Abstract
The complete genome of the green-sulfur eubacterium Chlorobium tepidum TLS was determined to be a single circular chromosome of 2,154,946 bp. This represents the first genome sequence from the phylum Chlorobia, whose members perform anoxygenic photosynthesis by the reductive tricarboxylic acid cycle. Genome comparisons have identified genes in C. tepidum that are highly conserved among photosynthetic species. Many of these have no assigned function and may play novel roles in photosynthesis or photobiology. Phylogenomic analysis reveals likely duplications of genes involved in biosynthetic pathways for photosynthesis and the metabolism of sulfur and nitrogen as well as strong similarities between metabolic processes in C. tepidum and many Archaeal species.
Chlorobium tepidum, a Gram-negative bacterium of the green-sulfur phylum (Chlorobia), was originally isolated from high-sulfide hot springs in New Zealand (1). Chlorobia are obligate anaerobic photolithoautotrophs and are widely distributed in aquatic environments, where anoxic layers containing reduced sulfur compounds are exposed to light. The genus Chlorobium includes six species of which C. tepidum is the only thermophile, growing optimally at 48°C.
The Chlorobia have unique mechanisms of photosynthesis relative to other phototrophs. They use multiple types of bacteriochlorophylls and carotenoids in unique structures known as chlorosomes for light harvesting. Photosynthetic reaction centers (RCs) are special enzymes that use light energy to drive electron transport reactions leading to the production of protonmotive force for ATP synthesis and/or reducing power (2). Plant chloroplasts and cyanobacteria use both Type I (Fe-S acceptors; Photosystem I) and Type II (quinone acceptors; Photosystem II) RCs to oxidize water, producing oxygen as waste. Many bacteria, including the photosynthetic species in the green nonsulfur bacteria and the α, β, and γ-subdivisions of the Proteobacteria, contain only Type II RCs. The Chlorobia and the Gram-positive heliobacteria uniquely possess only Type I homodimeric RCs (all other RCs are heterodimeric). Unlike plants and cyanobacteria, Chlorobia perform anoxygenic photosynthesis. In addition, instead of using the Calvin cycle, the Chlorobia perform autotrophic CO2 fixation via the reductive tricarboxylic acid (TCA) cycle, using electrons derived from hydrogen or reduced sulfur compounds (3). This cycle, which was discovered in the Chlorobia, is found only in a limited number of taxa. Because of these unique features, studies of the Chlorobia are important for understanding the evolution and mechanisms of photosynthesis and energy metabolism. It has even been proposed (4) that the ancestral photoautotroph was a green-sulfur bacterium. Here we report the determination and analysis of the complete genome of C. tepidum TLS, the type strain of this species.
Materials and Methods
Genome Sequencing.
C. tepidum genomic DNA was isolated as described (5). Cloning, sequencing, assembly, and genome closure were performed as described (6). The complete sequence has been assigned GenBank accession no. AE006470 and is available at http://www.tigr.org/tdb.
Genome Analysis.
An initial set of ORFs likely to encode proteins (CDS) was identified with glimmer (7); those shorter than 30 codons as well as some with overlaps were eliminated. Frame shifts and point mutations were corrected or annotated as “authentic.” Functional assignment, identification of membrane-spanning domains, determination of paralogous gene families, and identification of regions of unusual nucleotide composition were done as described (6). Phylogenomic analysis (8) was used to aid in functional predictions. Alignments and phylogenetic trees were generated as described (9).
Comparative Genomics.
All putative C. tepidum proteins were searched with FASTA3 (10) against the predicted proteomes of published complete organismal genomes, Porphyromonas gingivalis (R. D. Fleischmann, personal communication), and a set of complete plastid, mitochondrial, plasmid, and viral genomes. The results of these searches were used (i) for phylogenetic profile analysis (11), (ii) to identify putative lineage-specific duplications, those proteins with a top E-value score to another protein from C. tepidum, and (iii) to determine the number of best-scoring matches to different species.
Results and Discussion
Genome Features.
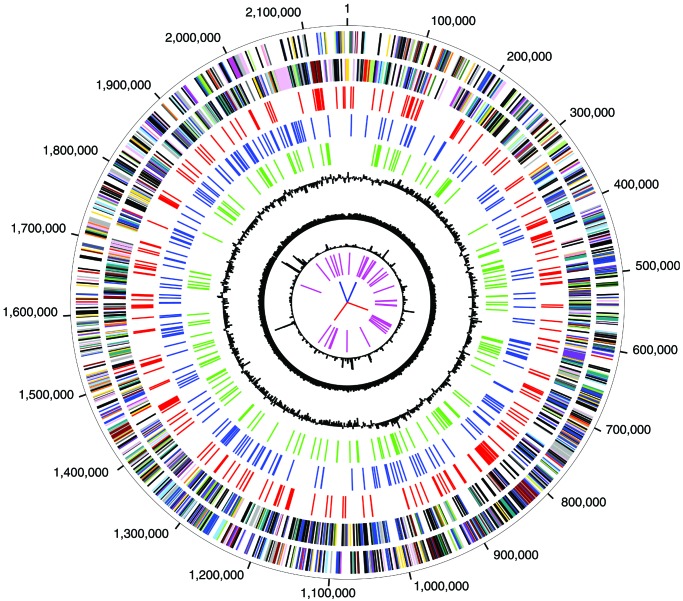
The genome sequence, which corresponds well with the published restriction map (12), is composed of a single, circular molecule of 2,154,946 bp (Figs. 1 and 4, the latter of which is published as supporting information on the PNAS web site, www.pnas.org). A region containing the likely origin of replication was identified based on GC skew (13), and base pair 1 was designated adjacent to the dnaN gene, which is in this region. General features of the genome and the 2,288 CDS are summarized in Tables 1 and 2, the latter of which is published as supporting information on the PNAS web site. Eight genomic regions with strongly atypical trinucleotide composition were identified (Fig. 1). Most of the CDS in these regions are hypothetical proteins. One hundred twenty-three genes possibly duplicated in the C. tepidum lineage were identified (Table 3, which is published as supporting information on the PNAS web site); their amplification suggests they may confer important evolutionary adaptations for this species.
