Abstract
Advanced and aggressive prostate cancer (PCa) depends on glutamine for survival and proliferation. We have previously shown that inhibition of glutaminase 1, which catalyzes the rate-limiting step of glutamine catabolism, achieves significant therapeutic effect; however, therapy resistance is inevitable. Here we report that while the glutamine carbon is critical to PCa survival, a parallel pathway of glutamine nitrogen catabolism that actively contributes to pyrimidine assembly is equally important for PCa cells. Importantly, we demonstrate a reciprocal feedback mechanism between glutamine carbon and nitrogen pathways which leads to therapy resistance when one of the two pathways is inhibited. Combination treatment to inhibit both pathways simultaneously yields better clinical outcome for advanced PCa patients.
Keywords: prostate cancer, therapeutic resistance, glutamine metabolism, GLS1, CAD
Introduction
Prostate cancer (PCa) is the most common non-cutaneous cancer in men, leading to ~30,000 deaths annually in the US1. Androgen deprivation therapy (ADT) remains the mainstay for patients with advanced and metastatic PCa. Although the treatment is efficacious initially, the disease will eventually recur as castration resistant PCa (CRPC). Second generation hormonal therapy drugs, such as abiraterone and enzalutamide, are useful for CRPC but resistance eventually develops2. Histologically, most cases relapse as adenocarcinoma, maintaining luminal differentiation including the expression of androgen receptor (AR) and prostate specific antigen (PSA). However, in up to 25% of the patients, the recurrent tumor after hormonal therapy shows neuroendocrine (NE) phenotype with suppressed AR signaling 3-5. Thus, developing therapeutic strategies independent of AR is a pressing unmet clinical need.
Rewired cellular metabolism is one of the most significant cancer hallmarks which is important to meet the needs of tumor cells’ uncontrolled proliferation6. In addition to the well-known Warburg effect which describes glucose flux primarily towards lactate, glutamine has also been implicated as a pleiotropic energy and building source in many solid cancer types7,8. Our recent publication has discovered that therapy-resistant PCa is extremely addicted to glutamine9. Thus, targeting glutamine metabolism is an attractive therapeutic strategy for androgen/AR-independent PCa.
As an anaplerotic nutrient that fuels the tricarboxylic acid (TCA) cycle, glutamine’s carbon backbone incorporates into α-ketoglutarate (α-KG) through glutaminase 1 (GLS1)-mediated catabolism10. Although our study has demonstrated therapeutic potential of inhibiting GLS1 in therapy-resistant PCa, experience in other tumor types suggests that monotherapy targeting GLS1 produces limited efficacy 11,12. Importantly, the nitrogen released during the process of glutamine carbon catabolism is used for the synthesis of nucleotides and other nitrogen-containing molecules, which are also critical for the proliferating cancer cells13-15. The coordinated roles of glutamine carbon and nitrogen in cancer cells’ energy production and biosynthesis suggest that targeting both arms of glutamine catabolism (nitrogen and carbon) may result in better therapeutic outcomes than targeting either pathway alone in therapy-resistant PCa.
In this study, we show that glutamine nitrogen participates in pyrimidine biosynthesis in therapy-resistant PCa. Importantly, we uncovered a mechanism of reciprocal regulation of glutamine carbon and nitrogen catabolism. Our results demonstrate that inhibition of glutamine carbon catabolism results in a compensatory increase of glutamine nitrogen utilization, thereby maintaining tumor cell survival. Combinatorial therapy targeting both pathways leads to near complete tumor regression which is a promising strategy for the treatment of advanced PCa.
Results
1. Glutamine carbon and nitrogen are both required by advanced PCa cells.
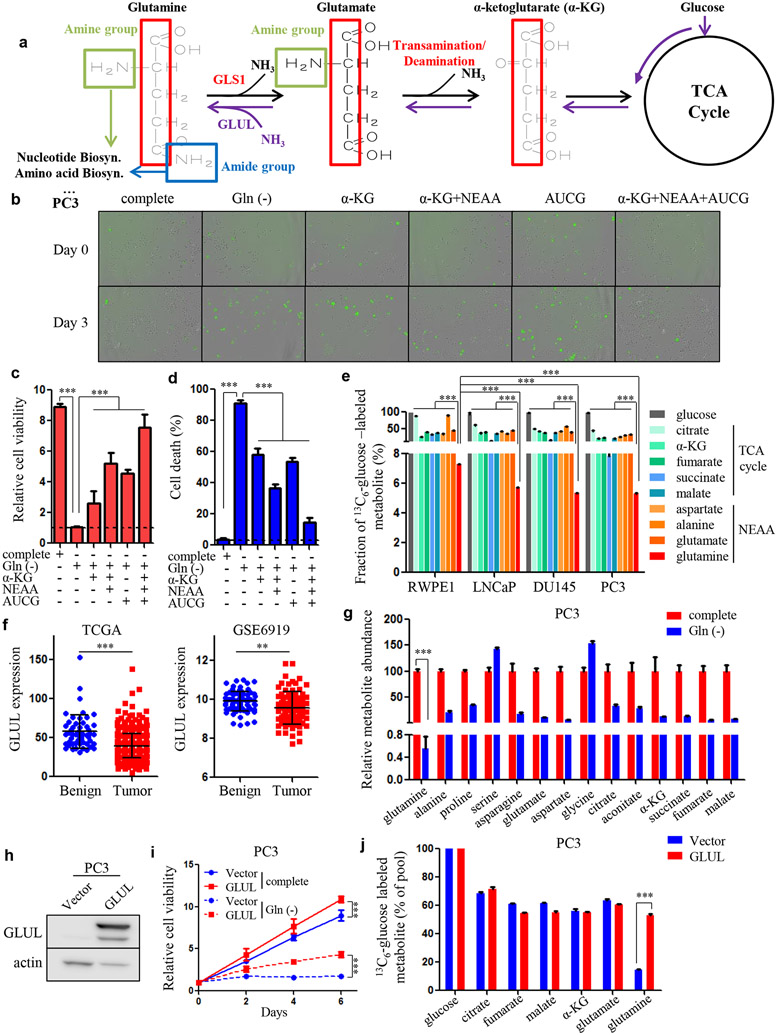
Glutamine contains carbon and nitrogen, which helps to maintains the TCA cycle activity and contributes to amino acid and nucleotide biosynthesis, respectively16 (Fig. 1a). Glutamine depletion led to dramatic cell growth arrest and cell death in two AR-negative, androgen-independent PCa cell lines, PC3 and DU145 (Fig. 1b-d and Supplementary Fig. S1a-c), consistent with our previous observation that advanced PCa cells are addicted to glutamine9. Supplementing the culture media with α-KG, the glutamine carbon inheritor, modestly rescued the cell death due to glutamine deprivation (Fig. 1b-d and Supplementary Fig. S1a-c). Importantly, this rescue effect was enhanced by the addition of non-essential amino acids (NEAAs, glutamine not included) (Fig. 1b-d and Supplementary Fig. S1a-c). In addition to the synthesis of NEAAs, glutamine nitrogen, particularly the amide group, also participates in the production of nucleotides 17. Indeed, supplementation of nucleosides in the culture media also showed a partial rescue effect under glutamine-deprived conditions (Fig. 1b-d and Supplementary Fig. S1a-c). Notably, combination treatment of α-KG, NEAAs and nucleosides achieved a synergistic rescue effect (Fig. 1b-d and Supplementary Fig. S1a-c). These results indicate that both glutamine carbon and nitrogen are needed by PCa cells for optimal survival and proliferation.
Figure 1.
Glutamine carbon and nitrogen are required equivalently for tumor survival in advanced PCa. A, Schematic overview showing glutamine carbon and nitrogen metabolism. B, Cellular viability and apoptosis in PC3 cells cultured with complete or glutamine-free medium and glutamine-free medium supplemented with dimethyl-α-ketoglutarate (α-KG, 2 mM) combined with/without NEAA solution (2 mM) and glutamine-free medium supplemented with nucleosides (0.1 mM of adenosine (A), guanosine (G), cytidine (C) and uridine (U)) alone or in combination with α-KG and NEAA solution. Green fluorescent dots indicate cellular apoptosis. C and D, Quantification of relative cell viability and the percentage of apoptotic cells in (B). E, Mass isotopomer analysis of 13C6-glucose-derived metabolites in RWPE1, LNCaP, DU145 and PC3 cells. Fractions of the 13C6-glucose-labeled metabolite of the total metabolic pool are shown. F, GLUL gene expression retrieved from the indicated datasets. G, Metabolite abundance in PC3 cells treated with complete medium or glutamine-free medium. H, Western blot showing GLUL overexpression in PC3 cells. I, Cellular viability determining the effect of GLUL overexpression in complete medium and under glutamine-deprived conditions in PC3 cells. J, Mass isotopomer analysis of 13C6-glucose-derived metabolites in PC3/Vector and PC3 with GLUL overexpressed cells. Data are depicted as mean ± s.d. * P < 0.05, ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t-test or ANOVA.
