Abstract
Transcriptional silencing is a heritable form of gene inactivation that involves the assembly of large regions of DNA into a specialized chromatin structure that inhibits transcription. This phenomenon is responsible for inhibiting transcription at silent mating-type loci, telomeres and rDNA repeats in both budding yeast Saccharomyces cerevisiae and fission yeast Schizosaccharomyces pombe, as well as at centromeres in fission yeast. Although transcriptional silencing in both S.cerevisiae and S.pombe involves modification of chromatin, no apparent amino acid sequence similarities have been reported between the proteins involved in establishment and maintenance of silent chromatin in these two distantly related yeasts. Silencing in S.cerevisiae is mediated by Sir2p-containing complexes, whereas silencing in S.pombe is mediated primarily by Swi6-containing complexes. The Swi6 complexes of S.pombe contain proteins closely related to their counterparts in higher eukaryotes, but have no apparent orthologs in S.cerevisiae. Silencing proteins from both yeasts are also actively involved in other chromosome-related nuclear functions, including DNA repair and the regulation of chromatin structure.
INTRODUCTION
In addition to the binding of gene-specific repressor proteins to DNA, eukaryotes have also evolved other specialized mechanisms to repress transcription. One such mechanism is transcriptional silencing. Transcriptional silencing involves the establishment of a repressive chromatin structure, called silent chromatin, that can persist throughout the cell cycle and that may be inherited by daughter cells following cell division.
Silencing has been observed in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, as well as in higher eukaryotes. In Drosophila, the heterochromatic structure concentrated at centromeric and telomeric regions, variably, but stably represses the expression of nearby genes, a phenomenon termed position-effect variegation (1). A second example is the Polycomb-mediated inactivation of homeotic genes, an event crucial for proper segmentation and differentiation in Drosophila (2). A classic example in mammals is X-chromosome inactivation, a process involving dosage compensation of X-linked genes through the transcriptional silencing of one of the X chromosomes in females (1,2). In both S.cerevisiae and S.pombe, heterochromatin-like silent chromatin is responsible for silencing at silent mating type loci (HM), telomeres and rDNA repeats, as well as S.pombe centromeres (3–5). Interestingly, transcriptional silencing has also been observed at sites adjacent to the P1 plasmid centromere in bacteria (6).
S.cerevisiae and S.pombe represent two attractive models for studying mechanisms of transcriptional silencing (7–19). Over the past few years considerable progress has been made toward the identification of novel factors involved in silencing in both yeasts. Recent discoveries have provided new insight into the role of S.cerevisiae Sir proteins in a broad range of cellular processes including DNA double-strand break (DSB) repair, aging, cell cycle control and meiotic checkpoint control. In this review, I discuss the trans-acting factors that orchestrate silencing in S.cerevisiae and S.pombe, and outline the similarities and differences between the silencing mechanisms of these two yeasts.
TRANSCRIPTIONAL SILENCING IN S.CEREVISIAE
Transcriptional silencing at the HML and HMR loci
Silencing at the HM loci is one of best-studied examples of transcriptional silencing. The two silent mating-type loci, HMR and HML, confer donor information to the MAT locus, and are responsible for the mating-type switch (20). The HM loci are flanked by ∼150 bp cis-acting elements, silencers E and I, both of which are located ∼1 kb from the genes they regulate. The silencers function to initiate assembly of the silent information regulator (Sir) complex. They contain binding sites for the origin replication complex (ORC), repressor and activator protein 1 (Rap1p), and ARS-binding factor (Abf1p) (13,21). Isolated binding sites for any of the silencer binding proteins, termed protosilencers, are unable to act as silencers on their own, but can function to enhance silencing by cooperating with intact, distant silencers (22). HMR-E is the best-characterized silencer. While mutations in any one of the protein-binding sites at the HMR-E locus does little to affect silencing (23,24), a combination of mutations within the binding sites for ORC, Rap1p and Abf1p causes severe defects in HMR silencing (24). The role of silencers is not limited to initiating silencing. Silencer I, for example, also serves as an insulator, separating active and inactive chromatin at the HML locus (25).
Silencing at the HM loci requires the Sir complex, which is composed of Sir1p, Sir2p, Sir3p and Sir4p (26). None of the SIR genes is essential for viability, but deletion of SIR2, SIR3 or SIR4 completely abolishes silencing, whereas disruption of SIR1 partially reduces silencing. The Sir complex is recruited by silencers and their associated proteins. Sir3p and Sir4p are recruited by Rap1p (27–29). Sir4p is also recruited by Sir1p, which binds to ORC (30). Interestingly, the function of Sir1p can be replaced by overexpression of ESC2 (establishment of silent chromatin 2) (31). Esc2p has been proposed to function in the recruitment or stabilization of Sir proteins to silent chromatin (31). Once bound to chromatin, the Sir complex is believed to spread over nucleosomes via interactions between Sir3p and Sir4p with the hypoacetylated N-terminal tails of histones H3 and H4 (32,33).
In addition to the establishment of silent chromatin, the maintenance and stable inheritance of the silenced state are also important for repression of the HM genes (34). Recent studies suggest that silencers are important for the maintenance of silencing during the cell cycle (35,36). In agreement with their function, Sir2p, Sir3p and Sir4p have been shown to be required for the maintenance of silent chromatin (34). Interestingly, a proline to arginine substitution at 898 in Sir3p results in a deficiency in the maintenance of silent chromatin, but not in the establishment of silencing (37). A recent study also suggests that the role of Sir1p, like Sir2p, Sir3p and Sir4p, may not be limited to the assembly of the silent chromatin at the HM loci, but may also play a role in maintaining silent chromatin throughout the cell cycle (38).
Silencing at the HM loci is also preferentially affected by a set of proteins that include Sum1p, Mga2p and Spt2p. Sum1p (suppressor of mar or sir) was initially identified in a screen for suppressors of the mating defects caused by deletion of SIR2 (39). Sum1p is a DNA-binding protein that represses expression of meiotic genes involved in sporulation (40). The SUM1-1 mutation suppresses silencing defects caused by deletion of SIR2, SIR3, SIR4, a specific mutation in RAP1, and mutations in the silencer HMR-E that are important for the binding of Abf1p, Rap1p and Orc1p (41–43). The SUM1-1 mutation also suppresses silencing defects caused by mutations in the N-terminal tails of the histones H3 and H4. How does the SUM1-1 mutation bypass the requirement for Sir proteins? Chromatin immunoprecipitation (Chip) and co-immunoprecipitation assays suggest that Sum1p normally does not associate with the HM loci. A threonine to isoleucine change at 988 in Sum1p appears to increase its affinity for ORC which may then be able to recruit Sum1-1p to the HM loci (44,45). Sum1-1p-mediated silencing requires Hst1p, a homolog of Sir2p, that normally forms a complex with Sum1p (40). It is likely that Hst1p plays a role similar to that of Sir2p in Sum1-1p-mediated silencing.
Mga2p and Spt23p are structurally- and functionally-related transcription factors that influence transcription by regulating chromatin structure. Deletion of MGA2 or SPT23 increases the silencing efficiency in the SUM1-1 strain and suppresses the silencing defects caused by deletion of SIR1 or mutations in the HMR silencer (46). These results suggest that MGA2- or SPT23-dependent activating functions compete with the silencing machinery at the silent HM loci. However, deletion of MGA2 or SPT23 causes further decrease in the silencing efficiency of a strain in which the HML silencer is mutated (46). This complex effect on silencing at HML suggests that the activating or repressing activities of MGA2 and SPT23 are context dependent.
The mechanism by which Sir-mediated heterochromatin represses transcription is not well understood. A recent study suggests that Sir-promoted heterochromatin does not impair the accessibility of RNA polymerase II machinery to DNA, suggesting that it may regulate gene expression by blocking a step downstream of pre-initiation complex recruitment (47).
Transcriptional silencing at the telomeres
Telomeres are protein–DNA complexes formed at the end of chromosomes that are important for chromosome end stability and proper organization of chromosomes within the nucleus (48–50). In S.cerevisiae, telomeric DNA consists of C1–3A/TG1–3 repeats that are ∼300 bp in length at the ends of chromosomes. These repeats are organized into a non-nucleosomal chromatin structure termed the telosome (51). The ends of S.cerevisiae chromosomes also contain a number of subtelomeric repeats including X repeats (52,53). In some organisms, such as S.cerevisiae, S.pombe, Drosophila and humans, marker genes placed near telomeres are repressed in a position-dependent manner, a phenomenon known as telomere position effect (TPE) (54–58). In addition, telomeric silencing is dependent on telomere length (59,60).
Compared with silencing at the HM loci, telomeric silencing is more sensitive to subtle changes in the levels of silencing proteins (61). The telomeres contain multiple Rap1p-binding sites that recruit the Sir complex (62). The interaction of Sir3p with Rap1p is competed by Rap1p-interacting factors 1 and 2 (Rif1p and Rif2p) which act as negative regulators of telomeric silencing and telomeric length (27,29,59,63–65). The inhibitory role of Rif proteins is counteracted by Ku which is also required for telomeric silencing, suggesting that Ku may play a role in the recruitment of Sir proteins (66, and see below). One-hybrid studies have shown that Sir proteins are integral components of the telosome (67). While the association of Sir4p with internal tracts of C1–3A/TG1–3 DNA is Sir3p independent, the Sir2p association is Sir3p dependent. The association of Sir proteins with internal tracts of C1–3A/TG1–3 DNA suggests that the Sir proteins may initially be recruited to the internal tracts of telomeric DNA (67). Once assembled on telomeres, Sir proteins propagate over the nucleosome to form a silent chromatin structure at the telomeres. The composition of the silent chromatin structure at telomeres is not uniform. The ‘core’ region contains all three Sir proteins whereas the ‘surrounding’ regions contain mainly Sir3p (60,68).
Orc1p, Abf1p and Sir1p are not required for the assembly of Sir proteins on the telomeres (34,61). However, the binding sites for these proteins have been identified in the core X element of subtelomeric X repeats. Moreover, these sites are important for silencing (69,70). The core X element acts as protosilencer that enhances silent chromatin propagation away from telomeres (70). These subtelomeric silencing elements are separated from the telomeric repeats by subtelomeric anti-silencing regions (STARs) that create a discontinuity in the propagation of silent chromatin (70).
Telomeres are localized to the nuclear periphery. The myosin-like nuclear pore complex proteins, Mlp1p and Mlp2p, have been proposed to tether telomeres to the nuclear periphery via their interactions with the chromosome end-binding heterodimeric protein, Ku, and the nuclear pore protein, Nyp145p (71). The proper maintenance of nuclear localization and structure of telomeres appears to be important for telomeric silencing. First, the HMR locus with a defective silencer can be silenced if tethered to the nuclear periphery, indicating that perinuclear localization aids in the establishment of silent chromatin (72). Secondly, deletion of YKU70/HDF1 or YKU80/HDF2, the genes encoding Ku, disrupts the perinuclear location of telomeres and telomeric silencing (73,74). Thirdly, deletion of MLP1 or MLP2 also disrupts the organization of silent telomeric chromatin and causes loss of telomeric silencing (71). Finally, the perinuclear localization of telomeres also appears to provide a storage site for Sir3p and Sir4p. Immunostaining studies reveal that Sir3p, Sir4p and Rap1p co-localize with subtelomeric DNA in foci near the nuclear periphery (75,76). Moreover, mutations disrupting telomeric silencing also disrupt the association of Sir proteins with subtelomeric DNA (28,75). Although perinuclear localization of telomeres favors silencing, the localization of telomeres to the nuclear periphery is not sufficient to cause telomeric silencing (77,78).
Silencing proteins are also involved in telomeric functions. Deletion of SIR3 or SIR4 causes shortening of telomeric repeats and mitotic instability of chromosomes (79). This finding, and the fact that there is no known regulatory function for telomeric silencing, suggest that the primary role of silencing proteins at telomeres is to maintain the stability and integrity of the chromosome ends, and that telomeric silencing is a secondary effect.
Transcriptional silencing at rDNA repeats
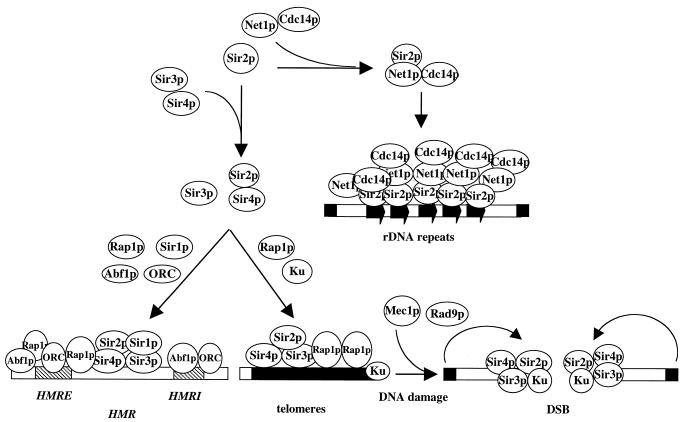
Saccharomyces cerevisiae ribosomal DNA (rDNA) is organized in tandem arrays of approximately 150 repeats that are separated by intergenic spacers (IGS). The rDNA repeats form a unique structure called the nucleolus (80) and are transcribed by RNA polymerase (Pol) I (81). Since rDNA is actively transcribed it was unexpected that Pol II genes would be repressed when inserted at this locus. This repression likely results as a consequence of the suppression of mitotic and meiotic recombination within rDNA repeats since silencing at rDNA repeats and suppression of recombination among rDNA repeats is closely linked and Sir2p dependent (82–84). Unlike silencing at the HM loci and telomeres, rDNA silencing requires a silencing complex termed RENT (regulator of nucleolar silencing and telophase exit). This complex consists of at least three components: Sir2p, Net1p (nucleolar silencing establishing factor and telophase regulator 1) and Cdc14p (85,86). Sir1p, Sir3p and Sir4p are not required for rDNA silencing and are not part of the RENT complex (83,85). However, Sir4p regulates the efficiency of rDNA silencing by modulating the distribution of a limited amount of Sir2p between telomeres and rDNA repeats (87). Normally, most Sir2p localizes to nucleolus, although a small fraction also localizes to telomeres (76).
