Abstract
Purpose:
Ring sideroblasts (RS) define the low-risk myelodysplastic neoplasm (MDS) subgroup with RS but may also reflect erythroid dysplasia in higher risk myeloid neoplasm. The benign behavior of MDS with RS (MDSRS+) is limited to SF3B1-mutated cases without additional high-risk genetic events, but one third of MDSRS+ carry no SF3B1 mutation, suggesting that different molecular mechanisms may underlie RS formation. We integrated genomic and transcriptomic analyses to evaluate whether transcriptome profiles may improve current risk stratification.
Experimental Design:
We studied a prospective cohort of MDSRS+ patients irrespective of World Health Organization (WHO) class with regard to somatic mutations, copy-number alterations, and bone marrow CD34+ cell transcriptomes to assess whether transcriptome profiles add to prognostication and provide input on disease classification.
Results:
SF3B1, SRSF2, or TP53 multihit mutations were found in 89% of MDSRS+ cases, and each mutation category was associated with distinct clinical outcome, gene expression, and alternative splicing profiles. Unsupervised clustering analysis identified three clusters with distinct hemopoietic stem and progenitor (HSPC) composition, which only partially overlapped with mutation groups. IPSS-M and the transcriptome-defined proportion of megakaryocyte/erythroid progenitors (MEP) independently predicted survival in multivariable analysis.
Conclusions:
These results provide essential input on the molecular basis of SF3B1-unmutated MDSRS+ and propose HSPC quantification as a prognostic marker in myeloid neoplasms with RS.
Translational Relevance.
MDS with ring sideroblasts (RS) and low myeloblast count is considered a lower-risk entity according to World Health Organization classifications, while the International Consensus Classification demands presence of SF3B1 mutation. Notably, RS are also found in SF3B1 wild-type (WT) myeloid neoplasms with or without increased blast, but molecular studies on these subtypes are lacking. We carried out a comprehensive evaluation of genomic and transcriptomic profiles of a prospective cohort of 129 MDSRS+ irrespective of classification. We identified SF3B1, SRSF2, or TP53multi-hit mutations as distinct disease-modifying entities in 89% of MDSRS+, and showed that outcome differed significantly between categories, highlighting the importance of identifying TP53multi-hit status also in MDSRS+. Transcriptome-defined hematopoietic progenitor signatures predicted survival independently from mutational categories and IPSS-M in multivariable analysis, proposing hematopoietic progenitor quantification as a valuable prognostic marker in myeloid neoplasms.
Introduction
Ring sideroblasts (RS) are erythroblasts with iron-loaded mitochondria visualized by Prussian blue staining (Perls’ reaction) as a perinuclear ring of blue granules (1) and constitute a hallmark of myelodysplastic syndrome (MDS) with RS (MDSRS+). Missense somatic mutations in the SF3B1 gene, a core component of the splicing factor machinery, are identified in approximately 80% of MDS-ring sideroblast with single/multilineage dysplasia (MDS-RS-SLD/MLD; refs. 2, 3) according to the 2016 World Health Organization (WHO) classification (4). The identification of SF3B1 mutations in MDSRS+ paved the way for characterization of the pathobiology of splicing factor mutated MDS (5–8) and iron metabolism dysregulation in RS (9, 10). On the basis of this evidence, the recent International Consensus Classification (ICC; ref. 11) as well as the WHO classification (12) recognize MDS with low blasts and SF3B1 mutation as a distinct nosological entity because of its well-defined molecular, clinical and biological characteristics (13). However, according to the 2022 WHO criteria, the detection of ≥15% RS may substitute for SF3B1 mutation, which makes the new systems diverge on wild-type SF3B1 MDS with low blasts and ring sideroblasts classification. This is relevant because the benign behavior of MDS-RS is limited to SF3B1-mutated cases (14). Indeed, RS can also be a marker of erythroid dysplasia in bone marrow (BM) smears of MDS with excess blasts, myeloproliferative neoplasms (MPN), MDS/MPN and acute myeloid leukemia (AML) as well as nonclonal and transitory conditions (e.g., exposure to isoniazid, linezolid, ethanol, or lead poisoning; ref. 15).
Although a strong association between RS and SF3B1 mutation has been confirmed throughout the spectrum of myeloid neoplasms, a large proportion of cases (up to 10% in MDS/MPN-RS and thrombocytosis; MDS/MPN-RS-T, 20% in MDS-RS-SLD/MLD, 90% in AML with RS) carry no SF3B1 mutation, suggesting that different molecular mechanisms may underlie RS formation (2, 16–20). The well-documented causative relationship between SF3B1 mutation and RS formation in MDS-RS-SLD/MLD is elicited by ABCB7 downregulation and mitochondrial iron accumulation, but little is known about the molecular pathophysiology of SF3B1-unmutated MDS-RS-SLD/MLD, including whether ABCB7 plays a role also in RS formation in SF3B1-unmutated MDS-RS-SLD/MLD (9, 10). Furthermore, a transcriptomic signature enriched for genes specific for erythroid and megakaryocytic progenitors has been to be associated with SF3B1-mutated MDS-RS-SLD/MLD and favorable prognosis in MDS, while no evidence is available on SF3B1-unmutated MDS-RS-SLD/MLD transcriptomics and molecular determinants of its widely variable clinical course (2, 7, 14).
The recently published Molecular International Prognostic Scoring System (IPSS-M) identified different risk groups based on previous revised-IPSS criteria plus comprehensive targeted DNA-sequencing data (DNA-seq; ref. 21). An important question for this study was therefore to assess whether IPSS-M compensates for the questions raised above, and whether transcriptomic profiling adds a distinct new dimension to the understanding and prognostication of MDSRS+.
We analyzed a large cohort of MDSRS+ irrespective of WHO categorization, and integrated somatic mutations, copy-number variation (CNV) and CD34+ BM mononuclear cells (MNC) transcriptome profiles at the time of diagnosis, assessing whether transcriptome profiles add to prognostication and provide input to disease classification.
Materials and Methods
Patients and samples
We studied a cohort of 129 MDSRS+ identified within a population of 834 myeloid neoplasms (682 MDS, 51 AML with myelodysplastic-related changes, 101 MDS/MPN) evaluated at Karolinska University Hospital in Stockholm between February 2004 and August 2020 for suspected MDS. Diagnostic procedures were performed according to the recommendations of the European LeukemiaNet and WHO organization (4, 13). All cases were reevaluated by a central pathologist (B. Sander). MNCs from BM were collected at diagnosis and cryopreserved at Karolinska Institutet MDS Biobank following standard operating procedure. RS were deemed present if percentage of RS was ≥ 5% of total nucleated erythroid cells in BM smears. Ten healthy individuals were included in the study as normal BM (NBM) controls for the comparative transcriptomic analysis. Diagnostic criteria were updated according to 2016 revision of WHO classification of hematopoietic tumors. Karolinska Institutet MDS Biobank was started on 2001 and received primary human BM samples from Stockholm area investigated for MDS or MDS/MPN. Considering the evolving classification of myeloid neoplasms over the last two decades, refractory anemia with excess blast in transformation (reclassified as AML since 2008) were also referred to the Biobank and therefore included into this study. For the same reason, cases with suspected MPN were not included in this study. Central pathology review according to the last WHO classification was carried out and diagnoses were confirmed at multiprofessional conference. Extensive clinical and survival information was available for all patients and retrieved from national electronic chart system. Cases that received any malignant hematologic-related treatment except from Erythropoiesis Stimulating Agents (ESA) prior biobanking were excluded from the study. Three additional MDS cases without RS (MDSRS−) were also included into the study as control in to validate results from digital sorting (Supplementary Table S2). Cryopreserved MNCs were thawed in RPMI1640 Glutamax + 20% FBS + 100 U/mL DNase I (Merck) with a median cell viability of 92% (range 80–95). This study was approved by the local ethics committee at Karolinska Institutet (Stockholm, Sweden). All the procedures followed were in accordance with the Helsinki Declaration of 1975, regularly revised up to the last 2013 version. All samples were collected after written informed consent was obtained from patients and healthy donors. SRSF2 and TP53MH MDSRS+ overall survival (OS) was compared with historical MDS controls without RS from two recent large studies evaluating clinical characteristics of SRSF2 and TP53MH-mutated myeloid neoplasms (22, 23), to explore the outcome of SF3B1-unmutated MDSRS+ stratified by RS amount.
Targeted capture DNA-seq
Genomic DNA was extracted from MNC following standard protocols for human tissue in all cases and controls and submitted for DNA-seq. A validated targeted sequencing panel was used to evaluate 152 genes recurrently mutated in MDS, as well as 1,118 genome-wide SNP probes for copy-number analysis, as previously reported (21, 23). Libraries were sequenced with paired-end Illumina HiSeq with a 100 or 125 bp read length, with a median coverage of 1,241× per sample (range, 338–1,956×). Raw sequence data were aligned to the human genome (GRCh37).
