Abstract
dsDNA in bacteriophages is highly stressed and exerts internal pressures of many atmospheres (1 atm = 101.3 kPa) on the capsid walls. We investigate the correlation between packaged DNA length in λ phage (78–100% of WT DNA) and capsid strength by using an atomic force microscope indentation technique. We show that phages with WT DNA are twice as strong as shorter genome mutants, which behave like empty capsids, regardless of high internal pressure. Our analytical model of DNA-filled capsid deformation shows that, because of DNA-hydrating water molecules, an osmotic pressure exists inside capsids that increases exponentially when the packaged DNA density is close to WT phage. This osmotic pressure raises the WT capsid strength and is approximately equal to the maximum breaking force of empty shells. This result suggests that the strength of the shells limits the maximal packaged genome length. Moreover, it implies an evolutionary optimization of WT phages allowing them to survive greater external mechanical stresses in nature.
Keywords: atomic force microscopy, viral capsids, bacteriophage, DNA hydration forces, osmotic pressure
The majority of viruses have spherical protein shells (capsids) with icosahedral symmetry, with radii varying between 10 and 100 nm, and with thicknesses of few nanometers, corresponding to a single protein layer. Viral capsids protect genomes that can be tens of micrometers in contour length. In prokaryotic viruses (bacteriophages), capsid proteins first assemble in empty capsids before the genome is actively packaged by a molecular motor that is part of the capsid (1, 2). Matching the capsid size and genome length is of great importance for efficient packaging and viral infectivity. For example, WT λ phage infects Escherichia coli cells and has its DNA (48,502 bp) contained in an icosahedral T = 7 capsid to which a flexible, noncontractile tail is attached. The mature capsid has an outer diameter of 63 nm and a shell thickness of between 1.8 and 4.1 nm (1, 2). λ phages can be packaged with DNA lengths in the range of 78–106% of WT DNA and remain infectious (3). If the genome is shorter than 78% of the WT-DNA, then the phage fails to infect. When the DNA is longer than 106% of WT length, packaging does not occur. It was recently shown that dsDNA inside many phages is highly stressed because of electrostatic repulsion and the bending energy of the packaged DNA chain, resulting in internal pressures of several tens of atmospheres (1 atm = 101.3 kPa) (4–13). This finding suggests that the infection is in part driven by the internal DNA pressure. Thus, if DNA is significantly shorter than WT, the internal pressure becomes too low and is therefore incapable of injecting enough DNA into the bacteria. On the other hand, if the DNA is longer than WT, the internal pressure might be too high, and the force that builds up in the capsid could exceed either the strength of the packaging motor (4, 14) or the maximum internal force that the capsid can withstand. Thus, the capsid size and strength might limit the extent to which the genome can be pressurized in the capsid.
In this work we investigate the physical coupling between packaged genome length and capsid size by measuring capsid deformation as a function of the externally applied force using atomic force microscope (AFM) tips. We compare the properties of empty λ phages, λ phages filled with WT DNA, and phages partially filled with DNA. We found that WT λ had a spring constant nearly double that of the other λ phages and could withstand forces twice as high before irreversible damage occurred. Using an analytical model, we show that only the packaging density of the WT λ phage provides a “DNA pressure” that remarkably makes the capsid stronger. These results seem to indicate that WT λ phages have optimized their dimensions with regard to internal pressure and capsid strength. Considering that the λ phage host (E. coli cell) often adheres to surfaces exposed to a significant shear stress (15) suggests an answer to why genetically modified mutants, which are identical in structure but differ in DNA length, are infectious but not abundant in nature. This behavior seems to mirror biological cells, in which osmotic pressure also creates strength against external mechanical deformation.
