Abstract
To assess the potential of protein function prediction in environmental genomics data, we analyzed shotgun sequences from four diverse and complex habitats. Using homology searches as well as customized gene neighborhood methods that incorporate intergenic and evolutionary distances, we inferred specific functions for 76% of the 1.4 million predicted ORFs in these samples (83% when nonspecific functions are considered). Surprisingly, these fractions are only slightly smaller than the corresponding ones in completely sequenced genomes (83% and 86%, respectively, by using the same methodology) and considerably higher than previously thought. For as many as 75,448 ORFs (5% of the total), only neighborhood methods can assign functions, illustrated here by a previously undescribed gene associated with the well characterized heme biosynthesis operon and a potential transcription factor that might regulate a coupling between fatty acid biosynthesis and degradation. Our results further suggest that, although functions can be inferred for most proteins on earth, many functions remain to be discovered in numerous small, rare protein families.
Keywords: fatty acid, heme, neighborhood, environmental genomics, metagenome annotation
Recent years have seen an explosion in the amount of shotgun sequence data gathered from diverse natural environments. Since 2004, almost 2 billion base pairs resulting from published large-scale metagenomics sequencing projects have been deposited [as of January of 2007 (1–8)], eclipsing the entire 764 Mbp of previously sequenced microbial genomes (9). Large-scale environmental sequencing efforts have the potential to considerably enhance our understanding of cellular processes, identify ubiquitous as well as unique biological functions in each environment, and close the gaps in our knowledge between genotype, phenotype, and environment. Until the identified ORFs are correctly annotated with biological functions, however, we are simply left with a vast amount of information but no contextual knowledge, analogous to the early days of genome sequencing.
Currently, characterizing an unknown sequence involves comparing it to sequences or protein domains of known function in public databases, usually by using BLAST (10) or other homology search tools (11). By applying BLAST-based annotation methods to newly sequenced genomes, functions can typically be assigned to ≈70% of the gene products (11–13). Unfortunately, these predictions have been estimated to include 13–15% database propagation errors (14) and are only possible if the unknown sequence has at least one BLAST hit. To complement homology-based function prediction, particularly in prokaryotes, additional information from genomic neighborhood (15, 16), phylogenetic profiles (17), gene coexpression (18), and gene fusion (19, 20) has been used and combined (18, 21). As yet, only the exploitation of genomic neighborhood (including gene fusions) is feasible in the context of metagenomic shotgun data.
In the first large-scale shotgun metagenomics projects from four diverse and complex environments [tropical surface water from the Sargasso Sea near Bermuda (2), farm soil from Minnesota (4), an acidophilic biofilm from an iron ore mine in northern California (1), and three samples from “whale fall” carcasses on the deep Pacific and Antarctic ocean floor (4)], functions have been predicted based on sequence similarity for only 27–48% of the 1.4 million genes in the different samples [see supporting information (SI) Table 1]. This implies that for the majority of proteins in the environment, functions remain unknown, and no attempt has yet been made to discover novel functionality. Furthermore, for each project, different methods, parameters, and even definitions of function were used, which are often not easily accessible to the community, making a comparison of the different samples difficult. To be able to comprehensively predict functions from various metagenomics samples and to get a consistent overview of function in different environments, we developed a sensitive prediction protocol that complements BLAST- and domain-based function predictions with newly developed and adapted gene neighborhood methods. Applying this protocol to the samples revealed a considerable predictive power, indicating that function can be inferred for most of the genes on earth; yet the majority of functions appear to reside in numerous rare, small protein families that remain largely unexplored.
Results and Discussion
An Operational Definition of Protein Function.
Biological function is a fuzzy term summarizing a complex concept applicable to different spatial scales (22, 23). At the molecular and (sub-)cellular level, an operational framework with clearly defined terms and thresholds is therefore required when attempting to quantify protein function. To infer specific function from existing database annotations by using homology, we require similarity to an environmental (partial) ORF >60 bits, corresponding roughly to an e-value of 10−8 in Uniref90 searches (4). This level of sequence similarity is rather strict in terms of homology identification but without further analysis may be insufficient to distinguish between paralogs and orthologs, thus not capturing all functional features such as enzyme substrate specificity. It is, however, sufficient to capture basic functionality.
