Abstract
The dopaminergic system, including five dopamine receptors (D1R to D5R), plays essential roles in the central nervous system (CNS); and ligands that activate dopamine receptors have been used to treat many neuropsychiatric disorders, including Parkinson’s Disease (PD) and schizophrenia. Here, we report cryo-EM structures of all five subtypes of human dopamine receptors in complex with G protein and bound to the pan-agonist, rotigotine, which is used to treat PD and restless legs syndrome. The structures reveal the basis of rotigotine recognition in different dopamine receptors. Structural analysis together with functional assays illuminate determinants of ligand polypharmacology and selectivity. The structures also uncover the mechanisms of dopamine receptor activation, unique structural features among the five receptor subtypes, and the basis of G protein coupling specificity. Our work provides a comprehensive set of structural templates for the rational design of specific ligands to treat CNS diseases targeting the dopaminergic system.
Subject terms: Cryoelectron microscopy, Molecular modelling
Introduction
Dopamine and the dopamine receptor system play critical roles in motor functions, cognition, and addiction.1–3 The action of dopaminergic system is mediated by five subtypes of dopamine receptors, a subfamily of G protein-coupled receptors (GPCRs). The dopamine receptors are divided into D1-like and D2-like groups. The D1-like group includes D1R and D5R, whereas the D2-like group includes D2R, D3R, and D4R. D1-like receptors are coupled to the stimulatory G proteins (Gs) and linked to the activation of adenylate cyclase. The D2-like receptors are coupled to the inhibitory subtypes of G proteins (Gi and Go) and linked to the inhibition of adenylate cyclase.4
Dopamine receptors are a prototypical class of drug targets for many central nervous system (CNS) diseases, including Parkinson’s disease,5 schizophrenia,6 and attention deficit hyperactivity disorder (ADHD).7 There are several dozens of dopaminergic drugs,8 with many of them having distinct properties of polypharmacology, which can act on multiple dopamine receptors or even other types of monoamine neurotransmitter receptors. However, understanding the polypharmacology of dopaminergic drugs remains a tremendous challenge due to the promiscuous binding of drugs to many different receptors with various pharmacology. Rotigotine, a drug for Parkinson’s Disease (PD) and restless legs syndrome (RLS), is a pan-agonist that activates all five dopamine receptors.9,10 The molecular basis for the pan-agonism of rotigotine to the dopaminergic receptor system is unclear.
To date, several structures of dopamine receptors have been reported, including active D1R, D2R, and D3R structures and inactive D2R, D3R, and D4R structures.11–18 No active-state structure of D4R or any state structure of D5R has been reported. The lack of the D5R structure and the active D4R structure impedes our understanding of the dopaminergic system. In addition, the basis of how different types of dopamine receptors bind ligands with similar or diverse affinity is not well understood, making it challenging to develop therapeutic agents with lower side effects. Here, we report cryo-electron microscopy (cryo-EM) structures of all five subtypes of dopamine receptors in complex with rotigotine and their cognate G protein subtypes, Gs or Gi. These structures reveal the basis for the pan-agonism of rotigotine and dopamine receptor polypharmacology, as well as a mechanism of dopamine receptor activation and G protein coupling selectivity. Together with mutagenesis and functional studies, our results provide important insights into the biology of dopaminergic system and templates for rational design of drugs treating CNS diseases.
Results and discussion
Cryo-EM structures of all the five dopamine receptors
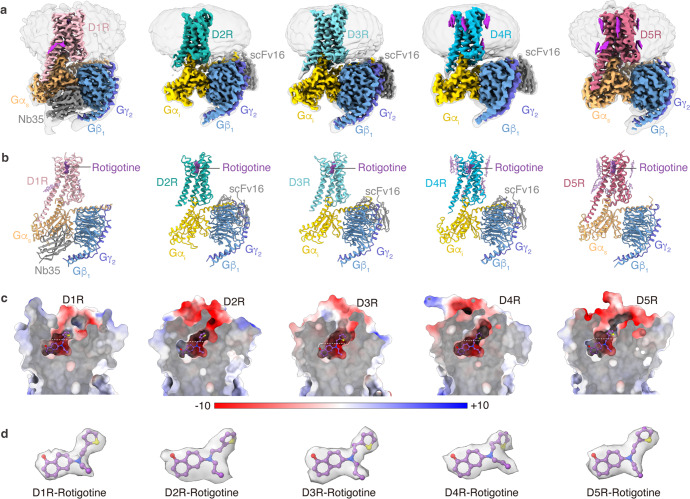
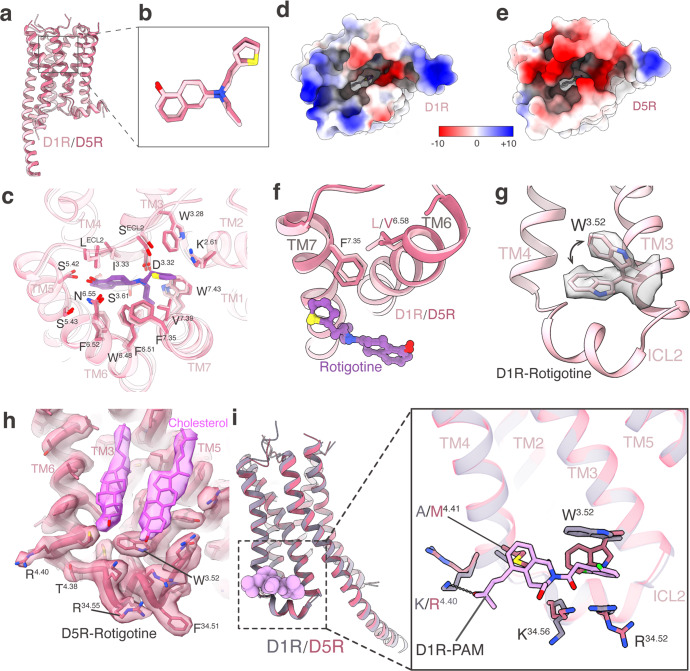
For cryo-EM studies, we used the wild-type (WT) human dopamine receptors for structural determination. To assist the expression and purification of the receptor–G protein complexes, we fused cytochrome b562 RIL (BRIL)19 and His tag at the N-termini of the receptors. To achieve the stable formation of the receptors with G proteins, D1R or D5R were expressed with the dominant-negative form of Gs (DNGs),20 and D2R or D3R were expressed with dominant-negative form of Gi (DNGi)21 in Trichoplusia ni insect cell. The D4R–Gi complex was not assembled stably, and the yield was low. To obtain the stable complex of D4R bound to Gi protein, we used another form of engineered Gi protein, which consists of αN and α5 helices of Gi and Ras domain of DNGs.22 In addition, NanoBiT tethering strategy was introduced to enhance the assembly of the D4R–Gi complex by fusing the LgBiT to the C-terminus of the receptor and fusing the SmBiT to the C-terminus of Gβ.23 For the D1R–Gs and D5R–Gs complex, nanobody35 (Nb35)24 was used to further stabilize the complexes. For the D2R–Gi, D3R–Gi, and D4R–Gi complexes, scFv1625 was used. The pan-agonist rotigotine and apyrase were added during purification to stabilize the complexes in the active states. The structures were determined at global resolutions of 3.2 Å (D1R–Gs), 3.0 Å (D2R–Gi), 2.7 Å (D3R–Gi), 3.2 Å (D4R–Gi), and 3.1 Å (D5R–Gs), respectively (Fig. 1; Supplementary information, Figs. S1, S2 and Table S1). The density maps of the five complexes allowed us to model the majority of the receptor residues, ligands, and G proteins, as well as a number of cholesterol molecules in D1R, D4R, and D5R (Fig. 1a, b, d). Several regions in the complexes were not observed in the EM maps, including the flexible N-terminus, a portion of extracellular loop 2 (ECL2), intracellular loop 3 (ICL3), C-terminus of each receptor, and the alpha-helical domains (AHDs) of Gα subunits (Fig. 1b). Although Nb35 was added during the purification of both D1R–Gs and D5R–Gs complexes, the density of Nb35 was not observed in the D5R–Gs complex (Fig. 1a; Supplementary information, Fig. S2m, n).
Fig. 1. Cryo-EM structures of D1R, D2R, D3R, D4R and D5R signaling complexes.
a, b The cryo-EM density maps (a) and models (b) of the D1R–Gs, D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes. c The ligand-binding pockets of the D1R–Gs, D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes. Electrostatic surface potential is colored by red (−10 kT/e), blue (+10 kT/e), and white (neutral). d The rotigotine structure in the D1R–Gs, D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes. The EM densities of rotigotine in the five structures are shown.
