Abstract
Prion diseases are fatal neurodegenerative disorders of humans and animals that are important because of their impact on public health and because they exemplify a novel mechanism of infectivity and biological information transfer. These diseases are caused by conformational conversion of a normal host glycoprotein (PrPC) into an infectious isoform (PrPSc) that is devoid of nucleic acid. This review focuses on the current understanding of prion diseases at the cell biological level. The characteristics of the diseases are introduced, and a brief history and description of the prion hypothesis are given. Information is then presented about the structure, expression, biosynthesis, and possible function of PrPC, as well as its posttranslational processing, cellular localization, and trafficking. The latest findings concerning PrPSc are then discussed, including cell culture systems used to generate this pathogenic isoform, the subcellular distribution of the protein, its membrane attachment, proteolytic processing, and its kinetics and sites of synthesis. Information is also provided on molecular models of the PrPC→PrPSc conversion reaction and the possible role of cellular chaperones. The review concludes with suggestions of several important avenues for future investigation.
Prion diseases, also called spongiform encephalopathies, are fatal neurodegenerative disorders that have attracted enormous attention not only for their unique biological features but also for their impact on public health. This group of diseases includes kuru, Creutzfeldt-Jakob disease (CJD), Gerstmann-Sträussler syndrome (GSS), and fatal familial insomnia (FFI) in human beings, as well as scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, and encephalopathies in mink, cats, mule deer, elk, and several exotic ungulates (61, 128). The primary symptom of the human disorders is dementia, usually accompanied by manifestations of motor dysfunction such as cerebellar ataxia, myoclonus, and pyramidal or extrapyramidal signs. FFI is also characterized by dysautonomia and sleep disturbances. The symptoms appear insidiously in middle to late adult life and last from months (CJD, FFI, and kuru) to years (GSS) prior to death. Neuropathologically, these disorders produce a characteristic spongiform degeneration of the brain, as well as deposition of amyloid plaques (most prominent in GSS and kuru). Prion diseases share important clinical, neuropathological and cell biological features with another, more common cerebral amyloidosis, Alzheimer’s disease (122).
Prion diseases have now become a subject of intense general interest because of the potential risks they pose to public health. An epidemic of BSE (“mad cow disease”) that began in Britain in 1986 has caused the deaths of almost 200,000 cattle, and a much larger number of animals have probably been infected (3, 129, 179). The epidemic arose as a result of a change in the rendering process used to prepare cattle feed from ruminant offal. Although a government-imposed feed ban in 1988 has significantly reduced the incidence of the disease, it has had a major economic and political impact throughout Europe. What is most troubling is the appearance since 1996 of about 40 cases of “new-variant” CJD in Britain and 1 case in France (33, 183). The clinical, neuropathological, molecular, and transmission characteristics of these cases distinguish them from sporadic CJD and strongly indicate that they were acquired by consumption of beef from cattle affected with BSE (19, 42, 183). Whether new-variant CJD will acquire epidemic proportions or be self-limited is difficult to predict at this point because of the long incubation time of the disease. Other examples of inadvertent spread of prion diseases include cases of CJD that resulted from contaminated dura mater grafts, corneal transplants, neurosurgical instruments, and cadaveric growth hormone (18). There is also debate about whether infection can be transmitted via blood transfusion or administration of blood-derived products (17).
MANIFESTATIONS OF PRION DISEASES AND THE PRION HYPOTHESIS
Three different manifestations of prion diseases are recognized: infectious, familial, and sporadic. Studies of the infectious etiology of these disorders and the chemical form of the transmissible agent have an interesting history. The infectious nature of scrapie, a neurological disease of sheep which is characterized by intense pruritus that causes the animals to scrape against walls and fences, was first demonstrated in 1939, when the disease was experimentally transmitted to goats by intraocular injection (46). In the 1950s, Carleton Gajdusek and his colleagues described a strange neurodegenerative disease of the Fore tribe of New Guinea called kuru, which was transmitted among the members of this group by ritual cannibalism (63). After William Hadlow pointed out the neuropathological similarities between kuru and scrapie (67), subsequent work demonstrated that kuru and CJD were transmissible to nonhuman primates, clearly establishing the infectious nature of these illnesses (62). There is a remarkable species specificity to the transmission of prion diseases, first pointed out by Pattison (117), that manifests itself in prolonged incubation times upon first passage between species. This effect is observable even between closely related species such as mice and hamsters (149).
The unusual properties of the infectious agent became the focus of attention beginning in the 1960s, and in the early 1980s Stanley Prusiner, building upon earlier suggestions (1, 66), proposed the prion hypothesis (127). This stated that the infectious agent in human and animal spongiform encephalopathies was composed exclusively of a single kind of protein molecule designated PrPSc without any encoding nucleic acid. Subsequent work has shown that PrPSc is, in fact, a conformationally altered form of normal, host-encoded membrane glycoprotein called PrPC (81, 128, 132). It was proposed that PrPSc impresses its abnormal conformation on PrPC, thereby generating additional molecules of PrPSc in an autocatalytic reaction (10, 39, 44, 60, 79). Prion diseases therefore exemplify a novel pathogenic mechanism based on a self-propagating change in protein conformation. The identification of proteins in yeast (92, 98) and filamentous fungi (45) that behave like prions makes it likely that inheritance of biological information via protein conformation will prove to be a quite general phenomenon in nature.
There is now a great deal of compelling evidence in favor of the prion hypothesis, which has been critically reviewed elsewhere (23, 36, 132). There is little question that PrP plays a key role in the disease process, and it has become increasingly difficult to explain all the existing data by a viral theory of pathogenesis. It is important to recognize, however, that perhaps the most definitive test of the protein-only model has yet to be successfully carried out: producing infectivity de novo in a test tube by experimental manipulation of recombinant or synthetic PrP. The absence of this evidence does not invalidate the prion hypothesis but may simply underscore the difficulty of reconstituting this remarkable molecular transformation.
Prion diseases are unique because they can arise by inheritance as well as by infection. About 10% of the cases of CJD and all cases of GSS and FFI are linked to germ line mutations in the PrP gene on chromosome 20 (116, 130, 186). The mutations are presumed to favor spontaneous conversion of the protein to the PrPSc state without the necessity for contact with exogenous infectious agent (39). Point mutations occur in the C-terminal half of the PrP molecule and are associated with either CJD, GSS, or FFI. Insertional mutations, which are associated with CJD, occur in the N-terminal half of the protein and consist of one to nine additional copies of an octapeptide repeat that is normally present at five copies. A polymorphism at codon 129, which can encode either methionine or valine, can profoundly influence the phenotypic characteristics of the disease caused by pathogenic mutations in other positions (64). Although they arise spontaneously as a result of a PrP mutation, familial prion diseases are transmissible to laboratory animals, thus demonstrating that the mutation has induced the formation of infectious prions.
Sporadic forms of prion disease, which include most cases of CJD, display no obvious infectious or genetic etiology. They may be attributable to spontaneous conversion of wild-type PrPC to the PrPSc state, or to the presence of as yet undetected somatic mutations in the protein that favor its conversion to PrPSc (39).
EXPERIMENTAL APPROACHES TO PRION DISEASES
Defining the mechanisms underlying the generation of PrPSc from PrPC has become one of the central issues in understanding the pathogenesis of prion diseases. This question has been addressed experimentally by a variety of approaches, including structural analysis of PrP isoforms by spectroscopic techniques (31, 55, 113, 137, 142), generation of mice carrying modified or ablated PrP genes (38, 152, 180), and development of model systems in which PrP peptides and purified proteins are used to reconstitute the production of PrPSc in vitro (7, 84, 88, 89).
One particularly useful approach has been a cell biological one: investigating the biosynthesis, posttranslational processing, cellular localization, and trafficking of PrPC and PrPSc. This kind of analysis, which involves techniques of cell culture, metabolic labeling, microscopy, and subcellular fractionation, has a number of advantages. First, it allows one to examine the properties of PrP in the setting of native cellular structures and cofactors that are likely to be critical in mediating the efficient conversion of PrPC into PrPSc. Second, a great deal is now known about the biology of membrane glycoproteins, and a number of sophisticated experimental techniques are available for studying them. Finally, a cell biological approach has been enormously fruitful in the study of a closely related but much more common neurodegenerative disorder, Alzheimer’s disease (153), and it is possible to gain insight from this work which informs our understanding of prion diseases.
In this review, I discuss what is known about the cell biology of PrP, focusing first on PrPC, then on PrPSc, and finally on the molecular nature of the PrPC→PrPSc conversion reaction.
CELL BIOLOGY OF PRPC
Expression and Function of PrPC
PrPC is normal cellular protein that is expressed in the neurons and glia of the brain and spinal cord, as well as in several peripheral tissues and in leukocytes (6, 25, 54, 73, 103, 108). PrP mRNA is first detectable in the brains of mice and chickens beginning early in embryogenesis, and its level increases as development proceeds (73, 103). In the adult central nervous system, PrP and its mRNA are widely distributed, with particular concentrations in neocortical and hippocampal neurons, cerebellar Purkinje cells, and spinal motor neurons (49, 90).
The normal function of PrPC remains unknown, although its localization on the cell surface would be consistent with roles in cell adhesion and recognition, ligand uptake, or transmembrane signaling. Defining the physiological role of PrPC may be relevant to understanding the disease state, since the protein may fail to perform its normal function when it is converted to the PrPSc isoform. Mice in which the endogenous PrP gene has been disrupted display no gross developmental or anatomical defects (21) but are reported in some studies to have electrophysiological and structural abnormalities in the hippocampus (40, 41, 43, 104, 182), loss of cerebellar Purkinje cells (144), alterations in circadian rhythm and sleep pattern (176), and changes in learning and memory (112). However, other investigators did not observe certain of these features (21, 78, 100); this may be due to differences between lines of mice in the region of the PrP gene that was replaced. In any case, how the abnormal phenotypes of PrP-null mice, when present, relate to the normal function of PrPC is unclear.
It has recently been suggested that PrPC may play a role in copper metabolism, since purified PrPC binds copper via its peptide repeat region and since the brains of PrP-null mice display a reduced content of membrane-associated copper and a decreased activity of copper-zinc superoxide dismutase (15, 16, 80). Our own analysis of the effect of copper ions on the cellular trafficking of PrPC (118) supports the hypothesis that PrPC may serve as an endocytic receptor for the uptake of copper from the extracellular milieu (see below).
Structure and Biosynthesis of PrPC
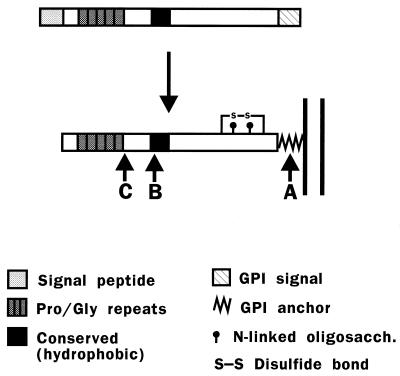
The mammalian PrP gene encodes a protein of approximately 250 amino acids that contains several distinct domains, including an N-terminal signal peptide, a series of five proline- and glycine-rich octapeptide repeats, a central hydrophobic segment that is highly conserved, and a C-terminal hydrophobic region that is a signal for addition of a glycosyl-phosphatidylinositol (GPI) anchor (Fig. 1).
FIG. 1.
Structure and posttranslational processing of PrP. (Top) Structure of the primary translation product of mammalian PrP. The five proline- and glycine-rich repeats in mouse PrP have the sequence P(Q/H)GG(T/G/S)WGQ. (Bottom) Structure of the mature protein. The GPI anchor attaches the polypeptide chain to the membrane (see Fig. 7 for a schematic of the core anchor structure). Arrows A and B indicate the positions of cleavage sites in PrPC, and arrow C indicates a cleavage site in PrPSc. Site A lies within the GPI anchor, between the diacylglycerol moiety and the ethanolamine residue that is attached to the C-terminal amino acid. Site B lies near position 110, and site C lies near position 89.
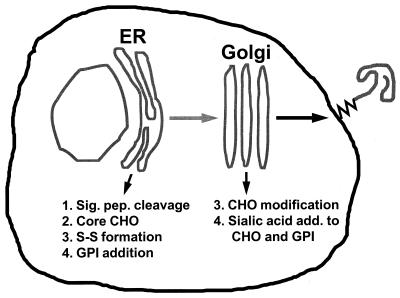
Like other membrane proteins, PrPC is synthesized in the rough endoplasmic reticulum (ER) and transits the Golgi on its way to the cell surface. During its biosynthesis, PrPC is subject to several kinds of posttranslational modifications (Fig. 2), including cleavage of the N-terminal signal peptide, addition of N-linked oligosaccharide chains at two sites, formation of a single disulfide bond, and attachment of the GPI anchor (68, 163, 177). The N-linked oligosaccharide chains added initially in the ER are of the high-mannose type and are sensitive to digestion by endoglycosidase H; these are subsequently modified in the Golgi to yield complex-type chains that contain sialic acid and are resistant to endoglycosidase H (26). The GPI anchor, which is added in the ER after cleavage of the C-terminal hydrophobic segment, has a core structure common to other glycolipid-anchored proteins, consisting of an ethanolamine residue amide-bonded to the C-terminal amino acid, three mannose residues, an unacetylated glucosamine residue, and a PI molecule which is embedded in the outer leaflet of the lipid bilayer (see Fig. 7). The GPI anchors of both PrPC and PrPSc are unusual because their cores are modified by the addition of sialic acid residues (161). Available evidence indicates that the oligosaccharide chains and GPI anchors of PrPC and PrPSc do not differ, although complete structures have been worked out only for PrPSc (57, 161).
