Abstract
Purpose:
Develop and deploy a robust discovery platform that encompasses heterogeneity, clinical annotation, and molecular characterization and overcomes the limited availability of prostate cancer models. This initiative builds on the rich MD Anderson (MDA) prostate cancer (PCa) patient-derived xenograft (PDX) resource to complement existing publicly available databases by addressing gaps in clinically annotated models reflecting the heterogeneity of potentially lethal and lethal prostate cancer.
Experimental Design:
We performed whole-genome, targeted, and RNA sequencing in representative samples of the same tumor from 44 PDXs derived from 38 patients linked to donor tumor metadata and corresponding organoids. The cohort includes models derived from different morphologic groups, disease states, and involved organ sites (including circulating tumor cells), as well as paired samples representing heterogeneity or stages before and after therapy.
Results:
The cohort recapitulates clinically reported alterations in prostate cancer genes, providing a data resource for clinical and molecular interrogation of suitable experimental models. Paired samples displayed conserved molecular alteration profiles, suggesting the relevance of other regulatory mechanisms (e.g., epigenomic) influenced by the microenvironment and/or treatment. Transcriptomically, models were grouped on the basis of morphologic classification. DNA damage response–associated mechanisms emerged as differentially regulated between adenocarcinoma and neuroendocrine prostate cancer in a cross-interrogation of PDX/patient datasets.
Conclusions:
We addressed the gap in clinically relevant prostate cancer models through comprehensive molecular characterization of MDA PCa PDXs, providing a discovery platform that integrates with patient data and benchmarked to therapeutically relevant consensus clinical groupings. This unique resource supports robust hypothesis generation and testing from basic, translational, and clinical perspectives.
Translational Relevance.
The discovery platform, founded on 44 comprehensively characterized MD Anderson prostate cancer patient-derived xenografts spanning the prostate cancer spectrum and linked to donor tumor metadata, recapitulates the molecular landscape of potentially lethal and lethal prostate cancer. This unique data inventory integrating genomic and transcriptomic analysis, complements publicly available datasets to enhance hypothesis generation and testing, enabling the informed selection of clinically annotated models suitable for experimentation. This rich resource supports clinical, translational, and basic research perspectives, with the aim to identify markers and/or drivers of progression and to prioritize targets for prosecution by combination/sequential treatment strategies.
Introduction
Metastatic prostate cancer that progresses after androgen ablation therapy [castration-resistant prostate cancer (CRPC)] remains incurable. Although recent large-scale genomic studies have identified deregulated pathways in metastatic CRPC (1, 2), our understanding of metastasis and therapy resistance has been hindered by a lack of models representing the clinical and biological complexity of prostate cancer. Development of patient-derived xenografts (PDX) has led to therapeutically relevant approaches (3–6), further driving the impetus to characterize and improve PDX utility to bridge the gap between experimental and clinical observations, complementing publicly available patient databases to enhance hypothesis generation and testing.
The MD Anderson (MDA) prostate cancer (PCa) PDX series is a collection of clinically annotated models representing the range of potentially lethal disease, namely therapy-naïve and therapy-resistant prostate cancer, encompassing clinical (classic and aggressive) and morphologic [e.g., adenocarcinoma (Ad), neuroendocrine carcinoma (NEPC)] variants from both primary sites and metastases, and reflecting the progression of the donor tumor (7, 8). In contrast with other PDX repositories, our program collects non–end-stage disease samples, including models from different areas of the same tumor, which represent prostate cancer heterogeneity, and those taken before and after therapy, which enable longitudinal studies.
Aiming to potentiate the utility and applicability of this PDX collection, we selected 44 MDA PCa PDX models derived from 38 patient tumors, reflecting different morphologic groups, treatment statuses, and organ sites of involvement, and performed whole-genome sequencing (WGS), targeted sequencing, and RNA sequencing (RNA-seq). The molecular and morphologic analyses of each PDX were performed in representative samples of the same tumor, facilitating the integration of the different approaches. Complementing patient data and recapitulating the genomic landscape found clinically, this comprehensive PDX platform provides a resource for clinical and molecular interrogation of models suitable for experimentation.
Materials and Methods
Animals
Experiments were conducted in accordance with standards of care and approved by the Institutional Animal Care and Use Committee at MD Anderson Cancer Center (MDACC).
PDX establishment and propagation
PDX propagation and harvesting was performed as described previously (8). Briefly, MDA PCa PDXs are established from tumor specimens obtained from patients with prostate cancer undergoing radical prostatectomy, cystoprostatectomy/pelvic exenteration, or resection/biopsy analysis of metastatic lesions. Tissues are evaluated and macrodissected by a pathologist for implantation in intact male CB17/Icr-Prkdcscid/IcrlcoCrl mice (RRID: IMSR_CRL:236; Charles River Laboratories). Grafts are implanted subcutaneously on the flank with supplemental Matrigel. Mice are monitored for initial growth up to 18 months after implantation. Tumors that grow are serially passaged without Matrigel. A line is considered established after five passages. At each passage, representative samples are stored in various forms for characterization and continued propagation. These PDXs are submitted to rigorous quality control. Tumor samples from later passages (>3) were used for sequencing.
MDA PCa PDX selection
We selected 44 PDXs derived from 38 patients with clinically annotated, potentially lethal, hormone-naïve prostate cancer or CRPC from primary or different metastatic sites. Reflecting the current clinical prostate cancer landscape, the cohort includes Ads (including ductal Ad), NEPCs, poorly differentiated carcinomas, and sarcomatoid carcinomas. Model selection prioritized reasonable propagation rate to allow for experimentation (Supplementary Table S1). Sample selection criteria: consistent morphology, high tumor content (>80%), and minimal areas of necrosis or fibrosis (<20%). PDX morphologic classification was performed and routinely reviewed by a clinical pathologist (P. Troncoso), verifying consistency with the tumor of origin. Each selected PDX tumor was divided into representative samples to perform next-generation sequencing (NGS) and formalin-fixed paraffin embedding, also reviewed by the clinical pathologist (P. Troncoso). NEPC classification was defined by morphology, lack of androgen receptor (AR), and at least one positive neuroendocrine marker by IHC (chromogranin A or synaptophysin). Double-negative (DN) carcinomas were defined by lack of both AR and NEPC markers.
DNA and RNA preparation
DNA and RNA were extracted from fresh frozen PDX tissue at the Biospecimen Extraction Facility (MDACC) using the QIAGEN QIAamp DNA Mini and RNeasy Kits, respectively.
NGS
We performed WGS, targeted sequencing [T200.1 panel, including 263 genes implicated in solid cancers (Supplementary Table S2)], and RNA-seq at the Advanced Technology Genomics Core (MDACC).
Targeted sequencing
Illumina-compatible indexed libraries were prepared from 200 ng of Biorupter Ultrasonicator (Diagenode) sheared gDNA using the KAPA Hyper Library Preparation Kit (Kapa Biosystems). Library size distribution was assessed using the 4200 TapeStation High Sensitivity D1000 ScreenTape (Agilent Technologies, RRID:SCR_018435). Libraries were then prepared for capture with seven cycles of preligation-mediated PCR. Amplified libraries were assessed as above and quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Equimolar quantities of libraries were then multiplexed, six libraries per pool. Targeted capture was performed using the SeqCap EZ Library T200.1 panel. Target-enriched library pools were amplified using six cycles of PCR and assessed as above for size distribution and quantity. Equimolar quantities of library pools were multiplexed, and the resultant combined pool was quantified by qPCR using the KAPA Library Quantification Kit (Kapa Biosystems), then sequenced in one lane of the NovaSeq6000 (RRID:SCR_016387) S2-Xp flow cell using the 100-nt paired-end format.
