Abstract
Global gene expression in yeast was examined in five different nutrient-limited steady states and in their corresponding starvation-induced stationary phases. The use of chemostats, with their ability to generate defined and reproducible physiological conditions, permitted the exclusion of the confounding variables that frequently complicate transcriptome analyses. This approach allowed us to dissect out effects on gene expression that are specific to particular physiological states. Thus, we discovered that a large number of ORFs involved in protein synthesis were activated under ammonium limitation, whereas the expression of ORFs concerned with energy and metabolism was enhanced by carbon limitation. Elevated transcription of genes in high-affinity glucose uptake, the trichloroacetic acid cycle, and oxidative phosphorylation were observed in glucose-limiting, but not glucose-abundant, conditions. In contrast, genes involved in gluconeogenesis and, interestingly, genes subject to nitrogen catabolite repression increased their transcription when ethanol was the carbon source, even though ammonium was in excess. This result suggests that up-regulation of genes sensitive to nitrogen catabolite repression may contribute anapleurotic intermediates in ethanol-grown cells. The different starvation conditions produced two general types of transcription profiles, with carbon-starved cells transcribing far fewer genes than cells starved for any of the other macronutrients. Nonetheless, each starvation condition induced its own peculiar set of genes, and only 17 genes were induced >5-fold by all five starvations. In all cases, analysis of the upstream sequences of clusters of coregulated genes identified motifs that may be recognized by transcription factors specific for controlling gene expression in each of the physiological conditions examined.
Keywords: chemostat culture, global gene expression, nutrient limitation, nutrient starvation
The availability of complete genome sequences for an increasing number of organisms has created a demand for techniques to facilitate the analysis of gene function that are as comprehensive as the genome sequences themselves. Thus, the concepts of transcriptome, proteome, and metabolome have evolved to represent, respectively, the complete set of mRNAs, proteins, and low-molecular-weight intermediates present in a cell (or cell type) under a given set of physiological, developmental, or pathological conditions (1). Among these different levels of functional genomic analysis, the transcriptome has attracted the most attention, because the technique of hybridization-array analysis is facile, high throughput, and truly comprehensive in scope. However, the comprehensive nature of transcriptome analysis is also its greatest weakness. The problem is to garner the relevant information from the huge amount of data obtained. There are two main ways of addressing this problem. The first is to carry out a large number of experiments and use data-mining techniques to extract the desired information from the mass of data. This solution is best exemplified by the compendium approach (2). The alternative is to carry out a much smaller number of experiments but to control experimental conditions such that all, or most, confounding variables are eliminated, and only relevant information is obtained.
This second approach is most readily pursued with microorganisms because, with these, the experimenter has complete control over the cells' physical and chemical environment. Even so, the usual design of experiments to investigate gene expression in microorganisms, involving the growth of cells in batch culture, presents considerable complexity to the investigator. This is because, during batch culture, the cells pass through five distinct phases of growth: lag, acceleration, exponential, deceleration, and stationary. Even during exponential growth, the growth rate may change due to phenomena such as diauxie. In any case, throughout the exponential phase, cell numbers increase, nutrient concentrations decrease, and the levels of excreted metabolites increase. To all of these confounding variables may be added the fact that the experimenter has no control over the growth rate achieved by the culture during the exponential phase, and the physiological or genetic phenomena under study may well affect growth rate. Recent studies have demonstrated that the impact on gene expression due to the reduced growth rate of the mcm1 mutants completely obscured the primary effects of the mutations on the transcription of specific sets of genes (3, 4).
All of these confounding variables and the complexity associated with them may be removed by the use of chemostat culture (5), because, at steady state, all culture parameters are held constant, including temperature, pH, dissolved oxygen, nutrients, excreted metabolites, cell numbers, and growth rate. By judicious medium design, any nutrient can be made to determine (or limit) the growth rate of the cells. Moreover, growth rate can be set at any value below the maximum achievable by the particular microbial strain used on the nutrient medium chosen by controlling the dilution rate of the culture. Despite these advantages, few previous studies have exploited chemostat culture in yeast transcriptome analysis. One work compared gene expression in yeast cultures grown aerobically and anaerobically (6) and another, the transition between growth on glucose to that on oleate (7). Here we present data on the nutrient control of gene expression in Saccharomyces cerevisiae using chemostat cultures. In all experiments, the same strain of yeast and chemically defined medium were used, and the same growth rate was maintained for each nutrient-limiting condition. Nutrient starvation was achieved by simply turning off the medium feed to the steady-state cultures. By controlling the culture parameters in this way, we have eliminated confounding variables and produced a considerable amount of information concerning the nutritional control of gene expression, as well as revealing some previously undescribed relationships between the different domains of metabolism within the yeast cell.
