Abstract
Huge amounts of genomic information are currently being generated. Therefore, biologists require structured, exhaustive and comparative databases. The PyloriGene database (http://genolist.pasteur.fr/PyloriGene) was developed to respond to these needs, by integrating and connecting the information generated during the sequencing of two distinct strains of Helicobacter pylori. This led to the need for a general annotation consensus, as the physical and functional annotations of the two strains differed significantly in some cases. A revised functional classification system was created to accommodate the existing data and to make it possible to classify coding sequences (CDS) into several functional categories to harmonize CDS classification. The annotation of the two complete genomes was revised in the light of new data, allowing us to reduce the percentage of hypothetical proteins from ∼40 to 33%. This resulted in the reassignment of functions for 108 CDS (∼7% of all CDS). Interestingly, the functions of only ∼13% of CDS (222 out of 1658 CDS) were annotated as a result of work done directly on H.pylori genes. Finally, comparison of the two published genomes revealed a significant amount of size variation between corresponding (orthologous) CDS. Most of these size variations were due to natural polymorphisms, although other sources of variation were identified, such as pseudogenes, new genes potentially regulated by slipped-strand mispairing mechanism, or frame-shifts. 113 of these differences were due to different start codon assignments, a common problem when constructing physical annotations.
INTRODUCTION
Helicobacter pylori, a Gram-negative, spiral bacterium with a relatively small genome (∼1.65 Mb), is responsible for one of the most common bacterial infections, affecting ∼50% of the human population. This bacterium was first cultivated in 1982 and was subsequently found to be the causative agent of gastritis, and of gastric and duodenal ulcers (1,2). Helicobacter pylori is also considered to be a major risk factor for adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma of the stomach. Thus, the sequencing of the H.pylori genome was considered to be a major advance in the understanding of its unique lifestyle and the associated diseases. The fact that this single bacterial species is associated with several clinical outcomes suggests that all H.pylori strains are not equally pathogenic and virulent. Indeed, the outcome of H.pylori infection appears to depend, at least partially, on strain diversity (3,4) although host and environmental factors are also important (5–7 and references therein).
The availability of two complete genomic sequences (8,9) confirmed the panmictic structure of H.pylori, which is characterized by a high mutation rate (microdiversity) and free recombination (10–13). However, the apparent macrodiversity of H.pylori populations revealed by typing techniques relying on nucleotide sequence variations could be explained by a high variability at the third codon position. Comparison of the two sequences revealed that genetic organization was similar in both strains. Besides the cag pathogenicity island, which is known to be a variable region, 6–7% of the annotated genes are strain specific. Half of these strain-specific genes are clustered in a hypervariable region, known as a ‘plasticity zone’.
The publication of the second H.pylori genome sequence improved the functional assignment of the coding sequences (CDS), particularly as several other genomes had been published in the meantime. Alm et al. (9) also revised the physical annotation by reducing the total number of CDS from 1590 to 1552 in strain 26695. Since this time, no further annotations have been made available in public databases. Therefore, we decided to create a database (http://genolist.pasteur.fr/PyloriGene) integrating the genomes of both strains.
The main objective of the PyloriGene database is to continuously update the annotation of both genomes whenever new genomes and new literature that might improve the functional annotation of CDS are published. Parallel analysis of the two published H.pylori genomes revealed some inconsistencies in the physical annotation, functional assignments and functional classification of CDS that needed to be harmonized by establishing a consensus. This resulted in the definition of a single list of functional categories for both genomes and the establishment of principles of annotation. The re-annotated database generated was also used to update the annotations of the H.pylori protein–protein interaction map (14). PyloriGene release 1.6 available since January 2003 reflects this current analysis and re-annotation of both H.pylori genomes.
MATERIALS AND METHODS
The genomic sequences of strains 26695 and J99 were recovered from GenBank (AE000511 and AE001439). The annotated CDS were systematically analyzed by use of Smith and Waterman scanning reports (15) against a non-redundant protein database run on a multi-processor Paracel GeneMatcher, and available at the PyloriGene Web site (http://genolist.pasteur.fr/PyloriGene/) for each CDS. When judged pertinent, manual analysis was done using BLAST analysis (16) and the Lasergene 99 software package (DNASTAR Inc.). In general, we followed the annotation procedures proposed by Bork et al. (17).
The CDS annotations of strains 26695 (http://www.tigr.org/tigr-scripts/CMR2/gene_table.spl?db=ghp) and J99 (http://scriabin.astrazeneca-boston.com/first.html) that were used for our analysis were the ones available in October 2001 in each respective database. We verified that these annotations had not changed by October 2002. This analysis does not reflect changes that might have occurred since this time in either database. Furthermore, the physical annotation of CDS in the PyloriGene database corresponds to the deposited coordinates at the time of retrieval of sequences from GenBank except for the few genes removed by TIGR for strain 26695 database.
We used the original nomenclature of each gene for the purpose of the PyloriGene database, i.e. HPxxxx and JHPxxxx for genes from strain 26695 and J99, respectively. We decided to use the list of corresponding CDS (orthologous CDS in the two strains) published by Doig et al. (18) because this is the generally accepted reference for H.pylori. In total, there are 1658 CDS from the two genomes, 1408 that are present in both strains, 87 CDS present only in strain J99 and 163 CDS present only in strain 26695.
RESULTS AND DISCUSSION
Structure of the PyloriGene database
The PyloriGene database was constructed by use of a generic database model (WWW server v3.1) called ‘GenoList’. GenoList was derived from the work performed on the Colibri (19) and SubtiList (20,21) databases (http://genolist.pasteur.fr), which are dedicated to the genomes of Escherichia coli and Bacillus subtilis, respectively. This model has progressed from a fragmented view of bacterial genomes (before 1997) to an original data structure designed for the representation, manipulation and maintenance of complete microbial genomes. The conceptual data schema was implemented as a relational database using the Sybase® SQL DataBase Management System. The PyloriGene database schema also uses dedicated tables making it possible to define relationships between both H.pylori strain genomes. Thus, genes found in both strains can be identified.
A user-oriented interface for browsing and querying the database has also been developed. It is accessible on the Internet through a Web server that runs dynamic HTML pages, generated by a CGI script written with the C/C++ programming languages. The interface is aimed at providing the user with easy access to the data of interest, based on the most frequently asked questions. For example, chromosome regions defined around a gene of interest can easily be drawn (or represented as customizable gene lists), through a few mouse clicks. Detailed information on every single gene can then be displayed in another frame, whilst keeping the gene list or the drawing present on the screen. Similarly, the most common queries can be accessed directly at any time (e.g. gene name, chromosome region, functional classification). A highly efficient tool is directly accessible to search for DNA or protein patterns. When searching for these patterns, constraints, such as the genome region to be searched or the location of start and stop codons, can be defined and the results can be analyzed immediately in the context of the chromosome environment. BLAST and Smith-Waterman analyses are also available, making it possible to search the H.pylori data for a given sequence, or providing pre-calculated and regularly updated reports comparing each H.pylori gene against a non-redundant protein databank. Finally, gene function and product information can be searched through a multicriteria search tool, taking advantage of the database capabilities provided by the relational data model. Gene and protein sequences are readily accessible from any point during a query process or during sequence analysis.
