Abstract
Genome-wide association studies (GWAS) have become an important method for mapping the genetic loci underlying complex phenotypic traits in many species. A crucial issue when performing GWAS is to control for the underlying population structure because not doing so can lead to spurious associations. Population structure is a particularly important issue in nonhuman species since it is often difficult to control for population structure during the study design phase, requiring population structure to be corrected statistically after the data have been collected. It has not yet been established if GWAS is a feasible approach in Saccharomyces cerevisiae, an important model organism and agricultural species. We thus performed an empirical study of statistical methods for controlling for population structure in GWAS using a set of 201 phenotypic traits measured in multiple resequenced strains of S. cerevisiae. We complemented our analysis of real data with an extensive set of simulations. Our main result is that a mixed linear model using the local ancestry of the strain as a covariate is effective at controlling for population structure, consistent with the mosaic structure of many S. cerevisiae strains. We further studied the evolutionary forces acting on the GWAS SNPs and found that SNPs associated with variation in phenotypic traits are enriched for low minor allele frequencies, consistent with the action of negative selection on these SNPs. Despite the effectiveness of local ancestry correction, GWAS remains challenging in highly structured populations, such as S. cerevisiae. Nonetheless, we found that, even after correcting for population structure, there is still sufficient statistical power to recover biologically meaningful associations.
Keywords: genome-wide association studies, Saccharomyces cerevisiae, mixed linear model, population structure, local ancestry
A key concern in biology is understanding the nature of the nucleotides underlying the variation in complex phenotypic traits among individuals. Recent breakthroughs in genotyping and sequencing technologies have facilitated an explosion of genome-wide association studies (GWAS) resulting in the discovery of many new associations of SNPs with complex traits, especially in humans (Hindorff et al. 2009). GWAS is also an increasingly important mapping approach in other nonhuman species, including plant species such as maize, rice, and Arabidopsis (reviewed in Brachi et al. 2011), mouse (Payseur and Place 2007), Drosophila (Mackay et al. 2012), and dog (Tsai et al. 2012). However, the feasibility of GWAS in Saccharomyces cerevisiae, an important model organism and agricultural species, has not yet been fully studied.
A critical issue that needs to be addressed in any GWAS is the presence of underlying population structure in the individuals studied, since population structure generally leads to spurious associations (Price et al. 2010). Accordingly, a number of statistical techniques have been proposed to correct for population structure, including genomic control, which corrects the GWAS test statistics by a variance inflation factor that is constant over all SNPs (Devlin and Roeder 1999; Reich and Goldstein 2001) and structured association (Pritchard et al. 2000b). An elegant approach is to use a linear model with ancestry components estimated from principal components analysis (Price et al. 2006) or mixture models (Pritchard et al. 2000a) as covariates. Other statistical approaches are also possible when there is pedigree structure underlying the data (Thornton and McPeek 2010).
Here we explored the feasibility of using GWAS to map complex traits in S. cerevisiae. We used whole-genome resequencing data from multiple S. cerevisiae strains (Liti et al. 2009) and data for 201 phenotypic traits for a significant fraction of these strains (Warringer et al. 2011). Previous population genetic studies of S. cerevisiae have made it clear that there is considerable population structure in this species (Liti et al. 2009; Schacherer et al. 2009; Elyashiv et al. 2010). We took an empirical approach to comparing different statistical methods for correcting for population structure in GWAS. Specifically, we assessed the performance of each statistical method by the shape of the quantile-quantile (Q-Q) plot of observed vs. expected P-values. Throughout our analysis we considered only statistical methods based on linear models since these allow the use of population structure data as covariates in a natural and flexible way. We also complemented our empirical study with an extensive set of simulations to assess the performance of the different GWAS methods.
Materials and Methods
Parameters for the program STRUCTURE
To study the population structure in the S. cerevisiae strains, we used the program STRUCTURE, which uses a Markov Chain Monte Carlo approach to assign individuals to ancestral populations and simultaneously infer the allele frequencies of the ancestral populations (Pritchard et al. 2000a; Falush et al. 2003). The parameters for the preliminary short run of STRUCTURE used for SNP selection (see Methods for SNP Selection) were default parameters except for PLOIDY = 1; BURNIN = 5000; NUMREPS = 5000; LINKAGE = 1; ADMBURNIN = 2500; and SITEBYSITE = 1. The long STRUCTURE runs used for the rest of the analysis used the same parameters except for BURNIN = 50,000; NUMREPS = 50,000; and ADMBURNIN = 25,000.
Methods for SNP selection
As described in the main text (in Selecting SNPs for population structure analysis), it is important to remove SNPs in high linkage disequilibrium (LD) because STRUCTURE does not model background LD. To do this, we implemented two procedures for selecting an independent set of SNPs: a “sliding-window” procedure and a “sequential” procedure.
Sliding-window procedure:
We implemented a sliding-window procedure in which we selected a small number of SNPs for each window of N consecutive SNPs. To select the SNPs, we calculated the LD, measured by D′2 for each pair of SNPs in the window. If the LD was above a threshold, the SNPs with more missing data were removed, with ties broken randomly. We tested window sizes of N = 10–100 consecutive SNPs in increments of 10 and D′2 thresholds of 0.1 to D = 0.9 in increments of 0.1. These parameters resulted in ∼1700–15,500 SNPs selected out of a total of 150,077 SNPs. Most windows contained one SNP, resulting in a roughly uniform distribution of SNPs across the genome. The window size N was the main determinant of the number of SNPs selected and the threshold D′2 had a relatively small effect on the choice of SNPs (data not shown). We chose a final window size of N = 30 as the number of SNPs chosen at this window size was approximately the average of all trials (data not shown). Instead of removing the SNP with more missing data, we also implemented several other procedures, including removing a random SNP, choosing the SNP with the higher minor allele frequency (MAF), and choosing the SNP that was more differentiated among the ancestral populations, as previously described (Liti et al. 2009). However, the set of SNPs selected was essentially unchanged (data not shown).
