Abstract
Transcription initiation by RNA polymerase II (RNA pol II) requires interaction between cis-acting promoter elements and trans-acting factors. The eukaryotic promoter consists of core elements, which include the TATA box and other DNA sequences that define transcription start sites, and regulatory elements, which either enhance or repress transcription in a gene-specific manner. The core promoter is the site for assembly of the transcription preinitiation complex, which includes RNA pol II and the general transcription fctors TBP, TFIIB, TFIIE, TFIIF, and TFIIH. Regulatory elements bind gene-specific factors, which affect the rate of transcription by interacting, either directly or indirectly, with components of the general transcriptional machinery. A third class of transcription factors, termed coactivators, is not required for basal transcription in vitro but often mediates activation by a broad spectrum of activators. Accordingly, coactivators are neither gene-specific nor general transcription factors, although gene-specific coactivators have been described in metazoan systems. Transcriptional repressors include both gene-specific and general factors. Similar to coactivators, general transcriptional repressors affect the expression of a broad spectrum of genes yet do not repress all genes. General repressors either act through the core transcriptional machinery or are histone related and presumably affect chromatin function. This review focuses on the global effectors of RNA polymerase II transcription in yeast, including the general transcription factors, the coactivators, and the general repressors. Emphasis is placed on the role that yeast genetics has played in identifying these factors and their associated functions.
This review presents an overview of the RNA polymerase II (RNA pol II) core transcriptional machinery. I discuss promoter elements and then review recent advances pertaining to three classes for transcription factors: general transcription factors (GTFs), transcriptional coactivators, and general transcriptional repressors. I focus on the transcriptional machinery from the yeast Saccharomyces cerevisiae, emphasizing the combined roles of yeast genetics and biochemistry in defining factors and their associated functions. However, this subject cannot be considered separately from the RNA pol II core transcriptional machinery from higher eukaryotic organisms, where results obtained with human, rat and Drosophila systems have often led the way. Although I emphasize the yeast system, I have attempted to integrate information from both yeast and metazoan systems whenever appropriate.
A breakthrough in understanding the mechanism of transcription initiation followed the discovery in the laboratory of Roeder that purified RNA pol II would selectively and accurately initiate transcription from template DNA when supplemented with a crude cell extract (529). This activity provided an assay for the fractionation and subsequent identification of the GTFs, defined as factors required for accurate, basal-level transcription initiation in vitro (311). Similar work defined analogous factors in rats, Drosophila, and yeast, suggesting that the GTFs are indeed “general” factors, required for expression of most, perhaps all, class II genes. Thus, the process of transcription initiation by RNA pol II is highly conserved among eukaryotic organisms, allowing for the experimental advantages offered by different organisms to be exploited to identify and define these factors.
S. cerevisiae has proven to be extraordinarily valuable in these studies. In 1987, Lue and Kornberg established an in vitro transcription system derived from yeast nuclei that would accurately initiate transcription from exogenous template DNA (295). A second in vitro transcription system was derived from yeast whole-cell extracts (541, 542). These systems have been instrumental not only for identifying the GTFs and their functions, but also for defining other transcription factors that influence the rate of transcription initiation.
A second advantage of yeast is the potential to exploit the power of classical and molecular genetic methods to investigate fundamental biological problems. An array of genetic selections has been developed to identify factors affecting RNA pol II transcription. In many cases, these studies have identified novel transcription factors or unexpected activities associated with these factors that had gone undetected by biochemical means. As a notable example, genetic selections for mutants unable to ferment sucrose (snf), for mutants that suppress promoter defects caused by insertion mutations (spt), and for mutants defective in mating-type switching (swi) converged, leading to the discovery of the SWI/SNF chromatin-remodeling complex that facilitates transcriptional activation (reviewed in references 51 and 537). Another important example is the genetic selection for suppressors of the conditional growth defect associated with truncation of the RNA pol II carboxy-terminal repeat domain (srb), which led to the discovery of the RNA pol II holoenzyme (reviewed in reference 261a).
The information available from the extensive collection of well-characterized yeast mutants and the complete sequence of the yeast genome are additional advantages to the yeast system. Accordingly, sequence information for proteins identified biochemically, either from yeast or from other organisms, can be compared to the yeast database. In many cases, a biochemically identified protein corresponds to the product of a gene identified in a genetic selection or screen. This combination of biochemistry and genetics often provides novel insight into protein function. Two remarkable examples are a histone acetyltransferase from Tetrahymena and a histone deacetylase from humans. Sequence analysis of these two proteins revealed similarity to the products of the genetically defined yeast GCN5 and RPD3 genes (38, 481). Although the biochemical function of neither gene had been defined, GCN5 was identified in a genetic selection for transcriptional coactivators whereas RPD3 was identified in a selection for transcriptional repressors. This combination of biochemistry and genetics led to the identification of Gcn5 and Rpd3 as histone acetyltransferase and histone deacetylase, respectively. Moreover, it provided a direct link between histone acetylation/deacetylation and transcriptional activation/repression (reviewed in reference 181).
PROMOTER STRUCTURE
Eukaryotic promoters can be divided into core elements and regulatory elements (reviewed in reference 456). Core promoter elements define the site for assembly of the transcription preinitiation complex (PIC) and include a TATA sequence, located upstream of the transcription start site, and an initiator sequence (Inr), encompassing the start site. Promoters can include a TATA box, an Inr sequence, or both of these control elements. A third core element, the downstream promoter element (DPE), was initially described in Drosophila and is located about 30 bp downstream of the start site (48). The DPE appears to function, in conjunction with the Inr element, as a TFIID binding site at TATA-less promoters.
Regulatory elements are gene-specific sequences that are located upstream of the core promoter and control the rate of transcription initiation; they include both upstream activation sequences (UAS) and upstream repression sequences (URS), which serve as binding sites for enhancers and repressors of transcription, respectively. In addition, poly(dA-dT) sequences are bidirectional upstream promoter elements that facilitate constitutive gene expression, not as UAS-like elements but apparently by forming a structure that is less stable to repressing nucleosomes. These elements are reviewed below.
TATA Elements
TATA elements in S. cerevisiae are typically located 40 to 120 bp upstream of the transcription initiation site. This is in contrast to other eukaryotes, including Schizosaccharomyces pombe, where the TATA element is almost always located at a fixed distance of 25 to 30 bp from the start site (reviewed in reference 456). The TATA sequence is the binding site for the TATA binding protein (TBP). TBP-TATA association nucleates the assembly of an approximately 4-MDa transcription preinitiation complex, a step that can be rate limiting for transcription initiation in vivo (257).
Mutational analysis and random selection for functional TATA elements defined TATAAA as the consensus TATA sequence in yeast (74, 441, 539). Many derivatives of this sequence also confer TATA function, albeit with diminished activity. One derivative, TGTAAA, eliminated TATA function and was used to select for TBP derivatives with altered binding specificity (454). TBPm3, described below, allowed transcription from TGTAAA promoters but not from certain other single-nucleotide derivatives of TATAAA (454). This mutant demonstrated the importance of specific interactions between the TATA element and TBP for efficient initiation. Functional analysis of mutated TATA elements revealed that yeast and human TBP have nearly identical TATA sequence requirements, underscoring the evolutionary conservation of the TBP-TATA interaction (539).
Some yeast promoters contain multiple TATA elements. For example, transcriptional analysis of site-directed mutations defined two functional TATA-like sequences within the CYC1 promoter (281). These two elements differed in sequence; one is denoted β-type, and the other is denoted α-type. Interestingly, when both elements were present, both were used equally to direct initiation within distinct but overlapping windows. However, if the same type (either β or α) was present at both sites, only the upstream element was used, directing initiation within the upstream window. These results were interpreted to mean that β- and α-type TATA elements are recognized by different factors of the transcriptional apparatus. However, TBP binds both consensus and nonconsensus TATA elements (178), suggesting that regulatory factors other than TBP might confer differential recognition of closely related TATA elements (281).
Yeast promoters lacking canonical TATA elements (TATA-less promoters) have also been identified. For example, the HIS3 promoter contains two TATA elements, one of which (TR) conforms to the canonical TATAAA sequence and is responsible for initiation at position +13 in response to activation by Gcn4. The other element (TC) does not resemble a consensus TBP binding site, directs initiation from position +1, and supports initiation in the absence of activators. Nonetheless, TC-directed transcription is TBP dependent in vivo (99). Interestingly, the relative utilization of TC and TR depends upon the overall level of transcription (223). TC is preferentially utilized at low levels of transcription, TC and TR are utilized equally well at moderate levels of transcription, and TR is preferentially utilized at high levels of transcription. These results suggest that transcription initiation from weak TATA elements is not mechanistically distinct from that mediated by canonical TATA elements but is determined instead by the overall level of transcription (223).
Transcription from TATA-less promoters remains TBP dependent. Accordingly, the term “TATA-less promoter” denotes relatively weak TBP-DNA affinity rather than a fundamentally distinct promoter element. Nonetheless, the rate-limiting step in PIC assembly at TATA-less promoters is unlikely to be TBP recruitment. Presumably, another component(s) of the core machinery recognizes a promoter structure other than TATA to nucleate PIC assembly. Indeed, TAFII60 and TAFII40 from human and Drosophila cells play a direct role in basal transcription by specifically binding the DPE of TATA-less promoters (47, 48).
Initiator Elements
Inr elements are DNA sequences encompassing transcription start sites. The fixed distance between TATA and the Inr element in eukaryotic organisms other than S. cerevisiae suggests that the Inr is determined simply by spacing from TATA. In contrast, the variable distance between TATA and the Inr in S. cerevisiae implies the existence of specific sequences that permit transcription initiation. Experiments to determine the relationship between TATA and the Inr established that the TATA element defines the window within which initiation can occur but that specific sequences within the window define the Inr element (179, 191, 281, 331, 403). Mutational analyses and surveys of start sites have defined preferred Inr sequences (146), yet there is no clearly defined Inr consensus sequence.
Yeast mutants that affect start site selection have been identified (11, 27, 147, 193, 216, 367). Specific mutations in TFIIB and the largest subunit of RNA pol II shift initiation downstream of normal (27, 367). However, in no case is the downstream site a “new” initiation site. Rather, these sites are normal, albeit minor initiation sites that generally conform to preferred Inr sequences. Thus, defects in TFIIB and RNA pol II do not alter the specificity of Inr element recognition but instead shift the window within which initiation can occur further downstream. Although the mechanism of Inr recognition is unclear, RNA pol II and TFIIB are key players in this process (284, 366).
The Inr element, as defined in higher eukaryotes, is not simply the DNA sequence encompassing the transcription start site. Rather, an Inr element was initially defined at the TATA-less promoter of the terminal deoxynucleotidyltransferase gene as a core promoter element, distinct from TATA, that can nucleate PIC assembly (442). Inr elements were subsequently identified at many promoters, both TATA-containing and TATA-less, and have been implicated in transcriptional control by directing accurate initiation in a TATA-independent manner (reviewed in reference 530). Proteins that bind Inr elements include CIF (241), YY1 (497), E2F (318), TFII-I, and USF (398, 399), as well as RNA pol II itself (12, 61). The CIF complex includes a homolog of Drosophila TAFII150 (241), which binds promoter DNA overlapping the Inr region (509) and has been implicated in differential recognition of two tandem Adh promoter elements (187). Furthermore, a recombinant TBP-TAFII150-TAFII250 subcomplex is minimally required for efficient utilization of Inr and downstream promoter elements in a reconstituted transcription system (507). Thus, it appears that TFIID can be recruited to a promoter by either of two distinct pathways, one involving TBP-TATA interaction and the other involving TAF-Inr interaction.
It is not clear whether Inr elements that function as distinct core promoter elements to nucleate PIC assembly exist in yeast. There are some intriguing prospects, though. For example, the GAL80 promoter has been reported to include both an Inr element and a TATA element, with these two elements directing initiation at distinct sites. The GAL80 Inr element is functionally portable, and a GAL80 Inr-binding protein has been detected (412). These results suggest that GAL80 transcription is driven by two independent pathways, one Inr dependent and the other TATA-dependent (412). This scenario is reminiscent of transcription at the HIS3 promoter (456). Thus, Inr elements that facilitate transcriptional control might be a universal feature of eukaryotic of RNA pol II transcription.
UAS and URS Elements
UAS elements are DNA sequences that function as binding sites for specific transcriptional activators. As such, UAS elements are analogous to metazoan enhancers, functioning in either orientation and at variable distances from the core promoter. An important functional distinction between enhancers and UAS elements is that UAS elements do not function when positioned downstream of the TATA box (168, 455). However, this has been reported for only a few genes and needs to be tested more thoroughly. Once associated with their cognate UAS elements, transcriptional activators facilitate assembly of the PIC, either by direct contact with GTFs or indirectly through coactivators, which in some cases mediate activator-GTF interactions. Consistent with its role in activation, deletion of a UAS element diminishes mRNA synthesis under activating conditions.
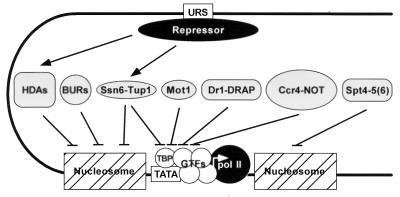
URS elements are binding sites for gene-specific transcriptional repressors. URS-repressor complexes can impair transcription by several different mechanisms, including interference with activator-UAS binding; interference with the activation domain of an activator-UAS complex; or by contact with the core transcriptional machinery, a process analogous to activation, albeit with opposite effects (reviewed in reference 233). URS-repressor complexes can also mediate repression indirectly by recruiting another complex that targets either the core transcriptional machinery or histones. For example, several URS-repressor complexes recruit the Ssn6-Tup1 complex, which appears to mediate repression by affecting histone function (214). Transcriptional repression associated with histone deacetylation is another example of this type of repression. In the best-characterized example in yeast, the GC-rich URS1 element binds the Ume6 repressor, which in turn recruits the Sin3-Rpd3 histone deacetylase complex (234). This process is analogous to transcriptional repression mediated by histone deacetylases in metazoan systems (reviewed in reference 359).
Poly(dA-dT) Elements
Homopolymeric dA-dT sequences are a common feature of yeast promoters and in several cases have been shown to be required for normal levels of transcription in vivo (reviewed in reference 224). Poly(dA-dT) sequences have distinct structural characteristics that impair nucleosome assembly or stability, which led to the proposal that poly(dA-dT) sequences function as promoter elements based on their intrinsic structure, rather than as conventional UAS elements to which sequence-specific transcription factors bind (75). However, a naturally occurring poly(dA-dT) sequence activated transcription in vitro, an effect that could be squelched by addition of a related oligonucleotide (294). This result argued for involvement of a poly(dA-dT)-specific transcription factor. Indeed, a poly(dA-dT)-binding protein, datin, has been identified (538).
The mechanism of poly(dA-dT)-mediated transcriptional activation has been investigated by using a combination of functional assays and probes of chromatin structure. A poly(dA-dT) sequence located upstream of the Gcn4 binding site in the HIS3 promoter stimulated Gcn4p-activated transcription in a length-dependent manner (224). Moreover, datin repressed, rather than stimulated, gene expression, and poly(dG-dC), which also affects nucleosome structure, functioned similarly to poly(dA-dT) (224). These results imply that poly(dA-dT) stimulates transcription as a consequence of its intrinsic structure, rather than as a conventional UAS element. This conclusion is supported and extended by the demonstration that a poly(dA-dT) element located adjacent to the Candida glabrata metal-dependent transcriptional activator gene, AMT1, plays a critical role in transcriptional autoactivation by causing a localized distortion of the nucleosomal DNA, allowing Amt1 to gain access to its cognate promoter element (565). Recently, a whole-genome analysis revealed that poly(dA-dT) tracts are abundant in S. cerevisiae and occur predominantly at unit nucleosomal length both upstream and downstream of open reading frames, leading to the proposal that such tracts modulate nucleosome positioning (382).
RNA POLYMERASE II
Overview
Yeast RNA pol II is composed of 12 subunits encoded by the RPB1 to RPB12 genes (543). There is extensive structural conservation among the subunits of eukaryotic RNA pol II. Indeed, six subunits of human RNA pol II can functionally replace their homologs in yeast (317). The two largest RNA pol II subunits, Rpb1 (∼200 kDa) and Rpb2 (∼150 kDa), are the most highly conserved subunits. Moreover, Rpb1 and Rpb2 are homologous to the β′ and β subunits, respectively, of bacterial RNA polymerase. Rpb3 is related to the α subunit of bacterial RNA polymerase based on partial amino acid sequence similarity, size similarity, identical subunit stoichiometry (two per molecule), and assembly defects associated with mutations in either subunit (543). None of the RNA pol II subunits appears to be closely related to the bacterial ς-subunit family, although structural and functional similarities between ς and certain GTFs have been identified (see below).
The Rpb1, Rpb2, Rpb3, and Rpb11 subunits of RNA pol II are homologous to subunits of RNA polymerases I and III. Moreover, five subunits, Rpb5, Rpb6, Rpb8, Rpb10, and Rpb12, are common to all three RNA polymerases. Only Rpb4, Rpb7, and Rpb9 are unique to RNA pol II. Thus, RNA polymerases are assembled from common as well as class-specific subunits. Ten of the yeast genes encoding RNA pol II subunits are essential for cell viability. Only the RPB4 and RPB9 genes are dispensable, although deletion of either gene confers conditional growth phenotypes.
The sequence similarity between Rpb1 and Rpb2 and the bacterial β′ and β subunits occurs in highly conserved domains, designated A to H in Rpb1, and A to I in Rpb2 (555). This structural similarity extends among the two largest subunits for all eukaryotic RNA pol II investigated. Not surprisingly, the structural similarity between Rpb1/β′ and Rpb2/β extends to functional similarity. Both Rpb1 and β′ are involved in DNA binding, whereas Rpb2 and β bind nucleotide substrates.
Many mutations in RPB1 and RPB2 have been isolated and characterized (reviewed in reference 6). Most amino acid replacements are located within the highly conserved domains. Specific mutations in RPB1 and RPB2 affect the accuracy of transcription initiation, demonstrating a role for these subunits in defining start site selection (11, 27, 193). Other mutations in RPB1 and RPB2 confer sensitivity to 6-azauracil (6-AU), a phenotype associated with transcription elongation defects, suggesting that both subunits are also involved in overcoming transcriptional arrest (7, 374). 6-AU-sensitive rpb1 mutants can be suppressed by overexpression of PPR2, the gene encoding the elongation factor SII (7). In the case of RPB2, 6-AU-sensitive alleles encode elongation-defective forms of RNA pol II (374).
The Rpb4 and Rpb7 subunits are functionally related. These two subunits can be dissociated from RNA pol II, and RNA pol II purified from a rpb4 null mutant lacks Rpb7 (122). This form of RNA pol II is indistinguishable from wild-type RNA pol II in an in vitro elongation assay but is inactive in promoter-directed transcription initiation. Furthermore, this form of RNA pol II could be complemented in vitro by an inactive RNA pol II with a defective form of Rpb1. These results demonstrate that Rpb4 and Rpb7 function in transcription initiation and suggest that they can shuttle between RNA pol II molecules (122). Interestingly, rpb4 mutants exhibit substantially impaired growth rates at elevated temperature or under conditions of nutritional deprivation, implicating Rpb4 in tolerance of RNA pol II to stress (80).
Similar mutational analyses are needed to define functions for the other RNA pol II subunits. Limited mutational analysis revealed that Rpb3 is involved in RNA pol II assembly (262). Mutations in RPB9 affect start site selection (147, 148, 216, 460), and in one case an rpb9 allele suppresses a TFIIB defect that affects start site selection (460). Thus, Rpb9, like Rpb1 and Rpb2, affects the accuracy of initiation, perhaps through interaction with TFIIB.
Carboxy-Terminal Repeat Domain
A unique feature of the largest RNA pol II subunit is the presence of tandem repeats of a heptapeptide sequence at its carboxy-terminus. This carboxy-terminal repeat domain (CTD) has the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser is highly conserved among eukaryotic organisms. Although the CTD is a ubiquitous feature of RNA pol II, the repeat length varies. For example, yeast Rpb1 includes 26 or 27 repeats, the C. elegans CTD has 34 repeats, the Drosophila CTD has 43 repeats, and the human CTD 52 repeats, suggesting that repeat length increases with increasing genome complexity.
Although the CTD is essential for cell viability, its function is not entirely clear. There are two forms of RNA pol II in vivo, designated IIO, which is extensively phosphorylated at the CTD, and IIA, which is not phosphorylated. The IIA form preferentially enters the PIC, whereas IIO is found in the elongating complex (reviewed in reference 103). Conversion of IIA to IIO occurs concomitant with or shortly after the transition from initiation to elongation and is accompanied by extensive CTD phosphorylation (292, 346). These results implicate the CTD in conversion of RNA pol II from a form involved in promoter recognition to an elongation-competent form. Nonetheless, a form of RNA pol II lacking the CTD (IIB) is able to initiate transcription from TATA-containing promoters in vitro, although not from TATA-less promoters (2, 40).
The kinase activity of TFIIH can mediate CTD phosphorylation (132, 293, 429), although other kinases, including Cdc2 (83), Ctk1 (274), the Srb10-Srb11 kinase-cyclin pair (285), and P-TEFb (310), have also been implicated in CTD phosphorylation. Drosophila P-TEFb (positive transcription elongation factor b) affects the transition from abortive to productive transcription elongation (310). The catalytic subunit of P-TEFb is homologous to PITALRE, a Cdc2-related protein kinase (564). Human P-TEFb associates with the Tat protein of human immunodeficiency virus type 1 to potentiate transcriptional elongation (307, 564). Thus, multiple kinases appear to mediate phosphorylation of the RNA pol II CTD. Whether these CTD kinases are gene specific or affect different steps in the transition from initiation to elongation remains to be determined.
A phosphatase responsible for dephosphorylation of the CTD has also been identified (66). CTD phosphatase activity is regulated by TFIIB and TFIIF (67). The RAP74 subunit of TFIIF stimulates CTD phosphatase activity, whereas TFIIB inhibits the stimulatory activity of TFIIF. Since the dephosphorylated form of RNA pol II (IIA) preferentially enters the PIC (103), these results suggest that the CTD phosphatase, TFIIF, and TFIIB interact to regulate RNA pol II recycling.
Although the CTD is essential for cell growth, all but 8 to 10 repeats can be deleted from yeast Rpb1 without loss of viability (345, 532). However, strains with a minimum number of CTD repeats exhibit a cold-sensitive growth defect, a phenotype that was exploited to isolate extragenic suppressors of CTD truncations (345). This selection identified SRB genes, which encode components of the SRB-mediator complex required for transcriptional activation (28). Thus, the CTD functions in transcriptional activation.
The CTD has also been implicated in pre-mRNA processing. A speculative model proposed that the negatively charged, hyperphosphorylated CTD of the IIO form of RNA pol II facilitates electrostatic interactions with positively charged regions of certain splicing factors (167). This model has received considerable experimental support. Splicing is inhibited in vitro (557) and in vivo (118) by CTD repeat polypeptides and in vitro by an anti-CTD antibody (557). Furthermore, proteins that might connect the spliceosome to RNA pol II via the CTD have been identified (450, 557). The CTD has also been implicated in 5′ capping of mRNA and 3′-end formation. The 5′-capping enzyme specifically binds the phosphorylated IIO form of RNA pol II, suggesting a mechanism for coupling of cap addition to RNA pol II transcription (79, 313, 556). A role for the CTD in 3′-end formation was discovered by the effects of CTD truncations on 3′ processing and poly(A) addition and substantiated by the association of cleavage and polyadenylation factors with the CTD (314). These results suggest that the CTD functions as a platform for the recruitment and assembly of factors involved in pre-mRNA processing (reviewed in reference 449).
RNA Polymerase II Holoenzymes
RNA pol II and the GTFs assemble in a defined order on promoter DNA in vitro (42, 301, 503). These results suggested stepwise assembly of the PIC. However, several GTFs were known to associate with RNA polymerase in the absence of DNA, hinting at the existence of an RNA pol II holoenzyme complex (96). Antibodies directed against SRB proteins provided direct evidence for a holoenzyme complex that includes a subset of the GTFs, as well as SRB proteins (261). Unlike core RNA pol II, holoenzyme responds to transcriptional activators in vitro. Holoenzyme was independently discovered based on the association of RNA pol II with mediator, the protein complex required for transcriptional activation (252). As described in the coactivator section below, mediator includes SRB proteins, MED proteins, and a subcomplex composed of Gal11, Sin4, Rgr1, and Med3. These two holoenzymes are comparable, although one complex is reported to include TFIIB, TFIIF, and TFIIH (261) whereas the other includes TFIIF as the only GTF (252). One holoenzyme preparation is also reported to include Srb8 to Srb11 (194, 285) and the SWI/SNF chromatin remodeling complex (535), whereas the other includes neither of these sets of proteins (57, 252, 330). Regardless of the distinction between these two holoenzyme preparations, a key point is that holoenzyme supports activated transcription with only TBP and other GTFs in reconstituted transcription systems. Transcriptional activation is mediator dependent but TAF independent. This is in contrast to metazoan transcription systems, where activation is TAF dependent (reviewed in reference 183).
There are at least two distinct forms of yeast RNA pol II holoenzyme (reviewed in reference 68). A second form was discovered by a purification strategy based on an RNA pol II affinity column immobilized through the CTD (435). Isolated proteins include TFIIB, TFIIF, TFIIS, and Gal11 but not SRB/mediator components (515). Novel components of this complex include Paf1 and Cdc73 (435, 515), as well as Ccr4 and Hpr1 (68). This form of the holoenzyme affects the expression of a different spectrum of genes and is therefore functionally distinct from the SRB/mediator-containing holoenzyme (436). The percentage of total RNA pol II contained in the Paf1-Cdc73-Ccr4-Hpr1 holoenzyme has not been reported, although this holoenzyme is less abundant than the SRB/mediator holoenzyme (227).
RNA pol II holoenzyme complexes have also been identified in mammalian cells (69, 302, 352). Like the yeast holoenzymes but distinct from core RNA pol II, these complexes are able to respond to transcriptional activators in vitro. In addition to the GTFs, a provocative array of proteins has been found in these enzyme preparations, including DNA repair proteins (302), splicing and polyadenylation factors (314), and even the breast cancer tumor suppressor BRCA1 (427).
GENERAL TRANSCRIPTION FACTORS
The GTFs include TBP, TFIIB, TFIIE, TFIIF, and TFIIH and were identified biochemically as factors required for accurate transcription initiation by RNA pol II from double-stranded DNA templates in vitro (reviewed in references 395 and 558). Fractionation of whole-cell extracts from yeast also identified five factors, designated a, b, d, e, and g, that are required for promoter-specific transcription by RNA pol II (420). The yeast factors are comparable in structure and function to the mammalian GTFs and are now designated by the mammalian nomenclature. These factors are reviewed below and summarized in Table 1.
TABLE 1.
Yeast general transcription factors
| Factora | Mass (kDa) | Gene(s) | Essential | Characteristics | Metazoan homolog(s) | Reference(s) |
|---|---|---|---|---|---|---|
| TBP (factor d) | 27 | SPT15 | Yes | Binds TATA element; nucleates PIC assembly; recruits TFIIB | TBP | 124, 177 |
| TFIIB (factor e) | 38 | SUA7 | Yes | Stabilizes TATA-TBP interaction; recruits RNA pol II-TFIIF; affects start site selection; zinc ribbon | TFIIB | 366 |
| TFIIF (factor g) | 82 | TFG1, SSU71 | Yes | Facilitates RNA pol II-promoter targeting; stimulates elongation; functional interaction with TFIIB | RAP74 | 196, 459 |
| 47 | TFG2 | Yes | ς factor homology; destabilizes nonspecific RNA pol II-DNA interactions | RAP30 | 196 | |
| 27 | TFG3, ANC1, SWP29, TAF30 | No | Common subunit of TFIID, TFIIF, and the SWI/SNF complex | AF-9, ENL | 55, 196, 531 | |
| TFIIE (factor a) | 66 | TFA1 | Yes | Recruits TFIIH; stimulates TFIIH catalytic activities; functions in promoter melting and clearance; zinc binding domain | TFIIE-α | 133 |
| 43 | TFA2 | Yes | TFIIE-β | 133 | ||
| TFIIHb (factor b) | 95 | SSL2, RAD25 | Yes | Functions in promoter melting and clearance; ATP-dependent DNA helicase (3′ → 5′); DNA-dependent ATPase; ATPase/helicase required for both transcription and NER | XPB, ERCC3 | 169 |
| 85 | RAD3 | Yes | ATP-dependent DNA helicase (5′ → 3′); DNA-dependent ATPase; ATPase/helicase required for NER but not transcription | XPD, ERCC2 | 22, 171 | |
| 73 | TFB1 | Yes | Required for NER | p62 | 160 | |
| 59 | TFB2 | Yes | Required for NER | p52 | 134 | |
| 50 | SSL1 | Yes | Required for NER; zinc binding domain | p44 | 528, 553 | |
| 47, 45 | CCL1 | Yes | TFIIK subcomplex with Kin28 | Cyclin H | 464 | |
| 37 | TFB4 | Yes | p34 | 134 | ||
| 32 | TFB3 | Yes | Zinc RING finger; links core-TFIIH with TFIIK; unlike Mat1, not a subunit of kinase/cyclin subcomplex | Mat1 | 134 | |
| 33 | KIN28 | Yes | TFIIK subcomplex with Ccl1 | MO15, Cdk7 | 136, 499 |
The initial designations of the yeast general transcription factors by Kornberg’s laboratory are denoted in parentheses.