Materials and Methods
Chemostat Culture and RNA Extraction. Diploid strain FY1679/ΔHO was used in all experiments. Five chemostats, each run at a dilution rate of 0.1 h-1, were set up as described (8) and run in duplicate. All five chemostats were maintained at 100% O2 saturation. To establish the starvation-induced stationary phases, the medium feed to each chemostat was switched off during steady state, and stationary-phase samples were taken after the cell density (OD600) had reached a constant value, and >90% of the cells had no buds (after 48 h). Cells were broken with a dismembranator (Braun, Melsungen, Germany) and total RNA extracted by using TRIzol reagent (3).
Target Labeling and Hybridization. Arrays, comprising PCR products for each of 6,200 yeast ORFs, spotted (in duplicate) on nylon membranes, were supplied by J. Hoheisel (Deutsches Krebsforschungszentrum, Heidelberg). Total yeast RNA (30-35 μg) was used to make first-strand cDNA target (9). An assessment of incorporation efficiency of the 33P-dCTP label was made before each hybridization. Prehybridization, hybridization, and washing were carried out according to published protocols (10).
Imaging and Data Analysis. Hybridization images were captured by using the storm 860 phosphorimager (Molecular Dynamics). Primary data were analyzed by using arrayvision 4.0 software (Imaging Research, St. Catherine's, ON, Canada). The expression level of an ORF was normalized as its fractional contribution to the total hybridization signal on a membrane. Blank spots were treated as controls. Twice the SD of the signal from the blank spots was used as the threshold to ensure that a signal was truly a result of hybridization (>95% probability) rather than due to background radiation. To assess reproducibility, duplicate experiments were carried out. Without filtering the data, the correlation coefficient was 0.96 for duplicates using the same filter and 0.94 for different filters. All of the raw data are presented at www.cs.man.ac.uk/cogeme/data, and full details of data analysis, including the normalization and statistical regimes, can be found in Supporting Text and Figs. 4-16, which are published as supporting information on the PNAS web site.
To determine the degree to which transcription of a particular ORF was regulated under a given nutrient limitation, the normalized value from that condition was divided by the corresponding value from the other nutrient-limited conditions, where the studied nutrient was in excess, and converted to log2. Positive values >1 and negative values <-1 under all of the comparisons then define up- and down-regulated ORFs under the studied nutrient limitation, with reference to the nutrient excess conditions. Regulated ORFs were placed into 16 functional categories as defined by the Munich Information Center for Protein Sequences Yeast Genome Database (http://mips.gsf.de/proj/yeast) (11).
The regulation of transcription across five different nutrient-limited conditions was compared by using self-organizing maps to detect clusters of genes with similar expression patterns. Clustering was performed by using genecluster software (http://www-genome.wi.mit.edu/cancer/software/software.html) (12). Identification of possible upstream regulatory sites in groups of coregulated genes was performed by using regulatory sequence analysis tools (http://rsat.ulb.ac.be/rsat). Possible regulatory sites were searched for in both the literature and the Saccharomyces cerevisiae Promoter Database (http://cgsigma.cshl.org/jian).
Results
Global Transcription Changes in Response to Different Nutrient Limitations and Starvations. Among the 6,280 ORFs arrayed on the filter, 1,297 of them (20.6% of the genome) were expressed at a level lower than a 2 × SD of the background in all of the steady states and starvation conditions. These ORFs were defined as not, or very lowly, expressed under these conditions. There were 1,038 ORFs (16.5% of the genome) that could be detected at a level higher than this threshold under all conditions. These ORFs may be regarded as housekeeping genes; 15% of them are classified as essential genes in the Munich Information Center for Protein Sequences database. This fraction is similar to the percentage of essential genes for the entire yeast genome (13), but this is the first indication that they may be required for stationary phase survival as well as mitotic growth. The number of ORFs expressed at a measurable level under each condition is summarized in Table 1.
Table 1. Number of ORFs expressed under different nutrient conditions.
| Limiting nutrient | SS | ST | Shut-off from SS to ST | Induced >5-fold at ST |
|---|---|---|---|---|
| Glucose | 3,903 (62.1) | 2,204 (35.0) | (27.1) | 268 |
| Ethanol | 3,551 (56.5) | 2,419 (38.5) | (18.0) | 68 |
| Ammonium | 4,361 (69.4) | 4,138 (65.8) | (3.6) | 171 |
| Phosphate | 3,446 (54.8) | 3,404 (54.1) | (0.7) | 231 |
| Sulphate | 2,488 (39.6) | 2,019 (32.1) | (7.5) | 85 |
| All conditions | 1,883 (29.9) | 1,346 (21.4) | — | 17 |
| <2 × SD in any condition | 1,646 (26.2) | 1,767 (28.1) | — | — |
SS, steady state; ST, starvation. Percentage of genome is included in brackets.
