Abstract
The post-transcriptional gene silencing in animals and plants is called RNA interference (RNAi). Guides for the sequence-specific degradation of mRNA are 21-nt small interfering RNAs (siRNAs) that are generated by Dicer-dependent cleavage from longer double-stranded RNAs (dsRNAs). To examine the relationship between the localization of dsRNA and the target cleavage of RNAi in human cells, we constructed five kinds of dsRNA expression vector that were controlled by tRNAVal or U6 promoter. Transcripts of tRNA-dsRNA were consistently localized in the cytoplasm and were efficiently processed by Dicer. In contrast, transcripts of tRNA-dsRNA were not processed in cells that expressed Dicer-directed ribozymes. In addition, transcripts of U6-dsRNA were basically localized in the nucleus and were not significantly processed, unless the transcripts of U6-dsRNAs possessed a microRNA-based loop motif: in the latter case, U6-dsRNAs with a microRNA-based loop were transported to the cytoplasm and were effectively processed. More over, tRNA-dsRNA directed against a mutant k-ras transcript cleaved its target mRNA efficiently in assays of RNAi not only in vitro with a cytoplasmic extract but also in vivo. Therefore, it appears that RNAi in human cells occur in the cytoplasm. Importantly, the same tRNA-dsRNA did not affect the degradation of the normal k-ras mRNA in vitro and in vivo. Our tRNA-dsRNA technology should be a powerful tool for studies of the mechanism of RNAi and the functions of various genes in mammalian cells with potential utility as a therapeutic agent.
INTRODUCTION
RNA interferance (RNAi) is a process in which double-strand RNA (dsRNA) induces a sequence-dependent degradation of a cognate mRNA (1,2). The natural roles of RNAi might include defense against viral infection (3–7) and regulation of the expression of cellular genes (8,9). Genetic and biological studies have revealed that RNAi is a very complex process that involves many different proteins with mostly unidentified functions (3–11).
It has been demonstrated in vitro that dsRNA targets mRNA for cleavage in lysates of early Drosophila embryos and in extracts of cultured Drosophila S2 cells (12–14), and such reactions in vitro require ATP (14). The molecular basis for the requirement for ATP is due, in part, to a requirement for ATP in the initial processing of long dsRNA into the 21–25-nt small interfering RNAs (siRNAs) that serve as guides for targeted cleavage (14–18).
Recent studies with synthetic RNA duplexes demonstrated that the siRNA duplex must have 2- or 3-nt overhanging 3′-ends for efficient cleavage of its target (16). Such 3′ overhangs are characteristic of the products of cleavage reactions catalyzed by RNase III, and, in cultured Drosophila S2 cells, cleavage of dsRNA into siRNAs requires a multidomain RNase III, known as Dicer (17). Subsequently, siRNAs seem to associate with a multicomponent nuclease, identified in Drosophila and designated RNAi-induced silencing complex (RISC), and then they guide this enzyme so that it catalyzes the sequence-specific degradation of mRNA (13,17,19).
RNAi provides a method for inactivating genes of interest and, thus, provides a powerful tool for studies of gene function in Caenorhabditis elegans, Drosophila melanogaster and plants. Specific inhibition of gene expression also can be achieved by the stable or inducible expression of dsRNA in animals and plants (11). Inactivation of genes by dsRNA has been achieved in mouse embryonal carcinoma (EC) cells and embryonic stem (ES) cells (20,21), but elicitation of RNAi using long dsRNAs has generally been less successful in cultured mammalian cells. Such failures can be explained most readily by the activation of dsRNA-dependent protein kinase (PKR) and 2′,5′-oligoadenylate synthetase (2′,5′-AS) which are activated by long dsRNA (>30 bp; 22,23).
It was reported recently that 21-nt synthetic siRNAs (24) and siRNAs that were transcribed by U6 or H1 promoter (25–32) specifically suppressed the expression of endogenous genes in several lines of mammalian cells. These 21-nt siRNA duplexes were able to evade non-specific reduction of mRNAs and these findings suggested that RNAi or an RNAi-related system might exist in mammals. Some mammalian homologs of RNAi-associated proteins, such as Dicer, eIF2C2 and WRN have, indeed, been identified (17,33–36). However, details of the characteristics and mechanisms of RNAi in human somatic cells remain to be determined.
In this study, we constructed five kinds of dsRNA expression vectors that were controlled by tRNAVal or widely used U6 promoter in order to examine the relationship between the localization and a target cleavage of RNAi in mammalian cells. Transcripts of tRNA-dsRNA were localized in the cytoplasm and were processed efficiently by Dicer. In addition, a mutant k-ras-directed tRNA-dsRNA efficiently cleaved the targeted mRNA in vitro and in vivo. In contrast, the tRNA-dsRNA did not affect the expression of normal k-ras in HeLa cells. Therefore, these results indicated that RNAi in mammalian cells occurred in the cytoplasm and our tRNA-dsRNA expression system should be a powerful tool for studying mechanism of RNAi and other gene functions in mammalian cells with potential utility as a therapeutic agent.
MATERIALS AND METHODS
Culture and transfection of cells
SW480 human colon cancer cells were cultured in L-15 medium (ICN Biomedicals, Inc., OH) supplemented with 10% fetal bovine serum (FBS). HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS. Transfections were performed with the Effectin™ reagent (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. dsRNA-expressing SW480 and dsRNA-expressing HeLa cells were selected by incubation with puromycin for two weeks.
