Abstract
Centrosomes consist of two centrioles surrounded by an amorphous pericentriolar matrix (PCM), but it is unknown how centrioles and PCM are connected. We show that the centrioles in Drosophila embryos that lack the centrosomal protein Centrosomin (Cnn) can recruit PCM components but cannot maintain a proper attachment to the PCM. As a result, the centrioles “rocket” around in the embryo and often lose their connection to the nucleus in interphase and to the spindle poles in mitosis. This leads to severe mitotic defects in embryos and to errors in centriole segregation in somatic cells. The Cnn-related protein CDK5RAP2 is linked to microcephaly in humans, but cnn mutant brains are of normal size, and we observe only subtle defects in the asymmetric divisions of mutant neuroblasts. We conclude that Cnn maintains the proper connection between the centrioles and the PCM; this connection is required for accurate centriole segregation in somatic cells but is not essential for the asymmetric division of neuroblasts.
Introduction
Centrosomes comprise a pair of centrioles and a surrounding pericentriolar matrix (PCM). They are the major microtubule (MT) organizing centers (MTOCs) in animal cells and are thought to play an important part in organizing many cell processes, including cell polarity, cell migration, and cell division (Doxsey et al., 2005). Centrioles are also required for the formation of cilia and flagella (Basto et al., 2006), and they have important roles in many developmental processes (Davis et al., 2006). Not surprisingly, centriole or centrosome dysfunction has been implicated in a wide variety of human genetic diseases (Badano et al., 2005).
Although much is known about the protein composition of the centrosome, it remains unclear how centrosome structure and organization are maintained. In flies, the centrosomal protein Centrosomin (Cnn) is required to recruit several proteins to the centrosome (Megraw et al., 1999, 2001; Vaizel-Ohayon and Schejter, 1999; Terada et al., 2003). In cnn mutant embryos, and in somatic cells lacking Cnn, the centrosomes fail to function as MTOCs during mitosis and anastral spindles assemble through a centrosome-independent pathway. This leads to dramatic mitotic defects in embryos (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999) but only to subtle mitotic defects in somatic cells (Megraw et al., 2001; Mahoney et al., 2006), presumably because centrosomes are not essential for cell division in somatic cells (Bettencourt-Dias et al., 2005; Basto et al., 2006).
Cnn is a member of a family of structurally related proteins that have been implicated in organizing MT arrays. In the yeast S. pombe, the Cnn-related protein Mto1 recruits the γ-tubulin complex to several types of MTOCs (Sawin et al., 2004; Venkatram et al., 2004). In human cells, the Cnn-related proteins CDK5RAP2 and Myomegalin/PDE4-DIP are concentrated at centrosomes, but their function is unknown (Verde et al., 2001; Bond et al., 2005). Mutations in the gene encoding CDK5RAP2, however, cause autosomal recessive primary microcephaly, in which the brain is small at birth and thereafter (Bond et al., 2005). The underlying cause of microcephaly is unknown, but it has been proposed that a failure of the centrosomes to function as efficient MTOCs in mitosis might lead to defects in asymmetric neuroblast (NB) divisions during fetal development (Basto et al., 2006; Bond and Woods, 2006; Fish et al., 2006). Here, we have used live confocal imaging to examine how Cnn functions to ensure the proper organization of the centrosome in flies, and to test whether Cnn is required for asymmetric divisions in larval NBs.
Results and discussion
Centrioles recruit PCM components and MTs but cannot maintain their connection to them in the absence of Cnn
In fixed embryos and somatic cells that lack Cnn, PCM components are barely detectable at the poles of the mitotic spindles (Megraw et al., 1999, 2001; Vaizel-Ohayon and Schejter, 1999). Centrioles are still present in cnn mutant cells (Megraw et al., 2001), but their function and positioning within the centrosome have not been analyzed. To understand better how Cnn normally recruits PCM components to the centrioles, we generated transgenic Drosophila lines expressing an mRFP-centriolar marker (either mRFP-Fzr or mRFP-PACT), together with one of three PCM markers fused to GFP: Aurora A–GFP, Grip75-GFP (a component of the γ-tubulin ring complex), and GFP- D-TACC. It has previously been shown that Cnn can interact with both the γ-tubulin ring complex and Aurora A (Terada et al., 2003), but we found no evidence for an interaction between Cnn and D-TACC in coimmunoprecipitation experiments (unpublished data).
