Abstract
The human body is composed of diverse cell types with distinct functions. While it is known that lineage specification depends on cell specific gene expression, which in turn is driven by promoters, enhancers, insulators and other cis-regulatory DNA sequences for each gene1–3, the relative roles of these regulatory elements in this process is not clear. We have previously developed a chromatin immunoprecipitation-based microarray method (ChIP-chip) to locate promoters, enhancers, and insulators in the human genome4–6. Here, we use the same approach to identify these elements in multiple cell types and investigated their roles in cell type-specific gene expression. We observed that chromatin state at promoters and CTCF-binding at insulators are largely invariant across diverse cell types. By contrast, enhancers are marked with highly cell type-specific histone modification patterns, strongly correlate to cell type-specific gene expression programs on a global scale, and are functionally active in a cell type-specific manner. Our results defined over 55,000 potential transcriptional enhancers in the human genome, significantly expanding the current catalog of human enhancers and highlighting the role of these elements in cell type-specific gene expression.
We performed ChIP-chip analysis as previously described5 to determine binding of CTCF (insulator-binding protein) and the coactivator p300, and patterns of histone modifications in five human cell lines: cervical carcinoma HeLa, immortalized lymphoblast GM06690 (GM), leukemia K562, embryonic stem cells (ES), and BMP4-induced ES cells (dES). We first investigated 1% of the human genome selected by the ENCODE Consortium7, using DNA microarrays consisting of 385,000 50-mer oligos that tile 30 million basepairs (bp) at 36bp resolution. We examined mono- and tri-methylation of histone H3 lysine 4 (H3K4me1, H3K4me3) and acetylation of histone H3 lysine 27 (H3K27ac) at well-annotated promoters, reasoning that the state of these histone modifications would vary in a cell type-specific manner. To our surprise, the chromatin signatures at promoters are remarkably similar across all cell types (Figure 1A). Quantitative comparison of ChIP-chip enrichment (see Supplementary Information) revealed highly-correlated histone modification patterns at promoters across all cell types, with an average Pearson correlation coefficient of 0.71 (Figure S1A). This observation also holds for the larger set of Gencode promoters (Figure S2).
Figure 1. Chromatin modifications at promoters are cell type-invariant while those at enhancers are cell type-specific.
We employed ChIP-chip to map histone modifications (H3K4me1, H3K4me3, and H3K27ac) in the ENCODE regions in five cell types (HeLa, GM, K562, ES, dES). (A) We performed k-means clustering on the chromatin modifications found +/− 5 kb from 414 promoters, and observe them to be generally invariant across cell types. (B) As in (A), but clustering on 1423 non-redundant enhancers predicted on the basis of chromatin signatures.
Next, we identified putative insulators in the ENCODE regions for these cell types based on CTCF binding, as mammalian insulators are generally understood to require CTCF to block promoter-enhancer interactions3. We observed nearly identical CTCF binding sites (Table S1, Figure S1E) and highly-correlated CTCF enrichment patterns across all five cell types (Figure S1B), providing experimental support for the mostly cell type invariant function of CTCF as suggested by DNase hypersensitivity mapping results8.
We then investigated transcriptional enhancers in the ENCODE regions, performing ChIP-chip in HeLa, K562, and GM cells to locate binding sites for the transcriptional coactivator protein p300 (Tables S2–S4) as p300 is known to localize at enhancers9. We observed highly cell type-specific histone modification patterns at distal p300 binding sites (Figure S1F), in sharp contrast to the similarities in histone modifications across cell types at promoters. We then employed our chromatin signature-based prediction method5 to identify additional enhancers in the ENCODE regions in these cell types (Figure 1B, Table S5–S9). In addition to the characteristic H3K4me1 enrichment, predicted enhancers are frequently marked by acetylation of H3K27, DNaseI hypersensitivity, and/or binding of transcription factors and coactivators, and many contain evolutionarily conserved sequences (Figure S3–S4, see Supplementary Information). Unlike promoters and insulators, but similar to p300 binding sites, the histone modification patterns at predicted enhancers are largely cell type-specific (Figure 1B, S1D), in agreement with observations that H3K4me1 is distributed in a cell type-specific manner10.
