Abstract
Previously, we presented evidence that it is possible to predict functional coupling between genes based on conservation of gene clusters between genomes. With the rapid increase in the availability of prokaryotic sequence data, it has become possible to verify and apply the technique. In this paper, we extend our characterization of the parameters that determine the utility of the approach, and we generalize the approach in a way that supports detection of common classes of functionally coupled genes (e.g., transport and signal transduction clusters). Now that the analysis includes over 30 complete or nearly complete genomes, it has become clear that this approach will play a significant role in supporting efforts to assign functionality to the remaining uncharacterized genes in sequenced genomes.
Gene clusters are known to be prominent features of bacterial chromosomes. Demerec and Hartman (1) postulated in 1959 that “regardless of how the gene clusters originated, natural selection must act to prevent their separation” and the “mere existence of such arrangements shows that they must be beneficial, conferring an evolutionary advantage on individuals and populations which exhibit them.” One of the most striking features of prokaryotic gene clusters is that typically they are composed of functionally related genes. For the past 40 years, there has been vigorous, ongoing discussion on the functional significance of gene arrangement on the chromosome, as well as the origin and mechanisms of maintenance of gene clusters (see, for example, refs. 2–5).
Here, we present a method that uses conserved gene clusters from a large number of genomes to predict functional coupling between genes in those genomes. This article further develops the approach that we previously reported (6) and uses this method to reconstruct several major metabolic and functional subsystems.
Methodology
The data presented below are computed via the WIT system (http://wit.mcs.anl.gov/WIT2/), developed by Overbeek et al. (7) at Argonne National Laboratory. WIT was designed and implemented to support genetic sequence analysis, metabolic reconstructions, and comparative analysis of sequenced genomes; it currently contains data from over 30 genomes, albeit a few of them are incomplete.
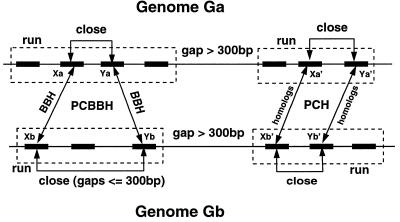
Our approach to detection of conserved clusters of genes is based on the following definitions: a set of genes occurring on a prokaryotic chromosome will be called a “run” if and only if they all occur on the same strand and the gaps between adjacent genes are 300 bp or less. Any pair of genes occurring within a single run is called “close.” Given two genes Xa and Xb from two genomes Ga and Gb, Xa and Xb are called a “bidirectional best hit (BBH)” if and only if recognizable similarity exists between them (in our case, we required fasta3 scores lower than 1.0 × 10−5), there is no gene Zb in Gb that is more similar than Xb is to Xa, and there is no gene Za in Ga that is more similar than Xa is to Xb. Genes (Xa, Ya) from Ga and (Xb, Yb) from Gb form a “pair of close bidirectional best hits (PCBBH)” if and only if Xa and Ya are close, Xb and Yb are close, Xa and Xb are a BBH, and Ya and Yb are a BBH. The notion of a PCBBH is illustrated graphically in Fig. 1.
Figure 1.
Illustration of the definitions of PCBBHs and “pairs of close homologs” (PCHs).
Computation of PCBBHs for 31 complete or nearly complete prokaryotic genomes established several critical points:
- 1.
We found 58,498 PCBBHs among the 31 genomes considered.
- 2.
As is typical of most forms of comparative evidence, the number of PCBBHs grows roughly as the square of the number of genomes (see Table 1).
- 3.
From the 31 complete or partial genomes, we were able to infer that approximately 35% of the genes assigned enzymatic functions from known pathways appeared in the same run with genes assigned other functions from the same pathway.
- 4.
A smaller percentage of genes showed inferred couplings that could not be confirmed as “real.” This set of coupled genes no doubt includes some “false positive” couplings, as well as pairs of genes that are indeed functionally related but whose connection has not yet been experimentally confirmed.
Table 1.
