Abstract
The regulation of segmentation gene expression is investigated by computational modeling using quantitative expression data. Previous tissue culture assays and transgene analyses raised the possibility that Hunchback (Hb) might function as both an activator and repressor of transcription. At low concentrations, Hb activates gene expression, whereas at high concentrations it mediates repression. Under the same experimental conditions, transcription factors encoded by other gap genes appear to function as dedicated repressors. Models based on dual regulation suggest that the Hb gradient can be sufficient for establishing the initial Kruppel (Kr) expression pattern in central regions of the precellular embryo. The subsequent refinement of the Kr pattern depends on the combination of Hb and the Giant (Gt) repressor. The dual-regulation models developed for Kr also explain some of the properties of the even-skipped (eve) stripe 3+7 enhancer. Computational simulations suggest that repression results from the dimerization of Hb monomers on the DNA template.
Keywords: computational model, Drosophila development, enhancer, dual transcriptional regulators, binding site
The segmentation of the Drosophila embryo depends on sequential spatial domains of gap gene expression, particularly Kruppel (Kr), knirps (kni), and giant (gt) in the presumptive thorax and abdomen. Classical genetic and molecular studies suggest that the maternal Bicoid (Bcd), Hunchback (Hb), and Caudal (Cad) gradients are essential for the establishment of these gap gene expression patterns (1–4). The subsequent refinement and maintenance of the gap patterns depend on extensive cross-regulatory interactions (5–8). However, the exact combinations of maternal morphogens used to control the gap system are still unclear (9).
There is considerable information about the establishment of the initial kni and gt expression patterns in the presumptive anterior and posterior abdomen, respectively. kni is regulated by the combination of the Bcd activator and the Hb gradient, which functions as a dedicated repressor in the context of the kni enhancer (1, 9–11). The regulation of gt is not as well defined, but appears to depend on the broad Bcd and Cad activator gradients; the anterior and posterior limits of the pattern are established by the Kr and Hb repressors, respectively (3, 6, 12). Mechanisms for the establishment of the central Kr expression pattern are very controversial.
A potentially important clue regarding Kr regulation is suggested by previous tissue culture assays. Kr regulatory sequences were attached to a chloramphenicol acetyltransferase (CAT) reporter gene and cotransfected with different concentrations of the Hb protein (13). Low levels of Hb were found to activate CAT expression, but surprisingly, high levels caused repression. Moreover, an artificial anterior–posterior (AP) gradient of Hb was found to be necessary and sufficient to establish a nearly normal Kr expression pattern in transgenic embryos in the absence of Bcd (1, 14). However, the exact positioning of the Kr pattern appears to depend on Bcd and the Torso terminal patterning pathway (15, 16). Additional complications regarding Kr regulation arise from the extensive cross-regulatory interactions that are thought to be important for the maintenance of the pattern, but might also contribute to the initial regulation (3). The difficulties in formulating an accurate quantitative model for Kr regulation were recently summarized (9).
The current study examines computational models for Kr regulation that account for both the establishment of the initial pattern during nuclear cleavage cycle 14 and the dynamic changes in the pattern (including the anterior shift in expression) observed during cellularization. The models are based on the following observations: the dual-regulatory activities of Hb in tissue culture assays (13) (Fig. 1A) and the analysis of antagonistic interactions between similarly distributed gradients (11, 17). Here, we present evidence that the Hb gradient can account for the initial Kr expression pattern and might contribute to the formation of even-skipped (eve) stripes 3 and 7.
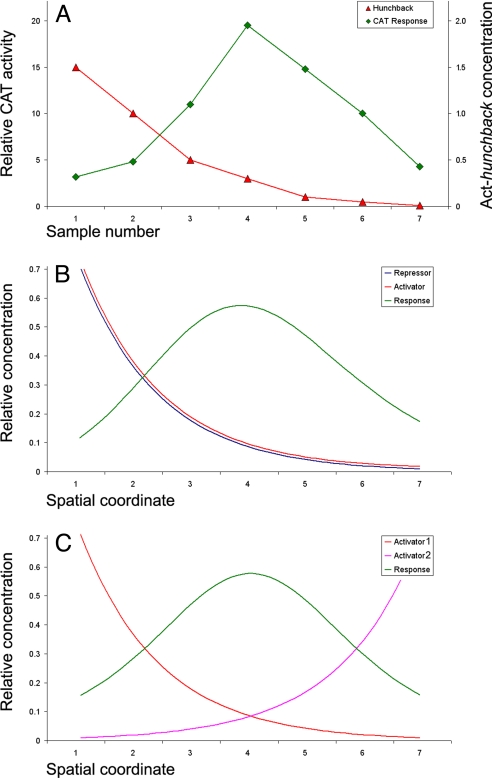
Fig. 1.