RNA sequencing
CD34+ cells (median 0.4 × 106, range 0.1–0.5 × 106) were isolated from the MNCs using AUTO-MACS with double-separation option (Miltenyi Biotec) and submitted for RNA extraction for all cases and controls. RNA was extracted with RNeasy Microkit (Qiagen) and treated with DNase, according to the manufacturer's instructions. RNA integrity number was estimated using Agilent RNA 6000 Pico (Agilent Technologies) and was greater than 6.5 for all the samples (median 8.2). The RNA-sequencing (RNA-seq) libraries were prepared from total RNA using SMARTer Stranded Total RNA-Seq Kit v2 Pico Input Mammalian with enzymatic ribosomal depletion (Takara Bio). Libraries were sequenced using the Novaseq 6,000 with paired-end 150-bp configuration. The reads were processed with nf-core/rnaseq pipeline (ref. 24; version 1.0), using human genome GRCh37 for STAR alignment. Uniquely mapped read pairs were counted using featureCounts (ref. 25; version 1.6.3) and used for gene expression analysis. Differential expression analysis was performed with DESeq2 (26) version 1.26. Genes with raw read counts <50 in less than 10% of samples were filtered out. Raw read counts were normalized by DESeq2 by applying variance stabilizing transformation and used for the downstream unsupervised clustering analysis after logarithmic transformation. The significance of differential expression among groups was calculated using likelihood ratio test including sequencing batch in the reduced model. Genes with false discovery rate Padj (FDR) < 0.001 were considered as significantly differentially expressed. ClusterProfiler (27), Reactome PA (28) version 3.14, and fgsea (29) version 3 R packages were used for Gene Ontology (GO), pathway and gene-set enrichment analyses. Differential splicing analysis was assessed using rMATS turbo (ref. 30; version 4.1) on BAM alignments, comparing disease subgroups against NBM controls. Splicing events with FDR < 0.0001 and absolute delta-PSI > 0.1 were considered statistically significant.
Flow cytometry and HSPC sorting
MNCs isolated from cryopreserved BM samples were washed twice with PBS (Thermo Fisher Scientific) with 5% FBS before staining. Cells were sorted on a FACS ARIA II Fusion (Becton Dickinson) at the WIRM FACS facility at the Karolinska Institutet, Flemingsberg (Huddinge, Sweden). All experiments included fluorescent-minus-one (FMO) and single-stained controls. A previously described strategy was used to define the HSPC populations (31).
Clonal hierarchy analysis
The bioinfomatic analysis of hierarchical rank of detected mutations was carried out with both DPClust (22, 32) and Pyclone (33). Mutations belonging to the largest clone were tagged as dominant and distinguished from other secondary mutations harbored by smaller subclones. Single-cell–derived hematopoietic stem and progenitor colonies from SF3B1-SRSF2 comutated patients were obtained as previously described (22). DNA from each single-cell–derived colony underwent droplet digital PCR (ddPCR) using mutation-specific primers, according to the manufacturer's instructions, as previously described (34).
Unsupervised clustering analysis
Consensus clustering with partitioning around medoids algorithm and binary distance measures was performed using genetic features, the latter considered as binary variable by means of ConsensusClusterPlus (version 1.50; refs. 35–37). The same methods with K-means algorithm and Euclidean distance was used to carry out unsupervised clustering analysis of scaled normalized gene expression counts (7).
Digital sorting on RNA-seq CD34+ MNC
To further explore composition of bulk CD34+ MNC, CIBERSORTx algorithm was used to provide a digital estimation of hemopoietic stem and progenitor cells (HSPC; ref. 38). Publicly available single-cell RNA-seq data representative of HSPC purified from CD34+ BM MNCs (39, 40) were used to create a signature matrix and quantify hemopoietic precursors using the following parameters: κ = 13, q-value = 10−5, number of barcode genes = 3–50, minimum expression = 1, replicates = 30, sampling = 0.5. Only genes shared between the single cell and MDSRS+ bulk RNA-seq data were selected. Normalized gene expression counts were therefore used to quantify hemopoietic precursors with the following settings: absolute mode, B-mode batch correction and 500 permutations. Only outputs with Monte Carlo sampling empirical P < 0.05 for the deconvolution were considered for further analysis.
Statistical analysis
Categorical variables were compared using χ2 test or Fisher exact test, in accordance with variable characteristics. Numerical variables were compared using Mann–Whitney U, Kruskal-Wallis one-way ANOVA or Spearman correlation test, in accordance with variable characteristics. For multiple test correction, P values were adjusted by the Benjamini–Hochberg FDR.
Survival was computed from study enrollment to the event of interest (e.g., death, AML progression) and accounted for right censoring at the time of disease-modifying treatments, including intensive chemotherapy and allogeneic stem cell transplantation. Survival probabilities were estimated using Kaplan–Meier methodology, and comparisons of survival across subgroups were conducted using the two-sided log-rank test. Multivariable models of OS were performed with Cox proportional hazards regression and optimized on the results of Akaike information criterion stepwise selection. HRs and 95% confidence intervals (CI) were reported for covariates, along with P values from the Wald test. All statistical analyses were performed with R 3.6.2 (https://www.r-project.org) software.
Data availability
Deidentified individual participant data are available in the Swedish National Data Service repository at https://doi.org/10.48723/zt59-8x04 upon request.
Results
Mutational landscape of MDSRS+
A total of 129 MDSRS+ cases were included in the study. Median age at sampling was 74 years (range 20–88), male/female ratio was 1.2, median OS was 5.5 years (interquartile range 0.9–9.5). Ninety-five patients (74%) had MDS without blast excess (EB), 23 (18%) MDS with EB, 9 (7%) MDS/MPN, and 2 AML. Detailed clinical and molecular features of MDSRS+ are reported in Table 1.
Table 1.
Clinical and hematologic features of the MDS with RS (MDSRS+), stratified by genetic classification.
| Total (N = 129) | SF3B1 mutated N = 82 (64%) | SRSF2 mutated N = 15 (11%) | TP53M H N = 13 (10%) | NOS N = 19 (15%) | P | |
|---|---|---|---|---|---|---|
| Age (years) | 0.426 | |||||
| Mean (range) | 72.7 (20–88) | 73.3 (44–88) | 69.6 (53–83) | 69 (20–86) | 72.7 (20–88) | — |
| Gender | — | — | — | — | — | 0.067 |
| Female | 58 (45.0%) | 39 (47.6%) | 2 (13.3%) | 7 (53.8%) | 10 (52.6%) | — |
| Male | 71 (55.0%) | 43 (52.4%) | 13 (86.7%) | 6 (46.2%) | 9 (47.4%) | — |
| Diagnosis according to the WHO 2016 classification | <0.001 | |||||
| MDS RS MLD | 62 (48.1%) | 45 (54.9%) | 7 (46.7%) | 4 (30.8%) | 6 (31.6%) | — |
| MDS RS SLD | 23 (17.8%) | 23 (28%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | — |
| MDS MPN RS T | 7 (5.4%) | 2 (2.4%) | 0 (0.0%) | 0 (0.0%) | 5 (26.3%) | — |
| MDS MLD | 8 (6.2%) | 0 (0.0%) | 4 (26.7%) | 1 (7.7%) | 3 (15.8%) | — |
| MDS EB 1 | 11 (8.5%) | 6 (7.3%) | 0 (0.0%) | 3 (23.1%) | 2 (10.5%) | — |
| MDS EB 2 | 12 (9.3%) | 5 (6.1%) | 2 (13.3%) | 5 (38.5%) | 0 (0.0%) | — |
| AML MRC | 2 (1.6%) | 0 (0.0%) | 1 (6.7%) | 0 (0.0%) | 1 (5.3%) | — |
| MDS del5q | 1 (0.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (5.3%) | — |
| MDS UNS | 1 (0.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (5.3%) | — |
| CMML 2 | 1 (0.8%) | 1 (1.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | — |
| MDS MPN | 1 (0.8%) | 0 (0.0%) | 1 (6.7%) | 0 (0.0%) | 0 (0.0%) | — |
| WBC (×109/L) | 0.369 | |||||
| Mean (range) | 6.4 (1.4–34.6) | 6.2 (1.4–32.8) | 6.9 (1.7–34.6) | 5.1 (1.9–8.9) | 7.6 (1.5–18.2) | — |
| ANC (×109/L) | 0.263 | |||||
| Mean (range) | 3.7 (0.3–21.1) | 3.6 (0.3–18.5) | 3.6(0.3–18.5) | 2.7 (0.8–5.6) | 4.8(0.3–12.2) | — |
| Monocytes (×109/L) | 0.419 | |||||
| Mean (range) | 0.7 (0–10) | 0.6 (0–4.9) | 1.1 (0.1–10) | 0.5 (0–1) | 1.0 (0.1–4.7) | — |
| HB (g/dL) | 0.012 | |||||
| Mean (range) | 10.0 (6–14.6) | 10 (6–13.6) | 10.8 (8–14.2) | 8.8 (6–10.9) | 9.8 (7–14.6) | — |
| Platelets (×109/L) | <0.001 | |||||
| Mean (range) | 258 (23–1234) | 272.3 (57–715) | 144.9 (25–261) | 152.4 (23–389) | 357.7 (36–1234) | |
| BM Blasts (%) | <0.001 | |||||
| Mean (range) | 3.6 (0.5–20) | 2.9 (0.5–19) | 5.3 (1.5–18) | 6.5 (1–15) | 3.6 (1–20) | — |
| BM RS (%) | <0.001 | |||||
| Mean (range) | 37.1 (5–88) | 43.8 (9–88) | 24.2(6–73) | 25.2 (6–52) | 26.8 (5–75) | |
| BM Cellularity (%) | 0.051 | |||||
| Mean (range) | 63 (10–100) | 60.2 (20–100) | 66 (30–100) | 74.6 (50–100) | 64.8 (10–100) | |
| BM Erythroid cell (%) | 0.166 | |||||
| Mean (range) | 31.9 (3–83) | 33.1 (7.5–83) | 25.7 (8.5–43) | 36.3 (13.5–65) | 28.2 (3–80.5) | |
| IPSS-M | <0.001 | |||||
| Very Low | 37 (29%) | 32 (39%) | 2 (13%) | 0 | 3 (16%) | — |
| Low | 51 (39%) | 36 (44%) | 4 (27%) | 2 (15%) | 9 (48%) | — |
| Intermediate low | 8 (6%) | 3 (4%) | 3 (20%) | 1 (8%) | 1 (5%) | — |
| Intermediate high | 5 (4%) | 1 (1%) | 3 (20%) | 1 (8%) | 0 | — |
| High | 15 (12%) | 8 (10%) | 2 (13%) | 0 (%) | 5 (26%) | — |
| Very high | 13 (10%) | 2 (2%) | 1 (7%) | 9 (69%) | 1 (5%) | — |
| mOS | <0.0001 | |||||
| Median (IQR) | 5.5 years (1.7–9.5) | 7.6 years (4–9.5) | 3.3 years (1–5.9) | 0.7 years (0.3–1.7) | 4.4 years (1.6–4.9) | — |
| Total SNV hits | 0.034 | |||||
| Mean (range) | 3 (0–11) | 2.8 (1–6) | 4.2 (1–11) | 2.7 (1–5) | 2.9 (0–9) | — |
| Total CNV hits | <0.001 | |||||
| Mean (range) | 1.1 (0–11) | 0.5 (0–8) | 0.3 (0–2) | 6.2 (2–11) | 0.6 (0–2) | — |
Abbreviations: AML MRC, acute myeloid neoplasms with myelodysplastic related changes; ANC, absolute neutrophil count; CMML 2, chronic myelomonocytic neoplasms type 2; HB, hemoglobin; IQR, interquartile range; MDS MPN RS T, MDS/myeloproliferative neoplasms with RS and thrombocytosis; MH, multi hit mutated; MLD, multi lineage dysplasia; mOS, median overall survival; NOS, not otherwise specified; SLD, single lineage dysplasia; SNV, single-nucleotide variant; UNS, unspecified; WBC, white blood cell.
