Abstract
LINE-1 (L1) is a highly successful autonomous non-LTR retrotransposon and a major force shaping mammalian genomes. Although there are about 600 000 L1 copies covering 23% of the rat genome, full-length rat L1s (L1Rn) with intact open reading frames (ORFs) representing functional master copies for retrotransposition have not been identified yet. In conjunction with studies to elucidate the role of L1 retrotransposons in tumorigenesis, we isolated and characterized 10 different cDNAs from transcribed full-length L1Rn elements in rat chloroleukemia (RCL) cells, each encoding intact ORF1 proteins (ORF1p). We identified the first functional L1Rn retrotransposon from this pool of cDNAs, determined its activity in HeLa cells and in the RCL cell line the cDNAs originated from and demonstrate that it is mobilized in the tumor cell line in which it is expressed. Furthermore, we generated monoclonal antibodies directed against L1Rn ORF1 and ORF2-encoded recombinant proteins, analyzed the expression of L1-encoded proteins and found ORF1p predominantly in the nucleus. Our results support the hypothesis that the reported explosive amplification of genomic L1Rn sequences after their transcriptional activation in RCL cells is based on L1 retrotransposition. Therefore, L1 activity might be one cause for genomic instability observed during the progression of leukemia.
INTRODUCTION
LINE-1s (Long Interspersed Nuclear Element-1s or L1s) represent a family of abundant non-long-terminal repeat (non-LTR) retrotransposons in mammalian genomes. More than 99.8% of L1s are retrotransposition-defective because they are 5′ truncated, contain internal rearrangements or harbor mutations within their open reading frames (ORFs) (1–3). However, the average human genome is estimated to contain 80–100 retrotransposition-competent (RC) L1s, and ∼10% of these elements are classified as highly active or ‘hot’ (4). By comparison, the mouse genome is estimated to contain at least 3000 active L1s (5,6). L1s have shaped mammalian genomes through a number of mechanisms (7,8). First, they have greatly expanded the genome both by their own retrotransposition and by providing the machinery necessary for the retrotransposition of other mobile elements, such as Alus and processed pseudogenes. Second, they have shuffled non-L1 sequences throughout the genome by a process termed transduction. Third, they have affected gene expression by several mechanisms.
The best-studied L1s are those found in mice and men. There are at least 599 000 copies of mouse L1 (L1Md) that are interspersed in all chromosomes, and together comprise ∼19% of the genomic DNA (9). RC L1Mds are ∼6500 nt in length. Full-length RC-L1s have a 5′ untranslated region (UTR) with internal promoter activity, two ORFs, a 3′ UTR that ends in an AATAAA polyadenylation signal and a poly A tail. The role of the ORF1-encoded protein (ORF1p) is poorly understood. ORF1p represents a basic protein that co-fractionates as a large ribonucleoprotein complex with L1 RNA in extracts of human and mouse cells (10–12), binds RNA and single-stranded (ss) DNA in vitro (13–15), and possesses nucleic acid chaperone activity (16). It was suggested that ORF1p is capable of protein–protein interactions (14). The ORF2 protein (ORF2p) provides the enzymatic activities of endonuclease (EN) (17) and reverse transcriptase (RT) (18) for a process termed ‘target-site primed reverse transcription’ (TPRT) (19,20) which is initiating the insertion of a new non-LTR retrotransposon copy into genomic DNA. For efficient retrotransposition, both functional L1-encoded proteins are required in cis and must be translated from the primary bicistronic transcript (21).
L1 is the only autonomous non-LTR retrotransposon in rodents. Approximately 600 000 copies, in variable stages of decay, comprise ∼23% of the rat genome (22). Considering the fraction of the genome it is occupying, L1 of Rattus norvegicus (L1Rn) is the most successful mammalian L1 retrotransposon identified so far. Following the mouse-rat split, L1 activity appears to have increased in rat. The 3′ UTR sequences define six rat-specific L1 subfamilies, represented by 150 000 copies that cover 12% of the rat genome. L1 copies accumulated over the same period in mouse cover only 10% of the genome. This higher accumulation of L1 copies could explain some of the size difference between the rat and mouse genome. Full-length L1Rn elements have a structural arrangement similar to that of L1Md and many elements are present as 7-kb full-length copies (Figure 1A) (23). The 5′ UTR of L1 in rodents is bipartite: there is a 5′ module that can occur as a tandem repeat and therefore is referred to as a monomer (24) and a 3′ module which is called the tether (25) that links the monomer to ORF1. Although the entire 5′ UTRs of the modern mouse and rat L1 subfamilies are not homologous, the regulatory property of the monomer was shown using transient expression assays (26). The 5′ one-third of murine ORF1s contains a hypervariable domain (length polymorphic region, LPR), which has uniquely been the site of repeated deletions and duplications, and in rat it is the locus of replacement base substitutions (25,27). The ancestral rat hypervariable domain contains a 21-bp sequence that is deleted in some modern L1Rn elements and a 66-bp sequence that is tandem-duplicated in some elements (28). The currently active L1 in rat is L1mlvi2 (or L1Rn) (29), but although evolution of L1Rn elements was intensively studied, structure and nucleotide sequence of a functional L1Rn element is unknown hitherto and there is only sparse information on expression of these elements.
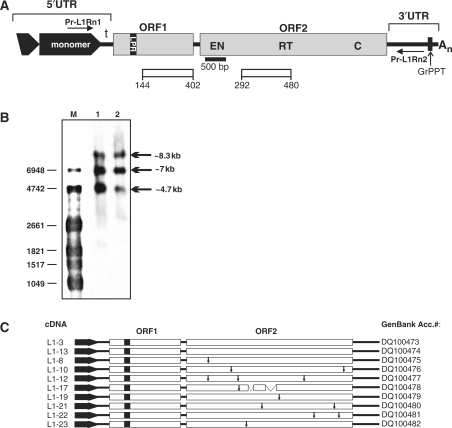
Figure 1.
Full-length L1Rn elements are transcribed in RCL cells. (A) Structure of L1Rn. A functional full-length L1Rn element is characterized by two ORFs flanked by 5′ and 3′ UTRs. The bipartite 5′ UTR consists of a monomer, which can be tandemly repeated, and a non-repeated tether (t). The most 5′ monomer is only partially duplicated (black truncated box with arrowhead) in all genomic rat elements identified so far. Horizontal arrows indicate the binding sites of the oligonucleotides Pr-L1Rn1 and Pr-L1Rn2 used to amplify cDNAs generated from full-length transcripts. The binding site of the 500-bp digoxigenin-labeled probe used to detect L1-specific transcripts is localized at the 5′ end of ORF2 (black bar). Open bars represent ORF1- and ORF2-encoded polypeptides against which monoclonal antibodies were raised. The polypeptides are covering amino acid positions 144–402 and 292–480 of ORF1p and ORF2p, respectively (accession no. DQ100480). LPR, length polymorphism region; GrPPT, G-rich polypurine tract; An, A-rich 3′ tract; EN, endonuclease; RT, reverse transcriptase; C, cysteine-rich motif. (B) L1Rn transcriptional products in RCL cells. PolyA+ RNA was isolated from RCL cells that had reached the maximum population density of ∼106 cells/ml (lane 2) and from cells that were UV-irradiated (lane 1). Two microgram of each RNA were separated by agarose gel electrophoresis and subjected to northern blot analysis using a 500-bp probe (Figure 1A). (C) Schematic structures of 10 L1Rn cDNAs synthesized from poly(A)+ RNA from UV-irradiated RCL cells. cDNAs are flanked by primer sequences Pr-L1Rn1 and Pr-L1Rn2. Names of the resulting cDNAs are listed on the left, while accession numbers are specified on the right. Termination codons within ORF2 sequences are indicated by vertical arrows. Two deletions in the ORF2-coding region of cDNA L1-17 covering 103 and 220 nts, respectively, are indicated by interrupted bars.
To date, transcriptional activation of L1Rn sequences by ∼10-fold was demonstrated in RCL cells reaching ∼50% of their maximal population density in suspension culture (30). Also, UV irradiation was shown to rapidly induce L1Rn transcription during fast exponential growth of these cells (31). L1Rn RNA could be detected in liver, kidney and skeletal muscle of the adult rat (23), in kidney tissue during the neonatal period (32) as well as in multipotent neural progenitor cells and neurons (33). Recently, L1Rn transcription was shown to be up-regulated in adult cardiac tissue after ischemia-reperfusion and the presence of L1-encoded proteins could be demonstrated (34).
Here we describe isolation and characterization of the first functional rat LINE-1 retrotransposon from a pool of 10 different cDNAs generated from actively transcribed endogenous full-length L1Rn elements which are all coding for intact ORF1ps in rat chloroleukemia (RCL) cells. We determined the retrotransposition activity of this active L1Rn element in HeLa cells as well as in the RCL cell line from which the cDNAs originated. This is the first report on a functional endogenous L1 element that is mobilized in the tumor cell line in which it was shown to be endogenously expressed. In order to investigate L1 translation and subcellular localization of L1-encoded proteins in rat cells, we generated monoclonal antibodies directed against recombinant ORF1p and ORF2p. Our experiments revealed that both L1-encoded proteins are expressed from endogenous elements in RCL cells. While endogenous ORF1 proteins appeared almost exclusively in the nucleus, transiently expressed ORF1p localized predominantly to the cytoplasm. Localization studies with ORF1p deletion mutants indicate that the N-terminal third of ORF1p has a function in cytoplasmic retention and targeting to cytoplasmic intermediate filaments. Our data demonstrating expression and retrotransposition of functional endogenous L1 elements in RCL cells support the idea that L1 activity might be at least one cause for the accumulation of molecular and chromosomal abnormalities leading to genomic instability which is characteristic for the progression of leukemia.
MATERIALS AND METHODS
Cell culture and UV irradiation
RCL cells growing in suspension culture (30) and HeLa S3 cells were kindly provided by K. Servomaa and by Dr A. Shatkin, respectively, and maintained in DMEM containing 10% FCS. Rat embryo fibroblasts (REFs) and mouse embryo fibroblasts (MEFs) from whole embryos were cultured in DMEM/10% FCS. Cells were transfected using FUGENE 6 lipid reagent (Roche). RCL cells were UV irradiated under following conditions: 312 nm (VL-215M; Bioblock Scientific), 4.65 mW/cm2, as measured with a VLH-3W radiometer equipped with a CX-312 filter.
Cloning of transcribed L1Rn sequences from RCL cells
Total and poly(A)+ RNA were prepared from control and irradiated RCL cells with the RNeasy and Oligotex mRNA kits (Qiagen), respectively. cDNA was synthesized from poly(A)+ RNA employing SuperScript™ II reverse transcriptase (Invitrogen) and oligo(dT) primer. About 6 kb long L1Rn sequences encompassing ORF1 and ORF2 were amplified from cDNA with the Taq/Pwo Proof Mix (AGS-Hybaid) and primers Pr-L1Rn1 (5′-GGA AGA GAC CAC CAA CAC TGC TCA C-3′, pos. 547 -571) and Pr-L1Rn2 (5′-GCT ACA TCC GGC TCC TGT CAG TCT GC-3′, pos. 6600 - 6576) specific for the monomeric region of the L1Rn 5′ UTR and for the 3′ UTR, respectively (accession no. M13100) (23,35). The PCR products were cloned into pBluescript II SK (+) (Stratagene). Ten recombinant plasmids containing L1Rn cDNA inserts were sequenced.
