Abstract
The formation of a transcriptionally active complex by RNA polymerase involves a series of short-lived structural intermediates where protein conformational changes are coupled to DNA wrapping and melting. We have used time-resolved KMnO4 and hydroxyl-radical X-ray footprinting to directly probe conformational signatures of these complexes at the T7A1 promoter. Here we demonstrate that DNA melting from m12 to m4 precedes the rate-limiting step in the pathway and takes place prior to the formation of full downstream contacts. In addition, on the wild-type promoter, we can detect the accumulation of a stable off-pathway intermediate that results from the absence of sequence-specific contacts with the melted non-consensus –10 region. Finally, the comparison of the results obtained at 37°C with those at 20°C reveals significant differences in the structure of the intermediates resulting in a different pathway for the formation of a transcriptionally active complex.
INTRODUCTION
The binding of RNA polymerase to promoter DNA is a key step in the regulation of gene expression. This process has been described as a multi-step pathway that begins with the formation of a competitor-sensitive ‘closed complex’, followed by isomerization into a competitor-resistant complex. This isomerization process is distinguished by an initial phase during which nucleation of DNA melting takes place and the promoter DNA becomes completely protected from cleavage by DNaseI or hydroxyl radicals (1–4), followed by a second step involving the complete melting of the transcription bubble and the formation of a transcriptionally active complex, also known as the ‘open complex’ (5). A series of conformational changes in both the protein and the DNA are necessary to obtain the active complex, and each of these presents a possible target for both regulation and for subtle modulation during evolution (6–8). A precise description of the pathway leading to the formation of a transcriptionally active complex is thus necessary in order to understand how regulatory mechanisms control RNA polymerase activity at the promoter.
At some strong promoters such as the A1 promoter of T7 phage (9,10) the Escherichia coli RNA polymerase holoenzyme (RNAP, made of six subunits, 2 α, β, β′, ω and σ) is able to recognize and bind specifically to the promoter, melt the DNA and initiate transcription in the absence of additional transcription factors. The study of binding of RNAP to these promoters has been used to define the sequence of events for this process as described above (3,11–14). The T7A1 promoter's wild-type (wt) sequence is characterized by the presence of an AT-rich UP element extending to –71, a near consensus –35 sequence (TTGACT instead of TTGACA) and a non-consensus –10 sequence (GATACT instead of TATAAT).
The description of structural intermediates in the process of macromolecular recognition remains a challenge due to the presence of short-lived, unstable complexes. We previously characterized the structural kinetics of binding of RNAP to the T7A1 promoter using time-resolved X-ray footprinting (14). The tens of millisecond temporal resolution of this technique allows the characterization of different intermediates not only by their extent of protection but also by their kinetic properties. In addition, the single nucleotide resolution from hydroxyl radical cleavage permits a structural interpretation of the footprinting patterns that facilitates the deduction of possible conformational changes in the intermediate complexes. The construction of a full structural model is further made possible, thanks to the considerable amount of existing biochemical data concerning the identification of specific DNA–protein interactions, with the caveat that some of the protection observed in intermediates could be due to short-lived interactions that are not present in the complex at equilibrium (15).
Our previous results described a kinetic pathway where an equilibrium is established among several, relatively unstable, intermediates preceding the energetic barrier of a rate-limiting step leading to the formation of a more stable complex (14). The protection from hydroxyl radical cleavage at a specific region thus can appear during at least two phases: the first, fast phase corresponding to the formation of protein–DNA contacts in the early intermediates in the pathway, and the second, slower phase in the kinetic profile corresponding to the formation of a stable complex where the DNA remains protected from cleavage in the same region.
In this work, the improved temporal resolution and thermal stability of the stopped-flow apparatus has allowed us to obtain a more detailed characterization of conformational changes taking place at 37°C and to determine how both temperature and the DNA sequence in the –10 region may affect the structure of intermediates. For the first time, time-resolved permanganate experiments at 37°C permit direct identification of intermediates in the pathway where different bases of the –10 sequence become accessible to modification. Here, we show that in the context of the wt promoter at 37°C DNA melting precedes the rate-limiting step and the formation of downstream contacts. In addition, the comparison of the intermediates formed under these different conditions strongly suggests that the presence of an off-pathway intermediate at 37°C results from the lack of specific contacts with the fork junction structure at the upstream end of the partially melted –10 region. The absence of this off-pathway intermediate at 20°C implies that its stability is dependent in part on a temperature dependent protein conformational change.
MATERIALS AND METHODS
DNA preparation
DNA fragments of 172 bp length containing wt T7A1 promoter and either radioactively or fluorescently labelled at the 5′−end were prepared by PCR using pDS1−A1220 plasmid DNA as a template and primer pairs T7A1−up(−91) for the non-template strand (NTS) and T7A1−down(+81) for the template strand (TS). One of these primers was end-labelled with 32P or Alexa Fluor 647 fluorescent dye (Invitrogen). The DNA fragments were purified by HPLC on a monoQ column using a MiLiChrom chromatograph (EcoNova, Novosibirsk, Russia). The T7A1 promoter mutant containing a consensus –10 region was obtained by ligation of commercially synthesized DNA oligonucleotides (IBA, Göttingen, Germany) followed by purification by HPLC and then amplified by PCR.
RNA polymerase purification
His-tag RNA polymerase was isolated from E. coli strain RL916 as described in our previous work (14).
Time-resolved X-ray footprinting experiments
Time-resolved hydroxyl radical footprinting experiments were performed at the ID10A white light undulator beamline at the European Synchrotron Radiation Facility (ESRF) (Grenoble, France) using a modified BioLogic stopped−flow machine (SFM−400) (Claix, France) (see Supplementary Figure 1). During a typical experiment, 13 µl of 50 nM fluorescently labeled DNA and 13 µl of 200 nM RNA polymerase in BBcac20 buffer (50 mM sodium cacodylate, pH 7.5; 20 mM NaCl; 6 mM MgCl2) were rapidly mixed inside the stopped-flow machine. After incubation of the reagents for a specific time (from 50 ms to 5 min), the sample was exposed to the X-ray beam by pushing the solution through a quartz capillary at a specific speed determining the exposure time, between ∼ 0.18 and 0.26 ms depending on the ring current. Each irradiated sample (final volume 100 µl) was mixed with 10 µl of 3 M sodium acetate (pH 5.0) containing carrier DNA (0.1 mg/ml) and with four volumes of cold ethanol to precipitate the DNA. The DNA fragments were then resolved on 8% denaturing polyacrylamide gel using an ALF Express II DNA analyzer (Amersham Pharmacia Biotech).
Time-resolved potassium permanganate footprinting experiments
Time-resolved permanganate footprinting experiments were performed in two steps. First, 10 µl of 50 nM DNA and 10 µl of 200 nM RNA polymerase in BBcac20 buffer were rapidly mixed inside the stopped−flow machine of our own construction (Supplementary Figure 5). The DNA fragment was radioactively labelled at the 5′−end of the TS (it was found that fluorescent labels do not withstand KMnO4 treatment). The reagents were incubated for a specific amount of time (from 1 s to 4 min), following which the sample was mixed with 20 µl of 60 mM KMnO4 for 0.2 s. The sample was then expelled into an Eppendorf tube containing 100 µl of 2× stop-KMnO4 solution (10% β-mercaptoethanol; 0.6 M NaAc, pH 5.0; 20 µg/ml carrier DNA) where the oxidation reaction is rapidly quenched. Each sample (final volume ∼200 µl) was mixed with four volumes of cold ethanol to precipitate the DNA. The DNA was cleaved at the modified thymines by reaction with piperidine: the pellet was dissolved in 90 µl of freshly prepared 10% piperidine and incubated at 90°C for 20 min. Ten microlitres of 5 M LiCl was then added and DNA fragments were precipitated with 400 µl of ethanol and subsequently resolved by gel electrophoresis. For quantification, gels were exposed to Fuji Imager plate BAS IIIS, which was scanned with Bas−1000 PhosphorImager instrument.
