Abstract
Endogenous small interfering RNAs (endo-siRNAs) have been discovered in many organisms, including mammals. In C. elegans, depletion of germline-enriched endo-siRNAs found in complex with the CSR-1 Argonaute protein causes sterility and defects in chromosome segregation in early embryos. We discovered that knockdown of either csr-1, the RNA-dependent RNA polymerase (RdRP) ego-1, or the dicer-related helicase drh-3, leads to defects in histone mRNA processing, resulting in severe depletion of core histone proteins. The maturation of replication-dependent histone mRNAs, unlike that of other mRNAs, requires processing of their 3′UTRs through an endonucleolytic cleavage guided by the U7 snRNA, which is lacking in C. elegans. We found that CSR-1-bound antisense endo-siRNAs match histone mRNAs and mRNA precursors. Consistently, we demonstrate that CSR-1 directly binds to histone mRNA in an ego-1-dependent manner using biotinylated 2′-O-methyl RNA oligonucleotides. Moreover, we demonstrate that increasing the dosage of histone genes rescues the lethality associated with depletion of CSR-1 and EGO-1. These results support a positive and direct effect of RNAi on histone gene expression.
Keywords: C. elegans , CSR-1, histone, mRNA processing, RNA interference
Introduction
C. elegans contains a large number of endogenous siRNAs, the majority of which are antisense to protein-coding mRNAs and are loaded by Argonaute (AGO) proteins (Ambros et al, 2003; Ruby et al, 2006; Pak and Fire, 2007; Claycomb et al, 2009; Gu et al, 2009). Endo-siRNAs produced by RNA-dependent RNA Polymerases (RdRPs) (Ruby et al, 2006; Aoki et al, 2007; Pak and Fire, 2007) are termed as 22G-RNAs and can be divided into classes based on the AGOs to which they bind (Claycomb et al, 2009; Gu et al, 2009). The Argonaute CSR-1 was shown to preferentially bind gene-specific endo-siRNAs (Claycomb et al, 2009). The synthesis of endo-siRNAs in the CSR-1 22G-RNA pathway is dependent on the RdRP EGO-1 (Smardon et al, 2000; Claycomb et al, 2009) and the dicer-related helicase DRH-3 (Duchaine et al, 2006; Nakamura et al, 2007). Depletion of EGO-1, CSR-1 or DRH-3 results in similar sterility and defects in chromosome segregation in early embryos (Smardon et al, 2000; Duchaine et al, 2006; Yigit et al, 2006; Nakamura et al, 2007; Claycomb et al, 2009; She et al, 2009). Based on the established model in S. pombe (Halic and Moazed, 2009; Lejeune and Allshire, 2011), it was proposed that the CSR-1 22G-RNA pathway aids in the proper compaction of the holocentric chromosomes of the worm (Claycomb et al, 2009). However, in fission yeast, siRNAs match pericentromeric repeat regions (Halic and Moazed, 2009), in contrast to C. elegans siRNAs, which were shown to primarily match protein-coding genomic regions (Claycomb et al, 2009), including histone genes (Grishok et al, 2008).
Here, we demonstrate that knockdown of csr-1, ego-1 or drh-3 leads to misprocessed histone mRNA, resulting in severe depletion of core histone proteins, and that expression of transgene-encoded histone transcripts rescues the lethality of the RNAi mutants. We propose that this depletion of core histones is responsible for the sterility and chromosome segregation defects of the CSR-1 RNAi pathway mutants. In most organisms, specific 3′UTR processing of histone mRNAs includes a cleavage by the Cleavage and Polyadenylation Specificity Factor (CPSF) subunit CPSF73 just downstream of a conserved stem-loop. This stem-loop is recognized by a conserved stem-loop binding protein (SLBP), while the histone downstream element (HDE) sequence is recognized by the U7 snRNA (Marzluff et al, 2008). Surprisingly, in C. elegans, U7 and its binding partner Lsm11, as well as the HDE sequence normally present at 3′UTRs of histone genes, are missing (Davila Lopez and Samuelsson, 2008). This opens the possibility for direct involvement of endo-siRNAs in histone mRNA 3′end processing. Consistent with this possibility, we show that CSR-1 interacts with 2′-O-methyl oligonucleotides that mimic the histone mRNA 3′UTR sequences in an ego-1-dependent, that is, siRNA-dependent, manner. We also show that CSR-1 interacts with histone mRNA in an ego-1-dependent manner. We conclude that CSR-1-dependent endo-siRNAs play a positive role in the regulation of core histone expression in C. elegans, although it remains to be determined whether CSR-1 is directly involved in cleaving histone mRNA precursors.
Results
CSR-1-bound siRNAs directly target histone genes
Previous studies reported no global changes in the expression of the genes targeted by the CSR-1 endogenous RNAi pathway in csr-1(tm892) mutant adults (Claycomb et al, 2009; Gu et al, 2009). Our analysis of the microarray data performed on csr-1(tm892) by Claycomb et al (2009) showed, as previously reported, that the majority (65%) of CSR-1 targets, defined by their antisense endo-siRNAs enriched in CSR-1-IP, had no change in gene expression in adults (Figure 1A and B). Interestingly, we found that the ratio of downregulated genes to upregulated genes among the CSR-1 targets was significantly (10-fold, P<0.0001) higher compared to that of the global gene expression profile (Figure 1A and B). However, most of the downregulated genes could be a consequence of comparing sterile to gravid adults. Nevertheless, downregulation of a few critical CSR-1-targeted germline genes could be causal in inducing the phenotypes observed in csr-1, ego-1 and drh-3 mutants. Among the downregulated genes, we found the majority (66%) of the histone genes to be depleted in csr-1(tm892) mutants (Figure 1C). This is significant because downregulation of just one core histone gene by RNAi has previously been shown to be sufficient to cause chromosome segregation and sterility phenotypes (Kodama et al, 2002; Sonnichsen et al, 2005). We hypothesize that since histones are essential for chromatin compaction, defects in their production could underlie the phenotypes observed in the RNAi mutants.
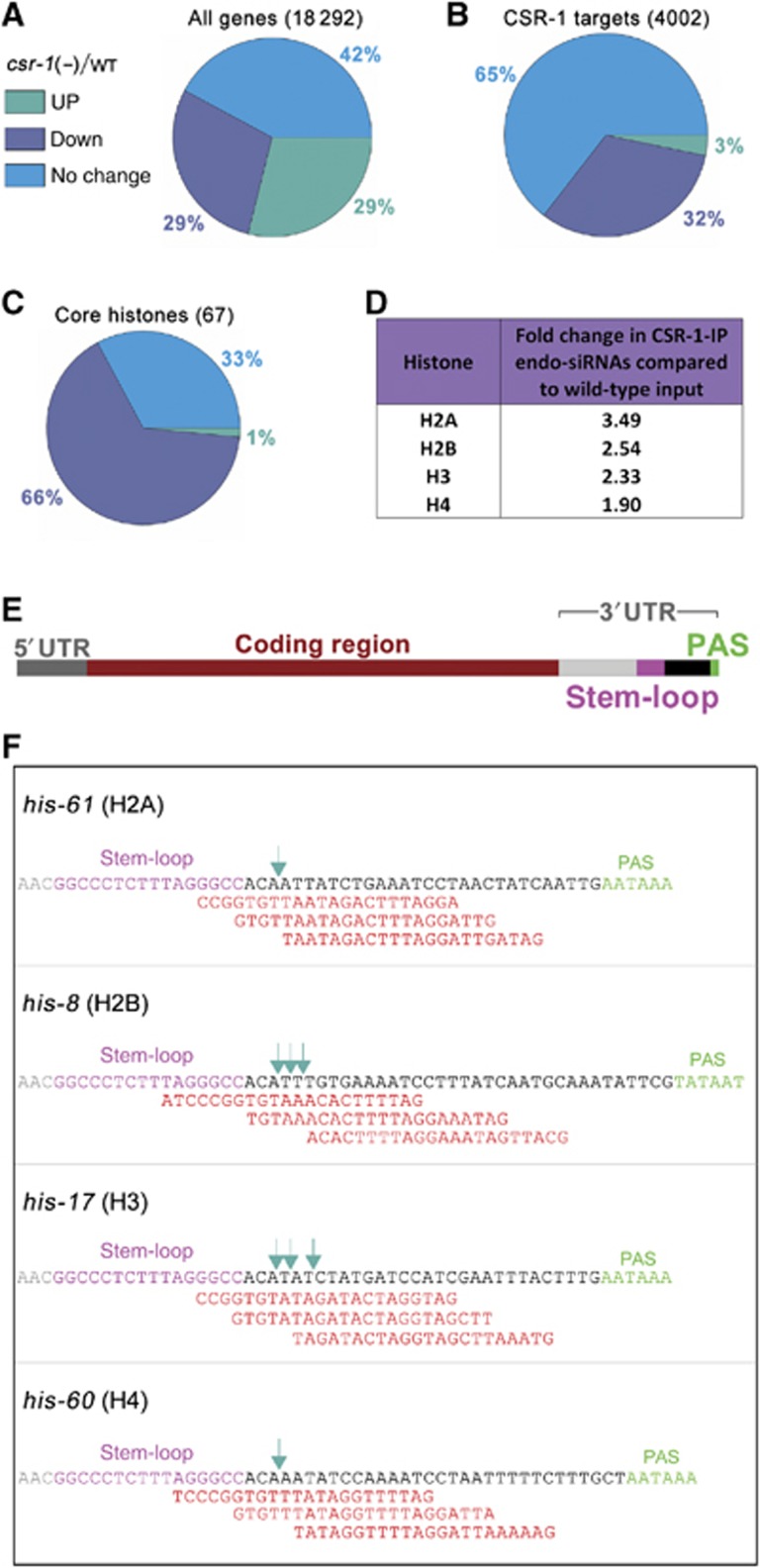
Figure 1.