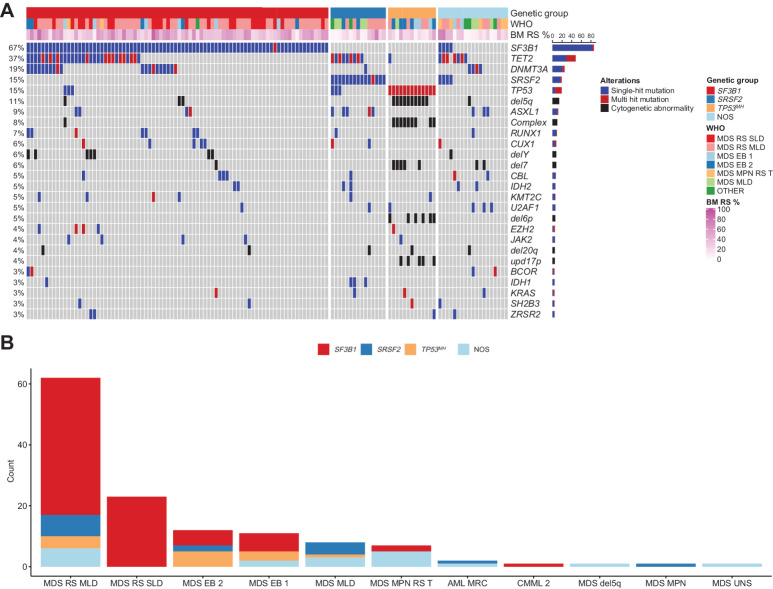
Overall, 392 somatic mutations were identified in the MDSRS+ population. SF3B1 was found to be the most recurrent mutated gene (67%) followed by TET2 (37%), DNMT3A (19%), SRSF2 (15%), and TP53 (15%). MDSRS+ were found to have ≥1, ≥2 or ≥3 recurrent somatic mutations in 97.7%, 79.2%, and 54.6% of cases, respectively. A total of 138 chromosomal alterations [96 unbalanced, 15 balanced aberrations and 27 copy-neutral loss of heterozygosity (LOH)] were identified in 54 (41.5%) MDSRS+ cases. Two MDS/MPN-RS-T cases did not harbor any clonal marker. NBM controls were all negative for clonal hematopoiesis with a tumor cell fraction cutoff of 1%. Multihit gene alterations (defined as having ≥2 distinct mutations in the same gene or one mutation associated with chromosomal alterations of the gene locus) were assessed by combining CNV analysis with somatic mutation profiling. TET2 and TP53 were the most recurrent genes with multi-hit (MH) mutations, with a prevalence of 19 of 48 (48%) TET2-mutated and 13 of 19 (68%) TP53-mutated cases, respectively.
Unsupervised analysis based on consensus clustering of genetic alternations was used to identify subsets with homogenous genetic profiles. We identified three major subgroups within MDSRS+, each driven by SF3B1, SRSF2, or TP53MH mutation, respectively (Fig. 1A; Table 1). Patients (11%) negative for SF3B1, SRSF2, and TP53MH mutation showed mutational patterns concordant with a recent publication and were classified as MDSRS+ not otherwise specified (NOS; Supplementary Fig. S1; ref. 20). TP53MH never occurred with SF3B1 or SRSF2, whereas the combination SF3B1-SRSF2 was found in 4 cases (3%). SF3B1-SRSF2 comutated cases were classified as NOS in downstream analysis and underwent additional clonal hierarchy studies. The analysis of mutations hierarchical ranks indicated that SF3B1 and SRSF2 mutations were comutated at the cellular level in two cases. Single-cell–derived hemopoietic and progenitor colony genotyping experiment confirmed coexistence of SF3B1K666T/SRSF2P95H and SF3B1K626S/SRSF2P95H mutations in these two cases. The other two comutated cases harbor SF3B1K700E mutation in the dominant clone, with a small secondary clone harboring a less common SRSF2 mutation outside the P95 hotspot (Supplementary Fig. S2A and S2B). The latter two cases had gene-expression and alternative splicing profiles similar but not identical to the SF3B1-only mutated cases, while the two cases with true concomitant SF3B1 and SRSF2 mutation showed a gene-expression and alternative splicing profiles with mixed features in-between single-mutated SF3B1 and SRSF2 cases. Given the current limited applicability into clinical practice of hierarchical clonal evaluation and transcriptomic profiling, we cautiously classified these four cases in the NOS subgroup, considering the limited power of any clinical outcome analysis carried out on 4 patients.
Figure 1.
Mutational landscape of MDSRS+. A, Mutation profile displaying clusters identified by unsupervised clustering analysis in MDSRS+. Genetic aberrations and somatic mutations with prevalence >2% are shown. Genes having ≥2 distinct mutations or one mutation associated with chromosomal alterations of the same gene locus are labeled as multihit and distinguished by single-mutation. Genetic subgroup, WHO subtypes, and BM RS percentage (BM RS %) are shown above the heatmap. B, Distribution of genetic classes across WHO categories of the MDSRS+. Complex, complex karyotype; del, deletion; mh, multihit; upd, uniparental disomy.
Association between mutation profile and clinical features
Mutations in DNMT3A, TET2, ASXL1 (usually referred to as DTA mutations) and loss of chromosome Y (-Y) – the most recurrent genetic aberrations in age-related clonal hematopoiesis (37, 41) – cooccurred with SF3B1 or SRSF2 in 53% and 75% of cases, respectively. Notably, TP53MH cases were depleted for DTA mutations or -Y (OR = 0.07, P < 0.001) but enriched for del5q (OR = 94.7, P < 0.001), del7q/-7 (OR = 32.9, P < 0.001), and complex karyotype (OR = 29.9, P < 0.001). No significant difference in age was found among the genetic subgroups.
The four genetic categories (i.e., SF3B1, SRSF2, TP53MH, and NOS) were variably distributed in WHO subcategories (Fig. 1B). MDSRS+SF3B1mutated accounted for 100% of MDS-RS-SLD (P = 0.01), whereas MDSRS+TP53MH were enriched in MDS-EB type 2 (MDS-EB-2, P = 0.01), but also observed in four cases with myeloblasts < 5% and RS ≥ 15%. When exploring genotype-phenotype associations irrespective of WHO classification, TP53 (both single and double) and RUNX1 mutations were found to be the strongest predicting factors of a blastic phenotype (BM blast count ≥5%, OR = 6.6 and 6.8, P < 0.001 and 0.01, respectively). Thrombocytosis was associated with JAK2 (OR = 25, P < 0.001), but no other independent molecular predictor of proliferative phenotype (e.g., leukocytosis, monocytosis) was identified in this cohort. In SRSF2 and TP53MH-mutated MDS, no survival difference was found when comparing cases with high (≥15%), low (5%–14%), RS and historical controls without RS (Supplementary Fig. S3).
