Abstract
Petunia inflata S-locus F-box (Pi SLF) is thought to function as a typical F-box protein in ubiquitin-mediated protein degradation and, along with Skp1, Cullin-1, and Rbx1, could compose an SCF complex mediating the degradation of nonself S-RNase but not self S-RNase. We isolated three P. inflata Skp1s (Pi SK1, -2, and -3), two Cullin-1s (Pi CUL1-C and -G), and an Rbx1 (Pi RBX1) cDNAs and found that Pi CUL1-G did not interact with Pi RBX1 and that none of the three Pi SKs interacted with Pi SLF2. We also isolated a RING-HC protein, S-RNase Binding Protein1 (Pi SBP1), almost identical to Petunia hybrida SBP1, which interacts with Pi SLFs, S-RNases, Pi CUL1-G, and an E2 ubiquitin-conjugating enzyme, suggesting that Pi CUL1-G, SBP1, and SLF may be components of a novel E3 ligase complex, with Pi SBP1 playing the roles of Skp1 and Rbx1. S-RNases interact more with nonself Pi SLFs than with self Pi SLFs, and Pi SLFs also interact more with nonself S-RNases than with self S-RNases. Bacterially expressed S1-, S2-, and S3-RNases are degraded by the 26S proteasomal pathway in a cell-free system, albeit not in an S-allele–specific manner. Native glycosylated S3-RNase is not degraded to any significant extent; however, deglycosylated S3-RNase is degraded as efficiently as the bacterially expressed S-RNases. Finally, S-RNases are ubiquitinated in pollen tube extracts, but whether this is mediated by the Pi SLF–containing E3 complex is unknown.
INTRODUCTION
Self-incompatibility (SI) is an intraspecific reproductive barrier that allows pistils of a plant to reject self pollen (genetically related pollen) but to accept nonself pollen (genetically unrelated pollen) for fertilization (de Nettancourt, 2001). SI is further classified into two major types, sporophytic SI (SSI) and gametophytic SI (GSI), based on how pollen behavior in SI interactions is determined. In the simplest cases, both SSI and GSI are controlled by a highly polymorphic locus, named the S-locus, which contains two separate genes: one encoding female specificity and the other encoding male specificity (Takayama and Isogai, 2005). For SSI, the pollen phenotype is determined by the S-genotype of the pollen parent, and for GSI, the pollen phenotype is determined by the S-genotype of the pollen itself. In GSI, matching of the pollen S-allele with either S-allele of the pistil results in the inhibition of pollen tube growth in the style. Only pollen carrying a different S-allele from both S-alleles of the pistil can grow down through the style to effect fertilization in the ovary.
One type of GSI mechanism that has been studied extensively is the S-RNase–based SI, which has been found in the Solanaceae, Scrophulariaceae, and Rosaceae (Kao and Tsukamoto, 2004). In these families, the S-RNase gene, a highly polymorphic gene first identified in Nicotiana alata (Solanaceae) (Anderson et al., 1986), controls the pistil specificity in SI (Lee et al., 1994; Murfett et al., 1994). The RNase activity of S-RNases is required for their function in rejecting self pollen (Huang et al., 1994), and results consistent with rRNA degradation being responsible for growth inhibition of self pollen tubes have been obtained (McClure et al., 1990). S-RNases are glycoproteins with various numbers of N-linked glycan chains; however, the carbohydrate moiety is not required for their function in SI (Karunanandaa et al., 1994). Thus, the recognition function of S-RNases appears to reside in the protein backbone.
The S-Locus F-box (SLF) gene, which controls pollen specificity, was identified recently. Lai et al. (2002) first reported the identification of Ah SLF (Antirrhinum hispanicum SLF) from sequencing a 63-kb region of the S2-locus of A. hispanicum (Scrophulariaceae) that contains the S2-RNase gene. Subsequently, SLF (also named SFB for S-Locus F-Box) was identified in several species of the genus Prunus (Rosaceae). For example, Pm SLF (P. mume SLF) was identified from sequencing a 62.5-kb S7-locus region (Entani et al., 2003), and Pd SFB (P. dulcis SFB) was identified from sequencing a 70-kb Sc-locus region of P. dulcis (Ushijima et al., 2003). In Petunia inflata (Solanaceae), Pi SLF was identified from sequence analysis of a 328-kb S-locus region that contains the S2-RNase gene (Wang et al., 2004). The role of Pi SLF in SI was demonstrated by introducing its S2-allele, Pi SLF2, into S1S1, S1S2, and S2S3 plants and showing that the Pi SLF2 transgene caused the breakdown of SI in pollen carrying the S1- or S3-allele but not in pollen carrying the S2-allele (Sijacic et al., 2004). The rationale for this approach was based on the well-documented phenomenon called competitive interaction, in which pollen carrying two different S-alleles (heteroallelic pollen), but not pollen carrying two copies of the same S-allele (homoallelic pollen), fails to function in SI. Interestingly, introducing the S2-allele of Ah SLF into Petunia hybrida S3S3 plants also caused the breakdown of SI in S3 pollen that inherited the transgene (Qiao et al., 2004a), even though Ah SLF2 is only ∼30% identical in amino acid sequence to Ph SLF3 and Pi SLF2. Several pollen-part mutants have been found to be associated with either truncation or deletion of SLF/SFB, strongly suggesting that this gene controls the pollen specificity in the Rosaceae (Ushijima et al., 2004; Sonneveld et al., 2005).
The finding that the pollen specificity gene encodes an F-box protein provides a clue to the biochemical mechanism of S-specific inhibition of pollen tube growth. Most F-box proteins are involved in ubiquitin-mediated protein degradation. This system uses E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes to catalyze the transfer of polyubiquitin chains to specific protein substrates for degradation by the 26S proteasome (Bai et al., 1996). One class of E3s is the RING-related class, which can be further divided into single subunit RING/U-box E3s and multisubunit E3s (Moon et al., 2004; Willems et al., 2004). SCF, a class of the multisubunit E3, consists of Skp1, Cullin-1, F-box protein, and Rbx1 (a RING-H2 protein) (Willems et al., 2004). Cullin-1 serves as the scaffold protein; its N- and C-terminal domains interact with Skp1 and Rbx1, respectively. The F-box domain of F-box proteins interacts with Skp1, and a separate domain interacts with specific protein substrates (Zheng et al., 2002). The substrate-interacting domains may contain WD40 repeats, Leu-rich repeats, other protein–protein interaction modules, or unrecognizable motifs (Cenciarelli et al., 1999; Moon et al., 2004). None of the SLFs characterized to date contains any known protein–protein interaction motifs in the C-terminal region (i.e., outside of the N-terminal F-box domain). Qiao et al. (2004b) have shown, by yeast two-hybrid, in vivo coimmunoprecipitation, and pull-down assays, that this C-terminal region of Ah SLF2 interacts with both its self and nonself S-RNases with no allelic specificity. Huang et al. (2006) recently used yeast two-hybrid screens to identify a Skp1-like protein that interacted with the F-box domain of Ah SLF2, suggesting that Ah SLF is likely to be a component of an SCF complex.
Using immunolocalization of pollinated pistils, Luu et al. (2000) showed that, in Solanum chacoense (Solanaceae), both self and nonself S-RNases were localized in the cytoplasm of the pollen tube. By contrast, Goldraij et al. (2006) reported that, in N. alata, both self and nonself S-RNases were sequestered in a vacuolar compartment of the pollen tube at an early stage (16 h after pollination) of pollen tube growth in the style. At a later stage (36 h after pollination), the compartment in the incompatible pollen tube was disrupted, presumably releasing S-RNases into the cytoplasm of the pollen tube to exert their cytotoxic function, whereas the compartment in the compatible pollen tube remained intact. However, Goldraij et al. (2006) could not rule out the possibility that some small amounts of self and nonself S-RNases taken up by the pollen tube were present in the cytoplasm. Thus, even if S-RNases are indeed compartmentalized after they are taken up by the pollen tube, it would appear that some mechanism should still be required to prevent nonself S-RNases from exerting their cytotoxic effect on pollen tubes.
A current model for the function of SLF is predicated on the assumption that SLF is a component of a canonical SCF complex (Qiao et al., 2004a, 2004b; Sijacic et al., 2004; Huang et al., 2006). This model predicts that an SLF interacts with its self and nonself S-RNases differently so that only nonself S-RNases are ubiquitinated and degraded by the 26S proteasome. As a first step toward testing the validity of this model in P. inflata, we set out to identify potential components of the putative SCFPi SLF complex. The results suggest that a Cullin-1 (named Pi CUL1-G), a RING finger protein (named Pi SBP1), and Pi SLF are likely to be components of a novel E3 ligase complex, with Pi SBP1 playing the roles of Skp1 and Rbx1 in the canonical SCF complex. The in vitro binding assay was also used to examine the interactions between S-RNases and Pi SLFs, and the results suggest that S-RNases interact with their nonself Pi SLFs to a greater extent than with their self Pi SLF and that Pi SLFs exhibit a similar binding property for S-RNases. Finally, we used pollen tube extracts and purified recombinant S1-, S2-, and S3-RNases, as well as both glycosylated and deglycosylated forms of S3-RNase purified from pistils, to show that all nonglycosylated S-RNases were degraded via the 26S proteasome pathway and that the recombinant S-RNases were ubiquitinated.
RESULTS
Isolation and Characterization of Three Skp1 Genes of P. inflata
To isolate Skp1 genes of P. inflata, we used cDNAs for ASK1 and ASK2, two well-characterized Arabidopsis thaliana Skp1s, as probes to screen an S1S1 pollen cDNA library under low-stringency hybridization conditions. Screening of 3 × 105 plaque-forming units (pfu) resulted in four independent clones, and sequencing revealed that they all corresponded to the same gene. The longest cDNA was 681 bp, with a 468-bp open reading frame. The deduced amino acid sequence was 80 and 83% identical to the amino acid sequences of ASK1 and ASK2, respectively, suggesting that this cDNA encodes a Skp1. The corresponding gene was thus named Pi SK1 (for P. inflata SKP1). Genomic gel blot analysis revealed that, under low-stringency hybridization conditions, Pi SK1 hybridized to at least three additional genomic fragments of P. inflata (data not shown). To isolate other Skp1 genes homologous with Pi SK1, we used the full-length Pi SK1 cDNA as a probe to screen 3 × 106 pfu of an S2S2 pollen cDNA library under low-stringency hybridization conditions. Twenty-two positive clones were isolated, and sequencing revealed that 6 encoded Pi SK1 and the other 16 corresponded to two Pi SK1 homologues. These two genes were named Pi SK2 and Pi SK3. Alignment of the amino acid sequences of Pi SK1, Pi SK2, and Pi SK3 with those of ASK1, ASK2, and a human SKP1 (Schulman et al., 2000) is shown in Supplemental Figure 1 online. Pair-wise amino acid sequence identities between these three P. inflata Skp1 proteins range from 90 to 92%, which is greater than the 79% amino acid sequence identity between ASK1 and ASK2.
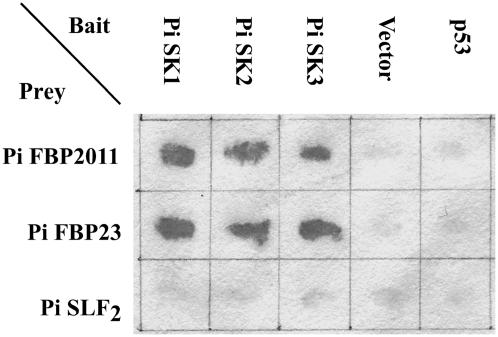
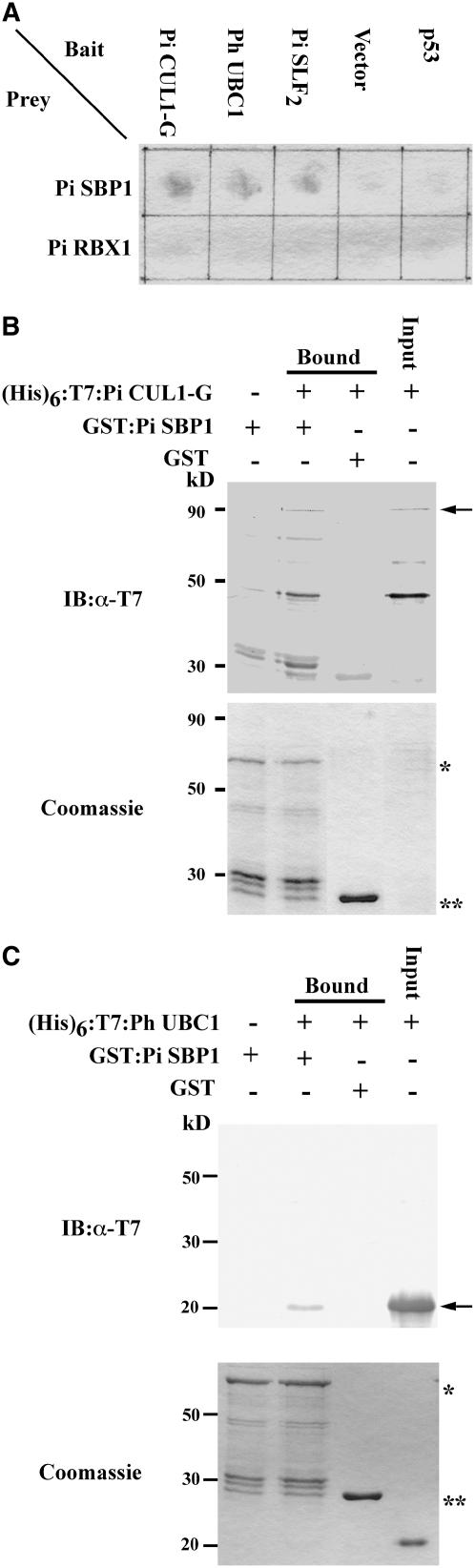
We next used the yeast two-hybrid assay to examine whether Pi SK1, Pi SK2, and Pi SK3 interact with Pi SLF. The coding sequences of Pi SK1, Pi SK2, and Pi SK3 were inserted into a yeast two-hybrid bait vector, pGBD-C1 (James et al., 1996), and the coding sequence of Pi SLF2, the product of the S2-allele of Pi SLF, was inserted into a prey vector, pGAD-C1 (James et al., 1996). No positive interactions were observed between any of these three Pi SKs and Pi SLF2 (Figure 1). The yeast two-hybrid assay was also performed using Pi SK1 and Pi SK2 in pGAD-C1 and Pi SLF1 (for S1-allele of Pi SLF) and Pi SLF2 in pGBD-C1. Again, no interactions were observed in any of the four possible combinations of Pi SKs and Pi SLFs (data not shown). To ascertain whether these three Pi SKs are bona fide Skp1s, we used pGBD-C1-Pi SK1 as bait to screen an S2S2 pollen prey library previously constructed in pGAD424 (Skirpan et al., 2001). Twenty independent colonies were isolated under high-stringency screening. PCR fingerprinting and sequencing revealed that these 20 clones represented seven different genes, and the deduced amino acid sequences of all of them contained an F-box domain at the N terminus. β-Galactosidase activity assays showed that all seven of these F-box proteins interacted strongly with Pi SK1, Pi SK2, and Pi SK3; the results for two of these F-box proteins, named Pi FBP23 and Pi FBP2011 (for P. inflata F-Box Protein 23 and 2011, respectively), are shown in Figure 1. The observation that all of the interacting proteins of Pi SK1 isolated from the yeast two-hybrid screen are F-box proteins suggests that Pi SK1 and its homologues, Pi SK2 and Pi SK3, are bona fide Skp1 proteins. None of the genes encoding these seven F-box proteins are likely linked to the S-locus, because no restriction fragment length polymorphism was observed when cDNAs for these genes were used as probes in genomic gel blot analysis of S1S1, S2S2, and S3S3 genotypes (data not shown).