Functional categories
To combine the data into a single database that directly links the corresponding CDS of the two strains, we decided to harmonize the functional categories for the following three reasons. First, each of the two H.pylori databases used different categories for the functional classification of genes (see Table S1 in Supplementary Material). Secondly, the annotations used did not necessarily place corresponding CDS into the same functional category, even when the same category was present in both classification schemes. For example, HP0220 or nifS was predicted to be an aminotransferase by AstraZeneca rather than a cysteine desulfurase. Some other examples include HP0384, HP0599, HP0748, HP0814, HP1264, HP1265, HP1560, etc. The divergent classification of orthologs sometimes reflects their multifunctional role, therefore, we decided to introduce a function making it possible to place any given CDS in more than one functional category (ex: NapA, GroES, FrxA, RdxA, FtsI or PBP3, etc., and see purine synthesis examples below). Finally, some of the categories and sub-categories seemed poorly suited in light of the biological characteristics of H.pylori.
The functional categories retained for the PyloriGene database were generally more similar to the TIGR classification established for strain 26695 (http://www.tigr.org/tigr-scripts/CMR2/gene_table.spl?db=ghp) than to that of AstraZeneca for strain J99 (http://scriabin.astrazeneca-boston.com/first.html). Therefore, the choice of functional category rearrangements affected mainly the classification of the CDS of strain J99. Some of these categories or sub-categories were deleted, renamed, split into several sub-categories or transferred into other categories (see Table S1). The three most divergent ‘AstraZeneca’ categories were ‘Cellular processes’, ‘DNA metabolism’ and ‘Translation’. All of the changes made are summarized in Table S1.
In the original annotations, a total of 404 pairs of corresponding CDS were placed in distinct functional groups. Most differences resulted either from divergence in the classification criteria (124 CDS) or from distinct organization of functional categories between the two annotation efforts (62 CDS). This latter group of CDS was re-classified manually one by one. From now on, we will only refer to the PyloriGene categories (designated numbers 1–17, see Table S1) in which all CDS were transferred (automatically and/or manually). The other differences were due to annotation conflicts (see Table 1).
Table 1. Summary of discrepancies in CDS annotations between strains 26695 and J99.
| Strain J99 | |||
|---|---|---|---|
| Strain 26695 | CDS with predicted function | Conserved hypothetical CDS | Hypothetical CDS specific to H.pylori |
| CDS with predicted function | 57 (33)a | 50b | 7 |
| Conserved hypothetical CDS | 16b | – | 12 |
| Hypothetical CDS specific to H.pylori | 29 | 71 | – |
aThese 57 CDS have a predicted but different proposed biological function in both annotation efforts. Of these 57 CDS, 33 were classified in different functional categories.
bAs an example, AstraZeneca proposed a biological function for these 16 CDS while TIGR considered them to be conserved hypothetical CDS. Inversely, 50 CDS with a function predicted by TIGR were considered to be conserved hypothetical by AstraZeneca.
For example, the annotation of strain 26695 classified most of the genes predicted to be involved in purine metabolism in sub-category 2.2, i.e. de novo synthesis (8). Knowledge of purine metabolism in enterobacteria (22), combined with the analysis of the J99 genome, led Doig et al. (18) to conclude that H.pylori does not have the capability to synthesize de novo purine ribonucleotides. Thus, most of these CDS were predicted to be involved exclusively in the salvage pathway and have been classified in sub-category 2.4. Indeed, sequence similarity analysis suggests that H.pylori only has a complete salvage pathway.
However, there is now biochemical evidence showing that H.pylori can sustain de novo purine synthesis (23). Furthermore, three CDS were annotated as homologs of the bifunctional PurB (HP1112/JHP1039), PurD (HP1218/JHP1140) and PurU (HP1434/JHP1327) proteins. These proteins have a role in de novo synthesis in other bacteria, suggesting that a complete, but as yet unidentified pathway, exists. Consequently, H.pylori may in fact be able to synthesize de novo purine ribonucleotide, so we classified any CDS predicted to be involved simultaneously in de novo nucleotide synthesis and the salvage pathway in both sub-categories: PurA (HP0255/JHP0239), GuaA (HP0409/JHP0972), GuaB (HP0829/JHP0768) and PurB (HP1112/JHP1039). The same rule is valid for some other CDS.
The analysis of sequence similarities also led to other incorrect predictions concerning the metabolic capabilities of H.pylori. For instance, genome analysis suggested that H.pylori did not have a complete traditional TCA cycle, but rather a branched anaerobic one working in a reductive mode. Recent studies, however, have demonstrated that H.pylori has an unusual, but complete, TCA cycle (24).
It is important to acknowledge the multifunctional role of some proteins, which is not compatible with restricting their classification to a single functional category. Failure to take this into account is reductionist and might be misleading, as it leads to the underestimation of metabolic capabilities and can mask the relationship between different metabolic pathways or protein networks in a given organism.
Functional assignments: principles of annotation
The study of H.pylori is relatively new and is highly focused in certain research areas (e.g. clinical, pathogenesis, etc.). Therefore, the annotation process is largely based on homology with other better-characterized organisms, which can raise some problems due to the relative genetic distance between H.pylori and E.coli or B.subtilis. Therefore, we decided to establish stringent annotation rules to raise the awareness of researchers to potentially biased annotations.
We created four main fields for each CDS. These were named ‘product’, ‘function’, ‘description’ and ‘comments’. These fields were completed according to our current knowledge about a gene and/or the biological function of the gene product, and about the current annotation status. Although we entirely revised the two genomes, our priority was the detailed annotation of genes of unknown function (see Table S2), of genes for which an annotation conflict existed (242 pairs of corresponding CDS, see summary in Table 1), or for genes that were relevant to the H.pylori protein–protein interaction map (14). Three situations were observed and different annotation rules were used for each of them.
The gene has been studied in H.pylori. In this case, we completed each major field with information from the H.pylori literature. References were attached to justify our comments (see http://genolist.pasteur.fr/PyloriGene). In some cases, when a CDS had not been characterized in detail in H.pylori, we also added references on better characterized homologs or from recent reviews, when available. In the release 1.6 of PyloriGene, ∼13% (222 out of 1658 CDS) of all genes correspond to genes that have been studied in H.pylori.
The gene has not been studied in H.pylori but homologs have been characterized in other organisms. As the gene product has not been studied in H.pylori, the function is predicted on the basis of sequence similarity. In general, we left the ‘function’ field empty, and completed the ‘product’, ‘description’ and ‘comments’ fields. This is a subjective choice that might be debatable. Thus, the name of the closest previously characterized homologous protein, preceded by the word ‘Predicted’, was entered into the ‘product’ field. In most cases, the closest homolog was from E.coli. The ‘comments’ field was used to summarize the function of the closest homologous protein. Any corresponding references were also cited to justify the annotation. These annotation rules were intended to emphasize that annotation based on similarity scores needs to be validated biologically in H.pylori.