Net1p is essential for rDNA silencing, and is responsible for recruiting Sir2p to the nucleolus (85). Net1p also interacts with Pol I and stimulates Pol I transcription presumably by tethering Pol I within the nucleolus (88). Net1p also regulates nucleolar structure. In addition to nucleolar functions, Net1p functions to regulate the cell cycle by sequestering Cdc14p (a protein phosphatase) in the nucleolus until anaphase (86,89). Once released, Cdc14p dephosphorylates nuclear and cytoplasmatic substrates that regulate the activity of the anaphase-promoting components, thus destroying the anaphase-promoting components (90,91).
The cis-acting elements responsible for silencing at rDNA repeats are not well understood. Sir2p is believed to be recruited to rDNA repeats by Net1p (85). How Net1p itself is recruited to rDNA repeats is unknown since no apparent DNA-binding motif has been identified in Net1p.
Competition among three silent loci
The local concentration of Sir proteins as well as the stoichiometry of Sir proteins in the Sir complex are important determinants in silencing efficiency due to competition among different silent loci for limited Sir proteins (92–97) (Fig. 1). Indeed, Sir3p is limiting at telomeres since overexpression of SIR3 extends telomeric silencing from 4 to 20 kb from telomeric repeats (60,68). Moreover, a rap1-12 mutation specifically reduces silencing at the HMR locus, but improves telomeric silencing, supporting the hypothesis that Rap1p plays a regulatory role in sequestering a limited amount of Sir proteins at telomeres (29,97).
Figure 1.
The assembly of Sir proteins at HMR (chromosome III), telomeres, rDNA repeats (chromosome XII) and a DNA DSB. Sir3p is loosely associated with Sir2p and Sir4p (156). Sir proteins are recruited by Rap1p, Abf1p and ORC to HMR, whereas Sir proteins are recruited by Ku and Rap1p to telomeres. Sir2p is recruited to rDNA repeats by Net1p. The hatched box represents silencer. The filled box represents the chromosome ends. The arrow represents rDNA repeats.
Sir2p is believed to be a limiting factor for rDNA silencing (87). Overexpression of SIR2 or disruption of the SIR3 and SIR4 genes enhances rDNA silencing, suggesting that telomeres and rDNA repeats compete for a limited amount of Sir2p (83). Similarly, overexpression of SIR4 enhances telomeric silencing, while decreasing rDNA silencing (87). This is likely due to the release of Sir2p from rDNA by overexpression of SIR4. In contrast, overexpression of the N-terminal 214 amino acids of Sir3p (Sir3N) improves silencing at telomeres, while relieving rDNA silencing (98). These effects on silencing coincide with displacement of a portion of Sir2p from the nucleolus (98). Moreover, expression of a C-terminal truncated Sir4p (Sir4C) strengthens silencing at rDNA repeats, but interferes with silencing at the HM loci and telomeres by titrating the HM and telomere Sir protein recruiting factors (87,99). Consistent with this notion, Sir3p and Sir4p are relocalized to nucleolus when Sir4C is overproduced (100). The relocalization of Sir3p to nucleolus appears to be mediated, at least in part, by Sir2p and two other uncharacterized proteins, Uth4p and Yh1023p (76). In contrast, relocalization of Sir proteins from telomeres to the HM loci seems to be mediated by Sir1p, since increasing Sir1p decreases telomeric silencing (97).
Chromatin-modulating factors and silencing histones
Histones that mediate the folding of DNA to form nucleosomes have direct roles in silencing. As noted above, the N-terminal regions of histones H3 and H4 that interact with Sir proteins are important for silencing at HM and telomeres (32,101–105). Moreover, histones H2A and H2B are important for rDNA silencing (106,107). For example, reducing histone H2A-H2B levels by deletion of histone genes HTA1-HTB1 perturbs local chromatin structure and abolishes silencing at rDNA repeats (106). Interestingly, Htz1p, a variant of histone H2A, is also required for efficient silencing at the HM loci and telomeres (31). Overexpression of HTZ1 increased silencing at the HMR locus, suggesting that silent chromatin at the HMR locus favors Htz1p. Indeed, Htz1p was present at the HMR locus (31).
Chromatin assembly factors
Yeast chromatin assembly factor I, yCAF-I, is one of the proteins that mediates assembly of histones onto the 146 bp of DNA that form a nucleosome (108). yCAF-I is a three-subunit complex (Cac1-3p) encoded by three genes, CAC1, CAC2 and CAC3. Deletion of any of these three genes reduces silencing at HM, telomeres and rDNA repeats because the silent chromatin state cannot be maintained (109,110). The contribution of yCAF-I to silencing can be altered by mutations in proliferating cell nuclear antigen (PCNA) (111). PCNA is believed to recruit yCAF-I to DNA by direct interaction (112). yCAF-I also plays a role in coupling chromatin assembly to DNA replication and repair by preferentially assembling nucleosomes onto newly replicated or repaired DNA (109,110,113,114).
Another yeast chromatin assembly factor, Asf1p (anti-silencer factor 1), was identified as a high-copy disruptor of telomeric silencing (115,116). Unlike the cacΔ mutation, the asf1Δ mutation displays only minor defects in silencing (115,116). However, the asf1Δ mutation exacerbates the silencing defect associated with the cacΔ mutation (111,117). Moreover, combination of the asf1Δ mutation with a mutation in the DNA polymerase processivity factor PCNA (pol30-8) prevents yCAF-I from contributing to telomeric silencing, and eliminates residual telomeric silencing (111). These genetic data suggest that Asf1p and yCAF-I have distinct, yet overlapping functions in the formation of silent chromatin. Importantly, a Drosophila homolog of Asf1p is present in the RCAF complex (replication-coupling assembly factor) that has been proposed to mediate chromatin assembly after DNA replication and repair (117). It would be interesting to know if Asf1p, like yCAF-I, also couples chromatin assembly to DNA replication and repair.
Histone regulators (Hir proteins) that control histone gene expression are also involved in silencing. Although deletion of HIR genes have no observable silencing defects at the HM loci and telomeres in wild-type or asf1Δ cells, they do exacerbate silencing at HM and telomeres in cacΔ cells (105,111). Moreover, Asf1p interacts with Hir proteins in vitro and in cellular extract (111). These biochemical and genetic studies suggest that Asp1p and Hir proteins function in silencing through the same genetic pathway. The contribution of Asf1p, Hir proteins and yCAF-I in silencing can be abolished by mutations in PCNA, suggesting that PCNA is involved in the silencing mediated through these silencing proteins (111).
SET domain proteins, which take their name from their founding members Su(v)ar 3-9, E(z) and trithorax, are involved in chromatin-mediated transcriptional regulation (118). SET domain proteins have been identified in S.cerevisiae and S.pombe. The S.cerevisiae SET domain protein, Set1p, is a regulator of chromatin structure, DNA repair and telomeric functions. Deletion of SET1 alleviates telomeric silencing (118), and increases the repair capacity of cells after DNA damage by, at least in part, alleviating the repression of repair genes (119). Interestingly, the silencing defect caused by the SET1 deletion can be specifically suppressed by the overexpression of genes encoding human (EZH2) or mouse (Ezh1) SET domain-containing proteins (120). In addition, the silencing defects can also partially be suppressed by disruption of the checkpoint gene MEC3, suggesting that Set1p and Mec3p may regulate telomeric silencing in opposite ways (121). Recently, it has been shown that Set1p complex contains a homolog of trithorax protein and specifically methylates histone 3 lysine4 (122).
Histone acetylation and deacetylation
It is well known that acetylation and deacetylation of core histones plays a role in regulation of gene transcription (123–125). The hyperacetylation of histones activates gene expression, whereas hypoacetylation represses gene expression. Consistent with this model, the transcriptionally silenced regions of genomes are generally hypoacetylated (33). However, in contradiction to this model, deletion of two S.cerevisiae histone deacetylase (HDAC) genes, hda1 and rpd3, increases repression at HM (126), the telomeric loci (127) and rDNA repeats (107). One possible interpretation is that the hda1Δ or the rpd3Δ mutations decreases specific nucleosome acetylation in heterochromatin required for its effects on transcription. For example, acetylation of lysine 12 of histone H4 is important for silencing at HM (128) and the rpd3Δ mutation decreases acetylation of histones H3 and H4 (127). Alternatively, the HDACs may modulate silencing efficiency by regulating the activity of silencing complexes. Rpd3p is a catalytic subunit of the HDAC complex that includes Sin3p, Sap30p, Sds3p and Pho23p (127,129–134). Mutations in Sin3p, Sds3p and Pho23p also enhance silencing at HM, telomeres and rDNA repeats.
Histone acetyltransferase (HAT) has also been shown to regulate transcriptional silencing, although silenced regions appear to differ in their sensitivity to acetylation status. For instance, two S.cerevisiae HAT genes, SAS2 and SAS3 (something about silencing), behave as both positive and negative regulators of transcriptional silencing at HM (135,136). SAS2 or SAS3 deletion suppresses silencing defects at the HMR locus. The effect of suppressing silencing at the HMR locus caused by deleting SAS2 or SAS3 may be an indirect consequence of disrupting telomeric silencing. In contrast, deleting SAS2 or SAS3 enhances silencing defects at the HML locus (135,136). The differential regulation of the HML and HMR loci suggests that silencing at the HML and HMR loci may be different and that the repression or activation functions of SAS2 and SAS3 are context dependent. Both Sas2p and Sas3p belong to the MYST family of proteins that includes HATs Esa1p (137) and Tip60 (138). Members of the MYST family also share homology with members of the GNAT superfamily including Gcn5p (139). Whereas recombinant Sas2p does not show HAT activity in vitro, Sas3p has been shown to have HAT activity (140). Sas3p is the catalytic subunit of a HAT complex and interacts with the Spt16p subunit of FACT (facilitates chromatin transcription), a complex implicated in the regulation of transcription and DNA replication (141).
It appears that Sas4p and Sas5p are functionally related to Sas2p and Sas3p. Like Sas2p and Sas3p, Sas4p and Sas5p are negative regulators of silencing at the HMR locus (142), but positive regulators of silencing at the HML locus and telomeres (143). It has been proposed that Sas4p and Sas5p may be components of the Sas2p-dependent acetylase complex or are targets of Sas2p-dependent acetylase (143). Sas4p is a novel protein, whereas Sas5p is a member of protein family that includes the uncharacterized S.pombe open reading frame YD67 and two human proteins encoded by the AF-9 and ENL genes (142). AF-9 and ENL are fusion partners for the human trithorax homolog MLL and are known to contribute to leukemogenesis (144). Both AF-9 and ENL have been shown to interact with hPc3, a member of the human Polycomb protein family that functions as a transcriptional repressor (145).
Role of Sir2p and its homologs in silencing
Sir2p is the only Sir protein that is required for silencing at all silent loci in S.cerevisiae. Consistent with its central role in silencing, Sir2p is also the only Sir protein that is evolutionarily conserved in organisms from bacteria to humans (146,147). The Sir2p family is characterized by a highly conserved approximately 200 amino acid core sequence that is required for silencing (146). The human core sequence can readily be substituted for the S.cerevisiae core sequence (148).
Sir2p and several Sir2-like proteins are nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylases, as well as weak NAD-dependent ADP-ribosyltransferases (149,150). The HDAC activity of these proteins is unique: it is NAD dependent, and not inhibited by trichostatin A (TSA), which inhibits the deacetylase activity of class I and class II HDACs (151,152).
The HDAC activity of Sir2p is absolutely required for silencing (33,151,153,154). Moreover, overexpression of SIR2 reduces histone acetylation (33). Surprisingly, deletion of SIR2 did not change the overall levels of histone acetylation, suggesting that the in vivo target of Sir2p deacetylation may not be limited to histones (154). This is supported by the finding that TAFI68, a component of human RNA polymerase I transcription factor, is deacetylated by murine mSir2A, and that this deacetylation impairs rDNA transcription in vitro (155).
Co-immunoprecipitation experiments have revealed that Sir4p associates with Sir2p tightly, but does not associate with Sir3p due to a regulatory domain in Sir4p that inhibits its interaction with Sir3p (26,62). This is consistent with recent biochemical characterization of Sir2p which showed that Sir2p exists in two complexes (156). One contains Sir4p, and has an NAD-dependent HDAC activity, whereas the other contains Net1p, and has a HDAC activity largely independent of NAD. It is also interesting that Sir3p is absent from both complexes.
Four Sir2p homologs, Hst1p, Hst2p, Hst3p and Hst4p, exist in S.cerevisiae. Among them, Hst1p and Hst2p are the two most well characterized. While deletion of HST1 does not affect silencing at the HM loci, telomeres or rDNA repeats, overexpression of HST1 partially suppresses the silencing defects at HMR caused by deletion of SIR2 (146,157). This suggests that Hst1p may also regulate chromatin structure (146,157). Indeed, Hst1p is important for Sum1p-mediated silencing in the absence of Sir proteins.
