Abstract
Ras plays a pivotal role in many cellular activities, and its subcellular compartmentalization provides spatial and temporal selectivity. Here we report a mode of spatial regulation of Ras signaling in the Golgi apparatus by two highly homologous proteins PAQR10 and PAQR11 of the progestin and AdipoQ receptors family. PAQR10 and PAQR11 are exclusively localized in the Golgi apparatus. Overexpression of PAQR10/PAQR11 stimulates basal and EGF-induced ERK phosphorylation and increases the expression of ERK target genes in a dose-dependent manner. Overexpression of PAQR10/PAQR11 markedly elevates Golgi localization of HRas, NRas and KRas4A, but not KRas4B. PAQR10 and PAQR11 can also interact with HRas, NRas and KRas4A, but not KRas4B. The increased Ras protein at the Golgi apparatus by overexpression of PAQR10/PAQR11 is in an active state. Consistently, knockdown of PAQR10 and PAQR11 reduces EGF-stimulated ERK phosphorylation and Ras activation at the Golgi apparatus. Intriguingly, PAQR10 and PAQR11 are able to interact with RasGRP1, a guanine nucleotide exchange protein of Ras, and increase Golgi localization of RasGRP1. The C1 domain of RasGRP1 is both necessary and sufficient for the interaction of RasGRP1 with PAQR10/PAQR11. The simulation of ERK phosphorylation by overexpressed PAQR10/PAQR11 is abrogated by downregulation of RasGRP1. Furthermore, differentiation of PC12 cells is significantly enhanced by overexpression of PAQR10/PAQR11. Collectively, this study uncovers a new paradigm of spatial regulation of Ras signaling in the Golgi apparatus by PAQR10 and PAQR11.
Keywords: Ras, Signal transduction, Golgi apparatus, compartmentalization, ERK
Introduction
Ras proteins are essential components of intracellular signaling cascades that regulate a variety of fundamental cellular activities, including proliferation, apoptosis, differentiation and senescence 1. Recently, compartmentalized signaling has become an important theme that imparts an increased complexity to signal transduction 2, 3. In addition to its main actions on the plasma membrane (PM), Ras-mediated signaling has been observed in endosomes 4, 5, endoplasmic reticulum (ER) 6, 7, Golgi apparatus 4, 8, 9 and mitochondria 10, 11. Scaffold proteins, guanine exchange factors (GEFs) and adapter proteins are implicated in the spatial regulation of Ras-mediated signaling on internal membranes. Ras signaling in endosomes is mediated by MP1 12, p14 13 and β-arrestin 14. Ras signaling in the Golgi apparatus involves RasGRP1 15 and PLCγ 16, whereas Ras to ERK signaling in ER can be mediated by RasGRFs 6.
Ras undergoes a dynamic trafficking within cells in a membrane-restricted manner 17, 18. There are four isoforms of Ras proteins: HRas, NRas, KRas4A and KRas4B. All four Ras isoforms share > 90% sequence homology. The major structural difference among Ras isoforms is located at the C-terminal 23-24aa hypervariable region (HVR), which contains the membrane-interacting and -anchoring sequences. On the other hand, the regions for effector interaction and guanine nucleotide binding are almost identical among Ras isoforms. After being synthesized in cytosolic ribosome, all four Ras isoforms undergo farnesylation in the cytosol at the cysteine of the C-terminal CAAX motif by farnesyl protein transferase, followed by proteolysis of the AAX sequence in ER. Farnesylation of Ras proteins only provides a weak signal for membrane binding, and a second motif in HVR is required for membrane association 19. In the case of HRas, NRas and KRas4A, the second membrane-anchoring signal consists of one or two palmitoylation groups (one for NRas and KRas4A, two for HRas) at cysteine(s) adjacent to the farnesylated cysteine. The palmitoylation of these Ras proteins occurs at the surface of the Golgi apparatus and the palmitoylated Ras traffics to the PM via a secretory pathway and inserts into the PM. On the other hand, the second membrane-targeting signal for KRas4B is comprised of a hexa-lysine polybasic sequence that electrostatically interacts with acidic lipid headgroup of the PM. Therefore, the difference in palmitoylation of Ras isoforms can explain why HRas, NRas and KRas4A, but not KRas4B, can be localized at the Golgi apparatus. In addition, palmitoylated Ras proteins undergo a dynamic de/re-acylation cycle and depalmitoylation detaches Ras from the PM, and the solely farnesylated Ras rapidly repartitions to other intracellular membranes, such as ER and Golgi apparatus 20. Ras proteins are repalmitoylated in the Golgi and then redirected back to the PM via secretory vesicles. During the dynamic compartmentalization of Ras within a cell, the Golgi pool of palmitoylated Ras can be considered as a trafficking intermediate. It is unknown at present why palmitoylated Ras has a relatively high concentration in the Golgi apparatus. It has been recognized that growth factor-induced activation of Ras signaling on the PM is rapid and transient, while Ras signaling in the Golgi apparatus is delayed and sustained 9, 20, 21. However, how Ras activity in the Golgi apparatus can be maintained and uncoupled from initial activation on the PM upon growth factor stimulation remains unclear 17.
The PAQR (progestin and AdipoQ receptors) family was first identified in 2005 22, characterized by an ancient seven-transmembrane motif. Evolutionary analysis reveals that PAQR family members exist throughout the eukaryotic kingdom, as well as in selected species of eubacteria 22. Eleven PAQR family members were discovered in humans with high conservation. The existence of PAQR family members in ancient species and the high conservation characteristics indicate that the proteins of this family may exert important biological functions. Recently, our laboratory identified the biological function of PAQR3 within the PAQR family and renamed it RKTG (Raf kinase trapping to Golgi) 23. We found that RKTG functions as a negative regulator of Ras to ERK signaling by sequestrating Raf kinase in the Golgi apparatus 23, 24, and such regulation is likely associated with the development of cancer in mouse models 24, 25. Adiponectin receptors (PAQR1 and PAQR2), in addition to their established role in glucose and lipid metabolism 26, were also found to regulate p38 26, JNK 27 and tumorigenesis 28, 29, 30. These findings, therefore, indicate that members of the PAQR family may regulate basic cell activities through MAPK signaling pathways.
In pilot studies, we analyzed the subcellular distribution of PAQR family members and observed that PAQR10 and PAQR11 were exclusively localized in the Golgi apparatus, similar to PAQR3/RKTG. Among the 11 PAQR members, PAQR10 and PAQR11 have the deepest evolutionary root, with a high sequence similarity to the bacterial hemolysin III-type proteins 22. PAQR11 displays a relatively ubiquitous distribution in a majority of tissues including lung, intestine, skeletal muscle, adipose and heart, while PAQR10 is highly expressed in the brain and testis 22. PAQR11 may have effects on macrophage differentiation 31, but the biological actions of PAQR10 and PAQR11 remain poorly resolved. In this study, using molecular and cellular approaches, we demonstrate that PAQR10 and PAQR11 can bind, mobilize and activate Ras in the Golgi apparatus, leading to activation of ERK signaling pathway in the Golgi apparatus, thereby identifying PAQR10 and PAQR11 as a new class of Ras modulators in the Golgi apparatus.
Results
PAQR10 and PAQR11 are localized to the Golgi apparatus
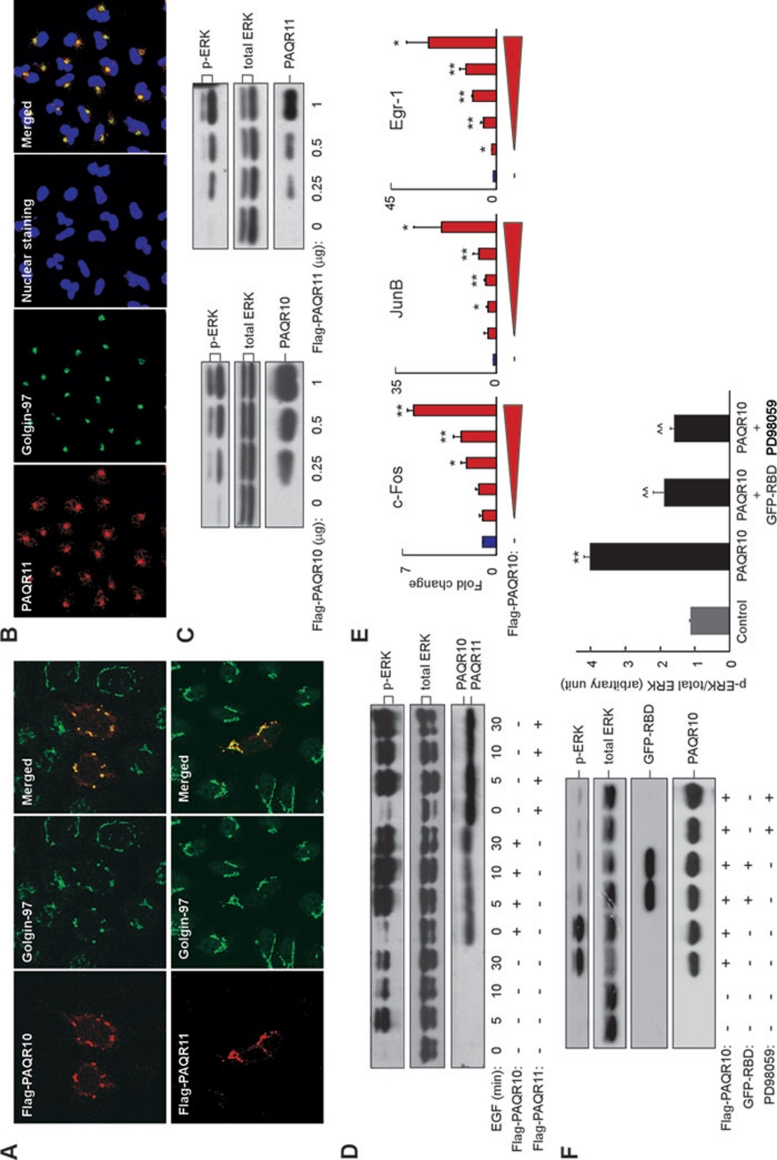
PAQR10 and PAQR11 are two highly homologous proteins within the PAQR family with a sequence homology of 83% at the amino acid level. Hydrophobicity analysis indicated that both PAQR10 and PAQR11 contain seven transmembrane domains (Supplementary information, Figure S1), similar to the topological structure of other members in PAQR family 22, 23, 26. To identify the potential cellular functions of these two genes, we first investigated subcellular distribution of PAQR10 and PAQR11. When transiently transfected into Hela cells, N-terminally-tagged human PAQR10 and PAQR11 were both restricted to the Golgi apparatus and colocalized with Golgin-97, a Golgi marker (Figure 1A). To further confirm that PAQR10 and PAQR11 are Golgi proteins, we generated polyclonal antibodies against PAQR10 and PAQR11, respectively (Supplementary information, Figure S2). Consistently, we found that the staining pattern of endogenous PAQR11 overlapped well with Golgin-97 in A549 cells, which express high levels of PAQR11 (Figure 1B).
