Abstract
The field covered in this review is new; the first sequence of a gene encoding the molecular chaperone Hsp70 and the first description of a chaperonin in the archaea were reported in 1991. These findings boosted research in other areas beyond the archaea that were directly relevant to bacteria and eukaryotes, for example, stress gene regulation, the structure-function relationship of the chaperonin complex, protein-based molecular phylogeny of organisms and eukaryotic-cell organelles, molecular biology and biochemistry of life in extreme environments, and stress tolerance at the cellular and molecular levels. In the last 8 years, archaeal stress genes and proteins belonging to the families Hsp70, Hsp60 (chaperonins), Hsp40(DnaJ), and small heat-shock proteins (sHsp) have been studied. The hsp70(dnaK), hsp40(dnaJ), and grpE genes (the chaperone machine) have been sequenced in seven, four, and two species, respectively, but their expression has been examined in detail only in the mesophilic methanogen Methanosarcina mazei S-6. The proteins possess markers typical of bacterial homologs but none of the signatures distinctive of eukaryotes. In contrast, gene expression and transcription initiation signals and factors are of the eucaryal type, which suggests a hybrid archaeal-bacterial complexion for the Hsp70 system. Another remarkable feature is that several archaeal species in different phylogenetic branches do not have the gene hsp70(dnaK), an evolutionary puzzle that raises the important question of what replaces the product of this gene, Hsp70(DnaK), in protein biogenesis and refolding and for stress resistance. Although archaea are prokaryotes like bacteria, their Hsp60 (chaperonin) family is of type (group) II, similar to that of the eukaryotic cytosol; however, unlike the latter, which has several different members, the archaeal chaperonin system usually includes only two (in some species one and in others possibly three) related subunits of ∼60 kDa. These form, in various combinations depending on the species, a large structure or chaperonin complex sometimes called the thermosome. This multimolecular assembly is similar to the bacterial chaperonin complex GroEL/S, but it is made of only the large, double-ring oligomers each with eight (or nine) subunits instead of seven as in the bacterial complex. Like Hsp70(DnaK), the archaeal chaperonin subunits are remarkable for their evolution, but for a different reason. Ubiquitous among archaea, the chaperonins show a pattern of recurrent gene duplication—hetero-oligomeric chaperonin complexes appear to have evolved several times independently. The stress response and stress tolerance in the archaea involve chaperones, chaperonins, other heat shock (stress) proteins including sHsp, thermoprotectants, the proteasome, as yet incompletely understood thermoresistant features of many molecules, and formation of multicellular structures. The latter structures include single- and mixed-species (bacterial-archaeal) types. Many questions remain unanswered, and the field offers extraordinary opportunities owing to the diversity, genetic makeup, and phylogenetic position of archaea and the variety of ecosystems they inhabit. Specific aspects that deserve investigation are elucidation of the mechanism of action of the chaperonin complex at different temperatures, identification of the partners and substitutes for the Hsp70 chaperone machine, analysis of protein folding and refolding in hyperthermophiles, and determination of the molecular mechanisms involved in stress gene regulation in archaeal species that thrive under widely different conditions (temperature, pH, osmolarity, and barometric pressure). These studies are now possible with uni- and multicellular archaeal models and are relevant to various areas of basic and applied research, including exploration and conquest of ecosystems inhospitable to humans and many mammals and plants.
The purpose of this review is to examine the information available on archaeal stress genes and proteins, particularly those of the Hsp70 and Hsp60 families, while critically discussing the data in comparison with what is known for the bacterial and eukaryotic equivalents. The aim was to treat the specific topics of the review embedded in the framework of closely related areas of science. Cross-fertilization between research with archaea and research with bacteria and eukaryotes is highlighted to show how the study of archaea has contributed, and will continue to contribute, to other fields, both basic and applied.
A deliberate effort has been made to simplify the text and make it readable to a general audience. Consequently, terms are explained and the data and theories are presented within a historical perspective. A minimal amount of overlapping between related sections distant from one another in the body of the review is included to enhance the flow, particularly when a later section expands on an earlier one.
A comprehensive search of printed literature and databases was attempted. Colleagues were consulted. The majority of the data are displayed in tables and figures, but only illustrative cases are explained in the text. Reviews rather than original reports are cited for topics related to but not strictly dealing with archaeal genes, proteins, or organisms, to reduce the number of references and save space while providing access to a wealth of published information.
Archaea have been found in a wide variety of ecosystems with very different characteristics, very hot or very cold, temperate, anoxic, oxygenated, etc. (9, 12, 50, 59, 178, 297). Thus, what represents a stressor for a species may be a condition required for the optimal growth of another species. The term “stressor,” therefore, must be understood in relation to a particular species or group of organisms that share similar living conditions, for example temperature. In this regard, organisms are classified into psychrophiles (optimal temperature for growth [OTG] 15°C or lower), psychrotolerant organisms (OTG, 20 to 30°C), mesophiles (OTG, 35 to 40°C), thermophiles (OTG, 50 to 70°C), and hyperthermophiles (OTG, 80°C or higher) (186). Temperatures higher or lower than the optimal may cause stress and induce a stress response. A temperature upshift causes a heat shock response (47, 90, 120, 128, 177, 252, 292), whereas a temperature downshift induces a cold stress or cold shock response (255, 275, 304). The latter is not dealt with in this review. Likewise, adaptation to high osmotic and barometric pressures and the response of the cell to their changes (33, 68, 77, 121, 138, 190, 201, 204, 232, 266, 271) are not treated in any detail.
We draw attention, though, to the formation of multicellular structures that result in improved cell resistance to physical, mechanical, and chemical stressors. These structures are of various types and have considerable potential for the biotechnology industry and for the exploration and conquest of inhospitable ecosystems, but their relevance to stress resistance is rarely discussed. We highlight the topics that future studies ought to address in relation to the proteins (and their genes) and other molecules that build the intercellular connective material to keep the cells together in a functional, three-dimensional arrangement.
STRESS, STRESS RESPONSE, STRESS GENES AND PROTEINS, HEAT SHOCK, MOLECULAR CHAPERONES, AND CHAPERONINS
Primer
A cell confronted with an abrupt change in its immediate surroundings suffers stress. The cause, or stressor, may be of various types, for example, physical (temperature elevation) or chemical (increase or decrease in pH, salinity, or oxygen concentration) (47, 113, 252, 292).
A key component of stress is protein denaturation (93, 96, 123, 124, 223, 293). Many proteins lose their native, functional configuration and tend to aggregate. The process may be reversible up to a degree, beyond which it becomes irreversible and generalized within the cell, which ultimately dies.
Another main component of stress is the down-regulation of many housekeeping genes, some of which are actually shut off. Whether this is all due to protein denaturation and represents just the breakdown of the cellular machinery or is an active, induced process by which genes are “told” to slow down or stop has not yet been elucidated. Perhaps both mechanisms, gene failure and regulated shutdown, participate.
Protein damage and gene down-regulation are part of the stress response. There is yet another important component of the stress response, i.e., activation of the stress genes (90, 120, 205, 208, 219, 253, 254, 279). The concentrations of the protein products of these genes increase in response to stressors, protecting the cell from the destructive effects of stress and enhancing post-stress recovery by promoting renaturation (refolding) of partially denatured proteins (93, 101, 223, 293).
Thus, stress inactivates or down-regulates many genes but activates others, whose function is to save the cell. Most stress genes also function in the absence of stress, namely, under normal physiological conditions. The proteins encoded by these genes play critical roles in physiological protein biogenesis. They assist in the folding, translocation, and assembly of other proteins (191, 214, 235, 236, 247). This is the reason why many stress proteins are also called molecular chaperones (72). They help other cellular proteins to (i) fold correctly during and after translation; (ii) migrate to the cell’s locale, where they will reside and function; and (iii) assemble into the quaternary structure that will make them useful to the cell when the proteins function as polymers.
Furthermore, some stress proteins participate in the degradation of other polypeptides, for example when these are denatured beyond recovery and could pose a serious threat to the cell if they aggregated (39, 92, 97, 98, 122, 123, 133).
In summary, stress proteins, particularly those that are molecular chaperones, aid and protect other cellular proteins from their birth on, but they also contribute to the elimination of polypeptides that are no longer useful and endanger cell viability.
It is important to bear in mind that not all stress proteins are chaperones and, vice versa, that not every molecular chaperone is a stress protein.
The wide spectrum of activities of stress proteins is not limited to the chaperoning of other proteins as described above. These activities also include other functions, for example, modulation of their own synthesis (6, 20, 90), regulation of the stress kinase JNK (85), association with enzymes (for purposes yet to be determined) (43), and participation in signal transduction pathways (175) and in rRNA processing (249). It is therefore clear that stress proteins are multifunctional and ubiquitous. They play their roles in all cells, cell compartments, and organelles and are said to be promiscuous because they interact with a great variety of other molecules.
This diversity of functions is reflected in the structural features of the stress proteins, which are composed of domains and motifs with specific roles. As we discuss below, characterization of these domains and motifs has helped in the classification of newly found genes and proteins, identification of stress proteins in their various anatomical locations, and determination of their evolutionary origins.
Stress versus Heat Shock
Stress genes and proteins are often named heat shock genes and proteins in today’s literature, for historical reasons. Because of this, they are represented by the acronyms hsp and Hsp, respectively.
hsp and Hsp were first observed in Drosophila exposed to a temperature higher than the optimum for growth (27°C) (reference 177 and references therein). The genes activated by the temperature upshift were called heat shock genes, and their products were called heat shock proteins. In this review, we use the terms “stress” and “heat shock” interchangeably to qualify the words response, gene, and protein, although we favor the use of “stress” rather than “heat shock” and reserve the latter for the specific instances in which the stressor is a temperature elevation.
Hsp (and their genes) are classified into groups or families according to their molecular mass in kilodaltons (Table 1). The proteins of the 55- to 64-kDa group, or Hsp60 family, are also called chaperonins and are included within the molecular chaperones, generally speaking. More specifically, the latter term is applied to the Hsp70 family. The genes and proteins belonging to the Hsp60 and Hsp70 families have been extensively studied in many bacterial and eukaryotic species.
TABLE 1.
Classification of Hsp into families according to their molecular mass
Phylogenetic Domains
The classification of all living cells into three main evolutionary lines, or phylogenetic domains, Bacteria (eubacteria), Archaea (formerly archaebacteria), and Eucarya (eukaryotes) (11, 297, 298, 300), is still useful despite its limitations and the challenges generated by new findings and contrasting theories (66, 67, 80, 103, 105, 106, 109–111, 188, 189, 192, 198, 234, 286, 298). It helps us to visualize how evolution produced what we see today and to track down genes from the past to the present.
The overwhelming majority of information available on stress genes and proteins comes from studies of bacteria (e.g., Escherichia coli and Bacillus subtilis) and eukaryotes (e.g., Drosophila melanogaster, Saccharomyces cerevisiae, plants, and mammals including humans). The study of stress genes and proteins in organisms of the phylogenetic domain Archaea began only a decade ago and is much less advanced than in the other two domains.
HSP70(DNAK) LOCUS
Structure and Organization
The terms hsp70 for the gene and Hsp70 for the protein are used for eukaryotes, while the same gene and protein are called dnaK and DnaK, respectively, in bacteria. We use hsp70 and Hsp70 in most cases, regardless of the origin, for simplicity and because these terms are more widely known than dnaK and DnaK, and also because archaea are not bacteria.
The first hsp70 gene identified by cloning and sequencing within the domain Archaea was reported in 1991 (182). The gene was found in the mesophilic methanogen Methanosarcina mazei S-6 (OTG, 37°C). Shortly thereafter, in 1992, a homolog was cloned and sequenced from another archaeon, Halobacterium (Haloarcula) marismortui (110). This organism is also mesophilic but belongs to a group, the extreme halophiles, different from that of M. mazei S-6.
For a while, the above genes were the only two archaeal hsp70 genes known. In 1994, two additional homologs were reported: one in another mesophilic extreme halophile, Halobacterium cutirubrum (OTG, 45°C), and the other in a thermophile, Thermoplasma acidophilum (OTG, 55°C) (111).
These findings seemed to indicate that the hsp70 gene was present in archaea, confirming the widely held notion that this gene is one of the most highly conserved, occurring in all organisms. This idea was challenged in 1996, when the sequencing of the whole genome of Methanococcus jannaschii (OTG, 85°C) revealed the absence of the hsp70 gene in this archaeon (31), a result that confirmed observations in other laboratories (47, 164).
More recently, in 1997, the sequencing of the thermophilic methanogen Methanobacterium thermoautotrophicum ΔH (OTG, 55°C) was published (263). The hsp70 gene is present in this methanogen, which in this regard is therefore similar to the mesophilic M. mazei S-6 but different from the hyperthermophilic methanogen M. jannaschii.
While the sequencing of the M. thermoautotrophicum ΔH was under way, another hsp70 gene was cloned and sequenced from a second Methanosarcina species, the thermophilic species Methanosarcina thermophila TM-1 (OTG, 50°C) (126). So by this time, it was clear that at least some mesophilic and thermophilic methanogens do have the gene but that some (perhaps all) hyperthermophiles do not.
After the discovery of an archaeal hsp70 gene in M. mazei S-6 in 1991, more sequencing up- and downstream of this gene revealed the other genes that accompany hsp70(dnaK) in bacteria: hsp40(dnaJ) and hsp23(grpE); these discoveries were made in 1993 (181) and 1994 (45), respectively.
It is pertinent to note here, as was done above for hsp70, that the hsp40 gene and its protein Hsp40 are named dnaJ and DnaJ, respectively, when they are from bacteria. Similarly, the bacterial hsp23 gene and its protein, Hsp23, are called grpE and GrpE, respectively.
We use the terms hsp40 and Hsp40 when referring to archaea, for the same reasons we use the terms hsp70 and Hsp70. However, for the hsp23 gene, we use the terms grpE and GrpE, because the alternative hsp23 and Hsp23 may be confusing since there are several different small heat shock proteins with a mass close to 23 kDa (76, 78, 156, 206, 227). Also, the archaeal GrpE molecule has a counterpart in bacteria and the eukaryotic organelles of bacterial origin but apparently not in the eukaryotic cytosol. The latter does not seem to have a GrpE protein but has some other alternative to exercise similar functions, although recent findings suggest that grpE homologs might also occur in the eukaryotic cytosol (211, 212).
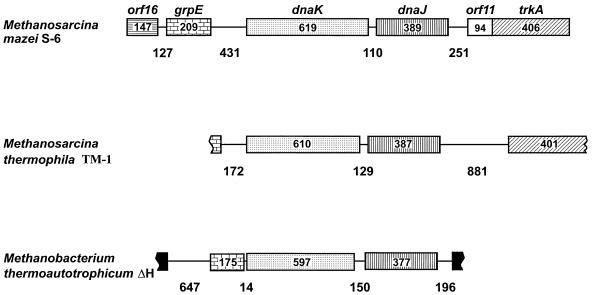
As a result of the sequencing of the M. mazei S-6 and M. thermophila TM-1 hsp70 chromosomal regions and the sequencing of the M. thermoautotrophicum ΔH genome, there are today three archaeal hsp70 loci whose structure and organization have been determined (Fig. 1) (31, 126, 179). The gene order 5′-grpE-hsp70-hsp40-3′ occurs in the three archaeal loci and is the same as that observed in many bacteria, particularly gram-positive bacteria (Fig. 2) (179). However, there are differences between the three archaeal loci. For example, they differ in the length of the 5′-grpE-hsp70-3′ and 5′-hsp40-next gene-3′ intergenic regions and in the gene that follows hsp40 downstream. This gene is the same in M. mazei S-6 and M. thermophila TM-1 but different in M. thermoautotrophicum ΔH.
FIG. 1.
The hsp70(dnaK) locus genes of the archaea for which sequences are available, including genes up- and downstream of hsp70(dnaK). The genes are represented by rectangular boxes from the 5′ to the 3′ end (left to right), with their names above their respective boxes in the locus on top [dnaK and dnaJ are used instead of hsp70(dnaK) and hsp40(dnaJ) for clarity]. The numbers within the boxes indicate the number of amino acids encoded. The lines joining the boxes represent the intergenic regions, with their lengths, in base pairs, shown underneath. The sequences of M. thermophila TM-1 grpE and trkA are still incomplete (what is available would encode 53 and 401 amino acids, respectively). Accession numbers and other details are provided in Tables 2, 5, and 10. Reprinted from references 126 and 179 with permission of the publishers.
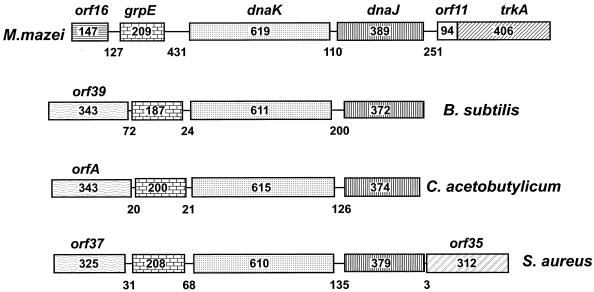
FIG. 2.
The hsp70(dnaK) locus genes of the archaeon M. mazei S-6 and three gram-positive bacteria: B. subtilis (M84964), C. acetobutylicum (M74569), and S. aureus (D30690). Symbols are the same as those described in the legend to Fig. 1. Modified from reference 179 with permission of the publisher.
The length of the intergenic region between hsp70 and hsp40 is conserved in the three loci, particularly in comparison with the other intergenic regions.
The meaning of these structural characteristics is not completely understood. They suggest, for example, that hsp70 and hsp40 may have evolved together, as a unit. This notion is also supported by the conservation of the homologous gene pair in many bacteria (see, for example, Fig. 2).
As discussed later in this review, there are indications that the hsp70 gene in archaea was received via lateral transfer from bacteria. Perhaps it was accompanied by hsp40 and grpE, since both are always present whenever hsp70 is, and the three genes appear next to each other in many bacteria. However, other structural characteristics and experimental results, to be discussed below (see “Occurrence of hsp70 in nature”), tend to make this notion less credible, at least in its simplest formulation. Analyses of the nucleotide sequences between the protein-coding regions of the genes do not reveal obvious similarities, except for the presence of putative archaea-type promoters and bacterial-type termination signals in the expected locations with regard to the translation start and stop codons, respectively (44, 181, 182). Other sequence features vary with the intergenic region and the species, but the regions upstream of hsp70 in two of the methanosarcinas possess a series of repeats and palindromes. They might be cis-acting signals, namely, binding sites for regulatory factors (168). In contrast, the region upstream of hsp70 in M. thermoautotrophicum ΔH is very short and lacks anything that might be a promoter or a cis-acting site.
No bacterial-type promoter sequences (52, 120, 219, 258, 259) are identifiable in these archaeal intergenic regions, nor are there bacterium-type regulatory elements, such as CIRCE (259, 310, 313) or ROSE (208, 209), that one can detect by sequence comparisons.
If one considers the high degree of conservation of these regulatory sequences among bacteria, it is reasonable to conclude that they do not occur in the three known archaeal loci and that regulation of the hsp70 locus genes in these organisms is mediated by factors different from those operating in bacteria. Thus, despite the similarities in organization between the archaeal and bacterial hsp70 loci, their mode of expression and regulatory mechanisms appear to be different.
Also remarkable is that the 5′-grpE-hsp70-3′ intergenic region and the distance between grpE and the next gene upstream in M. mazei S-6 and M. thermophila TM-1 are considerably longer than the equivalent regions in bacteria. The latter have their genes closer to one another, in agreement with their polycistronic mode of transcription and their being regulated as a unit, or operon. Instead, the structure of the archaeal loci in the two methanosarcinas shown in Fig. 2 does not suggest the bacterial modes of transcription and regulation but different ones (see also experimental data, given below).
Expression
Functional analyses of the hsp70 genes have been carried out for M. mazei S-6 and to a lesser extent for M. thermophila TM-1. No functional information exists for the other four archaeal hsp70 genes that have been cloned and sequenced thus far.
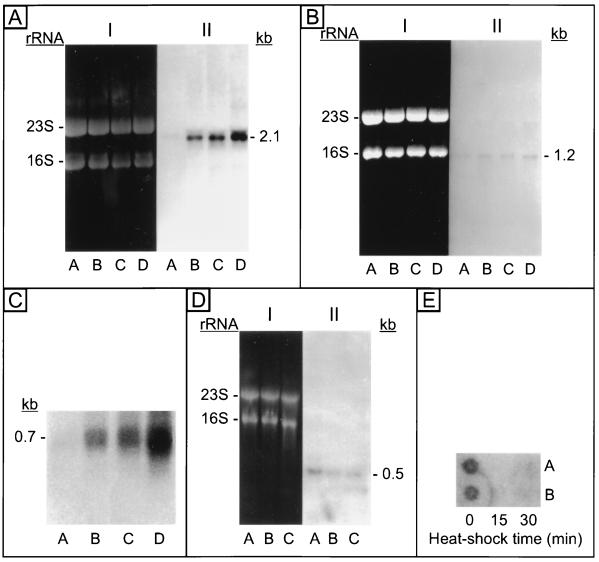
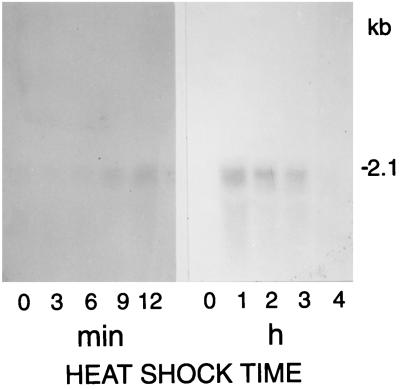
M. mazei S-6 hsp70(dnaK), hsp40(dnaJ), and grpE respond to heat shock by an increase in the production of their transcripts (Fig. 3) (42, 44), as one would expect for stress genes. The transcripts are monocistronic as in eukaryotes (302) and in contrast to bacteria (10, 95, 120, 131, 132, 294, 313). Likewise, the peak response in terms of transcript levels is reached after heat shocks longer than those that would induce a peak response in bacteria (Fig. 4) (164), also in agreement with what is observed in eukaryotes. The transcription initiation sites map to positions reminiscent of the eukaryotic initiation sites with respect to the promoter (Fig. 5) (42, 44, 179). Furthermore, the genes respond to temperatures ranging from 45 to 60°C (Fig. 6) (164) and to other stressors such as cadmium (Cd2+) (Fig. 7) (179) and ammonia (165) as expected for heat shock genes. Thus, the data show that the archaeal hsp70 locus genes are stress or heat shock genes but have mixed bacterial and eucaryal characteristics.
FIG. 3.
(A to D) Northern blots with M. mazei S-6 total RNA (10 μg/lane) showing an increase in the levels of transcripts of hsp70(dnaK) (A), hsp40(dnaJ) (B), and grpE (C), and a decrease in the level of the transcript of orf16 (D) in response to heat shock. (E) Dot blot showing a decrease in the level of the transcript of orf11-trkA in response to heat shock. Hybridizations were done in all cases with radiolabelled probes specific for the respective genes. In panels A, B, and D, I is the gel stained with ethidium bromide showing the RNAs, 23S and 16S while II is the corresponding Northern blot. Lanes: A, total RNA from M. mazei S-6 cells maintained at the optimal growth temperature of 37°C, i.e., non-heat-shocked cells; B and C or B to D, total RNA from cells heat shocked at 45°C for increasing time periods, from 15 to 60 min. The sizes of the transcripts in panels A to D are indicated in kilobases. Transcripts were detected for all the genes in non-heat-shocked cells. Heat shock caused an increase in the levels of the transcripts of hsp70, hsp40, and grpE. The reverse occurred for orf16, and orf11-trkA. The latter two genes overlap and are cotranscribed, whereas the other genes are transcribed monocystronically. Reprinted from references 42, 44, 49, and 184, with permission of the publishers.
FIG. 4.
Response of the M. mazei S-6 hsp70(dnaK) gene to heat shocks of various durations. Northern blots of total RNA (10 μg/lane) extracted from M. mazei S-6 cells before heat shock (lane 0 in both panels) or after a heat shock at 45°C for the times indicated in the horizontal axis, in minutes (min) or hours (h). Hybridization was done with a probe for dnaK. The size of the hybridization bands in kilobases is indicated to the right. Reprinted from reference 164 with permission of the publisher.
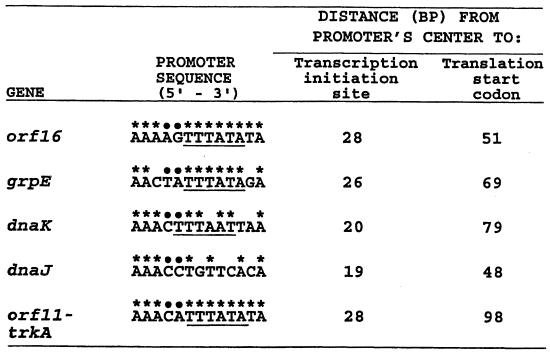
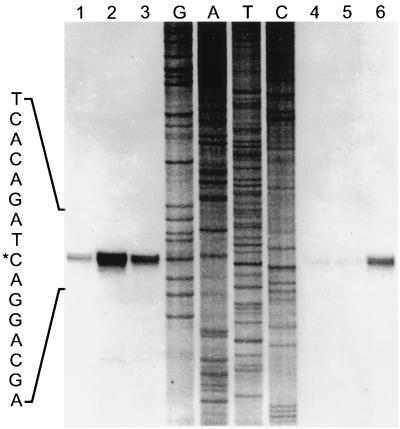
FIG. 5.
M. mazei S-6 promoters for the hsp70(dnaK) locus genes. Bases with asterisks are identical to those in the consensus sequence for promoters in methanogens, and bases with dots denote positions without base preference (25); underlined bases represent the archaeal box A (reference 312 and references therein). The consensus sequence was derived from comparative analysis of promoters for many non-heat-shock genes (25). There is no information on promoters for archaeal grpE, hsp70(dnaK), or hsp40(dnaJ), except that shown here and in Tables 2, 5, and 10. Therefore, no consensus sequence is available for these archaeal heat shock genes. Note that while the promoters for the non-heat-shock genes orf16 and orf11-trkA match the consensus 100%, the grpE, hsp70(dnaK), and hsp40(dnaJ) promoters do not match it to the same extent. Reprinted from reference 179 with permission of the publisher.
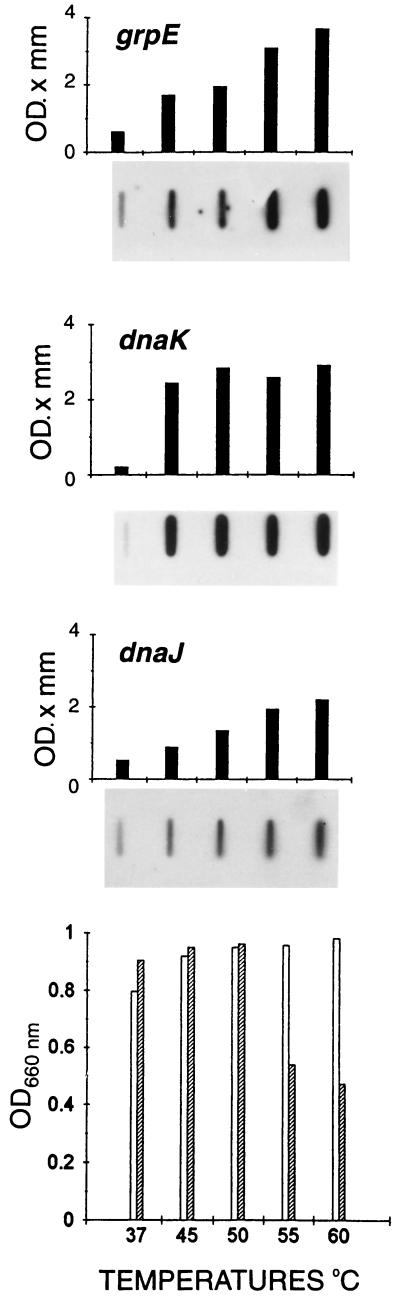
FIG. 6.
Response of the M mazei S-6 genes grpE, hsp70(dnaK), and hsp40(dnaJ) to heat shock at various temperatures demonstrated by slot-blotting with M. mazei S-6 RNA. The levels of mRNA for grpE, hsp70(dnaK), and hsp40(dnaJ) (top three panels) are represented by vertical bars expressed in the optical density (OD) × millimeter units given by the densitometer. The respective slot blots (10 μg of total RNA from S-6 cells per slot) are shown at the foot of the bars, while the heat shock temperatures are indicated in the horizontal axis at the bottom of the figure (°C). Hybridization was done with the respective labelled probes. The culture density is shown in the bottom panel. The OD660 was determined at time zero (open bars) and at 30 min (hatched bars) in cultures maintained at 37°C or heat shocked during this 30-min period at the temperatures indicated at the foot of the bars. Reprinted from reference 164 with permission of the publisher.
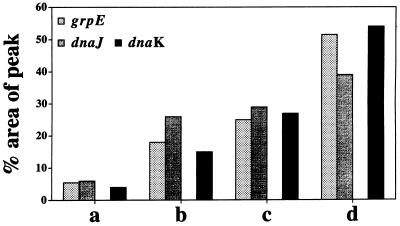
FIG. 7.
Response of the M. mazei S-6 genes grpE, hsp40(dnaJ), and hsp70(dnaK) to the stressors cadmium (Cd2+) and heat. The bars represent levels of mRNA determined by slot blotting with probes for the grpE, hsp40(dnaJ), and hsp70(dnaK) genes. The total RNAs were from cells grown at 37°C (i.e., the optimal temperature for growth of M. mazei S-6) in medium without Cd2+ (a) and in medium with 5 or 27 mM CdCl2 (b and c, respectively) and from cells grown in medium without Cd2+ but heat shocked at 45°C for 30 min (d). Note that the levels of the mRNAs from the three genes increased after heat shock by comparison with the levels before heat shock (constitutive or basal levels; compare a and d). Likewise, the presence of Cd2+ in the medium also induced an increase in the levels of the three mRNAs. This effect was more marked with 27 mM than with 5 mM CdCl2 (compare a with b and c; and compare b with c). Reprinted from reference 179 with permission of the publisher.