Evaluation of the data of time-resolved hydroxyl radical footprints
The processes of quantification and normalization of time-resolved hydroxyl radical footprints as well as the fitting of the resulting kinetic data to single and double exponential equations were carried out as described in our previous work (14). Representative plots of the raw output from the electrophoresis in the ALF sequencer and the result of the subsequent quantitation of each peak's area are found in Supplementary Figures 2 and 3.
Analysis of potassium permanganate footprinting data
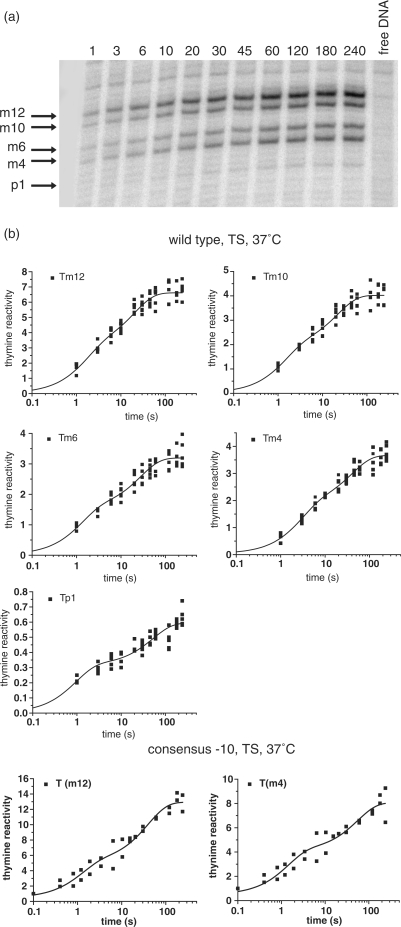
Band intensity profiles along each gel lane were determined using the MacBas software. For integration of the area of the peaks in each lane the lowest intensity points in the lane were used as the horizontal baseline and peaks were fit to a Lorenzian curve. The values for the peaks’ area were then normalized on the total amount of DNA material applied on corresponding gel lane. Normalized area values of the peaks in the lane of DNA treated with KMnO4 in the absence of RNAP (local background intensity) were then subtracted from the normalized values of corresponding peaks within each lane. The plots of the increase of KMnO4 accessibility at each thymine position were fit to single or double exponential equations and processed in the same way as in case of hydroxyl radicals footprints (see Supplementary Figure 4 and Table 1).
Table 1.
Rates of potassium permanganate reactivity
| Nucleotide Position | Apparent rate (s−1) |
Percentage of total amplitude |
||
|---|---|---|---|---|
| kA | kB | A | B | |
| Wild-type T7A1 promoter at 37°C | ||||
| m12 | 0.6 ± 0.2 | 0.049 ± 0.009 | 45 | 55 |
| m10 | 0.7 ± 0.2 | 0.046 ± 0.008 | 47 | 53 |
| m6 | 0.8 ± 0.2 | 0.036 ± 0.006 | 47 | 53 |
| m4 | 0.35 ± 0.07 | 0.025 ± 0.004 | 49 | 51 |
| p1 | 1.0 ± 0.2 | 0.019 ± 0.004 | 52 | 48 |
| Consensus –10 T7A1 promoter at 37°C | ||||
| m12 | 0.8 ± 0.3 | 0.025 ± 0.006 | 37 | 63 |
| m4 | 0.7 ± 0.2 | 0.018 ± 0.006 | 48 | 52 |
RESULTS
We used time-resolved hydroxyl radical and permanganate footprinting in order to characterize the effects of the –10 sequence and of temperature on the structure of intermediates in the process of open complex formation by RNA polymerase. We measured the rate of change in both permanganate reactivity and the extent of protection from hydroxyl radical cleavage of each base on the DNA after mixing RNA polymerase and an end-labelled DNA fragment containing the sequence for the T7A1 promoter in a stopped-flow apparatus. From the analysis of these results it is possible to distinguish several structural intermediates due to their unique structural and kinetic properties.
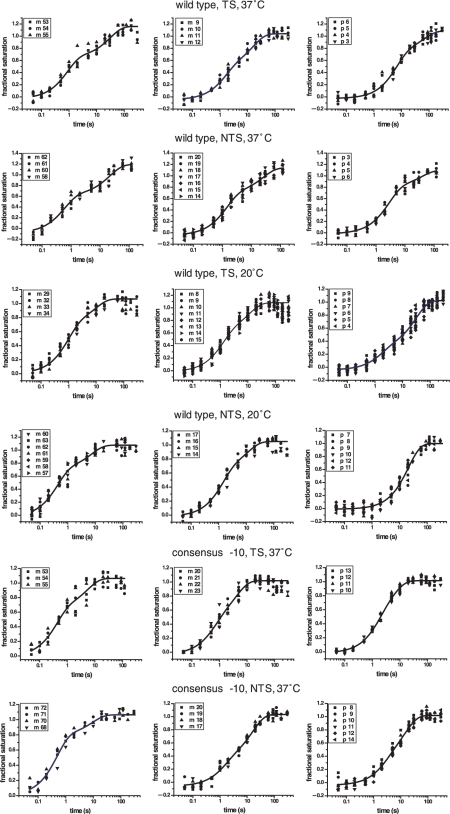
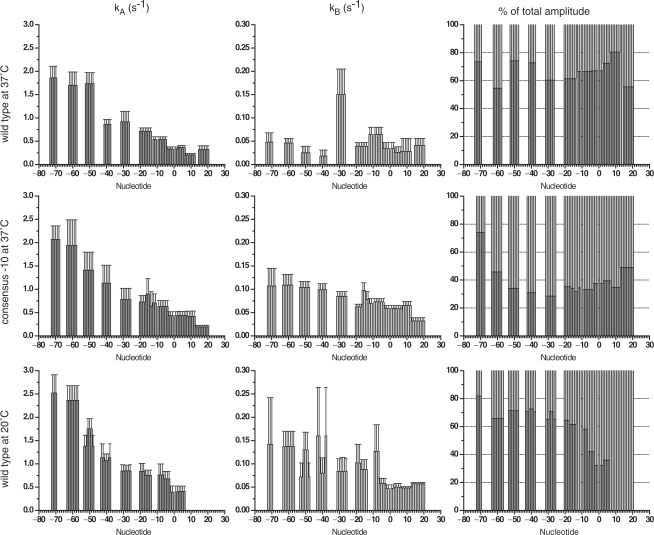
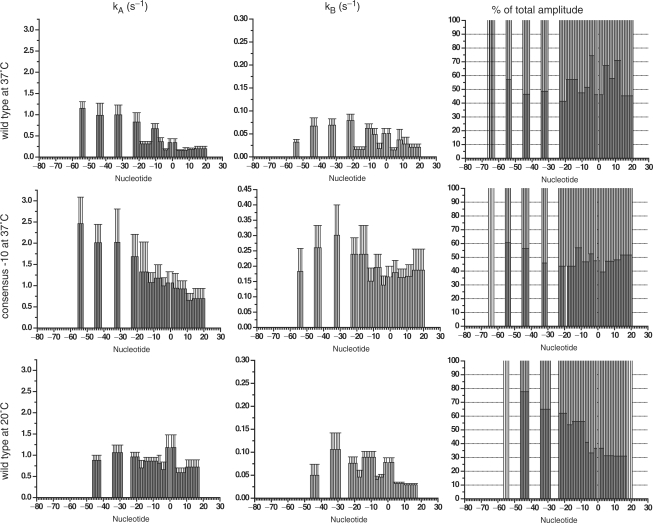
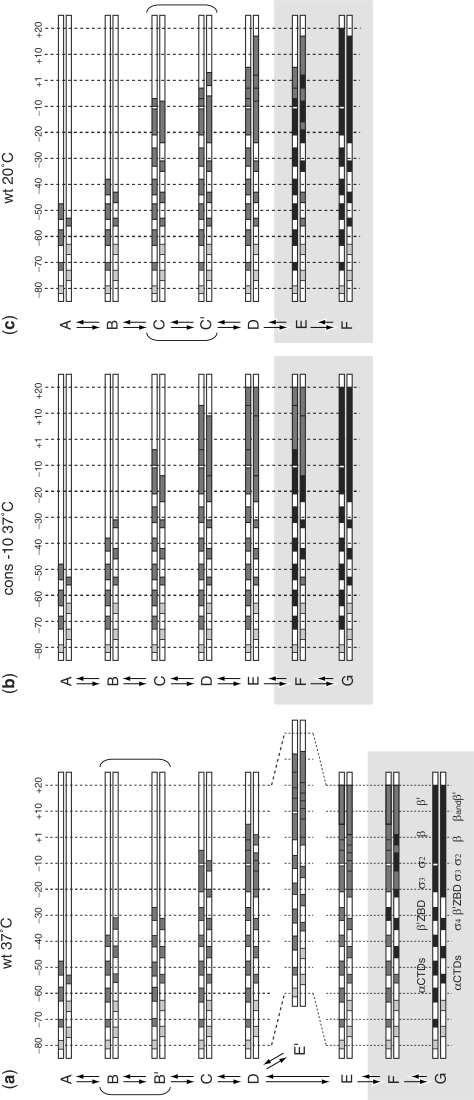
We recently built a new stopped-flow apparatus with an increased time resolution and a more stable and uniform temperature control resulting in improved data quality and reproducibility (see Supplementary Figure 1). The footprinting patterns obtained on the DNA at different mixing times were analysed as described (14). The raw data and the result of quantification are shown in Supplementary Figures 2–4 and Supplementary Table 1. A representative sample of plots of the change in the fraction of protection as a function of time in different regions of the promoter is shown in Figure 1. The rate of appearance of the protections at each site on the promoter is often best described by a double exponential (see Supplementary Table 2 for the results of the F-test statistical analysis and Supplementary Figure 4 for the comparison of the residuals). The values of the rate and amplitude of both phases depend on the position of the base in the promoter, the –10 region sequence and the temperature. Results are summarized in Figures 2 and 3. The extent of protection from hydroxyl radical cleavage of the DNA backbone is a function of solvent accessibility to these sites (16). The differences in the rates and amplitudes of appearance of protection allow us to assign each protection to a possible structural intermediate in the pathway (Figure 7). The protections of the different intermediates are described here and in Figures 5–7 in a sequential fashion; however we cannot exclude that those within a rapid equilibrium (shown within the parenthesis in Figure 6) could also be described by the presence of a branched pathway (14).
Figure 1.
