Abstract
In mammalian cells, the GW182 protein localizes to cytoplasmic bodies implicated in the regulation of messenger RNA (mRNA) stability, translation, and the RNA interference pathway. Many of these functions have also been assigned to analogous yeast cytoplasmic mRNA processing bodies. We have characterized the single Drosophila melanogaster homologue of the human GW182 protein family, which we have named Gawky (GW). Drosophila GW localizes to punctate, cytoplasmic foci in an RNA-dependent manner. Drosophila GW bodies (GWBs) appear to function analogously to human GWBs, as human GW182 colocalizes with GW when expressed in Drosophila cells. The RNA-induced silencing complex component Argonaute2 and orthologues of LSm4 and Xrn1 (Pacman) associated with 5′–3′ mRNA degradation localize to some GWBs. Reducing GW activity by mutation or antibody injection during syncytial embryo development leads to abnormal nuclear divisions, demonstrating an early requirement for GWB-mediated cytoplasmic mRNA regulation. This suggests that gw represents a previously unknown member of a small group of genes that need to be expressed zygotically during early embryo development.
Introduction
The GW182 protein is a critical component of cytoplasmic RNP bodies that have been shown to function in mRNA degradation, storage, and, recently, microRNA (miRNA)- and siRNA-based gene silencing (Eystathioy et al., 2003; Yang et al., 2004; Ding et al., 2005; Jakymiw et al., 2005; Liu et al., 2005a; Rehwinkel et al., 2005). GW182 was named for the presence of multiple glycine (G)–tryptophan (W) amino acid pairs in the N-terminal region of a 182-kD protein with a predicted C-terminal RNA recognition motif (RRM). It localizes into cytoplasmic GW bodies (GWBs; Eystathioy et al., 2002; Maris et al., 2005) that also contain factors involved in 5′–3′ mRNA decay, including the exonuclease XRN1, decapping enzymes DCP1 and DCP2, and the LSm1–7 decapping activator, pointing to a role for GWBs in regulating mRNA stability (Ingelfinger et al., 2002; Eystathioy et al., 2003; Cougot et al., 2004). These bodies may participate in additional roles in mRNA regulation, as they also contain the m7G cap–binding protein eIF4E and the eIF4E transporter but no other components of translation machinery (Andrei et al., 2005; Kedersha et al., 2005). Importantly, intact GWBs are required for the functioning of the RNAi pathway in human cells potentially via direct interaction between GW182 (and the related TNRC6B protein) and Argonaute1 (Ago1) and 2 (Ago2; Jakymiw et al., 2005; Liu et al., 2005a,b; Meister et al., 2005).
GWBs are thought to be analogous to Saccharomyces cerevisiae cytoplasmic processing bodies (PBs). They are involved in mRNA decapping and 5′–3′ exonucleolytic decay (Sheth and Parker, 2003), and their integrity depends on the presence of nontranslating mRNAs (Sheth and Parker, 2003; Cougot et al., 2004; Teixeira et al., 2005). Both PBs and GWBs dissociate when polysomes are stabilized with drugs such as cycloheximide (Sheth and Parker, 2003; Cougot et al., 2004; Teixeira et al., 2005). However, despite similar compositions, there are functional differences between GWBs and PBs. GWBs increase in size and number in proliferating cells (Yang et al., 2004), whereas PBs increase in size and number during growth limitation and increased cell density (Teixeira et al., 2005). GWBs and PBs also differ in their responses to stress, as PBs increase in size and number in response to environmental stress. This is likely caused by decreased translation initiation because this response can be reproduced using a temperature-sensitive allele of Prt1p, a subunit of the eIF3 complex (Teixeira et al., 2005). In stressed mammalian cells, stalled preinitiation complex mRNAs are first targeted to stress granules (SGs), which may function as triage sites where mRNAs are sorted for future degradation, storage, or reinitiation of translation. Observation of interactions between SGs and GWBs in live cells suggest that transcripts may be exported from SGs to GWBs for degradation (Kedersha et al., 2005).
We have characterized the role of gawky (gw), the Drosophila melanogaster orthologue of the human GW182 gene family. GW localizes to punctate structures in the cytoplasm of Drosophila embryos and cultured S2 cells. Drosophila GWBs are electron-dense nonmembrane-bound cytoplasmic foci. These structures are targeted by human GW182 and its paralogues TNRC6B and TNRC6C in Drosophila cells. Unlike what is seen in some mammalian cells, only some foci colocalize with the previously identified GWB components LSm4, the Drosophila Xrn1 orthologue Pacman (PCM), and AGO2 (Ingelfinger et al., 2002; Eystathioy et al., 2003; Kedersha et al., 2005; Liu et al., 2005a; Sen and Blau, 2005). There is a requirement for the zygotic expression of full-length Drosophila GW during early embryonic nuclear divisions. This suggests a critical role for GWB-based cytoplasmic RNA regulation in Drosophila beginning with early embryo development.
Results
Embryonic gw expression is required for early Drosophila development
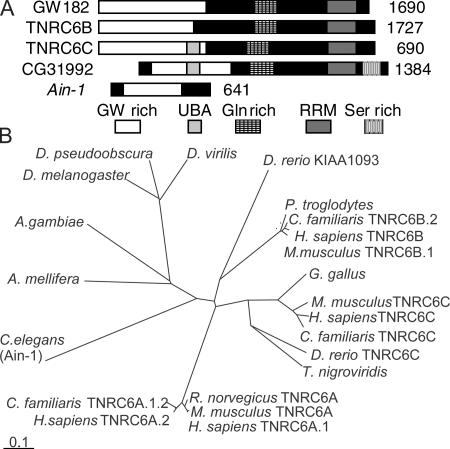
The gw1 mutation was isolated in a screen for recessive lethal zygotic mutations on the Drosophila fourth chromosome and mapped to a region predicted to contain a single gene, CG31992 (Adams et al., 2000). This gene encodes a 143-kD protein containing a C-terminal RRM domain and an N-terminal glycine- and tryptophan-rich region (20% G or W), which are features also found in the human GW182 protein (Eystathioy et al., 2002). There are three human GW-like proteins (Fig. 1 A). The Caenorhabditis elegans AIN-1 gene is also proposed to be part of this family, although it lacks an RRM domain (Ding et al., 2005). Although many vertebrate species have up to three GW-related proteins, invertebrates seem to have only a single form (Fig. 1 B).
Figure 1.
A comparison of the GW protein family. (A) The product of CG31992, the Drosophila GW protein (GenBank/EMBL/DDBJ accession no. AE003843), contains three regions that are common to all human GW182-related proteins: an N-terminal GW-rich region, a C-terminal RRM domain, and a glutamine-rich region. It has a predicted ubiquitin-associated domain (UBA) that is also found in TNRC6C and a C-terminal serine-rich region that is not found in human GW proteins. Drosophila GW is 17.8–20% identical and 24–28.3% similar to the human GW protein family. It is most similar to TNRC6C. C. elegans AIN-1 is also suggested to be a member of the GW protein family (Ding et al., 2005) because it is GW rich and contains one region of significant (24%) amino acid similarity. (B) Predicted evolutionary relationships between GW proteins from vertebrates and invertebrates. Bar, 0.1 amino acid substitutions per site.
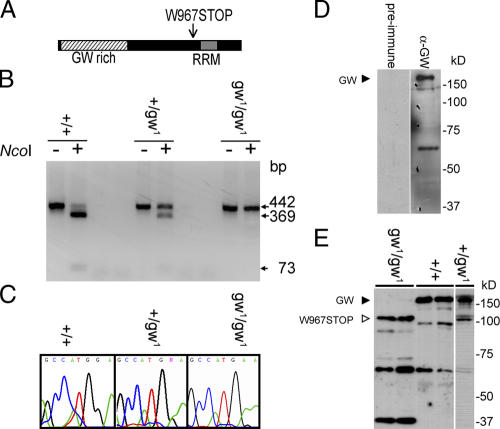
The mutant gw1 allele encodes a 100-kD truncated protein containing the GW-rich region but not the C-terminal RRM domain as a result of a nonsense mutation (Fig. 2 A). The location of this gene on chromosome four required an alternate approach to confirm the genotype of mutant embryos as a result of the lack of early developmental markers on this chromosome. We confirmed the presence of the mutation in individual embryos by PCR amplification of the region flanking the mutation (Fig. 2, B and C). We raised a polyclonal GW antibody that recognized a 160-kD protein (Fig. 2 D), which is within ∼10% of the predicted molecular mass of 143 kD. This antibody also recognized the 100-kD truncated GW protein in gw1 homozygotes. This truncated protein is also present in heterozygous adults, strongly suggesting that it is functionally inactive and has no dominant-negative effects (Fig. 2 E).