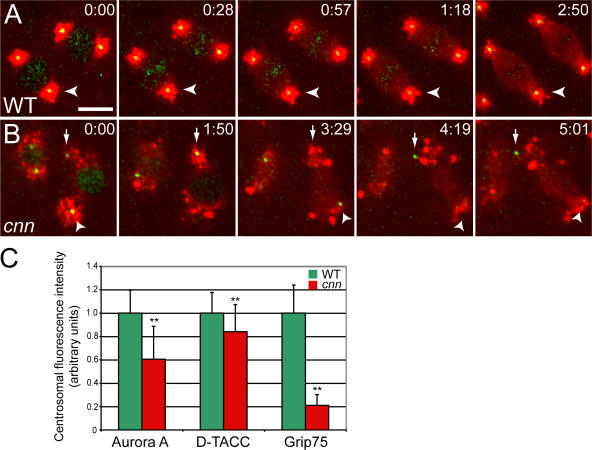
In wild-type (WT) syncytial embryos, centrioles recruited approximately equal amounts of PCM at all stages of the rapid mitotic cycles, and they remained well centered within the PCM throughout the cell cycle (Fig. 1; Fig. S1; and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). During interphase, the centrioles were always closely associated with the nuclear envelope, whereas in mitosis, they were always closely associated with the spindle poles (Videos 1 and 2). In embryos laid by cnn homozygous females (hereafter, cnn embryos), we were surprised to observe that the centrioles were associated with appreciable amounts of PCM, but they were often not properly centered within it (Fig. 1; Fig. S1; and Videos 1 and 2). In video recordings of cnn embryos, the centrioles appeared to be constantly nucleating PCM but seemed unable to maintain their connection to it. The centrioles often exhibited irregular, stochastic movements, leaving a trail of PCM behind them as they moved away. This PCM trail was most easily seen in cnn embryos expressing GFP–D-TACC (Video 1), as this protein was recruited in particularly large amounts to the centrioles, and large clusters of GFP–D-TACC often remained in the cytoplasm for some time after the centrioles had moved away. Smaller amounts of Aurora A–GFP and Grip75-GFP were recruited to the centrioles (Video 2), and so only small amounts of these proteins remained associated with the centrioles as they moved around the embryo. As a result of this abnormal centriole behavior, the centrioles in cnn embryos often lost their attachment to the nuclear envelope in interphase and to the spindle poles in mitosis. We refer to this behavior of the centrioles as “centriole rocketing” (see the following section).
Figure 1.
The centrioles in cnn embryos cannot maintain a proper connection to the PCM. (A and B) Still images from videos of WT and cnn syncytial embryos expressing the PCM marker GFP–D-TACC (pseudocolored red), and the centriole marker mRFP-Fzr (pseudocolored green; note that mRFP-Fzr is also concentrated in the nucleus in interphase). Time in min:s. (A) In WT embryos, the centrioles are always well centered within the PCM. (B) In cnn embryos, the centrioles are associated with PCM, but they “rocket” around within the cytoplasm and no longer maintain their proper connection to the PCM (see Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). (C) Quantification of the amount of PCM recruited to centrioles in WT and cnn embryos. Error bars represent SD (n = 45 for each marker and genotype). **, P < 0.001 (t test). Bar, 10 μm.
Previous studies suggested that centrosomes lacking Cnn fail to function as MTOCs during mitosis (Megraw et al., 1999, 2001; Vaizel-Ohayon and Schejter, 1999; Mahoney et al., 2006). We therefore examined whether the PCM organized by the centrioles in cnn embryos was capable of nucleating MTs. As shown in Fig. 2 (A–D) and Videos 3 and 4 (available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1), the centrosomes in cnn embryos organized astral MT arrays but seemed unable to maintain their connection with them. When the embryos entered mitosis, many nuclei were not associated with centrioles, and anastral spindles assembled around the mitotic chromatin (not depicted). Many nuclei, however, were close enough to a centriole for the astral MTs to contribute to spindle assembly (Fig. 2 D, arrow). Often, however, these centrioles failed to maintain their position at the spindle pole and either wandered around within the spindle (Fig. 2 D, red arrowhead) or lost their connection to the spindle altogether (Fig. 2 D, yellow arrowhead). We conclude that the dramatic mitotic defects observed in cnn embryos do not result from a failure of the centrioles to recruit PCM, or of the centrosomes to nucleate astral MTs, but instead result from the failure of the centrioles to maintain a stable connection to the PCM and MTs that they organize.
Figure 2.