Stranded mRNA
Stranded mRNA libraries were prepared using the KAPA Stranded mRNA-seq Kit. Briefly, 250 ng of total RNA was captured using magnetic Oligo-dT beads. Resultant PolyA RNA was fragmented using heat and magnesium. First-strand synthesis was performed using random priming followed by second-strand synthesis with the incorporation of dUTP into the second strand. The ends of the resulting double-stranded cDNA fragments were repaired, 5′-phosphorylated, 3′-A tailed, and Illumina-specific indexed adapters were ligated. The products were purified and enriched for full-length library with nine cycles of PCR. The strand marked with dUTP was not amplified, resulting in a strand-specific library. The libraries were quantified and assessed for size distribution as described above, then multiplexed, 48 libraries per pool. The library pool was quantified and sequenced as described above.
WGS
Illumina-compatible indexed libraries were prepared from 200 ng of Diagenode Biorupter Pico sheared DNA using the KAPA Hyper Library Preparation Kit (KAPA Biosystems, Inc.). Libraries were amplified by two cycles of PCR, then assessed for size distribution and quantified as described above. Equimolar quantities of the indexed libraries were multiplexed, 24 libraries per pool. The pool was quantified as described above. Then each pool was sequenced on the Illumina NovaSeq6000 S4 flow cell using the 150-nt paired-end format.
Data processing
NGS data were processed using an established in-house bioinformatics pipeline (9) at the MDACC Department of Genomic Medicine. Briefly, raw sequencing base call (BCL) files were first converted into FASTQ files using Illumina bcl2fastq2 conversion software v2.20. DNA sequences were then aligned to the combined human (hg19) and mouse (mm10) reference genome using the Burrows-Wheeler Aligner (RRID:SCR_010910). Picard toolset was used to convert the data into BAM format with duplicate reads (v1.112, http://broadinstitute.github.io/picard/, RRID:SCR_006525). Finally, the genome analysis toolkit (GATK, RRID:SCR_001876) was used to perform local realignments. Reads mapped to mm10 were filtered out (Supplementary Table S3) to keep HumanOnly (hg19) alignments. MuTect (RRID:SCR_000559) was used to identify somatic SNPs and small insertions and deletions (indel). ANNOVAR (RRID:SCR_012821) was applied to annotate each genetic variant with coding sequence change and allele frequency in control populations, including Exome Aggregation Consortium (ExAC, RRID:SCR_004068), Genome Aggregation Database (gnomAD, RRID:SCR_014964), the 1000 Genomes Project (RRID:SCR_008801), and NHLBI-ESP (RRID:SCR_012761) 6500 exomes.
The following filters were applied to select somatic mutations of good sequencing quality: total reads ≥20 in the tumor sample and ≥10 in the normal sample (matched germline sample or merged normal tissues); for SNVs, variant allele frequency (VAF) ≥0.02 in the tumor sample and ≤0.02 in the normal sample; for indels, VAF ≥0.05 in the tumor sample and not observed in normal tissue; length of indels ≤100 bp; exclusion of intronic and intergenic mutations; population allele frequency <0.01 in all four control databases [DNA allele changes observed from healthy populations or populations with some common/non-cancer diseases (e.g., diabetes)]. Mutations that are annotated as oncogenic or likely oncogenic in OncoKB (RRID:SCR_014782) or pathogenic or likely pathogenic in Sanger (RRID:SCR_011784) database were rescued even if they were found in control databases.
For the purpose of this publication, we focused on known prostate cancer–related genes (refs. 1, 2, 10; https://cancer.sanger.ac.uk/census).
SNPs and indels were determined from targeted sequencing data. For data representation in the article, we considered mutations annotated as driver mutations in cBioportal (e.g., OncoKB, Cancer Hotspots) and VAF ≥0.1.
Somatic copy-number variations (CNV) were identified from WGS data compared with a common normal consisting of 11 blood samples from patients with non-prostate cancer using HMMcopy (Bioconductor; RRID:SCR_006442). The gene-level copy number was assessed by calculating the mean value of the derived log2 scores of tumors versus normal reads for each gene. Copy-number status was further categorized using log2.mean cutoffs: ≥1.5, Amp.; 1–1.5, Gain; −2 to −1, Shal. Del.; ≤−2 Deep Del. In cases where no alterations were found for PTEN, TP53, or RB1, manual inspection was performed to determine transcript loss.
Gene fusions were assessed in both WGS and RNA-seq data. Seven bioinformatics software tools were used: three tools for WGS [brass (RRID:SCR_017091), delly (RRID:SCR_004603), lumpy (RRID:SCR_003253)] and four for RNA-seq [deFuse (RRID:SCR_003279), MapSplice (RRID:SCR_010844), TopHat-Fusion (RRID:SCR_011899), FusionMap (RRID:SCR_005242)]. We used Integrative Genomics Viewer (IGV; RRID:SCR_011793) for breakpoint fine mapping.
Organoids
Fresh PDX tissue was chopped, digested with collagenase II (R&D) on advanced DMEM/F-12 (Gibco) with 1% PenStrep (Sigma-Aldrich), 10 mmol/L HEPES (Gibco), and 1X GlutaMAX (Gibco) on an orbital shaker (37°C, 2 hours), and passed sequentially through a 100- and 70-μm strainer. After centrifugation, cells were incubated for 5 minutes with ammonium–chloride–potassium (ACK) lysis solution (Quality Biological). Cells were plated in PCM media for 2 to 4 days and then plated in Cultrex Basement Membrane Extract (Cell density: 1 × 106 in 100 μL) in PCM medium. For histology, organoids were fixed with 10% formalin ON, transferred to 70% ethanol, and paraffin embedded. Hematoxylin and eosin and IHC using anti-AR antibody (Agilent, M3562, RRID:AB_2060174) were performed using standard techniques (11).
PCM medium was prepared as in ref. 12 with these modifications: Noggin conditioned medium 10%, R-spondin 1 conditioned medium 5%. N-acetyl-L-cysteine and SB202190 were not added.
Organoid genetic modification
PDX-derived cells were transduced with lentiviral particles for GFP expression (Origene) following manufacturer's protocol (multiplicity of infection ≈ 20), plated, and treated with antibiotic for selection.
Drug testing
Organoids were treated with cisplatin, paclitaxel, cabazitaxel, bicalutamide (Casodex), enzalutamide, and/or niraparib (Selleckchem) or vehicle [ethanol (for cisplatin) or DMSO] for 96 hours in 96-well plates. Viability was evaluated with Promega CellTiter-Glo 3D Cell Viability Assay.
ARv7
IGV sashimi plots were used to identify putative ARv7 variants (≥5 reads). We confirmed the presence of ARv7 by IHC, using standard techniques (11) and anti-ARv7 antibody, clone RM7 (RevMab Biosciences, RRID:AB_2716436). Staining was evaluated by semiquantitative analysis of pattern, percentage of cells, and intensity on a scale of 1 to 3.
Analysis of Ad and NEPC morphologic groups
Differential gene expression between PDX groups was obtained from cBioPortal (RRID:SCR_014555) “group comparison” function. Volcano plot was created with vplot function ggplot2 package (RRID:SCR_014601) on R. Interactive plot (http://rpubs.com/nanselmino/1084846) was created and shared using the Plotly package on R. Ingenuity Pathway Analysis (IPA, QIAGEN, https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/; RRID:SCR_008653) was used to study differentially regulated canonical and metabolic pathways. Analysis was based on log2ratio with q-value cutoff <0.05. Benjamini–Hochberg multiple testing correction P value was performed.
Unsupervised clustering analysis of DNA damage response (DDR) genes was performed using ggplot2 and pheatmap (RRID:SCR_016418; https://rdrr.io/cran/pheatmap/) packages in R.
FGFR1 CpG methylation
Bisulfite pyrosequencing methylation analysis of the FGFR1 promoter region was performed at the MDACC Epigenomics Profiling Core on 1 μg DNA in 40 μL water/EB buffer. Average of all CpG sites within an assay is presented.
