Abstract
The phosphoinositide-3-OH-kinase-related kinases (PIKKs) are atypical protein kinases exclusive to eukaryotes. They mediate the cellular response to a range of stresses, including genome and RNA surveillance and availability of nutrients for growth. Orthologues of five out of the six PIKK family members are present in plant genomes. Recent studies in plant PIKKs have revealed features unique to, and in common with, other PIKKs. This review summarizes the basic knowledge of these proteins in mammals and yeast in comparison with what is known for Arabidopsis and other plants.
Keywords: phosphoinositide-3-OH-kinase-related kinases, TOR (target of rapamycin), ATM (ataxia-telangiectasia mutated), ATR (ATM and RAD3-related), TRRAP (transactivation/transformation-domain-associated protein)
The PIKK family
In 1985, it was observed that the addition of double-stranded DNA (dsDNA) to human cell extracts stimulated the phosphorylation of several proteins and, four years later, the protein kinase responsible was identified (Lees-Miller & Meek, 2003). DNA-dependent protein kinase catalytic subunit (DNA-PKcs), as it came to be known, was found to be a crucial mediator of DNA damage and repair. Ten years later, the gene responsible for the rare human disease ataxia-telangiectasia was identified and designated ataxia-telangiectasia mutated (ATM). The catalytic domain of the ATM sequence is highly similar to that of DNA-PKcs, and together, these proteins are most closely related to the phosphoinositide-3-OH-kinase (PI3K) catalytic domain, despite the fact that neither of them are known to phosphorylate lipids (for a review, see Kurz & Lees-Miller, 2004).
During the past few years, additional PI3K-related kinases (PIKKs) have been identified, and the family now has six known members (Abraham, 2004a). In addition to DNA-PKcs and ATM, the family includes ATM and RAD3-related (ATR), mammalian target of rapamycin (mTOR), suppressor of morphogenesis in genitalia (SMG1) and transactivation/transformation-domain-associated protein (TRRAP). These enzymes are referred to as atypical protein kinases (Manning et al, 2002). All family members except TRRAP have kinase activity towards Ser or Thr residues in proteins, and all but mTOR show specificity for Ser/Thr-Gln motifs. None of them show any lipid kinase activity. The kinase activity of these proteins in vitro is fully activated when using Mn2+ as a co-factor, as compared with classic protein kinases that usually use Mg2+ (Kim et al, 1999). It is not yet known whether this is biochemically significant, as DNA-PKcs can be activated without Mn2+ as long as its accessory proteins (Ku70/80) and dsDNA are bound. TRRAP retains most of the catalytic domain, but lacks the residues that are essential for binding ATP and does not show protein kinase activity (McMahon et al, 1998).
The general domain structure of the PIKKs is well conserved between organisms (Fig 1). PIKKs are defined by their carboxy-terminal PI3K catalytic domain and are flanked on the amino- and C-terminal sides by the FAT (FRAP, ATM and TRRAP) and FATC (FRAP, ATM and TRRAP C-terminal) domains, respectively. Together, these three domains account for approximately 5%–10% of the total size of the protein. The remaining sequence is almost exclusively made up of HEAT (Huntingtin, elongation factor 1A, protein phosphatase 2A Asubunit, TOR) repeats (Perry & Kleckner, 2003), except in DNA-PKcs in which the related ARM (Armadillo) repeats are found.
Figure 1.
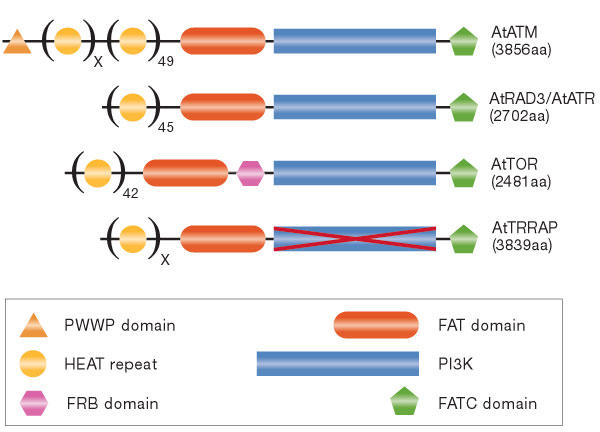
Domain structure of the phosphoinositide-3-OH-kinase-related kinase family members identified in Arabidopsis thaliana. Phosphoinositide-3-OH-kinase-related kinase (PIKK) family members are AtATM (genInfo identifier (gi) 11357182), AtRAD3/AtATR (gi 18422029), AtTOR (gi 22330143) and AtTRRAP (gi 22329206). HEAT (Huntingtin, elongation factor 1A, protein phosphatase 2A A-subunit, TOR) repeats, FAT (FRAP, ATM and TRRAP), PI3K (phosphoinositide 3-OH-kinase) and FATC (FRAP, ATM and TRRAP C-terminal) domains are characteristic of the proteins in this family. Other depicted domains are PWWP, named for its Pro-Trp-Trp-Pro sequence, and the FRB (FKBP12-rapamycin binding) domain. The crossed-out PI3K domain in AtTRRAP indicates its lack of catalytic activity as predicted by the absence of key catalytic residues. The figure is not to scale.
