Abstract
Despite the recent large-scale efforts dedicated to comprehensive phylogenetic analyses using mitochondrial and nuclear DNA sequences, several relationships among mammalian orders remain controversial. Here, we present an extensive application of retroposon (L1) insertion analysis to the phylogenetic relationships among almost all mammalian orders. In addition to demonstrating the validity of Glires, Euarchontoglires, Laurasiatheria, and Boreoeutheria, we demonstrate an interordinal clade that links Chiroptera, Carnivora, and Perissodactyla within Laurasiatheria. Re-examination of a large DNA sequence data set yielded results consistent with our conclusion. We propose a superordinal name “Pegasoferae” for this clade of Chiroptera + Perissodactyla + Carnivora + Pholidota. The presence of a single incongruent L1 locus generates a tree in which the group of Carnivora + Perissodactyla associates with Cetartiodactyla but not with Chiroptera. This result suggests that incomplete lineage sorting of an ancestral dimorphism occurred with regard to the presence or absence of retroposon alleles in a common ancestor of Scrotifera (Pegasoferae + Cetartiodactyla), which was followed by rapid divergence into the extant orders over an evolutionarily short period. Accordingly, Euungulata (Cetartiodactyla + Perissodactyla) and Fereuungulata (Carnivora + Pholidota + Perissodactyla + Cetartiodactyla) cannot be validated as natural groups. The interordinal mammalian relationships presented here provide a cornerstone for future studies in the reconstruction of mammalian classifications, including extinct species, on evolution of large genomic sequences and structure, and in developmental analysis of morphological diversification.
Keywords: intron, long interspersed element 1, mammalian phylogeny
Interordinal phylogenetic relationships of eutherian mammals have been well analyzed from both morphological and molecular points of view (1–5). Comprehensive analyses of large collections of DNA sequences mostly reject the conclusions from morphological analyses (6), and recent analyses of nuclear gene sequences suggest that 18 orders of extant eutherian mammals can be grouped into four major groups, namely, Euarchontoglires (Primates + Dermoptera + Scandentia + Rodentia + Lagomorpha), Laurasiatheria (Cetartiodactyla + Perissodactyla + Carnivora + Chiroptera + Pholidota + Eulipotyphla), Xenarthra, and Afrotheria (Proboscidea + Sirenia + Hyracoidea + Afrosoricida + Tubulidentata + Macroscelidea) (5). However, many of the interordinal relationships within each of the superorders remain unclear even after extensive molecular analysis (7).
Among these orders, the phylogenetic positions of Perissodactyla (odd-toed ungulates; horses and allies) and Chiroptera (bats) are two interesting issues that are not settled at present. Based on morphological data (2), Perissodactyla was originally grouped with other hoofed animals in the clade Ungulata, which includes Cetartiodactyla (whales, even-toed ungulates), Tubulidentata (aardvarks), Proboscidea (elephants), Sirenia (dugongs and manatees) and Hyracoidea (hyraxes). Subsequent molecular studies demonstrated that ungulates can be included in two groups, Afrotheria and Laurasiatheria (3–5). Although the monophyly of Laurasiatheria is strongly supported by nuclear gene sequence analyses, the relationships within Laurasiatheria, in which Perissodactyla is included, remain ambiguous. The existence of Fereuungulata (Carnivora + Pholidota + Perissodactyla + Cetartiodactyla) seems to be generally accepted, although there is no firm evidence from molecular data. Furthermore, it is of interest to determine whether either of the clades Euungulata (Perissodactyla + Cetartiodactyla) or Zooamata (Prissodactyla + Carnivora + Pholidota) is valid (8) with respect to the evolutionary origin of hooves in mammals.
The phylogenetic position of Chiroptera has been another controversial issue. Although many morphological studies place Chiroptera within the superorder Archonta (Primates + Dermoptera + Scandentia + Chiroptera) (1, 2), DNA sequence analysis rejects the clade and places the order within Laurasiatheria (5). However, the phylogenetic position of Chiroptera in Laurasiatheria frequently changes in molecular phylogenetic analyses of both mtDNA and nuclear gene data, depending on the combination of genes used in the analysis. Pumo et al. (9) presented strong evidence based on mitochondrial genome data that Chiroptera is closely related to Fereuungulata, and that report showed that Archonta can no longer be considered a natural group. Although those investigators place Chiroptera as a sister group to Fereuungulata, support for the monophyly of Fereuungulata is based on a bootstrap probability (BP) of ≈90%, and thus there is some uncertainty for this grouping. A recent analysis with a larger number of DNA sequences, however, appears to support the monophyly of Fereuungulata (7, 8). On the other hand, several groups (10, 11) have suggested the monophyly of Insectiphillia (Chiroptera + Eulipotyphla). Therefore, the phylogenetic positions of both Perissodactyla and Chiroptera are historically important issues to be settled.