Results and Discussion
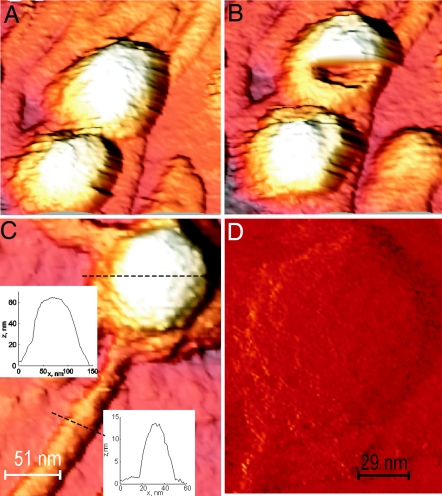
The mature heads of λ phage capsids are built of 415 copies of gpE protein (40 kDa) and 405–420 copies of gpD protein (11.4 kDa). gpE forms the actual shell and is clustered in hexamers and pentamers. gpD decorates the shell, and six trimers surround each hexamer (1, 2). We imaged empty and (partially) full λ phage capsids repeatedly and with high resolution by AFM in jumping mode (Fig. 1). The 173-nm tail, which is attached to one of the icosahedral fivefold vertices of the capsid (16), is easily dislodged during AFM imaging. Detachment of the tail is usually followed by the complete disintegration of the capsid. With high resolution imaging in jumping mode (17, 18), individual capsomers can be seen, presumably due to differences in height and lateral stiffness as a result of the gpD decoration (Fig. 1D).
Fig. 1.
High-resolution imaging of λ capsids (3D with shadow). (A) Intact λ phage. (B) Same phage as in A but after breakage. (C) Cross-sections of intact phage. (D) High-resolution normal force mode image of capsomers on capsid surface.
We determined the elastic and nonelastic response of intact mature λ capsids with different amounts of packaged DNA (0%, 78%, 94%, and 100% of 48,502-bp WT DNA) by nanoindentation using AFM cantilevers as described in ref. 17. Note that all different DNA length phages and the empty capsid have identical mature capsids (same diameter and capsid thickness) (R. W. Hendrix, personal communication, and refs. 19 and 20), where the empty shells have been emptied of their genetic content after maturation (see Experimental Procedures). We also confirmed that result with high-resolution cryoelectron microscopy by averaging several hundreds of phage particles together (Fig. 2). The packaged genome length (in every phage sample) was verified by extracting DNA from the capsids and determining its length with pulse-field gel electrophoresis. DNA length in each case corresponded to that of a single DNA length population of phage particles (37.7-, 45.7-, or 48.5-kb DNA). For each of the capsid conditions we recorded, the applied force as a function of piezo-related displacement of the sample toward the tip [force–distance (FZ) curves] for 10–15 individual objects. The typical features of the FZ curves are the following: (i) the moment the tip touches the virus shell (contact point), (ii) a linear reduction of the shell height with applied force, (iii) sudden nonlinear response of the shell (capsid breakage or collapse), and (iv) retraction of the tip (Fig. 3 A and B). Fig. 3C shows an example sequence of FZ curves of a shell. These curves reveal a repetitive linear response with no hysteresis or shifting of the contact point. Such response, which was seen for all capsids, indicates that the shells respond elastically without plastic deformation up to indentations of ≈25% of the capsid radius. After several indentations, however, the capsid starts to show nonlinear behavior and breaks irreversibly. This kind of material fatigue was also observed for φ29 proheads and cowpea chlorotic mottle virus capsids but typically, only after many more rounds of pushing (17, 18). λ phages thus seem more fragile. One possible explanation is the existence of the long tail in mature λ phages. They appear to break off rather easily, as can be concluded from many detached tails observed on the glass surface. Repetitive deformation of the capsids by the AFM tip might cause the tail to fall off quickly, hence inducing an irreversible defect. Irreversible shell collapse or breakage can also be induced by a single push in which the AFM cantilever flattens the capsid against the glass surface. We note here that, in all cases before deforming the phage capsid with an AFM tip, we confirmed that the tail was attached to the capsid by imaging the entire phage under very small forces (as shown in Fig. 1). As mentioned above, without tails, capsids were either partially or completely disintegrated, and those capsid parts had a significantly different, nonlinear deformation response compared with the intact shells. However, we also observed for a large number of phage particles that, upon AFM cantilever-induced deformation, the capsid would partially disintegrate while the tail would remain attached to the capsid remains (Fig. 1B). We always confirmed that the capsid was broken by imaging the surface after breaking the phage.
Fig. 2.
Two-dimensional projections of cryoelectron microscopy reconstructions of the 100%, 94%, and 78% DNA-filled and empty λ phage capsids. Several hundreds of particles with similar orientations were classified into groups and averaged together assuming icosahedral symmetry. Radial averages (data not shown) of these projections confirm that there is no difference in either capsid diameters or thickness.
Fig. 3.
