A fluorescence imaging-based forward genetic screen that detects components of endocytic recycling is used to identify BEX5/RabA1b as a regulator of protein trafficking to the plasma membrane.
Abstract
Constitutive endocytic recycling is a crucial mechanism allowing regulation of the activity of proteins at the plasma membrane and for rapid changes in their localization, as demonstrated in plants for PIN-FORMED (PIN) proteins, the auxin transporters. To identify novel molecular components of endocytic recycling, mainly exocytosis, we designed a PIN1-green fluorescent protein fluorescence imaging–based forward genetic screen for Arabidopsis thaliana mutants that showed increased intracellular accumulation of cargos in response to the trafficking inhibitor brefeldin A (BFA). We identified bex5 (for BFA-visualized exocytic trafficking defective), a novel dominant mutant carrying a missense mutation that disrupts a conserved sequence motif of the small GTPase, RAS GENES FROM RAT BRAINA1b. bex5 displays defects such as enhanced protein accumulation in abnormal BFA compartments, aberrant endosomes, and defective exocytosis and transcytosis. BEX5/RabA1b localizes to trans-Golgi network/early endosomes (TGN/EE) and acts on distinct trafficking processes like those regulated by GTP exchange factors on ADP-ribosylation factors GNOM-LIKE1 and HOPM INTERACTOR7/BFA-VISUALIZED ENDOCYTIC TRAFFICKING DEFECTIVE1, which regulate trafficking at the Golgi apparatus and TGN/EE, respectively. All together, this study identifies Arabidopsis BEX5/RabA1b as a novel regulator of protein trafficking from a TGN/EE compartment to the plasma membrane.
INTRODUCTION
Plants have a complex endomembrane system with specialized intracellular trafficking pathways. The endomembrane system of plants, although not yet entirely understood, is characterized by several well-defined anterograde and retrograde pathways (Paul and Frigerio, 2007; Bassham et al., 2008; Robinson et al., 2008; Irani and Russinova, 2009). The biosynthetic pathway begins with the translocation of the newly synthesized proteins into the endoplasmic reticulum lumen (Vitale and Denecke, 1999; Vitale and Galili, 2001). Here, they are folded and subsequently transported through the endoplasmic reticulum to the cis-cisternae of the Golgi apparatus (GA) and, further, through the following medium- and trans-cisternae (Matheson et al., 2006). From the trans-cisternae of the GA, the proteins flow to the trans-Golgi network/early endosome (TGN/EE), a critical sorting compartment that lies at the intersection of the secretory route with the endocytic route (Dettmer et al., 2006; Viotti et al., 2010). Hence, cargos that reach the TGN/EE are subsequently sorted either into the secretion route (cargos destined to the plasma membrane [PM] and/or the extracellular matrix) or into the degradation route (cargos delivered to the vacuole).
A typical feature of plant PM proteins is their ability to recycle between PM and endosomes (Geldner et al., 2001; Murphy et al., 2005; Kleine-Vehn and Friml, 2008). The endocytic recycling mechanism allows the regulation of the protein amount at the PM (Paciorek et al., 2005; Robert et al., 2010) as well as rapid changes in the polar distribution of the PIN-FORMED (PIN) auxin transporters (Petrásek et al., 2006) during organogenesis and embryogenesis and also during gravitropic and phototropic responses (Friml et al., 2002, 2003; Benková et al., 2003; Reinhardt et al., 2003; Heisler et al., 2005; Kleine-Vehn et al., 2008, 2010; Ding et al., 2011; Rakusová et al., 2011). Endocytic recycling typically begins with clathrin-mediated endocytosis at the PM (Dhonukshe et al., 2007) and trafficking to the TGN/EE, where the cargos are subsequently sorted, either for recycling (back to the PM) or degradation (to the vacuole). Thus, cargos destined for endocytic recycling, such as PIN1, pass the early endosomes positive for the GTP exchange factor on ADP-ribosylation factors (ARF-GEF) HOPM INTERACTOR7/BFA-VISUALIZED ENDOCYTIC TRAFFICKING DEFECTIVE1 (MIN7/BEN1) (Tanaka et al., 2009) and, presumably, are further transported to the GNOM-positive recycling endosomes (RE), from where they are then delivered to the PM by an ARF GEF GNOM-dependent trafficking mechanism (Geldner et al., 2003).
The molecular mechanisms that regulate the events of intracellular transport, such as vesicle formation, movement, tethering/docking, and fusion, are complex and poorly understood. However, large protein families, including small GTPases (such as Rab GTPases and ARF GTPases) and ARF-GEF activators of ARF GTPases have been found as crucial regulators of vesicle trafficking (Zerial and McBride, 2001; Rutherford and Moore, 2002; Vernoud et al., 2003; Anders and Jürgens, 2008; Nielsen et al., 2008; Woollard and Moore, 2008). In yeast and mammals, every step in vesicle trafficking on secretory, endocytic, and recycling routes is regulated by members of Rab and ARF GTPases. Hence, the evolutionarily conserved function of the small GTPases throughout eukaryotes, together with their increased number in more complex organisms such as plants and mammals, demonstrate the importance of these proteins for the regulation of intracellular vesicle trafficking.
The small GTPases function as molecular switches that are activated and inactivated by the binding and hydrolysis of GTP. They constitutively cycle between a membrane-associated and active GTP-bound form and a mainly cytosolic and inactive GDP-bound conformation (Vernoud et al., 2003; Grosshans et al., 2006). GDP–GTP cycling is crucial for intracellular transport. In the GTP-bound form, Rab GTPases interact with their downstream effectors and regulate diverse processes, such as vesicle budding, vesicle delivery, tethering of vesicles to the target compartment, and fusion at the level of different endosomal compartments (Zerial and McBride, 2001; Rutherford and Moore, 2002; Vernoud et al., 2003; Grosshans et al., 2006).
Rab GTPases represent the largest family of small GTPases. In the Arabidopsis thaliana genome, 57 members have been identified and, based on their sequence homology and similarities with the yeast and mammalian orthologs, classified in eight subfamilies or classes: RabA to RabH (Pereira-Leal and Seabra, 2000; Rutherford and Moore, 2002; Vernoud et al., 2003; Bassham et al., 2008). The best-represented subfamily is the RabA GTPases (Rutherford and Moore, 2002; Vernoud et al., 2003; Bassham et al., 2008). Characterization of dominant negative (DN) and constitutive active (CA) mutants and localization studies showed that RabA GTPase members play a role in post-Golgi protein trafficking (Ueda et al., 1996; Preuss et al., 2004; Chow et al., 2008).
In this study, we identify and characterize bex5 (for BFA-visualized exocytic trafficking defective), a novel dominant mutant defective in the exocytosis of polar and nonpolar PM proteins. We identified bex5 from a forward genetic screen designed to isolate regulators of PIN1 endocytic recycling. BEX5 encodes RabA1b (for RAS GENES FROM RAT BRAINA1b), a member of the large RabA GTPase class, which localizes to the TGN/EE compartment and plays a role in protein trafficking in Arabidopsis roots, presumably by regulating vesicle formation, budding, and trafficking from the TGN/EE to the PM.
RESULTS
Screen for Mutants with Altered PIN1-GFP Endocytic Recycling
To understand the mechanism of PIN1 endocytic recycling and to identify molecular components that regulate this process, we used a forward genetic approach. Complete inhibition of protein cycling might be lethal; therefore, a typical morphology-based screen for the identification of mutants defective in the endocytic recycling of the proteins might be ineffective. To overcome this limitation, we designed a PIN1-green fluorescent protein (GFP) fluorescence imaging-based screen and looked for mutants that showed subcellular defects in the brefeldin A (BFA)–induced intracellular accumulation of PIN1-GFP. The fungal toxin BFA has proven to be an excellent tool to investigate protein trafficking, as it inhibits intracellular trafficking, mainly secretion, and causes the accumulation of PM proteins, such as basal (rootward) localized PIN1, into large aggregates known as BFA compartments or BFA bodies (Geldner et al., 2001). Nonetheless, long-term BFA treatment leads to the gradual disappearance of PIN1 from the BFA compartment and its relocation to the apical (shootward) PM (Kleine-Vehn et al., 2008). Hence, we screened for mutants that, in contrast with the PIN1pro:PIN1-GFP control line, showed abnormal PIN1-GFP intracellular accumulation after a 16-h treatment with 50 µM BFA, indicating defective exocytosis. This approach is distinct from the previously reported PIN1pro:PIN1-GFP screens that were focused on PIN1-GFP aggregations in the absence of any treatments (pat mutants; Feraru et al., 2010; Zwiewka et al., 2011) or on short-term BFA treatments (ben mutants; Tanaka et al., 2009). By screening the progeny of 1200 M1 ethyl methanesulfonate–mutagenized PIN1pro:PIN1-GFP individuals, we obtained five nonallelic mutants (H. Tanaka and J. Friml, unpublished data) that were named bex1 to -5. Here, we identify and characterize the bex5 mutant.
bex5 Is Hypersensitive to BFA
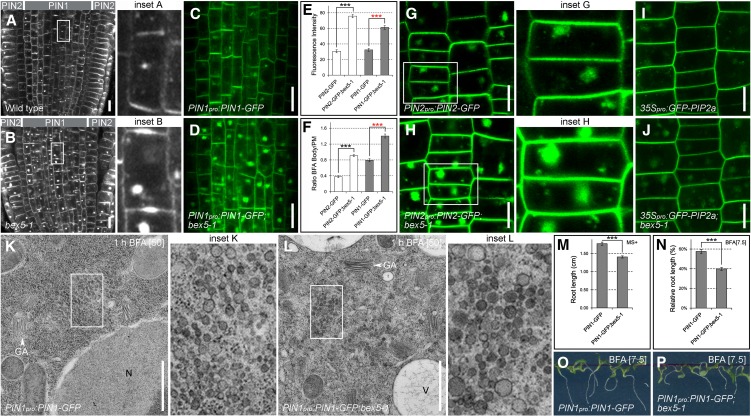
In the wild type, endogenous PIN1 or transgenic PIN1-GFP proteins showed intracellular endosomal agglomerations after a 1.5-h treatment with 25 µM BFA (Figures 1A to 1F; see Supplemental Figure 1A online). These BFA-induced PIN1 or PIN1-GFP agglomerations were enhanced in the bex5-1 mutant (Figures 1A to 1F; see Supplemental Figure 1A online). Similar to PIN1, the PIN2 and PIN2-GFP proteins displayed enhanced intracellular aggregation in the bex5-1 mutant following BFA treatment (Figures 1A, 1B, and 1E to 1H; see Supplemental Figure 1A online). Interestingly, other proteins, such as ARF1 (see Supplemental Figures 1B and 1C online), and nonpolar PM proteins, such as aquaporin GFP-PIP2a (Figures 1I and 1J; see Supplemental Figures 1F and 1G online) or PM ATPase (see Supplemental Figures 1D and 1E online), also showed enhanced BFA-induced intracellular accumulation in the PIN1pro:PIN1-GFP;bex5-1 mutant. These results show that the bex5-1 mutation enhances BFA-induced protein accumulation, indicating a more general defect in protein trafficking.
