Abstract
Transposable elements are often considered parasitic DNA sequences, able to invade the genome of their host thanks to their self-replicating ability. This colonization process has been extensively studied, both theoretically and experimentally, but their long-term coevolution with the genomes is still poorly understood. In this work, we aim to challenge previous population genetics models by considering features of transposable elements as quantitative, rather than discrete, variables. We also describe more realistic transposable element dynamics by accounting for the variability of the insertion effect, from deleterious to adaptive, as well as mutations leading to a loss of transposition activity and to nonautonomous copies. Individual-based simulations of the behavior of a transposable-element family over several thousand generations show different ways in which active or inactive copies can be maintained for a very long time. Results reveal an unexpected impact of genetic drift on the “junk DNA” content of the genome and strongly question the likelihood of the sustainable long-term stable transposition-selection equilibrium on which numerous previous works were based.
Keywords: genome evolution, molecular domestication, population genetics
Transposable elements (TEs) are among the major components of genomes. Their universality is usually attributed to their ability to invade genomes similarly to parasites (1–3); they provide no adaptive advantage to the individual carrying them and are thus often considered “junk DNA” (4).
It was rapidly acknowledged that the complexity of the evolutionary forces at work on TE sequences, including several levels of selection, need to be approached through specific population genetics models (5–7). Consequently, a number of theoretical studies have been carried out, some of which address general issues common to all TEs, whereas others are specific to a particular TE family and host species. All of these models brought insightful advances in the understanding of TE biology, showing that the invasion of parasitic DNA sequences in the genome of a species is realistic for a broad range of parameters. Although active replicative transposition enhances colonization, natural selection, as well as deletions, tends to limit the spread of “selfish” copies. Because the TE content of known genomes does not seem to show any general and dramatic increasing or decreasing trend, it has been supposed that, after a first-invasion stage, the appearance of new copies is roughly balanced by the losses, and TE families could be maintained over the long term through a so-called transposition–selection equilibrium (see refs. 8 and 9, and refs. therein).
However, the evolution of TEs after their initial colonization is still only partly understood, and theoretical models fail to grasp the complex dynamics, history, and insertion patterns of TE and TE-derived sequences. Indeed, most of these models do not account for polymorphism among TE sequences of the same family; TE evolution is thus reduced to the competition or coevolution between a “host” and a homogeneous group of “parasites.” However, data provided by sequencing programs generally demonstrate that the majority of TE-derived sequences correspond not to autonomous elements but rather to the relics of formerly active TE families. Some nonautonomous elements, such as MITEs (11, 12) or SINEs (13, 14), can even “superparasitize” the transposition machinery produced by autonomous copies and thus modify the dynamics of the whole system (15, 16), promoting, e.g., cyclic reinvasions of active copies (3, 17). Moreover, if the presence of TEs is known to be intrinsically deleterious (18–20), it is also thought that the insertion effect ranges from lethal insertions to neutral or even adaptive effects. More and more examples of TE copies fixed in the genome of a species without being degraded have been indeed evidenced. Such sequences sometimes show typical signs of positive selection (21) and seem to have escaped from their parasitic role; they are often said to be “domesticated” by the genome (22, 23). Finally, there is little doubt that genetic drift contributes to the structure of TE content in genomes, e.g., by promoting random departures from the expected equilibrium state or by influencing the efficiency of the elimination of deleterious insertions by natural selection.
Evolutionary geneticists, as well as molecular biologists involved in genome analyses, thus need more realistic (yet general) models accounting for the intragenomic evolution of TE sequences. Here, we propose to investigate, through stochastic simulations in finite-sized populations, the long-term coevolution of a TE family within its host genome. To do so, we relaxed the usual hypothesis that all copies from the same family must be identical, by introducing variability into both the effects of insertions on fitness and the production of transposition-related proteins. The possibility of mutations (i.e., modifications in the transposition efficiency of copies) leads to the exploration of long-term dynamics, far beyond the invasion stage. This model allows us to raise issues that could hardly be addressed by previous studies. Specifically, we will focus on investigating how the long-term maintenance of TEs in a genome can be reached, and whether the well documented transposition–selection equilibrium is the only way to reach it.
Results
The purpose of the model is to analyze the dynamic properties of a TE family over several thousand generations. Each TE copy is defined by its transposition (u) and deletion (v) rates, as well as by its impact on host fitness (s) and its transposition activity (a). The activity (i.e., the ability to produce functional transposition machinery) is thus separated from the transposition rate itself (the capacity to transpose given the presence of the transposition machinery) (Fig. 1). The activity can decrease with mutations, occurring at a rate m. More details about the model are provided in Material and Methods and in supporting information (SI) Figs. 5 and 6.