Indenting λ capsids. (A) FZ curves of full WT versus empty λ phage. Black dashed lines represent the fit of the linear part of the FZ curves. FZ curves for filled and empty capsids are solid, but the dashed parts show the areas when the capsid breaks, and there is a discontinuity in the curve. The capsid breaks stepwise, which is why there are several “jumps” in the FZ curves without any FZ data between these gaps. (B) The data in A displayed as observed capsid height vs. force. The deformation of the capsid is linear until a critical deformation is reached, after which a sudden decrease in capsid height can be observed. The dashed line is inserted to guide the eye, because there is no data recorded between these gaps. (C) A sequence of FZ curves performed on a phage capsid containing 94% of the WT genome.
We extracted the spring constant of the linear region and the force and indentation depth at which the shell starts to respond nonlinearly from the FZ curves of the various capsids (Figs. 4 and 5 and Table 1). It is clear from Figs. 4 and 5 and Table 1 that capsids containing WT DNA are almost twice as stiff and as strong as the others. The maximal indentation WT λ shells can withstand also shows some increase (≈25%), which might stem from a difference in deformation shape of the capsid due to the DNA “pressure” (i.e., less buckling and more flattening than the partially filled and empty capsids). Nevertheless, the actual failure of the shell is probably related to a critical protein–protein separation independent of the amount of DNA inside the capsid. Therefore, reaching the threshold of this separation might determine the tensile strength of the capsids.
Fig. 4.
Histogram of spring constants for WT, 94% and 78% filled, and empty phages. The data were statistically compared by using an unpaired t test to determine the statistical significance of the difference between the means within different groups. The calculated P values for the difference between the WT capsids and 78% and 94% filled capsids are 0.0027 and 0.00011, respectively (both below the common threshold value of 0.05 for statistical significance). The data for empty phages and the mutants shows no statistical significance, with P values of 0.406 and 0.336, respectively. The solid line is a fit to WT phage data, and the dashed line is a joint fit to 94% and 78% filled and empty phage data.
Fig. 5.
Capsid properties vs. DNA density in capsids. (A) Capsid spring constant versus genome length. (B) Breaking force versus genome length (experimental data shown by circles and model calculation with a line). (C) Deformation versus genome length. Error bars are shown as standard errors.
Table 1.
Spring constant of the linear region and the force and indentation depth (indent) at which the shell starts to respond nonlinearly
| Genome length, % | Spring constant k, N/m | k SD | k SE | Breaking force Fbreak, nN | Fbreak SD | Fbreak SE | Indent, nm | Indent SD | Indent SE |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.13 | 0.04 | 0.01 | 0.8 | 0.4 | 0.1 | 5.7 | 2 | 0.6 |
| 78 | 0.15 | 0.07 | 0.02 | 0.9 | 0.4 | 0.1 | 6.1 | 2 | 0.7 |
| 94 | 0.14 | 0.05 | 0.01 | 1.0 | 0.3 | 0.1 | 6.6 | 2 | 0.5 |
| 100 | 0.23 | 0.06 | 0.01 | 1.6 | 0.6 | 0.1 | 7.6 | 3 | 0.6 |
A continuum theory of elasticity for thin homogeneous shells (21) and finite element analysis can be used to describe the mechanical properties of empty capsids measured using FZ curves (17, 18). In the continuum model approximation, the spring constant, k, can be related to the Young's modulus, E, of the protein capsid with k = αE h2/r0, where h is the capsid thickness, r0 is the capsid radius, and α is the geometry-dependent proportionality factor. Using finite element simulations for empty λ phage capsids, we obtained a Young's modulus of 1.0 GPa (average ro, 29.5 nm; thickness of the gpE shell, ≈1.8 nm). From this simulation, we found the capsid geometry-dependent proportionality factor, α, to be around unity (17, 18, 21). This Young's modulus value is close to that reported for φ29 proheads, a virus that also keeps its genome under pressure (4, 17). The average force to break an empty λ capsid is 0.8 nN (Table 1), a value similar to that found for empty cowpea chlorotic mottle virus but half the breaking force of φ29 at loading rates comparable to these experiments (≈1.5 nN at 1,500 pN/s). Thus, even though φ29 proheads have the same thickness as the gpE shells of λ capsids, they are able withstand higher deformation forces. On the other hand, λ capsids are considerably more elastic than φ29 proheads (0.13 N/m vs. 0.31 N/m). As a result, the relative deformation before failure of both capsids is about the same (20–25%). Thus, the difference in strength seems to be caused primarily by the difference in rigidity of the capsids. This result indicates that the energy potential of the capsomer–capsomer interactions in λ phages is lower and that the mechanical properties of empty viral capsids seem to depend strongly on these local interactions (22). It should be noted, however, that failure of the λ capsids could also have been triggered by the loss of the capsid's tail, resulting in a lowering of the apparent maximal strength.