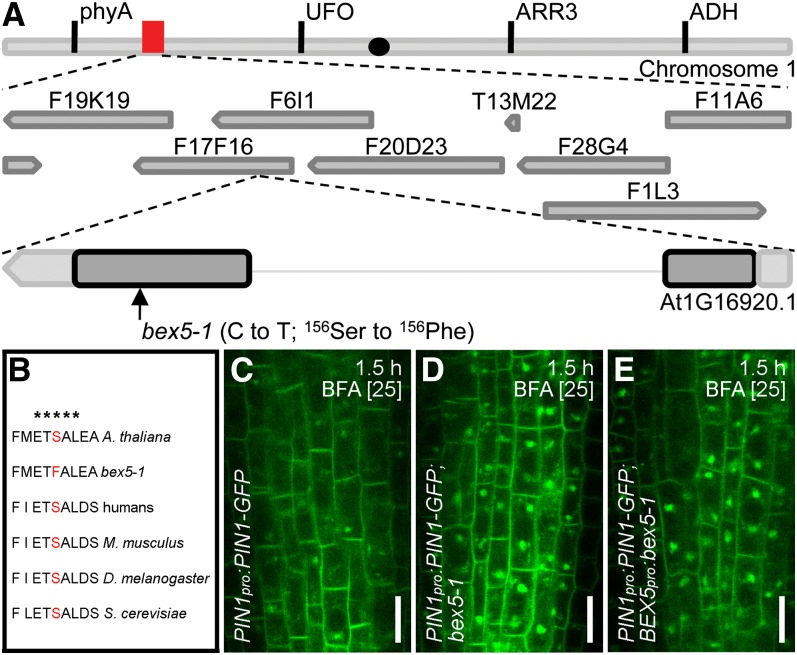
Figure 1.
bex5 Is Oversensitive to BFA.
(A) to (J) A 1.5-h treatment with 25 µM BFA causes enhanced intracellular protein accumulation in the bex5-1 mutant. Immunolocalization of PIN1 (vasculature) and PIN2 (epidermal and cortex cells) ([A] and [B]) and live imaging of PIN1-GFP (vasculature) ([C] and [D]) are shown in the wild-type (A), bex5-1 (B), PIN1pro:PIN1-GFP (C), and PIN1pro:PIN1-GFP;bex5-1 (D). PIN1-GFP and PIN2-GFP fluorescence measurements show enhanced accumulation into BFA bodies in the bex5-1 mutant vasculature and epidermal cells, respectively (n = 100 BFA bodies from 10 roots; red stars, t test P = 1.07E-53; black stars, t test P = 8.37E-29) (E). PIN1-GFP and PIN2-GFP accumulate into larger BFA bodies in the bex5-1 mutant vasculature and epidermal cells, respectively (n = 100 BFA bodies from 10 roots; red stars, t test P = 1.37E-52; black stars, t test P = 4.22E-29) (F). Live imaging of PIN2-GFP ([G] and [H]) and GFP-PIP2a ([I] and [J]) is shown in the epidermal cells of PIN2pro:PIN2-GFP (G), PIN2pro:PIN2-GFP;bex5-1 (H), 35Spro:GFP-PIP2a (I), and 35Spro:GFP-PIP2a;bex5-1. (J). Insets represent enlargements of the boxed regions from the corresponding images.
(K) and (L) The morphology of BFA bodies is impaired in the bex5-1 mutant. PIN1pro:PIN1-GFP;bex5-1 treated for 1 h with 50 µM BFA shows abnormal BFA bodies (L). While PIN1pro:PIN1-GFP displays 93.54% (n = 31 BFA bodies from five roots) typical BFA bodies (K), 61.9% (n = 42 BFA bodies from five roots) of the BFA bodies in the PIN1pro:PIN1-GFP;bex5-1 mutant are enlarged and less compact and the vesicle-like structures present in the BFA bodies have abnormal shapes, sizes, and composition (L). Insets represent enlargements of the boxed regions from the corresponding images. N, Nucleus; V, vacuole.
(M) Five-day-old PIN1pro:PIN1-GFP;bex5-1 has shorter roots than the control (n = 61; *** t test P = 4.41E-10). Seedlings were grown on MS medium with Suc (MS+).
(N) to (P) PIN1pro:PIN1-GFP;bex5-1 is oversensitive to BFA-mediated root growth inhibition. Relative root length is shown for 7-d-old PIN1pro:PIN1-GFP and PIN1pro:PIN1-GFP;bex5-1 germinated and grown on 7.5 µM BFA (n = 61; *** t test P = 1.84E-10) (N). When grown on 7.5 µM BFA, the roots of 7-d-old PIN1pro:PIN1-GFP;bex5-1 seedlings (P) are shorter than those of control seedlings (O).
Bars = 10 µm for (A) to (D) and (G) to (J) and 1 µm for (K) and (L). Error bars represent se. See also Supplemental Figure 1 online.
To further investigate the cause of bex5 hypersensitivity to BFA, we analyzed the cell ultrastructure by transmission electron microscopy (TEM). In PIN1pro:PIN1-GFP roots, a 1-h treatment with 50 µM BFA caused the heterogeneous aggregation of vesicle-like structures that became surrounded at their periphery by Golgi stacks (Figure 1K), as reported previously (Geldner et al., 2003; Grebe et al., 2003; Paciorek et al., 2005). We named this characteristic endosomal agglomeration a “typical BFA compartment,” as 94% of the total BFA compartments counted in the PIN1pro:PIN1-GFP roots (n = 31 BFA compartments) had this morphology. In contrast, in the PIN1pro:PIN1-GFP;bex5-1 mutant, BFA induced a different aggregation pattern (Figures 1K and 1L; see Supplemental Figure 1H online). Only 38% of the PIN1pro:PIN1-GFP;bex5-1 BFA-induced aggregates (n = 42 BFA-induced aggregates) displayed the typical BFA compartment morphology, while the others looked more enlarged, abnormally shaped, and less compact, and the vesicles from their composition displayed an altered morphology such as abnormal size (often larger than in the control), shape (distortion of the round shape probably due to vesicle fusion), and composition (more dense) (Figures 1K and 1L; see Supplemental Figure 1H online).
To test whether the impaired protein trafficking caused by the bex5 mutation affects overall plant development, we investigated the morphology of PIN1pro:PIN1-GFP;bex5 seedlings. PIN1pro:PIN1-GFP;bex5 seedlings had smaller cotyledons and shorter roots than PIN1pro:PIN1-GFP seedlings (Figure 1M; see Supplemental Figures 1I and 1J online), indicating that the BEX5 protein is important for plant growth and development. Previous studies have shown that BFA also affects root growth (Geldner et al., 2001, 2003). To investigate whether PIN1pro:PIN1-GFP;bex5-1 displays a different root growth sensitivity to BFA, we germinated and grew the mutant on medium supplemented with BFA. PIN1pro:PIN1-GFP;bex5-1 seedlings on BFA had shorter roots than control seedlings (Figures 1N to 1P; see Supplemental Figure 1K online), indicating that, not only at the subcellular level but also at the plant level, PIN1pro:PIN1-GFP;bex5-1 is hypersensitive to BFA. Together, these findings support the hypothesis that the bex5-1 mutant is defective in some BFA-sensitive trafficking processes.