Figure 1.
Yeast Two-Hybrid Assay of Interactions between Three Pi SKs and Three F-Box Proteins.
Pi FBP2011 and Pi FBP23 are F-box proteins that are likely encoded by genes unlinked to the S-locus. The bait vector is pGBD-C1, and the prey vector is pGAD-C1; p53 is a tumor suppressor and was used here as a negative control. Six colonies of yeast SFY526 coexpressing a pair of bait and prey proteins were streaked on filter paper for β-galactosidase activity assay. The paper was incubated in an X-Gal–containing solution (Breeden and Nasmyth, 1985) for 1 h at 30°C.
Pi SLF Does Not Interact with Arabidopsis Skp1 Proteins
The observation that Pi SLF2 did not interact with Pi SK1, Pi SK2, or Pi SK3 suggested that Skp1 might not be a component of the complex containing Pi SLF. To address this possibility, we first examined whether there is any Skp1 that interacts with Pi SLF2. Because the complete genome sequence of Petunia was not available, we tested the interactions of Pi SLF2 with the Arabidopsis Skp1s. The Arabidopsis genome sequence predicts the existence of 19 Skp1 genes, ASK1 through ASK19 (Farras et al., 2001; Gagne et al., 2002; Zhao et al., 2003), which, based on phylogenetic studies, have been classified into seven subgroups (Zhao et al., 2003). We chose one member from each subgroup (ASK1, -4, -5, -9, -11, -13, and -16) to test whether their encoded proteins interact with Pi SLF2. The yeast two-hybrid assay showed that none of these seven ASKs interacted with Pi SLF2 (data not shown). Because Skp1 interacts with F-box proteins through their F-box domain, we also used the yeast two-hybrid assay to examine whether any of these seven Arabidopsis ASKs interacts with Pi SLF2(FB), the F-box domain (amino acids 1 to 49) of Pi SLF2. Again, no interactions were observed (data not shown).
Identification of Pi SBP1 as an Interacting Partner of Pi SLF
To further examine the possibility that Pi SLF might interact with other Skp1(s) present in the pollen tube, we used Pi SLF2, Pi SLF2(FB), and Pi SLF2(CTD), which contains the C-terminal domain (amino acids 50 to 389), as baits to separately screen the S2S2 pollen prey library; 10, 13, and 7 positive colonies were isolated for pGBD-C1-Pi SLF2, pGBD-C1-Pi SLF2(FB), and pGBD-C1-Pi SLF2(CTD), respectively. Restriction digestion and sequence analysis revealed that the cDNAs contained in all of the positive clones corresponded to the same gene whose deduced amino acid sequence was 98% identical with that of Ph SBP1 (for P. hybrida S-RNase Binding Protein1). Ph SBP1 had previously been identified by Sims and Ordanic (2001) from yeast two-hybrid screens using as baits truncated P. hybrida S-RNases (containing the two hypervariable regions, HVa and HVb, and the conserved region C3 but missing the conserved regions C1, C2, C4, and C5; defined in Ioerger et al., 1991). A Ph SBP1 homologue was identified in S. chacoense using the same approach (O'Brien et al., 2004). Because the amino acid sequence of the protein found to interact with Pi SLF2 is almost completely identical to that of Ph SBP1, the protein was named Pi SBP1 (for P. inflata SBP1). An alignment of the amino acid sequences of Pi SBP1, Ph SBP1, and Sc SBP1 (for S. chacoense SBP1) is shown in Supplemental Figure 2 online. Amino acid sequence analysis by SMART showed that all three of these proteins have two protein–protein interaction domains: a coiled-coil region between amino acids 183 and 227, and a RING-HC domain between amino acids 289 and 323.
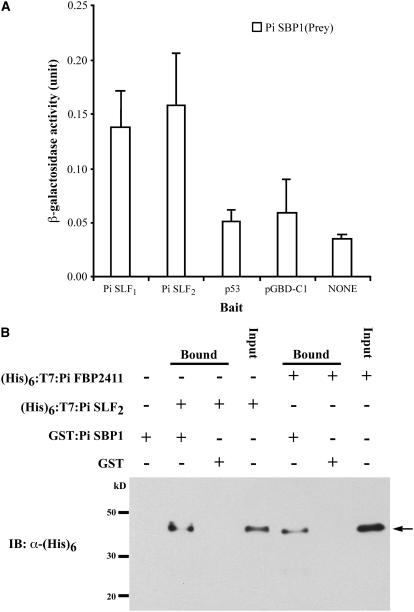
To determine whether Pi SLF1 also interacts with Pi SBP1, pGAD-C1-Pi SBP1 was separately cotransformed with pGBD-C1-Pi SLF1 and pGBD-C1-Pi SLF2 into yeast. The interactions of Pi SBP1 with Pi SLF1 and Pi SLF2 were quantified by an o-nitrophenyl-β-d-galactoside assay of the β-galactosidase activity produced by the colonies (Figure 2A). Comparable activities were detected for the interactions of Pi SBP1 with Pi SLF1 and Pi SLF2, whereas only background activities were detected in the negative controls.
Figure 2.
Analyses of Interactions of Pi SBP1 with Pi SLF1 and Pi SLF2.
(A) Yeast two-hybrid assay. Pi SBP1 (in prey vector pGAD-C1) along with three bait constructs containing Pi SLF1, Pi SLF2, or p53, and with the bait vector pGBD-C1, were separately cotransformed into the yeast strain SFY526, and the transformants were used for the assay. Pi SBP1 (in pGAD-C1) alone was also transformed into SFY526 (NONE). The β-galactosidase activities shown are mean values + sd measured from six independent yeast transformants.
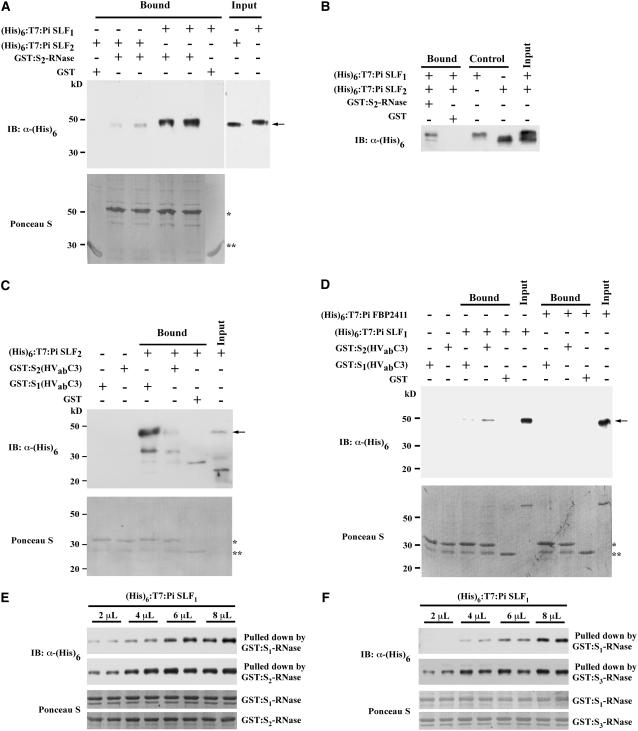
(B) In vitro binding assay. Pi SLF2 and Pi FBP2411 (an F-box protein likely encoded by a gene unlinked to the S-locus) were expressed as (His)6:T7:Pi SLF2 and (His)6:T7:Pi FBP2411, respectively, and the purified proteins were incubated separately with GST:Pi SBP1-bound Glutathione Sepharose 4 Fast Flow resin. As negative controls in this figure and in Figures 4B, 6B, and 6C, the same amount of GST:Pi SBP1-bound resin used in each binding assay was incubated without any (His)6:T7-tagged protein, and GST-bound resin was incubated with the (His)6:T7-tagged protein, as indicated, under the same conditions used in each binding assay. The bound proteins (arrow) were eluted and analyzed by immunoblotting (IB) using an anti-(His)6 antibody. Input lanes in this figure and in Figures 4B, 5C, 5D, 6B, and 6C contain a fraction of the (His)6:T7-tagged protein, as indicated, used in the binding assay.
The interaction between Pi SBP1 and Pi SLF2 was further confirmed by an in vitro binding assay. The coding sequence of Pi SBP1 was cloned into an expression vector, pGEX-5X-1, to produce a glutathione S-transferase (GST) fusion protein, GST:Pi SBP1, and the coding sequence of Pi SLF2 was cloned into another expression vector, pET28, to produce a (His)6- and T7-tagged protein, (His)6:T7:Pi SLF2. To determine whether Pi SBP1 interacts with other F-box proteins, we also cloned the coding sequence of Pi FBP2411 (one of the seven F-box proteins found to interact with Pi SK1) into pET28 to produce (His)6:T7:Pi FBP2411. As shown in Figure 2B, (His)6:T7:Pi SLF2 was detected only when both the GST:Pi SBP1-bound resin and this protein were present in the reaction mixture. Similarly, (His)6:T7:Pi FBP2411 was detected only when both the GST:Pi SBP1-bound resin and this protein were present in the reaction mixture. Thus, the interaction of Pi SBP1 with Pi SLF is not specific to the F-box protein involved in SI.
Genomic Complexity and Expression Pattern of Pi SBP1
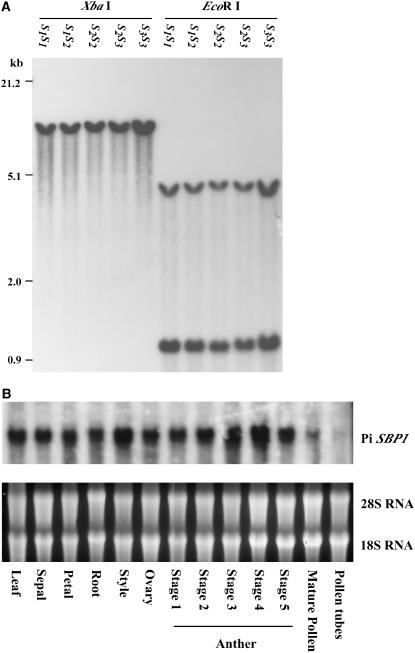
To determine whether Pi SBP1 exhibits S-specific restriction fragment length polymorphism, we used the full-length Pi SBP1 cDNA as a probe for genomic gel blot analysis of five different S-genotypes, S1S1, S1S2, S2S2, S2S3, and S3S3, of P. inflata (Figure 3A). The same hybridizing fragment was detected in XbaI digests of all of the genotypes (no XbaI recognition sequence is present in the Pi SBP1 cDNA), and the same two hybridizing fragments were detected in EcoRI digests of all of the genotypes (one EcoRI recognition sequence is present in the Pi SBP1 cDNA). Thus, it is likely that Pi SBP1 is a single-copy gene and unlinked to the S-locus. RNA gel blot analysis showed that Pi SBP1 was expressed in all of the tissues examined (Figure 3B). All of these results are similar to those reported for Ph SBP1 by Sims and Ordanic (2001). Interestingly, the relative expression level of Pi SBP1 in anthers of different developmental stages and in pollen/pollen tubes is very similar to that of Pi SLF (Sijacic et al., 2004). The expression level peaked in stage 3 and stage 4 anthers, after the completion of meiosis of pollen mother cells to produce microspores (Lee et al., 1996), and was reduced significantly in mature pollen and in vitro–cultured pollen tubes (Figure 3B).
Figure 3.
Genomic Hybridization and Expression Pattern of Pi SBP1.
(A) DNA gel blot analysis. Genomic DNA (15 μg) isolated from each S-genotype indicated was digested with EcoRI or XbaI, and the blot was hybridized with the full-length Pi SBP1 cDNA.
(B) RNA gel blot analysis. Each lane contained 20 μg of total RNA isolated from the tissue indicated, and the blot was hybridized with the full-length Pi SBP1 cDNA. The anther stages are defined by flower-bud size as described previously (Lee et al., 1996). Ethidium bromide staining of the gel used in blotting shows equal loading of the RNA samples.
Pi SBP1 Interacts with S-RNases but Not with an S-Like RNase
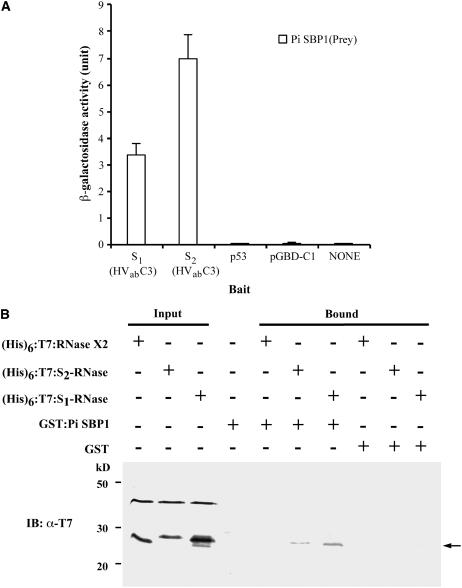
Because both Ph SBP1 and Sc SBP1 had been shown by the yeast two-hybrid assay to interact with truncated S-RNases containing the two hypervariable regions (HVa and HVb) and the conserved region C3 (Sims and Ordanic, 2001; O'Brien et al., 2004), we examined whether Pi SBP1 also interacts with the corresponding region of S1- and S2-RNases of P. inflata. The sequences for this region of S1-RNase (amino acid residues 47 to 97) and S2-RNase (amino acid residues 46 to 95) were used to make two bait constructs, pGBD-C1-S1(HVabC3) and pGBD-C1-S2(HVabC3), both of which were separately cotransformed with pGAD-C1-Pi SBP1 into yeast. The o-nitrophenyl-β-d-galactoside assay showed that Pi SBP1 interacted with the truncated S1- and S2-RNases (Figure 4A). Similar to what was reported by Sims and Ordanic (2001) for Ph SBP1 and by O'Brien et al. (2004) for Sc SBP1, when the mature S1- and S2-RNases were used as baits, no interaction with Pi SBP1 was detected by the yeast two-hybrid assay (data not shown). Hereafter, the terms S1-RNase, S2-RNase, and S3-RNase refer to the mature forms of these S-RNases without the leader peptide (i.e., amino acid residues 1 to 200 for S1-RNase, 1 to 199 for S2-RNase, and 1 to 200 for S3-RNase).