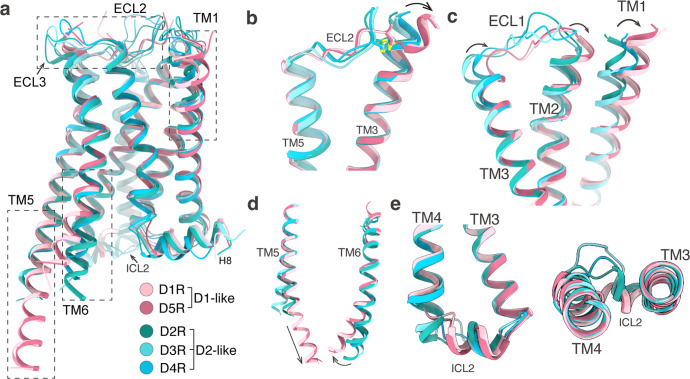
Overall, the five structures of the dopamine receptors exhibit similar backbone conformations (Fig. 2a). The seven transmembrane helical structures are highly overlapped, except for the extracellular sides of TM1–3 (Fig. 2b, c), the intracellular sides of TM5–6 (Fig. 2d), and ICL2 (Fig. 2e). Multiple cholesterol molecules were observed in the transmembrane domains (TMDs) of D1R, D4R, and D5R structures, but not in the D2R and D3R structures (Fig. 1a). The observation of cholesterols in D1R structure and the absence of cholesterols in D2R and D3R structures are consistent with the previously reported cryo-EM structures of D1R,11,17 D2R,11 and D3R.12 As the two members of D1-like dopamine receptor, D1R and D5R share almost identical backbone conformations, with a root-mean-square deviation (RMSD) of 0.48 Å as measured by the Cα atoms of the receptors (Fig. 2a). Our D1R–Gs complex is similar to the previously reported structures of D1R–Gs complexes solved by cryo-EM, with RMSD values ranging from 0.37 Å to 0.87 Å over the Cα atoms of the receptor part (Supplementary information, Fig. S3a).11,17,18,26 Nevertheless, the only X-ray structure of D1R–Gs complex27 shows a ~5 Å translocation at αN helix of Gαs subunit and Gβγ subunit from the cryo-EM structures, possibly due to crystal packing associated with the Gβγ subunits27 (Supplementary information, Fig. S3a). For the D2-like group, D2R, D3R, and D4R also exhibit very similar conformation in their TMDs, but show obvious differences in ECL2, ECL3, ICL2, and H8 helix (Fig. 2a). The D2R and D3R are more similar to each other than to D4R, with RMSD values of 0.51 Å between D2R and D3R, 0.73 Å between D2R and D4R, and 0.67 Å between D3R and D4R, respectively. This is consistent with the sequence identity between D2R and D3R (44%), which is higher than the sequence identity between D2R and D4R (31%) or between D3R and D4R (32%). The rotigotine-bound D2R–Gi structure, when compared with other cryo-EM structures of D2R–Gi protein complexes,11,13 showed conformational changes in both extracellular and intracellular regions, including TM6, TM7, ECL3, and Gi protein (Supplementary information, Fig. S3c–e). In contrast, our D3R–Gi structure shares very high similarity with the previously reported D3R–Gi structures (Supplementary information, Fig. S3f–h).12 The conformational change in the extracellular regions of D2R is possibly due to the adaptation of the receptor with the binding of bromocriptine, which contains bulkier branch groups relative to rotigotine (Supplementary information, Fig. S3d, e, g). The ligand-binding pockets of all five dopamine receptors are highly negatively charged, which allows the positively charged amine ligands to bind into their pockets (Fig. 1c). The differences between D1-like receptors and D2-like receptors were observed in the intracellular regions, including the intracellular ends of TM5/6 and ICL2 (Fig. 2d, e). The most notable difference exists in the conformation of TM5, wherein TM5 domains of the D1-like receptors are extended with three extra helical turns into the cytoplasmic side compared to the D2-like receptors (Fig. 2d). In addition, TM6 domains from the D1-like receptors move outwardly by ~7 Å compared to the D2-like receptors in the intracellular ends (Fig. 2d), consistent with their selective coupling of Gs and Gi subtypes.28 For ICL2, the ICL2 domains of the D1-like dopamine receptors are one more α-helical turn longer than those of the D2-like dopamine receptors (Fig. 2e).
Fig. 2. Structural feature comparison of all active-state dopamine receptors.
a Structural superposition of D1R, D2R, D3R, D4R, and D5R. b Structural alignment of ECL2, TM3, and TM5. c Structural alignment of ECL1 and TM1. d Structural alignment of TM5 and TM6. e Structural alignment of ICL2.
Rotigotine binds to orthosteric and extended binding pockets
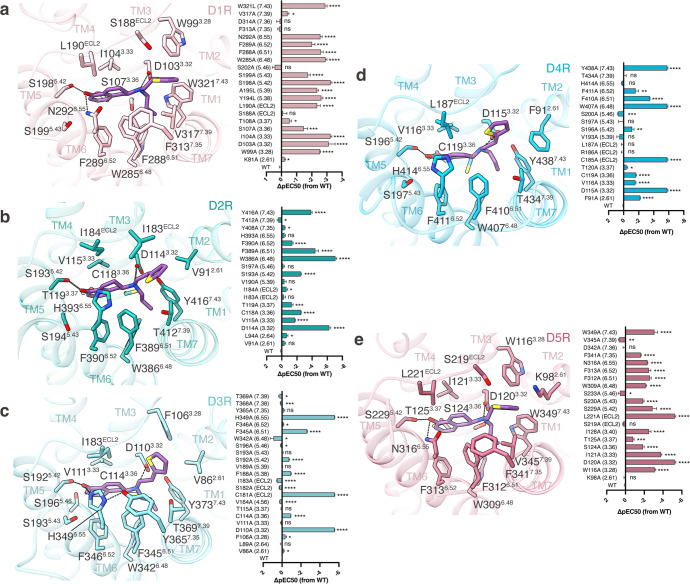
All five dopamine receptors harbor an open ligand-binding pocket within the top half of their TMDs which consists of an orthosteric binding pocket (OBP) and an extended binding pocket (EBP) (Fig. 1c). The OBP sits in the lower half of the entire pocket, reaching to the middle of the receptor transmembrane helix. The EBP opens upwardly, connecting the OBP and the extracellular space (Fig. 1c). The sequences of OBPs share higher similarities than those of EBPs in five dopamine receptors, as well as in aminergic receptors.11,12 Rotigotine binds to both OBPs and EBPs in all dopamine receptors. In the OBPs, rotigotine displays a nearly identical conformation in all structures (Figs. 1c and 3). The primary amine group from rotigotine forms charged interactions with the conserved D3.32 residue of the receptors, and the tetrahydronaphthalene group forms hydrophobic interactions with I3.33, F6.51 and F6.52 of the receptors. The amino-linked ethyl group inserts into a small hydrophobic pocket formed by the receptor residues W6.48, W/Y7.43, and F6.51 (Fig. 3). In the EBPs, rotigotine exhibits a conserved binding mode in D1R and D5R, while displays different binding modes among D2R, D3R, and D4R (Figs. 1d and 3). The hydroxyl group of rotigotine forms a hydrogen bond with S5.42, which is a conserved residue in all dopamine receptors, and forms another hydrogen bond with N6.55 of D1R and D5R (Fig. 3). However, the residue H6.55 in D2-like receptors shows different interaction patterns with rotigotine. The hydroxyl group of rotigotine only forms hydrogen bond with H6.55 in D3R, but not in D2R and D4R, although the H6.55 residue is conserved in D2-like receptors (Fig. 3b–d). Consequently, the H6.55A mutation only significantly affects the potency of rotigotine in D3R, rather than D2R and D4R. Comparison of the structures of D2R and D3R indicates that the extracellular ends of TM6 and TM7 in D2R moved more outwardly relative to those of D3R. These conformational differences prevent the H6.55 residue in D2R from forming hydrogen bond with the hydroxyl group of rotigotine (Fig. 3). Superposition of D4R and D3R structures shows that the extracellular end of TM6 in D4R moved more outwardly by ~2 Å than that of D3R when measured by the Cα atoms of Q6.58, resulting in the absence of hydrogen-bond interaction between rotigotine and H6.55 of D4R. Thus, the different interaction patterns between the hydroxyl group of rotigotine and D2-like receptors provide the basis of the higher affinity of rotigotine to D3R over D2R and D4R, consistent with the results of our functional assays (Fig. 3b–d; Supplementary information, Fig. S4 and Tables S3–S5). The thiophene group of rotigotine forms hydrophobic interactions with residues from the EBPs of the five dopamine receptors (Figs. 1c and 3). In both D1R and D5R structures, the thiophene group shares a nearly identical interaction mode between the two receptors owing to the conserved residues and conformations in the EBPs, in which the thiophene group forms hydrophobic interactions with receptor residues W3.28, F7.35, and V7.39 (Fig. 3a, e; Supplementary information, Fig. S5). On the other hand, in the structures of the D2-like receptors, the thiophene group of rotigotine displays different conformations due to the different shapes and topologies of the receptor EBPs (Fig. 3b–d; Supplementary information, Fig. S5). The D2R and D3R structures show that the thiophene group forms hydrophobic interactions with residues F3.28, V2.61, T7.39, Y7.35, and I/SECL2. Interestingly, the thiophene group displays a different interaction mode in D4R from D2R and D3R. In D4R, the thiophene group is pointed toward TM2 and forms hydrophobic interactions with the nonconserved residue F912.61, which corresponds to K2.61 in D1R/D5R and V2.61 in D2R/D3R (Fig. 3d). The potency of rotigotine to D4R was reduced by 100-fold with the mutation F91A, whereas the corresponding alanine mutation of residues 2.61 at other dopamine receptors had little effect on rotigotine binding (Fig. 3; Supplementary information, Fig. S4 and Tables S2–S6). These results support the unique interacting mode of rotigotine in D4R.