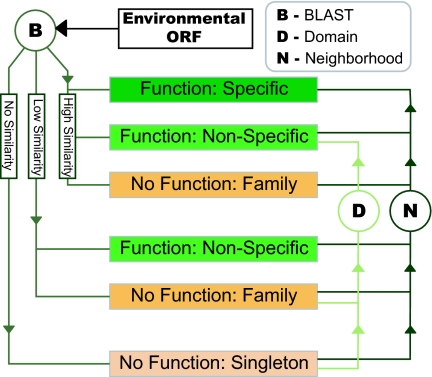
We used a hierarchical classification scheme, favoring manual annotation, to divide environmental ORFs and, for comparison, 124 prokaryotic proteomes into four categories based on the level of functional annotation possible: (i) those with strong similarity to, or in the genomic neighborhood of, a gene with specific functional annotation; (ii) those with strong similarity to genes with nonspecific functional information, weak but significant similarity to genes with any functional annotation, or in the genomic neighborhood of either of these; (iii) those with strong similarity to, or in the genomic neighborhood of, a gene of unknown function; (iv) those with neither similarity to sequences in annotated databases nor significant genomic neighborhood (Fig. 1).
Fig. 1.
Flow chart of function prediction procedure. By using homology to genes in the KEGG, COG, and UniRef90 databases, ORFs were divided into four categories based on the level of functional annotation possible; (i) specific functional annotation: ORFs similar to genes with specific functional information; (ii) nonspecific functional annotation: ORFs similar to genes that have been characterized at a general level or low similarity; (iii) no functional annotation but member of an existing family: ORFs with homologs in one of the databases but no functional information (e.g., “conserved hypothetical”); (iv) singletons: ORFs that have no significant similarity to known sequences. ORFs containing domains from the SMART and Pfam A databases were upgraded to having nonspecific annotation where applicable. Finally genomic neighborhood methods were used to infer functional links between ORFs and upgrade the functional annotation accordingly.
We used sequence similarity to infer functional information from the KEGG (24), COG (12), UniRef90 (25), SMART (26), and Pfam (27) databases (see Materials and Methods for parameter choices, benchmarks, and definitions of functional annotation). We used gene neighborhood evidence from the STRING database (21) and adapted existing gene neighborhood function prediction methods, based on intergenic distance and evolutionary conservation, for use in fragmented shotgun metagenomics data. First, we exploited the fact that intergenic distances tend to be shorter between genes of the same operon than between operons (28). Although several operon prediction methods have been introduced that are based solely on intergenic distances (28–31), they are species-specific, trained with experimentally verified transcript information (28), and/or require the context of a complete genome. Here, we calibrated directly on each sample to establish the likelihood of being functionally associated, given a positional distance within a read. Second, we used the fact that neighboring ORFs are more likely to be functionally associated if they are conserved over long evolutionary distances (15, 16, 32). We recorded multiple occurrences of neighboring genes, measured the sequence similarity of the respective neighborhoods to each other, and derived a metric based on evolutionary distance. We then combined these measures for intergenic and evolutionary distance to predict functional relationships between genes in the metagenomic data (see Materials and Methods).
Consistent Functional Characterization of ORFs in Four Environmental Data Sets.
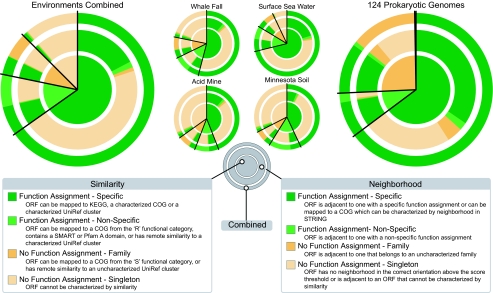
By combining homology searches and neighborhood methods, we were able to infer specific functional information for 76% of the 1.4 million predicted environmental ORFs and a more general level of functional information for another 7% (dark and light green segments respectively of the outermost ring in Fig. 2; see also SI Table 2). By using sequence similarity alone, a specific function can be inferred for almost two-thirds (65%) of the ORFs, and a general function for another 13% (inner circle Fig. 2). Neighborhood-based methods provide functional information for 30% of the ORFs (green segments in middle ring; Fig. 2), complementing similarity-based molecular characterizations with functional interactions. They also provide functional information for almost a quarter of the ORFs (75,448), where homology-based methods fail. This 30% of neighborhood-based predictions is considerably lower than the 56% achieved when the same methods are applied to the 124 prokaryotic genomes (SI Table 3). However, only 47% of the ORFs in the metagenomic data sets have a neighbor in the same transcription direction, as compared with 88% in completely sequenced genomes (SI Table 4), which implies that the predictive power of neighborhood methods is comparable in genomes and metagenomes. Indeed, the combined methods perform almost equally well in metagenomes (83% functional characterization) as in fully sequenced genomes (86%). Moreover, the metagenomic ORFs that cannot be characterized by similarity are significantly shorter than those that can (SI Fig. 5). Some of these may be fragmented ORFs that are too short to assign significant similarity; others may have resulted from erroneous ORF predictions. The latter would imply that the true fraction of gene products for which functions can be predicted is even higher. In either case, the quality of predictions should improve in the future because sequence coverage is likely to increase in metagenomics projects, allowing more reads to be assembled into longer contigs.