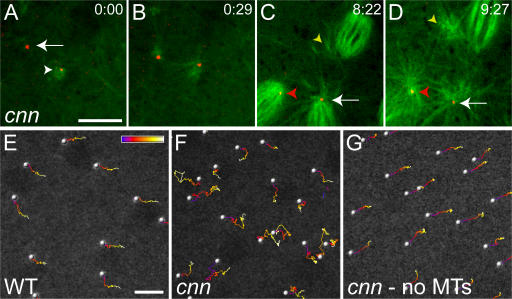
The centrioles associate with astral MTs in cnn embryos, and centriole rocketing is MT dependent. (A–D) The centrioles in a cnn embryo were visualized with mRFP-PACT (red) and the MTs with GFP–α-tubulin (green). (A) At the start of this time series (time in min:s), one centriole is associated with MTs (arrowhead), whereas another is not (arrow). The latter centriole starts to nucleate MTs from one side (B), and both centrioles exhibit “rocketing” movements as they recruit PCM and nucleate MTs asymmetrically around themselves (see Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). As this embryo enters mitosis (C), the centrioles continue to rocket around in the cytoplasm (Video 4). In this panel, one centriole is located at the pole of a mitotic spindle (arrow), and this pole is associated with astral MTs. Another centriole loses its connection to the spindle pole (red arrowhead) and starts to move around within the spindle; another centriole (that is not in focus, but whose position can be inferred from its associated astral MTs; yellow arrowhead) starts to migrate away from the spindle into the cytoplasm. By the end of mitosis (D), the latter centriole has completely lost contact with the spindle (yellow arrowhead); the other centriole has migrated back to the pole (red arrowhead). (E–G) Traces of centriole movement over time are shown for a WT embryo (E), a cnn embryo (F), and the same cnn embryo after colchicine injection (G). The position of each centriole over time is indicated by the color scale, which represents 150 s. See Video 5. Bars, 10 μm.
Centriole rocketing in cnn embryos is MT dependent
The centriole rocketing appeared to be driven by the asymmetric organization of the PCM and MTs around the centrioles (Videos 1–3). To test whether the rocketing was MT dependent, we injected the MT-depolymerizing drug colchicine into embryos. In WT embryos in late interphase, the centrioles had already migrated around the nuclei and, when we plotted their movement over time, the centrioles moved regularly across the embryo cortex (Fig. 2 E and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). This regular movement of the centrosomes and their associated nuclei across the cortex is driven by actin- and myosin-dependent cortical contractions, and it continued after colchicine injection (Video 5). In contrast, when we plotted centriole movement in cnn embryos in late interphase, we observed the rocketing behavior described previously (Fig. 2 F and Video 5). The rocketing ceased after colchicine injection, and the centrioles reverted to a regular movement across the embryo cortex (Fig. 2 G and Video 5). Thus, centriole rocketing in cnn embryos depends on intact MTs.
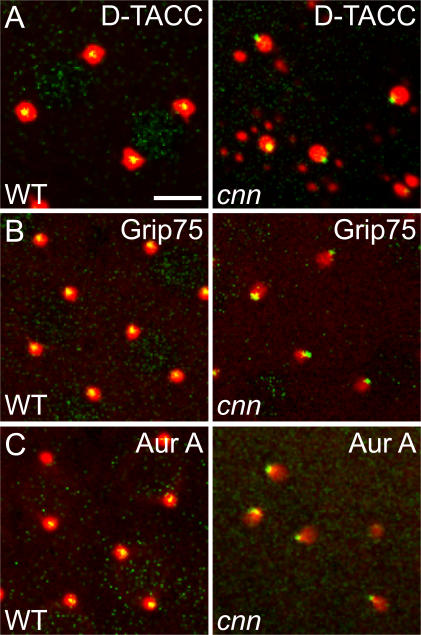
The injection of colchicine into cnn embryos also enabled the centrioles to remain associated with the PCM (Fig. 3), suggesting that it is the MT-dependent rocketing of the centrioles that ultimately breaks the link between the centrioles and the PCM in cnn embryos. Intriguingly, however, the injection of colchicine into cnn embryos did not correct the positioning defect of the centrioles within the PCM: whereas the centrioles were usually (>90%) well centered within the PCM in colchicine-injected WT embryos (between 50 and 100 centrioles observed with each of the three different PCM markers; Fig. 3 and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1), they were very rarely centered within the PCM in colchicine-injected cnn embryos (<10%) and were usually positioned at the very edge of the PCM (>100 centrioles observed with each of the three different PCM markers; Fig. 3 and Video 6).
Figure 3.
The centrioles are displaced to the edge of the PCM in cnn embryos. The WT and cnn embryos shown were injected with colchicine to depolymerize the MTs, and they express the centriole marker mRFP-Fzr (pseudocolored green) and the PCM markers GFP–D-TACC (A), Grip75-GFP (B), and Aurora A– GFP (C; all pseudocolored red). Note how the centrioles are displaced toward the edge of the PCM in cnn embryos (right). See Video 6 (available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). Bar, 10 μm.