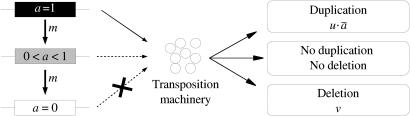
Fig. 1.
Scheme of the transposition model. Each copy i is defined by its specific activity 0 ≤ ai ≤ 1 (representing its capacity to produce a functional transposition machinery), transposition rate ui, and deletion rate vi. The activity can change with mutations, occuring with a rate m. Small circles and rectangles represent the transposition machinery and the copies inserted in the genome respectively (black, full activity; gray, partial activity; white, no activity).
Transposition Selection.
Active transposition stages correspond to situations in which the element is actively maintained in the genome because of its transposition ability. Transposition balances natural selection and mutational events, so the element can “survive” as a selfish DNA sequence.
Previous studies of TE dynamics have generally shown that the average copy number of a TE family is supposed to stabilize after a fairly long period, because of the balance between transposition and natural selection (7). In our model, when the adaptive insertion probability (ps>0) and the mutation rate (m) are 0 (or very low, i.e., at least 100× lower than the transposition rate), the conditions necessary for such an equilibrium state are achieved, and all of the parameters of the population are stabilized for a very long period (Fig. 2A). All or almost all TE copies remain deleterious and are maintained in the population only because of their “parasitic” ability; the average fitness of the population is lower than in a TE-free population (high genetic load for the population), and all copies remain highly transpositionally active. Even if they appear sporadically through rare mutational events, low-activity copies are rapidly eliminated by both deletions, preventing the fixation of the copy, and natural selection, driving its frequency to 0.
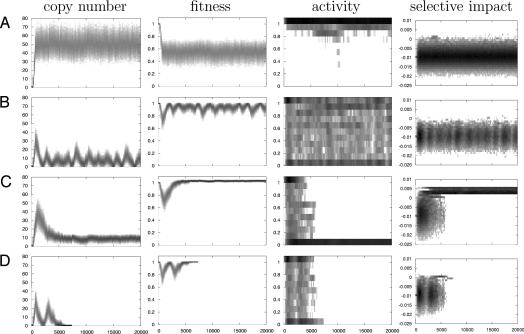
Fig. 2.
Description of the characteristic dynamics identified. (A) Equilibrium (obtained with the parameter set m = 10−5, σa = 0.1, ps>0 = 0.01%). (B) Cycles (m = 10−3, σa = 1, ps>0 = 0.01%). (C) Domestication (m = 10−3, σa = 1; ps>0 = 0.5%). (D) Loss (m = 10−3, σa = 1, ps>0 = 0.1%). The x axis represents the time in generations. First column, distribution of the copy number among the individuals of the population; second column, distribution of the fitnesses among the individuals of the population; third column, distribution of the activity among all of the elements present in the population; and fourth column, distribution of the selective impact among all elements. The gray scale corresponds to the distribution of the parameter of each generation (dark gray for high frequencies).
Mutations.
When activity mutations (i.e., mutations affecting the capacity to produce the transposition machinery without modifying the transposition ability itself) are introduced in the simulations, two “populations” of TEs may cohabit in the genome; nonautonomous copies settle, sometimes with a relatively high copy number, along with the regular fully functional elements (Fig. 2B and SI Fig. 7). Even if the model allows small step mutations, partially active elements do not seem to be able to maintain for a long time.
In large populations, this cohabitation between autonomous and nonautonomous copies can be maintained for a very long time (>20,000 generations, the upper limit of our simulations). As shown (3, 17), this situation leads to a succession of invasions and regressions for both autonomous and nonautonomous copies, very similar to known host–parasite demographic patterns. The resulting pattern is a cyclic invasion of TEs (Fig. 2B), appearing even at relatively low mutation rates (<100× less frequent than transpositions).
Variability of Selective Impact.
When the variability of the selective impact increases, transposition generates very deleterious copies, as well as neutral or slightly adaptive ones. Deleterious copies being rapidly eliminated, it appears that the main impact of the variability of the selective coefficients is due to the generation of “positive-effect” copies, which can settle and can be maintained in the genome for a very long time without being eliminated by natural selection. Despite a very low rate of appearance, such copies tend to accumulate in the genome and eventually impact the whole dynamics.