Sequential SNP selection procedure:
We implemented a sequential SNP selection procedure in which we identified LD blocks along a chromosome and kept one SNP per block. Starting with the first SNP in a chromosome (SNP A), we calculated LD between this SNP and the next SNP (SNP B). If SNP B was in high LD with SNP A (upper bound), the SNPs with less missing data were kept, with ties broken randomly. If SNP B was in LD with SNP A but not above a certain threshold (lower bound), the next SNP was considered and no change was made to SNP A. If SNP B was not in LD with SNP A, then we kept SNP A as the defining SNP for the previous LD block and let SNP B begin a new LD block. We varied the lower bound cutoff and the upper bound cutoff from D′2 = 0.05–0.95 in increments of 0.05 and found that the upper bound cutoff did not significantly impact the number of SNPs selected, while the lower bound cutoff had a significant impact, with more SNPs selected with a higher lower bound. This trend plateaued at a lower bound of 0.90 (given an upper bound of 0.95), with ∼5700 SNPs. We chose a D′2 cutoff for high LD of 0.95 and a D′2 cutoff for low LD of 0.16, resulting in a total of 3723 SNPs. We compared the distribution of SNPs chosen with a more relaxed lower bound of D′2 = 0.5 to the distribution of SNPs chosen with D′2 = 0.16 and found that both patterns were consistent (data not shown).
Methods for correcting for population structure in GWAS
A general linear model (LM) without covariates does not correct for population structure and therefore serves as a baseline statistical method for comparison with more sophisticated GWAS methods. Several other linear models are commonly used for population correction, including a general linear model with whole-genome ancestry estimates as covariates and a mixed linear model (MLM) with the kinship matrix as a random effect, both with and without whole-genome ancestry covariates. There are several existing programs that implement these methods in an efficient way for genome-scale studies, including TASSEL (Yu et al. 2006) and EMMAX (Kang et al. 2010). TASSEL performs both LM and MLM, while EMMAX performs only MLM. We implemented an LM both with and without covariates in R, and we used both EMMAX and TASSEL’s MLM algorithms. A summary of all methods used follows.
LM with covariates:
The following methods were used for LM with covariates:
Whole-genome ancestry covariates (obtained from STRUCTURE) implemented in R;
Local-ancestry covariates (obtained from STRUCTURE) implemented in R;
Both whole-genome ancestry and local ancestry covariates implemented in R.
MLM with covariates, implemented in both EMMAX and TASSEL:
The following methods were used for MLM with covariates, implemented in both EMMAX and TASSEL:
Kinship matrix only (kinship matrix computed by EMMAX-KIN from the EMMAX package);
Whole-genome ancestry covariates and kinship matrix, implemented in EMMAX and TASSEL;
Local ancestry covariates and kinship matrix, implemented in EMMAX;
Local ancestry covariates, whole-genome ancestry covariates, and kinship matrix, implemented in EMMAX.
We denote whole-genome ancestry covariates by Q, following the notation used in STRUCTURE (Pritchard et al. 2000a), local ancestry covariates as LA, and the kinship matrix as K. The statistical methods are summarized in Table 1.
Table 1. GWAS methods used in this study.
| Abbreviation | Statistical method | Covariates |
|---|---|---|
| R-LM | LM | None |
| R-Q | LM | Q |
| R-LA | LM | LA |
| R-LAQ | LM | LA + Q |
| EMMAX-K | MLM | K |
| EMMAX-QK | MLM | Q + K |
| TASSEL-K | MLM | K |
| TASSEL-QK | MLM | Q + K |
| EMMAX-KLA | MLM | LA + K |
| EMMAX-KLAQ | MLM | LA + K + Q |
Computing global ancestry
We estimated the global ancestry for each strain by running STRUCTURE with the 3723 SNPs selected by the sequential SNP selection method (see Methods for SNP selection). To determine the most likely number of ancestral populations, we first ran STRUCTURE (see short-run parameters in Parameters for the program STRUCTURE) by varying the value of K, the number of populations, between 3 and 8. Our results indicated that the most likely number of populations is six, consistent with previous studies (Liti et al. 2009; Schacherer et al. 2009). We performed a longer run of STRUCTURE using the linkage model with K = 6 to get a more refined analysis of the population structure (see long-run parameters in Parameters for the program STRUCTURE).
Computing local ancestry
STRUCTURE produces local ancestry estimates for each nucleotide based on a hidden Markov model (Falush et al. 2003). We removed SNPs in background LD using the sequential SNP selection scheme described above (see Methods for SNP selection). To verify the local ancestry estimates of STRUCTURE, we used the program WINPOP (Pasaniuc et al. 2009) to more carefully analyze the mosaic strains by using as ancestral populations the non-mosaic strains as determined by STRUCTURE, since we expect those lineages to be primarily of a single ancestry across the genome. For this purpose, the North American strains were grouped with the mosaic strains as previously determined by Liti et al. (2009), as they exhibit widespread mosaicism. We used a recombination rate of r = 3.5 × 10−6 following Ruderfer et al. (2006). While the number of generations since admixture is unknown, one estimate of the number of outcrossing events since the most recent common ancestor of two particular S. cerevisiae strains is 314 (Ruderfer et al. 2006), and the estimate from STRUCTURE was 28 generations. Therefore, we ran WINPOP for the following range of generations: 5, 10, 15, 28, 157, and 314. The first four values result in very similar patterns of ancestry while the last two values give very noisy results. We found that the local ancestry calls were significantly similar between WINPOP and STRUCTURE (data not shown) so we used the STRUCTURE local ancestry estimates for the remainder of the analysis.
Computing the kinship matrix
We computed a kinship matrix for use in the EMMAX and TASSEL MLMs using the EMMAX-KIN program. We used PLINK (Purcell et al. 2007) to convert .ped and .map files to EMMAX-readable .tped and .tmap files. These converted files were then input to EMMAX-KIN. We used the “identity-by-state” matrix option, following Zhao et al. (2007).