The gene has not been studied in any organisms. This third category includes conserved hypothetical CDS and H.pylori-specific CDS of unknown function. In both cases, the ‘product’ and ‘function’ fields were left empty. The ‘description’ field contained ‘Predicted coding region HPxxxx’ or ‘Predicted coding region HPxxxx with no homolog in the databases’ for conserved hypothetical CDS and H.pylori-specific hypothetical CDS, respectively. In some cases, when the CDS revealed a particular structure, such as sequences characteristic of secretion, a lipoprotein anchoring motif or putative transmembrane domains, the ‘description’ field was filled accordingly (e.g. ‘predicted lipoprotein’ or ‘integral membrane protein’).
After systematically re-analyzing the two genomes, the number of conserved CDS of unknown function decreased to 233 (15%) for strain 26695 and to 228 (15.3%) for strain J99. The number of H.pylori-specific CDS of unknown function also decreased, to 334 (21.5%) for strain 22695 and to 278 (18.5 %) for strain J99. This significant drop is mainly due to the publication of the sequence of the related pathogen, Campylobacter jejuni (25).
For these CDS annotations, the literature and new genome sequences must be constantly followed. These literature searches mainly concern H.pylori and related organisms, such as C.jejuni. However, literature on model organisms, such as E.coli and B.subtilis, is also examined to update the CDS of unknown function.
Examples of new functional annotations based on similarity searches and analysis of recent literature
Manual analysis allowed us to propose new putative and/or demonstrated functions for 95 CDS, to reassign the functions of 13 CDS (see Table S2) and to identify a new gene (HP0037.1/JHP0033.1) (26). This represents ∼7% of all CDS from the two strains.
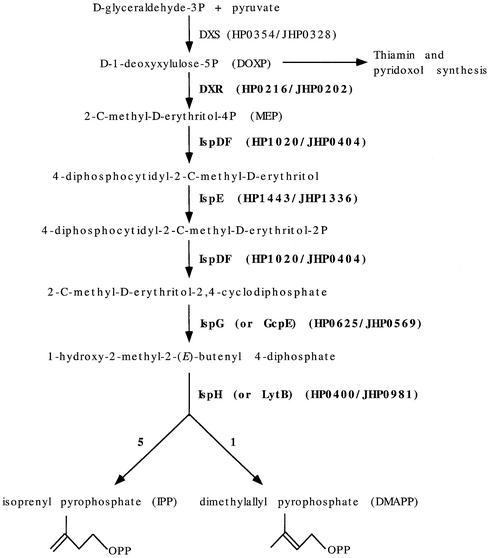
An example of conserved hypothetical CDS for which we were able to assign a function following systematic BLAST searches and literature reviews is isoprenoid synthesis. Isoprenoids comprise a highly diverse family of essential and secondary compounds. Their biosynthesis requires two ubiquitous building blocks: isoprenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP, see Fig. 1). In animals, fungi and some bacteria, IPP is synthesized via the mevalonate pathway. In chloroplasts, algae, cyanobacteria and other bacteria, including H.pylori, an alternative pathway, known as the deoxyxylulose-5-phosphate pathway, is used to synthesize IPP and DMAPP. A homolog of the protein involved in the first enzymatic step of this pathway was the only protein from this pathway to have been identified in H.pylori: deoxyxylulose-5-phosphate synthase (DXS) encoded by HP0354/JHP0328 (Fig. 1). Systematic BLAST searches and literature reviews led to the identification of homologs of the proteins involved in the second (DXR), third (IspD), fourth (IspE) and fifth (IspF) enzymatic steps described recently in E.coli (27–30). Interestingly, the IspD and IspF homologs are fused as a single polypeptide in H.pylori, and are encoded by HP1020/JHP0404. DXR and IspE are encoded by HP0216/JHP0202 and HP1443/JHP1336, respectively. Genomic distribution among organisms and subsequent studies showed that LytB (HP0400/JHP0981, previously annotated as being involved in penicillin tolerance) and GcpE (HP0625/JHP0569, previously annotated as being a protein of unknown function) are involved in the final steps of IPP and DMAPP synthesis, although their exact enzymatic activities remained unknown (31–33). Since then, the activities of GcpE and LytB have been characterized and the proteins have been renamed IspG and IspH, respectively (34,35).
Figure 1.
Identification of a complete deoxyxylulose-5-phosphate pathway for isoprenoid synthesis. The dexoxyxylulose-5-phosphate synthase (DXS) protein, for which a homolog had already been annotated in both H.pylori genomes (HP0354/JHP0328), is responsible for the first enzymatic step of the three distinct pathways that synthesize isoprenoid, thiamin and pyridoxol. Proteins involved in the subsequent enzymatic reactions had not been characterized biochemically in H.pylori or identified genetically in general. Therefore, the genome analysis data did not make it possible to draw any conclusions concerning the ability of H.pylori to synthesize de novo isoprenoid precursors, IPP and DMAPP. We were subsequently able to reconstitute the complete metabolic pathway for the synthesis of IPP and DMAPP by combining systematic sequence similarity searches and a review of recent literature. Note that the last step of IPP and DMAPP synthesis is catalyzed by the same protein (IspH) with a ratio of 5:1, respectively, in E.coli (35).
The analysis of the two genomes allowed other metabolic predictions to be made. For example, the biosynthesis of tyrosine and phenylalanine occurs via a common pathway up to the synthesis of chorismate (36). Homologs for all of the genes encoding proteins involved in the synthesis of chorismate have been identified in H.pylori. TyrA and PheA convert chorismate into tyrosine and phenylalanine precursors, respectively. A tyrA homolog (HP1380/JHP1294) has been annotated previously. However, no pheA homolog has been annotated in either strain. TyrA and PheA are bifunctional proteins. Both have an equivalent chorismate mutase domain that converts chorismate into prephenate, which is then converted into phenylpyruvate and 4-hydroxyphenylpyruvate by the second domain of PheA (prephenate dehydratase domain) and TyrA (prephenate dehydrogenase domain), respectively (36). Phenylpyruvate and 4-hydroxyphenylpyruvate are then converted into phenylalanine and tyrosine, respectively, by the aminotransferase TyrB.
The coding sequence HP0291/JHP0276 shows similarity to PheA and TyrA homologs, which is restricted exclusively to the chorismate mutase domain common to both proteins (36). HP0291/JHP0276 includes the entire chorismate mutase domain (PFAM01817) but lacks the prephenate dehydratase domain found in other organisms, which corresponds to the remaining two-thirds of PheA proteins. Interestingly, streptococci also seem to have PheA proteins restricted to the chorismate mutase domain. Therefore, these PheA variants are predicted to only synthesize prephenate. However, prephenate is an unstable precursor molecule that can be converted spontaneously into phenylpyruvate. Some phenylalanine auxotrophic E.coli without prephenate dehydratase activity compensate for their auxotrophy after prolonged culture due to this chemical conversion (37–39). Therefore, H.pylori might not need the prephenate dehydratase domain to synthesize phenylalanine, which would explain the contradictory results on H.pylori phenylalanine requirements and/or use (40,41). This functional assignment of HP0291/JHP0276 as pheA was only possible because BLAST searches were followed by manual assessment.
