Abstract
Formation of mRNA 3′ ends in eukaryotes requires the interaction of transacting factors with cis-acting signal elements on the RNA precursor by two distinct mechanisms, one for the cleavage of most replication-dependent histone transcripts and the other for cleavage and polyadenylation of the majority of eukaryotic mRNAs. Most of the basic factors have now been identified, as well as some of the key protein-protein and RNA-protein interactions. This processing can be regulated by changing the levels or activity of basic factors or by using activators and repressors, many of which are components of the splicing machinery. These regulatory mechanisms act during differentiation, progression through the cell cycle, or viral infections. Recent findings suggest that the association of cleavage/polyadenylation factors with the transcriptional complex via the carboxyl-terminal domain of the RNA polymerase II (Pol II) large subunit is the means by which the cell restricts polyadenylation to Pol II transcripts. The processing of 3′ ends is also important for transcription termination downstream of cleavage sites and for assembly of an export-competent mRNA. The progress of the last few years points to a remarkable coordination and cooperativity in the steps leading to the appearance of translatable mRNA in the cytoplasm.
Posttranscriptional cleavage of mRNA precursor is an essential step in mRNA maturation. Following cleavage, most eukaryotic mRNAs, with the exception of replication-dependent histone transcripts in some organisms, acquire a poly(A) tract at their 3′ ends. The process of 3′-end formation promotes transcription termination (101) and transport of the mRNA from the nucleus (215). The poly(A) tail, most probably by providing a binding site for poly(A) binding protein (105), also enhances the translation and stability of mRNA (149, 368, 399, 488).
Defects in mRNA 3′-end formation can profoundly alter cell viability, growth, and development. The essential nature of the yeast genes encoding components of the polyadenylation pathway emphasizes the importance of this process. In metazoan cells, in vivo depletion of one of the cleavage proteins, CstF-64, causes cell cycle arrest and ultimately apoptotic cell death (451). A failure to correctly modify the metazoan poly(A) polymerase during the cell cycle is thought to cause a lower growth rate and cell accumulation in the G0-G1 phase (516). The appearance of short GCG repeats in the gene encoding the PAB II polyadenylation factor is associated with oculopharyngeal muscular dystrophy (59). The formation of mRNA 3′ ends is a key regulatory step in the expression of many genes, and in some cases aberrant polyadenylation leads to disease. In humans, such defects cause thalassemias (203, 345) and a lysosomal storage disorder (161). Inappropriate polyadenylation may also contribute to the abnormal processing of the EAAT2 glutamate transporter transcripts observed in the brains of patients with sporadic amyotrophic lateral sclerosis (262). In this disease, the loss of functional EAAT2 correlates with motor neuron degeneration. Research into the fundamental mechanism of mRNA 3′-end formation and its regulation should lead to a better understanding of its crucial role in normal cell growth and development.
The past few years have brought astounding progress in our understanding of the biochemistry of mRNA 3′-end formation, its regulation, and its interaction with other aspects of mRNA synthesis. The factors which comprise the basic polyadenylation machinery have been identified, and the coding sequence of many, if not most, of the protein subunits has become available. The molecular mechanism by which several regulatory elements stimulate or inhibit polyadenylation has been dissected in exquisite detail, and the intimate involvement of splicing factors at these sites has been made clear. In addition, much information has accumulated on how the basic polyadenylation machinery is regulated to control the choice of poly(A) site or activity of the poly(A) polymerase. Finally, the coupling of transcription and mRNA 3′-end formation has been convincingly demonstrated in a variety of ways.
We have tried to provide sufficient background information that the reader can evaluate the developments in our understanding of mRNA 3′-end formation, primarily over the last 5 years. Due to space constraints, we are not able to give a more thorough historical account, and so we have focused on a limited number of examples to illustrate the paradigms emerging in the field. We ask the readers to refer to several recent reviews on constitutive and regulated polyadenylation for additional details (101, 139, 238, 473, 475a). Cytoplasmic polyadenylation and the role of the poly(A) tail in translation is covered in a review by Richter elsewhere in this issue (384a).
CLEAVAGE/POLYADENYLATION PATHWAY
RNA Sequences Which Specify Cleavage and Polyadenylation
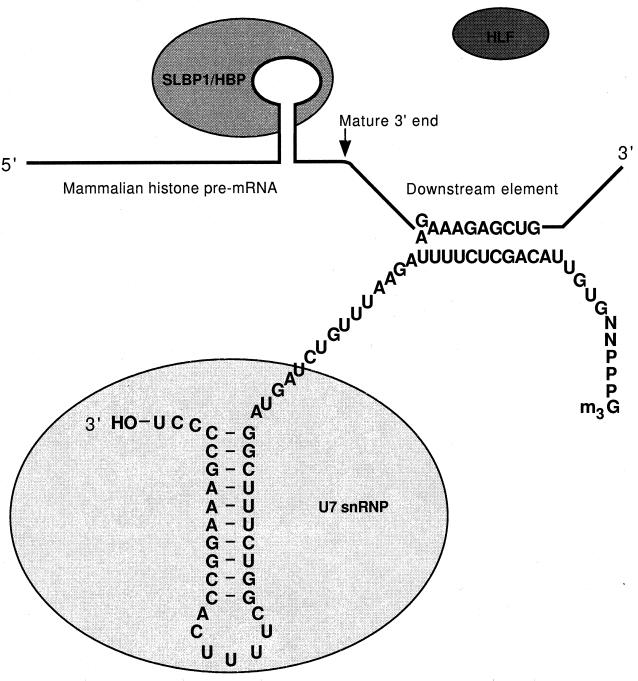
Sequences on the RNA precursor ultimately determine the processing efficiency in a given cellular environment. The cis-acting elements specifying cleavage and polyadenylation and the cleavage of replication-dependent histone pre-mRNA in animal cells are well defined. Research in the last few years has led to a much better understanding of the polyadenylation signals in yeasts.
Mammalian polyadenylation signals.
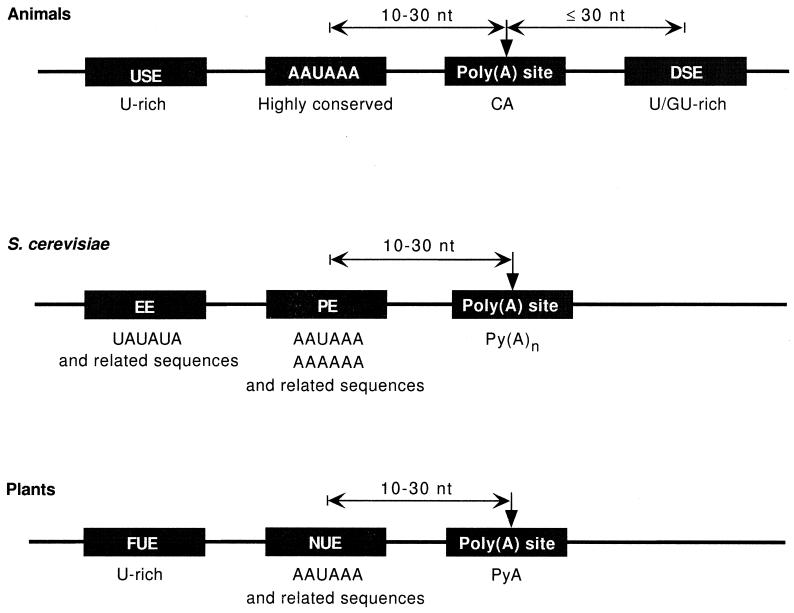
In mammalian cells, three elements define the core polyadenylation signal—the highly conserved hexanucleotide AAUAAA found 10 to 30 nucleotides upstream of the cleavage site, a less highly conserved U-rich or GU-rich element located downstream of the cleavage site, and the cleavage site itself, which becomes the point of poly(A) addition and is thus generally referred to as the poly(A) site (Fig. 1). Additional sequences outside of this core recruit regulatory factors or maintain the core signal in an open and accessible structure.
FIG. 1.
Schematic representation of poly(A) signals in animals, the yeast S. cerevisiae, and plants. USE, auxiliary upstream enhancer; DSE, downstream element; EE, efficiency element; PE, positioning element; FUE, far-upstream element; NUE, near-upstream element; nt, nucleotides. Adapted from reference 387.
(i) AAUAAA motif.
The consensus sequence AAUAAA was initially revealed by a comparison of nucleotide sequences preceding the poly(A) sites in several mRNAs (377) and has since been found in almost all polyadenylated mRNAs of animal cells (473). Extensive mutagenesis studies and the analysis of naturally occurring mutations have conclusively established that this hexanucleotide is essential for both cleavage and poly(A) addition (reviewed in references 286, 472, and 487). The sequence AAUAAA is one of the most highly conserved sequence elements known (374). The most frequent variant is AUUAAA, whose activity is comparable to that of the canonical sequence. Mutations of any other nucleotide strongly inhibit processing (422, 489, 496), although in some genes, these forms are the only hexanucleotide-like sequences upstream of the poly(A) site. Poly(A) sites with these variants, as well as those that have no discernible upstream AAUAAA element, i.e., no sequence differing by less than 2 nucleotides from the consensus, are usually associated with alternative (208, 421, 508) or tissue-specific (76, 476) polyadenylation.
(ii) Downstream elements.
The second element of the core polyadenylation signal is within approximately 30 nucleotides downstream of the poly(A) site. This downstream element (DSE) is more diffuse and poorly conserved, and two main types have been described, a U-rich element and a GU-rich element. The U-rich element is a short run of U residues (94, 162). The GU-rich type has the consensus YGUGUUYY (Y = pyrimidine) and has been found downstream of the poly(A) site in two-thirds of the 70 genes surveyed in a 1985 study (302). A polyadenylation signal may have only one DSE (94, 300) or may have both a U-rich element and a GU-rich element working together synergistically (162). However, some DSEs contain no matches to either the U- or GU-rich motif (431). Point mutations or small deletions in the DSE have only weak effects, and larger deletions are required to abolish function, which is in agreement with the idea that the DSE is poorly defined and possibly redundant (510). Nevertheless, the proximity of the DSE to the poly(A) site can affect the cleavage site position (280, 294) and the efficiency of cleavage (162, 300).
(iii) Poly(A) site.
The selection of the cleavage site is determined mainly by the distance between the upstream AAUAAA sequence and the DSE(s) (80). The local sequence surrounding the cleavage site is not conserved, although adenosine is found at the cleavage site of 70% of vertebrate mRNAs (422). Thus, the first nucleotide of the poly(A) tail in most mRNAs is probably template encoded, although this has been proven experimentally in vitro for only two poly(A) sites (317, 422). A study involving saturation mutagenesis demonstrated that the order of preference for the cleavage site nucleotide follows the order of A > U > C ≫ G (80). The pentultimate nucleotide is most often a C residue (in 59% of all genes analyzed) (422). Thus, a CA dinucleotide defines the poly(A) site for most genes.
(iv) Auxiliary sequences.
Other sequence elements can modulate the efficiency of 3′ processing in a positive or negative fashion. The molecular mechanisms by which some of these operate are described in later sections, and only the auxiliary sequences are discussed here. One class of enhancer sequence is located upstream of the AAUAAA element (USE) (Fig. 1), and has been found primarily in viral poly(A) sites, such as adenovirus E3 (43), adenovirus L1 (126), adenovirus L3 (372, 373), adenovirus L4 (431), Epstein-Barr virus DNA polymerase (427), simian virus 40 (SV40) late (74, 277, 410), ground squirrel hepatitis virus (392), and retroviruses such as human immunodeficiency virus type 1 (HIV-1) (65, 87, 127, 128, 466). These USEs are often U rich, but a consensus sequence has not emerged (Table 1).
TABLE 1.
Summary of enhancer elements for cleavage/polyadenylation in animal cells
| Gene | Motif | Binding protein(s)a | Reference(s) |
|---|---|---|---|
| USEs in virus genes | |||
| Adenovirus L1 | U-rich | 126 | |
| Adenovirus L3 | UUCUUUUU | hnRNP-C | 372, 373 |
| Adenovirus L4 | AUCUUUGUUGUC/AUCUCUGUGCUG | 431 | |
| Adenovirus E3 | U-rich | 43 | |
| EBV DNA polymerase | UUUGUA | 427 | |
| HBV PS1 region | AU-rich | 392 | |
| HBV transport element | 217 | ||
| HIV-1 U3 region | U-rich | CPSF-160 | 65, 87, 125, 127, 163, 164, 465 |
| HSV-TK transport element | hnRNP L | 217, 266 | |
| SV40 late | 3x AUUUGURA | U1 snRNP-A | 277, 410 |
| USEs in cellular genes | |||
| C2 complement | 42% U | PTB, CstF-64 | 319, 320 |
| Lamin B2 | 2–3 U tracts | 57 | |
| Histone H2a transport element | 217 | ||
| Downstream enhancers | |||
| SV40 late | GGGGGAGGUGUGGG | DSEF-1 | 26, 27, 81 |
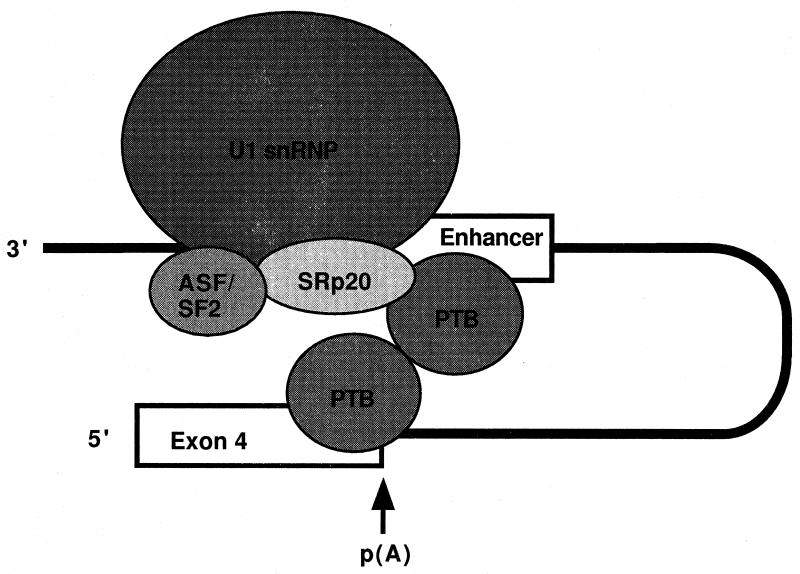
| Calcitonin/CGRP | CTCCGCTCCTCTTCCAGGTAAGAC | U1 snRNP, PTB, SR20 | 271 |
PTB, polypyrimidine tract-binding protein; hnRNP, heterogeneous nuclear ribonucleoprotein; DSEF-1, a member of the hnRNP H family of RNA-binding proteins; SR20, member of SR family of splicing factors.
While USEs are common among viruses, such USEs have recently been identified in the poly(A) sites of two cellular genes, one encoding the complement factor C2 (319, 320) and the other encoding lamin B2 (57). The polyadenylation of both mRNAs is enhanced by their respective USEs. As with viral USEs, they are also U rich (57, 320). An additional class of USEs, found in transcripts from the mouse histone H2a, herpes simplex virus thymidine kinase (HSV-TK) and hepatitis B virus genes, were originally identified by their ability to stimulate the transport of intronless mRNAs and subsequently shown to promote polyadenylation as well (217).
In contrast to USEs, it has been difficult to define an auxiliary downstream enhancer, primarily due to the diverse nature of the DSE and the ability of these elements to work additively. However, investigation of sequences downstream of the U-rich element of the SV40 late polyadenylation signal has led to the identification of a G-rich sequence that positively influences processing efficiency (27, 81). In another example, a pseudoexon sequence in calcitonin/CGRP transcripts has been shown to stimulate processing at an upstream poly(A) site (270, 271).
The sequences described above are thought to work by acting as recognition sites for factors which stabilize the polyadenylation complex. However, the last few years have brought an increasing appreciation of the influence of secondary structure on polyadenylation. In many lentiviruses, the RNA structure itself is the key feature. In the transcripts of human T-cell leukemia virus types 1 and 2 and bovine leukemia virus, the AAUAAA signal and the cleavage site are almost 300 nucleotides apart and a stem-loop in the intervening sequence brings the two sequences closer together (reviewed in reference 180). In HIV-1, the TAR stem-loop structure juxtaposes the USE and the AAUAAA (163). Secondary structure downstream of the murine immunoglobulin M secretory poly(A) site is critical for positively regulating the use of this site (361), while a stem-loop containing the HIV-1 core poly(A) signal (247) as well as base-pairing in hepatitis delta virus transcripts (211) reduces processing. The presentation of the adenovirus L4 AAUAAA in a loop whose stem is partially composed of the upstream enhancer is important for efficient processing (430). Sequences chosen for optimal function as a USE by selection amplification were found to keep the AAUAAA in an extensive open region (172, 173), and a similar function has been proposed for sequences beyond the DSE of several genes (81).
Negative regulatory elements have been described upstream of the U1A and the bovine papillomavirus late poly(A) sites (154, 177, 179) and downstream of the promoter-proximal HIV-1 poly(A) site (20). Two elements have been identified in Xenopus albumin mRNA that cause the poly(A) tail to be limited in vivo to about 20 nucleotides (117). These elements share a similar 8-nucleotide sequence, CUGARRAR (R = purine). Because the short tails are found on incompletely spliced RNAs, this regulation, which operates in both Xenopus and mouse cells, is thought to occur in the nucleus (382). In vitro analysis should clarify the mechanism of this interesting regulation.
Yeast polyadenylation signals.
Signals which direct mRNA 3′-end formation in the yeast Saccharomyces cerevisiae are somewhat different from those used in higher eukaryotes in both sequence and organization. Yeast polyadenylation signals are less highly conserved than are poly(A) signals in higher eukaryotes and are unexpectedly complicated. At least three elements are needed to make up a minimal yeast mRNA 3′-end region: (i) the UA-rich efficiency element and related sequences, functioning to activate the positioning element; (ii) the A-rich positioning element, which directs the position of the cleavage site; and (iii) the actual site of polyadenylation, PyAn (Fig. 1) (reviewed in reference 182). A recent analysis of 1,352 pre-mRNA 3′-end processing sites, corresponding to 861 different genes, has confirmed this organization of signal sequences (170).
(i) Efficiency element.
Efficiency elements are found at a variable distance upstream of the cleavage site and often contain alternating UA dinucleotides or U-rich stretches. Early comparisons of a number of yeast mRNA 3′ ends led to the proposal of a bipartite motif UAG ⋯ UAUGUA (395). Another sequence, UUUUUAUA, was also identified as an efficiency element in several yeast genes including GCN4, PHO5, and ADH1 (141, 181, 198, 199, 224). A subsequent study reduced these putative signals to a hexanucleotide, UAYRUA, with the sequence UAUAUA working best for mRNA 3′-end formation (223). The U residues at the first and fifth positions are the most critical nucleotides in this sequence. Furthermore, computer analysis has shown that more than half of the approximately 1,000 yeast nuclear genes examined contain UAUAUA sequences in their 3′ region (181). Thus, most yeast genes, e.g., GAL7 and MRP2 (1, 356), use UAUAUA as the efficiency element, while other genes, e.g., CYC1 and GCN4, appear to use related sequences, such as UAUUUA, UAUGUA, and UUUUUAUA (141, 181).
(ii) Positioning element.
The second element, the positioning element, directs cleavage to a position approximately 20 nucleotides downstream of this sequence. Deletion or mutation of the sequence UUAAGAAC in the 3′ region of CYC1 changes the location of the poly(A) site but not the overall mRNA processing level, indicating that it represents a signal distinct from the efficiency element (395). In addition to the sequence UUAAGAAC, several other A-rich sequences have been characterized as positioning elements, with AAUAAA and AAAAAA being the most efficient (183). Related motifs are also functional, except that sequences such as GAUAAA, GAAGAA, and GAAUAA are either inefficient or completely inactive, suggesting that a guanosine residue at the first position has an inhibitory effect on the signal function.
The positioning element can also contribute to the efficiency of processing, as seen from the fact that deletion of the AAUAAA motif in the 3′-end region of the ADH2 gene, the GAL7 gene, or a heterologous cauliflower mosaic virus gene resulted in a reduction in the use of wild-type poly(A) site (1, 222, 225). A single point mutation in the TRP4 gene altered the efficiency of processing as well as the selection of the poly(A) site (133). The positioning element normally resides between the efficiency element and the poly(A) site, but for the FBP1 gene, it is found upstream of a series of efficiency motifs (18).
(iii) Poly(A) site.
Mapping of the poly(A) sites of several yeast genes has shown that polyadenylation occurs most frequently at a Py(A)n sequence (Py = pyrimidine) (36, 195). In contrast to animal genes, in which a single poly(A) site is found downstream of AAUAAA, many yeast genes use a cluster of poly(A) sites downstream of the efficiency and positioning elements. When positioning elements are mutated, the poly(A) sites become scattered over a much broader region (17, 194, 396). A recent survey indicates that T-rich motifs are frequently found immediately before and after the poly(A) site, especially in genes with suboptimal efficiency or positioning elements (170). The importance of these flanking sequences has not been tested experimentally.
(iv) Additional properties of yeast polyadenylation signals.
Some yeast polyadenylation signals function in both orientations (17, 224, 356). This is in part due to the convergent transcription of closely packed genes. When this is not the case, it is likely that the sequence variability of the yeast signal motifs, the general TA richness of the 3′ ends of yeast genes (170), and the primary dependence on the UA-rich element, and not the positioning element, for efficient processing will increase the probability that an adequate signal is fortuitously found in the reverse orientation.
The efficiency and positioning elements are not only degenerate but also redundant, and most yeast polyadenylation signals are more complex than the minimal one presented in Fig. 1. The sequence redundancy provides an explanation why deletion or mutation of such motifs in several yeast genes has only slight or no effect. In the GCN4 gene, mutation of both copies of the UUUUAU sequence was necessary to reduce processing activity in vitro (141). The CYC1 gene employs multiple weaker elements (UUUAUA, UAUGUU, and UAUUUA) which act additively to constitute a strong signal (181). Deletion of an AAUAAA sequence upstream of the poly(A) site of the ADE8 transcript had no apparent effect, which may be due to the presence of two copies of the AAAAA sequence adjacent to AAUAAA (199).
The necessity for a specific sequence downstream of the cleavage site in yeast is not clear. A downstream sequence is required for efficient in vivo 3′-end processing of the ADH2 transcript (222). In many cases, deletion of all or nearly all downstream sequences has little or no effect on 3′-end formation (18, 141, 210, 356, 401), and in vitro substrates with as few as 7 to 10 nucleotides beyond the poly(A) site are efficiently cleaved (82, 401). The presence of mRNA secondary structure around signal sequences may be important in some yeast genes (222, 401), and a long-range interaction of 5′ and 3′ untranslated regions has been demonstrated for MFA2 mRNA (130). While the contribution of RNA conformation has not been rigorously investigated for any site, circular RNA substrates are not cleaved in vitro (443).
The polyadenylation signals of the fission yeast, Schizosaccharomyces pombe, have been characterized only for the ura4 gene (219). In this case, three elements were important: two site-determining elements upstream of the poly(A) sites and an efficiency element downstream of the poly(A) sites.
Plant polyadenylation signals.
The process of mRNA 3′-end formation in plants is poorly understood, but some information on important cis-acting elements is available (reviewed in references 220 and 387). At least three signals are required: the near-upstream element (NUE), the far-upstream element (FUE), and the cleavage site itself (Fig. 1). NUE is located about 10 to 30 nucleotides upstream of the cleavage site and presents in variant forms, from AAUAAA-like motifs to other related or unrelated sequences. FUE is usually U rich and is found approximately 100 nucleotides upstream of the cleavage site. Similar to other organisms, cleavage often occurs at a PyA dinucleotide. There are multiple cleavage sites in many genes, and use of a particular site is determined predominantly by the position of the NUE.
Comparison of polyadenylation signals in mammals, yeast, and plants.
A tripartite signal, composed of an A-rich sequence, a U-rich element, and a PyA cleavage site forms a common minimal polyadenylation signal in all eukaryotes (Fig. 1). In mammals, the hexanucleotide AAUAAA is highly conserved, present in a single copy, and absolutely necessary for 3′-end processing. In yeast, the A-rich motif sometimes serves only to position the poly(A) site and is often duplicated. The second set of sequence elements are U rich or UA rich and work in conjunction with the A-rich sequence. In mammals, these are most often present in single copy downstream of the cleavage site and are essential. When located upstream of the A-rich signal, they are stimulatory. In yeast and plants, this type of signal is often redundant and is usually found upstream of the cleavage site and most often upstream of the A-rich sequence as well. Cleavage occurs preferentially at CA in mammals and PyA in yeast and plants. No downstream signals have been clearly identified in yeast or plants. For the most part, the polyadenylation signal appears to be recognized as a one-dimensional string of nucleotides.
Cleavage/Polyadenylation Machinery
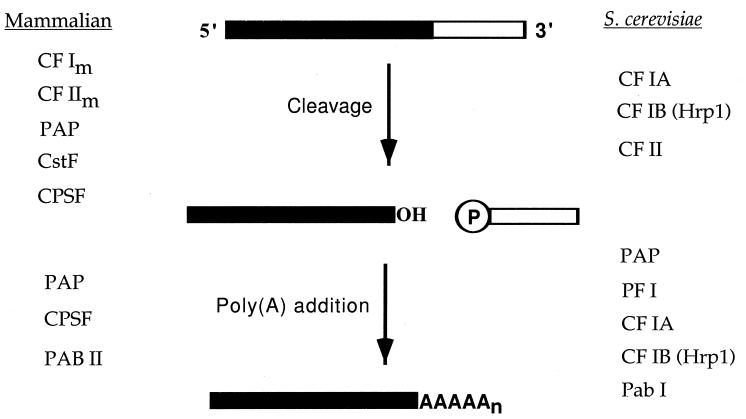
Multiple protein factors are required for the formation of mRNA 3′ ends and are generally conserved between yeast and mammals. The in vitro processing assays developed in mammalian cells by Moore and Sharp (315) and in yeast cells by Butler and Platt (68) have provided a successful approach for isolating these activities via fractionation of cell extracts and cloning the relevant genes and cDNAs from protein sequence. In mammals, cleavage/polyadenylation specificity factor (CPSF), cleavage-stimulatory factor (CstF), cleavage factors Im and IIm (CF Im and CF IIm), RNA polymerase II (pol II) and poly(A) polymerase (PAP) are involved in the cleavage step and CPSF, PAP, and poly(A)-binding protein II (PAB II) are involved in polyadenylation (Fig. 2). In yeast, cleavage requires CF IA CF IB, and CF II and polyadenylation uses CF IA, CF IB, polyadenylation factor I (PF I), Pab1, and Pap1 (Fig. 2). Yeast genetics and application of the two-hybrid screen in yeast cells has led to the discovery of some of the important yeast genes, and the recent completion of the S. cerevisiae genomic sequence has allowed the quick identification of several yeast proteins as homologues of mammalian CPSF subunits. In this section, we describe what is known about these factors (summarized in Tables 2 and 3 for mammal and yeast cell factors, respectively). At the end, we present a model for the complexes involved in each step of the reaction.
FIG. 2.
mRNA precursors are processed at their 3′ ends in a two-step reaction. A primary transcript is cleaved endonucleolytically at the poly(A) site, and this is followed by the addition of adenylate residues to the 3′ end of the upstream fragment to form a poly(A) tail. The factors responsible for each step of the reaction in mammals and in the yeast S. cerevisiae are indicated on each side.
TABLE 2.