FIG. 2.
Steps in the biosynthesis of PrPC. CHO, oligosaccharide; S-S, disulfide bond; Sig. pep., signal peptide.
FIG. 7.
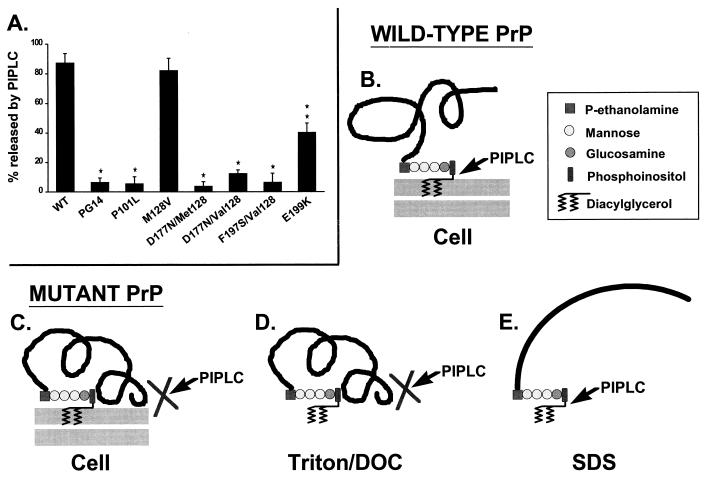
moPrPs carrying disease-related mutations are not released from the cell surface by PI-PLC. (A) CHO cells expressing wild-type (WT) and mutant PrPs were biotinylated with the membrane-impermeant reagent sulfo-biotin-X-NHS at 4°C and were then incubated with PI-PLC at 4°C prior to lysis. moPrP in the PI-PLC incubation media and cell lysates was immunoprecipitated, separated by SDS-polyacrylamide gel electrophoresis, and visualized by developing blots of the gel with horseradish peroxidase-streptavidin and enhanced chemiluminescence. PrP bands from three separate experiments were quantitated by densitometry, and the amount of PrP released by PI-PLC was plotted as a percentage of the total amount of PrP (medium plus cell lysate). Each bar represents the mean ± standard deviation. Values that are significantly different from wild-type moPrP by Student’s t test are indicated by single (P < 0.01) and double (P < 0.001) asterisks. All of the moPrPs carrying pathogenic mutations are less PI-PLC releasable than are wild-type and M128V moPrPs. E199K moPrP is more releasable than the other mutants, consistent with our observation that there are subtle biochemical differences among the mutant proteins. Phenotypes associated with the homologous human PrPs are given in the legend to Fig. 6. (B) Schematic of the membrane attachment of wild-type PrPC, which is can be completely released from cells by treatment with PI-PLC. The core structure of the GPI anchor, along with the site cleaved by PI-PLC, is indicated. (C to E) Schematics showing how PI-PLC is proposed to interact with mutant PrP. (C) On intact cells, the mutant protein adopts a PrPSc-like conformation that physically blocks access of PI-PLC to the GPI anchor. It is also possible that aggregation of mutant PrP molecules contributes to the inaccessibility of the anchor. (D) After extraction from the membrane by using nondenaturing detergents like Triton X-100 and deoxycholate (DOC), the abnormal conformation of the mutant protein is maintained and the anchor is still inaccessible to PI-PLC. (E) After denaturation in SDS, the conformation of the mutant protein is disrupted, and the anchor becomes susceptible to PI-PLC cleavage. Panel A is modified from reference 72 with permission of the publisher.
There is evidence that experimental alterations in N glycosylation modify the biosynthetic transport of PrPC. Mutation of both consensus sites for N glycosylation in rodent PrP or of the more N-terminal site alone causes the protein to misfold and accumulate in a compartment proximal to the mid-Golgi stack (97, 138). However, correct N glycosylation is not absolutely required for biosynthetic transport, since mutation of the C-terminal consensus site alone or synthesis of wild-type PrPC in the presence of the glycosylation inhibitor tunicamycin still allows a substantial number of molecules to be expressed on the cell surface (97). In fact, small amounts of unglycosylated wild-type PrPC are often detectable on the plasma membrane even in the absence of any inhibitors (26, 97).
Surprisingly, molecules with either one or both N-glycosylation consensus sites mutated exhibit several biochemical properties of PrPSc, and the same is true to a limited extent for wild-type PrPC molecules synthesized in the presence of tunicamycin (97). The conclusion from these studies is that wild-type PrPC has an intrinsic tendency to adopt some PrPSc-like features during its normal conformational maturation but that N-linked glycan chains protect against this change. This phenomenon may be related to the observation that different prion strains sometimes display distinctive patterns of glycosylation (42, 50, 87, 107, 114).
Posttranslational Cleavage of PrPC
Our own work (71) and that of others (12, 26, 27, 34, 168, 171) with transfected cell lines, as well as brain tissue and cerebrospinal fluid, has shown that PrPC undergoes two posttranslational cleavages as part of its normal metabolism. One cleavage (labeled A in Fig. 1) occurs within the GPI anchor and releases the polypeptide chain into the extracellular medium. The cellular location in which this cleavage occurs is not known, although for other GPI-anchored proteins which undergo a similar cleavage it has been speculated that a cell surface phospholipase is responsible (102). The second cleavage (labeled B in Fig. 1) is proteolytic and occurs within a segment of 16 hydrophobic amino acids that is completely conserved in all cloned PrP species. Our own data indicate that cleavage B occurs within an endocytic compartment of the cell (see the next section), although Taraboulos et al. (171) suggested that cholesterol-rich domains of the plasma membrane are involved. Both cleavages occur relatively slowly in comparison with the half-life of the protein, so that at steady state several different cleavage products in addition to the intact protein can be detected. The physiological significance of the cleavages is uncertain. The 10-kDa N-terminal fragment released by cleavage B could serve as a biologically active ligand, by analogy to polypeptide growth factors that are released by cleavage of membrane-bound precursors. Alternatively, if PrPC functions as a cell surface receptor, the cleavages may represent mechanisms of receptor down-regulation.
Subcellular Localization and Trafficking of PrPC
PrPC is a cell surface protein, most of which is found on the plasma membrane. Because it is attached exclusively by its GPI anchor, it can be released from the cell surface by treatment with the bacterial enzyme PI-specific phospholipase C (PI-PLC), which cleaves off the diacylglycerol portion of the anchor (see Fig. 7B) (13, 24, 94).
There is biochemical evidence that PrPC in neurons is axonally transported to nerve terminals (11), consistent with the localization of the protein in synaptic profiles as shown by immunoelectron microscopy (59, 145). Light microscopic immunocytochemistry shows that PrPC is concentrated primarily in synaptic fields of the olfactory bulb, limbic structures, and striato-nigral complex, with little staining of neuronal perikarya or fiber pathways, except for the olfactory nerve (145). These morphological data would suggest a role for PrPC in synaptic function, although additional studies are clearly necessary. For example, it will be important to determine the relative distribution of the protein in presynaptic and postsynaptic elements and whether it is present in synaptic vesicles. In addition, nothing is known about the pathways involved in biosynthetic targeting of PrPC in polarized cell types like neurons, which contain distinct axonal and somatodendritic surfaces.
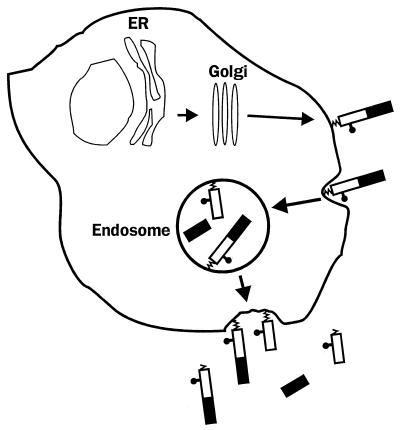
We have, however, learned a great deal about the subcellular trafficking of PrPC from studies of transfected cell lines that express the protein. Work from our laboratory has focused on the chicken version of PrPC, although the substantial homology between the avian and mammalian versions of the protein makes it likely that the conclusions we have reached are generally applicable. Our results demonstrate that PrPC does not remain on the cell surface after its delivery there but, rather, constitutively cycles between the plasma membrane and an endocytic compartment (Fig. 3) (155). This conclusion is based on a number of pieces of evidence, including the sensitivity of cleavage B to lysosomotropic amines, leupeptin, and brefeldin A; uptake of fluorescently labeled anti-PrP antibodies from the cell surface; and internalization and recycling of PrP molecules that have been labeled with membrane-impermeant iodination or biotinylation reagents. In cultured neuroblastoma cells, PrPC molecules cycle through the cell with a transit time of ∼60 min, and during each passage 1 to 5% of the molecules are cleaved at site B.
FIG. 3.
Cellular trafficking and cleavage of PrP. After reaching the cell surface, PrPC is internalized into an endocytic compartment from which most of the molecules are recycled intact to the cell surface. A small percentage of the endocytosed molecules are proteolytically cleaved (site B in Fig. 1), and the N- and C-terminal cleavage products are then externalized. Some of the membrane-anchored protein is released into the extracellular medium by cleavage within the GPI anchor (site A in Fig. 1). Reprinted from reference 155 with permission of the publisher.
This endocytic recycling pathway is of interest for two reasons. First, it may be the route along which certain steps in the conversion of PrPC to PrPSc take place (see below). Consistent with this proposal, we have found that sulfated glycans, which are potent antiscrapie therapeutic agents, dramatically stimulate endocytosis of PrPC, directing some of the molecules to late endosomes and/or lysosomes in which we have speculated that conversion to PrPSc may be inefficient (157).
The existence of a recycling pathway also suggests that one physiological function of PrPC might be to serve as a receptor for uptake of an extracellular ligand, by analogy to the receptors responsible for uptake of transferrin and low-density lipoprotein. One attractive candidate for such a ligand is the copper ion. As discussed above, there are several lines of evidence favoring a connection between PrPC and copper metabolism. We have hypothesized (118) that PrPC binds copper ions on the cell surface and then delivers them to an endocytic compartment within which the bound ions dissociate from PrPC and are transferred to other copper carrier proteins that move the ions into the cytosol; PrPC would then return to the cell surface to begin another cycle. Consistent with this model, we have found that copper ions at physiologically relevant concentrations rapidly and reversibly stimulate endocytosis of PrPC from the cell surface (118).
Clathrin-Coated Pits and Caveolae
Endocytosis of a number of cell surface receptors such as those for transferrin and low-density lipoprotein takes place in specialized infoldings of the plasma membrane called clathrin-coated pits (148). Clathrin is a large, oligomeric protein that assembles into lattice structures on the inner surface of the plasma membrane, thereby causing the membrane to invaginate and pinch off to form a clathrin-coated vesicle which can subsequently fuse with other intracellular organelles. We have found that clathrin-coated pits and vesicles are the morphological structures responsible for endocytic uptake of PrPC (154). This conclusion is based on immunogold localization of chicken PrPC in these organelles by electron microscopy; inhibition of PrPC internalization by incubation of cells in hypertonic sucrose, which disrupts clathrin lattices; and detection of PrPC in purified preparations of coated vesicles from brain tissue. The N-terminal half of the PrPC polypeptide chain is essential for efficient clathrin-mediated endocytosis (156). Deletions within this region diminish the internalization of PrPC measured biochemically and reduce the concentration of the protein in coated pits determined morphometrically.
Transmembrane receptors that are endocytosed in coated pits display in their cytoplasmic domains specific amino acid motifs, often containing critical tyrosine residues, that bind to clathrin via specialized adapter proteins (148). The involvement of clathrin-coated pits in endocytosis of PrPC is therefore surprising, since GPI-anchored proteins such as PrPC lack a cytoplasmic domain. Indeed, it has been speculated that other GPI-anchored proteins are excluded from coated pits and are internalized via other surface invaginations called caveolae (2). Caveolae, which are coated on their cytoplasmic surface with a protein called caveolin, are particularly abundant in fibroblasts, smooth muscle cells, and endothelial cells. They are postulated to be involved in several kinds of physiological processes, including transcytosis, transmembrane signaling, and cellular uptake of small molecules such as the vitamin folic acid. We have found, however, that caveolae are not responsible for internalization of PrPC in neuronal cells, since these cells lack morphologically recognizable caveolae, and do not express caveolin (154).
We have observed that when cells are extracted with Triton X-100 at 4°C, PrPC is recovered along with other GPI-anchored proteins in large, detergent-resistant complexes that contain cholesterol and sphingolipids, as well as signaling molecules such as Src-family tyrosine kinases and G-protein subunits (65). Complexes of similar composition have been prepared from a number of cells and tissues, and it has been speculated that they represent the biochemical equivalent of caveolae (32, 99). Our results indicate that this hypothesis cannot be true, however, since caveolae are not found in the neuronal cells from which we have extracted PrP-containing complexes. Although it is possible that the complexes are simply an artifact of detergent extraction, they may also correspond to specialized microdomains of the plasma membrane that exist in the intact cell. It has been proposed that such domains (or “rafts”) are involved in transmembrane signaling and polarized sorting of membrane proteins (158). It would be intriguing if PrP were involved in such processes in neurons.