Representative plots of the kinetics of protection. Increase in fractional saturation at different sites on the promoter on both the template and non template strands for the three conditions studied here, wt T7A1 at 37°C, wt T7A1 at 20°C and the consensus –10 mutant T7A1 promoter at 37°C. The goodness to fit to either a single or double exponential were determined by a visual analysis of the residuals and an F-test analysis (see Supplementary Table 2 online and Supplementary Figure 4 online for the complete set of the plots and the analysis of the residuals).
Figure 2.
Summary of the rates and amplitudes for the appearance of protection at each base on the non-template strand of the promoter region. The data obtained on the wild-type T7A1 promoter at 37°C, on the consensus –10 mutant of the T7A1 promoter at 37°C and on the wild-type T7A1 promoter at 20°C are shown in the first, second and third rows, respectively. The plots in the first column represent values for the rates within the fast phase of the kinetics, while in the second column the plots represent values for the rates within the slow phase. The plots in the third column show the percentage of the amplitudes of the two phases for each position on the promoter, in dark grey is the amplitude of the fast phase while in light grey is that of the slow phase.
Figure 3.
Summary of the rates and amplitudes for the appearance of protection at each base on the template strand of the promoter region. The data obtained on the wild-type T7A1 promoter at 37°C, on the consensus –10 mutant of the T7A1 promoter at 37°C and on the wild-type T7A1 promoter at 20°C are shown in the first, second and third rows, respectively. The plots in the first column represent the values for the rates within the fast phase of the kinetics, while in the second column the plots represent the values for the rates within the slow phase. The plots in the third column shown the percentage of the amplitude of the two phases for each position on the promoter, in dark grey is the amplitude of the fast phase while in light grey is that of the slow phase.
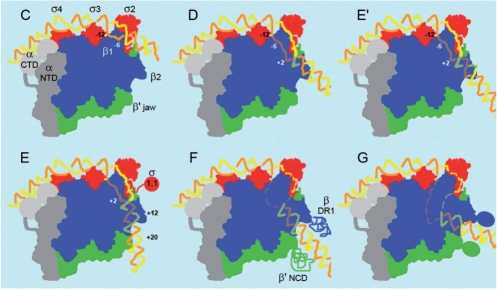
Figure 7.
Proposed structures for the key intermediates in the pathway of formation of a transcriptionally active complex on the wild type T7A1 promoter at 37°C. The model of the open complex proposed by Darst and co-workers from the crystal structure of the Thermus aquaticus RNAP was used as a starting point to create these images (17). α-CTDs and NTDs are shown in light and dark grey respectively. β and β′ are in blue and green, respectively, while sigma is in red. The template strand is in orange and the nontemplate strand in yellow. The shaded parts of the DNA are those that have entered the active site channel and the jaws and are therefore placed behind the β subunit. These intermediates correspond to those shown in Figure 5a, C through G. Following the formation of early complexes stabilized by the interaction of the α-CTDs with the UP-element and σ region 4 with the –35 region of the promoter (A, B and B′ not shown) the DNA is bent towards σ regions 3 and 2 where contacts are made with the spacer and the upstream end of the –10 region, C. The E′ intermediate corresponds to the off-pathway complex. While the pattern of protection in complexes E, F and G does not change, the extent of protection increases as the complex isomerizes into a transcriptionally active structure where the template strand is placed at the active site (G). Protection of the DNA down to p20 is likely due to an interaction with the β DR1 and β′ NCD and their subsequent folding stabilizing the transcriptionally active complex (3,54).
Figure 5.
Protection pattern of each of the intermediates identified by time-resolved X-ray footprinting. The identification of specific intermediates in the pathway was determined from the differences in the rates or in the relative amplitudes of the fast and slow phases of appearance of protection as a function of the position on the promoter. (a) Wild-type T7A1 promoter at 37°C. (b) Consensus –10 mutant of the T7A1 promoter at 37°C. (c) Wild-type T7A1 promoter at 20°C. Light grey boxes correspond to regions with weak protection; grey boxes correspond to the appearance of protection during the first, fast phase; black boxes correspond to the regions which further become protected in the slow phase of the kinetics. The RNAP domains interacting with specific promoter regions are shown for the final complex on the wt promoter. The complexes formed within the slow phase are within the light grey background.
Figure 6.