Figure 4.
Analyses of Interactions of Pi SBP1 with Full-Size and Truncated S1- and S2-RNases.
(A) Yeast two-hybrid assay. The assay was performed as described in the legend to Figure 2A. S1(HVabC3) and S2(HVabC3) are truncated S1- and S2-RNase, respectively, each containing the two hypervariable regions and the conserved C3 region. All of the negative controls are as described in the legend to Figure 2A.
(B) In vitro binding assay. The interactions between GST:Pi SBP1 and purified (His)6:T7-tagged S1-RNase, S2-RNase, and RNase X2 were analyzed as described in the legend to Figure 2B. The bound proteins (arrow) were detected by immunoblotting (IB) using an anti-T7 antibody. The band above the (His)6:T7-tagged protein in each input lane is an E. coli protein that copurified with the tagged protein.
The failure to detect any interaction between Pi SBP1 and S1- and S2-RNases by the yeast two-hybrid assay could be because S-RNase did not fold properly in the yeast cell, as it has eight conserved Cys residues involved in four intramolecular disulfide bonds (Ishimizu et al., 1996; Oxley and Bacic, 1996). Therefore, we used the in vitro binding assay, as described for Figure 2B, to reexamine the interaction. The coding sequences of S1- and S2-RNases were cloned into pET28 to produce (His)6:T7:S1-RNase and (His)6:T7:S2-RNase, respectively. As a control, the coding sequence (without the signal peptide; amino acid residues 1 to 193) of RNase X2 of P. inflata (Lee et al., 1992) was also cloned into pET28 to produce (His)6:T7:RNase X2. RNase X2, an S-like RNase, is 43% identical to S1- and S2-RNases, and it was used to ascertain whether any interaction observed with either S-RNase is specific to S-RNase. The results showed that Pi SBP1 interacted with both S1- and S2-RNases but not with RNase X2 (Figure 4B).
S-RNases Interact with Nonself Pi SLFs to a Greater Extent Than with Self Pi SLFs, and Pi SLFs Show a Similar Binding Property for S-RNases
The in vitro binding assay described above was used to examine whether a Pi SLF interacts with its self S-RNase and/or nonself S-RNases, and if it interacts with both, whether there is any difference in the extent of the interaction. The coding sequences of S1-, S2-, and S3-RNases were fused in-frame to the GST coding sequence in expression vector pGEX-5X-1 to produce GST:S1-RNase, GST:S2-RNase, and GST:S3-RNase, respectively. The coding sequence of RNase X2 (without the signal peptide) was similarly fused to the GST coding sequence to produce GST:RNase X2. The coding sequence of Pi SLF1 was cloned into pET28 to produce (His)6:T7:Pi SLF1. We first showed that both Pi SLF1 and Pi SLF2 interacted with S1- and S2-RNases but not with RNase X2 (see Supplemental Figure 3 online), suggesting that Pi SLFs specifically interact with S-RNases. To examine whether an S-RNase interacts with its self and nonself Pi SLFs to different extents, equal amounts of (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 were separately incubated with the same amount of resin-bound GST:S2-RNase, and the bound proteins were detected by an anti-(His)6 antibody. As shown in Figure 5A, the intensity of the (His)6:T7:Pi SLF1 band was much stronger than that of the (His)6:T7:Pi SLF2 band. The identity of these two bands was further confirmed by the anti-T7 antibody (data not shown). No binding was detected between GST and either (His)6:T7:Pi SLF1 or (His)6:T7:Pi SLF2 (Figure 5A). These results suggest that S2-RNase interacts with its nonself Pi SLF to a greater extent than with its self Pi SLF. To further confirm this finding, we performed the in vitro binding assay in a single reaction mixture containing GST:S2-RNase and equal amounts of (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 (Figure 5B). As these two (His)6:T7-tagged proteins can be clearly separated by SDS-PAGE (see control lanes), we were able to assess the relative intensity of these two protein bands. Consistent with the results shown in Figure 5A, the intensity of the (His)6:T7:Pi SLF1 band was much stronger than that of the (His)6:T7:Pi SLF2 band.
Figure 5.
In Vitro Binding Assay of Interactions of Pi SLFs with S1-, S2-, and S3-RNases.
(A) Interactions of Pi SLF1 and Pi SLF2 with S2-RNase. Equal amounts of purified (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 were assayed for their interactions with GST:S2-RNase, as described in the legend to Figure 2B, except that two independent reactions were performed to assess each interaction. Reaction mixtures containing GST-bound resin and the (His)6:T7-tagged protein, as indicated, were used as negative controls. Each input lane contains 10% of the amount of the (His)6:T7-tagged protein used in the binding reaction; note that the intensities of the (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 bands are approximately equal. Top panel, immunoblot (IB). The arrow indicates the (His)6:T7:Pi SLF1 band and the (His)6:T7:Pi SLF2 band detected by the anti-(His)6 tag antibody. Bottom panel, Ponceau S staining of a part of the blot shown in the top panel before immunoblotting to reveal equal amounts of GST:S2-RNase used in the binding assay. The single asterisk indicates the GST:S2-RNase band, and the double asterisks indicate the GST band.
(B) Competition between Pi SLF1 and Pi SLF2 for interactions with S2-RNase. Equal amounts of purified (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 were mixed and used with GST:S2-RNase in the binding assay. Purified (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 were loaded in the control lanes to mark their respective positions on the gel. The reaction mixture containing GST-bound resin and both (His)6:T7-tagged proteins served as a negative control. The input lane contains 10% of the amount of the mixture of the two (His)6:T7-tagged proteins used in the binding reaction; note that the intensities of the (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 bands are approximately equal. (His)6:T7:Pi SLF1 (top band) and (His)6:T7:Pi SLF2 (bottom band) were detected by the anti-(His)6 tag antibody.
(C) Interactions of Pi SLF2 with truncated S1- and S2-RNases. Purified (His)6:T7:Pi SLF2 was tested for its interaction with GST:S1(HVabC3) and GST:S2(HVabC3). The binding assay and negative control assays were performed as described in the legend to Figure 2B, except that GST:S1(HVabC3)-bound and GST:S2(HVabC3)-bound resins were used. The two truncated S-RNases are described in the legend to Figure 4A. Top panel, immunoblot. The arrow indicates the (His)6:T7:Pi SLF2 band detected by the anti-(His)6 tag antibody; the other bands with lower molecular masses cross-reacted with the antibody and were detected only after a long exposure of the blot. Bottom panel, Ponceau S staining of the blot shown in the top panel before immunoblotting; the single asterisk indicates the GST:S1(HVabC3) and GST:S2(HVabC3) bands, and the double asterisks indicate the GST band.
(D) Interactions of Pi SLF1 with truncated S1- and S2-RNases. Except for the use of (His)6:T7-tagged Pi SLF1 and Pi FBP2411, the binding assay and negative control assays were performed as described for (C). The top and bottom panels are as described for (C), except that the arrow indicates the (His)6:T7:Pi SLF1 and (His)6:T7:Pi FBP2411 bands.
(E) Binding differences between Pi SLF1 and S1- and S2-RNases. Different amounts of purified (His)6:T7:Pi SLF1 were used with equal amounts of GST:S1-RNase and GST:S2-RNase in separate binding assays, as described in the legend to Figure 2B. The top two panels show immunoblots against the anti-(His)6 tag antibody, and the bottom two panels show Ponceau S staining of the same blots before immunoblotting.
(F) Binding differences between Pi SLF1 and S1- and S3-RNases. The assays were performed as described for (E) except that a different batch of purified (His)6:T7:Pi SLF1 and a different GST-tagged nonself S-RNase, GST:S3-RNase, were used.
We next examined whether Pi SLFs have a similar binding property for S-RNases, using the truncated S1- and S2-RNases that had been shown to interact with Pi SBP1 (Figure 4A). Purified GST:S1(HVabC3) and GST:S2(HVabC3) fusion proteins were used along with (His)6:T7:Pi SLF2 in the in vitro binding assay. As shown in Figure 5C, (His)6:T7:Pi SLF2 interacted with both truncated S-RNases, but the intensity of the (His)6:T7:Pi SLF2 band was stronger in the binding reaction containing GST:S1(HVabC3) than in that containing GST:S2(HVabC3). Moreover, (His)6:T7:Pi SLF1 interacted with both truncated S-RNases, but the intensity of the (His)6:T7:Pi SLF1 band was stronger in the binding reaction containing GST:S2(HVabC3) than in that containing GST:S1(HVabC3) (Figure 5D). These results suggest that both (His)6:T7:Pi SLF1 and (His)6:T7:Pi SLF2 interact with their respective truncated nonself S-RNases to a greater extent than with their respective truncated self S-RNases.
To determine whether the interaction between S-RNase and Pi SLF is specific to Pi SLF, we examined whether (His)6:T7:Pi FBP2411 interacts with GST:S1(HVabC3) and GST:S2(HVabC3) in the in vitro binding assay. Pi FBP2411 was chosen because we had shown that both Pi FBP2411 and Pi SLF2 interacted with Pi SBP1 (Figure 2B). No interactions were observed between (His)6:T7:Pi FBP2411 and either of these two truncated S-RNases (Figure 5D), suggesting that S-RNases most likely interact specifically with Pi SLFs.
We performed additional in vitro binding assays to further assess the relative extent of binding between Pi SLF1 and S1-, S2-, and S3-RNases. In one assay, equal amounts of GST:S1-RNase and GST:S2-RNase were separately incubated with four different amounts of (His)6:T7:Pi SLF1, with each incubation in duplicate. Increasing binding of (His)6:T7:Pi SLF1 to both GST:S1-RNase and GST:S2-RNase was observed as the amount of (His)6:T7:Pi SLF1 was increased (Figure 5E). For all of the amounts examined, (His)6:T7:Pi SLF1 interacted with its nonself S-RNase, GST:S2-RNase, to a greater extent than with its self S-RNase, GST:S1-RNase. Most importantly, at the lowest amount used (2 μL), there was very little binding with GST:S1-RNase but significant binding with GST:S2-RNase. Another assay was performed similarly except that a different preparation of (His)6:T7:Pi SLF1 was used and that GST:S3-RNase, instead of GST:S2-RNase, was used as the nonself S-RNase. The results (Figure 5F) were similar to those shown in Figure 5E. For all of the amounts examined, (His)6:T7:Pi SLF1 interacted with its nonself S-RNase, GST:S3-RNase, to a greater extent than with its self S-RNase, GST:S1-RNase, and again at the lowest amount used (2 μL), there was very little binding of (His)6:T7:Pi SLF1 with GST:S1-RNase but significant binding with GST:S3-RNase.
Pi SBP1 Interacts with S-RNase and Pi SLF Differently
Because Pi SBP1 contains a coiled-coil region (amino acid residues 183 to 227; see Supplemental Figure 2 online) that could potentially be involved in protein–protein interactions, we examined whether this region is required for Pi SBP1 to interact with Pi SLF and S-RNase. The sequence for the coiled-coil region was deleted from the full-length Pi SBP1 cDNA to produce Pi SBP1(Δcoiled-coil), which was cloned into pGAD-C1. pGAD-C1-Pi SBP1(Δcoiled-coil) was separately cotransformed with pGBD-C1-S1(HVabC3), pGBD-C1-S2(HVabC3), and pGBD-C1-Pi SLF2 into yeast for the two-hybrid assay. The results showed that Pi SBP1(Δcoiled-coil) interacted with S1(HVabC3) and S2(HVabC3) but not with Pi SLF2 (see Supplemental Figure 4 online). Thus, the coiled-coil region of Pi SBP1 is either directly or indirectly required for its interaction with Pi SLF2 but is not required for its interaction with S-RNase, suggesting that S-RNase and Pi SLF interact differently with Pi SBP1.
Pi SBP1 Interacts with a Cullin-1 and an E2 Ubiquitin-Conjugating Enzyme
We isolated cDNAs for Cullin-1 and Rbx1 of P. inflata to examine whether they are components of the complex containing Pi SLF. A partial cullin-1 cDNA clone, previously isolated during our attempts to identify S-linked pollen-expressed genes by RNA differential display (J.A. Verica and T.-h. Kao, unpublished results; McCubbin et al., 2000), was used as a probe to screen 3 × 105 pfu of the S1S1 pollen cDNA library and 6 × 105 pfu of the S2S2 pollen cDNA library. Full-length cDNA clones for this gene, named Pi CUL1-C, were isolated from the S2S2 library, and full-length cDNA clones for another cullin-1 gene, named Pi CUL1-G, were isolated from both cDNA libraries. Pi CUL1-C and Pi CUL1-G shared 80% amino acid sequence identity, and among the five Arabidopsis Cullins (Moon et al., 2004), they are most similar to ATCUL1 (83 and 78% identity, respectively). The deduced amino acid sequences of Pi CUL1-C, Pi CUL1-G, ATCUL1, and a human CUL1 are aligned in Supplemental Figure 5 online. Because Pi CUL1-C and Pi CUL1-G are quite similar in sequence, we chose Pi CUL1-G for all subsequent studies. Screening of 6 × 105 pfu of the S2S2 pollen cDNA library using the full-length cDNA for an Arabidopsis Rbx1, At RBX1 (Lechner et al., 2002), as a probe resulted in the isolation of one class of cDNA clones. The corresponding gene was named Pi RBX1; its deduced amino acid sequence is 87% identical to that of At RBX1. An alignment of the amino acid sequences of Pi RBX1, At RBX1, and a human Rbx1 is shown in Supplemental Figure 6 online.