Transcriptomic landscape
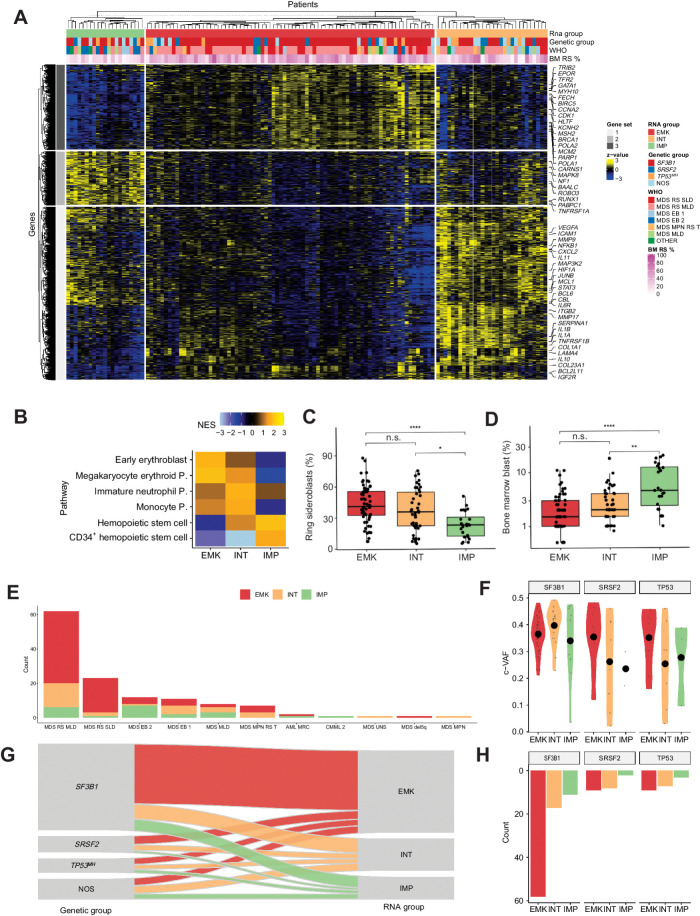
RNA-Seq analysis of CD34+ BM MNC was carried out in all 129 MDSRS+ and 10 NBM controls. Median sequencing coverage per patient was 67 million reads (Supplementary Tables S1 and S2). Overall, transcription analysis revealed a continuous spectrum of changes in transcriptome profiles (Supplementary Results; Supplementary Figs. S4–S7). Unsupervised consensus cluster analysis revealed three stable clusters (Fig. 2A; Supplementary Figures S8–S10). Gene-set enrichment analysis (GSEA) of differentially expressed genes (DEG) among the three groups detected a specific enrichment of genes expressed by immature myeloid progenitors (IMP) in the first cluster (hereafter referred to as IMP group), and erythroblast-megakaryocyte (EMK) precursors in the second cluster (EMK group). These results were in line with a previous transcriptome study in MDS (7). A third cluster was identified displaying intermediate molecular features between EMK and IMP, with a strong upregulation of genes involved in mature myelopoiesis. We referred to this third group as intermediate (INT) group (Fig. 2B; Supplementary Table S3).
Figure 2.
Transcriptomic landscape of MDSRS+. A, Heat map showing expression levels of differentially expressed genes in MDSRS+ across the three transcriptomic groups. Rows represent genes and samples are depicted in columns. Covariates display gene expression–based group, genetic group, WHO subtypes and BM RS %. B, GSEA on DEGs among the three clusters. Normalized enrichment score (NES) was computed considering the comparison between each group and NBM used as controls. P, progenitor. C–D, Comparison of BM RS (C) and blasts (D) distributions across the three groups derived from gene expression unsupervised clustering analysis. E, Frequency distribution of transcriptomic classes across WHO categories of the MDSRS+ cohort. F, Distribution of variant allele frequency (VAF) corrected by CNV/LOH (c-VAF) of SF3B1, SRSF2, and TP53MH, stratified by gene expression groups. Specifically, in case of deletion or LOH, VAF was halved to making it comparable with mutation without CNV/LOH. In case of multiple TP53 mutations, TP53MH c-VAF referred to the second-ranked TP53 mutation VAF. G, Sankey diagram showing the relationship between genomic and transcriptomic classification. Color labels distinguish cases according to the three gene expression groups. H, Absolute frequency distribution of SF3B1, SRSF2, and TP53MH mutation, stratified by gene expression group.
Relationship among clinical features, genetic, and transcriptome profiles
Compared with the EMK group, IMP cases had lower BM RS (median 22% vs. 40%, P < 0.001; Fig. 2C) and higher BM blast (median 5% vs. 1.5%, P = 0.002; Fig. 2D). IMP group was enriched in MDS-EB-2 and MDS with multilineage dysplasia (MDS-MLD, 50% and 62%, P < 0.001 and 0.4, respectively) and underrepresented in MDS-RS-SLD/MLD and MDS/MPN-RS-T (21% and 0, respectively, both P = 0.02; Fig. 2E). Consistently with transcriptomic analysis, INT signature was associated with an intermediate morphologic phenotype between EMK and IMP groups. No other significant differences were found in age, peripheral blood counts, serum ferritin, serum erythropoietin or BM myeloid/erythroid ratio across the three groups.
Subsequently, we explored the relationship between genomic and transcriptomic classification in the context of MDSRS+. Genetic class-defining allelic burden (median 0.35%; range, 0.02–0.48) did not differ across the genetic and transcriptomic subgroups (Fig. 2F). EMK-like signature was associated with SF3B1 mutation (OR 2.7, P = 0.01; Fig. 2G and H) and, interestingly, INT signature was associated with TP53MH mutation (OR 3.5, P = 0.02). IMP signature was not restricted to any of the genetic subgroups but was specifically associated with ASXL1 (OR = 11, P < 0.001), del7q/-7 (OR = 8.9, P = 0.004), and RUNX1 (OR = 6.4, P = 0.01) mutations.
However, none of the three transcriptomic signatures was highly specific for any mutation, suggesting that transcriptomic heterogeneity is influenced but not fully explained by genetic abnormalities. Therefore, we carried out supervised DEG analysis across the previously described genetically defined subgroups to get insight into the molecular pathophysiology of MDSRS+.
Genetically defined MDSRS+ subgroups display distinct gene expression and alternative splicing profiles
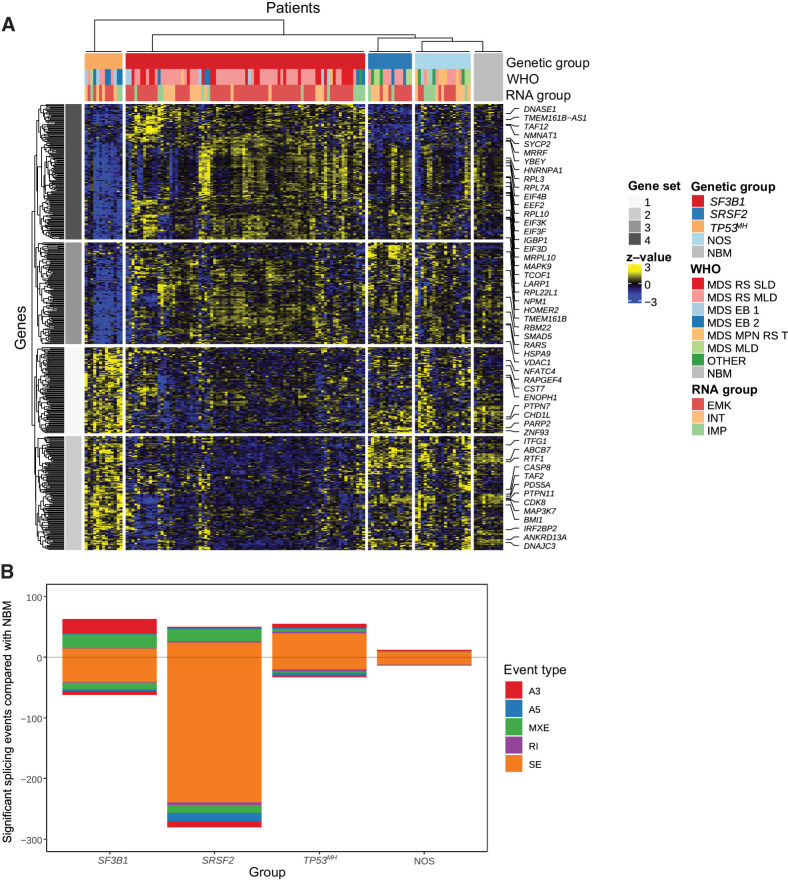
DEG analysis revealed distinct signatures among SF3B1, SRSF2, and TP53MH mutated MDSRS+ (Fig. 3A). ABCB7 – an iron mitochondrial transporter known to be downregulated in MDS-RS-SLD/MLD (6, 9, 10) – was found selectively downregulated in SF3B1mut MDSRS+ (Fig. 3A; Supplementary Fig. S11). In addition, SF3B1mut MDSRS+ also displayed significant downregulation of DNA/RNA polymerases (e.g., DNAJC3, PARP2, RTF1) and phosphatases involved in cellular signaling (e.g., PTPN11, ENOPH1), together with upregulation of genes involved in protein translation. Conversely, MDSRS+ harboring TP53MH mutation showed strong downregulation of genes involved in protein translation and upregulation of DNA/RNA polymerases and phosphatases, as compared with other MDSRS+ and NBM controls. ABCB7, phosphatases, DNA/RNA polymerases and protein translation gene expression was not significantly altered in SRSF2mut MDSRS+.
Figure 3.
Supervised transcriptome analysis according to MDSRS+ genomic classification. A, Heat map showing differentially expressed genes among SF3B1, SRSF2, and TP53MH MDSRS+ compared with NBM controls. Rows represent genes and columns represent samples. Genetic, WHO, and transcriptomic subtypes are shown as covariates. B, Types and numbers of splicing alterations significantly associated with each genetic group. Bars in the top half indicate alternative splicing events that were enriched in MDSRS+, bars in the bottom half indicate alternative splicing events enriched in the healthy donors used as controls, stratified by splicing alterations subtypes.