Northern blot analysis
Two microgram of poly(A)+ RNA from control and irradiated RCL cells were separated on standard formaldehyde-agarose gels, transferred to a nylon membrane (Hybond-N, Amersham Biosciences, Freiburg, Germany) by vacuum blotting in a Vacu-Blot System (Biometra, Göttingen, Germany), and fixed to the membrane by UV-crosslinking. A 500-bp digoxigenin-labeled double-stranded (ds) DNA probe specific for the EN-coding region of L1Rn (accession no. DQ100480) was generated by PCR using L1-21 (Figure 1A) as template, primers 5′-GGT CAG AAA AAG CAA GCT GGA GTA GCC-3′ (pos. 2111–2137) and 5′-AGG CCG TGG TGG TCC GAT AG-3′ (pos. 2610–2591), and digoxigenin-11-dUTP (Roche) at a molar ratio of 1:20 relative to dTTP. Hybridization was performed in ULTRAhyb ultrasensitive hybridization buffer (Ambion Europe Ltd.). The chemiluminescence-based detection of the digoxigenin moieties was performed as recommended by the manufacturer (Roche).
Retrotransposition reporter assay
Retrotransposition reporter plasmids pAD5/L1-3 and pAD6/L1-13 were generated by replacing the 5965-bp NotI/BstZ17I fragment of pJM101/L1.3 (36), which encompasses pos. 1-5965 of the human L1.3 element (37), by the 5860-bp NotI/EcoRV fragments of L1-3 and L1-13, respectively, comprising the entire particular cloned L1Rn-cDNA cassette. Antibiotic selection for transfected HeLa (0.4 mg/ml hygromycin) and RCL (0.7 mg/ml hygromycin) cells was started 1 and 3 days after transfection with FUGENE 6 lipid reagent, respectively. HeLa cells were selected for hygromycin resistance for 14 days, followed by G418 (1 mg/ml) selection for 11 days. Resulting colonies were stained with Giemsa solution. RCL cells growing in suspension were selected for hygromycin resistance for 29 days, followed by G418 (1.5 mg/ml) selection for at most 27 days. G418R RCL suspension cells were maintained and passaged in DMEM containing 10% FCS and 1.5 mg/ml G418. Live cell imaging was performed by trypan blue exclusion. 3′-junctions of L1-3 de novo retrotransposition events from the large background of HeLa genomic DNA were identified by EPTS/LM PCR (38) (Supplementary Data, Experimental Procedures).
Protein expression and purification
For expression of recombinant L1Rn ORF1p in Escherichia coli, the pET System (Merck Biosciences GmbH) was employed (Figure S6, A to C). Recombinant, 6xHis-tagged ORF1p, ORF1pΔ1–143, and ORF2pM292–480 were purified from soluble protein fractions and inclusion bodies via the cobalt IMAC resin Talon (BD Biosciences) technique, using gravity flow columns under native and denaturing conditions, as described in the user manual. Full-length recombinant Trx-6xHis-ORF1pΔ1–143, ORF1pΔ1–143-6xHis and ORF1p-6xHis, devoid of C- and N-terminal degradation products, were obtained by ssDNA-cellulose [heat-denatured calf thymus DNA (Sigma, Deisenhofen, Germany) covalently bound to CF-11 cellulose (Whatman, Göttingen, Germany)] chromatography. Briefly, to exchange buffer, protein samples were dialyzed against ssDNA-cellulose binding buffer (10 mM Tris–HCl, pH 7.6, 10 mM EDTA, 6 mM 2-mercaptoethanol, 6 M urea) and applied to a small column packed with ssDNA-cellulose. Bound protein was eluted with a 0–200 mM KCl gradient in binding buffer. The full-length products were detected in the fractions eluted at 150 mM KCl.
SDS–polyacrylamide gel electrophoresis and electroelution
Standard SDS–PAGE was carried out in Mini-Protean II dual slab cells (Bio-Rad, Munich, Germany). The separated proteins were stained with Coomassie brilliant blue or visualized by silver staining with the GelCode SilverSNAP stain kit (Perbio Science, Bonn, Germany). After preparative 9–15% gradient SDS–PAGE followed by copper staining, the desired protein bands were excised and electroeluted using the Model 422 Electro-Eluter (Bio-Rad), employing conditions recommended by the manufacturer.
Anti-ORF1p and -ORF2p mouse monoclonal antibodies
Recombinant Trx-6xHis-ORF1pΔ1–143 and -ORF2pM292–480 fusion proteins were expressed from pET32b-ORF1Δ1–143 and pET32b-ORF2M292–480 (Figure S6, A and F; Supplementary Data, Experimental Procedures), respectively, purified by electroelution from preparative SDS–polyacrylamide gradient gels and used for the immunization of BALB/c mice. Immunization and preparation of hybridoma cells were carried out as described in standard protocols. Myeloma P3X63-Ag8.653 cells were kindly provided by G. Hämmerling (DKFZ, Heidelberg, Germany) and maintained in RPMI-1640 (ICN Pharmaceuticals) supplemented with 15% FCS. For antibody production, hybridoma cells were grown in serum-free RPMI medium plus 10% HC-3 supplement, formulated on the basis of colostrum ultrafiltrate (BioConcept, Umkirch, Germany). Primary analysis and screening of hybridoma cells were performed employing the immuno-dot-blotting method. For that purpose, the tagged L1-encoded antigens used for immunization were immobilized on a nitrocellulose membrane (0.45 µm; Schleicher & Schuell, Dassel, Germany), which was fixed in a 96-well dot blot chamber (‘Minifold’, Schleicher & Schuell). To exclude hybridoma clones producing antibodies against Trx- and/or 6xHis tags, all selected clones in the primary analysis were then tested using total cell lysate of bacteria expressing the Trx-6xHis fusion protein from pET32b(+) (Novagen). Mice immunized with Trx-6xHis-ORF1pΔ1–143 and -ORF2pM292–480 fusion proteins were used to obtain hybridoma clones referred to as rG24 (isotype IgG2, κ light chain) and 2H9 (isotype IgM, κ light chain), respectively. The specificity of the monoclonal antibodies was further tested by immunofluorescence staining of transfected cells, immunoprecipitation, immunoblotting and/or mass spectrometry. Antibodies from supernatants of the hybridoma cells were used either directly or, depending on isotype, purified by affinity chromatography on protein G-Sepharose (Amersham Biosciences) or protein l-agarose (Perbio Science).
Indirect immunofluorescence
Adherent (REF and Hela) and suspension (RCL) cells were plated in a 6-well culture plate on untreated or poly-l-lysine (Sigma)-coated glass coverslips, respectively, at a density of 5 × 104 cells/well, fixed with 4% paraformaldehyde in PBS and permeabilized with 1% Triton X-100 in PBS. Immunostaining of cells was done as previously described (39). For in situ protein overlay, MEF cells were fixed with 4% paraformaldehyde in 1×PBS and permeabilized with 1% Triton X-100 in 1×PBS [as described for conventional immunostaining (39)], incubated in 1×PBS (pH 7.4) containing 1% bovine serum albumin (BSA) for 30 min, treated with a solution of recombinant ORF1p (50 µg/ml in 1% BSA/PBS, pH 7.4) for 30 min, washed three times with PBS (pH 7.4) and treated with 4% paraformaldehyde for 10 min. After washing and blocking in 1% BSA/PBS, cells were stained with anti-ORF1p (rG24) and affinity-purified polyclonal goat anti-vimentin antibodies (39). The cell specimens were examined on a Leica TCS SP2 confocal laser scanning microscope (CLSM) equipped with argon/krypton and neon ion lasers (Leica Lasertechnik, Heidelberg, Germany). In the case of double/triple labeling, the sequential mode of scanning was employed, capturing the images separately for each fluorescent dye used. To improve the signal-to-noise ratio, eight scans of one focal plane were averaged. Following image acquisition, the raw data were exported to the Huygens Essential software (version 2.7.2p0, Scientific Volume Imaging B.V., Hilversum, The Netherlands; http://www.svi.nl) and digital deconvolution was performed using the Maximum Likelihood Estimation (MLE) algorithm. The restored image data set was visualized and processed with the Imaris software package (version 4.1.3, Bitplane AG, Zürich, Switzerland; http://www.bitplane.com). Co-localization was analyzed with the ImarisColoc module integrated into the Imaris software package and the map of the co-localized voxels was saved as separate channel.
Immunoprecipitation of ORF1p and ORF2p
108 RCL cells were treated with 1 ml of lysis buffer (1% Triton X-100, 20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA) supplemented with Protease Inhibitor Cocktail Set III and briefly sonicated on ice. After pre-absorption with protein G-Sepharose or protein l-agarose (Pierce) for 1 h at 4°C, the cell lysate was supplemented with monoclonal anti-ORF1p (rG24) or anti-ORF2p (2H9) antibody at a ratio of 1 µg of antibody/100 µg of cellular protein, and gently shaken for 10–12 h at 4°C. Subsequently, 50 µl (packed bed volume) of protein G-Sepharose or protein l-agarose in lysis buffer were added and the suspension was incubated for another 4 h at 4°C. The matrix-bound immunocomplexes were pelleted by centrifugation and washed with lysis buffer five times for 10 min, before the immunocomplexes were eluted with SDS–PAGE sample buffer lacking 2-mercaptoethanol and analyzed by SDS–PAGE and silver staining.
Subcellular fractionation
Subcellular fractionation was performed using the ProteoExtract, Subcellular Proteome Extraction Kit (Calbiochem, Nottingham, UK). Protein fractions were enriched by acetone precipitation to use them for immunoblot analysis. Lamin B was used as a marker for nuclear and cytoskeletal matrix fractions.
Immunoblot analyses
RCL cell extracts and purified proteins were separated on SDS–polyacrylamide gels and electroblotted onto nitrocellulose membranes (Schleicher & Schuell). After protein transfer, membranes were blocked for 1 h in a 10% solution of non-fat milk powder in Tris-buffered saline (TBS, 10 mM Tris–HCl, pH7.6, 150 mM NaCl), and incubated with the respective primary antibody overnight at 4°C. To detect L1Rn-ORF2p, monoclonal 2H9 antibody was used in a 1:20 dilution in TBS/0.1% Tween 20 containing 1% milk powder, membranes were washed four times with TBS/0.1% Tween 20 and incubated with an HRP-conjugated, secondary anti-mouse IgM antibody (Dianova) at a dilution of 1:10 000 for 45 min. To detect L1Rn-ORF1p, primary monoclonal rG24 antibody was used at a dilution of 1:20, whereas the HRP-conjugated secondary anti-mouse IgG antibody was applied at dilutions of 1:10 000. After incubation with the secondary antibody, membranes were washed four times with TBS/0.1% Tween 20 and immunocomplexes were visualized using luminol-based ECL western blotting reagent (Amersham Biosciences). Anti-His Tag antibody G-18 (Santa Cruz Biotechnology), monoclonal anti-β-actin antibody (Sigma), and anti-α1-catenin antibody (Abcam) were used according to manufacturer's protocols. Digital gel images were captured with a Bio-Rad Fluor-S MultiImager and processed with Bio-Rad Quantity One Software (Bio-Rad) and Adobe Photoshop CS2 software (Adobe Systems Inc., Mountain View, CA, USA).