Figure 1.
PAQR10 and PAQR11 are localized in the Golgi apparatus and activate Ras/ERK signaling. (A) Ectopically expressed PAQR10 and PAQR11 are localized in the Golgi apparatus. Hela cells were transfected with Flag-tagged PAQR10 and PAQR11, followed by immunostaining and confocal analysis. The Golgi was labeled by Golgin-97. (B) Endogenous PAQR11 is localized in the Golgi. A549 cells were stained with affinity-purified, anti-PAQR11 antibody and the Golgi marker Golgin-97. (C) Dose-dependent activation of ERK phosphorylation by PAQR10 and PAQR11. HEK293T cells were transiently transfected with increasing amounts of Flag-tagged PAQR10 (left panel) or Flag-tagged PAQR11 (right panel). Twenty-four hours after transfection, the cells were serum-starved for 6 h and the cell lysate was used in immunoblotting, using antibodies against phosphorylated ERK, total ERK and Flag tag. The same experiment was carried out at least three times, and one of the representative data is shown. This is the same for most immunoblotting analyses throughout the study. (D) PAQR10 and PAQR11 enhance and prolong EGF-induced ERK signaling. HEK293T cells were transiently transfected with Flag-tagged PAQR10 and PAQR11. Twenty-four hours after transfection, cells were serum-starved for 6 h before treatment with EGF (100 ng/ml) for various lengths of time. Total cell lysate was used in immunoblotting. (E) PAQR10 stimulates expression of transcription factors downstream of ERK in a dose-dependent manner. HEK293T cells were transiently transfected with increasing amounts of Flag-PAQR10. Twenty-four hours after transfection, the cells were collected and real-time RT-PCR was used to determine the expression of cFos, junB, Egr-1, and actin. Fold change of each gene compared with actin was shown as mean ± SD for three independent experiments. The fold change of control group was set to 1. *P < 0.05 and **P < 0.01 in comparison with the control group. (F) PAQR10-mediated stimulation of ERK phosphorylation is inhibited by GFP-RBD and PD98059. HEK293T cells were transiently transfected with the plasmids or treated with 10 μM of PD98059 as indicated. Twenty-four hours after transfection, the cells were serum-starved for 6 h, and the total cell lysate was used in immunoblotting. The right panel represents pERK/total ERK ratio (mean ± SD) calculated from densitometer analyses of three independent experiments. **P < 0.01 as compared to the control group and ^^P < 0.01 as compared to PAQR10 alone.
Activation of ERK signaling by PAQR10/PAQR11
Members of the PAQR family have been implicated in the regulation of MAPK signaling pathway 23, 26, and so, we examined ERK signaling following graded overexpression of PAQR10/PAQR11. In HEK293T cells, overexpression of PAQR10 or PAQR11 dose-dependently increased the phosphorylation of ERK (Figure 1C). Overexpression of PAQR10 or PAQR11 also significantly enhanced and prolonged the phosphorylation of ERK upon EGF treatment (Figure 1D). As a control, another member of the PAQR family, AdipoR2 (PAQR2), had little effect on EGF-stimulated ERK phosphorylation (Supplementary information, Figure S3). We also examined the expression levels of a few target genes downstream of ERK signaling, including c-Fos, JunB and Egr-1 . Consistently, overexpression of PAQR10 elevated the expression levels of these genes in a dose-dependent manner (Figure 1E). As a control experiment, TGN38, another Golgi-localized protein 32, had no effect on the expression of these genes (Supplementary information, Figure S4). In addition, we found that PAQR10-activated ERK phosphorylation was abrogated by inhibitors that were specific for the Ras signaling cascade (Figure 1F). Green fluorescence protein-Ras-binding domain (GFP-RBD) is a RBD of Raf-1 fused with GFP and this protein could efficiently bind with Ras in the GTP-bound state and competitively inhibit Ras activity 9, 33, 34. PD98059 is a specific inhibitor of MEK. The PAQR10-induced transcription of Egr-1 was blocked by GFP-RBD and PD98059 (Supplementary information, Figure S5). These data, collectively, indicate that PAQR10 and PAQR11 are able to activate Ras/ERK signaling pathway.
PAQR10 and PAQR11 increase retention of HRas, NRas and KRas4A in the Golgi apparatus
Considering that PAQR10/PAQR11 are Golgi-localized proteins and may directly act on Ras signaling, we next investigated whether they can alter localization of Ras protein in the Golgi apparatus. In humans, there exists four Ras isoforms including HRas, NRas, KRas4A and KRas4B 35. These four isoforms have almost complete sequence conservation in the N-terminal region, with the Ras HVR in the C terminus enabling differential membrane targeting and localization 19, 36, 37. All Ras isoforms exhibit PM localization, whereas the extent of endomembranous localization varies with a different degree of affinity for the Golgi apparatus in the order of NRas ≥ HRas, KRas4A >> KRas4B 38.
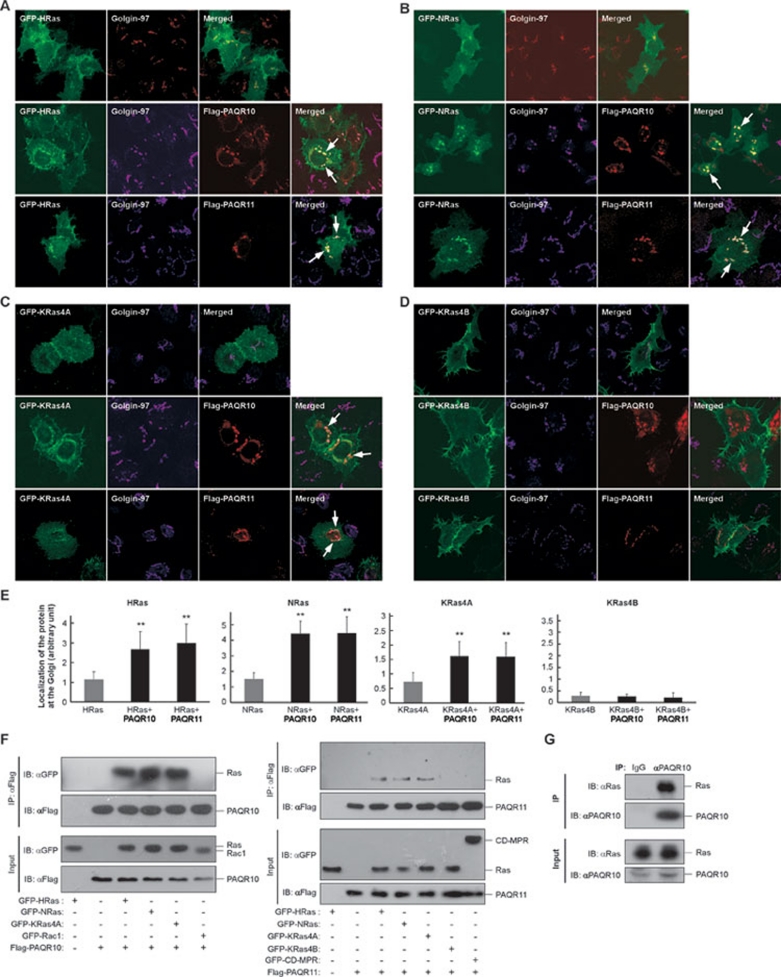
When HRas was overexpressed in Hela cells, it was largely localized on the PM with a little Golgi localization (Figure 2A), consistent with previous reports 4, 8. However, when coexpressed with Flag-tagged PAQR10 or PAQR11, the localization of HRas in the Golgi apparatus was significantly increased with a clear colocalization with PAQR10/PAQR11 and Golgin-97 (Figure 2A). Overexpression of PAQR10/PAQR11 also markedly increased Golgi localization of NRas and KRas4A (Figure 2B and 2C), but not KRas4B (Figure 2D). As a control experiment, PAQR10 had little colocalization with GFP (Supplementary information, Figure S6A), and another small G protein Rac1 (Supplementary information, Figure S6B). PAQR11 could not either increase Golgi localization of CD-MPR (Supplementary information, Figure S6C), which was previously reported to traffic through the Golgi apparatus 39. In MDCK cells, PAQR10 also elevated Golgi localization of HRas, NRas and KRas4A, but not KRas4B (Supplementary information, Figure S7). In addition, consistent with our findings, the amount of HRas on the PM was significantly reduced by PAQR10/PAQR11 overexpression, whereas the amount of HRas in the Golgi apparatus was significantly increased (Supplementary information, Figure S8). Collectively, these data indicate that PAQR10 and PAQR11 are able to significantly enhance localization of HRas, NRas and KRas4A in the Golgi apparatus (summarized in Figure 2E).