Another Sir2p homolog, Hst2p, is a major HDAC (153,154). While not required for the silencing at the HM loci, telomeres or rDNA repeats, overexpression of HST2 increases silencing at rDNA repeats, but decreases silencing at telomeres (146,157). Unlike Hst1p that is nuclear, Hst2p is cytoplasmic (158). These results led to a model which suggests that Hst2p and Sir2p might share a common ligand that is required for telomeric silencing (158). Overexpression of HST2 may sequester this ligand, thus releasing Sir2p, which can then relocalize to the nucleolus and enhance rDNA silencing (158). These studies suggest that in vivo, Hst1p, Hst2p and Hst4p may have roles distinct from Sir2p.
Role of Sir proteins in other cellular processes
Sir proteins and aging. In addition to their silencing functions, recent studies have led to the proposal that Sir proteins may be determinants of yeast life span and that Sir2p may serve to link silencing, metabolism and aging (159–161). The intracellular level of Sir2p correlates with cellular longevity, suggesting that Sir2p may be a limiting component of the molecular machinery involved in regulating cellular longevity (162). In addition, Sir3p and Sir4p are relocalized from telomeres to nucleolus in aging cells, and this relocalization, in the case of Sir3p, correlates with increased longevity (100). Sir3p and Sir4p are also relocalized to the nucleolus in a SIR4-42 mutant that displays increased longevity (99). These results suggest that increased levels of Sir proteins at the nucleolus may be crucial for longevity in yeast.
Sir2p has also been proposed to regulate the longevity of yeast cells by suppressing extrachromosomal rDNA circle (ERC) formation, as well as by a mechanism independent of ERC formation. The accumulation of ERCs in mother cells has been shown to be a cause of aging in S.cerevisiae (163). Sir2p suppresses the ERCs formation by two mechanisms. First, Sir2p directly suppresses homologous recombination at rDNA repeats (82). Shortened longevity caused by SIR2 deletion could be suppressed by FOB1 deletion that promotes rDNA recombination, suggesting that FOB1 and SIR2 may act in the same pathway (162). Secondly, Sir2p acts with Sir3p and Sir4p to repress transcription at the HM loci (162). As noted above, loss of silencing at the HM loci results in co-expression of the a and α genes, which increases recombination between rDNA repeats and as a consequence ERC formation (162). Calorie restriction (CR) also extends longevity in S.cerevisiae by reducing rDNA recombination and ERC formation (164,165). Interestingly, this effect requires Sir2p and Npt1p, an enzyme involved in the synthesis of NAD (164). It is likely that the elevated levels of NAD under CR increases the NAD-dependent HDAC activity of Sir2p and ultimately reduces ERC formation.
Sir proteins and DNA repair
Sir proteins have been implicated in Ku-dependent DNA DSB repair by non-homologous end joining (NHEJ) (73,166). Sir proteins and Ku form a complex (Ku/Sir complex) that associates with subtelomeric regions (167). The formation of this complex is likely mediated by interactions between Ku and Sir4p (166). Upon DNA breakage, the Ku/Sir complex is mobilized from a telomeric reservoir to the break sites and functions to facilitate DSB repair (167–169). This mobilization correlates with a loss in telomeric silencing (168). The redistribution of the Ku/Sir complex from the subtelomeric region to the sites of DSB requires the passage of damaged cells from the G1 to S phase of the cell cycle, and is dependent on the MEC1/RAD9 checkpoint pathway (167,169). However, the rad9Δ mutation does not affect telomeric silencing (170). These observations led to a suggestion that, in addition to participating in silencing of telomeric chromatin, Sir proteins may also promote NHEJ of DSB repair by creating a silent chromatin structure at the sites of DSBs (167–169). However, a direct role for Sir proteins in NHEJ has been disputed (171,172). The NHEJ defect caused by deletion of SIR may be a consequence of derepression of the HM loci which leads to co-expression of a and α gene in haploid cells, thus creating a pseudo-diploid state. In pseudo-diploids, homologous recombination of DNA damage is elevated, whereas NHEJ is reduced (173–175). Indeed, the absence of mating-type heterozygosity suppresses the defect in NHEJ caused by the deletion of SIR (171,172).
Role of Sir proteins in meiotic checkpoint control. As in the mitotic cell cycle, a meiotic checkpoint, referred to as the pachytene checkpoint, monitors recombination and synaptonemal complex (SC) formation during meiosis to ensure proper meiotic chromosome segregation (176). Zip1p, a component of the central region of the SC, is one of the pachytene checkpoint regulators. zip1 mutants are defective in the pachytene checkpoint and have reduced sporulation frequency and spore viability because of chromosome missegregation (177,178). Pch2p and Sir2p are also regulators of pachytene checkpoint function. However, unlike Zip1p, Pch2p and Sir2p are dispensable for normal meiosis. They are, however, required for checkpoint-induced pachytene arrest. Both Pch2p and Sir2p localize predominantly to nucleolus. The nucleolar localization of Pch2p is important for its checkpoint function and is Sir2p dependent (178). In contrast, the nucleolar localization of Sir2p is dependent on the nuclear protein Dot1p, which is involved in pachytene checkpoint control and silencing at the HM loci and telomeres (116). Pch2p can be relocalized to telomeres by overexpressing SIR4 or deleting rDNA repeats (178). Only when associated with telomeric heterochromatin is the mislocalized Pch2p able to provide some checkpoint function, suggesting that the heterochromatin established at rDNA repeats is important for Pch2p checkpoint function. Alternatively, Pch2p may sequester within the nucleolus an unknown protein that is required for the pachytene checkpoint function. Interestingly, Pch2p and Sir2p also repress meiotic recombination by excluding from the nucleolus meiosis-specific Hop1p that promotes meiotic recombination (178).
It is worth noting that Dot1p shows sequence similarity with S-adenosyl-l-methionine (SAM) methyltransferase and comparative modeling suggests that Dop1p contains a methyltransferase fold found in SAM methyltransferease (179). Based on these observations Dot1p has been proposed to be a histone methyltransferase (179).
TRANSCRIPTIONAL SILENCING IN S.POMBE
Transcriptional silencing at the silent mating-type loci
The mating-type region of S.pombe consists of three mating-type loci: mat1, mat2-P and mat3-M. The mat2-P locus is separated from the mat1 locus by a 15 kb interval designated the L region. The mat3-M locus is separated from the mat2-P locus by an 11 kb interval designated the K region (180). The mating-type of S.pombe is determined by the presence of either Plus (P) or Minus (M) information at the mat1 locus. mat2-P and mat3-M are donors of P or M information to the mat1 locus during mating-type switching (12). These donor loci are normally maintained in a transcriptionally silent state (181). The region subject to silencing at the mating-type region of S.pombe is much bigger than that of S.cerevisiae. It extends throughout a large 15 kb chromosomal domain that covers mat2-P, mat3-M, a ∼3 kb region at the mat2-P-proximal side of the L region, and the entire K region (182,183). In addition to being subjected to transcriptional suppression, mat2-P, mat3-M and the K region are refractory to meiotic recombination (184,185). The K region also controls the mating-type switching directionality, a non-random selection of switching information (186,187).
Three cis-acting elements responsible for silencing at the silent mating-type loci have been identified and characterized. One, designated REII, is located at the junction between mat2-P and the L region (188). Another element, termed the mat3-M element, lies within 500 bp of mat3-M (189). These two cis-acting elements were proposed to function together with the products of the esp1, esp2 and esp3 genes to suppress silencing at the donor loci (187). The third cis-acting element is located in the K region and bears strong homology to the dg and dh sequences of centromeric repeats (182). Deletion of the K region, REII or the Mat3-M element alone fully derepresses silencing at the donor loci. However, the combination of the REII element or the mat3-M proximal element deletion with mutations in some trans-acting factors (e.g. Swi6, Clr1, Clr2, Clr3 and Clr4) can cause pronounced effects on silencing at the donor loci. This suggests that the REII and mat3-M elements act in synergy with these trans-acting factors to suppress donor loci expression (183,187,189,190). In contrast to the K region, which has a global effect on silencing, the REII and mat3-M elements appear to control local expression around the donor loci (189,191). The REII element is a protosilencer. It is unable to promote silencing on its own, but can cooperate with centromeric-like sequences in the K region to enhance silencing stability (191). Interestingly, the REII element also serves as boundary elements of silent chromatin at the donor loci (191). In addition to the REII element, two 2 kb identical inverted repeats, termed IR-L and IR-R, may also serve as boundary elements of the silent chromatin at the donor loci (192).
Mutations in several trans-acting genes including clr1, clr2, clr4, clr6, swi6 and rik1, have been found to cause partial derepression at mat2-P and mat3-M (12,182,183,186,187,190,193–197). In addition to repressing transcription, these factors also inhibit meiotic recombination (12,182,183,186,187,190,193–197). Genetic data have shown that these genes act in the same pathway (183). Clr6 and Clr3 (cryptic loci regulator) are HDACs (196). Clr6 belongs to class I deacetylases and specifically deacetylates H3 at lysine 9 (198). Clr3 is a member of the class II HDAC family and specifically deacetylates histone H3 at lysine 14 (198). The deacetylase activity of Clr3 is critical to its silencing function (199).
Clr4 and Swi6 are chromo-domain proteins. Clr4 also contains a SET domain and is a member of the SU(VAR)3-9 protein family which includes human SUV39H1, murine Suv39h1 and Drosophila SU(VAR)3-9 (118,200–202). Members of this protein family have been identified as histone H3 methyltransferases that specifically methylate histone H3 at lysine 9 (199,203). These modifications create a binding site for heterochromatin-associated proteins like HP1 (204,205). The SET domain is required for methyltransferase activity (199). Similarly, Clr4 controls silent chromatin assembly at mat2-P, mat3-M and centromeres by specifically methylating H3 at lysine 9 (202). In addition to repressing gene repression, Clr4 also activates a number of genes including cdl3, a gene encoding a homolog of the Escherichia coli thermosensible glucokinase protein (202). Clr4 has also been implicated to function in switching directionality (202).
The chromo-domain protein Swi6 also contains chromo-shadow domains (206,207) and shares a high degree of homology with chromatin-associated proteins such as Drosophila HP1 and Polycomb proteins that function in assembly of transcriptionally inactive chromatin (207,208). Swi6 is the structural component of silent chromatin at mat2-P, mat3-M, centromeres and telomeres, and its localization to silent chromatin requires Clr4 (193,209). Swi6 is also critical to the maintenance and stable inheritance of silencing at mat2-P and mat3-M during mitosis and meiosis (193,209). In addition to functioning in silencing, Swi6 also regulates the efficiency of mating-type switching (210).
Rik1 contains β-propeller-like domains typically found within WD-40 domain proteins that participate in protein–protein interactions (211). A mutation in Rik1 completely abolishes methylation of H3 at lysine 9 and localization of Swi6 at both the silent mating-type region and centromeres (198). The WD-40 domain proteins have been suggested to function as chaperones that bring chromatin assembly factors, chromatin-remodeling factors, HATs and HDACs to histones (212). Thus, Rik1 may bring histone-modifying enzymes, such as Clr4, and silent chromatin-binding proteins, such as Swi6, to silent chromatin.
Another chromo-domain protein, Chp2, identified by the S.pombe sequencing project, also functions at silent mating-type loci (213,214). Chp2 is highly homologous to Swi6. Similarly to Swi6, Chp2 also participates in silencing at many silent loci (214). However, unlike Swi6, Chp2 is not absolutely required for efficient mating-type switching (214).
In addition to the previously described HDACs Clr3 and Clr6, S.pombe contains a third putative HDAC gene, hda1+ (also termed phd1+ or hoc1+) (215). Hda1 is 52% identical to S.cerevisiae Rpd3p and 58% identical to human HDAC1. Biochemical studies have revealed that although bacterially expressed Hda1 shows no HDAC activity, affinity-purified Hda1 does (216). Moreover, the deacetylase activity of the affinity-purified Hda1 complex is inhibited by TSA, a HDAC inhibitor, albeit at a relatively high concentration (IC50 value of 43 nM) (216). Since the Hda1 complex is not well characterized it is unclear if Hda1 or its associated proteins are responsible for HDAC activity. Similar to the effects observed from mutations in the S.cerevisiae Rpd3p and the Drosophila homolog of Rpd3p, removal of Hda1 enhances silencing at silent mating-type loci, centromeres and telomeres (215). Moreover, deletion of the hda1 leads to partial inhibition of cell growth at low concentrations of TSA, suggesting that Hda1 is one possible target for inhibition by TSA in vivo (215). In addition to being involved in silencing, Hda1 may also be involved in the early stages of meiotic cell cycle control by regulating the expression of a set of genes whose products function in the meiotic cell cycle (215,216).
Transcriptional silencing at centromeres
Unlike the centromeres of S.cerevisiae chromosomes, which are relatively small (∼125 bp), the centromeres in S.pombe are large (40–100 kb) and contain a 15 kb central region composed of a 4–7 kb central core domain (cnt) flanked by inverted repeats (imr). This central region is surrounded by 10–100 kb of repeated sequences (otr) containing highly repeated motifs (dg and dh) common to all centromeres (217). This arrangement resembles the arrangement at the centromeres of higher eukaryotes which also contain large arrays of repetitive satellite sequence (4,5). The centromeres of S.pombe, like those of human chromosomes, show characteristics of heterochromatin where the central region is weakly silenced and the outer flanking repeats are more strongly silenced. Similar to the situation at the silent mating-type loci, silencing and suppression of recombination at centromeres are also tightly linked (4,5,184,185).