Figure 1.
Circular representation of the C. tepidum genome. Circles 1 and 2: predicted protein-coding regions on the plus and minus strand, color-coded by putative role categories as in ref. 6. Circles 3, (red), 4 (blue), and 5 (green): proteins with top matches to proteins from Archaea, photosynthetic species, and C. tepidum, respectively. Other circles: 6, GC skew; 7, percent G+C; 8, χ2 value for trinucleotide composition in a 2,000-bp window; 9, tRNA genes; and 10, rRNA genes (blue) and sRNA genes (red).
Table 1.
General features of the C. tepidum genome
| Complete genome size, bp | 2,154,946 |
| Total no. of sequences | 36,670 |
| G + C percent | 56.5 |
| Total no. of CDS | 2,288 |
| Average CDS size, bp | 837 |
| Percent coding | 88.9 |
| No. of rRNA operons (16S-23S-5S) | 2 |
| No. of tRNA genes | 50 |
| No. of sRNA genes | 2 |
| Proteins similar to proteins of known function and role category | 1,217 |
| Proteins similar to proteins of known function but unknown role category | 98 |
| Conserved hypothetical proteins | 293 |
| Hypothetical proteins | 680 |
Comparative Genomics.
Analysis of the C. tepidum genome supports the hypothesis that the phyla Chlorobia and Cytophaga–Flavobacteria–Bacteroides (CFB) are related (14, 15). P. gingivalis, a member of the CFB phylum, groups with C. tepidum in most phylogenetic analyses of housekeeping genes (Table 4, which is published as supporting information on the PNAS web site), and its proteome is most similar to that of C. tepidum among species whose genomes are completely sequenced.
A high percentage (∼12%) of the C. tepidum proteins are most similar to proteins from Archaeal species (Fig. 5, which is published as supporting information on the PNAS web site); this value is greater than that for any bacterial species except Thermotoga maritima (16) and Aquifex aeolicus (17). The presence of so many Archaeal-like proteins can be explained by multiple scenarios including the loss or rapid rate of evolution of these genes in other bacteria (18) or past lateral gene transfer between the C. tepidum and Archaeal lineages. Because C. tepidum is apparently not deeply branching within the bacterial tree (15), for the gene loss/rapid evolution explanations to be correct, such events would have to have occurred in all of the earlier branching bacterial lineages. The lateral transfer possibility is consistent with the following observations: (i) some Archaeal species are much better represented in the top-scoring matches (Fig. 5B); (ii) the Archaeal-like proteins are biased toward role categories for which transfer is considered more likely (19) (e.g., central intermediary metabolism but not transcription; Fig. 5C); and (iii) many of the Archaeal-like genes branch in phylogenetic trees with genes from Archaeal species (see below). Distinguishing gene transfer from gene loss will require genomic analyses of other Chlorobia.
A number of proteins in C. tepidum that are most similar to those from photosynthetic species (213 in total; 110 and 63 to the cyanobacteria Synechocystis sp. PCC6803 and Nostoc sp. PCC7120, respectively; 31 to the plant Arabidopsis thaliana and 9 to proteins from chloroplast genomes). Those that have no assigned function (38 in total; see Table 5, which is published as supporting information on the PNAS web site) may have as yet undefined roles in photobiology. Some of the proteins most similar to A. thaliana lack strong homologs in the cyanobacterial genomes (12 for Nostoc and 13 for Synechocystis). These genes likely were encoded in the ancestor of plastids and cyanobacteria, and were transferred to the nucleus in plants but lost from one or both of the Synechocystis/Nostoc lineages. This hypothesis is supported by the finding that some not in Nostoc are in Synechocystis and vice versa.
Transcriptional Regulation and Translation.
The C. tepidum genome encodes few proteins for regulatory responses (Table 2). Only 20 likely transcription regulators could be identified. Few genes likely involved in environmental sensing and responses are present (two CheY-like response regulators, two DNA-binding response regulators, two sensor histidine kinases, and six sensor histidine kinase/response regulator proteins). This number of regulatory genes is ≈8-fold fewer than for the photoautotroph Synechocystis sp. PCC6803 (20). In addition, only five RNA polymerase sigma factors were identified—a vegetative RpoD subunit (CT1551), a member of the RpoN family (CT1193), and three Type III (ECF) factors (CT0502, CT0648, and CT0278). It is not known whether this small number of genes for regulatory responses is typical of the Chlorobia or has evolved by selective gene loss because of the relatively constant environment in which C. tepidum grows.