Overall observations of plant PIKKs
Three out of the six known PIKKs have been characterized to some degree in Arabidopsis: AtATM, AtATR and AtTOR. A fourth member, TRRAP, was noted in a previous study through analysis of model organism genomes (McMahon et al, 1998). In an attempt to find unique PIKK family members, we used BLAST software to search the Arabidopsis thaliana genome with the FAT, PI3K and FATC domains from human (Hs) ATM, with no success. Previous work did not find an orthologue to SMG1 in the Arabidopsis genome (Maquat, 2004), but we identified a likely candidate in rice (see below). No DNA-PKcs gene has been found in Arabidopsis (Tamura et al, 2002) or in other plant genomes and thus will not be discussed further here. It is interesting to note that there seems to be no duplication of the PIKK genes in plants in which numerous genome duplications have occurred. Sequencing of the A. thaliana genome has revealed at least three rounds of genome duplication, which is thought to provide a major resource for genome and gene evolution (Moore & Purugganan, 2005). Many plant genes exist as two or more copies; however, some genes were selectively lost during genome evolution after a duplication event. This seems to be true for the plant PIKKs and probably supports the idea that the PIKKs are ancient molecules with functions that were defined in an early eukaryotic ancestor and have not been expanded on with the genetic material provided by genome duplication.
Examination of the four PIKK family members found in A. thaliana and SMG1 in rice allows some comparison of overall features (Figs 1,2; supplementary Figs 1,2,3,4 online). First, analysis shows that in plants, as in mammals, TRRAP lacks several key catalytic residues, such as the DXXXXN motif present in all protein kinases. The other three PIKKs in Arabidopsis and the rice SMG1 homologue have these key residues, and as such are predicted to show protein kinase activity (Fig 2; supplementary Fig 5 online). Wortmannin—a potent inhibitor of PI3Ks and a relatively potent inhibitor of the PIKK family—functions by covalently modifying a lysine residue in the ATP-binding pocket of the kinase domain (Wymann et al, 1996). This lysine is conserved in the four plant PIKK family members that are predicted to have protein kinase activity, but not in TRRAP. This indicates that wortmannin may be effective as a PIKK inhibitor in plants and thus could be a powerful biochemical tool in the study of plant PIKK-regulated events.
Figure 2.
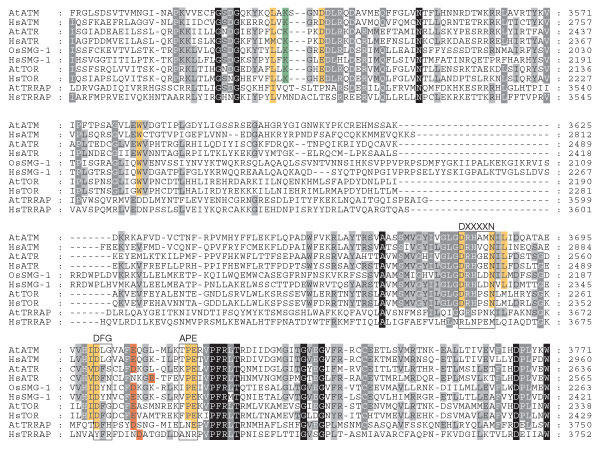
Alignment of the catalytic serine/threonine kinase domain of phosphoinositide-3-OH-kinase-related kinases common to both Arabidopsis and humans. Phosphoinositide-3-OH-kinase-related kinase (PIKK) family members include: ataxia-telangiectasia mutated (ATM; human gi 13878337, Arabidopsis gi 11357182); ATM and RAD3-related (ATR; human gi 4502325, Arabidopsis gi 18422029); target of rapamycin (TOR; human gi 4826730, Arabidopsis gi 22330143); and transactivation/transformation-domain-associated protein (TRRAP; human gi 4507691, Arabidopsis gi 22329206). Arabidopsis does not contain an obvious suppression of morphogenesis in genitalia 1 (SMG1) orthologue, but Oryza sativa does (gi 50918505), and is shown along with its human counterpart (gi 18765739). Residues in yellow are important for ATP binding, and are necessary for kinase activity. Of particular interest are the highlighted DXXXXN, 'DFG' and 'APE' motifs conserved in all active protein kinases. Acidic residues (Asp or Glu) in the putative activation loop are marked in red. The lysine in green is also involved in ATP binding and is covalently modified by the phosphoinositide-3-OH-kinase (PI3K) inhibitor, wortmannin. Sequence alignment was performed using ClustalX (Thompson et al, 1997).