If the mutation rate is very low and the rate of adaptive insertions high, all copies remain indefinitely active, and the accumulation of neutral or adaptive copies is not reversible. The copy number exponentially increases toward an uncontrolled invasion that stops only when the memory limit of the computer is reached (not shown). This is probably an unrealistic scenario, involving parameters beyond the reasonable biological range.
This pattern changes radically when mutations are introduced at the same time (Fig. 2 C and D, SI Fig. 8). Because the longer the time spent in the genome, the more likely the inactivation, the neutral or adaptive copies do not contribute significantly to the production of transposition machinery. Active transposition thus relies on newly inserted, often deleterious copies, that are likely to be rapidly eliminated by natural selection. The transposition rate tends to drop with the progressive loss of active copies, and the TE content of the genome freezes.
Depending on the probability of adaptive insertions, two different final states have been reached in our simulations. In the case where adaptive insertions are rare and their effect is small, most remaining copies are neutral or slightly deleterious. In the absence of transposition, they are rapidly lost by the combination of rare deletion events (generating polymorphism at fixed loci) and genetic drift (Fig. 2D). On the contrary, if the active transposition stage was long enough and the probability of adaptive insertions high enough for some large-effect beneficial copies to be fixed, natural selection can maintain them for a very long time (up to the end of the simulation; Fig. 2C). This last situation corresponds to the well known “molecular domestication” events, in which TE-derived sequences turn into regular “genes,” necessary for the survival of the organism. The simulations stress that, after a first decline due to invasion by autonomous and deleterious elements, the average fitness of the host increases and finally exceeds the initial level (i.e., the fitness of an individual without any copy); despite a transitional genetic load, invasion by a TE family has brought an adaptive potential to the population.
Genetic Drift and General Dynamic Properties.
Because the model is stochastic, the transition from one dynamics to another remains a random process, and there is no certainty concerning the state of the population at the end of a simulation. Nevertheless, despite the randomness brought by drift in small populations (Fig. 3), a general pattern can be described. If the mutation rate is very low, the equilibrium situation can be achieved. In all other cases, the copy number will decrease after an initial burst, because of mutant elements that will prevent the regular transposition process. The number of cycles occurring before the loss of transposition activity depends on the rate of adaptive insertions and on the population size. It can be reduced to a single cycle (in small populations), or cycles can be maintained until the end of the simulation when the effects of genetic drift are reduced (large populations). After loss of activity, a phase with a low copy number can be observed. The length of this phase depends on the population size and deletion rate (SI Fig. 8), but it is also highly influenced by the number of adaptive insertions that occurred when the element was able to transpose actively. Genetic drift will progressively eliminate these insertions, probably starting with the neutral ones. Significantly adaptive insertions can be maintained until the end of the simulations.
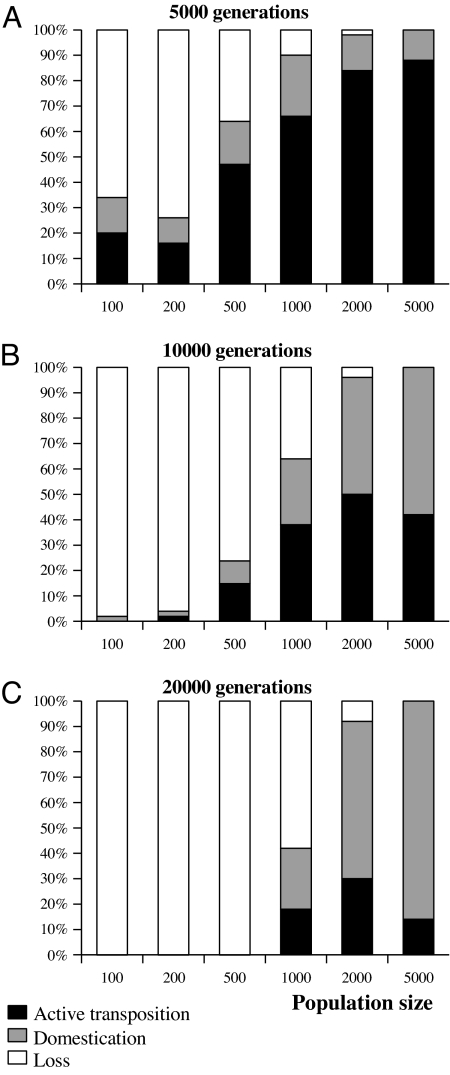
Fig. 3.
Influence of population size on TE evolution. The three histograms represent the percentage of each situation (active transposition, domestication, and loss) after a given number of generations (A, 5,000 generations; B, 10,000 generations; C, 20,000 generations) over 40 repetitions for each parameter set. Other parameters: m = 10−3, σa = 1, ps>0 = 0.1%.