The body of structural and functional data available at present suggests that the mechanism of transcription initiation for the archaeal hsp70 locus genes differs from those known to operate in bacteria (26, 27, 120, 132, 208–210, 242, 259, 310, 311, 313) and must involve factors which are not of a bacterial type, i.e., different from ς factors (180). These data, as well as the fact that all transcription initiation studies with archaeal systems (albeit none involving heat shock genes and practically all done with hyperthermophilic systems) have demonstrated transcription factors of the eucaryal type (16, 51, 94, 117, 118, 264, 274, 276, 312), force the prediction that initiation for the M. mazei S-6 hsp70, hsp40, and grpE genes involves eucaryal-type factors.
TATA-Binding Protein
The archaeal homologs of the eucaryal TATA-binding protein (TBP) and the transcription factor IIB (TFIIB) (aTFB and aTFA, respectively [117, 118]), have been identified and shown to be required for the transcription of archaeal, non-heat shock genes in vitro (57, 94, 117, 118, 276). There is no comparable information for archaeal stress genes, but one may hypothesize, based on the observations described in the previous section, that these genes will also require TBP and TFIIB as basal factors. Moreover, it is likely that other factors would also be necessary to induce the response to stressors and preferentially, or even specifically, start transcription of hsp70 and its teammates, hsp40 and grpE.
The tbp gene of M. mazei S-6 has been cloned and sequenced (51). The deduced amino acid sequence of the protein possesses some of the expected archaeal characteristics, but it also shows unique features. For example, like all archaeal TBPs known (reviewed in reference 264), the M. mazei S-6 protein is shorter than most eucaryal homologs, amounting to what is the C-terminal domain in eucaryal molecules. Also, the M. mazei S-6 protein is acidic, like the other known archaeal proteins, but it differs from them in that its N-terminal third is basic, not acidic. The direct, imperfect repeats found in all TBPs, archaeal and eucaryal, are also present in the M. mazei S-6 molecule. Repeats of approximately 42 amino acids separated by a spacing segment of 51 residues on average can be identified (51). The repeats are better conserved in the archaeal than in the eucaryal TBPs, and this is also true for the M. mazei S-6 homolog. A few archaeal TBPs have an acidic tail composed of a series of Glu residues in the C-terminal end. This acidic tail is not present in the M. mazei S-6 molecule.
The overall and regional characteristics of the M. mazei S-6 protein most probably determine its functional properties in what pertains to the binding to DNA at the promoter and to the potential interaction with other transcription factors, such as TFIIB and perhaps stress-specific factors (139a). These structure-function aspects of M. mazei S-6 TBP are being investigated at present. Purified TBP binds to the M. mazei S-6 hsp70 promoter, as demonstrated by the electrophoretic mobility shift assay (EMSA) (57a).
Research to determine how transcription initiation starts and proceeds under constitutive (basal) conditions and in response to stress (heat shock) is under way. In experiments with cell lysates from M. mazei S-6, it was demonstrated that TBP present in the lysates binds to the hsp70 promoter (51a). The phenomenon is observed with lysates from both unstressed and stressed cells. In the latter, a protein appears or increases in concentration or in its ability to bind DNA or TBP, which causes an additional shifted band in EMSA. The nature and role of the protein are under investigation. It might be a regulatory factor that binds near the hsp70 promoter.
ARCHAEAL HSP70 AND HSP70
The Gene
The salient characteristics of the archaeal genes sequenced thus far are described in Table 2. The promoters, terminators, and ribosome-binding sequences or sites (RBS) are putative, except for the M. mazei S-6 promoter, for which preliminary experimental evidence supports the promoter shown (Fig. 5).
TABLE 2.
Archaeal hsp70(dnaK) genesa
| Organism | Accession no. | Size (bp)/no. of amino acids encoded | Promoter/terminator/RBS | Other structures | Expression | Reference(s) |
|---|---|---|---|---|---|---|
| Halobacterium cutirubrum | L35530 | 1,887/628 | NR/NR/NRb | NR | NR | 111 |
| Haloarcula marismortui | M84006 | 1,909/635 | NR/NR/NR | NR | NR | 110 |
| Methanobacterium thermoautotrophicum ΔH | AE000894 | 1,791/596 | NR/NR/gaggtg (−8)c | Downstream repeats; stem-loops | NR | 263 |
| Methanosarcina mazei S-6 | X60265 | 1,857/619 | aaactttaattaa (−79)/inverted repeats/aggatataa (−5) | Up- and downstream repeats; stem-loops | Heat shock inducible | 42, 182 |
| Methanisarcina thermophila TM-1 | Y17862 | 1,833/610 | aacttttatcta (−60)/tctttttt (+38)/agtgaggataaa (−7) | Palindrome; distinctive features upstream | Heat shock inducible | 126, 164 |
| Thermoplasma acidophilum | L35529 | 1,785/595 | NR/NR/NR | NR | NR | 111 |
The Protein
The archaeal Hsp70 proteins are quite similar to each other and, most remarkably, equally so to proteins from gram-positive bacteria (Table 3). Among the archaeal proteins, the most similar pairs are those from the two methanosarcinas, the two extreme halophiles, and the two thermophiles (T. acidophilum and M. thermoautotrophicum ΔH) (see “Hsp70-based phylogenetic trees” below).
TABLE 3.
Comparison of the Hsp70(DnaK) amino-acid sequences from archaea and those most similar from bacteria
| Organism name | Accession no. | % Identity or similaritya
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| M. mazei S-6 | M. thermophila TM-1 | C. acetobutylicum | B. subtilis | M. thermoautotrophicum ΔH | H. cutirubrum | H. marismortui | T. acidophilum | ||
| Methanosarcina mazei S-6 | P27094 | 90.8 | 67.4 | 65.4 | 59.1 | 57.4 | 57.6 | 53.9 | |
| Methanosarcina thermophila TM-1 | Y17862 | 94.7 | 65.6 | 64.6 | 59.9 | 56.0 | 58.6 | 55.8 | |
| Clostridium acetobutylicum | P30721 | 76.1 | 74.8 | 67.7 | 59.7 | 52.9 | 53.5 | 55.0 | |
| Bacillus subtilis | P17820 | 75.4 | 74.4 | 75.4 | 58.9 | 54.6 | 56.3 | 55.0 | |
| Methanobacterium thermoautotrophicum ΔH | O27351 | 69.1 | 69.1 | 69.3 | 69.1 | 54.0 | 53.5 | 63.2 | |
| Halobacterium cutirubrum | P42372 | 66.2 | 64.4 | 63.6 | 64.8 | 64.9 | 74.6 | 48.0 | |
| Haloarcula marismortui | Q01100 | 65.9 | 66.8 | 63.6 | 66.8 | 64.3 | 82.2 | 49.5 | |
| Thermoplasma acidophilum | P50023 | 65.1 | 64.7 | 64.9 | 64.9 | 71.8 | 59.0 | 59.9 | |
Percent identity above and percent similarity (identity plus conservative substitutions) below the diagonal blank space.
The archaeal proteins all have the universal markers for Hsp70 and DnaK and the bacterial markers (Table 4). However, they do not have any of the markers typical of eucaryal molecules. Thus, the archaeal Hsp70 is of bacterial type in sequence and in structural features that reflect its function.
TABLE 4.
Archaeal Hsp70(DnaK) amino acid sequences deduced from cloned genes: motifs
| Motifa | Function | Reference(s) | Synonyma |
|---|---|---|---|
| Phosphate 1 | ATPase; nucleotide binding | 23 | Hsp70 family sig. (ATP α-phosphate) |
| Connect 1 | 23 | Hsp70 family sig. (6 aa) | |
| Phosphate 2 | 23 | Hsp70 family sig. | |
| Adenosine | 23 | None | |
| Connect 2 | 23 | None | |
| DnaK loop [N-29,b (A,S)-30, E-31, G-32, R-34, E-369] | GrpE binding | 28 | None |
| Residue E-171 | Hinge | 29 | None |
| T-12, T-13, D-367 | Interdomain communication | 265, 268 | None |
| Leucine zipper | Oligomerization (?) | 180a | None |
| Hsp70 family sigs. (6 & 7 aa) | ? | 239 | None |
| EEDKKRRERb (archaea, Gram+; not in Gram−) | ? | 142 | Hypercharge run |
| NLS (eukaryotic ct) | Nuclear localization signal | 239 | None |
| Eukaryotic (ct; ER) Hsp70 sig. | ? | 239 | None |
| EEVDb (eukaryotic Hsp70 COOH end) | DnaJ binding; regulatory | 81 | None |
| GDAWVb (mitochondria & alpha, beta, gamma proteobacteria) | ? | 91 | mt sig. Box A |
| Reference | Motif presentc
|
|||||
|---|---|---|---|---|---|---|
| M. mazei S-6 (182) | M. thermophila TM-1 (126) | M. thermoautotrophicum ΔH | T. acidophilum (111) | H. cutirubrum (111) | H. marismortui (110) | |
| 8 | Yes | Yes | Yes | Yes | Yes | Yes |
| 239 | Yes | Yes | Yes | Yes | Yes | Yes |
| 8 | Yes | Yes | Yes | Yes | Yes | Yes |
| NA | Yes | Yes | Yes | Yes | Yes | Yes |
| NA | Yes | Yes | Yes | Yes | Yes | Yes |
| NA | N-29 | N-29 | N-31 | N-31 | ||
| A-30 | A-30 | A-33 | S-30 | |||
| E-31 | E-31 | E-34 | E-31 | E-33 | E-33 | |
| G-32 | G-32 | G-35 | G-32 | G-34 | G-34 | |
| R-34 | R-34 | R-36 | ||||
| E-376 | E-376 | E-364 | E-362 | E-375 | E-375 | |
| NA | E-183 | E-183 | E-170 | E-168 | E-160 | E-162 |
| D-171 | D-171 | D-169 | D-174 | D-171 | D-171 | |
| NA | T-11 | T-11 | T-14 | T-11 | T-13 | T-13 |
| T-12 | T-12 | S-15 | S-12 | T-14 | T-14 | |
| D-367 | D-367 | D-367 | D-365 | D-366 | D-366 | |
| NA | Yes | Yes | Yes | Yes | Yes | Yes |
| NA | Yes | Yes | Yes | Yes | Yes | Yes |
| 142 | Yes | Yes | Yes | Yes | Yes | Yes |
| NA | No | No | No | No | No | No |
| NA | No | No | No | No | No | No |
| NA | E-611 | No | No | No | E-624 | E-626 |
| V-612 | D-625 | D-627 | ||||
| V-613 | V-626 | V-628 | ||||
| D-614 | D-627 | E-629 | ||||
| 91 | No | No | No | No | No | No |
Abbreviations: NA, not applicable; sig. or sigs., signature(s); aa, amino acid(s); NLS, nuclear localization signal; ct, cytosol; ER, endoplasmic reticulum; mt, mitochondria.
Amino acid single-letter symbols (followed by position number when pertinent).
References are given in parentheses. Accession numbers are the same as in Table 3.
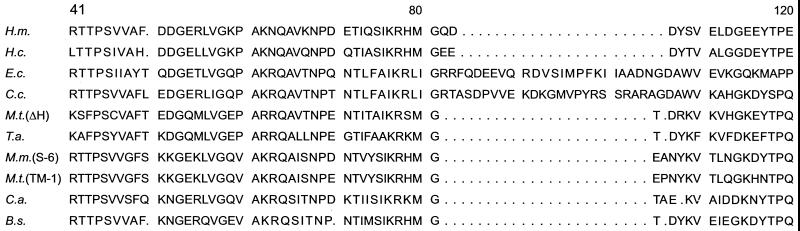
A remarkable feature that appears to be distinctive for the archaeal Hsp70 is the absence of a stretch of 23 to 25 amino acids in the N-terminal quadrant, which became evident when the sequences were aligned with those of proteins from gram-negative bacteria (Fig. 8) (182). This major marker is shared with the DnaK proteins from gram-positive bacteria and is not present in eukaryotic homologs (109–111).
FIG. 8.
Amino acid sequences (single-letter symbols) of six archaeal Hsp70(DnaK) proteins and of four bacterial homologs, two from gram-negative bacteria (E. coli and C. crescentus) and two from gram-positive bacteria (C. acetobutylicum and B. subtilis) between positions 41 and 120, aligned with the program PileUp (Genetics Computer Group, University of Wisconsin, Madison, Wis.). The absence of 23 residues in the proteins from the archaea and gram-positive bacteria compared with those from gram-negative bacteria is shown by dots. Organisms and accession numbers are as follows: H.m., Haloarcula (Halobacterium) marismortui (Q01100); H.c., Halobacterium cutirubrum (P42372); E.c., Escherichia coli (P04475); C.c., Caulobacter crescentus (P20442); M.t. (ΔH), Methanobacterium thermoautotrophicum ΔH (O27351); T.a., Thermoplasma acidophilum (P50023); M.m., Methanosarcina mazei S-6 (P27094); M.t. (TM-1), Methanosarcina thermophila TM-1 (Y17862); C.a., Clostridium acetobutylicum (P30721); B.s., Bacillus subtilis (P13343). Modified from references 179 and 182 with permission of the publishers.
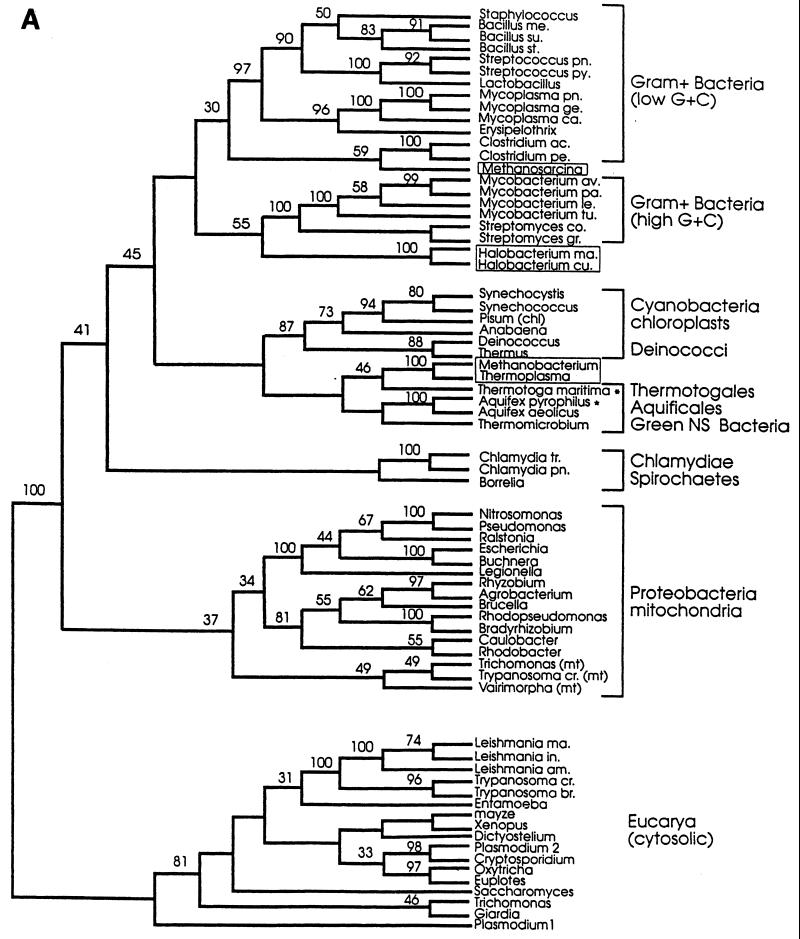
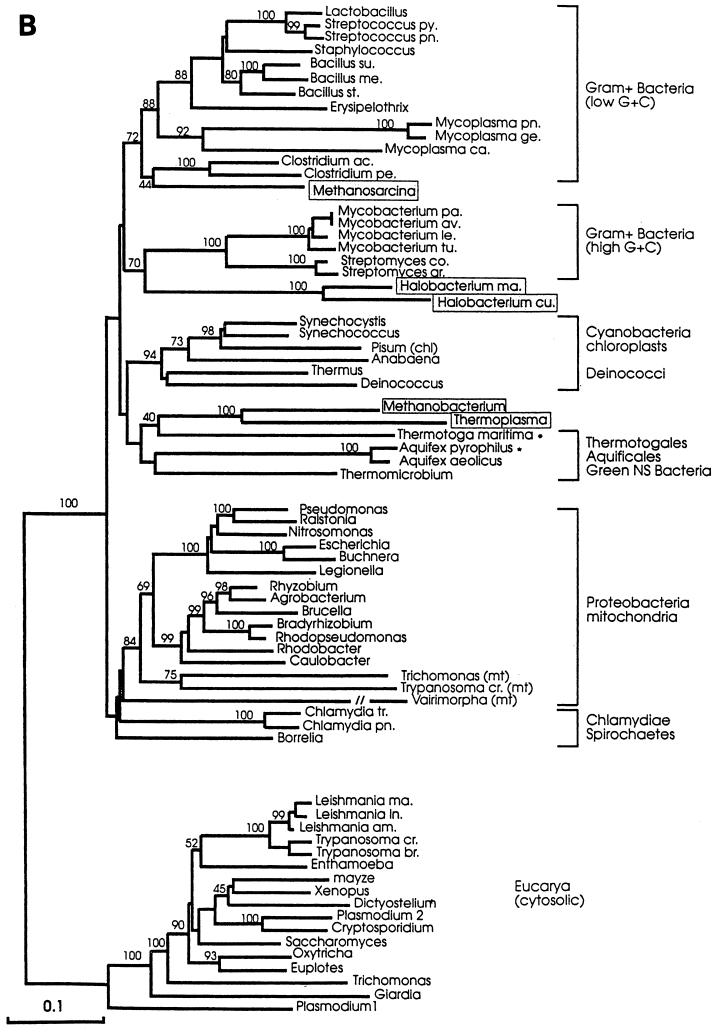
The evolutionary and functional significance of this sequence gap in archaea and gram-positive bacteria has not been elucidated. However, it has given support to a phylogenetic classification that places the archaea closer to gram-positive bacteria than to eukaryotes (105, 106, 109–111), in contrast to the classical 16S-18S rRNA-based tree (11, 299, 300). In the Hsp70-based tree, the extant gram-negative bacteria would have separated from their ancestors, i.e., the ancestors of today’s gram-positive bacteria, early in evolution. As this happened, or shortly thereafter, the gram-negative line acquired the 23 to 25 extra amino acids that characterize its Hsp70. Also, within the framework of this hypothesis, the eukaryotic nucleus would have arisen from a fusion of a primitive archaeon with a gram-negative ancestor. The gene that ultimately became established in the eukaryotic line was that which came from the bacterial partner.
These are speculations based on sequence comparisons and other data that are not completely satisfactory in view of all the information available today. Alternative explanations have been put forward and are discussed in some detail in subsequent sections of this review.
ARCHAEAL HSP40 AND HSP40
The Gene
The four archaeal hsp40(dnaJ) genes sequenced thus far are described in Table 5. They are remarkably similar to each other, as are the proteins they encode (see below).
TABLE 5.
Archaeal hsp40(dnaJ) genesa
| Organism | Accession no. | Size (bp)/no. of amino acids encoded | Promoter/terminator/RBS | Other structures | Expression | Reference(s) |
|---|---|---|---|---|---|---|
| Halobacterium cutirubrum | U93357 | 1,167/389 | NR/NR/NRb | NR | NR | 32 |
| Methanobacterium thermoautotrophicucm ΔH | AE000894 | 1,131/376 | acatttttttatt (−63)c/NR/aggtg (−9) | Up- and downstream repeats; stem-loops | NR | 263 |
| Methanosarcina mazei S-6 | X60265 | 1,167/389 | aaacctgttcaca (−100)/t-rich region/aacagggaatctg (−8) | Up- and downstream repeats; stem-loops | Heat shock inducible | 42, 181 |
| Methanosarcina thermophila TM-1 | AJ010152 | 1,167/388 | aaacctgcact (−55)/tcttttt (+30), t-rich region/atgacagggaa (−11) | Inverted repeat upstream; t-rich region downstream | NR | 126 |
The Protein
The four archaeal Hsp40(DnaJ) proteins known at present are similar to one another and to their bacterial homologs (Table 6). As is the case for the Hsp70, the most similar pair is that of the two proteins from methanosarcinas. The universal motifs and signatures that characterize the Hsp40 molecule, whether from eukaryotes or from bacteria, also occur in the archaeal homologs, except those that are distinctive for the eucaryal molecules (Table 7).
TABLE 6.
Comparison of the Hsp40(DnaJ) amino-acid sequences from archaea and those most similar from bacteria
| Organism | Accession no. | % Identity or similaritya
|
|||||
|---|---|---|---|---|---|---|---|
| M. mazei S-6 | M. thermophila TM-1 | B. subtilis | C. acetobutylicum | M. thermoautotrophicum ΔH | H. cutirubrum | ||
| Methanosarcina mazei S-6 | P35515 | 80.1 | 49.6 | 50.4 | 49.0 | 41.3 | |
| Methanosarcina thermophila TM-1 | AJ010152 | 84.7 | 46.7 | 48.9 | 50.7 | 39.2 | |
| Bacillus subtilis | P17631 | 59.9 | 56.8 | 50.5 | 49.0 | 41.5 | |
| Clostridium acetobutylicum | P30725 | 59.7 | 59.6 | 59.3 | 48.3 | 40.8 | |
| Methanobacterium thermoautotrophicum ΔH | O27352 | 56.4 | 57.5 | 59.7 | 57.7 | 43.7 | |
| Halobacterium cutirubrum | U93357 | 47.0 | 45.2 | 49.2 | 49.7 | 50.7 | |
Percent identity above and percent similarity (identity plus conservative substitutions) below the diagonal blank space.
TABLE 7.
Archaeal Hsp40(DnaJ) amino acid sequences deduced from cloned genes: motifsa
| Motif | Function | Reference(s) | Synonym | Reference(s) | Motif presentc
|
|||
|---|---|---|---|---|---|---|---|---|
| M. mazei S-6 (181) | M. thermophila TM-1 (126) | M. thermoautotrophicum ΔH | M. cutirubrum (32) | |||||
| J-domain | Regulates Hsp70 ATPase; acts as competitive inhibitor of DnaJ in protein refolding | 176, 272, 284 | N terminus | 37 | Yes | Yes | Yes | Yes |
| G-rich domain | Flexible spacer between domains; aids J-domain in stimulating Hsp70 ATPase; competitive inhibitor of DnaJ in protein refolding | 176, 272, 284 | None | NA | Yes | Yes | Yes | Yes |
| HPDb | Stimulates Hsp70 ATPase | 284 | None | NA | Yes | Yes | Yes | Yes |
| Zn finger (CXXCXGXG)b | Interacts with unfolded polypeptides and with denatured proteins | 176, 272 | None | NA | Yes | Yes | Yes | Yes |
| C terminus | Polypeptide (substrate) binding; possibly aids Zn fingers interact with protein substrates | 176, 272 | None | NA | Yes | Yes | Yes | Yes |
| HDELb | ER retention signal | 34, 107 | None | NA | No | No | No | No |
| CAAXb | Prenylation | 34, 35 | CaaX box | 34, 35 | No | No | No | No |
Abbreviations: NA, not applicable; ER, endoplasmic reticulum.
Amino acid single-letter symbols (X, any amino acid).
References are given in parentheses. Accession numbers are the same as in Table 6.
The Gly-rich domain of the H. cutirubrum Hsp40 is longer and has a higher percentage of Gly than those of the three molecules from methanogens (Table 8). The H. cutirubrum molecule also shows a different pattern of distribution of the CxxCxGxG motif from the molecules from the methanogens (Table 9). Motifs 1 and 2 (counting from the N to the C terminus) are separated by 9 amino acids, motifs 2 and 3 are separated by 18 amino acids, and motifs 3 and 4 are separated by 6 amino acids in the four molecules. However, motif 1 begins farther away from the N terminus in the H. cutirubrum molecule than in the others. In consequence, motif 4 is the closest to the C terminus in the molecule from the extreme halophile compared with those from the methanogens. The question remains open whether these seemingly unique features of the molecule from the extreme halophile reflect an adaptation to life under high-salinity conditions and/or to cope with salinity changes.
TABLE 8.
Archaeal Hsp40(DnaJ) amino acid sequences deduced from cloned genes: glycine-rich domaina
| Organism (reference) | Amino acid position
|
Total no. of:
|
% Gly | ||
|---|---|---|---|---|---|
| First | Last | Amino acids | Gly residues | ||
| M. mazei S-6 (181) | 70 | 114 | 45 | 12 | 26.67 |
| M. thermophila TM-1 (126) | 70 | 113 | 44 | 11 | 25.00 |
| M. thermoautotrophicum ΔH | 69 | 118 | 50 | 12 | 24.00 |
| H. cutirubrum (32) | 68 | 135 | 68 | 32 | 47.06 |
Accession numbers are the same as in Table 6.
TABLE 9.
Archaeal Hsp40(DnaJ) amino acid sequences deduced from cloned genes: CXXCXGXG motif
| Motif no.a | Organism (reference)d | Amino acid position
|
Sequence qualityb | |
|---|---|---|---|---|
| First | Last | |||
| 1 | M. mazei S-6 (181) | 144 | 151 | C—C-G-G (4/4) |
| M. thermophila TM-1 (126) | 143 | 150 | C—C-G-G (4/4) | |
| M. thermoautotrophicum ΔH | 148 | 155 | C—C-G-Rc (3/4) | |
| H. cutirubrum (32) | 165 | 172 | C—C-G-G (4/4) | |
| 2 | M. mazei S-6 (181) | 161 | 168 | C—C-G-G (4/4) |
| M. thermophila TM-1 (126) | 160 | 167 | C—C-G-G (4/4) | |
| M. thermoautotrophicum ΔH | 165 | 172 | C—C-G-G (4/4) | |
| H. cutirubrum (32) | 182 | 189 | C—C-G-G (4/4) | |
| 3 | M. mazei S-6 (181) | 187 | 194 | C—C-G-G (4/4) |
| M. thermophila TM-1 (126) | 186 | 194 | C—C-G-G (4/4) | |
| M. thermoautotrophicum ΔH | 191 | 198 | C—C-G-G (4/4) | |
| H. cutirubrum (32) | 208 | 215 | C—C-G-G (4/4) | |
| 4 | M. mazei S-6 (181) | 201 | 208 | C—C-G-G (4/4) |
| M. thermophila TM-1 (126) | 200 | 207 | C—C-G-G (4/4) | |
| M. thermoautotrophicum ΔH | 205 | 212 | C—C-G-G (4/4) | |
| H. cutirubrum (32) | 222 | 229 | C—C-G-G (4/4) | |
For all the organisms, motifs 1 and 2, 2 and 3, and 3 and 4 are separated by 9, 18, and 6 amino acids, respectively.
-, one amino acid and —, two amino acids. Numbers in parentheses indicate the number of amino acids matching the four consensus residues.
Deviation from the norm, R (Arg) instead of G (Gly).
Accession numbers are the same as in Table 6.
Motif 1 in the M. thermoautotrophicum ΔH molecule is, barring a sequencing error, aberrant in that the last residue is Arg (R) instead of Gly (G).
ARCHAEAL GRPE AND GRPE
The Gene
The two archaeal grpE genes whose sequences have been determined are described in Table 10. The genes differ considerably in length; the M. mazei S-6 gene encodes a molecule 35 amino acids longer than that encoded in the M. thermoautotrophicum ΔH homolog. This disparity confirms the poor degree of conservation of grpE and predicts that it will be very difficult to identify homologs in nature on the basis of sequence comparisons alone. The failure to detect GrpE in the eukaryotic-cell cytosol for example, may be due to its diversity. Methods other than structural analyses may be necessary to unveil the true spectrum of this molecule, as suggested by recent work (212) and by the data in Table 10 (see also below).
TABLE 10.
Archaeal grpE genesa
| Organism | Accession no. | Size (bp)/no. of amino acids encoded | Promoter/terminator/RBS | Other structures | Expression | Reference(s) |
|---|---|---|---|---|---|---|
| Methanobacterium thermoautotrophicum ΔH | AE000894 | 525/174 | aaatttttatata (−87)b/NRc/aggtg (−7) | Upstream repeats; stem-loops | NR | 263 |
| Methanosarcina mazei S-6 | X74353 | 630/209 | aactatttataga (−69)/inverted repeat (+76)/ atggg (−11) | Up- and downstream repeats; stem-loops | Heat shock inducible | 44, 45 |
The Protein
The amino acid sequence of GrpE is not as highly conserved as that of Hsp70 or even Hsp40 (Table 11). However, if discrete regions, for example regions I and II (294), are compared, the similarity increases (Table 12). These regions and the GrpE motifs (45) are shown in Fig. 9. The functions of these structural features have not been determined. It has been suggested that they might be important portions of the molecule, involved in the interaction of GrpE with the other members of the chaperone machine, Hsp70 and Hsp40 (45, 294).
TABLE 11.
Comparison of the GrpE amino acid sequences from archaea and those most similar from bacteria
| Organism | Accession no. | % Identity or similaritya
|
||||
|---|---|---|---|---|---|---|
| M. mazei S-6 | C. acetobutylicum | B. burgdorferi | B. subtilis | M. thermoautotrophicum ΔH | ||
| Methanosarcina mazei S-6 | P42367 | 34.0 | 31.4 | 28.6 | 27.0 | |
| Clostridium acetobutylicum | P30726 | 45.7 | 35.2 | 40.1 | 32.7 | |
| Borrelia burgdorferi | P28609 | 41.6 | 44.7 | 27.9 | 30.1 | |
| Bacillus subtilis | P15874 | 40.5 | 49.5 | 39.9 | 33.1 | |
| Methanobacterium thermoautotrophicum ΔH | O27350 | 39.1 | 47.4 | 42.2 | 50.6 | |
Percent identity above and percent similarity (identity plus conservative substitutions) below the diagonal blank space.