bex5 Is Defective in Protein Trafficking to the PM
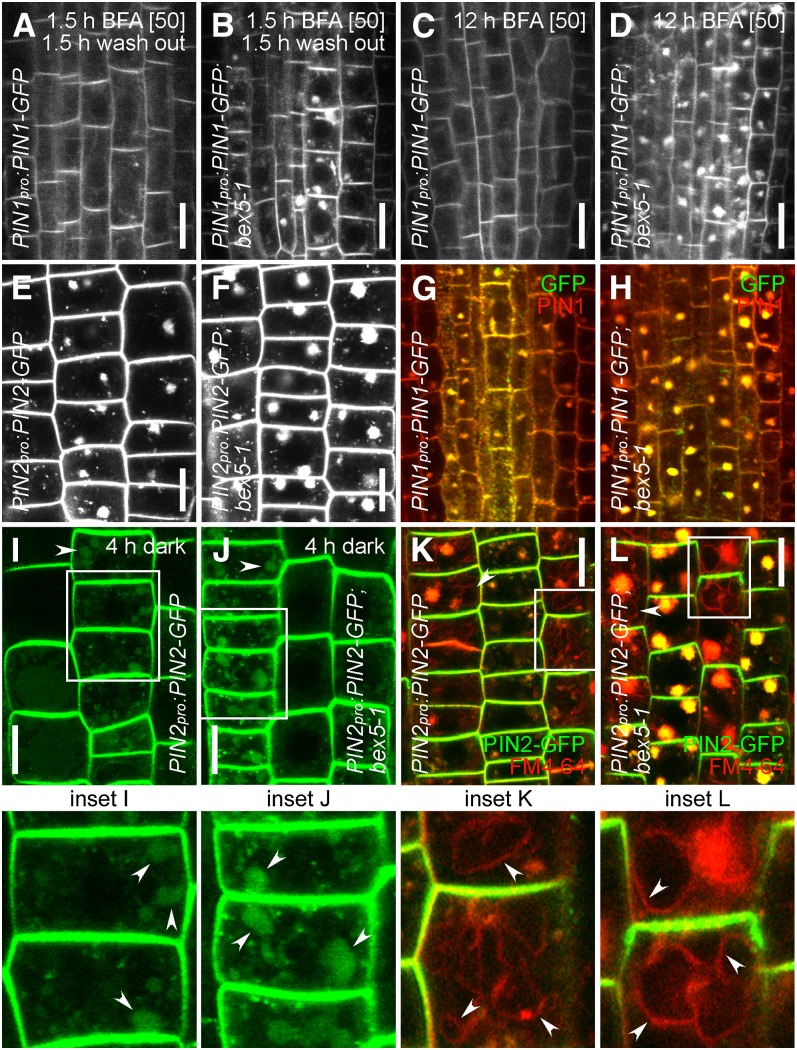
To test if the bex5 mutant is defective in exocytosis, we performed a 90-min treatment with 50 µM BFA followed by a 90-min wash out in Murashige and Skoog (MS) liquid medium. The intracellular agglomeration of PIN proteins caused by BFA treatment is reversible (Geldner et al., 2001; Kleine-Vehn et al., 2008); hence, we could observe almost complete disappearance of PIN1-GFP, PIN2, or GFP-PIP2a from the BFA compartments in the control cells treated with BFA followed by wash out (Figure 2A; see Supplemental Figures 2A and 2C online). In contrast, strong BFA-induced PIN1-GFP, PIN2, or GFP-PIP2a agglomerations persisted in the bex5-1 mutant after BFA removal (Figures 2A and 2B; see Supplemental Figures 2A to 2D online). This result suggests that exocytosis is defective in the PIN1pro:PIN1-GFP;bex5-1 mutant and indicates that BEX5 acts as a regulator of protein trafficking to the PM.
Figure 2.
bex5 Is Defective in Protein Trafficking to the PM.
(A) and (B) Exocytosis is defective in the bex5-1 mutant. A 1.5-h treatment with 50 µM BFA followed by a 1.5-h wash out with MS liquid medium led to almost complete disappearance of PIN1-GFP from the BFA bodies in the control vasculature cells (A). In contrast, strong persistent PIN1-GFP accumulation into BFA bodies could be observed in bex5-1vasculature cells (B).
(C) and (D) Transcytosis is defective in bex5-1. No intracellular PIN1-GFP accumulation could be observed in the wild-type vasculature cells treated for 12 h with 50 µM BFA (C). In contrast, pronounced intracellular PIN1-GFP accumulation persisted in the bex5-1 vasculature cells (D).
(E) to (H) A 0.5-h pretreatment with 50 µM cycloheximide followed by a 1.5-h cotreatment with 50 µM cycloheximide and 25 µM BFA does not interfere with the overaccumulation of PIN2-GFP ([E] and [F]) or PIN1-GFP ([G] and [H]) into abnormal BFA bodies in the bex5-1 mutant ([F] and [H]) as compared with the control ([E] and [G]). Live imaging of epidermal cells is shown in (E) and (F). Merged images show immunolocalization of PIN1 (red) and GFP (green) in the vasculature cells ([G] and [H]).
(I) to (L) Trafficking to the vacuole is not affected in the bex5-1 mutant. A 4-h dark treatment results in PIN2-GFP accumulation into vacuoles (arrowheads) of both wild-type (I) and bex5-1 (J) epidermal cells. Epidermal cells of the wild type (K) and bex5-1 (L) prestained with FM4-64 (4 µM) and treated with BFA (1.5 h; 25 µM) show overaccumulation of PIN2-GFP (green) and FM4-64 (red) into abnormal BFA bodies in bex5-1 (L) as compared with the control (K). Moreover, in both lines, FM4-64 also reaches and labels the tonoplast (arrowheads), indicating no defects in protein trafficking to the vacuoles in bex5-1. Insets represent enlargements of the boxed regions from the corresponding images.
Bars = 10 µm. See also Supplemental Figure 2 online.
During the long-term BFA treatment, PM proteins with a basal localization, such as PIN1, first agglomerate in the BFA compartments and then subsequently disappear from the BFA compartment and relocalize to the apical PM (Kleine-Vehn et al., 2008). This process involves protein translocation from the basal to the apical side of a polarized cell and is called transcytosis (Kleine-Vehn et al., 2008). No or very little PIN1-GFP agglomerations could be observed in the PIN1pro:PIN1-GFP root cells treated for 12 h with 50 µM BFA (Figure 2C; see Supplemental Figure 2E online). In contrast, PIN1pro:PIN1-GFP;bex5-1 showed strong intracellular PIN1-GFP accumulation (Figures 2C and 2D; see Supplemental Figures 2E and 2F online). The BFA-treated seedlings were briefly stained with propidium iodide to determine their cell viability (see Supplemental Figures 2E and 2F online). The observed impaired transcytosis indicates that the bex5-1 mutant is defective in PIN1-GFP trafficking from the BFA compartment to the apical side of the PM, which supports the hypothesis that the bex5-1 mutation affects protein trafficking to the PM.
Defective protein trafficking to the PM in the bex5 mutant may impair secretion and/or recycling. The bex5-1 mutant displayed BFA hypersensitivity also when we performed the BFA treatment in the presence of the protein synthesis inhibitor cycloheximide (Figures 2E to 2H), indicating that the PIN2-GFP (Figures 2E and 2F) or PIN1-GFP (Figures 2G and 2H) accumulation in the enlarged BFA bodies comes from a recycled but not newly synthesized pool of proteins.
To examine if the degradation pathway is impaired in the bex5-1 mutant, we performed a dark treatment experiment that allows the visualization of the vacuolar targeting of GFP-tagged proteins (Tamura et al., 2003; Feraru et al., 2010). A 4-h dark treatment followed by the observation of PIN2-GFP fluorescence revealed that in both the wild type and the bex5-1 mutant, PIN2-GFP accumulated into the vacuoles (Figures 2I and 2J). Furthermore, a 5-min staining with 2 µM FM4-64 followed by a 1.5-h treatment with 25 µM BFA showed the expected overaccumulation of FM4-64 and PIN2-GFP into abnormal BFA bodies in the bex5-1 mutant (Figures 2K and 2L). Moreover, in both PIN2pro:PIN2-GFP and PIN2pro:PIN2-GFP;bex5-1 seedlings, FM4-64 also labeled the tonoplast (Figures 2K and 2L). These two findings show that the bex5 mutation does not interfere with the trafficking of proteins from the TGN/EE to the vacuoles and support the hypothesis that recycling is defective in the bex5 mutant.
bex5 Displays Impaired Endosomal Compartments
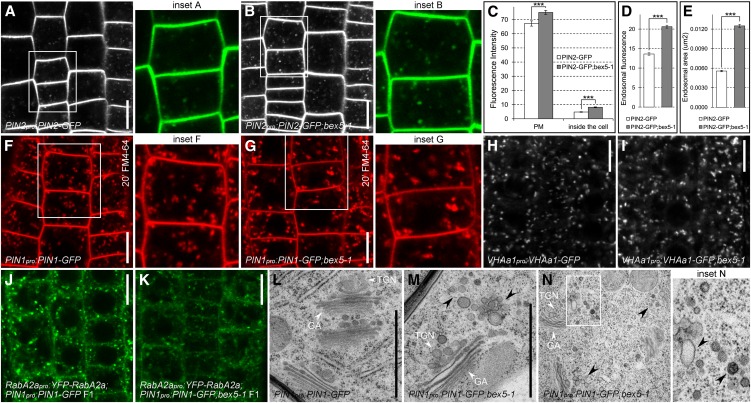
To gain further insight into trafficking defects in bex5, we analyzed the localization of the PIN2-GFP marker in the bex5 mutant. Live imaging of untreated bex5 mutant roots revealed an increased endosomal and PM PIN2-GFP signal (Figures 3A to 3D). PIN2-GFP endosomes in the mutant cells were enlarged, brighter, and sometimes appeared in aggregates (Figures 3A to 3E). Similarly, the uptake of the membrane-selective fluorescent dye FM4-64, commonly used to investigate endocytosis and vesicle trafficking in eukaryotic cells (Ueda et al., 2004; Tse et al., 2004; Dettmer et al., 2006; Chow et al., 2008; Jelínková et al., 2010), revealed slightly abnormal endosomal compartments in the bex5-1 mutant (Figures 3F and 3G; see Supplemental Figures 3A and 3B online). These findings, together with the abnormal morphology of the BFA compartment, indicate that the bex5-1 mutation affects endosomal compartments that are sensitive to BFA, such as the Golgi and/or the TGN/EE.
Figure 3.
bex5 Shows Abnormal Endosome Morphology.
(A) to (E) PIN2-GFP shows defective endosomes and increased PM accumulation in the bex5-1 mutant. Live imaging shows PIN2-GFP in the wild type (A) and bex5-1 (B). Measurements of PIN2-GFP fluorescence at the PM (n = 100 PMs from 10 roots; *** t test P = 6.49E-06) and inside the cell (n = 100 inner cell fluorescence from 10 roots; *** t test P = 1.04E-25) (C). Evaluation of PIN2-GFP endosomal fluorescence (D) (300 endosomes from 10 roots; *** t test P = 1.43E-42) and endosomal area (E) (300 endosomes from 10 roots; *** t test P = 1.50E-105) in the wild-type and bex5-1. Insets represent enlargements of the boxed regions from the corresponding images.