Glutamine is an NEAA and can be endogenously synthesized by cells in addition to being absorbed from outside of the cell. The de novo glutamine synthesis is mediated by glutamine synthetase (GS)/glutamate-ammonia ligase (GLUL), which catalyzes the condensation of glutamate and ammonia18 (Fig. 1a). Since glucose is the major donor of the glutamine carbon backbone, we performed 13C6-glucose isotopomer tracing to determine the degree of glutamine formation in benign prostatic epithelial cell line RWPE1 as well as AR-positive PCa cell line, LNCaP, and AR-negative cell lines, PC3 and DU145. Compared to the substantially labeled metabolites in the TCA cycle and other NEAAs, such as aspartate, alanine and glutamate, glucose was barely incorporated into glutamine across the tested cell lines (Fig. 1e), suggesting that glutamine anabolism is not active in prostate. We observed a significant block of conversion from glutamate (~50% glucose-labeled fraction) to glutamine (~7% glucose-labeled fraction) (Fig. 1e), suggesting that the activity of GLUL may be low. Consistent with this observation, the expression of the GLUL gene, which encodes the key enzyme for glutamine production, was expressed at relatively low levels in PCa in comparison to other cancers (Supplementary Fig. S1d), indicating that PCa does not carry out robust glutamine synthesis. Moreover, in comparing fractions of isotopomer-labeled glutamine in RWPE1 and other three PCa cell lines, we found that glucose-derived glutamine was lower in all tested PCa cells (LNCaP, PC3 and DU145) than in benign prostatic epithelial cells (RWPE1) (Fig. 1e), which was corroborated by two public datasets that also demonstrated lower GLUL gene expression in PCa tissues than in benign tissues (Fig. 1f). Consistent with these observations, glutamine deprivation caused a dramatic decrease in intracellular glutamine levels (about 200-fold) in PC3 and DU145 cells but had much less effect on other NEAAs and the TCA cycle intermediates (Fig. 1g and Supplementary Fig. S1e), indicating that unlike other metabolites, glutamine cannot be efficiently produced in PCa due to low expression of GLUL and exogenous source is critical for tumor cell survival.
We ectopically expressed GLUL in PC3 and DU145 cells (Fig. 1h and Supplementary Fig. S1f) and observed increased cellular viability and profound rescue effect under glutamine-depleted conditions (Fig. 1i and Supplementary Fig. S1g). We also found that overexpression of GLUL in the two cell lines dramatically increased the glucose-labeled glutamine proportions whereas other metabolites remained essentially unaffected (Fig. 1j and Supplementary Fig. S1h). Taken together, these results suggest that glutamine is a “conditional essential” amino acid in PCa and both carbon and nitrogen components are required for cell survival and proliferation.
2. Glutamine-nitrogen is enriched in pyrimidine nucleotides in advanced PCa cells.
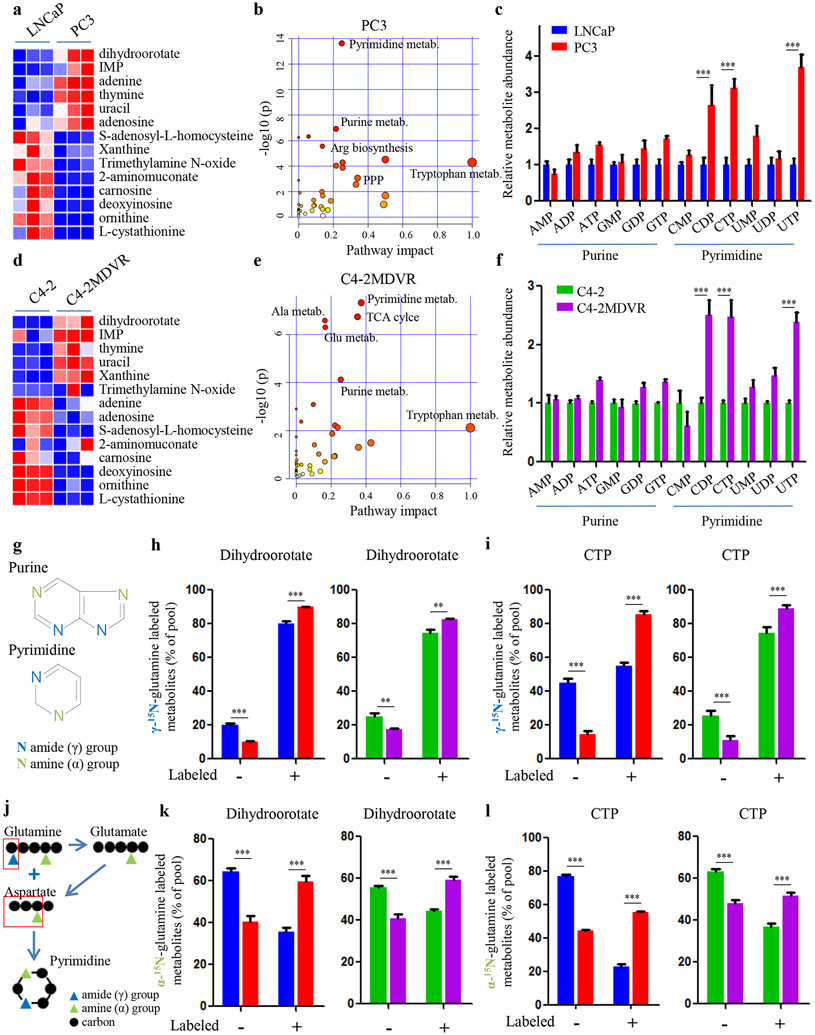
Due to an isoform switch of GLS1, glutamine carbon is more efficiently oxidized via the TCA cycle in therapy-resistant PCa than primary hormone-sensitive PCa to help tumor cells escape the inhibitory effect of ADT9. However, this process would generate excess glutamine nitrogen which is potentially toxic to tumor cells (Fig. 1a). To investigate how advanced PCa cells dispose of the accumulated nitrogen while utilizing glutamine carbon, we performed nitrogen-targeted metabolite profiling analyses to compare metabolic reprogramming between AR-positive, androgen-dependent LNCaP cells and androgen-independent, AR-null PC3 cells. Interestingly, in PC3 cells, nitrogen was significantly enriched in metabolites involved in nucleotide biosynthesis pathways such as dihydroorotate and IMP, the precursors for pyrimidine and purine synthesis, respectively (Fig. 2a). Pathway impact and enrichment analyses further demonstrated that pyrimidine and purine synthesis were among the top affected metabolic pathways in PC3 cells (Fig. 2b and Supplementary Fig. S2a). In addition, cellular nucleotides and their derivatives were increased in PC3 cells compared to LNCaP cells (Fig. 2c). This metabolic rewiring of nitrogen and its association with disease progression was also verified in an isogenic pair of cell lines, C4-2 (enzalutamide-sensitive) and C4-2MDVR (enzalutamide-resistant) (Fig. 2d-f and Supplementary Fig. S2b), suggesting that advanced PCa preferentially uses nitrogen to synthesize nucleotides.
Figure 2.
Glutamine nitrogen is largely enriched in nucleotide biosynthesis. A and D, Heat-map showing the comparison of nitrogen-contained metabolites in two paired PCa cell lines (LNCaP versus PC3 in A and C4-2 versus C4-2MDVR in D). B and E, Pathway impact analysis showing metabolic pathways that are significantly altered in advanced PCa cells (PC3 in B, C4-2MDVR in E). C and F, Metabolite abundance of nucleotides and derivatives in the indicated PCa cell lines. G, Chemical structure of purine and pyrimidine bases. H and I, Amide (γ)-15N-glutamine tracing analysis showing the incorporation of glutamine into dihydroorotate (H) and CTP (I) in the indicated PCa cell lines. J, Schematic overview of how glutamine amine (α) and amide (γ)-nitrogen contribute to the pyrimidine assembly. K and L, Amine (α)-15N-glutamine tracing analysis showing the incorporation of glutamine into dihydroorotate (K) and CTP (L) in the indicated PCa cell lines. Data are depicted as mean ± s.d. * P < 0.05, ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t-test.
The fact that glutamine deprivation of androgen-independent PCa cell lines (PC3 and DU145) dramatically reduced the levels of cellular nucleotides and their derivatives indicates that glutamine is indispensable for nucleotide generation (Supplementary Fig. S2c). To determine how glutamine nitrogen contributes to nucleotide biosynthesis in advanced PCa, we performed 15N-glutamine isotopomer tracing studies in PCa cell lines. Surprisingly, the amide nitrogen group, which directly participates in purine and pyrimidine synthesis (Fig. 2g), was minimally assimilated into the purine synthetic pathway in the more advanced PC3 and C4-2MDVR cells, as there was minimal labeling of IMP and ATP by glutamine (Supplementary Fig. S2d). In contrast, the labeled fractions of dihydroorotate and CTP were high in PC3 and C4-2MDVR cells (Fig. 2h,i), suggesting that advanced, therapy-resistant PCa preferentially utilizes glutamine amide-nitrogen for pyrimidine over purine production. In support of this conclusion, although we found that PC3 and C4-2MDVR cells had higher levels of purine and pyrimidine concentrations than LNCaP and C4-2 cells, the increases in the levels of pyrimidine nucleotides (CDP, CTP and UTP) were much more pronounced (Fig. 2c,f).
Unlike glutamine amide nitrogen, glutamine amine nitrogen does not directly contribute to pyrimidine synthesis but can potentially be integrated into pyrimidine rings after being transferred to aspartate through glutamate-aspartate transamination (Fig. 2j). Amine-15N-glutamine tracing studies showed that glutamate and aspartate were substantially labeled and the glutamine-derived fractions were significantly increased in PC3 and C4-2MDVR cells (Supplementary Fig. S2e). In line with the higher incorporation of glutamine amide-15N in dihydroorotate and CTP (Fig. 2h,i), we also found that advanced PCa cells assimilated more glutamine amine-15N into dihydroorotate and CTP synthesis than the AR-positive, hormone-sensitive LNCaP and C4-2 cells (Fig. 2k,l). Taken together, these findings demonstrate that advanced PCa cells utilize the released glutamine nitrogen for nucleotide, particularly pyrimidine, biosynthesis to keep pace with the hyper-activity of glutamine carbon oxidation towards the TCA cycle.