Retroposons propagate their copies via reverse transcription of their RNA intermediates in a host genome (12, 13). To date, no mechanism has been described for the reversal of retroposon integration, and it is highly unlikely that the same type of retroposon would integrate into the same genomic locus independently in different lineages. Because of these characteristics, retroposons are quite useful as nearly homoplasy-free phylogenetic markers (14, 15) and thus have been used to resolve phylogenetic relationships of various mammalian species, such as cetartiodactyls (16–18), primates (19–21), and afrotherians (22).
We performed in silico screening of annotated whole genomic data for long interspersed element 1 (L1) sequences present in introns for use in insertion analysis. L1s are retroposons, and a major long interspersed element (LINE) family distributed in all mammalian genomes (23). Because L1 sequences occupy a large portion (16%) of the human genome (24), L1s are one of the best-characterized LINEs and thus have been analyzed in detail (25). Mammalian L1s are classified into >50 subfamilies based on diagnostic motifs and nucleotides in their 3′ UTR region (26, 27). Additionally, although the full length of L1 is ≈6 kb, many L1 sequences in genomes are present as partial elements, the lengths of which vary by several hundred base pairs (28). This partial L1 is caused by truncation, in which synthesis of L1 cDNA from the 3′ UTR is terminated during the integration process. These characteristics of L1s make them almost completely free of homoplasy, because it is unlikely that insertions of a member of a certain L1 subfamily occurred independently in the same orthologous locus with the same truncated length in different lineages during evolution.
Comparative analysis of orthologous DNA sequences among mammalian orders has been recently performed on a large scale, and several retroposon-inserted loci that may be phylogenetically informative have been isolated (29–31). However, all of the data provided by these analyses are not sufficient to delineate certain interordinal relationships because of the small number of mammalian species compared. For example, Thomas et al. (29) provided a few loci in which retroposon insertions are shared by artiodactyls and carnivores but not by primates and rodents, but their phylogenetic implications are still ambiguous because retroposon presence or absence in other orders of Laurasiatheria, such as Perissodactyla, Chiroptera, and Eulipotyphla, is unknown.
In the present study, we performed a comprehensive comparison of many orthologous retroposon loci among all orders of eutherian mammals (except for Pholidota) to reconstruct interordinal mammalian phylogeny and especially to address the phylogenetic positions of Perissodactyla and Chiroptera. We demonstrate that these orders are closer than previously expected, forming a clade with Carnivora.
Results and Discussion
Validity of the L1 Insertion Method by Showing Further Evidence for Monophyly for Euarchontoglires, Laurasiatheria, and Boreoeutheria.
Because the divergence periods of the major orders of eutherian mammals occurred 70–110 million years ago, it is, in general, extremely difficult to obtain orthologous sequences by PCR from such distantly related mammalian species. To avoid that difficulty, we compared L1 insertions in PCR-amplified introns by using primers for the 3′ and 5′ flanking exons of four mammalian species (human, mouse, dog, and cow) according to the procedure shown in Fig. 1 (see Materials and Methods). By comparing L1 insertions at 192 loci among various mammals, we found 44 parsimony-informative loci that indicated interordinal phylogenetic relationships (Table 2, which is published as supporting information on the PNAS web site). Among them, nine loci support the monophyly of Euarchontoglires (Fig. 2), clearly rejecting other hypotheses (P < 0.001, calculated according to ref. 8). For three loci, we confirmed that L1 insertions were found in all five orders of Euarchontoglires (Table 2 and Fig. 4A, which are published as supporting information on the PNAS web site). This finding is evidence of retroposon insertions that confirms the monophyly of Euarchontoglires.
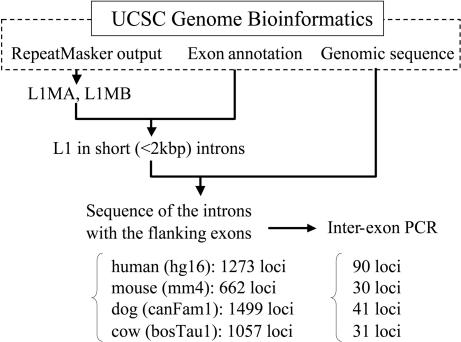
Fig. 1.
The in silico screening of genomic data for L1 sequences in introns performed in this study by using the databases available from University of California Santa Cruz Genome Bioinformatics (http://genome.ucsc.edu). The number of L1 insertions found and the number of loci used in interexonic PCR are shown for each species. The L1MA and L1MB are L1 subfamilies used for insertion comparison in this study. hg16, mm4, canFam1, and bosTau1 denote the database versions of human, mouse, dog, and cow, respectively.