Mammalian cleavage/polyadenylation
| Factor | Processing step | Subunits (kDa) | Sequence featuresa | Characteristics |
|---|---|---|---|---|
| CF Im | Cleavage | 68 | RBD in N terminus, P-rich in middle, SR protein homology in C terminus | Binds RNA with preference for polyadenylation precursor |
| 59 | —b | — | ||
| 25 | — | Binds RNA | ||
| CF IIm | Cleavage | Unknown | — | — |
| CstF | Cleavage | 77 | Eight HAT repeats | Bridges CstF-64 and CstF-55, interacts with CPSF-160 |
| 64 | RBD near N terminus, PG-rich C terminus, MEARA/G or LEPRG repeats | Binds GU- and U-rich downstream signals | ||
| 50 | Seven WD-40 repeats | — | ||
| CPSF | Cleavage & poly(A) addition | 160 | Divergent RBD, Rse1 homology | Binds to AAUAAA sequence; interacts with PAP and CstF-77 |
| 100 | 23% identity, 49% similarity to CPSF-73 | — | ||
| 73 | 23% identity, 49% similarity to CPSF-100 | — | ||
| 30 | Five CCCH zinc fingers, one CCHC zinc knuckle | Binds to poly(U) preferentially; binds CPSF-30 | ||
| PAP | Cleavage & poly(A) addition | 82/77 | Catalytic core near N terminus; C terminus contains RBS, bipartite NLS, ST-rich region | Catalyzes the poly(A) synthesis; nonspecific activity by itself; regulated by phosphorylation in ST-rich region |
| PAB II | Poly(A) elongation | 33 | Very acidic N terminus, very basic C terminus, RBD in middle region | Binds poly(A) and CPSF-30; responsible for processive elongation of poly(A) tail and control of tail length |
| RNA pol II (large subunit) | Cleavage | 200 | Phosphorylated CTD | Binds CPSF and CstF, substitutes for CP |
CP, creatine phosphate; CTD, C-terminal domain; RBD, RNA-binding domain; HAT repeats and WD-40 repeats, potential protein-protein interaction motifs.
—, unknown.
TABLE 3.
Yeast cleavage/polyadenylation
| Factor | Processing step | Subunits (kDa) | Gene | Essential | Sequence featuresa | Characteristics | Mammalian homologue (% identity) |
|---|---|---|---|---|---|---|---|
| CF IA | Cleavage and poly(A) addition | 76 | RNA14 | Yes | HAT repeat | Associates tightly with Rna15 | CstF-77 (24%) |
| 72 | PCF11 | Yes | Possible CTD-interaction domain, Q20 stretch, leucine zipper | Interacts with both Rna14 and Rna15 | |||
| 50 | EF1a or Clp1 | Yes | |||||
| 38 | RNA15 | Yes | RBD near N terminus, opa-like sequence | Binds U-rich sequences | CstF-64 (43% in RBD) | ||
| CF IB | Cleavage and poly(A) addition | 73 | HRP1 | Yes | Two RRMs | Interacts with Rna14 and Rna15; binds to UA repeats | hnRNP A/B/D |
| CF II | Cleavage and poly(A) addition | 150 | CFT1/YHH1 | Yes | —b | — | CPSF-160 (24%) |
| 105 | CFT2/YDH1 | Yes | — | RNA binding dependent on ATP, UA repeat and sequence at or beyond cleavage site | CPSF-100 (24%) | ||
| 100 | BRR5/YSHI | Yes | P-loop | — | CPSF-73 (53% in the first 500 amino acids) | ||
| 90 | PTA1 | Yes | — | — | — | ||
| PFI | Poly(A) addition | 58 | PFS1 | Yes | Zinc knuckle | — | — |
| 55 | FIP1 | Yes | Acidic N terminus and P-rich C terminus | Interacts with Pap1, Yth1, Rna14, and CF II | |||
| 53 | PFS2 | Yes | WD-40 repeats | ||||
| 24 | YTH1 | Yes | Five zinc finger repeats | Interacts with Fip1 | CPSF-30 (40%) | ||
| Pap1 | Poly(A) addition | 64 | PAP1 | Yes | Catalytic domain near N terminus; SpDs, C-terminal RBS | Catalyzes poly(A) synthesis; interacts directly with Fip1 | PAP (47%) |
| Pab I | Poly(A) addition | 70 | PAB1 | Yes | Four RBDs | Interacts with Rna15; controls the poly(A) tail length | PAB I (51%) |
| PAN | Poly(A) tail shortening | 127 | PAN2 | No | RNase D family | Catalytic subunit; 3′-5′ exoribonuclease; interacts with Pan3 and Pab1 | — |
| 76 | PAN3 | No | — | Interacts with Pan2 | — |
SpDs, specificity domains that interact with Fip1; RBS, RNA-binding site; HAT repeat and WD-40 repeat, potential protein-protein interaction motifs; RRMs, RNA recognition motifs, including RNP1 and RNP2 sequences.
—, unknown.
Mammalian cells.
(i) Cleavage/polyadenylation specificity factor.
In the late 1980s, it was discovered that AAUAAA-dependent processing required a factor termed cleavage and polyadenylation factor (CPF), specificity factor (SF), or polyadenylation factor 2 (PF2), and now called CPSF (96, 168, 453, 454). CPSF is required for both the cleavage and poly(A) addition reactions and, consistent with this function, recognizes AAUAAA, a signal also essential for both reactions. Gel retardation experiments showed that CPSF specifically binds AAUAAA-containing RNAs (32, 45, 237). RNA modification interference assays indicated that all six nucleotides of AAUAAA are necessary for binding (237) and that RNAs as short as 10 nucleotides can be bound specifically (491). CPSF thus appears to recognize only the AAUAAA sequence, independent of any secondary structure. The binding of purified CPSF is very weak but can be greatly enhanced by a cooperative interaction with CstF bound to the downstream signal sequence (167, 280, 485, 499). CPSF purified from calf thymus or HeLa cells is a large protein complex containing subunits of 160, 100, 70, and 30 kDa, referred to as CPSF-160, CPSF-100, CPSF-70, and CPSF-30, respectively (45, 328).
The sequence of the largest subunit (160 kDa) contains a possible bipartite nuclear localization signal (NLS) and sequences roughly similar to the RNP1 and RNP2 motifs found in many RNA-binding proteins (228, 329). The carboxyl end of CPSF-160 also has homology to the C terminus of Rse1, a yeast pre-mRNA-splicing protein (79). The cross-linking of CPSF-160 to RNA in the processing extract depends on AAUAAA (314), and recombinant CPSF-160 (rCPSF-160) alone binds preferentially to AAUAAA-containing RNAs (329), supporting the idea that this subunit is crucial for AAUAAA recognition. However, the specific binding of rCPSF-160 is less efficient than that observed with intact CPSF, suggesting that the participation of other CPSF subunits facilitates the recognition of AAUAAA. With HIV-1 pre-mRNAs, CPSF-160, as part of CPSF, can be cross-linked to the RNA in two places, at the AAUAAA and at the USE (164). CPSF-160 interacts specifically with the 77-kDa subunit of CstF and with PAP (329), which is consistent with the cooperative interactions of CPSF with CstF or PAP in forming stable complexes on the RNA precursor (reviewed in references 101, 238, and 473). Interestingly, rCPSF-160 inhibits the activity of PAP in nonspecific assays, implying that CPSF may facilitate both poly(A) synthesis and termination (329). The Drosophilia CPSF-160 is essential for viability (402).
The 100- and 73-kDa subunits of CPSF are closely related, with 23% identity and 49% similarity (227, 229). Antibodies raised against CPSF-100 coimmunoprecipitate all four subunits of CPSF, confirming their association as a complex (227). The functions of both CPSF-100 and CPSF-73 are unknown, but CPSF-100, as part of CPSF in extract, can be cross-linked to RNA by UV light (136), suggesting close contact with the precursor and perhaps a role in RNA binding.
The fourth subunit, CPSF-30, contains five CCCH zinc finger repeats followed by a CCHC zinc knuckle. Both types of motifs have been implicated in binding nucleic acid. In agreement with this sequence feature, CPSF-30 binds RNA polymers, with a distinct preference for poly(U). It has not always been detected in active CPSF preparations (164, 328) and may be less strongly associated with the other CPSF subunits under some conditions. However, it is coimmunoprecipitated with the other CPSF subunits (31, 227), and immunodepletion of this protein from extract or partially purified CPSF fractions inhibits cleavage and polyadenylation (31). The role of CPSF-30 is most probably to cooperate with CPSF-160 in the recognition of RNA substrates and, through an interaction with PAB II (85), to stabilize the polyadenylation complex.
The Drosophila clipper (clp) gene encodes a homolog of CPSF-30 which has five CCCH zinc finger motifs and two CCHC zinc knuckles. Members of this highly conserved family of proteins have been found in mouse, zebrafish, Caenorhabditis elegans, and S. cerevisiae (29). CLP is a nuclear protein that is posttranscriptionally regulated during development, and the zebrafish homolog, no arches, is essential for normal pharyngeal arch development (155). The yeast homolog, Yth1, is part of the PF I polyadenylation factor (31). The region containing the zinc finger motifs of CLP has endoribonucleolytic activity specific for RNA hairpins (28), and the C-terminal zinc knuckles confer to CLP a binding preference for RNA with G- and/or C-rich clusters (29). While these properties have not been reported for CPSF-30 and an association of CLP with other CPSF subunits has not been demonstrated, the remarkable conservation of the two proteins certainly raises the possibility that CPSF-30 is directly involved in cleavage at the poly(A) site.
(ii) Cleavage stimulation factor.
CstF is necessary for cleavage but not for poly(A) addition (168, 453), although it can stimulate poly(A) addition on substrates with a CstF binding site upstream of the AAUAAA hexanucleotide (319). Purification of CstF from HeLa cells showed that it consists of three polypeptides of 77, 64, and 50 kDa (167, 452). cDNAs for all three have been sequenced (447, 448, 450). CstF-77 and its yeast and Drosophila homologs have eight repeats very similar to the tetratricopeptide repeat (TPR)-like motifs found in the yeast Prp39 and Prp42 U1 small nuclear ribonucleoproteins (snRNP) (303, 438). The repeats in the CstF-77 family of proteins lack the highly conserved alanine and glycine residues of a TPR, and this new motif has been termed HAT (half a TPR) (370). Like TPR repeats, HAT repeats may mediate protein-protein interactions (101, 370). Consistent with this sequence feature, CstF-77 was shown to be the middle subunit bridging CstF-64 and CstF-55, with three of them arranged in a linear fashion (448). Its direct interaction with CPSF-160 probably contributes to the mutual stabilization of the CPSF-CstF-RNA complex (329). CstF-77 is homologous to the Drosophila suppressor of forked [su(f)] protein (448). Mutations in su(f) can enhance or suppress the effects of transposon insertion, probably through changes in polyadenylation.
CstF-64 contains a classical RNA-binding domain (RBD) close to its amino terminus. It is connected by a hinge region to an unusual proline- and glycine-rich region (40%) in the carboxy terminus (447). Embedded in the Pro-Gly-rich region is a domain of 12 repeats of MEAR(A/G) in the mouse and human genes and 11 repeats of LEPRG in the chicken gene (455). By a UV cross-linking assay with whole-cell extracts, CstF-64 was first implicated in AAUAAA-dependent binding (314, 497). However, the site of CstF-64 was subsequently mapped to the U-rich DSE of pre-mRNA (280). The AAUAAA dependence reflects the strong cooperative binding of CPSF to AAUAAA and of CstF to the DSE. RNA ligands selected by CstF resemble DSEs in sequence and function (42). Selection-amplification (SELEX) using the CstF-64 RBD confirmed that this region is sufficient to recognize GU- and U-rich sequences (449).
CstF-50 contains seven transducin, or WD-40, repeats, a motif which can mediate protein-protein interactions in other proteins (450).
(iii) Cleavage factors Im and IIm.
CF Im and CF IIm are required only for cleavage. (The designations CF I and CF II have been used for both the mammalian and yeast systems, but the factors are not homologous. The subscript “m” is included to differentiate the mammalian factor from the yeast one.) CF Im has been purified to near homogeneity. Three polypeptides of 25, 59, and 68 kDa and possibly a fourth one of 72 kDa copurify with CF Im activity (388, 389). The three smaller subunits can be UV cross-linked to RNA substrates (388). By gel retardation assays, purified CF Im has higher affinity for RNAs containing polyadenylation signal sequences than for unrelated RNAs. Furthermore, CF Im increases the stability of the CPSF-RNA complex, suggesting that this factor may also interact with CPSF and contribute to the overall stability of the 3′-end-processing complex (388). cDNAs encoding the 25- and 68-kDa subunits have been recently isolated (389). While CF Im-25 has no known motifs, CF Im-68 contains three distinct domains: an amino-terminal RNP-type RBD, a proline-rich region in the middle, and a carboxyl-terminal section consisting of alternating residues of opposite charge, with arginine residues alternating with glutamate, aspartate, and serine residues. This domain organization is strongly reminiscent of that found in the superfamily of RS-rich splicing factors. Most interestingly, recombinant CF Im-25 and CF Im-68 can be assembled in vitro and can replace purified CF Im in cleavage assays. Preliminary sequence data from studies of the CF Im-59 polypeptide reveal that it is similar to CF Im-68, suggesting either that it is a degradation product of CF Im-68 or that CF Im exists as heterodimers of CF Im-25-68 or CF Im-25-59 (389). Analysis of the kinetics of the cleavage reaction indicates that interaction of CF Im with RNA substrate may be an early step in the assembly of the 3′-end-processing complex, which facilitates the recruitment of other processing factors (389).
CF IIm has not been purified to homogeneity, and its function is not known.
(iv) RNA polymerase II.
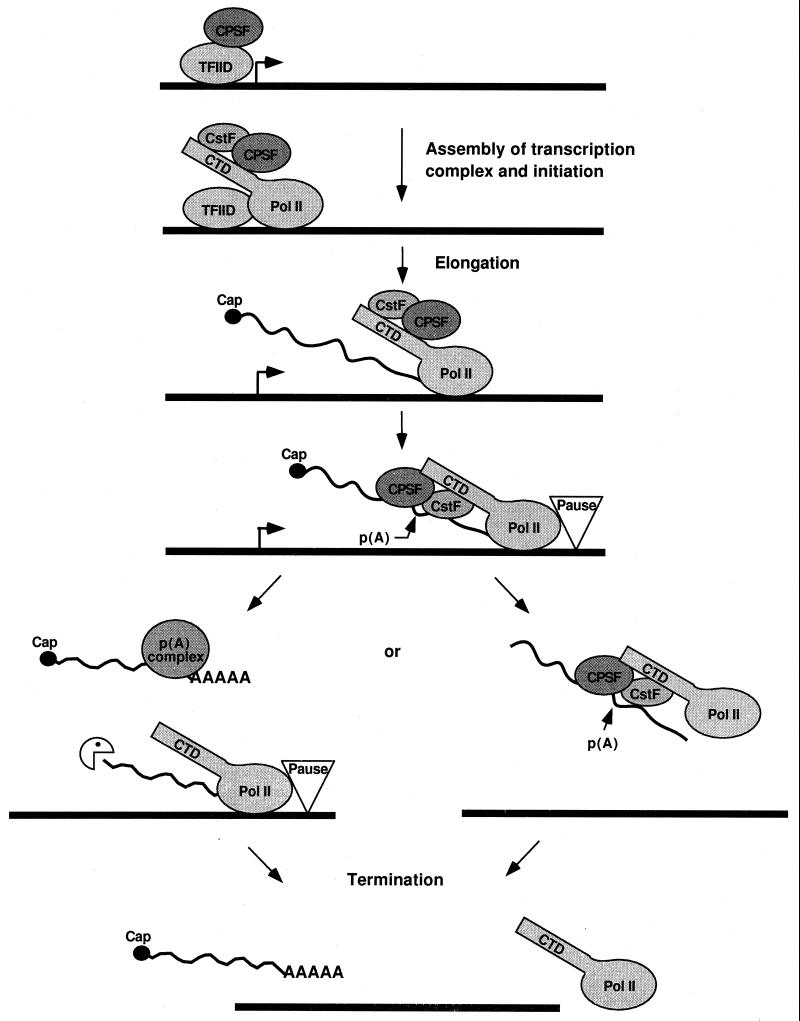
Pol II, through the conserved carboxyl-terminal domain (CTD) of its largest subunit, has properties which make it an authentic cleavage factor. The CTD is found in a hyperphosphorylated form in elongating transcriptional complexes (186). An understanding of a role for the CTD in the cleavage of pre-mRNAs evolved from a study of the function of creatine phosphate (CP) in this reaction (204). In the early in vitro work on cleavage and polyadenylation, ATP was shown to be necessary for cleavage of precursor mRNAs (316), and therefore CP was included to regenerate ATP through creatine phosphokinase activity in the extracts. However, other phosphocompounds can substitute for CP in the cleavage reaction, leading to the proposal that CP was acting as a mimic for a phosphoprotein (204, 205). This hypothesis was confirmed when a synthetic CTD, even without phosphorylation, or purified pol II was shown to direct the cleavage of polyadenylation precursor in the absence of CP or ATP (205). Pol II does not appear to be involved in the poly(A) addition step. The interaction of the CTD with CPSF and CstF (299) may stabilize the cleavage complex or, as proposed by Hirose and Manley (205), have allosteric effects, such as those found for CTD and the capping enzyme (443). This involvement of Pol II in cleavage is consistent with earlier studies showing that protein-encoding mRNAs transcribed by Pol I and Pol III were for the most part not polyadenylated (257, 429, 432) and with the recent demonstration of the colocalization of CstF and phosphorylated pol II in vivo (439). Other consequences of the coupling of transcription and mRNA 3′-end formation are explored in a later section.
(v) Poly(A) polymerase.
An activity which added adenosine residues to the 3′ ends of RNAs was discovered in the early 1960s, and at the time it was a reaction of unknown significance (reviewed in reference 135). It is now well established that the this activity, PAP, plays a key role in the 3′-end formation of mRNA in eukaryotic cells. The first PAP, purified to homogeneity from calf thymus, was a degradation product of 57 to 60 kDa (475). Cloning and expression of the bovine PAP cDNAs have identified at least two isoforms of PAP which are generated by alternative splicing (380, 475). The longest forms, PAP I (77 kDa) and PAP II (82 kDa), differ only at their C termini and are enzymatically active (380, 515). PAP II may be the predominant full-length species, since most cDNAs isolated from other animal cells encode PAP II-related isoforms (30, 460, 475). Several other short forms of PAP (PAPs III, V, and VI) encode truncated proteins that are enzymatically inactive, and their function is unknown (380, 515).
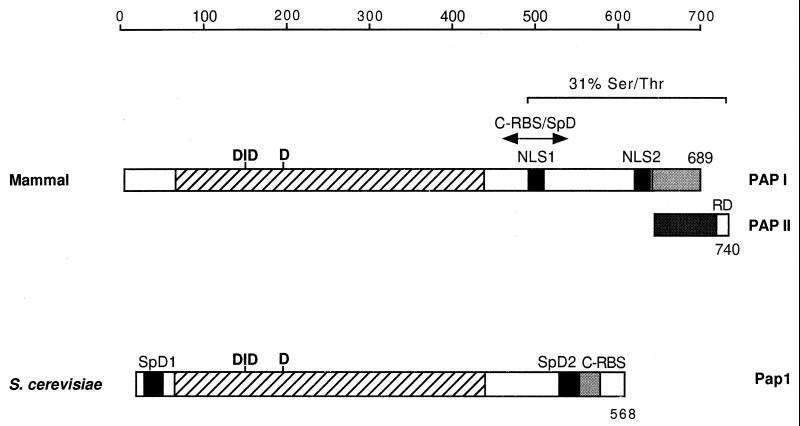
Analysis of PAP has indicated an organization of functional domains as illustrated in Fig. 3 (290, 291, 380, 381). The amino-terminal two-thirds of PAP is highly conserved in eukaryotes and contains a catalytic domain with homology to a family of nucleotidyltransferases including many DNA and RNA polymerases (207, 291, 507). The catalytic core of this family is characterized by a triad of conserved aspartate residues that are essential for activity. These three aspartate residues are located at positions 113, 115, and 167 in bovine PAP (291). A primer-binding domain (C-RBS in Fig. 3) is located between amino acids 488 and 508, in the carboxyl-terminal portion of PAP (291). It overlaps with a region (amino acids 493 to 538) needed for AAUAAA-dependent activity (460), which may interact with CPSF-160. This region also encompasses NLS-1, which is required, together with a second NLS (NLS-2) about 140 residues downstream of the first, for efficient localization of PAP to the nucleus (129). Besides the bipartite NLS, the carboxyl-terminal region is also rich in serine and threonine residues, which are the targets for multiple phosphorylations which regulate PAP activity (2, 102). The last 20 amino acids of PAP (amino acids 720 to 739) are involved in autoregulation of U1A transcripts and in the coupling of splicing and polyadenylation (177).
FIG. 3.
Schematic diagram of the bovine and yeast poly(A) polymerases. The scale at the top indicates the size in amino acids. The hatched bar indicates the conserved regions. DID and D mark the locations of the three aspartate residues essential for nucleotidyltransferase catalytic activity. Gray bars in different shades in the bovine PAP indicate the C terminus of PAP I and PAP II generated from alternative splicing. NLS, nuclear localization signal; RD, regulatory domain involved in inhibition by U1A and in coupling of splicing and polyadenylation; SpD, specificity domains; C-RBS, carboxyl-terminal RNA-binding site.
Under physiological conditions and in the absence of other factors, PAP has only a very low level of activity (472). The rate of poly(A) addition is significantly elevated when manganese is substituted for magnesium in the reaction, increasing the affinity of the enzyme for the primer (475). PAP is specific for the utilization of ATP (475) but has no specificity for RNA substrate. In AAUAAA-dependent polyadenylation, PAP is recruited to the processing complex by interaction with CPSF-160 (329) and is also required for cleavage of most pre-mRNAs.
(vi) Poly(A)-binding protein II.
CPSF and PAP suffice for poly(A) addition to a precleaved RNA substrate. However, rapid elongation and control of poly(A) tail length requires an additional factor, PAB II (44). The 33-kDa PAB II contains a very acidic amino-terminal domain, a very basic carboxyl-terminal domain, and a single RNP domain in its middle region. The protein tends to form oligomers and binds specifically to poly(A) and poly(G) (331, 474). In vitro assays indicate that PAB II binds directly to CPSF-30 (85). A Drosophila homolog is the product of the rox2 gene (60).
Yeast cells.
Fractionation of whole-yeast-cell extracts has identified five functionally distinct activities involved in cleavage and polyadenylation (82, 241). CF IA, IB, and II are sufficient for the cleavage reaction, while specific poly(A) addition requires CF IA and CF IB, Pap1, Pab1, and PF I (Fig. 1). All these factors have been purified to near homogeneity, and genes encoding most of the components have been cloned (Table 3). All the genes that have been cloned are essential. While the polyadenylation signals used by mammals and yeast are rather different in consensus sequence and organization, the factors which comprise the cleavage/polyadenylation apparatus in these two organisms exhibit surprising conservation.
At this point, it is important to mention several considerations about experimental procedures used in the yeast system, which, when not taken into account, can make some of the findings described below seem confusing. Immunodepletion or copurification is useful to indicate the involvement of a protein in cleavage or polyadenylation, but sometimes this may simply show that a complex exists between two factors. Mutations in proteins involved in 3′-end formation may show defects in cleavage or poly(A) addition or both. A defect in only one step of the reaction could mean that domains important for the other step have not been affected by the mutation. Furthermore, the levels of factors may be coordinately regulated in ways which are not clear. For example, temperature-sensitive mutations in the CF IA subunits Rna15 and Rna14 or in the poly(A) polymerase, Pap1, can all lead to reduced levels of Rna15 protein at 37°C (11). Thus, a mutation which presents defects in both steps may be affecting one of the steps only indirectly. Finally, even though an activity appears to be purified to homogeneity, there is always the caveat that a critical subunit is present in substoichiometric amounts or is not easily detectable by protein-staining methods. Nevertheless, the accumulation of biochemical and genetic data described below is leading to a much clearer understanding of the roles of the various proteins in cleavage and polyadenylation.
(i) Cleavage/polyadenylation factor IA.
CF I was originally identified as an activity needed for both the cleavage and poly(A) addition reactions (82), and further purification separated it into two components, CF IA and CF IB (241). CF IA consists of four polypeptides, Rna14 and Rna15 (241, 311), Pcf11 (13), and a 50-kDa polypeptide (241, 369). The first indication of the involvement of Rna14 and Rna15 in poly(A) mRNA metabolism came from the dramatic poly(A) tail shortening seen in strains harboring temperature-sensitive mutations in the RNA14 and RNA15 genes (312). These mutations are synergistically lethal with mutations in the PAP1 gene (311). Extracts from rna14 and rna15 mutants are defective in both cleavage and poly(A) addition, and fraction complementation assays suggested that Rna14 and Rna15 were components of CF I (311). Purification of CF I showed that Rna14 and Rna15 are indeed CF IA subunits (241). These two polypeptides are tightly associated, as indicated by a two-hybrid assay (239) and by coimmunoprecipitation as a heterodimer from whole-cell extract or CF I-containing fractions by antibodies against Rna15 (241). The 76-kDa Rna14 has sequence homology to mammalian CstF-77 (24% identity) (448).
Rna15, the 38-kDa subunit, is the yeast homologue of CstF-64 (448). It contains an RNA recognition motif (RRM)-type RBD in its amino-terminal region and can be UV cross-linked to substrate RNA (241, 312, 448). Its RNA-binding site in the processing complex is not known, and the existing clues are somewhat conflicting. Alone or as part of CF IA, Rna15 cross-links equally well to wild-type RNA and mutant RNA lacking the AU-rich efficiency element (308). Interestingly, the RBD found in Rna15 closely resembles that of mammalian CstF-64, and both show higher affinity for U-rich sequences (449). However, RNAs selected by Rna15-RBD affinity do not interact with CstF-64, and they bear some similarity to the upstream AU-rich efficiency element (449). The quite distinct RNA-binding preferences of these two closely related RBDs is consistent with the divergence in polyadenylation signal sequences between yeast and mammals. Downstream of the RBD, Rna15 has a stretch of glutamines and asparagines, similar to the opa sequences of Drosophila developmental genes and to those of several transcriptional regulators (312).
Pcf11 was identified as a 70-kDa protein which interacts with both Rna14 and Rna15 in a two-hybrid screen (13). The N terminus has some similarity to CTD-binding regions in other proteins such as Nrd1, a yeast heterogeneous nuclear RNP (hnRNP)-like protein which interacts with the mouse Pol II CTD in a two-hybrid assay and affects the elongation of Pol II transcripts containing an Nrd1-binding site (438, 439). Pcf11 also contains a striking stretch of 20 consecutive glutamine residues followed by the region responsible for the interaction with Rna14 and Rna15 (13). Extracts from pcf11 temperature-sensitive mutant strains are defective in both cleavage and poly(A) addition (13). Moreover, Pcf11-specific antibodies recognize the 70-kDa polypeptide of purified CF IA (240). Homology to Pcf11 has not been found in the mammalian system.
Microsequencing of internal peptides from the 50-kDa subunit revealed that it corresponded to elongation factor 1α (EF-1α), an essential GTP-binding protein functioning in translation elongation (240). Antibodies against Pcf11 bring down only the four subunits of CF IA from CF I-containing fractions, and the 50-kDa protein in the immunoprecipitate was recognized by EF-1α-specific antibodies (235, 514). However, a 50-kDa subunit has also been identified as a new protein called Clp1, which, like EF-1α, has a P-loop motif indicative of ATP-GTP binding (369). Thus, the identity of the 50-kDa subunit of CF IA needs to be further clarified.
(ii) Cleavage/polyadenylation factor IB.
Purified CF IB is a single polypeptide of 73 kDa (241) encoded by the HRP1/NAB4 gene (239). This gene was previously identified as a suppressor of a temperature-sensitive npl3 allele, a gene encoding a protein which is involved in mRNA export (201) and can be cross-linked to nuclear poly(A)+ RNA in yeast (308). Hrp1 is structurally related to the mammalian hnRNP A, B, and D proteins (239) and has two RRMs in its middle region, both containing RNP1 and RNP2 sequences (201). The last 50 amino acids of Hrp1 are rich in arginine and glycine, with potential RGG methylation sites. A similar domain of hnRNP A1 can mediate protein-protein interactions (97). Experimental data indicate that the UA-rich polyadenylation signal is the likely binding site for Hrp1. The UV-induced RNA cross-linking of recombinant Hrp1 and the endogenous protein in yeast extracts is greatly enhanced by the presence of this sequence (83, 239). A recent SELEX analysis has also shown that UAUAUA is a high-affinity binding site for Hrp1 (464). The closest counterpart to Hrp1 in the mammalian system, at least in terms of having an amino-terminal RBD and a function in cleavage, is CF Im-68.