This last observation was unexpected, and we are unaware of any other perturbation to the centrosome that results in this very specific displacement of the centrioles from the center of the PCM. This observation may have important implications for understanding how Cnn functions to maintain the link between the centrioles and the PCM. One interesting possibility is that the MT-dependent centriole rocketing we observe in cnn embryos may be mechanistically related to the actin-dependent rocketing of certain pathogenic bacteria (Borisy and Svitkina, 2000; Higgs and Pollard, 2001). These bacteria are coated with proteins that initially stimulate the polymerization of an actin “cloud” symmetrically around the surface of the bacteria. If the actin surrounding the bacteria is structurally weak, it can “fracture”, allowing the bacteria to move to the edge of the actin cloud and rocketing to begin (van Oudenaarden and Theriot, 1999; van der Gucht et al., 2005). Thus, we propose that the primary function of Cnn may be to mechanically strengthen the PCM: in the presence of Cnn, the PCM is structurally strong and the centrioles can maintain their position at the center of the PCM; in the absence of Cnn, the PCM is weakened and the centrioles move to the edge of the PCM. This then initiates centriole rocketing, although the exact mechanism of this MT-dependent rocketing remains unclear.
Centriole segregation is defective in cnn mutant somatic cells
Maintaining the proper connection between the centrioles and the PCM is clearly crucial in syncytial embryos, as a lack of Cnn results in catastrophic failures in mitosis. In contrast, somatic cells that lack Cnn have few mitotic defects, and cnn mutant flies are viable (Megraw et al., 2001; Mahoney et al., 2006). To test whether Cnn was required to maintain the proper connection between the centrioles and the PCM in somatic cells, we treated third instar larval brain cells with colchicine to depolymerize the MTs and then fixed and stained them to examine the distribution of the centrioles and the PCM. We found that hardly any PCM was detectable around the centrioles in cnn brain cells that had not been treated with colchicine (unpublished data). In cnn cells treated with colchicine, however, considerable amounts of PCM accumulated around the centrioles, but, as in cnn embryos, the centrioles were displaced from the center of the PCM (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1).
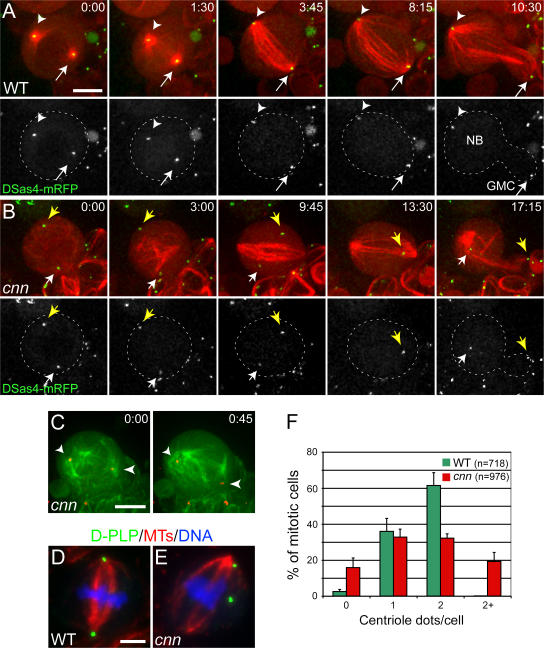
To further investigate whether the centrioles in cnn somatic cells behaved in the same way as the centrioles in cnn embryos, we examined living third instar larval NBs expressing the centriole marker DSas-4–mRFP and GFP–α-tubulin. In WT NBs entering mitosis, the centrioles were always centered within astral MT arrays, and the centrioles remained tightly associated with the poles of the spindle throughout mitosis (Fig. 4 A). In contrast, the centrioles in cnn NBs were often not associated with prominent astral MTs and exhibited irregular movements throughout the cell during mitosis. As a consequence, they were often abnormally displaced from the poles of the mitotic spindles (Fig. 4 B). Nevertheless, we could transiently detect astral MTs associated with some of the “rocketing” centrioles in some cnn NBs (Fig. 4 C, arrows; see the next section). In fixed larval cnn NBs, the centrioles were often randomly positioned around the cell (Fig. 4 E), and we noticed that 20–30% of brain cells had either too few or too many centrioles (Fig. 4 F). Taken together, these findings suggest that the centriole behavior is similar in cnn embryos and somatic cells; while these defects do not lead to dramatic errors in somatic cell division, they do lead to errors in centriole segregation. These findings support the hypothesis that centrioles have evolved the ability to recruit PCM to ensure the equal partitioning of the centrioles during cell division, rather than to ensure the efficient assembly of the mitotic spindle (Pickett-Heaps, 1969; Rieder et al., 2001).
Figure 4.