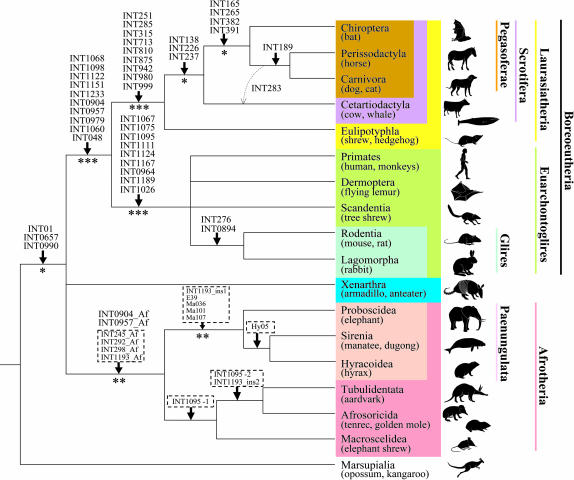
Fig. 2.
An interordinal mammalian phylogeny reconstructed by our retroposon insertion analysis. Downward arrows denote insertions of retroposons into each lineage. Locus INT283, denoted by a dashed arrow, supports the monophyly of Cetartiodactyla, Perissodactyla, and Carnivora. The loci surrounded by dashed lines in Afrotheria were identified in our previous study (22). Asterisks below the branches denote that the monophylies are statistically significant (∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
In addition, we found two loci, INT276 and INT0894, at which insertions of L1MA6 and MLT1A0 are shared in rodents and rabbit, indicating the monophyly of Glires (Figs. 2 and 4B). The positions of Rodentia and Lagomorpha by nuclear DNA and mtDNA analyses are still controversial (5, 32, 33), whereas morphological approaches support the monophyly of Glires (1, 2) with >10 morphological synapomorphic characters (34). Our retroposon data for two loci support the monophyly of Glires as well.
We discovered nine loci that indicate the monophyly of Laurasiatheria (Figs. 2 and 4C). Although the monophyly of Laurasiatheria is currently supported by a nuclear gene analysis (5), it is ambiguous with regard to mtDNA data because the phylogenetic position of the hedgehog lineage often comes to a root of eutherians (32). Our present data strongly support the result of nuclear DNA sequence analysis, confirming that the hedgehog is also within Laurasiatheria by one locus (INT713). The conflicting result from mtDNA sequence analysis is probably caused by the high substitution rates of mtDNA and analytical artifacts (35). We characterized many informative L1 loci supporting the monophyly of Laurasiatheria (P < 0.001; ref. 8), implying that this clade existed for a relatively long period. It is interesting to note that Laurasiatherian species are extremely diverse morphologically and that morphological studies found no synapomorphy for the clade, suggesting that future paleontological analysis of extinct species may recognize an ancestral Laurasiatherian species. The validity of Laurasiatheria clearly rejects the Archonta hypothesis proposed by morphological studies (1, 2).
We also found 10 loci that parsimoniously indicate the monophyly of Boreoeutheria (Euarchontoglires + Laurasiatheria; Figs. 2 and 4D). Morphological analyses have not suggested the monophyly of the clade (2, 6), whereas comprehensive analyses of a large number of nuclear DNA sequences support the clade (5). The monophyly of the group has not been supported by mtDNA analysis (32) because of the misleading positions of hedgehog and rodents, which tend to be regarded as basal lineages of Eutheria. In addition to recent molecular phylogenetics, our retroposon insertion data provide conclusive evidence for the monophyly of Boreoeutheria (P < 0.001; ref. 8).
We previously analyzed the Afrotherian clade in detail (22). In the present study, we isolated two additional loci in which AfroSINEs (Afrotheria-specific retroposons) are inserted in the genome of a common ancestor of Afrotheria. These six loci in total constitute convincing evidence for the Afrotheria clade (P < 0.01; ref.8 and Fig. 2).
Pegasoferae: An Unexpected Clade Within Laurasiatheria.
As described in the Introduction, the interordinal relationships within Laurasiatheria have not been resolved. We provide here three loci, which strongly indicate that Eulipotyphla diverged first in Laurasiatheria (P < 0.05; ref. 8 and Figs. 2 and 4E). Our L1 insertion patterns validate the monophyly of Scrotifera (Chiroptera + Carnivora + Pholidota + Perissodactyla + Cetartiodactyla) proposed by Waddell et al. (36). Remarkably, four loci (INT165, INT265, INT382, and INT391) indicate the monophyly of Carnivora, Perissodactyla, and Chiroptera, excluding Cetartiodactyla and Eulipotyphla (Fig. 3 and Table 2). It should be noted that these insertions are inconsistent with the Fereuungulata clade (36). The bootstrap values to support Fereuungulata (32, 37) are not high; thus, the validity remains controversial (7). The present analysis provides conclusive evidence that Chiroptera is nested deeply within Laurasiatheria, and we named this clade Pegasoferae (see below).
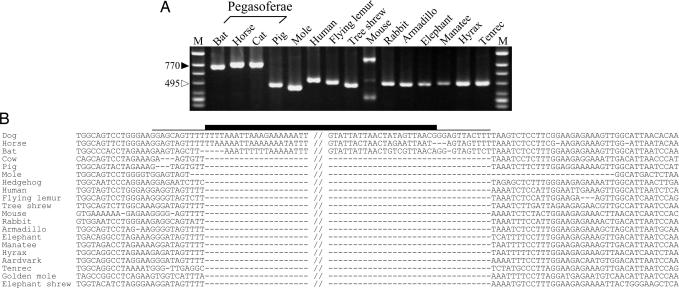
Fig. 3.
