Abstract
The nuclear envelope in Saccharomyces cerevisiae harbors two essential macromolecular protein assemblies: the nuclear pore complexes (NPCs) that enable nucleocytoplasmic transport, and the spindle pole bodies (SPBs) that mediate chromosome segregation. Previously, based on metazoan and budding yeast studies, we reported that reticulons and Yop1/DP1 play a role in the early steps of de novo NPC assembly. Here, we examined if Rtn1 and Yop1 are required for SPB function in S. cerevisiae. Electron microscopy of rtn1Δ yop1Δ cells revealed lobular abnormalities in SPB structure. Using an assay that monitors lateral expansion of the SPB central layer, we found that rtn1Δ yop1Δ SPBs had decreased connections to the NE compared to wild type, suggesting that SPBs are less stable in the NE. Furthermore, large budded rtn1Δ yop1Δ cells exhibited a high incidence of short mitotic spindles, which were frequently misoriented with respect to the mother–daughter axis. This correlated with cytoplasmic microtubule defects. We found that overexpression of the SPB insertion factors NDC1, MPS2, or BBP1 rescued the SPB defects observed in rtn1Δ yop1Δ cells. However, only overexpression of NDC1, which is also required for NPC biogenesis, rescued both the SPB and NPC associated defects. Rtn1 and Yop1 also physically interacted with Ndc1 and other NPC membrane proteins. We propose that NPC and SPB biogenesis are altered in cells lacking Rtn1 and Yop1 due to competition between these complexes for Ndc1, an essential common component of both NPCs and SPBs.
Keywords: reticulons, Yop1/DP1, Ndc1, spindle pole body, nuclear pore complex
THE nuclear envelope (NE), which physically separates the nucleoplasm from the cytoplasm, is a characteristic feature of all eukaryotic cells and structurally based upon two distinct yet connected membrane bilayers. These NE membranes harbor specialized functions, with the outer nuclear membrane (ONM) continuous with the endoplasmic reticulum (ER) and the inner nuclear envelope (INM) having a unique protein composition (Schirmer et al. 2003; Lusk et al. 2007; Antonin et al. 2011). However, specific connections between the ONM and INM are critical for cell function. For example, ONM protein–INM protein interactions that bridge the perinuclear space are required for nuclear positioning (Hiraoka and Dernburg 2009; Razafsky and Hodzic 2009). Moreover, the ONM and INM are specifically fused at sites of nuclear pores (Doucet and Hetzer 2010). The NE is further distinguished by the presence of large protein assemblies; for example, the nuclear pore complex (NPC) found in all eukaryotes and the spindle pole body (SPB) in the budding yeast Saccharomyces cerevisiae. A full understanding of the dynamics between the NE membranes and its different NE protein assemblies has not yet been achieved.
The NPCs in the NE are responsible for regulating the trafficking of macromolecules between the nucleoplasm and cytoplasm, and between the ONM and INM (Lusk et al. 2007; Tetenbaum-Novatt and Rout 2010). As >60 MDa proteinaceous complexes, the NPCs are assembled from ∼30 different proteins termed nucleoporins (Nups) or pore membrane proteins (Poms) with each Nup or Pom present in multiples of eightfold stoichiometry (8, 16, or 32 copies) (Alber et al. 2007). NPCs have structurally distinct modules: the nuclear basket filaments, the cytoplasmic filaments, the outer, central and lumenal rings, and a set of linker complexes. In the closed mitosis of S. cerevisiae and during metazoan interphase, all NPCs assemble de novo into an intact NE (D’Angelo et al. 2006; Alber et al. 2007; Antonin et al. 2008; Brohawn et al. 2008; Brohawn et al. 2009; Capelson et al. 2010; Talamas and Hetzer 2011). This NPC biogenesis mechanism requires a multistep process that is dependent on both ONM and INM events. The first steps of de novo NPC assembly require ONM/INM fusion and stabilization of the resulting highly curved pore membrane, a process that is not yet fully understood (D’Angelo et al. 2006; Antonin et al. 2008; Fernandez-Martinez and Rout 2009; Doucet and Hetzer 2010; Talamas and Hetzer 2011). Membrane-bending and curvature-stabilizing proteins, as well as potential changes in lipid composition, are likely required (Doucet and Hetzer 2010). Current models propose that the initial pore fusion event is mediated by NPC-associated Poms. In S. cerevisiae, this potentially includes Ndc1, Pom152, Pom34, and Pom33 (Madrid et al. 2006; Mansfeld et al. 2006; Antonin et al. 2008; Hetzer and Wente 2009; Onischenko et al. 2009; Chadrin et al. 2010; Doucet and Hetzer 2010). In addition, an early step in de novo NPC biogenesis requires the reticulons (Rtn) and Yop1/DP1 (Dawson et al. 2009; Chadrin et al. 2010), proteins in the outer membrane leaflet that act to stabilize/maintain membrane curvature (De Craene et al. 2006; Voeltz et al. 2006; Hu et al. 2008; West et al. 2011). After fusion of the INM and ONM, the Rtns and Yop1/DP1 are speculated to transiently localize at and stabilize the nascent pore (Dawson et al. 2009; Hetzer and Wente 2009). The subsequent recruitment of peripheral membrane Nups would maintain the curved pore membrane and provide a scaffold on which other Nups then assemble.
The S. cerevisiae SPB is the functional equivalent of the centrosome, nucleating both cytoplasmic microtubules involved in nuclear positioning and cytoplasmic transport as well as nuclear microtubules required for chromosome segregation (Byers and Goetsch 1975). Much like the NPC, the SPB is a modular structure and is formed by five subcomplexes: the γ-tubulin complex that nucleates microtubules, the linker proteins that connect the γ-tubulin complex to the cytoplasmic and nuclear face of the core SPB, the soluble core SPB/satellite components that form the foundation of the SPB and SPB precursor, the membrane anchors that tether the core SPB in the NE, and the half-bridge components that are important for SPB assembly (Jaspersen and Winey 2004). Duplication of the ∼0.5-GDa SPB begins with formation of a SPB precursor, known as the satellite, at the distal tip of the half-bridge. Continued expansion of the satellite by addition of soluble precursors, and expansion of the half-bridge, leads to the formation of a duplication plaque. The SPB is then inserted into a pore in the NE, allowing for assembly of nuclear components to create duplicated side-by-side SPBs (Byers and Goetsch 1974; Byers and Goetsch 1975; Adams and Kilmartin 1999; Jaspersen and Winey 2004; Winey and Bloom 2012). The membrane anchors and half-bridge components both play a role in this SPB insertion step (Winey et al. 1991, 1993; Schramm et al. 2000; Araki et al. 2006; Sezen et al. 2009; Witkin et al. 2010; Friederichs et al. 2011; Kupke et al. 2011; Winey and Bloom 2012). Unlike NPC assembly, SPB duplication is spatially and temporally restricted. The new SPB is assembled during late G1-phase, approximately 100 nm from the preexisting SPB (Byers and Goetsch 1975). However, although the exact mechanism of SPB insertion is unknown, its insertion into the NE is thought to require a pore membrane similar to that found at the NPC.
Interestingly, previous studies have revealed physical and/or functional links between the factors required for NPC and SPB assembly and integrity. One of the SPB membrane anchors is Ndc1, a conserved integral membrane protein that is also an essential NPC Pom and required for NPC assembly (Chial et al. 1998; Mansfeld et al. 2006; Stavru et al. 2006; Kind et al. 2009). Some NPC components are required for proper remodeling of SPB core components and regulation of SPB size (Niepel et al. 2005; Greenland et al. 2010), whereas the loss of other NPC components rescues SPB mutant assembly phenotypes (Chial et al. 1998; Sezen et al. 2009; Witkin et al. 2010). The exact mechanism by which all of these NPC components influence SPB assembly is not known. With the relationships between NPC and SPB biogenesis, we examined S. cerevisiae cells lacking Rtn1 and Yop1 for altered SPB structure and function. Indeed, we found perturbations in SPB integrity and NE attachment that were rescued by Ndc1 overproduction. Physical and genetic data indicated that Ndc1 function at NPCs is specifically altered in rtn1 null (Δ) yop1Δ cells. We propose that these observations reflect the known dual requirement for Ndc1 in both NPC and SPB assembly and pinpoint a role for Rtn1 and Yop1 in Ndc1 function at the NPC. These results also further implicate the role of Ndc1 in a common NPC and SPB biogenesis step that potentially requires NE membrane remodeling events for pore formation and complex insertion.
Materials and Methods
Yeast strains and plasmids
All strains and plasmids used in this study are listed in Supporting Information, Table S1 and Table S2. Strains denoted with SWY are derived from the BY4741 and BY4742 S288C lineage, whereas SLJ strains are derivatives of W303. Unless otherwise noted, yeast genetic techniques were performed by standard procedures described previously (Sherman et al. 1986), and yeast were transformed by the lithium acetate method (Ito et al. 1983). All strains were cultured in either rich (YPD: 1% yeast extract, 2% peptone, and 2% dextrose) or complete synthetic minimal (CSM) media lacking appropriate amino acids and supplemented with 2% dextrose. Kanamycin resistance (conferred by the KANR gene) was selected on medium containing 200 µg/ml G418 (US Biological). Yeast were serially diluted and spotted onto YPD to assay fitness and temperature sensitivity as previously described (Tran et al. 2007).
