Abstract
The cyclin-dependent protein kinase (CDK) encoded by CDC28 is the master regulator of cell division in the budding yeast Saccharomyces cerevisiae. By mechanisms that, for the most part, remain to be delineated, Cdc28 activity controls the timing of mitotic commitment, bud initiation, DNA replication, spindle formation, and chromosome separation. Environmental stimuli and progress through the cell cycle are monitored through checkpoint mechanisms that influence Cdc28 activity at key cell cycle stages. A vast body of information concerning how Cdc28 activity is timed and coordinated with various mitotic events has accrued. This article reviews that literature. Following an introduction to the properties of CDKs common to many eukaryotic species, the key influences on Cdc28 activity—cyclin-CKI binding and phosphorylation-dephosphorylation events—are examined. The processes controlling the abundance and activity of key Cdc28 regulators, especially transcriptional and proteolytic mechanisms, are then discussed in detail. Finally, the mechanisms by which environmental stimuli influence Cdc28 activity are summarized.
Saccharomyces cerevisiae possesses five cyclin-dependent protein kinases (CDKs) (Cdc28, Pho85, Kin28, Ssn3, and Ctk1), but Cdc28, the subject of this review, is the best studied by far. Cdc28 is the central coordinator of the major events of the yeast cell division cycle. Environmental effects that influence the decision to undergo cell division or the fidelity and rate of key mitotic events ultimately affect Cdc28 kinase activity. This review strives to provide a comprehensive survey of the published literature on how Cdc28 activity is generated and regulated. There have been many excellent shorter reviews of various aspects of this system in the last few years, and they provide an ideal general introduction to various aspects of the yeast cell cycle and opportunities for looking at specific topics in depth. The long-review format of Microbiological and Molecular Biology Reviews allows us to present a more exhaustive summary that we hope will be of use to our coworkers and will serve as a secondary source for those already familiar with basic yeast physiology. Discussion of the functions of the CDKs is kept to a minimum, except for the (numerous) instances when CDKs act as CDK regulators. Likewise, a discussion of the many homologous genes and gene products from other species is minimized or omitted; it is used mostly to help make sense of regulatory modes that are well worked out in other systems but not in S. cerevisiae. This compromise was necessary to limit what is already a voluminous topic, and we apologize to the many investigators whose work anticipated and inspired the parallel work in budding yeast but that we were unable to cite.
Instead of conducting a gene-by-gene summary or a walk through the cell cycle, we have chosen to organize the topics in this review by starting with a short description of key Cdc28 regulators (cyclins, CDK inhibitors [CKIs], the enigmatic Cks1, and phosphorylation of Cdc28) and then organizing the influences on these regulators by large-scale process starting with transcription and ending with proteolysis. Finally, the effects of environmental influences on these processes and regulators are discussed.
Nomenclature and Conventions
Many of the genes discussed have been identified by multiple laboratories over a long span of time and have consequently acquired multiple labels. To simplify the discussion, we use the gene names favored by the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces) and Proteome (www.proteome.com). Aliases for these genes can be found at the Saccharomyces Genome Database and Proteome Web sites and in Table 1. Table 1 also contains a short synopsis of the function of each gene and the positions of important domains discussed in the text. Standard S. cerevisiae genetic conventions are used throughout (dominant or wild-type genes and their mRNAs are in capital italics, recessive mutants are in lowercase italics, and Δ refers to a gene deletion or disruption; e.g. CLN3 is wild type, CLN3-1 is a dominant mutant allele, cln3 is a recessive mutant allele, and cln3Δ is a deletion). Superscripts are added to denote alleles with specific properties: cln3ts is a temperature-sensitive allele, and CLN3stab encodes a protein that is hyperstable relative to the wild-type protein. The protein products of a particular gene are in roman type (Cln3 is the gene product of CLN3 and Cln3-1 is the product of CLN3-1 allele). Genes under the transcriptional control of heterologous promoters are designated, e.g., GAL1p→CLN3, which indicates that the promoter element from the GAL1 gene is used to control expression of the open reading frame (ORF) for CLN3. Protein fusions are indicated with superscripts indicating the region of the protein that is present: Cln3404–488–β-Gal is a fusion of Cln3 residues 404 to 488 to β-galactosidase.
TABLE 1.
S. cerevisiae gene products that influence Cdc28 activity
| Gene products | No. of amino acids | Function | Reference(s) |
|---|---|---|---|
| Ace2 | 770 | Transcriptional activator of Sic1 and Rme1. | 4, 71, 133, 134 |
| Aos1 | 347 | With Uba2, acts as an E1 ligase for the ubiquitin-like Smt3. | 269 |
| Apc1 | 1,748 | Essential component of the APC. | 634 |
| Apc2, Rsi1 | 853 | Essential, cullin family component of the APC. | 305, 630, 633 |
| Apc4 | 652 | Essential component of the APC. | 633 |
| Apc5 | 685 | Essential component of the APC. | 633 |
| Apc9 | 265 | Nonessential component of the APC. | 633 |
| Apc11 | 165 | Essential, RING finger-containing component of the APC. | 633 |
| Apc13 | ? | 13-kDa component of the APC. | 633 |
| Bck2, Ctr7 | 851 | Positive factor in cyclin gene expression. | 128, 161 |
| Bub2, Pac7 | 306 | Required for cell cycle arrest in response to spindle failure. | 250, 551 |
| Cak1, Civ1, Mca28 | 368 | Protein kinase that phosphorylates and activates Cdc28. | 91, 111, 165, 275, 543, 560, 600 |
| Ccl1 | 393 | Cyclin activator of Kin28. | 545, 586 |
| Cdc14, Oaf3 | 551 | Dual-specificity protein phosphatase. Possible activator of the APC. | 500, 552, 604 |
| Cdc15 | 974 | Protein kinase. Possible activator of the APC. | 485, 500 |
| Cdc16 | 840 | Essential, TPR-containing component of the APC. Ten TPRs are contained in residues 359–392, 263–329, and 497–741. | 231, 232, 260, 291, 319, 417, 634 |
| Cdc20, Pac5 | 610 | Factor that targets Pds1 to the APC. | 256, 340, 343, 449, 482, 491, 503, 551, 599 |
| Cdc23 | 626 | Essential, TPR-containing component of the APC. Nine TPRs are contained in residues 215–248 and 295–569. | 260, 319, 449, 504, 513, 514, 632, 634 |
| Cdc26, Hit3, Scd26 | 124 | Nonessential component of the APC. | 20, 634 |
| Cdc27, Snb1 | 758 | TPR-containing component of the APC. Ten TPRs are contained in residues 154–187 and 438–709. | 232, 291, 319, 632, 634 |
| Cdc28, Cdk1, Srm5, Hsl5, Elm7 | 298 | Cyclin dependent protein kinase responsible for coordinating major cell cycle events. Inhibitory phosphorylations occurs on T18 and Y19, the PSTAIRE domain is in residues 52–58, the T-loop is at 161–179, CAK1 activates by phosphorylation at T169. | See text and Table 2 |
| Cdc34, Ubc3, Dna6 | 295 | E2 ubiquitin conjugating enzyme required for proteolysis of G1 regulators of Cdc28. SCF binding region in residues 171–209. | See text |
| Cdc37, Smo1 | 506 | Molecular chaperone needed for proper Cdc28-cyclin interaction. | 58, 194, 290 |
| Cdc4 | 779 | F box component of E3 ubiquitin ligase SCFCdc4 (with Cdc53 and Skp1) that recognizes Sic1, Far1, and Cdc6. Needed for Cdc34 essential function. F box is contained in residues 275–319, and WD-40 repeats are at 377–658. | See text |
| Cdc5, Pkx2, Msd2 | 705 | Protein kinase of the “polo” family. Activator of the APC. | 85, 222, 295, 503 |
| Cdc53 | 815 | “Cullin” component of E3 ubiquitin ligase (with Skp1 and either Cdc4 or Grr1). Needed for Cdc34 essential function. Residues 794–815 are required for Rub1 modification. | 27, 173, 320, 337, 368, 430, 612 |
| Cdc55 | 526 | B subunit of type 2A protein phosphatase that is important for Clb2-Cdc28 activation and has a role in the kinetochore/spindle checkpoint. | 230, 605 |
| Cdc6 | 513 | Required for DNA replication. Helps ensure single round of replication per cell division cycle. Inhibitor of Clb-Cdc28 complexes. | 68, 95, 123, 138, 141, 156, 338, 345, 437, 438, 548, 640–642 |
| Cdh1, Hct1 | 566 | Factor that targets Clb cyclins to the APC. | 483, 599 |
| Cks1 | 150 | Component of Cdc28-cyclin complexes. Possible assembly factor. May target Cdc28 complexes to the APC. May affect Cdc28 substrate specificity. May affect phosphorylation and activation/deactivation of Cdc28 complexes. | 42, 213, 549 |
| Clb1, Scb1 | 471 | Cyclin activator of Cdc28 at G2M. Destruction box at 35–43. | See text and Table 2 |
| Clb2 | 491 | Cyclin activator of Cdc28 at G2/M. Destruction box at 25–33. | See text and Table 2 |
| Clb3 | 427 | Cyclin activator of Cdc28 in S. Destruction box at 51–59. | See text and Table 2 |
| Clb4 | 460 | Cyclin activator of Cdc28 in S. Destruction box at 43–51. | See text and Table 2 |
| Clb5 | 435 | Cyclin activator of Cdc28 at Start. Destruction box at 56–64, acidic domain at 110–123. | See text and Table 2 |
| Clb6 | 380 | Cyclin activator of Cdc28 at Start. | See text and Table 2 |
| Cln1 | 546 | Cyclin activator of Cdc28 at Start. Cyclin box is at 20–127. | See text and Table 2 |
| Cln2, Daf3 | 545 | Cyclin activator of Cdc28 at Start. Cyclin box is at 20–127. | See text and Table 2 |
| Cln3, Daf1, Whi1, Fun10 | 580 | Cyclin activator of Cdc28 in G1. Cyclin box is at 106–206. | See text and Table 2 |
| Dbf2 | 572 | Protein kinase. Possible activator of the APC. | 272, 426, 570, 572, 573 |
| Dbf20 | 544 | Protein kinase. Possible activator of the APC. | 570, 572, 573 |
| Dig1, Rst1 | 452 | Repressor of Ste12. Links Kss1 and Fus3 to Ste12. | 101, 555 |
| Dig2, Rst2 | 323 | Repressor of Ste12. Links Kss1 and Fus3 to Ste12. | 101, 555 |
| Doc1, Hrc283, Apc10 | 283 | Nonessential component of the APC. | 257, 633 |
| Far1 | 830 | CKI specific for Cdc28-Cln complexes. CDK inhibitory domain is at 99–390. Activity and transcription is induced by mating pheromone. | See text |
| Far3 | 204 | Important for pheromone-mediated G1 arrest. | 249 |
| Fus3, Dac2 | 353 | Protein kinase of the MAPK family required for cell cycle arrest in response to mating pheromone. Also participates with Kss1 in the activation of pheromone dependent transcription. | 84, 151–154, 164, 187, 191, 192, 435, 555, 577 |
| Gin4, Erc47 | 1,142 | Protein kinase needed for full function of Clb2-Cdc28 complexes. | 2, 5 |
| Grr1, Sdc1, Cat80, Cot2, Toc1, Ssu2 | 1,151 | F box component of E3 ubiquitin ligase SCFGrr1 (with Cdc53 and Skp1) that recognizes Cln1 and Cln2. F box is contained in residues 317–362, and leucine-rich repeats are at 410–725. | 33, 97, 162, 182, 294, 333, 422, 430, 519, 589 |
| Hsc82 | 704 | Molecular chaperone of the Hsp90 family that may participate in Cdc28-cyclin complex formation. | 290 |
| Hsl1, Nik1 | 1,518 | Protein kinase homologous to S. pombe Cdr1 and A. nidulans Nim1 that negatively regulates Swe1. | 359 |
| Hsl7 | 827 | Negative regulator of Swe1. | 359 |
| Hsp82, Hsp83, Hsp90 | 708 | Heat-inducible molecular chaperone of the Hsp90 family that may participate in Cdc28-cyclin complex formation. | 290 |
| Kin28 | 306 | Cyclin-dependent kinase activated by Ccl1. Homologous to CDK activating kinases in other species, it does not activate Cdc28 but is a component of TFIID and acts as an RNA polymerase II CTD kinase. | 93, 172, 518, 586, 587 |
| Kss1 | 368 | Protein kinase of the MAPK family that participates with Fus3 in the activation of pheromone-dependent transcription. | 101–103, 151, 152, 192, 555 |
| Lte1, Msi2 | 1,435 | GTP/GDP exchange factor. May participate in activation of APC. | 284, 500, 501 |
| Mad1 | 749 | Part of complex bound to Cdc20 that prevents anaphase entry when the spindle is damaged. | 221, 256, 334 |
| Mad2 | 196 | Part of complex bound to Cdc20 that prevents anaphase entry when the spindle is damaged. | 221, 256, 334 |
| Mad3 | 515 | Part of complex bound to Cdc20 that prevents anaphase entry when the spindle is damaged. | 221, 256, 334 |
| Mbp1 | 833 | DNA binding component of MBF transcription factor. Important for Start-specific expression of Clb5 and Clb6. DNA binding domain is in residues 1–124, Swi6 binding domain is in residues 1–124, Swi6 binding domain is at 630–833. | 301, 553, 620 |
| Mcm1, Fun80 | 286 | Transcription factor important for expression of Clb1, Clb2, Cln3, Swi4, Swi5, Ace2, Far1, and Cdc6. Essential DNA binding and protein-protein interactions domains are within residues 17–97. | 4, 13, 65, 86, 90, 149, 170, 262, 312, 358, 362, 364, 373, 413, 447 |
| Mih1 | 474 | Protein phosphatase that dephosphorylates Cdc28 on Y19. Opposes the action of Swe1. | 469 |
| Nab3, Hmd1 | 802 | Inhibits processing of CLN3 mRNA. | 538 |
| Nap1 | 417 | Needed for full function of Clb2-Cdc28 complexes. Binds Clb2 and Gin4. | 5, 282 |
| Pcl1, Hcs26 | 279 | Cyclin activator of Pho85. | 166, 378, 414 |
| Pcl2, OrfD, Cln4 | 279 | Cyclin activation of Pho85. | 377, 378 |
| Pds1 | 373 | Needed for chromosomal separation in mitosis. Degraded by APC in a Cdc20-dependent fashion. | 96, 599, 623, 624 |
| Pho80, Tup7, Ags3 | 293 | Cyclin activator of Pho85. | 243, 274, 418, 584 |
| Pho81 | 1,178 | CKI specific for Pho80-Pho85 complexes. | 94, 105, 243, 415, 481 |
| Pho85, Ssg3 | 305 | Cyclin-dependent protein kinase activated by Pho80, Pcl1, and Pcl2. Involved in phosphate metabolism and bud emergence. | 166, 243, 251, 274, 377, 378, 418, 477, 556, 561, 564, 582 |
| Pph21 | 369 | Catalytic subunit of a type 2A protein phosphatase important for Clb2-Cdc28 activation. | 344, 466 |
| Pph22 | 377 | Catalytic subunit of a type 2A protein phosphatase important for Clb2-Cdc28 activation. | 344, 466 |
| Pph3 | 308 | Catalytic subunit of a type 2A protein phosphatase important for Clb2-Cdc28 activation. | 344, 466 |
| Prt1, Cdc63, Dna26 | 763 | eIF3η, a translation initiation factor that regulates the supply of 40S ribosomal subunits and their association with eIF-2–GTP–tRNAMet. Needed for efficient CLN3 translation. | 216, 217 |
| Rad53, Spk1, Mec2, Sad1 | 821 | Serine/threonine/tyrosine protein kinase with a checkpoint function in S and G2. | 3, 289, 475, 510, 539 |
| Ras1 | 306 | GTPase. Activator of adenylate cyclase and possibly needed for activation of the APC. | 279, 391 |
| Ras2, Ctn5, Glc5, Asc1 | 318 | GTPase. Activator of adenylate cyclase. | 31, 279, 391, 392, 598 |
| Rat1, Xrn2, Rsf11, Tap1, Hke1 | 1,006 | 5′,3′-Exo-RNase required for efficient nucleocytoplasmic RNA trafficking. Needed for efficient Start-specific gene expression. | 567 |
| Rme1, Csp1 | 300 | Positive factor in Cln2 expression. Negatively regulates early sporulation-specific genes. | 104, 567 |
| Rts1, Scs1 | 757 | B subunit of type 2A protein phosphatase that is important for Clb2-Cdc28 activation. | 167, 506, 638 |
| Rub1 | 76 | Ubiquitin-like protein that modifies Cdc53. | 320, 337 |
| Sap155 | 1,000 | Positive regulator of Sit4. | 357 |
| Sap185 | 1,058 | Positive regulator of Sit4. | 357 |
| Sap190 | 1,033 | Positive regulator of Sit4. | 357 |
| Sap4 | 818 | Positive regulator of Sit4. | 357 |
| Sic1, Sdb25, Byc1, Ric2 | 284 | CKI specific for Cdc28-Clb complexes. Domain conferring instability is at 28–161. CDK inhibitory domain is at 159–284. | See text |
| Sim1, Sag1 | 475 | Required for maintenance of Clb5-Cdc28 activity. Possibly involved in mRNA maturation. | 114 |
| Sis2, Hal3 | 562 | Positive regulator of Start-specific gene expression. | 127 |
| Sit4, Pph1 | 311 | Type 2A protein phosphatase needed for Start-specific gene expression. | 175, 426, 544 |
| Skp1, Mgo1 | 194 | E3 ubiquitin ligase (with Cdc53 and either Cdc4 or Grr1). Needed for Cdc34 essential function. | 27, 99, 173, 277, 333, 528 |
| Skn7, Pos9, Bry1 | 622 | Transcription factor capable of stimulating Cln1 and Cln2 expression independent of SBF. Heat shock factor domain (residues 87–150), coiled-coil domain (243–303), homology to bacterial two-component response regulators (378–497), Gln-rich domain (497–622). | 62, 63, 307, 388, 389 |
| Sln1, Ypd2, Nrp2 | 1,220 | Transmembrane histidine kinase that positively affects Mcm1 transcriptional activity. | 170, 361, 420, 445, 628 |
| Smt3 | 97 | Ubiquitin-like protein. Activated by Aos1-Uba2 and conjugated to substrates via Ubc9. | 268, 269, 484 |
| Spo12 | 173 | Required for sporulation. Possibly needed for activation of APC. | 297, 391, 426, 573 |
| Spo13 | 291 | Required for sporulation. Meiotic regulator of Cdc28 activity? | 297, 371 |
| Srp1, Scm1, Nbp70, Kap60 | 542 | Homolog of importin, the nuclear import receptor. Needed for Clb2 proteolysis at anaphase. | 36, 347, 625 |
| Ssd1, Srk1, Cla1, Rlt1, Mcs1, Ssl1 | 1,250 | RNA binding protein needed for efficient expression of Start-specific transcripts. | 113, 175, 544, 583 |
| Ste12 | 688 | Transcriptional activator of pheromone-responsive genes. | 135, 163, 293, 413, 522 |
| Ste7 | 515 | MAP kinase kinase that activates Fus3 and Kss1 in response to mating-pheromone exposure. | 164 |
| Swe1 | 819 | Protein kinase homolog of Wee1 that inactivates Clb2-Cdc28 complexes by phosphorylating Y19 of Cdc28. | 47, 359, 508 |
| Swi4, Art1 | 1,093 | DNA binding component of SBF transcription factor. Important for Start-specific expression of Cln1 and Cln2. DNA binding domain in residues 37–155, Swi6 binding domain at 1017–1093. | See text |
| Swi5, Ric1 | 709 | Transcription factor important for expression of Sic1, Cdc6, and Rme1. | See text |
| Swi6, Sds11, Psl8 | 803 | Regulatory component of SBF and MBF transcription factors important for Start-specific gene expression. Swi4 and Mbp1 binding domain in residues 663–787, leucine zipper domain at 585–612. | See text |
| Taf145, Taf130 | 1,066 | Component of TFIID that is specifically needed for Start-specific gene expression. | 603 |
| Taf90 | 798 | Component of TFIID that is needed for G2/M phase progression. | 19 |
| Tap42 | 366 | Positive regulator of Sit4, Pph21, and Pph22. | 126 |
| Tem1 | 245 | GTPase. Possibly needed for activation of APC. | 502 |
| Tor1, Drr1 | 2,470 | Phosphatidylinositol kinase needed for efficient translation of CLN3 mRNA. | 29, 233, 234, 311 |
| Tor2, Drr2 | 2,473 | Phosphatidylinositol kinase needed for efficient translation of CLN3 mRNA. | 29, 233, 234, 311 |
| Tpd3, Fun32 | 635 | “A” subunit of the type 2A protein phosphatase that is important for Clb2-Cdc28 activity. | 405, 592 |
| Tpk1, Sra3, Pka1 | 397 | cAMP-dependent protein kinase. | 79, 562 |
| Tpk2, Pka2 | 380 | cAMP-dependent protein kinase. | 562 |
| Tpk3, Pka3 | 398 | cAMP-dependent protein kinase. | 562 |
| Tsm1, Taf150 | 1,407 | Component of TFIID that is needed for G2/M phase progression. | 603 |
| Uba1 | 1,023 | E1 ubiquitin-activating enzyme. | 372 |
| Uba2, Pip2, Ual1 | 636 | With Aos1, acts as an E1 for the ubiquitin-like Smt3. | 132, 269 |
| Uba3 | 299 | With Ula1, acts as an E1 ligase for the ubiquitin-like Rub1. | 337 |
| Ubc4 | 147 | E2 ubiquitin conjugating enzyme that ubiquitinates short-lived and abnormal proteins. | 291, 493 |
| Ubc5 | 147 | E2 ubiquitin conjugating enzyme that ubiquitinates short-lived and abnormal proteins. | 493 |
| Ubc9 | 157 | E2-like enzyme that transfers the ubiquitin-like Rub1 to target proteins. | 40, 268, 484, 492 |
| Ubc11 | 156 | E2 enzyme with greatest similarity to metazoan E2-C, the ubiquitin conjugating enzyme associated with anaphase proteolysis. | 569 |
| Ubc12 | 188 | E2-like enzyme that transfers the ubiquitin-like Smt3 to target proteins. | 337 |
| Ubi4, Scd2 | 76 | Polyubiquitin. | 178, 423 |
| UbpX | Family of proteases that specifically remove ubiquitin from ubiquitin protein conjugates. | ||
| Ula1, Enr2, Lpa14 | 462 | With Uba3, acts as an E1 ligase for the ubiquitin-like Rub1. | 337 |
| Whi2 | 486 | Needed to down-regulate CLN1 and CLN2 expression in stationary phase. | 283, 393, 394, 451, 452, 478, 479, 537 |
| Xbp1 | 647 | Swi4-like transcriptional repressor that is responsive to stress. DNA binding domain from residues 346 to 384. | 363 |
| Ydj1, Mas5 | 406 | Molecular chaperone that is required for Cdc28-Cln2-dependent phosphorylation of Cdc28-Cln3 complexes. | 323, 622 |
| Zds1, Nrc1, Ces1, Ckm1, Oss1, Hst1, Bfr1, Rtg2S1, YM8156.15 | 915 | Involved in repression of SBF-mediated transcripts in G2. | 41, 359, 631 |
| Zds2, Ces4, Mcs1, YM8339.10 | 922 | Homolog of Zds1. | 41, 359, 631 |
General CDK Principles and IssuesCDKs.
As the name implies, the CDKs are protein kinases that are dependent for their activity on the binding of a cyclin subunit (for general reviews, see references 390 and 440). A large amount of useful information about CDKs and protein kinases in general can be obtained at www.sdsc.edu/Kinases/. The CDK catalytic subunits are generally recognized by a shared high degree of sequence identity with other members of the family (218), particularly in a domain near the N terminus known as the PSTAIRE motif. CDKs were first discovered during the genetic analyses of the cell cycles of budding (227, 351, 403) and fission (240, 410) yeasts, and, in landmark studies, a CDK was found to be a component of Xenopus mitosis promoting factor (MPF; known as maturation-promoting factor at that time) (144, 348). Eukaryotic cells generally possess multiple CDKs that are involved in a wide range of activities. For historical reasons, the CDK most involved in M phase initiation is called Cdc2 in most organisms (325), but is Cdc28 in S. cerevisiae. In the fungi, the other CDKs have names based on their phenotypes, but in most other systems the CDKs are labeled Cdk2, Cdk3, etc., based on their order of discovery in mouse or human systems. Cdk1 is seeing increased usage—it is equivalent to Cdc2.
CDKs are proline-directed kinases that phosphorylate serine or threonine in S/T-P motifs (321, 494), but individual CDK-cyclin complexes have more stringent substrate specificities (248, 296, 523, 524). The crystal structure of human Cdk2, critical for G1- and S-phase progression, has been solved (118) and has served as a model for other CDKs, including Cdc28. As observed for the catalytic core of other protein kinases, the Cdk2 structure is bilobed with an N terminus that is primarily β-sheet and a C terminus that is primarily α-helix. ATP binds in a cleft between the two lobes. Solitary CDK catalytic subunits have little or no protein kinase activity. Comparison of the monomeric Cdk2 structure with that of protein kinases that are active as monomers, such as the cyclic AMP (cAMP)-dependent protein kinase (299), indicates that Cdk2 lacks enzymatic activity because its N-terminal lobe is displaced relative to the C-terminal lobe—causing misalignment of key catalytic residues involved in phosphate transfer—and the protein substrate binding site is obstructed by the “T-loop” (see “Activation by phosphorylation”) (118).
Full activation of CDKs generally requires two events—cyclin binding and stimulatory phosphorylation. This activation is opposed by the binding of inhibitory proteins, the CKIs, and by inhibitory phosphorylation events as summarized below. Regulators of CDK activity are under complex transcriptional, translational, and proteolytic controls that vary from species to species. A common conserved feature is that the proteolytic controls are generally, although not exclusively (for example, see reference 89), mediated by a ubiquitin-dependent mechanism (236). In contrast to its regulators, the CDK catalytic subunits are usually stable and the regulation of their abundance has generally been of interest only in cells that are moving out of a prolonged stationary phase or during development.
Activation by cyclins.
Cyclins were discovered biochemically as proteins that appeared and disappeared in synchrony with early embryonic cleavage divisions in sea urchins (168) and genetically in yeast for their cell cycle effects (45, 82, 214, 402, 537). The realization in 1989 that cyclins were complexed with CDKs in diverse eukaryotes (46, 140, 193, 379, 617) marked the birth of the modern era in cell cycle research. Most organisms possess multiple cyclins and CDKs, and although cyclin-CDK interactions are specific, CDKs can be activated by multiple cyclins and cyclins can activate multiple CDKs (21, 496). (As yet, there is no example in S. cerevisiae of a cyclin activating more than one kinase, however [14].)
Cyclins are defined by their ability to bind and activate a CDK but are often recognized by the presence of a conserved domain, the “cyclin box” (300). This domain was first recognized based on sequence alignments with diverse cyclins. Now that the crystal structures of mammalian cyclins A (64, 263) and H have been solved (11, 12, 288), the cyclin box is recognized as a sequence element with a recognizable structural motif, the “cyclin fold,” consisting of five α-helices (407). Many, but apparently not all, cyclins possess a second cyclin fold that is often difficult to recognize due to low sequence conservation (198, 378). Interestingly, the cyclin fold is also found in the transcription factor TFIIB (26, 198, 406) and in the retinoblastoma tumor suppressor family (198, 287). The crystal structure of the human Cdk2-cyclin A complex has been solved (263). The principal intersubunit contacts are between a face of the cyclin A cyclin box domain and the PSTAIRE and T-loop regions of Cdk2. There are few differences between the structures of the bound and free forms of cyclin A, but the catalytic residues and the T-loop of the Cdk2 subunit undergo major conformational changes. These changes are presumably responsible for the 40,000-fold increase in protein kinase activity observed in vitro when cyclin A binds Cdk2 (98). Similar events likely occur upon activation of the other CDK-cyclin complexes.
Activation by phosphorylation.
Full activation of most CDK-cyclin complexes requires phosphorylation in the T loop at the position corresponding to T169 of Cdc28 (120, 142, 207, 521). In the crystal structure of the phosphorylated Cdk2-cyclin A complex, phosphorylation of Cdk2 T160 (equivalent to Cdc28 T169) results in additional movement of the T-loop, opening up the protein substrate binding region and increasing the number of contacts between the Cdk and the cyclin (472). The T-loop is a site for autophosphorylation in many protein kinases but not in CDKs. Phosphorylation at this position in a CDK requires a CDK-activating kinase (CAK). The first CAKs to be identified were purified from animal cells and are themselves CDKs, consisting of Cdk7 (177, 444, 520), cyclin H (180, 365) and, in some circumstances, a third protein, Mat1 (124, 179, 550). The Cdk7-cyclin H-Mat1 complex is also a component of the general transcription factor TFIIH (468, 489, 498), which phosphorylates the long carboxy-terminal domain (CTD) of RNA polymerase II and participates in transcription initiation and nucleotide excision repair. This pattern is not invariant, however, as results of the studies of CAK activity in S. cerevisiae made clear. The S. cerevisiae homologs of Cdk7, cyclin H, and Mat1 are Kin28, Ccl1 (586), and Rig2 (171), respectively, but although Kin28-Ccl1-Rig2 is a component of TFIIH and phosphorylates the CTD repeat of RNA polymerase II (172, 545, 587), Kin28-Ccl1-Rig2 does not possess CAK activity (93). As discussed below, the true CAK in S. cerevisiae is not a CDK and is not a component of TFIIH (see “Stimulatory phosphorylation on T169”) (165, 275, 560). Based on a very small sample, it appears that plants resemble the budding-yeast pattern (585) while Schizosaccharomyces pombe CAK has animal-like features (66, 116). These differing patterns seem to reveal an early evolutionary split in the manner in which eukaryotes handle CTD versus CDK phosphorylation events.
Inhibition by CKIs.
Opposing the action of the cyclins are the CKIs. These were first described genetically (83) and biochemically (380) in S. cerevisiae. Recognizable homologs of the yeast Cdc28 inhibitors have yet to be identified in metazoans. The mammalian CKIs (for a review, see reference 497) have received extensive attention due to their roles as tumor suppressors and developmental regulators. These CKIs are grouped into two major classes based on shared structural features and biochemical function. Members of the INK4 class are characterized by the presence of multiple 32- to 33-residue “ankyrin repeats” (49). These CKIs bind to and inhibit a small subset of CDKs (Cdk4 and Cdk6) (490), free or in complex with a cyclin, that are primarily responsible for promoting passage through G1. Crystal (594) and nuclear magnetic resonance spectroscopy (356) structures of two members of this class have been published. The budding-yeast Pho81, inhibitor of the Pho80-Pho85 cyclin-CDK complex, is structurally similar to the INK4 proteins (243, 416, 481). Members of the second class of mammalian CKIs are general CDK inhibitors and recognize both CDK and cyclin components. Analysis of this class is complicated by the observations that (i) the founding member, p21Cip1/Waf1, is a CDK-cyclin assembly factor at low concentrations—and thus a CDK activator—and inhibits only at higher concentrations (635) and (ii) that p21Cip1/Waf1 also binds and inhibits proliferating-cell nuclear antigen (637). No member of this class has been identified in budding yeast.
Inhibition by phosphorylation.
Phosphorylation on the CDK catalytic subunit at positions corresponding to T18 (306, 408) and Y19 (208) of Cdc28 inhibits the activity of CDKs (for a review, see reference 38). In vertebrate systems, both phosphorylations are needed for maximal CDK inhibition (306, 408). In fungal systems, tyrosine phosphorylation alone seems sufficient to meet known regulatory needs, although phosphorylation at the position corresponding to T18 has been detected (9, 119). The side chains of T18 and Y19 are near the ATP binding site, but the mechanism by which phosphorylation at these sites inhibits the CDK activity has not been established. Phosphorylation of Y15 in human Cdc2 (equivalent to Cdc28 Y19) does not significantly alter the Km for ATP (25). It is postulated that the positioning of the ATP γ phosphate or of active-site residues may be disrupted by Y19 phosphorylation, but it is also possible that interactions between the CDK and its protein substrates or other interacting factors are affected. Inhibitory phosphorylation on Cdc2 is associated with checkpoints preventing entry into M phase due to incomplete DNA synthesis (159), to unrepaired DNA damage (267, 286, 626), or to intrinsic cell cycle requirements such as cell size (147, 208). Tyrosine phosphorylation on other CDKs is also important for regulating other cell cycle transitions (258, 473, 557) but has been studied to a lesser extent. These phosphorylation events are controlled by multiple, dually specific protein kinases and phosphatases which, in turn, are under complex and not well-understood controls (for reviews, see references 38, 266, 330, and 568).
Cdc28
The first mutant allele of CDC28 was originally isolated in the early 1970s (226, 228, 229) and was quickly recognized as an important integrator of external controls on cell cycle events (225). The recognition that it encoded a protein kinase (351, 457) whose activity was cell cycle regulated (382, 615), that was activated by cyclins (214, 462, 614), and that was highly conserved in eukaryotic evolution (35, 240, 325) came over a 10-year span in the 1980s. Cdc28 is now recognized as the central component of an elaborate mechanism that controls the timing of events in the yeast cell cycle.
The gene encoding Cdc28 is essential. Most of the original cdc28 mutants arrest cell cycle progression at Start when shifted to restrictive conditions (225, 383, 455, 456), but alleles with other phenotypes are known. A few alleles, cdc28-1N being the most prominent, arrest predominantly in G2 (439, 541). This late cell cycle arrest can also be observed with many of the Start-arrest alleles when the restrictive conditions are applied to cells shortly after Start (458). Defects in postmating nuclear fusion (146), mitotic chromosome stability (125), mitochondrial DNA transmission (125), radiation sensitivity (304), spindle pole body separation (341), and meiosis (507) have also been attributed to defects in Cdc28 function.
Although its protein kinase activity is under multiple, complex controls, the abundance of the Cdc28 polypeptide is virtually unchanged throughout the cell cycle (382). Very little has been reported on environmental or cell cycle effects on Cdc28 transcription, but the protein product is stable (40) and naturally occurs in excess (75, 617). Constitutive overproduction of wild-type Cdc28 also seems to be tolerated relatively well by the cell (383, 454), and so transcriptional and translational regulation of Cdc28 has not been considered important. Virtually all of the controls on Cdc28 activity are manifested at the posttranslational level and are detailed in the remainder of this review.
The Other Budding-Yeast CDKs
It is often but erroneously stated that S. cerevisiae has a single CDK. It is appropriate at this point to emphasize that in addition to Cdc28, four other budding-yeast CDKs are known (Table 2). Three of these—Kin28 (93, 587), Ssn3 (309, 339), and Ctk1 (324), in association with their cyclin activators Ccl1 (545, 586), Ssn8 (309, 339), and Ctk2 (530), respectively—phosphorylate the carboxy-terminal repeat domain of RNA polymerase II and thus play a role in transcription. The remaining CDK, Pho85, is activated by a complex family of at least 10 cyclins (see reference 14 for a review). The functions of this CDK are still being delineated, but they include roles in the regulation of phosphate and glycogen metabolism. Of special interest for this review, the Pho85-Pcl1 and Pho85-Pcl2 complexes play a role in G1 passage. Both cyclin components are periodically expressed late in G1 (414, 578), and deletions of the genes encoding these cyclins or of PHO85 are synthetically lethal with deletions of CLN1 and CLN2 (166, 377), late G1 activators of CDC28 (see “G1 cyclins”). Another cyclin activator of Pho85, Pcl9 is periodically expressed at the M/G1 border and may play a role in cell cycle regulation as well (556). Pcl1, Pcl2, and Pcl9 belong to a subfamily of Pho85 cyclins (the other members are Clg1 and Pcl5) that play a role in the determination of bud site selection (556). As yet, there is no indication that any of these complexes regulate Cdc28 activity, and they are not discussed further in this review.
TABLE 2.
Functions of S. cerevisiae CDKs and their cyclin activators
| CDK | Cyclin | Functions and important properties | References |
|---|---|---|---|
| Cdc28 | Cln1 | Mediates glucose control of cell size at budding. All functions listed for Cln2. | 33, 112, 128, 129, 131, 184, 214, 411, 462, 533, 565, 578, 617 |
| Cln2 | Expressed at Start. Commits cell to mitotic division cycle (Start). Stimulates Sic1 degradation. Initiates localized growth leading to budding. Initiates SPB duplication. Represses pheromone-induced transcription. | 106, 112, 122, 128, 129, 131, 214, 328, 332, 411, 462, 533, 578, 617 | |
| Cln3 | Expressed throughout the cell cycle. Stimulates Start-specific transcription. Mediates cell size control. | 82, 107, 109, 129, 253, 327, 328, 396, 462, 534, 537, 578, 579, 621 | |
| Clb1 | Expressed at G2/M. Minor contributor to mitotic promoting factor. Most important cyclin for meiosis II. | 10, 160, 181, 196, 209, 460, 541 | |
| Clb2 | Expressed at G2/M. Major contributor to mitotic promoting factor. Promotes spindle elongation. Negatively regulates bud emergence. Promotes switch to depolarized bud growth. Represses SBF-mediated transcription. | 8, 10, 47, 160, 181, 209, 259, 332, 460, 540, 541 | |
| Clb3, Clb4 | Expressed in mid S to G2. Important for spindle formation. Can initiate S phase when Clb5 or Clb6 is lacking. | 160, 181, 209, 460, 487, 541 | |
| Clb5 | Expressed at Start. Important for S-phase initiation. Can stimulate SBF-regulated gene transcription. Prevents reinitiation on DNA replication origins that have already ’fired’. Has a possible role in spindle formation. Can fulfill essential Cln roles when overexpressed. | 34, 114, 160, 173, 310, 412, 486, 487, 519 | |
| Clb6 | Expressed at Start. Important for S-phase initiation. Represses Start-specific transcription. Has a possible role in spindle formation. Can fulfill essential Cln roles when overexpressed. | 34, 310, 486, 487 | |
| Pho85 | Clg1, Pcl1, Pcl2, Pcl5, Pcl9 | Roles in Start, bud emergence, and hyperpolarized growth (Δpcl1 Δpcl2 Δcln1 Δcln2 is lethal and fails to bud; Δclg1 Δpcl1 Δpcl2 Δpcl5 Δpcl9 has elongated buds and connected chains of cells). | 14, 166, 377, 378, 556 |
| Pho80 | Repressor of acid phosphatase transcription. | 243, 274, 326, 418, 481, 584 | |
| Pcl6, Pcl7 | Unknown function. | 14, 378 | |
| Pcl8, Pcl10 | Negative regulators of glycogen synthase 2. | 252, 378 | |
| Kin28 | Ccl1 | Phosphorylates carboxy terminal repeats on largest subunit of RNA polymerase II. Component of transcription factor TFIIH. | 93, 171, 172, 518, 545, 586, 587 |
| Ssn3a | Ssn8b | Phosphorylates carboxy terminal repeats on largest subunit of RNA polymerase II. Component of RNA polymerase II holoenzyme. | 309, 339, 542 |
| Ctk1 | Ctk2 | Phosphorylates carboxy terminal repeats on largest subunit of RNA polymerase II. | 324, 530 |
Ssn3 also known as Ume5, Srb10, and Are1.
Ssn8 also known as Sbr11 and Ume3.
CDC28 ACTIVATORS: CYCLINS AND CKS1
Historically, Cdc28 cyclins have been classified into two broad groups: the three G1 cyclins (Cln1 to Cln3) and the six B-type cyclins (Clb1 to Clb6). As the name implies, the G1 cyclins primarily regulate events during the cell cycle interval between mitosis and DNA replication. The yeast B-type cyclins receive their name from homology to the B cyclins of metazoans (546, 610) and are expressed in three successive waves from Start to M. With the exception of CLN3, all of the cyclin genes are paired, with both members of each pair possessing a common overall amino acid sequence and a similar pattern of transcription. Each of the cyclins confers a limited range of functions on Cdc28. The ranges overlap extensively, however, and this has considerably complicated the interpretation of investigations into their function. In this section, the structural and functional properties of the nine cyclin activators of Cdc28 are summarized. The mechanisms controlling cyclin abundance are discussed in later sections.
G1 Cyclins
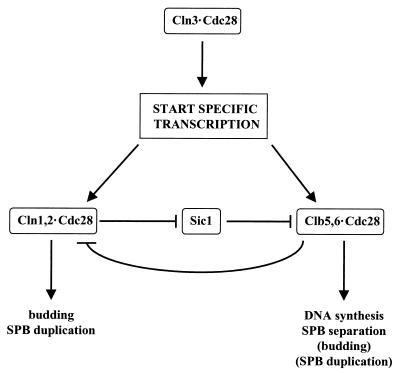
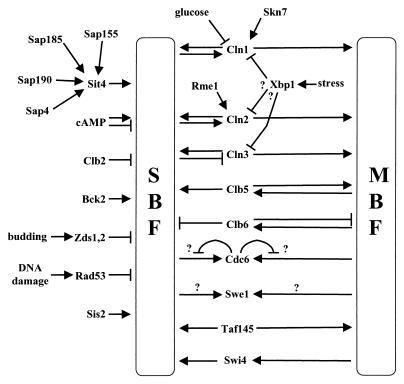
The three G1 cyclins constitute an essential gene family; i.e., the loss of any two CLN genes is tolerated, but at least one must be expressed or the cells arrest at Start (106, 214, 462). Despite this genetic overlap, the CLN gene products differ in their functions, properties, and regulation (an outline of the relationships among the key G1 regulators is given in Fig. 1).
FIG. 1.
Simplified outline of the relationships among major cell cycle regulators during the G1-to-S transition. Arrows indicates stimulatory interaction, lines ending in a “T” indicate inhibitory interactions.
Cln1 and Cln2.
CLN1 and CLN2 were originally identified as high-copy-number suppressors of cdc28-4ts mutations (214). These cyclins are 57% identical, but the homology rises to 72% identity in their N-terminal halves, which contain the cyclin box. The more divergent C termini contain determinants that destabilize the protein by a ubiquitin-dependent mechanism (see “Start proteolysis”). Using the crystal structure of human cyclin A as a guide for an extensive mutational analysis of CLN2, Huang et al. (253) have concluded that the cyclin box domains of Cln2 and cyclin A have similar structures, with the possible exception that helix 4 is missing or is unimportant for the function of Cln2. Genetic analyses (214, 462), coimmunoprecipitation experiments (578, 617), and in vitro reconstitutions (for Cln2 only) (122) show that Cln1 and Cln2 bind to Cdc28 and activate its protein kinase activity, presumably by a mechanism similar but not identical to that seen for the activation of human Cdk2 by cyclin A. Short of a crystal structure for any of the yeast Cdc28-cyclin complexes, genetic methods are being used to probe differences in Cdc28 recognition by different cyclins. Levine et al. have isolated Cdc28-csr mutants that are defective in Cln2 binding and kinase activity but do not affect Clb2 binding and activity (328). Cln3 binding is also diminished but not as dramatically as for Cln2. These mutations, K187E and Q188P, are in the T loop and identify a potential site of Cln2-Cdc28 interaction not seen in the crystal structure of the Cdk2-cyclin A complex. Loss of the C terminus also seems to destabilize the Cln1-Cdc28 interaction (33), indicating the presence of an interaction that is also not predicted by the existing crystal structure.