TFIIH is composed of core-TFIIH (Rad3, Ssl1, Tfb1 to Tfb4), plus Ssl2/Rad25 and the TFIIK kinase/cyclin subcomplex (Kin28, Ccl1).
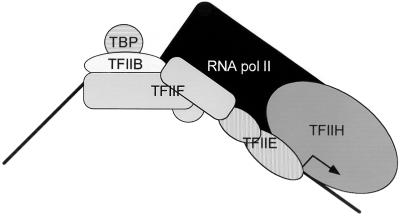
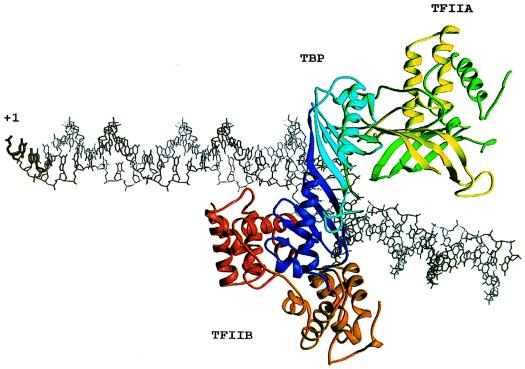
Order-of-addition experiments demonstrated that PIC assembly is nucleated in vitro by TBP binding to the TATA element followed by binding of TFIIB, RNA pol II-TFIIF, TFIIE, and TFIIH (42; reviewed in references 351 and 395). This scenario was challenged by the discovery of RNA pol II holoenzyme complexes, first in yeast and later in mammalian systems (see the RNA polymerase II section, above). These findings suggest that at least some GTFs associate with RNA pol II prior to promoter binding and have implications for the mechanisms of transcriptional regulation. Whether PIC assembly occurs in a stepwise manner or by holoenzyme recruitment, the order-of-addition experiments provided valuable information about GTF interactions within the PIC. A schematic representation of the PIC is presented in Fig. 1; a crystallographic representation of a DNA-TBP-TFIIA-TFIIB complex is presented in Fig. 2.
FIG. 1.
Schematic depiction of the transcription PIC. PIC assembly is nucleated by TBP binding to the TATA box, inducing a sharp bend in the DNA template, followed by association of TFIIB, RNA pol II/TFIIF, TFIIE, and TFIIH. Each pattern denotes a distinct general transcription factor. Subunit composition is indicated, except for TFIIH (9 subunits) and RNA pol II (12 subunits). Although PIC assembly can occur by stepwise addition of the general transcription factors (GTFs) in vitro, the discovery of RNA pol II holoenzyme complexes that include GTFs suggests that stepwise assembly might not occur in vivo.
FIG. 2.
Tertiary structure of a TATA-TBP-TFIIA-TFIIB complex. The amino- and carboxy-terminal direct repeats of TBP are light and dark blue, respectively. The amino- and carboxy-terminal repeats of core-TFIIB are red and orange, respectively. The Toa1 and Toa2 subunits of TFIIA are green and yellow, respectively. Reprinted from reference 351 with permission of the publisher.
TATA Binding Protein
Overview.
TBP is a universal transcription factor, required for initiation by all three eukaryotic RNA polymerases (reviewed in reference 198). TBP was identified as a subunit of TFIID, the large (∼750-kDa) multisubunit complex composed of TBP and TBP-associated factors (TAFs). Whereas TBP functions in basal level transcription, TFIID is required for response to transcriptional activators in metazoan in vitro transcription systems (reviewed in references 50, 378, and 508). Direct evidence that TBP plays a general role in eukaryotic transcription came from Comai et al., who found TBP to be an essential subunit of the RNA pol I transcription factor SL1 (93). TBP was also identified as a subunit of the RNA pol III general factor TFIIIB (215, 239, 290, 440, 473, 533). Furthermore, mutations in yeast TBP diminished expression by all three RNA polymerases in vivo (99, 426). Thus, TBP plays a requisite role in transcription initiation by all three RNA polymerases, functioning as a common subunit of SL1, TFIID, and TFIIIB.
Yeast TBP is a monomer of 27 kDa and is functionally interchangeable with mammalian TBP in in vitro transcription systems (43, 65, 178). This result demonstrated that GTF functions are conserved among eukaryotic organisms. Based on protein sequence information, the yeast gene encoding TBP was isolated (64, 177, 210, 424). Sequence analysis revealed identity to SPT15, which was identified in a genetic selection for suppressors of a Ty insertion in the HIS4 promoter (his4-917δ) (124). In the his4-917δ mutant, transcription initiates within the Ty δ element to produce abnormally long, nonfunctional transcripts. Mutations at the SPT15 locus suppressed his4-917δ by shifting initiation from the δ promoter element to the his4 promoter, resulting in a His+ phenotype. Furthermore, spt15 mutants are pleiotropic and deletion of SPT15 is lethal. These results demonstrated that TBP is an essential transcription factor that affects promoter recognition and is required for the expression of many, if not all, genes in vivo (124).
Sequence analysis of the deduced TBP amino acid sequence revealed two direct repeats encompassing the C-terminal two-thirds of the protein. Subsequent comparison with the phylogenetic series of TBP sequences demonstrated that the C-terminal direct repeats are highly conserved. Although the N-terminal domain is more divergent, it is conserved among vertebrate forms of TBP and regulates RNA pol III transcription at the U6 promoter (326). Unlike other DNA binding proteins, TBP recognizes its binding site through minor groove contacts (273, 448).
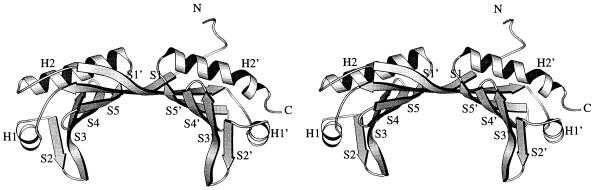
The crystal structure of Arabidopsis TBP revealed a remarkable structure containing a new DNA binding fold, resembling a molecular “saddle” that sits astride the DNA (342; reviewed in reference 340). The protein molecule includes two similar structural domains related by approximate twofold symmetry. Each α/β domain (∼90 amino acids) corresponds to each of the C-terminal direct repeats and is composed of two α-helices and five antiparallel β-sheets connected in the order S1-H1-S2-S3-S4-S5-H2 (Fig. 3) (343). The crystal structures of yeast and Arabidopsis TBP-TATA complexes demonstrated that the TBP saddle induces kinks at both ends of the 8-bp TATA element, bending the DNA 80° toward the major groove (246, 251). The DNA binding surface is a curved, anti-parallel β-sheet, and the convex seat of the saddle is potentially available for interaction with other factors.
FIG. 3.
Tertiary structure of TBP-2 from Arabidopsis thaliana. The three-dimensional structure is viewed perpendicular to the intramolecular twofold symmetry axis. The α-helices (H1, H2 and H1′, H2′) and β-sheets (S1 to S5 and S1′ to S5′) are labeled and can be correlated with the TBP amino acid replacement data summarized in Table 2. Reprinted from reference 342 with permission of the publisher.
TBP plays a critical role in the mechanism of transcriptional activation. This is implied by direct contact between the activation domains of many gene-specific activators and TBP (reviewed in reference 339). The best evidence that TBP plays a critical role in transcriptional activation comes from studies in yeast, where acidic activators enhance the kinetics of TBP recruitment (257). Consistent with this finding, tethering of TBP to a promoter by a heterologous DNA binding domain bypasses the need for a transcriptional activator (70, 254, 544).
Recruitment of TBP to the promoter is a slow and potentially rate-limiting step in transcriptional activation (257, 259, 286, 525). Complex formation between TBP and the TATA element has been proposed to occur by a two-step mechanism involving a slow TBP-TATA association followed by a rapid conformational change (209). Alternatively, the rate-limiting step in TBP-TATA complex formation has been proposed to be dissociation of TBP-TBP dimers. Human TBP has been reported to dimerize in solution, thereby blocking TBP-DNA association (88). TFIID also dimerizes in the absence of DNA, with dimer formation mediated by TBP-TBP association (474). Consistent with these findings, TBP from Arabidopsis crystallized as a dimer with the dimer interface overlapping the DNA binding domain (339, 342). Thus, TBP dimerization and TBP-DNA association appear to be mutually exclusive. Furthermore, TBP dimers dissociate slowly, suggesting that dimer dissociation dictates the kinetics of TBP-DNA binding (87). Kinetic analysis of yeast TBP-DNA interactions provides further support for the existence of TBP-TBP dimers that undergo slow dissociation at physiological concentrations (360). These results have important implications for the mechanisms by which certain activators and coactivators affect the rate of transcription.
A prediction based on TBP recruitment as a rate-limiting step in transcriptional activation is that TBP mutants that are specifically defective in activation yet have no effect on uninduced expression should be found. Indeed, several different genetic selections and screens have identified activation-defective TBP derivatives. These can be divided into functionally distinct classes. One class is defective in TBP-TATA interaction; a second is defective in TBP-TFIIA interaction; a third is defective in TBP-TFIIB interactions; and a fourth class is distinct from the other three. Each of these classes of TBP derivatives is reviewed below; a compilation of yeast TBP derivatives and their associated defects is presented in Table 2.
TABLE 2.
Yeast TBP derivatives
| Amino acid replacement(s) | Regiona | Functional defect | Reference(s) |
|---|---|---|---|
| N69S | S1 | Selectively increases transcription from weak promoters; does not alter TBP-TATA affinity | 29 |
| V71E | S1 | Diminished DNA binding | 387 |
| V71A | S1 | Activation defective; diminished DNA binding | 9 |
| R105C | S3 | Diminished DNA binding | 387 |
| P109A, P109Q | S3-S4 loop | Diminished DNA binding; promoter-specific activation defects | 9 |
| T112K | S4 | Diminished DNA binding | 387 |
| L114K | S4 | Activation defective; impaired interaction with VP16 | 248 |
| L114F | S4 | Altered DNA binding specificity | 10 |
| F116Y | S4-S5 loop | Diminished DNA binding | 9, 387 |
| S118L | S4-S5 loop | Activation defective; diminished DNA binding | 275 |
| K133L, K138L | H2 | Eliminates TFIIA binding; temperature-sensitive growth defect suppressed by high-copy-number BRF1/TDS4/PCF4 | 45, 46 |
| K133L, K145L | H2 | Eliminates TFIIA binding | 46 |
| K138T, Y139A (N2-1) | H2 | Activation defective; eliminates TFIIA binding | 447 |
| F148H | Inter-repeat strand | Activation defective; normal interaction with TATA, TFIIA, TFIIB and acidic activation domains | 446 |
| F148L | Inter-repeat strand | Activation defective; diminished DNA binding | 275 |
| T153I | Inter-repeat strand | Activation defective; normal interaction with TATA, TFIIA, TFIIB, and acidic activation domains | 446 |
| N159D | S1′ | Diminished DNA binding; promoter-specific activation defects | 9 |
| N159L | S1′ | Activation defective; diminished DNA binding | 275 |
| V161E | S1′ | Diminished DNA binding | 387 |
| V161A | S1′ | Activation defective; diminished DNA binding | 9, 275 |
| G174E | H1′ | Physical interaction with Spt3 | 123 |
| E186A | S2′-S3′ stirrup | Diminished TFIIB binding; activation responsive | 276 |
| E188A | S2′-S3′ stirrup | Diminished TFIIB binding; activation responsive | 276 |
| L189A | S2′-S3′ stirrup | Activation competent; deficient in forming a TBP-TFIIB complex | 276 |
| L189K | S2′-S3′ stirrup | Activation defective; deficient in forming a TBP-TFIIB complex | 248 |
| P191S | S2′-S3′ loop | Intragenic suppressor of L205F; corrects altered TATA binding specificity | 10 |
| I194F, V203T, L205V (TBPm3) | S3′ and S4′ | Altered DNA binding specificity | 454 |
| R196C | S3′ | Diminished DNA binding | 387 |
| V203K | S4′ | Diminished DNA binding | 387 |
| L205F | S4′ | Altered TATA binding specificity | 10 |
| F207Y | S4′-S5′ loop | Diminished DNA binding | 387 |
| K211L | S5′ | Activation defective | 248 |
| E236P | H2′ | Activation defective; normal interaction with TATA, TFIIA, TFIIB, and acidic activation domains | 446 |
| F237D | Immediately follows H2′ | Activation defective; normal interaction with TATA; impaired interaction with TFIIA and TFIIB | 446 |
| F237V | Downstream of H2′ | Intragenic suppressor of G174E | 123 |
The positions of the α-helices (H1, H2, H1′, and H2′) and β-sheets (S1 to S5 and S1′ and S5′) are indicated within the crystal structure of TBP shown in Fig. 2.
TBP-TATA interactions.
Several mutational studies of TBP have addressed the domains and residues that bind DNA. Random mutagenesis of SPT15 identified dominant mutations that diminished DNA binding (387). These included V71E, R105C, T112K, and F116Y single-residue derivatives. Each of these replacements occurs in the N-terminal direct repeat domain of TBP. Site-directed replacements of the analogous residues in the C-terminal repeat, V161E, R196C, V203K, and F207Y, also inhibited DNA binding, suggesting that DNA binding is partitioned between the two direct repeats (387). This prediction was confirmed by the crystal structure of the TATA-TBP complex (246, 251).
Regional mutagenesis, focusing on residues 190 to 205, identified a TBP derivative with altered TATA binding specificity. The TBPm3 derivative is the result of a triple amino acid replacement, I194F, V203T, and L205V, that supports transcription from promoters containing a TGTAAA promoter (454). Both the I194F and L205V replacements are required for this effect; V203T is dispensable but augments TGTAAA recognition. Although TBPm3 binds efficiently to the TATAAA sequence, it is unable to support cell growth. This mutant identified residues that interact directly with the TATA element, a conclusion confirmed by the TATA-TBP crystal structure. The TBPm3 derivative has been especially valuable for studying promoter-specific transcription using promoters containing a mutated TGTA element (see, e.g., reference 544).
Characterization of spt mutants also identified a TBP derivative with altered DNA binding specificity. The spt15-122 allele encodes an L205F replacement that enhances TBP binding to nonconsensus TATA elements (8). This accounts for the Spt phenotype of the spt15-122 mutant by shifting TBP recognition from the Ty promoter to the downstream his4 promoter. Based on this result, the analogous replacement, L114F, in the first repeat was constructed. L114F and L205F derivatives conferred nearly identical phenotypes (8). However, in vivo assays that utilize a complete set of point mutations in the TATAAA element found that L114F and L205F play distinct roles in TATA element recognition (10). The most notable distinction is that L114F enhances TATAAG recognition, whereas L205F enhances CATAAA recognition. These results support the premise that orientation of TBP with respect to the TATA element in the TBP-TATA crystal structure reflects the structure that occurs in vivo. These results also suggest that factors might exist to control transcriptional activation by affecting TBP recognition of nonconsensus TATA elements.
The concave surface of TBP interacts with the TATA element, and the convex surface interacts other proteins, including gene-specific activators. In an effort to understand the role of TBP in mediating transcriptional activation, a genetic selection for activation-defective derivatives of TBP that are normal for uninduced expression was devised (275). Presumably this selection would identify residues on the convex surface that are critical for protein-protein interactions that mediate activation. In this study, S118L, N159L, V161A, and F148L were identified as activation-defective TBP derivatives. Surprisingly, each of these positions, except F148, directly contacts DNA, and all four derivatives are defective for TATA binding. Each derivative interacts normally with TFIIB and GAL4-VP16. These results imply that the TBP-TATA interface is a critical determinant of transcriptional activation (275).
Activation-defective TBP mutants were identified in another, independent genetic screen. Random mutations in SPT15 were generated by error-prone PCR and subsequently screened for inositol auxotrophy (Ino−) or failure to grow on galactose medium (Gal−), phenotypes often associated with defects in transcriptional activation (9). Six different amino acid replacements at five unique positions, all within the repeat domains, were identified. Three of these replacements, V71A, F116Y, and V161A, were found at sites previously identified based on diminished DNA binding (387). Three replacements at two unique positions, P109A, P109Q, and N159D, led to mutants that were also defective in DNA binding and exhibited promoter-specific defects in activation. These results are consistent with those described above (275), strongly suggesting that transcriptional activators enhance the formation or stability of the TBP-TATA complex at certain promoters in vivo (9).
TBP-TFIIA interactions.
The core domain of TBP includes helices located at the ends of each direct repeat (H2, H2′) (Fig. 3). These structures are components of the convex surface of TBP, comprising the seat of the saddle, suggesting that the saddle mediates protein-protein interactions (251). Indeed, mutational analysis of basic residues within the H2 helix demonstrated that residues K133/K138 and K133/K145 are required for interaction with TFIIA, as defined by gel shift analysis (46).
In a genetic screen for TBP derivatives that cause a temperature-sensitive phenotype yet maintain normal pol III transcription, a double K138T/Y139A replacement within the H2 helix was found (447). This double mutant (designated N2-1) failed to interact with TFIIA and is defective for activation by Gal4, Gcn4, and Ace1. This result is consistent with the effect of the K133/K138 replacements (46) and demonstrates that the TBP-TFIIA interaction can be essential for activation in vivo (447).
Surprisingly, the crystal structure of a yeast TATA-TBP-TFIIA ternary complex revealed no direct contacts between TFIIA and the H2 helix (155, 475). The apparent discrepancy between this result and the effects of H2 replacements on TBP-TFIIA interaction might be accounted for by the forms of TFIIA used in the crystallographic studies. The crystal structures of two TATA-TBP-TFIIA ternary complexes have been solved. In both cases, crystals were generated with TFIIA derivatives deleted for large internal regions of the largest (Toa1) subunit (155, 475).
In a study of human TBP-TFIIA interactions, amino acids replacements in TBP (A86, N91, E93, R107 [yeast numbering system]) that are required for TBP-TFIIA interaction in vitro and for transcriptional activation in vivo were identified (39). In this case, the altered residues are located within the H1 helix and the S2 and S3 β-sheets that comprise one of the TBP stirrups (Fig. 3). These residues directly contact TFIIA (Fig. 2), thereby confirming the importance of the TFIIA-TBP contacts for transcriptional activation. These results predict that certain gene-specific transcriptional activators will stimulate TBP-TFIIA-promoter complex assembly by direct binding to TFIIA, a prediction borne out for the Zta and VP16 activators (259).
Until recently, no TBP mutants had been described that stimulated pol II transcription. In a hunt for TBP mutants that would enhance transcription from a weak, synthetic transcriptional activator, the TBP N69S derivative was identified (29). TBP N69S selectively increases transcription from genes with weak pol II promoters, including those lacking a functional TATA box. TBP N69S does not alter the affinity of TBP for DNA but appears to enhance TBP recruitment to the promoter. Since the L205K replacement disrupts TBP-TATA binding and the double L138T/Y139A replacement disrupts TBP-TFIIA interaction, it was suggested that the N69S effect is dependent upon TBP-TFIIA interaction but independent of TBP-TATA interaction (29). Although the mechanism by which N69S enhances promoter recruitment is unknown, this mutant strengthens the premise that TBP recruitment can be rate limiting for transcription initiation.
TBP-TFIIB interactions.
Site-directed mutations were generated in the C-terminal core domain of yeast TBP in an attempt to define residues critical for GAL4-VP16 activation (248). Three replacements, L114K, L189K, and K211L, selectively blocked activation with no effect on basal transcription in vitro. Each of these TBP derivatives was defective in GAL4-VP16-mediated recruitment of TFIIB to the promoter complex and one, L189K, disrupted TBP-TFIIB interaction. These results were interpreted to mean that GAL4-VP16 activation involves TBP recruitment, as well as stabilization or isomerization of an activation-specific TATA-TBP-TFIIB complex (248). A role for TBP-TFIIB interaction in transcriptional activation is consistent with activator-mediated recruitment of TFIIB to preinitiation complexes in vitro (82) and with a model invoking activator-dependent conformational changes in TBP-TFIIB interaction (discussed in reference 175).
A critical role for TBP-TFIIB interaction in transcriptional activation has additional support. Based on the crystal structure of the TATA-TBP-TFIIB ternary complex (341) and on TBP amino acid replacements that block ordered assembly of the PIC (477), amino acid replacements in TBP were made at positions known to directly interact with TFIIB (478). In this case, a human TBP E284R (yeast E186) derivative disrupted the interaction with TFIIB and blocked transcriptional activation in human cells. If the activation defect were due specifically to defective TBP-TFIIB interaction, an amino acid replacement in TFIIB at the position that interacts with TBP E284 might restore activation. Indeed, a TFIIB R169E replacement compensated for the activation defect caused by E284R, restoring activation by GAL4-VP16, GAL4-CTF, and GAL4-p53. In contrast, GAL4-Sp1 was insensitive to disruption of the TBP-TFIIB interaction, implying the selective use of the TBP-TFIIB interaction by activators in higher eukaryotes in vivo (478).
Interestingly, the opposite conclusion was reached in a study of yeast TBP, in this case performed in vivo. As described above, TBP derivatives that impaired TBP-TATA or TBP-TFIIA interactions were activation defective (275, 447). In contrast, TBP replacements (E186A, E188A, and L189A) that disrupted TBP-TFIIB interactions did not block transcriptional activation, leading to the conclusion that TBP-TFIIB interaction is not generally limiting for transcription in vivo (276). This conclusion is further supported by TFIIB derivatives that are defective for TBP-TFIIB-DNA complex formation yet support viability and respond to transcriptional activators (18, 81). To resolve the discrepancy between the effects of the L189A and L189K replacements on activation, it was suggested that the activation defect associated with L189K is not due to impaired TBP-TFIIB interaction but, rather, to impaired TBP-TATA interaction, since L189 contacts both TFIIB and template DNA (276).
In another study of human TBP, the effects of an array of single amino acid replacements on basal and activated transcription, as well as TBP-DNA and TBP-TFIIB binding, were assessed (39). This study included replacements of residues E284, E286, and L289, which are equivalent to residues E186, E188, and L189 in yeast. E284R, E286R, and L287E replacements blocked TBP-TFIIB interaction (39). However, these three mutants were defective in both activated and basal transcription, although the E286R replacement caused only a twofold drop in activated transcription in transient-transfection assays (39). This dramatic effect on TBP-TFIIB interaction, coupled with the subtle effect on activated transcription, is consistent with the conclusion that TBP-TFIIB interaction is not generally limiting for activation (276). Perhaps impaired TBP-TFIIB interactions might readily be compensated for by interaction of TFIIB other components of the PIC (39) or by TFIIB-DNA interactions (270).
The discrepancies among these studies on the role of TBP-TFIIB interaction in activated transcription is likely to be accounted for by the different experimental methods used. Some were performed in vitro, whereas other were done in vivo with either yeast or human cells. The identification and characterization of activation-defective TFIIB mutants might resolve these discrepancies and ultimately provide valuable insights into the role of TBP-TFIIB interactions in gene activation.
Other TBP derivatives.
In the same genetic selection that uncovered the activation-defective K138T/Y139A double replacement that affects TBP-TFIIA interaction (447), four activation-defective single replacements, F148H, T153I, E236P, and F237D, were found (446). These replacements lie on the convex surface of TBP. None affects TATA binding, and the F148H, T153I, and E236P replacements do not affect interaction with TFIIA, TFIIB, or glutathione S-transferase–VP16. These activation defects were at least partially rescued by artificial recruitment of the F148H, T153I, and F273D derivatives to the promoter. These results were interpreted as evidence for a second step in transcriptional activation in vivo, one involving recruitment of TBP to the promoter and the other involving activator interaction with another component of the PIC after TBP recruitment (446). This conclusion is consistent with a mutational analysis of human TBP, where activation-defective mutations were identified on the convex surface of TBP that did not affect interaction with either TFIIA or TFIIB (39).
Suppressors of TBP derivatives.
The extensive collection of well-defined TBP derivatives is a lucrative source of primary mutations for isolation of suppressors. In one study, dosage-dependent suppression of the temperature sensitivity associated with the K133L/K138L double mutant identified a TFIIB homolog that is a component of the RNA pol III transcriptional machinery (45). In another case, the same factor was isolated as a suppressor of the P65S replacement in TBP (86). Consistent with its role in RNA polymerase III transcription, the same factor was isolated as a dominant suppressor of a tRNA “A block” promoter mutation (291). The gene encoding this factor is designated BRF1/TDS4/PCF4 and encodes the 67-kDa subunit of TFIIB. The N-terminal half of Brf1/Tds4/Pcf4 is significantly similar to TFIIB, an observation that underscores the similarity of RNA pol II and pol III transcriptional systems and suggests that Brf1/Tds4/Pcf4 is a key factor distinguishing the polymerase specificity of a gene (45, 86).
A novel TBP replacement, P191S, was generated as an intragenic suppressor of the Spt phenotype associated with the L205F replacement encoded by spt15-122 (10). P191 is located in the S2′-S3′ stirrup and, like L205, contacts the DNA backbone at the A residue of the first base pair of the TATA element. This result is consistent with the relaxed specificity of TATA element recognition associated with L205F.
The spt15-21 allele encodes a G174E replacement within the H1′ helix. G174E does not affect TBP stability, DNA binding, basal transcription, or transcriptional activation (123). In an effort to further define the G174E defect, intragenic and extragenic suppressors were isolated and defined. All three intragenic suppressors encode a single residue replacement, F237V, which compensates for the Spt phenotype of G174E but does not confer an Spt phenotype on its own (123). F237 immediately follows the H2′ helix, which is in proximity to the H1′ helix. All extragenic suppressors of G174E were the result of recessive mutations in the same gene, SPT3. Mutations in SPT3, like mutations in SPT7, SPT8, and SPT15 but unlike all other spt mutations, suppress the his4-917δ allele. The spt3 suppressors of spt15 are allele specific, consistent with physical interaction between TBP and Spt3 (123). These results suggest that Spt3 forms a complex with TBP distinct from TFIID.
A novel gene, designated RTF1, was recently uncovered as a genomic suppressor of the TBP L205F derivative (452). Rtf1 is a nuclear protein of unknown function with no apparent similarity to other proteins. Characterization of rtf1 suppressor and null mutations suggests that Rtf1 regulates the DNA binding properties of TBP and might play a role in recognition of nonconsensus TATA elements in vivo.
TFIIB
Overview.
Yeast TFIIB is a monomer of 38 kDa encoded by the SUA7 gene. SUA7 was initially identified in a genetic selection for suppressors of a translational defect at the CYC1 locus (366). Recessive sua7 mutations shifted transcription start site selection downstream of normal such that the translational impediment was eliminated from the cyc1 transcript. This result demonstrated that TFIIB functions in transcription start site selection in vivo.
TFIIB enters the PIC after TBP and as a prerequisite for recruitment of RNA pol II (42). TFIIB interacts directly with TBP and RNA pol II, as well as with other GTFs, including the RAP30 (174) and RAP74 (128) subunits of TFIIF and the TAFII40 subunit of TFIID (161). TFIIB-RAP74 binding precludes TFIIB-RAP30 binding, implying a dynamic interaction between TFIIB and TFIIF during PIC assembly (128). TFIIB has also been implicated as the direct target of many gene-specific transcriptional activators, leading to the proposal that certain activators stimulate transcription by TFIIB recruitment (288, 392).
Sequence analysis of the human and yeast genes encoding TFIIB revealed several structural motifs, including a zinc binding motif near the N terminus and two imperfect repeats encompassing most of the C-terminal two-thirds of the molecule (173, 303, 366). The C-terminal region folds into a protease-resistant core (cTFIIB) that binds TBP, whereas the N-terminal region interacts with RNA pol II-TFIIF (18, 19, 44, 174, 202, 304, 547). The N- and C-terminal domains engage in an intramolecular interaction that undergoes an activator-induced conformational change, allowing assembly of the PIC (391).
A solution structure for human cTFIIB (17) and a crystal structure for a TATA-TBP-cTFIIB ternary complex have been determined (341; reviewed in reference 340). cTFIIB consists of two similar domains, each consisting of five α-helices, corresponding approximately to the two imperfect repeats. The α-helices defined by the nuclear magnetic resonance structure are termed A1 to E1 and A2 to E2 for the first and second repeats, respectively (17). Based on primary-structure analysis, TFIIB was classified as a member of a protein superfamily that includes the cyclin A and retinoblastoma proteins (159). Consistent with that proposal, the tertiary structure of cyclin A is arranged similarly to cTFIIB, including two imperfect repeats, each arranged as five α-helices (228).