Centriole segregation is abnormal in cnn mutant larval brain cells. (A and B) Time series (min:s) of living WT and cnn mutant NBs expressing the centriole marker DSas-4–mRFP (arrows; pseudocolored green) and GFP–α-tubulin (pseudocolored red). (A) In WT cells, the centrioles are always located at the poles of the mitotic spindle. (B) In cnn NBs, the centrioles move erratically around the cell and no longer maintain their proper connection to the poles of the spindle. Note that one of the centrioles temporarily moves out of the focal plane (13:30) but reappears by the end of mitosis. (C) Another cnn NB expressing the same markers as in A and B (colors are inverted). Both centrioles are able to nucleate astral MTs but fail to maintain their connection to them. (D and E) The distribution of MTs (red), centrioles (D-PLP; green), and DNA (blue) in fixed WT and cnn mutant cells. In cnn mutant cells (E), the centrioles are more randomly distributed in the cell and are sometimes of unequal size (which may reflect the clustering of some centrioles). (F) Bar chart showing the number of centrioles present in WT and cnn mutant mitotic brain cells. Error bars represent SD. Bars, 5 μm.
Asymmetric cell divisions are only mildly perturbed in cnn mutant NBs
The Cnn-related protein CDK5RAP2 has been implicated in human microcephaly (Bond et al., 2005), and several recent studies have shown that centrosomes exhibit an asymmetric behavior during the asymmetric divisions of male germline stem cells (GSCs) and larval neural stem cells (NBs) (Rebollo et al., 2007; Rusan and Peifer, 2007; Yamashita et al., 2007). During interphase in these cells, only one centrosome is initially associated with PCM and MTs, and this centrosome becomes anchored on one side of the cell (near the stem cell niche in GSCs, or near cortically localized cell polarity markers in NBs). When the second centrosome eventually associates with PCM and MTs, it localizes to the opposite side of the cell, thus ensuring that the forming mitotic spindle is correctly oriented relative to these positional cues.
This asymmetric centrosome behavior appears to be important in male GSCs, as the asymmetric division of these cells is dramatically perturbed in cnn mutants (Yamashita et al., 2003). Although cnn mutant NBs have defects in aligning their spindles with cortical determinants early in mitosis (Megraw et al., 2001), it is not clear that this ultimately leads to failures in asymmetric division: early mitotic spindle alignment defects are often corrected in these cells by the time the cells divide (Cai et al., 2001). To determine whether cnn mutant NBs ultimately divide asymmetrically, we analyzed living WT and cnn third instar larval NBs expressing only GFP–α-tubulin.
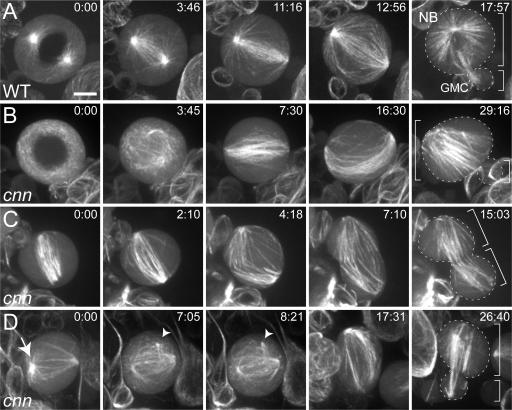
As reported previously (Rebollo et al., 2007; Rusan and Peifer, 2007), a single, anchored MTOC was usually visible in WT NBs before the entry into mitosis (not depicted). After nuclear envelope breakdown (NEB), however, both centrosomes nucleated prominent arrays of MTs, and spindle assembly occurred primarily by a centrosomal pathway (Fig. 5 A and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). As expected, the cells divided asymmetrically to produce a large NB and a small ganglion mother cell (GMC). In most cnn NBs, no prominent MTOC was detectable before NEB, and spindle assembly occurred largely by an acentrosomal pathway (Fig. 5 B and Video 8). Nonetheless, ∼95% of cnn NBs ultimately divided asymmetrically (n = 81; Fig. 5 B and Video 8), whereas ∼4% divided symmetrically (Fig. 5 C and Video 9) and ∼1% failed in cytokinesis (not depicted). Although this failure rate is modest, we believe it is considerably higher than in WT, as we observed only one symmetric division in >100 WT central brain NBs examined (unpublished data; Basto, R., and C.I. Dix, personal communication).
Figure 5.
Asymmetric cell divisions are mildly perturbed in cnn mutant NBs. Time series (min:s) of MT behavior in living third instar larval NBs. (A) In a WT NB, the centrosomes nucleate prominent arrays of astral MTs before NEB (0:00), and the spindle is assembled primarily from these MTs. This cell divides asymmetrically (17:57) to produce a large NB and a small GMC (cell areas are outlined; see Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1). (B) In this cnn mutant NB, astral MTs are undetectable, and the spindle appears to assemble around the chromatin; nevertheless, the cell divides asymmetrically (Video 8). (C) In this cnn mutant NB, some astral MTs are visible, but the cell ultimately divides symmetrically (Video 9). (D) In this cnn mutant NB, prominent astral MTs transiently form at one of the poles of the spindle (arrow), and a small, transient focus of MTs can be seen moving around in the cytoplasm (arrowheads; Video 10). Bar, 5 μm.