The plasmids pSW3673, pSW3674, pSW3675, and pSW3676 were generated by subcloning genomic DNA fragments containing the coding sequence, promoter and 3′-UTR into the SacI and SacII sites of pRS425. For MPS2, a 2.5-kb genomic fragment was isolated by PCR amplification with Klentaq-LA (Sigma) using primers 5′-TCGACCGCGGTGGTGGAAGGTTTCCTTGAG-3′ and 5′-CGCATCTGAGCTGTAACATGACTCGAGTCGA-3′. A 2.2-kb BBP1 genomic fragment was amplified with 5′-TCGACCGCGGCGTGCGATACGCAAATAGAA-3′ and 5′-CGGGAATTACAGCTCGTGTTCTCGAGTCGA-3′ and inserted into SacI and SacII sites of pRS425 (Christianson et al. 1992). Likewise, APQ12 and BRR6 were isolated in 1.6-kb and 1.9-kb PCR fragments, respectively using the primers 5′-TCGACCGCGGCGAATCCGTCAACGAGTTTT-3′, 5′-CAATGCTGCTGCTGTTGTTTCTCGAGTCGA-3′ and 5′-TCGACCGCGGTTAAAGAGGCAGGGAGAGCA-3′, 5′- TCCACAAGTTGGAAGTGCATCTCGAGTCGA-3′.
The plasmid pSW3594 [for amino (N)-terminal tagging with GFP] was generated by subcloning the GFP coding sequence into pSW3447 at HindIII and SalI using the oligos 5′-GCATAAGCTTATGAGTAAAGGAGAAGAACTTTTCACT-3′ and 5′-GTACGTCGACgtTTTGTATAGTTCATCCATGCCATG-3′. The GFP-TUB3 integration cassette was generated by PCR from this plasmid using the oligonucleotides 5′-GATCAGGTATCTCATAAAGTACATTAATCGACTAAGCAAGCGACTTGAGACAATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTGTCCC-3′ and 5′-CCAGCATGCATTACCTATTTGACAACCTGCTTGACCAACATTAATACTAATGACCTCTCTAGTGGATCTGATATCACCTA-3′. Integration of the GFP–TUB3–HIS5 cassette and excision of the HIS5 marker sequence were accomplished as previously described (Terry and Wente 2007).
Cell cycle arrest
Wild-type and rtn1Δ yop1Δ cells were arrested at different stages in the cell cycle by the addition of hydroxyurea (HU) (Sigma), nocodazole (Sigma), or α-factor (ZymoResearch) at a concentration of 200 mM, 2.5 μg/ml, or 5 μg/ml, respectively as described (Jacobs et al. 1988). Arrest was observed as 95% population synchronization by phase contrast microscopy. For HU arrest, early log phase (OD 0.2) cultures of wild type (YOL183) and rtn1Δ yop1Δ (SWY3811) cells were arrested in YPD for 3 hr at 30°. For indirect immunofluorescence, cells were fixed in 3.7% formaldehyde for 1.5 hr at room temperature and processed as described (Strawn et al. 2004) with mouse anti-α-tubulin (clone DM1A, 1:200, Sigma). Bound antibodies were detected by incubation with Alexa Fluor 594-conjugated goat anti-mouse IgG (1:300, Molecular Probes). Samples were washed and mounted for imaging in 90% glycerol and 1 mg/ml p-phenylenediamine, pH 8.0. All images were taken on a confocal microscope (LSM 510; Carl Zeiss) with a 63 × Plan-Apochromat 1.4 NA oil immersion lens at a zoom of 4. Fluorescence was acquired using a 543-nm laser and an LP560-nm-long pass filter. Images were processed with ImageJ (National Institutes of Health; Abramoff et al. 2004) and Adobe Creative Suite 4 (Adobe).
For nocodazole release experiments, cells were grown to an OD600 of 0.15 in YPD with 1% DMSO at 23° and arrested for 3.5 hr. Cells were washed two times with cold CSM, suspended in room temperature CSM and plated onto small CSM agarose pads on VALAP sealed slides. To visualize spindles in live cells, endogenously expressed GFP–Tub3 was used. Since Tub3 is a minor component of microtubules, we reasoned that tagging TUB3 would be less detrimental to microtubule function than tagging TUB1. Live cell results using GFP–Tub3 were consistent with immunofluorescence results stained for Tub1 (data not shown). For time-lapse microscopy, Z stacks of bright field and direct GFP–Tub3 epifluorescence were taken for individual cells every 5 min using a microscope (BX50; Olympus) equipped with a motorized stage (Model 999000, Ludl), a UPlanF1 100× NA 1.30 oil immersion objective, and digital charge coupled device camera (Orca-R2; Hamamatsu). Images were collected and scaled using Nikon Elements and processed with ImageJ or Photoshop 12.0 software.
To monitor spindle dynamics following α-factor arrest, cells were grown to an OD600 of 0.15 at 30° in YPD, pH 3.9, and then arrested for 2 hr at 30°. Cells were washed twice with equal volumes of YPD, pH 6.5, suspended in fresh YPD equal to the original volume and incubated at 30°. At 15-min intervals, cell samples were fixed for indirect immunofluorescence as described (Stage-Zimmermann et al. 2000) and mounted on slides. Asynchronous cell populations expressing endogenous GFP–Tub3 were also imaged using a microscope (BX50; Olympus) equipped with a motorized stage (Model 999000, Ludl), a UPlanF1 100× NA 1.30 oil immersion objective, and digital charge coupled device camera (Orca-R2; Hamamatsu). Images were collected and scaled using Nikon Elements and processed with ImageJ or Photoshop 12.0 software. Images of cells were scored by bud index and position of SPB or spindle within the cell. Large budded cells were counted and scored as having separate GFP-positive foci in mother and daughter bud (postmitosis), GFP-positive foci in mother and daughter bud connected by GFP-positive spindle (anaphase spindle), or GFP-positive foci connected by spindle sequestered the mother bud (pre-anaphase spindle). Pre-anaphase spindles were considered misaligned if the closest SPB within the cell was greater than 1 µm from the bud neck, or greater than 60° different than the mother bud axis.
GFP–Tub1/Spc42–mCherry images were acquired with a 100× 1.4 NA oil objective on an inverted Zeiss 200m equipped with a Yokagawa CSU-10 spinning disc. For GFP and mCherry, respectively, 488-nm excitation and 568-nm excitation were used and emission was collected through BP 500- to 550-nm and BP 590- to 650-nm filters, respectively, onto a Hamamatsu EMCCD (C9000-13). For each channel, a Z-stack was acquired using 0.6- or 0.7-μm spacing. Thirteen total slices were acquired and a maximum projection image was created using ImageJ (NIH).
Hydroxyurea survival
To assay recovery from arrest at early S-phase, 200 mM HU was added to wild-type (YOL183) and rtn1Δ yop1Δ (SWY3811) cells at an OD of 0.15 in YPD with 1% DMSO. Cells were incubated for 6 hr at 30° and washed in ddH2O, and equivalent cell counts were plated onto YPD agar. Cell survival was calculated after 3 days’ growth at 30° by the percentage of colonies formed from HU-arrested cultures vs. those treated with DMSO alone.
Immunoprecipitation
Lysates from Ndc1–TAP cells were prepared from mid-log-phase cultures using a bead beater (Biospec) as described (Bolger et al. 2008). Solubilized fractions were added to 25 μl of packed IgG-coated sepharose beads and incubated for 4 hr at 4°. Proteins bound to the sepharose beads were washed in wash buffer (0.05% Tween, 150 mM NaCl, 50 mM Tris–HCL ph6.5), eluted by boiling in SDS sample buffer, resolved by SDS–PAGE, and detected with rabbit affinity purified anti-GFP IgG [a gift of M. Linder, Cornell University, Ithaca, NY (1:2000) and horseradish peroxidase-conjugated donkey anti-rabbit antibodies (1:5000, GE Healthcare)].
For Yop1–3xFLAG, liquid nitrogen ground lysates were prepared from 200 OD600 mid-log-phase cells as described (Jaspersen et al. 2006) and 40-µl anti-Flag resin (Sigma-Aldrich) was added. After overnight incubation at 4°, beads were washed five times at 4° and resuspended with loading buffer. Samples were analyzed by SDS–PAGE followed by immunoblotting. The following primary antibody dilutions were used: 1:1000 anti-HA 3F 10 (Roche) and 1:1000 anti-FLAG M2 (Sigma-Aldrich). Alkaline phosphatase-conjugated secondary antibodies were used at 1:10,000 (Promega).
Membrane yeast two-hybrid system
Bait and prey constructs were generated by amplifying SFII–SFII fragments and directionally inserted into the SFII site of pBT3N or pBT3–STE or pPR3N. Plasmids were cotransformed into SLJ5572 (Dualsystem Biotech NMY51). Transformants were spotted onto SD–LEU–TRP and SD–LEU–TRP–HIS–ADE plates and grown for 2–3 days at 30°.
Superplaque assay and thin-section electron microscopy
Myc–Spc42 localization and spindle morphology were analyzed by indirect immunofluorescence microscopy as described (Jaspersen et al. 2002). Cells were examined with a Zeiss Axioimager using a 100× Zeiss Plan-Fluar lens (NA 1.45), and images were captured with a Hamamatsu Orca-ER digital camera and processed using ImageJ (NIH). Superplaque formation was assayed by high pressure freezing and freeze substitution (HPF/FS) electron microscopy (EM) as described (Castillo et al. 2002). Samples were frozen on the Leica EM-Pact (Wetzlar, Germany) at ∼2050 bar and then transferred under liquid nitrogen into 2% osmium tetroxide/0.1% uranyl acetate/acetone and transferred to the Leica AFS (Wetzlar, Germany). The freeze substitution protocol was as follows: −90° for 16 hr, up 4°/hr for 7 hr, −60° for 19 hr, up 4°/hr for 10 hr, −20° for 20 hr. Samples were removed from the AFS and placed in the refrigerator for 4 hr and then allowed to incubate at room temperature for 1 hr. Samples went through three changes of acetone over 1 hr and were removed from the planchettes. They were embedded in acetone/Epon mixtures to final 100% Epon over several days in a stepwise procedure as described (McDonald 1999). Serial thin sections (60 nm) were cut on a Leica UC6 (Wetzlar, Germany), stained with uranyl acetate and Sato’s lead and imaged on a FEI Technai Spirit (Hillsboro, OR).