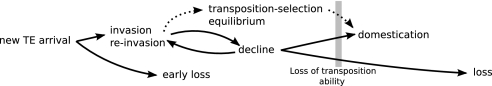
The polymorphism among the observed dynamics (Fig. 2, SI Fig. 9) can be explained by using a few general rules about the expected evolution of a TE family. Reaching a transposition–deletion equilibrium leads to the long-term maintenance of the family, but the conditions for its completion (e.g., low mutation rate) hardly correspond to realistic TEs. The most common dynamics is the occurrence of one (or several) invasion–regression cycles, generally followed by a domestication event, before the definitive loss of the family (Fig. 4). The general trend is an instability of the cycles when genetic drift increases; the larger the population size, the longer the maintenance of the active transposition stage.
Fig. 4.
Summary of the expected dynamics of a TE family in the genome of a species.
Discussion
TE Evolution.
TE dynamics are usually split into different phases. The first is necessarily the invasion of the population after the arrival of a new active TE sequence in the genome of a species, either by reactivation of a formerly inactive copy or, more likely, by horizontal transfer (24–26). In such a situation, an element has to be immediately active and transpositionally efficient; otherwise, its loss because of genetic drift and natural selection is likely (27). This invasion step has already been experimentally observed in model species such as Drosophila melanogaster, in both natural populations [e.g., the P element (28)] and laboratory experiments (e.g., refs. 29 and 30); some elements thus seem able to invade a new species efficiently, even if they come from a different one (31, 32).
The next steps of the dynamics cannot be observed directly, but further information may come from the genomic sequences. A rough analysis reveals that most TE-derived sequences seem to be nonfunctional. Moreover, the proportion of active copies is highly heterogeneous among species. Less than 20% of D. melanogaster's elements appear to be full-length (33, 34), but this figure is <5% (a single family) in Schizosaccharomyces pombe (35). In humans, only 1% of L1 elements are complete, and 1% of these full-length L1s are thought to be active, 100× fewer than in the mouse (36). More interestingly, such variability can be retrieved in a single genome. For instance, in D. melanogaster, >50% of the 27 roo copies identified by ref. 33 are complete, whereas no active copies have been found among the 16 R1 insertions or the 14 Opus elements.
Two hypotheses can be proposed to explain such heterogeneity. (i) The different TE families from the same genome and the TEs from the same family in different species are not in the same phase of their evolution; some of them could be invading, whereas others are in a decreasing dynamic, leading to different insertion patterns. (ii) Because slight differences in the parameter sets used in our simulations lead to distinct dynamics, the long-term evolution of a TE family can be significantly affected by the characteristics of the TE, of its host, or by specific TE–host interaction. Unfortunately, our knowledge of TE evolution is not complete enough to supply arguments in favor of one or the other hypothesis. Moreover, both hypotheses are nonexclusive, and insertion patterns might be explained by a combination of both processes.
TEs and Host Fitness.
One of the main original features of our model derives from the quantitative variability of the selective effect of the insertion sites. Traditionally, the insertion effect is modeled as constant (17, 27) or variable according to the genomic copy number (7, 37), but all copies of the genome potentially have the same impact. However, this is probably unrealistic. A part of the deleterious effect of the presence of a TE family may be shared by all copies of the family (e.g., genetic load due to ectopic recombinations) or by a subset of the genomic copies (e.g., selection against the transposition activity or against active copies only). Nevertheless, the direct effect of an insertion remains specific and depends on the precise locus in which the element is inserted (intergenic, exon, intron, promoter sequence, etc.).
The respective impacts of these selection mechanisms have not yet been quantified (9, 19), but a single TE insertion is known to lead to a wide range of consequences for fitness, from lethal mutations (38) to adaptive insertions (ref. 21, and see below). This variability has already led to specific models (39), in which several groups of insertion sites were defined (e.g., neutral or selected). This last model mainly generates an accumulation of fully active TEs in the neutral sites. We have not systematically reached such a conclusion in the present work, because TEs are also subject to mutations and can be progressively inactivated. In this study, the mean insertion effect has been fixed at 1% most of the time, which corresponds to the order of magnitude of the experimental measures of the selective impact of a TE in Drosophila (20, 40, 41). The average effect of the elements present in a genome at a given time is slightly lower, because the most deleterious ones are preferentially eliminated (e.g., Fig. 2C). The normal distribution of selective effects, with a peak in the slightly negative values, can be seen as the result of the combination of insertion effects, variable (from very deleterious to adaptive) but most often almost neutral, and deleterious genome-wide effects (e.g., ectopic recombinations) shared more or less equally among copies.