Construction of dsRNA expression plasmids
To construct vectors for expression of tRNA-dsRNA, we used the pPUR-tRNA plasmid that includes the chemically synthesized promoter for a human gene for tRNAVal (37) between the EcoRI and BamHI sites of pPUR (Clontech, CA). Chemically synthesized oligonucleotides encoding mutant k-ras-directed dsRNA that included loop 1 (5′-GAG CTC GGT AGT TGG AGC TGT TGG CGT AGG CAA GAG AAA ATC TTG CCT ACG CCA ACA GCT CCA ACT ACC GGT ACC-3′) were amplified as double-stranded sequences by PCR. After digestion with SacI and KpnI, the fragments were cloned downstream of the promoter of the tRNA gene in pPUR-tRNA. The construction of vectors for expression of dsRNA from the mouse U6 promoter has been described elsewhere (38,39) except for the insertion step of dsRNA sequences. Chemically synthesized oligonucleotides encoding k-ras-directed dsRNA that included loop 1 (5′-GAA TTC GGT AGT TGG AGC TGT TGG CGT AGG CAA GAG AAA ATC TTG CCT ACG CCA ACA GCT CCA ACT ACC TCT AGA-3′) or loop 2 (5′-GAA TTC GGT AGT TGG AGC TGT TGG CGT AGG CAA GAC TTC CTG TCA TCT TGC CTA CGC CAA CAG CTC CAA CTA CCC TCG AG-3′) were amplified as double-stranded sequences by PCR with specific up primer and down primer which contained EcoRI and XhoI linker sequences, respectively. After digestion with EcoRI and XhoI, the fragments were cloned downstream of the promoter of the mouse U6 gene. In the case of a human U6 promoter, we used pUC-hU6 (40). Chemically synthesized oligonucleotides encoding k-ras-directed dsRNA that included oop 1 (5′-GTC GAC GGT AGT TGG AGC TGT TGG CGT AGG CAA GAG AAA ATC TTG CCT ACG CCA ACA GCT CCA ACT ACC TCT AGA-3′) or loop 2 (5′-GTC GAC GGT AGT TGG AGC TGT TGG CGT AGG CAA GAC TTC CTG TCA TCT TGC CTA CGC CAA CAG CTC CAA CTA CCT CTA GA-3′) were amplified as double-stranded sequences by PCR with specific up primer and down primer which contained SalI and XbaI linker sequences, respectively. After digestion with SalI and XbaI, the fragments were cloned downstream of the promoter of the human U6 gene.
Construction of Dicer-directed ribozyme expression plasmids
Chemically synthesized oligonucleotides encoding Dicer-directed ribozyme sequence (5′-TCC CCG GTT CGA AAC CGG GCA CTA CAA AAA CCA ACT TTC AAA GAA AGC TGA TGA GGC CGA AAG GCC GAA ACC CAT TGG GGT ACC CCG GAT ATC TTT TTT-3′) with a pol III termination sequence (TTTTTT) were converted to double-stranded DNAs by PCR. After digestion with Csp45I and PstI, the fragments were cloned downstream of the tRNA promoter of pUC-dt (37,41,42). To generate poly(A)-connected ribozyme, we inserted a poly(A) sequence of 100 nt between the ribozyme and the pol III termination sequence (42–45).
Preparation of the nuclear fraction and the cytoplasmic fraction of cells
SW480 or HeLa cells were grown to ∼5 × 106 cells and were transfected with a tRNA-dsRNA or U6-dsRNA expression vector with the Effectin™ reagent (QIAGEN). Thirty-six hours after transfection, cells were harvested. For the preparation of the cytoplasmic fraction, collected cells were washed twice with PBS and then resuspended in digitonin lysis buffer (50 mM HEPES/KOH, pH 7.5, 50 mM potassium acetate, 8 mM MgCl2, 2 mM EGTA and 50 µg/ml digitonin) on ice for 10 min. The lysate was centrifuged at 1000× g and the supernatant was collected as the cytoplasmic fraction. The pellets were resuspended in NP-40 lysis buffer (20 mM Tris–HCl, pH 7.5, 50 mM KCl, 10 mM NaCl, 1 mM EDTA and 0.5% NP-40) and held on ice for 10 min and the resultant lysate was used as the nuclear fraction.
Northern blot analysis
Cytoplasmic RNA and nuclear RNA were extracted and purified from the cytoplasmic fraction and the nuclear fraction, respectively, with ISOGEN reagent (Wako, Osaka, Japan). Thirty micrograms of total RNA per lane were loaded on a 15% polyacrylamide gel. After electrophoresis, bands of RNA were transferred to a Hybond-N™ nylon membrane (Amersham Co., Buckinghamshire, UK). The membrane was probed with synthetic oligonucleotides that were complementary to the sequences of the k-ras gene. The synthetic probe was 32P-labeled by T4 polynucleotide kinase (Takara Shuzo Co., Kyoto, Japan).
RT–PCR analysis
RT–PCR was performed using an RNA PCR Kit ver. 2 (Takara, Kyoto, Japan) with dicer upstream (nucleotides 1–24) and downstream (nucleotides 435–459) primers or GADPH upstream (nucleotides 230–254) and downstream (nucleotides 442–466) primers as a control. The products of PCR were analyzed by electrophoresis on a 2% agarose gel.