Figure 2.
Characterization of the gw mutation and localization of the GW protein. (A) gw 1 is caused by a nonsense mutation of the tryptophan codon at position 967 to stop. (B) The gw 1 mutation causes the loss of an NcoI restriction site and allowed rapid embryo genotyping by PCR. (C) Mutations were confirmed by DNA sequencing. (D) A polyclonal antibody raised against the N-terminal region of GW recognizes a 160-kD band representing the endogenous protein. (E) The anti-GW antibody also recognizes a 100-kD truncated form of GW in gw1/gw1 embryos that is not present in wild-type embryos.
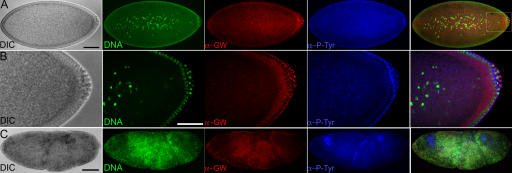
Heterozygous gw1/CiD parents produced embryos with disorganized internal structures 90– 130 min after egg deposition (AED; Fig. 3 C). These were found to be homozygous gw1 mutant, whereas embryos that developed normally were found to have at least one gw + allele (n = 200) by PCR. In early embryos, GW localizes to foci surrounding cortical nuclei (Fig. 3, A and B). Homozygous gw1 mutant embryos failed to cellularize, and DNA, GW, and membrane can be seen forming disorganized aggregates (Fig. 3 C).
Figure 3.
GW localization in normal Drosophila tissues and homozygous gw1 mutant embryos. (A–C) Embryos were fixed 90–130 min AED. (A) In normal embryos undergoing cellularization (differential interference contrast [DIC]), GW (α-GW) localized to foci surrounding the cortical nuclei (DNA). The plasma membrane is visualized using antiphosphotyrosine (α-P-Tyr). (B) The boxed area in A is shown magnified. Note the presence of brightly staining GW foci in the cytoplasm surrounding the nuclei. (C) In homozygous gw 1 mutant embryos, the DNA, anti-GW, and antiphosphotyrosine staining form disorganized aggregates. Bars, 100 μM.
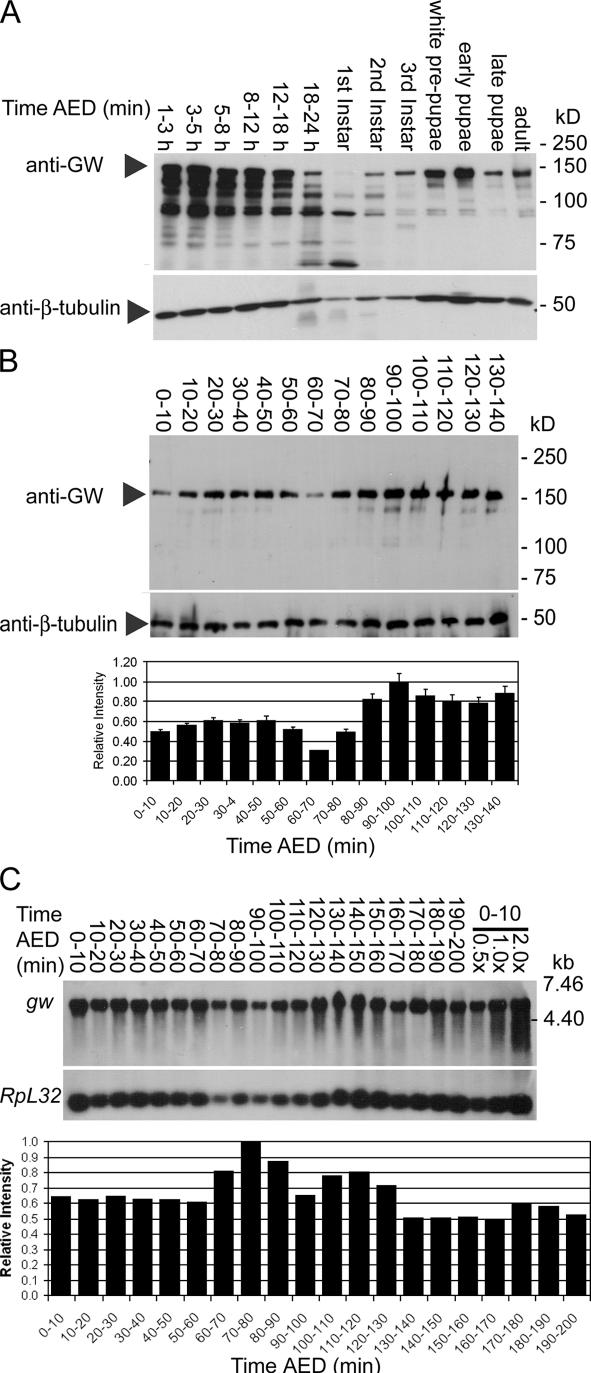
The highest relative levels of GW were found during early embryonic development and pupariation (Fig. 4 A). The presumptive maternal GW contribution to the embryo appears to be depleted by 60–70 min AED followed by an increase in GW levels starting at 80 min AED (Fig. 4 B). The activation of zygotic gw transcription was confirmed by Northern blotting. There is a significant maternal contribution of gw mRNA (Fig. 4 C). Corresponding to the increase observed in GW protein levels, the relative levels of gw mRNA increase at 80–90 min AED (Fig. 4 C).
Figure 4.
GW protein is expressed at varying levels during development. (A) Western blots showed high levels of GW protein during early embryonic development until ∼18 h and again during pupariation. (B) Relative GW protein levels are reduced at 60–70 min AED and subsequently increase at 70–80 min AED. Error bars represent the SD of the relative values obtained from three separate Western blots. (C) There is an increase in relative gw mRNA levels at ∼80–90 min AED compared with the mRNA encoding the RpL32 ribosomal protein. To confirm the accuracy of quantitation, the same sample was loaded at 0.5, 1.0, and 2.0× concentration.
Drosophila GWBs are homologous to human GWBs
GW localizes to punctate cytoplasmic bodies in Drosophila embryos (Fig. 3 C) and S2 cells (Fig. 5 and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1). In transmission EM sections, GWBs appeared as electron-dense nonmembrane-bound cytoplasmic particles (Fig. 6, A–C). Because of these similarities to human GWBs (Eystathioy et al., 2002), we tested the functional conservation between human GW182 and Drosophila GW. To assay GW in living cells, we created a transgenic cell line expressing a GW-GFP fusion that localized to cytoplasmic foci. GFP alone showed diffuse fluorescence throughout the cell (Fig. S1). Several key proteins found in GWBs/PBs, including PCM, the Drosophila Xrn1 homologue, AGO2, and a representative of the Drosophila LSm proteins, LSm4, colocalize with GW (Fig. 5, A–C). This colocalization is not caused by aggregation of the GFP tag, as FLAG-AGO2 costains with endogenous Drosophila GW. The association between AGO2 and Drosophila GW was further confirmed by coimmunoprecipitation of AGO2 with Drosophila GW (Fig. S2). Functional conservation with human GWBs is also suggested by the targeting of GFP-GW and RFP fusions of the human GW182 and its paralogues TNRC6B and TNRC6C in S2 cells (Fig. 5, D–F).
Figure 5.