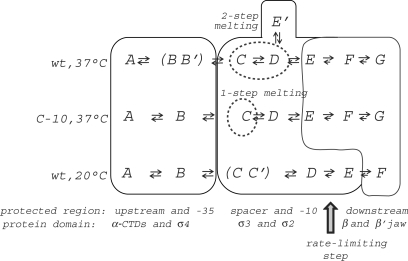
Summary of the intermediates identified in the pathway for RNAP binding to the wild type T7A1 promoter at 37°C, the consensus −10 T7A1 mutant (C-10) at 37°C and the wild type promoter at 20°C. The thin black line places the different intermediates in three groups depending on the nature of the main protein–DNA interactions involved in the intermediate structures. The intermediates within the parenthesis are in rapid equilibrium, while those within the dotted circles correspond to the DNA melting steps. The intermediates in each of the pathways can have different protection patterns even though they are identified by the same letter.
High-resolution characterization of the formation of intermediates on the wt T7A1 promoter at 37°C reveals the presence of a stable, off-pathway complex
A fast phase is observed during protection of all the nucleotides eventually protected in the open complex at the wt promoter at 37°C (Figures 1–3). Nucleotides at the upstream end of the promoter region are protected at a faster rate than those downstream. The stepwise decrease in the rate of the fast phase on passing from upstream to downstream defines the boundaries of each intermediate (Figure 5). In addition, differences in the amplitude of each phase provided additional criteria for the identification of intermediate structures (Figures 2 and 3). The earliest intermediates observed are characterized by the protection of the distal UP element sites by the α-C-Terminal Domain (CTD) (m55-m53 on the TS and m73-m48 on the non-NTS (Figure 5a, A intermediate); the numbering refers to the distance of the bases from the site of transcription initiation either upstream, m, or downstream, p). On the NTS, the protection extends over three sites within a 25-base region. At the m62-m58 site the amplitude of the fast phase is lower compared those on either side, indicating that this protection results from a less favorable interaction within the set of these early complexes in rapid equilibrium. However, the degree of protection at this site is greater than at the neighboring sites, indicating that a more stable interaction is formed in the final complex. This implies a rearrangement of the α-CTD domains during the process of RNAP binding to the promoter.
A set of protections are also observed near m80; however, the low signal to noise in this region did not allow for precise kinetic measurements. In addition, the weak protection on the TS at sites upstream of m55 indicates that the two strands are protected by the α-CTDs in an asymmetric fashion (the same weak protection is also observed in the other two data sets for the TS at 20°C and with the consensus –10 mutant).
The two sets of protections of the proximal UP element and the –35 sequence by the α -CTDs, σ region 4.2 and β′ (m45-m31 TS; m41-m27 NTS) appear at the same rate but with different amplitudes of the fast phase for the different regions, a signature of the presence of two intermediates in rapid equilibrium (Figure 5a, B intermediates).
The next step reveals protection at positions m23-m20 and m12-m9 on the TS and m20-m5 on the NTS (C intermediate). The continuous protection of the downstream half of the spacer region of the NTS (m20-m14) likely involves the σ non-conserved region (NCR) and σ region 3, whereas the protection of the –10 sequence is due to the σ region 2 protruding from the surface of the protein (3,17). In the following intermediate, protection extends to include the transcription start site (m19-m13, m8-m6, m2-p2 TS; m4-p6 NTS) (D intermediate) as the DNA enters the active site channel.
The protected regions m5-m3 and p3-p12 TS and p7-p12 NTS appear at a slower rate for both the first and second exponential phase. In addition, protection kinetics at these sites exhibit larger amplitudes for the fast phase compared to other protections either upstream or downstream. We propose that this larger amplitude results from the sum of two possible complexes being protected at these sites (intermediates E and E′ in Figure 5a). In the first complex, E, these contacts are formed at the same time as the protection of the downstream DNA (p13-p20 TS and NTS); in the second complex, E′, the same protection pattern appears in the absence of the downstream contacts. These results indicate that there is a branching of the pathway where a fraction of the intermediates, E′, is in a conformation that does not efficiently isomerize to form the downstream contacts. These contacts can form following a conversion of E′ back to the D intermediate and the slow rate of this conversion results in an accumulation of this off-pathway complex.
The rates of the second, slower phase correspond to isomerization steps leading to the formation of a stable complex and are very similar to rates measured for the formation of a transcriptionally active complex (12,18). In this second phase of the pathway two sets of rates can be distinguished at the different protection sites; the faster rate corresponding to the protection within the core promoter sequence (Figures 2 and 3). These differences in the rate of the second phase are more evident in the data set for the TS, where the upstream, spacer and downstream protection become stabilized at a slower rate than the core promoter (Figure 3). The bases at m31-m27 on the NTS are instead protected faster than the others in the second phase (Figure 2). This protection is likely caused by an interaction of the NTS with the β′ Zn+2 finger. A change in conformation of the DNA in the spacer region thus precedes the final isomerisation step, and could be coupled with the rearrangement of the α-CTDs described above. The absence of a higher rate for the equivalent site on the TS reflects the asymmetry resulting from a change in twist associated with the last steps of open complex formation, as the single-stranded TS is led deep into the core of the enzyme towards the active site. A similar rearrangement in this region was observed by Li and McClure and proposed by Davis and co-workers (3,19).
Time-resolved potassium permanganate reactivity measurements show that DNA melting precedes the rate-limiting step
The transcriptionally active complex formed by RNAP on the wt T7A1 promoter at 37°C in the presence of Mg2+ is characterized by DNA duplex unwinding from position m12 to position p2 (10). In order to determine the timing of DNA melting with respect to the appearance of protections from hydroxyl radical cleavage, we carried out time-resolved potassium permanganate (KMnO4) footprinting experiments. Potassium permanganate modification of thymines occurs when the unsaturated C5-C6 bond becomes accessible in the major groove due to significant distortion of the double helix disrupting base stacking (10). Our results clearly show that the permanganate signals resulting from modification of the thymines on the TS at m12, m10 and m6 appear with double exponential kinetics with similar rates to those measured for the appearance of protection of the NTS in the −10 region (C intermediate), while the modification of the thymine at m4 appears at a rate similar to the protections of the D intermediate (Figure 4 and Table 1). The reactivity of the thymine at m12 is almost twice that of the other bases within the –10 region. This reflects the key role of the fork junction structure for nucleation of DNA melting and for the positioning of the resulting bend in the double helix possibly increasing the accessibility to permanganate at this site (20). The reactivity of the thymine at p1 is significantly weaker than at other positions. The faster first phase in the appearance of reactivity at this base corresponds to a small change compared to the total reactivity observed at other bases (∼4%) and could reflect an early DNA conformational change caused by DNA wrapping upon formation of the first complexes. An additional increase in signal takes place within the slow phase of the kinetics. These results indicate that DNA melting from m12 to m4 takes place in at least two steps, that it precedes binding of downstream DNA within the jaws and that it is not the rate-limiting step in the formation of a transcriptionally active complex.
Figure 4.
Evolution of time-resolved potassium permanganate reactivity of the thymines on the template strand during formation of the RNAP–promoter complex. (a) Representative gel of the reactivity pattern obtained upon mixing RNAP on wt T7A1 promoter for different amounts of mixing time, in seconds, shown above each lane. (b) Plots of the increase in thymine modification as a function of time. The goodness to fit to either a single or double exponential was determined by a visual analysis of the residuals and an F-test analysis (see Supplementary Table 2 online and Supplementary Figure 4 online).
We have also carried out time-resolved permanganate experiments on the mutant T7A1 promoter where a consensus –10 sequence (C-10), TATAAT, replaces the wt −10, GATACT. On this promoter the signals at m12 and m4 appear at the same rate, DNA opening thus appears to take place more cooperatively due to the more stable interactions with the fork junction and the single-stranded NTS (20,21) (Figure 4 and Table 1).
