Abstract
Exonucleases are key enzymes involved in many aspects of cellular metabolism and maintenance and are essential to genome stability, acting to cleave DNA from free ends. Exonucleases can act as proofreaders during DNA polymerisation in DNA replication, to remove unusual DNA structures that arise from problems with DNA replication fork progression, and they can be directly involved in repairing damaged DNA. Several exonucleases have been recently discovered, with potentially critical roles in genome stability and ageing. Here we discuss how both intrinsic and extrinsic exonuclease activities contribute to the fidelity of DNA polymerases in DNA replication. The action of exonucleases in processing DNA intermediates during normal and aberrant DNA replication is then assessed, as is the importance of exonucleases in repair of double-strand breaks and interstrand crosslinks. Finally we examine how exonucleases are involved in maintenance of mitochondrial genome stability. Throughout the review, we assess how nuclease mutation or loss predisposes to a range of clinical diseases and particularly ageing.
Keywords: Exonuclease, Aging, Ageing, WRN, FAN1, FEN1, EXOG, EXDL2, Mitochondria, Proofreading, DNA repair, DNA replication
Introduction
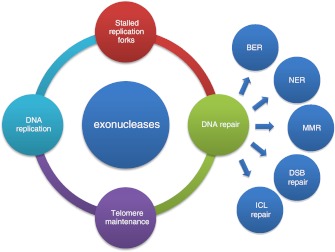
Maintenance of an intact genome is a key requisite both for evolutionary fitness and for health of the individual organism. While cells have evolved protective DNA packaging, such as eukaryotic chromatin, DNA is an active molecule involved in replication and transcription, and it can be modified by direct covalent changes such as CpG methylation during epigenetic regulation. It is also subject to mutation from both internal and external sources. Highly conserved surveillance and repair mechanisms have therefore evolved to keep pace with the daily insults that the genome undergoes. Most, if not all, of these mechanisms require cleavage of the DNA’s sugar–phosphate backbone in a controlled and accurate way by enzymes collectively called nucleases. Nucleases act in multiple pathways of DNA metabolism, including during the normal course of DNA replication, providing proofreading capacity to enhance polymerase fidelity, in mismatch repair (MMR), in tackling ends and unusual structures at stalled DNA replication forks, during repair of damaged bases (BER) or larger lesions (NER and recombinational repair) and in telomere maintenance (Fig. 1). Here, we describe the action of exonucleases that are most likely to be associated with DNA metabolism important in preventing premature ageing (Table 1).
Fig. 1.
DNA transactions involving exonucleases. Exonucleases are central to many processes of DNA metabolism. They are important in DNA replication in providing proofreading capacity both intrinsic to the replicative DNA polymerases δ and ε and extrinsic (e.g. ExoN) or autonomous proofreaders (e.g. TREX1 and 2). During the process of Okazaki fragment processing on the lagging strand, nucleases FEN1 and Dna2 (with RNaseH) are important for cleaving the primer and iDNA. Where replication forks stall or collapse, exonucleases are important in resolving aberrant DNA structures to permit fork restart. Such nucleases include the progeroid WRN protein and EXO1. Telomere maintenance also requires the action of exonucleases, possibly also using the same nucleases WRN and EXO1. DNA repair takes many forms depending on the lesion (on one or both strands); nucleases are involved in base excision repair (BER) (e.g. FEN1), in nucleotide excision repair, mismatch repair (e.g. EXO1) and in repair of DNA double-strand breaks (e.g. Mre11). Finally, repair of interstrand crosslinks (ICL) requires the action of a newly discovered nuclease FAN1, which is thought to be recruited by association with the Fanconi anaemia protein complex FANC ID (see “FAN1”)
Table 1.
Exonucleases discussed in this article
| Exonuclease | Polarity | Metabolic pathway | References |
|---|---|---|---|
| Polymerase δ, ε, γ | 3′-5′ | Replication (proofreading) | Hubscher et al. 2002; Tran et al. 1999; Longley et al. 2001 |
| ExoN | 3′-5′ | Unknown—proofreading? | Brown et al. 2002 |
| TREX1/TREX2 | 3′-5′ | Aberrant ssDNA removal/proofreading? | Chen et al. 2007; Yang et al. 2007 |
| p53 | 3′-5′ | Multiple; proofreading? | Mummenbrauer et al. 1996 |
| APE1/APE2a | 3′-5′ | BER, replication | Wilson 2003; Burkovics et al. 2006 |
| FEN1a | 5′-3′ | Okazaki fragment processing, BER | Harrington and Lieber, 1994 |
| XPG/ERCC5 | 5′-3′ | NER; transcription stabilisation | Habraken et al. 1994a, b |
| EXO1a | 5′-3′ | Multiple: MMR, DSBR, meiosis | Fiorentini et al. 1997; Lee and Wilson 1999; Tishkoff et al. 1998; Tsubouchi and Ogawa 2000 |
| WRNb | 3′-5′ | Multiple: DNA repair, telomeres | Cooper et al. 2000; Huang et al. 1998, 2000 |
| Dna2 | 3′-5′ | Replication—Okazaki fragment processing | Reviewed in Budd et al. (2009) |
| MRE11c/RAD50/NBS1 | 3′-5′ | DSBR; replication restart | Buis et al. 2008; D'Amours and Jackson 2002 |
| hRAD9 | 3′-5′ | Checkpoint | Bessho and Sancar 2000; Parker et al. 1998 |
| EXDL2 | 3′-5′d | Unknown—ICLR? | Smogorzewska et al. 2010 |
| FAN1 | 5′-3′ | ICLR? | Kratz et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010 |
| EXOGa | 5′-3′ | Multiple in mitochondria | Cymerman et al. 2008 |
BER base excision repair, DSBR double-strand DNA break repair, MMR mismatch repair, ICLR interstrand crosslink repair
aEXO1 also has (flap) endonucleolytic activity and 3′-5′ polarity, whilst the major activities of FEN1 and APE1 are as Flap endonucleases. EXOG also has endonucleolytic activity
bWRN has 3′-5′ exonuclease and a reported 5′-3′ activity in conjunction with Ku70/86
cMRE11 contains the exonuclease activity
dEXDL2 exonuclease activity is assumed by homology
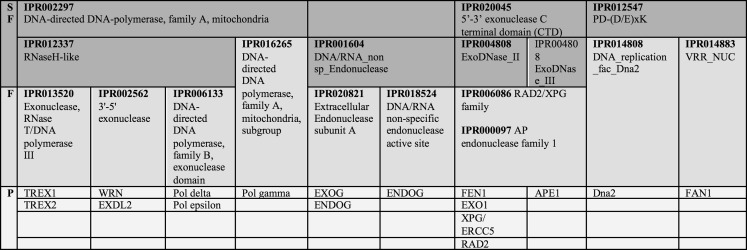
Nucleases are highly evolutionarily conserved and can be classified into families based on both sequence and functional homology. Of particular relevance to this review are the 5′-3′ exo C-terminal domain (CTD) superfamily and the RNaseH domain superfamily, the latter of which includes the DnaQ-like family and other 3′-5 exonucleases (Table 2). While endonucleases cleave DNA internally by cutting the phosphodiester backbone, exonucleases act biochemically to catalyse the removal of a single nucleotide monophosphate (dNMP) from the end of one strand of DNA. A critical characteristic of such nucleases is thus their ability to bind to the relevant DNA substrate, and most exonucleases do so in a non-sequence specific manner, though they generally show some degree of structure specificity. For example, the 5′-3′ exo CTD superfamily share a common helix hairpin helix (HhH) motif (Doherty et al. 1996; Thayer et al. 1995) that permits broad specificity DNA binding, probably through hydrogen bonding between the sugar–phosphate backbone of DNA and the protein backbone.
Table 2.
Relationship of nuclease superfamilies, families and individual proteins
Note that some proteins (e.g. Fen1) constitute their own family or subgroup
SF superfamily, F family, P protein (see also http://www.ebi.ac.uk/interpro/)
The functions of exonucleases are many and varied, reflected in the large number of independent nucleases found in many species. For example, of the DnaQ-like family (Table 2), humans possess 25 different family members, mice have 23, and other experimentally relevant organisms also have multiple DnaQ-like exonucleases (seven in fruit flies, eight in Arabidopsis and three in each of Schizosaccharomyces pombe and Saccharomyces cerevisiae).1 Within a single nuclease, there may reside multiple specificities: the human APE1 nuclease has both 3′-5′ exonuclease and abasic-site endonuclease activities, while FEN1 is both a structure-specific flap endonuclease and a 5′-3′ exonuclease. Similarly, the RAD2 domain of human EXO1 contains both 5′-3′ exonuclease and flap endonuclease activities (see Table 1 and references therein).
Nucleases may also be partially or fully redundant, depending upon the pathway, and such redundancy may complement functional losses. For example, in the yeast S. cerevisiae, 5′ end resection in double-strand DNA break repair can be catalysed by MRE11/Rad50/Xrs2, Exo1 or Rad27 (yeast FEN1) (Moreau et al. 2001). The general trend observed on nuclease mutation, however, is decreased viability and repair as more exonuclease activities are lost, suggesting overlapping but non identical roles.
Nucleases involved in DNA replication
Intrinsic proofreading activity of DNA polymerases
DNA replication is a highly accurate and precise process for copying the genome prior to segregation to daughter cells. Accumulation of errors occurring during DNA replication may contribute to both carcinogenesis and cellular ageing. Accuracy in the first instance depends on high fidelity of the replicative DNA polymerases, resulting from a combination both of a solvent-inaccessible polymerisation active site that allows 10,000-fold discrimination for correct over incorrect Watson–Crick base-pairing of substrate with template, and editing which involves a 40° shift of the new duplex DNA into the 3′-5′ exonuclease active site for proofreading (Franklin et al. 2001). This results in a base misincorporation rate during DNA replication in yeast of ~1 × 10−5 for DNA polymerase δ and approximately double that for DNA polymerase ε (Nick McElhinny et al. 2009). Note that human polymerase ε has a 5.5 fold higher fidelity of nucleotide incorporation (Korona et al. 2010) than its orthologue in yeast.
Replication fidelity rates vary slightly depending upon the base incorporated, suggesting that levels of individual nucleotides in the environment can modulate fidelity (Chen et al. 2000), thus replication stress occurs when nucleotide pools are imbalanced, e.g. following treatment with hydroxyurea. Translesion DNA polymerases can take over from faithful DNA polymerases in cases of DNA damage where correct templating is not possible, but these have solvent-accessible active sites and lack proofreading capacity, leading to very high error rates (Kunkel 2009) and accounting for the highly mutagenic effects of lesions that block the replicative polymerases.
