Abstract
Transcription initiation in eukaryotes is controlled by nucleoprotein complexes formed through cooperative interactions among multiple transcription regulatory proteins. These complexes may be assembled via stochastic collisions or defined pathways. We investigated the dynamics of Fos-Jun-NFAT1 complexes by using a multicolor fluorescence resonance energy transfer assay. Fos-Jun heterodimers can bind to AP-1 sites in two opposite orientations, only one of which is populated in mature Fos-Jun-NFAT1 complexes. We studied the reversal of Fos-Jun binding orientation in response to NFAT1 by measuring the efficiencies of energy transfer from donor fluorophores linked to opposite ends of an oligonucleotide to an acceptor fluorophore linked to one subunit of the heterodimer. The reorientation of Fos-Jun by NFAT1 was not inhibited by competitor oligonucleotides or heterodimers. The rate of Fos-Jun reorientation was faster than the rate of heterodimer dissociation at some binding sites. The facilitated reorientation of Fos-Jun heterodimers therefore can enhance the efficiency of Fos-Jun-NFAT1 complex formation. We also examined the influence of the preferred orientation of Fos-Jun binding on the stability and transcriptional activity of Fos-Jun-NFAT1 complexes. Complexes formed at sites where Fos-Jun favored the same binding orientation in the presence and absence of NFAT1 exhibited an 8-fold slower dissociation rate than complexes formed at sites where Fos-Jun favored the opposite binding orientation. Fos-Jun-NFAT1 complexes also exhibited greater transcription activation at promoter elements that favored the same orientation of Fos-Jun binding in the presence and absence of NFAT1. Thus, the orientation of heterodimer binding can influence both the dynamics and promoter selectivity of multiprotein transcription regulatory complexes.
Many transcription regulatory proteins form heterodimers that bind to palindromic DNA sequences. Such heterodimers can potentially bind their recognition sites in either of two opposite orientations (1–5). Heterodimers that bind in opposite orientations present different protein surfaces for interactions with adjacent DNA binding proteins. Thus, the orientation of heterodimer binding may influence interactions with other transcription factors. Opposite orientations of heterodimer binding can result in different transcriptional activities (6–9). The mechanisms whereby the orientation of heterodimer binding controls transcriptional activity have not been defined.
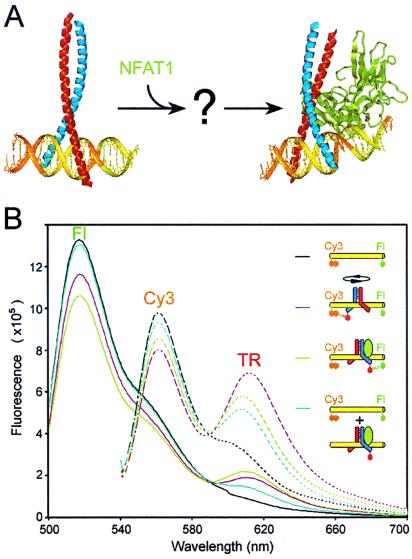
Fos and Jun family basic region–leucine zipper (bZIP) proteins regulate the expression of different genes in different cell types through cooperative interactions with other transcription factor families (10). Complexes formed by Fos and Jun with members of the NFAT family at composite regulatory elements induce cytokine gene expression in T cells activated by antigen presentation (11, 12). The x-ray crystal structures of DNA-bound complexes formed by the bZIP domains of Fos and Jun alone as well as by the DNA binding domains of Fos, Jun, and NFAT1 have been solved (Fig. 1A) (13, 14). In the Fos-Jun–AP-1 crystal, Fos-Jun heterodimers bind to the palindromic AP-1 sequence in both orientations (13). At different AP-1 sites, Fos-Jun heterodimers favor opposite binding orientations in solution (2–5). The contact interface between Fos-Jun and NFAT1 observed in the Fos-Jun-NFAT1–ARRE2 crystal can only be formed with heterodimers bound in one orientation (14). Heterodimers that are bound in the opposite orientation must be rotated by 180° about the dimer axis (1, 15). It is not known whether the heterodimer orientation is reversed by dissociation and rebinding of Fos-Jun or through reorientation of the heterodimer in association with DNA.
Figure 1.
Reversal of the orientation of Fos-Jun heterodimer binding by NFAT1. (A) The x-ray crystal structures of the bZIP domains of Fos (red)-Jun (blue) at the AP-1 site (only one of the two complexes is shown) (13) and Fos-Jun-NFAT1 (green) at the composite ARRE2 element (14) illustrate the structural barriers to Fos-Jun reorientation. Heterodimer reorientation may involve Fos-Jun dissociation from the composite binding site or a pathway that allows reversal of the binding orientation in association with DNA. (B) Multicolor fluorescence resonance energy transfer analysis of Fos-Jun binding, reorientation, and dissociation. The fluorescence emission spectra of a composite AP-1–NFAT site labeled with Cy3 and fluorescein (black), complexes formed by Fos-JunTR (magenta) and Fos-JunTR-NFAT1 (lime), and the dissociated components after incubation with genomic DNA (cyan) were determined after equilibration. The residual acceptor fluorescence of the dissociated complex was elicited by direct excitation. The solid lines show emission spectra obtained by excitation of fluorescein at 460 nm and the dashed lines of the same colors show emission spectra of the same complexes obtained by excitation of Cy3 at 530 nm. Fluorescence resonance energy transfer analysis of Fos-Jun binding, reorientation, and dissociation using single donor fluorophores on opposite ends of the oligonucleotide is shown in Fig. 5, which is published as supplemental material.