Dot blot overlay assay
Purified L1Rn-ORF1p was diluted in 1×PBS, pH7.4 at 0.66 µg/µl and applied onto a nitrocellulose membrane using a dot blot filtration apparatus (2 µg/dot). Protein samples were allowed to adsorb onto the membrane for 30 min before vacuum filtration. The membrane was incubated in blocking buffer (5% non-fat dry milk/1×TBS) for 30 min. The nitrocellulose membrane was cut into four squares each carrying one ORF1p dot. Subsequently, each membrane square was incubated in 2 ml blocking buffer supplemented with 5 µg/ml of purified vimentin, α-tubulin or β-actin, respectively. Cytoskeletal proteins bound to immobilized-ORF1p were detected employing primary antibodies specifically directed against each of the different cytoskeletal proteins and luminol-based ECL western blotting reagent. Antibodies used were goat anti-vimentin (39), mouse anti-β-actin/clone AC-40 (Sigma) and rat anti-α-tubulin/clone YOL 1/34 (Chemicon). Respective secondary antibodies were purchased from Santa Cruz Biotechnology. Recombinant mouse vimentin was produced in E. coli. Bovine α-tubulin and human β-actin were purchased from Cytoskeleton, Inc. (Denver, CO, USA).
In-silico analyses
Potential coiled-coil sequences in ORF1ps were searched for by accessing the MARCOIL server [http://www.isrec.isb-sib.ch/webmarcoil/webmarcoilC1.html; (40)]. Sequence alignments were calculated using the Clustal X (1.83) program (41). BLAST analysis of L1Rn sequences was performed using the L1base web portal [http://l1base.molgen.mpg.de/; (42)]. The collection of 377 L1Rn entries from this database was kindly provided by Tomasz Zemojtel (Max-Planck-Institute for Molecular Genetics, Berlin, Germany). Nuclear localization signals were predicted employing the WoLF PSORT (Protein Subcellular Localization Prediction) program [http://wolfpsort.seq.cbrc.jp; (43)] and PredictNLS server [http://www.rostlab.org/predictNLS/; (44)].
RESULTS
Isolation of cDNAs generated from polyadenylated full-length L1Rn RNAs
In order to identify functional L1Rn elements and characterize their expression, we were seeking for cell lines and/or growth conditions which are distinguished by an activation of L1Rn transcription. We chose RCL cells, since they were reported to increase transcription of endogenous L1s by ∼10-fold after they reached 50% of the maximum population density of the suspension culture, and by even more as a consequence of UV irradiation (30,31). Polyadenylated [poly(A)+] RNA was prepared from RCL cells that were growing under the aforementioned conditions activating L1 transcription, and submitted to northern blot analysis using a probe specific for the EN-coding region of L1 ORF2 (Figure 1A and B). Among the three detected transcriptional products, a predominant ∼7-kb band corresponds to full-length L1Rn transcripts. The probe also detected high-molecular-weight transcripts that migrated at 8.3 and ∼4.7-kb. The 8.3-kb transcript could be the consequence of transcriptional read-through of a full-length L1 element, while the latter might result either from transcription of distinct 5′-truncated L1Rn copies or from premature polyadenylation which has been shown to be conserved in mammalian L1s (45).
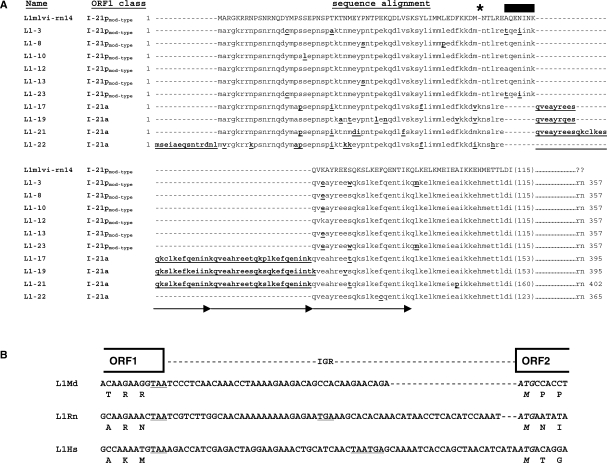
To identify and isolate full-length RC-L1s from rat, cDNA derived from RNA of UV- irradiated RCL cells was used as template for long-distance PCR employing the oligonucleotides Pr-L1Rn1 and Pr-L1Rn2 which are specific for the monomeric promoter region of the 5′ UTR (35) and for the 3′ UTR of L1Rn, respectively (Figure 1A). Ten resulting PCR products were cloned and sequenced (Figures 1C and S1), each consisting of ∼5.9 kb L1Rn sequences flanked by ∼640 nt 5′ UTR- and ∼260 nt 3′ UTR sequences. As a consequence of the chosen 5′ UTR-specific oligonucleotide Pr-L1Rn1, which binds position 156–181 of the functional L1Rn promoter region (26,35), the amplified 5′-UTR sequences are covering only the following 450–453 bp of the 608-bp promoter sequence (Figure S1). Although the most prominent sequence divergences were observed in the highly heterogeneous N-terminal third of ORF1p (Figures 2A and S2A), none of the 10 cloned L1Rn cDNA sequences carried any termination codon within ORF1. In contrast, only two isolated cDNAs, L1-3 and L1-13, represent typical mammalian L1-specifc bicistronic RNAs and are characterized by potentially functional ORF2p coding regions that are not interrupted by any termination codons, insertions and/or deletions (Figures 1C and S3). Based on the nomenclature of the heterogeneous ORF1 regions (25,29), we categorized our L1Rn sequences in the two classes I-21p and I-21a characterized by the presence (p) or absence (a) of a seven-amino acids insertion around position 64. Figure 2A shows an alignment of amino acid sequences of the hypervariable N-terminal third (N-1/3) of ORF1 proteins encoded by the cloned L1Rn cDNAs with the ORF1p sequence of L1mlvi2–rn14 (accession no. U87602), which is a member of the currently active L1 family in R. norvegicus (29). ORF1p sequences encoded by L1-3, -8, -10, -12, -13 and -23 are of the I-21pmod-type class, defining the modern, most recently emerged subfamily of L1Rn-encoded ORF1ps (25). Members of this family have the characteristic lysine deletion at position 59 and a 7-amino acid insertion in the N-1/3 region of ORF1p. Both deletion and insertion are absent in ORF1ps encoded by L1-17, -19, -21 and -22 (Figure 2A). L1-17-, -19- and -21-encoded ORF1ps include tandem repeats in the hypervariable N-terminal region. While L1-17 and -19 are coding for proteins carrying two slightly differing 22-amino acid tandem repeat insertions resulting from non-synonymous base substitutions, L1-21 is coding for a protein characterized by a 7-amino acid imperfect duplication in the first tandem repeat (Figure 2A). In sharp contrast to these I-21a class sequences, L1-22 is encoding an ORF1p lacking tandem repeats. L1-22 apparently has the potential for an alternative translational start site at nucleotide position -42, leading to an extension of ORF1p at its N-terminus by 14 amino acids (Figures 2A and S2A).
Figure 2.
Analyses of cDNA-encoded ORF1p classes and intergenic regions (IGR). (A) Alignment of the N-terminal third of the cDNA-encoded ORF1p sequences including the hypervariable region. Amino acid substitutions and insertions relative to the L1mlvi2-rn14 reference sequence (accession no. U87602) are in bold and underlined. Lysine deletion at position 59 (asterisk) and a seven-amino acids insertion (black bar) are hallmarks diagnostic for the I-21pmod-type class. Twenty-two-amino acids tandem repetitions are indicated by arrows. cDNA names and ORF1 classes to which the encoded protein sequences can be allocated, are listed. (B) Alignment of the non-coding IGR of the functional L1Rn-cDNA L1-3 with IGRs of functional L1Md (accession no. AY053455) and L1Hs (accession no. M80343) elements. The sequences start in the poorly conserved C-terminal regions of ORF1 and extend through the first ATG of ORF2. ORF1 stop codons as well as stop codons that are localized in the IGRs and are in frame with ORF1 are underlined; ATG start codons of each ORF2 are italicized.
Only 2 out of the 10 isolated L1Rn cDNA sequences, L1-3 and L1-13 (Figure 1C), have an intact ORF2 with coding capacity for a ∼150 kDa protein (1275 amino acids) including an N-terminal EN domain (residues 1–239), a central RT domain (residue 509–774) and a C-terminal zinc knuckle-like domain with a CX3CX7HC zinc-binding motif (residue 1131–1144). All three domains are highly conserved in mammalian L1-encoded ORF2 proteins (Figures 1A and S3). ORF2 regions of L1-8 and L1-23 are each disrupted by a termination codon (L1-8: 481CAG→ TAG; L1-23: 1372GAA→ TAA; Figures 1C and S3). The remaining six ORF2 sequences are characterized by termination codons and deletions (Figure 1C; accession numbers: DQ100476–DQ100481) [Nucleotide sequences of the L1Rn-specific cDNAs were deposited in the NCBI nucleotide database (GenBank accession numbers: DQ100473–DQ100482)]. The most prominent deletions covering 103 and 220 bp are found in the ORF2 region of L1-17 (Figures 1C and S1).
The intergenic non-coding spacer regions (IGR) between ORF1 and ORF2 of the different L1Rn cDNAs have lengths varying from 58 to 60 nt and are 99% identical to each other (Figures 1A, C and S5). The spacer sequences are rich in adenines and bear resemblance to those from active mouse and human L1s (Figures 2B and S5). In the case of the intact L1-3 cDNA, the 60-base inter-ORF spacer contains two in-frame stop codons and separates both intact L1 ORFs from each other.
Taken together, our data show that in RCL cells a multitude of different full-length L1Rn copies are actively transcribed and the resulting RNAs are polyadenylated. While each identified cDNA encodes an intact ORF1p, only two cDNAs, L1-3 and L1-13, are coding for intact and more conserved ORF2 proteins and could have the potential to actively retrotranspose.