Nineteen tRNA synthetases were identified, with those for asparagine and glutamine apparently being absent. Although a Type III glutamine synthetase (CT1411) is encoded, glutamine cannot be charged onto tRNAGln because of the missing GlnS enzyme. C. tepidum probably cannot synthesize the free amino acid asparagine, because no asparagine synthetase could be identified. C. tepidum thus possibly misacylates both tRNAGln and tRNAAsn with glutamic acid and aspartic acid, respectively, and then converts the misacylated products to the correctly charged products by transamidation with the GatABC enzyme (CT0268, CT2215, and CT1833) and glutamine. The use of GatABC for making both tRNAGln and tRNAAsn has not been observed previously for any bacterium (21).
DNA Repair and Oxidative Protection.
Because C. tepidum is an anaerobe, is sensitive to even brief oxygen exposure, and is exposed to only low levels of solar radiation, it might be expected to have limited capabilities for DNA repair and oxidative stress protection. However, a full suite of DNA repair genes is present including some components not found in most bacteria (22): two UvrAs (CT0527 and CT1689), an Archaeal-like family B DNA polymerase (CT2284), and a class II photolyase/blue-light receptor homolog (CT0511). The genome also encodes many proteins likely involved in oxidative damage protection (Table 2). These include superoxide dismutase (CT1211), rubredoxin oxygen oxidoreductase (CT2285) (23), and cytochrome bd quinol oxidase (CT1818 and CT1819). The presence of the latter two enzymes suggests that C. tepidum encounters oxygen; these enzymes may function in protecting oxygen-sensitive enzymes such as the photosynthetic apparatus and nitrogenase from damage (24, 25).
Light Harvesting and Electron Transfer.
All Chlorobia possess unique light-harvesting antennae known as chlorosomes (26), which contain bacteriochlorophyll c (BChl c) aggregates (27, 28) and are surrounded by a protein-stabilized, galactolipid monolayer. Genes had been identified for most of the 10 known chlorosome proteins (29) and for the BChl a binding protein FMO (CT1499), which likely attaches the chlorosome to the underlying RCs (Fig. 2) (30). Genome analysis allows the identification of CsmX (CT0652), a chlorosome envelope protein similar to CsmJ (29).
Figure 2.
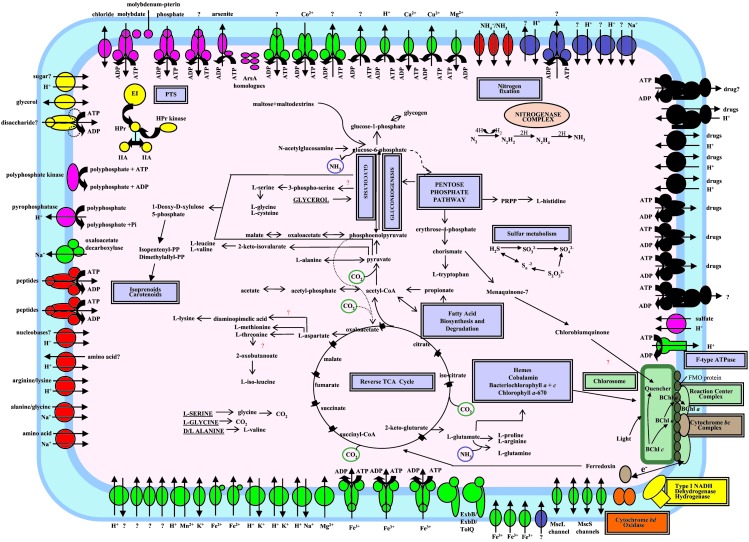
Overview of known and predicted metabolism (energy production and organic compounds) and transport in C. tepidum. Transporters are grouped by substrate specificity: inorganic cations (green), inorganic anions (pink), carbohydrates (yellow), amino acids/peptides/amines/purines and pyrimidines (red), drugs (black), other (blue), and uncertain specificity (??). Arrows indicate direction of transport. Energy coupling mechanisms are also shown: solutes transported by channel proteins (double-headed arrow); secondary transporters (two-arrowed lines indicating both the solute and the coupling ion); ATP-driven transporters (ATP hydrolysis reaction); and unknown energy coupling mechanism (single arrow).
Genome analysis reveals only single gene copies of the known RC subunits (PscABCD), which is consistent with experimental studies that suggest that the RC core is a homodimer of PscA subunits in Chlorobia (31). The photosynthetic electron transport chain of the Chlorobia has been assumed to resemble that surrounding PS I of plants, with electrons being transferred from the RC to highly reducing ferredoxins and ultimately by means of ferredoxin:NADP+ oxidoreductase (FNR) to NADP+. Three highly similar 8Fe-8S bacterial ferredoxins (CT1260, CT1261, and CT1736), all of which can act as soluble electron acceptors from C. tepidum RCs (32), were identified. Additionally, more than a dozen other Fe-S or flavin-containing electron transport proteins are present (Table 2). Eleven c-type cytochromes were present, but no homolog of cytochrome c1 or f was found. Although early experimental studies suggested the presence of an FNR-like activity in Chlorobium thiosulfatophilum (33), more recent attempts to isolate FNR from Chlorobium sp. Had failed. However, Seo and Sakurai (61) have recently shown that CT1512, which has sequence similarity to thioredoxin reductases, has FNR activity. The metabolic role of this enzyme could be to provide reduced pyridine nucleotides for gluconogenesis, although reduced ferrodoxins that are required to drive the reductive reactions of the reverse TCA cycle are expected to be the primary products of photosynthetic electron transport in C. tepidum.