Materials and Methods
Fluorescent Proteins and Oligonucleotides.
Proteins encompassing the bZIP domains of Fos (residues 137-200) and Jun (residues 255-318), with unique cysteine residues at positions 136 and 254, respectively, were expressed as hexahistidine fusion proteins in Escherichia coli and purified (2, 15). The bZIP domains were modified to contain unique cysteine residues through replacement of the native cysteines by serines and addition of cysteine residues on the amino terminal sides of the bZIP domains. The extended rel-homology region of NFAT1 (residues 396-692) and full-length Fos and Jun were expressed and purified as described (16). Jun was labeled by incubation with Texas red C2 maleimide (Molecular Probes) and purified by either nickel chelate affinity or size exclusion chromatography (4, 5). Complexes formed by the labeled and unlabeled proteins exhibited indistinguishable dissociation rates. Similar results were obtained by using proteins labeled at three different positions on the amino-terminal side of the bZIP domain. Thus, the position of the fluorescent label on the proteins did not influence the results. Oligonucleotides containing composite AP-1–NFAT sites were prepared with fluorescein Cy3 at their 5′ ends as described in supplemental text, which is published on the PNAS web site, www.pnas.org.
Multicolor Fluorescence Resonance Energy Transfer.
To favor heterodimer formation by the labeled proteins, JunTR was incubated with a 2-fold molar excess of unlabeled Fos. Heterodimers and oligonucleotides were mixed at equimolar (20 nM) concentrations in 20 mM Tris⋅Cl (pH 7.6), 50 mM NaCl, 5% glycerol, 5 mM DTT, 0.5 mg/ml BSA. NFAT1 was added to the final concentration (100 nM) that elicited the maximal rate of Fos-Jun reorientation. The AP-1 competitor oligonucleotide contained the same nucleotide sequence as the binding site with the exception for the NFAT recognition sequence. Competitor oligonucleotides were used at concentrations required to observe the maximal rates of Fos-Jun (500 nM AP-1) and Fos-Jun-NFAT1 (10 μM AP-1–NFAT) complex dissociation. Sonicated herring genomic DNA was used at 1 mg⋅ml−1.
The fluorescence of fluorescein and Cy3 were measured by alternate excitation at 460 nm and 530 nm and emission at 517 nm and 562 nm, respectively. Texas red emission was measured at 610 nm. The slight overlap of the emission spectra (see Fig. 1B) did not influence interpretation of the results. Because of the large change in distances (≈30 Å) caused by heterodimer reorientation and because of the relatively unconstrained mobilities of the fluorophores indicated by low anisotropies, the changes in the relative efficiencies of energy transfer from opposite ends of the oligonucleotides reflect primarily changes heterodimer orientations. The anisotropy of fluorescein emission was determined by measuring the vertical and horizontal components of fluorescence and correcting for the grating factor. The anisotropy measurements were performed under the same conditions as the energy transfer experiments with the exception that shorter oligonucleotides (31 bp) were used. Reagents were injected manually into a stirred cuvette with a mixing time of ≈2 s. All experiments were performed at 25°C.
In Vitro Transcription.
The templates used for in vitro transcription contained one composite AP-1–NFAT binding site upstream of the basal promoter from the c-fos gene and a G-less transcription unit. The promoters differed only at the base pairs at the ±6 positions flanking the AP-1 site and in the position of the NFAT site relative to the AP-1 site as shown in Fig. 4B. Each promoter was fused to two different transcription units that generated transcripts of 390 bp and 242 bp (the templates for in vitro transcription are described in detail in the supplemental text).
Figure 4.
Influence of Fos-Jun orientation preference on the synergy between Fos-Jun and NFAT1. (A) Comparison of the dissociation rates of Fos-Jun-NFAT1 complexes at binding sites containing single base pair substitutions. The changes in fluorescein (green) and Texas red fluorescence after addition of genomic DNA to Fos-Jun-NFAT1 complexes at the binding site shown in each graph are shown. The diagram in each graph indicates the preferred orientation of Fos-Jun binding in the absence of NFAT1 (4, 5). (B) Analysis of the transcriptional activities of Fos-Jun-NFAT1 complexes at regulatory elements with different orientation preferences of Fos-Jun binding and different relative positions of AP-1 and NFAT1 recognition sequences. The orientation preferences of Fos-Jun binding and the positions of the NFAT recognition sequences on the various promoters are shown above the autoradiogram. The green arrow indicates the orientation of Fos-Jun binding required for stable interaction with NFAT1. Each reaction contained two templates with the promoters indicated below the lanes linked to G-less transcription units of different lengths indicated to the right of the promoters. The transcription reactions were supplemented with the proteins indicated above the lanes. Quantitation of the efficiencies of transcription activation and the relative transcriptional activities of the promoters in the absence of NFAT1 are shown in Fig. 7, which is published as supplemental material.