Since the high degree of replicative DNA polymerase accuracy requires intrinsic proofreading activity whereby incorrectly incorporated bases are excised by a 3′-5′ exonuclease subunit of the polymerase (Hubscher et al. 2002), it is not surprising that mutation of the proofreading domains of either pol δ or pol ε increases the error rate for base substitution 12–13-fold in yeast (Nick McElhinny et al. 2009) or sevenfold for mutated human pol ε (Korona et al. 2010). Such loss of fidelity can have major biological consequences, as seen by the development of cancer in mice which have lost the proofreading function of polymerase δ in vivo (Goldsby et al. 2001). While there have yet been no studies directly linking nuclear DNA polymerase fidelity loss with ageing,2 it is notable that polymerase ε is required to prevent replicative senescence in budding yeast that lack telomerase activity (Deshpande et al. 2011). Moreover, instability of the nuclear genome can trigger apoptosis, neoplastic change or senescence, leading to organismal ageing.
Extrinsic proofreaders that act in concert with DNA polymerases
Proofreading during DNA synthesis does not necessarily have to proceed via intramolecular activity of intrinsic exonuclease domains of the DNA polymerase, since priming from a mismatch may be orders of magnitude slower than from paired bases (Huang et al. 1992), allowing time for intermolecular intervention. A number of exonucleases may supply extrinsic proofreading activity for DNA polymerases that do not possess their own capacity such as DNA polymerase α, which synthesises initiator DNA (iDNA) at the start of each new Okazaki fragment, and DNA polymerase β, important in repair. Yeast studies with DNA polymerases bearing point mutations in their exonuclease sites provide evidence that the proofreading component of polymerase δ can lend its exonuclease function to polymerase α to correct errors during lagging strand replication. Mutation of pol δ exonuclease prevented correction of the large number of errors generated by a mutant pol α (L868M which in vivo confers a mutator phenotype), while mutation of pol ε exonuclease had no effect (Nick McElhinny et al. 2006, 2009; Pavlov et al. 2006; Perrino and Loeb 1990).
The high error rate of non-proofreading DNA pol α may also be corrected in vivo by the action of ExoN, a 45.3-kDa 3′-5′ exonuclease with the ability to remove 3′ mismatched termini from double-stranded DNA (dsDNA), including the nucleotide analogues 1-beta-D-arabinofuranosylcytosine monophosphate (araC) and 9-beta-D-arabinofuranosyl-2-fluoroadenine 5′-monophosphate. ExoN is reported to interact with DNA pol α and may supply it with proofreading capability (Brown et al. 2002; Perrino and Loeb 1990; Skalski et al. 2000). ExoN upregulation by approximately sixfold in araC-resistant leukemia cells suggests that these cancer cells overcome drug treatment through efficient removal of drug-induced mismatches, and hence ExoN is likely to be a clinically relevant target in cancer (Skalski et al. 2000) and possibly age-related accumulation of genetic damage. It is important to note, however, that the genetic locus of ExoN has not yet been specified, so it might prove to be a known exonuclease, or possibly a novel isoform or duplication of one already described.
Autonomous proofreaders
Autonomous 3′-5′ exonucleases are, as their name suggests, nucleases that are not necessarily intrinsic to DNA polymerases. They may be fairly non-specific and may act with overlapping functionality depending upon the DNA polymerase involved and the type of proofreading required.
TREX1 and TREX2
The mammalian proteins TREX1 and TREX2 (three prime repair exonucleases) are small homodimeric exonucleases that cleave DNA non-processively from the 3′ end (Hoss et al. 1999; Mazur and Perrino 1999, 2001a, b). The TREX proteins make up the bulk of nuclear 3′-5′ exonuclease activity in mammals (Mazur and Perrino 2001a), suggesting that they serve an important role in metazoan nuclear genome maintenance, possibly acting as autonomous proofreaders (note that there are no TREX homologues in yeast). For example, TREX1 can remove mismatched DNA at a strand end (Mazur and Perrino 2001a), a required activity of a proofreader nuclease during DNA replication.
TREX1, whilst predominantly a cytoplasmic protein, is upregulated via the AP-1/Fos transcriptional pathway and relocated to the nucleus under conditions of genotoxic stress such as UV damage (Christmann et al. 2010). This may reflect both a role for TREX1 in DNA repair and in dealing with damage during DNA replication. However, TREX1−/− null mice show no increase in spontaneous mutation (although induced mutation has not been tested) or increase in cancer, but this might merely show that the nuclear function of TREX1 may be complemented by one of the other autonomous proofreaders, perhaps TREX2.
TREX1 in the cytoplasm appears to degrade single DNA strands arising from processing of aberrant replication intermediates that migrate to the endoplasmic reticulum (ER) after S-phase (Yang et al. 2007). Furthermore, it participates in Granzyme-A-mediated cell death in concert with the NM23-H1 nuclease (Chowdhury et al. 2006), resulting in DNA fragmentation characteristic of apoptosis. As loss-of-function mutations in the TREX1 gene cause the human disease Aicardi–Goutieres syndrome (a neurological syndrome that has symptomatic overlap with systemic lupus erythematosis—Crow et al. 2006; Lindahl et al. 2009), this suggests that non-degradation of ER/cytoplasmic DNA moieties might lead to immune dysfunction based upon an immune response to self-DNA, particularly if the DNA enters the secretory pathway via the ER. Notably, release of DNA from damaged cells is known to trigger inflammatory responses through ‘damage-associated molecular patterns’ (DAMPS) which are likely to trigger an innate immune response (Zhang et al. 2010), as such factors closely resemble ‘pathogen-associated molecular patterns’ (PAMPS) to which the innate immune system is tuned. Indeed, mice null for Trex1 only live for one-fifth as long as wild-type mice and develop inflammatory myocarditis (Morita et al. 2004), consistent with autoimmunity and/or premature ageing, since high levels of inflammation are associated with premature ageing (the ‘inflamm-aging’ hypothesis e.g. Franceschi et al. 2007). Removal of the aberrant cytoplasmic DNA by TREX1 probably mutes DNA damage signalling and checkpoint activation so that the inflammatory response cannot mount.
Defective TREX1 is associated not only with Aicardi–Goutieres syndrome but also with ataxia telangiectasia-like symptoms (Lindahl et al. 2009) and autosomal dominant retinal vasculopathy with cerebral leukodystrophy (Richards et al. 2007), caused by C-terminal truncation and accompanied by loss of perinuclear localisation (see Table 3). This gives rise to the onset of stroke, dementia, loss of visual acuity and other pathologies prematurely in middle age, with death occurring within 10 years of disease onset (Richards et al. 2007). Pathologies of this type become more common in old age; early onset might thus suggest defects in TREX1 or similar exonuclease activities.
Table 3.
Clinical features associated with exonuclease mutation
| Protein | Disease | Notes |
|---|---|---|
| Polymerase proofreader | Cancer | Lack of proofreading by replicative or repair polymerases leads to increased DNA mutation and cancer predisposition |
| TREX1 | Aicardi–Goutieres syndrome, lupus, Cree encephalitis | Deleterious DNA mutation leads to immune system dysfunction by disruption of the inflammatory response |
| p53 | Cancer | Most p53 mutations found in cancers are in the DNA-binding core region, which contains the exonuclease activity, suggesting that loss of the proofreading functionality promotes cancer via mutation (cf. polymerase proofreaders) |
| APE1 | Cancer | Deficient BER causes genome instability and predisposition to cancer |
| FEN1 | Cancer, autoimmunity, chronic inflammation, possibly trinucleotide repeat expansion disease | Mutator phenotype from nuclease mutation (compromised OFP and BER) can cause cancer; chronic inflammation resulting from FEN1 dysfunction promotes cancer progression. Trinucleotide repeat expansion on FEN1 mutation in yeast and mouse models; role in repeat expansion less clear in humans (Huntington’s disease triplet repeats stable in absence of FEN1 nuclease) though possibly implicated in e.g. Friedreich’s ataxia |
| XPG/ERCC5 | Xeroderma pigmentosum (XP), Cockayne syndrome (CS), trichothiodystrophy (TTD), cancer | Dysfunctional NER leading to sensitivity to sunlight (UV) and increased genome instability. Some mutations can cause symptoms of Cockayne syndrome or TTD (premature ageing, neural and physical defects, skin and hair abnormalities). Note that mutation of the related XPF (endonuclease) results in severe progeroid phenotypes |
| EXO1 | Cancer, immune deficiency | Cancer from compromised mismatch repair (HNPCC: human non-polyposis colon cancer). Compromised immunoglobulin production owing to lack of class switching and somatic hypermutation |
| WRN | Werner syndrome (WS) | Segmental adult-onset progeroid syndrome resulting from loss of the RecQ helicase/exonuclease WRN, which functions in multiple genome stability pathways (notably BER, replication restart and telomere maintenance); causes many symptoms associated with normal ageing to present early (for example cancer, atherosclerosis and heart disease) |
| DNA2 | None described | Defective DNA replication might cause genome instability and a rise in cancer predisposition |
| MRE11 (as part of MRN) | Nijmegen breakage syndrome (NBS), ataxia telangiectasia-like (AT-L) | A member of the MRN complex (with NBS1 and Rad50) and associates with ATM/ATR. Compromised DSBR and HJ resolution causes microcephaly and other physical defects, immunodeficiency, UV sensitivity and lymphoma predisposition |
| hRAD9 | None described | Loss of the RAD9 DNA damage checkpoint might cause aberrant cellular division/loss of apoptosis, presumably allowing cellular dysfunction and possible cancer predisposition |
| FAN1 | Fanconi’s anaemia (FA) | Nuclease associated with FA; symptoms are early cancer (mainly acute myelogenous leukaemia) and bone marrow failure. Also associated with congenital defects, including physical and developmental disabilities, and early death |
| EXDL2 | None described | Putative association with cancer (see FAN1); may have a mitochondrial DNA maintenance function |
| POLG | Premature ageing and multiple clinical features (see notes) | Mutation of mitochondrial polymerase gamma proofreading exonuclease leads to accelerated ageing symptoms in mice. The human mitochondrial genome is densely coded meaning that any base substitution errors from loss of proofreading have a high chance of disrupting mitochondrial genes and hence can cause neuromuscular disorders through disruption of ATP production, e.g. Leigh’s syndrome, NARP, MELAS (mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes), MERRF (myoclonic epilepsy with ragged red fibres), LHON (Leber’s hereditary optic neuropathy) |
| EXOG/ENDOG | None specifically associated (see notes) | DNA editing and processing defects give rise to mitochondrial DNA instability and may cause mitochondrial diseases as listed above (see POLG) |
Thus TREX1 has both nuclear and cytoplasmic roles, possibly as an autonomous proofreader in replication but also in removing potential inflammatory triggers. Similarly, human TREX2, which is 44% homologous to TREX1, shows punctate nuclear staining with some cytoplasmic localisation. It is cell cycle regulated, being downregulated during G2/M; like TREX1, its levels are highest in S-phase and lowest at mitosis (Chen et al. 2007). HeLa cells in which TREX2 has been knocked out show reduced cellular proliferation (Chen et al. 2007), suggesting an as yet undetermined role in proliferative DNA metabolism for TREX2 but consistent with action as an extrinsic proofreader.
p53
The tumour suppressor protein p53 has been suggested to possess 3′-5′ exonuclease activity (Mummenbrauer et al. 1996), in addition to its better-characterised roles as a transcriptional activator and an inducer of apoptosis (reviewed in Cox and Lane 1995; Green and Kroemer 2009; Vousden and Prives 2009). The 3′-5′ exonuclease activity of p53 is active on both single-stranded DNA (ssDNA) and dsDNA and has a preference for removing mismatches from replicating strand DNA, while paired bases inhibit the exonuclease activity (Huang et al. 1998). p53 may act by enhancing a further nuclease, AEN (apoptosis enhancing nuclease), to result in the DNA fragmentation seen in p53-dependent apoptosis (Kawase et al. 2008). However, p53 is found to co-localise with DNA synthesis during S-phase and might itself provide proofreading functionality for DNA polymerase α, since it enhances the replication fidelity of error-prone pol α but not of high-fidelity proofreading pol ε (Hollstein et al. 1996). Thus a novel way in which p53 might safeguard the genome is by increasing replication fidelity when necessary, for example under conditions of genomic stress following genotoxic damage. It is notable that over half of all human cancers have p53 mutations, frequently in the DNA binding and exonuclease domain of the protein.3 p53 is also known to be critical in establishing cell senescence, though it is likely that this is primarily through its transcriptional activation of p21 (El-Deiry et al. 1993; Noda et al. 1994) rather than through its nuclease activity.