An example of INT391 locus suggesting Pegasoferae clade. (A) An electrophoretic profile of PCR products of locus INT391, in which L1 is present in horse, cat, and bat, but not other mammals. The larger size of the mouse PCR product is caused by species-specific insertions of another retrotransposon (database position chr19:55196706–55197619 in mm7). M, size markers (øX174/HincII digest). (B) An alignment of locus INT391 sequences. Thick and thin lines denote the L1 and the direct repeat sequences, respectively, that were generated during integration. The central region of the inserted L1 sequence is omitted.
Reanalysis of the Nuclear Sequence Data of Murphy et al.
Murphy et al. (5) have made an important contribution to elucidating phylogenetic relationships of major mammalian lineages. Because the interordinal relationships in Laurasiatheria are still ambiguous (7), we here reanalyzed their nuclear sequence data about the laurasiatherian species. All 105 possible trees among Chiroptera, Eulipotyphla, Carnivora, Perissodactyla, and Cetartiodactyla, with Euarchontoglires as an outgroup, were examined. The maximum-likelihood (ML) tree for Murphy’s 10,666-site data set (which excludes indels and missing data) is Tree-1, as shown in Table 1, which supports the Chiroptera/Perissodactyla/Carnivora clade (although with only a 35% BP; Fig. 5, which is published as supporting information on the PNAS web site). This tree coincides with the tree reconstructed by our L1 analysis including the monophyly of Carnivora, Perissodactyla, and Chiroptera (Fig. 2). Although Tree-1 is the ML tree for the 10,666-site data set, other trees are also nearly equally likely from this data set (Table 1). The BPs for the Chiroptera + Carnivora + Perissodactyla clade were 35% and 12%, respectively, from the Murphy’s 10,666- and 16,671-site data (Fig. 6, which is published as supporting information on the PNAS web site), and those for Fereuungulata were 17% and 37% for these data (Figs. 5 and 6). Murphy et al. (5) suggested the Cetartiodactyla + Perissodactyla + Carnivora + Pholidota clade, but bootstrap support was only 59%. Although Baysesian posterior probability for this clade was 98%, it is well known that it tends to give overconfidence (8, 38). Thus, the data from Murphy et al. (5) do not contain enough information to settle the issue of the position of Chiroptera relative to Fereuungulata.
Table 1.
Comparison of alternative trees among Eulipotyphla (Eul), Chiroptera (Chi), Carnivora (Car), Cetartiodactyla (Cetart), and Perissodactyla (Per) with Euarchontoglires as an outgroup
| Tree topology | 10,666 sites (5) | 16,671 sites (5) | Mitochondrial amino acids | Mitochondrial codons |
|---|---|---|---|---|
| 1. (Eul,((Chi,(Per,Car)),Cetart)) | 〈−65,312.6〉 | −3.0 ± 8.1 | −23.8 ± 8.8 | −23.4 ± 9.2 |
| 2. (Eul,(Chi,(Cetart,(Per,Car)))) | −0.5 ± 2.2 | −0.5 ± 6.8 | 〈−44,796.2〉 | 〈−118,056.2〉 |
| 3. (Eul,(Chi,((Cetart,Per),Car))) | −7.1 ± 6.4 | 〈−101,887.3〉 | −3.2 ± 9.4 | −3.5 ± 7.9 |
| 4. (Eul,((Chi,Cetart),(Per,Car))) | −0.6 ± 2.2 | −3.2 ± 8.0 | −16.1 ± 10.7 | −12.8 ± 11.7 |
| 5. (Eul,((Chi,Car),(Cetart,Per))) | −4.3 ± 7.2 | −0.6 ± 3.9 | −20.7 ± 14.3 | −25.5 ± 13.6 |
| 6. (Eul,((Chi,(Cetart,Car)),Per)) | −8.6 ± 6.3 | −8.1 ± 7.0 | −26.7 ± 14.8 | −29.3 ± 13.7 |
| 7. (Eul,((Chi,(Cetart,Per)),Car)) | −6.2 ± 6.7 | −1.3 ± 3.6 | −24.4 ± 13.7 | −25.9 ± 13.6 |
| 8. (Eul,(((Chi,Car),Cetart),Per)) | −6.2 ± 6.9 | −8.8 ± 7.2 | −21.8 ± 15.6 | −30.7 ± 14.1 |
| 9. (Eul,(((Chi,Cetart),Per),Car)) | −7.7 ± 6.2 | −9.2 ± 6.7 | −21.8 ± 14.0 | −19.5 ± 14.6 |
| 10. (Eul,((Chi,Per),(Cetart,Car))) | −6.6 ± 6.4 | −2.4 ± 8.3 | −35.2 ± 12.6 | −33.3 ± 12.8 |
| 11. (Eul,(((Chi,Cetart),Car),Per)) | −8.3 ± 6.2 | −9.8 ± 7.1 | −20.3 ± 15.1 | −18.0 ± 15.0 |
| 12. (Eul,(((Chi,Car),Per),Cetart)) | −4.8 ± 6.0 | −7.9 ± 7.4 | −30.7 ± 13.0 | −35.0 ± 12.6 |
| 13. (Eul,(((Chi,Per),Car),Cetart)) | −5.6 ± 5.8 | −3.1 ± 8.7 | −36.4 ± 12.1 | −36.4 ± 12.3 |
| 14. (Eul,(((Chi,Per),Cetart),Car)) | −5.6 ± 6.7 | −3.3 ± 8.3 | −32.4 ± 13.4 | −34.5 ± 13.4 |
| 15. (Eul,(Chi,((Cetart,Car),Per))) | −9.1 ± 5.9 | −6.2 ± 5.1 | −10.8 ± 7.1 | −8.1 ± 6.4 |
The log-likelihood of the ML tree is given in angled brackets, and the differences in the log-likelihoods of alternative trees from that of the ML tree ± 1 SE were estimated by using the formula of Kishino and Hasegawa (53). Comparison of trees other than the Eulipotyphla-basal in Laurasiatheria and P values of several tests are given in Tables 3–6, which are published as supporting information on the PNAS web site.