CSR-1-associated siRNAs target replication-dependent core histone genes. (A–C) Gene expression profile of csr-1(tm892) adults compared to wild type. Pie charts showing the distribution of no change, up- (>1.5-fold) and downregulated (<1.5-fold) genes for the whole genome (A), CSR-1 targets (B) and core histones (C) (microarray data from Claycomb et al, 2009). The majority of all genes and CSR-1 target genes showed no change in gene expression in the csr-1 mutant, but core histone genes were predominantly down. (D) Fold change in total number of endo-siRNAs corresponding to each core histone in CSR-1-IP compared to wild-type input (deep sequencing data from Claycomb et al, 2009). Fold change is calculated from total endo-siRNA counts for all genes corresponding to each histone. (E) Schematic of a typical histone gene (to scale): an upstream 5′UTR followed by the coding region, typically without introns, and a downstream 3′UTR containing a stem-loop forming sequence (pink) followed by the poly(A) signal (PAS, green). (F) Endo-siRNAs (red) present in wild-type libraries, and not found in the ego-1 mutant library (Claycomb et al, 2009), mapping to the histone 3′UTR adjacent to the stem-loop (pink) for one example of each core histone gene [his-61 (H2A), his-8 (H2B), his-17 (H3) and his-60 (H4)] are shown. The dark green arrows indicate the previously detected 3′UTR ends of the mature histone mRNAs (Keall et al, 2007).
We observed that antisense endo-siRNAs targeting all core histone genes were enriched in CSR-1-IP libraries (Claycomb et al, 2009; Figure 1D), indicating a possible direct role in histone mRNA regulation by CSR-1 endo-siRNAs. The 3′ends of histone mRNAs were mapped to a few nucleotides downstream of the stem-loop (Keall et al, 2007; Figure 1F, green arrows), which is consistent with tiling microarray data (Spencer et al, 2011). However, polyadenylation signals (PAS) are also present at 3′UTRs of histone genes (Keall et al, 2007; Mangone et al, 2010; Figure 1E and F), and it has recently been discovered that a small portion of C. elegans and mammalian histone mRNAs are polyadenylated (Mangone et al, 2010; Jan et al, 2011; Shepard et al, 2011). Unprocessed histone mRNAs that accumulate in mutants deficient in processing are also known to be polyadenylated (Godfrey et al, 2006; Ideue et al, 2009). It has been proposed that the production of polyadenylated histone messages precedes the histone mRNA-specific processing (Mangone et al, 2010). Such a two-step mechanism for histone mRNA biogenesis would allow more inputs for potential regulation. The stem-loop and PAS are located in close proximity (∼30 nt) within 3′UTRs of C. elegans histone messages (Figure 1F). We therefore searched for endo-siRNAs antisense to these portions of 3′UTRs, which would have the potential to guide the cleavage reaction. Indeed, we found ego-1-dependent endo-siRNAs (Claycomb et al, 2009) that match 3′UTRs in antisense orientation opposite to the mapped 3′ends of mature histone mRNAs (Keall et al, 2007; Figure 1F). These analyses suggest the possibility of a direct role of the CSR-1 endo-siRNA pathway in histone mRNA biogenesis.
CSR-1 binds to histone mRNA in an ego-1-dependent manner
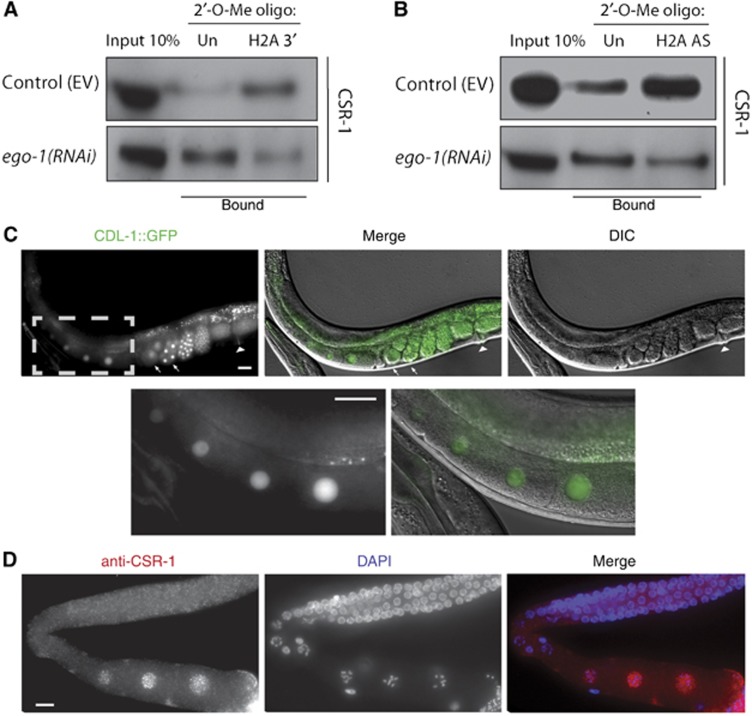
To confirm that CSR-1 binds to endo-siRNAs specific to the 3′UTR of histone genes, we designed a 34-nucleotide-long 2′-O-methyl RNA oligonucleotide identical to the region of the 3′UTR in between the stem-loop and the PAS of H2A pre-mRNA (Figure 1F, black). We incubated the biotinylated 2′-O-methyl RNA oligonucleotides with total worm lysates, performed a pull down with streptavidin beads according to established protocols (Hutvágner et al, 2004; Aoki et al, 2007), and investigated whether CSR-1 was present in the pulled-down material by western with anti-CSR-1 antibodies (Claycomb et al, 2009). We found an enrichment of CSR-1 in pull-downs using the H2A 3′UTR sequence, but not in experiments with an unrelated sequence (Figure 2A, top panel). This enrichment was lost upon ego-1(RNAi), indicating that the binding of CSR-1 to the H2A sequence was dependent on siRNAs produced by EGO-1 (Figure 2A, bottom panel). This experiment shows that CSR-1 is capable of binding to the 3′UTR of H2A pre-mRNA, and specifically to the region that is cleaved during its processing.
Figure 2.