We previously showed that mutations in DSas-4, which encodes the Drosophila homologue of the human microcephaly protein CenpJ/CPAP (Bond et al., 2005), also lead to defects in the asymmetric divisions of larval NBs. The defects were much more severe in DSas-4 mutants, which completely lack centrioles/ centrosomes (∼15% of NBs divided symmetrically, whereas ∼15% failed in cytokinesis; Basto et al., 2006). The much milder defects in asymmetric division that we observe in cnn NBs suggest that centrosomes are partially functional as MTOCs in cnn mutant somatic cells, consistent with our previous observations (Fig. 4 C and Fig. S2). Indeed, we frequently observed relatively well-focused astral MT arrays forming and disassembling in the cytoplasm, and these were often transiently associated with the spindle poles in cnn NBs (Fig. 5 D, arrow; and Video 10, available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1).
Taken together, our observations on cnn and DSas-4 mutant NBs reveal that, unlike the situation in male GSCs, the asymmetric behavior of the centrosomes is not essential for the accurate asymmetric division of larval NBs. Nevertheless, mutations in the Drosophila homologues of two of the three human centrosomal proteins implicated in microcephaly do lead to relatively subtle defects in NB divisions in flies. Drosophila cnn and DSas-4 mutants do not have small brains, suggesting that flies are able to compensate for defects in these divisions in a way that perhaps humans cannot.
Materials and methods
Fly stocks
Oregon R or yw flies were used as the WT stock, and yw flies as the parental stock for the generation of all transgenic lines. The cnnHK21 null allele has been described previously (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999), and the cnnf04547 allele, a piggyBac insertion in the middle of the cnn gene (Exelixis stock no. f04547), was obtained from the Exelixis Stock Centre (Harvard Medical School, Boston, MA). In Western blotting experiments, embryos laid by females homozygous for this allele had no detectable Cnn protein.
GFP and mRFP fusion proteins
GFP fusions to the following proteins were used in this study: Aurora A–GFP (Lau, J., personal communication), GFP–D-TACC (Lee et al., 2001), Grip75-GFP (Schnorrer et al., 2002), GFP–α-tubulin (Grieder et al., 2000), and DSas-4–GFP (Peel et al., 2007). We also generated fusions between mRFP and the full-length fzr cDNA, the PACT domain of D-PLP (Martinez-Campos et al., 2004), and DSas-4 (Basto, R., personal communication). Most of these fusions were subcloned into the pWR-Ubq transformation vector that drives the ubiquitous expression of the fusion protein at moderately high levels. For Grip75-GFP and GFP–α-tubulin, we used previously established lines in which the expression of these proteins is under the control of the UASp promoter (Rorth, 1998); we drove their expression in embryos using the maternal 67C α-tubulin–GAL4 promoter (Lee et al., 2001), and in brains using the 69B enhancer trap line (Brand and Perrimon, 1993).
Transgenic lines were generated using standard methods. Most studies were performed using the cnnHK21 allele, but we obtained similar results with either the cnnf04547 allele or transheterozygous combinations of the two alleles.
Live imaging of syncytial embryos
Live embryos expressing fluorescent fusion proteins were examined as described previously (Huang and Raff, 1999). The embryos were observed on an ERS spinning disc confocal system (PerkinElmer), mounted on an inverted microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) that was equipped with a charge-coupled device camera (Orca ER; Hamamatsu), using a 63×/1.25 NA objective. For each combination of GFP and mRFP fusions, 4–15 WT and cnn mutant syncytial blastoderm stage embryos were examined. For drug injections, live embryos were injected at the desired stage with 5 mg/ml colchicine (Sigma-Aldrich). All images were captured and made into videos using the Ultraview ERS software (PerkinElmer).
Live imaging of larval NBs
Third instar larval brains were prepared as described previously (Basto et al., 2006), and central brain NBs were followed by time-lapse confocal microscopy as described, with a 100×/1.3 NA objective. 12–20 focal planes spaced by 0.5 μm were acquired every 25 or 45 s (0.5–1 s/frame, respectively). All images shown are maximum intensity projections of z stacks at selected time points.
Immunofluorescence of larval brain preparations
Third instar larval brains were fixed and stained as described previously (Martinez-Campos et al., 2004). For drug treatment, dissected brains were incubated for 2 h at 25°C in PBS containing 1 μg/ml colchicine before fixation. The following primary antibodies were used: rabbit anti–D-PLP at 1–2 μg/ml (Martinez-Campos et al., 2004); mouse anti–α-tubulin at 1:1,000 (DM1α; Sigma-Aldrich), mouse anti–γ-tubulin at 1:1,000 (GTU88; Sigma-Aldrich), and mouse anti–phospho-Histone H3 at 1:2,000 (Abcam). All secondary antibodies coupled to the appropriate fluorophore (Alexa 488 or 568; Invitrogen) were used at 1:1,000 in PBT. Fixed preparations were examined on a widefield upright microscope (Axioskop II; Carl Zeiss MicroImaging, Inc.), equipped with a camera (CoolSnap HQ; Photometrics) and MetaMorph software (Molecular Devices), using a 100×/1.3 NA objective. Images of fixed brain cells are all maximum intensity projections of optical sections acquired at 0.1– 0.2-μm intervals.