Recombinant Hrp1 can fully replace the yeast CF IB in both the reconstituted cleavage and poly(A) addition assays (239). Synergistic-lethal-interaction assays and two-hybrid analysis indicated that Hrp1 interacts in vivo with Rna14 and Rna15 but not with Pap1 (239), consistent with its copurification with CF IA. Hrp1 shuttles between the nucleus and the cytoplasm (239), a property facilitated by Hmt1-catalyzed arginine methylation (202, 423). A recent study has reported that Hrp1 is not essential for cleavage of pre-RNAs and instead may regulate cleavage site utilization (308). In this study, pre-mRNA substrates were cleaved at additional sites as well as the normal cleavage site in the absence of Hrp1, and Hrp1 acted in a concentration-dependent manner to suppress the use of the alternate sites. This discrepancy may be attributable to relative concentrations of factors in the reconstituted assays, to differences in the composition of CF II used in the two studies, or to other experimental procedures.
(iii) Cleavage factor II.
CF II has been purified by taking advantage of its ability to reconstitute the cleavage reaction in the presence of purified CF IA and CF IB (512). It contains four polypeptides, Cft1/Yhh1 (150 kDa) (442, 512), Cft2/Ydh1 (105 kDa), Brr5/Ysh1 (100 kDa), and Pta1 (90 kDa) (513). Cft1/Yhh1 was first identified by its sequence homology to mammalian CPSF-160 (24% identity and 51% similarity) (442). Depletion of extract with antibodies to Cft1/Yhh1 abolished both cleavage and poly(A) addition, consistent with CF II also being part of PF I (see below). However, addition of a CF II-containing fraction restored cleavage activity but not poly(A) addition (442). Cft1/Yhh1-specific antibodies recognize the 150-kDa component of purified CF II (512) and precipitate only four subunits (Cft1/Yhh1, Cft2/Ydh1, Brr5/Ysh1, and Pta1) from partially purified preparations (513).
Cft2/Ydh1, the 105-kDa subunit of CF II, has significant homology to CPSF-100 (24% identity and 43% similarity) (512). Cft2/Ydh1, as part of CF II, can be UV cross-linked to wild-type full-length pre-mRNA substrate but not to wild-type precleaved RNA or mutated substrate that lacks a (UA)6 efficiency element, suggesting that Cft2/Ydh1 may recognize the efficiency element and/or poly(A) site (512). The cross-linking of Cft2/Ydh1 to RNA is also ATP dependent.
The third subunit (100 kDa) of CF II is Brr5/Ysh1 (512), a yeast homologue of mammalian CPSF-73 with 23% identity and 48% similarity through its entire length and 53% identity in the first 500 amino acids (78, 229). Brr5/Ysh1 was identified by this homology and as a mutation which gave a cold-sensitive defect in the in vivo splicing of mRNA. The smallest subunit of CF II has been recently identified as a protein encoded by PTA1, an essential gene affecting pre-tRNA processing (342, 513). In one report, extracts from brr5/ysh1 and pta1 mutant strains were shown to be deficient in poly(A) addition but not cleavage (78, 369), whereas a recent study found that these extracts were defective in both steps (513). The cleavage defect could be rescued by CF II. These discrepancies may be due to differences in extract preparation or culture conditions, which could influence the concentration or stability of proteins in the mutant extracts.
A protein corresponding to Yth1, the yeast CPSF-30 homolog, was not detected by silver staining in the purified preparation of Zhao et al. (512). It has been reported that Yth1 can be detected by immunoblotting in partially purified CF II (unpublished results cited in reference 308) and that point mutations in the second zinc finger cause reduced cleavage activity in vitro (309). Interestingly, the entire CF II complex copurified with PF I (see below) (369), suggesting that CF II also plays a role in poly(A) addition. The involvement of CF II in both cleavage and poly(A) addition and the sequence homology of three of its subunits to those of CPSF support the idea that CF II is the functional homolog of this mammalian factor.
(iv) Polyadenylation factor I.
PF I was originally identified as an activity which supported poly(A) addition but not cleavage (82). A multiprotein complex from yeast containing PF I activity has been recently purified by restoration of polyadenylation activity to extracts with mutated Fip1 subunit of PF I (369). In addition to Fip1, this complex contained Pap1 (see below), Yth1, all four subunits of CF II, and two uncharacterized proteins, Pfs1 (58 kDa) and Pfs2 (53 kDa). The PF I-Pap1-CF II association is also indicated by coimmunoprecipitation experiments with yeast whole-cell extracts, since all CF II components as well as Pap1 and Fip1 were precipitated specifically by antibodies against Pap1 or Fip1 (513). It is not clear whether CF II in extract and cells exists in two forms (free and associated with PF I-specific subunits) or whether it is separated from PF I activity only by chromatography.
Fip1 (for “Factor interacting with Pap1”) was identified as a protein that interacts with Pap1 in a two-hybrid screen (371). It has a predicted molecular mass of 35 kDa but migrates as 55 kDa on sodium dodecyl sulfate-polyacrylamide gels. Fip1 has some sequence homology to CPSF-160 (329) and contains a very acidic amino terminus and a proline-rich region at the carboxyl terminus. This proline-rich domain is also found in several other proteins, including CstF-77 (448), the 70K U1 snRNP (379), and Pab1 (398). This domain in Fip1 is not required for cell viability (371), although truncation of Fip1 beyond the proline-rich region causes temperature-sensitive growth. Extract from this strain is defective in poly(A) addition but efficient in cleavage. Biochemical analysis with in vitro translation products demonstrated that Fip1 interacts directly with Pap1, Yth1, and Rna14, although the binding to Rna14 is weak (31, 371). Fip1 alone can alter the processivity of Pap1 (517). These properties suggest that it is the functional homolog of CPSF-160 in the specific poly(A) addition reaction and plays a central role in the assembly of the yeast polyadenylation machinery.
Like Cft1/Yhh1 and Brr5/Ysh1, Yth1 (for “yeast thirty-kilodalton homolog”) was also isolated by sequence similarity to a CPSF subunit. It has 40% identity and 60.5% similarity to CPSF-30 (31). However, Yth1 lacks the C-terminal zinc knuckle motifs found in the metazoan proteins (31). Yeast strains containing the yth1-1 mutant, which is truncated at its carboxyl terminus to leave only four zinc finger motifs, are temperature sensitive for growth. Extracts prepared from this mutant are normal in cleavage but deficient in poly(A) addition. While Yth1 in a highly purified PF I preparation cannot be detected by silver staining, its presence as a PF I subunit was confirmed by immunoblot analysis (369).
The two additional proteins, p58 and p53, that copurified with PF I are encoded by the essential PFS1 and PFS2 genes (unpublished data cited in reference 238). Pfs1 has a zinc knuckle, and Pfs2 has WD-40 repeats.
(v) Poly(A) polymerase.
Yeast Pap1 was the first factor of the yeast 3′-end processing machinery to be purified, and its gene, PAP1, was the first to be identified. PAP1 was cloned both by sequencing of the purified 64-kDa protein and by complementation of a temperature-sensitive pap1 allele (263, 351). The yeast and mammalian Pap1 proteins are 47% identical within the first 400 amino acids, a region thought to comprise the catalytic domain and include the nucleotidyltransferase active site (291) (Fig. 3). The carboxyl-terminal regions are not conserved. Monoclonal antibodies which recognize epitopes in the amino- and carboxyl-terminal regions of Pap1 do not recognize the mammalian enzyme (242). An RNA-binding site at the carboxyl-terminus (C-RBS) is thought to interact in part with the phosphate backbone of polynucleotides and is essential for processive activity (517). At least two other contacts with the RNA substrate exist in addition to C-RBS. While their location on the enzyme is not known, one is thought to recognize the last 3 nucleotides of the RNA primer and to help the enzyme discriminate against deoxyribonucleotide substrates, and another base-specific site is proposed to interact with the primer 12 to 14 nucleotides upstream of the 3′ end (517).
Mutational analysis has identified two specificity domains, SpD1 and SpD2, located at either end of Pap1 as indicated in Fig. 3 (517, 518). These are probably necessary to recruit Pap1 to the polyadenylation machinery by interaction with specificity factors and to regulate its activity. In agreement with this idea, deletion of SpD1 eliminates the activity of the enzyme in association with the polyadenylation factors but has no effect on its ability to extend an oligo(A) primer. Furthermore, both SpD domains interact with the Fip1 subunit of PF I by two-hybrid analysis. Interestingly, SpD2 partially overlaps with the C-RBS. The presence of recombinant Fip1 increases the Km of Pap1 for RNA 50-fold, prevents the cross-linking of Pap1 to RNA, and results in a shift to a distributive mode of action, consistent with a direct interaction of Fip1 at SpD2 (517). It is interesting that a specificity domain of the mammalian PAP also overlaps with a carboxyl-terminal primer-binding site (291, 460).
Unlike the mammalian system, the yeast Pap1 is not required for efficient cleavage of precursor in vitro. However, a mutation in Pap1 conferring temperature-sensitive growth can influence the choice of poly(A) site in the ACT1 transcript (284).
(vi) Poly(A)-binding protein I and poly(A) nuclease.
Yeast Pab1 is the major RNP associated with the poly(A) tails of mRNA in both the nucleus and cytoplasm (3, 420, 446). Amino acid sequence analysis of this 70-kDa polypeptide revealed four RRM-type RBDs at its amino-terminal region and a proline-rich carboxyl terminus (67). Pab1 is important for translation initiation (456–458) and for deadenylation-dependent mRNA turnover (71). Recent studies have found that it is also involved in mRNA poly(A) tail formation. Pab1 cofractionates with CF IA (310) and interacts with Rna15 in two-hybrid assays and by coimmunoprecipitation (12). Cells bearing a pab1 mutation conferring temperature-sensitive growth show aberrantly long poly(A) tails in vivo (400) and in vitro (12, 310). Addition of anti-Pab1 antibodies to a processing extract results in an elongated poly(A) tail but has no effect on the cleavage reaction (12). In a reconstituted-poly(A) addition assay, Pab1 was further confirmed to function in limiting the length of poly(A) tail but was not required for cleavage (239).
Pab1 acts in concert with a poly(A)-specific nuclease (PAN) to affect the poly(A) tail length (120, 274). PAN is composed of at least two subunits, Pan2 (127 kDa) and Pan3 (76 kDa), both encoded by nonessential genes (50, 64). Deletion of the PAN2 and/or PAN3 gene resulted in a similar increase in mRNA poly(A) tail lengths in vivo and in the loss of Pab1-stimulated PAN activity in yeast extracts (50, 64). Pan2 and Pan3 directly interact with each other, as shown by coimmunoprecipitation and two-hybrid analysis (64). Pan2, which interacts with Pab1 (50), is likely to be the catalytic subunit of the PAN complex, since it is a member of the RNase D family of 3′-5′ exoribonucleases (306, 321). The proline-rich C terminus of Pab1 is necessary for PAN activity in vitro (285). Recently identified deadenylating nucleases from Xenopus and human also belong to the RNase D family (249). Deadenylating nuclease is localized in both the nucleus and cytoplasm and, unlike PAN, is inhibited by PAB I, the mammalian homolog of the yeast Pab1. A recent study with polyadenylation extracts found that PAN matures newly synthesized poly(A) tails to defined poly(A) tail lengths of 50 to 90 nucleotides (63). In vivo, this process is rapid and appears to precede translation and mRNA degradation. However, it is not clear whether PAN-dependent deadenylation occurs in the nucleus as an integral step of the 3′-end-processing reaction or, instead, as an early cytoplasmic mRNA maturation event.
Pab1 appears to play two roles in regulating tail length: (i) suppression of the activity of Pap1 by limiting its access to the RNA substrate (517) and (ii) recruitment of PAN. These negative effects of Pab1 are somewhat alleviated by Pbp1, a protein isolated by two-hybrid analysis as a Pab1-interactor (285). In extracts from a strain lacking Pbp1, the tails are shorter, and its proposed function is to help regulate poly(A) tail length by suppressing the activity of Pab1 or perhaps the association of PAN with Pab1.
(vii) Proteins with possible auxiliary or regulatory roles in yeast polyadenylation.
Other proteins which are not components of constitutive polyadenylation factors can influence the efficiency or accuracy of processing in yeast. Some of these gene products suggest interactions of polyadenylation with other cell processes or ways of regulating the activity or specificity of the reaction. For example, temperature-sensitive mutations in the essential ESS1/PTF1 gene lead to increased readthrough of certain poly(A) sites and a decrease in the level of total poly(A)+ RNA in the cell (187). This gene encodes a peptidylprolyl-cis/trans-isomerase (PPIase), an enzyme thought to function in protein folding. While in vitro work is necessary to localize the defect to transcription termination or cleavage/polyadenylation, the authors noted that PPIases associate with the phosphorylated C-terminal domain of mammalian RNA pol II (55) and with the U4/U6.U5 tri-snRNP splicing factor (209, 459) and are proposed to promote disassociation of the HIV-1 capsid core (56). These findings led to speculation that the yeast PPIase activity might be involved in the assembly of polyadenylation or transcription complexes or their disassembly at termination sites, a topic discussed later in this review.
Ref2, an RNA-binding protein, is important for the efficient processing of weak poly(A) sites (393). Disruption of the gene encoding Fir1/Pip1 (122, 394), a nonessential protein which interacts with many nuclear proteins such as Pap1, Ref2, Sir4, topoisomerase 1, and lamin, exacerbates the defective processing seen in cells lacking Ref2 (394). The yeast Pap1 was found to interact specifically with two essential proteins, Ufd1 and Uba2, by two-hybrid analysis in vivo and by coimmunoprecipitation in vitro (122). Ufd1 has been implicated in ubiquitin-mediated protein degradation (230), and Uba2 is involved in the conjugation of the ubiquitin-like protein, Smt3, to other proteins (231). Depletion of these proteins from cells affects the efficiency of processing in vitro (122).
For another class of proteins, a direct connection to polyadenylation is less obvious. Mutations in several proteins involved in nucleocytoplasmic transport cause the appearance of longer poly(A) tails and transcripts which extend beyond the normal poly(A) sites, and these are discussed below. A deletion of SSM4, a gene of unknown function (283), or overexpression of SSM1, encoding a ribosomal protein (360), can suppress temperature-sensitive mutations in RNA14. The SSM4 deletion also restores proper ACT1 poly(A) site selection in an rna14-3 mutant (284). STS1, a gene implicated in nuclear targeting, protein transport, rRNA stability, and chromosome segregation, suppresses the mRNA-processing defect of rna15-2 by restoring normal levels of the Rna15 protein (11). Mutations in RET1, encoding an RNA pol III subunit, or RRP6, a gene important for 5.8S rRNA 3′-end formation, can partially rescue the growth defect of a temperature-sensitive pap1-1 mutation (61, 62), and a mutation in LCP5, a gene involved in 18S rRNA maturation, is synthetically lethal in combination with the pap1-7 allele (490). The mechanism by which these effects are mediated is not clear. Some of the mutations may simply make the cell less dependent on a poly(A) tail for translation or may worsen translation problems due to polyadenylation defects, or they may affect polyadenylation indirectly by affecting nuclear import and export of processing factors and mature mRNA.
A 5′-3′ exonucleolytic activity degrades the 3′-cleavage products of both the yeast and mammalian reactions. However, this activity is not required for in vitro processing of polyadenylation precursor, since it is not present when the reaction is reconstituted from purified factors. Possible additional roles in transcription termination (discussed below) or in recycling polyadenylation factors by destroying their binding sites have not been investigated. The identity of this exonuclease is not known. The activity is magnesium dependent, and in yeast it may be provided by Rat1, a nuclear exoribonuclease which has been implicated in mRNA transport (10).
Steps in Processing: Assembly, Cleavage, and Polyadenylation
Mammals.
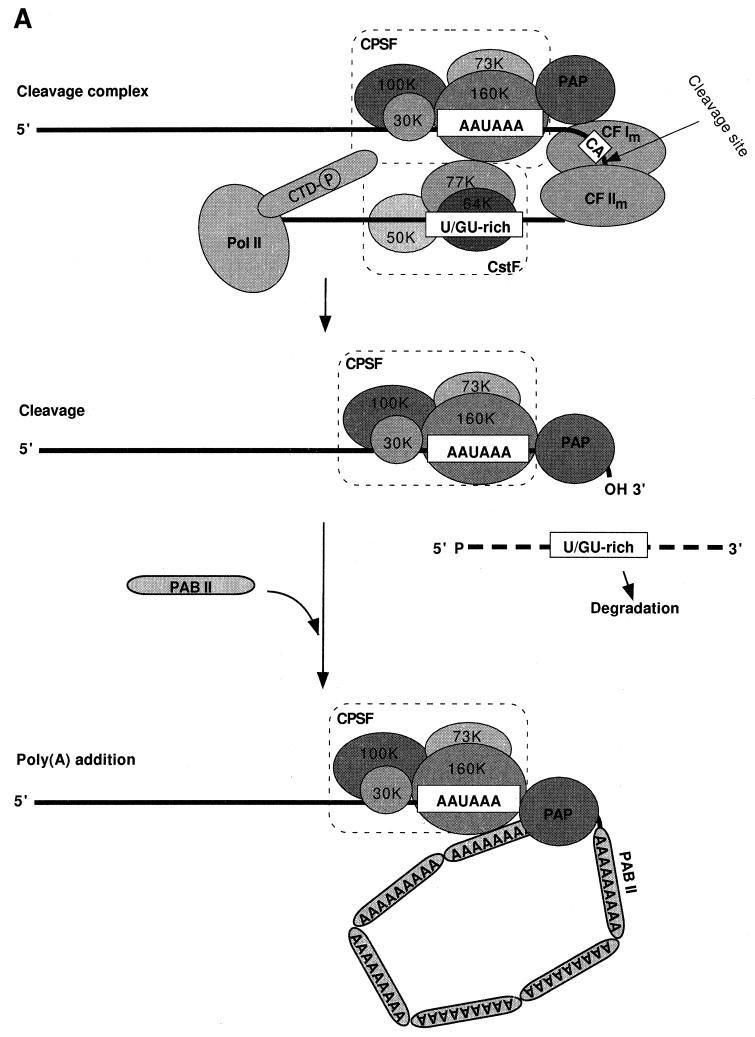
The mutually cooperative interactions of CPSF, CstF, CF Im, PAP, and PAB II in catalyzing accurate and efficient cleavage and polyadenylation have been well documented (167, 329, 388, 471, 499; for a review, see reference 473). The following model of mRNA 3′-end formation in mammalian cells (Fig. 4A) is derived from these numerous studies. The initiating step in assembly of a functional cleavage/polyadenylation complex is probably the recognition of signals on the precursor by CPSF and CstF in a process assisted by CF Im. CPSF binds to AAUAAA through CPSF-160 with the help of CPSF-30 and possibly CPSF-100, and CstF binds to the DSE via CstF-64. The individual interactions of CPSF and CstF with their cognate sequences are weak but are stabilized by the cross-factor interaction of CPSF-160 and CstF-77. A final component of the initiation complex is pol II (205). While it is not known when the precise cleavage site is chosen, the CPSF-CstF interactions define the region in which it must lie. The formation of a cleavage-competent complex requires the additional recruitment of CF IIm and PAP. The contacts of CF Im and CF IIm with the other factors and RNA are not known, but PAP at this point probably interacts with CPSF-160.
FIG. 4.
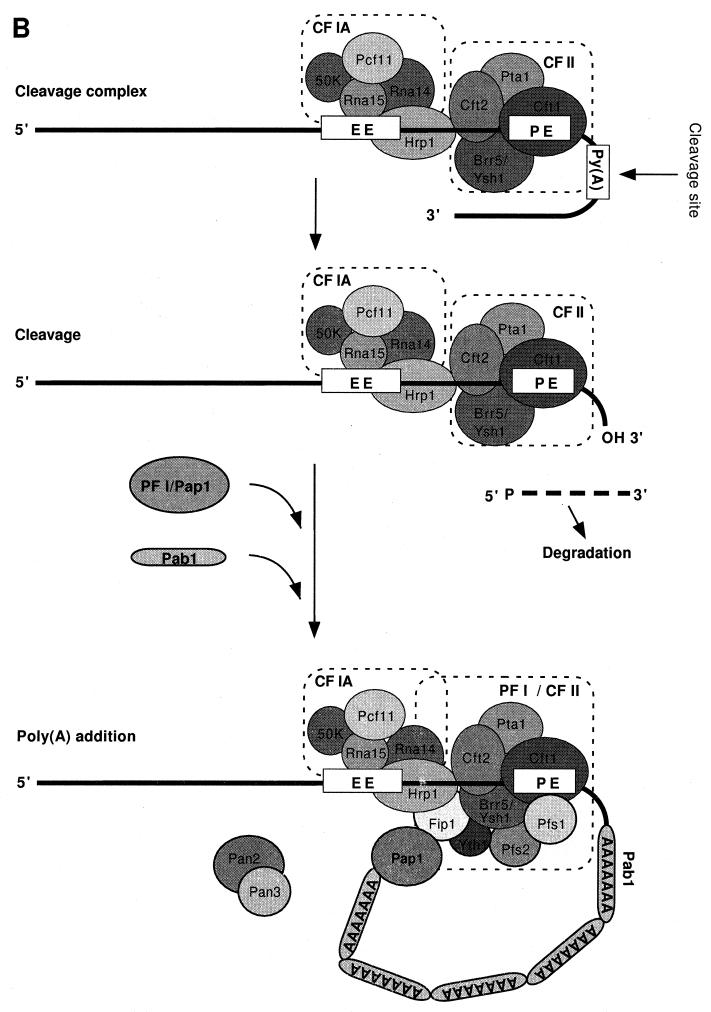
Schematic representation of the mammalian (A) and yeast (B) mRNA 3′-end-processing complexes. (A) The mammalian cleavage complex assembles through a cooperative binding of CPSF at the AAUAAA signal and CstF at the U- or GU-rich sequence. CPSF-160 directly interacts with CstF-77 and PAP. The arrangement of CF Im and CF IIm is not known. After the cleavage step, CPSF and PAP remain bound to the cleaved RNA and elongate the poly(A) tail in the presence of PAB II. (B) CF IA, Hrp1, and CF II are sufficient for the cleavage step in yeast. Poly(A) tail synthesis requires the addition of Pap1, PF I and Pab1. The Pan2/Pan3 deadenylase helps to regulate the poly(A) tail length.
The assembly of all of these factors may be sufficient to enable cleavage to occur. There is no indication of a reorganization of the complex before catalysis, as occurs during the splicing reaction (436). However, based on the information that reconstitution of polyadenylation in vitro requires only CPSF and PAP, significant changes probably take place after cleavage, with release and degradation of the 3′-cleavage product and dissociation of the CstF, pol II, CF Im, and CF IIm cleavage factors. The situation may be different in the cell, with some factors remaining in the processing complex. This possibility is supported by evidence that CstF can stimulate polyadenylation if a CstF-binding site is present upstream of the AAUAAA element (319).
Assembly and cleavage are followed by polyadenylation. The interaction with CPSF serves to recruit PAP to the AAUAAA-containing substrate. A complex of CPSF and PAP is adequate for a slow polymerization of an adenosine tract (44), but full processive activity requires PAB II. A biphasic poly(A) addition is observed in vitro, with synthesis being slow for the first 10 nucleotides until a binding site for PAB II has been created. The rapid elongation of the poly(A) tail to the final 250 adenosine residues found on nuclear poly(A)+ mRNA is promoted by the cooperative interactions of CPSF, PAP, and PAB II (471). When the tails reach this length, processive elongation switches to a slow and distributive mode again (471). PAB II is necessary to terminate poly(A) addition at this point, but the mechanism is not understood. This termination is probably coupled with release of CPSF and PAP from the finished product.
Yeast.
Since many of the yeast factors have only recently been characterized, less is known about how they interact with the RNA precursor and with each other. The available data and the homology to mammalian factors do allow us to construct a working model for assembly and processing in this organism (Fig. 4B). By analogy to the mammalian system, CF II and CF IA are probably prime players in the initiation of complex assembly. Their binding sites are not known, but from the structure of the core polyadenylation site in yeast, they are most probably located upstream of the cleavage site and may correspond to the UA-rich efficiency element and the A-rich positioning element. Extrapolating from the similarity of the best A-rich motifs (AAAAAA and AAAUAA) to the CPSF-binding site, we have placed CF II at this position. However, the cross-linking of Cft2 from highly purified CF II is dependent on the UA-rich sequence and the sequence at or downstream of the cleavage site, a finding which is not in accord with this model. Rna15 of CF IA shows some preference for U-rich RNAs, and CF IA may participate in recognition of the UA-rich motif or U tracts flanking the cleavage site. In yeast, Hrp1 (CF IB) appears to stabilize the assembly of the cleavage complex at the authentic poly(A) site (308). Recent findings suggest that it interacts with the UA-rich element as well (83, 239, 464) and, in doing so, can prevent the cross-linking of Cft2/Ydh1. The positioning of factors in the model is obviously provisional, and further experiments must be done to establish the architecture of the processing complex. A possible sequence of events is that CF II initially identifies the poly(A) site by interacting with both the UA-rich and A-rich elements and that the subsequent recruitment of Hrp1 and CF IA results in a reorganization of these contacts. Moreover, the prevalence of adenosine and uridine residues in the yeast 3′ untranslated sequence may provide abundant contact points for both Rna15 and Hrp1 if they are generally positioned in the vicinity of the cleavage site by CF II.
CF IA, Hrp1, and CF II are sufficient for accurate cleavage in vitro (239). Polyadenylation requires CF IA, Hrp1, Pap1, Pab1, and PF I (239). PF I is a complex of CF II plus PF I-specific subunits (369). The Yth1 and Pfs1 subunits of PF I probably provide additional contacts with the cleaved RNA which are not shown in the model in Fig. 4B. Direct interaction with the Fip1 subunit of PF I incorporates the catalytic subunit, Pap1, into the polyadenylation holoenzyme (371). Like the mammalian PAB II, Pab1 is necessary to limit the size of the poly(A) tail (12, 239, 310), probably helped in yeast by the opposing action of PAN (64). Findings such as the copurification of Pab1 with CF I over many columns (239, 310) and the depletion of CF I activity by immunoprecipitation of Pap1 from extracts (242) suggest that in the cell, all of the factors may be preassembled into a cleavage/polyadenylation complex which can be dissected apart in vitro.
The redundancy of signals at many yeast poly(A) sites suggests a need for proteins which interact with each of these elements and contribute to the stability of the processing complex. RNA-binding proteins which have been implicated directly or indirectly in 3′-end formation in yeast but are not part of the basic machinery (e.g., Ref2 and nuclear transport proteins such as Nab2, Grs1, and Npl3) may supply additional contacts necessary for the most efficient utilization of a yeast site. The hnRNP family of proteins may play a similar role in mammalian polyadenylation.
Perhaps the best function of the models presented above for cleavage and polyadenylation in mammals and yeast is to point out how much we do not understand about this process. For example, PAP is the obvious catalytic subunit for polyadenylation, but what is the catalytic subunit for cleavage? Why are there not obvious counterparts for some of the proteins (such as Pcf11, Pta1, Fip1, and Hrp1 in yeast and CF Im in mammals) in the two types of organisms? Surely the missing pieces are not all located in the uncharacterized CF IIm. Have some of the essential yeast proteins assumed auxiliary roles that have yet to be identified in the mammalian process? Phosphatase treatment of extract blocks processing (95), raising the unexplored issue of whether cycles of phosphorylation will be used, as in splicing (492), for transiting from a cleavage to a poly(A) addition complex or for dissociation of factors after poly(A) addition. How important is it to balance the levels of these factors in the cell? The amounts of Rna14, Rna15, and Pap1 appear to be coordinately regulated in yeast (11), and overexpression of Rna14 and Rna15 in yeast is not tolerated unless the two CF IA subunits are coexpressed (53). In chicken cells, a 10-fold increase or decrease in the CstF-64 concentration did not markedly affect cell growth and did not induce changes in the level of CstF-77 (451, 455) whereas slight overexpression of PAP reduced the growth rate significantly (516).
Polyadenylation in Other Organisms
Polyadenylation in prokaryotes has been recently reviewed by Sarkar (406), and the topic is only briefly summarized here. Two different poly(A) polymerases have been identified in Escherichia coli, (69, 70). Neither bears resemblance at the amino acid sequence level to the eukaryotic poly(A) polymerases, although PAP1 does have a nucleotidyltransferase catalytic site (291). Neither enzyme is essential for bacterial growth, but a poly(A) tract is thought to promote the degradation of mRNA. The association of the S1 ribosomal protein with poly(A) suggests an additional role for this modification in translation.