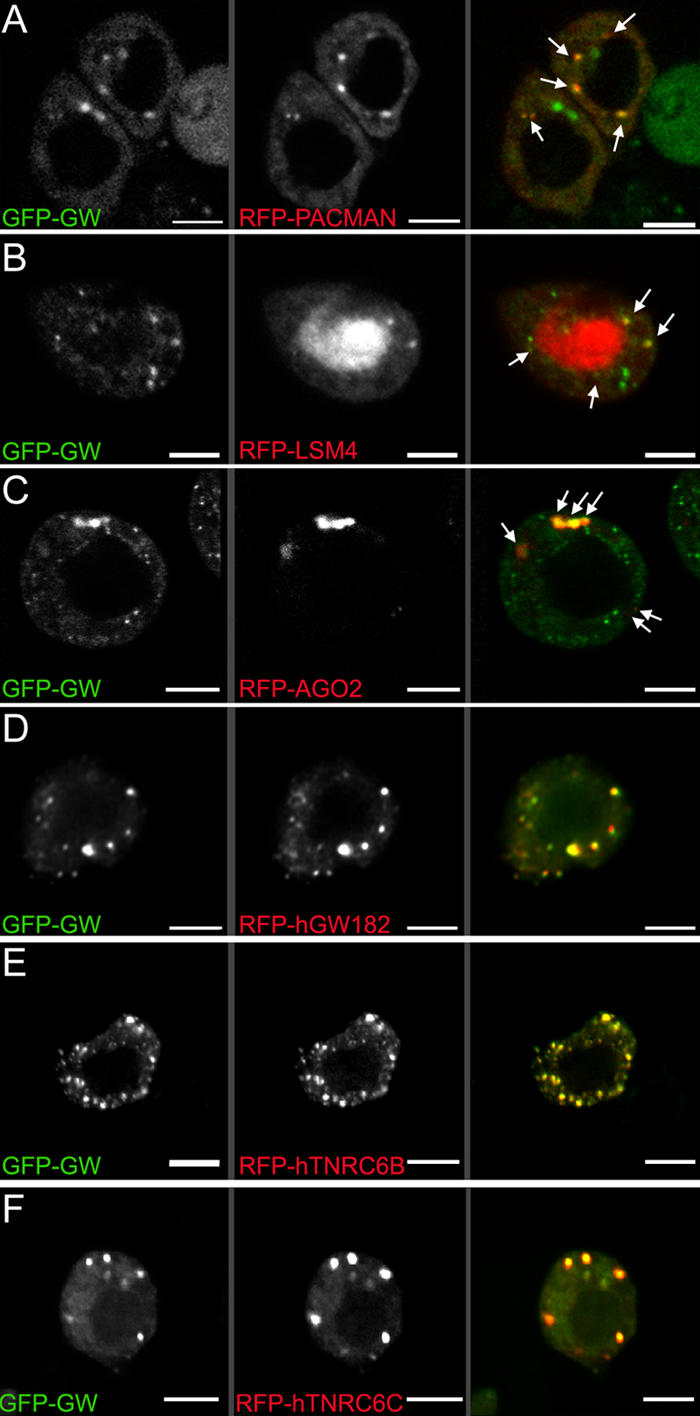
Colocalization of GW with markers associated with GWBs/PBs in Drosophila S2 cells. (A) A C-terminal fusion of RFP to PCM localized to discrete cytoplasmic foci. Several of these (arrows) colocalized with a GFP-GW fusion protein. (B) Another human GWB component, LSm4, localized to the nucleus (middle), but some signal was also detected in cytoplasmic foci (arrows). Some, but not all, Drosophila GWBs colocalized with the LSm4 foci. (C) AGO2, a RISC component, also colocalized with some cytoplasmic Drosophila GWBs (arrows). Notably, the cytoplasmic bodies containing GFP-GW and RFP-AGO2 were consistently larger than those containing only GFP-GW. (D–F) Protein fusions between RFP and the three major human GW182 family proteins transfected into Drosophila S2 cells were found in the same structures as Drosophila GW. The expression of human GW182 could not be detected without a coincident RNAi knockdown of endogenous GW. Bars, 5 μM.
Figure 6.
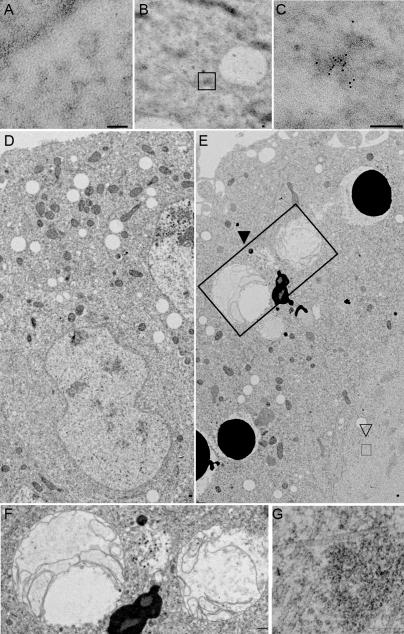
Ultrastructural analysis of Drosophila GWBs and the effect of GW loss on embryos. (A) Thin sections of embryos do not show appreciable immunogold localization when preimmune serum is used. (B and C) Sections stained with α-GW antibodies show appreciable immunogold signal in irregular, electron-dense structures. These are not membrane bound or associated with any other known cytoplasmic structure. Boxed area in B represents a single structure; a representative example is shown at higher magnification in C. (D) Thin sections of wild-type 3-h embryos show characteristic structures (including nuclei) surrounded by a distinct bilayer membrane, which is continuous with the rough endoplasmic reticulum, as well as mitochondria. (E) Homozygous gw 1 3-h mutant embryos have few recognizable nuclei and darkly staining membrane-bound vesicles, presumably corresponding to yolk particles in the embryo cortex, from which they are usually excluded at this later stage of development. Large multivesicular bodies (closed arrowhead and large box) are seen and are shown in higher magnification in F. (G) A higher magnification of the aggregates of filamentous structures indicated by the open arrowhead and small box in E. Bars, 0.2 μm.
Both human GW182 and Drosophila GW contain an RRM domain within the C-terminal of the protein (Fig. 1). Concomitant with a requirement for intact RNA for the formation of GWBs and PBs (Liu et al., 2005b; Teixeira et al., 2005), we have shown a requirement for intact RNA for the formation of Drosophila GWBs. After RNase treatment, only 15% of cells had localized GWBs compared with 97% of untreated cells (Fig. 7).
Figure 7.
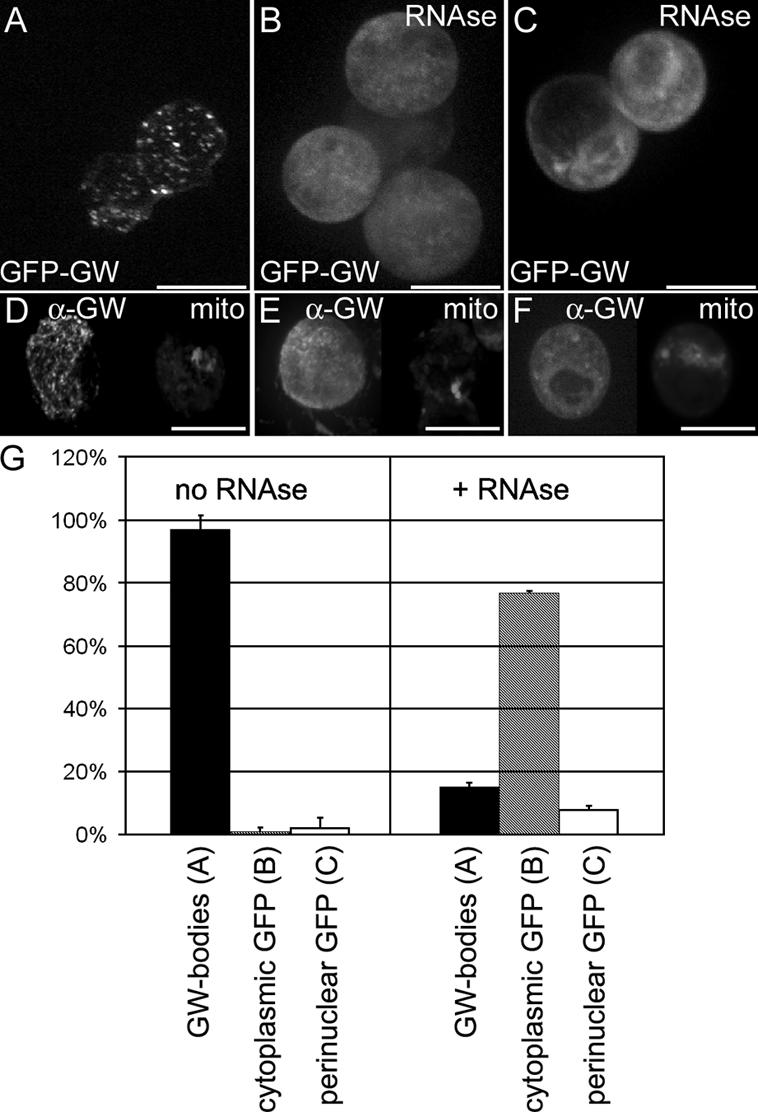
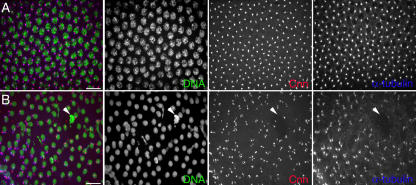
Cytoplasmic GWBs require the presence of intact RNA. (A) Drosophila GWBs were detected in S2 cells expressing a GFP-GW protein fusion. (B) 5 min after RNase treatment, punctate GWBs were no longer present, and the GFP-GW signal became diffuse throughout the cytoplasm. (C) In 10% of RNase-treated and 4% of untreated cells, an alternate perinuclear pattern of GFP-GW was seen. Each image represents a maximum projection of a three-dimensional stack of confocal images encompassing the entire cell. (D–F) Endogenous GW (α-GW) and mitochondria (Mitotracker) was also observed to ensure that the RNase treatment did not cause general organelle breakdown. (G) Quantification of the number of cells displaying each of the patterns of GFP-GW (A–C) with or without RNase treatment (n = 320). Error bars represent the SD from three separate experiments. Bars (A–C), 10 μm; (D–F) 20 μM.