Predicting new functional assignments raised several issues. A large number of CDS had different predicted functions in the two genome annotations (57 CDS of which only 33 resulted in different functional classifications, see Table 1). This can be explained by the fact that in the time between the release of the first and second sequences, new sequence data and new functional characterizations of CDS became available, generally improving predictions of function for the annotation performed by AstraZeneca (9,18). The same is valid for the 45 hypothetical CDS for which AstraZeneca predicted a function in the meantime (Table 1).
Interestingly, although the publication of the second genome resulted in an improved annotation of both genomes, a small subset of CDS (57 CDS, see Table 1) had a predicted function assigned in strain 26695 that became hypothetical CDS in strain J99 (9,18). The level of sequence similarity of these CDS was too low to be considered as significant by these authors. The fact that H.pylori is distantly related to model organisms is often reflected by poor sequence conservation, in particular for small CDS. The annotation of 26695 by TIGR might have benefited from in-house sequences that were not publicly released at that time. Given our current knowledge of the physiology of H.pylori, most of these functional assignments were probably correct. Some of these CDS have subsequently been studied in H.pylori and their functional assignment confirmed. For example, HP0333 was annotated as a homolog of the Haemophilus influenza DrpA (42), whereas JHP0316 was deemed to be a conserved hypothetical CDS. Since then, two groups have shown that HP0333/JHP0316 indeed encodes a DrpA homolog involved in the natural transformation in H.pylori (43,44).
Physical annotation of the genomes
During our analysis of CDS pairs, several orphan CDS were found to actually have counterparts. A previously orphan CDS from strain J99 (JHP0533) was found to have a counterpart in strain 26695 that was not physically annotated (HP0595.1 and reported by SWISS-PROT; accession number P57798). Moreover, JHP1322 appeared to be split into two CDS in strain 26695 (a previously not annotated CDS, HP1439.1 and HP1439), due to a premature stop codon. Two orphan CDS in strain J99 (JHP0896 and JHP1306) were found to correspond to larger CDS in strain 26695 (HP1412 and HP0963, respectively). Similarly, three orphan CDS in strain 26695 (HP0678, HP0992 and HP1018) were found to correspond to three larger CDS in strain J99 (JHP0620, JHP0939 and JHP0405, respectively). Two orphan CDS (HP1369 and HP1370) were found to have a corresponding CDS previously annotated as a single orphan CDS, JHP1284. Finally, although the analysis by Doig et al. (18) included a secE homolog, which was not initially annotated physically either by Tomb et al. (8) or Alm et al. (9), this gene was never included in AstraZeneca’s database. Their identification of a secE homolog was recently confirmed in silico by Médigue et al. (45). The H.pylori secE homolog (HP1203.1 or JHP1126.1) has been included in the PyloriGene database.
Genetic diversity
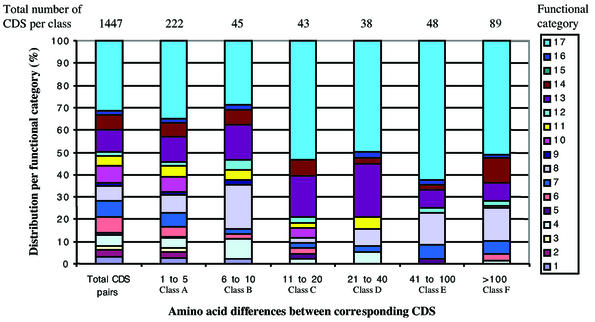
CDS length variation analysis. Systematic manual examination of the two genomes revealed a large number of size differences between corresponding CDS. Helicobacter pylori is a highly diverse species at the nucleotide level (9–11). Indeed, 485 of the 1447 pairs of corresponding CDS differ in length. This prompted us to investigate the sources of this apparent genetic diversity. We analyzed the polymorphism of the predicted proteins and classified them into six different groups based on the difference in the total amino acid length (Table 2): class A (1–5 amino acid difference between corresponding proteins), class B (6–10 amino acids), class C (11–20 amino acids), class D (21–40 amino acids), class E (41–100 amino acids) and class F (>100 amino acids). In each of the six classes defined above, we analyzed the distribution of the polymorphic proteins in the different functional categories and compared this distribution to the overall distribution of the 1447 pairs of proteins (Fig. 2). When a CDS was classified in more than one functional category, the corresponding assignments were ordered. For the purpose of this analysis, we only took the first functional category into account.
Table 2. Summary of CDS pairs that differ in length classified per functional category.
| Total number of CDS pairs | Amino acid differences | Total number of CDS pairs with length variation | ||||||
|---|---|---|---|---|---|---|---|---|
| Functional categories | 1–5 | 6–10 | 11–20 | 21–40 | 41–100 | >100 | ||
| 1. Amino acid biosynthesis | 44 | 6 | 1 | 0 | 0 | 0 | 0 | 7 |
| 2. Purine, pyrimidines, nucleosides, and nucleotides | 44 | 6 | 0 | 0 | 0 | 0 | 0 | 6 |
| 3. Fatty acid and phospholipid metabolism | 27 | 4 | 0 | 0 | 0 | 0 | 0 | 4 |
| 4. Biosynthesis of cofactors, prosthetic groups, and carriers | 73 | 10 | 4 | 1 | 2 | 0 | 1 | 18 |
| 5. Central intermediary metabolism | 16 | 1 | 0 | 1 | 0 | 1 | 0 | 3 |
| 6. Energy metabolism | 100 | 10 | 1 | 1 | 0 | 0 | 3 | 15 |
| 7. Transport and binding proteins | 105 | 14 | 1 | 1 | 1 | 3 | 5 | 25 |
| 8. DNA metabolism | 97 | 18 | 9 | 1 | 3 | 7 | 13 | 51 |
| 9. Transcription | 19 | 3 | 1 | 0 | 0 | 0 | 0 | 4 |
| 10. Protein synthesis | 108 | 15 | 0 | 2 | 0 | 0 | 0 | 17 |
| 11. Protein fate | 66 | 11 | 2 | 1 | 2 | 0 | 1 | 17 |
| 12. Regulatory functions | 28 | 4 | 2 | 1 | 0 | 1 | 2 | 10 |
| 13. Cell envelope | 142 | 24 | 7 | 8 | 9 | 4 | 8 | 60 |
| 14. Cellular processes | 98 | 14 | 3 | 3 | 1 | 1 | 10 | 32 |
| 15. Other categories | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16. Unknown | 26 | 4 | 1 | 0 | 1 | 1 | 1 | 8 |
| 17. Hypothetical | 454 | 78 | 13 | 23 | 19 | 30 | 45 | 208 |
| Total | 1447 | 222 | 45 | 43 | 38 | 48 | 89 | 485 |
Figure 2.