Fig. 3. Rotigotine recognition at all dopamine receptors.
a–e Left, detailed interaction between rotigotine and D1R (a), D2R (b), D3R (c), D4R (d) or D5R (e). Right, effects of mutations of the ligand-binding pocket residues of D1R (a), D2R (b), D3R (c), D4R (d) or D5R (e) on changes in ΔpEC50 in response to stimulation of rotigotine, evaluated using a GloSensor cAMP assay. All data are presented as means ± SEM of three independent experiments for the WT and mutants (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, not significant (two-tailed paired t-test).
Since the OBP sequences of dopamine receptors are highly conserved, many ligands were developed using the same chemical scaffold. Compounds catechol and ergoline are two classes of prototypical ligands of dopamine receptors and many other aminergic receptors. Rotigotine does not belong to either catechol or ergoline class of ligands. To uncover the differences of the rotigotine binding modes from the catechol agonists, we analyzed the detailed interactions by comparing the structures of dopamine receptor D1R bound to rotigotine and dopamine (a prototypical catechol ligand),26 revealing six sets of intermolecular interactions between the bound ligands and the receptor. Among the six sets of interactions, three sets are similar, and the other three sets are different (Supplementary information, Fig. S5c–e). The three similar sets of interactions include the conserved salt bridge formed by the primary amine group with the D1033.32 residue, the hydrophobic interactions of the tetrahydronaphthalene group of rotigotine and the benzene group of dopamine with hydrophobic residues 6.51, 6.52, and 3.33, and the hydrogen-bond interactions of the 5-hydroxyl group of rotigotine and dopamine with polar residues S1985.42, S1995.43, and N2926.55 (Supplementary information, Fig. S5c, d). Mutations of these residues to Ala significantly reduced the potencies of both rotigotine and dopamine to D1R by similar degrees (Supplementary information, Table S2). The three different sets of interactions include extra hydrogen bonds of the 4-hydroxyl group of dopamine with residues S2025.46 and T1083.37, while the corresponding interactions cannot be formed by rotigotine, because rotigotine does not have the corresponding hydroxyl group as in dopamine (Supplementary information, Fig. S5c–e). Correspondingly, S202A or T108A mutations in all dopamine receptors reduced the dopamine potencies by over 1000-fold, but hardly affected the potencies of rotigotine (S202A) or mildly reduced the potencies of rotigotine by 20-fold in all dopamine receptors (T108A) (Supplementary information, Tables S2–S6). In addition, the propyl group in rotigotine makes extra hydrophobic contacts with residues F2886.51, V3177.39 and W3217.43, which are absent in dopamine-bound D1R structure. The last different set of interactions was observed in the EBPs, because dopamine lacks a branch group like the thiophene group in rotigotine, which forms additional hydrophobic contacts with the EBPs (Supplementary information, Fig. S5c–e). Correspondingly, mutations of EBP residues in all five dopamine receptors affected the potencies of rotigotine more significantly than those of dopamine, which is mostly bound within the OBP (Supplementary information, Table S2).
Two cryo-EM structures of D1R–Gs complex bound to non-catechol agonists, including tavapadon and PW0464, were recently reported.17,18 Superposition of D1R structures bound to rotigotine and non-catechol agonists suggests major differences in their binding to the EBP of D1R. In rotigotine-bound D1R, the thiophen ring interacts mainly with hydrophobic residues such as W993.28 and F3137.35 in EBP. However, in addition to a similar set of hydrophobic interactions, the pyrimidinedione groups of tavapadon and PW0464 form extra polar contacts with residues K812.61, C186ECL2 and S188ECL2 compared to rotigotine (Supplementary information, Fig. S5f), which may account for their higher potencies toward D1R. Noticeable differences were also observed in the topologies of ECL2 in rotigotine-, tavapadon- and PW0464-bound D1R structures, indicating the plasticity in the conformation of D1R EBP when bound to ligands with distinct chemical scaffolds.
Rotigotine polypharmacology
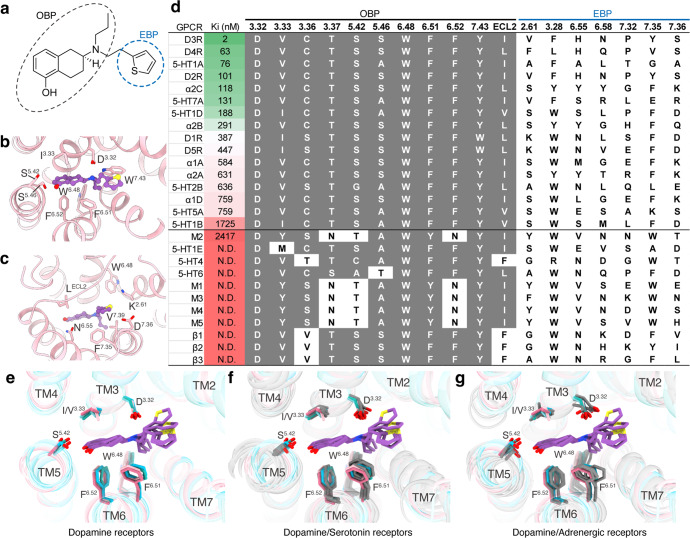
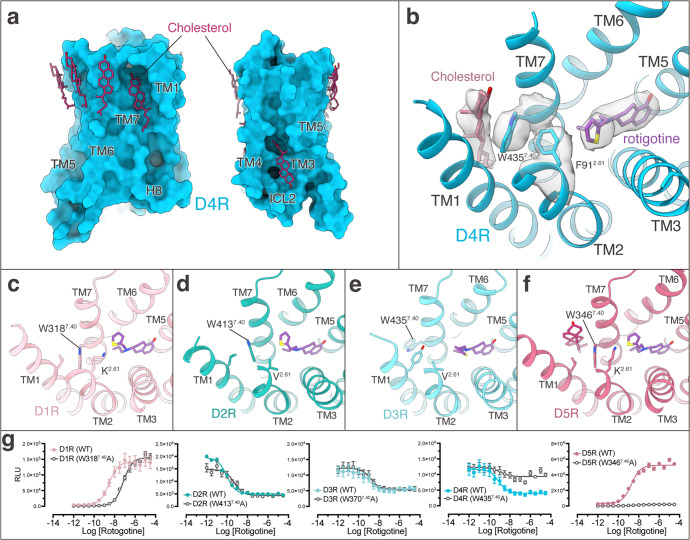
Rotigotine has been reported to activate all dopamine receptors, as well as several types of aminergic receptors.29 To reveal the polypharmacological profile of rotigotine, we screened the binding activity of rotigotine to over 300 GPCRs (Supplementary information, Table S7). The results showed that rotigotine exhibited high affinities to dopamine receptors, serotonin receptors, and adrenergic receptors and unexpectedly displayed agonist activities at somatostatin receptors, adenosine receptors, opioid receptors, and melatonin receptors (Fig. 4; Supplementary information, Table S7). To illustrate the basis of the promiscuous binding of rotigotine, we aligned the sequences of the ligand-binding pockets of aminergic receptors. We found that the high affinity of rotigotine is highly related to the conserved sequences of OBP in many monoamine receptors (Fig. 4d). Structure comparisons of the rotigotine-bound dopamine receptors with serotonin receptors and adrenergic receptors revealed highly overlapped conformations shared by the conserved OBP residues, including D3.32, I/V3.33, F6.51, F6.52, and W6.48 (Fig. 4e–g). Mutations of these conserved OBP residues in dopamine receptors greatly affect rotigotine binding (Supplementary information, Tables S2–S6), indicating that the polypharmacology of rotigotine is mainly attributed to the conserved OBP.
Fig. 4. Polypharmacological profile of rotigotine.
a The chemical structure of rotigotine. b The interaction of D1R OBP with rotigotine. c The interaction of D1R EBP with rotigotine. d The affinities (Ki) of rotigotine to different GPCRs as indicated by radioligand competition binding assays and the alignment of OBP-EBP residues. Receptors are listed in order of decreasing rotigotine affinity. e Structural superposition of five dopamine receptors and bound rotigotine. f Structural superposition of five rotigotine-bound dopamine receptors compared with serotonin receptors (gray). 5-HT1A (PDB: 7E2Y), 5-HT1B (PDB: 6G79), 5-HT1D (PDB: 7E32), 5-HT2B (PDB: 6DRY), and 5-HT5A (PDB: 7X5H). g Structural superposition of five rotigotine-bound dopamine receptors compared with adrenergic receptors (gray). α2A (PDB: 6KUY), α2B (PDB: 6K41), and α2C (PDB: 6KUW).