Responses to dual regulators and opposing gradients. (A) In vitro response (units of CAT activity) of a Kr-CAT fusion gene to different concentrations of Hb expression vector (μg of plasmid). Data are based on a published analysis (13). (B and C) Predicted spatial pattern produced by the antagonistic activities of two identical gradients (B) or two opposing activator gradients (C). The predicted response seen in B is very similar to that observed for the regulation of Kr by Hb.
Results
There are at least three models for the establishment of the initial Kr expression pattern in central regions of the early embryo. First, the Bcd activator and Hb repressor gradients could regulate Kr, in a manner similar to the establishment of the early kni pattern (1, 18). However, available quantitative simulations are inconsistent with this model (9, 11). Second, opposing Bcd and Cad gradients (Fig. 1C) originating from the anterior and posterior poles could work in a synergistic fashion to activate Kr in central regions where the two gradients overlap. The simplest version of this model is excluded by the observation that neither bcd nor cad mutants eliminate Kr expression (14, 18). Finally, the Kr pattern may be defined solely by the Hb gradient, acting as a concentration-dependent activator and repressor as discussed (1, 2, 13, 14). Dual regulation by Hb is an extension of the recent analysis of antagonistic interactions between gradients establishing dorsal–ventral patterning (11, 17) (Fig. 1B).
Dual-Regulation Models and Mechanisms.
There are numerous precedents for dual regulation by a single sequence-specific transcription factor. For example, the human folate receptor gene is activated by a combination of Sp1 and Ets transcription factors (19). However, at high concentrations Sp1 represses transcription by blocking Ets binding to neighboring sites. The Dorsal gradient mediates both transcriptional activation and repression. It directly silences the expression of target genes that contain regulatory sequences with linked Dorsal binding sites and “AT elements.” The proteins that bind the AT elements interact with Dorsal, and the resulting protein complex recruits the Groucho corepressor protein (20, 21). Pax5 activates gene expression in B lymphocytes but represses expression in erythrocytes, myeloid cells, and T lymphocytes by interacting with alternative coregulators (22). Interactions between the Pax6, Orthodenticle, and Prospero activators result in mutually exclusive expression of rhodopsin genes in fly retina cells (23, 24). Finally, the promyelocytic leukemia zinc finger protein (PLZF) requires dimerization for its repressor function. A specific amino acid substitution converts the repressor into a dedicated activator, presumably by disrupting dimerization (25).
Hb contains two zinc-finger domains; the central domain mediates DNA binding, whereas the C-terminal domain is responsible for dimerization (DZF) (26). Whereas mutations in the DNA-binding domain disrupt expression of both Kr and kni, mutations in the DZF domain affect only Kr, suggesting that dimerization may be selectively required for Kr regulation (27).
Based on the preceding information, several dual-regulation models for the Hb gradient were constructed and tested. The simplest model, “dual-B,” was derived from antagonistic interactions of two identical gradients [see Fig. 1B and supporting information (SI) Text] (11, 17):
In this model, the response P to a dual regulator X is mediated by two binding sites; one is an activator site (binding constant KA) and the other is a repressor site (binding constant KR). The second model, “dual-P” involves dimerization of Hb on a series of equivalent binding sites (see SI Text and SI Figs. 6 and 7). In the case of two linked Hb sites, activation is observed only when one of the two sites is occupied. There is no activation if both sites are occupied (e.g., at high concentrations of Hb). The third model, “dual-C” invokes the concerted action of two activators, X and Y, originating from the anterior and posterior poles, respectively (see Figs. 1C, 2A, and 3A and SI Text).
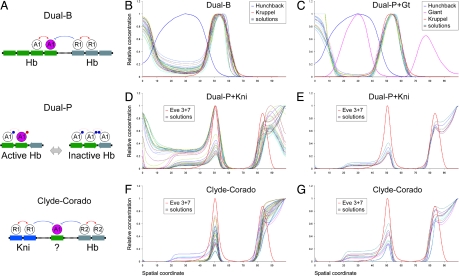
Fig. 2.