Distribution of corresponding CDS among the different functional categories according to variations in amino acid length. The total number of CDS pairs (1447 CDS) is based on the list of orthologous CDS published by Doig et al. (18) plus the new corresponding CDS identified during our analysis. These CDS were classified in the different functional categories used for the PyloriGene database, taking into account our latest analysis of the two genomes presented in this study. Out of the 1447 pairs of CDS, 485 showed a difference in amino acid length. We classified these CDS according to the amino acid length difference (classes A–F) and to their distribution in the functional categories. The total number of CDS per class is indicated above each class.
The class A, and to some extent class B, proteins were found in all functional categories, indicating that these size variations are mostly due to natural variability. When corresponding proteins differed by >10 amino acids, several functional categories were over-represented. These categories include ‘DNA metabolism, PyloriGene category 8’, ‘Cell envelope, PyloriGene category 13’, ‘Cellular processes, PyloriGene category 14’ and particularly ‘Hypothetical, PyloriGene category 17’. The genes that were over-represented were shown or predicted to be regulated by slipped-strand mispairing mechanisms (genes coding for DNA restriction–modification systems or for enzymes involved in LPS biosynthesis) and/or subject to antigenic variation (adhesins, outer membrane proteins, virulence factors) or to genetic decay (DNA restriction–modification systems) (46–52). In addition, the high percentage of CDS with large amino acid length differences in category 17 (hypothetical proteins), is certainly related to the lack of information on those hypothetical CDS, potentially resulting in incorrect start codon assignments or undetected sequencing errors that might generate premature stop codons, or the artificial splitting or merging of genes.
A similar analysis was performed by Alm and Trust (53). Although these authors used the Astra functional classification, they also observed a bias in length variation towards certain functional categories (cell envelope, DNA restriction, modification, recombination and repair, cellular processes and hypothetical proteins). Similarly to our analysis, the average length variation in these latter categories was >10 amino acids.
Source of CDS length variation. To assess the extent to which each source of polymorphism was responsible for the length variation between corresponding CDS, we individually analyzed all CDS pairs and classified them according to the source of variation, which could be multiple (Table 3; more detailed analysis for each pair of CDS is available as Table S3). Most of the CDS (201 out of 485 CDS) differed in amino acid length exclusively due to amino acid insertions and/or deletions (classes 1, 2c and 3b and a combination of these; see Table 3). However, 113 out of these 485 CDS differed in predicted size partially due to different start codon assignments and in 91 cases this was the sole difference. We analyzed the conservation of sequence similarity of the upstream region to determine whether it was a potential coding sequence or an intergenic region (Table S3 for more details). The start codons of 30 pairs of corresponding CDS with different lengths between the two sequenced strains have already been revised by (i) SWISS-PROT for 19 CDS (54), (ii) by the PIR database for four CDS [although we do not agree with one of the revised start codons; entry D64590; see (55)] and (iii) by researchers for seven CDS (49,56,57). For another 17 CDS, we found that one of the assigned start codons was not conserved in both strains, thus potentially solving the start codon assignment problem. These corrections led to corresponding CDS with the same size for 41 CDS out of 47 (Table S3).
Table 3. Source of CDS size variation between strains 26695 and J99.
| Source of variation | Number of CDSa |
|---|---|
| 1. Insertions and/or deletions | 184 (152) |
| 2. Different start codon | |
| 2a. Distinct assignment of start codon | 113 (91) |
| 2b. Nucleotide substitution or frame-shiftb | 14 (10) |
| 2c. Apparent natural polymorphismc | 23 (12) |
| 3. Different stop codon | |
| 3a. Nucleotide substitution or frame-shiftb | 50 (40) |
| 3b. Apparent natural polymorphismc | 41 (24) |
| 4. Intragenic frameshift | 57 |
| 5. Slipped-strand mispairing mechanism | 28 (22) |
| 6. Pseudogenes and others | 27 |
| Totald | 537 (435) |
| Number of CDS with a variation in amino acid length | 485 |
aValues in parenthesis represent CDS that differ in size exclusively due to the source of variation indicated in the left column.
bNucleotide substitutions that disrupt the reading frame by introducing a premature stop codon.
cInsertions and/or deletions of one or more amino acids restricted to the N- or C-terminus.
dCertain orthologous proteins have more than one source of size variation.
For the remaining 44 CDS, start codon assignment was not straightforward. However, in some cases we could suggest a better choice for one of the strains. Ultimately, the assessment of the correct start codon requires experimental data. Even approaches such as the one used during the analysis of the genome sequence of the enterohemorragic E.coli O157:H7 strain (58), in which it was decided to choose alternative start codons to conform to the genome of E.coli K-12, might generate incorrect assignments. Indeed, we found an example of this in H.pylori for HP1415 and JHP1310, which encode homologs of tRNA delta(2)-isopentenylpyrophosphate transferase (MiaA). These CDS were predicted to be 266 and 277 amino acids long, respectively. Conforming JHP1310 to HP1415 or vice-versa would have resulted in incorrect predicted proteins. Both predicted proteins lack an ATP-binding site that appears to be important for the in vivo regulation of MiaA activity (59). Based on the presence of other potential start codons upstream from the predicted ones, we suggest that the protein is in fact 311 amino acids long in 26695 and 312 amino acids long in J99 and thus includes the previously missing ATP-binding domain.
The characterization of H.pylori anti-sigma 28 factor FlgM (HP1122) also highlighted the problem of alternative start codon annotations (56,57). Indeed, the HP1122 homolog in strain J99, JHP1051, was predicted to start at a different AUG codon, thus producing a protein that is nine amino acids shorter than the HP1122 product. If an upstream alternative UUG start codon was attributed to JHP1051, the two anti-sigma 28 factors were identical in length (78 amino acids). This appears to be also the case for the FlgM protein of three additional H.pylori strains (57). Indeed, H.pylori is predicted to have a different percentage of alternative start codons (10.4% of UUG and 6.7% of GUG) (9) to E.coli K-12 (60). One might speculate that variations of initiation codons might affect the efficiency of translation and result in subtle regulatory mechanism of protein expression in H.pylori strains.
The problem of start codon assignment has been reported for many sequenced genomes and probably reflects the limitations of the existing predictive algorithms. This problem is not restricted to corresponding CDS with different sizes and might also affect the rest of the CDS of the two genomes both for corresponding CDS of the same size and for strain-specific CDS.
Interestingly, corresponding CDS of different size due to the start codon assignment problems are distributed uniformly among the functional categories, as is the overall distribution of CDS. Therefore, the over-representation of CDS of different size within the hypothetical CDS category cannot be attributed to different start codon assignment. Alternat ively, size differences might be due to frame-shifts. These might result from undetected sequencing errors or natural mechanisms such as slipped-strand mispairing, antigenic variation and gene decay. Natural mechanisms should be observed randomly in both strains, whereas undetected sequencing errors might be more common in one strain than in the other.