Figure 2.
Colocalization and interaction of PAQR10 and PAQR11 with Ras. (A-D) Analyses of colocalization of PAQR10/PAQR11 with different forms of Ras: HRas (A), NRas (B), KRas4A (C) and KRas4B (D). Hela cells were transfected with GFP-fused Ras plasmids as indicated with or without Flag-tagged PAQR10 or Flag-tagged PAQR11, following by immunostaining and confocal analysis. The arrows indicate apparent colocalization signals in the Golgi apparatus. (E) Quantitation for data of A to D. The relative signal intensity of different Ras proteins at the Golgi apparatus was quantified and shown as mean ± SD. **P < 0.01 as compared to the group with Ras expressed alone. (F) Interaction of PAQR10/PAQR11 with HRas, NRas and KRas4A. HEK293T cells were transiently transfected with GFP-fused HRas, NRas, KRas4A, KRas4B, Rac1 and CD-MPR, with Flag-tagged PAQR10 (left panel) or Flag-tagged PAQR11 (right panel) as indicated. Twenty-four hours after transfection, cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) with the antibodies as indicated. (G) Interaction of endogenous PAQR10 with endogenous Ras. Total cell lysate from Hela cells was subjected to IP with an anti-PAQR10 antibody, followed by IB with antibodies against pan-Ras and PAQR10. Rabbit IgG was used as a negative control.
Interaction of PAQR10 and PAQR11 with Ras
To provide evidence that PAQR10/PAQR11 can form a complex with Ras, we employed co-immunoprecipitation (Co-IP) assays to analyze the in vivo interaction between the proteins. We found that immunoprecipitation of both PAQR10 and PAQR11 could pull down HRas, NRas and KRas4A, but not KRas4B (Figure 2F). Inversely, immunoprecipitation of HRas, NRas and KRas4A, but not KRas4B, was able to pull down PAQR10 and PAQR11 (Supplementary information, Figure S9). Consistent with the colocalization data (Supplementary information, Figure S6), Rac1 and CD-MPR had no interaction with PAQR10/PAQR11 (Figure 2F and Supplementary information, Figure S9). Furthermore, we found that endogenous Ras could also interact with endogenous PAQR10 in Hela cells (Figure 2G). These data, collectively, indicate that PAQR10 and PAQR11 are able to physically associate with multiple forms of Ras.
Activation of Ras by PAQR10 and PAQR11 in the Golgi apparatus
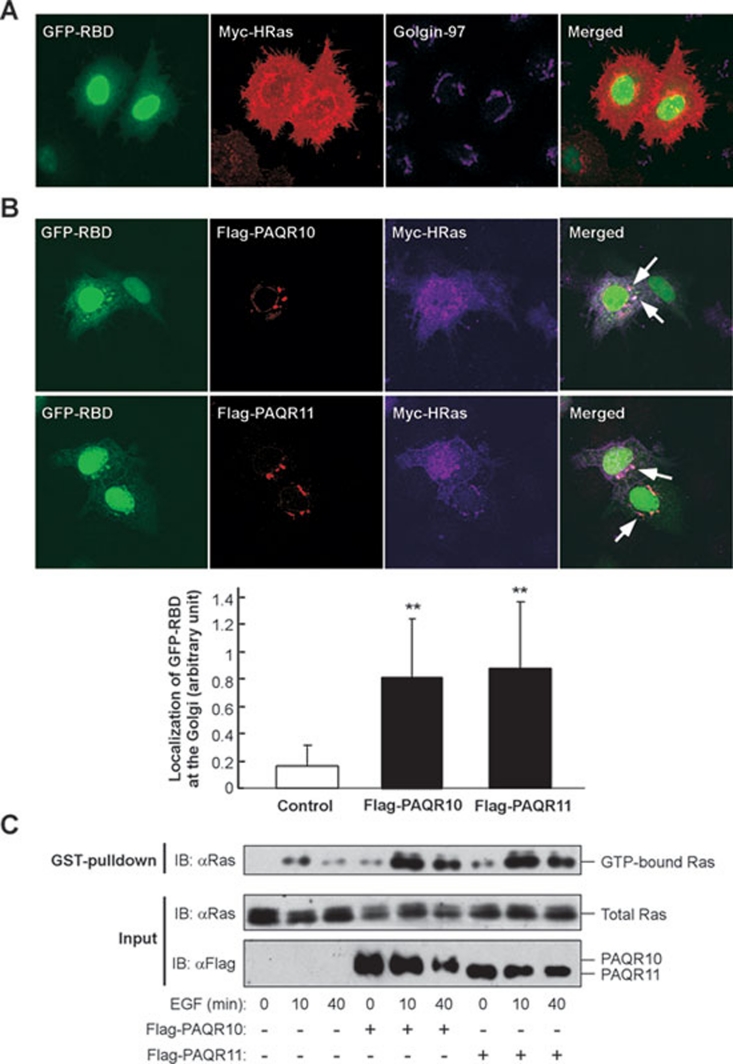
As PAQR10 and PAQR11 can interact with Ras, enhance localization of Ras in the Golgi apparatus and elevate ERK signaling cascade, we hypothesized that the Golgi-localized Ras mediated by PAQR10/PAQR11 should be in an active form. To address this issue, we employed a previously well-characterized in situ marker of activated Ras, GFP-RBD, a RBD of Raf-1 fused with GFP 9, 33. Under unstimulated condition, GFP-RBD was primarily localized in the nucleus and not associated with any intracellular structures, including the PM (Figure 3A), consistent with previous reports 9, 33. However, when GFP-RBD was coexpressed with PAQR10 or PAQR11, we observed evident colocalization of GFP-RBD with PAQR10/PAQR11 and HRas (Figure 3B), indicating that the Ras proteins mobilized onto the Golgi apparatus by PAQR10/PAQR11 are in an active state. Furthermore, we found that overexpression of both PAQR10 and PAQR11 significantly increased GTP-bound Ras, under unstimulated conditions and after EGF treatment (Figure 3C), further confirming that the PAQR10/PAQR11-mediated Golgi-localized Ras protein is active.
Figure 3.
Activation of Ras in the Golgi apparatus by PAQR10 and PAQR11. (A) Ras is in an inactive state in unstimulated cells. Hela cells were transfected with GFP-RBD and Myc-tagged HRas and serum-starved overnight before fixation and confocal analysis. Note that there is no colocalization of GFP-RBD with HRas. (B) PAQR10 and PAQR11 activate Ras in the Golgi apparatus. Hela cells were co-transfected with the plasmids as indicated, and used for immunostaining and confocal analysis. The arrow indicates increased activation of Ras in the Golgi apparatus by PAQR10 and PAQR11 as revealed by GFP-RBD colocalized with PAQR10/PAQR11 and HRas. The relative signal intensity of GFP-RBD in the Golgi apparatus was quantified and shown in the lower panel as mean ± SD. **P < 0.01 as compared to the group without PAQR10/PAQR11 expression. (C) PAQR10/PAQR11 overexpression increases activation of Ras upon EGF stimulation. HEK293T cells were transfected with the plasmids as indicated. Twenty-four hours after transfection, cells were serum-starved for 6 h before treatment with EGF (100 ng/ml) for various lengths of time, and the cell lysate was used in GST pull-down assay with GST-RBD, followed by immunoblotting with the antibodies as indicated.