Identifying significant associations
We used two methods for multiple hypothesis testing correction. First, we used the Holm correction, which is a family-wise error rate (FWER) method, similar to the Bonferroni correction. Second, we used the Benjamini–Hochberg correction, which is a false discovery rate (FDR) correction. We used a threshold of 0.05 in both cases.
To identify genes corresponding to statistically significant SNPs, we used two methods. First, given a significant SNP, if the SNP fell within a gene, we counted the gene as significant. Otherwise, we treated both flanking genes as significant. Second, given a significant SNP, we considered the entire linkage block containing the SNP and all genes that overlapped the linkage block as significant. Linkage blocks were determined with a procedure similar to the sequential SNP selection method: starting with the SNP (SNP A), we computed pairwise LD between the SNP and the next SNP (SNP B). If LDs between SNP A and SNP B were above the threshold D′2 > 0.95 (this threshold was chosen to be consistent with that discussed in Methods for SNP selection), then we considered SNP B to be part of the LD block. We did the same for the SNPs before SNP A as well. We preferred the second method since the manner in which the SNPs were chosen to represent each LD block involved some randomness.
Simulations
In the first set of simulations, we chose one known phenotype (specifically, maltose 2% growth rate) from Warringer et al. (2011). We randomly chose a certain number of causal SNPs, n, from the 3723 SNPs used in our analysis. For each of the strains containing the major allele of each of the causal SNPs, we added a certain fixed effect, V, to the phenotypic value for the trait. We set n to [3, 20, 100] and we set V to [1, 3] to explore a wide range of genetic architectures. In addition, we performed one simulation with n = 3 and V = 10 to simulate the case of a few causal SNPs of large effect. After all the effects attributed to these causal SNPs were assigned, we normalized the phenotypic data by subtracting the mean and dividing by the standard deviation across all strains. The mean and variance for the original maltose 2% growth rate trait were 0.9347 and 0.5781, respectively.
For the second set of simulations, we used genotypic data from Schacherer et al. (2009), which contains a larger set of 63 strains. To make the simulations comparable to our previous analyses using the strains from Liti et al. (2009), we selected SNPs from the set of all 101,343 SNPs reported by Schacherer et al. (2009) using the same SNP selection procedure that we used to select SNPs from the strains from Liti et al. (2009). This procedure selected a total of 12,916 SNPs. To compute the local and global ancestry covariates, we ran STRUCTURE on this set of SNPs for K = 6 ancestral populations, which is consistent with the number of populations described by Schacherer et al. (2009) and also with our previous analysis of the strains from Liti et al. (2009). For the mixed linear models, we generated an identity-by-state kinship matrix based on the 12,916 SNPs selected using the EMMAX-KIN program. To simulate the phenotypic data, we first let phenotypes for all 63 strains be 0. Then we proceeded as with the first set of simulations by randomly selecting n causal SNPs and adding to the phenotypic values the phenotypic effect V. As before, we let n = [3, 20, 100] and V = [1, 3], with an additional simulation of n = 3 and V = 10 to simulate the case of a few causal SNPs with a very large effect.
For both types of simulations, 200 iterations were run for each pair of parameters, n and V. For each of these simulations, we ran all GWAS methods analyzed in our previous analyses. To compute the receiver operating characteristic (ROC) curves, for each pair of parameters and each simulation, we determined the ranks of the n planted SNPs and then took the average ranks over all simulated data sets. That is, we took the average rank of the highest-ranked planted SNP, and then we took the average rank of the next highest-ranked planted SNP, etc. Finally, we plotted these average ranks in the ROC curves.
Results
We obtained whole-genome resequencing data for 38 strains of S. cerevisiae (Liti et al. 2009) and processed the raw data as previously described (Chen et al. 2010). We obtained phenotypic data for 35 of the strains in 67 different environmental conditions (Warringer et al. 2011). There were three measurements per environment—growth rate (population doubling time), adaptation (proliferation lag), and efficiency (population density change)—resulting in 201 total phenotypes for each strain.
Selecting SNPs for population structure analysis
We investigated the population structure of the 35 S. cerevisiae strains using the program STRUCTURE (Pritchard et al. 2000a; Falush et al. 2003). An important issue that has not received much attention in the literature is how to select an appropriate set of SNPs when running STRUCTURE (Miclaus et al. 2009). STRUCTURE requires SNPs that are somewhat linked but not so tightly linked that there is significant background LD among the SNPs (i.e., LD present in the ancestral populations prior to admixture) (Falush et al. 2003). Indeed, we found that the results of STRUCTURE were sensitive to the choice of SNPs (Materials and Methods and Supporting Information, Figure S1).
We thus performed a comparison of SNP spacing produced by the two SNP selection procedures, sliding-window selection and sequential selection (Materials and Methods). We found that SNPs selected by the sliding-window procedure were more uniformly distributed across the genome while sequentially selected SNPs tended to form clusters (Figure S2). Sequential SNPs were chosen in a manner consistent with the recombination landscape of the genome (Mancera et al. 2008), with fewer SNPs chosen around the centromeres where recombination is low and more SNPs chosen around recombination hotspots, while windowed SNPs were more uniform across the genome (Figure S2). Thus we performed all further analyses with SNPs selected by the sequential method. When we set the LD cutoff to D′2 = 0.16, the sequential procedure resulted in a set of 3723 SNPs. To confirm the clustering result of STRUCTURE, we performed principal components analysis using the 3723 SNPs and found that the non-admixed strains found by STRUCTURE cluster together (Figure S3).