Fig. 2.
Protein function prediction in genomes and metagenomes. Many proteins can be functionally characterized in both data sets. The degree of functional characterization for four metagenomic data sets is shown on the left and for 124 prokaryotic genomes on the right. The inner pie chart represents the level of functional characterization possible by using the homology-based approach. The middle ring shows the level of functional characterization possible by using neighborhood methods. The outer ring summarizes the combined level of characterization possible. Surprisingly, it implies that most metagenomic ORFs (83% of the data) can be functionally characterized, similar to the level possible in fully sequenced genomes.
In the original reports of the metagenomics data sets, specific functions were assigned to 27–48% of the predicted gene products (1, 2, 4), indicating marked differences in the function prediction protocols caused by various technical issues such as the stringency of BLAST cutoffs, the choice of functional databases, and variations in gene calling (a comparison is presented in SI Table 1; for an expanded comparison see ref. 9). Because our benchmarks and manual confirmations of parameter settings show a negligible false-positive rate (see Materials and Methods), we believe that the near doubling in functional assignments is not caused by a looser function definition or more spurious assignments but is due to better utilization of existing functional information. The latter uncovers marked trends such as overrepresentation at the gene, family, or pathway level, in line with earlier studies (4) (SI Table 5). For example, we find that bacterial chemotaxis, flagellar assembly, and type III secretion genes are 3-fold more frequent in the genomes than the metagenomes (dominated by the surface sea water data set), perhaps because of the futility of bacterial motility in strong ocean currents. On the other hand, genes involved in amino acid metabolism as well as in the biosynthesis of nucleotides, carbohydrates, and lipids are significantly underrepresented in the genomes as compared with the metagenomes, perhaps because of the bias toward sequencing obligate pathogens, which tend to acquire these compounds from their hosts.
Comparison of Environmental Samples.
Among the four environments, the fraction of functional assignments differs considerably as it does between organisms (Fig. 2 and SI Figs. 6 and 7). In the surface sea water, specific functions are inferable for 82% of ORFs (dark green sections in Fig. 2); the corresponding fraction in whale fall is 66% and in soil only 53%. These differences can be partially attributed to inherent differences in the sequence data: for example, the average read length of the sea water data is longer than that of soil [818 bp vs. 673 bp after quality filtering (2, 4)] and 60% of the sea water reads can be assembled into longer contigs compared with <1% in soil (33). Also, environments have been previously characterized to different degrees, and for some environments, complete genome sequences are available that closely resemble those from the environment [e.g., SAR11 as a frequent ocean bacterium (34)]. This means not only more gene context in a certain environment but also more BLAST assignments for short fragmented ORFs and hence more reliable gene predictions. Finally, a major fraction of the acid mine sample is comprised of Archaea, which are generally less functionally characterized than bacteria, thus lowering our functional understanding of the sample. Nevertheless, we believe that most differences between the environments are caused by multiple effects linked to genuine diversity in phylogeny and lifestyle. For example, genomes of species in the sea water samples are smaller than in soil, with a higher fraction of essential, well characterized genes (33), but they also evolve faster (35), which should make homology searches less sensitive. Farm soil might supply the most stressors to microbial life because of its high population density, microhabitats, and physical and systemic perturbations (e.g., temperature, nutrient availability, and pH) (36), leading to a broad repertoire of stress-response phenotypes with hitherto uncharacterized functions. Similarly, the unusual ecological niche created by a deep-sea whale carcass, with its extreme conditions of darkness, cold, and high pressure, lead to highly specialized microbial adaptations such as barotolerance and temperature-induced lipid fluidity (37) that do not resemble those in other environments or genomes.