The C. tepidum genome contains an apparent operon, ndhCHJKAIGEFDB (CT0766–CT0776), encoding 11 subunits of a type I NADH dehydrogenase (NDH-1). Similar to Synechocystis sp. PCC6803 (20), no homologs of the Escherichia coli proteins NuoEFG were found. However, genes encoding an Ni-Fe uptake hydrogenase (CT0777) and an associated b-cytochrome (CT0778) may also be part of this operon, suggesting a structural and functional relationship between NDH-1 and the uptake hydrogenase. Similar suggestions were made concerning the bidirectional Ni-Fe hydrogenase and the NDH-1 complex of cyanobacteria (34). The presence of NDH-1 suggests that reduced pyridine nucleotides for gluconeogenesis may also be generated by reverse electron transport as in purple bacteria (35).
CO2 Fixation.
For the autotrophic fixation of CO2, C. tepidum utilizes the reverse (reductive) TCA cycle (36) (Fig. 2). In this process, all but three steps are catalyzed in reverse by the same enzymes that carry out the forward (oxidative) TCA cycle. Two of the reverse cycle-specific steps are catalyzed by the ferredoxin-dependent enzymes pyruvate synthase and 2-oxoglutarate synthase, which are encoded by an NifJ homolog (CT1628) and a complex of CT0163/CT0162, respectively (ref. 37; F. R. Tabita, personal communication). Citrate lyase (CL), which catalyzes the conversion of citrate to acetyl CoA and oxaloacetate, is the third enzyme specific to this cycle. The CL of C. tepidum is unique among prokaryotes in that it is an ATP-dependent enzyme, much like CLs of eukaryotes (36, 38). Eukaryotic CLs are composed of three domains forming either one (animals) or two (plants/fungi) proteins. Each domain corresponds to distinct single-domain proteins that function in the oxidative TCA cycle in many species. As with other bacteria, C. tepidum encodes each single-domain protein (CT0380, CT0269, CT1834) but unlike any other bacterium for which genome data is available additionally possesses a plant/fungal-type CL composed of two polypeptides (CT1088 and CT1089) (see Fig. 6, which is published as supporting information on the PNAS web site). Phylogenetic analysis reveals that each domain of CT1088/CT1089 is most closely related to the corresponding domain of plant CLs.
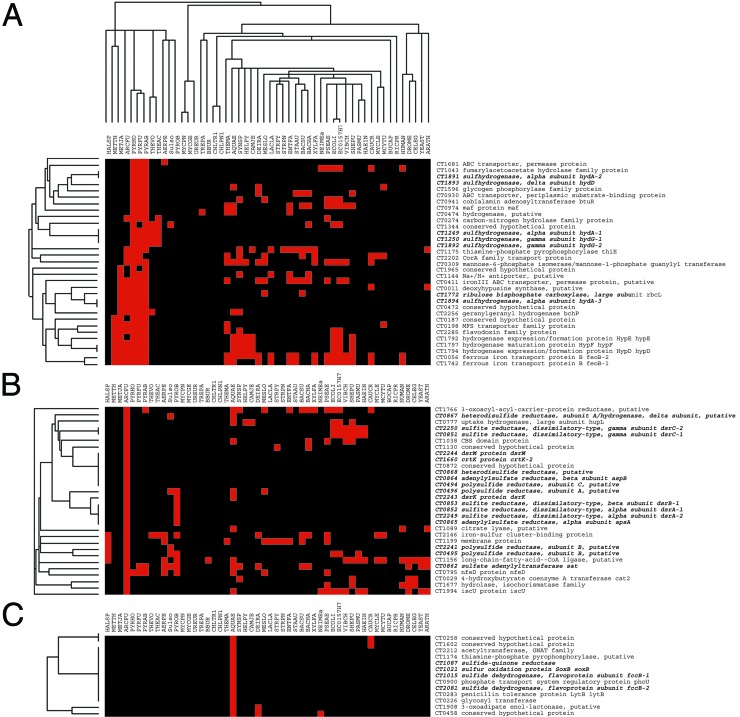
CO2 assimilation in higher plants, algae, and most characterized autotrophic prokaryotes relies on the ribulose-1,5-bis-phosphate carboxylase (RubisCO). Low levels of RubisCO activity were initially reported for C. thiosulfatophilum (39, 40). The subsequent discovery of the reverse TCA cycle led to the conclusion that RubisCO activity is not present in Chlorobium sp. However, C. tepidum encodes an ortholog of the large-subunit of RubisCO (RbcL CT1772), although no homolog of the small subunit is detectable, a pattern also observed in some other bacteria and Archaea (41). The RbcLs from these species form a distinctive class (type IV) within the RbcL family (41). The purified C. tepidum RbcL has no detectable CO2 fixation activity, and strains with the rbcL gene deleted showed phenotypes consistent with possible defects in sulfur metabolism (41). Consistent with the gene knockout results, phylogenetic profile analysis reveals that the C. tepidum RbcL has a species distribution pattern similar to many sulfur metabolism genes (Fig. 3).
Figure 3.