CSR-1 interacts with histone mRNA and shows a nuclear localization in maturing oocytes, similarly to CDL-1::GFP. (A) Western blot analysis with anti-CSR-1 antibodies (Claycomb et al, 2009) of pull-down experiments using biotinylated 2′-O-methyl oligonucleotides. ‘Un’ indicates an unrelated sequence used as a negative control where ‘H2A 3′’ is the sense strand of the 3′UTR of H2A in between the stem-loop and PAS (Figure 1F, black sequence). The top panel shows enrichment of CSR-1 using the H2A sequence but not the unrelated sequence. The bottom panel shows the loss of this enrichment upon knockdown of ego-1 by RNAi. (B) The same as in (A), only using biotinylated 2′-O-methyl oligonucleotides antisense (AS) to the coding region of H2A. The top panel shows enrichment of CSR-1 using the H2A antisense sequence, but not the unrelated sequence. The bottom panel shows loss of this enrichment upon knockdown of ego-1 by RNAi. (C) CDL-1::GFP is expressed in the germline and early embryos. The arrows show embryos in the first two cell divisions while the arrowhead indicates the vulva. The grey dotted box shows the enlarged region in the top panel to better visualize expression in the nuclei of maturing oocytes. (D) Immunostaining of dissected adult worm gonads with anti-CSR-1 antibodies (pink) (Aoki et al, 2007) and DNA (DAPI, blue). The signal is strongest in the maturing oocytes. All scale bars represent 20 μm. Figure source data can be found with the Supplementary data.
To validate the interaction of CSR-1 with the H2A mRNA sequence further, we designed 2′-O-methyl RNA oligonucleotides antisense to the coding region of H2A and repeated the pull-down experiment (see Materials and methods for details). The antisense H2A oligos were also able to pull down CSR-1 in an ego-1-dependent manner (Figure 2B), indicating that CSR-1 interacts with the histone mRNA through its bound siRNAs. We confirmed that H2A mRNA was similarly enriched in both the control (empty vector, EV) and ego-1(RNAi) pull-down samples by RT–qPCR (Supplementary Figure S1A). We also confirmed that the depletion of CSR-1 protein in the pull down upon knockdown of ego-1 is not due to lower levels of CSR-1 overall (Supplementary Figure S1B). These results demonstrate that CSR-1 interacts with histone mRNA through the base pairing of its bound siRNAs. CSR-1 may then facilitate cleavage of the 3′UTR downstream of the stem-loop or it may act to recruit other cleavage factors for the proper processing of the histone mRNA.
CDL-1 (SLBP) and CSR-1 are strongly enriched in the nuclei of maturing oocytes
One factor necessary for the processing, stabilization and efficient translation of histone mRNA is SLBP (Marzluff et al, 2008). Downregulation of C. elegans SLBP (CDL-1) by RNAi causes a severe depletion of histones as well as sterility in adults and chromosome segregation defects in early embryos (Kodama et al, 2002; Pettitt et al, 2002). To better understand the connection between CDL-1 function and its localization, we created transgenic worms expressing CDL-1 with a C-terminal GFP tag using its endogenous promoter. A previous study described strong expression of a GFP reporter driven by the cdl-1 (i.e., R06F6.1) promoter in the dividing cells of embryo, larva, and adult (Pettitt et al, 2002). However, germline expression was not reported, probably due to transgene silencing in this tissue. We observed expression of the CDL-1::GFP translational fusion protein in adult germline and embryos (Figure 2C). Notably, there is a strong enrichment of CDL-1::GFP in the nuclei of developing oocytes (Figure 2C, enlarged region). This is consistent with findings in other animals, as SLBP has been shown to be present in maturing oocytes in mammals and frogs (Allard et al, 2005; Gorgoni et al, 2005; Thelie et al, 2012) and to be required maternally in flies (Sullivan et al, 2001). Interestingly, CSR-1 is present in the peri-nuclear P-granules in the distal germline (Claycomb et al, 2009; Updike and Strome, 2009), but becomes nuclear in maturing oocytes (Claycomb et al, 2009). Immunostaining of dissected adult germlines with anti-CSR-1 antibodies (Aoki et al, 2007) revealed that CSR-1 is enriched in oocyte nuclei at the same stage as CDL-1::GFP (Figure 2D). This signal is specific to CSR-1 as it is lost in the csr-1(tm892) null mutant (Supplementary Figure S2A and B). Since CSR-1 directly binds to histone mRNAs, we propose that it may have a role, along with CDL-1, in histone mRNA processing in oocytes to ensure that early embryos undergoing rapid cell divisions, before the onset of zygotic transcription, are supplied with histone proteins and mRNA. In early embryos, CDL-1 is likely to be involved in histone mRNA translation, as has been found in other systems (Sullivan et al, 2001; Allard et al, 2005).
Interestingly, some phenotypes of the RdRP mutant ego-1 can be related to histone biogenesis defects. The germline defects of ego-1 mutants have been extensively characterized and include a remarkable extension of the transition zone (Smardon et al, 2000) (transition from mitosis to meiosis), enriched in nuclei in premeiotic S-phase (Kimble and Crittenden, 2005). This phenotype is consistent with histone biogenesis defects, which were shown to result in S-phase extension and cell-cycle delay in cultured cells (Wagner et al, 2005; Ideue et al, 2009).
In summary, both phenotypic and expression data are consistent with the role of RNAi in histone biogenesis in the germline.
Knockdown of RNAi pathway components leads to a depletion in total histone mRNA and an accumulation of unprocessed histone mRNA
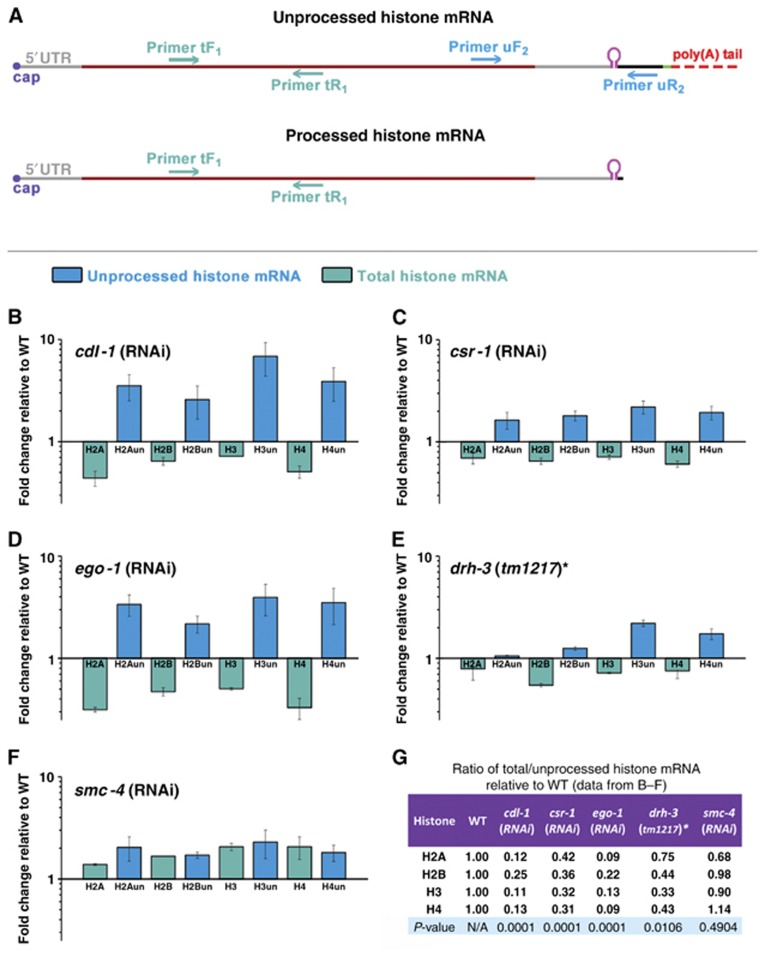
Although cdl-1 has been shown to be required for proper histone expression in C. elegans (Kodama et al, 2002; Pettitt et al, 2002), accumulation of unprocessed histone mRNA has not been previously reported for the CDL-1 knockdown (Keall et al, 2007). To measure the level of histone mRNA misprocessing, we designed primers (Supplementary Table S1) to specifically detect total histone mRNA (primers tF1, tR1) and unprocessed histone mRNA (primers uF2, uR2) for all core histone messages (Figure 3A). To test whether misprocessing of histone mRNA could lead to the defects observed in cdl-1(RNAi), we measured the levels of unprocessed and total histone mRNA in young adults, at a stage prior to the emergence of the sterility and chromosome segregation phenotype. Indeed, we found increased levels of the unprocessed mRNA and a depletion of total mRNA for all the core histones in cdl-1(RNAi) compared to wild type (empty vector, EV) (Figure 3B). To evaluate the role of endogenous RNAi in the processing of histone mRNA, we quantified the level of misprocessing after the knockdown of endogenous RNAi components. We observed a signature very similar to that of cdl-1(RNAi) in csr-1(RNAi) and ego-1(RNAi) (Figure 3C and D), suggesting that these RNAi factors regulate histone mRNA processing. We did not achieve efficient knockdown of DRH-3 by RNAi; therefore, we performed the mRNA analysis in the drh-3(tm1217) null mutant and found similar misprocessing (Figure 3E). We also confirmed that all the RNAi treatments resulted in sterility of mature adults. To verify that the sterility we observed in the RNAi knockdown was due to histone mRNA depletion, and not vice versa, we used RNAi against SMC-4, a subunit of the mitotic condensin complex whose knockdown also results in sterility and chromosome segregation defects, specifically in anaphase bridging (Hagstrom et al, 2002). We did not observe any histone mRNA depletion in smc-4(RNAi) (Figure 3F). This signature of mRNA misprocessing of all core histones, as quantified by the ratio of the change in total mRNA to the change in unprocessed mRNA, was statistically significant for cdl-1(RNAi), csr-1(RNAi), ego-1(RNAi) and drh-3(tm1217), but not for smc-4(RNAi) compared to wild type (P-values are provided in Figure 3G). This suggests that misprocessing of replication-dependent histone mRNA can cause the defects observed after the knockdown of RNAi pathway components.