Predicting Functional Novelty: In-Depth Analysis of Two Neighborhood-Based Findings.
Whereas homology-based methods require additional analysis to identify novel functions (e.g., via novel subgroups in a characterized sequence family), neighborhood methods can directly provide novel functional associations. Novelty can be obtained either by (i) seeing unexpected functional coupling of known genes or (ii) assigning unknown genes to known processes. The first is evident in the fact that there are as many as 5,851 pairs of neighboring COGs unique to metagenomes, even though these COGs occur individually in the 124 prokaryotic genomes, implying many novel functional interactions. These frequently include enzymes involved in amino acid biosynthesis with novel links to numerous protein degradation and regulatory proteins, probably reflecting the different nutritional constraints (SI Table 6). The second can be seen in the 75,448 ORFs (5% of the total) that are solely characterized by neighborhood. Here, we provide detailed functional annotation for two families: a previously uncharacterized gene family associated with a well known pathway (heme biosynthesis) and a transcription factor, unique to the Sargasso Sea data set, that potentially regulates the coupling of two opposing processes (fatty acid biosynthesis and degradation). These and other functional predictions, including annotations for nearly half a million previously uncharacterized proteins, are available online (www.bork.embl.de/Docu/harrington).
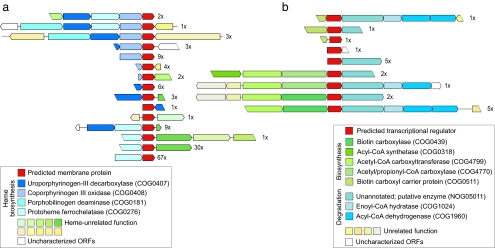
Neighborhood information can help characterize a gene family if members of that gene family occur next to different genes belonging to the same pathway in different species. By using such a query, we discovered members of a large uncharacterized gene family (COG1981) with several hundred ORFs in the surface sea water and whale fall samples, adjacent to various enzymes from the well studied heme biosynthesis pathway (Fig. 3a). Heme feeds into the synthesis of both cytochromes and chlorophyll and thus plays a key role in enzymatic reactions, energy production, and metabolic regulation (38). In addition, it functions as a prosthetic group to proteins involved in bacterial stress response, oxidative damage, and virulence (39). Sequence analysis of the uncharacterized family reveals that it comprises hydrophobic, putative membrane-associated proteins that are unlikely to have enzymatic functions. They might thus be implicated as scaffolding proteins in tethering the pathway to the membrane and/or enabling sufficient substrate fluxes.
Fig. 3.
Prediction of function in previously uncharacterized gene families by using genomic neighborhood. Whereas homology-based approaches quantify the known functions, neighborhood approaches reveal functional novelty, even in conjunction with well known processes. (a) A putative transmembrane protein belonging to an uncharacterized COG (COG1981 shown in red) that consistently cooccurs with members of the well characterized heme biosynthesis pathway (colored blue). The putative membrane-associated protein occurs on 174 distinct contigs in the surface sea water and whale fall data sets that can be grouped into at least 15 unique operon arrangements, strongly suggesting a role in this process. (b) A predicted putative regulator, shown in red, that links fatty acid biosynthesis (upstream, colored green) with fatty acid degradation (downstream, colored blue), a functional link not seen in fully sequenced genomes. The regulator appears on 20 distinct contigs in the sea water, of which there are at least five unique operon arrangements.
Whereas the heme-associated gene family had previously been observed in fully sequenced genomes, another family of 20 members was found exclusively in the surface sea water samples by using our clustering procedure (see Materials and Methods). Even though no homology could be found by using our automated methods, detailed analysis revealed weak but significant similarity to a family of helix–turn–helix (HTH) transcription factors. An examination of its neighboring genes implies that this family is found in a variety of species, the most closely related being Actinobacteria. As the genes are on various contigs with differing gene orders, we could assign it to an entire operon that additionally contains three downstream genes consistently occurring in the same orientation. The first downstream gene of unknown function (NOG05011) has been observed in completely sequenced genomes; in-depth sequence and secondary structure analyses suggest an enzymatic function (data not shown). The second and third genes of this potential operon (COG1024 and COG1960) catalyze successive steps of the β-oxidation of fatty acids (usually involved in degradation) (38, 40). Interestingly, this invariant operon, apparently controlled by the newly predicted transcriptional regulator, frequently occurs downstream of various genes involved in fatty acids biosynthesis (Fig. 3b). Thus, context-based methods predict a coupling between fatty acid degradation and biosynthesis, whereby the previously undescribed gene might provide the regulation of this link. It is intriguing to speculate that this coupling of two antagonistic processes is an adaptation to repeatedly changing environmental conditions. For instance, strongly regulated circadian rhythms are followed by several marine bacteria (41). These bacteria actively migrate to different depths in a periodic fashion to balance the efficient usage of light for energy against the danger of DNA damage (42, 43). Energy storage during the light-dependent phase by biosynthesis of fatty acid and energy release in the light-independent phase could thus be a regulated switch during locomotion from light to dark and vice versa.