Patient datasets
The Cancer Genome Atlas (TCGA)–prostate adenocarcinoma (PRAD; ref. 13) and Stand Up To Cancer (SU2C; ref. 10) datasets, containing data from primary prostate cancer or CRPC samples, respectively, were analyzed on cBioPortal.
Statistical analysis
Differences between NEPC and Ad were assessed with Fisher exact test (mutations and CNVs) using “R” (RRID:SCR_001905). Comparison of mRNA expression between groups was analyzed by t test (two groups) or one-way ANOVA (>2 groups). Spearman Pearson method was used for correlation analyses.
Data availability
Data are available in cBioPortal [Prostate Cancer MDA PCa PDX (MD Anderson; https://www.cbioportal.org/study/summary?id=prad_msk_mdanderson_2023)] and dbGaP (https://www.ncbi.nlm.nih.gov/gap/; RRID:SCR_002709), accession number phs003420.v1.p1. MDA PCa PDXs are available through a material transfer agreement. Contact e-mail: pcapdxprogram@mdanderson.org.
Results
The MDA PCa PDX program is an accessible, dynamic collection, representing the evolving landscape of prostate cancer progression under conventional and novel therapies (androgen deprivation, chemotherapy, and targeted therapies). To date, we have developed two cell lines and 150 PDXs derived from 90 patients with prostate cancer (8). The establishment of conditions to grow prostate cancer cells as organoids (refs. 14, 15; Supplementary Fig. S1A and S1B) has enabled the use of PDXs for in vitro experimentation, that is, genetic editing (Supplementary Fig. S1C) and drug testing (Supplementary Fig. S1D and S1E), widening the disease spectrum represented by the available prostate cancer cell lines.
We performed WGS (30X coverage), exome-targeted sequencing (400X coverage) for genes implicated in solid cancers (T200.1 panel; Supplementary Table S2), and RNA-seq of 44 MDA PCa PDX models derived from 38 patient tumors, reflecting the various morphologic groups, treatment statuses, organ sites of involvement, and racial distribution (Fig. 1A; Supplementary Table S1). Clustering based on transcriptomic data resulted in groups consistent with morphologic classification (Fig. 1B).
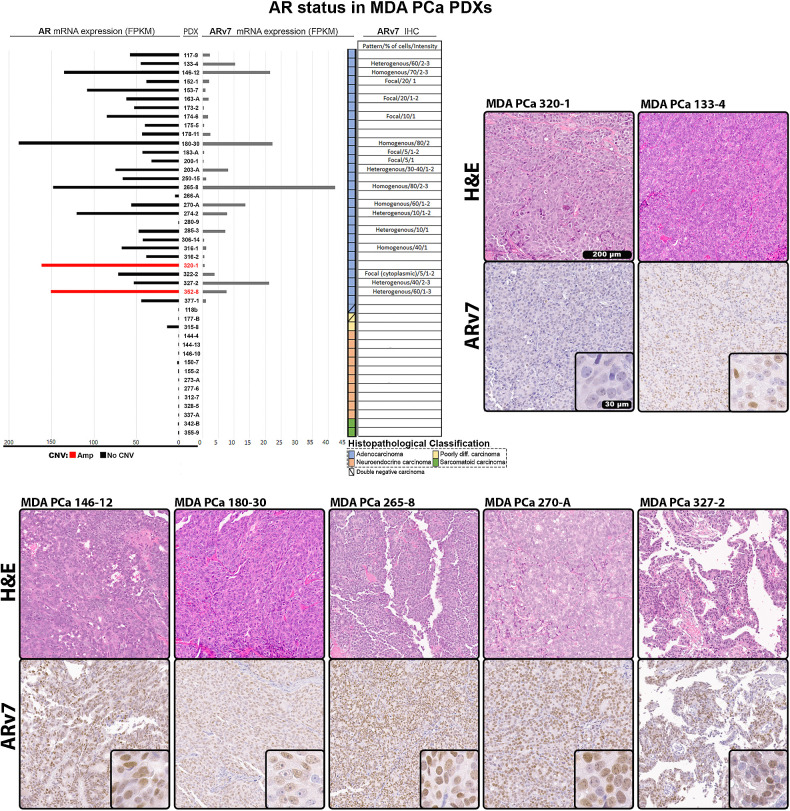
Figure 1.
Morphologic and clinical distribution of the MDA PCa PDX cohort. A, Morphologic classification, site of tumor collection, and previous treatment of the 44 PDXs (from 38 patients with prostate cancer) used for genomic analysis [WGS, targeted sequencing (T200.1 panel; 263 genes), and RNA-seq]. Note: second- or next-generation androgen receptor signaling inhibition (ARSI) comprises abiraterone acetate, enzalutamide, and apalutamide. B, Heat map showing transcriptomic data covariance clustering. Highlighted and colored are longitudinal and heterogenous samples from the same patient, respectively. CTC, circulating tumor cells; diff, differentiated.
Genomic alterations of prostate cancer–associated genes
Mutations and CNVs frequently observed in prostate cancer were found in our cohort (Fig. 2A). PDXs presented chromosome 8 copy-number gains (8q) and losses (8p; Supplementary Fig. S2A), characteristic of prostate cancer (16). Prostate cancer is characterized by deletions (10, 13), and our PDXs showed a prevalence of deep deletions, mainly phosphatase and tensin homolog (PTEN) and retinoblastoma 1 (RB1; Supplementary Fig. S2B). Thus, critical aspects of clinical prostate cancer are reflected in the PDXs. In some cases, when comparing the CNV profile from WGS with T200 and RNA-seq reads (Supplementary Fig. S4), we found lack of expression (full/partial), consistent with deletion in a portion of the gene.
Figure 2.
Landscape of genomic alterations of known prostate cancer–associated genes in MDA PCa PDXs. A, Oncoplot depicting SNP/indel (allele frequency > 0.1) and CNV of known prostate cancer–associated genes identified by targeted sequencing (T200.1 panel) and WGS, respectively, in the 44 MDA PCa PDXs studied. Highlighted and colored are longitudinal and heterogenous samples from the same patient, respectively. B, Mutation (mut), CNV, and mRNA status of genes frequently altered in prostate cancer. Highlighted in purple are the models with no relevant alterations in the genes analyzed. *, Full or partial copy-number/transcript loss determined by manual inspection for TP53, RB1, and PTEN; +, found on DNA only, together with presence of an interstitial loss between the genes in the copy-number profile; Amp, amplification.
Several fusions previously identified in human prostate cancer (1, 8, 17–19) were detected in our PDXs at both the DNA and RNA levels (Supplementary Table S4; Supplementary Fig. S3). We found transmembrane serine protease 2–ETS-related gene (TMPRSS2-ERG) fusion in 13 models, but only 10 correlated with increased ERG expression (Supplementary Fig. S3A). Lack of correlation was consistent with low/null AR, a known positive TMPRSS2 regulator. Although in 144-13 the fusion was detected only at the DNA level, CNV profiles showed that 144-4 (TMPRSS2-ERG detected in DNA and RNA) and 144-13, both derived from the same tumor, have an interstitial loss between TMPRSS2 and ERG (Supplementary Fig. S3B).
Fusions with other members of the ETS family have also been found in prostate cancer (19). We identified TMPRSS2-ETV4 fusion with increased ETV4 expression in 2 PDXs (Fig. 3A). ETV1 was found in four other fusions (ETV1-FOXA1, FOXA1-ETV1, ACSL3-ETV1, ETV1-ACSL3; Supplementary Table S4), but no changes in the expression of these genes were detected. The biological consequences of these fusions remain to be elucidated. We also identified SLC45A3-ELK4 fusion in 11 Ad models at the RNA level, consistent with being reported as a posttranscriptional fusion in prostate cancer (refs. 20, 21; Fig. 3A).
Figure 3.