The number of genes expressed in each nutrient-starved stationary phase was compared with that expressed in its corresponding steady state. As summarized in Table 1, transcripts for fewer ORFs were detected in the stationary phase than in growing cells, in agreement with previous studies (14, 15). Furthermore, the transcription profiles of the five stationary phases may be divided into two classes, according to the number of ORFs expressed. Class 1 comprises carbon-starved (glucose and ethanol) stationary phases and is characterized by their being far fewer genes transcribed than in steady state. Class 2 contains all noncarbon-starved (ammonium, phosphate, and sulfate) stationary phases. In this class, the transcription pattern in each stationary phase was qualitatively similar to that in its corresponding steady state. These results imply that deprivation of a carbon source results in a major reduction in gene activity that may be crucial for maintaining cell viability over prolonged periods without growth.
Comparing each dataset with all other limiting conditions, where the studied nutrient was abundant, we were able to identify ORFs that were up- or down-regulated under each nutrient limitation. We initially tested this approach by comparing the dataset from the phosphate-limited steady state with those from phosphate-abundant conditions (C, N, and S limitations). As might have been expected, PHO5 and PHO84 were the two most up-regulated genes in phosphate-limited cells. Several additional genes, including PHO11, PHO12, PHM6, and SPL2, were activated in response to phosphate limitation (see Table 4, which is published as supporting information on the PNAS web site, for complete list). These data suggested that the cross comparison to the nutrient-excess cultures was an effective way of assessing the regulatory impact of specific nutrient limitations.
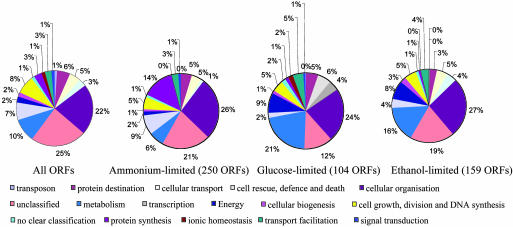
We then identified ORFs up-regulated under each of the nutrient limitations. For instance, there were 250 and 104 ORFs up-regulated under glucose and ammonium limitations, respectively. These ORFs were then categorized according to their functions as defined by the Munich Information Center for Protein Sequences database. As compared to the distribution of all ORFs and those up-regulated under the glucose-limiting condition, a larger number of ORFs activated under nitrogen limitation fall into the protein synthesis class (Fig. 1). In contrast, the transcription of more ORFs from the metabolism and energy categories was increased when glucose was limiting. Similarly, as compared with either glucose-limiting or -abundant (N-, P-, or S-limiting) conditions, 159 ORFs were found up-regulated with ethanol as carbon source. These ORFs have a similar distribution of functions to those up-regulated under glucose limitation (Fig. 1), suggesting that carbon limitation has a similar global impact on expression of genes from the different functional categories, irrespective of the identity of the carbon source.
Fig. 1.
Functional categories of all ORFs and of up-regulated ORFs. The percentage of these ORFs in each of the 16 functional categories is shown.
Genes Encoding Enzymes in the Trichloroacetic Acid (TCA) and Glyoxylate Cycles Were Activated by Glucose and Ethanol Limitation, Respectively. Among the 250 ORFs that showed increased transcript levels when glucose was limiting, 76 were up-regulated >3-fold. Nearly half of these (33 ORFs) fall into the metabolism functional category, which prompted us to study these ORFs in more detail by fitting them into the pathways of central carbon metabolism, by using the Kyoto Encyclopedia of Genes and Genomes database (www.genome.ad.jp/kegg/metabolism.html). The expression of two genes encoding high-affinity hexose transporters (HXT6 and HXT7) and all three genes specifying glucose kinases (HXK1, HXK2, and GLK1) was up-regulated under glucose limitation. In addition, genes encoding enzymes in the pentose phosphate pathway and the TCA cycle increased their transcript levels in response to glucose limitation. Fig. 2 shows that almost every step in the TCA cycle has one or more genes up-regulated in the glucose-limited steady state. Furthermore, the two glutamate-dehydrogenase genes (GDH1 and GDH3) were shown to be up-regulated. Gdh3p accounts for <0.5% of the total GDH-NADP activity of cells grown with ammonia as the sole nitrogen source (16) and may have a sensing role in nitrogen regulation (17). Its up-regulation on glucose limitation suggests that it may act in both carbon and nitrogen regulation in response to the depletion of glutamate, an essential intermediate for amino acid and nucleotide biosynthesis.
