Abstract
An often limiting factor for studying protein folding by single-molecule fluorescence resonance energy transfer (FRET) is the ability to site-specifically introduce a photostable organic FRET donor (D) and a complementary acceptor (A) into a polypeptide chain. Using alternating-laser excitation and chymotrypsin inhibitor 2 as a model, we show that chemical labeling of a unique cysteine, followed by enzymatic modification of a reactive glutamine in an N-terminally appended substrate sequence recognition tag for transglutaminase (TGase) affords stoichiometrically D-/A-labeled protein suitable for single-molecule FRET experiments. Thermodynamic data indicate that neither the presence of the TGase tag nor D/A labeling perturbs protein stability. As the N terminus in proteins is typically solvent accessible, a TGase tag can (in principle) be appended to any protein of interest by genetic engineering. Two-step chemical/enzymatic labeling may thus represent a simple, low-cost, and widely available strategy for D/A labeling of proteins for FRET-based single-molecule protein folding studies, even for non-protein-experts laboratories.
Keywords: protein labeling, transglutaminase, fluorescence resonance energy transfer, single-molecule spectroscopy, alternating laser excitation, fluorescence-aided molecular sorting
Fluorescence resonance energy transfer between a single donor (D) and a single acceptor (A) (single-pair or sp-FRET) is a sensitive method for monitoring conformational dynamics in biomolecules (Selvin 2000; Weiss 2000; Deniz et al. 2001; Ha 2001, 2004; Haran 2003). The FRET efficiency (E) depends on the D/A distance (R) according to E = [1 ± (R/R0)6]−1. The Förster radius R0 is a characteristic constant that corresponds to R at which E = 50% (Clegg 1992). For typical single-molecule fluorophores, R0 ~ 4–6 nm, allowing the use of sp-FRET as a distance ruler in the 2–8 nm range (Stryer and Haugland 1967).
As cysteines (Cys) are underrepresented in proteins, D/A labeling can be achieved by sequential modification of two engineered Cys in a polypeptide chain. However, unless the dye accessibility of the thiol side chains differs drastically, the first added fluorophore can react with either of the two Cys, resulting in mixtures of D-/A-labeled molecules and their dye-permutated A/D analogs. Unless such mixtures can be chromatographically separated (Ratner et al. 2002), this may lead to sample heterogeneity, as the photophysical properties of the conjugated fluorophore may depend on local environment (charge, pH, or hydrophobicity) (Moerner and Orrit 1999; Brasselet and Moerner 2000).
Methodologies to improve labeling specificity include total chemical synthesis of proteins (Jia et al. 1999; Deniz et al. 2000; Talaga et al. 2000); specific chemical modification/labeling of certain N-terminal residues (Gaertner et al. 1992; Geoghegan and Stroh 1992; Zhang and Tam 1996; Schuler and Pannell 2002; Tolbert and Wong 2002); segmental labeling/assembly of proteins using intein, chemical, or chemoenzymatic ligation (Kaiser et al. 1989; Dawson et al. 1994; Liu and Tam 1994; Muir et al. 1998);the use of protein–protein interactions to protect a Cys in a protein–protein interface (Jäger et al. 2005); in vitro transcription/translation with tRNAs aminoacylated with non-canonical amino acids (Kiga et al. 2002; Olejnik et al. 2005); or (most elegantly, but not yet generally available) the incorporation of nonnatural amino acid analogs in vitro (or in vivo) using genetically engineered orthogonal tRNA/tRNA synthetase pairs (Noren et al. 1989; Wang et al. 2002; Chin et al. 2003).
Enzymatic post-translational modification of proteins is an alternative approach toward site-specific labeling. Transglutaminase (TGase) catalyzes the acyl transfer reaction between the γ-carboxyamide group of an acceptor glutamine (Gln) residue and a primary amine donor (Folk 1980). Although the sequence requirements that render a Gln into a TGase substrate are not fully understood, tissue TGase seems to exhibit a very stringent sequence specificity and structural requirements for the amine acceptor side around the Gln residues (Gorman and Folk 1980a, b, Gorman and Folk 1981, 1984). Only clearly solvent-exposed Glns in a flexible environment are recognized as substrates (Coussons et al. 1992a,b; Matsumura et al. 1996). The substrate specificity toward the amine donor seems to be more relaxed; both natural primary amines and nonbiological substrates are recognized (Lorand et al. 1979; Gorman and Folk 1980a; Sato et al. 1996, 2000; Sato 2002; Tanaka et al. 2005). Studies with chimeric proteins, in which a short substrate recognition tag for tissue TGase (Folk 1980) (sequence Pro-Lys-Pro-Gln-Gln-Phe-Met) was appended at the N or the C terminus of the protein, allowed site-specific labeling with a single fluorophore (Sato et al. 1996; Taki et al. 2004) or derivatization with synthetic polymers (Sato 2002). These studies also showed that the first of the two Glns (Gln4) in the TGase tag used is the primary acceptor for the incoming amine.