Functional Prediction vs. Functional Diversity.
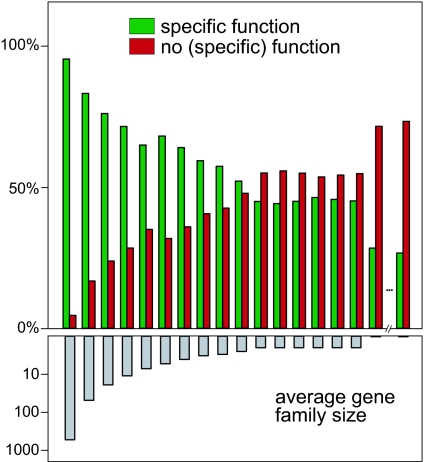
As more environments are explored, we expect that core protein functions (for example, translational machinery) will be seen repeatedly and will dominate every sample. Novel, rare, and perhaps environment-specific functions, on the other hand, might not be classifiable because they are not yet captured by the experimental studies that underlie most current knowledge about biological function. To reconcile our gene-centric view of the data with a function-based one, we performed an all-against-all similarity search of all predicted ORFs in all four environments, clustered the results into gene families, and recorded their functional status according to our operational definition (see Fig. 4 and Materials and Methods). We find that specific functional knowledge is indeed heavily skewed toward large families: functionally characterized families make up 89% of the largest families (200 or more members), whereas uncharacterized ones make up 72% of the smallest families (three or fewer members). Thus, although most of the proteins in the environmental samples can be functionally characterized because they belong to well studied large gene families, numerous distinct, rare functions remain to be identified. Because these are likely to be adaptations to specific environmental constraints, they should have the potential for exploitation in biotechnology and medicine. Of all of the families (including singletons), functions can be assigned for only 32%, but this fraction contains 85% of all of the proteins studied here. If singletons are disregarded, the fraction of characterizable proteins in the complex environments studied increases further, from 72% to 79%. Although these remain qualitative assignments of low resolution (i.e., substrate specificity or cellular roles are often not specified), even general molecular classifications such as “dehydrogenase” imply some basic functional understanding, and more than a quarter of these are further complemented by associations to other genes predicted by the neighborhood method. Despite this remarkably high coverage, our functional knowledge about the proteins on earth can be further increased by deeper sequencing that generates longer assemblies and less-fragmented ORFs. This should improve gene predictions and reduce the number of uncharacterized singletons that are skewed toward short ORFs. Moreover, longer contigs would allow the application of indirect neighborhood methods (that is, operon membership), which we have not considered here. This huge potential to functionally characterize the vast majority of proteins in current and upcoming complex samples calls for strategies to capture functional novelty, for example by experimental procedures that enrich in those many small and rare families of unknown functions, analogous to normalizations of EST libraries introduced in the early 1990s (2). Coupled with systematic biochemical screens, a census of the repertoire of protein functions on earth (at least at the low level of resolution currently used in sequence annotation) might thus be feasible in the very near future.
Fig. 4.
Dependence of functional characterization on family size. Colored bars in this histogram of gene families binned by size represent the proportion of families with specific functional annotation (if >20% of the members were classified as such; green) and no specific annotation (a combination of nonspecific and no functional annotation; red). Gray bars indicate average gene family size in that bin. Only two of 174,124 bins containing singletons are shown for clarity. Most large gene families have a known function, whereas many small families remain uncharacterized.
Materials and Methods
Sequence Data and Similarity Searches.