The ‘off-pathway’ intermediate is not present during open complex formation on the T7A1 consensus –10 mutant
The identity of the bases within the –10 region plays an important role at different steps during formation of a transcriptionally active complex (20,22–24). The wt T7A1 promoter contains an imperfect –10 sequence (GATACT instead of TATAAT). The presence of guanosine at position –12 is especially detrimental to the stability of the intermediates leading to the open complex (22,25). In order to determine how the lack of optimal contacts with this region may influence the structure and stability of intermediates in this process we have carried out time-resolved X-ray footprinting experiments on a mutant T7A1 promoter with a consensus –10 sequence.
The rates of appearance of protection for the different intermediates decrease from upstream to downstream; however, the differences are not as distinct as for the wt promoter (Figures 2 and 3). The most probable interpretation of these results is that, in the presence of the consensus –10 sequence, the energetic barriers between the intermediates are lower than for the wt promoter all along the pathway.
The protection at m20-m4 NTS appears at the same rate as the protection of nucleotides at positions m31-m26 on NTS in the C intermediate, in the absence of a B′ intermediate, reflecting a more cooperative conformational change involving contacts with both the spacer and –10 regions. In addition, on the TS, the C intermediate is protected continuously from m23 to m14 by interactions with β′ and σ region 3, while on the wt promoter the protection due to σ is lacking. This could be directly due to the presence of the m12 non-consensus guanosine on the wt promoter, while the more extended protection of the D intermediate compared to wt can be attributed to the substitution of the m8 cytosine by the consensus adenine within the context of the already single-stranded DNA.
The amplitudes for the fast phase on both strands show a different pattern than that observed on the wt promoter. Indeed, in the C-10 data sets there is no large difference in the values for the amplitudes between the nucleotide positions m5−m3 and p3−p12 TS and p7−p12 NTS (intermediate E′ on wt) and those of surrounding sites. The absence of the ‘off pathway’ intermediate indicates that its formation is favored on the wt promoter by the decreased stability of the interactions of σ regions 3 and 2 with the non-consensus fork junction sequence and the single-stranded NTS (20,26,27).
Time-resolved footprinting at 20°C reveals a temperature-dependent conformational change inhibiting protection of downstream DNA within the fast phase
While changing the –10 sequence to its consensus form should increase the stability of the intermediates by favoring specific interactions of the protein side chains with this region of the promoter, a decrease in temperature could affect the rate of melting of the double helix and thus have an opposite effect on those intermediates in the pathway that are dependent on the presence of single stranded DNA. We compared the time-resolved hydroxyl radical footprints obtained at 20°C with those at 37°C in an effort to identify a conformational change responsible for the strong temperature effects previously observed at this promoter (10,12).
The formation of a transcriptionally active complex by RNAP at the T7A1 promoter shows a steep temperature dependence between 20°C and 37°C (10). At 20°C, only 10% of the complexes are transcriptionally active even though the DNA is melted from m12 to m4. In these experimental conditions, the reactivity to permanganate oxidation of the thymines at p2 (NTS) and at p1 (TS) are about 40% and 10%, respectively compared to their values at 37°C. The reactivity at these bases increases with increasing temperatures and the reactivity at p1 (TS) correlates with the appearance of transcription activity (10).
Differences in the rates of the first phase of the NTS protections clearly distinguish different intermediates, however, on the TS all the rates are more or less within the experimental error. As observed for the consensus –10 promoter, the protection at m31-m26 appears at the same rate as the protection downstream, at m20-m7, suggesting that there is a decrease in a kinetic barrier during the formation of contacts with the –10 region compared to the wt promoter at 37°C. This can be attributed to the specific recognition of double-stranded DNA by the σ subunit (22) and a decrease in the flexibility of the still double-stranded DNA reducing the number of possible orientations of the double helix.
The amplitudes of the first phase show a very different pattern than that observed at 37°C. These differences in amplitude allow us to propose the existence of two complexes in rapid equilibrium whose protection extends to m7-m3 NTS and m8-m6, m2-p3 TS (C and C′ in Figure 5c). These intermediates precede a conformational change resulting in the downstream extension of the protection on the TS (D). The notable difference compared to the other two data sets is that the protection downstream of p6 on the NTS is best described by a single exponential whose rate corresponds to the second, slow phase. Thus, at lower temperatures the conformational change resulting in burial of downstream DNA within the jaws becomes rate limiting at this promoter.
DISCUSSION
A1 is one of the first T7 phage genes to be expressed upon infection. Its transcription occurs in the absence of specific phage proteins; it is thus a naturally strong promoter that can successfully compete for the pool of the host's RNA polymerase enzymes. The strength of this promoter results from two main properties: the upstream contacts between the AT-rich UP element and the α-CTDs assuring the rapid recruitment and stabilization of the enzyme on the DNA and the less-than-perfect interactions with the non-consensus –10 sequence resulting in efficient promoter escape (28,29). Between these two events a series of isomerisations steps contribute to the wrapping and bending of the DNA on the surface of the enzyme and result in DNA melting and the burial of the TS deep within the enzyme to reach the active site (17,30). Previous biochemical characterization of this process has identified the specific protein–DNA interactions necessary for stable promoter binding and DNA melting (5). Here, we provide the first description of the melting process within the context of the promoter-binding pathway.
At the T7A1 promoter, specific binding by RNAP is mediated by early contacts of the α -CTDs and σ region 4 with the upstream, AT-rich UP element and the –35 region, respectively, in all three cases studied here: at 37°C and 20°C on the wt promoter and on a consensus –10 T7A1 mutant promoter at 37°C (Figure 6). The extent of protection observed in the following isomerization steps, however, depends on the cooperativity between spacer distortion and formation of stable interactions of σ regions 3 and 2 with the fork junction and the –10 region that bend the DNA towards the active site channel and induce DNA melting from m12 to m4 prior to the entry of downstream DNA within the jaws. Depending on the temperature or promoter sequence, different structural intermediates can thus accumulate before the downstream DNA becomes buried within the jaws of the enzyme formed by the downstream β lobe and the non-conserved or dispensable domains of β′ and β. At 37°C on the wt promoter, one of these intermediates appears to be ‘off-pathway’, possibly resulting from the lack of specific contacts with the melted –10 region. Finally, these results show that, while at 37°C the rate-limiting step in the pathway does not entail a significant change in the extent of DNA protection, at 20°C the downstream NTS becomes protected only within this slow conformational change, suggesting the presence of a different rate-limiting step. At the T7A1 promoter, the rate-limiting step is independent of the sequence of the –10 region and mainly involves protein conformational changes within the core enzyme placing the template strand at the active site (Figure 7).