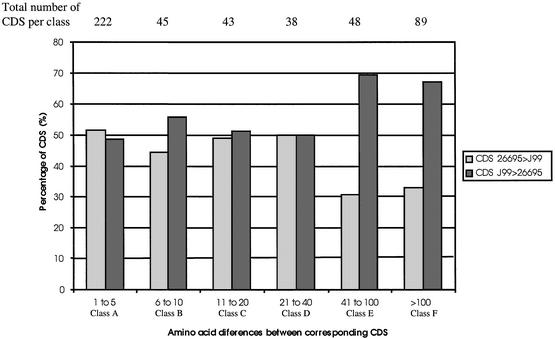
CDS length bias between the two sequenced strains. For each of the 485 pairs of corresponding CDS, we determined which strain had the longest protein (Fig. 3). Interestingly, there is no significant difference for the first four classes (classes A–D, differences <41 amino acids), suggesting that there is no bias for the different sources of variation in these classes. This result was expected at least for CDS in classes A and B, as these CDS are distributed in all the functional categories like the overall distribution (Fig. 2). CDS from classes C and D did not seem to be systematically longer in either one of the strains. Therefore, the over representation of hypothetical CDS in classes C and D (Fig. 2) might reflect either non-essential proteins that have undergone substantial gene decay or intrinsically variable proteins such as antigenic factors. Accordingly, we identified two new CDS that could be subject to slipped-strand mispairing due to a poly A track (HP0368 and HP0991) and several pseudogenes (HP0667/HP0668/HP0669; HP0732/HP0733; HP0764/HP0765; HP0937/HP0938; HP0964/HP0965). Most of these pseudogenes were classified as hypothetical CDS, although we noticed that the HP0667/HP0668/HP0669 cluster had decayed from an ancestral type III restriction–modification system similar to the endonuclease and methylase LlaGI from Lactococcus lactis (61).
Figure 3.
Bias in CDS size variation between the two sequenced genomes of H.pylori. For each pair of corresponding CDS that varied in size (organized in the same classes A–F as in Figure 2), we determined which strain had the longer CDS.
However, for differences exceeding 41 amino acids, there is a clear bias towards longer proteins in strain J99. This suggests that some of these differences might be due to a higher frequency of sequencing errors for the genome of strain 26695, even though 44 CDS were smaller in strain J99 than in strain 26695, indicating that some differences are due to natural variability. This result was confirmed by restricting our analysis to the 116 CDS that differ in size due to frame-shifts. Seventy percent of the CDS were longer in strain J99. Two examples described in the literature illustrate these conclusions (26,62).
CONCLUSIONS AND FUTURE PERSPECTIVES
Researchers are constantly faced with problems concerning the update of the annotation of genomic sequences whilst consulting databases. Most databases are static, keeping the original annotations unchanged. Indeed, the H.pylori databases of TIGR and AstraZeneca have been improved very little since the publication of the respective sequences. However, the huge amount of information generated has opened new fields of research that have produced enormous amounts of new data and hypotheses. Unfortunately, this new scientific knowledge is too fragmented and is not yet exploited efficiently. Although databases such as SWISS-PROT are very informative and updated frequently, for researchers in certain fields like that of H.pylori, a specialized database should become a reference, such as is the case for the SubtiList database for the B.subtilis community (21).
Another challenging issue faced by researchers results from the fact that H.pylori research is highly focused on clinically relevant fields (drug resistance, epidemiology, serology, etc.). This is reflected by the fact that most annotations are based on sequence similarity rather than on work done on H.pylori. Discrepancies in functional annotations were often attributed to the fact that annotations are mostly done automatically by sequence similarity searches. Combining automatic BLAST searches with continuous update of new published information can bring an added value and increase the accuracy of functional assignment predictions. This exhaustive approach seems effective as our efforts to assign functions to previously hypothetical CDS reduced the number of these CDS from ∼40% to ∼33% for each strain. This percentage is similar to that of other organisms such as E.coli K-12 (http://genolist.pasteur.fr/Colibri/) and B.subtilis (http://genolist.pasteur.fr/SubtiList/).
Part of our analysis was also aimed at comparing CDS polymorphisms as H.pylori was the first organism for which two complete genome sequences were published. This raised the question of the threshold to be used when considering two corresponding CDS as orthologs, and consequently, to determine which are strain-specific. We decided to use the list of corresponding CDS published by Doig et al. (18) as this list is widely considered to be the reference for H.pylori. The definition of a strain-specific CDS might vary according to the algorithm used. A recent approach, using an improved algorithm applied to H.pylori (63), resulted in a revised number of strain-specific CDS. The differences were minor compared with the previous set of CDS (18) and were unlikely to have affected the outcome of our analysis. However, this is an important issue, as strain-specific genes might be involved in the different clinical outcomes of H.pylori infection and/or adaptation to different niches in the human host (64,65).
The analysis presented in this manuscript emphasized the two conflicting objectives of the functional annotation: to provide as much information as possible whilst avoiding erroneous physical and functional assignments. The best way of constructing such a highly informative database inevitably requires exhaustive manual annotation by experts. Obviously, errors occur due to the massive amount of information gathered. Therefore, we encourage reports on incorrect information and the communication of recent work that might improve the annotation of H.pylori genomes. Technical problems and suggestions for the improvement of the WWW server are also welcomed.
Besides improving the annotation of H.pylori genomes, our future goals for the PyloriGene database include providing information on the distribution of CDS in the highly diverse H.pylori population according to the geographical origins of strains and their associated diseases. This information can be obtained from whole-genome microarray-based technology (66,67), and from the specific analysis of certain important loci such as the cag pathogenicity island. Finally, we would like to integrate and connect the database with other information such as the essential or conditional nature of each CDS or protein expression 2D-PAGE profiles (http://www.mpiib-berlin.mpg.de/2D-PAGE) (68–72).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank L. M. Jones for transferring the data from the 4th Dimension® DataBase Management Systems (DBMS) to the mainframe computer using the Sybase® SQL DBMS. We would also like to thank P. Legrain and R. Ferrero for critical review of the manuscript. We would like to thank Hybrigenics Inc. for financial support, in particular to I.G.B. and J.-C.E. I.G.B. is also a recipient of a post-doctoral fellowship from the Fundação para a Ciência e a Tecnologia, Ministry of Science and Technology, Portugal (SFRH/BPD/1567/2000).