Preparation of figures
Individual images were imported into Photoshop 7.0 (Adobe) and adjusted to use the full range of pixel intensities. In panels for some figures, pixel intensities were adjusted using the “curves” control panel, and an unsharp mask and despeckle filter were applied to the whole image. In all cases, the images from control and experimental embryos were adjusted in the same way.
Quantification of the PCM recruitment around the centrioles in syncytial embryos
To quantify the amount of PCM recruited to centrioles, individual images were imported into MetaMorph. The centrosome was circled, and the integrated intensity was calculated for independent centrosomes after subtraction of cytoplasmic background fluorescence. The integrated intensity per pixel area was determined from at least three different WT and cnn embryos per centrosomal marker. A total of 45 centrosomes were scored for each marker. Error bars represent the SD. The data were analyzed for statistical significance using a two-tailed t test.
Quantification of centriole number in fixed larval brain cells
To quantify the number of centriole dots per mitotic cell, fixed preparations of third instar larval brains were stained with anti–D-PLP and anti–phospho-Histone H3 antibodies. We examined a minimum of 70 mitotic (phospho–Histone H3 positive) cells per larval brain from at least six different brains. A total of 718 mitotic cells from WT and 976 mitotic cells from homozygous cnnHK21 mutants were scored. Error bars represent the SD.
Centriole tracking
Semiautomated tracking software (Imaris 4.5.2; Bitplane AG) was used to identify and track DSas-4–GFP trajectories over time (150 time frames; 1 s/frame). Image segmentation was performed to convert pixel intensities above a given threshold into computerized spots, and this method was applied equally to all the images in the time series. Semiautomatic track building was based on autoregressive motion algorithms.
Online supplemental material
Fig. S1 shows the localization of Aurora A–GFP, Grip75-GFP, and centrioles in WT and cnn embryos. Fig. S2 shows the centriole positioning defects in cnn larval brain cells after colchicine treatment. 10 additional videos are also included, showing the behavior of centrioles and GFP-D-TACC in WT and cnn embryos (Video 1); centrioles and Aurora A–GFP or Grip75-GFP in WT and cnn embryos (Video 2); centrioles and GFP– α-tubulin in a cnn embryo (Videos 3 and 4); centrioles in WT and cnn embryos before and after colchicine injection (Video 5); centrioles and PCM in WT and cnn embryos after colchicine injection (Video 6); a WT larval NB expressing GFP–α-tubulin, dividing asymmetrically (Video 7); a cnn larval NB expressing GFP–α-tubulin, dividing asymmetrically (Video 8); a cnn larval NB expressing GFP–α-tubulin, dividing symmetrically (Video 9); a cnn larval NB expressing GFP–α-tubulin with prominent astral MTs, and a small focus of MTs moving around the cell (Video 10). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200704081/DC1.
Supplementary Material
Acknowledgments
We thank R. Basto and C.I. Dix for sharing unpublished data, C. Lehner for the full-length fzr cDNA and the Bloomington and Harvard stock centres for stocks. We also thank members of the Raff laboratory for valuable discussions and comments on the manuscript and A. Sossick for help with microscopy and image analysis.
This work was funded by a Research Fellowship from Cancer Research UK (J.W. Raff) and a PhD studentship from Fundação para a Ciência e a Tecnologia, Portugal (E.P. Lucas).
Abbreviations used in this paper: Cnn, centrosomin; GSC, germline stem cell; MT, microtubule; MTOC, MT organizing center; NB, neuroblast; NEB, nuclear envelope breakdown; PCM, pericentriolar matrix; WT, wild-type.