D1-like receptors
The two D1-like dopamine receptors, D1R and D5R, exhibit highly conserved sequence homology, particularly at the orthosteric binding pocket (Fig. 4d). To further explore the remaining differences in ligand affinity for D1R and D5R, we performed structural superimposition of rotigotine-bound D1R and D5R. The rotigotine-bound D1R and D5R structures revealed that D1R and D5R share almost identical conformations in their ligand-binding pockets (Fig. 5a–c). However, both dopamine and rotigotine have higher potencies to D5R over D1R (Supplementary information, Fig. S5a and Tables S2, S6). The cAMP accumulation assays showed that the potency of dopamine is ~10-fold higher on D5R (pEC50 = 9.82) than on D1R (pEC50 = 8.86) and that rotigotine is also ~10-fold more potent on D5R (pEC50 = 9.25) than on D1R (pEC50 = 8.49) (Supplementary information, Tables S2 and S6). Comparison of their ligand-binding pockets and electrostatic surface showed that D5R has more negative charges on the extracellular surface than D1R (Fig. 5d, e). These findings indicate that positively charged ligands, such as dopamine and rotigotine, would prefer to enrich within the pocket of D5R. In addition, we performed a docking study of rotigotine into the ligand-binding pockets of D1R and D5R, which revealed that rotigotine had a better docking score on D5R (–7.09) than on D1R (–4.85), consistent with our functional studies. Further comparison of the residue pair F7.35–L6.58 of D1R with the F7.35–V6.58 of D5R showed subtle difference in the rotigotine binding modes between these two receptors. Residue F7.35 in EBPs forms hydrophobic interactions with the thiophene group of rotigotine in both D1R and D5R. However, the side chain of F7.35 in D5R is slightly closer to the thiophene group of rotigotine than in D1R (Fig. 5f). Thus, the F3417.35A mutation in D5R showed a greater effect on rotigotine binding (ΔpEC50 = –1.63) than the corresponding F3137.35A mutation in D1R (ΔpEC50 = 0.16) (Supplementary information, Tables S2, S6).
Fig. 5. Comparison of D1R and D5R in rotigotine binding and PAM binding.
a Structural superposition of D1R–Gs and D5R–Gs complexes when receptors were aligned. b Comparison of rotigotine binding poses in D1R and D5R structures. c Structural comparison of rotigotine recognition between D1R and D5R. d, e Rotigotine-binding pockets of D1R (d) and D5R (e) viewed from the extracellular side. Electrostatic surface potential is colored by red (−10 kT/e), blue (+10 kT/e), and white (neutral). f Comparison of TM6 and TM7 residues for rotigotine recognition between D1R and D5R. g The side chain of W3.52 residue shows two alternative conformations in the D1R–Gs–rotigotine structure. h The unique conformation of W3.52 residue is stabilized by a cholesterol molecule in the D5R–Gs–rotigotine structure. i Comparison of TM3, TM4 and ICL2 residues for compound LY3154207 recognition between D1R and D5R.
In addition to orthosteric agonists, positive allosteric modulators (PAMs) represent a promising strategy for discovering D1R- and D5R-targeting drugs with high selectivity and low side effect. LY3154207 is the first clinical PAM of D1R with a high level of selectivity.30 To explore the PAM selectivity between D1R and D5R, we compared the structures of the LY3154207-bound D1R with the rotigotine-bound D1R and D5R. A notable difference is that A4.41 in D1R is replaced by M4.41 in D5R, resulting in a steric hindrance for D5R to bind LY3154207 (Fig. 5i). In addition, W3.52 of D1R was found to adopt two alternative conformations in the rotigotine-bound D1R structure (Fig. 5g). One of the conformations, with the side chain in the up configuration, could adapt to the LY3154207 binding.26 The other conformation, with the side chain in the down configuration, would prevent LY3154207 binding (Fig. 5g, i). In the D5R structure, residue W3.52 only has one conformation in its down configuration, which would prevent D5R from binding to LY3154207. The unique conformation of W3.52 in D5R is further stabilized by a cholesterol molecule (Fig. 5h), which is not observed in the corresponding site of D1R. Thus, the structures determined here provide a framework for understanding the mechanism of PAM selectivity and could assist in the design and optimization of D1R-selective therapeutic modulators.
D2-like receptors
Despite that the three D2-like receptors share relatively high sequence conservation and that all of them couple to Gi protein, they play different physiological functions and show different affinities to various ligands.4 The variety of the D2R structures and the conservation of the D3R structures (Supplementary information, Fig. S3c–h) are consistent with the notion that D2R is more dynamic than D3R.12 Since no other active-state D4R structure is currently available, we compared the structures of D4R with D2R and D3R. We found that there were multiple cholesterol molecules surrounding the D4R TMD but not in D2R and D3R (Fig. 1a). Remarkably, a clear cholesterol molecule locates between TM1 and TM7 in D4R. This cholesterol forms hydrophobic interactions with W4357.40, which is further stabilized by the pocket residue F912.61 (Fig. 6a, b). Interestingly, this cholesterol is only found in the D4R structure and the 2.61 residue is not conserved in other dopamine receptors (Fig. 6b–f). Consistently, mutation of residue 2.61 to Ala significantly reduced the potency of rotigotine on D4R but not on other dopamine receptors (Supplementary information, Tables S2–S6), suggesting that the CHL–W7.40–F912.61 interaction network is important for ligand binding in D4R. A similar cholesterol-interacting network has also been observed in the 5-HT1A receptor–ligand complexes.31
Fig. 6. The binding of rotigotine in D4R is regulated by cholesterol.
a Cholesterol molecules at the surface of D4R. b A cholesterol molecule is located between TM1 and TM7 of D4R and stabilizes rotigotine binding through residues W4357.40 and F912.61. c–f Structural comparison of the TM1–TM7 region and residue 2.61 of D1R (c), D2R (d), D3R (e), and D5R (f) show differences from those of D4R. g Concentration response of WT and W7.40A mutant of all five dopamine receptors stimulated by rotigotine.
Activation of the dopamine receptors
The availability of all five dopamine receptor structures allowed us to examine the common features of dopamine receptor activation mechanism. The activation of D4 and D5 receptors displays similar characteristics to the previously reported activation of D1R, D2R, and D3R.11,12 For all five dopamine receptors, the binding of rotigotine leads to the downward movement of the “toggle switch” residue W6.48, which further induces conformational changes in the PIF, DRY and NPxxY motifs. These conformational changes eventually cause the outward movement of TM6, allowing the α5 helix of G protein to insert into the intracellular pocket of the TMD. Within the D1-like receptors, the active structures of both D1R and D5R share nearly identical conformations in the “toggle switch” residue, PIF, DRY, and NPxxY motifs, suggesting a potentially common mechanism of rotigotine-induced activation (Supplementary information, Fig. S5g). Within the D2-like receptors, all motifs related to receptor activation are conserved between D2R and D3R, and the motif residues share similar conformational changes between the inactive and active structures. However, the D4R structure shows different conformational changes from D2R and D3R (Supplementary information, Fig. S6). In particular, the intracellular end of TM3 in the active D4R undergoes a 3-Å inward translocation from the inactive state. This translocation is not observed in D2R or D3R upon inactive-to-active transition (Supplementary information, Fig. S6), revealing a unique feature of D4R activation.
Previous studies reported that the catechol agonists can activate the aminergic receptor through a hydrogen bond with S5.46, which induces an inward movement of residue P5.50 and the rearrangement of the PIF motif and TM6.32,33 In all dopamine receptor structures, rotigotine does not form the same hydrogen bond with the S5.46 as dopamine (a catechol ligand with two hydroxyl groups), because rotigotine contains only one hydroxyl group, which forms hydrogen bonds with S5.42 and N6.55 (Supplementary information, Fig. S5c, d). However, rotigotine, despite not forming hydrogen bond with S5.46, can still activate dopamine receptors with similar efficiency as dopamine (Supplementary information, Tables S2–S6). To reveal the different activation mechanisms of rotigotine and catechol agonists, we compared the structures of rotigotine-bound D1R and dopamine-bound D1R.11 We found that the closest distance between residue W6.48 and rotigotine is 3.4 Å, but for dopamine this distance is 5.3 Å. This distance difference may cause the different strength of interactions of rotigotine and dopamine with W6.48, therefore affecting their activation of D1R. Consistent with these observations, W6.48A mutation in D1R leads to a greater reduction of G protein signaling induced by rotigotine compared to that induced by dopamine, although rotigotine exhibits a higher potency for the mutated receptor (Supplementary information, Table S2). These results suggest that dopamine activates receptors through both the residues S5.46 and W6.48, whereas rotigotine activates receptors mainly through residue W6.48. Our results indicate that there are different mechanisms of catechol and non-catechol agonists-induced dopamine receptor activation.
To illustrate the agonism and antagonism of dopamine receptors, we analyzed all available dopamine receptor structures and focused on the interactions of rotigotine with residue W6.48, as well as the interactions of residue W6.48 with the PIF motif. We found the activation of the respective receptor is highly related to the distance between residues W6.48 and I3.40. Specifically, all activated dopamine receptors display shorter W6.48–I3.40 distances (< 5 Å) than all inactive dopamine receptors (> 6 Å), except for the antagonist L745870-bound D4R structure, where the W6.48–I3.40 distance is 4.9 Å. However, the structural activation analysis showed that this antagonist-bound D4R structure exhibited 83% activation34 (Supplementary information, Table S8). The typical antagonists of dopamine receptors could prevent the W6.48–I3.40 interaction by inserting deeply into the OBP, whereas agonists only bind to the upper half of receptors. These results suggest the importance of the W6.48–I3.40 interaction for receptor activation and further reveal the basis of agonism and antagonism of dopamine receptors.