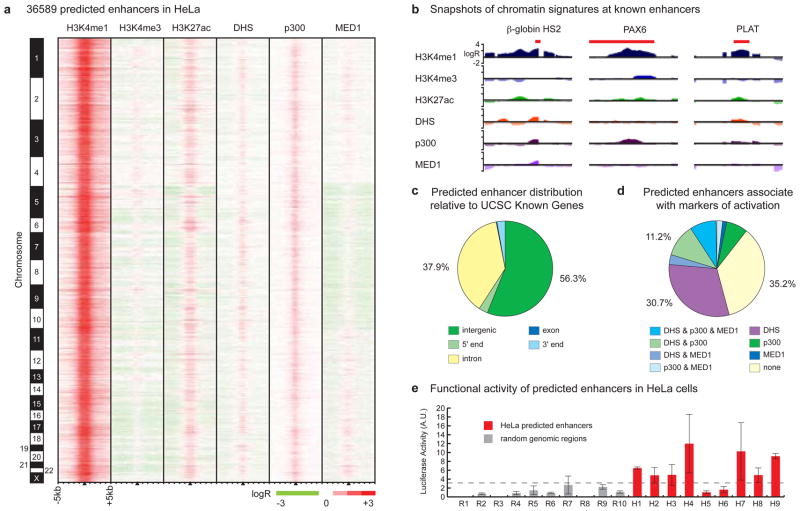
These results suggest that enhancers are the most variable class of transcriptional regulatory element between cell types and are likely of primary importance in driving cell type-specific patterns of gene expression. Knowledge of enhancers is therefore critical for understanding mechanisms that control cell type-specific gene expression, yet our incomplete knowledge of enhancers in the human genome has confined previous studies of gene regulatory networks mainly to promoters. To identify enhancers on a genome-wide scale and facilitate global analysis of gene regulatory mechanisms, we performed ChIP-chip throughout the entire human genome as described6, mapping enrichment patterns of H3K4me1 and H3K4me3 in HeLa cells. Using previously described chromatin signatures for enhancers5, we predicted 36589 enhancers in the HeLa genome (Figure 2A, Table S10, see Supplementary Information). This method correctly located several previously characterized enhancers, including the β-globin HS2 enhancer11 and distal enhancers for the PAX612 and PLAT (t-PA)13 genes (Figure 2B). Most predicted enhancers are distal to promoters (Figure 2C), exhibit strong evolutionary conservation (see Supplementary Information), and are marked by histone acetylation (H3K27ac), binding of coactivator proteins (p300, MED1), or DNaseI hypersensitivity (DHS) (Figure 2A, 2D) (see Supplementary Information). We verified the functional potential of predicted HeLa enhancers using luciferase reporter assays as described5 (see Methods). Of nine predicted enhancers that we evaluated, seven (78%) were active in reporter assays (Figure 2E, Table S11), with median activity significantly different from random genomic regions (p = 3.25 × 10−4). These results support the suitability of using chromatin signatures to identify genomic regions with enhancer function.
Figure 2. Genome-wide enhancer predictions in human cells.