The trans-acting factors Swi6, Clr1, Clr2, Clr3, Clr4, Rik1, Chp2 and Hda1, which mediate silencing at mat2-P and mat3-M, are also required for silencing at centromeres (5,196,214,215). In particular, Clr4, Rik1 and Swi6 are required for silencing within the outer repeats of centromeres, but not within the central core region. Silencing at centromeres also involves additional centromere-specific factors. The centromere-specific factor Chp1 is a chromo-domain protein identified by the S.pombe sequencing project (213,214). Chp1 is responsible for silencing at the outer repeats of centromeres (218). Like Swi6, Chp1 is localized to the flanking outer repeats, an association that requires Rik1 and Clr4 (218). An additional 12 csp (centromere: suppressor of position effect) genes have been identified genetically. Mutations in these genes have been shown to alleviate silencing in the outer repeats of centromeres, but not at the mating-type region (219). In contrast to the previously described factors, two other factors, Mis6 and Mis12, are required for silencing within the central core and associate with the central region of centromeres. It is unclear how Mis6 and Mis12 are assembled on centromeres (218). Silencing at centromeres may also be regulated by components of the proteasome 19S, suggesting that some components of centromeric silent chromatin may be regulated by proteolytic degradation (220).
The importance of trans-acting factors for silencing generally correlates with their importance for centromere function. First, mutations in Clr4, Clr6, Rik1 and Swi6 result in increased chromosome loss (5,194,196). In addition, many of these mutants show sensitivity to microtubule-destabilizing drugs, suggesting that these silencing proteins interact with microtubules at the kinetochore, a protein–DNA complex at centromeres that is critical for proper chromosome segregation (194). Secondly, Chp1 shows a genetic interaction with α-tubulin and is required for proper chromosome segregation (213). Finally, consistent with defective centromere structure and function, csp mutants display sensitivity to spindle-destabilizing drugs and elevated chromosome loss rates (219). These results suggest that heterochromatin at centromeres may be crucial to centromere function in S.pombe.
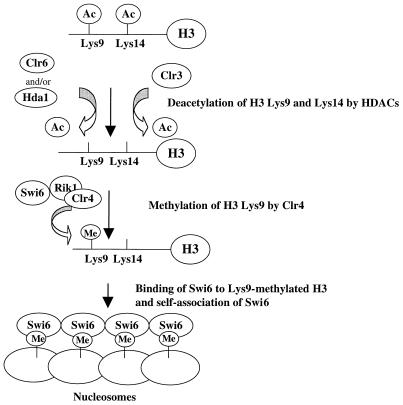
Compared with S.cerevisiae, little is known about the assembly of silent chromatin at mat2-P, mat3-M and centromeres in S.pombe. Recent biochemical studies suggested that histone deacetylases and methylases act in a sequential process (Fig. 2) (198). First, Clr3 and Clr6 deacetylate histone H3 at lysine 14 and lysine 9, respectively. This deacetylation is then followed by methylation of histone H3 at lysine 9 by Clr4 which establishes a ‘histone code’ that can be recognized by Swi6 (221,222). The binding of Swi6 to lysine 9-methylated histone H3 and self-association of Swi6 along nucleosomes result in the establishment of a heterochamatin-like structure at silent loci in S.pombe (198).
Figure 2.
Model for the assembly of silent chromatin in S.pombe. This model is adapted from Nakayama et al. (198). Ac represents the acetyl group on histones H3 lysine 9 and lysine 14; Me represents the methyl group on histone H3 lysine 9. Genetic data suggest that Swi6, Rik1 and Clr4 likely form a complex. A physical association between these proteins has yet to be demonstrated.
Transcriptional silencing at telomeres and rDNA repeats
Silencing has also been observed at telomeres and rDNA repeats in S.pombe, both of which are characterized by repeated sequences (56,214). S.pombe rDNA is organized in tandem arrays of approximately 70–100 repeats in two large domains (500–1000 kb long) (223). These rDNA repeats are located at both ends of chromosome 3, adjacent to the telomeres (223,224). Similarly, S.pombe telomeres consist of ∼300 bp of G-rich repeats arranged in tandem arrays. The arrangement of telomeric complexes at the telomeric repeats are apparently different in S.cerevisiae and S.pombe. Whereas S.cerevisiae Rap1p binds telomeric DNA directly, S.pombe Rap1-like protein may be recruited by the TTAGGG Repeat Factor (TRF)-like protein (225).
The cis-acting elements and trans-acting factors responsible for silencing at rDNA repeats and telomeres are not well characterized. Recently, it has been shown that the four chromo-domain proteins, Ch1p, Ch2p, Swi6 and Clr4, are involved in silencing at rDNA repeats (214). Consistent with its role in rDNA silencing, Clr4 is localized in the nucleolus during interphase (226). In addition to these chromo-domain proteins, other S.pombe trans-acting factors including Clr3 are also important for rDNA silencing (214). Mutations in clr1, clr2, clr3, clr4, clr6, swi6 and rik1 that affect silencing at mat2-P, mat3-M, centromeres and in some cases rDNA, also affect telomeric silencing (5,196). In addition, telomeric silencing is specifically affected by mutations in taz1, rat1 and lot2 (227). The telomere-binding protein Taz1 is a member of the TRF family of proteins that are characterized by two domains: a TRF homology (TRFH) domain and a Myb-like domain (225). The TRF family proteins are telomere-associated proteins that negatively regulate telomere length (225). S.pombe Taz1 plays other roles at telomeres. First, Taz1 appears to suppress recombination at telomeres (228). Secondly, Taz1p is required for proper telomere aggregation at the spindle pole body which is crucial for the telomere-led movement of nuclei, chromosome segregation and recombination during meiotic prophase (227). The factors encoded by lot2 and rat1 have yet to be characterized (227).
Sir homologs in S.pombe
There are three S.pombe members of the Sir2p family: spSir2, spHst2 and spHst4 (158). Among them, spHst4 is the best characterized. SpHst4 is most closely related in sequence to S.cerevisiae Hst3p and Hst4p, and also appears to be functionally related to Hst3p and Hst4p (229). Overexpression of sphst4 rescues temperature sensitivity and silencing defects caused by deletion of HST3 and HST4 (229). Moreover, sphst4Δ cells exhibit phenotypes (e.g. growth and morphological defects) similar to those of S.cerevisiae hst3 and hst4 mutants. Finally, like Hst4p, spHst4 is concentrated at the nucleolus (229). Thus, the role in the maintenance of chromatin structure and integrity is likely conserved for these Hst proteins.
SpHst4 shares many phenotypes with S.pombe trans-acting factors including Clr4, Clr6, Rik1 and Swi6. Deletion of sphst4 affects silencing at centromeres and telomeres, but not at the silent mating-type loci (229). In addition to silencing, spHst4 is important for centromere function. The sphst4Δ cells display increased chromosome loss rates, fragmented DNA and are sensitive to the microtubule-destabilizing drug thiabendazole (TBZ) (229). However, sphst4Δ cells display growth and morphology defects, and severely fragmented DNA that is not observed in clr4, clr6, rik1 and swi6 mutants. One possible explanation is that spHst4 may function with other silencing factors at some loci, while functioning independently at others.
SILENCING PROTEINS INVOLVED IN OTHER CELLULAR PROCESSES
DNA repair proteins and telomeric silencing
Although telomeric silencing and DNA repair are controlled by two different cellular pathways, recent studies suggest that DNA repair is related to chromatin structure modulation and silencing. As noted above, the chromatin assembly factor yCAF-I, which couples DNA repair to chromatin assembly, is involved in silencing. Another example comes from studies on Rad6p, an E2 ubiqutin-conjugating enzyme. Rad6p appears to have multiple functions. It is involved in DNA postreplication repair, sporulation, recombination, degradation of proteins by the N-end rule pathway, and silencing (106,230). Ubiquitin-conjugating activity of Rad6p is required for these biological functions (230–232).
It has been proposed that the DNA repair activity and silencing activity of Rad6p may involve ubiquitination of different substrates (233). Moreover, ubiquitination of these different substrates by Rad6p may require a different partner protein (233). The hypothesis that Rad6p ubiquitinates a protein involved in silencing is supported by the finding that deletion of UBP3, encoding a deubiquitinating enzyme that interacts with Sir4p, enhances silencing at telomeres and HM loci (210). It appears that the role of controlling chromatin structure by S.cerevisiae Rad6p is conserved in S.pombe. Rhp6, a homolog of S.cerevisiae Rad6p, has been shown to regulate the expression of silent mating-type loci by modulating chromatin structure (234). Moreover, the silencing defect at mat2-P and mat3-M, and changes in chromatin structure brought on by rhp6 mutations, are switching dependent. Thus, Rhp6 was proposed to be required for reestablishment of chromatin structure which is disrupted during mating-type switching (234).
Another DNA repair protein involved in silencing is the chromosome end-binding protein, Ku. Ku functions in DNA DSB repair by the NHEJ pathway (235), telomere maintenance (73,236–238) and telomeric silencing (73). Recent genetic studies suggest that Ku may play a direct role in telomeric silencing by overcoming the negative effects of Rif1p and Rifp2p, thus helping to recruit the Sir complex to telomeres (66).
Although S.pombe Ku70 (Pku70) is also required for DSB repair by NHEJ and maintenance of telomere length, it does not play a role in telomeric silencing (239). Furthermore, in contrast to S.cerevisiae Ku that is concentrated at telomeres, S.pombe ku70 is distributed throughout nuclei. Consistent with these results, S.cerevisiae Ku associates with Sir proteins (e.g. Sir3p and Sir4p), whereas no apparent homologs of Sir3p and Sir4p exist in S.pombe.
DNA repair checkpoint proteins and silencing
Studies of DNA repair checkpoint proteins have demonstrated that there is a connection between checkpoint control and silencing. DNA repair checkpoint control involves a constant surveillance of the state of DNA. When DNA is damaged, DNA repair checkpoint proteins sense the DNA damage, transduce a signal to the DNA repair machinery, and suspend cell cycle progression until the damaged DNA is repaired. Several proteins required for this checkpoint control are also required for telomere function and telomeric silencing.
One of the checkpoint proteins involved in telomeric silencing is Mec3p. Deletion of MEC3 leads to increased telomere length and increased telomeric silencing (121). Mec3p physically interacts with Set1p, a protein involved in the regulation of chromatin structure, DNA repair and telomeric functions. This association links Mec3p to chromatin structure modulation. Interestingly, Mec3p antagonizes Set1p in telomere function and telomeric silencing (121). The SET domain of Set1p participates in telomeric silencing in a MEC3-dependent manner, whereas the non-SET part of Set1p participates in telomeric silencing in a MEC3-independent manner (121).
Another checkpoint protein involved in silencing is Mec1p. Mec1p is a protein kinase that is related to human ATM (Ataxia Telangiectasia) protein kinase family (240). The kinase domain of Mec1p has been shown to be important for DNA damage repair and telomere length maintenance (240), and has been implicated in telomeric silencing (241,242). The transcriptional silencing and telomere length maintenance functions of Mec1p are separable. It has been proposed that Mec1p-dependent phosphorylation of silencing proteins is necessary for the telomeric silencing activity of these proteins (241,242). Interestingly, a silencing defect associated with a mec1 mutation could be suppressed by a mutation in SML1 (suppressor of mec1 lethality) (241). SML1 encodes the ribonucleotide reductase inhibitor, the removal of which by Mec1p and Rad53p kinases is required to ensure DNA replication during cell growth and after DNA damage (243). This may explain, at least in part, why no defect in telomeric silencing was observed in the mec1Δ mutant, since this strain also carries a deletion of SML1 (241). Alternatively, the silencing defects associated with overexpression of MEC1 could be suppressed by overexpressing Scs2p (suppressor of choline sensitivity) which was originally identified as a suppressor of the inositol auxotrophy of cse1 (choline sensitive) and ire15 (inosito requiring) genes (244). It is likely that Scs2p regulates telomeric silencing via a pathway different from that used by Mec1p (242). A similar role for Mec1p in telomeric silencing has also been proposed for Dun1p.
The checkpoint protein Rad3, a S.pombe homolog of S.cerevisiae Mec1p, also plays a role in the control of telomere integrity, suggesting that the link between checkpoint proteins and maintenance of telomeres is conserved in S.pombe (245). However, the role of Rad3 in silencing remains to be addressed. While one report has shown that deletion of rad3+ does not affect telomeric silencing (239), another study revealed that the rad3-136 mutation has a moderate effect on telomeric silencing (245).
A connection between checkpoint proteins and telomeric silencing also comes from studies on the checkpoint proteins Mrc1p (Mediator of the DNA replication checkpoint) and Rad53p. Both of these proteins function in the S-phase checkpoint pathway and are required for telomeric silencing (246). Rad53p is an essential Ser/Thr/Tyr protein kinase. This kinase domain of Rad53p seems to be involved in both telomeric silencing and telomere length maintenance since inactivation of the kinase activity of Rad53p increases telomeric silencing and shortening of the telomere (170). Transcriptional silencing and telomere length maintenance represent two separable functions of Rad53p. Recent studies suggest that Rad53p may directly regulate chromatin assembly by releasing Asf1p (Anti-silencing function) in response to DNA damage and DNA replication blockage. Rad53p was found to physically associate with Asf1p, a chromatin assembly factor that has been implicated in deposition of acetylated H3 and H4 during DNA replication and repair (117). The Rad53p–Asf1p association is disrupted in response to replication blocks and DNA damage, and this disruption correlates with Rad53p phosphorylation by Mec1p (246,247). The biological relevance of this association is provided by the findings that a temperature-sensitive phenotype of the mrc1-1rad53-21 mutant and the hydroxyurea (HU; which inhibits DNA replication) sensitivity of rad53-21 mutant can be suppressed by ASF1 overexpression (246). These results suggest that Asf1p physically links Rad53p to chromatin assembly.