TABLE 12.
Comparison of the GrpE amino acid sequences from M. mazei S-6 and M. thermoautotrophicum ΔH: entire molecule, and regions I and II
FIG. 9.
Amino acid sequences (single-letter symbols) of the archaeal GrpE proteins from M. mazei S-6 (M.m. S-6; P42367) and M. thermoautotrophicum ΔH (M.t. ΔH; O27350) aligned with the program PileUp. Regions I and II (294), in that order from the N terminus, are underlined. Motifs 1, 2, and 3 (45), also from the N to the C terminus, are shaded, with their respective consensus sequences on top (light and dark shades show hydrophobic and hydrophilic residues, respectively). The M. thermoautotrophicum ΔH molecule is shorter than the M. mazei S-6 protein, with amino acids missing at the beginning and the end (tildes) and inside (dots).
OCCURRENCE OF HSP70 IN NATURE
The Archaeal Puzzle
The absence of the hsp70 gene in some archaeal species has been noted since the early 1990s (47) and was also found later, when it could not be detected in the hyperthermophiles Methanothermus fervidus, Sulfolobus sp., and M. jannaschii or in the mesophile Methanospirillum hungateii (47, 164). These reports, however, were based on negative results obtained by Northern, Southern, and Western blots with heterologous probes. Consequently, they could not be taken as proof of the absence of the gene.
A definitive confirmation came in 1996, when the sequencing of the M. jannaschii genome did indeed reveal that this organism does not contain hsp70 or the other two genes of the chaperone machine triad, hsp40 and grpE (31). Although this finding helped to give credence to previous negative results obtained by blotting procedures with heterologous nucleic acid and antibody probes and to reaffirm the idea that some organisms may indeed lack hsp70, it raised questions about the earlier finding of the gene in M. mazei S-6. This organism is a methanogen like M. jannaschii. Why is it, then, that the former contains hsp70 while the latter does not? Was the reported M. mazei S-6 gene real or artifactual?
There were additional data confirming the occurrence of hsp70 in other methanosarcinas, different from M. mazei S-6, from before the M. jannaschii genome had been sequenced (42). However, once again, these data had been obtained by Northern and Southern blots with a probe for the M. mazei S-6 gene, and the possibility of nonspecific hybridizations could not be ruled out.
The situation was finally clarified when the full genome sequence of another methanogen, M. thermoautotrophicum ΔH was reported in 1997 (263). Like M. mazei S-6, this methanogen contains hsp70, as well as hsp40 and grpE (Fig. 1).
As things stand today, it is clear that hsp70 occurs in some but not all methanogens. It also occurs in extreme halophiles, but it is not known whether there are organisms in this group that lack the gene—this remains to be demonstrated. The gene does not occur in any of the extreme thermoacidophilic archaea investigated up until now. This had been suggested, as mentioned above, by results obtained by blotting procedures (47, 164) and was confirmed for Archaeoglobus fulgidus (151) and other species by whole-genome sequencing (Table 13). In addition, a search for the hsp70(dnaK) relative hsc66, found in Escherichia coli and other bacteria (260), in the genomes of A. fulgidus, Pyrococcus horikoshii, M. jannaschii, and M. thermoautotrophicum did not reveal its presence (180a).
TABLE 13.
Occurrence, or lack thereof, of the hsp70(dnaK) gene among archaea and representatives of thermophilic and hyperthermophilic bacteria
| Organism | OTG (°C) | hsp70 (dnaK) present | Genome size (Mb) | Demonstrated bya: | Reference(s) |
|---|---|---|---|---|---|
| Archaea | |||||
| Methanosarcina mazei S-6 | 37 | Yes | 2.8 | S, N, W, seq. | 42, 44, 182 |
| Methanosarcina mazei JC3 | 37 | Yes | NDb | N | 42 |
| Methanosarcina mazei LYC | 37 | Yes | ND | N | 42 |
| Methanosarcina sp. strain JVC | 37 | Yes | ND | N | 42 |
| Methanosarcina acetivorans C2A | 37 | Yes | 2.7 | N | 42 |
| Methanosarcina barkeri | 37 | Yes | 2.7 | S | 10 |
| Methanosarcina thermophila TM-1 | 50 | Yes | 2.7 | S, N, seq. | 126, 164 |
| Methanospirillum hungateii | 37 | No | ND | S | 164 |
| Methanobacterium thermoautotrophicum ΔH | 65 | Yes | 1.7 | seq. | 263 |
| Methanococcus voltae | 37 | No | ND | S, W | 119 |
| Methanococcus vannielii | 37 | No | ND | S, P | 99 |
| Methanococcus jannaschii | 85 | No | 1.7 | S, seq. | 31, 164 |
| Methanothermus fervidus | 85 | No | ND | S, P | 99, 164 |
| Methanopyrus kandleri | 100 | No | ND | S, P | 99 |
| Haloarcula marismortui | 45 | Yes | ND | seq. | 110 |
| Halobacterium cutirubrum | 45 | Yes | ND | seq. | 111 |
| Halobacterium halobium | 45 | Yes | ND | S, P | 99 |
| Thermoplasma acidophilum | 55 | Yes | 1.7 | seq., P | 99, 111 |
| Sulfolobus solfataricus | 70 | No | 3.1 | S, P | 99 |
| Sulfolobus sp. | 70 | No | ND | S | 164 |
| Archaeoglobus fulgidus | 83 | No | 2.2 | seq., P | 99, 151 |
| Desulfurococcus mobilis | 85 | No | ND | S, P | 99 |
| Thermococcus tenax | 88 | No | ND | S, P | 99 |
| Pyrococcus furiosus | 100 | No | 2.0 | seq. | 293a |
| Pyrococcus horikoshii | 100 | No | 1.7 | seq. | 144, 145 |
| Pyrococcus woesei | 100 | No | ND | S, P | 99 |
| Pyrococcus abyssi | 100 | No | 1.8 | seq. | P. abyssi; website |
| Pyrobaculum aerophilum | 100 | No | 2.2 | seq. | 79, 79a |
| Aeropyrum pernix K1 | 100 | No | 1.7 | seq. | 143 |
| Bacteria | |||||
| Thermus thermophilus | 70 | Yes | ND | seq. | 220 |
| Thermomicrobium roseum | 70 | Yes | ND | seq. | 108 |
| Thermotoga maritima | 80 | Yes | ND | seq. | 99, 213 |
| Aquifex aeolicus | 83 | Yes | ND | seq. | 58 |
| Aquifex pyrophilus | 83 | Yes | ND | seq. | 99 |
S, N, and W, Southern, Northern, and Western blotting, respectively; P, PCR; seq., sequencing of gene or genome.
ND, not determined.
Several important conclusions may be derived from the data available at present: (i) the absence of hsp70 seems to be a characteristic of archaeal species that live at very high temperatures (hyperthermophiles); (ii) in sharp contrast, no hyperthermophilic bacterium has been found yet that lacks the gene; (iii) hsp70 is scattered among methanogenic archaea that are either mesophiles or thermophiles, like M. mazei S-6 and M. thermophila TM-1, but is absent in other methanogens; (iv) whenever hsp70 was present in a genome, hsp40 and grpE were also found if enough sequencing was done; (v) conversely, genome sequencing has demonstrated that if the hsp70 gene is absent, hsp40 and grpE are also absent; and (vi) the gene has been found in two extreme halophiles, but there are no reports of full-genome sequences for this group of organisms, and so it is not possible to be certain about the situation with them. Do they all have hsp70, and also hsp40 and grpE? We know that at least one of them, H. cutirubum, has hsp40 in addition to hsp70 (32). It may be argued that archaea did not have the genes to begin with and that some of them received the genes via lateral gene transfer. However, the observations listed above, particularly (iv) and (v), and other data challenge the lateral-gene-transfer hypothesis, at least in its simplest form.
One must assume that the three genes jumped as a block, or unit, from a bacterium into an archaeon, particularly the pair hsp70 and hsp40. This implies that the unit also carried the intergenic regions. In agreement with the hypothesis, the proteins encoded by the genes, particularly Hsp70 and Hsp40, are of a bacterial type. Against this hypothesis stands the fact that there are no signal sequences of the bacterial type in the intergenic regions. They are typically archaeal. Hence, more data are needed to determine the origin of the archaeal hsp70 locus genes, i.e., archaeal or bacterial, or prearchaeal or prebacterial; and if the origin is archaeal or prearchaeal, more research is necessary to elucidate the mechanism by which these genes came to be in today’s species that have them.
Hsp70-Based Phylogenetic Trees
The Hsp70 molecule lends itself to comparative analyses for making phylogenetic trees. It is widely distributed among organisms of the domains Bacteria and Eucarya, and it also occurs among the archaea. The molecule is long enough to allow for many mutations to be detected and for useful alignments, since about 500 residues are conserved in a molecule which on average is a little over 600 amino acids in length. Furthermore, Hsp70 has segments that are highly conserved and others that are less so, which allows the detection of variations while maintaining alignable portions and the identification of structural markers that can easily be seen in all members of the family.
Several groups of investigators have used Hsp70 to make phylogenetic trees (22, 99, 105–111, 142, 239). Most of these do not agree with the classical tree based on comparisons of 16-18S rRNA sequences (11, 299, 300). In the rRNA-based tree, archaea and eucarya have a common line up to a point at which they diverge. The archaeal-eucaryal and the bacterial lines are shown to branch off earlier from a primitive, common line. Hsp70-based trees do not support the archaea-eucarya sisterhood or the monophyletic character of archaea suggested by the rRNA-based tree. Some Hsp70 trees suggest a close relationship between archaea and gram-positive bacteria on one side and between eucarya and gram-negative bacteria on the other (105, 106).
The reliability of the trees has been questioned lately. This applies to both rRNA- and protein-based trees (66, 67, 80, 99, 105, 106, 188, 189, 192, 286, 298).
Evidence showing that lateral gene transfer events are more frequent and widespread than was previously realized has been accumulating in the last couple of years (1, 4, 66, 67, 213, 298). Thus, finding that the Hsp70 proteins, for example, of two organisms are very similar does not necessarily mean that the organisms are phylogenetically close. It only means that their Hsp70 molecules are close to each other and have a common ancestor. It does not prove that the ancestor molecule was present in a common ancestor of the two organisms. One of the two organisms may have received its Hsp70 via lateral gene transfer and thus mistakenly appears to be a close relative of the donor’s ancestral lineage.
In summary, by studying amino acid sequences, one can follow the natural history of genes and their occurrence in nature, namely, their itinerary, as it were, along the series of organisms in which they are found.
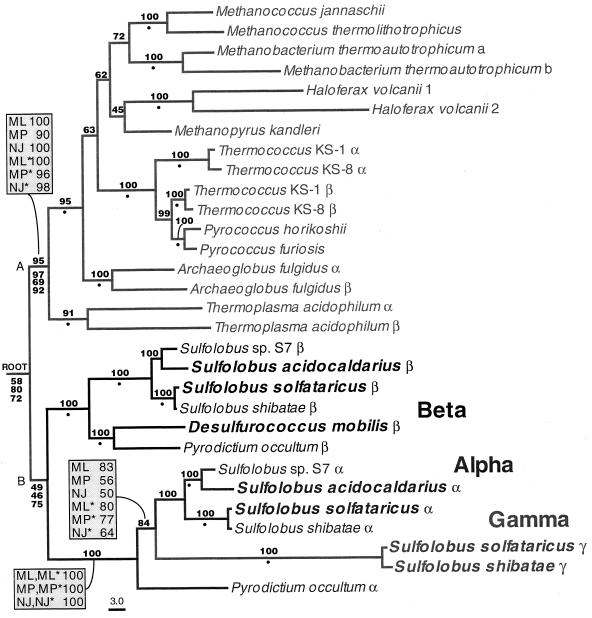
A recent study addresses these points, taking advantage of the fact that considerably more Hsp70 sequences are known now than when previous studies were carried out (99). A systematic search for hsp70 among archaea was performed, and the gene was cloned from Aquifex pyrophilus and Thermotoga maritima. These two bacterial species represent the deepest offshoots in the rRNA-based tree. The gene was not found in 8 of the 10 archaea investigated (Table 13). Alignments of 70 Hsp70 sequences, including the 2 new ones from A. pyrophilus and T. maritima, confirmed the previous observations that the M. mazei S-6 protein clusters with the proteins from the low-G+C gram-positive bacteria while the proteins from the extreme halophiles cluster with the proteins from the high-G+C gram-positive bacteria (Fig. 10). Remarkably, the Hsp70 proteins from T. acidophilum and M. thermoautotrophicum ΔH clustered together (Table 3), along with those from the Aquifexales, Thermotogales, and green nonsulfur bacteria (Fig. 10). The two archaeal Hsp70 proteins in this group appeared to have an ancestor in common with T. maritima. In brief, the Hsp70 from the archaeal species T. acidophilum and M. thermoautotrophicum ΔH did not cluster with the proteins from gram-positive species, as suggested by others, but with bacteria unrelated to the gram positive ones. Another unexpected finding was that the Hsp70 from T. maritima did not have the 23-amino-acid insert characteristic, it was believed, of gram-negative bacteria. Thus, T. maritima Hsp70 possesses a structural feature (i.e., a 23-amino-acid gap in its N-terminal quadrant [Fig. 8]) that is assumed to be distinctive for gram-positive bacteria and archaea, despite the fact that this organism is not a gram-positive bacterium or an archaeon.
FIG. 10.
Maximum-parsimony (A) and evolutionary-distance (B) phylogenetic trees based on Hsp70(DnaK) sequences. Both trees show essentially the same clustering of the archaeal molecules with those from gram-positive bacteria and a group formed by the Aquifexales, Thermotogales, and green nonsulfur bacteria. Numbers represent bootstrap confidence levels calculated from 100 bootstraps (only those that were 30% or higher are shown). Asterisks indicate the newly described genes-proteins (see reference 99). Abbreviations: ac, acetobutylicum; am, amazonensis; av, avium; br, brucei; ca, capricolum; chl, chloroplasts; co, coelicolor; cr, cruzis; cu, cutirubrum; ge, genitalium; gr, griseus; in, infantum; le, leprae; ma, marismortui or major (Leishmania); me, megaterium; mt, mitochondria; NS, nonsulfur; pa, paratuberculosis; pe, perfringens; pn, pneumonia; py, pyogenes; st, stearothermophilus; su, subtilis; tr, trachomatis; and tu, tuberculosis. Reprinted from reference 99 with permission of the American Society for Microbiology.
There are several possibilities to explain these observations. For example, if one assumes that the eucarya and archaea had a common ancestor that contained hsp70, it is possible that both received the gene but some archaea lost it afterwards. A second possibility is that there was no hsp70 in the common ancestor and the gene was acquired after the three lineages separated, with some archaea being excluded. A third possibility is that there was a common ancestor which contained hsp70 and that the three lineages received the gene vertically but the archaeal lineage lost it very early (the species that have the gene today received it via lateral transfer). Finally, if one disregards the common-ancestor idea, another possibility is that the archaea never had the gene while the bacteria and eucarya had it from the beginning. Here, also, archaea that have the gene today must have acquired it by lateral transfer.
The above possibilities and others one might easily think of are more or less improbable depending on (i) how one interprets available data from other studies; (ii) what molecule(s) and criteria were used to generate these data; (iii) what methods were applied to obtain, study, and statistically validate the data; and (iv) what classification scheme was adopted as a master scaffolding to compare with the Hsp70-based tree.
In any case, all the possibilities mentioned share an important characteristic: they stimulate research in this fascinating area of biology and evolution. Molecular phylogeny and detailed analyses of proteins and other macromolecules have already demonstrated their enormous value as tools for research. They are instrumental in uncovering relationships between organisms, the origins of the eukaryotic cell components, the functional meaning of structural motifs, and the role of domains in large proteins of eukaryotes whose ancestors are smaller proteins in more primitive organisms.
FUNCTIONS OF ARCHAEAL MOLECULAR CHAPERONES
Biochemistry
There is little information on the functions of the archaeal Hsp70, Hsp40, and GrpE molecules, as assessed in vitro or in vivo. Since they are so similar in sequence and/or structural features to the homologs from bacteria (Tables 4 and 7; Fig. 9), it may be assumed that both groups of proteins have the same functions and participate in the same mechanisms as molecular chaperones and regulators of their own synthesis.
For example, the bacterial Hsp70(DnaK) is an ATPase and binds ATP and unfolded polypeptides (substrate) (191, 214, 236). Hsp70(DnaK)-ATP has a lower affinity for substrate than Hsp70(DnaK)-ADP. Thus, ATP hydrolysis, which is enhanced by Hsp40(DnaJ) via interaction of its J domain with at least two sites on the Hsp70(DnaK) molecule (87, 270), promotes substrate binding, and the polypeptide is maintained in an extended form, avoiding aggregation. Interaction with GrpE, or nucleotide exchange factor, regenerates the Hsp70(DnaK)-ATP complex, lowers the affinity for the substrate, and releases it (the polypeptide may then be taken by the chaperonin system for final folding (see “Chaperonins” below). Hsp40(DnaJ) is thought to also bind the substrate, before Hsp70(DnaK) does, and to tag the polypeptide so that the Hsp70(DnaK)-ATP complex “sees” it and binds it (176, 191, 236, 272). Also, E. coli DnaK interacts with ribosome-bound trigger factor (62). There is no experimental information on whether the archaeal Hsp70 system operates like that of bacteria and, if so, to what extent the details described above are similar or dissimilar. This is an area that requires investigation and deserves to be explored. It has the potential for revealing how a bacterial-like molecular machine works in a cell whose genome bears eucaryal-like features and probably encodes accessory (regulatory, auxiliary) factors of the eucaryal type while lacking the complementary Hsp60 system of the bacterial type.
Regulation: More Archaeal Puzzles
It would be of particular interest to explore whether a self-regulating circuit similar to that described for E. coli (references 6, 20, 90, and 259 and references therein), or some variation of it, also operates in archaea. Hsp70(DnaK) in some bacteria participates along with Hsp40(DnaJ) and perhaps also GrpE in the degradation of ς32 as a way to down-regulate hsp70(dnaK) transcription. How much of this mechanism operates in archaea? We know that archaea do not have ς factors, and so regulatory circuits for Hsp70(DnaK) synthesis cannot include this factor. Does it include another?
We also know that in eukaryotes, the Hsp70 protein intervenes to prevent trimerization of the heat shock factor (HSF) and thus precludes induction of the hsp70 gene (205, 253). Archaea do not have an identifiable HSF, or heat shock element (75), in the hsp70 promoter region (180a). How, then, is the archaeal hsp70 gene regulated? Does Hsp70 participate in this process? If so, how? Does it interact with an archaeal equivalent of the eucaryal HSF or with another kind of regulator?
These are but some of the fascinating questions posed by recent research with the archaeal hsp70 locus genes. The answers to these questions will shed light on the details of the transcription initiation machinery for stress-inducible genes in archaea and will help us to understand the evolution and principles of the transcription mechanisms in the three phylogenetic domains, not just in the Archaea.
CHAPERONINS
Chaperonin Systems I and II
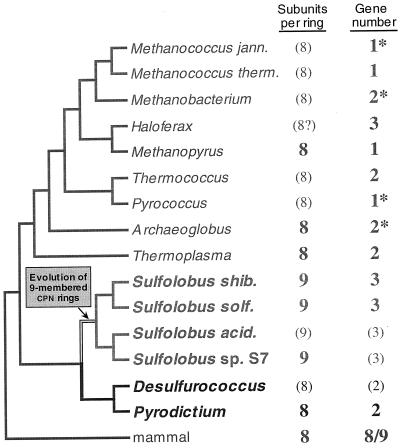
One of the most striking features of archaea is that although they are prokaryotes, they do not have a chaperonin (Hsp60) system like that of the other prokaryotes, the bacteria, but instead have a eukaryotic type of chaperonin complex. No exception to this rule has yet been reported; all archaea investigated have a chaperonin system which resembles that of the eukaryotic cytosol.
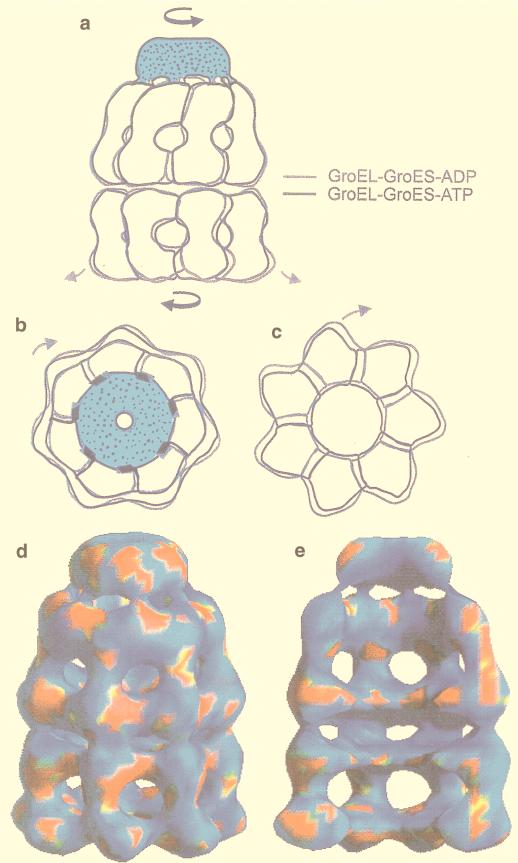
The bacteria have the “bacterial” type (group) I chaperonins, i.e., the genes/proteins groEL/GroEL and groES/GroES (191, 214, 235, 247). A three-dimensional (3-D) view of the barrel-like GroEL/S complex is shown in Fig. 11. What makes the lack of this chaperonin system in archaea very intriguing is that these organisms, at least some of them, have an Hsp70(DnaK) molecular chaperone machine very similar to the bacterial homolog, as described in previous sections. The absence of type I chaperonins and the presence of the Hsp70(DnaK) chaperone machine in a single organism is perplexing if one considers that in bacteria the two systems coexist and seemingly evolved together to interact with each other. The Hsp70(DnaK) system acts early in protein biogenesis to avoid aggregation of nascent polypeptides during translation or immediately thereafter. Subsequently, the polypeptide reaches the GroEL/S system for final folding (191, 214, 235, 247). Now we know that only a minority of polypeptides require the GroEL/S machinery for correct folding (214), but there is no doubt that the coordinated action between the Hsp70(DnaK) and GroEL/S systems is an important physiologic characteristic of the bacterial cell.
FIG. 11.
The bacterial chaperonin complex GroEL/S, and its allosteric changes upon interaction with nucleotide phosphate, which is a major player in chaperonin action. The barrel shape of the complex is apparent, with one base flat and the other convex due to GroES (dotted shading in panels a and b). Also apparent are the two stacked GroEL rings that form the barrel, the subunits of the rings, the domains of the subunits, and the windows between the intermediate domains. GroES looks like a lid, occluding one of the bases of the barrel. The figure also shows the morphologic changes that the complex undergoes when it passes from the ADP to the ATP-bound stages. The structural differences between GroEL-GroES complexes in ATP and ADP were determined by cryoelectron microscopy and computer-assisted image reconstruction. The upper part of the figure (a to c) illustrates domain movements between GroEL/S-ADP and GroEL/S-ATP complexes (gray and black outlines, respectively). The complexes are viewed from the side (a; GroES is dotted-shaded), from above (b; cis apical and equatorial domains surrounding the dotted-shaded GroES), and from below (c; transapical domains). The comparison showed small twists of the subunit domains, particularly in the apical domains of the lower ring. The lower part of the figure represents the GroEL/S-ATP complex as a whole viewed from the side (d) and the same complex cut open to expose the inner cavity (e). The complex is color coded to display the significance map of the differences between the ADP- and ATP-bound stages; i.e., the different colors show significance differences from t tests between the two structures. Regions with significant change (P ≪ 0.0005) are red, and regions with no significant change are blue. The comparison demonstrated that there were domain movements throughout the complex. The main regions of differences (red) observed in GroEL were the ends of the apical and equatorial domains and the hinge regions. There was a localized region of significance at the interring contact 2 (between the equatorial massess, on the outside surface of the structure). The pinwheel pattern of variation in GroES suggested that its subunits were being twisted by the change in the orientation of the apical domain of GroEL. Reprinted from reference 246 with permission of the publisher.
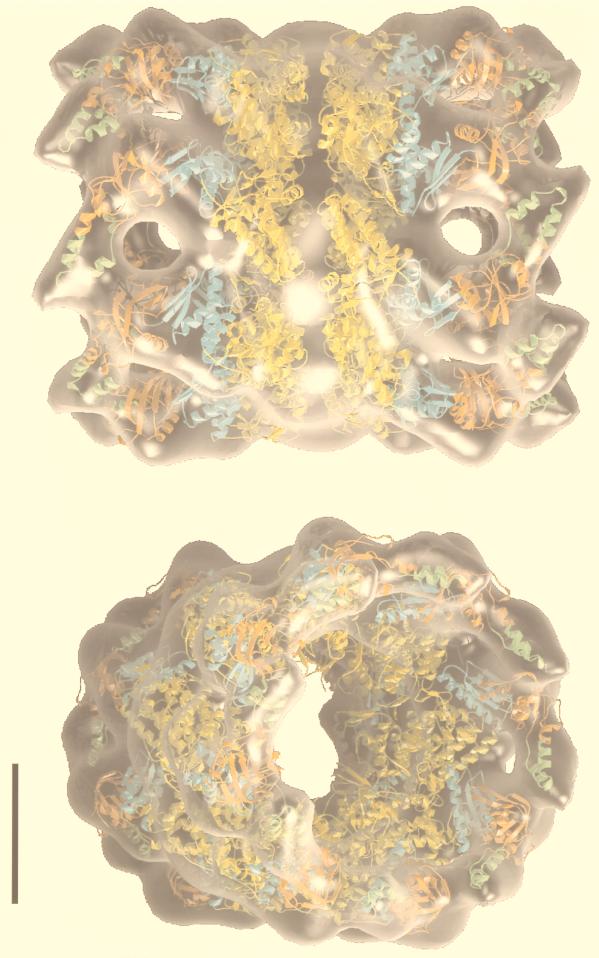
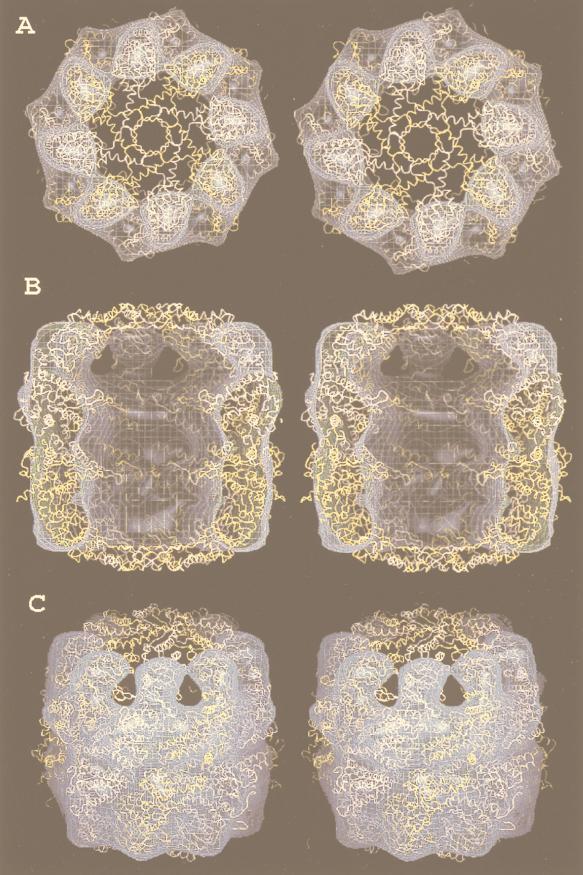
In contrast, archaea, even those that have the hsp70 locus genes [i.e., those that possess the components of the molecular chaperone machine Hsp70(DnaK), Hsp40(DnaJ), and GrpE], do not have the chaperonin type I but the type (group) II system. An example of an archaeal chaperonin complex is shown in Fig. 12. It has also a cylindrical shape like the GroEL/S bacterial complex, but both ends are flat; there is no equivalent of GroES in archaea. In this regard, the archaeal complex resembles that of the eukaryotic cytosol, which is also called TCP-1 (tailless complex polypeptide 1), CCT (chaperonin-containing TCP-1), or TRiC (TCP-1 ring complex) (245, 295). A 3-D reconstruction of ATP-bound CCT is shown in Fig. 13, in stereoview for 3-D visualization with appropriate glasses. In this figure, the X-ray structure of an archaeal chaperonin has been superposed on CCT to demonstrate how the two complexes match with each other.
FIG. 12.
An example of archaeal chaperonin complex. The cylindrical, barrel-like shape is apparent in the top panel, but in contrast to the bacterial GroEL/S complex (Fig. 11), both bases are flat (there is no GroES homolog here). The figure is a semitransparent surface representation of the electron-tomographic 3-D reconstruction of the α-only thermosome showing the complex in an open conformation with a composite atomic model fitted into it. The atomic model was derived from the crystal structures of the intermediate (blue) and equatorial (yellow) domains of the cis-ring of GroEL/S and the apical domain (orange and light green) of the thermosomal α subunit. The complex is viewed from the side (top) and at 60° with respect to the x-y plane (bottom). The black scale bar (bottom left) corresponds to 5 nm. Reprinted from reference 217 with permission of the publisher.
FIG. 13.