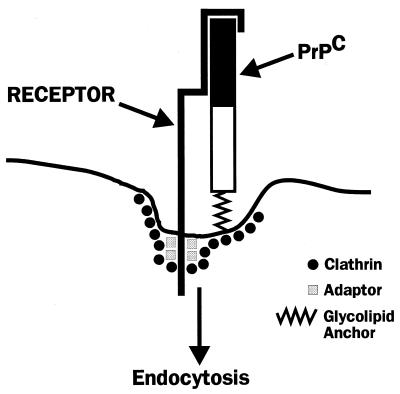
A Putative Receptor for PrPC
To explain the association of PrPC with coated pits, we have postulated the existence of a PrPC receptor, i.e., a transmembrane protein that has a coated-pit localization signal in its cytoplasmic domain and whose extracellular domain binds the N-terminal portion of PrPC (Fig. 4) (70). Binding of copper ions may enhance the affinity of PrPC for this putative receptor, in line with the stimulatory effect of the metal on endocytosis of PrPC (118). Identification of a PrPC receptor is now of great importance, since it is likely to provide further clues to the normal function of PrPC and may allow the design of therapeutic strategies for blocking endocytic uptake of PrPC, thereby inhibiting prion replication. Such a receptor might also be involved in the conversion of PrPC into PrPSc or in the initial uptake of PrPSc-containing prion particles into cells.
FIG. 4.
Hypothetical model for a PrPC receptor. The cytoplasmic domain of the receptor contains signals for interacting with adapter molecules and clathrin, which are components of coated pits. The extracellular domain of the receptor binds to the N-terminal part of the PrPC molecule (solid rectangle), a region that is essential for efficient endocytosis. Reprinted from reference 70 with permission of the publisher.
We have begun to search for a PrPC receptor by identifying cell surface binding sites for radioiodinated bacterial fusion proteins incorporating segments of the PrP sequence (157). Thus far, we have detected saturable and specific binding sites on the surfaces of cultured cells that have an affinity of 70 to 240 nM, with ∼1 million sites per cell. Binding is seen only with fusion proteins incorporating the N-terminal half of the PrP sequence, consistent with the importance of this region in endocytic targeting. Many but not all of the detected binding sites are glycosaminoglycan (GAG) molecules, based on their biochemical properties. Whether either the GAG or non-GAG sites play a role in endocytosis of PrPC remains to be determined.
CELL BIOLOGY OF PRPSC
Cell Culture Models of Prion Formation
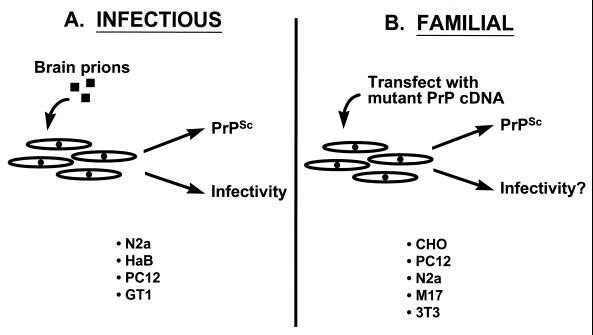
Experimental analysis of conventional viral diseases depends on the ability to grow the infectious agent in cultured cells, and the same has been true for prion diseases. Several cell types are thought to produce PrPSc following prion infection in vivo, including neurons, astrocytes, and lymphoreticular cells (9, 49, 53, 134). However, only some neuronally derived cell lines appear to be susceptible to infection with scrapie prions in vitro, including N2a mouse neuroblastoma cells (22, 133), PC12 rat pheochromocytoma cells (140), spontaneously immortalized hamster brain (HaB) cells (172), and T-antigen-immortalized hypothalamic neurons (GT1 cells) (146) (Fig. 5A). Once infected, these cells continuously produce low levels of PrPSc which can be recognized by its protease resistance and detergent insolubility in biochemical experiments and by its infectivity in animal bioassays. Surprisingly, the infected cells display no obvious cytopathology, with the exception of the GT1 cells, a subpopulation of which appears to undergo apoptosis (146). Scrapie-infected N2a cells show alterations in bradykinin-mediated responses and in membrane fluidity, although these abnormalities do not seem to affect the growth or viability of the cells (91, 184). Indeed, all the infected lines can be cloned and maintained in culture for many passages.
FIG. 5.
Cell culture systems for generation of PrPSc. (A) To model the infectious manifestation of prion diseases, several kinds of cultured cells have been infected with prions purified from rodent brain. (B) To model familial prion diseases, cultured cells have been transfected to express PrP molecules carrying disease-specific mutations. N2a and M17 are, respectively, mouse and human neuroblastoma cells; HaB are spontaneously immortalized hamster brain cells; PC12 are rat pheochromocytoma cells; GT1 are T-antigen-immortalized hypothalamic neurons; CHO are Chinese hamster ovary cells; and 3T3 are transformed mouse fibroblasts.
Scrapie-infected cultured cells provide a model of infectious forms of prion diseases, but until recently there had been no comparable model of familial forms (Fig. 5B). In principle, it should be possible to model familial prion diseases by expression in transfected cells of PrP molecules carrying pathogenic mutations. By comparing cells expressing mutant PrPs with those that have been infected with exogenous prions, it should be possible to directly contrast the familial and infectious manifestations of prion disease at the cellular level. It is important to keep in mind that the mechanisms underlying PrPSc formation may be different for the infectious and genetic forms. In the first case, exogenous PrPSc serves as a catalyst for conversion of endogenous PrPC to the PrPSc state, while in the second case, mutant PrP is spontaneously transformed to PrPSc.
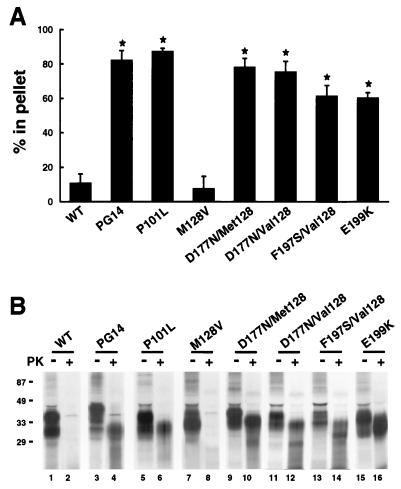
During the last several years, my laboratory has developed a model of familial prion diseases by constructing stably transfected lines of Chinese hamster ovary (CHO) cells that express murine homologues of mutant PrPs associated with all three familial prion diseases of humans (47, 93–97). We have found that each of seven different mouse PrPs (moPrPs) carrying a pathogenic mutation displays biochemical properties of PrPSc. As shown in Fig. 6, these properties include detergent insolubility, manifested by sedimentation at 265,000 × g from Triton-deoxycholate lysates, and protease resistance, manifested by production of an N-terminally truncated core of 27 to 30 kDa after treatment with proteinase K. Wild-type moPrP and moPrP carrying a mutation (M128V) homologous to a nonpathogenic polymorphism of human PrP do not display these properties. We have also noted subtle biochemical differences among the mutant moPrPs in their glycosylation pattern, proteinase K cleavage site, and degree of detergent insolubility, which may relate to the different clinical and neuropathological phenotypes associated with each protein. In support of the relevance of the cell culture model to an in vivo system, we have recently demonstrated that a mutant PrP expressed in transgenic mice acquires PrPSc-like properties identical to those seen in CHO cells and also produces clinical neurological dysfunction and neuropathology in the animals (38).
FIG. 6.
MoPrPs carrying disease-related mutations are detergent-insoluble and protease-resistant when expressed in cultured CHO cells. (A) CHO cells expressing wild-type (WT) and mutant moPrPs were labeled with [35S]methionine for 20 min and then chased for 3 h. Detergent lysates of the cells were centrifuged first at 16,000 × g for 5 min and then at 265,000 × g for 40 min. moPrP in the supernatants and pellets from the second centrifugation was immunoprecipitated and analyzed by SDS-polyacrylamide gel electrophoresis. PrP-specific bands were quantitated with a PhosphorImager, and the percentage of PrP in the pellet was calculated. Each bar represents the mean ± standard deviation of values from three experiments. Values that are significantly different from wild-type moPrP by Student’s t test (P < 0.001) are indicated by an asterisk. MoPrPs carrying disease-related mutations sediment (i.e., are detergent insoluble), while WT and M128V moPrPs remain largely in the supernatant. Human homologues of the mutant moPrPs analyzed here are associated with the following phenotypes: PG14 (9-octapeptide insertion), CJD variant; P101L, GSS; M128V, normal; D177N/Met128, FFI; D177N/Val128, CJD; F197S/Val128, GSS; E199K, CJD. (B) CHO cells expressing each moPrP were labeled for 3 h with [35S]methionine and chased for 4 h. Proteins in cell lysates were either digested at 37°C for 10 min with 3.3 μg of proteinase K per ml (+ lanes) or were untreated (− lanes) prior to recovery of moPrP by immunoprecipitation. Five times as many cell equivalents were loaded in the + lanes as in the − lanes. Molecular mass markers are in kilodaltons. moPrPs carrying pathogenic mutations yield a protease-resistant fragment of 27 to 30 kDa, while WT and M128V moPrPs are completely degraded. In separate experiments, we have shown that the PrP 27- to 30-kDa fragments are N-terminally truncated after the octapeptide repeats, the same region within which PrPSc from infected brain is cleaved (96). Modified from reference 72 with permission of the publisher.
There are now several other cultured cell systems that have been used to analyze the metabolism of mutant PrP molecules. We have recently shown that mutant moPrPs synthesized in PC12 cells have the same biochemical properties as those produced in CHO cells (37). Priola and Chesebro (126) have reported that PrP molecules carrying CJD-linked octapeptide insertions are partially protease resistant and detergent insoluble when expressed in N2a neuroblastoma cells and 3T3 fibroblasts. Interestingly, these properties were more pronounced for molecules with longer insertions than for those with shorter insertions and for proteins expressed in 3T3 cells than for those expressed in N2a cells. In 3T3 cells, PrP molecules with longer insertions were also more resistant to PI-PLC release, although in N2a cells, the mutant proteins were released as readily as wild-type PrP. In M17 human neuroblastoma cells, PrP molecules with a D178N mutation (linked to either FFI or CJD, depending on the presence of methionine or valine, respectively, at codon 129) were degraded prior to reaching the cell surface, a defect that was particularly severe for unglycosylated forms (120). The mutant proteins were protease sensitive and PI-PLC releasable. Finally, the GSS-linked Q217R mutation caused inhibition of surface transport and synthesis of forms lacking a GPI anchor and also increased the protease resistance and detergent insolubility of the protein in M17 cells (159). Most of these defects were reversed by culturing cells at 24°C, consistent with misfolding of the mutant protein.
Taken together, these studies of cultured cells make it clear that pathogenic mutations induce significant alterations in the metabolism of PrP. One effect of the mutation is that the protein acquires PrPSc-like biochemical properties, although the extent to which this occurs can vary with the cell type and with the mutation being expressed. It thus seems likely that these culture systems are modeling important features of the PrPC→PrPSc conversion process. However, it is important to point out that mutant PrPs synthesized in cultured cells differ in at least one respect from PrPSc isolated from the brains of humans and animals: the cell-derived PrPs have a considerably lower degree of protease resistance (95, 126, 159). It remains to be determined whether this difference reflects a variation in protease resistance that is often seen in scrapie isolates from different sources (8) or whether it results from the inefficiency of cultured cells in carrying out some critical step in the conversion process. Clearly, the definitive test of whether mutant PrPs synthesized in cultured cells represent authentic PrPSc will be to determine whether these proteins are infectious in animal bioassays.
Subcellular Localization of PrPSc
The subcellular distribution of PrPSc has been difficult to determine, primarily because this form of the protein displays poor immunoreactivity unless treated with denaturing agents that have a deleterious effect on cell morphology. Immunofluorescence studies of scrapie-infected N2a cells suggest that some PrPSc molecules reside intracellularly, colocalizing with Golgi markers in some clones but not in others (172). Electron microscopic studies of both N2a cells and brain tissue show that PrPSc also colocalizes with late endosomal and lysosomal markers (4, 106). It is also clear that some PrPSc molecules, both mutant and infectious, are present on the cell surface, as shown by immunogold staining for electron microscopy and by labeling of intact cells with membrane-impermeant probes (28, 83, 95, 165). The tentative conclusion from these studies is that PrPSc may be widely distributed within infected cells, although improved methods for in situ detection of PrPSc are clearly needed to establish the relative proportions of the protein in these multiple locations.