Transcription reactions were carried out in 40 mM Hepes (pH 7.6), 30 mM KCl, 5 mM MgCl2, 1 mM DTT, 4% glycerol, and 1% polyvinyl alcohol. Full-length Fos-Jun heterodimers (0.8 μM) and NFAT1 (396-692) (1 μM) were incubated with an equimolar mixture of the test templates (10 μg/ml each) and the AdML190 control template (2 μg/ml) at 30°C for 5 min. A total of 3 μM of an oligonucleotide competitor containing the composite AP-1–NFAT binding site was added. After 1-min incubation at 30°C, 50 μg of nuclear extract protein from Namalwa cells (16) was added. This extract contained low endogenous Fos-Jun and NFAT1 activities. After a 5-min incubation at 30°C, ATP and CTP (500 μM each), 3′O-methyl-GTP (100 μM), UTP (10 μM), [α32P]UTP (15 μCi), and creatine phosphate (10 mM) were added. After 30 min at 30°C, 100 units of RNase T1 were added and the incubation was continued for 30 min at 30°C. The nucleic acids were purified by phenol/chloroform extraction and ethanol precipitation. The transcripts were separated by denaturing PAGE and quantitated by PhosphorImager analysis.
Results and Discussion
Multicolor Fluorescence Resonance Energy Transfer Analysis of Fos-Jun-NFAT1 Complexes.
To investigate the reorientation of Fos-Jun by NFAT1 and to characterize the dynamics of Fos-Jun-NFAT1 complexes, a multicolor fluorescence resonance energy transfer assay was developed. In this approach, oligonucleotides containing a composite AP-1–NFAT binding site in the center were labeled with two different donor fluorophores (fluorescein and Cy3) on opposite ends of the duplex. The oligonucleotides were incubated with heterodimers formed by the bZIP domains of Fos and Jun. One subunit of the heterodimer was labeled with a fluorophore (Texas red) that could accept energy from either donor. The efficiency of energy transfer between each pair of fluorophores is a function of the distance between the fluorophores and the relative directions of their dipole axes (17, 18). Changes in the orientation of heterodimer binding thereby could be detected as reciprocal changes in the efficiencies of energy transfer from the two donor fluorophores. This assay allows independent determination of changes in the number and orientations of complexes.
The fluorescence emission spectra of Fos-JunTR and Fos-JunTR-NFAT1 complexes at a composite binding site demonstrated heterodimer reorientation (Fig. 1B). Binding of Fos-JunTR heterodimers to the labeled oligonucleotides resulted in energy transfer from both donors to the acceptor. Addition of NFAT1 caused an increase in energy transfer from one end (Fl), and a simultaneous decrease in energy transfer from the opposite end (Cy3), consistent with a shift in heterodimer orientation that brings the acceptor fluorophore on Jun in closer proximity to the fluorescein donor on the right end of the oligonucleotide. When the same complexes were formed on oligonucleotides where the positions of the donor fluorophores were exchanged, NFAT1 caused the converse changes in fluorescence emissions (compare Fl and Cy3 in Figs. 2B and 3A). The effect of NFAT1 on the fluorescence emissions was specific for composite AP-1–NFAT sites where Fos-Jun and NFAT1 exhibited cooperative binding and was not observed at sites where Fos-Jun and NFAT1 bind independently of each other (15) (data not shown). We therefore interpret reciprocal changes in fluorescence emissions that are reversed by exchange of the fluorophores on opposite ends of the oligonucleotide to reflect heterodimer reorientation.
Figure 2.