Increase of the number of PCBBHs with the number of genomes
| No. of genomes | No. of PCBBHs with scores >0.1 |
|---|---|
| 4 | 998 |
| 8 | 4859 |
| 16 | 12570 |
| 24 | 23144 |
| 31 | 58498 |
The question of whether gene clusters are widely present in the Archaea is worth a comment. Our computation shows that there are 2,504 PCBBHs among Methanococcus jannaschii, Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum, and Pyrococcus horikoshii. The number of PCBBHs for the first four sequenced bacterial genomes—Haemophilus influenzae, Mycoplasma genitalium, Synechocystis sp., and Helicobacter pylori—equals 1,616. However, when Haemophilus influenzae, Escherichia coli, Bacillus subtilis, and Synechocystis sp. are used, we find 2,981 PCBBHs. Finally, if one considers PCBBHs among the four organisms Haemophilus influenzae, Helicobacter pylori, Methanococcus jannaschii, and Archaeoglobus fulgidus, one sees the following numbers of PCBBHs: 262 PCBBHs between the two archaeal genomes, 329 between the two bacterial genomes, and 132 between an archaeal and a bacterial genome. Hence, it seems likely that gene clusters also play an important role in the Archaea. However, we have far too little data on the Archaea to make an accurate estimate that takes into account the size of the genomes and phylogenetic distance between the organisms.
Motivating the Definition of a PCBBH.
The significance of the coupling information provided by PCBBHs will be covered in detail below. First, we motivate the definitions above. The essential questions are as follows:
- 1.
Is it important that we restrict our attention to genes clustered on the same strand? We know that occasionally divergent genes are coregulated (8), that horizontal transfer may be a dominant theme, and that either of these considerations might lead to a situation in which mere proximity might convey information relating to functional coupling, without regard to strand.
- 2.
Assuming that we do restrict our attention to genes clustered on the same strand, what maximum gap size should be used in the definition of a PCBBH?
To address these questions, we performed a number of computations. First, we restricted our attention to a set of ORFs that we have reason to believe are functionally coupled, and that occur within 10,000 bp of one another. For this set, we tabulated the number of ORFs that occurred on the same strand, the number of ORFs on the same strand with no intervening genes on the opposite strand, the number of ORFs on opposite strands, and the number on opposite strands that were immediately adjacent. To estimate the frequency of occurrence of functionally coupled ORFs in a run as a result of chance alone, we performed one more experiment. We repeatedly took the same set of ORFs (along with their function assignments), randomly shuffled the set of locations, and recomputed the values above. The results of both computations are shown in Table 2. Table 2 suggests that co-occurrence of functionally related ORFs on the same strand is of primary significance. The results also suggest that, although there appear to be more cases of divergent genes with coupled functions than expected from chance, their frequency is nearly two orders of magnitude smaller than that of same-strand ORFs with coupled functions; hence, we shall not examine divergent pairs further in this paper.
Table 2.
Locations of functionally related ORFs on the chromosome
| Strand | Real data
|
“Shuffled” data
|
|
|---|---|---|---|
| Functionally related ORFs | Mean no. of functionally related ORFs | SD | |
| Same strand | 10,968 | 445 | 23 |
| Same strand, no intervening genes | 10,583 | 273 | 24 |
| Opposite strand | 349 | 256 | 18 |
| Divergent genes | 43 | 19 | 5 |
Having argued that the essence of the phenomenon we are observing is co-occurrence in runs of genes on the same strand, we next ask: what is the range of gaps that occur between genes in such runs? To answer this question, we examined the gaps for the 10,583 cases used to construct Table 2. The average maximum gap between pairs of related genes was 94 bp, with a standard deviation of 194 bp; after we trimmed 50 obvious outliers, the average reduces to 91 bp, with a standard deviation of 136 bp. As suggested by the fact that the standard deviation is significantly larger than the mean, the gap distribution is rather skewed; nevertheless, two standard deviations above the mean still provides a reasonable cutoff for the maximum allowed gap, showing that our initial assumption of a 300 bp maximum gap in a “run” used in ref. 6 was not too far off.