The yeast two-hybrid assay showed that there were no interactions between Pi CUL1-G and Pi RBX1 (Figure 6A) or between Pi RBX1 and Pi SLF2 (Figure 6A). The findings that Pi RBX1 did not interact with either Pi CUL1-G or Pi SLF2, and that Pi SBP1 interacted with Pi SLF2 (Figures 2 and 6A), raised the possibility that Pi SBP1, but not Pi RBX1, is the RING protein that brings E2 into the complex containing Pi SLF. To address this possibility, we examined whether Pi SBP1 interacts with Pi CUL1-G and Ph UBC1, an E2 of P. hybrida. The yeast two-hybrid assay showed that Pi SBP1 interacted with both Pi CUL1-G and Ph UBC1 (Figure 6A). These interactions were further examined by the in vitro binding assay. The coding sequences of Pi CUL1-G and Ph UBC1 were cloned into pET28 to produce (His)6:T7:Pi CUL1-G and (His)6:T7:Ph UBC1, respectively. The results of the binding assay showed that GST:Pi SBP1 interacted with both (His)6:T7:Pi CUL1-G (Figure 6B) and (His)6:T7:Ph UBC1 (Figure 6C). The interactions of Pi SBP1 with a Cullin-1 and an E2 conjugating enzyme suggest that in the complex containing Pi SLF, Pi SBP1 plays the role of Rbx1 in the canonical SCF complex.
Figure 6.
Interactions of Pi SBP1 with Pi CUL1-G and Ph UBC1.
(A) Yeast two-hybrid assay showing that Pi SBP1, but not Pi RBX1, interacts with Pi CUL1-G, Ph UBC1 (an E2 ubiquitin-conjugating enzyme of P. hybrida), and Pi SLF2. The β-galactosidase activity assay was performed as described in the legend to Figure 1 except that the filter paper was incubated for 5 h at 30°C.
(B) In vitro binding assay of the interaction between Pi SBP1 and Pi CUL1-G. Purified (His)6:T7:Pi CUL1-G was tested for its interaction with GST:Pi SBP1 in the binding assay as described in the legend to Figure 2B. Top panel, immunoblot (IB). The bound (His)6:T7:Pi CUL1-G was detected by the anti-T7 tag antibody and is indicated with an arrow. The other bands with lower molecular masses cross-reacted with the antibody and were detected only after a long exposure of the blot. Bottom panel, Coomassie blue staining of a duplicate gel of that used in immunoblotting. The asterisk indicates the GST:Pi SBP1 band, and the double asterisks indicate the GST band.
(C) In vitro binding assay of the interaction between Pi SBP1 and Ph UBC1. Purified (His)6:T7:Ph UBC1 was used along with GST:Pi SBP1 in the assay as described in the legend to Figure 2B. The bound (His)6:T7:Ph UBC1 was detected by an anti-T7 tag antibody. Top panel, immunoblot. The arrow indicates the (His)6:T7:Ph UBC1 band. Bottom panel, Coomassie blue staining of a duplicate gel of that used in immunoblotting. The asterisk indicates the GST:Pi SBP1 band, and the double asterisks indicate the GST band.
S-RNases Are Degraded via the 26S Proteasomal Pathway in a Non–S-Specific Manner in Pollen Tube Extracts
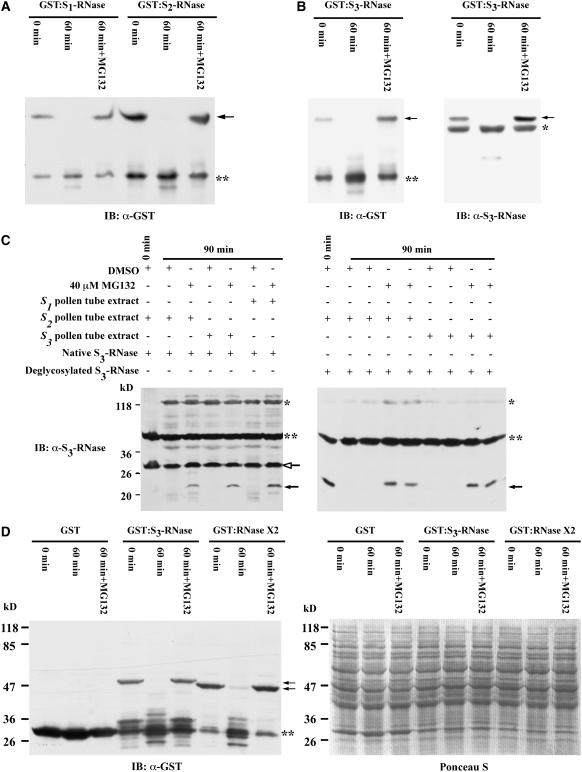
To examine whether pollen tubes contain a ubiquitin-26S proteasome pathway that degrades S-RNases, we developed a cell-free system using extracts of in vitro–germinated pollen tubes and purified GST:S-RNases or native S-RNases. First, GST:S1-RNase and GST:S2-RNase were incubated separately with extracts of in vitro–germinated S2 pollen tubes at 30°C for 1 h in the absence or presence of MG132 (40 μM), a specific 26S proteasome inhibitor (Lee and Goldberg, 1998; Smalle and Vierstra, 2004). The proteins were then separated by SDS-PAGE, and an anti-GST antibody was used to detect GST:S1-RNase and GST:S2-RNase by protein gel blot analysis (Figure 7A). Neither GST:S1-RNase nor GST:S2-RNase was detectable after 1 h of incubation in the absence of MG132. However, in the presence of MG132, the amount of each protein after 1 h of incubation was approximately the same as that before the incubation.
Figure 7.
Degradation Assay of Bacterially Expressed GST, GST:S1-RNase, GST:S2-RNase, GST:S3-RNase, and GST:RNase X2, as Well as S3-RNase Purified from Pistils, by Extracts of S1, S2, or S3 Pollen Tubes.
(A) GST:S1-RNase and GST:S2-RNase. Purified GST fusion proteins (0.3 μg each) were incubated separately with extracts of in vitro–germinated S2 pollen tubes in either the absence or presence of 40 μM MG132 (a specific inhibitor of the 26S proteasome) for 1 h at 30°C. An anti-GST antibody was used to detect GST:S1-RNase and GST:S2-RNase (arrow) as well as GST (double asterisks). IB, immunoblot.
(B) GST:S3-RNase. Purified GST:S3-RNase (0.3 μg) was used in the assay as described for (A), except that both anti-GST and anti-S3-RNase antibodies were used to detect GST:S3-RNase. The arrow indicates GST:S3-RNase, the single asterisk indicates a cross-reacting protein present in the reaction mixture, and the double asterisks indicate GST.
(C) Native glycosylated S3-RNase and its deglycosylated form. S3-RNase (0.1 μg) purified from pistils of the S3S3 genotype (left panel) and an equal amount of purified deglycosylated S3-RNase (right panel) were incubated separately with extracts of S1, S2, and S3 pollen tubes in either the absence or presence of MG132 (40 μM) for 90 min at 30°C. The anti-S3-RNase antibody was used to detect both glycosylated (open-headed arrow) and deglycosylated (closed arrows) S3-RNase. Single and double asterisks indicate cross-reacting proteins in the reaction mixture. A longer exposure time was used for the blot shown in the left panel (5 min) than that shown in the right panel (30 s).
(D) GST:S3-RNase, GST:RNase X2, and GST. Each purified protein (0.3 μg) was used in the assay as described for (A). Left panel, immunoblot. The arrows indicate GST:S3-RNase or GST:RNase X2, and the double asterisks indicate GST. Right panel, Ponceau S staining of the blot shown in the left panel before immunoblotting.
The disappearance of both GST:S-RNase bands, as assayed by the anti-GST antibody, could be attributable to the removal of the GST tag from the fusion proteins by proteolytic cleavage. To rule out this possibility, we performed an additional degradation assay using both the anti-GST antibody and an anti-S3-RNase antibody to examine the fate of GST:S3-RNase in S2 pollen tube extract. As shown in Figure 7B, the GST:S3-RNase band, detected by both antibodies before incubation, was not detectable by either antibody after 1 h of incubation in the absence of MG132, confirming that the S-RNase part of the fusion protein was degraded. As observed in Figure 7A, the disappearance of the GST:S3-RNase band was prevented when MG132 was included in the extract. To further confirm these results, we also performed degradation assays on GST:S1-RNase and GST:S2-RNase using S1 pollen tube extract. The same results were obtained (data not shown).
Because native S-RNases are glycosylated but the bacterially expressed GST:S-RNases are not, we examined whether S3-RNase purified from pistils of the S3S3 genotype was also degraded in pollen tube extracts. The purified native S3-RNase was separately incubated with S1, S2, and S3 pollen tube extracts, and the fate of the protein was analyzed by protein gel blotting using the anti-S3-RNase antibody. The results (Figure 7C, left panel) showed that the native S3-RNase was not degraded to a significant extent, if at all, after 90 min of incubation in the absence of MG132. However, when MG132 was present, a faint band (closed arrow) was detected in all three pollen tube extracts after the same length of incubation. Because this protein band was not detectable before the incubation and had a lower molecular mass than the native S3-RNase band, we hypothesized that it might correspond to the deglycosylated S3-RNase. That is, a small amount of S3-RNase might have been deglycosylated and then degraded during the incubation in the pollen tube extracts without MG132.
To test this hypothesis, S3-RNase purified from the pistils was treated with Peptide:N-Glycosidase F (PNGase F) to remove the N-linked glycan chain attached to Asn-29 (Karunanandaa et al., 1994) and then incubated with S2 or S3 pollen tube extract. The results of the degradation assay are shown in Figure 7C, right panel. The native S3-RNase was completely deglycosylated by PNGase F (0-min lane), and the deglycosylated S3-RNase band was virtually nondetectable after 90 min of incubation in either S2 or S3 pollen tube extract in the absence of MG132. However, the intensity of the deglycosylated S3-RNase band after 90 min of incubation in the presence of MG132 was comparable to that at 0 min.
When a GST fusion protein is produced in Escherichia coli, the GST moiety of some fusion protein molecules is cleaved off around the Factor Xa cleavage site. This was also the case for GST:S1-RNase, GST:S2-RNase, and GST:S3-RNase (Figures 7A and 7B). Under the assay conditions in which nonglycosylated S-RNases were completely degraded, GST molecules released from these recombinant S-RNases were not degraded. To further confirm that the degradation of nonglycosylated S-RNases in our in vitro system was not caused by general proteolytic cleavage, and to assess whether the degradation was specific to S-RNases, recombinant GST and GST:RNase X2 were examined, along with GST:S3-RNase, in S2 pollen tube extract. As shown in Figure 7D, GST was not degraded, whereas GST:RNase X2 was degraded in a similar 26S proteasome–dependent manner as GST:S3-RNase. The findings that neither GST nor the native glycosylated S3-RNase was degraded suggest that the degradation of nonglycosylated S-RNases was not caused by general proteases present in the pollen tube extracts. However, the degradation observed in our in vitro system was not S-specific, nor was it specific to S-RNases.
S-RNases Are Ubiquitinated by Pollen Tube Extracts in a Non–S-Specific Manner
Because the bacterially expressed nonglycosylated GST:S-RNases were degraded to similar extents in pollen tube extracts as the deglycosylated native S3-RNase, we decided to use GST:S2-RNase and GST:S3-RNase to examine whether the degradation of S-RNase resulted from ubiquitination. Recombinant GST and GST:RNase X2 were used as controls to assess whether ubiquitination was restricted to the protein moiety (but not to the GST tag) and was specific to S-RNases. Using GST-tagged S-RNases and RNase X2 also allowed us to simplify the isolation of their ubiquitinated forms, if any, by affinity purification from among many other ubiquitinated proteins in pollen tube extracts (see Supplemental Figure 7 online). The ubiquitination assay was performed in a similar manner to the degradation assay except for the following modifications. First, 30 μg of ubiquitin or (His)6:ubiquitin was added to S2 pollen tube extract before incubation. Second, after incubation, GST:S2-RNase, GST:S3-RNase, GST:RNase X2, GST, and any products derived from them were isolated by Glutathione Sepharose 4 Fast Flow resin. The bound proteins were eluted and separated on two duplicated reducing SDS-polyacrylamide gels, and protein gel blotting was performed separately using the anti-GST antibody and an anti-ubiquitin or anti-(His)6 antibody.
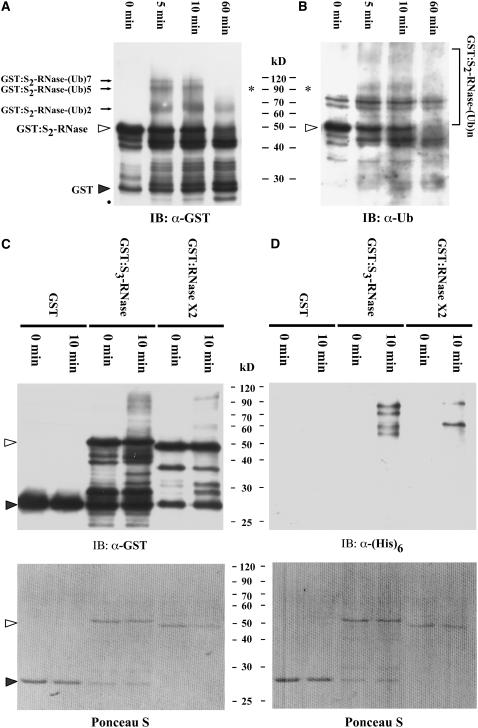
As shown in Figure 8A, after 5 min of incubation in the S2 pollen tube extract at 30°C, the anti-GST antibody detected three discrete protein bands with molecular masses greater than that of GST:S2-RNase, and all of the bands remained clearly visible after 10 min of incubation. One of these bands (Figures 8A and 8B, asterisks) was also detected by the anti-ubiquitin antibody at the 5- and 10-min time points (Figure 8B). Based on the estimated molecular masses of the three bands (67, 93, and 110 kD) detected by the anti-GST antibody, we attributed them to be GST:S2-RNase (50 kD) conjugated with two, five, and seven ubiquitins (with total molecular masses of 67, 92.5, and 109.5 kD, respectively). After 1 h of incubation, the two bands with higher molecular masses had disappeared, but not the one possibly corresponding to GST:S2-RNase conjugated to two ubiquitins. This ubiquitinated GST:S2-RNase might have escaped degradation because the number of conjugated ubiquitin subunits was fewer than the minimum of four required for recognition by the 26S proteasome (Thrower et al., 2000). Moreover, the intensity of a protein band (Figure 8A, black dot) with a lower molecular mass than GST:S2-RNase became more and more visible from the 5-min time point onward. This band could correspond to degradative product(s) of GST:S2-RNase.
Figure 8.