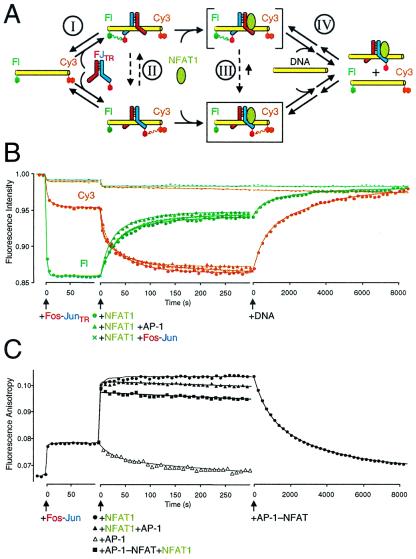
Analysis of the dynamics of the Fos-Jun-NFAT1 complex. (A) Alternate pathways of Fos-Jun heterodimer reorientation (I, II, III, and IV). The dashed lines indicate hypothetical reorientation of the heterodimer on DNA. The complex enclosed in a box represents the Fos-Jun-NFAT1 complex observed in the crystal (14). The complex in brackets represents a putative intermediate during Fos-Jun reorientation. (B) Kinetics of Fos-Jun heterodimer binding, reorientation by NFAT1, and Fos-Jun-NFAT1 complex disassembly analyzed by multicolor fluorescence resonance energy transfer. The fluorescein (green) and Cy3 (orange) emissions after the addition of Fos-JunTR, NFAT1, and genomic DNA are shown as a function of time. The effects of addition of NFAT1 alone (●), addition of NFAT1 followed by AP-1 oligonucleotide competitor (2 μM) (▴), and addition of NFAT1 followed by unlabeled Fos-Jun competitor (200 nM) (X) on fluorescence emissions are superimposed in the graph. The fluorescence emissions of complexes formed by unlabeled Fos-Jun heterodimers under the same conditions are shown as a control (−). The fluorescence emissions were corrected for dilution and were expressed relative to the emissions from the oligonucleotide before protein addition. (C) Analysis of intermediates during Fos-Jun-NFAT1 complex formation by fluorescence anisotropy. The anisotropy of fluorescein linked to the oligonucleotide was monitored during the same transitions examined in B. The increased anisotropy reflects the reduction in fluorophore mobility caused by protein binding to the oligonucleotide. The effects of addition of NFAT1 alone (●), addition of NFAT1 followed by AP-1 oligonucleotide competitor (10 μM) (▴), addition of NFAT1 (2 μM) after AP-1–NFAT oligonucleotide competitor (10 μM) (■), and addition of AP-1 oligonucleotide competitor (10 μM) alone (▵) are superimposed in the graph. Dissociation of Fos-Jun-NFAT1 complexes was measured after addition of AP-1–NFAT oligonucleotide (10 μM final concentration) after heterodimer reorientation. Fluorescence resonance energy transfer analysis of Fos-Jun binding, reorientation, and dissociation using single donor fluorophores on opposite ends of the oligonucleotide is shown in Fig. 6, which is published as supplemental material.
Pathway of Fos-Jun Heterodimer Reorientation by NFAT1.
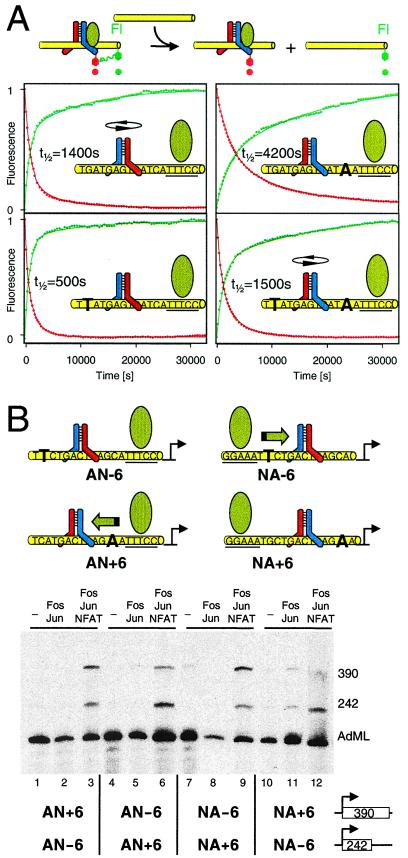
The changes in fluorescence emissions allow real-time analysis of Fos-Jun-NFAT1 complex dynamics (Fig. 2). There are many possible pathways of Fos-Jun reorientation by NFAT1 (upper left to lower right in Fig. 2A): I. Fos-Jun may dissociate and rebind the oligonucleotide. II. Fos-Jun may reorient on DNA before NFAT1 binding. III. Fos-Jun may form transient intermediates with NFAT1 and reorient in association with the composite binding site. IV. NFAT1 may displace the incorrectly oriented Fos-Jun, and a new heterodimer may bind to the preformed NFAT1 complex in the correct orientation. Addition of Fos-JunTR to the oligonucleotide labeled on both ends caused rapid quenching of both donor fluorophores (Fig. 2B), consistent with fast binding by the heterodimer. Addition of NFAT1 to the Fos-JunTR complex caused a gradual increase in energy transfer from Cy3 and a decrease in energy transfer from fluorescein, consistent with a shift in heterodimer orientation that brings the acceptor fluorophore in closer proximity to the Cy3 donor on the right end of the oligonucleotide. There was little effect of NFAT1 on the fluorescence of complexes formed by unlabeled Fos-Jun (Fig. 2B) or JunTR homodimers, confirming that the reciprocal changes in fluorescence emissions specifically reflect heterodimer reorientation.