It is important to note that we are dealing with data that suffer from many sources of error and uncertainty. For example, the actual starting positions for ORFs in the collection are often inaccurate, and, in many cases, short genes were missed in the initial analysis of each genome. The use of “partial” genomes, having a generally lower quality of sequence data, numerous frameshifts, and frequent truncated genes, also makes analysis more difficult—although we emphasize that our method itself appears to be largely insensitive to these problems and that we are grateful for the enormous wealth of data that such partial genomes represent. Taken together, these figures and observations would perhaps support a slightly larger threshold than the 300 bp value used in our definition of a run; however, overall the basic definitions used accurately capture a useful characterization of the notion “pair of close bidirectional best hits.”
Motivating the Score of a PCBBH.
The significance of the evidence for functional coupling provided by a PCBBH depends on a number of factors, the most important of which is the phylogenetic distance between the organisms. In phylogenetically close organisms, there is a significant probability that two pairs of nearby genes will form a PCBBH as a result of chance alone, presumably because whatever processes are rearranging the gene order have not yet had enough time to act. By the same token, in phylogenetically distant organisms, it is rather unlikely that two pairs of genes would form a PCBBH as a result of chance alone. To reflect the importance of the phylogenetic distance between the genomes in deciding whether the observed linkage of their genes is due to chance, we developed the following simple scoring mechanism: the score of a PCBBH is given by the phylogenetic distance between organism Ga and organism Gb in the 16S rRNA tree (9), regardless of the physical distance between the ORFs in either run, or the degree of similarity of either BBH. We give some representative phylogenetic distances in Table 3. A number of other scoring functions were explored, but none appeared to display a significant advantage over this simple scheme.
Table 3.
PCBBH scores based on phylogenetic distances between pairs of organisms
| Pair of organisms | Phylogenetic distance |
|---|---|
| N. gonorrhoea, N. meningitidis | 0.01 |
| M. genitalium, M. pneumoniae | 0.01 |
| E. coli, H. influenzae | 0.21 |
| M. genitalium, B. subtilis | 0.41 |
| M. genitalium, Synechocystis sp. | 0.80 |
| M. genitalium, E. coli | 0.88 |
| M. genitalium, P. furiosus | 1.57 |
N., Neisseria; M., Mycoplasma; E., Escherichia; H., Haemophilus; P., Pyrococcus.
The rate at which the number of PCBBHs grows as a function of the number of genomes present in the analysis is worth considering. Our current data points are shown in Table 1. Because there is large variability between genomes in terms of size, number of contigs, accuracy with which genes have been identified, and so forth, one would expect only a rough correspondence to be evident from these values. Nevertheless, it does appear that the number of PCBBHs increases as the square of the number of genomes.
A generalization of PCBBHs was proposed by W. Pearson (personal communication). There is no need to insist that the pairs of genes be BBHs. We can also define the concept of “pairs of close homologs” (PCHs) as follows: genes (X′a, Y′a) from Ga and (X′b, Y′b) from Gb form a PCH if and only if X′a and Y′a are close, X′b and Y′b are close, X′a and X′b are recognizably similar, and Y′a and Y′b are recognizably similar. Here, we will consider two genes to be recognizably similar if their gene products produce fasta3 scores lower than 1.0 × 10−5. We use a scoring scheme analogous to the one described for PCBBHs to evaluate the connections between PCHs, except that if Ga and Gb are the same genome, we assign an arbitrary “same-genome score” (“same-genome” pairs cannot occur for PCBBHs by definition, but for PCHs they are possible). Unlike PCBBHs from two very close genomes for which contiguity is completely uninformative in the vast majority of cases, PCHs allow recognition of gene clusters that play similar (but usually not identical) roles (such as two transport cassettes containing pairs of homologs) in the same or similar organisms. The arbitrary “same-genome score” should, we believe, have a value that is high enough to rank such instances as significant. In ref. 6, we found that PCBBHs with score above 0.1 were significant, and PCBBHs with scores above 1.0 were highly significant; choosing a “same genome” score of 0.5 seems a reasonable first approximation. With this choice, we have 103,449 PCHs with scores greater than 0.1, as compared with 58,498 PCBBHs; of these PCHs, approximately 20% represent “same-genome” pairs. This generalization to PCHs has allowed us to detect broad categories of functionally coupled proteins for which BBHs proved to represent a too restrictive criterion for homology; two examples are transport cassettes and signal transduction operons.