Fig. 2.
Genes of central carbon metabolism activated under glucose-limited (red) and ethanol-limited (green) conditions. Boxes are drawn around the sections of the metabolic chart that include the glyoxylate and TCA cycles, respectively, to highlight the gluconeogenic and glucose-catabolic regions of metabolism. Question marks next to gene names indicate uncertainty over their involvement in that step of the pathway.
Among the 159 ORFs induced under ethanol limitation, 52 were activated >3-fold as compared to cells grown under all conditions where glucose was the carbon source. In the list of the TCA-cycle-related genes activated under glucose limitation, only two (CIT1 and GDH3, blue in Fig. 2) were further up-regulated, three (LPD1, IDP1, and GDH1) down-regulated, and the rest remained unchanged when ethanol was the carbon source. Instead, genes encoding proteins involved in the pathways leading from ethanol to: (i) the synthesis of acetyl-CoA and its transport into mitochondria; (ii) the glyoxylate cycle, which is responsible for gluconeogenesis and succinate replenishment for the TCA cycle; and (iii) the export of α-ketoglutarate out of the mitochondria for glutamate and lysine biosynthesis were all shown to be activated (genes in green in Fig. 2). Moreover, genes encoding subunits of respiratory complexes were specifically up-regulated in ethanol-limited, as opposed to glucose-limited, conditions, indicating that the oxidative phosphorylation pathway is further activated in ethanol-limited cells.
More interestingly, a few genes under nitrogen catabolite repression increased their transcription in the ethanol-limited, as compared to the aerobic glucose-limited, steady state. These included MEP2, PUT4, DAL2, DAL4, and DAL5. It was expected that these genes would be repressed by the excess of ammonium in the carbon-limited culture (18). Indeed, this is true for the glucose-limited steady state. That these genes are induced under ethanol-limitation suggests there is cross-talk between carbon and nitrogen metabolism, probably through some key regulators controlling both these domains of metabolism (see Discussion).
Genes Commonly Activated at Starvation Conditions. About 250 ORFs were found to be down-regulated in all five starvation-induced stationary phases. Twelve percent have no known function, and the majority of the remainder fall into three functional categories: cellular organization (75), transcription (35), and protein synthesis (40). In the transcription category, most encode proteins required for synthesizing and processing rRNAs (8) or encode RNA polymerases II and III (2) or proteins involved in transcription activation and DNA binding (6), including histones (8). Within the protein synthesis category, the majority of these ORFs encode ribosomal proteins (19), have products involved in translation initiation and elongation (11), or specify aminoacyl-tRNA-synthases (8). When cells enter stationary phase, their protein synthetic rate decreases to <0.3% of that observed in exponentially growing cells (15). Our data show that they concomitantly down-regulate the expression of genes encoding components of the transcriptional and translational apparatus, although there may still be excess translational capacity in stationary-phase cells (19).
In contrast to the general shut-down of transcription on starvation, a few genes are up-regulated in the nutrient-starved stationary phases that we examined. The number of ORFs induced in response to different nutrient starvations, compared with their preceding nutrient-limited steady state, is summarized in Table 1. Little difference in their distribution among the functional groups was found, and ≈25% of these genes have no recognized functions. Details of the top 50 ORFs activated under each of the starvation conditions are listed in Tables 5-9, which are published as supporting information on the PNAS web site.
There are just 17 ORFs that are commonly up-regulated >5-fold in all five starvation conditions (Table 2). Only one of them (TBF1) is essential for vegetative growth. This group is of special interest, because it may provide some clues as to the ways yeast cells adapt to a nutrient-starved stationary phase from an actively growing state. Until now, this adaptation has been very poorly characterized, as demonstrated by the fact that 11 of these 17 genes have no known functions.
Table 2. Genes commonly induced > 5-fold in all starvation conditions.