To determine whether Fos-Jun dissociates from the composite binding site during heterodimer reorientation, unlabeled AP-1 site competitor oligonucleotide was added together with NFAT1 to Fos-JunTR bound to the composite element (Fig. 2B). Surprisingly, the reciprocal changes in energy transfer were observed in the presence of a 100-fold molar excess of AP-1 site oligonucleotide competitor. Heterodimer reorientation was observed at the highest oligonucleotide concentrations tested (1,000-fold molar excess). Reorientation also was not affected by the presence of a 10-fold molar excess of unlabeled Fos-Jun heterodimers (Fig. 2B; higher concentrations of Fos-Jun affected fluorescence emissions through nonspecific DNA binding). The effects of the competitors on the shift in fluorescence were less than 10%, corresponding to the experimental variation in the absence of competitors. Thus, more than 90% of the heterodimers that were reoriented by NFAT1 in the absence of competitors also were reoriented in the presence of AP-1 or Fos-Jun competitors. We define dissociation operationally as the loss of the preferential interaction between a protein and a DNA oligonucleotide. Consequently, these results demonstrate that Fos-Jun remains associated with the oligonucleotide during heterodimer reorientation.
The intermediates during Fos-Jun heterodimer reorientation were investigated further by measuring the anisotropy of fluorescein to monitor complex formation and stability. The shift in anisotropy caused by NFAT1 binding was much faster than the rate of reorientation observed by energy transfer under the same experimental conditions (Fig. 2 B and C). These results indicate that NFAT1 binding to sites where Fos-Jun was bound in the reverse orientation resulted in the formation of transient intermediates (indicated by a bracket in Fig. 2A) whose reorientation was rate-limiting in formation of mature Fos-Jun-NFAT1 complexes. The kinetics of reorientation was not affected by the concentrations of Fos-Jun or the binding site oligonucleotide (data not shown), consistent with a unimolecular reorientation mechanism. The intermediates formed on NFAT1 binding to Fos-Jun complexes were stable in the presence of AP-1 oligonucleotide competitors (Fig. 2C). To confirm that no heterodimer reorientation occurred before competitor addition and to verify that the difference in nucleotide sequences between the AP-1 competitor and the AP-1–NFAT binding site did not affect the result, we added AP-1–NFAT competitor to the reaction before NFAT1 (Fig. 2C). The slightly smaller shift in anisotropy reflects competition for NFAT1 by the added oligonucleotide. The Fos-Jun-NFAT1 complexes that formed in the presence of oligonucleotide competitor were more stable than complexes formed by either Fos-Jun or NFAT1 alone (t1/2 = 120 s and 35 s, respectively). Thus, NFAT1 and Fos-Jun mutually stabilized binding by each other during heterodimer reorientation. The gradual decrease in anisotropy in the presence of competitor oligonucleotides is consistent with the dissociation rate of mature Fos-Jun-NFAT1 complexes. The changes in anisotropy and fluorescence energy transfer in the presence of competitor oligonucleotides demonstrate that both Fos-Jun and NFAT1 were specifically bound to their recognition sites before heterodimer reorientation because complexes formed on oligonucleotides lacking these recognition sites had high dissociation rates (t1/2 < 2 s). These results reveal the existence of intermediates in which NFAT1 co-occupies the composite element with Fos-Jun heterodimers bound in the disfavored orientation. The results do not exclude reorientation of some complexes through other pathways, but indicate that the majority of complexes are reoriented through pathway III.
Kinetics of Fos-Jun Reorientation at Different Binding Sites.
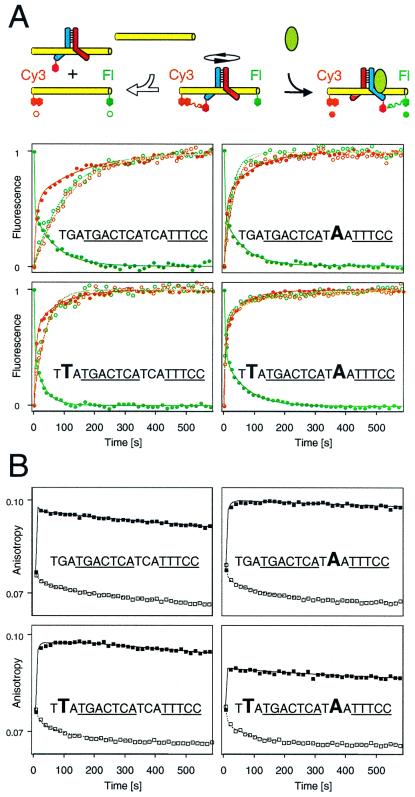
To investigate the relationship between Fos-Jun reorientation and dissociation, we compared their kinetics at different AP-1 sites (Fig. 3). At binding sites with slow dissociation rates, Fos-Jun reorientation was faster than dissociation (Fig. 3A Upper Left). At such sites, Fos-Jun dissociation can be excluded as a step in the reorientation pathway on kinetic grounds. Base substitutions outside the core AP-1 recognition element increased the rate of Fos-Jun dissociation up to 10-fold, but had small effects on Fos-Jun reorientation (Fig. 3A Lower Right). The kinetics of Fos-Jun reorientation were bi-phasic, consistent with multiple intermediates or pathways of reorientation. Neither of the rate coefficients for Fos-Jun reorientation correlated with the rates of dissociation at different binding sites. Thus, heterodimer dissociation is unlikely to be the rate-limiting step in reorientation of these Fos-Jun complexes.