AR status in MDA PCa PDXs. Expression of AR and ARv7 evaluated from RNA-seq data in MDA PCa PDXs. AR bars in the chart are depicted in red for AR amplification or black for no CNV. Table and representative images show IHC for ARv7 performed in samples from the 44 MDA PCa PDX models. Scale bar: 200 μm and 30 μm in magnified section. Blank spaces in the table correspond to negative staining. Staining was evaluated by semiquantitative analysis of pattern, percentage of cells, and intensity on a scale of 1 to 3. FPKM, fragments per kilobase of transcript per million mapped reads.
Prostate cancer drivers
We found oncogenic molecular alterations in PTEN, RB1, TP53, and AR in most PDXs (Fig. 2B). 265-8 had high AR expression in the absence of alterations, consistent with AR enhancer region amplification (ref. 22; Supplementary Fig. S3C), highlighting the importance of this regulatory region. However, other models presenting this amplification did not exhibit increased AR expression (Fig. 2B). Cases like 306-14 and 273-A exhibited low/null PTEN or RB1 expression, respectively, which was not associated with deep deletion or mutation (Fig. 2B; Supplementary Figs. S2B and S4).
Consistent with clinical findings, 9% of PDXs lacked alterations in prostate cancer driver genes (Fig. 2B, highlighted). In these models, we looked at other reported cancer genes, including ETS family gene fusions (1, 23) and CDK12 alterations (Supplementary Fig. S3D). We found four PDXs with CDK12 alterations in the cohort. 174-6 and 322-2 presented two CDK12 mutations each, likely resulting in biallelic loss. In addition, examination of CNV profiles showed that 117-9 and 328-5 exhibit typical small focal gains associated with CDK12 loss (24) but lack biallelic mutations (Supplementary Fig. S5A). 117-9 exhibited loss of the last two CDK12 exons (RNA-seq and CNV profiles), likely due to structural variation (Supplementary Fig. S5B). 328-5 had a monoallelic missense mutation in the kinase domain that is not reported as a driver mutation per OncoKB. RNA-seq showed that monoallelic mutations represent approximately 80% of transcripts, suggesting inactivation of the wild-type allele (Supplementary Fig. S5C). Moreover, there was an aberrantly spliced exon in approximately 20% of reads, suggesting that the nonmutated allele harbors aberrant splicing, which leads to biallelic inactivation, depicting different mechanisms of CDK12 inactivation.
Only 133-4 and 316-2 exhibited known AR oncogenic mutations (H875Y and T878A, respectively), while 320-1 and 352-8 had AR amplification with concomitant high expression. 265-8 had high AR expression explained by AR enhancer region amplification (Supplementary Fig. S3C). Nonetheless, 180-30 had the highest mRNA levels in absence of alterations, implicating transcriptional regulation of AR beyond specific alterations at the gene locus.
We also evaluated the best-characterized AR splice variant, ARv7 (a constitutively active variant and proposed prostate cancer driver; ref. 25), analyzing the inclusion/exclusion of the cryptic exon 3 on RNA-seq reads, and confirming the presence of ARv7 by IHC (Fig. 3). Immunostaining was consistent with mRNA levels, with 146-12, 180-30, 265-8, 270-A, and 327-2 having the highest ARv7 expression. It remains unknown whether ARv7 could be a prostate cancer driver by itself (25).
Prostate cancer heterogeneity
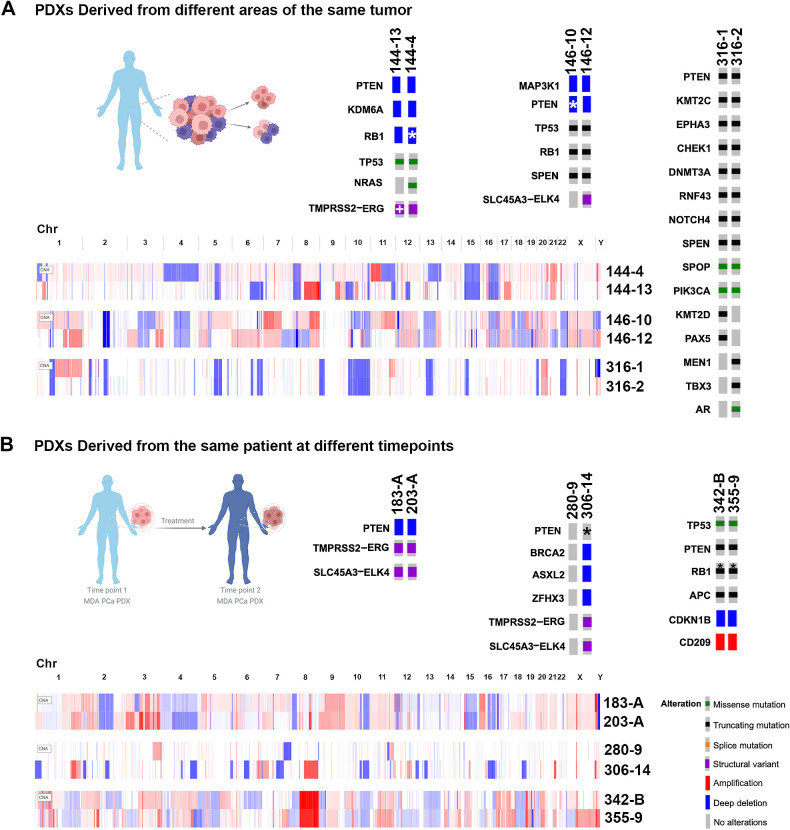
Different PDXs were derived from different areas of the same tumor or from CTCs obtained at the same time (144, 146, 316; Supplementary Tables S2, S4, and S5). CNV profiles evidenced similarities shared between sample pairs in certain chromosome regions, as well as specific alterations for each model, reflecting heterogeneity (Fig. 4A).
Figure 4.
Alterations in MDA PCa PDX pairs derived from the same patient tumor and longitudinal samples. A, Oncoprints depicting alterations in models derived from two areas of the same tumor. The following pairs are represented: 144-13 and 144-4; 146-10 and 146-12; 316-1 and 316-2. B, Oncoprints depicting alterations in longitudinal samples. The following pairs are represented: 183-A and 203-A; 280-9 and 306-14; 342-B and 355-9. Note: oncoprints show only one alteration when multiple hits are present. *, Full or partial copy-number/transcript loss determined by manual inspection for TP53, RB1, and PTEN; TMPRSS2-ERG fusion in MDA PCa 144-13 was determined by the presence of an interstitial loss between the genes in the copy-number profile. (Patient schemes created with BioRender.com.)
144-13 and 144-4 were derived from different areas of a single primary NEPC. Both were AR null and shared genomic alterations in known prostate cancer drivers (PTEN and KDM6A deep deletion and TP53 mutation), consistent with null/almost null expression (Fig. 4A; Supplementary Table S5), and a deletion in the same RB1 region (Supplementary Fig. S4). Interestingly, 144-4 had an oncogenic mutation in NRAS (Q61K; Fig. 4A; Supplementary Table S5), seldomly implicated in prostate cancer (26).
146-10 and 146-12 were derived from different areas of a single primary mixed Ad-NEPC and subsequently established as pure NEPC (146-10) and Ad (146-12). As expected, the NEPC 146-10 did not express AR or its target, ZBTB16 (Supplementary Table S5). Despite differences in morphology, both shared TP53 and RB1 mutations and PTEN deletion (Fig. 4A; Supplementary Table S5) among other genomic alterations, including a mutation in SPEN, a hormone-inducible transcription repressor, implicated in prostate cancer (27).
316-1 and 316-2 were developed from a bone metastasis and CTCs, respectively, obtained at the same time from the same patient. Despite their different sites of origin, both established as Ads, exhibiting similar transcriptomic profiles (Fig. 1B). They shared mutations in some prostate cancer driver genes, including PTEN, negative regulator of Wnt pathway RNF43 (16), SPOP, and SPEN. However, they also presented mutations specific to each model, probably due to the influence of the microenvironment. 316-1 had PAX5 (28) and KMT2D (29) mutations, while 316-2 had TBX3 (30), MEN1, and AR mutations (Fig. 4A; Supplementary Table S5). Although both presented mutations in NOTCH4 and a second mutation in RNF43, the specific alterations were different in each model.