DNA replication proteins and silencing
Several lines of evidence suggest that DNA replication is linked to silencing. First, the establishment of silencing requires passage of yeast cells through S phase (111,117). Secondly, silencers contain the DNA replication initiation factor (ORC) binding site and the Orc1p–Sir1p interaction is thought to be important for the recruitment of the Sir complex in the HM loci (30,248). Thirdly, mutations that disrupt silencing have been identified in DNA replication proteins and DNA replication-related proteins such as DNA polymerase α (Pol α, required for both priming and DNA replication) (107,249), DNA helicase Dna2p, PCNA (required for DNA replication and nucleotide excision repair) (250), Rfc1p (PCNA loading factor) and the two replication-coupled chromatin assembly factors yCAF-1 and Asf1p (107,109,113,116,251). In addition, mutations that are capable of suppressing silencing defects are found in DNA replication proteins including DNA polymerase ɛ (Pol ɛ, a protein complex required for chromosomal replication and DNA repair) (252), the replication initiation factor Cdc45, PCNA and RF-C (replication factor C, a protein also involved in DNA repair) (253).
However, it appears that DNA replication is not the requisite S phase event for establishment of silencing. First, the requirement for the ORC-binding site and initiation of DNA replication can be bypassed by tethering Sir1p to the silencer, suggesting that DNA replication at the silent site is not required for silencing (254). Secondly, the functions of ORC in replication and silencing are genetically separable, indicating that some replication proteins may have dual roles (255,256). Recent studies have shown that while the establishment of silencing is cell cycle dependent, it can occur independent of DNA replication (257,258). The finding that silencing can be established in the absence of DNA replication raised an interesting question as to how the DNA replication proteins involved in silencing can be uncoupled from DNA replication. It is likely that DNA replication proteins may aid in the establishment of silencing, as well as the inheritance and maintenance of silent chromatin.
A role for DNA replication proteins in transcriptional silencing has recently been demonstrated in S.pombe (234,259,260). For example, a mutation in S.pombe Pol α (also named Swi7) suppresses repression at the mating-type region and centromeres (259,260). Moreover, CHIP assays have revealed that the Pol α mutant is defective in Swi6 localization at the mating-type region, centromeres and telomeres. Pol α interacts directly with Swi6, suggesting that Pol α may affect Swi6 localization directly. These data imply that Pol α may participate directly in silent chromatin assembly through its interaction with Swi6. Alternatively, mutations in Pol α might impair nucleosome assembly by S.pombe CAF-1 (259,260). It is worthy to note that like other replication proteins in S.cerevisiae, the silencing activity of Pol α is separable from its replication activity (259,260).
CONCLUSION
Transcriptional silencing is one of several molecular mechanisms in the cell that is dependent on the establishment and maintenance of repressed or silent chromatin. Other mechanisms include recombination, chromosome maintenance and segregation, nuclear organization and possibly DNA repair. With this in mind, it is not surprising to see that many silencing proteins from S.cerevisiae and S.pombe are also involved in several chromosome-related nuclear functions. Conversely, many proteins that are directly or indirectly involved in chromatin structure modulation also regulate silencing.
Silent chromatin in the budding yeast S.cerevisiae and the fission yeast S.pombe are governed by sets of different cis-acting elements and trans-acting factors, suggesting that silent chromatin in these two yeasts may be structurally and biochemically different. Whereas Sir proteins are essential for transcriptional silencing and constitute the ‘core’ component of the silencing machinery in S.cerevisiae, the chromodomain protein Swi6 and chromatin modifying proteins Clr3 and Clr4 are crucial for transcriptional silencing in S.pombe. The absence of homologs of these S.pombe silencing factors in S.cerevisiae is likely due to co-elimination of functionally linked genes that encode chromodomain proteins and other chromatin-associated proteins (261). The S.cerevisiae Sir proteins that have no apparent orthologs in S.pombe and higher eukaryotes have likely been functionally displaced by the chromatin-associated proteins that promote silencing in S.pombe. Remarkably, Sir proteins have diverse functions, including DNA DSB repair, regulation of the cell cycle and meiosis, and aging. Many of the proteins that function as the regulators of silent chromatin structure in S.cerevisiae have their homologs in S.pombe (Table 1). However, some of these S.pombe homologs (e.g. Pku70) are not involved in silencing.
Table 1. Conservation of silencing proteins in S.cerevisiae and S.pombe.
| S.cerevisiae | Functions in S.cerevisiae in addition to silencing | Homolog in S.pombea | Silencing function in S.pombe |
|---|---|---|---|
| Sir1p | None | ||
| Sir2p | NAD-dependent HDAC; aging; DNA repair; Pachytene checkpoints; telomere structure; mating-type switching | spSir2 (158) | |
| Sir3p | DNA repair; aging; telomere structure | None | |
| Sir4p | DNA repair; aging; telomere structure | None | |
| Hst1-4p | Homolog of Sir2p, cell cycle; chromosome stability | SpHst2, spHst4 (158) | + |
| Orc1p | DNA replication | Orp1p (264) | |
| Abf1p | Gene activation | SPCC663.05Cb | |
| Rap1p | Gene activation and repression; telomere function; replication | SPBC1778.02 | |
| Sum1p | Transcriptional repression | Sum1 (265) | |
| Rif1p, Rif2p | Regulator of telomere structure | None | |
| Sas4p | Regulator of chromatin structure | None | |
| Sas5p | Regulator of chromatin structure | YD67 (142) | |
| Mga2p, Spt23 | Regulator of chromatin structure; transcription activation | SPAC26H5.05 | |
| Scs2p | Synthesis or processing of phospholipids | Unknown | |
| Hda1p | HDAC | Hda1 (214), Clr3 (196) | + |
| Rpd3p | HDAC | Clr6 (196) | + |
| Sds3p | Component of Rpd3 HDAC complex | Unknown | |
| Sin3p | Component of Rpd3 HDAC complex | Pst1 (266) | |
| Pho23p | Component of Rpd3 HDAC complex | Png1, Png2 (267) | |
| Sas2p, Sas3p | Histone acetylase homolog | SpSas (135) | |
| yCAF-I | Chromatin assembling; DNA repair | Unknown | |
| Asf1p, Asf2p | Chromatin assembling | SPCC663.05C | |
| Set1p, Set2pc | DNA repair; regulation of chromatin structure | Clr4, SP CC306.04C | + |
| Rad6p | DNA repair; sporation; recombination; ubiquitination | Rhp6 (234) | + |
| Ku70, Ku80 | DNA repair; telomere function | Pku70 (239) | |
| Mec3p | DNA damage checkpoint; telomere functions | Hus1(268) | |
| Mec1p | DNA damage checkpoint; telomere functions | Rad3 (245) | |
| Mrc1p | DNA damage checkpoint | SPAC694.06C | |
| Rad53p | DNA damage checkpoint; telomere functions | Cds1 (269) | |
| Pch2p | DNA damage checkpoint | None | |
| Zip1p | DNA damage checkpoint | None | |
| Dot1p | DNA damage checkpoint | None | |
| PCNA | DNA replication and repair | Pcn1 (270) | |
| Rfc1p (RF-C) | DNA replication and repair checkpoints | Rfc1 (271) | |
| Pol2p (scPolɛ) | DNA replication and checkpoints | spPolɛ (272) | |
| Pol1p (scPolα) | DNA replication | Polα (spPolα) (257,258) | + |
| Net1p | Cell cycle; nucleolar structure; rDNA synthesis | Unknown | |
| None | Swi6 | + | |
| None | Rik1 | + |
aGenes encoding putative S.pombe homolog of S.cerevisiae silencing protein are listed by their access numbers, which are accessible at http://www.sanger.ac.uk/cgi-bin/yeastpub/comp_class_search.p.
bSPAC694.06C is probably the S.pombe ortholog of Mrc1p (V.Wood, personal communication).
cSet2p contains the SET domain and cysteine-rich regions shared by Clr4 and SUV39H1 (199).
Despite these significant differences, silent chromatin in these two yeasts shares a number of similarities between them and with the heterochromatin structure of higher eukaryotes. For example, silent chromatin is predominantly composed of repetitive DNA and contains a low density of genes. It is also relatively inaccessible to DNA-modifying reagents, replicates late during S phase, and is inherited during the cell cycle (234,262,263). Finally, many S.cerevisiae and S.pombe silencing proteins have functional orthologs in higher eukaryotes. Therefore, the study of transcriptional silencing in these two yeasts should provide insight into the mechanisms of transcriptional silencing in higher eukaryotes.
ACKNOWLEGEMENTS
I thank Dr N. C. Martin for encouragement, Drs A. J. S. Klar, S. J. Elledge, D. S. Gross, H. Nojima, M. P. Longhese and V. Wood for kindly answering my inquires, members of the Sanger Center for analysis and annotation of S.pombe genomic DNA, and Drs J. A. Bogan, V. Russanova and E. McGillicuddy for comments and discussion.
REFERENCES
- 1.Heard E., Clerc,P. and Avner,P. (1997) X-chromosome inactivation in mammals. Annu. Rev. Genet., 31, 571–610. [DOI] [PubMed] [Google Scholar]
- 2.Hennig W. (1999) Heterochromatin. Chromosoma, 108, 1–9. [DOI] [PubMed] [Google Scholar]
- 3.Steiner N.C. and Clarke,L. (1994) A novel epigenetic effect can alter centromere function in fission yeast. Cell, 79, 865–874. [DOI] [PubMed] [Google Scholar]
- 4.Allshire R.C., Javerzat,J.P., Redhead,N.J. and Cranston,G. (1994) Position effect variegation at fission yeast centromeres. Cell, 76, 157–169. [DOI] [PubMed] [Google Scholar]
- 5.Allshire R.C., Nimmo,E.R., Ekwall,K., Javerzat,J.P. and Cranston,G. (1995) Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev., 9, 218–233. [DOI] [PubMed] [Google Scholar]
- 6.Rodionov O., Lobocka,M. and Yarmolinsky,M. (1999) Silencing of genes flanking the P1 plasmid centromere. Science, 283, 546–549. [DOI] [PubMed] [Google Scholar]
- 7.Loo S. and Rine,J. (1995) Silencing and heritable domains of gene expression. Annu. Rev. Cell. Dev. Biol., 11, 519–548. [DOI] [PubMed] [Google Scholar]
- 8.Allshire R.C. (1996) Transcriptional silencing in the fission yeast: a manifestation of higher order chromosome structure and functions. In Russo,V.E.A., Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 443–466.
- 9.Grunstein M. (1997) Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol., 9, 383–387. [DOI] [PubMed] [Google Scholar]
- 10.Kamakaka R.T. (1997) Silencers and locus control regions: opposite sides of the same coin. Trends Biochem. Sci., 22, 124–128. [DOI] [PubMed] [Google Scholar]
- 11.Sherman J.M. and Pillus,L. (1997) An uncertain silence. Trends Genet., 13, 308–313. [DOI] [PubMed] [Google Scholar]
- 12.Klar A.J. (2001) Differentiated parental DNA chain causes stem cell pattern of cell-type switching in Schizosaccharomyces pombe. In Marshak,D.R., Gadner,R.L. and Gottlieb,D. (eds), Stem Cell Biology. Cold Spring Harbor Laboratory Press, NY, pp. 17–35.