As a rule, classic protein kinases are made catalytically competent by phosphorylation in the middle of their activation loops, loosely defined by the 'DFG' and 'APE' boundaries. Mutation of the appropriate Ser, Thr or Tyr to an Asp or Glu in an activation loop can activate these protein kinases to varying extents. Plant calcium-dependent protein kinases are not phosphorylated in their activation loops and have an Asp or Glu residue in the equivalent position of most activation-loop Ser/Thr phosphorylation sites (Harper et al, 2004). The putative activation loops (as defined by DFG–APE; Fig 2) of the Arabidopsis and human PIKKs, with the exception of HsDNA-PKcs, do not contain a Ser, Thr or Tyr residue at this position. However, most retain an Asp or Glu in the middle of this loop. This highlights the potential for these kinases to be considered in an activated state and thus regulated by alternative means. This is not unlike the first protein kinase to be characterized, phosphorylase kinase, which has a Glu residue in its activation loop. When this is mutated to Ser, catalytic efficiency decreases 20-fold (Nolen et al, 2004).
The presumed site of protein–protein interaction for the PIKK family—the HEAT repeats—is rather difficult to define precisely because the sequence motif and connecting loop length vary considerably. However, a study of HEAT repeats in AtATM, AtRAD3 (ATR) and AtTOR found that the PIKKs should all have the same number of HEAT repeats as their mammalian counterparts, with the exception of AtATM, in which 800 extra amino acids were not examined for HEAT repeats (Perry & Kleckner, 2003).
ATM
The ATM protein is a key component in the regulation of the cellular response to ionizing radiation (IR). Under DNA-damaging conditions such as double-stranded breaks (DSBs), HsATM becomes phosphorylated and an autophosphorylation event on Ser1981 is vital for enzyme activation. Protein phosphatase 2A (PP2A) has been shown recently to control the phosphorylation status of HsATM, which leads to the possibility that the key activator of ATM may not be the initiation of autophosphorylation, but a decrease in dephosphorylation (Goodarzi et al, 2004). This is achieved by the rapid dissociation of PP2A from HsATM in response to DSBs. Once activated, ATM phosphorylates some effector proteins, such as p53, directly and others indirectly, as it also activates several protein kinases, such as Chk1 and Chk2 (for a review, see Kurz & Lees-Miller, 2004).
The ATM homologue in A. thaliana (AtATM) was identified originally through expressed sequence tag (EST) data and confirmed with BAC sequencing before the Arabidopsis genome was published (Garcia et al, 2000). Comparisons with the human ATM sequence reveal some similarities and differences (Fig 2; supplementary Fig 1 online). The AtATM protein is 17% identical to its human counterpart across the entire length of the protein (22% similar), and the FAT–PI3K–FATC region is 34% identical (54% similar). Whereas the other members of the PIKK family in Arabidopsis are of similar length to their human counterparts, the AtATM protein is 800 amino acids longer than HsATM. These extra residues are not found in any other ATM orthologue (including the orthologue from Oryza sativa (rice) and are located at the extreme N-terminus. Part of this extra sequence comprises a PWWP domain (Garcia et al, 2000), which binds to DNA and specifically localizes proteins to chromatin (Qiu et al, 2002). The PWWP domain of AtATM aligns with the PWWP domains of several other proteins (supplementary Fig 6 online), including a human methyltransferase that binds to DNA through its PWWP domain. DNA binding is thought to involve an extremely basic surface in the PWWP motif and this is maintained in the Arabidopsis protein.