Membrane Attachment of PrPSc
The precise mechanism by which PrPSc is attached to cell membranes has been a matter of uncertainty. Chemical analysis of the purified protein demonstrates that PrPSc, like PrPC, possesses a C-terminal GPI anchor (161). Unlike PrPC, however, PrPSc is not releasable from brain membranes with PI-PLC (24, 141, 164). The lack of release is not due to sequestration in the lumen of a vesicular compartment, since membrane-bound PrPSc is accessible to biotinylation and protease digestion. After surface labeling, PrPSc in scrapie-infected N2a cells also cannot be released by incubation of intact cells with PI-PLC (95). The GPI anchor of purified and denatured PrPSc is cleavable by PI-PLC (160), suggesting that the anchor is not modified, for example by acylation of the inositol ring, so as to make it intrinsically PI-PLC resistant. Taken together, all of these results argue that PrPSc is associated with membranes in a way that is different from PrPC, although exactly how they differ is unclear.
Our analysis of mutant PrPs in cultured CHO cells has shed new light on the issue of the membrane attachment of PrPSc (94). We found that PrPs carrying pathogenic mutations are glycosylated and expressed on the cell surface but that like PrPSc from scrapie-infected brain, the mutant proteins are not released from the membrane by treatment with PI-PLC (Fig. 7A). This property does not result from absence of a GPI anchor structure, since the mutant PrPs metabolically incorporate the anchor precursors [3H]ethanolamine, [3H]palmitate, and [3H]stearate. Although we originally postulated that mutant PrPs possessed a secondary mechanism of membrane attachment in addition to their GPI anchors (94), more recent evidence suggests that the mutant molecules are resistant to PI-PLC-mediated release because their GPI anchors become physically inaccessible to the phospholipase as part of their conversion to the PrPSc state (110) (Fig. 7C to E). This conclusion is based on the failure of PI-PLC to quantitatively remove [3H]palmitate label from the proteins or to render them hydrophilic by Triton X-114 phase partitioning or phenyl-Sepharose chromatography, suggesting that the GPI anchor is retained. Resistance to cleavage is observed when PI-PLC is applied to intact cells (Fig. 7C), as well as when treatment is carried out after lysis in nondenaturing buffers (Fig. 7D). However, denaturation in sodium dodecyl sulfate (SDS) renders the GPI anchor of the mutant PrPs susceptible to cleavage, indicating that PI-PLC resistance depends on a native structure of the protein (Fig. 7E). We now view PI-PLC resistance as being an operational property analogous to protease resistance and postulate that it reflects an alteration in the structure of PrP attendant upon conversion to the PrPSc state. We hypothesize that the same structural changes that renders the polypeptide chain of PrPSc partially inaccessible to proteinase K may also render it inaccessible to PI-PLC. This effect presumably involves a change in the conformation of the polypeptide chain, but it may also be related to aggregation or polymerization of the protein.
Although most PrPC molecules are attached to the cell membrane exclusively by their GPI anchor, it has been reported that there exists a subpopulation that displays a transmembrane orientation. This conclusion was originally based on analysis of PrP molecules synthesized in vitro from synthetic mRNA by using either wheat germ extract or rabbit reticulocyte lysate in the presence of canine pancreatic microsomes (75, 101). To explain the existence of transmembrane forms, Lingappa and colleagues proposed that a 24-amino-acid region of PrP (the “stop transfer effector”) induces a pause in translocation of the polypeptide chain and that cytosolic factors determine whether this intermediate is either converted into a stable transmembrane species or fully translocated into the ER lumen (51, 185). In subsequent work, these authors postulated the existence of two transmembrane species of PrP with the same membrane-spanning segment (residues 112 to 135) but with opposite orientations of the polypeptide chain (76, 77). It was suggested that an increase in the proportion of one of these species was induced by the presence of a disease-causing mutation in PrP and that this form was a cause of neurodegeneration. However, a difficulty with all of these studies is that the percentage of the transmembrane forms is usually quite low, and in addition it is not clear whether the GPI anchor has been properly attached, particularly in the in vitro translation system.
Kinetics of PrPSc Production
The kinetics of PrPSc production have been examined both in scrapie-infected N2a and HaB cells and in CHO cells expressing mutant PrPs. Protease-resistant PrP in infected cells is generated only during the chase period after pulse-labeling, with half-maximal accumulation requiring several hours (13, 14, 28). This result is consistent with the idea that transformation of PrPC into PrPSc is a posttranslational event. Once formed, PrPSc appears to be metabolically stable for periods as long as 24 to 48 h, in contrast to PrPC, which turns over with a half-life of 4 to 6 h (13, 14, 28). Only a minority of PrPC molecules are converted to PrPSc in infected cells, with the remainder being degraded by pathways similar to those found in uninfected cells; these pathways have not been clearly identified, but they may involve acidic compartments such as endosomes and lysosomes (171).
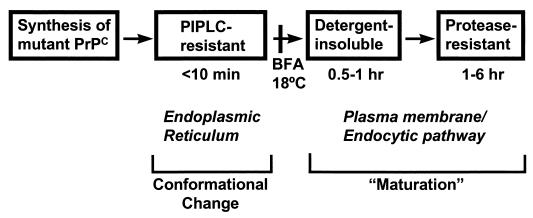
We have used transfected CHO cells to identify intermediate biochemical steps in the conversion of mutant PrPs to the PrPSc state (47). Our strategy was to measure the kinetics with which three PrPSc-related properties (PI-PLC resistance, detergent insolubility, and protease resistance) develop in pulse-chase labeling experiments. This has allowed us to define three steps in the conversion process (Fig. 8). The earliest biochemical change we could detect in mutant PrP, one that was observable within minutes of pulse-labeling the cells, was the acquisition of PI-PLC resistance, a property that was revealed by partitioning of the phospholipase-treated protein into the detergent phase of Triton X-114 lysates or by its binding to phenyl-Sepharose. The second step is acquisition of detergent insolubility, which is not maximal until after 1 h of chase (96), arguing that it occurs after the acquisition of PI-PLC resistance. Detergent insolubility presumably reflects aggregation of PrP molecules, and by sucrose gradient fractionation we were able to detect aggregates ranging in size from 4S (monomeric) to over 20S (more than 30 PrP molecules). The third step is acquisition of protease resistance, which is not maximal until several hours after labeling (96). We have shown that mutant PrP is detergent insoluble and protease sensitive at early times after pulse-labeling, directly demonstrating that acquisition of detergent insolubility and protease resistance are temporally distinct steps. We have hypothesized that the fundamental conformational change that underlies the conversion of mutant PrPC into PrPSc is reflected in the acquisition of PI-PLC resistance. In this view, detergent insolubility and protease resistance are secondary properties that develop some time after the initial molecular conversion and reflect further “maturation” of mutant PrP molecules, perhaps by aggregation or polymerization.
FIG. 8.
Scheme for transformation of mutant PrPs to a PrPSc state. Mutant PrPs are initially synthesized in the PrPC state and acquire PrPSc properties in a stepwise fashion as they traverse different cellular compartments. PI-PLC resistance, which develops in the ER, reflects folding of the polypeptide chain into the PrPSc conformation. Detergent insolubility and protease resistance, which develop upon arrival at the plasma membrane or along an endocytic pathway, result from intermolecular aggregation (“maturation”). The times given underneath the boxes indicate when after pulse-labeling the corresponding property is detected. Addition of brefeldin A (BFA) to cells or incubation at 18°C, treatments which block the movement of proteins beyond the Golgi apparatus, inhibit the acquisition of detergent insolubility and protease resistance but not PI-PLC resistance. Adapted from reference 47 with permission of the publisher.
Although a PI-PLC-resistant intermediate underlies the conversion of mutant PrPs to the PrPSc state, such an intermediate may not play a role in PrPSc formation from wild-type PrPC after infection with exogenous prions. Consistent with this idea, there is evidence that PrPSc in infected neuroblastoma cells is synthesized from a precursor that is PI-PLC sensitive (28). However, much work remains to be done in characterizing the molecular intermediates in PrPSc formation in infected cells.
Subcellular Site of PrPSc Formation
There is surprisingly little information available about how extracellular PrPSc is taken up by cells during the initial stage of infection. If this uptake were to occur via an endocytic mechanism, PrPSc may interact with PrPC on the plasma membrane or in endosomes and key events in the conversion process may take place in these locations. In fact, several pieces of evidence suggest that an endocytic pathway is involved in the generation of PrPSc in scrapie-infected N2a and HaB cells. First, this idea is consistent with the localization of at least some PrPSc molecules in endosomes and lysosomes. Second, surface iodination of infected cells results in incorporation of radiolabel into PrPSc after a chase period, arguing that PrP molecules transit the plasma membrane prior to conversion into PrPSc (28). Third, treatment of cells with PI-PLC or proteases inhibits the production of PrPSc, presumably by removal of the PrPC precursor from the cell surface (14, 28). Consistent with this last result, generation of PrPSc is also inhibited by treatment of cells with the fungal metabolite brefeldin A, which blocks surface delivery of membrane and secretory proteins (169). These results do not distinguish between the plasma membrane or endosomes as the relevant sites for PrPSc production.
Recent studies suggest that detergent-resistant subdomains of the plasma membrane (rafts [158] [discussed above]) may be involved in the formation of PrPSc. Consistent with this idea, both PrPC and PrPSc are found in raft domains isolated biochemically (65, 111, 171, 178). In addition, pharmacological depletion of cellular cholesterol, which is known to disrupt rafts, inhibits PrPSc formation (171). Finally, artificially constructed transmembrane forms of PrP, which are excluded from rafts, are poor substrates for conversion to PrPSc (85). Since PrPSc is defined by its protease resistance in all of these studies, the results suggest that rafts are involved in the acquisition of this particular property but do not address whether other steps of PrPSc formation take place in these domains.
Our kinetic studies of mutant PrPs synthesized in CHO cells suggest that individual steps in the formation of PrPSc may take place in at least two different cellular locations (Fig. 8). Since mutant PrPs become PI-PLC resistant within minutes of synthesis in pulse-labeling experiments, this early step must take place in the ER. Consistent with this conclusion, acquisition of PI-PLC resistance is not affected by treatment of cells with brefeldin A or by incubation at 18°C, manipulations which block the exit of proteins beyond the Golgi (47). In contrast, detergent insolubility and protease resistance, which do not develop until later times of chase and are reduced by brefeldin A and 18°C incubation, are likely to be acquired after arrival of the protein at the cell surface, either on the plasma membrane itself or in endocytic compartments. We have evidence that raft domains might be involved in the acquisition of detergent insolubility and protease resistance, since these two properties are never acquired by chimeric forms of mutant PrP that contain a transmembrane domain in place of a GPI anchor (14a).
Thus, the results of our studies of cells expressing mutant PrPs are consistent with experiments on scrapie-infected cells, but with the additional finding that the very earliest step in PrPSc synthesis, formation of a PI-PLC-resistant, protease-sensitive intermediate, takes place in the ER rather than on the plasma membrane or in endosomes. Further work is required to determine whether an early ER intermediate is also generated in infected cells. If so, this would presumably require that external PrPSc somehow gain access to the lumen of the ER in order to interact with newly synthesized PrPC.
The idea that the generation of PrPSc from mutant PrPs may begin in the ER is novel, and it is theoretically appealing because of the well-known role of this organelle in protein folding. It is also reasonable to suggest that ER chaperones are good candidates for the hypothetical cellular cofactors that are widely thought to play an important regulatory role in prion synthesis (see below).
Posttranslational Cleavage of PrPSc
PrPSc in cultured cells and brain undergoes a proteolytic cleavage that removes a portion of the N terminus (29, 34, 169). This cleavage occurs at a site (labeled C in Fig. 1) distinct from the one at which PrPC is cleaved (labeled B). Cleavage at site C ensues within 1 h after formation of PrPSc and occurs within the region attacked by proteinase K to yield PrP 27-30. It has been pointed out that cleavage of PrPC site B occurs within an amyloidogenic and neurotoxic region of the polypeptide chain whereas cleavage of PrPSc at site C leaves this segment intact, thereby implicating N-terminal trimming in regulation of the pathogenicity of PrP (34). Sensitivity to lysosomotropic amines and protease inhibitors indicates that cleavage of PrPSc occurs in endosomes or lysosomes, compartments that may play a role in the generation of this isoform, as discussed above. However, N-terminal trimming is not an essential step in the formation of PrPSc, since PrPSc is still produced in the presence of lysosomotropic amines that inhibit the trimming (29, 169).
In samples from many patients with GSS, PrPSc fragments smaller than 27 to 30 kDa, which are generated by additional cleavages, have been observed (115, 121, 166, 167). A 7- to 8-kDa fragment is produced by cleavage near residues 80 and 150, and an 11-kDa fragment is produced by cleavage near residues 50 and 150; the termini of these fragments are usually heterogeneous. In some cases, these smaller fragments are major constituents of the amyloid plaques that are characteristic of GSS (121, 166, 167), and it is reasonable to propose that PrP molecules carrying GSS-linked mutations are metabolized in a distinctive way to generate these amyloidogenic products.
A small percentage of PrPSc molecules also appear to undergo C-terminal proteolytic cleavage at a position 3 residues from the GPI attachment site (160); this cleavage would appear to be distinct from cleavage A in PrPC (Fig. 1), which occurs within the anchor structure itself.