The archaeal and eukaryotic chaperonin complexes resemble each other; both look like a barrel with flat bases. The structure of the archaeal complex (thermosome) from Thermoplasma acidophilum as determined by X-ray crystallography (yellow ribbon) is shown superposed on the 3-D reconstruction of CCT bound to ATP generated by cryoelectron microscopy and computer-assisted image processing (blue). The stereoview pairs (which produce single three-dimensional images when viewed with appropriate glasses) are as follows: (A) a base, or end, of the chaperonin complex seen along the longitudinal axis; i.e., the barrel-like chaperonin complex may be imagined to be standing upright on one of its bases and viewed from above to see the inner cylindrical cavity; (B) the complex split open in half; i.e., the barrel has been cut through the sagittal plane, and the half-cut structure is seen from a line of view perpendicular to the longitudinal axis of the barrel, into the inner cavity; and (C) the whole barrel seen from the side as if it were standing on one of its bases, slightly tilted against the longitudinal axis toward the observer to expose the other base. Note that the fitting is between an asymmetric (CCT-ATP) and a symmetric (thermosome) complex. The fitting in the CCT-ATP complex is excellent for the ATP ring but not as good in the apo-ring. The slight mismatch of the apo-rings is consistent with the fact that the apical domains in the apo-ring of CCT do not point toward the cavity but contact each other around the circumference of the ring whereas the apical domains of the thermosome protrude toward the central cavity. Reprinted from reference 170 with permission of the publisher.
It should be noted here that the organelles of the eukaryotic cell, e.g., mitochondria and chloroplasts (which are the descendants of endosymbiotic bacteria) (1, 73, 91, 146, 229, 243, 244, 256, 296), do have the type I chaperonins. Notably, however, the genes coding for the organellar chaperonins are located in the nucleus—to where, it is believed, they migrated from the genome of the primitive bacterium after the endosymbiotic event (1, 19, 296). In fact, comparative analyses of the amino acid sequences of chaperonins and chaperones have helped considerably in establishing the origins of the components of the eukaryotic cell (discussed in other sections of this review).
Structure-Function
The chaperonin systems of the members of the Bacteria, Archaea, and Eucarya are relatively large multimeric rings that form barrel-like structures visible under the electron microscope (Fig. 11 to 13). As a consequence, they have been characterized morphologically and functionally by combining two or more of the following procedures: electron microscopy, molecular genetics, biochemistry, and crystallography. A sample of the results of structural studies of archaea is given in Table 14.
TABLE 14.
Examples of multimeric complexes formed by some archaeal chaperonins and related proteinsa
| Organism | Protein or complexb | Methodb | Detailsb | Reference(s) |
|---|---|---|---|---|
| Methanococcus jannaschii | sHsp | Expression in E. coli, X-ray crystallography | Oligomeric complexes, 24-mer, octahedron, Vm 22 Å3/Da | 148, 149 |
| Thermoplasma acidophilum | Thermosome | Overexpression in E. coli, crystallization, transmission EM, cryo-EM, electron tomography | Two stacked, 8-membered rings of alternating α and β subunits, α-α and β-β pairs, spherical; OD = 164 by 158; lid domain blocks central cavity; 75% of surface area of α and β are solvent exposed; ID = 54–86; chamber volumes = 325,000 Å3/assembled complex and 130,000 Å3/ring; hydrophobic, access through side windows; nonintact proteins only | 65, 216, 289, 290 |
| Methanopyrus kandleri | Thermosome | Transmission EM | Two stacked, 8-membered rings; homo-oligomers; OD = 145 by 136, ID = 43 | 3 |
| Methanococcus thermolithotrophicus | MTTS | EM | Barrel-like structure; forms filaments in vitro | 84 |
| Sulfolobus sp. strain 7 | Chaperonin | EM | Barrel-like structure | 207 |
| Sulfolobus shibatae | Chaperonin (TF55) | EM, circular dicroism spectroscopy, PAGE, spectrophotometry | Two stacked, 9-membered rings; 9-fold symmetry; 2 subunits; open and closed complexes; forms filaments | 234, 280, 281, 307 |
| Sulfolobus solfataricus | Chaperonin (TF55) | EM | Two stacked rings; 9-fold symmetry; 2 subunits; OD = 160 by 175, ID: = 45; crystallization | 71, 154, 187 |
| Pyrodictium occultum | Chaperonin (thermosome) | EM | Two stacked, 8-membered hetero-oligomeric rings; OD = 160 by 165, ID = 155; crystals | 230, 231 |
| Pyrodictium brockii | Chaperonin (thermosome) | EM | Barrel-like structure; OD = 160 by 155, ID = 165 | 231 |
| Thermococcus strain KS-1 | Chaperonin | EM | Forms homo-oligomeric rings in vitro | 308 |
See also Fig. 15.
Abbreviations: Vm, volume to mass ratio; EM, electron microscopy; OD, outer diameter (angstroms); ID, inner diameter (angstroms); PAGE, polyacrylamide gel electrophoresis; MTTS, Methanococcus thermolithotrophicus chaperonin.
The bacterial type (or group) I chaperonin system consists of two protein components, GroEL and GroES, of approximately 60 and 10 kDa, respectively (this system is classified within the Hsp60 family because of the size of GroEL) (235, 247). The two proteins form homoheptameric rings. GroEL builds a barrel-like complex or cage, with two stacked rings (14 subunits in all). Each subunit has three domains, equatorial, intermediate, and apical. The equatorial domain is at the base of the ring, the apical domain forms the contour of the ring toward the base of the barrel, and the intermediate domain connects the other two. Since the intermediate domain is thinner than the other domains it leaves open spaces or windows that connect the outside with the inner cavity of the barrel (Fig. 11). GroES, instead, is a single ring (seven subunits) and completes the functional chaperonin by serving as a lid to occlude one of the two ends of the barrel formed by the GroEL rings (Fig. 11).
CCT, the chaperonin of the eukaryotic cytosol, is the paradigm of the type (or group) II system. It has a structure similar to that of the GroEL/S complex but is built with eight different subunits in the 50- to 68-kDa range per ring (heteropolymeric ring) (170, 171, 235, 245, 246, 273, 295). In addition to the morphologic differences, type I and II chaperonins differ in functional aspects. For example, while GroEL binds a number of different polypeptide substrates (although it may not actively fold all the substrates it can bind), CCT may be more selective. It is able to bind and mediate the ATP-dependent folding of actin and tubulin, but more data are needed to determine the range of substrates that CCT can bind and fold (it might turn out to be wider than is currently believed).
The archaeal chaperonin system (152) has been studied mostly in thermophiles and extreme thermophiles but there is also some information on the homolog of one extreme halophilic species (Tables 14, 15, and 16 and references therein). It consists of either one or two subunits, depending on the species, although recent findings suggest the existence of a third subunit (see “Evolution” below). The subunits are similar to each other in organisms that have more than one subunit and have been given different names (e.g., α and β, a and b, or 1 and 2). They form complexes with the same general design in all species. Detailed features of the archaeal chaperonin complex as seen by electron microscopy were first reported in 1991 (230). The structure was described as a “large cylindrical complex” present in relatively large quantities in the cytoplasm of the extreme thermophilic archaeon Pyrodictium occultum. Salient characteristics were its thermostable ATPase activity and its accumulation after heat shock. The component protein, TF55, of a chaperonin complex from Sulfolobus shibatae was also purified, and its encoding gene was cloned and sequenced in 1991 (281). TF55 formed oligomeric complexes of two stacked rings similar to the bacterial GroEL complex. However, the amino acid sequence of TF55 was found to be more similar to that of the eukaryotic cytosolic TCP-1 (now known to be a component of CCT, as mentioned above) than to that of GroEL. This finding helped to assign possible functions to TCP-1 and boosted research on the eukaryotic cytosolic chaperonin.
TABLE 15.
A sample of archaeal hsp60 (chaperonin) genesa
| Organism | Chaperonin (accession no.) | Size (bp) | Promoter |
|---|---|---|---|
| Archaeoglobus fulgidus | thsA/Cpn-α (AE000950) | 1,635 | tatata (−49)b, tgcaa (−19) |
| thsB/Cpn-β (AF035826) | 1,635 | cttata (−49), tgcaa (−19) | |
| Desulforococcus strain SY | HHSP (S79557) | 1,635 | aggagg (−15) |
| Haloferax volcanii | cct1 (AF010470) | 1,683 | tttata (−26), cgaa (−34) |
| cct2 (AF010469) | 1,674 | tttata (−26), cgaa (−34) | |
| cct3 (AF029873) | Partial seq. | tttata (−26), cgaa (−36) | |
| Methanobacterium thermoautotrophicum ΔH | Chaperonin (α-subunit) (MT0794) | 1,617 | NR |
| Chaperonin (MT0218) | 1,659 | NR | |
| Methanococcus jannaschii | Chaperonin (U67542) | 1,626 | NR |
| Methanococcus thermolithotrophicus | MTTS (AB015435) | 1,632 | tttatata (−75) |
| Methanopyrus kandleri | Thermosome (Z50745) | 1,635 | tttaaata (−60), atgc (−42) |
| Pyrobaculum aerophilum | TF55 (PA306e) | NR | NR |
| TF56 (PA307e) | NR | NR | |
| TF55 (PA27e) | NR | NR | |
| Pyrococcus kodakaraensis KOD1 | cpkA (AB018432) | 1,647 | NR |
| cpkB (D29672) | 1,641 | NR | |
| Pyrococcus abyssi | α-Subunit-thsA (PAB2410) | 1,650 | NR |
| Sulfolobus shibatae | TF55-β (X63834) | 1,659 | tttata (−40) |
| TF56-α (L34691) | 1,680 | tttata (−40) | |
| Sulfolobus sp. strain 7 | α-Subunit (AB001085) | 1,680 | ttttatata (−37) and tgc (−18) |
| β-Subunit (AB001086) | 1,659 | ttttatata (−37) and tgc (−18) | |
| Thermococcus strain KS-1 | α-Subunit (AB001080) | 1,638 | NR |
| β-Subunit (AB001081) | 1,647 | NR | |
| Thermoplasma acidophilum | α-Subunit (Z46649) | 1,638 | tttata (−40) |
| β-Subunit (Z46650) | 1,632 | ttatata (−33), tttatttttta (−21) | |
| Aeropyrum pernix K1 | Thermosome subunit (AEP0907) | 1,671 | NR |
| Thermosome subunit (AEP2072) | 1,665 | NR |
| Terminator | RBS | Remarks | Reference(s) |
|---|---|---|---|
| NRc | aggtg (−9) | Enhanced Box A elementd | 74, 151 |
| NR | aggtg (−9) | Enhanced Box A element | |
| tttga–t-rich-9–tcaaa (+18) | NR | 140 | |
| NR | NR | Induced by heat and hypotonic shocks; enhanced Box A element | 163, 277 |
| NR | NR | Induced by heat and hypotonic shocks; enhanced Box A element | |
| NR | NR | Induced by heat shock | |
| NR | NR | 263 | |
| NR | NR | ||
| NR | NR | 31 | |
| t-rich region (+20) | NR | Recombinant | 84 |
| c-rich region | aggtgat (+18) | 3 | |
| NR | NR | 79 | |
| NR | NR | ||
| NR | NR | ||
| NR | NR | 137, 305 | |
| NR | NR | ||
| NR | NR | P. abyssi website | |
| ttttttt (+7) | aggtg (−8) | Enhanced Box A element | 141, 281 |
| ttttttt (+6) | tgaggt (−4), ggtg (−12) | Enhanced Box A element | |
| NR | ggtg (−6) | 207 | |
| NR | ggtg (−9) | ||
| NR | NR | Recombinant | 308 |
| NR | NR | Recombinant | |
| t-rich region (+6 to +38) | atgcg (−22), aggtgat (−10), caggttcc (+16), tccat (−12) | 288 | |
| ttttt and aaaaa (+14), attta-9-taaat (+35) | tccat (−12) | ||
| NR | NR | 143 | |
| NR | NR |
(−) and (+) refer to position of center of sequence upstream from translation start codon or downstream from translation stop codon, respectively.
NR, not reported.
Conserved sequences as an extension of Box A and between Box A and Box B.
Identification number used in genome project.
TABLE 16.
Examples of archaeal Hsp60 proteins (chaperonins) that have been studied experimentally
| Organism | Protein or structured | Length (amino acids)/ mass (kDa)a | Function/structure | Remarks | Reference(s) |
|---|---|---|---|---|---|
| Archaeoglobus fulgidus | ThsA/Cpn-α | 545/59.0 | NRb | Heat shock-induced | 74 |
| ThsB/Cpn-β | 545/59.7 | NR | increase | ||
| Desulforococcus strain SY | HHSP | NR | 140 | ||
| Haloferax volcanii | Cct1 | 560/59.0 | Two ATP-binding sites, including GDGTTc | 163 | |
| Cct2 | 557/59.0 | Two ATP-binding sites, including GDGTT | |||
| Methanococcus jannaschii | Chaperonin | NR/60.0 | ATP-binding site (GDGTT) | Refolding activity | 161 |
| Methanopyrus kandleri | Thermosome | 545/59.5 | ATP-binding site (GDGTT) | NH4+-dependent ATPase activity | 2, 3 |
| Methanococcus thermolithotrophicus | MTTS | 544/NR | ATP-binding site (GDGTT); unique GGX (M,D,S)c repeats in C terminus | ATPase activity; refolding activity; forms filaments; specific nucleotide requirements | 84 |
| Pyrococcus kodakaraensis KOD1 | CpkA | 549/59.2 | ATP-binding site (GDGTT) | Enhances solubility of peptide | 137, 305 |
| CpkB | 546/59.1 | ATP-binding site (GDGTT) | Prevents thermal denaturation; enhances thermostability | ||
| Pyrodictium brockii | ATPase complex | NR/56 and 59 | NR | Heat shock-induced increase | 230 |
| Pyrodictium occultum | TF55/ATPase complex | NR/56 and 59 | NR | Heat shock-induced increase; ATPase activity | 202, 230 |
| Sulfolobus shibatae | TF55-β | 552/59.7 | No conserved ATP-binding site | Heat-shock induced increase; ATPase activity; binds unfolded protein; conformational changes; forms filaments | 141, 234, 280, 307 |
| TF56-α | 560/59.9 | ATP-binding site (GDGTT) | |||
| Sulfolobus solfataricus | α subunit | NR/NR | NR | Associated with aminopeptidase activity; RNA binding; involved in rRNA processing; specific endonuclease activity; refolding activity | 43, 101, 154, 187, 249 |
| β subunit | NR/NR | NR | |||
| Sulfolobus sp. strain 7 | α subunit | 559/60.2 | NR | No ATPase activity; involved in refolding; able to form homo-oligomeric complexes in vitro | 207, 309 |
| β subunit | 552/59.9 | NR | |||
| Thermococcus strain KS-1 | α subunit | 545/59.1 | ATP-binding site (GDGTT), G-Mc motif in C-terminus | ATPase activity; able to form homo-oligomeric complexes in vitro; involved in refolding | 308 |
| β subunit | 548/59.2 | ATP-binding site (GDGTT) | |||
| Thermoplasma acidophilum | α subunit | 545/58.2 | ATP-binding site (GDGTT) | Alternating α and β units; binds unfolded protein; ATPase activity | 65, 153, 216, 289, 290 |
| β subunit | 543/58.5 | ATP-binding site (GDGTT) |
Calculated from the deduced amino acid sequence.
NR, not reported.
Amino acid single-letter symbols.
HHSP, hyperthermophilic heat shock protein; MTTS, Methanococcus thermolithotrophicus thermosome.
The archaeal chaperonin subunits form homo- or heteropolymeric rings (Table 14) and two rings stacked end to end build a barrel (called a thermosome in some species) that is assumed to be a peptide-folding cage (2, 3, 153, 216, 217, 308, 309). The archaeal barrel does not have the detachable lid characteristic of the bacterial folding chamber (Fig. 11), because archaea lack a homolog of the bacterial small-subunit GroES. Instead, archaea possess one or two large subunits, depending on the species, of approximately 60 kDa—hence the assignment of this system to the Hsp60 family. The subunit(s) is equivalent to the eukaryotic TCP-1 molecule. Recent results (5) indicate that a third gene homolog exists in some organisms and that its expression product would be a third chaperonin subunit. If this protein were in fact present in the cytoplasm, it could account for the ninefold symmetry of the chaperonin complexes observed in the Sulfolobus species that have the third gene (Table 14) (see “Evolution” below).
One of the best-studied archaeal chaperonin complexes is the thermosome from T. acidophilum (65). It is composed of two rings, each with two alternating, different subunits (named α and β) of ∼58 kDa. Electron microscopy and crystallography showed a spheroid or short cylinder with dome-shaped ends instead of the bacterium-like barrel that has one open end and one closed end. The peculiar appearance of the archaeal complex might have been caused by the crystallization conditions. Also, the structure might have been “fixed” while the folding cage was closed, as one would expect it to be during peptide folding, according to what is known for the bacterial GroEL/S system (235, 247, 251). In the bacterial system, the barrel would be alternatively open (acceptor stage, when the polypeptide needing assistance for folding enters the barrel) or closed (folding stage). In the latter stage, the polypeptide would encounter a hydrophilic environment inside the barrel and would bury its hydrophobic residues to stay in solution. The closed stage occurs when the GroES ring attaches to one of the ends of the barrel and covers it as if it were a lid. At this point, the chaperonin complex viewed from the side looks like a bullet, with one end flat and the other convex with the GroES ring (Fig. 11). Instead, the T. acidophilum thermosome appeared more as a sphere than as a barrel (65). The two ends of the double-ringed barrel were not flat. In this archaeal thermosome, protruding structures from the inner surface of the rings themselves might act as a built-in lid.
The functional inferences from these and other structural and morphodynamic analyses (235, 246) are that while the GroEL barrel is closed, the inside of its wall changes from hydrophobic to hydrophilic, owing to conformational changes induced by the binding of ATP and the GroES ring to the GroEL barrel (Fig. 11). Thus, the prisoner polypeptide finds no partner to aggregate with but does find conditions conducive to correct folding. The next move would be the opening of the barrel by nucleotide-driven release of the GroES ring and the exit of the folded polypeptide. If folding is incomplete, the same polypeptide would reenter the barrel and another cycle would begin. Recycling would continue until folding was complete.
These functional inferences may not apply entirely to the archaeal chaperonin complex. As pointed out above, its structure is different from that of its bacterial counterpart. Also, the latter may receive polypeptides from the Hsp70(DnaK) chaperone machine and seems prepared for this interaction. However, archaea that have the Hsp70(DnaK) protein, like T. acidophilum [it is not yet known if this organism also has Hsp40(DnaJ) and GrpE], have a eucarya-like chaperonin system, which may require a different mechanism for interaction with the chaperone machine. In fact, the eukaryotic CCT does not seem to interact with the Hsp70 system and also differs in other aspects from the bacterial GroEL/S complex (152, 170, 171, 245, 273, 295). It remains to be seen how the two systems interact, if at all, in the archaea, and how they work. This is one area in which research with archaeal systems may provide crucial insights into protein folding and refolding mechanisms pertinent to all species, not just the archaea.
The chaperone system of the extreme halophile Haloferax volcanii consists of at least two, perhaps three, components (162, 163, 277). Two genes have been sequenced, and a third has been inferred from a partial sequence that encompasses a putative promoter region (Table 15). These genes have been named cct1, cct2, and cct3, which emphasizes their similarity to the eukaryotic CCT system. However, in contrast to the eukaryotic cct genes, which are not heat shock inducible, the archaeal cct homologs are induced by heat and hypotonic shocks (the latter osmotic shock is a stressor for H. volcanii, since this organism is adapted to live in hypersaline environments).
Evolution
The evolution of Hsp70(DnaK) in archaea is puzzling, as discussed elsewhere in this review, and challenges the imagination of evolutionary and molecular biologists and taxonomists. Likewise, the evolution of the archaeal chaperonin subunits presents a rather interesting panorama in which gene duplication is a salient feature (5). Archaeal chaperonin subunits in a given organism tend to be more similar to each other than to their homologs from other archaea. For example, the α subunit of Thermoplasma acidophilum is more closely related to the β subunit of this organism than to the chaperonin subunits of other archaea (Fig. 14). Also intriguing is the variation in the number of subunits among archaeal species, which may affect the structural configuration (and perhaps functionality) of their chaperonin complexes (Fig. 15). A few archaeal species have only one subunit, others have two, and still others may have three. Indeed, beside the genes encoding the two well-known chaperonins, some archaeal species have another gene, which encodes a third subunit named γ (5). In this work, seven new archaeal genes encoding chaperonin subunits were identified by cloning and sequencing in the archaeotae Sulfolobus solfataricus, Sulfolobus acidocaldarius, Sulfolobus shibatae, and Desulfurococcus mobilis. The relationship of these genes to each other and to those from other archaea are shown in Fig. 14 and 15. In the latter figure, known and predicted subunit genes are listed along with the known and inferred symmetries of the respective chaperonin complexes. Type (or group) II chaperonin systems with multiple subunit species appear to have arisen many times independently during archaeal evolution. It remains to be determined how such differences in the number of subunits in a given organism might affect the function and/or mechanism of action of the chaperonin complex.
FIG. 14.
Phylogeny of archaeal chaperonins as illustrated by a maximum-likelihood tree constructed with the chaperonin amino acid sequences. The two major branches, euryarchaeota (top half; light lines) and crenarchaeota (bottom half; dark lines), are shown. Percent support values are given above each node. Inset boxes indicate support for nodes of particular interest—values were derived from various tree reconstruction methods: maximum-likelihood (ML), maximum-parsimony (MP), and neighboring-joining distance (NJ). The influence of site-by-site rate variation on the support for these nodes was also tested; support values from analyses in which fastest-evolving sites were removed are labeled with an asterisk. The position of the eukaryote outgroup root (ROOT) was determined from additional phylogenetic analyses. Support values for nodes A, B, and ROOT are given in the order ML, MP, and NJ from top to bottom. The names of the species from which new chaperonin genes (not listed in Table 15) were cloned and sequenced are shown in boldface. The scale bar indicates 3.0 substitutions per 100 amino acid sites. The data suggest that among euryarchaeota, linage-specific gene duplications occurred in M. thermoautotrophicum, H. volcanii, A. fulgidus, T. acidophilum, and the Pyrococcus/Thermococcus clade, and that among crenarchaeota, the α and β genes arose from a duplication pre-dating the separation of Sulfolobus and Pyrodictium but the α and γ paralogs of Sulfolobus resulted from a duplication subsequent to that separation. Reprinted from reference 5 (in which sources of sequences, methods, and other details are given) with permission of the publisher.
FIG. 15.
A proposal for the evolution of chaperonin gene number and chaperonin complex symmetry. The cladogram on the left displays the archaeal relationships based on the phylogenetic analysis of the chaperonin amino acid sequences shown in Fig. 14. The number of known or predicted subunits per chaperonin ring and of known or predicted chaperonin genes in each archaeal species are listed in the two columns on the right. Subunits per ring: boldface values indicate known subunit stoichiometry from electron microscopic studies (see also Table 14); values within parentheses are predicted. Gene number: boldface values indicate known gene numbers from sequence data; asterisks indicate total gene numbers confirmed by complete genome sequence; values within parentheses are predicted. The data show that the eight-membered chaperonin ring is widespread among euryarchaeota (see also Table 14), as is the case in eukaryotes (295), but that the situation may be different in Sulfolobus. It would appear from gene sequence data that a transition from eight- to nine-membered chaperonin (CPN) rings (inset) occurred in an ancestor of the extant Sulfolobus during crenarchaeal evolution. This suggestion awaits confirmation from experiments that would demonstrate the existence of a third, distinct protein that is functional in vivo along with the other two subunits. Reprinted from reference 5 (in which source of sequences, methods, and other details are given) with permission of the publisher.
Expression
Expression of genes encoding archaeal chaperonin subunits has been studied in extreme thermophiles to determine the basal levels in unstressed cells and to measure the effect of heat shock (Tables 15 and 16). For example, Sulfolobus shibatae (OTG, 70°C) was heat shocked at 85 or 90°C, and the levels of mRNA for the α and β subunit genes were assessed (141). The levels of the two mRNAs increased in the heat-shocked cells. Extremely stressful conditions caused by exposing the cells to 90°C or higher temperatures stopped all detectable synthesis of all cellular proteins, except that of the two chaperonin subunits. The role that these subunits and the assembled chaperonin complex play in cell survival or in protein biogenesis in general in stressed and unstressed cells has not yet been fully elucidated. Studies in this area may help in the development of methods to protect cells from extreme environments, and they will have practical implications for the design of strategies to explore and exploit such environments. However, these studies must await the development of usable vectors, transformation protocols, mutants, and other tools necessary to carry out the required experiments.
Studies of the expression of the two fully sequenced hsp60 genes, cct1 and cct2, in the extreme halophile H. volcanii have shown that they are inducible by heat and hypotonic shocks (163, 277). The amount of mRNA detected by Northern blotting increased when the cells were heat shocked at 60°C (the OTG for H. volcanii is 45°C) for different periods ranging from 5 to 75 min each. The first clear increase in mRNA was observed after a heat shock of 30 min, and the peak response occurred after the longest heat shock tested, i.e., 75 min. In this regard, the response of H. volcanii hsp60 genes is similar to that of the hsp70 locus genes in the mesophilic methanogen M. mazei S-6 (Fig. 4 and 6). A common feature of these archaeal stress genes is that the duration of the heat shock that induces the highest increase in the amounts of gene products is considerably longer than that which causes a peak response in bacteria. Likewise, the response was detectable in H. volcanii when the cells were heat shocked for 45 min at various temperatures, from 45 to 65°C. This pattern is also similar to that observed for the hsp70 locus genes in M. mazei S-6 (Fig. 6).
Thus, in terms of the duration of heat shock that induces a peak response and of the magnitude of temperature upshift that induces a measurable response, the two archaeal species follow a similar pattern, which differs from that of most bacteria. The latter reach a peak response with a heat shock of about 15 min, and longer heat exposures inhibit the response (10, 95, 120, 259, 292). Similarly, heat shocks at temperatures that would cause a clear response in archaea would not have this effect in bacteria or would kill the cells, even if the OTG for the bacteria and archaeal species under comparison were the same.
Regulation
There is limited information on the molecular mechanisms of transcription initiation and regulation of the chaperone genes in hyperthermophilic archaea, except for some structural and culture data (Tables 15 and 16).
Some information is available for the cct genes in the extreme halophile H. volcanii (222, 277). As mentioned above, this organism responds to heat shock by an increase in the production of mRNA for cct1 and cct2. A study focused on cct1 showed that the heat shock response depends on signals located in a DNA region of about 200 bp upstream of the gene. In this DNA segment, a putative promoter was identified with the sequence 5′-TTTATA-3′ centered 25 bp upstream of the transcription initiation site. Deletion mutagenesis indicated that basal and heat-induced transcriptions required sequence elements very close to the promoter and on both sides of it.
Interestingly, three TBP (tbp) and six TFIIB (tfIIb) genes were found in H. volcanii (222). One of these genes, named tfb2, seems to be induced by heat (278). These observations are noteworthy. Only a single copy of the tbp gene has been found among the archaea studied thus far; however, these did not include an extreme halophile, and so it is not yet clear whether a multiplicity of tbp and tfIIb is a characteristic of all extreme halophiles or is unique to H. volcanii. Extreme halophiles harbor large extrachromosomal elements (172), which may account for the gene multiplicity as suggested by the H. volcanii case, since in this organism some of the tbp and tfIIb genes were indeed located in extrachromosomal DNA.
It has been suggested that in H. volcanii a specific TBP-TFIIB pair might be involved in the selective induction of the cct1 gene in response to heat shock (222). This hypothesis remains to be proven. However, the information available at present indicates that transcription initiation and regulation of the hsp60 genes in H. volcanii must be mediated by a mechanism different from that operating in M. mazei S-6 for the hsp70 locus genes. Moreover, these two gene groups are evolutionarily quite diverse: the hsp60 system is of the eukaryotic (archaeal) type, whereas the hsp70 locus genes are of the bacterial type, very close to the homologs from gram-positive bacteria. While the former may be aboriginal to the archaea and is perhaps the ancestor of the eukaryotic cytosolic CCT, the hsp70 system of M. mazei S-6 and other archaea may not be genuinely archaeal but, rather, may be foreign.
ORIGINS: VERTICAL VERSUS LATERAL
Studies of archaeal stress genes and proteins have provided insights into many areas of biology beyond microbiology. One recent example is the demonstration that while some archaea have the hsp70 gene, others do not, and those that have it probably received it via lateral transfer (99, 164). This finding highlighted the role of lateral gene transfer in determining the genome contents of the cells in nature and thus their behavior and evolution.
Along the same lines, comparative analyses of the amino acid sequences of archaeal chaperones and chaperonins among themselves and with homologs from bacteria and the various components of the eukaryotic cell enhanced our understanding of the origins of the mitochondria, plastids, and other eukaryotic-cell organelles such as the hydrogenosome (73, 91, 107, 146, 167, 229, 243, 244, 256, 296). The impact of these contributions is reflected, for example, in the way in which phylogenetic trees based on Hsp70 and other proteins are interpreted (32, 99, 103–111, 142, 229, 239). Other illustrations of this effect are the classification schemes of cells and organisms we see today in the literature, which display not only the gram-positive bacteria, gram-negative bacteria, cyanobacteria, archaea, etc., but also the mitochondria, chloroplasts, and cytosol as life units with a history of their own (Fig. 10). This notion originated well before the archaea entered the scene, but received considerable input from studies of stress genes and proteins since 1991, when the first archaeal hsp70 gene and the first archaeal chaperonin were sequenced (152, 182). Subsequent comparative analyses of Hsp70(DnaK), Hsp40(DnaJ), GrpE, and Hsp60 types I and II from organisms belonging to the domains Archaea, Bacteria, and Eucarya have revealed the distribution and the origins of these proteins in nature (Table 17).
TABLE 17.