For thin-section EM (TEM) of SPBs, early log-phase cultures of parental (BY4724) and rtn1Δ yop1Δ yeast strains (SWY3811) grown in YPD were processed to preserve and stain dense protein and membrane structures as previously described (Dawson et al. 2009). Grids were examined on a CM-12 120-keV electron microscope (FEI, Hillsboro, OR). Images were acquired with an Advantage HR or MegaPlus ES 4.0 camera (Advanced Microscopy Techniques, Danvers, MA) and processed with ImageJ and Photoshop 12.0 software.
Results
Rtn1 and Yop1 are required for normal spindle pole body morphology
In S. cerevisiae lacking Rtn1 and Yop1, NPCs are clustered in a limited NE region and NPC assembly is altered (Dawson et al. 2009). Based on connections between SPB and NPC assembly (Chial et al. 1998; Adams and Kilmartin 1999; Jaspersen and Winey 2004; Sezen et al. 2009; Witkin et al. 2010), we speculated that the rtn1Δ yop1Δ mutant cells might have SPB perturbations. Using TEM, SPB morphology was assessed in rtn1Δ yop1Δ cells. In wild-type cells, SPBs were embedded in the NE with the documented laminar structure of central, inner, and outer plaques (Figure 1A). Nuclear microtubules organized from the inner plaque were also apparent. However, in the micrographs from rtn1Δ yop1Δ cells, the SPBs had strikingly altered morphology (Figure 1, B–E, and Figure S1). SPBs appeared to have unusually separated laminar structure with atypical plaque densities as well as peripheral lobular densities adjacent to the central plaque (Figure 1, B–C, and Figure S1). Of the 15 SPBs identified by this method, 12 exhibited this altered SPB morphology. As illustrated in Figure 1E, the aberrant SPB morphologies in the rtn1Δ yop1Δ cells were distinct from mutants with defects in SPB membrane components wherein the SPB structural perturbations typically include half bridge instability or an inability to insert the newly duplicated SPB into the NE, both of which result in a monopolar mitotic spindle (Jaspersen and Winey 2004). Moreover, to date, there are no reports of SPB structural alterations in other NPC clustering mutants (e.g., nup133Δ and nup120Δ); however, others have documented shorter spindles in nup120Δ cells (Aitchison et al. 1995).
Figure 1 .
SPBs have abnormal morphology and colocalize with NPC clusters in rtn1Δ yop1Δ cells. (A–D) Parental wild-type (A) or rtn1Δ yop1Δ (B–D) cells were grown to early log phase at 23° and processed for TEM. Scale bar, 100 nm. Arrowheads point to SPBs, arrows point to NPCs, stars indicate abnormal lobular structures on SPBs. (E) Scheme of SPBs from wild-type, SPB-insertion mutants, and rtn1Δ yop1Δ cells. cMTs, cytoplasmic microtubules; nMTs, nuclear microtubules; OP, outer plaque; IP, inner plaque; CP, central plaque; HB, half-bridge; DP, duplication plaque/uninserted SPB; L, lobular abnormalities. (F) Parental wild-type, rtn1Δ yop1Δ, nup133Δ, and nup120Δ cells expressing endogenously tagged Nic96–mCherry and Bbp1–GFP were grown to early log phase at 25°. Representative DIC and direct fluorescence microscopy images are shown. Scale bar, 2 μm. (G) Quantitative analysis of Bbp1–GFP and Nic96–mCherry colocalization. Cells were scored for presence of a Bbp1 foci within the Nic96 cluster (SWY4950, n = 882; SWY5033, n = 602; SWY4971, n = 571). Error bars represent standard error.
The rtn1Δ yop1Δ TEM micrographs also revealed a prevalence of NPCs clustering near the aberrant SPB structures (Figure 1C). Others have reported NPC localization near SPBs in the NE in both wild-type and NPC clustering strains (Heath et al. 1995; Winey et al. 1997; Adams and Kilmartin 1999; Schramm et al. 2000). To gain a further understanding of their distributions in the NE, colocalization of SPBs and NPC clusters was assayed in rtn1Δ yop1Δ cells. For direct comparison, the same analysis was conducted in nup133Δ and nup120Δ cells that also have clustered NPCs (Heath et al. 1995; Pemberton et al. 1995). Strains expressing chromosomally integrated BBP1–GFP (encoding a SPB component; Schramm et al. 2000) and NIC96–mCherry (encoding a Nup; Grandi et al. 1993) were analyzed by direct fluorescence microscopy (Figure 1F). As determined by the association of Bbp1–GFP foci with a Nic96–mCherry cluster, the SPBs localized coincident with NPC clusters at a frequency of 57.2 and 48.8%, respectively, for the nup133Δ and nup120Δ cells. In wild-type cells NPCs do not cluster and the Bbp1–GFP foci were found on the Nic96–mCherry-labeled NE rim. Strikingly, in rtn1Δ yop1Δ cells, the colocalization of NPC clusters with SPBs increased significantly to 86.0% of cells (Figure 1G). Taken together, the rtn1Δ yop1Δ mutant resulted in both SPB morphology defects that were distinct from other known NPC clustering mutants and an increased coincidence of NPC clusters near SPBs.
Since SPBs were associated with NPC clusters in 57.2% of nup133Δ cells, we speculated that this mutant could be used to determine if Rtn1 is enriched at SPBs. For this, nup133Δ RTN1–GFP cells expressing SPC42–MCHERRY (encoding a SPB component) were analyzed by direct fluorescence confocal microscopy (Figure S2). In cells where the Spc42–mCherry foci were clearly distinct from the Rtn1–GFP/NPC cluster, no coincident Rtn1–GFP intensity was observed at the Spc42–mCherry foci. Although this did not eliminate the possibility that Rtn1 and Yop1 colocalize with SPBs, it suggests that any association is below the detection limit of this method.
SPB superplaques in rtn1Δ yop1Δ cells are unstable in the NE
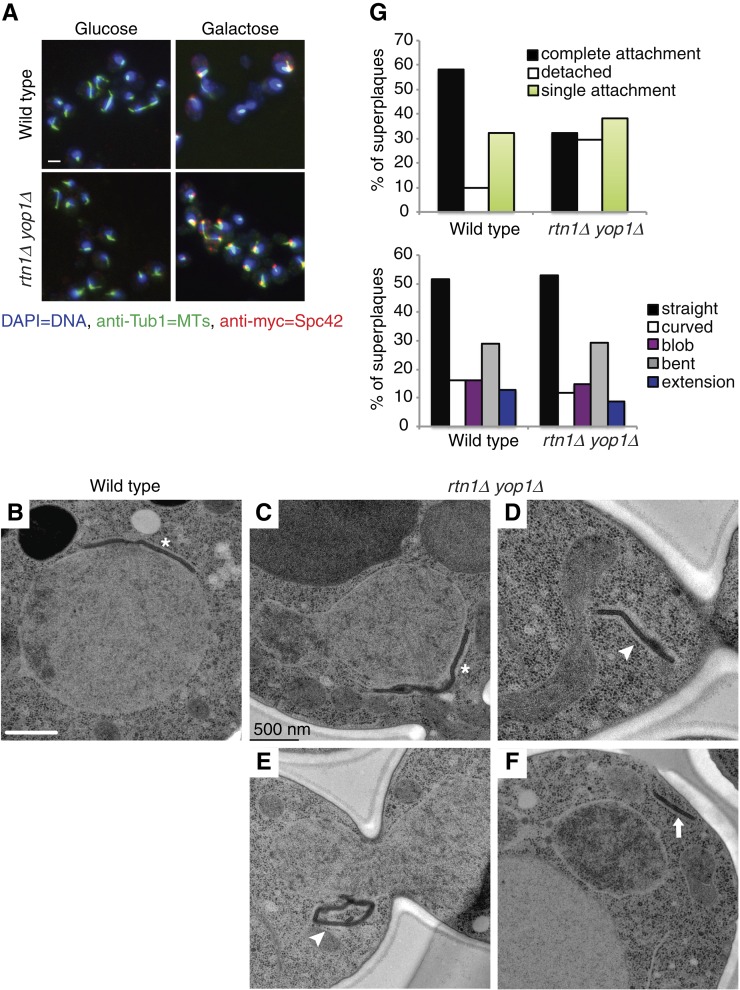
When the SPB component Spc42 is overproduced, the excess protein is incorporated into the central plaque of the SPB. This results in a lateral expansion of the SPB to form a structure termed the superplaque (Donaldson and Kilmartin 1996). Others have found that many of the same molecular and regulatory events required for SPB duplication are also required for superplaque formation (Donaldson and Kilmartin 1996; Castillo et al. 2002; Jaspersen and Winey 2004). To further test SPB structural integrity and connections of the SPB to the NE, we examined the ability of rtn1Δ yop1Δ cells to stably maintain superplaque attachment. Using a galactose-inducible myc-SPC42, superplaque formation was induced in wild-type and rtn1Δ yop1Δ cells. By indirect immunofluorescence, as compared to superplaques in wild-type cells, the rtn1Δ yop1Δ superplaques were more variable in size. In addition, an increased proportion was extended away from the microtubules and DNA (Figure 2A). Examination of superplaques by TEM revealed that 29% of the rtn1Δ yop1Δ superplaques were completely disconnected from the NE, compared to 10% in wild-type cells (Figure 2, B–G). Interestingly, the overall laminar structure of the superplaques in rtn1Δ yop1Δ cells was not significantly altered, with >50% of these structures showing a straight-layered structure similar to the SPB central plaque (Figure 2, B–G). These data suggested that Rtn1 and Yop1 play a role in stable attachment of SPB structures to the NE.
Figure 2 .