DNA melting precedes rate-limiting conformational changes
These results provide the first description of the melting process by RNA polymerase within the context of the structural intermediates in the promoter-binding pathway. Two alternative models have been proposed for DNA melting in which separation of the two strands in the –10 region and at the transcription start site takes place either in a concerted fashion or in two separate steps (5,31). A two-step DNA melting process is observed with several mutant protein and/or promoter complexes (15,32–34) and on a set of naturally strong promoters, such as the wt tyrT promoter that, although it has a consensus –10 sequence, also has a stable, GC-rich, discriminator region upstream of the transcription start site providing a thermodynamic barrier to propagation of the transcription bubble (35). In all these cases, a complex with a short bubble is formed at equilibrium and propagation of the melted region from upstream to downstream is favoured by the addition of nucleotides and the initiation of the transcription process. The results described here obtained by time-resolved permanganate footprinting show that, at the T7A1 promoter, DNA melting occurs in at least two steps, from m12 to m4 within the fast phase followed by propagation to p1 within the slow phase. In addition, within the fast phase we can differentiate two sets of rates on the wt promoter, from m12 to m6 and then at m4, while at the C-10 promoter melting occurs simultaneously at m12 and at m4 (Figure 4 and Table 1). The formation of the first bubble at the wt T7A1 promoter is associated with a slower conformational change from the C to the D intermediate as compared to what is observed at the C-10 promoter. In these intermediates two sets of contacts are formed: the first by σ region 3 and 2 upstream of, and with, the –10 region (m19-m13, m8-m6 TS) and the second by σ region 1.2 and the β subunit with the discriminator region and with downstream double-stranded DNA (m2-p2 TS and m4-p6 NTS)(17,36,37), in agreement with the proposed role of these interactions in the strand separation process (33,38). It is interesting to note that this two-step melting process initiating outside of the RNAP catalytic cleft resembles the mechanism observed for DNA melting by σ54-RNAP (39).
This DNA-melting isomerization step at T7A1 occurs at a similar rate as the change in the protein's intrinsic fluorescence measured by Johnson and Chester for the binding of RNAP to the T7A1 promoter and is thus likely associated with a significant protein conformational change (12). The nature of this conformational change remains to be determined; however, it has been proposed that the presence of stable contacts between σ region 2.2 and the single-stranded DNA in the C-10 mutant may be transmitted via the β′ coiled coil to other domains of the enzyme favouring the conformational change necessary for subsequent isomerisation (40,41). This could result in the opening of the active-center cleft of the enzyme necessary to allow entry of double-stranded DNA downstream of the melted –10 region (Figure 7 complex D) (42). This step has recently been shown to be the target of antibiotic activity by specific binding to the clamp switch region (43,44). The accumulation of these relatively unstable structures prior to the rate-limiting step may help explain the efficacy of these drugs.
Recent results by deHaseth and co-workers (45) from measurements of the change in fluorescence of 2-aminopurine substituted at different sites of the –10 region of a consensus promoter also provide evidence for DNA melting following RNAP binding and preceding the formation of a stable complex. Moreover, the change in fluorescence appears at the same rate from m11 to m4, in agreement with what we observe here on the C-10 promoter by KMnO4 reactivity. The observation that melting of the DNA occurs within the early steps in the pathway contrasts with previously proposed models for the formation of a transcriptionally active complex that placed DNA melting within the last step (1,2,46–48). One possible explanation for this discrepancy is that previous studies of DNA melting were often carried out by trapping the intermediates at low temperatures assuming that the basic sequence of events is not affected by this parameter (1,2,10,46).
The non-linearity of the promoter recognition process
On the wt T7A1 promoter, the lack of stable contacts between the σ subunit of RNAP and the DNA at the non-consensus –10 region results in a less cooperative DNA binding and melting process compared to the C-10 promoter, as shown by the presence of additional intermediates and by the kinetics of DNA melting (Figures 4 and 6). In the consensus-10 mutant both the C and D intermediates show a more extended protection of the fork junction and downstream of p1, respectively, evidence of a larger, more stable, interaction surface compared to the wt promoter. The instability of these contacts on the wt promoter allows for the formation of an off-pathway complex (E′). The protection pattern of E′ is similar in extent to that of the D intermediate on the C-10 promoter (Figure 5). In the light of structural models that have been proposed for the open complex, the position of the downstream boundary of protection near p12 suggests that these protections result from the entry of downstream DNA within the active-center cleft and an interaction of the DNA with the β′ jaw and the downstream lobe of the β subunit, also known as β2 or lobe 1 of the β pincer (Figure 7) (17,36,38,49). We propose that the difference between these two complexes, E′ on wt and D on C-10, is due to a different orientation of the DNA within the jaws such that contacts further downstream are not accessible from the E′ complex, possibly leading to the formation of a ‘moribund’ complex (50).
This effect could also be coupled to the presence of σ region 1.1 within the jaws of the enzyme competing for the entry of the DNA into the active site channel (36). It was shown that the more stable interactions, formed between the protein and the DNA in a consensus –10 region, favours the complex in which σ region 1.1 has been ejected from the jaws (51,52). In the next step, the extension of the protection from p13 to p20 in the E intermediate is dependent on an interaction with the β′ G non-conserved domain (NCD) and possibly the β dispensable region 1 (DR1) (13,53,54) (Figure 7). Saecker and co-workers (13,54) have proposed that the folding of the β′ G and G′ domains upon interaction with downstream DNA follows σ1.1 ejection and is involved in the rate-limiting step at the λPR promoter. This induced fit is likely to play a role also in this context, in the isomerizations of the second, slower phase during the stabilization of the transcriptionally active complex (see below).
Work by Shimamoto and co-workers (50) has provided evidence for the presence of a branched pathway within different promoter contexts, showing that the stability of the off-pathway intermediates leading to transcriptionally ‘moribund’ complexes may be affected by environmental parameters such as pH and salt concentration. In addition, the possible regulatory role of the presence of these structures has been underlined by the effect of the greA and greB factors on their stability and the probability of accumulation of stalled initiation complexes (55,56). Here, we directly identify a step in the recognition pathway that may lead to the formation of such a branched pathway.
The rate-limiting step at 37°C involves a re-orientation of the DNA within the open complex
On the wt promoter the slow rate of appearance of protection in the second phase corresponds to the stabilization of the complex; its value is similar to that of the rate-limiting step in the appearance of the transcriptionally active complex as measured by abortive initiation assay (18). This rate-limiting step, however, is not accompanied by a significant change in the protection pattern or to a change in protein fluorescence (12). The same is observed for the C-10 mutant, the sequence of the –10 region is thus not significantly affecting this conformational change. We propose that unstable contacts of the enzyme with the DNA downstream from the transcription start site induce additional conformational changes to take place within the active site of the enzyme and the downstream jaws that reorient the DNA at the active site, as suggested by the weak increase in permanganate reactivity at p1. This internal rearrangement of the DNA within the active site channel could be coupled to the closing of the clamp and the folding of the conserved G and G′ β′ domains and of the non-conserved β′ NCD and β DR1 regions upon binding to downstream DNA (11,38,57) (Figure 7). This is consistent with a lack of change in the protein's fluorescence in this step since only one of the nine tryptophans of β′ is found within this region.
An open complex can form in the absence of these downstream interactions, as shown above and for some RNAP mutants (34,58); however, their presence is necessary for subsequent steps in the transcription process. The closing of the downstream clamp and the interaction of non-conserved elements of the E. coli enzyme with the DNA must not be too stable, or sequence specific, in order to allow for a rotation of the enzyme along the groove during transcription elongation (59); however, they are necessary to constrain downstream DNA in the correct orientation resulting in propagation of the transcription bubble (33,34) and for increased stability of both the transcription initiation and elongation complexes (38,54,60). In addition, the presence of this slow isomerization step requires that the RNAP–promoter interactions formed up to this point stabilize the complex long enough for it to arrive to completion. We propose that this increases the specificity of selection of the transcription initiation site, especially in the light of results showing that multiple potential core promoter sequences may be found upstream of coding regions in E. coli (61).