We analyzed published microbial shotgun sequence data from four environmental samples, totaling 1,438,944 genes: 1,086,400 genes from tropical surface water from the Sargasso Sea (2), 183,586 genes from farm soil from Minnesota (4), 122,146 genes from isolated whale fall carcasses (4), and 46,862 genes from an acidophilic biofilm from an iron ore mine (1). In parallel, we analyzed 344,619 genes from 124 prokaryotic genomes from the STRING database (21) (SI Table 7). Analyses were carried out at three different levels of stringency, the figures reported here use a bit score cutoff of 60 bits for orthology assignment (a prerequisite to predict specific functions) and 40 bits for homology assignment (for details of parameter exploration see SI Text). To map functionally characterized domains to metagenomic ORFs, we scanned the HMM profile signatures from Pfam (27) and SMART (26) against the metagenomic sequences by using HMMER (http://hmmer.wustl.edu/) software and applied the corresponding family-specific cutoffs.
Gene Family Analysis.
We grouped genes from all four environmental data sets into 206,217 gene families by first constructing a single-linkage graph of an all-against-all BLAST (60-bit cutoff), with nodes representing proteins, and edges representing BLAST hits between proteins weighted by BLAST bit scores. This graph was then clustered by using Markov chain linkage clustering with an inflation value of 1.1 (44, 45) (SI Table 8).
Function Prediction Using Sequence Similarity.
ORFs were assigned to KEGG pathways and COGs by using the method described by Tringe et al. (4) using a 60 bit cutoff. For the 124 prokaryotic genomes, the KEGG and COG assignments from the STRING database were used. ORFs were also compared against the UniRef90 database, divided into functionally characterized and uncharacterized clusters (see SI Text) and annotated with domains from the SMART and Pfam databases. These annotations were combined in a hierarchical manner, favoring manually annotated databases, placing each ORF into one of the above categories. By definition, any ORF that mapped to KEGG was considered to have a specific function assigned. Of the remaining ORFs, those that mapped to a COG were considered to have a specific function assigned, with the exception of those in functional classes “R” and “S,” which were considered to have nonspecific and no function assigned, respectively. The remaining ORFs were considered to have specific functional annotation if they had strong similarity (>60 bits) to functionally characterized UniRef90 clusters, nonspecific functional annotation if they contain a domain from the SMART or Pfam A database or have remote similarity (>40 bits) to functionally characterized UniRef90 clusters. All other ORFs were considered to have no function assigned, those with similarity to uncharacterized UniRef90 clusters were considered to be part of a family, and the rest singletons.
Function Prediction Using Genomic Neighborhood.
Using the contig positions of the ORFs in each data set, we constructed a list of pair-wise neighborhoods. For this analysis, we considered only codirectionally transcribed genes (for the treatment of overlapping genes, see SI Text). To investigate the conservation of neighborhoods, we constructed a graph for each set of homologous neighborhoods. An edge was placed between two neighborhoods if there were BLAST hits >60 bits between both pairs of genes, except in cases where a gene from one neighborhood hit both genes in the other. This graph was then used to construct clusters of neighborhoods representing a conserved gene pair. To estimate the evolutionary distance over which a neighborhood is conserved, we adapted a weighting scheme used for multiple sequence alignment (46) to derive a score with the property that it will be low for small clusters of closely related sequences and large for clusters with distantly related sequences. For each metagenomic data set, we then constructed a benchmark set of pair-wise neighborhoods where both genes have a KEGG mapping. At each intergenic and evolutionary distance within the benchmark set, we determined the proportion of neighborhoods that map to the same KEGG pathway. This relationship was then interpolated and used to derive a value P for each neighborhood in the data set, corresponding to the probability that a pair of genes in a neighborhood is functionally related (SI Figs. 6 and 8–11 and SI Table 2). We also applied this method to individual organisms (SI Figs. 7 and 12 and SI Table 3) to assess the effect of species-specific genome architectures on the method. It is clear that the relationship between intergenic and evolutionary distance and P is highly species-specific. The vast majority of P values exceed the random expectation (16%, the probability that a random pair of genes map to the same KEGG pathway). To ensure that we were dealing with high quality predictions, we considered a pair of genes to be functionally linked only if the P value was >0.4 [found to have an accuracy approaching 70% at the level of functional modules (47)]. For the ORFs that map to COGs, additional neighborhood information was taken from the STRING database (see SI Text).
