Abstract
The olfactory neuroepithelium serves as a sensory organ for odors and forms part of the nasal mucosal barrier. Olfactory sensory neurons are surrounded and supported by epithelial cells. Among them, microvillous cells (MVCs) are strategically positioned at the apical surface, but their specific functions are enigmatic and their relationship to the other specialized epithelial cells, particularly the solitary chemosensory cell family, is unclear. Here, we establish that the family of MVCs comprises tuft cells and ionocytes in both mice and humans. Integrating analysis of the respiratory and olfactory epithelia, we define the distinct receptor expression of TRPM5+ tuft-MVCs compared to Gɑ-gustducinhigh respiratory tuft cells and characterize a previously undescribed population of glandular DCLK1+ tuft cells. To establish how allergen sensing by tuft-MVCs might direct olfactory mucosal responses, we employed an integrated single-cell transcriptional and protein analysis. Inhalation of Alternaria induced mucosal epithelial effector molecules including Chil4, and a distinct pathway leading to proliferation of the quiescent olfactory horizontal basal stem cell (HBC) pool, both triggered in the absence of olfactory apoptosis. While the Chil4 pathway was dependent on STAT6 signaling and innate lymphocytes, neither were required for HBC proliferation. Alternaria- and ATP-elicited HBC proliferation was dependent on TRPM5+tuft-MVCs, identifying these specialized epithelial cells as regulators of olfactory stem cell responses. Together our data provide high resolution characterization of nasal tuft cell heterogeneity and uncover the functional capacity of TRPM5+ tuft-MVCs to direct the olfactory mucosal response to allergens.
One Sentence Summary
TRPM5+ microvillous cells in the olfactory neuroepithelium are tuft cells that regulate olfactory stem cell proliferation.
INTRODUCTION:
The nose contains two functionally distinct but anatomically overlapping mucosal compartments: the respiratory and olfactory epithelia. The olfactory neuroepithelium is a unique organ comprised of olfactory sensory neurons (OSNs) surrounded by specialized epithelial cells. OSNs have a limited lifespan and continuously regenerate from a population of proliferating progenitor cells called globose basal cells, specific to the olfactory mucosa (1). A second type of stem cells specific to the olfactory mucosa, the horizontal basal stem cell (HBCs) are quiescent and serve as a reserve population, but can be activated by profound damage with loss of neuroepithelial structure to repopulate both the OSNs and the olfactory epithelial cells (1–4). The nasal olfactory and respiratory epithelia overlay a dense network of submucosal glands and interspersed immune cells. Thus, the nose represents a site of closely interacting epithelial cells, OSNs, stem cells, and secretory glands at the first entry of inhaled air.
Sustentacular cells and two distinct subsets of microvillous cells (MVCs) are specialized olfactory epithelial cell types with emerging functions in mucosal immune responses (5, 6). The smaller, pear-shaped MVCs lining the apical surface of the olfactory neuroepithelium are distinguished by the expression of the Transient Receptor Potential Cation Channel Subfamily M Member 5 (TRPM5), a taste receptor-linked calcium-activated monovalent cation channel (5, 7). Another subset of MVCs, with a broad cell body and a slender cytoplasmic process extending to the basement membrane, are distinguished by expression of both the type 3 IP3 receptor (IP3R3) and the 5’ exonucleotidase CD73, and a lack of expression of TRPM5 (5, 8–11). Both TRPM5+ and TRPM5− MVCs derive from a common c-kit+ progenitor but differ in their differentiation trajectory (10, 12). TRPM5− MVCs are implicated as a source of the neuropeptide Y, capable of directing the activation and proliferation of globose basal cells (11, 13), while deletion of TRPM5+ MVC leads to impaired olfactory-guided behaviors after extended exposure to strong odorants (14).
TRPM5+ MVCs also express the acetylcholine-producing enzyme choline acetyltransferase (ChAT) (15). TRPM5 and ChAT notably also mark tracheal tuft (also known as brush) cells (16–18), intestinal tuft cells (19, 20), and the related nasal solitary chemosensory cells (SCCs) (21–23), which suggests shared activation pathways between tuft cells and TRPM5+ MVCs. TRPM5+ MVC development depends on Pou2f3 (24), a transcription factor also required for tuft cell development in all mucosal compartments (25–27). However, other defining features of tuft cells such as the expression of taste receptor type I and II families and the taste signaling G protein Gα gustducin (28, 29) are low or absent in TRPM5+ MVCs (5, 7, 24, 30). Our previous studies demonstrated that ChAT-eGFP+ nasal epithelial cells with morphological and transcriptional features of TRPM5+ MVCs are directly activated by allergens and respond by generating cysteinyl leukotrienes CysLTs (31). Intestinal and airway tuft cells are also the dominant epithelial source of IL-25 (31–33). Tuft cell activation – by parasites, or metabolites derived from microbiota – directs the activation of type 2 innate lymphoid cells (ILC2) leading to IL-13-driven type 2 inflammation and stem cell activation (34). Whether TRPM5+ MVCs, like tuft cells, engage with ILC2s to drive inflammation or stem cell proliferation in the olfactory mucosa after allergen inhalation is not known.
Here, we applied single-cell sequencing and histological approaches to demonstrate that TRPM5+ MVC, SCCs and a third “intermediate” population of ChAT-eGFP+ nasal epithelial cells all belong to the tuft cell family. The three tuft cell subsets share a core transcriptional profile including Trpm5, Pou2f3, Chat, Avil, Il25 and Ltc4s. The distinct population of TRPM5− MVCs corresponds transcriptionally to pulmonary ionocytes (expressing Cftr, Foxi1, Ascl3, Coch). By generating a single-cell atlas of the mouse nasal mucosa (containing 63,125 cells), we found that Alternaria inhalation caused a marked proliferation of HBCs in the olfactory mucosa, even in the absence of profound damage to the OSNs or accessory epithelial cells. Strikingly, both allergen- and ATP-induced stem cell proliferation were substantially reduced after genetic deletion of tuft cells using Pou2f3−/− mice. Genetic ablation of lymphocytes (in Il7r−/− mice) and IL-4/IL-13 receptor signaling (in Stat6−/− mice) did not affect allergen-induced stem cell proliferation, indicating that tuft cells are part of a neuroepithelial-intrinsic circuit that drives activation of typically quiescent HBC stem cells. Together, our data define extensive heterogeneity of sensory neuroepithelial cell-types in the nasal mucosa and delineate a previously undescribed role for tuft-MVCs in the control of stem cell proliferation.
RESULTS:
Mouse olfactory TRPM5+ MVCs are tuft cells and TRPM5− MVCs are ionocytes.
To define the transcriptional profile of mouse olfactory and nasal respiratory chemosensory ChAT-eGFP+ cells, we separated olfactory from respiratory mucosa along the anterior portion of the olfactory turbinates and obtained single-cell suspensions from each compartment (Fig. S1A). The olfactory mucosa contained the majority of ChAT-eGFP+ cells assessed by flow cytometry (FACS) (69%) compared to 31% derived from the respiratory preparation (Fig. S1B–F). ChAT-eGFP+ cells were abundant in the olfactory mucosa (22% of EpCAMhigh cells) but were also surprisingly prevalent in the respiratory mucosa (10% of EpCAMhigh cells and 1% of live cells) (Fig. S1B–F). We confirmed that most ChAT-eGFP FSClow cells were derived from the olfactory mucosa while rare FSChigh were mostly respiratory (Fig. S1G–H) consistent with our previous findings that FSClow ChAT-eGFP+ cells correspond to olfactory MVCs and FSChigh ChAT-eGFP+ correspond to respiratory nasal SCCs (31).
We then compared the ChAT-eGFP+ cells to EpCAMhigh cells from each of olfactory and respiratory epithelia by scRNAseq (Fig. 1A). To ensure sufficient SCC numbers, we enriched for the scarce elongated FSChigh ChAT-eGFP+ cells (Fig. S1I). Unsupervised clustering identified a subset of TRPM5+ epithelial cells which expressed known tuft cell markers and clustered distinctly from all other EpCAMhigh cells from the respiratory and olfactory mucosa (Fig. 1A). Confirming their identity, transcriptome-wide analysis using a random forest classifier mapped 94% of the cells in the TRPM5+ epithelial cell cluster (itself 44.1% of all cells) to tracheal tuft cells (35) (Fig. 1B). Besides tuft cells, we identified a second population of abundant differentiated epithelial cells NEGATIVE for Chat and Trpm5 but POSITIVE for Ascl3 and Nt5e (CD73), consistent with descriptions of TRPM5− MVCs (10, 11, 36). We mapped 96% of them to pulmonary ionocytes (Fig. 1B), unambiguously identifying them as nasal ionocytes.
Figure 1. Tuft cells and ionocytes comprise the two MVC types of the olfactory epithelium in mice and humans.
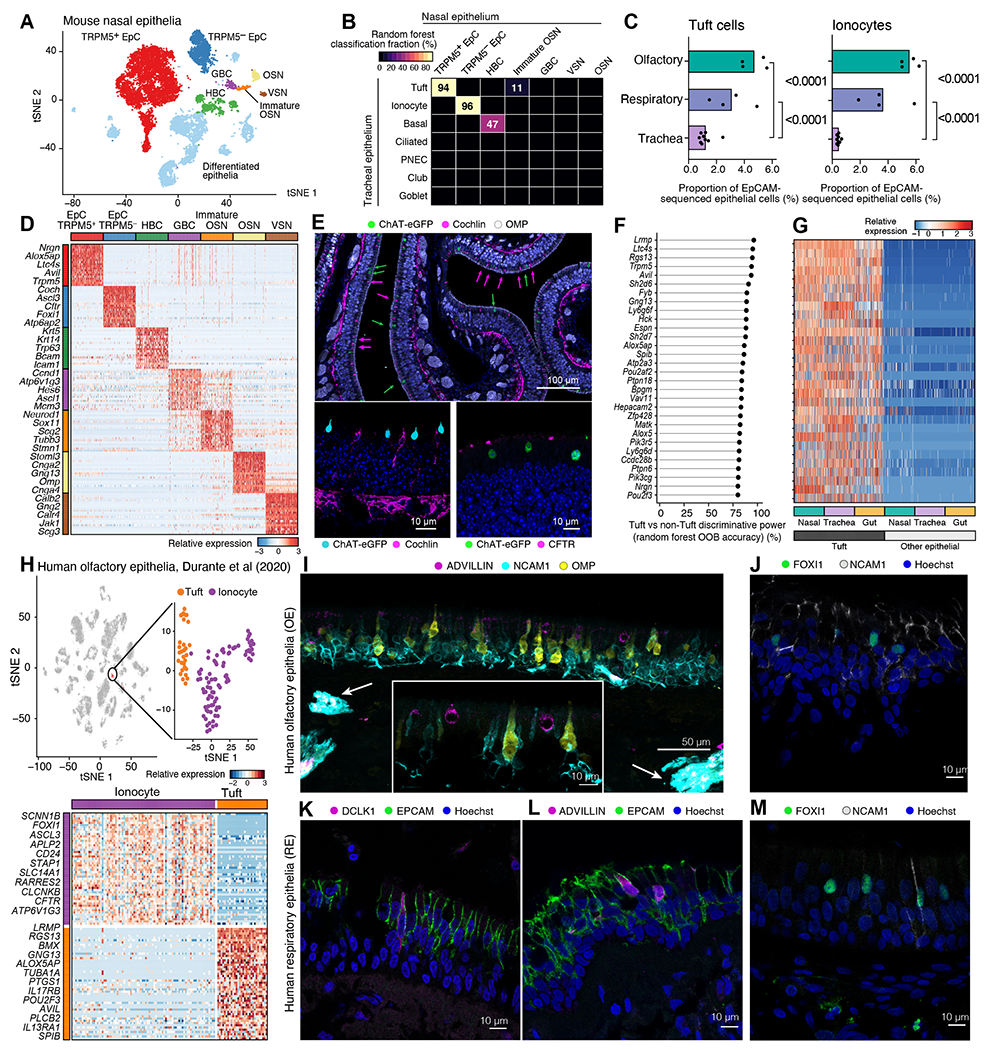
(A-G) The nasal respiratory and olfactory epithelium of ChAT-eGFP mice was assessed after enrichment for ChAT-eGFP cells (as in S1A). (A) tSNE embedding of 13,052 cells (n=4 mice from two independent experiments), colored and labeled by unsupervised clustering. (B) Heatmap shows the fraction of cells (color bar) of each nasal epithelial cell type (columns, types as in A) classified as each tracheal cell type (rows) using a random forest. (C) Fraction (x-axis) of tuft cells (top) and ionocytes (bottom) in all epithelial cells from each mouse (points), in each tissue, from EpCAM+ cells in naïve nasal mucosa (dataset introduced in Fig. 3, p-values: Wald test). (D) Relative expression (row-wise z-score of log2(TPM+1)), (color bar) of top 50 genes (rows) specific to each cell type (FDR<0.05) in our dataset; two selected genes are shown. (E) Cross-section of olfactory epithelium of ChAT-eGFP mice stained for ChAT-eGFP (tuft cells), cochlin and CFTR (ionocytes), OMP (OSNs, top image). Hoechst (blue) is used to mark nuclei in all images. Green arrows point at tuft cells, magenta arrows point at ionocytes in the top image. (F) Top 30 pan-tissue tuft cell marker genes (y-axis) ranked by out of bag (OOB) accuracy (x-axis) from random forest classification. (G) Relative expression of top tuft cell marker genes (ordered as in F) in tuft and other epithelial cells across three tissues. (H-M) Single-cell sequencing (H) and histology (I-M) of human nasal mucosa. (H) tSNE embeddings (top) of 3,528 olfactory epithelial, immune and neuronal cells (40) showing new re-analysis of a cluster of 116 MVCs containing tuft cells and ionocytes (inset), and heatmap (bottom) of differentially expressed genes (FDR<0.05, rows) between the MVC subtypes (40). Selected marker genes are shown. (I-M) Immunofluorescence of olfactory (I, J) or respiratory epithelium (K-M) from superior turbinates of controls without sinus disease (I, J, M) or of patients with CRS (n=3) (K, L). Arrows in I indicate olfactory axon bundles in the submucosa.
Tuft cells and ionocytes appeared markedly more abundant in the nose than their reported frequency in the trachea (35). To characterize their actual distribution, we evaluated the numbers of tuft cells and ionocytes in a non-enriched scRNAseq dataset from the nose (introduced in Fig. 3) and compared to published tracheal scRNAseq data (35). We found that tuft cells comprise 4.7% of EpCAM+ cells from the olfactory epithelium, 3.7-fold more common than in the trachea (Fig. 1C). Ionocytes comprised 5.4% of EpCAM+ cells from the olfactory epithelium, 12.3-fold more common than in the trachea (Fig. 1C).
Figure 3. Single-cell analysis of intercellular communication and compositional shifts during allergic inflammation in the mouse nasal mucosa.