Here we combine traditional chemical thiol labeling with enzymatic modification of an engineered, reactive Gln by TGase to obtain highly pure, stoichiometrically D-/A-labeled protein from recombinant sources, suitable for single-molecule FRET studies.
Results and Discussion
Model system and proposed labeling scheme
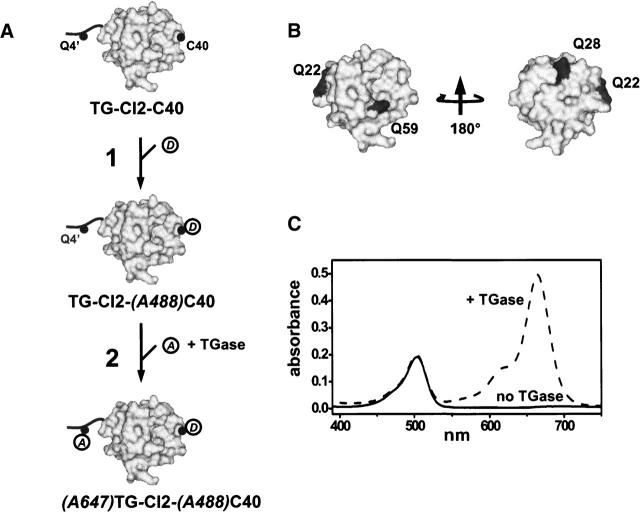
Chymotrypsin inhibitor 2 (CI2) (Fig. 1A) is a 64-residue, single-domain serine protease inhibitor and has served as a model system for experimental and theoretical protein folding studies (Daggett et al. 1996; Ladurner et al. 1998; Deniz et al. 2000; Kazmirski et al. 2001). A unique Cys residue (Cys40) was introduced at a solvent-exposed position into CI2 for site-specific labeling with Alexa488 maleimide (A488 hereafter). A substrate sequence-recognition tag (Pro-Lys-Pro-Gln-Gln-Phe) for tissue TGase was appended at the N terminus of CI2. TGase will catalyze the acyl transfer reaction between the primary amine of Alexa647 (A647 hereafter) cadaverine (A647–(CH2)6–NH2) and the γ-carboxyamine moiety of the reactive Gln (Gln4) in the TGase tag (Sato et al. 1996; Taki et al. 2004). The three internal Glns (Gln22, Gln28, Gln 59) in CI2 (Fig. 1B), although partially solvent-exposed, are embedded in a rigid tertiary structure context and should not be recognized as substrates for tissue TGase.
Figure 1.
(A) Labeling scheme of Chymotrypsin inhibitor 2 (CI2), represented in surface-accessible area mode (gray). The unique Cys40 is indicated by a solid black dot and labeled (C40). The added flexible N-terminal TGase tag is depicted by a solid black line with the reactive Gln indicated by a solid black dot and labeled (Q4′). A488-maleimide (A488) and A647-cadaverine (A647) are shown as open circles. (B) Location and solvent accessibility of the internal Glns (dark gray) in CI2 (light gray). (C) Labeling specificity of TGase. Absorbance spectra obtained with CI2-(A488)C40 (solid line) or TG-CI2-(A488)C40 (dotted line) after incubation with a 10-fold stoichiometric excess of A647 in the presence of TGase. Unreacted A647 was removed by filtration prior to acquisition of spectra. For comparison, spectra are normalized to the absorbance at 488 nm.
Labeling nomenclature
In the following, the Cys40 variant of CI2 without the N-terminal TGase tag is dubbed CI2-C40, and the tagged variant is abbreviated TG-CI2-C40. CI2, singly labeled with A488 at Cys40, is named CI2-(A488)C40 (no TGase tag) or TG-CI2-(A488)C40 (with TGase tag). D-/A-labeled CI2 is abbreviated (A647)TG-CI2-(A488)C40, with A647 attached to Gln4 in the appended N-terminal TGase tag and A488 conjugated to Cys40.
Labeling specificity of TGase
A prerequisite for the proposed labeling scheme (Fig. 1A) is that TGase modifies the solvent-exposed, reactive Gln in the N-terminal TGase tag (Gln4) but none of the semiexposed intrinsic Glns (Fig. 1B). To verify that only Gln4 in the appended N-terminal TGase tag is reactive, we incubated CI2-(A488)C40 or TG-CI2-(A488)C40 with a 10-fold stoichiometric excess of A647. The extent of A647 labeling was estimated from absorption spectra taken with the protein solution after removal of non-reacted A647 (extensive washing on a 3-kDa-cutoff membrane until the absorbance of the flow through at 647 nm was negligible). As TGase does not contribute to absorption in the visible range, it was not chromatographically removed at this stage. As shown in Figure 1C, TG-CI2- (A488) C40 exhibited two absorption bands with maxima at 488 and 647 nm (typical for A488 and A647, respectively). Only the A488 peak was detectable for CI2-(A488)C40, lacking the N-terminal TGase tag. We conclude that TGase labeling is specific for Gln4 in the flexible TGase tag; none of the three semiexposed intrinsic Glns is recognized as a substrate for TGase.