REFERENCES
- 1.Warren J.R. and Marshall,B.J. (1983) Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet, i, 1273–1275. [PubMed] [Google Scholar]
- 2.Marshall B.J. and Warren,J.R. (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet, 1, 1311–1315. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M.J., Perez-Perez,G.I., Kleanthous,H., Cover,T.L., Peek,R.M., Chyou,P.H., Stemmermann,G.N. and Nomura,A. (1995) Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res., 55, 2111–2115. [PubMed] [Google Scholar]
- 4.Censini S., Lange,C., Xiang,Z., Crabtree,J.E., Ghiara,P., Borodovsky,M., Rappuoli,R. and Covacci,A. (1996) cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl Acad. Sci. USA, 93, 14648–14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Omar E.M., Carrington,M., Chow,W.H., McColl,K.E., Bream,J.H., Young,H.A., Herrera,J., Lissowska,J., Yuan,C.C., Rothman,N. et al. (2000) Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature, 404, 398–402. [DOI] [PubMed] [Google Scholar]
- 6.Machado J.C., Pharoah,P., Sousa,S., Carvalho,R., Oliveira,C., Figueiredo,C., Amorim,A., Seruca,R., Caldas,C., Carneiro,F. et al. (2001) Interleukin 1B and interleukin 1RN polymorphisms are associated with increased risk of gastric carcinoma. Gastroenterology, 121, 823–829. [DOI] [PubMed] [Google Scholar]
- 7.Botterweck A.A., van den Brandt,P.A. and Goldbohm,R.A. (2000) Vitamins, carotenoids, dietary fiber and the risk of gastric carcinoma: results from a prospective study after 6.3 years of follow-up. Cancer, 88, 737–748. [PubMed] [Google Scholar]
- 8.Tomb J.F., White,O., Kerlavage,A.R., Clayton,R.A., Sutton,G.G., Fleischmann,R.D., Ketchum,K.A., Klenk,H.P., Gill,S., Dougherty,B.A. et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature, 388, 539–547. [DOI] [PubMed] [Google Scholar]
- 9.Alm R.A., Ling,L.S., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Noonan,B., Guild,B.C., deJonge,B.L. et al. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 10.Suerbaum S. (2000) Genetic variability within Helicobacter pylori. Int. J. Med. Microbiol., 290, 175–181. [DOI] [PubMed] [Google Scholar]
- 11.Suerbaum S., Smith,J.M., Bapumia,K., Morelli,G., Smith,N.H., Kunstmann,E., Dyrek,I. and Achtman,M. (1998) Free recombination within Helicobacter pylori. Proc. Natl Acad. Sci. USA, 95, 12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G., Humayun,M.Z. and Taylor,D.E. (1999) Mutation as an origin of genetic variability in Helicobacter pylori. Trends Microbiol., 7, 488–493. [DOI] [PubMed] [Google Scholar]
- 13.Bjorkholm B., Sjolund,M., Falk,P.G., Berg,O.G., Engstrand,L. and Andersson,D.I. (2001) Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl Acad. Sci. USA, 98, 14607–14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rain J.C., Selig,L., De Reuse,H., Battaglia,V., Reverdy,C., Simon,S., Lenzen,G., Petel,F., Wojcik,J., Schachter,V. et al. (2001) The protein–protein interaction map of Helicobacter pylori. Nature, 409, 211–215. [DOI] [PubMed] [Google Scholar]
- 15.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 17.Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genes to genomes and back. J. Mol. Biol., 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 18.Doig P., de Jonge,B.L., Alm,R.A., Brown,E.D., Uria-Nickelsen,M., Noonan,B., Mills,S.D., Tummino,P., Carmel,G., Guild,B.C. et al. (1999) Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol. Mol. Biol. Rev., 63, 675–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Médigue C., Viari,A., Hénaut,A. and Danchin,A. (1993) Colibri: a functional database for the Escherichia coli genome. Microbiological Rev., 57, 623–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moszer I., Glaser,P. and Danchin,A. (1995) SubtiList: a relational database for the Bacillus subtilis genome. Microbiology, 141, 261–268. [DOI] [PubMed] [Google Scholar]
- 21.Moszer I., Jones,L.M., Moreira,S., Fabry,C. and Danchin,A. (2002) SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res., 30, 62–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zalkin H. and Nygaard,P. (1996) Biosynthesis of purine nucleotides. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella. Cellular and Molecular Biology. Second edition. ASM Press, Washington DC, pp. 561–579.
- 23.Mendz G.L. (2001) Nucleotide metabolism. In Mobley,H.L.T., Mendz,G.L. and Hazell,S.L. (eds), Helicobacter pylori. Physiology and Genetics. ASM Press, Washington DC, pp. 147–158. [PubMed]
- 24.Kather B., Stingl,K., van der Rest,M.E., Altendorf,K. and Molenaar,D. (2000) Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J. Bacteriol., 182, 3204–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkhill J., Wren,B.W., Mungall,K., Ketley,J.M., Churcher,C., Basham,D., Chillingworth,T., Davies,R.M., Feltwell,T., Holroyd,S. et al. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature, 403, 665–668. [DOI] [PubMed] [Google Scholar]
- 26.Hofreuter D., Odenbreit,S. and Haas,R. (2001) Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol., 41, 379–391. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi S., Kuzuyama,T., Watanabe,H. and Seto,H. (1998) A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl Acad. Sci. USA, 95, 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herz S., Wungsintaweekul,J., Schuhr,C.A., Hecht,S., Luttgen,H., Sagner,S., Fellermeier,M., Eisenreich,W., Zenk,M.H., Bacher,A. et al. (2000) Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2,4-cyclodiphosphate. Proc. Natl Acad. Sci. USA, 97, 2486–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luttgen H., Rohdich,F., Herz,S., Wungsintaweekul,J., Hecht,S., Schuhr,C.A., Fellermeier,M., Sagner,S., Zenk,M.H., Bacher,A. et al. (2000) Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Proc. Natl Acad. Sci. USA, 97, 1062–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohdich F., Wungsintaweekul,J., Fellermeier,M., Sagner,S., Herz,S., Kis,K., Eisenreich,W., Bacher,A. and Zenk,M.H. (1999) Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl Acad. Sci. USA, 96, 11758–11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altincicek B., Kollas,A.K., Sanderbrand,S., Wiesner,J., Hintz,M., Beck,E. and Jomaa,H. (2001) GcpE is involved in the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J. Bacteriol., 183, 2411–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altincicek B., Kollas,A., Eberl,M., Wiesner,J., Sanderbrand,S., Hintz,M., Beck,E. and Jomaa,H. (2001) LytB, a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett., 499, 37–40. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham F.X. Jr, Lafond,T.P. and Gantt,E. (2000) Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol., 182, 5841–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hecht S., Eisenreich,W., Adam,P., Amslinger,S., Kis,K., Bacher,A., Arigoni,D. and Rohdich,F. (2001) Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl Acad. Sci. USA, 98, 14837–14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohdich F., Hecht,S., Gartner,K., Adam,P., Krieger,C., Amslinger,S., Arigoni,D., Bacher,A. and Eisenreich,W. (2002) Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl Acad. Sci. USA, 99, 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pittard A.J. (1996) Biosynthesis of the aromatic amino acids. In Neidhardt,F.C. (ed.), Escherichia coli and Salmonella. Cellular and Molecular Biology. Second Edition. ASM Press, Washington DC, pp. 458–484.