Ubiquitination Assay of GST, GST:S2-RNase, GST:S3-RNase, and GST:RNase X2 by Extracts of S2 Pollen Tubes.
(A) and (B) Time course of ubiquitination and degradation of GST:S2-RNase. GST:S2-RNase (0.5 μg) was used in each reaction containing ubiquitin, and the reaction was stopped at different time points as indicated. The anti-GST antibody was used in the protein gel blot shown in (A), and an anti-ubiquitin antibody was used in the blot shown in (B). The open triangles indicate GST:S2-RNase, and the closed triangle indicates GST. The black dot indicates the degradative products of GST:S2-RNase. The closed arrows indicate three ubiquitinated forms of GST:S2-RNase with two, five, and seven ubiquitin subunits, and the asterisks indicate GST:S2-RNase conjugated with five ubiquitin subunits that was detected by both anti-GST and anti-ubiquitin antibodies. IB, immunoblot.
(C) and (D) Ubiquitination assay of GST, GST:S3-RNase, and GST:RNase X2. Purified GST, GST:S3-RNase, and GST:RNase X2 (0.5 μg each) were used in the assay as described for (A) and (B), except that the reaction contained (His)6:ubiquitin and was analyzed only at the 10-min time point. Each reaction mixture was then divided equally and electrophoresed on two duplicate gels. The transferred membranes were immunoblotted with the anti-GST antibody (C) and the anti-(His)6 antibody (D). Top panels, immunoblot. Bottom panels, Ponceau S staining of the blots shown in the top panels before immunoblotting.
(His)6:ubiquitin was used in the ubiquitination assay of GST, GST:S3-RNase, and GST:RNase X2 in the S2 pollen tube extract (Figures 8C and 8D). Using (His)6:ubiquitin allowed us to detect the ubiquitinated proteins with the anti-(His)6 antibody (Figure 8D) and to compare the results with those obtained with the use of the anti-ubiquitin antibody (Figure 8B). The results of GST:S3-RNase and GST:RNase X2 were similar to those of GST:S2-RNase: after 10 min of incubation in the pollen tube extract, discrete bands with molecular masses greater than those of the respective GST fusion proteins were detected by both the anti-GST antibody (Figure 8C) and the anti-(His)6 antibody (Figure 8D). By contrast, no ubiquitinated forms of GST were detected, confirming that ubiquitination of the GST fusion proteins occurred exclusively in the proteins that were fused to GST. However, similar to the results of the degradation assay, the ubiquitination observed in our in vitro system was not S-specific, nor was it specific to S-RNases.
DISCUSSION
The recent identification of the SLF gene as the pollen S-gene has opened up opportunities to investigate the biochemical mechanism of S-RNase–based SI. As the RNase activity of S-RNases is required for their function in SI (Huang et al., 1994) and rRNA of pollen tubes may be degraded after incompatible pollination (McClure et al., 1990), S-RNases are thought to act as cytotoxic molecules to inhibit the growth of self pollen tubes through RNA degradation. Both Luu et al. (2000) and Goldraij et al. (2006) have shown that uptake of S-RNases by pollen tubes is not S-allele–specific. Before the identification of SLF as the pollen S-gene, one of the models proposed that the pollen S-allele product encodes a cytosolic RNase inhibitor, which specifically inhibits the RNase activity of all nonself S-RNases and renders them nonfunctional inside a pollen tube (Kao and McCubbin, 1996; Golz et al., 2001). Because most F-box proteins are involved in ubiquitin-mediated protein degradation, this model has now been modified to state that a pollen S-allele product mediates the degradation of its nonself S-RNases, but not that of its self S-RNase, inside a pollen tube (Qiao et al., 2004b; Sijacic et al., 2004; Huang et al., 2006). According to this model, pollen S-allele products regulate the stability, rather than the RNase activity, of S-RNases. In this work, we have begun to address the biochemical role of the SLF of P. inflata.
The Complex That Contains Pi SLF Is Not a Canonical SCF Complex
If Pi SLF functions as a conventional F-box protein, it would be expected to be a component of an SCF complex. In this work, we have obtained several lines of evidence to suggest that the Pi SLF–containing complex is a not a canonical SCF complex and instead is a novel E3 ligase complex containing a Cullin-1 protein (Pi CUL1-G), a RING-HC protein (Pi SBP1), and Pi SLF but not containing Skp1 or Rbx1. First, we isolated cDNAs for three Skp1s of P. inflata and showed that none of them interact with Pi SLF2 in the yeast two-hybrid assay (Figure 1) or the in vitro binding assay (data not shown). When Pi SK1, one of these Skp1s, was used as bait in yeast two-hybrid library screening, all seven classes of prey proteins identified were F-box proteins, suggesting that Pi SK1, and most likely its homologues, Pi SK2 and Pi SK3, are bona fide Skp1s. Second, the Arabidopsis genome is predicted to encode 19 Skp1s, but none of the 7 Skp1s representing all seven subgroups interact with Pi SLF2 in the yeast two-hybrid assay. Third, Pi CUL1-G interacts with Pi SBP1 but not with Pi RBX1 (Figures 6A and 6B). Fourth, Pi SLF1 and Pi SLF2 interact with Pi SBP1 (Figure 2) but not with Pi RBX1 (Figure 6A). Fifth, Pi SBP1 interacts with an E2 conjugating enzyme (Figure 6C). Interestingly, the expression of both Pi SLF and Pi SBP1 peaks during pollen development and declines significantly in mature pollen and in vitro–germinated pollen tubes (Figure 3B) (Sijacic et al., 2004). It is possible that the protein produced in developing microspores is retained in mature pollen and pollen tubes.
We have concluded that, in the Pi SLF–containing complex, Pi SBP1 likely plays the roles of Skp1 and Rbx1 of a canonical SCF complex. This is because Pi SBP1, like Skp1, bridges the Cullin-1 component (Pi CUL1-G) and an F-box protein (Pi SLF) and, like Rbx1, interacts with the Cullin-1 component and E2. Pi SBP1 (335 amino acids) is approximately three times the size of Pi RBX1 (116 amino acids), so it could potentially interact with more proteins than does Pi RBX1. Variants of the SCF complex have been reported that contain some but not all of the typical components (Willems et al., 2004). For example, an SCF-like complex in humans consists of Skp1, an F-box protein (Ebi), a RING-HC protein (Siah-1), and an adaptor protein (SIP) that bridges Siah-1 and Skp1 (Matsuzawa and Reed, 2001). In this complex, Siah-1 plays the roles of Cullin-1 and Rbx1, and interestingly, Siah-1 (298 amino acids) is also larger than Rbx1. Another example is the E2F1 transcription factor, which can be ubiquitinated by multiple ROC-Cullin ligases that do not contain Skp1 (Ohta and Xiong, 2001).
Moreover, in Drosophila melanogaster, Sina, a RING-HC protein, forms an E3 ligase complex with Phyllopod (Phyl) and Ebi to regulate the degradation of a transcription repressor protein, Tramtrack (Ttk) (Li et al., 2002). In this complex, Ebi interacts with Sina and the substrate, Ttk. Sina and Phyl can mediate the degradation of Ttk, but Ebi promotes a more efficient degradation of Ttk, perhaps by stabilizing the Sina-Phyl-Ttk complex (Boulton et al., 2000; Li et al., 2002). Because SBP1 could potentially function as a single-subunit E3, the finding by Sims and Ordanic (2001) that Ph SBP1 interacted with S-RNases posed a conundrum concerning why two E3s (Ph SBP1 and the then predicted SLF-containing SCF complex) would be involved in the ubiquitination of S-RNases (McClure, 2004). Our finding that Pi SBP1 is a component of a novel E3 ligase complex containing Pi SLF and that Pi SBP1 likely assumes the roles of Skp1 and Rbx1 of a canonical SCF complex would provide a solution to this conundrum. Moreover, because Pi SBP1 alone interacts with S-RNases, the Pi CUL1-G-Pi SBP1-Pi SLF complex may function in a similar manner as the Sina-Phyl-Ebi complex in that Pi CUL1-G-Pi SBP1 could mediate the basal degradation of all S-RNases and Pi SLF could promote the efficient degradation of specific S-RNases (see below).
Our finding that Skp1 is not a component of the Pi SLF–containing complex contradicts the results for Ah SLF of A. hispanicum. Qiao et al. (2004b) used an anti-Ah SLF2 antibody to pull down protein complexes that contained Ah SLF2 from mixtures of pollen and stylar proteins under both compatible and incompatible conditions. They found proteins that cross-reacted with an anti-ASK1 antibody or with an anti-ATCUL1 antibody. Huang et al. (2006) subsequently used yeast two-hybrid screens to identify a Skp1-like protein, named Ah SSK1, that interacted with the F-box domain of Ah SLF2. Thus, the Ah SLF–containing complex of A. hispanicum may have a different subunit composition from that of the Pi SLF–containing complex of P. inflata. In this context, it is interesting that the SLF of the Solanaceae functions differently at the mechanistic level from the SLF/SFB of the Rosaceae, as all of the pollen-part self-compatible mutants characterized to date in the Solanaceae resulted from duplication, not deletion, of the pollen S-allele (SLF) (Golz et al., 1999, 2001), whereas those in the Rosaceae resulted from defects (deletion or frame-shift mutations) in SLF/SFB (Ushijima et al., 2004; Sonneveld et al., 2005). Moreover, in the Solanaceae, pollen carrying two different functional S-alleles (i.e., SLF alleles) fails to function in SI as a result of competitive interaction (Golz et al., 2001; Sijacic et al., 2004), whereas in sour cherry (Prunus cerasus) of the Rosaceae, such heteroallelic pollen has been shown to function normally in SI (Hauck et al., 2006). Thus, it appears that even though all three of these families use F-box proteins and S-RNases as the male and female determinants, respectively, the biochemical mechanisms of SI are likely to have diverged.
S-RNases Are Degraded via a Ubiquitin-26S Proteasomal Pathway
If the Pi SLF–containing E3 ligase complex is involved in the degradation of S-RNases in pollen tubes, one would expect the degradation to be via the ubiquitin-mediated 26S proteasomal pathway. Previously, Qiao et al. (2004b) addressed the role of this protein degradation pathway in SI of Antirrhinum by examining the effect of 26S proteasome inhibitors on the in vitro growth of pollen tubes in the presence of either compatible or incompatible stylar extracts. They found that the inhibitors had no effect on the growth inhibition of pollen tubes by incompatible stylar extracts but inhibited the growth of pollen tubes in compatible stylar extracts by ∼50%. Qiao et al. (2004b) further showed that protease inhibitors had no significant effect on the growth of compatible pollen tubes; thus, they concluded that protein degradation by the 26S proteasome is required for the growth of compatible pollen tubes. The ubiquitin-mediated 26S proteasomal pathway is thought to be involved in many developmental processes. For example, >5% of the Arabidopsis proteome is related to this pathway (Smalle and Vierstra, 2004). Thus, inhibitors of the 26S proteasome will likely affect a plethora of biochemical events during pollen tube growth, complicating the interpretation of results.
Another approach to address the role of the 26S proteasome in SI is to compare the amounts of S-RNase in pistils after compatible pollination with those after incompatible pollination at various times after pollination, up to the time when the growth of incompatible pollen tubes is inhibited in the style. For example, Qiao et al. (2004b) pollinated S2S5 pistils of Antirrhirum with compatible or incompatible pollen, isolated total proteins from pistils collected at eight different time points (up to 60 h) after compatible or incompatible pollination, and determined the amounts of S2-RNase. They detected a lower amount of S2-RNase in compatibly pollinated pistils than in incompatibly pollinated pistils at 36 h after pollination, and they interpreted the results to mean that S2-RNase was degraded in compatible pollen tubes. Goldraij et al. (2006) used a similar approach to measure the amounts of SC10-RNase of N. alata in SC10SC10 pistils that had been pollinated with SC10 or S105 pollen, but they did not detect any significant difference in the amounts of SC10-RNase between compatibly and incompatibly pollinated pistils up to 48 h after pollination. Because S-RNase is abundant in the style and it is very likely that not all of the S-RNase molecules are taken up by pollen tubes, it is difficult, if not impossible, to know precisely how much of the total amount determined from pollinated pistils is contributed by the S-RNase inside pollen tubes. This may be the reason why Qiao et al. (2004b) and Goldraij et al. (2006) reached opposite conclusions about whether S-RNase is degraded in compatible pollen tubes.
Goldraij et al. (2006) also dissected pollen tubes from SA2SA2 pistils pollinated with compatible or incompatible pollen and determined the amounts of SA2-RNase in soluble and membrane fractions of both compatible and incompatible pollen tubes. They concluded that there was no significant difference in the amounts of SA2-RNase between the soluble or membrane fractions of compatible and incompatible pollen tubes. Accurate determination of the amount of S-RNase inside pollen tubes by this approach requires the isolation of intact pollen tubes completely free of contaminating style tissue, which is abundant in S-RNase. This may be technically difficult, as the pollen tube preparations of Goldraij et al. (2006) also contained transmitting tract cells and extracellular matrix material of the style tissue. Another potential problem of using pollinated pistils or dissected pollen tubes to examine the fate of S-RNase is that if the great majority of S-RNase taken up by a pollen tube is indeed sequestered in a vacuolar compartment and not subject to degradation, as suggested by Goldraij et al. (2006), it would be difficult to accurately assess the degradation of a small amount of S-RNase among a large amount of sequestered S-RNase.
Here, we have developed a cell-free system to examine whether S-RNases are degraded in compatible pollen tubes, and if so, whether this is mediated by the ubiquitin/26S proteasomal pathway. The system involves the use of extracts of in vitro–germinated pollen tubes and exogenously added GST:S-RNases or native S-RNases purified from pistils. As the ubiquitin-mediated 26S proteasome degradation machinery is active in pollen tube extracts, we were able to use this cell-free system to show that the bacterially expressed GST:S1-RNase, GST:S2-RNase, GST:S3-RNase, and GST-RNase X2, but not GST alone, were degraded by S2 pollen tube extracts (Figures 7A, 7B, and 7D). Moreover, we found that the degradation was dependent of the activity of the 26S proteasome, as MG132, a specific inhibitor of the 26S proteasome, completely inhibited the degradation (Figures 7A, 7B, and 7D). We also showed that GST:S2-RNase, GST:S3-RNase, and GST-RNase X2, but not GST alone, were rapidly ubiquitinated in this cell-free system [with the addition of ubiquitin or (His)6:ubiquitin] (Figure 8). The time course of ubiquitination of GST:S2-RNase suggested that the molecules conjugated with four or more ubiquitins were subsequently degraded (Figure 8A and 8B). The use of the native glycosylated S3-RNase in the degradation assay led us to the discovery that deglycosylation was required for the degradation of the native S3-RNase (Figure 7C). It will be interesting to determine why this is the case in our in vitro system and whether this is also true in vivo. One possible explanation is that the N-linked glycan chain of the native S3-RNase might mask the Lys residue(s) essential for ubiquitination.