Zygotic expression of full-length GW is required for early Drosophila development
Syncytial Drosophila embryos undergo 14 synchronous nuclear cycles (NCs) during early development before they cellularize (Foe and Alberts, 1983). In homozygous gw1 embryos, defects in nuclear spacing and morphology were observed beginning at approximately NC10, as they migrate to the embryo cortex. Mutant embryos had fewer cortical nuclei, and these had irregular spacing (Fig. 8). These nuclei had abnormally positioned centrosomes (Fig. 8 B), and examination of the ultrastructure of 2-h AED gw1 mutant embryos showed larger than normal nuclei and an abnormal clearing of the embryo cortex. By 3 h AED, no recognizable nuclei were found, and large multivesicular bodies and homogeneous patches devoid of organelles were seen (Fig. 6, D–G). Higher magnification of the homogenous regions showed that they were composed of filamentous elements (Fig. 6 G), which may represent large tubulin aggregates.
Figure 8.
Lack of functional GW protein leads to nuclear breakdown caused by defects in mitosis. (A) In wild-type embryos of the same age, a regular array of nuclei, each with a pair of centrosomes, can been seen immediately below the embryo cortex. (B) In homozygous gw 1 embryos 90 min AED stained with anticentrosomin (Cnn, red) antibody and antitubulin (blue), severe defects are observed after NC10. Fewer nuclei (PicoGreen) are seen, and the majority of these have improperly localized centrosomes. Large, brightly staining DNA aggregates are also seen (arrowheads). Maximum projection of 125 slices that are 10 μm deep. Bars, 5 μM.
Homozygous gw1 mutant embryos that do not express full-length GW are extremely fragile as a result of what appears to be abnormal cellularization (Fig. 3 C). Thus, we examined the localization of chromatin in live embryos expressing histone-GFP, which can be used to track chromatin dynamics after NC10 (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1; Clarkson and Saint, 1999). In homozygous gw1 embryos, fewer nuclei reached the cortex at NC10, and the majority of those that did could not successfully complete subsequent mitosis (Video 2). The remaining GFP-labeled chromatin could be seen fusing into large aggregates within the cytoplasm, which is similar to the pattern observed with DNA staining of fixed embryos (Figs. 3 C and 8 B).
Loss of functional GW can be linked to defects in chromosome separation
The rapid degradation of internal structures that occurs in homozygous gw1 embryos made linking specific effects to the loss of gw function difficult. Therefore, we interfered with GW function in a localized manner by injecting anti-GW antibody into live embryos. Loss of GW function occurs in a graded manner starting closest to the injection site. When GW antibody was injected into histone-GFP–expressing embryos, the chromosomes failed to successfully separate during mitosis similar to what is seen in gw1/gw1 embryos (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1). As the effect of the anti-GW antibody diffused anteriorly, additional nuclei were observed failing to separate with each NC. In both anti-GW–injected and gw1 mutant embryos, the chromatin was observed forming ring-shaped patterns that broke apart with time. Additionally, one to two NCs after injection, the nuclei were no longer anchored at the cortex as they moved freely within the embryonic cytoplasm (Video 3).
Live embryos expressing GFP fusions that selectively mark the spindles (tubulin), pseudocleavage furrows (actin), or nuclei (nuclear localization sequence) were treated in a similar fashion. The pseudocleavage furrows act as barriers between adjacent spindles and regress during late anaphase and telophase (Sullivan and Theurkauf, 1995). These can be monitored by following the actin network that forms apical caps over the cortical nuclei that correspondingly divides with each NC (Warn et al., 1984; Warn, 1986). As each nucleus enters prophase, the centrosomes normally migrate to opposite poles, and the apical actin caps reorganize into the pseudocleavage furrows. Subsequently, the nuclear envelope is broken down, and the spindle poles begin to separate during chromosome separation (Karr and Alberts, 1986; Sullivan and Theurkauf, 1995; Foe et al., 2000). A tubulin-GFP fusion faithfully marks the localization of the spindles during embryonic nuclear divisions (Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1). The defects in tubulin localization induced by anti-GW injection (Video 5) are similar to those detected in fixed gw mutant embryos by indirect immunofluorescence using antibodies to tubulin or centrosomin (Fig. 8). In both cases, nuclei were often observed with an abnormal number of spindles, which subsequently broke down to form large tubulin aggregates (Video 5). The dynamics of actin reorganization during the cell cycle in wild-type embryos can be seen using an actin-GFP fusion (Video 6). Blocking GW function by antibody injection at NC10 causes a stabilization of actin in the hexagonal pattern that is associated with pseudocleavage furrows beginning at the site of injection (Fig. 9, A–F; and Video 7). The stabilized actin configuration was seen even after 30 min following injection (Fig. 9 F and Video 7) but eventually breaks down into a large aggregate (Video 7).
Figure 9.
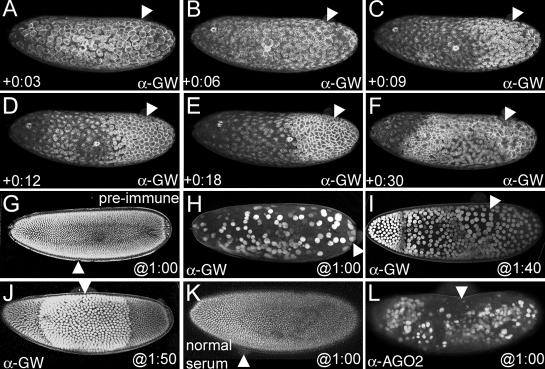
Loss of GW causes breakdown of the cortical cytoskeleton and nuclear expansion. (A) An embryo at NC10 immediately after injection with anti-GW antibody. Actin surrounds each dividing nucleus, and no obvious differences in this pattern are observed at the site of injection (arrowheads in all panels). (B) At 6 min after injection, the majority of the actin has formed apical “caps” over the interphase nuclei. However, at the anti-GW injection site, actin remains in the honeycomb pattern indicative of mitotic nuclei. (C) This stabilization of actin into the pseudocleavage furrows spreads from the site of injection. (D and E) 10–15 min after injection, the stabilized actin network elongates, and the region of stabilized actin enlarges with time. Areas more distant from the injection site (arrowheads) still form interphase caps for one more NC. (F) By 30 min after injection, the majority of the actin cytoskeleton is in a stabilized pattern, and structures nearest the injection site are beginning to break down. (G) An embryo after NC14 (2 h 10 min AED) expressing an NLS-GFP fusion that localizes to the nuclei injected with guinea pig preimmune serum at 1 h AED. No significant alterations in the morphology or spacing of the nuclei are seen, and posterior pole cells are observed. (H) Injection of anti-GW antibody at 1 h AED produces embryos with ∼200 enlarged nuclei (approximately four times normal size) at 2 h 10 min AED. These nuclei migrate to the cortex but are not anchored there and subsequently move freely within the embryo cytoplasm. (I) Anti-GW injection at 1 h 40 min AED shows a stepwise nuclear enlargement phenotype that is greatest proximal to the injection site at 2 h 10 min AED. Measurements of the nuclear diameter reveal that they are on average two (anterior), four, and eight times larger than those found in wild-type stage 14 embryos. (J) Injection at 1:50 produces a region of nuclei twice the normal size proximal to the injection site at 2 h 10 min AED. (K) Injection of normal rabbit serum into embryos 1 h AED has no effect on nuclear-GFP localization at 2 h 10 min AED. (L) Injection of polyclonal anti-AGO2 antibody into 1-h AED embryos produces a similar phenotype to the injection of anti-GW shown in H.