| ORF (gene) | Function | Interaction |
|---|---|---|
| YBL049w | Unknown | CAP2, YIP1 |
| YBL075c (SSA3) | Heat-shock protein | CDC25 |
| YBL078c (AUT7) | Microtubule-binding protein | APG7, AUT1, AUT2 |
| YDL169c (UGX2) | Unknown | |
| YDL218w | Unknown | Rna15p |
| YGR201c | Unknown | |
| YGR236c (SPG1) | Unknown | |
| YHR096c (HXT5) | Hexose transporter | |
| YHR097c | Unknown | |
| YIL055c | Unknown | |
| YIL057c | Unknown | |
| YJL144w | Unknown | |
| YML058w (SML1) | Ribonucleotide reductase inhibitor | DDC2, RNR1 |
| YMR107w | Unknown | |
| YPL128c (TBF1) | TTAGGG repeat-binding factor | YKL090W |
| YPL186c | Unknown | |
| YPL223c (GRE1) | Induced by osmotic stress |
Among the six genes with known functions, SML1 encodes an inhibitor of ribonucleotide reductase (20), which catalyzes a rate-limiting step in DNA precursor synthesis (21). AUT7 is predicted to play a role in the Cvt pathway, during active growth, and in the autophagic uptake of proteins during starvation (22). Their up-regulation indicates that DNA synthesis is inhibited and bulk protein degradation activated in nutrient-starved cells. Ssa3p is a cytosolic member of the Hsp70 family that is induced by a variety of stress conditions, including heat shock and nutrient starvation (23). It is known that cytosolic members of the Hsp70 family are involved in the transport of cytosolic contents into the yeast vacuole (24). The specific up-regulation of SSA3 suggests it may play a dominant role as a chaperone in nutrient-starved cells. Hxt5p is a hexose transporter, but deletion of HXT5 has no clear phenotype (13). That HXT5 is strongly induced under all five nutrient-starved stationary phases in the absence or presence of extracellular glucose suggests that its expression is controlled by the reduction or cessation of growth, a fact confirmed by the chemostat studies of Verwaal et al. (25).
Among the ORFs with no clear function, GRE1, SPG1, UGX2, YDL218w, YIL055c, YHR097c, and YPL186c are genes whose transcripts accumulate in response to any combination of stress conditions (26). The universal induction of these genes under different nutrient-starved stationary phases and different stresses indicates that nutrient starvation is another kind of stress condition. Previous studies have indicated that YBL049w encodes a putative N-myristoylprotein (27), and that specific N-myristoylproteins contribute to stationary-phase survival, suggesting that these genes may play an important role in cell survival during stationary phase.
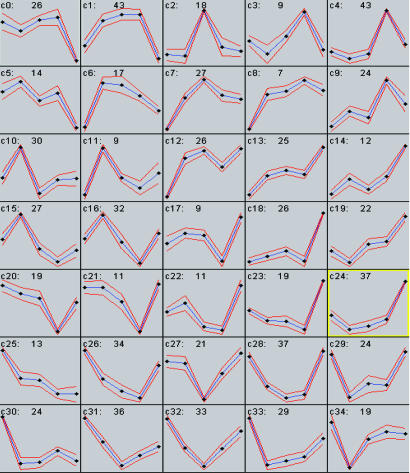
Clustering Coregulated Genes and Promoter Analysis. Only ORFs whose expression was up- or down-regulated >2-fold across different nutrient limitation conditions were subjected to clustering (see Materials and Methods). As shown in Fig. 3, the 18 ORFs in cluster c2 have their peak expression levels in the phosphate-limited steady state and comprise mostly PHO genes. The most significant oligonucleotide motifs identified for this cluster are CACGT(G/C) (Table 3), the core sequences bound by the transcription factor Pho4p, which is required for expression of phosphate pathway genes on phosphate limitation (28).
Fig. 3.
Clustering of ORFs up-regulated >2-fold across five different nutrient-limited conditions. The y axis stands for relative expression level across glucose-, ammonium-, phosphate-, sulfate-, and ethanol-limited conditions (x axis, left to right). The blue lines represent the average expression pattern of each cluster; the red lines indicate variation around the average pattern. Each cell is labeled with the cluster name and the number of ORFs that it contains.
Table 3. Overrepresented motifs in ORF upstream regions.
| Cluster | Oligonucleotide and dyad motifs, significance |
|---|---|
| c0 | ccaga 3.4, catca 3.3, gcaccc 3.0 (AFT1), cctn14ggc 1.2, ctcn15aga 1.1 |
| c1 | ccaca 2.5, aaccc 1.9, ccgaa 1.4, gtgn1gca 3.4, cgcn1tgc 2.2, accn5ggt 1.98 (ZAP1) |
| c28 | ggcca 5.5, cgggc 4.7, gcgac 4.2, cgcn11cac 3.0, cggn17cgc 2.8, ctgn7gcc 2.1 |
| c29 | ccgcc 4.3, cgggc 3.2, ctccc 2.5, gcgn8cca 3.5, tggn20caa 2.4, gggn19cca 1.9 |
| c30 | cccct 5.0 (MSN2/4), ccccc 2.8, cccccc 2.8, agcn9agc 2.3, cccn7gcc 2.0 |
| c18 | catcc 3.2, gatga 1.1, tcan18tca 6.6, atcn12atc 2.9, gatn20tga 2.1 |
| c23 | tccca 1.4, ctcn15cag 1.3 |
| c24 | gccca 3.9, ccgcc 3.3, aatgg 2.0, ccgn5ccg 2.0 (CAT8), ccan5ccg 1.4 (CAT8) |
| c15, c16 | ccaga 3.3, ggcca 2.3, agatg 1.6, tggn8gaa 2.5, atgn3ggg 1.4, atgn2tgg 1.2 |
| c4 | ctgga 3.4, ggcca 2.8, caagg 2.7, ccan8ttc 1.8, ccan15agg 1.2, caan11tgg 1.1 |
| c2 | acgtg 6.4 (PHO4), acgtgc 2.6, acgtgg 2.0 |
The significance index, listed beside the sequences, is the -log10 transform of the E value; higher values are associated with the most significant patterns. A significance of 1 indicates that, if random sequences were submitted to the program, such a level of overrepresentation would be expected every 10 trials (http://rsat.ulb.ac.be/rsat). Only the top three motifs are listed for each category if significance is >1.