Downregulation of PAQR10/PAQR11 affects Ras signaling in the Golgi apparatus
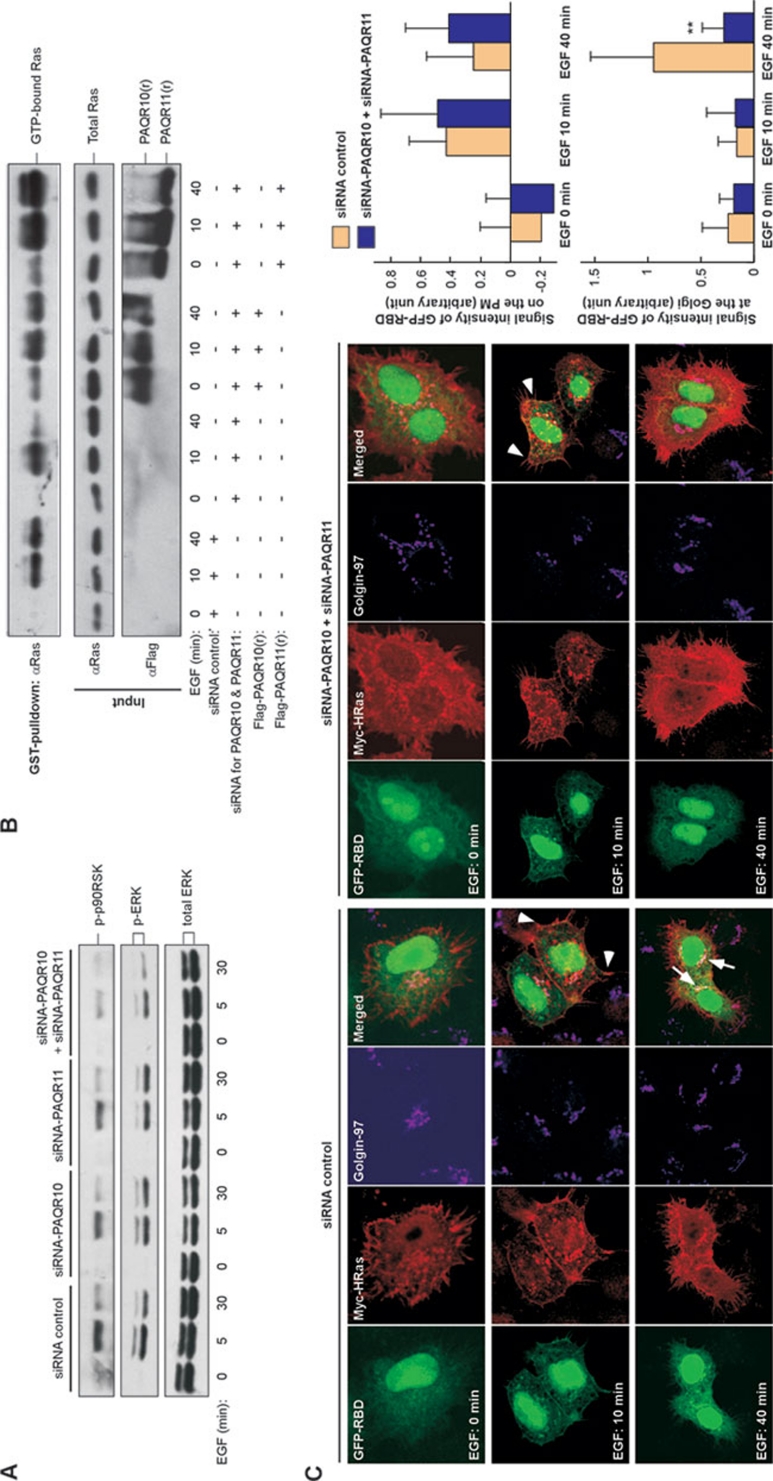
To further explore the functional significance of PAQR10/PAQR11 on Ras/ERK signaling, we employed a siRNA strategy to knockdown endogenous PAQR10 and PAQR11. We first confirmed the efficiency of siRNA to silence the expression of PAQR10 and PAQR11, shown by about a 50% decrease at the protein level (Supplementary information, Figure S10). Hela cells were transfected with the siRNA oligos, followed by EGF stimulation. Consistent with our observation from the overexpression experiments (Figure 1C and 1D), the phosphorylation levels of ERK and p90RSK, a cytosolic target of ERK 40, were markedly reduced when both PAQR10 and PAQR11 were silenced (Figure 4A). The phosphorylation levels of ERK or p90RSK was not significantly affected when only PAQR10 or PAQR11 was silenced (Figure 4A), likely due to the functional redundancy of these two proteins. We next carried out a GST pull-down assay to analyze the activation of endogenous Ras (Figure 4B). As expected, EGF was able to increase the amount of GTP-bound Ras. When PAQR10 and PAQR11 were both silenced, GTP-bound Ras was significantly reduced at 40 min upon EGF stimulation. To confirm that the silencing effect of PAQR10/PAQR11 siRNA was specific, we constructed siRNA-resistant expression plasmids PAQR10(r) and PAQR11(r), respectively. We found that the abrogation of Ras activation by PAQR10/11 siRNAs was rescued by PAQR10(r) and PAQR11(r) (Figure 4B). We also investigated in situ Ras activation using GFP-RBD. When PAQR10 and PAQR11 were both silenced, EGF-mediated rapid activation of Ras on the PM was not affected, but the delayed activation of Ras in the Golgi apparatus was largely diminished (Figure 4C). Taken together, these results indicate that endogenous PAQR10 and PAQR11 are involved in the regulation of the Ras activity in the Golgi apparatus.
Figure 4.
Downregulation of PAQR10/PAQR11 reduces ERK signaling. (A) Silencing of PAQR10 and PAQR11 affects EGF-induced ERK signaling. Hela cells were transfected with synthesized siRNA as indicated. Forty-eight hours after transfection, the cells were serum-starved overnight and treated with EGF (100 ng/ml). Total cell lysate was used in immunoblotting with the antibodies as indicated. (B) Downregulated Ras activity by PAQR10/PAQR11 siRNA is abrogated by PAQR10/PAQR11 constructs (PAQR10(r) and PAQR11(r)) that are refractory to RNAi interference. Hela cells were co-transfected with synthesized siRNA and the plasmid as indicated. Forty-eight hours after transfection, the cells were serum-starved overnight and treated with EGF (100 ng/ml). Total cell lysate was used in a GST pull-down assay with GST-RBD, followed by immunoblotting with the antibodies as indicated. (C) Silencing of PAQR10/PAQR11 affects activation of Ras in the Golgi after EGF stimulation. Hela cells were co-transfected with synthesized siRNA, GFP-RBD and Myc-HRas as indicated. Forty-eight hours after transfection, the cells were serum-starved overnight and treated with EGF (100 ng/ml) for the indicated times before immunostaining and confocal analysis. The arrowhead indicates activated Ras on the plasma membrane and the arrow indicates activated Ras in the Golgi. The relative signal intensity of GFP-RBD at the plasma membrane (PM) and the Golgi apparatus was quantified respectively and shown in the right panels. **P < 0.01 between control group and PAQR10/PAQR11 siRNA group.
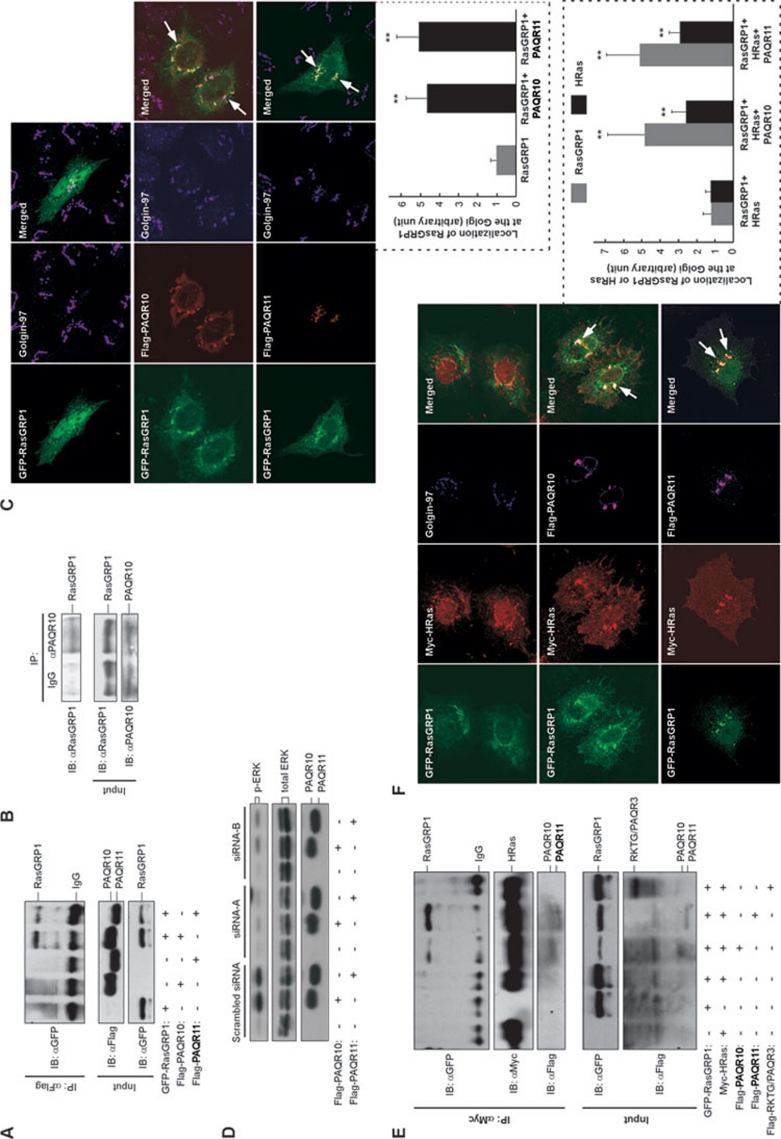
RasGRP1 is involved in PAQR10/PAQR11-mediated activation of Ras in the Golgi apparatus
How can Ras be activated by PAQR10 and PAQR11 in the Golgi apparatus? Classically, Ras activation requires guanine nucleotide exchange factors (GEFs) that facilitate GDP to GTP exchange on Ras. Previously, a Ras GEF, RasGRP1, has been characterized to translocate to the Golgi apparatus upon extracellular stimuli and activates Ras in the Golgi 15, 16. We first analyzed whether RasGRP1 could interact with PAQR10/PAQR11 by a co-IP assay. When PAQR10/PAQR11 and RasGRP1 were coexpressed, both PAQR10 and PAQR11 could be successfully co-purified with RasGRP1 (Figure 5A). We also found that endogenous PAQR10 could interact with endogenous RasGRP1 (Figure 5B). Considering that PAQR10/PAQR11 physically associates with RasGRP1, we hypothesized that PAQR10/PAQR11 could also increase Golgi localization of RasGRP1. Under the condition of serum starvation, RasGRP1 was predominantly diffusely distributed in the cytoplasm and nucleus with a small portion localized to the Golgi (Figure 5C). However, when PAQR10 or PAQR11 was coexpressed, the localization of RasGRP1 in the Golgi compartment was profoundly increased under unstimulated condition (Figure 5C). To further examine whether RasGRP1 is required for PAQR10/PAQR11 to activate Ras in the Golgi apparatus, we employed a siRNA strategy to knock down endogenous RasGRP1. Three siRNA oligos were designed and tested (Supplementary information, Figure S11), and two of the oligos (siRNA-A and siRNA-B) provided efficient silencing of RasGRP1 and were used to transfect HEK293T cells. We found that the PAQR10/PAQR11-induced ERK phosphorylation was inhibited by RasGRP1-specific siRNAs (Figure 5D). In Hela cells, RasGRP1-specific siRNA could also abrogate PAQR10/PAQR11-mediated accumulation of GFP-RBD in the Golgi apparatus (Supplementary information, Figure S12), further indicating that RasGRP1 is implicated in PAQR10/PAQR11-mediated activation of Ras in the Golgi apparatus.