We end this section with a fact that is relevant to understanding the underlying phenomena producing an unexpectedly high number of PCBBHs and PCHs: consider all pairs of runs ℛa and ℛb from organisms that have a phylogenetic distance greater than 0.1 in the rRNA tree such that they each contain at least three bidirectional best hits. Then about 88% of the time the order of the corresponding genes is exactly preserved: of the 3,821 such pairs of runs in our data, only 473 contained permutations of the gene order.
Significance of the PCBBH and PCH Scores.
At this time, on the average only half of the gene functions in newly sequenced genomes can be predicted on the basis of sequence analysis. Finding new approaches to establish the functions of such “hypothetical” proteins is one of the major goals of our current research. We have found hundreds of instances in which a hypothetical protein is paired with a protein of known function via one or more PCBBHs or PCHs. The central question is: how meaningful are such predicted couplings? In this section, we explore this question by examining predicted couplings between proteins of known function.
Suppose that two genes X and Y from a single run occur in one or more PCBBHs. Then, by the “BBH coupling score,” we mean the sum of the scores of all the PCBBHs containing X and Y. Similarly, by the “coupling score” we mean the sum of the scores of the PCHs containing X and Y. In other words, to gain an estimate of whether two genes in a run are functionally coupled, we propose simply to add up the scores for the relevant PCBBHs or PCHs.
Once we have defined the notion of BBH coupling score, it becomes possible to form clusters of genes that are coupled at some level exceeding a specified threshold. Basically, one starts with a gene, finds those genes with which it has high coupling scores, adds those genes (and the corresponding genes from related genomes) to the emerging set, and repeats this procedure until no new genes can be added to the set. (Details of this approach are given as algorithm 1, which is published as supplemental material on the PNAS web site, www.pnas.org.)
Results
Below, we show the results of applying algorithm 1 to reconstruct two common metabolic pathways: purine biosynthesis and glycolysis. (A number of additional examples of reconstructed metabolic pathways and functional subsystems, as well as signal-transduction pathways and metabolite transport, are presented in the supplemental material on the PNAS web site.) The utility of the algorithm was evaluated by asking three questions:
- 1.
How much of the functional coupling implied by the pathway could be determined directly from the PCBBHs?
- 2.
How often could the entire pathway be derived directly from just the PCBBHs?
- 3.
How many spurious (“false positive”) functional couplings were predicted?
To answer these questions, we present our results for the de novo purine biosynthetic pathway and the glycolytic pathway. We have tabulated all couplings between genes known to be related to the particular pathway, as well as to other genes that have no obvious connections to it. These latter genes are candidates for “false positive” results, and a detailed analysis of some of these “false positive” couplings is presented.
De Novo Purine Biosynthesis.
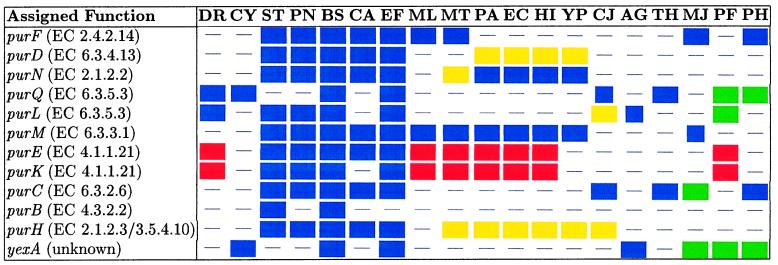
Below, we present a reconstruction of the de novo purine biosynthetic pathway (12–14) from PCBBHs. Table 4 shows the inferred clustering of genes from a number of genomes. Each row depicts a set of bidirectional best hits associated with the function defined in the leftmost column, and each column represents one or more gene clusters from a single genome (distinct colors indicate distinct clusters). Dashes represent enzymes that are not present in PCBBHs in the given organism. So, for example, the Deinococcus radiodurans (DR) genome has two gene clusters—purEK and purQL—from this pathway.
Table 4.