We also identified clusters of genes whose transcription responded to changes of carbon source. Motifs overrepresented in their upstream regions and possible binding transcription factors are summarized in Table 3. In cluster c30, in which ORFs were highly expressed only under glucose limitation, the most overrepresented motif, CCCCT, is the so-called stress-response element. This is the binding site for Msn2/4p transcriptional activator (29) and is responsible for reprogramming gene expression on glucose exhaustion for ethanol-sustained growth (30). Surprisingly, in clusters c28 and c29, where the transcription of ORFs was elevated when glucose was limiting or ethanol was used as the carbon source, CCCCR(A/G), the consensus sequence to which the transcriptional repressor Mig1p binds, was not found to be the most over-represented. Instead, CGGGC was found to be significant for both clusters. No putative transcription factor in SCPD was found to bind this sequence. A search of their promoter sequences revealed that 40 of 61 ORFs contain the CCCCR sequence in their promoters, suggesting that Mig1p is responsible for repression of most, but not all, of the glucose-repressed genes.
Among the clusters in which ORFs were expressed only during ethanol limitation, overrepresented motifs targeted by known transcription factors were extracted from promoters of cluster c24. GGAGA (significance = 0.61, not shown in Table 3) and CCGN5CCG are the consensus sequences to which Adr1p and Cat8p bind, respectively. ADR1 encodes a transcription factor involved in the regulation of ADH2 and peroxisomal genes (31). Cat8p is required for derepression of genes involved in gluconeogenesis (32) and acts on the carbon-source response. Indeed, this cluster consists primarily of genes involved in ethanol utilization, the glyoxylate cycle, and gluconeogenesis.
Cluster c1 contained ORFs that were highly induced when glucose was abundant in the medium. HXT1, encoding a low-affinity glucose transporter expressed only at high glucose concentrations (33), was clustered with a number of genes with known functions, including MIG2, ADH4, ROM1, and PPQ1. Promoter analysis revealed a known dyad sequence, ACCN5GGT, to which Zap1p binds. Zap1p is a zinc-responsive transcriptional activator that regulates genes involved in zinc uptake (34). The transcription of at least eight genes in this cluster, including YNL254c, YOL154w, YGL258w, YOR387c, ZRT3, MNT2, ADH4, and YPL250c (ICY2), has been shown to be significantly up-regulated by zinc deficiency (35). This may indicate that cells grown under conditions of glucose excess are zinc-deficient. In addition, a few genes involved in iron uptake and its subsequent utilization were grouped in the c0 cluster with genes highly induced when glucose (rather than ethanol) was the carbon source. This result suggests that the group of iron-associated genes, including FTH1, FET3, FRE3, FIT2, and FIT3, are activated by glycolysis, irrespective of the cells' respiratory status.
Discussion
Most studies on the yeast transcriptome have been performed by using batch culture systems. In such cultures, the environment is continuously changing, and it is difficult to study the effects of individual physiological parameters on cell growth and metabolism or to make reproducible comparisons among exponential phases. Furthermore, secondary effects, such as growth-rate-dependent factors, tend to obscure the identification of genes really pertinent to a particular experimental treatment, mutation, or physiological condition (3, 4). In contrast, the steady states obtained in chemostat culture are well defined and highly reproducible. In this paper, we have examined the effects of different nutrient limitations on the yeast transcriptome by using chemostat cultures run at the same dilution (growth) rate.