The L1-3 cDNA is retrotransposition-competent in cultured cells
We next determined whether the non-interrupted ORFs in L1-3 and L1-13 encode functional proteins, which are able to facilitate L1 retrotransposition. For that purpose, we used a cell culture assay that accurately detects authentic retrotransposition events (46,47). In this assay, L1 retrotransposition does not require an intact 5′ UTR if an exogenous promoter is provided. We replaced the human L1.3 sequence of pJM101/L1.3 covering the 5′UTR, both ORFs and the 5′ part of the 3′UTR (37) by the L1-3 and L1-13 cDNA sequences (Figure 1C) which are lacking 155 nt of the 5′ end of the L1Rn-specific promoter sequence (35). In the resulting retrotransposition reporter constructs, pAD5/L1-3 and pAD6/L1-13 (Figure 3A), transcription of the L1Rn cDNAs was under control of the cytomegalovirus (CMV) immediate early promoter. While the control construct pJM101/L1RP (48) as well as pAD5/L1-3 generated G418R foci in HeLa cells, both the EN-defective L1RP-mutant expressed by pJM101/L1RPH230A and pAD6/L1-13 failed to generate G418R foci (Figure 3B). pAD5/L1-3 generated retrotransposition events at frequencies of 30% relative to pJM101/L1RP, while pAD6/L1-13 did not retrotranspose at all (Figure 3B, Table 1). As both ORFs encoded by L1-13 are not destroyed by stop-codons or deletions, we conclude that single point mutations within the ORFs might have inactivated the retrotransposition capacity of the L1-13 element. cDNA sequences of the active L1-3 and the inactive L1-13 element vary in 9, 2 and 5 nucleotides in their 5′-UTR-, IGR and 3′-UTR sequences, respectively. Point mutations within the coding regions are resulting in 6 and 20 amino acid substitutions in ORF1p and ORF2p, respectively (Figures S2A and S3). However, the three blocks of amino acids, REKG (pos. 257–260), ARR (pos. 282–284) and YPAKLS (pos. 304–309), which are essential for L1 retrotransposition (46), localized close to the C-terminal end of L1-ORF1p and are highly conserved among rat, mouse, rabbit and human are also conserved between L1-3 and L1-13.
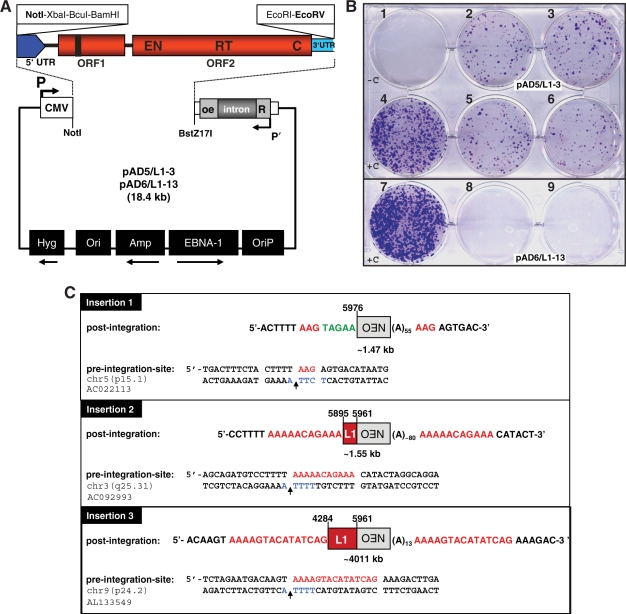
Figure 3.
The L1Rn element L1-3 retrotransposes at high frequency in HeLa cells. (A) L1Rn retrotransposition reporter constructs. The human L1.3 sequence of pJM101/L1.3 (flanked by NotI and BstZ17I sites) was replaced by the L1-3 and L1-13 cDNAs (flanked by NotI and EcoRV sites), respectively. The resulting reporter constructs are pAD5/L1-3 and pAD6/L1-13. (B) Results of the retrotransposition reporter assay. G418R foci were fixed to 6-well dishes and stained with Giemsa solution. At the timepoint of transfection with pAD5/L1-3 (well # 2, 3, 5, 6), pAD6/L1-13 (well # 8, 9) and the control constructs pJM101/L1RP (+C, well # 4, 7) and pJM101/L1RPH230A (–C, well # 1), respectively, each well was containing 2 × 105 HeLa cells. The negative control construct pJM101/L1RPH230A is expressing an EN-defective allele of L1RP. (C) Structures of three L1-3-derived genomic de novo insertions. Both post-integration sites and pre-integration sequences are shown. The nucleotide position of the 5′ truncation within the L1 reporter cassette is indicated. Numbering corresponds to the nucleotide position in the active L1-3 element (accession no. DQ100473). Insertion lengths range from 1.47 to 4.01 kb. All insertions are demonstrating the structural hallmarks of retrotransposition by TPRT as they are truncated at their 5′ ends, end in a poly(A)+ tail, and are flanked by short TSDs (red) of 3–15 bp. All three insertions integrated into sequences (blue) that are preferred sites for mammalian L1 integration in vivo (as denoted by the vertical arrows). The L1 endonuclease cleavage site on the bottom strand is indicated in blue. Notably, insertion #1 is truncated 15 nucleotides upstream of the 3′ end of the mneoI indicator cassette (pos. 5976). This insertion also resulted in five extra nucleotides (indicated in green) at the left junction between L1-mneo and genomic DNA.
Table 1.
Retrotransposition frequencies of tested L1 reporter constructs
| Construct | N | Retrotransposition frequency (×10−6) | Experimental range (×10−6) | Wild-type activity (%) |
|---|---|---|---|---|
| pJM101/L1RP | 3 | 2837 | 2470–3090 | 100 |
| pAD/L1-3 | 8 | 853 | 690–1000 | 30.1 |
| pAD/L1-13 | 8 | 0 | 0 | 0 |
| pJM101/L1RPH230A | 3 | 1.3 | 0–4 | 0.05 |
N = number of individual transfection experiments.
Next, we examined the genomic structures generated by retrotransposition events that derived from the active L1-3 element. Analysis of pre- and post-integration sites of three pAD5/L1-3-derived insertions isolated from HeLa cells (Figure 3C) revealed structural hallmarks of mammalian L1 retrotransposition, like 5′ truncations, target site duplications (TSDs), poly(A)+ tails at their 3′ ends, a 5′-TTTT/A-3′ consensus target sequence and, in one case, the insertion of five extra nucleotides between TSD and 5′-end of the L1 sequence (49,50). Taken together, structures of the post-integration sites indicate that they result from conventional TPRT as described for human L1 retrotransposition (19).
To determine whether the RC L1-3 cDNA is also able to retrotranspose in RCL cells the RNA was isolated from, cells were transfected separately and in triplicate with the L1Rn reporter constructs (pAD5/L1-3, pAD6/L1-13), and with the control constructs (pJM101/L1RPH230A, pJM101/L1RP) and selected for de novo retrotransposition by growth in G418-containing medium for up to 27 days (Figure 4A). Whereas RCL cells transfected with the negative control constructs pJM101/L1RPH230A and pAD6/L1-13 were completely eliminated after 25 days of G418-selection, G418R cells did arise after transfection with the active L1 constructs pAD5/L1-3 and pJM101/L1RP and grew to a maximum density of 1.3–1.5 × 106 cells/ml (Figure 4A). To test whether the G418R RCL suspension cultures that were derived from pAD5/L1-3- and pJM101/L1RP-transfected cells, are still able to proliferate, growth curves of each of the six different cultures were determined (Figure 4B). The observed exponential growth of the six G418R RCL cultures to a maximum density of ∼1.5 × 106 cells/ml is consistent with stable integration of intact neoR genes in each of the cultures.
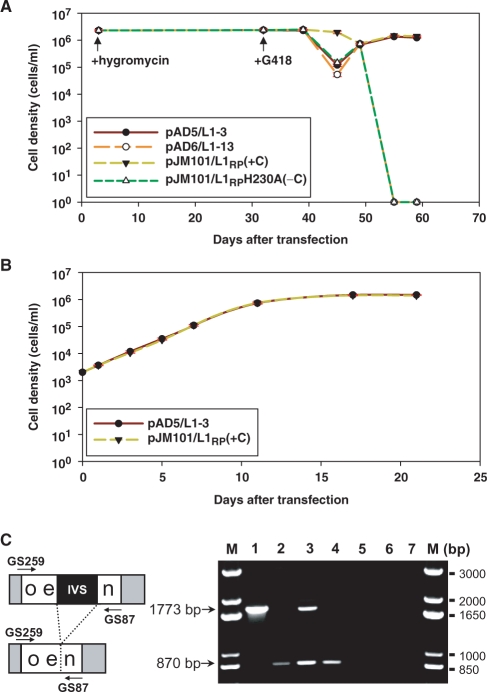
Figure 4.
Functional L1Rn elements retrotranspose in RCL suspension cells. (A) Growth curves of differently transfected RCL cells. Cells were transfected with pJM101/L1RP, pJM101/L1RPH230A, pAD5/L1-3 and pAD6/L1-13, separately, and were grown in hygromycin-containing medium for the following 29 days. Transfection experiments were performed in triplicate. In each of these experiments identical numbers of hygromycin-resistant cells (HygR) expressing the different L1 retrotransposition reporter constructs were then plated in medium containing G418 separately, and selected for G418 resistance for another 28 days. Live cells were counted by trypan blue exclusion. Transfection of RCL cells with pAD6/L1-13 or with the negative control construct pJM101/L1RPH230A led to the elimination of RCL cells after 18-25 days of G418 selection. By contrast, transfection with pJM101/L1RP and pAD5/L1-3 was leading to G418R cells that continued to grow at a density of 1.3–1.5 × 106 cells/ml. Each curve represents arithmetic means of cell densities resulting from three independent transfection experiments performed with the same reporter plasmid. Error bars were calculated but are barely visible because the courses of each of the three growth curves are almost identical. (B) When cell densities of pJM101/L1RP- and pAD5/L1-3-transfected suspension cultures reached their maximum after 26–30 days of G418 selection in all six independent transfection experiments, neoR-cells of each suspension culture were harvested to inoculate G418-containing medium at a density of 2 × 103 cells/ml in order to monitor growth curves of the differently transfected RCL cells. Rapid exponential growth without cell loss is demonstrating proliferation of pJM101/L1RP- and pAD5/L1-3-transfected and neoR-selected RCL cells in all three independent experiments. Error bars were calculated but are not visible as the courses of all six growth curves are almost identical. (C) PCR assay for correct splicing of the artificial intron in the neoR gene after retrotransposition from the reporter plasmids pJM101/L1RP and pAD5/L1-3. Genomic DNA was extracted from the differentially transfected RCL cells after 28 days of G418 selection, and used as template for PCR (lanes 3 and 4). This strategy allows distinction of the spliced and reverse-transcribed form of the neoR gene (870 bp PCR product) from the original unspliced form (1773 bp PCR product) expressed from the reporter construct and confirmed integration into the genome via authentic retrotransposition (80,81). PCR was performed on pAD5/L1-3 plasmid DNA mixed with genomic RCL DNA (lane 1), pSV2neo (BD Biosciences) mixed with genomic RCL DNA (lane 2), genomic DNA from pJM101/L1RP–transfected RCL cells (lane 3), and genomic DNA from pAD5/L1-3 –transfected RCL cells (lane 4); lanes 5 and 6, negative control genomic DNAs from untransfected RCL and HeLa cells, respectively; lane 7, minus template control; lane M, 1-kb Plus DNA ladder (Invitrogen).