Alternative splicing analysis found that splicing factor mutated MDSRS+ had a significantly higher number of alternative splicing events than TP53MH subgroup when compared with NBM controls (Fig. 3B; Supplementary Fig. S12). Consistently with previous reports (6, 9), SF3B1mut MDSRS+ had a specific increase of A3´ splice site events, whereas SRSF2mut had a substantial decrease in exon skipping events. Missplicing events affecting ABCB7 and TMEM14C, already described in MDS-RS-SLD/MLD (6, 42), were confirmed to be restricted to the SF3B1mut subgroup, together with other genes involved in DNA homeostasis (CHD1 L and PDS5A) and in protein translation (MAT2B and TCOF1). Results from alternative splicing analysis applied to the transcriptomic stratification groups mostly reflected different distribution of splicing factor mutations across the groups; redundancy with genotyping data limited the translational application of these findings in following analyses. Overall, distinct patterns of differentially expressed genes and aberrant splicing events were found in the genetically defined subgroups, providing additional evidence that MDSRS+ genetic classification unveils distinct disease entities.
CD34+ transcriptome profile reflects HSPC composition in MDSRS+
Considering that GSEA and pathway analyses were significant for transmembrane signaling activity involved in hematopoietic precursors differentiation (as previously shown in Fig. 2B; Supplementary Figs. S6 and S7), and transcriptomic heterogeneity was only partly explained by genetic abnormalities (Fig. 2G), we hypothesized that the BM CD34+ transcriptome may reflect composition in hemopoietic stem and progenitor cells (HSPC). To explore this hypothesis and better characterize the relationship between gene and hemopoietic precursor signatures, we used single-cell transcriptomic–based deconvolution to dissect distinct HSPCs in the BM CD34+ transcriptome. This technique allowed accurate HSPC quantification by integrating quantitative evaluation of a restricted gene-set specific for each HSPC subtype. Granulocyte/macrophage progenitors (GMP), megakaryocyte/erythroid progenitors (MEP), and hemopoietic stem cells (HSC) were the most represented estimated cell subtypes and accounted for 39%, 35%, and 6% of the total imputed CD34+ cell population in MDSRS+ (Fig. 4A; Supplemental Results and Supplementary Figs. S13–S17). Among the most represented subpopulations, cells with MEP-like signature showed the highest variability across the transcriptomic groups (median value was 45%, 37%, and 13% in EMK, INT, and IMP group, respectively, P < 0.0001; Fig. 4B), with IMP cases displaying very low value of MEP compared with other MDSRS+ and NBM controls. Results from RNA deconvolution were validated using multiparameter flow cytometry (R = 0.9, P 0.0001; Fig. 4C; Supplementary Fig. S15) in a subset of five cases already included into this study. MEP was also the only estimated population that retained statistical significance when multinomial logistic regression model was built to predict results from transcriptome unsupervised clustering analysis (P < 0.001). Cases with BM blasts ≥ 10% displayed significantly low MEP levels compared with other cases, with very high variance especially in cases without blast excess (<5%, Supplementary Fig. S18). Importantly, MEP-like signature was independent from myeloid/erythroid ratio evaluated on total BM cellularity. These results suggest that MDSRS+ transcriptome reflect heterogeneity of the HSPC compartment, simplistically summarized as a decreasing gradient in MEP-like signature across the EMK, INT, and IMP subgroups and not mimicked by standard morphologic features.
Figure 4.
![Figure 4. Digital sorting analysis carried out on bulk CD34+ MNC transcriptome from MDSRS+ cases. A, Digital sorting analysis, gated for HSPCs. Mature cells were gated out to focus only on HSPC compartment (results including mature cells are shown in Supplementary Fig. S13); CMP, common myeloid progenitors; GMP, granulocyte–monocyte progenitors; HSC, hematopoietic stem cells; MLP, multi-lymphoid progenitors; MPP, multipotent progenitors; MEP, megakaryocyte-erythroid progenitors). Cases (x-axis) are sorted by decreasing estimated MEP abundance, showing enriched IMP signature in cases with MEP depletion on right side of the plots. B, Relative distribution of estimated MEP in the three transcriptomic groups and (NBM). C, Scatter plot comparing results from multiparameter flow (x-axis) and in silico cytometry (y-axis). MEP, GMP, and HSC were quantified in the CD34+ BM MNCs by means of the two techniques, showing a highly concordance [correlation line and 95% confidence interval (CI) are represented in dark and light gray, respectively, Pearson correlation coefficient = 0.9, P < 0.001].](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/93b4/10570683/ee11fff227ea/4256fig4.jpg)
Digital sorting analysis carried out on bulk CD34+ MNC transcriptome from MDSRS+ cases. A, Digital sorting analysis, gated for HSPCs. Mature cells were gated out to focus only on HSPC compartment (results including mature cells are shown in Supplementary Fig. S13); CMP, common myeloid progenitors; GMP, granulocyte–monocyte progenitors; HSC, hematopoietic stem cells; MLP, multi-lymphoid progenitors; MPP, multipotent progenitors; MEP, megakaryocyte-erythroid progenitors). Cases (x-axis) are sorted by decreasing estimated MEP abundance, showing enriched IMP signature in cases with MEP depletion on right side of the plots. B, Relative distribution of estimated MEP in the three transcriptomic groups and (NBM). C, Scatter plot comparing results from multiparameter flow (x-axis) and in silico cytometry (y-axis). MEP, GMP, and HSC were quantified in the CD34+ BM MNCs by means of the two techniques, showing a highly concordance [correlation line and 95% confidence interval (CI) are represented in dark and light gray, respectively, Pearson correlation coefficient = 0.9, P < 0.001].
Clinical relevance of combined genomic–transcriptomic profiling
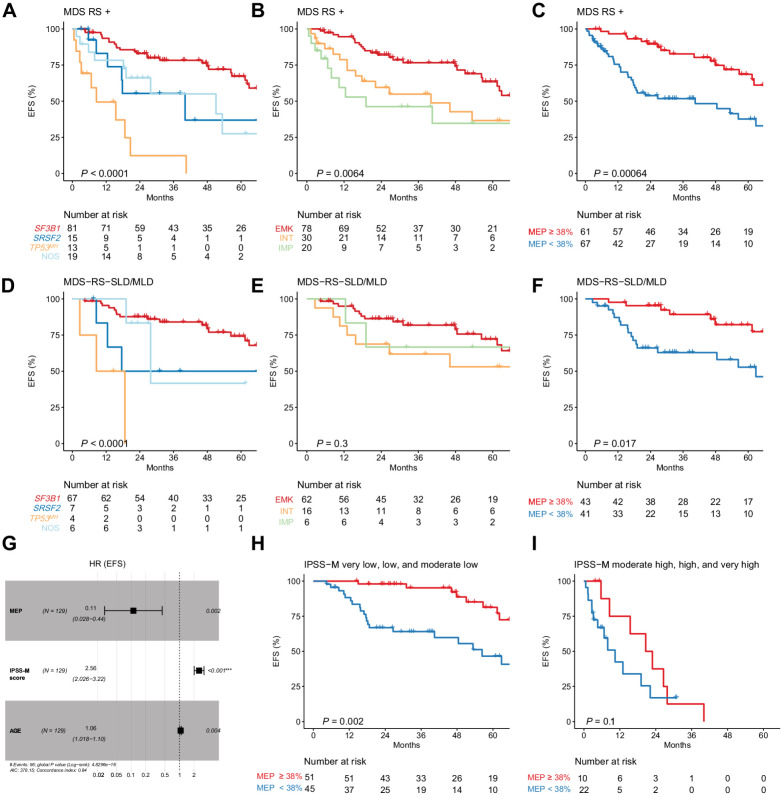
Finally, we explored the predictive value of multiomics characterization on patient outcome. In univariate analysis, age, blast, complete blood count, cytogenetic risk, and IPSS-M score were associated with distinct OS and EFS, together with genetic and transcriptomic classification and estimated MEP proportion (Fig. 5A and B; Supplementary Fig. S19A and S19B, and Supplementary Tables S4 and S5).
Figure 5.
Prognostic effect of genomic and transcriptomic analyses on MDSRS+ outcome. EFS stratified by genomic (A), transcriptomic classification (B), and estimated MEP percentage (C) in all MDSRS+ (A–C) and MDS-RS-SLD/MLD only (D–F). Multivariable Cox proportional hazard model for EFS in all MDSRS+ including age, IPSS-M score, and estimated MEP percentage as continuous variables (G). EFS stratified by estimated MEP percentage and IPSS-M risk category (H and I) (full representation of the six IPSS-M categories shown in Supplementary Fig. S21A).
Given the reduced applicability in clinical practice of continuous variables as a prognostic factor, we applied maximally selected rank statistics method and found that 38% was the most significant MEP cut-off level in survival univariate analysis (both OS and EFS). MEP ≥ 38% predicted EMK profile with the accuracy of 70% (positive predictive value 64%, negative predictive value 81%) and was consistently associated with longer OS and EFS (Supplementary Fig. S19C and Fig. 5C, respectively; and Supplementary Fig. S20).
Prognostic effect of genetic classification and MEP was also confirmed when only MDS-RS-SLD/MLD were considered (Fig. 5D–F; Supplementary Fig. S19D–S19F). MEP depletion was found to be an independent prognostic factor when the new IPSS-M was included in the multivariable analysis (Fig. 5G–I; Supplementary Figs. S19G–S19I and S21). Similar results were found when we restricted our analysis to the MDS with low blast and SF3B1 mutation, as defined according to the 2022 WHO and ICC new classifications (Supplementary Fig. S22).
Discussion
In this study we explored the clinical relevance of combined genomic/transcriptomic profiling in MDSRS+. To study the association of a RS phenotype with other clinical features in an unbiased manner, we included any MDS cases with ring sideroblast (i.e., ≥5%) without limitation to MDS specific subclasses as defined by recent classifications of myeloid neoplasm. Mutations in SF3B1, SRSF2 or TP53MH were found to be the most relevant drivers in 89% of all MDSRS+. Unsupervised clustering of genetic alterations confirmed that these three genotypes cooccur very rarely. Indeed, TP53MH never cooccurred with SF3B1 or SRSF2 mutation, prompting future studies evaluating RNA splicing impairment in TP53MH-mutant cells. Coexistence of SF3B1/SRSF2 mutation was very rare (3%) and, when cooccurring in the same cell, involved a permissive combination of less common alleles with reduced effects on RNA splicing (43).