Cln1 and Cln2 and their associated protein kinase activities are maximal at Start (578, 617), suggesting a role in commitment to the mitotic division process, a suggestion that has received abundant genetic support. Although individual gene knockouts do not have dramatic phenotypes, double cln1Δ cln2Δ mutant cells grow slowly, are aberrantly shaped (214), and have greatly delayed times of bud emergence and DNA synthesis initiation (129, 533). Hyperstable alleles of Cln2, on the other hand, accelerate passage through Start (214). Following Start, yeast cells initiate DNA replication, bud formation, and spindle pole body duplication. Cln-Cdc28 complexes stimulate DNA synthesis indirectly by accelerating the proteolysis of the Clb-Cdc28 inhibitor Sic1 (see “Sic1”), but the mechanisms by which bud formation and spindle pole body duplication are stimulated by Cln-Cdc28 complexes have not been delineated. In addition to the Start functions, Cln1-Cdc28 and Cln2-Cdc28 are specifically able to repress pheromone-inducible transcription, a function not shared with Cln3-Cdc28 or the Clb-Cdc28 complexes (411). Despite their similarity, some functional differences between these cyclins have been noted. For example, extended overproduction of Cln2 but not Cln1 is lethal in some strain backgrounds (462) and Cln1 but not Cln2 modulates an increase in cell size at budding in response to glucose (184, 565).
Cln3.
In many ways, Cln3 is the oddest of the Cdc28 cyclins. It does not have a close yeast homolog and has only ∼20 to 25% identity to its namesakes, Cln1 and Cln2 (107, 396), and actually has greater overall sequence similarity to Clb5 and Clb6. Sequence similarity is highest in the cyclin box region. Activation of Cdc28 protein kinase activity probably occurs in a manner similar to activation by Cln1 and Cln2, but the strength of the cyclin-CDK interaction and specific protein-protein contacts no doubt differ. Cross and Blake have isolated a mutant Cdc28, Cdc28-5r83, that binds Cln1 but not Cln3 (109), providing an entrée to the genetic analysis of differences in Cdc28 activation by the G1 cyclins.
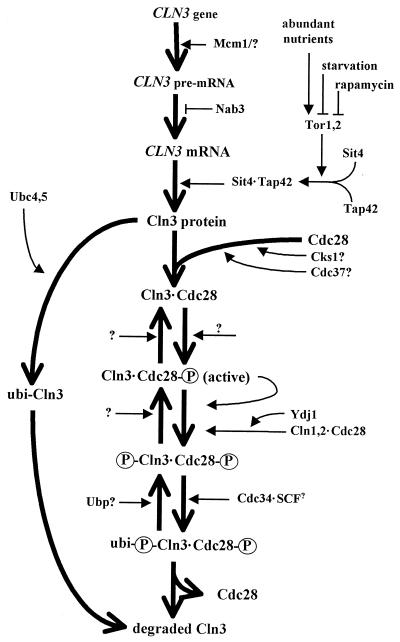
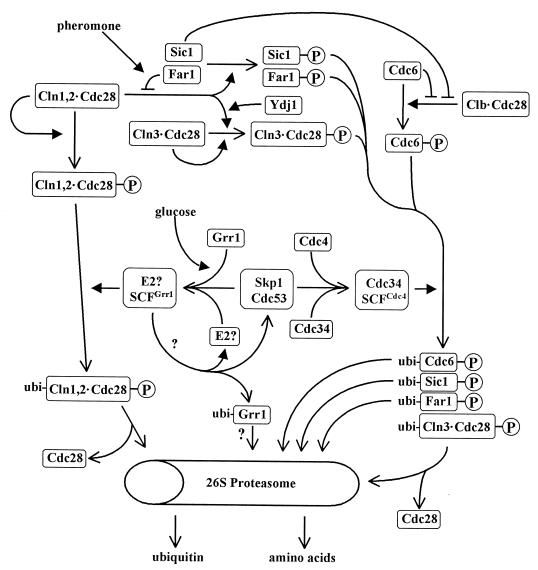
Unlike the other cyclins, CLN3 transcription is not strongly periodic with respect to the cell cycle, but there is a small rise at the M/G1 border over its basal levels (see “M/early-G1-specific transcription”) and protein levels exhibit moderate periodicities in amplitude (109, 578). CLN3 mutants have the strongest phenotypes of the G1 cyclins, and, fittingly, CLN3 is the only cyclin discovered by classical genetic methods, having been originally identified as WHI1-1 (now CLN3-1) by its small-cell phenotype (82, 537) and as DAF1-1 (now CLN3-2) by its resistance to mating pheromone (107). Both of these dominant mutations remove the C-terminal one-third of CLN3, which, like Cln1 and Cln2, contains a determinant that makes Cln3 a target for rapid turnover (see “Start Proteolysis”) (109, 579). In addition to reducing Cln3 turnover rates, C-terminal truncations appear to reduce the ability of Cln3 to activate Cdc28 (109, 621), but this reduction is more than overcome by the increase in Cln3 stability, which accounts for the small size of the CLN3stab cells. Cells with CLN3 deleted are enlarged and have an extended G1 period but have an overall normal growth rate due to compensation in other parts of the cell cycle (107, 129, 396, 534). Despite the prominence of the phenotypic effects relative to cln1Δ and cln2Δ mutants, Cln3 is estimated to be 5- to 100-fold less abundant and the specific activity of the associated protein kinase activity (with histone H1 as a substrate) is 2- to 20-fold lower than the corresponding values for Cln1 or Cln2 (327, 578). These results and others support the hypothesis that Cln3-Cdc28 plays a unique role in G1 as an activator of CLN1 and CLN2 transcription (see “Control of SBF and MBF activity by Cln-Cdc28”). Known and suspected influences on Cln3 activity are outlined in Fig. 2 and discussed throughout the review.
FIG. 2.
Birth, life, and death of Cln3. An outline of processes influencing the synthesis, activation, and destruction of Cln3 is shown. Heavy, open arrowheads indicate transitions involving CLN3 and its gene product. Lighter, solid arrowheads denote cellular components and environmental influences that positively regulate the indicates tep. T-shaped lines denote cellular components and environmental influences that negatively regulate the indicated step. The circled “P” indicates a phosphorylated protein, “ubi-” indicates a ubiquitinated protein. Indicated relationships may be indirect, and some steps are speculative. See the text for details.
B-Type Cyclins
The six B-type cyclins are commonly subdivided into three distinct pairs based on sequence homology and transcriptional regulation. As with the G1 cyclins, the functions of the members of this family are complex and partially overlapping.
Clb5 and Clb6.
Clb5 and Clb6 are 50% identical. Of the six Clb proteins, these two have the least similarity to metazoan B-type cyclins, with Clb5 being more divergent than Clb6. For both proteins, the cyclin homology is in the C-terminal half of the protein. Clb5 can directly activate Cdc28 protein kinase activity in vitro (173, 519), and both Clb5 and Clb6 interact with Cdc28 in a two-hybrid assay in vivo (310). Clb5 possesses a mitotic destruction box (160, 310, 487) that may accelerate Clb5 proteolysis during mitosis (see “Anaphase proteolysis”), but Clb6 does not (310, 487). Clb5 also possesses a highly acidic domain that is not shared with Clb6.
The CLB5 and CLB6 genes are coexpressed with CLN1 and CLN2 (160, 310, 487) and could, in a sense, be classified as G1 cyclins. Consistent with such a classification, Δcln1 Δcln2 Δclb5 Δclb6 cells are inviable (487). Furthermore, overexpression of CLB5 (160, 487) or CLB6 (34) suppresses the cln1Δ cln2Δ cln3Δ lethality. No other CLB gene has this ability (160, 329). Under normal conditions, however, Clb5 and Clb6 do not carry out most Start functions, since they are kept in an inactive state by Sic1 until after Cln1-Cdc28 and Cln2-Cdc28 activities have appeared (486) (see “Sic1”).
The primary roles for Clb5 and Clb6 are to initiate S phase in a timely fashion (486) (see reference 566 for a review), prevent reinitiation on replication origins that have already “fired” (114), and negatively regulate Cln-Cdc28 activity (34). Consistent with these roles, cells lacking Clb5 have an extended S phase (160, 310, 487) and a clb5Δ clb6Δ double mutant has a long S-phase initiation delay, but once initiated, the S phase is of normal length (310, 487). CLB6 knockouts have reduced G1 times and small cells, indicative of an early Start transition, while overexpression of CLB6 represses the transcription of both CLN2 and CLB5 (34). Clb5, on the other hand, does not seem to have this repressive effect on transcription and, when overexpressed, stimulates at least some Start-specific transcripts (412). Both Clb5 and Clb6 seem to have a negative effect on formation of Cln2-Cdc28 complexes that is independent of the transcriptional effects, however, since S-phase-arrested cells lacking either Clb5 or Clb6 have levels of Cln2-Cdc28 complexes that are 1.5 to 2 times that of wild-type cells (34). Analyses of multiple CLB and CLN knockouts indicate that both Clb5 and Clb6 may play a role in spindle formation as well (487), but Clb5 and Clb6 are not sufficient to form the bipolar spindles needed for mitosis (10, 181, 460).
Clb3 and Clb4.
CLB3 and CLB4 were originally identified by high-copy-number suppression of the G2-arresting cdc28-1N mutation (541), degenerate PCR (181, 460, 541), and low-stringency hybridization (460). The C-terminal 276 residues of both proteins contain the region most homologous to cyclin B and are 62% identical to each other. Destruction box consensus regions are found within the less homologous amino termini (see “Anaphase proteolysis”). Like CLB5, CLB3 has a highly acidic domain. CLB3 and CLB4 transcripts arise near the beginning of S phase (after the CLN1 and CLN2 peak) and remain high until late anaphase (160, 181, 460). The associated protein kinase activity has a similar periodicity (209). Measurements of absolute levels of protein kinase activity in asynchronous cells indicate that Clb3-Cdc28 constitutes the majority (67%) of all Cdc28 activity in asynchronous log phase cultures. Clb4-Cdc28 is a minor component. This abundance is not reflected phenotypically, though, since clb3Δ, clb4Δ, and clb3Δ clb4Δ mutants have no obvious mitotic phenotypes (181, 460, 487). The clb3Δ clb4Δ clb5Δ triple mutant, however, cannot make spindles and is inviable. The clb3Δ clb4Δ clb5Δ clb6Δ mutant, also inviable, has difficulty initiating S phase (487). Given the timing of their appearance, it appears that Clb5 and Clb6 are normally involved in S-phase initiation, although Clb3 and Clb4 can fill in if necessary. Clb3 and Clb4 appear to play a role in spindle formation that cannot be fulfilled by Clb5 and Clb6 but can be accomplished by Clb1 and Clb2, which appear later (10, 460).
Clb1 and Clb2.
The CLB1 and CLB2 genes were cloned along with CLB3 and CLB4 as high-copy-number suppressors of the G2-arresting cdc28-1N mutation (541), degenerate PCR (181, 196, 460, 541), and low-stringency hybridization (460). CLB2 and CLB5 are adjacent genes transcribed convergently. CLB1 and CLB6 are arranged similarly—a fortunate circumstance that facilitated the cloning of both CLB5 and CLB6 (310, 487). This arrangement is apparently an evolutionary holdover, reflecting two successive duplications of a primordial CLB gene. There is no indication that CLB2 and CLB5 or CLB1 and CLB6 are regulated coordinately at the genetic level. The C-terminal 276 residues of both proteins contain the region most homologous to cyclin B and are 78% identical to each other (62% identical overall) but only 40 to 44% identical to the analogous region of Clb3 and Clb4. Destruction box consensus regions are found within the less homologous amino termini (see “Anaphase proteolysis”). As previously observed for cyclin B in Xenopus oocyte lysates (395), Clb2 mutants lacking the destruction box have difficulty exiting M phase (196, 540), indicating that CDK activation and inactivation are needed for proper cell cycle advancement.
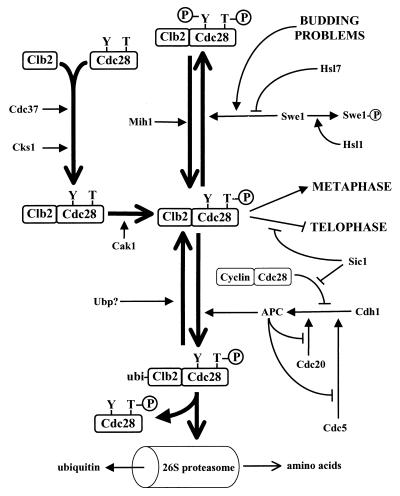
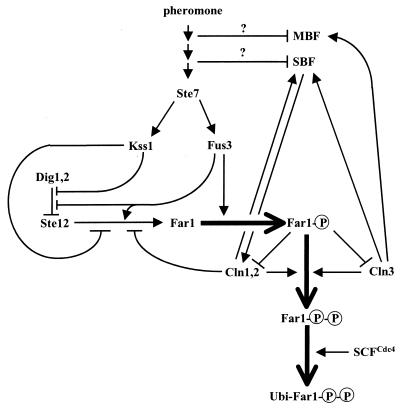
CLB1 and CLB2 transcripts are strongly periodic, peaking about 10 min before anaphase (181, 196, 460, 541). The associated protein kinase activity has a similar periodicity (209, 540). Measurements of absolute levels of protein kinase activity indicate that Clb2-Cdc28 constitutes the majority (85%) of Cdc28 activity in mitotically arrested cells. Clb1-Cdc28 is a minor component (209). Phenotypically, CLB2 is the most important of the CLB genes. Deletion mutants of clb2 are somewhat larger than normal, and the cultures have a high percentage of budded G2-phase cells (160, 181, 460, 541). Double-mutant combinations of clb2Δ with clb1Δ or clb3Δ are lethal (the clb2Δ clb4Δ and clb2Δ clb5Δ combinations are viable). In contrast, clb1Δ has no obvious mitotic phenotype (but see “Meiosis”) and even the triple clb1Δ clb3Δ clb4Δ mutant has only a mild mitotic defect (10, 160, 181, 196, 460, 541). The inviable combinations that include clb2Δ arrest prior to mitosis and indicate that Clb2-Cdc28 constitutes the yeast MPF with some assistance from Clb1-Cdc28. Consistent with this, Clb2-Cdc28 is important for spindle elongation (332). Clb2 also negatively regulates SBF-promoted transcription (see “Repression of SBF activity by Clb2-Cdc28 complexes”) (10) and bud emergence (47, 196, 332, 540) but promotes the switch from tip-directed growth to isotropic growth in buds (332). When inappropriately expressed, Clb2 can activate DNA synthesis (8, 259) but not budding (8). Key events surrounding Clb2 metabolism are diagrammed in Fig. 3.
FIG. 3.
Processes centered around Clb2 activation, regulation, and destruction. Conventions and caveats are as in Fig. 2.
Cks1
Cks1, the budding-yeast homolog of the Schizosaccharomyces pombe p13Suc1 protein (60, 239), is essential for proper Cdc28 function (213), but the nature of this function has been mysterious and controversial. Cks1 binds to many, but not all, CDK-cyclin complexes with high specificity, an activity that has been exploited as a tool to purify CDKs (317). Recent biochemical data argue strongly for a role as a CDK-cyclin assembly factor (173, 519, 595), but this does not preclude additional functions for this small protein. The budding-yeast gene was originally cloned along with CLN1 and CLN2 as a high-copy-number suppressor of cdc28ts mutants (213). It is highly conserved but has an extended C-terminal tail containing a 16-residue polyglutamine tract not found in its human or fission yeast counterparts (213, 461). Cks1 abundance does not vary with the cell cycle (213). Mutants lacking CKS1 arrest at Start (213), at G2, or in a mixture of G1 and G2 states (549) depending upon how Cks1 function is eliminated. Studies in other systems indicate that at least part of the essential function of Cks1 is its interaction with a CDK, since mutations in either CKS1 (606) or CDC28 (143) homologs that reduce Cks1-Cdk binding are lethal. Cks1 is not needed for CDK catalytic function per se, however, since cks1ts cells at the restrictive temperature possess high levels of Cdc28 protein kinase activity (549) and purified human Cdc2-cyclin B complexes lacking the human Cks1 homolog retain full protein kinase activity (316). In vitro, Cks1 is required to reconstitute active Cdc28-Cln2 (173, 519, 595) but is not needed for Cdc28-Clb5 activity (173), supporting a role for Cks1 as an assembly factor in vivo for at least some CDK-cyclin complexes. If this is the only role of Cks1, the G2 arrest phenotype of cks1 (549) predicts that the M-phase Cdc28 complexes may also require Cks1 for their assembly. A test of this hypothesis has not yet been reported.
Overexpression of Cks1 delays G2 progression (461), indicating that Cks1 may do more than simply promote Cdc28-cyclin assembly. Studies on Cks1 homologs in other systems have suggested other potential functions, including narrowing of the CDK substrate specificity (316), inhibition of CDK dephosphorylation on phosphotyrosine (see “Phosphorylation of Cdc28”) (145, 429), inhibition of CDK activation following phosphotyrosine hydrolysis (265), and inhibition of CDK activity following mitosis (387, 429). Compensating for the lack of hard information on Cks1 function, there is abundant structural data on the Cks1 protein. The crystal structures of the S. pombe p13Suc1 homolog (50, 157), the human CksHs1 (24) and CksHs2 (425) homologs, and the human CksHs1-Cdk2 complex (51) have been solved. The free Cks1 can undergo dramatic conformational changes and exists as monomers, dimers, or hexamers. Only the monomer is capable of binding CDKs, however (606), and the relevance of the multimeric forms is not clear. Watson et al. have proposed that regulated oligomerization of Cks1 may control its association with Cdk complexes (606). The crystal structure has also revealed the presence of an “anion-binding site” capable of interacting with phosphate and sulfate (50, 157, 425) that might target Cdc2 complexes to other phosphoproteins (51, 429). Sudakin et al. suggest that one such target is the APC, the complex responsible for ubiquitinating A and B cyclins at anaphase (see “Anaphase Proteolysis”) (536). The phosphorylated APC binds to Cks proteins, most probably through the anion binding site, and these investigators have speculated that this might be important for Cdk-cyclin B degradation at anaphase.
CDC28 INHIBITORS: CKIS
Far1
FAR1 was originally discovered as a gene required for mating-pheromone-induced cell cycle arrest but not needed for induction of pheromone-responsive genes (83). The gene product was initially reported to be an inhibitor of Cln1-Cdc28 and Cln2-Cdc28 protein kinase activity (436, 577) and later to have activity against Cln3-Cdc28 complexes as well (264), but it was not able to inhibit Clb5-Cdc28 and Clb2-Cdc28 in vitro (436). The biochemical nature of Far1 activity has recently been called into question by Gartner et al., who found that Far1 did not reduce the specific activity of immunoprecipitated Cln2-Cdc28 from mating-pheromone-treated cells although Far1 was present in the Cln2-Cdc28 immunoprecipitate (191). Gartner et al. have argued that the previous results may be an artifact of overproduction of Far1, Cln2, or both, but they did not provide data that supported an alternative mechanism for Far1 action. These newer results are difficult to reconcile with the previous findings in this field and indicate that much of the biochemistry in this area may need to be reevaluated. In this review, we will still consider Far1 to be a specific inhibitor of Cln-Cdc28 complexes with the caveat that its substrate specificity and possibly its mechanism of action may undergo considerable revision in the near future.
If the traditional mode of action for Far1 is upheld, Far1 probably inhibits by substrate exclusion, since Cln-Cdc28 activity is regained when Far1 is washed off the complexes (ruling out irreversible modification or disruption) (436) and since Far1 can be phosphorylated in Cln-Cdc28-Far1 complexes (making allosteric change to an inactive form of Cdc28 unlikely) (577). The Cln-Cdc28 binding and inhibitory activity has been mapped to residues 99 to 390. (Note that the original sequence analysis missed the first 150 bases of the coding sequence [376]). The positions in this review have been corrected for that difference.) The sequence of this region does not show any homology to other CKIs. The N terminus confers regulated instability on Far1 (376) (see “Start Proteolysis”), and the C terminus plays a separate role, not yet related to Cdc28 regulation, in mating and bud site selection (83, 87, 139, 590), which is not discussed further in this review.
Far1 is regulated at multiple levels. Its transcription is cell cycle regulated, with a peak near the M/G1 transition (375) (see “M/early-G1-specific transcription”), suggesting that Far1 may have a cell cycle function independent of its role in mating. Consistent with this, Far1 is found bound to Cln1-Cdc28 and Cln2-Cdc28 complexes in cells unexposed to pheromone and far1Δ strains have a reduced G1 phase relative to the wild type (376), indicating that Far1 acts constitutively to moderate Cln activity at Start. FAR1 is not expressed in diploids and is presumably under Mata-Matα repression (83). Mating pheromone induces additional Far1 transcription (83), and this induction is necessary but not sufficient for pheromone-induced cell cycle arrest (84, 375). The protein product is predominantly nuclear (as a green fluorescence protein fusion) (235). It is stable in G1 but is degraded rapidly following Start (375) (see “Start Proteolysis”).
Far1 is unique among the known CKIs, in that its inhibitory activity is apparently enhanced by an inducible, posttranslational modification. Activation of the pheromone response pathway stimulates increased association of Far1 with all three Cln-Cdc28 complexes (435, 577). This is not simply due to increased FAR1 expression, since overexpression of FAR1 in the absence of pheromone does not stop cell cycle progress and has only weak effects in a cln1 CLN2 cln3 strain (84, 375, 413). Furthermore, wild-type Far1 from cells not treated with pheromone or produced from bacterial expression systems apparently has little or no inhibitory activity in vitro, although Far1 from mating-pheromone-treated cells or a constitutive Far1 allele (Far1-22S87P) produced in bacteria is fully active (436). Attention has focused on phosphorylation by Fus3, a mitogen-activated protein (MAP) kinase homolog, as the required activator of Far1 because (i) Fus3 is at the base of the protein kinase cascade that responds to mating pheromone; (ii) Fus3 interacts with Far1 in a two-hybrid assay; (iii) overexpression of FAR1 suppresses the sterility of fus3ts mutants, but FUS3 overproduction does not suppress far1Δ mutations; (iv) Fus3 isolated from pheromone-treated but not untreated cells phosphorylates Far1 in vitro; (v) phosphorylation of Far1 is lost in any mutation which inactivates the pheromone response pathway; (vi) mutants of Fus3 which are defective in cell cycle arrest but not mating-inducible transcription do not phosphorylate Far1; and (vii) Far1 associates with Cln-Cdc28 complexes in pheromone-treated FUS3+ but not fus3 cells (154, 164, 435, 577). Despite this evidence, there has been no report of Fus3 activation of Far1 inhibitory activity in vitro. The threonine at position 306 may be the phosphorylation site for whatever kinase, probably Fus3, is critical to Far1 activation, since the Far1-T306A mutant protein lacks the pheromone-induced sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) mobility shift seen with wild type Far1, does not bind to Cln2-Cdc28 complexes, and is inactive in effecting a pheromone-mediated cell cycle arrest (191).
The inability to reconstitute an in vitro assay of Cln-Cdc28 inhibition by wild-type Far1 has complicated the determination of the substrate specificity of Far1. The original genetic analysis was interpreted to suggest that Far1 would be active only against Cln2-Cdc28 complexes (83). This rested primarily on the observation that while far1Δ mutants were unable to stop dividing in the presence of mating pheromone, double mutants combining far1Δ with cln2Δ were pheromone sensitive. Double or triple mutants combining far1Δ with cln1Δ, cln3Δ, or cln1Δ and cln3Δ are pheromone resistant (83). The implication of this result is that Far1 is the only activity holding Cln2-Cdc28 in check and that other mechanisms would be needed to inhibit Cln1-Cdc28 and Cln3-Cdc28, but it does not prevent Far1 from having activity on Cln1-Cdc28 or Cln3-Cdc28. In accord with a broader specificity for Far1, Tyers and Futcher found that Far1 binds all three Cln-Cdc28 complexes (577). After mating-pheromone addition, the binding to Cln1-Cdc28 and Cln2-Cdc28 rises within minutes from an already appreciable basal rate. In contrast, the binding to Cln3-Cdc28 is not detectable until after an hour of pheromone exposure. The binding correlated with partial inhibition of Cln1-Cdc28 and Cln2-Cdc28 activity, but changes in Cln3-Cdc28 activity, which are difficult to measure, were not detected in this study. Using a different approach to enhance Cln3-Cdc28 activity (Cln3 overproduction instead of inhibition of Cdc34-dependent ubiquitination) Jeoung et al. have found that Cln3-Cdc28 activity is down-regulated 10-fold in a FAR1- and mating-pheromone pathway-dependent manner (264). The time course of inhibition of Cln3-Cdc28 after pheromone addition was considerably longer than that seen for Cln1-Cdc28 and Cln2-Cdc28—occurring on the order of a cell cycle instead of in minutes—in accord with the kinetics of Far1 binding (577), suggesting that a cell cycle-regulated process was necessary to activate Far1 for Cln3-Cdc28 inhibition. Supporting this conjecture, Cln3-Cdc28 activity was not inhibited by pheromone addition in cells blocked in G2/M by nocodazole (264). The biochemical nature of this cell cycle-dependent activation is not clear, but stabilization and overexpression of Far1 restores Cln3-Cdc28 inhibition in G2/M, indicating a potential role for proteolysis. The putative inhibition of Cln3-Cdc28 complexes by Far1 is generally very sensitive to the relative levels of Cln3 and Far1. Mutations or genetic constructs that increase Cln3 production or stability are mating-pheromone resistant (112, 131, 214, 235, 264, 396, 578), a resistance that can be counteracted by overproduction or stabilization of Far1 (235, 264). Similar overproduction or stability of Cln2 or Cln1 produces cells that are considerably less resistant to mating pheromone, which is more readily reversed by higher Far1 levels (112, 214, 235, 436). The data give the impression that Far1 is a potent and rapidly acting inhibitor of Cln1-Cdc28 and Cln2-Cdc28 complexes, but is less potent and acts more slowly against Cln3-Cdc28. This impression will have to be reevaluated in the light of the data of Gartner et al. (191).
Peter and Herskowitz have shown that a bacterially produced Far1 mutant, Far1-22S87P, inhibits Cln1-Cdc28 and Cln2-Cdc28 complexes in vitro (436). This mutation eliminates a Cdc28 phosphorylation site and results in a hyperstable gene product in vivo (see “Start proteolysis”) (235). Expression of FAR1-22S87P results in constitutive, pheromone-independent cell cycle arrest and seems to cause arrest in cell cycle intervals past Start. Truncations that remove the first 50 amino acids of Far1 also hyperstabilize the protein and, when expressed in yeast, can cause cells to arrest (still in a pheromone-dependent manner) in a budded state (376). These results open up the possibility that hyperstabilized Far1 has additional targets that Far1+ does not. These targets are apparently not Clb5-Cdc28 and Clb2-Cdc28 if the in vitro assays are reliable indicators of in vivo function (436).
Sic1
Sic1 is an inhibitor of Cdc28-Clb complexes (380, 486) and thus has an activity complementary to that of Far1. Originally discovered as a tight-binding Cdc28 substrate in immunoprecipitated Cdc28 complexes (457), it was later shown to have CDK-inhibitory activity (380), the first biochemical demonstration of CKI activity. Like Far1, Sic1 inhibitory activity is due to its ability to exclude substrates from the Cdc28 active site. Cdc28-Clb5 binding activity has been mapped to the C-terminal half of Sic1 (596). This domain has weak similarity to the inhibitory domain of Rum1, an S. pombe CKI that has many functional and regulatory parallels with Sic1 (476). There is no noticeable resemblance to mammalian CKIs, but two sequences at the extreme C terminus of Sic1 (but not found in Rum1) match the ZRXL motif that has been proposed to be a CDK-cyclin recognition motif (1). Sic1 protein expression is limited to the G1 phase (137, 382). This pattern of expression is due to periodic transcription peaking at the G1/M-phase border (see “M/early-G1-specific transcription”) and to Cln-Cdc28-dependent proteolysis at Start (see “Start proteolysis”). An N-terminal domain is sufficient and necessary for Sic1 ubiquitination in vitro (596) and thus may regulate Sic1 stability in vivo.
SIC1 is a nonessential gene (137, 409), but sic1Δ cultures contain a high percentage of cells permanently arrested in G2 (409). Two major functions have been assigned to Sic1, and either function could account for the dying cells. The first is to prevent premature S-phase initiation until after Cln-Cdc28 levels have risen sufficiently to complete bud initiation and spindle pole body duplication (486). This function is carried out by inhibiting Clb5-Cdc28 and Clb6-Cdc28 complexes until Sic1 is destroyed, which is, in turn, initiated by Cln-Cdc28-dependent phosphorylation of Sic1 (see “Start proteolysis”). Sic1 destruction is the only essential function of the CLN genes, i.e., a cln1Δ cln2Δ cln3Δ sic1Δ mutant is viable (161, 480, 576). The second major function is to assist in the down-regulation of Clb-Cdc28 activity in late anaphase to telophase (137, 571). The exact role of Sic1 in this process is not clear; it may inhibit a fraction of Clb-Cdc28 activity that is not accessible to proteolysis at anaphase, or it may help down-regulate Clb-Cdc28 so that anaphase proteolysis can be activated (see “Anaphase proteolysis”) (7).
Cdc6?
Cdc6 also has characteristics that indicate that it may act as a Cdc28 CKI. It is better known for its role in ensuring single rounds of DNA replication during a cell cycle (reviewed in reference 566). In addition to or as part of this function, Elsasser et al. have reported that Cdc6 can bind and inhibit Clb-Cdc28 complexes (156), a function that may explain the G2 delay seen in Cdc6 overexpressers (34, 68). This inhibitory activity of Cdc6 is enhanced by Clb5 and Clb6 by both transcriptional and posttranscriptional means (34). The Cdc6-Clb-Cdc28 interaction appears weaker than the Sic1-Clb-Cdc28 interaction, since Sic1 can displace Cdc6 from Clb-Cdc28 complexes (156). Like Sic1, Cdc6 is an unstable protein and is destroyed at the G1/S border (438). Its destruction is dependent upon the same ubiquitination system that degrades Sic1 (see “Start proteolysis”) (141, 437), but the role of phosphorylation in initiating Cdc6 degradation has not been established. Cdc6 is phosphorylated by Clb-Cdc28 complexes (156, 437), not Cln-Cdc28 complexes (156), so if Cdc28-dependent phosphorylation is a required prerequisite for Cdc6 proteolysis, Sic1 destruction would precede Cdc6 turnover. CKI activity by Cdc6 may be a means of fine-tuning Clb5 and Clb6 activity in a highly localized manner at the origin of replication.
PHOSPHORYLATION OF CDC28
Stimulatory Phosphorylation on T169
The major site of phosphorylation on Cdc28 is at T169 (93). Consistent with the requirement for phosphorylation at the equivalent position in the CDKs of other organisms, the nonphosphorylatable Cdc28-T169A mutant cannot be activated in vitro (122) or support cell division in vivo (342). Three groups independently identified the S. cerevisiae CAK that phosphorylates T169 by purifying Cdc28-activating activity (one group finding that yeast CAK was stably associated with Cdc28 [560]) and cloning its gene, CAK1 (165, 275, 560). CAK1 has also been identified by its synthetic lethality with the sit4 protein phosphatase (543) and the kin28 CDK (560), as a suppressor of the sporulation defect of the smk1 MAP kinase (600), and as a mutant that gives a cdc34-like phenotype (91). Sequence comparisons indicate that, within the protein kinase family, Cak1 is most closely related to the CDKs but is a very distant relative. It has some unusual sequence features, the most striking being the lack of the almost invariant GxGxxG motif involved in nucleotide binding in most other protein kinases (Drosophila NinaC and Yersinia YPKA are other exceptions [91]). In addition, a mutant allele of Cak1 in which arginine replaces an invariant lysine (K31R) thought to be involved in phosphate transfer during catalysis can still support vegetative growth (600). This mutation would cripple most other protein kinases. Also, unexpectedly for a CDK-activating kinase, Cak1 is clearly active as a monomeric protein; i.e., no cyclin subunit is required for its activity (165, 275, 560).
Both monomeric CDKs and CDK-cyclin complexes act as substrates for Cak1, although only the CDK-cyclin complexes gain kinase activity upon phosphorylation by Cak1 (165, 275, 560). Cdc28 activation by Cak1 is essential for viability, and, accordingly, CAK1 is an essential gene. Immunodepletion (165, 275) and assays of extracts from cakts mutants (560) indicate that Cak1 is the predominant, if not the only, protein kinase capable of phosphorylating and activating Cdc28. Cdc28 activation is also the only essential function of Cak1. Cross and Levine have identified mutant derivatives of CDC28-T169E that no longer require Cak1 for activation. In these multiply mutant CDC28 backgrounds, CAK1 is no longer essential but the cak1Δ derivative had a slow-growth phenotype relative to the CAK1+ control, indicative of another, nonessential function (111). This other role has not been identified. Unlike CAKs from other species, Cak1 is unable to phosphorylate the carboxy-terminal repeat domain of RNA polymerase II (275). At least one cak1 allele (civ1-4) is synthetically lethal with kin28ts mutations and CAK1 overexpression suppresses kin28ts alleles (560), opening the possibility that Cak1 acts on Kin28 and possibly other yeast CDKs in addition to Cdc28. Sutton and Freiman, however, saw no effect on generalized transcription or acid phosphatase secretion in a cak1-22 strain at the restrictive temperature, as would have been expected if Kin28 or Pho85 activity were inhibited (543). Cak1 is required for spore wall morphogenesis during the later stages of meiosis (600), but this defect is not expected to lead to mitotic growth defects.
CAK1 mutant phenotypes are complex. Temperature-sensitive alleles of cak1 arrest cell division at multiple stages, with the fraction of cells found at each particular stage being determined by the cak1 allele (91, 165, 275, 543, 560, 600). The civ1-4 allele of CAK1 isolated by Thuret et al. (560) arrests predominantly in G1, a C-terminal truncation isolated by Chun and Goebl (91) has a cdc34-like phenotype (pre-S arrest with multiple buds), and the cak1-1 and cak1-22 alleles of Kaldis et al. (275) and Sutton and Freiman (543) arrest predominantly in G2. These varied phenotypes are consistent with a failure to fully activate Cdc28 protein kinase activity, with individual Cdc28-cyclin complexes being differentially affected in an allele-specific manner. As a consequence, CAK1 genetic interactions are also complex. The cak1-1 allele of Kaldis et al. is suppressed by overexpression of CLB2, but not CLN2, and is synthetically lethal with clb2Δ and with a cln1Δ cln3Δ double mutation (275, 543).
Do cyclical changes in the T169 phosphorylation state of Cdc28 play a role in cell cycle entry or progression? There is evidence that dephosphorylation of T169 plays a role in anaphase in Xenopus (349). In an attempt to simulate the constitutive phosphorylation of T169 in S. cerevisiae, Lim et al. (342) replaced T169 with glutamate. The Cdc28-T169E protein has weak but noticeable kinase activity relative to the wild type, and its expression allows the growth of cdc28-1N (mitosis-defective) but not cdc28-4 (Start-defective) mutant strains. Clb2 coimmunoprecipitates with Cdc28-T169E, but Cln2 does not. The multiply mutant, Cak1-independent Cdc28-T169E derivatives constructed by Cross and Levine promote almost normal growth and behavior even when complementing a cdc28 deletion, but, despite binding Cln2 more efficiently, they also had a severe defect in Cln2-associated protein kinase activity measured in vitro (111). These results suggest that G1 cyclins might be more efficient activators of unphosphorylated Cdc28, while the B-type cyclins require phosphorylated T169 to be fully active. In any case, there does not appear to be a strong requirement for periodic phosphorylation/dephosphorylation of Cdc28 T169 in the usual laboratory physiological tests.
There is also no indication that Cdc28 T169 phosphorylation is periodic. There is little bulk change in the Cdc28 phosphorylation state in response to G1 arrest by starvation or mating-pheromone exposure (212) or during the cell cycle (9), but these studies would have missed changes that affected only the small fraction of Cdc28 bound to a particular Cln or Clb (615, 617). Cak1 activity appears to be constitutive throughout the cell cycle (165, 543), but studies on its regulation are still at an early stage. Cak1 autophosphorylates (543), but the significance of this activity is unclear since the autophosphorylation is severely reduced in the cak1-K31R allele (543), which has an otherwise wild-type phenotype (543, 600). Proteins appear to associate with Cak1 in immunoprecipitates in substoichiometric amounts, but their identities or functions are unknown (543). The flip side of the coin, a protein phosphatase analogous to the mammalian KAP (CDK-associated phosphatase) (443) that would dephosphorylate phospho-T169, has not been identified. Sit4 is a potential candidate, since sit4 alleles are defective in cell cycle events associated with G1 CDK activation (175), but the synthetic lethality of sit4 cak1 strains is difficult to explain in this context (543).
Inhibitory Phosphorylation on Y19 and T18
Y19.
Y19 is clearly phosphorylated in Cdc28, reaching maximal levels in S and G2 and being undetectable in M and G1 (9, 525). Different groups using different approaches have reported widely varying extents of Cdc28 inactivation by Y19 phosphorylation, ranging from inconsequential to substantial (47, 331, 341, 344, 525). Despite these uncertainties, it is generally thought that tyrosine phosphorylation of Cdc28 delays entry into mitosis. Consistent with this and indicative of a specific effect at mitosis, Cdc28-Y19E (which partially mimics the permanently phosphorylated form) retains near-wild-type protein kinase activity in anti-Clb2 immunoprecipitates and supports DNA replication, bud emergence, and spindle pole body duplication but does not support spindle pole body separation or nuclear division (341). Phosphorylation on Y19 increases when cells are UV irradiated (195) or arrested by hydroxyurea (9), consistent with a role for Y19 phosphorylation in an S-phase or DNA damage checkpoint. Surprisingly, however, cells expressing the unphosphorylatable CDC28-Y19F allele do not differ from the wild type with respect to arrest due to DNA damage or incomplete DNA replication (9, 525), precluding an essential role in the checkpoint coupling DNA metabolism with mitosis. On the other hand, Y19 phosphorylation is an essential part of a different checkpoint system used to delay nuclear division when bud formation (polarized growth) is prevented or delayed (331). In addition, Y19 phosphorylation may play a role in mitotic exit (see “Cdc20, Cdh1, and regulation of anaphase proteolysis”) (384, 605).
Swe1.
Despite monitoring a different cell cycle checkpoint, the proximal regulators of Cdc28 Y19 phosphorylation are homologous to those originally described in S. pombe. SWE1, which phosphorylates Y19, encodes a protein kinase homologous to S. pombe wee1 (47, 471). The action of Swe1 is cyclin specific, since it phosphorylates Cdc28-Clb2 but not Cdc28-Cln2 or Cdc28-Cln3 (47). The basis for this specificity has not been determined but may reside in a recognition site in Clb2, a conformational change in Cdc28 induced by Clb2 but not Cln cyclins, or the occlusion of a recognition site on Cdc28 by the Cln cyclins but not by Clb2.
Overexpression of SWE1 causes a premitotic cell cycle arrest that resembles the phenotypes that are generated by cdc28-Y19E. The SWE1 overexpression phenotypes are suppressed by CDC28-Y19F (341). The arrested cells have either a short mitotic spindle (47) or duplicated but unseparated spindle pole bodies (341), indicating that the Cln-Cdc28 complexes are not inhibited. DNA replication is unaffected by SWE1 overexpression (47), indicating either that the Clb5 or Clb6 complex with Cdc28 is not inhibited or not phosphorylated by Swe1 or that tyrosine-phosphorylated Cdc28 possesses sufficient activity to promote S phase but not mitosis. That Cdc28 phosphorylated on Y19 might retain kinase activity is suggested by the similar phenotypes of the cdc28-Y19E mutant.
Deletion of SWE1 generates a phenotype identical to that of cells carrying the CDC28-Y19F allele—suppression of the nuclear division delay caused by inhibition of bud formation (508)—but has no other overt effects (47). The effect on nuclear division delay is very sensitive to SWE1 gene copy number. SWE1/swe1 heterozygotes have a shorter delay than do SWE1/SWE1 homozygotes (508). This observation indicates that the periodic transcription of SWE1—the SWE1 promoter possesses an SCB box (see “Start-specific transcription”) and its transcription peaks at about the time of Start with kinetics similar to CLN2 and CLB5 (341, 359, 508)—might play an important functional role. Replacement of the normal SWE1 promoter with a weak constitutive promoter, however, has no effect on nuclear division kinetics in response to inhibited bud emergence (508), indicating that the biologically relevant regulation of Swe1 levels or activity for the budding checkpoint, if it occurs, is posttranscriptional.
Swe1 is negatively regulated by Hsl1 (359), a protein kinase related to the Cdr1/Nim1 protein kinase of S. pombe. Mutational loss of Hsl1 function produces a G2 delay that is suppressed by deletion of swe1 or by replacement of CDC28 with the CDC28-Y19F allele, consistent with Hsl1 being a negative regulator of Swe1. Another gene, Hsl7, was identified in the same screen with Hsl1 and has very similar genetic properties. Its function has not been further defined (359).
Mih1.
The action of Swe1 is opposed by Mih1 (469), the S. cerevisiae homolog of the protein phosphatase encoded by the S. pombe cdc25+ gene (470). Deletion of MIH1 either has no effect (508) or causes a slight acceleration of cell cycle progression (469) under normal growth conditions. When polarized cell growth is inhibited, however, the subsequent mitotic delay is exacerbated in a strain lacking MIH1 (508) and suppressed when MIH1 is overexpressed (331). As with SWE1, the mitotic delay response to bud emergence failure is very sensitive to the level of MIH1 expression, with a greater delay seen in mih1/MIH1 heterozygotes than in MIH1/MIH1 homozygotes (508). An mih1 deletion mutant is also supersensitive to expression of the S. pombe wee1+ gene (469), a sensitivity suppressed by the CDC28-Y19F allele (525). These phenotypes are consistent with the presumed role of Mih1 in the dephosphorylation of Cdc28 on Y19 in response to successful passage through a checkpoint regulating bud emergence. They have also been used to argue that the mitotic delay seen when bud emergence is inhibited cannot be due to inhibition of Mih1 activity (508) and have focused attention on Swe1 as the likely target of the bud emergence checkpoint pathway.
T18.
Very little has been reported on T18 phosphorylation in S. cerevisiae Cdc28. Amon et al. (9) detected weak phosphorylation at this position but did not find a phenotype associated with a CDC28-T18A,Y19F mutant. These studies were carried out before the connection between bud emergence and Y19 phosphorylation was recognized (331), however, and so a revisit of this issue may be in order. There is no data on protein kinases and phosphatases that regulate T18 phosphorylation of Cdc28.