At 20°C a protein conformational change becomes rate limiting for the entry of downstream DNA within the jaws
In the data sets at 20°C, we observe the accumulation of three different intermediates (C, C′ and D in Figure 5c) prior to the rate-limiting step. The C and C′ complexes are distinguished by their decreased amplitude for the fast phase indicating that at this temperature the formation of the sharp bend of the DNA at the upstream end of the –10 region becomes rate limiting within the fast phase. This bend, which is favoured by the nucleation of DNA melting, is required for the DNA downstream of m8 NTS to become protected by its entry into the active-center cleft. In the following steps, the downstream end of the NTS, at p7-p20, becomes protected only within the second slow phase, in contrast to what is observed at 37°C (Figures 2 and 5c). While protection of the TS at p7-p13 is likely due to an interaction with β′, β DR1 and the downstream β lobe, the NTS down to p14 only cross-links to β′ (38,49), suggesting that the short footprint is due to β′ not being correctly folded onto the DNA (54).
Johnson and Chester (12) have shown that the rate of change of the protein's intrinsic fluorescence during T7A1 promoter binding exhibits a non-linear temperature dependence with a break point at 28°C, after which the isomerization rate decreases more steeply as a function of temperature. As described above, at 37°C this protein conformational change, associated with the propagation of DNA melting, is found within the fast phase of the kinetics of protection (C and D complexes). The extrapolation of the data from Johnson and Chester to 20°C would bring the value for this protein isomerizations to about 0.03 s–1 (12) which is similar to the rate we measure for the appearance of the downstream protection on the NTS at 20°C, 0.048 ± 0.002 s–1 for p7-p12 and 0.056 ± 0.003 s–1 for p13-p20 (Figure 2 and Supplementary Table 3). It is possible that as the temperature is decreased DNA melting occurs later in the pathway, only after the downstream DNA has completely entered the jaws (11). DNA melting at low temperatures is inhibited by the presence of the downstream β lobe (see below) (33). The decrease in the rate of the protein conformational change relieving this inhibition may in turn result from the lack of an interaction of the σ subunit with single-stranded DNA. Finally, the decreased flexibility of double-stranded DNA compared to a melted bubble and the different sequence requirement for the formation of stable contacts of the σ subunit with the still double-stranded DNA (22) may explain the absence of the off-pathway complex under these conditions.
More than one pathway for the formation of a transcriptionally active complex
Two different models have been proposed for the mechanism of DNA melting during open complex formation, the capture of single-stranded DNA by the protein, in particular by σ region 2 (34), and the active destabilization of the double helix by the protein (11,33). Our results suggest that, while at the T7A1 promoter at 37°C one can invoke a model for the formation of an open complex where wrapping and bending of the DNA on the surface of the protein destabilizes the double helix sufficiently for capture by σ, a more active role of the protein in DNA melting may become significant as the temperature is decreased, particularly at non-consensus promoters such as this one, with a lower AT content in the –10 region. At lower temperatures, the entry of the still double-stranded DNA within the active site channel and the jaws may distort the double helix at the –10 region enough to destabilize it and nucleate opening, but a significant protein conformational change may be required for this to take place. A mutant of RNAP where the downstream β lobe, including the DR1, has been deleted (Δ186-433) can form an open complex even at 0°C, albeit with a smaller bubble and a shortened footprint in the absence of nucleotides (58). This supports the idea that as the temperature is decreased a protein conformational change, dependent on the presence of this domain, becomes rate limiting, and eventually inhibiting, for DNA melting. Severinov and coworkers (58) proposed that this conformational change is dependent on a ‘two-stroke’ coupling of the upstream and downstream β lobes (β1 and β2 in Figure 7), the first interacting with the upstream end of the –10 region and the second with the downstream DNA. As described above, at 20°C, the downstream β lobe protection of the TS appears within the fast phase, supporting the possibility that an interaction, or a steric clash, with this domain at 20°C may be responsible for the decrease in the isomerisation rate for the conformational change leading to burial of downstream DNA and a change in protein fluorescence. It remains to be determined whether the presence of the downstream β lobe may affect the stability of σ region 1.1 within the jaws.
Thus, the differences between the pathways at 37°C and 20°C are not merely due to a decrease in the rates but also to the presence of different structural intermediates, as the double helix becomes more stable at lower temperatures a change in the mechanism for open complex formation takes place.
At the λPR promoter, the extent of protection of downstream DNA increases upon isomerizations of the closed complex (I1) to the open complex at 17°C (3). Significant changes are also observed in the protections upstream of the promoter, as it is observed here, suggesting that in both cases there is a rearrangement of upstream contacts upon open complex formation as the downstream contacts become more stable (3). However, on the T7A1 promoter the interactions of α-CTDs with a near-consensus UP element, absent in the λPR promoter, stabilize the early complexes with a short footprint that then expands downstream in a series of step-wise wrapping and bending steps. In the absence of the UP element, the stability of the early complexes is dependent on the wrapping of the DNA concomitant with the formation of downstream contacts and the entry of double-stranded DNA into the jaws, as it was also observed for the lacUV5 promoter (3,4,32,62). Whether DNA melting precedes the rate-limiting step in these specific promoter contexts at 37°C remains to be determined. However, recent results obtained on a consensus promoter lacking an UP element suggest that this may be the case (45).
The dependence of the mechanism of formation of a transcriptionally active complex on both the temperature and promoter sequence described here explains the differences that are observed between these results and the previous models where the open complex was assumed to be associated with the last step of the pathway mainly from the extrapolation of low temperature results to the temperatures for optimal growth (1,2,10,46). The presence of multiple possible pathways within the same promoter context endows robustness and specificity on the mechanism of formation of a transcriptionally active complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Agence Nationale pour la Recherche (grant JC05-53151 to B.S.); DAAD/PROCOPE (grant D/0628177 to A.R., H.H. and E.Z.); and Egide PHC PROCOPE (grant 14852YG to B.S. and M.B.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the staff of the ID10 beamline at the ESRF for their support, particularly Federico Zontone and Anders Madsen. We thank Anne Olliver, John Herrick and Andrew Travers for critical reading of the manuscript.