The putative positive effect of TEs has long been suggested, because the mutational activity of transposition, despite the numerous deleterious insertions, is likely to promote genetic diversity and speed up the adaptation process (18, 42, 43). Recent results have shown that these molecular domestication events (i.e., integration of an adaptive TE-derived sequence in the host genome) seem even more frequent than previously thought, and a significant proportion of genes, including promoters and regulatory sequences, indeed appears to derive from former TE insertions (21–23, 44–46). However, domestication remains rather a consequence than a cause of the presence of TE in genomes, and they represent (very good) examples of exaptations at the molecular level (47, 48).
Model Parameters.
In our model, the maximum transposition rate has been generally set to u = 2 · 10−2 events per copy and per generation, which is rather high compared with known transposition rates for various types of elements, generally around or below 10−3 (49–52). However, it is worth noting that the generation of nonautonomous copies will reduce the average activity ā in the genome, and consequently the simulated transposition rate u · ā will drop as well. This is consistent with the idea of a decrease of the transposition rate after the invasion, often modeled by a “self-regulation” parameter (e.g., refs. 7 and 27), and would mean that most of the time, the transposition rate evaluated from natural populations is smaller than the maximal activity the TE family had at the beginning of the invasion.
Like transposition, the deletion rate that was chosen (v = 10−3) also appears overestimated compared with the measured rates. The aim of choosing high-parameter values was to get realistic simulation results while sparing computer resources. Indeed, it is generally expected that lowering all parameters at the same time gives a similar, but slower, dynamic pattern (see, e.g., ref. 54). This becomes less straightforward when considering a complex model, especially when genetic drift is involved; however, additional simulations showed convincingly that very similar results are obtained when the transposition rate and the selective impact of elements are both reduced by the same order of magnitude, being much closer to what they might be, e.g., in mammals (SI Fig. 10).
The mutation model implemented in the simulations is particularly simple. Only one kind of mutation is allowed, turning functional copies into less autonomous ones, up to nonautonomous copies still able to be transactivated. Additionally less, “deletions” can play the role of large mutations that totally cancels out all of the effects of the copy. A more realistic model would probably include mutations that destroy both activity and mobility of the elements or mutations preventing the transposition without affecting the production of transposition-related proteins, etc.
Very few data concerning the mutation rate of TEs are available, and the rates of appearance of nonautonomous elements from autonomous copies are probably overestimated in the model. Moreover, in some cases, such as in human, nonautonomous SINEs are not derived from full-length LINE elements. Nevertheless, the current choice remains particularly convenient, because (i) a low number of parameters for the mutational model reduces the amount of arbitrarily set values; (ii) the appearance of nonautonomous copies by mutation allows the initialization of simulations with simple populations (active copies only); (iii) contrary to other kinds of defective copies, nonautonomous elements have a tremendous impact on long-term evolution, because they can maintain by “parasitic” self-duplication; and (iv) by turning, in each generation, a small part of low-activity copies into nonautonomous ones, the current mutation model introduces a slight advantage for the nonautonomous population of copies. This slight advantage is far from being unrealistic, because nonautonomous copies, generally smaller and not mobilizing the cellular machinery, are likely to be less deleterious than full-length ones (55). It has been shown (17) that this somewhat unrealistic but simple mutation-driven advantage has very similar effects compared with a more complex selection-based advantage model.
Genetic Drift.
One of the main advantages of a stochastic implementation of the model is to explore the impact of genetic drift on TE dynamics. Some of the stages of the TE life cycle appear to be particularly sensitive to these stochastic effects [e.g., the very beginning of the invasion (27)]. The particularity of such a multicopy system comes from the fact that the drift is determined by the “copy population size,” which depends on the number of individuals in the population, the copy number in each genome, and the fitness of the host. Simulations show that the end of each invasion cycle (when the number of functional copies is low) can lead to loss of transposition activity instead of another invasion, and that loss frequency decreases when population size increases (Fig. 3).
However, this result is rather unexpected. It is generally admitted that, on average, large population sizes lead to less deleterious junk DNA content because of a higher efficiency in purifying selection. Even if there is indeed such a trend within the tree of life (56), it does not always appear as clearly as expected (e.g., ref. 57), and other mechanisms, not yet precisely identified, are probably involved in the evolution of genome size. Our model, by evidencing that active junk DNA can suffer from the consequences of genetic drift exactly the same as genes, might provide an alternative hypothesis to explain why, in some cases, a small population size is not systematically associated with large proportions of TEs.