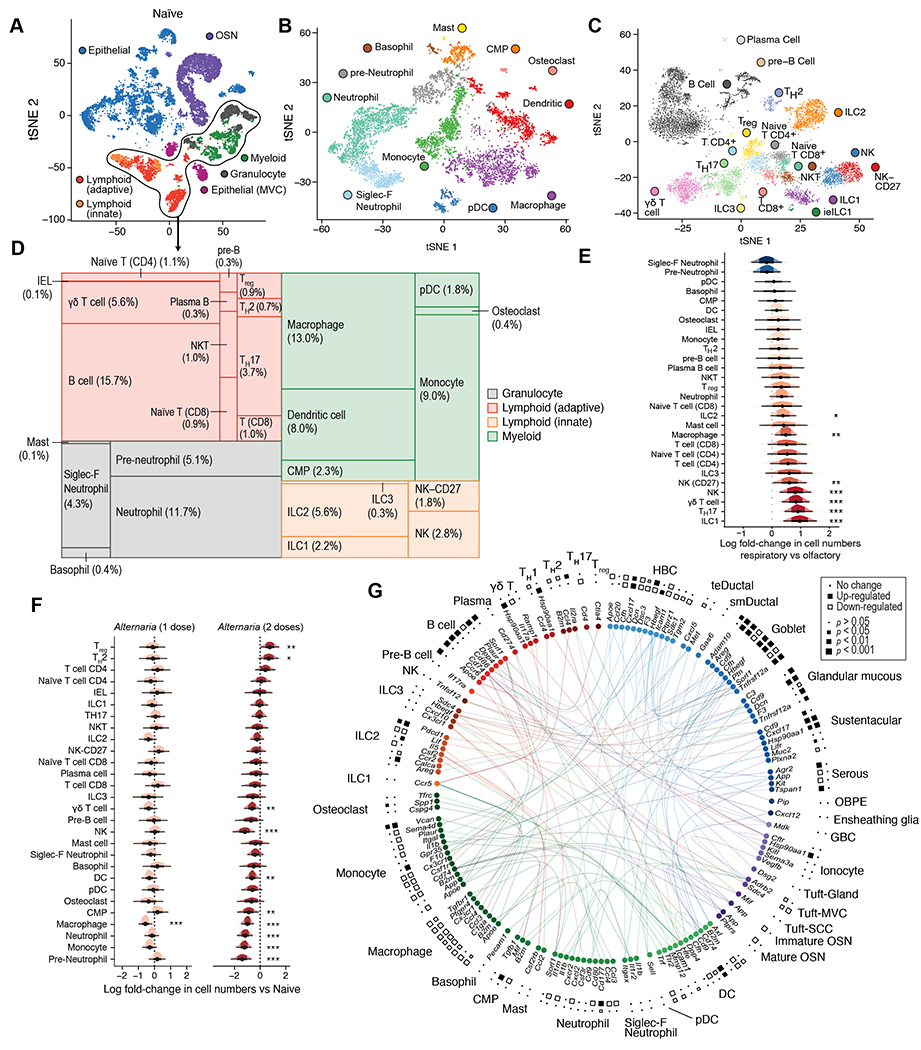
WT mice were assessed before Alternaria inhalation (n=4) (A-E) or after one (n=3) or two (n=2) doses of inhaled Alternaria as in Fig. S5A (F-G). (A) tSNE embedding of 19,275 cells from nasal mucosa of naïve mice assessed by scRNAseq, cell type lineages identified by unsupervised clustering are shown (color legend). (B-C) tSNE embeddings of 7,107 myeloid (B) and 7,213 lymphoid (C) cells (points). Cells are colored by the cell types (inset legend). (D) Tree map plot shows the cell-type composition (size of rectangles) of the immune compartment in naïve mice. (E-F) Estimated effect (log fold-change, x axis) of tissue location (E) and one (orange) and two (red) doses of inhaled Alternaria (F) on the proportions of each immune cell type. Density histograms show posterior distribution of Dirichlet Multinomial regression. Black dot: point estimate, thick bar: 66% credible interval, thin bar: 95% credible interval (CI) * 90%, ** 95%, *** 99% CI does not include 0. (G) Circle plot displays differentially expressed receptor ligand interactions between cell types (color) after Alternaria inhalation. Squares denote significance (size) of up (black) or down (white) regulation after one dose (inner ring) or two doses (outer ring) of Alternaria (see legend). Data are from two independent experiments with 1-2 mice/group.
The large population of nasal tuft cells identified here expressed the canonical markers of tracheal and intestinal tuft cells: the actin-binding protein advillin Avil, Trpm5, and eicosanoid biosynthetic enzymes Alox5ap and Ltc4s (Fig. 1D and Data File S2, S3). Nasal ionocytes (TRPM5− MVCs) expressed high levels of Cftr, Coch and Foxi1 (Fig. 1D, Data File S3). Immunofluorescence of cross sections confirmed that both cochlin+ and CFTR+ ionocyte-like epithelial cells and ChAT-eGFP+/TRPM5+ MVCs were abundant in the olfactory epithelium (Fig. 1E) and ionocytes are found in lower numbers in the respiratory epithelium (Fig. S2A). The remainder of the EpCAMhigh cells were comprised of differentiated epithelial cells, basal cells, and a minority of olfactory marker protein (OMP+) OSNs, Fig. 1A, D). To validate the separation of olfactory and respiratory mucosa, we used aquaporin 4 (Aqp4) as a distinguishing marker between OSNs and vomeronasal sensory neurons (VSNs) (37) (Fig. S2B–D). Although the vomeronasal organ (VNO) is an olfactory organ, it is located in the anterior part of the nose surrounded by respiratory epithelium (Fig. S1A). We found nearly all Aqp4+ neurons in our single cell preparations were derived from the respiratory portion (Fig. S2B), where the Aqp4+ VSNs are located (Fig. S2C–D), validating our separation of tissue origin.
TRPM5+/ChAT+ cells in the olfactory and respiratory mucosa of the nose are historically defined as distinct epithelial subsets – olfactory MVCs and respiratory SCCs, largely based on the absence of taste receptors and transduction machinery in MVCs (7, 38). However, we found that olfactory and respiratory Trpm5+/Chat+ cells shared a core transcriptional profile defined by transcripts involved in taste transduction (Trpm5, Gng13), calcium signaling molecules (Lrmp, Pik3cg, Hck, Vav1, Matk, Pik3r5), the transcription factors Spib, Pou2f3 and Pou2af2 and the CysLT biosynthetic enzymes Alox5, Alox5ap and Ltc4s (Fig. 1F). This core effector profile was shared by tuft cells in the trachea (35) and intestine (39), as defined by our previous single-cell analyses (Fig. 1G). Thus, tuft cells in all mucosal compartments are distinguished by the expression of the necessary machinery to rapidly generate Ca2+-signaling dependent mediators – eicosanoids and acetylcholine.
Human olfactory TRPM5+ MVCs are tuft cells and TRPM5− MVCs are ionocytes.
To determine whether human MVCs subdivide into tuft cells and ionocytes, we reanalyzed two published human scRNA-seq data sets. The Durante et al (40) dataset includes samples from the olfactory cleft while the Ordovas-Montanes et al. (41) contains samples derived exclusively from the respiratory mucosa of patients with chronic rhinosinusitis (CRS) with or without polyposis. Focused analysis of the cluster of 116 cells annotated by Durante et al. as MVCs identified two subsets (Fig. 1H). One subset (of 30 cells) was highly similar to mouse tuft cells, with distinct expression of the pan-tuft cell markers AVIL and LRMP, eicosanoid pathway components ALOX5AP and PTGS1, the IL-25 receptor IL17RB, and the tuft cell transcription factor POU2F3. In the Ordovas-Montanes dataset (41), 13 of the 19,196 cells profiled were nasal tuft cells. They were identifiable by their specific expression of pan-tuft cell markers AVIL, LRMP, RGS13, taste transduction protein PLCB2, and eicosanoid transcripts ALOX5 and PTGS1 (Fig. S2E, F). Finally, we confirmed that human olfactory TRPM5+MVCs express ALOX5, ALOX5AP, and LTC4S, required for CysLT generation (Fig. S2G) providing a functional connection to mouse tuft cells.
The second subset (of 86 cells) of MVCs in the Durante et al. olfactory-enriched dataset was highly similar to mouse TRPM5− MVCs (Fig. 1H, Data File S4) (40).We also identified 79 ionocytes in the human respiratory epithelium dataset (41) (Fig. S2E, Data File S4). Human nasal ionocytes from both datasets shared the transcriptional profile of human pulmonary ionocytes and mouse pulmonary and nasal ionocytes with specific expression of the ion channels CFTR, CLCNKB, the transcription factors FOXI1 and ASCL3, and the vacuolar ATP-ase ATP6V1G3. To locate human tuft cells and ionocytes in olfactory biopsies, we used neural cell adhesion molecule 1 (NCAM1) (42), which is expressed at high mRNA levels in both mature and immature human OSNs (Fig. S2G). In the olfactory neuroepithelium, marked by NCAM1 and OMP, advillin+ cells were interspersed between OSNs (Fig. 1I). These olfactory tuft cells had globular shape like olfactory TRPM5+ MVCs suggesting a shared morphology (Fig. 1I, inset). FOXI1+ ionocytes were also found in the human olfactory mucosa scattered between NCAM1+ OSNs (Fig. 1J). In the respiratory human epithelium, the adivillin- and DCLK1-positive tuft cells were spindle shaped (Fig. 1K–L). We also identified FOXI1+ ionocytes in the respiratory epithelium of human biopsies (Fig. 1M). In summary, we confirmed that the human tuft cells and ionocytes represent two distinct subsets of the previously described human MVCs in the olfactory mucosa. As in mice, olfactory and respiratory tuft cells share a common transcriptional profile, as is the case for ionocytes.
Three subsets of mouse nasal tuft cells correspond to MVCs, SCCs, and a population of glandular tuft cells.
Our single-cell analysis showed all Trpm5+/Chat+ nasal epithelial cells cluster together and share the tuft cell transcriptional profile. To reconcile this with the literature that defined olfactory MVCs as distinct from respiratory SCCs and other chemosensory cells, we examined their heterogeneity. Unsupervised clustering of the tuft cells identified three subgroups (Fig. 2A), all expressing the pan tuft cell signature defined by Nadjsombati et al. (32) (Fig. S3A) but each defined by distinct markers (Fig. 2B left, Data File S5) and G protein-coupled receptors (Fig. 2B right). We interpreted the dominant population of tuft cells as TRPM5+ MVCs (Fig. 2A) since they were largely derived from the olfactory mucosa (Fig. S3B–C) and did not express the wide range of taste receptors that defines SCCs (Fig. 2B). TRPM5+ MVCs were defined by the high and ubiquitous expression of tuft cell markers Trpm5, Pou2f3, and Chat (all original markers of TRPM5+ MVCs (7, 15, 24, 28)) and the pan-tuft marker Avil (Fig. 2B–C, S3D–E). ChAT-eGFP+ cells were abundant with a globular body and a single very short process ending with short brushes at the apical surface, identical to TRPM5+ MVCs (38) (Fig. S3F–G). These TRPM5+ MVCs expressed the taste receptor associated protein Gɑ-gustducin Gnat3 (Fig. 2C) at low levels in ~50% of these cells (Fig. S3E), consistent with other reports (30). They also expressed high levels of eicosanoid biosynthetic enzymes Ptgs1, Hpgds, Alox5, Alox5ap and Ltc4s (Fig. 2C, S3H) and the tuft cell cytokine Il25 (Fig. 2B–C, S3H). Comparison of tuft-MVCs to the tracheal tuft cell subsets using a machine learning classifier showed highest similarity to previously described eicosanoid-enriched tuft-2 cells (35) (Fig. 2D). Since TRPM5+MVCs shared the location and morphology of MVCs and the core transcriptional profile of tuft cells, we refer to them as tuft-MVCs.
Figure 2. Single-cell analysis identifies nasal tuft cell heterogeneity in mice.
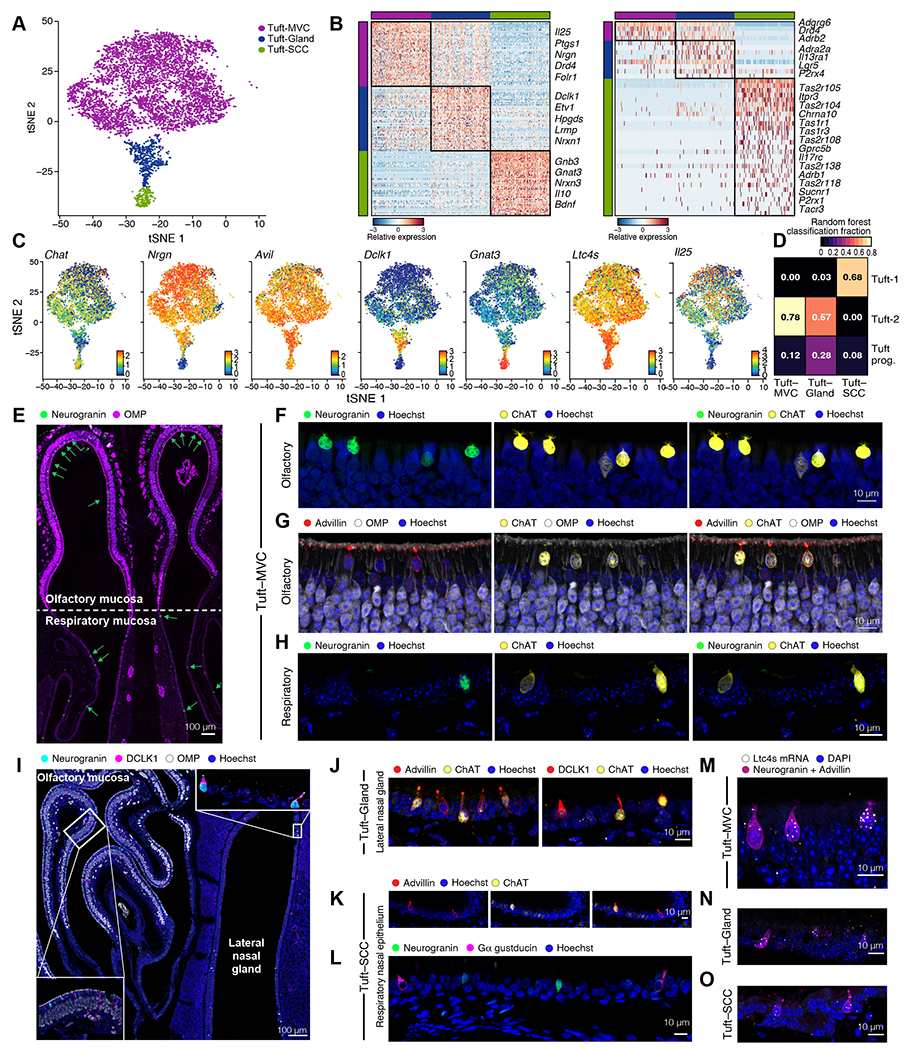
(A) tSNE embedding of 5,594 tuft cells derived from ChAT-eGFP mice colored by unsupervised clustering. (B) Heatmaps show the expression level (row-wise z-score of log2(TPM+1), color bar) of marker (left) and G protein-coupled receptor genes (right) specifically expressed (FDR<0.01) by each subset (color bars). (C) tSNE plots display expression in log2(TPM) + 1 (color bar) for shared tuft cell markers and subtype markers. (D) Heatmap shows the fraction of cells (color bar) of each nasal tuft cell sub-type (columns, types as in A) classified as each tracheal (35) tuft cell sub-type (rows) using a random forest. (E-L) Immunolocalization of tuft cell subsets. (E) Cross section through the anterior nasal cavity at the level of the nasal septum (Fig. S1A T1) captures the olfactory epithelium (top) and respiratory epithelium (bottom) stained for OMP (OSNs) and neurogranin (tuft-MVCs). Arrows point at neurogranin positive cells. (F-H) Tuft-MVC expression of ChAT-eGFP, neurogranin (F, H) and advillin (G) in the olfactory (F-G) and respiratory (H) epithelium. (I) Cross section through the posterior section of the nasal cavity captures the olfactory epithelium (top left), respiratory epithelium (bottom center) and LNG (right) (Fig. S1A T4). Inserts show high magnification of the olfactory mucosa (lower left) and the epithelium lining the LNG (top right). (J) Expression of ChAT-eGFP and advillin (left) or DCLK1 (right) in tuft cells lining the epithelium of the LNG. (K-L) Tuft-SCC expression of ChAT-eGFP and advillin (K), neurogranin and Gα gustducin (L). Hoechst was used for nuclear staining. (M-O) In situ hybridization with RNAscope probe for Ltc4s. Tuft cells were distinguished based on immunoreactivity for advillin and neurogranin. Representative images of tuft-MVC (M), tuft-Gland (N) and tuft-SCC (O) Nuclear staining with DAPI.