MOLECULAR NATURE OF THE PRPC→PRPSC CONVERSION REACTION
Structural Changes and Physical Interactions
All available evidence indicates that conversion of PrPC into PrPSc is conformational rather than covalent and that the two isoforms have the same primary amino acid sequence and probably the same posttranslational additions (132, 162). The conformational change involves a substantial increase in the amount of β-sheet structure of the protein, with possibly a small decrease in the amount of α-helix. Circular dichroism and infrared spectroscopy indicate that PrPC contains approximately 42% α-helix and 3% β-sheet, compared to 30% α-helix and 43% β-sheet for PrPSc (31, 113, 142). A tertiary structure of PrPC, based on nuclear magnetic resonance spectroscopy analysis of recombinant PrP produced in bacteria, includes a long, flexible N-terminal tail (residues 23 to 121), three α-helices, and two small, anti-parallel β-strands that flank the first α-helix (55, 136, 137). Although a tertiary structure of PrPSc has not yet been obtained, the current evidence suggests that generation of this isoform involves primarily changes in the N-terminal half of the protein, including folding of a portion of the N-terminal tail from residues 90 to 121 (and possibly part of the first α-helix) into β-sheet (119). Clearly, a key challenge in the field now is to obtain a complete structure for PrPSc by spectroscopic and crystallographic techniques and to specify at the atomic level how this structure is produced from the PrPC conformation.
It is thought that during prion infection, a highly specific physical interaction that is responsible for generating new molecules of PrPSc occurs between PrPC and PrPSc. This conclusion is based on several kinds of evidence. First, and perhaps most compelling, mice that do not synthesize PrPC because their endogenous PrP gene has been ablated are completely resistant to prion infection (20, 143). Second, expression of genetically engineered forms of PrP in transgenic mice and cultured cells profoundly alters their susceptibility to prion infection (131, 149, 150). For example, mice are not normally susceptible to prions derived from hamsters but expression of a hamster PrP transgene in mice renders them susceptible to infection.
Additional studies have provided evidence about which parts of the PrPC substrate are critical for the proposed interaction with PrPSc. The N-terminal 66-amino-acid stretch is not required for the conversion, nor is a 36-amino-acid segment that includes helix 1 and the second β-strand (58, 109, 139); at least one of the N-linked oligosaccharide chains is also dispensable (50, 170). Within the central region of the protein, the degree of amino acid homology between infectious PrPSc and endogenous PrPC substrate profoundly influences the efficiency of the conversion process (150, 151, 175); mismatch at a single amino acid residue can prevent the formation of PrPSc (125). In addition, nonhomologous PrP can interfere with the conversion of homologous PrP in cells expressing both proteins (124). The exquisite molecular specificity suggested by these experiments is thought to underlie the strong barrier against interspecies transmission of prions.
A final line of evidence favoring a direct interaction between PrPC and PrPSc comes from in vitro conversion systems in which these two isoforms are mixed in a test tube reaction. In theory, reconstitution of PrPSc production in vitro by using purified components would provide the most direct way to define the molecular requirements of the conversion reaction. Moreover, if infectivity could be recovered, it would provide the most unambiguous demonstration of the prion hypothesis. A widely used cell-free conversion system involves mixing PrPSc that has been purified from scrapie-infected brain tissue with metabolically labeled PrPC that has been immunoprecipitated from cultured cells and scoring the amount of radioactive PrP that becomes protease resistant (7, 88, 89, 135). This system mimics several features of prion propagation in vivo, including species specificity and strain specificity. However, the conversion requires a large excess of PrPSc over PrPC (>50-fold), the inverse of the ratio that probably exists during initiation of prion infection in vivo, and thus far it has not been possible to demonstrate infectivity of the converted product. It is possible that cellular factors that are absent in this purified system account in part for its poor stoichiometric efficiency.
Models of PrPSc Formation
There is currently a vigorous debate about the mechanistic details of the PrPC→PrPSc conversion process. One model (nucleated polymerization) suggests that spontaneous conversion of PrPC monomers to the PrPSc state is stabilized by addition of the monomers to the ends of a preexisting polymer composed of PrPSc (30, 44, 60, 69). This process is envisaged as a nucleated polymerization similar to actin or tubulin polymerization, with the barrier between PrPC and PrPSc being overcome by incorporation of subunits into an ordered polymer. A second model (template assistance) postulates that a relatively small number of PrPC and PrPSc molecules oligomerize, possibly along with a hypothetical molecular chaperone (5, 82). The PrPSc component then serves as a catalyst or template that impresses its conformation on the PrPC substrate or on a partially converted intermediate designated PrP*. In this case, the kinetic barrier between PrPC and PrPSc is surmounted by the catalytic action of PrPSc, possibly in conjunction with the chaperone. These two models, which are not mutually exclusive, are applicable to familial as well as infectious manifestations of prion diseases, since the presence of a pathogenic mutation presumably favors the spontaneous formation of either PrPSc nuclei or PrP*.
A piece of information that will be crucial in deciding between these two models is an understanding of the physical state of PrPSc inside cells. Molecules of PrPSc have a strong tendency to aggregate and polymerize, as evidenced by their sedimentation in detergent lysates, their presence in the form of extracellular fibrils in amyloid plaques, and their incorporation into artificial rods following limited proteolysis in the presence of detergent (48, 105). However, ordered PrPSc polymers have not been observed intracellularly by ultrastructural techniques, although this could be because they are too small or irregular in appearance to be resolved. To definitively ascertain the oligomeric state of PrPSc within cells, it will be necessary to trap and isolate a putative PrPC-PrPSc conversion intermediate by biochemical methods and to develop improved methods for visualizing PrPSc within cells.
Role of Molecular Chaperones in the Formation of PrPSc
Chaperones are proteins that facilitate the folding of polypeptides during their biosynthesis and transport into organelles and that help prevent protein aggregation during conditions of cellular stress such as heat shock (74). Chaperones are thought to act by binding to their substrates, sometimes in an ATP-dependent manner, and preventing the formation of unproductive folding intermediates. Chaperones are found in several cellular compartments, although the ER is the only organelle through which PrP passes during its cellular trafficking that contains known chaperone molecules. There is currently a great deal of discussion and excitement in the prion field about the possibility that chaperones play a critical role in the generation of PrPSc (81, 181). This idea is appealing from a theoretical point of view, since PrPSc formation involves changes in protein folding and possibly intermolecular aggregation, processes in which chaperones are known to play a role.
There are a number of lines of experimental evidence in favor of chaperone involvement in prion phenomena. First, there are studies of transmission patterns in transgenic mice (175). Human prions propagate poorly in mice carrying a human PrP transgene, but this effect is alleviated either by eliminating the expression of endogenous mouse PrP or by modifying the human PrP transgene to include N- and C-terminal segments from the mouse PrP gene. These results and others (86) have been interpreted to imply the existence of cellular chaperones, collectively referred to as protein X, that interact in a species-specific way with the C terminus of PrPC during prion propagation. If such chaperones were also cell specific, this might explain the selective propagation of prions in distinct neuronal populations and in particular types of peripheral cells such as those of the lymphoreticular system. Evidence from scrapie-infected neuroblastoma cells also implicates chaperones in prion biogenesis. Several “chemical chaperones” such as glycerol and dimethyl sulfoxide, which stabilize protein conformation, inhibit PrPSc production in infected cells (173). Moreover, these cells display alterations in the induction and localization of several cytoplasmic heat shock proteins which function as molecular chaperones (174). Finally, there is evidence that PrP can bind to two chaperones, mammalian Hsp60 and bacterial GroEL, in a yeast two-hybrid system and in a glutathione S-transferase–agarose pull-down assay (56).
The most direct evidence that chaperones can affect generation of PrPSc is provided by experiments in which the yeast chaperone Hsp104 and the bacterial chaperone GroEL have been shown to enhance the formation of PrPSc in a cell-free conversion system (51). Hsp104 was chosen for these experiments because it interacts both genetically and biochemically with Sup35, a subunit of a translation termination factor in yeast that can exist in a prion-like state referred to as [PSI+] (35, 147). Although the effects of Hsp104 and GroEL represent an important demonstration of the principle that chaperones can modulate the PrPC→PrPSc conversion process, the two chaperones chosen would not normally come into contact with PrP in a mammalian cell. It will thus be crucial to identify the chaperones with which mammalian PrP interacts in vivo and to see if alterations in the amount or activity of these molecules affect the production of PrPSc. Candidates might include known ER chaperones, other as yet unidentified chaperone proteins, or, conceivably, nonprotein molecules with which PrP is known to interact, such as glycosaminoglycans (123).
CONCLUSION AND PERSPECTIVES
The work reviewed here has begun to provide a detailed picture of how cells synthesize, process, and target PrPC and PrPSc. PrPC is synthesized and matures along the secretory pathway much like other membrane glycoproteins. One important feature is the addition of a GPI anchor, which serves to attach the polypeptide chain to the lipid bilayer without the necessity for a transmembrane domain. Once it reaches the plasma membrane, PrPC is constitutively endocytosed via clathrin-coated vesicles, and some of the molecules are proteolytically cleaved in a highly conserved domain before recycling to the cell surface. Efficient endocytosis depends on structural features in the N-terminal half of the PrP molecule and may be mediated by a transmembrane PrPC receptor.
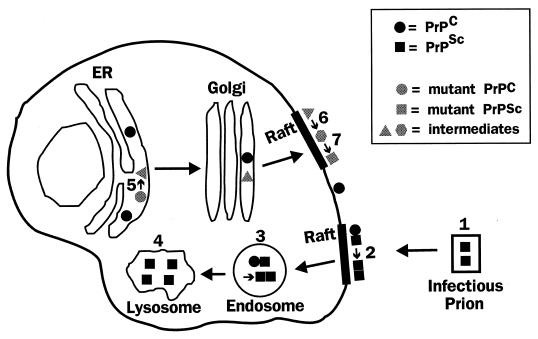
The cellular pathways involved in the formation of PrPSc are summarized in Fig. 9. PrPSc is produced from the PrPC precursor as a result of a conformational change involving increased β-sheet structure in the N-terminal half of the polypeptide chain. In the infectious manifestation, this structural alteration is induced by a sequence-specific physical interaction between exogenous PrPSc and endogenous PrPC, a process that may take place in detergent-resistant rafts on the plasma membrane or in endocytic organelles, although other locations have not been ruled out. Once generated, PrPSc is metabolically stable and becomes localized partly but not exclusively in intracellular organelles, perhaps endosomes and lysosomes, which may be the sites where the N terminus of the protein is proteolytically cleaved.
FIG. 9.
Model of the cellular pathways involved in generation of PrPSc. In the infectious manifestation of prion diseases, extracellular PrPSc in the form of a prion particle (labeled 1) interacts with PrPC on the cell surface, possibly in detergent-resistant rafts, catalyzing its conversion to PrPSc (step 2). Conversion may also occur after uptake of the proteins into an endosomal compartment (step 3). Once formed, some PrPSc accumulates in lysosomes (step 4), although the protein is probably found in a number of other cellular locations as well. In familial prion disorders, mutant PrPC is converted spontaneously to the PrPSc state via a series of biochemical intermediates, the earliest of which is a PI-PLC-resistant form generated in the ER (step 5). Mutant PrP molecules are subsequently delivered to the cell surface, where they become detergent insoluble (step 6) and then protease resistant (step 7), possibly in raft domains. Steps 6 and 7 could also occur in endocytic organelles.
In familial forms of prion diseases, conversion of PrPC to the PrPSc conformation is induced by a mutation in the amino acid sequence of the protein. This transformation is accompanied by stepwise acquisition of several biochemical properties characteristic of PrPSc. The earliest recognizable change is acquisition of PI-PLC resistance, which takes place in the ER and may register the initial conformational alteration of the protein. Detergent insolubility and protease resistance develop later, possibly in raft domains on the plasma membrane or in endocytic compartments.
Several important avenues for future investigation are suggested by the cell biological results summarized here. First, it will be critical to define the mechanism by which PrPC molecules are concentrated in coated pits and vesicles, to determine whether a specific surface receptor is really responsible, and, if so, to isolate and clone it. Further exploration of the interaction between PrPC and copper ions at the cellular level is also warranted. These investigations are likely to provide important clues to the normal function of PrPC, loss of which could play some role in the disease process. Second, we will want to understand the individual steps along the pathway from PrPC to PrPSc, isolating and characterizing the earliest conversion intermediates biochemically and determining where in the cell they are generated. This information will help distinguish between the nucleated-polymerization and template assistance models of PrPSc formation. Third, it will be important to identify other cellular molecules that undoubtedly play a crucial in the conversion process. These species may be already identified molecular chaperones, or they may be novel proteins in cellular compartments through which PrP trafficks. Adding purified cofactors to in vitro conversion systems or using subcellular fractions that retain these components may make it possible to greatly enhance the efficiency of PrPSc production and possibly the generation of infectious prions. Each of these cell biological approaches will allow us to develop a more detailed understanding of the pathogenesis of prion diseases and will ultimately provide the basis for the development of rational therapeutic modalities.
ACKNOWLEDGMENTS
Work in my laboratory is supported by grants from the National Institutes of Health, the American Health Assistance Foundation, and the Alzheimer’s Association.