3. Glutamine carbon is associated with glutamine nitrogen in enhanced pyrimidine synthesis in advanced PCa cells.
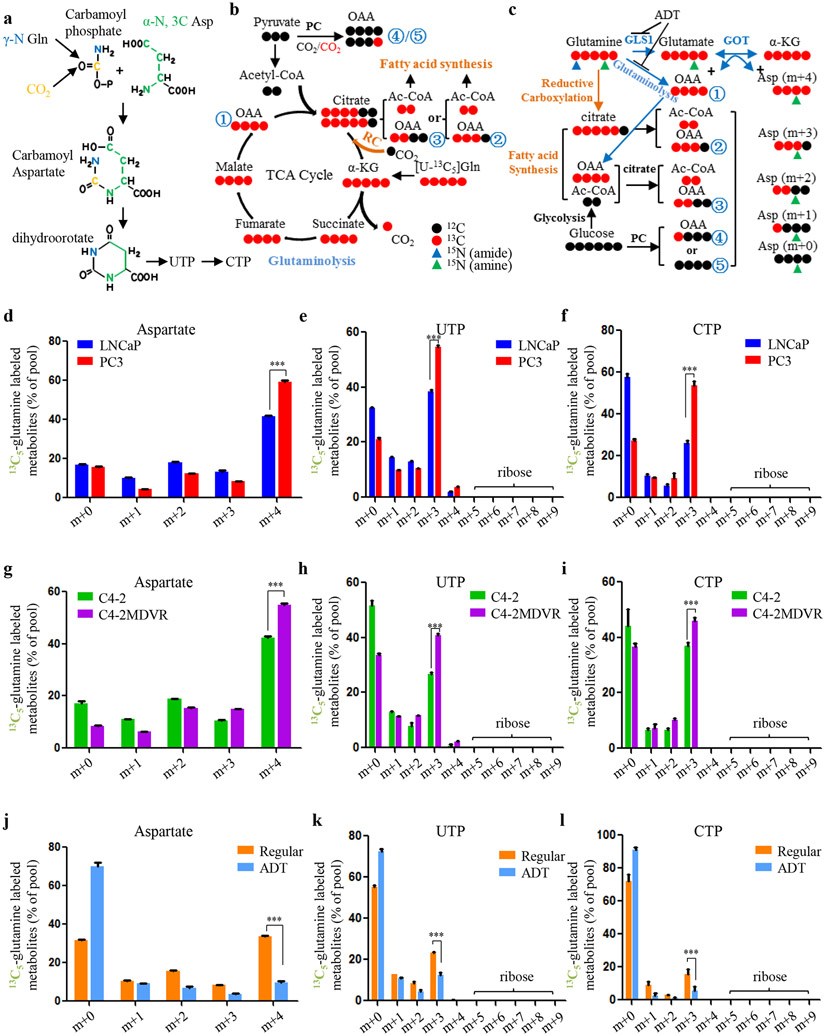
In addition to the requirement of glutamine nitrogen, glutamine carbon is also an important contributor for nucleotide biosynthesis. Generally, glutamine passages its carbon to aspartate which is the direct precursor of dihydroorotate (m+3) (Fig. 3a). This carbon transfer is usually mediated by the generation of oxaloacetic acid (OAA). Isotopomer tracing flow shows that glutaminolysis is the pathway for m+4 OAA generation while the reductive carboxylation pathway is able to produce m+3 and m+2 OAA as the by-products split from citrate (Fig. 3b). Moreover, m+1 and m+0 OAA can be traced from glycolysis depending on whether the assimilated carbon dioxide (CO2) is derived from glutamine or not (Fig. 3b). Accordingly, m+4, m+3, m+2, m+1 and m+0 aspartate are formed through transamination by which OAA accepts amine-nitrogen from glutamate while retaining the carbon backbone from glutamine/glutamate (Fig. 3c). To determine whether glutamine carbon catabolism is associated with its nitrogen catabolic rates when advanced PCa cells actively synthesize pyrimidine, we traced the 13C5-glutamine isotope-labeled intermediates in pyrimidine biosynthesis. Across all the tested cell line models, the proportion of m+4 aspartate was the largest among the aspartate pool, suggesting that glutaminolysis is the dominant pathway for aspartate synthesis in PCa (Fig. 3d, g). In comparing the metabolic differences between cell lines representing different stages of the disease, we found that the more advanced PC3 and C4-2MDVR cells had more m+4 aspartate proportions than LNCaP and C4-2 cells (Fig. 3d, g). Accordingly, we observed significantly increased levels of glutamine-aspartate-derived pyrimidine nucleotides (m+3 UTP and m+3 CTP) in advanced PCa cells (Fig. 3e, f, h, i). Conversely, when glutaminolysis, catalyzed by the rate-limiting enzyme GLS1, was suppressed by ADT9, the glutamine-derived aspartate was dramatically decreased which led to significant reduction of m+3 UTP and m+3 CTP (Fig. 3j-l). These findings, together with our previously published data, indicate that while advanced PCa avidly consumes glutamine carbon, the intermediates (such as OAA and aspartate) can coordinate with the released glutamine nitrogen to participate in pyrimidine biosynthesis.
Figure 3.
Glutamine carbon is associated with glutamine nitrogen in building pyrimidine rings in PCa. A, Schematic overview of the synthesis of pyrimidine. B and C, Schematic overview of glutamine carbon flux toward related metabolites. Gln, glutamine. Asp, aspartate. OAA, oxaloacetic acid. α-KG, α-ketoglutarate. Ac-CoA, acetyl-CoA. PC, pyruvate carboxylase. GLS1, glutaminase. GOT, glutamic oxaloacetic transaminase. RC, reductive carboxylation. ADT, androgen deprivation therapy. D-I, Fractions of 13C5-glutamine derived isotopologues in aspartate (D and G), UTP (E and H) and CTP (F and I) in the indicated PCa cell lines. J-L, Fractions of 13C5-glutamine derived isotopologues in aspartate (J), UTP (K) and CTP (L) in LNCaP cells treated in regular medium or charcoal-stripped medium (ADT). Data are depicted as mean ± s.d. * P < 0.05, ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t-test.
Purine bases normally obtain carbon from glycine and the one-carbon unit, which is frequently generated via conversion between serine and glycine (Supplementary Fig. S3a). As an indispensable component for purine assembly, glycine is synthesized from serine, which is originally generated by glucose through phophoglycerate. To this end, the carbon backbone of glycine is largely derived from glucose, independent of glutamine, although the nitrogen part can be acquired via glutamine metabolism (Supplementary Fig. S3b). Indeed, glutamine carbon inefficiently labeled serine, glycine and ATP in all tested cell lines (Supplementary Fig. S3c-e). Additionally, no differences in isotope fractions were observed between paired cell lines representing different stages of PCa (Supplementary Fig. S3c-e). Moreover, inhibition of glutamine metabolism by ADT showed little effect on the carbon constitution of the metabolite pools of serine, glycine and ATP (Supplementary Fig. S3f-h). These observations suggest that glutamine may not be the major carbon contributor to purine synthesis in PCa.