(A) We predict 36589 enhancers in HeLa cells based on chromatin signatures for H3K4me1 and H3K4me3 as determined by ChIP-chip using genome-wide tiling microarrays and condensed enhancer microarrays (see Supplementary Information). Enhancer predictions are located at the center of 10 kb windows as indicated by black triangles, and ordered by genomic position. Enrichment data are shown for histone modifications (H3K4me1, H3K4me3, and H3K27ac), DNaseI hypersensitivity (DHS), and binding of p300 and MED1. (B) ChIP-chip enrichment profiles at several known enhancers (indicated in red) recovered by prediction: β-globin HS2 (chr11:5258371-5258665)11, PAX6 (chr11:31630500-31635000)12, PLAT (chr8:42191500-42192400)13 (5 kb windows centered on enhancer predictions; images generated in part at the UCSC Genome Browser). (C) Most enhancers have intergenic (56.3%) or intronic (37.9%) localization relative to UCSC Known Gene 5′-ends. (D) Most enhancers (64.8%) are significantly marked by DNaseI hypersensitivity, binding of p300, binding of MED1, or some combination thereof. (E) 7 of 9 enhancers predicted in HeLa cells were active in reporter assays (red bars) as compared to none of the random fragments selected as controls (gray), where activity is defined as relative luciferase value greater than 2.33 standard deviations (p = 0.01) above the median random activity (gray dashed line). Error bars represent standard deviation. Regions of ~1–2kb in size were randomly selected for validation in reporter assays based on histone modification patterns as in (A), overlap with features in (D), and sequence features amenable to cloning via PCR (see Supplementary Information).
We evaluated the predicted enhancers for conserved motif-like sequences using several hundred shuffled TRANSFAC motifs across 10 mammals in a phylogenetic framework that tolerates motif movement, partial motif loss, and sequencing or alignment discrepancies (see Methods). Predicted enhancers showed conservation for 4.3% of instances (at Branch-Length-Score > 50%, see Methods), substantially greater than for the remaining intergenic regions (2.9%, p < 1 × 10−100) and even promoter regions (3.9%, p = 1 × 10−57). Additionally, testing a list of 123 unique TRANSFAC motifs as described14 (see Supplementary Information), we found that 67 (54%) are over-conserved and 39 (32%) are enriched in predicted enhancers (Table S12). We also performed de novo motif discovery in enhancer regions using multiple alignments of 10 mammalian genomes (see Methods), revealing 41 enhancer motifs, of which 19 match known transcription factor motifs while 22 are novel (Table S13). These motifs show conservation rates between 7% and 22% in enhancers (median 9.3%), compared to only 1.1% for control shuffled motifs of identical composition. Furthermore, over 90% of these motifs appear to be unique to enhancers, as only 4 motifs are enriched in promoter regions and 12 are in fact depleted in promoters (Table S13), indicating that predicted enhancers contain unique regulatory sequences that may be specific to enhancer function.
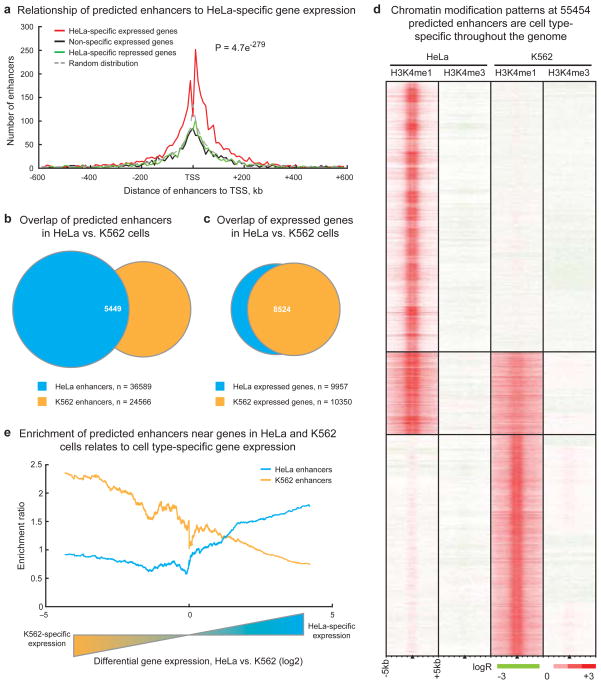
To investigate the association of predicted enhancers with HeLa-specific gene expression, we used Shannon entropy15 to rank genes by the specificity of their expression levels in HeLa as compared to three other cell lines (K562, GM06990, IMR90) (Figure S5, see Supplementary Information), then plotted the distribution of enhancers around genes within insulator-delineated domains (as defined by CTCF binding sites in Figure S6, see Supplementary Information). Predicted enhancers are strikingly enriched near HeLa-specific expressed genes (Figure 3A), particularly within 200 kb of promoters. We observed a 1.83-fold enrichment (p = 4.71 × 10−279) of predicted enhancers around HeLa-specific expressed genes relative to random (see Supplementary Information) and significant depletion of enhancers around non-specific expressed genes (p = 5.43 × 10−15) and HeLa-specific repressed (p = 4.63 × 10−2) genes.