REFERENCES
- 1.Alper T, Cramp W A, Haig D A, Clarke M C. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764–766. doi: 10.1038/214764a0. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R G W. Plasmalemmal caveolae and GPI-anchored membrane proteins. Curr Opin Cell Biol. 1993;5:647–652. doi: 10.1016/0955-0674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 3.Anderson R M, Donnelly C A, Ferguson N M, Woolhouse M E J, Watt C J, Udy H J, MaWhinney S, Dunstan S P, Southwood M E J, Wilesmith J W, Ryan J B M, Hoinville L J, Hillerton J E, Austin A R, Wells G A H. Transmission dynamics and epidemiology of BSE in British cattle. Nature. 1996;382:779–788. doi: 10.1038/382779a0. [DOI] [PubMed] [Google Scholar]
- 4.Arnold J E, Tipler C, Laszlo L, Hope J, Landon M, Mayer R J. The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. J Pathol. 1995;176:403–411. doi: 10.1002/path.1711760412. [DOI] [PubMed] [Google Scholar]
- 5.Bamborough P, Wille H, Telling G C, Yehiely F, Prusiner S B, Cohen F E. Prion protein structure and scrapie replication: theoretical, spectroscopic, and genetic investigations. Cold Spring Harbor Symp Quant Biol. 1996;61:495–509. [PubMed] [Google Scholar]
- 6.Bendheim P E, Brown H R, Rudelli R D, Scala L J, Goller N L, Wen G Y, Kascsak R J, Cashman N R, Bolton D C. Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology. 1992;42:149–156. doi: 10.1212/wnl.42.1.149. [DOI] [PubMed] [Google Scholar]
- 7.Bessen R A, Kocisko D A, Raymond G J, Nandan S, Lansbury P T, Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 8.Bessen R A, Marsh R F. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blättler T, Brandner S, Raeber A J, Klein M A, Volgtländer T, Weissmann C, Aguzzi A. PrP-expressing tissue required for transfer of scrapie infectivity from spleen to brain. Nature. 1997;389:69–73. doi: 10.1038/37981. [DOI] [PubMed] [Google Scholar]
- 10.Bolton D C, Bendheim P E. A modified host protein model of scrapie. In: Brown F, editor. Ciba Foundation Symposium. 135. Novel infectious agents and the central nervous system. Chichester, United Kingdom: John Wiley & Sons; 1988. pp. 164–181. [DOI] [PubMed] [Google Scholar]
- 11.Borchelt D R, Koliatsos V E, Guarnieri M, Pardo C A, Sisodia S S, Price D L. Rapid anterograde axonal transport of the cellular prion glycoprotein in the peripheral and central nervous systems. J Biol Chem. 1994;269:14711–14714. [PubMed] [Google Scholar]
- 12.Borchelt D R, Rogers M, Stahl N, Telling G, Prusiner S B. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology. 1993;3:319–329. doi: 10.1093/glycob/3.4.319. [DOI] [PubMed] [Google Scholar]
- 13.Borchelt D R, Scott M, Taraboulos A, Stahl N, Prusiner S B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borchelt D R, Taraboulos A, Prusiner S B. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J Biol Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- 14a.Brewer, J. A., and D. A. Harris. Unpublished data.
- 15.Brown D R, Qin K F, Herms J W, Madlung A, Manson J, Strome R, Fraser P E, Kruck T, Vonbohlen A, Schulzschaeffer W, Giese A, Westaway D, Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 16.Brown D R, Schulzschaeffer W J, Schmidt B, Kretzschmar H A. Prion protein-deficient cells show altered response to oxidative stress due to decreased SOD-1 activity. Exp Neurol. 1997;146:104–112. doi: 10.1006/exnr.1997.6505. [DOI] [PubMed] [Google Scholar]
- 17.Brown P. Donor pool size and the risk of blood-borne Creutzfeldt-Jakob disease. Transfusion. 1998;38:312–315. doi: 10.1046/j.1537-2995.1998.38398222878.x. [DOI] [PubMed] [Google Scholar]
- 18.Brown P, Preece M A, Will R G. “Friendly fire” in medicine: hormones, homografts, and Creutzfeldt-Jakob disease. Lancet. 1992;340:24–27. doi: 10.1016/0140-6736(92)92431-e. [DOI] [PubMed] [Google Scholar]
- 19.Bruce M E, Will R G, Ironside J W, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock C J. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 20.Büeler H, Aguzzi A, Sailer A, Greiner R A, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 21.Büeler H, Fischer M, Lang Y, Fluethmann H, Lipp H-P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Normal development and behavior of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 22.Butler D A, Scott M R D, Bockman J M, Borchelt D R, Taraboulos A, Hsiao K K, Kingsbury D T, Prusiner S B. Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caughey B, Chesebro B. Prion protein and the transmissible spongiform encephalopathies. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 24.Caughey B, Neary K, Buller R, Ernst D, Perry L L, Chesebro B, Race R E. Normal and scrapie-associated forms of prion protein differ in their sensitivities to phospholipase and proteases in intact neuroblastoma cells. J Virol. 1990;64:1093–1101. doi: 10.1128/jvi.64.3.1093-1101.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caughey B, Race R E, Chesebro B. Detection of prion protein mRNA in normal and scrapie-infected tissues and cell lines. J Gen Virol. 1988;69:711–716. doi: 10.1099/0022-1317-69-3-711. [DOI] [PubMed] [Google Scholar]
- 26.Caughey B, Race R E, Ernst D, Buchmeier M J, Chesebro B. Prion protein biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J Virol. 1989;63:175–181. doi: 10.1128/jvi.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caughey B, Race R E, Vogel M, Buchmeier M J, Chesebro B. In vitro expression in eukaryotic cells of a prion protein gene cloned from scrapie-infected mouse brain. Proc Natl Acad Sci USA. 1988;85:4657–4661. doi: 10.1073/pnas.85.13.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caughey B, Raymond G J. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 29.Caughey B, Raymond G J, Ernst D, Race R E. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caughey B, Raymond G J, Kocisko D A, Lansbury P T. Scrapie infectivity correlates with converting activity, protease resistance, and aggregation of scrapie-associated prion protein in guanidine denaturation studies. J Virol. 1997;71:4107–4110. doi: 10.1128/jvi.71.5.4107-4110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caughey B W, Dong A, Bhat K S, Ernst D, Hayes S F, Caughey W S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 32.Chang W-J, Ying Y-S, Rothberg K G, Hooper N M, Turner A J, Gambliel H A, De Gunzburg J, Mumby S M, Gilman A G, Anderson R G W. Purification and characterization of smooth muscle caveolae. J Cell Biol. 1994;126:127–138. doi: 10.1083/jcb.126.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chazot G, Broussolle E, Lapras C, Blattler T, Aguzzi A, Kopp N. New variant of Creutzfeldt-Jakob disease in a 26-year-old French man. Lancet. 1996;347:1181. doi: 10.1016/s0140-6736(96)90638-8. [DOI] [PubMed] [Google Scholar]
- 34.Chen S G, Teplow D B, Parchi P, Teller J K, Gambetti P, Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 35.Chernoff Y O, Lindquist S L, Ono B-I, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 36.Chesebro B. BSE and prions: uncertainties about the agent. Science. 1998;279:42–43. doi: 10.1126/science.279.5347.42. [DOI] [PubMed] [Google Scholar]
- 37.Chiesa, R., and D. A. Harris. Unpublished data.
- 38.Chiesa R, Piccardo P, Ghetti B, Harris D A. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- 39.Cohen F E, Pan K M, Huang Z, Baldwin M, Fletterick R J, Prusiner S B. Structural clues to prion replication. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 40.Colling S B, Collinge J, Jefferys J G R. Hippocampal slices from prion protein null mice: disrupted Ca2+-activated K+ currents. Neurosci Lett. 1996;209:49–52. doi: 10.1016/0304-3940(96)12596-9. [DOI] [PubMed] [Google Scholar]
- 41.Colling S B, Khana M, Collinge J, Jefferys J G R. Mossy fibre reorganization in the hippocampus of prion protein null mice. Brain Res. 1997;755:28–35. doi: 10.1016/s0006-8993(97)00087-5. [DOI] [PubMed] [Google Scholar]
- 42.Collinge J, Sidle K C L, Meads J, Ironside J, Hill A F. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 43.Collinge J, Whittington M A, Sidle K C, Smith C J, Palmer M S, Clarke A R, Jefferys J G. Prion protein is necessary for normal synaptic function. Nature. 1994;370:295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 44.Come J H, Fraser P E, Lansbury P T. A kinetic model for amyloid formation in the prion diseases: importance of seeding. Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuillé J, Chelle P L. Experimental transmission of trembling to the goat. CR Seances Acad Sci. 1939;208:1058–1060. [Google Scholar]
- 47.Daude N, Lehmann S, Harris D A. Identification of intermediate steps in the conversion of a mutant prion protein to a scrapie-like form in cultured cells. J Biol Chem. 1997;272:11604–11612. doi: 10.1074/jbc.272.17.11604. [DOI] [PubMed] [Google Scholar]
- 48.DeArmond S J, McKinley M P, Barry R A, Braunfeld M B, McColloch J R, Prusiner S B. Identification of prion amyloid filaments in scrapie-infected brain. Cell. 1985;41:221–235. doi: 10.1016/0092-8674(85)90076-5. [DOI] [PubMed] [Google Scholar]
- 49.DeArmond S J, Mobley W C, DeMott D L, Barry R A, Beckstead J H, Prusiner S B. Changes in the localization of brain prion proteins during scrapie infection. Neurology. 1987;37:1271–1280. doi: 10.1212/wnl.37.8.1271. [DOI] [PubMed] [Google Scholar]
- 50.DeArmond S J, Sanchez H, Yehiely F, Qiu Y, Ninchakcasey A, Daggett V, Camerino A P, Cayetano J, Rogers M, Groth D, Torchia M, Tremblay P, Scott M R, Cohen F E, Prusiner S B. Selective neuronal targeting in prion disease. Neuron. 1997;19:1337–1348. doi: 10.1016/s0896-6273(00)80424-9. [DOI] [PubMed] [Google Scholar]
- 51.DebBurman S K, Raymond G J, Caughey B, Lindquist S. Chaperone-supervised conversion of prion protein to its protease-resistant form. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Fea K A, Nakahara D H, Calayag M C, Yost C S, Mirels L F, Prusiner S B, Lingappa V R. Determinants of carboxyl-terminal domain translocation during prion protein biogenesis. J Biol Chem. 1994;269:16810–16820. [PubMed] [Google Scholar]
- 53.Diedrich J F, Bendheim P E, Kim Y S, Carp R I, Haase A T. Scrapie-associated prion protein accumulates in astrocytes during scrapie infection. Proc Natl Acad Sci USA. 1991;88:375–379. doi: 10.1073/pnas.88.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dodelet V C, Cashman N R. Prion protein expression in human leukocyte differentiation. Blood. 1998;91:1556–1561. [PubMed] [Google Scholar]
- 55.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Structure of the recombinant full-length hamster prion protein PrP(29-231): the N terminus is highly flexible. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker E L. Prion protein PrPC interacts with molecular chaperones of the Hsp60 family. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Endo T, Groth D, Prusiner S B, Kobata A. Diversity of oligosaccharide structures linked to asparagines of the scrapie prion protein. Biochemistry. 1989;28:8380–8388. doi: 10.1021/bi00447a017. [DOI] [PubMed] [Google Scholar]