(F) to (K) Live imaging of FM4-64 (20 min; 4 µM) ([F] and [G]), VHAa1-GFP ([H] and [I]), and YFP-RabA2a ([J] and [K]) localization in the bex5-1 mutant ([G], [I], and [K]) as compared with the control ([F], [H], and [J]). Insets represent enlargements of the boxed regions from the corresponding images.
(L) to (N) PIN1pro:PIN1-GFP (L) and PIN1pro:PIN1-GFP;bex5-1 ([M] and [N]) cell ultrastructure. PIN1pro:PIN1-GFP;bex5-1 shows abnormal endosomal/vesicle clusters in the neighborhood of the GA-TGN/EE complex (black arrowheads) ([M] and [N]). Note the impaired morphology of the vesicles, such as size, shape, and composition (black arrowheads). Inset N represents an enlargement of the boxed region from the corresponding image.
Bars = 10 µm for (A), (B), and (F) to (K) and 1 µm for (L) and (M). Error bars represent se. See also Supplemental Figure 3 online.
Thus, we investigated the localization of the Golgi marker Sec21 (Movafeghi et al., 1999). By performing immunolocalization experiments, we found that Sec21 did not show altered localization in the bex5-1 mutant (see Supplemental Figures 3C and 3D online), indicating that the GA morphology may not be affected by the bex5-1 mutation. As the TGN/EE is another endosomal compartment that is a target of BFA, we investigated the localization of commonly used TGN/EE markers such as VHAa1 (Dettmer et al., 2006), clathrin light chain (CLC; Konopka et al., 2008), and RabA2a (Chow et al., 2008). Live imaging of VHAa1-GFP and CLC-GFP in the bex5-1 mutant revealed no dramatic or quantifiable defects in the localization of these proteins, although sometimes VHAa1-GFP and CLC-GFP endosomes appeared slightly enlarged and brighter (Figures 3H and 3I; see Supplemental Figures 3E and 3F online). Notably, we found that RabA2a GTPase, a protein defining a TGN compartment that contributes to cell plate formation (Chow et al., 2008), displayed an impaired localization in the bex5-1 mutant (Figures 3J and 3K). These results suggest that the bex5-1 mutant is impaired in a process that takes place at a TGN/EE compartment.
TEM performed on roots of untreated seedlings confirmed that bex5-1 endosomes have a defective morphology (Figures 3L to 3N; see Supplemental Figures 3G to 3J online). The typical morphology of the GA-TGN/EE complex observed in the PIN1pro:PIN1-GFP root cells was impaired in the root cells of the bex5-1 mutant (Figures 3L to 3N; see Supplemental Figures 3G to 3J online). The predominant defect in the mutant was characterized by endosomal/vesicle-like clusters present in the neighborhood of GA-TGN/EE, mainly at one of its sides. The vesicles had a more heterogeneous size and generally larger size and were often fused and abnormally shaped (Figures 3L to 3N; see Supplemental Figures 3G to 3J online). The abnormal accumulation of aberrant vesicles in the vicinity of the TGN/EE suggests that bex5-1 is defective in vesicle budding and/or trafficking and supports a role of BEX5 in protein/vesicle trafficking from a TGN/EE compartment to the PM.
BEX5 Has Different Subcellular Functions than GNL1 and BEN1
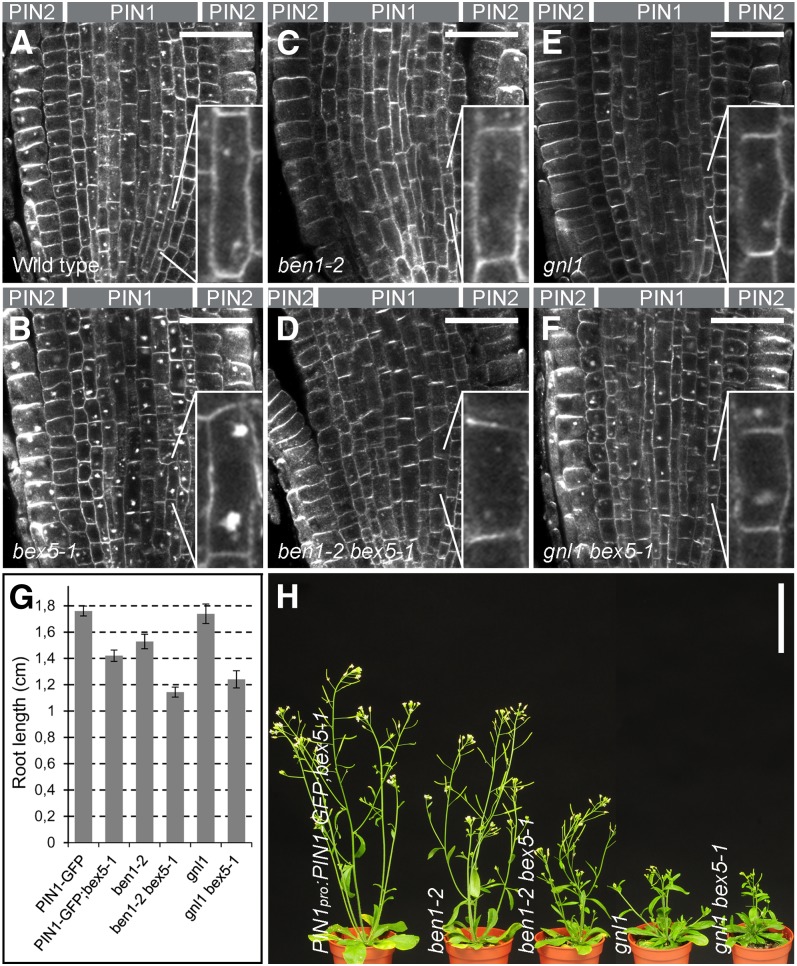
Next, we investigated the site of BEX5 action in protein trafficking. For this purpose, we analyzed the genetic relationship between bex5 and ben1, a mutant defective in protein trafficking from the TGN/EE to the RE (Tanaka et al., 2009), and gnl1, a mutant impaired at the GA (Richter et al., 2007; Teh and Moore, 2007). Both ben1 and gnl1 are distinct from bex5, as they showed a dramatically reduced intracellular accumulation of PIN1 or PIN2 following a 1.5-h treatment with 25 µM BFA (Figures 4A to 4F), a phenotype largely opposite to that of bex5. When crossed into the bex5-1 mutant background, ben1 showed complete and gnl1 showed partial epistasis to bex5-1 (Figures 4A to 4F; see Supplemental Figures 4A to 4C online). Thus, the root cells of the double ben1-2 bex5-1 or gnl1 bex5-1 mutants showed less BFA-induced accumulation of PIN1, PIN1-GFP, or PIN2 protein than the single bex5-1 mutant (Figures 4A to 4F; see Supplemental Figures 4A to 4C online). This suggests that BEX5 and the large ARF-GEFs BEN1 and GNL1 act at different steps along the intracellular trafficking pathway and that, in a subcellular membrane flow, BEX5 acts after BEN1 or GNL1. However, both double mutants showed additive effects regarding the root length (Figure 4G) and overall plant growth and development of the aerial part (Figure 4H), consistent with the notion that combined partial defects at two instances of the protein-trafficking pathways have a greater impact on plant growth than a single defect. Overall, these results are consistent with the finding that bex5, ben1, and gnl1 disrupt intracellular protein trafficking at distinct steps, with bex5 impairing protein trafficking in the post-TGN/EE pathway.
Figure 4.
BEX5 Acts Downstream of BEN1 and GNL1.
(A) to (F) Immunolocalization of PIN1 and PIN2 in the wild type (A), bex5-1 (B), ben1-2 (C), ben1-2;bex5-1 (D), gnl1 (E), and gnl1;bex5-1 (F) treated for 1.5 h with 25 µM BFA shows that gnl1 and ben1-2 mutations are partly (gnl1) or completely (ben1-2) epistatic to the bex5-1 mutation. Note that the BFA subcellular hypersensitivity of the bex5-1 mutant cannot be rescued by the genetic interaction with ben1-2 or gnl1. Insets represent magnifications of the indicated cells. Median sections of the root tips are shown.
(G) and (H) ben1-2 bex5-1 and gnl1 bex5-1 double mutants show additive effects with respect to root growth (G) and overall plant morphology (H). ben1-2 bex5-1 and gnl1 bex5-1 have shorter roots (n = 45) (G) and reduced overall plant growth (H) compared with the single bex5-1, ben1-2, or gnl1 mutant.
Bars = 30 µm for (A) to (F) and 5 cm for (H). Error bars represent se. See also Supplemental Figure 4 online.
BEX5 Encodes a Small GTP Binding Protein, RabA1b
We identified bex5-1 as a dominant mutant (see Supplemental Figures 5A to 5C online). Using 291 recombinants selected for the wild-type subcellular phenotype, we cloned bex5-1 by map-based cloning to a region of 236 kb on the left arm of chromosome 1 (Figure 5A). Because of a low recombination rate in this chromosome region, we chose not to further narrow down the interval but to use a candidate approach for gene identification. We sequenced several candidates and identified a point mutation that causes amino acid substitution S156F in a conserved GTP binding site in the open reading frame of RabA1b (Rutherford and Moore, 2002; Vernoud et al., 2003; Bassham et al., 2008) (Figure 5B).
Figure 5.
BEX5 Encodes RabA1b.
(A) Schematic representation of the chromosomal localization of BEX5/RabA1b. The organization of chromosome 5, the approximate position of BACs (red box), and the organization of BEX5 with the approximate position of the bex5-1 mutation are depicted in the scheme. The black dot indicates the approximate position of the centromer. ADH, alcohol dehydrogenase; ARR3, ARABIDOPSIS RESPONSE REGULATOR3; UFO, UNUSUAL FLORAL ORGANS.
(B) Alignment of several RabA1b amino acids from different model organisms. bex5-1 shows a point mutation that causes an amino acid substitution (Ser-156 to Phe) in the GTP binding site ETSAL, which is conserved among different eukaryotes. The asterisks indicate the conserved ETSAL GTP binding site.