Previously characterized plant poly(A) polymerases are chloroplast species (118, 259) which are likely to be related to the bacterial enzymes. A plant homolog of PAP has appeared in the genome database, and homologs of the other factors are likely to follow. A major hindrance to further elucidation of this process in plants has been the lack of an in vitro system.
Vaccinia virus is a DNA virus which replicates in the cytoplasm of mammalian cells. Accordingly, it encodes its own enzymes for mRNA transcription, capping, and polyadenylation. The catalytic subunit of the heterodimeric vaccinia virus PAP (VP55) shows no sequence homology to either the eukaryotic or prokaryotic enzymes except for sharing the nucleotidyltransferase active site (291). Biochemical and crystallographic studies have provided very interesting insights into how this polymerase works (for a recent review, see reference 158). VP55 recognizes uridylate residues at the ends of viral transcripts and processively synthesizes an oligo(A) of approximately 35 nucleotides. At this point, it can no longer stably interact with the RNA, and so it shifts to a distributive mode of activity. The association of the second subunit, VP39, which is also the 5′ cap methylase, restores contact with the substrate and allows processive synthesis to continue.
FORMATION OF THE 3′ ENDS OF HISTONE MRNAS
In most higher eukaryotes, the majority of histone mRNAs are not polyadenylated and instead possess a strong stem-loop at the 3′ end of the RNA. In plants (248, 445) and in lower eukaryotes, such as yeast (144) and protozoans (378), the 3′ ends of histone mRNAs are polyadenylated. In vertebrates, two distinct sets of histone genes, one coding for replication-dependent, nonpolyadenylated mRNAs (413) and the other coding for replication-independent, polyadenylated histone mRNAs (480, 486), have emerged. Interestingly, histone genes containing both polyadenylation and nonpolyadenylation signals have also been identified (76, 86, 104, 245, 288, 479).
The formation of the 3′ ends of histone mRNAs, like polyadenylation, also involves an endonucleolytic cleavage of pre-mRNA (159, 366) and transcription termination downstream of the cleavage site (92, 176). The cis-acting sequences defining the cleavage site of histone mRNA precursor in higher eukaryotes consist of a highly conserved stem-loop structure 5′ to the cleavage site and a loosely conserved purine-rich sequence 3′ to the cleavage site, referred to as the histone downstream element (HDE) (Fig. 5) (for reviews, see references 325 and 413). In addition, the nucleotides surrounding the cleavage site, between the two cis-acting sequences are conserved amongst vertebrate histone genes, with cleavage occurring preferentially after an A (153). The stem-loop structure contributes to the efficiency of 3′-end formation but is not absolutely required (112, 322, 349, 441). Like the poly(A) tail, it fulfills additional roles in transport, translation, and stability (131, 156, 493).
FIG. 5.
Factors involved in cleavage of the histone mRNA precursor. SLBP1/HBP, stem-loop binding protein 1/hairpin-binding factor; HLF, heat-labile factor. Reproduced with modifications from reference 324 with permission.
At least three trans-acting factors are involved in histone 3′ end processing in metazoans (Fig. 5). The U7 snRNP is anchored to the HDE via the 5′ end of its RNA component (112, 304, 323). This snRNP is composed of the core snRNP proteins B, DD′, E, F, and G, the U7 snRNA, and two U7-specific proteins of 14 and 50 kDa (433). The essential HDE-U7 interaction serves as a molecular guide, defining the exact position of the cleavage site (52, 89, 407, 408). Disruption of this base pairing between the HDE and U7 can severely affect 3′-end formation (52, 409).
The stem-loop binding protein (SLBP1), also called the hairpin-binding factor (HBF), interacts with the hairpin loop and stabilizes the association of the U7 snRNP (131, 131a, 435, 469, 481). It remains associated with the mature histone mRNA on polysomes (189). SLPB1/HBFs from human, mouse and Xenopus laevis are conserved, but a second protein from Xenopus, SLBP2, and a Caenorhabditis elegans protein have significant homology to the others only in a central RBD (290, 478, 481). SLBP2 is bound to stored histone mRNAs in Xenopus oocytes and is thought to be involved in translation repression (478). A third factor required for 3′-end processing is a two-component heat labile factor (160). In the in vitro histone cleavage reaction, the 3′-end product is degraded to shorter forms by a 5′-3′ exonuclease (477). It has been proposed that this activity plays a role in transcription termination or in disengaging the U7 snRNP by destroying its binding site. The endonuclease responsible for the specific cleavage has yet to be identified. Since this reaction has not been reconstituted in vitro from purified components, other factors may also be required. The currently known components (SLBP1/HBF and U7 snRNP) show no relationship to those which form the polyadenylation machinery, some of which also await identification.
INTERRELATIONSHIP OF MRNA 3′-END FORMATION AND OTHER NUCLEAR PROCESSES
In vitro studies have clearly demonstrated that the reactions of transcription, capping, splicing, and mRNA 3′-end formation can take place independently. However, recent work indicates that in vivo, the cell creates the equivalent of an assembly line which couples all of these processes on each RNA polymerase II transcript (335).
Transcription by RNA Polymerase II
The necessity of a functional polyadenylation signal for efficient transcription termination downstream of poly(A) sites has long been acknowledged (for reviews, see references 375 and 473), and new findings bearing on the mechanism underlying this requirement are presented in this section. Factors necessary for mRNA 3′-end processing are found at sites of RNA synthesis in the nucleus (412). However, it has only recently been appreciated that an interaction of pol II with the polyadenylation machinery may begin at the promoter itself. This association could be maintained as transcription proceeds across the gene such that some of the factors are actually delivered to the polyadenylation signal by the transcriptional complex. The role of pol II extends beyond that of a delivery service—its presence is important for efficient cleavage of the precursor. The assembly of the processing complex and perhaps execution of the cleavage step is then thought to alter the pol II, leading to transcription termination farther downstream (reviewed in reference 437).
Transcription initiation.
The first indication of an early and direct linkage of transcription and polyadenylation factors came from a study examining the ability of pol II lacking a functional CTD to transcribe a reporter plasmid in mammalian cells (299). When transcription was dependent on polymerases containing a severely truncated CTD, splicing, polyadenylation, and transcription termination were all inhibited. These results established that in the absence a CTD, termination was aberrant even in the presence of a functional poly(A) signal. Furthermore, the cleavage/polyadenylation factors CPSF and CstF specifically bound to CTD affinity columns and copurified with pol II in a high-molecular-mass complex.
CPSF stably associates with TFIID, a factor important for assembly of preinitiation complexes at pol II promoters (115). It is also found in preinitiation complexes formed in vitro and then in complexes with pol II when the transcriptional complex is allowed to enter the elongation phase, leading to the proposal that CPSF is recruited to the promoter by TFIID and then transferred to pol II during initiation. The study by Hirose and Manley (205), discussed above in the section on the cleavage/polyadenylation machinery, subsequently demonstrated that the pol II CTD was also a cleavage factor, extending the role of pol II beyond simply bringing essential components of the processing machinery to the polyadenylation site. An interesting corollary of these findings is that the efficiency of polyadenylation might vary depending on the promoter and how well polyadenylation factors are loaded onto the CTD.
Transcription termination downstream of poly(A) sites.
Transcription termination downstream of 3′-end-processing sites is important in reducing nonproductive transcription and in avoiding transcriptional interference at downstream promoters (140, 175, 376) and chromosomal elements such as centromeres (194) and origins of replication (84). It can also affect the utilization of an upstream poly(A) site and thus can influence the amount of mRNA derived from that gene (46, 143). In eukaryotes, the efficiency of transcription termination generally correlates with the strength of the polyadenylation signal and the presence of a termination site or region located downstream of the poly(A) site (22, 138, 175, 221, 305, 476; see references 375 and 473 for earlier references). In some cases, this termination site pauses the elongating polymerase (46, 140, 143).
A recent study with mammalian cells has shown that a splice acceptor site, through its enhancement of 3′-end processing as part of terminal-exon definition, also contributes indirectly to efficient termination (133a). We have already discussed what constitutes a strong, functional polyadenylation signal. We focus here on how cleavage/polyadenylation factors contribute to termination, what other factors might be necessary for this event, and what creates a pause site, and we provide models to explain how the polyadenylation process might cause pausing to change to transcription termination, as defined by the release of the nascent RNA and the RNA polymerase from the template.
(i) Role of cleavage/polyadenylation factors.
Direct evidence for the dependence of termination on cleavage/polyadenylation factors came from an elegant study which examined transcription termination in several yeast strains defective in 3′-end processing (47). Transcription run-on analysis was used to measure the density of RNA polymerases along a specific DNA template. This density normally shows a dramatic decrease beyond the transcription termination sites found downstream of poly(A) sites. Temperature-sensitive mutations in the Rna14p, Rna15p, and Pcf11p subunits of CF IA resulted in increased transcriptional readthrough by pol II at the nonpermissive temperature. In contrast, mutants with mutations in the polyadenylation-specific proteins Pap1p, Fip1p, and Yth1p exhibited wild-type levels of termination, even at the nonpermissive temperature.
(ii) Other factors with possible roles in termination.
(a) Processivity of pol II . Other factors, some identified and others not yet identified, are likely to be important in influencing termination. Elongation factors that dissociate or are modified after transcribing through the polyadenylation site could reduce the processivity of pol II and thus contribute to the termination event (268). Comprehensive reviews that focus on elongation have been recently published (424, 425, 462), and therefore these factors are not addressed fully here. The phosphorylation of the CTD may be important for transcription elongation (186) and enhances its function in the cleavage reaction (205), implying that kinases and phosphatases may be crucial in regulating the sensitivity of pol II to transcription termination sites. Several CTD-specific kinases and phosphatases have been identified (for reviews, see references 72, 186, 424, 425, and 437).
(b) Pausing and release. Pausing of pol II downstream of the poly(A) addition site may be an obligatory step leading to termination (45, 139, 142, 375). Therefore, factors that cause the enzyme to stop without necessarily releasing the transcript can also be considered termination factors. In fact, this two-step mechanism of pause and release is utilized by the other eukaryotic polymerases, pol I and pol III, as necessary components of termination (289, 383). Two types of signals control pol II pausing. Intrinsic signals are encoded by the template and act in an ill-defined manner to cause the polymerase to alter the rate of transcript elongation (462). These have been found downstream of poly(A) sites (46, 143, 347).
In addition to the intrinsic pausing elements, pol II may pause as a result of protein roadblocks (60, 61, 89, 90, 106). For example, the MAZ protein-binding site was mapped to the termination region in the intergenic sequence of two closely spaced mammalian genes, C2 and factor B (21, 22). Mutational analysis of the MAZ-binding site, RNA-binding studies, and poly(A) competition assays demonstrated that binding of MAZ to the termination region was necessary but not sufficient for efficient 3′-end formation. In this case, protein binding induces pausing of pol II downstream of the polyadenylation site and promotes termination. Unfortunately, the presence of a universal protein-DNA interaction at termination sites has not been found. Thus, in contrast to pol I, which utilizes a specific protein to facilitate the termination of rRNA transcription, and pol III, which utilizes a specific termination sequence, pol II appears to be much more versatile in its response to both sequence and protein factors to facilitate pausing and termination of transcription.
The release of the nascent RNA from the ternary transcription complex is the hallmark of the termination event. The specific events responsible for this release are not understood, but some release factors have been identified. The best understood is factor 2, a component of N-TEF (for “negative-transcription elongation factor”) (501), whose activity has been implicated in premature termination before the poly(A) site is reached (37, 236, 462). Factor 2 belongs to the Swi2/Snf2 superfamily (264), a class of factors with ATPase activity involved in many aspects of DNA-protein interactions (353). A model for transcript release by factor 2 states that the protein utilizes its ATPase activity to disrupt the interaction between pol II and the template in a manner similar to that of other Swi2/Snf2 family members (502). Factor 2 may play a comparable role in transcription termination downstream of poly(A) sites.
The conservation of mechanisms related to transcription between the eukaryotes and prokaryotes suggests that many of the components of the transcriptional apparatus are functionally conserved. Thus, it was surprising, based on sequence homology, that the bacterial transcription termination factor rho does not have a eukaryotic homologue (for reviews, see references 367 and 384). However, the possibility remains that a rho-like helicase activity may be utilized by eukaryotic cells. For example, purified rho factor added to an in vitro transcription reaction mixture can arrest pol II as well as cause the release of transcript from pol II paused at a protein roadblock (251a, 500). The requirement for ATP hydrolysis indicates that the rho protein was acting on pol II transcripts in a manner analogous to its role in rho-dependent termination in bacteria. A eukaryotic factor performing a rho-like function has not been identified, but there is no shortage of known ATP-dependent RNA:DNA helicases that could, in principle, provide this function (142).
A new type of transcription termination factor has recently been discovered. At the nonpermissive temperature, a mutation in Grs1, a putative tRNA synthetase in yeast, causes transcription readthrough of the termination signals associated with yeast poly(A) sites in vivo and in vitro (281). Processing extracts made from this strain are not defective for cleavage and polyadenylation, making this the first factor associated with transcription termination in yeast which is not a component of the cleavage/polyadenylation machinery.
Discussion.
The RNA factory of the nucleus may restrict polyadenylation to pol II transcripts through the association of cleavage/polyadenylation factors with the transcriptional complex and by the requirement of the pol II CTD for efficient processing. This linkage of transcription with polyadenylation also ensures that the transcript is not terminated before its polyadenylation is guaranteed. How, then, is transcription termination coupled to pre-mRNA processing? One model (Fig. 6) proposes that cleavage of the precursor occurs while the polymerase is still engaged in transcription (107, 375). After cleavage, a 5′-3′ RNA exonuclease acting on the uncapped 3′ cleavage product would participate in the release of pol II. A recent electron microscopy study of nascent pol II transcripts suggests that this mechanism does not account for polymerase release downstream of all poly(A) sites (345a).
FIG. 6.
Interrelationship of transcription and mRNA 3′-end formation. See the text for details.
It is now evident that cleavage/polyadenylation factors in yeast are indeed a prerequisite for efficient termination (47). These factors will probably play a similar role in metazoan cells, a hypothesis which could perhaps be tested now that cell lines with regulatable CstF-64 are available (451). A fundamental unanswered question is whether cleavage at the poly(A) site is a prerequisite for termination or if assembly of the complex is sufficient to cause termination (Fig. 6). A variation of the model presented above might be that cleavage releases the CTD and, in doing so, somehow creates a termination-competent pol II or causes a paused pol II to disengage from the DNA template. Resolution of this issue may require the development of an in vitro coupled transcription-polyadenylation assay.
There are also cases which do not fit neatly into the models discussed above. For a yeast gene with a complex set of polyadenylation signals, a reduction in polyadenylation due to deletions of subsets of these signals did not always correlate with a decrease in termination efficiency (18), leading to a hypothesis that assembly of a partial but nonfunctional processing complex might be sufficient to cause termination. When constructs containing tandem poly(A) signals were introduced into mouse cells, the termination efficiency was determined by the upstream signal, even if it was the weaker one (412).
The requirement for cleavage factors for transcription termination in yeast (47) suggests that as in mammals, an early association of cleavage factors with the transcriptional complex might occur in this organism. However, a role for yeast pol II in the reconstituted cleavage reaction has not been established. Deletion of the CTD of yeast pol II perturbs the 3′-end processing of CUP1 transcripts but not that of CYC1 (303a). Furthermore, when a poly(A) site is transcribed in vivo by pol I or pol III in yeast, the transcripts are polyadenylated, suggesting that pol II is not always necessary for processing (260, 267). Differences between yeast and metazoans have also been noted in the pol II-directed transcription of the U-type snRNA genes. In metazoans, formation of the 3′ ends of these transcripts requires special snRNA promoter elements, suggesting that the transcriptional complexes on these genes are different from those on genes encoding mRNAs (91, 151). This is not the case in yeast (352, 419). In addition, when the processing site on a yeast U2 gene is deleted, these transcripts are now polyadenylated (260).
Splicing
A temporal relationship between splicing and polyadenylation in metazoans has been inferred from observations that 3′-end formation often occurs before removal of introns in vivo (34, 148, 213, 336, 483). While this order is not obligatory in vivo, especially with introns transcribed long before the poly(A) site is reached (35, 40, 41, 255), single-intron RNA molecules which are first spliced in vitro are not subsequently polyadenylated (338). Much evidence has accumulated in support of a physical connection between the splicing and the polyadenylation machinery. Depending on the context, splicing factors can stimulate or inhibit cleavage and poly(A) addition. Moreover, some of the interactions between splicing and polyadenylation factors appear to have evolved into regulatory mechanisms, and there are several cases in which the cell uses splicing factors to influence the decision of how or when to polyadenylate an mRNA precursor. In this section, we discuss studies in which the molecular mechanism coupling splicing and polyadenylation has been explored in some detail. More general aspects of how these interactions come into play during alternative mRNA processing are addressed later.
Activation of polyadenylation by splicing factors.
(i) Recognition of constitutive 3′-terminal exons.
Recent work has shown that splicing factors can interact across exons as well as across introns. This mechanism, called exon definition, is an attractive one for marking off exons on complex mRNA precursors (39, 48). A need for a functional linkage between splicing and polyadenylation was then suggested by the question of how to recognize terminal exons, i.e., ones at the 5′ and 3′ ends of the precursor. Proteins bound to the 5′ cap structure are thought to stabilize splicing factors on a downstream 5′ splice site (106, 258). Likewise, an interaction of splicing and polyadenylation factors could bridge the last exon, which begins with a 3′ splice site and ends with a poly(A) site (Fig. 7).
FIG. 7.
Exon definition and processing lead to formation of an export-competent mRNP. CBP, cap-binding proteins; U1, U2, the U1 and U2 snRNPs; SR, members of the serine- and arginine-rich family of splicing factors; U2AF, U2 snRNP auxiliary factor; SF-U1A, snRNP-free U1A complex; CPC, cleavage/polyadenylation complex; PAB II, poly(A)-binding protein II. The identity of factors directly contacting splicing factors during the bridging of terminal exons is not certain. This figure is adapted from a similar one in the review by Berget (39).
This model is supported by numerous studies showing that an intron enhances the utilization of a downstream poly(A) site in vivo and in proportion to the efficiency with which the intron is spliced in vivo (4, 16, 88, 214, 265, 332, 333, 348, 416) and in vitro (340). A functional 5′ splice site upstream of the terminal exon is not necessary for stimulation of polyadenylation in vitro (108, 340, 482), and mutation of the 5′ splice site in vivo had no effect on polyadenylation of β-globin transcripts (16) and less of an effect than a 3′ splice site mutation for SV40 late transcripts (88). These findings imply that the critical interactions of splicing and polyadenylation factors extend only across the terminal exon.
In some cases, these interactions are mutually beneficial for both polyadenylation and splicing. For example, mutation of the AAUAAA polyadenylation signal impairs the in vitro splicing of proximal but not distal exons (337) and the poly(A) site strength has a proportional effect on the efficiency of the splicing reaction in vivo (416). However, the relationship is not always reciprocal. For the human triose-phosphate isomerase gene, the efficiency of polyadenylation in vivo did not affect the efficiency at which the final intron was removed (334). Furthermore, 3′ splice signals found in some terminal introns may be better suited for coupling with polyadenylation sites (16). In agreement with the lack of introns in histone genes, a cooperative interaction was not observed between splicing and the histone cleavage mechanism, although insertion of an intron into the histone pre-mRNA did activate a cryptic polyadenylation site (348).
The physiological significance of exon definition in determining the relative processing efficiencies of two poly(A) sites is also illustrated by the need to balance the amount of α-globin mRNA made from four genes with that of β-globin made from two genes in erythrocytes. The last intron of β-globin pre-mRNAs is much more effective at enhancing polyadenylation than is the equivalent intron of α-globin transcripts, suggesting that 3′-end formation may play a role in maintaining the equimolar amounts of these mRNAs found in erythrocytes (16).
The cross talk of splicing and polyadenylation across a terminal exon is likely to involve factors such as U2AF, which are associated with early recognition of the 3′ splice site (39). However, examination of factors which bind to polyadenylation enhancer sequences suggests that polyadenylation proteins may not directly contact 3′-splice-site factors but instead may interact with ones normally involved in recognition of the 5′ splice site, such as U1 snRNP. Early studies have shown that antibodies against the Sm epitope of U snRNA-binding proteins or against U1 snRNP inhibit polyadenylation in vitro (316) and that antibodies against poly(A) polymerase coimmunoprecipitate U1 but not other snRNPs (381). More recent work has demonstrated a direct interaction of U1 components with regions near the poly(A) site.
For example, the stimulation of polyadenylation by the USE in the SV40 late polyadenylation signal is attributed to the U1A protein binding to this site (277) and to stabilization of the interaction of CPSF with substrate by direct contact with the CPSF 160-kDa subunit (278). U1A does not have to be associated with U1 RNA for this activation, since recombinant U1A has a similar effect (278). In fact, the cellular component recognizing the USE may be a naturally occurring snRNP-free U1A, in complex with several proteins, one of which is an essential splicing factor, the polypyrimidine tract-binding protein-associated factor (276). In support of this idea, antibodies against a U1A epitope which is masked when U1A is bound to U1 RNA inhibit polyadenylation in vitro on substrates containing only a poly(A) site (343) and inhibit both splicing and polyadenylation in a coupled reaction (276). This epitope is near the RRM of U1A, which binds to the second stem-loop of U1 (279). These data suggest that this RRM of U1A may contact the USE, and they bring into question an involvement of U1 snRNP at this enhancer element. Furthermore, this novel U1A complex may be a special adaptation for recognition of 3′-terminal exons.
Nevertheless, the U1 snRNA does interact with RNA in the vicinity of some poly(A) sites, suggesting that U1 snRNP may help form the bridge between splicing factors at the 3′ splice site and the polyadenylation-processing complex. The U1 snRNA in cell extract can be cross-linked to regions in the adenovirus L3 site and the SV40 late site upstream of the AAUAAA signal (482). These regions have some complementarity to the U1 5′ end. The presence of a 3′ splice site on the RNA substrate enhanced both U1 snRNA cross-linking and polyadenylation. Interestingly, deletion of the U1 snRNA-binding site did not prevent the stimulation of polyadenylation conferred by the 3′ splice site. While this result implies a minor role for the base pairing of U1 snRNA in the recognition of terminal exons, other activities of U1 snRNP, such as its interaction with the 3′ end of introns (152) and its interaction with U2 snRNP (119), also do not require its 5′ end.
(ii) Recognition of alternative 3′-terminal exons.
In the alternative processing of the pre-mRNA for calcitonin/calcitonin gene-related peptide (CGRP), the use of exon 4 as the terminal exon in calcitonin-specific processing requires a sequence located within the intron 168 nucleotides downstream of the exon 4 poly(A) site (273). The calcitonin enhancer stimulates the in vitro polyadenylation of precursor containing the exon 4 poly(A) site or heterologous poly(A) sites. It contains a polypyrimidine stretch and a CAG (characteristic of 3′ splice sites), followed immediately by a 5′ splice donor site, which binds the U1 snRNP, alternative splicing factor/splicing factor 2 (ASF/SF2), and the SR protein SRp20 (270, 272) (Fig. 6). Polypyrimidine tract-binding protein (PTB), which binds the polypyrimidine tract of the intron enhancer, has been implicated in the exclusion of several alternative exons (23, 77, 169, 326, 341, 355). The action of the calcitonin/CGRP intronic enhancer requires U1 snRNP binding and is enhanced by PTB and Srp20 (271, 272). In this case, the binding of PTB may block access of the essential splicing factor U2AF to the enhancer pyrimidine tract and thus may interrupt productive recognition of the enhancer as a splice site (271).
PTB may also play a more direct role in activating polyadenylation. It has been shown to stimulate cleavage, but not poly(A) addition, by binding to an enhancer element upstream of the C2 complement poly(A) site in reactions with purified cleavage/polyadenylation factors (319). In the calcitonin/CGRP transcript, an additional PTB-binding site was found close to the AAUAAA hexanucleotide of exon 4 (271). The importance of this pyrimidine tract has not been explored, but PTB does oligomerize (344), an interaction proposed to bring the entire enhancer complex closer to the poly(A) site (271) (Fig. 8). While it is not known which splicing or polyadenylation factors are in direct contact, these interactions stabilize the binding of CstF to the exon 4 poly(A) site and represent a novel means of defining a 3′-terminal exon (271).
FIG. 8.
Calcitonin/CGRP intron enhancer complex. Adapted from reference 271.
The calcitonin/CGRP polyadenylation enhancer, which resembles a pseudo-exon, functions downstream of the calcitonin exon 4 poly(A) site. While this is not the configuration of processing signals found in 3′-terminal exons, it does suggest that splicing factors such as U1 snRNP, SR proteins, and PTB may play a role in recognition of 3′-terminal exons. This arrangement is also reminiscent of the coupling of polyadenylation and transplicing in trypanosomes. In these protozoans, the downstream acceptor splice site with an accompanying polypyrimidine tract, and not AAUAAA-like consensus polyadenylation signals, determines the position of the poly(A) site in the intergenic regions of polycistronic pre-mRNAs (252, 296, 414). Damaging the spliced leader RNA by oligonucleotide/RNase H treatment of a U2-like region simultaneously inhibits trans-splicing and polyadenylation (461), suggesting that assembly of a fully functional splicing complex is necessary to promote polyadenylation. A similar interaction may occur in the polycistronic precursors of C. elegans, in which trans-splicing to the second open reading frame is dependent on the upstream AAUAAA polyadenylation signal (49).
Inhibition of polyadenylation by splicing factors.
As described below, the U1 snRNP or its components in certain contexts can also inhibit polyadenylation.
(i) Inhibition by a downstream splice donor site.
A scenario in which splicing factors downregulate the use of a poly(A) site can be found in the transcripts of HIV-1. Transcription of integrated viral DNA results in a precursor with duplicated poly(A) sites, but polyadenylation at the promoter-proximal poly(A) site is suppressed. The factors influencing the alternative polyadenylation of this precursor are reviewed later in the section on that topic, and here we describe only the involvement of splicing factors. A recent study has shown that the promoter-proximal poly(A) site is repressed by interaction of the U1 snRNP with a splice donor site almost 200 nucleotides downstream, which is used in processing of viral mRNAs (20). Recognition of a poly(A) site at this position is possible if the splice site is mutated or moved further away or if the HIV-1 poly(A) site is replaced with a more efficient one (19). The mechanism for this inhibition has not yet been worked out; it may involve direct or indirect interactions of U1 snRNP to destabilize polyadenylation factors or interaction of U1 snRNP with cap-binding proteins to define the first exon, thus blocking access to the poly(A) site (258). This is rather different from U1 snRNP action at the calcitonin/CGRP intronic enhancer and probably reflects the fact that in HIV-1, this U1 snRNP binding site is active in splicing.
(ii) Inhibition by an upstream splice donor site.
A strong U1 snRNP interaction upstream of a poly(A) site can inhibit polyadenylation. For example, a poly(A) site, even a strong one, inserted into an intron is not recognized (4, 58, 256). However, mutation of the splicing signals activates the inserted poly(A) site, suggesting that assembly of the splicing complex across the intron excludes the polyadenylation machinery. It has also been demonstrated that insertion of a bona fide 5′ splice site between the 3′ splice site and the poly(A) site abolishes the coupling of the two reactions in vitro and in vivo, perhaps by making the RNA look more like an internal exon (339, 482).
This arrangement of a U1 snRNP-binding site upstream of a poly(A) site occurs naturally in the transcripts of papillomaviruses. Polyadenylation of the late mRNAs of bovine papillomavirus type 1 in undifferentiated epithelial cells is repressed by the U1 snRNP bound to a cryptic but consensus 5′ splice site upstream of the late poly(A) site (154). The human papillomavirus type 16 has four overlapping motifs in a similar region which have partial 5′-splice-site homology (154). Surprisingly, this inhibition is mediated by direct contact of the U1 70K protein with PAP (178). This result suggests that the downregulation is mediated by direct protein-protein interaction at a late step in 3′-end formation, not only by interference with definition of the 3′-terminal exon. It is not known what relieves this block in terminally differentiated epithelial cells.
The mammalian U1A protein also downregulates the level of its mRNA by inhibition of PAP (177, 179). This mechanism requires an RNA structure bound by two U1A proteins and, like the bovine papillomavirus type 1 example described above, does not target the initial step of polyadenylation complex assembly. Instead, U1A binding inhibits poly(A) addition by a direct interaction between a site in the last 20 amino acids of PAP (Fig. 3) and a 17-amino-acid domain of U1A which is similar in sequence to repeats in the U1 70K inhibitory region (178). If multiple copies of this U1A peptide are brought together by conjugation with BSA, this conjugate is sufficient to inhibit PAP and, interestingly, to uncouple splicing and polyadenylation in vitro without affecting the processing of RNA with only splice sites or poly(A) sites.