Deletion of reticulons affects superplaque formation. Parental (SLJ1433) and rtn1Δ yop1Δ (SLJ3828) were grown overnight in YEP + 2% raffinose at 30° until they were in early log phase then divided into two cultures. To one culture, glucose was added to a final concentration of 2% while the other was treated with 2% galactose to induce expression of myc-SPC42. After 4 hr of continued growth at 30°, cultures where harvested and examined by indirect immunofluorescence microscopy and by EM. (A) Microtubules (green) and myc-Spc42 (red) were labeled using anti-Tub1 and anti-myc antibodies, respectively. DNA (blue) was visualized using DAPI. Only when galactose was added were Spc42 plaques observed. Bar, 5 µm. (B–F) Superplaque structures in parental (B) and rtn1Δ yop1Δ (C–F) were further examined by EM and characterized by shape and attachment to the NE. Asterisks indicate SPB superplaques with complete attachment, arrowheads at superplaques with single attachment, and arrows at superplaques completely detached from nucleus. Scale bar, 500 nm. (G) Superplaque structures were quantified in 31 wildtype and 34 rtn1Δ yop1Δ nuclei.
Cells lacking Rtn1 and Yop1 have defects in the mitotic spindle
The observation that SPB morphology is altered in rtn1Δ yop1Δ cells indicated that SPB function might also be impaired. To assay SPB function, we used a variety of cellular arrest factors to examine SPBs and spindles at distinct stages in the cell cycle. SPB remodeling occurs throughout the cell cycle, starting with duplication of a new SPB in late G1-phase and then growth of the SPB core through exchange of subunits in S-phase and G2/M. SPB size decreases as cells exit mitosis, presumably through the removal of core subunits (Byers and Goetsch 1975; Yoder et al. 2003). Therefore, SPBs in wild-type cells arrested with HU or nocodazole in S-phase or G2/M, respectively, undergo a lateral expansion and increase the overall size. In contrast, the SPBs in wild-type cells arrested in G1-phase using α-factor are contracted in size.
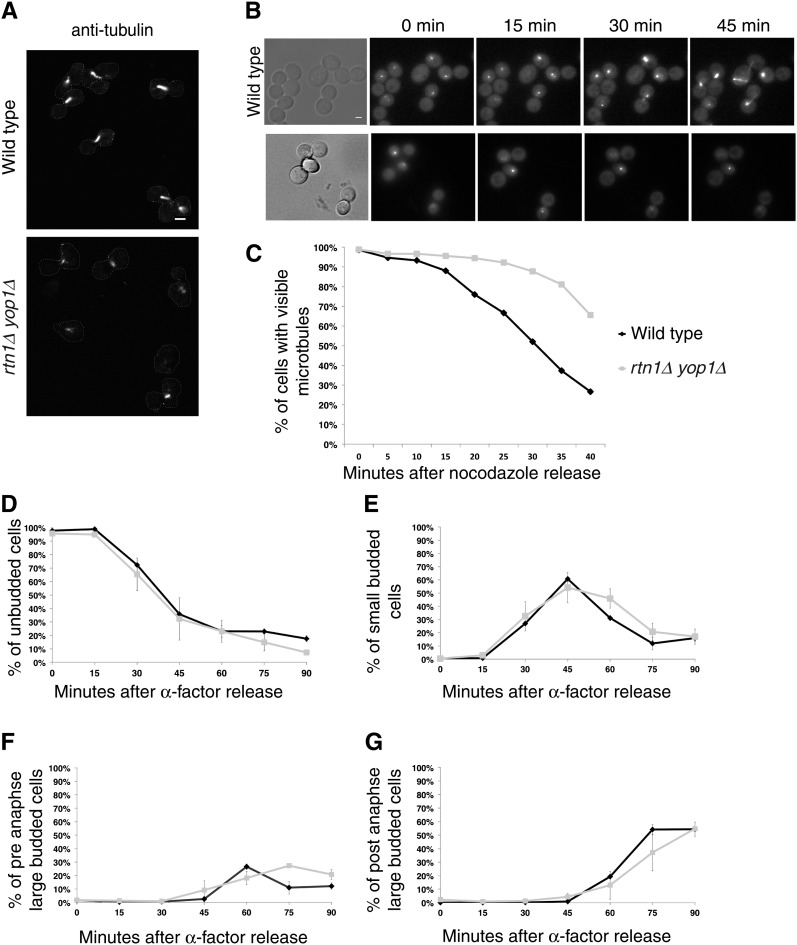
Microtubule structure of wild-type and rtn1Δ yop1Δ cells in arrested and released cells was observed using indirect immunofluorescence for anti-α-tubulin or direct fluorescence microscopy of GFP–Tub3 to determine if there were defects in the microtubule cytoskeleton. As reported (Miller and Rose 1998), in wild-type cells with α-factor treatment, the late G1 arrest point in wild-type cells was characterized by frequent alignment of the SPB with the shmoo extension and astral microtubules that extend into the shmoo. However, the α-factor arrested microtubules of rtn1Δ yop1Δ cells appeared to have a minor spindle positioning defect (Table 1). SPBs were more frequently misoriented away from the shmoo in rtn1Δ yop1Δ cells compared to wild type, 12.6 and 7.4%, respectively. This suggests a possible impairment of cytoplasmic microtubules. Further analysis of this phenotype by treatment of cells with HU, which results in a S-phase arrest in wild-type cells with a short bar-like spindle positioned at the bud neck, revealed additional defects in rtn1Δ yop1Δ cells (Figure 3A). A single bright focus of GFP–Tub3 fluorescence was observed in the mother cells of HU-arrested rtn1Δ yop1Δ cells (Figure 3A), suggesting that loss of RTN1 and YOP1 function is associated not only with a defect in nucleation of cytoplasmic microtubules needed for spindle positioning but also with a defect in the formation of a bipolar spindle. Furthermore, prolonging HU treatment of rtn1Δ yop1Δ cells for up to 6 hr did not increase the percentage of cells with wild-type short spindles (data not shown).
Table 1 . rtn1Δ yop1Δ cells have mild SPB positioning defects upon α-factor arrest.
| Wild type | rtn1Δ yop1Δ | |
|---|---|---|
| Microtubules positioned in shmoo | 335 (92.6%) | 384 (87.3%) |
| Microtubules positioned away from shmoo | 27 (7.4%) | 56 (12.6%) |
| Total | 362 | 440 |
Parental (YOL183) or rtn1Δ yop1Δ (SWY3811) cells expressing GFP-Tub3 arrested with α-factor. Cells were fixed to preserve GFP fluorescence and imaged and scored based on proximity of SPB and microtubules to the shmoo; P-value= 0.00012.
Figure 3 .
Mitotic arrest leads to collapsed spindles and reduced microtubule function in rtn1Δ yop1Δ cells. (A) Microtubules in parental wild-type (YOL183) or rtn1Δ yop1Δ (SWY3811) cells arrested with 200 mM HU were detected by indirect anti-tubulin immunofluorescence and laser scanning confocal microscopy. Scale Bar, 2 μm. (B) Direct fluorescence of GFP–Tub3 was visualized following nocodazole or α-factor arrest in GFP–Tub3 (SWY4617) or rtn1Δ yop1Δ GFP–Tub3 (SW4935) cells. Scale bar, 2 μm. (C) Time-lapse images were scored for release from nocodazole arrest as the percentage of cells exhibiting of microtubule re-polymerization. (D, E, F, and G) Time-lapse images were scored for release from α-factor arrest based on bud index and position of SPBs within the cells.
To determine if rtn1Δ yop1Δ mutants have a defect in spindle formation, we treated cells with nocodazole, which inhibits spindle formation, and assessed the ability of the spindle to repolymerize following removal of the nocodazole. Wild-type and rtn1Δ yop1Δ GFP-Tub3 cells were arrested in G2/M with nocodazole. Time-course imaging on agarose pads was conducted of individual cells following release. Wild-type cells showed repolymerization of microtubules by 15 min after nocodazole washout. However, repolymerization in rtn1Δ yop1Δ cells was delayed until ∼30 min (Figure 3, B and C). This significant delay in rtn1Δ yop1Δ cells was not due to growth defects since release from α-factor arrest was not delayed in rtn1Δ yop1Δ cells compared to wild type (Figure 3, D–G). We concluded that rtn1Δ yop1Δ cells have altered microtubule dynamics.
Because cytoplasmic microtubules are critical for spindle positioning along the mother–daughter axis, we speculated that rtn1Δ yop1Δ cells were defective in nucleation or maintenance of cytoplasmic microtubules (Hoepfner et al. 2002; Moore et al. 2009; Winey and Bloom 2012). To further analyze the microtubules of rtn1yop1Δ, we imaged cells expressing GFP–Tub1 and Tub4–mCherry by live-cell microscopy. The GFP–Tub1 localization results were consistent with the GFP–Tub3 data; however, the cytoplasmic microtubules were more easily observed with GFP–Tub1 (Figure 4A). From these images, we found that short spindles nucleated cytoplasmic microtubules that went toward the bud. Strikingly, as the spindles elongated, cytoplasmic microtubules were present less frequently in the rtn1Δ yop1Δ cells (52.4% compared to 83.7% in wild type). To determine if rtn1Δ yop1Δ cells were deficient in cytoplasmic microtubules nucleation, TEM micrographs of cells under HPF/FS conditions were analyzed. Similar to our other TEM observations (Figure 1, B–D), rtn1Δ yop1Δ SPBs were frequently flanked by NPCs (12 of 17) and associated with some type of detached NE structure (12 of 17) (Figure 4, B and C). Also, rtn1Δ yop1Δ SPBs often lacked visible cytoplasmic microtubules (8 of 17) compared to wild type (1 of 10); however, all were associated with nuclear microtubules. Taken together, we concluded that rtn1Δ yop1Δ cells have defects in nuclear positioning caused by insufficient cytoplasmic microtubules.