Labeling stoichiometry
Labeling stoichiometry was confirmed by matrix-assisted laser-desorption ionization (MALDI) mass spectrometry. A summary of the mass spectrometric analysis is given in Table 1. Raw mass spectra are provided as supplementary material.
Table 1.
Probing of labeling specificity and labeling stoichiometry by matrix-assisted-laser-desorption-ionization mass spectrometry
| Protein/fluorophore | Theoretical mass (Da) | Experimental mass (Da) |
| A488a | 720.20 | |
| A647b | 911.0c | |
| CI2-C40 | 7245.5d | 7235.6 |
| TG-CI2-C40 | 7971.4e | 7960.5 |
| TG-CI2-(A488)C40 | 8691.6 | 8679.4 |
| (A647)TG-CI2- (A488)C40 | 9602.6 | 9619.0 |
aAlexa-488 maleimide (http://www.probes.com).
bAlexa-647 cadaverine (A647-NH-CO-(CH2)5-NH2 (http://www.probes.com).
cEstimated from A647-succinimidylester (1020 Da) (http://www.idtdna.com).
dMolecular weight of CI2-C40 with the initiation-Met (sequence: MKTE…).
eMolecular weight of TG-CI2-C40 without the initiation-Met (sequence: PKPQQFMKT...).
Single-molecule FRET
Alternating-laser excitation (ALEX) is a novel single-molecule spectroscopic technique that allows fluorescence-aided molecular sorting (FAMS) of freely diffusing molecules by calculation of two ratiometric values (Kapanidis et al. 2004, 2005; Jäger et al. 2005; Lee et al. 2005). The traditional FRET efficiency E = FAemDexc/(FAemDem+ γF Dem Dexc) reports on the D/A distance; the distance-independent stoichiometry ratio S = FDexc/(FDexc + FAexc) reports on the relative D/A stoichiometry. FDemDexc is the D-excitation-based D emission, FAemDexc is the D-excitation-based A emission, FDexc is a sum of D-excitation-based emissions, FAexc is a sum of A-excitation-based emissions, and γ is a detection correction factor. Previous studies showed that γ ~1 (Ha et al. 1999), and γ = 1 was assumed in this study.
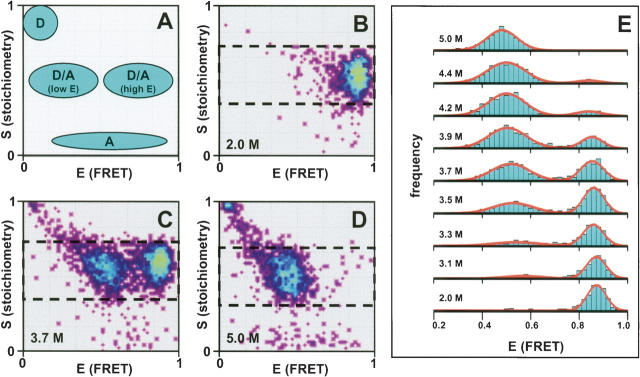
The expected location of D-only, A-only, and D/A sub-populations in a 2D E–S histogram is schematically depicted in Figure 2A. Experimental E–S histograms obtained with (A647)TG-CI2-(A488)C40 are depicted in Figure 2, B–D. At (or below) 2.0 M GdnCl, CI2 is folded and a single subpopulation with mean E ~0.9 and mean S ~0.55 is observed (Fig. 2B). A high E ratio indicates that the interdye distance is much smaller than the Förster radius (R0) for the A488/A647 dye pair (~56 ) (http://www.probes.com), consistent with a Cα –Cα distance of 28 Å between residue Cys40 and the N-terminal methionine in CI2 lacking the TGase tag (Radisky and Koshland 2002). Essentially no A488-only or A647-only subpopulations are visible, indicating that unreacted TG-CI2-(A488)C40 has been removed by ion-exchange chromatography (IEX) (Fig. 1A, step 2) and that fluorophore photobleaching during sample preparation/data acquisition is negligible. Addition of denaturant leads to unfolding of CI2, as indicated by the emergence of a second subpopulation with lower mean E (E ~0.45, corresponding to a D/A distance of ~55 Å assuming the experimentally determined R0 = 56 Å ) (Fig. 2C,D). At 3.6 M GdnCl (midpoint of unfolding), native and unfolded subpopulations are approximately equally populated. Complete unfolding is observed at (or above) 5 M GdnCl.
Figure 2.
Single-molecule FRET. (A) Schematic 2D E–S histogram showing the expected location of A488-only CI2, A647-only CI2, and A647/A488-labeled CI2 (folded protein, high-E; denatured protein, lower E). (B–D) Measured E–S histogram for A647/A488-labeled CI2 at 2 M GdnCl (B), 3.7 M GdnCl (midpoint of folding) (C), and 5 M GdnCl (D). The area of the E–S histograms selected for projection of the 1D E histograms (shown in E) is indicated by a dashed black box. (E) 1D E histograms of A647/A488-labeled CI2 at various concentrations of GdnCl. The solid red lines are fits of the data to Gaussian functions.