- 13.Lustig A.J. (1998) Mechanisms of silencing in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev., 8, 233–239. [DOI] [PubMed] [Google Scholar]
- 14.Guarente L. (1999) Diverse and dynamic functions of the Sir silencing complex. Nature Genet., 23, 281–285. [DOI] [PubMed] [Google Scholar]
- 15.Roeder G.S. and Bailis,J.M. (2000) The pachytene checkpoint. Trends Genet., 16, 395–403. [DOI] [PubMed] [Google Scholar]
- 16.Gottschling D.E. (2000) Gene silencing: two faces of SIR2. Curr. Biol., 10, R708–R711. [DOI] [PubMed] [Google Scholar]
- 17.Grewal S.I. (2000) Transcriptional silencing in fission yeast. J. Cell Physiol., 184, 311–318. [DOI] [PubMed] [Google Scholar]
- 18.Gartenberg M.R. (2000) The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr. Opin. Microbiol., 3, 132–137. [DOI] [PubMed] [Google Scholar]
- 19.Moazed D. (2001) Common themes in mechanisms of gene silencing. Mol. Cell, 8, 489–498. [DOI] [PubMed] [Google Scholar]
- 20.Haber J.E. (1998) Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet., 32, 561–599. [DOI] [PubMed] [Google Scholar]
- 21.Loo S., Fox,C.A., Rine,J., Kobayashi,R., Stillman,B. and Bell,S. (1995) The origin recognition complex in silencing, cell cycle progression and DNA replication. Mol. Biol. Cell, 6, 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boscheron C., Maillet,L., Marcand,S., Tsai-Pflugfelder,M., Gasser,S.M. and Gilson,E. (1996) Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J., 15, 2184–2195. [PMC free article] [PubMed] [Google Scholar]
- 23.Brand A.H., Micklem,G. and Nasmyth,K. (1987) A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell, 51, 709–719. [DOI] [PubMed] [Google Scholar]
- 24.Kimmerly W., Buchman,A., Kornberg,R. and Rine,J. (1988) Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J., 7, 2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bi X., Braunstein,M., Shei,G.J. and Broach,J.R. (1999) The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl Acad. Sci. USA, 96, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moazed D., Kistler,A., Axelrod,A., Rine,J. and Johnson,A.D. (1997) Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl Acad. Sci. USA, 94, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moretti P., Freeman,K., Coodly,L. and Shore,D. (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev., 8, 2257–2269. [DOI] [PubMed] [Google Scholar]
- 28.Cockell M., Palladino,F., Laroche,T., Kyrion,G., Liu,C., Lustig,A.J. and Gasser,S.M. (1995) The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol., 129, 909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buck S.W. and Shore,D. (1995) Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev., 9, 370–384. [DOI] [PubMed] [Google Scholar]
- 30.Triolo T. and Sternglanz,R. (1996) Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature, 381, 251–253. [DOI] [PubMed] [Google Scholar]
- 31.Dhillon N. and Kamakaka,R.T. (2000) A histone variant, Htz1p and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell, 6, 769–780. [DOI] [PubMed] [Google Scholar]
- 32.Hecht A., Laroche,T., Strahl-Bolsinger,S., Gasser,S.M. and Grunstein,M. (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- 33.Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- 34.Pillus L. and Rine,J. (1989) Epigenetic inheritance of transcriptional states in Saccharomyces cerevisiae. Cell, 59, 637–647. [DOI] [PubMed] [Google Scholar]
- 35.Bi X. and Broach,J.R. (1997) DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol. Cell. Biol., 17, 7077–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng T.H. and Gartenberg,M.R. (2000) Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev., 14, 452–463. [PMC free article] [PubMed] [Google Scholar]
- 37.Enomoto S., Johnston,S.D. and Berman,J. (2000) Identification of a novel allele of SIR3 defective in the maintenance, but not the establishment, of silencing in Saccharomyces cerevisiae. Genetics, 155, 523–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner K.A. and Fox,C.A. (2001) The Sir1 protein’s association with a silenced chromosome domain. Genes Dev., 15, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klar A.J., Kakar,S.N., Ivy,J.M., Hicks,J.B., Livi,G.P. and Miglio,L.M. (1985) SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics, 111, 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie J., Pierce,M., Gailus-Durner,V., Wagner,M., Winter,E. and Vershon,A.K. (1999) Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J., 18, 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livi G.P., Hicks,J.B. and Klar,A.J. (1990) The sum1-1 mutation affects silent mating-type gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurenson P. and Rine,J. (1991) SUM1-1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics, 129, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chi M.H. and Shore,D. (1996) SUM1-1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol., 16, 4281–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rusche L.N. and Rine,J. (2001) Conversion of a gene-specific repressor to a regional silencer. Genes Dev., 15, 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutton A., Heller,R.C., Landry,J., Choy,J.S., Sirko,A. and Sternglanz,R. (2001) A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol., 21, 3514–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dula M.L. and Holmes,S.G. (2000) MGA2 and SPT23 are modifiers of transcriptional silencing in yeast. Genetics, 156, 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekinger E.A. and Gross,D.S. (2001) Silenced chromatin is permissive to activator binding and PIC recruitment. Cell, 105, 403–414. [DOI] [PubMed] [Google Scholar]
- 48.Bryan T.M. and Cech,T.R. (1999) Telomerase and the maintenance of chromosome ends. Curr. Opin. Cell Biol., 11, 318–324. [DOI] [PubMed] [Google Scholar]
- 49.Zakian V.A. (1996) Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet., 30, 141–172. [DOI] [PubMed] [Google Scholar]
- 50.Chikashige Y., Ding,D.Q., Imai,Y., Yamamoto,M., Haraguchi,T. and Hiraoka,Y. (1997) Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J., 16, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright J.H., Gottschling,D.E. and Zakian,V.A. (1992) Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev., 6, 197–210. [DOI] [PubMed] [Google Scholar]
- 52.Louis E.J., Naumova,E.S., Lee,A., Naumov,G. and Haber,J.E. (1994) The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics, 136, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pryde F.E., Huckle,T.C. and Louis,E.J. (1995) Sequence analysis of the right end of chromosome XV in Saccharomyces cerevisiae: an insight into the structural and functional significance of sub-telomeric repeat sequences. Yeast, 11, 371–382. [DOI] [PubMed] [Google Scholar]
- 54.Levis R., Hazelrigg,T. and Rubin,G.M. (1985) Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science, 229, 558–561. [DOI] [PubMed] [Google Scholar]
- 55.Gottschling D.E., Aparicio,O.M., Billington,B.L. and Zakian,V.A. (1990) Position effect at Saccharomyces cerevisiae telomeres: reversible repression of Pol II transcription. Cell, 63, 751–762. [DOI] [PubMed] [Google Scholar]
- 56.Nimmo E.R., Cranston,G. and Allshire,R.C. (1994) Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J., 13, 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horn D. and Cross,G.A. (1995) A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell, 83, 555–561. [DOI] [PubMed] [Google Scholar]
- 58.Baur J.A., Zou,Y., Shay,J.W. and Wright,W.E. (2001) Telomere position effect in human cells. Science, 292, 2075–2077. [DOI] [PubMed] [Google Scholar]
- 59.Kyrion G., Liu,K., Liu,C. and Lustig,A.J. (1993) RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev., 7, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 60.Renauld H., Aparicio,O.M., Zierath,P.D., Billington,B.L., Chhablani,S.K. and Gottschling,D.E. (1993) Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength and by SIR3 dosage. Genes Dev., 7, 1133–1145. [DOI] [PubMed] [Google Scholar]
- 61.Aparicio O.M., Billington,B.L. and Gottschling,D.E. (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in Saccharomyces cerevisiae. Cell, 66, 1279–1287. [DOI] [PubMed] [Google Scholar]
- 62.Strahl-Bolsinger S., Hecht,A., Luo,K. and Grunstein,M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev., 11, 83–93. [DOI] [PubMed] [Google Scholar]
- 63.Hardy C.F., Sussel,L. and Shore,D. (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev., 6, 801–814. [DOI] [PubMed] [Google Scholar]
- 64.Wotton D. and Shore,D. (1997) A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev., 11, 748–760. [DOI] [PubMed] [Google Scholar]
- 65.Marcand S., Wotton,D., Gilson,E. and Shore,D. (1997) Rap1p and telomere length regulation in yeast. Ciba Found. Symp., 211, 76–93. [DOI] [PubMed] [Google Scholar]
- 66.Mishra K. and Shore,D. (1999) Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr. Biol., 9, 1123–1126. [DOI] [PubMed] [Google Scholar]
- 67.Bourns B.D., Alexander,M.K., Smith,A.M. and Zakian,V.A. (1998) Sir proteins, Rif proteins and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol., 18, 5600–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hecht A., Strahl-Bolsinger,S. and Grunstein,M. (1996) Spreading of transcriptional repressor Sir3 from telomeric heterochromatin. Nature, 383, 92–96. [DOI] [PubMed] [Google Scholar]
- 69.Pryde F.E. and Louis,E.J. (1999) Limitations of silencing at native yeast telomeres. EMBO J., 18, 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lebrun E., Revardel,E., Boscheron,C., Li,R., Gilson,E. and Fourel,G. (2001) Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics, 158, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galy V., Olivo-Marin,J.C., Scherthan,H., Doye,V., Rascalou,N. and Nehrbass,U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [DOI] [PubMed] [Google Scholar]
- 72.Andrulis E.D., Neiman,A.M., Zappulla,D.C. and Sternglanz,R. (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature, 394, 592–595. [DOI] [PubMed] [Google Scholar]
- 73.Boulton S.J. and Jackson,S.P. (1998) Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J., 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laroche T., Martin,S.G., Gotta,M., Gorham,H.C., Pryde,F.E., Louis,E.J. and Gasser,S.M. (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol., 8, 653–656. [DOI] [PubMed] [Google Scholar]
- 75.Gotta M., Laroche,T., Formenton,A., Maillet,L., Scherthan,H. and Gasser,S.M. (1996) The clustering of telomeres and colocalization with Rap1, Sir3 and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol., 134, 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gotta M., Strahl-Bolsinger,S., Renauld,H., Laroche,T., Kennedy,B.K., Grunstein,M. and Gasser,S.M. (1997) Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J., 16, 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cockell M. and Gasser,S.M. (1999) Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev., 9, 199–205. [DOI] [PubMed] [Google Scholar]
- 78.Tham W., Wyithe,J.S., Ferrigno,P.K., Silver,P.A. and Zakian,V.A. (2001) Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell, 8, 189–199. [DOI] [PubMed] [Google Scholar]
- 79.Palladino F., Laroche,T., Gilson,E., Axelrod,A., Pillus,L. and Gasser,S.M. (1993) SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell, 75, 543–555. [DOI] [PubMed] [Google Scholar]
- 80.Shaw P.J. and Jordan,E.G. (1995) The nucleolus. Annu. Rev. Cell. Dev. Biol., 11, 93–121. [DOI] [PubMed] [Google Scholar]
- 81.Dammann R., Lucchini,R., Koller,T. and Sogo,J.M. (1995) Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol., 15, 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gottlieb S. and Esposito,R.E. (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell, 56, 771–776. [DOI] [PubMed] [Google Scholar]
- 83.Smith J.S. and Boeke,J.D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev., 11, 241–254. [DOI] [PubMed] [Google Scholar]
- 84.Fritze C.E., Verschueren,K., Strich,R. and Easton Esposito,R. (1997) Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J., 16, 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Straight A.F., Shou,W., Dowd,G.J., Turck,C.W., Deshaies,R.J., Johnson,A.D. and Moazed,D. (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell, 97, 245–256. [DOI] [PubMed] [Google Scholar]
- 86.Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W., Jang,J., Charbonneau,H. and Deshaies,R.J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- 87.Smith J.S., Brachmann,C.B., Pillus,L. and Boeke,J.D. (1998) Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by SIR4p. Genetics, 149, 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shou W., Sakamoto,K.M., Keener,J., Morimoto,K.W., Traverso,E.E., Azzam,R., Hoppe,G.J., Feldman,R.M., DeModena,J., Moazed,D. et al. (2001) Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell, 8, 45–55. [DOI] [PubMed] [Google Scholar]
- 89.Visintin R., Hwang,E.S. and Amon,A. (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature, 398, 818–823. [DOI] [PubMed] [Google Scholar]
- 90.Schwab M., Lutum,A.S. and Seufert,W. (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell, 90, 683–693. [DOI] [PubMed] [Google Scholar]
- 91.Visintin R., Prinz,S. and Amon,A. (1997) Cdc20 and Cdh1: a family of substrate-specific activators of apc-dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- 92.Ivy J.M., Klar,A.J. and Hicks,J.B. (1986) Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sussel L., Vannier,D. and Shore,D. (1993) Epigenetic switching of transcriptional states: cis- and trans-acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 3919–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stavenhagen J.B. and Zakian,V.A. (1994) Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes Dev., 8, 1411–1422. [DOI] [PubMed] [Google Scholar]
- 95.Lustig A.J., Liu,C., Zhang,C. and Hanish,J.P. (1996) Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2483–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maillet L., Boscheron,C., Gotta,M., Marcand,S., Gilson,E. and Gasser,S.M. (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev., 10, 1796–1811. [DOI] [PubMed] [Google Scholar]
- 97.Marcand S., Buck,S.W., Moretti,P., Gilson,E. and Shore,D. (1996) Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap1 protein. Genes Dev., 10, 1297–1309. [DOI] [PubMed] [Google Scholar]
- 98.Gotta M., Palladino,F. and Gasser,S.M. (1998) Functional characterization of the N terminus of Sir3p. Mol. Cell. Biol., 18, 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kennedy B.K., Austriaco,N.R.,Jr, Zhang,J. and Guarente,L. (1995) Mutation in the silencing gene SIR4 can delay aging in Saccharomyces cerevisiae. Cell, 80, 485–496. [DOI] [PubMed] [Google Scholar]
- 100.Kennedy B.K., Gotta,M., Sinclair,D.A., Mills,K., McNabb,D.S., Murthy,M., Pak,S.M., Laroche,T., Gasser,S.M. and Guarente,L. (1997) Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell, 89, 381–391. [DOI] [PubMed] [Google Scholar]
- 101.Johnson L.M., Kayne,P.S., Kahn,E.S. and Grunstein,M. (1990) Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 87, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Megee P.C., Morgan,B.A., Mittman,B.A. and Smith,M.M. (1990) Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science, 247, 841–845. [DOI] [PubMed] [Google Scholar]
- 103.Park E.C. and Szostak,J.W. (1990) Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol., 10, 4932–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thompson J.S., Ling,X. and Grunstein,M. (1994) Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature, 369, 245–247. [DOI] [PubMed] [Google Scholar]