Figure 4 .
rtn1Δ yop1Δ cells have defects in cytoplasmic microtubules. (A) Asynchronous cultures of parental wild-type (SLJ3996) or rtn1Δ yop1Δ (SLJ3994) cells expressing GFP–Tub1 and Tub4–mCherry were grown to early log phase and imaged. Cells were analyzed for the presence or absence of cytoplasmic microtubules and length of spindles. Arrows point to duplicated SPBs in large budded cells. Single asterisk indicates a cell with duplicated poles and cytoplasmic microtubules that go toward bud and mother. The double asterisk indicates a cell with spindle elongation in the mother. (B and C) Asynchronous rtn1Δ yop1Δ cells were processed by HPF/FS and imaged by EM. Black arrows point to SPBs. Asterisk indicates NPC in close proximity to SPB. Arrowheads point to nuclear and cytoplasmic microtubules. White arrows point to electron-dense structure present in the nucleoplasm associated with nuclear microtubules (B) and to an electron dense structure resembling the satellite (C). Scale bar, 100 nm.
Rtn1 and Yop1 affect proper spindle function
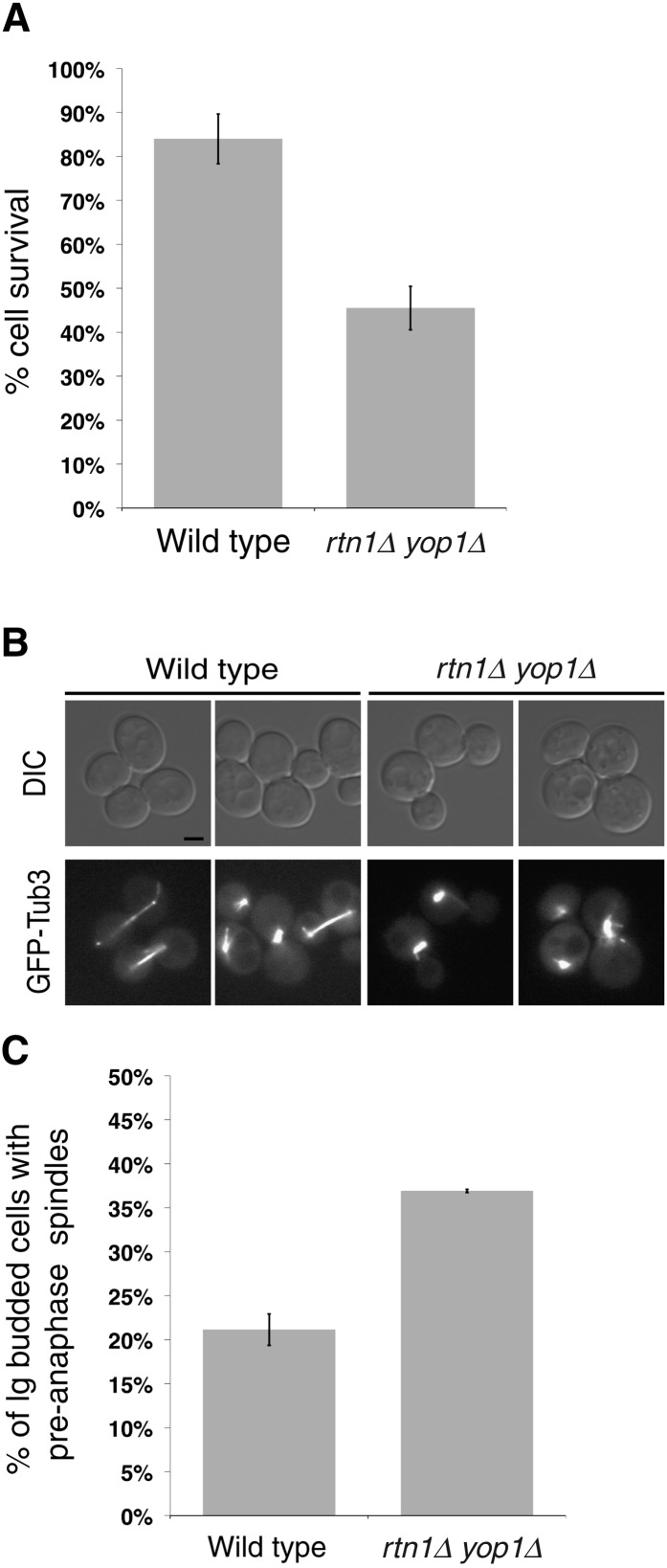
Since rtn1Δ yop1Δ cells exhibit spindle defects during HU arrest and following release from G2/M, cell-viability assays were performed to determine if these defects in spindle morphology result in compromised spindle function, chromosome segregation errors, and ultimately cell death. The rtn1Δ yop1Δ cells were arrested with HU for 6 hr, released into the cell cycle, and then plated on YPD plates. Compared to wild type, rtn1Δ yop1Δ cells had 50% reduced viability after HU treatment (Figure 5A). Overall, these results suggested that when arrested in S-phase, rtn1Δ yop1Δ cells are vulnerable to reduced spindle integrity, resulting in increased cell death.
Figure 5 .
rtn1Δ yop1Δ cells exhibit functional defects in spindle positioning. (A) Parental wild-type (YOL183) and rtn1Δ yop1Δ (SWY3811) cells were arrested with 200 mM HU. Cell viability following HU arrest was measured by colony formation after 3 days growth. (B) Live-cell direct fluorescence microscopy was conducted with GFP–Tub3 and rtn1Δ yop1Δ GFP–Tub3 cells grown to early log phase at 23°. Scale bar, 2 μm. (C) Bud index was scored in DIC images of parental GFP–Tub3 (SWY4616, n = 423) and rtn1Δ yop1Δ GFP–Tub3 (SWY4877, n = 750).
We also speculated that rtn1Δ yop1Δ cells would exhibit defects in SPB function in untreated cells. GFP–Tub3 was used to observe the spindles in an asynchronously growing population of rtn1Δ yop1Δ cells. There was no increase in the number of rtn1Δ yop1Δ cells with extra SPBs or evidence of nonfunctional SPBs that did not nucleate microtubules (Figure 4B and data not shown). However, the overall rtn1Δ yop1Δ population harbored an increase in large budded cells with pre-anaphase spindles (spindles of <2 μm) (Figure 5, B and C). Furthermore, when compared to wild type, the pre-anaphase spindles in rtn1Δ yop1Δ cells were more frequently misaligned within the mother bud (Figure 6B). Thus, rtn1Δ yop1Δ cells exhibited poor spindle function in asynchronous cells, likely due to reduced SPB integrity and the defects in the cytoplasmic microtubules.
Figure 6 .
Overexpression of SPB insertion factors rescues rtn1Δ yop1Δ defect. Parental wild-type GFP–Tub3 and rtn1Δ yop1Δ GFP–Tub3 cells transformed with plasmids expressing NDC1, RTN1, POM152, BBP1, MPS2, or empty vector were grown to midlog phase at 30° and visualized by live-cell direct fluorescence microscopy. (A) Cells were scored for bud index by quantification of DIC images and cell-cycle position by spindle stage (parental + pRS315, n = 1251; + pRS425; n = 1483; SWY4877 + pRS315, n = 409; +pRSS425; n = 2372; + pNDC1; n = 2073; + pRTN1, n = 2095; + pPOM15; n = 904; + pBBP1, n = 792; + pMPS2, n = 2475). (B) Large budded cells with pre-anaphase spindles were further characterized by orientation of their spindle. Error bars indicate standard error. The asterisk and double asterisk denotes statistical significance (P-value < 0.04, P-value <0.01, respectively).
Overexpression of SPB insertion factors specifically rescues rtn1Δ yop1Δ spindle defects
Previously, we demonstrated that NPC clustering in the rtn1Δ yop1Δ cells is rescued by the overexpression of NDC1 or POM152 (Dawson et al. 2009). Pom152 and Ndc1 interact in a complex in the NPC, and they have partially overlapping roles in NPC assembly (Madrid et al. 2006). To determine if altered NPC assembly/function was indirectly affecting SPBs, the shortened misaligned spindles phenotype was assessed by live-cell microscopy in rtn1Δ yop1Δ GFP–TUB3 cells overexpressing NDC1 or POM152. Compared to empty vector, overexpression of NDC1 rescued both of the SPB defects observed in rtn1Δ yop1Δ cells, as reflected by reduced numbers of large budded cells with short spindles (Figure 6A) and wild-type levels of properly oriented pre-anaphase spindles (Figure 6B). In contrast, overexpression of POM152 did not have the same effect on spindle defects in rtn1Δ yop1Δ cells (Figure 6, A and B), and the decrease in the average percentage of short or misaligned spindles was not significant (P-values of 0.20 and 0.13, respectively).
Since overexpression of POM152 inhibits wild-type cell growth (Wozniak et al. 1994), it is of note that decreased growth rate was not observed in rtn1Δ yop1Δ cells (Figure S3). Importantly, overexpression of NDC1 rescued the mild growth defect of rtn1Δ yop1Δ cells whereas POM152 overexpression did not (Figure S3), suggesting that the compromised growth of rtn1Δ yop1Δ cells reflects the reduced fidelity of SPB function. Overall, overexpression of either NDC1 or POM152 rescued NPC clustering in rtn1Δ yop1Δ cells (Dawson et al. 2009); however, only NDC1 overexpression rescued the rtn1Δ yop1Δ spindle defect. Thus, simply rescuing the NPC clustering defect did not rescue the SPB defect, suggesting the rtn1Δ yop1Δ effect was not an indirect overall NPC perturbation impact.