Preliminary analysis of the phenotypic data
We started by computing the variance of each phenotype across all strains. We also measured the degree of population structure underlying each phenotype by performing GWAS using a general linear model with no correction for population structure (Materials and Methods). One empirical way to ascertain the existence of population structure in GWAS is to plot the P-values produced by a statistical method as a Q-Q plot. A standard result states that, under the null hypothesis (in this case, the hypothesis of no population structure), the P-values are uniformly distributed. Since population structure is expected to produce spurious associations when in fact there are none, in the presence of population structure the observed P-values will tend to be smaller than expected. We defined the amount of population structure remaining after statistical correction by taking the mean squared distance (MSD) between the points generated by the Q-Q plot and the corresponding point on the line Y = X (i.e., the expected P-values).
The top 10 conditions that are most genetically structured as measured by the MSD statistic are: rapamycin 0.5 µg/ml adaptation; rapamycin 1 µg/ml adaptation; pH 3.5 adaptation; LiCl 150 mM efficiency; CuCl2 0.75 mM rate; CuCl2 0.375 mM rate;KCl 1.45 M rate; CoCl2 0.015 mM adaptation; maltose 2% rate; and LiCl 225 mM efficiency. The average phenotypic variance for these 10 conditions (0.77) was significantly higher than the average phenotypic variance for all 201 conditions (0.30; P-value 0.0036, 100,000 bootstrap replicates). This suggests that conditions with high levels of population structure (at the genotype level) also have more varied phenotypes. We repeated the analysis with a different measure of population stratification, the variance inflation factor (Devlin and Roeder 1999), instead of the MSD statistic and obtained a nearly identical result (data not shown).
There are a total of 67 types of environments, for example, environments with various levels of pH or glucose concentration. Five environment types were significantly structured compared to background (Table 2). We conclude that it is important to account for population structure when performing GWAS in S. cerevisiae.
Table 2. Environment types that are significantly structured compared to the background.
| Environment | P-value |
|---|---|
| Rapamycin | 0.0006 |
| CuCl2 | 0.0024 |
| LiCl | 0.026 |
| Kcl | 0.084 |
| pH | 0.095 |
The level of population structure was measured by the MSD statistic from the Q-Q plot produced by a general linear model.
Comparison of statistical methods for GWAS
To assess the performance of different statistical methods for correcting for population structure, we carried out GWAS analysis for all 201 phenotypes using the methods listed in Table 1 (Materials and Methods). We found a wide range of significant SNPs called by the different statistical methods, highlighting the importance of considering different statistical methods in GWAS (Table 3). We combined the best scoring method (according to the MSD statistic) for each phenotype into a meta-statistical method that we will refer to as the “BEST” method.
Table 3. SNPs found significant across all phenotypes after correction for FWER (Holm) and FDR (Benjamini-Hochberg), both at a threshold of 0.05.
| Including duplicates |
Unique SNPs |
|||
|---|---|---|---|---|
| Method | Holm | BH | Holm | BH |
| R-LM | 389 | 2662 | 99 | 1200 |
| R-Q | 249 | 1024 | 106 | 415 |
| EMMAX-K | 235 | 353 | 31 | 57 |
| EMMAX-QK | 68 | 97 | 14 | 29 |
| R-LA | 431 | 2154 | 220 | 891 |
| R-LAQ | 191 | 1261 | 107 | 723 |
| EMMAX-KLA | 637 | 1477 | 65 | 171 |
| EMMAX-KLAQ | 90 | 144 | 17 | 44 |
| BEST | 162 | 359 | 63 | 177 |
The multiple testing corrections were applied to each condition separately, not to all 201 phenotypes all together. SNPs that are found to be significant in multiple phenotypes are included multiple times under the “Including duplicates” but only once under the “Unique SNPs.” BH, Benjamini-Hochberg.
Some SNPs were found to be statistically significant in multiple phenotypes, and we call these “pleiotropic SNPs.” When removing pleiotropic SNPs, we still found a wide range of numbers of SNPs found by the different statistical methods (Table 3). The number of GWAS loci detected under the Holm correction, a commonly used Bonferroni-type multiple testing correction, was small (fewer than two per phenotype for the general linear model). This is consistent with the power of the study based on the number of strains, and we discuss this point at greater length in the Discussion.
We compared the P-values produced by the different statistical methods (Table 4) and found a strong Pearson correlation between the MLM methods implemented in EMMAX and TASSEL when only the kinship matrix K was used (R = 0.82). However, this correlation was much lower with the addition of the global ancestry covariate Q (R = 0.25). Additionally, we found that the methods R-LA and R-LAQ both exhibited relatively low correlations to the other methods. Surprisingly, R-LM, which used no covariates for correction, had a fairly high correlation with EMMAX-K. We are not sure why the EMMAX and TASSEL implementations of the MLM differ. To the best of our knowledge, the only difference between the two programs is the implementation of the compression method, and this might be the reason for the different results.
Table 4. Correlation coefficients between the P-values produced by different GWAS methods.