(C) to (E) BEX5pro:bex5-1 T1 seedlings treated for 1.5 h with 25 µM BFA (E) display similar abnormal PIN1-GFP intracellular agglomerations to bex5-1 (D). Note that the wild type displays weaker BFA-induced PIN1-GFP intracellular accumulation (C). Live imaging of vasculature cells is shown.
Bars = 10 µm. See also Supplemental Figures 5 and 6 online.
To test whether the mutation in RabA1b is responsible for the defective protein trafficking, we analyzed two knockout T-DNA insertion alleles, bex5-2 and bex5-3 (see Supplemental Figure 5D online). These mutants did not show similar phenotypes to bex5-1 regarding the root growth hypersensitivity to BFA (see Supplemental Figure 5E online) or exocytosis defects (see Supplemental Figures 5F to 5I online), although the bex5-2 mutant showed mild defects in subcellular BFA sensitivity (see Supplemental Figures 5F to 5I online). These results suggest functional redundancy with other RabA1 GTPases or GTPases from the RabA class.
To confirm that the bex5-1 mutation in RabA1b is responsible for the observed protein-trafficking defects, we expressed the bex5-1 dominant mutation (genomic DNA amplified from the bex5-1 mutant) under the control of both the native and the strong constitutive cauliflower mosaic virus 35S promoter and subsequently transformed these constructs into the PIN1pro:PIN1-GFP transgenic line. Similar to the bex5-1 mutant, transgenic BEX5pro:bex5-1 (Figures 5C to 5E) or 35Spro:bex5-1 (see Supplemental Figures 6A to 6C online) showed high intracellular PIN1-GFP agglomerations after a 1.5-h treatment with 25 µM BFA. In contrast, week and sporadic BFA compartments could be observed in the PIN1pro:PIN1-GFP control (Figures 5C to 5E; see Supplemental Figures 6A to 6C online). Similar retention of PIN1-GFP into abnormal BFA bodies in bex5-1, BEX5pro:bex5-1, and 35Spro:bex5-1 confirms that the bex5-1 mutation causes the observed protein-trafficking defects. Surprisingly, overexpression of the wild-type clone (35Spro:GFP-RabA1b) in the wild-type background also resembles the bex5-1 subcellular phenotype (see Supplemental Figures 6D to 6I online). This indicates that wild-type 35Spro:GFP-RabA1b does not suppress the bex5-1 phenotype but generates a dominant phenotype.
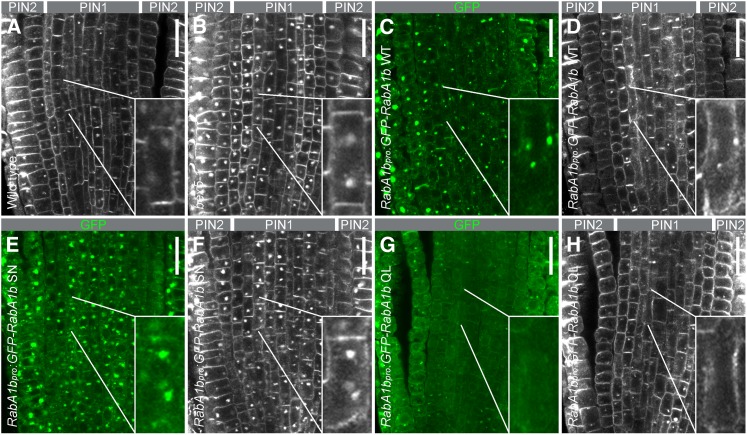
To investigate the nature of the bex5-1 mutation, we generated GTP binding and GTP hydrolysis RabA1b mutants by creating mutations in the conserved sequence motifs that are well known to either stabilize the GDP-bound (SN) or GTP-bound (QL) form (Chow et al., 2008). Thus, we generated RabA1bpro:GFP-RabA1b S27N and RabA1bpro:GFP-RabA1b Q72L mutants known to act as DN and CA versions, respectively (Chow et al., 2008). Immunolocalization of PIN1 and PIN2 proteins in the roots of RabA1bpro:GFP-RabA1b S27N and RabA1bpro:GFP-RabA1b Q72L treated for 1.5 h with 25 µM BFA showed that DN RabA1bpro:GFP-RabA1b S27N but not CA RabA1bpro:GFP-RabA1b Q72L displayed BFA oversensitivity similarly to the bex5-1 mutant (Figure 6). These results show that bex5-1, carrying the S156F mutation in the ETSAL conserved sequence motif of RabA1b small GTPase, acts as a DN mutant and identifies this mutation as a potential target of DN manipulations in small GTPases.
Figure 6.
bex5 Is a DN Mutant.
Immunolocalization of PIN1 (gray pseudocolor in the vasculature cells) ([A], [B], [D], [F], and [H]), PIN2 (gray pseudocolor in the epidermal and cortex cells) ([A], [B], [D], [F], and [H]), and GFP (green color) ([C], [E], and [G]) in the wild type (A), bex5-1 (B), RabA1bpro:GFP-RabA1b WT ([C] and [D]), RabA1bpro:GFP-RabA1b SN ([E] and [F]), and RabA1bpro:GFP-RabA1b QL ([G] and [H]) treated for 1.5 h with 25 µM BFA shows abnormal overaccumulation of PIN1 and PIN2 in RabA1bpro:GFP-RabA1b SN (F) and in the bex5-1 mutant (B). Insets represent magnifications of the indicated cells. Median sections of the root tips are shown. Bars = 20 µm.
BEX5/RabA1b Localizes to a TGN/EE Compartment
Next, we investigated the subcellular localization of BEX5/RabA1b by generating transgenic RabA1bpro:GFP-RabA1b and RabA1bpro:RFP-RabA1b in the wild-type background. Both transgenic constructs showed the same pattern of localization throughout the root, with higher occurrence in the root meristem, specifically in the root cap (Figure 7A; for RabA1bpro:GFP-RabA1b). At the subcellular level, red fluorescent protein (RFP)-RabA1b (Figure 7B) or GFP-RabA1b (Figure 7C) labeled punctate structures resembling endosomes that were sensitive to BFA (Figure 7D). In addition, RFP-RabA1b and GFP-RabA1b showed faint localization at the PM (Figures 7A to 7C) and, notably, they strongly labeled the cell plates during cell division (Figures 7A and 7B).
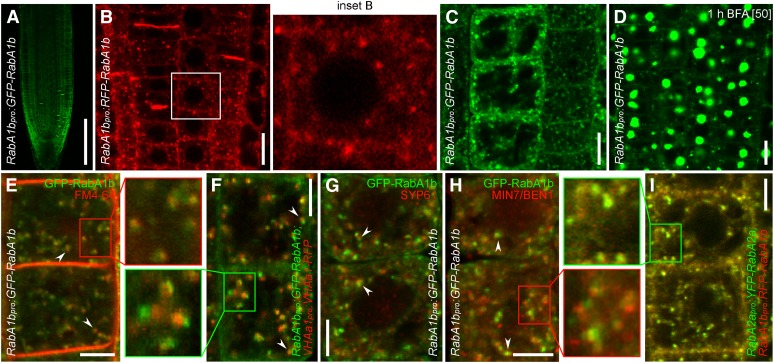
Figure 7.
BEX5 Localizes at Domains of the TGN/EE.
(A) RabA1bpro:GFP-RabA1b shows localization throughout the root, with a higher occurrence in the root cap.
(B) and (C) RFP-RabA1b (B) and GFP-RabA1b (C) show predominant localization at the endosomes and cell plate and faint localization at the PM. Inset B represents an enlargement of the boxed region from the corresponding image.
(D) GFP-RabA1b endosomes are sensitive to BFA (1-h treatment with 50 µM BFA).
(E) to (I) RabA1b localizes at a TGN/EE compartment. GFP-RabA1b ([E] to [H]) or RFP-RabA1b (I) endosomes partly (arrowheads) ([E] to [H]) or completely (I) overlap with TGN/EE markers such as FM4-64 (5-min treatment with 4 µM; red) (E), VHAa1-RFP (F), SYP61 (G), BEN1/MIN7 (H), and YFP-RabA2a (I). Live imaging is shown in (E), (F), and (I) and immunolocalization is shown in (G) and (H). Insets represent enlargements of the boxed regions from the corresponding images.
Bars = 100 µm for (A), 10 µm for (B) to (D), and 5 µm for (E) to (I). See also Supplemental Figure 7 online.
To address the identity of the BEX5/RabA1b compartments, we performed colocalization experiments and found that GFP-RabA1b endosomes showed partial colocalization with early accumulation (5 min after treatment) of the lipophilic styryl dye FM4-64 (Figure 7E; see Supplemental Figure 7A online). It has been shown that FM4-64 labels endosomal compartments of the endocytic pathway (Tse et al., 2004; Ueda et al., 2004; Dettmer et al., 2006) and that the early site of FM4-64 accumulation corresponds to the TGN/EE (Dettmer et al., 2006; Chow et al., 2008). The partial colocalization of GFP-RabA1b endosomes with early FM4-64–labeled endosomes indicates that BEX5/RabA1b localizes at a TGN/EE compartment. Moreover, GFP-RabA1b showed partial or complete colocalization with other known TGN/EE markers, such as VHAa1-RFP (Figure 7F; see Supplemental Figure 7B online), SYP61 (Figure 7G; see Supplemental Figure 7C online), MIN7/BEN1 (Figure 7H; see Supplemental Figure 7D online), and YFP-RabA2a (Figure 7I; see Supplemental Figure 7E online) (Sanderfoot et al., 2001; von der Fecht-Bartenbach et al., 2007; Tanaka et al., 2009). In contrast, GFP-RabA1b did not colocalize with Golgi markers Sec21 (see Supplemental Figure 7F online) and N-ST-GFP (see Supplemental Figure 7G online) (Grebe et al., 2003), although it often showed localization in their immediate proximity (see Supplemental Figures 7F and 7G online). Together, these results show that BEX5/RabA1b localizes at a TGN/EE compartment and further support the notion that BEX5/RabA1b plays a role in protein trafficking from this compartment to the PM.