Western blot analysis
SW480 or HeLa cells that had been transfected with individual dsRNA-expression vectors were harvested. Proteins were resolved by SDS–PAGE (10% polyacrylamide) and transferred to a polyvinylidene difluoride (PVDF) membrane (Funakoshi Co., Tokyo, Japan) by electroblotting. Immune complexes were visualized with ECL kit (Amersham Co., Buckinghamshire, UK) using specific polyclonal antibodies against K-Ras (UBI, CA) and Actin (Santa Cruz, CA).
Assay of RNAi in vitro
For cleavage of a target RNA, we synthesized a mutant k-ras template DNA (70 nt) and a normal k-ras template DNA (70 nt) using an automated DNA synthesizer. For preparation of k-ras mRNA substrates, we amplified template DNAs by PCR using a k-ras-specific up primer that included the T7 promoter sequence and a k-ras-specific down primer. The amplified k-ras DNA templates were transcribed by T7 polymerase. Transcribed normal and mutant k-ras mRNA substrates were purified by PAGE. These mRNAs were 32P-labeled by T4 polynucleotide kinase. For detection of the cleavage of the target mRNA, target mRNA (5–10 nM) was incubated with a lysate of SW480 cells that expressed tRNA-dsRNA under standard conditions (14) for 2 h at 25°C. In the case of siRNA targeted to k-ras mRNA, 100 nM siRNA was incubated with target RNA (5–10 nM) in a lysate of SW480 cells under standard conditions (14) for 2 h at 25°C.
Detection of rates of cell proliferation
The rate of proliferation of each line of cells was measured with a Cell Proliferation Kit II (Roche Ltd, Switzerland) according to the manufacturer’s instructions.
RESULTS
Construction of two kinds of dsRNA-expression plasmid with pol III promoters
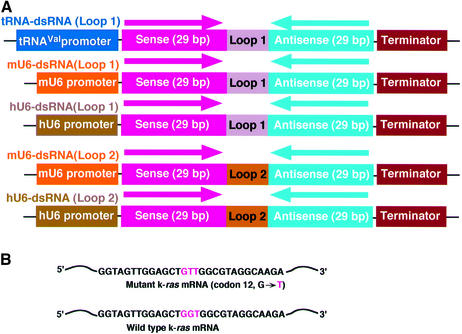
It has not yet been determined whether a long dsRNA is processed by Dicer in the nucleus or the cytoplasm. Therefore, we used two kinds of pol III promoter (the promoter of a gene for human tRNAVal, and mouse U6 or human U6 promoter) for expression of dsRNAs in mammalian cells. If designed appropriately, transcripts generated with the promoter of a human gene for tRNAVal are transported efficiently to the cytoplasm and we demonstrated previously that the cytoplasmic localization was important in improving ribozyme activities in vivo (41,46,47). In contrast, small RNAs transcribed under the control of the U6 promoter remained localized in the nucleus (37). Thus, we attached the gene for dsRNA (an extended stem–loop RNA; Fig. 1A) to the 3′ end of the promoter of a gene for tRNAVal (tRNA-dsRNA), to the 3′ end of the mouse U6 promoter (mU6-dsRNA) or to the 3′ end of the human U6 promoter (hU6-dsRNA).
Figure 1.
Construction of dsRNA expression plasmids. (A) The plasmid for expression of tRNA-dsRNA included the promoter sequence from a human gene for tRNAVal and a terminator sequence. The plasmid for expression of mouse U6- or human U6-dsRNA included a mouse or a human U6 promoter, respectively. The Loop 1 sequence (5′-GAAAA-3′) or loop2 sequence [miR-23 loop sequence (48): 5′-CUUCCUGUCA-3′] was inserted between a k-ras sense-strand sequence (29 bp) and an antisense-strand sequence (29 bp). (B) Sequences of mutant and normal k-ras mRNAs that were targeted by dsRNA. The mutant k-ras gene had a point mutation (codon 12 GGT→GTT).
We constructed dsRNA expression plasmids targeted to the mRNA for a mutant of K-Ras with a point mutation in codon 12 of the k-ras gene (Fig. 1B). The length of the double-stranded region within the dsRNA was kept at 29 bp because long dsRNAs (>30mers) induce non-specific reduction of mRNAs (22,23). In the case of U6-based constructs, we used two kinds of loop motif for stem–loop RNAs. One is a loop motif that consists of 5 nt (5′-GAAAA-3′), namely Loop 1 (Fig. 1A). The other is a microRNA (human mir-23; 48) loop motif, namely Loop 2 (Fig. 1A). It is believed that precursor microRNAs are transported to the cytoplasm and the processed microRNAs act post-transcriptional gene silencing (48–54).