Figure 5.
RasGRP1 is involved in PAQR10/PAQR11-mediated activation of Ras in the Golgi. (A) Interaction of PAQR10/PAQR11 with RasGRP1. HEK293T cells were transiently transfected with the plasmids as indicated. Twenty-four hours after transfection, cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) with the antibodies as indicated. (B) Interaction of endogenous PAQR10 with endogenous RasGRP1. Total cell lysate of Hela cells was subjected to IP with an anti-PAQR10 antibody, followed by IB with an antibody against human RasGRP1. Rabbit IgG was used as a negative control. (C) PAQR10/PAQR11 increases localization of RasGRP1 in the Golgi apparatus. Hela cells were transfected with GFP-fused RasGRP1 with or without Flag-tagged PAQR10/PAQR11, followed by immunostaining and confocal analysis. The arrow indicates colocalization of RasGRP1 with PAQR10 or PAQR11 in the Golgi. The relative signal intensity of RasGRP1 in the Golgi apparatus was quantified and shown in the lower panel (mean ± SD). **P < 0.01 as compared to the group with RasGRP1 expressed alone. (D) Silencing of RasGRP1 affects PAQR10/PAQR11-induced ERK phosphorylation. HEK293T cells were transfected with synthesized siRNA (a scrambled siRNA and two siRNAs for RasGRP1) and Flag-tagged PAQR10 or PAQR11 as indicated. Seventy-two hours after transfection, cells were serum-starved for 6 h and total cell lysate was used in immunoblotting with antibodies as indicated. (E) Overexpression of PAQR10/PAQR11 increases the protein interaction between HRas and RasGRP1. HEK293T cells were transiently transfected with the plasmids as indicated. Twenty-four hours after transfection, cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) with the antibodies as indicated. (F) Overexpression PAQR10/PAQR11 increases colocalization of HRas and RasGRP1 in the Golgi simultaneously. Hela cells were transfected with the plasmids as indicated, followed by immunostaining and confocal analysis. The arrow indicates colocalization of RasGRP1, HRas and PAQR10/PAQR11 in the Golgi. The relative signal intensity of RasGRP1 and HRas at the Golgi apparatus was quantified and shown in the right panel (mean ± SD). **P < 0.01 as compared to the group without PAQR10/PAQR11 expression, respectively.
We next analyzed whether PAQR10/PAQR11 had an effect on the interaction of RasGRP1 with Ras. By a co-IP assay, we found that the interaction of RasGRP1 with HRas was barely detectable in the absence of PAQR10/PAQR11 overexpression (Figure 5E). However, when PAQR10 or PAQR11 was coexpressed, the interaction of RasGRP1 with HRas was significantly elevated (Figure 5E). As a control, the interaction of RasGRP1 with HRas was not elevated by RKTG/PAQR3 (Figure 5E), another Golgi-localized protein 23. In agreement, overexpression of PAQR10 or PAQR11 profoundly increased Golgi localization of RasGRP1 and HRas, simultaneously (Figure 5F). Interestingly, the increased Golgi localization of HRas by PAQR10/11, and the interaction between PAQR10/11 and HRas were independent of RasGRP1, as knockdown of RasGRP1 had no effect on these events (Supplementary information, Figure S13). Collectively, these data indicate that PAQR10/PAQR11 can enhance the interaction of RasGRP1 with Ras in the Golgi apparatus, in which Ras is activated by RasGRP1.
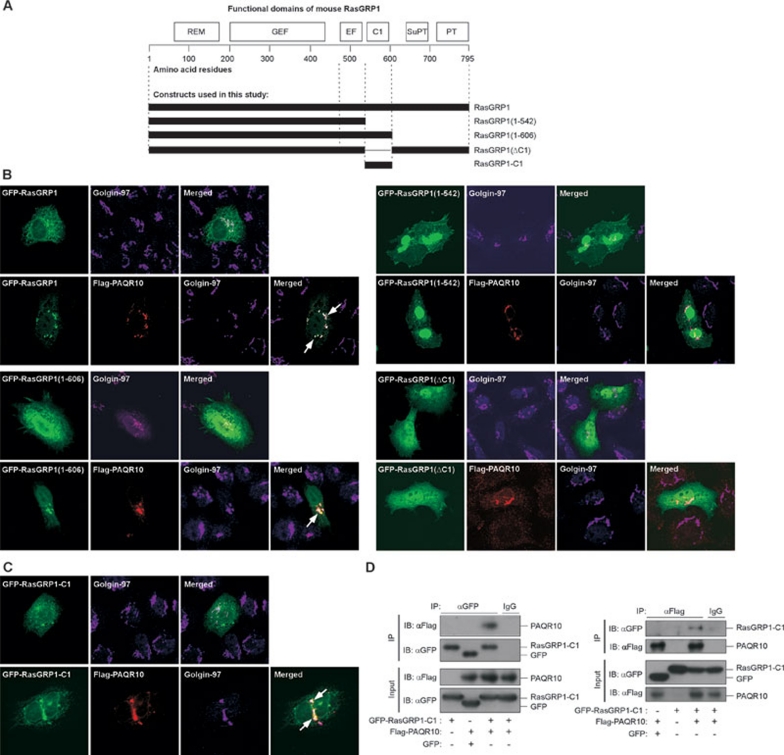
We next tried to identify the domain(s) necessary for the interaction of RasGRP1 with PAQR10 using different deletion or truncation forms of RasGRP1 15, 41 (Figure 6A). PAQR10 could significantly increase Golgi localization of the full-length RasGRP1, as well as its mutant RasGRP1(1-606) that had a deletion of the C-terminus containing SuPT and PT domains (Figure 6B). In contrast, PAQR10 could not increase Golgi localization of RasGRP1(1-542) that lacks the C1, SuPT and PT domains (Figure 6B). Based on these observations, we hypothesized that the C1 domain might be critical for the interaction of PAQR10 with RasGRP1. To address this hypothesis, we generated another RasGRP1 mutant that had a deletion of the C1 domain (RasGRP1ΔC1). We observed that PAQR10 could not elevate Golgi localization of this mutant (Figure 6B). Furthermore, we constructed a truncated version of RasGRP1, containing only its C1 domain (RasGRP1-C1). Interestingly, we found that PAQR10 could significantly increase Golgi localization of RasGRP1-C1 (Figure 6C). These results, therefore, indicate that the C1 domain of RasGRP1 is both necessary and sufficient for mobilization of RasGRP1 to the Golgi apparatus by PAQR10. To further confirm these results, we performed co-IP assays to analyze the interaction between PAQR10 and RasGRP1-C1. By two-way co-IP assays, we found that PAQR10 could interact with the C1 domain of RasGRP1 (Figure 6D). In summary, we demonstrated that the C1 domain of RasGRP1 is essential for the interaction of RasGRP1 with PAQR10, and such interaction is implicated in tethering RasGRP1 to the Golgi apparatus by PAQR10.
Figure 6.