Studies of AtATM have shown that it, like its human counterpart, is expressed ubiquitously and is involved in the response to DNA damage. AtATM mutants are partially sterile due to various meiotic defects, and are hypersensitive to IR as well as being deficient at inducing transcription of the genes that are involved in the repair of DSBs (Garcia et al, 2003). Recently, it has been observed that AtATM and AtATR (see the next section) are each responsible for a subset of histone H2AX phosphorylation events in response to IR (Friesner et al, 2005). Phosphorylation of H2AX at the site of DSBs is one of the earliest known events in response to this insult. By monitoring the foci formed through the phosphorylated H2AX using immunocytochemistry, it is possible to quantify the number of DSBs in each cell. Approximately 90% of this foci formation is dependent on AtATM. Interestingly, observations made in the same study also indicate that plants have roughly threefold less sensitivity to IR, and that the increased resistance is probably due to a greater resistance to damage as opposed to a greater DNA damage tolerance.
RAD3/ATR
ATR was originally discovered when analysing genes with sequence similarity to ATM in Saccharomyces cerevisiae and Schizosaccharomyces pombe, and was found to be related to the RAD3 protein of S. pombe. Whereas the loss of ATM function causes serious disease in humans, the null mutation of ATR is embryonic-lethal in mice. ATR can also complement some of the function lost in ATM mutants, which indicates that the functions of the two proteins overlap to some extent. ATR has been found to be activated by replication protein A (RPA)-bound DNA. Because RPA normally binds to single-stranded DNA (ssDNA) in DNA replication (as well as in DNA repair), this indicates a role for ATR in the normal progression of the cell cycle and possibly explains the embryonic lethality of ATR-null mutants (for a review, see Shechter et al, 2004).
The Arabidopsis homologue AtATR is 23% identical (42% similar) to HsATR and has no significant extra or missing residues (Fig 2, supplementary Fig 2 online; Culligan et al, 2004). Whereas ATR is necessary for development in mammals, this is not the case in plants. ATR-null mutants in Arabidopsis show no discernible phenotype under standard growth conditions. Cell-cycle arrest by IR at G2 is altered slightly in atr plants; however, aphidicolin treatment, which prevents DNA replication and normally leads to G2 arrest, does not arrest the null plants at G2. This shows that in plants, as in mammals and yeast, ATR is involved in the G2 checkpoint response. One main difference observed in plants is that hydroxyurea leads to G1 arrest, as opposed to the G2 arrest observed in mammals and yeast. This indicates a checkpoint response in plants that is absent from other eukaryotes (Culligan et al, 2004). Approximately 10% of the IR-induced foci formation in Arabidopsis cells is dependent on ATR, and in mutants that are defective in both ATM and ATR, no IR-induced H2AX phosphorylation occurs. Because of the non-lethal phenotype of atr plants, and the lack of a plant DNA-PK, the substrates and roles of ATM and ATR may be more specifically elucidated in plants. Providing these targets are found to be conserved, plants offer an ideal model to study DNA damage and repair in eukaryotes.
TOR
The TOR protein is now recognized as one of the primary controls of growth in eukaryotic cells. In mammalian cells, the activity and function of TOR—and thus the activity of its targets—is controlled by various factors including amino-acid levels, growth factors, ATP availability, hypoxia, polyphosphate and phosphatidic acid (Martin & Hall, 2005). In this way, cell growth is coupled tightly to nutrient availability. Mammals and yeast have rapamycinsensitive and -insensitive TOR complexes. in which rapamycin functions as a TOR inhibitor by forming a complex with the protein FK506 binding-protein 12 (FKBP12) that then binds TOR (Fig 3). The rapamycinsensitive complex contains mTOR, regulatory associated protein of mTOR (Raptor) and GβL (G protein β-subunit like), and the equivalent proteins TOR, KOG (kontroller of growth) and LST8 (lethal with sec thirteen 8) in S. cerevisiae. The mammalian complex of mTOR, Rictor and GβL (TOR, AVO1–3 (adheres voraciously to TOR2) and LST8 in S. cerevisiae) is rapamycin-insensitive due to its inability to bind rapamycin–FKBP12. The complex containing Rictor (mammals) or AVO1–3 (yeast) regulates the actin cytoskeleton (Jacinto et al, 2004). The two targets of the TOR–Raptor complex that are best characterized are the ribosomal protein S6 kinase 1 (S6K1) and the transcriptional repressor 4E-BP1. As the name suggests, S6K1 was characterized as an S6 protein kinase, but recently discovered targets of S6K, such as upstream binding factor (UBF), eukaryotic initiation factor 4B (eIF4B) and S6K Aly/Ref-like target (SKAR) have shed new light on the role of this enzyme. In mammals, phosphorylation that is dependent on mTOR causes the activation of S6K1 and the inhibition of 4E-BP1, together upregulating both transcriptional machinery synthesis and general protein synthesis (for a review, see Martin & Hall, 2005).