DISCUSSION
A Fluorescence Imaging–Based Forward Genetic Screen Identifies bex5, a Novel Dominant Mutant of the Small RabA1 GTPases
Recently, several fluorescence imaging–based forward genetic screens have been used to identify regulators of crucial subcellular trafficking pathways and processes, such as enodocytosis, and vacuole biogenesis and function (Tanaka et al., 2009; Feraru et al., 2010; Zwiewka et al., 2011). The design of the screens allowed the identification of regulators impaired in essential cellular processes, which would be difficult to isolate in classical, plant morphology–based genetic screens because they lack dramatic morphological phenotypes, presumably due to functional redundancy. Here, we used a similar strategy and identified a GTPase from the previously uncharacterized RabA1 class.
The Arabidopsis genome contains 26 RabA GTPases that are divided into six classes, based on their sequence homology: RabA1 to RabA6 (Rutherford and Moore, 2002; Vernoud et al., 2003; Bassham et al., 2008). RabA GTPases are a large family of vesicle-trafficking regulators; however, knowledge about the more specific, cellular function of many members of this family is still lacking (Rutherford and Moore, 2002; Vernoud et al., 2003; Nielsen et al., 2008; Woollard and Moore, 2008). BEX5, identified in this study, encodes RabA1b, one of the nine previously uncharacterized members of the RabA1 subclass.
A widely used approach to investigate the function of Rab GTPases is to create mutations in their conserved sequence motifs. Thus, mutations in the conserved sequence motifs GxxxxGKS/T, WDTAGQE, and GNKxD have been generated previously to create DN or CA mutants impaired in nucleotide binding or hydrolysis (Rutherford and Moore, 2002). Here, we identified bex5-1 as a dominant mutant that shows an amino acid substitution in a GTP conserved sequence motif, ETSAK/L, that has not been used previously to generate Rab GTPase dominant mutants (Rutherford and Moore, 2002). Hence, we show that mutations in the conserved ETSAL sequence motif are sufficient to alter the function of RabA1 GTPases, while the S156F mutation in the ETSAL conserved sequence motif shows that Ser-156 is important for the function of RabA1b and, in addition, causes a phenotype that is similar to that caused by a classical DN mutation in the GxxxxGKS/T motif. Thus, the PIN1-GFP fluorescence imaging–based forward genetic screen presented here successfully identified bex5, a RabA1b GTPase mutant that behaves in a DN manner.
BEX5/RabA1b Is a Regulator of TGN-Mediated Protein Trafficking
The bex5-1 mutant shows enhanced BFA-induced intracellular protein accumulation in Arabidopsis root cells and is defective in the exocytosis and transcytosis of PM proteins. The results from TEM performed on BFA-treated roots are consistent with the oversensitivity of bex5-1 to BFA. Abnormally sized and shaped vesicles can be observed in the composition of the BFA bodies, and presumably, this is the cause of abnormally enlarged and less compact BFA bodies in the bex5-1 mutant.
In the absence of BFA, the bex5-1 mutant shows impaired endosome morphology and enhanced PIN2-GFP fluorescence at the PM. The impaired intracellular labeling of PIN2-GFP and FM4-64 indicates the presence of defective endosomal compartments in the bex5 mutant. Moreover, defective localization of YFP-RabA2a, which defines a domain of the TGN/EE required for cell plate formation (Chow et al., 2008), shows that bex5 is defective in a cellular process that occurs at the TGN/EE. This finding is supported by the analysis of bex5-1 cell ultrastructure, which revealed abnormal accumulation of misshaped, missized, and misfused vesicles in the proximity of the GA-TGN/EE complex. The alteration of the GA-TGN/EE complex (Kang et al., 2011) morphology indicates that the bex5 mutation may interfere with the formation of secretory vesicles at the TGN and suggests a role for BEX5 in the formation and budding of secretory vesicles.
Another question we addressed in this study is whether the protein-trafficking defects in bex5 come from secretion and/or recycling. The abnormal protein overaccumulation in the bex5 mutant is independent of protein biosynthesis. Furthermore, when treated with BFA, the double mutant ben1-2 bex5-1 shows that ben1-2, which is defective in trafficking from the TGN/EE to the RE (Tanaka et al., 2009), is completely epistatic to bex5-1. In contrast, the gnl1 mutant, which is defective in the maintenance of GA integrity (Richter et al., 2007; Teh and Moore, 2007), has only a partial epistatic effect to the bex5-1 mutant. Together, these results indicate that in the bex5 mutant a substantial pool of the PM proteins available for TGN-mediated trafficking to the PM largely comes from endocytic recycling. However, a secretion defect in the bex5-1 mutant is not completely excluded. Moreover, our results show that the mutation in BEX5 results in abnormal TGN/EE complexes in which proteins accumulate, presumably due to a defect in the formation, budding, and transport of secretory vesicles from the TGN/EE to the PM. This is in accordance with the localization of RabA1b at the RabA2a domain of the TGN/EE. Hence, we show that BEX5 is a novel regulator of protein trafficking that probably specifically regulates vesicle formation, budding, and transport from the TGN/EE compartment to the PM.
RabA1 GTPases Have an Evolutionarily Conserved Function among Eukaryotes
The large expansion of the RabA GTPase family in plants, as exemplified by Arabidopsis, shows the complex organization of the plant endomembrane system and the importance of these proteins for the regulation of vesicle trafficking. Arabidopsis RabA GTPases are similar to mammalian Rab11 (Rab11A, Rab11B, and Rab25) and Saccharomyces YEAST PROTEIN TRANSPORT (YPT; Saccharomyces pombae YPT3 or Saccharomyces cerevisiae YPT31 and YPT32) (Rutherford and Moore, 2002; Vernoud et al., 2003). Yeast YPT3 and YPT31/32 play a role in protein exit from the yeast trans-Golgi (Jedd et al., 1997) and in protein recycling through this compartment (Chen et al., 2005), while the mammalian putative ortholog Rab11 functions at the RE in regulating the recycling of PM proteins (Ullrich et al., 1996; Green et al., 1997; Ren et al., 1998; Prekeris et al., 2000).
In plants, members of RabA GTPases have also been associated with post-Golgi endomembranes (Ueda et al., 1996; Inaba et al., 2002; Preuss et al., 2004; Chow et al., 2008). So far, there is no information about the function of Arabidopsis RabA1 proteins. However, the analyses of similar regulators from other plant species are consistent with our identified role of BEX5/RabA1b in regulating exocytosis. Thus, tomato (Solanum lycopersicum) SlRab11a and rice (Oryza sativa) OsRab11 were suggested to regulate protein trafficking from the TGN/EE to the PM (Heo et al., 2005; Rehman et al., 2008), while tobacco (Nicotiana tabacum) NtRab11 plays a role in exocytosis (de Graaf et al., 2005).
The analysis of the T-DNA insertion mutants and the localization of BEX5/RabA1b suggest that some members of RabA GTPases might act redundantly. In contrast with bex5-1, the knockout bex5-2 and bex5-3 mutants did not show or showed only mild root and subcellular BFA hypersensitivity or defective trafficking processes. This result suggests a redundant function with closely related RabA GTPase members (Preuss et al., 2004). This hypothesis is strongly supported by the localization and function of other members of the RabA GTPase family (Chow et al., 2008; Geldner et al., 2009; Kang et al., 2011). It has been shown that RabA members such as RabA1e, RabA1g, RabA2a to -d, RabA3, and RabA4b GTPases are localized at the TGN/EE (Chow et al., 2008; Geldner et al., 2009; Kang et al., 2011). In addition, they are abundant at the cell plate (Chow et al., 2008; Geldner et al., 2009). This similar localization pattern to RabA1b indicates that other RabA members, such as RabA2 and RabA3, might have a similar function at the TGN/EE level to RabA1b/BEX5; however, this was not explored in detail because the investigation of RabA2 and RabA3 GTPases, for example, focused mainly on their role in cytokinesis (Chow et al., 2008), whereas we investigated BEX5-mediated trafficking from the TGN/EE to the PM. By identifying and characterizing BEX5/RabA1b as a regulator of protein trafficking from the TGN/EE to the PM, we show that Rab11-like Rab GTPases have an evolutionarily conserved function among different eukaryotes.
METHODS
Plant Material and Growth Conditions
PIN1pro:PIN1-GFP (Benková et al., 2003), PIN2pro:PIN2-GFP (Xu and Scheres, 2005), VHAa1pro:VHAa1-GFP (Dettmer et al., 2006), VHAa1pro:VHAa1-RFP (von der Fecht-Bartenbach et al., 2007), RabA2apro:YFP-RabA2a (Chow et al., 2008), 35Spro:GFP-PIP2a (Cutler et al., 2000), 35Spro:CLC-GFP (Konopka et al., 2008), and 35Spro:N-ST-YFP (Grebe et al., 2003) marker lines and the ben1-2 (Tanaka et al., 2009) mutant in Arabidopsis thaliana have been described previously. The seeds were stratified for 2 d in the dark at 4°C, and the seedlings were germinated and grown vertically in Petri dishes containing 0.8% agar and 0.5× MS medium with 1% Suc (pH 5.9) mainly at 18°C, long-day photoperiod but also at 22°C, continuous illumination. gnl1 (Salk_030701), bex5-2 (SAIL_875_F08), and bex5-3 (GABI_726E08) alleles were identified from available T-DNA collections (Alonso et al., 2003).