Footprinting (277) and cross-linking (102, 269) experiments demonstrated that TFIIB binds beneath and to one face of the TATA-TBP complex, a result confirmed by the crystal structure of the TATA-TBP-TFIIB ternary complex (Fig. 2) (341). A basic region within the D1 and E1 helices of the first repeat of cTFIIB interacts with the acidic C-terminal stirrup of TBP. Residues within the C2 and E2 helices of the second repeat contact DNA, upstream of the TATA box. Interestingly, helices D2 and E2 form a helix-turn-helix motif, which has been proposed to constitute a sequence-specific DNA binding domain (270). Accordingly, TFIIB, like TBP, is a sequence-specific GTF and the template sequence bound by TFIIB defines a novel promoter element (270). Photo-cross-linking studies demonstrated that other GTFs also directly bind promoter DNA (102, 249, 269, 390), raising the possibility that additional GTFs recognize promoter elements in a sequence-specific manner.
The N-terminal region was not included in either the crystal or solution structures of human TFIIB (17, 341). However, a solution structure for the metal binding domain of Pyrococcus TFIIB demonstrated that the N-terminal region forms a zinc ribbon (563). The zinc ribbon and core domain flank the most phylogenetically conserved region of the protein. This region plays an important but undefined role in transcription start site selection in vivo (18, 355, 367).
Start site selection.
The effects of sua7 on start site selection are not limited to the CYC1 gene but also affect other genes, including ADH1 (27, 366). Interestingly, the start site shift is always downstream of normal. Furthermore, the downstream start sites are never “new” sites but represent enhanced initiation at minor sites. Therefore, TFIIB defects shift the window within which certain start sites are recognized rather than altering the specificity of start site recognition. Also, the downstream shift is not a consequence of alternative TATA usage. For example, none of the sua7 suppressors compensate for Ty element insertions at the HIS4 or LYS2 loci, which is the basis for isolation of the spt class of suppressors (536).
Neither the mechanism of start site selection nor the basis of the sua7-induced downstream shift is understood. Nonetheless, RNA pol II is known to be an integral component of this process. In addition to sua7, selection for suppressors of the cyc1 translational defect yielded sua8 mutants. Moreover, sua7 and sua8 suppressors confer identical downstream start site shifts (27). The sua8 suppressors are allelic to RPB1, the gene encoding the largest subunit of RNA pol II. Furthermore, double sua7 sua8 mutants are inviable (synthetic lethality) and sua7/SUA7+ sua8/SUA8+ heterozygous diploids display sua phenotypes (nonallelic noncomplementation). These results imply that accurate start site selection involves interaction between TFIIB and the Rpb1 subunit of RNA pol II. This interpretation is consistent with results of biochemical studies. Pairwise replacement of RNA pol II and TFIIB from S. cerevisiae by their counterparts from S. pombe was both necessary and sufficient to shift start sites from the pattern characteristic of S. cerevisiae to that of S. pombe (284).
The sequences of the sua7 suppressor alleles were determined in an effort to define the role of TFIIB in start site selection. Each of four independent sua7 alleles encodes a single amino acid replacement: E62K, R78C, or R78S (355, 367). E62 and R78 lie within the phylogenetically conserved region of TFIIB, immediately downstream of the zinc ribbon. The opposite charge of E62 and R78 and the identical effects of replacements at these two positions on both start site selection and cold sensitivity imply that E62 and R78 interact, perhaps forming an ion pair. Consistent with this hypothesis, the inviability of an R78E replacement is suppressed by an E62R replacement, confirming a functional, if not direct, interaction between E62 and R78 (367). Although this interaction is clearly important for start site selection, it cannot be essential, since mutants expressing either E62K or R78C not only are viable but also exhibit only modestly impaired growth at 30°C. Also, residues in addition to E62 and R78 can affect start site selection, as demonstrated by the downstream shift associated with an R64E replacement (18).
What is the role of TFIIB in start site selection? The N-terminal region of TFIIB is critical for interaction with RNA pol II-TFIIF (19, 44), and the TFIIB R78C derivative binds RNA pol II with ∼100-fold-diminished affinity (53). The downstream start site shift therefore correlates with diminished TFIIB-RNA pol II interaction. Based on two-dimensional electron crystallographic data, the distance between TFIIB and the RNA pol II catalytic site is ∼110 Å, corresponding to 32 bp of B-form DNA (280). This is the approximate distance between the TATA box and start site for most RNA pol II promoters. Template DNA has been proposed to follow a linear path from the TATA box to the RNA pol II active site and that longer distances between TATA and start sites would result from RNA pol II scanning further downstream (280). Consistent with this proposal, RNA pol II forms an open promoter complex at a fixed distance of approximately 20 bp from the TATA box, regardless of the distance between TATA and start sites (158). To account for the variable distances between TATA and start sites in yeast, RNA pol II was proposed to reach downstream sites by template scanning (158). It was recently proposed that the mechanism of start site selection involves arrest of the scanning polymerase by specific promoter sequences in a manner analogous to arrest of the elongating polymerase (16). Accordingly, altered forms of TFIIB, Rpb1, and other factors that affect start site selection might allow RNA pol II to adopt conformations either more or less compatible with sequences at the start sites. Although reasonable, this model does not address whether or how RNA pol II would clear the promoter to scan for downstream start sites. It is also not clear why scanning might occur in S. cerevisiae yet transcription seems to occur at a fixed distance from TATA in other eukaryotes.
Human TFIIB does not functionally replace yeast TFIIB in vivo (433). Since TFIIB is a determinant of accurate start site selection (366), the differential spacing between TATA and start sites in human (25 to 30 bp) and yeast (40 to 120 bp) could account for the incompatibility of human TFIIB in yeast. By constructing chimeric human-yeast TFIIB hybrids, a species-specific region of TFIIB was identified within a solvent-exposed region of the first repeat cTFIIB (433). Interestingly, mutations within this region impair gene-specific transcriptional activation yet have only subtle effects on start site selection. Thus, the functional distinction between yeast and human TFIIB does not appear to be a consequence of the role of TFIIB in start site selection but is more likely to be accounted for by the differential responses to transcriptional activators (432, 433).
The SUA7 gene was also uncovered in another genetic selection. Mutations in eight different genes, designated soh1 to soh8, were isolated as suppressors of the temperature-sensitive growth defect of an hpr1 hyperrecombination mutant of S. cerevisiae (127). Two of these genes encode components of the general transcriptional machinery. SOH2 and SOH4 are identical to RPB2 and SUA7, respectively (126). Furthermore, SOH1 encodes a novel protein that interacts with components of both the DNA repair and transcriptional machinery. These results imply a link between transcription and recombination (126). A transcription-recombination link is further supported by other genetic results. Mutations in GCR3, whose product plays an undefined role in glycolytic gene expression, and in SRB2, which encodes a component of the SRB/mediator complex, also suppress the temperature-sensitive growth defect of hpr1Δ mutants (369, 495). Also, mutations in SPT4 and SPT6, members of the histone class of SPT genes (see below), confer a hyperrecombination phenotype (300). These results are consistent with earlier observations that link transcription with recombination (242, 482, 512), although the molecular basis of this process is unresolved.
Suppressors of TFIIB derivatives.
In an effort to identify factors that interact with TFIIB, the cold-sensitive phenotype of the sua7-1 mutant was exploited to isolate extragenic suppressors. Cold sensitivity is often associated with defects in assembly of multisubunit complexes (reviewed in reference 180); therefore, suppressors of cold sensitivity seemed likely to identify other components of the PIC or perhaps factors that facilitate PIC assembly.
Mutations in two genes, designated SSU71 and SSU73, were isolated as suppressors of sua7-1. In addition to suppression of cold sensitivity, the ssu71 suppressors caused heat lethality, but only in combination with sua7-1. This effect demonstrated a functional relationship between SSU71 and SUA7 and provided a phenotype for cloning SSU71. Sequence analysis of SSU71 identified an 82-kDa protein with sequence similarity to mammalian RAP74, the largest subunit of TFIIF. Subsequent comparison of the Ssu71-deduced amino acid sequence with that of Tfg1, the largest subunit of yeast TFIIF, revealed that Ssu71 and Tfg1 are identical. Whereas sua7-1 shifts start site selection downstream of normal, the ssu71 suppressors compensate for this effect, partially restoring the normal initiation pattern (459). Thus, the largest subunit of TFIIF genetically interacts with TFIIB, an interaction that can influence start site selection.
The ssu73-1 suppressor of sua7-1 is allelic to RPB9 (460). Like the ssu71 suppressors, ssu73-1 compensates for the downstream start site shift associated with sua7-1. The ssu73-1 allele encodes a nonsense mutation immediately following the second of two metal binding motifs within Rpb9. This motif is predicted to form a zinc ribbon that would be disrupted in the truncated Rpb9 derivative (460). Interestingly, the sua7-1-encoded E62K replacement lies near the end of the TFIIB zinc ribbon motif, suggesting that TFIIB and Rpb9 interact either with each other or with the DNA template via their zinc ribbon structures. In contrast to the ssu71 suppressors, which do not appear to affect start site selection in a SUA7 wild-type background (459), the ssu73 suppressor shifts initiation upstream of normal in both SUA7 wild-type and mutant backgrounds (460). This suggests that the Ssu71 (Tfg1) subunit of TFIIF and Rpb9 affect start site selection by different mechanisms.
A role for Rpb9 in start site selection was discovered in two independent studies. In one case, a selection for mutations that affect the spacing between the TATA element and start sites identified a gene designated shi (145), which is allelic to RPB9 (147). In the other study, transcription in the absence of Rpb9 shifted initiation upstream of normal both in vitro and in vivo (216). This effect was attributed solely to Rpb9 because the start site shift was rescued by recombinant Rpb9 and because RNA pol II isolated from an rpb9 null mutant was intact, lacking only the Rpb9 subunit (216). Thus, three independent studies demonstrate a role for Rpb9 in start site selection.
In contrast to SSU71 and SSU73, a mutation in the SSU72 gene was identified as an enhancer rather than a suppressor of the sua7-1 defect (459). Whereas a sua7-1 mutant is cold sensitive but not heat sensitive, a double sua7-1 ssu72-1 mutant is both cold and heat sensitive; moreover, the heat-sensitive phenotype is dependent upon both sua7 and ssu72. The ssu72-1 mutation dramatically enhances the downstream start site shift associated with sua7-1, an effect that can be rescued by either wild-type SUA7 or SSU72. SSU72 is an essential gene, encoding a novel protein of 206 amino acids with unknown function. Although a mammalian counterpart of Ssu72 has not been found in cell-free transcription assays, a human Ssu72 homolog (64% similar) exists in the databases.
The SUB1 gene was also identified as a suppressor of a TFIIB defect. In this case, high-copy-number expression of SUB1 suppressed the cold-sensitive growth defect of TFIIB R78H and E62G derivatives (258). Sub1 directly interacts with TFIIB in vitro and inhibits formation of the TATA-TBP-TFIIB promoter complex. SUB1 is identical to TSP1 and encodes a homolog of the mammalian PC4 transcriptional coactivator (195, 258).
TFIIF
The subunits of TFIIF were identified as RAP30 and RAP74 based on affinity of these two proteins for RNA pol II (52). RAP30 and RAP74 were required for accurate transcription initiation from several promoters, defining the RAP30-RAP74 complex as a general initiation factor that binds RNA pol II (141). Although TFIIF was not initially identified as one of the four HeLa cell chromatographic fractions (TFIIA, TFIIB, TFIID, and TFIIE) required for accurate initiation by RNA pol II (418), further purification resolved the TFIIE fraction into two factors, TFIIE and TFIIF, both of which are essential for initiation (142). Purification of the TFIIF fraction defined two subunits, identical to RAP30 and RAP74 (139). TFIIF was also identified in human cells as factor FC (253), in rat cells as factor βγ (95), and in Drosophila cells as factor 5 (377). In each case, these factors were required for specific initiation of transcription in vitro.
TFIIF has several characteristics reminiscent of bacterial ς factors. These include tight binding of TFIIF to RNA polymerase; suppression of nonspecific binding of RNA pol II to DNA; and stabilization of the PIC (96, 166). Both subunits exhibit limited sequence similarity to bacterial ς factors (149, 315, 444, 459, 552). It is not clear whether these structural similarities reflect functional similarities. However, human RAP30-RAP74 binds E. coli RNA polymerase and can be displaced by ς70 (315). Moreover, polymerase binding is attributed to region 2.1 of ς70, which is the region of similarity between ς70 and RAP30 (315). Thus, RAP30 appears to be partially analogous to bacterial ς70.
TFIIF probably does not play a significant role in promoter selectivity but contributes to PIC stability. Photo-cross-linking studies have defined the topology of a TBP-TFIIB-TFIIF-RNA pol II-TFIIE promoter complex. RAP74 and RAP30 bind promoter DNA between the TATA box and start site, a region where TFIIE and RNA pol II also cross-link (102, 249, 390). RAP74 also binds DNA upstream of TATA, inducing a conformational change that affects the position of RNA pol II relative to the DNA template (144).
In addition to its role in initiation, TFIIF functions in transcriptional elongation by suppressing transient pausing of the polymerase (24, 32, 142, 225, 377). Although RAP30 and RAP74 were initially thought to function exclusively in initiation and in elongation, respectively, both subunits are now known to function in both processes (476).
Yeast TFIIF (factor g) stably associates with RNA polymerase, is required for specific initiation at all promoters tested, and is composed of three subunits with apparent molecular masses of 105, 54, and 30 kDa (197). The genes encoding the two largest subunits of TFIIF were cloned based on the partial sequence of the purified proteins (196). Analysis of the deduced amino acid sequences of TFG1 and TFG2 revealed 50 and 51% similarity to human RAP74 and RAP30, respectively, thereby confirming the relationship between factor g and TFIIF. TFG1 and TFG2 are present in single copy, and both are essential for cell viability (196, 459). TFG3 encodes the 30-kDa subunit and is identical to ANC1, a gene originally identified based on genetic interaction with ACT1 (531). Tfg3 is less tightly associated with the complex than are Tfg1 and Tfg2 and has no counterpart in mammalian TFIIF, although Tfg3 is similar in sequence to the leukemogenic proteins ENL and AF-9 (55). Tfg3 is found in RNA pol II holoenzyme complexes (252, 261, 515), probably as a component of TFIIF. Interestingly, Tfg3 is identical to the TAFII30 subunit of the yeast TFIID complex (371) and to the Swp29 subunit of the SWI/SNF chromatin-remodeling complex (55). As such, Tfg3 establishes a connection between basal and regulatory components of the transcriptional machinery (55, 196). Tfg3 is the only yeast GTF that is not essential for cell viability (196), perhaps because yeast contains a Tfg3 homolog (YOR213c), which is a subunit of the RSC chromatin-remodeling complex (57).
The only gene encoding a TFIIF subunit to be identified in a genetic selection for transcription factors is TFG1. As described in the TFIIB section (above), recessive mutations in the SSU71 gene were isolated as suppressors of a TFIIB defect (sua7) that altered transcription start site selection (459). Sequence analysis revealed that Ssu71 is homologous to RAP74 and identical to Tfg1 (196, 459). Thus, TFIIF functionally interacts with TFIIB. Although ssu71 suppressor alleles compensate for the downstream start site shift caused by the TFIIB defect, it remains to be determined whether TFIIF plays a direct role in the accuracy of initiation and whether Tfg2 and Tfg3 are involved in this process.
TFIIE
Order-of-addition experiments demonstrated that TFIIE enters the PIC after RNA pol II and prior to TFIIH (42, 140). TFIIE interacts directly with the unphosphorylated form of RNA pol II (IIA), with both subunits of TFIIF, and with TFIIH (142, 312). TFIIE has also been implicated as the direct target of certain gene-specific transcriptional activators (416, 562). Functions attributed to TFIIE include recruitment of TFIIH to the PIC, stimulation of TFIIH-dependent phosphorylation of the RNA pol II CTD, and stimulation of TFIIH-dependent ATP hydrolysis (293, 347, 348).
TFIIE was purified to homogeneity from HeLa cell nuclear extracts (220, 350). Biochemical analyses revealed that human TFIIE is composed of 56-kDa (TFIIE-α) and 34-kDa (TFIIE-β) subunits that form an α2β2 heterotetramer. However, two-dimensional crystallography of a TFIIE-RNA pol II complex suggests that yeast TFIIE exits as an αβ dimer (280). The human genes encoding both subunits were cloned based on the partial sequences of the purified proteins (349, 363, 457). Both subunits are highly charged, with pI values of 4.5 and 9.5 for TFIIE-α and TFIIE-β, respectively. Neither subunit is closely related to any other proteins in the databases, although both subunits include defined structural motifs. These include a zinc ribbon motif (C-X2-C-X21-C-X2-C) and protein kinase consensus sequences in TFIIE-α and a consensus nucleotide binding site in TFIIE-β. However, no enzymatic activities have been demonstrated for TFIIE, including DNA-dependent ATPase, topoisomerase, or helicase activities (350; reviewed in reference 558). Both subunits exhibit limited sequence similarities to bacterial sigma factor. Whether these structural similarities are functionally significant remains to be determined.
Yeast factor a is required for accurate transcription in vitro (420). Characterization of purified factor a identified two subunits with apparent molecular masses of 66 and 43 kDa, suggesting that factor a might be the yeast counterpart of either TFIIE or TFIIF (419). The relationship between factor a and metazoan GTFs was clarified after cloning of the yeast genes encoding the two subunits. These genes, designated TFA1 and TFA2, were cloned based on partial sequences of purified factor a (133). TFA1 and TFA2 are present in single copy in the yeast genome, and both genes are essential for cell viability. The deduced amino acid sequences revealed that Tfa1 and Tfa2 are homologous to TFIIE-α and TFIIE-β, respectively, exhibiting 52 and 53% sequence similarity. Like their metazoan counterparts, Tfa1 is acidic (pI = 4.1) and Tfa2 is basic (pI = 10.4). Thus, factor a is the yeast counterpart of TFIIE. In contrast to metazoan TFIIE, neither the kinase nor nucleotide binding motifs are present in Tfa1 or Tfa2, suggesting that these motifs might not be functionally relevant.
Like human TFIIE-α, the sequence of Tfa1 includes a zinc ribbon motif (C-X2-C-X21-C-X2-C), suggesting that this motif plays an important, albeit undefined role in TFIIE function. A similar motif is present in TFIIB and in the elongation factor TFIIS, which is able to bind single- and double-stranded DNA (380). This suggests that TFIIB and TFIIE might interact to form or stabilize melted DNA in the initiator region (351). Indeed, yeast TFIIE binds single-stranded DNA (266), a result which could account for the dispensability of TFIIE for transcription initiation from premelted template DNA (208, 354, 480).
TFIIE is functionally linked to TFIIH. This was elegantly demonstrated by the inability of S. cerevisiae TFIIE to functionally replace the S. pombe counterpart of TFIIE in a reconstituted transcription system, unless they are exchanged as a TFIIE-TFIIH pair (284). Still, TFIIE is the least well understood of the GTFs. So far, yeast genetics has provided few clues; neither TFA1 nor TFA2 has turned up in any of the genetic selections designed to identify transcription factors. Structure-function analysis suggests that TFIIE might act as a checkpoint for formation of the PIC via its control of TFIIH recruitment and activities (347). Furthermore, two-dimensional crystallography of a TFIIE-RNA pol II complex suggested that TFIIE promotes a conformational switch at the active center upon RNA pol II-DNA interaction (280).
Mutational analysis of the TFA1-encoded subunit of yeast TFIIE identified two functionally distinct domains. Substitutions of cysteine residues that comprise the zinc ribbon motif in the N-terminal half of Tfa1 confer growth defects at elevated temperature (266, 487), whereas deletions within the C-terminal portion confer growth defects at reduced temperature (266, 411). Either depletion of Tfa1 (266) or growth of tfa1 conditional mutants at the nonpermissive temperature (411, 487) rapidly diminished the steady-state levels of poly(A)+ RNA, establishing that TFIIE is essential for RNA pol II-mediated transcription in vivo. However, analysis of specific mRNA species revealed a promoter-specific dependence for TFIIE (411, 487), an effect consistent with the differential dependence of mammalian promoters upon TFIIE (208, 358). It is not clear why some promoters are more dependent upon TFIIE than others. TFIIE dependence has been reported to correlate with the presence of a promoter TATA element (407, 411), although another study argues against that correlation (487). Interestingly, the Gal11 component of the SRB/mediator physically interacts with both subunits of TFIIE and Gal11-mediated stimulation of transcription is TFIIE dependent (407, 409).
TFIIH
The GTFs TFIIB, TFIID, TFIIE, and TFIIF, along with TFIIA, were sufficient for accurate transcription initiation by RNA pol II in vitro. It was not until these factors were more extensively purified or replaced by recombinant factors that the requirement for an additional factor, TFIIH, was discovered in mammalian cells. TFIIH was identified from rat liver cells (factor δ) (97), human cells (TFIIH, BTF2) (140, 157), and yeast (factor b) (131). The functional analogy of factor δ, factor b, BTF2, and TFIIH was suggested by similar polypeptide compositions and supported by immunological cross-reactivities and common subunit activities.
TFIIH is the only GTF with known enzymatic activities, which include DNA-dependent ATPase (97, 401), ATP-dependent DNA helicase (422, 430), and CTD kinase (132, 293, 429) activities. In addition to its fundamental role in transcription, TFIIH functions as an essential component in nucleotide excision repair (NER) and has been implicated in mammalian cell cycle progression. Consistent with these multiple functions, TFIIH is the most complex of the GTFs, consisting of nine subunits with a total mass of approximately 500 kDa, comparable to the mass of RNA pol II.
TFIIH performs critical roles at both initiation and postinitiation stages of transcription. Formation of an open promoter complex by RNA pol II requires ATP-dependent DNA helicase activity (229, 523). Since TFIIE, TFIIH, and ATP hydrolysis are dispensable for initiation from supercoiled promoter DNA (357, 358, 493) or from a premelted template (208, 354, 480), open-complex formation appears to be mediated by the TFIIH DNA helicase activity. Although another study implicated TFIIE and TFIIH in promoter clearance rather than open-complex formation (162), subsequent studies strongly support a role for ATP, TFIIE, and TFIIH in open-complex formation (120, 207).
TFIIH also regulates the transition from transcription initiation to elongation, presumably mediated by the CTD kinase. This was suggested by the observation that RNA pol II enters the PIC in the unphosphorylated IIA form and is converted to the phosphorylated IIO form by CTD kinase upon promoter clearance (104, 346). Presumably, phosphorylation of the CTD causes a conformational change within the PIC, resulting in disruption of the CTD-TBP interaction and promoter clearance (496). Recently, TFIIH was also shown to promote the transition from very early elongation complexes to stable elongation complexes (119). Thus, TFIIH performs multiple roles in transcription, affecting steps before, during, and immediately after initiation (119).
A role for TFIIH in NER was suggested by its subunit composition (reviewed in reference 466). The largest subunit of human TFIIH, p89, is identical to the DNA excision repair protein ERCC3 (excision repair cross-complement), which complements the DNA repair deficiency associated with the XPB gene defect in xeroderma pigmentosum patients (422). Additional subunits of TFIIH were subsequently identified as components of the NER complex (116, 217, 421, 528). A direct role for TFIIH in NER was established by the ability of purified TFIIH to rescue the repair deficiency of mammalian and yeast TFIIH mutants (116, 505, 527). The discovery that TFIIH plays a dual role in transcription and DNA repair nicely accounts for earlier observations suggesting that transcriptionally active genes are preferentially repaired (30, 322). The Rad2 and Rad4 proteins of the DNA repair machinery, although not components of TFIIH, interact directly with TFIIH subunits, suggesting a mechanism for preferential targeting of repair proteins to actively transcribing genes (21, 22).
The yeast homolog of TFIIH, factor b, was initially identified as a three-subunit complex of 85, 73, and 55 kDa that, along with TBP, would restore transcriptional activity to heat-inactivated nuclear extracts (131). The 73-kDa subunit, encoded by the TFB1 gene, was subsequently used to purify a five-subunit core-TFIIH complex of 85-, 73-, 55-, 50-, and 38-kDa polypeptides (135). This complex resembled the subunit composition of human BTF2 and rat δ, further supporting the premise that factor b is the yeast counterpart of mammalian TFIIH. Although core-TFIIH was able to replace heat-inactivated factor b in a crude in vitro transcription system, it was nonfunctional in a highly purified, reconstituted system. This in turn provided an assay for the isolation of holo-TFIIH, composed of the five-subunit core-TFIIH, Ssl2, and two additional subunits of 47/45 and 33 kDa that comprise a subcomplex denoted TFIIK (465). Core-TFIIH and holo-TFIIH were distinguished in three functional assays: (i) CTD kinase activity, (ii) promoter-specific transcription in heat-treated nuclear extracts, and (iii) promoter-specific transcription from reconstituted components. Holo-TFIIH is active in all three, whereas core-TFIIH is functional only in the heat-treated extract. Therefore, core-TFIIH and TFIIK are both essential transcription factors.
The observation that TFIIH exists in multiple forms and physically interacts with components of the NER machinery suggested that the form of TFIIH involved in NER is different from that required for transcription. Indeed, the TFIIK subcomplex is dispensable for NER. NER-proficient TFIIH includes core-TFIIH, Ssl2, and all the other known NER proteins, including Rad1, Rad2, Rad4, Rad10, and Rad14. The structural distinction between holo-TFIIH and the repairosome suggested a mechanism for transcription-coupled NER: when associated with RNA pol II at the promoter, core-TFIIH would bind more tightly to TFIIK, but in the presence of DNA damage, a conformational change would displace TFIIK in favor of NER proteins (467). Although this is an appealing model, other data argues that NER components do not exist in a preassembled repairosome but instead assemble sequentially at the site of DNA damage, with only Rad1-Rad10-Rad14 existing as a complex (172). In this case, NER complex assembly would be similar in yeast and prokaryotes (reviewed in reference 414).
It now appears that all subunits of TFIIH have been identified and that their corresponding genes have been cloned (134). Notable features of specific subunits are summarized below and in Table 1.
The 85-kDa subunit of factor b was identified as Rad3, a 5′→3′ DNA helicase shown previously to function in NER (135). Rad3 is homologous to the DNA excision repair protein ERCC2, which complements the DNA repair deficiency of the xeroderma pigmentosum XPD gene defect. Immunodepletion of Rad3 inhibited transcription in vitro (135), and a rad3(Ts) mutant exhibited dramatically reduced mRNA synthesis at the restrictive temperature in vivo (171). A rad3 mutation (rad3-21) that presumably eliminates ATPase/helicase activity and affects NER had no effect on transcription. Thus, Rad3 appears to have dual functions, one in repair and the other in transcription, with the ATPase/helicase activity required for NER but not for transcription (135, 171).
The 50-kDa subunit of factor b was identified as the product of the SSL1 gene (135). SSL1 was identified previously in a genetic selection for suppressors of an artificial stem-loop structure in the transcribed leader region of the his4 gene (553). Although it is not clear how ssl1 suppresses the his4 defect, a role for Ssl1 in DNA repair was suggested by the increased UV sensitivity of ssl1 mutants (553).
SSL2 was identified in the same genetic selection that uncovered SSL1 (169). Sequence analysis of the cloned gene revealed that SSL2 is the yeast homolog of human XPB, a 3′→5′ DNA helicase. Moreover, deletion of the Ssl2 C terminus conferred UV sensitivity. SSL2 was cloned independently based on hybridization with a human ERCC3 probe and designated RAD25 (356). Although none of the five subunits of core-TFIIH cross-reacted with anti-Ssl2 antibody, Ssl2 was detected at earlier stages of factor b purification. Moreover, antisera to both Rad3 and Tfb1 coimmunoprecipitated Ssl2 (135), and Ssl2 interacts with Rad3 in vitro (22). Therefore, Ssl2 associates with core-TFIIH, but is not a subunit of the core complex. This contrasts with human ERCC3, which not only is a component of TFIIH but, as noted above, provided the initial evidence for the dual role of TFIIH in transcription and NER.
The TFIIK subcomplex of TFIIH includes the KIN28-encoded protein kinase and the CCL1-encoded cyclin H homolog (136, 464). Kin28 is a member of the p34/cdc2/Cdc28 family of cyclin-dependent protein kinases and is homologous to MO15/cdk7, the catalytic subunit of human TFIIH (400). A kin28(Ts) mutant displays a rapid decline in CTD phosphorylation at the restrictive temperature, demonstrating that the Kin28 subunit of TFIIH is a principal effector of CTD phosphorylation in vivo (498). The cyclin H homolog of TFIIK was identified based on ccl1 suppression of a kin28 mutation; physical interaction between Kin28 and Ccl1 was established by using the yeast two-hybrid system (498). The 47- and 45-kDa subunits of TFIIK were subsequently identified as the products of CCL1, confirming that TFIIK is a Cdk-cyclin dimer, composed of the Kin28 kinase and either the 47- or 45-kDa form of cyclin H (464). The human Cdk7-cyclin H subcomplex of TFIIH includes a third subunit, Mat1 (1). This heterotrimeric complex comprises the Cdk-activating kinase, denoted CAK (see below). Yeast TFIIH includes a homolog of Mat1, denoted Tfb3, but unlike Mat1, Tfb3 is found in the core-TFIIH complex rather than in TFIIK (134). Tfb3 and Kin28 physically interact, suggesting that Tfb3 links core-TFIIH with TFIIK.