Type 2A Phosphatases
Four genes—PPH21, PPH22, PPH3, and SIT4—encode the catalytic subunits of type 2A serine/threonine protein phosphatases in S. cerevisiae. SIT4 is an essential gene with a role in CLN transcription (175) discussed below (see “Phosphatase requirement for Start-specific transcription”). PPH21, PPH22, and PPH3 constitute an essential gene family (466). Strains temperature sensitive for Pph activity arrest in G2, an arrest that is suppressed by CLB2 overexpression (344). Deletion of mih1 potentiates the severity of pph21Δ pph22Δ PPH3 phenotypes, and this effect is suppressed by the CDC28-Y19F mutation. While this data suggests a role for Pph activity in the dephosphorylation of Cdc28 Y19, additional results indicate that Pph activity is needed for full activation of the Clb2-Cdc28Y19F mutant as well as the wild type. This indicates that the Pph phosphatases play a role in Clb2-Cdc28 activation that is unrelated to the phosphorylation on Y19, but as yet this role is undefined (344).
Further support for a role in Clb2-Cdc28 activation comes from an analysis of the regulatory subunits of the Pph phosphatases. Type 2A phosphatases typically have a heterotrimeric structure consisting of the catalytic (or C) components mentioned above and A and B regulatory subunits. Rts1 and Cdc55 are B subunits that are essential at high and low temperatures, respectively (167, 230, 505). Tpd3 is an A subunit that is essential at both high and low temperatures—it is viable but has a poor growth phenotype at 23°C—and is needed for binding of Rts1 (and probably Cdc55) to the catalytic subunit (592). Both rts1Δ and cdc55Δ have cell cycle progression defects that can be suppressed by CLB2 overexpression (506, 605) and, in the case of cdc55Δ, by expression of CDC28-Y19F (605).
TRANSCRIPTION
Four waves of cell cycle-specific transcription affecting Cdc28 activity are generally recognized. The best studied, by far, is the transcriptional activation of genes at Start (CLN1, CLN2, CLB5, and CLB6). Also receiving increasing attention is a wave occurring at the M/G1 border (CLN3, CDC6, SIC1, and FAR1). Less well studied are the waves occurring during S (CLB3 and CLB4) and in G2 (CLB1 and CLB2). As key mechanisms in the “clock” that regulates cell cycle events, it should be no surprise that the activities of the factors controlling each wave of transcription would be partially dependent, in a domino-like fashion, on Cdc28 activity generated in a previous transcriptional wave. Not all of the dominoes have been identified; virtually nothing is known about the factors regulating CLB3 and CLB4 transcription, for example. Completing the identification and the characterization of the interrelationships among these factors remains a major challenge in this field. Additionally, the falling of individual dominoes is not inevitable, since environmental influences and cell cycle checkpoints modulate each step through a variety of signal transduction pathways. Characterizing and quantifying these various inputs is also a critical task. In the following, the current state of knowledge of transcriptional regulation—as it pertains to control of Cdc28 activity—is summarized. Given the current status of knowledge and the dependence of one transcriptional wave on products generated during an earlier wave, it makes sense to begin this discussion with the G2-phase transcription.
G2-Phase Transcription
Genes that are important for Cdc28 regulation and that are coordinately expressed at G2 include CLB1 and CLB2 (181, 196, 460, 541), SWI5 (400) and ACE2 (133) (both transcription factors involved in the next transcriptional wave), and CDC5 (4) and CDC20 (503) (both implicated in the regulation of the anaphase proteolysis [see “Cdc20, Cdh1, and regulation of anaphase proteolysis”). SWI6 mRNA, encoding a transcription factor component involved in Start, also shows a modest peak of accumulation in G2 (54). The periodic transcription of CLB1, CLB2, SWI5, ACE2, and CDC5 results largely, if not entirely, from the activities of Mcm1 and SFF (Swi five factor) (4, 358, 362). Regulators of SWI6 and CDC20 transcription have not been studied. Key relationships are depicted in Fig. 4.
FIG. 4.
Relationships among genes transcribed in the G2 wave of cell cycle-dependent transcription and their regulators. Transcription factors are outlined with diamond-shaped boxes. Proteins with other activities have rectangular outlines. Other conventions are as in Fig. 2.
Mcm1.
Mcm1 is an essential, acidic transcription factor containing multiple polyglutamine stretches. It was originally identified genetically by the inability of mcm1 mutants to stably maintain yeast minichromosomes (364) and has since been shown to be involved in the transcriptional activation and repression of multiple genes involved in diverse processes. It is best known in the context of controlling mating-type-specific genes (6, 37, 150, 262, 280, 428), but its essential nature indicated the existence of other functional roles, particularly in the G2/M transition (4, 150). The diversity of its activities rests in its ability to interact with different coregulators at the promoters of the various genes that Mcm1 influences (136). Virtually all of the important functions of Mcm1, including DNA binding and protein-protein interactions, are contained within residues 17 to 97 (65, 90, 447), but CLB1 expression (and not CLB2 or SWI5 expression) also depends upon residues in the Mcm1 C terminus (4). The amino-terminal domain, known as the MADS box, is similar to that of other transcriptional activators, including the mammalian serum response factor (574), which all bind a similar DNA element, CCw6GG (where w is A or T) called the Mcm1 cell cycle element (MCE). By analyzing random DNA fragments bound by Mcm1, Wynne and Treisman (619) defined the more degenerate MCE listed in Table 3.
TABLE 3.
Transcription factors important for synthesis of CDK regulators
| Transcription factor | DNA element recognized | Recognition motifa | Target genes | Reference(s) |
|---|---|---|---|---|
| SBF (Swi4-Swi6) | SCB | mrCGAAA | CLN1, CLN2, CDC6?, SWE1? | 17, 53, 110, 130, 301, 353, 397, 399, 414, 427, 532 |
| MBF (Mbp1-Swi6) | MCB | ACGCGT | CLB5, CLB6, SWI4, CDC6?, SWE1? | 186, 204, 301, 354, 374, 487 |
| Xbp1 | GcCTGArGmgr | CLN1, CLN3? | 363 | |
| Mcm1 | MCE | dCCywwwnnrG | ACE2, CDC5, CLB1, CLB2, SWI5, CDC20?, SWI6? | 4, 312, 358, 362, 619 |
| Mcm1 | ECB | TTwCCCnnwnAGGAAA | CDC6, CLN3, FAR1, SWI4, | 373, 413 |
| SFF | SFRE | AnGTmAACAA | CLB1, CLB2, SWI5, ACE2?, CDC5?, CDC20?, SWI6? | 4, 358, 362 |
| Swi5, Ace2 | tGCTGGT | CDC6, RME1, SIC1, | 44, 134, 298, 438, 567, 574 | |
| Ste12 | PRE | ATGAAACA | FAR1, CLN2?, CLN3? | 83, 308, 375, 396, 413, 575, 591, 617 |
m is C or A; n is any base; r is A or G; w is A or T; y is T or C. Lowercases letters indicate positions where there is not a stringent requirement for a particular base.
SFF.
MCEs are found in the promoters of CLB1, CLB2, and SWI5 (4, 312, 358, 362), and each of these genes, as well as ACE2, requires Mcm1 for its expression. Mutational analysis of the promoter region of SWI5 indicate that nucleotides flanking but outside the MCE are necessary for SWI5 transcription (358). Furthermore, mutations in this flanking sequence still permit it to be bound by Mcm1, but higher-molecular-weight complexes seen in band shift experiments are eliminated. On the basis of these results, Lydall et al. postulated the existence of SFF as a necessary cofactor for Mcm1-dependent transcriptional activation of SWI5 (358). Additional putative SFF binding sites (SFREs [see Table 3 for a consensus]) have been found in the CLB2 and CLB1 promoters (4, 362), and, for CLB2 at least, the MCE/SFRE motif is required for gene expression. SFF still awaits genetic identification, but it is known not to be Ste12, Matα1, or Matα2, cofactors required for Mcm1 regulation of genes specifying mating type (358).
Control of Mcm1-SFF activity.
Little is known about how Mcm1-SFF induces periodic transcription of the genes it regulates. As assayed by in vivo footprinting, the entire complex is bound to CLB2 and SWI5 promoter elements throughout the cell cycle (4), and so neither Mcm1 nor SFF is thought to be periodically expressed. Analysis of MCE and SFRE point mutations indicate that Mcm1 is required for SFF binding but SFF is not needed for Mcm1 binding to DNA (358), leading to a model in which Mcm1 recruits SFRE to the promoter and SFRE is responsible for the transcriptional activation. Consistent with this model, but not exclusive to it, fusion of the Mcm1 DNA binding domain to the strong transcriptional activation domain of the herpes simplex virus VP16 protein (Mcm1DBD-VP16TAD) results in constitutive SWI5, CLB1, and CLB2 expression (4). How G2-specific Mcm1-SFF activity would be generated is not clear, but Amon et al. have shown that the expression of CLB1, CLB2, and SWI5 is dependent upon Clb2-Cdc28 activity in a strain lacking functional Clb1, Clb3, and Clb4 (10). This indicates that these G2-expressed genes may be involved in a positive feedback loop, but it is not clear how the loop is activated. The best candidates for this role are Clb3-Cdc28 and Clb4-Cdc28, which are transcribed before CLB1 and CLB2 (181, 209, 460) but have not been tested for this function. Clb5-Cdc28 and Clb6-Cdc28 do not suffice (10). Mcm1 is an obvious target for Clb-Cdc28 phosphorylation—it has three potential CDK phosphorylation sites—and is known to be phosphorylated in vivo (313), but no evidence for functional CDK-specific phosphorylation has been described.
Two other proteins, Nap1 and Gin4, have been identified that may participate in the putative feedback loop activating CLB2 transcription. Nap1 was identified as a Clb2 binding protein by affinity chromatography and was also found to be a Cdc28-Clb2 substrate in vitro (281). Mutations in GIN4, which encodes a protein kinase, were found in a visual screen of mutagenized clb1Δ clb3Δ clb4Δ cells that had an elongated bud morphology (5). Subsequent analyses indicate that Nap1 binds to Gin4 and promotes Clb2-dependent phosphorylation and activation of the Gin4 protein kinase. Both Nap1 and Gin4 are needed to allow Cdc28-Clb2 to carry out a subset of its mitotic functions, particularly those dealing with bud morphogenesis but including the ability of Cdc28-Clb2 to amplify its own production (5, 282).
M/Early-G1-Specific Transcription
At about the time of telophase, the transcription of six genes important for CDK regulation during G1 phase—CLN3 (373), SWI4 (54, 175, 373), RME1 (an inhibitor of sporulation-specific transcription and activator of Start-specific transcription) (567), CDC6 (373, 438, 642), FAR1 (413), and SIC1 (137, 486)—is stimulated. CLN3, SWI4 and probably FAR1 are controlled by an Mcm1-containing factor that acts at a site called the early cell cycle box (ECB) (373). RME1 and SIC1 are controlled by a different pair of transcription factors, Swi5 and Ace2 (44, 298, 567, 571). CDC6 transcription is activated by both ECB- and Swi5-dependent mechanisms (and along with SWI4, a Swi6-dependent mechanism to be discussed below [see “Start-specific transcription”]) (373, 438).
The genes transcribed at this period of the cell cycle and the factors that control their expression form a major link in the chain of events that make up the cell cycle clock. Swi5 and Ace2 levels are determined, in part, during G2, as discussed above, and their transcriptional activities are stimulated by the reduction in mitotic Cdc28-cyclin activity that occurs in anaphase (see “Anaphase Proteolysis”). The transcription of G2-specific genes and of CLN3, SWI4, and RME1 is dependent upon Mcm1 activity, and, although the manner in which Mcm1 activity is controlled has not been deciphered, it is expected that a dependence on Cdc28-Clb activity will be found. It is also expected that transcriptional activity at this time of the cell cycle will be dependent upon external factors, since decisions relating to continued cycles of cell proliferation will have to be made. As is evident from the following discussion, progress in this area is in its early stages and complete regulatory pathways have not been delineated. Finally, as we shall see in the next section (“Start-specific transcription”), the gene products produced at this stage of the cell cycle have a decisive influence on the activity and timing of the next wave of transcription. Key relationships are summarized in Fig. 5.
FIG. 5.
Relationships among genes transcribed in the M/G1 wave of cell cycle-dependent transcription and their regulators. Conventions are as in Fig. 4.
ECB.
It has been widely reported that CLN3 transcription is invariant due to a relatively high basal level and small amplitude compared to genes like CLN1 and CLN2 (175, 249, 396, 411, 578, 617), but McInerny et al. (373) point out that the observed four- to fivefold variation in CLN3 mRNA levels, peaking in late M to early G1, may be significant since the rate of G1 progression and overall cell size are sensitive to less than twofold changes in CLN3 gene expression (107, 108, 396). This reevaluation of CLN3 transcriptional periodicity was prompted by studies of the transcription of SWI4 (373), a transcription factor important for CLN1 and CLN2 expression (399, 414). SWI4 mRNA shows a pronounced periodicity, varying 10-fold in abundance with a peak in early G1 (54, 175). Three MCB motifs (see “Start-specific transcription”), involved in late G1 transcriptional activation, are part of the SWI4 promoter. Their deletion reduces SWI4 transcription 10-fold, but the residual transcription is still periodic (varying 8-fold in amplitude) with a peak in early G1 (186). Most of the residual promoter activity maps to a sequence 3′ to the MCB elements that matches the Mcm1 consensus binding site and that can bind Mcm1 (373). Expression of the Mcm1DBD-VP16TAD chimera (see “Control of Mcm1-SFF activity”) results in constitutive expression of the SWI4 gene. Despite accounting for only a small fraction (<10%) of total SWI4 gene expression, this transcription is significant, since mutation of the Mcm1 binding site in the SWI4 promoter delays SWI4 mRNA production and concomitantly increases the mean cell size. Site-directed mutagenesis was used to define the boundaries of this element, indicating that residues outside of the Mcm1 consensus recognition site were also important for its function. Computer probing of the yeast genome with this extended sequence indicated the presence of similar elements in the promoters of CLN3, FAR1, and CDC6 (373). These genes, like SWI4, are transcribed in late M and early G1 (68, 375, 642), and DNA fragments containing the common sequence element from each of these promoters were able to confer late M/early G1 expression on a reporter gene (373). As observed for SWI4, constitutive expression of Mcm1DBD-VP16TAD leads to constitutive expression of CDC6, CLN3 (373), and FAR1 (413); FAR1 expression, at least, is dependent upon Mcm1 (413), supporting a role for Mcm1 in ECB-dependent regulation of multiple genes. Comparison of all these related sequence elements led to the designation of this sequence as the early cell cycle box (ECB) with the consensus listed in Table 3 (373).
ECB regulation.
As with the G2 expression discussed above, it is not understood how Mcm1 activity at ECB promoter elements generates periodic transcription or how that periodicity is confined to a cell cycle interval different from the timing conferred by the Mcm1-SFF complex. A logical mechanism would include activation by Clb1-Cdc28 or Clb2-Cdc28. There is weak evidence for this possibility: FAR1 transcription is reduced considerably in cells arrested at G2/M by shifting a clb1 clb2ts clb3 clb4 strain to the restrictive temperature (413), and CLB6 overexpression enhances the transcription of CDC6 (34). In neither case has it been shown that the effect of the Clb-Cdc28 kinase is through an Mcm1-dependent mechanism, and in the latter example, it is not clear that other Clbs can substitute for Clb6.
It is possible that an additional factor interacts with Mcm1 at the ECB. Most of the ECB-regulated genes are known to be controlled by other transcription factors, but there is little evidence to indicate that any of them work with Mcm1 to confer M/G1 timing. The best candidate, Swi6, is a transcription factor important for Start-specific transcription and is required for most but not all of the periodicity of SWI4 gene expression at Start (186). The SWI4 periodicity that remains in the swi6 mutant peaks 30 min after the normal late G1 peak of expression, and so Swi6 would seem to be required for M/G1 timing as well as for Start. Ste12 is needed for maximal FAR1 transcription (83), but the reduction in FAR1 expression seen in ste12 mutants occurs primarily in late G1 (10-fold reduction) and only modestly at M (2-fold reduction), indicating that Ste12 is not a required cofactor for ECB-directed gene expression (413). Swi5 is needed for CDC6 transcription (438), but there is no evidence for an interaction with Mcm1. There is also no indication of a cofactor needed for CLN3 periodicity, and McInerny et al. (373) have suggested that Mcm1 may be acting alone, since the ECB in the CLN3 promoter, which is sufficient for periodic transcription, is entirely protected by Mcm1 in in vitro binding experiments (312).
The transcriptional activity of Mcm1 is influenced posttranscriptionally by a number of factors relevant to CDK regulation. Chen and Tye showed that decreased glycolytic flux suppressed the plasmid loss phenotype of mcm1-1 mutations and increased the transcription of a reporter gene driven by an MCE element (86). Since mcm1-1 plasmid loss is due to the failure to adequately transcribe CDC6 (373), this suggests that rates of glycolysis could control the rates of transcription from ECBs, a result of obvious importance for controlling the rate of passage through early G1 in response to cellular metabolism. Unfortunately, decreased glycolysis has the opposite effect on transcription rates and plasmid stability in a wild-type MCM1 background (373). Altered cell cycle phase distributions (as a cause of altered Mcm1 activity) was not controlled for, and so the interpretation of these experiments is not straightforward. Mcm1 activity is also affected by Sln1, a transmembrane histidine kinase similar to the bacterial two-component sensors (420), that regulates a MAP kinase cascade involved in protecting the cell against hyperosmotic conditions (59, 361). Deletion of SLN1 severely reduces Mcm1 activity, while a hyperactivated allele increases Mcm1 activity (628). This effect on Mcm1 is independent of the downstream MAP kinase pathway, since deletion of the MAP kinase gene (HOG1) or of the gene encoding the upstream activator (SSK1) of the MAP kinase kinase kinase component (Ssk2 and Ssk22) had no effect on Sln1 regulation of Mcm1 activity (170, 628). It is not clear whether the Ypd1 intermediary in the Sln1-to-Ssk1 phosphorelay system is required for this effect on Mcm1 activity. Kuo et al. have reported that mcm1 mutants confer reduced or enhanced survival to high-salt conditions and that exposure to high salt induces a new Mcm1 isoform (probably a new phosphorylated species) (313). Anderson and Lopes found effects of carbon source regulation and SLN1 on MCM1-dependent transcription of the gene encoding phosphatidylinositol synthase (13). Another connection with carbon source regulation was found by Yu and Fassler (629), who showed that Mcm1 is negatively regulated by GAL11, a subcomponent of the “mediator of transcriptional regulation” component of the RNA polymerase holoenzyme (336), which has been associated with transcriptional regulation by carbon source. No direct connection between any of these influences on Mcm1 activity and transcriptional regulation of SWI4, CLN3, or CDC6 or G1 progression has yet been made. Nonetheless, these results outline a possible link between environmental and metabolic conditions through the transcriptional machinery that influences SWI4 and CLN3 levels in early G1 and ultimately the timing of Start.
After the peak of transcription caused by the ECB, SWI4 transcription continues to rise under the influence of Start-specific promoter elements (see “Start-specific transcription”), but CLN3 and CDC6 transcription decline to basal levels. The mechanism responsible for shutting off ECB activity has not been determined, although, for CLN3 at least, it appears to require the action of Swi4 and Swi6—either directly or through transcriptional induction of Start-specific genes (399). It should be pointed out that the relevance of ECB-controlled transcription of CLN3 to G1 control has not been demonstrated directly and remains controversial.
Swi5 and Ace2.
The RME1 and SIC1 genes do not possess recognizable ECBs or MCEs in their upstream regions. Their transcriptional periodicity has been ascribed to the action of Swi5 and Ace2 (44, 298, 567, 571), which are also required for M/G1-phase peak expression of CDC6 (438). Swi5 and Ace2 both possess two C2H2 and one C2HC zinc finger motif. They are 37% identical overall and 83% identical in the region of the zinc fingers (71). The residues predicted, on the basis of comparison to the crystal structure of the related Zif268-DNA complex (433), to make contact with DNA are identical (133). Not surprisingly, both recognize identical DNA sequence motifs (Table 3). Despite these and other similar modes of regulation, Swi5 and Ace2 function differently in vivo. For example, Ace2 but not Swi5 activates CTS1 transcription and Swi5 but not Ace2 activates HO transcription (133). This differential activity on these particular promoters is in large part due to interactions with different positive and negative cofactors that recognize neighboring promoter elements (134).
Differential behavior of Swi5 and Ace2 is also observed with regard to RME1 and SIC1 transcription. Expression of RME1 is reduced in a swi5Δ background but is more strongly affected by ace2Δ. Deletion of both ace2 and swi5 nearly abolishes RME1 expression (567). For SIC1, Swi5 activity is clearly predominant, accounting for 50% of total expression and most of its periodicity (like CLN3, SIC1 is transcribed at an appreciable basal level throughout the cell cycle) (298, 571), but Ace2 is responsible for the residual periodic activity. Even on the same promoter, these transcription factors have different physical behaviors. In vitro gel band shift experiments indicate that a single Swi5 molecule binds the SIC1 promoter while two molecules of Ace2 leave a larger footprint in the same region (298). These different behaviors affect the relative timing of the transcriptional peaks dependent on Swi5 versus Ace2. Toyn et al. noticed that SIC1 mRNA levels peaked earlier than those of the Ace2-driven CTS1 gene but that the residual SIC1 periodicity in a swi5Δ strain coincided with CTS1 expression (571), indicating that Swi5-driven transcription initiates earlier than Ace2-driven transcription. In cells synchronized in telophase by arrest due to overexpression of CLB2 or by shift to the restrictive temperature in a dbf2ts strain, SIC1 transcription was reduced fourfold in a swi5Δ strain but CTS1 was not expressed. A similar effect of swi5Δ on SIC1 expression in cells synchronized in mitosis by cdc15 arrest was observed by Knapp et al. (298).
The finding that SIC1 is expressed in a SWI5-dependent manner when the level of Clb2-Cdc28 kinase activity is high is somewhat surprising given the way that Swi5 is regulated. As discussed above (see “G2-phase transcription”), SWI5 is transcribed in an Mcm1-SFF-dependent fashion during S, G2, and M. Once produced, the Swi5 protein remains in the cytoplasm until mitosis, when it rapidly enters the nucleus, allowing Swi5-dependent transcription to initiate (398). Nuclear entry of Swi5 can be made constitutive by mutating three serines—S522, S646, and S664—to alanines, with partial nuclear localization seen in single and double mutants. These three serines fit the consensus for CDK phosphorylation, are phosphorylated by Cdc28 complexes in vitro, have reduced or no phosphorylation in vivo in a cdc28-4ts mutant at the restrictive temperature, and are dephosphorylated after release from mitotic arrest as Swi5 enters the nucleus. This, as well as the observation that Swi5 is cytoplasmic when Cdc28-Clb activity levels are high and nuclear when Cdc28-Clb levels are low, fits nicely with a model in which Cdc28-Clb phosphorylation of Swi5 prevents nuclear entry of Swi5 (386). Thus, it seemed surprising that SIC1 and a Swi4-Swi6-independent allele of HO (whose transcription is also dependent upon Swi5) can be transcribed in a Swi5-dependent fashion when Cdc28-Clb is active (298, 398, 571). Apparently, small amounts of Swi5 “leak” into the nucleus in sufficient quantities to have an effect on transcription, despite the exclusionary signal coming from Cdc28. This leakage can be detected with hyperstable alleles of Swi5 (386, 398, 554).
Nuclear leakage of Swi5 seems to be tightly controlled. In studies on the Swi4-Swi6-independent, Swi5-dependent HO construct, there was no detectable HO transcription in cells arrested in late telophase by growth of a cdc14ts mutant or of a cdc14ts cdc15ts double mutant at the restrictive temperature but there was substantial HO transcription at the cdc15ts arrest point (398). Both CDC14 and CDC15 have been implicated in the control of Clb degradation (see “Proteolysis”) (260, 500). Mutations in CDC15, in particular, allow a substantial but incomplete fraction of Cdc28-Clb activity to be degraded (260). Apparently this reduction in activity is sufficient to allow enough Swi5 to enter the nucleus to permit transcription. Ace2, whose transcription and nuclear localization behave similarly to those of Swi5 (133), is apparently not permitted to enter the nucleus until completion of telophase, explaining the discrepancy in the timing of SIC1 transcription driven by Swi5 and Ace2 (571). Toyn et al. have proposed that the leakage of Swi5 into the nucleus initiates a positive feedback loop that is important for telophase and cytokinesis (571). By this model, Sic1 produced by the limited amount of nuclear Swi5 can inhibit residual Cdc28-Clb complexes not destroyed by the APC (260) (see “Anaphase proteolysis”), further reducing Swi5 phosphorylation and increasing its nuclear localization. This might also allow Ace2 to enter the nucleus.
Start-Specific Transcription
Control of Cdc28 activity at Start is perhaps the most critical event in the life cycle of a yeast cell, since this determines whether the cell will commit itself to a round of mitotic division. Phase-specific transcription at this point in the cell cycle is a key event, critical for the production of major Cdc28 regulators that include Cln1, Cln2 (617), Clb5 (160, 310, 487), and Clb6 (310, 487). The transcription of other important Cdc28 regulators such as Cdc6 (68, 438, 641), Swi4 (54), and Swe1 (359) is also strongly influenced by events at Start. This expression is generally ascribed to the action of two related transcription factors, SBF and MBF, which are responsible for most of the periodic, late-G1-specific mRNA production in freely dividing cells. SBF and MBF clearly do not work alone, however, and the list of factors that influence or work in parallel with SBF and MBF is growing. The roles and relationships of many of these factors are poorly defined. As the key commitment step of the mitotic cycle, transcription at Start is also affected by many environmental influences, which bring in many more regulators (summarized in Fig. 6). In this section, the factors involved in Start-specific transcription and the controls over their activity in dividing cells are discussed. Environmental events that alter this regulation are discussed separately (see “Perturbations to the normal cell cycle”).
FIG. 6.
Relationships among genes transcribed at Start and their regulators. Conventions are as in Fig. 4.
SBF.
SBF, the dominant factor controlling the expression of CLN1 and CLN2 (399, 414), was originally discovered as the factor controlling Start-specific transcription of HO (the mating-type switching endonuclease). Promoter deletion studies had identified eight copies of a CACGAAAA consensus sequence in the HO upstream region, which conferred late-G1-specific transcription to HO or to a reporter (397). Mutants defective in 10 SWI genes required for normal HO expression (55, 211, 529) were tested for their effects on transcription driven from, and transcription factor binding to, the CACGAAAA element. Only two, SWI4 and SWI6, whose gene products bind as a complex (18, 446, 509), were found to be specifically required (15, 55). The CACGAAAA element is known as the SCB (Swi4-Swi6-dependent cell cycle box [formerly the CCB]), and the Swi4-Swi6 complex is known as the SBF (SCB binding factor [formerly the CCBF]). Detailed site-specific mutagenesis studies, coupled with the discovery of additional SBF-regulated genes, have resulted in the more degenerate consensus listed in Table 3 (17, 427). The DNA sequence specificity of SBF is somewhat plastic, however, since binding of SBF to DNA sequences lacking a recognizable SCB has been reported (547).
MBF.
The existence of MBF, the dominant factor controlling the expression of CLB5 and CLB6, was first suspected from studies on late-G1-specific transcription of genes needed for DNA replication (204, 354, 374, 419, 611). The sequence ACGCGT, usually in multiple copies, was found in the upstream region of these genes and shown to be able to confer Start-specific transcription to normally aperiodic reporters (204, 354, 374). This sequence is an MluI restriction site, hence the name MluI cell cycle box (MCB). The transcription factor that binds is called MCB binding factor (MBF [also DSC1]). MBF binding to MCBs and the periodic expression conferred by MCBs is abolished in swi6 but not swi4 mutants (130, 353, 597), indicating that Swi6 is a component of MBF. This was confirmed when Koch et al. (301) purified MBF and found that it consisted of Swi6 and a 120-kDa protein. The gene encoding the 120-kDa protein, MBP1, was cloned on the basis of its homology to SWI4 (30% overall identity at the amino acid level) and a partial peptide sequence (301).
Swi4, Mbp1, and Swi6 structure.
Although both Swi4 and Mbp1 form complexes with Swi6, they are capable of binding specific DNA sequences on their own (18, 301, 446, 509), but in neither case has the binding been shown to be due to the full-length protein and not a C-terminal truncation (52). Swi4 and Mbp1 possess similar DNA binding domains in their N termini (16, 301, 446) that are related to real and potential transcription factors in other fungi, including cdc10, res1, and res2 of S. pombe, and may be distantly related to the E2F family in vertebrates (52). The X-ray crystal structure of the DNA binding domain of Mbp1 has been determined and consists of a six-stranded β-barrel packed against a bundle of four α-helices (553, 620). The structure is related to that of a number of helix-turn-helix DNA binding proteins, particularly the Escherichia coli catabolite activator protein, histone H5, and hepatocyte nuclear factor 3γ. Mbp1 is apparently modified in vivo, since it migrates differently on SDS-PAGE when it is prepared from yeast cells from when it is prepared from a reticulocyte lysate (301). The nature or function of this modification is not known.
Swi6 does not have a recognizable DNA binding domain and is incapable of binding DNA unless complexed with Swi4 or Mbp1 (223, 301, 446, 509). Complex formation between Swi6 and Swi4 or Mbp1 requires the C termini of both partners (18, 446, 509, 512). Swi6 also has a leucine zipper-like domain that promotes Swi4-Swi6 complex formation and binding to SCBs (509).
The most striking feature of this group of proteins is that each possesses four full and one partial “ankyrin repeats” in their central portion. The middle two and last partial repeat are less recognizable than the first and fourth repeats (16, 49, 52). Ankyrin repeats are generally 33 amino acid residues in length, highly degenerate, and found in hundreds of proteins (49). They were thought to mediate protein-protein interactions, a hypothesis substantiated by the solution of the X-ray crystal structure of the ankyrin repeat-containing human 53BP2 in complex with the p53 tumor suppressor (205). Nuclear magnetic resonance spectroscopy (276) and crystal (594) structures of the ankyrin repeat domains of mammalian CKIs are similar to that of 53BP2, but co-crystal structures with CDKs have not yet been reported. The ankyrin repeats in Swi4 and Swi6 are not needed for complex formation (18), but the repeats in Swi6 are important for the SCB binding activity of SBF, perhaps suggesting a more subtle role in proper subunit alignment (509). The ankyrin repeat domain of Swi4, whether it is part of the native protein or an isolated fragment, binds Cdc28-Clb2 complexes (512), an activity with potentially important regulatory consequences for both Swi4 and Cdc28-Clb2 (it is not known whether Swi4 acts as a CKI). Swi4 is phosphorylated in the bound complex (10). The ankyrin repeat domain in Mbp1 also binds Cdc28-Clb2 complexes but only when the C terminus of Mbp1 is removed (512). Whether this reflects an in vivo interaction that could be regulated by conformational changes in Mbp1 remains to be determined. The Swi6 ankyrin repeat domain interacts only weakly with Cdc28-Clb2 complexes and only in the context of the full-length protein.
Swi4, Mbp1, and Swi6 genetic interactions.
Since Swi6 is a component of both SBF and MBF, one might expect that swi6 mutants would have the severest transcriptional defects, but this is not what is observed. Phenotypic analyses of cells possessing null and temperature-sensitive mutations in SWI4, SWI6, and MBP1 give the impression that Swi4 plays the dominant role in essential Start-specific transcription, with Swi6 and Mbp1 playing increasingly less important, subsidiary roles. Mutants with null mutations in swi4 are viable (15, 55, 529) but slow growing, with enlarged and misshapen cells that resemble cln1Δ cln2Δ double mutants. The slow-growth phenotype is exacerbated when combined with a cln3Δ mutation and is cured by heterologous expression of CLN2 (399, 414). In some strains, swi4Δ is temperature sensitive for growth, producing a cdc28-like arrest at the restrictive temperature, and homozygosity for swi4Δ is lethal in diploids (414). Mutants with null mutations in swi6 are also enlarged and misshapen, but temperature-sensitive growth has not been reported (56). Deletion of MBP1 produces no obvious growth defects—no delay in bud emergence or DNA synthesis—in haploids or homozygous diploids (301). Double mutants containing swi4 and swi6 or mbp1 arrest just prior to Start and are inviable (55, 301, 399). The mbp1Δ swi6Δ double mutant, however, is viable, with a phenotype resembling that of the swi6Δ single mutant (301). SWI4 overexpression suppresses the swi6 phenotypes (56), but SWI6 overexpression has no effect on swi4 mutants (16). Understanding the differences between the expected and observed phenotypes requires closer inspection of the functions of SBF and MBF and the roles that the individual components contribute to the overall function.
Swi4, Swi6, and Mbp1 mutant effects on transcription.
Appreciation of the complexity of Swi4, Swi6, and Mbp1 function has come from studies of the effects of null mutations on the transcription of individual genes. Despite their structural similarities and the presence of a common component, the roles of SBF and MBF in promoting periodic transcription appear to be quite different. SBF is required for full transcriptional activation and periodicity at the promoters it controls, while MBF often imparts periodicity to promoters that have a strong basal expression, acting as both an activator at Start and a repressor at other times. For example, expression of SCB-driven HO is almost entirely eliminated in swi4 backgrounds (54, 55). Strains lacking mbp1, however, produce normal amounts of mRNA from MCB-driven genes like TMP1 or POL1, but their periodic transcription is completely abolished (301). Like mbp1Δ strains, swi6Δ strains have normal levels of transcripts from some MCB-driven genes, like RNR1, TMP1, and POL1, but have lost the ability to regulate them in a cell-cycle-specific manner (130, 353, 597). mRNA levels from the MCB-driven CDC9 and from tandem synthetic MCBs, however, are substantially diminished in a swi6Δ background (130, 353, 597). Similarly, transcription from the SCB-driven HO promoter is reduced 100-fold by a swi6 mutation (55). These results indicate that Swi6 is required for periodic modulation of some promoters and for overall activation of transcription at others.
The separation between SBF-controlled SCBs and MBF-controlled MCBs is not absolute. Swi4 binds MCBs and Mbp1 binds SCBs in vitro (301, 427); SCBs compete for the factors that bind MCBs (130); and MCBs compete with SCBs for Swi4 binding (446). The transcriptional activity of a promoter driven by multiple synthetic MCBs in tandem is reduced two- to eightfold in a swi4 background (597). Deletion of a distal MCB element in the TMP1 promoter decreases expression fivefold whether the strain is mbp1 or wild type, indicating the activity of another transcription factor, possibly SBF, at this MCB (301). Not all MCBs are activated by SBF, however. Transcription of MCB-driven RNR1 is unaffected in a swi4 mutant (130), and although transcription complex formation on MCB elements in one study of the TMP1 promoter was reduced in a swi4 background, none of the complexes that formed in the SWI4+ background supershifted in gel retardation assays when anti-Swi4 antibodies were added (427). Others, however, have reported that protein complex formation on TMP1 MCBs or on synthetic MCBs was not affected in swi4 mutants but was abolished in mbp1Δ strains (130, 301, 353, 597). In any case, these results indicate that Start-specific transcription cannot readily be assigned to particular transcription factors on the basis of the presence of consensus promoter elements in an upstream region. For this reason, the effects of swi4, swi6, and mpb1 mutants on the transcription of CLN1, CLN2, CLB5, SWI4, and CDC6 are discussed individually. Little has been reported about CLB6 and SWE1 transcription beyond the timing of their peak transcription at Start, and so these genes are not discussed further.
(i) CLN1 transcription.
There are two near matches to the SCB consensus and three matches or near matches to the MCB consensus in the CLN1 upstream region (414), suggesting the possible involvement of both SBF and MBF in CLN1 gene expression. Consistent with this possibility, transcription of CLN1 mRNA levels is severely reduced in a swi4 background (399, 414) and is undetectable in a swi4 mbp1 double-mutant background (301) (Table 4). There is no effect on CLN1 mRNA levels in an mbp1 SWI4+ background (301), indicating that the contribution of MBF is probably negligible when SBF is functional. Contrary to expectations, loss of Swi6 activity, which should lead to loss of both SBF and MBF function, has a relatively modest effect on CLN1 transcript levels (130, 399). These results indicate that both Swi4 and Mbp1 can have appreciable transcriptional activity on the CLN1 promoter in the absence of Swi6.
TABLE 4.
Transcript levels of CLN1 and CLN2 mRNA in SBF and MBF component mutants
| Mutant background | Transcript level (% of wild-type level) ofa:
|
|
|---|---|---|
| CLN1 | CLN2 | |
| swi4 | 12 (399) | 20–30 (110, 399, 532) |
| mbp1 | 100 (301) | 100 (110, 301) |
| swi6 | 35 (399) | 53 (399) |
| swi4 mbp1 | NDb (301) | ND (301) |
| swi4 swi6 | 3 (399) | 8 (399) |
References are given in parentheses.
ND, not detected.
These genetic analyses are complicated by the ability of Swi4, Swi6, and Mbp1 to function in what may be unnatural ways when the activity of any one is altered or eliminated. This becomes even more of an issue when the question of how each factor contributes to the timing of transcription is approached. Mutants lacking Swi4 and Swi6 function, in particular, have altered cell cycle phase transitions and are difficult to synchronize. Genetic manipulations that are used to subvert these difficulties may introduce artifacts that prevent straightforward interpretations. With these considerations in mind, it appears that the cell cycle timing of CLN1 transcription cannot simply be assigned to any of the known SBF and MBF components. Near-normal timing of CLN1 mRNA periodicity has been observed in swi6 (53, 353; but see reference 130), swi4 (53), and mbp1 (301) mutants. The experiments with swi4 (53), in which a suppressor of the swi4 transcriptional defect and constitutive expression of SWI4 were used, show the lengths to which it is sometimes necessary to go to obtain a reasonably synchronous population of cells. A more complete understanding of how transcriptional periodicity is generated will probably require the development of an in vitro assay system to complement the genetics.
Close examination of the promoter elements responsible for CLN1 transcription led to an unexpected result. A DNA fragment carrying only the SCB-containing region is unable to promote the transcription of a reporter gene, but the MCB-containing sequence activates transcription efficiently with normal cell cycle kinetics (427). Transcription driven from the MCB element is sensitive to mutations in swi4 and swi6, but mbp1 mutations have only a minor effect. Furthermore, protein complexes bind the MCB element in extracts from mbp1 strains but not when made from swi4 or swi6 strains, and these complexes react with anti-Swi4 and anti-Swi6 antibodies. No complexes that bound the SCB element were found. Finally, point mutations in the MCB elements reduce transcription from the entire promoter fourfold. Additional point mutations that also eliminated the SCB reduce transcription another twofold (427). These results indicate that much of CLN1 transcription is dependent upon SBF acting on MCB elements, with minor participation, at best, by MBF and the SCBs. The residual transcription from promoter constructs lacking recognizable SCB and MCB elements, however, was still periodic with normal Start-specific expression (427).
(ii) CLN2 transcription.
There are three matches to the SCB consensus (110, 399, 414, 532) and three matches to the MCB “core,” two of which overlap the SCB consensus (532) in the CLN2 upstream region, once again indicating the possible involvement of both SBF and MBF in CLN2 gene expression. Analyses of mutations in SBF and MBF components (Table 4) lead to conclusions similar to those obtained with CLN1 with respect to effects on transcript levels (110, 130, 301, 399, 532) and cell cycle timing (110, 130, 301, 353, 386, 399, 532). Detailed analysis of the promoter region once again led to surprising results that differed from expectations and from what was seen with CLN1. Combinations of point mutations and deletions that eliminate the CLN2 SCBs and MCBs reduce expression to 8 to 25% of control levels, indicating that the bulk of CLN2 transcription was promoted from the SCBs with a minor contribution by the MCBs. The expression that remained, however, was still periodic with normal cell cycle timing. In a swi4 background, this remaining periodicity was eliminated, indicating that Swi4 was acting on sequences not recognizable as SCBs or MCBs (110, 532). Unlike SCBs or MCBs, this other element has appreciable promoter activity during a cdc28 arrest that increases further upon return to permissive conditions in a manner that is dependent upon new protein synthesis. This element is inactive during arrest at Start by pheromone treatment and is not active during heat shock (532). Confusingly, it has been reported to be inactive during Cln depletion (532) and to show dramatic increases in activity when Cln or Cdc28 activity is eliminated (533). Efforts to identify the sequence element conferring this periodicity have been unsuccessful (110, 532) but have revealed additional promoter complexities. The picture we seem to be left with is that transcriptional periodicity, involving Swi4 and possibly involving Mbp1, is conferred by the SCBs and MCBs, while Swi4 also acts in parallel on non-SCB, non-MCB-containing sequences.
(iii) CLB5 transcription.
Transcriptional control of CLB5 seems less complex, but it has not been as intensively studied as that of CLN1 and CLN2. CLB5 appears to be controlled solely by MBF, with no SBF involvement. There are five MCB-like sequences in the CLB5 upstream region and one SCB in the coding sequence. The SCB is probably not functional, but neither the MCBs nor the SCB has been tested in a mutational analysis. Mutations in swi4 have no effect on CLB5 mRNA levels or periodicity (160, 487). Mutants defective in swi6 lose CLB5 periodicity, as would be expected for MBF involvement, and Swi6 but not Swi4 was found in complexes that bind the region containing four of the five MCBs (487). No major change in CLB5 expression in cells from asynchronous cultures was observed in mbp1 mutants, but the periodicity of CLB5 in cells synchronized by centrifugal elutriation was entirely lost (301). Curiously, CLB5 periodicity is retained in mbp1 mutants when Cln deprivation and resynthesis, as opposed to elutriation, is used to synchronize the cells. This behavior is not seen with TMP1 and POL1 expression, which loses all periodicity in mbp1 backgrounds, no matter which synchrony protocol is used, and has yet to be explained. The factors responsible for the basal expression of CLB5 in mbp1 mutants have also not been identified. Swi4 does not seem to be involved, since the asynchronous levels of CLB5 mRNA do not change in swi4ts mbp1 double mutants at the restrictive temperature compared to mbp1 single mutants or to the wild type (301).
(iv) SWI4 transcription.
Transcription of SWI4 initiates at the M/G1 border, as discussed above, but the bulk of SWI4 gene expression is dependent upon three MCB elements, which are responsible for the late-G1 peak of SWI4 transcription (186). Loss of Swi6 activity has no effect on the overall level of SWI4 transcription from the wild-type promoter sequence, but the amplitude of the periodicity is greatly reduced, implicating a Swi6-containing complex, which has been assumed to be MBF, in the control of periodic SWI4 transcription. Since this work was done in a background in which SWI4 was overexpressed (to promote greater cell cycle synchrony in the swi6 background) and since recent data have shown that SBF can operate at MCB elements (427), it remains to be proven that MBF, and not SBF or some other factor, is normally active in SWI4 gene expression. The second of the three MCB elements in the SWI4 promoter is an exact match for the MCB consensus (the others are somewhat degenerate). Deletion of this central, “perfect” MCB has no effect on SWI4 promoter activity, but deletion of all three reduces transcription 10-fold. The remaining transcription is dependent upon the ECB, as described above (see “M/early-G1-specific transcription”). In a swi6 background, deletion of the central MCB actually increases promoter activity three- to fivefold. This could indicate the existence of negative regulators active at this promoter but could also have resulted from the inadvertent creation of a new, more powerful promoter element (186).