REFERENCES
- 1.Roe JH, Burgess RR, Record M., Jr. Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J. Mol. Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 2.Buc H, McClure WR. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 3.Davis CA, Bingman CA, Landick R, Record MT, Saecker RM. Real-time footprinting of DNA in the first kinetically significant intermediate in open complex formation by Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 2007;104:7833–7838. doi: 10.1073/pnas.0609888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spassky A, Kirkegaard K, Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985;24:2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- 5.Sclavi B. In: RNA Polymerases as Molecular Motors. Buc H, Strick T, editors. Cambridge, UK: Royal Society of Chemistry Book Publishing; 2008. [Google Scholar]
- 6.Hochschild A, Dove S. Protein-protein contacts that activate and repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 7.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 8.Travers A, Muskhelishvili G. DNA supercoiling – a global transcriptional regulator for enterobacterial growth? Nat. Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 9.Kadesch TR, Rosenberg S, Chamberlin MJ. Binding of Escherichia coli RNA polymerase holoenzyme to bacteriophage T7 DNA. Measurements of binding at bacteriophage T7 promoter A1 using a template competition assay. J. Mol. Biol. 1982;155:1–29. doi: 10.1016/0022-2836(82)90489-2. [DOI] [PubMed] [Google Scholar]
- 10.Zaychikov E, Denissova L, Meier T, Gotte M, Heumann H. Influence of Mg2+ and temperature on formation of the transcription bubble. J. Biol. Chem. 1997;272:2259–2267. doi: 10.1074/jbc.272.4.2259. [DOI] [PubMed] [Google Scholar]
- 11.Craig ML, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Saecker RM, Record MT., Jr. DNA footprints of the two kinetically significant intermediates in formation of an RNA polymerase-promoter open complex: evidence that interactions with start site and downstream DNA induce sequential conformational changes in polymerase and DNA. J. Mol. Biol. 1998;283:741–756. doi: 10.1006/jmbi.1998.2129. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RS, Chester RE. Stopped-flow kinetic analysis of the interaction of Escherichia coli RNA polymerase with the bacteriophage T7 A1 promoter. J. Mol. Biol. 1998;283:353–370. doi: 10.1006/jmbi.1998.2101. [DOI] [PubMed] [Google Scholar]
- 13.Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Capp MW, Record TM. Kinetic studies and structural models of the association of E. coli [sigma]70 RNA polymerase with the [lambda]PR promoter: large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 14.Sclavi B, Zaychikov E, Rogozina A, Walther F, Buckle M, Heumann H. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc. Natl Acad. Sci. USA. 2005;102:4706–4711. doi: 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brodolin K, Zenkin N, Severinov K. Remodeling of the [sigma]70 subunit non-template DNA strand contacts during the final step of transcription initiation. J. Mol. Biol. 2005;350:930–937. doi: 10.1016/j.jmb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 16.Balasubramanian B, Pogozelski W, Tullius T. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl Acad. Sci. USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 18.Johnson R, Bowers M, Eaton Q. Preparation and characterization of N-(1-pyrenyl)iodoacetamide-labeled Escherichia coli RNA polymerase. Biochemistry. 1991;30:189–198. doi: 10.1021/bi00215a028. [DOI] [PubMed] [Google Scholar]
- 19.Li XY, McClure WR. Characterization of the closed complex intermediate formed during transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 1998;273:23549–23557. doi: 10.1074/jbc.273.36.23549. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc. Natl Acad. Sci. USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenton M, Gralla J. Effect of DNA bases and backbone on sigma70 holoenzyme binding and isomerization using fork junction probes. Nucleic Acids Res. 2003;31:2745–2750. doi: 10.1093/nar/gkg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton MS, Gralla JD. Function of the bacterial TATAAT -10 element as single-stranded DNA during RNA polymerase isomerization. Proc. Natl Acad. Sci. USA. 2001;98:9020–9025. doi: 10.1073/pnas.161085798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKane M, Malone C, Gussin G. Mutations at position -10 in the lambda PR promoter primarily affect conversion of the initial closed complex (RPc) to a stable, closed intermediate (RPi) Biochemistry. 2001;40:2023–2031. doi: 10.1021/bi0019085. [DOI] [PubMed] [Google Scholar]
- 24.Heyduk E, Kuznedelov K, Severinov K, Heyduk T. A consensus adenine at position -11 of the nontemplate strand of bacterial promoter is important for nucleation of promoter melting. J. Biol. Chem. 2006;281:12362–12369. doi: 10.1074/jbc.M601364200. [DOI] [PubMed] [Google Scholar]
- 25.Roberts CW, Roberts JW. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 26.Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth PL. Different roles for basic and aromatic amino acids in conserved region 2 of Escherichia coli sigma(70) in the nucleation and maintenance of the single-stranded DNA bubble in open RNA polymerase-promoter complexes. J. Biol. Chem. 2001;276:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- 27.Fenton MS, Lee SJ, Gralla JD. Escherichia coli promoter opening and -10 recognition: mutational analysis of sigma70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vo N, Hsu L, Kane C, Chamberlin M. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- 29.Strainic MG, Jr, Sullivan JJ, Velevis A, deHaseth PL. Promoter recognition by Escherichia coli RNA polymerase: effects of the UP element on open complex formation and promoter clearance. Biochemistry. 1998;37:18074–18080. doi: 10.1021/bi9813431. [DOI] [PubMed] [Google Scholar]
- 30.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 31.Helmann JD, deHaseth PL. Protein-nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 32.Davis CA, Capp MW, Record MT, Saecker RM. The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc. Natl Acad. Sci. USA. 2005;102:285–290. doi: 10.1073/pnas.0405779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nechaev S, Chlenov M, Severinov K. Dissection of two hallmarks of the open promoter complex by mutation in an RNA polymerase core subunit. J. Biol. Chem. 2000;275:25516–25522. doi: 10.1074/jbc.M002511200. [DOI] [PubMed] [Google Scholar]
- 34.Young BA, Gruber TM, Gross CA. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- 35.Auner H, Buckle M, Deufel A, Kutateladze T, Lazarus L, Mavathur R, Muskhelishvili G, Pemberton I, Schneider R, Travers A. Mechanism of transcriptional activation by FIS: role of core promoter structure and DNA topology. J. Mol. Biol. 2003;331:331–344. doi: 10.1016/s0022-2836(03)00727-7. [DOI] [PubMed] [Google Scholar]
- 36.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 37.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 38.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst S. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 39.Wigneshweraraj S, Bose D, Burrows P, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, et al. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol. Microbiol. 2008;68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- 40.Arthur T, Burgess R. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta' subunit. J. Biol. Chem. 1998;273:31381–31387. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- 41.Young B, Anthony L, Gruber T, Arthur T, Heyduk E, Lu C, Sharp M, Heyduk T, Burgess R, Gross C. A coiled-coil from the RNA polymerase beta' subunit allosterically induces selective nontemplate strand binding by sigma(70) Cell. 2001;105:935–944. doi: 10.1016/s0092-8674(01)00398-1. [DOI] [PubMed] [Google Scholar]
- 42.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 43.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, et al. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belogurov G, Vassylyeva M, Sevostyanova A, Appleman J, Xiang A, Lira R, Webber S, Klyuyev S, Nudler E, Artsimovitch I, et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;457:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroeder L, Gries T, Saecker R, Record MJ, Harris M, DeHaseth P. Evidence for a tyrosine-adenine stacking interaction and for a short-lived open intermediate subsequent to initial binding of Escherichia coli RNA polymerase to promoter DNA. J. Mol. Biol. 2009;385:339–349. doi: 10.1016/j.jmb.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.deHaseth PL, Zupancic ML, Record MT. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hook-Barnard I, Hinton D. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul. Syst. Bio. 2007;1:275–293. [PMC free article] [PubMed] [Google Scholar]
- 49.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 50.Susa M, Sen R, Shimamoto N. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 2002;277:15407–15412. doi: 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- 51.Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- 52.Wilson C, Dombroski AJ. Region 1 of sigma70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
- 53.Chlenov M, Masuda S, Murakami K, Nikiforov V, Darst S, Mustaev A. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase beta' subunit. J. Mol. Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 54.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT. Solute probes of conformational changes in open complex (RPo) formation by Escherichia coli RNA polymerase at the lambdaPR promoter: evidence for unmasking of the active site in the isomerization step and for large-scale coupled folding in the subsequent conversion to RPo. Biochemistry. 2006;45:2161–2177. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Susa M, Kubori T, Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol. Microbiol. 2006;59:1807–1817. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reppas N, Wade J, Church G, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 57.Kontur WS, Saecker RM, Davis CA, Capp MW, Record MT. Solute probes of conformational changes in open complex (RPo) formation by Escherichia coli RNA polymerase at the lambdaPR promoter: evidence for unmasking of the active site in the isomerization step and for large-scale coupled folding in the subsequent conversion to RPo. J. Mol. Biol. 2006;45:2161–2177. doi: 10.1021/bi051835v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Severinov K, Darst SA. A mutant RNA polymerase that forms unusual open promoter complexes. Proc. Natl Acad. Sci. USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakata-Sogawa K, Shimamoto N. RNA polymerase can track a DNA groove during promoter search. Proc. Natl Acad. Sci. USA. 2004;101:14731–14735. doi: 10.1073/pnas.0406441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Artsimovitch I, Svetlov V, Murakami K, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 61.Huerta A, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 62.Amouyal M, Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J. Mol. Biol. 1987;195:795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.