Injecting anti-GW or anti-AGO2 into embryos causes similar defects in nuclear division
The number and size of nuclei can be monitored in developing embryos expressing an NLS-GFP fusion (Fig. 9, G–J). The effect of the blocking of GW function on nuclear proliferation was assayed by injecting antibody at interphase of NC13 and observing the resulting effects at the time when NC14 would have occurred in wild-type embryos (130 min AED; Fig. 9 G). When anti-GW was injected at any point before NC9, significantly fewer nuclei are observed at the embryo periphery (Fig. 9 H). These nuclei were on average 8–10 times greater in diameter than stage 14 nuclei of control injected embryos (Fig. 9, G and H). When anti-GW was injected later, a graded response was observed. In embryos injected at 1 h 40 min AED, three distinct regions of enlarged nuclei were seen with a distinct boundary between nuclei that was eight and four times greater in size as well as between nuclei that was four times and twice the size farther from the site of injection (Fig. 9 I and Video 8, available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1). Embryos injected at later time points (1 h 50 min) showed nuclei twice the normal size in the area proximal to the injection point, whereas the diameter and number of nuclei in the anterior and posterior were similar to wild type. Additionally, in these embryos, the posterior pole cells developed normally (Fig. 9, G and J). This graded response to a presumptive gradient of anti-GW activity could be correlated to the number of nuclear divisions that elapsed between the time of injection and 130 min AED. A video of a live embryo expressing NLS-GFP injected with anti-GW antibody at NC10 shows that with subsequent three mitotic cycles, a corresponding increase in nuclear size could be observed beginning at the site of injection and progressing anteriorly (Video 8). Finally, because AGO2 and GW colocalize in some Drosophila GWBs (Fig. 5 C and Fig. S2), we also tested the effect of injection of anti-AGO2 antibody using a similar assay (Fig. 9, K and L). In all cases (n = 12), the injection of anti-AGO2 at 1 h AED produced an effect similar to the injection of anti-GW at the same time (Fig. 9, H and L).
Discussion
Drosophila GWBs are similar to yeast PBs and human GWBs
Our results confirm that GW is homologous to human GW182 and that Drosophila GWBs are analogous to human GWBs and yeast PBs. GW localizes to rapidly moving (Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1) and electron-dense, nonmembrane-bound cytoplasmic structures (Fig. 6, B and C). Colocalization of GW to homologues of known GWB or PB components LSm4, AGO2, and PCM (Xrn1) shows that Drosophila GWBs are of similar composition to PBs and GWBs. Another similarity between GWBs and PBs is that Drosophila GWBs also require intact RNA to maintain their integrity (Fig. 7). Functionally, human and Drosophila GW homologues are all targeted to the same foci when coexpressed in S2 cells (Fig. 5, D–F). However, not all Drosophila GWBs contain the mRNA decay enzymes LSm4 and PCM or AGO2 associated with GWBs or PBs. There is an apparent lack of interdependence in functions of the nonsense-mediated decay, RNAi, and miRNA pathways in Drosophila S2 cells, as the depletion of proteins involved in one pathway did not affect the function of another (Rehwinkel et al., 2005). Thus, the variable composition of Drosophila GWBs provides evidence that there may be distinct functions for these cytoplasmic structures. It may be possible to discern functionally distinct classes of GWBs by analyzing relative localizations of other mRNA-processing proteins as they become known.
GW is required for early Drosophila embryonic development
There have been several exhaustive screens to identify zygotically transcribed genes that affect Drosophila precellular embryonic development (Merrill et al., 1988; Wieschaus and Sweeton, 1988). Currently, a total of seven genes are thought to be expressed before the cellular blastoderm stage (Merrill et al., 1988; Wieschaus and Sweeton, 1988). However, these screens focused on the X chromosome and autosomes two and three, but not four (Merrill et al., 1988). We propose that gw represents an additional zygotically expressed gene required for successful completion of the early embryo development in Drosophila. The reduction in GW protein observed at 60–70 min AED (Fig. 4 B) suggests that maternally supplied GW is depleted. This would be subsequently replenished by zygotic gw transcription, as shown by rising mRNA levels beginning at 70–80 min AED (Fig. 4 C), a time of rapid nuclear division that culminates in the cellularization and subsequent gastrulation steps of embryo development (Foe, 1989). Notably, increased levels of Drosophila GW expression are also observed during pupal development (Fig. 4 A), which is another time of rapid cell proliferation (Milan et al., 1996). The increase in GW expression during periods of rapid cell division is consistent with elevated GW182 levels observed in proliferating human cells (Yang et al., 2004).
What is the role of GWBs in early embryonic development?
The function of GWBs described in mammalian cells suggests a potential role for these structures in Drosophila development. In many organisms, siRNA and miRNA, which are produced by Dicer-mediated cleavage of longer double-stranded or hairpin RNA precursors, regulate several developmental functions (for review see Jaronczyk et al., 2005). For both siRNA and miRNA activity, the RNA-induced silencing complex (RISC) binds and selectively suppresses or degrades complementary target mRNA (Dykxhoorn et al., 2003; Finnegan and Matzke, 2003; Bartel, 2004; Nolan and Cogoni, 2004). Several recent studies have identified a link between GWBs and the RNAi pathway. RISC components Ago1–4 localize to GWBs (Liu et al., 2005b; Sen and Blau, 2005), as do reporter mRNAs targeted for miRNA-mediated translational repression (Liu et al., 2005b). In addition, intact GWBs are required for siRNA silencing (Jakymiw et al., 2005; Liu et al., 2005b). The effects of miRNA expression on Drosophila development were characterized in a screen of 46 embryonically expressed miRNAs. Injection of antisense RNA to block these miRNAs into 30-min AED embryos revealed 25 miRNAs with visible phenotypes affecting a variety of developmental processes. Blocking miR-9 resulted in several severe defects, including nuclear division and migration, actin cytoskeleton formation, and cellularization (Leaman et al., 2005). A role for components of the RNAi machinery in the timing of heterochromatin formation and accurate chromosome separation has been reported in Schizosaccharomyces pombe (Volpe et al., 2003; Carmichael et al., 2004) and the trypanosome Trypanosoma brucei (Durand-Dubief and Bastin, 2003). Drosophila Ago2 mutants show several defects in early embryogenesis, including defects in centromeres, nuclear division, nuclear migration, and germ cell migration. However, homozygous Ago2 mutants are, for the most part, fertile and viable (Deshpande et al., 2005). Therefore, cytoplasmic-based RISC-mediated miRNA may have an effect on the control of timing of protein reorganization associated with cytoskeletal and mitotic events during early development.
The putative C. elegans GW protein orthologue Ain-l localizes to cytoplasmic foci with a composition similar to PBs and GWBs and forms complexes with ALG-1 (argonaute-like gene) Dicer-1 and miRNAs. However, C. elegans Ain-1 and RNAi components dicer-1, alg-1, and alg-2 function in the heterochronic pathway that regulates developmental timing in many postembryonic cell lineages (Grishok et al., 2001; Ding et al., 2005), while xrn1 is required in embryogenesis for ventral epithelial closure (Newbury and Woollard, 2004).
The phenotypes associated with blocking Drosophila GW function suggest that functional GWBs are required for the completion of nuclear divisions during early embryonic development. These effects, although similar to Drosophila Ago2 mutants, are far more severe. Injection of anti-AGO2 antibody into early embryos caused a reduction in number and enlargement in the size of the embryonic nuclei detected by NLS-GFP (Fig. 9 L). The more severe defects resulting from GW depletion may be caused by the nature of the Ago2 mutation, which does not completely block protein function (Deshpande et al., 2005), or may be the consequence of additional functions of Drosophila GWBs (which are not related to AGO2) and, by extension, RISC function.