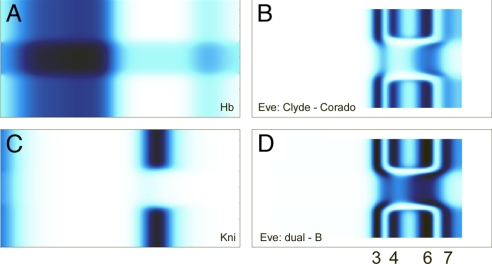
Dual-regulation models agree with the Kr and Eve patterns. (A) Graphical representation of models for dual regulation. (Top) The dual-B model assumes that Hb activates transcription when bound to some sites (in green), but represses transcription from others (in gray). (Middle) The dual-P model assumes that Hb binds to a specific arrangement of sites, and occupancy of all of the sites leads to the masking of the Hb activation domain. The exposed activation domain is shown in red, blocked is in blue. (Bottom) The Clyde-Corado model assumes ubiquitous activation along with localized repression by Hb and Kni. (B) Top 40 solutions for Kr regulation based on the dual-B model. The blue line shows the Hb gradient. (C) Top 40 solutions for Kr regulation based on the dual-P model, combined with Gt repression. (D) Top 40 solutions for eve stripe 3 regulation based on the dual-P model for Hb and Kni repression. (E) Some of the solutions for eve stripe 3 regulation also display the stripe 7 pattern. High levels of expression in the posterior pole are caused by the absence of the torso terminal system. (F) Top 40 solutions for eve stripe 3 regulation based on the Clyde-Corado model (see ref. 34). (G) Some of the solutions also display stripe 7.
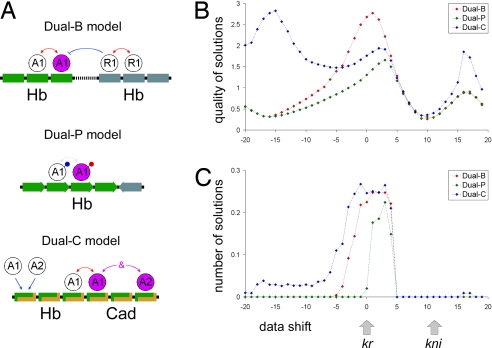
Fig. 3.
Positional cues for Kr. (A) Summary of three different dual-regulation models (see also Fig. 1A). (B and C) Positional cue test. Computer simulations were done to examine the consequences of changing the positions of the Kr expression pattern along the AP axis. In most cases, both the solution quality (B) and the number of good solutions (C) are maximal when Kr is located in its normal position (see Methods). None of the dual-regulation solutions match the endogenous Kni expression pattern (see the gray arrows). Thus, dual regulation by Hb can explain the Kr pattern, but not Kni expression.
Formation and Maintenance of the Kr Expression Pattern.
Dual regulation by the Hb gradient accurately predicts the normal Kr expression pattern in precellular embryos (Fig. 2 B and C). Each of the three models was used to produce computer simulations with four to five parameters, including the number of Hb binding sites and their binding affinities. All three models provide good data-to-model agreements, with correlations between the model and quantitative Kr expression data varying in the range of 0.95 to 0.99 (see SI Table 1). The dual-C model (opposing X and Y activator gradients) produced slightly lower correlations than the dual-B and dual-P models. The dual-B model (distinct Hb activator and repressor sites) produced strong correlations with the expression data, but required considerably higher binding constants for Hb activator sites than repressor sites (see SI Table 1 and SI Fig. 8). The dual-P model (dimerization at linked sites) provided the optimal performance, producing comparable binding constants for different Hb sites (low parameter skew).
Each of the tested models contain relatively few parameters, but it is still possible that the strong correlations with the quantitative expression data were achieved by chance. To address this potential limitation, the behavior of the Hb gradient (input) was explored after shifting the Kr patterns (output) along the AP axis (Fig. 3). The dual-B and dual-P models produced the best solutions only when the Kr pattern was fixed within its normal expression limits (shift = 0). In contrast, the dual-C model produced the best solutions outside the native Kr position (15% shift along the AP axis; Fig. 3 B and C). Nevertheless, all three models produced the largest number of accurate solutions around the native Kr position (shift = 0; Fig. 3C). Interestingly, when the Kr pattern was shifted to the location of the normal kni expression pattern (arrows in Fig. 3B), there was a minimum in both the quality and quantity of solutions. This finding is in agreement with evidence that the role of Hb in the regulation of kni is very different from its regulation of Kr (27). Overall, the computer simulations suggest that dual regulation by Hb is sufficient to account for the formation of the initial Kr pattern. Differences between the current attempt to model Kr regulation and preceding work (9) could be attributed to differences in assumptions about the detailed molecular mechanisms.