- 37.Davis B.D. (1953) Autocatalytic growth of a mutant due to accumulation of an unstable phenylalanine precursor. Science, 118, 251–252. [DOI] [PubMed] [Google Scholar]
- 38.Katagiri M. and Sato,R. (1953) Accumulation of phenylalanine by a phenylalanineless mutant of Escherichia coli. Science, 118, 250–251. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds S. (1950) The metabolism of phenylalalnine and tyrosine in mutant strains of Escherichia coli. J. Biol. Chem., 185, 755–762. [PubMed] [Google Scholar]
- 40.Reynolds D.J. and Penn,C.W. (1994) Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology, 140, 2649–2656. [DOI] [PubMed] [Google Scholar]
- 41.Mendz G.L. and Hazell,S.L. (1995) Aminoacid utilization by Helicobacter pylori. Int. J. Biochem. Cell Biol., 27, 1085–1093. [DOI] [PubMed] [Google Scholar]
- 42.Karudapuram S. and Barcak,G.J. (1997) The Haemophilus influenzae dprABC genes constitute a competence-inducible operon that requires the product of the tfoX (sxy) gene for transcriptional activation. J. Bacteriol., 179, 4815–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smeets L.C., Bijlsma,J.J., Kuipers,E.J., Vandenbroucke-Grauls,C.M. and Kusters,J.G. (2000) The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol. Med. Microbiol., 27, 99–102. [DOI] [PubMed] [Google Scholar]
- 44.Ando T., Israel,D.A., Kusugami,K. and Blaser,M.J. (1999) HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol., 181, 5572–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Médigue C., Wong,B.C., Lin,M.C., Bocs,S. and Danchin,A. (2002) The secE gene of Helicobacter pylori. J. Bacteriol., 184, 2837–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pride D.T., Meinersmann,R.J. and Blaser,M.J. (2001) Allelic variation within Helicobacter pylori babA and babB. Infect. Immun., 69, 1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin L.F., Posfai,J., Roberts,R.J. and Kong,H. (2001) Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl Acad. Sci. USA, 98, 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Appelmelk B.J., Martino,M.C., Veenhof,E., Monteiro,M.A., Maaskant,J.J., Negrini,R., Lindh,F., Perry,M., Del Giudice,G. and Vandenbroucke-Grauls,C.M. (2000) Phase variation in H type I and Lewis a epitopes of Helicobacter pylori lipopolysaccharide. Infect. Immun., 68, 5928–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alm R.A., Bina,J., Andrews,B.M., Doig,P., Hancock,R.E. and Trust,T.J. (2000) Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun., 68, 4155–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Appelmelk B.J., Martin,S.L., Monteiro,M.A., Clayton,C.A., McColm,A.A., Zheng,P., Verboom,T., Maaskant,J.J., van den Eijnden,D.H., Hokke,C.H. et al. (1999) Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in alpha3-fucosyltransferase genes. Infect. Immun., 67, 5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Covacci A., Censini,S., Bugnoli,M., Petracca,R., Burroni,D., Macchia,G., Massone,A., Papini,E., Xiang,Z., Figura,N. et al. (1993) Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl Acad. Sci. USA, 90, 5791–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atherton J.C., Cao,P., Peek,R.M.,Jr, Tummuru,M.K., Blaser,M.J. and Cover,T.L. (1995) Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem., 270, 17771–17777. [DOI] [PubMed] [Google Scholar]
- 53.Alm R.A. and Trust,T.J. (1999) Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J. Mol. Med., 77, 834–846. [DOI] [PubMed] [Google Scholar]
- 54.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGarvey P.B., Huang,H., Barker,W.C., Orcutt,B.C., Garavelli,J.S., Srinivasarao,G.Y., Yeh,L.S., Xiao,C. and Wu,C.H. (2000) PIR: a new resource for bioinformatics. Bioinformatics, 16, 290–291. [DOI] [PubMed] [Google Scholar]
- 56.Colland F., Rain,J.C., Gounon,P., Labigne,A., Legrain,P. and De Reuse,H. (2001) Identification of the Helicobacter pylori anti-sigma28 factor. Mol. Microbiol., 41, 477–487. [DOI] [PubMed] [Google Scholar]
- 57.Josenhans C., Niehus,E., Amersbach,S., Horster,A., Betz,C., Drescher,B., Hughes,K.T. and Suerbaum,S. (2002) Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol., 43, 307–322. [DOI] [PubMed] [Google Scholar]
- 58.Perna N.T., Plunkett,G.,III, Burland,V., Mau,B., Glasner,J.D., Rose,D.J., Mayhew,G.F., Evans,P.S., Gregor,J., Kirkpatrick,H.A. et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature, 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 59.Leung H.C., Chen,Y. and Winkler,M.E. (1997) Regulation of substrate recognition by the MiaA tRNA prenyltransferase modification enzyme of Escherichia coli K-12. J. Biol. Chem., 272, 13073–13083. [DOI] [PubMed] [Google Scholar]
- 60.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 61.Madsen A. and Josephsen,J. (2001) The LlaGI restriction and modification system of Lactococcus lactis W10 consists of only one single polypeptide. FEMS Microbiol. Lett., 200, 91–96. [DOI] [PubMed] [Google Scholar]
- 62.Chirica L.C., Elleby,B. and Lindskog,S. (2001) Cloning, expression and some properties of alpha-carbonic anhydrase from Helicobacter pylori. Biochim. Biophys. Acta, 1544, 55–63. [DOI] [PubMed] [Google Scholar]
- 63.Janssen P.J., Audit,B. and Ouzounis,C.A. (2001) Strain-specific genes of Helicobacter pylori: distribution, function and dynamics. Nucleic Acids Res., 29, 4395–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Israel D.A., Salama,N., Krishna,U., Rieger,U.M., Atherton,J.C., Falkow,S. and Peek,R.M.,Jr (2001) Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl Acad. Sci. USA, 98, 14625–14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kersulyte D., Chalkauskas,H. and Berg,D.E. (1999) Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol., 31, 31–43. [DOI] [PubMed] [Google Scholar]
- 66.Thiberge J.-M. and Labigne,A. (2000) Use of DNA chips to study the biodiversity of H. pylori clinical isolates from different geographical origins. Gut, 47 (Suppl. 1), A1.11086475 [Google Scholar]
- 67.Salama N., Guillemin,K., McDaniel,T.K., Sherlock,G., Tompkins,L. and Falkow,S. (2000) A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl Acad. Sci. USA, 97, 14668–14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sabarth N., Lamer,S., Zimny-Arndt,U., Jungblut,P.R., Meyer,T.F. and Bumann,D. (2002) Identification of surface-exposed proteins of Helicobacter pylori by selective biotinylation, affinity purification and two-dimensional gel electrophoresis. J. Biol. Chem., 277, 27896–27902. [DOI] [PubMed] [Google Scholar]
- 69.Haas G., Karaali,G., Ebermayer,K., Metzger,W.G., Lamer,S., Zimny-Arndt,U., Diescher,S., Goebel,U.B., Vogt,K., Roznowski,A.B. et al. (2002) Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics, 2, 313–324. [DOI] [PubMed] [Google Scholar]
- 70.Jungblut P.R., Bumann,D., Haas,G., Zimny-Arndt,U., Holland,P., Lamer,S., Siejak,F., Aebischer,A. and Meyer,T.F. (2000) Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol., 36, 710–725. [DOI] [PubMed] [Google Scholar]
- 71.Bumann D., Aksu,S., Wendland,M., Janek,K., Zimny-Arndt,U., Sabarth,N., Meyer,T.F. and Jungblut,P.R. (2002) Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun., 70, 3396–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bumann D., Meyer,T.F. and Jungblut,P.R. (2001) Proteome analysis of the common human pathogen Helicobacter pylori. Proteomics, 1, 473–479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.