Equilibrium.
Interestingly, one of the less-supported dynamics remains the long-term equilibrium state. Indeed, it seems very unlikely to maintain a constant copy number in a genome, despite mutations, evolution of the regulation system, and insertions into neutral or adaptive loci. Equilibria can be obtained only with weak (or no) mutation rates, low adaptive insertion rates, and low amplitude of mutations. These conditions contrast with what is generally known from real TEs, which are particularly unstable in the genomes and frequently subject to recombinations, deletions, and inactivations [e.g., because of the insertion of another element in it (58, 59)]. Surprisingly, despite its unlikelihood, the equilibrium state is generally chosen as a reference from which TE features (such as the transposition rate or selective impact) can be computed, which is probably erroneous. This point underlines the need to acquire better definitions of the basic dynamics properties of these selfish DNA sequences.
Materials and Methods
Transposition.
Each TE copy i is characterized by its (i) transposition rate ui, (ii) deletion rate vi, (iii) activity ai, and (iv) impact on the fitness of the individual si. A transposition event corresponds to the duplication of an existing element. The new copy will be identical to the template, except for its position, which is uniformly sampled in the genome, and its selective impact (see below). A deletion consists of the total disappearance of a copy from a genomic location. The activity ai is the production capacity of “transposition machinery” (the proteins required for the transposition process). ai = 0 represents a copy that is not able to produce any protein, but unlike defective elements (i.e., those that are unable to transpose), such a copy is still able to move because of the transposition machinery produced by other sequences in the genome and will be called “nonautonomous.” On the other hand, fully efficient elements are characterized by ai = 1; intermediate values correspond to partially active copies. This transposition machinery is shared among all copies of the genome, whatever their respective activities (Fig. 1). Thus, each individual is defined by its overall transposition activity ā, which is the arithmetic mean of the activities of the n elements present in its genome (i.e., ā = Σai/n). Therefore, each TE of the genome has a probability of being duplicated of ā · u ≤ u. Deletions are supposed to be transposition-unrelated, and the deletion probability is always v, whatever the transposition activity in the genome.
Selection.
Individual fitnesses are calculated relative to a TE-free individual, whose fitness is w = 1. Each TE copy i has an additive selective coefficient si, and the total fitness of an individual is w = 1 + Σsi. If si < 0, then the copy i is deleterious, whereas si > 0 corresponds to an adaptive insertion. In the (rare) case in which there are many deleterious elements, so that w < 0, w is artificially set to 0 (a nonviable or sterile individual). The selection coefficient is supposed to result from a combination of insertion effects (e.g., the disruption of a coding sequence), and other deleterious consequences, such as ectopic recombinations. For each new insertion (i.e., for each transposition event), a selection effect is randomly determined by sampling in a Gaussian distribution of mean μs (fixed at μs = −0.01) and variance σs2. Several variance values have been used in the present work and were chosen according to the corresponding probability of adaptive mutations (SI Fig. 6).
Mutation.
The activity ai of copies does not change except through mutation. Mutation is a transposition-independent process and is considered as occurring only during meiosis. Each time a new gamete is computed, a TE copy has a probability m of being affected by a mutation. Here, only mutations that decrease the activity of the element will be considered. If an element mutates, its activity is decreased by a value δa, sampled in a truncated Gaussian distribution with a variance σa2 (with δa>0, i.e., the activity cannot increase). If the new activity is <0, then it is set to 0, and the element is considered as definitely nonautonomous.
Reproduction.
N diploid individuals (N/2 males and N/2 females), with a genome of five chromosomes of 100 cM, constitute a panmictic population. A nonoverlapping generation consists in the creation of N offspring (as many males as females) from the previous population. For each offspring, two parents (one male and one female) are randomly chosen; a parent j with fitness wj having a probability pj = wj/(N · w̄) of being chosen (where w̄ = Σwj/N is the mean fitness of the population). A haploid gamete is then generated from the genome of each parent. The number of crossovers is evaluated by using a Poisson process, and their positions are determined according to a uniform distribution. Then, for each pair of chromosomes, one homologue is randomly chosen and copied in the gamete until the next crossover, where the template chromosome is swapped. Mutations occur during gametogenesis. Both gametes are merged in a new diploid genome, and transposition occurs at this stage; the average transposition activity of the genome is determined, and TEs are duplicated or deleted after the transposition and deletion probabilities detailed above. Finally, the fitness of the individual is computed from its current genome.
Simulation Procedure.