Figure 3.
Knockdown of CSR-1 RNAi pathway components results in the distinct signature of core histone mRNA misprocessing. (A) Schematic diagram of unprocessed and processed histone mRNAs showing the primers used to detect total (unprocessed and processed) mRNA (tF1 and tR1) and unprocessed mRNA only (uF2 and uR2) by RT–qPCR in young adult worms. Primer sequences are provided in Supplementary Table S1. (B–E) Total mRNA (green) of core histones decreased and unprocessed mRNA (blue) increased in cdl-1, csr-1 and ego-1 RNAi and drh-3(tm1217) as compared to appropriate wild-type controls. *DA1316 is a congenic wild-type strain for drh-3(tm1217). (F) smc-4(RNAi) showed no decrease in total histone mRNA. (G) The ratio of the change in total to the change in unprocessed histone mRNA levels for all core histones was significantly lower in cdl-1 (P<0.0001), csr-1 (P<0.0001) and ego-1 (P<0.0001) RNAi and drh-3(tm1217) (P=0.01) when compared to wild type. This effect was not seen in smc-4(RNAi) (P=0.49). Error bars represent the standard deviation from the mean from three independent experiments.
Histone proteins are severely depleted in the knockdown of RNAi pathway components
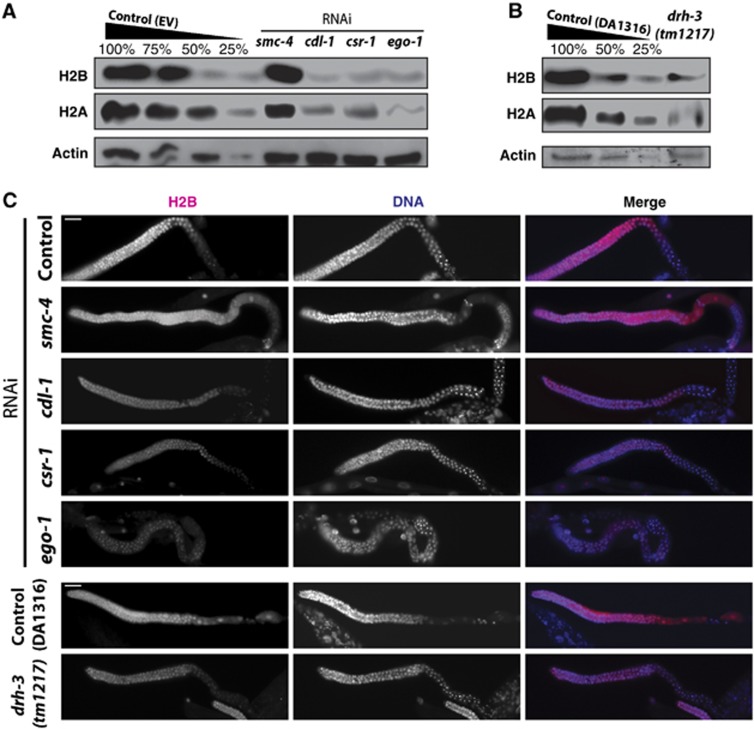
To evaluate the depletion of core histone proteins, we performed western blot analyses using extracts of worms depleted of csr-1 and ego-1 by RNAi as well as extracts of drh-3(tm1217) null mutants. Extracts from cdl-1(RNAi) and smc-4(RNAi) were included as positive and negative controls, respectively. Consistent with our mRNA results, the levels of H2A (50–75% depletion compared to wild type) and H2B histones (∼50% compared to wild type) were dramatically reduced in csr-1 and ego-1(RNAi) and in drh-3(tm1217), similar to the depletion observed in cdl-1(RNAi) worms (Figure 4A and B). H2A and H2B levels were not reduced in smc-4(RNAi) (Figure 4A). In a different approach, we stained dissected gonads from young adult worms treated with csr-1 and ego-1 RNAi and drh3(tm1217) mutants with anti-H2B antibodies to confirm that the depletion takes place in the germline and therefore should influence the histone pool in early embryos. As expected from our western blot results, we detected a severe depletion of H2B in the cdl-1, csr-1 and ego-1 RNAi and drh-3(tm1217), but not in smc-4(RNAi) (Figure 4C).
Figure 4.
Knockdown of CSR-1 RNAi pathway components results in severe depletion in H2A and H2B proteins. (A) Western blot results with total young adult worm lysates: core histones H2A and H2B showed severe depletion in cdl-1, csr-1 and ego-1 RNAi, but not in smc-4 RNAi when compared to empty vector (EV) control. Actin is shown as a loading control. (B) Same as (A) for the drh-3(tm1217) null mutant with its congenic wild type (DA1316) as the control. (C) Histone H2B immunostaining of dissected gonads from young adults (pink) shown with DNA (DAPI, blue). H2B was depleted in cdl-1, csr-1 and ego-1 RNAi, but not in smc-4 RNAi, consistent with western blotting. Similarly, H2B was depleted in drh-3(tm1217), but not in the control (DA1316). Scale bar is 20 μm. Figure source data can be found with the Supplementary data.
Although depletion of just one core histone has been shown to be sufficient to cause sterility and chromosome segregation phenotypes (Kodama et al, 2002; Sonnichsen et al, 2005), we wanted to determine whether H3 and H4 were also affected. Our mRNA results indicate that all four histones should be similarly affected (Figure 3). We found, by using the same biological samples for westerns, that one antibody specific to H4 reveals a severe depletion (∼75%) in csr-1(RNAi) and ego-1(RNAi) samples while another shows no difference in H4 levels under the knockdown conditions (Supplementary Figure S3, compare A and B). There is no apparent change in H3 protein levels (Supplementary Figure S3B). Thus, it appears that using more sensitive antibodies may preclude the detection of any difference in protein levels due to saturation of the signal.
Since RNAi targeting histone genes in C. elegans was shown to cause chromosome segregation defects (Kodama et al, 2002; Sonnichsen et al, 2005), our findings of severe histone depletion in csr-1 and ego-1 RNAi and drh-3(tm1217) point to it as the most likely cause of their published cell division phenotypes (Duchaine et al, 2006; Yigit et al, 2006; Nakamura et al, 2007; Claycomb et al, 2009).
A higher level of expression of histone proteins is also needed during DNA endoreduplication in the intestinal cells of C. elegans (McGhee, 2007). Consistently, we find by immunostaining with anti-CSR-1 antibodies (Aoki et al, 2007) that CSR-1 is present in the intestinal nuclei of adult worms (Supplementary Figure S2A). Previous studies reported a lower level of CSR-1 protein expression in somatic tissues compared to the germline, although csr-1 mRNA specific to the long isoform of the protein is produced in the soma at high levels (Claycomb et al, 2009). Our results are consistent with these findings. Furthermore, we find that upon csr-1(RNAi), H2B is depleted in intestinal nuclei, confirming that the dependence of histone production on the RNAi pathway is not limited to the germline (Supplementary Figure S4).