Progression through the cell cycle is controlled by Cdks, which include p34/cdc2/Cdc28. Cdk activation requires phosphorylation, catalyzed by CAK. The identification of the CAK subunits Cdk7 and cyclin H as subunits of TFIIH suggested that in addition to its roles in transcription and repair, TFIIH functions in cell cycle control (137, 431, 437). Although this might be the case in mammalian cells, Kin28 neither regulates phosphorylation of the yeast cell cycle kinase Cdc28 nor has CAK activity in vitro (84). Rather, the kinase activity of CAK in S. cerevisiae is encoded by CAK1/CIV1, a protein that is active as a monomer and is not a component of TFIIH (125, 236, 485). Thus, TFIIH does not control cell cycle progression by regulating Cdk activity in S. cerevisiae; rather, S. cerevisiae appears to have evolved two protein kinases, Kin28 and Cak1, to carry out the functions of Cdk7 in vertebrates.
TRANSCRIPTIONAL COACTIVATORS
Transcriptional coactivators, also known as mediators or adapters, are required for transcriptional activation. Coactivators are distinct from GTFs in that they are dispensable for basal-level transcription in vitro and distinct from activators in that most do not directly bind DNA and none appears to bind DNA in a sequence-specific manner. In some cases, coactivators appear to bridge the interaction between gene-specific activator proteins and GTFs, whereas in other cases, coactivators facilitate chromatin remodeling. Several functionally distinct classes of coactivators have been described. These include the TAF components of TFIID, the SRB/mediator complex that associates with RNA pol II, TFIIA, SAGA and related complexes that catalyze nucleosomal histone acetylation, and the SWI/SNF and related chromatin-remodeling complexes. Additional coactivators have been described in mammalian systems, including the general cofactor designated USA (upstream stimulatory activity). Each of these classes of coactivators is reviewed here. A schematic summary of coactivators and their functions is presented in Fig. 4.
FIG. 4.
Schematic summary of yeast coactivators and their activities. Transcriptional activation by a UAS-activator complex can occur by direct interaction between an acidic activator protein and components of the core transcriptional machinery or can be indirect, mediated by coactivators that interact either with components of the core transcriptional machinery (TFIIA, TAFs, SRB/mediator) or with nucleosomes (SWI/SNF, HATs). TFIIA interacts with the core transcriptional machinery through TBP, functioning as either an antirepressor or coactivator. TAFs also interact with TBP as components of the TFIID complex. Although initially thought to be requisite coactivators of transcription, TAFs now appear to be required for activation of only a subset of genes with noncanonical TATA elements. SRB/mediator is a component of an RNA pol II holoenzyme complex, interacting with RNA pol II through the CTD of Rpb1. The SRB/mediator includes subunits that function in transcriptional repression as well as activation. Several HATs have been identified in yeast, including the Gcn5-containing SAGA complex. HATs appear to mediate transcriptional activation by acetylation of nucleosomal histones, resulting in chromatin remodeling. The SWI/SNF complex also appears to facilitate chromatin remodeling, in this case by promoting nucleosome displacement in an ATP-dependent manner.
Additional coactivators that are either gene or cell type specific have been described in metazoan systems. These include the thyroid hormone receptor-associated proteins (143) and the B-cell-specific coactivator OCA-B/OBF-1/Bob-1 (250). Although certainly relevant to the mechanisms of transcriptional activation, these cofactors are beyond the scope of this review and are not included here.
TBP-Associated Factors
TBP is a universal transcription factor, required by all three RNA polymerases (reviewed in reference 198). In each case, TBP is associated with a distinct set of factors, which are defined by either copurification or coimmunoprecipitation with TBP. Four distinct TBP complexes have been described in metazoan systems. TFIID is specific for RNA pol II and includes TBP plus 8 to 11 polypeptides. SL1 (human) is specific for RNA pol I and consists of TBP plus three TAFs. TFIIIB is specific for RNA pol III and is composed of TBP plus at least two additional polypeptides. SNAPC is another RNA pol III TBP complex, required for transcription of certain small nuclear RNA (snRNA) genes. This section is restricted to RNA pol II-specific TAFs, with emphasis on the yeast system. An excellent review of the biochemistry and structural biology of TFIID was published recently (50).
Although TBP is sufficient for promoter recognition and subsequent assembly of other factors into a functional PIC (43), transcriptional activation in metazoan systems is observed only when the PIC is assembled with the multisubunit TFIID complex (204, 379). This observation led to the discovery of the TAFs and the hypothesis that TAFs are requisite mediators of transcriptional activation (reviewed in reference 378). Consistent with this hypothesis, certain TAFs directly contact activator proteins whereas other TAFs directly bind either GTFs or promoter DNA (48, 71, 161, 203, 486, 507). Thus, TAFs were proposed to function in transcriptional activation by relaying information from activators to the core transcriptional machinery.
In contrast to human and Drosophila TBP, yeast TBP was initially thought to exist in a monomeric form, resulting in interchangeable use of the terms “TBP” and “TFIID” (43, 211). However, mutations in the class of SPT genes that includes SPT15 (TBP) confer a related set of pleiotropic phenotypes and the product of one of these genes, SPT3, physically interacts with TBP (123). These results suggested that at least a portion of TBP is complexed with other factors in vivo. Indeed, under native conditions, the majority of cellular TBP chromatographed as a large complex and immunoprecipitation of TBP resulted in copurification of nine polypeptides (373). One of these proteins was identified as Brf1, a subunit of the RNA pol III-specific factor TFIIIB, and the purified complex contains TFIIIB activity in vitro (373). Another protein was identified as Mot1, which functions in RNA pol II transcription, apparently by displacing TBP from DNA (372). These results established the existence of yeast TAFs, defined simply as proteins stably associated with TBP.
The polypeptide composition of an immunopurified TBP-TAF preparation suggested that in addition to TFIIIB and Mot1, the yeast counterpart of TFIID was included. Indeed, the TBP-TAF complex functioned as a coactivator, conferring transcriptional activation by RNA pol II in vitro (371). The genes encoding three of these TAFs, TAF130, TAF90, and TAF60, were cloned based on partial sequence of the purified proteins; a fourth gene, TSM1, had been cloned previously based on physical linkage to the MAT locus (385). Sequence analysis revealed that all four proteins are homologous to Drosophila and/or human TFIID components.
Yeast TFIID was also identified by affinity chromatography with TBP as the ligand (388). Consistent with metazoan TFIID, and similar to the immunopurified TBP-TAF preparation described above (371), this complex was required for activated transcription in vitro. The genes for two components of this complex, TAF145 and TAF90, encode homologs of higher eukaryote TAFII250 and TAFII80, respectively, and are identical to yeast TAF130 and TAF90, described independently (371). These studies established that yeast TBP exists in distinct complexes that are the counterparts of higher eukaryote TFIID and TFIIIB and in a complex with Mot1. In a separate study, yeast TBP was found in a distinct complex (TBP-Rrn6-Rrn7-Rrn11) that is functionally related to the mammalian RNA pol I transcription factor SL1, although there are no obvious sequence similarities between the RRN proteins and SL1 subunits (287).
A computer search of the yeast genome database identified additional TAF genes based on sequence similarity to known human and Drosophila TAFs (329). Each of these proteins coimmunoprecipitates with TBP, establishing that they are bona fide TAFs. Furthermore, all are present in a complex with TAFII145/TAFII130, which is thought to be the scaffold for TFIID assembly. In total, 12 TAF subunits of yeast TFIID have been identified (255, 256, 329, 371, 388, 517). With the exception of TFG3, all of the genes encoding these subunits are essential for cell viability. Also, with only one exception, all metazoan TAFs have a homolog in yeast. The exception is TAFII110, which is required for activation by the glutamine-rich activator Sp1; interestingly, glutamine-rich activators do not function in yeast, perhaps due to the absence of a TAFII110 homolog (329). A summary of yeast TFIID subunits is presented in Table 3.
TABLE 3.
TAF subunits of yeast TFIID
| Factor | Mass (kDa) | Gene(s) | Essential | Characteristics | Metazoan homolog(s) | Reference(s) |
|---|---|---|---|---|---|---|
| TAFII150 | 155 | TSM1 | Yes | dTAFII150 | 371, 385 | |
| TAFII145/TAFII130 | 121 | TAF130 | Yes | Scaffold for assembly of TFIID; directly contacts TBP | hTAFII250 (CCG1), dTAFII230 | 371, 385 |
| TAFII90 | 89 | TAF90 | Yes | dTAFII80 binds TFIIEα and TFIIFα | hTAFII100, dTAFII80 | 371, 388 |
| TAFII67 | 67 | TAF67 | Yes | hTAFII55 | 329 | |
| TAFII61/TAFII68 | 61 | TAF61 | Yes | Structural similarity to histone H2B | hTAFII15/20, dTAFII30α (p28/p22) | 329, 517 |
| TAFII60 | 58 | TAF60 | Yes | Structural similarity to histone H4 | hTAFII80, dTAFII60 | 371 |
| TAFII47 | 40 | TAF47 | Yes | None | 517 | |
| TAFII40 | 41 | TAF40 | Yes | dTAFII40 binds TFIIB | hTAFII28, dTAFII30β | 255, 329 |
| TAFII30 | 27 | TAF30, ANC1, TFG3, SWP29 | No | Subunit common to TFIID, TFIIF, and the SWI/SNF complex | AF-9, ENL | 55, 196, 531 |
| TAFII23/TAFII25 | 23 | TAF25 | Yes | hTAFII30 | 256, 329 | |
| TAFII19 | 19 | TAF19, FUN81 | hTAFII18 | 329 | ||
| TAFII17 | 17 | TAF17 | Yes | Structural similarity to histone H3 | hTAFII32, dTAFII40/42 | 329 |
Immunodepletion of yeast TFIID by TAF-specific antibodies blocks in vitro transcription by RNA pol II but not by RNA pol I or RNA pol III (255). Furthermore, the degree of inhibition parallels the degree of TAF depletion. These results confirm that TFIID is indeed RNA pol II specific and that TFIID is the principal, if not sole, TBP-TAF complex specific to RNA pol II in yeast. As noted above, TAFII30 is identical to the Tfg3 subunit of TFIIF and to the Swp29 subunit of SWI/SNF and is a component of the RNA pol II holoenzyme (55, 196). The TFG3-encoded TAFII30 subunit therefore establishes a connection between the core and coactivator components of the transcriptional machinery.
It is interesting that none of the genes encoding TAF subunits of TFIID was uncovered in any of the extensive genetic selection schemes designed to identify transcription factors. Only TAF30/TFG3/SWP29/ANC1 turned up in a genetic selection, in this case based on a search for cytoskeletal components that interact with actin (actin noncomplementing) (531). Although MOT1 was identified genetically (105, 364) and the Mot1 protein exists in a complex with TBP (372), the Mot1-TBP complex is distinct from TFIID (see the Mot1 section, below). Thus, yeast genetics provided no insight into TFIID function.
The structural and functional conservation of TFIID in human, Drosophila and yeast and the essential nature of the yeast genes encoding yeast TFIID clearly point to a critical role for TAFs in cell physiology. Nonetheless, the function of TAFs remains unclear. It had generally been assumed that TAFs were requisite coactivators of transcription, acting as the targets of gene-specific activator proteins (reviewed in reference 378). The identification of the yeast genes encoding TFIID allowed that premise to be tested directly. Quite unexpectedly, depletion or inactivation of several TAFs, including TAFII145/TAFII130, the scaffold for TFIID assembly and the only TAF known to directly bind TBP, did not compromise transcriptional activation in vivo (328, 517). The only exception was reduced activation from promoters lacking canonical TATA elements (328). This result cannot be explained by functional redundancy, since the yeast genome includes only a single homolog for each of the metazoan TFIID subunits (329). Thus, in stark contrast to the TFIID requirement for activation in metazoan in vitro transcription systems, TAFs are not generally required for activation in yeast (reviewed in references 183 and 479).
What, then, is the essential function of TAFs? One possibility is that TAFs are in fact essential coactivators but only for transcription of a subset of genes. This possibility is supported by the failure of a taf90(Ts) mutant to progress through the G2/M phase of the cell cycle at the restrictive temperature (5). Furthermore, a taf145(Ts) mutation blocks transcription of G1/S cyclin genes at the restrictive temperature (518). A specialized role for TAFs in transcription of cell cycle-specific genes is further supported by diminished levels of TAFs in the stationary (G0) phase whereas GTF levels are unaffected (518). Indeed, a role for TAFs in the regulation of cell cycle-specific genes was suggested by the identification of mammalian TAFII250 as the product of the CCG1 gene, which is required for passage through the G1 phase of the cell cycle (406). Consistent with this observation, a CCG1 mutant is defective in transcriptional activation of the cell cycle-regulated cyclin A promoter but not the c-fos promoter (519). The effects of TAF mutations have also been described in Drosophila, where altered forms of TAFII60 and TAFII110 diminished the transcription of Bicoid-dependent genes in the developing embryo (417). Taken together, TAFs appear to be critical coactivators of a subset of genes but are not required for the activation of all, or even most, RNA pol II-dependent genes.
The mechanism by which TAFs facilitate expression of specific genes is unknown. The TAFII250-dependent transcription of cyclin A is dependent upon the ATF activator protein (520). However, yeast TAFII145/TAFII130 (TAFII250 homolog) was found to be a core promoter selectivity factor, indicating that TAFII145/TAFII130 facilitates expression of specific genes through the core promoter, rather than by activator-TAFII145/TAFII130 interaction (434). This discrepancy between might be accounted for by different mechanisms of TAFII145- and TAFII250-mediated transcriptional activation in yeast and metazoan systems. It is intriguing, however, that TAFII250-dependent promoters lack canonical TATA boxes (520), and core promoter elements that confers TAFII145/TAFII130 dependence do not include the TATA box (434). These results support a role for TAFII250 and TAFII145/TAFII130 in mediating activation from promoters lacking consensus TATA sequences, a conclusion consistent with the earlier observation that TAFII145/TAFII130 is important for transcription from promoters lacking consensus TATA sequences (328).
Enzymatic activities have been reported for one subunit of TFIID. Human TAFII250 is a bipartite protein kinase with specificity for the RAP74 subunit of TFIIF (112). TAFII250 is also a histone acetyltransferase (HAT) (327). Thus, TAFII250 appears to play multiple roles in transcriptional activation, including phosphorylation of TFIIF and acetylation of histones. The yeast homolog of TAFII250, TAFII145/TAFII130, also has HAT activity (327). Sequence alignment between human TAFII250 and yeast TAFII145/TAFII130 revealed that neither the kinase domain nor TFIIF interaction domain is conserved in the yeast protein, suggesting that the TAFII250 kinase plays a promoter-specific role in transcriptional activation in higher eukaryotes (112).
Recent insights have been made into the structural organization of TFIID (reviewed in reference 206). Sequence analysis of TAFs revealed structural similarity to histones: Drosophila TAFII42 (dTAFII42) and human TAFII31 (hTAFII31) resemble histone H3; dTAFII62 and hTAFII80 resemble H4; and dTAFII30α and hTAFII20 resemble H2B (an H2A homolog has not been identified) (50). These similarities in primary structure extend to the tertiary and quaternary structures. The N-terminal portions of dTAFII42 and dTAFII62 form canonical histone folds that mediate the formation of a heterotetrameric (dTAFII42-dTAFII62)2 complex resembling the (H3-H4)2 tetrameric core of the histone octamer (545). Moreover, the structural relevance of the octamer-like structure is supported by biochemical studies (205, 333). These results establish the existence of a histone octamer-like structure within TFIID. However, the recent crystal structure of the nucleosome core particle defined arginine side chains as a predominant feature of the histone-fold-DNA interaction, yet these arginine residues are not conserved in the TAFs (296). Apparently, if these TAFs do bind DNA, the mode of interaction is likely to be different from that in the nucleosome (296). Whether an octamer-like structure is a component of TFIID in yeast is not known. However, yeast homologs of dTAFII62/hTAFII80 (TAFII60), dTAFII42/hTAFII31 (TAFII17) and dTAFII30α/hTAFII20 (TAFII61/TAFII67) have been identified (Table 3). In each case, the sequence similarity includes the histone-fold motif.
SRB/Mediator
SRB/mediator is a multisubunit complex isolated from yeast based on its requirement for transcriptional activation by RNA pol II in a purified system (reviewed in reference 28). Evidence for a transcriptional mediator came from squelching experiments, defined by the ability of one activator to inhibit transcription by another activator (138, 244). This effect could not be rescued by excess GTFs but was reversed by a partially purified yeast fraction. These results were interpreted as evidence for an intermediary molecule that would mediate the interaction between activators and components of the core machinery. Purified SRB/mediator is functionally defined by three activities: (i) stimulation of basal transcription in a highly purified system; (ii) response to transcriptional activators in vitro; and (iii) stimulation of phosphorylation of the RNA pol II CTD by the TFIIH kinase (252, 330). In contrast to TAFs, which appear to function as coactivators in a gene-specific manner, SRB/mediator appears to play a more general role in transcriptional activation.
Unlike the TAF components of TFIID, many of the SRB/mediator components were identified in genetic selections for mutations that affect transcription. One selection was based on suppression of the cold-sensitive growth phenotype associated with truncations of the RNA pol II CTD (345). Mutations in nine different genes, designated SRB2 and SRB4 to SRB11 (suppressor of RNA polymerase B) were identified. The products of the SRB2, SRB4, SRB5, SRB6, and SRB7 genes were found to be mediator subunits. This result suggested that SRB/mediator function is manifest through the CTD. The other SRB/mediator subunits include the products of the GAL11, SIN4, RGR1, and ROX3 genes, all identified based on mutations that affect transcription, and the products of the MED genes, most encoding novel SRB/mediator components. These factors are reviewed below and summarized in Table 4.
TABLE 4.
Yeast SRB/mediator subunitsa
| Factor | Mass (kDa) | Gene(s) | Essential | Metazoan homologs | Reference(s) |
|---|---|---|---|---|---|
| Srb2 | 23 | SRB2 | No | 252, 260, 261, 483 | |
| Srb4 | 78 | SRB4 | Yes | 252, 261, 483, 484 | |
| Srb5 | 34 | SRB5 | No | 252, 261, 483 | |
| Srb6 | 14 | SRB6 | Yes | 252, 261, 483, 484 | |
| Srb7 | 16 | SRB7 | Yes | Yes | 194, 283 |
| Srb8 | 167 | SRB8, SSN5, ARE2 | No | 194, 283 | |
| Srb9 | 160 | SRB9, SSN2, UME2 | No | 194, 283 | |
| Srb10 | 63 | SRB10, SSN3, UME5, ARE1 | No | 265, 285, 461, 516 | |
| Srb11 | 36 | SRB11, SSN8, UME3 | No | 98, 265, 285 | |
| Gal11 | 38 | GAL11, SPT13, SDS4, RAR3 | No | 130, 252, 261 | |
| Sin4 | 111 | SIN4, SSN4, TSF3 | No | 72, 230, 282 | |
| Rgr1 | 123 | RGR1 | Yes | Yes | 230, 282 |
| Rox3b | 25 | ROX3, SSN7 | Yes | 170, 396 | |
| Med1 | 64 | MED1 | No | Cited in 330 | |
| Med2 | 48 | MED2 | No | 330 | |
| Med3, Pgd1, Hrs1 | 47 | PGD1, HRS1 | No | 330 | |
| Med4 | 32 | MED4 | Yes | 330 | |
| Med6 | 33 | MED6, MTR32 | Yes | Yes | 278 |
| Med7 | 32 | MED7 | Yes | 330 | |
| Med8 | 25 | MED8 | Yes | 330 |
Srb2, Srb4, Srb5, Srb6, and Srb7 are subunits of the SRB/mediator as described by both the Kornberg and Young laboratories. Srb8 to Srb11 are components of the SRB/mediator defined by Young and coworkers (194) but not by Kornberg and coworkers (330). The SWI/SNF complex has also been reported by Young and colleagues to be associated with the holoenzyme (535) but is not included in the SRB/mediator defined by Kornberg and coworkers (330). Sug1 was identified as a suppressor of a gal4 mutation (468) and proposed to be a component of SRB/mediator (252, 261, 469). More recently, Sug1 has been defined as a subunit of the 26S proteosome (402) and is no longer included as a subunit of the SRB/mediator (330).
Rox3 was initially reported to be identical to Med8 (170). Subsequent analysis of Med8 revealed two distinct polypeptides, one of which is Rox3; however, the Med8 designation was retained for the other (330). Accordingly, Rox3 and Med8 now refer to distinct polypeptides that are both components of SRB/mediator.
The SRB2 gene is not essential for cell viability, although srb2 deletion mutants exhibit the same phenotypes as CTD truncations, including slow growth, inositol auxotrophy, and heat and cold sensitivity (260). Furthermore, the dominant SRB2-1 mutation exhibited allele-specific suppression of CTD truncation mutations. Srb2 physically associates with the PIC and directly binds TBP. These results established the potential for suppressors of CTD defects to identify novel transcription factors. Moreover, Srb2 revealed a functional link between the CTD and TBP (260).
The SRB4 and SRB6 genes are essential for cell viability (483). Like SRB2, SRB5 is not essential for cell viability, but deletion of SRB5 confers slow growth, as well as heat and cold sensitivity. Srb5 is a component of the PIC and is required for efficient transcription initiation. Srb2, Srb4, Srb5, and Srb6 physically associate with RNA pol II as components of a 1.2-MDa holoenzyme complex (483).
Not all of the cellular RNA pol II is found in the holoenzyme form (252, 261). This raised the question whether holoenzyme is a general requirement for transcription initiation. This was addressed in vivo by using a conditional srb4 mutant. A temperature shift to 37°C caused rapid growth arrest of the srb4 and srb6 temperature-sensitive mutants. Concomitant with the temperature shift, the levels of total mRNA, as well as of specific mRNAs, rapidly declined. These results established a general requirement for SRB proteins in RNA pol II transcription and implied that the holoenzyme is the form of RNA pol II recruited to most promoters in vivo (484).
The SRB7 to SRB11 genes have also been defined. Like mutations in SRB2, SRB4, SRB5, and SRB6, the suppressor mutations in SRB7 to SRB11 specifically compensate for the conditional phenotypes associated with the CTD truncation mutation but not for other rpb1 alleles, implying that all nine SRB gene products are functionally related to the CTD. SRB7, SRB8, and SRB9 are novel genes (194). SRB7 is essential for cell viability, whereas deletion of SRB8 and SRB9 confers similar heat- and cold-sensitive growth defects as well as cell flocculation. SRB10 and SRB11 encode a kinase-cyclin pair (285). All nine SRB proteins are reported to be components of one form of the holoenzyme and can be dissociated from the holoenzyme as a complex that also includes TFIIF and Gal11 (194). This complex stimulates transcriptional activation, confirming its mediator function. The SWI/SNF chromatin remodeling complex has also been found as a component of the holoenzyme (535). On the other hand, an independent holoenzyme preparation includes neither Srb8 to Srb11 nor SWI/SNF components (283, 330). Perhaps more than one form of the SRB/mediator complex exists in yeast cells.
A mutant lacking the SRB10-SRB11-encoded kinase-cyclin pair is defective in response to galactose induction and is deficient in CTD phosphorylation (285). This suggests that the Srb10 kinase is involved in CTD phosphorylation and that CTD phosphorylation is regulated in a cell cycle-dependent manner and plays a role in the response to transcriptional regulators in vivo (285).
The Gal11 component of the RNA pol II holoenzyme was identified in a genetic selection for factors required for full expression of galactose-inducible genes (344). Although not essential for cell viability, gal11 mutants are pleiotropic, indicating that Gal11 function is not limited to GAL gene expression (343). Indeed, gal11 was also identified in genetic selections for the spt (spt13) and snf genes (130, 500) and in a screen for factors (sds4) that affect transcriptional silencing at the HMR mating-type locus (462). A GAL11 mutation, GAL11P, was also identified as a potentiator of the Gal4-AH weak activator (200). These results suggested that Gal11 is a general coactivator of transcription (343), although Gal11 (Spt13/Sds4) has also been implicated in transcriptional repression (130, 462). Gal11 was found subsequently to copurify as an SRB/mediator component of the holoenzyme (252).
In a reconstituted transcription system Gal11 enhances basal transcription and facilitates activation by many, but not all, gene-specific activators (408). The gene-specific function of Gal11 appears to be defined by the core promoter. TATA-containing genes are under control of Gal11, whereas genes with noncanonical TATA elements are unaffected (410). Moreover, the gene-specific function of Gal11 is dependent upon TFIIE (407, 409), which also affects transcription in a promoter-dependent manner (266, 411, 487). A role for Gal11 as a TATA element-specific factor implies that transcription initiation at TATA-containing and TATA-less promoters is mechanistically distinct (410). By contrast, transcription directed by the HIS3 TR (TATA containing) and TC (TATA-less) promoter elements does not appear to be mechanistically distinct; rather, differential utilization of TR and TC appears to be a function of the overall level of transcription (223).
The GAL11P allele encodes a derivative of Gal11 with a single amino acid change (N342I); it has enhanced affinity for the dimerization domain, rather than the activation domain, of Gal4. This result suggests that N342I creates an artificial target for the Gal4 dimerization domain and that a single activator-holoenzyme contact is sufficient to recruit the holoenzyme to the promoter, resulting in activation (20). This conclusion is consistent with TBP tethering experiments, demonstrating that artificial recruitment of TBP to the promoter is sufficient for activation, bypassing the need for an activation domain (70, 254, 544). Experiments involving artificial recruitment of Gal11 were extended to determine the effect of holoenzyme recruitment on chromatin remodeling. Recruitment of the holoenzyme to the PHO5 promoter by fusing the DNA binding domain of the Pho4 activator to either Gal11 or Srb2, both holoenzyme components, resulted in displacement of four positioned nucleosomes from the PHO5 promoter (150). This result demonstrated that recruitment of the RNA pol II holoenzyme is sufficient for chromatin remodeling.
The phenotypes of gal11 mutants are similar to those of sin4 and rgr1 mutants, including diminished transcription of the GAL, Ty, and MATα genes. These results suggested that Sin4 and Rgr1 might be SRB/mediator components. Indeed, the presence of Sin4 and Rgr1 in the holoenzyme was confirmed by immunoblot analysis and microsequencing, respectively (282). Furthermore, Gal11, Sin4, Rgr1, and a 50-kDa polypeptide were found in a subcomplex of the SRB/mediator, thereby accounting for the similar phenotypes associated with gal11, sin4, and rgr1 mutations. Recently, the 50-kDa polypeptide was defined as Med3, the product of the HRS1/PGD1 gene (330). Mutation in HRS1/PGD1 causes transcriptional defects similar to mutations in GAL11 and SIN4 (368). Moreover, all four genes can exert both positive and negative effects on gene expression (129, 230, 232, 368, 463).
Several of the SRB/mediator components were also identified in a genetic selection designed to uncover factors that function in glucose repression (reviewed in reference 62). Mutations in the SSN family of genes were isolated as suppressors of mutations in SNF1, which encodes a protein kinase required for release from glucose repression. Accordingly, at least some SSN genes should encode transcriptional repressors. Indeed, Ssn1 is identical to Mig1 (501), which mediates glucose repression, and Ssn6, in association with Tup1, is a general transcriptional repressor (243). A link between the SSN and SRB genetics systems came from the characterization of SSN3 and SSN8, which are identical to SRB10 and SRB11, respectively (265, 285). SSN3/SRB10 and SSN8/SRB11 are also identical to UME5 and UME3, respectively, which encode important regulators of meiosis-specific genes (98, 461). Four other SSN genes are also identical to genes variously reported to encode SRB/mediator components: SSN2 = SRB9, SSN4 = SIN4, SSN5 = SRB8, and SSN7 = ROX3 (443). Despite the discrepancies regarding the composition of SRB/mediator in different laboratories (57, 194, 252, 261, 285, 330, 535), both complexes include transcriptional repressors: Srb8 to Srb11 in one case (194) and Sin4, Rgr1 and Rox3 in the other (170, 282). Thus, SRB/mediator components appear to confer both positive and negative effects on gene expression, suggesting that the SRB/mediator of “activation” might be more appropriately termed the SRB/mediator of transcriptional “regulation” (282).