Molecular chaperones and chaperonins in the three phylogenetic domains
| Protein in bacteriaa | Equivalent protein in:
|
|
|---|---|---|
| Archaeaa | Eucaryaa | |
| GroEL (Hsp60) | Nob | mt, chl: Hsp60 (Rubisco subunit binding protein); ct, ER: No |
| GroES (Hsp10) | No | mt, chl: Yes; ct, ER: No |
| No | TF55; thermosome subunits | ct: TRiC (CCT; TCP-1) subunits; mt, chl, ER: No |
| G+, DnaK(Hsp70) | Hsp70(DnaK)c; No-hyp. | No |
| G−, DnaK(Hsp70) | No | mt, chl: Hsp70; ct, ER: Hsp70 para.f |
| DnaJ(Hsp40) | Hsp40(DnaJ)c; No-hyp. | ct, mt, chl, ER: Yes |
| GrpE | GrpE; No-hyp. | mt, chl: Yes; ct, ER: No |
| G−, HptG | No | ct, ER: Hsp90 para.; mt, chl: No |
Abbreviations: mt, mitochondria; chl, chloroplast; ct, cytosol; ER, endoplasmic reticulum; G+ and G−, gram-positive and -negative type of DnaK, respectively. No-hyp., not yet investigated, or investigated but not yet found in hyperthermophiles; para., paralogous.
No, not yet investigated or not well characterized, or investigated but not yet found.
Protein similar to homologs in gram-positive bacteria but transcription initiation mechanism similar to that of eucarya.
The main features revealed by the data are as follows: (i) archaea have chaperonin II as the eukaryotic cystosol and Hsp70(DnaK) like that of the gram-positive bacteria; (ii) the eukaryotic organelles and gram-negative bacteria have the same Hsp70(DnaK); (iii) the type I chaperonin system is present in bacteria and organelles; (iv) hyperthermophilic archaea have the type II chaperonin system but no Hsp70(DnaK), Hsp40(DnaJ), or GrpE; and (v) GrpE is present in bacteria and in some archaeal species but has not yet been identified in eukaryotes outside the mitochondria, although recent work suggests otherwise (211, 212).
OTHER ARCHAEAL STRESS GENES AND PROTEINS
A number of genes and proteins have been found, in a variety of archaeal species, that may be considered pertinent to the stress response because of one or more of the following observations: (i) the amount of protein and/or the amount of mRNA increased during or after stress; (ii) the protein was present at higher concentrations than usual during a period of stress tolerance; (iii) the protein assisted polypeptide folding in vitro; and (iv) the sequence was similar to that of a known stress gene or protein. An illustrative list of these proteins is shown in Table 18. An interesting group comprises the small heat shock proteins (sHsp) (76, 78, 177, 206, 227), with a molecular mass around or below 30 kDa (Table 1). A subgroup is composed of proteins with peptidyl-prolyl cis-trans isomerase (PPIase) activity, which catalyze the isomerization of the peptidyl-prolyl bond and thus play an important role in protein biogenesis in bacteria and eukaryotes (78). PPIases can be distinguished from one another because they bind to and are inhibited by different immunosuppressive agents (this is the reason why PPIases are sometimes called immunophilins). Some PPIases bind cyclosporin and are termed cyclophilins, while others bind FK506. Examples of both types have been found in the archaea. A cyclophilin homolog was identified in H. cutirubrum (135), but most of the other PPIase gene homologs found in the archaea encode proteins of the FK506-binding type (83, 136). The functions and mechanisms of action of the archaeal PPIase homologs remain to be determined.
TABLE 18.
Other examples of archaeal stress or stress-related genes and proteins
| Protein | Mass (kDa) | Organism | Presumed function | Inducer | Reference(s) |
|---|---|---|---|---|---|
| Superoxide dismutase | 20 | Halobacterium halobium | Scavenger of oxygen free radicals | Heat | 17 |
| Crx protein trio | 40.8, 42.3, and 42.9 | Methanobacterium bryantii | Copper or general resistance | Copper | 147 |
| Betaine transporter | NRa | Methanosarcina thermophila TM-1 | Maintenance of internal ionic balance | Osmotic stress | 233 |
| Inositol compounds | NR | Methanococcus igneus, Pyrococcus woesei, Pyrococcus furiosus | Maintenance of internal ionic balance | Osmotic stress | 40, 193, 257 |
| TrkA | 44.1 | Methanosarcina mazei S-6 | Maintenance of internal K+ balance | Ammonia | 164 |
| DNA repair system | NR | Pyrococcus AL585 | DNA repair | UV light | 24 |
| Prefoldin | 14–23b | Methanococcus jannaschii | Protein folding | NR | 287 |
| sHsp | NR | Methanococcus jannaschii | RNA stabilization, thermotolerance | NR | 148 |
| ClpB | NR | Methanosarcina acetivorans | Growth and survival at high temperatures; involved in proteolysis | NR | 98 |
| Bacterioruberin | NR | Haloferax mediterranei | Maintenance of internal ionic balance | Osmotic stress | 68 |
| Several, unidentified | 22, 25, 40, 70, 55, and 90 | Thermococcus peptonophilus | General stress protection | Heat and pressure | 33 |
| PPIase | 19.4–31c | Halobacterium cutirubrum | Acceleration of rate-limiting step in protein folding | NR | 31, 83, 135, 136 |
| 16 or 42c | Methanococcus thermolithotrophicus | ||||
| NR | Methanococcus jannaschii | ||||
| 17.6 | Thermococcus sp. KS-1 | ||||
| Two, unidentified | 90 and 150 | Pyrococcus ES4 | General stress protection | Cold shock | 129 |
| Several, unidentified | 20, 22, 29, 30, 31, 32, 36, 42, and 88 | Sulfolobus acidocaldarius | General stress protection | P starvation | 221 |
| 17, unidentified | Mainly in the groups 21–28, 44–45, and 75–105 | Haloferax volcanii | General stress protection | Hyposalinity | 55 |
| Proteasome | 24 and 22d | Methanosarcina thermophila TM-1 | Protein degradation | NR | 195, 196, 248, 263 |
| 25.8 and 22.3d | Thermoplasma acidophilum | ||||
| NR | Methanobacterium thermoautotrophicum ΔH |
NR, not reported.
Six subunits within the indicated size range.
Depends on the method used.
α- and β-subunits, respectively.
Recently, a disulfide oxidoreductase that contains two thioredoxin fold units from the hyperthermophilic archaeon has been characterized (238). This protein falls within a large group of enzymes that share the sequence motif CXXC and comprises various subgroups such as the protein disulfide isomerase (PDIase), thioredoxin, glutaredoxin, and DsbA families (76, 238). The archaeal enzyme had the capacity to catalyze dithiol oxidation and reduced disulfide bridges. The crystal structure revealed similarities to the eukaryotic PDIase.
Other putative stress genes have been identified by computer analysis in sequenced archaeal genomes; a sample of these is presented in Table 19. These genes would encode proteins with sequences similar to known stress or stress related proteins. As in other fields of biology, the availability of whole-genome sequences has helped archaeal research and will continue to do so. We have already discussed how critical were the contributions made by the sequencing of the M. jannaschii and M. thermoautotrophicum ΔH genomes to the clarification of the distribution of hsp70 among archaea and its possible origin. This is just one example from a long list. The future is very promising. For example, an important area that will benefit from the availability of genome sequences is the study of the factors involved in the regulation of the archaeal stress genes. Also, it would be interesting to identify the whole set of genes that are active during stress and to determine which ones are essential for protein biogenesis under physiological conditions in the hyperthermophiles that lack the Hsp70(DnaK) chaperone machine.
TABLE 19.
Some stress related gene and protein homologs identified in sequenced archaeal genomes
| Organism | Protein | Identification no. | Reference(s) |
|---|---|---|---|
| Methanococcus jannaschii | Heat shock protein X | MJ1682 | 31 |
| Heat shock protein 31 | MJ0285 | ||
| DNA repair protein 45 | MJ0869 | ||
| DNA repair protein RAD51 | MJ0254 | ||
| DNA repair protein RAD2 | MJ1444 | ||
| PPIase | MJ0278 | ||
| PPIase | MJ0825 | ||
| Proteasome α-subunit | MJ0591 | ||
| Proteasome β-subunit | MJ1237 | ||
| Survival protein | MJ0559 | ||
| Methanobacterium thermoautotrophicum ΔH | Heat shock protein X | MTH569 | 263 |
| Heat shock-related protein X | MTH1817 | ||
| Heat shock protein class I | MTH859 | ||
| DNA repair protein rad2 | MTH1633 | ||
| DNA repair protein rad51 | MTH1693 | ||
| DNA repair protein radA | MTH541 | ||
| DNA repair protein rad32 | MTH1383 | ||
| PPIase | MTH1125 | ||
| PPIase B | MTH1338 | ||
| Proteasome, α subunit | MTH686 | ||
| Proteasome, β subunit | MTH1202 | ||
| Survival protein (SurE) | MTH1435 | ||
| Archaeoglobus fulgidus | Heat shock protein (htpx) | AF0235 | 151 |
| Small heat shock protein (hsp20-1) | AF1296 | ||
| sHsp (hsp20-2) | AF1971 | ||
| Proteasome, α subunit | AF0490 | ||
| Proteasome, β subunit | AF0481 | ||
| Pyrobaculum aerophilum | Heat shock protein | PA95 | 79 |
| Heat shock protein | PA305 | ||
| DNA repair protein rad2 | PA289 | ||
| DNA repair protein (XRCC1) | PA290 | ||
| β-Type proteasome | PA403 | ||
| Proteasome β | PA408 | ||
| Pyrococcus horikoshii OT3 | Thermophilic factor | PH0017 | 144, 145 |
| DNA damage-inducible protein | PH1807 | ||
| DNA repair protein | PH0263 | ||
| DNA repair protein, putative | PH1704 | ||
| Proteasome, β subunit, putative | PH0245 | ||
| Proteasome, β-subunit precursor | PH1402 | ||
| Proteasome, α subunit, putative | PH1553 | ||
| Pyrococcus abyssi | Heat shock protein (htpX) | PAB0758 | P. abyssi website |
| Heat shock protein (htpX) | PAB1974 | ||
| sHsp (hsp20) | PAB2072 | ||
| Aeropyrum pernix K1 | Heat shock protein | APE0754 | 143 |
| Heat shock protein (htpX) | APE1045 |
STRESSORS
As pointed out above, cell stress has become known, for historical reasons, as the heat shock response (reference 177 and references therein). While this name acknowledges the pioneers who discovered the reaction of a cell to a temperature elevation, it does not reflect the entire scope of the field.
It is true that the archetypal stressor used by most investigators to test genes and measure the stress response of a cell is heat. Nonetheless, there are many other stressors, at least as important as heat from the ecological, biotechnological, and medical viewpoints (177, 250, 252, 292). Certain stressors are specific to some organisms that live in ecosystems very different from those inhabited by terrestrial mammals such as humans. Some archaea do, indeed, thrive in extreme environments, under conditions of temperature, pressure, pH, or salt concentration which would stress or even kill human cells. “Extremophilic” archaea offer a unique opportunity to study how a cell copes with agents and conditions that would not be tolerated by other cells. The information may one day prove useful, for example, to design means of improving the resistance of human or plant cells to stressors that cause disease or impair crops.
Research in this particular area has included the testing of a range of stressors that are mostly pertinent, in each case, to the environment in which the organism lives, as illustrated by the examples listed in Table 20.
TABLE 20.
Examples of stressors, other than heat, tested with archaeal cells
| Stressor | Organism | Reference(s) |
|---|---|---|
| Hyperosmolarity | Pyrococcus furiosus, Methanosarcina thermophila TM-1, Methanosarcina mazei S-6, Haloferax volcanii, Methanococcus igneus, Methanococcus thermolithotrophicus | 40, 41, 77, 165a, 193, 204, 233 |
| Hypoosmolarity | Haloferax mediterranei, Haloferax volcanii | 68, 77, 163, 204 |
| Pressure | Pyrococcus strain ES4, Pyrococcus strain ES1, Methanococcus thermolithotrophicus, Methanococcus jannaschii, Thermococcus peptonophilus | 33, 130, 138, 200, 232 |
| Ethanol | Methanococcus voltae | 119 |
| UV light | Sulfolobus acidocaldarius, Pyrococcus furiosus | 64, 301 |
| Copper | Methanobacterium bryantii | 147 |
| Heavy metals | Methanosarcina mazei S-6 | 179 |
| H2O2 | Methanococcus voltae | 119 |
| Casamino Acid/Fe2+ starvation | Metallosphaera sedula | 114, 225 |
| pH | Pyrococcus furiosus | 36 |
| Ammonia | Methanosarcina mazei S-6, Methanosarcina thermophila TM-1 | 165, 165a |
| P starvation | Sulfolobus acidocaldarius | 221 |
STRATEGIES AND METHODS USED TO STUDY THE STRESS RESPONSE, GENES, AND PROTEINS IN THE ARCHAEA
Now that we have become acquainted with the archaeal organisms and the stress genes, proteins, and stressors pertinent to these organisms, we can summarize the methods that were applied as a practical guide for future research. The strategies and procedures were varied, as shown in Table 21. They included most of the methods routinely applied to the study of bacterial and eukaryotic cells but with the modifications necessary to deal with the special problems posed by some archaeal organisms. Examples of these special problems are growth at very high temperature and pressure, anaerobiosis required to preserve enzymatic activity, high resistance to cell disruption with the resulting difficulty in obtaining good yields of macromolecules relatively undamaged or unfragmented, and other equally serious technical obstacles that make research with some archaea quite challenging.
TABLE 21.
Strategies and methods used to study the stress response, genes, and proteins in archaea
| Method | Stressor/conditionsa | Gene or protein and organismb | Reference(s) |
|---|---|---|---|
| Culture | |||
| Viability, acquired stress tolerance | Heat Pressure Osmotic stress Irradiation P starvation |
PES4, SSH, MSE MJA, PES4, PES1, TPE MTP, HXV PFU SAC |
33, 64, 68, 114, 129, 130, 201, 204, 221, 225, 233, 279, 281, 282 |
| Growth | Continuous culturec | MSE | 114, 255 |
| Morphology | Heat/starvation Ammonia |
MSE (membrane potential effects) MMA, MTP (cell shape) |
165a, 255 |
| DNA | |||
| Southern blot as only identifier |
hsp70: MAC, MBA, MHU, HHA pho: SAC |
10, 42, 99, 164, 221 | |
| Sequencing of gene |
hsp60: AFU, DSY, HXV, MTH, MJA, MTL, MKA, PAE, PKOD, SSH, SSP7, TKS, TAC hsp70: HCU, HMA, MTH, MMA, MTP, TAC HCU (hsp40), MMA (grpE, hsp40, trkA), MTP (hsp40), PAL585 (dinF), MBR (crx) Otherd: MJA, AFU, PAE, PHO, MTH |
3, 24, 31, 32, 45, 79, 84, 105, 110, 111, 126, 140, 141, 143, 144, 147, 151, 163, 181–183, 207, 263, 281, 288, 305, 308 | |
| Stability of DNA | Heat | PFU | 224 |
| DNA repair | UV light | SAC, PFU | 64, 301 |
| DNA (plasmid) topology | Heat and cold | Sulfolobus species | 173, 174 |
| RNA | |||
| Stress related increase in transcript (Northern/slot blot) | Heat Heat, ammonia, metal Heat and ammonia Copper |
hsp60: SSH, HXV hsp70: MMA, MTPd Others: MMA (hsp40, grpE, trkA) MBR (crx) |
42, 44, 126, 141, 147, 163, 165, 277 |
| Transcriptionally active regions of genome | Heat and osmotic stress | HXV | 77, 283 |
| Protein | |||
| 1-D electrophoresis (unidentified proteins) | Osmotic stress Pressure Heat shock Copper Starvation Cold |
HME TPE, PES4 POC, TPE, MSE, SSH MBR MSE PES4 |
33, 68, 114, 129, 130, 141, 147, 225, 230 |
| 1-D electrophoresis, radiolabelled | Osmotic stress Heat Oxidative stress Ethanol |
HXV (several), HME, MTP (betaine) SSH (Hsp60e), MVO (11f), HMA (4), SAC (4) HXV MVO MVO (11f), SAC (none) |
55, 68, 110, 119, 139, 141, 204, 233, 234, 279, 282 |
| 2-D electrophoresis (unidentified proteins) | Heat P starvation |
HXV, MSE SAC |
114, 204, 221 |
| 2-D electrophoresis, radiolabelled | Pressure | MTL | 55, 138, 204, 221 |
| Osmotic stress | HXV | ||
| P starvation | SAC | ||
| Heat | HVO | ||
| Isoelectric focusing | SSO(Hsp60) | 154 | |
| Stress-accumulation determined by NMR | Osmotic stress Heat |
PFU, MIG, PWO (inositol compounds), PFU, MIG (inositol) | 40, 41, 193, 257 |
| Cross reaction with antibodies for stress proteins | Hsp60: ABR, TTE, POC, SMA, MSE, SAC, PAB, HBU, DAM, AFU, TAC | 114, 141, 225, 230, 290 | |
| Peptide analysis/sequencing | Hsp60: SSO, TAC, SSP7, MKA Crx: MBR |
3, 147, 154, 207, 289 | |
| Purification of protein | Hsp60: SSH, TAC, MKA, PKOD, SSO, POC, SSP7, MJA, MTL, TKS sHsp: MJA Crx: MBR |
3, 65, 84, 101, 147, 148, 154, 161, 187, 207, 216, 230, 234, 281, 289, 290, 305, 308 | |
| Structure | See Table 14 | Hsp60: SSH, TAC, MKA, MTL, POC, SSP7, PBR, SSO, TKS sHsp: MJA |
3, 65, 84, 148–150, 154, 187, 207, 216, 230, 231, 234, 279, 280, 281, 289, 290, 307, 308 |
| Conformational changes | Heat or ATP | Hsp60: SSH, SSO | 101, 154, 170, 171, 234, 246, 307 |
| Stress accumulation (Western blotting/activity gels) | Hsp60: POC, MSE, AFU Hsp70: MMA Superoxide dismutase: HHA Crx: MBR |
17, 42, 74, 114, 147, 230 | |
| Analytical gel filtration and ultrafiltration; spectral analysis | Recombinant proteins tested in vitro | Hsp60 (α and β subunits) | 202 |
| Refolding experiments | Hsp60: SSP7, SSH, SSO, MTL, PKOD, TKS, MJA | 84, 101, 161, 207, 234, 305, 308 | |
| ATP-binding site in sequence | Hsp60: TAC, MKA, MJA, SSH, PKOD, HXV, MTL, TKS | 3, 65, 84, 141, 161, 163, 289, 305, 308 | |
| ATPase activity as indicator of chaperonin action | Hsp 60: PKOD, SSO, SSH, SSP7, POC, TAC, MKA (negative result), MJA, MTL, TKS | 3, 65, 84, 101, 154, 161, 207, 230, 234, 281, 289, 305, 308 | |
| Stabilization of proteins | Pressure Heat |
MIG, MJA (protease) POC, MJA (protease), PKOD, MJA (Hsp60) |
121, 161, 200, 230, 305 |
| Recombinant expression of stress proteins in E. coli | TAC, MJA, PKOD, PAL585 (dinF), HXV, MTL, TKS, AFU | 24, 74, 84, 148, 163, 216, 290, 305, 308 | |
| Binding of denatured proteins | SSH, TAC, SSO, PKOD, MJA | 101, 161, 234, 289, 305 | |
| Proteasome function | TAC | 248 | |
| Thermostability of protein | MJA, POC | 200, 230 | |
| Reassembly | Hsp60: SSO, SSH, TAC | 141, 154, 234, 290 | |
| Filament formation | Hsp60: MTL, SSH | 84, 280, 307 | |
| RNA binding and processing | Hsp60: SSO | 249 | |
| Protein modification | P starvation | SAC (phosphorylation) SSO (methylation) |
13, 221 |
Blanks in this column indicate absence of information.
Organisms (in capitals) are: ABR, Acidianus brierleyi; AFU, Archaeoglobus fulgidus; DAM, Desulfurolobus ambivalens; DMO, Desulforococcus mobilis; DSY, Desulforococcus strain SY; HBU, Hyperthermus butyricus; HCU, Halobacterium cutirubrum; HHA, Halobacterium halobium; HMA, Haloarcula marismortui; HME, Haloferax mediterranei; HXV, Haloferax volcanii; MAC, Methanosarcina acetivorans; MBA, Methanosarcina barkeri; MBR, Methanobacterium bryantii; MFE, Methanothermus fervidus; MHU; Methanospirillum hungateii; MIG, Methanococcus igneus; MJA, Methanococcus jannaschii; MMA, Methanosarcina mazei S-6; MKA, Methanopyrus kandleri; MSE, Metallosphaera sedula; MTH, Methanobacterium thermoautotrophicum ΔH; MTL, Methanococcus thermolithotrophicus; MTP, Methanosarcina thermophila TM-1; MVO, Methanococcus voltae; PAB, Pyrodictium abyssi; PAE, Pyrobaculum aerophilum; PAL585, Pyrococcus strain IFREMER585; PBR, Pyrodictium brockii; PES1, Pyrococcus strain ES1; PES4, Pyrococcus strain ES4; PFU, Pyrococcus furiosus; PHO, Pyrococcus horikoshii OT3; POC, Pyrodictium occultum; PKOD, Pyrococcus st. KOD; PWO, Pyrococcus woesei; SAC, Sulfolobus acidocaldarius; SMA, Staphylothermus marimus; SSP7, Sulfolobus Sp. strain 7; SSH, Sulfolobus shibatae; SSO, Sulfolobus solfataricus; TAC, Thermoplasma acidophilum; TPE, Thermococcus peptonophilus; TKS, Thermococcus strain KS-1; TTE, Thermoproteus tenax. DNA (genes) and RNA (transcripts) are shown in italics.
All others are batch experiments.
Part of whole-genome projects.
Protein (first letter in capital and rest in lower case); Hsp60/hsp60, heat shock protein 60/gene belonging to the chaperonins; Hsp70/hsp70, heat shock protein 70/gene belonging to the Hsp70(DnaK) chaperone system; Crx/crx, copper-responsive protein/gene.
Number of induced proteins within parentheses.
STRESS TOLERANCE
Questions
It is difficult to think of a cell or organism that will not be exposed to stressors and suffer stress many times during its life. Does a cell learn anything from stress? Does it learn how to better cope with a second aggression by the same stressor or a similar one? Can a cell develop acquired resistance to a stressor or groups of similar stressors after a first aggression? How about archaea? Are they different from the better-studied bacteria and eucarya?
Research to answer these questions is important because it will not only uncover mechanisms of cell resistance but it will also provide the basis for practical applications aimed at the fortification of cells to enhance survival and recovery. The implications for the biotechnology industry, medicine, planetary exploration, and other endeavors of interest to mankind are obvious and need not be mentioned here.
Cells are equipped to deal with a first encounter with stressors as explained in preceding sections. In addition, there is experimental evidence indicating that cells do acquire higher levels of resistance to a second encounter, although this enhanced resistance (stress tolerance) is of relatively short duration (47, 114, 128–130, 203, 250, 261, 279, 282).
Typically, stress tolerance is defined by measuring cell survival after each of two consecutive stresses. The first is induced by a nonlethal dose of stressor, but the cells are challenged the second time with a dose that would be lethal for nonpretreated cells. If the cells survive the lethal dose, it is concluded that they have developed stress tolerance.
What makes the cell more resistant to stressors? What is the mechanism of stress tolerance?
We mentioned at the beginning of this review that a key component of stress is protein denaturation, followed by aggregation and ultimately cell death. Therefore, the central point of defense for the cell is to prevent protein denaturation and aggregation or to reverse these processes while they are still reversible. The cell has several means of achieving these goals, i.e., stress (heat shock) proteins, molecular chaperones, and chaperonins, as explained throughout this review. Moreover, cells have other molecules and mechanisms to deal with stress. These are thermoprotectants, formation of multicellular structures, and other factors such as intrinsic thermoresistance of the proteins themselves.
Thermoprotectants
A variety of nonprotein molecules also play a role in protecting the cell from the effects of stressors, and some of these molecules may play a role akin to that of the chaperone proteins (41, 122, 128, 190).
Two thermoprotectants have been found in hyperthermophilic archaea: di-myo-inositol phosphate (DIP) and cyclic diphosphoglycerate (cDPG) (128). High concentrations of both DIP and cDPG have been measured in Methanothermus fervidus and Methanopyrus kandleri (122, 128). DIP has been found in A. fulgidus, Pyrodictium occultum, Methanoccus igneus, Pyrococcus woesei, and Pyrococcus furiosus (40, 193, 257).
cDPG and DIP were tested in vitro and found to stabilize certain proteins, but not others, to make them resistant to high temperatures (128). These compounds are believed to be important for thermotolerance in organisms living at high temperatures and for their survival under stress. It is assumed that the enzymes needed to synthesize DIP, cDPG, and other molecules necessary to withstand stress must also be active during heat shock, perhaps due to built-in thermoresistant features (36, 56, 215, 254). These features are not yet completely understood, although clues that might explain why some proteins are more resistant to heat than others are beginning to emerge (115, 156). However, these enzymes may be considered stress proteins, since they would fall within the broad definition of these molecules that includes the characteristics of being present or increased, and active, during and immediately after stress.
Trehalose also plays a role in protein folding and in stress tolerance, at least in some organisms (reference 261 and references therein). The disaccharide has been found in a few archaeal species, although its functions have not been elucidated (61, 194). Likewise, osmolytes that seem to play a role in osmotic adaptation and in the response to osmotic stress have been identified in archaeal cells (54, 68, 122, 190, 233, 266).
Multicellular Structures
Another line of defense of some archaea against mechanical, physical, and chemical stressors is the cell wall (18, 50, 100, 157, 158, 228, 262, 267), which may be supplemented by the formation of supracellular structures. These may be flat or globular and are named biofilm and packet or granule, respectively (48, 134, 197, 306). They may be composed of a single archaeal species, as illustrated in Fig. 16 (197), or of several different organisms, including archaeal and bacterial species (43, 134, 185). An example of the latter mixed microbial structure or consortium, taken from a bioreactor processing organic substrates, is shown in Fig. 17 (180a). Methanosarcinas and other methanogens are found in these granular consortia. In addition, other archaea, for example A. fulgidus, M. jannaschii, and Thermococcus litoralis, can build up biofilms (166, 241). A. fulgidus maintained at the OTG (83°C) does not form a biofilm, but it does so when it is exposed to higher temperatures.
FIG. 16.
Archaeal multicellular structures. M. mazei S-6 packets (A) and lamina (B) are displayed, along with the single-cell morphotype (C) for comparison (197). The diameter of the single cells is ∼3 μm, and the magnification factor is the same for the three panels. The photographs were taken with phase-contrast optics of wet samples from live cultures between the glass slide and coverslip (180a).
FIG. 17.
Archaea-bacterial multicellular structures shown in thin histological sections. (A) Cross section of a granule (multicellular consortium) from a thermophilic (50°C), anaerobic, methanogenic bioreactor. Visible are the cortex and medulla (48, 134) and a large island of methanosarcina cells and packets (arrows) (180a). Hematoxylin-eosin stain. Magnification, ×736. (B) Another section of the same granule in which the presence of Methanosarcina thermophila TM-1 (optimal temperature for growth, 50°C) is demonstrated with a antibody probe for TM-1 by immunofluorescence (180a). Magnification, ×3,680.
M. mazei S-6 packets are considerably more resistant than single cells to mechanical and chemical stressors (180a). Also, induction of a detectable heat shock response in packets, assessed in terms of an increase in the level of the mRNA from the hsp70 locus genes, requires higher temperatures and longer heat shocks than does induction of the response in single cells. The results suggest that the cells inside the packets are protected from environmental changes by comparison with single cells, as illustrated in Fig. 18 (44).
FIG. 18.
Heat resistance of archaeal multicellular structures compared with the single-cell phenotype. Primer extension mapping of the transcription initiation site for M. mazei S-6 grpE was performed. A radiolabelled oligonucleotide primer complementary to bases 57 through 77 within the grpE coding region was used with 10 μg of total RNA from single cells (lanes 1 to 3) or packets (lanes 4 to 6) per test. Single cells and packets were grown at 37°C (lanes 1 and 4) or heat shocked at 45°C for 30 (lanes 2 and 5) or 60 (lanes 3 and 6) min. The primer-extended products were electrophoresed in a 6% acrylamide sequencing gel in parallel with the products of a sequencing reaction that was done with the same primer and the dideoxy chain termination method (lanes G, A, T, and C). These lanes show the complementary (antisense) strand sequence. The coding (sense) strand sequence and the initiation site (asterisk) are shown on the left. Reprinted from reference 44 with permission of the American Society for Microbiology.
The archaeal multicellular structures have an intercellular connective material made of acidic polysaccharides and proteins (86, 157, 158). There is some information about the biochemistry of these components, but more research is needed to elucidate their role, thermoresistance, insulating properties, and other characteristics that might achieve cell protection without interfering with the influx of nutrients and the efflux of catabolites. The bioreactor granules, for example, have a net of microtubes crisscrossing the interior and the cortex, in which the connective material is abundant, as shown in Fig. 19 (185). These tubes may provide a circulation network to ensure that the cells in the interior of the granules, even those in the deepest zones, receive nutrients and find a way to discharge what they produce. It is likely that the granules have evolved because of the frequent modifications in temperature, pH, and other conditions that occur in the ecosystems in which they are found and are better equipped than the solitary cells to withstand environmental changes.
FIG. 19.
Superficial, histological thin section of a granule like that shown in Fig. 17, passing through the cortex. Visible are circular openings which represent cross sections of the tubes that crisscross the cortex and enable communication between different zones of the granule (180a, 185). Hematoxylin-eosin stain. Magnification, ×800.
Other Factors
The cell surely possesses other means in addition to all those already discussed to protect itself and withstand environmental changes. For example, there must be several genes and proteins that are required to produce the known chaperones and chaperonins, the thermoprotectants, and the materials to form a biofilm or granule, which must be active during stress. Some of these proteins must be enzymes and other cofactors of various types, with built-in structural features that must make them more stable than the rest under stress conditions (13, 36, 56, 102, 115, 156, 215, 224, 254). Among this group of intrinsically stress-resistant molecules are probably the chaperones and chaperonins themselves.