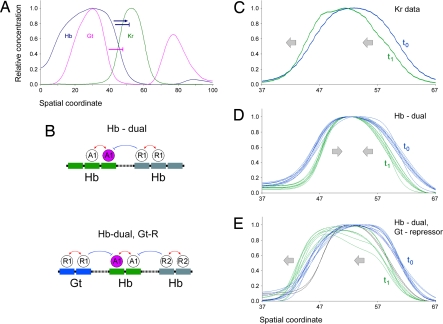
Cross-repressive interactions with other gap proteins are important for the refinement and maintenance of the Kr pattern in older embryos (5–8). Previous quantitative measurements of gene expression and dynamic modeling of the segmentation gene network led to a surprising observation: the gap gene expression patterns shift into anterior regions during cellularization (28–30). Based exclusively on Hb, the dual-regulation models failed to reproduce this anterior shift in the Kr pattern (Fig. 4D, compare with C). However, the shift is observed when the dual-regulation models are combined with known cross-repressive interactions with the Gt protein (5, 6) (Fig. 4E). Thus, the initial Kr pattern might be established solely by the dual-regulatory activities of the Hb gradient. Subsequent repression by Gt is required for the refinement of the pattern, including the anterior shift, during cellularization.
Fig. 4.
Dynamics of Kr pattern supports role for the Gt repressor. (A) The blue arrow and blue T-arrow show dual Hb activities on the Kr expression pattern. Repression by Gt (purple T-arrow) is required for maintaining the late Krl pattern. (B) Displayed are the dual-regulation model (dual-B) (Upper) and the same model with Gt repression (Lower). (C) Temporal changes (anterior shift) in the Kr expression pattern during nuclear cleavage cycle 14, from stage 14.4 to 14.6. (D) Blue lines show solutions for stage 14.4 based on the dual-regulation model. Green lines show behavior of these solutions using data from stage 14.6. In the absence of Gt repression, the Kr pattern narrows, which is not in agreement with the observed dynamics of the Kr pattern (see C). (E) In the presence of Gt repression, most solutions from one temporal stage are also valid for the other stage.
Formation of Eve Stripes.
The dual-regulatory activities of the Hb gradient might not be limited to Kr. The eve stripe 3+7 enhancer contains a series of closely linked Hb and Kni binding sites. There is evidence that the enhancer is activated by one or more ubiquitously distributed transcriptional activators, including components of the JAK–Stat pathway (31, 32). The borders of the stripes are established by Hb and Kni, which were considered to function as dedicated repressors (33, 34) (“Clyde-Corado” model; Fig. 2 A, F, and G). According to this model, the Hb repressor gradient establishes the anterior border of eve stripe 3 and the posterior border of stripe 7 (there is a second source of Hb in posterior regions) (35). The posterior border of eve stripe 3 and the anterior border of stripe 7 are limited by the Kni repressor.
Computer simulations produced very similar results for both the Clyde-Corado and dual-regulation models. Tests were performed only for eve stripe 3, although we also monitored the presence or absence of stripe 7 (Fig. 2 D–G). In this fairly stringent test, both models produced accurate solutions for the limits of stripe 3 (see SI Table 2). According to the Clyde-Corado model, differences in the positioning of eve stripes 3+7 and 4+6 are explained by different binding affinities in the two enhancers: Hb binds more strongly to the eve 4+6 enhancer than the eve 3+7 enhancer, whereas Kni binds stronger to 3+7 than 4+6. To compare the dual-regulation and the Clyde-Corado mechanisms further, variable Hb and Kni binding constants were analyzed in the eve stripe 3 and stripe 4 enhancers by using both models (see SI Table 3). The predicted balance between the Hb and the Kni repression activities was very similar for the dual-regulation and the Clyde-Corado models: stronger repression of stripe 4 by Hb and stronger repression of stripe 3 by Kni (see SI Fig. 9). The distributions of Hb and Kni binding sites in the two enhancers were determined previously (33, 34).