G protein coupling of dopamine receptors
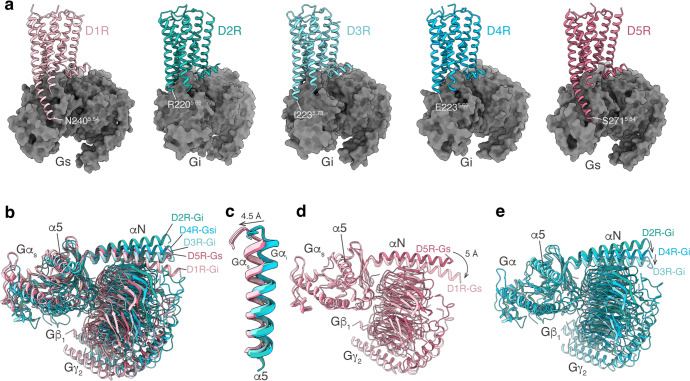
The interactions of the five subtypes of dopamine receptors with the respective G proteins display a relatively conserved mode as other aminergic receptors, with remarkable features that fit the TM5–TM6 switches for Gs and Gi/o selectivity.28 For the Gs-coupled D1R and D5R, their TM5 domains are extended into the cytoplasmic sides and end at residue 5.84, forming extensive interactions with the Gαs-Ras domain. In contrast, for the Gi-coupled D2R, D3R and D4R, their TM5 domains are not extended as those of D1R and D5R, which end at residue 5.69 (D2R and D4R) or 5.73 (D3R) (Fig. 7a). The TM5–Ras interactions are absent between the D2-like receptors and the Gi protein (Fig. 7a). This Gi- and Gs-coupling selectivity is consistent with the determinants of TM5–TM6 switches for the Gi and Gs selectivity as originally revealed in serotonin receptors.28 In addition to the differences in the receptor structures, several differences were also observed in the orientation of the G proteins between Gi and Gs complexes, with the α5 helices of Gαs subunit showing a 4.5-Å translocation from the Gi complexes (Fig. 7). At the ICL2 of receptors, the 34.51 residue is conserved as a hydrophobic residue and forms hydrophobic interactions with G protein by inserting its side chain into the cleft between αN and α5 of G protein (Supplementary information, Fig. S7a–f). The hydrophobic interactions between residue 34.51 and Gα cleft are conserved in all dopamine receptors and many other GPCRs.31,35–41 On the other hand, the rest of ICL2, which is not conserved in sequence, forms different interactions with the G proteins among the five dopamine receptors (Supplementary information, Fig. S7). Specifically, D1R residue E13234.54 forms unique polar interactions with Gs residue H41 (Supplementary information, Fig. S7b); D2R residue Y14634.57 forms unique polar interactions with Gi residue E28 (Supplementary information, Fig. S7d); and D3R residue H14034.55 forms unique polar interactions with the main chain of Gi residue A31 (Supplementary information, Fig. S7e). Together, these structural observations reveal common and unique features that determine G protein coupling specificity of dopamine receptors.
Fig. 7. G protein coupling of dopamine receptors.
a The structures of the D1R–Gs, D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes. b Comparison of the G protein conformations among the structures of D1R–Gs, D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes. c Structural comparison focused on the α5 helix of the Gα subunit bound to dopamine receptors. d Comparison of the G protein conformations among the structures of D1R–Gs and D5R–Gs complexes. e Comparison of the G protein conformations among the structures of D2R–Gi, D3R–Gi, and D4R–Gi complexes.
Concluding remarks
Here, we report the cryo-EM structures of all five dopamine receptors, D1R, D2R, D3R, D4R, and D5R, in complex with G proteins, among which the D4R and D5R structures are the first set of their active structures. The structures reveal a universal binding mode of the pan-agonist rotigotine in all five dopamine receptors and the specific intermolecular interactions that define the recognition of rotigotine by each of dopamine receptors. Structural and sequence comparisons indicate that the conserved OBP is the basis for promiscuous binding of rotigotine in all five dopamine receptors as well as in many other monoamine receptors, including receptor subtypes for serotonin, adrenergic amines, histamine, and muscarinic amines. Rotigotine is mainly prescribed for PD and RLS, with potential anti-depression effects from its cross reactivity of the serotonin receptor 5-HT1A (Fig. 4d). The structures and the binding results from this study therefore provide a rational basis for understanding the profound polypharmacology of rotigotine and its therapeutic effects.
The structures of all five dopamine receptors reveal a highly similar OBP where rotigotine binds, thus posing a great challenge to the design of subtype-specific orthosteric agonists. As an adjunct, one alternative strategy is to design subtype-specific allosteric modulators, such as LY315420, which is a D1R-specific PAM. Our structures reveal that D1R residues A4.41 and W3.52 are key to the selective agonism of LY3154207 on D1R. These structural observations provide critical insights into the PAM selectivity of LY315420 for D1R over D5R, and the basis for designing next generation PAMs targeting dopamine receptors.
The five dopamine receptor structures also reveal differential roles of cholesterol in dopamine signaling. Specifically, a number of cholesterol molecules are found in D1R, D4R and D5R, but not in D2R and D3R. Most of these cholesterol molecules are found to surround the extracellular half of the TMDs, in analogous to cholesterol molecules in the structures of 5-HT1A31 and class B GPCRs such as CRFR1, CRFR2,42 and PTH1R.43 The regulatory roles of cholesterol have been shown to be important in 5-HT1A31 and CRFR1/2.42 In this study, we also showed that cholesterol is involved in ligand binding. For example, the D4R structure reveals the mechanism of cholesterol-mediated regulation of ligand binding in D4R through an interaction network formed by a cholesterol molecule at the cleft of TM1 and TM7 (Fig. 6b). The location of this cholesterol in D4R is nearly identical to that in 5-HT1A, suggesting the conserved role of cholesterol in ligand binding in different monoamine GPCRs.
Through structural comparisons, we also uncovered conserved activation mechanisms of dopamine receptors and the detailed conformational changes during activation, as well as the basis of agonism and antagonism. We have additionally analyzed the selectivity and the unique features of all five dopamine receptors in G protein coupling. Together, our work presents the structural genomics of the human dopamine receptor system and provides structural templates for the development of selective or non-selective agonists, antagonists, and allosteric modulators of dopamine receptors, with potential significance for the treatment of CNS diseases.
Materials and methods
Cell lines
Spodoptera frugiperda (Sf9) and Trichoplusia ni (Hi5) cells were grown in ESF 921 medium (Expression Systems) at 27 °C and 120 rpm. HEKT cells were grown in a humidified 37 °C incubator with 5% CO2 using media supplemented with 100 IU/mL penicillin and 100 mg/mL streptomycin (Invitrogen). The human HEK293T cells were maintained in DMEM (VWR) containing 10% fetal bovine serum (FBS, VWR).
Constructs
The human WT D1R, D2R, D3R, D4R, or D5R (Supplementary information, Fig. S8) was cloned into the pFastBac (Thermo Fisher Scientific) vector using ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd). An N-terminal haemagglutinin (HA) signal sequence followed by a FLAG tag and a His tag was fused with the receptor proteins to facilitate expression and purification. A fragment of β2AR N-terminal tail region was fused in D1R, and BRIL was fused in D2R, D3R, D4R, and D5R as the fusion proteins. For the G proteins, dominant-negative (DN) mutations were induced in Gα subunits to decrease the affinity of nucleotide binding to the heterotrimer Gαβγ complex. For the D1R–Gs and D5R–Gs complexes, a mini-G format of Gαs was used. The mini-Gαs was generated by deleting the alpha-helical domain of Gαs and introducing stabilizing mutations under the previously reported sequence.44,45 Two DN mutations G226A and A366S were also introduced into the mini-Gαs.46 For the D2R–Gi and D3R–Gi complexes, a DN form of Gαi (DNGαi) was constructed by site-directed mutagenesis to incorporate mutations S47N, G203A, E245A, and A326S.20 For the D4R–Gi complex, a form of Gαsi construct with α5 helices from Gi and Ras-AHD domains from Gαs was used to obtain well-performed purifications. All these formats of Gα subunits, including mini-DNGαs, DNGαs, DNGαi, and DNGαsi as well as human Gβ1, Gγ2, and a single-chain antibody scFv1625,47 were cloned into the pFastBac vector.
Complex expression and purification
For the D1R–Gs complex, the recombinant baculoviruses of D1R, miniGαs, Gβ1 and Gγ2 were prepared individually following the manufacturer’s instructions about the Bac-to-Bac baculovirus expression system (Thermo Fisher Scientific). Prior to protein expression, Sf9 cell cultures were grown to cell density at ~4 × 106 cells/mL in ESF 921 serum-free medium (Expression Systems). Subsequently, the Sf9 cells were co-infected with the four types of baculoviruses prepared above at the ratio of 1:1:1:1. After infection for 48 h, the cultures were harvested and frozen at –80 °C for further usage. Before purification of the D1R–Gs signaling complex, the stabilizing nanobody, Nb35, was prepared through the previously described method,48 fast-frozen by liquid nitrogen and stored at –80 °C. For the purification of rotigotine–D1R–miniGs complex, cell pellet of 1 L culture was thawed at room temperature. The pellet was then resuspended in buffer containing 20 mM HEPES, pH 7.3, 75 mM NaCl, 5 mM CaCl2, 5 mM MgCl2, 10% Glycerol, 0.3 mM TCEP, protease inhibitor cocktail (Bimake, 1 mL/100 mL suspension). The protein complex was assembled on membrane by adding 100 μM rotigotine (TargetMol) and 10 μg/mL Nb35, which was added to stabilize the signaling complex. After incubation for half an hour, the suspension was treated with apyrase (25 mU/mL, NEB) and incubated for another 1 h at room temperature. The membrane in suspension was then solubilized by 0.5% Lauryl Maltose Neopentyl Glycol (LMNG, Anatrace), 0.1% (w/v) cholesteryl hemisuccinate TRIS salt (CHS, Anatrace), 0.025% (w/v) digitonin (Biosynth). The membrane was solubilized for 3 h at 4 °C before separation by ultracentrifugation at 100,000× g (Ti45, Beckman) for 45 min. The isolated supernatant was incubated for 2 h at 4 °C with pre-equilibrated FLAG resin (Smart-Lifesciences). Detergents were directly exchanged upon FLAG resin by two washing steps in buffer containing 20 mM HEPES, pH 7.3, 100 mM NaCl, 0.3 mM TCEP, 20 μM rotigotine, and supplemented with different detergents: first 0.01% LMNG, 0.002% CHS, 0.025% digitonin, then 0.015% LMNG, 0.005% glyco-diosgenin (GDN), 0.004% CHS, 0.025% digitonin for 10 column volumes, each. The protein complex was then eluted in buffer containing 20 mM HEPES, pH 7.3, 100 mM NaCl, 0.3 mM TCEP, 20 μM rotigotine, 0.015% LMNG, 0.005% GDN, 0.004% CHS, 0.025% digitonin, 200 μg/mL FLAG peptide. The eluted protein was concentrated to 0.5 mL by centrifugal filters with a 100 kDa molecular weight cut-off (Thermo Fisher Scientific) and then loaded onto a Superdex 200 10/300 GL Increase column (GE Healthcare). The separation column was pre-equilibrated and ran in buffer containing 20 mM HEPES, pH 7.3, 100 mM NaCl, 0.3 mM TCEP, 20 μM rotigotine, 0.00075% LMNG, 0.00025% GDN, 0.0002% CHS, 0.025% digitonin. Fractions of monomeric complex were collected and concentrated for electron microscopy experiments.