| R-LM | R-Q | EMMAX-K | EMMAX-QK | EMMAX-KLA | EMMAX-KLAQ | TASSEL-K | TASSEL-QK | R-LA | R-LAQ | |
|---|---|---|---|---|---|---|---|---|---|---|
| R-LM | 1 | 0.1684 | 0.7341 | 0.2024 | 0.0753 | 0.0936 | 0.6397 | 0.1460 | 0.0850 | 0.0566 |
| R-Q | 1 | 0.2540 | 0.2847 | 0.1686 | 0.0557 | 0.3073 | 0.8103 | 0.2886 | 0.0989 | |
| EMMAX-K | 1 | 0.3338 | 0.1374 | 0.1241 | 0.8153 | 0.2603 | 0.0977 | 0.0619 | ||
| EMMAX-QK | 1 | 0.2493 | 0.4173 | 0.2888 | 0.2542 | 0.1190 | 0.0492 | |||
| EMMAX-KLA | 1 | 0.2683 | 0.1084 | 0.1289 | 0.2208 | 0.1027 | ||||
| EMMAX-KLAQ | 1 | 0.0869 | 0.0408 | 0.0849 | 0.0738 | |||||
| TASSEL-K | 1 | 0.3425 | 0.1039 | 0.0630 | ||||||
| TASSEL-QK | 1 | 0.2476 | 0.0947 | |||||||
| R-LA | 1 | 0.2654 | ||||||||
| R-LAQ | 1 |
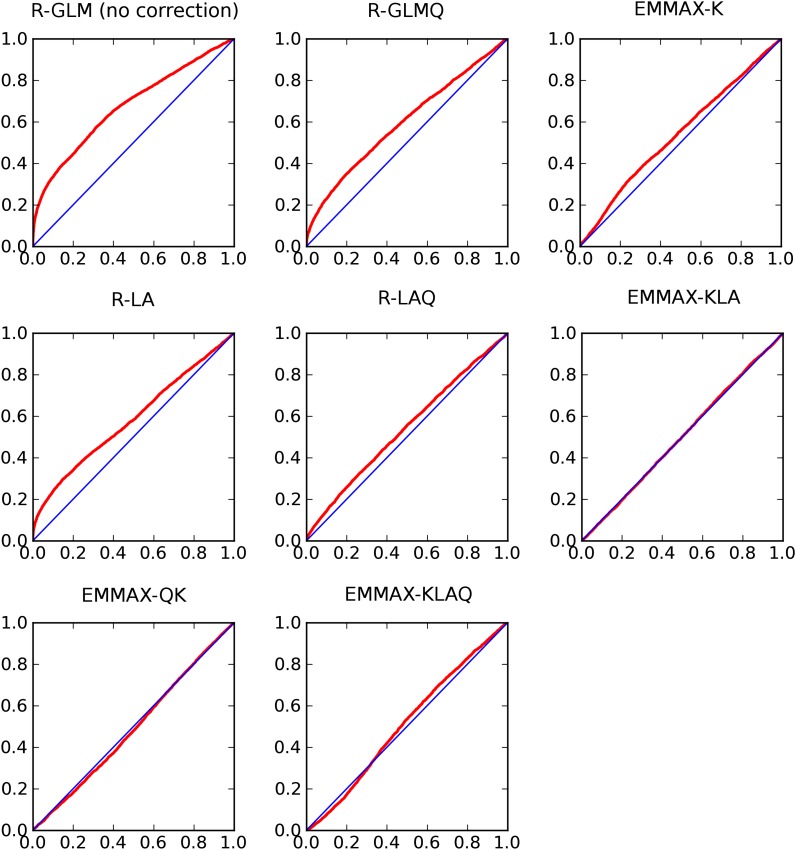
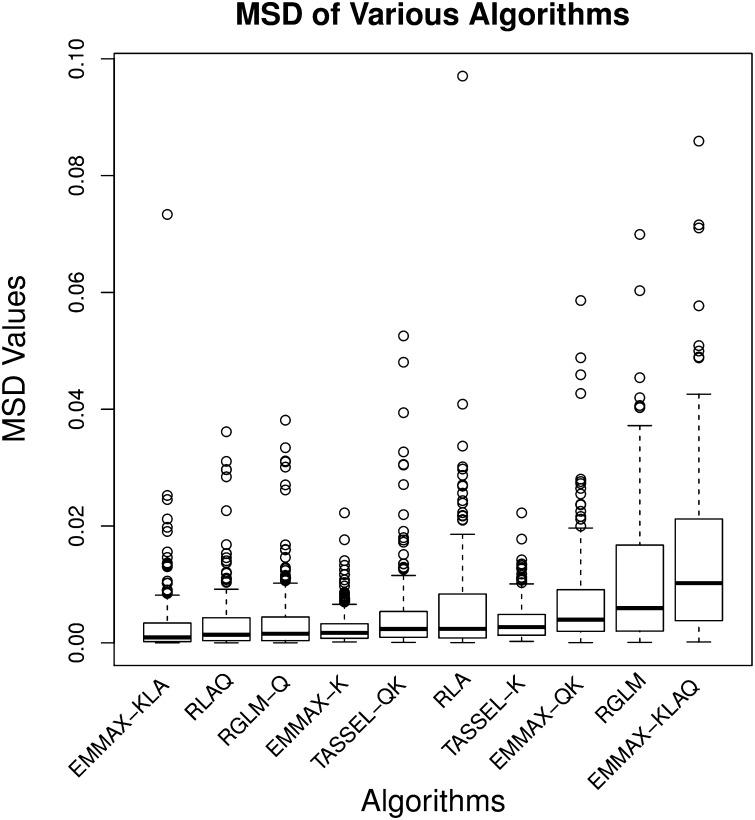
For each GWAS method and phenotype, we generated a Q-Q plot (Figure 1). Using the MSD statistic, we found that the EMMAX-KLA method was the most effective overall at correcting for population structure, followed by R-LAQ (Table 5; Table S1; Figure 2). Our data highlight the importance of correcting for the local ancestry, not just the global ancestry, of a strain when performing GWAS.
Figure 1.
Q-Q plots for the different algorithms for correcting for population structure. For each plot, the Y-axis shows the expected P-value and the X-axis shows the observed P-value.
Table 5. Statistical method and phenotypes where it performed best.
| Statistical method | No. of phenotypes where method performed best |
|---|---|
| R-LM | 11 |
| R-Q | 22 |
| R-LA | 14 |
| R-LAQ | 49 |
| EMMAX-K | 36 |
| EMMAX-QK | 5 |
| EMMAX-KLA | 62 |
| EMMAX-KLAQ | 2 |
Boldface type indicates the method which performed the best over all conditions.
Figure 2.
Mean Square Distance (MSD) score for each algorithm tested.
The results from the GWAS analysis reported above used the set of 3723 SNPs produced by our sequential SNP selection procedure (Materials and Methods). While removing SNPs in high LD is not required for GWAS, it reduces the multiple hypothesis testing burden while retaining most of the statistical power to detect associations; a similar strategy is used when selecting tag SNPs in human GWAS. To test the robustness of our choice of SNPs for the GWAS analysis, we selected an expanded set of 15,812 SNPs by choosing 1 SNP for each window of 10 SNPs. We then performed GWAS with the linear model with no covariates for the six copper-tolerance phenotypes. While using the expanded set of SNPs resulted in more SNPs declared significant, the end result was similar because of the higher multiple testing burden for the expanded set of SNPs (data not shown).