Drosophila GWBs may coordinate developmental posttranscriptional mRNA regulation
Drosophila GW is expressed throughout development and is required for the viability of cultured Drosophila cells (Boutros et al., 2004). Our data suggest that one function of GWBs is to coordinate the regulation of embryonic development in a posttranscriptional fashion. Subsets of eukaryotic mRNAs involved in the same cellular processes are often associated with specific RNA-binding proteins, depending on growth conditions (Keene and Tenenbaum, 2002; Nakahara et al., 2005). In one proposed model, RNP particles like GWBs coordinately regulate mRNAs encoding functionally related proteins, which is analogous to the operon-based coordination of prokaryotic gene expression (Keene and Lager, 2005). Thus, mRNAs with similar cis-elements would be recognized and trafficked by a common RNP to collectively regulate their translation or degradation (Takizawa et al., 2000; Tenenbaum et al., 2000, 2002; Keene, 2001; Keene and Tenenbaum, 2002; Penalva et al., 2004). Our data provide evidence that Drosophila GWBs mirror human GWB composition and function, providing an excellent model for genetic dissection of the potential role of GWBs in regulating mRNAs during development.
Materials and methods
Expression of fluorescent fusions in S2 cells
The gw open reading frame and 3′ untranslated region were amplified from cDNA LD47780 with primers 5′gw, CGCAGACGTCTTATGCGTGAAGCCC and 3′gw, TGCGGACGTCGACATATACATACATATGTATG and were cloned into pZero Blunt (Invitrogen) to make pZBgw. A GFP-GW fusion was expressed in S2 cells by recloning gw from pZBgw into the AatII site of pP(GS[hsEGFP3′]) (Schotta and Reuter, 2000) to make pPGFPgw. Approximately 106 cells were transfected with 1.6 μg pPGFPgw and 0.1 μg pCoHygro using 7 μl Cellfectin (Invitrogen), and stably transformed cells were selected using 300 μg/ml hygromycin. The pcm open reading frame was amplified from the LD22664 cDNA with 5′PCM, CACCATGGGCGTTCCCAAGTTCTTTC and 3′PCM, AGTTGGATGCGGGGAGTCGGG primers and cloned into pENTR/D (Invitrogen) to make pENTRpcm. It was then recombined into pAWR (provided by T. Murphy, Carnegie Institute, Troy, MI) to create a C-terminal RFP fusion under control of the actin5C promoter. The Drosophila LSm4 homologue (CG33677) was amplified from the RE35747 cDNA with 5′LSM, CACCATGCTGCCACTTTC and 3′LSM, CGATCCGAAGAACTATTTCCTATT primers, cloned into pENTR/D, and recombined into pAWR as described above. cDNAs of human GW182 and GW182-related proteins, which were provided by E. Chan (University of Floirda, Gainesville, FL), were also recombined into pAWR as described above. The AGO2 open reading frame was amplified from the REO4347 cDNA (Hammond et al., 2001) using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGGAAAAAAAGATAAGAACA-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGACAAAGTACATGGGGTT-3′, recombined into pDONR221 (Invitrogen), and recombined into pAWR. Double-stranded gw RNA was made using primers specific to the 3′ untranslated region of gw: 5′gw, RNAi TAATACGACTCACTATAGGGAAGATCAATTACCAGTTCCA and 3′gw, RNAi TAATACGACTCACTATAGGGACATATACATACATATGTATG, allowing direct synthesis of double-stranded RNA from the PCR product using the Megascript in vitro transcription system (Ambion).
Drosophila stocks
The gw1 mutant was identified during an ethylmethylsulfonate mutagenesis screen for recessive lethal loci located on chromosome four. This mutation was mapped to the 102C region, and only two nucleotide changes were identified: causing W967stop in gw1 and N144I in the N-terminal region of CG1838 (myoglianin). However, CG1838 contains a conserved proteolytic cleavage site, which would remove N144 from the mature protein (Lo and Frasch, 1999). The HS-GFP-GW strain was generated by transferring pPGFPGW into the pP(GS[w +, hsEGFP3′]) vector (Schotta and Reuter, 2000) and germline transformation of y1w1118;ry506 Sb1 P(ry t7.2 = Δ2−3) 99B/TM6 embryos (Spradling and Rubin, 1982). The histone-GFP;gw1/ciD strain carries the histone2AvD-GFP fusion (Clarkson and Saint, 1999). All other fly strains were obtained from the Bloomington Drosophila Stock Centre.
Production of an anti-GW antibody and immunolocalization
The 5′ XhoI fragment of pZBgw encoding the first 1,061 amino acids of GW was subcloned into pRSETA (Invitrogen), and recombinant protein was purified on Ni nitrilotriacetic acid agarose (QIAGEN), repurified by SDS-PAGE, electroeluted from polyacrylamide (Waterborg and Matthews, 1994), and injected into Hartley guinea pigs (Charles River Laboratories). Western blot analysis confirmed reactivity with the initial 100-kD recombinant protein, the endogenous 160-kD GW protein in embryos and S2 cells, as well as a 200-kD GFP-GW fusion. GW antibody was affinity purified using 100 μg of fusion protein bound to a 1-ml HiTrap N-hydroxysuccinimide–activated high performance column (GE Healthcare) and eluted using Immunopure gentle elution buffer (Pierce Chemical Co.). The eluted antibody was concentrated to 15 μg/μl using an ultrafiltration unit (centricon-10; Millipore) in a Tris, pH 8.0, and 50% glycerol solution. Anti-GW serum recognized cytoplasmic foci colocalizing with GFP-GW in stably transformed S2 cells fixed with 2% PFA (Fehon et al., 1990), whereas no specific signal was seen with the preimmune serum. Drosophila embryos were fixed as described previously (Rothwell and Sullivan, 2000), rehydrated in 1× PBS, and treated for 30 min with 10 μg/ml DNase-free RNase (Sigma Aldrich). The following primary antibodies were used: mouse anti–α-tubulin (1:100; Sigma-Aldrich), anti-actin (1:100; Sigma- Aldrich), rabbit anticentrosomin (1:100; a gift from T. Kaufman, Indiana University, Bloomington, IN), and antiphosphotyrosine (1:1,000; Cell Signaling). All secondary antibodies were AlexaFluor-conjugated 488, 546, or 647 (Invitrogen) used at 1:2,000. DNA was stained using PicoGreen (1:1,000; Invitrogen). All imaging was performed at 25°C. Confocal images were obtained using a spinning disk confocal system (Ultraview ERS; PerkinElmer) mated with a camera (Orca AG; Hamamatsu) and a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) with a 63× NA 1.4 plan-Apochromat lens.
Western blot analysis
Extracts were prepared in 2.5× SDS gel sample buffer (157 mM Tris-HCL, 0.025% bromophenol blue, 5% SDS, 25% glycerol, and 50 mM DTT), immediately heated to 98°C, and centrifuged for 5 min at 12,000 g. Approximately 200 μg of protein per 1 μl of sample buffer (embryos, larvae, and pupae) or one adult per 8 μl SDS sample buffer was loaded in each lane (Laemmli, 1970). Protein loading was standardized using E7 anti–β-tubulin monoclonal antibody (Developmental Studies Hybridoma Bank). Early developmental extracts contained five visually staged embryos in 25 μl of gel sample buffer for each time point, and the equivalent protein from one embryo was loaded per lane. Proteins were fractionated on 6% polyacrylamide gels, transferred to nitrocellulose, and incubated with anti-GW serum (1:1,000) and 1 μg/ml E7 anti–β-tubulin monoclonal antibody. This was followed by HRP-conjugated anti–guinea pig or anti–mouse secondary antibodies (1:50,000; Jackson ImmunoResearch Laboratories) and detected using Super Signal West Pico Chemiluminescent Substrate (Pierce Chemical Co.).
Northern blot analysis
Equal amounts of total RNA extracted from staged embryo TRIzol (Invitrogen) were separated on a 1.2% agarose gel (0.67% formaldehyde) and transferred to BrightStar-Plus Membrane (Ambion) using 10× SSC and 120 mJ UV cross-linked for 45 s. Blots were hybridized to digoxygenin- labeled antisense (1:5,000) gw RNAs that were in vitro transcribed using T7 RNA polymerase (New England Biolabs, Inc.) from the LD47780 cDNA-cut EagI and RpL32 (loading control) RNA probes T3 transcribed from RH03940 cut with EcoRI overnight at 68°C in 3 M urea, 5× SSC, 0.1% (wt/vol) N-lauroylsarcosine, 0.02% (wt/vol) SDS, 0.5% milk powder, and 0.2 mg/ml sonicated salmon sperm DNA. The membrane was then washed for 15 min with 0.1× SSC and 0.1% SDS, washed for 15 min with 2× SSC and 0.1% SDS, blocked for 30 min (0.1 M maleic acid, 0.15 M NaCl, 1% acelyated BSA, and 0.1% Tween 20, pH 7.5), and incubated with sheep antidigoxygenin-HRP (1:10,000) for 1 min (Roche). This was followed by two 15-min washes with blocking buffer and detection using North2South chemiluminescent substrate (Pierce Chemical Co.).