- 105.Kaufman P.D., Cohen,J.L. and Osley,M.A. (1998) Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol., 18, 4793–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bryk M., Banerjee,M., Murphy,M., Knudsen,K.E., Garfinkel,D.J. and Curcio,M.J. (1997) Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev., 11, 255–269. [DOI] [PubMed] [Google Scholar]
- 107.Smith J.S., Caputo,E. and Boeke,J.D. (1999) A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol., 19, 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 109.Enomoto S. and Berman,J. (1998) Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev., 12, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Monson E.K., de Bruin,D. and Zakian,V.A. (1997) The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc. Natl Acad. Sci. USA, 94, 13081–13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sharp J.A., Fouts,E.T., Krawitz,D.C. and Kaufman,P.D. (2001) Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol., 11, 463–473. [DOI] [PubMed] [Google Scholar]
- 112.Shibahara K. and Stillman,B. (1999) Replication-dependent marking of DNA by PCNA facilitates Caf-1-coupled inheritance of chromatin. Cell, 96, 575–585. [DOI] [PubMed] [Google Scholar]
- 113.Kaufman P.D., Kobayashi,R. and Stillman,B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev., 11, 345–357. [DOI] [PubMed] [Google Scholar]
- 114.Kamakaka R.T., Bulger,M., Kaufman,P.D., Stillman,B. and Kadonaga,J.T. (1996) Postreplicative chromatin assembly by Drosophila and human chromatin assembly factor I. Mol. Cell. Biol., 16, 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Le S., Davis,C., Konopka,J.B. and Sternglanz,R. (1997) Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast, 13, 1029–1042. [DOI] [PubMed] [Google Scholar]
- 116.Singer M.S., Kahana,A., Wolf,A.J., Meisinger,L.L., Peterson,S.E., Goggin,C., Mahowald,M. and Gottschling,D.E. (1998) Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics, 150, 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tyler J.K., Adams,C.R., Chen,S.R., Kobayashi,R., Kamakaka,R.T. and Kadonaga,J.T. (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature, 402, 555–560. [DOI] [PubMed] [Google Scholar]
- 118.Jenuwein T., Laible,G., Dorn,R. and Reuter,G. (1998) SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci., 54, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schramke V., Neecke,H., Brevet,V., Corda,Y., Lucchini,G., Longhese,M.P., Gilson,E. and Geli,V. (2001) The set1Δ mutation unveils a novel signaling pathway relayed by the Rad53-dependent hyperphosphorylation of replication protein A that leads to transcriptional activation of repair genes. Genes Dev., 15, 1845–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laible G., Wolf,A., Dorn,R., Reuter,G., Nislow,C., Lebersorger,A., Popkin,D., Pillus,L. and Jenuwein,T. (1997) Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at Saccharomyces cerevisiae telomeres. EMBO J., 16, 3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Corda Y., Schramke,V., Longhese,M.P., Smokvina,T., Paciotti,V., Brevet,V., Gilson,E. and Geli,V. (1999) Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nature Genet., 21, 204–208. [DOI] [PubMed] [Google Scholar]
- 122.Roguev A., Schaft,D., Shevchenko,A., Pijnappel,W.W., Wilm,M., Aasland,R. and Stewart,A.F. (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J., 20, 7137–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- 124.Ng H.H. and Bird,A. (2000) Histone deacetylases: silencers for hire. Trends Biochem. Sci., 25, 121–126. [DOI] [PubMed] [Google Scholar]
- 125.Kuo M.H. and Allis,C.D. (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays, 20, 615–626. [DOI] [PubMed] [Google Scholar]
- 126.Vannier D., Balderes,D. and Shore,D. (1996) Evidence that the transcriptional regulators SIN3 and RPD3 and a novel gene (SDS3) with similar functions, are involved in transcriptional silencing in S. cerevisiae. Genetics, 144, 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rundlett S.E., Carmen,A.A., Kobayashi,R., Bavykin,S., Turner,B.M. and Grunstein,M. (1996) Hda1 and Rpd3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl Acad. Sci. USA, 93, 14503–14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kasten M.M., Dorland,S. and Stillman,D.J. (1997) A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol. Cell. Biol., 17, 4852–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Laherty C.D., Billin,A.N., Lavinsky,R.M., Yochum,G.S., Bush,A.C., Sun,J.M., Mullen,T.M., Davie,J.R., Rose,D.W., Glass,C.K. et al. (1998) SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell, 2, 33–42. [DOI] [PubMed] [Google Scholar]
- 131.Zhang Y., Sun,Z.W., Iratni,R., Erdjument-Bromage,H., Tempst,P., Hampsey,M. and Reinberg,D. (1998) SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell, 1, 1021–1031. [DOI] [PubMed] [Google Scholar]
- 132.Bernstein B.E., Tong,J.K. and Schreiber,S.L. (2000) Genomewide studies of histone deacetylase function in yeast. Proc. Natl Acad. Sci. USA, 97, 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lechner T., Carrozza,M.J., Yu,Y., Grant,P.A., Eberharter,A., Vannier,D., Brosch,G., Stillman,D.J., Shore,D. and Workman,J.L. (2000) Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3·Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem., 275, 40961–40966. [DOI] [PubMed] [Google Scholar]
- 134.Loewith R., Smith,J.S., Meijer,M., Williams,T.J., Bachman,N., Boeke,J.D. and Young,D. (2001) Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem., 276, 24068–24074. [DOI] [PubMed] [Google Scholar]
- 135.Reifsnyder C., Lowell,J., Clarke,A. and Pillus,L. (1996) Yeast Sas silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nature Genet., 14, 42–49. [DOI] [PubMed] [Google Scholar]
- 136.Ehrenhofer-Murray A.E., Rivier,D.H. and Rine,J. (1997) The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics, 145, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Smith E.R., Eisen,A., Gu,W., Sattah,M., Pannuti,A., Zhou,J., Cook,R.G., Lucchesi,J.C. and Allis,C.D. (1998) ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl Acad. Sci. USA, 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kimura A. and Horikoshi,M. (1998) Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells, 3, 789–800. [DOI] [PubMed] [Google Scholar]
- 139.Neuwald A.F. and Landsman,D. (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci., 22, 154–155. [DOI] [PubMed] [Google Scholar]
- 140.Takechi S. and Nakayama,T. (1999) Sas3 is a histone acetyltransferase and requires a zinc finger motif. Biochem. Biophys. Res. Commun., 266, 405–410. [DOI] [PubMed] [Google Scholar]
- 141.John S., Howe,L., Tafrov,S.T., Grant,P.A., Sternglanz,R. and Workman,J.L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing hat complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev., 14, 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 142.Xu E.Y., Kim,S., Replogle,K., Rine,J. and Rivier,D.H. (1999) Identification of SAS4 and SAS5, two genes that regulate silencing in Saccharomyces cerevisiae. Genetics, 153, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu E.Y., Kim,S. and Rivier,D.H. (1999) SAS4 and SAS5 are locus-specific regulators of silencing in Saccharomyces cerevisiae. Genetics, 153, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tenen D.G., Hromas,R., Licht,J.D. and Zhang,D.E. (1997) Transcription factors, normal myeloid development and leukemia. Blood, 90, 489–519. [PubMed] [Google Scholar]
- 145.Garcia-Cuellar M.P., Zilles,O., Schreiner,S.A., Birke,M., Winkler,T.H. and Slany,R.K. (2001) The ENL moiety of the childhood leukemia-associated MLL-ENL oncoprotein recruits human Polycomb 3. Oncogene, 20, 411–419. [DOI] [PubMed] [Google Scholar]
- 146.Brachmann C.B., Sherman,J.M., Devine,S.E., Cameron,E.E., Pillus,L. and Boeke,J.D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes Dev., 9, 2888–2902. [DOI] [PubMed] [Google Scholar]
- 147.Frye R.A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun., 273, 793–798. [DOI] [PubMed] [Google Scholar]
- 148.Sherman J.M., Stone,E.M., Freeman-Cook,L.L., Brachmann,C.B., Boeke,J.D. and Pillus,L. (1999) The conserved core of a human SIR2 homologue functions in yeast silencing. Mol. Biol. Cell, 10, 3045–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tanny J.C., Dowd,G.J., Huang,J., Hilz,H. and Moazed,D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell, 99, 735–745. [DOI] [PubMed] [Google Scholar]
- 150.Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: SIR2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun., 260, 273–279. [DOI] [PubMed] [Google Scholar]
- 151.Imai S., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- 152.Yoshida M., Horinouchi,S. and Beppu,T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays, 17, 423–430. [DOI] [PubMed] [Google Scholar]
- 153.Landry J., Sutton,A., Tafrov,S.T., Heller,R.C., Stebbins,J., Pillus,L. and Sternglanz,R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA, 97, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Smith J.S., Brachmann,C.B., Celic,I., Kenna,M.A., Muhammad,S., Starai,V.J., Avalos,J.L., Escalante-Semerena,J.C., Grubmeyer,C., Wolberger,C. et al. (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA, 97, 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Muth V., Nadaud,S., Grummt,I. and Voit,R. (2001) Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J., 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ghidelli S., Donze,D., Dhillon,N. and Kamakaka,R.T. (2001) Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J., 20, 4522–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Derbyshire M.K., Weinstock,K.G. and Strathern,J.N. (1996) HST1, a new member of the SIR2 family of genes. Yeast, 12, 631–640. [DOI] [PubMed] [Google Scholar]
- 158.Perrod S., Cockell,M.M., Laroche,T., Renauld,H., Ducrest,A.L., Bonnard,C. and Gasser,S.M. (2001) A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J., 20, 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guarente L. (2001) SIR2 and aging—the exception that proves the rule. Trends Genet., 17, 391–392. [DOI] [PubMed] [Google Scholar]
- 160.Guarente L. (2000) Sir2 links chromatin silencing, metabolism and aging. Genes Dev., 14, 1021–1026. [PubMed] [Google Scholar]
- 161.Defossez P.A., Lin,S.J. and McNabb,D.S. (2001) Sound silencing: the Sir2 protein and cellular senescence. Bioessays, 23, 327–332. [DOI] [PubMed] [Google Scholar]
- 162.Kaeberlein M., McVey,M. and Guarente,L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev., 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 164.Lin S.J., Defossez,P.A. and Guarente,L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science, 289, 2126–2128. [DOI] [PubMed] [Google Scholar]
- 165.Jiang J.C., Jaruga,E., Repnevskaya,M.V. and Jazwinski,S.M. (2000) An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J., 14, 2135–2137. [DOI] [PubMed] [Google Scholar]
- 166.Tsukamoto Y., Kato,J. and Ikeda,H. (1997) Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature, 388, 900–903. [DOI] [PubMed] [Google Scholar]
- 167.Martin S.G., Laroche,T., Suka,N., Grunstein,M. and Gasser,S.M. (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell, 97, 621–633. [DOI] [PubMed] [Google Scholar]
- 168.McAinsh A.D., Scott-Drew,S., Murray,J.A. and Jackson,S.P. (1999) DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol., 9, 963–966. [DOI] [PubMed] [Google Scholar]
- 169.Mills K.D., Sinclair,D.A. and Guarente,L. (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell, 97, 609–620. [DOI] [PubMed] [Google Scholar]
- 170.Longhese M.P., Paciotti,V., Neecke,H. and Lucchini,G. (2000) Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics, 155, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee S.E., Paques,F., Sylvan,J. and Haber,J.E. (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol., 9, 767–770. [DOI] [PubMed] [Google Scholar]
- 172.Astrom S.U., Okamura,S.M. and Rine,J. (1999) Yeast cell-type regulation of DNA repair. Nature, 397, 310. [DOI] [PubMed] [Google Scholar]
- 173.Heude M. and Fabre,F. (1993) a/α-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics, 133, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schild D. (1995) Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics, 140, 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fasullo M., Bennett,T. and Dave,P. (1999) Expression of Saccharomyces cerevisiae MATa and MATa enhances the HO endonuclease-stimulation of chromosomal rearrangements directed by his3 recombinational substrates. Mutat. Res., 433, 33–44. [DOI] [PubMed] [Google Scholar]
- 176.Sym M., Engebrecht,J.A. and Roeder,G.S. (1993) Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell, 7 2, 365–378. [DOI] [PubMed] [Google Scholar]
- 177.San-Segundo P.A. and Roeder,G.S. (2000) Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell, 11, 3601–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.San-Segundo P.A. and Roeder,G.S. (1999) Pch2 links chromatin silencing to meiotic checkpoint control. Cell, 97, 313–324. [DOI] [PubMed] [Google Scholar]
- 179.Dlakic M. (2001) Chromatin silencing protein and pachytene checkpoint regulator Dot1p has a methyltransferase fold. Trends Biochem. Sci., 26, 405–407. [DOI] [PubMed] [Google Scholar]
- 180.Beach D.H. and Klar,A.J. (1984) Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J., 3, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Egel R. (1981) Mating-type switching and mitotic crossing-over at the mating-type locus in fission yeast. Cold Spring Harbor Symp. Quant. Biol., 45, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 182.Grewal S.I. and Klar,A.J. (1997) A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics, 146, 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Thon G., Cohen,A. and Klar,A.J. (1994) Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics, 138, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Klar A.J. and Miglio,L.M. (1986) Initiation of meiotic recombination by double-strand DNA breaks in Schizosaccharomyces pombe. Cell, 46, 725–731. [DOI] [PubMed] [Google Scholar]
- 185.Egel R., Willer,M. and Nielsen,O. (1989) Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive,pleotropic mutant rik1. Curr. Genet., 15, 407–410. [Google Scholar]
- 186.Grewal S.I. and Klar,A.J. (1996) Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell, 86, 95–101. [DOI] [PubMed] [Google Scholar]
- 187.Thon G. and Friis,T. (1997) Epigenetic inheritance of transcriptional silencing and switching competence in fission yeast. Genetics, 145, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ekwall K., Nielsen,O. and Ruusala,T. (1991) Repression of a mating type cassette in the fission yeast by four DNA elements. Yeast, 7, 745–755. [DOI] [PubMed] [Google Scholar]
- 189.Thon G., Bjerling,K.P. and Nielsen,I.S. (1999) Localization and properties of a silencing element near the mat3-M mating-type cassette of Schizosaccharomyces pombe. Genetics, 151, 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ayoub N., Goldshmidt,I. and Cohen,A. (1999) Position effect variegation at the mating-type locus of fission yeast: a cis-acting element inhibits covariegated expression of genes in the silent and expressed domains. Genetics, 152, 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ayoub N., Goldshmidt,I., Lyakhovetsky,R. and Cohen,A. (2000) A fission yeast repression element cooperates with centromere-like sequences and defines a mat silent domain boundary. Genetics, 156, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Noma K., Allis,C.D. and Grewal,S.I. (2001) Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science, 293, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 193.Ekwall K., Nimmo,E.R., Javerzat,J.P., Borgstrom,B., Egel,R., Cranston,G. and Allshire,R. (1996) Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci., 109, 2637–2648. [DOI] [PubMed] [Google Scholar]