(v) CDC6 transcription.
By using different protocols to achieve cell cycle synchrony, different groups reported that CDC6 transcription peaks at different times, either at the M/G1 border (642), as discussed above (see M/early-G1-specific transcription), or at Start (68, 641). Piatti et al. investigated this discrepancy and found that CDC6 transcript levels peak at Start only in cells that have not recently transited mitosis (438). In rapidly dividing cells, CDC6 transcription peaks at M/G1. They rationalized this complex pattern of transcription on the basis of the instability of the Cdc6 protein and its involvement in promoting S-phase initiation. In rapidly dividing cells, CDC6 mRNA is produced at the M/G1 border, so that Cdc6 protein is available for initiation of DNA replication. In cells with a long G1 phase—such as small daughter cells or cells arrested in G0 by pheromone or starvation—Cdc6 levels decline, due to its instability, and must be replenished by renewed transcription at Start. Piatti et al. further showed that the Start transcription of CDC6 was dependent upon Swi6, since swi6 mutants had constitutively high levels of CDC6 mRNA, indicating a role for either SBF or MBF, but did not further characterize this transcriptional pattern (438).
The suppression of Start-specific transcription of CDC6 by prior M/G1-dependent transcription can be contrasted with the pattern of SWI4 transcription, which appears to be a superposition of M/G1 and Start-specific transcription. Transcription of the HO endonuclease gene, required for mating-type switching, provides an example of yet another variation. HO transcription requires both Swi5 and SBF (55, 211, 529), yet HO transcripts are seen only at Start (401). In this case, transcription at the M/G1 border is suppressed and a factor known to operate at M/G1 is required for Start-specific transcription. It is not clear how the transcription factors interrelate to produce these varied effects or which other gene products are needed to generate these transcriptional patterns.
Transcriptional control of SBF and MBF activity.
Part of the timing of SBF activity, during free-running cell cycles, results from the periodic transcription of SWI4 (373), but this plays only a minor role in controlling SBF. Constitutive production of Swi4 reduces the amplitude of HO, CLN1, and CLN2 mRNA periodicity—by increasing expression during the troughs without affecting the peak height—but does not eliminate it (53, 54). SWI6 mRNA abundance varies modestly during the cell cycle with a pattern similar to that of SWI5, but constitutive production of Swi6 has no effect on HO periodicity (54). Studies of MBP1 transcription as a function of cell cycle position have not been reported, but the MCB binding activity of MBF does not change during the cell cycle (130), indicating that periodic variation in MBP1 mRNA levels should have little importance for controlling Start-specific transcription.
Control of SBF and MBF activity by Cln-Cdc28.
The best-known controls exerted on SBF and MBF activity are by Cdc28 complexes. SCB- and MCB-containing promoters lose activity in cdc28ts strains at the restrictive temperature or when CLN gene products are depleted (55, 112, 131, 273, 399, 578). Expression of any one of the three CLN genes or of CLB5 can restore SCB- and MCB-driven gene expression to a strain lacking cln function—an effect that, for the CLN1 and CLN2 promoters at least, is dependent upon Swi4 and Swi6 (399, 487). These results led to a model of SBF activation incorporating a positive feedback loop. It was proposed that low levels of SBF activity led to the production of Cln1 and Cln2 (and Swi4), which activated Cdc28, leading to increased SBF activity and more Cln1, Cln2, and Swi4 production (112, 131, 399, 414). CLN1 and CLN2 transcriptional activation, however, can occur in the absence of de novo protein synthesis (366). In addition, careful measurements of the timing of Cln production showed that under the laboratory growth conditions used in these studies, this positive feedback loop does not have time to operate (129, 533). The currently accepted model of SBF activation at Start dispenses with the positive feedback loop and assumes that Cln3-Cdc28 is the only CDK involved in activating SBF in normal cycling cells. Of course, there may exist conditions (in a cln3Δ strain, to give an unnatural example, or high glucose concentrations [184]) when a positive feedback loop becomes operative.
As a stimulator of Start-specific transcription, Cln3-Cdc28 seems particularly well adapted. It is more effective than Cln1-Cdc28 or Cln2-Cdc28 complexes (112, 327, 578), despite having considerably lower specific activity as a protein kinase (using histone H1 as a substrate), and is less able to promote budding (327, 533, 578). How Cln3-Cdc28 carries out this function is not understood, however. Since it is a protein kinase, attention has focused on its potential to phosphorylate SBF and MBF components, particularly Swi6, but no conclusive evidence confirming this role has been presented. Both Swi4 and Swi6 are phosphoproteins in vivo (10, 511, 547), but the kinases responsible for their phosphorylation have not been identified. The only function ascribed to any of these phosphorylations is for that of S160 in Swi6, which is part of a nuclear localization signal and fits the consensus for a CDK substrate.
Dephosphorylation of phospho-S160 occurs in G1 and correlates with Swi6 nuclear localization; the phosphorylated form is found during S, G2, and M and correlates with a cytoplasmic localization of Swi6 (511, 547). Cdc28 complexes do not appear to be primarily responsible for this phosphorylation (511), however, unlike the situation described above for Swi5. In addition, the nuclear localization of Swi6 is not necessary for the proper timing of SBF-dependent gene expression, although it does promote peak transcript levels (511).
A transcriptionally inactive form of SBF is found bound to SCB elements early in G1 and becomes active at Start in a Cln3-Cdc28-dependent manner (223, 302, 547). SBF activation coincides with an altered mobility of the SBF-SCB complex, but neither the cause nor the function of the mobility shift is known. Both the inactive and active forms of SBF are sensitive to phosphatase, indicating a potential requirement for a protein kinase in allowing SBF to bind the SCB, but this gives no information about the need for phosphorylation in the transition to the transcriptionally active state (547). In studies on the SBF- and MBF-related Cdc10-Res1 transcription factor from S. pombe, Beach and coworkers demonstrated a requirement for Cdc2 (the Cdc28 homolog), Puc1 (a Cln3-like cyclin activator of Cdc2), and the Ran1 protein kinase in Cdc10-Res1 activation (77, 100). This acts through a transient association of Puc1 (probably in complex with Cdc2 and Ran1) with Cdc10 (the Swi4 homolog) followed by Cdc2- and Ran1-dependent phosphorylation of Cdc10, dissociation of Puc1, and association of Cdc10 with Res1 (77, 100). It is not clear how much of this model is conserved in S. cerevisiae, since the SBF complex already seems to be preformed and promoter bound prior to the requirement for Cln3-Cdc28 (223, 302, 547).
Repression of SBF activity by Clb-Cdc28 complexes.
Clb-Cdc28 complexes participate in the complementary aspect of SBF periodicity—turning off transcription in late G2 and M when SBF dissociates from the promoter by a process that is dependent on the activity of Clb2-Cdc28 (302). Clb3, Clb4, Clb5, and Clb6 complexes with Cdc28 have considerably less activity in repressing Cln transcription in the post-Start phase of the cell cycle (10). Clb1-Cdc28 complexes have not been directly tested in this regard. During G1, overexpression of CLB6 (but not CLB5 [other CLB genes have not been tested]) inhibits the expression of both SBF- and MBF-dependent genes, consistent with the acceleration of Start that is observed in clb6 mutants (34).
The repressive activity of Clb2-Cdc28 in G2/M is dominant to activation by Cln-Cdc28 complexes (10), a feature that explains the failure of artificially maintained high levels of Cln3-Cdc28 to sustain high-level expression of SBF-controlled transcripts after Start (578). This is part of a system that ensures that CLN1 and CLN2 expression is kept low until the Clb2- Cdc28 complexes are destroyed during mitosis. The mechanism by which Clb2-Cdc28 represses SBF is not known. In vitro SCBs can be bound by SBF isolated from cells at any stage of the cell cycle (547) but are occupied by SBF in vivo only in G1 and S (302), indicating that biochemical events associated with repression are not readily maintained after cell lysis. A large fraction of cellular Swi4 is found in association with Clb2-Cdc28; therefore, repression may occur by a direct sequestration mechanism (10). Dissociation of these complexes in vitro may explain the ability to detect DNA binding activity from all cell cycle stages. Immunoprecipitated Clb2-Cdc28-Swi4 complexes phosphorylate the Swi4 component in vitro, but the in vivo relevance of this phosphorylation is not known. As noted above, activation of SBF coincides with altered mobility of SBF-SCB complexes in gel shift assays. This altered mobility persists well after SBF shutoff and could conceivably reflect a mechanism by which SBF activity is turned off soon after it is activated (547).
In summary, periodic transcription by SBF is controlled by its passage through three distinct states as the cell cycle progresses. From telophase to Start, it is bound to the promoter but is transcriptionally inactive. In a process requiring Cln-Cdc28 activity, especially Cln3-Cdc28, SBF is converted at Start into an active transcription factor. Finally, from G2 to M, SBF leaves the promoter in a Clb-Cdc28-dependent (particularly Clb2-Cdc28) fashion. Additional controls that affect peak transcription rates and fine-tune the timing of activation derive from cell cycle-regulated transcription of Swi4 and cell cycle-regulated nuclear localization of Swi6. Less is known about controls over MBF activity. MBF-driven promoters are not repressed by Clb-Cdc28 complexes in the post-Start phase of the cell cycle (10). This may provide a means for the cell to maintain the expression of genes needed for DNA synthesis or repair during an S-phase checkpoint when the initial stages of budding have already been completed and Cln-Cdc28 activity is no longer needed. During normal cell cycle progression, though, MBF activity is turned off with kinetics similar to that of SBF, but it is not known how this is accomplished. Contradictory reports on whether MBF DNA binding activity is constitutive or periodic (130, 354) have been published, and little has been reported beyond that.
Phosphatase requirement for Start-specific transcription.
SIT4 , the gene encoding the catalytic subunit of one of the yeast type 2A protein phosphatases (23), is required for transcription of Swi4- and SBF-dependent genes (175). Due to the pleiotropic nature of sit4 mutations (23, 190), the multitude and complexity of the feedback loops in this system, and the lack of a useful in vitro analytical system, the role of SIT4 in Start-specific transcription is not understood, even though the biochemical function of its gene product is known. The analyses to date have relied heavily on the phenotypes of multiple mutant constructions with sit4 and other genes which are also pleiotropic and whose functions are poorly understood. Depending upon the genetic background, sit4Δ mutations either arrest in G1 or have a slow-growing phenotype with an extended G1 phase (544). This difference in sit4Δ behavior has been traced to allelic variations in SSD1, which encodes an RNA binding protein (583) and has also turned up as a suppressor of mutations in many seemingly unrelated genes (see the list compiled at Proteome [190]). The ssd1-d alleles originally described by Sutton et al. (544) as conferring essentiality to SIT4 are impaired versions of the apparently fully functional SSD1-v allele. Loss of Ssd1 functionality potentiates the phenotypic severity of mutants defective in Cln1 and Cln2 expression, especially swi4 (113) and sit4 (175, 544), and is lethal in a cln1Δ cln2Δ background (113), but otherwise it causes rather subtle phenotypes. Overexpression of CLN2 allows DNA replication—but not budding—to initiate in sit4 ssd1 mutants (175). Surprisingly, PCL1 overexpression also suppresses the essential sit4ts ssd1 cell cycle defects (126) in a Pho85-dependent manner. These results are difficult to reconcile, given the current status of knowledge concerning Pcl1 and Cln function, particularly with respect to budding, but indicate that Sit4 plays an important, if ill-defined role in setting the level of SBF transcriptional activity at Start. They also indicate a complex interplay between Cln-Cdc28 and Pcl-Pho85 activities in early cell cycle events.
Sit4 levels remain steady during the cell cycle (544), but Sit4 physically associates with five other proteins, Sap155, Sap185, Sap190, Sap4 (Sap4 association has not been demonstrated but is strongly suggested), and Tap42, in separate complexes that are dependent upon cell cycle position (126, 357). Association with a Sap is required for Sit4 function, and overexpression of any SAP gene suppresses sit4ts mutations but not a sit4 deletion, suggesting that the Saps are positive regulators of Sit4 activity or are the downstream effectors of Sit4 function (357). Single sap knockouts have distinct, mild phenotypes relating to G1 progression, suggesting some specialization but with considerable overlap. Combinations of multiple sap deletions cause more severe phenotypes, the most severe of which resemble the sit4 knockouts (357). Sit4-Sap complexes are not found in G1-phase cells but appear at about the time of the G1/S transition, suggestive of a role for a Sit4-catalyzed dephosphorylation event in Start-specific transcription (544). The basis for G1/S timing of Sit4-Sap association or the identity of potential Sit4 substrates is not known.
Tap42 plays a role analogous to the Saps but is also found associated with other yeast type 2A protein phosphatases (Pph21 and Pph22) (126). The association of Tap42 with Sit4 is sensitive to the nutritional status of the medium. Sit4-Tap42 complexes are present in greatest abundance during growth in rich media and are present in reduced amounts in nutritionally limited media. TAP42 is essential for G1 progression. In high copy number, TAP42 suppresses temperature-sensitive mutations in sit4, pph21, and pph22, indicating that it is a positive regulator of type 2A phosphatase function. Di Como and Arndt have described indirect evidence suggesting that Sit4-Tap42 may be involved in the control of CLN3 translation (see “Translation”) (126).
Sis2 has also been implicated in Start-specific transcription on the basis of genetic interactions between overexpression and deletion alleles of SIS2 with mutations in SIT4 and SWI4 (127). A sis2Δ strain is salt sensitive (176) but has no other obvious phenotypes. The sis2Δ sit4Δ double mutant, however, is inviable (127). SIS2 overexpression suppresses the growth defects due to very low expression or activity of Sit4 but cannot suppress a sit4Δ mutation. Overexpression of SIS2 in a swi4Δ strain is toxic, but the toxicity is relieved by heterologous expression of CLN2. These results suggest that Sis2 stimulates Swi4-dependent transcription and depresses Swi4-independent transcription. The SIS2 gene product has a highly acidic domain (residues 496 to 553 are 88% Glu or Asp), which is required for the sit4 suppression activity of Sis2. Di Como et al. have argued that Sis2 may be involved in chromatin interactions on the basis of the nuclear localization of Sis2 (which conflicts with a cytoplasmic location found by Ferrando et al. [176]) and sensitivity of cells with reduced histone expression to Sis2 overproduction (127). There are two other ORFs in yeast (YOR54c and YKL088w) with substantial sequence homology to SIS2 (190), but there are no reports of their mutant phenotypes or genetic interactions.
Other factors.
(i) Bck2.
While Cln3 has received the greatest attention as an upstream activator of SBF and MBF, it is clear that there must be a parallel mechanism, since cln3Δ strains have normal growth rates (107, 396). A likely candidate for a component of this parallel pathway is BCK2, whose deletion and overexpression phenotypes have many similarities to that of CLN3. The double bck2Δ cln3Δ knockout is lethal or very slow growing depending upon the background, and this poor growth phenotype is suppressed by expression of CLN1 or CLN2 from heterologous reporters (128, 161). BCK2 overexpression increases the expression of CLN1, CLN2, PCL1, and CLB5 in a Swi4- and Swi6-independent fashion (but even greater induction is seen in SWI4+ SWI6+ backgrounds) (128) and is the likely explanation for the ability of high-copy-number BCK2 plasmids to suppress a cln1 cln2 cln3 triple mutant (161). Curiously, CLB2 transcription is also stimulated by BCK2 overexpression (128). Di Como et al. have suggested that Sit4 may act in the same pathway with Bck2 (128). cln3 sit4 double mutations cause defects that are more severe than those of sit4 or cln3 single mutations (175), but bck2 sit4 double-mutant defects are no more deleterious than either of the single-mutant defects (128). sit4 mutants clearly have more severe phenotypes relative to Start-specific transcription than either bck2Δ or cln3Δ mutants (128, 175, 544), suggesting that Sit4 may function in both pathways or at a point common to both pathways.
(ii) Skn7.
Overexpression of SKN7 suppresses the lethality of a swi4ts swi6Δ mbp1Δ triple mutant by restoring the expression of CLN1 and CLN2 (389). For CLN2, this restoration is dependent upon promoter MCB and SCB elements. Skn7 does not bind MCB and SCB elements directly, however, and may act through another factor. Deletion of skn7 potentiates the temperature sensitivity of a swi4ts Δswi6 strain. These results indicate that Skn7 plays a role independent of SBF in the promotion of Start-specific transcription and may act under conditions when SBF activity is compromised. It apparently acts in competition with SBF, since CLN1 and CLN2 expression is enhanced in an SKN7 overproducer when SBF activity is compromised. Skn7 is homologous to the receiver domain of bacterial “two-component” signal transduction systems (62, 63). Structurally, Skn7 consists of a heat shock factor (HSF) domain, a coiled-coil domain, a region homologous to bacterial response regulators, and a Gln-rich domain (63, 389). Skn7 seems to play no role in the heat shock response, but skn7Δ mutants are sensitive to oxidative conditions. It is not known whether CLN expression is influenced by Skn7 under these conditions (307, 388).
(iii) Rme1.
Another factor that stimulates CLN2 expression is Rme1, which is best known for its role as a repressor of IME1 transcription, the transcriptional activator required for meiosis and sporulation (385). Toone et al. found that RME1 overexpression increases CLN2 but not CLN1 transcription and can suppress the temperature sensitivity of a swi6Δ swi4ts strain in a fashion that requires CLN2 (567). Deletion of rme1 potentiates the severity of SBF mutants and causes a 30% decrease in CLN2 expression but has no effect on CLN1 expression. The CLN2 promoter has two matches to the Rme1 recognition element RRE motif (the Rme1 binding site [104]) near the MCB/SCB region. Rme1 binds this region in vitro but is unable to act at the MCB or SCB elements themselves. As mentioned above, RME1 is periodically expressed in the cell cycle with an M/G1 peak (567). In addition, RME1 is repressed by the Mata1-Matα1 repressor and is therefore not expressed in diploids, which may explain why swi4Δ mutations are lethal in diploids (414).
(iv) Taf145, Taf90, and Tsm1.
Taf145 is one of many components of TFIID, the TATA box binding general transcription factor needed for the expression of many activator-directed promoters (reviewed in reference 70). Mutations in Taf145, but not in other TFIID components, specifically arrest cell cycle progression in G1 (602). The transcription of CLN1, CLN2, CLB5, and CLB6, but not CLN3 or the bulk of cellular transcription, is lost in taf145ts mutants at the restrictive temperature, indicating a relatively specific effect of Taf145 on Start-specific gene expression. The mechanism by which Taf145 is involved is unknown. Walker et al. argue that the action of Taf145 is direct, based primarily on the rapid rate (less than 30 min) at which Start-specific transcripts are lost after the shift of a taf145ts strain to the restrictive temperature and on the effect of Taf145ts inactivation in strains arrested in G1 (603). The upstream activation site (UAS) of CLN2 and CLB5 can convey Taf145 dependence to a heterologous reporter, but it has not been established whether a specific DNA sequence element or a particular transcription factor component bound to the promoter is recognized by Taf145 (603). Two other components of TFIID, Taf90 and Tsm1, are required for G2/M progression. The mechanism for this is completely unknown, but it does not seem to involve decreases in the transcription of CLB2 (19, 603).
(v) Zds1 and Zds2.
Zds1 and Zds2 have been implicated in SBF repression during G2, since deletion of zds1 leads to increased expression of SWE1 and CLN2 in G2 and delayed expression at Start (359). CLB5 expression is relatively mildly affected by zds1, so the effect of Zds1 seems specific to the two SBF-driven genes tested. The function of ZDS2 was not examined in this regard, but Zds1 and Zds2 are 48% identical and a zds1Δ zds2Δ double mutation has much more severe effects than either single mutation, indicating that the two genes have a common function. Mutations in ZDS1 and ZDS2 are highly pleiotropic, a result that is responsible for its acronym (zillions of different screens [see references 41 and 631 for a listing]). The Zds1 protein interacts with the Cdc42 GTPase-activating protein and has been localized to presumptive bud sites (41), suggesting the possibility that Zds1 and Zds2 control Start-specific transcription in response to bud morphogenesis.
Cell size control.
Most cells possess an intrinsic cell size characteristic of the species. The maintenance of a specific size requires the coordination of cell growth with cell division. For S. cerevisiae, much of this coordination is exerted at Start (for a review, see reference 448). Cln3 is assumed to convey cell size information to the cell cycle machinery through its effect on SBF (82, 107, 109, 112, 129, 131, 396, 399, 537, 578). This is based primarily on the correspondence between Cln3 dosage and cell size—increased Cln3 dosage (through multiple gene copies, fusion to stronger promoters, or alleles that produce a hyperstable gene product) leads to small cell size at Start, and decreased dosage leads to increased cell size (82, 107, 109, 396, 537). There has been no demonstration of an effect of cell size on the amount, activity, or concentration of Cln3, however; consequently, no mechanism linking cell size to Start activity has been delineated. The critical factor may reside in difficult-to-measure quantities such as the ratio of Cln3-Cdc28 activity to DNA, cell volume, or cell surface area. The possibility also remains that Cln3 conveys only a basal signal to SBF and MBF that is modified in a Cln3-independent manner by as yet unrecognized cell size signals. In this view, mutations that increase Cln3 activity would increase the basal rate and act to decrease the amount of signal needed from the independent source for Start initiation.
Cell size is regulated by nutritional conditions. When grown on poor nutritional sources, yeast cells are smaller on average than when grown on rich media (270, 350, 380). The size increase at which budding initiates when cells are shifted to glucose has been particularly well studied. This shift causes a delay in the expression of multiple Start-expressed genes, including CLN1, CLN2, and SWI4 (184), and a specific repression of CLN1 transcription (184, 565). The repression of CLN1 transcripts is responsible for much but not all of the cell size increase at budding (184). This repression requires SBF and is mediated through a region of the promoter containing the MCB elements. CLN2 transcription, which is controlled by SBF acting at the SCB and MCB elements, is not normally repressed by glucose, but a derivative in which the SCBs were eliminated (but the MCBs were retained) became glucose repressible (184). These results indicate that when SBF activates transcription through MCB elements, it can be regulated quite differently (gaining glucose repression) from when it acts through SCB elements.
The mechanism by which cell size regulation of CLN transcription occurs has not been defined, but the Ras-activated cAMP-dependent protein kinase pathway, which communicates the nutritional status of the growth media to the cell and is essential for growth (see references 495, 558, and 616 for reviews), has been implicated. High levels of cAMP delay the time of passage through Start (32) and severely inhibit the transcription of Start-related transcripts, including CLN1, CLN2, CLB5, and SWI4 (32, 565). (The differences in effect between these studies and the studies with glucose [184, 565]) may just be a matter of degree: CLN1 transcription was most severely affected by exogenous cAMP addition.) CLN3 mRNA levels rise in response to added cAMP (32, 565), indicating that the effect of glucose and cAMP on Start transcription must be mediated by controls that act independently of CLN3 transcription. There is a complex relationship between cAMP and Start-specific transcription. The delay in passage through Start caused by increased cAMP is seen only in small cells—there is no effect in cells large enough to initiate budding (32). Furthermore, cAMP is actually required for Cln3-stimulated transcription of CLN1 and CLN2 in larger cells (254). It is not known how cAMP mediates these varied effects.
POSTTRANSCRIPTIONAL PROCESSING
RNA Processing
CLN3 mRNA processing.
Sugimoto et al. isolated NAB3 as a high-copy-number suppressor of the mating defect conferred by the hyperstable CLN3-2 (538). Nab3 had previously been identified as an essential nuclear protein that binds poly (A)-containing RNA in vivo (613). Slow depletion of Nab3 leads to the accumulation of unspliced pre-mRNAs, suggesting that Nab3 is required for an early step in mRNA processing. The Nab3 protein has a domain (residues 327 to 400) conserved in other RNA and single-stranded-DNA binding proteins. In addition, there is a highly acidic domain (residues 36 to 158 are 17% Asp and 41% Glu) and a Gln-Pro-rich domain (residues 568 to 785 are 28% Gln and 20% Pro) (538, 613). Overexpression of NAB3 reduces the accumulation of CLN3 transcripts but has only modest effects on the levels of CLN1, CLN2, and ACT1 (an intron-containing mRNA that encodes actin) transcripts. NAB3 overexpression also retards cell cycle progression in cln1 cln2 CLN3 strains but has no effect on cln1 CLN2 cln3 or CLN1 cln2 cln3 strains. By swapping promoter sequences, Sugimoto et al. showed that the NAB3 overexpression exerted its effects on the CLN3 coding region or 3′-flanking sequences and not on the CLN3 promoter (538). There is no evidence that the CLN3 mRNA is spliced, so the role of NAB3 in CLN3 function is still undefined.
Late-G1 transcripts.
As discussed above (see “Phosphatase requirement for Start-specific transcription”), impaired function of Ssd1 RNA binding protein potentiates the phenotypic severity of mutants defective in CLN1 and CLN2 expression, especially swi4 (113) and sit4 (175, 544). Toone et al. (567) have found that temperature-sensitive mutations in RAT1, which encodes a 5′,3′-exo-RNase required for efficient nucleocytoplasmic RNA trafficking, are lethal in a swi4 mutant background and that moderate overexpression of SWI4, CLN2, PCL1, PCL2, and RME1 suppresses the temperature sensitivity of rat1ts alleles. These results indicate a specific but as yet uncharacterized defect in the processing of mRNAs required for Start.
Maturation of CLB5 mRNA?
Dahmann et al. identified mutations in SIM1 in a hunt for mutants that would rereplicate their DNA without undergoing an intervening mitosis (114). SIM1 is required for the maintenance of Clb5-Cdc28 activity, but sim1 alleles do not affect CLB5 mRNA levels. The C terminus of Sim1 is 66% identical to Nca3 and 67% identical to the product of an uncharacterized ORF, YKR042w. NCA3 is involved in the maturation of mitochondrial transcripts (434), raising the possibility that SIM1 plays a role in CLB5 mRNA processing.
Translation
Nutritional deprivation slows or arrests passage through G1 while having little effect on other cell cycle phases (reviewed in references 448). One means of transmitting starvation signals is through the protein synthetic apparatus. Such a mechanism seems logical, since translation places heavy demands upon the cell’s material and energetic resources and is absolutely required for passage through G1 and G2 (69). G1 is particularly sensitive to treatments which limit translation rates (448), a result which has long been interpreted to imply that steps leading up to S-phase initiation are dependent upon the accumulation of a critical, labile protein (499). Lodish has suggested that mRNAs which are inefficiently translated are disproportionately affected by treatments that decrease the concentration of functional translation initiation complexes (346), adding a means of enhancing the relationship between levels of a critical protein and translation rates. The CLN genes are obvious candidates for Pardee’s critical, labile protein (see “Start proteolysis”), but Cln2 (and Cln1?) is stabilized by cycloheximide treatment (366), leaving Cln3 as the most likely target of a translationally based regulatory system controlling G1 passage. Recent results have provided strong support that Cln3 is the critical G1 target by showing that Cln3 levels are particularly sensitive to translation initiation rates by Lodish’s mechanism.
Gallego et al. found that Cln3 protein levels decrease sharply in the first two hours following nitrogen starvation (189). This decrease is due in large part to an eightfold decrease in the rate at which CLN3 mRNA is translated. During the same time interval, bulk cellular protein synthesis decreases only twofold, indicating that CLN3 translation is particularly sensitive to the loss of nitrogen. Translational repression requires the 5′ untranslated region of the CLN3 mRNA (189). This region was studied in greater detail by Polymenis and Schmidt who found that CLN3 transcription initiated 364 bp prior to the CLN3 start codon, a long leader sequence for S. cerevisiae. Within this 5′ untranslated region is a 3-amino-acid ORF (starting at −315 relative to the CLN3 ATG). Mutating the ATG of this upstream ORF to a noninitiating TTG had a strong negative effect on CLN3 translation when cells were deprived of a good source of nitrogen or carbon. Interestingly, the upstream ORF had a mild positive effect on CLN3 translation when cells were grown in rich medium at low cell densities (442). Unlike the well-studied control of GCN4 translation by upstream ORFs (241), translation of the CLN3 ORF does not seem to occur by reinitiation but probably involves a leaky scanning mechanism in which the ATG of the upstream ORF is occasionally bypassed, allowing initiation at the CLN3 ATG (442). In any event, the efficiency of CLN3 translation was not affected by 3-aminotriazole or amino acid starvation (conditions that increase GCN4 translation efficiency), indicating that CLN3 translational efficiency is not regulated by the same system that controls GCN4 translation.
Barbet et al. (29) have suggested that a pathway involving the putative phosphatidylinositol kinases, Tor1 and Tor2, is involved in CLN3 translational regulation. Inhibition of Tor1 and Tor2, by mutation or by the immunosuppressant rapamycin, generates a G0-like response (76, 233, 234, 311, 527, 639) and reduces the translation rate of bulk yeast proteins by 90% (29). The G0-like response is prevented when Cln3 levels are maintained by expression of hyperstabilized CLN3 alleles or expression of a CLN3 mRNA possessing the 5′ untranslated region from UBI4 (translation of the polyubiquitin-encoding UBI4 mRNA is not inhibited by rapamycin or starvation [178, 423]) but not by simple overexpression of wild-type CLN3 (29). This pathway may involve the Tor1- and Tor2-stimulated association of Sit4 with Tap42, since this association is also inhibited by rapamycin, an effect not seen in strains carrying alleles of TOR1 or TOR2 that confer rapamycin resistance (126). The association of Sit4 with certain Tap42 mutant proteins, the growth of cells containing those TAP42 alleles, and the growth of cells overexpressing Sit4 are resistant to rapamycin treatment. As mentioned above, the Tap42-Sit4 complex is sensitive to the nutritional status of the medium, with complex levels high in rich media and reduced in nutritionally limited cells (see “Phosphatase requirement for Start-specific transcription”). The tap42 mutants have greatly reduced levels of polysomes but possess large numbers of monomeric ribosomes, consistent with a role for Tap42 in protein translation initiation. Adding to this pathway, Berset et al. have found that nutritional deprivation or interruption of the Tor-dependent signal transduction pathway induces the degradation of eIF4G, a factor required for initiation of translation of capped mRNAs (39). These results suggest that nutritional signals favoring growth are mediated by the Tor1 and Tor2 proteins, which act somehow to inhibit the degradation of eIF4G and increase the association of Tap42 with Sit4 which then, presumably, promotes the translation of CLN3 and the subsequent stimulation of Start-specific transcription as discussed earlier.
Gallego et al. (189) have argued against the importance of this pathway as a specific mechanism controlling the nutritional response on the basis that translational rates are severely reduced by rapamycin treatment whereas down-regulation of the Cln3 translation rate occurs when overall translation rates are still quite high. They also point out that cln3Δ strains still undergo arrest due to nutritional starvation, so that other factors must still be at work. The existence of additional mechanisms is also suggested by work on CDC63. CDC63 encodes eIF3η, a translation initiation factor that regulates the supply of 40S ribosomal subunits and their association with eIF-2–GTP–tRNAMet (219). Generalized protein synthesis is decreased in mutant cdc63 cells at the restrictive temperature, and monosomes accumulate, but the cells continue to enlarge and are mating competent, suggesting that the translation of specific Start mRNAs is particularly affected (57, 216, 217). The G1 arrest phenotype of cdc63 mutants is not suppressed by overexpression of hyperstabilized Cln3 (442). This is unlike the cell cycle arrest caused by rapamycin, which is suppressed by hyperstable Cln3 (29), and indicates that other mRNAs needed for Start are also affected. Furthermore, the mutation eliminating translation of the short ORF upstream of the gene encoding Cln3 does not prevent normal arrest and retention of high levels of viability upon entering stationary phase although it does suppress the G1 arrest caused by rapamycin (cells were still rapamycin sensitive but arrested in a manner that was not cell cycle specific).
Protein Folding
Comparison of the crystal structure of the human Cdk2-cyclin A complex with the structures of the free components indicates that Cdk2 undergoes large structural changes to form the heterodimer with its cyclin activator (64, 118, 263). It is likely that Cdc28 also undergoes similar structural rearrangements. In vivo, formation of at least some Cdc28-cyclin complexes does not appear to be spontaneous but requires one or more assembly factors. One of these factors appears to be Cdc37, encoded by a gene that was originally identified in a screen for mutants that displayed a Start arrest phenotype similar to that of Cdc28 (456). Cdc37 is a highly conserved protein that is associated with many different protein kinases in diverse species. It acts as a molecular chaperone, capable of binding and stabilizing proteins—especially protein kinases—in partially folded or mature but unstable states (290). Consistent with this role, cdc37ts mutants have reduced levels of Cdc28-Cln2 and Cdc28-Clb2 complexes at the restrictive temperature in vivo (194). Furthermore, Cdc37 is apparently required for the activation of Cdc28 by Cln2 and Clb2 in vitro (122). These results suggest that Cdc37 may act to stabilize the inactive Cdc28 in a conformation that is able to bind and be activated by a cyclin. In many of its interactions in other systems, the involvement of a member of the 90-kDa heat shock protein (HSP90) family has also been implicated. The yeast HSP90 homologs Hsc82 and Hsp82 may fulfill this function in yeast since they are partially redundant in function with Cdc37 but are unable to replace it (290).
Yaglom et al. have reported that another molecular chaperone, Ydj1 (a homolog of the bacterial DnaJ gene product), is needed for the phosphorylation and degradation of Cln3 (622) (see Proteolysis). Ydj1 binds the C terminus of Cln3 when fused to β-galactosidase (Cln3404–488–β-gal), possibly affecting the conformation of this domain in the normal protein, but not affecting the ability of Cln3 to associate with and activate Cdc28. They speculate that increased production of Ydj1 and other Hsp70 family members during heat shock or other stress could accelerate the degradation of Cln3, resulting in the G1 arrest or delay associated with such stresses, but this has yet to be tested explicitly. Interestingly, overexpression of a human Ydj1 homolog (Dnj3) in yeast increases Cln3 levels, possibly by dominantly interfering with yeast Ydj1 function with respect to Cln3 (148).
PROTEOLYSIS
If the activity of a periodically expressed protein is to be dependent upon its concentration, the protein must have short half-lives relative to cell cycle phases. Not surprisingly, many regulators of Cdc28 activity are unstable proteins. Mutations that stabilize these proteins usually have noticeable phenotypic consequences. Stabilization of Clns makes cells insensitive to pheromone exposure and nutritional limitation, reduces the cell size, and shortens the G1 phase (82, 107, 129, 214, 322, 396, 537, 579). Far1 stabilization delays or prevents cell cycle progression (235, 376). Sic1 stabilization arrests the cell cycle between Start and the initiation of DNA synthesis (486, 595). Clb stabilization results in delayed mitotic exit (196, 463, 540) or in premature S-phase initiation (8, 259). The mechanisms influencing this instability fall into two major classes that we will call Start proteolysis and anaphase proteolysis. In both cases, the proteolysis is ubiquitin mediated. These two classes of proteolysis are regulated by different strategies. In Start proteolysis, the ubiquitination machinery is constitutively active. The substrates to be proteolyzed must be activated for destruction by phosphorylation, which is usually dependent upon Cdc28 (a summary of the interactions is given in Fig. 7). Start proteolysis is not specific to Start, but some of the major substrates undergoing destruction by this process are first marked for instability by the Cdc28 complexes that become active late in G1, and the effect is a rapid disappearance of these proteins at Start. In anaphase proteolysis, the ubiquitination machinery is cell cycle regulated—it is turned off from S phase to mitosis and then activated at anaphase—but the substrates appear to be “constitutively” active for degradation.
FIG. 7.
Relationships between SCF components and their substrates. Cln3 ubiquitination is shown to be catalyzed by SCFCdc4 for convenience; this has yet to be shown experimentally. Conventions are as in Fig. 2.
Ubiquitination Machinery
Ubiquitination of proteins is a highly conserved mechanism for targeted cellular proteolysis (for reviews, see references 92, 246, and 593). Ubiquitin marks proteins for degradation when it is covalently attached by an isopeptide linkage to the ɛ-amino group of lysine residues in the target polypeptide. Once attached, K48 of the ubiquitin moiety is usually a target for further ubiquitination. Sequential iterations of this process lead to the multiubiquitinated proteins that are the primary target for degradation by the 26S proteasome—a large, multisubunit complex of proteases with an evolutionarily conserved core structure (210, 352). The attachment of ubiquitin to the target protein requires a series of ubiquitin transfers. First, free ubiquitin is activated in an ATP-dependent manner by covalent attachment of the ubiquitin C terminus to an E1 (ubiquitin-activating) enzyme via a thioester bond. The ubiquitin is then transferred to an E2 (ubiquitin-conjugating) enzyme, to which it is also attached as a thioester. From the E2 enzyme, the ubiquitin can be transferred to the target protein. As is discussed in greater detail below, the ubiquitination reactions involved in Start and anaphase proteolysis require an E3 or ubiquitin ligase. At a minimum, the E3 enzymes interact with the substrate and with an E2 to confer substrate specificity to the ubiquitination reaction. In addition, some E3 enzymes actively participate in the catalytic process and form thioester intermediates with ubiquitin.
In S. cerevisiae, ubiquitin is encoded by four genes (UBI1 to UBI4), which produce polyproteins from which mature ubiquitin is cleaved. The E1 enzyme is encoded by UBA1 (372). On the basis of homology to known E2 enzymes and, in many cases, biochemical characterization, 13 genes encoding potential E2 enzymes have been recognized. These include CDC34 and RAD6 but are generally known as UBC genes. Recently, a number of ubiquitin-like proteins have been identified in yeast and other eukaryotic systems (reviewed in reference 245). Two of these, Smt3 and Rub1, play potential roles in Cdc28 regulation, as described below. In many respects, Smt3 and Rub1 behave like ubiquitin. They are activated by specific E1-like enzymes (Table 5) (132, 269, 320, 337) and then transferred via thioester linkages to specific E2 enzymes (Table 5) (268, 337, 484) and then to target proteins. The E1 enzymes for both Smt3 and Rub1 are heterodimers in which one component (Aos1 and Ula1) structurally resembles the N terminus of the ubiquitin E1 (269, 320, 337) and the other component (Uba2 and Uba3) resembles the C terminus of the ubiquitin E1 (132, 269, 337). The E2 enzymes are structurally similar to the E2 enzymes that transfer ubiquitin, and until recently they were thought to be involved in ubiquitin transfer. The functional significance of Smt3 or Rub1 modification is not yet known.
TABLE 5.
Proteins involved in transfer of ubiquitin or ubiquitin-like polypeptidesa
| Ubiquitin-like polypeptide | Protein involved in transfer
|
Relevant target(s) | ||
|---|---|---|---|---|
| E1 | E2 | E3 | ||
| Ubiquitin | Uba1 | Cdc34, Rad6, and up to 9 others | SCF, APC | Cyclins, Sic1, Far1, Cdc6 |
| Smt3 | Aos1-Uba2 | Ubc9 | ? | ? |
| Rub1 | Ula1-Uba3 | Ubc12 | ? | Cdc53 |
See Table 1 for references.
Start Proteolysis
Ubiquitin dependence.
Cln1 (33, 462), Cln2 (33, 121, 322, 474, 612, 617), Cln3 (33, 106, 109, 579, 621), Cdc6 (141, 438), Far1 (235), and Sic1 (27, 137, 480, 486) are unstable proteins. The reported half-lives for Cln1 (33), Cln2 (33, 121, 322, 474, 612, 617), Cln3 (33, 579, 621), and Sic1 (27) are in the range of 3 to 10 min; the half-life for Cdc6 is 13 to 14 min (141); and the half-life for Far1 is about 30 min (235, 375, 376). Only the cyclin half-lives have been determined by pulse-chase analysis at wild-type expression levels (33, 121, 322); the other half-life determinations relied on the use of galactose-inducible controllers to shut off continued synthesis of the protein. This technique is relatively simple to perform, and its use is often required to allow quantitative detection of CDK regulators, which are present at very low abundances. It has the disadvantage, however, that the abnormally high level of protein produced will be degraded by a nonphysiological mechanism (one that recognizes excess subunits free of their normal binding partners, for example) or will overwhelm the normal degradative mechanism. The pulse-chase mechanism also has the disadvantage that starvation conditions must often be used, which may dramatically affect cell cycle events. So far, no great differences in half-lives have been noticed when both pulse-chase with the endogenous promoter and galactose shutoff have been applied to the same protein (33, 121, 322, 474, 579, 612, 617, 621), but more detailed studies of mechanism may be more sensitive to technique. Another complication to measuring destruction rates of cell cycle regulators is that the half-lives of these proteins may vary with the cell cycle position or method of synchrony (their instability is often stimulated by Cdc28-dependent phosphorylation, as discussed below), factors which are sometimes not controlled for. Despite these caveats, the instability of all these proteins is generally accepted.
This instability is also generally accepted to be ubiquitin dependent, an assumption that rests on a large number of experimental results that vary in quality depending upon the substrate. Detection of ubiquitinated intermediates has been claimed for Cln2 (612), Cln3–β-galactosidase fusions (621), and Sic1 in vivo (519); in vitro ubiquitination has been demonstrated for Cln2 (121, 519), Far1 (235), and Sic1 (173, 519, 595, 596); and inhibition of enzymes needed for ubiquitination or ubiquitin-dependent proteolysis stabilizes Cln1 (33, 43), Cln2 (33, 43, 121, 612), Cln3 (189, 621), Cdc6 (141, 437), Far1 (235, 375), and Sic1 (27, 480, 486).
Sequences required in cis: PEST sequences.
PEST regions—defined as hydrophilic sequences containing at least one proline, one acidic residue, and a serine or a threonine bounded by basic residues but not containing any basic residues within the region of interest—are thought to mark proteins for rapid turnover (453, 465), and algorithms have been developed for finding and scoring such regions in proteins (one is available at www.at.embnet.org/embnet/tools/bio/PESTfind/). According to Rechsteiner and Rogers, scores should be greater than +5 to be considered interesting (453). All the major G1 regulators of Cdc28 have regions meeting these criteria (Table 6). For comparison, the highest PEST score for Cdc28, a stable protein, is 2.78.
TABLE 6.
PEST sequences in CDK regulators
| Gene product | Residues | Sequence | PEST scorea |
|---|---|---|---|
| Sic1 | 197–213 | RSQESEDEEDIIINPVR | 5.25 |
| Far1 | 351–365 | KMATTDPFDLSDDEK | 6.63 |
| Cln1 | 248–274 | HISSSPQSTGLDGDTTTMDEDEELNSK | 14.12 |
| Cln2 | 376–404 | KLTISTPSCSFENSNSTSIPSPASSSQSH | 6.99 |
| Cln3 | 445–471 | KDSISPPFAFTPTSSSSSPSPFNSPYK | 7.39 |
| 471–484 | KTSSSMTTPDSASH | 10.65 | |
| Cdc6 | 32–46 | KLQFTDVTPESSPEK | 5.15 |
Scores were derived from the PESTfind program (www.at.embnet.org/embnet/tools/bio/PESTfind/).