We repeated our analyses using the variance inflation factor (VIF) (Devlin and Roeder 1999) as a measure of the degree of population stratification instead of the MSD statistic. We note that the mean of the test statistics has been proposed as a good estimator for the variance inflation factor (Reich and Goldstein 2001; Devlin et al. 2004), so our MSD statistic has some basis in formal statistical theory. Overall, EMMAX-KLA still performed better than the other GWAS methods under the VIF statistic (data not shown). We also checked the genes found statistically significant by the “BEST” method where we now have two “BEST” methods: “BEST-VIF” and “BEST-MSD.” After FDR multiple testing correction, “BEST-MSD” found 58 conditions with significant genes while “BEST-VIF” found 48. Under about half the conditions, there were identical sets of significant genes. We observed very similar results for the Holm correction (data not shown). Thus our use of the MSD statistic did not affect our overall conclusions.
Functional analysis of GWAS SNPs
For the remaining analyses, we took the best correction method for each phenotype and used the P-values generated by that method. We refer to this meta-statistical method as the “BEST” method. To investigate the functional significance of the statistically significant SNPs, we examined the fraction of significant SNPs contained in genes. We found that 75% of SNPs called statistically significant under the FDR correction were in genes compared to 63% of all SNPs used in our analysis (P-value < 1e-4).
We also examined the biological functions of the GWAS SNPs using the GO term enrichment program FuncAssociate (Berriz et al. 2003). We found several interesting enriched functions, including biotin biosynthesis for the phenotype pH 3.5 adaptation (Fisher’s exact test, P-value 0.001). Because of the small number of SNPs, most functions did not reach statistical significance. Nonetheless, among the uncorrected P-values, we observed many suggestive functions, such as glucoside transport for the phenotype, glucose 8% efficiency (Fisher’s exact test, uncorrected P-value 0.003), and oligosaccharide metabolic process for the phenotype glucose 0.5% rate (Fisher’s exact test, uncorrected P-value 0.003). Although these functional results are preliminary, they suggest that more highly powered GWAS in S. cerevisiae may be able to elucidate important biochemical pathways.
Next we followed up on several associations previously detected by Cubillos et al. (2011) and Warringer et al. (2011). These researchers studied four broad phenotypes and their associated genes: copper tolerance, associated with CUP1/2; NaCl and LiCl tolerance, associated with ENA1/2/5; galactose growth, associated with GAL1/2/3; and maltose growth, associated with MAL31/32/33. To determine if the GWAS methods that we tested discovered the previously published associations, for each condition, gene, and GWAS method, we searched for all SNPs that were nominally significant at a P-value threshold of 0.05 in the vicinity of the relevant gene(s).
We computed how many relevant SNPs that each GWAS method found for all four reported associations and the percentage of SNPs found compared to the total number of nominally significant SNPs found by each algorithm (data not shown). The closest SNP to CUP1 and CUP2 (chr08:214751) was found by EMMAX-QK and LM-Q, while the next closest SNP (chr08:221695) was found by EMMAX-KLAQ and LM. For NaCl tolerance, four SNPs that fell within the ENA1 gene were discovered by several GWAS methods. Similarly for LiCl tolerance, four SNPs were discovered, three of which fell within ENA1, and one of which fell 1743 bp downstream of ENA5 (chr04:525679). The GWAS methods combined also discovered four SNPs associated with the maltose growth environments, all of which were located within MAL31. No significant SNPs were found in or near GAL1/2/3 by any of the GWAS methods only because there were few SNPs in our set near these genes (data not shown). We conclude that the GWAS methods are often able to recover previously known associations.
Evolutionary analysis of GWAS SNPs
It is also important to understand the nature of the evolutionary forces acting on SNPs that affect phenotypic variation. To address this issue, we considered the distributions of minor allele frequencies of the 3723 SNPs used in our analysis and all intergenic SNPs and compared them to the distribution of minor allele frequencies of the SNPs that were found to be statistically significant by the GWAS methods. We did not attempt to root the SNPs to obtain derived allele frequencies, similar to a previous study (Chen et al. 2010).
Overall, significantly associated SNPs were highly enriched for rare alleles compared to either the 3723 SNPs used in our analysis or all intergenic SNPs (Table 6). These results were robust whether we used an FWER or an FDR multiple testing correction method and whether we considered nonpleiotopic or pleiotropic SNPs. Note that in general GWAS methods have more statistical power for SNPs with higher MAF, so our tests were conservative because they showed that GWAS SNPs were nonetheless enriched in lower MAF. Also, the strain sampling and SNP selection procedures should not bias our result because all sets of SNPs should be equally affected.
Table 6. Minor allele frequencies for nonpleoitopic and pleiotropic SNPs found by the BEST method.
| Type of SNP | Multiple testing correction | P-value vs. GWAS | P-value vs. intergenic | Mean MAF | No. of SNPs |
|---|---|---|---|---|---|
| Nonpleiotropic | Holm | 1.676e-15 | 0.0088 | 0.1329 | 63 |
| FDR | 1.251e-17 | 4.975e-85 | 0.1227 | 359 | |
| Pleiotropic | Holm | 0.0028 | 7.692e-07 | 0.0898 | 14 |
| FDR | 1.005e-12 | 0.0125 | 0.1228 | 47 |
The BEST method refers to the best method of correction for each condition separately. The P-values are from one-sided Wilcoxon tests. By comparison, the mean MAF for intergenic SNPs was 0.1684, and the mean MAF for GWAS SNPs was 0.1839.