Live imaging of S2 cells and embryos
S2 cells were imaged in cell media (Perbio) in coverglass chambers (Lab-Tek). Visually staged embryos were prepared under Halocarbon 700 oil (Sigma-Aldrich) on coverslips as described previously (Johansen and Johansen, 2000), injected with 0.25 ng affinity-purified anti-GW antibody, guinea pig preimmune serum, or affinity-purified rabbit anti-Ago2 (ab5072; Abcam), and diluted in 1× PBS. Approximately 100–150 pl of antibody solution was injected, determined by estimation of the size of the liquid drops (Kennerdell and Carthew, 1998). RNase treatment of the cells expressing GFP-GW was performed as described previously (Sen and Blau, 2005). Mitochondria were stained using 100 nm Mitotracker red CMXRos (Invitrogen). All imaging was performed at 25°C. Time-lapse confocal images were obtained using a spinning disk confocal system (Ultraview ERS; PerkinElmer) mated to a camera (Orca AG; Hamamatsu) and a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) with a 20× NA 0.75 plan-Apochromat lens. 30–40 optical sections at a resolution of 672 × 512 with 2 × 2 binning were collected every 10 s. A maximum projection of each time point was generated, and uncompressed AVI videos were exported using Ultraview software (PerkinElmer). Each video was converted to QuickTime format using QuickTime Pro software (Apple). Still images of GFP-expressing cells and embryos were obtained using a confocal microscope (LSM 510; Carl Zeiss MicroImaging, Inc.) and software using a 63× NA 1.4 plan-Apochromat and 20× NA 0.75 plan-Apochromat lenses, respectively.
EM
Drosophila embryos were fixed 8–12 h AED using high pressure freezing (McDonald and Morphew, 1993) and embedded in LR white resin (London Resin Company). 70-nm thin sections were contrast stained with uranyl acetate and incubated with 1:25 anti-GW antibody or 1:25 preimmune serum followed by donkey anti–guinea pig IgG conjugated to 6 nM gold (1:25; Jackson ImmunoResearch Laboratories). Embryos were collected, aged for 1–3 h, dechorionated in 50% bleach, and fixed in heptane (Sigma-Aldrich) saturated with 25% glutaraldehyde (Sigma-Aldrich) in 50 mM sodium cocadylate, pH 7.0, for 20 min at 25°C. Mutant embryos were selected via direct phenotypic observation of nuclear morphology after staining with PicoGreen (Invitrogen), hand devitinellized under heptane, postfixed in 1% osmium tetroxide (EM Sciences), and embedded in Epon resin (McDonald et al., 2000). Thin sections were stained with lead citrate and uranyl acetate before sectioning and were imaged using a transmission electron microscope (TEM2000; Philips), digital camera (MegaView III; Soft Imaging System), and analySIS software (Soft Imaging System).
Genotype verification of single embryo
The gw1/gw1 genotype was confirmed by genomic PCR with the following primers: 5′outside (intron 6), TGTAACAGGCAGAAGGAAGCGTTTCCGACCAT and 3′outside (exon 9), GGCAGTCAATCCTGGCGGGGGACCTCGAGACG followed by a second nested PCR reaction with 5′inside (intron 6), CCATCTGTCCGTATGAACTTCGAG and 3′inside (exon 9), TCCGAAGTCGCGGTACATTGTTGA using 50 μl PCR Supermix (Invitrogen). The stop mutation (TGG to TGA) in gw1 disrupts an NcoI recognition sequence and was initially identified by digesting purified PCR products (Qiaquick; QIAGEN) with NcoI. Mutants were verified by DNA sequencing.
Immunoprecipitation of Drosophila GW-associated proteins
Approximately 107 S2 cells were transfected with 10 μg of the plasmid HSFLAG-Ago2. 48 h after transfection, cells were heat shocked for 40 min at 37°C and allowed to recover for 40 min at 25°C. Cells were lysed in 2 ml radioimmunoprecipitation buffer (1% sodium deoxycholate, 1% NP-40, 0.2% SDS, 150 mM NaCl, 50 mM Tris, pH 7.4, complete EDTA-free protease inhibitors [Roche], and 1 mM PMSF). The extract was incubated with 6 μl anti-GW antibody for 30 min and incubated for 2 h in the presence of 40 μl protein A–Sepharose beads (GE Healthcare) at 4°C. After washing, bound proteins were eluted with 2× SDS gel sample buffer, fractionated on a 6% low bisacrylamide (118:1) polyacrylamide gel, and transferred to nitrocellulose. Flag-AGO2 was detected with mouse anti-Flag M2 antibody (1:100; Sigma-Aldrich).
Online supplemental material
Video 1 shows chromatin organization during early development of a wild-type Drosophila embryo injected with guinea pig preimmune serum. Video 2 shows an abnormal pattern of chromosome division in a homozygous gw1 mutant expressing histone-GFP. Video 3 shows histone-GFP–expressing embryos after the localized depletion of GW function by antibody injection at the anterior pole during interphase of NC10. Video 4 shows localization of the spindles during early development in living Drosophila embryos. Video 5 shows anti-GW antibody injection into the posterior pole of tubulin-GFP–expressing embryos at NC10. Video 6 shows the dynamic pattern of actin localization monitored using the actin-binding domain of moesin-GFP expressed in live embryos. Video 7 shows that blocking GW function by anti-GW antibody injection at NC10 into moesin-GFP–expressing embryos leads to stabilization and then breakdown of the cortical actin network. Video 8 shows that injection of anti-GW antibody into embryos expressing NLS-GFP at NC10 causes progressive nuclear enlargement of the posterior pole and regional nuclear enlargement with each subsequent NC. Video 9 presents the visualization of Drosophila GWBs in living S2 cells. Fig. S1 shows GFP-GW and GFP expression in S2 cells. Fig. S2 shows Flag-AGO2 colocalized and associated with endogenous Drosophila GW in S2 cells, and Fig. S3 shows RNAi knockdown of gw mRNA phenocopies of the gw1 mutation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200512103/DC1.
Supplementary Material
Acknowledgments
We wish to acknowledge the technical assistance of Michelle Hamm, Janine Stienky, Honey Chan, Eden Foley, Wayne Rickoll, and Ed Chan and useful suggestions from Hua Deng, Marvin Fritzler, Shelagh Campbell, Ellen Homola, and Gary Eitzen.
M.D. Schneider was funded by a studentship, S. Chaker was supported by a Heritage Youth Research Scholarship, T.C. Hobman was funded by a Scientist award, and A.J. Simmonds was supported by a Scholar award from the Alberta Heritage Foundation for Medical Research. A.J. Simmonds also received a New Investigator award from the Canadian Institutes for Health Research (CIHR). The laboratory of A.J. Simmonds was supported by an operating grant and Rx&D Health Research Foundation special research allowance from the CIHR. The laboratories of T.C. Hobman and J. Locke were also supported by CIHR.
S.C. Hughes' present address is Department of Cell Biology, Faculty of Medicine and Dentistry, University of Alberta, Edmonton, Alberta T6G H7, Canada.
Abbreviations used in this paper: AED, after egg deposition; GW, Gawky; GWB, GW body; miRNA, microRNA; NC, nuclear cycle; PB, processing body; PCM, Pacman; RISC, RNA-induced silencing complex; RRM, RNA recognition motif; SG, stress granule.