We initiated this analysis by comparing gene expression in the phosphate-limited steady state with all four other steady states in which phosphate was abundant. Fourteen genes, including PHO5 and PHO84, were activated >3-fold in response to phosphate limitation. The transcription of about half of these ORFs has also been shown to be activated in phosphate-limiting medium by Ogawa et al. (36) and vice versa. There may be several reasons for the discrepancy. First, our experiment was performed by using a chemostat, whereas theirs used batch growth. For reasons discussed above, transcriptome data may be obscured by constant changes of environment in a batch culture. Second, during our analysis, spots were included in the dataset only if their intensity was greater than the threshold (2 × SD of background). As a result, some genes that were activated, in terms of the change in their absolute expression levels, were excluded from the phosphate-regulated dataset. Examples include PHO89 and PHM2, both of which were up-regulated >5-fold. Finally, phosphate concentrations may differ in both abundant and limiting conditions between the two studies.
Under glucose limitation, ORFs encoding TCA cycle enzymes were switched on (Fig. 2), as compared to their state in glucose-abundant conditions, indicating that both TCA cycle genes and those involved in oxidative phosphorylation are derepressed to provide intermediates and ATP needed for cellular growth. Compared with glucose-limiting conditions, the expression of TCA cycle genes remained at about the same level under ethanol limitation. However, genes in the glyoxylate cycle were massively induced, which may indicate that cytosolic acetyl-CoA synthesized from ethanol may be sufficient for ATP generation in mitochondria but may not provide enough TCA cycle intermediates for glutamate synthesis and gluconeogenesis. As a result, the glyoxylate cycle was induced to convert ethanol into acetate to replenish malate used for gluconeogenesis. Moreover, in addition to GDH1 and GDH3, the expression of GLT1 (responsible for converting ammonium and glutamine into glutamate) was induced (Fig. 2), suggesting that cells grown on ethanol may experience glutamate deficiency. This might explain why some of the nitrogen catabolite repression genes, such as MEP2, AGP2, PUT4, DAL4, and DAL5, were up-regulated when cells were grown under ethanol-limiting conditions, even though there was excess ammonium in the medium.
The nitrogen catabolite repression genes are activated by the partially redundant GATA factors Gln3p and Gat1p, which are retained in the cytoplasm by Ure2p in the presence of a good nitrogen source (37). Ure2p is itself a phosphoprotein, and a recent study (38) has shown that its phosphorylation state responds not to nitrogen availability but rather to the carbon source. Ure2p is dephosphorylated when cells grown on a fermentable energy source (glucose) are transferred to a nonfermentable energy source (ethanol or acetate). Dephosphorylation of Ure2p releases Gat1p, which is translocated into the nucleus and induces a specific set of genes involved in the anapleurotic pathways that replenish TCA cycle intermediates (38).
Nutrient starvation elicited dramatic changes in yeast's global gene expression pattern. Among the five conditions, carbon starvations have a greater quantitative impact on gene transcription, suggesting that genes involved in carbon metabolism make up more of the genome than those responsible for the metabolism of any other macronutrient. Alternatively, yeast cells might be unable to support as many general functions in the absence of a carbon source compared to when other macronutrients are absent, due to the fact that recycled carbon must be used as a source of energy as well as anabolites. Although different sets of ORFs were activated under different nutrient-induced starvations (see Tables 5-9), it will be interesting to extend the starvation experiment to assess the regulation of these genes during and after exhaustion of a particular nutrient. Nevertheless, those 17 genes that were commonly induced in all five conditions examined are of special interest, and the functions of most of them are not yet precisely characterized. Previous studies have revealed that 10 of these ORFs are induced by a variety of stresses, highlighting the overlapping response of cells to stress and nutrient deprivation and suggesting that starvation itself is a kind of stress. However, subsequent promoter analysis failed to identify the stress-response element in the upstream regions of these 17 ORFs, suggesting other mechanisms are regulating cells' entry into stationary phase.
The data presented (obtained from experiments with carefully controlled nutrient-limited steady states and defined nutrient-starved stationary phases) have provided much useful information about the nutrient control of gene expression in S. cerevisiae. These data suggest a number of hypotheses about the regulation of yeast metabolic pathways and the interrelationships among them. These hypotheses now need to be tested by more specific experiments.
Supplementary Material
Acknowledgments
We thank Jorg Hoheisel for providing the filters. This work was supported by a EUROFAN contract from the European Community and by Biotechnology and Biological Sciences Research Council grants (from the Engineering and Biological Systems Committee and the Consortium for the Functional Genomics of Microbial Eukaryotes consortium of the Investigating Gene Function Initiative) (to S.G.O.). J.W. is the grateful recipient of a scholarship from the University of Manchester Institute of Science and Technology Department, and K.P. thanks SachetPack Limited for her studentship.
Abbreviation: TCA, trichloroacetic acid.