In addition to the SRB proteins and the Gal11 subcomplex, SRB/mediator includes polypeptides designated MED proteins (283). Most of these proteins have now been identified (330). As mentioned above, Med3 is encoded by the previously identified HRS1/PGD1 genes (34, 415); is found in a subcomplex of the mediator that includes Gal11, Sin4, and Rgr1 (330); and functions as both a positive and negative regulator of transcription (368).
Med6 is a novel protein encoded by the essential MED6 gene (278). In contrast to TAFs, which are required for activation of only a subset of genes, Med6 is required for the activation of many, although not all, genes. Consistent with a general role for Med6 in transcription, a med6(Ts) mutant displays a broad array of phenotypes. Med6 has no effect on uninduced transcription. In this sense, Med6 is different from the SRB proteins, which can affect uninduced transcription, and different from the Gal11 subcomplex, which affects both activation and repression of transcription (278). Homologs of Med6 have been identified in Caenorhabditis elegans and humans, suggesting that Med6 is a universal transcription factor.
Med8 was initially reported to be identical to Rox3 (170). Subsequent analysis of Med8 revealed two distinct polypeptides, one of which is Rox3; Med8 now refers to the other polypeptide (330). Accordingly, Rox3 and Med8 are distinct polypeptides, both of which are components of SRB/mediator. Like components of the Gal11 subcomplex, Rox3 has been implicated in both activation and repression of transcription in vivo (reviewed in reference 170). Thus, Rox3, like the Gal11 subcomplex, offers strong support for the premise that the SRB/mediator affects transcriptional regulation in vivo.
TFIIA
TFIIA was initially identified as a GTF based on its requirement for specific transcription in vitro (311, 389). More recent studies established that TFIIA is dispensable for TBP-directed initiation but stimulates transcription in a TFIID-directed system (100, 106, 187, 353, 420, 458, 551). This differential effect has been attributed to TFIIA-mediated displacement of transcriptional repressors such as Dr1-DRAP1/NC2, PC3/Dr2 (topoisomerase I), HMG1, and Mot1 from the TFIID complex (14, 152, 221, 320, 324). TFIIA associates with the PIC through interactions with TBP (42) and stabilizes TBP-TATA box binding (219). TFIIA also interacts with specific transcriptional activators (353, 551), TAFII110 (550), and the coactivators PC4 and HMG2 (153, 438). Moreover, TFIIA is required to overcome a rate-limiting step during formation of an open promoter complex (525). Thus, TFIIA is dispensable for accurate initiation but plays an important role in transcriptional activation, functioning as either an antirepressor or a coactivator (238, 298).
The yeast homolog of metazoan TFIIA was identified by complementation of a mammalian in vitro transcription system (176). Yeast TFIIA activity copurified as two polypeptides with apparent molecular masses of 32 and 13.5 kDa (383). The genes encoding both subunits were cloned based on partial sequence of the purified subunits and designated TOA1 and TOA2 (384). Both TOA1 and TOA2 are essential for cell viability, underscoring the functional importance of TFIIA. Structural analysis of Toa1 and Toa2 defined domains of the two subunits that are required for subunit association, TBP-DNA interactions, and transcriptional activity (238). Neither TOA1 nor TOA2 has been identified in a genetic selection or screen.
Similar to the TAF subunits of TFIID, TFIIA is dispensable for activated transcription in vitro (252, 261). However, TBP mutants defective in TFIIA binding are activation-defective in vivo (447). This apparent discrepancy might be explained if TBP-TFIIA interaction blocks the effects of general transcriptional repressors, absent from the in vitro systems, that either promote TBP-TATA dissociation (e.g., Mot1 [13–15]) or impair TBP-TATA association (e.g., Ydr1-Bur6 [148, 163, 247, 375]).
Human TFIIA and Drosophila TFIIA are composed of three subunits with apparent molecular masses of approximately 35/30, 19/20, and 12/13 kDa. In both organisms, the two larger subunits are encoded by the same gene and appear to be posttranslationally modified forms of a precursor protein (107, 297, 550). The N-terminal 54 amino acids and the C-terminal 76 amino acids of the precursor protein exhibit structural similarity to the TOA1-encoded subunit of yeast TFIIA. Furthermore, the nonconserved central region of Toa1 is dispensable for function (238). The smallest subunit of human TFIIA is homologous to the TOA2-encoded subunit of yeast TFIIA (353, 458). Thus, the three subunits of metazoan TFIIA are encoded by two genes that are homologous to yeast TOA1 and TOA2.
Crystal structures for two forms of yeast TFIIA-TBP-DNA ternary complexes have been solved (155, 475). In both cases, the structures were determined with the smallest form of TFIIA that retained biological function. Accordingly, the largest subunit of TFIIA had the dispensable, central region of the polypeptide deleted. Two major structural elements were identified: a six-stranded β-sandwich and a four-helix bundle. The C termini of both subunits contribute three strands to the β-sandwich, and the N termini of each subunit contribute two helices to each helical bundle. TFIIA associates with the side of TBP-DNA opposite to TFIIB (Figure 2). Unlike TFIIB, which binds both upstream and downstream of the TATA box, TFIIA is located exclusively upstream of TATA and is unlikely to contact other general factors that bind downstream of TATA.
Histone Acetyltransferases
Based on selective inhibition of activated, but not basal, transcription by the acidic activation domain of GAL4-VP16, the existence of transcriptional adapter molecules that would bridge the interaction between activators and the core transcriptional machinery was proposed (25). The toxic effect of GAL4-VP16 overexpression was exploited to select for adapter-defective mutants (26). Five genes, ADA1 to ADA3, GCN5, and ADA5, were identified (26, 213, 308, 309, 365, 394). GCN5 was initially identified based on its requirement for full activation by the Gcn4 transcriptional activator (156). ADA2, ADA3, and GCN5 were required for full activation by a subset of transcriptional activators, and Ada2 binds the activation domains of VP16 and Gcn4 (23, 439). Presumably the ADA proteins and Gcn5 are directly recruited to promoter DNA by gene-specific activators.
Mutations in these genes cause a similar array of pleiotropic phenotypes, and double ada gcn5 deletion mutants exhibit phenotypes no more severe than single mutants (309, 365). These observations suggest that Gcn5 and the ADA proteins are functionally related, operating in a common pathway. Moreover, the Ada2, Ada3, and Gcn5 proteins physically interact with each other, both in vitro (212) and in vivo (58), suggesting the existence of an ADA-Gcn5 complex.
Although all of the evidence pointed to a role for the ADA and Gcn5 proteins in transcription, no specific function had been assigned to any of these proteins. A breakthrough occurred following purification of HAT from the Tetrahymena (37). Sequence analysis of the cloned HAT gene revealed homology to Gcn5, which was subsequently shown to have HAT activity (38). This was a key discovery because, in addition to defining an activity for Gcn5, it provided a direct link between histone acetylation and transcriptional activation.
The ADA5 gene is identical to SPT20, an important discovery because it connects the ADA and SPT genetic systems (308, 394). Furthermore, Spt20/Ada5 physically associates with Spt3, Spt7, and Spt8 (165, 393). This suggested that Gcn5 functions within a large complex that includes both SPT and ADA proteins.
The existence of a Gcn5 HAT complex was also suggested by the ability of recombinant Gcn5 to acetylate free histones but not histones assembled into nucleosomes (267, 548). Perhaps other components of the putative complex were required for recognition of nucleosomes. Consistent with this notion, both HAT activity and interaction with Ada2 are required for Gcn5 function in vivo (59). A recent collaboration among several laboratories succeeded in identifying four distinct nucleosomal HAT complexes (165). Two of these complexes, with apparent molecular masses of 1.8 and 0.8 MDa, included Gcn5 and Ada2. The genetic relationship among the GCN5, ADA, and SPT genes provided a clue to the identity of the other components. Specifically, the 1.8-MDa complex copurified with Gcn5, Ada2, Spt3, Spt7, and Spt20/Ada5, and the integrity of the complex was dependent upon intact GCN5, ADA2, ADA3, SPT7, and SPT20/ADA5 genes. This complex has been named SAGA (Spt-Ada-Gcn5-Acetyltransferase) and links nucleosomal histone acetylation with transcriptional activation associated with ADA and SPT proteins. SAGA is probably identical to two other ADA-containing complexes described recently (213, 413). Other nucleosomal HAT complexes have been described, including a 200-kDa complex that is different from the four HAT complexes described above (413), and a 170-kDa complex that includes Gcn5 (404).
These results establish that one class of transcriptional coactivator functions by acetylation of nucleosomal histones. Presumably, acetylation weakens histone-DNA interactions, thereby relieving the repressive effects of chromatin (540). This is a satisfying result because a correlation between histone acetylation and gene activation was recognized more than three decades ago (4). However, the cause-and-effect relationship had not been established. Does histone acetylation facilitate transcriptional activation, or does activation promote acetylation? The initial identification of the Gcn5 HAT based on two distinct genetic selections for transcriptional coactivators strongly supports the premise that acetylation promotes activation. Thus, SAGA and related HAT complexes appear to function as transcriptional coactivators by facilitating the removal or repositioning of nucleosomes.
Chromatin-Remodeling Complexes
Other complexes that facilitate transcriptional activation by affecting nucleosome structure yet do not catalyze histone acetylation have been described. These include yeast, human, and Drosophila SWI/SNF complexes and Drosophila NURF. Each of these complexes promotes nucleosome disruption or displacement in an ATP-dependent manner. Reviews describing these complexes have been published recently (51, 362). Here I review briefly the yeast complexes and discuss their relationship to the TFIID, SRB/mediator, and HAT complexes.
Yeast SWI/SNF is the most well-characterized of the remodeling complexes. The composition and function of the SWI/SNF complex was unraveled by linking disparate genetic systems (reviewed in reference 537). The initial set of SWI genes were identified in a screen for defects in mating-type switching (451), whereas SNF genes were identified based on diminished expression of SUC2 (336). A connection between the SNF and SWI systems was made when SNF2 and SWI2 were found to be identical. The link to chromatin function was made by characterization of suppressors of snf and swi mutations, defining ssn and sin genes, respectively. SSN20 and SIN2 turned out to be identical to SPT6 (85, 338) and HHT1 (264), respectively. The connection to SPT6 and HHT1 was revealing because SPT6, along with SPT4, SPT5, SPT11/HTA1, SPT12/HTB1, and SPT16/CDC68, is a member of the SPT class of genes that either encode histones or affect chromatin function, and HHT1 encodes histone H3. The identities of these genes and their effects on gene expression led to a model for the function of the SNF/SWI and SPT/SIN genes. Accordingly, SPT/SIN proteins repress transcription by formation of inactive chromatin whereas SWI/SNF proteins overcome chromatin repression (537). This model received direct support from defective chromatin remodeling at the SUC2 promoter in swi2/snf2 and snf5 mutants (201).
The SWI/SNF complex has been purified from yeast as a 2-MDa, 11-subunit complex (56, 101). Subunits include Swi1, Swi2/Snf2, Swi3, Snf5, Snf6, Snf11, Swp29, Swp59, Swp61, Swp73, and Swp82 (51). Swi2/Snf2 is the best-characterized component and, as a DNA-dependent ATPase, is the only subunit with known enzymatic activity. As noted above, the Swp29 subunit is identical to the Tfg3 subunit of TFIIF and to the TAFII30 subunit of TFIID (55). Thus, Swp29 is a functional link between the TFIID, TFIIF, and SWI/SNF complexes. The SWI/SNF complex has also been reported to be a component of the RNA pol II holoenzyme (535). This could endow the holoenzyme with the ability to promote PIC assembly by disrupting nucleosomal DNA. Although an appealing concept, SWI/SNF is not found in an independent preparation of RNA pol II holoenzyme (57, 330).
The yeast SWI/SNF complex has been extensively purified, and a number of its biochemical properties have been reported. SWI/SNF is a high-affinity DNA binding complex with properties similar to those of proteins containing HMG-box domains (381). Interestingly, SWI/SNF binds synthetic four-way junction DNA, which mimics the structure where DNA enters and exits the nucleosome, a property that has implications for the mechanism of chromatin remodeling (381).
In addition to HO, SUC2, and Ty, activation of the ADH1, ADH2, INO1, and STA1 promoters is SWI/SNF dependent (361, 554). However, many promoters are not SWI/SNF dependent for activation. As examples, PHO5, URA3, LYS2, CLN1, CLN2, CLN3, and HSC26 are SWI/SNF independent (361, 370). Why are some promoter SWI/SNF dependent while others are not? One possible explanation is that SWI/SNF is required only where critical promoter elements are contained within positioned nucleosomes. This would account for the effect of SWI/SNF on SUC2 (201). However, activation of PHO5 involves displacement of four positioned nucleosomes from the promoter region, yet PHO5 activation is SWI/SNF independent (150). An alternative explanation for SWI/SNF-independent activation is dependence of those promoters on alternative chromatin remodeling complexes or other coactivators. Indeed, there is functional overlap between SWI/SNF, SAGA, and SRB/mediator complexes (370, 393).
Functional overlap between coactivators is further supported by the identity of the SWI7, SWI8, and SWI9 genes. Mutations in these genes were identified in a screen for mutants defective in HO gene expression (33) and are therefore related the SWI genes encoding SWI/SNF components (451). However, SWI7, SWI8, and SWI9 do not encode SWI/SNF subunits but are identical to ADA3, ADA2, and GCN5, respectively (370). Moreover, swi7, swi8, and swi9 mutants are phenotypically similar to swi/snf mutants and are defective in the expression of a common set of genes (370). These results suggest that SWI/SNF and SAGA complexes work in concert to alter chromatin structure.
Based on homology to components of the SWI/SNF complex, a second chromatin remodeling complex, termed RSC (remodels the structure of chromatin), was isolated from yeast (57). Like SWI/SNF, RSC is a DNA-dependent ATPase whose activity is stimulated by both free and nucleosomal DNA. RSC is a 15-subunit complex that includes several SWI/SNF-related polypeptides: Sth1, Rsc6, and Rsc8 are homologous to Swi2/Snf2, Swp73, and Swi3, respectively (57); and Sfh1 is homologous to Snf5 (60). RSC is approximately 10-fold more abundant than SWI/SNF; it is present at several thousand molecules per cell, and, unlike SWI/SNF, at least certain genes encoding RSC subunits are essential for cell viability. However, sth1 mutations do not affect SUC2 and GAL10 expression and are not suppressed by histone gene mutations (117). Indeed, there is currently no evidence that RSC plays a direct role in transcription. Instead, mutations in SFH1 (60) and STH1 (117) cause cell cycle arrest in the G2/M phase of the cell cycle and Sfh1 is specifically phosphorylated in G1 (60). These results suggest that RSC and SWI/SNF are functionally distinct, with RSC playing a role in cell cycle progression.
SWI/SNF- and RSC-mediated chromatin remodeling is not specific to yeast. A SWI/SNF-like complex has been isolated from human cells (218, 268, 524, 526), and homologs of SWI/SNF components have been found in other organisms (237). Human SWI/SNF can also affect transcriptional elongation, as demonstrated by the SWI/SNF requirement to overcome nucleosome-enhanced transcriptional pausing on the hsp70 gene (36).
Other nucleosome-remodeling factors have also been described. These include NURF and CHRAC, both isolated from Drosophila (492, 506). NURF was identified based on GAGA-dependent formation of nuclease hypersensitive sites within an array of nucleosomes in vitro (492). NURF includes a SWI2/SNF2 homolog, ISWI (imitation SWI), but is otherwise distinct from SWI/SNF. A yeast complex homologous to NURF has not been described, but yeast homologs of the other NURF subunits have been found (491). CHRAC includes the ISWI subunit of NURF and facilitates the accessibility of DNA in chromatin, as well as chromatin assembly (506). Thus, eukaryotes contain multiple chromatin-remodeling complexes, some of which play a general role as transcriptional coactivators in organisms from yeast to humans.
Other Coactivators
As human GTFs were more extensively purified, in vitro transcription systems lost the ability to respond to transcriptional activators. This led to the identification of an additional class of cofactors, designated USA (upstream stimulatory activity) (320). USA includes both positive and negative effectors of transcription, hence the designation “cofactor” rather than “coactivator.” These cofactors interact with the PIC to repress transcription in the absence of activators or to stimulate transcription in the presence of activators (320). USA stimulates transcription in the presence of both TFIID and RNA pol II holoenzyme (69). Thus, TAFs, USA, and SRB/mediator appear to have overlapping but distinct functions in transcriptional activation.
Several independent USA components have been identified (reviewed in reference 235). One of these, PC4, dramatically stimulates activation and interacts directly with various activator domains and DNA-TBP-TFIIA complexes, demonstrating that PC4 mediates functional interaction between upstream activators and the PIC (154). However, the mechanism by which this interaction stimulates transcription has not been resolved.
Recently, a yeast homolog of human PC4 was found, both biochemically and genetically (195, 258). During the purification of yeast TFIIF, a contaminating polypeptide, p43, diminished the response to activators when purified away (195). When added back, p43 stimulated transcription, even in the presence of SRB/mediator. The gene encoding p43, designated TSP1, was cloned and found to encode a homolog of human PC4. Like PC4, Tsp1 interacts with both a transcriptional activator and a GTF, in this case TFIIB. Furthermore, Tsp1 phosphorylation regulates these interactions. TSP1 was also identified as the SUB1 gene, in this case as a high-copy-number suppressor of a TFIIB defect (258). Interestingly, high-copy-number SUB1 is allele specific in its suppression of TFIIB defects, compensating for the growth defects associated with amino acid replacements at positions E62 and R78, two positions involved in start site selection (258, 367). These results suggest functional overlap between the mechanisms affecting the accuracy and the rate of transcription initiation.
Other cofactors have also been defined. PC1 is poly(ADP-ribose) polymerase (321). Transcriptional activation by poly(ADP-ribose) polymerase requires the amino-terminal DNA binding domain, but not the carboxyl-terminal catalytic region. PC3/Dr2 is topoisomerase I and functions in both repression of uninduced transcription and stimulation of activated transcription (263, 324). The nonhistone chromosomal protein HMG2 was also identified as a transcriptional coactivator (438). In this case, the HMG box alone is sufficient for coactivator function, leading to the proposal that this “architectural” protein functions as a coactivator by stabilizing an activated form of the PIC (438). It is interesting that PC1, PC3, PC4, and HMG2 are all nonsequence-specific DNA binding proteins, suggesting that these cofactors function by affecting the accessibility of RNA pol II to chromatin (235).
In summary, multiple, functionally distinct classes of transcriptional coactivators have been identified. Is there a functional relationship among these factors, or does each function independently at specific genes? Several results indicate functional overlap. First, many of the genes encoding these factors are not essential for cell viability. In some cases, this can be accounted for by more than one gene encoding a specific activity. However, analysis of the complete yeast genome sequence revealed that many of these factors are encoded by unique genes. Second, a search for mutations that confer lethality in combination with defects in subunits of the SAGA complex identified genes encoding components of the SWI/SNF complex and SRB/mediator (393). Furthermore, a genetic selection similar to that which identified components of the SWI/SNF complex also uncovered components of the SAGA HAT complex (370). These results suggest that SAGA, SWI/SNF, and SRB/mediator, although functionally distinct, overlap in their roles as coactivators of gene expression.
GENERAL TRANSCRIPTIONAL REPRESSORS
General transcriptional repressors are genetically defined by mutations that cause increased transcription of multiple, functionally unrelated genes. In this sense, general transcriptional repressors are comparable to transcriptional coactivators, albeit with opposite effects. In contrast to the GTFs, most of which were initially identified biochemically, many of the general repressors were uncovered in genetic selections for mutations that enhanced transcription, in the absence of either a gene-specific transcriptional activator or its cognate UAS promoter element. An important and surprising feature of the general repressors is that many of them also function in transcriptional activation.
Two classes of general repressors have been recognized. One class operates through core promoter elements and includes factors that affect TBP function. The second class is functionally related to chromatin and includes histones, histone-related proteins, and histone deacetylases. Both classes are reviewed here, with an emphasis on the genetic schemes that were instrumental in identifying these factors. The yeast general transcriptional repressors are summarized in Table 5. A schematic summary of the general transcriptional repressors and their functions is presented in Fig. 5.
TABLE 5.
Summary of yeast general transcriptional repressors
| Factor | Mass (kDa) | Gene(s) | Essential | Characteristics | Reference(s) |
|---|---|---|---|---|---|
| Mot1 | 210 | MOT1, BUR3 | Yes | ATP-dependent dissociation of TBP from DNA; functions in both activation and repression of transcription; functionally related to Spt3, TFIIA, and NOT complex | 14, 90, 105, 299, 375 |
| Ccr4a | 95 | CCR4 | No | Component of Ccr4-NOT complex; implicated in both transcriptional activation and repression | 289 |
| Dbf2 | 66 | DBF2 | No | Component of Ccr4-NOT complex; implicated in both transcriptional activation and repression | 289 |
| Caf1 | 50 | POP2, CAF1 | No | Component of Ccr4-NOT complex; implicated in both transcriptional activation and repression | 289 |
| Not1 | 240 | NOT1, CDC39 | Yes | Component of Ccr4-NOT complex; represses transcription from TATA-less promoters; functionally related to Mot1 and Spt3 | 90–92, 289 |
| Not2 | 22 | NOT2, CDC36 | No | Component of Ccr4-NOT complex | 90–92, 289 |
| Not3 | 94 | NOT3 | No | Component of Ccr4-NOT complex | 90–92, 289 |
| Not4 | 65 | NOT4, MOT2, CCL1, SIG1 | No | Component of Ccr4-NOT complex | 90–92, 289 |
| Not5 | NOT5 | No | Component of Ccr4-NOT complex | 89 | |
| Bur1 | 74 | BUR1, SGV1 | Yes | Cdc28-related protein kinase | 376 |
| Bur2 | 46 | BUR2 | No | 376 | |
| Bur4 | BUR4 | 376 | |||
| Histone H3 (Bur5) | 15 | HHT1, BUR5, SIN2 | No | Nucleosome subunit | 376 |
| Bur6 | 16 | BUR6, NCB1 | Yes | Homologous to mammalian DRAP1/NC2α; associated with Ydr1/Ncb1; blocks TBP association with TFIIA, TFIIB? | 148, 163, 247, 375 |
| Ydr1/Ncb2 | 17 | YDR1, NCB2 | Yes | Homologous to mammalian Dr1/NC2β; associated with Bur6; blocks TBP association with TFIIA, TFIIB? | 148, 163, 247 |
| Spt4 | 11 | SPT4 | No | Physically associated with Spt5; human homolog; functions in chromatin remodeling? | 188, 300, 472 |
| Spt5 | 116 | SPT5 | Yes | Physically associated with Spt4; functions in chromatin remodeling? | 471, 472 |
| Spt6 | 168 | SPT6, SSN20, CRE2 | Yes | Functionally related to SPT4 and SPT5; might function in either nucleosome assembly or chromatin reorganization | 31, 85, 338, 470, 471 |
| Sin4 | 111 | SIN4, TSF3 | No | Component of SRB/mediator; functions in both activation and repression of transcription; affects chromatin organization | 72, 230, 231, 282 |
| Rpd3 | 49 | RPD3, SDI2, SDS6 | No | HDA; interacts with Sin3; homologous to Hda1, Hos1p, Hos2, and Hos3 | 234, 240, 405, 510, 511 |
| Sin3 | 175 | SIN3, UME4, RPD1, SDI1, CPE1, SDS16 | No | Component of Rpd3 HDA complex | 234, 453, 504, 511, 521, 522 |
| Ssn6 | 107 | SSN6, CYC8 | No | Component of Ssn6-Tup1 complex; does not bind DNA; recruited to promoter by DNA-repressor complexes; contains 10 TPR repeats | 243, 425, 490, 534 |
| Tup1 | 78 | TUP1, CYC9, FLK1, ROX4, SFL2, UMR7 | No | Component of Ssn6-Tup1 complex; does not bind DNA; recruited to promoter by DNA-repressor complexes; directly interacts with histones H3 and H4; contains 7 WD repeats | 214, 243, 279, 534 |
Ccr4 was initially identified as a transcriptional coactivator required for glucose derepression (289).
FIG. 5.
Schematic summary of general transcriptional repressors and their activities. Comparable to coactivators, general repressors can interact either with the core transcriptional machinery or with nucleosomes. Mot1, Dr1-DRAP1 (NC2), and the Ccr4-Not complex confer transcriptional repression by interaction with components of the core machinery. Mot1 interacts directly with TBP and promotes TATA-TBP dissociation in an ATP-dependent manner. Dr1-DRAP1 also interacts directly with TBP but, in contrast to Mot1, represses transcription by blocking TBP interaction with TFIIA and TFIIB rather than by displacing TBP from DNA. The Ccr4-NOT complex also targets the core machinery. Whereas Mot1 promotes TBP-DNA dissociation, the Ccr4/NOT complex has been proposed to negatively regulate the activity of factors (e.g., TFIIA) that facilitate TBP-TATA association. In contradistinction to HATs (Fig. 4), HDA complexes repress transcription by deacetylation of histones or other factors, presumably allowing reestablishment of repressive chromatin structures. HDAs do not bind DNA directly but are targeted by URS-repressor complexes. Ssn6-Tup1 is also targeted by URS-repressor complexes and was recently reported to interact with histones H3 and H4. Thus, HDAs and Ssn6/Tup1 are similar in their modes of transcriptional repression, although Ssn6/Tup1 is not an HDA. The BUR proteins, including Bur1, Bur2, Bur4, and Bur5, appear to mediate repression by affecting chromatin structure. (BUR5 is identical to HHT1/SIN2, which encodes histone H3.) The Spt4-Spt5 complex also regulates transcription by affecting the chromatin structure. Recently, a human Spt4-Spt5 complex, denoted DSIF, was identified as a transcription elongation factor. Spt6 is functionally related to Spt4 and Spt5 but does not appear to be a component of the Spt4-Spt5 complex. An important characteristic of several general transcriptional repressors is that they can also function in transcriptional activation.
Mot1
Mot1 was identified both biochemically and genetically. ADI (ATP-dependent inhibitor), was identified as a factor in yeast nuclear extracts that inhibited TBP binding to DNA in an ATP-dependent manner (13). ADI-mediated TBP displacement was not promoter specific and could be counteracted by TFIIA and to a lesser extent by TFIIB. Sequence analysis identified ADI as the product of the MOT1 gene (14), which was identified in a screen for mutants with enhanced basal expression of several unrelated genes (105, 364). Consistent with a functional relationship among Mot1, TBP, and TFIIA, overexpression of either TBP or TFIIA suppressed the growth defect associated with a dominant negative MOT1 allele (14). Mot1 is a member of the Snf2/Swi2 family of ATPases, but in contrast to Snf2/Swi2 (272), the Mot1 ATPase is not stimulated by DNA (15).
Mot1 was also identified as a 170-kDa protein that bound TBP in a complex distinct from TFIID (372). In mammalian cell extracts, the majority of TBP exists in an alternative form of TFIID, denoted B-TFIID (489). In contrast to TFIID, B-TFIID does not respond to transcriptional activators (489) and possesses ATPase activity (488). Recently, a TAFII170-kDa subunit of human B-TFIID was cloned. Sequence analysis revealed structural similarity to yeast Mot1 (502). These results provide direct support for a physical interaction between Mot1 and TBP and suggest that the yeast TBP-Mot1 complex is the counterpart of mammalian B-TFIID. This form of Mot1 is likely to be the transcriptional corepressor that mediates repression by Leu3 in yeast (514).
MOT1 was also identified in a genetic screen for factors that functionally interact with Spt3. Specifically, a mot1 allele conferred synthetically lethality in combination with an spt3 disruption (299). Surprisingly, mot1 and spt3 mutations cause similar phenotypes, including suppression of his4-912δ and diminished levels of certain other transcripts. A mot1 mutation also markedly diminished transcription from TATA-less elements in the HIS3 and HIS4 promoters (90). Thus, Mot1 can function as either an activator or a repressor of transcription. Furthermore, mutation in the TOA1 gene, encoding the larger subunit of TFIIA, resulted in lethality in combination with either mot1 or spt3, and TOA1 or TOA2 overexpression suppressed spt3 phenotypes (299). These results led to the proposal that Mot1, Spt3, and TFIIA regulate TBP-DNA interactions: Mot1 by displacing TBP from nonfunctional TATA boxes, and Spt3 and TFIIA by enhancing TBP association with functional TATA boxes (299). As reviewed in the next section, promoters regulated by Mot1 and Spt3 are also regulated by NOT proteins, albeit with opposite effects.
Ccr4-NOT Complex
As reviewed in the section on TATA elements (above), the HIS3 promoter includes two functionally distinct TATA elements, TC and TR, that are differentially utilized in constitutive and Gcn4-activated HIS3 transcription (223). The functional distinction between TC and TR was exploited to identify negative regulators of transcription. By selecting for strains with increased expression of HIS3 in a gcn4 mutant background, mutants that specifically enhanced expression from the TC-dependent +1 site were isolated. Mutations in four different genes, designated NOT1 through NOT4 (negative on TATA), were initially identified (91, 92); a fifth gene, NOT5, was recently identified from the same selection (89).