dsRNA transcribed under the control of the tRNAVal promoter was processed by a Dicer-complex in the cytoplasm
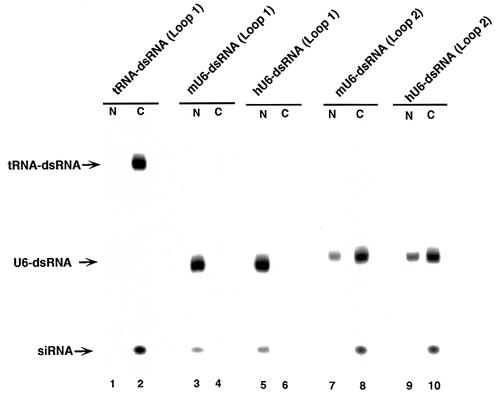
In RNAi, long dsRNAs are first processed to short RNA duplexes of predominantly 21 and 22 nt in length and with staggered 3′ ends in an RNase III-like reaction (17). To examine whether tRNA-dsRNA and U6-dsRNA might be processed by an RNase III complex in mammalian cells, we performed northern blotting analysis with a k-ras mRNA-specific probe. SW480 cells were transfected with plasmids that encoded dsRNA under the control of the tRNAVal or U6 promoter. Forty-eight hours after transfection, cells were collected and separated into cytoplasmic and nuclear fractions. Total RNA in each fraction was isolated and fractionated on a 15% polyacrylamide gel. As shown in Figure 2, in cells that expressed tRNA-dsRNA, processed siRNAs were detected in the cytoplasmic fraction and not in the nuclear fraction. Moreover, the sequences of processed siRNAs were confirmed by cloning and sequencing of the siRNAs (data not shown). In contrast, in cells that expressed either mU6- or hU6-dsRNA (Loop 1), predominantly unprocessed precursor dsRNAs were detected in the nucleus and the nuclear fraction contained very little siRNAs in cells that expressed either mU6- or hU6-dsRNA (Loop 1). However, mU6-dsRNAs (Loop 2) and hU6-dsRNAs (Loop 2) were transported to the cytoplasm and were processed efficiently (Fig. 2). A mammalian Dicer has been detected mostly in the cytoplasm by immunostaining in situ (20), so it seems likely that tRNA-dsRNA, mU6-dsRNAs (Loop 2) and hU6-dsRNAs (Loop 2) that had been transported to the cytoplasm were processed by the Dicer-like RNase III complex.
Figure 2.
Detection of precursor dsRNAs and siRNAs. The presence of precursor dsRNAs and siRNAs was analyzed by northern blotting analysis. Plasmids encoding tRNA-dsRNAs or U6-dsRNAs were introduced into SW480 cells. After 48 h, the cells were collected and divided into cytoplasmic (C) and nuclear (N) fraction. Total RNA in each fraction was isolated and fractionated on a 15% polyacrylamide gel. Northern blotting analysis was performed as in the text. (Lane 1, nuclear fraction of cells that expressed tRNA-dsRNA; lane 2, cytoplasmic fraction of cells that expressed tRNA-dsRNA; lane 3, nuclear fraction of cells that expressed mU6-dsRNA (Loop 1); lane 4, cytoplasmic fraction of cells that expressed mU6-dsRNA (Loop 1); lane 5, nuclear fraction of cells that expressed hU6-dsRNA (Loop 1); lane 6, cytoplasmic fraction of cells that expressed hU6-dsRNA (Loop 1); lane 7, nuclear fraction of cells that expressed mU6-dsRNA (Loop 2); lane 8, cytoplasmic fraction of cells that expressed mU6-dsRNA (Loop 2); lane 9, nuclear fraction of cells that expressed hU6-dsRNA (Loop 2); and lane 10, cytoplasmic fraction of cells that expressed hU6-dsRNA (Loop 2).
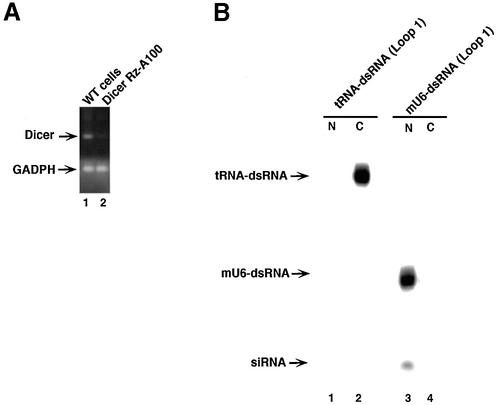
To confirm whether tRNA-dsRNAs are processed by Dicer, we constructed a poly(A)-connected Dicer-directed ribozyme (Dicer-RzA100) expression plasmid (42–44). Then this plasmid was introduced into HeLa cells stably. Stable cell lines were obtained by neomycin selection. Next, to examine suppression of expression of dicer gene by Dicer-RzA100, we performed the RT–PCR analysis with specific primers for dicer mRNA. As shown in Figure 3A, the level of dicer mRNA in cells that expressed Dicer-RzA100 was reduced compared with that in WT-HeLa cells. The level of GADPH mRNA as a control did not alter in both cell lines. These results indicated that the Dicer-RzA100 cleaved dicer mRNAs specifically.
Figure 3.
Effect of Dicer-ribozymes on processing of dsRNAs. (A) The levels of expression of dicer genes in cells that expressed poly(A)-connected Dicer-ribozyme (Dicer-RzA100). The dicer mRNA was detected by RT–PCR with primers specific for the dicer gene (see Materials and Methods). GADPH is an endogenous control. (B) Detection of precursor dsRNAs and siRNAs in cells that expressed the Dicer-RzA100. Lane 1, nuclear fraction of cells that expressed tRNA-dsRNA and Dicer-RzA100; lane 2, cytoplasmic fraction of cells that expressed tRNA-dsRNA and Dicer-RzA100; lane 3, nuclear fraction of cells that expressed U6-dsRNA and Dicer-RzA100; and lane 4, cytoplasmic fraction of cells that expressed U6-dsRNA and Dicer-RzA100.