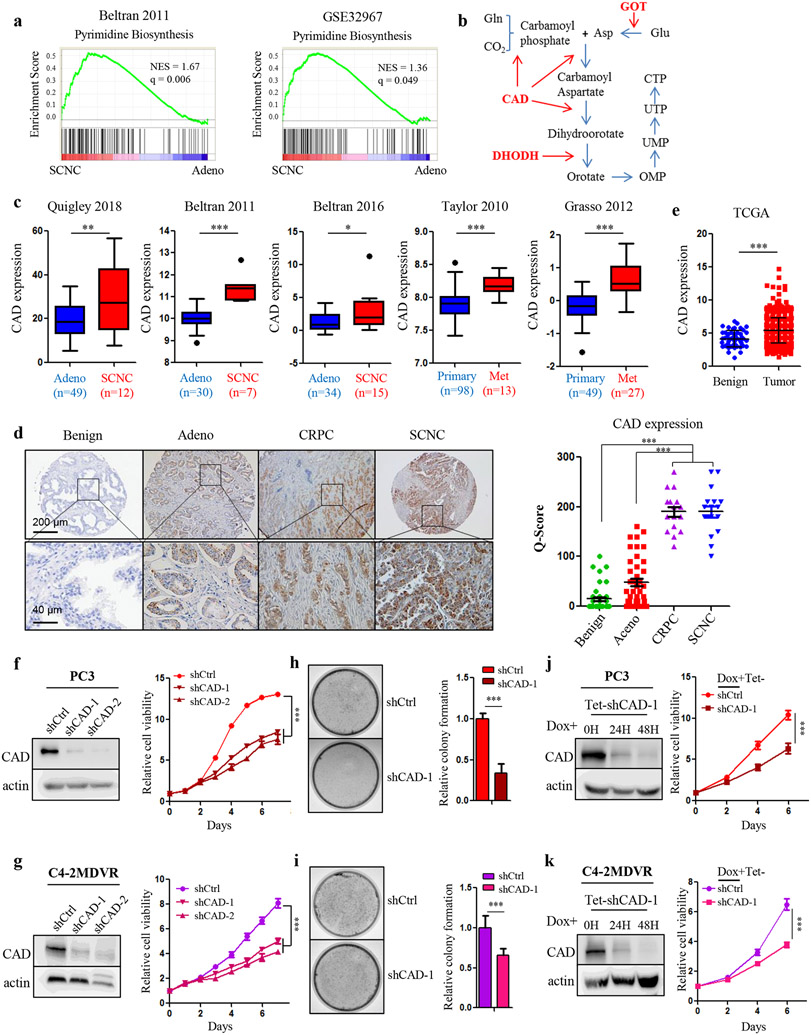
4. CAD is the key enzyme for pyrimidine synthesis in advanced PCa.
The above findings clearly demonstrate that the addiction of advanced PCa to glutamine leads to not only hyper activity of the TCA cycle, but also enhanced pyrimidine biosynthesis in tumor cells. Bioinformatics analyses of public datasets of human PCa specimens confirmed that genes belonging to pyrimidine synthesis pathway were significantly enriched in small cell neuroendocrine prostate cancer (SCNC), the most aggressive histologic variant of PCa often seen in terminal stages of the disease after hormonal therapy (Fig. 4a). There are three dominant enzymes in this anabolic pathway, carbamoyl-phosphate synthetase 2, aspartate transcarbamylase and dihydroorotase (CAD), dihydroorotate dehydrogenase (DHODH) and glutamic oxaloacetic transaminase (GOT). CAD is responsible for the first three steps of pyrimidine synthesis while DHODH catalyzes the subsequent reaction converting dihydroorotate to orotate (Fig. 4b). GOT is the key enzyme for aspartate generation catalyzing the transfer of an amine-group nitrogen between glutamate and aspartate (Fig. 4b). To determine if some or all of the pyrimidine biosynthesis enzymes play a role in advanced PCa, we queried publicly available datasets to compare gene expression between primary adenocarcinoma and therapy-resistant tumors. In all the cohorts analyzed, CAD was universally upregulated in advanced PCa, including SCNC and metastatic PCa, in comparison to primary adenocarcinoma samples 19-23 (Fig. 4c). In contrast, the expression of DHODH and GOT did not differ between hormone-naïve and therapy-resistant PCa in these datasets (Supplementary Fig. S4a-c). To confirm these results, we examined CAD expression by immunohistochemical (IHC) staining using tissue microarrays (TMAs) including benign prostate, primary PCa, castration-resistant PCa (CRPC) and SCNC. We found that CAD-positive cells were rarely present in benign tissues (Fig. 4d), which was consistent with the findings from the Cancer Genome Atlas (TCGA) dataset showing an overall lower expression of CAD in benign tissues (Fig. 4e). In contrast, CAD expression was slightly elevated in adenocarcinoma, and significantly enriched in CRPC and SCNC (Fig. 4d). These results strongly support those of bioinformatics analyses and together, suggest that CAD is associated with disease progression of human PCa (Fig. 4b).
Figure 4.
CAD plays a predominant role in advanced PCa. A, GSEA of pyrimidine biosynthesis signatures in SCNC and adenocarcinoma samples from Beltran 2011 and GSE32967 datasets. Adeno, adenocarcinoma. SCNC, small cell neuroendocrine prostate cancer. B. Schematic overview showing the process and the key enzymes of pyrimidine synthesis. Gln, glutamine. Glu, glutamate. Asp, aspartate. CAD, carbamoyl-phosphate synthetase 2, aspartate transcarbamylase and dihydroorotase. DHODH, dihydroorotate dehydrogenase. GOT, glutamic oxaloacetic transaminase. C, Bioinformatics analysis showing CAD gene expression in a variety of study cohorts. D, Representative images of CAD IHC staining on the TMAs including benign prostate tissue (n = 40), primary prostate adenocarcinoma (n = 40), CRPC (n = 16) and SCNC (n = 17). CRPC, castration-resistant prostate cancer. A plot of Quick-score quantifying CAD expression (the intensity of staining x the percentage of staining) in each group. P values were calculated using unpaired t-test. Scale bars are shown as indicated. E, Bioinformatics analysis showing CAD gene expression in benign samples and tumor samples from the TCGA dataset. F and G, Western blot showing two sets of shRNAs targeting CAD in PC3 (F) and C4-2MDVR (G) cells. Cellular viability was measured in the corresponding cell lines. H and I, Colony formation assay determining tumorigenic capability in the indicated cell lines in PC3 (H) and C4-2MDVR (I) cells. J and K, Western blot showing CAD protein levels in PC3 (J) and C4-2MDVR (K) cells transduced with shRNAs against CAD in an inducible vector system at various time points. Cellular viability was measured in the corresponding cell lines. Doxycycline (100 ng/ml) was added at Day 0. Data are depicted as mean ± s.d. * P < 0.05, ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t-test or ANOVA.
We next performed experiments to study the function of CAD in PCa. Ectopic expression of short-hairpin RNAs (shRNAs) targeting CAD reduced cellular proliferation of PC3 and C4-2MDVR cells whereas knocking down DHODH or GOT1 had little impact on PCa cells (Fig. 4f,g and Supplementary Fig. S4d,e). Similar results were obtained by colony formation assays (Fig. 4h,i and Supplementary Fig. S4f,g). To confirm the results obtained through constitutive expression of shRNAs, we employed a doxycycline-inducible shRNA knockdown model to study loss-of-function phenotypes. Similarly, cellular proliferation decreased dramatically upon doxycycline treatment targeting CAD (Fig. 4j,k). Importantly, CAD deficiency did not show any significant impact on the benign prostatic epithelial cell line RWPE1 (Supplementary Fig. S4h). These results suggest that CAD is required in advanced PCa.
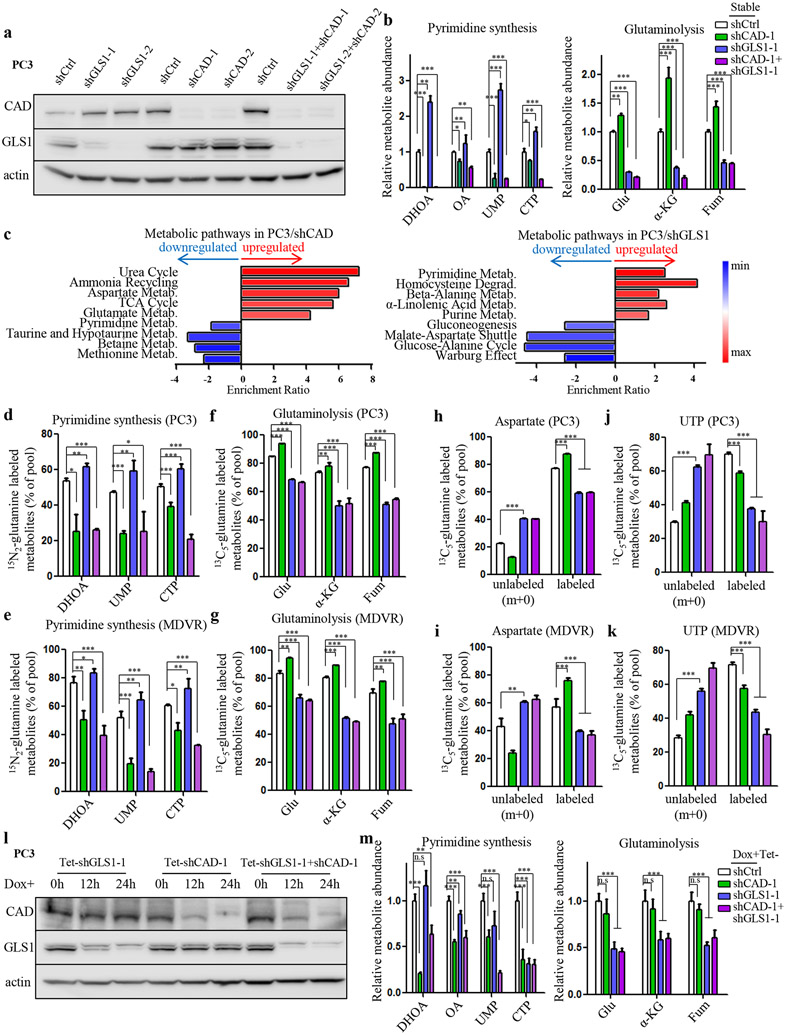
5. Reciprocal regulation between glutamine carbon and nitrogen pathways in PCa.
Although acute inhibition of either CAD or GLS1, key enzymes for glutamine nitrogen and carbon catabolism, respectively, can achieve significant tumor suppression in advanced PCa, the inhibitory effect gradually diminished over time (Supplementary Fig. S5a). This observation raised the possibility that tumor cells may acquire resistance through compensatory mechanisms after either pathway is inhibited. Given that glutamine carbon and nitrogen pathways both promote tumor proliferation and are tightly connected, we hypothesized that there may be a reciprocal regulation of the two pathways in PCa cells so that inhibition of one pathway will eventually lead to the activation of the other, resulting in therapeutic resistance. To determine whether the cellular metabolism underwent reprogramming in those genetically modified cells that are resistant to either GLS1 or CAD inhibition, we performed metabolite profiling experiments in cells with single knockdown of GLS1 or CAD, respectively and their combination knockdown in PC3 and C4-2MDVR cells (Fig. 5a and Supplementary Fig. S5b). As expected, the downstream products of CAD (dihydroorotate, orotate, UMP and CTP) and GLS1 (glutamate, α-KG and fumarate), respectively, were markedly reduced when CAD and GLS1 were knocked down (Fig. 5b and Supplementary Fig. S5c). In the meantime, many other metabolic pathways and metabolites were upregulated. For instance, loss of CAD activated ammonia assimilation pathways, such as urea cycle and ammonia recycling (Fig. 5c), largely because pyrimidine synthesis pathway can also function to scavenge the accumulated ammonia13. Surprisingly, loss of CAD also led to significantly enhanced glutaminolysis-related pathways including glutamate metabolism, aspartate metabolism and the TCA cycle (Fig. 5c), as well as increased levels of specific intermediate (glutamate, α-KG and fumarate) (Fig. 5b and Supplementary Fig. S5c). Conversely, suppressing GLS1 elevated pyrimidine synthesis activity (Fig 5b,c and Supplementary Fig. S5c). The reciprocal regulation of glutamine carbon and nitrogen catabolism was abolished when both enzymes were repressed (Fig. 5b and Supplementary Fig. S5c). Interestingly, while there was a moderate elevation of intracellular glutamine concentration in cells deficient in either GLS1 or CAD, the increase was much more dramatic in cells in which both enzymes had been knocked down (Supplementary Fig. S5d,e), indicating that inhibition of both pathways maximally prevents glutamine from being utilized by the tumor cells.
Figure 5.