This unexpected insight suggests that PAP, which is known to be part of the cleavage complex, may provide a direct link to splicing factors during recognition of the 3′-terminal exon. When the base pairing of U1 snRNA upstream of a poly(A) site is not strong, as in the examples described by Wassarman and Steitz (482), would a similar interaction of PAP and U1 70K contribute to the recognition of 3′-terminal exons? If so, such a mechanism would require a reorganization of the splicing/polyadenylation complex on the last exon following cleavage so that the PAP would be free to synthesize the poly(A) tail.
Role of hnRNP and cap-binding proteins.
The hnRNP proteins A, B, F, H, and I (PTB) influence the efficiency or pattern of splicing by interacting with splicing enhancers or silencers (75, 93, 121, 171, 307) and perhaps with the nuclear cap-binding protein complex (157). The critical role of PTB in the calcitonin/CGRP polyadenylation enhancer (271) and the recent identification of the auxiliary polyadenylation factor DSEF-1 as hnRNP H′, a variant of hnRNP H (26), suggests that we can anticipate a greater involvement of hnRNP proteins in modulating the assembly of the polyadenylation complex and thus in contributing to terminal-exon definition. DSEF-1, a 50-kDa nuclear protein, binds specifically to a G-rich element downstream of the SV40 late poly(A) site and increases the cross-linking of CstF to the polyadenylation precursor (26, 27). hnRNP C interacts with the adenovirus L3 polyadenylation enhancer element (372) and with U-rich sequences downstream of several poly(A) sites (440, 495, 498), but in at least one case, competition for hnRNP C binding with poly(U) did not affect polyadenylation (440).
Intronless transcripts such as the HSV-TK, hepatitis B virus, and mouse histone H2a mRNAs have upstream elements which stimulate polyadenylation and nuclear transport (212, 216–218, 266). The important motifs in these elements have not been finely mapped, and it is not known whether splicing factors will be involved in recognition of this type of polyadenylation enhancer. The HSV-TK element has a stretch [AG(G/G)UGAGA] with significant homology to the 5′-splice-site consensus, but a GU-to-AU point mutation does not prevent the element from promoting cytoplasmic accumulation of intronless transcripts (266). However, it is interesting that the hnRNP L protein interacts specifically with the HSV-TK sequence (266) and with PTB (184).
The presence of a 5′ m7GpppG cap on the polyadenylation precursor enhances the efficiency of cleavage of these RNAs in mammalian extracts (108, 166). This effect is due to stabilization of the cleavage complex by the cap-binding proteins CBP20 and CBP80 (147), and the poly(A) addition step is not affected. The interaction is not direct, but the intermediate is not known. It may involve the same family of factors, possibly hnRNPs F and H (157), which mediate the cap-dependent stimulation of splicing.
Summary.
In summary, the effect of splicing factors on polyadenylation in vertebrates often depends on the position of actual or cryptic splice donor or acceptor sites in relation to the poly(A) site and how well these sites match consensus splicing signals. In the natural situation of a poly(A) site as part of a terminal exon, the two processes cooperate and the presence of a 3′ splice site activates polyadenylation (Figure 7). In certain contexts, the placement of a strong 5′ splice site downstream or immediately upstream of the poly(A) site and the recruitment of splicing factors to this 5′ splice site tends to inhibit polyadenylation or at least prevent its stimulation by a 3′ splice site. In the case of calcitonin/CGRP, the intron enhancer demonstrates how, in a special context, a 5′ splice site can stimulate polyadenylation and suggests how interactions resembling those at a 5′ splice site can promote the recognition of 3′-terminal exons.
The available data indicate that the coupling of splicing and polyadenylation at the last exon will probably involve interactions of factors such as U1 snRNP, PTB, SR proteins, the U1A snRNP-free complex, CPSF, and PAP in a configuration that remains to be elucidated. Many of the elements that have been grouped as polyadenylation upstream enhancer elements (Table 1) are probably there for the purpose of stabilizing this terminal exon complex. When the 3′ splice site upstream of a poly(A) site is weak, as is found in some alternatively processed transcripts, additional interactions mediated by splicing factors bound to exonic enhancers may also be enlisted.
It is not known whether remnants of this linkage of splicing and polyadenylation will be found in yeast. The remaining introns in yeast genes tend to be at the 5′ end of transcripts and thus are not likely to be ones originally in need of a poly(A) site for recognition. In fact, natural or artificially inserted intron sequences located 200 bp or further downstream of the 5′ end of a yeast transcript are not spliced out efficiently (33, 506). Furthermore, poly(A) sites placed in yeast introns predominate over splicing (222, 393). However, at least one yeast polyadenylation factor, Brr5/Ysh1, was discovered in a screen for cold-sensitive defects in splicing (78). Efforts at exhaustive two-hybrid screening are also providing tantalizing leads: Pcf11 interacts with Lsm8, a protein with similarity to snRNA-associated proteins of the Sm family (151), and a protein encoded by YMR117c interacts with Fir1 and the U2 snRNP-associated splicing factor, Prp11 (90). Additional relationships may yet emerge.
Transport
As discussed above, mRNA synthesis involves a cooperative interaction of factors involved in transcription, splicing, and polyadenylation. The next step in mRNA utilization requires its transport from the nucleus to the cytoplasm. Recent progress in the field of nuclear trafficking has demonstrated that each major class of RNAs (tRNA, rRNA, mRNA, and U snRNAs) uses a distinct, saturable export pathway (226, 444). The mRNA precursor becomes rapidly associated with numerous RNA-binding proteins known as hnRNP proteins (132). Many of these proteins have been shown to shuttle between the nucleus and cytoplasm (364), leading to the notion that export signals on this type of protein mediate the transport of mRNA out of the nucleus.
mRNA 3′-end formation facilitates efficient transport.
Several observations have led to the hypothesis that transport factors are deposited onto the RNA during splicing and 3′-end formation. Some intron-containing genes are poorly expressed as cDNAs, a phenomon attributed to both a decrease in stability of transcripts in the nucleus and a defect in nuclear transport (254, 396). Cytoplasmic accumulation is greatly improved if an intron or certain cis-acting elements found in intronless genes are included (16, 66, 216, 332). Like 3′-terminal splice sites, these transport elements also facilitate polyadenylation, leading to the proposal that enhancement of transport is a consequence of more efficient polyadenylation (216, 217). In support of this conclusion, the absence of a polyadenylation signal resulted in retention of pol II reporter transcripts in the yeast nucleus (269), and the inhibition of polyadenylation of intronless transcripts by the influenza virus NS1 protein prevents their exit from the nucleus (330). The positioning of a transcribed poly(A) tract at the end of the mRNA by ribozyme cleavage still resulted in poor transport, arguing against a poly(A)-induced increase in stability as the sole reason for increased RNA levels in the cytoplasm (215).
In a similar vein, nonpolyadenylated histone mRNAs are not efficiently exported if the ends are produced by ribozyme cleavage (134) or if the stem-loop structure at the end is mutated so as to reduce the binding of SLBP1/HBF (493). These findings linking mRNA 3′-end formation with export are consistent with the emerging view that the early steps in mobilizing an mRNA for transport actually occur in the nucleoplasm and not at the nuclear pore (350).
Genetic analysis in yeast has also pointed to coordination between polyadenylation and mRNA transport. Npl3/Nop3 is a yeast hnRNP with two RNA recognition motifs and a C-terminal domain containing RGG repeats (54, 391). It shuttles between the nucleus and cytoplasm in a transcription-dependent manner but is found predominantly in the nucleus (146) in association with polyadenylated RNA (391, 494). Mutations in Npl3/Nop3 interfere with mRNA export (253, 391, 428), nuclear import of proteins (54), and the late steps of rRNA maturation (390), and they can lead to poly(A) tails which are longer than normal (428). The mRNA transport defects of the npl3-1 allele can be suppressed by extragenic mutations in the RNA15 and HRP1 genes (201), and other NPL3 alleles are synthetically lethal with a temperature-sensitive allele of RNA14 (250). Recent work from the laboratory of Cole has shown that certain alleles of RNA14, RNA15 and PAP1 cause defects in both mRNA export and in vitro processing (185).
Possible mechanisms for coupling 3′-end formation and transport.
The stimulation of cytoplasmic accumulation of mRNA by splicing could be attributed in part to the enhancement of polyadenylation, as discussed above. Splicing also serves another important function through the removal of splicing factors which act as nuclear retention factors (444). Polyadenylation may play a similar role, in that the presence of certain polyadenylation factors bound to the unprocessed mRNA may prevent interaction with the transport machinery or assembly of an export-competent mRNP. This possibility has not received much attention, although it has been reported that the yeast CF IA subunit Rna15 does not exit the nucleus (200).
The polyadenylation apparatus may also play a direct role in facilitating transport by leading to the acquisition of transport factors. Perhaps the clearest link to transport has come from the discovery that some RNA-binding proteins involved in polyadenylation also have the property of shuttling. These are Hrp1/Nab4 (239) in yeast and PAB II (73, 85) in mammals. The metazoan PAB I, which was originally thought to be only cytoplasmic, does accumulates in nuclei upon transcription inhibition (7). The yeast homolog, Pab1, may also shuttle, since it has both nuclear and cytoplasmic functions. However, depletion of Pab1 from the cell or inhibition of its function through a temperature-sensitive allele does not cause mRNA accumulation in the nucleus (234), and perhaps its role in transport is redundant. Experiments directly addressing the role of Hrp1/Nab4 and PAB II in transport have not been performed.
Because of the importance of the poly(A) tail to RNA translation and turnover, leakage of a significant amount of poly(A)− mRNAs into the cytoplasm could be undesirable to the cell. The recruitment of specific proteins onto the mRNA as part of the polyadenylation process could mark an RNA as mature and ready for transport and translation. Like Npl3/Nop3, the shuttling of Hrp1/Nab4 requires ongoing transcription pol II (423). If the only way for Hrp1 to get on the mRNA is through the polyadenylation of nascent transcripts, it is reasonable that transcription is a prerequisite for its exit from the nucleus.
There are several ways in which such a monitoring system could work at a molecular level if some 3′-end formation proteins, such as Hrp1 and SLBP1/HBF, have a second function in facilitating transport. These polyadenylation proteins, and perhaps others yet to be identified, could possess nuclear export signals and thus contribute to mRNP transport by interacting directly with nuclear export receptors. Analysis of the transport of the large Balbiani ring mRNP particles of Chironomus tentans by electron microscopy indicates that both the 5′ and 3′ ends of the RNA appear to contact the basket of the nuclear pore complex, even though the 5′ end is the first part to cross the nuclear membrane into the cytoplasm (114). If this is the case for all mRNPs, interactions of the pore with the poly(A) tail and/or proteins bound to it may also help correctly orient the particle for translocation.
Another mechanism for how polyadenylation proteins might participate in transport is that they interact with proteins containing export signals, such as Gle1/Rss1 (123, 327), and in doing so, recruit them to the mRNP. Alternatively, proteins such as Hrp1 could stabilize the RNA binding of export effectors such as Npl3/Nop3, the cap-binding proteins, or the recently identified Mex67 (417), which has a Rev-like nuclear export signal and binds poly(A)+ RNA.
Mutations in several classes of genes cause mRNA export defects as well as an increase in the length of the poly(A) tails by about 10 to 20 nucleotides, similar to that seen in the absence of the yeast poly(A) nuclease (63). These include ACC1, MTR2, MTR3, MTR4 (232), NAB2, NPL3/NOP3 (428), PRP20 (150), and RNA1 (365). Some of these mutations (ones in PRP20, MTR3, MTR4, and PRP20) also produce extended RNA polymerase II transcripts (150, 232), suggestive of additional defects in transcription termination. A subgroup of these proteins are associated with RNA: Mtr2, Mtr4, Nab2, and Npl3/Nop3. Mtr2 interacts with the mRNA-binding protein Mex67 and with nucleopores (405). Mtr4 is a member of the DEAD-box helicase family (261). Nab2, like the Npl3/Nop3 discussed above, is a yeast hnRNP. It has an RGG box and seven repeats of a CCCH zinc-binding motif, shuttles from the nucleus to cytoplasm, and is bound to poly(A)+ RNA (references 8 and 15 and unpublished data cited in reference 494). A second class includes proteins generally involved in nuclear trafficking (Prp20 and Rna1) or in synthesis of nuclear membrane components (Acc1). Rna1 is the yeast Ran GTPase-activating protein, and Prp20 is the guanine nucleotide exchange factor which helps Ran release GDP (98, 113, 318). ACC1 encodes acetyl coenzyme A carboxylase, the rate-limiting enzyme of de novo fatty acid synthesis (411). The third class (Mtr3) is a nucleolar protein (233).
So far, these proteins have not copurified with any of the basic polyadenylation factors and have not been shown to have any effect on the in vitro polyadenylation reaction. Mutants which exhibit atypically long poly(A) tails often exhibit global defects in rRNA and tRNA processing and in splicing of pre-mRNA (470). The simplest explanation for the longer tails is that trimming of the poly(A) occurs in the cytoplasm (150). Recent work by Brown and Sachs (63) suggests that this answer may not suffice. This study showed that the combination of the PAP and the poly(A) nuclease in processing extract produces poly(A) tracts of the correct length, and upon longer incubations in vitro, the tails of the processed substrate tend to shorten. If this in vitro situation is an accurate representation of conditions in the nucleus, a longer stint in the nucleus due to the export block would not be expected to give longer tails on mRNA. Instead, the longer tails and the extended transcripts might reflect factors being titrated by the poly(A)+ RNA accumulating in the nucleus or factors not being able to recycle if assembly of an export complex involving proteins such as Npl3/Nop3 and Nab2 is prevented. For some mutations, especially ones in general nuclear trafficking factors such as the Ran1 complexes or in proteins causing alterations of the nuclear envelope such as Acc1, the increase in poly(A) size may be a secondary effect of concomitant blocks in protein import.
Finally, the job of some 3′-end-formation factors may extend beyond the nucleus and into the cytoplasm. In this case, they may not play a direct role in transport but, by arriving in the cytoplasm with the mRNP, are immediately available for other functions in translation and mRNA turnover. In addition, the release and recycling of export factors may depend on interactions of these factors with the translational machinery. The most clear-cut examples of this possibility are the SLBP1/HBF of histone mRNA processing, PTB, and the Pab1 of yeast. SLBP1/HBF is thought to be important for regulating both the translation and stability of histone mRNAs (131, 478). PTB, a positive effector of polyadenylation, is a nuclear shuttling protein (364) and participates in translation regulation and translation initiation at internal ribosome entry sites (463). The ICP27 protein of HSV-1 has activities similar to those of PTB. It stimulates the polyadenylation of HSV late mRNAs and enhances the binding of the 64-kDa subunit of CstF to these poly(A) sites (188, 301), but it represses the splicing of host mRNAs (404). It also binds to and mediates the transport of intronless HSV-1 transcripts (403). As discussed above, Pab1p is necessary in yeast for the termination of poly(A) synthesis, a function assumed by PAB II in mammalian cells. Its role in promoting the initiation of translation has been firmly established in both organisms (399). While the shuttling ability of yeast Pab1p has not been proven experimentally, it seems reasonable that the Pab1p deposited onto the poly(A) tail in the nucleus stays put during and after transport.
The RRM-type RNA-binding domains of Hrp1 have the greatest sequence similarity to those of hnRNP A1, hnRNP A2/B1, and hnRNP D/AUF1 (239). AUF1/hnRNP D is a protein which binds the AU-rich degradation signal of short-lived mRNAs (511) and has been implicated in regulating mRNA editing (14) and cytoplasmic mRNA stability (124, 244). In fact, recent work has shown that mRNAs normally targeted for degradation by the nonsense-mediated decay pathway are stabilized in Hrp1-defective yeast strains (354). Thus, Hrp1, like SLBP1/HBF and AUF1/hnRNP D, is likely to have multiple functions, acting in 3′-end formation, transport, and mRNA turnover.
REGULATION OF MRNA 3′-END FORMATION
General Themes
Regulation of mRNA 3′-end formation is one of the many ways in which cells can vary the amount and type of mRNA derived from a particular transcriptional unit. A recent review by Edwalds-Gilbert et al. (139) provided a thorough compilation of genes known to be subject to this form of alternative processing. At the level of 3′ end formation, two types of choices can be made: (i) whether to process the transcript and (ii) where on the transcript to place the 3′ end. These choices can be influenced by the growth conditions of the cell, its position in the cell cycle, or its differentiated state. The first situation involves only one processing site, which is used with different efficiencies. In this case, transcripts which are not polyadenylated, or cleaved in the case of histone mRNAs, will be degraded or not transported efficiently into the cytoplasm, and the amount of protein expressed from that gene will decrease. In the second type of processing choice, the cell is faced with more than one polyadenylation site, and utilization of one or the other will change the coding sequence or 3′ untranslated region in the final product. This type of transcript falls into three categories (Fig. 9): ones with tandem arrays of poly(A) sites within a single 3′ untranslated region, ones with alternative 3′ terminal exons, and ones with a composite exon whose 3′ end is formed by either a 5′ splice site or a poly(A) site (139). In this section, we focus on a few examples which point to the molecular mechanisms governing these choices.
FIG. 9.
Types of alternative polyadenylation choices include multiple poly(A) sites in the 3′ untranslated region (3′UTR) (A), a choice in defining the end of an exon by a 5′ splice site (5′SS) or a poly(A) site (B), and a choice of two different 3′-terminal exons (C). Adapted from reference 139.
Two mechanisms are commonly used to execute these decisions. First, the efficiency with which the processing complex assembles on a site can be regulated. Many studies have shown that the efficiency with which a poly(A) site is used correlates with the stability of the 3′-end-processing complex formed on that site, which in turn depends on the strength of the core polyadenylation signals (163, 372, 373, 485). Many regulated poly(A) sites are weak because the signal sequences are poor matches to the consensus sequence or are in an unfavorable sequence context (139). As discussed above, a stable polyadenylation complex is the consequence of multiple, cooperative interactions among the basic polyadenylation factors, an arrangement which affords many targets for regulation. Taking all of this into consideration, the efficiency at which the processing complex forms will be a combined effect of the concentration of polyadenylation factors in the cell and the additional stabilizing influence of other factors recruited onto polyadenylation enhancer elements adjacent to the regulated site. Second, regulation can also be achieved by directly influencing the activity of processing enzymes by posttranslational modifications or by interaction with repressor proteins.
In several examples of alternative polyadenylation, important cis-acting regulatory sequences and the factors which bind them have been identified. As discussed in the section on the coupling of splicing and polyadenylation, the critical trans-acting components are often ubiquitous splicing factors or a protein such as PTB, involved in alternative processing in many types of cells. Tissue-specific or transcript-specific factors have not been discovered. Also, little is known about the molecular switches which control the levels, activity, or localization of basic polyadenylation and splicing factors, so that they now provide regulatory functions. Given the coupling of transcription and processing, it is conceivable that part of the decision may occur at the promoter. Furthermore, the choice to use a particular poly(A) site may be made rapidly following transcription through it, within a limited window of opportunity, as was found for the alternative splicing of one of the alpha-tropomyosin exons (385). If regulatory modifications are made in discrete locales in the nucleus, corresponding to the space occupied by the gene producing the regulated transcripts, it may be difficult to detect changes in processing activity in cell extracts. Nevertheless, in the cases described below, there has been significant progress in understanding how changing the activity or concentration of factors involved in mRNA 3′-end formation can influence the utilization of a 3′-end-processing site.
Regulatory Mechanisms in the Selection of Alternate 3′-Terminal Exons
A common form of alternative polyadenylation results in the inclusion or exclusion of a 3′-terminal exon located within a multiexon transcript. To understand how this choice is made, we must consider what makes the alternative 3′-terminal exon visible (or invisible) to the processing machinery. From our knowledge of the mechanism of last exon recognition, it is clear that activities in the regulated environment will most probably alter the association of factors with either the 3′ splice site or the poly(A) site. In some cases, the primary event is at the level of splicing, in some it is at polyadenylation, and in others it is at both steps. Often, weak polyadenylation or 3′ splicing signals make inclusion of the regulated exon unfavorable, and such suboptimal signals can even be necessary for regulation.
Alternative polyadenylation of calcitonin/CGRP transcripts.
The mammalian calcitonin/CGRP transcriptional unit provides an insightful example into how these choices are made. This gene has six exons and is alternatively processed in a tissue-specific manner (Fig. 10A). In thyroid cells and most other cell types, the first four exons are almost always spliced together and polyadenylated at exon 4 to give mRNAs encoding calcitonin (397). In neuronal cells, exon 3 is spliced instead to exon 5 nearly 100% of the time and polyadenylated at exon 6 (9). Translation of these mRNAs gives a neuropeptide, CGRP. Dissection of the mechanism of this regulated processing has been facilitated by the availability of cell lines which make predominantly one or the other form of mRNA.
FIG. 10.
(A) Organization of exons and introns in calcitonin/CGRP pre-mRNA and the structure of the primary mRNA products produced in thyroid cells or neurons. (B) Alternative processing choices in the immunoglobulin M (IgM) heavy-chain (μ) precursor. (C) Structure of the integrated proviral DNA of the HIV-1 retrovirus and the primary transcript. The transcription initiation site is indicated by the arrow. USE, upstream polyadenylation enhancer; LTR, long-terminal repeat sequence; TAR, TAT-binding site; MSD, major splice donor used by all spliced transcripts. The promoter proximal and promoter-distal poly(A) sites and alternative splice donor (◊) and acceptor (▵) are also indicated. For ease of representation, the elements are not in correct size scale with respect to each other.
Several elements on the calcitonin/CGRP precursor affect the recognition of exon 4. The exon 4 poly(A) site lacks a strong GU- or U-rich sequence and is processed poorly in vitro (468). The intronic enhancer discussed in the section on splicing/polyadenylation coupling is essential for inclusion of exon 4 because the splicing factors assembled on it stabilize the polyadenylation complex at this weak poly(A) site (271). The exon 4 splice acceptor site has a noncanonical uridine or cytidine residue as the branch point and a short pyrimidine tract (6, 111, 505). Recognition of this deficient 3′ splice site in calcitonin-producing cells is probably assisted by the presence of internal exon 4 sequences. One is a region in the first 45 nucleotides of exon 4 (109, 111) which forms part of a stem-loop structure whose duplex nature is important for usage of the site in vitro (100). Two additional elements are farther downstream, and both are necessary to fully prevent the skipping of exon 4 (467, 509). The first element of this pair resembles the Drosophila doublesex exonic enhancer critical for female-specific splicing, and the second is purine rich (467), suggesting that Tra or Tra2 homologs and members of the SR protein family might be involved in their recognition (5). From what we understand about 3′ exon definition, all of the above interactions could influence the use of the exon 4 poly(A) site.
If the strength of exon 4 recognition in calcitonin-producing cells is a composite of how well the different exonic and intronic elements are recognized, how is the cellular environment regulated to exclude exon 4 in neuronal cells? At present, there are some enticing leads but no clear answer to the question. If the exonic or intronic elements affecting exon 4 are mutated, exon 3 can be spliced to exon 5 in calcitonin-producing cells (273, 467), implying that the relevant event is tissue-specific activation and/or repression of exon 4 rather than exon 5. The first element in exon 4 binds a 66-kDa protein from HeLa cells, which make the calcitonin-specific splice, and extracts from CGRP-producing cells inhibit both the binding of this protein and the inclusion of exon 4 in vitro (110, 111), possibly through the association of two brain-specific polypeptides of 41 and 43 kDa (386). An SR protein, SRp20, is necessary for assembly of the active intron enhancer complex, and this protein is limiting in cells which preferentially exclude exon 4 (272). Exon 4 inclusion also increases when the PTB level is increased experimentally in vivo (271). It is not known if the PTB level or binding affinity is normally depressed in neuronal cells, but neuron-specific isoforms have been reported (23, 77). However, constructs containing a substitution of the first exon element with the equivalent region of exon 3, deletion of both of the downstream exon enhancers, or mutations which partially disrupted the intron enhancer were still regulated appropriately in neuronal cells; i.e., the use of exon 4 was downregulated (273, 505). A reasonable explanation is that more than one site is a target for regulation, with this regulation occurring either by negatively acting neuronal factors or by lowered amounts of positive factors which bind to these sites. The cis- and trans-acting factors contributing to inclusion or exclusion of exon 4 are summarized in Table 4.
TABLE 4.
Known and potential factors contributing to alternative polyadenylation in calcitonin/CGRP, immunoglobulin heavy chain, and HIV-1 transcripts
| Calcitonin/CGRP exon 4 inclusion |
| Exclusion |
| Weak 3′ splice site (nonconsensus BP, short Py tract) |
| Weak core p(A) signal (poor DSE) |
| Limiting PTB and SRp20 in neuronal cells |
| Repressor in neuronal cells |
| Inclusion |
| Intron polyadenylation enhancer and sufficient PTB and SRp20 |
| Two exonic splicing enhancers |
| Stem-loop containing 3′ splice site and first part of exon |
| Possible positive effector acting in first part of exon |
| Use of secretory poly(A) site in IgM heavy chain transcripts |
| Suppression in immature B cells |
| Weak core p(A) signal |
| Limiting CstF |
| U1 snRNP on functional upstream 5′ splice site |
| Possible repressor of secretory site and activator of membrane site |
| Activation in plasma cells |
| Increase in CstF activity |
| Weak 5′ splice site |
| Possible activator binding at distal DSE |
| Increased transcription termination just downstream of secretory p(A) site |
| HIV-1 |
| Suppression of the proximal p(A) site |
| U1 snRNP on downstream 5′ splice site |
| Lack of USE |
| Cap structure instead of 3′ splice site upstream of p(A) site |
| Incorporation of p(A) core signal into stem loop |
| Activation of the distal p(A) site |
| USE stabilizing CPSF interaction |
| TAR stem-loop juxtaposing USE and AAUAAA |
| 3′ splice site upstream of p(A) site |
Another example of the need for multiple factors to effectively bridge the alternate 3′ exon is found in Drosophila doublesex transcripts. On this precursor RNA, recognition of the weak 3′ splice site in the female specific exon 4 requires cooperative binding of the tra, tra-2, and SR proteins to enhancer sequences located in this exon between the 3′ splice site and the poly(A) site (75). This enhancer element is also important for efficient use of the poly(A) site (193).
Alternative polyadenylation of immunoglobulin transcripts.
The switch from the membrane-bound to the secreted forms of the immunoglobulin heavy chain is regulated during B-cell development by differential splicing and poly(A) site selection of the primary transcripts, and since the discovery of the phenomenon in 1980, it has been one of the most intensively studied examples of this type of alternative processing (reviewed in reference 139). Two poly(A) sites, each forming part of a different 3′-terminal exon, are present on the primary transcript, with the first being found on mRNA encoding secreted immunoglobulins and the second being used on mRNAs producing the membrane-bound form (Fig. 10B). In immature B cells, the use of an internal splice donor site in exon 4 removes the secretory poly(A) site, and the two forms of mRNA are produced in approximately equal abundance. In terminally differentiated plasma cells, the secreted form is predominant. All of the immunoglobulin genes which undergo a membrane-to-secreted switch have a similar gene structure and are thought to be regulated by the same mechanism (2, 139).
In the processing of calcitonin/CGRP transcripts, the choice was whether to use the regulated exon as the 3′-terminal exon. With the immunoglobulin transcripts, the processing pattern is determined by whether the alternative exon 4 is seen as an internal exon flanked by a 3′ splice acceptor site and 5′ donor site or instead as a 3′-terminal exon defined by the same 3′ acceptor site but ending with the secretory poly(A) site. The competing processing sites on exon 4 both have suboptimal signals (295, 359), and it has been proposed that small changes in the efficiency of either the splicing or polyadenylation reactions could tip the balance in favor of the secreted poly(A) site (358).