- 194.Ekwall K., Javerzat,J.P., Lorentz,A., Schmidt,H., Cranston,G. and Allshire,R. (1995) The chromodomain protein Swi6: a key component at fission yeast centromeres. Science, 269, 1429–1431. [DOI] [PubMed] [Google Scholar]
- 195.Thon G. and Klar,A.J. (1992) The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics, 131, 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Grewal S.I., Bonaduce,M.J. and Klar,A.J. (1998) Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics, 150, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ekwall K. and Ruusala,T. (1994) Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics, 136, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nakayama J., Rice,J.C., Strahl,B.D., Allis,C.D. and Grewal,S.I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science, 292, 110–113. [DOI] [PubMed] [Google Scholar]
- 199.Rea S., Eisenhaber,F., O’Carroll,D., Strahl,B.D., Sun,Z.W., Schmid,M., Opravil,S., Mechtler,K., Ponting,C.P., Allis,C.D. and Jenuwein T. (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature, 406, 593–599. [DOI] [PubMed] [Google Scholar]
- 200.Koonin E.V., Zhou,S. and Lucchesi,J.C. (1995) The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res., 23, 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cavalli G. and Paro,R. (1998) Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr. Opin. Cell Biol., 10, 354–360. [DOI] [PubMed] [Google Scholar]
- 202.Ivanova A.V., Bonaduce,M.J., Ivanov,S.V. and Klar,A.J. (1998) The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet., 19, 192–195. [DOI] [PubMed] [Google Scholar]
- 203.O’Carroll D., Scherthan,H., Peters,A.H., Opravil,S., Haynes,A.R., Laible,G., Rea,S., Schmid,M., Lebersorger,A., Jerratsch,M. et al. (2000) Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol., 20, 9423–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Jenuwein T. (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol., 11, 266–273. [DOI] [PubMed] [Google Scholar]
- 205.Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 206.Aasland R. and Stewart,A.F. (1995) The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res., 23, 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lorentz A., Ostermann,K., Fleck,O. and Schmidt,H. (1994) Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from drosophila and mammals. Gene, 143, 139–143. [DOI] [PubMed] [Google Scholar]
- 208.Paro R. and Hogness,D.S. (1991) The polycomb protein shares a homologous domain with a heterochromatin-associated protein of drosophila. Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nakayama J., Klar,A.J. and Grewal,S.I. (2000) A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell, 101, 307–317. [DOI] [PubMed] [Google Scholar]
- 210.Moazed D. and Johnson,D. (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in Saccharomyces cerevisiae. Cell, 86, 667–677. [DOI] [PubMed] [Google Scholar]
- 211.Neuwald A.F. and Poleksic,A. (2000) PSI-BLAST searches using hidden markov models of structural repeats: prediction of an unusual sliding DNA clamp and of β-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res., 28, 3570–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- 213.Doe C.L., Wang,G., Chow,C., Fricker,M.D., Singh,P.B. and Mellor,E.J. (1998) The fission yeast chromo domain encoding gene chp1+ is required for chromosome segregation and shows a genetic interaction with α-tubulin. Nucleic Acids Res., 26, 4222–4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Thon G. and Verhein-Hansen,J. (2000) Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics, 155, 551–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Olsson T.G., Ekwall,K., Allshire,R.C., Sunnerhagen,P., Partridge,J.F. and Richardson,W.A. (1998) Genetic characterisation of hda1+, a putative fission yeast histone deacetylase gene. Nucleic Acids Res., 26, 3247–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kim Y.B., Honda,A., Yoshida,M. and Horinouchi,S. (1998) phd1+, a histone deacetylase gene of Schizosaccharomyces pombe, is required for the meiotic cell cycle and resistance to trichostatin a. FEBS Lett., 436, 193–196. [DOI] [PubMed] [Google Scholar]
- 217.Takahashi K., Murakami,S., Chikashige,Y., Funabiki,H., Niwa,O. and Yanagida,M. (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell, 3, 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Partridge J.F., Borgstrom,B. and Allshire,R.C. (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev., 14, 783–791. [PMC free article] [PubMed] [Google Scholar]
- 219.Ekwall K., Cranston,G. and Allshire,R.C. (1999) Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics, 153, 1153–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Javerzat J.P., McGurk,G., Cranston,G., Barreau,C., Bernard,P., Gordon,C. and Allshire,R. (1999) Defects in components of the proteasome enhance transcriptional silencing at fission yeast centromeres and impair chromosome segregation. Mol. Cell. Biol., 19, 5155–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Turner B.M. (2000) Histone acetylation and an epigenetic code. Bioessays, 22, 836–845. [DOI] [PubMed] [Google Scholar]
- 222.Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 223.Hoheisel J.D., Maier,E., Mott,R., McCarthy,L., Grigoriev,A.V., Schalkwyk,L.C., Nizetic,D., Francis,F. and Lehrach,H. (1993) High resolution cosmid and P1 maps spanning the 14 MB genome of the fission yeast Schizosaccharomyces pombe. Cell, 73, 109–120. [DOI] [PubMed] [Google Scholar]
- 224.Schaak J., Mao,J. and Soll,D. (1982) The 5.8S RNA gene sequence and the ribosomal repeat of Schizosaccharomyces pombe. Nucleic Acids Res., 10, 2851–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Li B., Oestreich,S. and de Lange,T. (2000) Identification of human Rap1: implications for telomere evolution. Cell, 101, 471–483. [DOI] [PubMed] [Google Scholar]
- 226.Sawin K.E. and Nurse,P. (1996) Identification of fission yeast nuclear markers using random polypeptide fusions with green fluorescent protein. Proc. Natl Acad. Sci. USA, 93, 15146–15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Nimmo E.R., Pidoux,A.L., Perry,P.E. and Allshire,R.C. (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature, 392, 825–828. [DOI] [PubMed] [Google Scholar]
- 228.Nakamura T.M., Cooper,J.P. and Cech,T.R. (1998) Two modes of survival of fission yeast without telomerase. Science, 282, 493–496. [DOI] [PubMed] [Google Scholar]
- 229.Freeman-Cook L.L., Sherman,J.M., Brachmann,C.B., Allshire,R.C., Boeke,J.D. and Pillus,L. (1999) The Schizosaccharomyces pombe hst4+ gene is a SIR2 homologue with silencing and centromeric functions. Mol. Biol. Cell, 10, 3171–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Huang H., Kahana,A., Gottschling,D.E., Prakash,L. and Liebman,S.W. (1997) The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6693–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sung P., Berleth,E., Pickart,C., Prakash,S. and Prakash,L. (1991) Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the n-end-recognizing e3 enzyme. EMBO J., 10, 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sung P., Prakash,S. and Prakash,L. (1990) Mutation of cysteine-88 in the Saccharomyces cerevisiae Rad6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl Acad. Sci. USA, 87, 2695–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Freiberg G., Mesecar,A.D., Huang,H., Hong,J.Y. and Liebman,S.W. (2000) Characterization of novel rad6/ubc2 ubiquitin-conjugating enzyme mutants in yeast. Curr. Genet., 37, 221–233. [DOI] [PubMed] [Google Scholar]
- 234.Singh J., Goel,V. and Klar,A.J. (1998) A novel function of the DNA repair gene rhp6 in mating-type silencing by chromatin remodeling in fission yeast. Mol. Cell. Biol., 18, 5511–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Critchlow S.E. and Jackson,S.P. (1998) DNA end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 236.Gravel S., Larrivee,M., Labrecque,P. and Wellinger,R.J. (1998) Yeast Ku as a regulator of chromosomal DNA end structure. Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 237.Polotnianka R.M., Li,J. and Lustig,A.J. (1998) The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol., 8, 831–834. [DOI] [PubMed] [Google Scholar]
- 238.Nugent C.I., Bosco,G., Ross,L.O., Evans,S.K., Salinger,A.P., Moore,J.K., Haber,J.E. and Lundblad,V. (1998) Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol., 8, 657–660. [DOI] [PubMed] [Google Scholar]
- 239.Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mallory J.C. and Petes,T.D. (2000) Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human atm protein kinase. Proc. Natl Acad. Sci. USA, 97, 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Craven R.J. and Petes,T.D. (2000) Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 2378–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Craven R.J. and Petes,T.D. (2001) The Saccharomyces cerevisiae suppressor of choline sensitivity (SCS2) gene is a multicopy suppressor of mec1 telomeric silencing defects. Genetics, 158, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhao X., Chabes,A., Domkin,V., Thelander,L. and Rothstein,R. (2001) The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J., 20, 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kagiwada S., Hosaka,K., Murata,M., Nikawa,J. and Takatsuki,A. (1998) The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J. Bacteriol., 180, 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Matsuura A., Naito,T. and Ishikawa,F. (1999) Genetic control of telomere integrity in Schizosaccharomyces pombe: rad3+ and tel1+ are parts of two regulatory networks independent of the downstream protein kinases chk1+ and cds1+. Genetics, 152, 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hu F., Alcasabas,A.A. and Elledge,S.J. (2001) Asf1 links Rad53 to control of chromatin assembly. Genes Dev., 15, 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Emili A., Schieltz,D.M., Yates,J.R.,3rd and Hartwell,L.H. (2001) Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor asf1. Mol. Cell, 7, 13–20. [DOI] [PubMed] [Google Scholar]
- 248.Gardner K.A., Rine,J. and Fox,C.A. (1999) A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics, 151, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Adams Martin, A., Dionne,I., Wellinger,R.J. and Holm,C. (2000) The function of DNA polymerase α at telomeric g tails is important for telomere homeostasis. Mol. Cell. Biol., 20, 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shivji K.K., Kenny,M.K. and Wood,R.D. (1992) Proliferating cell nuclear antigen is required for DNA excision repair. Cell, 69, 367–374. [DOI] [PubMed] [Google Scholar]
- 251.Zhang Z., Shibahara,K. and Stillman,B. (2000) Pcna connects DNA replication to epigenetic inheritance in yeast. Nature, 408, 221–225. [DOI] [PubMed] [Google Scholar]
- 252.Wang Z., Wu,X. and Friedberg,E.C. (1993) DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase ɛ and is influenced by DNA polymerases α and δ in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ehrenhofer-Murray A.E., Kamakaka,R.T. and Rine,J. (1999) A role for the replication proteins PCNA, RF-C, polymerase ɛ and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics, 153, 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fox C.A., Ehrenhofer-Murray,A.E., Loo,S. and Rine,J. (1997) The origin recognition complex, Sir1 and the S phase requirement for silencing. Science, 276, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 255.Dillin A. and Rine,J. (1997) Separable functions of Orc5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics, 147, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fox C.A., Loo,S., Dillin,A. and Rine,J. (1995) The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev., 9, 911–924. [DOI] [PubMed] [Google Scholar]
- 257.Kirchmaier A.L. and Rine,J. (2001) DNA replication-independent silencing in Saccharomyces cerevisiae. Science, 291, 646–650. [DOI] [PubMed] [Google Scholar]
- 258.Li Y.C., Cheng,T.H. and Gartenberg,M.R. (2001) Establishment of transcriptional silencing in the absence of DNA replication. Science, 291, 650–653. [DOI] [PubMed] [Google Scholar]
- 259.Nakayama J., Allshire,R.C., Klar,A.J. and Grewal,S.I. (2001) A role for DNA polymerase α in epigenetic control of transcriptional silencing in fission yeast. EMBO J., 20, 2857–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ahmed S., Saini,S., Arora,S. and Singh,J. (2001) Chromodomain protein Swi6-mediated role of DNA polymerase α in establishment of silencing in fission yeast. J. Biol. Chem., 1, 1. [DOI] [PubMed] [Google Scholar]
- 261.Aravind L., Watanabe,H., Lipman,D.J. and Koonin,E.V. (2000) Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA, 97, 11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Singh J. and Klar,A.J. (1992) Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev., 6, 186–196. [DOI] [PubMed] [Google Scholar]
- 263.Stevenson J.B. and Gottschling,D.E. (1999) Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev., 13, 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Grallert B. and Nurse,P. (1996) The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev., 10, 2644–2654. [DOI] [PubMed] [Google Scholar]
- 265.Humphrey T. and Enoch,T. (1998) Sum1, a highly conserved WD-repeat protein, suppresses S-M checkpoint mutants and inhibits the osmotic stress cell cycle response in fission yeast. Genetics, 148, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Dang V.D., Benedik,M.J., Ekwall,K., Choi,J., Allshire,R.C. and Levin,H.L. (1999) A new member of the Sin3 family of corepressors is essential for cell viability and required for retroelement propagation in fission yeast. Mol. Cell. Biol., 19, 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Loewith R., Meijer,M., Lees-Miller,S.P., Riabowol,K. and Young,D. (2000) Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol., 20, 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Caspari T., Dahlen,M., Kanter-Smoler,G., Lindsay,H.D., Hofmann,K., Papadimitriou,K., Sunnerhagen,P. and Carr,A.M. (2000) Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol., 20, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Walworth N., Davey,S. and Beach,D. (1993) Fission yeast Chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature, 363, 368–371. [DOI] [PubMed] [Google Scholar]
- 270.Waseem N.H., Labib,K., Nurse,P. and Lane,D.P. (1992) Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J., 11, 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Tanaka H., Tanaka,K., Murakami,H. and Okayama,H. (1999) Fission yeast Cdc24 is a replication factor C- and proliferating cell nuclear antigen-interacting factor essential for S-phase completion. Mol. Cell. Biol., 19, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.D’Urso G. and Nurse,P. (1997) Schizosaccharomyces pombe cdc20+ encodes DNA polymerase ɛ and is required for chromosomal replication but not for the S phase checkpoint. Proc. Natl Acad. Sci. USA, 94, 12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]