Recently, the effect of packaged DNA length on the internal pressure in λ phage capsids has been investigated both theoretically (4–6, 8, 9, 12) and experimentally (7, 10, 11, 13, 14). It was found that internal DNA pressure increases monotonically and significantly as the DNA packaging progresses to completion. Eventually, internal pressures reach tens of atmospheres in the λ capsid (10). We have investigated the effect of DNA internal pressure on capsid strength in response to an external deformation (Fig. 4 and Table 1). The surprising difference in strength and elasticity between the WT (100% DNA-filled) capsids and the partially filled ones (78% and 94%) exists despite the fact that DNA in the partially filled capsids still exerts high internal pressure on the capsid walls [≈20 and ≈40 atm, respectively (6, 14)]. In fact, the partially filled shells responded to external deformation like an empty capsid. Only WT phage with an internal pressure of ≈60 atm is approximately twice as stiff and, thus, twice as resilient to external forces.
Could this unexpected behavior be the result of DNA hydration forces when the capsids are deformed? It has been shown that the interaction of water-soluble molecules can be described through the interaction of these molecules with the water (23). By using the osmotic stress method, hydration forces were measured between phospholipid bilayers (24), between polysaccharides (24), and between DNA double helices (25). Condensed DNA was seen to be packed into a lattice with well defined interaxial spacing, d, that decreases with the concentration of the added polymer (osmotic pressure). The force of DNA compression, given by the polymer's osmotic pressure, is equal and opposite to the interhelical repulsion at a given spacing between DNA strands (25). It was found that, at separations of <35 Å, the repulsive force between DNA molecules varies exponentially and cannot be described by the electrostatic theory. These forces were termed “hydration forces.” The hydration force represents the work of the removal of water from the vicinity of the surface of the DNA molecule. Likewise, x-ray diffraction studies on phages (26–28) showed that the interhelical distance between packaged DNA strands in bacteriophage heads is in the same range of distances measured by using the osmotic stress method. From both results, an empirical relation was determined for the resulting internal pressure of DNA, P(d), on the walls of the phage capsid (6, 12, 25):
where Π(d) is the osmotic pressure, and F0 and c are constants that depend on the nature, charge, and concentration of ions present in the solution. For the ionic conditions of our experiment (10 mM MgSO4), F0 = 1.2 × 104 pN/nm2 and c = 0.30 nm (12, 25, 29). As shown in ref. 12, the relationship between the interaxial spacing, d; the genome length, L; and the capsid volume, V, can be reasonably described by
 |
Now if the volume of a capsid is changed because of AFM indentation, DNA cannot leave the capsid, whereas water can. As a result, the change in capsid volume requires dehydration of DNA, which increases the DNA–DNA hydration force exponentially. To determine whether this force was significant in our AFM measurements, we needed to compare the additional work due to the hydration forces with the work of deforming empty capsids.