In addition, the murine immunoglobulin M secretory site has an unusual organization of polyadenylation signals, with a consensus AAUAAA downstream of an AU-rich region (AUAAAAAUUAG). This AU-rich region on its own can function inefficiently as an upstream signal, and two downstream GU-rich regions which are both necessary for full polyadenylation activity (363). The distal downstream element must be present as part of a stem-loop structure in order to enhance polyadenylation by stabilizing the CPSF-CstF complex (363). The arrangement of an AU-rich region adjacent to a consensus hexanucleotide and a stem-loop similarly positioned downstream of the cleavage site is evolutionarily conserved in the immunoglobulin M secretory sites and may be important for regulation of this site.
Enhancer elements favoring one or the other pattern of processing have not been identified in the immunoglobulin transcripts (139), and several studies have indicated that immunoglobulin-specific sequences are not necessary. Immunoglobulin genes with substitutions of the regulated processing sites or nonimmunoglobulin test genes designed to have an immunoglobulin-like structure are appropriately regulated in cell lines and in transgenic mice as long as the competing splice and polyadenylation sites are balanced in their efficiencies (357, 358, 418). No differences have been detected in the splicing efficiencies of B cells at different stages of development or in comparison to non-B-cell types (139). Other findings point instead to a mechanism in which changes in the amounts or activities of polyadenylation factors suffice to mediate the processing regulation.
Weak poly(A) sites, regardless of sequence composition, are used more efficiently in plasma cells than in other cell types (295). The switch to usage of the secreted site parallels an increase in the binding but not a change in the abundance or mobility of the 64-kDa subunit of the CstF cleavage factor (137) and the 100-kDa subunit of CPSF (136). Consistent with the idea that CstF activity is limiting in primary B cells, a 10-fold overexpression of CstF-64 in a chicken B-cell line is sufficient to switch heavy-chain RNA expression to the secreted poly(A) site in vivo, and supplying more CstF in a reconstituted in vitro reaction can increase the use of this site (455). This study also showed that the membrane site is two- to threefold more efficiently processed than the secreted site in the in vitro system and has a higher affinity for the CstF cleavage factor (455). These results implicate CstF and complex assembly as a primary target in determining poly(A) site choice.
On the other hand, a fivefold increase in CstF p64 levels seen when primary resting B cells are stimulated to proliferate by CD40 ligand exposure (a G0-to-S-phase transition) is not adequate to elicit the shift to secreted mRNAs, suggesting that other lymphokine-induced factors will also be important (292). The distal GU-rich element of the murine immunoglobulin M secreted site is necessary for the binding of a 30-kDa protein whose induction correlates with the appearance of secretory mRNAs (362). It is not yet known if this protein is the trans-acting factor responsible for the more efficient polyadenylation complex formation mediated by its binding site. In addition, a 50- to 55-kDa protein binds more strongly to the membrane poly(A) site under conditions which inhibit the accumulation of the secreted mRNAs (362). Regulation may involve both positive and negative factors—an activity has also been found in nuclear extracts from B-cell tumors which specifically destabilizes polyadenylation complexes on the secretory poly(A) site (504). Furthermore, sequences upstream of exon 4 are necessary to fully achieve normal ratios of secreted to membrane mRNAs in plasma cells (2), emphasizing that we still do not understand all that contributes to the fine-tuning of this regulation. The known possibilities are summarized in Table 4.
Other mechanisms can also contribute to the preferred use of an upstream poly(A) site. In plasma cells, transcription termination increases between the secretory poly(A) site and the membrane exons of the immunoglobulin secretory poly(A) site, but not that of other isoforms (reviewed in reference 139). However, from what we know of the dependence of transcription termination on polyadenylation, this may simply be due to an increase in use of the secretory site. In another interesting example, the exon-intron architecture of the alternatively processed alpha-thyroid hormone receptor mRNAs resembles that of immunoglobulin transcripts. However, the selection of the upstream poly(A) site in these mRNAs did not correlate with the differentiated state of the B-cell lines tested but, instead, correlated with the levels of antisense RNA naturally transcribed from an overlapping gene (191).
Utilization of duplicate poly(A) sites in HIV-1 transcripts.
Duplication of retroviral sequences during the process of integration results in one poly(A) signal immediately downstream of the viral promoter and a second, identical one more appropriately positioned at the end of the viral genome (Fig. 10C). During transcription of the viral genes, the first site is ignored and the second is used to produce the viral transcripts. Several mechanisms contribute to this pattern of polyadenylation (Table 4). First, the core HIV-1 viral poly(A) signal is weakened because its incorporation into a stem-loop structure causes suboptimal binding of polyadenylation factors (116, 246, 247). It is recognized efficiently in vivo and in vitro only in conjunction with an enhancer element present upstream of the promoter-distal but not the promoter-proximal poly(A) site (174, 465). This auxiliary element binds the 160-kDa subunit of CPSF and facilitates CPSF interaction at the AAUAAA (164). Functional introns upstream of distal poly(A) site also stimulate its use (416).
Taking all of this into consideration, the promoter-proximal poly(A) site is not favored, because of its inherent weakness and the lack of an upstream USE. The closeness of this poly(A) site to the promoter was also found to be detrimental (87, 484), due to nuclear instability of short polyadenylated transcripts (415). Moreover, the site is very effectively squelched by the strong splice donor found downstream of it (19), an arrangement which we have discussed above.
Cell Cycle Regulation of Factors Involved in 3′-End Formation
Histone pre-mRNA cleavage factors.
Precise control mechanisms are required to produce new histones in a time frame within the cell cycle coincident with the replication of the genome. This is essential to ensure that newly synthesized DNA is efficiently packaged into nucleosomes. Thus, the vast majority of histones are regulated in a cell cycle-dependent manner (197). This regulation is achieved mainly at the histone mRNA level through stringent control of histone gene transcription, mRNA 3′-end formation, and mRNA degradation (reviewed in references 196, 293, 325, 346, and 413).
Posttranscriptional events, including 3′-end processing and mRNA stability, contribute significantly to this cell cycle regulation (190, 346, 503). The heat-labile component of the histone 3′-end-processing complex becomes limiting in G0- and G1-arrested mouse cells (160, 206, 275), and maximum 3′-end processing coincides with DNA synthesis in the S phase of the cell cycle. The levels of U7 snRNP do not appear to vary during the cell cycle in human cells (51). Studies with serum-starved murine cells showed that the 5′ end of U7 is protected from micrococcal nuclease digestion in G0 cells (206). This protection is removed when cells are released from G0 by the addition of serum, suggesting that binding of a protein factor or secondary structure prevents the interaction of U7 with the histone downstream element in the G0 state. However, a similar protection of the 5′ end of U7 was not observed in human cells (51).
Cleavage/polyadenylation factors.
The synthesis and utilization of polyadenylated mRNAs in mammalian cells fluctuates during the mitotic cell cycle, with a large increase as cells move into S phase (38), and a general repression of transcription and translation is seen during M, when nuclear breakdown, chromosome condensation, and chromosome segregation occur (101). Early studies reported a concomitant increase in polyadenylation activity as cells entered S phase (99, 192), and the advent of antibodies against many of the cleavage/polyadenylation factors has made possible an initial probe into the molecular basis of this increase. Martincic et al. (292) have recently shown that the amount of the CstF 64-kDa subunit increases fivefold during the G0-to-S-phase transition when NIH 3T6 fibroblasts or primary B cells are stimulated to proliferate. In that study, the levels of CstF-50 and CF Im-25 rose about twofold in B cells but not fibroblasts, while the levels of CstF-77, CPSF-100, and CPSF-30 remained constant. However, the Drosophila homolog of CstF-77, Su(f), accumulates in mitotically active cells compared to nondividing cells (24), suggesting that cell cycle regulation may differ depending on the organism.
Differences in the expression of CstF-64 have been recently discovered in the germ cells of the mouse testis (476). A 70-kDa form replaces the 64-kDa protein in meiotic spermatocytes. The CstF-64 gene maps to the X chromosome in both humans and mice. Thus, the 64-kDa form may be absent from spermatocytes because of X chromosome inactivation during meiosis, implying that the 70-kDa species is expressed from a homologous autosomal gene. This variant CstF subunit may facilitate the processing of the many testis-specific transcripts which lack a canonical AAUAAA (476).
The phosphorylated state of poly(A) polymerase changes during Xenopus oocyte maturation and early postfertilization development (30) and in mammals as cells progress through the cell cycle (102). The mammalian PAP is hyperphosphorylated during M phase by the p34cdc2-cyclin B complex at multiple consensus and nonconsensus sites in its carboxyl-terminal serine- and threonine-rich region (102, 103). The observation that the hyperphosphorylated forms of PAP do not accumulate at sites of transcription (412) is consistent with the inhibition of the catalytic activity of PAP by this extensive modification (102, 103). The S. cerevisiae PAP is phosphorylated and ubiquitinated at a point in the cell cycle corresponding to late S/G2, but the modified species disappear at M phase (313). The effects of these modifications on PAP activity are not known. The change in timing of the modification from that of the mammalian enzyme may relate to differences in how S. cerevisiae progresses through the cell cycle and to the fact that the nucleus remains intact in M phase.
The expression of mRNAs from some of the S. cerevisiae genes encoding cleavage/polyadenylation factors has been examined across the cell cycle (90). This analysis did not reveal any significant periodic fluctuations in the levels of mRNAs encoding Rna15, Pcf11, Ysh1, Yth1, Fip1, Pap1, or Pab1.
Alterations of the Cleavage/Polyadenylation Machinery during Viral Infection
Viruses often alter the polyadenylation capacity of infected cells, sometimes as a means to differentially express viral genes during the course of viral replication and in other cases as a means to suppress the synthesis of cellular mRNAs in favor of viral ones. For example, during early infection by adenovirus, transcripts made from the adenovirus major late transcriptional unit are polyadenylated predominantly at the promoter-proximal L1 poly(A) site, while late in infection, a switch occurs to more distal sites. Sequences flanking the core L1 poly(A) site are responsible for this regulation (372), and in vitro analysis has shown that they stabilize the binding of purified CPSF to this site (165). These investigators propose that the L1 sequences, like the HIV-1 auxiliary element (164), may provide an additional contact site for CPSF-160 or simply place the core poly(A) site in a more favorable structure. However, late in infection the positive effect of these auxiliary elements cannot compensate for a substantial decline in CstF activity, and the stronger downstream sites prevail (287, 372). However, if cells in the late phase of infection are reinfected with virus, transcripts derived from the superinfected virus (or transfected reporter constructs) show a polyadenylation pattern typical of the early stage of infection (145). This finding has brought about the speculation that there could be localized modification or depletion of CstF in late viral transcription centers (125).
In other types of viral infections, viral proteins appear to play a more direct role in affecting polyadenylation efficiency. The ICP27/IE63 protein of HSV-1 is necessary for the switch from early- to late-gene expression. While an interaction of ICP27/IE63 with polyadenylation factors has yet to be demonstrated, this viral protein enhances processing at the weak poly(A) sites of late transcripts by increasing the binding of CstF-64 to these sites (301). The Epstein-Barr virus early proteins SM and M, which are homologs of HSV ICP27, are important for a posttranscriptional increase in mRNAs encoding the Epstein-Barr virus DNA polymerase and may function by stimulating the polyadenylation of these transcripts, which are weakened by a noncanonical hexanucleotide, UAUAAA (243).
Influenza virus uses a complex regulatory network to ensure the optimal temporal production and utilization of viral mRNAs. Some of the negative effects on cellular mRNAs are mediated by the abundant NS1 viral protein (reviewed in reference 251). This RNA-binding protein inhibits pre-mRNA splicing and nuclear export of polyadenylated mRNAs. By binding to double-stranded RNA produced during viral replication, it blocks the translation inhibition caused by double-stranded RNA-activated protein kinase. NS1 also inhibits 3′-end processing of cellular pre-mRNAs in vivo, and these unprocessed RNAs accumulate in the nucleus (330, 426). The reason for this inhibition, surprisingly, turns out to be a direct interaction of NS1 with the 30-kDa subunit of CPSF, an association which prevents the binding of CPSF to precursor RNA and blocks both cleavage and polyadenylation in vitro (330). The polyadenylation of any cellular mRNAs which do get processed is prevented because NS1 also directly inhibits PAB II function in tail elongation (85). Influenza virus circumvents these blocks and achieves the polyadenylation of its own mRNAs through reiterative copying of virally encoded uridine stretches by the viral transcriptase (251). The selection of CPSF-30 for negative regulation by a viral protein and the importance of the zebrafish homolog to proper development (155) may foretell its use as a target in other regulated polyadenylation events.
Alternative mRNA Polyadenylation in Yeast
While many yeast genes have multiple poly(A) sites, regulated alternative polyadenylation had been observed only for transcripts from the CBP1 gene (297). Two sizes of mRNAs are produced from this gene, and the induction of respiration by growth on glycerol results in the preferential accumulation of the shorter one, which is polyadenylated at multiple sites in a region near the middle of the coding sequence (298). The increased abundance of the short transcripts is not due to a change in stability of the two mRNAs or to transcription termination between the two poly(A) signals (434). Unexpectedly, the same carbon source regulation was recently shown to operate on the transcripts of other yeast genes (AEP2/ATP13, RNA14, and SIR1) which produce long mRNAs as well as truncated ones that end within the coding sequence (434). A consensus sequence which might specify this regulation could not be perceived by examination of the sequences surrounding the different poly(A) sites.
The molecular mechanism underlying this regulation has not been identified, but it may involve the Hrp1 cleavage/polyadenylation factor. It was recently found that inaccurate cleavage can occur upstream of a normal poly(A) site when this protein is not present in processing reactions (308), leading to the speculation that limiting amounts of Hrp1 may provoke the shift in polyadenylation sites described above. On the other hand, changes in levels of other processing factors may also be involved, since limiting the amounts of active Rna14, Rna15, or Pap1 by temperature-sensitive mutations favors the use of the more distal poly(A) sites in ACT1 and RNA14 transcripts (282, 284). Interestingly, a similar regulation has been reported for the Drosophila homolog of Rna14, Su(f), in that accumulation of truncated Su(f) transcripts requires wild-type Su(f) protein (25).
CONCLUSIONS AND PERSPECTIVES
Regarding the basic polyadenylation mechanism, more research is needed to define the architecture of the processing complex, i.e., how the factors interact with each other and with the RNA substrate and how this changes as the cleavage complex is converted to a poly(A) addition complex. The future will probably bring additional understanding of the dynamic interplay of processes involved in mRNA synthesis and utilization. Models of how transcription termination and 3′-end formation are coupled need to be tested, and the contacts with splicing and transport factors should be probed in greater detail. Several unique mechanisms to elicit alternative polyadenylation have been identified. The recent review by Edwald-Gilbert et al. (139) described 33 known genes in which the choice of poly(A) site was regulated at different developmental stages or in different tissues, and there are probably many more such genes. How many different ways will the cell have devised to accomplish this alternative processing? Certain players continue to make appearances at regulated sites, and common themes should continue to manifest with future research. A major challenge will be to determine how the cellular environment modulates the activity or levels of these regulatory factors. All of these endeavors will no doubt be aided by the rapid accumulation of genomic sequence information, by the growing sophistication of tools to analyze this huge database, and by the increasingly efficient methods to determine genome-wide expression patterns and protein-protein interactions.
ACKNOWLEDGMENTS
We are grateful to G. Carmichael, G. Gilmartin, S. Gross, S. Helmling, M. Henry, C. Milcarek, and A. Zhelkovsky for valuable comments on the manuscript and to our colleagues who provided information prior to publication.
REFERENCES
- 1.Abe A, Hiraoka Y, Fukasawa T. Signal sequence for generation of mRNA 3′ ends in the Saccharomyces cerevisiae GAL7 gene. EMBO J. 1990;9:3691–3697. doi: 10.1002/j.1460-2075.1990.tb07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abuodeh R, Wei H, Yuan D. Effect of upstream RNA processing on selection of μS versus μM poly(A) sites. Nucleic Acids Res. 1998;26:5417–5424. doi: 10.1093/nar/26.23.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam S A, Nakagawa T, Swanson M S, Woodruff T K, Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adami G, Nevins J R. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 1988;7:2107–2116. doi: 10.1002/j.1460-2075.1988.tb03050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams M D, Rudner D Z, Rio D C. Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339. doi: 10.1016/s0955-0674(96)80006-8. [DOI] [PubMed] [Google Scholar]
- 6.Adema G J, van Hulst K L, Baas P D. Uridine branch acceptor is a cis-acting element involved in regulation of the alternative processing of calcitonin/CGRP-1 pre-mRNA. Nucleic Acids Res. 1990;18:5365–5373. doi: 10.1093/nar/18.18.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afonina E, Stauber R, Pavlakis G N. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 8.Aitchison J D, Blobel G, Rout M P. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- 9.Amara S G, Jonas V, Rosenfeld M G, Ong E S, Evans R M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- 10.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 11.Amrani N, Dufour M E, Bonneaud N, Lacroute F. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15-2 mutation in Saccharomyces cerevisiae. Mol Gen Genet. 1996;252:552–562. doi: 10.1007/BF02172401. [DOI] [PubMed] [Google Scholar]
- 12.Amrani N, Minet M, LeGouar M, Lacroute F, Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amrani N, Minet M, Wyers F, Dufour M, Aggerbeck L, Lacroute F. PCF11 encodes a third protein component of the yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anant S, Giannoni F, Antic D, DeMaria C T, Keene J D, Brewer G, Davidson N O. AU-rich RNA binding proteins Hel-N1 and AUF1 bind apolipoprotein B mRNA and inhibit posttranscriptional C to U editing. Nucleic Acids Symp Ser. 1997;36:115–118. [Google Scholar]
- 15.Anderson J, Wilson S, Datar K, Swanson M. Nab2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antoniou M, Geraghty F, Hurst J, Grosveld F. Efficient 3′-end formation of human beta-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res. 1998;26:721–729. doi: 10.1093/nar/26.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranda A, Perez-Ortin J E, Moore C, del Olmo M. The yeast FBP1 poly(A) signal functions in both orientations and overlaps with a gene promoter. Nucleic Acids Res. 1998;26:4588–4596. doi: 10.1093/nar/26.20.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aranda A, Perez-Ortin J E, Moore C, del Olmo M L. Transcription termination downstream of the Saccharomyces cerevisiae FBP1 poly(A) site does not depend on efficient 3′end processing. RNA. 1998;4:303–318. [PMC free article] [PubMed] [Google Scholar]
- 19.Ashe M P, Griffin P, James W, Proudfoot N J. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 20.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashfield R, Enriquez-Harris P, Proudfoot N J. Transcription termination between the closely linked human complement genes C2 and factor B: common termination factor C2 and c-myc? EMBO J. 1991;10:4197–4207. doi: 10.1002/j.1460-2075.1991.tb04998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashfield R, Patel A J, Bossone S A, Brown H, Campbell R D, Marcu K B, Proudfoot N J. MAZ-dependent termination between closely spaced human complement genes. EMBO J. 1994;13:5656–5667. doi: 10.1002/j.1460-2075.1994.tb06904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 24.Audibert A, Juge F, Simonelig M. The suppressor of forked protein of Drosophila, a homologue of the human 77K protein required for mRNA 3′-end formation, accumulates in mitotically-active cells. Mech Dev. 1998;72:53–63. doi: 10.1016/s0925-4773(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 25.Audibert A, Simonelig M. Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc Natl Acad Sci USA. 1998;95:14302–14307. doi: 10.1073/pnas.95.24.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagga P S, Arhin G K, Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bagga P S, Ford L P, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai C, Tolias P P. Cleavage of RNA hairpins mediated by a developmentally regulated CCCH zinc finger protein. Mol Cell Biol. 1996;16:6661–6667. doi: 10.1128/mcb.16.12.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai C, Tolias P P. Drosophila clipper/CPSF 30K is a post-transcriptionally regulated nuclear protein that binds RNA containing GC clusters. Nucleic Acids Res. 1998;26:1597–1604. doi: 10.1093/nar/26.7.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ballantyne S, Bilger A, Astrom J, Virtanen A, Wickens M. Poly (A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA. 1995;1:64–78. [PMC free article] [PubMed] [Google Scholar]
- 31.Barabino S, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 32.Bardwell V J, Wickens M, Bienroth S, Keller W, Sproat B S. Site-directed ribose methylation identifies 2′-OH groups in polyadenylation substrates critical for AAUAAA recognition and poly(A) addition. Cell. 1991;65:125–133. doi: 10.1016/0092-8674(91)90414-t. [DOI] [PubMed] [Google Scholar]
- 33.Barta I, Iggo R. Autoregulation of expression of the yeast Dbp2p ‘DEAD-box’ protein is mediated by sequences in the conserved DBP2 intron. EMBO J. 1995;14:3800–3808. doi: 10.1002/j.1460-2075.1995.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauren G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 36.Bennetzen J L, Hall B D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase I. J Biol Chem. 1982;257:3018–3025. [PubMed] [Google Scholar]
- 37.Bentley D. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 38.Benz E W, Jr, Getz M J, Wells D J, Moses H L. Nuclear RNA polymerase activities and poly(A)-containing mRNA accumulation in cultured AKR mouse embryo cells stimulated to proliferate. Exp Cell Res. 1977;108:157–165. [PubMed] [Google Scholar]
- 39.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 40.Berget S M, Berk A J, Harrison T, Sharp P A. Spliced segments at the 5′ termini of adenovirus-2 late mRNA: a role for heterogeneous nuclear RNA in mammalian cells. Cold Spring Harbor Symp Quant Biol. 1978;42:523–529. doi: 10.1101/sqb.1978.042.01.055. [DOI] [PubMed] [Google Scholar]
- 41.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 42.Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′ end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 43.Bhat B M, Wold W S. A small deletion distant from a splice or polyadenylation site dramatically alters pre-mRNA processing in region E3 of adenovirus. J Virol. 1987;61:3938–3945. doi: 10.1128/jvi.61.12.3938-3945.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–19776. [PubMed] [Google Scholar]
- 46.Birse C E, Lee B A, Hansen K, Proudfoot N J. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Birse C E, Minvielle-Sebastia L, Lee B A, Keller W, Proudfoot N J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 48.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 49.Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. Bioessays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 50.Boeck R, Tarum S, Rieger M, Deardorff J A, Muller-Auer S, Sachs A B. The yeast Pan2 proteins is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 51.Bond U, Yario T A. The steady state levels and structure of the U7 snRNP are constant during the human cell cycle: lack of cell cycle regulation of histone mRNA 3′ end formation. Cell Mol Biol Res. 1994;40:27–34. [PubMed] [Google Scholar]
- 52.Bond U M, Yario T A, Steitz J A. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 53.Bonneaud N, Minvielle-Sebastia L, Cullin C, Lacroute F. Cellular localization of RNA14p and RNA15p, two yeast proteins involved in mRNA stability. J Cell Sci. 1994;107:913–921. doi: 10.1242/jcs.107.4.913. [DOI] [PubMed] [Google Scholar]
- 54.Bossie M, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourquin J P, Stagljar I, Meier P, Moosmann P, Silke J, Baechi T, Georgiev O, Schaffner W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braaten D, Franke E K, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brackenridge S, Ashe H L, Giacca M, Proudfoot N J. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 1998;25:2326–2335. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brady H A, Wold W S. Competition between splicing and polyadenylation reactions determines which adenovirus region E3 mRNAs are synthesized. Mol Cell Biol. 1988;8:3291–3297. doi: 10.1128/mcb.8.8.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brais B, Bouchard J P, Xie Y G, Rochefort D L, Chretien N, Tome F M, Lafreniere R G, Rommens J M, Uyama E, Nohira O, Blumen S, Korczyn A D, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau G A, Korcyn A D. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 60.Brand S F, Pichoff S, Noselli S, Bourbon H-M. Novel Drosophila melanogaster genes encoding RRM-type RNA-binding proteins identified by a degenerate PCR strategy. Gene. 1995;154:187–192. doi: 10.1016/0378-1119(94)00840-o. [DOI] [PubMed] [Google Scholar]
- 61.Briggs M W, Burkard K T, Butler J S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 62.Briggs M W, Butler J S. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics. 1996;143:1149–1161. doi: 10.1093/genetics/143.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown C E, Sachs A B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown C E, Tarun S Z, Boeck R, Sachs A B. PAN3 encodes a subunit of the Pab1-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown P H, Tiley L S, Cullen B R. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeats requires flanking U3-specific sequences. J Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buchman A R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;256:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 68.Butler J S, Platt T. RNA processing generates the mature 3′ ends of yeast CYC1 mRNA in vitro. Science. 1988;242:1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- 69.Cao G J, Pogliano J, Sarkar N. Identification of the coding region for a second poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:11580–11585. doi: 10.1073/pnas.93.21.11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao G J, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 72.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 73.Carmo-Fonseca, M. 1999. Unpublished data.