The high allelic burden of SF3B1, SRSF2, and TP53MH mutations suggests that mutations in these three genes may play a driving role in pathogenesis of MDSRS+, while comutation patterns contribute to further shape the clinical phenotype of the disease, as suggested by previous reports on myeloid neoplasms (19, 22). Survival analysis based on the resulting genetic stratification confirmed that the benign behavior of MDS-LB-RS is limited to case with SF3B1 mutation but without concomitant high-risk cytogenetic aberrations or mutations, arguing against the accuracy of RS as a surrogate marker for SF3B1 mutation in the recent WHO diagnostic criteria (12). In addition, TP53MH and SRSF2 MDSRS+ survival was similar to historical controls (i.e., MDS and MDS/MPN) with RS < 5% (22, 23), and no survival difference was found between having RS between 5% and 14% or ≥ 15% in SF3B1wt cases (as it was already known for MDSSF3B1+). Our results support the classification of SF3B1wt MDS-LB-RS cases within MDS-NOS irrespective of the number of RS, and confirm that the definition of MDS-LB-RS should be only limited to the cases with SF3B1 mutation, as proposed by new ICC 2022 classification of myeloid neoplasms (11). Further studies evaluating clinical characteristics and outcome of the rare cases with concomitant SF3B1/SRSF2 mutations are warranted, giving the relevant implication on prognostic stratification and the current lack of evidences on this rare molecular condition.
Transcriptome analysis of CD34+ enriched BM cells revealed that transcriptomic heterogeneity was mainly led by genes involved in inflammation, transmembrane signaling and extracellular matrix organization, highlighting the interaction between bone marrow microenvironment and HSPC in MDSRS+. Consistent with a previous study (7), we identified a gene-expression signature characterized by upregulation of genes involved in erythrocyte-megakaryocytic differentiation (i.e., EMK signature). Compared with previous report – that distinguished only two groups – we found one additional stratification in the non-EMK group, where we distinguished IMP from INT signature. Both INT and IMP signatures included genes upregulated in myeloid progenitors, while INT signature displayed a specific upregulation of genes involved in interleukin signaling and inflammation.
Given the differential expression pattern involving genes related to hematopoietic precursor differentiation, we hypothesized that signatures captured by bulk CD34+ MNC RNA-seq reflected a distinct HSPC composition. This hypothesis was confirmed by digital sorting and showed that EMK group had higher proportion of MEP as compared IMP/INT cases and NBM controls that was independent from myeloid/erythroid ratio evaluated on total BM cellularity. To translate these findings into a clinically useful tool, we explored the prognostic value of MEP proportion on survival analysis and found that low MEP predicted poor survival, with 38% threshold as the optimal cut-off value. Multivariable survival analysis showed that MEP proportion and IPSS-M score independently contributed to survival prediction. These results are consistent with previous studies evaluating CD34+ BM MNCs by flow cytometry analysis (44) and gene array (45) reporting MEP decrease in high-risk MDS, and were found to be significant even when the IPSS-M risk score was taken into account, in particular in low/moderate-low risk classes, where clinical decision making may be a challenge. Overall, our results suggest that MEP (Lin−CD34+CD38+CD123−CD45RA−) quantification by diagnostic flow cytometry analysis on BM aspirate at diagnosis should be explored as a surrogate marker for high-risk transcriptomic profile in larger prospective trials aiming to improve MDS prognostication.
Furthermore, results from differential gene expression and alternative splicing analyses supported distinction of SF3B1mut from SRSF2mut and TP53MH MDSRS+ and suggests that RS development has a distinct pathobiology depending on the genetic driver. Indeed, SF3B1mut and SRSF2mut MDSRS+ displayed distinct iron and mitochondrial homeostasis regulators among differentially expressed and differentially spliced genes. Impaired expression of ABCB7 and TMEM14C was found to be restricted to SF3B1mut MDSRS+ (42), whereas CASP8 and ESWR1 – both known to be involved in mitochondrial homeostasis (46, 47) – were specifically misspliced in SRSF2mut cases. Conversely, TP53MH MDSRS+ exhibited a selective dysregulation of genes involved in protein synthesis, suggesting that mitochondrial iron overload in these cases might be consequent to defective iron incorporation into hemoglobin rather than increased mitochondrial iron uptake. Further studies to explore the interconnection between clonal hematopoiesis pathophysiology and iron metabolism may elucidate the molecular mechanisms leading to impaired iron cellular balance in distinct genetically-defined MDSRS+ subtypes.
In conclusion, integrated genomic-transcriptomic analysis supports the distinction of SF3B1mut, SRSF2mut, TP53MH, and NOS MDSRS+ and introduces HSPC transcriptomes as a novel and independent prognostic variable. Our results provide essential input on the molecular basis of SF3B1-unmutated MDSRS+, and support SF3B1mut MDSRS+ as a stand-alone category in the myeloid neoplasms classifications.
Supplementary Material
Supplementary Results
Mutation profiles of clusters identified by unsupervised clustering of genetic aberrations and somatic mutations. WHO subtypes and bone marrow ring sideroblasts percentage (BM RS %) are shown above the oncoplot.
Clonal hierarchy analysis of SF3B1-SRSF2 co-mutated cases. A. Results from the hierarchical rank analysis of mutations detected by DNAseq in the 4 cases harboring both SF3B1 and SRSF2 mutation using Pyclone. The analysis was also performed using DPClust, which provided converging results (data not shown). Tumor cell fraction (axes) for each detected clone and its 95% confidence interval (dot size) is represented in a scatter-plot, using different color according to the cluster driving mutation (SF3B1, red; SRSF2, blue; other drivers, gray). SF3B1 dominancy in MDS392 and MDS 640, together with the SRSF2 large clone size are suggestive of both splicing factor mutations in the same clone. On the contrary, MDS694 and MDS965 had a dominant SF3B1 clone associated with a very little and probably independent SRSF2 secondary clone. B. Results from single-cell derived colony-forming unit (CFU) genotyping confirming the concurrent double splicing factor mutations within the same clone in patient MDS382 and MDS640. CFU experiment was not carried out for the other two cases because of the very low probability of SF3B1/SRSF2 double positive clone identification, as already suggested by current data in the literature related to the presence of SF3B1K700E mutation (ref. 1).
Overall survival of MDSRS+ with SRSF2 and TP53MH genotype stratified by RS burden. Cases (colored Kaplan-Meier curves) were compared to historical controls (gray curve) with similar clinical and molecular features and no RS (i.e. RS<5%) from two large studies evaluating clinical characteristics of SRSF2 and TP53MH -mutated MDS, respectively (ref. 2,3). SF3B1 mutated MDSRS+ were not considered for this analysis because of the small number of cases with RS between 5 and 15% (2 out of 82 SF3B1-mutated ones) and consolidated evidences in the literature showing no effect of RS burden on SF3B1-mutated MDS outcome (ref. 4,5); sample size also limited further analysis in the MDSRS+ NOS category.
UMAP plot of MDSRS+ bone marrow CD34 mononuclear cells transcriptomes. Each point is one patient, overlaid with results from unsupervised clustering.
Cumulative proportion of principal components derived from principal component analysis. The first 14 explain 2/3 of the total variability.
– Results from Gene Ontology enrichment analysis (molecular function dataset) for the top 250 contributors of principal component 1 (PC1)
Results from Gene Ontology enrichment analysis (molecular function dataset) for the top 250 contributors of principal component 2 (PC2)
Unsupervised consensus clustering analysis on gene expression data. Consensus clustering applied to the MDSRS+ cohort revealed 3 transcriptomic profiles, named EMK, INT and IMP according to the results from gene set enrichment analysis.
Item consensus for K=3 showing stable item-consensus values across the whole cohort.
Optimal number of clusters defined by average silhouette width method.
ABCB7 expression throughout the spectrum of MDSRS+ genotypes. ABCB7 downregulation is restricted to SF3B1-mutant MDSRS+.
Scatter plots comparing mean PSI values between the MDSRS+ and healthy donors used as normal bone marrow controls (NBM), stratified by genetic classification. Significantly associated events (FDR < 0.0001) are color coded by splicing alterations subtypes.
Digital sorting analysis on bulk CD34+ MNC transcriptome from MDSRS+ cases. Plots refer to unfiltered data, i.e. including hematopoietic stem/progenitor and mature hematopoietic cells. CMP, common myeloid progenitors; EryP, erythroid precursors; GMP, granulocyte–monocyte progenitors; Gran, granulocytes; HSC, hematopoietic stem cells; Lymph, lymphocytes; MK, megakaryocytes; MLP, multi-lymphoid progenitors; MPP, multipotent progenitors; MEP, megakaryocyte-erythroid progenitors). Cases (x-axis) are sorted by decreasing estimated MEP abundance, showing enriched IMP signature in cases with MEP depletion on right-side of the plots.
The proportion of estimated erythroblast-like cells correlate positively with the percentage of RS in BM smears.