Reciprocal regulation between glutamine carbon and nitrogen metabolism in PCa. A, Western blot showing knockdown of either GLS1 or CAD or in combination mediated by shRNAs in PC3 cells. B, Metabolite profiling results showing levels of representative metabolites in pyrimidine synthesis and glutaminolysis upon indicated treatment in PC3 cells. C, Pathway enrichment analysis showing upregulated metabolic pathways in CAD-deficient (left panel) and GLS1-deficient (right panel) PC3 cells. D and E, 15N2-glutamine tracing analysis showing the incorporation of glutamine into CAD downstream metabolites in PC3 (D) and C4-2MDVR (E) cells. F and G, 13C5-glutamine tracing analysis showing the incorporation of glutamine into GLS1 downstream metabolites in PC3 (F) and C4-2MDVR (G) cells. H and I, 13C5-glutamine tracing analysis showing the fraction of glutamine-labeled and glutamine-unlabeled aspartate in PC3 (H) and C4-2MDVR (I) cells. J and K, 13C5-glutamine tracing analysis showing the fraction of glutamine-labeled and glutamine-unlabeled UTP in PC3 (J) and C4-2MDVR (K) cells. L, Western blot showing doxycycline-induced knockdown (100 ng/ml) of either GLS1 or CAD or in combination mediated by shRNAs in PC3 cells. M, Metabolite profiling results showing levels of representative metabolites in pyrimidine synthesis and glutaminolysis upon indicated treatment in PC3 cells. DHOA, dihydroorotate. OA, orotate. Glu, glutamate. α-KG, α-ketoglutarate. Fum, fumarate. n.s, not significant. Data are depicted as mean ± s.d. * P < 0.05, ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t-test or ANOVA.
To determine whether the CAD-GLS1 balance affects glutamine flux distributions, we performed isotopomer analysis with 13C5/15N2-glutamine. In agreement with the metabolite profiling results above, inhibition on CAD markedly decreased the 15N labeling for dihydroorotate, UMP and CTP but increased 13C fractions of glutamate, α-KG and fumarate (Fig. 5d,e). Similarly, loss of GLS1 reduced the glutamine carbon flux towards its downstream products while increasing 15N incorporation for the pyrimidine synthesis pathway (Fig. 5f,). These isotopomer analyses suggest that blocking one pathway (carbon or nitrogen) of glutamine metabolism causes a detour of its metabolic flux to the other. 13C5-glutamine tracing analyses also revealed that glutamine-derived aspartate, particularly the m+4 aspartate, decreased dramatically upon loss of GLS1 (Fig. 5h,i and Supplementary Fig. S5f,g). Accordingly, significant reduction of glutamine-labeled carbon was observed in UTP (Fig. 5j,k and Supplementary Fig. S5h,i). In contrast, the fractions of m+0 aspartate and the resultant m+0 UTP were markedly elevated (Fig. 5h-k), indicating that although glutamine carbon is no longer able to contribute to the building of pyrimidine base due to GLS1 deficiency, other non-glutamine carbon sources were engaged in the process to keep pace with the loss-of-GLS1-facilitated glutamine nitrogen incorporation rate for pyrimidine synthesis. Additionally, as the direct precursor of pyrimidine bases, although aspartate carbon production from glutamine was induced by CAD suppression (Fig. 5h,i and Supplementary Fig. S5f,g), its pyrimidine product UTP, particularly the m+3 UTP, was not increased accordingly (Fig. 5j,k and Supplementary Fig. S5h,i). This was because loss of CAD hindered glutamine/aspartate-derived carbon flux into pyrimidine synthesis and decreased the overall levels of UMP and CTP (Fig. 5b and Supplementary Fig. S5c). Taken together, our observations demonstrate that CAD-GLS1 balance governs glutamine fate and a reciprocal regulation exists between glutamine carbon and nitrogen catabolism in PCa.
The metabolic shift between glutamine carbon and nitrogen enables tumor cells to continue using glutamine when either GLS1 or CAD is inhibited. However, since significant tumor cell inhibition was observed in advanced PCa upon short-term downregulation of either enzyme alone, we wondered if long-term treatment is required to trigger this compensatory mechanism. Consistent with this hypothesis, doxycycline-induced acute knockdown of GLS1 or CAD reduced their respective downstream metabolites; however, the other glutamine catabolism branch was not activated as shown by the relatively unchanged levels of the downstream products (Fig. 5l,m and Supplementary Fig. S5j,k). Similar to stable shRNA treatment, both pathways were inhibited when the two enzymes were knocked down simultaneously (Fig. 5l,m and Supplementary Fig. S5j,k). Collectively, the above results indicate that although monotherapy that inhibits one branch of glutamine metabolism can be effective initially, tumor cells will eventually develop resistance by activating the other metabolic branch through a compensatory mechanism.
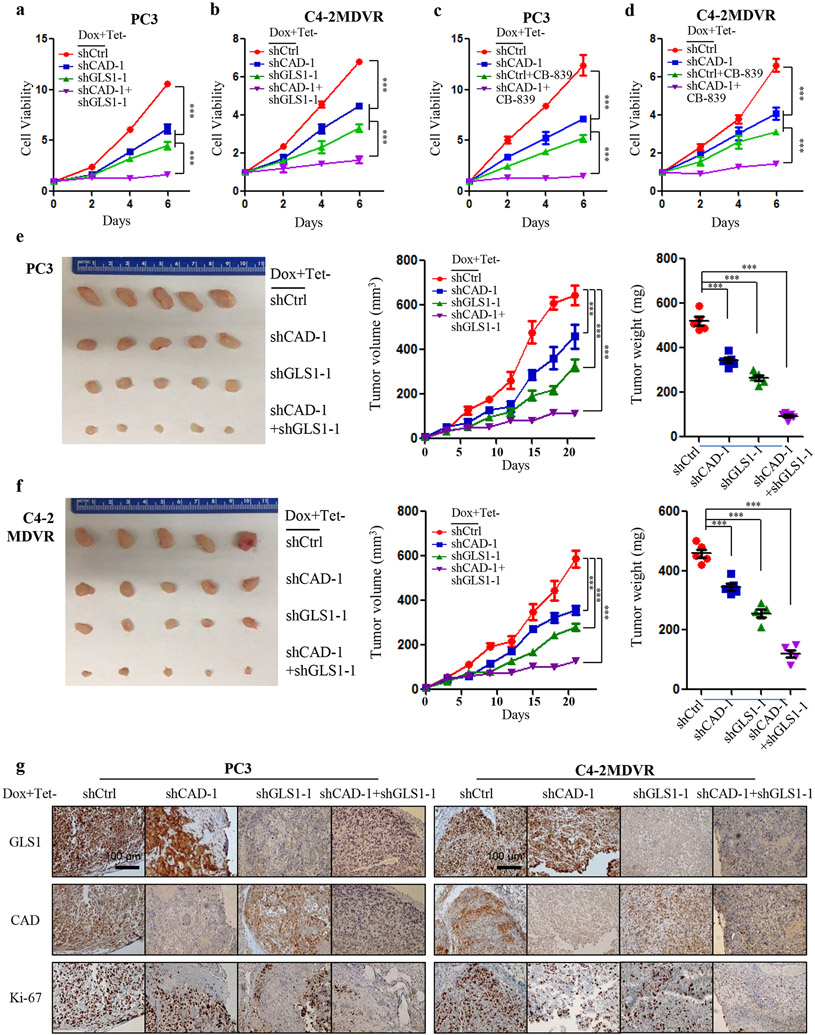
6. Combined targeting of both branches of glutamine catabolism achieves superior inhibitory effects in advanced PCa cells.
Since the reciprocal regulation of the two branches of glutamine catabolism network is an important resistance mechanism after glutamine carbon- or nitrogen-targeted monotherapy, we tested whether simultaneously targeting both pathways in advanced PCa could achieve a synergistic effect. Doxycycline-induced reduction of GLS1 or CAD displayed modest activity in inhibiting PC3 and C4-2MDVR cells (Fig. 6a,b). However, their combined knockdown led to a more profound inhibition of cell viability (Fig. 6a,b). The combination of CAD knockdown together with CB-839, a selective GLS1 inhibitor, also achieved significant inhibitory effects in advanced PCa cells (Fig. 6c,d). To demonstrate if the combinatorial effect can be observed in vivo, we subcutaneously transplanted PC3 and C4-2MDVR cells into immuno-compromised mice and fed mice with doxycycline chow to induce functional knockdown of GLS1 and CAD. Similar to the in vitro findings, tumor volumes and weights were marginally decreased in CAD knockdown group, substantially reduced in GLS1 knockdown group and the strongest inhibition was observed in the group receiving double knockdown (Fig. 6e,f). The in vivo effects on the protein levels of the two enzymes after doxycycline addition were confirmed by IHC staining (Fig. 6g). IHC study also showed that the decrease in the expression of the proliferative marker Ki-67 was more pronounced in the group with dual knockdown (Fig. 6g). Collectively, our data demonstrates that simultaneous targeting of glutamine carbon and nitrogen pathways is a more efficacious treatment approach.
Figure 6.
Synergistic effect of CAD and GLS1 inhibtion. A and B, Cellular viability of scramble control, CAD/GLS1 knockdown alone or in combination upon doxycycline induction (100 ng/ml) in PC3 (A) and C4-2MDVR (B) cells. C and D, Cellular viability of scramble control, CAD knockdown/CB-839 (500 nM) alone, or in combination upon doxycycline induction (100 ng/ml) in PC3 (C) and C4-2MDVR (D) cells. E and F, Representative images, relative tumor growth rate and tumor weights showing the effect of knockdown of CAD/GLS1 alone or dual knockdown in PC3 (E) and C4-2MDVR (F) xenograft tumors compared to that of the intact tumors. The concentration of doxycycline is 200 mg per kg of diet. G, Representative images of IHC staining showing GLS1, CAD and Ki-67 protein expression in the indicated xenograft tumors. Scale bars are shown as indicated. Data are depicted as mean ± s.d. * P < 0.05, ** P < 0.01 and *** P < 0.001 by two-tailed Student’s t-test or ANOVA.