Misexpression of Hb in the ventral mesoderm distorts gap gene expression and eve stripes 3–7 (34) (schematic of the Hb misexpression is shown in Fig. 5A). eve stripes 4 and 5 are lost in the ventral mesoderm, while the stripe 3 pattern contains “arms” and stripe 7 contains a “bulge.” These unusual 2D expression patterns were “reverse-modeled” to compare the performance of the dual-regulation and Clyde-Corado models (Fig. 5 B and D). The expression patterns were reproduced by using Hb 1D data (29), Snail 1D data (17) and the solutions for eve stripes described above (see SI Tables 2 and 3 and Fig. 2 D–G). Both the dual-regulation and Clyde-Corado models accurately produced the observed stripe 3 arms and stripe 7 bulge (Fig. 5 B and D).
Fig. 5.
Reverse modeling of abnormal eve patterns. (A) Misexpression of Hb in the presumptive mesoderm (see ref. 34). In silico representation using Snail data from the DvEx database. (B and D). Both the Clyde-Corado and dual-regulation models can reproduce the altered pattern of eve expression resulting from Hb misexpression, including the stripe 3 arms and stripe 7 bulge. (C) Repression of kni by ectopic Hb in silico.
Discussion
Hb is highly conserved in a variety of insect systems where it is essential for initiating AP patterning and segmentation. Most previous efforts to understand the potent patterning activity of the Hb gradient assumed that it functions as a dedicated repressor, like other gap proteins such as Kr, Kni, and Gt. However, there is evidence that Hb can function as both an activator and repressor (1, 13, 14). These diverse regulatory activities can be explained, at least in part, by the protein structure. Hb is a zinc finger protein with two zinc finger domains. While the central zinc finger domain is involved in DNA binding, the C-terminal zinc finger (DZF) mediates dimerization (26). Dimerization of Hb on the DNA template might provide the basis for Hb-mediated transcriptional repression, as we discuss below.
Regulation of Kr.
It has been argued that Kr is activated by Bcd and repressed by components of the Torso terminal patterning system (16, 36). This model is based on the observation that the Kr expression pattern is expanded in bcd + torso-like mutants (bcd−, tsl−); the strength of the anterior expansion is proportional to the number of copies of the Hb gene (up to four times). However, an alternative explanation is that the Hb gradient is sufficient for the central domain of Kr expression in precellular embryos. In bcd mutants, the only source of Hb is provided by maternal transcripts. These levels are considerably lower than those produced from zygotic transcripts induced by the Bcd gradient. Even four copies of the maternal product might be sufficient for activation, but not repression, of Kr expression.
The dual-regulation model provides a quantitative explanation for the dynamic pattern of Kr expression. As early as nuclear cleavage cycle 10–11, Hb alone is sufficient for the formation of the initial Kr pattern. Later, and throughout nuclear cycle 14, the anterior border of the Kr expression pattern is maintained by Hb dual regulation, along with repression by Gt. Thus, the dual-regulation model can account for the anterior shift in the Kr expression pattern (Fig. 4 C and E). There is no need to invoke an unknown or additional component of the segmentation network. However, there is still the unresolved question of how much Hb is needed for Kr repression vs. activation. Quantitative expression data (29) shows that Kr expression reaches 50% of the peak levels in the region of the embryo containing ≈50% of the peak levels of the Hb gradient. Removing Gt repression from the dual-regulation models (see SI Fig. 10) suggests that peak levels of Hb can permit 50–80% of peak Kr expression.
The misexpression of Hb using the sna enhancer delivered up to 60% of the peak levels of the endogenous Hb gradient, but nonetheless had no detectable effect on Kr expression (S. Small, personal communication). These levels may be insufficient to achieve repression, or ectopic Hb was delivered too late, after the establishment of the initial Kr pattern.
The current analysis raises the possibility that the Hb gradient is sufficient for establishing the initial Kr expression pattern. This model is not incompatible with the more traditional view that Kr is regulated by one or more broadly distributed activators, along with spatially localized repressors (including Hb) that define the anterior and posterior borders of the pattern. Indeed, this principle is one of the most broadly used mechanisms for establishing localized patterns of segmentation gene expression in the early embryo. However, as discussed earlier, the available information suggests that this mechanism may not be sufficient to account for the initial Kr expression pattern. Perhaps dual regulation contributes to this pattern or works in a partially redundant manner with a different mode of regulation.