- 74.Carswell S, Alwine J C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 76.Challoner P B, Moss S B, Groudine M. Expression of replication-dependent histone genes in avian spermatids involves an alternate pathway of mRNA 3′-end formation. Mol Cell Biol. 1989;9:902–913. doi: 10.1128/mcb.9.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan R C, Black D L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chanfeau G, Noble S M, Guthrie C. An essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF) Science. 1996;274:1511–1514. doi: 10.1126/science.274.5292.1511. [DOI] [PubMed] [Google Scholar]
- 79.Chen E J, Frand A R, Chitouras E, Kaiser C A. A link between secretion and pre-mRNA processing defects in Saccharomyces cerevisiae and the identification of a novel splicing gene, RSE1. Mol Cell Biol. 1998;18:7139–7146. doi: 10.1128/mcb.18.12.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen F, MacDonald C C, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen F, Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J, Moore C L. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen S, Hyman L E. A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 1998;26:4965–4974. doi: 10.1093/nar/26.21.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen S, Reger R, Miller C, Hyman L E. Transcriptional terminators of RNA polymerase II are associated with yeast replication origins. Nucleic Acids Res. 1996;24:2885–2893. doi: 10.1093/nar/24.15.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Z, Li Y, Krug R. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′ end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng G H, Nandi A, Clerk S, Skoultchi A I. Different 3′-end processing produces two independently regulated mRNAs from a single H1 histone gene. Proc Natl Acad Sci USA. 1989;86:7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cherrington J, Ganem D. Regulation of polyadenylation in HIV: contributions of promoter proximity and upstream sequences. EMBO J. 1992;11:1513–1524. doi: 10.1002/j.1460-2075.1992.tb05196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chiou H C, Dabrowski C, Alwine J C. Simian virus 40 late mRNA leader sequences involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J Virol. 1991;65:6677–6685. doi: 10.1128/jvi.65.12.6677-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cho D C, Scharl E C, Steitz J A. Decreasing the distance between the two conserved sequence elements of histone pre-messenger RNA interferes with 3′ processing in vitro. RNA. 1995;1:905–914. [PMC free article] [PubMed] [Google Scholar]
- 90.Cho R J, Campbell M J, Winzeler E A, Steinmetz L, Conway A, Wodicka L, Wolfsberg T G, Gabrielian A E, Landsman D, Lockhart D J, Davis R W. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 91.Cho R J, Fromont-Racine M, Wodicka L, Feierbach B, Stearns T, Legrain P, Lockhart D J, Davis R W. Parallel analysis of genetic selections using whole genome oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:3752–3757. doi: 10.1073/pnas.95.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chodchoy N, Pandey N B, Marzluff W F. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol Cell Biol. 1991;11:497–509. doi: 10.1128/mcb.11.1.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chou M Y, Rooke N, Turck C W, Black D L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chou Z F, Chen F, Wilusz J. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 1994;22:2525–2531. doi: 10.1093/nar/22.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chrislip K M, Hengst-Zhang J A, Jacob S T. Polyadenylation of SV40 late pre-mRNA is dependent on phosphorylation of an essential component associated with the 3′ end processing machinery. Gene Expression. 1991;1:197–206. [PMC free article] [PubMed] [Google Scholar]
- 96.Christofori G, Keller W. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988;54:875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- 97.Cobianchi F, Biamonti G, Maconi M, Riva S. Human hnRNP protein A1: a model polypeptide for a structural and genetic investigation of a broad family of RNA binding proteins. Genetica. 1994;94:101–114. doi: 10.1007/BF01443425. [DOI] [PubMed] [Google Scholar]
- 98.Cole C N, Hammell C M. Nucleocytoplasmic transport: driving and directing transport. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- 99.Coleman M S, Hutton J J, Bollum F J. Terminal riboadenylate transferase in human lymphocytes. Nature. 1974;248:407–409. doi: 10.1038/248407a0. [DOI] [PubMed] [Google Scholar]
- 100.Coleman T P, Roesser J R. RNA secondary structure: an important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochemistry. 1998;37:15941–15950. doi: 10.1021/bi9808058. [DOI] [PubMed] [Google Scholar]
- 101.Colgan D, Manley J. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 102.Colgan D F, Murthy K G, Prives C, Manley J L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 103.Colgan D F, Murthy K G, Zhao W, Prives C, Manley J L. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Collart D, Romain P L, Huebner K, Pockwinse S, Pilapil S, Cannizzaro L A, Lian J B, Croce C M, Stein J L, Stein G S. A human histone H2B.1 variant gene, located on chromosome 1, utilizes alternative 3′ end processing. J Cell Biochem. 1992;50:374–385. doi: 10.1002/jcb.240500406. [DOI] [PubMed] [Google Scholar]
- 105.Coller J M, Gray N K, Wickens M P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colot H V, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 107.Connelly S, Manley J L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 108.Cooke C, Alwine J C. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cote G J, Nguyen I N, Berget S M, Gagel R F. Calcitonin exon sequences influence alternative RNA processing. Mol Endocrinol. 1990;4:1744–1749. doi: 10.1210/mend-4-11-1744. [DOI] [PubMed] [Google Scholar]
- 110.Cote G J, Nguyen I N, Lips C J, Berget S M, Gagel R F. Validation of an in vitro RNA processing system for CT/CGRP precursor mRNA. Nucleic Acids Res. 1991;19:3601–3606. doi: 10.1093/nar/19.13.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cote G J, Stolow D T, Peleg S, Berget S M, Gagel R F. Identification of exon sequences and an exon binding protein involved in alternative RNA splicing of calcitonin/CGRP. Nucleic Acids Res. 1992;20:2361–2366. doi: 10.1093/nar/20.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel M L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. EMBO J. 1988;7:801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dahlberg J E, Lund E. Functions of the GTPase Ran in RNA export from the nucleus. Curr Opin Cell Biol. 1998;10:400–408. doi: 10.1016/s0955-0674(98)80017-3. [DOI] [PubMed] [Google Scholar]
- 114.Daneholt B. A look at messenger RNP moving through the nuclear pore. Cell. 1997;88:585–588. doi: 10.1016/s0092-8674(00)81900-5. [DOI] [PubMed] [Google Scholar]
- 115.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 116.Das A T, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Das Gupta J, Gu H, Chernokalskaya E, Gao X, Schoenberg D R. Identification of two cis-acting elements that independently regulate the length of poly(A) on Xenopus albumin pre-mRNA. RNA. 1998;4:766–776. doi: 10.1017/s1355838298971837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Das Gupta J, Li Q S, Thomson A B, Hunt A G. Characterization of a cDNA encoding a novel plant poly(A) polymerase. Plant Mol Biol. 1998;37:729–734. doi: 10.1023/a:1006000213403. [DOI] [PubMed] [Google Scholar]
- 119.Daugeron M C, Tazi J, Jeanteur P, Brunel C, Cathala G. U1-U2 snRNPs interaction induced by an RNA complementary to the 5′ end sequence of U1 snRNA. Nucleic Acids Res. 1992;20:3625–3630. doi: 10.1093/nar/20.14.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deardorff J A, Sachs A B. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 121.Del Gatto-Konczak F, Olive M, Gesnel M C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.del Olmo M, Mizrahi N, Gross S, Moore C. The Uba2 and Ufd1 proteins of S. cerevisiae interact with poly(A) polymerase and affect the polyadenylation activity of extracts. Mol Gen Genet. 1997;255:209–218. doi: 10.1007/s004380050491. [DOI] [PubMed] [Google Scholar]
- 123.Del Priore V, Snay C A, Bahr A, Cole C N. The product of the Saccharomyces cerevisiae RSS1 gene, identified as a high-copy suppressor of the rat7-1 temperature-sensitive allele of the RAT7/NUP159 nucleoporin, is required for efficient mRNA export. Mol Biol Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 125.DeZazzo J D, Falck-Pedersen E, Imperiale M J. Sequences regulating temporal poly(A) site switching in the adenovirus major late transcription unit. Mol Cell Biol. 1991;11:5977–5984. doi: 10.1128/mcb.11.12.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.DeZazzo J D, Imperiale M J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.DeZazzo J D, Scott J M, Imperiale M J. Relative roles of signals upstream of AAUAAA and promoter proximity in regulation of human immunodeficiency virus type 1 mRNA 3′ end formation. Mol Cell Biol. 1992;12:5555–5562. doi: 10.1128/mcb.12.12.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.DeZazzo J K, Kilpatrick J E, Imperiale M J. Involvement of a long terminal repeat U3 sequence overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol Cell Biol. 1991;11:1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 130.Doktycz M J, Larimer F W, Pastrnak M, Stevens A. Comparative analyses of the secondary structures of synthetic and intracellular yeast MFA2 mRNAs. Proc Natl Acad Sci USA. 1998;95:14614–14621. doi: 10.1073/pnas.95.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dominski Z, Sumerel J, Hanson R J, Marzluff W F. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- 131a.Dominski Z, Zheng L-X, Sanchez R, Marzluff W F. Stem-loop binding protein facilitates formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 133.Duvel K, Egli C M, Braus G H. A single point mutation in the yeast TRP4 gene affects efficiency of mRNA 3′ end processing and alters selection of the poly(A) site. Nucleic Acids Res. 1999;27:1289–1295. doi: 10.1093/nar/27.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133a.Dye M J, Proudfoot N J. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–378. doi: 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 134.Eckner R, Ellmeier W, Birnstiel M. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Edmonds M, Winters M A. Polyadenylate polymerases. Prog Nucleic Acids Res Mol Biol. 1976;17:149–179. doi: 10.1016/s0079-6603(08)60069-0. [DOI] [PubMed] [Google Scholar]
- 136.Edwalds-Gilbert G, Milcarek C. The binding of a subunit of the general polyadenylation factor cleavage-polyadenylation specificity factor (CPSF) to polyadenylation sites changes during B cell development. Nucleic Acids Symp Ser. 1995;33:229–233. [PubMed] [Google Scholar]
- 137.Edwalds-Gilbert G, Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol Cell Biol. 1995;15:6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Edwalds-Gilbert G, Prescott J, Falck-Pedersen E. 3′ RNA processing efficiency plays a primary role in generating termination-competent RNA polymerase II elongation complexes. Mol Cell Biol. 1993;13:3472–3480. doi: 10.1128/mcb.13.6.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Edwalds-Gilbert G, Veraldi K L, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eggermont J, Proudfoot N J. Poly(A) signals and transcriptional pause sites combine to prevent interference between RNA polymerase II promoters. EMBO J. 1993;12:2539–2548. doi: 10.1002/j.1460-2075.1993.tb05909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Egli C, Springer C, Braus G. A complex unidirectional signal element mediates GCN4 mRNA 3′ end formation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2466–2473. doi: 10.1128/mcb.15.5.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eisen A, Lucchesi J C. Unraveling the role of helicases in transcription. Bioessays. 1998;8:634–641. doi: 10.1002/(SICI)1521-1878(199808)20:8<634::AID-BIES6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 143.Enriquez-Harris P, Levitt N, Briggs D, Proudfoot N J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991;10:1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fahrner K, Yarger J, Hereford L. Yeast histone mRNA is polyadenylated. Nucleic Acids Res. 1980;8:5725–5737. doi: 10.1093/nar/8.23.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Falck-Pedersen E, Logan J. Regulation of poly(A) site selection in adenovirus. J Virol. 1989;63:532–541. doi: 10.1128/jvi.63.2.532-541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins D, Silver P. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Flaherty S M, Fortes P, Izaurralde E, Mattaj I W, Gilmartin G M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ford J P, Hsu M T. Transcription pattern of in vivo-labeled late simian virus 40 RNA: equimolar transcription beyond the mRNA 3′ terminus. J Virol. 1978;28:795–801. doi: 10.1128/jvi.28.3.795-801.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ford L P, Bagga P S, Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol Cell Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 151.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 152.Fu X D, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci USA. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Furger A, Schaller A, Schumperli D. Functional importance of conserved nucleotides at the histone RNA 3′ processing site. RNA. 1998;4:246–256. [PMC free article] [PubMed] [Google Scholar]
- 154.Furth P A, Choe W T, Rex J H, Byrne J C, Baker C C. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 156.Gallie D R, Lewis N J, Marzluff W F. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gamberi C, Izaurralde E, Beisel C, Mattaj I W. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol Cell Biol. 1997;17:2587–2597. doi: 10.1128/mcb.17.5.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gershon P. mRNA 3′ end formation by vaccinia virus: mechanism of action of a heterodimeric poly(A) polymerase. Semin Virol. 1998;8:343–350. [Google Scholar]
- 159.Gick O, Kramer A, Keller W, Birnstiel M L. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gick O, Kramer A, Vasserot A, Birnstiel M L. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gieselmann V, Polete A, Kreysing J, von Figura K. Arylsulfatase A pseudodeficiency: loss of a polyadenylation signal and N-glycosylation site. Proc Natl Acad Sci USA. 1989;86:9436–9440. doi: 10.1073/pnas.86.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gil A, Proudfoot N J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 163.Gilmartin G M, Fleming E S, Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 165.Gilmartin G M, Hung S L, DeZazzo J D, Fleming E S, Imperiale M J. Sequences regulating poly(A) site selection within the adenovirus major late transcription unit influence the interaction of constitutive processing factors with the pre-mRNA. J Virol. 1996;70:1775–1783. doi: 10.1128/jvi.70.3.1775-1783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gilmartin G M, McDevitt M A, Nevins J R. Multiple factors are required for specific RNA cleavage at a poly(A) addition site. Genes Dev. 1988;2:578–587. doi: 10.1101/gad.2.5.578. [DOI] [PubMed] [Google Scholar]
- 167.Gilmartin G M, Nevins J R. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol Cell Biol. 1991;11:2432–2438. doi: 10.1128/mcb.11.5.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gilmartin G M, Nevins J R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2189. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 169.Gooding C, Roberts G C, Smith C W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 170.Graber J H, Cantor C R, Mohr S C, Smith T F. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Grabowski P J. Splicing regulation in neurons: tinkering with cell-specific control. Cell. 1998;92:709–712. doi: 10.1016/s0092-8674(00)81399-9. [DOI] [PubMed] [Google Scholar]
- 172.Graveley B R, Fleming E S, Gilmartin G M. Restoration of both structure and function to a defective poly(A) site by in vitro selection. J Biol Chem. 1996;271:33654–33663. doi: 10.1074/jbc.271.52.33654. [DOI] [PubMed] [Google Scholar]
- 173.Graveley B R, Fleming E S, Gilmartin G M. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Graveley B R, Gilmartin G M. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol. 1996;70:1612–1617. doi: 10.1128/jvi.70.3.1612-1617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Greger I H, Proudfoot N J. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gu X, Marzluff W F. 3′ Processing and termination of mouse histone transcripts synthesized in vitro by RNA polymerase II. Nucleic Acids Res. 1996;24:3797–3805. doi: 10.1093/nar/24.19.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gunderson S I, Beyer K, Martin G, Keller W, Boelens W C, Mattaj L W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 178.Gunderson S I, Polycarpou-Schwarz M, Mattaj I W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 179.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 180.Guntaka R V. Transcription termination and polyadenylation in retroviruses. Microbiol Rev. 1993;57:511–521. doi: 10.1128/mr.57.3.511-521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guo Z, Russo P, Yun D-F, Buttler S, Sherman F. Redundant 3′ end forming signals for the yeast CYC1 mRNA. Proc Natl Acad Sci USA. 1995;92:4211–4214. doi: 10.1073/pnas.92.10.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Guo Z, Sherman F. 3′-end forming signals of yeast mRNA. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 183.Guo Z, Sherman F. 3′-end-forming signals of yeast mRNA. Mol Cell Biol. 1995;15:5983–5990. doi: 10.1128/mcb.15.11.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hahm B, Cho O H, Kim J E, Kim Y K, Kim J H, Oh Y L, Jang S K. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 1998;425:401–406. doi: 10.1016/s0014-5793(98)00269-5. [DOI] [PubMed] [Google Scholar]
- 185.Hammell, C., and C. Cole. 1999. Unpublished data.
- 186.Hampsey M H. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol Mol Biol Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hani J, Schelbert B, Bernhardt A, Domdey H, Fischer G, Wiebauer K, Rahfeld J U. Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3′-end formation of a pre-mRNA in Saccharomyces cerevisiae. J Biol Chem. 1999;274:108–116. doi: 10.1074/jbc.274.1.108. [DOI] [PubMed] [Google Scholar]
- 188.Hann L E, Cook W J, Uprichard S L, Knipe D M, Coen D M. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J Virol. 1998;72:7709–7714. doi: 10.1128/jvi.72.10.7709-7714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hanson R J, Sun J, Willis D G, Marzluff W F. Efficient extraction and partial purification of the polyribosome-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry. 1996;35:2146–2156. doi: 10.1021/bi9521856. [DOI] [PubMed] [Google Scholar]
- 190.Harris M E, Bohni R, Schneiderman M H, Ramamurthy L, Schumperli D, Marzluff W F. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hastings M L, Milcarek C, Martincic K, Peterson M L, Munroe S H. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–4300. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hauser H, Knippers R, Schafer K P. Increased rate of RNA-polyadenylation. An early response in concanavalin A activated lymphocytes. Exp Cell Res. 1978;111:175–184. doi: 10.1016/0014-4827(78)90247-1. [DOI] [PubMed] [Google Scholar]
- 193.Hedley M L, Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- 194.Hegemann J H, Fleig U N. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- 195.Heidmann S, Schindewolf C, Stumpf G, Domdey H. Flexibility and interchangeability of polyadenylation signals in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4633–4642. doi: 10.1128/mcb.14.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 197.Heintz N, Sive H L, Roeder R G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Henikoff S, Cohen E H. Sequences responsible for transcription termination on a gene segment in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1515–1520. doi: 10.1128/mcb.4.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Henikoff S, Kelly J D, Cohen E H. Transcription terminates in yeast distal to a control sequence. Cell. 1983;33:607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- 200.Henry, M. 1999. Unpublished data.
- 201.Henry M, Borland C, Bossie M, Silver P. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Henry M, Silver P. A novel methyltransferase (Hmt1p) modifies poly(A)+ RNA-binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Higgs D R, Goodburn S, Lamb J, Clegg J B, Weatherall D J, Proudfoot N J. α-Thalassemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- 204.Hirose Y, Manley J L. Creatine phosphate, not ATP, is required for 3′ end cleavage of mammalian pre-mRNA in vitro. J Biol Chem. 1997;272:29636–29642. doi: 10.1074/jbc.272.47.29636. [DOI] [PubMed] [Google Scholar]
- 205.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 206.Hoffmann I, Birnstiel M L. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature. 1990;346:665–668. doi: 10.1038/346665a0. [DOI] [PubMed] [Google Scholar]
- 207.Holm L, Sander C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- 208.Hook A G, Kellems R E. Localization and sequence analysis of poly(A) sites generating multiple dihydrofolate reductase mRNAs. J Biol Chem. 1988;263:2337–2343. [PubMed] [Google Scholar]
- 209.Horowitz D S, Kobayashi R, Krainer A R. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- 210.Hou W, Russnak R, Platt T. Poly(A) site selection in the yeast Ty retroelement requires an upstream region and sequence specific titratable factors in vitro. EMBO J. 1994;13:446–452. doi: 10.1002/j.1460-2075.1994.tb06279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Hsieh S Y, Yang P Y, Ou J T, Chu C M, Liaw Y F. Polyadenylation of the mRNA of hepatitis delta virus is dependent on the structure of the nascent RNA and regulated by the small or large delta antigen. Nucleic Acids Res. 1994;22:391–396. doi: 10.1093/nar/22.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Huang J, van der Ploeg L H. Maturation of polycistronic pre-mRNA in Trypanosoma brucei: analysis of trans splicing and poly(A) addition at nascent RNA transcripts from the hsp70 locus. Mol Cell Biol. 1991;11:3180–3190. doi: 10.1128/mcb.11.6.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huang M T, Gorman C M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang Y, Carmichael G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Huang Y, Carmichael G G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Huang Y, Wimler K M, Carmichael G G. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Huang Z M, Yen T S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Humphrey T, Birse C E, Proudfoot N J. RNA 3′ end signals of the S. pombe ura4 gene comprise a site determining and efficiency element. EMBO J. 1994;13:2441–2451. doi: 10.1002/j.1460-2075.1994.tb06529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hunt A G. Messenger RNA 3′ end formation in plants. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:47–60. [Google Scholar]
- 221.Hyman L E, Moore C L. Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol Cell Biol. 1993;13:5159–5167. doi: 10.1128/mcb.13.9.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hyman L E, Seiler S H, Whoriskey J, Moore C L. Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3′-end formation. Mol Cell Biol. 1991;11:2004–2012. doi: 10.1128/mcb.11.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Irniger S, Braus G H. Saturation mutagenesis of a polyadenylation signal reveals a hexanucleotide element essential for mRNA 3′ end formation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:257–261. doi: 10.1073/pnas.91.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Irniger S, Egli C M, Braus G H. Different classes of polyadenylation sites in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3060–3069. doi: 10.1128/mcb.11.6.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Irniger S, Sanfacon H, Egli C M, Braus G. Different sequence elements are required for function of the cauliflower mosaic virus polyadenylation site in Saccharomyces cerevisiae compared with plants. Mol Cell Biol. 1992;12:2322–2330. doi: 10.1128/mcb.12.5.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 227.Jenny A, Hauri H-P, Keller W. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol Cell Biol. 1994;14:8183–8190. doi: 10.1128/mcb.14.12.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jenny A, Keller W. Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res. 1995;23:2629–2635. doi: 10.1093/nar/23.14.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jenny A, Minvielle-Sebastia L, Preker P J, Keller W. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science. 1996;274:1514–1517. doi: 10.1126/science.274.5292.1514. [DOI] [PubMed] [Google Scholar]
- 230.Johnson E, Ma P, Ota I, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 231.Johnson E, Schwienhorst I, Dohmen R J, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff A M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kadowaki T, Schneiter R, Hitomi M, Tartakoff A M. Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1103–1110. doi: 10.1091/mbc.6.9.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kadowaki T, Zhao Y, Tartakoff A M. A conditional yeast mutant deficient in mRNA transport from nucleus to cytoplasm. Proc Natl Acad Sci USA. 1992;89:2312–2316. doi: 10.1073/pnas.89.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kamath A, Chakraburtty K. Protein synthesis in yeast. J Biol Chem. 1986;261:12593–12595. [PubMed] [Google Scholar]
- 236.Kane C M. Transcript elongation and gene regulation in eukaryotes. New York, N.Y: Raven Press; 1994. [Google Scholar]
- 237.Keller W, Bienroth S, Lang K M, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 1991;10:4241–4249. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′ end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 239.Kessler M, Henry M, Gross S, Shen E, Zhao J, Silver P, Moore C. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kessler, M., and C. Moore. Unpublished data.
- 241.Kessler M M, Zhao J, Moore C L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 242.Kessler M M, Zhelkovsky A M, Skvorak A, Moore C L. Monoclonal antibodies to yeast poly(A) polymerase (PAP) provide evidence for asociation of PAP with cleavage factor I. Biochemistry. 1995;34:1750–1759. doi: 10.1021/bi00005a032. [DOI] [PubMed] [Google Scholar]
- 243.Key S C, Yoshizaki T, Pagano J S. The Epstein-Barr virus (EBV) SM protein enhances pre-mRNA processing of the EBV DNA polymerase transcript. J Virol. 1998;72:8485–8492. doi: 10.1128/jvi.72.11.8485-8492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kiledjian M, DeMaria C T, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kirsh A L, Groudine M, Challoner P B. Polyadenylation and U7 snRNP-mediated cleavage: alternative modes of RNA 3′ processing in two avian histone H1 genes. Genes Dev. 1989;3:2172–2179. doi: 10.1101/gad.3.12b.2172. [DOI] [PubMed] [Google Scholar]
- 246.Klasens B I, Das A T, Berkhout B. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 1998;26:1870–1876. doi: 10.1093/nar/26.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Klasens B I F, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by localRNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Konig A, et al. Cell specific expression of a plant histone 2A. Plant Cell. 1991;3:656–657. doi: 10.1105/tpc.3.7.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Korner C G, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lacroute, F. Unpublished results cited in reference 201.
- 251.Lamb R, Krug R. Orthomyxoviridae: the viruses and their replication. In: Fields B N, editor. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1353–1395. [Google Scholar]
- 251a.Lang W H, Platt T, Reeder R H. Escherichia coli rho factor induces release of yeast RNA polymerase II but not polymerase I or III. Proc Natl Acad Sci USA. 1998;95:4900–4905. doi: 10.1073/pnas.95.9.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.LeBowitz J H, Smith H Q, Rusche L, Beverley S M. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 253.Lee M, Henry M, Silver P. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 254.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 255.LeMaire M F, Thummel C S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Levitt N, Briggs D, Gil A, Proudfoot N. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 257.Lewis E D, Manley J L. Polyadenylylation of an mRNA precursor occurs independently of transcription by RNA polymerase II in vivo. Proc Natl Acad Sci USA. 1986;83:8555–8559. doi: 10.1073/pnas.83.22.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 259.Li Q S, Gupta J D, Hunt A G. Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J Biol Chem. 1998;273:17539–17543. doi: 10.1074/jbc.273.28.17539. [DOI] [PubMed] [Google Scholar]
- 260.Liang S, Briggs M W, Butler J S. Regulation of tRNA suppressor activity by an intron-encoded polyadenylation signal. RNA. 1997;3:648–659. [PMC free article] [PubMed] [Google Scholar]
- 261.Liang S, Hitomi M, Hu Y H, Liu Y, Tartakoff A M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lin C L, Bristol L A, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein J D. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 263.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 264.Liu M, Xie Z, Price D H. A human RNA polymerase II transcription termination factor is a Swi2/Snf2 family member. J Biol Chem. 1998;273:25541–25544. doi: 10.1074/jbc.273.40.25541. [DOI] [PubMed] [Google Scholar]
- 265.Liu X, Mertz J. Sequence of the polypyrimidine tract of the 3′-terminal 3′-splicing signal can affect intron-dependent pre-mRNA processing in vivo. Nucleic Acids Res. 1996;24:1765–1773. doi: 10.1093/nar/24.9.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Liu X, Mertz J E. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 267.Lo H-J, Huang H K, Donahue T F. RNA polymerase I-promoted HIS4 expression yields uncapped, polyadenylated mRNA that is unstable and inefficiently translated in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:665–675. doi: 10.1128/mcb.18.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Logan J, Falck-Pedersen E, Darnell J E, Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci USA. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Long R M, Elliott D J, Stutz F, Rosbash M, Singer R H. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 270.Lou H, Gagel R F, Berget S M. An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev. 1996;10:208–219. doi: 10.1101/gad.10.2.208. [DOI] [PubMed] [Google Scholar]
- 271.Lou H, Helfman D M, Gagel R F, Berget S M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lou H, Neugebauer K M, Gagel R F, Berget S M. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18:4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lou H, Yang Y, Cote G J, Berget S M, Gagel R F. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol Cell Biol. 1995;15:7135–7142. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lowell J E, Rudner D Z, Sachs A B. 3′-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992;6:2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- 275.Luscher B, Schumperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lutz C, Cooke C, O’Connor J P, Kobayashi R, Alwine J. The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine tract binding protein associated splicing factor. RNA. 1999;4:1493–1499. doi: 10.1017/s1355838298981183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lutz C S, Alwine J C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 278.Lutz C S, Murthy K G, Schek N, O’Connor J P, Manley J L, Alwine J C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 279.Lutz-Freyermuth C, Query C C, Keene J D. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc Natl Acad Sci USA. 1990;87:6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.MacDonald C C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Magrath, C., and L. Hyman. A mutation in Grs1, a glycine tRNA synthetase, affects mRNA 3′ end formation in S. cerevisiae. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 282.Mandart E. Effects of mutations in the Saccharomyces cerevisiae RNA14 gene on the abundance and polyadenylation of its transcripts. Mol Gen Genet. 1998;258:16–25. doi: 10.1007/s004380050702. [DOI] [PubMed] [Google Scholar]
- 283.Mandart E, Dufour M, Lacroute F. Inactivation of SSM4, a new Saccharomyces cerevisiae gene, suppresses mRNA instability due to rna14 mutations. Mol Gen Genet. 1994;245:323–333. doi: 10.1007/BF00290112. [DOI] [PubMed] [Google Scholar]
- 284.Mandart E, Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol Cell Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Mangus D A, Amrani N, Jacobson A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Manley J L. Polyadenylation of mRNA precursors. Biochim Biophys Acta. 1988;950:1–12. doi: 10.1016/0167-4781(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 287.Mann K P, Weiss E A, Nevins J R. Alternative poly(A) site utilization during adenovirus infection coincides with a decrease in the activity of a poly(A) site-processing factor. Mol Cell Biol. 1993;13:2411–2419. doi: 10.1128/mcb.13.4.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Mannironi C, Bonner W M, Hatch C L. H2A.X, a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3′ processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Maraia R, Kenan D, Keene J. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Martin F, Schaller A, Eglite S, Schumperli D, Muller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 292.Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze M T, Milcarek C. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc Natl Acad Sci USA. 1998;95:11095–11100. doi: 10.1073/pnas.95.19.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Marzluff W F. Histone 3′ ends: essential and regulatory functions. Gene Expression. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 294.Mason P J, Elkington J A, Lloyd M M, Jones M B, Williams J G. Mutations downstream of the polyadenylation site of a Xenopus beta-globin mRNA affect the position but not the efficiency of 3′ processing. Cell. 1986;46:263–270. doi: 10.1016/0092-8674(86)90743-9. [DOI] [PubMed] [Google Scholar]
- 295.Matis S A, Martincic K, Milcarek C. B-lineage regulated polyadenylation occurs on weak poly(A) sites regardless of sequence composition at the cleavage and downstream regions. Nucleic Acids Res. 1996;24:4684–4692. doi: 10.1093/nar/24.23.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Matthews K R, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 297.Mayer S A, Dieckmann C L. The yeast CBP1 gene produces two differentially regulated transcripts by alternative 3′-end formation. Mol Cell Biol. 1989;9:4161–4169. doi: 10.1128/mcb.9.10.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Mayer S A, Dieckmann C L. Yeast CBP1 mRNA 3′ end formation is regulated during the induction of mitochondrial function. Mol Cell Biol. 1991;11:813–821. doi: 10.1128/mcb.11.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 300.McDevitt M A, Hart R P, Wong W W, Nevins J R. Sequence capable of restoring poly(A) site function define two distinct downstream element. EMBO J. 1986;5:2907–2931. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly (A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.McLauchlan J, Gaffney D, Whitton J L, Clements J B. The consensus sequence YGTGTTYY located downstream from the AAUAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.McLean M R, Rymond B C. Yeast pre-mRNA splicing requires a pair of U1 snRNP-associated tetratricopeptide repeat proteins. Mol Cell Biol. 1998;18:353–360. doi: 10.1128/mcb.18.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303a.McNeil J B, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Mehlin H, Daneholt B, Skoglund U. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell. 1992;69:605–613. doi: 10.1016/0092-8674(92)90224-z. [DOI] [PubMed] [Google Scholar]
- 305.Meijer D, Hermans A, von Lindern M, van Agthoven T, de Klein A, Mackenbach P, Grootegoed A, Talarico D, Della Valle G, Grosveld G. Molecular characterization of the testis specific c-abl mRNA in mouse. EMBO J. 1987;6:4041–4048. doi: 10.1002/j.1460-2075.1987.tb02749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Mian I S. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Min H, Chan R C, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 308.Minvielle-Sebastia L, Beyer K, Krecic A M, Hector R E, Swanson M S, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Minvielle-Sebastia, L., and W. Keller. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol., in press. [DOI] [PubMed]
- 310.Minvielle-Sebastia L, Preker P, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Natl Acad Sci USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Minvielle-Sebastia L, Preker P J, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 312.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate: sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Mizrahi, N., and C. Moore. 1999. Unpublished data.