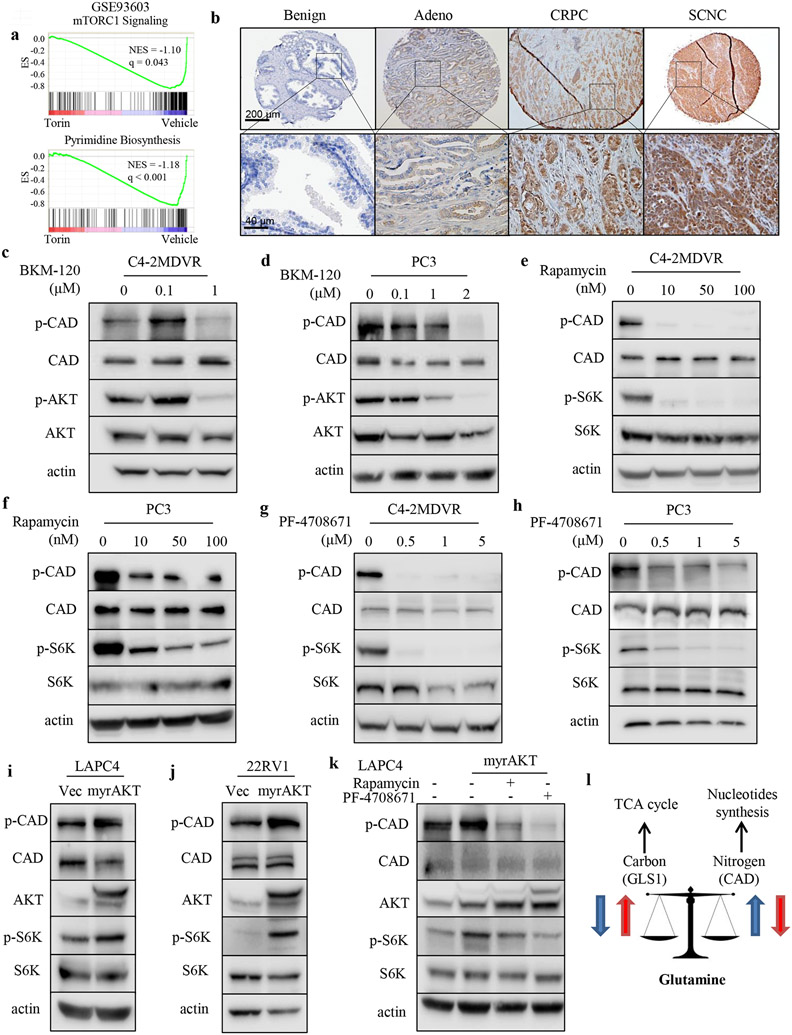
7. CAD is phosphorylated by PI3K-AKT pathway in advanced PCa.
CAD can be phosphorylated by mammalian target of rapamycin (mTOR)-S6K axis on Ser1859 24,25. Given that alterations of the PI3K-phosphatase and tensin homolog (PTEN)-mTOR pathway are extremely frequent in PCa (including both CRPC and SCNC)23,26, we hypothesized that activation of PI3K-AKT-mTOR pathway may phosphorylate and activate CAD. In addition to the upregulated gene signature of pyrimidine synthesis in advanced PCa (Fig. 4a), gene set enrichment analysis (GSEA) revealed that mTOR signaling pathway activity was also enriched in both SCNC and metastatic PCa (Supplementary Fig. S6a). Furthermore, inhibition of mTOR with Torin1 significantly suppressed the expression of genes involved in pyrimidine synthesis (Fig. 7a), suggesting that pyrimidine synthesis is dependent on mTOR signaling. Indeed, IHC staining of human PCa TMAs also displayed pronounced expression of CADS1859 in CRPC and SCNC where mTOR pathway is more frequently activated in comparison to benign prostate tissue and primary adenocarcinoma (Fig. 7b and Supplementary Fig. S6b). In PCa, activation of PI3K-AKT-mTOR pathway is often trigged by PTEN deletion 27,28. Consistent with this hypothesis, PTEN deficient PCa cell models PC3 and C4-2MDVR (Supplementary Fig. S6c) had significantly upregulated levels of CAD phosphorylation (CADS1859) as well as increased phosph-AKT and phosph-S6K levels, in comparison to PTEN wide-type (WT) cell models LAPC4 and 22RV1 (Supplementary Fig. S6c). Treatment with the PI3K inhibitor BKM-120 reduced CADS1859 phosphorylation without changing total CAD levels in PC3 and C4-2MDVR cells (Fig. 7c, d). Specific inhibitors for downstream mediators of the PI3K pathway, rapamycin (an inhibitor of mTOR) and PF-4708671 (an inhibitor of S6K1), also significantly inhibited CADS1859 phosphorylation (Fig. 7e, h). We overexpressed myristoylated AKT29 in PTEN WT cell lines LAPC4 and 22RV1 where the endogenous phospho-AKT levels are low and observed increased CAD phosphorylation along with activation of S6K (Fig. 7i,j). Targeting the cells with rapamycin or PF-4708671 abrogated the myristoylated AKT-induced CAD phosphorylation (Fig. 7k). Together, these findings suggest that loss of PTEN stimulates CAD phosphorylation mediated by PI3K-AKT-mTOR-S6K signaling axis which contributes to increased pyrimidine synthesis in advanced PCa.
Figure 7.
PI3K-AKT signaling phosphorylates CAD in advanced PCa. A, GSEA of “mTORC1 Signaling” and “Pyrimidine Biosynthesis” gene sets in the comparison of cells treated with torin or vehicle. B, Representative images of phosphorylated CADS1859 IHC staining on the TMAs including benign prostate tissue (n = 40), primary prostate adenocarcinoma (n = 40), CRPC (n = 16) and SCNC (n = 17). Adeno, adenocarcinoma. CRPC, castration-resistant prostate cancer. SCNC, small neuroendocrine prostate cancer. Scale bars are shown as indicated. C and D, Western blot assessment of phosphorylated CADS1859, total CAD, phosphorylated AKTS473 and total AKT protein levels after treatment with the PI3K inhibitor BKM-120 for 48 hours in C4-2MDVR (C) and PC3 (D) cells. E and F, Western blot assessment of phosphorylated CADS1859, total CAD, phosphorylated S6KT389 and total S6 protein levels after treatment with the mTOR inhibitor rapamycin for 48 hours in C4-2MDVR (E) and PC3 (F) cells. G and H, Western blot assessment of phosphorylated CADS1859, total CAD, phosphorylated S6KT389 and total S6 protein levels after treatment with the S6K inhibitor PF-4708671 for 48 hours in C4-2MDVR (G) and PC3 (H) cells. I and J, Western blot assessment of phosphorylated CADS1859, total CAD, total AKT, phosphorylated S6KT389 and total S6 protein levels after infection of myristoylated AKT in LAPC4 (I) and 22RV1 (J) cells. K, Western blot assessment of phosphorylated CADS1859, total CAD, total AKT, phosphorylated S6KT389 and total S6 protein levels after infection of myristoylated AKT together with the PI3K inhibitor (BKM-120) or the mTOR inhibitor (rapamycin) treatment in LAPC4 cells. L, Diagram of proposed mechanism for the crosstalk within glutamine metabolic network.
Discussion
Our recent study showed that advanced PCa is addicted to glutamine, a process that is regulated by AR9. Targeting GLS1, the rate-limiting enzyme for glutamine catabolism, is likely a more specific and efficacious approach than AR-directed therapy for advanced PCa. However, acquired treatment resistance is still inevitable. Here, we identify activation of nitrogen metabolism following GLS1 inhibition as an important mechanism of therapy resistance and reveal a reciprocal regulation of the glutamine carbon and nitrogen pathways governed by the balance of GLS1 and CAD, key enzymes for the two pathways, respectively. Our study shows that glutamine carbon and nitrogen pathways coordinate with each other in advanced PCa to fuel tumor cell proliferation. Inhibition of one pathway will eventually activate the other through a mechanism of reciprocal regulation, leading to therapy failure. We therefore propose a combinatorial therapeutic strategy by targeting both pathways of glutamine catabolism to improve therapeutic efficacy (Fig. 7l).
Glutamine carbon and nitrogen catabolism is a well-coordinated process. While glutamine carbon is catabolized to fuel the TCA cycle, the released nitrogen, which is potentially cytotoxic if allowed to accumulate, is used to synthesize numerous nitrogenous metabolites13. Although the two groups of glutamine nitrogen have functional overlap in constituting intermediates, the alpha (amine) nitrogen is primarily transferred to generate other NEAAs while the gamma (amide) nitrogen participates in building nucleotide bases16,18. Our metabolomics data reveal that the nitrogen-containing metabolites are largely enriched in the nucleotide biosynthesis pathway in advanced PCa. Indeed, increased de novo nucleotide synthesis is necessary for DNA replication and RNA production to support energetics and generate macromolecules which are particularly important for tumor cells14,30. In line with this notion, our data demonstrates that PCa is also nucleotide-dependent since supplementing nucleosides rescues the inhibitory effect caused by depletion of glutamine, the most important nitrogen source for nucleotide rings. Therefore, together with our previous discoveries, we have dissected two critical metabolic pathways that are equally important for the glutamine addiction of therapy-resistant PCa.