Phylogenetic (PG) profile analysis of C. tepidum. C. tepidum proteins were clustered according to their distribution patterns across species for which complete genomes are available, using the single-linkage clustering algorithm with column weighting of CLUSTER (http://rana.lbl.gov). A protein was considered present (and is shown in red) if there is a match with an E-value ≤ 10−15. Clusters were viewed with TREEVIEW (http://rana.lbl.gov). (A) PG profile of C. tepidum's large subunit RubisCO homolog showing its grouping with some genes involved in S metabolism. (B and C) PG profile of many of C. tepidum's other S metabolism genes.
Organic Molecule Metabolism.
Genome analysis suggests that C. tepidum has very limited capabilities for the assimilation of organic compounds (Fig. 2), which is consistent with experiments that found that only acetate and pyruvate stimulate phototrophic growth (1). The inability to use glucose correlates with the apparent absence of a glucokinase and glucose transporter. A putative operon consisting of genes for a maltodextrin glycosyltransferase (CT0977), a putative carbohydrate kinase (CT0976), and a putative sugar transporter (CT0975) was identified. Although the substrate specificities of these proteins are unclear, C. tepidum might transport and utilize malto-oligosaccharides, the products of which could then be fed into glycolysis and the pentose phosphate pathway as glucose-6-phosphate. Maltose for this pathway could potentially be generated by an α-amylase-like protein (CT0755).
The genome encodes some but not all components of a phosphotransferase system (PTS) [Enzyme I (CT0204), HPr (CT2211), 2 Enzyme IIAs (CT0235, CT0629), and a HPr serine kinase (CT1633) are present but a membrane-bound Enzyme II PTS transporter is not]. The C. tepidum EIIAs might function with a novel PTS Enzyme II. Alternatively, these PTS components may serve regulatory rather than transport functions. Consistent with this hypothesis, one of the EIIAs is similar to EIIANtr, which may function in coordinating nitrogen and carbon metabolism in E. coli (42).
N and S Metabolism.
The primary sources of nitrogen seem to be NH4 and N2 (which is reduced to yield NH4). C. tepidum possesses three probable ammonia/ammonium AmtB transporters (CT0498, CT0986, and CT0133). The nitrogen fixation genes are present in a single region of the genome. The genes in this cluster have nearly the same arrangement as the Nif cluster of the Archaeon Methanobacterium thermoautotrophicum (see Fig. 7, which is published as supporting information on the PNAS web site) and most are most closely related to genes from Archaea (not shown), suggesting that the entire cluster may have been laterally transferred between these two lineages.
C. tepidum can use both sulfide and thiosulfate as sources of sulfur and as electron donors for photosynthesis (1) (see Fig. 8A, which is published as supporting information on the PNAS web site). The genes involved in sulfur oxidation are mainly localized in four clusters in the genome. One cluster includes most of the sox genes, which are common in other bacteria and can be used to oxidize thiosulfate. However, the cluster is missing genes encoding SoxC and SoxD, which have been shown to function as a sulfur dehydrogenase in other species (43). Three genes encoding flavoprotein reductases related to sulfide-quinone oxidoreductase are present; these enzymes may function in oxidizing sulfide to form polysulfides (Fig. 8A). The further oxidation of sulfide to sulfate probably takes place in the cytoplasm by using dissimilatory sulfite reductase (DSR), APS reductase, and ATP sulfurylase (Fig. 8A). A sulfate exporter is not yet identified in the genome, although a likely sulfate/thiosulfate importer is present. Many sulfur metabolism genes have been duplicated in the C. tepidum lineage (Table 3), including two nearly identical copies of a DSR-encoding operon (Fig. 8B). Finally, two operons encoding components of a hydrogenase/sulfur reductase similar to those of Archaea (44) are present.
Reduced inorganic sulfur compounds can be used as electron donors for phototrophic or lithotrophic growth by Archaea and bacteria. Sulfate is the major oxidation product whether the dissimilatory sulfur oxidation occurs under oxic or anoxic conditions. In Archaea, sulfide and thiosulfate are aerobically oxidized by members of the order Sulfolobales. Phylogenetic profile analysis indicates that the C. tepidum sulfur oxidation genes are more extensively shared with the anaerobic, sulfate-reducing Archaeon Archaeoglobus fulgidus than the aerobic Sulfolabales (Fig. 3). The unusual crenarchaeon Pyrobaculum aerophilum contains homologs of the genes encoding ATP sulfurylase, two subunits of the APS reductase (AprA and AprB), and a sulfite:acceptor oxidoreductase (45). It is not clear whether these genes function in assimilatory or dissimilatory sulfur metabolism. Both the aprA and aprB genes are apparently defective, possibly explaining the sensitivity of P. aerophilum to reduced sulfur compounds (45). However, the presence of genes encoding PAPS reductase and two thiosulfate:sulfur transferases suggests that wild-type P. aerophilum might be able to oxidize thiosulfate.
Other Transport Functions.
C. tepidum has an extensive repertoire of transporters for inorganic compounds (Fig. 2) with almost 50% of transporters being predicted to function in metal ion homeostasis, a similar percentage to that for Synechocystis sp. PCC6803 (46). C. tepidum encodes six homologs of ArsA, a protein that couples arsenite efflux to ATP hydrolysis (47). Chloroflexus aurantiacus, a green nonsulfur bacterium that also contains chlorosomes, also encodes multiple ArsA homologs, many of which seem to be orthologs of the C. tepidum proteins (see Fig. 9, which is published as supporting information on the PNAS web site). In both species, some of these genes are located in close proximity to the csmA gene, encoding the major protein of the chlorosome envelope. We propose that these ArsA homologs may play a role in energizing transport across or into the chlorosome envelope.