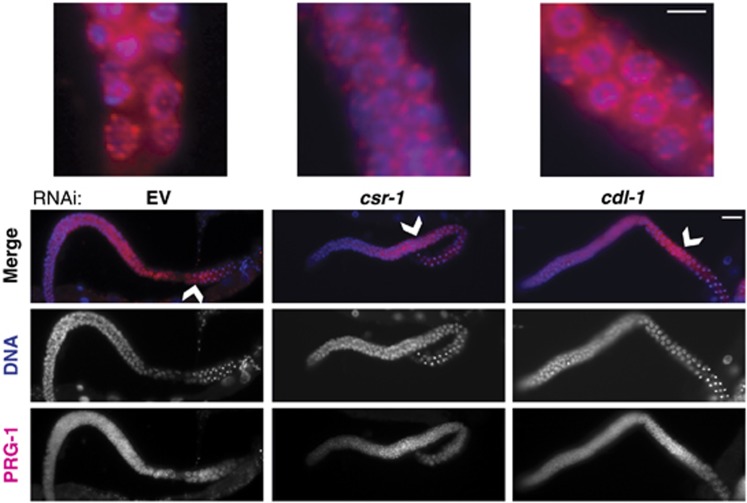
P-granule formation is not affected by the knockdown of CDL-1
In addition to sterility and chromosome segregation defects, the endogenous RNAi pathway components CSR-1, EGO-1 and DRH-3 have also been shown to affect P-granule formation in the C. elegans gonad (Updike and Strome, 2009). Next, we asked whether sterility in cdl-1(RNAi) correlated with the disruption of P-granule morphology as well, by staining for PRG-1, a PIWI protein found in P-granules (Batista et al, 2008). We did not observe any defects in P-granule morphology in dissected gonads from cdl-1(RNAi) young adults compared to csr-1(RNAi) (Figure 5). Since CSR-1-bound endo-siRNAs produced by EGO-1 regulate additional germline genes (Maniar and Fire, 2011) while CDL-1 has a specific and conserved role in histone mRNA processing, these results indicate that histone mRNA misprocessing can cause sterility in cdl-1(RNAi) without affecting P-granule morphology. Therefore, histone mRNA misprocessing could be sufficient to cause sterility in the RNAi pathway mutants.
Figure 5.
csr-1(RNAi) P-granule defects are not related to histone misregulation. PRG-1 immunostaining of dissected gonads from young adults (pink) shown with DNA (DAPI, blue) to visualize P-granule morphology in csr-1 and cdl-1 RNAi (anti-PRG-1 is from Batista et al, 2008). Knockdown of cdl-1(RNAi) does not affect P-granule morphology despite sterility, indicating that the P-granule phenotype in csr-1 is independent of the histone processing defect. Scale bar is 20 μm. Arrowheads indicate enlarged regions, scale bar is 5 μm.
Overexpression of transgenic histones rescues the sterility caused by the knockdown of RNAi pathway components
Next, we tested whether the sterility of RNAi mutants can be rescued by increased histone production from transgenes. We chose two pairs of histone genes: H2A/H2B and H3/H4 (his-59-62 locus) arranged in head-to-head orientation and designed constructs for rescue experiments. The constructs included promoters, coding regions and truncated 3′UTRs of histone genes ending after the conserved stem-loop (see Materials and methods for experimental details). Our results described above suggest that the processing of histone mRNAs in C. elegans is dependent on the RNAi pathway. We hypothesized that endo-siRNAs antisense to 3′UTRs of histone genes and mapping downstream of the stem-loop (Figure 1F) regulate histone mRNA-specific processing, which includes a cleavage between the stem-loop and PAS. Since an increase in unprocessed histone mRNA resulted in the depletion of total mRNA (Figure 3) and histone protein levels (Figure 4), we hypothesized that the removal of the 3′UTR downstream of the stem-loop, including the PAS, should stimulate stem-loop-dependent histone expression. Therefore, we tested whether the constructs containing the stem-loop but lacking the PAS in the 3′UTRs would bypass the requirement of RNAi for histone production.
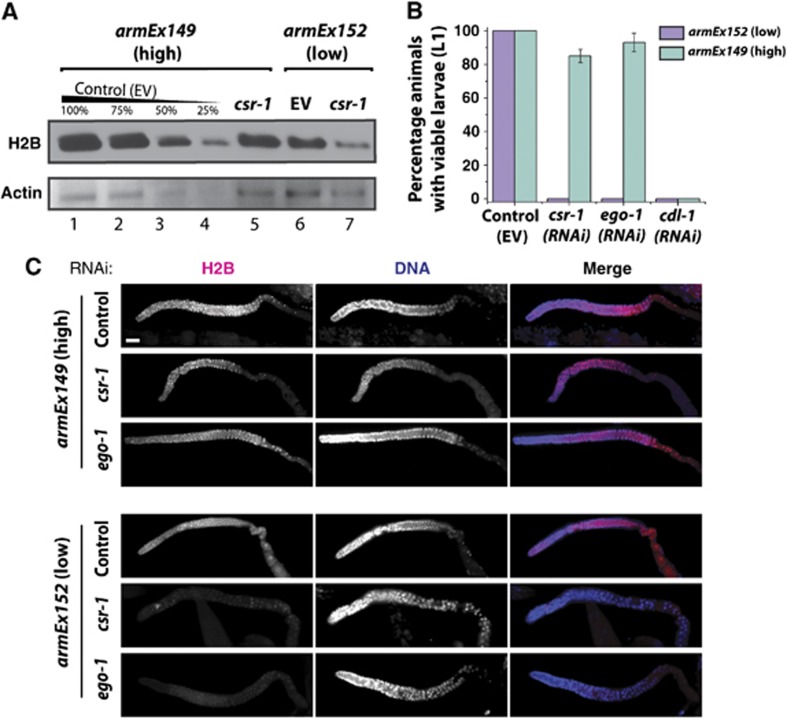
We examined the expression levels of the transgene-encoded histone mRNAs in several transgenic lines and selected high (armEx149) and low (armEx152) expressing lines for further experiments. There was a notable increase in the total levels of histone proteins in the high expressing armEx149 line compared to the low expressing control armEx152 line (Figure 6A, compare lanes 1 and 6, Figure 7B, compare lanes 1 and 3). Next, we downregulated csr-1, ego-1 and cdl-1 by RNAi in high- and low-expressing transgenic lines. We initiated RNAi by feeding young larva (L1) and observed survival of their progeny. csr-1(RNAi), ego-1(RNAi) and cdl-1(RNAi) induced penetrant embryonic lethality in the low-expressing transgenic control line (Figure 6B, purple bars, P<0.0001). However, csr-1(RNAi) and ego-1(RNAi) failed to induce significant embryonic lethality in the strain expressing high levels of transgenic histone mRNAs (Figure 6B, green bars). cdl-1(RNAi) lethality was not rescued in this transgenic background, consistent with the requirement for the SLBP in nuclear export and translation of histone mRNAs (Allard et al, 2005; Marzluff et al, 2008). Consistently, H2B protein levels are not reduced in the germlines of young adult worms treated with csr-1(RNAi) or ego-1(RNAi) in the high-expressing line but are depleted in the low-expressing line (Figure 6C). The resistance of the armEx152 line to csr-1(RNAi) and ego-1(RNAi) is not the result of a general insensitivity to RNAi in this strain (see Supplementary Figure S5A and B).
Figure 6.
Expression of core histones from a transgenic array rescues the sterility phenotype arising from the knockdown of CSR-1 RNAi pathway components. (A) Western blot results with young adult worm lysates: total histone H2B protein levels were reduced in both the low-expressing transgenic line armEx152 and high-expressing line armEx149 upon csr-1(RNAi) compared to control RNAi; however, total H2B levels were higher in the armEx149, accounting for a considerable persistence of H2B expression in csr-1(RNAi)-treated animals. Actin is shown as a loading control. (B) Knockdown of csr-1 or ego-1 by RNAi, but not the control treatment (empty vector, EV), caused lethality in the progeny of the treated worms in the low-expressing transgenic line armEx152 (purple) (n>60). However, in the high-expressing line armEx149 (green), knockdown of csr-1 or ego-1 by RNAi did not cause lethality since greater than 80% of RNAi-treated hermaphrodites produced viable broods (n>60, P<0.0001). Knockdown of cdl-1 by RNAi in either transgenic line resulted in progeny lethality (n>60). The standard error of the proportion for the animals with viable progeny for csr-1(RNAi) is ±4.85% and for ego-1(RNAi) is ±4.34% with n=60. (C) Histone H2B immunostaining of dissected gonads from young adults (pink) shown with DNA (DAPI, blue). H2B is not depleted in csr-1(RNAi) or ego-1(RNAi) in armEx149 (high transgene expression) compared to empty vector control, but is depleted in csr-1(RNAi) and ego-1(RNAi) in armEx152 (low transgene expression), consistent with the western blotting shown in Figure 6A. Scale bar is 20 μm. Figure source data can be found with the Supplementary data.