- 58.Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 59.Fournier J-G, Escaig-Haye F, Billette de Villemeur T, Robain O. Ultrastructural localization of cellular prion protein (PrPC) in synaptic boutons of normal hamster hippocampus. C R Acad Sci Paris. 1995;318:339–344. [PubMed] [Google Scholar]
- 60.Gajdusek D C. Spontaneous generation of infectious nucleating amyloids in the transmissible and nontransmissible cerebral amyloidoses. Mol Neurobiol. 1994;8:1–13. doi: 10.1007/BF02778003. [DOI] [PubMed] [Google Scholar]
- 61.Gajdusek D C. Infectious amyloids: subacute spongiform encephalopathies as transmissible cerebral amyloidoses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2851–2900. [Google Scholar]
- 62.Gajdusek D C, Gibbs C J, Alpers M. Experimental transmission of a kuru-like syndrome to chimpanzees. Nature. 1966;209:794–796. doi: 10.1038/209794a0. [DOI] [PubMed] [Google Scholar]
- 63.Gajdusek D C, Zigas V. Degenerative disease of the central nervous system in New Guinea: the endemic occurrence of “kuru” in the native population. N Engl J Med. 1957;257:974–978. doi: 10.1056/NEJM195711142572005. [DOI] [PubMed] [Google Scholar]
- 64.Goldfarb L G, Petersen R B, Tabaton M, Brown P, LeBlanc A C, Montagna P, Cortelli P, Julien J, Vital C, Pendelbury W W, Haltia M, Wills P R, Hauw J J, McKeever P E, Monari L, Schrank B, Swergold G D, Autilio-Gambetti L, Gajdusek D C, Lugaresi E, Gambetti P. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: disease phenotype determined by a DNA polymorphism. Science. 1992;258:806–808. doi: 10.1126/science.1439789. [DOI] [PubMed] [Google Scholar]
- 65.Gorodinsky A, Harris D A. Glycolipid-anchored proteins in neuroblastoma cells form detergent-resistant complexes without caveolin. J Cell Biol. 1995;129:619–627. doi: 10.1083/jcb.129.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Griffith J S. Self-replication and scrapie. Nature. 1967;215:1043–1044. doi: 10.1038/2151043a0. [DOI] [PubMed] [Google Scholar]
- 67.Hadlow W J. Scrapie and kuru. Lancet. 1959;ii:289–290. [Google Scholar]
- 68.Haraguchi T, Fisher S, Olofsson S, Endo T, Groth D, Tarentino A, Borchelt D R, Teplow D, Hood L, Burlingame A, Lycke E, Kobata A, Prusiner S B. Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch Biochem Biophys. 1989;274:1–13. doi: 10.1016/0003-9861(89)90409-8. [DOI] [PubMed] [Google Scholar]
- 69.Harper J D, Lansbury P T. Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 70.Harris D A, Gorodinsky A, Lehmann S, Moulder K, Shyng S-L. Cell biology of the prion protein. Curr Top Microbiol Immunol. 1996;207:77–93. doi: 10.1007/978-3-642-60983-1_7. [DOI] [PubMed] [Google Scholar]
- 71.Harris D A, Huber M T, van Dijken P, Shyng S-L, Chait B T, Wang R. Processing of a cellular prion protein: identification of N- and C-terminal cleavage sites. Biochemistry. 1993;32:1009–1016. doi: 10.1021/bi00055a003. [DOI] [PubMed] [Google Scholar]
- 72.Harris D A, Lehmann S. Mutant prion proteins acquire PrPSc-like properties in cultured cells: an experimental model of familial prion diseases. In: Iqbal K, Winblad B, Nishimura T, Takeda M, Wisniewski H M, editors. Alzheimer’s disease: biology, diagnosis and therapeutics. Chichester, United Kingdom: John Wiley & Sons; 1997. pp. 631–643. [Google Scholar]
- 73.Harris D A, Lele P, Snider W D. Localization of the mRNA for a chicken prion protein by in situ hybridization. Proc Natl Acad Sci USA. 1993;90:4309–4313. doi: 10.1073/pnas.90.9.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 75.Hay B, Prusiner S B, Lingappa V R. Evidence for a secretory form of the cellular prion protein. Biochemistry. 1987;26:8110–8115. doi: 10.1021/bi00399a014. [DOI] [PubMed] [Google Scholar]
- 76.Hegde R S, Mastrianni J A, Scott M R, Defea K A, Tremblay P, Torchia M, Dearmond S J, Prusiner S B, Lingappa V R. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 77.Hegde R S, Voigt S, Lingappa V R. Regulation of protein topology by trans-acting factors at the endoplasmic reticulum. Mol Cell. 1998;2:85–91. doi: 10.1016/s1097-2765(00)80116-1. [DOI] [PubMed] [Google Scholar]
- 78.Herms J W, Kretzschmar H A, Titz S, Keller B U. Patch-clamp analysis of synaptic transmission to cerebellar Purkinje cells of prion protein knockout mice. Eur J Neurosci. 1995;7:2508–2512. doi: 10.1111/j.1460-9568.1995.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 79.Hope J, Morton L J, Farquhar C F, Multhaup G, Beyreuther K, Kimberlin R H. The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP) EMBO J. 1986;5:2591–2597. doi: 10.1002/j.1460-2075.1986.tb04539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hornshaw M P, McDermott J R, Candy J M. Copper binding to the N-terminal tandem repeat regions of mammalian and avian prion protein. Biochem Biophys Res Commun. 1995;207:621–629. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 81.Horwich A L, Weissman J S. Deadly conformation—protein misfolding in prion disease. Cell. 1997;89:499–510. doi: 10.1016/s0092-8674(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 82.Huang Z, Prusiner S B, Cohen F E. Structures of prion proteins and conformational models for prion diseases. Curr Top Microbiol Immunol. 1996;207:49–67. doi: 10.1007/978-3-642-60983-1_5. [DOI] [PubMed] [Google Scholar]
- 83.Jeffrey M, Goodsir C M, Bruce M E, McBride P A, Scott J R, Halliday W G. Infection specific prion protein (PrP) accumulates on neuronal plasmalemma in scrapie infected mice. Neurosci Lett. 1992;147:106–109. doi: 10.1016/0304-3940(92)90785-6. [DOI] [PubMed] [Google Scholar]
- 84.Kaneko K, Peretz D, Pan K M, Blochberger T C, Wille H, Gabizon R, Griffith O H, Cohen F E, Baldwin M A, Prusiner S B. Prion protein (PrP) synthetic peptides induce cellular PrP to acquire properties of the scrapie isoform. Proc Natl Acad Sci USA. 1995;92:11160–11164. doi: 10.1073/pnas.92.24.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen F E, Prusiner S B. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kascsak R J, Rubenstein R, Merz P, Carp R I, Robakis N K, Wisniewski H M, Diringer H. Immunological comparison of scrapie-associated fibrils isolated from animals infected with four different scrapie strains. J Virol. 1986;59:676–683. doi: 10.1128/jvi.59.3.676-683.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Cell-free formation of protease-resistant prion protein. Nature. 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 89.Kocisko D A, Priola S A, Raymond G J, Chesebro B, Lansbury P T, Jr, Caughey B. Species specificity in the cell-free conversion of prion protein to protease-resistant forms: a model for the scrapie species barrier. Proc Natl Acad Sci USA. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kretzschmar H A, Prusiner S B, Stowring L E, DeArmond S J. Scrapie prion proteins are synthesized in neurons. Am J Pathol. 1986;122:1–5. [PMC free article] [PubMed] [Google Scholar]
- 91.Kristensson K, Feuerstein B, Taraboulos A, Hyun W C, Prusiner S B, DeArmond S J. Scrapie prions alter receptor-mediated calcium responses in cultured cells. Neurology. 1993;43:2335–2341. doi: 10.1212/wnl.43.11.2335. [DOI] [PubMed] [Google Scholar]
- 92.Kushnirov V V, Ter-Avanesyan M D. Structure and replication of yeast prions. Cell. 1998;94:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 93.Lehmann S, Daude N, Harris D A. A wild-type prion protein does not acquire properties of the scrapie isoform when coexpressed with a mutant prion protein in cultured cells. Mol Brain Res. 1997;52:139–145. doi: 10.1016/s0169-328x(97)00231-3. [DOI] [PubMed] [Google Scholar]
- 94.Lehmann S, Harris D A. A mutant prion protein displays an aberrant membrane association when expressed in cultured cells. J Biol Chem. 1995;270:24589–24597. doi: 10.1074/jbc.270.41.24589. [DOI] [PubMed] [Google Scholar]
- 95.Lehmann S, Harris D A. Mutant and infectious prion proteins display common biochemical properties in cultured cells. J Biol Chem. 1996;271:1633–1637. doi: 10.1074/jbc.271.3.1633. [DOI] [PubMed] [Google Scholar]
- 96.Lehmann S, Harris D A. Two mutant prion proteins expressed in cultured cells acquire biochemical properties reminiscent of the scrapie isoform. Proc Natl Acad Sci USA. 1996;93:5610–5614. doi: 10.1073/pnas.93.11.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lehmann S, Harris D A. Blockade of glycosylation promotes acquisition of scrapie-like properties by the prion protein in cultured cells. J Biol Chem. 1997;272:21479–21487. doi: 10.1074/jbc.272.34.21479. [DOI] [PubMed] [Google Scholar]
- 98.Lindquist S. Mad cows meet Psi-chotic yeast: the expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 99.Lisanti M P, Scherer P E, Vidugiriene J, Tang Z, Hermanowski-Vosatka A, Tu Y-H, Cook R F, Sargiacomo M. Characterization of caveolin-rich membrane domains isolated from an endothelial-rich source: implications for human disease. J Cell Biol. 1994;126:111–126. doi: 10.1083/jcb.126.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lledo P M, Tremblay P, Dearmond S J, Prusiner S B, Nicoll R A. Mice deficient for prion protein exhibit normal neuronal excitability and synaptic transmission in the hippocampus. Proc Natl Acad Sci USA. 1996;93:2403–2407. doi: 10.1073/pnas.93.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lopez C D, Yost C S, Prusiner S B, Myers R M, Lingappa V R. Unusual topogenic sequence directs prion protein biogenesis. Science. 1990;248:226–229. doi: 10.1126/science.1970195. [DOI] [PubMed] [Google Scholar]
- 102.Low M G. The glycosyl-phosphatidyl anchor of membrane proteins. Biochim Biophys Acta. 1989;988:427–454. doi: 10.1016/0304-4157(89)90014-2. [DOI] [PubMed] [Google Scholar]
- 103.Manson J, West J D, Thomson V, McBride P, Kaufman M H, Hope J. The prion protein gene: a role in mouse embryogenesis? Development. 1992;115:117–122. doi: 10.1242/dev.115.1.117. [DOI] [PubMed] [Google Scholar]
- 104.Manson J C, Hope J, Clarke A R, Johnston A, Black C, MacLeod N. PrP gene dosage and long term potentiation. Neurodegeneration. 1995;4:113–114. doi: 10.1006/neur.1995.0014. [DOI] [PubMed] [Google Scholar]
- 105.McKinley M P, Meyer R K, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner S B. Scrapie prion rod formation in vitro requires both detergent extraction and limited proteolysis. J Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKinley M P, Taraboulos A, Kenaga L, Serban D, Stieber A, DeArmond S J, Prusiner S B, Gonatas N. Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab Investig. 1991;65:622–630. [PubMed] [Google Scholar]
- 107.Monari L, Chen S G, Brown P, Parchi P, Petersen R B, Mikol J, Gray F, Cortelli P, Montagna P, Ghetti B, Goldfarb L, Gajdusek D C, Lugaresi E, Gambetti P, Autilio-Gambetti L. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: different prion proteins determined by a DNA polymorphism. Proc Natl Acad Sci USA. 1994;91:2839–2842. doi: 10.1073/pnas.91.7.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moser M, Colello R J, Pott U, Oesch B. Developmental expression of the prion protein gene in glial cells. Neuron. 1995;14:509–517. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 109.Muramoto T, Scott M, Cohen F E, Prusiner S B. Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Narwa, R., and D. A. Harris. Unpublished data.