Are PEST sequences determinants of protein stability for Cdc28 regulators? For this group of proteins, the evidence indicates that PEST regions are often contained within an extended domain that destabilizes their host protein but that they are not in themselves sufficient to confer such instability. This is most clearly seen in the studies on Cln2, where deletion of little more than the PEST region (Cln2Δ373–409) produces a protein that is significantly more stable than the wild type. Additional stability is achieved by deletions that remove the entire carboxy terminus (Cln2Δ371–545), indicating that the PEST region alone is not sufficient to confer normal Cln2 degradative rates. These conclusions are supported by the converse experiment, in which transfer of Cln2368–545 confers instability to a heterologous protein but a smaller region containing little more than the PEST domain (Cln2368–409) does not. As far as they go, studies of Cln1, Cln3, and Cdc6 present a similar story. The C-terminal region of Cln3 contains two regions with significant PEST scores (Table 6) and three other PEST-like sequences (621). Large C-terminal truncations significantly stabilize Cln3 (109, 579, 621), while smaller deletions in this region produce intermediate levels of stability (621). Cln1Δ266–546 (33) and Cdc6Δ2–47 (141), both of which lack more than just the PEST sequences (the Cln1 mutant removes only a portion of its PEST region), are more stable than the corresponding wild-type proteins. Cln3404–580 confers instability to a heterologous protein (621), but Cdc61–47 does not (141).
Although both Far1 and Sic1 contain PEST regions, studies of the cis-acting factors that determine stability in these proteins have taken a different direction that highlight non-PEST determinants. Proteins with a graded series of N-terminal deletions up to residue 30 of Far1 have progressively longer half-lives in vivo (376) and thus behave similarly to the C-terminal deletion series of Cln3 (621). Like the Cdc6 N terminus (141), this region is not sufficient to confer instability when attached to a heterologous protein (376). No similar study of the in vivo stability of Sic1 derivatives has been published, but a study of a series of truncations has shown that the N-terminal 27 amino acids and the C terminus containing the PEST region (residues 162 to 284) are not important for Sic1 ubiquitination in an in vitro assay with purified recombinant proteins that is thought to accurately reflect the in vivo mechanism (596). Although the regions that have been identified as conferring instability in both Far1 and Sic1 do not score highly by the rules originally proposed by Rogers et al. (465), they are relatively rich in serine, threonine, and proline, as are the non-PEST regions of the G1 cyclins that also influence instability. It may be that yeasts use a modified version of the PEST motif that will become more defined as substrate recognition motifs are further refined experimentally.
The recent development of an efficient in vitro assay for the ubiquitination of Sic1 with purified recombinant components (173, 519) is a major advance that will promote our understanding of the substrate recognition process in Start proteolysis. The primary difficulty with in vivo approaches is the frequent need to overproduce the substrate being studied so that accurate quantitation can be obtained. All of these substrates form multisubunit complexes; therefore, there is a real danger that overproduction will result in excess protein that is recognized as excess and degraded by mechanisms that are not part of the normal regulatory pathway. This possibility may explain the biphasic degradation observed in the studies of the turnover of Cln2 deletion. The initial degradation rates of both Cln2Δ373–409 and Cln2Δ371–545 were changed relatively little compared with wild-type protein but showed more pronounced stability at longer times (474). These experiments relied on galactose-induced overexpression of the various Cln2 derivatives to generate detectable protein, and so it is likely that the biphasic response represents two different types of proteolysis—a rapid PEST-independent turnover (of free Cln2?) and a slower PEST-dependent turnover (of Cln2 in complex with Cdc28?). The defined in vitro ubiquitination system will bypass these difficulties by eliminating the alternative mechanism.
Requirement for substrate phosphorylation by Cdc28.
Due to their composition, PEST and PEST-like sequences are expected to be rich in the (T/S)-P motif that is the minimal consensus Cdk phosphorylation site, and, as it happens, phosphorylation by Cdc28 appears to be an important prerequisite for Cln, Far1, and Sic1 instability. This is well documented for Sic1, which is phosphorylated on multiple sites in vitro by Cln-Cdc28 and Clb-Cdc28 complexes (380, 457, 486, 519, 595) and is phosphorylated in vivo in a fashion that is primarily CLN dependent (480, 595). At least some of the in vivo phosphorylated sites correspond to sites phosphorylated by Cdc28 in vitro (595). Mutation of three of these sites (T5, T33, and S76) to nonphosphorylatable residues results in a form of Sic1 that has a much longer half-life than the wild type in vivo and is poorly ubiquitinated in vitro in the ubiquitination assay dependent upon purified recombinant components mentioned above. Phosphorylation at additional sites also promotes Sic1 ubiquitination (595). Previous or concomitant phosphorylation of wild-type Sic1 by active Cdc28 complexes is necessary for in vitro ubiquitination when using either this highly purified system (173, 519) or a cruder system involving fractionated yeast lysates and recombinant proteins (596). Finally, inactivation of Cdc28 leads to Sic1 accumulation (137, 382, 486) and inhibition of the machinery responsible for Start proteolysis results in the accumulation of a phosphorylated form of Sic1 in vivo (137, 480, 486, 595). Together, these data and the data cited previously indicate that phosphorylation of Sic1 by Cdc28 complexes induces the ubiquitination and degradation of Sic1. This mechanism ensures that Sic1 inhibition of Clb-Cdc28 complexes remains in effect until after Cln-Cdc28 activity has become established. In fact, stimulation of Sic1 ubiquitination by phosphorylation is thought to be the only essential function of the Cln cyclins, since otherwise lethal cln1 cln2 cln3 triple mutations are suppressed by deletion of SIC1 (161, 480, 576).
A similar story is seen with Far1. Far1 is stable in pre-Start G1, becomes hyperphosphorylated at Start, and is then degraded with a 30-min half-life (375, 376). It is stabilized in cdc28ts mutants at the restrictive temperature and can be phosphorylated in vitro in Cln-Cdc28 immunoprecipitates but not when the extracts are taken from temperature-sensitive cdc28 mutants (435, 577). This phosphorylation is required for in vitro ubiquitination (235). Mutation of a CDK consensus phosphorylation site, S87, to proline (Far1-22) increases the half-life of the mutant protein and eliminates its ability to be ubiquitinated in in vitro assays (235). Cln2-Cdc28 phosphorylates S87 in wild-type Far1 in vitro, but very little phosphate is incorporated into the Far1-22 protein, indicating that S87 is the major in vitro site of Cln2-Cdc28 or that phosphorylation at other sites depends upon S87. In vivo analysis is complicated by Fus3-dependent phosphorylation, whose sites overlap with the Cdc28 site(s) (435). Phosphorylation by Fus3 is thought to enhance the association of Far1 with Cln-Cdc28 complexes (see “Far1”), but its effect on Far1 turnover is unknown.
Ascertaining the role of Cdc28 in Cln2 stability has been complicated due to apparently different requirements for degradation of free Cln2 and degradation of Cln2 in complex with Cdc28. The direct experiment—showing an effect of Cdc28 inactivation on Cln2 stability—has resulted in only modest (twofold) (121, 322) or undetectable (436, 474) changes in the Cln2 half-life. A series of indirect experiments have strongly suggested a requirement for phosphorylation by Cdc28, however. Ubiquitination of Cln2 is promoted in crude in vitro ubiquitination reactions by Cdc28 and by phosphorylation (121). When Start proteolysis is inhibited in vivo, Cln2 accumulates in a hyperphosphorylated state (33, 612). The bulk of Cln2 phosphorylation in vivo is dependent upon Cdc28 activity (322). Cln2 mutants in which alanines replace CDK consensus phosphorylation sites are hyperstabilized relative to wild-type Cln2 (322). As was also seen with Sic1 (595), mutation of multiple phosphorylation sites results in enhanced Cln2 stability (322). There has been no direct demonstration that any of these putative Cln2 phosphorylation sites are, in fact, phosphorylated by Cdc28, but a hyperstable Cln2 mutant (Cln24T3S) lacking all seven CDK consensus sites is poorly phosphorylated (322) and ubiquitinated (612) in vivo. Given this evidence, why is there so little apparent dependence of Cln2 stability on Cdc28 activity in vivo? Lanker et al. have suggested that only Cln2 in Cln2-Cdc28 complexes is subject to phosphorylation-dependent turnover whereas free Cln2 is degraded by a slightly slower, Cdc28-independent process (322). In support of this interpretation, they have found that Cln2 mutants defective in Cdc28 binding (Cln2-Δxs) are poorly phosphorylated in vivo and in vitro and are moderately stabilized relative to the wild type but are not further stabilized when additionally mutated in all seven consensus CDK phosphorylation sites (Cln2-Δxs4T3S). They argue that Cln2 binds thermolabile Cdc28 poorly at the restrictive temperature and that therefore the in vivo studies of Cdc28 dependence involving cdc28ts alleles measured only the slower, Cdc28-independent turnover of Cln2. The development of an in vitro ubiquitination assay with purified components should further clarify these issues.
Consistent with a role for Cdc28 activity in determining Cln3 stability, and unlike the situation seen for Cln2, Cln3 stability increases in cdc28ts mutants at the restrictive temperature (109, 189, 621). Furthermore, when Cln3 proteolysis is inhibited in a CDC28+ background, Cln3 accumulates in a hyperphosphorylated form (578). A systematic analysis of Cln3 phosphorylation has not been undertaken, but S468, which fits the Cdk substrate consensus, seems to be an important site (621). Mutation of S468 to alanine stabilizes a Cln3404–488–β-galactosidase construct and decreases the abundance of the phosphorylated form in vivo. A similar analysis with the Cln3404–580–β-galactosidase fusion has a lesser effect on the stability and abundance of the phosphorylated form. The longer construct apparently contains additional degradation signals that make up for the loss of S468 phosphorylation. As with Cln2, but perhaps not as strongly, association between Cln3 and Cdc28 seems to be important for Cln3 turnover, since Cln3 is more stable in a cdc28-5r83 background (Cdc28-5r83 appears to be specifically defective in Cln3 binding [109]).
E2: Cdc34.
CDC34 encodes the only essential ubiquitin-conjugating enzyme (190, 201). Under restrictive conditions, cdc34ts mutants are multiply budded but mononucleate, with unreplicated DNA and duplicated but unseparated spindle pole bodies (72, 73). This phenotype is also observed in cells depleted of Clb function (486) and in cells expressing hyperstable SIC1 alleles (595). The distinctive phenotype of cdc34ts mutations is clearly due to the failure of these cells to degrade Sic1. Sic1 accumulates in cdc34ts mutants at the restrictive temperature (480, 486), but, more dramatically, sic1Δ cdc34ts double mutants replicate their DNA and arrest in G2 with a single large bud (381, 486), indicating that Cdc34 is not needed for its pre-S-phase function in the absence of Sic1. Cdc34 almost certainly acts directly on Sic1. Efficient in vitro ubiquitination of Sic1, dependent upon Cdc34 and prior phosphorylation by a Cdc28-cyclin complex, has been described (595, 596) and refined to the point that only purified recombinant proteins can recapitulate the entire reaction (173, 519). Other E2 enzymes such as Rad6 and Ubc4 cannot utilize Sic1 as a substrate (596).
In addition to Sic1, it has been found that Far1, Cdc6, and the Clns appear to be Cdc34 substrates. In vitro, Cdc34-dependent ubiquitination has been reported for Cln2 (121, 519) and Far1 (235) and shift of a cdc34ts strain to the restrictive temperature results in the stabilization of Cln1 (43), Cln2 (43, 121, 612), Cln3 (189, 579, 621), and Far1 (235, 375). Cln3 may also have a Cdc34-independent mode of degradation. Yaglom et al. (621) have found that in addition to being stabilized in cdc34 mutants, a Cln3–β-galactosidase fusion is also stabilized in ubc4 ubc5 double mutants but is not stabilized by mutations in five other ubiquitin-conjugating enzyme mutants. Ubc4 and Ubc5 share responsibility for degrading a variety of short-lived and abnormal proteins (493) and may thus be involved in a mechanism by which improperly folded or unassociated cyclin is removed from the cell. It is not clear, however, whether involvement of Ubc4 and Ubc5 in Cln3 degradation is an artifact of the use of the Cln3–β-galactosidase fusion or is a real component of Cln3 metabolism in vivo. In addition, the molecular chaperone Ydj1, known for its involvement in the turnover of many short-lived and abnormal proteins (323) and mentioned above for its role in Cln3 phosphorylation (see “Protein folding”), is required for rapid turnover of Cln3 (622).
Blondel and Mann contend that Cln1 and Cln2 are not direct substrates of Cdc34 in vivo (43). This is based on their observations that although Cln1 and Cln2 degradation is slowed in strains with defective Cdc34 activity, it is also slowed in strains with low Clb1 to Clb4 activity but is normal in a cdc34 sic1Δ double mutant. They argue that Cln1 and Cln2 degradation is actually dependent upon Clb-Cdc28 activity and that since Sic1 is stabilized in cdc34 mutants, Clb-Cdc28 activity will be inhibited and the lack of active Clb-Cdc28 results in stabilization of Cln1 and Cln2. These results have not been reconciled with the Cdc34 dependence of Cln2 ubiquitination in in vitro systems (121, 519), but it should be noted that in vitro Cln2 ubiquitination has been achieved only systems with unfractionated yeast lysates and not in a fully defined system (519).
Blondel and Mann also found that Cln1 and Cln2 are stabilized in a ubc9 mutant (43) which has also been implicated in Clb2 and Clb5 degradation (492). As mentioned above (see “Ubiquitination machinery”), Ubc9 is an E2 enzyme for the ubiquitin-like protein Smt3 (268, 484). SMT3 and all the genes required for Smt3 transfer to target proteins are essential and have similar terminal phenotypes (132, 269). It is not yet known which substrates Smt3 is transferred to, what effect it has on those substrates, or what role they play in cyclin turnover. There is no evidence that modification by Smt3 induces proteasome recognition or protein turnover.
E3: SCF.
Although Cdc34 can transfer activated ubiquitin to artificial substrates in vitro (201), its cell cycle function in vivo requires the assistance of a member of the SCF family of ubiquitin ligases for proper substrate recognition. SCF family members have two common subunits, Skp1 and Cdc53 (the S and the C in SCF) but differ in a third subunit, known as the F box protein (the F in SCF) (27), which determines the substrate specificity of the complex as a whole (430, 519). Skp1 was originally identified as the product of a high-copy-number suppressor of cdc4ts mutations (27), as a yeast homolog of a human cyclin F-binding protein (27) and cyclin A-cdk2-associated protein (636), and as a component of the yeast kinetochore (99, 528). (Only the Start proteolysis functions of Skp1 are covered in this review, but see reference 277 for its function in the kinetochore.) CDC53 was isolated as an essential gene that conferred a cdc34-like phenotype when mutated, indicative of its role in Sic1 degradation (368), and independently as the gene encoding a protein found in association with Cln2 (612). The F box was originally discovered as a sequence motif of about 44 amino acids that was shared by the yeast Cdc4 protein and two human proteins, cyclin F (which gave the F box its name) and Skp2, which were involved in a series of genetic interactions (27). Database searches revealed the presence of this motif in many other proteins, including 17 from S. cerevisiae (27, 431), 2 of which—Cdc4 and Grr1—play important roles in Cdc28 regulation. Cdc4 and Grr1 are discussed more fully below (see “SCFCdc4 recognizes Sic1, Far1, and Cdc6” and “SCFGrr1 recognizes Cln1 and Cln2”).
Cdc34 binds to an SCF through residues just distal to the Cdc34 catalytic core (369). A slightly larger region encompassing this domain (Cdc34171–244) had previously been shown to be unnecessary for ubiquitin acceptance activity but was required for substrate recognition in vitro and cell cycle progression in vivo (450). It appears to act as a portable recognition determinant since its addition to Rad6, a nonessential E2 enzyme, allows Rad6-Cdc34171–244 to suppress the lethality of cdc34Δ mutants (303, 517), presumably by allowing Rad6 to bind the SCF and recognize Cdc34 substrates. Mutations in residues 109 to 113 also eliminate Cdc34 function in vivo but not E3-independent ubiquitination in vitro, and they may define another site of interaction with the SCF (441).
Cdc53, classified as a “cullin” due to its homology to a group of Caenorhabditis elegans proteins (292), acts as a modular bridge linking Cdc34 to the other SCF components (430, 519). Deletions in the C-terminal half of Cdc53 abolish its binding to Cdc34 in coimmunoprecipitation assays while retaining its interactions with Skp1 and F-box proteins. Conversely, deletions in the N-terminal half of Cdc53 abolish its interactions with Skp1 and F-box proteins while retaining Cdc34 binding (430). Mathias et al. have suggested that Cdc4 may also interact directly with Cdc34, since overexpression of Cdc34 but not Cdc53 significantly increases the abundance of overexpressed Cdc4 (369). These interactions have not been observed in coimmunoprecipitation assays, however (430, 519).
A substantial fraction of cellular Cdc53 is modified by the ubiquitin-like Rub1 protein (320, 337), which is attached to Cdc53 via a linkage resistant to reducing agents and denaturing conditions (337, 430, 612) and not requiring a cysteine (430)—probably an isopeptide linkage. Cdc53 appears to be the major Rub1 substrate in the cell (320, 337). Rub1 modification requires the C terminus of Cdc53, which is highly conserved among the cullins (320) but which is not required for the interaction with Cdc34 (430). Rub1 modification of Cdc53 is also dependent upon the presence of functional Skp1 (320). Despite its prominence, the role of Rub1 modification is subtle, at best. It does not affect the stability of Cdc53, since both Cdc53 and Rub1-Cdc53 conjugates have similar half-lives (320). CDC53Δ794–815 mutants and mutants lacking Rub1 or enzymes specifically involved in Rub1 conjugation (Table 5) have no discernible phenotypes on their own but are synthetically lethal or potentiate the severity of mutations in cdc34, cdc53, skp1, or cdc4 (320, 337), suggesting that Rub1 may help stabilize Cdc34-SCF complexes. Overexpression of either Cdc53 or Cdc34 in cells unable to attach Rub1 to Cdc53 is also detrimental to growth (320), however, which seems to indicate a role for Rub1 in maintaining a narrow range of Cdc34-SCF activity. Indeed, Lammer et al. noted that both over- and underexpression of CDC34 and CDC53 increase the amount of Rub1-Cdc53 conjugates relative to total cellular Cdc53 (320). Conditions have yet to be found in which Rub1 modification plays an important physiological role, however.
In addition to stimulating Rub1 modification of Cdc53 (320), Skp1 links Cdc53 to at least three F-box proteins—Cdc4 (173, 519), Grr1 (333, 519), and Met30 (430). This binding is mutually exclusive (only one F-box protein per SCF complex) (333, 430, 519) and requires the F-box motif (27, 294, 333, 430). Different SCF complexes are distinguished in print by superscript F-box components (i.e., SCFCdc4, SCFGrr1, and SCFMet30). Skp1 is also found in complex with Ctf13, an F-box protein found in the kinetochore (99, 528), independently of Cdc53 (277). These results indicate that the F box is a protein-protein interaction domain recognized by Skp1. Other regions of the F-box protein may contribute to the stability of the SCF. Grr1 has twelve 26-residue leucine-rich repeats (28, 182), some or all of which are necessary for interaction with Skp1 in two-hybrid assays although not in coimmunoprecipitation experiments (333). Both Cdc4 (627) and Met30 (559) have multiple WD-40 repeats (185, 404), of which at least some are needed for maximal interaction with the SCF (430). Differential interactions with different F-box proteins may explain some of the mutant skp1 phenotypes—some skp1 mutants arrest in G1 with a multibudded morphology like that of cdc34, cdc53, and cdc4, while others arrest in G2 or as a mixed population of G1 and G2 cells (27, 99). A direct demonstration that mutant skp1 proteins fail to bind distinct subsets of F-box proteins has not been reported, however.
Some E3 enzymes participate in the ubiquitination reaction by forming thioester intermediates. Does the SCF play such a role? Mutational elimination of all the cysteines in Skp1 and Cdc53 and four of the eight cysteines in Cdc4 does not affect the essential function of this SCF (430). This still leaves an opening for a catalytic contribution, but the expectation is that the primary function of the SCF is its ability to recognize substrate and not to participate directly in ubiquitin transfer.
SCFCdc4 recognizes Sic1, Far1, and Cdc6.
Different SCF complexes have different substrate specificities that are determined by the F-box component. Mutants with defects in SCFCdc4 components accumulate or have low turnover rates for Sic1 (27, 480, 486), Far1 (375), and Cdc6 (141, 437). Each of the SCFCdc4 components is also required for in vitro ubiquitination of Sic1 (173, 519) and Far1 (235). With the exception of certain skp1 alleles mentioned above, mutants with mutations in SCFCdc4 components generally arrest in G1 with a multibudded phenotype (27, 73, 88, 99, 224, 368), which can be partially suppressed by deletion of sic1 (resulting in a G2 arrest for the double mutant) (27, 486). Mutations in other F-box proteins do not generate these phenotypes, suggesting that the recognition of Far1, Cdc6, and especially Sic1 is ultimately mediated by Cdc4, the unique SCF component. In support of this, Sic1 can be coimmunoprecipitated with purified Cdc4 (173, 519) but not with any other SCF component (519). Skp1 enhances the Cdc4-Sic1 association (173, 519), but it is not known whether this is due to Skp1 stabilizing an altered conformation of Cdc4 or whether Skp1 makes weak Sic1 contacts that help stabilize a tertiary complex. The eight WD-40 repeats of Cdc4, which are needed to fully stabilize the Cdc4-Skp1 interaction (430), are also thought to mediate substrate recognition. Deletion of the last three repeats abolishes in vitro binding to Sic1 (519), and a region containing little more than the WD-40 repeats (and lacking the F box) interacts with Cdc6 in two-hybrid assays in vivo (141).
It is not yet known what SCFCdc4 sees in its substrates. Two regions of the N terminus of Cdc6 (residues 1 to 18 and, more robustly, 17 to 47) interact with Cdc4 in two-hybrid assays (141), indicating that recognition sites exist within these sequences. Sic1Δ1–27 and Sic1Δ159–284 are both efficiently ubiquitinated in vitro, but further truncations from the N terminus abolish ubiquitination. Additional truncations from the C terminus reduce ubiquitination, but even Sic11–105 still retains some ubiquitin acceptance activity in the in vitro assay (596). A Cdc4 recognition site presumably exists within this domain. Similar analysis has not been reported for Far1, but as discussed above, the N terminus is important for Far1 turnover (376). These regions of Cdc6, Sic1, and Far1 are very basic (their pIs range from 9.6 to 12.1) and rich in proline. It is not clear that these are the characteristics recognized by Cdc4, but they are characteristics that surround CDK phosphorylation sites (248), and for Sic1, phosphorylation by Cdc28 is clearly a prerequisite for SCFCdc4 recognition. The in vitro ubiquitination reactions (173, 519, 596) and the coimmunoprecipitation with Cdc4 (173, 519) fail when Sic1 is not phosphorylated. In vivo, Sic1 turnover is dependent upon Cln-Cdc28 activity (see “Requirement for substrate phosphorylation by Cdc28”), but in vitro Cln2-Cdc28, Clb5-Cdc28, and Clb2-Cdc28 complexes are all capable of carrying out the requisite phosphorylation (173, 519, 596). (This discrepancy between in vitro and in vivo results is probably the result of the use of excess Clb-Cdc28 complexes in vitro over what would normally be present in late G1 in vivo when degradation of Sic1 becomes important.) In addition to being stable in vivo and failing to be ubiquitinated in vitro (595), the Sic1-T5G,T33A,S76A mutant lacking multiple Cdc28 phosphorylation sites does not coimmunoprecipitate with Cdc4 (173). Once Sic1 is phosphorylated, there is no further requirement for Cdc28 complexes and phospho-Sic1 can be ubiquitinated by Cdc34-SCFCdc4 whether it is free or bound to Cdc28-Clb5 (173, 519). Far1 ubiquitination appears to have a similar dependence upon Cdc28 phosphorylation. A crude, in vitro Far1 ubiquitination system involving yeast lysates requires active Cdc28, and the Far1-22 mutant lacking a Cdc28 phosphorylation site was poorly ubiquitinated (235). Ubiquitination of Cdc6 has not been shown to be dependent upon prior phosphorylation, but Cdc6 can be phosphorylated by Clb-Cdc28 complexes and the N-terminal domain of Cdc6—which contains an important instability determinant—contains four CDK consensus phosphorylation sites, indicating that Cdc28 phosphorylation could play a role. On the other hand, Cdc6 is unstable in mating-pheromone-arrested cells (141), conditions that severely reduce Cdc28 protein kinase activity and that lead to stabilization of Sic1 (382, 486). Whether Cdc6 is phosphorylated by another kinase or has a phosphorylation-independent association with Cdc4 are open questions.
SCFGrr1 recognizes Cln1 and Cln2.
Turnover of Cln2 is slowed in grr1 (33), cdc53 (612), and some skp1 (27) mutants, indicating that SCFGrr1 mediates Cln2 ubiquitination. Consistent with this, Cln1—whose stability is also increased in grr1 mutants [33])—and Cln2 bind Grr1 in vitro (519). As was seen for the recognition of Sic1 by Cdc4 and consistent with the dependence of Cln1 and Cln2 stability on Cdc28 activity, Cln1 and Cln2 must be phosphorylated to interact with SCFGrr1 in vitro (519, 612) and phosphorylated Cln1 and Cln2 accumulate in grr1 mutants (33). There is little if any overlap between SCFGrr1 and SCFCdc4 activities—SCFCdc4 binds Cln1 and Cln2 weakly and does not ubiquitinate them in vitro, and SCFGrr1 does not bind Sic1 (519). Consistent with this, Patton et al. have reported that Cln2 is not stabilized in cdc4 mutants (430), but Blondel and Mann report that it is (43). The latter group argue, as with Cdc34 discussed above (see “E2: Cdc34”), that Cdc4 involvement is indirect since deletion of sic1 stabilizes Cln1 and Cln2 in a cdc4 background. However, the discrepancy between these two experimental results has not been resolved.
Unlike CDC4 and the other SCF components, GRR1 is not essential, but grr1Δ mutants are highly pleiotropic with defects in morphology (28, 97, 182, 422), divalent cation transport (97), hexose transport (421, 422, 589), and glucose repression (28, 182, 183). In studies of the role of Grr1 in glucose repression, Li and Johnston noticed that the association between Grr1 and Skp1 increased fourfold when cells are grown in glucose instead of raffinose (333). This is the first indication that SCF complexes might be regulated by environmental conditions. This particular control may act through a negative element in the C terminus of Grr1, since Grr1Δ920–1151 interacts with Skp1 equally well in raffinose and glucose (333). Li and Johnston have proposed that this may be a mechanism tying the carbon source to the rate of G1 progression. In this scheme, growth in glucose would increase the levels of SCFGrr1, leading to increased rates of Cln1 and Cln2 degradation. This would delay passage through Start and contribute to the increase in average cell size that is observed in glucose-grown cells (270, 350, 580). Li and Johnston further hypothesize that F boxes compete for limited Cdc53-Skp1 complexes. The evidence supporting this proposition is weak—it is based on the ability of Grr1 and, more dramatically, GrrΔ920–1151 overexpressers to exacerbate the cell cycle defects of some skp1 alleles (333)—but the possibility of competition between F-box proteins suggests that it may be no accident that the stability of CDK inhibitors and that of activators are controlled by different SCF complexes. Li and Johnston also noticed an inverse correlation between Grr1 levels and the ability of Grr1 to interact with Skp1 (333). Since Grr1 is ubiquitinated by Cdc34, it is possible that SCFGrr1 self-regulates its levels by promoting its own ubiquitination and destruction.
The work on the glucose repression phenotype of grr1 mutants returns us to the question of the E2 enzyme involved. Grr1, Skp1, and Cdc53 are required for the induction of glucose transporters, but Cdc34 is not (333). If SCFGrr1-mediated ubiquitination plays a role in glucose repression, it would seem to involve an E2 enzyme other than Cdc34, casting additional doubt on Cdc34 involvement in other SCFGrr1-mediated processes such as Cln1 and Cln2 degradation. Furthermore, although Far1 and Sic1 can be ubiquitinated in highly defined in vitro reactions dependent upon added Cdc34-SCFCdc4 (173, 235, 519), Cln1 and Cln2 could not be ubiquitinated in comparable reactions dependent upon Cdc34-SCFGrr1 or Cdc34-SCFCdc4, even though binding of Cln1 and Cln2 to these complexes is detectable (519). If Cdc34 is not the E2 enzyme involved in SCFGrr1-mediated ubiquitinations, it would imply that the F-box component would determine not only the substrate to be ubiquitinated but also the E2 to be used. This still would not explain why partially fractionated yeast lysates can support a Cdc34-dependent ubiquitination of Cln1 and Cln2 (121, 519), but the in vivo relevance of this reaction still needs to be established. Also unexplained is the dependence of Cln1 and Cln2 degradation on Clb-Cdc28 activity (43).
Other Cdc28 regulators unstable at Start.
Cln3 is not stabilized in grr1Δ backgrounds (33), but it coimmunoprecipitates with Cdc53 (612) and its degradation is slowed in cdc34 and cdc28 mutant backgrounds (189). These results indicate that Cln3 is not recognized by SCFGrr1 but leaves interactions with other SCFs untested. Clb5 is partially stabilized in skp1 mutants (27), which also indicates a role for an SCF complex in the turnover of this B-type cyclin. Swi5 is also extremely unstable during the G1 phase (398, 400). A region rich in S/T-P motifs confers the instability (554), making Swi5 another candidate for SCF-mediated degradation.
Anaphase Proteolysis
Ubiquitin dependence.
Clb protein levels are periodic, with maximum accumulations occurring in the post-G1 phase of the cell cycle and sharp declines occurring in anaphase (8, 196, 540). The Clb2, Clb3, and Clb5 half-lives are 1 to 2 min during the G1 phase when its proteolysis is active. Throughout S and G2, Clb2 and Clb3 are stable (8, 259) but Clb5 continues to turn over; however, it has a longer half-life, of 10 to 15 min (492). As with the Start proteolysis substrates, the Clbs are thought to be proteolyzed via ubiquitinated intermediates, but there is no direct evidence that any Clb is ubiquitinated in vivo in yeast. In vitro ubiquitination with crude cell lysates has been demonstrated for Clb2 and Clb3 (632, 634), however, and inhibition of enzymes needed for ubiquitination or ubiquitination-dependent proteolysis stabilize Clb2 (197, 257, 260, 492, 634), Clb3 (197, 259), and Clb5 (259, 492).
Sequences required in cis: destruction boxes.
The destruction box was originally defined for mitotic cyclins of other species such as RxxLxyzxN, where y is N, D, or E for B-type cyclins and V for A-type cyclins and z is either I, V, or L (199). Sequences matching the destruction box consensus for B-type cyclins have been identified near the N terminus of Clb1, Clb2, Clb3, and Clb4. Clb5 has a more divergent destruction box that is a closer match to the cyclin A consensus (Table 7). Clb6 does not have a significant match to either consensus (487). Overexpression of constructs lacking the N-terminal 152 amino acids of Clb1 or the destruction box of Clb2 (Clb2Δ24–34) results in cell cycle arrest in telophase due to the accumulation of these cyclins (196, 540). The G1-phase half-life of Clb2 rises from less than 1 min for the wild type to more than 10 min when the destruction box is deleted (8) and the ability to be ubiquitinated in an in vitro assay is lost (634). Single point mutations in individual destruction box residues increase the Clb2 half-lives moderately. Deletion of the Clb5 destruction box also results in partial stabilization of this cyclin (259, 260).
TABLE 7.
Putative destruction box sequences in selected yeast APC substrates
| Substrate (residues) | Destruction box sequenceb | Reference(s) |
|---|---|---|
| Clb1 (35–43) | RTILGNVTN | 181 |
| Clb2 (25–33) | RLALNNVTN | 181 |
| Clb3 (51–59) | RVALSRVTN | 181 |
| Clb4 (43–51) | RVALGDVTS | 181 |
| Clb5 (56–64) | RALTDVPVN | 160, 310, 487 |
| Cdc5 (17–25) | RSKLVHTPI | 503 |
| Cdc5 (61–69) | RKKLSALCK | 503 |
| Cdc20 (17–25) | RSVLSIASP | 449, 503 |
| Cdc20 (60–68) | RPSLQASAN | 449, 503 |
| Pds1 (85–93) | RLPLAAKDN | 96, 623 |
| Cyclin A consensusa | RxALGVzxN | 199 |
| Cyclin B consensusa | RxALGyzxN | 199 |
x is any amino acid; y is N, D, or E; and z is either I, V, or L.
Boldface indicates conserved residues in yeast which match those of metazoan consensus sequences.
E2?
The E2 involved in anaphase proteolysis has yet to be unambiguously identified in yeast. Ubc11 (569) is the closest yeast homolog to E2-C, the metazoan ubiquitin-conjugating enzyme involved in anaphase cyclin proteolysis (22). Unfortunately, Ubc11 cannot replace E2-C in in vitro ubiquitination assays and deletion of UBC11 has no effect on Clb2 turnover in vivo (569). Ubc4 can replace E2-C in in vitro cyclin B ubiquitination assays (291), but the only in vivo evidence supporting a role for Ubc4 in anaphase proteolysis is the observation that double mutants combining ubc4 and cdc23, a component of the anaphase-promoting complex (see “E3: APC”) are inviable (260). Other experiments are negative: extracts from ubc4Δ mutants retain the ability to ubiquitinate Clb2 in vitro (632), and ubc4Δ mutants have no effect on Clb2 turnover in vivo (569). Double ubc4 ubc11 mutants are no more defective for growth, mitotic progression, or Clb2 turnover than are single ubc4 mutants (569). Ubc5 closely resembles Ubc4 in sequence and shares with Ubc4 an essential function (they are required for the degradation of short-lived and abnormal proteins) (493), but ubc11 ubc5 mutants also show no defects in Clb2 turnover (569). The ubc4 ubc5 double mutant and the ubc4 ubc5 ubc11 triple mutant do not appear to have been tested for defects in anaphase proteolysis, however. Ubc9 has also been advanced as a candidate for the APC ubiquitin-conjugating enzyme, since ubc9 mutants arrest in late G2 and Clb2 is stabilized (492). As noted above, Ubc9 is not a true ubiquitin-conjugating enzyme but an Smt3-conjugating enzyme (268). In any case, the biochemical evidence is negative, since the in vitro ubiquitination assay is not affected in extracts from ubc9 mutants (632). It is possible that multiple ubiquitin-conjugating enzymes function with the APC, making genetic analyses of this function less than straightforward.
E3: APC.
Genetic screens for mutants defective in Clb2 proteolysis or colethal with Sic1 identified alleles of APC1, CDC26 (634), CDC16, CDC23 (260), DOC1 (257), and APC2 (305). The protein products of these five genes and that of another gene, CDC27 (319), associate into a larger 20S or 36S particle (257, 634) known as the cyclosome or anaphase-promoting complex (APC) (291, 535). Additional components—Apc4, Apc5, Apc9, Apc10, Apc11, and Apc13—have recently been identified by mass spectrometric analysis of the proteins that coimmunoprecipitated with some of the genetically identified APC components (633) or by homology to components of the human APC (630). The APC was initially identified as the E3 for cyclins A and B in Xenopus and clam oocytes (291, 535). Consistent with this role for the yeast APC, extracts from cdc16, cdc23 (632), apc1, cdc26 (634), apc2 (305, 633), and apc11 (633) are defective in ubiquitination of Clb2 and Clb3 in vitro. Clb2 ubiquitination is reduced, but not eliminated, in extracts from cdc27 strains, consistent with the lesser effect that cdc27 mutants have on Clb2 stability (632).
Mutations in APC component genes generally result in similar but not identical phenotypes. Most of these genes are essential—arresting as large-budded cells with a G2 DNA content and a single nucleus with a short mitotic spindle (73, 232, 259, 305, 515, 630, 633, 634). These phenotypes are consistent with a failure to degrade proteins whose removal is essential for anaphase. Deletions of cdc26 (20, 634), apc9 (633), and doc1 (257) result in viable phenotypes, however. The cdc26Δ strains are temperature sensitive, with an arrest phenotype similar to that of the other APC genes (20, 634). Cdc26 amounts increase 10-fold at the restrictive temperature (634); therefore, the primary role of Cdc26 may be to stabilize the APC under extreme conditions. Deletion mutants of apc9 have delayed progression through mitosis (633), while doc1Δ mutants are slow growing and accumulate cells with large buds (257)—both phenotypes suggestive of an important though not essential role for these proteins in APC function. APC13 mutant phenotypes have not been reported.
Other than being required for anaphase proteolysis, there is little information on the biochemical functions for individual APC components. As might be expected for components of a high-molecular-weight particle, most of the identifiable sequence motifs in APC proteins are associated with protein-protein interactions. Multiple, highly degenerate, 34-amino-acid TPR (tetratricopeptide repeat) domains are found in Cdc16, Cdc23, and Cdc27 (242, 513). The X-ray crystal structure of the TPR repeat of protein phosphatase 5 show that each repeat unit consists of two α-helices lying side to side in an antiparallel fashion. Different repeats stack atop one another to create an amphipathic channel that is expected to be involved in interprotein interactions (117). At least some TPRs are required for APC function, since TPR mutations of CDC23 are lethal and reduce the stability of associations between Cdc23 and other APC components (514, 515). The three TPR proteins and Apc1 are present in at least two copies per APC (319, 634), but the exact stoichiometry of these or any other components has not yet been ascertained. Cdc27 association with the APC is stabilized by Apc9, since APCs prepared from apc9Δ mutants lack Cdc27 (633). Similarly, Cdc26 seems to stabilize the association of Cdc16, Apc9, and Cdc27 (633). Apc11 contains a RING motif (633). This motif (C3HC4) in other proteins is capable of binding two Zn2+ ions and is also thought to mediate protein-protein interactions (48). Apc2, like Cdc53, is a cullin family member (305, 630, 633) and, by analogy, may be the link with the E2 component. As described above (see “E2?”), the E2 for the yeast APC is not yet known, but Apc2 does not associate with Cdc34 (630), the E2 that binds Cdc53.
Cdc20, Cdh1, and regulation of anaphase proteolysis.
In a manner that, on the surface, seems analogous to the use of multiple F-box proteins to recognize different groups of substrates for Start proteolysis by the SCF, the APC requires additional factors to destabilize different targets. These factors are Cdc20 and Cdh1, two related proteins that—like the F-box proteins, Cdc4, and Met30—contain WD-40 motifs (483, 491, 599). CDC20 alleles were first isolated in the initial screen for cell division cycle mutants (226). Its terminal arrest point is just prior to anaphase (73, 424, 491). Cdh1 was identified through its homology to Cdc20 (599) and its ability to act as a high-copy-number suppressor of cdc20 mutants (483). Unlike Cdc20, Cdh1 is not essential, but cdh1Δ strains have a slow-growth phenotype (483, 599) and an extended stay in late anaphase (599). Critical clues to their function derived from the studies of APC activity during the cell cycle, and particularly the observation that the APC is active and Clb2 is extremely unstable in pre-Start G1 cells (8). Inactivation of Cdh1 (483, 599) but not Cdc20 (551, 599) stabilizes Clb2 in these early-G1 cells. Conversely, Pds1, whose APC-dependent degradation is needed for chromosome separation (96, 623, 624), is stabilized in cdc20 but not cdh1Δ mutants (599). In S phase, Clb2 (8) and Pds1 (96) proteolysis is turned off but overexpression of Cdh1 accelerates Clb2 turnover (483, 599) without affecting Pds1 stability (599) while Cdc20 overexpression accelerates Pds1 turnover without affecting Clb2 (483, 503, 599). In all cases, the accelerated degradation is dependent upon Cdc23 (483, 599) and, for Clb2 at least, the presence of a functional destruction box (483), indicative of APC involvement. These results indicate that Cdh1 allows the APC to target Clb2 whereas Cdc20 is specific for Pds1.
The situation seems to change as cells approach anaphase. When cells are arrested at the G2/M border with nocodazole, overexpression of Cdc20 induces slow but clearly increased proteolysis of Clb2 whereas overexpression of Cdh1 induces a similar change in Pds1 turnover rates (599). This seems to derive in part from a dependence of Cdh1 activity on prior Cdc20 function and in part from the mechanism by which cell cycle progress is blocked by nocodazole. The Cdh1 dependence on Cdc20 function is seen by the inability of terminally arrested cdc20 and cdc20 pds1 mutants to induce Clb2 proteolysis (340, 503). This explains why Pds1 is degraded just before chromosome separation (anaphase A) while Clb2 proteolysis begins after the sister chromatids have separated (anaphase B or telophase) (503). Nocodazole is a microtubule poison, whose action invokes the spindle checkpoint preventing G2/M progression (for reviews, see references 203 and 220). A number of gene products required for this checkpoint have been identified (250, 334, 608). Of these, three—Mad1, Mad2, and Mad3—form a complex with Cdc20 (256). Mad complex formation with Cdc20 is required for the checkpoint function, since Cdc20 mutants that disrupt this binding fail to activate the spindle checkpoint (256, see also reference 482). It is not clear how the Mad complex affects Cdc20 activity, since there does not seem to be a change in the Mad-Cdc20 interaction when the checkpoint is activated. Mad1 becomes hyperphosphorylated in many (221) but not all (169) cases in which the checkpoint is activated, but it is not clear what role this plays. In any case, overexpression of Cdc20 and Cdh1 seems to affect the stoichiometric balance required to make the checkpoint work and pushes the cell into anaphase (256, 503, 599). The mechanism by which these factors function is unclear, however. Unlike the work with the F-box proteins, there has been no demonstration of direct physical interactions between Cdh1 or Cdc20 and any APC substrate. Lim et al. have reported that Cdc20 and the APC component Cdc23 coimmunoprecipitate (340), but neither factor was found in purified APCs (633), so the significance of this result is unclear.
How Cdh1 activity is made to be dependent upon Cdc20 activity is also unclear. The BUB2 gene, originally isolated for its role in the spindle checkpoint (250), may be important for this process. Double cdc20 bub2 mutants arrest with reduced mitotic Cdc28 activity and a multibudded phenotype that may result from prolonged G1 cyclin expression (551). Its biochemical role is uncertain, however. Since Cdh1 overexpression can override this dependence (483, 599), stoichiometric balances once again seem to be important. Change in cellular concentration is clearly an important feature of Cdc20 regulation. Cdc20 levels are strongly periodic, accumulating in the nucleus as Pds1 levels start to decline in early anaphase (503) but then declining sharply at cytokinesis along with Clb2 (503). This pattern is partly due to a CLB2-like transcriptional pattern (with the exception that CDC20, unlike Clb2, is induced by mating pheromone) (449) and partly due to changes in Cdc20 stability (449, 503). At all times Cdc20 has a short half-life (∼3 min), but it is even more unstable in G1 (449). APC mutations cause significant increases in Cdc20 stability in all parts of the cell cycle (449, 503), but the half-life is still short—indicating that Cdc20 stability is controlled by both APC-dependent and -independent mechanisms (449). Furthermore, a destruction box sequence (Table 7) near the N terminus contributes to the G1 instability (449, 503) but not to the instability at other cell cycle stages (449), suggesting that three distinct mechanisms affect Cdc20 turnover. This work also reveals that there is little or no Cdc20 in G1 cells but that Cdc20 is required for rapid Pds1 turnover during this cell cycle phase (599). Either an undetectable level of Cdc20 remains in G1 cells or Cdc20 is involved in the establishment but not the maintenance of Pds1-directed APC activity (503).