- 314.Moore C, Chen J, Whoriksey J. Two proteins crosslinked to RNA containing the adenovirus L3 polyadenylation site require the AAUAAA sequence for binding. EMBO J. 1988;7:3159–3169. doi: 10.1002/j.1460-2075.1988.tb03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Moore C, Sharp P. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell. 1985;41:845–855. doi: 10.1016/s0092-8674(85)80065-9. [DOI] [PubMed] [Google Scholar]
- 316.Moore C, Sharp P. Site-specific polyadenylation in a cell-free system. Cell. 1984;36:581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- 317.Moore C, Skolnik-David H, Sharp P. Analysis of RNA cleavage at the adenovirus-2 L3 polyadenylation site. EMBO J. 1986;5:1929–1938. doi: 10.1002/j.1460-2075.1986.tb04446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Moore M S. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- 319.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Moreira A, Wollerton J, Monks J, Proudfoot N J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Moser M J, Holley W R, Chatterjee A, Mian I S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mowry K L, Oh R, Steitz J A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3′-end processing reaction. Mol Cell Biol. 1989;9:3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Mowry K L, Steitz J A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 324.Mowry K L, Steitz J A. snRNP mediators of 3′ end processing: functional fossils? Trends Biochem Sci. 1988;13:447–451. doi: 10.1016/0968-0004(88)90220-4. [DOI] [PubMed] [Google Scholar]
- 325.Muller B, Schumperli D. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin Cell Dev Biol. 1997;8:567–576. doi: 10.1006/scdb.1997.0182. [DOI] [PubMed] [Google Scholar]
- 326.Mulligan G J, Guo W, Wormsley S, Helfman D M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 327.Murphy R, Wente S R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 328.Murthy K G, Manley J L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 329.Murthy K G K, Manley J L. The 160 kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′ end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 330.Nemeroff M E, Barabino S M, Li Y, Keller W, Krug R M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 331.Nemeth A, Krause S, Blank D, Jenny A, Jeno P, Lusting A, Wahle E. Isolation of genomic and cDNA clones encoding bovine poly(A) binding protein II. Nucleic Acids Res. 1995;23:4034–4041. doi: 10.1093/nar/23.20.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Nesic D, Cheng J, Maquat L E. Sequences within the last intron function in RNA 3′-end formation in cultured cells. Mol Cell Biol. 1993;13:3359–3369. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Nesic D, Maquat L E. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev. 1994;8:363–375. doi: 10.1101/gad.8.3.363. [DOI] [PubMed] [Google Scholar]
- 334.Nesic D, Zhang J, Maquat L E. Lack of an effect of the efficiency of RNA 3′-end formation on the efficiency of removal of either the final or the penultimate intron in intact cells. Mol Cell Biol. 1995;15:488–496. doi: 10.1128/mcb.15.1.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Neugebauer K, Roth M. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 336.Nevins J R, Darnell J E. Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978;15:1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- 337.Niwa M, Berget S M. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 338.Niwa M, Berget S M. Polyadenylation precedes splicing in vitro. Gene Expression. 1991;1:5–14. [PMC free article] [PubMed] [Google Scholar]
- 339.Niwa M, MacDonald C C, Berget S M. Are vertebrate exons scanned during splice-site selection? Nature. 1992;360:277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- 340.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 341.Norton P A. Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res. 1994;22:3854–3860. doi: 10.1093/nar/22.19.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.O’Conner J, Peebles C. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992;12:3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.O’Connor J P, Alwine J C, Lutz C S. Identification of a novel, non-snRNP protein complex containing U1A protein. RNA. 1997;3:1444–1455. [PMC free article] [PubMed] [Google Scholar]
- 344.Oh Y L, Hahm B, Kim Y K, Lee H K, Lee J W, Song O, Tsukiyama-Kohara K, Kohara M, Nomoto A, Jang S K. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem J. 1998;331:169–175. doi: 10.1042/bj3310169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Orkin S H, Cheng T, Antonarakis E, Kazazian H H. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human β-globin gene. EMBO J. 1985;4:453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345a.Osheim Y N, Proudfoot N J, Beyer A L. EM visualization of transcription by RNA polymerase II: downstream termination requires a poly(A) signal but not transcript cleavage. Mol Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- 346.Osley M A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 347.Oyen O, Myklebust F, Scott J D, Cadd G G, McKnight G S, Hansson V, Jahnsen T. Subunits of cyclic adenosine 3′,5′-monophosphate-dependent protein kinase show differential and distinct expression patterns during germ cell differentiation: alternative polyadenylation in germ cells gives rise to unique smaller-sized mRNA species. Biol Reprod. 1990;43:46–54. doi: 10.1095/biolreprod43.1.46. [DOI] [PubMed] [Google Scholar]
- 348.Pandey N B, Chodchoy N, Liu T J, Marzluff W F. Introns in histone genes alter the distribution of 3′ ends. Nucleic Acids Res. 1990;18:3161–3170. doi: 10.1093/nar/18.11.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Pandey N B, Williams A S, Sun J H, Brown V D, Bond U, Marzluff W F. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol Cell Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Pante N, Jarmolowski A, Izaurralde E, Sauder U, Baschong W, Mattaj I W. Visualizing nuclear export of different classes of RNA by electron microscopy. RNA. 1997;3:498–513. [PMC free article] [PubMed] [Google Scholar]
- 351.Patel D, Butler J S. A conditional defect in mRNA 3′-end processing caused by a mutation in the gene for poly(A) polymerase. Mol Cell Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Patterson B, Guthrie C. An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell. 1987;49:613–624. doi: 10.1016/0092-8674(87)90537-x. [DOI] [PubMed] [Google Scholar]
- 353.Pazin M, Kadonaga J. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;40:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 354.Peltz, S. 1999. Unpublished data.
- 355.Perez I, Lin C H, McAfee J G, Patton J G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 356.Peterson J A, Myers A M. Functional analysis of mRNA 3′ end formation signals in the convergent and overlapping transcription units of the S. cerevisiae genes RHO1 and MRP2. Nucleic Acids Res. 1993;21:5500–5508. doi: 10.1093/nar/21.23.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Peterson M L. Balanced efficiencies of splicing and cleavage-polyadenylation are required for mu-s and mu-m mRNA regulation. Gene Expression. 1992;2:319–327. [PMC free article] [PubMed] [Google Scholar]
- 358.Peterson M L. Regulated immunoglobulin (Ig) RNA processing does not require specific cis-acting sequences: non-Ig RNA can be alternatively processed in B cells and plasma cells. Mol Cell Biol. 1994;14:7891–7898. doi: 10.1128/mcb.14.12.7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Peterson M L, Perry R P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Pettijean A, Bonneaud N, Lacroute F. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eukaryotic homolog of the eubacterial and archaebacterial L1 ribosomal proteins. Mol Cell Biol. 1995;15:5071–5081. doi: 10.1128/mcb.15.9.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Phillips C, Kyriakopoulou C B, Virtanen A. Identification of a stem-loop structure important for polyadenylation at the murine IgM secretory poly(A) site. Nucleic Acids Res. 1999;27:429–438. doi: 10.1093/nar/27.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Phillips C, Schimpl A, Dietrich-Goetz W, Clements J B, Virtanen A. Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur J Immunol. 1996;26:3144–3152. doi: 10.1002/eji.1830261247. [DOI] [PubMed] [Google Scholar]
- 363.Phillips C, Virtanen A. The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res. 1997;25:2344–2351. doi: 10.1093/nar/25.12.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 365.Piper P W, Aamand J L. Yeast mutation thought to arrest mRNA transport markedly increased the length of the 3′ poly(A) on polyadenylated RNA. J Mol Biol. 1989;208:697–700. doi: 10.1016/0022-2836(89)90159-9. [DOI] [PubMed] [Google Scholar]
- 366.Pironcheva G, Russev G. 3′ processing of histone H4 precursor mRNA requires the presence of a small nuclear RNP particle. Int J Biochem. 1991;23:1043–1047. doi: 10.1016/0020-711x(91)90143-b. [DOI] [PubMed] [Google Scholar]
- 367.Platt T. Rho and RNA: models for recognition and response. Mol Microbiol. 1994;11:983–990. doi: 10.1111/j.1365-2958.1994.tb00376.x. [DOI] [PubMed] [Google Scholar]
- 368.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 369.Preker P, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Preker P J, Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- 371.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 372.Prescott J, Falck-Pedersen E. Sequence elements upstream of the 3′ cleavage site confer substrate strength to the adenovirus L1 and L3 polyadenylation sites. Mol Cell Biol. 1994;14:4682–4693. doi: 10.1128/mcb.14.7.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Prescott J C, Falck-Pedersen E. Varied poly(A) site efficiency in the adenovirus major late transcription unit. J Biol Chem. 1992;267:8175–8181. [PubMed] [Google Scholar]
- 374.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 375.Proudfoot N J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- 376.Proudfoot N J. Transcriptional interference and termination between duplicated alpha-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 377.Proudfoot N J, Brownlee G G. 3′ non coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 378.Puerta C, Martin J, Alonso C, Lopez M C. Isolation and characterization of the gene encoding histone H2A from Trypanosoma cruzi. Mol Biochem Parasitol. 1994;64:1–10. doi: 10.1016/0166-6851(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 379.Query C C, Bentley R C, Keene J D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 380.Raabe T, Bollum F J, Manley J L. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- 381.Raju V S, Jacob S T. Association of poly(A) polymerase with U1 RNA. J Biol Chem. 1988;263:11067–11070. [PubMed] [Google Scholar]
- 382.Rao M N, Chernokalskaya E, Schoenberg D R. Regulated nuclear polyadenylation of Xenopus albumin pre-mRNA. Nucleic Acids Res. 1996;24:4078–4083. doi: 10.1093/nar/24.20.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Reeder R H, Lang W H. Terminating transcription in eukaryotes: lessons learned from RNA polymerase I. Trends Biochem Sci. 1997;22:473–477. doi: 10.1016/s0968-0004(97)01133-x. [DOI] [PubMed] [Google Scholar]
- 384.Richardson J P. Transcription termination. Crit Rev Biochem Mol Biol. 1993;28:1–30. doi: 10.3109/10409239309082571. [DOI] [PubMed] [Google Scholar]
- 384a.Richter J. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Roberts G C, Gooding C, Mak H Y, Proudfoot N J, Smith C W J. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Roesser J R, Liittschwager K, Leff S E. Regulation of tissue-specific splicing of the calcitonin/calcitonin gene-related peptide gene by RNA-binding proteins. J Biol Chem. 1993;268:8366–8375. [PubMed] [Google Scholar]
- 387.Rothnie H M. Plant mRNA 3′-end formation. Plant Mol Biol. 1996;32:43–61. doi: 10.1007/BF00039376. [DOI] [PubMed] [Google Scholar]
- 388.Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 389.Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to splicesomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 390.Russell I, Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Russell I, Tollervey D. Yeast Nop3p has structural and functional similarities to mammalian pre-mRNA binding proteins. Eur J Cell Biol. 1995;66:293–301. [PubMed] [Google Scholar]
- 392.Russnak R, Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–767. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 393.Russnak R, Nehrke K W, Platt T. REF2 encodes an RNA binding protein directly involved in yeast mRNA 3′ end formation. Mol Cell Biol. 1994;15:1689–1697. doi: 10.1128/mcb.15.3.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Russnak R, Pereira S, Platt T. RNA binding analysis of yeast Ref2 and its two-hybrid interaction with a new gene product Fir1. Gene Expression. 1997;6:241–258. [PMC free article] [PubMed] [Google Scholar]
- 395.Russo P, Li W-Z, Hampsey D M, Zaret K S, Sherman F. Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991;10:563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Ryu W S, Mertz J E. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Sabate M I, Stolarsky L S, Polak J M, Bloom S R, Varndell I M, Ghatei M A, Evans R M, Rosenfeld M G. Regulation of neuroendocrine gene expression by alternative RNA processing. Colocalization of calcitonin and calcitonin gene-related peptide in thyroid C-cells. J Biol Chem. 1985;260:2589–2592. [PubMed] [Google Scholar]
- 398.Sachs A, Bond M, Kornberg R. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 399.Sachs A, Sarnow P, Hentze M. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 400.Sachs A B, Deardorff J A. Tranlation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 401.Sadhale P P, Platt T. Unusual aspects of in vitro RNA processing in the 3′ regions of the GAL1, GAL7, and GAL10 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4262–4270. doi: 10.1128/mcb.12.10.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Salinas C A, Sinclair D A, O’Hare K, Brock H W. Characterization of a Drosophila homologue of the 160-kDa subunit of the cleavage and polyadenylation specificity factor CPSF. Mol Gen Genet. 1998;257:672–680. doi: 10.1007/s004380050696. [DOI] [PubMed] [Google Scholar]
- 403.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Sandri-Goldin R M. Properties of an HSV-1 regulatory protein that appears to impair host cell splicing. Infect Agents Dis. 1994;3:59–67. [PubMed] [Google Scholar]
- 405.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu Rev Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- 407.Scharl E C, Steitz J A. Length suppression in histone messenger RNA 3′-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc Natl Acad Sci USA. 1996;93:14659–14664. doi: 10.1073/pnas.93.25.14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Scharl E C, Steitz J A. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Schaufele F, Gilmartin G M, Bannwarth W, Birnstiel M L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- 410.Schek N, Cooke C, Alwine J C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Schneiter R, Hitomi M, Ivessa A S, Fasch E V, Kohlwein S D, Tartakoff A M. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Schul W, van Driel R, de Jong L. A subset of poly(A) polymerase is concentrated at sites of RNA synthesis and is associated with domains enriched in splicing factors and poly(A) RNA. Exp Cell Res. 1998;238:1–12. doi: 10.1006/excr.1997.3808. [DOI] [PubMed] [Google Scholar]
- 413.Schumperli D. Multilevel regulation of replication-dependent histone genes. Trends Genet. 1988;4:187–191. doi: 10.1016/0168-9525(88)90074-1. [DOI] [PubMed] [Google Scholar]
- 414.Schurch N, Hehl A, Vassella E, Braun R, Roditi I. Accurate polyadenylation of procyclin mRNAs in Trypanosoma brucei is determined by pyrimidine-rich elements in the intergenic regions. Mol Cell Biol. 1994;14:3668–3675. doi: 10.1128/mcb.14.6.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Scott J M, Imperiale M J. Promoter-proximal poly(A) sites are processed efficiently, but the RNA products are unstable in the nucleus. Mol Cell Biol. 1997;17:2127–2135. doi: 10.1128/mcb.17.4.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Scott J M, Imperiale M J. Reciprocal effects of splicing and polyadenylation on human immunodeficiency virus type 1 pre-mRNA processing. Virology. 1996;224:498–509. doi: 10.1006/viro.1996.0556. [DOI] [PubMed] [Google Scholar]
- 417.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Seipelt R L, Spear B T, Snow E C, Peterson M L. A nonimmunoglobulin transgene and the endogenous immunoglobulin mu gene are coordinately regulated by alternative RNA processing during B-cell maturation. Mol Cell Biol. 1998;18:1042–1048. doi: 10.1128/mcb.18.2.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Seraphin B, Abovich N, Rosbash M. Genetic depletion indicates a late role for U5 snRNP during in vitro spliceosome assembly. Nucleic Acids Res. 1991;19:3857–3860. doi: 10.1093/nar/19.14.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Setyono B, Greenburg J R. Proteins associated with poly(A) and other regions of mRNA and hnRNA molecules as investigated by crosslinking. Cell. 1981;24:775–783. doi: 10.1016/0092-8674(81)90103-3. [DOI] [PubMed] [Google Scholar]
- 421.Setzer D R, Mcgrogan M, Nunberg J H, Schimke R T. Size heterogeneity in the 3′ end of dihydrofolate reductase messenger RNAs in mouse cells. Cell. 1980;22:361–370. doi: 10.1016/0092-8674(80)90346-3. [DOI] [PubMed] [Google Scholar]
- 422.Sheets M D, Ogg S C, Wickens M P. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Shen E, Henry N, Weiss V, Valentini S, Silver P, Lee M. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 425.Shilatifard A. The RNA polymerase II general elongation complex. J Biol Chem. 1998;379:27–31. doi: 10.1515/bchm.1998.379.1.27. [DOI] [PubMed] [Google Scholar]
- 426.Shimizu K, Iguchi A, Gomyou R, Ono Y. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology. 1999;254:213–219. doi: 10.1006/viro.1998.9555. [DOI] [PubMed] [Google Scholar]
- 427.Silver Key S C, Pagano J S. A noncanonical poly(A) signal, UAUAAA, and flanking elements in Epstein-Barr virus DNA polymerase mRNA function in cleavage and polyadenylation assays. Virology. 1997;234:147–159. doi: 10.1006/viro.1997.8647. [DOI] [PubMed] [Google Scholar]
- 428.Singleton D, Shoping C, Hitomi C, Kumagai C, Tartakoff A. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- 429.Sisodia S S, Sollner-Webb B, Cleveland D W. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Sittler A, Gallinaro H, Jacob M. The secondary structure of the adenovirus-2 L4 polyadenylation domain: evidence for a hairpin structure exposing the AAUAAA signal in its loop. J Mol Biol. 1995;248:525–540. doi: 10.1006/jmbi.1995.0240. [DOI] [PubMed] [Google Scholar]
- 431.Sittler A, Gallinaro H, Jacob M. Upstream and downstream cis-acting elements for cleavage at the L4 polyadenylation site of adenovirus-2. Nucleic Acids Res. 1994;22:222–231. doi: 10.1093/nar/22.2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Smale S T, Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol Cell Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Smith H O, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel M L. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci USA. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Sparks K A, Dieckmann C L. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Spycher C, Streit A, Stefanovic B, Albrecht D, Koning T H, Schumperli D. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 437.Steinmetz E. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 438.Steinmetz E J, Brow D A. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci USA. 1998;95:6699–6704. doi: 10.1073/pnas.95.12.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Steinmetz E J, Brow D A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Stolow D T, Berget S M. UV cross-linking of polypeptides associated with 3′-terminal exons. Mol Cell Biol. 1990;10:5937–5944. doi: 10.1128/mcb.10.11.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Streit A, Koning T W, Soldati D, Melin L, Schumperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Stumpf G, Domdey H. Dependence of yeast pre-mRNA 3′-end processing on Cft1: a sequence homolog of the mammalian AAUAAA binding factor. Science. 1996;274:1517–1520. doi: 10.1126/science.274.5292.1517. [DOI] [PubMed] [Google Scholar]
- 443.Stumpf G, Goppelt A, Domdey H. Pre-mRNA topology is important for 3′-end formation in Saccharomyces cerevisiae and mammals. Mol Cell Biol. 1996;16:2204–2213. doi: 10.1128/mcb.16.5.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 445.Sundas A, Tandre K, Kvarnheden A, Engstrom P. cDNA sequence and expression of an intron-containing histone H2A gene from Norway spruce, Picea abies. Plant Mol Biol. 1993;21:595–605. doi: 10.1007/BF00014543. [DOI] [PubMed] [Google Scholar]
- 446.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Takagaki Y, MacDonald C, Shenk T, Manley J. The human 64-kDa polyadenylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc Natl Acad Sci USA. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Takagaki Y, Manley J. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 449.Takagaki Y, Manley J. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Takagaki Y, Manley J L. A human polyadenylation factor is a G protein β-subunit homologue. J Biol Chem. 1992;267:23471–23474. [PubMed] [Google Scholar]
- 451.Takagaki Y, Manley J L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 452.Takagaki Y, Manley J L, MacDonald C C, Wilusz J, Shenk T. A multicomponent complex is required for the AAUAAA-dependent crosslinking of a 64-kilodalton protein to polyadenylation substrates. Genes Dev. 1990;4:2112–2120. doi: 10.1128/mcb.10.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Takagaki Y, Ryner L C, Manley J L. Four factors are required for 3′ end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–1724. doi: 10.1101/gad.3.11.1711. [DOI] [PubMed] [Google Scholar]
- 454.Takagaki Y, Ryner L C, Manley J L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA processing and polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 455.Takagaki Y, Seipelt R L, Peterson M L, Manley J L. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 456.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 457.Tarun S Z, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 458.Tarun S Z, Wells S E, Deardoff J A, Sachs A B. Translation initiation factor eIF-4G mediates in vivo poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Teigelkamp S, Achsel T, Mundt C, Gothel S F, Cronshagen U, Lane W S, Marahiel M, Luhrmann R. The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA. 1998;4:127–141. [PMC free article] [PubMed] [Google Scholar]
- 460.Thuresson A-C, Aetrom J, Astrom A, Gronvik K-O, Virtanen A. Multiple forms of poly(A) polymerases in human cells. Proc Natl Acad Sci USA. 1994;91:979–983. doi: 10.1073/pnas.91.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Ullu E, Tschudi C. 2′-O-methyl RNA oligonucleotides identify two functional elements in the trypanosome spliced leader ribonucleoprotein particle. J Biol Chem. 1993;268:13068–13073. [PubMed] [Google Scholar]
- 462.Uptain S M, Kane C M, Chamberlin M J. Mechanisms of transcript elongation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 463.Valcarcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 464.Valentini S, Weiss V, Silver P. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA. 1999;5:272–280. doi: 10.1017/s1355838299981633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Valsamakis A, Zeichner S, Carswell S, Alwine J C. The human immunodeficiency virus type 1 polyadenylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylation. Proc Natl Acad Sci USA. 1991;88:2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.van Oers C C, Adema G J, Zandberg H, Moen T C, Baas P D. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol Cell Biol. 1994;14:951–960. doi: 10.1128/mcb.14.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.van Oers C C, Bakker L, Baas P D. The exon 4 poly(A) site of the human calcitonin/CGRP-I pre-mRNA is a weak site in vitro. Biochim Biophys Acta. 1994;1218:55–63. doi: 10.1016/0167-4781(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 469.Vasserot A P, Schaufele F J, Birnstiel M L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci USA. 1989;86:4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 471.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 472.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 473.Wahle E, Kuhn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog Nucleic Acid Res Mol Biol. 1997;57:41–70. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- 474.Wahle E, Lusting A, Jeno P, Maurer P. Mammalian poly(A)-binding protein II. J Biol Chem. 1993;268:2937–2945. [PubMed] [Google Scholar]
- 475.Wahle E, Martin G, Schiltz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475a.Wahle, E., and U. Rüegsegger. 3′ end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., in press. [DOI] [PubMed]
- 476.Wallace, A., B. Dass, S. Ravnik, V. Tonk, N. Jenkins, D. Gilbert, N. Copelant, and C. MacDonald. Two distinct forms of the 64,000 Mr protein of the Cleavage Stimulation Factor are expressed in mouse male germ cells. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 477.Walther T N, Wittop Koning T H, Schumperli D, Muller B. A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA. 1998;4:1034–1046. doi: 10.1017/s1355838298971771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Wang Z F, Ingledue T C, Dominski Z, Sanchez R, Marzluff W F. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol. 1999;19:835–845. doi: 10.1128/mcb.19.1.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Wang Z F, Sirotkin A M, Buchold G M, Skoultchi A I, Marzluff W F. The mouse histone H1 genes: gene organization and differential regulation. J Mol Biol. 1997;271:124–138. doi: 10.1006/jmbi.1997.1166. [DOI] [PubMed] [Google Scholar]
- 480.Wang Z F, Tisovec R, Debry R W, Frey M R, Matera A G, Marzluff W F. Characterization of the 55-kb mouse histone gene cluster on chromosome 3. Genome Res. 1996;6:702–714. doi: 10.1101/gr.6.8.702. [DOI] [PubMed] [Google Scholar]
- 481.Wang Z F, Whitfield M L, Ingledue T C, 3rd, Dominski Z, Marzluff W F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 482.Wassarman K M, Steitz J A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- 483.Weber J, Blanchard J M, Ginsberg H, Darnell J E., Jr Order of polyadenylic acid addition and splicing events in early adenovirus mRNA formation. J Virol. 1980;33:286–291. doi: 10.1128/jvi.33.1.286-291.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Weichs an der Glon C, Ashe M, Eggermont J, Proudfoot N J. Tat-dependent occlusion of the HIV poly(A) site. EMBO J. 1993;12:2119–2128. doi: 10.1002/j.1460-2075.1993.tb05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Weiss E A, Gilmartin G M, Nevins J R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Wells D, Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci USA. 1985;82:2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 488.Wickens M, Anderson P, Jackson R J. Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 489.Wickens M, Stephenson P. Role of the conserved AAUAAA sequence: four point mutants prevent messenger RNA 3′ end formation. Science. 1984;226:1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- 490.Wiederkehr T, Pretot R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Wigley P L, Sheets M D, Zarkower D A, Whitmer M E, Wickens M. Polyadenylation of mRNA: minimal substrates and a requirement for the 2′ hydroxyl of the U in AAUAAA. Mol Cell Biol. 1990;10:1705–1713. doi: 10.1128/mcb.10.4.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Will C L, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 493.Williams A S, Ingledue T C, 3rd, Kay B K, Marzluff W F. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Wilson S, Datar K, Paddy M, Swedlow J, Swanson M. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Wilusz J, Feig D I, Shenk T. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol Cell Biol. 1988;8:4477–4483. doi: 10.1128/mcb.8.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Wilusz J, Pettine S, Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989;17:3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Wilusz J, Shenk T. A 64kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- 498.Wilusz J, Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990;10:6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Wilusz J, Shenk T, Takagaki Y, Manley J. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol Cell Biol. 1990;10:1244–1248. doi: 10.1128/mcb.10.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Wu S-Y, Platt T. Transcriptional arrest of yeast RNA polymerase II by Escherichia coli Rho protein in vitro. Proc Natl Acad Sci USA. 1993;90:6606–6610. doi: 10.1073/pnas.90.14.6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Xie Z, Price D H. Drosophila factor 2, an RNA polymerase II transcript release factor, has DNA-dependent ATPase activity. J Biol Chem. 1997;272:31902–31907. doi: 10.1074/jbc.272.50.31902. [DOI] [PubMed] [Google Scholar]
- 502.Xie Z, Price D H. Unusual nucleic acid binding properties of factor 2, an RNA polymerase II transcript release factor. J Biol Chem. 1998;273:3771–3777. doi: 10.1074/jbc.273.6.3771. [DOI] [PubMed] [Google Scholar]
- 503.Xu H X, Johnson L, Grunstein M. Coding and noncoding sequences at the 3′ end of yeast histone H2B mRNA confer cell cycle regulation. Mol Cell Biol. 1990;10:2687–2694. doi: 10.1128/mcb.10.6.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Yan D H, Weiss E A, Nevins J R. Identification of an activity in B-cell extracts that selectively impairs the formation of an immunoglobulin mu s poly(A) site processing complex. Mol Cell Biol. 1995;15:1901–1906. doi: 10.1128/mcb.15.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Yeakley J M, Hedjran F, Morfin J P, Merillat N, Rosenfeld M G, Emeson R B. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol Cell Biol. 1993;13:5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Yoshimatsu T, Nagawa F. Effect of artificially inserted intron on gene expression in Saccharomyces cerevisiae. DNA Cell Biol. 1994;13:51–58. doi: 10.1089/dna.1994.13.51. [DOI] [PubMed] [Google Scholar]
- 507.Yue D, Maizels N, Weiner A M. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archael hyperthermophile Sulfolobus shibatae. RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- 508.Yue X, Connolly T, Futcher B, Beach D. Human D-type cyclin. Cell. 1991;65:691–699. doi: 10.1016/0092-8674(91)90100-d. [DOI] [PubMed] [Google Scholar]
- 509.Zandberg H, Moen T C, Baas P D. Cooperation of 5′ and 3′ processing sites as well as intron and exon sequences in calcitonin exon recognition. Nucleic Acids Res. 1995;23:248–255. [PMC free article] [PubMed] [Google Scholar]
- 510.Zarkower D, Wickens M. A functionally redundant downstream sequence in SV40 late pre-mRNA is required for mRNA 3′ end formation and for assembly of a precleavage complex in vitro. J Biol Chem. 1988;263:5780–5788. [PubMed] [Google Scholar]
- 511.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Zhao J, Kessler M, Moore C. Cleavage factor II of S. cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J Biol Chem. 1997;272:10831–10838. doi: 10.1074/jbc.272.16.10831. [DOI] [PubMed] [Google Scholar]
- 513.Zhao, J., M. M. Kessler, and C. L. Moore. Unpublished data.
- 514.Zhao, J., and C. Moore. Unpublished data.
- 515.Zhao W, Manley J L. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Zhao W, Manley J L. Deregulation of poly(A) polymerase interferes with cell growth. Mol Cell Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Zhelkovsky A, Helmling S, Moore C. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol Cell Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Zhelkovsky A M, Kessler M M, Moore C L. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase. J Biol Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]