Figure 2E depicts 1D E histograms obtained with (A647)TG-CI2-(A488)C40 at denaturant concentrations close to or within the unfolding transition region (2–5 M GdnCl), where folded and unfolded conformers coexist. The FAMS capabilities of ALEX were exploited to construct these histograms by projection of only the region of the 2D E–S histogram onto the E-axis that contained photons from D/A-labeled molecules (selected area indicated by dashed black boxes in Fig. 2B–D; Kapanidis et al. 2004, 2005; Jäger et al. 2005; Lee et al. 2005). This option eliminates the contaminating D-only (and A-only) contribution to the 1D FRET efficiency E histogram, which becomes more pronounced in the presence of high concentrations of denaturant (Deniz et al. 1999; Schuler et al. 2002; McCarney et al. 2005). Although not required here, ALEX-FAMS also allows accurate mean E determinations for distances outside the dynamic FRET range (E < 0.1, due to D/A colocalization). For clarity, only the 0.2 < E < 1.0 range is shown in Figure 2E. The solid lines represent fits of the E histograms to Gaussian functions.
The disappearance of the high-E subpopulation under strongly denaturing conditions is further single-molecule evidence that the internal Glns in CI2 (Fig. 1B) are not recognized as substrates by TGase. Assuming that the sequence bracketed by Cys40 and any of the three internal Glns can be approximated by a random coil in >5 M denaturant (Kazmirski et al. 2001), the root-mean-square value of the distance r between the Cα atoms of Cys40 (attachment site for A488) and an internal Gln (potential acceptor site for A647) is given by 〈r2〉 01/2 = n1/2 · l (where n is the number of residue separating the two fluorophore chromophores and l is the bond length). With l = 3.8 Å (the length of a peptide bond in the trans-configuration), n = 12 (Gln28), 18 (Gln28), or 19 (Gln59) and R0 = 56 Å for the A647/A488 dye pair, this would lead to r = 13.1–16.5 Å, corresponding to a mean FRET efficiency E of ~1, even under conditions in which CI2 is fully denatured.
Thermodynamic analysis
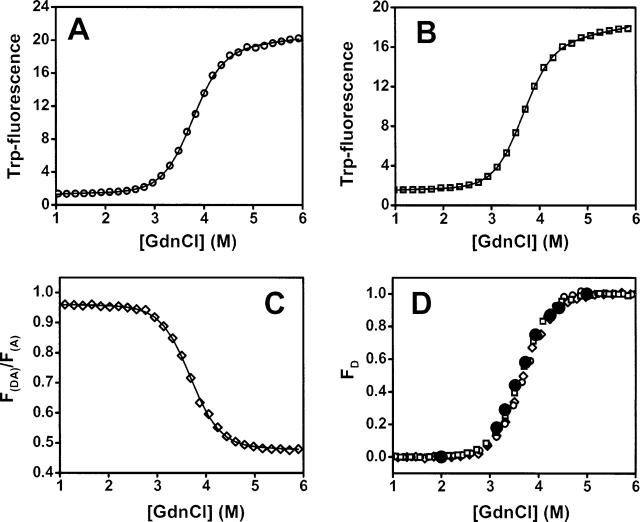
For TGase labeling to be useful for protein folding studies, it needs to be shown that the presence of the moderately hydrophobic N-terminal TGase tag (Pro-Lys-Pro-Gln-Gln-Phe) does not significantly perturb the folding energy landscape of CI2. To investigate this, we studied the equilibrium unfolding of CI2-C40 (no TGase tag) and TG-CI2-C40 (TGase-tagged) by monitoring the change in intrinsic Trp fluorescence (Fig. 3A,B). The effect of D/A labeling on CI2 stability was addressed by monitoring the unfolding of (A647)TG-CI2-(A488)C40 via A488-sensitized A647 emission (F(DA)/F(A)) as described elsewhere (Jäger et al. 2005; Fig. 3C). All three samples unfolded cooperatively and normalized denaturation curves are essentially super-imposable (Fig. 3D). Fitting the denaturation curves to a two-state unfolding model (Santoro and Bolen 1988) yielded the following thermodynamic parameters (ΔGN-D is the free energy of folding; meq is the unfolding cooperativity): (1) CI2-C40: ΔGN-D = 31.0 ± 0.6 kJ/mol, meq = 8.2 ± 0.2 kJ/mol M 1; (2) TG-CI2-C40: ΔGN-D = 30.4 ± 0.4 kJ/mol, meq = 8.4 ± 0.1 kJ/mol M− 1; (3) (A647)TG-CI2-(A488)C40: ΔGN-D = 30.2 ± 0.8 kJ/mol, meq = 8.2 ± 0.2 kJ/mol M−1.
Figure 3.
Equilibrium denaturation curves of (A) CI2-C40, (B) TG-CI2-C40, and (C) (A647) TG-CI2-(A488)C40 (measured by ensemble FRET). (D) Normalized curves are shown in A–C (same symbols used). The black-filled dots are data obtained from fitting the single-molecule FRET histograms shown in Figure 2E.