In contrast to Cdc20, Cdh1 levels are constant throughout the cell cycle (449, 503), so a mechanism other than changes in abundance must explain changes in Cdh1-dependent proteolytic activity. An attractive but speculative possibility is that Cdh1 is regulated by Cdc28. Cdc28 activity—whether it is complexed with Cln1, Cln2, or any of the Clb cyclins—represses Clb proteolysis (7, 8, 129), and inhibition of Cdc28 at any point in the cell cycle results in increased Clb turnover. This behavior seemed to result from changes in the activity of the APC since extracts from G1-arrested cells are capable of ubiquitinating added Clb2 in vitro in an APC-dependent fashion, but extracts from cells arrested in other parts of the cell cycle have no Clb ubiquitination activity (632). Pds1 degradation, however, is not inhibited by Cdc28 activity (503). This difference between the sensitivity of Pds1 and Clb2 turnover to Cdc28 activity could be readily achieved if Cdh1 but not Cdc20 activity was sensitive to Cdc28. Consistent with this possibility, but far short of confirming it, Cdh1 isolated from early-G1 cells migrates more rapidly on SDS-PAGE gels than when it is isolated from other cell cycle phases (449) as if it were in a hypophosphorylated state in G1 relative to the remainder of the cell cycle.
If this mechanism keeps Clb2 proteolysis off throughout S and G2, what activates Clb2 turnover in late anaphase? In metazoan systems, Cdk activity stimulates APC activity and mitotic cyclin ubiquitination in vivo (174, 318, 355, 395) and in vitro (237, 291, 535). This is in contrast to the inhibition of Clb2 turnover by Cdc28 activity observed in yeast (7, 8, 129). There is some genetic support for a specific role of Clb2 in APC activation, however. The clb2 cdc23ts mutant combination is lethal, a synthetic lethality specific for CLB2 (260). Mutant combinations of cdc23ts with deletions in the other CLB genes, either singly or in combinations such as clb5 clb6 and clb1 clb3 clb4 are viable, indicating that Clb2 may have a specific function related to APC activation not possessed by any of the other Cdc28 complexes (260). Inappropriate expression of CLB2 outside of metaphase, however, does not activate anaphase proteolysis (7), and so it is clear that there must be more to this putative “switch.” There is evidence that Sic1 directly relieves the Cdc28-mediated repression of anaphase proteolysis (7, 137). Sic1 is synthesized near the start of anaphase proteolysis (137, 298, 486, 571), and thus its appearance is timed to mediate reduction in Clb-Cdc28 kinase activity. Consistent with such a role, mutant sic1Δ cells have a delayed anaphase-to-telophase transition (571). Double-mutant combinations between sic1 and cdh1 (483, 599) and some APC components (305) are also lethal. The difficulty with this model is that some reduction in Clb-Cdc28 activity would seem to be required to activate SIC1 transcription (see “Swi5 and Ace2”).
Another mechanism for initiating Clb2 turnover might involve phosphorylation of Cdc28 on tyrosine. When spindles are damaged in cdc55 mutants (Cdc55 is a WD-40 repeat-containing, regulatory B subunit of protein phosphatase 2A), sister chromatid separation occurs without the checkpoint delay (384, 605). Clb2 is not degraded under these conditions, but Cdc28 accumulates in an inactive form with high levels of phosphotyrosine. CDC28-Y19F cdc55Δ double mutants regain the spindle checkpoint (384). Inactivation of the APC also suppresses the loss of the spindle checkpoint (605). These results suggest that the appearance of separated sister chromatids (or some other correlative event) stimulates the phosphorylation of Cdc28 on Y19 as a means of inactivating Clb2-Cdc28 when APC function is abrogated. Once again, this cannot be the only mechanism, because the CDC28-Y19F mutant has no gross cell cycle defects (9, 525) and therefore must be capable of entering and completing anaphase.
Yet another means of activating Clb2 proteolysis is provided by Cdc5 (295), a protein kinase of the “polo” family (named from its Drosophila homolog [for reviews, see references 200 and 202). Mutants defective in Cdc5 arrest late in anaphase with low Pds1 but elevated Clb2 levels (85, 503). Clb2 ubiquitination in extracts from cdc5 is reduced (85). Overexpression of Cdc5 does not affect Pds1 levels (85) but induces declines in Clb2 levels (85, 503) in a manner that is APC dependent but not Sic1 dependent (503). In vitro Clb2 ubiquitination is stimulated in extracts from Cdc5-overproducing cells but not if those extracts are defective for Cdh1 (85). Together, these results indicate that Cdc5 plays a specific role in the stimulation of Cdh1-dependent APC activity or in the relief of Cdc28-dependent inhibition of Cdh1-dependent APC activity. Cdc5 activity seems to be controlled similarly to that of Clb2 and Cdc20. Cdc5 protein (222, 503) and protein kinase activity levels (85) are strongly periodic due to cell cycle dependent transcription (295) and proteolysis (85, 503). Cdc5 proteolysis is dependent upon Cdh1-dependent APC activity in G1 and requires the presence of two destruction boxes in the Cdc5 N terminus (85, 503) (Table 7).
Cdc5 and Sic1 have been tied together in a complex genetic web with a host of additional factors. These include Cdc15 (485, 500), Dbf2 (137, 272, 426), and Dbf20 (570, 573) (all protein kinases), Cdc14 (604) and Sit4 (426) (both protein phosphatases), Lte1 (a GDP/GTP exchange factor) (501), Tem1 (502) and Ras1 (both GTPases) (391), and Spo12 (an activator of Dbf2 and Dbf20) (391, 426, 573). These are all good components for a signal transduction pathway or two, but they have not yet been organized into a coherent scheme. The behavior of Cdc15 is particularly interesting. Mutant cdc15 strains are often used to arrest cells at a point before anaphase proteolysis occurs (195, 259, 540, 632), but Irniger et al. have found that a subclass of Clb2 is degraded at a cdc15 arrest point and that proteolysis of the remainder requires passage through the cdc15 arrest point (260). This mutant seems to be arrested at a step in which Clb2-Cdc28 has been partially deactivated, perhaps enough to allow partial activation of Cdh1-dependent APC activity by one of the mechanisms described above.
Role for nuclear import?
Mutants with defects in SRP1, an essential gene with a G2 arrest phenotype, also have defects in Clb2 turnover (347). SRP1 encodes the yeast homolog of importin, the nuclear import receptor (36, 347, 625). srp1 mutant extracts are able to ubiquitinate Clb2 in vitro; therefore, Srp1 may be needed to activate the APC or to deliver substrates or other components to the APC. The role of transport of cyclins between the cytoplasm and nucleus during mitosis (215, 618) is a developing story in non-yeast systems but has been little studied in S. cerevisiae.
The other Clbs.
Proteolysis of the other Clb genes has, in general, been ignored. In the few studies in which they have been examined Clb1, Clb3, and Clb4 behave similarly to Clb2. Clb3 is stabilized in APC and proteasome mutants (197, 259) and is ubiquitinated in vitro (632) similarly to Clb2. Clb1 is dependent upon Cdh1 for its destruction (483). Clb5 seems to be controlled by a combination of Start and anaphase proteolysis with features all its own that keeps Clb5 moderately unstable at all points in the cell cycle. Clb5 stability is increased in skp1 mutants (27), indicating participation by SCF, but unlike the Clns, it is not found in complex with Cdc53 (612). It is not recognized by Cdh1 (483) but is degraded along with Pds1 (503), and its stability is partially dependent upon APC components (259), Ubc9 (492), and its destruction box (195). Clb6, which lacks a destruction box, has not been studied at all.
PERTURBATIONS TO THE NORMAL CELL CYCLE
DNA Damage and Other Checkpoints
Successful duplication of a cell requires the coordination of many complex processes. Due to environmental influences or even just to random errors or delays in synthesis, transport or assembly, some of these processes can be delayed relative to others. For this reason, the cell must have mechanisms to monitor cell cycle events and, in the event of a problem, be able to halt or delay some steps until coordination can be reestablished. These coordination mechanisms are called checkpoints (607). Some of the best-studied checkpoints are invoked in response to DNA damage, incomplete DNA replication, the failure to duplicate the spindle pole body, and the failure to assemble a proper mitotic spindle (see references 80, 155, 158, and 432 for reviews). Checkpoints that operate in S, G2, or M arrest cell division with high Cdc28 activity levels (9, 256, 335, 525, 534, 608). This was initially surprising because studies of other systems, particularly S. pombe, indicated that DNA damage and replication checkpoints operated through inhibition of CDK activity by phosphorylation on tyrosine (see references 155 and 158 for reviews). As discussed above (see “Inhibitory phosphorylation on Y19 and T18”), S. cerevisiae has redirected this pathway to monitor budding defects. S. cerevisiae is unusual in that it builds spindles very early in its cell cycle and so initiates the early steps of mitosis around the time of S-phase initiation. This difference may require the maintenance of high Cdk activity to preserve the spindle and may have forced S. cerevisiae to find ways to halt cell cycle progression in response to delays or errors in the nuclear cycle that would not reduce Cdk activity. Therefore, as far as Cdc28 regulation is concerned, the function of a checkpoint operating in S, G2, or M has to be to prevent the destruction of the Clb cyclins. The critical role that Cdc20 plays in the spindle damage checkpoint has already been discussed (see “Cdc20, Cdh1, and regulation of anaphase proteolysis”). This pathway is also important for at least some responses to DNA damage (256, 343) but not for the checkpoint monitoring the completion of DNA synthesis (256).
Another mechanism may operate through Cks1. Allen et al. (3) have found that cks1 mutations can suppress the hydroxyurea sensitivity of mutations in rad53, which encodes a Ser/Thr/Tyr protein kinase required for the DNA damage checkpoint. Cks1 has a proposed role in targeting Cdc28-cyclin complexes to the APC (535); therefore, inhibition of Cks1 binding to either the APC or Cdc28 could also reduce Clb turnover and maintain Cdc28 protein kinase activity.
DNA damage in G1.
Checkpoint operation in G1 must prevent Cdc28 activation. The mechanisms by which this occurs are only beginning to be worked out. Treatment with a DNA-damaging agent such as methyl methanesulfonate (MMS) in G1 delays the appearance of CLN1 and CLN2 mRNAs and represses their transcription even in cells that have already passed Start (but not entered S) (510). Cells with defects in the DNA damage checkpoint pathway, such as mec1 and rad53 mutants (a “PI” kinase and a Ser/Thr/Tyr protein kinase, respectively), initially respond to MMS treatment like wild-type cells but recover much more quickly. Overexpression of a SWI4 allele lacking the Swi6 binding domain or deletion of SWI6 generates a response that resembles the mec1 and rad53 mutations, indicating that Mec1 and Rad53 may operate through Swi6. The Swi6-Rad53 interaction appears to be direct, since Rad53 immunoprecipitated from MMS-treated but not Rad53 from control cells can phosphorylate Swi6 in vitro. Furthermore, Swi6 is phosphorylated on the same peptides in vivo in an MMS- and rad53-dependent fashion (510). Ho et al. (244) have also implicated Swi6 as the most proximal element in the DNA damage response. They found that Swi6, Swi4, and the casein kinase Hrr25 are needed for the induction of DNA damage response genes such as RNR2 and RNR3. Hrr25 binds to and phosphorylates Swi6 in vitro. It is not known what role Hrr25 plays in the response of CLN1 and CLN2.
Transcription of CLN3 and CDC6 is reduced by ∼50% during treatment with MMS, but CLN3 is hyperinduced following MMS removal (when damage control is still needed and CLN1 and CLN2 mRNA levels remain low) while CDC6 returns to basal levels (510). These results indicate that the ECB may be regulated by DNA damage as well and could be a factor in the initial transcriptional response of CLN1 and CLN2. Another mechanism, possibly involving the phosphorylation of Swi6 by either Hrr25 or Rad53, must keep CLN1 and CLN2 transcription low during the period of CLN3 hyperexpression.
Defects in bud formation are also handled at a G1 checkpoint. The G1 cyclins are particularly adept at promoting bud formation, while Clb2 activity inhibits bud initiation. When proper bud formation is perturbed, such as by osmotic stress, the cell must respond by enhancing Cln-Cdc28 kinase activity but keeping Clb-Cdc28 activity turned off. In this case, phosphorylation on Y19 seems to be the preferred mode of action (see “Inhibitory phosphorylation on Y19 and T18”) (331).
Stress
Exposure to elevated temperature results in a transient decline in the transcription of CLN1, CLN2 and SWI4 but not CLN3 and results in a pause at Start (467). This effect has been at least partially traced to changes in Cln3 activity but does not involve Far1, Fus3, or the cAMP-dependent protein kinase pathway (467). Yaglom et al. have hypothesized that this could be mediated by destabilization of Cln3 through induction of stress-related proteins such as Ydj1 (see “Protein folding”), but they have provided no evidence that this actually occurs (622).
A possible component of a stress-induced transcriptional repression system has been identified by Mai and Breeden. Two sites in the CLN1 promoter (at −434 and −450) that were originally thought to be SBF binding sites (414) but were later shown to lack UAS activity (427) are recognized by Xbp1 (363). Xbp1 has a DNA binding domain that is 40% identical to that of Swi4 and Mbp1. It recognizes a consensus sequence which has an XhoI restriction endonuclease site as its core (giving Xbp1 its name, i.e., XhoI site binding protein [Table 3]) and does not bind sites known to function as SCBs or MCBs. The promoter for XBP1 contains a number of elements (five stress response elements [STREs], one AP-1 recognition element, and one heat shock element) that confer response to stress, and, correspondingly, XBP1 transcription is induced by heat shock, glucose starvation, oxidative stress, high osmolarity, and DNA damage. CLN1 transcription declines in a manner that mirrors XBP1 induction in time-dependent and dose-dependent fashions. Artificial production of Xbp1 from the GAL1 promoter leads to repression of CLN1 transcription and generates a slow-growth, large-cell phenotype resulting from an increase in G1 length. These observations make XBP1 a good candidate for a factor that would mediate the CLN1 stress response, but unfortunately the CLN1 responses in wild-type and xbp1Δ strains are identical (363). XBP1 is also induced by some agents, such as diamide, which do not affect CLN1 transcription. Furthermore, CLN2 and CLN3 mRNA levels also decline when Xbp1 is induced artificially, although the CLN2 promoter does not have a recognizable Xbp1 binding consensus and CLN3 transcription is not affected by the heat shock response (CLN3 has a recognizable Xbp1 consensus binding site at −415). These results indicate that if XBP1 is involved in the stress response of the G1 cyclins, the mechanism is complex. Structurally, Xbp1 has some interesting features in addition to the Swi4/Mbp1-like DNA binding domain. There are three homopolymer domains (one of which is 42% identical to the N terminus of the Swi1 transcription factor) and a region that is 40% identical to the C-terminal domain of testis-specific histone H1, a region which (in histone H1) can efficiently condense chromatin (285, 609) and could play a role in the transcriptional repression activity of Xbp1.
Nutritional Limitation
Nutritional limitation, commonly achieved through reductions in the concentration or changes in the species of a metabolizable carbon- or nitrogen-containing compound, causes an overall increase in cell doubling times. When an essential nutrient is completely lacking, yeast cells arrest in a state commonly, but with some controversy, called G0. Most of the increase in the doubling time of cells growing in poor media is due to an expansion of G1, with little changes in the duration of the S to M portion of the cell cycle (81, 271), indicating that changes in Cln-Cdc28 activity should play a dominant role in this response. In fact, down-regulation of Cln2-Cdc28 and Cln1-Cdc28 is required for a proper starvation response, since cells expressing CLN1 from a mild constitutive promoter or carrying hyperstable alleles of CLN2 do not properly arrest in response to starvation signals (189, 214). Part of the response is transcriptional. Levels of mRNAs from SBF- and MBF-driven genes and some but not all components of TFIID decrease 2 to 4 h after the cells are deprived of a source of metabolizable nitrogen (189, 603). This response is fairly specific since CLN3, SWI6, and MBP1 transcript levels do not change and SIC1 mRNA levels actually increase (189). Silljé et al. contend that the absolute amounts of CLN1 and CLN2 transcripts are unaffected during conditions of nutritional limitation but that since cell cycle times increase, the relative amounts of SBF- and MBF-driven mRNAs in an asynchronous population of cells will decline (516). Their experiments involving a synchronous fed-batch system support this contention. By varying the nutritional content of their media, Silljé et al. produced populations of cells with G1 periods that ranged from 40 to 580 min. No matter how long the G1 period, CLN1 and CLN2 mRNAs did not appear until immediately prior to bud emergence. The SWI4 mRNA peaks at variable times before the CLN1 and CLN2 peaks, indicating that the nutritional response signals pass, at least in part, through SWI4 (516).
Part of this transcriptional response to starvation is due to reductions in the levels of Cln3 protein and can be suppressed by constitutively expressing CLN3 from a heterologous promoter (189). As discussed previously, reduced translational efficiency of CLN3 mRNA (see “Translation”) and increased Cln3 proteolysis (see “Proteolysis”) both play a role in this effect. Hyperstable CLN3 strains respond to nutritional signals, however, although with a poorer efficiency (214, 396, 537), indicating that the nutritional response may be only partially due to changes in Cln3-Cdc28 activity. There are also indications that Cln1, Cln2, and Clb5 proteolysis is also accelerated in starved cells (189).
Ras-cAMP pathway.
The upstream controls are still poorly worked out, due in part to the multiplicity of pathways involved and the corresponding complexity of the genetics involved in their analysis. Possible components of a translational control mechanism for Cln3 have been discussed earlier (see “Translation”). The Ras-cAMP pathway has long been assumed to be involved, since mutations which reduce the activity of the cAMP-dependent protein kinases (Tpk1, Tpk2, or Tpk3) cause cell cycle arrest with the characteristics of G0 cells (61, 278, 370, 464, 563) while mutations which superactivate the Tpks make cells resistant to starvation-induced G1 arrest (79, 279, 563). On the other hand, Markwardt et al. (367) have shown that cells with constitutively high Tpk activity down-regulate CLN2 transcription and cease dividing during nitrogen starvation, like wild-type cells, but do not uniformly arrest in G1. In addition, cells that constitutively express a very low level of Tpk activity are still able to show a normal G1 arrest response to nutritional limitation (78). Markwardt et al. contend that the active Tpks prevent cells from storing sufficient nutrients to complete a cell cycle in response to nitrogen starvation and that the cAMP pathway does not directly control the G1 cyclin response (367).
Whi2.
Mutations in whi2 were isolated based on their small-cell phenotype (537). These mutations are defective in their ability to properly undergo stationary-phase arrest (283, 479, 537), a defect that is due in part to their failure to down-regulate CLN1 and CLN2 expression (451). Ectopic low-level expression of CLN1 in cells nearing stationary phase phenocopies many but not all (they are not as sensitive to heat shock, for example) aspects of whi2 mutants. Higher-level CLN1 expression causes an elongated cell phenotype, but these cells are still not as sensitive as whi2 cells are to heat shock. The WHI2 gene has been sequenced (283), but nothing is known of its function. Like the CLN1 and CLN2 genes, WHI2 transcription is down-regulated as cells approach stationary phase; this down-regulation is not affected when Whi2 itself is nonfunctional (whi2 transcript levels decline normally as whi2 cells approach stationary phase) (393). The whi2 phenotype is observed only when the oxygen saturation levels in the culture are above 40% (452). The ssd-d2 mutation found naturally in W303a genetic backgrounds promotes an earlier decline of CLN1, CLN2, and CLN3 mRNA levels and has an uncharacterized interaction with whi2 (451).
Pheromone-Mediated Cell Cycle Arrest
Model.
Exposure to mating pheromone allows yeast cells to complete their current cell cycle but then forces an arrest at Start. This response is required for mating to be successful (459). Arrest at Start is achieved by shutting down Cln-Cdc28 activity while allowing cells that have already activated Clb-Cdc28 complexes at the time of exposure to complete their program. This is accomplished, in part, by repressing the transcription of Start-specific genes and inducing the synthesis and activation of Far1 (see “Cdc28 inhibitors: CKIs”). These effects are mediated through the pheromone response pathway that leads through a serpentine surface receptor, a heterotrimeric G protein, a Rho-family GTPase, and a protein kinase cascade that has two MAP kinases, Fus3 and Kss1, at its base (for reviews, see references 30, 238, 315, and 526). Fus3 and Kss1 are both capable of activating the induction of pheromone-responsive mRNAs, but Fus3 alone has been held responsible for the cell cycle arrest response (103, 151–153, 188). The transcriptional response is thought to result from the phosphorylation of the zinc finger transcription factor Ste12 and two repressors of Ste12, Dig1 and Dig2, by either Fus3 or Kss1 (30, 101, 154, 555). The cell cycle arrest response results, at least in part, from the Fus3 phosphorylation-dependent activation of Far1 and its inhibition of Cln-Cdc28 activity (summarized in Fig. 8).
FIG. 8.
Model for the pheromone response, focusing on regulation of Cln-Cdc28 activity. Conventions are as in Fig. 2.
Pheromone-induced transcription of Cdc28 regulators.
FAR1 is rapidly induced after pheromone exposure. This induction and a large fraction of basal G1 transcription is dependent upon Ste12 (83, 413), a zinc finger transcription factor that activates the expression of many genes required for the mating process. Three Ste12 consensus binding sites (PREs) have been identified in the near-upstream region of FAR1 (83) but seem to be inactive, since deletion of sequences more distal confers low-level, constitutive transcription on FAR1 that is not pheromone inducible (375). This upstream region has not been explored for Ste12 binding activity. The basal activity of Ste12 allows significant expression of FAR1 in vegetatively growing haploids but, in response to pheromone, Ste12-dependent FAR1 transcription increases four- to fivefold. In addition to FAR1, PREs have been noted in the UASs of CLN2 (617) and CLN3 (396). CLN3 mRNA levels increase in response to pheromone but CLN2 transcripts decline (617). There has been no demonstration that Ste12 is involved in the regulation of either gene. The apparent CLN3 induction may be due to a failure to shut off ECB-dependent transcription, which is sensitive to the activity of SBF (399).
Expression of pheromone-inducible genes is antagonized by Cln1-Cdc28 and Cln2-Cdc28, but not by Cln3-Cdc28 or Clb-Cdc28 (411). Levine et al. have isolated a set of cdc28 alleles that affect this function (328). A subset of these alleles was, surprisingly, defective in protein kinase activity and in Cln binding but still retained the ability to repress pheromone-inducible transcription. This does not seem to be due to a dominant negative effect since at least one other kinase-inactive allele of Cdc28 is not able to repress pheromone-inducible transcription. Furthermore, two other Cdc28 mutations, I56T and L61S, in the PSTAIRE helix retain efficient Cln2 binding and display Cln2-dependent protein kinase activity in vitro but eliminate repression of pheromone-inducible transcription (328). These results indicate that the role of Cdc28 in repression of pheromone-induced transcription is not due to its catalytic activity, but an alternative mechanism has not been described.
Repression of Start-specific transcripts.
In response to mating factor, CLN1, CLN2, and CLB6 mRNA levels decline (617). The effects on CLB5 have not been reported, but since CLB5 overexpression can suppress a cln1Δ cln2Δ cln3Δ triple mutant, activate SBF, and suppress mating-pheromone arrest (160, 487), reduced CLB5 transcription is expected. There is no evidence for a specific repression mechanism, and most of the effort in this area has gone toward understanding how the normal cell cycle activation of Start mRNAs is abrogated when cells respond to pheromone. There does not seem to be a single overriding mechanism, but a collection of reinforcing influences keeps Start-specific transcription from initiating. These influences work on both the SBF-dependent and -independent transcription of Start-expressed genes (112, 588).
Inhibition of Cln-Cdc28 activity.
Cln1 and Cln2 protein levels decline rapidly following pheromone exposure (617). The reduction in Cln1 levels is due to the rapid loss of mRNA coupled to the natural instability of Cln1. CLN2 transcript levels reportedly decline much more slowly (617), and when CLN2 mRNA is produced constitutively with the CLN3 promoter, Cln2 protein levels still decline rapidly in response to pheromone in a Far1-dependent manner (588). These results suggest that CLN2 translational efficiency might be reduced or that Cln2 turnover rates increase. In support of the latter mechanism, Cln2 is stable in a cdc28ts background at the restrictive temperature but is rapidly lost upon pheromone addition (617). Using stronger promoters to drive CLN2 expression, other groups did not see this effect (436, 474), and there has been no further characterization of this potential pheromone-induced proteolytic system. The protein kinase activity associated with Cln2 declines even more rapidly than the protein levels (436, 578). This has long been thought to be due to the direct action of Far1 on the Cln-Cdc28 complex, but as discussed above, this has been questioned by a recent publication (see “Far1”) (191).
Cln3.
Regulation of Cln3-Cdc28 activity seems to be a key event. Wild-type strains carrying CLN3stab alleles or cdc34ts strains overexpressing CLN3+ at the permissive temperature are resistant to pheromone-induced cell cycle arrest (112, 131, 214, 396, 578). A similar effect can be seen with CLN2stab alleles but only at lower pheromone concentrations (112, 214). The controls on Cln3 are not fully understood, however. CLN3 transcription is stimulated somewhat by pheromone (617), but Cln3 protein levels decline twofold (264). Reports of pheromonal effects on Cln3-Cdc28 activity have been contradictory (264, 577), but it is now considered likely that Cln3 activity is repressed by pheromone. Simply reducing Cln3-Cdc28 cannot be the whole story, however, since cln3Δ strains still express CLN1 and CLN2 and are pheromone sensitive. Since Start-dependent transcription is dependent upon the activity of any Cln-Cdc28 complex, the direct inhibition of all the Cln-Cdc28 complexes by Far1 could explain the pheromone sensitivity of transcription, but now the finding by Gartner et al. (191) that Cln2-Cdc28 activity is not reduced by Far1 indicates that there may be more going on.
Fus3 and Kss1.
The signal transduction pathway leading from receptor to CDK is not discussed in this review, but some issues relating to the terminal elements of the protein kinase cascade, Fus3 and Kss1, are pertinent. Fus3 and Kss1 are similar MAP kinases with 55% identity. Both are activated in response to pheromone stimulation by phosphorylation on T180 and Y182 by the MAPK kinase Ste7 (164, 192). It is widely reported that Fus3 alone is responsible for the cell cycle arrest response whereas both Fus3 and Kss1 participate in the induction of pheromone-specific transcripts. This result is strain and/or laboratory dependent. In the initial results of Elion et al., fus3 mutants were defective in cell cycle arrest due to pheromone (153). Induction of pheromone-responsive transcripts could still occur in either fus3Δ or kss1Δ mutants but not in fus3Δ kss1Δ double mutants (152). Others, however, have reported that pheromone resistance is not seen or is weak in fus3 strains and that the resistance is considerably stronger in combination with kss1Δ mutations (249, 577). Many laboratory strains are apparently naturally kss1 (152), which may be one source of some of the conflicting results, but this clearly does not explain all the differences, and it is likely that additional modifiers of Fus3 or Kss1 activity remain to be found.
Whether it is Fus3 alone or Fus3 and Kss1 in parallel, these protein kinases are thought to act directly on Ste12 and Far1. Both Ste12 and Far1 are substrates for Fus3 in vitro (154), but direct stimulation of the activity of either substrate has not been demonstrated. Ste12 transcriptional activity is repressed by Dig1 and Dig2, which are also Fus3 and Kss1 substrates in vitro and promote a physical interaction between Ste12 and Fus3 and Kss1 (101, 555). It is hypothesized that phosphorylation of Ste2, Dig1, and Dig2 is needed for full activation of pheromone-inducible transcripts. As discussed above, Fus3 but not Kss1 is needed to activate Far1 inhibitory activity (see “Far1”). With this in mind, the phenotype of a cln3Δ fus3Δ CLN1+ CLN2+ KSS1+ strain is of interest. This strain is pheromone sensitive (152), indicating that Fus3 is not needed when Cln3 is absent and that there must be a parallel route (independent of Far1?) that delivers the mating-pheromone signal to inhibit Cln1-Cdc28 and Cln2-Cdc28. This also implies that the only essential function of Fus3 in the mating-pheromone response is to inhibit Cln3-Cdc28 function and that there is no other activity capable of doing this.
Far3.
The genetic analyses of Chang and Herskowitz (83), Elion et al. (152, 153), and Tyers (576) argue for the existence of at least one additional pheromone-induced inhibitor, and Horecka and Sprague seem to have found one (249). Working in a genetic background in which fus3 mutations do not confer pheromone resistance, they found that mutants with mutations in far3 are pheromone resistant but still allow Ste12-controlled transcription (249). Through the examination of double-mutant combinations, Horecka and Sprague found that far3Δ mutations increase the pheromone resistance of far1Δ and fus3Δ mutants and that while far3Δ cln1Δ and far3Δ cln2Δ double mutants remained pheromone resistant, far3Δ cln1Δ cln2Δ triple mutants and far3Δ cln3Δ double mutants were pheromone sensitive. Far3 is transcriptionally regulated either during the cell cycle or in response to pheromone. It does not seem to affect the transcription of any of the CLN genes, nor does it affect the turnover of their gene products (249), so its function is still mysterious.
Return to the model.
As described above, a cln1Δ cln2Δ cln3Δ sic1Δ mutant is viable (161, 480, 576). Despite lacking all apparent G1 controls on Cdc28, this strain arrests division in response to pheromone. This result is completely unexpected, given the emphasis of the past work on pheromone control of Cln activity, and indicates that attention should be directed at pheromonal control of Clb5 and Clb6 activity. With Sic1 eliminated, the major controls on these B cyclins are transcriptional and proteolytic. The transcriptional controls on CLN1 and CLN2 share many elements with those of CLB5 and CLB6 and could be a common target for regulation by the pheromone response system; therefore, we have included input from Fus3 and Kss1 into the Start-specific transcriptional in the model diagram, although there are no direct data supporting this function. In this fashion, instead of overriding Start, the information from the pheromone response pathway is integrated with the other inputs that govern Start-specific gene expression. This may be necessary because the cell may need a certain level of Cln-Cdc28 activity to promote the directed growth that allows mating partners to contact each other (261, 360, 488), just as it needs Cln-Cdc28 activity at Start to initiate polarized growth in the bud (332). However, the Cln-Cdc28 activity would have to be kept below the level that would promote entry into the mitotic cell cycle. Thus, we should not expect that the activity of the various gene products in this model should be on or off. Such major differences clearly do not occur in this system. There is a constant basal level of activity of Fus3 and Ste12 before pheromonal exposure, and after exposure the activity will vary as a function of pheromone concentration, length of exposure, and cell cycle position. This is an analog, not a digital, system, with multiple input values setting a range of output responses.
Meiosis
Sporulation, the result of meiosis in yeast, initiates in cells starved for nitrogen while growing on a poor carbon source (see reference 314 for a review). In haploid cells, these conditions down-regulate the activity of Cln-Cdc28 and the cells enter stationary phase. In a diploid heterozygous for mating type, Cln-Cdc28 is presumably similarly regulated, but then the DNA replicates and undergoes meiotic recombination, the homologs pair and separate (meiosis I), the sister chromatids separate (meiosis II), and four haploid nuclei are encapsulated into individual spores within the body of the mother cell (the ascus). The mechanics of many of these steps superficially resemble the events of mitosis, and so an involvement of Clb-Cdc28 is expected. Indeed, the B cyclins were originally discovered (168, 546) and many of their properties were deduced in the meiotic cells of metazoans. Therefore, it is somewhat surprising that relatively little is known about Cdc28 regulation during meiosis.
Cdc28 is clearly required for sporulation (507). At restrictive temperatures, a cdc28ts mutant arrests at pachytene prior to meiosis I. At semipermissive temperatures, many cdc28ts mutations efficiently form dyads instead of tetrads. The spores within the dyads are mostly viable and diploid. Genetic analyses indicate that these spores arise from a failure to carry out meiosis II, indicating that meiosis II is particularly sensitive to reduced levels of Cdc28 activity. Dyads are also frequently seen in sporulating cultures of diploids lacking Clb1 function and predominate in clb1Δ clb3Δ, clb1Δ clb4Δ, and clb1Δ clb3Δ clb4Δ homozygotes (115, 209). These dyads also arise from a failure to carry out meiosis II (115). Electron microscopic observations of the sporulating cdc28ts/cdc28ts diploids suggest that dyad formation results from the failure to form a spindle during meiosis II. The SPBs duplicate but do not separate (507). In mitosis, clb1Δ clb2Δ clb3Δ clb4Δ strains also fail to make a spindle (10, 181). These results indicate that the B cyclins have similar functions, relative to spindle formation, in meiosis II to their functions in mitosis but that spindle formation in meiosis I seems to be controlled quite differently. A requirement for Clb5 at an undetermined stage of meiosis has also been reported (160).
At the regulatory level, there are major differences in cyclin function. In contrast to mitosis, in which Clb2 has the dominant role, there is little CLB2 expression in meiosis (209) and clb2Δ mutants have no discernible phenotype (115, 209). Clb1 seems to be playing the dominant role, with support from Clb3 and Clb4. As sporulation progresses, Clb4 protein and its associated protein kinase activity appear first, followed by Clb1 and then Clb3. All three proteins disappear rapidly following meiosis II (209). No changes in activity were observed between meiosis I and meiosis II, but the investigators did not believe that the cells were sufficiently synchronized to permit changes to be detected.
SPO13 has received attention as a potential meiosis-specific regulator of Cdc28 activity (371). Like the cdc28ts mutants under semipermissive conditions and the clb1Δ clb3Δ clb4Δ strains, diploids lacking spo13 activity also produce primarily dyads when sporulated, but the chromosomal segregation that occurs during the single meiotic division is a mixture of reductional and equational divisions (with each chromosome pair in a cell seeming to independently choose which type of separation it will use) (206, 247, 255, 297, 601). Normally, SPO13 is transcribed early in meiosis (67, 531, 542). Overexpression of SPO13 in meiosis delays meiosis I and can suppress the dyad-forming tendency of cdc28ts strains (371). SPO13 overexpression in mitosis arrests the cell cycle with high levels of Cdc28 protein kinase activity, but with a spindle morphology and elongated bud phenotype that suggests that Clb1-Cdc28 and Clb2-Cdc28 kinase is not activated. McCarroll and Esposito (371) have proposed that Spo13 delays meiosis I to allow the chromosomes to be prepared for reductional division and that it later promotes entry into meiosis II. It is not yet clear just how Spo13 and Cdc28 functions are related, but taken together, these results imply that Cdc28 and Spo13 but not Clb1 to Clb4 are needed for meiosis I whereas Cdc28 and Clb1 are required for meiosis II.
REVIEW
With a process as complex as cell division, it is easy to get lost in the details of the mechanics. Even when limited to just the controls placed on the activity of a single (but crucial) component like Cdc28, the complexity can overwhelm. Stepping back from the detail a bit, we can see that two processes—transcription and proteolysis—play dominant roles. While the Cdc28 polypeptide itself seems to play a mostly passive role, the regulators that activate and inhibit Cdc28 are produced by transcriptional waves and removed by specific proteolytic systems timed to match key cell cycle events.
A newborn yeast cell contains a stock of Cln3-Cdc28, Sic1, SBF, and MBF that will take it through most of G1 to Start. This interval is primarily one of growth, but when the cell attains a size appropriate for the growing conditions, Cln3-Cdc28 activates SBF and MBF to transcribe four new cyclins Cln1, Cln2, Clb5, and Clb6, which form complexes with Cdc28. The activity of the Clb5-Cdc28 and Clb6-Cdc28 complexes is kept in check by Sic1, but the rising activity of Cln1-Cdc28 and Cln2-Cdc28 induces budding, SPB duplication, inhibition of Cdh1-dependent APC activity, and the phosphorylation-, ubiquitin-, and SCF-dependent destruction of Sic1 (as well as its own destruction). The loss of Sic1 frees Clb5-Cdc28 and Clb6-Cdc28, allowing it to initiate S phase and SPB separation. A new wave of cyclin synthesis (that of Clb3 and Clb4) begins, which allows the spindle to mature and may induce the synthesis of yet another set of cyclins (Clb1 and Clb2) and a pair of proteolytic regulators, Cdc20 and Cdc5. Clb1 and, particularly, Clb2 complexes with Cdc28 to initiate the early events of mitosis and inhibition of SBF activity. Cdc20 stimulates the APC-mediated breakdown of Pds1, allowing sister chromatid separation, and, along with Cdc5, counteracts the Cdc28-mediated inhibition of Cdh1-activated APC activity. Destruction of Clb1, Clb2, Cdc20, and Cdc5 follows immediately, leading to telophase, Swi5/Ace2-mediated transcription of Sic1, and ECB-mediated increases in Cln3 and Swi4 transcription. This returns the cell to the early-G1 state.
Environmental influences and checkpoint regulators act at several positions to slow or stop these processes. Mating pheromone stimulates the transcription and activation of Far1 to prevent the rise in Cln-Cdc28 activity at Start. Problems with budding act through Swe1 to phosphorylate and inhibit Cdc28 activity. DNA damage and spindle problems act through Cdc20 to prevent the APC-activated proteolysis of Pds1 and Clb proteins. An ever-growing list of additional factors act in diverse ways through these elements to convey other signals to moderate various steps in the cell division process.
CONCLUSION AND FUTURE DIRECTIONS
As studies on the cell cycle of S. cerevisiae continue, we can expect that more details about known components will be revealed and that the influence of new factors will be uncovered. Interactions based initially on genetic data are increasingly being replaced and enriched by biochemical descriptions—a process that is expected to accelerate now that the complete sequence of the yeast genome is known and the ability to produce cellular components and reproduce cellular processes in vitro is improved. At the time this review was being written, the extremely rapid progress in understanding the control of cell cycle-dependent proteolysis was particularly striking. It also seems that a dramatically improved understanding of the connections between Cdc28 activity and checkpoint mechanisms is about to be elucidated. This still leaves some major scientific problems to be solved, however. Despite a long and extensive literature, there does not yet exist a good biochemical description of how periodic transcription is produced. With the exception of pheromonal control, the way in which passage through G1 is controlled by external events is still fairly murky and size control is pretty much a mystery. Despite expectations to the contrary, what we know about Cdc28 regulators has yet to make much of an impact on the studies of yeast alternative life-styles, such as meiosis, invasive and pseudohyphal growth, and stationary phase. Another realm, intentionally avoided in the writing of this article, is the downstream role of Cdc28. How do changes in its partners and its activity actually carry out the events of cell division? Much of the work on Cdc28 function and Cdc28 substrates has been limited to Cdc28 regulators, but progress on downstream events is gaining speed and deserves its own review. The writing, assembly, and illustration of this review also indicated the inadequacy of the written language and two-dimensional figures to describe complex phenomena with multiple inputs and outputs that change with time. The pathways that have already been delineated contain numerous feedback and feed forward loops (the whole process is a loop) that act in negative and positive fashions and often intersect. It is difficult to convey, and usually not possible to know, the relative importance of a given influence under a particular set of conditions. In the end, mathematical modeling of these processes (for example, see reference 581) may be the only way that the entire process can be assembled and the full picture can be viewed.
ACKNOWLEDGMENTS
We thank Marilyn Duncan and Jennifer Williams for critical comments on the manuscript, James Shine and Marcia Ballard for technical assistance, and three anonymous reviewers for wading through the original manuscript and making many critical and helpful comments.