Functional couplings between the genes of the purine biosynthetic pathway
The colors label genes occuring in corresponding runs in each organism. DR, Deinococcus radiodurans; CY, Synechocystis sp.; ST, Streptococcus pyogenes; PN, Streptococcus pneumoniae; BS, Bacillus subtilis; CA, Clostridium acetobutylicum; EF, Enterococcus faecalis; ML, Mycobacterium leprae; MT, Mycobacterium tuberculosis; PA, Pseudomonas aeruginosa; EC, Escherichia coli; HI, Haemophilus influenzae; YP, Yersinia pestis; CJ, Campylobacter jejuni; AG, Archaeoglobus fulgidus; TH, Methanobacterium thermoautotrophicum; MJ, Methanococcus jannaschii; PF, Pyrococcus furiosus; PH, Pyrococcus horikoshii.
As one can see in Table 4, there is a substantial difference in the organization of pur genes in Gram-positive and Gram-negative bacteria. In the low G + C Gram-positive group (Bacillus subtilis, Enterococcus faecalis, Streptococcus pneumoniae, Streptococcus pyogenes, and Clostridium acetobutylicum), pur genes form tight clusters containing most of the genes related to purine biosynthesis. In Gram-negative organisms belonging to the gamma subdivision of the Proteobacteria (Pseudomonas aeruginosa, Escherichia coli, Haemophilus influenzae, and Yersinia pestis), pur genes form three highly preserved clusters: purEK, described in Escherichia coli (10, 11), purMN, and purHD. In Deinococcus radiodurans, Mycobacterium leprae, Mycobacterium tuberculosis, and Synechocystis sp., as well as in all the archaeobacterial genomes under consideration, pur genes are instead gathered into short clusters scattered about the genome.
The coupling scores between the distinct functional roles in the pathway are shown in Table 5. The values represent the strongest coupling between the designated functions. Almost all of the enzymes of purine biosynthesis are connected by PCBBH coupling scores above 0.3.
Table 5.
Connection matrix between the genes of the purine biosynthetic pathway
| Gene names | purF | purD | purN | purQ | purL | purM | purE | purC | purK | purB | purH | Unknown yexA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| purF | — | 0.81 | 1.82 | 0.10 | 0.48 | 8.72 | 0.81 | 1.82 | 0.10 | 0 | 1.33 | 0.10 |
| purD | 0.81 | — | 0.81 | 0.10 | 0.10 | 0.81 | 1.33 | 0.81 | 0.34 | 0.20 | 4.28 | 0.10 |
| purN | 1.82 | 0.81 | — | 0.10 | 0.48 | 4.26 | 0.81 | 1.82 | 0.10 | 0 | 1.77 | 0.10 |
| purQ | 0.10 | 0.10 | 0.10 | — | 3.75 | 0.10 | 0.10 | 3.95 | 0.10 | 0 | 0.10 | 2.98 |
| purL | 0.48 | 0.10 | 0.48 | 3.75 | — | 0.48 | 0.10 | 0.48 | 0.10 | 0 | 1.75 | 2.56 |
| purM | 8.72 | 0.81 | 4.26 | 0.10 | 0.48 | — | 0.81 | 1.82 | 0.10 | 0 | 1.33 | 0.10 |
| purE | 0.81 | 1.33 | 0.81 | 0.10 | 0.10 | 0.81 | — | 0.81 | 13.3 | 0.37 | 0.81 | 0.10 |
| purC | 1.82 | 0.81 | 1.82 | 3.95 | 0.48 | 1.82 | 0.81 | — | 0.10 | 0 | 1.33 | 1.41 |
| purK | 0.10 | 0.34 | 0.10 | 0.10 | 0.10 | 0.10 | 13.3 | 0.10 | — | 0 | 0.10 | 0.10 |
| purB | 0 | 0.20 | 0 | 0 | 0 | 0 | 0.37 | 0 | 0 | — | 0 | 0 |
| purH | 1.33 | 4.28 | 1.77 | 0.10 | 1.75 | 1.33 | 0.81 | 1.33 | 0.10 | 0 | — | 0.10 |
| Unknown yexA | 0.10 | 0.10 | 0.10 | 2.98 | 2.56 | 0.10 | 0.10 | 1.41 | 0.10 | 0 | 0.10 | — |
Our analysis predicted only one connection outside the known purine biosynthetic pathway that may be interpreted as a false positive result: a set of seven bidirectional best hits that were all assigned the function “hypothetical cytosolic protein” (yexA homolog). Homologous proteins were found in Bacillus subtilis, Synechocystis sp., Enterococcus faecalis, and the archaeal genomes Archaeoglobus fulgidus, Methanococcus jannaschii, Pyrococcus furiosus, and Pyrococcus horikoshii. It has strong PCBBH scores with the purL, purQ, and purC gene products. We hope that future laboratory experiments will confirm or reject the functional connection of this protein to the purine biosynthetic cluster.