References
- Adams, M.D., S.E. Celniker, R.A. Holt, C.A. Evans, J.D. Gocayne, P.G. Amanatides, S.E. Scherer, P.W. Li, R.A. Hoskins, R.F. Galle, et al. 2000. The genome sequence of Drosophila melanogaster. Science. 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- Andrei, M.A., D. Ingelfinger, R. Heintzmann, T. Achsel, R. Rivera-Pomar, and R. Luhrmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 11:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. [DOI] [PubMed] [Google Scholar]
- Boutros, M., A.A. Kiger, S. Armknecht, K. Kerr, M. Hild, B. Koch, S.A. Haas, H.F. Consortium, R. Paro, and N. Perrimon. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 303:832–835. [DOI] [PubMed] [Google Scholar]
- Carmichael, J.B., P. Provost, K. Ekwall, and T.C. Hobman. 2004. ago1 and dcr1, two core components of the RNA interference pathway, functionally diverge from rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol. Biol. Cell. 15:1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson, M., and R. Saint. 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol. 18:457–462. [DOI] [PubMed] [Google Scholar]
- Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun, and P. Schedl. 2005. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 19:1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., A. Spencer, K. Morita, and M. Han. 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 19:437–447. [DOI] [PubMed] [Google Scholar]
- Durand-Dubief, M., and P. Bastin. 2003. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 1:2. [DOI] [PMC free article] [PubMed]
- Dykxhoorn, D.M., C.D. Novina, and P.A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 4:457–467. [DOI] [PubMed] [Google Scholar]
- Eystathioy, T., E.K. Chan, S.A. Tenenbaum, J.D. Keene, K. Griffith, and M.J. Fritzler. 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 13:1338–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy, T., A. Jakymiw, E.K. Chan, B. Seraphin, N. Cougot, and M.J. Fritzler. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 9:1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon, R.G., P.J. Kooh, I. Rebay, C.L. Regan, T. Xu, M.A. Muskavitch, and S. Artavanis-Tsakonas. 1990. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 61:523–534. [DOI] [PubMed] [Google Scholar]
- Finnegan, E.J., and M.A. Matzke. 2003. The small RNA world. J. Cell Sci. 116:4689–4693. [DOI] [PubMed] [Google Scholar]
- Foe, V.E. 1989. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 107:1–22. [PubMed] [Google Scholar]
- Foe, V.E., and B.M. Alberts. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61:31–70. [DOI] [PubMed] [Google Scholar]
- Foe, V.E., C.M. Field, and G.M. Odell. 2000. Microtubules and mitotic cycle phase modulate spatiotemporal distributions of F-actin and myosin II in Drosophila syncytial blastoderm embryos. Development. 127:1767–1787. [DOI] [PubMed] [Google Scholar]
- Grishok, A., A.E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D.L. Baillie, A. Fire, G. Ruvkun, and C.C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 106:23–34. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., S. Boettcher, A.A. Caudy, R. Kobayashi, and G.J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 293:1146–1150. [DOI] [PubMed] [Google Scholar]
- Ingelfinger, D., D.J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jakymiw, A., S. Lian, T. Eystathioy, S. Li, M. Satoh, J.C. Hamel, M.J. Fritzler, and E.K. Chan. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 7:1267–1274. [DOI] [PubMed] [Google Scholar]
- Jaronczyk, K., J.B. Carmichael, and T.C. Hobman. 2005. Exploring the functions of RNA interference pathway proteins: some functions are more RISCy than others? Biochem. J. 387:561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, K., and J. Johansen. 2000. Studying nuclear organization in embryos using antibody tools. In Methods in Molecular Biology: Drosophila Cytogenetics Protocols. Vol. 247. D.S. Henderson, editor. Humana Press, Totowa, NJ. 215–234. [DOI] [PubMed]
- Karr, T.L., and B.M. Alberts. 1986. Organization of the cytoskeleton in early Drosophila embryos. J. Cell Biol. 102:1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M.J. Fitzler, D. Scheuner, R.J. Kaufman, D.E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA. 98:7018–7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D., and S.A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 9:1161–1167. [DOI] [PubMed] [Google Scholar]
- Keene, J.D., and P.J. Lager. 2005. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 13:327–337. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R., and R.W. Carthew. 1998. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 95:1017–1026. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Leaman, D., P.Y. Chen, J. Fak, A. Yalcin, M. Pearce, U. Unnerstall, D.S. Marks, C. Sander, T. Tuschl, and U. Gaul. 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 121:1097–1108. [DOI] [PubMed] [Google Scholar]
- Liu, J., F.V. Rivas, J. Wohlschlegel, J.R. Yates 3rd, R. Parker, and G.J. Hannon. 2005. a. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., M.A. Valencia-Sanchez, G.J. Hannon, and R. Parker. 2005. b. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, P.C., and M. Frasch. 1999. Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-beta superfamily. Mech. Dev. 86:171–175. [DOI] [PubMed] [Google Scholar]
- Maris, C., C. Dominguez, and F.H. Allain. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- McDonald, K., and M.K. Morphew. 1993. Improved preservation of ultrastructure in difficult-to-fix organisms by high pressure freezing and freeze substitution: I. Drosophila melanogaster and Strongylocentrotus purpuratus embryos. Microsc. Res. Tech. 24:465–473. [DOI] [PubMed] [Google Scholar]
- McDonald, K.L., D.J. Sharp, and W. Rickoll. 2000. Preparation of thin sections of Drosophila for examination by transmission electron microscopy. In Drosophila Protocols. W. Sullivan, M. Ashburner, and R.S. Hawley, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 245–272.
- Meister, G., M. Landthaler, L. Peters, P.Y. Chen, H. Urlaub, R. Luhrmann, and T. Tuschl. 2005. Identification of novel argonaute-associated proteins. Curr. Biol. 15:2149–2155. [DOI] [PubMed] [Google Scholar]
- Merrill, P.T., D. Sweeton, and E. Wieschaus. 1988. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 104:495–509. [DOI] [PubMed] [Google Scholar]
- Milan, M., S. Campuzano, and A. Garcia-Bellido. 1996. Cell cycling and patterned cell proliferation in the Drosophila wing during metamorphosis. Proc. Natl. Acad. Sci. USA. 93:11687–11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara, K., K. Kim, C. Sciulli, S.R. Dowd, J.S. Minden, and R.W. Carthew. 2005. Targets of microRNA regulation in the Drosophila oocyte proteome. Proc. Natl. Acad. Sci. USA. 102:12023–12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury, S., and A. Woollard. 2004. The 5′-3′ exoribonuclease xrn-1 is essential for ventral epithelial enclosure during C. elegans embryogenesis. RNA. 10:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, T., and C. Cogoni. 2004. The long hand of the small RNAs reaches into several levels of gene regulation. Biochem. Cell Biol. 82:472–481. [DOI] [PubMed] [Google Scholar]
- Penalva, L.O., S.A. Tenenbaum, and J.D. Keene. 2004. Gene expression analysis of messenger RNP complexes. Methods Mol. Biol. 257:125–134. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J., I. Behm-Ansmant, D. Gatfield, and E. Izaurralde. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 11:1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell, W.F., and W. Sullivan. 2000. Fluorescent analysis of Drosophila embryos. In Drosophila Protocols. W. Sullivan, M. Ashburner, and R.S. Hawley, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 141–157.
- Schotta, G., and G. Reuter. 2000. Controlled expression of tagged proteins in Drosophila using a new modular P-element vector system. Mol. Gen. Genet. 262:916–920. [DOI] [PubMed] [Google Scholar]
- Sen, G.L., and H.M. Blau. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633–636. [DOI] [PubMed] [Google Scholar]
- Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A.C., and G.M. Rubin. 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 218:341–347. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., and W.E. Theurkauf. 1995. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 7:18–22. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., J.L. DeRisi, J.E. Wilhelm, and R.D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 290:341–344. [DOI] [PubMed] [Google Scholar]
- Teixeira, D., U. Sheth, M.A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum, S.A., C.C. Carson, P.J. Lager, and J.D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA. 97:14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum, S.A., P.J. Lager, C.C. Carson, and J.D. Keene. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 26:191–198. [DOI] [PubMed] [Google Scholar]
- Volpe, T., V. Schramke, G.L. Hamilton, S.A. White, G. Teng, R.A. Martienssen, and R.C. Allshire. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11:137–146. [DOI] [PubMed] [Google Scholar]
- Warn, R.M. 1986. The cytoskeleton of the early Drosophila embryo. J. Cell Sci. Suppl. 5:311–328. [DOI] [PubMed] [Google Scholar]
- Warn, R.M., R. Magrath, and S. Webb. 1984. Distribution of F-actin during cleavage of the Drosophila syncytial blastoderm. J. Cell Biol. 98:156–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg, J.H., and H.R. Matthews. 1994. The electrophoretic elution of proteins from polyacrylamide gels. Methods Mol. Biol. 32:169–175. [DOI] [PubMed] [Google Scholar]
- Wieschaus, E., and D. Sweeton. 1988. Requirements for X-linked zygotic gene activity during cellularization of early Drosophila embryos. Development. 104:483–493. [DOI] [PubMed] [Google Scholar]
- Yang, Z., A. Jakymiw, M.R. Wood, T. Eystathioy, R.L. Rubin, M.J. Fritzler, and E.K. Chan. 2004. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 117:5567–5578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.