Figure 7.
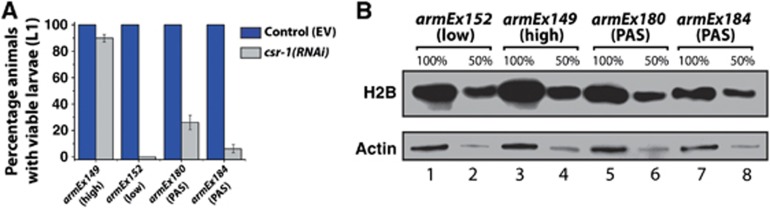
Histone transgenes containing the PAS sequence and no siRNA sites between the stem-loop and PAS do not produce high levels of histone proteins and do not rescue csr-1(RNAi) lethality. (A) Knockdown of csr-1 by RNAi fails to cause lethality in armEx149 (>80% viability, as in Figure 6B) but leads to lethality in armEx152 (0% as in Figure 6B). Introduction of the PAS sequence results in a significant decrease in viability after csr-1(RNAi) in armEx180 (26%, P-value<0.001) and armEx184 (6%, P-value<0.001) compared to armEx149. The standard error of the proportion for the animals with viable progeny upon csr-1(RNAi) is ±3.87% in armEx149, ±0% in armEx152, ±5.59% in armEx180 and ±2.81% in armEx184 with n=60. (B) Western blot analysis of young adult worm lysates for armEx149, armEx152, armEx180, and armEx184, showing an increase in total H2B in armEx149 as compared to the other three. In all, 100 and 50% amounts are shown to give relative abundance. Actin is shown as a loading control. Figure source data can be found with the Supplementary data.
Although the total levels of histone mRNA and histone proteins decreased in the high-expressing line treated with csr-1(RNAi) (Figure 6A, compare lanes 1 and 5; Supplementary Figure S6A), the expression level of transgene-encoded messages did not change under these conditions (Supplementary Figure S6B), nor were transgene-encoded histone mRNAs polyadenylated (Supplementary Figure S6C). Due to the overexpression of transgenic histones from the array, we found a significant difference in the expression of histone proteins in the rescued transgenic line treated with csr-1(RNAi) compared to the line with low transgene expression, as confirmed by western (Figure 6A, compare lanes 5 and 7) and immunofluorescence analyses for csr-1(RNAi) and ego-1(RNAi) (Figure 6C). This difference in histone protein levels in the germlines of rescued versus sensitive transgenic worms under csr-1(RNAi) and ego-1(RNAi) conditions is consistent with either the survival or the lethality of their progeny shown in Figure 6B.
To confirm that the lack of the PAS in the transgenic array contributed to RNAi-independent histone biogenesis, we re-introduced a PAS sequence into the 3′UTRs of H2A and H2B genes on the transgenic array. This was done so that the plasmid backbone sequence was present in place of sequences normally targeted by siRNAs. We selected two lines expressing the transgene at a higher levels than armEx152 (Supplementary Figure S7) and tested whether these arrays were able to rescue the lethality phenotype caused by csr-1(RNAi). We found that the lines containing the PAS showed significantly lower levels of rescue compared to the 90% rescue in the armEx149 line (Figure 7A, compare with 26% rescue in armEx180, and 6% in armEx184, P-value<0.001). We note that these lines had a higher level of rescue compared to armEx152, consistent with some level of histone expression in the germline. However, this increase is minimal as it was not possible to see any difference in histone protein levels between armEx152 and armEx180 by western (Figure 7B). Therefore, although the transgenes containing the PAS express mRNA (Supplementary Figure S7), this does not translate into a significant increase in total histone protein, which explains the lack of rescue in these lines. The caveat of these experiments is that none of the transgenic lines designed to express histone mRNA with the stem-loop and PAS separated by plasmid sequences produced mRNA at levels similar to that of armEx149 (Supplementary Figure S7). The stability of unprocessed histone mRNAs appears to be lower compared to that of properly processed ones; note the decrease in total mRNA levels in samples containing abundant unprocessed transcripts (Figure 3B–E). This increased destabilization of unprocessed histone transcripts has been observed in the maternal mutant embryos of SLBP in flies, indicating an active degradation process during oogenesis (Sullivan et al, 2001). Since the C. elegans SLBP homologue is enriched in oocytes, it is likely that a similar degradation mechanism of unprocessed transcripts takes place in the C. elegans germline as well. Therefore, it may not be possible to generate transgenic lines with high expression of RNAs mimicking the unprocessed histone mRNA precursors.
We conclude that histone messages containing both the stem-loop and the PAS, but no siRNA sites between them, appear to be expressed and utilized poorly, which suggests the need for an RNAi-dependent processing step.
Discussion
In most organisms, maturation of mRNAs coding for replication-dependent core histones, unlike other mRNAs, requires the processing of their 3′UTRs through an endonucleolytic cleavage between the conserved stem-loop and the specific element recognized by U7 snRNA (Marzluff et al, 2008). Although the vast majority of mature histone mRNAs are not polyadenylated, recent deep sequencing studies discovered polyadenylated histone mRNAs in C. elegans (Mangone et al, 2010; Jan et al, 2011) and in mammalian cells (Shepard et al, 2011). Hence, it was proposed that addition of polyA precedes histone mRNA-specific cleavage events (Mangone et al, 2010). Since nematodes lack the U7 snRNA (Davila Lopez and Samuelsson, 2008), it is possible that endo-siRNAs assume its role in guiding histone mRNA processing in C. elegans either by directly cleaving the transcript through the slicer activity of CSR-1 (Aoki et al, 2007) or by recruiting other conserved cleavage factors specific to 3′end processing. We have shown that CSR-1 directly interacts with histone mRNAs in an ego-1-dependent manner and that the knockdown of CSR-1 leads to histone mRNA misprocessing that results in severe histone protein depletion. We further demonstrate that the overexpression of transgenic histone proteins rescues the lethality caused by csr-1 and ego-1 RNAi. Together, these results indicate that an increase in core histone expression is sufficient to rescue the lethality of the CSR-1 pathway mutants. Therefore, we propose that it is the lack of adequate histone production that leads to the published phenotypes in these RNAi mutants (Smardon et al, 2000; Duchaine et al, 2006; Yigit et al, 2006; Nakamura et al, 2007; Claycomb et al, 2009; She et al, 2009).
Higher levels of histone expression in C. elegans correlate well with the presence of actively dividing cells, with almost all histone expression in adult worms taking place in the germline (Keall et al, 2007). We observe the highest levels of CDL-1 (SLBP) expression in the oocytes and early embryos, consistent with the need for histone production during early development. Importantly, CSR-1 is also highly enriched in the oocytes and its depletion in the germline phenocopies the depletion of CDL-1.
As mentioned earlier, one possibility for the function of RNAi in histone processing is a direct role of CSR-1 in the endonucleolytic cleavage of histone 3′UTRs since it has been shown to possess endonuclease cleavage activity in vitro (Aoki et al, 2007). Our findings of the endo-siRNA-dependent binding of CSR-1 to a 2′-O-methyl RNA oligonucleotide identical to the region of the 3′UTR in between the stem-loop and the PAS strongly support this possibility. However, we cannot exclude that the RNAi machinery serves to recruit other cleavage factors to process the histone mRNA. Overall, our results and the existing published data (Mangone et al, 2010; Jan et al, 2011) are consistent with a sequential model of histone mRNA processing where the messages are initially polyadenylated, used by the RdRP EGO-1 as templates for antisense endo-siRNA production, and finally cleaved by an endonuclease, guided by ego-1-dependent endo-siRNAs matching the 3′UTR between the stem-loop and the PAS.