Figure 3.
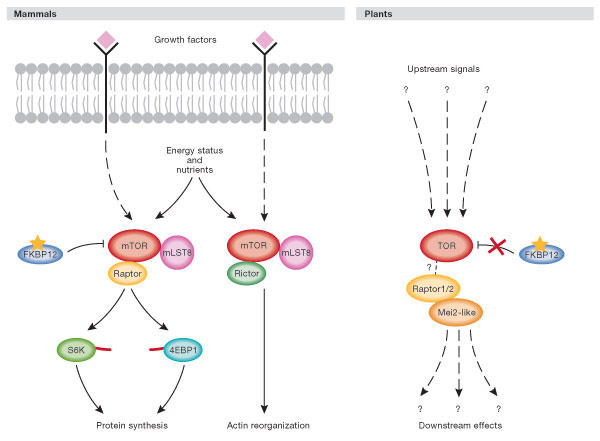
Model of target of rapamycin signalling in mammalian and plant cells. In mammalian cells, target of rapamycin (TOR) receives growth factor, nutrient and energy status signals. Mammalian TOR (mTOR), like yeast TOR, resides in two complexes—a rapamycin-sensitive and an insensitive form—that each have common and distinct protein components. Many of the components upstream of mTOR that receive and interpret the growth factor, nutrient and energy status inputs have been defined, but are not shown here for clarity (see Anderson & Hanson, 2005; Deprost et al, 2005; Martin & Hall, 2005). Rapamycin sensitivity of the mTOR–Raptor complex is conferred through the binding of rapamycin (shown as a yellow star) to FKBP12. The protein mLST8 is also known as GβL. S6K and 4EBP1 are known to dock with the mTOR complex through a short amino-acid sequence called the TOR signalling (TOS) motif that is illustrated as a red line on each protein. In plants, the upstream signalling events and downstream readouts of TOR have yet to be defined at the molecular level. Although rapamycin binds plant FKBP12, this complex fails to interact with TOR and plants are consequently rapamycin insensitive (illustrated by an X between plant TOR and FKBP12). The interaction between the plant Raptor protein and TOR has yet to be shown formally, and a dashed line is therefore used to connect them. The recently identified plant Raptor-binding protein Mei2-like is shown and is probably a TOR substrate.
Arabidopsis and rice have a single TOR gene. AtTOR is a 2,481 amino-acid protein (compared with 2,549 amino acids in HsTOR) that is 39% identical (57% similar) to its human counterpart, making it the most highly conserved member of the PIKK family between humans and plants (Fig 2; supplementary Fig 3 online; Menand et al, 2002). The FKBP12/rapamycin-binding (FRB) domain is situated between the FAT and the PI3K domains, as in HsTOR, and is nearly 62% identical to the FRB of HsTOR (supplementary Fig 3 online). Despite the fact that this domain is conserved, A. thaliana and all other plants examined so far have been found to be rapamycin insensitive. This is due to the inability of plant FKBP12 to mediate complex formation with plant TOR (Xu et al, 1998). The two mammalian interactors of the rapamycinsensitive HsTOR complex, Raptor and GβL, have orthologues in plants (Templeton & Moorhead, 2004), whereas Rictor/AVO1–3 seems not to be present in plants (R. Loewith and J. M. Mulet, personal communication). Deprost et al (2005) have identified two Raptor genes in Arabidopsis (AtRaptor1 and AtRaptor2) and rice, and have shown that T-DNA knockout of Raptor1 leads to seed abortion and arrest of embryo development at a stage earlier than a TOR knockout.