To verify that the observed G418 resistance of pAD5/L1-3- and pJM101/L1RP- transfected RCL cells is a consequence of retrotransposition events launched from these plasmids, genomic DNA was extracted from G418R RCL cells after 28 days of G418 selection. Using the genomic DNAs as templates, PCR was performed employing neoR gene-specific primers flanking the intron in the neoR gene to determine whether the intron was spliced from the retrotransposition indicator cassette. The presence of an 870-bp PCR product amplified from genomic DNA, which was isolated from pAD5/L1-3- and pJM101/L1RP -transfected cells (Figure 4C) clearly indicates that the spliced functional neoR cassette was retrotransposed together with the respective L1 element. The presence of an additional 1773-bp PCR product in the case of pJM101/L1RP -transfected cells, which is indicative for the unspliced neoR cassette, is suggesting that the reporter plasmid either integrated into the genome or is still present episomally. Taken together, the data demonstrate authentic retrotransposition from pAD5/L1-3 and pJM101/L1RP in RCL cells. These results are also indicating that the functional genomic L1-3 element the isolated cDNA is derived from is actively retrotransposing in the RCL tumor cell line as well.
The L1Rn cDNAs we had identified could not be attributed unambiguously to defined L1 copies annotated in the present R. norvegicus genome database. This is not really surprising as it is known that only 50–60% of functional L1 sequences are fixed in mammalian genomes (4,51,52). This means that our particular active L1-3 sequence might not be annotated in the published rat genome sequence, simply because it does not cover all existing allele variations. Alternatively, we might not find our particular L1 sequence because the presently available sequence ‘draft’ is covering only ∼90% of the rat genome (22). Nonetheless, we screened L1base, a database containing L1Rn elements with intact ORFs (42), by a BLAST analysis with the L1-3 nucleotide sequence in order to identify additional potentially functional genomic L1Rn copies. Of the 377 full-length L1s with intact ORFs listed in L1base, we identified eight elements (Figure S4A) that differ from L1-3-encoded ORF1p and ORF2p only in ≤3 residues and ≤23 residues, respectively (Figure S4B).
Transiently expressed ORF1p appears in the cytoplasm with a speckled pattern and is localized to the nucleus
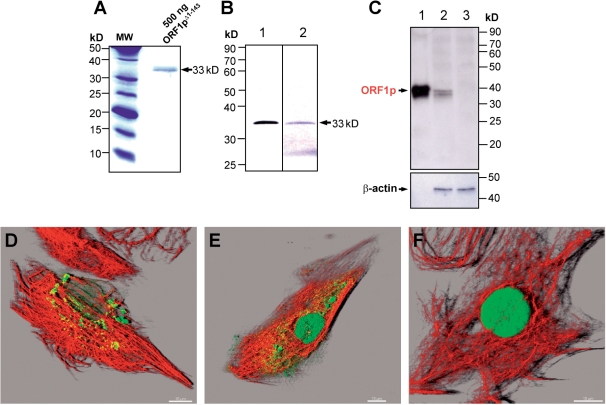
If the identified L1Rn mRNAs, which are coding for both functional ORFs are actively retrotransposing in RCL cells, the presence of both functional proteins, ORF1p and ORF2p, is an essential prerequisite (46). In order to be able to study the L1Rn-encoded protein machinery, we raised monoclonal antibodies against proteins encoded by intact L1 ORFs. To generate antibodies that are able to react with both major variants of intact ORF1p, we immunized mice with the recombinant, L1-21-encoded ORF1pΔ1–143 polypeptide (∼33 kDa) lacking the hypervariable N-terminal region (Figures 5A and S2A). We selected one hybridoma clone named rG24, which is producing IgG specifically recognizing an epitope in the His-tagged ORF1pΔ1–143 (Figure 5B) that is expected to be conserved among the different classes of L1Rn ORF1ps. To confirm the specificity of this antibody, we transiently transfected REFs with the L1Rn-ORF1 expression constructs pORF1-IRESpuro and pORF1Δ1–143-IRESpuro (Figure S6, D and E) and with the empty expression vector pIRESpuro (BD Biosciences). We chose REF cells because they do not contain any detectable amounts of RNA expressed from endogenous genomic full-length L1Rn elements (Tolstonog and Schumann, data not shown). Subsequently, transfected cells were tested for specific detection of recombinant ORF1-encoded proteins by immunoblot analysis (Figure 5C) and immunofluorescence staining with rG24 IgG (Figure 5D–F). The molecular weight of the polypeptide (∼33 kDa) detected with the rG24 antibody in pORF1Δ1–143-IRESpuro-transfected, but not in pIRESpuro-transfected REF cells (Figure 5C) is consistent with the theoretical mass of the ORF1Δ1–143 protein expressed in these cells. pORF1-IRESpuro-transfected cells exhibited predominantly cytoplasmic staining with the rG24 IgG (Figure 5D and E), indicating that the antibody is specifically interacting with the recombinant ORF1p. The absence of detectable staining in non-transfected as well as pIRESpuro-transfected REF cells (data not shown) is consistent with the lack of any traceable L1 RNA in these cells, indicates that expression of endogenous L1-encoded ORF1ps is either negligible or not existing, and supports the specificity of the rG24 antibody. Concurrently, transfection of REF cells gave us also the opportunity to study the intracellular localization of transiently expressed ORF1p and ORF1pΔ1–143 (Figure 5D–F). Co-localization analyses of deconvoluted immunofluorescence images of REF cells transiently transfected with pORF1-IRESpuro, demonstrate that ORF1p is predominantly cytoplasmic forming large aggregates with most of them being associated with vimentin filaments (Figure 5D). A small subset of transiently transfected cells (<1%) was characterized by cytoplasmic as well as nuclear localization of ORF1p (Figure 5E). These observations are in line with a previous report demonstrating predominant cytoplasmic aggregation of transiently expressed L1Hs ORF1p but also translocation of epitope-tagged L1Hs ORF1p into the nucleus (53,54). As demonstrated for human ORF1p (10) and recombinant mouse ORF1p (55), the overexpressed L1Rn ORF1 protein is probably able to form stable multimeric complexes, which cannot enter the nucleus (53). However, an exiguous pool of monomeric ORF1p may exist in the cytoplasm, at least when overexpressed, which are small enough to be transported into the nucleus (53). By contrast, ORF1pΔ1–143 was distinctly localized to the nucleus in all transfected cells (Figure 5F), suggesting that the N-terminal third of ORF1p may either be essential for the formation of multimers (10), which cannot passively enter the nucleus, or have a function in cytoplasmic retention as recently demonstrated for the human L1 ORF1p (54). The nuclear localization of the N-terminally truncated version of ORF1p could be explained by the putative bipartite nuclear localization signal KKEHMETTLDIENQKKR (residues 150–166 in ORF1p encoded by L1-21) and KRKK (residues 204–207) predicted in the central third of L1Rn ORF1p (Figure S2B) by the WoLF PSORT server (43). Our data are also consistent with the recent identification of a sequence within L1Hs ORF1p that was suggested to be responsible for the nuclear localization of truncated L1Hs ORF1p fragments (54).
Figure 5.
Monoclonal anti-ORF1pΔ1–143 antibodies (rG24) specifically recognize ORF1-encoded proteins. (A) Recombinant 33-kDa ORF1pΔ1–143 was used for the generation of monoclonal antibodies. MW, molecular weight marker. (B) Immunoblot detection of 10 ng of purified His-tagged ORF1pΔ1–143 with rG24 (lane 1) and anti-His-tag antibodies (lane 2). (C) Immunoblot analysis examining the specificity of the generated monoclonal anti-ORF1p antibody. Sixty microgram whole cell extracts from pORF1pΔ1–143-IRESpuro (lane 2)- or pIRESpuro-transfected (lane 3) REF cells were loaded on a 12% SDS–PAA gel. In contrast to the parental empty expression vector pIRESpuro, pORF1pΔ1–143-IRESpuro is expressing the N-terminally truncated 33-kDa L1Rn-ORF1 protein, which is detected by the rG24 antibody. As a loading control the membrane was stripped and incubated with an anti-β-actin antibody; lane 1, 10 ng purified ORF1pΔ1–143 protein; (D–F) Confocal images of REF cells transfected with ORF1p- (D, E) and ORF1pΔ1–143- (F) expressing constructs. ORF1-encoded proteins and vimentin were stained using monoclonal rG24 (green) and polyclonal anti-vimentin antibodies (red), respectively. The co-localization channel (yellow) was generated using the ImarisColoc module. Cytoplasmic ORF1p aggregates co-localize with vimentin filaments partially. Scale bar—10 µm.
Multimerization properties of L1Rn ORF1p
Several lines of evidence indicate that L1Rn ORF1ps have the inherent propensity to form oligo/multimers: (i) human, rabbit and mouse L1 ORF1p have the potential for leucine zipper- and coiled-coil formation (3,55,56) and in this respect as well as due to their nucleic acid binding activity (16) are quite similar to the intermediate filament (IF) proteins (57), and (ii) recombinant L1Rn ORF1p tends to aggregate under non-denaturating conditions forming filament-like structures that can be prevented only by the supplementation of protein solutions with urea or guanidinium chloride (Kirilyuk, Tolstonog and Traub, data not shown).
Applying the computer program MARCOIL which can predict the coiled-coil domain-characteristic heptad motif (abcdefg)n (40), we found that 84 amino acid residues in the conservative central third of L1Rn ORF1p follow a heptad motif (Figure S2B; I-21p class and I-21a class). An additional shorter heptad motif was detected in the N-terminal third of both ORF1p classes, although the presence of consecutive hydrophobic residues in positions 76 and 77 of I-21a class ORF1p introduces some ambiguity (‘stutter’) in assigning residues to the heptad positions. The in silico predicted polymerization potential of L1Rn ORF1p was supported by electron microscopy (Figure S7) and the in situ overlay procedure (Figure 6A–C). In this experiment, paraformaldehyde (PFA)-fixed and Triton X-100-permeabilized MEF were incubated with recombinant L1-21-encoded ORF1p, whose binding to cellular structures was stabilized by subsequent fixation with PFA and detected with the rG24 antibody (Figure 6A). Cytoskeletal vimentin filaments as the most prominent structure in MEFs were decorated with anti-vimentin antibody (Figure 6B). Under these conditions, ORF1p forms filamentous structures (Figure 6A) which partially (∼15% pixels) co-aligned with the vimentin filament network as demonstrated by co-localization analysis of the deconvoluted images (Figure 6C). This result points to a possible direct interaction between ORF1ps and cytoplasmic IFs or IF-associated proteins.
Figure 6.