Genetic evidence supports a functional relationship among the NOT proteins (92). First, not suppressors enhance expression of the same spectrum of functionally unrelated genes. Second, suppression, including allele-specific suppression, occurs among various combinations of not mutations. Third, Not1-Not2, Not1-Not4, Not1-Not5, Not3-Not4, and Not3-Not5 interact in two-hybrid assays. Moreover, Not1 purifies as a large complex that includes the other NOT proteins.
Three of the NOT genes were identified previously in other mutant hunts. NOT1 and NOT2 are identical to CDC39 and CDC36, respectively, and NOT4 is identical to MOT2, which was identified in separate genetic selections for suppressors of mutations in the STE4 and STE11 genes (54, 222). Mutations in each of the NOT genes caused constitutive expression of specific genes, consistent with their roles as global repressors of transcription (54, 91, 92, 222). Although NOT2 to NOT5 can be deleted without loss of cell viability, NOT1 is an essential gene, implying that the NOT repressor complex is critical for cell viability.
The NOT repressors are functionally distinct from the Ssn6-Tup1 repressor complex since ssn6 and tup1 mutations do not affect HIS3 basal transcription from either the +1 or +13 sites (92). Also, the NOT repressors appear to be distinct from general chromatin-based repressors, since loss of histone H4 function, or defects in the histone-related protein Spt6, preferentially enhance HIS3 transcription from the +13 site, rather than +1 (92). These results suggest that NOT proteins are global transcriptional repressors that target the general transcriptional machinery, preferentially affecting basal, rather than activated, transcription (92).
Mutations in the NOT genes and in MOT1 exert opposite effects on transcription from TATA-less promoters. Whereas not mutations increase transcription, mot1 mutations decrease transcription (90). In addition, genetic interactions among the MOT1, SPT3, and NOT genes were found. Based on these interactions and the opposite effects of mot1 and not mutations on gene expression from TATA-less promoters, it was proposed that the Mot1, Spt3, and NOT proteins functionally interact to regulate the distribution of limiting TBP on weak and strong promoter elements. Accordingly, Mot1 promotes TBP-TATA dissociation from canonical TATA elements whereas NOT proteins negatively regulate the activity of factors such as Spt3 (and presumably TFIIA) that promote TBP-TATA association. This is similar to the model proposed for the functions of Mot1, Spt3, and TFIIA at specific promoters (299). A fundamental distinction between the two models is whether Mot1 stimulates the displacement of TBP from nonfunctional TATA sequences, thereby acting as an activator (299), or from canonical TATA elements, thereby acting as a repressor (90).
Most recently, NOT proteins were identified as components of a 1.2-MDa Ccr4 complex (289). Ccr4 is a global transcription factor, affecting the expression of genes involved in nonfermentative growth, cell wall integrity, and ion sensitivity. Although identified based on its requirement for activation of ADH2 (108, 109), Ccr4 has also been implicated in transcriptional repression (316, 423). Ccr4 exists in a large multisubunit complex that includes Caf1 and Dbf2, a cell cycle-regulated protein kinase, as well as several other polypeptides (115). Four of these polypeptides have now been identified as Not1 to Not4 (289). Moreover, mutations in NOT genes affected many of the same genes and functions affected by mutations in CCR4, CAF1, and DBF2. Accordingly, this complex is designated Ccr4-NOT and can affect transcription in either a positive or negative manner.
BUR Proteins
The effects of Mot1 and NOT proteins on differential TATA usage demonstrate that core promoter elements can be derepressed in vivo and that the repressed state must be maintained by general transcriptional repressors. A genetic selection was developed to identify such factors based on suppression of the phenotype associated with deletion of the UAS element from the SUC2 promoter (376). Genes identified in this selection were designated BUR (bypass of UAS requirement). All bur mutants exhibit multiple pleiotropic phenotypes, indicating that BUR genes affect more than SUC2 expression. Several of the BUR genes are identical to previously isolated SPT genes, a result that was not unexpected since mutations in the histone class of SPT genes were known to partially bypass the SUC2 UAS requirement. In addition, six new genes, denoted BUR1 to BUR6, were defined.
The BUR proteins are likely to repress transcription by two different mechanisms. The bur3 and bur6 mutants have common phenotypes, and neither suppresses a deletion of the SNF5 gene, which encodes a component of the SWI/SNF chromatin-remodeling complex that is required for SUC2 activation. Therefore, Bur3 and Bur6 were suggested to repress SUC2 by a chromatin-independent mechanism, perhaps by interacting with GTFs (376). Indeed, BUR3 is identical to MOT1 and BUR6 encodes the yeast homolog of DRAP1/NC2α (see the section on Dr1-DRAP1/NC2, below). Thus, both Bur3 and Bur6 are global repressors of transcription that target TBP, a conclusion consistent with the similar phenotypes of bur3 and bur6 mutants (376).
The bur1, bur2, bur4, and bur5 suppressors cause a common set of pleiotropic phenotypes and, in contrast to bur3 and bur6, suppress a snf5Δ mutation (375, 376). BUR1 (SGV1) encodes a Cdc28-related protein kinase, implicating protein phosphorylation in repression of the SUC2 basal promoter. The substrate(s) for this kinase has not been identified, although several general factors that affect transcription, including histone H4, the Rpb1 subunit of RNA pol II, and the largest subunit of TFIIF, are known to be phosphorylated. BUR5 is identical to HHT1/SIN2, which encodes histone H3. These results suggest that this group of BUR genes is functionally related to chromatin (376). Characterization of the BUR2 and BUR4 genes has not yet been reported.
Defects in several other genes are also capable of suppressing the effects of UAS deletions. Depletion of histone H4 suppresses UAS deletions within the CYC1, GAL1, and PHO5 promoters (184, 185). Also, mutations in SPT5 (472), SPT6 (85, 338), SPT16 (305), SUD1/SPT10 (334, 546), and HTA1-HTB1 (114) enhance expression from promoters that lack UAS elements (reviewed in reference 376). These studies strengthen the conclusion that histones and histone-related proteins can function through core promoter elements to maintain genes in a repressed state.
Dr1-DRAP1/NC2
The human Dr1-DRAP1 complex, which is identical to the NC2 complex, represses transcription by blocking the association of TBP with TFIIA and TFIIB (221, 319). Dr1/NC2β directly binds TBP (221, 319), whereas DRAP1/NC2α is a corepressor that enhances Dr1/NC2β activity (247, 325). cDNAs encoding human Dr1 (221) and DRAP1 (164, 245, 325) have been isolated and sequenced. Both proteins include histone-fold motifs that appear to mediate the Dr1-DRAP1 interaction (325).
Yeast contains homologs of both Dr1 and DRAP1. The yeast counterpart of DRAP1/NC2α, encoded by the BUR6/NCB1 gene, was identified by sequence analysis of the yeast genome (163, 247) and also in two separate genetic selections. In one case, a mutation in BUR6 was identified in the selection for bypass suppressors of the SUC2 UAS deletion (375, 376). In the other case, a mutation in NCB1 was found as a suppressor of a mutation in SRB4, which encodes a subunit of the SRB/mediator complex (148). These results are consistent with the proposed role of DRAP1 in transcriptional repression, since both selections were designed to uncover negative effectors of gene expression.
A yeast homolog of Dr1/NC2β, encoded by the YDR1/NCB2 gene, was also identified by analysis of the yeast genome sequence (148, 163, 247). Consistent with its role as a global negative regulator of transcription, high-copy-number expression of YDR1 causes diminished mRNA accumulation and a slow-growth phenotype (247). Moreover, the growth phenotype is partially suppressed by overexpression of TBP (247), a result consistent with the reversal of human Dr1-mediated repression by TBP overexpression (549). Both the YDR1/NCB2 and BUR6/NCB1 genes are essential for cell viability. Taken together, these results confirm that Dr1-DRAP1/NC2 functions as a general transcriptional repressor that targets TBP and that this function is conserved among eukaryotic organisms.
The bur6/ncb1 suppressor of srb4 functionally links repression of the core transcriptional apparatus with the SRB/mediator of gene expression (148). This connection is also supported by results from an independent genetic selection. In this case, mutations that relieved repression at the GAL1 and GAL10 promoter were sought (73). Recessive mutations in six different genes, TSF1 to TSF6, were identified. Mutations in all six genes caused multiple pleiotropic effects, indicating that they encode global rather than GAL-gene-specific transcription factors. To date, only the identification of TSF3 has been reported (72). Similar to the bur mutations and consistent with the notion that TSF3 encodes a global transcriptional repressor, tsf3 mutations enhance expression from UAS-less promoters (72, 231). TSF3 is identical to SIN4, a component of the SRB/mediator. This provides independent support for a connection between the SRB/mediator and general transcriptional repressors that function through core promoter elements.
Spt4-Spt6
The SPT4, SPT5, and SPT6 genes are members of the SPT class of genes that encode either histones or proteins that affect chromatin function (536). Mutations in these three genes confer similar pleiotropic phenotypes, and Spt5 and Spt6 physically interact (472). Although SPT4 is not essential for cell viability (306), both SPT5 and SPT6 (SPT6 = SSN20 = CRE2) are essential genes (470, 471). Similar to deletion of HTA1-HTB1 (201, 264), one of two gene pairs encoding histones H2A and H2B, mutations in SPT4, SPT5, and SPT6 suppress defects in components of the SWI/SNF chromatin-remodeling complex (537). Since hta1-htb1-mediated suppression occurs by altering chromatin structure (201), Spt4, Spt5, and Spt6 were implicated in transcriptional regulation by affecting chromatin function (472). Indeed, Spt6 interacts directly with histones to control chromatin structure in vivo (31). Accordingly, Spt6 was proposed to function either in nucleosome assembly or as a histone acceptor/donor during chromatin reorganization (31).
Spt4, Spt5, and Spt6 homologs have been identified in humans, suggesting that their function is conserved among eukaryotic organisms (76–78, 188, 445). Recently, a human Spt4-Spt5 complex, denoted DSIF, was identified as an elongation factor that negatively and positively regulates RNA pol II processivity (513). Genetic and biochemical evidence is consistent with roles for a yeast Spt4-Spt5 complex in regulation of transcriptional elongation (189). Moreover, mutations in SPT5 can exert both positive and negative effects on transcription (94, 189, 472). Based on the genetic connection to chromatin, the Spt4-Spt5 complex might be an effector of transcriptional elongation that modulates chromatin structure to either repress or stimulate gene expression. The relationship of Spt6 to the Spt4-Spt5 complex is not yet clear.
Histone Deacetylases
A corollary to the relationship between histone acetylation and gene activation (see the section on histone acetyltransferases, above) implies that histone deacetylases (HDAs) would function as transcriptional repressors. Although this appears reasonable, until recently there was no direct experimental support. (HDAs, like HATs, have historically been difficult to purify.) Recently, the HDA inhibitor trapoxin was used to develop an HDA affinity matrix (481). This led to the purification of human HDA and subsequent isolation of the corresponding gene. Similar to the story for HAT, sequence analysis provided a link between histone deacetylation and transcriptional repression. Human HDA was found to be homologous to the product of the yeast RPD3 gene, identified in a genetic selection for transcriptional repressors (510). No biochemical function had been assigned to Rpd3. Thus, biochemistry revealed a function for the Rpd3 protein, whereas genetics provided a direct connection between histone deacetylation and transcriptional repression.
Yeast genetics suggested that HDAs function within multisubunit complexes. The original genetic selection that yielded RPD3 also identified SIN3 (RPD1) (510). Mutations in SIN3 and RPD3 cause the same array of phenotypes, and rpd1 rpd3 double mutants are phenotypically indistinguishable from single mutants. These results implied that Sin3 and Rpd3 are functionally related and operate either in the same pathway or in the same complex. Mutations in SIN3 were isolated in multiple genetic selections (SIN3 = RPD1 = UME4 = SDS16) based on relief of transcriptional repression of functionally unrelated genes (453, 504, 511, 521). Sin3 does not bind directly to DNA, but when tethered to a promoter as a LexA-Sin3 fusion, it represses gene expression (522). Also, LexA-Sin3-mediated repression is RPD3 dependent. Taken together, these results suggested a model for HDA-mediated transcriptional repression involving Sin3 as a corepressor, bridging gene-specific repressors to HDAs.
Direct support for this model comes from characterization of Ume6-mediated transcriptional repression, which is SIN3 and RPD3 dependent (234). Furthermore, Sin3 and Rpd3 physically interact (234). A 2-MDa complex containing both Sin3 and Rpd3 has since been identified (240). Other multisubunit HDA complexes have also been identified in yeast (63). One complex (HDB; 600 kDa) includes Rpd3, whereas the other (HDA; 350 kDa) includes Hda1, which is an Rpd3 homolog (405). Also, three new yeast genes, designated HOS1, HOS2, and HOS3, were identified based on sequence similarity to RPD3 and HDA1 (405). Mutations in all of these genes increase the acetylation of histones H3 and H4, suggesting that each affects histone acetylation in vivo (405). The components and functions of the yeast HDA complexes are likely to be highly conserved. Indeed, several novel components of mammalian HDA complexes, including SAP18 and SAP30, have functional counterparts in yeast (559, 560).
Large complexes containing Sin3 and Rpd3 have also been identified in mammalian cells (3, 190, 192, 271, 332, 559). These complexes mediate repression by unliganded nuclear hormone receptors or by Mad-Max, in each case dependent upon Sin3 and Rpd3. The beauty of these results is that they suggest a simple model to account for the genetic switch mediated by Mad-Max or the hormone receptors: Mad-Max or unliganded receptor complexes bind DNA and recruit the Sin3-Rpd3 complex, resulting in repression by histone deacetylation. In the case of the hormone receptors, the interaction with Sin3-Rpd3 is mediated by the corepressor N-CoR or SMRT (3, 192, 332). The switch to activation occurs when Myc-Max replaces Mad-Max, or upon receptor-hormone binding, resulting in replacement of the Sin3-Rpd3 HDA complex with a HAT complex that catalyzes histone acetylation. In essence, the model invokes targeted histone acetylation and deacetylation as a toggle between transcriptional activation and repression. A caveat to this model is that Sin3-mediated repression is not completely Rpd3 dependent (234, 271), which might be explained by redundant HDAs. Similarly to the Gcn5 dependence upon HAT activity (59), it was recently reported that Rpd3-mediated repression requires HDA activity (234a).
Ssn6-Tup1
The SSN6 gene was found as a suppressor of mutations in the SNF1 gene, which encodes a protein kinase required for expression of glucose-repressible genes (336, 425, 490). TUP1 was identified based on inappropriate expression of mating-specific genes (279). A functional connection between SSN6 and TUP1 was made based on similar pleiotropic phenotypes associated with mutations in both genes. Specifically, ssn6 and tup1 mutants exhibit constitutive invertase expression, are mating and sporulation defective, and are flocculent in liquid growth medium (337). Like ssn6, tup1 also suppresses snf1 and snf2 defects (337). Furthermore, SSN6 and TUP1 are identical to CYC8 and CYC9, respectively, which were identified based on enhanced expression of CYC7-encoded iso-2-cytochrome c conferred by mutations at these loci (397). This provided an early link between Ssn6/Cyc8 and Tup1/Cyc9 and suggested that these proteins might function as transcriptional repressors.
Repression of a-specific genes in α cells is mediated by the α2-Mcm1 transcriptional repressor complex. However, α2-Mcm1 association with operator DNA was not sufficient for repression. Rather, α2-Mcm1-mediated repression required Ssn6 and Tup1 (243). In addition, Mig1-mediated repression in response to glucose (335), Rox1-mediated repression in response to oxygen (566), and repression of DNA damage-inducible genes (561) require Ssn6 and Tup1. Thus, Ssn6 and Tup1 are general transcriptional repressors, affecting the expression of many functionally unrelated genes.
Ssn6 and Tup1 function within a complex (534). However, neither Ssn6 nor Tup1 directly binds DNA. The Ssn6-Tup1 complex is recruited to promoter DNA by gene-specific repressors, such as α2-Mcm1 (243). This requirement can be bypassed by tethering either Ssn6 or Tup1 to promoter DNA as LexA-Ssn6 or LexA-Tup1 fusion proteins through a LexA operator (243, 494). Although LexA-Ssn6-mediated repression is Tup1 dependent (243), LexA-Tup1 repression is not Ssn6-dependent (494). Thus, Ssn6 and Tup1 play distinct roles within a general repressor complex that is recruited to promoter DNA by gene-specific repressors.
The role of Ssn6-Tup1 as a general transcriptional repressor is reminiscent of the role of the Rpd3-Sin3 HDA complex in transcriptional repression. Both complexes actively repress transcription, but neither complex binds directly to promoter DNA. Interestingly, Tup1 interacts directly with the amino-terminal tails of histones H3 and H4, and this interaction is required for Tup1 function (121). Furthermore, mutations in the amino-terminal tails of H3 and H4 overcome Ssn6-Tup1-mediated repression (214). These results suggest that Ssn6-Tup1 represses transcription by affecting chromatin. Indeed, ssn6 and tup1 mutations alter SUC2 chromatin structure (151). Moreover, this effect can be suppressed by swi1 mutations, suggesting an interplay between the SWI/SNF chromatin-remodeling complex and the Ssn6-Tup1 repressor complex (151). Whether there is functional redundancy between Ssn6-Tup1 and HDA complexes and the extent to which transcriptional repression mediated by these complexes might be overcome by chromatin-remodeling complexes, remain to be determined.
Components of the core transcriptional machinery have also been implicated in the mechanism of Ssn6-Tup1-mediated repression. Several components of the SRB/mediator complex, including Rox3, Sin4, and Srb8 to Srb11, have been genetically identified as components of the Ssn6-Tup1 repression pathway in vivo (73, 265, 396, 443, 516). Furthermore, the Ssn6-Tup1 complex can repress transcription in vitro by using naked DNA templates (199, 386). These results suggest that Ssn6-Tup1 confers repression by more than a single mechanism, in one case by affecting chromatin structure and in the other case by interacting with components of the core transcriptional machinery.
CONCLUSIONS AND PERSPECTIVES
The convergence of genetics and biochemistry has provided a much clearer picture of the mechanisms involved in transcription initiation by RNA pol II. The inability of purified RNA pol II to accurately initiate transcription in vitro provided the initial assay for the GTFs (311, 529). Genetic studies have confirmed that GTFs do indeed play a general role in transcription initiation in vivo and in many cases identified either new factors or functions that had gone undetected biochemically. The crystal structures of TBP (342), TATA-TBP (246, 251), TATA-TBP-TFIIA (155, 475), TATA-TBP-TFIIB (341), a TAF heterotetramer (205), and, most recently, the nucleosome core particle (296) have been especially revealing. This multifaceted approach involving biochemistry, genetics, and structural biology will to continue to yield extraordinary insight into the organization of the PIC and the mechanisms of transcriptional activation.
It is interesting that most of the yeast GTFs have not been found genetically. Among the GTFs, only TBP (SPT15) (124) and the larger subunit of TFIIF (TFG1/SSU71) (459) turned up in genetic selections designed specifically to identify transcription factors. Although other GTF subunits were identified genetically, in no case was the selection set up with this in mind. As examples, sua7, ssl1, and ssl2 were found in genetic selections designed to uncover translation initiation factors (169, 182, 553) and several genes encoding TFIIH subunits were identified based on NER phenotypes. Moreover, among the disparate genetic selections for transcription factors, none has uncovered a gene encoding a TAF subunit of TFIID. In contrast, many of the coactivators other than TAFs and most of the general transcriptional repressors have been found genetically. Indeed, many of these factors would have gone undetected if not for these genetic selections. Presumably, the underrepresentation of genetically identified GTF and TAF genes is a consequence of the essential nature of these genes—only alleles that retain at least partial function can be uncovered for essential genes, whereas any allele that confers a phenotype can be uncovered for nonessential genes.
An emerging theme from the study of eukaryotic transcription factors is the existence of multiple forms of functionally distinct transcription complexes (reviewed in references 41 and 68). In yeast, there are at least two distinct forms of the holoenzyme, distinguished by their mediator composition and the spectrum of genes whose expression is affected (252, 261, 435). Multiple forms of TFIID have also been identified. A second form of TFIID (B-TFIID) was identified in mammalian cells (488, 489) and was recently reported to include a homolog of Mot1 (502). Accordingly, B-TFIID might be the mammalian counterpart of the yeast TBP-Mot1 complex (372, 514). Other TBP-TAF complexes have been described that respond to specific classes of activators (35, 226, 323), are cell cycle specific (428, 520), or are cell type specific (113, 186). Multiple forms of coactivators and repressors have also been described, including distinct HAT (165, 413) and HDA (405) complexes. Although functionally distinct, many of these complexes are redundant with other transcription factor complexes (393).
Recently, two fundamental premises of RNA pol II transcription have been challenged. It had been generally assumed that the core promoter was a generic element affecting only the accuracy of transcription initiation and that regulation was conferred by enhancer or repressor elements, independent of the core promoter. However, recent studies define specific interactions between TAFs and core promoter elements (47, 48). Moreover, yeast TAFII145 was identified as a core promoter-selectivity factor, suggesting that core promoter-TAFII145 interactions, rather than activator-TAFII145 interactions, mediate gene expression (434). Second, the concept of TAFs as requisite coactivators of transcription is no longer valid. TAFs are not generally required for activation of yeast genes (328, 517) but, instead, are required for the expression of specific genes, including those involved in cell cycle progression (5, 518). It will be interesting to learn whether other general factors, including GTFs and SRB/mediator components, might be dispensable for activation of certain genes.
Another theme to emerge from the characterization of global transcription factors is their ability to mediate either activation or repression. For example, SRB/mediator includes factors that affect either activation or repression (129, 230, 232, 368, 463). Similarly, most general repressors can also mediate activation. The recent identification of the Ccr4-NOT complex is a case in point, where Ccr4 was initially identified as a transcriptional coactivator and several Ccr4-NOT components either activate or repress the transcription of specific genes (289). This is also evident in the effect of histone deacetylation on telomeric silencing—rather than facilitating genomic silencing, Rpd3 counteracts silencing in both Drosophila and yeast (111). Clearly, much remains to be learned regarding the mechanisms by which these factors mediate activation and repression.
Now that all of the GTFs have been identified in both metazoan and yeast systems, the immediate challenge is to identify how these factors interact within the PIC and how PIC assembly responds to transcriptional regulators. Another challenge is to identify the components and associated activities of the various coactivator and repressor complexes. Ultimately, the spectrum of genes affected by these complexes must be defined. Yeast will remain a seminal organism in these studies, continuing to offer the power of classical and molecular genetic methods. These studies will be facilitated by the complete sequence of the yeast genome, allowing for factors, identified either genetically or biochemically, to be immediately defined. Moreover, DNA microarray analysis offers an unprecedented opportunity to define the effects of single-gene mutations on the expression of virtually every gene of S. cerevisiae (110).
ACKNOWLEDGMENTS
I am grateful to D. Reinberg, D. S. Gross, and members of my laboratory for valuable comments on the manuscript; to S. K. Burley and D. Reinberg for their generous gift of GTF figures; and to my colleagues who provided information prior to publication.
The research in my laboratory is supported by NIH grant GM-39484.