To examine whether reduction of Dicer affects the processing of the tRNA-dsRNA, we performed northern blot analysis using total RNAs from cells that expressed the tRNA-dsRNA. As shown in Figure 3B, in the case of a total RNA from cells that expressed the Dicer-RzA100, siRNAs that are generated from tRNA-dsRNAs were not observed in both nucleus and cytoplasm. Therefore, these results suggest that tRNA-dsRNAs are processed by Dicer in the cytoplasm. In contrast, reduction of Dicer did not affect processing of mU6-dsRNA (Loop 1). Thus, it is possible that the processing of mU6-dsRNA (Loop 1) is dealt with another RNase III-like enzyme.
Degradation of target mRNA in vitro in a cell extract that contained tRNA-dsRNA, U6-dsRNA or synthetic siRNAs
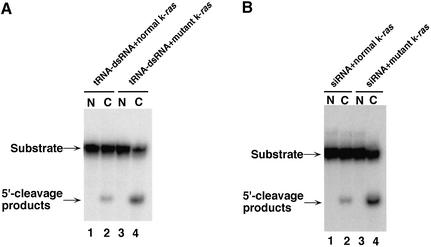
To examine the cell compartment in which degradation of a target mRNA by dsRNA-mediated gene silencing occurs, we performed assays of RNAi in vitro using cell extracts that contained transcripts of tRNA-dsRNA. In these assays, we used mutant and normal k-ras partial mRNAs, which had been transcribed in vitro by T7 polymerase, as substrates. For cleavage of the target mRNA, each substrate was incubated for 2 h at 25°C with an extract of SW480 cells that had been transfected with the tRNA-dsRNA expression vector. The 5′-cleavage products were resolved on sequencing gels. As shown in Figure 4A, the mutant k-ras mRNA substrate was cleaved in the cytoplasmic fraction of cell extracts that contained tRNA-dsRNA (lane 4). In contrast, in the nuclear fraction of the cell extracts, the substrate was not cleaved (lane 3). These results support the earlier reports that the RISC is included in a ribosomal fraction (14,17,19).
Figure 4.
Detection of dsRNA- and siRNA-mediated cleavage of mutant k-ras mRNA. (A) Cleavage of mutant k-ras mRNA by cell extracts that contained tRNA-dsRNA. In vitro RNAi: the assay of RNAi in vitro was performed as described in the text. Lane 1, nuclear fraction (N) of cells that expressed tRNA-dsRNA and normal k-ras RNA; lane 2, cytoplasmic fraction (C) of cells that expressed tRNA-dsRNA and normal k-ras mRNA; Lane 3, nuclear fraction of cells that expressed tRNA-dsRNA and mutant k-ras mRNA; and lane 4, cytoplasmic fraction of cells that expressed tRNA-dsRNA and mutant k-ras mRNA. (B) Cleavage of mutant k-ras mRNA mediated by k-ras-directed siRNAs and extracts of SW480 cells. Lane 1, nuclear fraction of SW480 cells and normal k-ras mRNA; lane 2, cytoplasmic fraction of SW480 cells and normal k-ras mRNA; lane 3, nuclear fraction of SW480 cells and mutant k-ras mRNA; and lane 4, cytoplasmic fraction of SW480 cells and mutant k-ras mRNA.
In contrast to these results, in the case of the normal k-ras mRNA substrate, significantly smaller amounts of 5′-cleavage products were detected and these were found only in the cytoplasmic fraction of cell extracts that contained transcripts of tRNA-dsRNA (Fig. 4A, lane 2). These results are in accord with the report that synthesized siRNAs with one mismatched base pair in the middle of the siRNA was reduced in efficiency of cleavage of a target RNA in an assay of RNAi in vitro with lysates of D.melanogaster (55). We confirmed that the siRNA generated from the tRNA-dsRNA formed a mismatched base pair with normal k-ras mRNA in the middle of the siRNA by sequencing analysis (data not shown). We obtained similar results with synthesized siRNAs targeted against the mutant k-ras mRNA (Fig. 4B; 36). Thus, our results suggested that degradation of a target mRNA by dsRNA-mediated gene silencing occurs in the cytoplasm and that siRNAs are capable of recognizing one mismatched base pair in a middle position.