We identified a previously undescribed marker of tuft-MVCs: Nrgn, encoding neurogranin, a postsynaptic neuronal protein in the calpacitin family (43) (Fig. 2B–C), and validated its expression at the protein level (Fig. 2E–F) and absence in tuft cell-deficient Pou2f3−/− mice (Fig. S4A–B). Nearly all ChAT-eGFP+ olfactory epithelial cells were positive for neurogranin (Fig. 2F) and advillin (Fig. 2G) but negative for Gɑ gustducin protein (Fig. S4C–E). Scattered rare neurogranin+ cells were also detectable in the respiratory epithelium (Fig. 2E, H, S4B, D–E). Surprisingly, tuft-MVCs had very low levels of both Dclk1 and the encoded DCLK1 protein, commonly used to identify tuft cells in the intestine and trachea (27, 44) (Fig. 2C, I). In the olfactory epithelium, DCLK1 marked epithelial cells with ductal morphology (Fig. 2I inset), indicating that DCLK1 is not a tuft cell-specific marker in the nose.
A second, rarer population of tuft cells was exclusively derived from the respiratory epithelium (Fig. 2A, S3B–C), and strongly enriched for transcripts that define the elongated nasal respiratory SCCs, including type 1 (Tas1r1, Tas1r3) and type 2 (Tas2r104, Tas2r105, Tas2r108, Tas2r118, and Tas2r138) taste receptors, the succinate receptor Sucnr1 and ubiquitous and high expression of gustducin components Gnat3 and Gnb3 (21, 28, 45)(Fig. 2B–C, S3E). These cells had a spindle-shaped morphology (Fig. S3F) and were also enriched for the core tuft cell genes (Fig. 2C, S3A, D–E, H). Because of this morphology and marker expression characteristic of nasal respiratory SCCs, combined with the transcriptional profile of tuft cells, we refer to them as tuft-SCCs. The tuft-SCCs were most similar to the taste receptor-enriched tracheal tuft-1 cells (Fig. 2D). Histologically, tuft-SCCs were largely negative for neurogranin (Fig. S4D–E), but were marked by ChAT-eGFP, advillin and Gɑ-gustducin (Fig. 2K–L, Fig. S4E–F). Tuft-SCCs highly expressed neuropeptide bone-derived neurotrophic factor (Bdnf) and Il10 (Fig. 2B) suggesting a possible immunomodulatory role.
Finally, we identified a transcriptionally intermediate population of tuft cells (Fig. 2A–C) that expressed high levels of Dclk1 and was enriched for Il13ra1 (Fig. 2B), responsible for type 2 inflammation-induced tuft cell proliferation in the intestine (34). They were also enriched for Ly6d and Ly6e, transcripts associated with breast and prostate glandular cancers (46) (Data File S5). We identified a distinct population of tuft cells in the lateral nasal gland (LNG) – a large serous glandular structure unique to the murine nasal cavity (Fig. 2I, Fig. S1A T3–T4). These ChAT+/advillin+ tuft cells were also positive for neurogranin, Gɑ-gustducin and DCLK1, and thus correspond to this intermediate tuft cell population (Fig. 2C, I–J, S3E, S4G–H). Based on their transcriptional profile and their anatomical location in the LNG we refer to them as tuft-gland. Deconvolution (47) of bulk RNAseq of sorted elongated (FSChigh) and globular (FSClow) tuft cells from olfactory and respiratory epithelia suggested that tuft-gland cells are likely of intermediate morphology, since they were equally represented in elongated (FSChigh) and globular (FSClow) gates (Fig. S4I). We also characterized the vomeronasal organ (VNO) ChAT-eGFP+ cells as positive for Gα-gustducin (48) but negative for neurogranin and DCLK1, making them most similar to tuft-SCCs, despite their proximity to OMP+ neurons (Fig. S4J–L). Finally, we confirmed the ubiquitous expression of Ltc4s mRNA in all subsets of nasal tuft cells: tuft-MVCs in the olfactory mucosa, tuft-gland in the LNG and tuft-SCCs in the respiratory epithelium (Fig. 2M–O).
Together, these data define three nasal tuft cell subsets with common and distinct transcriptional, morphological, and anatomical features. All three shared expression of Trpm5, Chat, Il25 and the transcripts of the eicosanoid generating cascade Alox5, Alox5ap, Ltc4s, Ptgs1, and Hpgds, but could be distinguished based on their expression of neurogranin (tuft-MVC and tuft-gland), protein expression of Gɑ-gustducin (tuft-SCC and tuft-gland) and DCLK1 (only tuft-gland). Each group had distinct receptor expression of taste and succinate receptors as well as expression of adrenergic receptors. Adrb2, Adra2a, and Adrb1 were enriched in tuft-MVC, tuft-gland, and tuft-SCC subtypes respectively (Fig. 2B, right), raising the possibility of distinct interactions with norepinephrine secreting sympathetic neurons.
Characterization of the mouse nasal mucosal immune system during homeostasis and in response to inhaled allergen
We next sought to map the nasal mucosal immune and neuroepithelial system at homeostasis and during response to inhaled allergens. We performed scRNA-seq at homeostasis (n=4 mice) and after nasal inhalation of the mold allergen Alternaria alternata in the innate phase (36h after a single inhalation, n=3) and adaptive phase (36h after 2 inhalations given 6 days apart, n=2) (Fig. S5A). We included equal numbers of CD45+ immune cells and EpCAM+ cells (both EpCAMhigh and EpCAMintermediate), expanding FACS gating to capture OSNs, and again sampled cells from olfactory and respiratory mucosa (Fig. S5B). In total, we profiled 50,448 high-quality epithelial and immune cells and OSNs (Fig. 3A). Unsupervised cluster analysis detected 11 subtypes of myeloid (Fig. 3B, S5C) and 17 subtypes of lymphoid cells (Fig. 3C, S5D).
In the myeloid compartment at homeostasis, macrophages were most abundant (13%) followed by tissue-resident conventional neutrophils (11.7%), an additional group (4.3%) of the recently described Siglec F+ neutrophils (49) (Fig. 3B, D). Siglec F+ neutrophils and neutrophil precursors were highly abundant in the olfactory mucosa (Fig. 3D, S5E) (49). Lymphocytes were concentrated in the nasal respiratory portion, likely due to the nasal lymphoid tissue there (Fig. 3E). The ILC2 subset accounted for 5 and 6% of the hematopoietic cell pool in the olfactory and respiratory epithelia, respectively (Fig. S5E–F, Data File S6). We recovered all canonical T helper subsets (TH) except for TH1 cells and observed a population of γδ T cells which expressed high levels of Il17a and Rorc (Fig. S5D) as did TH17 cells. Both Il17a-expressing subsets were respiratory-enriched, as were ILC1, and NK cells (Fig. 3E, S5D–F).
Alternaria inhalation triggered rapid recruitment of eosinophils and ILC2 expansion in the nose when assessed by flow cytometry (Fig. S6A), similar to the immune response in the lung (44, 50, 51). Interestingly, we also found a moderate increase in Siglec F+ neutrophils (Fig. S6A) (49). Analyzing fewer cells, our scRNA-seq data was not statistically powered to detect all of these changes (Fig. 3F). One day after a single inhalation, the proportions of immune cell subsets were relatively stable (Fig. 3F, S6A–B). During the adaptive phase of inflammation (36h after the second dose of Alternaria), TH2 cells and regulatory T cells doubled (Fig. 3F) while the relative fraction of almost all myeloid cells decreased (Fig. 3F, Data File S6), likely a reflection of their stable absolute numbers.
Next, we defined cell-type specific (Data File S7) and Alternaria-induced genes (Fig. S6C–E, Data File S8). As expected, Alternaria drove a type 2-polarized innate immune response 24h after inhalation with type 2 effector cytokines Il5 and Il13 specifically upregulated by ILC2s (Data File S8), while TH2 cells showed the strongest transcriptional response to a second Alternaria dose (Fig. S6E). Finally, we characterized the intercellular signaling induced by the aeroallergen among the diverse cell types of the nasal mucosa (Fig. 3G). We mapped all receptor-ligand gene pairs – annotated in either CellPhoneDB 2.0 (52) or the FANTOM5 (53) databases – that showed differential expression in response to Alternaria (Data File S9). In addition to the classical cytokines Il5 and Il13, additional effector signals associated with type 2 inflammation was upregulated by ILC2s including Calca, (Fig. 3G) and Il17rb (Data File S8). Together, these data characterize the specific cell types and pathways constituting the type 2 polarized response to Alternaria in the nasal mucosa.
Mapping the subtypes of epithelial cells in the mouse nasal mucosa.
We next defined the diversity of olfactory epithelial cells (Fig. 4A–J, S7, and Data File S7). Sustentacular cells, tuft-MVCs, ionocytes, ductal cells and globose basal cells (Fig. S7A) were predominantly derived from the olfactory mucosa (Fig. 4B, Fig. 1A, 1D, S7A). (4, 54, 55). The epithelial stem cells of the respiratory mucosa and olfactory HBCs were transcriptionally highly similar despite their distinct morphology (S7B–C), but we identified distinct markers including the respiratory-enriched Krt15 (Fig. S7D).
Figure 4. Mapping the subtypes of secretory epithelial cells in the mouse nasal mucosa.
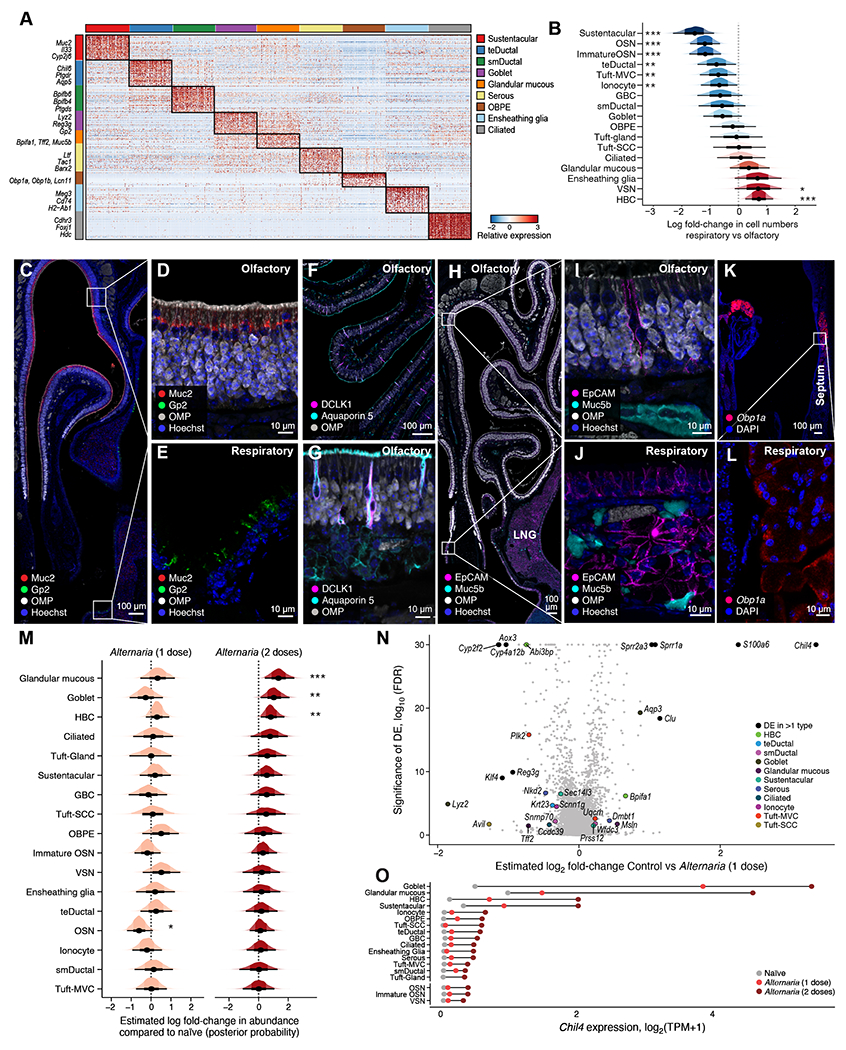
WT mice were assessed before Alternaria inhalation (n=4) (A-L) or after one (n=3) or two (n=2) doses of inhaled Alternaria (M-O) as in Fig. S5A. (A) Heatmap shows the expression level (row-wise z-score of log2(TPM+1), color bar) of top 30 marker genes (rows) specifically expressed (FDR<0.001) by each non-sensory epithelial subset (color bars). (B) Estimated effect (log fold-change, x axis) of tissue location on proportions of each epithelial cell type. Density histograms show posterior distribution of Dirichlet Multinomial regression. Black dot: point estimate, thick bar: 66% CI, thin bar: 95% CI. * 90%, ** 95%, *** 99% CI does not include 0. (C-L) Immunolocalization of epithelial cell subsets in the olfactory and respiratory nasal mucosa. (C) Cross section through the anterior section of the nose (Fig. S1A, T1) immunolabeled with OMP (olfactory epithelium), Muc2 (sustentacular) and GP2 (goblet cells). Closeup of the olfactory (D, top inset of C) and respiratory epithelium (E, bottom inset of C) identifies Muc2+ sustentacular and GP2+ goblet cells. (F) Olfactory turbinates marked by OMP and DCLK1 in combination with Aquaporin 5 (ductal cells) (G) Closeup of F. (H) Cross section through the posterior portion of the nose with olfactory turbinates (top) (Fig. S1A, T4), respiratory epithelium (bottom) and LNG (lower right) demonstrating the distribution of Muc5b in the olfactory (labeled with OMP) and respiratory mucosa. (I) Closeup of olfactory epithelium and submucosa from H. (J) Closeup of respiratory mucosa of the nasal septum from H with epithelium overlying the submucosal glands. (K) Cross section through the anterior portion of the nose (Fig. S1A, T1) with septum on the right and lateral turbinates on the left demonstrating the expression of Obp1a mRNA detected by RNA scope ISH. (L) Closeup of Obp1a mRNA expression in the septal submucosal gland from K. (M) Estimated effect (log fold-change, x axis) of one (left) and two (right) doses of inhaled Alternaria on proportions of epithelial cell subsets, visualized as in B. (N) Volcano plot shows the relationship between differential expression (log2 fold change, x axis) and significance (y axis) of genes (points) within epithelial subsets (color legend) after one dose of inhaled Alternaria. (O) Expression level (x-axis) of chitinase-like protein 4 (Chil4), before and after inhalation of Alternaria (color legend). Data are from two independent experiments with 1-2 mice/group.