Simulation study of S. cerevisiae GWAS
To complement our GWAS analysis on real data, we performed two sets of simulations over a wide range of genetic architectures: one set based on the existing phenotypic data from Warringer et al. (2011) and one set with purely simulated phenotypes (Materials and Methods). To determine the performance of each GWAS method, we plotted ROC curves to compare the average ranks of the randomly chosen causal SNPs (Materials and Methods). In general, all the GWAS methods performed better in the simulations with simpler genetic architectures (i.e., fewer causal SNPs of larger effect). All methods performed close to random in the simulations involving 100 SNPs (Figure S4), but they performed much better with three SNPs and large phenotypic value (Figure S5). The purely simulated phenotypes generally performed better than the phenotypes based in the existing phenotype (Figure S6), perhaps due to the larger sample size in the analyses involving the data set of Schacherer et al. (2009). These simulation results are consistent with the recent results of Connelly and Akey (2012).
In most cases, EMMAX-K performed the best in terms of the ROC curves, having a lower rate of false positives. To address why the methods based on local ancestry appear to be performing worse according to the ROC curves, we computed the variance of the ranks in the simulations. We observed that the methods based on local ancestry have higher variance (data not shown), presumably because when randomly selecting SNPs to be causal SNPs, some of the SNPs chosen were correlated with local ancestry so their effect was inadvertently corrected by the GWAS method. This raises an important broader point: while we have concentrated on trying to reduce false positive associations so far, any such method will necessarily also reduce the power of the statistical approach. Nonetheless, we have shown above that the statistical power of our local ancestry approach is not extremely low because it still is able to recover biologically meaningful associations previously published in Cubillos et al. (2011) and Warringer et al. (2011).
Discussion
GWAS have proven to be a highly effective way to map the genes underlying complex phenotypic traits in many species. In all applications of GWAS, it is crucial to control for underlying population structure, since it can cause spurious associations. Here we have performed an empirical study of statistical methods for correcting for population structure when performing GWAS in the important model organism, S. cerevisiae. Our main results are that GWAS is indeed a feasible approach in S. cerevisiae and that it is important to take into account the local ancestry of an S. cerevisiae strain when performing GWAS. At a practical level, the EMMAX mixed linear model implementation (Kang et al. 2010) using an identity-by-state kinship matrix as a random effect and local ancestry inferred by STRUCTURE (Falush et al. 2003) as a fixed effect performed best in our experiments. Importantly, our work also shows that existing methods for detecting local ancestry, such as STRUCTURE (Falush et al. 2003) and WINPOP (Pasaniuc et al. 2009), are effective in S. cerevisiae, at least for the purposes of GWAS. Nonetheless, the demographic history of S. cerevisiae is complex (Liti et al. 2009; Schacherer et al. 2009) and properly modeling it will probably require more specialized statistical methods than the methods designed for the comparatively simpler cases of recent punctate admixture in human populations, particularly Latinos and African-Americans (Verdu and Rosenberg 2011).
There are many differences between performing GWAS in S. cerevisiae and humans. First, the burden of multiple hypothesis testing correction is much lower in S. cerevisiae because it has a much smaller genome size. Our analysis used only 3723 SNPs compared to the ∼500,000 typically used in human GWAS studies. If a simple Bonferroni-type correction is used, we would expect an S. cerevisiae GWAS to be far more powerful than a comparable GWAS in humans. Second, the extent of linkage disequilibrium is much less in S. cerevisiae, so GWAS in S. cerevisiae is more likely to pinpoint the actual causal variant than in humans, where it is more likely to find an association with a tag SNP. The S. cerevisiae genome is also much more gene-rich than the human genome, so each significant SNP is easier to link to a putative causal gene than in the human case. Third, since it is relatively cost-effective to fully resequence S. cerevisiae genomes, we were able to use whole-genome resequencing data compared to the SNP genotyping chips still typically used in human GWAS studies (although continued decreases in sequencing cost may make whole-genome resequencing for humans feasible at some point in the future). Thus GWAS in S. cerevisiae can in principle test causal SNPs for association rather than tag SNPs. It has previously been shown that the power to detect association is much higher when testing the causal SNPs than when testing a tag SNP (Ohashi and Tokunaga 2001). Fourth, with S. cerevisiae it is possible to perform replicate phenotypic measurements to reduce the environmental noise. For all of these reasons, we believe that the power of GWAS in S. cerevisiae mitigates the relatively small sample sizes of individuals used in our study. We also note that it is possible to study many environmental conditions in S. cerevisiae, such as drug treatments, which would be impossible or unethical to do in humans.
S. cerevisiae is an important model organism for many aspects of molecular biology. Recent work on mapping complex traits in this species using recombinant inbred lines has yielded many important insights (Ehrenreich et al. 2009). In addition to its use as a model organism, S. cerevisiae is also an important agricultural species in its own right. Thus we hope that the statistical methods for GWAS investigated here will lead to further advances in our understanding of the genotype–phenotype map in this important species. Our comparisons to previous mapping results in S. cerevisiae (Cubillos et al. 2011; Warringer et al. 2011) are promising in this regard. A recent study of GWAS in S. cerevisiae also found similar results to our study (Connelly and Akey 2012). In particular, they showed through simulations on the same set of S. cereivisae strains that GWAS in S. cerevisiae is generally difficult because of the complex population structure but is feasible for Mendelian trait and cis QTL mapping. One difference is that Connelly and Akey (2012) stressed the difficulties of GWAS in S. cerevisiae whereas we have stressed the relative utility of using local ancestry corrections in S. cerevisiae GWAS, while continuing to acknowledge the overall difficulty of using GWAS methods in this species. Nonetheless, our improved GWAS performance on the larger set of S. cerevisiae strains from Schacherer et al. (2009) suggests that increased sampling and sequencing of strains will improve GWAS results in the future. Such studies will be facilitated by the small size of the S. cerevisiae genome (∼12 Mb), the decreasing cost of DNA sequencing, and the relative tractability of high-throughput phenotyping in yeast (Ohya et al. 2005).