Dockland et al. (1) proposed from their structural study of λ phage that capsid proteins of λ do not seem to interact directly with the packaged DNA. Therefore, in our model we describe the force of capsid deformation as a sum of the force required to deform an empty capsid and the force required to deform the DNA inside the capsid. From a thermodynamic perspective, the work (w) exerted by the external force, F, on the capsid is given by
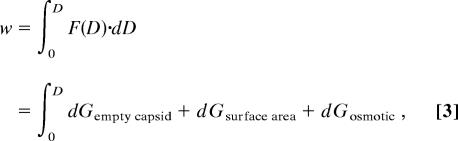 |
where D is distance of deformation in the z direction, dGempty capsid is the work of deformation of an empty shell (bending and stretching), dGsurface area is the work of capsid area change due to capsid stretching induced by the packaged DNA, and dGosmotic is the work induced by osmotic pressure change due to the capsid volume modification. The work required for small mechanical deformations of the capsid shell is
 |
where dA is a change in the capsid surface area due to the deformation distance dD and parameters k1 and k2 are material-specific constants for the capsid proteins. k1 corresponds to kempty (0.13 N/m) the spring constant for an empty λ capsid (Table 1). The deformation dD also gives rise to a volume change, dV, which in turn leads to an osmotic work (dGosmotic = −Πosmotic·dV). The force of deformation F(D) can thus be expressed as
We set the capsid area constant (dA/dD = 0), a value that, considering the Young's modulus of the capsids, is expected to be small. We show in supporting information (SI) Text (see also SI Figs. 6 and 7) that, independent of the deformed capsid shape (oblate or a truncated sphere depending on the assumed AFM cantilever tip shape and size), (dV/dD) ≈ [(π r0)/2]·D. Thus the total deformation force F(D) can be expressed as:
where Πosmotic (c0·V0/V) is the osmotic pressure of DNA in the deformed capsid with the volume changed from V0 to V, c0 is DNA concentration in the undeformed capsid, and c0·V0/V is the DNA concentration in the deformed capsid with volume change V0/V ≈ 1 + [3/(16·r02)]·D2 (derived from Eqs. 9 and 10 in SI Text). This volume change before a capsid breaks is almost negligible for small deformations (in our case, ΔV ≈ 1% at maximum deformation Dmax = 7.6 nm). Therefore, the osmotic pressure Πosmotic (c0·V0/V) ≈ Πosmotic(c0) ≈ constant during the entire deformation before the capsid breaks, which explains why we observe a linear deformation for all DNA-filled λ capsids even though the DNA is under high pressure.
By setting kempty·D = 0.7 nN in Eq. 6 (the lower estimate of the breaking force for empty capsids) and by using the maximal indentation averaged over all measured values (D = 6.5 nm), we can calculate the maximal force a capsid can withstand as a function of DNA length (solid line in Fig. 5B) by using V0 = 87,114 nm3 and r0 = 29.5 nm. Although it shows a small overestimation, the theoretical prediction in Fig. 5B shows good agreement between the calculated values and the measured data, in particular when one considers that there are no fitting parameters used in this model. The overestimation of the data can be explained by the fact that in our model, we have set the capsid area constant. However, some change in the area upon deformation should be expected, which would imply a smaller change in the volume and thus a smaller contribution from the volume-dependent osmotic force term to the total force F(D). Eq. 6 thus provides an upper limit of the deformation force.
These results show that the packaging density of WT DNA (or close to its length) in λ phage is capable of providing a significant internal support to the mechanical strength of the capsid with the help of DNA hydration forces. We suggest that it is not coincidence, but rather the evolutionary energy optimization by nature that has selected and optimized the WT DNA phages, which can survive external mechanical stresses in the environment that are twice as high, in comparison to its mutants. Such an assumption is supported by the fact that E. coli cells which are infected by λ are often found colonizing the surfaces experiencing high shear forces due to the laminar liquid flows (15). Because the phage needs, in turn, to adhere to the E. coli cell surface to infect, it will also experience this mechanical stress. Indeed, practical experience shows that phages have a limited life due to mechanical breakage induced by the shear forces in the solution (30). From our own experience, WT λ phages seem to lose their infectivity more slowly than their shorter genome mutants, which we confirmed by titering WT phage and its 37.7-kb mutant before and after 2 weeks of storage at 4°C.
Finally, we performed AFM indentation experiments on the WT λ phage in 1 mM spermine tetrahydrochloride. Spermine is a tetravalent cation that is known to cause DNA condensation by introducing attractive interactions between the DNA strands (24). Capsid deformation response was again linear, but the capsid breaking force was similar to the empty capsid (our unpublished data). Indeed, we have shown earlier that 1 mM spermine will significantly reduce forces between packaged DNA strands within the capsid, because spermine cations permeate the capsid (7, 10). Rau et al. (24) showed that hydration forces between DNA strands will significantly decrease in the presence of polyvalent ions such as spermine. Therefore, this experimental observation confirms our above model interpretation that the osmotic force inside WT phage under standard buffer conditions will contribute to the capsid's strength against external mechanical deformation.