The C1 domain of RasGRP1 is involved in its interaction with PAQR10. (A) Schematic representation of the RasGRP1 proteins used in the experiments. REM, Ras exchange motif; GEF, guanine nucleotide exchange domain that catalyzes GTP loading of Ras GTPases; EF, EF-hands, calcium binding motif; C1, phorbol esters/diacylglycerol binding domain; PT and SuPT, the plasma membrane targeting regulatory domains. (B, C) Colocalization of PAQR10 with different deletion mutants of RasGRP1. Hela cells were cotransfected with different GFP-fused RasGRP1 mutants with or without Flag-tagged PAQR10, followed by immunostaining and confocal analysis. The arrow indicates colocalization of RasGRP1 mutants with PAQR10 in the Golgi apparatus. (D) Interaction of PAQR10 with the C1 domain of RasGRP1. HEK293T cells were transiently transfected with the plasmids as indicated. Twenty-four hours after transfection, the cell lysate was used in immunoprecipitation (IP) and immunoblotting (IB) with the antibodies as indicated.
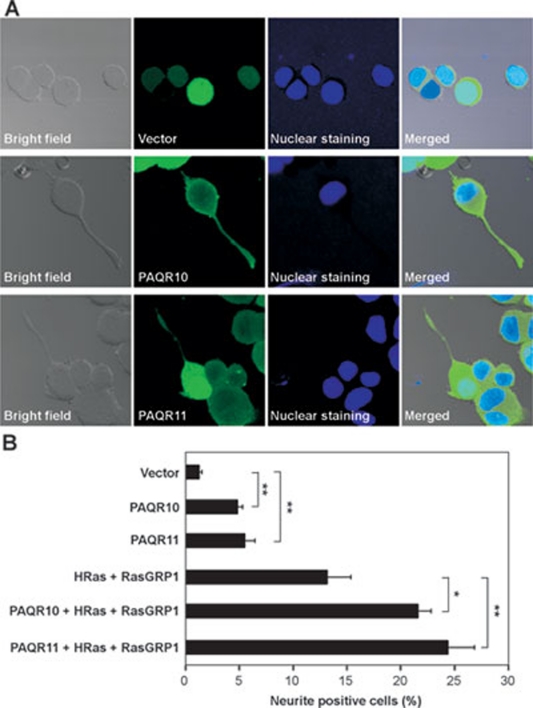
PAQR10 and PAQR11 potentiate differentiation of PC12 cells
We next used PC12 cell as a cellular model to investigate the potential biological activity of PAQR10/11. It has been reported that activation of Golgi-localized Ras signaling is able to promote differentiation of PC12 cells 16. When PAQR10 or PAQR11 was expressed in PC12 cells, either of them could significantly stimulate neurite outgrowth (Figure 7), a marker of PC12 cell differentiation. Furthermore, we found that PC12 cell differentiation induced by coexpression of HRas and RasGRP1 was significantly enhanced by PAQR10 or PAQR11 overexpression (Figure 7B), further supporting our model that PAQR10/11 could activate a sustained Ras signaling in the Golgi apparatus.
Figure 7.
PAQR10 and PAQR11 potentiate PC12 cell differentiation. PC12 cells were co-transfected with pEGFP and excess amount of other plasmids as indicated, and neurite outgrowth was analyzed 48 h later. Representative images are shown in A and the statistical analysis is shown in B. *P < 0.05 and **P < 0.01 as compared to the group without PAQR10/PAQR11 expression.
Discussion
In this study, we demonstrate for the first time that PAQR10 and PAQR11, the two Golgi-localized transmembrane proteins, can interact with Ras and enhance its retention and activity in the Golgi apparatus. Furthermore, localization of RasGRP1 in the Golgi apparatus and the interaction between Ras and RasGPR1 are elevated by PAQR10/PAQR11. The differentiation of PC12 cells stimulated by Ras and RasGRP1 is also enhanced by overexpression of PAQR10 and PAQR11. These findings, therefore, reveal a new paradigm through which Ras signaling is regulated in the Golgi apparatus via the coordinating activities of the PAQR10/PAQR11 proteins.
It has been recognized that Ras proteins (except for KRas4B) undergoes a dynamic trafficking within a cell via de/re-palmitoylation. Depalmitoylation detaches Ras from the PM and allows Ras to repartition to other intracellular membrane such as ER and Golgi apparatus. Ras proteins are repalmitoylated in the Golgi apparatus and then retrograde to the PM via the secretory pathway. Given its unique role in Ras palmitoylation, Golgi apparatus is considered as a trafficking intermediate for Ras proteins. However, it was unknown why palmitoylated Ras has a relatively high concentration in the Golgi apparatus. Furthermore, it was unclear how Ras activity in the Golgi apparatus can be maintained and uncoupled from initial activation on the PM upon stimulation by growth factors on cell surface. Our studies indicate that PAQR10 and PAQR11 are implicated in both tethering Ras to the Golgi and maintaining Ras activity in this organelle. Overexpression of PAQR10 and PAQR11 markedly increased Golgi localization of palmitoylation-regulated Ras isoforms (Figure 2). PAQR10 and PAQR11 also interacted with these Ras proteins by co-IP assays (Figure 2). Therefore, PAQR10/11 may directly or indirectly serve as a scaffold protein to increase retention of Ras proteins in the Golgi apparatus. On the other hand, PAQR10/11 may also tether activated Ras to the Golgi apparatus after Ras is detached from the PM via rapid depalmitoylation upon growth factor stimulation. This idea is supported by our finding that EGF-stimulated and delayed Ras activity in the Golgi apparatus was abrogated by downregulation of PAQR10/PAQR11 (Figure 4C).
It has been known that growth factor-induced activation of Ras signaling on the PM is rapid and transient, while Ras activity in the Golgi is delayed and sustained 9, 20, 21, and the increased Ras activity in the Golgi via PAQR10/PAQR11 may give rise to a sustained Ras signaling. This concept is supported by our finding that PC12 differentiation, a process that mainly relies on sustained Ras signaling 16, 42, was significantly enhanced by overexpression of PAQR10 and PAQR11 (Figure 7). At present, it is unclear how PARQ10 and PAQR11 themselves are regulated. EGF treatment appeared to have no effect on the expression of PAQR10 and PAQR11 (Supplementary information, Figure S14). It would be important to investigate the regulation of PAQR10 and PAQR11 in future, as elucidation of such regulation may uncover how Golgi-restricted Ras signaling is modulated.
Golgi-specific activation of Ras signaling involves RasGRP1, a GEF that is regulated by calcium and diacylglycerol (DAG) 15, 16, 43, 44. Overexpression of RasGRP1 leads to Ras activation in the Golgi apparatus 15, and such activation is found to be dependent on PLCγ 16. Based on these studies, it was proposed that the signaling cascade of “receptors/PLCγ/DAG-Ca2+/RasGRP1” is mainly implicated in the activation of Ras signaling in the Golgi apparatus 45. Consistent with previous reports, we demonstrate that RasGRP1 is also involved in PAQR10/11-mediated activation of Ras signaling in the Golgi apparatus (Figures 5 and 6). Our structural studies indicate that the C1 domain of RasGRP1 is both necessary and sufficient for the interaction of RasGRP1 with PAQR10 (Figure 6). The C1 domain was originally identified as the binding site for DAG and phorbol ester 46. Recently, it was found that the C1 domain may serve as a protein interaction module that regulates cellular localization of the C1 domain-containing proteins, such as their localization in the Golgi apparatus 47. Therefore, we propose that the C1 domain of RasGRP1 may play an essential role in PAQR10/11-mediated activation of Ras signaling in the Golgi apparatus.
Collectively, our data demonstrate a new paradigm of regulation of Ras signaling in the Golgi apparatus by PAQR10 and PAQR11. PAQR10/PAQR11 interacts directly or indirectly with RasGRP1 and Ras proteins, increases localization of these two proteins in the Golgi apparatus, in which Ras is activated through elevated interaction between RasGRP1 and Ras, thereby leading to activation of Golgi-restricted Ras signaling cascade. At present, it is unknown whether PAQR10 and PAQR11 have other biological activities in addition to their roles in Golgi-localized Ras signaling. Furthermore, it is noteworthy that PAQR10 and PAQR11 share certain degree of sequence homology with other members of the PAQR family 22. PAQR1 and PAQR2 are cell surface receptors for adiponectin and play important roles in glucose and lipid metabolism 26. PAQR3 is a Golgi-localized protein that negatively regulates Ras to ERK signaling by sequestrating Raf kinase to the Golgi apparatus 23, 24. However, replacing the N-terminal sequence of PAQR10 with that of PAQR3 abrogated Golgi localization and Ras activation (Y Chen, unpublished data), indicating that such sequence is indispensable for the unique activity of PAQR10. We postulate that the functional difference among PAQR members is dependent on their structural difference and cellular localization. Our findings that PAQR10 and PAQR11 are implicated in modulating Golgi-localized Ras signaling, therefore, have provided additional evidence that PAQR family with the ancient seven-transmembranal motif does have important biological activities.