For the D2R–Gi, D3R–Gi, D4R–Gi complexes, the D2R/D3R/D4R, DNGαi/ DNGαsi, Gβ1, Gγ2, and scFv16 were co-expressed in Hi5 insect cells using the Bac-to-Bac Baculovirus Expression System (Invitrogen). The D5R, mini-DNGαs, Gβ1, Gγ2 were also co-expressed in Hi5 insect cells. In addition, the D1R, DNGαs, Gβ1, Gγ2 were co-expressed in Sf9 insect cells. Cell cultures were grown in ESF 921 medium (Expression Systems) to a density of 3 × 106 cell/mL and then infected with the different types of baculoviruses. Cell culture was collected by centrifugation 48 h post infection and stored at –80 °C until use.
For the purification of D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes, cell pellets were lysed by homogenization in 20 mM HEPES, pH 7.4, 20 mM KCl and 10 mM MgCl2 supplemented with Protease Inhibitor Cocktail (Bimake). The sample was centrifuged at 65,000× g for 30 min, then the membranes were re-suspended in 20 mM HEPES, pH 7.4, 100 mM NaCl, 20 mM KCl, 10 mM MgCl2, 5 mM CaCl2, 25 mU/mL Apyrase (Sigma) and 10 µM rotigotine. After incubation at room temperature for 1 h, the membranes were solubilized by addition of 0.5% (w/v) DDM (Anatrace) and 0.1% (w/v) CHS (Anatrace) for 2 h at 4 °C. The supernatant was cleared by centrifugation and incubated with TALON (Clontech) resin overnight. After binding, the resin was washed with 20 column volumes of 20 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) LMNG (Anatrace), 0.002% (w/v) CHS, 25 mM imidazole and 10 µM rotigotine. The complex was eluted with 5 column volumes of 20 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM MgCl2, 0.01% (w/v) LMNG, 0.002% (w/v) CHS, 250 mM imidazole and 10 µM rotigotine. The protein was then concentrated and loaded onto a Superdex 200 Increase 10/300 column (GE Healthcare) pre-equilibrated with buffer containing 20 mM HEPES, pH 7.4, 100 mM NaCl, 0.00075% (w/v) LMNG, 0.00025% (w/v) GDN (Anatrace), 0.0002% (w/v) CHS and 10 µM rotigotine. The fractions for the monomeric complex were collected and concentrated for electron microscopy experiments.
Cryo-EM grid preparation and data collection
For the preparation of cryo-EM grids, 3 μL of the purified complexes at 20 mg/mL for the D2R–rotigotine–Gi complex, 17 mg/mL for the D3R–rotigotine–Gi complex, 13 mg/mL for the D4R–rotigotine–Gi complex, 20 mg/mL for the D5R–rotigotine–Gs complex and 15 mg/mL for the D1R–rotigotine–Gs complex were applied onto a glow-discharged holey carbon grid (Quantifoil R1.2/1.3). Grids were plunge-frozen in liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific). Frozen grids were transferred to liquid nitrogen and stored for data acquisition. For the D1R–Gs complex, D3R–Gi complex, D4R–Gi complex, and D5R–Gs complex, automatic data collection was performed on a Titan Krios equipped with a Gatan K3 direct electron detector in the Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences (Shanghai, China). Cryo-EM imaging was performed, and micrographs were recorded in counting mode at a dose rate of ~8.0 e/Å2/s with a defocus ranging from –1.0 μm to –3.0 μm using the SerialEM software.49 The total exposure time was 8 s and 40 frames were recorded per micrograph. A total of 6301, 5156, 3562 and 4746 movies were collected for D1R–Gs complex, D3R–Gi complex, D4R–Gi complex, and D5R–Gs complex, respectively. For the D2R–Gi complex, automatic data collection was performed on a Titan Krios at 300 kV using Gatan K2 Summit detector in the Center of Cryo-Electron Microscopy, Zhejiang University (Hangzhou, China). Cryo-EM imaging was performed, and micrographs were recorded in counting mode at a dose rate of ~8.0 e/Å2/s with a defocus ranging from –1.0 μm to –3.0 μm using the SerialEM software.49 The total exposure time was 8 s and 40 frames were recorded per micrograph. A total of 5324 movies were collected for D2R–Gi complex.
Image processing and map construction
Dose-fractionated image stacks were aligned using MotionCor2.1.50 Contrast transfer function (CTF) parameters for each micrograph were estimated by Gctf.51 Cryo-EM data processing was performed using RELION-3.0-beta2.52
For the D1R–Gs complex, particle selections for 2D and 3D classifications were performed on a binned dataset with a pixel size of 1.60 Å. Automated particle picking yielded 2,685,434 particles that were subjected to reference-free 2D classification to discard poorly defined particles, producing 2,145,182 particles. After 6 rounds of 3D classification, a well-defined subset containing 448,516 particles was used to obtain the final map using a pixel size of 0.80 Å. Further refinement produced a final map with an indicated global resolution of 3.2 Å at a Fourier shell correlation (FSC) of 0.143.
For the D2R–Gi complex, particle selections for 2D and 3D classifications were performed on a binned dataset with a pixel size of 2.09 Å. Automated particle picking yielded 7,064,860 particles that were subjected to reference-free 2D classification to discard poorly defined particles. After 2 rounds of 3D classification, two well-defined subsets were selected. The selected subsets were subsequently subjected to 2 rounds of 3D classification with a mask on the receptor. One subset showing the high-quality receptor density was selected, producing 140,237 particles. The selected subset was subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 3.0 Å at an FSC of 0.143.
For the D3R–Gi complex, particle selections for 2D and 3D classifications were performed on a binned dataset with a pixel size of 2.142 Å. Automated particle picking yielded 8,770,602 particles that were subjected to reference-free 2D classification to discard poorly defined particles. After 3 rounds of 3D classification, three well-defined subsets with 1,786,008 particles were selected and subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 2.7 Å at an FSC of 0.143.
For the D4R–Gi complex, particle selections for 2D and 3D classifications were performed on a binned dataset with a pixel size of 2.16 Å. Automated particle picking yielded 4,333,829 particles that were subjected to reference-free 2D classification to discard poorly defined particles. After 3D classification, two well-defined subsets were selected and subsequently subjected to 3D classification with a mask on the receptor. Two subsets showing the high-quality receptor density were selected, producing 471,638 particles. The selected subsets were subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 3.2 Å at an FSC of 0.143.
For the D5R–Gs complex, particle selections for 2D and 3D classifications were performed on a binned dataset with a pixel size of 2.142 Å. Automated particle picking yielded 7,900,346 particles that were subjected to reference-free 2D classification to discard poorly defined particles. After 2 rounds of 3D classification, one well-defined subset was selected and subsequently subjected to additional 4 rounds of 3D classification. Four subsets showing the high-quality receptor density were selected, producing 2,652,297 particles. The selected subsets were subsequently subjected to 3D refinement, CTF refinement, and Bayesian polishing. The final refinement generated a map with an indicated global resolution of 3.1 Å at an FSC of 0.143. Local resolution was determined using the Bsoft53 package with half maps as input maps.
Model building and refinement
The structure of the D1R–Gs–Apomorphine complex (PDB: 7JVQ) was used as the initial model for model rebuilding and refinement against the electron microscopy maps of D1R–Gs–rotigotine and D5R–Gs–rotigotine complexes. The structure of the D2R–Gi–Bromocriptine complex (PDB: 7JVR) was used as the initial model for model rebuilding and refinement against the electron microscopy maps of D2R–Gi–rotigotine complexes. The structure of the D3R–Gi–PD128907 complex (PDB: 7CMV) was used as the initial model for model rebuilding and refinement against the electron microscopy maps of the D3R–Gi–rotigotine and the D4R–Gi–rotigotine complexes. The model was docked into the electron microscopy density map using Chimera,54 followed by iterative manual adjustment and rebuilding in COOT55 and ISOLDE.56 Real space and reciprocal space refinements were performed using Phenix programs.57 The model statistics were validated using MolProbity.58 Structural figures were prepared in Chimera, ChimeraX59 and PyMOL (https://pymol.org/2/). The final refinement statistics are provided in Supplementary information, Table S1.