REFERENCES
- 1.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G., Jr Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akada R, Yamamoto J, Yamashita I. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol Gen Genet. 1997;254:267–274. doi: 10.1007/s004380050415. [DOI] [PubMed] [Google Scholar]
- 3.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 4.Althoefer H, Schleiffer A, Wassman K, Nordheim A, Ammerer G. Mcm1 is required to coordinate G2-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5917–5928. doi: 10.1128/mcb.15.11.5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altman R, Kellog D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ammerer G. Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Dev. 1990;4:299–312. doi: 10.1101/gad.4.2.299. [DOI] [PubMed] [Google Scholar]
- 7.Amon A. Regulation of B-type cyclin proteolysis by Cdc28-associated kinases in budding yeast. EMBO J. 1997;16:2693–2702. doi: 10.1093/emboj/16.10.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- 9.Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- 10.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 11.Andersen G, Busso D, Poterszman A, Hwang J R, Wurtz J M, Ripp R, Thierry J C, Egly J M, Moras D. The structure of cyclin H: common mode of kinase activation and specific features. EMBO J. 1997;16:958–967. doi: 10.1093/emboj/16.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen G, Poterszman A, Egly J M, Moras D, Thierry J C. The crystal structure of human cyclin H. FEBS Lett. 1996;397:65–69. doi: 10.1016/s0014-5793(96)01143-x. [DOI] [PubMed] [Google Scholar]
- 13.Anderson M S, Lopes J M. Carbon source regulation of PIS1 gene expression in Saccharomyces cerevisiae involves the MCM1 gene and the two-component regulatory gene, SLN1. J Biol Chem. 1996;271:26596–26601. doi: 10.1074/jbc.271.43.26596. [DOI] [PubMed] [Google Scholar]
- 14.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 15.Andrews B J, Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989;57:21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- 16.Andrews B J, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- 17.Andrews B J, Moore L. Mutational analysis of a DNA sequence involved in linking gene expression to the cell cycle. Biochem Cell Biol. 1992;70:1073–1080. doi: 10.1139/o92-152. [DOI] [PubMed] [Google Scholar]
- 18.Andrews B J, Moore L A. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc Natl Acad Sci USA. 1992;89:11852–11856. doi: 10.1073/pnas.89.24.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apone L M, Virbasius C-M A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 20.Araki H, Awane K, Ogawa N, Oshima Y. The CDC26 gene of Saccharomyces cerevisiae is required for cell growth only at high temperature. Mol Gen Genet. 1992;231:329–331. doi: 10.1007/BF00279807. [DOI] [PubMed] [Google Scholar]
- 21.Arellano M, Moreno S. Regulation of CDK/cyclin complexes during the cell cycle. Int J Biochem Cell Biol. 1997;29:559–573. doi: 10.1016/s1357-2725(96)00178-1. [DOI] [PubMed] [Google Scholar]
- 22.Aristarkhov A, Eytan E, Moghe A, Admon A, Hershko A, Ruderman J V. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc Natl Acad Sci USA. 1996;93:4294–4299. doi: 10.1073/pnas.93.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arndt K T, Styles C A, Fink G R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 24.Arvai A S, Bourne Y, Hickey M J, Tainer J A. Crystal structure of the human cell cycle protein CksHs1: single domain fold with similarity to kinase N-lobe domain. J Mol Biol. 1995;249:835–842. doi: 10.1006/jmbi.1995.0341. [DOI] [PubMed] [Google Scholar]
- 25.Atherton-Fessler S, Parker L L, Geahlen R L, Piwnica-Worms H. Mechanisms of p34cdc2 regulation. Mol Cell Biol. 1993;13:1675–1685. doi: 10.1128/mcb.13.3.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 27.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 28.Bailey R B, Woodward A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- 29.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bardwell L, Cook J G, Inouye C J, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- 31.Baroni M D, Martegani E, Monti P, Alberghina L. Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2715–2723. doi: 10.1128/mcb.9.6.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baroni M D, Monti P, Marconi G, Alberghina L. cAMP-mediated increase in the critical cell size required for the G1 to S transition in Saccharomyces cerevisiae. Exp Cell Res. 1992;201:299–306. doi: 10.1016/0014-4827(92)90277-f. [DOI] [PubMed] [Google Scholar]
- 33.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 34.Basco R D, Segal M D, Reed S I. Negative regulation of G1 and G2 by S-phase cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5030–5042. doi: 10.1128/mcb.15.9.5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beach D, Durkacz B, Nurse P. Functionally homologous cell cycle control genes in fission yeast and budding yeast. Nature. 1982;300:706–709. doi: 10.1038/300706a0. [DOI] [PubMed] [Google Scholar]
- 36.Belanger K D, Kenna M A, Wei S, Davis L I. Genetic and physical interactions between Srp1p and nuclear pore complex proteins Nup1p and Nup2p. J Cell Biol. 1994;126:619–630. doi: 10.1083/jcb.126.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bender A, Sprague G F., Jr MATα1 protein, a yeast transcriptional activator, binds synergistically with a second protein to a set of cell-type specific genes. Cell. 1987;50:681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- 38.Berry L D, Gould K L. Regulation of Cdc2 activity by phosphorylation at T14/Y15. Prog Cell Cycle Res. 1996;2:99–105. doi: 10.1007/978-1-4615-5873-6_10. [DOI] [PubMed] [Google Scholar]
- 39.Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Betting J, Seufert W. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J Biol Chem. 1996;271:25790–25796. doi: 10.1074/jbc.271.42.25790. [DOI] [PubMed] [Google Scholar]
- 41.Bi E, Pringle J R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birck C, Vachette P, Welch M, Swarén P, Samama J-P. Is the function of the cdc2 kinase subunit proteins tuned by their propensities to oligomerize? Conformational states in solution of the cdc2 kinase partners p13suc1 and p9cksphy. Biochemistry. 1996;35:5577–5585. doi: 10.1021/bi952199b. [DOI] [PubMed] [Google Scholar]
- 43.Blondel M, Mann C. G2 cyclins are required for the degradation of G1 cyclins in yeast. Nature. 1996;384:279–282. doi: 10.1038/384279a0. [DOI] [PubMed] [Google Scholar]
- 44.Bobola N, Jansen R-P, Shin T H, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 45.Booher R, Beach D. Interaction between cdc13+ and cdc2+ in the control of mitosis in fission yeast; dissociation of the G1 and G2 roles of the cdc2+ protein kinase. EMBO J. 1987;6:3441–3447. doi: 10.1002/j.1460-2075.1987.tb02667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Booher R N, Alfa C E, Hyams J S, Beach D H. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- 47.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borden K L, Freemont P S. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 49.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins Struct Funct Genet. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 50.Bourne Y, Arvai A S, Bernstein S L, Watson M H, Reed S I, Endicott J E, Noble M E, Johnson L N, Tainer J A. Crystal structure of the cell cycle-regulatory protein suc1 reveals a β-hinge conformational switch. Proc Natl Acad Sci USA. 1995;92:10232–10236. doi: 10.1073/pnas.92.22.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourne Y, Watson M H, Hickey M J, Holmes W, Rocque W, Reed S I, Tainer J A. Crystal structure and mutational analysis of the human CDK2 kinase complex with cell cycle-regulatory protein CksHs1. Cell. 1996;84:863–874. doi: 10.1016/s0092-8674(00)81065-x. [DOI] [PubMed] [Google Scholar]
- 52.Breeden L. Start-specific transcription in yeast. Curr Top Microbiol Immunol. 1996;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- 53.Breeden L, Mikesell G. Three independent forms of regulation affect expression of HO, CLN1 and CLN2 during the cell cycle of Saccharomyces cerevisiae. Genetics. 1994;138:1015–1024. doi: 10.1093/genetics/138.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Breeden L, Mikesell G E. Cell cycle-specific expression of the SWI4 transcription factor is required for the cell cycle regulation of HO transcription. Genes Dev. 1991;5:1183–1190. doi: 10.1101/gad.5.7.1183. [DOI] [PubMed] [Google Scholar]
- 55.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting factors. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 56.Breeden L, Nasmyth K. Similarity between cell-cycle genes of budding yeast and fission yeast and the Notch gene of Drosophila. Nature. 1987;329:651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- 57.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Breter H-J, Ferguson J, Peterson T A, Reed S I. Isolation and transcriptional characterization of three genes which function at Start, the controlling event of the Saccharomyces cerevisiae cell division cycle: CDC36, CDC37, and CDC39. Mol Cell Biol. 1983;3:881–891. doi: 10.1128/mcb.3.5.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 60.Brizuela L, Draetta G, Beach D. p13suc1 acts in the fission yeast cell division cycle as a component of the p34cdc2 protein kinase. EMBO J. 1987;6:3507–3514. doi: 10.1002/j.1460-2075.1987.tb02676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The Saccharomyces cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- 62.Brown J L, Bussey H, Stewart R C. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown J L, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown N R, Noble M E M, Edicott J A, Garman E F, Waklatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson L N. The crystal structure of cyclin A. Structure. 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 65.Bruhn L, Hwang-Shum J-J, Sprague G F., Jr The N-terminal 96 residues of Mcm1, a regulator of cell type-specific genes in Saccharomyces cerevisiae, are sufficient for DNA binding, transcription activation, and interaction with α1. Mol Cell Biol. 1992;12:3563–3572. doi: 10.1128/mcb.12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buck V, Russell P, Millar J B. Identification of a cdk-activating kinase in fission yeast. EMBO J. 1995;14:6173–6183. doi: 10.1002/j.1460-2075.1995.tb00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buckingham L E, Wang H-T, Elder R T, McCarroll R M, Slater M R, Esposito R E. Nucleotide sequence and promoter analysis of SPO13: a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:9406–9410. doi: 10.1073/pnas.87.23.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bueno A, Russell P. Dual functions of CDC6: a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burke D J, Church D. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3691–3698. doi: 10.1128/mcb.11.7.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 71.Butler G, Thiele D J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol Cell Biol. 1991;11:476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Byers B. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. Alfred Benzon Symp. 1981;16:119–133. [Google Scholar]
- 73.Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- 74.Reference deleted.
- 75.Byers B, Sowder L. Gene expression in the yeast cell cycle. J Cell Biol. 1980;87:6. [Google Scholar]
- 76.Cafferkey R, Young P R, McLaughlin M M, Bergsma D J, Koltin Y, Sathe G M, Faucette L, Eng W K, Johnson R K, Livi G P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin toxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caligiuri M, Connolly T, Beach D. Ran1 functions to control the Cdc10/Sct1 complex through Puc1. Mol Biol Cell. 1997;8:1117–1128. doi: 10.1091/mbc.8.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cameron S, Levin L, Zoller M, Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat-shock resistance in S. cerevisiae. Cell. 1988;53:555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- 79.Cannon J F, Tatchell K. Characterization of Saccharomyces cerevisiae genes encoding subunits of cAMP-dependent protein kinase. Mol Cell Biol. 1987;7:2653–2663. doi: 10.1128/mcb.7.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carr A M. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- 81.Carter B L A, Jagadish M N. The relationship between cell size and cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1978;112:15–24. doi: 10.1016/0014-4827(78)90520-7. [DOI] [PubMed] [Google Scholar]
- 82.Carter B L A, Sudbery P E. Small-sized mutants of Saccharomyces cerevisiae. Genetics. 1980;96:561–566. doi: 10.1093/genetics/96.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 84.Chang F, Herskowitz I. Phosphorylation of FAR1 in response to α-factor: a possible requirement for cell-cycle arrest. Mol Biol Cell. 1992;3:445–450. doi: 10.1091/mbc.3.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Charles J F, Jaspersen S L, Tinker-Kulberg R L, Hwang L, Szidon A, Morgan D O. The polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- 86.Chen Y, Tye B-K. The yeast Mcm1 protein is regulated posttranscriptionally by the flux of glycolysis. Mol Cell Biol. 1995;15:4631–4639. doi: 10.1128/mcb.15.8.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chenevert J, Valtz N, Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1297. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi W-J, Clark M W, Chen J X, Jong A Y. The CDC4 gene product is associated with the yeast nuclear skeleton. Biochem Biophys Res Commun. 1990;172:1324–1330. doi: 10.1016/0006-291x(90)91594-i. [DOI] [PubMed] [Google Scholar]
- 89.Choi Y H, Lee S J, Nguyen P, Jang J S, Lee J, Wu M L, Takano E, Maki M, Henkart P A, Trepel J B. Regulation of cyclin D1 by calpain protease. J Biol Chem. 1997;272:28479–28484. doi: 10.1074/jbc.272.45.28479. [DOI] [PubMed] [Google Scholar]
- 90.Christ C, Tye B-K. Functional domains of the yeast transcription/replication factor Mcm1. Genes Dev. 1991;5:751–763. doi: 10.1101/gad.5.5.751. [DOI] [PubMed] [Google Scholar]
- 91.Chun K T, Goebl M G. Mutational analysis of Cak1p, an essential protein kinase that regulates cell cycle progression. Mol Gen Genet. 1997;256:365–375. doi: 10.1007/s004380050580. [DOI] [PubMed] [Google Scholar]
- 92.Ciechanover A, Schwartz A L. The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc Natl Acad Sci USA. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cismowski M J, Laff G M, Solomon M J, Reed S I. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Coche T, Prozzi D, Legrain M, Hilger F, Vandenhaute J. Nucleotide sequence of the PHO81 gene involved in the regulation of the repressible acid phosphatase gene in Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:2176. doi: 10.1093/nar/18.8.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 96.Cohen-Fix O, Peters J-M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 97.Conklin D S, Kung C, Culbertson M R. The COT2 gene is required for glucose-dependent divalent cation transport in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2041–2049. doi: 10.1128/mcb.13.4.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Connell-Crowley L, Solomon M J, Wei N, Harper J W. Phosphorylation independent activation of human cyclin-dependent kinase 2 by cyclin A in vitro. Mol Biol Cell. 1993;4:79–92. doi: 10.1091/mbc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Connolly T, Caligiuri M, Beach D. The Cdc2 protein kinase controls Cdc10/Sct1 complex formation. Mol Biol Cell. 1997;8:1105–1115. doi: 10.1091/mbc.8.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cook J G, Bardwell L, Kron S J, Thorner J. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 1996;10:2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- 102.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 103.Courchesne W E, Kunisawa R, Thorner J. A putative protein kinase overcomes pheromone-induced arrest of cell cycling in S. cerevisiae. Cell. 1989;58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 104.Covitz P A, Mitchell A P. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993;7:1598–1608. doi: 10.1101/gad.7.8.1598. [DOI] [PubMed] [Google Scholar]
- 105.Creasy C, Madden S L, Bergman L W. Molecular analysis of the PHO81 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:1975–1982. doi: 10.1093/nar/21.8.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cross F R. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheromone signalling pathway. Mol Cell Biol. 1990;10:6482–6490. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cross F R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–7684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cross F R. Further characterization of a size control gene in Saccharomyces cerevisiae. J Cell Sci Suppl. 1989;12:117–127. doi: 10.1242/jcs.1989.supplement_12.10. [DOI] [PubMed] [Google Scholar]
- 109.Cross F R, Blake C M. The yeast Cln3 protein is an unstable activator of Cdc28. Mol Cell Biol. 1993;13:3266–3271. doi: 10.1128/mcb.13.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cross F R, Hoek M, McKinney J D, Tinkelenberg A H. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cross F R, Levine K. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol Cell Biol. 1998;18:2923–2931. doi: 10.1128/mcb.18.5.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 113.Cvrcková F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dahmann C, Diffley J F X, Nasmyth K A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 115.Dahmann C, Futcher B. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics. 1995;140:957–963. doi: 10.1093/genetics/140.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Damagnez V, Makela T P, Cottarel G. Schizosaccharomyces pombe Mop1-Mcs2 is related to mammalian CAK. EMBO J. 1995;14:6164–6172. doi: 10.1002/j.1460-2075.1995.tb00307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Das A K, Cohen P W, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.de Bondt H L, Rosenblatt J, Jancarik J, Jones H D, Morgan D O, Kim S-H. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 119.Den Haese G J, Walworth N, Carr A M, Gould K L. The Wee1 protein kinase regulates T14 phosphorylation of fission yeast Cdc2. Mol Biol Cell. 1995;6:371–385. doi: 10.1091/mbc.6.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deshaies R J, Kirschner M. G1 cyclin-dependent activation of p34CDC28 (Cdc28p) in vitro. Proc Natl Acad Sci USA. 1995;92:1182–1186. doi: 10.1073/pnas.92.4.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Detweiler C S, Li J J. Cdc6p establishes and maintains a state of replication competence during G1 phase. J Cell Sci. 1997;110:753–763. doi: 10.1242/jcs.110.6.753. [DOI] [PubMed] [Google Scholar]
- 124.Devault A, Martinez A M, Fesquet D, Labbé J-C, Morin N, Tassan J P, Nigg E A, Cavadore J C, Dorée M. MAT1 (‘menage a trois’): a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 1995;14:5027–5036. doi: 10.1002/j.1460-2075.1995.tb00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Devin A B, Prosvirova T Y, Peshekhonov V T, Chepurnaya O V, Smirnova M E, Koltovaya N A, Troitskaya E N, Arman I P. The start gene CDC28 and the genetic stability of yeast. Yeast. 1990;6:231–243. doi: 10.1002/yea.320060308. [DOI] [PubMed] [Google Scholar]
- 126.Di Como C J, Arndt K T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 127.Di Como C J, Bose R, Arndt K T. Overexpression of SIS2, which contains an extremely acidic region, increases the expression of SWI4, CLN1 and CLN2 in sit4 mutants. Genetics. 1995;139:95–107. doi: 10.1093/genetics/139.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Di Como C J, Chang H, Arndt K T. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dirick L, Moll T, Auer H, Nasmyth K. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature. 1992;357:508–513. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- 131.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 132.Dohmen R J, Stappen R, McGrath J P, Forrová H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 133.Dohrmann P R, Butler G, Tamai K, Dorland S, Greene J R, Thiele D J, Stillman D J. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control HO and chitinase transcription. Genes Dev. 1992;6:93–104. doi: 10.1101/gad.6.1.93. [DOI] [PubMed] [Google Scholar]
- 134.Dohrmann P R, Voth W P, Stillman D J. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol Cell Biol. 1996;16:1746–1758. doi: 10.1128/mcb.16.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dolan J, Kirkman C, Fields S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc Natl Acad Sci USA. 1989;86:5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dolan J W, Fields S. Cell-type-specific transcription in yeast. Biochim Biophys Acta. 1991;1088:155–169. doi: 10.1016/0167-4781(91)90051-m. [DOI] [PubMed] [Google Scholar]
- 137.Donovan J D, Toyn J H, Johnson A L, Johnston L H. p40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- 138.Donovan S, Harwood J, Drury L S, Diffley J F X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dorer R, Pryciak P M, Hartwell L H. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J Cell Biol. 1995;131:845–861. doi: 10.1083/jcb.131.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Draetta G, Luca F, Westendorf J, Brizuela L, Ruderman J, Beach D. cdc2 protein kinase is complexed with both cyclin A and B: evidence for proteolytic inactivation of MPF. Cell. 1989;56:829–838. doi: 10.1016/0092-8674(89)90687-9. [DOI] [PubMed] [Google Scholar]
- 141.Drury L S, Perkins G, Diffley J F X. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ducommun B, Brambilla P, Félix M-A, Franza J, Karsenti B R, E, Draetta G. cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 1991;10:3311–3319. doi: 10.1002/j.1460-2075.1991.tb04895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ducommun P, Brambilla P, Draetta G. Mutations at sites involved in Suc1 binding inactivate Cdc2. Mol Cell Biol. 1991;11:6177–6184. doi: 10.1128/mcb.11.12.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dunphy W G, Brizuela L, Beach D, Newport J. The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- 145.Dunphy W G, Newport J W. Fission yeast p13 blocks mitotic activation and tyrosine dephosphorylation of the Xenopus cdc2 protein kinase. Cell. 1989;58:181–191. doi: 10.1016/0092-8674(89)90414-5. [DOI] [PubMed] [Google Scholar]
- 146.Dutcher S K, Hartwell L H. The role of S. cerevisiae cell division cycle genes in nuclear fusion. Genetics. 1982;100:175–184. doi: 10.1093/genetics/100.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Edgar B A, O’Farrell P H. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Edwards M C, Liegeois N, Horecka J, DePinho R A, Sprague G F, Jr, Tyers M, Elledge S J. Human CPR (cell cycle progression restoration) genes impart a Far− phenotype on yeast cells. Genetics. 1997;147:1063–1076. doi: 10.1093/genetics/147.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Elble R, Tye B-K. Chromosome loss, hyperrecombination, and cell cycle arrest in a yeast mcm1 mutant. Mol Cell Biol. 1992;3:971–980. doi: 10.1091/mbc.3.9.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Elble R, Tye B K. Both activation and repression of a-mating type-specific genes in yeast require transcription factor Mcm1. Proc Natl Acad Sci USA. 1991;88:10966–10970. doi: 10.1073/pnas.88.23.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Elion E A, Brill J A, Fink G R. Functional redundancy in the yeast cell cycle: FUS3 and KSS1 have both overlapping and unique functions. Cold Spring Harbor Symp Quant Biol. 1991;61:41–49. doi: 10.1101/sqb.1991.056.01.007. [DOI] [PubMed] [Google Scholar]
- 152.Elion E A, Brill J A, Fink G R. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc Natl Acad Sci USA. 1991;88:9392–9396. doi: 10.1073/pnas.88.21.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elion E A, Grisafi P L, Fink G R. FUS3 encodes a cdc2+/CDC28-related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 154.Elion E A, Satterberg B, Kranz J E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 156.Elsasser S, Lou F, Wang B, Campbell J L, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Endicott J A, Noble M E, Garman E F, Brown N, Rasmussen B, Nurse P, Johnson L N. The crystal structure of p13suc1, a p34cdc2-interacting cell cycle control protein. EMBO J. 1995;14:1004–1014. doi: 10.1002/j.1460-2075.1995.tb07081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Enoch T, Norbury C. Cellular responses to DNA damage: cell-cycle checkpoints, apoptosis and the roles of p53 and ATM. Trends Biochem Sci. 1995;20:426–430. doi: 10.1016/s0968-0004(00)89093-3. [DOI] [PubMed] [Google Scholar]
- 159.Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- 160.Epstein C B, Cross F R. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 161.Epstein C B, Cross F R. Genes that can bypass the CLN requirement for Saccharomyces cerevisiae cell cycle START. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Erickson J R, Johnston M. Suppressors reveal two classes of glucose repression genes in the yeast Saccharomyces cerevisiae. Genetics. 1994;136:1271–1278. doi: 10.1093/genetics/136.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- 164.Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 165.Espinoza F H, Farrell A, Erdjument-Bromage H, Tempst P, Morgan D O. A cyclin-dependent kinase activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science. 1996;273:1714–1717. doi: 10.1126/science.273.5282.1714. [DOI] [PubMed] [Google Scholar]
- 166.Espinoza F H, Ogas J, Herskowitz I, Morgan D O. Cell cycle control by a complex of the cyclin HCS26 (PCL1) and the kinase PHO85. Science. 1994;266:1388–1391. doi: 10.1126/science.7973730. [DOI] [PubMed] [Google Scholar]
- 167.Evangelista C C, Rodriguez T, Limbach M P, Zitomer S R. ROX3 and RTS1 function in the global stress response pathway in Baker’s yeast. Genetics. 1996;142:1083–1093. doi: 10.1093/genetics/142.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Evans T, Rosenthal E T, Youngblom J, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- 169.Farr K A, Hoyt M A. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fassler J S, Gray W M, Malone C L, Tao W, Lin H, Deschenes R J. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through Hog1 osmosensing pathway. J Biol Chem. 1997;272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- 171.Faye G, Simon M, Valay J G, Fesquet D, Facca C. Rig2, a RING finger protein that interacts with the Kin28/Ccl1 CTD kinase in yeast. Mol Gen Genet. 1997;255:460–466. doi: 10.1007/s004380050518. [DOI] [PubMed] [Google Scholar]
- 172.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 173.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 174.Félix M-A, Labbé J-C, Dorée M, Hunt T, Karsenti E. Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature. 1990;346:379–382. doi: 10.1038/346379a0. [DOI] [PubMed] [Google Scholar]
- 175.Fernandez-Sarabia M J, Sutton A, Zhong T, Arndt K T. SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev. 1992;6:2417–2428. doi: 10.1101/gad.6.12a.2417. [DOI] [PubMed] [Google Scholar]
- 176.Ferrando A, Kron S J, Rios G, Fink G R, Serrano R. Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol Cell Biol. 1995;15:5470–5481. doi: 10.1128/mcb.15.10.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fesquet D, Labbé J-C, Derancourt J, Capony J-P, Galas S, Girard F, Lorca T, Shuttleworth J, Dorée M, Cavadore J-C. The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 1993;12:3111–3121. doi: 10.1002/j.1460-2075.1993.tb05980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Finley D, Özkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperature, starvation, and stress. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 179.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 180.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 181.Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Flick J S, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Flick J S, Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Flick K, Chapman-Shimshoni D, Stuart D, Guaderrama M, Wittenberg C. Regulation of cell size by glucose is exerted via repression of the CLN1 promoter. Mol Cell Biol. 1998;18:2492–2501. doi: 10.1128/mcb.18.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fong H K, Hurley J B, Hopkins R S, Miake-Lye R, Johnson M S, Doolittle R F, Simon M I. Repetitive segmental structure of the transducin beta subunit: homology with the CDC4 gene and identification of related mRNAs. Proc Natl Acad Sci USA. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Foster R, Mikesell G E, Breeden L. Multiple SWI6-dependent cis-acting elements control SWI4 transcription through the cell cycle. Mol Cell Biol. 1993;13:3792–3801. doi: 10.1128/mcb.13.6.3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fujimura H-A. The DAC2/FUS3 protein kinase is not essential for transcriptional activation of the mating pheromone response pathway in Saccharomyces cerevisiae. Mol Gen Genet. 1992;235:450–452. doi: 10.1007/BF00279392. [DOI] [PubMed] [Google Scholar]
- 188.Fujimura H-A. Identification and characterization of a mutation affecting the division arrest signaling of the pheromone response pathway in Saccharomyces cerevisiae. Genetics. 1990;124:275–282. doi: 10.1093/genetics/124.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gallego C, Garí E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Garrels, J. Yeast Protein Database. http://www.proteome.com.
- 191.Gartner A, Jovanovic A, Jeoung D I, Bourlat S, Cross F R, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 193.Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller J L. Cyclin is a component of maturation-promoting factor from Xenopus. Cell. 1990;60:487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- 194.Gerber M R, Farrell A, Deshaies R J, Herskowitz I, Morgan D O. Cdc37 is required for association of the protein kinase Cdc28 with G1 and mitotic cyclins. Proc Natl Acad Sci USA. 1995;92:4651–4655. doi: 10.1073/pnas.92.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Germain D, Hendley J, Futcher B. DNA damage inhibits proteolysis of the B-type cyclin Clb5 in S. cerevisiae. J Cell Sci. 1997;110:1813–1820. doi: 10.1242/jcs.110.15.1813. [DOI] [PubMed] [Google Scholar]
- 196.Ghiara J B, Richardson H E, Sugimoto K, Henze M, Lew D J, Wittenberg C, Reed S I. A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell. 1991;65:163–174. doi: 10.1016/0092-8674(91)90417-w. [DOI] [PubMed] [Google Scholar]
- 197.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutant arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 198.Gibson T J, Thompson J D, Blocker A, Kouzarides T. Evidence for a protein domain superfamily shared by the cyclins, TFIIB and RB/p107. Nucleic Acids Res. 1994;22:946–952. doi: 10.1093/nar/22.6.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 200.Glover D M, Ohkura H, Tavares A. Polo kinase: the choreographer of the mitotic stage? J Biol Chem. 1996;135:1681–1684. doi: 10.1083/jcb.135.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 202.Golsteyn R M, Lane H A, Mundt K E, Arnaud L, Nigg E A. The family of polo-like kinases. Prog Cell Cycle Res. 1996;2:107–114. doi: 10.1007/978-1-4615-5873-6_11. [DOI] [PubMed] [Google Scholar]
- 203.Gorbsky G J. Cell cycle checkpoints: arresting progress in mitosis. Bioessays. 1997;19:193–197. doi: 10.1002/bies.950190303. [DOI] [PubMed] [Google Scholar]
- 204.Gordon C B, Campbell J L. A cell cycle-responsive transcriptional control element and a negative control element in the gene encoding DNA polymerase α in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:6058–6062. doi: 10.1073/pnas.88.14.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gorina S, Pavletich N P. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 206.Gottlieb S, Wagstaff J, Esposito R E. Evidence for two pathways of meiotic recombination in yeast. Proc Natl Acad Sci USA. 1989;86:7072–7076. doi: 10.1073/pnas.86.18.7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gould K L, Moreno S, Owen D J, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gould K L, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- 209.Grandin N, Reed S I. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik H D, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 211.Haber J E, Garvik B. A new gene affecting the efficiency of mating type interconversion in homothallic strains of S. cerevisiae. Genetics. 1977;87:33–50. doi: 10.1093/genetics/87.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hadwiger J A, Reed S I. Invariant phosphorylation of the Saccharomyces cerevisiae Cdc28 protein kinase. Mol Cell Biol. 1988;8:2976–2979. doi: 10.1128/mcb.8.7.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hadwiger J A, Wittenberg C, Mendenhall M D, Reed S I. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol Cell Biol. 1989;9:2034–2041. doi: 10.1128/mcb.9.5.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hadwiger J A, Wittenberg C, Richardson H E, de Barros Lopes M, Reed S I. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hanic-Joyce P J, Johnston G C, Singer R A. Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp Cell Res. 1987;172:134–145. doi: 10.1016/0014-4827(87)90100-5. [DOI] [PubMed] [Google Scholar]
- 217.Hanic-Joyce P J, Singer R A, Johnston G C. Molecular characterization of the yeast PRT1 gene in which mutations affect translation initiation and regulation of cell proliferation. J Biol Chem. 1987;262:2845–2851. [PubMed] [Google Scholar]
- 218.Hanks S K, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 219.Hannig E M. Protein synthesis in eukaryotic organisms: new insights into the function of translation initiation factor eIF-3. Bioessays. 1995;17:915–919. doi: 10.1002/bies.950171103. [DOI] [PubMed] [Google Scholar]
- 220.Hardwick K G. The spindle checkpoint. Trends Genet. 1998;14:1–4. doi: 10.1016/S0168-9525(97)01340-1. [DOI] [PubMed] [Google Scholar]
- 221.Hardwick K G, Murray A W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hardy C F J, Pautz A. A novel role for Cdc5p in DNA replication. Mol Cell Biol. 1996;16:6775–6782. doi: 10.1128/mcb.16.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Harrington L A, Andrews B J. Binding to the yeast Swi4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hartwell L H. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 225.Hartwell L H. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hartwell L H. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hartwell L H, Culotti J, Pringle J, Reid B. Genetic control of the cell division cycle in yeast: a model. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 228.Hartwell L H, Culotti J, Reid B. Genetic control of the cell division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hartwell L H, Mortimer R K, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Healy A M, Zolnierowicz S, Stapleton A E, Goebl M, DePaoli R A, Pringle J R. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol. 1991;11:5767–5780. doi: 10.1128/mcb.11.11.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Heichman K A, Roberts J M. CDC16 controls initiation at chromosome replication origins. Mol Cell. 1998;1:457–463. doi: 10.1016/s1097-2765(00)80046-5. [DOI] [PubMed] [Google Scholar]
- 232.Heichman K A, Roberts J M. The yeast CDC16 and CDC27 genes restrict DNA replication to once per cell cycle. Cell. 1996;85:39–48. doi: 10.1016/s0092-8674(00)81080-6. [DOI] [PubMed] [Google Scholar]
- 233.Heitman J, Movva N R, Hall M N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 234.Helliwell S B, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall M N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologs in yeast. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R J, Peter M. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 237.Hershko A, Ganoth D, Sudakin V, Dahan A, Cohen L H, Luca F C, Ruderman J V, Eytan E. Components of a system that ligates cyclin to ubiquitin and their regulation by the protein kinase cdc2. J Biol Chem. 1994;269:4940–4946. [PubMed] [Google Scholar]
- 238.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 239.Hindley J, Phear G, Stein M, Beach D. Suc1+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol Cell Biol. 1987;7:504–511. doi: 10.1128/mcb.7.1.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hindley J, Phear G A. Sequence of the cell division gene cdc2 from Schizosaccharomyces pombe: pattern of splicing and homology to protein kinases. Gene. 1984;31:129–134. doi: 10.1016/0378-1119(84)90203-8. [DOI] [PubMed] [Google Scholar]
- 241.Hinnebusch A G. Translational control of GCN4: gene specific regulation by phosphorylation of eIF2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1996. pp. 199–244. [Google Scholar]
- 242.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein muc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 243.Hirst K, Fisher F, McAndrew P C, Goding C R. The transcription factor, the Cdk, its cyclin and their regulator: directing the transcriptional response to a nutritional signal. EMBO J. 1994;13:5410–5420. doi: 10.1002/j.1460-2075.1994.tb06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hochstrasser M. There’s the rub: a novel ubiquitin-like modification linked to cell cycle regulation. Genes Dev. 1998;12:901–907. doi: 10.1101/gad.12.7.901. [DOI] [PubMed] [Google Scholar]
- 246.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 247.Hollingsworth N M, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Holmes J K, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 249.Horecka J, Sprague G F., Jr Identification and characterization of FAR3, a gene required for pheromone-mediated G1 arrest in Saccharomyces cerevisiae. Genetics. 1996;144:905–921. doi: 10.1093/genetics/144.3.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hoyt M A, Totis L, Robert B T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 251.Huang D, Farkas I, Roach P. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Huang D, Moffat J, Wilson W A, Moore L, Cheng C, Roach P J, Andrews B. Cyclin partners determine pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by pcl8 and pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Huang K N, Odinsky S A, Cross F R. Structure-function analysis of the Saccharomyces cerevisiae G1 cyclin Cln2. Mol Cell Biol. 1997;17:4654–4666. doi: 10.1128/mcb.17.8.4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Hubler L, Bradshaw-Rouse J, Heideman W. Connections between the ras-cyclic AMP pathway and G1 cyclin expression in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:6274–6282. doi: 10.1128/mcb.13.10.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Hugerat Y, Simchen G. Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics. 1993;135:297–308. doi: 10.1093/genetics/135.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hwang L H, Lau L F, Smith D L, Mistrot C A, Hardwick K G, Hwang E S, Amon A, Murray A W. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 257.Hwang L H, Murray A W. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol Biol Cell. 1997;8:1877–1887. doi: 10.1091/mbc.8.10.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-beta in cells lacking the CDK inhibitor p15. Nature. 1997;387:417–422. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 259.Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- 260.Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–277. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 261.Jackson C L, Hartwell L H. Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 1990;63:1039–1051. doi: 10.1016/0092-8674(90)90507-b. [DOI] [PubMed] [Google Scholar]
- 262.Jarvis E E, Clark K L, Sprague G F. The yeast transcription activator PRTF, homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev. 1989;3:939–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- 263.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 264.Jeoung D-I, Oehlen L J W M, Cross F R. Cln3-associated kinase activity in Saccharomyces cerevisiae is regulated by the mating factor pathway. Mol Cell Biol. 1998;18:433–441. doi: 10.1128/mcb.18.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Jessus C, Ducommun B, Beach D. Direct activation of cdc2 with phosphatase: identification of p13suc1-sensitive and insensitive steps. FEBS Lett. 1990;266:4–8. doi: 10.1016/0014-5793(90)90002-c. [DOI] [PubMed] [Google Scholar]
- 266.Jessus C, Ozon R, editors. Function and regulation of cdc25 protein phosphatase through mitosis and meiosis. Vol. 1. New York, N.Y: Plenum Press; 1995. [DOI] [PubMed] [Google Scholar]
- 267.Jin P, Gu Y, Morgan D O. Role of inhibitory CDC phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Johnson E S, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 269.Johnson E S, Schwienhorst I, Dohmen R J, Blobel G. The ubiquitin-like Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Johnston G C, Ehrhardt C W, Lörincz A T, Carter B L A. Regulation of cell size in the yeast Saccharomyces cerevisiae. J Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Johnston G C, Singer R A, Sharrow S O, Slater M L. Cell division in the yeast Saccharomyces cerevisiae growing at different rates. J Gen Microbiol. 1980;118:479–484. [Google Scholar]
- 272.Johnston L H, Eberly S L, Chapman J W, Araki H, Sugino A. The product of the Saccharomyces cerevisiae cell cycle gene DBF2 has homology with protein kinases and is periodically expressed in the cell cycle. Mol Cell Biol. 1990;10:1358–1366. doi: 10.1128/mcb.10.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Johnston L H, Thomas A P. The isolation of new DNA synthesis mutants in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;186:439–444. doi: 10.1007/BF00729466. [DOI] [PubMed] [Google Scholar]
- 274.Kaffman A, Herskowitz I, Tjian R, O’Shea E K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 275.Kaldis P, Sutton A, Solomon M J. The Cdk-activating kinase (CAK) from budding yeast. Cell. 1996;86:553–564. doi: 10.1016/s0092-8674(00)80129-4. [DOI] [PubMed] [Google Scholar]
- 276.Kalus W, Baumgartner R, Renner C, Noegel A, Chan F K M, Winoto A, Holak T A. NMR structural characterization of the CDK inhibitor p19INK4d. FEBS Lett. 1997;401:127–132. doi: 10.1016/s0014-5793(96)01465-2. [DOI] [PubMed] [Google Scholar]
- 277.Kaplan K B, Hyman A A, Sorger P K. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 278.Kataoka T, Broek D, Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985;43:493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- 279.Kataoka T, Powers S, McGill C, Fasonao O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 280.Keleher C A, Goutte C, Johnson A D. The yeast cell-type-specific repressor α2 acts cooperatively with a non-cell type-specific protein. Cell. 1988;53:927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- 281.Kellog D R, Kikuchi A, Fujii-Nakata T, Turck C W, Murray A W. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kellog D R, Murray A W. NAP1 acts with Clb2 to perform mitotic functions and to suppress polar bud growth in budding yeast. J Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kelly D E, Trevethick J, Mountain H, Sudbery P E. Transcript characterisation, gene disruption and nucleotide sequence of the Saccharomyces cerevisiae WHI2 gene. Gene. 1988;66:205–213. doi: 10.1016/0378-1119(88)90357-5. [DOI] [PubMed] [Google Scholar]
- 284.Keng T, Clark M W, Storms R K, Fortin N, Zhong W, Ouellette B F F, Barton A B, Kaback D B, Bussey H. LTE1 of Saccharomyces cerevisiae is a 1435 codon open reading frame that has sequence similarities to guanine nucleotide releasing factors. Yeast. 1994;10:953–958. doi: 10.1002/yea.320100710. [DOI] [PubMed] [Google Scholar]
- 285.Khadake J R, Rao M R. Condensation of DNA and chromatin by an SPKK-containing octapeptide repeat motif present in the C-terminus of histone H1. Biochemistry. 1997;36:1041–1051. doi: 10.1021/bi961617p. [DOI] [PubMed] [Google Scholar]
- 286.Kharbanda S, Saleem A, Datta R, Yuan Z, Weichselbaum R, Kufe D. Ionizing radiation induces rapid tyrosine phosphorylation of p34cdc2. Cancer Res. 1994;54:1412–1414. [PubMed] [Google Scholar]
- 287.Kim H-Y, Cho Y. Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box. Nat Struct Biol. 1997;4:390–395. doi: 10.1038/nsb0597-390. [DOI] [PubMed] [Google Scholar]
- 288.Kim K K, Chamberlin H M, Morgan D O, Kim S-H. Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase. Nat Struct Biol. 1996;3:849–855. doi: 10.1038/nsb1096-849. [DOI] [PubMed] [Google Scholar]
- 289.Kim S, Weinert T A. Characterization of the checkpoint gene RAD53/MEC2 in Saccharomyces cerevisiae. Yeast. 1997;13:735–745. doi: 10.1002/(SICI)1097-0061(19970630)13:8<735::AID-YEA136>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 290.Kimura Y, Rutherford S L, Miyata Y, Yahara I, Freeman B C, Yue L, Morimoto R I, Lindquist S. Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev. 1997;11:1775–1785. doi: 10.1101/gad.11.14.1775. [DOI] [PubMed] [Google Scholar]
- 291.King R W, Peters J-M, Tugendreich S, Rolfe M, Hieter P, Kirschner M W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 292.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 293.Kirkman-Crreia C, Stroke I L, Fields S. Functional domains of the yeast STE12 protein, a pheromone-responsive transcriptional activator. Mol Cell Biol. 1993;13:3765–3772. doi: 10.1128/mcb.13.6.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kishi T, Seno T, Yamao F. Grr1 functions in the ubiquitin pathway in Saccharomyces cerevisiae through association with Skp1. Mol Gen Genet. 1998;257:143–148. doi: 10.1007/s004380050633. [DOI] [PubMed] [Google Scholar]
- 295.Kitada K, Johnson A L, Johnston L H, Sugino A. A multicopy suppressor of the Saccharomyces cerevisiae G1 cell cycle gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kitagawa M, Higashi H, Jung H-K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J-Y, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 297.Klapholz S, Esposito R E. Recombination and chromosome segregation during the single division meiosis in spo12-1 and spo13-1 diploids. Genetics. 1980;96:589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Knapp D, Bhoite L, Stillman D J, Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol Cell Biol. 1996;16:5701–5707. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Knighton D R, Zheng J, Ten Eyck L F, Ashford V A, Xuong N-h, Taylor S S, Sowadski J M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 300.Kobayashi H, Stewart E, Poon R, Adamczewski J P, Gannon J, Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol Biol Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 302.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 303.Kolman C J, Toth J, Gonda D K. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Koltovaya N A, Arman I P, Devin A B. Mutations of the CDC28 gene and the radiation sensitivity of Saccharomyces cerevisiae. Yeast. 1998;14:133–146. doi: 10.1002/(SICI)1097-0061(19980130)14:2<133::AID-YEA206>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 305.Kramer K M, Fesquet D, Johnson A L, Johnston L H. Budding yeast RSI1/APC2, a novel gene necessary for initiation of anaphase, encodes an APC subunit. EMBO J. 1998;17:498–506. doi: 10.1093/emboj/17.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Krek W, Nigg E A. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 1991;10:3331–3341. doi: 10.1002/j.1460-2075.1991.tb04897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Krems B, Charizanis C, Entian K-D. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- 308.Kronstad J W, Holly J A, MacKay V L. A yeast operator overlaps an upstream activation site. Cell. 1987;50:369–377. doi: 10.1016/0092-8674(87)90491-0. [DOI] [PubMed] [Google Scholar]
- 309.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kühne C, Linder P. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 1993;12:3437–3447. doi: 10.1002/j.1460-2075.1993.tb06018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 312.Kuo M, Grayhack E. A library of yeast genomic MCM1 binding sites contains genes involved in cell cycle control, cell wall and membrane structure, and metabolism. Mol Cell Biol. 1994;14:348–359. doi: 10.1128/mcb.14.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Kuo M-H, Nadeau E T, Grayhack E J. Multiple phosphorylated forms of the Saccharomyces cerevisiae Mcm1 protein include an isoform induced in response to high salt concentration. Mol Cell Biol. 1997;17:819–832. doi: 10.1128/mcb.17.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kupiec M, Byers B, Esposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 315.Kurjan J. Pheromone response in yeast. Annu Rev Biochem. 1992;61:1097–1129. doi: 10.1146/annurev.bi.61.070192.005313. [DOI] [PubMed] [Google Scholar]
- 316.Kusubata M, Tokui T, Matsuoka Y, Okumura E, Tachibana K, Hisanaga S-I, Kishimoto T, Yasuda H, Kamijo M, Ohba Y, Tsujimura K, Yatani R, Inagaki M. p13suc1 suppresses the catalytic function of p34cdc2 kinase for intermediate filament proteins, in vitro. J Biol Chem. 1992;267:20937–20942. [PubMed] [Google Scholar]
- 317.Labbé J-C, Cavadore J-C, Dorée M. M phase-specific cdc2 kinase: preparation from starfish oocytes and properties. Methods Enzymol. 1991;200:291–301. doi: 10.1016/0076-6879(91)00147-o. [DOI] [PubMed] [Google Scholar]