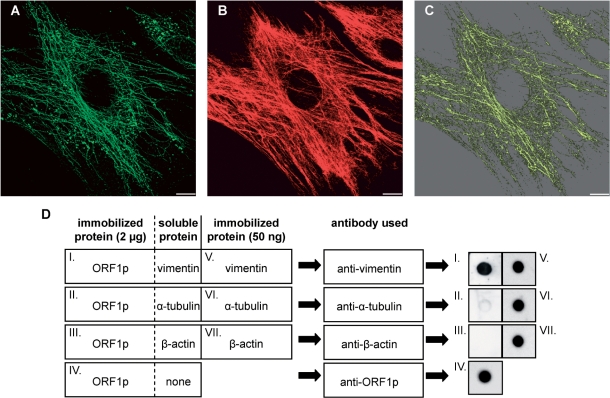
L1Rn ORF1p is able to form filaments that co-align with vimentin filaments. (A and B) In situ overlay of recombinant L1-21 ORF1p onto PFA-fixed and Triton X-100-extracted MEFs. Immunofluorescence images of L1Rn ORF1p (A) and vimentin (B) are shown in green and red colors, respectively. Each panel represents the 3D reconstruction of xy confocal sections. (C) ORF1p and vimentin filaments co-align partially. The co-localization channel (yellow) was generated using the ImarisColoc module. Scale bar—10 µm. (D) Dot blot overlay assay demonstrating interaction of ORF1p with vimentin. ORF1p was immobilized onto a nitrocellulose membrane (dots I–IV). Each ORF1p dot contained 2 μg of protein. After blocking, immobilized ORF1p was overlaid with an excess of soluble vimentin, α-tubulin or β-actin, respectively. Incubation was carried out for 60 min. In order to control for successful immobilization and immunoblotting procedure, 50 ng of soluble vimentin, α-tubulin and β-actin were immobilized on the nitrocellulose membrane in parallel (dots V–VII). Membranes were washed subsequently and submitted to immunoblot analyses with anti-vimentin, anti-α-tubulin or anti-β-actin antibodies, respectively, due to the schematic shown. In contrast to α-tubulin and β-actin, only vimentin can be detected with its specific antibody on the nitrocellulose membrane, after overlaying ORF1p dots with the respective IF protein (dots I–III). This is indicating that only vimentin is binding to ORF1p.
In order to confirm the interaction between vimentin and L1Rn ORF1p, we evaluated the affinity of recombinant ORF1p for several cytoskeletal proteins by far-western analysis (58). Dot blot overlay assays (Figure 6D) revealed a high affinity of L1Rn ORF1p for the cytoplasmic protein vimentin. In contrast, no binding could be observed for the cytoskeletal proteins α-tubulin and β-actin.
L1Rn ORF1- and ORF2-encoded proteins are expressed from endogenous elements in RCL cells
If the rG24 antibody is specifically recognizing L1Rn ORF1p and if the ORF1 regions of the identified full-length L1 RNA sequences are translated in RCL cells where L1 retrotransposition takes place, we would expect to be able to detect members of the two intact ORF1p classes, I-21p and I-21a, by immunoprecipitation with our antibody. Indeed, in silver-stained polyacrylamide gels we could visualize two polypeptides with estimated molecular weights of ∼40 and ∼46 kDa (Figure 7A) which correspond to the theoretical molecular masses of the two classes of ORF1p, I-21pmod-type and I-21a, calculated from both genomic ORF1 sequences (25,29) and isolated cDNA sequences (Figure 2A). As a next step, we wanted to determine the distribution of endogenous ORF1p in RCL cells where the expressed and tagged L1-3 cDNA was shown to retrotranspose. Confocal microscopy of RCL cells immunostained with the rG24 IgG localized the vast majority of ORF1p to the nucleus and only relatively low or barely detectable amounts to the cytoplasm (Figure 7B). To confirm nuclear localization of ORF1p, we next analyzed subcellular fractions of RCL cells for the presence of L1 ORF1p by immunoblotting with the rG24 antibody (Figure 7D). As expected ORF1-encoded proteins were absent from the cytoplasmic fraction and could be visualized predominantly in the nuclear fraction. Minor amounts of ORF1-encoded proteins could also be detected as faint bands in the cytoskeletal fraction which is consistent with the weak cytoplasmic immunostaining of RCL cells (Figure 7B). Endogenous ORF1 proteins as part of the cytoskeletal fraction are also in support of the proposed interaction between vimentin and ORF1p demonstrated in immunofluorescence experiments (Figure 6A–C) and dot blot overlay assays (Figure 6D). Molecular weights of the detected proteins correspond to the ORF1p classes, I-21p and I-21a, and are consistent with those verified by immunoprecipitation (Figure 7A).
Figure 7.
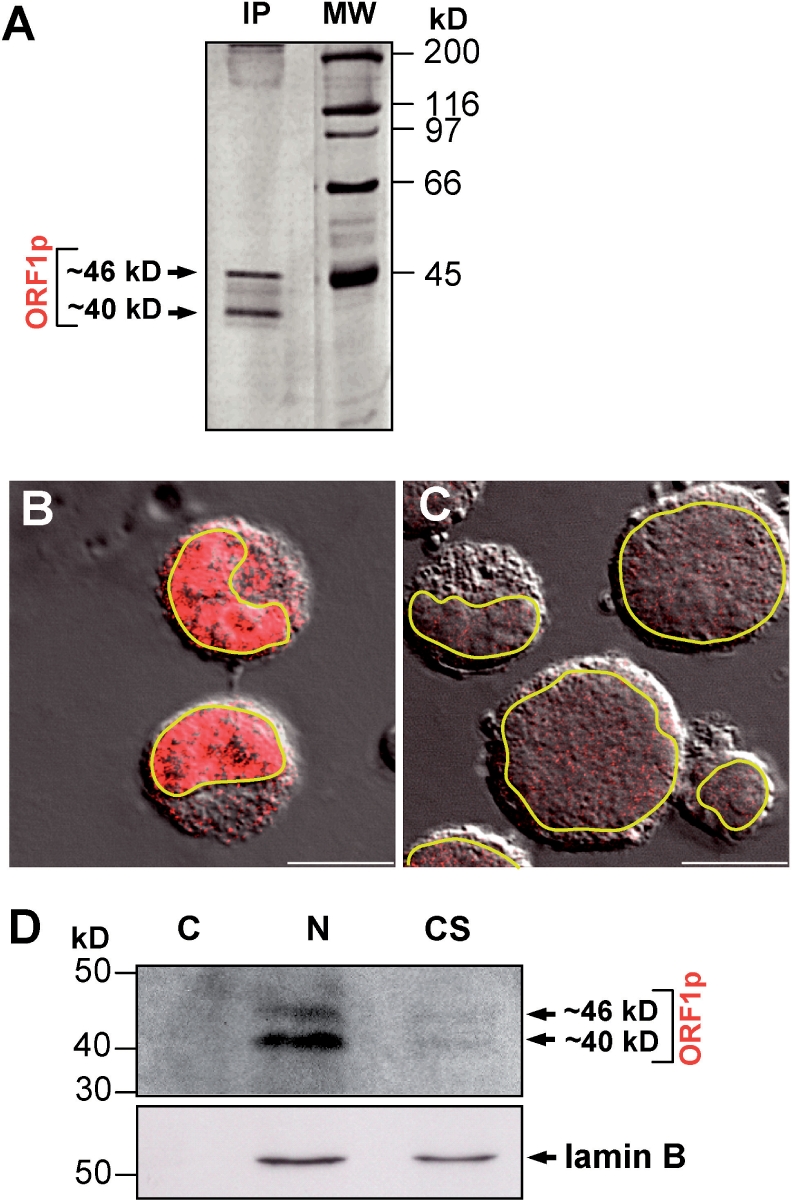
Endogenous L1Rn ORF1-encoded proteins are predominantly localized to the nucleus of RCL cells. (A) Silver staining of L1Rn ORF1-encoded proteins immunoprecipitated from RCL cell lysates with RG24 IgG. Molecular weights of the two proteins of ∼46 and ∼40 kDa correspond to the theoretical masses of L1Rn-ORF1 protein classes I-21a and I-21p, respectively. (B) Confocal images of RCL cells immunostained with the rG24 antibody (red). (C) Secondary antibody control. Confocal images were merged with a differential interference contrast (DIC) micrograph to show the outlines and nuclei of cells. Outlines of nuclear envelopes are emphasized in yellow. Scale bar—10 µm. (D) Immunoblot analysis of subcellular fractions of RCL cells. Three different protein fractions (C, cytosolic; N, nuclear; CS, cytoskeletal matrix proteins) were separated by SDS–PAGE and blotted onto nitrocellulose. The membrane was incubated consecutively with anti-ORF1p- antibody (upper panel) and with an antibody directed against the nucleus-specific laminB protein (lower panel), which shows efficient separation of the subcellular fractions.
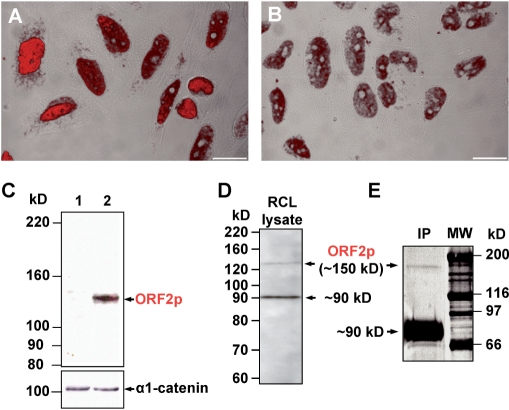
In order to generate mouse monoclonal antibodies specifically recognizing L1Rn ORF2p, the N-terminal fragment of the RT domain covering residues Met292 to 480 of ORF2p (Figure 1A, accession no. DQ100473) was expressed from pET32b-ORF2M292–480 (Figure S6F) in bacteria. The resulting ORF2M292–480–Trx-6xHis fusion protein was used as antigen for the generation of monoclonal anti-ORF2p antibodies. We identified one hybridoma cell clone (referred to as 2H9) producing IgM antibodies, which specifically recognize recombinant ORF2pM292–480 in bacterial lysates (data not shown). To test this antibody for its ability to detect native ORF2p in situ, we transfected the ORF2p expression construct pEF6-ORF2 (Figure S6G) into HeLa cells and immunostained them with the 2H9 IgM (Figure 8A). While HeLa cells that were transfected with the empty vector displayed only weak background immunoreactivity (Figure 8B) that may be explained by endogenous human ORF2 expression (59), transfection with pEF6-ORF2 resulted after only 48 h in a much more intense nuclear immunostaining. Detection of endogenous human L1 ORF2p with the 2H9 IgM could be possible as the antibody is directed against the RT domain which is highly conserved among human and murine L1s (Figure S3). To further confirm the specificity of the 2H9 antibody, we transiently transfected REF cells with ORF2p-expressing pEF6-ORF2 and its empty parental vector pET6V5-His, and tested for specific detection of L1Rn ORF2-encoded proteins by immunoblot analysis (Figure 8C). A ∼150-kDa protein was detected only in extracts from REF cells transfected with the ORF2p expressing plasmid, but not in those from cells transfected with the empty vector. This is strongly suggesting that the anti ORF2p IgM antibody is specifically recognizing L1Rn-ORF2 encoded proteins.
Figure 8.