In-silico cytometry data validation. A) Experimental workflow and multi parameter flow cytometry gating strategy used to sort Lin+ and Lin- from CD34+ MNC and to explore HSPC composition. B) Scatter plot comparing results from multi-parameter-flow (x-axis) and in-silico cytometry (y-axis) partitioned by megakaryocyte/erythroid progenitors (MEP), granulocyte/macrophage progenitors (GMP) and hemopoietic stem cell (HSC), showing significant concordances for MEP and GMP (Pearson correlation coefficient 0.94 and 0.87, P 0.02 and 0.05, respectively; correlation line and 95% confidence interval are represented in dark and light gray, respectively).
EMK and IMP signature validation on HSPC sorted populations. Differential gene expression analysis across MDSRS+ with EMK profile, MDSRS- with IMP profile and NBM (3 cases each) according to Lin marker expression. Differential expressed genes (row) between EMK and IMP groups were selected. Each row represents a gene, and each column represents a sample. Design comparison (case) and sorted population (condition) are shown as covariates.
Gene set enrichment analysis on differentially expressed genes in LIN- CD34+ cells. Three MDS with RSEMK were compared to 3 MDS without RSIMP and displayed an enrichment of genes expressed in early erythroblast/megakaryocyte progenitor genes and a depletion of genes expressed in myeloid progenitors.
Distribution of estimated MEP proportion according to bone marrow blast percentage. P-value for each pairwise comparison is reported in the upper part of the plot.
Prognostic effect of genomic and transcriptomic analyses on MDSRS+ outcome. Overall survival (OS) stratified by genomic (A), transcriptomic classification (B) and estimated MEP percentage (C) in all MDSRS+ (A-C) and MDS-RS-SLD/MLD only (D-F). Multivariable Cox proportional hazard model for OS in all MDSRS+ including age, IPSS-mol score and estimated MEP percentage as continuous variables (G). OS stratified by estimated MEP percentage and IPSS-mol risk category (full representation of the 6 IPSS-mol categories shown in Supplemental Figure 21B).
MDSRS+ event free survival (EFS) stratified by estimated MEP percentage in the four genetic subgroups (A-D).
MDSRS+ event free survival (EFS, A) and overall survival (OS, B) stratified by estimated MEP percentage and IPSS-mol risk category.
Transcriptomic impact on SF3B1 mutant MDS and low blasts outcome. Overall survival (OS) and Event Free Survival (EFS) stratified by transcriptomic classification (A-B) and estimated MEP percentage (C-D), respectively, in SF3B1 mutant MDS and low blasts, as defined according to ICC and WHO 2022 classification.
RNA Sequencing metrics.
Demographic and clinical features of cases and controls included into the study.
List of differentially expressed genes associated with EMK, INT and IMP profile.
Variables significant for survival analysis in univariate analysis
Treatment. Number of cases that underwent to each treatment category and the associated hazard-ratio for death is reported for each genetic subgroup and for the whole cohort.
Acknowledgments
This study was supported by the Swedish Cancer Society (Cancerfonden, 150269, 190200 and 210340), the Cancer Research Foundations of Radiumhemmets (Radiumhemmets Forskningsfonder, 151103), the Swedish Research Council (Vetenskapsrådet, 521–2013–3577, Associazione Italiana per la Ricerca sul Cancro (Milan, Italy, 22796- 21267- 20125). SER is financially supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab. We would like to acknowledge the MedH Flow Cytometry core facility at Karolinska Institutet for providing cell analysis services and technical expertise, National Genomics Infrastructure in Stockholm for providing sequencing service, SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science (project sens2019530) for access to the UPPMAX computational infrastructure.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
E. Bernard reports personal fees from Pfizer and grants from Edward P Evans Foundation outside the submitted work. S.E. Reinsbach reports grants from Knut and Alice Wallenberg Foundation during the conduct of the study. E. Papaemmanuil reports grants from MDS Foundation outside the submitted work; is a founder, equity holders and holds fiduciary roles in Isabl Inc.; is also a Scientific Advisor for Tensixteen Bio and accordingly has shares in the company; and has received travel funding from Illumina. E. Hellström-Lindberg reports grants from The Swedish Wallenberg Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
G. Todisco: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Creignou: Data curation, formal analysis. E. Bernard: Resources, data curation, formal analysis, methodology. A. Björklund: Data curation, formal analysis, investigation. P.L. Moura: Data curation, formal analysis, investigation. B. Tesi: Data curation, formal analysis, investigation. T. Mortera Blanco: Data curation, formal analysis, investigation. B. Sander: Data curation, formal analysis, investigation. M. Jansson: Data curation, formal analysis, investigation. G. Walldin: Data curation. I. Barbosa: Data curation, formal analysis, investigation. S.E. Reinsbach: Data curation, formal analysis, investigation. I.J. Hofman: Data curation, formal analysis, investigation. C. Nilsson: Data curation, formal analysis. T. Yoshizato: Data curation, formal analysis, investigation. M. Dimitriou: Data curation, formal analysis, investigation. D. Chang: Data curation. S. Olafsdottir: Data curation. S. Venckute Larsson: Data curation. M. Tobiasson: Data curation, formal analysis. L. Malcovati: Supervision, funding acquisition, methodology. P. Woll: Supervision, methodology. S.W. Jacobsen: Supervision, methodology. E. Papaemmanuil: Supervision, methodology. E. Hellström-Lindberg: Conceptualization, resources, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Cazzola M, Invernizzi R. Ring sideroblasts and sideroblastic anemias. Haematologica 2011;96:789–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jädersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015;126:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011;365:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 5. Pellagatti A, Armstrong RN, Steeples V, Sharma E, Repapi E, Singh S, et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: Dysregulated genes/pathways and clinical associations. Blood 2018;132:1225–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shiozawa Y, Malcovati L, Gallì A, Sato-Otsubo A, Kataoka K, Sato Y, et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat Commun 2018;9:3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiozawa Y, Malcovati L, Gallì A, Pellagatti A, Karimi M, Sato-Otsubo A, et al. Gene expression and risk of leukemic transformation in myelodysplasia. Blood 2017;130:2642–53. [DOI] [PubMed] [Google Scholar]
- 8. Beauchamp EM, Leventhal M, Bernard E, Hoppe ER, Todisco G, Creignou M, et al. ZBTB33 is mutated in clonal hematopoiesis and myelodysplastic syndromes and impacts RNA splicing. Blood cancer Discov 2021;2:500–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dolatshad H, Pellagatti A, Liberante FG, Llorian M, Repapi E, Steeples V, et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 2016;30:2322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nikpour M, Scharenberg C, Liu A, Conte S, Karimi M, Mortera-Blanco T, et al. The transporter ABCB7 is a mediator of the phenotype of acquired refractory anemia with ring sideroblasts. Leukemia 2013;27:889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood 2022;140:1200–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia 2022;36:1703–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malcovati L, Hellström-Lindberg E, Bowen D, Adès L, Cermak J, Del Cañizo C, et al. Diagnosis and treatment of primary myelodysplastic syndromes in adults: recommendations from the European LeukemiaNet. Blood 2013;122:2943–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malcovati L, Stevenson K, Papaemmanuil E, Neuberg D, Bejar R, Boultwood J, et al. SF3B1-mutant MDS as a distinct disease subtype: a proposal from the international working group for the prognosis of MDS. Blood 2020;136:157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willekens C, Dumezy F, Boyer T, Renneville A, Rossignol J, Berthon C, et al. Linezolid induces ring sideroblasts. Haematologica 2013;98:e138–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Montalban-Bravo G, Kanagal-Shamanna R, Darbaniyan F, Siddiqui MT, Sasaki K, Wei Y, et al. Clinical, genomic, and transcriptomic differences between myelodysplastic syndrome/myeloproliferative neoplasm with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T) and myelodysplastic syndrome with ring sideroblasts (MDS-RS). Am J Hematol 2021;96:E246–9. [DOI] [PubMed] [Google Scholar]
- 17. Berger G, Gerritsen M, Yi G, Koorenhof-Scheele TN, Kroeze LI, Stevens-Kroef M, et al. Ring sideroblasts in AML are associated with adverse risk characteristics and have a distinct gene expression pattern. Blood Adv 2019;3:3111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malcovati L, Della Porta MG, Pietra D, Boveri E, Pellagatti A, Gallì A, et al. Molecular and clinical features of refractory anemia with ringed sideroblasts associated with marked thrombocytosis. Blood 2009;114:3538–45. [DOI] [PubMed] [Google Scholar]
- 19. Malcovati L, Papaemmanuil E, Ambaglio I, Elena C, Gallì A, Della Porta MG, et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014;124:1513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huber S, Haferlach T, Meggendorfer M, Hutter S, Hoermann G, Summerer I, et al. Mutations in spliceosome genes in myelodysplastic neoplasms and their association to ring sideroblasts. Leukemia 2023;37:500–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Ossa JEA, Nannya Y, et al. Molecular international prognostic scoring system for myelodysplastic syndromes. NEJM Evid; 2022. Available from: https://evidence.nejm.org/doi/full/10.1056/EVIDoa2200008 [DOI] [PubMed]
- 22. Todisco G, Creignou M, Gallì A, Guglielmelli P, Rumi E, Roncador M, et al. Co-mutation pattern, clonal hierarchy, and clone size concur to determine disease phenotype of SRSF2 P95-mutated neoplasms. Leukemia 2021;35:2371–81. [DOI] [PubMed] [Google Scholar]
- 23. Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat Med 2020;26:1549–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020;38:276–8. [DOI] [PubMed] [Google Scholar]
- 25. Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst 2016;12:477–9. [DOI] [PubMed] [Google Scholar]
- 29. Sergushichev AA. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. Biorxiv; 2016. Available from: https://www.biorxiv.org/content/10.1101/060012v1
- 30. Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 2014;111:E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woll PS, Kjällquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell 2014;25:794–808. [DOI] [PubMed] [Google Scholar]
- 32. Salcedo A, Tarabichi M, Espiritu SMG, Deshwar AG, David M, Wilson NM, et al. A community effort to create standards for evaluating tumor subclonal reconstruction. Nat Biotechnol 2020;38:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods 2014;11:396–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hofman IJF, Mortera-Blanco T, Moura PL, Vestlund J, Larsson SV, Elvarsdottir EM, et al. The extent of residual WT HSPCs is associated with the degree of anemia in patients with SF3B1-mutated MDS-RS. Blood Adv 2022;6:475–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagata Y, Zhao R, Awada H, Kerr CM, Mirzaev I, Kongkiatkamon S, et al. Machine learning demonstrates that somatic mutations imprint invariant morphologic features in myelodysplastic syndromes. Blood 2020;136:2249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gallì A, Todisco G, Catamo E, Sala C, Elena C, Pozzi S, et al. Relationship between clone metrics and clinical outcome in clonal cytopenia. Blood 2021;138:965–76. [DOI] [PubMed] [Google Scholar]
- 38. Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 2019;37:773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun 2019;10:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oetjen KA, Lindblad KE, Goswami M, Gui G, Dagur PK, Lai C, et al. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 2018;3:e124928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Malcovati L, Gallì A, Travaglino E, Ambaglio I, Rizzo E, Molteni E, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood 2017;129:3371–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Clough CA, Pangallo J, Sarchi M, Ilagan JO, North K, Bergantinos R, et al. Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 2022;139:2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taylor J, Mi X, North K, Binder M, Penson A, Lasho T, et al. Single-cell genomics reveals the genetic and molecular bases for escape from mutational epistasis in myeloid neoplasms. Blood 2020;136:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Will B, Zhou L, Vogler TO, Ben-Neriah S, Schinke C, Tamari R, et al. Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood 2012;120:2076–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Coelho-Silva JL, Silveira DRA, Pereira-Martins DA, Rojas CAO, Lucena-Araujo AR, Rego EM, et al. Molecular-based score inspired on metabolic signature improves prognostic stratification for myelodysplastic syndrome. Sci Rep 2021;11:1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fritsch M, Günther SD, Schwarzer R, Albert MC, Schorn F, Werthenbach JP, et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 2019;575:683–7. [DOI] [PubMed] [Google Scholar]
- 47. Park JH, Kang HJ, Lee YK, Kang H, Kim J, Chung JH, et al. Inactivation of EWS reduces PGC-1α protein stability and mitochondrial homeostasis. Proc Natl Acad Sci U S A 2015;112:6074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Results
Mutation profiles of clusters identified by unsupervised clustering of genetic aberrations and somatic mutations. WHO subtypes and bone marrow ring sideroblasts percentage (BM RS %) are shown above the oncoplot.