Current work in mammals indicates that signalling through mTOR/Raptor requires docking to the TOR complex through the TOR signalling (TOS) motif identified in S6K1 and 4E-BP1. Although plants have two S6K proteins, they do not have a TOS motif in their N-terminus or the recently identified S6K C-terminal motif RSPRR necessary for the mTOR-dependent suppression of S6K activation (Schalm et al, 2005). Like yeast, there is no apparent 4E-BP1 orthologue in plants although functionally similar proteins do exist (Cosentino et al, 2000). The best candidate for a TOR substrate in plants is the Mei2-like protein, which has been shown to interact with one of the Arabidopsis Raptor proteins (Anderson & Hanson, 2005). In S. pombe, Mei2 interacts with the Raptor protein Mip1, and it is thought to be a substrate for TOR due to its phosphorylation under high nutrient conditions, when TOR is active. Under nutrient-poor conditions, S. pombe Mei2 accumulates in the nucleus as an unphosphorylated protein that binds a non-coding RNA molecule through one of its three RNA-recognition motifs (RRMs). These RRMs are conserved in the plant version of Mei2. Curiously, the Raptor-binding protein Mei2 does not exist in budding yeast or metazoans. One final mystery is that in mammals, functional TOR is expressed in all tissues, whereas in plants, TOR has been found only in non-differentiated, rapidly growing meristematic cells. This leads to questions about nutrient regulation of protein synthesis in mature tissues (Menand et al, 2002).
TRRAP
The only catalytically inactive member of the PIKK family, TRRAP, seems to have lost its kinase activity and functions solely in a protein scaffolding role (McMahon et al, 1998). Initially purified from human cells as an interacting protein of c-Myc, it is now known to be in a complex with many different histone acetyltransferase (HAT) proteins. TRRAP, like the rest of its PIKK brethren, has also been found to regulate mitotic checkpoints, and genome-wide analysis indicates involvement in the cytoskeleton, cell adhesion, protein turnover, metabolism and signal transduction (for a review, see Herceg & Wang, 2005).
The TRRAP orthologue in Arabidopsis has 24% identity (45% similarity) to the human protein and is approximately the same length. It is also missing many of the key ATP-binding residues from the catalytic domain, indicating that it, too, is devoid of kinase activity (Fig 2; supplementary Fig 4 online). Human TRRAP contains many N-terminal HEAT repeats, like other PIKKs, but the exact number has yet to be defined. Because AtTRRAP differs in size to HsTRRAP by only nine amino acids, it probably has a similar number of HEAT repeats. There does not seem to be a TRRAP orthologue in the current version of the Oryza sativa genome sequence.
SMG1
SMG1 is the most recent member of the PIKK family to be identified. In the other PIKK family members, the kinase domains are no more than 200 amino acids from the C-terminal FATC domain, but in HsSMG1 and SMG1 orthologues, an additional 1,000 amino acids are inserted between the kinase domain and the FATC domain. The most highly characterized role of SMG1 is in nonsense-mediated decay (NMD). This is a process by which messenger RNAs with premature termination codons are degraded rapidly to ensure that potentially deleterious truncated proteins are not made. The possible roles of SMG1 now include stress-induced signalling, after it was discovered that HsSMG1 phosphorylates Ser15 of p53 with higher specific activity than ATM, which itself is a kinase for this site (for a review, see Abraham, 2004b).
Database searches for an Arabidopsis orthologue of SMG1 were unsuccessful (Maquat, 2004); however, when the search was broadened to other plant genomes, a potential homologue was found in O. sativa that is 15% identical (32% similar) to HsSMG1 over the entire length of the protein (for comparison, Caenorhabditis elegans SMG-1 is 14% identical (29% similar) to HsSMG1; see supplementary Fig 5 online). The Arabidopsis genome has been searched for orthologues to other proteins known to be involved in the NMD pathway in eukaryotes, namely SMG1–7 (Maquat, 2004). All eukaryotes so far examined, including plants, contain orthologues of SMG2–4 (also called Upf1–3), and although there are no proteins with high similarity to Arabidopsis SMG5–7, there are two proteins with homology to the tetratricopeptide (TPR) repeats found in the N-terminus of SMG5–7 in metazoans (Maquat, 2004). Recently, these repeats have been shown to function like 14-3-3 proteins in their ability to bind phosphoserine peptides, and in the case of human SMG7, to specifically bind to phosphorylated Upf1 (Fukuhara et al, 2005). It is not yet known if the plant TPR-containing proteins have a similar function.