The monoclonal anti-ORF2pM292–480 antibody 2H9 detects endogenous ORF2-encoded proteins. (A and B) Confocal images of HeLa cells transfected with either ORF2p-expressing pEF6-rORF2 (A) or the parental empty vector pEF6V5-His (B) and stained with the 2H9 IgM (red). (C) Immunoblot analysis examining the specificity of the generated monoclonal antiORF2p antibody. Fifty micrograms of whole cell extract from pEF6-rORF2 (lane 2)- or pEF6V5-His-transfected (lane 1) REF cells were loaded on a 6% SDS–PAA gel. In contrast to the parental empty expression vector pEF6V5-His, pEF6-rORF2 is expressing intact L1Rn ORF2p. As a loading control the membrane was stripped and incubated with an anti-α1-catenin antibody (lower panel). (D) Immunoblot analysis of RCL cell extract with monoclonal 2H9 antibody reveals two bands representing 150- and 90-kDa polypeptides. The 150-kDa band corresponds to the theoretical molecular mass of L1Rn ORF2p. (E) Silver staining of L1Rn ORF2-encoded proteins immunoprecipitated from RCL cell lysates with 2H9 IgM. Two polypeptides with apparent molecular masses of 150 and 90 kDa were detected and are consistent with the proteins detected by immunoblot analyses. Scale bar—10 µm.
Next, we wanted to evaluate whether we were able to detect ORF2 proteins in RCL cells. This would be expected, if actively transcribed endogenous L1Rn elements are mobilized in these cells, as suggested by the L1 retrotransposition reporter assays (Figure 4). Immunoblot analysis of RCL cell lysates with the 2H9 IgM uncovered two bands with estimated molecular masses of ∼150 and ∼90 kDa (Figure 8D). Detection of a protein with an approximate molecular mass of 150 kDa is consistent with the translation of L1mlvi2-ORF2p which is encompassing 1275 amino acids and is also encoded by the functional L1-3 element (accession no. DQ100473, Figure S3). Also, the weak signal intensity of the ∼150-kDa band (Figure 8D and E) would be consistent with a reduction of the translation rate of ORF2p relative to ORF1p which was reported to be the consequence of an unconventional termination/reinitiation mechanism during translation of human L1 elements (60). In order to test the possibility that the detected 90-kDa band could be caused by a cross-reacting protein species not related to L1 which is found in RCL but not in REF cells, we immunoprecipitated RCL cell lysates with the 2H9 IgM (Figure 8E) and subjected the ∼90-kDa polypeptide to tandem mass spectrometry (MS/MS) analyses. The obtained results revealed indeed that the 90-kDa protein recognized by the 2H9 antibody is the cellular Heat Shock Protein 90 (HSP90) (data not shown) which is involved in processing and presentation of cellular peptides (61).
Taken together, we were able to verify the presence of ORF1- and ORF2-encoded proteins in rat cells by applying monoclonal antibodies directed against recombinant L1Rn-encoded proteins. Molecular weights of the proteins detected with the anti-ORF1p IgM antibody, and the identification of a ∼150-kDa polypeptide applying the anti-ORF2p IgM antibody are consistent with the presence of functional L1 proteins.
DISCUSSION
Transcription of both functional and inactive endogenous full-length L1s coincides with L1 retrotransposition in RCL cells
The recently published genome sequence of R. norvegicus (22) shows that the rat L1 element is the most successful member of the mammalian L1 family identified until now. Although previous extensive studies of different structures of rat L1s were focusing on 5′UTR sequences and ORF1-coding regions (28), functional L1s whose protein machinery is responsible for the success of these mobile elements had not been identified so far. Therefore, we set out to identify functional and actively transcribed endogenous L1Rn elements, and to generate monoclonal antibodies to identify L1Rn-encoded proteins. Furthermore, we explored whether functional endogenous L1 elements are mobilized in a given tumor cell line which could support genome instability generally attributed to tumor cells.
In RCL cells that were grown under conditions supporting transcriptional activation of L1Rn (30,31,62), we detected significant amounts of L1-specific transcriptional products of three different sizes (Figure 1B). We were able to generate 10 L1 cDNAs from RNA isolated from UV-irradiated RCL cells, each covering nearly full-length L1 sequences. The fact that we isolated almost full-length cDNAs from RCL cells is suggesting that they are derived from the putative ∼7-kb full-length transcripts and/or the putative ∼8.3-kb read-through products. Six isolated L1Rn cDNAs, including the functional L1-3 sequence, are derived from the phylogenetically youngest family of L1Rn elements, the I-21pmod-type, and only four originate from the older I-21a class (25,29). While each of the cDNAs had coding capacity for intact ORF1ps, only two of them also included non-interrupted ORF2 coding regions, and therefore had the potential to represent RC-L1s. Applying a retrotransposition reporter assay in HeLa as well as in RCL cells, we tested the latter two cDNAs for their potential to retrotranspose. We demonstrated that only the L1-3 cDNA is functional and the transposition rate in HeLa cells amounted to 30% relative to the highly active human L1 element, L1RP. De novo insertions carry the hallmarks of human L1 retrotransposition initiated by TPRT (19). We further show that the tagged L1-3 cDNA can retrotranspose in the RCL tumor cell line the endogenous L1-3 element was originally expressed in and from which its RNA was isolated. Therefore, we conclude that the untagged, endogenously expressed L1-3 RNA is also retrotransposing in RCL cells. This conclusion is supported by a previous publication presenting evidence for transcriptional activation of endogenous L1Rn elements in RCL cells and subsequent amplification of genomic L1 sequences ∼24 h later (30). Activation of endogenous L1 expression is also observed in chronic myeloid leukemia (CML) cells isolated from CML patients (63), and it was shown to be caused by hypomethylation of L1 promoter sequences. It was suggested that such a reactivation of retroelements might contribute to genomic instability in cancer (64). Furthermore, it was hypothesized that ongoing L1 retrotransposition in CML cell lines might be an explanation for the observation that the transition of CML from the chronic phase to the blast crisis is characterized by the accumulation of molecular and chromosomal abnormalities (65).
Translation and subcellular localization of ORF1p
Similar to L1Hs, the L1Rn transcript is unusual because a 60-bp intergenic spacer containing two in-frame stop codons separates ORF1 and ORF2 (Figure 2B). It is likely that L1Rn-encoded ORF1p and ORF2p are translated from one full-length, bicistronic L1 mRNA by an unconventional termination/reinitiation mechanism (45,60,66–69). Also, the fact that our rG24 antibody reactive to native L1Rn ORF1p has only identified ∼40- and ∼46-kDa products argues against the synthesis of an ORF1p-ORF2p fusion protein (3). Consistently, immunoblot analyses of RCL cell extracts with the anti-ORF2p antibody led to the detection of a ∼150 kDa protein in RCL cells (Figure 8D). This again is in accordance with the detection of an L1 ORF2 protein with a similar molecular weight in human cells (59).
Our experiments indicate that both L1Rn ORF1p classes, I-21a and I-21p, are present in RCL cells (Figure 7A, B and D) The results are suggesting that there is no obvious bias in expression activity of any of the two classes, and that there is a highly heterogeneous group of ORF1ps expressed in these cells. Because 9 out of 10 isolated close-to-full-length cDNAs are retrotransposition-incompetent but are coding for intact ORF1ps, it can be concluded that the vast majority of the detected cellular ORF1ps is not part of functional RNP complexes. The heterogeneous pool of ORF1p visualized in the nuclei of RCL cells could also include defective proteins, which are encoded by mutated L1 ORF1 copies. Also, a fraction of the detected ∼40-kDa ORF1ps may be the result of expression of copies of the non-autonomous HAL1 element that is derived from L1 by deletion of its ORF2 (70). There are about 7500 HAL1 copies in the rat genome, and it was concluded that the currently identified HAL1-like elements (RNHAL1) still propagate (22). RNHAL1 contains only an ORF1, and the 5′ 2600 bases are 98% identical to the L1 currently active in rat (29). The repeated origin and high copy number of HAL1s suggest that the ORF1 product, which binds strongly to its messenger RNA (71), may render this transcript a superior target for L1-mediated reverse transcription. Taken together, the majority of these heterogeneous, endogenous ORF1p may not be able to be part of functional L1 RNPs, because of the absence of an intact ORF2 protein, or because they are not able to multimerize or interact with their full-length RNA due to mutations. These monomeric ORF1ps may be small enough to be transported into the nucleus and cause the predominant nuclear staining of RCL cells observed in Figure 7B. Accumulation of both the vast majority of endogenous ORF1p and a fraction of transiently expressed ORF1p in the nuclei of RCL and few REF cells (Figures 7B and 5E), respectively, could be explained by two predicted NLS in the conserved C-terminal two-thirds of both L1Rn-ORF1p classes (Figures 2A and S2B). The presence of an NLS in ORF1p is supported by the exclusive nuclear localization of the transiently expressed, N-terminally truncated ORF1pΔ1–143 in REF cells (Figure 5F). As, by contrast, transiently expressed ORF1p was found predominantly in the cytoplasm in an aggregated form (Figure 5D and E), the N-terminal third of ORF1p may either be essential for the formation of multimers (10) which cannot be transported into the nucleus, or have a function in cytoplasmic retention as recently demonstrated for the human L1 ORF1p (54).
Although endogenous ORF1p patterns in mouse and humans were described to be predominantly cytoplasmic (10,12), more recent publications are reporting that L1 ORF1p is also migrating into the nucleus: (i) Transiently expressed functional human ORF1p was detected mainly in the cytoplasm and in nucleolar sites in human osteosarcoma and MCF-7 cells (53,72). (ii) Immunohistochemical analyses revealed both cytoplasmic and nuclear localization of rat ORF1p in cardiomyocytes (34); (iii) Tagged human L1 ORF1p was visualized in the nucleus of human 143 Btk- and 293T cells (54).
Also, the ORF1-encoded Gag protein of TART, a LINE-like non-LTR retrotransposon from Drosophila, is found predominantly in the nucleus where it has a role in telomeric localization of TART (73). Although the precise function of ORF1p in L1 retrotransposition remains unclear, the nucleic acid chaperone activity of ORF1p proposed to contribute to reverse transcription by TPRT (56) would require the presence of functional ORF1p in the nucleus.
The interaction between ORF1p and vimentin observed under in vitro conditions raises the possibility that cytoskeletal proteins, such as IF proteins, might be important for the intracellular localization of ORF1p (Figure 6). The localization of RNA encoded by the LINE-like retrotransposon Doc in the region of the cytoskeleton of Drosophila oocytes (74) would be consistent with a role of IFs in intracellular localization of retrotransposon RNPs. Two possible roles of an interaction of ORF1p with intermediate filaments could be envisioned:
One of the steps of the L1 retrotransposition cycle that could be affected by this interaction is the transport of cytoplasmic RNP complex particles to the nucleus. Since intermediate filaments directly connect nuclear and plasma membranes (75), they could serve as a guide to direct RNPs toward the nucleus. ORF1p, as a major part of the L1 RNP and because of its affinity for vimentin, could consequently serve as an anchor molecule and allow the two-dimensional movement of RNPs along IFs through the cytoplasm to the nuclear membrane. Such a guided movement of RNPs would be significantly more efficient than undirected diffusion. A similar role in the guided movement of viral particles from the cytoplasm to the nucleus was proposed for the recently reported interaction between human immunodeficiency virus type 1 (HIV-1) Vif protein and vimentin (76). Theiler's murine encephalomyelitis (TMEV) capsid protein was also shown to interact with vimentin (77) but the function of this interaction is unknown.