Glycolysis.
Our analysis detected two clusters produced by genes encoding glycolytic enzymes, shown in Fig. 2. Both clusters were found only in the bacterial genomes. The first cluster includes tpi (triosephosphate isomerase, EC 5.3.1.1), gap (glyceraldehyde 3-phosphate dehydrogenase, EC 1.2.1.12), pgk (phosphoglycerate kinase, EC 2.7.2.3), pgm (2,3-bisphosphoglycerate-independent phosphoglycerate mutase, EC 5.4.2.1), eno (enolase, EC 4.2.1.11), and a hypothetical protein. These results agree well with the limited data on clustering of glycolytic enzymes (15–17). The hypothetical protein, which is functionally connected to most of the enzymes in this cluster, is most probably a transcriptional regulator. It is highly homologous to a hypothetical transcriptional regulator from Bacillus megaterium (sp|P35168) and contains a weak signature for the deoR family of transcriptional regulators.
Figure 2.
Functional clusters in the glycolysis pathway; BB, Borrelia burgdorferi; DR, Deinococcus radiodurans; CA, Clostridium acetobutylicum; BS, Bacillus subtilis; EF, Enterococcus faecalis; MP, Mycoplasma pneumoniae; MG, Mycoplasma genitalium; ML, Mycobacterium leprae; MT, Mycobacterium tuberculosis; CJ, Campylobacter jejuni; TP, Treponema pallidum; HP, Helicobacter pylori; ST, Streptococcus pyogenes; PN, Streptococcus pneumoniae.
The second glycolytic cluster contains pfk (phosphofructokinase, EC 2.7.1.11) and pyk (pyruvate kinase, EC 2.7.1.40)—the only two glycolytic enzymes that do not participate in gluconeogenesis.
This cluster was previously described in the literature (18–20), where it was suggested that both genes constitute an operon. Our analysis shows functional relationship between this cluster and the alpha chain of DNA polymerase III (dnaE). Although it is possible that there is a connection between glycolysis and replication, we currently consider the presence of dnaE (EC 2.7.7.7) in this cluster to be a “false positive” result. Tables 6 and 7 show the connection matrices for both glycolytic clusters.
Table 6.
Connection matrix of the glycolytic enzymes at 0.4 threshold (cluster 1)
| EC no. | 5.3.1.1 | 1.2.1.12 | 2.7.2.3 | 5.4.2.1 | 4.2.1.11 | Unknown |
|---|---|---|---|---|---|---|
| 5.3.1.1 | — | 2.96 | 3.38 | 1.85 | 0.81 | 0.42 |
| 1.2.1.12 | 2.96 | — | 9.30 | 0.42 | 0.81 | |
| 2.7.2.3 | 3.38 | 9.30 | — | 0.38 | 0.81 | 0.42 |
| 5.4.2.1 | 1.85 | 0.38 | — | 0.38 | ||
| 4.2.1.11 | 0.81 | 0.42 | 0.81 | 0.38 | — | 0.42 |
| Unknown | 0.42 | 0.81 | 0.42 | 0.42 | — |
Table 7.
Connection matrix of the glycolytic enzymes at 0.4 threshold (cluster 2)
| EC no. | 2.7.1.11 | 2.7.1.40 | 2.7.7.7 |
|---|---|---|---|
| 2.7.1.11 | — | 2.79 | 1.44 |
| 2.7.1.40 | 2.79 | — | 0.94 |
| 2.7.7.7 | 1.44 | 0.94 | — |
A More Systematic Exploration of Functional Couplings.
The simple algorithm alluded to in the preceding section can be used very effectively to gain insights into the roles of specific genes. Two questions immediately arise:
- 1.
How much of known metabolism can be deduced from gene clusters?