Overall, these results show that known prostate cancer gene alterations are highly preserved within heterogeneous pairs derived from the same tumor, highlighting their relevance as main disease drivers. Nevertheless, our results do not fully explain the differences between models in each pair, emphasizing the need to consider these drivers in the context of other specific alterations and design specific treatment strategies.
Longitudinal studies
By establishing PDXs from non–end-stage samples obtained from the same patient at different times during disease progression, we developed three longitudinal pairs: 183/203, 280/306, and 342/355 (Supplementary Tables S5 and S6).
In 183/203, 183-A was derived from a treatment-naïve Ad bone biopsy. The patient was placed on leuprolide [luteinizing hormone-releasing hormone (LHRH) agonist], and 1 year later, 203-A was derived from a second bone biopsy. These models displayed a conserved CNV profile with no major differences in molecular alterations (Fig. 4B; Supplementary Table S5).
The longitudinal pair 280/306 is a unique case, as 280-9 was obtained from a therapy-naïve lymph node metastasis with no molecular alterations in the best-known prostate cancer drivers (Fig. 2B), but with low/null AR expression (Supplementary Tables S5 and S6). The patient was placed on antiandrogen therapy and cabozantinib. One year later, we developed 306-14, which was derived from the prostate of the same patient and displayed several alterations not detected in 280-9 (Fig. 4B; Supplementary Tables S5 and S6). These models exhibited distinct CNVs and TMPRSS2-ERG status and did not share a single somatic variant on T200, suggesting that they either arose from distinct primaries or diverged early during tumorigenesis.
Sarcomatoid carcinomas 342 and 355 were developed from samples obtained 5 months apart from the prostate or local extension to bladder. These models presented identical mutations and similar CNV profiles (Fig. 4B; Supplementary Table S5).
Overall, our results show that longitudinal models from the same site mainly conserve the same molecular alteration profile, indicating that treatment does not define the prevalence of specific genomic alterations. It remains to be determined whether specific niches favor molecular profiles, conferring treatment resistance.
Molecular comparison between the largest morphologic groups
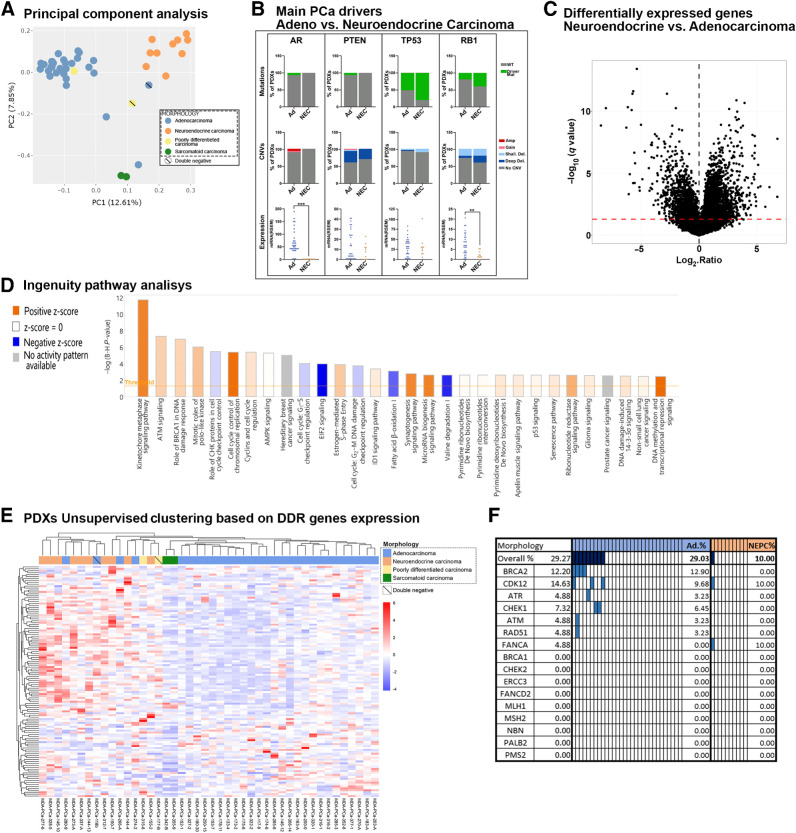
On the basis of transcriptomic profiles, principal component analysis (PCA) segregates groups according to morphologic classification (Fig. 5A). Similar to previous reports (10), four models [three Ad (118b, 266-A, 280-9) and one poorly differentiated carcinoma (177-B)] were localized in a region between both major clusters, Ad and NEPC. This segregation correlates with low (266-A) or lack of (118b, 280-9, 177-B) AR expression (Supplementary Fig. S6A). 118b, 177-B, and 280-9 fit the molecular definition of DN prostate cancer (negative for AR and neuroendocrine markers; ref. 31), reported to emerge following AR signaling inhibition (31). However, being a naïve case, 280-9 is not considered as a DN, requiring further investigation into whether 280-9 represents a rare case within this group (32). Of note, independent of their morphologic classification as poorly differentiated carcinomas, 315-8 clustered within the Ad group, whereas 177-B and 118b (both DN) both clustered closer to NEPC. This discrepancy could be related to the fact that, although low, 315-8 did express AR (Supplementary Fig. S6A).
Figure 5.
Molecular comparison between largest morphologic groups, Ad and NEPC. A, PCA of RNA-seq data. Scatter plot displays the first two principal components (PC). Each point represents a sample. Samples with similar gene expression profiles are clustered together. Samples are colored on the basis of morphologic groups. B, Percentage distribution of driver mutations or CNVs and expression of AR, PTEN, TP53, and RB1 in Ad and NEC (NEPC). P = 9 × 10−5 for RB1 and P = 4 × 10−9 for AR mRNA expression. C, Volcano plots depicting differential expression of each gene between Ad and NEPC PDX groups. Red dotted line represents a q-value of 0.05 to indicate significance threshold. D, Bar chart showing top 30 most significant “canonical pathways” identified from IPA based on differentially expressed genes (NEPC/Ad). The orange and blue bars indicate predicted pathway activation (z-score ≥ 2) or inhibition (z-score ≤ −2). For gray bars, no activity prediction can currently be made. White bars indicate pathways with z-scores at or very close to 0 or those that are ineligible for analysis because there are fewer than four molecules in the dataset associated with the pathway (z-score = NaN). E, Nonsupervised clustering based on the expression of DDR-associated genes in MDA PCa PDXs. Top column depicts model morphology. F, Genomic status of main DDR genes in Ad and NEPC MDa PCa PDXs. Colored rectangles indicate presence of driver genomic alterations.
We assessed mutation frequency, CNVs, and mRNA expression of AR, PTEN, TP53, and RB1 between Ad and NEPC (10, 33, 34). RB1 and AR expression were significantly lower in NEPC (Fig. 5B). Although no significant genomic differences were detected (probably due to cohort size), some trends were evident in the groups, including AR mutations and amplifications observed only in Ad, as well as higher frequency of TP53 mutations and RB1 mutations and deep deletions in NEPC (Fig. 5B). When considering CNVs and mutations together, RB1-altered models were more frequent in NEPC than in Ad (P = 0.0002).