Supplementary Material
Acknowledgments
We thank the P.B. group for helpful discussions. E.D.H. was supported by the European Community's FP6 Marie Curie Fellowship for Early Stage Training (E-STAR) under contract number MEST-CT-2004-504640. This work was supported by the European Union 6th Framework Program (Contract No. LSHG-CT-2004-503567).
Abbreviations
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- COG
Clusters of Orthologous Groups.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702636104/DC1.
References
- 1.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 3.Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 4.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, et al. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 5.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 6.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia Martin H, Ivanova N, Kunin V, Warnecke F, Barry KW, McHardy AC, Yeates C, He S, Salamov AA, Szeto E, et al. Nat Biotechnol. 2006;24:1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Raes J, Harrington ED, Singh AH, Bork P. Curr Opin Struct Biol. 2007;17:362–369. doi: 10.1016/j.sbi.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Bork P, Koonin EV. Nat Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 12.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huynen MA, Snel B, von Mering C, Bork P. Curr Opin Cell Biol. 2003;15:191–198. doi: 10.1016/s0955-0674(03)00009-7. [DOI] [PubMed] [Google Scholar]
- 14.Brenner SE. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 15.Dandekar T, Snel B, Huynen M, Bork P. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 16.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Proc Natl Acad Sci USA. 1999;96:4285. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 19.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Science. 1999;285:751. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 20.Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 21.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. Nucleic Acids Res. 2005;33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bork P, Dandekar T, Diaz-Lazcoz Y, Eisenhaber F, Huynen M, Yuan Y. J Mol Biol. 1998;283:707–725. doi: 10.1006/jmbi.1998.2144. [DOI] [PubMed] [Google Scholar]
- 23.Bork P, Serrano L. Cell. 2005;121:507–509. doi: 10.1016/j.cell.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, et al. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Proc Natl Acad Sci USA. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price MN, Huang KH, Alm EJ, Arkin AP. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuda S, Katayama T, Kawashima S, Goto S, Kanehisa M. Nucleic Acids Res. 2006;34:D358–D362. doi: 10.1093/nar/gkj037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan Y, Moult J. Proteins. 2006;64:615–628. doi: 10.1002/prot.21021. [DOI] [PubMed] [Google Scholar]
- 32.Korbel JO, Jensen LJ, von Mering C, Bork P. Nat Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 33.Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, et al. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 35.von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Science. 2007;315:1126–1130. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- 36.Torsvik V, Ovreas L. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- 37.Yayanos AA. Annu Rev Microbiol. 1995;49:777–805. doi: 10.1146/annurev.mi.49.100195.004021. [DOI] [PubMed] [Google Scholar]
- 38.Michal G. Biochemical Pathways: An Atlas of Biochemistry and Molecular Biology. New York: Wiley; 1999. [Google Scholar]
- 39.Frankenberg N, Moser J, Jahn D. Appl Microbiol Biotechnol. 2003;63:115–127. doi: 10.1007/s00253-003-1432-2. [DOI] [PubMed] [Google Scholar]
- 40.Yang XY, Schulz H, Elzinga M, Yang SY. Biochemistry. 1991;30:6788–6795. doi: 10.1021/bi00241a023. [DOI] [PubMed] [Google Scholar]
- 41.Lakin-Thomas PL, Brody S. Annu Rev Microbiol. 2004;58:489–519. doi: 10.1146/annurev.micro.58.030603.123744. [DOI] [PubMed] [Google Scholar]
- 42.Alexandre G, Greer-Phillips S, Zhulin IB. FEMS Microbiol Rev. 2004;28:113–126. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Bebout BM, Garcia-Pichel F. Appl Environ Microbiol. 1995;61:4215–4222. doi: 10.1128/aem.61.12.4215-4222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enright AJ, Van Dongen S, Ouzounis CA. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Dongen S. A Cluster Algorithm for Graphs. Amsterdam: National Research Institute for Mathematics and Computer Science in The Netherlands; 2000. [Google Scholar]
- 46.Gerstein M, Sonnhammer EL, Chothia C. J Mol Biol. 1994;236:1067–1078. doi: 10.1016/0022-2836(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 47.von Mering C, Zdobnov EM, Tsoka S, Ciccarelli FD, Pereira-Leal JB, Ouzounis CA, Bork P. Proc Natl Acad Sci USA. 2003;100:15428–15433. doi: 10.1073/pnas.2136809100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.