- 2.
How many hypothetical proteins can be coupled to functional subsystems?
To explore these questions systematically, we developed the approach presented in the next section.
Identifying Corresponding Genes from Different Organisms.
We begin by forming sets of genes that we call “role groups.” A role group is a set of genes such that the set contains at most one gene from an organism, each gene in the set is a bidirectional best hit with at least two other genes in the set, the set is “connected” in the sense that one could not split it without separating two bidirectional best hits, and the set contains no pair of genes Xa and Xb from organisms Ga and Gb, respectively, such that Xa is a bidirectional best hit with Yb from Gb and Yb is not Xb. The last condition is especially important in cases with a large number of paralogs, where our ability to accurately identify corresponding genes from distinct organisms is limited. We will call these sets “role groups,” because we are attempting to isolate genes from different organisms that play identical roles in each organism (and again, we emphasize that our ability to accurately compute such groupings is limited).
These role groups are related to the much better known clusters of orthologous genes (COGs) developed by R. L. Tatusov et al. (21). COGs play an invaluable role in attempting to characterize families of proteins. They tend to be much larger groupings than the role groups; that is, COGs are often the union of a set of role groups. COGs represent an attempt to group proteins at the level of abstraction appropriate to assigning function; role groups attempt to identify corresponding genes in distinct organisms. In this sense, COGs have a much more clearly defensible conceptual basis (and require more judgment to curate). Both COGs and role groups have extremely interesting properties and utility. The current WIT system has over 5,200 identified role groups.
Connecting Role Groups.
After computing role groups for a set of organisms, one can compute connections between specific groups based on coupling scores (or coupling BBH scores) as follows. Let X and Y be genes from a single organism such that the coupling score between X and Y is Sc. Then if X is from one role group Rx and Y is from another group Ry, (X, Y) is said to be a connection at score Sc between Rx and Ry. If Rx and Ry are connected by two or more such connections with scores greater than or equal to some threshold T, Rx and Ry are said to be connected at threshold T. That is, one can compute a set of connections between role groups imposed by the coupling scores between genes in the groups. Among other things, these connections between role groups can be used to infer functional couplings between genes that do not occur in gene clusters.
Clustering Role Groups.
Our first attempt to cluster role groups was based on the approach we used for algorithm 1. We then devised a better approach that computes all connections between role groups at a threshold of 0.1, orders the connections based on the maximum connection score, and allows a knowledgeable biologist or biochemist to decide whether to add a new role group to an existing cluster of connected groups or to terminate the search; once a group has been added to a cluster, it is removed from further consideration, ensuring that each group occurs in one and only one cluster. (For details, see algorithm 2 in the supplemental material on the PNAS web site.)
There were 7,464 connections between the role groups maintained in WIT2; 343 clusters of role groups were produced. Each such cluster represents a working hypothesis of the composition of a functional subsystem in some set of organisms.
Conclusion
The availability of multiple genomes provides an opportunity to gain new insights into the processes that drive the dispersion and formation of chromosomal gene clusters. The results obtained with the method described above confirm that conserved gene clusters accurately convey functional coupling between the genes present in them. We have supported this by anecdotal evidence, with further examples being available in the supplemental material on the PNAS web site. The importance of simultaneous analysis of a large number of genomes for the reconstruction of functional subsystems using functional coupling is illustrated by the following calculation.
Three parameters determine the utility of this class of data: the percentage of genes that occur within clusters, the average size of a cluster, and the size of the real subsystems. From the experiments described in Methodology, we found that the percentage of genes assigned to a pathway that occur within the same run with at least one other gene from the same pathway is approximately 35%. We consider 3 genes to be a conservative underestimate of the average size of a gene cluster, because it was the median size of “same pathway” clusters found in the experiments of Methodology.
Consider a subsystem composed of 5 genes. How many genomes containing this subsystem will be needed before the coupling between two specific genes Gx and Gy in the subsystem might be revealed via PCBBHs? Under the assumptions that a given gene will occur in a run in 35% of the genomes and that the average length of a gene cluster is 3 genes, one expects to see 1 co-occurrence of Gx and Gy in a run (i.e., a single pair) in 6 genomes, about 2 co-occurrences in 11 genomes (that is, 11 genomes is the smallest number for which one expects to first see a PCBBH containing Gx and Gy), and about 3 co-occurrences in 17 genomes.