All of the results from the degradation and ubiquitination assays together suggest that S-RNases are degraded via the ubiquitin-mediated 26S proteasomal pathway. However, for the S1, S2, and S3 pollen tubes tested, no S-specific degradation was observed because both self and nonself S-RNases were degraded in their extracts. It is possible that our in vitro system cannot duplicate the S-specific degradation that exists in vivo. For example, as stated above, Pi CUL1-G-Pi SBP1 might act as an E3 ligase complex to mediate the nonspecific degradation of all S-RNases, and an allelic variant of Pi SLF might promote the degradation of its nonself S-RNases to confer allelic specificity to the E3 ligase complex containing Pi CUL1-G, Pi SBP1, and Pi SLF. If this scenario is true and if the Pi CUL1-G-Pi SBP1 complex predominates over the Pi CUL1-G-Pi SBP1-Pi SLF complex in the pollen tube extracts (e.g., because of the dissociation of Pi SLF from the complex during extraction), we would expect to see the degradation of both self and nonself S-RNases. In vivo, the Pi CUL1-G-Pi SBP1-Pi SLF complex would predominate, resulting in specific degradation of nonself S-RNases. That is, two different E3 ligase complexes may operate in the ubiquitin-26S proteasome pathway to regulate the amount of S-RNases in the cytoplasm of the pollen tube: one for specific degradation of nonself S-RNases, and the other for nonspecific degradation of all S-RNases.
RNase X2 was also ubiquitinated and degraded in our in vitro system (Figures 7D and 8C). This is not unexpected given that ubiquitin-mediated protein degradation is involved in regulating the stability of many proteins. Also, many different E3 ligases are used in this process; for example, ∼1300 genes of the Arabidopsis genome are annotated as encoding components of E3 ligases (Smalle and Vierstra, 2004). Moreover, our results showed that many proteins were ubiquitinated in pollen tube extracts (see Supplemental Figure 7 online). Because RNase X2 did not interact with Pi SLFs (see Supplemental Figure 3 online) or Pi SBP1 (Figure 4B), it would seem likely that an E3 ligase different from the Pi SLF–containing complex mediated the ubiquitination and degradation of RNase X2 in the pollen tube extract. However, we cannot rule out the possibility that the ubiquitination and degradation of S-RNases in our in vitro system also were not mediated by the Pi SLF–containing complex.
Biochemical Model for SI
Two different versions of the inhibitor model had been put forward to explain the S-specific inhibition of pollen tube growth before the identification of the pollen determinant (Golz et al., 2001; Luu et al., 2001). Both models predict that the pollen S-allele products contain an allele-specific domain, but they differ in how the RNase activity of nonself S-RNases is inhibited in the pollen tube. One version proposes that the pollen S-allele products also contain an RNase inhibition domain, whereas the other invokes a general RNase inhibitor as being responsible for the inhibition of RNase activity. Both models predict that the interaction between the matching S-allele–specificity domains of a pollen S-allele product and its cognate S-RNase is thermodynamically more favorable than the nonspecific interaction between the RNase catalytic domain of S-RNases and either the RNase inhibition domain of the pollen S-allele products or the general RNase inhibitor (Kao and Tsukamoto, 2004). As a result, in the case of self interaction, the RNase activity of self S-RNase would not be inhibited either by its cognate pollen S-allele product or by the general RNase inhibitor. On the contrary, in the case of nonself interaction, the RNase activity of any nonself S-RNase would be inhibited through the binding of the RNase catalytic domain with either the RNase inhibition domain of its noncognate pollen S-allele product or the general RNase inhibitor.
Although the prediction of preferential binding between a pollen S-allele product and its cognate S-RNase would seem logical, it is at odds with the competitive interaction phenomenon observed in the Solanaceae (Luu et al., 2001; Takayama and Isogai, 2005). For example, according to this prediction, if a heteroallelic pollen grain carrying S1- and S2-alleles is used to pollinate an S1S2 pistil, the pollen S1-allele product would preferentially interact with S1-RNase and the pollen S2-allele product would preferentially interact with S2-RNase inside the pollen tube. Consequently, neither S-RNase would be inhibited and both would exert their cytotoxic effects on the pollen tube to inhibit its growth. This outcome is precisely the opposite of the SI behavior of heteroallelic S1S2 pollen, which fails to function in SI and is rejected by pistils of any S-genotype. To explain competitive interaction, Luu et al. (2001) proposed that the active form of the pollen S-allele product is a homotetramer. When two different S-alleles are carried by a pollen grain, the products mainly form heterotetramers, which do not efficiently interact with any S-RNase. The general inhibitor would then bind any S-RNase to inhibit its RNase activity, and as a result, heteroallelic pollen would be compatible with pistils of any S-genotype. To date, no biochemical evidence supporting this model has been reported.
Goldraij et al. (2006) recently proposed an alternative model based on their findings of subcellular localizations of S-RNases in compatible and incompatible pollen tubes and of the requirement of several non–S-specific proteins for SI. The model predicts that both self and nonself S-RNases are sequestered in a vacuolar compartment in compatible pollen tubes throughout pollen tube growth in the style, whereas in incompatible pollen tubes this compartment is disrupted at later stages of pollen tube growth, releasing both self and nonself S-RNases into the cytoplasm. Because a non–S-specific protein, named HT-B, is required for SI (McClure et al., 1999) and because it appears to be preferentially degraded in compatible pollen tubes (Goldraij et al., 2006), this model also predicts that HT-B is responsible for the disruption of the compartment in the incompatible pollen tube. According to this model, the interactions between an SLF and its self and nonself S-RNases do not result in specific degradation of nonself S-RNases; rather, they indirectly regulate the stability of HT-B. In compatible pollen tubes, the interaction between an SLF and its nonself S-RNase would lead to the degradation of HT-B by a hypothetical protease, whereas in incompatible pollen tubes, the interaction between an SLF and its self S-RNase would result in the stabilization of HT-B and thus disruption of the compartment. The proposed role of HT-B and the proposed regulation of its stability have yet to be demonstrated experimentally. Moreover, this model predicts that heteroallelic pollen will be incompatible with pistils of any S-genotype. For example, when heteroallelic pollen containing both SLF1 and SLF2 is used to pollinate an S1S1 pistil, based on this model, the interaction between SLF1 and S1-RNase would result in the stabilization of HT-B and the subsequent breakdown of the compartment to release S1-RNase into the cytoplasm. However, as stated above, in the Solanaceae, heteroallelic pollen is compatible with pistils of any S-genotype as a result of the phenomenon of competitive interaction.
Although Goldraij et al. (2006) did not detect S-RNase in the cytoplasm of compatible pollen tubes or degradation of S-RNase, they could not rule out the possibility of the existence of some S-RNase in the cytoplasm and of the degradation of small amounts of S-RNase. Also, SLF appears to be localized in the cytoplasm of pollen tubes, as revealed by immunolocalization of Ah SLF (Wang and Xue, 2005) and by green fluorescent protein (GFP) fluorescence of Pi SLF2:GFP in transgenic pollen tubes germinated in vitro (unpublished data). Therefore, if the stability of HT-B is indeed controlled by the interactions of SLF with its self and nonself S-RNase, as proposed by Goldraij et al. (2006), it would seem that some S-RNase should be present in the cytoplasm for their interactions with SLF to take place.
Here, we propose a new model (Figure 9) to explain the biochemical basis of S-specific inhibition of pollen tubes based on our findings that Pi SLF1 and Pi SLF2 interact with their respective nonself S-RNases (S2- and S3-RNase for PiSLF1 and S1-RNase for PiSLF2) to a greater extent than with their respective self S-RNases (S1-RNase for Pi SLF1 and S2-RNase for Pi SLF2). This model also takes into account the possible existence of an S-specific and a general degradation mechanism for S-RNases in the pollen tube. As the significance of the deglycosylation of S-RNase for its degradation in vivo is not known, this model does not address this aspect. Although the physiological concentration of Pi SLF in the cytoplasm of the pollen tube is unknown, the level is likely to be quite low. This is based on RNA gel blot analysis of the Pi SLF transcript (Sijacic et al., 2004) and protein blot analysis of the levels of Pi SLF2:FLAG and Pi SLF2:GFP proteins expressed by the Pi SLF2 promoter in transgenic plants (unpublished data). Our model thus makes an assumption that the physiological concentration of Pi SLF in the cytoplasm of the pollen tube is in the range in which Pi SLF was found to show significant binding differences between self and nonself S-RNases in our binding assays (Figures 5E and 5F).
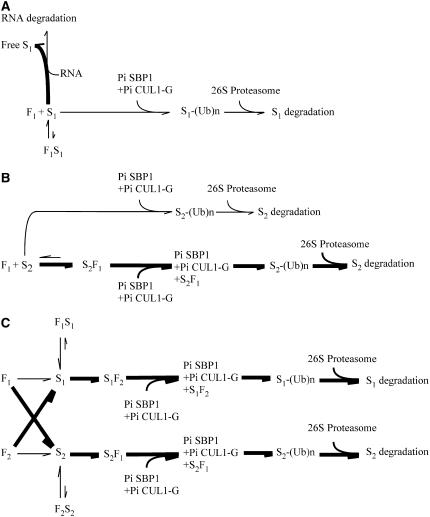
Figure 9.
Model for the Biochemical Mechanism of S-Specific Inhibition of Pollen Tube Growth.
F1 and F2 indicate Pi SLF1 and Pi SLF2, respectively; S1 and S2 indicate S1-RNase and S2-RNase, respectively; S1-(Ub)n and S2-(Ub)n indicate S1-RNase and S2-RNase conjugated with ubiquitins, respectively. The interactions between a Pi SLF and its self and nonself S-RNases are depicted differently (e.g., F1S1 for self interaction and S2F1 for nonself interaction) to emphasize that self and nonself interactions likely involve different domains of both Pi SLF and S-RNase. Thick lines denote the predominant reactions.
(A) Incompatible pollination. Pi SLF1 produced in the S1 pollen tube interacts weakly with its self S-RNase, S1-RNase, and thus most of the S1-RNase molecules taken up by the pollen tube and located in the cytoplasm are free to degrade RNA. The small amount of S1-RNase molecules that interact with Pi SLF1 are not degraded via the S-specific degradation mechanism. Some degradation of S1-RNase occurs via the non–S-specific degradation machinery mediated by Pi CUL-G-Pi SBP1; this could account for the basal degradation of S1-RNase observed in our in vitro system.
(B) Compatible pollination. Pi SLF1 interacts strongly with its nonself S-RNase, S2-RNase, and the interaction results in the degradation of S2-RNase via the S-specific degradation mechanism mediated by the E3 ligase complex containing Pi SBP1, Pi CUL1-G, and Pi SLF1. Thus, most of the S2-RNase molecules taken up by the pollen tube and located in the cytoplasm are ubiquitinated and degraded. A small number of S2-RNase molecules that do not interact with Pi SLF1 are degraded by the non–S-specific mechanism, as in incompatible pollination (A).
(C) Competitive interaction. When Pi SLF1 and Pi SLF2 are produced in the same pollen tube, Pi SLF1 preferentially interacts with S2-RNase and Pi SLF2 preferentially interacts with S1-RNase. Thus, other than having two Pi SLFs and two S-RNases in the same pollen tube, the scenarios described for (B) apply here. For simplicity, the basal degradation pathways, as shown in (A) and (B), are not depicted.
The model also predicts that the outcomes of the interactions of a Pi SLF with its self and nonself S-RNases are different. This could be accomplished if a Pi SLF interacts with its self and nonself S-RNases through different domains on both Pi SLF and S-RNase. The interaction with the self S-RNase might mask the ubiquitination site or might result in ubiquitination sites different from those on nonself S-RNases. The positions of the Lys residues to which polyubiquitin chains are attached play an important role in the degradation of protein substrates by the 26S proteasome. For example, the 6 N-terminal Lys residues of Sic1, the S-phase CDK inhibitor of yeast, contribute to the major degradation signal, whereas the other 14 Lys residues, when changed to other amino acids, do not affect the degradation of Sic1 (Petroski and Deshaies, 2003). For solanaceous S-RNases, there is only one Lys residue (in the C4 region) that is absolutely conserved. Qin et al. (2005) showed that replacing this Lys of S11-RNase of S. chacoense with Arg did not affect the function of S11-RNase in SI, suggesting that this Lys alone is not the target for ubiquitination. S1-, S2-, and S3-RNases of P. inflata contain 17, 18, and 19 additional Lys residues, respectively; thus, a more comprehensive and systematic approach would be required to determine which Lys residues are involved in ubiquitination. Therefore, we speculate that even when a Pi SLF binds its self S-RNase (in the context of the Pi CUL1-G-Pi SBP1-Pi SLF complex), this interaction would not result in degradation of the self S-RNase.
Our model addresses three different scenarios: incompatible pollination, compatible pollination, and competitive interaction, and all of the interactions depicted occur in the cytoplasm of the pollen tube. Figure 9A shows incompatible pollination, represented by selfing of an S1S1 plant. Because Pi SLF1 interacts weakly with S1-RNase, most of the S1-RNase molecules would be free to degrade RNA, resulting in growth inhibition of the S1 pollen tube. The small amount of S1-RNase molecules that do interact with Pi SLF1 in the context of the Pi CUL1-G-Pi SBP1-Pi SLF1 complex would not result in its degradation, as discussed above. However, the Pi CUL1-G-Pi SBP1 complex would mediate basal-level degradation of S1-RNase; this could account for the observed degradation of self S-RNase in our in vitro system.
In the case of compatible pollination, represented by pollination of an S2S2 plant by pollen from an S1S1 plant (Figure 9B), because Pi SLF1 interacts strongly with S2-RNase, most of the S2-RNase molecules in the cytoplasm would interact with Pi SLF1 and be assembled into an E3 ligase complex with Pi SBP1 and Pi CUL1-G. As a result, these S2-RNase molecules would be ubiquitinated and degraded. The small amount of free S2-RNase molecules that do not interact with Pi SLF1 would also be degraded via the basal degradation machinery mediated by Pi CUL1-G-Pi SBP1.