APE1 and APE2
A novel class of proteins that may also act as autonomous proofreaders are the AP endonucleases. There are four types of AP endonucleases, based upon the sites of incision: class I and class II incise such that 3′-hydroxyl and 5′-phosphate ends arise. Class III and class IV instead generate the reverse—a 3′-phosphate and a 5′-hydroxyl (Myles and Sancar 1989). Human APE1 belongs to the most common class II. Both APE1 and APE2 (Hadi and Wilson 2000) have been shown to possess specific, non-processive but nevertheless possibly important abilities to cleave 3′-termini from DNA. APE1 removes nucleotides from matched, mismatched and beta-l-dioxolane-cytidine nicked DNA (Chou and Cheng 2002, 2003), with a marked preference for 3′-mismatched and nicked substrates. APE2 also has a preference for removal of mismatched bases (Burkovics et al. 2006). It remains to be determined whether and how much these proteins contribute to proofreading during replication or whether they utilise this functionality only within the context of DNA repair.
Replication fidelity is enhanced through nucleases acting in mismatch repair
In addition to proofreading by intrinsic or extrinsic exonucleases, a further mechanism that can increase replication fidelity by up to 1,000-fold involves the mismatch repair (MMR) machinery. Errors in mismatch repair are linked to microsatellite instability and cancer. Eukaryotic MMR requires not only the well-characterised MSH (MutS homologue) proteins which survey the genome for both mismatches and small insertion/deletion loops but also exonucleases such as EXO1 (see Hsieh and Yamane 2008 and references therein).
Eukaryotic EXO1 is a member of the Rad2/FEN1 family of nucleases (Lee and Wilson 1999) (Table 2) and is one of four nucleases that may be involved in MMR, although it is not (as originally reported) an orthologue of the Escherichia coli MMR protein EXO1 (Genschel et al. 2002; Tishkoff et al. 1997, 1998). There are some reports that loss of EXO1 gives rise to one form of the MMR-deficient syndrome human nonpolyposis colon cancer, HNPCC (Wu et al. 2001), but a direct association has been questioned (Thompson et al. 2004). Eukaryotic EXO1 is involved in various DNA metabolic pathways other than MMR, including meiosis (Fiorentini et al. 1997) and recombination (Tsubouchi and Ogawa 2000). EXO1 may also play a role in resecting ends at broken replication forks to prevent generation of recombinogenic intermediates.
Human EXO1 has 5′-3′ activity on both dsDNA and ssDNA, although its activity is greater on 3′-overhang and blunt duplex substrates, including those containing gaps or nicks than on 5′ overhang duplex or ssDNA (Lee and Wilson 1999). It can also degrade RNA in vitro. EXO1 is also involved in 3′-nick directed repair, suggesting either that it might have a cryptic 3′-5′ activity or that it is required to activate another as yet uncharacterised 3′-5′ nuclease (Genschel et al. 2002). EXO1 has been shown to resect DNA in vitro, with DNA affinity increased by the RecQ helicase BLM and loading and processivity by the MRN complex and RPA (Nimonkar et al. 2011). In yeast, the flap endonuclease activity of EXO1 has been suggested to be a functional backup for the major Okazaki fragment processing flap endonuclease FEN1 (see “FEN1”), again showing the overlapping redundancies of many cellular nucleases. Moreover human EXO1 can complement its yeast homologue (Qiu et al. 1999).
Notably, it has recently been shown that a SNP in the EXO1 promoter leading to higher levels of EXO1 expression is enriched in female centenarians (Nebel et al. 2009), suggesting a possible role for EXO1 activity in prevention of premature ageing, possibly by reducing cancer risk. By contrast, deletion of the nuclease activity of EXO1 in short-lived late-generation telomerase-deficient mice rescues lifespan (Schaetzlein et al. 2007). This rather unexpected finding may result from repression of the cyclin-kinase inhibitor p21, which leads to the increased lifespan in terc−/− mice, as observed in mice lacking other components of MMR such as PMS2 (Siegl-Cachedenier et al. 2007) and which directly phenocopies p21−/− Terc−/− mice (Choudhury et al. 2007). Thus it is possible that rescue of longevity upon EXO1 loss in telomerase null mice was effected by suppression of the DNA damage response checkpoint—perhaps via lack of EXO1 end-processing and recruitment of downstream response elements—to allow proliferation in cells with uncapped telomeres and other DNA damage. Taken together, these data might suggest that upregulation of EXO1 is only useful for long lifespan in the presence of robust telomeres and DNA proofreading/repair.
Processing DNA intermediates at DNA replication forks
During the maturation step of DNA replication, Okazaki fragments on the lagging strand of the replication fork must be processed to remove both the RNA primers generated by primase and the iDNA synthesised by error-prone DNA pol α, prior to ligation into high molecular weight DNA. Several models of Okazaki fragment processing (OFP) have been proposed (reviewed in Budd et al. 2009), all requiring exonucleases, particularly the flap endonuclease/exonuclease 1, FEN1. The DNA flaps and similar structures arising during OFP are likely also to be formed during DNA lesion processing, and the nucleases important in OFP have also been shown to play roles in DNA repair.
FEN1
FEN1 is a structure-specific endo/exonuclease originally cloned from mouse cells (Harrington and Lieber 1994) and subsequently humans (Hiraoka et al. 1995) and is essential for DNA replication in vitro (Waga et al. 1994). The FEN1 homologue in budding yeast is encoded by the rad27 gene. It acts during Okazaki fragment processing in DNA replication to remove the terminal deoxyribonucleotide (or ribonucleotide) by endonucleolytic cleavage at the elbow of DNA junctions bearing a 5′ flap generated by incoming DNA pol δ on the lagging strand (Budd et al. 2009). In addition, FEN1 can act exonucleolytically in a 5′-3′ direction on flap substrates.
Recruitment of FEN1 to sites of exonuclease action involves direct binding to the sliding clamp PCNA through a conserved PCNA-interacting motif (PIP, Cox 1997; Warbrick et al. 1997), the interaction positioning FEN1 to act preferentially as an exonuclease rather than an endonuclease (Hosfield et al. 1998a, b). The mode of action has been elucidated by X-ray crystallography, showing that association with PCNA allows flexibility around a hinge region in FEN1 that brings the enzyme’s nuclease active site in direct contact with the DNA substrate (Sakurai et al. 2005).
Furthermore, FEN1 has recently been crystallised both alone (Sakurai et al. 2008; Tsutakawa et al. 2011) and in association with the Rad9–Rad1–Hus1 (9-1-1) DNA damage checkpoint complex (Dore et al. 2009). Analysis of the structures of FEN1 (Tsutakawa et al. 2011) and EXO1 (Orans et al. 2011), which were solved at similar times, suggest a unified mechanism for this type of nuclease involving DNA bending and two nucleotide ‘fraying’ of one strand (Orans et al. 2011).
Mutation of FEN1’s nuclease active site (E160D) makes cells more susceptible to chemically induced cancers (Xu et al. 2011). Loss of only one copy of FEN1 increases colon cancer incidence in mice heterozygous for mutation of the Apc tumour suppressor, with Fen1 halpoinsufficiency leading to rapid tumour progression (Kucherlapati et al. 2002). FEN1 also shows gap endonuclease (GEN) activity, which is critical for the resolution of stalled replication forks following DNA damage (Zheng et al. 2005). Notably, these authors showed that a mutation in FEN1 that abolished GEN activity (E178A) removed FEN1’s ability to rescue camptothecin and UV sensitivity in a yeast Fen1 mutant.4 Moreover, transgenic mice bearing a point mutation in FEN1 that abolished the majority of 5′ exonuclease and GEN activity developed significant inflammation and autoimmunity, together with high cancer incidence (Zheng et al. 2007). An inability to recruit FEN1 to sites of action through mutation-critical aromatic residues within its PCNA-interacting peptide (F343A F344A) (Warbrick et al. 1997) also results in loss of DNA processing and aneuploid cancers (Zheng et al. 2011).
Mutational studies in yeast have suggested that FEN1 restrains recombination between short DNA sequences (Negritto et al. 2001) such as the flaps arising during strand displacement DNA synthesis on the lagging strand of the replication fork or during DNA repair. These findings are also consistent with the triplet repeat expansion phenotype observed on FEN1/Rad27 mutation (Liu et al. 2004; White et al. 1999) and with its involvement in long patch BER along with its partner PCNA (Frosina et al. 1996). (Note that FEN1 is important for removal of oxidative damage in both the nucleus (Asagoshi et al. 2010) and in mitochondria (Liu et al. 2008)). Mutation of pol ε in a Rad27 null background led to elevated +1 frameshifts on homonucleotide runs (Kirchner et al. 2000) suggesting a role for FEN1 either in mismatch repair or proofreading, in addition to its roles in OFP and BER. Notably, mutation of FEN1 leads to decreased yeast lifespan (Hoopes et al. 2002). Hence loss of function of FEN1 results in a mutator phenotype that predisposes to cancer and signs of premature ageing.
WRN
WRN is a multifunctional replication/repair RecQ-helicase family member (Brosh et al. 2001b) that in deficiency causes the premature ageing Werner’s syndrome, an adult-onset progeria phenocopying early many normal ageing diseases such as atherosclerosis, cancer and cataracts. WRN association with FEN1 may be important in Okazaki fragment processing during DNA replication as FEN1’s nuclease activity is stimulated by binding to the helicase domain of WRN (Brosh et al. 2002a).
Unlike other RecQ helicases, in addition to helicase activity, WRN has a well-characterised 3′-5′ exonuclease domain (Huang et al. 2000) which is highly conserved and of the DnaQ exonuclease family (Perry et al. 2006) (Table 2). In Drosophila, and other invertebrate and plant species, the exonuclease appears as an autonomous protein (Boubriak et al. 2009; Cox et al. 2007; Plchova et al. 2003; Saunders et al. 2008; Sekelsky et al. 1999).