It has been reported recently that Sulfolobus acidocaldarius, a thermoacidophilic archaeon, changes its motility according to the ambient temperature (169). The findings suggested that motility changes were a response to heat stress and were a mechanism for escaping lethal temperatures. This is an interesting working hypothesis that ought to be tested by comparing archaeal species from various habitats with different temperatures.
Lastly, some conditions in the environment may increase stress tolerance in organisms that live in extreme ecosystems, for example deep down on the ocean floor. It has been reported that hydrostatic pressure enhances thermotolerance and protein stability (121, 128–130, 138, 159, 200, 201, 232). For instance, Pyrococcus strain ES4 (OTG, 99°C) acquired thermotolerance to a heat shock at 105°C (a lethal temperature for ES4) after exposure to 102°C for 90 min (130). Cells grown at low pressure (3 MPa) also acquired thermotolerance when they were transferred to a culture at 22 MPa. This thermotolerance lasted for a considerable time after the cells were returned to 3 MPa, suggesting that hyperpressure had set in motion events that were meant to persist.
PERSPECTIVES, OPEN QUESTIONS, AREAS FOR EXPLORATION, AND APPLICATIONS
Biochemistry and Function of Archaeal Chaperones and Chaperonins
The functions of the archaeal Hsp70, Hsp40, and GrpE (the Hsp70 machine) proteins have not been studied in vitro or in vivo to any significant extent; they are certainly much less extensively studied than their homologs from the bacteria and eucarya. It is not known to what extent the archaeal proteins resemble those from the other two domains in their interaction with each other and with substrates, nucleotides, ions, cochaperones, auxiliary factors, and regulatory molecules.
There are several aspects that are unique to the archaea and whose elucidation will enhance our understanding of protein biogenesis and molecular chaperoning in general. For example, is prefoldin (89, 287) critical for protein folding in archaea, and does it require another complementary chaperonin system? Does the archaeal Hsp70 machine interact with chaperonins that have yet to be identified? Does the Hsp70 machine interact with the type (or group) II chaperonins in archaea? If so, how? What is known about interactions between the Hsp70 machine and the chaperonins has been learned mostly from studies in bacteria. These have type I chaperonins which differ from those of type II. Along the same line, another question is pertinent: What, if anything, replaces the Hsp70 machine in the archaeal species that lack it? Is there an archaeal equivalent of the Hsc66 protein in some bacterial species in addition to Hsp70(DnaK) (260), which performs at least some of the functions typical of the latter? The answers to these questions will most probably shed light on the protein-folding problem in general. If a substitute for Hsp70(DnaK) were found in archaea, it would probably be the ancestor of a eucaryal homolog, whose functions could then be better understood by studying both in parallel.
All the archaeal species that lack the Hsp70 machine and that have been examined to identify chaperonins have been found to possess the type II system. This raises the following question: does the archaeal chaperonin system (thermosome) require interaction with a second system that would play a role similar to that of the Hsp70 machine? Data from eukaryotic models suggest that the cytosolic chaperonin system (type II) does not interact with the Hsp70 machine. It is not safe, however, to conclude that the same is true for the archaea, or at least for all archaea. Although the archaeal and eukaryotic chaperonin complexes (e.g., CCT and the thermosome), with their respective subunits, are similar and are both classified as type II, they do show structural differences (Fig. 13). Hence, differences in the function and mechanism of action between the thermosome and CCT are to be expected. What are their respective, preferred substrates? Do they both interact with prefoldin?
It would also be of interest, for example, to determine the substrates for the archaeal Hsp70 machine and how this machine recognizes the substrates. Differences by comparison with the bacterial machine will probably be found, since differences in binding properties between the cytosolic and mitochondrial Hsp70 molecules, for example, have been demonstrated (7).
Elucidation of the mechanism of polypeptide folding in vitro by the archaeal chaperonin complex should be pursued. For example, additional data ought to be obtained on the open stage of the thermosome, polypeptide capture, and release. Cryoelectron tomography of a thermosome formed only by recombinant α subunits expressed in E. coli revealed the 3-D structure of the open state (Fig. 12) (217). This experimental model of a homopolymeric complex was about 18 nm long and had a diameter of approximately 15 nm. The central channel was calculated to have a volume probably too large to influence the selection of which substrate would enter the cavity solely based on size. It seems unlikely that such a large cavity would have the plasticity to discriminate between substrates that differ in size, particularly when the differences are not pronounced and when the substrates are very large or very small. In contrast, the estimated volume of the closed chamber was small and would accommodate substrates (polypeptides) of up to 50 kDa at most, which is about the same as that suggested for the bacterial chaperonin complex (251). Several questions remain unanswered. For instance, is prefoldin involved in substrate selection and/or presentation to the thermosome and in the closing of the folding chamber? Experiments ought to be done to elucidate how a folding cavity without a removable lid passes from an acceptor to a folding phase. Moreover, a comparison between the chaperonins of hyperthermophiles and those of mesophiles and psychrophiles should provide insights into the evolution of the system and its modifications to cope with different environments.
An area in need of development pertains to in vivo studies of chaperone and chaperonin function and mechanism of action. In this regard, the most rewarding work will be the standardization of transformation vectors and protocols and the identification and generation of mutants. For some archaeal groups, like the methanogens, the methods necessary for molecular genetic manipulations are still scarce (46, 199, 285).
Very little is known also about archaeal stress proteins that are not members of the Hsp70 and Hsp60 families and that may or may not be chaperones but still play a role in the stress response or in stress tolerance. Small heat shock proteins (sHsp) (149, 150) are good candidates for study. Also, experiments with bacterial models are revealing novel genes that respond to environmental stressors (21, 112). Do archaea have similar genes? Comparative genomics will help initiate experimental research, which will probably unveil antistress mechanisms that are as important as those attributed to the better-known chaperones and chaperonins and should help our understanding of stress resistance in eukaryotes. It is also possible that these novel mechanisms will be important during physiologic differentiation and during the development of supracellular structures in response to stress.
Regulation
Regulation of stress genes in the archaea is an open field and a very intriguing one. In brief, the hsp70 locus genes produce proteins with bacterial characteristics, betraying their origin, but they have to function within cells containing a genome in which the transcription machinery is of the archaeal (eucarya-like) type. This suggests a hybrid heritage. It remains to be established when, and how, the hsp70 locus genes came to be located in the archaea that have them and how are they regulated. It is known that the transcription initiation factors in archaea are homologs of some of the eucaryal transcription factors (16, 264, 276). However, it is unclear whether the regulatory signals and factors are of bacterial or eucaryal type or whether they have novel, archaeal, characteristics (180). These points remain to be elucidated and are relevant to basic and applied sciences. For example, stress proteins are expressed as a reaction to pollutants (21, 112). Moreover, heat shock gene promoters have been used, along with reporter genes, to detect stressors of relevance to public health (15, 69, 70, 269). Can archaeal heat shock gene promoters be used similarly? Will these archaeal promoters function inside eukaryotic cells?
Likewise, very little is known about posttranscriptional regulation. The information available comes from genome analyses, but experimental data are scarce. For instance, there are very few data on transcription termination of archaeal stress genes. Examination of sequences and computer analyses suggest that the formation of an mRNA hairpin may be the device used for termination by some genes (42, 44, 45, 49, 50, 179, 181–184). However, a recent computer analysis of available archaeal genomes suggested that hairpin formation was not the mechanism used by most genes (291).
Cell Differentiation, Development, and Adaptation
Heat shock genes in many organisms are expressed under normal, physiological conditions. This basal or constitutive expression is low level by comparison with that which occurs in response to stressors. Furthermore, many heat shock genes respond to normal, physiological signals related to cell differentiation and tissue and organism development (95, 205, 240, 253). This aspect of stress gene regulation has not been studied in the archaea to any significant extent. Preliminary data suggest that the hsp70(dnaK) locus genes in M. mazei S-6 are constitutively expressed at levels that vary with the stage of the developmental cycle, which includes the formation of packets, single cells, and lamina (180a).
The lamina morphotype is a flat structure, typically with a thickness of one cell (although “old” lamina may be thicker and encompass two or three cells, vertically) and abundant intercellular connective material. It resembles a biofilm formed by many bacteria and some archaea, as discussed above (see “Multicellular structures”). However, the lamina morphotype has peculiar characteristics that distinguish it from other biofilms. For instance, a lamina does not adhere to glass or plastic surfaces. It sits on the bottom of the culture flask, or floats when the culture is stirred or when there is methane trapped within it that provides buoyancy. Packets have a different anatomy. They are compact globular structures, with the cells present inside a layer of intercellular material, and may reach a diameter equivalent to 10 cells or more. These globular masses are very resistant to disruption by chemical and mechanical means and do not allow antibiotics to reach the cells inside (180a; see also reference 40 for possible applications of packet formation). Why is it that M. mazei S-6 has evolved the capacity to form laminas and packets? Do these structures represent developmental stages different from the single-cell phenotype? Did the capacity to form packets (the most resistant of the three phenotypes to stressors) evolve as a survival mechanism to cope with harmful environmental changes? Research addressing these questions will provide information useful to our understanding of the evolution of histogenesis and its advantages for survival.
Voids To Be Filled: Proteases and Auxiliary Factors
There are many molecules and molecular families directly or indirectly related to stress and/or molecular chaperoning and protein management in the bacteria and eucarya that have not yet been identified or well characterized in archaea. A few examples follow.
The Clp system occurs in bacteria and consists of a family of ATP-dependent proteases that are relatively well studied in E. coli (96–98) and Bacillus subtilis (92). Proteases do, indeed, play a critical role in maintaining the physiological array of proteins and their optimal levels by, among other mechanisms, eliminating those that are damaged and whose accumulation inside the cell would be deleterious (60, 96–98, 123, 124). It is not known whether the archaea harbor a system resembling the bacterium-type Clp, or any other that might take its place, but with unique, archaeal characteristics. A suggestion in this direction is provided by a report of positive hybridization obtained with a probe for Clp and DNA from Methanosarcina acetivorans (98).
The proteasome is another protease system (14, 39, 60, 167, 314). It is a large structure with a central cavity or chamber, formed by stacked rings constituted of several protein subunits. Superficially, the proteasome resembles the thermosome, but it differs from the latter in several important aspects and has different functions. The proteasome digests polypeptides inside its central chamber rather than promoting their correct folding as the thermosome is thought to do. Although a proteasome has been identified in a couple of archaeal species (167, 195, 196), little is known about its function, mechanism of action, and diversity according to ecosystem or phylogenetic branch. Experiments done with T. acidophilum have demonstrated that proteasome inhibition does not affect cell viability under physiologic conditions but causes growth arrest in heat-shocked cultures (248). The data indicated that the proteasome plays a role in the survival of T. acidophilum under stress.
The group of sHsp is widespread, diverse, and important for bacteria and eukaryotes (76, 78, 177, 206, 227). Some information is beginning to emerge about sHsp in the archaea. For example, an sHsp of 16.5 kDa from M. jannaschii has been purified and crystallized (148, 149). The folding unit was found to consist of nine β-strands in two sheets, two short helices, and one short β-strand, with one of the β-strands coming from an adjacent subunit. Twenty-four monomers formed a large (∼400-kDa) spherical complex with octahedral symmetry (Fig. 20). The sphere appeared to enclose a cavity with a volume of 140,000 Å3, i.e., about half that of the estimated volume of the inner space in the GroEL barrel. In vitro studies showed that the M. jannaschii sHsp protected other proteins from heat denaturation and prevented their aggregation (150). Thus, this archaeal protein seems to play a role similar to that of the sHsp homologs in organisms of the other two phylogenetic domains.
FIG. 20.
Space-filling model of the multimeric complex formed by an sHsp from the hyperthermophilic methanogenic archaeaon M. jannaschii. The model was derived from crystallographic analysis at 2.9-Å resolution, and each atom is represented by a small sphere. The whole structure consists of 24 identical protein (the sHsp) subunits (16.5 kDa each) arranged in regular octahedral symmetry with a total molecular mass of about 400 kDa. Twelve dimers are represented by different colors, with monomers in each dimer shown by different shading of the same color. The complex is a hollow sphere with eight triangular and six square openings or windows that connect the inner cavity with the outside. The view in the figure is along the axis that aligns two triangular windows, one in front, closest to the observer, and the other on the back of the sphere, farthest from the observer. Reprinted from reference 149 with permission of the publisher.
Among the sHsp of E. coli, one has been identified very recently that binds nucleic acids (160). Although its role in vivo was not elucidated, it was deemed important because of its abundance. Do archaea have homologs that also bind nucleic acids (and assist them in some way) rather than polypeptides?
On the other side of the spectrum, the occurrence and role of high-molecular-weight (90 kDa or higher) (Table 1) (30, 53, 93, 175, 177, 218, 236, 293) stress proteins and chaperones in the archaea have not been determined. This is another area of archaeal research open for exploration that will benefit from genomics and that in turn will help the study of chaperone machines different from the Hsp70(DnaK) and chaperonin systems.
Cooperation between chaperones and chaperonins seem to be an important physiological characteristic of the cell, as mentioned in previous sections of this review. This idea is being extended to encompass cooperation with regulatory or auxiliary factors, or cofactors (38, 62, 82, 88, 127). Examples of the latter are Hip (for “Hsc-70 interacting protein”) and Hop (for “Hsp70/Hsp90-organizing protein”); the latter is also named p60, IEF-SSP-3521, extendin, or Sti1 (38, 127). These factors occur in eukaryotes; hence, it is justified to suspect that they (or their primitive ancestors) might also be present in the archaea, or at least in some of them.
Beyond Chaperones and Chaperonins
There are other important areas for investigation that extend beyond chaperones and chaperonins. One pertains to all the other stress genes and proteins alluded to in previous sections. A second topic that also deserves investigation and promises practical dividends is stress resistance. Archaea are particularly interesting in this regard because they encompass an array of species that inhabit a wide variety of ecosystems, some very similar to the optimal conditions for human cells and some that are extreme by comparison, with all gradations in between (9, 11, 12, 59, 178, 186, 232, 297). To what extent do protein biogenesis under physiological conditions and molecular chaperoning under stress differ in species that live in disparate ecosystems? How different must two ecosystems be in terms of temperature, pH, salinity, or pressure to require differences in the chaperoning mechanisms of the organisms that thrive in them? What are the sensors for temperature stress, for example, in psychrophilic compared with thermophilic species? All these questions point to specific aspects of the stress response in archaea that merit investigation. Again, genome analyses are helping our knowledge to progress. Some experimental data on the thermostability of DNA and some proteins are already available (13, 36, 56, 101, 102, 173, 174, 215, 224, 254), and they should also enhance the implementation of projects to clarify more completely how life is sustained in extreme environments.
Similarly, thermoprotectants are being identified (128), as are molecules that may be seen as insulants, like histones and small, nonhistone DNA-binding proteins (226, 237). Progress in this area should improve our understanding of the mechanisms used by the archaea, particularly those that live in extreme environments, to pack DNA and to unpack it to allow gene transcription.
Studies of the thermoprotectants made by the archaea followed by laboratory synthesis of similar compounds with desired properties should provide a means of increasing stress resistance to a variety of stressors in useful organisms. These improved organisms could then be used in a variety of activities such as decontamination of toxic wastes and planetary exploration.
Stress resistance and survival depend to a great extent on the properties of the cell envelope. Some archaea have an S-layer with protective functions, allowing the passage of water and other molecules (50, 158, 262) [and other articles in the same issue], (267). The characteristics of the S-layer proteins of M. mazei S-6 seem to have evolved to provide rapid adaptation to environmental changes and to promote the formation of multicellular structures when necessary (50, 262). This field, encompassing stress resistance mechanisms mediated by cell surface molecules and intercellular connective material, will probably undergo major advances in the near future if enough attention is directed to it. Firstly, it will be necessary to understand the mechanism responsible for the formation of multicellular structures in response to environmental signals. Second, the proteins and other molecules involved will have to be characterized and their genes will have to be studied to determine what induces them and how they are regulated. Finally, manipulation of these genes ought to provide means of engineering multicellular structures as needed, for instance, in bioreactors with violent flow and drastic, unpredictable oscillations in temperature or pH. Moreover, learning about the formation of multicellular structures in the archaea will help in the development of methods to prevent the formation of or to destroy undesirable biofilms, such as those involved in biofouling and in disease.
Along these lines, the prospects are promising for multicellular structures with several archaeal and bacterial species in a food web, like the granular consortia of methanogenic bioreactors. How are these consortia formed at the molecular level? What brings the various species together, and what keeps them close to each other? What is the composition of the extracellular connective material? When and how do the genes responsible for the synthesis of the extracellular material become activated?
If one considers the complex architecture of the multicellular structures of M. mazei S-6 (197) and even more so that of the granular consortia (48, 185), it is easy to imagine that the building process must involve a complicated mechanism with stress sensors, genes, and molecules of various kinds. The process must include not only the synthesis and export of the building blocks for the intercellular connective material but also the construction of the microtubes that crisscross the granules, with the participation, most probably of extracellular proteases.
CONCLUSION
Inferences from the data and hypotheses have been enunciated throughout the text, mostly at the end of each subsection. In brief, there is considerable information on the archaeal Hsp70 family of chaperones in what pertains to their genes but very little in what pertains to their function. Virtually no experimental data are available that will elucidate what these molecules do and how, when, and where they will do it. Since the proteins are very similar to bacterial homologs whose function have been extensively studied in vitro and in vivo, one may be tempted to extrapolate the data and assume that the archaeal molecules do the same things in the same way as the bacterial chaperones. This may be true but only to a degree. The intracellular environment and the batteries of cochaperones and auxiliary molecules that interact with the Hsp70 team may be different in archaea from in bacteria. This is certainly an area of biochemistry that deserves investigation; it promises to unveil details of molecular interactions that studies with bacteria cannot reveal and that may be useful to our understanding of comparable interactions in the eukaryotic cell.
In contrast to Hsp70, the functions of the archaeal Hsp60 molecules have been studied more extensively than their genes. Actually, initial studies of archaeal chaperonin molecules helped research on the chaperonins of the eukaryotic-cell cytosol. However, the bulk of our information comes from in vitro studies. We must wait for in vivo experiments to show the real-life functions of the archaeal chaperonins and their mechanisms of action inside a living cell.
sHsp are beginning to emerge as important players in cell physiology and stress resistance. A few have been identified in archaea that are PPIases, and another has been crystallized. Studies with archaeal sHsp should enhance our understanding of the chaperone functions of these proteins, their interaction and cooperation with other chaperone and chaperonin systems in the cell, and their role in the assembly of the cytoskeleton. Archaea do not possess a cytoskeleton, in contrast to eukaryotes, but several reports (43, 84, 280, 307) suggest that they might have a primitive version of it. This hypothesis merits further investigation. The fact that archaea do have sHsp and chaperonins known to associate with tubulin and actin in eukaryotes is another incentive to search for cytoskeletal ancestors in the archaea.
The study of archaeal and mixed (archaeal-bacterial) supracellular structures has been rewarding so far, to the extent that it has revealed complex formations with regional heterogeneity and hints of organization as if they were primitive tissues and has shown that these structures are more resistant to stressors than are the single-celled morphotypes. These findings ought to serve as the starting point for investigations of the molecular and genetic mechanisms involved in this primeval type of histogenesis and their role in stress resistance.
ACKNOWLEDGMENTS
We thank Andrea De Biase for assistance, Anthony Hickey for his help with the tabulation of data, Tammi Stock for initial word processing, the members of the Photo-Art unit of this Center for the graphic and art works, and our collaborators throughout the years (too many to mention here, but the names of most of them appear in the list of references) for their contributions. We also thank our colleagues who provided photographs for figures, as follows: Helen Saibil (Birkbeck College, University of London, London, United Kingdom) for Fig. 11; Wolfgang Baumeister and Michael Nitsch (Max Plack Institute for Biochemistry, Martinsried, Germany) for Fig. 12; José M. Valpuesta (Centro Nacional de Biotecnología, Universidad Autónoma de Madrid, Madrid, Spain) for Fig. 13; J. M. Archibald, J. M. Logsdon, and W. Ford Doolittle (Department of Biochemistry, Dalhousie University, Halifax, Canada) for Fig. 14 and 15; and R. Kim, S.-H. Kim, and K. K. Kim (Lawrence Berkeley National Laboratory and Department of Chemistry, University of California at Berkeley, Berkeley, Calif.) for Fig. 20. Our gratitude also goes to John M. Archibald, José M. Valpuesta, Wolfgang Baumeister, and Helen Saibil for critically reading the manuscript, and for their suggestions.
Work in the A.J.L.M. and E.C.M. laboratories was supported by grants from NYSERDA, the U.S. Department of Energy (DOE), and the National Science Foundation (NSF). M.L. was supported by the Danish Technical Science Council.
REFERENCES
- 1.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Pontén T, Alsmark U C M, Podowski R M, Näslund A K, Eriksson A-S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 2.Andrä S, Frey G, Jaenicke R, Stetter K O. The thermosome from Methanopyrus kandleri possesses an NH4+-dependent ATPase activity. Eur J Biochem. 1998;255:93–99. doi: 10.1046/j.1432-1327.1998.2550093.x. [DOI] [PubMed] [Google Scholar]
- 3.Andrä S, Frey G, Nitsch M, Baumeister W, Stetter K O. Purification and structural characterization of the thermosome from the hyperthermophilic archaeum Methanopyrus kandleri. FEBS Lett. 1996;379:127–131. doi: 10.1016/0014-5793(95)01493-4. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L, Tatusov R L, Wolf Y I, Walker D R, Koonin E V. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 5.Archibald J M, Logsdon Jr J M, Doolittle W F. Recurrent paralogy in the evolution of archaeal chaperonins. Curr Biol. 1999;9:1053–1056. doi: 10.1016/s0960-9822(99)80457-6. [DOI] [PubMed] [Google Scholar]
- 6.Arsène F, Tomoyasu T, Mogk A, Schirra C, Schulze-Specking A, Bukau B. Role of region C in regulation of the heat shock gene-specific sigma factor of Escherichia coli, ς32. J Bacteriol. 1999;181:3552–3561. doi: 10.1128/jb.181.11.3552-3561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artigues A, Crawford D L, Iriarte A, Martinez-Carrion M. Divergent Hsc70 binding properties of mitochondrial and cytosolic aspartate aminotransferase. J Biol Chem. 1998;273:33130–33134. doi: 10.1074/jbc.273.50.33130. [DOI] [PubMed] [Google Scholar]
- 8.Bairoch A. Prosite: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardwell J C A, Craig E A. Major heat shock gene of Drosophila and the Escherichia coli heat-inducible dnaK gene are homologous. Proc Natl Acad Sci USA. 1984;81:848–852. doi: 10.1073/pnas.81.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baross J A, Holden J F. Overview of hyperthermophiles and their heat-shock proteins. Adv Protein Chem. 1996;48:1–34. doi: 10.1016/s0065-3233(08)60360-5. [DOI] [PubMed] [Google Scholar]
- 13.Baumann H, Knapp S, Ladenstein R, Härd T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Struct Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 14.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 15.Belkin, S., Y. Levi, S. Dunkan, and D. Touati. Death by disinfection: molecular approaches to understanding bacterial sensitivity and resistance to free chlorine. In E. Rosenberg (ed.), Microbial ecology and infectious diseases, in press. American Society for Microbiology, Washington, D.C.
- 16.Bell S D, Jackson S P. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 17.Bergonia G B, Salin M L. Elevation of superoxide dismutase in Halobacterium halobium by heat shock. J Bacteriol. 1991;173:5582–5584. doi: 10.1128/jb.173.17.5582-5584.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beveridge T J, Stewart M, Doyle R J, Sprott G D. Unusual stability of the Methanospirillum hungatei sheath. J Bacteriol. 1985;162:728–737. doi: 10.1128/jb.162.2.728-737.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchard J L, Schmidt G W. Pervasive migration of organellar DNA to the nucleus in plants. J Mol Evol. 1995;41:397–406. doi: 10.1007/BF00160310. [DOI] [PubMed] [Google Scholar]
- 20.Blaszczak A, Georgopolous C, Liberek K. On the mechanism of FtsH-dependent degradation of the ς32 transcriptional regulator of Escherichia coli and the role of the DnaK chaperone machine. Mol Microbiol. 1999;31:157–166. doi: 10.1046/j.1365-2958.1999.01155.x. [DOI] [PubMed] [Google Scholar]
- 21.Blom A, Harder W, Matin A. Unique and overlapping pollutant stress proteins of Escherichia coli. Appl Environ Microbiol. 1992;58:331–334. doi: 10.1128/aem.58.1.331-334.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borchiellini C, Boury-Esnault N, Vacelet J, Le Parco Y. Phylogenetic analysis of the Hsp70 sequences reveals the monophyly of metazoa and specific phylogenetic relationships between animals and fungi. Mol Biol Evol. 1998;15:647–655. doi: 10.1093/oxfordjournals.molbev.a025968. [DOI] [PubMed] [Google Scholar]
- 23.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouyoub A, Barbier G, Querellou J, Forterre P. A putative SOS repair gene (dinF-like) in a hyperthermophilic archaeon. Gene. 1995;167:147–149. doi: 10.1016/0378-1119(95)00651-6. [DOI] [PubMed] [Google Scholar]
- 25.Brown J W, Daniels E J, Reeve J N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16:287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- 26.Bucca G, Ferina G, Puglia A M, Smith C P. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol Microbiol. 1995;17:663–674. doi: 10.1111/j.1365-2958.1995.mmi_17040663.x. [DOI] [PubMed] [Google Scholar]
- 27.Bucca G, Hindle Z, Smith C P. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J Bacteriol. 1997;179:5999–6004. doi: 10.1128/jb.179.19.5999-6004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchberger A, Schröder H, Büttner M, Valencia A, Bukau B. A conserved loop in the ATPase domain of the DnaK chaperone is essential for stable binding of GrpE. Struct Biol. 1994;1:95–101. doi: 10.1038/nsb0294-95. [DOI] [PubMed] [Google Scholar]
- 29.Buchberger A, Valencia A, McMacken R, Sander C, Bukau B. The chaperone function of DnaK requires the coupling of ATPase activity with substrate binding through residue E171. EMBO J. 1994;13:1687–1695. doi: 10.1002/j.1460-2075.1994.tb06433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchner J. Hsp90 & Co.—a holding for folding. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 31.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Goeghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 32.Bustard K, Gupta R S. The sequences of heat shock protein 40 (DnaJ) homologs provide evidence for a close evolutionary relationship between the Deinococcus-Thermus group and cyanobacteria. J Mol Evol. 1997;45:193–205. doi: 10.1007/pl00006219. [DOI] [PubMed] [Google Scholar]
- 33.Canganella F, Gonzalez J M, Yanagibayashi M, Horikoshi K. Pressure and temperature effects on growth and viability of the hyperthermophilic archaeon Thermococcus peptonophilus. Arch Microbiol. 1997;168:1–7. doi: 10.1007/s002030050462. [DOI] [PubMed] [Google Scholar]
- 34.Caplan A J, Douglas M G. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caplan A J, Tsai J, Casey P J, Douglas M G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J Biol Chem. 1992;267:18890–18895. [PubMed] [Google Scholar]
- 36.Cavagnero S, Zhou Z H, Adams M W W, Chan S I. Response of rubredoxin from Pyrococcus furiosus to environmental changes: implications for the origin of hyperthermostability. Biochemistry. 1995;34:9865–9873. doi: 10.1021/bi00031a007. [DOI] [PubMed] [Google Scholar]
- 37.Cheetham M E, Brion J P, Anderton B H. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem J. 1992;284:469–476. doi: 10.1042/bj2840469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S, Smith D F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and Hsp90 chaperone machinery. J Biol Chem. 1998;273:35194–35200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- 39.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciulla R A, Burgraff S, Stetter K O, Roberts M F. Occurrence and role of di-myo-inositol-1,1′-phosphate in Methanococcus igneus. Appl Environ Microbiol. 1994;60:3660–3664. doi: 10.1128/aem.60.10.3660-3664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciulla R A, Roberts M F. Effects of osmotic stress on Methanococcus thermolithotrophicus: 13C-edited 1H-NMR studies on osmolyte turnover. Biochim Biophys Acta. 1999;1427:193–204. doi: 10.1016/s0304-4165(99)00033-1. [DOI] [PubMed] [Google Scholar]
- 42.Clarens M, Macario A J L, Conway de Macario E. The archaeal dnaK-dnaJ gene cluster: organization and expression in the methanogen Methanosarcina mazei. J Mol Biol. 1995;250:191–201. doi: 10.1006/jmbi.1995.0370. [DOI] [PubMed] [Google Scholar]
- 43.Condo I, Ruggero D, Reinhardt R, Londei P. A novel aminopeptidase associated with the 60kDa chaperonin in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 1998;29:775–785. doi: 10.1046/j.1365-2958.1998.00971.x. [DOI] [PubMed] [Google Scholar]
- 44.Conway de Macario E, Clarens M, Macario A J L. Archaeal grpE: transcription in two different morphologic stages of Methanosarcina mazei and comparison with dnaK and dnaJ. J Bacteriol. 1995;177:544–550. doi: 10.1128/jb.177.3.544-550.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway de Macario E, Dugan C B, Macario A J L. Identification of a grpE heat-shock gene homolog in the archaeon Methanosarcina mazei. J Mol Biol. 1994;240:95–101. doi: 10.1006/jmbi.1994.1422. [DOI] [PubMed] [Google Scholar]
- 46.Conway de Macario E, Guerrini M, Dugan C B, Macario A J L. Integration of foreign DNA in an intergenic region of the archaeon Methanosarcina mazei without effect on transcription of adjacent genes. J Mol Biol. 1996;262:12–20. doi: 10.1006/jmbi.1996.0494. [DOI] [PubMed] [Google Scholar]
- 47.Conway de Macario E, Macario A J L. Heat-shock response in Archaea. Trends Biotechnol. 1994;12:512–518. doi: 10.1016/0167-7799(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 48.Conway de Macario E, Macario A J L. Diversity, dynamics and topographic arrangement of microorganisms are essential parameters that identify a microbial consortium. In: Priest F G, Ramos-Cormenzana A, Tindall B J, editors. Bacterial diversity and systematics. New York, N.Y: Plenum Press; 1994. pp. 161–171. [Google Scholar]
- 49.Conway de Macario E, Macario A J L. Transcription of the archaeal trkA homolog in Methanosarcina mazei S-6. J Bacteriol. 1995;177:6077–6082. doi: 10.1128/jb.177.21.6077-6082.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway de Macario E, Macario A J L. S-layer and ABC transporters in methanogenic archaea. FEMS Microbiol Rev. 1997;20:59–64. [Google Scholar]
- 51.Conway de Macario E, Thomsen J, Hausner W, Macario A J L. Keystone Symposium. Archaea: bridging the gap between Bacteria and Eukarya. 1999. Eucaryal and archaeal features of the Methanosarcina mazei S-6 TATA-binding protein (TBP) p. 31. [Google Scholar]
- 51a.Conway de Macario, E., and A. J. L. Macario. Unpublished data.