Tetrapyrroles: Hemes, Chlorophylls, and Cobalamins.
C. tepidum synthesizes at least eight tetrapyrroles from a uroporphyrinogen III backbone (48) including three chlorophylls (BChl a, BChl c, and Chl aPD), four hemes, and cobalamin. Based on genetic studies in Rhodobacter species (49) and the genome analysis, a complete pathway for chlorophyll synthesis in C. tepidum can be proposed (see Fig. 10, which is published as supporting information on the PNAS web site). Orthologs of each Rhodobacter sp. enzyme for the conversion of protoporphyrin IX into BChl a were identified, suggesting that the synthesis of BChl a proceeds as in Rhodobacter sp. The assigned functions of CT1610/BchG-1, CT1958/BchM, and CT1421/BchF-1 have been confirmed by complementation of Rhodobacter capsulatus mutants (50). The Chl aPD synthase is likely CT1270, which is a close homolog of the chlorophyll synthase of plants and cyanobacteria.
Before genome sequencing, little was known about BChl c synthesis. Only three steps are likely catalyzed by the same enzymes as for BChl a synthesis (see Fig. 10). The presence of three close homologs of the BchH subunit of the Mg chelatase (CT1295, CT1955, and CT1957) may allow for the formation of Mg chelatase isoenzymes sensitive to the three Chl end products. The remaining enzymes for BChl c biosynthesis were identified as paralogs of other genes for tetrapyrrole biosynthesis. The C-20 methylation is possibly catalyzed by CT1763, a paralog of the precorrin 2 methyl-transferase CbiL (CT0388). Of two paralogs of BchF, CT1776 likely modifies BChl c and CT1421 BChl a, because the latter is encoded in an apparent operon with genes specific for BChl a synthesis (bchX and bchC). Analysis of several BchE paralogs (Table 3, see below) suggests that CT1697 is the most likely enzyme for formation of the isocyclic ring of BChl c. The BChl c synthase is CT1992; it groups in phylogenetic trees with the putative BChl c synthase of C. aurantiacus, and a null mutant cannot synthesize Bchl c (51).
When cells are grown at low light intensities, increased alkylation of the C8 and C12 substituents of BChl c leads to a red-shift in the antenna pigment absorption and presumably better light energy utilization. The carbon for the alkylation comes from S-adenosyl methionine (SAM) (52). This alkylation activity is probably catalyzed by proteins in the BchE/P-methylase family, because proteins in this family contain SAM-binding motifs, require SAM for activity (53), and have undergone extensive duplication in C. tepidum (Table 3). C. tepidum encodes nine members of this family including two paralogs each of BchE (CT1959 and CT1697) and HemN (CT2010 and CT0372). The other five (CT1903, CT1320, CT1502, CT1777, and CT0072) are good candidates for the multiple alkylation activities.
Although vitamin B12 is routinely added to the growth medium for C. tepidum, the genome predicts a complete set of enzymes for the de novo synthesis of 5′deoxyadenosyl cobalamin. Two types of cobalt chelatases are found: CbiK (CT0389) and a possible ATP-dependent chelatase CobN-CobT-CobS-CobN (CT0418, CT0420, CT0421, and CT0422). The ATP-dependent chelatase genes are clustered with a putative TonB-dependent outer membrane receptor for vitamin B12, suggesting that this enzyme may insert cobalt into vitamin B12 scavenged from the environment, whereas CbiK is used during de novo biosynthesis. Four C. tepidum proteins contain two Cbi domains: CbiGF, CbiHC, CbiJD, and CbiET (CT0385, CT0387, CT0384, and CT0386). In most other respects, the biosynthesis is similar to the oxygen-independent pathway of Salmonella typhimurium (54). The dependence of Chl synthesis on vitamin B12 (53) suggests that such reactions existed before the evolution of photosynthesis.
Isoprenoids: Carotenoids, Quinones, and Undecaprenol.
C. tepidum synthesizes the precursors of all isoprenoid compounds from pyruvate and glyceraldehyde 3-phosphate (Fig. 2) by using the deoxyxylulose phosphate pathway (55). All presently known enzymes of this pathway were identified (see Fig. 11, which is published as supporting information on the PNAS web site), as were the gcpE and lytB genes that have recently been implicated in the synthesis of isoprenoid precursors (56). Many of the enzymes for the synthesis of C. tepidum's five major carotenoids (57) could tentatively be identified (Fig. 11), but no lycopene cyclase was identified. Possible candidates include the multiple homologs of CrtD/CrtI (CT0649, CT0807, CT1414, and CT0180). C. tepidum synthesizes two major quinones, menaquinone-7 and chlorobiumquinone (58). Homologs of all enzymes for the conversion of chorismate into menaquinone in E. coli (59) were identified. The 1′-keto group on the isoprenoid chain of chlorobiumquinone may be introduced by one of the BchE paralogs (possibly CT1903), because this chemical moiety is similar to the 131-keto group found in the isocyclic ring of chlorophyll.
Conclusions
The complete genome of C. tepidum reveals an unusual combination of genes for energy metabolism, including a CL that resembles those of plants, many genes most similar to those from Archaea (RubisCO large-subunit, sulfhydrogenase, ATP synthase, and nitrogenase homologs), and multiple duplications of genes for photosynthesis and sulfur metabolism. Further genome analysis and experimental work should help provide insights into the evolution of photosynthesis and other pathways of energy metabolism. Importantly, hypotheses generated from in silico analyses such as those described here can now be readily tested experimentally, because conditions for efficient natural transformation of C. tepidum have recently been developed (60).