Also depicted in Figure 3D are data points obtained from Gaussian fitting of the histogram series shown in Figure 2E (solid black circles). The fraction of denatured protein (FD) at each GdnCl concentration was calculated from the integrated area of the folded (high-E) and denatured (low-E) subpopulations. The single molecule data reproduce the ensemble data that were obtained by assuming the validity of a two-state unfolding scenario. We conclude that neither the presence of the N-terminal TGase tag nor labeling of the tagged protein with single A647/A488 FRET-fluorophore pair significantly perturbs the folding thermodynamics of CI2.
Concluding remarks
Using CI2 as a model, we have shown that chemical labeling of a unique Cys, followed by site-specific enzymatic modification of a reactive Gln in an N-terminally TGase substrate recognition tag, affords site-specifically D/A-labeled CI2 for FRET-based single-molecule studies. A unique Cys in CI2 avoids sample heterogeneity due to the formation of A-/D-labeled and D-/A-labeled molecules, which is possible in conventional sequential labeling of two Cys in a polypeptide chain (Ratner et al. 2002). Mass spectrometry in conjunction with fluorescence-aided single molecular sorting verified the high substrate specificity of tissue TGase. In agreement with previous studies (Sato et al. 1996; Taki et al. 2004), only the solvent-exposed Gln in the flexible N-terminally appended TGase tag was labeled; none of the semiburied internal Glns has been modified. Even though this result may not be representative for other proteins, the frequency of Gln residues (3.7%) is below the statistically expected value, and in most cases, it should be possible to replace reactive internal Glns with near-isosteric substitutes (e.g., Asn, Glu) without compromising protein stability or function. Equilibrium unfolding experiments revealed that the TGase tag (although slightly hydrophobic) does not significantly affect protein stability. While this again may not be representative for all proteins, it should be mentioned that similar observations were made with Acyl-CoA binding protein (Kragelund et al. 1999) and Protein L (Scalley et al. 1997; M. Jäger and S. Weiss, unpubl.). Two-step chemical/enzymatic labeling of proteins may thus represent an attractive alternative to more costly or labor-intensive labeling strategies by affording a simple and widely accessible method for site-specific labeling of proteins for single-molecule FRET studies, even for non-protein-experts’s laboratories.
Materials and methods
Alexa488 maleimide (A488) and Alexa647 cadaverine (A647) were purchased from Molecular Probes. Bovine pig liver trans-glutaminase (TGase) was from Sigma. An expression plasmid for CI2 (Glu26Ala/Lys53Arg double mutant) was a gift from Dr. Daniel Koshland (UC Berkeley). Two variants of CI2 are discussed in this study. Variant 1 has a unique Cys at position 40 (dubbed CI2-C40), while variant 2 carries an additional N-terminal TGase substrate sequence recognition tag (dubbed TG-CI2-C40).
Protein expression and purification of CI2-C40 and TG-CI2-C40 were performed as described elsewhere (Radisky and Koshland 2002; Radisky et al. 2003). Labeling of CI2-C40 or TG-CI2-C40 with A488 was performed in 20 mM sodium phosphate (pH 7.0, dye:protein ratio = 2:1) for 4 h at 25° C in the dark. Unreacted dye was removed from the solution by repeated concentration/dilution (in 5 mM Tris at pH 8.0) of the protein solution on a Centricon YM3 membrane (Millipore) until the absorbance of the flowthrough fraction at 488 nm (emission maximum of A488) was negligible. Purification of CI2-(A488)C40 from trace amounts of unreacted protein was accomplished by ion-exchange chromatography (IEX) using a MonoQ 5/5-column (Amersham-Pharmacia). A linear salt gradient (0–500 mM sodium chloride in 5 mM Tris at pH 8.0, 45 min, 2 mL/min) was used [nonlabeled CI2 did not bind to the MonoQ-resin under these conditions; elution of CI2-(A488)C40 occurred at ~80 mM NaCl]. Enzymatic labeling of TG-CI2-(A488)C40 (10 μM) with A647 was performed in 100 mM Tris (pH 8.0), 1 mM A647, 10 mM CaCl, and 0.5 unit of TGase for 24 h at 25° C in the dark in a total volume of 1 mL. Unreacted A647 was removed by gel filtration, and (A647)TG-CI2-(A488)C40 was purified from unreacted TG-CI2-(A488)C40 and TGase by IEX [(A647)TG-CI2-(A488)C40 eluted at ~180 mM NaCl]. Yields of double-labeled CI2 were consistently >80%, as judged from the integrated peak areas of A488-labeled and A647/A488-labeled CI2 (based on excitation at 488 nm).
For molecular weight determinations using matrix-assisted laser desorption-ionization mass spectrometry (MALDI), protein samples were extensively washed with doubly distilled water on a Centricon YM3 membrane (Millipore) prior to data acquisition.