Our data analysis also shows that, in the WT phage, the osmotic force contribution to maximum deformation force (≈0.8 nN) is nearly equal to the breaking force of an empty capsid. This result suggests that the mechanical strength of the capsid sets the upper limit to the amount of DNA that can be packaged (i.e., the maximal internal force due to the osmotic pressure). Although there is no lower limit of DNA length that can be packaged into λ phage (as long as phage does not have to infect the cells and multiply), there is, however, an upper limit (106% of the WT genome), which could be related to the strength of the capsids (3). This balance between the internal DNA pressure and the capsid strength also has been discussed in the context of osmotic shock experiments (31). Therefore, we expect the packaging density of DNA to be optimized to the capsid strength and to be the key parameter in phage stability and infectivity.
Experimental Procedures
Bacteriophage Strain and Preparation of Phage Stock.
WT bacteriophage λ cI857, with a genome length of 48.5 kb, and its shorter mutant, with a genome length of 45.7 kb, were produced by thermal induction of lysogenic E. coli strains AE1 and AE2 derived from S2773 and S2739 strains (generously provided by Stanley Brown, Copenhagen University, Copenhagen, Denmark). The AE1 and AE2 strains were modified to grow without LamB protein expressed on its surface to increase the yield of phage induced in the cell. The culture was then lysed by temperature induction. λ phage λb221 with a length of 37.7 kb was extracted from single plaques. Phage purification details are described elsewhere (7). All phage samples were purified by CsCl equilibrium centrifugation and dialyzed from CsCl against TM buffer (10 mM MgSO4/50 mM Tris·HCl, pH 7.4). The final titer was ≈1012 virions per milliliter, which was determined by plaque assay (32).
Empty phage particles were prepared by incubating WT phage with its extracted LamB receptor for 1 h at 37°C, which causes all phages to eject their DNA. The ejected DNA was digested by DNase I, and the empty phage “ghosts” were purified by filter centrifugation.
Preparation of LamB λ Phage Receptor.
The receptor was the LamB protein purified from pop 154, a strain of E. coli K12 in which the lamB gene has been transduced from Shigella sonnei 3070 (33, 34). This protein has been shown to cause complete in vitro ejection of DNA from λ at 37°C in the absence of the added solvents required with the WT E. coli receptor (35, 36). Purified LamB was solubilized from the outer membrane with the detergent octyl polyoxyethylene.
AFM.
All AFM experiments were performed with AFM (Nanotec Electronica, Madrid, Spain) operated in jumping mode with WSxM software. A detailed description of the method and apparatus can be found elsewhere (37). Both imaging and FZ curves were done in buffer conditions at room temperature. The AFM sample was prepared as is described by Ivanovska et al. (17). The virus particles were deposited for adsorption on preliminary cleaned, hydrophobic glass coverslips. Rectangular gold-coated cantilevers (Olympus, Tokyo, Japan) were used with a tip apex nominal value of <20 nm. The cantilevers' spring constants were calibrated by using the method described in ref. 38 and were found to be 0.07 N/m, varying by ≈15% in a wafer unit.
Supplementary Material
Acknowledgments
We thank William M. Gelbart, Charles M. Knobler, Christoph F. Schmidt, and Michael Feiss for valuable discussions and advice; Gabriel Lander and John E. Johnson for taking EM images of λ capsids; Stanley Brown for providing the bacterial strain for phage growth; and Caroline Fraysse (University of Basel, Basel, Switzerland) for providing the strain for LamB production. This work was supported by the Laser Center at Vrije University Amsterdam and the Swedish Research Council (to A.E.), and by grants from the Dutch Organization for Fundamental Research of Matter and the Netherlands Organization for Scientific Research (to G.W.).
Abbreviations
- AFM
atomic force microscope
- FZ
force–distance.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703166104/DC1.