Proper targeting of Ndc1 to SPBs occurs by its association with other SPB insertion factors at the NE (Winey et al. 1991; Schramm et al. 2000; Kupke et al. 2011). Bbp1 and Mps2 are SPB-specific proteins that interact with Ndc1 and play roles in SPB insertion and stability (Winey et al. 1991; Muñoz-Centeno et al. 1999; Schramm et al. 2000). We hypothesized that overexpressing BBP1 or MPS2 would rescue the rtn1Δ yop1Δ spindle defects but not the NPC clustering defect. By examining GFP–Tub3, we found that SPB defects were rescued in rtn1Δ yop1Δ cells overexpressing BBP1 or MPS2 (Figure 6, A and B). For BBP1 overexpression, the numbers of large budded cells that had not completed mitosis (31% vs. 50% for rtn1Δ yop1Δ alone) and the proportion with misoriented anaphase spindles (17% vs. 28% for rtn1Δ yop1Δ alone) were clearly reduced. Likewise, in the population of cells overexpressing MPS2, there were fewer large budded cells that had not completed mitosis (34%) and a lower proportion with misoriented anaphase spindles (13%). Indeed, the spindle defect rescue levels in the BBP1 and MPS2 experiments were similar to that found with overexpressing NDC1. However, NPC clusters were still present in rtn1Δ yop1Δ cells overexpressing BBP1 or MPS2 (data not shown). Thus, rescue of the rtn1Δ yop1Δ spindle defects by overexpression of SPB anchoring components was specific. These results indicated that the NPC and SPB defects are separable and both potentially the result of defects or insufficiencies in NE membrane proteins.
We speculated that the underlying cause for the rtn1Δ yop1Δ mutant phenotypes might be a perturbation in the function of shared SPB and NPC component(s). Ndc1 has roles at both SPBs and NPCs (Winey et al. 1993; Chial et al. 1998; Lau et al. 2004). Two other NE membrane proteins, Brr6 and Apq12, have also been linked to both NPC biogenesis and SPB insertion (Scarcelli et al. 2007; Hodge et al. 2010; Schneiter and Cole 2010; Tamm et al. 2011). To test for specificity, BRR6 and APQ12 overexpression was analyzed. Overproduction of neither Brr6 nor Apq12 altered the SPB or NPC defects in rtn1Δ yop1Δ cells (data not shown). Thus, the rtn1Δ yop1Δ cells had NPC and SPB defects that are separate from the lipid homeostasis defects and membrane fluidity function associated with BRR6 and APQ12. Moreover, NDC1 overexpression was unique in rescuing both the SPB and NPC defects.
High osmolarity reduces NPC clustering but not spindle defects of rtn1Δ yop1Δ cells
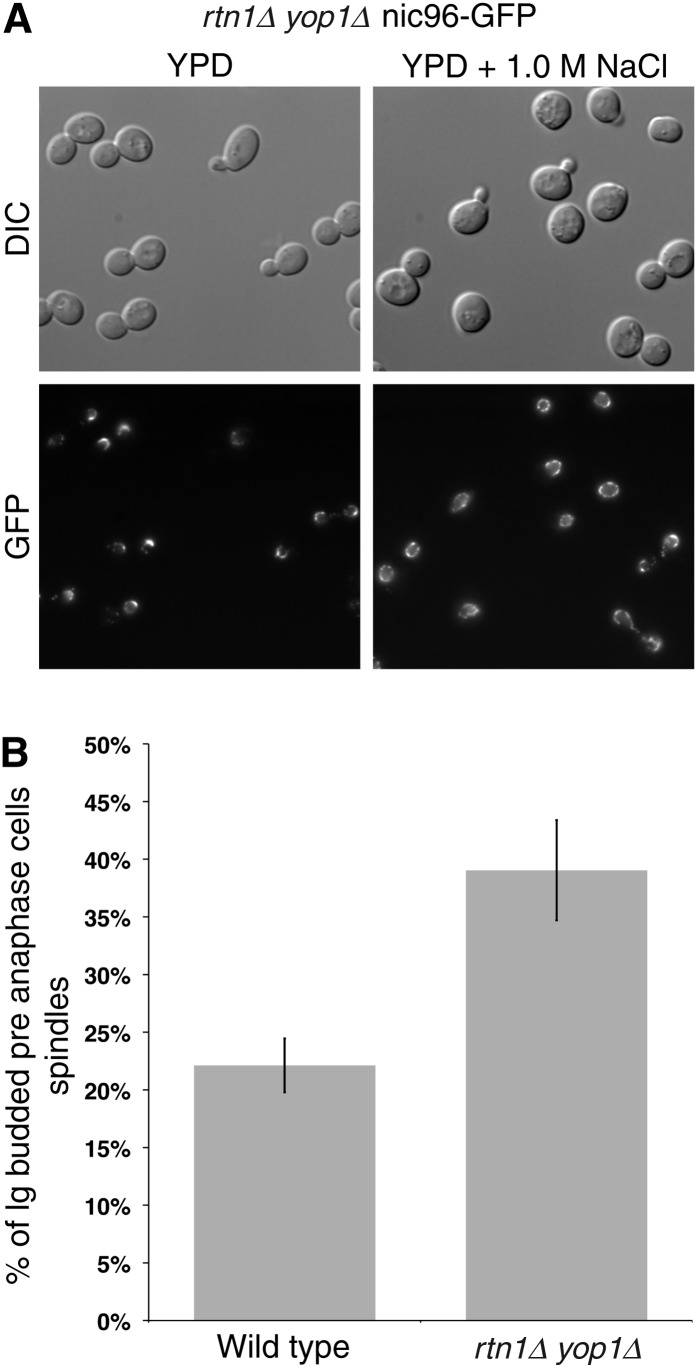
To further test the functional separation of NPC and SPB defects in cells, experiments were conducted after growth of cells in high osmolarity media (1 M NaCl). Strikingly, the percentage of rtn1Δ yop1Δ cells with distinct NPC clusters was reduced in high osmolarity media from 71 to 22% (Figure 7A). This differed from a previous report for the nup120Δ clustering mutant wherein high osmolarity rescues growth and nucleocytoplasmic transport defects but not NPC clustering (Heath et al. 1995). However, while growth of rtn1Δ yop1Δ cells in high osmolarity (1 M NaCl) rescued NPC clustering, it did not rescue the observed SPB defects (Figure 7B). These results again highlighted differential NPC and SPB effects in the rtn1Δ yop1Δ cells. Previous work has shown that high osmolarity results in increased RTN2 expression, which could compensate for the loss of Rtn1 and Yop1 at NPCs (De Craene et al. 2006; Romero-Santacreu et al. 2009).
Figure 7 .
Growth in high osmolarity only reduces NPC clusters in rtn1Δ yop1Δ cells. (A) Asynchronous cultures of rtn1Δ yop1Δ nic96–GFP cells (SWY4725) were grown to log phase at 23° in YPD. After shifting to YPD alone (control) or YPD + 1.0 M NaCl, cells were grown at 23° for an additional 5 hr and imaged. (B) Asynchronous cultures of parental and rtn1Δ yop1Δ cells endogenously expressing GFP–TUB3 (SWY4616 and SWY4877, respectively) were grown to log phase at 23° in YPD. After shifting to YPD + 1.0 M NaCl, cells were grown at 23° for an additional 5 hr and imaged. Cells were scored for bud index by quantification of DIC images and cell-cycle position by spindle stage (SWY4616, n = 171; SWY4877, n = 233). P-value = 0.041.
Rtn1 and Yop1 interact with Ndc1
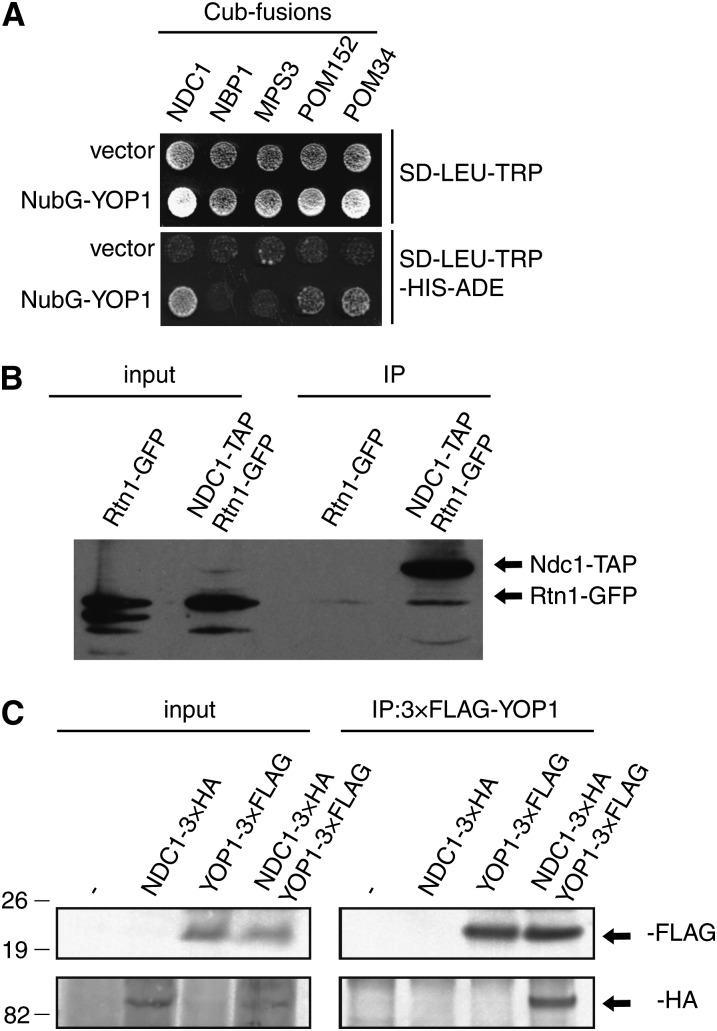
Based on the genetic and functional connections, we investigated whether Rtn1 and/or Yop1 physically interact with integral membrane proteins of the NPC and/or SPB. Rtn1 and Yop1 interact by co-immunoprecipitation (Voeltz et al. 2006). Furthermore, based on a published large-scale split ubiquitin-based two hybrid screen, Yop1 interacts with both Pom33 and Pom34 (Miller et al. 2005). Using the split ubiquitin two-hybrid assay, we used a candidate approach to identify other possible Yop1 interaction partners. Remarkably, Pom34, Pom152, and Ndc1 were all positive for interaction with Yop1. However, Yop1 did not interact with either Nbp1 or Mps3, two proteins involved in SPB insertion, using this system (Figure 8A) (Araki et al. 2006; Friederichs et al. 2011).