Olfactory Cyp2g1+ cells were comprised of two major subsets with distinct characteristics: first, sustentacular cells were marked by Cyp2g1, Muc2 and Il33; and while a second Cyp2g1+ population negative for Muc2 expressed Aqp4 and Aqp5, markers of Bowman’s gland ductal cells (56) (Fig. 4A, C–G, S7E, F). We confirmed that MUC2 was also highly enriched in human sustentacular cells by reanalyzing data published by Durante et al (40) (Data File S4). The Bowman’s gland ductal cells were also unexpectedly positive for DCKL1 (Fig. 4F–G), previously considered a tuft cell marker. Since they were positioned between OSNs and epithelial cells, we refer to them as transepithelial ductal cells. They were enriched for the PGD2 receptor DP1 (Ptgdr) (Fig. 4A), associated with mucus secretion in humans (57). We identified a new population of submucosal ductal cells distinct in their expression of several members of the bactericidal/permeability-increasing protein (BPI) fold family members – Bpifb3, Bpifb4 and Bpifb6 (Fig. 4A, G, Data File S7), and high expression of the SARS-CoV2 entry molecule ACE2 (Fig. S7G–I). Notably, BPI proteins have been linked to regulation of viral replication (58).
We identified two additional types of mucous cells: Gp2+ goblet cells and Muc5b+/Tff2+ glandular mucous cells (Fig. 4B). Gp2+ goblet cells were abundant and scattered in the respiratory epithelium (Fig. 4C, E) and LNG in the posterior portion of the nose (Fig. S7J–K). The Muc5b+ glandular mucous cells specifically expressed the trefoil factor Tff2, secreted in concert with mucins and involved in epithelial restitution (39), as well as Bpifa1 (SPLUNC1), important for nasal epithelial defense (59)(Fig. 4A). We used Muc5b protein as a marker of glandular mucous cells and were found in 1) the acinar cells in the Bowman’s glands of the olfactory epithelium (Fig. 4H–I); 2) respiratory submucosal glands (Fig. 4H, J) and 3) acinar cells in the LNG (Fig. 4H). We detected only low Muc5b expression in Gp2+ goblet cells on the luminal surface of the respiratory epithelium and LNG (Fig. S7K–L). Consistent with this wide distribution of mucin-expressing cells, we find multiple Periodic Acid Schiff positive cells in the respiratory mucosa and Bowman’s glands (Fig. S7M).
Finally, we identified a population of epithelial cells marked by odorant binding protein (OBP)-encoding genes: Obp1a, Obp1b, Obp2a (Fig. 4A, S7N–O). OBPs reversibly bind odorants for delivery to the OSNs (60). These cells were detected in the submucosal area of the LNG based on high Obp1a expression by in situ hybridization (Fig. S7N), consistent with scRNA-seq (Fig. S7O), and similar to previous studies in rats (60). In addition, we found high Obp1a expression in the septal submucosal glands (Fig. 4K–L).
We then analyzed the responses of the olfactory and respiratory mucosa to aeroallergen inhalation of Alternaria. We identified a 1.4- and 3.9-fold increase in Muc5b glandular mucous cells in the epithelium after one and two Alternaria doses respectively, and a 2.9-fold increase in Gp2 goblet cells after two Alternaria doses (Fig. 4M, S8A–B, Data File S6). Differential expression analysis distinguished between cell-type specific and cell type-independent effects, identifying Muc5b+ glandular mucous and Gp2+ goblet cells as the most strongly responsive cell-types, and chitinase-like protein 4 (Chil4) as the gene most strongly up-regulated across multiple cell types (Fig. 4N–O, S8C, D). Chil4 increases in lung type 2 inflammation (61), in the nasal mucosa in an Il13 overexpressing system (62), and was recently shown to form crystals thought to amplify type 2 inflammation (63). Additionally, S100a6, Sprr1a and Sprr2a3 were induced by Alternaria inhalation (Fig. 4N). This was notable as these transcripts are prominently upregulated when the olfactory neuroepithelium is regenerated after full ablation (3), suggesting that the allergen Alternaria induces IL13-dependent remodeling (Chil4) but also a distinct olfactory injury-triggered neuroregeneration program (S100a6, Sprr1a and Sprr2a3).
Allergen inhalation induces horizontal basal stem cell proliferation.
Inhalation of the mold allergen Aspergillus leads to rapid OSN and epithelial cell death assessed histologically (64). To determine whether Alternaria causes similar disruption of neuroepithelial structure, we assessed the integrity of olfactory and respiratory mucosa by histology. Notably, the multilayered organization of the olfactory neuroepithelium – with OMP+ OSNs overlying 1-2 layers of β-tubulin+ (TUJ1+) immature olfactory sensory neurons (ISNs) and a single layer of proliferating Ki67+ globose basal cells – was preserved after Alternaria (Fig. 5A, S9A). In addition, neither the thickness of the OMP layer (Fig. 5A, S9B), nor the proportion of OMP+ OSNs and sustentacular cells detected in scRNAseq showed any significant change (Data File S6), together implying there was no profound OSN loss. Assessment using TUNEL showed clear evidence of apoptotic cells in respiratory but not in olfactory epithelia after a single Alternaria inhalation (Fig. S9C–D). We did not detect induction of necrosis or apoptosis-associated transcriptional programs in OSNs, while a minor signal was induced in HBCs after a single challenge and in sustentacular cells only after two challenges (Fig. S9E). Together, these data show that Alternaria inhalation does not cause substantial olfactory cell death.
Figure 5. Induction of HBC proliferation by allergen inhalation.
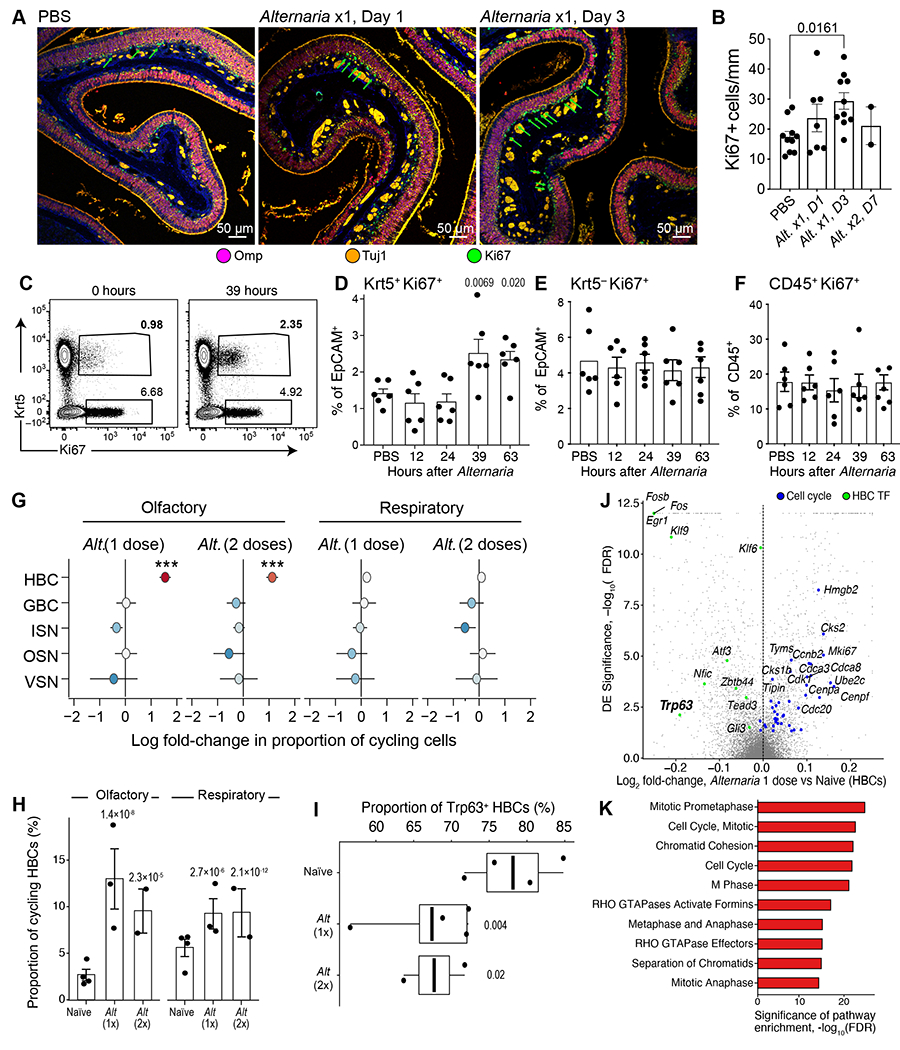
WT mice were assessed before Alternaria inhalation (n=4) (A-L) or after one (n=3) or two (n=2) doses of inhaled Alternaria (M-O) as in Fig. S5A. (A) Histological assessment of the effect of Alternaria inhalation (right hand panels) on overall morphology, and on OSNs (OMP, purple), proliferating cells (Ki67, green) and ISNs (Tuj1, Orange) in the olfactory neuroepithelium compared to PBS controls (left). (B) Histological quantification of proliferating (Ki67+) cells at baseline and after Alternaria assessed at days 1 and 3 after single inhalation (Alt x1 D1, D3), or a day after 2 inhalations (Alt x2, D7). (C-F) Flow cytometry analysis of Ki67 expression after a single Alternaria inhalation (30μg) at the indicated time points. Representative flow cytometry plot demonstrating the expression of Ki67 among CD45−EpCAM+ cells before and 39h after Alternaria inhalation (C). (D-F) Quantitation of the percent of Keratin5+Ki67+ (D) and Keratin5−Ki67+ (E) among EpCAM+ cells and the proportion of Ki67+ cells among CD45+ immune cells (F). Data in B, D-F are means ± SEM from 3 independent experiments, each dot is a separate mouse, p-values: Wald test. (G-H) Cell-type identity of Alternaria-induced proliferating cells from scRNA-seq data. (G) Estimated regression coefficients from Bayesian Dirichlet Multinomial regression (Methods) modeling the fold-change in proliferative state (x-axis) for each cell type (y-axis) after inhalation of Alternaria in the olfactory (left) and respiratory (right) nasal mucosa. Colored dot: point estimate, bar: standard deviation of the posterior, *** 99% CI does not include 0. (H) Estimated proportion (y-axis) of proliferating HBCs (Methods) in each mouse (dots) naïve mucosa and after inhalation of Alternaria (x-axis) assessed by scRNAseq. Bars show the mean and error bars show the SEM. P-values: Wald test. (I-K) Transcriptional activation of HBCs by Alternaria from scRNA-seq data. (I) Proportion of Trp63+ HBCs (J) Volcano plot shows DE (y axis shows −log10(FDR) and effect size (x axis) for HBCs after 1 dose of Alternaria. Cell-cycle and HBC-enriched transcription factors (FDR<0.001) are highlighted (color legend, top). (K) Statistical significance (x axis, −log10(FDR)) of top-ranked Reactome pathways enriched among up-regulated genes in HBCs. Data are from two independent experiments with 1-2 mice/group.
The most notable change was along the basal layer of the olfactory neuroepithelium where the number of Ki67+ cells visibly increased after Alternaria inhalation (Fig. 5A, B). Conversely, in the respiratory epithelium, the number of Ki67+ cells, which was much lower at baseline (reflecting the lack of globose basal cells), did not significantly increase after 1 or 2 Alternaria challenges (Fig. S9F–G). By immunofluorescence, we found that the olfactory Ki67+ cells are aligned at the basement membrane and partially co-localize with Keratin 5 (Krt5), suggesting they are HBCs and not globose basal cells, the cycling stem cell population in the nose (Fig. S10A). We confirmed using FACS that Krt5+ HBCs proliferate after Alternaria inhalation, while there was no significant change in the numbers of proliferating immune or non-immune cells including Ki67+Kit+ globose basal cells (Fig. 5C–F, S10B–D). Finally, we also found proliferation gene expression programs were strongly and specifically induced in HBCs in the olfactory epithelium. There was a 6.1-fold increase in the fraction of proliferating Krt5+ cells in the olfactory epithelium and only a 1.8-fold increase in the respiratory epithelium (Fig. 5G–H, S10E–F).
The transcription factor p63 (Trp63) promotes olfactory stem cell self-renewal by inhibiting HBC differentiation (65, 66) and its loss precedes HBC proliferation (67). Consistent with aeroallergen-induced HBC activation, we found that Trp63 expression in HBCs was markedly reduced after Alternaria (Fig. 5I). Alternaria inhalation also down-regulated the Notch signaling pathway, particularly Jag2 (FDR<0.0001) and Notch2 (FDR=0.09, Data File S8), consistent with known mechanisms of olfactory HBC activation (55, 68). Unsupervised analysis consistently showed genes upregulated in HBCs were dominated by cell-cycle programs (Fig. 5J), and pathway analysis demonstrated that proliferation programs were the most significantly enriched (Fig. 5K). Together, these data demonstrate that inhalation of Alternaria induced proliferation of normally quiescent HBCs within 36-72h but without apoptosis of OSNs or sustentacular cells.
Allergen-induced olfactory stem cell plasticity.
To define the plasticity of the olfactory stem cell niche in the setting of allergen exposure, we performed ‘pseudotime’ analysis (69) of epithelial and neuronal cells from the olfactory epithelium (Fig. 6A, S11A–C). The inferred trajectory of cellular states (Fig. 6A) suggested that HBCs give rise to globose basal cells, which in turn differentiate along a major pathway to OSNs, and a minor pathway to MVCs (identified here as tuft cells and ionocytes, Fig. 1), consistent with the results of Fletcher et al. (70). Early genes whose expression significantly varied (FDR<0.001) with progression through neurogenesis included canonical HBCs markers such as Krt5 and Trp63 (65), while genes at the latest stages included markers of mature OSNs such as Omp, Chga encoding chromogranin A, and Olfm1 for olfactomedin 1 (Fig. 6B, S11A–C, Data File S10). As expected, globose basal cell markers Ascl1 and Kit were transiently activated during intermediate stages (Fig. 6B). Early in the fate branch toward OSNs, we observed up-regulation of a module of genes involved in regulation of cell cytoskeleton, including stathmin-1 (Stmn1), alpha (Tuba1a, Tuba1b) and beta-tubulin (Tubb5). Cells that were specifically induced by Alternaria inhalation were positive for the trio of markers Mki67/Krt5/Stmn1 but not for Mki67 and Tubb3 (Fig. 6C, S11D). We validated that stathmin-1 marks a subset of early precursors using immunofluorescence and that very few proliferating cells were positive for the ISN marker TUJ1 (Fig. S12A, B), but some were positive for stathmin-1 (Fig. 6D). Lastly, histology revealed that Alternaria induced an elongation in the morphology of HBCs (Fig. 6D), consistent with our observation of change in forward and side scatter by FACS (Fig. S10D) indicative of the previously identified ‘morphological switch from a flattened quiescent phenotype to a pyramidal, proliferative phenotype’ (71) of activated HBCs.
Figure 6. Mold and house dust mite allergens direct distinct olfactory stem cell activation programs.