Despite the important role of GLS1 in many cancers, therapy resistance following GLS1 inhibition has been observed11. We believe that GLS1-targeted monotherapy suffers from the following shortcomings: 1. The role of GLS1 appears to be context-dependent since in certain tumor types, GLS1 acts as a tumor suppressor instead of being a canonical tumor promoter 31,32. Importantly, even in the same tumor type, the response to GLS1 inhibition in vivo may differ from that in vitro, which heightens the challenge of GLS1-targeted therapy in clinical applications11; 2. Metabolic interaction of cancer cells with tumor microenvironment (TME), such as stromal cells, can also mitigate the effect of targeting GLS1 alone since TME can send paracrine signals conferring cancer cells resistance to therapies in nutrient-deprived conditions12,33; 3. Multiple cellular salvage responses to GLS1 inhibition may enable tumor cells to adapt to the GLS1 functional deficiency11,31,34. Within the glutamine catabolism network, given the tight connection of glutamine carbon and nitrogen pathways, it is likely that the metabolic flux detours to one pathway when the other is blocked. A recent study showed that the ratio of GLS1 to phosphoribosyl pyrophosphate amidotransferase (PPAT), the committed enzyme for the purine synthesis, governs the shift between glutamine carbon and nitrogen which significantly affects the efficiency of malignant transformation32. Therefore, inhibiting GLS1 alone is unlikely sufficient in the long run. Our metabolomics analyses elucidate a metabolic cross-talk between GLS1-CAD-governed glutamine carbon and nitrogen network that enables metabolic flux to shift towards pyrimidine synthesis when GLS1 is inhibited, and vice versa. This tumor-protective mechanism enables tumor cells to adapt to the hostile environment following therapy and more importantly, resulting in therapy resistance.
Combination therapy has become a common strategy in cancer treatment. The goal for the combinatorial strategy is to impose additional suppression on the adaptively upregulated pathway induced by the initial treatment. For instance, after GLS1 inhibitor (CB-839) treatment, pancreatic ductal adenocarcinoma (PDAC) cells activate adaptive metabolic networks such as redox pathways and fatty acid oxidation, allowing the tumor cells to utilize available nutrients to sustain cellular proliferation. Combining inhibitors to these compensatory pathways (e.g., gamma-glutamylcysteine inhibitor buthionine-sulfoximine (BSO), and carnitine palmitotyl-transferase inhibitor etomoxir) with CB-839 abrogates the regrowth of PDAC cells that have developed resistance to CB-83911. Additionally, in ovarian cancer, simultaneously disrupting glutamine anabolism (GLUL as the target) in cancer-associated-fibroblasts (CAFs) and glutamine catabolism (GLS1 as the target) in cancer cells proves deleterious to tumor growth12. Similarly, findings from our current study also illustrate a remarkable synergistic effect of simultaneously targeting glutamine nitrogen and carbon pathways by cutting off the feedback loop of glutamine metabolic connections. This synthetic lethal approach could achieve more desirable therapeutic outcomes for patients with therapy-resistant PCa.
Reciprocal regulation is a common phenomenon as a protective and survival mechanism for cancer cells to cope with the hostile environment in order to escape anti-cancer treatment. It occurs on multiple levels including transcription, translation, epigenetics, signal transduction and metabolism 35. An example in the PCa context is the cross-talk and reciprocal regulation between AR and AKT, two critical oncogenic signaling pathways, to maintain tumor cell survival36,37. Activation of the AKT pathway following AR suppression prompted researchers to develop AKT-directed drugs to overcome hormonal therapy resistance. Unfortunately, up to now, single AKT inhibition or dual targeting of AR and AKT have not achieved the desired outcome26. Our previous work has shown GLS1 to be regulated by the AR pathway and the current work shows CAD to be regulated by the AKT signaling pathways. In comparison to inhibition of the upstream AR and AKT pathways for which the tumor can easily develop bypass mechanisms, inhibition of the downstream metabolic enzymes GLS1 and CAD, which directly fuel tumor growth, may lead to more specific therapeutic effects that are difficult for the tumor to overcome. Since cellular metabolism is the final common mechanism to promote tumor proliferation, targeting metabolic vulnerability may prove to be superior than targeting signaling pathways.
Methods
Cell lines, plasmids and reagents
Human PCa cell lines, LNCaP, C4-2, PC3, DU145 and immortal prostatic epithelial cell line, RWPE1 were purchased from ATCC. C4-2MDVR was provided by Allen Gao from UC Davis38. LNCaP, C4-2, C4-2MDVR, PC3 and DU145 cells were culture in DMEM medium supplemented with 10% fetal bovine serum (FBS) and 1% streptomycin and penicillin. RWPE1 cells were maintained in keratinocyte serum-free medium (SFM). For ADT treatment, RPMI-1640 (no phenol red) supplemented with 5% charcoal-stripped FBS was applied. Dimethyl-α-KG and nucleosides (AGCU) were purchased from Sigma. GLUL plasmid was purchased from Origene (RC204039). Myristoylated AKT plasmid was given by Jung Wook Park from Duke University29. BKM-120, rapamycin and PF-4708671 were purchase from MedChem Express (HY-70063), Cell Signaling Technology (9904S) and Selleckchem (S2163).
Cell viability and apoptosis assay
Cells were seeded in 96-well plate with the density of 1000 cells/well. Medium was changed on the following day with desired treatment. Cell confluence was monitored by the Incucyte equipment. For cell apoptosis assay, Incucyte Cytotox Green Dye (4633) was added to the cell culture medium and green fluorescence will be shown when cells become unhealthy. For the doxycycline-induced cell models, 500ng/ml doxycycline (Sigma, 24390) was added when cells were seeded.
Mass spectrometry and isotopomer analysis
Cells were seeded in 6-well plates. Fresh regular medium (for metabolite profiling) or medium containing corresponding isotope (tracing analysis) was added to replace the original culture conditions. After 24 hours incubation, medium was aspirated and 80% pre-chilled HPLC grade methanol was added to each well. Adherent cells were then scraped into the solvent and the supernatant was extracted after cell pellets and debris were spun down. Solvent was further dried with a speed vacuum and dry pellets were collected. Samples were submitted to the core facility of Duke University for mass spectrometry analysis. DMEM without glutamine medium (Gibco, 11960) and DMEM without glucose medium (Gibco, 11966) were used as the corresponding nutrient-deficient medium. 13C5-glutamine (Cambridge, CLM-1822-H-PK), 15N2-glutamine (Cambridge, NLM-1328-PK), alpha-15N-glutamine (Cambridge, NLM-1016-PK), amide-15N-glutamine (Cambridge, NLM-557-PK) and 13C6-glucose (Cambridge, CLM-1396-1) were supplemented accordingly. The raw data of metabolite intensity was further processed by Metaboanalyst online tool.
Tissue microarrays
Several TMAs were constructed and reported previously4,9. Benign and adenocarcinoma TMAs were built with prostatectomy specimens including PCa and the adjacent benign tissue from (n = 40) patients. CRPC TMAs were obtained through transurethral resection for those PCa patients who received hormonal therapy, rather than prostatectomy, but finally had urinary obstruction due to tumor recurrence. SCNC TMAs were constructed from (n = 17) primary SCNC cases. AR was uniformly positive in adenocarcinoma samples and negative in SCNC samples. All samples were collected from patients with informed consent, and all related procedures were performed with the approval of the internal review and ethics boards of Duke University.
Immunohistochemical staining
All TMA or xenograft sections were deparaffinized, rehydrated, and boiled in citrate buffer (pH=6.0) for 40 minutes. Antibodies were incubated for 1 hour followed by incubation of Dako EnVision+System-HRP secondary antibody. Diaminobenzidine (DAB) was then applied for visualization. Quick-score system was performed in all the IHC staining tissues as reported4,9. The intensity of staining is multiplied by the percentage of staining to derive a composite score (a range from 0 to 300). Primary antibodies used are as following: CAD (Thermo Fisher, PA5-21352, 1:100 dilution), p-CAD S1859 (Cell Signaling, 67235, 1:100 dilution), GLS1 (Abcam, ab156876, 1:800 dilution), Ki-67 (Thermo Fisher, RM-9106, 1:300 dilution).
Gene silencing
Plasmids expressing shRNAs targeting human CAD (TRCN0000045910, TRCN0000045908), human DHODH (TRCN0000025839, TRCN0000025868), human GOT1 (TRCN0000034784, TRCN0000034785), human GLS1 (TRCN0000051135, TRCN0000051136) were purchased from Sigma. ShRNAs that had the best knockdown efficiency targeting CAD and GLS1 were subsequently cloned to pLKO-Tet-On vector (Addgene) to generate the doxycycline-inducible knockdown system. Lentiviral particles were produced by packaging shRNA plasmids with the packaging vectors, pMDL, pVSVg and pRev, using the standard calcium phosphate transfection method. Stable cell lines were selected by constant puromycin addition. Western blot was used to verify the knockdown efficiency.
Immunoblotting
Whole cell lysates were prepared with phosphatase and protease inhibitor cocktail addition and loaded to the SDS-PAGE gel with equal amount of total protein. After electrophoresis, proteins were transferred to polyvinylidene difuoride transfer membrane (PVDF) followed by blocked in 5% non-fat milk and incubated with primary antibodies. After washing with TBST (TBS with 0.1% Tween), corresponding secondary antibodies conjugated with HRP were used to incubate the membranes. Samples were developed by Chemiluminescent Substrate (Thermo Fisher) and exposed by Odyssey Imaging Systems (LI-COR). Primary antibodies used are as following: CAD (Santa Cruz, sc-376072, 1:200 dilution), GLUL (Santa Cruz, sc-74430, 1:200 dilution), DHODH (Santa Cruz, sc-166348, 1:200 dilution), GOT1(Santa Cruz, sc-515641, 1:200 dilution), GLS1 (Abcam, ab156876, 1:1000 dilution), p-CAD S1859 (Cell Signaling, 67235, 1:1000 dilution), PTEN (Cell Signaling, 9559, 1:1000 dilution), total AKT (Cell Signaling, 2920, 1:1000 dilution), p-AKTS473 (Cell Signaling, 9271, 1:1000 dilution), total S6K (Cell Signaling, 2708, 1:1000 dilution), p-S6KT389 (Cell Signaling, 9234, 1:1000 dilution), actin (Santa Cruz, sc-47778, 1:200 dilution).