- 111.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem. 1996;272:6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 112.Nishida N, Katamine S, Shigematsu K, Nakatani A, Sakamoto N, Hasegawa S, Nakaoke R, Atarashi R, Kataoka Y, Miyamoto T. Prion protein is necessary for latent learning and long-term memory retention. Cell Mol Neurobiol. 1997;17:537–545. doi: 10.1023/A:1026315006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen S G, Farlow M, Dickson D W, Sima A A F, Trojanowski J Q, Petersen R B, Gambetti P. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 115.Parchi P, Chen S G, Brown P, Zou W, Capellari S, Budka H, Hainfellner J, Reyes P F, Golden G T, Hauw J J, Gajdusek D C, Gambetti P. Different patterns of truncated prion protein fragments correlated with distinct phenotypes in P101L Gerstmann-Sträussler-Scheinker disease. Proc Natl Acad Sci USA. 1998;95:8322–8327. doi: 10.1073/pnas.95.14.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parchi P, Gambetti P. Human prion diseases. Curr Opin Neurol. 1995;8:286–293. doi: 10.1097/00019052-199508000-00007. [DOI] [PubMed] [Google Scholar]
- 117.Pattison I H. The relative susceptibility of sheep, goats and mice to two types of the goat scrapie agent. Res Vet Sci. 1966;7:207–212. [PubMed] [Google Scholar]
- 118.Pauly P C, Harris D A. Copper stimulates endocytosis of the prion protein. J Biol Chem. 1998;273:33107–33110. doi: 10.1074/jbc.273.50.33107. [DOI] [PubMed] [Google Scholar]
- 119.Peretz D, Williamson R A, Matsunaga Y, Serban H, Pinilla C, Bastidas R B, Rozenshteyn R, James T L, Houghten R A, Cohen F E, Prusiner S B, Burton D R. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 120.Petersen R B, Parchi P, Richardson S L, Urig C B, Gambetti P. Effect of the D178N mutation and the codon 129 polymorphism on the metabolism of the prion protein. J Biol Chem. 1996;271:12661–12668. doi: 10.1074/jbc.271.21.12661. [DOI] [PubMed] [Google Scholar]
- 121.Piccardo P, Ghetti B, Dickson D W, Vinters H V, Giaccone G, Bugiani O, Tagliavini F, Young K, Dlouhy S R, Seiler C, Jones C K, Lazzarini A, Golbe L I, Zimmerman T R, Perlman S L, McLachlan D C, Stgeorgehyslop P H, Lennox A. Gerstmann-Sträussler-Scheinker disease (PRNP P102L): amyloid deposits are best recognized by antibodies directed to epitopes in PrP region 90-165. J Neuropathol Exp Neurol. 1995;54:790–801. doi: 10.1097/00005072-199511000-00006. [DOI] [PubMed] [Google Scholar]
- 122.Price D L, Borchelt D R, Sisodia S S. Alzheimer disease and the prion protein disorders: amyloid β-protein and prion protein amyloidoses. Proc Natl Acad Sci USA. 1993;90:6381–6384. doi: 10.1073/pnas.90.14.6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Priola S A, Caughey B. Inhibition of scrapie-associated PrP accumulation: probing the role of glycosaminoglycans in amyloidogenesis. Mol Neurobiol. 1994;8:113–120. doi: 10.1007/BF02780661. [DOI] [PubMed] [Google Scholar]
- 124.Priola S A, Caughey B, Race R E, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Priola S A, Chesebro B. A single hamster PrP amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Priola S A, Chesebro B. Abnormal properties of prion protein with insertional mutations in different cell types. J Biol Chem. 1998;273:11980–11985. doi: 10.1074/jbc.273.19.11980. [DOI] [PubMed] [Google Scholar]
- 127.Prusiner S B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 128.Prusiner S B. Prions. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2901–2950. [Google Scholar]
- 129.Prusiner S B. Prion diseases and the BSE crisis. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 130.Prusiner S B, Hsiao K K. Human prion diseases. Ann Neurol. 1994;35:385–395. doi: 10.1002/ana.410350404. [DOI] [PubMed] [Google Scholar]
- 131.Prusiner S B, Scott M, Foster D, Pan K M, Groth D, Mirenda C, Torchia M, Yang S L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 132.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 133.Race R E, Caughey B, Graham K, Ernst D, Chesebro B. Analyses of frequency of infection, specific infectivity, and prion protein biosynthesis in scrapie-infected neuroblastoma cell clones. J Virol. 1988;62:2845–2849. doi: 10.1128/jvi.62.8.2845-2849.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Race R E, Ernst D. Detection of proteinase K-resistant prion protein and infectivity in mouse spleen by 2 weeks after scrapie agent inoculation. J Gen Virol. 1992;73:3319–3323. doi: 10.1099/0022-1317-73-12-3319. [DOI] [PubMed] [Google Scholar]
- 135.Raymond G J, Hope J, Kocisko D A, Priola S A, Raymond L D, Bossers A, Ironside J, Will R G, Chen S G, Petersen R B, Gambetti P, Rubenstein R, Smits M A, Lansbury P T, Caughey B. Molecular assessment of the potential transmissibilities of BSE and scrapie to humans. Nature. 1997;388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 136.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain PrP(121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 137.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 138.Rogers M, Taraboulos A, Scott M, Groth D, Prusiner S B. Intracellular accumulation of the cellular prion protein after mutagenesis of its Asn-linked glycosylation sites. Glycobiology. 1990;1:101–109. doi: 10.1093/glycob/1.1.101. [DOI] [PubMed] [Google Scholar]
- 139.Rogers M, Yehiely F, Scott M, Prusiner S B. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc Natl Acad Sci USA. 1993;90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rubinstein R, Deng H, Scalici C L, Papini M C. Alterations in neurotransmitter-related enzyme activity in scrapie-infected PC12 cells. J Gen Virol. 1991;72:1279–1285. doi: 10.1099/0022-1317-72-6-1279. [DOI] [PubMed] [Google Scholar]
- 141.Safar J, Ceroni M, Gajdusek D C, Gibbs C J., Jr Differences in the membrane interaction of scrapie amyloid precursor proteins in normal and scrapie- or Creutzfeldt-Jakob disease-infected brains. J Infect Dis. 1991;163:488–494. doi: 10.1093/infdis/163.3.488. [DOI] [PubMed] [Google Scholar]
- 142.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 143.Sailer A, Büeler H, Fischer M, Aguzzi A, Weissmann C. No propagation of prions in mice devoid of PrP. Cell. 1994;77:967–968. doi: 10.1016/0092-8674(94)90436-7. [DOI] [PubMed] [Google Scholar]
- 144.Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, Sugimoto T, Nakatani A, Kataoka Y, Houtani T, Shirabe S, Okada H, Hasegawa S, Miyamoto T, Noda T. Loss of cerebellar Purkinje cells in aged mice homozygous for a disrupted PrP gene. Nature. 1996;380:528–531. doi: 10.1038/380528a0. [DOI] [PubMed] [Google Scholar]
- 145.Salès N, Rodolfo K, Hässig R, Faucheux B, Di Giamberardino L, Moya K L. Cellular prion protein localization in rodent and primate brain. Eur J Neurosci. 1998;10:2464–2471. doi: 10.1046/j.1460-9568.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- 146.Schätzl H M, Laszlo L, Holtzman D M, Tatzelt J, Dearmond S J, Weiner R I, Mobley W C, Prusiner S B. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J Virol. 1997;71:8821–8831. doi: 10.1128/jvi.71.11.8821-8831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schirmer E C, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schmid S L. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 149.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 150.Scott M, Groth D, Foster D, Torchia M, Yang S L, DeArmond S J, Prusiner S B. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 151.Scott M R, Köhler R, Foster D, Prusiner S B. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Scott M R D, Telling G C, Prusiner S B. Transgenetics and gene targeting in studies of prion diseases. Curr Top Microbiol Immunol. 1996;207:95–123. doi: 10.1007/978-3-642-60983-1_8. [DOI] [PubMed] [Google Scholar]
- 153.Selkoe D J. Cell biology of the β-amyloid precursor protein and the genetics of Alzheimer’s disease. Cold Spring Harbor Symp Quant Biol. 1996;61:587–596. [PubMed] [Google Scholar]
- 154.Shyng S L, Heuser J E, Harris D A. A glycolipid-anchored prion protein is endocytosed via clathrin-coated pits. J Cell Biol. 1994;125:1239–1250. doi: 10.1083/jcb.125.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shyng S L, Huber M T, Harris D A. A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J Biol Chem. 1993;268:15922–15928. [PubMed] [Google Scholar]
- 156.Shyng S L, Moulder K L, Lesko A, Harris D A. The N-terminal domain of a glycolipid-anchored prion protein is essential for its endocytosis via clathrin-coated pits. J Biol Chem. 1995;270:14793–14800. doi: 10.1074/jbc.270.24.14793. [DOI] [PubMed] [Google Scholar]
- 157.Shyng S-L, Lehmann S, Moulder K L, Harris D A. Sulfated glycans stimulate endocytosis of the cellular isoform of the prion protein, PrPC, in cultured cells. J Biol Chem. 1995;270:30221–30229. doi: 10.1074/jbc.270.50.30221. [DOI] [PubMed] [Google Scholar]
- 158.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 159.Singh N, Zanusso G, Chen S G, Fujioka H, Richardson S, Gambetti P, Petersen R B. Prion protein aggregation reverted by low temperature in transfected cells carrying a prion protein gene mutation. J Biol Chem. 1997;272:28461–28470. doi: 10.1074/jbc.272.45.28461. [DOI] [PubMed] [Google Scholar]
- 160.Stahl N, Baldwin M A, Burlingame A L, Prusiner S B. Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry. 1990;29:8879–8884. doi: 10.1021/bi00490a001. [DOI] [PubMed] [Google Scholar]
- 161.Stahl N, Baldwin M A, Hecker R, Pan K-M, Burlingame A L, Prusiner S B. Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry. 1992;31:5043–5053. doi: 10.1021/bi00136a600. [DOI] [PubMed] [Google Scholar]
- 162.Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson G W, Burlingame A L, Prusiner S B. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 163.Stahl N, Borchelt D R, Hsiao K, Prusiner S B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–249. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 164.Stahl N, Borchelt D R, Prusiner S B. Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry. 1990;29:5405–5412. doi: 10.1021/bi00474a028. [DOI] [PubMed] [Google Scholar]
- 165.Stahl N, Borchelt D R, Prusiner S B. Glycolipid anchors of the cellular and scrapie prion proteins. In: Turner A J, editor. Molecular and cell biology of membrane proteins: glycolipid anchors of cell-surface proteins. London, United Kingdom: Ellis Horwood; 1990. pp. 189–216. [Google Scholar]
- 166.Tagliavini F, Prelli F, Ghiso J, Bugiani O, Serban D, Prusiner S B, Farlow M R, Ghetti B, Frangione B. Amyloid protein of Gerstmann-Sträussler-Scheinker disease (Indiana kindred) is an 11 kd fragment of prion protein with an N-terminal glycine at codon 58. EMBO J. 1991;10:513–519. doi: 10.1002/j.1460-2075.1991.tb07977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tagliavini F, Prelli F, Porro M, Rossi G, Giaccone G, Farlow M R, Dlouhy S R, Ghetti B, Bugiani O, Frangione B. Amyloid fibrils in Gerstmann-Sträussler-Scheinker disease (Indiana and Swedish kindreds) express only PrP peptides encoded by the mutant allele. Cell. 1994;79:695–703. doi: 10.1016/0092-8674(94)90554-1. [DOI] [PubMed] [Google Scholar]
- 168.Tagliavini F, Prelli F, Porro M, Salmona M, Bugiani O, Frangione B. A soluble form of prion protein in human cerebrospinal fluid: implications for prion-related encephalopathies. Biochem Biophys Res Commun. 1992;184:1398–1404. doi: 10.1016/s0006-291x(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 169.Taraboulos A, Raeber A J, Borchelt D R, Serban D, Prusiner S B. Synthesis and trafficking of prion proteins in cultured cells. Mol Biol Cell. 1992;3:851–863. doi: 10.1091/mbc.3.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Taraboulos A, Rogers M, Borchelt D R, McKinley M P, Scott M, Serban D, Prusiner S B. Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc Natl Acad Sci USA. 1990;87:8262–8266. doi: 10.1073/pnas.87.21.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner S B, Avraham D, Laszlo L, Prusiner S B. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Taraboulos A, Serban D, Prusiner S B. Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J Cell Biol. 1990;110:2117–2132. doi: 10.1083/jcb.110.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tatzelt J, Prusiner S B, Welch W J. Chemical chaperones interfere with the formation of the scrapie prion protein. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- 174.Tatzelt J, Zuo J, Voellmy R, Scott M, Hartl U, Prusiner S B, Welch W J. Scrapie prions selectively modify the stress response in neuroblastoma cells. Proc Natl Acad Sci USA. 1995;92:2944–2948. doi: 10.1073/pnas.92.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Telling G C, Scott M, Mastriani J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 176.Tobler I, Gaus S E, Deboer T, Achermann P, Fischer M, Rulicke T, Moser M, Oesch B, McBride P A, Manson J C. Altered circadian activity rhythms and sleep in mice devoid of prion protein. Nature. 1996;380:639–642. doi: 10.1038/380639a0. [DOI] [PubMed] [Google Scholar]
- 177.Turk E, Teplow D B, Hood L E, Prusiner S B. Purification and properties of the cellular and scrapie hamster prion proteins. Eur J Biochem. 1988;176:21–30. doi: 10.1111/j.1432-1033.1988.tb14246.x. [DOI] [PubMed] [Google Scholar]
- 178.Vey M, Pilkuhn S, Wille H, Nixon R, Dearmond S J, Smart E J, Anderson R G W, Taraboulos A, Prusiner S B. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Weissmann C, Aguzzi A. Bovine spongiform encephalopathy and early onset variant Creutzfeldt-Jakob disease. Curr Opin Neurobiol. 1997;7:695–700. doi: 10.1016/s0959-4388(97)80091-8. [DOI] [PubMed] [Google Scholar]
- 180.Weissmann C, Raeber A J, Shmerling D, Cozzio A, Flechsig E, Aguzzi A. The use of genetically modified mice in prion research. In: Harris D A, editor. Prions: molecular and cellular biology. Wymondham, United Kingdom: Horizon Scientific Press; 1999. pp. 87–106. [Google Scholar]
- 181.Welch W J, Gambetti P. Chaperoning brain diseases. Nature. 1998;392:23–24. doi: 10.1038/32049. [DOI] [PubMed] [Google Scholar]
- 182.Whittington M A, Sidle K C, Gowland I, Meads J, Hill A F, Palmer M S, Jefferys J G, Collinge J. Rescue of neurophysiological phenotype seen in PrP null mice by transgene encoding human prion protein. Nat Genet. 1995;9:197–201. doi: 10.1038/ng0295-197. [DOI] [PubMed] [Google Scholar]
- 183.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 184.Wong K, Qiu Y, Hyun W, Nixon R, Vancleff J, Sanchezsalazar J, Prusiner S B, DeArmond S J. Decreased receptor-mediated calcium response in prion-infected cells correlates with decreased membrane fluidity and IP3 release. Neurology. 1996;47:741–750. doi: 10.1212/wnl.47.3.741. [DOI] [PubMed] [Google Scholar]
- 185.Yost C S, Lopez C D, Prusiner S B, Myers R M, Lingappa V R. Non-hydrophobic extracytoplasmic determinant of stop transfer in the prion protein. Nature. 1990;343:669–672. doi: 10.1038/343669a0. [DOI] [PubMed] [Google Scholar]
- 186.Young K, Piccardo P, Dlouhy S, Bugiani O, Tagliavini F, Ghetti B. The human genetic prion diseases. In: Harris D A, editor. Prions: molecular and cellular biology. Wymondham, United Kingdom: Horizon Scientific Press; 1999. pp. 139–175. [Google Scholar]