While WRN exonuclease acts on both single-stranded (Machwe et al. 2006) and duplex DNA with overhanging ends, it is also active on substrates that resemble either DNA replication intermediates or structures found during the processing of DNA damage (Brosh et al. 2002b). However, in vitro without other factors, WRN cannot cleave blunt-ended duplex DNA. In DNA replication, WRN is implicated in processing unusual DNA structures that, if left unprocessed, would result in replication fork regression and formation of recombinogenic intermediates including Holliday junctions (HJs). This is supported by the accumulation of stalled replication forks in cells lacking WRN protein (Rodriguez-Lopez et al. 2002; Sidorova et al. 2008), and it is likely that this reflects a need for the nuclease activity of WRN, since many of the defects observed in WS cells, including reduced proliferative capacity, low S-phase fraction and drug sensitivity, can be corrected by ectopic expression of a bacterial Holliday junction resolvase (Rodriguez-Lopez et al. 2007). Recruitment of WRN to stalled replication forks may occur through binding to PCNA via a classical PIP (Lebel et al. 1999; Rodriguez-Lopez et al. 2003).
Thus WRN may be involved in directing the appropriate pathway during recombinational repair, for example in response to collapsed replication forks or other sources of double-strand breaks (Plchova et al. 2003; Rodriguez-Lopez et al. 2002; Saintigny et al. 2002; Swanson et al. 2004). It may, like other RecQs such as yeast Rqh1 (in combination with EXO1 or the cross-over junction nuclease MUS81-EME1) (Doe et al. 2002), direct non-cross over pathways, perhaps by removing a potentially invasive DNA strand arising from replication fork stalling or dissociation of leading and lagging strand polymerases. Indeed, Rqh1 null yeast are hyperrecombinant, while ectopic expression of human WRN in such cells represses recombination (Yamagata et al. 1998). That this suppression of recombination results from the DNA exonuclease activity of WRN is supported by the finding that fruit flies deficient in the orthologue of human WRN exonuclease, DmWRNexo, show extremely high levels of mitotic recombination (Saunders et al. 2008) and marked developmental abnormalities (R. Lasala et al., manuscript in preparation). The DNA-end binding Ku70/86 dimer specifically and strongly stimulates WRN’s exonuclease activity (Comai and Li 2004; Li and Comai 2002; Li et al. 2004, 2005), changing its specificity to allow degradation of blunt and even 5′-protruding ends (Cooper et al. 2000), suggesting a particular role for WRN during Ku-directed repair such as non-homologous end joining (NHEJ) or in an alternative pathway to degrade free ends at broken replication forks.
WRN exonuclease is implicated in other DNA metabolic pathways in addition to its likely role as an accessory protein at the replication fork. For example, WRN may play an important role at the telomere, including telomeric D-loop resolution (Opresko et al. 2004). It is likely that this requires intramolecular co-operation between the intrinsic helicase and the exonuclease activities of WRN such that the helicase unwinds unusual DNA structures (including G4 quadruplexes which mimic telomeric structures) and is followed by exonuclease activity to process the resulting ends. Indeed, a K577M point mutation within the helicase domain has a dominant negative effect on any subsequent exonuclease activity and prevents lagging strand Okazaki fragment processing at the telomere (Crabbe et al. 2004). (Note that WRN also binds to telomere proteins POT1 and TRF2 (Opresko et al. 2002, 2005)). Interestingly, WRN also interacts physically with and stimulates EXO1 (Aggarwal et al. 2010; Sharma et al. 2003), a nuclease that is implicated both in Okazaki fragment processing and in telomere processing. This interaction may be important for either WRN or EXO1, or both, to process stalled or regressed replication forks and to maintain the T and D loops at telomeres. Loss of telomere integrity drives replicative senescence and ageing, and prolonged replication stress can similarly result in ‘deep’ senescence and contribute to organismal ageing.
WRN exonuclease can also function as an extrinsic proofreader for DNA polymerase β during base excision repair (BER) (Harrigan et al. 2006). Studies based upon the X-ray crystal structure of human WRN exonuclease domain give a firm assignment of editing functionality (Perry et al. 2006), akin to end-processing activities of other members of the DnaQ family. Moreover, deletion of the exonuclease domain results in cells that are hypersensitive to DNA damage (Kashino et al. 2005). Confusingly, p53, itself a nuclease, strongly inhibits the exonuclease activity of WRN (Brosh et al. 2001a), highlighting the complex synergistic/antagonistic interplay between cellular nucleases. Since loss of WRN causes the segmental progeroid Werner’s syndrome (Huang et al. 1998; Yu et al. 1996), this provides a direct evidential link between genome instability and ageing.
Dna2
Dna2 is a combined helicase/nuclease that interacts with FEN1 and is likely to be involved in Okazaki fragment processing, as yeast cells with a mutant allele of Dna2 (dna2-1) can only synthesize short stretches of DNA under restrictive conditions (Budd et al. 2009). It is likely that Dna2 is recruited to process Okazaki fragments under conditions when long flaps arise during pol δ-mediated strand displacement of the primer DNA and iDNA, as these flaps become coated with RPA and resistant to cleavage by FEN1. Indeed, recruitment of Dna2 is likely to occur through direct interaction with RPA. Exonuclease cleavage of the flap DNA by Dna2 eventually results in flaps short enough to be processed by FEN1 (reviewed in Kao and Bambara 2003; MacNeill 2001; Budd et al. 2009).
In addition to a helicase domain in the C-terminus, Dna2 possesses 3′-5′ exonuclease activity with further 5′ flap endo-exonuclease activity (Fortini et al. 2011) present in the amino terminal half of the protein. The gene is itself essential with deletion mutants being inviable, and yeast cells mutant for both Fen1 and Dna2 are inviable (dna2-1 rad27Δ), while Fen1 overexpression can complement Dna2-1 phenotypes (reviewed in Budd and Campbell 2009), although Dna2 stimulates Fen1’s activity (Kao et al. 2004). Consistent with a role in preventing ageing, presumably by ensuring genome stability especially of the repetitive ribosomal DNA (rDNA) or at the telomeres, Dna2 mutants in yeast have a short lifespan (Weitao et al. 2003a, b), and similarly, Caenorhabditis elegans lacking worm Dna2 show compromised genome maintenance and shortened lifespan (Lee et al. 2011).
The human homologue of Dna2 is also a helicase/nuclease, which does not have an obvious nuclear localisation signal but in deficiency causes nuclear genome instability (Duxin et al. 2009) and co-localises with DNA (nucleoids) in the mitochondrion. It interacts with and stimulates mitochondrial DNA polymerase γ, and functions with FEN1 in mitochondrial long-patch BER (Copeland and Longley 2008; Zheng et al. 2008). In vitro reconstituted DSB repair complexes show that in the nucleus, end resection proceeds via two pathways (Nimonkar et al. 2011), one of which involves human DNA2 nuclease activity and the RecQ helicase activity of BLM, stimulated by RPA. RPA also ensures that DNA2 exonuclease activity occurs in the correct 5′-3′ polarity. The MRN complex is found to stimulate activity by recruiting BLM to the site of resection (Nimonkar et al. 2011). DNA2 therefore probably suppresses genome instability via both DNA repair (BER, DSB repair) and recombination pathways.
Nucleases in nucleotide excision repair and transcription
Bulky lesions such as UV-induced pyrimidine dimers that prevent replication fork progression or transcription elongation are generally removed through the nucleotide excision repair (NER) pathway, involving many proteins of the Xeroderma pigmentosum (XP) complementation group. Nucleases including XPG5 are required to make incisions either side of the lesion, hence mutation of the incision activity of XPG gives rise to symptoms of XP (Tian et al. 2004), conferring sensitivity to UV damage and consequently great susceptibility to epithelial cancers via defects in nucleotide excision repair (NER) (Table 3). By contrast, total absence of XPG protein causes symptoms of Cockayne’s syndrome (Arenas-Sordo Mde et al. 2006), with developmental defects and premature ageing. These phenotypes are thought to arise through loss of XPG’s role in stabilising transcription, rather than its activity in DNA repair (Friedberg and Wood 2007). Hence XPG mutation can give rise to various symptoms of both CS and XP including premature ageing (Friedberg and Wood 2007; Wijnhoven et al. 2007). In addition to its incision activity (O'Donovan et al. 1994; Habraken et al. 1994b), XPG belongs to the FEN1/RAD2 family because of its conserved 5′-3′ exonuclease activity (Habraken et al. 1994a, see Table 2). Of note, XPG has recently been shown to interact with the WRN helicase/exonuclease (Trego et al. 2011) via the C-terminal domain of both. XPG co-localises with WRN during S-phase and stimulates its helicase activity (Trego et al. 2011). Whether this association is important in preventing premature ageing is not yet clear.
Exonucleases involved in double-strand DNA break repair
Mre11
The first nuclease recruited in repair of DNA double-stranded breaks is MRE11 (Stracker et al. 2004), a component of the MRN complex (Mre11–Rad50–Nbs1) which plays a key role early in the DNA damage response (DDR) (Berkovich et al. 2007; Lisby et al. 2004; Mirzoeva and Petrini 2001). MRN detects and localises rapidly to DSBs after damage and secures and shields the frayed DNA ends (Williams and Tainer 2005) whilst activating checkpoint signalling via ATM (Lee and Paull 2007; Williams and Tainer 2005). Two Mre11 and two Rad50 molecules form a heterotetramer that interacts with Nbs1 (Xrs2 in yeast) and can bind both ends of a DSB, although Mre11 can itself form homomultimers (reviewed in Budd and Campbell 2009). Yeast and human Mre11 have multiple nuclease activities (Furuse et al. 1998; Moreau et al. 1999; Paull and Gellert 1998) and in addition have helicase-like activities in that they can cause both strand-annealing (de Jager et al. 2001) and strand dissociation.
Mre11 has 3′-5′ dsDNA exonuclease activity in vitro, as well as dsDNA and ssDNA endonuclease activity, but is specific for blunt or 3′-recessed ends, or, very weakly, for hairpin structures (D'Amours and Jackson 2002). Whilst the 3′-5′ exonuclease activity is necessary for DSB repair (Paull and Gellert 1998), experiments on S. cerevisiae mutants show that functions of Mre11 are separable and distinct, with the N-terminal nuclease domain required for DSB repair and the C-terminal dsDNA-binding domain needed for meiotic functions such as chromatin modification (Furuse et al. 1998).