REFERENCES
- 1.Adamczewski J P, Rossignol M, Tassan J P, Nigg E A, Moncollin V, Egly J M. MAT1, cdk7 and cyclin H form a kinase complex which is UV light-sensitive upon association with TFIIH. EMBO J. 1996;15:1877–1884. [PMC free article] [PubMed] [Google Scholar]
- 2.Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 3.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 4.Allfrey V G, Faulkner R, Mirsky A E. Acetylation and methylation of histones and their possible role in regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apone L M, Virbasius C M A, Reese J C, Green M R. Yeast TAF(II)90 is required for cell-cycle progression through G(2)/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 6.Archambault J, Friesen J D. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archambault J, Lacroute F, Ruet A, Friesen J D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arndt K M, Ricupero S L, Eisenmann D M, Winston F. Biochemical and genetic characterization of a yeast TFIID mutant that alters transcription in vivo and DNA binding in vitro. Mol Cell Biol. 1992;12:2372–2382. doi: 10.1128/mcb.12.5.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arndt K M, Wobbe C R, Ricupero-Hovasse S, Struhl K, Winston F. Equivalent mutations in the two repeats of yeast TATA-binding protein confer distinct TATA recognition specificities. Mol Cell Biol. 1994;14:3719–3728. doi: 10.1128/mcb.14.6.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt K T, Styles C A, Fink G R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 12.Aso T, Conaway J W, Conaway R C. Role of core promoter structure in assembly of the RNA polymerase II preinitiation complex—a common pathway for formation of preinitiation intermediates at many TATA and TATA-less promoters. J Biol Chem. 1994;269:26575–26583. [PubMed] [Google Scholar]
- 13.Auble D T, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 14.Auble D T, Hansen K E, Mueller C G F, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 15.Auble D T, Wang D, Post K W, Hahn S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Mol Cell Biol. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awrey D E, Weilbaecher R G, Hemming S A, Orlicky S M, Kane C M, Edwards A M. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J Biol Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- 17.Bagby S, Kim S J, Maldonado E, Tong K I, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 18.Bangur C S, Pardee T S, Ponticelli A S. Mutational analysis of the D1/E1 core helices and the conserved N-terminal region of yeast transcription factor IIB (TFIIB): identification of an N-terminal mutant that stabilizes TATA-binding protein-TFIIB-DNA complexes. Mol Cell Biol. 1997;17:6784–6793. doi: 10.1128/mcb.17.12.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barberis A, Muller C W, Harrison S C, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 21.Bardwell A J, Bardwell L, Iyer N, Svejstrup J Q, Feaver W J, Kornberg R D, Friedberg E C. Yeast nucleotide excision repair proteins Rad2 and Rad4 interact with RNA polymerase II basal transcription factor b (TFIIH) Mol Cell Biol. 1994;14:3569–3576. doi: 10.1128/mcb.14.6.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardwell L, Bardwell A J, Feaver W J, Svejstrup J Q, Kornberg R D, Friedberg E C. Yeast RAD3 protein binds directly to both SSL2 and SSL1 proteins: implications for the structure and function of transcription/repair factor b. Proc Natl Acad Sci USA. 1994;91:3926–3930. doi: 10.1073/pnas.91.9.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barlev N A, Candau R, Wang L A, Darpino P, Silverman N, Berger S L. Characterization of physical interactions of the putative transcriptional adapter, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- 24.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berger S L, Cress W D, Cress A, Triezenberg S J, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adapters. Cell. 1990;61:1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- 26.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adapter required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 27.Berroteran R W, Ware D E, Hampsey M. The sua8 suppressors of Saccharomyces cerevisiae encode replacements of conserved residues within the largest subunit of RNA polymerase II and affect transcription start site selection similarly to sua7 (TFIIB) mutations. Mol Cell Biol. 1994;14:226–237. doi: 10.1128/mcb.14.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjorklund S, Kim Y J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 29.Blair W S, Cullen B R. A yeast TATA-binding protein mutant that selectively enhances gene expression from weak RNA polymerase II promoters. Mol Cell Biol. 1997;17:2888–2896. doi: 10.1128/mcb.17.5.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohr V A, Smith C A, Okumoto D S, Hanawalt P C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 31.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 32.Bradsher J N, Tan S, McLaury H J, Conaway J W, Conaway R C. RNA polymerase II transcription factor SIII. II. Functional properties and role in RNA chain elongation. J Biol Chem. 1993;268:25594–25603. [PubMed] [Google Scholar]
- 33.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 34.Brohl S, Lisowsky T, Riemen G, Michaelis G. A new nuclear suppressor system for a mitochondrial RNA polymerase mutant identifies an unusual zinc-finger protein and a polyglutamine domain protein in Saccharomyces cerevisiae. Yeast. 1994;10:719–731. doi: 10.1002/yea.320100604. [DOI] [PubMed] [Google Scholar]
- 35.Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly J M, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 37.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 39.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 40.Buermeyer A B, Strasheim L A, McMahon S L, Farnham P J. Identification of cis-acting elements that can obviate a requirement for the C-terminal domain of RNA polymerase II. J Biol Chem. 1995;270:6798–6807. doi: 10.1074/jbc.270.12.6798. [DOI] [PubMed] [Google Scholar]
- 41.Buratowski S. Multiple TATA-binding factors come back into style. Cell. 1997;91:13–15. doi: 10.1016/s0092-8674(01)80004-0. [DOI] [PubMed] [Google Scholar]
- 42.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 43.Buratowski S, Hahn S, Sharp P A, Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988;334:37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- 44.Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 46.Buratowski S, Zhou H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science. 1992;255:1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- 47.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 49.Burley S K. The TATA box binding protein. Curr Opin Struct Biol. 1996;6:69–75. doi: 10.1016/s0959-440x(96)80097-2. [DOI] [PubMed] [Google Scholar]
- 50.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 51.Burns L G, Peterson C L. Protein complexes for remodeling chromatin. Biochim Biophys Acta. 1997;1350:159–168. doi: 10.1016/s0167-4781(96)00162-5. [DOI] [PubMed] [Google Scholar]
- 52.Burton Z F, Killeen M, Sopta M, Ortolan L G, Greenblatt J. RAP30/74: a general initiation factor that binds to RNA polymerase II. Mol Cell Biol. 1988;8:1602–1613. doi: 10.1128/mcb.8.4.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bushnell D A, Bamdad C, Kornberg R D. A minimal set of RNA polymerase II transcription protein interactions. J Biol Chem. 1996;271:20170–20174. doi: 10.1074/jbc.271.33.20170. [DOI] [PubMed] [Google Scholar]
- 54.Cade R M, Errede B. MOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3139–3149. doi: 10.1128/mcb.14.5.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cairns B R, Henry N L, Kornberg R D. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cairns B R, Lorch Y, Li Y, Zhang M C, Lacomis L, Erdjumentbromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 58.Candau R, Berger S L. Structural and functional analysis of yeast putative adapters—evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 59.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carcamo J, Buckbinder L, Reinberg D. The initiator directs the assembly of a transcription factor IID-dependent transcription complex. Proc Natl Acad Sci USA. 1991;88:8052–8056. doi: 10.1073/pnas.88.18.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 63.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 64.Cavallini B, Faus I, Matthes H, Chipoulet J M, Winsor B, Egly J M, Chambon P. Cloning of the gene encoding the yeast protein BTF1Y, which can substitute for the human TATA box-binding factor. Proc Natl Acad Sci USA. 1989;86:9803–9807. doi: 10.1073/pnas.86.24.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavallini B, Huet J, Plassat J L, Sentenac A, Egly J M, Chambon P. A yeast activity can substitute for the HeLa cell TATA box factor. Nature. 1988;334:77–80. doi: 10.1038/334077a0. [DOI] [PubMed] [Google Scholar]
- 66.Chambers R S, Dahmus M E. Purification and characterization of a phosphatase from HeLa cells which dephosphorylates the C-terminal domain of RNA polymerase II. J Biol Chem. 1994;269:26243–26248. [PubMed] [Google Scholar]
- 67.Chambers R S, Wang B Q, Burton Z F, Dahmus M E. The activity of COOH-terminal domain phosphatase is regulated by a docking site on RNA polymerase II and by the general transcription factors IIF and IIB. J Biol Chem. 1995;270:14962–14969. doi: 10.1074/jbc.270.25.14962. [DOI] [PubMed] [Google Scholar]
- 68.Chang M, Jaehning J A. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 70.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 71.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 72.Chen S, West R W, Jr, Johnson S L, Gans H, Kruger B, Ma J. TSF3, a global regulatory protein that silences transcription of yeast GAL genes, also mediates repression by alpha 2 repressor and is identical to SIN4. Mol Cell Biol. 1993;13:831–840. doi: 10.1128/mcb.13.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S, West R W, Jr, Ma J, Johnson S L, Gans H, Woldehawariat G. TSF1 to TSF6, required for silencing the Saccharomyces cerevisiae GAL genes, are global regulatory genes. Genetics. 1993;134:701–716. doi: 10.1093/genetics/134.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W, Struhl K. Saturation mutagenesis of a yeast his3 “TATA element”: genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci USA. 1988;85:2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen W, Tabor S, Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987;50:1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- 76.Chiang P W, Fogel E, Jackson C L, Lieuallen K, Lennon G, Qu X, Wang S Q, Kurnit D M. Isolation, sequencing, and mapping of the human homologue of the yeast transcription factor, SPT5. Genomics. 1996;38:421–424. doi: 10.1006/geno.1996.0646. [DOI] [PubMed] [Google Scholar]
- 77.Chiang P W, Wang S, Smithivas P, Song W J, Ramamoorthy S, Hillman J, Puett S, Van Keuren M L, Crombez E, Kumar A, Glover T W, Miller D E, Tsai C H, Blackburn C C, Chen X N, Sun Z, Cheng J F, Korenberg J R, Kurnit D M. Identification and analysis of the human and murine putative chromatin structure regulator SUPT6H and Supt6h. Genomics. 1996;34:328–333. doi: 10.1006/geno.1996.0294. [DOI] [PubMed] [Google Scholar]
- 78.Chiang P W, Wang S Q, Smithivas P, Song W J, Crombez E, Akhtar A, Im R, Greenfield J, Ramamoorthy S, Van Keuren M, Blackburn C C, Tsai C H, Kurnit D M. Isolation and characterization of the human and mouse homologues (SUPT4H and Supt4h) of the yeast SPT4 gene. Genomics. 1996;34:368–375. doi: 10.1006/geno.1996.0299. [DOI] [PubMed] [Google Scholar]
- 79.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the Rna polymerase Ii carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choder M, Young R A. A portion of RNA polymerase II molecules has a component essential for stress responses and stress survival. Mol Cell Biol. 1993;13:6984–6991. doi: 10.1128/mcb.13.11.6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chou S, Struhl K. Transcriptional activation by TFIIB mutants that are severely impaired in interaction with promoter DNA and acidic activation domains. Mol Cell Biol. 1997;17:6794–6802. doi: 10.1128/mcb.17.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choy B, Green M R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993;366:531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 83.Cisek L J, Corden J L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- 84.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clark-Adams C D, Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 87.Coleman R A, Pugh B F. Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding. Proc Natl Acad Sci USA. 1997;94:7221–7226. doi: 10.1073/pnas.94.14.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coleman R A, Taggart A K P, Benjamin L R, Pugh B F. Dimerization of the TATA binding protein. J Biol Chem. 1995;270:13842–13849. doi: 10.1074/jbc.270.23.13842. [DOI] [PubMed] [Google Scholar]
- 89.Collart, M. A. Personal communication.
- 90.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Collart M A, Struhl K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Collart M A, Struhl K. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 93.Comai L, Tanese N, Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 94.Compagnone-Post P A, Osley M A. Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae. Genetics. 1996;143:1543–1554. doi: 10.1093/genetics/143.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conaway J W, Conaway R C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J Biol Chem. 1989;264:2357–2362. [PubMed] [Google Scholar]
- 96.Conaway R C, Conaway J W. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–690. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 97.Conaway R C, Conaway J W. An RNA polymerase II transcription factor has an associated DNA-dependent ATPase (dATPase) activity strongly stimulated by the TATA region of promoters. Proc Natl Acad Sci USA. 1989;86:7356–7360. doi: 10.1073/pnas.86.19.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cooper K F, Mallory M J, Smith J B, Strich R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 100.Cortes P, Flores O, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: purification and analysis of transcription factor IIA and identification of transcription factor IIJ. Mol Cell Biol. 1992;12:413–421. doi: 10.1128/mcb.12.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 102.Coulombe B, Li J, Greenblatt J. Topological localization of the human transcription factors IIA, IIB, TATA box-binding protein, and RNA polymerase II-associated protein 30 on a class II promoter. J Biol Chem. 1994;269:19962–19967. [PubMed] [Google Scholar]
- 103.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 104.Dahmus M E. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog Nucleic Acid Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- 105.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DeJong J, Bernstein R, Roeder R G. Human general transcription factor TFIIA: Characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.DeJong J, Roeder R G. A single cDNA, hTFIIA/alpha, encodes both the p35 and p19 subunits of human TFIIA. Genes Dev. 1993;7:2220–2234. doi: 10.1101/gad.7.11.2220. [DOI] [PubMed] [Google Scholar]
- 108.Denis C L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984;108:833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Denis C L, Draper M P, Liu H Y, Malvar T, Vallari R C, Cook W J. The yeast CCR4 protein is neither regulated by nor associated with the SPT6 and SPT10 proteins and forms a functionally distinct complex from that of the SNF/SWI transcription factors. Genetics. 1994;138:1005–1013. doi: 10.1093/genetics/138.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 111.De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 112.Dikstein R, Ruppert S, Tjian R. TAF(II)250 is a bipartite protein kinase that phosphorylates the basal transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 113.Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- 114.Dollard C, Ricupero-Hovasse S L, Natsoulis G, Boeke J D, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Draper M P, Liu H Y, Nelsbach A H, Mosley S P, Denis C L. CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol Cell Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Drapkin R, Reardon J T, Ansari A, Huang J C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 117.Du, J., I. Nasir, and B. C. Laurent. Personal communication.
- 118.Du L, Warren S L. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dvir A, Conaway R C, Conaway J W. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc Natl Acad Sci USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dvir A, Garrett K P, Chalut C, Egly J M, Conaway J W, Conaway R C. A role for ATP and TFIIH in activation of the RNA polymerase II preinitiation complex prior to transcription initiation. J Biol Chem. 1996;271:7245–7248. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 121.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 122.Edwards A M, Kane C M, Young R A, Kornberg R D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 123.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 124.Eisenmann D M, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–91. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 125.Espinoza F H, Farrell A, Erdjumentbromage H, Tempst P, Morgan D O. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 126.Fan H Y, Cheng K K, Klein H L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 Delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fan H Y, Klein H L. Characterization of mutations that suppress the temperature-sensitive growth of the hpr1 delta mutant of Saccharomyces cerevisiae. Genetics. 1994;137:945–956. doi: 10.1093/genetics/137.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fang S M, Burton Z F. RNA polymerase II-associated protein (RAP) 74 binds transcription factor (TF) IIB and blocks TFIIB-RAP30 binding. J Biol Chem. 1996;271:11703–11709. doi: 10.1074/jbc.271.20.11703. [DOI] [PubMed] [Google Scholar]
- 129.Fassler J S, Gray W, Lee J P, Yu G Y, Gingerich G. The Saccharomyces cerevisiae SPT14 gene is essential for normal expression of the yeast transposon, Ty, as well as for expression of the HIS4 gene and several genes in the mating pathway. Mol Gen Genet. 1991;230:310–320. doi: 10.1007/BF00290682. [DOI] [PubMed] [Google Scholar]
- 130.Fassler J S, Winston F. The Saccharomyces cerevisiae SPT13/GAL11 gene has both positive and negative regulatory roles in transcription. Mol Cell Biol. 1989;9:5602–5609. doi: 10.1128/mcb.9.12.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Feaver W J, Gileadi O, Kornberg R D. Purification and characterization of yeast RNA polymerase II transcription factor b. J Biol Chem. 1991;266:19000–19005. [PubMed] [Google Scholar]
- 132.Feaver W J, Gileadi O, Li Y, Kornberg R D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 133.Feaver W J, Henry N L, Bushnell D A, Sayre M H, Brickner J H, Gileadi O, Kornberg R D. Yeast TFIIE—cloning, expression, and homology to vertebrate proteins. J Biol Chem. 1994;269:27549–27553. [PubMed] [Google Scholar]
- 134.Feaver W J, Henry N L, Wang Z, Wu X, Svejstrup J Q, Bushnell D A, Friedberg E C, Kornberg R D. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J Biol Chem. 1997;272:19319–19327. doi: 10.1074/jbc.272.31.19319. [DOI] [PubMed] [Google Scholar]
- 135.Feaver W J, Svejstrup J Q, Bardwell L, Bardwell A J, Buratowski S, Gulyas K D, Donahue T F, Friedberg E C, Kornberg R D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 136.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 137.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 138.Flanagan P M, Kelleher R J, Sayre M H, Tschochner H, Kornberg R D. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 139.Flores O, Ha I, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 140.Flores O, Lu H, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 141.Flores O, Maldonado E, Burton Z, Greenblatt J, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. RNA polymerase II-associating protein 30 is an essential component of transcription factor IIF. J Biol Chem. 1988;263:10812–10816. [PubMed] [Google Scholar]
- 142.Flores O, Maldonado E, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem. 1989;264:8913–8921. [PubMed] [Google Scholar]
- 143.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Forget D, Robert F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Furter-Graves E M, Furter R, Hall B D. SHI, a new yeast gene affecting the spacing between TATA and transcription initiation sites. Mol Cell Biol. 1991;11:4121–4127. doi: 10.1128/mcb.11.8.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Furter-Graves E M, Hall B D. DNA sequence elements required for transcription initiation of the Schizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol Gen Genet. 1990;223:407–416. doi: 10.1007/BF00264447. [DOI] [PubMed] [Google Scholar]
- 147.Furter-Graves E M, Hall B D, Furter R. Role of a small RNA pol II subunit in TATA to transcription start site spacing. Nucleic Acids Res. 1994;22:4932–4936. doi: 10.1093/nar/22.23.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Garrett K P, Serizawa H, Hanley J P, Bradsher J N, Tsuboi A, Arai N, Yokota T, Arai K, Conaway R C, Conaway J W. The carboxyl terminus of RAP30 is similar in sequence to region 4 of bacterial sigma factors and is required for function. J Biol Chem. 1992;267:23942–23949. [PubMed] [Google Scholar]
- 150.Gaudreau L, Schmid A, Blaschke D, Ptashne M, Horz W. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell. 1997;89:55–62. doi: 10.1016/s0092-8674(00)80182-8. [DOI] [PubMed] [Google Scholar]
- 151.Gavin I M, Simpson R T. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 153.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 154.Ge H, Zhao Y M, Chait B T, Roeder R G. Phosphorylation negatively regulates the function of coactivator PC4. Proc Natl Acad Sci USA. 1994;91:12691–12695. doi: 10.1073/pnas.91.26.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Geiger J H, Hahn S, Lee S, Sigler P B. Crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 156.Georgakopoulos T, Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gerard M, Fischer L, Moncollin V, Chipoulet J M, Chambon P, Egly J M. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J Biol Chem. 1991;266:20940–20945. [PubMed] [Google Scholar]
- 158.Giardina C, Lis J T. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 159.Gibson T J, Thompson J D, Blocker A, Kouzarides T. Evidence for a protein domain superfamily shared by the cyclins, TFIIB and RB/p107. Nucleic Acids Res. 1994;22:946–952. doi: 10.1093/nar/22.6.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gileadi O, Feaver W J, Kornberg R D. Cloning of a subunit of yeast RNA polymerase II transcription factor b and CTD kinase. Science. 1992;257:1389–1392. doi: 10.1126/science.1445600. [DOI] [PubMed] [Google Scholar]
- 161.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 162.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 163.Goppelt A, Meisterernst M. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4450–4455. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 165.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 166.Greenblatt J. RNA polymerase-associated transcription factors. Trends Biochem Sci. 1991;16:408–411. doi: 10.1016/0968-0004(91)90165-r. [DOI] [PubMed] [Google Scholar]
- 167.Greenleaf A L. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- 168.Guarente L, Hoar E. Upstream activation sites of the CYC1 gene of Saccharomyces cerevisiae are active when inverted but not when placed downstream of the “TATA box.”. Proc Natl Acad Sci USA. 1984;81:7860–7864. doi: 10.1073/pnas.81.24.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gulyas K D, Donahue T F. SSL2, a suppressor of a stem-loop mutation in the HIS4 leader encodes the yeast homolog of human ERCC-3. Cell. 1992;69:1031–1042. doi: 10.1016/0092-8674(92)90621-i. [DOI] [PubMed] [Google Scholar]
- 170.Gustafsson C M, Myers L C, Li Y, Redd M J, Lui M, Erdjumentbromage H, Tempst P, Kornberg R D. Identification of Rox3 as a component of mediator and RNA polymerase II holoenzyme. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 171.Guzder S N, Qiu H, Sommers C H, Sung P, Prakash L, Prakash S. DNA repair gene RAD3 of S. cerevisiae is essential for transcription by RNA polymerase II. Nature. 1994;367:91–94. doi: 10.1038/367091a0. [DOI] [PubMed] [Google Scholar]
- 172.Guzder S N, Sung P, Prakash L, Prakash S. Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J Biol Chem. 1996;271:8903–8910. doi: 10.1074/jbc.271.15.8903. [DOI] [PubMed] [Google Scholar]
- 173.Ha I, Lane W S, Reinberg D. Cloning of a human gene encoding the general transcription initiation factor IIB. Nature. 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 174.Ha I, Roberts S, Maldonado E, Sun X, Kim L U, Green M, Reinberg D. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 175.Hahn S. Efficiency in activation. Nature. 1993;363:672–673. doi: 10.1038/363672a0. [DOI] [PubMed] [Google Scholar]
- 176.Hahn S, Buratowski S, Sharp P A, Guarente L. Identification of a yeast protein homologous in function to the mammalian general transcription factor, TFIIA. EMBO J. 1989;8:3379–3382. doi: 10.1002/j.1460-2075.1989.tb08501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hahn S, Buratowski S, Sharp P A, Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 178.Hahn S, Buratowski S, Sharp P A, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hahn S, Hoar E T, Guarente L. Each of three “TATA elements” specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 181.Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- 182.Hampsey M, Na J G, Pinto I, Ware D E, Berroteran R W. Extragenic suppressors of a translation initiation defect in the cyc1 gene of Saccharomyces cerevisiae. Biochimie. 1991;73:1445–55. doi: 10.1016/0300-9084(91)90177-3. [DOI] [PubMed] [Google Scholar]
- 183.Hampsey M, Reinberg D. Why are TAFs essential? Curr Biol. 1997;7:44–46. doi: 10.1016/s0960-9822(06)00018-2. [DOI] [PubMed] [Google Scholar]
- 184.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 185.Han M, Kim U J, Kayne P, Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988;7:2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hansen S K, Takada S, Jacobson R H, Lis J T, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- 187.Hansen S K, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 188.Hartzog G A, Basrai M A, Ricupero-Hovasse S L, Hieter P, Winston F. Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2848–2856. doi: 10.1128/mcb.16.6.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 191.Healy A M, Helser T L, Zitomer R S. Sequences required for transcriptional initiation of the Saccharomyces cerevisiae CYC7 genes. Mol Cell Biol. 1987;7:3785–3791. doi: 10.1128/mcb.7.10.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 193.Hekmatpanah D S, Young R A. Mutations in a conserved region of RNA polymerase II influence the accuracy of mRNA start site selection. Mol Cell Biol. 1991;11:5781–5791. doi: 10.1128/mcb.11.11.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hengartner C J, Thompson C M, Zhang J H, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 195.Henry N L, Bushnell D A, Kornberg R D. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem. 1996;271:21842–21847. doi: 10.1074/jbc.271.36.21842. [DOI] [PubMed] [Google Scholar]
- 196.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 197.Henry N L, Sayre M H, Kornberg R D. Purification and characterization of yeast RNA polymerase II general initiation factor g. J Biol Chem. 1992;267:23388–23392. [PubMed] [Google Scholar]
- 198.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 199.Herschbach B M, Arnaud M B, Johnson A D. Transcriptional repression directed by the yeast alpha 2 protein in vitro. Nature. 1994;370:309–311. doi: 10.1038/370309a0. [DOI] [PubMed] [Google Scholar]
- 200.Himmelfarb H J, Pearlberg J, Last D H, Ptashne M. GAL11P: a yeast mutation that potentiates the effect of weak GAL4-derived activators. Cell. 1990;63:1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- 201.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 202.Hisatake K, Roeder R G, Horikoshi M. Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature. 1993;363:744–747. doi: 10.1038/363744a0. [DOI] [PubMed] [Google Scholar]
- 203.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 204.Hoffmann A, Sinn E, Yamamoto T, Wang J, Roy A, Horikoshi M, Roeder R G. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID) Nature. 1990;346:387–390. doi: 10.1038/346387a0. [DOI] [PubMed] [Google Scholar]
- 205.Hoffmann A, Chiang C M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. A histone octamer-like structure within TFIID. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 206.Hoffmann A, Oelgeschlager T, Roeder R G. Considerations of transcriptional control mechanisms: do TFIID-core promoter complexes recapitulate nucleosome-like functions? Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Holstege F C, van der Vliet P C, Timmers H T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 208.Holstege F C P, Tantin D, Carey M, Vandervliet P C, Timmers H T M. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hoopes B C, LeBlanc J F, Hawley D K. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J Biol Chem. 1992;267:11539–11547. [PubMed] [Google Scholar]
- 210.Horikoshi M, Wang C K, Fujii H, Cromlish J A, Weil P A, Roeder R G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989;341:299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- 211.Horikoshi M, Wang C K, Fujii H, Cromlish J A, Weil P A, Roeder R G. Purification of a yeast TATA box-binding protein that exhibits human transcription factor IID activity. Proc Natl Acad Sci USA. 1989;86:4843–4847. doi: 10.1073/pnas.86.13.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Horiuchi J, Silverman N, Marcus G A, Guarente L. ADA3, a putative transcriptional adapter, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Horiuchi J, Silverman N, Pina B, Marcus G A, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huang L, Zhang W, Roth S Y. Amino termini of histones H3 and H4 are required for α1-α2 repression in yeast. Mol Cell Biol. 1997;17:6555–6562. doi: 10.1128/mcb.17.11.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huet J, Sentenac A. The TATA-binding protein participates in TFIIIB assembly on tRNA genes. Nucleic Acids Res. 1992;20:6451–6454. doi: 10.1093/nar/20.24.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hull M W, Mckune K, Woychik N A. RNA polymerase II subunit RPB9 is required for accurate start site selection. Genes Dev. 1995;9:481–490. doi: 10.1101/gad.9.4.481. [DOI] [PubMed] [Google Scholar]
- 217.Humbert S, van Vuuren H, Lutz Y, Hoeijmakers J H, Egly J M, Moncollin V. p44 and p34 subunits of the Btf2/Tfiih transcription factor have homologies with Ssl1, a yeast protein involved in Dna repair. EMBO J. 1994;13:2393–2398. doi: 10.1002/j.1460-2075.1994.tb06523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 219.Imbalzano A N, Zaret K S, Kingston R E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 220.Inostroza J, Flores O, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of general transcription factor IIE. J Biol Chem. 1991;266:9304–9308. [PubMed] [Google Scholar]
- 221.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 222.Irie K, Yamaguchi K, Kawase K, Matsumoto K. The yeast MOT2 gene encodes a putative zinc finger protein that serves as a global negative regulator affecting expression of several categories of genes, including mating-pheromone-responsive genes. Mol Cell Biol. 1994;14:3150–3157. doi: 10.1128/mcb.14.5.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Iyer V, Struhl K. Mechanism of differential utilization of the his3 T-R and T-C TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 226.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAF(II)30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 227.Jaehning, J. A. Personal communication.
- 228.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 229.Jiang Y, Triezenberg S J, Gralla J D. Defective transcriptional activation by diverse VP16 mutants associated with a common inability to form open promoter complexes. J Biol Chem. 1994;269:5505–5508. [PubMed] [Google Scholar]
- 230.Jiang Y W, Dohrmann P R, Stillman D J. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Jiang Y W, Stillman D J. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4503–4514. doi: 10.1128/mcb.12.10.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jiang Y W, Stillman D J. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 234.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 234a.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kaiser K, Meisterernst M. The human general co-factors. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 236.Kaldis P, Sutton A, Solomon M J. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 237.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 238.Kang J J, Auble D T, Ranish J A, Hahn S. Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kassavetis G A, Joazeiro C A, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 240.Kasten M M, Dorland S, Stillman D J. A large complex containing the yeast Sin3p and Rpd3 transcriptional regulators. Mol Cell Biol. 1997;17:4852–4856. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kaufmann J, Verrijzer C P, Shao J, Smale S T. CIF, an essential cofactor for TFIID-dependent initiator function. Genes Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- 242.Keil R L, Roeder G S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 243.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 244.Kelleher R J, Flanagan P M, Kornberg R D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 245.Kim J, Parvin J D, Shykind B M, Sharp P A. A negative cofactor containing Dr1/p19 modulates transcription with TFIIA in a promoter-specific fashion. J Biol Chem. 1996;271:18405–18412. doi: 10.1074/jbc.271.31.18405. [DOI] [PubMed] [Google Scholar]
- 246.Kim J L, Nikolov D B, Burley S K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 247.Kim S, Na J G, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kim T K, Hashimoto S, Kelleher R J, Flanagan P M, Kornberg R D, Horikoshi M, Roeder R G. Effects of activation-defective TBP mutations on transcription initiation in yeast. Nature. 1994;369:252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- 249.Kim T K, Lagrange T, Wang Y H, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kim U, Qin X F, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder R G. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 251.Kim Y, Geiger J H, Hahn S, Sigler P B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 252.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 253.Kitajima S, Tanaka Y, Kawaguchi T, Nagaoka T, Weissman S M, Yasukochi Y. A heteromeric transcription factor required for mammalian RNA polymerase II. Nucleic Acids Res. 1990;18:4843–4849. doi: 10.1093/nar/18.16.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 255.Klebanow E R, Poon D, Zhou S, Weil P A. Cloning and characterization of an essential Saccharomyces cerevisiae gene, TAF40, an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1997;272:9436–9442. doi: 10.1074/jbc.272.14.9436. [DOI] [PubMed] [Google Scholar]
- 256.Klebanow E R, Poon D, Zhou S, Weil P A. Isolation and characterization of TAF25, an essential yeast gene that encodes an RNA polymerase II-specific TATA-binding protein-associated factor. J Biol Chem. 1996;271:13706–13715. doi: 10.1074/jbc.271.23.13706. [DOI] [PubMed] [Google Scholar]
- 257.Klein C, Struhl K. Increased recruitment of TATA-binding protein to the promoter by transcriptional activation domains in vivo. Science. 1994;266:280–282. doi: 10.1126/science.7939664. [DOI] [PubMed] [Google Scholar]
- 258.Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 259.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Koleske A J, Buratowski S, Nonet M, Young R A. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- 261.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 261a.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 262.Kolodziej P A, Young R A. Mutations in the three largest subunits of yeast RNA polymerase II that affect enzyme assembly. Mol Cell Biol. 1991;11:4669–4678. doi: 10.1128/mcb.11.9.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kretzschmar M, Meisterernst M, Roeder R G. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 265.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSM8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kuldell N H, Buratowski S. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol Cell Biol. 1997;17:5288–5298. doi: 10.1128/mcb.17.9.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 268.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 269.Lagrange T, Kim T K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lagrange T, Kim T-K, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 272.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 273.Lee D K, Horikoshi M, Roeder R G. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–50. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- 274.Lee J M, Greenleaf A L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 275.Lee M, Struhl K. Mutations on the DNA-binding surface of TATA-binding protein can specifically impair the response to acidic activators in vivo. Mol Cell Biol. 1995;15:5461–5469. doi: 10.1128/mcb.15.10.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lee M, Struhl K. A severly defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lee S, Hahn S. Model for binding of transcription factor TFIIB to the TBP-DNA complex. Nature. 1995;376:609–612. doi: 10.1038/376609a0. [DOI] [PubMed] [Google Scholar]
- 278.Lee Y C, Min S, Gim B S, Kim Y-J. A transcriptional mediator protein that is required for activation of many RNA polymerase promoters and is conserved from yeast to humans. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lemontt J F, Fugit D R, MacKay V L. Pleiotropic mutations at the TUP1 locus that affect the expression of mating-type-dependent functions in Saccharomyces cerevisiae. Genetics. 1980;94:899–920. doi: 10.1093/genetics/94.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Leuther K K, Bushnell D A, Kornberg R D. Two-dimensional crystallography of TFIIB- and IIE-RNA polymerase II complexes: implications for start site selection and initiation complex formation. Cell. 1996;85:773–779. doi: 10.1016/s0092-8674(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 281.Li W Z, Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Li Y, Bjorklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Li Y, Bjorklund S, Kim Y J, Kornberg R D. Yeast RNA polymerase II holoenzyme. Methods Enzymol. 1996;273:172–175. doi: 10.1016/s0076-6879(96)73017-3. [DOI] [PubMed] [Google Scholar]
- 284.Li Y, Flanagan P M, Tschochner H, Kornberg R D. RNA polymerase II initiation factor interactions and transcription start site selection. Science. 1994;263:805–807. doi: 10.1126/science.8303296. [DOI] [PubMed] [Google Scholar]
- 285.Liao S M, Zhang J H, Jeffrey D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, Vanvuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 286.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 287.Lin C W, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder R H. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6436–6443. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 289.Liu H-Y, Badarinarayana V, Audino D C, Rappsilber J, Mann M, Denis C L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lobo S M, Tanaka M, Sullivan M L, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 291.Lopez-De-Leon A, Librizzi M, Puglia K, Willis I M. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 292.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Lu H, Zawel L, Fisher L, Egly J M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 294.Lue N F, Buchman A R, Kornberg R D. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element. Proc Natl Acad Sci USA. 1989;86:486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lue N F, Kornberg R D. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 297.Ma D, Watanabe H, Mermelstein F, Admon A, Oguri K, Sun X, Wada T, Imai T, Shiroya T, Reinberg D, Handa H. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 1993;7:2246–2257. doi: 10.1101/gad.7.11.2246. [DOI] [PubMed] [Google Scholar]
- 298.Ma D M, Olave I, Merino A, Reinberg D. Separation of the transcriptional coactivator and antirepression functions of transcription factor IIA. Proc Natl Acad Sci USA. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Malagon F, Aguilera A. Differential intrachromosomal hyper-recombination phenotype of spt4 and spt6 mutants of S. cerevisiae. Curr Genet. 1996;30:101–106. doi: 10.1007/s002940050107. [DOI] [PubMed] [Google Scholar]
- 301.Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 303.Malik S, Hisatake K, Sumimoto H, Horikoshi M, Roeder R G. Sequence of general transcription factor TFIIB and relationships to other initiation factors. Proc Natl Acad Sci USA. 1991;88:9553–9557. doi: 10.1073/pnas.88.21.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Malik S, Lee D K, Roeder R G. Potential RNA polymerase II-induced interactions of transcription factor TFIIB. Mol Cell Biol. 1993;13:6253–6259. doi: 10.1128/mcb.13.10.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Malone E A, Clark C D, Chiang A, Winston F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5710–5717. doi: 10.1128/mcb.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Malone E A, Fassler J S, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- 307.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Marcus G A, Silverman N, Berger S L, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Marshall N F, Peng J, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 311.Matsui T, Segall J, Weil P A, Roeder R G. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- 312.Maxon M E, Tjian R. Transcriptional activity of transcription factor IIE is dependent on zinc binding. Proc Natl Acad Sci USA. 1994;91:9529–9533. doi: 10.1073/pnas.91.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Program A E, Shuman S, Bentley D L. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G H, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 315.McCracken S, Greenblatt J. Related RNA polymerase-binding regions in human RAP30/74 and Escherichia coli sigma 70. Science. 1991;253:900–902. doi: 10.1126/science.1652156. [DOI] [PubMed] [Google Scholar]
- 316.McKenzie E A, Kent N A, Dowell S J, Moreno F, Bird L E, Mellor J. The centromere and promoter factor 1, CPF1, of Saccharomyces cerevisiae modulates gene activity through a family of factors including SPT21, RPD1 (SIN3), RPD3 and CCR4. Mol Gen Genet. 1993;240:374–386. doi: 10.1007/BF00280389. [DOI] [PubMed] [Google Scholar]
- 317.McKune K, Moore P A, Hull M W, Woychik N A. Six human RNA polymerase subunits functionally substitute for their yeast counterparts. Mol Cell Biol. 1995;15:6895–6900. doi: 10.1128/mcb.15.12.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. The HIP1 binding site is required for growth regulation of the dihydrofolate reductase gene promoter. Mol Cell Biol. 1992;12:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 320.Meisterernst M, Roy A L, Lieu H M, Roeder R G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 321.Meisterernst M, Stelzer G, Roeder R G. Poly(ADP-ribose) polymerase enhances activator-dependent transcription in vitro. Proc Natl Acad Sci USA. 1997;94:2261–2265. doi: 10.1073/pnas.94.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mellon I, Hanawalt P C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 323.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. Cloning and characterization of hTAF(II)18, hTAF(II)20 and hTAF(II)28: three subunits of the human transcription factor TFIID. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 325.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 326.Mittal V, Hernandez N. Role for the amino-terminal region of human TBP in U6 snRNA transcription. Science. 1997;275:1136–1140. doi: 10.1126/science.275.5303.1136. [DOI] [PubMed] [Google Scholar]
- 327.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owenhughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 328.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 329.Moqtaderi Z, Yale J D, Struhl K, Buratowski S. Yeast homologues of higher eukaryotic TFIID subunits. Proc Natl Acad Sci USA. 1996;93:14654–14658. doi: 10.1073/pnas.93.25.14654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Nagawa F, Fink G R. The relationship between the “TATA” sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 333.Nakatani Y, Bagby S, Ikura M. The histone folds in transcription factor TFIID. J Biol Chem. 1996;271:6575–6578. doi: 10.1074/jbc.271.12.6575. [DOI] [PubMed] [Google Scholar]
- 334.Natsoulis G, Winston F, Boeke J D. The SPT10 and SPT21 genes of Saccharomyces cerevisiae. Genetics. 1994;136:93–105. doi: 10.1093/genetics/136.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Neigeborn L, Carlson M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics. 1987;115:247–253. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Neigeborn L, Celenza J L, Carlson M. SSN20 is an essential gene with mutant alleles that suppress defects in SUC2 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:672–678. doi: 10.1128/mcb.7.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Nikolov D B, Burley S K. 2.1 A resolution refined structure of a TATA box-binding protein (TBP) Nat Struct Biol. 1994;1:621–637. doi: 10.1038/nsb0994-621. [DOI] [PubMed] [Google Scholar]
- 340.Nikolov D B, Burley S K. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 342.Nikolov D B, Hu S H, Lin J, Gasch A, Hoffmann A, Horikoshi M, Chua N H, Roeder R G, Burley S K. Crystal structure of TFIID TATA-box binding protein. Nature. 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 343.Nishizawa M, Suzuki Y, Nogi Y, Matsumoto K, Fukasawa T. Yeast Gal11 protein mediates the transcriptional activation signal of two different transacting factors, Gal4 and general regulatory factor I/repressor/activator site binding protein 1/translation upstream factor. Proc Natl Acad Sci USA. 1990;87:5373–5377. doi: 10.1073/pnas.87.14.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Nogi Y, Fukasawa T. A novel mutation that affects utilization of galactose in Saccharomyces cerevisiae. Curr Genet. 1980;2:115–120. doi: 10.1007/BF00420623. [DOI] [PubMed] [Google Scholar]
- 345.Nonet M L, Young R A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.O’Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 347.Ohkuma Y, Hashimoto S, Wang C K, Horikoshi M, Roeder R G. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol Cell Biol. 1995;15:4856–4866. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Ohkuma Y, Roeder R G. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 349.Ohkuma Y, Sumimoto H, Hoffmann A, Shimasaki S, Horikoshi M, Roeder R G. Structural motifs and potential sigma homologies in the large subunit of human general transcription factor TFIIE. Nature. 1991;354:398–401. doi: 10.1038/354398a0. [DOI] [PubMed] [Google Scholar]
- 350.Ohkuma Y, Sumimoto H, Horikoshi M, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II: purification and characterization of general transcription factor TFIIE. Proc Natl Acad Sci USA. 1990;87:9163–9167. doi: 10.1073/pnas.87.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Orphanides G, LaGrange T, Reinberg D. The general initiation factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 352.Ossipow V, Tassan J-P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription initiation. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 353.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 354.Pan G H, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 355.Pappas, A. T., and M. Hampsey. Unpublished results.