Thermodynamic parameters were obtained by fitting the data to a two-state unfolding model (Santoro and Bolen 1988) as described in detail elsewhere (Jäger et al. 2005).
Single-molecule measurements were performed as described elsewhere (Jäger et al. 2005) with minor modifications. A water-immersion inverted fluorescence microscope (Olympus IX17, 60 × 1.2 NA water-immersion objective, 100 μm pinhole), modified to allow alternating-laser excitation (ALEX) using a two-laser excitation source (488 nm Ar+-laser, 634 nm diode-laser) was used. All measurements were carried out in 20 mM sodium phosphate (pH 6.3) and 100 μg/mL bovine serum albumin. Protein concentration was 40 pM. The A488-excitation power was 100 μW, and the A647-excitation power was 45 μW. Only bursts containing >180 photons were retained for further analysis.
Acknowledgments
This work was funded by NIH grant no. 1R01-GM65382 (to S.W.) and the NSF. The Center for Biophotonics, an NSF Science and Technology Center, is managed by the University of California, Davis, under Cooperative Agreement no. PHY0120999. E.N. is supported by the Human Frontier Science Program (HFSP).
Abbreviations
A, FRET acceptor
A488, Alexa488 maleimide
A647, Alexa647 cadaverine
ALEX, alternating-laser excitation
CI2, chymotrypsin inhibitor 2
D, FRET donor
E, FRET efficiency
FAMS, fluorescence-aided molecular sorting
FRET, fluorescence resonance energy transfer
IEX, ion-exchange chromatography
MW, molecular weight
R0, Förster radius
S, ALEX-ratio S
TGase, transglutaminase
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051851506.
Supplemental material: see www.proteinscience.org
References
- Brasselet, S. and Moerner, W.E. 2000. Fluorescence behaviour of single-molecule pH-sensors. Single Mol. 1: 17–23. [Google Scholar]
- Chin, J.W., Cropp, T.A., Anderson, J.C., Mukherji, M., Zhang, Z., and Schultz, P.G. 2003. An expanded eukaryotic genetic code. Science 301: 964–967. [DOI] [PubMed] [Google Scholar]
- Clegg, R.M. 1992. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211: 353–388. [DOI] [PubMed] [Google Scholar]
- Coussons, P.J., Price, N.C., Kelly, S.M., and Fothergill-Gilmore, L.A. 1992a. The modification of bovine β-casein using transglutaminase purified from guinea pig liver. Biochem. Soc. Trans. 20: 48S. [DOI] [PubMed] [Google Scholar]
- Coussons, P.J., Price, N.C., Kelly, S.M., Smith, B., and Sawyer, L. 1992b. Transglutaminase catalyses the modification of glutamine side chains in the C-terminal region of bovine β-lactoglobulin. Biochem. J. 283 (Pt. 3): 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett, V., Li, A., Itzhaki, L.S., Otzen, D.E., and Fersht, A.R. 1996. Structure of the transition state for folding of a protein derived from experiment and simulation. J. Mol. Biol. 257: 430–440. [DOI] [PubMed] [Google Scholar]
- Dawson, P.E., Muir, T.W., Clark-Lewis, I., and Kent, S.B. 1994. Synthesis of proteins by native chemical ligation. Science 266: 776–779. [DOI] [PubMed] [Google Scholar]
- Deniz, A.A., Dahan, M., Grunwell, J.R., Ha, T., Faulhaber, A.E., Chemla, D.S., Weiss, S., and Schultz, P.G. 1999. Single-pair fluorescence energy transfer on freely diffusing molecules: Observation of Foerster distance dependence and subpopulations. Proc. Natl. Acad. Sci. 96: 3670–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz, A.A., Laurence, T.A., Beligere, G.S., Dahan, M., Martin, A.B., Chemla, D.S., Dawson, P.E., Schultz, P.G., and Weiss, S. 2000. Single-molecule protein folding: Diffusion fluorescence energy transfer studies of the denaturation of chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. 97: 5179–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz, A.A., Laurence, T.A., Dahan, M., Chemla, D.S., Schultz, P.G., and Weiss, S. 2001. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 52: 233–253. [DOI] [PubMed] [Google Scholar]
- Folk, J.E. 1980. Transglutaminases. Annu. Rev. Biochem. 49: 517–531. [DOI] [PubMed] [Google Scholar]
- Gaertner, H.F., Rose, K., Cotton, R., Timms, D., Camble, R., and Offord, R.E. 1992. Construction of protein analogues by site-specific condensation of unprotected fragments. Bioconjug. Chem. 3: 262–268. [DOI] [PubMed] [Google Scholar]
- Geoghegan, K.F. and Stroh, J.G. 1992. Site-directed conjugation of non-peptide groups to peptides and proteins via perjodate oxidation of a 2-amino alcohol. Application to modification at N-terminal serine. Bioconjugate Chem. 3: 138–146. [DOI] [PubMed] [Google Scholar]