These simple calculations reveal an important characteristic of gene clusters: functional clustering could only be detected once we had access to about 10–15 genomes containing the functional subsystem of interest. This property has caused the utility of preserved chromosomal gene clusters to be undervalued while only a limited number of genomes were sequenced. However, given the availability of hundreds of genomes (which we certainly expect within the next few years), this class of data may well offer a very precise description of the functional coupling between genetic subsystems in prokaryotic genomes.
Supplementary Material
Acknowledgments
This work was supported in part by U. S. Department of Energy, under Contract W-31-109-Eng-38.
ABBREVIATIONS
- BBH
bidirectional best hit
- PCBBH
pair of close bidirectional best hits
- PCH
pair of close homologs
- COGs
clusters of orthologous genes
References
- 1.Demerec M E, Hartman P. Annu Rev Microbiol. 1959;13:377–406. [Google Scholar]
- 2.Lawrence J G, Roth J R. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence J G. Trends Microbiol. 1997;5:355–359. doi: 10.1016/S0966-842X(97)01110-4. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro J A. Trends Genet. 1997;13:98–104. doi: 10.1016/s0168-9525(97)01058-5. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal T. BioEssays. 1998;20:480–487. doi: 10.1002/(SICI)1521-1878(199806)20:6<480::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Overbeek R, Fonstein M, D’Souza M, Pusch G D, Maltsev N. In Silico Biol. 1998;1:0009. http://www.bioinfo.de/isb/1998/01/0009/ ( http://www.bioinfo.de/isb/1998/01/0009/). ). [PubMed] [Google Scholar]
- 7.Overbeek R, Larsen N, Smith W, Maltsev N, Selkov E. Gene. 1997;191:GC1–GC9. doi: 10.1016/s0378-1119(96)00854-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Smith T F. Microbial Comp Genomics. 1998;3:133–140. doi: 10.1089/omi.1.1998.3.133. [DOI] [PubMed] [Google Scholar]
- 9.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. J Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiedeman A A, Keyhani J, Kamholz J, Daum H A, Gots J S, Smith J M. J Bacteriol. 1989;171:205–212. doi: 10.1128/jb.171.1.205-212.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe W, Sampei G, Aiba A, Mizobuchi K. J Bacteriol. 1989;171:198–204. doi: 10.1128/jb.171.1.198-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebbole D J, Zalkin H. J Bacteriol. 1989;171:2136–2141. doi: 10.1128/jb.171.4.2136-2141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zalkin H, Dixon J E. Prog Nucleic Acid Res Mol Biol. 1992;42:259–287. doi: 10.1016/s0079-6603(08)60578-4. [DOI] [PubMed] [Google Scholar]
- 14.Smith J L. Curr Opin Struct Biol. 1995;5:752–757. doi: 10.1016/0959-440x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 15.Gebbia J A, Backenson P B, Coleman J L, Anda P, Benach J L. Gene. 1997;188:221–228. doi: 10.1016/s0378-1119(96)00811-6. [DOI] [PubMed] [Google Scholar]
- 16.Leyva-Vazquez M A, Setlow P. J Bacteriol. 1994;176:3903–3910. doi: 10.1128/jb.176.13.3903-3910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mier P D, Cotton D W. Nature (London) 1966;209:1022–1023. doi: 10.1038/2091022b0. [DOI] [PubMed] [Google Scholar]
- 18.Branny P, De La Torre F, Garel J R. J Bacteriol. 1996;178:4727–4730. doi: 10.1128/jb.178.15.4727-4730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belouski E, Watson D E, Bennett G N. Curr Microbiol. 1998;37:17–22. doi: 10.1007/s002849900330. [DOI] [PubMed] [Google Scholar]
- 20.Sakai H, Ohta T. Eur J Biochem. 1993;211:851–859. doi: 10.1111/j.1432-1033.1993.tb17618.x. [DOI] [PubMed] [Google Scholar]
- 21.Tatusov R L, Koonin E V, Lipman D J. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.