Figure 3. Chromatin modifications at enhancers are globally related to cell type-specific gene expression.
(A) Enhancer localization relative to genes that are HeLa-specific expressed compared to K562, GM06990, and IMR90 cells (red), non-specific expressed (green), HeLa-specific repressed (black), and a random distribution (dashed grey). Predicted enhancers are enriched around HeLa-specific expressed genes within insulator-defined domains and depleted in domains of ubiquitous or non-expressed genes (p-value reflects significance of enhancer enrichment in domains of HeLa-specific expressed genes, see Supplementary Information). (B) Most enhancers predicted in HeLa and K562 cells are cell-type specific while (C) most genes in HeLa and K562 cells are not specifically expressed; n = integer number of enhancers or genes in each set. (D) Chromatin modification patterns are cell type-specific at the majority of 55454 enhancers predicted in HeLa and K562 cells. (E) Comparison of enhancer enrichment and differential gene expression between HeLa cells and K562 cells revealed that HeLa enhancers are enriched near HeLa-specific expressed genes (blue line) while K562 enhancers are enriched near K562-specific expressed genes (orange line).
To more directly investigate the relationship between chromatin modification patterns at enhancers and cell type-specific gene expression, we expanded our global analysis to another cell type. We performed genome-wide ChIP-chip for H3K4me1 and H3K4me3 in K562 cells and identified 24566 putative enhancers in this cell type using our chromatin signature-based enhancer prediction method (Table S14) (see Supplementary Information). Consistent with results in the ENCODE regions, the vast majority of enhancers predicted in K562 and HeLa cells are unique to either cell type (Figure 3B) even though most expressed genes are common between the cell types (Figure 3C). Chromatin modification profiles at predicted enhancers throughout the genome are highly cell type-specific (Figure 3D), with a Pearson correlation coefficient of −0.32. Furthermore, these differences seem to have regulatory implications, as domains with HeLa-specific expressed genes are enriched in HeLa enhancers but depleted in K562 enhancers, and vice-versa (Figure 3E) (see Supplementary Information). These observations hold across all five cell types in the ENCODE regions (see Supplementary Information). To assess the cell type-specificity of enhancer activity, we cloned enhancers predicted specifically in K562 cells (and not in HeLa cells) and subjected them to reporter assays in HeLa cells as described above. Of nine K562-specific enhancers tested, only two (22%) were active in HeLa cells (Figure S7), and the median activity of the K-562 specific enhancers was not significantly different from random (p = 0.11), suggesting that the enhancer chromatin signature is a reliable marker of cell type-specific enhancer function.
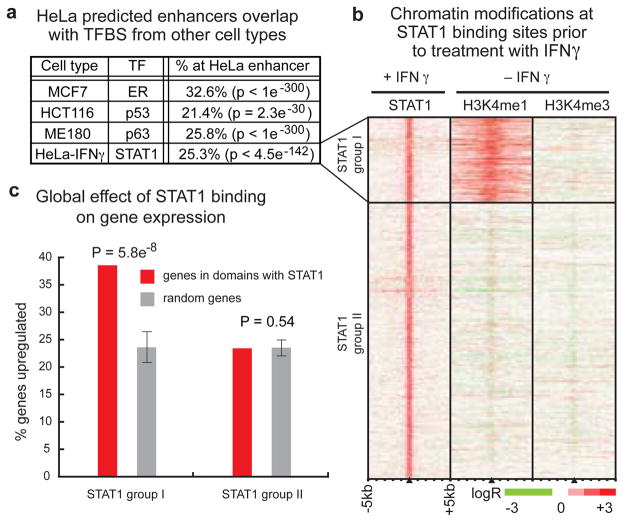
Though most enhancers are cell type-specific, the presence of predicted enhancers shared by HeLa and K562 (Figure 3B, 3D) suggests that some enhancers may be active in multiple cell types or conditions. We compared the HeLa enhancer predictions with the results of several genome-wide studies of binding sites for sequence-specific transcription factors in different cell types, namely estrogen receptor16 (ER), p5317, and p6318 in MCF7, HCT116, and ME180 cells, respectively. Interestingly, significant percentages of binding sites for each transcription factor (from 21.4% to 32.6%) overlap with predicted enhancers in HeLa cells (Figure 4A, Table S15), in sharp contrast to a significant depletion of the repressor NRSF/REST19 at predicted enhancers and minimal overlap with CTCF-binding sites (see Supplementary Information).