- Gorman, J.J. and Folk, J.E. 1980a. Structural features of glutamine substrates for human plasma factor XIIIa (activated blood coagulation factor XIII). J. Biol. Chem. 255: 419–427. [PubMed] [Google Scholar]
- ———. 1980b. Transglutaminase amine substrates for photochemical labeling and cleavable cross-linking of proteins. J. Biol. Chem. 255: 1175–1180. [PubMed] [Google Scholar]
- ———. 1981. Structural features of glutamine substrates for transglutaminases. Specificities of human plasma factor XIIIa and the guinea pig liver enzyme toward synthetic peptides. J. Biol. Chem. 256: 2712–2715. [PubMed] [Google Scholar]
- ———. 1984. Structural features of glutamine substrates for transglutaminases. Role of extended interactions in the specificity of human plasma factor XIIIa and of the guinea pig liver enzyme. J. Biol. Chem. 259: 9007–9010. [PubMed] [Google Scholar]
- Ha, T. 2001. Single-molecule fluorescence energy transfer. Methods 25: 78–86. [DOI] [PubMed] [Google Scholar]
- ——— . 2004. Structural dynamics and processing of nucleic acids revealed by single-molecule spectroscopy. Biochemistry 43: 4055–4063. [DOI] [PubMed] [Google Scholar]
- Ha, T., Ting, A.Y., Liang, J., Caldwell, W.B., Deniz, A.A., Chemla, D.S., Schultz, P.G., and Weiss, S. 1999. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. 96: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran, G. 2003. Single-molecule fluorescence spectroscopy of biomolecular folding. J. Phys. Condens. Matter 15: R1291–R1317. [Google Scholar]
- Jäger, M., Michalet, X., and Weiss, S. 2005. Protein–protein interactions as a tool for site-specific labeling of proteins. Protein Sci. 14: 2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., Talaga, D.S., Lau, W.L., Lu, H.S.M., DeGrado, W.F., and Hochstrasser, R.M. 1999. Folding dynamics of single GCN-4 peptides by fluorescence resonant energy transfer confocal microscopy. Chem. Phys. 247: 69–83. [Google Scholar]
- Kaiser, E.T., Mihara, H., Laforet, G.A., Kelly, J.W., Walters, L., Findeis, M.A., and Sasaki, T. 1989. Peptide and protein synthesis by segment synthesis-condensation. Science 243: 187–192. [DOI] [PubMed] [Google Scholar]
- Kapanidis, A.N., Lee, N.K., Laurence, T.A., Doose, S., Margeat, E., and Weiss, S. 2004. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser-excitation of single molecules. Proc. Natl. Acad. Sci. 101: 8936–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis, A.N., Laurence, T.A., Lee, N.K., Margeat, E., Kong, X., and Weiss, S. 2005. Alternating-laser excitation of single molecules. Acc. Chem. Res. 38: 523–533. [DOI] [PubMed] [Google Scholar]
- Kazmirski, S.L., Wong, K.B., Freund, S.M., Tan, Y.J., Fersht, A.R., and Daggett, V. 2001. Protein folding from a highly disordered denatured state: The folding pathway of chymotrypsin inhibitor 2 at atomic resolution. Proc. Natl. Acad. Sci. 98: 4349–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiga, D., Sakamoto, K., Kodama, K., Kigawa, T., Matsuda, T., Yabuki, T., Shirouzu, M., Harada, Y., Nakayama, H., Takio, K., et al. 2002. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl. Acad. Sci. 99: 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragelund, B.B., Knudsen, J., and Poulsen, F.M. 1999. Acylcoenzyme A binding protein (ACBP). Biochim. Biophys. Acta 1441: 150–161. [DOI] [PubMed] [Google Scholar]
- Ladurner, A.G., Itzhaki, L.S., Daggett, V., and Fersht, A.R. 1998. Synergy between simulation and experiment in describing the energy landscape of protein folding. Proc. Natl. Acad. Sci. 95: 8473–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, N.K., Kapanidis, A.N., Wang, Y., Michalet, X., Mukhopadhyay, J., Ebright, R.H., and Weiss, S. 2005. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88: 2939–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.F. and Tam, J.P. 1994. Peptide segment ligation strategy without use of protecting groups. Proc. Natl. Acad. Sci. 91: 6584–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorand, L., Parameswaran, K.N., Stenberg, P., Tong, Y.S., Velasco, P.T., Jonsson, N.A., Mikiver, L., and Moses, P. 1979. Specificity of guinea pig liver transglutaminase for amine substrates. Biochemistry 18: 1756–1765. [DOI] [PubMed] [Google Scholar]
- Matsumura, Y., Chanyongvorakul, Y., Kumazawa, Y., Ohtsuka, T., and Mori, T. 1996. Enhanced susceptibility to transglutaminase reaction of α-lactalbumin in the molten globule state. Biochim. Biophys. Acta 1292: 69–76. [DOI] [PubMed] [Google Scholar]
- McCarney, E.R., Werner, J.H., Bernstein, S.L., Ruczinski, I., Makarov, D.E., Goodwin, P.M., and Plaxco, K.W. 2005. Site-specific dimensions across a highly denatured protein; a single molecule study. J. Mol. Biol. 352: 672–682. [DOI] [PubMed] [Google Scholar]