Materials and Methods
Plasmids
The full-length human PAQR10 cDNA (GenBank entry NM_198403), PAQR11 cDNA (GenBank entry NM_012329) and adiponectin receptor 2 (GenBank entry NM_024551.2) were isolated from HEK293T cells by RT-PCR and confirmed by DNA sequencing. PAQR10, PAQR11 and AdipoR2 were subcloned into the mammalian expression vector pRc/CMV (Invitrogen, Carlsbad, CA, USA) with a Flag epitope tag added to the N-termini by a PCR-based method. Construction of PAQR10/PAQR11 refractory to RNA interference was managed by processing a same-sense mutation using a PCR-based method. The untagged PAQR10 and PAQR11 used in the differentiation of PC12 cells were subcloned into pCS2+ plasmids. HRas (GenBank entry NM_005343), NRas (GenBank entry, NM_002524) and KRas4A (GenBank entry, NM_033360.2) were cloned into pEGFP-C1 (Clontech, Mountain View, CA, USA) to fuse with an enhanced green fluorescence protein at the N-terminus. CD-MPR (GenBank entry NM_002355) was cloned into pEGFP-N1 (Clontech) to fuse with an enhanced green fluorescence protein at the C-terminus. The full-length RasGRP1 (GenBank entry NM_011246) were isolated from mouse brain cDNA by RT-PCR and confirmed by DNA sequencing. RasGRP1 cDNA was subcloned into pEGFP-C1 and pcDNA3.1(+) vectors. The deletion and truncation mutants of RasGRP1 were generated by a PCR-based method and confirmed by DNA sequencing. The RBD of Raf1 was cloned into the pEGFP-C1 (Clontech), as well as the pGEX-4T1 (Clontech) to generate GST-fused RBD. The HA-tagged TGN38 was kindly provided by Dr Juan S Bonifacino (National Institute of Child Health and Human Development, Bethesda, MD, USA).
Cell culture, cell transfection and confocal microscopy
HEK293T, Hela and A549 cells were cultured in DMEM containing 10% FBS. Transient transfection was performed with the polyethylenimine method for HEK293T and Hela cells 48. The methods for cell fixation, immunofluorescence staining and confocal analyses were described previously 23. Quantification of proteins localized in the Golgi apparatus and on the PM was performed using LSM images by Zeiss Confocal Microscopy Software and Physiology Software Rel. 3.2 as previously described 9, 49. The fluorescent signal in the Golgi apparatus was defined by staining with a Golgi marker, Golgin-97. We regarded the dispersed green signals in the cytoplasm as the background fluorescence. The fluorescence signal of the cytosol was subtracted from the signal in the Golgi apparatus and used to measure the fluorescence intensity of the Golgi apparatus. The images presented in the figures were captured using standardized setting and exposure times. More than 100 cells were observed in three independent experiments, and at least 20 cells were randomly chosen and quantified for each experimental group. The experiment of PC12 cell differentiation was performed as previously described 9. In brief, PC12 cells were cultured in RPMI1640 medium containing 10% horse serum and 5% FBS, and co-transfected with pEGFP vector and five-fold excess of other plasmids. Neurite outgrowth was measured 48 h later and quantified as the number of transected cells (scored by expression of GFP), with at least one neurite with a length equal or longer than the cell body diameter.
Antibodies, immunoblotting and immunoprecipitation
The antibodies were purchased as follows: phospho-ERK1/2 and phospho-p90RSK (Ser-380) from Cell Signaling Technology (Danvers, MA, USA); monoclonal and polyclonal anti-Flag antibodies from Sigma-Aldrich (St Louis, MO, USA); antibodies against Myc, tubulin, RasGRP1 and total ERK 1/2 from Santa Cruz Biotechnology (Santa Cruz, CA, USA); pan-Ras antibody from BD Bioscience Transduction Laboratories (Lexington, KY, USA); Golgin-97 monoclonal antibody, Alexa Fluor 488 donkey anti-mouse IgG, Alexa Fluor 546 goat anti-mouse, rabbit IgG and Hoechst 33342 from Molecular Probes (Eugene, OR, USA); and Cy5-labeled goat anti-mouse IgG from GE Healthcare (Chalfont St Giles, UK). The polyclonal PAQR10 and PAQR11 antibodies were generated by rabbit immunization with a His-tagged fusion protein that contains three hydrophilic regions of human PAQR10 and PAQR11, respectively. For the PAQR10 antibody, a fusion peptide with amino acid residues 1-69, 87-111 and 234-246 were used. For PAQR11 antibody, fusion peptide with amino acid residues 1-60, 85-101 and 221-238 were used. The antibodies were affinity-purified against recombinant GST-fused PAQR10 and PAQR11 polypeptides coupled to Glutathione Sepharose 4B according to the manufacturer's instructions (GE Healthcare). Both immunoblotting and immunoprecipitation assays were previously described by us 23.
RNA interference
We used both chemical-synthesized oligos and lentivirus-generated shRNA to interfere with the expression of endogenous genes. Chemical oligos targeting PAQR10 and PAQR11, as well as a nonspecific oligo used as control, were synthesized by GeneChem Co Ltd (Shanghai, China). Three targeted sequences for human RasGRP1 are chosen as follows (shown as the targeting sense sequence: RasGRP1-A: 5′-CCUGAUUGACACAACUCAATT-3′, RasGPR1-B: 5′-GGGAUGAGAUCACAGCCUATT-3′, RasGPR1-C: 5′-GCCCAGUCUUGGUCAGAAATT-3′. Three targeted sequences for PAQR10 and PAQR11 each are chosen as follows: 5′-GGAUGGUGGAACACUGUAU-3′, 5′-GCUUCUCUGCUACGUCGUA-3′, and 5′-GUGCUCAGUAGCCAAACAA-3′ for PAQR10; 5′-AGAUAACAGCAUGGAUUUAA-3′, 5′-AGGACAGUGGAGCAUUGUU-3′, and 5′-GCCACUUAAGGACAGUGGA-3′ for PAQR11; 5′-UUCUCCGAACGUGUCACGU-3′ for the nonspecific oligo. To achieve better silencing, mixtures of three oligonucleotides were used to transfect cells for PAQR10 and PAQR11. For lentivirus-mediated RNA interference, FG12 lentiviral vector, which has an independent open reading frame of GFP, was used to produce small, double-stranded shRNA to silence target gene expression; information about this vector system has been described in detail by Qin et al 50. The targeting sequences are 5′-CTTCTCTGCTACGTCGTA-3′ for PAQR10; 5′-AGATAACAGCATGGATTTA-3′ for PAQR11; 5′-TTCTCCGAACGTGTCACGT-3′ for nonspecific control. The virus was harvested from the culture medium 48 h after transfection, concentrated by centrifugation, and used to infect cells. The infection efficiency was measured by counting GFP-positive cells with a >70% infection efficiency.
Ras activity assay
To detect the GTP-bound Ras, cells were collected and lysed in buffer containing 20 mM HEPES (pH 7.4), 1% NP40, 150 mM NaCl, 5 mM MgCl2 and 10% glycerol. After centrifugation, the supernatant were collected and incubated with 20 μg of GST-RBD bound to GSH-beads (Amersham Biosciences, Sweden) at 4 °C for overnight. The beads were then collected and washed by lysis buffer. The amount of precipitated Ras proteins was analyzed by immunoblotting.
RNA isolation and real-time PCR
RNA isolation, reverse transcription, RT-PCR and real-time PCR were carried out as described 51. The sequence of primers for RT-PCR are as follows: 5′-GATGTAAAGACTGCGGGATG-3′ and 5′-TTCTGACCAAGACTGGGCTA-3′ for human RasGRP1; 5′-GCACTCTTCCAGCCTTTCCTG-3′ and 5′-GGAGTACTTGCGCTCAGGAGGAGC-3′ for human GAPDH. The sequence of primers for real-time PCR are as follows: 5′-CCAGGGCTGGCGTTGTGAAG-3′ and 5′-CTTGGAGTGTATCAGTCAGC-3′ for human c-Fos gene; 5′-CAGCACCTTCAACCCTCAGG-3′ and 5′-GTAACTGGTCTCCACCAGCA-3′ for human Egr-1 gene; 5′-CAGCTACTTTTCTGGTCAGG-3′ and 5′-GTGTAGGCGTCGTCGTGATC-3′ for human JunB gene; 5′-CAGGATGGTGGAACACTGTC-3′ and 5′-ACGGAAGCCATAATCCAGAC-3′ for human PAQR10; 5′-CGCCATTTGGAAATACCTTT-3′ and 5′-TCCCAAGTGCCATAATTGAA-3′ for human PAQR11; 5′-GATCATTGCTCCTCCTGAGC-3′ and 5′-ACTCCTGCTTGCTGATCCAC-3′ for human actin.
Statistical analysis
Student's t-test is used for all the statistical analysis.
Acknowledgments
We thank Drs Natlie G Ahn (University of Colorado), Piero Crespo (Consejo Superior de Investigaciones Científicas), Xosé R Bustelo (University of Salamanca), Robert J Kay (University of British Columbia) and Juan S Bonifacino (National Institute of Child Health and Human Development) for generously providing the plasmids. This work was supported by research grants from Ministry of Science and Technology of China (2007CB947100 to YC and 2010CB529506 to YP and ZW), National Natural Science Foundation of China (30830037 and 81021002 to YC and 30971660 to YP), Chinese Academy of Sciences (KSCX2-EW-R-08 to YC), and Shanghai Institutes for Biological Sciences (2009KIP207 to YP).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Hydrophobicity analysis of PAQR10 and PAQR11.
Characterization of PAQR10/11 antibodies.
PAQR10 but not AdipoR2 can activate ERK signaling.