Figure 4. Chromatin modifications are associated with increased regulatory response of transcription factor binding sites at enhancers.
(A) Predicted enhancers in steady-state HeLa cells overlap with significant fractions of transcription factor binding sites (ER, p53, p63) in diverse cell types (MCF7, HCT116, ME180), as well as with STAT1 binding sites in HeLa cells treated with the cytokine interferon-gamma (HeLa-IFNγ) (TFBS = Transcription factor binding sites, TF = Transcription Factor). (B) Hundreds of STAT1 binding sites after treatment (+IFNγ) are marked by the enhancer chromatin signature in HeLa cells even prior to treatment (−IFNγ). (C) In HeLa cells treated with IFNγ (upper panel), gene expression is significantly (p = 5.8 × 10−8) more likely to be induced by STAT1 binding at sites with the enhancer chromatin signature (red, STAT1 group I) than by STAT1 binding at other distal sites (red, STAT1 group II) relative to a random distribution (gray). Error bars represent standard deviation.
To examine the potential role of enhancers in regulating inducible gene expression, we treated HeLa cells with the cytokine interferon-gamma (IFNγ) and identified binding sites for the transcription factor STAT1 throughout the genome using ChIP-chip. STAT1 generally binds its target DNA sequences only after IFNγ induction20 with a small fraction of binding possible prior to induction21. In IFNγ-treated HeLa cells, we identified 1969 STAT1 binding sites (Table S16), with 85.8% of STAT1 binding sites occurring distal to Known Gene 5′-ends. Comparison of these distal STAT1 binding sites with recent ChIP-seq analysis of STAT1 binding in uninduced HeLa cells21 shows only 6.5% of IFNγ-induced STAT1 binding sites are occupied by STAT1 prior to induction. We observed that 429 distal STAT1 binding sites overlapped enhancers predicted in HeLa cells prior to induction (Figure 4A, Table S15). The H3K4me1 enhancer chromatin signature is present prior to induction at these STAT1 binding sites, which we designated as STAT1 group I, while no evidence of this signature is visible at the remaining 1260 distal STAT1 binding sites, designated STAT1 group II (Figure 4B). Intriguingly, we observed significant relative induction of expression of genes in the domains of STAT1 group I binding sites after just 30 minutes of IFNγ-induction, while induction levels remained relatively unchanged for genes in the domains of other distal STAT1 group II binding sites during this time (Figure 4C). These findings suggest that an enhancer chromatin signature confers increased regulatory responsiveness to a STAT1 binding site, in agreement with our previous discovery of functional enhancers in HeLa cells that were marked by the enhancer chromatin signature but were not active until they were bound by STAT15.
Our findings offer the first genome-wide evaluation of the relationship between chromatin modifications at transcriptional enhancers and global programs of cell type-specific gene expression. We determined over 55,000 potential enhancers in the human genome and showed that the chromatin modifications at the enhancers correlate with cell type-specific gene expression and functional enhancer activity. Perhaps the most intriguing observation is the large number of enhancers identified from the investigation of just two cell lines. Since enhancers are mostly cell type-specific, our data suggest the existence of a vast number of enhancers in the human genome, on the order of 105–106, that are used to drive specific gene expression programs in the 200 cell types of the human body. Future experiments with diverse cell types and experimental conditions will be necessary to comprehensively identify these regulatory elements and understand their roles in the specific gene expression program of each cell type.