The model described above has been implemented by using a simulation software named DyET (available on request from the authors). The initial state of the simulations is a population of size N; where N is complete, fully active elements are distributed at one locus (i.e., one per individual on average). This is quite an artificial initial state, but the main goal of the present work remains the analysis of long-term evolution. We thus prevent the early loss of the element, which would be highly likely if a more realistic initial state were used (e.g., only a few copies spread through the population) (27). Simulations are run for 20,000 generations, except if one of the following conditions is encountered: (i) the TE is totally lost (i.e., there are no more elements in the population), and (ii) there are too many elements in the system (the upper limit has been set to 2.106 elements in the whole population, i.e., 1,000 per genome).
Supplementary Material
Acknowledgments
We thank Malcolm Eden for review of the text. This work was supported by the Centre National de la Recherche Scientifique [UPR 9034 and GDR 2157 (entitled “Les éléments transposables: du génome aux populations”)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705238104/DC1.
References
- 1.Doolittle W, Sapienza C. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 2.Orgel LE, Crick FHC. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 3.Le Rouzic A, Dupas S, Capy P. Gene. 2007;390:214–220. doi: 10.1016/j.gene.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 4.Wong GK, Passey DA, Huang Y-Z, Yang Z, Yu J. Genome Res. 2000;10:1672–1678. doi: 10.1101/gr.148900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickey DA. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brookfield J. J Theor Biol. 1982;94:281–299. doi: 10.1016/0022-5193(82)90313-7. [DOI] [PubMed] [Google Scholar]
- 7.Charlesworth B, Charlesworth D. Genet Res Camb. 1983;42:1–27. [Google Scholar]
- 8.Charlesworth B, Sniegowski P, Stephan W. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 9.Le Rouzic A, Deceliere G. Genet Res Camb. 2005;85:171–181. doi: 10.1017/S0016672305007585. [DOI] [PubMed] [Google Scholar]
- 10.Quesneville H, Anxolabéhère D. Genetica. 1997;100:295–307. [PubMed] [Google Scholar]
- 11.Feschotte C, Mouchès C. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 12.Santiago N, Herraiz C, Goni J, Messeguer X, Casacuberta J. Mol Biol Evol. 2002;19:2285–2293. doi: 10.1093/oxfordjournals.molbev.a004052. [DOI] [PubMed] [Google Scholar]
- 13.Weiner A. Curr Opin Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- 14.Vassetzky N, Ten O, Kramerov D. Gene. 2003;319:149–160. doi: 10.1016/s0378-1119(03)00805-9. [DOI] [PubMed] [Google Scholar]
- 15.Tachida H, Iizuka M. Genetics. 1993;133:1023–1030. doi: 10.1093/genetics/133.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tachida H. Genetics. 1996;143:1033–1042. doi: 10.1093/genetics/143.2.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Rouzic A, Capy P. Genetics. 2006;174:785–793. doi: 10.1534/genetics.105.052241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackay TFC. Genet Res Camb. 1986;48:77–87. [Google Scholar]
- 19.Nuzhdin SV. Genetica. 1999;107:129–137. [PubMed] [Google Scholar]
- 20.Pasyukova E, Nuzhdin S, Morozova T, Mackay T. J Hered. 2004;95:284–290. doi: 10.1093/jhered/esh050. [DOI] [PubMed] [Google Scholar]
- 21.Schlenke T, Begun D. Proc Natl Acad Sci USA. 2004;101:1626–1631. doi: 10.1073/pnas.0303793101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller W, McDonald J, Nouaud D, Anxolabéhère D. Genetica. 1999;107:197–207. [PubMed] [Google Scholar]
- 23.Quesneville H, Nouaud D, Anxolabéhère D. Mol Biol Evol. 2005;22:741–746. doi: 10.1093/molbev/msi064. [DOI] [PubMed] [Google Scholar]
- 24.Kidwell MG. Curr Opin Genet Dev. 1992;2:868–873. doi: 10.1016/s0959-437x(05)80109-1. [DOI] [PubMed] [Google Scholar]