Forward Genetic Screen and Mapping
To identify bex mutants, 35,000 M2 PIN1pro:PIN1-GFP seedlings descended from 1200 0.3% ethyl methanesulfonate–mutagenized M1 plants were analyzed with the epifluorescence microscope. The bex5-1 mutant on chromosome 1 was mapped between F19K19 (5.722 Mb) simple sequence-length polymorphism and F28G4_FspBI (5.958 Mb) cleaved-amplified polymorphic sequence markers. A total of 291 recombinants from an F2 cross between bex5-1 (Columbia background) and Landsberg erecta were used. For Columbia/Landsberg erecta polymorphism information, the Monsanto Arabidopsis Polymorphism and the Ler Sequence Collection (Cereon Genomics) were used, and for information regarding single-nucleotide polymorphisms and insertions/deletions, The Arabidopsis Information Resource (http://www.Arabidopsis.org) was used.
Drug Treatments
Five-day-old seedlings were incubated for the indicated time points in MS medium containing one of the following drugs: BFA (Invitrogen; 25 or 50 μM in DMSO solvent), cycloheximide (Sigma; 50 μM in DMSO solvent), propidium iodide (Sigma; 1 mg/mL in water diluted 1:100), and FM4-64 (Invitrogen; 2 or 4 µM in water solvent); or they were germinated for 5 or 7 d on MS medium supplemented with BFA at 5 or 7.5 μM.
Quantitative Analysis of BFA Bodies
For the evaluation of bex5 oversensitivity to BFA, the fluorescence intensity of PIN2-GFP or PIN1-GFP was measured in a defined region of 1416 E-4 (PIN2-GFP) and 7080 E-5 (PIN1-GFP) for BFA bodies and in a defined region of 5015 E-4 (PIN2-GFP) and 2537 E-4 (PIN1-GFP) for PMs by using the corresponding functions of ImageJ 1.41 software (http://rsbweb.nih.gov/ij/). One hundred BFA bodies and 100 PMs (1 BFA body and 1 PM per cell; 10 cells per root) from 10 root tips were evaluated. To evaluate the area of the BFA bodies, 150 bodies from 10 root tips per line (∼15 BFA bodies per root) were measured. To examine the ultrastructure of the BFA bodies, the BFA bodies from 32 PIN1pro:PIN1-GFP cells (five root tips) and 38 PIN1pro:PIN1-GFP;bex5 cells (five root tips) were counted on TEM images.
Quantitative Evaluation of PM Fluorescence and/or Endosomal Area
To evaluate PIN2-GFP fluorescence at the PM and in the endosomes, the fluorescence of 100 PMs and the total fluorescence from the interior of 100 cells were measured. Ten cells per root and a total of 10 root tips were evaluated per line. Additionally, the fluorescence and area of PIN2-GFP endosomes were gauged in 300 endosomes from 10 root tips per line (30 endosomes per root from ∼10 cells).
Protein Immunolocalizations
Immunolocalizations were performed on 4- to 5-d-old seedlings using the Intavis in situ pro robot, as described by Sauer et al. (2006). The following dilutions were used for primary antibodies: anti-PIN1 rabbit (Paciorek et al., 2005), 1:1000; anti-PIN2 rabbit (Paciorek et al., 2005), 1:1000; anti-ARF1 rabbit (Pimpl et al., 2000), 1:1000; anti-MIN7/BEN1 rabbit (Nomura et al., 2006), 1:1000; anti-Sec21 rabbit (Movafeghi et al., 1999), 1:1000; anti-SYP61 rabbit (Sanderfoot et al., 2001), 1:500; anti-PM–ATPase mouse (Geldner et al., 2001), 1:1000; and anti-GFP mouse (Roche), 1:1000. For the secondary antibodies, Alexa488 anti-mouse (Invitrogen), 1:600 and Cy3 anti-mouse (Sigma), 1:600 were used.
TEM on Roots
Root tips of 4-d-old seedlings of PIN1pro:PIN1-GFP and PIN1pro:PIN1-GFP;bex5-1 untreated and treated for 1 h with 50 µM BFA were excised and processed as described by Tanaka et al. (2009).
Genotyping and RT-PCR
bex5-1 (bex5-1_BglII_FP, 5′-AGAATCGCTCTGCTTCATGGAAAGAT-3′, and bex5-1_RP, 5′-TCAATTTGAGCAGCACCCGAG-3′, product size is 211 bp; BglII cuts wild-type sequence), bex5-2 (bex5-2_FP, 5′-AATGTATGTCATGGCGTGAGTC-3′, and bex5-2_RP, 5′-CTCCCATTCACTCCAAAACTG-3′), bex5-3 (bex5-3_FP, 5′-AGTTGACTGCATTTGACCACC-3′, and bex5-3_RP, 5′-TGAACGTCGGCATTTTAGATC-3′), ben1-2 (ben1-2_FP, 5′-TGGAAAGTGAAATTGGTGAGC-3′, and ben1-2_RP, 5′-CAAGGATTCTTCTCTGCATGG-3′), and gnl1 (gnl1_FP, 5′-TCACGTTAGATCCCGTACTGG-3′, and gnl1_RP, 5′-AGCAGTTTCCAAATTGCCTC-3′) were genotyped using the stated primers.
For RT-PCR, total RNA was isolated from 6-d-old seedlings using the Qiagen RNeasy Plant Mini kit. The cDNA was synthesized with the Reverse Transcription System (Promega) from 1 µg of total RNA that was previously decontaminated with an Ambion TURBO DNA-free kit. The total length of BEX5 was amplified using BEX5_FP (5′-ATGGCAGGGTACAGAGTGGAA-3′) and BEX5_RP (5′-ATTTGAGCAGCACCCGAGTTT-3′) primers. Tubulin (Tub_FP, 5′-ACTCGTTGGGAGGAGGAACT-3′, and Tub_RP, 5′-ACACCAGACATAGTAGCAGAAATCAAG-3′) was used as a control. The PCR underwent 25 cycles.
Cloning and Transformation
To create RabA1bpro:GFP-RabA1b and RabA1bpro:RFP-RabA1b, the promoter was introduced into pDONR P4-P1R (attB4_ RabA1b_prom_FP, 5′-GGGGACAACTTTGTATAGAAAAGTTGTTATGACCATTTCGCTGACCGTA-3′, and attB1r_ RabA1b_prom_RP, 5′-GGGGACTGCTTTTTTGTACAAACTTGTCTCCGCCGTCTCCAGGTGGTT-3′) and the gene into pDONR P2R-P3 (attB2r_ RabA1b_FP, 5′-GGGGACAGCTTTCTTGTACAAAGTGGTTATGGCAGGGTACAGAGTGGAA-3′, and attB3_ RabA1b_stop_RP, 5′-GGGGACAACTTTGTATAATAAAGTTGTTCAATTTGAGCAGCACCCGAG-3′). Then, by multisite Gateway recombination, these fragments were introduced with pEN-L1-F-L2 or pEN-L1-R-L2 into pB7m34GW,0 (Karimi et al., 2002; http://www.psb.ugent.be/gateway). To create 35Spro:GFP-RabA1b, we introduced the entry clone containing RabA1b together with pEN-L4-2-R1 and pEN-L1-F-L2 into pK7m34GW,0. All constructs were transformed into the Columbia wild type.
To make 35Spro:bex5-1, the mutated form of BEX5 was amplified from bex5-1 DNA using attB1_bex5-1_FP, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTATGGCAGGGTACAGAGTGGAA-3′, and attB2_bex5-1_stop_RP, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAATTTGAGCAGCACCCGAG-3′. The fragment was introduced into pDONR221, then into pB7WG2,0, and transformed into the PIN1pro:PIN1-GFP transgenic line. To generate BEX5pro:bex5-1, the BEX5 promoter was amplified together with the mutated gene from the bex5-1 mutant DNA using attB1_BEX5_prom_FP, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTTATGACCATTTCGCTGACCGTA-3′, and attB2_bex5-1_stop_RP. The fragment was introduced into pDONR221, then into pB7FWG,0, and further transformed into the PIN1pro:PIN1-GFP transgenic line.
S27N and Q72L mutant versions of RabA1bpro:GFP-RabA1b were generated by PCR mutagenesis of RabA1bpro:GFP-RabA1b in pGWB1, which contained 2.5 kb of the 5′ flanking and 1.0 kb of the 3′ flanking sequences, using SN f, 5′-GAGTTGGCAAAAACAATCTCCTTTC-3′, SN r, 5′-GAAAGGAGATTGTTTTTGCCAACTC-3′, QL f, 5′-TTTGGGACACTGCTGGTCTGGAGAG-3′, and QL r, 5′-CTCTCCAGACCAGCAGTGTCCCAAA-3′. The original RabA1bpro:GFP-RabA1b fragment was made by the technique of fluorescent tagging of full-length proteins (Tian et al., 2004). Primer sequences used for the amplification of genomic fragments were RabA1b-1, 5′-CACCCATTTCGCTGACCGTAGATAACCCAGT-3′, RabA1b-2, 5′-CCGGAACTCACTCACTTGGATTTGGTA-3′, RabA1b-3, 5′-ACCACCTGGAGACGGCGGAGATGGTGAGCAAGGGCGAGGAGCTGT-3′, RabA1b-4, 5′-TCCTCGCCCTTGCTCACCATCTCCGCCGTCTCCAGGTGGTTGCTT-3′, RabA1b-5, 5′-TGTACAAGGGAGGTAGTGGCATGGCAGGGTACAGAGTGGAAGATG-3′, and RabA1b-6, 5′-CCTGCCATGCCACTACCTCCCTTGTACAGCTCGTCCATGCCGTGA-3′.
Confocal laser scanning microscopy images were taken with an Olympus FV10 ASW apparatus, and the most representative images are shown. Collages were assembled in Photoshop 7.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BEX5/RabA1b (At1G16920), PIN1 (At1g73590), PIN2 (At5G57090), PIP2a (At3G53420), VHAa1 (At2g28520), RabA2a (At1G09630), CLC (At2G40060), N-ST (AJ243198), BEN1 (At3G43300), ARF1 (M95166), SEC21 (AL023094), SYP61 (AF355754), and GNL1 (At5G39500).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. bex5 Shows Enhanced Sensitivity to BFA.
Supplemental Figure 2. bex5 Is Defective in Exocytosis and Transcytosis of PM Proteins.