Clonogenic assay,
As described previously5, single cell suspension was performed by trypsinization. Cells were seeded into 60 mm culture dishes with a density of 1000 cells/dish. Cells were incubated for two weeks without medium change. After medium was aspirated and cells were washed by PBS, 10% neutral buffered formalin was used to fix cells followed by 0.01% (w/v) crystal violet staining for at least one hour. Images were captured by Chemidoc (Bio-rad).
Bioinformatics analysis of gene and metabolite expression
Data cohorts were retrieved from the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and cBioPortal. Gene set enrichment analysis (GSEA) was performed by using GSEA software. Molecular signatures were downloaded from MSigDB collections. Heat-map presentation of metabolite intensity was generated by Morpheus (Broad Institute). Metabolic pathway enrichment analysis was achieved by MetaboAnalyst software.
Animal study
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Duke University (A057-19-03). Mice were randomized into one of the four group: 1) scramble control; 2) CAD knockdown; 3) GLS1 knockdown and 4) CAD and GLS1 double knockdown. Sample size was determined by G-Power software to obtain 90% power with a type I error of 0.01. 107 doxycycline-inducible tumor cells with either targeting GLS1 or CAD or in combination were subcutaneously injected into the two flanks of immune-compromised mice (Jackson Laboratories). When tumor were palpable, mice were fed with doxycycline diet (Bio-Serv) for the induction of suppression on corresponding proteins. Tumor volume was monitored by caliper measurement three times a week. Tumor weights were recorded at the endpoint and tumors were harvested, fixed in 4% formaldehyde and embedded in paraffin. Hematoxylin and eosin staining and IHC staining were performed for histological analysis.
Statistics
All quantitative results are displayed as the mean ± SD. The statistical difference between two groups was compared using a Mann–Whitney U test or a Student t test. If more than two groups were compared, ANOVA was used. Statistical analysis was determined using Prism 5 software (GraphPad Software, Inc.). A p value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by DOD-W81XWH-19-1-0411 (JH), DOD-W81XWH2110034 (LX), K99-K99CA237618 (XG), Prostate Cancer Foundation Movember Valor Challenge Award (2018).
Footnotes
Conflict of interest:
The authors declare no potential conflicts of interest.
Competing interest
JH is a consultant for or owns shares in the following companies: Kingmed, MoreHealth, OptraScan, Genetron, Omnitura, Vetonco, York Biotechnology, Genecode, VIVA Biotechnology and Sisu Pharma.
References
- 1.Rawla P Epidemiology of Prostate Cancer. World J Oncol 10, 63–89, doi: 10.14740/wjon1191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swami U, McFarland TR, Nussenzveig R & Agarwal N Advanced Prostate Cancer: Treatment Advances and Future Directions. Trends Cancer 6, 702–715, doi: 10.1016/j.trecan.2020.04.010 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Spetsieris N, Boukovala M, Patsakis G, Alafis I & Efstathiou E Neuroendocrine and Aggressive-Variant Prostate Cancer. Cancers 12, doi:ARTN 3792 10.3390/cancers12123792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y He Y Butler W Xu L Chang Y Lei K et al. Targeting cellular heterogeneity with CXCR2 blockade for the treatment of therapy-resistant prostate cancer. Sci Transl Med 11, doi: 10.1126/scitranslmed.aax0428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yin Y Xu L Chang Y Zeng T Chen X Wang A et al. N-Myc promotes therapeutic resistance development of neuroendocrine prostate cancer by differentially regulating miR-421/ATM pathway. Mol Cancer 18, 11, doi: 10.1186/s12943-019-0941-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergers G & Fendt SM The metabolism of cancer cells during metastasis. Nature reviews. Cancer, doi: 10.1038/s41568-020-00320-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park JH, Pyun WY & Park HW Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets. Cells-Basel 9, doi:ARTN 2308 10.3390/cells9102308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo HC, Yu YC, Sung Y & Han JM Glutamine reliance in cell metabolism. Exp Mol Med 52, 1496–1516, doi: 10.1038/s12276-020-00504-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L Yin Y Li Y Chen X Chang Y Zhang H et al. A glutaminase isoform switch drives therapeutic resistance and disease progression of prostate cancer. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2012748118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altman BJ, Stine ZE & Dang CV From Krebs to clinic: glutamine metabolism to cancer therapy. Nature Reviews Cancer 16, 619–634, doi: 10.1038/nrc.2016.71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biancur DE Paulo JA Malachowska B Rey MQDR Sousa CM Wang X et al. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat Commun 8, doi:ARTN 15965 10.1038/ncomms15965 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L Achreja A Yeung T Mangala L Jiang D Han C et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab 24, 685–700, doi: 10.1016/j.cmet.2016.10.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y Bai C Ruan Y Liu M Chu Q Yang C et al. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat Commun 10, doi:ARTN 201 10.1038/s41467-018-08033-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane AN & Fan TWM Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res 43, 2466–2485, doi: 10.1093/nar/gkv047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oizel K Tait-Mulder J Fernandez-de-Cossio-Diaz J Pietzke M Brunton H Lilla S et al. Formate induces a metabolic switch in nucleotide and energy metabolism. Cell Death Dis 11, doi:ARTN 310 10.1038/s41419-020-2523-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bott AJ, Maimouni S & Zong WX The Pleiotropic Effects of Glutamine Metabolism in Cancer. Cancers 11, doi:ARTN 770 10.3390/cancers11060770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Pavlova NN & Thompson CB Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. Embo J 36, 1302–1315, doi: 10.15252/embj.201696151 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cluntun AA, Lukey MJ, Cerione RA & Locasale JW Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer 3, 169–180, doi: 10.1016/j.trecan.2017.01.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beltran H Prandi D Mosquera JM Benelli M Puca L Cyrta J et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22, 298–305, doi: 10.1038/nm.4045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltran H Rickman DS Park K Chae SS Sboner A MacDonald TY et al. Molecular Characterization of Neuroendocrine Prostate Cancer and Identification of New Drug Targets. Cancer Discov 1, 487–495, doi: 10.1158/2159-8290.Cd-11-0130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasso CS Wu Y Robinson D Cao X Dhanasekaran SM Khan AP et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243, doi: 10.1038/nature11125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quigley DA Dang HX Zhao SG Lloyd P Aggarwal R Alumkal JJ et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 174, 758-+, doi: 10.1016/j.cell.2018.06.039 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor BS Schultz N Hieronymus H Gopalan A Xiao Y Carver B et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 18, 11–22, doi: 10.1016/j.ccr.2010.05.026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Sahra I, Howell JJ, Asara JM & Manning BD Stimulation of de Novo Pyrimidine Synthesis by Growth Signaling Through mTOR and S6K1. Science 339, 1323–1328, doi: 10.1126/science.1228792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robitaille AM Christen S Shimobayashi M Cornu M Fava LL Moes S et al. Quantitative Phosphoproteomics Reveal mTORC1 Activates de Novo Pyrimidine Synthesis. Science 339, 1320–1323, doi: 10.1126/science.1228771 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Crumbaker M, Khoja L & Joshua AM AR Signaling and the PI3K Pathway in Prostate Cancer. Cancers 9, doi:ARTN 34 10.3390/cancers9040034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M Zhang J Sampieri K, Clohessy JG Mendez L Gonzalez-Billalabeitia E et al. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet 50, 206-+, doi: 10.1038/s41588-017-0027-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamaspishvili T Berman DM Ross AE Scher HI Marzo AMD Squire JA et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol 15, 222–234, doi: 10.1038/nrurol.2018.9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JW Lee JK Phillips JW Huang P Cheng D Huang J et al. Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. P Natl Acad Sci USA 113, 4482–4487, doi: 10.1073/pnas.1603645113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X Yang K Wu Q Kim LJY Morton AR Gimple RC et al. Targeting pyrimidine synthesis accentuates molecular therapy response in glioblastoma stem cells. Sci Transl Med 11, doi:ARTN eaau4972 10.1126/scitranslmed.aau4972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson SM Papagiannakopoulos T Olenchock BA Heyman JE Keibler MA Luengo A et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 23, 517–528, doi: 10.1016/j.cmet.2016.01.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodama M Oshikawa K Shimizu H, Yoshioka S Takahashi M Izumi Y et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat Commun 11, doi:ARTN 1320 10.1038/s41467-020-15136-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavlova NN & Thompson CB The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47, doi: 10.1016/j.cmet.2015.12.006 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z Liu F Fan N Zhou C Li D Macvicar T et al. Targeting Glutaminolysis: New Perspectives to Understand Cancer Development and Novel Strategies for Potential Target Therapies. Front Oncol 10, doi:ARTN 589508 10.3389/fonc.2020.589508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Ma R, Wu YS, Zhai YS & Li SS Reciprocal Regulation of Metabolic Reprogramming and Epigenetic Modifications in Cancer. Front Genet 9, doi:ARTN 394 10.3389/fgene.2018.00394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carver BS Chapinski C Wongvipat J Hieronymus H Chen Y Chandarlapaty S et al. Reciprocal Feedback Regulation of PI3K and Androgen Receptor Signaling in PTEN-Deficient Prostate Cancer. Cancer Cell 19, 575–586, doi: 10.1016/j.ccr.2011.04.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulholland DJ Tran LM Li Y Cai H Morim A Wang S et al. Cell Autonomous Role of PTEN in Regulating Castration-Resistant Prostate Cancer Growth. Cancer Cell 19, 792–804, doi: 10.1016/j.ccr.2011.05.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C Lou W Zhu Y Nadiminty N Schwartz CT Evans CP et al. Niclosamide inhibits androgen receptor variants expression and overcomes enzalutamide resistance in castration-resistant prostate cancer. Clin Cancer Res 20, 3198–3210, doi: 10.1158/1078-0432.CCR-13-3296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.