We further compared the transcriptomes of major groups (Fig. 5C). IPA indicated differences at the metabolic level, mainly through downregulation of fatty acid β-oxidation and amino acid degradation in NEPC (Fig. 5D; Supplementary Fig. S6B). Major differences were found in cell cycle–associated pathways, mainly those related to DDR, which were upregulated in NEPC (Fig. 5D). As numerous studies reported impairment of DDR mechanisms in prostate cancer (35), we further examined DDR-associated gene expression (36) by nonsupervised clustering, resulting in groups concordant with morphologic classification (Fig. 5E). Consistent with the augmented expression of DDR genes in NEPC, these samples also exhibited elevated levels of MYCN and/or AURKA (Supplementary Fig. S6C), not associated with gene amplification (37). Frequent genomic alterations known to generate deficiencies in DDR (35, 38) were predominant in Ad PDX samples (Fig. 5F). The same profile was observed in the human prostate cancer SU2C dataset when samples were ranked on the basis of NEPC score and AR score (Supplementary Fig. S6D).
Exploiting the applicability of the PDX platform
Our group has a longstanding focus on the role of the FGF axis in advanced prostate cancer and bone progression (6, 39–41). We have previously shown that blockade of FGFR using the tyrosine kinase receptor inhibitor dovitinib has clinical activity in a subset of patients with CRPC skeletal metastases (6), emphasizing the need to stratify patients who will benefit from this targeted treatment.
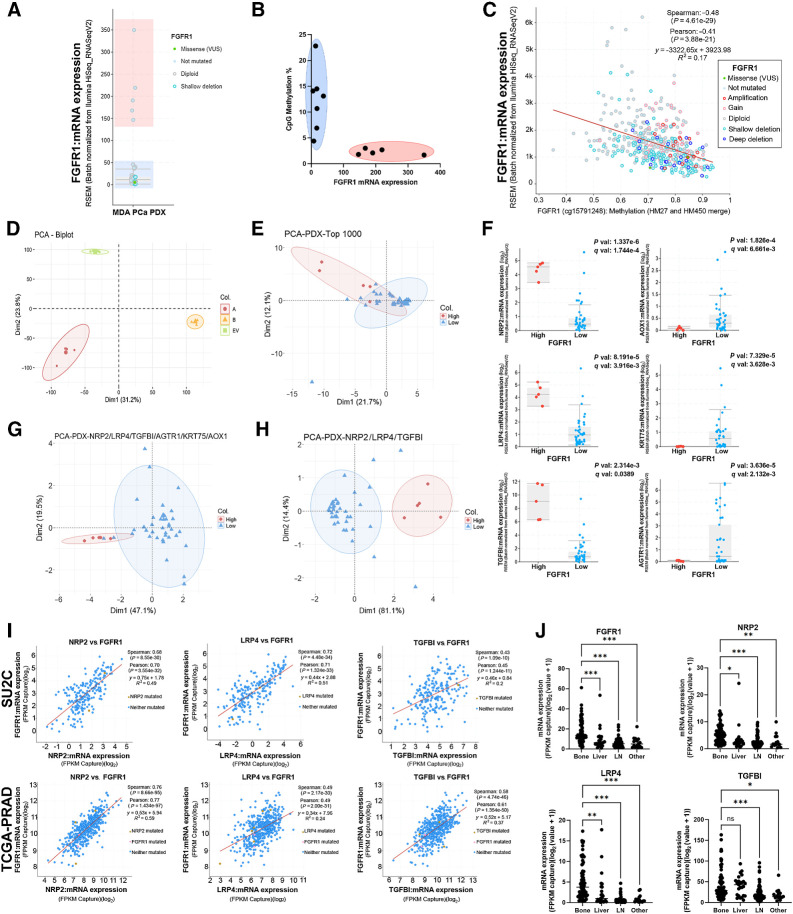
FGFR1 is the most prevalent FGFR in prostate cancer (6, 40). In line with clinical observations (40), elevated FGFR1 levels were not associated with genomic alterations in PDXs (Fig. 6A), suggesting the involvement of other regulatory mechanisms (e.g., epigenetics). Comparing PDX models with different FGFR1 levels assessed by RNA-seq, we found that CpG methylation at the FGFR1 promoter was associated with low expression (Fig. 6B). Accordingly, in TCGA-PRAD dataset (13), promoter methylation inversely correlates with FGFR1 expression (Fig. 6C), linking PDX findings to clinical observations.
Figure 6.
Exploiting the applicability of the PDX platform. A,FGFR1 mRNA expression assessed by RNA-seq in MDA PCa PDXs. Groups with low or high FGFR1 levels are highlighted in blue or red, respectively. B,FGFR1 promoter CpG methylation percentage versus mRNA levels in PDX models with high (red) or low (blue) FGFR1 expression. C,FGFR1 expression and promoter methylation in TCGA-PRAD. Red line depicts lineal regression. Spearman and Pearson methods were applied to assess correlation. D, PCA in PC3 cells expressing FGFR1 isoforms alpha (A), beta (B), or empty vector (EV). E, PCA of MDA PCa PDXs based on top 1,000 genes in dimension 2 with q-value <0.05 and logFoldChange >|0.2| from RNA-seq expression of PC3 cells expressing FGFR1 or controls. F, mRNA expression comparison for genes used in E that are differentially expressed between PDX models with high and low FGFR1 levels. Statistical significance was assessed via t test. G, PCA of MDA PCa PDXs based on genes from F. H, PCA of MDA PCa PDXs based on three genes from F that have high expression in the high-FGFR1 group. I, Correlation between FGFR1 and selected genes in SU2C and TCGA-PRAD. Red line depicts lineal regression. Spearman and Pearson methods were applied to assess correlation. J, Expression of FGFR1 and selected genes by metastatic site in SU2C. One-way ANOVA was used to assess statistical significance. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
Considering that the specific downstream targets of FGFR1 in prostate cancer remain to be defined, we explored RNA-seq data from PC3 cells overexpressing FGFR1 (alpha and beta isoforms, relevant for progression and bone dissemination; ref. 41) or empty vector (EV). PCA revealed a clear segregation between groups, with dimension 2 displaying the major separation between FGFR1 and EV (Fig. 6D). From the top 1,000 genes ranked on the basis of their contribution to dimension 2, we filtered those with q-value <0.05 and logFoldChange >|0.2|. The resulting 75 genes poorly separated models based on FGFR1 levels in PCA (Fig. 6E). Then, we kept the six genes that were differentially expressed between models with high and low FGFR1 levels (Fig. 6F), which improved the segregation between PDX samples (Fig. 6G). Three (NRP2, LRP4, TGFBI) out of these six genes displayed high expression in PDXs with high FGFR1 (Fig. 6F). Interestingly, the levels of these three genes accurately segregated models according to FGFR1 expression (Fig. 6H), providing confidence in the mechanistic association. To trace a parallelism with patient datasets, we corroborated the positive correlation between the expression of these genes and FGFR1 in SU2C (10) and TCGA-PRAD (ref. 13; Fig. 6I).
Furthermore, given the relevance of FGFR1 in prostate cancer bone progression, we detected its increased expression in bone metastases compared with other metastatic sites (Fig. 6J). NRP2, LRP4, and TGFBI were also prevalent in bone metastases, further supporting their link to FGFR1 (Fig. 6J).
Altogether, these results illustrate the utility of this queryable PDX platform for cross-interrogation with experimental and clinical data.
Discussion
PDXs have addressed the lack of models representing the prostate cancer spectrum and provided clinical relevance (3–6, 13, 14). Large-scale genomic studies of prostate cancer molecular alterations highlight the importance of in-depth characterization to improve model utility in tackling clinical gaps, linking molecular profiling to corresponding therapeutically relevant consensus clinical groups.
In this work, we performed a comprehensive characterization of a PDX cohort, integrating multiple approaches (WGS, targeted sequencing, RNA-seq) together with immunohistopathologic profiling within the same sample, spanning the clinical range of prostate cancer, including models derived from different areas of the same tumor or at different timepoints before and after therapy. This approach allows us to study alterations in the context of a larger and clinically annotated PDX collection, benchmarked to consensus clinical groupings and suitable for experimentation. Frequent prostate cancer genomic alterations were identified in our cohort, evidencing its clinical relevance to answer specific biological questions. Consistent with the clinic, 91% of PDXs presented oncogenic alterations in AR, RB1, TP53, or PTEN.