- 25.Silva J, Loreto E, Clark J. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 26.Sanchez-Gracia A, Maside X, Charlesworth B. Trends Genet. 2005;21:200–203. doi: 10.1016/j.tig.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Le Rouzic A, Capy P. Genetics. 2005;169:1033–1043. doi: 10.1534/genetics.104.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anxolabéhère D, Kidwell MG, Periquet G. Mol Biol Evol. 1988;5:252–269. doi: 10.1093/oxfordjournals.molbev.a040491. [DOI] [PubMed] [Google Scholar]
- 29.Kimura K, Kidwell MG. Genet Res Camb. 1994;63:27–38. doi: 10.1017/s0016672300032055. [DOI] [PubMed] [Google Scholar]
- 30.Biémont C. J Mol Evol. 1994;39:466–472. doi: 10.1007/BF00173415. [DOI] [PubMed] [Google Scholar]
- 31.Daniels SB, Chovnick A, Kidwell MG. Genetics. 1989;121:281–291. doi: 10.1093/genetics/121.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montchamp-Moreau C. Evolution (Cambridge, UK) 1990;44:194–203. doi: 10.1111/j.1558-5646.1990.tb04289.x. [DOI] [PubMed] [Google Scholar]
- 33.Bartolomé C, Maside X, Charlesworth B. Mol Biol Evol. 2002;19:926–937. doi: 10.1093/oxfordjournals.molbev.a004150. [DOI] [PubMed] [Google Scholar]
- 34.Quesneville H, Nouaud D, Anxolabéhère D. J Mol Evol. 2003;57:S50–S59. doi: 10.1007/s00239-003-0007-2. [DOI] [PubMed] [Google Scholar]
- 35.Bowen N, Jordan I, Epstein J, Wood V, Levin H. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prak E, Kazazian H., Jr Nat Rev Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- 37.Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. Genet Res Camb. 1988;52:223–235. doi: 10.1017/s0016672300027695. [DOI] [PubMed] [Google Scholar]
- 38.Mackay T, Lyman R, Jackson M. Genetics. 1992;130:315–332. doi: 10.1093/genetics/130.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlesworth B. Genet Res Camb. 1991;57:127–134. doi: 10.1017/s0016672300029190. [DOI] [PubMed] [Google Scholar]
- 40.Eanes WF, Wesley C, Hey J, Houle D. Genet Res Camb. 1988;52:17–26. [Google Scholar]
- 41.Houle D, Nuzhdin SV. Genet Res Camb. 2004;83:7–18. doi: 10.1017/s0016672303006505. [DOI] [PubMed] [Google Scholar]
- 42.McClintock B. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 43.Mackay TFC. Genetics. 1985;111:351–374. doi: 10.1093/genetics/111.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal A, Eastman QM, Schatz DG. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 45.Casacuberta E, Pardue M. Proc Natl Acad Sci USA. 2003;100:3363–3368. doi: 10.1073/pnas.0230353100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jordan I, Rogozin I, Glazko G, Koonin E. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 47.Gould SJ. The Structure of Evolutionary Theory. Cambridge, MA: Belknap Press of Harvard Univ Press; 2002. [Google Scholar]
- 48.Brookfield J. Nat Rev Genet. 2005;6:128–136. doi: 10.1038/nrg1524. [DOI] [PubMed] [Google Scholar]
- 49.Charlesworth B, Lapid A, Canada D. Genet Res Camb. 1992;60:103–114. doi: 10.1017/s0016672300030792. [DOI] [PubMed] [Google Scholar]
- 50.Nuzhdin SV, Mackay TFC. Mol Biol Evol. 1995;12:180–181. doi: 10.1093/oxfordjournals.molbev.a040188. [DOI] [PubMed] [Google Scholar]
- 51.Suh D, Choi E, Yamazaki T, Harada K. Mol Biol Evol. 1995;12:748–758. doi: 10.1093/oxfordjournals.molbev.a040253. [DOI] [PubMed] [Google Scholar]
- 52.Maside X, Assimacopoulos S, Charlesworth B. Genet Res Camb. 2000;75:275–284. doi: 10.1017/s0016672399004474. [DOI] [PubMed] [Google Scholar]
- 53.Eggleston W, Johnson-Schlitz D, Engels W. Nature. 1988;331:368–370. doi: 10.1038/331368a0. [DOI] [PubMed] [Google Scholar]
- 54.Dolgin E, Charlesworth B. Genetics. 2006;174:817–827. doi: 10.1534/genetics.106.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrov D, Aminetzach Y, Davis J, Bensasson D, Hirsh A. Mol Biol Evol. 2003;20:880–892. doi: 10.1093/molbev/msg102. [DOI] [PubMed] [Google Scholar]
- 56.Lynch M, Conery J. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 57.Vinogradov A. Curr Opin Genet Dev. 2004;14:620–626. doi: 10.1016/j.gde.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 58.San Miguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 59.Hua-Van A, Daviere J, Kaper F, Langin T, Daboussi M. Curr Genet. 2000;37:339–347. doi: 10.1007/s002940050537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.