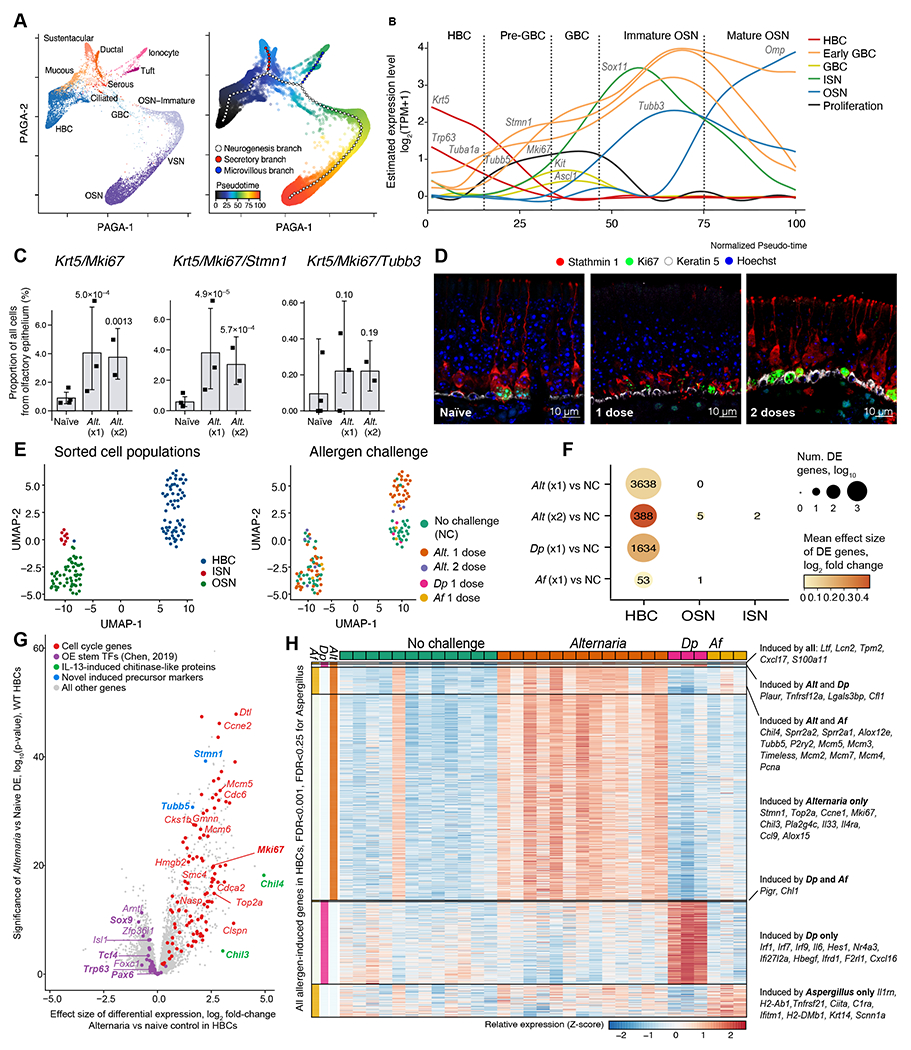
WT mice were given one or two inhalations of Alternaria and assessed by scRNAseq (as in Fig. S5A) (A-C) or by histology (D). (A) PAGA embeddings of 17,318 olfactory epithelial cells (points) colored by cell-type (left) and by pseudo-time (color legend, right). Trajectory (large points) and branches (color legend) were fit using elastic principal graphs. (B) Smoothed expression level (y-axis) of classical (Krt5, Bcam, Trp63, Kit, Ascl1, Omp, Tubb3) and novel (Tub1a, Tubb5, Stmn1) markers of intermediate stages of neurogenesis (color legend) along the pseudo-time course (x-axis) from HBCs to mature OSNs. Stages (dotted lines) are shown as a guide. (C) Mean proportion of cells in each mouse (point) in which each set of genes is detected by scRNAseq. Error bars: 95% CI, p-values: Wald test on logistic mixed model. (D) Immunofluorescence for Stathmin 1, Ki67 and Krt 5 in the olfactory epithelium of naïve mice (left) and after inhalation of one (center) and two (right) Alternaria doses. (E-H) WT mice were given a single inhalation of Alternaria (Alt), or Aspergillus fumigatus (Af) or Dermatophagoides pteronyssinus (Dp). Bulk RNAseq was performed on sorted HBCs (EpCAMintBCAM+), ISNs (EpCAMintNCAM1+) and OSNs (EpCAMlow/-NCAM1+) a day after allergen challenge. (E) Uniform Manifold Approximation and Projection (UMAP) embedding of bulk RNA-seq profiles of 130 FACS-sorted populations (color legend, left panel). (F) Dot plot shows the number (dot size) and mean fold-change (dot color) of DE genes in each population (x-axis) after each allergen challenge (y-axis) compared to no challenge (NC). (G) Volcano plot shows the differentially expressed genes in HBCs after 1 dose of Alternaria. (H) Heatmap shows the expression (row-wise Z-score, color bar) in FACS-sorted BCAM+ HBCs from each mouse (columns) of 1373 genes (rows) up-regulated (FDR<0.001 for Alternaria and Dp and FDR<0.25 for Aspergillus) grouped into categories where they are induced (color bars, left), selected genes are labeled.
Next, using a combination of basal cell adhesion molecule (BCAM), a marker of stem cells (72), NCAM1, a marker of OSNs and ISNs, and EpCAM, we identified the distinct developmental stages of OSNs by flow cytometry. We sorted EpCAMintBCAM+, EpCAMintNCAM1+, and EpCAMlow NCAM1+ and identified them as HBCs, ISNs and OSNs, respectively using histological markers (Fig. S12 C, D) and transcriptional analysis by bulk RNAseq (Fig. 6E and S12E). We then analyzed the transcriptional and compositional plasticity of each subset after inhalation of three different allergens: Alternaria, Aspergillus fumigatus (Af) or Dermatophagoides pteronyssinus (Dp) in WT mice. There were no significant compositional changes by flow cytometry 39h after single inhalation of the allergens (Fig. S12F–G) but BCAM+ stem cells increased three days after a single Alternaria inhalation and even further after 2 challenges (Fig. S12H). Transcriptionally, a single inhalation of each of these allergens caused significant changes in HBCs (Fig. 6F). Consistent with the scRNAseq, OSN and ISN gene expression profiles were not profoundly affected by Alternaria or Af inhalation, while Dp OSNs were not assessed (Fig. 6F). Up-regulated genes in HBCs from Alternaria challenged mice were prominently composed of cell cycle-related transcripts (Fig. 6G). Along with Trp63, Chen et al. (73) identify 38 HBC stemness transcription factors necessary for maintaining HBC stemness, three of which (Pax6, Tcf4 and Sox9) are also identified by Fletcher (65), and all of these were down-regulated (FDR < 0.5) in HBCs by Alternaria (Fig. 6G). Notch pathway signaling was also downregulated, including Notch1, Notch2, Jag1, and Jag2 (all FDR<0.0001, Data File S11), and Dll1 (FDR<0.05, Data File S11).
We then compared the genes induced in HBCs by Alternaria, Af and Dp, to determine common transcriptional programs and those specific to each allergen (Fig. 6H, Data File S11). Transcripts induced by all three allergens include the monocyte chemoattractant Cxcl17 and the antimicrobial peptides Lcn2 and Ltf, shown to increase in the epithelium in the setting of allergic inflammation (74). Alternaria led to the most robust induction of HBC proliferation, with stem cell activation markers Mki67, Top2a and Stmn1, all specifically upregulated. In addition, Alternaria specifically induced the chemokine Ccl9, the cytokine Il33, and Il4ra, consistent with strong type 2 response (Fig. 6G–H, S13A). Like Alternaria, Af caused an increase in HBC expression of cell cycle markers including Pcna, Mcm3, Mcm5 as well as the type 2-associated transcripts Chil4 and Alox12e, and the olfactory neuroregeneration-associated transcripts Sprr2a1 and Sprr2a2 but less strongly than Alternaria (Fig. 6H, S13C). Dp inhalation was associated with a robust interferon response and induction of NFκB signaling but without the robust proliferative response induced by Alternaria (Fig. 6H, S13B). In summary, we find that inhalation of several allergens causes downstream activation of HBCs without profound effects on OSNs. While some pathways, notably HBC proliferation, are shared with Af, Alternaria elicits the most profound transcriptional responses, inducing three distinct programs: stem cell proliferation, neuroregeneration (Sprr2a1 and Sprr2a2) and IL-13-dependent type 2 inflammatory (Chil4, Alox12e) programs.
Allergen-induced stem cell proliferation depends on tuft cells, not type 2 inflammatory pathways.
We next sought to define the mechanism by which allergens induce stem cell activation. In the olfactory neuroepithelium, stem cells are located beneath several layers of OSNs, 60-100μm below the luminal epithelial layer, which consists of TRPM5+ tuft-MVCs, sustentacular cells, and TRPM5− MVCs (ionocytes) (Fig. S7F). Since tuft-MVCs are directly activated by Alternaria (31), and their apical location in the olfactory epithelium positions them at the site of interaction with the airstream (Fig. 2E–G), we hypothesized that tuft-MVCs mediate the link between allergen and olfactory stem cells. To test this, we assessed the response to a single inhalation of Alternaria in Pou2f3−/− mice that lack tuft-MVCs (24) (Fig. 7A). Alternaria-induced stem cell proliferation, assessed as percent and number of proliferating Krt5+ epithelial cells, was reduced in tuft cell-deficient Pou2f3−/− mice (Fig. 7B–C, S14A–B, Data File S12). Proliferation of Krt5− epithelial cells, the number of Krt5+ stem cells at homeostasis or after allergen inhalation was unchanged by either allergen inhalation or tuft cell deficiency (Fig. S14C, D). Alternaria-induced nasal eosinophil and immune cell recruitment and proliferation were partially reduced in Pou2f3−/− mice (Fig. 7D, S14E, F). Finally, the proportions of BCAM+ stem cells, ISNs and OSNs were unchanged by Alternaria inhalation or deletion of tuft cells (Fig. S14G).
Figure 7. Allergen-induced stem cell proliferation is independent of type 2 inflammatory pathways and dependent on tuft cells.
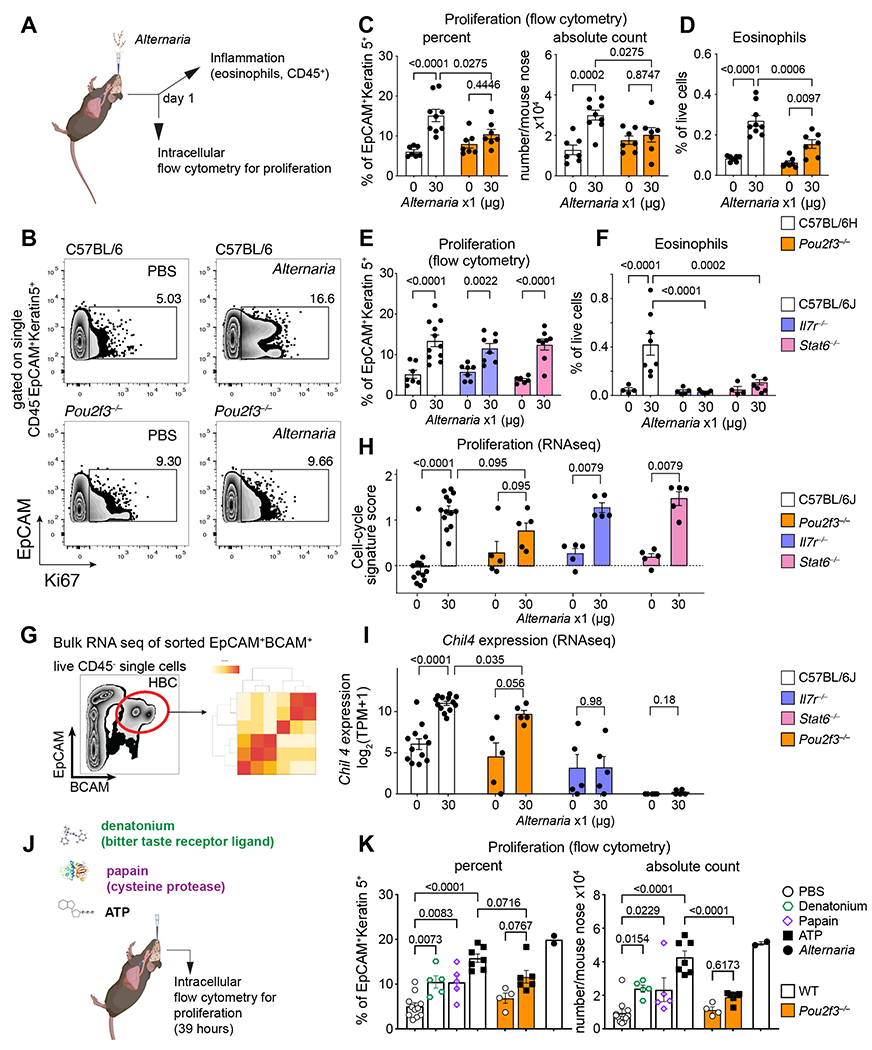
(A-I) Mice of the indicated genotypes were given a single dose of Alternaria intranasally and the nasal mucosa was assessed a day later for stem cell proliferation defined by Ki67 expression of Krt5+ cells (B, C, E) and eosinophil infiltration (D, F) by flow cytometry and for transcriptional reprograming by bulk RNAseq of sorted EpCAM+BCAM+ stem cells (G-I). RNAseq quantification of a validated cell-cycle signature score (Methods) (H), and the IL-13-dependent transcript Chil4 (I). (J-K) WT or Pou2f3−/− mice were given a single intranasal inhalation of denatonium, papain, ATP or Alternaria (color legend, right) and the percent and number of proliferating Krt5+ cells were assessed by intranuclear staining for Ki67 by flow cytometry after 39h. Data are means ± SEM from three independent experiments, each dot is a separate mouse. P-values for FACS quantification in C-F and K are from one-way ANOVA with Sidak’s multiple comparison correction. All other p-values (H-I) are Mann-Whitney U-test.
To determine whether the early olfactory proliferation and remodeling programs are dependent on innate lymphocytes or IL-13 signaling, we assessed the Alternaria response in Il7r−/− mice, deficient in innate and adaptive lymphocytes (75), and Stat6−/− mice, deficient in IL-4/IL-13 receptor signaling (76). Unexpectedly, we found that Alternaria-induced proliferation of Krt5+ stem cells was preserved in both Il7r−/− or Stat6−/− mice (Fig. 7E and S15A–C), indicating that innate lymphocytes and IL-13 signaling are not required for nasal stem cell proliferation. Consistent with previous findings in the lungs (50, 51), Alternaria-elicited eosinophil recruitment (Fig. 7F), nasal inflammation (Fig. S15D), and immune cell proliferation (Fig. S15E) were all ablated in both Il7r−/− and in Stat6−/− mice, and lymphocytes were strongly reduced in Il7r−/− mice (Fig. S15F).
Consistent with the flow cytometry data, RNAseq of sorted BCAM+ stem cells indicated both Mki67 (Fig. S15G) and a proliferation gene signature (77) were equally up-regulated in WT, Stat6−/− and Il7r−/− mice, but were reduced in Pou2f3−/− mice (Fig. 7G–H). An Alternaria signature score, computed based on the expression of up-regulated genes from scRNAseq profiles of HBCs (Data File S7), was also induced in Stat6−/− and Il7r−/− mice as in WT mice and reduced in Pou2f3−/− mice (Fig. S15H). However, transcriptional programs that are dependent on IL-13 signaling and innate lymphocytes such as Chil4 and the related chitinase Chil3 were completely abolished in Stat6−/−, strongly reduced in Il7r−/−, and only modestly affected in Pou2f3−/− mice (Fig. 7I, S15I). Together, these data demonstrate that nasal tuft cells direct allergen-induced stem cell proliferation independent of innate immune cells and IL-13 signaling.