Although removal of the nuclease activities of Mre11 in yeast results in only slight radiation sensitivity (Moreau et al. 1999), Mre11 mutation in mammals results in more severe phenotypes. Inherited hypomorphs cause ataxia telangiectasia-like symptoms such as extreme sensitivity to IR and cerebellar degeneration (Stewart et al. 1999, see Table 3), even though the nuclease-dead Mre11 mutant retains the ability to associate with the other members of the MRN complex and to activate ATM (Buis et al. 2008). This suggests that Mre11 nuclease activity may be more important in mammals than in yeast, and this importance is not based upon the DNA damage checkpoint acting through ATM but occurs via other roles of Mre11. It is possible that Mre11 is involved in preventing replicative senescence caused by telomere attrition, since in yeast it has been implicated in telomere maintenance acting as a block to replicative senescence (Joseph et al. 2010). Additionally, defective DSB repair has been reported in the progeroid Hutchinson Gilford progeria syndrome (HGPS), with a delay in localization of Mre11 to sites of DSBs suggesting that defects in Mre11 targeting may be associated with premature ageing characteristic of this syndrome (Constantinescu et al. 2010). Hence Mre11 nuclease activity may be necessary to prevent premature ageing.
hRAD9
The hRAD9 protein was discovered by homology to the S. pombe rad9 protein that is involved in the early DNA damage response checkpoint. Modified hRAD9, along with hRAD1 and hHus1, form the human 9-1-1 complex (Volkmer and Karnitz 1999), a heterotrimeric clamp complex structurally (Xu et al. 2009) and functionally analogous to PCNA that is loaded onto damaged DNA tracts (Burtelow et al. 2000; Carr 2002) by an RF-C like complex and is hyperphosphorylated in a damage-dependent manner, for example by ATM after ionising radiation (IR) (reviewed in Cox and Kearsey 2009). Such phosphorylation is required for IR-induced G1/S checkpoint activation (Chen et al. 2001a) but also after hydroxyurea-induced replication stress or UV-induced DNA damage. hRAD9 has 3′-5′ exonuclease activity (Bessho and Sancar 2000); as hRAD9 quickly localises to double-strand breaks, this suggests that the nuclease activity is important during early DNA processing. The 9-1-1 complex can also bind to and stimulate FEN1 nuclease activity (Wang et al. 2004) and also interacts with and enhances the function of DNA polymerase β, although not pol δ nor pol α (Toueille et al. 2004). hRad9 alone (not as part of the 9-1-1 complex) is also implicated in numerous other pathways; for example in ribonucleotide synthesis via stimulation of the carbamoyl phosphate synthetase activity of CAD (Lindsey-Boltz et al. 2004) and in apoptosis by interaction with members of the Bcl-2 family of proteins (Komatsu et al. 2000). Mouse rad9 nulls are not viable (Hopkins et al. 2004) presumably because Rad9 has so many different roles that there cannot be full complementation of all its activities. The exact role(s) of hRad9 nuclease activity has not yet been elucidated, nor its link with ageing, but in the nematode worm C. elegans, the 9-1-1 complex is implicated in telomere maintenance (Meier et al. 2006). Whether this is the case in higher organisms with an overt effect on ageing has yet to be determined.
Interstrand crosslink (ICL) repair
Interstrand crosslinks (ICLs) are chemical links between the bases (or backbones) on opposite strands of duplex DNA. They block progress of the replication fork, causing stalling; ICLs must therefore be removed in order for cells to proliferate and for their long-term survival. Deficiencies in ICL repair give rise to Fanconi’s anaemia (FA), a genome instability syndrome characterized by increased sensitivity to agents that promote ICLs in DNA (e.g. mitomycin C), and FA patients show many gross phenotypic abnormalities (Auerbach 1995), with greatly elevated predisposition to cancer (Alter 1996) associated with marked chromosomal instability. The FA protein core complex monoubiquitinates the ID complex comprising FANCI and FANCD2 (Garcia-Higuera et al. 2001), which localises to BRCA-containing nuclear DNA repair foci along with the recombination protein RAD51 (Taniguchi et al. 2002) and promotes removal of the DNA tract containing the crosslink.
FAN1
Short hairpin (shRNAi) screens to identify novel factors that confer resistance to ICL-inducing mitomycin C very recently identified FAN1 (Fanconi-associated nuclease 1) as a novel but important exonuclease involved in the ICL repair pathway (Smogorzewska et al. 2010). Furthermore, FAN1 was independently described as a putative nuclease that interacts with FANCD2. Absence of FAN1 was found to increase cellular sensitivity to ICL-inducing agents such as mitomycin C and cisplatin (Liu et al. 2010; MacKay et al. 2010).
FAN1 is a protein containing an N-terminal ubiquitin-binding domain and a C-terminal VRR_nuc nuclease domain (ancient lineage, see Table 2) and localises to interstrand crosslinks (ICL) along with monoubiquitinated FANCD2 (Kratz et al. 2010; Liu et al. 2010), a process requiring its ubiquitin-binding domain. Under mitomycin C treatment, FAN1 co-localises with FANCD2 at sites of damaged DNA, but since FAN1 depletion does not affect the ability of FANCD2 to become monoubiquitinated or localise to damage, FAN1 is likely to act downstream of, and may be recruited to sites of damage by, monoubiquitinated FANCD2.
Purified FAN1 is a structure-specific endonuclease capable of cleaving branched structures such as replication fork-like structures, splayed DNA arms and 5′ or 3′ flaps (Liu et al. 2010), but not duplex DNA. Like FEN1, FAN1 is also a 5′-3′ exonuclease as well as a flap endonuclease. Monoubiquitination of FANCD2—and thus the concomitant recruitment of FAN1 to damage foci—is required for the ‘unhooking’ step in removal of the ICL (Knipscheer et al. 2009). During removal, dsDNA excisions generating double-strand breaks are made either side of the crosslink in order to remove the crosslinked portion of the DNA. At least one of these incisions is catalysed by MUS81-EME1, a heterodimeric endonuclease (Ciccia et al. 2003) that can cleave branched structures and resolve Holliday junctions (Chen et al. 2001b). It is possible that MUS81-EME1 may also make the second incision, though the excision repair factor XPF-ERCC1 is another candidate (Fekairi et al. 2009). Alternatively, the structure arising from the first incision may resemble a 5′ flap, which could act as a substrate for the endonuclease activity of FAN1. While there is no difference in the rate of formation of cisplatin-induced or mitomycin C-induced DSBs in a FAN1-depleted background, their disappearance (removal) is slower than in controls (Kratz et al. 2010; MacKay et al. 2010), suggesting that FAN1 is not required to introduce DSBs, but rather is involved in their repair. Since DSB repair often occurs via homologous recombination (HR), this strongly implies that the FAN1 nuclease is involved in DNA processing during the latter stages of HR, but does not preclude the possibility that FAN1 may act in other as yet undiscovered pathways. The VRR_nuc domain is associated with the PD-(D/E)XK nuclease superfamily (including type III restriction enzymes and the replication/repair helicase/nuclease DNA2). The multiple nucleolytic abilities of FAN1 imply a multifunctional capacity similar to ExoG or Mre11. Whether the primary role of FAN1 is indeed in ICL repair (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010) or lies in other DNA transactions such as removal of flaps at stalled replication forks remains to be determined.
EXDL2
The genetic screen that isolated FAN1 (Smogorzewska et al. 2010) also identified a further putative exonuclease, EXDL2, which has sequence homology to the 3′-5′ exonuclease domain of WRN helicase/exonuclease, a DnaQ family nuclease (see “WRN”). While there is little published information concerning EXDL2, bioinformatics analysis shows that it has the DEDDy motif conserved in WRN and many related exonucleases. Various different isoforms of EXDL2 are reported; there is also a possible association with cancer.6 RNAi depletion of EXDL2 has no effect on ubiquitination of the Fanconi’s ID complex, leading Smogorzewska and colleagues (Smogorzewska et al. 2010) to suggest that EXDL2 may act in an ICL repair pathway either downstream of, or parallel to, the FA repair complex. In Drosophila, flies with an insertional mutation in the EXDL2 orthologue, encoded by CG6744, show slightly elevated rates of chromosomal recombination (Cox et al. 2007), implicating EXDL2 in nuclear DNA metabolism consistent with a role in repair. Intriguingly, EXDL2 in early Xenopus development has been localised by in situ hybridisation to the mitochondrial cloud (Cuykendall and Houston 2010), suggesting that it may also be involved in mitochondrial DNA metabolism, consistent with bioinformatics predictions of mitochondrial localisation of at least some of the isoforms of human EXDL2 (Mason and Cox, unpublished). The similarity between EXDL2 and WRN and its possible involvement in not only nuclear genome stability, perhaps in the ICL repair pathway, but also in mitochondrial DNA (mtDNA) maintenance make EXDL2 an interesting candidate for further research into possible links with ageing.
Mitochondrial nucleases
Mitochondria have their own highly conserved DNA replication and maintenance apparatus, which includes mitochondrial-specific nucleases. In humans, the circular mitochondrial genome is only 16.569 kb and encodes some components of the electron transport chain together with rRNAs and transfer RNAs (tRNAs) required for mitochondrial mRNA translation (the majority of gene products used in the mitochondria are, however, encoded by nuclear genes and the proteins subsequently imported across the mitochondrial membranes).
The mitochondrial genome has a mode of DNA replication unlike that of the nuclear genome, involving large regions of single-stranded DNA and use of several possible origins that may act bidirectionally or unidirectionally (Falkenberg et al. 2007; Yasukawa et al. 2009) and mediated by the single mitochondrial DNA polymerase γ, POLG. DNA repair pathways within mitochondria include MMR, BER and possibly DSB repair. Interestingly, nuclear repair factors including the nuclease FEN1 are implicated in maintaining mtDNA integrity (Kalifa et al. 2009). Accumulation of mitochondrial genome mutations and deletions has been reported with increasing chronological age, resulting in mitochondria with defects in metabolism, leading to the mitochondrial hypothesis of ageing (Harman 1972; Miquel et al. 1980). Links between telomere attrition resulting in mtDNA damage and loss of mitochondrial function have recently been drawn (Passos et al. 2010; Sahin and Depinho 2010). Thus maintenance of mtDNA integrity may be critical in preventing premature ageing, but until recently, little has been reported on the importance of nucleases in this process.
Proofreading nuclease of mitochondrial DNA POLG
As is the case for the nuclear genome, mitochondrial DNA requires high-fidelity replication, which is conducted by POLG. The importance of exonucleolytic proofreading during mtDNA replication is clear from the phenotype of mice lacking mitochondrial polymerase proofreading through mutation of POLG, which show early-onset of many age-related phenotypes (Trifunovic et al. 2004).7 Mice carrying high levels of mitochondrial DNA mutation show sarcopenia and mitochondrial dysfunction in skeletal muscle (Hiona et al. 2010), although this can be blunted by endurance exercise (Safdar et al. 2011). They also show an accelerated age-related loss of retinal function (Kong et al. 2011). Interestingly, this high mitochondrial mutation load alters the ability of hematopoietic stem cell lineages to undergo appropriate differentiation, leading to defects such as anaemia and lymphopenia similar to those caused by the premature senescence of this compartment (Norddahl et al. 2011). Hence loss of mtDNA integrity via loss of the proofreading function of polymerase γ is strongly associated with ageing.
EXOG/EndoG
In addition to the intrinsic proofreading nuclease activity of POLG, in S. cerevisiae the endo/exonuclease Nucp1 (Dake et al. 1988; Vincent et al. 1988) is the major nuclease in yeast mitochondria and is involved in a number of functions including mitochondrial DNA metabolism, recombination and cell death (Buttner et al. 2007; Zassenhaus and Denniger 1994). Nucp1 can degrade ssRNA and has DNA endonuclease activity (on both ssDNA and dsDNA) and 5′-3′ exonuclease activity (on dsDNA). The mammalian homologue, EndoG, was discovered as a nuclease involved in apoptosis (Li et al. 2001), where it is translocated to the nucleus and degrades chromatin in a caspase-independent manner. Active EndoG outside the mitochondria quickly induces cell death (Schafer et al. 2004). Like its yeast homologue, human EndoG is required for normal cellular proliferation (Huang et al. 2006).