Radioligand binding assays
Binding assays were performed using membranes from HEK293T (ATCC CRL-11268) cells transiently expressing WT dopamine receptors. For D1R and D5R, binding assays were set up in 96-well plates in standard binding buffer (50 mM HEPES, 50 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, pH 7.4). Saturation binding assays with 0.5–5 nM [3H]-SCH23390 (Perkin-Elmer) in standard binding buffer were performed to determine Kd and Bmax, whereas 10 μM final concentration of Butaclamol was used to define nonspecific binding. For D2R, D3R, and D4R, binding assays were set up in 96-well plates in standard binding buffer (50 mM Tris, 0.1 mM EDTA, 10 mM MgCl2, 0.1% (w/v) BSA, pH 7.40). Saturation binding assays with 0.5–5 nM [3H]Methylspiperone (Perkin-Elmer) in standard binding buffer were performed to determine Kd and Bmax, whereas 10 μM final concentration of Chlorpromazine was used to define nonspecific binding. All reactions were incubated for 2 h at room temperature in the dark and terminated by rapid vacuum filtration onto chilled 0.3% PEI-soaked GF/A filters (Perkin-Elmer) followed by three quick washes with cold washing buffer (50 mM Tris HCl, pH 7.40). Radioactivity counts were determined using a Wallac Trilux MicroBeta counter (Perkin-Elmer). Results were analyzed using GraphPad Prism 8.4 (Graphpad Software Inc., San Diego, CA) using “One site -- Total and nonspecific binding”. Competition assays were performed similarly to saturation binding assays except that various concentrations of competitor were premixed with [3H]-SCH23390 or [3H]Methylspiperone (Perkin-Elmer) near the pre-determined Kd and then incubated for 2 h at room temperature in the dark with membranes from HEK293T (ATCC CRL-11268) cells transiently expressing WT receptors. Results were analyzed using GraphPad Prism 8.4 (Graphpad Software Inc., San Diego, CA) using “One site – Fit Ki”.
Gs-mediated Gs-cAMP accumulation assay
For receptors D1R and D5R, Gs-mediated Gs-cAMP accumulation assays were performed with HEK293T (ATCC CRL-11268) cells transiently expressing human D1R or D5R WT or mutants along with the cAMP biosensor GloSensor-22F (Promega). Cells were seeded (20,000 cells/35 μL/well) into 384-well white clear-bottom, tissue culture plates in DMEM containing 1% (v/v) dialyzed FBS. Next day, 3× drug dilutions were diluted in HBSS, 20 mM N-(2-hydroxyethyl) piperazine-N′-ethanesulfonic acid (HEPES), 0.3% (w/v) bovine serum albumin (BSA), 0.03% (w/v) ascorbic acid, pH 7.4. Medium was decanted from 384-well plates and 20 μL of drug buffer (HBSS, 20 mM HEPES, pH 7.4) containing GloSensor reagent was added per well and allowed to equilibrate for at least 15 min at room temperature. Cells were then treated with 10 μL per well of 3× drug using a FLIPR (Molecular Devices). After 15 min, Gs-cAMP accumulation was read on a TriLux Microbeta (PerkinElmer) plate counter. For receptors D2R, D3R, and D4R, Gs-mediated Gs-cAMP accumulation assays were performed as above, except in the inhibition mode, a final concentration of 100 nM isoproterenol was added to the cells 15 min prior to the addition of the drug. Data were analyzed using the sigmoidal log(agonist) vs dose response or sigmoidal log(inhibitor) vs dose response function built into GraphPad Prism 8.4.
Surface expression analysis
Surface expression determination of WT receptors and mutants was performed using HEK293T cells (ATCC CRL-11268) maintained in DMEM containing 10% (v/v) dialyzed FBS, 1 IU/mL Penicillin G, and 100 μg/mL Streptomycin. Cells were passed to 6-well plates (Genesee Scientific, Cat# 25-106MP) and transfected using TransIT (Mirus Bio) and 0.4 μg of the given receptor. After at least 24 h, transfected cells were plated in poly-l-lysine-coated 96-well white clear-bottom cell culture plates (Greiner Bio-One) in plating media (DMEM containing 1% (v/v) dialyzed FBS, 1 IU/mL Penicillin G, and 100 μg/mL Streptomycin) at a density of 20,000 cells in 200 μL per well and incubated overnight. The following day, the medium was aspirated and cells were washed twice with 200 μL of 1× phosphate buffered saline (PBS). Then 100 μL of 1× PBS containing 5% (w/v) BSA was added to each well and incubated at room temperature. After 30 min, 100 μL of 1:10,000 anti-HA HRP conjugate (Sigma-Aldrich, Cat# A8592) was added to each well. After an additional 30 min, the medium was aspirated and cells were washed twice with 200 μL of 1× PBS. Chemiluminescence was observed by the addition of 50 μL of HRP substrate (Thermo Fisher Scientific, Cat# 37069) and counted using a Wallac Trilux MicroBeta counter (Perkin-Elmer). Chemiluminescence values were normalized to WT receptor and graphed as a percentage of WT using Graphpad Prism 8 (Graphpad Software Inc., San Diego, CA). Part of the surface expression data has been published in our previous paper.11
PRESTO-Tango GPCRome screening
Screening of the compounds in the PRESTO-Tango GPCRome was performed as previously described60 with slight modifications. First, HTLA cells were plated in poly-l-lysine-coated 384-well white plates in DMEM containing 1% dialyzed FBS for 6 h. Next, the cells were transfected with 20 ng per well PRESTO-Tango receptor DNAs overnight. The cells were then treated with 10 µM rotigotine without changing the medium and incubated for another 24 h. Each target was designed to have four wells for basal and four wells for sample. The remaining steps of the PRESTO-Tango protocol were followed. The results were plotted as fold change in the average basal signaling activity against individual receptors in GraphPad (v.9.0). Selective receptors were repeated as a full dose–response assay to confirm activity.
Molecular docking
The D1R and D5R cryo-EM structures were used for docking. The receptors were separated from the complex and prepared in the protein preparation wizard of Schrödinger, Maestro. We first assigned bond orders and add hydrogens to the protein. Meanwhile, disulfide bonds were created and residue het states were defined using Epik at pH = 7.0 ± 2.0. PROPKA was then applied to assign residue protonation states. The grid files for docking were generated according to the ligand-binding pocket. At last, rotigotine was docked to the grid files in standard precision of the glide program. The docking score was the score for the best-matching ligand pose.
Supplementary information
Acknowledgements
The cryo-EM data were collected at the Center of Cryo-Electron Microscopy, Shanghai Institute of Materia Medica, the Center of Cryo-Electron Microscopy, Zhejiang University, and the Cryo-Electron Microscopy Facility, Zhejiang University Medical Center/Liangzhu laboratory. This work was partially supported by the National Key R&D Programs of China (2018YFA0507002), Shanghai Municipal Science and Technology Major Project (2019SHZDZX02 and XDB08020303) to H.E.X.; the National Key Basic Research Program of China (2019YFA0508800), the Key R&D Projects of Zhejiang Province (2021C03039) and Fundamental Research Funds for the Central Universities (2019XZZX001-01-06) to Yan Z.; the Zhejiang Province Natural Science Fund for Excellent Young Scholars (LR22C050002) and the National Natural Science Foundation of China (32100959) to C.M.; the National Natural Science Foundation of China (31770796) and the National Science and Technology Major Project (2018ZX09711002) to Y.J.; grants from the NIMH Psychoactive Drug Screening Program to X.-P.H., Y.L., B.L.R., and RO1MH112205 to B.E.K. and B.L.R. The Special Research Assistant Project of Chinese Academy of Sciences to Youwen Z.
Author contributions
P.X. and S.H. designed the expression constructs, purified the complexes, and prepared protein samples for the D2R–Gi, D3R–Gi, D4R–Gi, and D5R–Gs complexes for cryo-EM data collection. P.X. and S.H. performed cryo-EM grid preparation, data acquisition, structure determination, and prepared the draft of the manuscript and figures. Youwen Z. designed the constructs, prepared the protein samples, conducted cryo-EM data collection and structure determination of D1R–Gs, and participated in the preparation of supplementary figures and manuscript editing. C.M. screened the cryo-EM conditions, prepared the cryo-EM grids, collected cryo-EM images, processed the EM data of the D2R–Gi complex, and participated in the preparation of supplementary figures. P.X. built the models and refined the structures. Yumu Z. and H.L. participated in the sample preparation and screening of the D4R–Gi and the D5R–Gs complexes. Y.W. participated in the sample preparation and screening of the D1R–Gs complex. B.E.K., X.-P.H., and Y.-F.L. performed cAMP, GPCRome, Tango, and radioligand binding assays. B.E.K. compiled assay data and participated in the preparation of the manuscript. X.H. performed the docking studies. W.Y. designed the Gα constructs used for the generation of the D4R–Gi complex. Y.J. participated in the funding acquisition. Yan Z. supervised C.M. and participated in manuscript editing. B.L.R. supervised pharmacological and mutagenesis experiments and participated in manuscript writing. H.E.X. conceived and supervised the project and wrote the manuscript with P.X.