References
- Badano, J.L., T.M. Teslovich, and N. Katsanis. 2005. The centrosome in human genetic disease. Nat. Rev. Genet. 6:194–205. [DOI] [PubMed] [Google Scholar]
- Basto, R., J. Lau, T. Vinogradova, A. Gardiol, C.G. Woods, A. Khodjakov, and J.W. Raff. 2006. Flies without centrioles. Cell. 125:1375–1386. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., A. Rodrigues-Martins, L. Carpenter, M. Riparbelli, L. Lehmann, M.K. Gatt, N. Carmo, F. Balloux, G. Callaini, and D.M. Glover. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. [DOI] [PubMed] [Google Scholar]
- Bond, J., and C.G. Woods. 2006. Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 18:95–101. [DOI] [PubMed] [Google Scholar]
- Bond, J., E. Roberts, K. Springell, S.B. Lizarraga, S. Scott, J. Higgins, D.J. Hampshire, E.E. Morrison, G.F. Leal, E.O. Silva, et al. 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355. [DOI] [PubMed] [Google Scholar]
- Borisy, G.G., and T.M. Svitkina. 2000. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12:104–112. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. [DOI] [PubMed] [Google Scholar]
- Cai, Y., W. Chia, and X. Yang. 2001. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J. 20:1704–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, E.E., M. Brueckner, and N. Katsanis. 2006. The emerging complexity of the vertebrate cilium: new functional roles for an ancient organelle. Dev. Cell. 11:9–19. [DOI] [PubMed] [Google Scholar]
- Doxsey, S., D. McCollum, and W. Theurkauf. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21:411–434. [DOI] [PubMed] [Google Scholar]
- Fish, J.L., Y. Kosodo, W. Enard, S. Paabo, and W.B. Huttner. 2006. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA. 103:10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder, N.C., M. de Cuevas, and A.C. Spradling. 2000. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 127:4253–4264. [DOI] [PubMed] [Google Scholar]
- Higgs, H.N., and T.D. Pollard. 2001. Regulation of actin filament network formation through ARP2/3 complex: activation by a diverse array of proteins. Annu. Rev. Biochem. 70:649–676. [DOI] [PubMed] [Google Scholar]
- Huang, J., and J.W. Raff. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.J., F. Gergely, K. Jeffers, S.Y. Peak-Chew, and J.W. Raff. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649. [DOI] [PubMed] [Google Scholar]
- Mahoney, N.M., G. Goshima, A.D. Douglass, and R.D. Vale. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569. [DOI] [PubMed] [Google Scholar]
- Martinez-Campos, M., R. Basto, J. Baker, M. Kernan, and J.W. Raff. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T.L., K. Li, L.R. Kao, and T.C. Kaufman. 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 126:2829–2839. [DOI] [PubMed] [Google Scholar]
- Megraw, T.L., L.R. Kao, and T.C. Kaufman. 2001. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11:116–120. [DOI] [PubMed] [Google Scholar]
- Peel, N., N.R. Stevens, R. Basto, and J.W. Raff. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17:834–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps, J. 1969. The evolution of the mitotic apparatus: an attempt at comparative ultrastuctural plant cytology in dividing plant cells. Cytobios. 3:257–280. [Google Scholar]
- Rebollo, E., P. Sampaio, J. Januschke, S. Llamazares, H. Varmark, and C. Gonzalez. 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 12:467–474. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., S. Faruki, and A. Khodjakov. 2001. The centrosome in vertebrates: more than a microtubule-organizing center. Trends Cell Biol. 11:413–419. [DOI] [PubMed] [Google Scholar]
- Rorth, P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78:113–118. [DOI] [PubMed] [Google Scholar]
- Rusan, N.M., and M. Peifer. 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin, K.E., P.C. Lourenco, and H.A. Snaith. 2004. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14:763–775. [DOI] [PubMed] [Google Scholar]
- Schnorrer, F., S. Luschnig, I. Koch, and C. Nusslein-Volhard. 2002. Gamma-tubulin37C and gamma-tubulin ring complex protein 75 are essential for bicoid RNA localization during Drosophila oogenesis. Dev. Cell. 3:685–696. [DOI] [PubMed] [Google Scholar]
- Terada, Y., Y. Uetake, and R. Kuriyama. 2003. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 162:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaizel-Ohayon, D., and E.D. Schejter. 1999. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9:889–898. [DOI] [PubMed] [Google Scholar]
- van der Gucht, J., E. Paluch, J. Plastino, and C. Sykes. 2005. Stress release drives symmetry breaking for actin-based movement. Proc. Natl. Acad. Sci. USA. 102:7847–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oudenaarden, A., and J.A. Theriot. 1999. Cooperative symmetry-breaking by actin polymerization in a model for cell motility. Nat. Cell Biol. 1:493–499. [DOI] [PubMed] [Google Scholar]
- Venkatram, S., J.J. Tasto, A. Feoktistova, J.L. Jennings, A.J. Link, and K.L. Gould. 2004. Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol. Biol. Cell. 15:2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde, I., G. Pahlke, M. Salanova, G. Zhang, S. Wang, D. Coletti, J. Onuffer, S.L. Jin, and M. Conti. 2001. Myomegalin is a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 276:11189–11198. [DOI] [PubMed] [Google Scholar]
- Yamashita, Y.M., D.L. Jones, and M.T. Fuller. 2003. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 301:1547–1550. [DOI] [PubMed] [Google Scholar]
- Yamashita, Y.M., A.P. Mahowald, J.R. Perlin, and M.T. Fuller. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 315:518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.