References
- 1.Dokland T, Murialdo H. J Mol Biol. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- 2.Yang F, Forrer P, Dauter Z, Conway JF, Cheng N, Cerritelli ME, Steven AC, Pluckthun A, Wlodawer A. Nat Struct Biol. 2000;7:230–237. doi: 10.1038/73347. [DOI] [PubMed] [Google Scholar]
- 3.Feiss M, Fisher RA, Crayton MA, Egner C. Virology. 1977;77:281–293. doi: 10.1016/0042-6822(77)90425-1. [DOI] [PubMed] [Google Scholar]
- 4.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 5.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. Proc Natl Acad Sci USA. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Biophys J. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Proc Natl Acad Sci USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purohit PK, Kondev J, Phillips R. Proc Natl Acad Sci USA. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purohit PK, Kodnev J, Phillips R. J Mech Phys Solids. 2003;51:2239–2257. [Google Scholar]
- 10.Evilevitch A, Castelnovo M, Knobler CM, Gelbart WM. J Phys Chem B. 2004;108:6838–6843. [Google Scholar]
- 11.Evilevitch A, Gober JW, Phillips M, Knobler CK, Gelbart WM. Biophys J. 2005;88:751–756. doi: 10.1529/biophysj.104.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Biophys J. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löf D, Schillén K, Jönsson B, Evilevitch A. J Mol Biol. 2007;368:55–65. doi: 10.1016/j.jmb.2007.01.076. [DOI] [PubMed] [Google Scholar]
- 14.Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. Virology. 2006;348:430–436. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas WE, Trintchina E, Forero M, Vogel V, Sokurenko EV. Cell. 2002;109:913–923. doi: 10.1016/s0092-8674(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix RW, Roberts JW, Stahl FW, Weisberg RA. Lambda II. Woodbury, NY: Cold Spring Harbor Lab Press; 1983. [Google Scholar]
- 17.Ivanovska IL, de Pablo PJ, Ibarra B, Sgalari G, MacKintosh FC, Carrascosa JL, Schmidt CF, Wuite GJ. Proc Natl Acad Sci USA. 2004;101:7600–7605. doi: 10.1073/pnas.0308198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJ, Schmidt CF. Proc Natl Acad Sci USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellenberger E, Edgar RS. In: The Bacteriophage Lambda. Hershey AD, editor. Woodbury, NY: Cold Spring Harbor Lab Press; 1971. pp. 271–295. [Google Scholar]
- 20.Hendrix RW, Tsui L. Proc Natl Acad Sci USA. 1978;75:136–139. doi: 10.1073/pnas.75.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau LD, Lifshitz EM. Theory of Elasticity. New York: Pergamon; 1986. [Google Scholar]
- 22.Zandi R, Reguera D. Phys Rev E. 2005;72:021917. doi: 10.1103/PhysRevE.72.021917. [DOI] [PubMed] [Google Scholar]
- 23.Leikin S, Parsegian VA, Rau DC, Rand RP. Annu Rev Phys Chem. 1993;44:369–395. doi: 10.1146/annurev.pc.44.100193.002101. [DOI] [PubMed] [Google Scholar]
- 24.Rau DC, Parsegian VA. Biophys J. 1992;61:260–271. doi: 10.1016/S0006-3495(92)81832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rau DC, Lee B, Parsegian VA. Proc Natl Acad Sci USA. 1984;81:2621–2625. doi: 10.1073/pnas.81.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Earnshaw WC, Harrison SC. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 27.Earnshaw WC, King J, Harrison SC, Eiserling FA. Cell. 1978;14:559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- 28.Earnshaw WC, Casjens SR. Cell. 1980;21:319–331. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 29.Parsegian VA, Rand RP, Fuller NL, Rau DC. Methods Enzymol. 1986;127:400–416. doi: 10.1016/0076-6879(86)27032-9. [DOI] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Woodbury, NY: Cold Spring Harbor Lab Press; 1983. [Google Scholar]
- 31.Cordova A, Deserno M, Gelbart WM, Ben-Shaul A. Biophys J. 2003;85:70–74. doi: 10.1016/S0006-3495(03)74455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silhavy TJ. Experiments with Gene Fusions. Woodbury, NY: Cold Spring Harbor Lab Press; 1984. [Google Scholar]
- 33.Roa M, Scandella D. Virology. 1976;72:182–194. doi: 10.1016/0042-6822(76)90322-6. [DOI] [PubMed] [Google Scholar]
- 34.Graff A, Sauer M, Van Gelder P, Meier W. Proc Natl Acad Sci USA. 2002;99:5064–5068. doi: 10.1073/pnas.062654499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Randall-Hazelbauer L, Schwartz M. J Bacteriol. 1973;116:1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roa M. FEMS Microbiol Lett. 1981;11:257–262. [Google Scholar]
- 37.de Pablo PJ, Colchero J, Gomez-Herrero J, Barro AM. Appl Phys Lett. 1998;73:3300–3302. [Google Scholar]
- 38.Sader JE, Chon JWM, Mulvaney P. Rev Sci Instrum. 1999;70:3967–3969. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.