- 52.Cowing D W, Bardwell J C A, Craig E A, Woolford C, Hendrix R W, Gross C A. Consensus sequence for Escherichia coli heat shock gene promoters. Proc Natl Acad Sci USA. 1985;82:2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Csermely P, Schnaider T, Sôti C, Prohászka Z, Nardai G. The 90 kDa molecular chaperone family: structure, function and clinical applications. Pharmacol Ther. 1998;79:129–168. doi: 10.1016/s0163-7258(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 54.Da Costa M S, Santos H, Galinski E A. An overview of the role and diversity of compatible solutes in bacteria and archaea. Adv Biochem Eng Biotechnol. 1998;61:118–153. doi: 10.1007/BFb0102291. [DOI] [PubMed] [Google Scholar]
- 55.Daniels C J, McKee A H Z, Doolittle W F. Archaebacterial heat-shock proteins. EMBO J. 1984;3:745–749. doi: 10.1002/j.1460-2075.1984.tb01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danson M J, Hough D W. Structure, function and stability of enzymes from the archaea. Trends Microbiol. 1998;6:307–314. doi: 10.1016/s0966-842x(98)01316-x. [DOI] [PubMed] [Google Scholar]
- 57.Darcy T J, Hausner W, Awery D E, Edwards A M, Thomm M, Reeve J N. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J Bacteriol. 1999;181:4424–4429. doi: 10.1128/jb.181.14.4424-4429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57a.De Biase, A., J. Thomsen, W. Hausner, M. W. Thomm, A. J. L. Macario, and E. Conway de Macario. Unpublished data.
- 58.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;329:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 59.DeLong F. Everything in moderation: archaea as ‘non-extremophiles’. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 60.DeMartino G N, Slaughter C A. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 61.Desmarais D, Jablonski P, Fedarko N S, Roberts M F. 2-sulfotrehalose, a novel osmolyte in haloalkaliphilic archaea. J Bacteriol. 1997;179:3146–3153. doi: 10.1128/jb.179.10.3146-3153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 63.Diaz-Perez S V, Alatriste-Mondragon F, Hernandez R, Birren B, Gunsalus R P. Bacterial artificial chromosome (BAC) library as a tool for physical mapping of the archaeon Methanosarcina thermophila TM-1. Microb Comp Genomics. 1997;2:275–286. doi: 10.1089/omi.1.1997.2.275. [DOI] [PubMed] [Google Scholar]
- 64.DiRuggiero J, Santangelo N, Nackerdien Z, Ravel J, Robb F T. Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1997;179:4643–4645. doi: 10.1128/jb.179.14.4643-4645.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ditzel L, Löwe J, Stock D, Stetter K O, Huber H, Huber R, Steinbacher S. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 66.Doolittle W F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 67.Doolittle W F, Logsdon J M. Archaeal genomics: do archaea have a mixed heritage? Curr Biol. 1998;8:R209–R211. doi: 10.1016/s0960-9822(98)70127-7. [DOI] [PubMed] [Google Scholar]
- 68.D’Souza S E, Altekar W, D’Souza S F. Adaptive response of Haloferax mediterranei to low concentrations of NaCl (<20%) in the growth medium. Arch Microbiol. 1997;168:68–71. doi: 10.1007/s002030050471. [DOI] [PubMed] [Google Scholar]
- 69.Dukan S, Dadon S, Smulksi D R, Belkin S. Hypochlorous acid activates the heat shock and soxRS systems of Escherichia coli. Appl Environ Microbiol. 1996;62:4003–4008. doi: 10.1128/aem.62.11.4003-4008.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dyk T K V, Mararian W R, Konstantinov K B, Young R M, Dhurjarti P S, Larossa R A. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminiscence gene fusions. Appl Environ Microbiol. 1994;60:1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellis M J, Knapp S, Koeck P J B, Fakoor-Biniaz Z, Landenstein R, Hebert H. Two-dimensional crystallization of the chaperonin TF55 from the hyperthermophilic archaeon Sulfolobus solfataricus. J Struct Biol. 1998;123:30–36. doi: 10.1006/jsbi.1998.4002. [DOI] [PubMed] [Google Scholar]
- 72.Ellis R J, van der Vies S M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- 73.Embley T M, Hirt R P. Early branching eukaryotes? Curr Opin Genet Dev. 1998;8:624–629. doi: 10.1016/s0959-437x(98)80029-4. [DOI] [PubMed] [Google Scholar]
- 74.Emmerhoff O J, Klenk H P, Birkeland N K. Characterization and sequence comparison of temperature-regulated chaperonins from the hyperthermophilic archaeon Archaeoglobus fulgidus. Gene. 1998;215:431–438. doi: 10.1016/s0378-1119(98)00314-x. [DOI] [PubMed] [Google Scholar]
- 75.Fernandes M, O’Brien T, Lis J T. Structure and regulation of heat shock gene promoters. In: Morimoto R I, Tisieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 375–393. [Google Scholar]
- 76.Ferrari D M, Söling H-D. The protein disulphide-isomerase family: unravelling a string of folds. Biochem J. 1999;339:1–10. [PMC free article] [PubMed] [Google Scholar]
- 77.Ferrer C, Mojica F J M, Juez G, Rodriguez-Valera F. Differentially transcribed regions of Haloferax volcanii genome depending on the medium salinity. J Bacteriol. 1996;178:309–313. doi: 10.1128/jb.178.1.309-313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fisher G, Tradler T, Zarnt T. The mode of action of peptidyl cis/trans isomerases in vivo: binding vs. catalysis. FEBS Lett. 1998;426:17–20. doi: 10.1016/s0014-5793(98)00242-7. [DOI] [PubMed] [Google Scholar]
- 79.Fitz-Gibbon S, Choi A J, Miller J H, Stetter K O, Simon M I, Swanson R, Kim U-J. A fosmid-based genomic map and identification of 474 genes of the hyperthermophilic archaeon Pyrobaculum aerophilum. Extremophiles. 1997;1:36–51. doi: 10.1007/s007920050013. [DOI] [PubMed] [Google Scholar]
- 79a.Fitz-Gibbon, S. Personal communication.
- 80.Forterre P. Protein versus rRNA: problems in rooting the universal tree of life. ASM News. 1997;63:89–95. [Google Scholar]
- 81.Freeman B C, Myers M P, Schumacher R, Morimoto R I. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frydman J, Höhfeld J. Chaperones get in touch: the Hip-Hop connection. Trends Biochem Sci. 1997;22:87–92. doi: 10.1016/s0968-0004(97)01005-0. [DOI] [PubMed] [Google Scholar]
- 83.Furutani M, Iida T, Yamano S, Kamino K, Maruyama T. Biochemical and genetic characterization of an FK506-sensitive peptidyl prolyl cis-trans isomerase from a thermophilic archaeon, Methanococcus thermolithotrophicus. J Bacteriol. 1998;180:388–394. doi: 10.1128/jb.180.2.388-394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Furutani M, Iida T, Yoshida T, Maruyama R. Group II chaperonin in a thermophilic methanogen, Methanococcus thermolithotrophicus. J Biol Chem. 1998;273:28399–28407. doi: 10.1074/jbc.273.43.28399. [DOI] [PubMed] [Google Scholar]
- 85.Gabai V L, Meriin A B, Yaglom J A, Vollcoh V Z, Sherman M Y. Role of Hsp70 in regulation of stress-kinase JNK: implications in apoptosis and aging. FEBS Lett. 1998;438:1–4. doi: 10.1016/s0014-5793(98)01242-3. [DOI] [PubMed] [Google Scholar]
- 86.Garberi J C, Macario A J L, Conway de Macario E. Antigenic mosaic of Methanosarcinaceae: partial characterization of Methanosarcina barkeri 227 surface antigens by monoclonal antibodies. J Bacteriol. 1985;16:1–16. doi: 10.1128/jb.164.1.1-6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gässler C S, Buchberger A, Laufen T, Mayer M P, Schröder H, Valencia A, Bukau B. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc Natl Acad Sci USA. 1998;95:15229–15234. doi: 10.1073/pnas.95.26.15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gebauer M, Melki R, Gehring U. The chaperone cofactor Hop/p60 interacts with the cytosolic chaperonin-containing TCP-1 and affects its nucleotide exchange and protein folding activities. J Biol Chem. 1998;273:29475–29480. doi: 10.1074/jbc.273.45.29475. [DOI] [PubMed] [Google Scholar]
- 89.Geissler S, Siegers K, Schiebel E. A novel protein complex promoting formation of functional α- and γ-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 91.Germot A, Philippe H, Le Guyader H. Presence of a mitochondrial-type 70-kDa heat shock protein in Trichomonas vaginalis suggests a very early mitochondrial endosymbiosis in eukaryotes. Proc Natl Acad Sci USA. 1996;93:14614–14617. doi: 10.1073/pnas.93.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gerth U, Krüger E, Derré L, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic components of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 93.Glover J R, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 94.Gohl H P, Gröndahl B, Thomm M. Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 1995;23:3837–3841. doi: 10.1093/nar/23.19.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gomes S L, Gober J W, Shapiro L. Expression of the Caulobacter heat-shock gene dnaK is developmentally controlled during growth at normal temperatures. J Bacteriol. 1990;172:3051–3059. doi: 10.1128/jb.172.6.3051-3059.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gottesman S. Protecting the neighborhood: extreme measures. Proc Natl Acad Sci USA. 1998;95:2731–2732. doi: 10.1073/pnas.95.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gottesman S, Roche E, Zhou Y, Sauer R T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick J S, Dalrymple B, Kuramitsu H, Shiroza T, Foster T, Clark W P, Ross B, Squires C L, Maurizi M R. Conservation of the regulatory subunit for the ClpATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci USA. 1990;87:3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gribaldo S, Lumia V, Creti R, Conway de Macario E, Sanangelantoni A, Cammarano P. Discontinuous occurrence of the hsp70(dnaK) gene among archaea and sequence features of HSP70 suggest a novel outlook on phylogenies inferred from this protein. J Bacteriol. 1999;181:434–443. doi: 10.1128/jb.181.2.434-443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grogan D W. Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can J Microbiol. 1996;42:1163–1171. [Google Scholar]
- 101.Guagliardi A, Cerchia L, Bartolucci S, Rossi M. The chaperonin from the archaeon Sulfolobus solfataricus promotes correct refolding and prevents thermal denaturation in vitro. Protein Sci. 1994;3:1436–1443. doi: 10.1002/pro.5560030910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guagliardi A, Napoli A, Rossi M, Ciaramella M. Annealing of complementary DNA strands above the melting point of the duplex promoted by an archaeal protein. J Mol Biol. 1997;267:841–848. doi: 10.1006/jmbi.1996.0873. [DOI] [PubMed] [Google Scholar]
- 103.Gupta R S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 104.Gupta R S. Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- 105.Gupta R S. What are archaebacteria: life’s third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Mol Microbiol. 1998;29:695–707. doi: 10.1046/j.1365-2958.1998.00978.x. [DOI] [PubMed] [Google Scholar]
- 106.Gupta R S. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gupta R S, Aitken K, Falah M, Singh B. Cloning of Giardia lamblia heat shock protein HSP70 homologs: implications regarding origin of eukaryotic cells and endoplasmic reticulum. Proc Natl Acad Sci USA. 1994;91:2895–2899. doi: 10.1073/pnas.91.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gupta R S, Bustard K, Falah M, Singh D. Sequencing of heat shock protein 70 (DnaK) homologs from Deinococcus proteolyticus and Thermomicrobium roseum and their integration in the protein-based phylogeny of prokaryotes. J Bacteriol. 1997;179:345–357. doi: 10.1128/jb.179.2.345-357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gupta R S, Golding G B. The origin of the eukaryotic cell. Trends Biochem Sci. 1996;21:166–171. [PubMed] [Google Scholar]
- 110.Gupta R S, Singh B. Cloning of the Hsp70 gene from Halobacterium marismortui: relatedness of archaebacterial Hsp70 to its eubacterial homologs and a model for the evolution of the Hsp70 gene. J Bacteriol. 1992;174:4594–4605. doi: 10.1128/jb.174.14.4594-4605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gupta R S, Singh B. Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus. Curr Biol. 1994;4:1104–1114. doi: 10.1016/s0960-9822(00)00249-9. [DOI] [PubMed] [Google Scholar]
- 112.Guzzo, J., C. Diorio, D. C. Alexander, and M. S. DuBow. Toward understanding metal stress in environmental microbial flora. In C. R. Bell, P. Johnson-Green, and M. Brylinsky (ed.), Microbial biosystems. New frontiers, in press. Acadia University Press, Halifax, Canada.
- 113.Hall H K, Karem K L, Foster J W. Molecular responses of microbes to environmental pH stress. Adv Microbiol Physiol. 1995;37:229–272. doi: 10.1016/s0065-2911(08)60147-2. [DOI] [PubMed] [Google Scholar]
- 114.Han C J, Park S H, Kelly R M. Acquired thermotolerance and stress phase growth of the extremely thermoacidophilic archaeon Metallosphaera sedula in continuous culture. Appl Environ Microbiol. 1997;63:2391–2396. doi: 10.1128/aem.63.6.2391-2396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haney P J, Badger J H, Buldak G L, Reich C I, Woese C R, Olsen G J. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc Natl Acad Sci USA. 1999;96:3578–3583. doi: 10.1073/pnas.96.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harrison C J, Hayer-Hartl M, Di Liberto M, Hartl F U, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431. [DOI] [PubMed] [Google Scholar]
- 117.Hausner W, Thomm M. Purification and characterization of a general transcription factor, aTFB, from the archaeon Methanococcus thermolithotrophicus. J Biol Chem. 1993;268:24047–24052. [PubMed] [Google Scholar]
- 118.Hausner W, Wettach J, Hethke C, Thomm M. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J Biol Chem. 1996;217:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 119.Hebert A M, Kropinski A M, Jarrell K F. Heat shock response of the archaebacterium Methanococcus voltae. J Bacteriol. 1991;173:3224–3227. doi: 10.1128/jb.173.10.3224-3227.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 121.Hei D J, Clark D S. Pressure stabilization of proteins from extreme thermophiles. Appl Environ Microbiol. 1994;60:932–939. doi: 10.1128/aem.60.3.932-939.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hensel R, König H. Thermoadaptation of methanogenic bacteria by intracellular ion concentration. FEMS Microbiol Lett. 1988;49:75–79. [Google Scholar]
- 123.Herman C, D’Ari R. Proteolysis and chaperones: the destruction/reconstruction dilemma. Curr Opin Microbiol. 1998;1:204–209. doi: 10.1016/s1369-5274(98)80012-x. [DOI] [PubMed] [Google Scholar]
- 124.Herman C, Thévenet D, Bouloc P, Walker G C, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hirt R P, Healy B, Vossbrick C R, Canning E U, Embley T M. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: molecular evidence that microsporidia once contained mitochondria. Curr Biol. 1997;7:995–998. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 126.Hofman-Bang J P, Lange M, Conway de Macario E, Macario A J L, Ahring B K. The genes coding for the hsp70(dnaK) molecular chaperone machine occur in the moderate thermophilic archaeon Methanosarcina thermophila TM-1. Gene. 1999;238:387–395. doi: 10.1016/s0378-1119(99)00343-1. [DOI] [PubMed] [Google Scholar]
- 127.Höhfeld J, Minami Y, Hartl F U. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 128.Holden, J., M. W. W. Adams, and J. A. Baross. Heat-shock response in hyperthermophilic microorganisms. In C. R. Bell, P. Johnson-Green, and M. Brylinsky (ed.), Microbial biosystems. New frontiers, in press. Acadia University Press, Halifax, Canada.
- 129.Holden J F, Baross J A. Enhanced thermotolerance and temperature-induced changes in protein composition in the hyperthermophilic archaeon ES4. J Bacteriol. 1993;175:2839–2843. doi: 10.1128/jb.175.10.2839-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Holden J F, Baross J A. Enhanced thermotolerance by hydrostatic pressure in the deep-sea hyperthermophile Pyrococcus strain ES4. FEMS Microbiol Ecol. 1995;18:27–34. [Google Scholar]
- 131.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Homuth G, Mogk A, Schumann W. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol Microbiol. 1999;32:1183–1197. doi: 10.1046/j.1365-2958.1999.01428.x. [DOI] [PubMed] [Google Scholar]
- 133.Hoskins J R, Pak M, Maurizi M R, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Howgrave-Graham A R, Macario A J L, Wallis F M. Quantitative analysis and mapping of micro-organisms in anaerobic digester granules using a combination of transmission electron microscopy with immunotechnology. J Appl Microbiol. 1997;83:587–595. [Google Scholar]
- 135.Iida T, Furutani M, Iwabuchi T, Maruyama T. Gene for a cyclophilin-type peptidyl-prolyl cis-trans isomerase from a halophilic archaeum, Halobacterium cutirubrum. Gene. 1997;204:139–144. doi: 10.1016/s0378-1119(97)00534-9. [DOI] [PubMed] [Google Scholar]
- 136.Iida T, Furutani M, Nishida F, Maruyama T. FKBP-type peptidyl-prolyl cis-trans isomerase from a sulfur-dependent hyperthermophilic archaeon, Thermococcus sp. KS-1. Gene. 1998;222:249–255. doi: 10.1016/s0378-1119(98)00484-3. [DOI] [PubMed] [Google Scholar]
- 137.Izumi M, Fujiwara S, Takagi M, Kanaya S, Imanaka T. Isolation and characterization of a second subunit of molecular chaperonin from Pyrococcus kodakaraensis KOD1: analysis of an ATPase-deficient mutant enzyme. Appl Environ Microbiol. 1999;65:1801–1805. doi: 10.1128/aem.65.4.1801-1805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jaenicke R, Bernhardt G, Lüdemann H D, Stetter K O. Pressure-induced alterations in the protein pattern of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Appl Environ Microbiol. 1988;54:2375–2380. doi: 10.1128/aem.54.10.2375-2380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jerez C A. The heat shock response in meso- and thermoacidophilic chemolitotrophic bacteria. FEMS Microbiol Lett. 1988;56:289–294. [Google Scholar]
- 139a.Kaczanowski, S., P. Zielenkiewicz, J. Thomsen, W. Hausner, M. W. Thomm, A. J. L. Macario, and E. Conway de Macario. Unpublished data.
- 140.Kagawa Y, Ohta T, Abe Y, Endo H, Yodha M, Kato N, Endo I, Hamamoto T, Ichida M, Hoaki T, Maruyama T. Gene of heat shock protein of sulfur-dependent archaeal hyperthermophile Desulfurococcus. Biochem Biophys Res Commun. 1995;214:730–736. doi: 10.1006/bbrc.1995.2346. [DOI] [PubMed] [Google Scholar]
- 141.Kagawa H K, Osipiuk J, Maltsev N, Overbeek R, Quaite-Randall E, Joachimiak A, Trent J D. The 60 kDa heat shock proteins in the hyperthermophilic archaeon Sulfolobus shibatae. J Mol Biol. 1995;253:712–725. doi: 10.1006/jmbi.1995.0585. [DOI] [PubMed] [Google Scholar]
- 142.Karlin S, Brocchieri L. Heat shock protein 70 family: multiple sequence comparisons, function, and evolution. J Mol Evol. 1998;47:565–577. doi: 10.1007/pl00006413. [DOI] [PubMed] [Google Scholar]
- 143.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Nakazawa H, Takamiya M, Masuda S, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kubota K, Nakamura Y, Nomura N, Sako Y, Kikuchi H. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 144.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Yoshizawa T, Nakamura Y, Robb F, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–75. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 145.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Yoshizawa T, Nakamura Y, Robb F, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3 (supplement) DNA Res. 1998;5(Suppl.):147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 146.Keeling P J. A kingdom’s progress: archezoa and the origin of eukaryotes. Bioessays. 1998;20:87–95. [Google Scholar]
- 147.Kim B K, Pihl T D, Reeve J N, Daniels L. Purification of the copper response extracellular proteins secreted by the copper-resistant methanogen Methanobacterium bryantii BKYH and cloning, sequencing, and transcription of the genes encoding these proteins. J Bacteriol. 1995;177:7178–7185. doi: 10.1128/jb.177.24.7178-7185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kim K K, Yokota H, Santoso S, Lerner D, Kim R, Kim S H. Purification, crystallization, and preliminary x-ray crystallographic data analysis of small heat shock protein homolog from Methanococcus jannaschii, a hyperthermophile. J Struct Biol. 1998;121:76–80. doi: 10.1006/jsbi.1998.3969. [DOI] [PubMed] [Google Scholar]
- 149.Kim K K, Kim R, Kim S-H. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 150.Kim R, Kim K K, Yokota H, Kim S-H. Small heat shock protein of Methanococcus jannaschii, a hyperthermophile. Proc Natl Acad Sci USA. 1998;95:9129–9133. doi: 10.1073/pnas.95.16.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Klenk H-P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 152.Klumpp M, Baumeister W. The thermosome: archetype of group II chaperonins. FEBS Lett. 1998;430:73–77. doi: 10.1016/s0014-5793(98)00541-9. [DOI] [PubMed] [Google Scholar]
- 153.Klumpp M, Baumeister W, Essen L O. Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell. 1997;91:263–270. doi: 10.1016/s0092-8674(00)80408-0. [DOI] [PubMed] [Google Scholar]
- 154.Knapp S, Schmidt-Krey I, Hebert H, Bergman T, Jörnvall H, Ladenstein R. The molecular chaperonin TF55 from the thermophilic archaeon Sulfolobus solfataricus. J Mol Biol. 1994;242:397–407. doi: 10.1006/jmbi.1994.1590. [DOI] [PubMed] [Google Scholar]
- 155.Koeck P J B, Kagawa H K, Ellis M J, Herbert H, Trent J D. Two-dimensional crystals of reconstituted β-subunits of the chaperonin TF55 from Sulfolobus shibatae. Biochem Biophys Acta. 1998;1429:40–44. doi: 10.1016/s0167-4838(98)00218-0. [DOI] [PubMed] [Google Scholar]
- 156.Kojoh K, Matsuzawa H, Wakagi T. Zinc and an N-terminal extra stretch of the ferredoxin from a thermoacidophilic archaeon stabilize the molecule at high temperature. Eur J Biochem. 1999;264:85–91. doi: 10.1046/j.1432-1327.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- 157.König H. Archaeobacterial cell envelopes. Can J Microbiol. 1988;34:395–406. [Google Scholar]
- 158.König H, Hartmann E, Kärcher U. Pathways and principles of the biosynthesis of methanobacterial cell wall polymers. Syst Appl Microbiol. 1994;16:510–517. [Google Scholar]
- 159.Konisky J, Michels P, Clark D S. Pressure stabilization is not a general property of thermophilic enzymes: the adenylate kinases of Methanococcus voltae, Methanococcus maripaludis, Methanococcus thermolithotrophicus, and Methanmococcus jannaschii. Appl Environ Microbiol. 1995;61:2762–2764. doi: 10.1128/aem.61.7.2762-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Korber P, Zander T, Herschlag D, Bardwell J C A. A new heat shock protein that binds nucleic acids. J Biol Chem. 1999;274:249–256. doi: 10.1074/jbc.274.1.249. [DOI] [PubMed] [Google Scholar]
- 161.Kowalski J M, Kelly R M, Konisky J, Clark D S, Wittrup K D. Purification and functional characterization of a chaperone from Methanococcus jannaschii. Syst Appl Microbiol. 1998;21:173–178. doi: 10.1016/S0723-2020(98)80021-0. [DOI] [PubMed] [Google Scholar]
- 162.Kuo Y-P, Thompson D K, Daniels C J. Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C.: American Society for Microbiology; 1994. Characterization and regulation of a heat shock gene from the Archaeon Haloferax volcanii; p. 264. [Google Scholar]
- 163.Kuo Y-P, Thompson D K, St. Jean A, Charlebois R L, Daniels C J. Characterization of two heat shock genes from Haloferax volcanii: a model system for transcription regulation in the Archaea. J Bacteriol. 1997;179:6318–6324. doi: 10.1128/jb.179.20.6318-6324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lange M, Macario A J L, Ahring B K, Conway de Macario E. Heat-shock response in Methanosarcina mazei S-6. Curr Microbiol. 1997;35:116–121. doi: 10.1007/s002849900222. [DOI] [PubMed] [Google Scholar]
- 165.Lange M, Macario A J L, Ahring B K, Conway de Macario E. Increased transcripts of the dnaK locus genes in Methanosarcina mazei S-6 exposed to supraoptimal concentrations of ammonia. FEMS Microbiol Lett. 1997;152:379–384. [Google Scholar]
- 165a.Lange, M. Unpublished results.
- 166.LaPaglia C, Hartzell P L. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl Environ Microbiol. 1997;63:3158–3163. doi: 10.1128/aem.63.8.3158-3163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Leibovitz D, Koch Y, Fridkin M, Pitzer F, Zwickl P, Dantes A, Baumeister W, Amsterdam A. Archaebacterial and eukaryotic proteasomes prefer different sites in cleaving gonadotropin-releasing hormone. J Biol Chem. 1995;270:11029–11032. doi: 10.1074/jbc.270.19.11029. [DOI] [PubMed] [Google Scholar]
- 168.Leigh J A. Transcriptional regulation in Archaea. Curr Opin Microbiol. 1999;2:131–134. doi: 10.1016/S1369-5274(99)80023-X. [DOI] [PubMed] [Google Scholar]
- 169.Lewus P, Ford R M. Temperature-sensitive motility of Sulfolobus acidocaldarius influences population distribution in extreme environments. J Bacteriol. 1999;181:4020–4025. doi: 10.1128/jb.181.13.4020-4025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Llorca O, Smyth M G, Carrascosa J L, Willison K R, Radermacher M, Steinbacher S, Valpuesta J M. 3D reconstruction of the ATP-bound form of CCT reveals the asymetric folding conformation of a type II chaperonin. Nat Struct Biol. 1999;6:639–642. doi: 10.1038/10689. [DOI] [PubMed] [Google Scholar]
- 171.Llorca O, Smyth M G, Marco S, Carrascosa J L, Willison K R, Valpuesta J M. ATP binding induces large conformational changes in the apical and equatorial domains of the eukaryotic chaperonin containing TCP-1 complex. J Biol Chem. 1998;273:10091–10094. doi: 10.1074/jbc.273.17.10091. [DOI] [PubMed] [Google Scholar]
- 172.López-Garcia P, Amils R, Anton J. Sizing chromosomes and megaplasmids in haloarchaea. Microbiology. 1996;142:1423–1428. doi: 10.1099/13500872-142-6-1423. [DOI] [PubMed] [Google Scholar]
- 173.López-Garcia P, Forterre P. DNA topology in hyperthermophilic archaea: reference states and their variation with growth phase, growth temperature, and temperature stresses. Mol Microbiol. 1997;22:1267–1279. doi: 10.1046/j.1365-2958.1997.3051668.x. [DOI] [PubMed] [Google Scholar]
- 174.Lopez-Garcia P, Forterre P. Control of DNA topology during thermal stress in hyperthermophilic archaea: DNA topoisomerase levels, activities and induced thermotolerance during heat and cold shock in Sulfolobus. Mol Microbiol. 1999;33:766–777. doi: 10.1046/j.1365-2958.1999.01524.x. [DOI] [PubMed] [Google Scholar]
- 175.Louvion J-F, Abbas-Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lu Z, Cyr D M. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem. 1998;273:5970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 177.Macario A J L. Heat-shock proteins and molecular chaperones: implications for pathogenesis, diagnostics, and therapeutics. Intl J Clin Lab Res. 1995;25:59–70. doi: 10.1007/BF02592359. [DOI] [PubMed] [Google Scholar]
- 178.Macario A J L, Conway de Macario E. Quantitative immunologic analysis of the methanogenic flora of digestors reveals a considerable diversity. Appl Environ Microbiol. 1988;54:79–86. doi: 10.1128/aem.54.1.79-86.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Macario A J L, Conway de Macario E. Stress genes: an introductory overview. Stress. 1997;1:123–134. doi: 10.3109/10253899709001102. [DOI] [PubMed] [Google Scholar]
- 180.Macario, A. J. L., and E. Conway de Macario. Transcription initiation of stress (heat-shock) genes in archaea. In C. R. Bell, P. Johnson-Green, and M. Brylinsky (ed.), Microbial biosystems. New frontiers, in press. Halifax, Acadia University Press, Halifax, Canada.
- 180a.Macario, A. J. L., and E. Conway de Macario. Unpublished data.