Figure 8 .
Rtn1 and Yop1 interact with Ndc1 and NPC components. (A) Split ubiquitin yeast two-hybrid vectors containing a LEU2 marker and the C-terminal region of ubiquitin (Cub) fused to NDC1, NBP1, MPS3, POM152, or POM34 (baits) were expressed in SLJ5572 and tested for their ability to interact with the N-terminal region of ubiquitin (NubG) fused to Yop1 or the N-terminal region of ubiquitin alone in a TRP1 vector (preys). Interaction of bait and prey proteins leads to cleavage of the split ubiquitin and release of a transcription factor, which activates reporter genes such as HIS3 and ADE2. (B) Lysates were prepared from wild-type, Ndc1–TAP Rtn1–GFP, and Rtn1–GFP cells and immunoprecipitated with IgG-coated sepharose beads. Analysis of cell lysates and immunoprecipitated proteins by western blotting with anti-GFP antibodies showed that Ndc1–TAP binds to Rtn1–GFP. (C) Lysates were prepared from wild-type, Ndc1–3xHA, Yop1–3xFLAG, and Ndc1–3xHA Yop1–3xFLAG cells and immunoprecipitated with anti-FLAG antibodies. Analysis of cell lysates and immunoprecipitated proteins by immunoblotting with anti-FLAG and anti-HA antibodies showed that Ndc1–3xHA binds to Yop1–3xFLAG. Positions of molecular mass markers (kilodaltons) are indicated to the left.
Using immunoprecipitation assays, we further examined the interaction between Ndc1 and Rtn1. Lysates of yeast cells exogenously expressing NDC1–TAP and RTN1–GFP were incubated with IgG-sepharose beads. By immunoblotting analysis, Rtn1–GFP was co-isolated with Ndc1–TAP (Figure 8B). Similarly, lysates of yeast cells exogenously expressing Ndc1–3xHA and Yop1–3XFLAG were incubated anti-FLAG affinity matrix and bound samples were analyzed by immunoblotting. As shown, Yop1–3xFLAG and Ndc1–3xHA were co-isolated (Figure 8C). Overall, these data showed that Rtn1 and Yop1 physically interact with Ndc1 and other membrane components of the NPC.
Discussion
Previously, we defined a role for Rtn1 and Yop1 in nuclear pore and NPC biogenesis (Dawson et al. 2009). Building on this, here we demonstrate novel functions of Rtn1 and Yop1 at the NE by discovering links to SPB morphology and microtubule dynamics. We conclude that the lack of Rtn1 and Yop1 perturbs Ndc1 function, an essential factor required for both SPB and NPC assembly. This is based on a complementary set of genetic, cell biological, and biochemical data. We find that rtn1Δ yop1Δ cells have structural and functional defects in SPBs, in the SPB-associated microtubule spindles and cytoplasmic microtubules, and in SPB superplaque formation. Overproduction of either Ndc1 or components involved in anchoring the SPB to the NE rescues the SPB defects in rtn1Δ yop1Δ cells. Furthermore, although increasing Ndc1 levels also rescues the NPC defects in rtn1Δ yop1Δ cells, overproducing NPC-specific or SPB-specific components rescues the defects only in their respective complex. Interestingly, Rtn1 and/or Yop1 physically interact with Ndc1. We conclude that Rtn1 and Yop1 facilitate proper Ndc1 function in the NE at NPCs and SPBs.
Together with our prior work, rtn1Δ yop1Δ mutants have clear defects in the structure of both NPCs and SPBs. In addition to the NPC clusters, the NE in rtn1Δ yop1Δ cells also has partial NPC-like structures present on only the INM or ONM surface (Dawson et al. 2009). Interestingly, the aberrant lobular SPB structures in rtn1yop1Δ cells are not similar to other reported SPB morphological defects (Figure 1). The rtn1Δ yop1Δ mutant cells also have altered spindle function, indicative of defects in SPB migration due to insufficient or defective cytoplasmic microtubules (Figures 3, 4, and 5). Although gross defects in insertion, such as monopolar spindles, are not observed, our data do suggest that the connections of the SPB to the NE are altered. Upon SPC42 overexpression, a greater proportion of the superplaques in rtn1Δ yop1Δ cells are partially or fully disconnected from the NE (Figure 2). We speculate that both the NPC and SPB defects in rtn1Δ yop1Δ cells reflect decreased stability of the respective structure/complex in the NE.
Ndc1 is to date the only known factor common to both NPCs and SPBs. Based on the work here, we propose that Rtn1 and Yop1 are also common effectors of both NPCs and SPBs. We have previously shown that Rtn1 and Yop1 colocalize to NPC clusters in nup133Δ cells (Dawson et al. 2009); however, there is no evidence of physical association of Rtn1 and Yop1 with SPBs. General changes to the lipid and protein composition of the NE are one of several possibilities by which the absence of Rtn1 and Yop1 could affect NPC and SPB stability. Alternatively, several pieces of evidence indicate that the rtn1Δ yop1Δ effect is directly perturbing NPCs and/or SPBs. The SPB is associated with the NPC clusters in rtn1Δ yop1Δ cells to a greater extent than it is in other NPC clustering mutants nup133Δ and nup120Δ (Figure 1, F and G). Furthermore, the gene specificity in the overexpression suppression analysis is intriguing and indicates that the rtn1yop1Δ defects are possibly not due to a general perturbation in NPC or the NE. Overexpression of POM152 rescues the NPC clustering defect but does not rescue the SPB defects in rtn1Δ yop1Δ mutants. Likewise, overexpression of MPS2 or BBP1 results in rescue of spindle defects, but not NPC clustering. Interestingly, these multicopy suppressors of the rtn1Δ yop1Δ phenotypes are physical or genetic interactors of Ndc1/NDC1. Moreover, elevated Ndc1 levels rescue both the SPB and NPC defects in the rtn1Δ yop1Δ mutant. Based on these genetic data and the physical interaction between Ndc1 and Rtn1/Yop1, we speculate that Ndc1 function is potentially controlled by Rtn1 and/or Yop1.
Others have provided key data supporting a role for Rtns and Yop1/DP1 in stabilizing membrane curvature. Lipid reconstitution assays in the presence of purified Yop1 result in the formation of stable membrane tubules (Hu et al. 2008), and in rtn1Δ rtn2Δ yop1Δ cells the ER structure is specifically altered (West et al. 2011). However, whereas all tubular ER is dramatically altered in rtn1Δ rtn2Δ yop1Δ cells, the overall structural properties of the NE are not altered. We speculate that the rtn1Δ yop1Δ defects in NPCs and SPBs are due to highly localized or highly temporal defects in stabilizing membrane structures at NPCs and/or SPBs. Moreover, the Rtns and Yop1/DP1 could serve to facilitate the function of other proteins directly involved in the respective membrane association of NPCs and SPBs (see below). During NPC assembly, both positive and negative membrane curvature are predicted to occur for the INM and ONM to fuse (Antonin 2009). The Rtns and Yop1/DP1 are proposed to function in the NE and stabilize the highly curved nuclear pore membrane during these early NPC biogenesis steps (Dawson et al. 2009). The physical interactions between Rtn1 and Yop1 with Ndc1 (Figure 5, B and C) and other membrane components of the NPC (Figure 5A and Chadrin et al. 2010) provide a plausible mechanism by which these proteins might be colocalized/recruited to nuclear pore membranes.
Our working model for how Rtn1 and/or Yop1 mediate NPC biogenesis extends directly to two alternative scenarios for how Rtn1 and/or Yop1 might affect SPB assembly. SPBs also require membrane curvature maintenance, with specific membrane changes required during SPB duplication and migration. First, it is possible that Rtn1 and Yop1 function with Ndc1 at both NPCs and SPBs. Loss of Rtn1 and Yop1 might result in the need for increased levels of Ndc1 at both complexes to allow proper function. As such, both NPCs and SPBs are defective or not correctly assembled without additional Ndc1. Second, alternatively, it is possible that Rtn1 and Yop1 function with Ndc1 only at the NPC. In this case, in the absence of Rtn1 and Yop1, increased levels of Ndc1 are sequestered by NPCs and potentially titrated away from SPBs. It is possible that overexpression of MPS2 or BBP1 rescues the SPB in rtn1Δ yop1Δ cells due to Mps2 and Bbp1 having overlapping functions with Ndc1 at the SPB or due to physical interactions between these proteins resulting in Ndc1 being more efficiently targeted away from the NPC to the SPB. This second model places NPC and SPB assembly as acting antagonistically in terms of Ndc1 function.
It has been previously suggested that a feedback mechanism exists in response to defects in SPB duplication, with this resulting in antagonistic roles of the NPC and SPB complexes (Witkin et al. 2010). Many SPB assembly mutants, including ndc1-1 and mps2-1, are suppressed by specific deletions in genes encoding NPC components (Chial et al. 1998; Sezen et al. 2009; Witkin et al. 2010; Friederichs et al. 2011). Interestingly, proper Ndc1 levels are critical for cell survival, as illustrated by its haplo-insufficiency and overexpression phenotypes leading to defects in SPB duplication (Chial et al. 1999). Our data, along with these studies, support a model of competition between SPBs and NPCs for a common limiting component, Ndc1. Since Ndc1 is thought to be targeted to SPBs and NPCs through specific physical interactions with other membrane proteins (Onischenko et al. 2009), loss of POM152 or POM34 could result in a shift of Ndc1 recruitment to SPBs, which might aid in SPB assembly. Such a model of Ndc1 altered recruitment would suggest that competition for Ndc1 leads to antagonism of SPBs and NPCs.