Efficiency and specificity of RNAi by tRNA-dsRNA in colon cancer cells
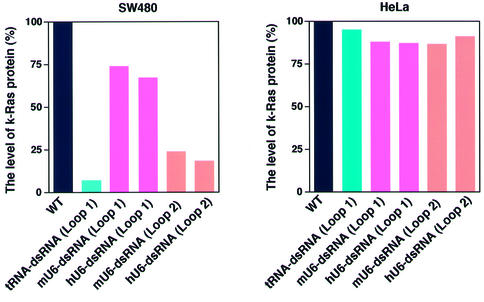
To examine the efficiency and specificity of RNAi by tRNA-dsRNA in human colon cancer SW480 cells, we introduced tRNA-dsRNAs and four kinds of U6-dsRNAs expression plasmids into SW480 and HeLa cells. The mutant k-ras gene was expressed in SW480 cells (56). We used HeLa cells that expressed a normal k-ras gene as controls. We generated stable lines of cells that expressed tRNA-dsRNAs or each U6-dsRNA by selection in the presence of puromycin. We examined levels of K-Ras protein in cells that expressed tRNA-dsRNA or each U6-dsRNA by western blotting with K-Ras-specific antibodies. For quantitation, intensities of bands were analyzed by densitometry using the NIH Image Analysis. As shown in Figure 5, the level of K-Ras protein in SW480 cells that expressed tRNA-dsRNA, mU6-dsRNA (Loop 2) or hU6-dsRNA (Loop 2) was significantly lower than that in wild-type SW480 cells, whereas the level of K-Ras protein in SW480 cells that expressed mU6-dsRNA (Loop 1) or hU6-dsRNA (Loop 1) was reduced only slightly compared with that in wild-type SW480 cells. The level of actin, chosen as an endogenous control, remained constant in these cell lines. Moreover, in HeLa cells that expressed tRNA-dsRNA, the level of K-Ras protein was similar to that in wild-type cells and in cells that expressed respective U6-dsRNA. These results demonstrated clearly both the efficiency and specificity of the tRNA-dsRNA in human cancer cells.
Figure 5.
Efficiency and specificity of RNAi by tRNA-dsRNA in vivo. The level of K-Ras protein in cells expressed that tRNA-dsRNAs or U6-dsRNAs was analyzed by western blotting analysis with specific k-Ras antibodies. Mutant k-ras gene is expressed in SW480 cells. In contrast, normal k-ras gene is expressed in HeLa cells. For quantitation, intensities of bands were analyzed by densitometry using the NIH Image Analysis.
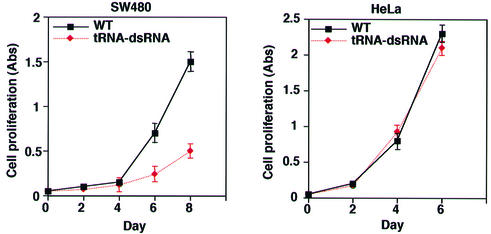
To examine the phenotype of cells that expressed tRNA-dsRNA, we analyzed the rates of proliferation of various lines of cells. As shown in Figure 6, SW480 cells that expressed tRNA-dsRNA proliferated significantly more slowly than wild-type SW480 cells. In contrast, the rate of proliferation in HeLa cells that expressed tRNA-dsRNA was the same as that of wild-type SW480 cells. These results indicated that the reduced rate of proliferation of SW480 cells that expressed tRNA-dsRNA was correlated with the reduction in the level of K-Ras protein in the cells. Thus, our tRNA-dsRNA targeted to the mutant k-ras gene appears to have potential utility as a therapeutic agent.
Figure 6.
Effect of proliferation of cells by tRNA-dsRNA. Rates of proliferation of cells that expressed tRNA-dsRNA. Rates of proliferation were determined as described in the text. Values are means ±SD of results from three replicates in each case. SW480 cells that expressed tRNA-dsRNA proliferated significantly more slowly than wild-type SW480 cells. In contrast, the rate of proliferation in HeLa cells that expressed tRNA-dsRNA was the same as that of wild-type SW480 cells.
DISCUSSION
The discovery that dsRNA could induce gene silencing in animals and plants has raised the possibility that RNAi might be a nearly universal mechanism of gene silencing. An outline of the processes involoved in RNAi and several components have been identified in C.elegans, D.melanogaster and plants, but details of the mechanism and many of the necessary participants in RNAi in mammalian cells remain unclear. In this study, to clarify the relationship between the localization and the target specificity of RNAi in mammalian cells, we constructed two kinds of dsRNA expression plasmid, namely, tRNA-dsRNA and U6-dsRNA expression plasmids. We demonstrated that tRNA-dsRNA which was localized in the cytoplasm was efficiently processed by the RNase III complex. An initial step in RNAi is the cleavage by Dicer, which is localized in the cytoplasm (20), of long dsRNAs. Although short dsRNAs are cleaved less effectively by Dicer in vitro (16,17), our tRNA-dsRNA was processed with significant efficiency in mammalian cells (Fig. 2). In addition, U6-dsRNAs that had a microRNA-based loop motif were transported to the cytoplasm and were processed. Although transcripts from the U6 promoter are generally localized in the nucleus, the microRNA-loop motif promotes the transport of dsRNAs to the cytoplasm. Thus, cytoplasmic localization of dsRNAs is important for processing by Dicer.
The degradation step of mRNA is a very interesting aspect of RNAi-mediated gene silencing. Although two groups proposed that an RNA-directed RNA polymerase (RdRP) chain reaction with siRNA amplifies the interference caused by a small amount of ‘trigger’ dsRNA in C.elegans (57), the mechanism is unclear in mammalian cells because an RdRP homolog does not exist in mammalian cells and siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways (58). In this study, we demonstrated that a cytoplasmic fraction from cells which expressed tRNA-dsRNA had mRNA degradation activity (Fig. 4A). In addition, synthetic siRNAs mixed with the cytoplasmic fraction also had mRNA degradation activity (Fig. 4B; 36). Moreover, it was recently reported that the level of HIV RRE-containing mRNA in the nucleus was not affected by siRNAs (59). Thus, it is likely that a siRNA-associated silencing complex (SASC) including Dicer or -Slicer- is localized in the cytoplasm of mammalian cells.