Conclusion
The PIKK family of proteins has a key role in many of the stress responses of the eukaryotic cell, and thus has attracted much attention in the mammalian research community. The fact that five out of the six PIKKs are present in plants supports the idea that the PIKKs are a group of ancient eukaryotic protein kinases the function of which is to carry out key cellular responses to a variety of stresses. By studying similarities and differences in the pathways and proteins involved in plants and other eukaryotes, especially with regard to DNA-PKcs and SMG1, it is hoped that a better understanding of the roles of these protein kinases will be found.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Fig 1
Supplementary Fig 2
Supplementary Fig 3
Supplementary Fig 4
Supplementary Fig 5
Supplementary Fig 6


Acknowledgments
We thank S.P. Lees-Miller for helpful suggestions for the manuscript. G.B.G.M. is funded by the Natural Sciences and Engineering Research Council of Canada. G.W.T. is supported by Natural Sciences Engineering Research Council of Canada and Alberta Ingenuity. We apologize to authors for not referencing original works because of space limitations.
References
- Abraham RT (2004) PI 3-kinase related kinases: 'big' players in stress-induced signaling pathways. DNA Repair (Amst) 3: 883–887 [DOI] [PubMed] [Google Scholar]
- Abraham RT (2004) The ATM-related kinase, hSMG-1, bridges genome and RNA surveillance pathways. DNA Repair (Amst) 3: 919–925 [DOI] [PubMed] [Google Scholar]
- Anderson GH, Hanson MR (2005) The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth. BMC Plant Biol 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosentino GP, Schmelzle T, Haghighat A, Helliwell SB, Hall MN, Sonenberg N (2000) Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol Cell Biol 20: 4604–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan K, Tissier A, Britt A (2004) ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Truong HN, Robaglia C, Meyer C (2005) An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun 326: 844–850 [DOI] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB (2005) Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell 16: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E (2005) SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell 17: 537–547 [DOI] [PubMed] [Google Scholar]
- Garcia V, Salanoubat M, Choisne N, Tissier A (2000) An ATM homologue from Arabidopsis thaliana: complete genomic organisation and expression analysis. Nucleic Acids Res 28: 1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A (2003) AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK (2004) Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J 23: 4451–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A (2004) Decoding Ca(2+) signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Herceg Z, Wang ZQ (2005) Rendez-vous at mitosis: TRRAPed in the chromatin. Cell Cycle 4: 383–387 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB (1999) Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274: 37538–37543 [DOI] [PubMed] [Google Scholar]
- Kurz EU, Lees-Miller SP (2004) DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 3: 889–900 [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Meek K (2003) Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85: 1161–1173 [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298: 1912–1934 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: a comparative analysis of different species. Curr Genomics 5: 175–190 [Google Scholar]
- Martin DE, Hall MN (2005) The expanding TOR signaling network. Curr Opin Cell Biol 17: 158–166 [DOI] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94: 363–374 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell 15: 661–675 [DOI] [PubMed] [Google Scholar]
- Perry J, Kleckner N (2003) The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X (2002) The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol 9: 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalm SS, Tee AR, Blenis J (2005) Characterization of a conserved C-terminal motif (RSPRR) in ribosomal protein S6 kinase 1 required for its mammalian target of rapamycin-dependent regulation. J Biol Chem 280: 11101–11106 [DOI] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J (2004) Regulation of DNA replication by ATR: signaling in response to DNA intermediates. DNA Repair (Amst) 3: 901–908 [DOI] [PubMed] [Google Scholar]
- Tamura K, Adachi Y, Chiba K, Oguchi K, Takahashi H (2002) Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: evidence for a role in the repair of DNA doublestrand breaks. Plant J 29: 771–781 [DOI] [PubMed] [Google Scholar]
- Templeton GW, Moorhead GB (2004) A renaissance of metabolite sensing and signaling: from modular domains to riboswitches. Plant Cell 16: 2252–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G (1996) Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol 16: 1722–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Liang S, Kudla J, Luan S (1998) Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J 15: 511–519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig 1
Supplementary Fig 2
Supplementary Fig 3
Supplementary Fig 4
Supplementary Fig 5
Supplementary Fig 6