Evidence indicates that this antagonism between NPCs and SPBs is regulated within the cell. Inhibition of Pom34 translation by the Smy2–Eap1–Scp160–Asc1 (SESA) network is sufficient to rescue the temperature-sensitive insertion defects of mps2-2 cells (Sezen et al. 2009). It is intriguing to consider that linking SPB and NPC assembly/function by such a mechanism might allow control of nuclear pore formation and number during specific cell-cycle stages and restrict SPB duplication in the G1-phase of the cell cycle.
Supplementary Material
Acknowledgments
We are grateful to Martin Hetzer, Brian Slaughter, Jay Unruh, and members of the Wente and Jaspersen laboratories for helpful discussions. Electron microscopy work was supported by the Vanderbilt Ingram Cancer Center Support Grant (P30 CA068485) through the use of the Vanderbilt University Medical Center Cell Imaging Shared Resource (supported by National Institutes of Health (NIH) grants CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126) This work was supported by the Stowers Institute for Medical Research (to S.L.J), the American Cancer Society (RSG-11-030-01-CSM) (to S. L. J), NIH (5R01 GM057438-13) (to S. R. W.), and National Science Foundation (Graduate Research Fellowship 2011100772) (to A.K.C.).
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Abramoff M., Magelhaes P., Ram S., 2004. Image processing with ImageJ, pp. 36–42 Biophotonics International [Google Scholar]
- Adams I. R., Kilmartin J. V., 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145: 809–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison J. D., Blobel G., Rout M. P., 1995. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 131: 1659–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Antonin W., 2009. Nuclear envelope: membrane bending for pore formation? Curr. Biol. 19: R410–R412 [DOI] [PubMed] [Google Scholar]
- Antonin W., Ellenberg J., Dultz E., 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582: 2004–2016 [DOI] [PubMed] [Google Scholar]
- Antonin W., Ungricht R., Kutay U., 2011. Traversing the NPC along the pore membrane: targeting of membrane proteins to the INM. Nucleus 2: 87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y., Lau C. K., Maekawa H., Jaspersen S. L., Giddings T. H., et al. , 2006. The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p. Mol. Biol. Cell 17: 1959–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger T. A., Folkmann A. W., Tran E. J., Wente S. R., 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Leksa N. C., Spear E. D., Rajashankar K. R., Schwartz T. U., 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Partridge J. R., Whittle J. R., Schwartz T. U., 2009. The nuclear pore complex has entered the atomic age. Structure 17: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L., 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb. Symp. Quant. Biol. 38: 123–131 [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L., 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124: 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Doucet C., Hetzer M. W., 2010. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb. Symp. Quant. Biol. 75: 585–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo A. R., Meehl J. B., Morgan G., Schutz-Geschwender A., Winey M., 2002. The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J. Cell Biol. 156: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadrin A., Hess B., San Roman M., Gatti X., Lombard B., et al. , 2010. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 189: 795–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H. J., Rout M. P., Giddings T. H., Winey M., 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143: 1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H. J., Giddings T. H., Siewert E. A., Hoyt M. A., Winey M., 1999. Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication gene, NDC1, leads to aneuploidy and polyploidy. Proc. Natl. Acad. Sci. USA 96: 10200–10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- D’Angelo M. A., Anderson D. J., Richard E., Hetzer M. W., 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science 312: 440–443 [DOI] [PubMed] [Google Scholar]
- Dawson T. R., Lazarus M. D., Hetzer M. W., Wente S. R., 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene J. O., Coleman J., Estrada de Martin P., Pypaert M., Anderson S., et al. , 2006. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol. Biol. Cell 17: 3009–3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. D., Kilmartin J. V., 1996. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J. Cell Biol. 132: 887–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C. M., Hetzer M. W., 2010. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma 119: 469–477 [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J., Rout M. P., 2009. Nuclear pore complex biogenesis. Curr. Opin. Cell Biol. 21: 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederichs J. M., Ghosh S., Smoyer C. J., McCroskey S., Miller B. D., et al. , 2011. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 7: e1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E. C., 1993. Purification of NSP1 reveals complex formation with ’GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 12: 3061–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenland K. B., Ding H., Costanzo M., Boone C., Davis T. N., 2010. Identification of Saccharomyces cerevisiae spindle pole body remodeling factors. PLoS ONE 5: e15426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath C. V., Copeland C. S., Amberg D. C., Del Priore V., Snyder M., et al. , 1995. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J. Cell Biol. 131: 1677–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. W., Wente S. R., 2009. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Dernburg A. F., 2009. The SUN rises on meiotic chromosome dynamics. Dev. Cell 17: 598–605 [DOI] [PubMed] [Google Scholar]
- Hodge C. A., Choudhary V., Wolyniak M. J., Scarcelli J. J., Schneiter R., et al. , 2010. Integral membrane proteins Brr6 and Apq12 link assembly of the nuclear pore complex to lipid homeostasis in the endoplasmic reticulum. J. Cell Sci. 123: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schaerer F., Brachat A., Wach A., Philippsen P., 2002. Reorientation of mispositioned spindles in short astral microtubule mutant spc72Delta is dependent on spindle pole body outer plaque and Kar3 motor protein. Mol. Biol. Cell 13: 1366–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Voss C., Shemesh T., Li Z., et al. , 2008. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A., 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R., 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107: 1409–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M., 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20: 1–28 [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Giddings T. H., Winey M., 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S. L., Martin A. E., Glazko G., Giddings T. H., Morgan G., et al. , 2006. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J. Cell Biol. 174: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind B., Koehler K., Lorenz M., Huebner A., 2009. The nuclear pore complex protein ALADIN is anchored via NDC1 but not via POM121 and GP210 in the nuclear envelope. Biochem. Biophys. Res. Commun. 390: 205–210 [DOI] [PubMed] [Google Scholar]
- Kupke T., Di Cecco L., Müller H. M., Neuner A., Adolf F., et al. , 2011. Targeting of Nbp1 to the inner nuclear membrane is essential for spindle pole body duplication. EMBO J. 30: 3337–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C. K., Giddings T. H., Winey M., 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly. Eukaryot. Cell 3: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk C. P., Blobel G., King M. C., 2007. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8: 414–420 [DOI] [PubMed] [Google Scholar]
- Madrid A. S., Mancuso J., Cande W. Z., Weis K., 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function. J. Cell Biol. 173: 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L. A., Panté N., Mall M., et al. , 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 22: 93–103 [DOI] [PubMed] [Google Scholar]
- McDonald K., 1999. High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 117: 77–97 [DOI] [PubMed] [Google Scholar]
- Miller J. P., Lo R. S., Ben-Hur A., Desmarais C., Stagljar I., et al. , 2005. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. USA 102: 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Rose M. D., 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140: 377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. K., Stuchell-Brereton M. D., Cooper J. A., 2009. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil. Cytoskeleton 66: 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Centeno M. C., McBratney S., Monterrosa A., Byers B., Mann C., et al. , 1999. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell 10: 2393–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B. T., Rout M. P., 2005. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 170: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E., Stanton L. H., Madrid A. S., Kieselbach T., Weis K., 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L. F., Rout M. P., Blobel G., 1995. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafsky D., Hodzic D., 2009. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J. Cell Biol. 186: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Santacreu L., Moreno J., Pérez-Ortín J. E., Alepuz P., 2009. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15: 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli J. J., Hodge C. A., Cole C. N., 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J. Cell Biol. 178: 799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E. C., Florens L., Guan T., Yates J. R., Gerace L., 2003. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301: 1380–1382 [DOI] [PubMed] [Google Scholar]
- Schneiter R., Cole C. N., 2010. Integrating complex functions: coordination of nuclear pore complex assembly and membrane expansion of the nuclear envelope requires a family of integral membrane proteins. Nucleus 1: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm C., Elliott S., Shevchenko A., Schiebel E., 2000. The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezen B., Seedorf M., Schiebel E., 2009. The SESA network links duplication of the yeast centrosome with the protein translation machinery. Genes Dev. 23: 1559–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., R Fink G., B Hicks J., 1986. Methods in Yeast Genetics: Laboratory Course Manual for Methods in Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Stage-Zimmermann T., Schmidt U., Silver P. A., 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11: 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F., Hülsmann B. B., Spang A., Hartmann E., Cordes V. C., et al. , 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173: 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R., 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6: 197–206 [DOI] [PubMed] [Google Scholar]
- Talamas J. A., Hetzer M. W., 2011. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J. Cell Biol. 194: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm T., Grallert A., Grossman E. P., Alvarez-Tabares I., Stevens F. E., et al. , 2011. Brr6 drives the Schizosaccharomyces pombe spindle pole body nuclear envelope insertion/extrusion cycle. J. Cell Biol. 195: 467–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R., 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178: 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetenbaum-Novatt J., Rout M. P., 2010. The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harb. Symp. Quant. Biol. 75: 567–584 [DOI] [PubMed] [Google Scholar]
- Tran E. J., Zhou Y., Corbett A. H., Wente S. R., 2007. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28: 850–859 [DOI] [PubMed] [Google Scholar]
- Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A., 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124: 573–586 [DOI] [PubMed] [Google Scholar]
- West M., Zurek N., Hoenger A., Voeltz G. K., 2011. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193: 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Bloom K., 2012. Mitotic spindle form and function. Genetics 190: 1197–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B., 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114: 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Hoyt M. A., Chan C., Goetsch L., Botstein D., et al. , 1993. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122: 743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Yarar D., Giddings T. H., Mastronarde D. N., 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell 8: 2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin K. L., Friederichs J. M., Cohen-Fix O., Jaspersen S. L., 2010. Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae. Genetics 186: 867–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R. W., Blobel G., Rout M. P., 1994. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 125: 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder T. J., Pearson C. G., Bloom K., Davis T. N., 2003. The Saccharomyces cerevisiae spindle pole body is a dynamic structure. Mol. Biol. Cell 14: 3494–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.