
Data Collection for: |
Meinhardt and de Boer (2001) Proc. Natl. Acad. Sci. USA 98 (25), 14202-14207. (10.1073/pnas.251216598) |
Overview: Pattern Formation in E. Coli: A Model for the Pole-to-Pole Oscillations of Min Proteins and the Localization of the Division Site
How a bacterium finds its center in order to localize the division machinery is a long standing question. The preparation for division starts with the assembly of a polymeric ring of the tubulin-like GTPase FtsZ (Z ring). In E. coli, this ring is localized to the center by the actions of the MinC, MinD, and MinE proteins. MinC inhibits the initiation of the Z ring. MinC colocalizes with MinD. In wild-type (WT) cells, MinC/D forms a polar pattern that oscillates between the poles, keeping the center free for initiation of cell division. Thus, virtually all of MinC/D dynamically assembles on the membrane in the shape of a test tube covering the membrane from one pole up to approximately midcell (see
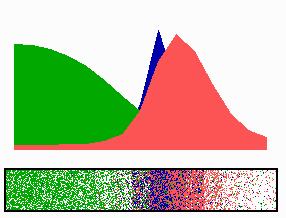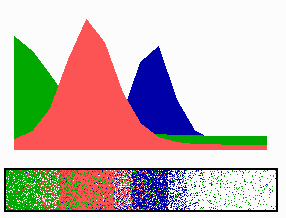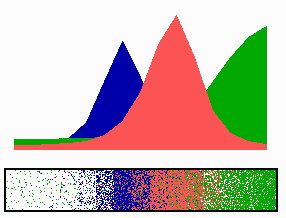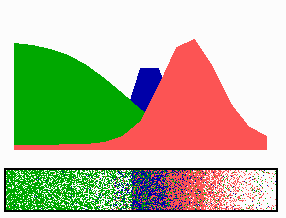
time-lapse fluorescence micrographs). Most of MinE accumulates at the rim of this tube, in the shape of a ring (the E ring). The rim of the MinC/D tube and associated E ring move from a central position to the cell pole until both the tube and ring vanish. Meanwhile, a new MinC/D tube and associated E ring form in the opposite cell half, and the process repeats, resulting in a pole-to-pole oscillation cycle of the division inhibitor. A full cycle takes about 50 s. The panel below shows a schematic drawing of the MinC/D (green) and MinE (red) localization cycles. The animations show a typical computer simulation using our model to describe the dynamic behaviour of these proteins. 

In the model, the signal for septum formation (the Z ring, blue) is generated by a pattern-forming process that becomes localized to the membrane at the cell center because of the pole-to-pole oscillation of the inhibitor of Z-ring assembly (MinC/MinD, green). A local high concentration of a substance (MinE, red) at the membrane is generated by a pattern formation reaction that depends on membrane-bound MinD. Membrane-associated MinE displaces membrane-bound MinD. Because local MinE assembly depends on membrane-bound MinD, the removal of MinD causes the local high MinE concentration to destabilize itself and shift into a neighboring position with higher MinD concentrations. This results in a traveling wave of MinE that "peels" MinD off the membrane as it moves toward a cell pole. Meanwhile, MinD reassembles on the membrane in the opposite cell half. This attracts a new MinE activation that, somewhat later, leads to the wave-like removal of MinD from the membrane in that half of the cell as well. The result is a pole-to-pole oscillation of MinD and associated MinC. On time average, the MinC/MinD concentration is highest at the poles, forcing the FtsZ pattern to the center. The model accounts for the experimental observations on Min protein dynamics in live cells (1-7).
The following simulations illustrate the elementary steps:
References