In addition, there are many other studies of GWAS in other model organisms that are similar to our study, including studies in mice (Payseur and Place 2007), Arabidopsis (Zhao et al. 2007), maize and rice (Brachi et al. 2011), tomato (Ranc et al. 2012), dog (Tsai et al. 2012), and Drosophila melanogaster (Mackay et al. 2012). Recent admixture is a pervasive phenomenon in many species. For example, there is strong evidence of non-African admixture in the DPGP D. melanogaster lines from Africa (J. Pool, unpublished results). GWAS in admixed human populations is also an important current research problem, and a very interesting goal for the future will be to combine admixture mapping with association mapping (Seldin et al. 2011; Shriner et al. 2011). Thus we believe that our results will also be useful for GWAS analyses in humans and other model systems as well.
Supplementary Material
Acknowledgments
We thank Steve Buyske for comments on the manuscript, Jonathan Flowers for advice on running the STRUCTURE program, David Gould for assistance with running the WINPOP program, and Jonas Warringer for giving us the yeast phenotype data. This work was partially funded by the National Institutes of Health (R00HG004515 to K.C.C.).
Footnotes
Communicating editor: S. Sen
Literature Cited
- Berriz G., King O., Bryant B., Sander C., Roth F., 2003. Characterizing gene sets with FuncAssociate. Bioinformatics 19: 2502–2504 [DOI] [PubMed] [Google Scholar]
- Brachi B., Morris G., Borevitz J., 2011. Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., van Nimwegen E., Rajewsky N., Siegal M., 2010. Correlating gene expression variation with cis-regulatory polymorphism in Saccharomyces cerevisiae. Genome Biol. Evol. 2: 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C., Akey J., 2012. On the prospects of whole-genome association mapping in Saccharomyces cerevisiae. Genetics 191: 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F., Billi E., Zorgo E., Parts L., Fargier P., et al. , 2011. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol. Ecol. 20: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Devlin B., Roeder K., 1999. Genomic control for association studies. Biometrics 55: 997–1004 [DOI] [PubMed] [Google Scholar]
- Devlin B., Bacanu S., Roeder K., 2004. Genomic control to the extreme. Nat. Genet. 36: 1129–1130 [DOI] [PubMed] [Google Scholar]
- Ehrenreich I., Gerke J., Kruglyak L., 2009. Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the BYxRM cross. Cold Spring Harb. Symp. Quant. Biol. 74: 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyashiv E., Bullaughey K., Sattath S., Rinott Y., Przeworski M., et al. , 2010. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res. 20: 1558–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff L., Sethupathy P., Junkins H., Ramos E., Mehta J., et al. , 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Sul J., Service S., Zaitlen N., Kong S., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S., 1979. A simple sequentially rejective multiple test procedure. Scand. J. Statist. 6(2): 65–70 [Google Scholar]
- Liti G., Carter D., Moses A., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T., Richards S., Stone E., Barbadilla A., Ayroles J., et al. , 2012. The Drosophila melanogaster Genetic Reference Panel. Nature 482: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miclaus K., Wolfinger R., Czika W., 2009. SNP selection and multidimensional scaling to quantify population structure. Genet. Epidemiol. 33: 488–496 [DOI] [PubMed] [Google Scholar]
- Ohashi J., Tokunaga K., 2001. The power of genome-wide association studies of complex disease genes: statistical limitations of indirect approaches using SNP markers. J. Hum. Genet. 46: 478–482 [DOI] [PubMed] [Google Scholar]
- Ohya Y., Sese J., Yukawa M., Sano F., Nakatani Y., et al. , 2005. High-dimensional and large-scale phenotyping of yeast mutants. Proc. Natl. Acad. Sci. USA 102: 19015–19020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B., Sankararaman S., Kimmel G., Halperin E., 2009. Inference of locus-specific ancestry in closely related populations. Bioinformatics 25: i213–i221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B., Place M., 2007. Prospects for association mapping in classical inbred mouse strains. Genetics 175: 1999–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A., Patterson N., Plenge R., Weinblatt M., Shadick N., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909 [DOI] [PubMed] [Google Scholar]
- Price A., Zaitlen N., Reich D., Patterson N., 2010. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 11: 459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J., Stephens M., Donnelly P., 2000a Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J., Stephens M., Rosenberg N., Donnelly P., 2000b Association mapping in structured populations. Am. J. Hum. Genet. 67: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranc N., Munos S., Xu J., Le Paslier M., Chauveau A., et al. , 2012. Genome-wide association mapping in tomato (Solanum lycopersicum) is possible using genome admixture of Solanum lycopersicum var. cerasiforme. G3: Genes, Genomes, and Genetics 2: 853–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Goldstein D., 2001. Detecting association in a case-control study while correcting for population stratification. Genet. Epidemiol. 20: 4–16 [DOI] [PubMed] [Google Scholar]
- Ruderfer D., Pratt S., Seidel H., Kruglyak L., 2006. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38: 1077–1081 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J., Ruderfer D., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin M., Pasaniuc B., Price A., 2011. New approaches to disease mapping in admixed populations. Nat. Rev. Genet. 12: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D., Adeyemo A., Rotimi C., 2011. Joint ancestry and association testing in admixed individuals. PLOS Comput. Biol. 7: e1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T., McPeek M., 2010. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am. J. Hum. Genet. 86: 172–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K., Noorai R., Starr-Moss A., Quignon P., Rinz C., et al. , 2012. Genome-wide association studies for multiple diseases of the German Shepherd dog. Mamm. Genome 23: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu P., Rosenberg N., 2011. A general mechanistic model for admixture histories of hybrid populations. Genetics 189: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J., Zorgo E., Cubillos F., Zia A., Gjuvsland A., et al. , 2011. Trait variation in yeast is defined by population history. PLoS Genet. 7: e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W., Vroh B., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhao K., Aranzana M., Kim S., Lister C., Shindo C., et al. , 2007. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.