Figure 3.
Effects of base substitutions flanking the AP-1 site on Fos-Jun reorientation and dissociation. (A) Comparison of the kinetics of Fos-Jun reorientation by NFAT1 (●) and Fos-Jun dissociation (○) at the binding sites shown in the graphs. The changes in fluorescein (green) and Cy3 (orange) fluorescence after addition of NFAT1 or genomic DNA to Fos-Jun heterodimers bound to the various sites were normalized to facilitate comparison of the rates. The AP-1 and NFAT recognition sequences are underlined, and base substitutions are shown in large type. (B) Comparison of the stabilities of Fos-Jun alone (□) and intermediates formed by Fos-Jun and NFAT1 (■) at different binding sites. The anisotropy of fluorescein was monitored after addition of AP-1–NFAT competitor oligonucleotide (□) or NFAT1 (2 μM final concentration) immediately after AP-1–NFAT competitor oligonucleotide (10 μM final concentration) (■) to Fos-Jun heterodimers bound to the oligonucleotide sequences shown in each graph linked to fluorescein.
To investigate the effects of these base substitutions on the stabilities of intermediates in the reorientation pathway, we measured the anisotropy of fluorescein in Fos-June NFAT1 complexes formed in the presence of AP-1–NFAT competitor oligonucleotides (Fig. 3B). NFAT1 caused a rapid and stable increase in anisotropy at all binding sites, consistent with formation of reorientation intermediates in which Fos-Jun and NFAT1 mutually stabilized binding by each other. There was little dissociation of these intermediates at any of the binding sites in the presence of competitor oligonucleotides in the time required for complete dissociation of Fos-Jun in the absence of NFAT1. The smaller anisotropy change at one binding site (Fig. 3B Lower Right) was not observed when NFAT1 was added before oligonucleotide competitor and likely was caused by the rapid dissociation of Fos-Jun from this site before NFAT1 binding. Thus, even at binding sites where the rate of Fos-Jun dissociation in the absence of NFAT1 was similar to the rate of heterodimer reorientation, less than 5% of the Fos-Jun heterodimers dissociated during reorientation in the presence of NFAT1. These results indicate that the intermediates formed by NFAT1 with Fos-Jun bound in the disfavored orientation stabilize Fos-Jun binding during heterodimer reorientation at all binding sites tested.
Role of Electrostatic Interactions in Fos-Jun Reorientation.
The contrasting effects of flanking base pair substitutions on the rates of Fos-Jun reorientation and on heterodimer dissociation suggest that interactions with flanking sequences are not altered during the rate-limiting step of Fos-Jun reorientation. In contrast, substitution of base pairs or amino acid residues that make direct contacts in the Fos-Jun–AP-1 crystal structure had parallel effects on the kinetics of reorientation and dissociation (data not shown). Base pairs flanking the AP-1 site influence the orientation of Fos-Jun heterodimer binding through electrostatic interactions (5). These results are consistent with the hypothesis that base-specific contacts to the core AP-1 recognition site are disrupted during reorientation, but that electrostatic interactions with flanking DNA sequences are maintained in the transition state and may stabilize intermediates in the reorientation pathway.
To investigate the role of electrostatic interactions during Fos-Jun reorientation, we examined reorientation in the presence of different concentrations of MgCl2. Fos-Jun reorientation in the presence of competitor oligonucleotides was observed at all MgCl2 concentrations tested (up to 10 mM). The efficiency of reorientation was reduced by MgCl2 concentrations above the physiological range. We previously have shown that high concentrations of MgCl2 attenuate electrostatic interactions between Fos-Jun and flanking DNA sequences (5). These results are consistent with stabilization of reorientation intermediates by electrostatic interactions with flanking DNA sequences. Electrostatic interactions also have been shown to contribute to other pathways that enhance the rate and efficiency of nucleoprotein complex formation including facilitated diffusion and intermolecular transfer (19). Thus, electrostatic interactions are important determinants of the dynamics of nucleoprotein complexes.
Effects of Heterodimer Orientation Preference on Fos-Jun-NFAT1 Complex Stability.