Regulation of Eve.
The Hb gradient plays multiple roles in the regulation of eve expression. Hb helps activate eve stripe 2 expression, and point mutations in the Hb binding sites in the minimal stripe 2 enhancer lead to a severe reduction in the activities of an otherwise normal stripe2–lacZ fusion gene (37). According to previous models, Hb differentially represses the eve stripe 3+7 and 4+6 enhancers. Relatively high levels of Hb are required for the repression of the 3+7 enhancer, whereas lower levels are sufficient to repress the 4+6 enhancer (32, 38). It has been suggested that ubiquitous components of the JAK–Stat pathway (e.g., dStat) participate in the activation of both enhancers in the early embryo (31, 32).
It is possible that the eve stripe 3+7 enhancer is subject to dual regulation by the Hb gradient. The dual-regulation and Clyde-Corado models performed equally well in a variety of computer simulations (see Figs. 2 D–G and 5 B and D). Indeed, the two models are not mutually exclusive and it is conceivable that a combination is used for the regulation of stripes 3 and 7. As discussed for the regulation of the initial Kr pattern, dual regulation might provide “back-up” or better precision for the expression patterns produced primarily by the Clyde-Corado mechanism whereby Hb and Kni function as dedicated repressors to define the stripe borders.
We prefer one specific form of the dual regulation model, the dimerization on DNA model (dual-P model; SI Fig. 6). The key feature of this model is that concentration-dependent activation or repression by the Hb gradient depends on a series of equivalent Hb binding sites in the Kr and eve 3+7 enhancers (see Fig. 2A and SI Fig. 6A). At low Hb concentrations, the bound Hb monomers function as activators, whereas at high concentrations Hb forms dimers that either repress transcription or block activation. Activation by Hb monomers may be the result of exposed peptide interfaces that recruit transcriptional coactivator complexes. In contrast, at high density, Hb monomers bound at neighboring sites are able to interact with one another, perhaps through the DZF domain (27), and sequester the coactivator recruitment interfaces. A critical test of this model would require the mutagenesis of alternating Hb binding sites in otherwise normal Kr and eve 3+7 enhancers. Such enhancers should exhibit only activation, not repression, by the Hb gradient.
Dual Regulation and the Conserved Role of Hb in Evolution.
The dual activities of the Hb regulatory gradient are consistent with its conserved activity as a critical determinant of segmentation in a variety of insects, and possibly other arthropods and ecdysozoans (39–41). It has been well established that the bcd gene is not conserved outside of the Drosophilids, but Hb gradients have been implicated in the segmentation of a broad spectrum of insects, including the grasshopper and flour beetle with short germband modes of development, and the parasitic wasp Nasonia, which exhibits a long germband mode of development (39, 42). The ability of Hb to function as both an activator and a repressor provides an explanation for its potent patterning activity in different insect embryos. Although the mechanistic details may be different, it is interesting to note that another regulatory morphogen with long-range patterning activity, the Dorsal gradient in Drosophila, also functions as both a transcriptional activator and repressor (20, 21).
Methods
Quantitative gene expression data were downloaded from the FlyEx database (29) and refined by interpolation as described in SI Text and SI Fig. 11. The input data for the Kr models were obtained for nuclear cleavage cycle 14.2, and the output data were produced for cleavage cycle 14.4. For the eve models, the input data were obtained for cycle 14.4 and the output data were produced for cycle 14.6. Quantitative data used in the current study reflect rather late, “mature” Kr pattern (cycle 14). However, the position of the earlier Kr pattern (cycles 11–13), recently assessed by Jaeger et al. (9) is consistent with the mature pattern.
The Metropolis–Hastings algorithm (SI Fig. 12) was used for fitting parameters (30, 43), based on the correlation, r, between the model and data as the objective function. Probability of acceptance was calculated from the likelihood ratio between the current (r0) and the proposed states (r1). The proposed state was accepted if the likelihood ratio produced a number greater than a random number U, derived from a uniform distribution:
An additional parameter α was sometimes used to optimize performance. At α = 1 the algorithm works as Metropolis–Hastings based on r, at α > 1 the algorithm exaggerates good solutions, at α < 1 the algorithm is able to cross deeper and longer valleys. In all fitting tests, the search was run for 300 steps per seed point for no more than 1,000 independent seed points.