We then used differential gene expression of sorted BCAM+ stem cells to further characterize the aspects of the Alternaria-induced transcriptional programs that depend on tuft cells (Pou2f3−/−), innate lymphocytes (in Il7r−/−) or IL-4/IL-13 receptor signaling (Stat6−/−). Of the 741 genes induced in stem cells by Alternaria, 305 were impaired (significant negative interaction term FDR<0.5) by at least one genotype (Fig. S16A, Data File S11). Of these, the majority (238 genes) were impaired only in tuft cell-deficient mice, and of these, 114 were cell-cycle associated, including Mki67. Conversely, Chil4 induction was reduced in both Il7r−/− and Stat6−/− mice consistent with its dependence on ILC2-derived IL-13. A further panel of IL-13-dependent transcripts were reduced only in Stat6−/− mice including other chitinase-like proteins Chil3, Chia, and the 15- lipoxygenase Alox15 (78), while the macrophage chemoattractant Cxcl17 and the asthma-associated transcript Tmem45a (79) were both specifically decreased in Il7r−/− mice (Fig. S16A). Together, these data define the dependence of specific innate epithelial effector molecules on the nasal mucosal immune system and demonstrate the necessity of tuft cells for allergen-induced olfactory stem cell proliferation.
Finally, we tested whether tuft cell activation is sufficient for stem cell proliferation. We gave mice a single inhalation of ATP, denatonium, or the cysteine protease papain, and assessed stem cell proliferation (Fig. 7J). ATP elicits calcium flux in ChAT+ MVCs (15) and CysLTs from unfractionated nasal ChAT+ tuft cells (31), ~95% of which we now show are tuft-MVCs. Denatonium, a Tas2r ligand, triggers calcium flux in a subset of ChAT+ MVCs (15). While Tas2rs are particularly highly expressed in tuft-SCCs, we also detected expression of Tas2r108, encoding a denatonium receptor (80, 81), and Tas2r138 in tuft-MVCs using both single-cell (Fig. S16B) and bulk RNAseq of sorted tuft cells from the olfactory mucosa (Fig. S16C). Expression of all Tas2rs was specific to tuft cells in the respiratory and olfactory mucosa (Fig. S16B–C). Papain is hypothesized to mimic the effect of cysteine proteases in allergens and is a putative protease receptor ligand (82). Two potential papain receptor candidates (F2r and F2rl1) were expressed by both tuft-SCCs and tuft-MVCs (Fig. S16B–C). Inhalation of denatonium and of papain induced a modest increase in the percent of Ki67+/Krt5+ cells (Fig. 7K, left) and a more marked change in the number of Ki67+/Krt5+ cells (Fig. 7K, right). ATP induced the most robust proliferation of stem cells, comparable with Alternaria assessed in parallel (Fig. 7K). Since ATP is a pluripotent epithelial activator, and the purinergic receptors P2ry2 and P2rx4 are broadly expressed by both tuft and non-tuft epithelial cells (Fig. S16B–C), we tested whether its effect on proliferation was reduced in tuft cell-deficient Pou2f3−/− mice. While not completely ablated, the absolute count of ATP-elicited proliferating stem cells was reduced in Pou2f3−/− mice (Fig. 7K), demonstrating that tuft cell activation is also sufficient to induce stem cell proliferation.
Taken together, these findings reshape our understanding of the component cell types of the nasal mucosa (Fig. S17) during homeostasis and in response to aeroallergen. Each compartment of the murine nasal epithelium – respiratory, vomeronasal, olfactory, and lateral nasal gland – differs in its cellular composition and the molecular markers required to dissect its heterogeneity of structure and function.
DISCUSSION:
Tuft cells are solitary chemosensory epithelial cells scattered in most mucosal surfaces where they activate neural and immune circuits to promote inflammation and epithelial remodeling. TRPM5+ MVCs were previously considered distinct from the rest of the chemosensory tuft family because of their distinct morphology, low expression of taste receptors and lack of intimate connections with sensory neurons (38). We demonstrate that the olfactory TRPM5+ MVCs share the core transcriptional profile of the solitary chemosensory tuft cell family and specifically its pro-inflammatory mediator cassette including transcripts for the cytokine IL-25 and the CysLT and prostaglandin generating enzyme Alox5, Alox5ap, Ltc4s, Ptgs1 and Hpgds. Finally, we identify a function of TRPM5+MVCs as drivers of allergen-induced non-apoptotic olfactory stem cell proliferation. Distinct from the function of other mucosal tuft cells as initiators of an ILC2-IL13 loop of epithelial stem cell activation (34), tuft-MVCs direct stem cell proliferation independent of innate lymphocytes and STAT6 signaling. This establishes a function of olfactory TRPM5+ tuft MVCs as drivers of neuroepithelial-intrinsic stem cell proliferation.
Tuft cells in most mucosal compartments were defined for their role in directing type 2 inflammation and downstream stem cell activation through a tuft cell-ILC2-IL-13-driven circuit (27, 34). Here we found a more limited role for nasal tuft cells as immune regulators, especially when compared to the degree of inflammatory cell reduction we observed in STAT6-deficient and lymphocyte-depleted mice. Our data indicate that a distinct role of nasal tuft-MVCs is the regulation of stem cell proliferation independent of inflammatory cascades, different from their pro-inflammatory role in other compartments. We considered the possibility that the multilayered organization of the olfactory epithelium might be responsible for the distinct role of tuft-MVCs here. A relay of TRPM5+ MVC (tuft-MVC)-derived acetylcholine signaling to IP3R3+ MVCs (ionocytes) to direct HBC proliferation was recently suggested (83). They also found a moderate reduction in HBC proliferation in Pou2f3−/− mice, albeit only after repeated inhalation of a mixture of odorants. Our more robust findings might be explained by the potent direct activation of tuft-MVCs by allergens (31, 33). Interestingly, tuft cell deletion in the intestine is also associated with a significant reduction in the baseline number of proliferating epithelial cells (84). Thus, a role for tuft cells in directing proliferation of epithelial cells has been suggested before in some compartments. Conversely, hyperplastic tuft cells found in the lung in the recovery phase after viral infections and bleomycin injury are dispensable for stem cell proliferation (85, 86). Interestingly, these tuft cells arise from Krt5+ stem cells but their development is independent of IL-13 signaling (85, 86). It is therefore likely that both the tuft cell activating signals and the specific cellular and mediator milieu of the distinct mucosal compartments where tuft cells reside determines their specific functions.
Our analysis of the olfactory and respiratory response to the mold allergen Alternaria identified the quiescent olfactory HBCs (55, 68) as an unexpected early responder to allergen inhalation. Given the absence of pronounced necrosis or apoptosis, the Alternaria-induced activation we demonstrate here provides a physiological model of olfactory HBC activation. HBC proliferation is also induced by inhalation of other allergens, specifically Aspergillus and the protease papain, suggesting that this is a shared allergen response pathway. Activation of HBCs is known to depend on the transcription factor p63 (Trp63), which promotes olfactory stem cell self-renewal by inhibiting differentiation (65, 66), and its down-regulation is required prior to HBC proliferation (67). Our analysis of Trp63 and Notch signaling using single-cell and bulk transcriptomics showed that along with HBC-specific down-regulation of Trp63, Alternaria inhalation also induced down-regulation of the Notch signaling pathway, consistent with the mechanisms of olfactory HBC activation described by the Schwob group (55, 68).
Our analysis of HBCs from Alternaria-challenged Pou2f3−/−, Il7r−/−, and Stat6−/− mice identified specific aspects of the integrated response contributed by tuft cells, lymphocytes, and IL4/IL-13 receptor signaling, respectively. Perhaps surprisingly, we found that the loss of tuft cells had the most profound impact on the transcriptional response, likely due to their role as direct, early sensors of allergens. Several critical epithelial effectors involved in barrier defense, which are also all implicated in asthma, including the protective signaling proteins SPLUNC1 (Bpifa1 (87, 88)) and Amphiregulin (Areg (89)), and anti-fungal chitinase-like proteins (Chia, Chil3 (90)), were all impaired in Stat6−/−, but not Il7r−/− or Pou2f3−/−, indicating their dependence on a non-lymphoid source of IL-13. Our data implicates mast cells and basophils as this potential source, as their expression of Il13 is highest apart from ILC2s. The set of Alternaria-induced genes impaired in both Stat6−/− and Il7r−/− mice was extremely small, which suggests that the direct regulation of HBCs by ILC2-derived IL-13 is a relatively minor pathway. Conversely, the program impaired exclusively in Il7r−/− was more extensive – indicating that lymphocytes, likely innate, regulate HBCs using IL-13-independent mechanisms – and included key modulators of the nasal immune response such as Cxcl17, recently implicated in macrophage-mediated airway remodeling (91).
Classically grouped together with TRPM5+ MVCs, olfactory TRPM5− MVCs are instead characterized by high expression of Cftr, Foxi1, Ascl3 and Itpr3, in addition to a transcriptome which we show here is almost identical to pulmonary ionocytes (35). This is in line with prior observations based on immunohistochemistry identification of CFTR- and ITPR3-expressing cells as MVCs (36). We do note, however, that ~5% of TRPM5+ tuft-MVCs also expressed Itpr3, indicating they could play a role in deficient neuroregenerative phenotypes observed in Itpr3−/− mice (11). Interestingly, although different in morphology, ionocytes from the olfactory and respiratory mucosa did not differ significantly in transcriptional makeup.
Although we identified tuft cells as the critical link between allergen sensing and HBC proliferation, we did not delineate the downstream signaling pathways that constitute this link. After allergen exposure, we found that the olfactory epithelium is reprogrammed in an IL-4/13-dependent fashion with induction of classical IL-13-dependent transcripts and goblet cell metaplasia, although the fate of the proliferating HBCs remains unclear. Pseudotime trajectories did not show any disruption of normal neurogenesis trajectories, but we are yet to define the fate of the proliferating stem cells. One intriguing possibility that will require future studies is whether tuft cells might induce a proliferative state in basal cells that is not associated with regeneration but may instead be part of an aberrant response to allergen sensing by the epithelium, which could be pathological rather than beneficial.
In sum, we provide extensive characterization of the rare subsets of nasal epithelial cells, identifying MVCs as belonging to the tuft cell and ionocyte families. We provide genetic evidence that olfactory tuft cells play an unexpected role in regulating the proliferation of the stem cell compartment, independent of classic inflammatory loops. This identifies a direct link between allergen sensing by tuft cells and activation of a reserve neural stem cell population. Together, these data deepen our understanding of the extensive epithelial heterogeneity in the nose and demonstrate that the functional capacity of tuft cells extends beyond triggering inflammation, as they are also regulators of tissue homeostasis and transformation.
MATERIALS AND METHODS:
Study design.
The aim of this study was to determine how olfactory TRPM5+ MVCs relate to the rest of the chemosensory family and define their function in the olfactory mucosa. The integrated response to allergens in the olfactory and respiratory mucosa was assessed in mice in vivo in mice with specific genetic deletion of tuft cells, immune cells and immune cell signaling. Mice were randomly assigned to treatment groups after matching for sex and age. All experimental replications are specifically described in the relevant figure legends (Table S1).
Mouse models.
Mice were maintained at the Brigham and Women’s Hospital specific pathogen-free animal facility, in accordance with the protocols approved by the Mass General Brigham (MGB) IACUC committee. C57BL/6 wild type (indicated as C57BL/6H) mice were originally from Charles River Laboratories, ChAT-eGFP from Jackson Laboratories and Pou2f3−/− (24) were all bred in house in parallel. Stat6−/−, Il7r−/− and age and sex-matched WT C57BL/6J control mice were from Jackson Laboratories. All mice were between 11 and 36 weeks old and included both sexes.
Human samples
Subjects between the ages of 18 and 75 years were recruited at the time of elective sinus surgery, and sinus or superior turbinate tissue was collected after subjects provided written informed consent. The local institutional review board – MGB IRB – approved the study.
Aeroallergen and tuft cell ligand challenge protocols.
For all in vivo allergen challenge experiments, mice were given intranasal inhalations of Alternaria, Dp or Af culture filtrate, ATPγS, denatonium benzoate or papain after sedation. Each substance was applied in a 20μl PBS solution to the tip of the nose or 10μl in each nostril.
Histochemistry, immunofluorescence, and quantitative assessment of cell numbers
Snouts were collected as described previously(31). All samples were stained with primary antibodies overnight at 4°C, after target retrieval with DAKO or EDTA target retrieval solution and incubated with secondary antibodies for 2 hours at room temperature unless otherwise indicated. Specific details can be found in the Supplementary Materials. Samples for in situ hybridization were fixed in DEPC-treated 4% PFA and decalcified with DEPC-treated EDTA solution and processed with RNAscope® Fluorescent Multiplex Reagent Ki, post-fixed with PFA and incubated with a probe to Lct4s or Obp1a after hydrogen peroxide and Protease III.
Single-cell suspension preparation for FACS and RNA seq.
To separate mouse respiratory from olfactory mucosa, the tissue overlying the proximal part of the nasal septal and lateral wall, together with VNO, was separated from olfactory epithelium, covering the distal portions of nasal cavity, along the anterior portion of the olfactory turbinate. The isolated olfactory and respiratory mucosa were processed as described previously (31). Single-cell suspensions from ChAT-eGFP mice were used for scRNAseq experiments at homoeostasis with specific enrichment of tuft cells (Figs. 1–2) or without enrichment of tuft cells (Figs 3–5). Single-cell suspensions for scRNAseq were collected in PBS and 0.04% BSA (w/v), run through 10X controller and individually bar-coded libraries were generated. Pooled library samples were sequenced on a NovaSeq S1 (Illumina) in partnership with Dana Farber Cancer Institute (DFCI) Molecular Biology Core Facilities (Figs. 3–6).
HBC proliferation in cell suspensions of the whole nasal mucosa was assessed with extracellular staining for CD45, Kit and EpCAM, followed by assessment of the intracellular markers Krt5 and Ki-67 after fixation and permeabilization with a Transcription Factor Buffer Set. HBC, ISN and OSNs were identified in whole nose single cell suspensions with antibodies against NCAM1, CD45, BCAM and EpCAM incubated at room temperature for 40 min. One thousand cells of each subset were sorted into TCL buffer with 1% 2-mercaptoethanol. Bulk RNA sequencing was at the Broad Institute Technology Labs using low-input eukaryotic Smart-seq 2. Smart-seq2 libraries were sequenced on an Illumina NextSeq500 using a High Output kit to generate 2 x 25 bp reads.
Dimensionality reduction and cell type identification
De-multiplexing, alignment to the mm10 transcriptome and UMI-collapsing were performed using the CellRanger toolkit (version 1.0.1, 10X Genomics). Quality filtering, variable gene selection and clustering we performed as described previously (39), and we interpreted clusters using known markers (Data File S2), merging clusters expressing the same known markers, and removing doublets and contaminating fibroblast populations. After comparing all cell type clusters to each other, marker genes (Data File S3, S5, S7) were defined as described previously (39). To identify proliferation all cells were scored for a previously defined cell-cycle signature gene set defined from scRNA-seq data (77) as described above, and the distribution of scores was clustered into two groups using an unsupervised mixture of gaussians (Mclust package in R (92)). The proportion of the cells in the high-expression cluster was reported as the proportion of proliferating cells. To analyze cell-cell signaling we combined CellPhoneDB (52) and FANTOM5 (53) interaction databases as described previously (93). For plotting purposes (Fig. 3G), we identified receptors or ligands differentially expressed after Alternaria (FDR < 0.05 and absolute log2 fold-change > 0.1). Circle plot visualization was produced using the ‘edgebundleR’ R package. For full details, see Supplemental Methods.
Identification of human tuft cell subsets.