- 181.Macario A J L, Dugan C B, Clarens M, Conway de Macario E. dnaJ in Archaea. Nucleic Acids Res. 1993;21:2773. doi: 10.1093/nar/21.11.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Macario A J L, Dugan C B, Conway de Macario E. A dnaK homolog in the archaebacterium Methanosarcina mazei S6. Gene. 1991;108:133–137. doi: 10.1016/0378-1119(91)90498-z. [DOI] [PubMed] [Google Scholar]
- 183.Macario A J L, Dugan C B, Conway de Macario E. An archaeal trkA homolog near dnaK and dnaJ. Biochim Biophys Acta. 1993;1216:495–498. doi: 10.1016/0167-4781(93)90022-6. [DOI] [PubMed] [Google Scholar]
- 184.Macario A J L, Simon V H, Conway de Macario E. An archaeal gene upstream of grpE different from eubacterial counterparts. Biochim Biophys Acta Gene Struct Expression. 1995;1264:173–177. doi: 10.1016/0167-4781(95)00163-b. [DOI] [PubMed] [Google Scholar]
- 185.Macario A J L, Visser F A, van Lier J B, Conway de Macario E. Topography of methanogenic subpopulations in a microbial consortium adapting to thermophilic conditions. J Gen Microbiol. 1991;137:2179–2189. [Google Scholar]
- 186.Madigan M T, Martinko J M, Parker J. Brock biology of microorganisms. 8th ed. Upper Saddle River, N.J: Prentice-Hall, Inc.; 1997. [Google Scholar]
- 187.Marco S, Ureña D, Carrascosa J L, Waldman T, Peters J, Hegerl R, Pfeiffer G, Sack-Kongehl H, Baumeister W. The molecular chaperone TF55. FEBS Lett. 1994;341:152–155. doi: 10.1016/0014-5793(94)80447-8. [DOI] [PubMed] [Google Scholar]
- 188.Margulis L. Archaeal-eubacterial mergers in the origin of Eukarya: phylogenetic classification of life. Proc Natl Acad Sci USA. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Martin W F. Is something wrong with the tree of life? Bioessays. 1996;18:523–527. [Google Scholar]
- 190.Martin D D, Ciulla R A, Roberts M F. Osmoadaptation in archea. Appl Environ Microbiol. 1999;65:1815–1825. doi: 10.1128/aem.65.5.1815-1825.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Martin J, Hartl F U. Chaperone-assisted protein folding. Curr Opin Struct Biol. 1997;7:41–52. doi: 10.1016/s0959-440x(97)80006-1. [DOI] [PubMed] [Google Scholar]
- 192.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 193.Martins L O, Santos H. Accumulation of mannoglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl Environ Microbiol. 1995;61:3299–3303. doi: 10.1128/aem.61.9.3299-3303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Maruta K, Mitsuzumi H, Nakada T, Kubota M, Chaen H, Fukuda S, Sugimoto T, Kurimoto M. Cloning and sequencing of a cluster of genes encoding novel enzymes of trehalose biosynthesis from the thermophilic archaebacterium Sulfolobus acidocaldarius. Biochim Biophys Acta. 1996;1992:177–181. doi: 10.1016/s0304-4165(96)00082-7. [DOI] [PubMed] [Google Scholar]
- 195.Maupin-Furlow J A, Aldrich H C, Ferry J G. Biochemical characterization of the 20S proteasome from the methanoarchaeon Methanosarcina thermophila. J Bacteriol. 1998;180:1480–1487. doi: 10.1128/jb.180.6.1480-1487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Maupin-Furlow J A, Ferry J G. A proteasome from the methanogenic archaeon Methanosarcina thermophila. J Biol Chem. 1995;270:28617–28622. doi: 10.1074/jbc.270.48.28617. [DOI] [PubMed] [Google Scholar]
- 197.Mayerhofer L E, Macario A J L, Conway de Macario E. Lamina, a novel multicellular form of Methanosarcina mazei S-6. J Bacteriol. 1992;174:3009–3014. doi: 10.1128/jb.174.1.309-314.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mayr E. Two empires or three? Proc Natl Acad Sci USA. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Metcalf W W, Zhang J K, Wolfe R S. An anaerobic, intrachamber incubator for growth of Methanosarcina spp. on methanol-containing solid media. Appl Environ Microbiol. 1998;64:768–770. doi: 10.1128/aem.64.2.768-770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Michels P C, Clark D S. Pressure-enhanced activity and stability of a hyperthermophilic protease from a deep-sea methanogen. Appl Environ Microbiol. 1997;63:3985–3991. doi: 10.1128/aem.63.10.3985-3991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Miller J F, Shah N N, Nelson C M, Ludlow J M, Clark D S. Pressure and temperature effects on growth and methane production of the extreme thermophile Methanococcus jannaschii. Appl Environ Microbiol. 1988;54:3039–3042. doi: 10.1128/aem.54.12.3039-3042.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Minuth T, Frey G, Lindner P, Rachel R, Stetter K O, Jaenicke R. Recombinant homo- and hetero-oligomers of an ultrastable chaperonin from the archaeon Pyrodictium occultum show chaperone activity in vitro. Eur J Biochem. 1998;258:837–845. doi: 10.1046/j.1432-1327.1998.2580837.x. [DOI] [PubMed] [Google Scholar]
- 203.Mohsenzadeh S, Saupe-Thies W, Steier G, Schroeder T, Fracella F, Ruoff P, Rensing L. Temperature adaptation of house keeping and heat shock gene expression in Neurospora crassa. Fungal Genet Biol. 1998;25:31–43. doi: 10.1006/fgbi.1998.1081. [DOI] [PubMed] [Google Scholar]
- 204.Mojica F J M, Cisneros E, Ferrer C, Rodriguez-Valera F, Juez G. Osmotically induced response in representatives of halophilic prokaryotes: the bacterium Halomonas elongata and the archaeon Haloferax volcanii. J Bacteriol. 1997;179:5471–5481. doi: 10.1128/jb.179.17.5471-5481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Morimoto R I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 206.Münchbach M, Nocker A, Narberhaus F. Multiple small heat shock proteins in rhizobia. J Bacteriol. 1998;181:83–90. doi: 10.1128/jb.181.1.83-90.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nakamura N, Taguchi H, Ishii N, Yoshida M, Suzuki M, Endo I, Miura K, Yodha M. Purification and molecular cloning of the group II chaperonin from the acidothermophilic archaeon Sulfolobus sp. strain 7. Biochem Biophys Res Commun. 1997;236:727–732. doi: 10.1006/bbrc.1997.6916. [DOI] [PubMed] [Google Scholar]
- 208.Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 209.Narberhaus F, Käser R, Nocker A, Hennecke H. A novel DNA element that controls bacterial heat shock gene expression. Mol Microbiol. 1998;28:315–323. doi: 10.1046/j.1365-2958.1998.00794.x. [DOI] [PubMed] [Google Scholar]
- 210.Narberhaus F, Kowarik M, Beck C, Hennecke H. Promoter selectivity of the Bradyrhizobium japonicum RpoH transcription factors in vivo and in vitro. J Bacteriol. 1998;180:2395–2401. doi: 10.1128/jb.180.9.2395-2401.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Naylor D J, Hoogenraad N J, Høj P B. Isolation and characterization of cDNA encoding rat mitochondrial GrpE, a stress-inducible nucleotide-exchange factor of ubiquitous appearance in mammalian organs. FEBS Lett. 1996;396:181–188. doi: 10.1016/0014-5793(96)01100-3. [DOI] [PubMed] [Google Scholar]
- 212.Naylor D J, Stines A P, Hoogenraad N J, Høj P B. Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the co-chaperones of specific Hsp70 members. J Biol Chem. 1998;273:21169–21177. doi: 10.1074/jbc.273.33.21169. [DOI] [PubMed] [Google Scholar]
- 213.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, White O, Salzberg S L, Smith H O, Venter J C, Fraser C M. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 214.Netzer W J, Hartl F U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 215.Niehaus F, Frey B, Antranikian G. Cloning and characterisation of a thermostable α-DNA polymerase from the hyperthermophilic archaeon Thermococcus sp. TY. Gene. 1997;204:153–158. doi: 10.1016/s0378-1119(97)00536-2. [DOI] [PubMed] [Google Scholar]
- 216.Nitsch M, Klumpp M, Lupas A, Baumeister W. The thermosome: alternating α and β-subunits within the chaperonin of the archaeon Thermoplasma acidophilum. J Mol Biol. 1997;267:142–149. doi: 10.1006/jmbi.1996.0849. [DOI] [PubMed] [Google Scholar]
- 217.Nitsch M, Walz J, Typke D, Klumpp M, Essen L O, Baumeister W. Group II chaperonin in an open conformation examined by electron tomography. Nat Struct Biol. 1998;5:855–857. doi: 10.1038/2296. [DOI] [PubMed] [Google Scholar]
- 218.Oh H J, Easton D, Murawski M, Kaneko Y, Subjeck J R. The chaperoning activity of hsp110. J Biol Chem. 1999;274:15712–15718. doi: 10.1074/jbc.274.22.15712. [DOI] [PubMed] [Google Scholar]
- 219.Ohta T, Honda K, Saito K, Hayashi H, Tano H, Hamamoto T, Kagawa Y. Heat shock promoter of thermophilic chaperonin operon. Biochem Biophys Res Commun. 1993;191:550–557. doi: 10.1006/bbrc.1993.1253. [DOI] [PubMed] [Google Scholar]
- 220.Osipiuk J, Joachimiak A. Cloning, sequencing, and expression of dnaK-operon proteins from the thermophilic bacterium Thermus thermophilus. Biochim Biophys Acta. 1997;1353:253–265. doi: 10.1016/s0167-4781(97)00071-7. [DOI] [PubMed] [Google Scholar]
- 221.Osorio G, Jerez C A. Adaptive response of the archaeon Sulfolobus acidocaldarius BC65 to phosphate starvation. Microbiology. 1996;142:1531–1536. doi: 10.1099/13500872-142-6-1531. [DOI] [PubMed] [Google Scholar]
- 222.Palmer J R, Thompson D K, Ray W C, Daniels C J. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C.: American Society for Microbiology; 1997. Occurrence of multiple TATA binding protein and TFIIB eucaryal-like transcription factors in the archaeon Haloferax volcanii and evidence for their differential regulation; p. 330. [Google Scholar]
- 223.Parsell D A, Lindquist S. The function of heat shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 224.Peak M J, Robb F T, Peak J G. Extreme resistance to thermally induced DNA backbone breaks in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1995;177:6316–6318. doi: 10.1128/jb.177.21.6316-6318.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Peeples T L, Kelly R M. Bioenergetic response of the extreme thermophilic Metallosphaera sedula to thermal and nutritional stresses. Appl Environ Microbiol. 1997;61:2314–2321. doi: 10.1128/aem.61.6.2314-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pereira S L, Reeve J N. Histones and nucleosomes in Archaea and Eukarya: a comparative analysis. Extremophiles. 1998;2:141–148. doi: 10.1007/s007920050053. [DOI] [PubMed] [Google Scholar]
- 227.Perng M D, Cairns L, van den Ijssel P, Hutcheson A M, Quinlan R A. Intermediate filament interactions can be altered by HSP27 and αB-crystallin. J Cell Sci. 1999;112:2099–2112. doi: 10.1242/jcs.112.13.2099. [DOI] [PubMed] [Google Scholar]
- 228.Peters J, Baumeister W, Lupas A. Hyperthermostable surface layer protein tetrabrachion from the archaebacterium Staphylothermus marinus: evidence for the presence of a right-handed coiled coil derived from the primary structure. J Mol Biol. 1996;257:1031–1041. doi: 10.1006/jmbi.1996.0221. [DOI] [PubMed] [Google Scholar]
- 229.Peyretaillade E, Broussolle V, Peyret P, Méténier G, Gouy M, Vivarès C P. Microsporidia, amitochondrial protists, possess a 70-kDa heat shock protein gene of mitochondrial evolutionary origin. Mol Biol Evol. 1998;15:683–689. doi: 10.1093/oxfordjournals.molbev.a025971. [DOI] [PubMed] [Google Scholar]
- 230.Phipps B M, Hoffmann A, Stetter K O, Baumeister W. A novel ATPase complex selectively accumulated upon heat shock is a major cellular component of thermophilic archaebacteria. EMBO J. 1991;10:1711–1722. doi: 10.1002/j.1460-2075.1991.tb07695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Phipps B M, Typke D, Hegerl R, Volker S, Hoffmann A, Stetter K O, Baumeister W. Structure of a molecular chaperone from a thermophilic archaebacterium. Nature. 1993;361:475–477. [Google Scholar]
- 232.Pledger R J, Crump B C, Baross J A. A barophilic response by two hyperthermophilic hydrothermal vent Archaea: upward shift in the optimal temperature and acceleration of growth rate at supra-optimal temperatures by elevated pressure. FEMS Microbiol Ecol. 1994;14:233–242. [Google Scholar]
- 233.Proctor L M, Lai R, Gunsalus R P. The methanogenic archaeon Methanosarcina thermophila TM-1 possesses a high-affinity betaine transporter involved in osmotic adaption. Appl Environ Microbiol. 1997;63:2252–2257. doi: 10.1128/aem.63.6.2252-2257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Quaite-Randall E, Trent J D, Josephs R, Joachimiak A. Conformational cycle of the archaeosome, a TCP1-like chaperonin from Sulfolobus shibatae. J Biol Chem. 1995;270:28818–28823. doi: 10.1074/jbc.270.48.28818. [DOI] [PubMed] [Google Scholar]
- 235.Ranson N A, White H E, Saibil H R. Chaperonins. Biochem J. 1998;333:233–242. doi: 10.1042/bj3330233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Rassow J, von Ahsen O, Bömer U, Pfanner N. Molecular chaperones: towards a characterization of the heat-shock protein 70 family. Trends Cell Biol. 1997;7:129–133. doi: 10.1016/S0962-8924(96)10056-8. [DOI] [PubMed] [Google Scholar]
- 237.Reeve J N, Sandman K, Daniels C J. Archaeal histones, nucleosomes, and transcription initiation. Cell. 1997;89:999–1002. doi: 10.1016/s0092-8674(00)80286-x. [DOI] [PubMed] [Google Scholar]
- 238.Ren B, Tibbelin G, de Pascale D, Rossi M, Bartolucci S, Ladenstein R. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units. Nat Struct Biol. 1998;5:602–611. doi: 10.1038/862. [DOI] [PubMed] [Google Scholar]
- 239.Rensing S A, Maier U G. Phylogenetic analysis of the stress-70 protein family. J Mol Evol. 1994;39:80–86. doi: 10.1007/BF00178252. [DOI] [PubMed] [Google Scholar]
- 240.Rensing L, Monnerjahn C, Meyer U. Differential stress gene expression during the development of Neurospora crassa and other fungi. FEMS Microbiol Lett. 1998;168:159–166. doi: 10.1111/j.1574-6968.1998.tb13268.x. [DOI] [PubMed] [Google Scholar]
- 241.Rinker K D, Kelly R M. Growth physiology of the hyperthermophilic archaeon Thermococcus litoralis: development of a sulfur-free defined medium, characterization of an exopolysaccharide, and evidence for biofilm formation. Appl Environ Microbiol. 1996;62:4478–4485. doi: 10.1128/aem.62.12.4478-4485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Roberts R C, Toochinda C, Avedissian M, Baldini R L, Gomes S L, Shapiro L. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J Bacteriol. 1996;178:1829–1841. doi: 10.1128/jb.178.7.1829-1841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Roger A J, Clark C G, Doolittle W F. A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis. Proc Natl Acad Sci USA. 1996;93:14618–14622. doi: 10.1073/pnas.93.25.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Roger A J, Svärd S G, Tovar J, Clark C G, Smith M W, Gillin F D, Sogin M L. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rommelaere H, Van Troys M, Gao Y, Melki R, Cowan N J, Vandekerckhove J, Ampe C. Eukaryotic cytosolic chaperonin contains t-complex polypeptide 1 and seven related subunits. Proc Natl Acad Sci USA. 1993;90:11975–11979. doi: 10.1073/pnas.90.24.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Roseman A M, Chen S, White H, Braig K, Saibil H R. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 1996;87:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 247.Ruddon R W, Bedows E. Assisted protein folding. J Biol Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125. [DOI] [PubMed] [Google Scholar]
- 248.Ruepp A, Eckerskorn C, Bogyo M, Baumeister W. Proteasome function is dispensable under normal but not under heat shock conditions in Thermoplasma acidophilum. FEBS Lett. 1998;425:87–90. doi: 10.1016/s0014-5793(98)00205-1. [DOI] [PubMed] [Google Scholar]
- 249.Ruggero D, Ciammaruconi A, Londei P. The chaperonin of the archaeon Sulfolobus solfataricus is an RNA-binding protein that participates in ribosomal RNA processing. EMBO J. 1998;17:3471–3477. doi: 10.1093/emboj/17.12.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sabehat A, Weiss D, Lurie S. Heat-shock proteins and cross-tolerance in plants. Physiol Plant. 1998;103:437–431. [Google Scholar]
- 251.Sakikawa C, Taguchi H, Makino Y, Yoshida M. On the maximum size of proteins to stay and fold in the cavity of GroEL underneath GroES. J Biol Chem. 1999;274:21251–21256. doi: 10.1074/jbc.274.30.21251. [DOI] [PubMed] [Google Scholar]
- 252.Sanders B M. Stress proteins in aquatic organisms: an environmental perspective. Crit Rev Toxicol. 1993;23:49–75. doi: 10.3109/10408449309104074. [DOI] [PubMed] [Google Scholar]
- 253.Satyal S H, Chen D, Fox S G, Kramer J M, Morimoto R I. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Scandurra R, Consalvi V, Chiaraluce R, Politi L, Engel P C. Protein thermostability in extremophiles. Biochimie. 1998;80:933–941. doi: 10.1016/s0300-9084(00)88890-2. [DOI] [PubMed] [Google Scholar]
- 255.Schindler T, Graumann P L, Perl D, Ma S, Schmid F X, Marahiel M A. The family of cold shock proteins of Bacillus subtilis. J Biol Chem. 1999;274:3407–3413. doi: 10.1074/jbc.274.6.3407. [DOI] [PubMed] [Google Scholar]
- 256.Schlicher T, Soll J. Chloroplastic isoforms of DnaJ and GrpE in pea. Plant Mol Biol. 1997;33:181–185. doi: 10.1023/a:1005784115363. [DOI] [PubMed] [Google Scholar]
- 257.Scholz S, Sonnenbichler J, Schäfer W, Hensel R. Di-myo-inositol-1,1′-phosphate: a new inositol phosphate isolated from Pyrococcus woesei. FEBS Lett. 1992;306:239–242. doi: 10.1016/0014-5793(92)81008-a. [DOI] [PubMed] [Google Scholar]
- 258.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Segal G, Ron E Z. Regulation and organization of the groE and dnaK operons in eubacteria. FEMS Microbiol Lett. 1996;138:1–10. doi: 10.1111/j.1574-6968.1996.tb08126.x. [DOI] [PubMed] [Google Scholar]
- 260.Silberg J J, Hoff K G, Vickery L E. The Hsc66-Hsc20 chaperone system in Escherichia coli: chaperone activity and interactions with the DnaK-DnaJ-GrpE system. J Bacteriol. 1998;180:6617–6624. doi: 10.1128/jb.180.24.6617-6624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Singer M A, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 262.Sleytr U B. I. Basic and applied S-layer research: an overview. FEMS Microbiol Rev. 1997;20:5–12. [Google Scholar]
- 263.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakeley D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Soppa J. Transcription initiation in Archaea: facts, factors and future aspects. Mol Microbiol. 1999;31:1295–1305. doi: 10.1046/j.1365-2958.1999.01273.x. [DOI] [PubMed] [Google Scholar]
- 265.Sousa M C, McKay D B. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry. 1998;37:15392–15399. doi: 10.1021/bi981510x. [DOI] [PubMed] [Google Scholar]
- 266.Sowers K R, Gunsalus R P. Halotolerance in Methanosarcina spp.: role of Nɛ-acetyl-β-lysine, α-glutamate, glycine betaine, and K+ as compatible solutes for osmotic adaptation. Appl Environ Microbiol. 1995;61:4382–4388. doi: 10.1128/aem.61.12.4382-4388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sprott G D, Beveridge T J. Microscopy. In: Ferry J G, editor. Methanogenesis. New York, N.Y: Chapman & Hall; 1993. pp. 81–127. [Google Scholar]
- 268.Sriram M, Osipiuk J, Freeman B C, Miromoto R I, Joachimiak A. Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure. 1997;5:403–414. doi: 10.1016/s0969-2126(97)00197-4. [DOI] [PubMed] [Google Scholar]
- 269.Stringham E G, Candido E P M. Transgenic hsp16-lacZ strains of the soil nematode Caenorhabditis elegans as biological monitors of environmental stress. Environ Toxicol Chem. 1994;13:1211–1220. [Google Scholar]
- 270.Suh W-C, Burkholder W F, Lu C Z, Zhao X, Gottesman M E, Gross C A. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 1998;45:15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Summit M, Scott B, Nielson K, Mathur E, Baross J. Pressure enhances thermal stability of DNA polymerase from three thermophilic organisms. Extremophiles. 1998;2:339–345. doi: 10.1007/s007920050077. [DOI] [PubMed] [Google Scholar]
- 272.Szabo A, Korszun R, Hartl F U, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- 273.Szpikowska B K, Swiderek K M, Sherman M A, Mas M T. MgATP binding to the nucleotide-binding domains of the eukaryotic cytoplasmic chaperonin induces conformational changes in the putative substrate-binding domains. Protein Sci. 1998;7:1524–1530. doi: 10.1002/pro.5560070705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tan S, Richmond T J. Eukaryotic transcription factors. Curr Opin Struct Biol. 1998;8:41–48. doi: 10.1016/s0959-440x(98)80008-0. [DOI] [PubMed] [Google Scholar]
- 275.Thomas T, Cavicchioli R. Archaeal cold-adapted proteins: structural and evolutionary analysis of the elongation factor 2 proteins from psychrophilic, mesophilic and thermophilic methanogens. FEBS Lett. 1998;439:281–286. doi: 10.1016/s0014-5793(98)01375-1. [DOI] [PubMed] [Google Scholar]
- 276.Thomm M. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 277.Thompson D K, Daniels C J. Heat shock inducibility of an archaeal TATA-like promoter is controlled by adjacent sequence elements. Mol Microbiol. 1998;27:541–551. doi: 10.1046/j.1365-2958.1998.00698.x. [DOI] [PubMed] [Google Scholar]
- 278.Thompson D K, Palmer J R, Daniels C J. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol Microbiol. 1999;33:1081–1092. doi: 10.1046/j.1365-2958.1999.01551.x. [DOI] [PubMed] [Google Scholar]
- 279.Trent J D, Gabrielsen M, Jensen B, Neuhard J, Olsen J. Acquired thermotolerance and heat shock proteins in thermophiles from the three phylogenetic domains. J Bacteriol. 1994;176:6148–6152. doi: 10.1128/jb.176.19.6148-6152.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Trent J D, Kagawa H K, Yaoi T, Olle E, Zaluzec N J. Chaperonin filaments: the archaeal cytoskeleton? Proc Natl Acad Sci USA. 1997;95:5383–5388. doi: 10.1073/pnas.94.10.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Trent J D, Nimmesgern E, Wall J S, Hartl F U, Horwich A L. A molecular chaperone from a thermophilic archaebacterium is related to the eukaryotic protein t-complex polypeptide-1. Nature. 1991;354:490–493. doi: 10.1038/354490a0. [DOI] [PubMed] [Google Scholar]
- 282.Trent J D, Osipiuk J, Pinkau T. Acquired thermotolerance and heat shock in the extremely thermophilic archaebacterium Sulfolobus sp. strain B12. J Bacteriol. 1990;172:1478–1484. doi: 10.1128/jb.172.3.1478-1484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Trieselmann B A, Charlebois R L. Transcriptionally active regions in the genome of the archaeabacterium Haloferax volcanii. J Bacteriol. 1992;174:30–34. doi: 10.1128/jb.174.1.30-34.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Tsai J, Douglas M G. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271:9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 285.Tumbula D L, Whitman W B. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol Microbiol. 1999;33:1–7. doi: 10.1046/j.1365-2958.1999.01463.x. [DOI] [PubMed] [Google Scholar]
- 286.Ueda K, Seki T, Kudo T, Yoshida T, Kataoka M. Two distinct mechanisms cause heterogeneity of 16S rRNA. J Bacteriol. 1999;181:78–82. doi: 10.1128/jb.181.1.78-82.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Vainberg I E, Lewis S A, Remmelaere H, Ampe C, Vandekerckhove J, Klein H L, Cowan N J. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- 288.Waldmann T, Lupas A, Kellermann J, Peters J, Baumeister W. Primary structure of the thermososme from Thermoplasma acidophilum. Biol Chem Hoppe-Seyler. 1995;376:119–126. doi: 10.1515/bchm3.1995.376.2.119. [DOI] [PubMed] [Google Scholar]
- 289.Waldmann T, Nimmesgern E, Nitsch M, Peters J, Pfeifer G, Müller S, Kellermann J, Engel A, Hartl F U, Baumeister W. The thermosome of Thermoplasma acidophilum and its relationship to the eukaryotic chaperonin TRiC. Eur J Biochem. 1995;227:848–856. doi: 10.1111/j.1432-1033.1995.tb20210.x. [DOI] [PubMed] [Google Scholar]
- 290.Waldmann T, Nitsch M, Klumpp M, Baumeister W. Expression of an archaeal chaperonin in E. coli: formation of homo- (α,β) and hetero-oligomeric (α+β) thermosome complexes. FEBS Lett. 1995;376:67–73. doi: 10.1016/0014-5793(95)01248-8. [DOI] [PubMed] [Google Scholar]
- 291.Washio T, Sasayama J, Tomita M. Analysis of complete genomes suggests that many prokaryotes do not rely on hairpin formation in transcription termination. Nucleic Acids Res. 1998;26:5456–5463. doi: 10.1093/nar/26.23.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Watson K. Microbial stress proteins. Adv Microb Physiol. 1990;31:183–223. doi: 10.1016/s0065-2911(08)60122-8. [DOI] [PubMed] [Google Scholar]
- 293.Weber-Ban E U, Reid B G, Miranker A D, Horwich A L. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 293a.Weiss, R. Personal communication.
- 294.Wetzstein M, Völker U, Dedio J, Löbau S, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Willison K R. Composition and function of the eukaryotic cytosolic chaperonin-containing TCP-1. In: Bukau B, editor. Molecular chaperones and folding catalysis. Sydney, Australia: Harwood Academic Publishers; 1999. pp. 555–571. [Google Scholar]
- 296.Wimmer B, Lottspeich F, van der Klei I, Veenhuis M, Gietl C. The glyoxysomal and plastid molecular chaperones (70-kDa heat shock protein) of watermelon cotyledons are encoded by a single gene. Proc Natl Acad Sci USA. 1997;94:13624–13629. doi: 10.1073/pnas.94.25.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Woese C R. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Woese C R. A manifesto for microbisal genomics. Curr Biol. 1998;8:R781–R783. doi: 10.1016/s0960-9822(07)00498-8. [DOI] [PubMed] [Google Scholar]
- 300.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wood E R, Ghané F, Grogan D W. Genetic responses of the thermophilic archaeon Sulfolobus acidocaldarius to short-wave length UV light. J Bacteriol. 1997;179:5693–5698. doi: 10.1128/jb.179.18.5693-5698.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wu B, Hunt C, Morimoto R I. Structure and expression of the human gene encoding major heat shock protein HSP70. Mol Cell Biol. 1985;5:330–341. doi: 10.1128/mcb.5.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Wu B, Wawrzynow A, Zylicz M, Georgopoulos C. Structure-function analysis of the Escherichia coli GrpE heat shock protein. EMBO J. 1996;15:4806–4816. [PMC free article] [PubMed] [Google Scholar]
- 304.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 305.Yan Z, Fujiwara S, Kohda K, Takagi M, Imanaka T. In vitro stabilization and in vivo solubilization of foreign proteins by the β subunit of a chaperonin from the hyperthermophilic archaeaon Pyrococcus sp. strain KOD1. Appl Environ Microbiol. 1997;63:785–789. doi: 10.1128/aem.63.2.785-789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Yao R, Macario A J L, Conway de Macario E. Immunochemical differences among Methanosarcina mazei S-6 morphologic forms. J Bacteriol. 1992;174:4683–4688. doi: 10.1128/jb.174.14.4683-4688.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Yaoi T, Kagawa H K, Trent J D. Chaperonin filaments: their formation and evaluation of methods for studying them. Arch Biochem Biophys. 1998;356:55–62. doi: 10.1006/abbi.1998.0758. [DOI] [PubMed] [Google Scholar]
- 308.Yoshida T, Yohda M, Iida T, Maruyama T, Taguchi H, Yazaki K, Ohta T, Odaka M, Endo I, Kagawa Y. Structural and functional characterization of homo-oligomeric complexes of α and β chaperonin subunits from the hyperthermophilic archaeum Thermococcus strain KS-1. J Mol Biol. 1997;273:635–645. doi: 10.1006/jmbi.1997.1337. [DOI] [PubMed] [Google Scholar]
- 309.Yoshida T, Yohda M, Suzuki M, Yazaki K, Miura K, Endo I. Characterization of homo-oligomeric complexes of α and β chaperonin subunits from the acidothermophilic archaeon, Sulfolobus sp. strain 7. Biochem Biophys Res Commun. 1998;242:640–647. doi: 10.1006/bbrc.1997.8026. [DOI] [PubMed] [Google Scholar]
- 310.Yuan G, Wong S L. Regulation of groE expression in Bacillus subtilis: the involvement of the ςA-like promoter and the roles of the inverted repeat sequence (CIRCE) J Bacteriol. 1995;177:5427–5433. doi: 10.1128/jb.177.19.5427-5433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Yuan G, Wong S L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Zillig W, Palm P, Klenk H-P, Langer D, Hüdephol U, Hain J, Lazendörfer M, Holz I. Transcription in archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea (archaebacteria). Amsterdam, The Netherlands: Elsevier; 1993. pp. 367–391. [Google Scholar]
- 313.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Zwickel P, Ng D, Woo K M, Klenk H-P, Goldberg A L. An archaebacterial ATPase, homologous to ATPases in the eukaryotic 26 S proteasome, activates protein breakdown by 20 S proteasomes. J Biol Chem. 1999;274:26008–26014. doi: 10.1074/jbc.274.37.26008. [DOI] [PubMed] [Google Scholar]