In the shifting tests (Fig. 3) only the Kr (output) data were shifted along the AP axis, while the input data (Hb) were in its original position. For each Kr data shift 1,000 seed points were explored. Best qualities are plotted in Fig. 3B, and the number of solutions exceeding r = 0.95 are plotted in Fig. 3C.
In pseudodynamics tests (Fig. 4), the model solutions (parameters) derived from fitting Kr at the stage 14.2 → 14.4 (Hb → Kr) were taken to test the same model, but using the data from the later stages (14.4 → 14.6).
All scripts and programs used in this study are available from D.P. on request (dxp@berkeley.edu).
Supplementary Material
ACKNOWLEDGMENTS.
We thank members of M.S.L.'s laboratory and the Center for Integrative Genomics for critical comments and stimulating discussions. This work was supported by National Institutes of Health Grant GM34431.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711941105/DC1.
References
- 1.Hulskamp M, Pfeifle C, Tautz D. A morphogenetic gradient of hunchback protein organizes the expression of the gap genes Kruppel and knirps in the early Drosophila embryo. Nature. 1990;346:577–580. doi: 10.1038/346577a0. [DOI] [PubMed] [Google Scholar]
- 2.Hulskamp M, Tautz D. Gap genes and gradients: The logic behind the gaps. BioEssays. 1991;13:261–268. doi: 10.1002/bies.950130602. [DOI] [PubMed] [Google Scholar]
- 3.Kraut R, Levine M. Spatial regulation of the gap gene giant during Drosophila development. Development. 1991;111:601–609. doi: 10.1242/dev.111.2.601. [DOI] [PubMed] [Google Scholar]
- 4.Rivera-Pomar R, Jackle H. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 1996;12:478–483. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 5.Pankratz MJ, Hoch M, Seifert E, Jackle H. Kruppel requirement for knirps enhancement reflects overlapping gap gene activities in the Drosophila embryo. Nature. 1989;341:337–340. doi: 10.1038/341337a0. [DOI] [PubMed] [Google Scholar]
- 6.Kraut R, Levine M. Mutually repressive interactions between the gap genes giant and Kruppel define middle body regions of the Drosophila embryo. Development. 1991;111:611–621. doi: 10.1242/dev.111.2.611. [DOI] [PubMed] [Google Scholar]
- 7.Jaeger J, et al. Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster. Genetics. 2004;167:1721–1737. doi: 10.1534/genetics.104.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkins T, Jaeger JJ, Reinitz J, Glass L. Reverse engineering the gap gene network of Drosophila melanogaster. PLoS Comput Biol. 2006;2:e51. doi: 10.1371/journal.pcbi.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaeger J, Sharp DH, Reinitz J. Known maternal gradients are not sufficient for the establishment of gap domains in Drosophila melanogaster. Mech Dev. 2007;124:108–128. doi: 10.1016/j.mod.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu X, Vasisht V, Kosman D, Reinitz J, Small S. Thoracic patterning by the Drosophila gap gene hunchback. Dev Biol. 2001;237:79–92. doi: 10.1006/dbio.2001.0355. [DOI] [PubMed] [Google Scholar]
- 11.Zinzen RP, Papatsenko D. Enhancer responses to similarly distributed antagonistic gradients in development. PLoS Comput Biol. 2007;3:e84. doi: 10.1371/journal.pcbi.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivera-Pomar R, Lu X, Perrimon N, Taubert H, Jackle H. Activation of posterior gap gene expression in the Drosophila blastoderm. Nature. 1995;376:253–256. doi: 10.1038/376253a0. [DOI] [PubMed] [Google Scholar]
- 13.Zuo P, et al. Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev. 1991;5:254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]
- 14.Schulz C, Tautz D. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development. 1994;120:3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- 15.Casanova J. Developmental evolution: Torso, a story with different ends? Curr Biol. 2005;15:R968–R970. doi: 10.1016/j.cub.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Brent AE, Yucel G, Small S, Desplan C. Permissive and instructive anterior patterning rely on mRNA localization in the wasp embryo. Science. 2007;315:1841–1843. doi: 10.1126/science.1137528. [DOI] [PubMed] [Google Scholar]
- 17.Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 18.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69:237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 19.Kelley KM, Wang H, Ratnam M. Dual regulation of ets-activated gene expression by SP1. Gene. 2003;307:87–97. doi: 10.1016/s0378-1119(03)00445-1. [DOI] [PubMed] [Google Scholar]