Supplementary Material
Acknowledgments
We acknowledge J. Buchoff, M. Heaney, M. Holmes, D. Kosack, B. Lee, V. Sapiro, and J. Shao for IT support; T. Dickinson, S. Angiuoli, M. Ermoleva, S. Salzberg, M. Eisen, M-I. Benito, R. Roberts, J. Posfai, C. Dunn, C. Friedrich, C. Dahl, and anonymous reviewers for comments and assistance with genome analysis; and the TIGR seqcore, informatics, and IT groups for support. This project was supported by the U.S. Department of Energy, Office of Biological Energy Research, Co-Operative Agreement DE-FC02-95ER61962 and DE-FG02-94ER20137 (to D.A.B.).
Abbreviations
- RC
reaction center
- TCA
tricarboxylic acid
- BChl
bacteriochlorophyll
- CL
citrate lyase
- FNR
ferredoxin:NADP+ oxidoreductase
- RubisCO
ribulose-1,5-bisphosphate carboxylase
- CDS
coding sequence
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE006470).
References
- 1.Wahlund T M, Woese C R, Castenholz R W, Madigan M T. Arch Microbiol. 1991;156:81–90. [Google Scholar]
- 2.Golbeck J H. Proc Natl Acad Sci USA. 1993;90:1642–1646. doi: 10.1073/pnas.90.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans M C, Buchanan B B, Arnon D I. Proc Natl Acad Sci USA. 1966;55:928–934. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavalier-Smith T. J Mol Evol. 2001;53:555–595. doi: 10.1007/s002390010245. [DOI] [PubMed] [Google Scholar]
- 5.Gruber T M, Bryant D A. Arch Microbiol. 1998;170:285–296. doi: 10.1007/s002030050644. [DOI] [PubMed] [Google Scholar]
- 6.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, et al. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 7.Salzberg S L, Delcher A L, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisen J A. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- 9.Salzberg S L, White O, Peterson J, Eisen J A. Science. 2001;292:1903–1906. doi: 10.1126/science.1061036. [DOI] [PubMed] [Google Scholar]
- 10.Pearson W R. Methods Mol Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrini M, Marcotte E M, Thompson M J, Eisenberg D, Yeates T O. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naterstad K, Kølsto A B, Sirevåg R. J Bacteriol. 1995;177:5480–5484. doi: 10.1128/jb.177.19.5480-5484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 14.Gruber T M, Eisen J A, Gish K, Bryant D A. FEMS Microbiol Lett. 1998;162:53–60. doi: 10.1111/j.1574-6968.1998.tb12978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woese C. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, et al. Nature (London) 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L, Tatusov R L, Wolf Y I, Walker D R, Koonin E V. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 18.Logsdon J M, Faguy D M. Curr Biol. 1999;9:R747–R751. doi: 10.1016/s0960-9822(99)80474-6. [DOI] [PubMed] [Google Scholar]
- 19.Jain R, Rivera M C, Lake J A. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 21.Ibba M, Soll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 22.Eisen J A, Hanawalt P C. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frazao C, Silva G, Gomes C M, Matias P, Coelho R, Sieker L, Macedo S, Liu M Y, Oliveira S, Teixeira M, et al. Nat Struct Biol. 2000;7:1041–1045. doi: 10.1038/80961. [DOI] [PubMed] [Google Scholar]
- 24.Poole R K, Hill S. Biosci Rep. 1997;17:303–317. doi: 10.1023/a:1027336712748. [DOI] [PubMed] [Google Scholar]
- 25.Lemos R S, Gomes C M, Santana M, LeGall J, Xavier A V, Teixeira M. FEBS Lett. 2001;496:40–43. doi: 10.1016/s0014-5793(01)02399-7. [DOI] [PubMed] [Google Scholar]
- 26.Vassilieva E V, Frigaard N-U, Bryant D A. Spectrum. 2000;13:7–13. [Google Scholar]
- 27.Steensgaard D B, van Walree C A, Permentier H, Baneras L, Borrego C M, Garcia-Gil J, Aartsma T J, Amesz J, Holzwarth A R. Biochim Biophys Acta. 2000;1457:71–80. doi: 10.1016/s0005-2728(99)00112-7. [DOI] [PubMed] [Google Scholar]
- 28.van Rossum B J, Steensgaard D B, Mulder F M, Boender G J, Schaffner K, Holzwarth A R, deGroot H J. Biochemistry. 2001;40:1587–1595. doi: 10.1021/bi0017529. [DOI] [PubMed] [Google Scholar]
- 29.Vassilieva E V, Antonkine M L, Zybailov B L, Yang F, Jakobs C U, Golbeck J H, Bryant D A. Biochemistry. 2001;40:464–473. doi: 10.1021/bi001917d. [DOI] [PubMed] [Google Scholar]
- 30.Blankenship R E, Miller M, Olson J M. In: Anoxygenic Photosynthetic Bacteria. Blankenship R E, Miller M, Bauer C E, editors. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 399–435. [Google Scholar]