In the dataset derived by Durante et al (40) the authors defined a subset of “Olfactory Microvillar Cells”. We restricted our analysis to this subset and reanalyzed these cells to compare them to murine nasal microvillous cells. For the Ordovas-Montañes dataset (41), no cluster of microvillous cells had been previously identified. To identify them we clustered the whole dataset and identified a cluster of 92 cells that were enriched for known microvillous cell markers (Foxi1, Cftr, Trpm5). We then re-clustered this subset and identified two sub-groups that expressed tuft and ionocyte markers respectively.
Pseudo-time analysis of olfactory neurogenesis.
We isolated the 17,318 CD45− epithelial and neuronal cells (Fig. 6A, left) to examine the progression through neurogenesis. The Partition-Based Graph Abstraction (PAGA) algorithm (69) was used to project cells into a low dimensional manifold, after defining unsupervised clusters generated using the Leiden algorithm (94), implemented with scanpy (95). Elastic principal graphs (96) were then used to fit a branching tree through the PAGA co-ordinate space. Spurious single-node branches were removed, producing a tree with a main branch from HBCs to mature OSNs, and two branches which were interpreted as a secretory and microvillous cell branch (Fig. 6A, right), consistent with earlier lineage tracing data (70). The node within the HBC cluster was manually selected as the root node. Genes significantly varying along the pseudo-time path we identified by fitting generalized additive models (97) to the expression level of each gene (Fig. S11B–C). Genes were ranked by the pseudo-time coordinate of their first non-zero fitted value in order to identify early markers of neurogenesis (Fig. S11C).
Bulk RNAseq processing and deconvolution-based cell type composition analysis
Computational pipelines for RNA seq analysis were implemented as described elsewhere (35, 39). Paired-end reads were mapped to the UCSC mm10 mouse transcriptome using Bowtie (98). Cell type proportions were estimated using CIBERSORTx (47) with default parameters. We tested for compositional changes using a Dirichlet Multinomial Mixture (DMM) model fit using the “brms” package in R (99) (see Supplement).
Statistical analyses.
For all statistical analyses of non-sequencing data, analysis was performed with GraphPad Prism software (version 10). For all in vivo experiments with ≧3 group comparisons, the overall significance was determined using a one-way analysis of variance (ANOVA), and pairwise comparison was performed with Sidak’s test to account for multiple comparisons. For experiments involving two categories (i.e genotype and treatment), a two-way ANOVA and Wald test was used to confirm a significant interaction effect. A value of P < 0.05 was considered significant. Sample sizes were not predetermined by statistical methods.
Supplementary Material
Figure S1: Isolation of single-cell suspensions from nasal respiratory and olfactory epithelium.
Figure S3: Three morphologically distinct types of nasal tuft cells share a common molecular profile.
Figure S2: Anatomical and molecular characterization of olfactory epithelial and neuronal subtypes in mice and humans.
Figure S5: Single-cell mapping of immune cell subtypes in the naive nasal mucosa.
Figure S6: Effects of allergen inhalation on gene expression and cell type proportions in the nasal immune system.
Figure S4: Neurogranin acts as a novel tuft cell marker of tuft-MVCs and tuft-gland cells.
Figure S7: Expression of SARS-CoV-2 receptor ACE2 in epithelial cells and marker genes of differentiated mucosal subtypes in naïve mice.
Figure S9: Alternaria inhalation is not associated with significant olfactory damage.
Figure S10: Alternaria inhalation induces HBC proliferation in the absence of additional induction of globose basal cell proliferation.
Figure S11: Pseudo-time analysis identifies novel marker genes for stages of olfactory neurogenesis.
Figure S12. Characterization of HBC and OSN populations in the olfactory mucosa.
Figure S13: Transcriptional programs induced in HBCs by multiple aeroallergens.
Figure S14: Tuft cells are required for allergen-induced HBC proliferation.
Figure S15: Allergen-induced HBC proliferation is independent of type 2 inflammatory pathways.
Figure S16: Characterization of HBC transcriptional responses to allergen and receptor expression by nasal epithelial cells.
Figure S17. Diverse neuroepithelial cell types comprise distinct regions of the murine nose.
Figure S8: Effects of allergic inflammation on gene expression and cell type proportions in the nasal epithelium.
Table S1. Reproducibility checklist
Data File S3. Molecular signatures for all cell types in the nasal epithelium.
Data File S2. Known marker gene sets used for interpretation of cell clusters.
Data File S4. Marker genes for human nasal tuft cells and ionocytes.
Data File S5. Molecular signatures for novel subsets of nasal tuft cells.
Data File S6. Cell type composition dynamics during allergic inflammation.
Data File S7. Molecular signatures for epithelial and immune cell subsets in the nose.
Data File S8. Cell type-specific gene expression dynamics after allergic inflammation.
Data File S10. Gene expression dynamics during olfactory neurogenesis.
Data File S12. Additional statistical analysis of genotype interaction with treatment effect of Alternaria inhalation by two-way ANOVA.
Data File S9. Modulation of cell-cell signaling during allergic inflammation.
Data File S11. Allergen-induced inflammation-dependent and independent transcriptional programs in the olfactory epithelium by bulk RNA seq.
Data File S1. Raw data points underlying all digital Figures and Supplementary Figures.
Acknowledgments:
We thank Adam Chicoine, Juning Case, Gerald Watts, Zhu Zhu, and Kevin Wei from the Brigham and Women’s Hospital Center for Cellular Profiling for their invaluable assistance with cell isolation and single-cell RNA sequencing. We gratefully acknowledge Leslie Gaffney for help with arranging figures, and Daniel Montoro for technical advice. We are also thankful to Dr. Lisa Goodrich at the Department of Neurobiology at Harvard Medical School for providing reagents and infrastructure for in situ hybridization. The authors gratefully acknowledge the MicRoN (Microscopy Resources on the North Quad) Core for their support & assistance in this work. Schematic diagrams were created using BioRender.
Funding:
This work was supported by grants from the National Institutes of Health grant K08 AI132723 (to LGB), 1R21AI154345 (to LGB and ALH), 5T32AI007306 (to JAB, SU), R01AI078908, R37AI052353, R01AI136041, R01HL136209 (to JAB), U19 AI095219 (to NAB, JAB), R01AI134989 (to NAB), AAAAI Foundation Faculty Development Award and Joycelyn C. Austen Fund for Career Development of Women Physician Scientists (to LGB), the Parker B. Francis Fellowship (to ALH) and a generous donation by the Vinik family (to LGB).
Footnotes
Competing interests: Authors declare that they have no competing interests.
Data and materials availability:
All raw and processed sequencing data is deposited in the GEO online database (GSE245074). No custom code was used as part of this analysis. Standard code required to reproduce the main steps of the analysis will be made available for research purposes on reasonable request. All other data needed to support the conclusions of the paper are present in the paper or the Supplementary Materials.
References
- 1.Schwob JE, Huard JM, Luskin MB, Youngentob SL, Retroviral lineage studies of the rat olfactory epithelium. Chem Senses 19, 671–682 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Herrick DB, Guo Z, Jang W, Schnittke N, Schwob JE, Canonical Notch Signaling Directs the Fate of Differentiating Neurocompetent Progenitors in the Mammalian Olfactory Epithelium. J Neurosci 38, 5022–5037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Fletcher RB, Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Stem Cell 21, 775–790.e779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein BJ, Goss GM, Hatzistergos KE, Rangel EB, Seidler B, Saur D, Hare JM, Adult c-Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J Comp Neurol 523, 15–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen A, Finger TE, Is TrpM5 a reliable marker for chemosensory cells? Multiple types of microvillous cells in the main olfactory epithelium of mice. BMC Neurosci 9, 159–112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baxter BD, Larson ED, Merle L, Feinstein P, Polese AG, Bubak AN, Niemeyer CS, Hassell J Jr., Shepherd D, Ramakrishnan VR, Nagel MA, Restrepo D, Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium. BMC Genomics 22, 224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin W, Ezekwe EA Jr., Zhao Z, Liman ER, Restrepo D, TRPM5-expressing microvillous cells in the main olfactory epithelium. BMC Neurosci 9, 114 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegg CC, Jia C, Chick WS, Restrepo D, Hansen A, Microvillous cells expressing IP3 receptor type 3 in the olfactory epithelium of mice. Eur J Neurosci 32, 1632–1645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfister S, Dietrich MG, Sidler C, Fritschy JM, Knuesel I, Elsaesser R, Characterization and turnover of CD73/IP(3)R3-positive microvillar cells in the adult mouse olfactory epithelium. Chem Senses 37, 859–868 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Weng PL, Vinjamuri M, Ovitt CE, Ascl3 transcription factor marks a distinct progenitor lineage for non-neuronal support cells in the olfactory epithelium. Sci Rep 6, 38199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia C, Hayoz S, Hutch CR, Iqbal TR, Pooley AE, Hegg CC, An IP3R3- and NPY-expressing microvillous cell mediates tissue homeostasis and regeneration in the mouse olfactory epithelium. PLoS One 8, e58668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goss GM, Chaudhari N, Hare JM, Nwojo R, Seidler B, Saur D, Goldstein BJ, Differentiation potential of individual olfactory c-Kit+ progenitors determined via multicolor lineage tracing. Dev Neurobiol 76, 241--251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia C, Hegg CC, NPY mediates ATP-induced neuroproliferation in adult mouse olfactory epithelium. Neurobiol Dis 38, 405–413 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemons K, Fu Z, Aoude I, Ogura T, Sun J, Chang J, Mbonu K, Matsumoto I, Arakawa H, Lin W, Lack of TRPM5-Expressing Microvillous Cells in Mouse Main Olfactory Epithelium Leads to Impaired Odor-Evoked Responses and Olfactory-Guided Behavior in a Challenging Chemical Environment. eNeuro 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura T, Szebenyi SA, Krosnowski K, Sathyanesan A, Jackson J, Lin W, Cholinergic microvillous cells in the mouse main olfactory epithelium and effect of acetylcholine on olfactory sensory neurons and supporting cells. J Neurophysiol 106, 1274–1287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasteva G, Canning BJ, Hartmann P, Veres TZ, Papadakis T, Muhlfeld C, Schliecker K, Tallini YN, Braun A, Hackstein H, Baal N, Weihe E, Schutz B, Kotlikoff M, Ibanez-Tallon I, Kummer W, Cholinergic chemosensory cells in the trachea regulate breathing. Proc Natl Acad Sci 108, 9478–9483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaske S, Krasteva G, Konig P, Kummer W, Hofmann T, Gudermann T, Chubanov V, TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells. BMC Neurosci 8, 49 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perniss A, Liu S, Boonen B, Keshavarz M, Ruppert A-L, Timm T, Pfeil U, Soultanova A, Kusumakshi S, Delventhal L, Aydin Ö, Pyrski M, Deckmann K, Hain T, Schmidt N, Ewers C, Günther A, Lochnit G, Chubanov V, Gudermann T, Oberwinkler J, Klein J, Mikoshiba K, Leinders-Zufall T, Offermanns S, Schütz B, Boehm U, Zufall F, Bufe B, Kummer W, Chemosensory Cell-Derived Acetylcholine Drives Tracheal Mucociliary Clearance in Response to Virulence-Associated Formyl Peptides. Immunity 52, 683–699.e611 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Bezencon C, Furholz A, Raymond F, Mansourian R, Metairon S, Le Coutre J, Damak S, Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J Comp Neurol 509, 514–525 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Schutz B, Jurastow I, Bader S, Ringer C, von Engelhardt J, Chubanov V, Gudermann T, Diener M, Kummer W, Krasteva-Christ G, Weihe E, Chemical coding and chemosensory properties of cholinergic brush cells in the mouse gastrointestinal and biliary tract. Front Physiol 6, 87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulbransen BD, Clapp TR, Finger TE, Kinnamon SC, Nasal Solitary Chemoreceptor Cell Responses to Bitter and Trigeminal Stimulants In Vitro. J Neurophysiol 99, 2929–2937 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W, Ogura T, Margolskee RF, Finger TE, Restrepo D, TRPM5-expressing solitary chemosensory cells respond to odorous irritants. J Neurophysiol 99, 1451–1460 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Saunders CJ, Christensen M, Finger TE, Tizzano M, Cholinergic neurotransmission links solitary chemosensory cells to nasal inflammation. Proc Natl Acad Sci U S A 111, 6075–6080 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi T, Yamashita J, Ohmoto M, Aoude I, Ogura T, Luo W, Bachmanov AA, Lin W, Matsumoto I, Hirota J, Skn-1a/Pou2f3 is required for the generation of Trpm5-expressing microvillous cells in the mouse main olfactory epithelium. BMC Neurosci 15, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita J, Ohmoto M, Yamaguchi T, Matsumoto I, Hirota J, Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS One 12, e0189340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohmoto M, Yamaguchi T, Yamashita J, Bachmanov AA, Hirota J, Matsumoto I, Pou2f3/Skn-1a is necessary for the generation or differentiation of solitary chemosensory cells in the anterior nasal cavity. Biosci Biotechnol Biochem 77, 2154–2156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P, Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finger TE, Böttger B, Hansen A, Anderson KT, Alimohammadi H, Silver WL, Solitary Chemoreceptor Cells in the Nasal Cavity Serve as Sentinels of Respiration. Proc Natl Acad Sci 100, 8981–8986 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merigo F, Benati D, Tizzano M, Osculati F, Sbarbati A, alpha-Gustducin immunoreactivity in the airways. Cell Tissue Res 319, 211–219 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Genovese F, Tizzano M, Microvillous Cells in the Olfactory Epithelium Express Elements of the Solitary Chemosensory Cell Transduction Signaling Cascade. PLoS one 13, e0202754–0202715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ualiyeva S, Hallen N, Kanaoka Y, Ledderose C, Matsumoto I, Junger WG, Barrett NA, Bankova LG, Airway brush cells generate cysteinyl leukotrienes through the ATP sensor P2Y2. Sci Immunol 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Gowda GAN, Fontana MF, Erle DJ, Anderson MS, Locksley RM, Raftery D, von Moltke J, Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49, 33–41.e37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ualiyeva S, Lemire E, Aviles EC, Wong C, Boyd AA, Lai J, Liu T, Matsumoto I, Barrett NA, Boyce JA, Haber AL, Bankova LG, Tuft cell–produced cysteinyl leukotrienes and IL-25 synergistically initiate lung type 2 inflammation. Sci Immunol 6, eabj0474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Moltke J, Ji M, Liang H-E, Locksley RM, Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature 529, 221–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, Rogel N, Burgin G, Tsankov AM, Waghray A, Slyper M, Waldman J, Nguyen L, Dionne D, Rozenblatt-Rosen O, Tata PR, Mou H, Shivaraju M, Bihler H, Mense M, Tearney GJ, Rowe SM, Engelhardt JF, Regev A, Rajagopal J, A Revised Airway Epithelial Hierarchy Includes CFTR-expressing Ionocytes. Nature 560, 319–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister S, Weber T, Härtig W, Schwerdel C, Elsaesser R, Knuesel I, Fritschy J-M, Novel role of cystic fibrosis transmembrane conductance regulator in maintaining adult mouse olfactory neuronal homeostasis. The Journal of comparative neurology 523, 406–430 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Ablimit A, Aoki T, Matsuzaki T, Suzuki T, Hagiwara H, Takami S, Takata K, Immunolocalization of water channel aquaporins in the vomeronasal organ of the rat: expression of AQP4 in neuronal sensory cells. Chem Senses 33, 481–488 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Tizzano M, Finger TE, Chemosensors in the nose: guardians of the airways. Physiology (Bethesda) 28, 51–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A, A Single-cell Survey of the Small Intestinal Epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durante MA, Kurtenbach S, Sargi ZB, Harbour JW, Choi R, Kurtenbach