The mitochondrial genome analysis suggested Tree-2: (Eulipotyphla, (Chiroptera, (Cetartiodactyla, (Perissodactyla, Carnivora)))) as the most likely tree (Tables 5 and 6), irrespective of whether the analysis was performed at the amino acid or codon level; moreover, that mitochondrial analysis suggested that Tree-1 is unlikely, even though it was suggested by our L1 insertion analysis. Tree-1 was rejected with the most conservative test (39), although it was marginally at the 5% level (Tables 5 and 6). However, the monophyly of Fereuungulata, excluding Chiroptera as an outgroup, was supported with only 87% and 84% BP from the amino acid and codon analyses, respectively (Fig. 7, which is published as supporting information on the PNAS web site), and the position of Chiroptera in Laurasiatheria also remained uncertain from the mitochondrial genome analyses.
Characterization of an Inconsistent Locus.
In addition to the four loci that support the Pegasoferae clade, we isolated one L1 locus (INT283) that indicates the monophyly of Carnivora, Perissodactyla, and Cetartiodactyla, supporting Fereuungulata. The probability of homoplasy can be considered extremely low for L1 insertions, and the same target-site duplications (that are produced in the process of integration) were clearly observed for both patterns of the insertions (Figs. 3B and 4F). Thus, this inconsistency cannot be explained by homoplasy, and both of the incongruent trees are convincing gene trees (40). If clear homoplasy-free genetic markers show incongruent trees, it can be interpreted that lineage sorting of polymorphism in a common ancestor of the species was incomplete. Namely, the L1 insertion at locus INT283 occurred in the genome of a common ancestor of Scrotifera, and the ancestral dimorphism of alleles containing or lacking L1 had been retained in the population during the divergence of at least three lineages such as Cetartiodactyla, Chiroptera, and a group of Carnivora + Perissodactyla, followed by random fixation of the alleles. Accordingly, the time period in which the divergences occurred is relatively shorter than the coalescence time of the ancestral population (22, 40). Waddell et al. (8) proposed a likelihood analysis to estimate significance for the lineage supported by retroposon method. According to the estimation, the clade supported by four loci is statistically significant (P = 0.025; ref. 8) despite the existence of one incongruent locus. Therefore, it seems likely that the monophyly of Carnivora, Perissodactyla, and Chiroptera reflect the true species’ relationships, and Fereuungulata (including Carnivora, Perissodactyla and Chiroptera) supported by one locus can no longer be considered a robust clade.
Phylogenetic Positions of Perissodactyla and Chiroptera.
For one locus (INT189), the L1 insertion pattern indicated the monophyly of Carnivora and Perissodactyla (Figs. 2 and 4G). Although this clade, named Zooamata (41), is favored by recent molecular analyses of nuclear DNA, BP supports for the clade are relatively low in many cases (5). Although Schwartz et al. (30) provided one locus in which L1 is present in Carnivora and Perissodactyla but not in Cetartiodactyla, it is not enough to suggest the monophyly of Zooamata because it remains unknown whether the L1 is present or absent in the ortholog of Chiroptera. INT189 demonstrates the close relationship between Perissodactyla and Carnivora.
Regarding the phylogenetic position of Perissodactyla, an alternative hypothesis of Euungulata composed of Cetartiodactyla and Perissodactyla has also been proposed based on molecular analyses of concatenated nuclear DNA and mtDNA sequences (8). However, we did not isolate a locus that supports Euungulata, and instead, the five loci described above (INT165, INT265, INT382, INT391, and INT283) are obviously incompatible with this hypothesis. Therefore, Euungulata cannot be validated.