The production of the polyadenylated histone pre-mRNA intermediate could help in the recruitment of factors required for further processing or, alternatively, could attract proteins promoting pre-mRNA degradation in the oocytes, such that only properly processed messages ready for translation are deposited into the zygote and early embryo.
It is possible that regulation of histone genes by CSR-1-bound endo-siRNAs includes both transcriptional control and a direct role in histone mRNA processing. The transcriptional role could be served by siRNAs antisense to the coding regions while the processing role fits siRNAs matching the 3′UTR sequences in the model described earlier. Also, there are two isoforms of the CSR-1 protein (Claycomb et al, 2009), consistent with its diverse functions.
In conclusion, our study has revealed a novel positive role of RNAi in histone mRNA processing and demonstrated that the severe developmental phenotypes of the RNAi pathway mutants in C. elegans are due to insufficient production of core histone proteins.
Materials and methods
C. elegans strains
Worms were cultured according to Brenner (1974). Strains used in this study are GR1373: eri-1(mg366) IV, WM216: avr-14(ad1302) drh-3(tm1217) I/hT2 [bli-4(e937) let-?(q782) qIs48(myo-2::gfp; pes-10::gfp; ges-1::gfp)] (I; III); avr-15(ad1051) glc-1(pk54) V, DA1316: avr-14(ad1302) I; avr-15(ad1051) glc-1(pk54) V, AGK482: unc-119(ed3) III; eri-1(mg366) IV, AGK458: unc-119(ed3) III; eri-1(mg366) IV; armEx149(his-59,60,61,62; unc-119(+)), AGK461: unc-119(ed3) III; eri-1(mg366) IV; armEx152(his-59,60,61,62; unc-119(+)), AGK516: unc-119(ed3) III; eri-1(mg366) IV; armEx180(his-59,60,61(PAS),62(PAS); unc-119(+)), AGK520: unc-119(ed3) III; eri-1(mg366) IV; armEx184(his-59,60,61(PAS),62(PAS); unc-119(+)), and AGK537: unc-119(ed3) III; armEx199(pcdl-1::CDL-1::GFP; unc-119(+)).
Bioinformatics
Gene expression profile for all genes, CSR-1 targets and core histones in csr-1(tm892) compared to wild type (Figure 1A–C) was constructed from the published microarray data (Claycomb et al, 2009). CSR-1 targets (identified in Claycomb et al, 2009) were classified based on endo-siRNA enrichment in CSR-1-IP small RNA library compared to wild-type input library (Figure 1D). Endo-siRNA sequences used in Figure 1F are from Claycomb et al (2009).
RNA interference
RNAi feeding experiments were all done at 20°C in eri-1(mg366) sensitized background. RNAi was done by feeding on large NGM plates (150 mm diameter) in order to obtain sufficient amount of worms for RNA extraction, staining and western blotting from the same experiment. We used the following clones from the C. elegans feeding library (Kamath et al, 2001): cdl-1 (JA: R06F6.1), csr-1 (JA: F20D12.1), ego-1 (JA: F26A3.3), smc-4 (JA: F35G12.8), and pos-1 (JA:F52E1.1). Bacteria containing each RNAi clone were cultured in Luria Broth (LB) containing 50 μg/ml ampicillin at 37°C for 8–14 h in 350 ml. The cultures were then spun at 4000, r.p.m. (Beckman J2-21) and resuspended in 3.5 ml of LB. In all, 1 ml of the concentrated RNAi food was then spread on a large NGM plate containing 1 mM IPTG and 50 μg/ml ampicillin and left to dry. To perform the RNAi, ∼20 000 synchronized L1 worms were placed on each plate of the RNAi media and allowed to grow for 60–70 h at 20°C. Worms were collected as young adults prior to the formation of eggs for each type of analysis: western blotting, RNA extraction and immunostaining.
RNA extraction and quantitative RT–PCR
A phenol-chloroform extraction and ethanol precipitation were performed to extract RNA using TRI Reagent (MRC), followed by a DNase (Ambion) treatment at 37°C for 1 h. cDNA was prepared using RevertAid (Fermentas) reverse transcriptase, oligo dTs and random hexamers at 42°C for 1 h. qPCR was performed on cDNA on the Eppendorf Mastercycler ep realplex thermal cycler using Quantifast SYBR Green PCR Kit (Qiagen). Thermocycling was done for 50 cycles in a 25-μl reaction containing 12.5 μl SYBR Green, 0.15 μl of 100 μM forward and reverse primers (Supplementary Table S1), 5 μl cDNA and 7.2 μl dH2O. Primers were designed to amplify either processed or unprocessed cDNA based on the respective 3′UTRs for each histone type. Each experiment was performed in triplicate for each biological replica using Actin as a normalizing standard. mRNA expression levels were analysed using the comparative Ct method.
Western blotting
Synchronized worm populations were sonicated in 1 M TSE solution to ensure extraction of all histone proteins at 20% output for 30 s on, 30 s off for eight rounds using the Branson 500 Sonic Dismembrator. Samples were then centrifuged to remove cell debris at 4°C, 12 000 r.p.m. (Eppendorf AG 5424) and total protein content was quantified using the Bradford Dye Reagent (Bio-Rad). Equal amounts of total lysate for each sample were then resolved via SDS–PAGE on a 12% gel (Invitrogen NuPAGE Bis-Tris) and transferred onto a 2-μm nitrocellulose membrane (Thermo Scientific) at 100 mA for 30 min. The membrane was blocked using 3% (w/v) BSA in PBS with 0.01% Tween for 1 h, followed by overnight incubation at 4°C with one of the following antibodies: anti-H2B (abcam ab1790), anti-H2A (abcam ab13923), anti-H3 (Millipore 05-928), anti-H4 (Millipore 07-108 or abcam ab7311), anti-CSR-1 (Claycomb et al, 2009) and anti-actin (Millipore MAB1501R) diluted 1:2000 in PBST-3% BSA. The membrane was then washed three times with PBST and incubated for 1 h at room temperature with either anti-rabbit or anti-mouse HRP-conjugated secondary antibody (Perkin-Elmer) diluted 1:5000 in PBST-3% BSA and visualized by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) using a Series 2000A Film Processor (TIBA). Results shown are representative of at least three biological replicates.
Immunostaining
Young adult worms were handpicked and placed in PBS (137 mM NaCl, 10 mM Phosphate, 2.7 mM KCl, pH 7.4) with 0.01% Tween and 25 mM sodium azide in a glass petri dish. Worms were dissected using a scalpel and fixed with 2% paraformaldehyde for 1 h at room temperature and post-fixed in ice-cold methanol for 5 min. Worms were transferred to glass tubes and blocked with 3% (w/v) BSA in PBS. Blocked worms were then incubated overnight at 4°C with anti-H2B antibody (abcam ab1790) diluted 1:8000 or anti-PRG-1 (Batista et al, 2008) diluted 1:8000 or anti-CSR-1 (Aoki et al, 2007) diluted 1:4000, washed three times with PBST and incubated for 1 h at room temperature with anti-rabbit Alexa Fluor 555 (Molecular Probes) diluted 1:300. Worms were then mounted on polylysine slides (LabScientific, Inc) using Mounting solution with DAPI (Vector Laboratories) and all images were taken at × 20 on Zeiss AxioImager Z1 with 50 ms exposure for red channels, and 10 ms exposure for blue channels. Results shown are representative of at least three biological replicates.
Transgene plasmid construction
DNA sequence containing the promoter, coding region and 3′UTR (including the stem-loop) of his-61 and his-62 genes was obtained from the PCR performed on C. elegans N2 genomic DNA with
5′seqGCCGCGGTCGACATAATTGTGGCCCTAAAGAGGGCCGTTGGGTTCGGTTGGGTTTTTAAGAAAAAAGTTC forward primer and
5′seqGATCAAGGATCCATATTTGTGGCCCTAAAGAGGGCCGTTGGGTTCGGTTGGTTAGTTTTCAGCCAATGGC reverse primer.
pPD95.75 (Addgene) plasmid and the his-61,62 PCR product were digested with BamHI and SalI restriction enzymes and ligated to form pPD95.75w/his-61,62 plasmid. DNA sequence containing the promoter, coding region and 3′UTR (including the stem-loop) of his-59 and his-60 genes was obtained from the PCR performed on C. elegans N2 genomic DNA with
5′seqGCCGCCGGTACCATATTTGTGGCCCATAAGAGGGCCGTTGGGTTCGGTAATTGGTTTTGATGAAAACAG forward primer and
5′seqAAGAGTGGGCCCTAAATTGTGGCCCTAAAGAGGGCCGTTGGGTTCGGTGAGTTTTGAGTTGAAGCTTAC reverse primer.
pPD95.75w/his-61,62 plasmid and the his-59,60 PCR product were digested with KpnI and ApaI restriction enzymes and ligated to form the final plasmid pPD95.75w/his-59,60,61,62.