Methods Summary
HeLa, K562, and IMR90 cells were obtained from ATCC. GM06990 cells were acquired from Coriell. All were cultured under recommended conditions. Passage 32 H1 cells were cultured as described22 with/without 200ng/ml BMP4 for 6 days (RND systems). Chromatin preparation, ChIP, DNA purification, and LM-PCR were performed as described using commercially available and custom antibodies, and ChIP samples were hybridized to tiling microarrays and to custom condensed enhancer microarrays (NimbleGen Systems, Inc.) as described5,6. DNase-chip was performed and the data analyzed as described23. Cloning and reporter assays were performed as described5. Data were normalized as described5 and ChIP-chip targets for CTCF, p300, MED1, and STAT1 were selected with the Mpeak program. We used MA2C24 to normalize and call peaks on Nimblegen HD2 arrays. Enhancers were predicted and K-means clustering, intersection analysis, and evolutionary conservation analysis were performed as described5. Motif analysis was performed as described25. Gene expression was analyzed using HGU133 Plus 2.0 microarrays (Affymetrix) as described5. Specificity of expression was determined using a function of Shannon entropy15. We use the MAS5 algorithm from the Bioconductor R package to generate gene expression Present/Absent calls. Detailed methods may be found in the Supplementary Information. Supplementary data for the microarray experiments has been formatted for viewing in the UCSC genome browser via http://bioinformatics-renlab.ucsd.edu/enhancer
Supplementary Material
Coordinates are listed in hg17 for 729 non-redundant CTCF binding sites identified in HeLa, GM, K562, ES, and dES cells (see Supplementary Information).
Coordinates are listed in hg17 for 36589 enhancers predicted in HeLa cells based on chromatin signatures (see Supplementary Information).
Coordinates (hg17) and primers used to amplify regions containing predicted enhancers in HeLa (H1-H9) and K562 (K1-K9) cells for cloning and reporter assays, as well as random regions selected as controls (R1–R10).
Enrichment of motifs in enhancers was analyzed as described 25,28. Over-conservation and Enrichment are calculated as the excess conservation and overabundance, respectively, of a motif in enhancers or promoters relative to that expected for a random motif of identical composition. All significance values are expressed as Z-scores, corresponding to the number of standard deviations away from the mean of a normal distribution.
Known Match score represents the shared information content between novel and known motif28. Over-conservation is calculated as the excess conservation of a motif in enhancers or promoters relative to that expected for a random motif of identical composition. Enrichment is calculated as the over-abundance of a motif in enhancers or promoters relative to that expected for a random motif of identical composition. Enhancer-Specific motifs are those lacking significant promoter enrichment. All significance values are expressed as Z-scores, corresponding to the number of standard deviations away from the mean of a normal distribution.
Coordinates are listed in hg17 for 24566 enhancers predicted in K562 cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for each HeLa predicted enhancer with notation of overlap with experimentally determined transcription factor binding sites (see Supplementary Information).
Coordinates are listed in hg17 for 1969 STAT binding sites as determined by ChIP-chip.
Coordinates are listed in hg17 for p300 binding sites identified in HeLa cells (see Supplementary Information).
Coordinates are listed in hg17 for p300 binding sites identified in GM cells (see Supplementary Information).
Coordinates are listed in hg17 for p300 binding sites identified in K562 cells (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in HeLa cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in GM cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in K562 cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in ES cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in dES cells based on chromatin signatures (see Supplementary Information).
Acknowledgments
We thank members of the Ren lab for comments. This work was supported by funding from American Cancer Society (RDH), LICR (BR), NHGRI (BR), NCI (BR), and CIRM (BR).