According to the molecular studies to date, including ours, Cetartiodactyla and Perissodactyla are included in Laurasiatheria, whereas other ungulates are in Afrotheria. Regarding Afrotheria, we have suggested that Tubulidentata is not close to the clade of Proboscidea + Sirenia + Hyracoidea (22). Furthermore, in the present study, we provided five loci that reject the monophyly of Cetartiodactyla and Perissodactyla. Therefore, the superordinal clade Ungulata proposed by morphological studies is polyphyletic in eutherian mammals, and the multiple morphological synapomorphic characters for the clade, such as those described by Shoshani and McKenna (2), must have independently evolved in each lineage.
Conclusion
Together with a recent retroposon analysis by Kriegs et al. (42), we here confirm the three major superordinal clades of eutherian mammals (Euarchontoglires, Laurasiatheria, and Afrotheria) and the Boreoeutheria clade (Euarchontoglires + Laurasiatheria). Addressing relationships within Laurasiatheria, the first divergence of Eulipotyphla is strongly supported, indicating the monophyly of Scrotifera. For intrarelationships in Scrotifera, the monophyly of Carnivora, Perissodactyla, and Chiroptera is supported by four loci.
We also show that the extensive sequence analyses conducted to date do not provide enough resolution regarding interordinal relationships in Laurasiatheria. We reanalyzed the data from Murphy et al. (5) with the ML method, and a subset of the data (excluding indels and missing sites data) gave the L1 tree in Fig. 2 as the ML tree although the support of the Chiroptera + Perissodactyla + Carnivora clade from the sequence analysis had only a 35% BP. However, our L1 insertion analysis was able to resolve this ambiguity, and we found an unexpected clade comprising Chiroptera, Perissodactyla, Carnivora, and Pholidota. Although Pholidota was not included in the present work, many other molecular studies (3–5, 8, 37) gave strong evidence for the Carnivora + Pholidota grouping. We propose the name Pegasoferae for this clade, because Pegasus refers to the flying horse (Chiroptera + Perissodactyla) in Greek mythology and Ferae refers to the monophyletic clade of Carnivora + Pholidota (41, 43). Interestingly, Waddell and Shelley (44) also obtained the Pegasoferae clade in the Bayesian maximum posterior probability tree (figure 10 of ref. 44) of their concatenated sequences of nuclear and mitochondrial genes, the data for which are mostly independent of Murphy et al. (5). Waddell and Shelley, however, did not give much attention to this finding.
The interordinal mammalian phylogeny presented here will contribute to the reconstruction of mammalian classifications and elucidation of the history of their diversification, including placement of extinct species. Furthermore, the discovery of an interordinal clade that links Chiroptera, Carnivora, and Perissodactyla will be useful for shedding light on the origin of each flying and hoofed mammal and the developmental mechanism of their morphological specialization from the inclusive viewpoints of paleontology, anatomy, and developmental biology.
Materials and Methods
Strategy for the L1 Insertion Method.
We collected all L1s present in short (<2 kb) introns from the annotated genomic database of four mammalian species (human, mouse, dog, and cow) according to the computational procedure shown in Fig. 1. First, we obtained annotated data files of the gene exon/intron structures (refFlat) and repetitive elements (chr*_rmsk; output of repeatmasker) for each species, human (data version hg16), mouse (mm4), dog (canFam1), and cow (bosTau1), from University of California Santa Cruz Genome Bioinformatics (http://genome.ucsc.edu) (45). The data for the L1MA and L1MB subfamilies were extracted from the annotation files on repetitive elements because these subfamilies have been suggested to have propagated 50–150 million years ago, thereby covering the divergence period of the major eutherian orders (24, 46). By comparing the annotation data of genes with those of L1, information on L1s that are present in introns <2 kb was collected. Next, we extracted the sequences of the introns and the 3′ and 5′ flanking exons from whole genomic sequence databases. Thus, we obtained 1,273, 662, 1,499, and 1,057 intronic loci, which included members of L1MA or L1MB subfamilies, from human, mouse, dog, and cow databases, respectively. For mouse, we also screened for MLT sequences, namely mammalian LTR-retrotransposons, in short introns.
To determine the presence or absence of L1 sequences among mammalian species, we selected 90, 30, 41, and 31 L1 loci from human, mouse, dog, and cow, respectively, to perform interexonic PCR. The species samples and databases that we used in this study are shown in Table 7, which is published as supporting information on the PNAS web site. The primer sequences used for each locus are shown in Table 8, which is published as supporting information on the PNAS web site. All PCR products were sequenced, and the presence or absence of L1 was confirmed as shown in Table 2. The sequences determined in this study have been deposited in GenBank (accession nos. AB258671–AB258977).
Reanalysis of a Large Gene Sequence Data Set.