NFAT1 can overcome the intrinsic orientation preference of Fos-Jun and reorient the heterodimer at all composite sites examined (1, 15). To investigate whether the preferred orientation of heterodimer binding affected the properties of Fos-Jun-NFAT1 complexes subsequent to reorientation, we examined the effects of heterodimer orientation preference on the stabilities and transcriptional activities of Fos-Jun-NFAT1 complexes. First, we compared the dissociation rates of Fos-Jun-NFAT1 complexes at binding sites that contained base substitutions that have opposite effects on the preferred orientation of heterodimer binding (4, 5) (Fig. 4A). The dissociation rates were determined by measuring the increase in donor emission and the decrease in acceptor emission resulting from the loss of energy transfer on complex dissociation. Base substitutions that shift the orientation of Fos-Jun binding in the same direction as the interaction with NFAT1 (4, 5, 15) increased the stability of Fos-Jun-NFAT1 complexes (Fig. 4A Upper Right). Conversely, symmetry-related base substitutions that shift the orientation preference of Fos-Jun heterodimers in the opposite direction reduced the stability of the complexes (Fig. 4A Lower Left). Fos-Jun heterodimers exhibited similar rates of dissociation at these sites in the absence of NFAT1 (t1/2 = 38 s vs. t1/2 = 36 s, Fig. 3A). Symmetrical base substitutions on both sides of the AP-1 site had little effect on the rate of Fos-Jun-NFAT1 dissociation (Fig. 4A Lower Right). The differences in orientation preference caused by these base substitutions affected the stability of Fos-Jun-NFAT1 complexes regardless of the order of Fos-Jun and NFAT1 binding. The base substitutions flanking the AP-1 site had the same effect on the dissociation rates of Fos-Jun-NFAT1 complexes formed by unlabeled proteins (data not shown). Thus, the orientation preference of Fos-Jun heterodimer binding modulated the stability of Fos-Jun-NFAT1 complexes at composite binding sites.
Because the orientation of heterodimer binding influences the stability of Fos-Jun-NFAT1 complexes, it may seem surprising that intermediates in which Fos-Jun is bound in the opposite orientation do not dissociate during heterodimer reorientation (Fig. 3). However, when heterodimer reorientation is faster than dissociation of the intermediates, the fraction of complexes that dissociate before reorientation is small. Fos-Jun heterodimers can reorient during polyacrylamide gel electrophoresis (2), but it is not known whether reorientation in the absence of NFAT1 requires heterodimer dissociation. NFAT1 may facilitate Fos-Jun reorientation either directly by accelerating reorientation or indirectly by stabilizing intermediates in the reorientation pathway.
Influence of Heterodimer Orientation Preference on the Transcriptional Activities of Fos-Jun-NFAT1 Complexes.
To investigate the functional effects of the preferred orientation of Fos-Jun heterodimer binding, we compared the transcriptional activities of Fos-Jun-NFAT1 complexes at regulatory elements where heterodimers favored opposite binding orientations in the absence of NFAT1 (Fig. 4B, quantitation shown in Fig. 7A, which is published as supplemental material). Two templates containing AP1-NFAT regulatory elements with opposite orientation preferences for Fos-Jun binding (4, 5) were transcribed in the same reaction. Fos-Jun and NFAT1 activated transcription of both templates synergistically (Fig. 4B). The efficiency of transcription activation by Fos-Jun and NFAT1 was higher for the template where the orientation preference of Fos-Jun binding favored the interaction with NFAT1 (Fig. 4B, green arrow). This difference in transcriptional activities between the two templates was not observed in the absence of Fos-Jun and NFAT1, and Fos-Jun alone exhibited little transcription activation (Fig. 7B). Exchange of the promoters between the two transcription units reversed the relative transcriptional activities of the two templates (Fig. 4B, compare lanes 3 and 6). Thus, the orientation preference of Fos-Jun heterodimer binding controlled their relative transcriptional activities.
To confirm that the influence of the preferred orientation of Fos-Jun binding on transcription activation at these promoters was determined by their interaction with NFAT1, we examined the effects of the same base substitutions on transcription activation at promoters that contained the NFAT recognition element on the opposite side of the AP-1 site (Fig. 4B, lanes 7–12). The effects of the base substitutions were reversed at these promoters, indicating that the orientation preference of heterodimer binding affected transcription activation specifically through the interaction with NFAT1. Consequently, the orientation preference of heterodimer binding can influence the synergistic activation of transcription by Fos-Jun and NFAT1 at composite regulatory elements.
Conclusions
The reorientation of Fos-Jun heterodimers in association with NFAT1 and DNA indicates that transcription regulatory protein complexes are more dynamic than generally assumed. The dynamics of transcription regulatory protein complexes provides the potential for control of the rate, specificity, and duration of transcription activation. Multicolor fluorescence resonance energy transfer analysis of the dynamics of Fos-Jun-NFAT1 complexes revealed that Fos-Jun could reorient while remaining bound to DNA. Heterodimer reorientation is a novel mechanism for the facilitated assembly of nucleoprotein complexes. The order of Fos-Jun and NFAT1 binding to composite regulatory elements in the cell is unknown. However, because Fos and Jun are constitutively nuclear whereas NFAT1 is translocated into the nucleus after activation (20, 21), it is likely that Fos-Jun bind to some composite regulatory elements before NFAT1.