RNAi has been shown to be a powerful tool for studies of gene function in C.elegans, D.melanogaster and plants. However, in mammalian cells, a long dsRNA causes the non-specific reduction in expression of many genes. Thus, it was believed initially that RNAi could not be used for gene inactivation in mammalian cells. However, Tuschle’s group demonstrated that siRNA could specifically suppress the expression of a target gene specifically (24). Exploitation of RNAi in mammalian cells requires evasion of non-specific reduction of mRNAs. In addition, since the putative SASC is located in the cytoplasm, it is important that dsRNA transcripts be localized in the cytoplasm. If properly designed, tRNA-dsRNAs (with a short hairpin structure) can be transported to the cytoplasm and can escape the non-specific reduction of mRNAs. Indeed, we found that PKR was not activated in cells that expressed tRNA-dsRNA (data not shown).
Although our U6-dsRNA with a general loop motif that consisted of five nucleotides was not transported to the cytoplasm and we could detect only marginal RNAi by our U6-dsRNA (Figs 2 and 5), transcripts of U6-dsRNAs (Loop 2) that had a microRNA-based loop motif were transported to the cytoplasm and were processed by Dicer (Fig. 2). In addition, they induced RNAi-mediated gene silencing (Fig. 5).
When alternative conditions were used, other U6-dsRNAs prepared by several independent groups (length of dsRNA and the size and the sequence of the hairpin-loop in these constructs were different from those of our U6-dsRNA) could support RNAi in mammalian cells (25–30). In one case, a microRNA motif was used as the loop motif of dsRNA and the efficacy could have been enhanced by a microRNA pathway (32). Thus, the length and the nucleotide sequence of the loop in hairpin types of dsRNAs are important for construction of effective hairpin types of dsRNAs. Now we are examining effects of both different length and structure of the loop on the localization and RNAi-mediated gene silencing by hairpin types of dsRNAs in detail.
Taken together, our results indicate that RNAi in mammalian cells occurs in the cytoplasm and is very specific in mammalian cells. Our tRNA-dsRNAs should be powerful tools for studies of the mechanism of RNAi and the functions of specific genes in mammalian cells, and they might also be useful as therapeutic agents.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by grants from the Ministry of Economy, Trade and Industry (METI) of Japan, and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Culture (MEXT) of Japan.
REFERENCES
- 1.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans.Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery M.K., Xu,S. and Fire,A. (1998) RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA, 95, 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montgomery M.K. and Fire,A. (1998) Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet., 14, 255–258. [DOI] [PubMed] [Google Scholar]
- 4.Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G. and Zamore,P.D. (2002) RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev., 12, 225–232. [DOI] [PubMed] [Google Scholar]
- 6.Li W.X. and Ding,S.W. (2001) Viral suppressors of RNA silencing. Curr. Opin. Biotechnol., 12, 150–154. [DOI] [PubMed] [Google Scholar]
- 7.Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- 8.Smardon A., Spoerke,J., Stacey,S., Klein,M., Mackin,N. and Maine,E. (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans.Curr. Biol., 10, 169–178. [DOI] [PubMed] [Google Scholar]
- 9.Aravin A.A., Naumova,N.M., Tulin,A.V., Vagin,V.V., Rozovsky,Y.M. and Gvozdev,V.A. (2001) Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D.melanogaster germline. Curr. Biol., 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein E., Denli,A.M. and Hannon,G.J. (2001) The rest is silence. RNA, 7, 1509–1521. [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001) Post-transcriptional gene silencing by double-stranded RNA. Nature Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- 12.Tuschl T., Zamore,P.D., Lehmann,R., Bartel,D.P. and Sharp,P.A. (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev., 13, 3191–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond S.M., Bernstein,E., Beach,D. and Hannon,G.J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 14.Zamore P., Tuschl,T., Sharp,P. and Bartel,D. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton A.J. and Baulcombe,D.C. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science, 286, 950–952. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 18.Tuschl T. (2001) RNA interference and small interfering RNAs. Chembiochem., 2, 239–254. [DOI] [PubMed] [Google Scholar]
- 19.Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute 2, a link between genetic and biochemical analysis of RNAi. Science, 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 20.Billy E., Brondani,V., Zhang,H., Müller,U. and Filipowicz,W. (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA, 98, 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paddison P.J., Caudy,A.A. and Hannon,G.J. (2002) Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manche L., Green,S.R., Schmedt,C. and Mathews,M.B. (1992) Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12, 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minks M.A., West,D.K., Benvin,S. and Baglioni,C. (1979) Structural requirements of double-stranded RNA for the activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells. J. Biol. Chem., 254, 10180–10183. [PubMed] [Google Scholar]
- 24.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 25.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 26.Miyagishi M. and Taira,K. (2002) U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 27.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, 500–505. [DOI] [PubMed] [Google Scholar]