The in-depth analysis presented here contextualizes specific alterations with comprehensive knowledge of the genomic-transcriptomic scenario within each model and in comparison with the whole PDX cohort. This was illustrated by capturing CDK12 deficiencies that would have been missed in a nonintegrative analysis; detecting promoter amplification that could regulate ARv7; and defining a novel FGFR1 prostate cancer signature that could serve as an indicator of pathway activation. Altogether, these findings reinforce the utility of this PDX collection, which complements patient datasets, helping to define and select specific models to functionalize clinical observations and test hypotheses.
Prostate cancer is increasingly recognized as a heterogeneous disease (42, 43), leading to a lack of uniform therapy responses, underscoring the need to integrate our knowledge of prostate cancer biology into clinical application. For samples derived from different areas of the same tumor, we found that genomic alterations were mainly conserved, but some pairs exhibited differences in their transcriptomic profiles (144 and 146) and even distinct morphologies (146-10 NEPC; 146-12 Ad). This suggests that other regulatory mechanisms (e.g., epigenetics) could be implicated in tumor heterogeneity.
Longitudinal samples represent a precious subset derived from particular cases exhibiting early emergence of resistance, including metastatic hormone-naïve models. Although biased and limited in number, each of these unique pairs serves as a valuable source for understanding disease progression. We found preserved alteration profiles in longitudinal models, suggesting that, for tumors derived from the same site, treatment does not select for particular alterations and that driver alterations fail to explain treatment resistance. Furthermore, transcriptomic profiles also appear to be conserved (Fig. 1B). Notably, regardless of treatment status and collection site of the tumor of origin, our PDX models are all grown subcutaneously in untreated mice. Thus, the effect of treatment pressure or specific niches on these phenotypes remains to be addressed.
As seen clinically, Ad and NEPC are the main morphologic groups in our cohort. Comparison between these groups did not reveal significant differences in the frequency of AR, PTEN, TP53, and RB1 genomic alterations, despite observing some trends consistent with prior studies in human samples (10, 33, 34). The lack of significance could be a consequence of cohort size and of the fact that the models were selected to represent a wide range of the disease. Considered together, genomic alterations in RB1 were significantly more frequent in NEPC than in Ad (Fig. 5B). Furthermore, transcriptomic profiles were consistent with histomorphologic classification (Figs. 1B and 6A), and notably, NEPC presented increased DDR-associated processes compared with Ad. In addition to cell cycle modulation, the top altered DDR-associated categories involved BRCA, ATM, and CHEK signaling, suggesting that homologous recombination could be implicated. The inositol phosphate synthesis pathway, involved in DNA repair regulation (44), was also augmented in NEPC. Moreover, NEPC had higher expression of the main DDR genes, consistent with increased expression of AURKA and MYCN (37, 45), while driver mutations on these genes were more frequent in Ad. A GR-MYCN-CDK5-RB1-E2F1 mechanism was associated with enzalutamide-induced NEPC differentiation, impaired by PARP inhibition (45, 46). Thus, cross-interrogation of PDXs and patient datasets indicates the relevance of DDR functionality in NEPC. At the metabolic level, results also show that processes like fatty acid β-oxidation and amino acid degradation are downregulated in NEPC (Supplementary Fig. S6A). Accordingly, we previously reported that a metabolic shift occurs in an Ad PDX during CRPC relapse (11).
Recently, prostate cancer organoid development has been shown to recapitulate features of human disease (47). The application of this technology to PDXs serves as a complimentary approach to perform in vitro experiments on clinically relevant well-characterized models, expanding their usefulness to address scientifically meaningful questions in the field.
In summary, the integration of PDXs, organoids, human donor tumors, and their molecular characterization, provides a comprehensive way to study prostate cancer pathogenesis, envisage treatment response and resistance mechanisms, and accelerate the discovery of effective therapies.
Supplementary Material
MDA PCa PDX-derived organoids
General copy number profile and association with expresion for PTEN and RB1
Fusion genes expression levels. CNV details on TMPRSS2-ERG fusion and AR enhancer region. Putative PCa drivers in models lacking alterations in four major genes
Detail of manually curated full or partial copy number/ transcript loss
CDK12 loss of function in MDA PCa PDX
Comparison between adenocarcinoma and neuroendocrine carcinoma
Clinical and morphological features of 44 MDA PCa PDX
T200.1 panel: genes implicated in the pathogenesis of solid cancers
Percentage of reads matching with mouse DNA
Fusions identified in MDA PCa PDX cohort
Clinical, morphological and molecular features of MDA PCa PDXs pairs derived from the same patient
STR Profiling of samples derived from the same patient
Acknowledgments
We thank Sarah E. Townsend for editing the article. We thank the J. William Fulbright Foreign Scholarship Board for awarding a Fulbright grant to G. Gueron. We acknowledge the Biospecimen Extraction Facility (MDACC) for sample processing and DNA and RNA extraction. The Advanced Technology Genomics Core is funded by NCI Grant P30CA016672. This work was supported by the Prostate Cancer Foundation, Cancer Center Prostate Cancer SPORE (NIH/NCI P50 CA140388; P. Troncoso), David H. Koch Center for Applied Research in Genitourinary Cancers at MD Anderson (Houston, TX), and NIH/NCI U01 CA224044 (N.M. Navone, Y. Chen, A. Futreal), P50CA092629 (MSKCC SPORE), P30CA008748 (MSKCC Cancer Center Grant), STARR Cancer Consortium Grant, Geoffrey Beene Cancer Research Center (Y. Chen).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
S.K. Subudhi reports grants, personal fees, and non-financial support from Bristol Myers Squibb and Merck; personal fees from Amgen, Baird, Boxer Capital, Breaking Data, Dendreon, Bayer, and Portage; grants and personal fees from AstraZeneca, Janssen, Pfizer, and Regeneron; personal fees and non-financial support from Dava Oncology outside the submitted work. A. Futreal reports personal fees from Scorpion Therapeutics outside the submitted work. Y. Chen reports other support from ORIC Pharmaceuticals, grants from Foghorn Therapeutics, and personal fees from Fog Pharmaceuticals and Belharra Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
N. Anselmino: Data curation, formal analysis, validation, investigation, visualization, writing–original draft, project administration, writing–review and editing. E. Labanca: Data curation, formal analysis, validation, investigation, visualization, writing–original draft, project administration, writing–review and editing. P.D.A. Shepherd: Resources, data curation, investigation, methodology. J. Dong: Resources, methodology. J. Yang: Resources, investigation, methodology. X. Song: Data curation, software. S. Nandakumar: Data curation. R. Kundra: Resources, data curation, software, visualization. C. Lee: Investigation. N. Schultz: Resources, validation, visualization. J. Zhang: Data curation, software. J.C. Araujo: Resources, investigation. A.M. Aparicio: Resources, investigation. S.K. Subudhi: Resources, investigation. P.G. Corn: Resources, investigation. L.L. Pisters: Resources, investigation. J.F. Ward: Resources, investigation. J.W. Davis: Resources, investigation. E.S. Vazquez: Visualization, writing–original draft, writing–review and editing. G. Gueron: Visualization, writing–review and editing. C.J. Logothetis: Conceptualization, resources, funding acquisition. A. Futreal: Data curation, software, funding acquisition. P. Troncoso: Conceptualization, resources, data curation. Y. Chen: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, project administration, writing–review and editing. N.M. Navone: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;174:758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Varkaris A, Corn PG, Parikh NU, Efstathiou E, Song JH, Lee YC, et al. Integrating murine and clinical trials with cabozantinib to understand roles of MET and VEGFR2 as targets for growth inhibition of prostate cancer. Clin Cancer Res 2016;22:107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 2017;547:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dai X, Gan W, Li X, Wang S, Zhang W, Huang L, et al. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med 2017;23:1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wan X, Corn PG, Yang J, Palanisamy N, Starbuck MW, Efstathiou E, et al. Prostate cancer cell-stromal cell crosstalk via FGFR1 mediates antitumor activity of dovitinib in bone metastases. Sci Transl Med 2014;6:252ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Navone NM, van Weerden WM, Vessella RL, Williams ED, Wang Y, Isaacs JT, et al. Movember GAP1 PDX project: an international collection of serially transplantable prostate cancer patient-derived xenograft (PDX) models. Prostate 2018;78:1262–82. [DOI] [PubMed] [Google Scholar]