A Golgi-specific protein TGN38 does not affect ERK signaling.
PAQR10-mediated stimulation of Egr-1 gene expression is inhibited by GFP-RBD and MEK inhibitor PD98059.
Control experiments for confocal studies.
PAQR10 increases Golgi localization of HRas, NRas, KRas4A but no KRas4B in the Golgi apparatus in MDCK cells.
The distribution of HRas on the plasma membrane and Golgi apparatus is affected by PAQR10/PAQR11 overexpression.
Interaction of PAQR10/PAQR11 with HRas, NRas and KRas4A.
Analysis of PAQR10/PAQR11 siRNA efficiency.
Analysis of RasGRP1 siRNA efficiency.
Silencing of RasGRP1 affects activation of Ras in the Golgi apparatus by PAQR10/PAQR11 overexpression.
Knockdown of RasGRP1 does not affect PAQR10/PAQR11-induced Golgi localization of HRas and the interaction between HRas and PAQR10/PAQR11.
EGF treatment has no effect on the expression of PAQR10 and PAQR11.
References
- Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- Philips MR. Teaching resources. Imaging signal transduction in living cells with fluorescent proteins. Sci STKE. 2005;2005:tr28. doi: 10.1126/stke.3142005tr28. [DOI] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Jiang X, Sorkin A. Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol Biol Cell. 2002;13:1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arozarena I, Matallanas D, Berciano MT, et al. Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol Cell Biol. 2004;24:1516–1530. doi: 10.1128/MCB.24.4.1516-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobering AK, Romeo MJ, Vay HA, Levin DE. A novel Ras inhibitor, Eri1, engages yeast Ras at the endoplasmic reticulum. Mol Cell Biol. 2003;23:4983–4990. doi: 10.1128/MCB.23.14.4983-4990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolloni A, Prior IA, Lindsay M, Parton RG, Hancock JF. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol Cell Biol. 2000;20:2475–2487. doi: 10.1128/mcb.20.7.2475-2487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu VK, Bivona T, Hach A, et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- Rebollo A, Perez-Sala D, Martinez AC. Bcl-2 differentially targets K-, N-, and H-Ras to mitochondria in IL-2 supplemented or deprived cells: implications in prevention of apoptosis. Oncogene. 1999;18:4930–4939. doi: 10.1038/sj.onc.1202875. [DOI] [PubMed] [Google Scholar]
- Bivona TG, Quatela SE, Bodemann BO, et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell. 2006;21:481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Schaeffer HJ, Catling AD, Eblen ST, et al. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Roudabush FL, Choy EW, et al. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci USA. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caloca MJ, Zugaza JL, Bustelo XR. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J Biol Chem. 2003;278:33465–33473. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- Bivona TG, Perez De Castro I, Ahearn IM, et al. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Bastiaens PI. Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Curr Opin Cell Biol. 2006;18:351–357. doi: 10.1016/j.ceb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Henis YI, Hancock JF, Prior IA. Ras acylation, compartmentalization and signaling nanoclusters. Mol Membr Biol. 2009;26:80–92. doi: 10.1080/09687680802649582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Peyker A, Rocks O, Bastiaens PI. Imaging activation of two Ras isoforms simultaneously in a single cell. Chembiochem. 2005;6:78–85. doi: 10.1002/cbic.200400280. [DOI] [PubMed] [Google Scholar]
- Tang YT, Hu T, Arterburn M, et al. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol. 2005;61:372–380. doi: 10.1007/s00239-004-0375-2. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie X, Ding Q, et al. Spatial regulation of Raf kinase signaling by RKTG. Proc Natl Acad Sci USA. 2007;104:14348–14353. doi: 10.1073/pnas.0701298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Feng L, He J, et al. RKTG sequesters B-Raf to the Golgi apparatus and inhibits the proliferation and tumorigenicity of human malignant melanoma cells. Carcinogenesis. 2008;29:1157–1163. doi: 10.1093/carcin/bgn119. [DOI] [PubMed] [Google Scholar]
- Xie X, Zhang Y, Jiang Y, et al. Suppressive function of RKTG on chemical carcinogen-induced skin carcinogenesis in mouse. Carcinogenesis. 2008;29:1632–1638. doi: 10.1093/carcin/bgn139. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Kamon J, Ito Y, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423:762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- Luo XH, Guo LJ, Yuan LQ, et al. Adiponectin stimulates human osteoblasts proliferation and differentiation via the MAPK signaling pathway. Exp Cell Res. 2005;309:99–109. doi: 10.1016/j.yexcr.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Petridou ET, Mitsiades N, Gialamas S, et al. Circulating adiponectin levels and expression of adiponectin receptors in relation to lung cancer: two case-control studies. Oncology. 2007;73:261–269. doi: 10.1159/000127424. [DOI] [PubMed] [Google Scholar]
- Arditi JD, Venihaki M, Karalis KP, Chrousos GP. Antiproliferative effect of adiponectin on MCF7 breast cancer cells: a potential hormonal link between obesity and cancer. Horm Metab Res. 2007;39:9–13. doi: 10.1055/s-2007-956518. [DOI] [PubMed] [Google Scholar]
- Williams CJ, Mitsiades N, Sozopoulos E, et al. Adiponectin receptor expression is elevated in colorectal carcinomas but not in gastrointestinal stromal tumors. Endocr Relat Cancer. 2008;15:289–299. doi: 10.1677/ERC-07-0197. [DOI] [PubMed] [Google Scholar]
- Rehli M, Krause SW, Schwarzfischer L, Kreutz M, Andreesen R. Molecular cloning of a novel macrophage maturation-associated transcript encoding a protein with several potential transmembrane domains. Biochem Biophys Res Commun. 1995;217:661–667. doi: 10.1006/bbrc.1995.2825. [DOI] [PubMed] [Google Scholar]
- Wilde A, Reaves B, Banting G. Epitope mapping of two isoforms of a trans Golgi network specific integral membrane protein TGN38/41. FEBS Lett. 1992;313:235–238. doi: 10.1016/0014-5793(92)81199-v. [DOI] [PubMed] [Google Scholar]
- Bondeva T, Balla A, Varnai P, Balla T. Structural determinants of Ras-Raf interaction analyzed in live cells. Mol Biol Cell. 2002;13:2323–2333. doi: 10.1091/mbc.E02-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: 'it ain't over 'til it's over'. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- Mitin N, Rossman KL, Der CJ. Signaling interplay in Ras superfamily function. Curr Biol. 2005;15:R563–574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64:2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Grubb JH, Sly WS. The overexpressed human 46-kDa mannose 6-phosphate receptor mediates endocytosis and sorting of beta-glucuronidase. Proc Natl Acad Sci USA. 1990;87:8036–8040. doi: 10.1073/pnas.87.20.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- Beaulieu N, Zahedi B, Goulding RE, et al. Regulation of RasGRP1 by B cell antigen receptor requires cooperativity between three domains controlling translocation to the plasma membrane. Mol Biol Cell. 2007;18:3156–3168. doi: 10.1091/mbc.E06-10-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Bottorff DA, Chan EY, et al. RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Springett GM, Toki S, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quatela SE, Philips MR. Ras signaling on the Golgi. Curr Opin Cell Biol. 2006;18:162–167. doi: 10.1016/j.ceb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Ono Y, Fujii T, Igarashi K, et al. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci USA. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kazanietz MG. p23/Tmp21 differentially targets the Rac-GAP beta2-chimaerin and protein kinase C via their C1 domains. Mol Biol Cell. 2010;21:1398–1408. doi: 10.1091/mbc.E09-08-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Han R, Wang Z, Chen Y.Regulation of adiponectin receptors in hepatocytes by the peroxisome proliferator-activated receptor-gamma agonist rosiglitazone Diabetologia 2006491303–1310.16609881 [Google Scholar]
- Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Jin T, Wang Z, Chen Y. Catalase potentiates retinoic acid-induced THP-1 monocyte differentiation into macrophage through inhibition of peroxisome proliferator-activated receptor gamma. J Leukoc Biol. 2007;81:1568–1576. doi: 10.1189/jlb.1106672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hydrophobicity analysis of PAQR10 and PAQR11.
Characterization of PAQR10/11 antibodies.
PAQR10 but not AdipoR2 can activate ERK signaling.
A Golgi-specific protein TGN38 does not affect ERK signaling.
PAQR10-mediated stimulation of Egr-1 gene expression is inhibited by GFP-RBD and MEK inhibitor PD98059.
Control experiments for confocal studies.
PAQR10 increases Golgi localization of HRas, NRas, KRas4A but no KRas4B in the Golgi apparatus in MDCK cells.
The distribution of HRas on the plasma membrane and Golgi apparatus is affected by PAQR10/PAQR11 overexpression.
Interaction of PAQR10/PAQR11 with HRas, NRas and KRas4A.
Analysis of PAQR10/PAQR11 siRNA efficiency.
Analysis of RasGRP1 siRNA efficiency.
Silencing of RasGRP1 affects activation of Ras in the Golgi apparatus by PAQR10/PAQR11 overexpression.
Knockdown of RasGRP1 does not affect PAQR10/PAQR11-induced Golgi localization of HRas and the interaction between HRas and PAQR10/PAQR11.
EGF treatment has no effect on the expression of PAQR10 and PAQR11.