- 20.Jiang J, Rushlow CA, Zhou Q, Small S, Levine M. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 1992;11:3147–3154. doi: 10.1002/j.1460-2075.1992.tb05387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ratnaparkhi GS, Jia S, Courey AJ. Uncoupling dorsal-mediated activation from dorsal-mediated repression in the Drosophila embryo. Development. 2006;133:4409–4414. doi: 10.1242/dev.02643. [DOI] [PubMed] [Google Scholar]
- 22.Fuxa M, Skok JA. Transcriptional regulation in early B cell development. Curr Opin Immunol. 2007;19:129–136. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Papatsenko D, Nazina A, Desplan C. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech Dev. 2001;101:143–153. doi: 10.1016/s0925-4773(00)00581-5. [DOI] [PubMed] [Google Scholar]
- 24.Tahayato A, et al. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- 25.Melnick A, et al. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol Cell Biol. 2000;20:6550–6567. doi: 10.1128/mcb.20.17.6550-6567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarty AS, Kleiger G, Eisenberg D, Smale ST. Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell. 2003;11:459–470. doi: 10.1016/s1097-2765(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 27.Hulskamp M, Lukowitz W, Beermann A, Glaser G, Tautz D. Differential regulation of target genes by different alleles of the segmentation gene hunchback in Drosophila. Genetics. 1994;138:125–134. doi: 10.1093/genetics/138.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myasnikova E, Samsonova A, Kozlov K, Samsonova M, Reinitz J. Registration of the expression patterns of Drosophila segmentation genes by two independent methods. Bioinformatics. 2001;17:3–12. doi: 10.1093/bioinformatics/17.1.3. [DOI] [PubMed] [Google Scholar]
- 29.Poustelnikova E, Pisarev A, Blagov M, Samsonova M, Reinitz J. A database for management of gene expression data in situ. Bioinformatics. 2004;20:2212–2221. doi: 10.1093/bioinformatics/bth222. [DOI] [PubMed] [Google Scholar]
- 30.Jaeger J, et al. Dynamic control of positional information in the early Drosophila embryo. Nature. 2004;430:368–371. doi: 10.1038/nature02678. [DOI] [PubMed] [Google Scholar]
- 31.Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 32.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 33.Lifanov AP, Makeev VJ, Nazina AG, Papatsenko DA. Homotypic regulatory clusters in Drosophila. Genome Res. 2003;13:579–588. doi: 10.1101/gr.668403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clyde DE, Corado MS, Wu X, Pare A, Papatsenko D, Small S. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 35.Margolis JS, et al. Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development. 1995;121:3067–3077. doi: 10.1242/dev.121.9.3067. [DOI] [PubMed] [Google Scholar]
- 36.Li WX. Functions and mechanisms of receptor tyrosine kinase Torso signaling: Lessons from Drosophila embryonic terminal development. Dev Dyn. 2005;232:656–672. doi: 10.1002/dvdy.20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff C, Sommer R, Schroder R, Glaser G, Tautz D. Conserved and divergent expression aspects of the Drosophila segmentation gene hunchback in the short germ band embryo of the flour beetle Tribolium. Development. 1995;121:4227–4236. doi: 10.1242/dev.121.12.4227. [DOI] [PubMed] [Google Scholar]
- 40.Schroder R. The genes orthodenticle and hunchback substitute for bicoid in the beetle Tribolium. Nature. 2003;422:621–625. doi: 10.1038/nature01536. [DOI] [PubMed] [Google Scholar]
- 41.Kerner P, Zelada Gonzalez F, Le Gouar M, Ledent V, Arendt D, Vervoort M. The expression of a hunchback ortholog in the polychaete annelid Platynereis dumerilii suggests an ancestral role in mesoderm development and neurogenesis. Dev Genes Evol. 2006;216:821–828. doi: 10.1007/s00427-006-0100-9. [DOI] [PubMed] [Google Scholar]
- 42.Pultz MA, et al. A major role for zygotic hunchback in patterning the Nasonia embryo. Development. 2005;132:3705–3715. doi: 10.1242/dev.01939. [DOI] [PubMed] [Google Scholar]
- 43.Hoffman JG, Metropolis N, Gardiner V. Study of tumor cell populations by Monte Carlo methods. Science. 1955;122:465–466. doi: 10.1126/science.122.3167.465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.