Alternatively, vimentin may interact with L1 ORF1p as one component of stress granules (54). This would suggest a mechanism whereby the host cell may attenuate the potential mutagenic effects of retrotransposition by sequestering L1 RNPs and possibly targeting them for degradation (54). This hypothesis is supported by the finding that RNP particles of variable composition (stress granules) are associated with vimentin in Ptk1 and HeLa cells (78).
Unfortunately, ORF2p localization studies in RCL cells cannot be performed with our monoclonal 2H9 antibody raised against recombinant L1Rn ORF2p, because mass spectrometry revealed that this antibody is not only recognizing the L1Rn ORF2-encoded 150-kDa protein but is also cross-reacting with the cellular HSP90 protein. Nevertheless, this antibody is applicable for the study of recombinant L1Rn ORF2p transiently expressed in transfected cells (Figure 8C). In silico analyses of the L1-3-encoded ORF2p sequence on the PredictNLS server (44) revealed that the conservative amino acid sequence LRDKDR (residues 48–53, accession no. AAY88216) which is localized within the EN domain, is a putative NLS. Its functionality in the context of the EN domain is supported by the nuclear localization of transiently expressed full-length L1Rn ORF2p in HeLa cells (Figure 8A). The observation that transiently expressed full-length L1Hs ORF2p is mainly accumulated in the cytoplasm of HeLa cells (59) and 143B osteosarcoma cells (53) could be explained by differences between the putative NLS sequences of L1Rn-EN (LRDKDR) and the respective corresponding region in L1Hs-EN (LTDKDT) (Figure S3). Based on analyses on the PredictNLS server, the latter sequence motif is not consistent with a putative NLS function.
Applications
The functional L1Rn element L1-3 has the potential to serve as a tool in transgenic research. In recent years, transgenic rats were increasingly favored as model organisms to study cardiovascular diseases and brain physiology. Also, the laboratory rat is the principle model in the initial stages of drug development because their organ sizes allows more detailed investigations, which are problematic in mice (79). In transgenic rats, a tagged functional L1Rn element that is under control of a tissue-specific promoter will be useful as insertional mutagen or as a gene-trap tool helping to dissect genetic mechanisms of pathologic processes. Furthermore, the ORF1p-specific monoclonal antibody described in this work will enable cell-biological studies on L1 ORF1p with broad application to rat tumor models and will help expand our knowledge about L1 biology and the role of L1s in disease development and tumorigenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Annemarie Scherbarth and Ulrike Traub for providing tissue culture cells, Klaus Boller for crucial advice in immunofluorescence imaging, Heike Strobel and Katharina Grikscheit for excellent technical support, and Johannes Löwer for helpful discussions. We are indebted to Alexandre Zougman (Max-Planck Institute for Biochemistry, Martinsried) for performing NanoLC-ESI MS/MS. This work was supported in part by grants SCHU 1014/5-1 and 1014/5-2 from the Deutsche Forschungsgemeinschaft and grant Az.10.01.1.104 from the Fritz-Thyssen-Stiftung (to G.G.S.). Funding to pay the Open Access publication charges for this article was provided by the Paul-Ehrlich-Institut.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. EMBO J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 4.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nat. Genet. 1998;20:288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- 6.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- 8.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 9.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 10.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 11.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 12.Martin SL. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohjoh H, Singer MF. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J. Biol. Chem. 2003;278:8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- 15.Martin SL, Cruceanu M, Branciforte D, Wai-Lun LP, Kwok SC, Hodges RS, Williams MC. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 2005;348:549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 17.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 18.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 19.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 23.D'Ambrosio E, Waitzkin SD, Witney FR, Salemme A, Furano AV. Structure of the highly repeated, long interspersed DNA family (LINE or L1Rn) of the rat. Mol. Cell. Biol. 1986;6:411–424. doi: 10.1128/mcb.6.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padgett RW, Hutchison C.A., III, Edgell MH. The F-type 5' motif of mouse L1 elements: a major class of L1 termini similar to the A-type in organization but unrelated in sequence. Nucleic Acids Res. 1988;16:739–749. doi: 10.1093/nar/16.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabot EL, Angeletti B, Usdin K, Furano AV. Rapid evolution of a young L1 (LINE-1) clade in recently speciated Rattus taxa. J. Mol. Evol. 1997;45:412–423. doi: 10.1007/pl00006246. [DOI] [PubMed] [Google Scholar]
- 26.Nur I, Pascale E, Furano AV. The left end of rat L1 (L1Rn, long interspersed repeated) DNA which is a CpG island can function as a promoter. Nucleic Acids Res. 1988;16:9233–9251. doi: 10.1093/nar/16.19.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxton JA, Martin SL. Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome. J. Mol. Biol. 1998;280:611–622. doi: 10.1006/jmbi.1998.1899. [DOI] [PubMed] [Google Scholar]
- 28.Furano AV. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:255–294. doi: 10.1016/s0079-6603(00)64007-2. [DOI] [PubMed] [Google Scholar]
- 29.Hayward BE, Zavanelli M, Furano AV. Recombination creates novel L1 (LINE-1) elements in Rattus norvegicus. Genetics. 1997;146:641–654. doi: 10.1093/genetics/146.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Servomaa K, Rytomaa T. Suicidal death of rat chloroleukaemia cells by activation of the long interspersed repetitive DNA element (L1Rn) Cell Tissue Kinet. 1988;21:33–43. doi: 10.1111/j.1365-2184.1988.tb00769.x. [DOI] [PubMed] [Google Scholar]
- 31.Servomaa K, Rytomaa T. UV light and ionizing radiations cause programmed death of rat chloroleukaemia cells by inducing retropositions of a mobile DNA element (L1Rn) Int. J. Radiat. Biol. 1990;57:331–343. doi: 10.1080/09553009014552441. [DOI] [PubMed] [Google Scholar]
- 32.Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proc. Natl Acad. Sci. USA. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 34.Lucchinetti E, Feng J, Silva R, Tolstonog GV, Schaub MC, Schumann GG, Zaugg M. Inhibition of LINE-1 expression in the heart decreases ischemic damage by activation of Akt/PKB signaling. Physiol Genomics. 2006;25:314–324. doi: 10.1152/physiolgenomics.00251.2005. [DOI] [PubMed] [Google Scholar]
- 35.Furano AV, Robb SM, Robb FT. The structure of the regulatory region of the rat L1 (L1Rn, long interspersed repeated) DNA family of transposable elements. Nucleic Acids Res. 1988;16:9215–9231. doi: 10.1093/nar/16.19.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 37.Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt M, Hoffmann G, Wissler M, Lemke N, Mussig A, Glimm H, Williams DA, Ragg S, Hesemann CU, et al. Detection and direct genomic sequencing of multiple rare unknown flanking DNA in highly complex samples. Hum. Gene Ther. 2001;12:743–749. doi: 10.1089/104303401750148649. [DOI] [PubMed] [Google Scholar]
- 39.Hartig R, Huang Y, Janetzko A, Shoeman R, Grub S, Traub P. Binding of fluorescence- and gold-labeled oligodeoxyribonucleotides to cytoplasmic intermediate filaments in epithelial and fibroblast cells. Exp. Cell Res. 1997;233:169–186. doi: 10.1006/excr.1997.3543. [DOI] [PubMed] [Google Scholar]
- 40.Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- 41.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penzkofer T, Dandekar T, Zemojtel T. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res. 2005;33:D498–D500. doi: 10.1093/nar/gki044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 46.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 47.Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 48.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 49.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 50.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 51.Boissinot S, Chevret P, Furano AV. L1 (LINE–1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 52.Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH., Jr. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 2004;13:1041–1048. doi: 10.1093/hmg/ddh118. [DOI] [PubMed] [Google Scholar]
- 54.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNAi RISC. Mol Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin SL, Branciforte D, Keller D, Bain DL. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl Acad. Sci. USA. 2003;100:13815–13820. doi: 10.1073/pnas.2336221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin SL. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J. Biomed. Biotechnol. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traub P. Intermediate filaments and gene regulation. Physiol. Chem. Phys. Med. NMR. 1995;27:377–400. [PubMed] [Google Scholar]
- 58.Hall RA. Studying protein-protein interactions via blot overlay or far western blot. Methods Mol. Biol. 2004;261:167–174. doi: 10.1385/1-59259-762-9:167. [DOI] [PubMed] [Google Scholar]
- 59.Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, et al. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 60.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu. Rev. Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- 62.Luokkamäki M, Servomaa K, Rytomaa T. Onset of chromatin fragmentation in chloroma cell apoptosis is highly sensitive to UV and begins at non-B DNA conformation. Int. J. Radiat. Biol. 1993;63:207–213. doi: 10.1080/09553009314550271. [DOI] [PubMed] [Google Scholar]
- 63.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 64.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 65.Shet AS, Jahagirdar BN, Verfaillie CM. Chronic myelogenous leukemia: mechanisms underlying disease progression. Leukemia. 2002;16:1402–1411. doi: 10.1038/sj.leu.2402577. [DOI] [PubMed] [Google Scholar]
- 66.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr. Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 67.Kojima KK, Matsumoto T, Fujiwara H. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol. Cell Biol. 2005;25:7675–7686. doi: 10.1128/MCB.25.17.7675-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McMillan JP, Singer MF. Translation of the human LINE-1 element, L1Hs. Proc. Natl Acad. Sci. USA. 1993;90:11533–11537. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 71.Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rashkova S, Karam SE, Pardue ML. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc. Natl Acad. Sci. USA. 2002;99:3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao D, Bownes M. The RNA product of the Doc retrotransposon is localized on the Drosophila oocyte cytoskeleton. Mol. Gen. Genet. 1998;257:497–504. doi: 10.1007/s004380050675. [DOI] [PubMed] [Google Scholar]
- 75.Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 76.Karczewski MK, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nedellec P, Vicart P, Laurent-Winter C, Martinat C, Prevost MC, Brahic M. Interaction of Theiler's virus with intermediate filaments of infected cells. J. Virol. 1998;72:9553–9560. doi: 10.1128/jvi.72.12.9553-9560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olink-Coux M, Arcangeletti C, Pinardi F, Minisini R, Huesca M, Chezzi C, Scherrer K. Cytolocation of prosome antigens on intermediate filament subnetworks of cytokeratin, vimentin and desmin type. J. Cell Sci. 1994;107(Pt 3):353–366. [PubMed] [Google Scholar]
- 79.Tesson L, Cozzi J, Menoret S, Remy S, Usal C, Fraichard A, Anegon I. Transgenic modifications of the rat genome. Transgenic Res. 2005;14:531–546. doi: 10.1007/s11248-005-5077-z. [DOI] [PubMed] [Google Scholar]
- 80.Ostertag EM, DeBerardinis RJ, Goodier JL, Zhang Y, Yang N, Gerton GL, Kazazian HH., Jr A mouse model of human L1 retrotransposition. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 81.Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.