- 31.Buttner M, Xie D L, Nelson H, Pinther W, Hauska G, Nelson N. Biochim Biophys Acta. 1992;1101:154–156. [PubMed] [Google Scholar]
- 32.Seo D, Tomioka A, Kusumoto N, Kamo M, Enami I, Sakurai H. Biochim Biophys Acta. 2001;1503:377–384. doi: 10.1016/s0005-2728(00)00245-0. [DOI] [PubMed] [Google Scholar]
- 33.Buchanan B B, Evans M C. Biochim Biophys Acta. 1969;180:123–129. doi: 10.1016/0005-2728(69)90199-6. [DOI] [PubMed] [Google Scholar]
- 34.Appel J, Schulz R. Biochim Biophys Acta. 1996;1298:141–147. doi: 10.1016/s0167-4838(96)00176-8. [DOI] [PubMed] [Google Scholar]
- 35.Zannoni D. In: Anoxygenic Photosynthetic Bacteria. Blankenship R E, Miller M, Bauer C E, editors. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 949–971. [Google Scholar]
- 36.Wahlund T M, Tabita F R. J Bacteriol. 1997;179:4859–4867. doi: 10.1128/jb.179.15.4859-4867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon K S, Hille R, Hemann C, Tabita F R. J Biol Chem. 1999;274:29772–29778. doi: 10.1074/jbc.274.42.29772. [DOI] [PubMed] [Google Scholar]
- 38.Kanao T, Fukui T, Atomi H, Imanaka T. Eur J Biochem. 2001;268:1670–1678. [PubMed] [Google Scholar]
- 39.Buchanan B B, Sirevåg R. Arch Microbiol. 1976;109:15–19. doi: 10.1007/BF00425107. [DOI] [PubMed] [Google Scholar]
- 40.Tabita F R, McFadden B A, Pfennig N. Biochim Biophys Acta. 1974;341:187–194. doi: 10.1016/0005-2744(74)90079-5. [DOI] [PubMed] [Google Scholar]
- 41.Hanson T E, Tabita F R. Proc Natl Acad Sci USA. 2001;98:4397–4402. doi: 10.1073/pnas.081610398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabus R, Reizer J, Paulsen I, Saier M H., Jr J Biol Chem. 1999;274:26185–26191. doi: 10.1074/jbc.274.37.26185. [DOI] [PubMed] [Google Scholar]
- 43.Friedrich C G, Rother D, Bardischewsky F, Quentmeier A, Fischer J. Appl Environ Microbiol. 2001;67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams M W, Holden J F, Menon A L, Schut G J, Grunden A M, Hou C, Hutchins A M, Jenney F E, Jr, Kim C, Ma K, et al. J Bacteriol. 2001;183:716–724. doi: 10.1128/JB.183.2.716-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitz-Gibbon S T, Ladner H, Kim U J, Stetter K O, Simon M I, Miller J H. Proc Natl Acad Sci USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulsen I T, Nguyen L, Sliwinski M K, Rabus R, Saier M H., Jr J Mol Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 47.Zhou T, Radaev S, Rosen B P, Gatti D L. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avissar Y J, Ormerod J G, Beale S I. Arch Microbiol. 1989;151:513–519. doi: 10.1007/BF00454867. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki J Y, Bollivar D W, Bauer C E. Annu Rev Genet. 1997;31:61–89. doi: 10.1146/annurev.genet.31.1.61. [DOI] [PubMed] [Google Scholar]
- 50.Xiong J, Fischer W M, Inoue K, Nakahara M, Bauer C E. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 51. Frigaard, N.-U., Voigt, G. D. & Bryant, D. A. (2002) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 52.Senge M O, Smith K M. In: Anoxygenic Photosynthetic Bacteria. Blankenship R E, Miller M, Bauer C E, editors. Dordrecht, The Netherlands: Kluwer Academic; 1995. pp. 137–151. [Google Scholar]
- 53.Gough S P, Petersen B O, Duus J O. Proc Natl Acad Sci USA. 2000;97:6908–6913. doi: 10.1073/pnas.97.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth J R, Lawrence J G, Bobik T A. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 55.Eisenreich W, Rohdich F, Bacher A. Trends Plant Sci. 2001;6:78–84. doi: 10.1016/s1360-1385(00)01812-4. [DOI] [PubMed] [Google Scholar]
- 56.Hintz M, Reichenberg A, Altincicek B, Bahr U, Gschwind R M, Kollas A K, Beck E, Wiesner J, Eberl M, Jomaa H. FEBS Lett. 2001;509:317–322. doi: 10.1016/s0014-5793(01)03191-x. [DOI] [PubMed] [Google Scholar]
- 57.Takaichi S, Wang Z Y, Umetsu M, Nozawa T, Shimada K, Madigan M T. Arch Microbiol. 1997;168:270–276. doi: 10.1007/s002030050498. [DOI] [PubMed] [Google Scholar]
- 58.Frigaard N-U, Matsuura K, Hirota M, Miller M, Cox R P. Photosynth Res. 1998;58:81–90. [Google Scholar]
- 59.Meganathan R. Vitam Horm. 2001;61:173–218. doi: 10.1016/s0083-6729(01)61006-9. [DOI] [PubMed] [Google Scholar]
- 60.Frigaard N U, Bryant D A. Appl Environ Microbiol. 2001;67:2538–2544. doi: 10.1128/AEM.67.6.2538-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seo D, Sakura H. Biochim Biophys Acta. 2002;1597:123–132. doi: 10.1016/s0167-4838(02)00269-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.