- 356.Park E, Guzder S N, Koken M H, Jaspers-Dekker I, Weeda G, Hoeijmakers J H, Prakash S, Prakash L. RAD25 (SSL2), the yeast homolog of the human xeroderma pigmentosum group B DNA repair gene, is essential for viability. Proc Natl Acad Sci USA. 1992;89:11416–11420. doi: 10.1073/pnas.89.23.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Parvin J D, Sharp P A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 358.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 359.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 360.Perez-Howard G M, Weil P A, Beechem J M. Yeast TATA binding protein interaction with DNA: fluorescence determination of oligomeric state, equilibrium binding, on-rate, and dissociation kinetics. Biochemistry. 1995;34:8005–8017. doi: 10.1021/bi00025a006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 362.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine. Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 363.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Structure and functional properties of human general transcription factor IIE. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 364.Piatti S, Tazzi R, Pizzagalli A, Plevani P, Lucchini G. Control of DNA synthesis genes in budding yeast: involvement of the transcriptional modulator MOT1 in the expression of the DNA polymerase alpha gene. Chromosoma. 1992;102:S107–S113. doi: 10.1007/BF02451793. [DOI] [PubMed] [Google Scholar]
- 365.Pina B, Berger S, Marcus G A, Silverman N, Agapite J, Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Pinto I, Ware D E, Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992;68:977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 367.Pinto I, Wu W-H, Na J G, Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994;269:30569–30573. [PubMed] [Google Scholar]
- 368.Piraut J I, Chavez S, Aguilera A. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics. 1997;147:1585–1594. doi: 10.1093/genetics/147.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Piraut J I, Aguilera A. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics. 1996;143:1533–1542. doi: 10.1093/genetics/143.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Poon D, Bai Y, Campbell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Poon D, Campbell A M, Bai Y, Weil P A. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 373.Poon D, Weil P A. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- 374.Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Pugh B F, Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992;267:679–682. [PubMed] [Google Scholar]
- 379.Pugh B F, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 380.Qian X, Jeon C, Yoon H, Agarwal K, Weiss M A. Structure of a new nucleic-acid-binding motif in eukaryotic transcriptional elongation factor TFIIS. Nature. 1993;365:277–279. doi: 10.1038/365277a0. [DOI] [PubMed] [Google Scholar]
- 381.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 382.Raghavan S, Burma P K, Brahmachari S K. Positional preferences of polypurine/polypyrimidine tracts in Saccharomyces cerevisiae genome: implications for cis regulation of gene expression. J Mol Evol. 1997;45:485–498. doi: 10.1007/pl00006253. [DOI] [PubMed] [Google Scholar]
- 383.Ranish J A, Hahn S. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J Biol Chem. 1991;266:19320–19327. [PubMed] [Google Scholar]
- 384.Ranish J A, Lane W S, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 385.Ray B L, White C I, Haber J E. The TSM1 gene of Saccharomyces cerevisiae overlaps the MAT locus. Curr Genet. 1991;20:25–31. doi: 10.1007/BF00312761. [DOI] [PubMed] [Google Scholar]
- 386.Redd M J, Arnaud M B, Johnson A D. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 387.Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–57. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- 388.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAF(II)s in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 389.Reinberg D, Horikoshi M, Roeder R G. Factors involved in specific transcription in mammalian RNA polymerase II. Functional analysis of initiation factors IIA and IID and identification of a new factor operating at sequences downstream of the initiation site. J Biol Chem. 1987;262:3322–3330. [PubMed] [Google Scholar]
- 390.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 391.Roberts S G E, Green M R. Activator-induced conformational change in general transcription factor TFIIB. Nature. 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 392.Roberts S G E, Ha I, Maldonado E, Reinberg D, Green M R. Interaction between acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 393.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcnr proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 396.Rosenblum-Vos L S, Rhodes L, Evangelista C C, Jr, Boayke K A, Zitomer R S. The ROX3 gene encodes an essential nuclear protein involved in CYC7 gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5639–5647. doi: 10.1128/mcb.11.11.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Rothstein R J, Sherman F. Genes affecting the expression of cytochrome c in yeast: genetic mapping and genetic interactions. Genetics. 1980;94:871–889. doi: 10.1093/genetics/94.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Roy A L, Du H, Gregor P D, Novina C D, Martinez E, Roeder R G. Cloning of an Inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Roy A L, Malik S, Meisterernst M, Roeder R G. An alternative pathway for transcription initiation involving TFII-I. Nature. 1993;365:355–359. doi: 10.1038/365355a0. [DOI] [PubMed] [Google Scholar]
- 400.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J M. The MO15 cell cycle kinase is associated with the TFIIH transcription DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 401.Roy R, Schaeffer L, Humbert S, Vermeulen W, Weeda G, Egly J M. The DNA-dependent ATPase activity associated with the class II basic transcription factor BTF2/TFIIH. J Biol Chem. 1994;269:9826–9832. [PubMed] [Google Scholar]
- 402.Rubin D M, Coux O, Wefes I, Hengartner C, Young R A, Goldberg A L, Finley D. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature. 1996;379:655–657. doi: 10.1038/379655a0. [DOI] [PubMed] [Google Scholar]
- 403.Rudolph H, Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci USA. 1987;84:1340–1344. doi: 10.1073/pnas.84.5.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Ruiz-Garcia A B, Sendra R, Pamblanco M, Tordera V. Gcn5p is involved in the acetylation of histone H3 in nucleosomes. FEBS Lett. 1997;403:186–190. doi: 10.1016/s0014-5793(97)00049-5. [DOI] [PubMed] [Google Scholar]
- 405.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Ruppert S, Wang E H, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 407.Sakurai H, Fukasawa T. Yeast gal11 and transcription factor IIE function through a common pathway in transcriptional regulation. J Biol Chem. 1997;272:32663–32669. doi: 10.1074/jbc.272.51.32663. [DOI] [PubMed] [Google Scholar]
- 408.Sakurai H, Hiraoka Y, Fukasawa T. Yeast GAL11 protein is a distinctive type transcription factor that enhances basal transcription in vitro. Proc Natl Acad Sci USA. 1993;90:8382–8386. doi: 10.1073/pnas.90.18.8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Sakurai H, Kim Y J, Ohishi T, Kornberg R D, Fukasawa T. The yeast GAL11 protein binds to the transcription factor IIE through GAL11 regions essential for its in vivo function. Proc Natl Acad Sci USA. 1996;93:9488–9492. doi: 10.1073/pnas.93.18.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Sakurai H, Ohishi T, Fukasawa T. Core promoter elements are essential as selective determinants for function of the yeast transcription factor GAL11. FEBS Lett. 1996;398:113–119. doi: 10.1016/s0014-5793(96)01219-7. [DOI] [PubMed] [Google Scholar]
- 411.Sakurai H, Ohishi T, Fukasawa T. Promoter structure-dependent functioning of the general transcription factor IIE in Saccharomyces cerevisiae. J Biol Chem. 1997;272:15936–15942. doi: 10.1074/jbc.272.25.15936. [DOI] [PubMed] [Google Scholar]
- 412.Sakurai H, Ohishi T, Fukasawa T. Two alternative pathways of transcription initiation in the yeast negative regulatory gene GAL80. Mol Cell Biol. 1994;14:6819–6828. doi: 10.1128/mcb.14.10.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Saleh A, Lang V, Cook R, Brandl C J. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- 414.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 415.Santosrosa H, Clever B, Heyer W D, Aguilera A. The yeast HRS1 gene encodes a polyglutamine-rich nuclear protein required for spontaneous and hpr1-induced deletions between direct repeats. Genetics. 1996;142:705–716. doi: 10.1093/genetics/142.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 417.Sauer F, Wassarman D A, Rubin G M, Tjian R. TAF(II)s mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 418.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Sayre M H, Tschochner H, Kornberg R D. Purification and properties of Saccharomyces cerevisiae RNA polymerase II general initiation factor a. J Biol Chem. 1992;267:23383–23387. [PubMed] [Google Scholar]
- 420.Sayre M H, Tschochner H, Kornberg R D. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- 421.Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers J H, Egly J M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H, Chambon P, Egly J M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 423.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Schmidt M C, Kao C C, Pei R, Berk A J. Yeast TATA-box transcription factor gene. Proc Natl Acad Sci USA. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Schultz J, Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Schultz M C, Reeder R H, Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 427.Scully R, Anderson S F, Chao D M, Wei W, Ye L, Young R A, Livingston D M, Parvin J D. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 429.Serizawa H, Conaway R C, Conaway J W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor delta from rat liver. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Serizawa H, Conaway R C, Conaway J W. Multifunctional RNA polymerase II initiation factor delta from rat liver. Relationship between carboxyl-terminal domain kinase, ATPase, and DNA helicase activities. J Biol Chem. 1993;268:17300–17308. [PubMed] [Google Scholar]
- 431.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 432.Shaw S P, Carson D J, Dorsey M J, Ma J. Mutational studies of yeast transcription factor IIB in vivo reveal a functional surface important for gene activation. Proc Natl Acad Sci USA. 1997;94:2427–2432. doi: 10.1073/pnas.94.6.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Shaw S P, Wingfield J, Dorsey M J, Ma J. Identifying a species-specific region of yeast TFIIB in vivo. Mol Cell Biol. 1996;16:3651–3657. doi: 10.1128/mcb.16.7.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Shen W-C, Walker S S, Green M R. Yeast TAFII145 functions as a core promoter-selectivity factor, not a general co-activator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 435.Shi X, Chang M, Wolf A J, Chang C H, Frazer-Abel A A, Wade P A, Burton Z F, Jaehning J A. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Shi X M, Finkelstein A, Wolf A J, Wade P A, Burton Z F, Jaehning J A. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 438.Shykind B M, Kim J, Sharp P A. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 439.Silverman N, Agapite J, Guarente L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci USA. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Simmen K A, Bernues J, Lewis J D, Mattaj I W. Cofractionation of the TATA-binding protein with the RNA polymerase III transcription factor TFIIIB. Nucleic Acids Res. 1992;20:5889–5898. doi: 10.1093/nar/20.22.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Singer V L, Wobbe C R, Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990;4:636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- 442.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 443.Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Sopta M, Burton Z F, Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989;341:410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- 445.Stachora A A, Schafer R E, Pohlmeier M, Maier G, Ponstingl H. Human Supt5h protein, a putative modulator of chromatin structure, is reversibly phosphorylated in mitosis. FEBS Lett. 1997;409:74–78. doi: 10.1016/s0014-5793(97)00486-9. [DOI] [PubMed] [Google Scholar]
- 446.Stargell L A, Struhl K. A new class of activation-defective TATA-binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Stargell L A, Struhl K. The TBP-TFIIA interaction in the response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 448.Starr D B, Hawley D K. TFIID binds in the minor groove of the TATA box. Cell. 1991;67:1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- 449.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 450.Steinmetz E J, Brow D A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 452.Stolinski L A, Eisenmann D M, Arndt K M. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Strubin M, Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 455.Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci USA. 1984;81:7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 457.Sumimoto H, Ohkuma Y, Sinn E, Kato H, Shimasaki S, Horikoshi M, Roeder R G. Conserved sequence motifs in the small subunit of human general transcription factor TFIIE. Nature. 1991;354:401–404. doi: 10.1038/354401a0. [DOI] [PubMed] [Google Scholar]
- 458.Sun X Q, Ma D M, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: A role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 459.Sun Z W, Hampsey M. Identification of the gene (SSU71/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Sun Z W, Tessmer A, Hampsey M. Functional interaction between TFIIB and the Rpb9 (Ssu73) subunit of RNA polymerase II in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2560–2566. doi: 10.1093/nar/24.13.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Sussel L, Vannier D, Shore D. Suppressors of defective silencing in yeast: effects on transcriptional repression at the HMR locus, cell growth and telomere structure. Genetics. 1995;141:873–888. doi: 10.1093/genetics/141.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Suzuki Y, Nogi Y, Abe A, Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4991–4999. doi: 10.1128/mcb.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Svejstrup J Q, Feaver W J, Kornberg R D. Subunits of yeast RNA polymerase II transcription factor TFIIH encoded by the CCL1 gene. J Biol Chem. 1996;271:643–645. doi: 10.1074/jbc.271.2.643. [DOI] [PubMed] [Google Scholar]
- 465.Svejstrup J Q, Feaver W J, Lapointe J, Kornberg R D. RNA polymerase transcription factor IIH holoenzyme from yeast. J Biol Chem. 1994;269:28044–28048. [PubMed] [Google Scholar]
- 466.Svejstrup J Q, Vichi P, Egly J M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 467.Svejstrup J Q, Wang Z G, Feaver W J, Wu X H, Bushnell D A, Donahue T F, Friedberg E C, Kornberg R D. Different forms of TFIIH for transcription and DNA repair: Holo-TFIIH and a nucleotide excision repairosome. Cell. 1995;80:21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- 468.Swaffield J C, Bromberg J F, Johnston S A. Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activation domain in GAL4. Nature. 1992;357:698–700. doi: 10.1038/357698a0. [DOI] [PubMed] [Google Scholar]
- 469.Swaffield J C, Melcher K, Johnston S A. A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature. 1995;374:88–91. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- 470.Swanson M S, Carlson M, Winston F. SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol Cell Biol. 1990;10:4935–4941. doi: 10.1128/mcb.10.9.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:4286. doi: 10.1128/mcb.11.8.4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Swanson M S, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Taggart A K, Fisher T S, Pugh B F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 474.Taggart A K, Pugh B F. Dimerization of TFIID when not bound to DNA. Science. 1996;272:1331–1333. doi: 10.1126/science.272.5266.1331. [DOI] [PubMed] [Google Scholar]
- 475.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 476.Tan S Y, Aso T, Conaway R C, Conaway J W. Roles far both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 477.Tang H, Sun X Q, Reinberg D, Ebright R H. Protein-protein interactions in eukaryotic transcription initiation: structure of the preinitiation complex. Proc Natl Acad Sci USA. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Tansey W P, Herr W. Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science. 1997;275:829–831. doi: 10.1126/science.275.5301.829. [DOI] [PubMed] [Google Scholar]
- 479.Tansey W P, Herr W. TAFs: guilt by association? Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 480.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 481.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 482.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 483.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 484.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Thuret J Y, Valay J G, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 486.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAF(II)40 and TAF(II)60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 487.Tijerina P, Sayre M H. A debilitating mutation in transcription factor IIE with differential effects on gene expression in yeast. J Biol Chem. 1998;273:1107–1113. doi: 10.1074/jbc.273.2.1107. [DOI] [PubMed] [Google Scholar]
- 488.Timmers H T M, Meyers R E, Sharp P A. Composition of transcription factor B-TFIID. Proc Natl Acad Sci USA. 1992;89:8140–8144. doi: 10.1073/pnas.89.17.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Timmers H T M, Sharp P A. The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev. 1991;5:1946–1956. doi: 10.1101/gad.5.11.1946. [DOI] [PubMed] [Google Scholar]
- 490.Trumbly R J. Isolation of Saccharomyces cerevisiae mutants constitutive for invertase synthesis. J Bacteriol. 1986;166:1123–1127. doi: 10.1128/jb.166.3.1123-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 492.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 493.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 494.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 495.Uemura H, Pandit S, Jigami Y, Sternglanz R. Mutations in GCR3, a gene involved in the expression of glycolytic genes in Saccharomyces cerevisiae, suppress the temperature-sensitive growth of hpr1 mutants. Genetics. 1996;142:1095–1103. doi: 10.1093/genetics/142.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Specific interaction between the nonphosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- 497.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 498.Valay J G, Simon M, Dubois M F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 499.Valay J G, Simon M, Faye G. The kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- 500.Vallier L G, Carlson M. New SNF genes, GAL11 and GRR1 affect SUC2 expression in Saccharomyces cerevisiae. Genetics. 1991;129:675–684. doi: 10.1093/genetics/129.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Vallier L G, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137:49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.van der Knaap J A, Borst J W, van der Vliet P C, Gentz R, Timmers H T M. Cloning of the cDNA for the TATA-binding protein-associated factorII170 subunit of transcription factor B-TFIID reveals homology to global transcription regulators in yeast and Drosophila. Proc Natl Acad Sci USA. 1997;94:11827–11832. doi: 10.1073/pnas.94.22.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Van Dyke M W, Roeder R G, Sawadogo M. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 504.Vannier D, Balderes D, Shore D. Evidence that the transcriptional regulators SIN3 and RPD3, and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics. 1996;144:1343–1353. doi: 10.1093/genetics/144.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.van Vuuren A J, Vermeulen W, Ma L, Weeda G, Appeldoorn E, Jaspers N G, van der Eb A J, Bootsma D, Hoeijmakers J H, Humbert S, et al. Correction of xeroderma pigmentosum repair defect by basal transcription factor BTF2 (TFIIH) EMBO J. 1994;13:1645–1653. doi: 10.1002/j.1460-2075.1994.tb06428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 507.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 508.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 509.Verrijzer C P, Yokomori K, Chen J L, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 510.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 513.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Wade P A, Jaehning J A. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol Cell Biol. 1996;16:1641–1648. doi: 10.1128/mcb.16.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Wade P A, Werel W, Fentzke R C, Thompson N E, Leykam J F, Burgess R R, Jaehning J A, Burton Z F. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expression Purif. 1996;8:85–90. doi: 10.1006/prep.1996.0077. [DOI] [PubMed] [Google Scholar]
- 516.Wahi M, Johnson A D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAF(II)s. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 518.Walker S S, Shen W-C, Green M R. Yeast TAFII145 is required for transcription of G1/S cyclin genes and is regulated by the cellular growth rate. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 519.Wang E H, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 520.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Wang H, Clark I, Nicholson P R, Herskowitz I, Stillman D J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990;10:5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Wang H, Stillman D J. Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993;13:1805–1814. doi: 10.1128/mcb.13.3.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Wang W, Carey M, Gralla J D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–453. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 524.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 525.Wang W, Gralla J D, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- 526.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 527.Wang Z, Svejstrup J Q, Feaver W J, Wu X, Kornberg R D, Friedberg E C. Transcription factor b (TFIIH) is required during nucleotide-excision repair in yeast. Nature. 1994;368:74–76. doi: 10.1038/368074a0. [DOI] [PubMed] [Google Scholar]
- 528.Wang Z G, Buratowski S, Svejstrup J Q, Feaver W J, Wu X H, Kornberg R D, Donahue T F, Friedberg E C. The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol Cell Biol. 1995;15:2288–2293. doi: 10.1128/mcb.15.4.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Weil P A, Luse D S, Segall J, Roeder R G. Selective and accurate initiation of transcription at the Ad2 major late promoter in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979;18:469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- 530.Weis L, Reinberg D. Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J. 1992;6:3300–3309. doi: 10.1096/fasebj.6.14.1426767. [DOI] [PubMed] [Google Scholar]
- 531.Welch M D, Vinh D B, Okamura H H, Drubin D G. Screens for extragenic mutations that fail to complement act1 alleles identify genes that are important for actin function in Saccharomyces cerevisiae. Genetics. 1993;135:265–274. doi: 10.1093/genetics/135.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.West M L, Corden J L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.White R J, Jackson S P. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell. 1992;71:1041–1053. doi: 10.1016/0092-8674(92)90398-v. [DOI] [PubMed] [Google Scholar]
- 534.Williams F E, Varanasi U, Trumbly R J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 536.Winston F. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones and other transcription factors of yeast. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1271–1293. [Google Scholar]
- 537.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 538.Winter E, Varshavsky A. A DNA binding protein that recognizes oligo(dA) · oligo(dT) tracts. EMBO J. 1989;8:1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Wobbe C R, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 541.Woontner M, Jaehning J A. Accurate initiation by RNA polymerase II in a whole cell extract from Saccharomyces cerevisiae. J Biol Chem. 1990;265:8979–8982. [PubMed] [Google Scholar]
- 542.Woontner M, Wade P A, Bonner J, Jaehning J A. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:4555–4560. doi: 10.1128/mcb.11.9.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Woychik N A, Young R A. Exploring RNA polymerase II structure and function. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 227–242. [Google Scholar]
- 544.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 546.Yamashita I. Isolation and characterization of the SUD1 gene, which encodes a global repressor of core promoter activity in Saccharomyces cerevisiae. Mol Gen Genet. 1993;241:616–626. doi: 10.1007/BF00279904. [DOI] [PubMed] [Google Scholar]
- 547.Yamashita S, Hisatake K, Kokubo T, Doi K, Roeder R G, Horikoshi M, Nakatani Y. Transcription factor TFIIB sites important for interaction with promoter-bound TFIID. Science. 1993;261:463–466. doi: 10.1126/science.8332911. [DOI] [PubMed] [Google Scholar]
- 548.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 549.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr(1) reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 550.Yokomori K, Admon A, Goodrich J A, Chen J L, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- 551.Yokomori K, Zeidler M P, Chen J L, Verrijzer C P, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 552.Yonaha M, Aso T, Kobayashi Y, Vasavada H, Yasukochi Y, Weissman S M, Kitajima S. Domain structure of a human general transcription initiation factor, TFIIF. Nucleic Acids Res. 1993;21:273–279. doi: 10.1093/nar/21.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Yoon H, Miller S P, Pabich E K, Donahue T F. SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 1992;6:2463–2477. doi: 10.1101/gad.6.12b.2463. [DOI] [PubMed] [Google Scholar]
- 554.Yoshimoto H, Yamashita I. The GAM1/SNF2 gene of Saccharomyces cerevisiae encodes a highly charged nuclear protein required for transcription of the STA1 gene. Mol Gen Genet. 1991;228:270–280. doi: 10.1007/BF00282476. [DOI] [PubMed] [Google Scholar]
- 555.Young R A. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 556.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: A multi-step process. Prog Nucleic Acid Res. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 559.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 560.Zhang, Y., Z.-W. Sun, M. Hampsey, and D. Reinberg. Unpublished results.
- 561.Zhou Z, Elledge S J. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics. 1992;131:851–866. doi: 10.1093/genetics/131.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Zhu A H, Kuziora M A. Homeodomain interaction with the beta subunit of the general transcription factor TFIIE. J Biol Chem. 1996;271:20993–20996. doi: 10.1074/jbc.271.35.20993. [DOI] [PubMed] [Google Scholar]
- 563.Zhu W L, Zeng Q D, Colangelo C M, Lewis L M, Summers M F, Scott R A. The N-terminal domain of TFIIB from Pyrococcus furiosus forms a zinc ribbon. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 564.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Zhu Z, Thiele D J. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell. 1996;87:459–470. doi: 10.1016/s0092-8674(00)81366-5. [DOI] [PubMed] [Google Scholar]
- 566.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]