The results of both competition studies and kinetic analysis of Fos-Jun reorientation by NFAT1 support the existence of a facilitated pathway for reversal of Fos-Jun binding in association with DNA. Yet, steric considerations demand that interactions between the basic regions and the nucleotide bases must be severed during reorientation. Our data does not allow direct determination of the extent of the separation between the proteins and DNA during reorientation. However, the distinct effects of flanking base pair substitutions on dissociation and reorientation rates suggest that the influence of these base pairs on Fos-Jun interactions with DNA is not altered during reorientation. The reorientation pathway may be conceptually related to inelastic collisions between macromolecules that are predicted to facilitate protein–nucleic acid associations (19). However, in contrast to generic facilitated diffusion mechanisms, the intermediates during Fos-Jun reorientation are stabilized by NFAT1 and therefore are unaffected by the presence of competitors.
Fos-Jun heterodimers bind to different AP-1 sites with different orientation preferences (2–5). The orientation preference of heterodimer binding can influence both the stability and the transcriptional activity of Fos-Jun-NFAT1 complexes. Single base pair substitutions in flanking DNA sequences caused 8-fold differences in dissociation rates and 2-fold differences in relative transcriptional activities. These same single base pair substitutions caused a 4:1 ratio between the two orientations of Fos-Jun binding in the absence of NFAT1 (4, 5, 22). Thus, even modest differences in orientation preference can influence the promoter selectivity of Fos-Jun heterodimers. Heterodimers with stronger orientation preferences exhibited larger differences in the relative transcriptional activities of promoters that favored opposite binding orientations (22).
The orientation-dependent transcriptional synergy between Fos-Jun and NFAT1 demonstrates that transcription can be regulated through control of the orientation of heterodimer binding. The orientation of heterodimer binding at different regulatory elements can vary due to the recognition of flanking DNA sequences or asymmetric base pairs in nonconsensus binding sites as well as through interactions with other DNA binding proteins (1, 4, 5, 15). Because the orientation of heterodimer binding can be reversed without the requirement for dissociation and reassembly of the complex, it can provide a mechanism for rapidly reversible control of transcriptional activity.
Supplementary Material
Acknowledgments
We thank members of the Kerppola laboratory for valuable suggestions and constructive criticisms of the manuscript. V.R.R.-C. was supported by a Rackham Merit Fellowship.
Abbreviation
- bZIP
basic region–leucine zipper
References
- 1.Chen L, Oakley M G, Glover J N, Jain J, Dervan P B, Hogan P G, Rao A, Verdine G L. Curr Biol. 1995;5:882–889. doi: 10.1016/s0960-9822(95)00178-3. [DOI] [PubMed] [Google Scholar]
- 2.Leonard D A, Rajaram N, Kerppola T K. Proc Natl Acad Sci USA. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajaram N, Kerppola T K. EMBO J. 1997;16:2917–2925. doi: 10.1093/emboj/16.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonard D A, Kerppola T K. Nat Struct Biol. 1998;5:877–881. doi: 10.1038/2316. [DOI] [PubMed] [Google Scholar]
- 5.Ramirez-Carrozzi V R, Kerppola T K. J Mol Biol. 2001;305:411–427. doi: 10.1006/jmbi.2000.4286. [DOI] [PubMed] [Google Scholar]
- 6.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Nature (London) 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 7.Schrader M, Nayeri S, Kahlen J P, Muller K M, Carlberg C. Mol Cell Biol. 1995;15:1154–1161. doi: 10.1128/mcb.15.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chytil M, Peterson B R, Erlanson D A, Verdine G L. Proc Natl Acad Sci USA. 1998;95:14076–14081. doi: 10.1073/pnas.95.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falvo J V, Parekh B S, Lin H, C, Fraenkel E, Maniatis T. Mol Cell Biol. 2000;20:4814–4825. doi: 10.1128/mcb.20.13.4814-4825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinenov Y, Kerppola T K. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree G R. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 12.Kiani A, Rao A, Aramburu J. Immunity. 2000;12:359–372. doi: 10.1016/s1074-7613(00)80188-0. [DOI] [PubMed] [Google Scholar]
- 13.Glover J N, Harrison S C. Nature (London) 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Glover J N M, Hogan P G, Rao A, Harrison S C. Nature (London) 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 15.Diebold R J, Rajaram N, Leonard D A, Kerppola T K. Proc Natl Acad Sci USA. 1998;95:7915–7920. doi: 10.1073/pnas.95.14.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, et al. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 17.Förster T. Ann Physik. 1948;2:55–75. [Google Scholar]
- 18.Stryer L, Haugland R P. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berg O G, Winter R B, von Hippel P H. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 20.Luo C, Shaw K T-Y, Raghavan A, Aramburu J, Garcia-Cozar F, Perrino B A, Hogan P G, Rao A. Proc Natl Acad Sci USA. 1996;93:8907–8912. doi: 10.1073/pnas.93.17.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, McKeon F. Nature (London) 1999;398:256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- 22.Ramirez-Carrozzi, V. R. & Kerppola, T. K. (2001) J. Biol. Chem., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.