Interestingly, mammalian EndoG acts only as an endonuclease; unlike the multifunctional Nuc1p, it notably lacks the 5′-3′ exonuclease activity used to generate gaps in dsDNA during recombination and repair. Recently, a novel EndoG paralogue named EXOG was discovered in mitochondria, which is thought to have arisen during ancestral gene duplication (Cymerman et al. 2008). Human EXOG homodimer possesses the 5′-3′ exonuclease activity that is missing in EndoG, as well as its own endonuclease functionality with some preference towards ssDNA, and it can nick supercoiled DNA. Thus EXOG has the activity necessary to generate ssDNA gaps. Together, mammalian EndoG and EXOG recapitulate all the functions of yeast Nuc1p. EndoG has been reported to lie in the mitochondrial inter-membrane space away from the DNA (Ohsato et al. 2002), whilst EXOG is located in the inner membrane where the mitochondrial DNA attaches (Cymerman et al. 2008), suggesting a functional as well as a physical split, with EndoG responsible for the cell death function performed by Nuc1p in yeast and EXOG involved in cellular proliferation. Both exonic (Cymerman et al. 2008) and intronic SNPs have been reported in the ExoG gene; the latter is associated with type 2 diabetes (Moritani et al. 2007), a disease linked with age.
Conclusions
Many different mechanisms have been proposed to account for cellular and organismal ageing, including the adverse impact of oxidative damage on proteins, lipids and DNA, genome instability arising from defects in maintenance mechanisms, mitochondrial dysfunction, defects in autophagy, aberrant metabolic signalling through the IGF1 axis and mTOR, degradation of the stem cell compartment and telomere attrition. That these mechanisms are important in ageing has been demonstrated in several recent experimental models. For example, Jaskelioff and colleagues have shown that short-term telomerase reactivation can restore normal tissue integrity and function even to aged tissues in mice (Jaskelioff et al. 2011), demonstrating the importance of correct telomere maintenance (which may involve not only telomerase but also nucleases including EXO1 and WRN) in preventing tissue homeostasis collapse and ageing. Telomere integrity is also thought to be important in preventing premature ageing of stem cells populations (Sahin and DePinho 2010). Inhibition of the IGF-1/mTOR axis through rapamycin administration significantly increased lifespan in adult mice (Harrison et al. 2009), verifying the importance of IGF1-mTOR in ageing, while nutrient restriction had a similar impact in increasing primate lifespan (Colman et al. 2009; reviewed in Cox (2009); Cox and Mattison 2009).
It would at first sight seem obvious that deficiency of exonucleases, whose normal role is to promote genome stability, would result in premature cell senescence through accumulation of DNA damage. The exonucleases discussed above do show the importance of correct genome maintenance in preventing the onset of premature ageing or age-associated disease such as diabetes and cancer. However, in addition to the direct role in DNA stability, there appears to be much more complex interplay between nucleases and other pathways of ageing, with significant cross-talk between metabolic signalling and DNA damage. Perhaps nowhere is this exemplified more clearly than in the recent description of a severe progeroid phenotype resulting from mutation of the nucleotide excision repair nuclease, XPF, with a patient showing not only signs of both CS and XP (deficient global genomic NER and transcription coupled NER) but also severe effects on liver, blood, muscle and neurological systems (Niedernhofer et al. 2006). Mice engineered to lack ERCC1 similarly showed profound systemic progeroid symptoms and died before sexual maturity. These results suggest that XPF-ERCC1 are involved not only in NER but also in other critical systems, perhaps including interstrand crosslink repair. Perhaps most interestingly, in this mouse model, the IGF1 nutrient response axis was altered, presumably in response to the presence of unrepaired DNA damage (Niedernhofer et al. 2006). Hence ageing may be triggered by initial loss of a single nuclease activity which impacts on multiple downstream regulatory pathways resulting in the loss of tissue homeostasis characteristic of whole organismal ageing.
The very tight association between mitochondrial genome maintenance defects and ageing strongly supports the idea that precise processing of mtDNA is critical to stave off ageing. Nucleases including POLG and EXOG are important to the cell to prevent premature accumulation of mtDNA mutations. Multiple nucleases have evolved to fulfill the functions required for each separate pathway, although there is often redundancy and overlap in functionality, shown by the dearth of human diseases that arise from a nuclease deficiency. However, it is suggestive of the importance of exonucleases that the diseases that do arise from their deficiency are systemic—premature ageing, genome-wide destabilization and general inflammation are all hallmark phenotypes. It is highly probable that a combination of mitochondrial nucleases, together with nucleases acting on the genomic DNA within the nucleus (particularly the progeroid-associated WRN), play a central role in preventing accumulation of damage that would otherwise give rise to a senescent phenotype. Such nucleases therefore present promising targets for further study and in developing agents either to promote precocious senescence (e.g. in cancer therapy) or anti-ageing therapies based on supporting or augmenting the role of these nucleases.
Acknowledgements
The authors thank Sophie Riddell for careful reading of the manuscript. We gratefully acknowledge the financial support of the Economic and Social Research Council of Great Britain (ESRC) [ES/G037086/1] under the New Dynamics of Ageing programme, and the Biotechnology and Biological Sciences Research Council of Great Britain (BBSRC) [BB/E016995/1].
Footnotes
See http://supfam.cs.bris.ac.uk/SUPERFAMILY/, which also includes detailed phylogeny.
However, loss of fidelity of the mitochondrial replicative DNA polymerase γ is strongly associated with premature ageing phenotypes in mice—see later.
See http://p53.free.fr.
Note that the related human GEN1 (yeast Yen1) appears to act exclusively as an endonuclease (Ip et al. 2008, Nature 456:357–361) and so is not discussed in this review.
also known as ERCC5
(although the absolute cause of this phenotype has been questioned—see Kraytsberg et al. (2009))
Contributor Information
Penelope A. Mason, Email: penelope.mason@bioch.ox.ac.uk
Lynne S. Cox, Phone: +44-1865-613243, Email: lynne.cox@bioch.ox.ac.uk
References
- Aggarwal M, Sommers JA, Morris C, Brosh RM., Jr Delineation of WRN helicase function with EXO1 in the replicational stress response. DNA Repair (Amst) 2010;9:765–776. doi: 10.1016/j.dnarep.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP. Fanconi's anemia and malignancies. Am J Hematol. 1996;53:99–110. doi: 10.1002/(SICI)1096-8652(199610)53:2<99::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Arenas-Sordo Mde L, Hernandez-Zamora E, Montoya-Perez LA, Aldape-Barrios BC. Cockayne's syndrome: a case report. Literature review. Med Oral Patol Oral Cir Bucal. 2006;11:E236–E238. [PubMed] [Google Scholar]
- Asagoshi K, Tano K, Chastain PD, 2nd, Adachi N, Sonoda E, Kikuchi K, Koyama H, Nagata K, Kaufman DG, Takeda S, Wilson SH, Watanabe M, Swenberg JA, Nakamura J. FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol Cancer Res. 2010;8:204–215. doi: 10.1158/1541-7786.MCR-09-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia. Dermatol Clin. 1995;13:41–49. [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Bessho T, Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J Biol Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- Boubriak I, Mason PA, Clancy DJ, Dockray J, Saunders RD, Cox LS. DmWRNexo is a 3′-5′ exonuclease: phenotypic and biochemical characterization of mutants of the Drosophila orthologue of human WRN exonuclease. Biogerontology. 2009;10:267–277. doi: 10.1007/s10522-008-9181-3. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Karmakar P, Sommers JA, Yang Q, Wang XW, Spillare EA, Harris CC, Bohr VA. p53 modulates the exonuclease activity of Werner syndrome protein. J Biol Chem. 2001;276:35093–35102. doi: 10.1074/jbc.M103332200. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr, Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J Biol Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- Brown KR, Weatherdon KL, Galligan CL, Skalski V. A nuclear 3′-5′ exonuclease proofreads for the exonuclease-deficient DNA polymerase alpha. DNA Repair (Amst) 2002;1:795–810. doi: 10.1016/S1568-7864(02)00115-5. [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL. Interplay of Mre11 nuclease with Dna2 plus Sgs1 in Rad51-dependent recombinational repair. PLoS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Cox LS, Campbell JL. Coordination of nucleases and helicases during DNA replication and double-strand break repair. In: Cox LS, editor. Molecular themes in DNA replication. Cambridge: Royal Society of Chemistry; 2009. pp. 112–155. [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtelow MA, Kaufmann SH, Karnitz LM. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J Biol Chem. 2000;275:26343–26348. doi: 10.1074/jbc.M001244200. [DOI] [PubMed] [Google Scholar]
- Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, Sigrist S, Madeo F. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Carr AM. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair (Amst) 2002;1:983–994. doi: 10.1016/S1568-7864(02)00165-9. [DOI] [PubMed] [Google Scholar]
- Chen X, Zuo S, Kelman Z, O'Donnell M, Hurwitz J, Goodman MF. Fidelity of eucaryotic DNA polymerase delta holoenzyme from Schizosaccharomyces pombe. J Biol Chem. 2000;275:17677–17682. doi: 10.1074/jbc.M910278199. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Lin YT, Lieberman HB, Chen G, Lee EY. ATM-dependent phosphorylation of human Rad9 is required for ionizing radiation-induced checkpoint activation. J Biol Chem. 2001;276:16580–16586. doi: 10.1074/jbc.M008871200. [DOI] [PubMed] [Google Scholar]
- Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8:1117–1127. doi: 10.1016/S1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Ma SM, Dumitrache LC, Hasty P. Biochemical and cellular characteristics of the 3′→5′ exonuclease TREX2. Nucleic Acids Res. 2007;35:2682–2694. doi: 10.1093/nar/gkm151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415:655–659. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- Chou KM, Cheng YC. The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J Biol Chem. 2003;278:18289–18296. doi: 10.1074/jbc.M212143200. [DOI] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, Lee HW, Zglinicki T, Ganser A, Schirmacher P, Nakauchi H, Rudolph KL. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Christmann M, Tomicic MT, Aasland D, Berdelle N, Kaina B. Three prime exonuclease I (TREX1) is Fos/AP-1 regulated by genotoxic stress and protects against ultraviolet light and benzo(a)pyrene-induced DNA damage. Nucleic Acids Res. 2010;38:6418–6432. doi: 10.1093/nar/gkq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L, Li B. The Werner syndrome protein at the crossroads of DNA repair and apoptosis. Mech Ageing Dev. 2004;125:521–528. doi: 10.1016/j.mad.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Constantinescu D, Csoka AB, Navara CS, Schatten GP. Defective DSB repair correlates with abnormal nuclear morphology and is improved with FTI treatment in Hutchinson-Gilford progeria syndrome fibroblasts. Exp Cell Res. 2010;316:2747–2759. doi: 10.1016/j.yexcr.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- Copeland WC, Longley MJ. DNA2 resolves expanding flap in mitochondrial base excision repair. Mol Cell. 2008;32:457–458. doi: 10.1016/j.molcel.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS. Multiple pathways control cell growth and transformation: overlapping and independent activities of p53 and p21Cip1/WAF1/Sdi1. J Pathol. 1997;183:134–140. doi: 10.1002/(SICI)1096-9896(199710)183:2<134::AID-PATH960>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Cox LS. Live fast, die young: new lessons in mammalian longevity. Rejuvenation Res. 2009;12:283–288. doi: 10.1089/rej.2009.0894. [DOI] [PubMed] [Google Scholar]