Clonal hierarchy analysis of SF3B1-SRSF2 co-mutated cases. A. Results from the hierarchical rank analysis of mutations detected by DNAseq in the 4 cases harboring both SF3B1 and SRSF2 mutation using Pyclone. The analysis was also performed using DPClust, which provided converging results (data not shown). Tumor cell fraction (axes) for each detected clone and its 95% confidence interval (dot size) is represented in a scatter-plot, using different color according to the cluster driving mutation (SF3B1, red; SRSF2, blue; other drivers, gray). SF3B1 dominancy in MDS392 and MDS 640, together with the SRSF2 large clone size are suggestive of both splicing factor mutations in the same clone. On the contrary, MDS694 and MDS965 had a dominant SF3B1 clone associated with a very little and probably independent SRSF2 secondary clone. B. Results from single-cell derived colony-forming unit (CFU) genotyping confirming the concurrent double splicing factor mutations within the same clone in patient MDS382 and MDS640. CFU experiment was not carried out for the other two cases because of the very low probability of SF3B1/SRSF2 double positive clone identification, as already suggested by current data in the literature related to the presence of SF3B1K700E mutation (ref. 1).
Overall survival of MDSRS+ with SRSF2 and TP53MH genotype stratified by RS burden. Cases (colored Kaplan-Meier curves) were compared to historical controls (gray curve) with similar clinical and molecular features and no RS (i.e. RS<5%) from two large studies evaluating clinical characteristics of SRSF2 and TP53MH -mutated MDS, respectively (ref. 2,3). SF3B1 mutated MDSRS+ were not considered for this analysis because of the small number of cases with RS between 5 and 15% (2 out of 82 SF3B1-mutated ones) and consolidated evidences in the literature showing no effect of RS burden on SF3B1-mutated MDS outcome (ref. 4,5); sample size also limited further analysis in the MDSRS+ NOS category.
UMAP plot of MDSRS+ bone marrow CD34 mononuclear cells transcriptomes. Each point is one patient, overlaid with results from unsupervised clustering.
Cumulative proportion of principal components derived from principal component analysis. The first 14 explain 2/3 of the total variability.
– Results from Gene Ontology enrichment analysis (molecular function dataset) for the top 250 contributors of principal component 1 (PC1)
Results from Gene Ontology enrichment analysis (molecular function dataset) for the top 250 contributors of principal component 2 (PC2)
Unsupervised consensus clustering analysis on gene expression data. Consensus clustering applied to the MDSRS+ cohort revealed 3 transcriptomic profiles, named EMK, INT and IMP according to the results from gene set enrichment analysis.
Item consensus for K=3 showing stable item-consensus values across the whole cohort.
Optimal number of clusters defined by average silhouette width method.
ABCB7 expression throughout the spectrum of MDSRS+ genotypes. ABCB7 downregulation is restricted to SF3B1-mutant MDSRS+.
Scatter plots comparing mean PSI values between the MDSRS+ and healthy donors used as normal bone marrow controls (NBM), stratified by genetic classification. Significantly associated events (FDR < 0.0001) are color coded by splicing alterations subtypes.
Digital sorting analysis on bulk CD34+ MNC transcriptome from MDSRS+ cases. Plots refer to unfiltered data, i.e. including hematopoietic stem/progenitor and mature hematopoietic cells. CMP, common myeloid progenitors; EryP, erythroid precursors; GMP, granulocyte–monocyte progenitors; Gran, granulocytes; HSC, hematopoietic stem cells; Lymph, lymphocytes; MK, megakaryocytes; MLP, multi-lymphoid progenitors; MPP, multipotent progenitors; MEP, megakaryocyte-erythroid progenitors). Cases (x-axis) are sorted by decreasing estimated MEP abundance, showing enriched IMP signature in cases with MEP depletion on right-side of the plots.
The proportion of estimated erythroblast-like cells correlate positively with the percentage of RS in BM smears.
In-silico cytometry data validation. A) Experimental workflow and multi parameter flow cytometry gating strategy used to sort Lin+ and Lin- from CD34+ MNC and to explore HSPC composition. B) Scatter plot comparing results from multi-parameter-flow (x-axis) and in-silico cytometry (y-axis) partitioned by megakaryocyte/erythroid progenitors (MEP), granulocyte/macrophage progenitors (GMP) and hemopoietic stem cell (HSC), showing significant concordances for MEP and GMP (Pearson correlation coefficient 0.94 and 0.87, P 0.02 and 0.05, respectively; correlation line and 95% confidence interval are represented in dark and light gray, respectively).
EMK and IMP signature validation on HSPC sorted populations. Differential gene expression analysis across MDSRS+ with EMK profile, MDSRS- with IMP profile and NBM (3 cases each) according to Lin marker expression. Differential expressed genes (row) between EMK and IMP groups were selected. Each row represents a gene, and each column represents a sample. Design comparison (case) and sorted population (condition) are shown as covariates.
Gene set enrichment analysis on differentially expressed genes in LIN- CD34+ cells. Three MDS with RSEMK were compared to 3 MDS without RSIMP and displayed an enrichment of genes expressed in early erythroblast/megakaryocyte progenitor genes and a depletion of genes expressed in myeloid progenitors.
Distribution of estimated MEP proportion according to bone marrow blast percentage. P-value for each pairwise comparison is reported in the upper part of the plot.
Prognostic effect of genomic and transcriptomic analyses on MDSRS+ outcome. Overall survival (OS) stratified by genomic (A), transcriptomic classification (B) and estimated MEP percentage (C) in all MDSRS+ (A-C) and MDS-RS-SLD/MLD only (D-F). Multivariable Cox proportional hazard model for OS in all MDSRS+ including age, IPSS-mol score and estimated MEP percentage as continuous variables (G). OS stratified by estimated MEP percentage and IPSS-mol risk category (full representation of the 6 IPSS-mol categories shown in Supplemental Figure 21B).
MDSRS+ event free survival (EFS) stratified by estimated MEP percentage in the four genetic subgroups (A-D).
MDSRS+ event free survival (EFS, A) and overall survival (OS, B) stratified by estimated MEP percentage and IPSS-mol risk category.
Transcriptomic impact on SF3B1 mutant MDS and low blasts outcome. Overall survival (OS) and Event Free Survival (EFS) stratified by transcriptomic classification (A-B) and estimated MEP percentage (C-D), respectively, in SF3B1 mutant MDS and low blasts, as defined according to ICC and WHO 2022 classification.
RNA Sequencing metrics.
Demographic and clinical features of cases and controls included into the study.
List of differentially expressed genes associated with EMK, INT and IMP profile.
Variables significant for survival analysis in univariate analysis
Treatment. Number of cases that underwent to each treatment category and the associated hazard-ratio for death is reported for each genetic subgroup and for the whole cohort.
Data Availability Statement
Deidentified individual participant data are available in the Swedish National Data Service repository at https://doi.org/10.48723/zt59-8x04 upon request.