S, Goss GM, Matsunami H, Goldstein BJ, Single-cell Analysis of Olfactory Neurogenesis and Differentiation in Adult Humans. Nature Neuroscience 23, 323–326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, Wadsworth MH, Hughes TK, Kazer SW, Yoshimoto E, Cahill KN, Bhattacharyya N, Katz HR, Berger B, Laidlaw TM, Boyce JA, Barrett NA, Shalek AK, Allergic Inflammatory Memory in Human Respiratory Epithelial Progenitor Cells. Nature 560, 649–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holbrook EH, Wu E, Curry WT, Lin DT, Schwob JE, Immunohistochemical characterization of human olfactory tissue. The Laryngoscope 121, 1687--1701 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerendasy DD, Sutcliffe JG, RC3/neurogranin, a postsynaptic calpacitin for setting the response threshold to calcium influxes. Mol Neurobiol 15, 131–163 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Bankova LG, Dwyer DF, Yoshimoto E, Ualiyeva S, McGinty JW, Raff H, von Moltke J, Kanaoka Y, Frank Austen K, Barrett NA, The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Sci Immunol 3, eaat9453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perniss A, Boonen B, Tonack S, Thiel M, Poharkar K, Alnouri MW, Keshavarz M, Papadakis T, Wiegand S, Pfeil U, Richter K, Althaus M, Oberwinkler J, Schutz B, Boehm U, Offermanns S, Leinders-Zufall T, Zufall F, Kummer W, A succinate/SUCNR1-brush cell defense program in the tracheal epithelium. Sci Adv 9, eadg8842 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barros-Silva JD, Linn DE, Steiner I, Guo G, Ali A, Pakula H, Ashton G, Peset I, Brown M, Clarke NW, Bronson RT, Yuan GC, Orkin SH, Li Z, Baena E, Single-Cell Analysis Identifies LY6D as a Marker Linking Castration-Resistant Prostate Luminal Cells to Prostate Progenitors and Cancer. Cell Rep 25, 3504–3518 e3506 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D, Diehn M, Alizadeh AA, Determining cell type abundance and expression from bulk tissues with digital cytometry. Nature Biotechnology 37, 773--782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gulbransen BD, Finger TE, Solitary chemoreceptor cell proliferation in adult nasal epithelium. J Neurocytol 34, 117–122 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogawa K, Asano K, Yotsumoto S, Yamane T, Arita M, Hayashi Y, Harada H, Makino-Okamura C, Fukuyama H, Kondo K, Yamasoba T, Tanaka M, Frontline Science: Conversion of neutrophils into atypical Ly6G(+) SiglecF(+) immune cells with neurosupportive potential in olfactory neuroepithelium. J Leukoc Biol 109, 481–496 (2021). [DOI] [PubMed] [Google Scholar]
- 50.Doherty TA, Khorram N, Chang JE, Kim HK, Rosenthal P, Croft M, Broide DH, STAT6 regulates natural helper cell proliferation during lung inflammation initiated by Alternaria. Am J Physiol Lung Cell Mol Physiol 303, L577–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kouzaki H, Iijima K, Kobayashi T, O’Grady SM, Kita H, The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. Journal of immunology (Baltimore, Md. : 1950) 186, 4375–4387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polanski K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA, Single-cell Reconstruction of the Early Maternal–fetal Interface in Humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, Forrest ARR, A draft network of ligand-receptor-mediated multicellular signalling in human. Nature Communications 6, 7866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krolewski RC, Packard A, Schwob JE, Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. J Comp Neurol 521, 833–859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwob JE, Jang W, Holbrook EH, Lin B, Herrick DB, Peterson JN, Coleman JH, Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J Comp Neurol 525, 1034--1054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solbu TT, Holen T, Aquaporin pathways and mucin secretion of Bowman’s glands might protect the olfactory mucosa. Chemical Senses 37, 35 -- 46 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Oguma T, Asano K, Ishizaka A. a., Role of prostaglandin D2 and its receptors in the pathophysiology of asthma. Allergology International 57, 307–312 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Evans AS, Lennemann NJ, Coyne CB, BPIFB3 interacts with ARFGAP1 and TMED9 to regulate non-canonical autophagy and RNA virus infection. Journal of Cell Science 134, jcs251835 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akram KM, Moyo NA, Leeming GH, Bingle L, Jasim S, Hussain S, Schorlemmer A, Kipar A, Digard P, Tripp RA, Shohet RV, Bingle CD, Stewart JP, An innate defense peptide BPIFA1/SPLUNC1 restricts influenza A virus infection. Mucosal Immunol 11, 71–81 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Pevsner J, Hwang PM, Sklar PB, Venable JC, Snyder SH, Odorant-binding protein and its mRNA are localized to lateral nasal gland implying a carrier function. Proc Natl Acad Sci 85, 2383–2387 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sutherland TE, Logan N, Rckerl D, Humbles AA, Allan SM, Papayannopoulos V, Stockinger B, Maizels RM, Allen JE, Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nature Immunology 15, 1116–1125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saraswathula A, Liu MM, Kulaga H, Lane AP, Chronic interleukin-13 expression in mouse olfactory mucosa results in regional aneuronal epithelium. Int Forum Allergy Rhinol 13, 230–241 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heyndrickx I, Deswarte K, Verstraete K, Verschueren KHG, Smole U, Aegerter H, Dansercoer A, Hammad H, Savvides SN, Lambrecht BN, Ym1 protein crystals promote type 2 immunity. bioRxiv, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Epstein VA, Bryce PJ, Conley DB, Kern RC, Robinson AM, Intranasal Aspergillus fumigatus exposure induces eosinophilic inflammation and olfactory sensory neuron cell death in mice. Otolaryngol Head Neck Surg 138, 334–339 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, Ngai J, p63 regulates olfactory stem cell self-renewal and differentiation. Neuron 72, 748 -- 759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Packard A, Schnittke N, Romano RA, Sinha S, Schwob JE, Np63 Regulates Stem Cell Dynamics in the Mammalian Olfactory Epithelium. Journal of Neuroscience 31, 8748--8759 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI, Jang W, Schwob JE, Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc Natl Acad Sci 112, E5068–E5077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herrick DB, Lin B, Peterson J, Schnittke N, Schwob JE, Notch1 maintains dormancy of olfactory horizontal basal cells, a reserve neural stem cell. Proc Natl Acad Sci 114, E5589–E5598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Gttgens B, Rajewsky N, Simon L, Theis FJ, PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biology 20, 59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Deconstructing Olfactory Stem Cell Trajectories at Single-Cell Resolution. Cell Stem Cell 20, 817–830.e818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brann JH, Ellis DP, Ku BS, Spinazzi EF, Firestein S, Injury in aged animals robustly activates quiescent olfactory neural stem cells. Frontiers in Neuroscience 9, 367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Hallen NR, Lee M, Samuchiwal S, Ye Q, Buchheit KM, Maxfield AZ, Roditi RE, Bergmark RW, Bhattacharyya N, Ryan T, Gakpo D, Raychaudhuri S, Dwyer D, Laidlaw TM, Boyce JA, Gutierrez-Arcelus M, Barrett NA, Type 2 Inflammation Drives an Airway Basal Stem Cell Program Through Insulin Receptor Substrate Signaling. J Allergy Clin Immunol, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen M, Reed RR, Lane AP, Chronic Inflammation Directs an Olfactory Stem Cell Functional Switch from Neuroregeneration to Immune Defense. Cell Stem Cell 25, 501–513.e505 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dittrich AM, Krokowski M, Meyer HA, Quarcoo D, Avagyan A, Ahrens B, Kube SM, Witzenrath M, Loddenkemper C, Cowland JB, Hamelmann E, Lipocalin2 protects against airway inflammation and hyperresponsiveness in a murine model of allergic airway disease. Clin Exp Allergy 40, 1689–1700 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL, Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med 180, 1955–1960 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaplan MH, Schindler U, Smiley ST, Grusby MJ, Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 4, 313–319 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A, Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Research 25, 1860–1872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown CD, Kilty I, Yeadon M, Jenkinson S, Regulation of 15-lipoxygenase isozymes and mucin secretion by cytokines in cultured normal human bronchial epithelial cells. Inflamm Res 50, 321–326 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, Erle DJ, Yamamoto KR, Fahy JV, Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A 104, 15858–15863 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hollenhorst MI, Kumar P, Zimmer M, Salah A, Maxeiner S, Elhawy MI, Evers SB, Flockerzi V, Gudermann T, Chubanov V, Boehm U, Krasteva-Christ G, Taste Receptor Activation in Tracheal Brush Cells by Denatonium Modulates ENaC Channels via Ca(2+), cAMP and ACh. Cells 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ, T2Rs function as bitter taste receptors. Cell 100, 703–711 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Liang G, Barker T, Xie Z, Charles N, Rivera J, Druey KM, Naive T cells sense the cysteine protease allergen papain through protease-activated receptor 2 and propel TH2 immunity. J Allergy Clin Immunol 129, 1377–1386 e1313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lemons K, Fu Z, Ogura T, Lin W, TRPM5-expressing Microvillous Cells Regulate Region-specific Cell Proliferation and Apoptosis During Chemical Exposure. Neuroscience 434, 171–190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A, Chen X, May R, Houchen CW, Fox JG, Gershon MD, Quante M, Wang TC, Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 124, 1283–1295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barr J, Gentile ME, Lee S, Kotas ME, Holcomb M. Fernanda de Mello Costa N. P., Jaquish A, Palashikar G, Soewignjo M, McDaniel M, Matsumoto I, Margolskee R, Von Moltke J, Cohen NA, Sun X, Vaughan AE, Injury-induced pulmonary tuft cells are heterogenous, arise independent of key Type 2 cytokines, and are dispensable for dysplastic repair. Elife 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rane CK, Jackson SR, Pastore CF, Zhao G, Weiner AI, Patel NN, Herbert DR, Cohen NA, Vaughan AE, Development of solitary chemosensory cells in the distal lung after severe influenza injury. Am J Physiol Lung Cell Mol Physiol 316, L1141–L1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schaefer N, Li X, Seibold MA, Jarjour NN, Denlinger LC, Castro M, Coverstone AM, Teague WG, Boomer J, Bleecker ER, Meyers DA, Moore WC, Hawkins GA, Fahy J, Phillips BR, Mauger DT, Dakhama A, Gellatly S, Pavelka N, Berman R, Di YP, Wenzel SE, Chu HW, The effect of BPIFA1/SPLUNC1 genetic variation on its expression and function in asthmatic airway epithelium. JCI Insight 4, e127237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu T, Huang J, Moore PJ, Little MS, Walton WG, Fellner RC, Alexis NE, Di YP, Redinbo MR, Tilley SL, Tarran R, Identification of BPIFA1/SPLUNC1 as an epithelium-derived smooth muscle relaxing factor. Nature Communications 8, 14118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Enomoto Y, Orihara K, Takamasu T, Matsuda A, Gon Y, Saito H, Ra C, Okayama Y, Tissue remodeling induced by hypersecreted epidermal growth factor and amphiregulin in the airway after an acute asthma attack. J Allergy Clin Immunol 124, 913–920.e917 (2009). [DOI] [PubMed] [Google Scholar]
- 90.Zhu Y, Yan X, Zhai C, Yang L, Li M, Association between risk of asthma and gene polymorphisms in CHI3L1 and CHIA: a systematic meta-analysis. BMC Pulmonary Medicine 17, 193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu K, Kamimoto K, Zhang Y, Yang K, Keeler SP, Gerovac BJ, Agapov EV, Austin SP, Yantis J, Gissy KA, Byers DE, Alexander-Brett J, Hoffmann CM, Wallace M, Hughes ME, Crouch EC, Morris SA, Holtzman MJ, Basal-epithelial stem cells cross an alarmin checkpoint for post-viral lung disease. Journal of Clinical Investigation 131, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scrucca L, Fop M, Murphy TB, Raftery AE, mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. The R Journal 8, 289–317 (2016). [PMC free article] [PubMed] [Google Scholar]
- 93.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani A-C, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A, Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Traag VA, Waltman L, van Eck NJ, From Louvain to Leiden: guaranteeing well-connected communities. Scientific Reports 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biology 19, 1–5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Albergante L, Mirkes E, Bac J, Chen H, Martin A, Faure L, Barillot E, Pinello L, Gorban A, Zinovyev A, Robust and Scalable Learning of Complex Intrinsic Dataset Geometry via ElPiGraph. Entropy 22, 296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wood SN, Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. Journal of the Royal Statistical Society B 73, 3–36 (2011). [Google Scholar]
- 98.Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bürkner P-C, brms: An R Package for Bayesian Multilevel Models Using Stan. Journal of Statistical Software 80, 1–28 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Isolation of single-cell suspensions from nasal respiratory and olfactory epithelium.
Figure S3: Three morphologically distinct types of nasal tuft cells share a common molecular profile.
Figure S2: Anatomical and molecular characterization of olfactory epithelial and neuronal subtypes in mice and humans.
Figure S5: Single-cell mapping of immune cell subtypes in the naive nasal mucosa.
Figure S6: Effects of allergen inhalation on gene expression and cell type proportions in the nasal immune system.
Figure S4: Neurogranin acts as a novel tuft cell marker of tuft-MVCs and tuft-gland cells.
Figure S7: Expression of SARS-CoV-2 receptor ACE2 in epithelial cells and marker genes of differentiated mucosal subtypes in naïve mice.
Figure S9: Alternaria inhalation is not associated with significant olfactory damage.
Figure S10: Alternaria inhalation induces HBC proliferation in the absence of additional induction of globose basal cell proliferation.
Figure S11: Pseudo-time analysis identifies novel marker genes for stages of olfactory neurogenesis.
Figure S12. Characterization of HBC and OSN populations in the olfactory mucosa.
Figure S13: Transcriptional programs induced in HBCs by multiple aeroallergens.
Figure S14: Tuft cells are required for allergen-induced HBC proliferation.
Figure S15: Allergen-induced HBC proliferation is independent of type 2 inflammatory pathways.
Figure S16: Characterization of HBC transcriptional responses to allergen and receptor expression by nasal epithelial cells.
Figure S17. Diverse neuroepithelial cell types comprise distinct regions of the murine nose.
Figure S8: Effects of allergic inflammation on gene expression and cell type proportions in the nasal epithelium.
Table S1. Reproducibility checklist
Data File S3. Molecular signatures for all cell types in the nasal epithelium.
Data File S2. Known marker gene sets used for interpretation of cell clusters.
Data File S4. Marker genes for human nasal tuft cells and ionocytes.
Data File S5. Molecular signatures for novel subsets of nasal tuft cells.
Data File S6. Cell type composition dynamics during allergic inflammation.
Data File S7. Molecular signatures for epithelial and immune cell subsets in the nose.
Data File S8. Cell type-specific gene expression dynamics after allergic inflammation.
Data File S10. Gene expression dynamics during olfactory neurogenesis.
Data File S12. Additional statistical analysis of genotype interaction with treatment effect of Alternaria inhalation by two-way ANOVA.
Data File S9. Modulation of cell-cell signaling during allergic inflammation.
Data File S11. Allergen-induced inflammation-dependent and independent transcriptional programs in the olfactory epithelium by bulk RNA seq.
Data File S1. Raw data points underlying all digital Figures and Supplementary Figures.
Data Availability Statement
All raw and processed sequencing data is deposited in the GEO online database (GSE245074). No custom code was used as part of this analysis. Standard code required to reproduce the main steps of the analysis will be made available for research purposes on reasonable request. All other data needed to support the conclusions of the paper are present in the paper or the Supplementary Materials.