Supplemental Figure 3. bex5 Shows Abnormal Endosomal Compartments.
Supplemental Figure 4. gnl1 Is Epistatic to bex5.
Supplemental Figure 5. bex5 Is a Dominant RabA1b Mutant.
Supplemental Figure 6. BEX5 Encodes RabA1b.
Supplemental Figure 7. BEX5 Localizes at a TGN/EE Compartment.
Supplementary Material
Acknowledgments
We thank Sebastian Y. Bednarek, Eva Benková, Sheng Y. He, Christian Luschnig, Wolfgang Michalke, Ian Moore, Natasha V. Raikhel, David G. Robinson, Ben Scheres, Karin Schumacher, and Chris R. Somerville for providing published material; the Nottingham Arabidopsis Stock Centre and GABI-Kat for T-DNA–mutated seeds; Wilson Ardiles-Diaz, Carina Braeckman, and Karel Spruyt for sequencing, plant transformation, and plant photographs, respectively; and Annick Bleys for help with preparing the manuscript. This work was supported by the Odysseus program of the Research Foundation-Flanders and KörberStiftung (to J.F.), by projects CZ.1.07/2.3.00/20.0043 and CZ.1.05/1.1.00/02.0068 of the Central European Institute of Technology (to J.F.), and by a MEXT Grant-in-Aid for Scientific Research on Priority Areas (Grant 23012026 to H.T.).
AUTHOR CONTRIBUTIONS
E.F. designed and performed most of the experiments. M.I.F., R.A., R.D.R., T.P., H.T., and A.N. performed experiments and analyzed data. E.F. and J.F. analyzed the data, discussed the results, and wrote the article. All authors corrected and approved the article.
Glossary
- PM
plasma membrane
- BFA
brefeldin A
- GA
Golgi apparatus
- TGN/EE
trans-Golgi network/early endosome
- RE
recycling endosome
- DN
dominant negative
- CA
constitutive active
- TEM
transmission electron microscopy
- MS
Murashige and Skoog
References
- Alonso J.M.,, et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anders N., Jürgens G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65: 3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D.C., Brandizzi F., Otegui M.S., Sanderfoot A.A. (2008). The secretory system of Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0099, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Chen S.H., Chen S., Tokarev A.A., Liu F., Jedd G., Segev N. (2005). Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol. Biol. Cell 16: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.-M., Neto H., Foucart C., Moore I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf B.H.J., Cheung A.Y., Andreyeva T., Levasseur K., Kieliszewski M., Wu H.M. (2005). Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell 17: 2564–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.-D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.-D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Ding Z., Galván-Ampudia C.S., Demarsy E., Łangowski Ł., Kleine-Vehn J., Fan Y., Morita M.T., Tasaka M., Fankhauser C., Offringa R., Friml J. (2011). Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Feraru E., Paciorek T., Feraru M.I., Zwiewka M., De Groodt R., De Rycke R., Kleine-Vehn J., Friml J. (2010). The AP-3β adaptin mediates the biogenesis and function of lytic vacuoles in Arabidopsis. Plant Cell 22: 2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J., Wiśniewska J., Benková E., Mendgen K., Palme K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Geldner N., Dénervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.-D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.-D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428 [DOI] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Green E.G., Ramm E., Riley N.M., Spiro D.J., Goldenring J.R., Wessling-Resnick M. (1997). Rab11 is associated with transferrin-containing recycling compartments in K562 cells. Biochem. Biophys. Res. Commun. 239: 612–616 [DOI] [PubMed] [Google Scholar]
- Grosshans B.L., Ortiz D., Novick P. (2006). Rabs and their effectors: Achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA 103: 11821–11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Heo J.B., Rho H.S., Kim S.W., Hwang S.M., Kwon H.J., Nahm M.Y., Bang W.Y., Bahk J.D. (2005). OsGAP1 functions as a positive regulator of OsRab11-mediated TGN to PM or vacuole trafficking. Plant Cell Physiol. 46: 2005–2018 [DOI] [PubMed] [Google Scholar]
- Inaba T., Nagano Y., Nagasaki T., Sasaki Y. (2002). Distinct localization of two closely related Ypt3/Rab11 proteins on the trafficking pathway in higher plants. J. Biol. Chem. 277: 9183–9188 [DOI] [PubMed] [Google Scholar]
- Irani N.G., Russinova E. (2009). Receptor endocytosis and signaling in plants. Curr. Opin. Plant Biol. 12: 653–659 [DOI] [PubMed] [Google Scholar]
- Jedd G., Mulholland J., Segev N. (1997). Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J. Cell Biol. 137: 563–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelínková A., Malínská K., Simon S., Kleine-Vehn J., Parezová M., Pejchar P., Kubeš M., Martinec J., Friml J., Zazímalová E., Petrásek J. (2010). Probing plant membranes with FM dyes: Tracking, dragging or blocking? Plant J. 61: 883–892 [DOI] [PubMed] [Google Scholar]
- Kang B.-H., Nielsen E., Preuss M.L., Mastronarde D., Staehelin L.A. (2011). Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J., Ding Z., Jones A.R., Tasaka M., Morita M.T., Friml J. (2010). Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl. Acad. Sci. USA 107: 22344–22349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Konopka C.A., Backues S.K., Bednarek S.Y. (2008). Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 20: 1363–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson L.A., Hanton S.L., Brandizzi F. (2006). Traffic between the plant endoplasmic reticulum and Golgi apparatus: To the Golgi and beyond. Curr. Opin. Plant Biol. 9: 601–609 [DOI] [PubMed] [Google Scholar]
- Movafeghi A., Happel N., Pimpl P., Tai G.-H., Robinson D.G. (1999). Arabidopsis Sec21p and Sec23p homologs. Probable coat proteins of plant COP-coated vesicles. Plant Physiol. 119: 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.S., Bandyopadhyay A., Holstein S.E., Peer W.A. (2005). Endocytotic cycling of PM proteins. Annu. Rev. Plant Biol. 56: 221–251 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Cheung A.Y., Ueda T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223 [DOI] [PubMed] [Google Scholar]
- Paciorek T., Zazímalová E., Ruthardt N., Petrásek J., Stierhof Y.-D., Kleine-Vehn J., Morris D.A., Emans N., Jürgens G., Geldner N., Friml J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256 [DOI] [PubMed] [Google Scholar]
- Paul M.J., Frigerio L. (2007). Coated vesicles in plant cells. Semin. Cell Dev. Biol. 18: 471–478 [DOI] [PubMed] [Google Scholar]
- Pereira-Leal J.B., Seabra M.C. (2000). The mammalian Rab family of small GTPases: Definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301: 1077–1087 [DOI] [PubMed] [Google Scholar]
- Petrásek J.,, et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pimpl P., Movafeghi A., Coughlan S., Denecke J., Hillmer S., Robinson D.G. (2000). In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Scheller R.H. (2000). A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell 6: 1437–1448 [DOI] [PubMed] [Google Scholar]
- Preuss M.L., Serna J., Falbel T.G., Bednarek S.Y., Nielsen E. (2004). The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell 16: 1589–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakusová H., Gallego-Bartolomé J., Vanstraelen M., Robert H.S., Alabadí D., Blázquez M.A., Benková E., Friml J. (2011). Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 67: 817–826 [DOI] [PubMed] [Google Scholar]
- Rehman R.U., Stigliano E., Lycett G.W., Sticher L., Sbano F., Faraco M., Dalessandro G., Di Sansebastiano G.-P. (2008). Tomato Rab11a characterization evidenced a difference between SYP121-dependent and SYP122-dependent exocytosis. Plant Cell Physiol. 49: 751–766 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.-R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Ren M., Xu G., Zeng J., De Lemos-Chiarandini C., Adesnik M., Sabatini D.D. (1998). Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95: 6187–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.-D., Rios G., Koncz C., Robinson D.G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492 [DOI] [PubMed] [Google Scholar]
- Robert S.,, et al. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Jiang L., Schumacher K. (2008). The endosomal system of plants: Charting new and familiar territories. Plant Physiol. 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5: 518–528 [DOI] [PubMed] [Google Scholar]
- Sanderfoot A.A., Kovaleva V., Bassham D.C., Raikhel N.V. (2001). Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol. Biol. Cell 12: 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Paciorek T., Benková E., Friml J. (2006). Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Ono E., Tanaka Y., Nagatani A., Higashi S.-I., Watanabe M., Nishimura M., Hara-Nishimura I. (2003). Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397 [DOI] [PubMed] [Google Scholar]
- Teh O.K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496 [DOI] [PubMed] [Google Scholar]
- Tian G.W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y.C., Mo B., Hillmer S., Zhao M., Lo S.W., Robinson D.G., Jiang L. (2004). Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell 16: 672–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Anai T., Tsukaya H., Hirata A., Uchimiya H. (1996). Characterization and subcellular localization of a small GTP-binding protein (Ara-4) from Arabidopsis: Conditional expression under control of the promoter of the gene for heat-shock protein HSP81-1. Mol. Gen. Genet. 250: 533–539 [DOI] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Ullrich O., Reinsch S., Urbé S., Zerial M., Parton R.G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V., Horton A.C., Yang Z., Nielsen E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131: 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C., et al. (2010). Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Denecke J. (1999). The endoplasmic reticulum: Gateway of the secretory pathway. Plant Cell 11: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Galili G. (2001). The endomembrane system and the problem of protein sorting. Plant Physiol. 125: 115–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Fecht-Bartenbach J., Bogner M., Krebs M., Stierhof Y.-D., Schumacher K., Ludewig U. (2007). Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J. 50: 466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard A.A.D., Moore I. (2008). The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 11: 610–619 [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2: 107–117 [DOI] [PubMed] [Google Scholar]
- Zwiewka M., Feraru E., Möller B., Hwang I., Feraru M.I., Kleine-Vehn J., Weijers D., Friml J. (2011). The AP-3 adaptor complex is required for vacuolar function in Arabidopsis. Cell Res. 21: 1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.