The data set of Murphy et al. (5), consisting primarily of nuclear gene sequences, was reanalyzed with particular attention to the relationships within Laurasiatheria by using Euarchontoglires as an outgroup. We used sequences from the following 17 species: (i) Chiroptera, phyllostomid microbat, free tailed bat, and rousette fruit bat; (ii) Perissodactyla, horse and rhinoceros; (iii) Cetartiodactyla, dolphin, hippopotamus, and pig; (iv) Carnivora, cat and caniform, (v) Eulipotyphla, mole and shrew; and (vi) outgroup (Euarchontoglires), sciurid, mouse, pika, strepsirrhine, and human. The data were analyzed in two different ways, either including or excluding indels and missing data sites, with 16,671 or 10,666 sites, respectively. The baseml program in the paml package (version 3.14; ref. 47) was used to analyze the nucleotide sequences with the GTR + Γ model (48, 49).
We also analyzed concatenated 12 protein-encoding gene sequences in the same strand of mtDNA of 22 species from Chiroptera, Perissodactyla, Cetartiodactyla, Carnivora, Eulipotyphla, and Euarchontoglires (listed in Table 9, which is published as supporting information on the PNAS web site). Positions with gaps and regions of ambiguous alignment were excluded. The total number of remaining sites was 10,737 (3,579 codons). The codeml program in the paml package (version 3.14; ref. 47) was used to analyze the sequences both at the amino acid level with the mtREV + Γ model (49, 50) and at the codon level with the codon-substitution + Γ model (51, 52).
Supplementary Material
Acknowledgments
We thank Ueno Zoological Gardens (Tokyo) for tissue samples of Pteropus dasymallus, Dasypus novemcinctus, and Myrmecophaga tridacctyla; Ying Cao (The Institute of Statistical Mathematics) for the alignment of mtDNA sequences; and Prof. Dan Graur (University of Houston, Houston) for grammatically revising the name of the clade “Pegasoferae,” which was originally suggested by M.H. This work was supported by research grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N.O.).
Abbreviations
- LI
long interspersed element 1
- ML
maximum likelihood
- BP
bootstrap probability.
Footnotes
References
- 1.Novacek M. J. Nature. 1992;356:121–125. doi: 10.1038/356121a0. [DOI] [PubMed] [Google Scholar]
- 2.Shoshani J., McKenna M. C. Mol. Phylogenet. Evol. 1998;9:572–584. doi: 10.1006/mpev.1998.0520. [DOI] [PubMed] [Google Scholar]
- 3.Madsen O., Scally M., Douady C. J., Kao D. J., DeBry R. W., Adkins R., Amrine H. M., Stanhope M. J., de Jong W. W., Springer M. S. Nature. 2001;409:610–614. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- 4.Murphy W. J., Eizirik E., Johnson W. E., Zhang Y. P., Ryder O. A., O’Brien S. J. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 5.Murphy W. J., Eizirik E., O’Brien S. J., Madsen O., Scally M., Douady C. J., Teeling E., Ryder O. A., Stanhope M. J., de Jong W. W., et al. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 6.Novacek M. J. Curr. Biol. 2001;11:R573–R575. doi: 10.1016/s0960-9822(01)00347-5. [DOI] [PubMed] [Google Scholar]
- 7.Murphy W. J., Pevzner P. A., O’Brien S. J. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Waddell P. J., Kishino H., Ota R. Genome Inform. Ser. Workshop Genome Inform. 2001;12:141–154. [Google Scholar]
- 9.Pumo D. E., Finamore P. S., Franek W. R., Phillips C. J., Tarzami S., Balzarano D. J. Mol. Evol. 1998;47:709–717. doi: 10.1007/pl00006430. [DOI] [PubMed] [Google Scholar]
- 10.Mouchaty S. K., Gullberg A., Janke A., Arnason U. Mol. Biol. Evol. 2000;17:60–67. doi: 10.1093/oxfordjournals.molbev.a026238. [DOI] [PubMed] [Google Scholar]
- 11.Nikaido M., Kawai K., Cao Y., Harada M., Tomita S., Okada N., Hasegawa M. J. Mol. Evol. 2001;53:508–516. doi: 10.1007/s002390010241. [DOI] [PubMed] [Google Scholar]
- 12.Rogers J. H. Int. Rev. Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- 13.Weiner A. M., Deininger P. L., Efstratiadis A. Annu. Rev. Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 14.Shedlock A. M., Okada N. BioEssays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 15.Rokas A., Holland P. W. Trends Ecol. Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 16.Shimamura M., Yasue H., Ohshima K., Abe H., Kato H., Kishiro T., Goto M., Munechika I., Okada N. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido M., Rooney A. P., Okada N. Proc. Natl. Acad. Sci. USA. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido M., Matsuno F., Hamilton H., Brownell R. L., Jr., Cao Y., Ding W., Zuoyan Z., Shedlock A. M., Fordyce R. E., Hasegawa M., et al. Proc. Natl. Acad. Sci. USA. 2001;98:7384–7389. doi: 10.1073/pnas.121139198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitz J., Ohme M., Zischler H. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salem A. H., Ray D. A., Xing J., Callinan P. A., Myers J. S., Hedges D. J., Garber R. K., Witherspoon D. J., Jorde L. B., Batzer M. A. Proc. Natl. Acad. Sci. USA. 2003;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roos C., Schmitz J., Zischler H. Proc. Natl. Acad. Sci. USA. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishihara H., Satta Y., Nikaido M., Thewissen J. G., Stanhope M. J., Okada N. Mol. Biol. Evol. 2005;22:1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- 23.Fanning T. G., Singer M. F. Biochim. Biophys. Acta. 1987;910:203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- 24.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 25.Moran J. V., Gilbert N. In: Mobile DNA II. Craig N. L., Craigie R., Gellert M., Lambowitz A. M., editors. Washington, DC: Am. Soc. Microbiol.; 2002. pp. 836–869. [Google Scholar]
- 26.Smit A. F., Toth G., Riggs A. D., Jurka J. J. Mol. Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- 27.Hardies S. C., Wang L., Zhou L., Zhao Y., Casavant N. C., Huang S. Mol. Biol. Evol. 2000;17:616–628. doi: 10.1093/oxfordjournals.molbev.a026340. [DOI] [PubMed] [Google Scholar]
- 28.Sassaman D. M., Dombroski B. A., Moran J. V., Kimberland M. L., Naas T. P., DeBerardinis R. J., Gabriel A., Swergold G. D., Kazazian H. H., Jr. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 29.Thomas J. W., Touchman J. W., Blakesley R. W., Bouffard G. G., Beckstrom-Sternberg S. M., Margulies E. H., Blanchette M., Siepel A. C., Thomas P. J., McDowell J. C., et al. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz S., Elnitski L., Li M., Weirauch M., Riemer C., Smit A., Green E. D., Hardison R. C., Miller W. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bashir A., Ye C., Price A. L., Bafna V. Genome Res. 2005;15:998–1006. doi: 10.1101/gr.3493405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnason U., Adegoke J. A., Bodin K., Born E. W., Esa Y. B., Gullberg A., Nilsson M., Short R. V., Xu X., Janke A. Proc. Natl. Acad. Sci. USA. 2002;99:8151–8156. doi: 10.1073/pnas.102164299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Misawa K., Janke A. Mol. Phylogenet. Evol. 2003;28:320–327. doi: 10.1016/s1055-7903(03)00079-4. [DOI] [PubMed] [Google Scholar]
- 34.Novacek M. J., Wyss A. R. Cladistics. 1986;2:257–287. doi: 10.1111/j.1096-0031.1986.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 35.Nikaido M., Cao Y., Harada M., Okada N., Hasegawa M. Mol. Phylogenet. Evol. 2003;28:276–284. doi: 10.1016/s1055-7903(03)00120-9. [DOI] [PubMed] [Google Scholar]
- 36.Waddell P. J., Cao Y., Hauf J., Hasegawa M. Syst. Biol. 1999;48:31–53. doi: 10.1080/106351599260427. [DOI] [PubMed] [Google Scholar]
- 37.Amrine-Madsen H., Koepfli K. P., Wayne R. K., Springer M. S. Mol. Phylogenet. Evol. 2003;28:225–240. doi: 10.1016/s1055-7903(03)00118-0. [DOI] [PubMed] [Google Scholar]
- 38.Shimodaira H., Hasegawa M. In: Statistical Methods in Molecular Evolution. Nielsen R., editor. New York: Springer; 2005. pp. 463–493. [Google Scholar]
- 39.Shimodaira H., Hasegawa M. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- 40.Shedlock A. M., Takahashi K., Okada N. Trends Ecol. Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Waddell P. J., Okada N., Hasegawa M. Syst. Biol. 1999;48:1–5. [PubMed] [Google Scholar]
- 42.Kriegs J. O., Churakov G., Kiefmann M., Jordan U., Brosius J., Schmitz J. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna M. C., Bell S. K. Classification of Mammals Above Species Level. New York: Columbia Univ. Press; 1997. [Google Scholar]
- 44.Waddell P. J., Shelley S. Mol. Phylogenet. Evol. 2003;28:197–224. doi: 10.1016/s1055-7903(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 45.Karolchik D., Baertsch R., Diekhans M., Furey T. S., Hinrichs A., Lu Y. T., Roskin K. M., Schwartz M., Sugnet C. W., Thomas D. J., et al. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Springer M. S., Murphy W. J., Eizirik E., O’Brien S. J. Proc. Natl. Acad. Sci. USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez F., Oliver J. L., Marin A., Medina J. R. J. Theor. Biol. 1990;142:485–501. doi: 10.1016/s0022-5193(05)80104-3. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z. Trends Ecol. Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 50.Adachi J., Hasegawa M. J. Mol. Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- 51.Goldman N., Yang Z. Mol. Biol. Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 52.Yang Z., Nielsen R., Hasegawa M. Mol. Biol. Evol. 1998;15:1600–1611. doi: 10.1093/oxfordjournals.molbev.a025888. [DOI] [PubMed] [Google Scholar]
- 53.Kishino H., Hasegawa M. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.