Competitive interaction is represented by pollination of an S1S2 plant with pollen from a transgenic S1S1 plant that carries a single copy of a Pi SLF2 transgene (Figure 9C). The diagram shown is also applicable to the case of self-pollination of a tetraploid plant with the S1S1S2S2 genotype. S1 pollen that carries the transgene (or S1S2 pollen of the tetraploid plant) produces both Pi SLF1 and Pi SLF2 and is compatible with the S1S2 pistil (or the S1S1S2S2 pistil of the tetraploid plant). According to our model, Pi SLF1 would interact to a greater extent with S2-RNase and Pi SLF2 would interact to a greater extent with S1-RNase, and as explained in Figure 9B, both S-RNases would be degraded and would be unable to exert their cytotoxic effects. Again, Pi CUL1-G-Pi SBP1 would mediate the basal-level degradation of any free S1- and S2-RNase molecules (data not shown).
It would be interesting to reconstitute the Pi CUL1-G-Pi SBP1 complex and the Pi CUL1-G-Pi SBP1-Pi SLF1 (or Pi SLF2) complex in vitro and then compare their interactions with self and nonself S-RNases. The reconstituted complexes could also be used in conjunction with purified E1, E2, ubiquitin, and self or nonself S-RNases to determine whether self and/or nonself S-RNases is (are) ubiquitinated. Ultimately, all of the results obtained in vitro will have to be confirmed by in vivo approaches. If, in vivo, the Pi CUL1-G-Pi SBP1-Pi SLF complex indeed mediates the specific degradation of nonself S-RNases via the preferential binding of a Pi SLF with its nonself S-RNases, it will be interesting to investigate the biochemical basis for the binding difference between a Pi SLF and its self and nonself S-RNases. For example, is the binding difference attributable to a difference in the binding affinity, a difference in the binding stoichiometry, or other reasons?
METHODS
Plant Material
The five S-genotypes of Petunia inflata used in this study, S1S1, S1S2, S2S2, S2S3, and S3S3, have been described by Ai et al. (1990).
cDNA Library Screening
The S1 and S2 pollen cDNA libraries of P. inflata, constructed in λZAPII vector (Stratagene), were described by Skirpan et al. (2001). The ASK1 and ASK2 cDNA clones of Arabidopsis thaliana were radiolabeled with 32P using the Ready-To-Go DNA Labeling kit (GE Healthcare) and used as probes for screening the S1 pollen cDNA library as described by Mu et al. (1994), except for the following modifications. The nitrocellulose membranes (Millipore) were prehybridized in 10% (w/v) dextran sulfate, 1 M NaCl, 1% (w/v) SDS, and sonicated salmon sperm DNA (200 μg/mL) for 2 h, hybridized in the same buffer plus the 32P-labeled ASK1 and ASK2 cDNA probes overnight, and washed in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS twice for 20 min each and then in 1× SSC and 0.1% SDS for 30 min. All of the manipulations were performed at 52°C, allowing ASK1 and ASK2 cDNA probes to cross-hybridize with the homologous cDNAs of P. inflata. cDNAs of the positive clones were excised from the λZAPII vector and recirculated to form pBluescript SK− phagemid DNA according to the manufacturer's protocol (Stratagene). Plasmid DNA was purified with the NucleoSpin Plasmid kit (Clontech) using the procedure recommended by the manufacturer. The cDNA library screenings using Pi SK1, cullin-1, and At RBX1 cDNA probes were performed similarly.
Yeast Two-Hybrid Protein–Protein Interaction Assay
The coding sequences for Pi SK1, Pi SK2, Pi SK3, Pi SLF1, Pi SLF2, Pi SLF2(FB), Pi SLF2(CTD), Pi CUL1-G, and Ph UBC1 were cloned in-frame to the coding sequence of the GAL4 binding domain (BD) in pGBD-C1. The coding sequences for seven Arabidopsis ASKs (ASK1, ASK4, ASK5, ASK9, ASK11, ASK13 and ASK16), Pi SK1, Pi SK2, Pi SLF1, Pi SLF2, Pi RBX1, Pi SBP1, and Pi SBP1(Δcoiled-coil) were fused in-frame to the GAL4 activation domain (AD) in pGAD-C1. To test the interaction between a pair of proteins, the corresponding pGAD-C1 and pGBD-C1 constructs were cotransformed into Saccharomyces cerevisiae SFY526 (Clontech), and the transformants were plated out on synthetic dropout (SD) medium without Trp and Leu to select cells in which both BD and AD fusion proteins coexpressed. Six independent transformants were streaked together as a dot on the filter paper and then assayed using the X-Gal filter-lift method (Breeden and Nasmyth, 1985). For the β-galactosidase activity quantitative assay, six independent colonies were inoculated separately in 5 mL of SD medium lacking Leu and Trp. The cultures growing at the mid-log phase were used for β-galactosidase activity assay according to Miller (1972). Relative β-galactosidase activities were calculated using the methods described by Skirpan et al. (2001).
Yeast Two-Hybrid Library Screening
Pi SK1, Pi SLF2, Pi SLF2(FB), and Pi SLF2(CTD) were used as baits in the library screening. Yeast HF7C (Clontech) cells were transformed with 0.1 μg of each bait construct in pGBD-C1, and the transformants were subsequently transformed with 500 μg of DNA isolated from the S2 pollen yeast two-hybrid library (in pGAD424) constructed previously by Skirpan et al. (2001). Colonies that produced interacting proteins were selected on sd medium without Leu, Trp, and His but with 3-aminotriazole (10 mM) when Pi SK1 was used as bait. The positive colonies were further confirmed by X-Gal filter-lift assay as described above. To recover the prey plasmid from each positive colony, plasmid DNA was isolated and transformed into Escherichia coli HB101, and the transformants were plated out on Leu−Amp+ M9 agar medium. Plasmid DNA was purified from the E. coli cultures by the NucleoSpin Plasmid kit and sequenced using GAL4 AD forward primer (5′-TACCACTACAATGGATG-3′) and reverse primer (5′-TGAGATGGTGCACGATG-3′). Yeast colonies (6 × 106, 3 × 106, 1.4 × 106, and 1.7 × 106) were screened using Pi SK1, Pi SLF2, Pi SLF2(FB), and Pi SLF2(CTD) as baits, respectively.
DNA Sequence Analysis
All DNA sequencing was performed at the Nucleic Acid Facility of Pennsylvania State University. Nucleotide sequences were assembled and analyzed using DNA Strider 1.2.1. Database searches were run on the BLASTx program at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/). Alignments of amino acid sequences were performed using ClustalW (http://www.ebi.ac.uk/clustalw/); gonnet250 protein weight matrix was selected, and the gap opening and extension parameters were 10 and 0.05, respectively. Alignments were shaded using Boxshade version 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The protein interaction motifs were detected by SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1).
DNA and RNA Gel Blot Analyses
Genomic DNA was purified from young leaves of S1S1, S1S2, S2S2, S2S3, and S3S3 P. inflata plants using Plant DNAzol reagent (Invitrogen) according to the manufacturer's procedure. Genomic DNA (15 μg) from each tissue was digested overnight by EcoRI or XbaI; the digests were separated on a 0.7% (w/v) agarose gel and transferred to a charged nylon membrane, Biodyne B (Pall). Total RNA was purified from different tissues of S2S2 plants using Trizol reagent (Invitrogen), and 20 μg from each tissue was electrophoresed on a 1% (w/v) agarose/formaldehyde gel and blotted onto a Biodyne B membrane as described previously (Sijacic et al., 2004). The probe was 32P-labeled Pi SBP1 cDNA, which was obtained by PCR amplification of the yeast two-hybrid clone of Pi SBP1 using the GAL4 AD forward and reverse primers as described above and by radiolabeling with 32P using the Ready-To-Go DNA Labeling kit. Prehybridization, hybridization, and washing of the membranes were performed as described by Skirpan et al. (2001).
In Vitro Binding Assays
The coding sequences of Pi SLF1, Pi SLF2, Pi FBP2411, S1-RNase, S2-RNase, RNase X2, Pi CUL1-G, Ph UBC1, and Pi SLF2(CTD) were cloned separately in-frame behind the sequence for the (His)6:T7 tag in vector pET28 (Novagen). The coding sequences of Pi SBP1, S1-RNase, S2-RNase, S3-RNase, RNase X2, S1(HVabC3), S2(HVabC3), and S3(HVabC3) were cloned separately as in-frame fusions to the GST coding sequence in vector pGEX-5X-1 (GE Healthcare). The recombinant proteins were expressed in BL21 Codon Plus E. coli (Stratagene) and purified as described by Skirpan et al. (2001). To examine the binding between a GST fusion protein and a (His)6:T7 fusion protein, the GST fusion protein (0.5 to 1.0 μg) bound to 30 μL of Glutathione Sepharose 4 Fast Flow resin (GE Healthcare) was incubated with the (His)6:T7 fusion protein (0.5 to 1 μg) in 500 μL of the binding buffer (50 mM Tris-Cl, pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 0.01% Nonidet P-40) for 1 h at 25°C. After binding, the resin was washed three times with binding buffer, and the bound proteins were eluted by boiling in 30 μL of 2× SDS reducing sample buffer for 5 min and separated by SDS-PAGE. The presence of bound proteins was analyzed by protein gel blotting as described below. For the assays that used different amounts of (His)6:T7:Pi SLF1 to bind its self and nonself S-RNases, equal amounts of GST:S1-RNase and GST:S2-RNase (or GST:S3-RNase) were bound separately to 250 μL of Glutathione Sepharose 4 Fast Flow resin, and the resin with either bound GST:S1-RNase or bound GST:S2-RNase (or GST:S3-RNase) was divided equally into eight aliquots. Each aliquot was equilibrated with 500 μL of the binding buffer, and to each two aliquots were added 2, 4, 6, or 8 μL of a stock of purified (His)6:T7:Pi SLF1. The binding reaction was performed at 25°C for 90 min, and analysis of the bound proteins was performed as described above.
Purification of S3-RNase from Pistils
S3-RNase was purified from pistils of a P. inflata plant of S3S3 genotype as described previously (Lee et al., 1994). Briefly, 30 pistils were collected and ground with 1 mL of extraction buffer (50 mM Tris-HCl, pH 8.5, 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM CaCl2, and 1 mM DTT). After centrifugation at 12,000g for 10 min to remove tissue debris, the supernatant was filtered through a 0.45-μm MILLEX-GV filter (Millipore) and the filtrate was chromatographed on a Mono-S column (HR 5/5) equilibrated with 50 mM sodium phosphate, pH 6.0, using a fast protein liquid chromatograph (GE Healthcare). The bound proteins were eluted with a linear gradient of 0 to 500 mM NaCl in the same buffer at a flow rate of 0.5 mL/min. The eluted proteins were monitored at A280 with the sensitivity of the detector set to 0.1 absorbance unit full scale. The fractions containing S3-RNase were determined by SDS-PAGE and confirmed by immunoblotting using an anti-S3-RNase antibody. Protein concentrations were determined using a Bio-Rad protein assay kit with BSA as the standard.
Protein Gel Blot Analysis
Proteins were resolved on 10% polyacrylamide gels and blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore). To visualize the amount of proteins loaded in each lane, a duplicate gel was stained with Coomassie Brilliant Blue R 250 or the membrane was stained with Ponceau S before immunoblotting. The primary antibodies used were an anti-T7 tag monoclonal antibody (1:10,000; Novagen), an anti-(His)6 tag antibody (1:2000; Novagen), an anti-GST antibody (1:200; Oncogene Research Products), and an affinity-purified anti-S3-RNase antibody (1:1000). The antiserum for S3-RNase had been raised previously in rabbits against a synthetic peptide, DGDKFVSFSLKDRIV (corresponding to amino acids 48 to 62 of S3-RNase in the hypervariable region HVa). The monospecific antibody against S3-RNase was purified using GST:S3(HVabC3), which encodes amino acids 47 to 97 of S3-RNase, according to the procedure of Bar-Peled and Raikhel (1996). After the blots had been incubated with the secondary antibody of peroxidase-linked sheep anti-mouse IgG (1:10,000; GE Healthcare), the immunoreactive proteins were visualized with the Supersignal West Pico Chemiluminescent Substrate kit (Pierce). Alternatively, the immunoreactive proteins were detected with a Bio-Rad AP-Conjugate Substrate kit after the blots had been incubated with alkaline phosphatase–conjugated goat anti-mouse IgG (1:5000; Calbiochem).
Preparation of Pollen Tube Extracts
Fresh pollen was collected from S1S1, S2S2, and S3S3 plants and germinated separately in vitro for 3 h in the pollen germination medium described by Lee et al. (1996). Pollen tubes were harvested by centrifugation at 16,000g for 1 min and quickly frozen in liquid nitrogen before storage at −80°C. The pellets were homogenized in a 1.5-mL microfuge tube with a pestle and extracted with 500 μL of pollen tube extraction buffer (50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM DTT, and 1 mM PMSF). After centrifugation at 20,000g at 4°C for 30 min, the supernatants were divided into aliquots and stored at −80°C. The total protein concentration of each extract was determined by the Bio-Rad protein assay kit.
In Vitro Degradation and Ubiquitination Assays
For the degradation assay of bacterially expressed proteins, 0.3 μg of GST, GST:S1-RNase, GST:S2-RNase, GST:S3-RNase, or GST:RNase X2 was incubated with 10 μg of total pollen tube extracts (quantified based on the protein concentration) in a final volume of 30 μL of ubiquitin reaction buffer (50 mM Tris-HCl, pH 7.4, 2 mM ATP, 2 mM DTT, 5 mM MgCl2, ∼4 μg of creatine phosphokinase [Calbiochem], 10 mM creatine phosphate [Calbiochem], and 1 mM PMSF) for 1 h at 30°C. The reaction was stopped by the addition of 7 μL of 5× SDS reducing sample buffer, and the mixture was heated at 95°C for 5 min. The proteins were resolved on two duplicate 10% reducing SDS-polyacrylamide gels and then transferred to PVDF membranes. S3-RNase (0.1 μg), purified from pistils, and its deglycosylated form (0.1 μg) were similarly analyzed for degradation except that the incubation time was 1.5 h. The deglycosylation reaction was performed at 37°C using PNGase-F (New England Biolabs) according to the manufacturer's protocol.