To introduce the PAS sequence into the above plasmid, the QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used with the following primer pairs:
5′CAACTTGGAAATGAAATTTTATTGCATGCCTGCAGGTCG and
5′GTTGAACCTTTACTTTAAAATAACGTACGGACGTCCAGC for his-61
5′CCCCGGGATTGGCCAAAGGAAATAAAGGTATGTTTCGAATG and
5′GGGGCCCTAACCGGTTTCCTTTATTTCCATACAAAGCTTAC for his-62.
pPD95.75w/his-59,60,61,62 or pPD95.75w/his-59,60,61(PAS),62(PAS) was injected into AGK482: eri-1(mg366)IV; unc-119(ed3)III at 1 ng/μl, together with N2 genomic DNA (30 ng/μl) and pMM016B [unc-119(+)] (1 ng/μl). Transgenic lines were generated based on the rescue of unc-119 mutant phenotype. In the rescued lines, transgene histone expression was quantified by RT-qPCR.
CDL-1::GFP was also created using the pPD95.75 (Addgene) plasmid. The endogenous promoter and coding region were amplified from N2 genomic DNA using the following primers:
5′CATCCCGGGGACGTCCATCTCAACTCGCAATC forward
5′GTAGGTACCCTCTTGTCATCGTCATCTTTATAATCGTGCGACGACATCTTGGAGAAG reverse, including a FLAG tag in frame.
The 3.5-kb insert was then digested with SmaI and KpnI restriction enzymes and ligated into a similarly digested pPD95.75. The resultant CDL-1::GFP plasmid was co-bombarded with the plasmid pMM016b (AddGene) for unc-119(ed3)III rescue using microparticle bombardment with a PDS-1000 Hepta Apparatus (Bio-Rad). Transgenic lines were selected by unc-119 phenotypic rescue. The germline expressing transgenic line containing transgene armEx199 is called AGK537.
Statistics
Statistical significance between all genes and CSR-1 targets in Figure 1A and B was calculated based on the Z-test of two proportions. Statistical significance for the ratio of the change in total to the change in unprocessed histone mRNA levels for all core histones relative to wild type in Figure 3G was performed based on one sample t-test with the hypothetical mean of 1. Statistical significance in Figures 6B and 7A was calculated based on z-test of two proportions. Standard error of proportion was calculated to give the error bars for rescue experiments.
2′-O-methyl RNA pull-down experiments
Pull-down experiments were conducted according to Hutvágner et al (2004). The only modification to the existing protocol was to incubate two antisense oligonucleotides for the pull down of the mRNA as opposed to a direct complement to the desired small RNA itself. The results were analysed by western blot. The following oligonucleotides were used:
H2A UTR: 5′GGCCACAAUUAUCUGAAAUCCUAACUAUCAAUUG
Unrelated sequence: 5′UCUUUACGCUAACAACUUGGAAAUGAAAUAAGCU
H2A antisense 1: 5′UCUUGGCUUUGCCUCCCUUUCCACGUCCAGACAU
H2A antisense 2: 5′AUUGGAGUCCGGCUCUUGAUGAGCGGGACUUGGC.
Supplementary Material
Acknowledgments
We thank X Zhou for help in generating transgenic lines and I Greenwald for a generous offer of technical assistance by her staff. We thank members of the Grishok lab and J Canman for helpful discussions; S Nicholis for technical assistance; S Tisdale and L Pellizzoni for sharing antibodies and helpful suggestions; P Sharp and C Sarkar for critical reading of the manuscript; W Gu for deep sequencing data sets and suggestions; H Tabara for sharing CSR-1 antibodies; C Mello, D Conte and P Batista for strains and reagents. Some strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by National Institutes of Health National Center for Research Resources. This work was supported by NIH Director’s New Innovator Award 1DP2OD006412-01 to AG and long-term NIH Training Grant (DK07328) to DA.
Author contributions: AG conceived the project; DA, SP, and AG designed the experiments; DA performed pull-down experiments, CDL-1::GFP construction, western blotting, and immunostaining; SP performed bioinformatics; DA and SP performed RNAi experiments; SP and YS performed RT–qPCR and histone transgene construction for rescue experiments; DA, SP, YS, and AG analysed the data; DA, SP, and AG wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allard P, Yang Q, Marzluff WF, Clarke HJ (2005) The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Dev Biol 286: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H (2007) In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26: 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D Jr, Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC (2008) PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, Conte D Jr, Mello CC (2009) The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila Lopez M, Samuelsson T (2008) Early evolution of histone mRNA 3′ end processing. RNA 14: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR 3rd, Mello CC (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ (2006) U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 12: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Andrews S, Schaller A, Schumperli D, Gray NK, Muller B (2005) The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA 11: 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Hoersch S, Sharp PA (2008) RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc Natl Acad Sci USA 105: 20386–20391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, Chen CC, Chaves DA, Duan S, Kasschau KD, Fahlgren N, Yates JR 3rd, Mitani S, Carrington JC, Mello CC (2009) Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D (2009) 22G-RNAs in transposon silencing and centromere function. Mol Cell 36: 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, Simard MJ, Mello CC, Zamore PD (2004) Sequence-specific inhibition of small RNA function. PLoS Biol 2: E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Hino K, Kitao S, Yokoi T, Hirose T (2009) Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA 15: 1578–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP (2011) Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keall R, Whitelaw S, Pettitt J, Muller B (2007) Histone gene expression and histone mRNA 3′ end structure in Caenorhabditis elegans. BMC Mol Biol 8: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL (2005) Germline proliferation and its control. WormBook Aug 15: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y, Rothman JH, Sugimoto A, Yamamoto M (2002) The stem-loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development 129: 187–196 [DOI] [PubMed] [Google Scholar]
- Lejeune E, Allshire RC (2011) Common ground: small RNA programming and chromatin modifications. Curr Opin Cell Biol 23: 258–265 [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, Attie O, Chen K, Salehi-Ashtiani K, Vidal M, Harkins TT, Bouffard P, Suzuki Y, Sugano S, Kohara Y, Rajewsky N et al. (2010) The landscape of C. elegans 3′UTRs. Science 329: 432–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ (2011) EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol 21: 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ (2008) Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 9: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD (2007) The C. elegans intestine. WormBook Mar 27: 1–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Ando R, Nakazawa T, Yudazono T, Tsutsumi N, Hatanaka N, Ohgake T, Hanaoka F, Eki T (2007) Dicer-related drh-3 gene functions in germ-line development by maintenance of chromosomal integrity in Caenorhabditis elegans. Genes Cells 12: 997–1010 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Pettitt J, Crombie C, Schumperli D, Muller B (2002) The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J Cell Sci 115: 857–866 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP (2006) Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- She X, Xu X, Fedotov A, Kelly WG, Maine EM (2009) Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet 5: e1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y (2011) Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol 10: 169–178 [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R et al. (2005) Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434: 462–469 [DOI] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, Reinke V, Petrella L, Strome S, Von Stetina SE, Katz M, Shaham S, Ratsch G, Miller DM 3rd (2011) A spatial and temporal map of C. elegans gene expression. Genome Res 21: 325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ (2001) Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev 15: 173–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelie A, Pascal G, Angulo L, Perreau C, Papillier P, Dalbies-Tran R (2012) An oocyte-preferential histone mRNA stem-loop-binding protein like is expressed in several mammalian species. Mol Reprod Dev 79: 380–391 [DOI] [PubMed] [Google Scholar]
- Updike DL, Strome S (2009) A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 183: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Berkow A, Marzluff WF (2005) Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem Soc Trans 33: 471–473 [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC (2006) Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.