- 8. Palanisamy N, Yang J, Shepherd PDA, Li-Ning-Tapia EM, Labanca E, Manyam G, et al. The MD Anderson prostate cancer patient-derived xenograft series (MDA PCa PDX) captures the molecular landscape of prostate cancer and facilitates marker-driven therapy development. Clin Cancer Res 2020;26:4933–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rokita JL, Rathi KS, Cardenas MF, Upton KA, Jayaseelan J, Cross KL, et al. Genomic profiling of childhood tumor patient-derived xenograft models to enable rational clinical trial design. Cell Rep 2019;29:1675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Labanca E, Bizzotto J, Sanchis P, Anselmino N, Yang J, Shepherd PDA, et al. Prostate cancer castrate resistant progression usage of non-canonical androgen receptor signaling and ketone body fuel. Oncogene 2021;40:6284–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc 2016;11:347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014;159:176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang F, Xu D, Wang S, Wong CK, Martinez-Fundichely A, Lee CJ, et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 2022;376:eabe1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;44:685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature 2009;458:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Attard G, Clark J, Ambroisine L, Mills IG, Fisher G, Flohr P, et al. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer 2008;99:314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res 2009;69:2734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. SLC45A3-ELK4 chimera in prostate cancer: spotlight on cis-splicing. Cancer Discov 2012;2:582–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takeda DY, Spisák S, Seo JH, Bell C, O'Connor E, Korthauer K, et al. A somatically acquired enhancer of the androgen receptor is a noncoding driver in advanced prostate cancer. Cell 2018;174:422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol 2009;6:429–39. [DOI] [PubMed] [Google Scholar]
- 24. Wu Y-M, Cieślik M, Lonigro RJ, Vats P, Reimers MA, Cao X, et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 2018;173:1770–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ware KE, Garcia-Blanco MA, Armstrong AJ, Dehm SM. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocr Relat Cancer 2014;21:T87–T103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moul JW, Friedrichs PA, Lance RS, Theune SM, Chang EH. Infrequent RAS oncogene mutations in human prostate cancer. Prostate 1992;20:327–38. [DOI] [PubMed] [Google Scholar]
- 27. Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 29. Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015;21:1190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fischer K, Pflugfelder GO. Putative breast cancer driver mutations in TBX3 cause impaired transcriptional repression. Front Oncol 2015;5:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 2017;32:474–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cackowski FC, Kumar-Sinha C, Mehra R, Wu Y-M, Robinson DR, Alumkal JJ, et al. Double-negative prostate cancer masquerading as a squamous cancer of unknown primary: a clinicopathologic and genomic sequencing-based case study. JCO Precis Oncol 2020;4:PO:20.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aparicio AM, Shen L, Tapia ELN, Lu J-F, Chen H-C, Zhang J, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res 2016;22:1520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lozano R, Castro E, Aragón IM, Cendón Y, Cattrini C, López-Casas PP, et al. Genetic aberrations in DNA repair pathways: a cornerstone of precision oncology in prostate cancer. Br J Cancer 2021;124:552–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nombela P, Lozano R, Aytes A, Mateo J, Olmos D, Castro E. BRCA2 and other DDR genes in prostate cancer. Cancers 2019;11:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang W, Liu B, Wu W, Li L, Broom BM, Basourakos SP, et al. Targeting the MYCN-PARP-DNA damage response pathway in neuroendocrine prostate cancer. Clin Cancer Res 2018;24:696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mateo J, Cheng HH, Beltran H, Dolling D, Xu W, Pritchard CC, et al. Clinical outcome of prostate cancer patients with germline DNA repair mutations: retrospective analysis from an international study. Eur Urol 2018;73:687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li ZG, Mathew P, Yang J, Starbuck MW, Zurita AJ, Liu J, et al. Androgen receptor-negative human prostate cancer cells induce osteogenesis in mice through FGF9-mediated mechanisms. J Clin Invest 2008;118:2697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Labanca E, Vazquez ES, Corn PG, Roberts JM, Wang F, Logothetis CJ, et al. Fibroblast growth factors signaling in bone metastasis. Endocr Relat Cancer 2020;27:R255–R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Labanca E, Yang J, Shepherd PDA, Wan X, Starbuck MW, Guerra LD, et al. Fibroblast growth factor receptor 1 drives the metastatic progression of prostate cancer. Eur Urol Oncol 2022;5:164–75. [DOI] [PubMed] [Google Scholar]
- 42. Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol 2021;18:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berglund E, Maaskola J, Schultz N, Friedrich S, Marklund M, Bergenstråhle J, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun 2018;9:2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tsui MM, York JD. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv Enzyme Regul 2010;50:324–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Merkens L, Sailer V, Lessel D, Janzen E, Greimeier S, Kirfel J, et al. Aggressive variants of prostate cancer: underlying mechanisms of neuroendocrine transdifferentiation. J Exp Clin Cancer Res 2022;41:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu B, Li L, Yang G, Geng C, Luo Y, Wu W, et al. PARP inhibition suppresses GR-MYCN-CDK5-RB1-E2F1 signaling and neuroendocrine differentiation in castration-resistant prostate cancer. Clin Cancer Res 2019;25:6839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karkampouna S, La Manna F, Benjak A, Kiener M, De Menna M, Zoni E, et al. Patient-derived xenografts and organoids model therapy response in prostate cancer. Nat Commun 2021;12:1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MDA PCa PDX-derived organoids
General copy number profile and association with expresion for PTEN and RB1
Fusion genes expression levels. CNV details on TMPRSS2-ERG fusion and AR enhancer region. Putative PCa drivers in models lacking alterations in four major genes
Detail of manually curated full or partial copy number/ transcript loss
CDK12 loss of function in MDA PCa PDX
Comparison between adenocarcinoma and neuroendocrine carcinoma
Clinical and morphological features of 44 MDA PCa PDX
T200.1 panel: genes implicated in the pathogenesis of solid cancers
Percentage of reads matching with mouse DNA
Fusions identified in MDA PCa PDX cohort
Clinical, morphological and molecular features of MDA PCa PDXs pairs derived from the same patient
STR Profiling of samples derived from the same patient
Data Availability Statement
Data are available in cBioPortal [Prostate Cancer MDA PCa PDX (MD Anderson; https://www.cbioportal.org/study/summary?id=prad_msk_mdanderson_2023)] and dbGaP (https://www.ncbi.nlm.nih.gov/gap/; RRID:SCR_002709), accession number phs003420.v1.p1. MDA PCa PDXs are available through a material transfer agreement. Contact e-mail: pcapdxprogram@mdanderson.org.



![Figure 1. Morphologic and clinical distribution of the MDA PCa PDX cohort. A, Morphologic classification, site of tumor collection, and previous treatment of the 44 PDXs (from 38 patients with prostate cancer) used for genomic analysis [WGS, targeted sequencing (T200.1 panel; 263 genes), and RNA-seq]. Note: second- or next-generation androgen receptor signaling inhibition (ARSI) comprises abiraterone acetate, enzalutamide, and apalutamide. B, Heat map showing transcriptomic data covariance clustering. Highlighted and colored are longitudinal and heterogenous samples from the same patient, respectively. CTC, circulating tumor cells; diff, differentiated.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/cd61/11094415/6b44cd44dd97/2272fig1.jpg)