- Moerner, W.E. and Orrit, M. 1999. Illuminating single molecules in condensed matter. Science 283: 1670–1676. [DOI] [PubMed] [Google Scholar]
- Muir, T.W., Sondhi, D., and Cole, P.A. 1998. Expressed protein ligation: A general method for protein engineering. Proc. Natl. Acad. Sci. 95: 6705–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren, C.J., Anthony-Cahill, S.J., Griffith, M.C., and Schultz, P.G. 1989. A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244: 182–188. [DOI] [PubMed] [Google Scholar]
- Olejnik, J., Gite, S., Mamaev, S., and Rothschild, K.J. 2005. N-terminal labeling of proteins using initiator tRNA. Methods 36: 252–260. [DOI] [PubMed] [Google Scholar]
- Radisky, E.S. and Koshland Jr., D.E. 2002. A clogged gutter mechanism for protease inhibitors. Proc. Natl. Acad. Sci. 99: 10316–10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky, E.S., King, D.S., Kwan, G., and Koshland Jr., D.E. 2003. The role of the protein core in the inhibitory power of the classical serine protease inhibitor, chymotrypsin inhibitor 2. Biochemistry 42: 6484–6492. [DOI] [PubMed] [Google Scholar]
- Ratner, V., Kahana, E., Eichler, M., and Haas, E. 2002. A general strategy for site-specific double labeling of globular proteins for kinetic FRET studies. Bioconjugate Chem. 13: 1163–1170. [DOI] [PubMed] [Google Scholar]
- Santoro, M.M. and Bolen, D.W. 1988. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27: 8063–8068. [DOI] [PubMed] [Google Scholar]
- Sato, H. 2002. Enzymatic procedure for site-specific pegylation of proteins. Adv. Drug Deliv. Rev. 54: 487–504. [DOI] [PubMed] [Google Scholar]
- Sato, H., Ikeda, M., Suzuki, K., and Hirayama, K. 1996. Site-specific modification of interleukin-2 by the combined use of genetic engineering techniques and transglutaminase. Biochemistry 35: 13072–13080. [DOI] [PubMed] [Google Scholar]
- Sato, H., Yamada, N., Shimba, N., and Takahara, Y. 2000. Unique substrate specificities of two adjacent glutamine residues in EAQQIVM for transglutaminase: Identification and characterization of the reaction products by electrospray ionization tandem mass spectrometry. Anal. Biochem. 281: 68–76. [DOI] [PubMed] [Google Scholar]
- Scalley, M.L., Yi, Q., Gu, H., McCormack, A., Yates III, J.R., and Baker, D. 1997. Kinetics of folding of the IgG binding domain of peptostreptococcal protein L. Biochemistry 36: 3373–3382. [DOI] [PubMed] [Google Scholar]
- Schuler, B. and Pannell, L.K. 2002. Specific labeling of polypeptides at amino-terminal cysteine residues using Cy5-benzyl thioester. Bioconjugate Chem. 13: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Schuler, B., Lipman, E.A., and Eaton, W.A. 2002. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature 419: 743–747. [DOI] [PubMed] [Google Scholar]
- Selvin, P.R. 2000. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 7: 730–734. [DOI] [PubMed] [Google Scholar]
- Stryer, L. and Haugland, R.P. 1967. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. 58: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki, M., Shiota, M., and Taira, K. 2004. Transglutaminase-mediated N-and C-terminal fluorescein labeling of a protein can support the native activity of the modified protein. Protein Eng. Des. Sel. 17: 119–126. [DOI] [PubMed] [Google Scholar]
- Talaga, D.S., Lau, W.L., Roder, H., Tang, J., Jia, Y., DeGrado, W.F., and Hochstrasser, R.M. 2000. Dynamics and folding of single two-stranded coiled-coil peptides studied by fluorescent energy transfer confocal microscopy. Proc. Natl. Acad. Sci. 97: 13021–13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Kamiya, N., and Nagamune, T. 2005. N-terminal glycine-specific protein conjugation catalyzed by microbial transglutaminase. FEBS Lett. 579: 2092–2096. [DOI] [PubMed] [Google Scholar]
- Tolbert, T.J. and Wong, C.H. 2002. New methods for proteomic research: Preparation of proteins with N-terminal cysteines for labeling and conjugation. Angew. Chem. Int. Ed. 41: 2171–2174. [DOI] [PubMed] [Google Scholar]
- Wang, L., Zhang, Z., Brock, A., and Schultz, P.G. 2002. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. 100: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S. 2000. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 7: 724–729. [DOI] [PubMed] [Google Scholar]
- Zhang, L. and Tam, J.P. 1996. Thiazolidine formation as a general and site-specific conjugation method for synthetic peptides and proteins. Anal. Biochem. 233: 87–93. [DOI] [PubMed] [Google Scholar]