- Cox LS, Kearsey S. Ring structures and six-fold symmetry in DNA replication. In: Cox LS, editor. Molecular themes in DNA replication. Cambridge: Royal Society of Chemistry; 2009. pp. 47–85. [Google Scholar]
- Cox LS, Lane DP. Tumour suppressors, kinases and clamps: how p53 regulates the cell cycle in response to DNA damage. Bioessays. 1995;17:501–508. doi: 10.1002/bies.950170606. [DOI] [PubMed] [Google Scholar]
- Cox LS, Mattison JA. Increasing longevity through caloric restriction or rapamycin feeding in mammals: common mechanisms for common outcomes? Aging Cell. 2009;8:607–613. doi: 10.1111/j.1474-9726.2009.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS, Clancy DJ, Boubriak I, Saunders RD. Modeling Werner Syndrome in Drosophila melanogaster: hyper-recombination in flies lacking WRN-like exonuclease. Ann N Y Acad Sci. 2007;1119:274–288. doi: 10.1196/annals.1404.009. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi–Goutieres syndrome at the AGS1 locus. Nat Genet. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW. Identification of germ plasm-associated transcripts by microarray analysis of Xenopus vegetal cortex RNA. Dev Dyn. 2010;239:1838–1848. doi: 10.1002/dvdy.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymerman IA, Chung I, Beckmann BM, Bujnicki JM, Meiss G. EXOG, a novel paralog of endonuclease G in higher eukaryotes. Nucleic Acids Res. 2008;36:1369–1379. doi: 10.1093/nar/gkm1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dake E, Hofmann TJ, McIntire S, Hudson A, Zassenhaus HP. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1988;263:7691–7702. [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, Gent DC. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Ivanova IG, Raykov V, Xue Y, Maringele L. Polymerase epsilon is required to maintain replicative senescence. Mol Cell Biol. 2011;31:1637–1645. doi: 10.1128/MCB.00144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP. The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9–rad1–hus1 DNA damage checkpoint complex—implications for clamp loading and regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini P, Huang KN, Tishkoff DX, Kolodner RD, Symington LS. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini BK, Pokharel S, Polaczek P, Balakrishnan L, Bambara RA, Campbell JL. Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J Biol Chem. 2011;286:23763–23770. doi: 10.1074/jbc.M111.243071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panourgia MP, Invidia L, Celani L, Scurti M, Cevenini E, Castellani GC, Salvioli S. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/S0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Wood RD. New insights into the combined Cockayne/xeroderma pigmentosum complex: human XPG protein can function in transcription factor stability. Mol Cell. 2007;26:162–164. doi: 10.1016/j.molcel.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, Lane DP, Abbondandolo A, Dogliotti E. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/S1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y, Sung P, Prakash L, Prakash S. A conserved 5′ to 3′ exonuclease activity in the yeast and human nucleotide excision repair proteins RAD2 and XPG. J Biol Chem. 1994;269:31342–31345. [PubMed] [Google Scholar]
- Habraken Y, Sung P, Prakash L, Prakash S. Human xeroderma pigmentosum group G gene encodes a DNA endonuclease. Nucleic Acids Res. 1994;22:3312–3316. doi: 10.1093/nar/22.16.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi MZ, Wilson DM., 3rd Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen. 2000;36:312–324. doi: 10.1002/1098-2280(2000)36:4<312::AID-EM7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Harrigan JA, Wilson DM, 3rd, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase beta. Nucleic Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Lieber MR. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 1994;13:1235–1246. doi: 10.1002/j.1460-2075.1994.tb06373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiona A, Sanz A, Kujoth GC, Pamplona R, Seo AY, Hofer T, Someya S, Miyakawa T, Nakayama C, Samhan-Arias AK, Servais S, Barger JL, Portero-Otin M, Tanokura M, Prolla TA, Leeuwenburgh C. Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One. 2010;5:e11468. doi: 10.1371/journal.pone.0011468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka LR, Harrington JJ, Gerhard DS, Lieber MR, Hsieh CL. Sequence of human FEN-1, a structure-specific endonuclease, and chromosomal localization of the gene (FEN1) in mouse and human. Genomics. 1995;25:220–225. doi: 10.1016/0888-7543(95)80129-A. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Shomer B, Greenblatt M, Soussi T, Hovig E, Montesano R, Harris CC. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 1996;24:141–146. doi: 10.1093/nar/24.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes LL, Budd M, Choe W, Weitao T, Campbell JL. Mutations in DNA replication genes reduce yeast life span. Mol Cell Biol. 2002;22:4136–4146. doi: 10.1128/MCB.22.12.4136-4146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Frank G, Weng Y, Tainer JA, Shen B. Newly discovered archaebacterial flap endonucleases show a structure-specific mechanism for DNA substrate binding and catalysis resembling human flap endonuclease-1. J Biol Chem. 1998;273:27154–27161. doi: 10.1074/jbc.273.42.27154. [DOI] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/S0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MM, Arnheim N, Goodman MF. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 1992;20:4567–4573. doi: 10.1093/nar/20.17.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′ → 5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′ → 5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KJ, Ku CC, Lehman IR. Endonuclease G: a role for the enzyme in recombination and cellular proliferation. Proc Natl Acad Sci U S A. 2006;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadinanos J, Horner JW, Maratos-Flier E, Depinho RA. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph IS, Kumari A, Bhattacharyya MK, Gao H, Li B, Lustig AJ. An mre11 mutation that promotes telomere recombination and an efficient bypass of senescence. Genetics. 2010;185:761–770. doi: 10.1534/genetics.110.117598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L, Beutner G, Phadnis N, Sheu SS, Sia EA. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst) 2009;8:1242–1249. doi: 10.1016/j.dnarep.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HI, Bambara RA. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit Rev Biochem Mol Biol. 2003;38:433–452. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- Kao HI, Veeraraghavan J, Polaczek P, Campbell JL, Bambara RA. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J Biol Chem. 2004;279:15014–15024. doi: 10.1074/jbc.M313216200. [DOI] [PubMed] [Google Scholar]
- Kashino G, Kodama S, Suzuki K, Matsumoto T, Watanabe M. Exogenous expression of exonuclease domain-deleted WRN interferes with the repair of radiation-induced DNA damages. J Radiat Res (Tokyo) 2005;46:407–414. doi: 10.1269/jrr.46.407. [DOI] [PubMed] [Google Scholar]
- Kawase T, Ichikawa H, Ohta T, Nozaki N, Tashiro F, Ohki R, Taya Y. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene. 2008;27:3797–3810. doi: 10.1038/onc.2008.32. [DOI] [PubMed] [Google Scholar]
- Kirchner JM, Tran H, Resnick MA. A DNA polymerase epsilon mutant that specifically causes +1 frameshift mutations within homonucleotide runs in yeast. Genetics. 2000;155:1623–1632. doi: 10.1093/genetics/155.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Miyashita T, Hang H, Hopkins KM, Zheng W, Cuddeback S, Yamada M, Lieberman HB, Wang HG. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol. 2000;2:1–6. doi: 10.1038/71316. [DOI] [PubMed] [Google Scholar]
- Kong YX, Bergen N, Trounce IA, Bui BV, Chrysostomou V, Waugh H, Vingrys A, Crowston JG. Increase in mitochondrial DNA mutations impairs retinal function and renders the retina vulnerable to injury. Aging Cell. 2011;10:572–583. doi: 10.1111/j.1474-9726.2011.00690.x. [DOI] [PubMed] [Google Scholar]
- Korona DA, Lecompte KG, Pursell ZF. The high fidelity and unique error signature of human DNA polymerase epsilon. Nucleic Acids Res. 2010;39:1763–1773. doi: 10.1093/nar/gkq1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Simon DK, Turnbull DM, Khrapko K. Do mtDNA deletions drive premature aging in mtDNA mutator mice? Aging Cell. 2009;8:502–506. doi: 10.1111/j.1474-9726.2009.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati M, Yang K, Kuraguchi M, Zhao J, Lia M, Heyer J, Kane MF, Fan K, Russell R, Brown AM, Kneitz B, Edelmann W, Kolodner RD, Lipkin M, Kucherlapati R. Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc Natl Acad Sci U S A. 2002;99:9924–9929. doi: 10.1073/pnas.152321699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J Biol Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- Lee BI, Wilson DM., 3rd The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- Lee MH, Hollis SE, Yoo BH, Nykamp K. Caenorhabditis elegans DNA-2 helicase/endonuclease plays a vital role in maintaining genome stability, morphogenesis, and life span. Biochem Biophys Res Commun. 2011;407:495–500. doi: 10.1016/j.bbrc.2011.03.045. [DOI] [PubMed] [Google Scholar]
- Li B, Comai L. Displacement of DNA-PKcs from DNA ends by the Werner syndrome protein. Nucleic Acids Res. 2002;30:3653–3661. doi: 10.1093/nar/gkf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- Li B, Conway N, Navarro S, Comai L. A conserved and species-specific functional interaction between the Werner syndrome-like exonuclease atWEX and the Ku heterodimer in Arabidopsis. Nucleic Acids Res. 2005;33:6861–6867. doi: 10.1093/nar/gki984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE, Yang YG, Robins P. Biochemical properties of mammalian TREX1 and its association with DNA replication and inherited inflammatory disease. Biochem Soc Trans. 2009;37:535–538. doi: 10.1042/BST0370535. [DOI] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Wauson EM, Graves LM, Sancar A. The human Rad9 checkpoint protein stimulates the carbamoyl phosphate synthetase activity of the multifunctional protein CAD. Nucleic Acids Res. 2004;32:4524–4530. doi: 10.1093/nar/gkh789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol Cell Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, 3rd, Shen B, Demple B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008;28:4975–4987. doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Orren DK. Length-dependent degradation of single-stranded 3′ ends by the Werner syndrome protein (WRN): implications for spatial orientation and coordinated 3′ to 5′ movement of its ATPase/helicase and exonuclease domains. BMC Mol Biol. 2006;7:6. doi: 10.1186/1471-2199-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, Lilley DM, Rouse J. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill SA. DNA replication: partners in the Okazaki two-step. Curr Biol. 2001;11:R842–R844. doi: 10.1016/S0960-9822(01)00500-0. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′→5′ exonucleases. J Biol Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′→5′ exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3′→5′ exonuclease genes. J Biol Chem. 2001;276:14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- Meier B, Clejan I, Liu Y, Lowden M, Gartner A, Hodgkin J, Ahmed S. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2006;2:e18. doi: 10.1371/journal.pgen.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel J, Economos AC, Fleming J, Johnson JE., Jr Mitochondrial role in cell aging. Exp Gerontol. 1980;15:575–591. doi: 10.1016/0531-5565(80)90010-8. [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JH. DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3′→5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritani M, Nomura K, Tanahashi T, Osabe D, Fujita Y, Shinohara S, Yamaguchi Y, Keshavarz P, Kudo E, Nakamura N, Yoshikawa T, Ichiishi E, Takata Y, Yasui N, Shiota H, Kunika K, Inoue H, Itakura M. Genetic association of single nucleotide polymorphisms in endonuclease G-like 1 gene with type 2 diabetes in a Japanese population. Diabetologia. 2007;50:1218–1227. doi: 10.1007/s00125-007-0631-2. [DOI] [PubMed] [Google Scholar]
- Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/S0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- Myles GM, Sancar A. DNA repair. Chem Res Toxicol. 1989;2:197–226. doi: 10.1021/tx00010a001. [DOI] [PubMed] [Google Scholar]
- Nebel A, Flachsbart F, Till A, Caliebe A, Blanche H, Arlt A, Hasler R, Jacobs G, Kleindorp R, Franke A, Shen B, Nikolaus S, Krawczak M, Rosenstiel P, Schreiber S. A functional EXO1 promoter variant is associated with prolonged life expectancy in centenarians. Mech Ageing Dev. 2009;130:691–699. doi: 10.1016/j.mad.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Negritto MC, Qiu J, Ratay DO, Shen B, Bailis AM. Novel function of Rad27 (FEN-1) in restricting short-sequence recombination. Mol Cell Biol. 2001;21:2349–2358. doi: 10.1128/MCB.21.7.2349-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–962. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Pursell ZF, Kunkel TA. Mechanisms for high fidelity DNA replication. In: Cox LS, editor. Molecular themes in DNA replication. Cambridge: Royal Society of Chemistry; 2009. pp. 86–111. [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, Leeuwen W, Theil AF, Vermeulen W, Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211:90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, Sigvardsson M, Bryder D. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- O'Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- Ohsato T, Ishihara N, Muta T, Umeda S, Ikeda S, Mihara K, Hamasaki N, Kang D. Mammalian mitochondrial endonuclease G. Digestion of R-loops and localization in intermembrane space. Eur J Biochem. 2002;269:5765–5770. doi: 10.1046/j.1432-1033.2002.03238.x. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker AE, Weyer I, Laus MC, Oostveen I, Yon J, Verhasselt P, Luyten WH. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, Miwa S, Olijslagers S, Hallinan J, Wipat A, Saretzki G, Rudolph KL, Kirkwood TB, Zglinicki T. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/S1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Loeb LA. Hydrolysis of 3′-terminal mispairs in vitro by the 3′-5′ exonuclease of DNA polymerase delta permits subsequent extension by DNA polymerase alpha. Biochemistry. 1990;29:5226–5231. doi: 10.1021/bi00474a002. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- Plchova H, Hartung F, Puchta H. Biochemical characterization of an exonuclease from Arabidopsis thaliana reveals similarities to the DNA exonuclease of the human Werner syndrome protein. J Biol Chem. 2003;278:44128–44138. doi: 10.1074/jbc.M303891200. [DOI] [PubMed] [Google Scholar]
- Qiu J, Qian Y, Chen V, Guan MX, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- Richards A, Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-Labarca ML, Terwindt GM, Kasai Y, McLellan M, Grand MG, Vanmolkot KR, Vries B, Wan J, Kane MJ, Mamsa H, Schafer R, Stam AH, Haan J, Jong PT, Storimans CW, Schooneveld MJ, Oosterhuis JA, Gschwendter A, Dichgans M, Kotschet KE, Hodgkinson S, Hardy TA, Delatycki MB, Hajj-Ali RA, Kothari PH, Nelson SF, Frants RR, Baloh RW, Ferrari MD, Atkinson JP. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech Ageing Dev. 2003;124:167–174. doi: 10.1016/S0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Whitby MC, Borer CM, Bachler MA, Cox LS. Correction of proliferation and drug sensitivity defects in the progeroid Werner's Syndrome by Holliday junction resolution. Rejuvenation Res. 2007;10:27–40. doi: 10.1089/rej.2006.0503. [DOI] [PubMed] [Google Scholar]
- Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, Prolla TA, Tarnopolsky MA. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in Werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Morioka H, Hakoshima T. Crystallization and preliminary crystallographic analysis of the catalytic domain of human flap endonuclease 1 in complex with a nicked DNA product: use of a DPCS kit for efficient protein-DNA complex crystallization. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:39–43. doi: 10.1107/S1744309107065372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RD, Boubriak I, Clancy DJ, Cox LS. Identification and characterization of a Drosophila ortholog of WRN exonuclease that is required to maintain genome integrity. Aging Cell. 2008;7:418–425. doi: 10.1111/j.1474-9726.2008.00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Kodandaramireddy NR, Ju Z, Lechel A, Stepczynska A, Lilli DR, Clark AB, Rudolph C, Kuhnel F, Wei K, Schlegelberger B, Schirmacher P, Kunkel TA, Greenberg RA, Edelmann W, Rudolph KL. Exonuclease-1 deletion impairs DNA damage signaling and prolongs lifespan of telomere-dysfunctional mice. Cell. 2007;130:863–877. doi: 10.1016/j.cell.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer P, Scholz SR, Gimadutdinow O, Cymerman IA, Bujnicki JM, Ruiz-Carrillo A, Pingoud A, Meiss G. Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J Mol Biol. 2004;338:217–228. doi: 10.1016/j.jmb.2004.02.069. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J Biol Chem. 2003;278:23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl-Cachedenier I, Munoz P, Flores JM, Klatt P, Blasco MA. Deficient mismatch repair improves organismal fitness and survival of mice with dysfunctional telomeres. Genes Dev. 2007;21:2234–2247. doi: 10.1101/gad.430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalski V, Brown KR, Choi BY, Lin ZY, Chen S. A 3′-5′ exonuclease in human leukemia cells: implications for resistance to 1-beta-D-arabinofuranosylcytosine and 9-beta-D-arabinofuranosyl-2-fluoroadenine 5′-monophosphate. J Biol Chem. 2000;275:25814–25819. doi: 10.1074/jbc.M001460200. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/S0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Theunissen JW, Morales M, Petrini JH. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair (Amst) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Swanson C, Saintigny Y, Emond MJ, Monnat RJ., Jr The Werner syndrome protein has separable recombination and survival functions. DNA Repair (Amst) 2004;3:475–482. doi: 10.1016/j.dnarep.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D'Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson E, Meldrum CJ, Crooks R, McPhillips M, Thomas L, Spigelman AD, Scott RJ. Hereditary non-polyposis colorectal cancer and the role of hPMS2 and hEXO1 mutations. Clin Genet. 2004;65:215–225. doi: 10.1111/j.1399-0004.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- Tian M, Jones DA, Smith M, Shinkura R, Alt FW. Deficiency in the nuclease activity of xeroderma pigmentosum G in mice leads to hypersensitivity to UV irradiation. Mol Cell Biol. 2004;24:2237–2242. doi: 10.1128/MCB.24.6.2237-2242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci U S A. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Amin NS, Viars CS, Arden KC, Kolodner RD. Identification of a human gene encoding a homologue of Saccharomyces cerevisiae EXO1, an exonuclease implicated in mismatch repair and recombination. Cancer Res. 1998;58:5027–5031. [PubMed] [Google Scholar]
- Toueille M, El-Andaloussi N, Frouin I, Freire R, Funk D, Shevelev I, Friedrich-Heineken E, Villani G, Hottiger MO, Hubscher U. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Gordenin DA, Resnick MA. The 3′→5′ exonucleases of DNA polymerases delta and epsilon and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trego KS, Chernikova SB, Davalos AR, Perry JJ, Finger LD, Ng C, Tsai MS, Yannone SM, Tainer JA, Campisi J, Cooper PK. The DNA repair endonuclease XPG interacts directly and functionally with the WRN helicase defective in Werner syndrome. Cell Cycle. 2011;10:1998–2007. doi: 10.4161/cc.10.12.15878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, Cooper PK, Grasby JA, Tainer JA. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent RD, Hofmann TJ, Zassenhaus HP. Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:3297–3312. doi: 10.1093/nar/16.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer E, Karnitz LM. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- Wang W, Brandt P, Rossi ML, Lindsey-Boltz L, Podust V, Fanning E, Sancar A, Bambara RA. The human Rad9-Rad1-Hus1 checkpoint complex stimulates flap endonuclease 1. Proc Natl Acad Sci U S A. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair. Oncogene. 1997;14:2313–2321. doi: 10.1038/sj.onc.1201072. [DOI] [PubMed] [Google Scholar]
- Weitao T, Budd M, Campbell JL. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- Weitao T, Budd M, Hoopes LL, Campbell JL. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J Biol Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- White PJ, Borts RH, Hirst MC. Stability of the human fragile X (CGG)(n) triplet repeat array in Saccharomyces cerevisiae deficient in aspects of DNA metabolism. Mol Cell Biol. 1999;19:5675–5684. doi: 10.1128/mcb.19.8.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnhoven SW, Hoogervorst EM, Waard H, Horst GT, Steeg H. Tissue specific mutagenic and carcinogenic responses in NER defective mouse models. Mutat Res. 2007;614:77–94. doi: 10.1016/j.mrfmmm.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Williams RS, Tainer JA. A nanomachine for making ends meet: MRN is a flexing scaffold for the repair of DNA double-strand breaks. Mol Cell. 2005;19:724–726. doi: 10.1016/j.molcel.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Wilson DM., 3rd Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol. 2003;330:1027–1037. doi: 10.1016/S0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- Wu Y, Berends MJ, Post JG, Mensink RG, Verlind E, Sluis T, Kempinga C, Sijmons RH, Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM. Germline mutations of EXO1 gene in patients with hereditary nonpolyposis colorectal cancer (HNPCC) and atypical HNPCC forms. Gastroenterology. 2001;120:1580–1587. doi: 10.1053/gast.2001.25117. [DOI] [PubMed] [Google Scholar]
- Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human rad9–hus1–rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zheng L, Dai H, Zhou M, Hua Y, Shen B. Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice. Oncogene. 2011;30:1072–1081. doi: 10.1038/onc.2010.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci U S A. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zassenhaus HP, Denniger G. Analysis of the role of the NUC1 endo/exonuclease in yeast mitochondrial DNA recombination. Curr Genet. 1994;25:142–149. doi: 10.1007/BF00309540. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen B. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, Tsark W, Huang Q, Kernstine K, Zhang X, Lin D, Shen B. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med. 2007;13:812–819. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Hegde ML, Zhou M, Guo Z, Wu X, Wu J, Su L, Zhong X, Mitra S, Huang Q, Kernstine KH, Pfeifer GP, Shen B. Fen1 mutations that specifically disrupt its interaction with PCNA cause aneuploidy-associated cancer. Cell Res. 2011;21:1052–1067. doi: 10.1038/cr.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]