Footnotes
Author contributions: RDH, NDH, GCH and BR designed the experiments; RDH, NDH, LFH, ZY, LKL, RKS, CWC, HL, and XZ conducted the ChIP-chip experiments; GCH and KAC analyzed the ChIP-chip data; GCH predicted enhancers; RDH and LKL conducted the reporter assays; JEA, RS and JAT provided hES cells and expression data; PK, AS and MK analyzed the transcription factor motifs; GEC performed and analyzed the DNaseI-chip experiments; NDH, GCH, RDH and BR wrote the manuscript.
Microarray data have been submitted to the GEO repository under accession numbers GSE14083, GSE8098, GSE7872, and GSE7118.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions
References
- 1.Heintzman ND, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nightingale KP, O’Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 4.Kim TH, et al. A high-Resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 6.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.ENCODE Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science (New York NY) 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 8.Xi H, et al. Identification and characterization of cell type-Specific and ubiquitous chromatin regulatory structures in the human genome. PLoS genetics. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Koch CM, et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome research. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King DC, et al. Evaluation of regulatory potential and conservation scores for detecting cis-Regulatory modules in aligned mammalian genome sequences. Genome research. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinjan DA, et al. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- 13.Wolf AT, Medcalf RL, Jern C. The t-PA -7351C>T enhancer polymorphism decreases Sp1 and Sp3 protein binding affinity and transcriptional responsiveness to retinoic acid. Blood. 2005;105:1060–1067. doi: 10.1182/blood-2003-12-4383. [DOI] [PubMed] [Google Scholar]
- 14.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schug J, et al. Promoter features related to tissue specificity as measured by Shannon entropy. Genome biology. 2005;6:R33. doi: 10.1186/gb-2005-6-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 17.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 18.Yang A, et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science (New York, N Y) 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 20.Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science (New York, N Y) 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 21.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig TE, et al. Feeder-independent culture of human embryonic stem cells. Nature methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 23.Crawford GE, et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nature methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song JS, et al. Model-based analysis of two-color arrays (MA2C) Genome biology. 2007;8:R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome research. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 28.Stark A, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coordinates are listed in hg17 for 729 non-redundant CTCF binding sites identified in HeLa, GM, K562, ES, and dES cells (see Supplementary Information).
Coordinates are listed in hg17 for 36589 enhancers predicted in HeLa cells based on chromatin signatures (see Supplementary Information).
Coordinates (hg17) and primers used to amplify regions containing predicted enhancers in HeLa (H1-H9) and K562 (K1-K9) cells for cloning and reporter assays, as well as random regions selected as controls (R1–R10).
Enrichment of motifs in enhancers was analyzed as described 25,28. Over-conservation and Enrichment are calculated as the excess conservation and overabundance, respectively, of a motif in enhancers or promoters relative to that expected for a random motif of identical composition. All significance values are expressed as Z-scores, corresponding to the number of standard deviations away from the mean of a normal distribution.
Known Match score represents the shared information content between novel and known motif28. Over-conservation is calculated as the excess conservation of a motif in enhancers or promoters relative to that expected for a random motif of identical composition. Enrichment is calculated as the over-abundance of a motif in enhancers or promoters relative to that expected for a random motif of identical composition. Enhancer-Specific motifs are those lacking significant promoter enrichment. All significance values are expressed as Z-scores, corresponding to the number of standard deviations away from the mean of a normal distribution.
Coordinates are listed in hg17 for 24566 enhancers predicted in K562 cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for each HeLa predicted enhancer with notation of overlap with experimentally determined transcription factor binding sites (see Supplementary Information).
Coordinates are listed in hg17 for 1969 STAT binding sites as determined by ChIP-chip.
Coordinates are listed in hg17 for p300 binding sites identified in HeLa cells (see Supplementary Information).
Coordinates are listed in hg17 for p300 binding sites identified in GM cells (see Supplementary Information).
Coordinates are listed in hg17 for p300 binding sites identified in K562 cells (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in HeLa cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in GM cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in K562 cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in ES cells based on chromatin signatures (see Supplementary Information).
Coordinates are listed in hg17 for enhancers predicted in dES cells based on chromatin signatures (see Supplementary Information).