- 28.Paul C.P., Good,P.D., Winer,I. and Engelke,D.R. (2002) Effective expression of small interfering RNA in human cells. Nat. Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 29.Paddison P.J., Caudy,A.A., Bernstein,E., Hannon,G.J. and Conklin D.S. (2002) Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev., 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sui G., Soohoo,C., Affar,E.B., Gay,F., Shi,Y., Forrester,W.C. and Shi,Y. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J.Y., DeRuiter,S.L. and Turner,D.L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mcmanus M.T., Petersen,C.P., Haines,B.B., Chen,J. and Sharp,P.A. (2002) Gene silencing using micro-RNA designed hairpins. RNA, 8, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabara H., Sarkissian,M., Kelly,W.G., Fleenor,J., Grishok,A., Timmons,L., Fire,A. and Mello,C.C. (1999) The rde-1 gene, RNA interference and transposon silencing in C. elegans. Cell, 99, 123–132. [DOI] [PubMed] [Google Scholar]
- 34.Ketting R.F., Haverkamp,T.H., van Luenen,H.G. and Plasterk,R.H. (1999) Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell, 99, 133–141. [DOI] [PubMed] [Google Scholar]
- 35.Martinez J., Patkaniowska,A., Urlaub,H., Luhrmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 36.Hutvagner G. and Zamore,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- 37.Koseki S., Tanabe,T., Tani,K., Asano,S., Shioda,T., Nagai,Y., Shimada,T., Ohkawa,J. and Taira,K. (1999) Factors governing the activity in vivo of ribozymes transcribed by RNA polymerase III. J. Virol., 73, 1868–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plehn-Dujowich D. and Altman,S. (1998) Effective inhibition of influenza virus production in cultured cells by external guide sequences and ribonuclease P. Proc. Natl Acad. Sci. USA, 95, 7327–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato Y., Kuwabara,T., Warashina,M., Toda,H. and Taira,K. (2001) Relationships between the activities in vitro and in vivo of various kinds of ribozyme and their intracellular localization in mammalian cells. J. Biol. Chem., 276, 15378–15385. [DOI] [PubMed] [Google Scholar]
- 40.Bertrand E., Castanotto,D., Zhou,C., Carbonnelle,C., Lee,N.S., Good,P., Chatterjee,S., Grange,T., Pictet,R., Kohn,D., Engelke,D. and Rossi,J.J. (1997) The expression cassette determines the functional activity of ribozymes in mammalian cells by controlling their intracellular localization. RNA, 3, 75–88. [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki H., Eckner,R., Yao,T.P., Taira,K., Chiu,R., Livingston,D.M. and K.K. Yokoyama. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature, 393, 284–289. [DOI] [PubMed] [Google Scholar]
- 42.Kawasaki H. and Taira,K. (2002) Identification of genes by hybrid-ribozymes that couple cleavage activity with the unwinding activity of an endogenous RNA helicase. EMBO rep., 3, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Kawasaki H., Ohnuki,R., Suyama,E. and Taira,K. (2002) Identification of genes that function in the TNF-α-mediated apoptotic pathway using randomized hybrid ribozyme libraries. Nat. Biotechnol., 20, 376–380. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki H. and Taira,K. (2002) A functional gene discovery in the Fas-mediated pathway to apoptosis by analysis of transiently expressed randomized hybrid-ribozyme libraries. Nucleic Acids Res., 30, 3609–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warashina M., Kuwabara,T., Kato,Y., Sano,M. and Taira,K. (2001) RNA–protein hybrid ribozymes that efficiently cleave any mRNA independently of the structure of the target RNA. Proc. Natl Acad. Sci. USA, 98, 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuwabara T., Warashina,M., Koseki,S., Sano,M., Ohkawa,J., Nakayama,K. and Taira,K. (2001) Significantly higher activity of a cytoplasmic hammerhead ribozyme than a corresponding nuclear counterpart: engineered tRNAs with an extended 3′ end can be exported efficiently and specifically to the cytoplasm in mammalian cells. Nucleic Acids Res., 29, 2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuwabara T., Warashina,M., Sano,M., Tang,H., Wong-Staal,F., Munekata,E. and Taira,K. (2001) Recognition of engineered tRNAs with an extended 3′ end by Exportin-t (Xpo-t) and transport of tRNA-attached ribozymes to the cytoplasm in somatic cells. Biomacromol., 2, 1229–1242. [DOI] [PubMed] [Google Scholar]
- 48.Lagos-Quintana M., Rauhut, R, Yalcin,A., Meyer,J., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 49.Lee Y., Jeon,K., Lee,J.T., Kim,S. and Kim,V.N. (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J., 21, 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grishok A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–24. [DOI] [PubMed] [Google Scholar]
- 51.Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- 52.Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 53.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 54.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charroux,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elbashir S.M., Martinez,J., Patkaniowska,A., Lendeckel,W. and Tuschl,T. (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J., 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell C.E., Belinsky,S.A. and Lechner,J.F., (1995) Detection and quantitation of mutant K-ras codon 12 restriction fragments by capillary electrophoresis. Anal. Biochem., 224, 148–153. [DOI] [PubMed] [Google Scholar]
- 57.Sijen T., Fleenor,J., Simmer,F., Thijssen,K.L., Parrish,S., Timmons,L., Plasterk,R.H. and Fire,A. (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell, 107, 465–476. [DOI] [PubMed] [Google Scholar]
- 58.Schwarz D.S., Hutvagner,G., Haley,B. and Zamore,P.D. (2002) Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell, 10, 537–548. [DOI] [PubMed] [Google Scholar]
- 59.Zeng Y. and Cullen,B.R. (2002) RNA interference in human cells is restricted to the cytoplasm. RNA, 8, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]