For the ubiquitination assay, 0.5 μg of GST:S2-RNase, GST:S3-RNase, GST:RNase X2, or GST was incubated with 5 μg of total S2 pollen tube extracts and 1 μg/μL ubiquitin (Boston Biochem) or (His)6:ubiquitin in a final reaction volume of 30 μL of ubiquitin reaction buffer at 30°C. A (His)6:ubiquitin construct was made by digesting pET26Ub (Gohara et al., 1999) with NdeI and BamHI to release the DNA fragment encoding S. cerevisiae ubiquitin and cloning the fragment at the NdeI and BamHI sites in pET28. The recombinant ubiquitin protein was expressed and purified as described above. At various times during the assay, the reaction was stopped, and GST, GST-fused proteins, and their respective ubiquitinated forms were purified by binding to the Glutathione Sepharose 4 Fast Flow resin in GST binding buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10 mM EDTA, 1 mM DTT, 0.4% [w/v] Triton X-100, and 1 mM PMSF) for 30 min at room temperature. The resin was washed three times in GST washing buffer (50 mM Tris-HCl, pH 7.4, 500 mM NaCl, 10 mM EDTA, 1 mM DTT, and 0.4% [w/v] Triton X-100), and the bound proteins were eluted by heating at 95°C for 5 min in 30 μL of 2.5× SDS reducing sample buffer. The eluted proteins were separated on 10% reducing SDS-polyacrylamide gels and transferred to a PVDF membrane for immunoblotting.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Pi SBP1 (DQ250022), Pi SK1 (DQ250014), Pi SK2 (DQ250015), Pi SK3 (DQ250013), Pi CUL1-C (DQ250016), Pi CUL1-G (DQ250017), Pi RBX1 (DQ250021), Pi FBP23 (DQ250018), Pi FBP2011 (DQ250019), and Pi FBP2411 (DQ250020). The accession numbers for the sequence data used in this article are as follows: ASK1 (AAM45019), ASK2 (AAC14445), ATCUL1 (CAC85264) and At RBX1 (Q940X7) from Arabidopsis thaliana; Ph SBP1 (AAF28357) from Petunia hybrida; Sc SBP1 (AAS76633) from Solanum chacoense; Skp1 (AAH09839), CUL1 (NP_003583), and Rbx1 (NP_055063) from Homo sapiens; and Pi SLF1 (AAS79484), Pi SLF2 (AAS79485), S1-RNase (AAA33726), S2-RNase (AAG21384), S3-RNase (AAA33727), and RNase X2 (S28611) from Petunia inflata.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Amino Acid Sequences of Pi SK1, Pi SK2, and Pi SK3 of P. inflata, ASK1 and ASK2 of Arabidopsis, and a Skp1 of Human.
Supplemental Figure 2. Alignment of the Deduced Amino Acid Sequences of Pi SBP1 of P. inflata, Ph SBP1 of P. hybrida, and Sc SBP1 of S. chacoense.
Supplemental Figure 3. In Vitro Binding Assay of Interactions of Pi SLFs with S1-RNase, S2-RNase, and RNase X2.
Supplemental Figure 4. Role of the Coiled-Coil Region of Pi SBP1 in Interactions with Pi SLF2 and Truncated S-RNases.
Supplemental Figure 5. Alignment of Amino Acid Sequences of Pi CUL1-C and Pi CUL1-G of P. inflata, ATCUL1 of Arabidopsis, and CUL1 of Human.
Supplemental Figure 6. Alignment of Amino Acid Sequences of Pi RBX1 of P. inflata, At RBX1 of Arabidopsis, and an Rbx1 of Human.
Supplemental Figure 7. Ubiquitination Assay of Total Protein in S2 Pollen Tube Extract.
Supplementary Material
Acknowledgments
We thank Hong Ma for providing cDNA clones for ASK1 and ASK2, Richard Vierstra for providing seven ASK cDNA clones in yeast two-hybrid vector pBI771, Thomas Sims for providing Ph UBC1, and Craig Cameron for providing pET26Ub. We also thank Peter Dowd, Paja Sijacic, Andrea Skirpan, and Yan Wang for valuable comments, Anthony Omeis for greenhouse management, and Jiong Wang for routine laboratory assistance. This work was supported by Grants IOB-0235176 and IOB-0543201 from the National Science Foundation to T.-h.K.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Teh-hui Kao (txk3@psu.edu).
Online version contains Web-only data.
References
- Ai, Y., Singh, A., Coleman, C.E., Ioerger, T.R., Kheyr-Pour, A., and Kao, T.-h. (1990). Self-incompatibility in Petunia inflata: Isolation and characterization of cDNAs encoding three S-allele-associated proteins. Sex. Plant Reprod. 3 130–138. [Google Scholar]
- Anderson, M.A., et al. (1986). Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321 38–44. [Google Scholar]
- Bai, C., Sen, P., Hofman, K., Ma, L., Goebel, M., Harper, W., and Elledge, S. (1996). Skp1 connects cell cycle regulation to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86 263–274. [DOI] [PubMed] [Google Scholar]
- Bar-Peled, M., and Raikhel, N.V. (1996). A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 241 140–142. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., Brook, A., Staehling-Hampton, K., Heitzler, P., and Dyson, N. (2000). A role for Ebi in neuronal cell cycle control. EMBO J. 19 5376–5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden, L., and Nasmyth, K. (1985). Regulation of the yeast HO gene. Cold Spring Harb. Symp. Quant. Biol. 50 643–650. [DOI] [PubMed] [Google Scholar]
- Cenciarelli, C., Chiaur, D.S., Guardavaccaro, D., Parks, W., Vidal, M., and Pagano, M. (1999). Identification of a family of human F-box proteins. Curr. Biol. 9 1177–1179. [DOI] [PubMed] [Google Scholar]
- de Nettancourt, D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. (Berlin: Springer-Verlag).
- Entani, T., Zwano, M., Shiba, H., Che, F.S., Isogai, A., and Takayama, S. (2003). Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8 203–213. [DOI] [PubMed] [Google Scholar]
- Farras, R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1–SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohara, D.W., Ha, C.S., Ghosh, S.K.B., Arnold, J.J., Wisniewski, T.J., and Cameron, C.E. (1999). Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17 128–138. [DOI] [PubMed] [Google Scholar]
- Goldraij, A., Kondo, K., Lee, C.B., Hancock, C.N., Sivaguru, M., Vazquez-Santana, S., Kim, S., Phillips, T.E., Cruz-Garcia, F., and McClure, B. (2006). Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439 805–810. [DOI] [PubMed] [Google Scholar]
- Golz, J.F., Oh, H.-Y., Su, V., Kusaba, M., and Newbigin, E. (2001). Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc. Natl. Acad. Sci. USA 98 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F., Su, V., Clarke, A.E., and Newbigin, E. (1999). A molecular description of mutations affecting the pollen components of the Nicotiana alata S locus. Genetics 152 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, N.R., Yamane, H., Tao, R., and Iezzoni, A.F. (2006). Accumulation of non-functional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Zhao, L., Yang, Q., and Xue, Y. (2006). AhSSK1, a novel SKP1-like protein that interacts with the S-locus F-box protein SLF. Plant J. 46 780–793. [DOI] [PubMed] [Google Scholar]
- Huang, S., Lee, H.-S., Karunanandaa, B., and Kao, T.-h. (1994). Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger, T.R., Gohlke, J.R., Xu, B., and Kao, T.-h. (1991). Primary structural features of the self-incompatibility protein in Solanaceae. Sex. Plant Reprod. 4 81–87. [Google Scholar]
- Ishimizu, T., Norioka, S., Kanai, M., Clarke, A.E., and Sakiyama, F. (1996). Location of cysteine and cystine residues in S-ribonucleases associated with gametophytic self-incompatibility. Eur. J. Biochem. 242 627–635. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, T.-h., and McCubbin, A.G. (1996). How flowering plants discriminate between self and non-self pollen to prevent inbreeding. Proc. Natl. Acad. Sci. USA 93 12059–12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, T.-h., and Tsukamoto, T. (2004). The molecular and genetic bases of S-RNase-based self-incompatibility. Plant Cell 16 (suppl.), S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanandaa, B., Huang, S., and Kao, T.-h. (1994). Carbohydrate moiety of the Petunia inflata S3 protein is not required for self-incompatibility interactions between pollen and pistil. Plant Cell 6 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G., and Xue, Y. (2002). An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50 29–42. [DOI] [PubMed] [Google Scholar]
- Lechner, E., Xie, D., Grava, S., Pigaglio, E., Planchais, S., Murray, J.A., Parmentier, Y., Mutterer, J., Dubreucq, B., Shen, W.H., and Genschik, P. (2002). The AtRbx1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J. Biol. Chem. 277 50069–50080. [DOI] [PubMed] [Google Scholar]
- Lee, D.H., and Goldberg, A.L. (1998). Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 8 397–403. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., Huang, S., and Kao, T.-h. (1994). S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367 560–563. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., Karunanandaa, B., McCubbin, A., Gilroy, S., and Kao, T.-h. (1996). PRK1, a receptor-like kinase of Petunia inflata, is essential for postmeiotic development of pollen. Plant J. 9 613–624. [Google Scholar]
- Lee, H.-S., Singh, A., and Kao, T.-h. (1992). RNase X2, a pistil-specific ribonuclease from Petunia inflata, shares sequence similarity with solanaceous S proteins. Plant Mol. Biol. 6 1131–1141. [DOI] [PubMed] [Google Scholar]
- Li, S., Xu, C., and Carthew, R.W. (2002). Phyllopod acts as an adaptor protein to link the Sina ubiquitin ligase to the substrate protein Tramtrack. Mol. Cell. Biol. 22 6854–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.-T., Qin, X., Laublin, G., Yang, Q., Morse, D., and Cappadocia, M. (2001). Rejection of S-heteroallelic pollen by a dual-specific S-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics 159 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, D.-T., Qin, X., Morse, D., and Cappadocia, M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407 649–651. [DOI] [PubMed] [Google Scholar]
- Matsuzawa, S., and Reed, J.C. (2001). Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 response. Mol. Cell 7 915–926. [DOI] [PubMed] [Google Scholar]
- McClure, B. (2004). S-RNase and SLF determine S-haplotype-specific pollen recognition and rejection. Plant Cell 16 2840–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B.A., Gray, J.E., Anderson, M.A., and Clarke, A.E. (1990). Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347 757–760. [Google Scholar]
- McClure, B.A., Mou, B., Canevascini, S., and Bernatzky, R. (1999). A small asparagine-rich protein required for S-allele-specific pollen rejection in Nicotiana. Proc. Natl. Acad. Sci. USA 96 13548–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin, A.G., Wang, X., and Kao, T.-h. (2000). Identification of self-incompatibility (S-) locus linked pollen cDNA markers in Petunia inflata. Genome 43 619–627. [PubMed] [Google Scholar]
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, J.H., Lee, H.S., and Kao, T.-h. (1994). Characterization of a pollen expressed receptor like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell 6 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfett, J., Atherton, T.L., Mou, B., Gasser, C.S., and McClure, B.A. (1994). S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature 367 563–566. [DOI] [PubMed] [Google Scholar]
- O'Brien, M., Major, G., Chantha, S., and Matton, D.P. (2004). Isolation of S-RNase binding proteins from Solanum chacoense: Identification of an SBP1 (RING finger protein) orthologue. Sex. Plant Reprod. 17 81–88. [Google Scholar]
- Ohta, T., and Xiong, Y. (2001). Phosphorylation- and SKP1-independent in vitro ubiquitination of E2F1 by multiple ROC-Cullin ligase. Cancer Res. 61 1347–1353. [PubMed] [Google Scholar]
- Oxley, D., and Bacic, A. (1996). Disulphide bonding in a stylar self-incompatibility ribonuclease of Nicotiana alata. Eur. J. Biochem. 242 75–80. [DOI] [PubMed] [Google Scholar]
- Petroski, M.D., and Deshaies, R.J. (2003). Context of multiubiquitin chain attachment influences the rate of Sic1 degradation. Mol. Cell 11 1435–1444. [DOI] [PubMed] [Google Scholar]
- Qiao, H., Wang, F., Zhao, L., Zhou, J., Lai, Z., Zhang, Y., Robbins, T.P., and Xue, Y. (2004. a). The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, H., Wang, H., Zhao, L., Zhou, J., Huang, J., Zhang, Y., and Xue, Y. (2004. b). The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, X., Soulard J., Laublin, G., Morse, D., and Cappadocia, M. (2005). Molecular analysis of the conserved C4 region of the S11-RNase of Solanum chacoense. Planta 221 531–537. [DOI] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408 381–386. [DOI] [PubMed] [Google Scholar]
- Sijacic, P., Wang, X., Skirpan, A.L., Wang, Y., Dowd, P.E., McCubbin, A.G., Huang, S., and Kao, T.-h. (2004). Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429 302–305. [DOI] [PubMed] [Google Scholar]
- Sims, T.L., and Ordanic, M. (2001). Identification of a S-ribonuclease-binding protein in Petunia hybrida. Plant Mol. Biol. 47 771–783. [DOI] [PubMed] [Google Scholar]
- Skirpan, A.L., McCubbin, A.G., Ishimizu, T., Wang, X., Hu, Y., Dowd, P.E., Ma, H., and Kao, T.-h. (2001). Isolation and characterization of kinase interacting protein 1, a pollen protein that interacts with the kinase domain of PRK1, a receptor-like kinase of Petunia. Plant Physiol. 126 1480–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., Kenneth, R., Tobutt, K.R., Simon, P., Vaughan, S.P., and Robbins, T.P. (2005). Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S., and Isogai, A. (2005). Self-incompatibility in plants. Annu. Rev. Plant Biol. 56 467–489. [DOI] [PubMed] [Google Scholar]
- Thrower, J.S., Hoffman, L., Rechsteiner, M., and Pickart, C.M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Dandekar, A.M., Gradziel, T.M., Tao, R., and Hirano, H. (2003). Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., Yamane, H., Watari, A., Kakehi, E., Ikeda, K., Hauck, N.R., Iezzoni, A.F., and Tao, R. (2004). The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39 573–586. [DOI] [PubMed] [Google Scholar]
- Wang, H.-Y., and Xue, Y.-B. (2005). Subcellular localization of the S-locus F-box protein AhSLF2 in pollen and pollen tubes of self-incompatible Antirrhinum. J. Integr. Plant Biol. 47 76–83. [Google Scholar]
- Wang, Y., Tsukamoto, T., Yi, K.-w., Wang, X., Huang, S., McCubbin, A.G., and Kao, T.-h. (2004). Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S2-RNase. Plant Mol. Biol. 54 727–742. [DOI] [PubMed] [Google Scholar]
- Willems, A.R., Schwab, M., and Tyers, M. (2004). A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695 133–170. [DOI] [PubMed] [Google Scholar]
- Zhao, D., Ni, W., Feng, B., Han, T., Petrasek, M.G., and Ma, H. (2003). Members of the Arabidopsis SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol. 133 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-FboxSkp2 ubiquitin-protein ligase complex. Nature 416 703–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.