Data availability
Density maps and structure coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with accession codes EMD-35683 and 8IRR for the D1R–Gs–rotigotine complex; EMD-35684 and 8IRS for the D2R–Gi–rotigotine complex; EMD-35685 and 8IRT for the D3R–Gi–rotigotine complex; EMD-35686 and 8IRU for the D4R–Gi–rotigotine complex; EMD-35687 and 8IRV for the D5R–Gs–rotigotine complex.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Peiyu Xu, Sijie Huang, Brian E. Krumm, Youwen Zhuang, Chunyou Mao.
Contributor Information
Yan Zhang, Email: zhang_yan@zju.edu.cn.
Bryan L. Roth, Email: bryan_roth@med.unc.edu
H. Eric Xu, Email: eric.xu@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00808-0.
References
- 1.Robbins TW. Dopamine and cognition. Curr. Opin. Neurol. 2003;16:S1–S2. doi: 10.1097/00019052-200312002-00001. [DOI] [PubMed] [Google Scholar]
- 2.Volkow ND, et al. Association between decline in brain dopamine activity with age and cognitive and motor impairment in healthy individuals. Am. J. Psychiatry. 1998;155:344–349. doi: 10.1176/ajp.155.3.344. [DOI] [PubMed] [Google Scholar]
- 3.Koob GF. Dopamine, addiction and reward. Semin. Neurosci. 1992;4:139–148. [Google Scholar]
- 4.Neve, K. A. & Neve, R. L. Molecular biology of dopamine receptors. In: Neve, K. A. & Neve, R. L. (eds) The Dopamine Receptors. The Receptors. (Humana Press, Totowa, 1997).
- 5.Bonuccelli U, Del Dotto P, Rascol O. Role of dopamine receptor agonists in the treatment of early Parkinson’s disease. Parkinsonism Relat. Disord. 2009;15:S44–S53. doi: 10.1016/S1353-8020(09)70835-1. [DOI] [PubMed] [Google Scholar]
- 6.Martel JC, Gatti McArthur S. Dopamine receptor subtypes, physiology and pharmacology: new ligands and concepts in schizophrenia. Front. Pharmacol. 2020;11:1003. doi: 10.3389/fphar.2020.01003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faraone SV, Biederman J. Neurobiology of attention-deficit hyperactivity disorder. Biol. Psychiatry. 1998;44:951–958. doi: 10.1016/s0006-3223(98)00240-6. [DOI] [PubMed] [Google Scholar]
- 8.Mao Q, Qin W-Z, Zhang A, Ye N. Recent advances in dopaminergic strategies for the treatment of Parkinson’s disease. Acta Pharmacol. Sin. 2020;41:471–482. doi: 10.1038/s41401-020-0365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reynolds NA, Wellington K, Easthope SE. Rotigotine. CNS Drugs. 2005;19:973–981. doi: 10.2165/00023210-200519110-00006. [DOI] [PubMed] [Google Scholar]
- 10.Bogan RK. From bench to bedside: an overview of rotigotine for the treatment of restless legs syndrome. Clin. Ther. 2014;36:436–455. doi: 10.1016/j.clinthera.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang Y, et al. Structural insights into the human D1 and D2 dopamine receptor signaling complexes. Cell. 2021;184:931–942.e18. doi: 10.1016/j.cell.2021.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu P, et al. Structures of the human dopamine D3 receptor-Gi complexes. Mol. Cell. 2021;81:1147–1159.e4. doi: 10.1016/j.molcel.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Yin J, et al. Structure of a D2 dopamine receptor–G-protein complex in a lipid membrane. Nature. 2020;584:125–129. doi: 10.1038/s41586-020-2379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Im D, et al. Structure of the dopamine D2 receptor in complex with the antipsychotic drug spiperone. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-20221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, et al. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science. 2017;358:381–386. doi: 10.1126/science.aan5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao P, et al. Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes. Cell. 2021;184:943–956.e18. doi: 10.1016/j.cell.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng X, et al. Ligand recognition and biased agonism of the D1 dopamine receptor. Nat. Commun. 2022;13:3186. doi: 10.1038/s41467-022-30929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y-L, et al. Dominant negative G proteins enhance formation and purification of agonist-GPCR-G protein complexes for structure determination. ACS Pharmacol. Transl. Sci. 2018;1:12–20. doi: 10.1021/acsptsci.8b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu P, et al. The structural basis of the dominant negative phenotype of the Gαi1β1γ2 G203A/A326S heterotrimer. Acta Pharmacol. Sin. 2016;37:1259–1272. doi: 10.1038/aps.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nehmé R, et al. Mini-G proteins: novel tools for studying GPCRs in their active conformation. PLoS One. 2017;12:e0175642. doi: 10.1371/journal.pone.0175642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan J, et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun. 2020;11:4121. doi: 10.1038/s41467-020-17933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda S, et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 2018;9:1–9. doi: 10.1038/s41467-018-06002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang Y, et al. Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor. Cell Res. 2021;31:593–596. doi: 10.1038/s41422-021-00482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun B, et al. Crystal structure of dopamine D1 receptor in complex with G protein and a non-catechol agonist. Nat. Commun. 2021;12:3305. doi: 10.1038/s41467-021-23519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, et al. GPCRs steer Gi and Gs selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors. Mol. Cell. 2022;82:2681–2695.e6. doi: 10.1016/j.molcel.2022.05.031. [DOI] [PubMed] [Google Scholar]
- 29.Reichmann H, et al. Ergoline and non-ergoline derivatives in the treatment of Parkinson’s disease. J. Neurol. 2006;253:IV36–IV38. doi: 10.1007/s00415-006-4009-z. [DOI] [PubMed] [Google Scholar]
- 30.Hao J, et al. Synthesis and pharmacological characterization of 2-(2,6-dichlorophenyl)-1-((1S,3R)-5-(3-hydroxy-3-methylbutyl)-3-(hydroxymethyl)-1-methyl-3,4-dihydroisoquinolin-2(1H)-yl)ethan-1-one (LY3154207), a potent, subtype selective, and orally available positive allosteric modulator of the human dopamine D1 Receptor. J. Med. Chem. 2019;62:8711–8732. doi: 10.1021/acs.jmedchem.9b01234. [DOI] [PubMed] [Google Scholar]
- 31.Xu P, et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature. 2021;592:469–473. doi: 10.1038/s41586-021-03376-8. [DOI] [PubMed] [Google Scholar]
- 32.Weis WI, Kobilka BK. The molecular basis of G protein–coupled receptor activation. Ann. Rev. Biochem. 2018;87:897. doi: 10.1146/annurev-biochem-060614-033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manglik A, Kruse AC. Structural basis for G protein-coupled receptor activation. Biochemistry. 2017;56:5628–5634. doi: 10.1021/acs.biochem.7b00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Cao C, He L, Wang X, Zhang XC. Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870. Elife. 2019;8:e48822. doi: 10.7554/eLife.48822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu P, et al. Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119. Nat. Struct. Mol. Biol. 2022;29:863–870. doi: 10.1038/s41594-022-00816-5. [DOI] [PubMed] [Google Scholar]
- 36.Huang S, et al. Structural basis for recognition of anti-migraine drug lasmiditan by the serotonin receptor 5-HT1F-G protein complex. Cell Res. 2021;31:1036–1038. doi: 10.1038/s41422-021-00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen SGF, et al. Crystal structure of the beta(2) adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan J, et al. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature. 2022;609:854–859. doi: 10.1038/s41586-022-05173-3. [DOI] [PubMed] [Google Scholar]
- 39.Duan J, et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature. 2021;598:688–692. doi: 10.1038/s41586-021-03924-2. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, et al. Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor. Nat. Commun. 2021;12:5064. doi: 10.1038/s41467-021-25364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xing C, et al. Cryo-EM structure of the human cannabinoid receptor CB2-Gi signaling complex. Cell. 2020;180:645–654.e13. doi: 10.1016/j.cell.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma S, et al. Molecular basis for hormone recognition and activation of corticotropin-releasing factor receptors. Mol. Cell. 2020;77:669–680.e4. doi: 10.1016/j.molcel.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Zhao LH, et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science. 2019;364:148–153. doi: 10.1126/science.aav7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter B, Nehme R, Warne T, Leslie AG, Tate CG. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Nafria J, Lee Y, Bai X, Carpenter B, Tate CG. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. Elife. 2018;7:e35946. doi: 10.7554/eLife.35946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P, et al. The structural basis of the dominant negative phenotype of the Galphai1beta1gamma2 G203A/A326S heterotrimer. Acta Pharmacol. Sin. 2016;37:1259–1272. doi: 10.1038/aps.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeda S, et al. Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 2018;9:3712. doi: 10.1038/s41467-018-06002-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardon E, et al. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heymann JB. Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 2018;27:159–171. doi: 10.1002/pro.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Croll TI. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallor. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2020;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015;22:362–369. doi: 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Density maps and structure coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with accession codes EMD-35683 and 8IRR for the D1R–Gs–rotigotine complex; EMD-35684 and 8IRS for the D2R–Gi–rotigotine complex; EMD-35685 and 8IRT for the D3R–Gi–rotigotine complex; EMD-35686 and 8IRU for the D4R–Gi–rotigotine complex; EMD-35687 and 8IRV for the D5R–Gs–rotigotine complex.