- 318.Lahav-Baratz S, Sudakin V, Ruderman J V, Hershko A. Reversible phosphorylation controls the activity of cyclosome associated cyclin-ubiquitin ligase. Proc Natl Acad Sci USA. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Lamb J R, Michaud W A, Sikorski R S, Hieter P A. Cdc16p, Cdc23p, and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Langan T A, Gautier J, Lohka M, Hollingsworth R, Moreno S, Nurse P, Maller J, Sclafani R A. Mammalian growth-associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol Cell Biol. 1989;9:3860–3868. doi: 10.1128/mcb.9.9.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Lanker S, Valdivieso H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 323.Lee D H, Sherman M Y, Goldberg A L. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lee J M, Greenleaf A L. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Exp. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- 325.Lee M G, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 326.Lemire J M, Willcocks T, Halvorson H O, Bostian K A. Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1985;8:2131–2141. doi: 10.1128/mcb.5.8.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Levine K, Huang K, Cross F R. Saccharomyces cerevisiae G1 cyclins differ in their intrinsic functional specificities. Mol Cell Biol. 1996;16:6794–6803. doi: 10.1128/mcb.16.12.6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Levine K, Oehlen L J W M, Cross F R. Isolation and characterization of new alleles of the cyclin-dependent kinase gene CDC28 with cyclin-specific functional and biochemical defects. Mol Cell Biol. 1998;18:290–302. doi: 10.1128/mcb.18.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 330.Lew D J, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 331.Lew D J, Reed S I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Li R, Murray A W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 335.Li X, Cai M. Inactivation of the cyclin-dependent kinase Cdc28 abrogates cell cycle arrest induced by DNA damage and disassembly of mitotic spindles in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2723–2734. doi: 10.1128/mcb.17.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Li Y, Bjorklund S, Jiang Y W, Kim Y-J, Lane W S, Stillman D J, Kornberg R D. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 339.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 340.Lim H H, Goh P Y, Surana U. Cdc20 is essential for the cyclosome-mediated proteolysis of both Pds1 and Clb2 during M phase in budding yeast. Curr Biol. 1998;8:231–234. doi: 10.1016/s0960-9822(98)70088-0. [DOI] [PubMed] [Google Scholar]
- 341.Lim H H, Goh P-Y, Surana U. Spindle poly body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28. Mol Cell Biol. 1996;16:6385–6397. doi: 10.1128/mcb.16.11.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Lim H H, Loy C J, Zaman S, Surana U. Dephosphorylation of threonine 169 of Cdc28 is not required for exit from mitosis but may be necessary for Start in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4573–4583. doi: 10.1128/mcb.16.8.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Lim H H, Surana U. Cdc20, a β-transducin homologue, links RAD9-mediated G2/M checkpoint control to mitosis in Saccharomyces cerevisiae. Mol Gen Genet. 1996;253:138–148. doi: 10.1007/s004380050306. [DOI] [PubMed] [Google Scholar]
- 344.Lin F C, Arndt K. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 1995;14:2745–2759. doi: 10.1002/j.1460-2075.1995.tb07275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Lisziewicz J, Godany A, Agoston D V, Kuntzel H. Cloning and characterization of the Saccharomyces cerevisiae CDC6 gene. Nucleic Acids Res. 1988;16:11507–11520. doi: 10.1093/nar/16.24.11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Lodish H F. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 347.Loeb J D J, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G R. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Lohka M J, Hayes M K, Maller J L. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc Natl Acad Sci USA. 1988;85:3009–3013. doi: 10.1073/pnas.85.9.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Lorca T, Labbé J-C, Devault A, Fesquet D, Capony J P, Cavadore J C, Le Bouffant F, Dorée M. Dephosphorylation of cdc2 on threonine 161 is required for cdc2 inactivation and normal anaphase. EMBO J. 1992;11:2381–2390. doi: 10.1002/j.1460-2075.1992.tb05302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Lörincz A, Carter B L A. Control of cell size at bud initiation in Saccharomyces cerevisiae. J Gen Microbiol. 1979;113:287–295. [Google Scholar]
- 351.Lörincz A T, Reed S I. Primary structure homology between the product of yeast cell division control gene CDC28 and vertebrate oncogenes. Nature. 1984;307:183–185. doi: 10.1038/307183a0. [DOI] [PubMed] [Google Scholar]
- 352.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 353.Lowndes N F, Johnson A L, Breeden L, Johnston L H. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature. 1992;357:505–508. doi: 10.1038/357505a0. [DOI] [PubMed] [Google Scholar]
- 354.Lowndes N F, Johnson A L, Johnston L H. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans factor. Nature. 1991;350:247–250. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- 355.Luca F C, Shibuya E K, Dohrmann C E, Ruderman J V. Both cyclin AΔ60 and BΔ97 are stable and arrest cells in M-phase, but only cyclin BΔ97 turns on cyclin destruction. EMBO J. 1991;10:4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Luh F Y, Archer S J, Domaille P J, Smith B O, Owen D, Brotherton D H, Raine A R C, Xu X, Brizuela L, Brenner S L, Laue E D. Structure of the cyclin-dependent kinase inhibitor p19Ink4d. Nature. 1997;389:999–1003. doi: 10.1038/40202. [DOI] [PubMed] [Google Scholar]
- 357.Luke M M, Della Seta F, Di Como C J, Kobayashi R, Arndt K T. The SAPs, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol. 1996;16:2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Lydall D, Ammerer G, Nasmyth K. A new role for MCM1 in yeast: cell cycle regulation of SWI5 transcription. Genes Dev. 1991;5:2405–2419. doi: 10.1101/gad.5.12b.2405. [DOI] [PubMed] [Google Scholar]
- 359.Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 360.Madden K, Snyder M. Specification of sites for polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol Cell Biol. 1992;3:1025–1035. doi: 10.1091/mbc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 362.Maher M, Cong F, Kindelberger D, Nasmyth K, Dalton S. Cell cycle-regulated transcription of the CLB2 gene is dependent on Mcm1 and a ternary complex factor. Mol Cell Biol. 1995;15:3129–3137. doi: 10.1128/mcb.15.6.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Mai B, Breeden L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol. 1997;17:6491–6501. doi: 10.1128/mcb.17.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Maine G, Sinha P, Tye B-K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Mäkelä T P, Tassan J-P, Nigg E A, Frutiger S, Hughes G J, Weinberg R A. A cyclin associated with the CDK-activating kinase MO15. Nature. 1994;371:254–257. doi: 10.1038/371254a0. [DOI] [PubMed] [Google Scholar]
- 366.Marini N J, Reed S I. Direct induction of G1-specific transcripts following reactivation of the Cdc28 kinase in the absence of de novo protein synthesis. Genes Dev. 1992;6:557–567. doi: 10.1101/gad.6.4.557. [DOI] [PubMed] [Google Scholar]
- 367.Markwardt D D, Garrett J M, Eberhardy S, Heideman W. Activation of the Ras/cyclic AMP pathway in the yeast Saccharomyces cerevisiae does not prevent G1 arrest in response to nitrogen starvation. J Bacteriol. 1995;177:6761–6765. doi: 10.1128/jb.177.23.6761-6765.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Mathias N, Johnson S L, Winey M, Adams A E M, Goetsch L, Pringle J R, Byers B, Goebl M G. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Mathias N, Steussy C N, Goebl M G. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- 370.Matsumoto K, Uno I, Oshima Y, Ishikawa T. Isolation and characterization of yeast mutants defective in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1982;79:2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.McCarroll R M, Esposito R E. SPO13 negatively regulates the progression of mitotic and meiotic nuclear division in Saccharomyces cerevisiae. Genetics. 1994;138:47–60. doi: 10.1093/genetics/138.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.McGrath J P, Jentsch S, Varshavsky A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.McInerny C J, Partridge J F, Mikesell G E, Creemer D P, Breeden L L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- 374.McIntosh E M, Atkinson T, Storms R K, Smith M. Characterization of a short, cis-acting DNA sequence which conveys cell cycle stage-dependent transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:329–337. doi: 10.1128/mcb.11.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.McKinney J D, Chang F, Heintz N, Cross F R. Negative regulation of FAR1 at the Start of the yeast cell cycle. Genes Dev. 1993;7:833–843. doi: 10.1101/gad.7.5.833. [DOI] [PubMed] [Google Scholar]
- 376.McKinney J D, Cross F R. FAR1 and the G1 phase specificity of cell cycle arrest by mating factor in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2509–2516. doi: 10.1128/mcb.15.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Measday V, Moore L, Ogas J, Tyers M, Andrews B. The PCL2 (ORFD)-PHO85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science. 1994;266:1391–1395. doi: 10.1126/science.7973731. [DOI] [PubMed] [Google Scholar]
- 378.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B A. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Meijer L, Arion D, Golsteyn R, Pines J, Brizuela L, Hunt T, Beach D. Cyclin is a component of the sea urchin egg M-phase specific histone H1 kinase. EMBO J. 1989;8:2275–2282. doi: 10.1002/j.1460-2075.1989.tb08353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Mendenhall M D. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 381.Mendenhall M D, Al-jumaily W, Nugroho T T. The Cdc28 inhibitor p40Sic1. Prog Cell Cycle Res. 1995;1:173–185. doi: 10.1007/978-1-4615-1809-9_14. [DOI] [PubMed] [Google Scholar]
- 382.Mendenhall M D, Jones C A, Reed S I. Dual regulation of the yeast CDC28-p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell. 1987;50:927–935. doi: 10.1016/0092-8674(87)90519-8. [DOI] [PubMed] [Google Scholar]
- 383.Mendenhall M D, Richardson H E, Reed S I. Dominant negative protein kinase mutations that confer a G1 arrest phenotype. Proc Natl Acad Sci USA. 1988;85:4426–4430. doi: 10.1073/pnas.85.12.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Minshull J, Straight A, Rudner A D, Dernburg A F, Belmont A, Murray A W. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- 385.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 387.Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- 388.Morgan B A, Banks G R, Toone W M, Raitt D, Kuge S, Johnston L H. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Morgan B A, Bouquin N, Merrill G F, Johnston L H. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 1995;14:5679–5689. doi: 10.1002/j.1460-2075.1995.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 391.Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 1995;270:1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- 392.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Mountain H A, Sudbery P E. Regulation of the Saccharomyces cerevisiae WHI2 gene. J Gen Microbiol. 1990;136:727–732. doi: 10.1099/00221287-136-4-727. [DOI] [PubMed] [Google Scholar]
- 394.Mountain H A, Sudbery P E. The relationship of growth rate and catabolite repression with WHI2 expression and cell size in Saccharomyces cerevisiae. J Gen Microbiol. 1990;136:733–737. doi: 10.1099/00221287-136-4-733. [DOI] [PubMed] [Google Scholar]
- 395.Murray A W, Solomon M J, Kirschner M W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 396.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Nasmyth K. A repetitive DNA sequence that confers cell cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell. 1985;42:225–235. doi: 10.1016/s0092-8674(85)80118-5. [DOI] [PubMed] [Google Scholar]
- 398.Nasmyth K, Adolf G, Lydall D, Seddon A. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- 399.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 400.Nasmyth K, Seddon A, Ammerer G. Cell cycle regulation of SWI5 is required for mother-cell-specific HO transcription in yeast. Cell. 1987;49:549–558. doi: 10.1016/0092-8674(87)90457-0. [DOI] [PubMed] [Google Scholar]
- 401.Nasmyth K A. Molecular analysis of a cell lineage. Nature. 1983;302:670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- 402.Nasmyth K A, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- 403.Nasmyth K A, Reed S I. The isolation of genes by complementation in yeast: the molecular cloning of a cell cycle gene. Proc Natl Acad Sci USA. 1980;77:2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Neer E J, Schmidt C J, Nambudripad R, Smith T F. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 405.Nickels J T, Broach J R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 406.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 407.Noble M E M, Endicott J A, Brown N R, Johnson L N. The cyclin box fold: protein recognition in cell-cycle and transcription control. Trends Biochem Sci. 1997;22:482–487. doi: 10.1016/s0968-0004(97)01144-4. [DOI] [PubMed] [Google Scholar]
- 408.Norbury C, Blow J, Nurse P. Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. EMBO J. 1991;10:3321–3329. doi: 10.1002/j.1460-2075.1991.tb04896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Nugroho T T, Mendenhall M D. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol Cell Biol. 1994;14:3320–3328. doi: 10.1128/mcb.14.5.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980;96:627–637. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Oehlen L J W M, Cross F R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 412.Oehlen L J W M, Jeoung D-I, Cross F R. Cyclin-specific START events and the G1-phase specificity of arrest by mating factor in budding yeast. Mol Gen Genet. 1998;258:183–198. doi: 10.1007/s004380050722. [DOI] [PubMed] [Google Scholar]
- 413.Oehlen L J W M, McKinney J D, Cross F R. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol Cell Biol. 1996;16:2830–2837. doi: 10.1128/mcb.16.6.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Ogas J, Andrews B J, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 415.Ogawa N, Noguchi K-I, Sawai H, Yamashita Y, Yompakdee C, Oshima Y. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Ogawa N, Noguchi K-I, Yamashita Y, Yashuhara T, Hayashi N, Yoshida K, Oshima Y. Promoter analysis of the PHO81 gene encoding a 134 kDa protein bearing ankyrin repeats in the phosphatase regulon of Saccharomyces cerevisiae. Mol Gen Genet. 1993;238:444–454. doi: 10.1007/BF00292004. [DOI] [PubMed] [Google Scholar]
- 417.Ollendorff V, Donoghue D J. The serine/threonine phosphatase PP5 interacts with CDC16 and CDC27, two tetratricopeptide repeat-containing subunits of the anaphase- promoting complex. J Biol Chem. 1997;272:32011–32018. doi: 10.1074/jbc.272.51.32011. [DOI] [PubMed] [Google Scholar]
- 418.O’Neill E M, Kaffman A, Jolly E R, O’Shea E K. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 419.Ord R W, McIntosh E M, Lee L, Poon P P, Storms R K. Multiple elements regulate expression of the cell cycle-regulated thymidylate synthase gene of Saccharomyces cerevisiae. Curr Genet. 1988;14:363–373. [Google Scholar]
- 420.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 421.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Özcan S, Schulte F, Freidel K, Weber A, Ciriacy M. Glucose uptake and metabolism in grr1/cat80 mutants of Saccharomyces cerevisiae. Eur J Biochem. 1994;224:605–611. doi: 10.1111/j.1432-1033.1994.00605.x. [DOI] [PubMed] [Google Scholar]
- 423.Özkaynak E, Finley D, Solomon M J, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Palmer R E, Koval M, Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989;109:3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Parge H E, Arvai A S, Murtari D J, Reed S I, Tainer J A. Human CksHs2 atomic structure: a role for its hexameric assembly in cell cycle control. Science. 1993;262:387–395. doi: 10.1126/science.8211159. [DOI] [PubMed] [Google Scholar]
- 426.Parkes V, Johnston L H. SPO12 and SIT4 suppress mutations in DBF2, which encodes a protein kinase that is periodically expressed. Nucleic Acids Res. 1992;20:5617–5623. doi: 10.1093/nar/20.21.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Partridge J F, Mikesell G E, Breeden L L. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J Biol Chem. 1997;272:9071–9077. doi: 10.1074/jbc.272.14.9071. [DOI] [PubMed] [Google Scholar]
- 428.Passmore S, Maine G, Elble R, Christ C, Tye B-K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J Mol Biol. 1988;204:593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- 429.Patra D, Dunphy W G. Xe-p9, a Xenopus Suc1/Csk homolog, has multiple essential roles in cell cycle control. Genes Dev. 1996;10:1503–1515. doi: 10.1101/gad.10.12.1503. [DOI] [PubMed] [Google Scholar]
- 430.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 432.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 433.Pavletich N P, Pabo C O. Zinc finger-DNA-recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 434.Pelissier P, Camougrand N, Velours G, Guerin M. NCA3, a gene involved in the mitochondrial expression of subunits 6 and 8 of the F0-F1 ATP synthase of S. cerevisiae. Curr Genet. 1995;5:409–416. doi: 10.1007/BF00311209. [DOI] [PubMed] [Google Scholar]
- 435.Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- 436.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 437.Piatti S, Böhm T, Cocker J H, Diffley J F X, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 438.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a “reductional” anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Piggot J R, Rai B, Carter B L A. A bifunctional gene product involved in two phases of the yeast cell cycle. Nature. 1982;298:391–393. doi: 10.1038/298391a0. [DOI] [PubMed] [Google Scholar]
- 440.Pines J. Cyclins and cyclin-dependent kinases: a biochemical view. Biochem J. 1995;308:697–711. doi: 10.1042/bj3080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Pitluk Z W, McDonough M, Sangan P, Gonda D K. Novel CDC34 (UBC3) ubiquitin-conjugating enzyme mutants obtained by charge-to-alanine scanning mutagenesis. Mol Cell Biol. 1995;15:1210–1219. doi: 10.1128/mcb.15.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Polymenis M, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Poon R Y C, Hunter T. Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science. 1995;270:90–93. doi: 10.1126/science.270.5233.90. [DOI] [PubMed] [Google Scholar]
- 444.Poon R Y C, Yamashita K, Adamczewski J P, Hunt T, Shuttleworth J. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 1993;12:3123–3132. doi: 10.1002/j.1460-2075.1993.tb05981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 446.Primig M, Sockanathan S, Auer H, Nasmyth K. Anatomy of a transcription factor important for the Start of the cell cycle in Saccharomyces cerevisiae. Nature. 1992;358:593–597. doi: 10.1038/358593a0. [DOI] [PubMed] [Google Scholar]
- 447.Primig M, Winkler H, Ammerer G. The DNA binding and oligomerization domain of Mcm1 is sufficient for its interaction with other regulatory proteins. EMBO J. 1991;10:4209–4218. doi: 10.1002/j.1460-2075.1991.tb04999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Pringle J R, Hartwell L H. The Saccharomyces cerevisiae cell cycle. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 97–142. [Google Scholar]
- 449.Prinz S, Hwang E S, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- 450.Ptak C, Prendergast J A, Hodgins R, Kay C M, Chau V, Ellison M J. Functional and physical characterization of the cell cycle ubiquitin-conjugating enzyme CDC34 (UBC3). Identification of a functional determinant within the tail that facilitates CDC34 self-association. J Biol Chem. 1994;269:26539–26545. [PubMed] [Google Scholar]
- 451.Radcliffe P, Trevethick J, Tyers M, Sudbery P. Deregulation of CLN1 and CLN2 in the Saccharomyces cerevisiae whi2 mutant. Yeast. 1997;13:707–715. doi: 10.1002/(SICI)1097-0061(19970630)13:8<707::AID-YEA130>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 452.Rahman D R J, Sudbery P E, Kelly S, Marison I W. The effect of dissolved oxygen concentration of the growth physiology of Saccharomyces cerevisiae whi2 mutants. J Gen Microbiol. 1988;134:2241–2248. doi: 10.1099/00221287-134-8-2241. [DOI] [PubMed] [Google Scholar]
- 453.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 454.Reed S. Genetic and molecular analysis of division control in Saccharomyces. In: Nurse P, Streiblova E, editors. The microbial cell cycle. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 89–108. [Google Scholar]
- 455.Reed S I. The selection of amber mutations in genes required for completion of start, the controlling event of the cell division cycle of S. cerevisiae. Genetics. 1980;95:579–588. doi: 10.1093/genetics/95.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Reed S I. The selection of S. cerevisiae mutants defective in the Start event of cell division. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Reed S I, Hadwiger J A, Lörincz A T. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc Natl Acad Sci USA. 1985;82:4055–4059. doi: 10.1073/pnas.82.12.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Reed S I, Wittenberg C. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:5697–5701. doi: 10.1073/pnas.87.15.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Reid B J, Hartwell L H. Regulation of mating in the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1977;75:355–365. doi: 10.1083/jcb.75.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Richardson H E, Lew D J, Henze M, Sugimoto K, Reed S I. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- 461.Richardson H E, Stueland C S, Thomas J, Russell P, Reed S I. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 1990;4:1332–1344. doi: 10.1101/gad.4.8.1332. [DOI] [PubMed] [Google Scholar]
- 462.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 463.Richter-Ruoff B, Wolf D H. Proteasome and cell cycle. Evidence for a regulatory role of the protease on mitotic cyclins in yeast. FEBS Lett. 1993;336:34–36. doi: 10.1016/0014-5793(93)81603-w. [DOI] [PubMed] [Google Scholar]
- 464.Robinson L C, Gibbs J B, Marshall M S, Sigal I S, Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987;235:1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- 465.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 466.Ronne H, Carlberg M, Hu G-Z, Nehlin J O. Protein phosphatase 2A in S. cerevisiae: effects on cell growth and bud morphogenesis. Mol Cell Biol. 1991;11:4876–4884. doi: 10.1128/mcb.11.10.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Rowley A, Johnston G C, Butler B, Werner-Washburne M, Singer R A. Heat shock-mediated cell cycle blockage and G1 cyclin expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1034–1041. doi: 10.1128/mcb.13.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Roy R, Adamczeski J P, Seroz T, Vermeulen W, Tassan J-P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J-M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 469.Russell P, Moreno S, Reed S I. Conservation of mitotic controls in fission and budding yeasts. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- 470.Russell P, Nurse P. cdc25+ functions as an inducer in the mititoc control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- 471.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 472.Russo A A, Jeffrey P D, Pavletich N P. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 473.Saha P, Eichbaum Q, Silberman E D, Mayer B J, Dutta A. p21CIP1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4338–4345. doi: 10.1128/mcb.17.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Salama S R, Hendricks K B, Thorner J. G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol Cell Biol. 1994;14:7953–7966. doi: 10.1128/mcb.14.12.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 476.Sanchez-Diaz A, Gonzalez I, Arellano M, Moreno S. The cdk inhibitors p25rum1 and p40SIC1 are functional homologues that play similar roles in the regulation of the cell cycle in fission and budding yeast. J Cell Sci. 1998;111:843–851. doi: 10.1242/jcs.111.6.843. [DOI] [PubMed] [Google Scholar]
- 477.Santos R C, Waters N C, Creasy C L, Bergman L W. Structure-function relationships of the yeast cyclin-dependent kinase Pho85. Mol Cell Biol. 1995;15:5482–5491. doi: 10.1128/mcb.15.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Saul D J, Sudbery P E. Molecular cloning of WHI2, a gene involved in the regulation of cell-proliferation in Saccharomyces cerevisiae. J Gen Microbiol. 1985;131:1797–1806. doi: 10.1099/00221287-131-7-1797. [DOI] [PubMed] [Google Scholar]
- 479.Saul D J, Walton E F, Sudbery P E, Carter B L A. Saccharomyces cerevisiae whi2 mutants in stationary phase retain the properties of exponentially growing cells. J Gen Microbiol. 1985;131:2245–2251. [Google Scholar]
- 480.Schneider B L, Yang Q-H, Futcher A B. Linkage of replication to Start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 481.Schneider K R, Smith R L, O’Shea E K. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 482.Schott E J, Hoyt M A. Dominant alleles of Saccharomyces cerevisiae CDC20 reveal its role in promoting anaphase. Genetics. 1998;148:599–610. doi: 10.1093/genetics/148.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Schwab M, Lutum A S, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 484.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Schweitzer B, Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991;7:265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- 486.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 487.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 488.Segall J E. Polarization of yeast cells in spatial gradients of à mating factor. Proc Natl Acad Sci USA. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 490.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 491.Sethi N, Monteagudo M C, Koshland D, Hogan E, Burke D J. The CDC20 gene product of Saccharomyces cerevisiae, a β-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol Cell Biol. 1991;11:5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 493.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Shenoy S, Choi J-K, Bagrodia S, Copeland T D, Maller J M, Shalloway D. Purified maturation promoting factor phosphorylates pp60c-src at sites phosphorylated during fibroblast mitosis. Cell. 1989;57:763–774. doi: 10.1016/0092-8674(89)90791-5. [DOI] [PubMed] [Google Scholar]
- 495.Sherlock G, Rosamond J. Starting to cycle: G1 controls regulating cell division in budding yeast. J Gen Microbiol. 1993;139:2531–2541. doi: 10.1099/00221287-139-11-2531. [DOI] [PubMed] [Google Scholar]
- 496.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 497.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 498.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 499.Shilo B, Riddle V G, Pardee A B. Protein turnover and cell-cycle initiation in yeast. Exp Cell Res. 1979;123:221–227. doi: 10.1016/0014-4827(79)90462-2. [DOI] [PubMed] [Google Scholar]
- 500.Shirayama M, Matsui Y, Toh-e A. Dominant mutant alleles of yeast protein kinase gene CDC15 suppress the lte1 defect in termination of M phase and genetically interact with CDC14. Mol Gen Genet. 1996;251:176–185. doi: 10.1007/BF02172916. [DOI] [PubMed] [Google Scholar]
- 501.Shirayama M, Matsui Y, Toh-e A. Isolation of a CDC25 family gene, MSI2/LTE1, a multicopy suppressor of ira1. Yeast. 1994;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- 502.Shirayama M, Matsui Y, Toh-e A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Shpungin S, Liberzon A, Bangio H, Yona E, Katcoff D J. Association of yeast SIN1 with the tetratrico peptide repeats of CDC23. Proc Natl Acad Sci USA. 1996;93:8274–8277. doi: 10.1073/pnas.93.16.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Shu Y, Hallberg R L. SCS1, a multicopy suppressor of hsp60-ts mutant alleles, does not encode a mitochondrially targeted protein. Mol Cell Biol. 1995;15:5618–5626. doi: 10.1128/mcb.15.10.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Shu Y, Yang H, Hallberg E, Hallberg R. Molecular analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol Cell Biol. 1997;17:3242–3253. doi: 10.1128/mcb.17.6.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Shuster E O, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1980;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Sia R A L, Herald H A, Lew D J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Sidorova J, Breeden L. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1069–1077. doi: 10.1128/mcb.13.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Sidorova J M, Breeden L L. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Sidorova J M, Mikesell G E, Breeden L L. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Siegmund R F, Nasmyth K. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol Cell Biol. 1996;16:2647–2655. doi: 10.1128/mcb.16.6.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Sikorski R S, Boguski M S, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 514.Sikorski R S, Michaud W A, Hieter P. p62cdc23 of Saccharomyces cerevisiae: a nuclear tetratricopeptide repeat protein with two mutable domains. Mol Cell Biol. 1993;13:1212–1221. doi: 10.1128/mcb.13.2.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Sikorski R S, Michaud W A, Wootton J C, Boguski M S, Connelly C, Hieter P. TPR proteins as essential components of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1991;56:663–673. doi: 10.1101/sqb.1991.056.01.075. [DOI] [PubMed] [Google Scholar]
- 516.Silljé H H W, ter Schure E G, Rommens A J M, Huls P G, Woldringh C L, Verkleij A J, Boonstra J, Verrips C T. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Silver E T, Gwozd T J, Ptak C, Goebl M, Ellison M J. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function, and evolution of the E2s. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Simon M, Séraphin B, Faye G. KIN28, a yeast split gene coding for a putative protein kinase homologous to CDC28. EMBO J. 1986;5:2697–2701. doi: 10.1002/j.1460-2075.1986.tb04553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 520.Solomon M J, Harper J W, Shuttleworth J. CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J. 1993;12:3133–3142. doi: 10.1002/j.1460-2075.1993.tb05982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Solomon M J, Lee T, Kirschner M W. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Song O, Dolan J W, Yuan Y O, Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991;5:741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- 523.Songyang Z, Blechner S, Hoagland N, Hoekstra M F, Piwnica-Worms H, Cantley L C. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 524.Songyang Z, Lu K P, Kwon Y T, Tsai L-H, Filhol O, Cochet C, Brickey D A, Soderling T R, Bartleson C, Graves D J, DeMaggio A J, Hoekstra M F, Blenis J, Hunter T, Cantley L C. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Sorger P K, Murray A W. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature. 1992;355:365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- 526.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 527.Stan R, McLaughlin M M, Cafferkey R, Johnson R K, Rosenberg M, Livi G P. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- 528.Stemmann O, Lechner J. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- 529.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 530.Sterner D E, Lee J M, Hardin S E, Greenleaf A L. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Stuart D, Wittenberg C. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 534.Stueland C S, Lew D J, Cismowski M J, Reed S I. Full activation of p34CDC28 histone H1 kinase activity is unable to promote entry into mitosis in checkpoint-arrested cells of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3744–3755. doi: 10.1128/mcb.13.6.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca F C, Ruderman J V, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Sudakin V, Shteinberg M, Ganoth D, Hershko J, Hershko A. Binding of activated cyclosome to p13suc1. Use for affinity purification. J Biol Chem. 1997;272:18051–18059. doi: 10.1074/jbc.272.29.18051. [DOI] [PubMed] [Google Scholar]
- 537.Sudbery P E, Goodey A R, Carter B L A. Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature. 1980;288:401–404. doi: 10.1038/288401a0. [DOI] [PubMed] [Google Scholar]
- 538.Sugimoto K, Matsumoto K, Kornberg R D, Reed S I, Wittenberg C. Dosage suppressors of the dominant G1 cyclin mutant CLN3-2: identification of a yeast gene encoding a putative RNA/ssDNA binding protein. Mol Gen Genet. 1995;248:712–718. doi: 10.1007/BF02191711. [DOI] [PubMed] [Google Scholar]
- 539.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 540.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Surana U, Robitsch H, Schuster T, Fitch I, Futcher A B, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 542.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Sutton A, Freiman R. The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics. 1997;147:57–71. doi: 10.1093/genetics/147.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Svejstrup J Q, Feaver W J, Kornberg R D. Subunits of yeast RNA polymerase II transcription factor TFIIH encoded by the CCL1 gene. J Biol Chem. 1996;271:643–645. doi: 10.1074/jbc.271.2.643. [DOI] [PubMed] [Google Scholar]
- 546.Swenson K I, Farrell K M, Ruderman J V. The clam embryo protein cyclin A induces entry into M phase and the resumption of meiosis in Xenopus oocytes. Cell. 1986;47:861–870. doi: 10.1016/0092-8674(86)90801-9. [DOI] [PubMed] [Google Scholar]
- 547.Taba M R M, Muroff I, Lydall D, Tebb G, Nasymth K. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- 548.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 549.Tang Y, Reed S I. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993;7:822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]
- 550.Tassan J-P, Jacquenoud M, Fry A M, Frutiger S, Hughes G J, Nigg E A. In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 1995;14:5608–5617. doi: 10.1002/j.1460-2075.1995.tb00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Tavormina P A, Burke D J. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 1998;148:1701–1713. doi: 10.1093/genetics/148.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Taylor G S, Liu Y, Baskerville C, Charbonneau H. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J Biol Chem. 1997;272:24054–24063. doi: 10.1074/jbc.272.38.24054. [DOI] [PubMed] [Google Scholar]
- 553.Taylor I A, Treiber M K, Olivi L, Smerdon S J. The X-ray structure of the DNA-binding domain from the Saccharomyces cerevisiae cell-cycle transcription factor Mbp1 at 2.1 Å resolution. J Mol Biol. 1997;272:1–8. doi: 10.1006/jmbi.1997.1229. [DOI] [PubMed] [Google Scholar]
- 554.Tebb G, Moll T, Dowzer C, Nasmyth K. SWI5 instability may be necessary but is not sufficient for asymmetric HO expression in yeast. Genes Dev. 1993;7:517–528. doi: 10.1101/gad.7.3.517. [DOI] [PubMed] [Google Scholar]
- 555.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Regulation of the mating pheromone and invasive growth responses in yeast by two MAP kinase substrates. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 556.Tennyson C N, Lee J, Andrews B J. A role for the Pcl9-Pho85 cyclin-cdk complex at the M/G1 boundary in Saccharomyces cerevisiae. Mol Microbiol. 1998;28:69–79. doi: 10.1046/j.1365-2958.1998.00773.x. [DOI] [PubMed] [Google Scholar]
- 557.Terada Y, Tatsuka M, Jinno S, Okayama H. Requirement for tyrosine phosphorylation of Cdk4 in G1 arrest induced by ultraviolet irradiation. Nature. 1995;376:358–362. doi: 10.1038/376358a0. [DOI] [PubMed] [Google Scholar]
- 558.Thevelein J M. The RAS-adenylate cyclase pathway and cell cycle control in Saccharomyces cerevisiae. Antonie Leeuwenhoek. 1992;62:109–130. doi: 10.1007/BF00584466. [DOI] [PubMed] [Google Scholar]
- 559.Thomas D, Kuras L, Barbey R, Cherest H, Blaiseau P L, Surdin-Kerjan Y. Met30p, a yeast transcriptional inhibitor that responds to S-adenosylmethionine, is an essential protein with WD40 repeats. Mol Cell Biol. 1995;15:6526–6534. doi: 10.1128/mcb.15.12.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Thuret J-Y, Valay J-G, Faye G, Mann C. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell. 1996;86:565–576. doi: 10.1016/s0092-8674(00)80130-0. [DOI] [PubMed] [Google Scholar]
- 561.Timblin B K, Tatchell K, Bergman L W. Deletion of the gene encoding the cyclin-dependent protein kinase Pho85 alters glycogen metabolism in Saccharomyces cerevisiae. Genetics. 1996;143:57–66. doi: 10.1093/genetics/143.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Toda T, Cameron S, Sass P, Zoller M, Wigler M. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell. 1987;50:277–287. doi: 10.1016/0092-8674(87)90223-6. [DOI] [PubMed] [Google Scholar]
- 563.Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- 564.Toh-e A, Tanaka K, Uesono Y, Wickner R. PHO85, a negative regulator of the PHO system, is a homolog of a protein kinase, CDC28, of S. cerevisiae. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 565.Tokiwa G, Tyers M, Volpe T, Futcher B. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature. 1994;371:342–345. doi: 10.1038/371342a0. [DOI] [PubMed] [Google Scholar]
- 566.Toone W M, Aerne B L, Morgan B A, Johnston L H. Getting started: regulating the initiation of DNA replication in yeast. Annu Rev Microbiol. 1997;51:125–149. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- 567.Toone W M, Johnson A L, Banks G R, Toyn J H, Stuart D, Wittenberg C, Johnston L H. Rme1, a negative regulator of meiosis is also a positive activator of G1 cyclin gene expression. EMBO J. 1995;14:5824–5832. doi: 10.1002/j.1460-2075.1995.tb00270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Tourret J, McKeon F. Tyrosine kinases wee1 and mik1 as effectors of DNA replication checkpoint control. Prog Cell Cycle Res. 1996;2:91–97. doi: 10.1007/978-1-4615-5873-6_9. [DOI] [PubMed] [Google Scholar]
- 569.Townsley F, Ruderman J. Functional analysis of the Saccharomyces cerevisiae UBC11 gene. Yeast. 1998;14:747–757. doi: 10.1002/(SICI)1097-0061(19980615)14:8<747::AID-YEA271>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 570.Toyn J H, Araki H, Sugino A, Johnston L H. The cell-cycle-regulated budding yeast gene DBF2, encoding a putative protein kinase, has a homologue that is not under cell-cycle control. Gene. 1991;104:63–70. doi: 10.1016/0378-1119(91)90465-n. [DOI] [PubMed] [Google Scholar]
- 571.Toyn J H, Johnson A L, Donovan J D, Toone W M, Johnston L H. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics. 1996;145:85–96. doi: 10.1093/genetics/145.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Toyn J H, Johnston L H. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Toyn J H, Johnston L H. Spo12 is a limiting factor that interacts with the cell cycle protein kinases Dbf2 and Dbf20, which are involved in mitotic chromatid disjunction. Genetics. 1993;135:963–971. doi: 10.1093/genetics/135.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Treisman R, Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992;2:221–226. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- 575.Trueheart J, Boeke J D, Fink G R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Tyers M. The cyclin-dependent kinase inhibitor p40SIC1 imposes the requirement for Cln G1 cyclin function at Start. Proc Natl Acad Sci USA. 1996;93:7772–7776. doi: 10.1073/pnas.93.15.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13:5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Tyson C B, Lord P G, Wheals A E. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Tyson J J, Novak B, Odell G M, Chen K, Thron C D. Chemical kinetic theory: understanding cell-cycle regulation. Trends Biochem Sci. 1996;21:89–96. [PubMed] [Google Scholar]
- 582.Uesono Y, Tanaka K, Toh-e A. Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and characterization of PHO85. Nucleic Acids Res. 1987;15:10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Uesono Y, Toh-e A, Kikuchi Y. Ssd1p of Saccharomyces cerevisiae associates with RNA. J Biol Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- 584.Uesono Y, Tokai M, Tanaka K, Toh-e A. Negative regulators of the PHO system of Saccharomyces cerevisiae: characterization of PHO80 and PHO85. Mol Gen Genet. 1992;231:426–432. doi: 10.1007/BF00292712. [DOI] [PubMed] [Google Scholar]
- 585.Umeda M, Bhalerao R P, Schell J, Uchimiya H, Koncz C. A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998;95:5021–5026. doi: 10.1073/pnas.95.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Valay J G, Simon M, Faye G. The Kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J Mol Biol. 1993;234:307–310. doi: 10.1006/jmbi.1993.1587. [DOI] [PubMed] [Google Scholar]
- 587.Valay J-G, Simon M, Dubois M-F, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- 588.Valdivieso M H, Sugimoto K, Jahng K, Fernandes P M, Wittenberg C. FAR1 is required for posttranscriptional regulation of CLN2 gene expression in response to mating pheromone. Mol Cell Biol. 1993;13:1013–1022. doi: 10.1128/mcb.13.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Vallier L G, Coons D, Bisson L F, Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994;136:1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating hormones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Van Arsdell S W, Thorner J. The yeast repeated element Sigma contains a hormone-inducible promoter. Mol Cell Biol. 1987;7:749–759. doi: 10.1128/mcb.7.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.van Zyl W, Huang W D, Sneddon A A, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach J R. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Gen Genet. 1992;12:4946–4959. doi: 10.1128/mcb.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Varshavsky A. The ubiquitin system. Trends Biochem Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- 594.Venkataramani R, Swaminathan K, Marmorstein R. Crystal structgure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations. Nat Struct Biol. 1998;5:74–81. doi: 10.1038/nsb0198-74. [DOI] [PubMed] [Google Scholar]
- 595.Verma R, Annan R S, Huddleston M J, Carr S A, Reynard G, Deshaies R J. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- 596.Verma R, Feldman R M R, Deshaies R J. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol Biol Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Verma R, Smiley J, Andrews B, Campbell J L. Regulation of the yeast DNA replication genes through the MluI cell cycle box is dependent on SWI6. Proc Natl Acad Sci USA. 1992;89:9479–9483. doi: 10.1073/pnas.89.20.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 600.Wagner M, Pierce M, Winter E. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 1997;16:1305–1317. doi: 10.1093/emboj/16.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Wagstaff J E, Klapholz S, Esposito R E. Meiosis in haploid yeast. Proc Natl Acad Sci USA. 1982;79:2986–2990. doi: 10.1073/pnas.79.9.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;382:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 603.Walker S S, Shen W-C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 604.Wan J, Xu H, Grunstein M. CDC14 of Saccharomyces cerevisiae. J Biol Chem. 1992;267:11274–11280. [PubMed] [Google Scholar]
- 605.Wang Y, Burke D J. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:620–626. doi: 10.1128/mcb.17.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Watson M H, Bourne Y, Arvai A S, Hickey M J, Santiago A, Bernstein S L, Tainer J A, Reed S I. A mutation in the human cyclin-dependent kinase interacting protein, CksHs2, interferes with cyclin-dependent kinase binding and biological function, but preserves protein structure and assembly. J Mol Biol. 1996;261:646–657. doi: 10.1006/jmbi.1996.0490. [DOI] [PubMed] [Google Scholar]
- 607.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 608.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Wellman S E. Carboxyl-terminal peptides from histone H1 variants: DNA binding characteristics and solution conformation. Biopolymers. 1996;39:491–501. doi: 10.1002/(SICI)1097-0282(199610)39:4%3C491::AID-BIP2%3E3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 610.Westendorf J M, Swenson K I, Ruderman J V. The role of cyclin B in meiosis. J Cell Biol. 1989;108:1431–1444. doi: 10.1083/jcb.108.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.White J H M, Green S R, Barker D G, Dumas L B, Johnston L H. The CDC8 transcript is cell cycle regulated in yeast and is expressed with CDC9 and CDC21 at a point preceding histone transcription. Exp Cell Res. 1987;171:223–231. doi: 10.1016/0014-4827(87)90265-5. [DOI] [PubMed] [Google Scholar]
- 612.Willems A R, Lanker S, Patton E E, Craig K L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 613.Wilson S M, Datar K V, Paddy M R, Swedlow J R, Swanson M S. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Wittenberg C, Reed S I. Conservation of function and regulation with the Cdc28/cdc2 protein kinase family: characterization of the human Cdc2Hs protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:4064–4068. doi: 10.1128/mcb.9.9.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Wittenberg C, Reed S I. Control of the yeast cell cycle is associated with assembly/disassembly of the Cdc28 protein kinase complex. Cell. 1988;54:1061–1072. doi: 10.1016/0092-8674(88)90121-3. [DOI] [PubMed] [Google Scholar]
- 616.Wittenberg C, Reed S I. Plugging it in: signaling circuits and the yeast cell cycle. Curr Opin Cell Biol. 1996;8:223–230. doi: 10.1016/s0955-0674(96)80069-x. [DOI] [PubMed] [Google Scholar]
- 617.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae. Cell cycle, periodicity, regulation by mating pheromone, and assocation with the p34cdc28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 618.Wu L, Osmani S A, Mirabito P M. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J Cell Biol. 1998;141:1575–1587. doi: 10.1083/jcb.141.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Wynne J, Treisman SRF and MCM1 have related but distinct DNA binding specificities. Nucleic Acids Res. 1992;20:3297–3303. doi: 10.1093/nar/20.13.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Xu R M, Koch C, Liu Y, Horton R, Knapp D, Nasmyth K, Cheng X. Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis. Structure. 1997;5:349–358. doi: 10.1016/s0969-2126(97)00192-5. [DOI] [PubMed] [Google Scholar]
- 621.Yaglom J, Linskens M H K, Sadis S, Rubin D M, Futcher B, Finley D. p34cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Yaglom J A, Goldberg A L, Finley D, Sherman M Y. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol. 1996;16:3679–3684. doi: 10.1128/mcb.16.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Yano R, Oakes M, Yamaghishi M, Dodd J A, Nomura M. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5640–5451. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Ye X S, Fincher R R, Tang A, Osmani S A. The G2/M DNA damage checkpoint inhibits mitosis through Tyr15 phosphorylation of p34cdc2 in Aspergillus nidulans. EMBO J. 1997;16:182–192. doi: 10.1093/emboj/16.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Yochem J, Byers B. Structural comparison of the yeast cell division cycle gene CDC4 and a related pseudogene. J Mol Biol. 1987;195:233–245. doi: 10.1016/0022-2836(87)90646-2. [DOI] [PubMed] [Google Scholar]
- 628.Yu G, Deschenes R J, Fassler J S. The essential transcription factor, Mcm1, is a downstream target of Sln1, a yeast “two-component” regulator. J Biol Chem. 1995;270:8739–8743. doi: 10.1074/jbc.270.15.8739. [DOI] [PubMed] [Google Scholar]
- 629.Yu G, Fassler J S. SPT13 (GAL11) of Saccharomyces cerevisiae negatively regulates activity of Mcm1 transcription factor in Ty1 elements. Mol Cell Biol. 1993;13:63–71. doi: 10.1128/mcb.13.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Yu H, Peters J M, King R W, Page A M, Hieter P, Kirschner M W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- 631.Yu Y, Jiang Y W, Wellinger J, Carlson K, Roberts J M, Stillman D J. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol Cell Biol. 1996;16:5254–5263. doi: 10.1128/mcb.16.10.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Zachariae W, Shevchenko A, Andrews P D, Ciosk R, Galova M, Stark M J, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 634.Zachariae W, Shin T H, Galova M, Overmaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]
- 635.Zhang H, Hannon G J, Beach D. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 636.Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S-phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 637.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Zhao Y, Boguslawski G, Zitomer R S, DePaoli-Roach A A. Saccharomyces cerevisiae homologs of mammalian B and B′ subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J Biol Chem. 1997;272:8256–8262. doi: 10.1074/jbc.272.13.8256. [DOI] [PubMed] [Google Scholar]
- 639.Zheng X-F, Fiorentino D, Chen J, Crabtree G R, Schreiber S L. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 640.Zhou C, Huang S-H, Jong A Y. Molecular cloning of Saccharomyces cerevisiae CDC6 gene. Isolation, identification, and sequence analysis. J Biol Chem. 1989;264:9022–9029. [PubMed] [Google Scholar]
- 641.Zhou C, Jong A. CDC6 mRNA fluctuates periodically in the yeast cell cycle. J Biol Chem. 1990;265:19904–19909. [PubMed] [Google Scholar]
- 642.Zwerschke W, Rottjakob H-W, Küntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]