References
- 1.Oliver, S. (2000) Nature 403, 601-603. [DOI] [PubMed] [Google Scholar]
- 2.Hughes, T. R., Marton, M. J., Jones, A. R., Roberts, C. J., Stoughton, R., Armour, C. D., Bennett, H. A., Coffey, E., Dai, H., He, Y. D., et al. (2000) Cell 102, 109-126. [DOI] [PubMed] [Google Scholar]
- 3.Hayes, A., Zhang, N., Wu, J., Butler, P. R., Hauser, N. C., Hoheisel, J. D., Lim, F. L., Sharrocks, A. D. & Oliver, S. G. (2002) Methods 26, 281-290. [DOI] [PubMed] [Google Scholar]
- 4.Lim, F. L., Hayes, A., West, A. G., Pic, A., Darieva, Z., Morgan, B. A., Oliver, S. G. & Sharrocks, A. D. (2002) Mol. Cell. Biol. 23, 450-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirt, S. J. (1975) Principles of Microbe and Cell Cultivation (Blackwell Scientific, Oxford, U.K.).
- 6.ter Linde, J. J., Liang, H., Davis, R. W., Steensma, H. Y., van Dijken, J. P. & Pronk, J. T. (1999) J. Bacteriol. 181, 7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kal, A. J., van Zonneveld, A. J., Benes, V., van den, B. M., Koerkamp, M. G., Albermann, K., Strack, N., Ruijter, J. M., Richter, A., Dujon, B., et al. (1999) Mol. Biol. Cell 10, 1859-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baganz, F., Hayes, A., Marren, D., Gardner, D. C. & Oliver, S. G. (1997) Yeast 13, 1563-1573. [DOI] [PubMed] [Google Scholar]
- 9.Hauser, N. C., Vingron, M., Scheideler, M., Krems, B., Hellmuth, K., Entian, K. D. & Hoheisel, J. D. (1998) Yeast 14, 1209-1221. [DOI] [PubMed] [Google Scholar]
- 10.Engler-Blum, G., Meier, M., Frank, J. & Muller, G. A. (1993) Anal. Biochem. 210, 235-244. [DOI] [PubMed] [Google Scholar]
- 11.Mewes, H. W., Frishman, D., Gruber, C., Geier, B., Haase, D., Kaps, A., Lemcke, K., Mannhaupt, G., Pfeiffer, F., Schuller, C., et al. (2000) Nucleic Acids Res. 28, 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamayo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E. S. & Golub, T. R. (1999) Proc. Natl. Acad. Sci. USA 96, 2907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 14.Werner-Washburne, M., Braun, E., Johnston, G. C. & Singer, R. A. (1993) Microbiol. Rev. 57, 383-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner-Washburne, M., Braun, E. L., Crawford, M. E. & Peck, V. M. (1996) Mol. Microbiol. 19, 1159-1166. [DOI] [PubMed] [Google Scholar]
- 16.Avendano, A., Deluna, A., Olivera, H., Valenzuela, L. & Gonzalez, A. (1997) J. Bacteriol. 179, 5594-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson, B. M., James, C. M. & Walmsley, R. M. (1996) Microbiology 142, 1667-1673. [DOI] [PubMed] [Google Scholar]
- 18.ter Schure, E. G., van Riel, N. A. & Verrips, C. T. (2000) FEMS Microbiol. Rev. 24, 67-83. [DOI] [PubMed] [Google Scholar]
- 19.Dickinson, J. R. & Schweizer, M. (1999) The Metabolism and Molecular Physiology of Saccharomyces cerevisiae (Taylor and Francis, London).
- 20.Zhao, X., Georgieva, B., Chabes, A., Domkin, V., Ippel, J. H., Schleucher, J., Wijmenga, S., Thelander, L. & Rothstein, R. (2000) Mol. Cell. Biol. 20, 9076-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichard, P. (1988) Annu. Rev. Biochem. 57, 349-374. [DOI] [PubMed] [Google Scholar]
- 22.Thumm, M., Egner, R., Koch, B., Schlumpberger, M., Straub, M., Veenhuis, M. & Wolf, D. H. (1994) FEBS Lett. 349, 275-280. [DOI] [PubMed] [Google Scholar]
- 23.Werner-Washburne, M., Becker, J., Kosic-Smithers, J. & Craig, E. A. (1989) J. Bacteriol. 171, 2680-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horst, M., Knecht, E. C. & Schu, P. V. (1999) Mol. Biol. Cell 10, 2879-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verwaal, R., Paalman, J. W., Hogenkamp, A., Verkleij, A. J., Verrips, C. T. & Boonstra, J. (2002) Yeast 19, 1029-1038. [DOI] [PubMed] [Google Scholar]
- 26.Garay-Arroyo, A. & Covarrubias, A. A. (1999) Yeast 15, 879-892. [DOI] [PubMed] [Google Scholar]
- 27.Ashrafi, K., Farazi, T. A. & Gordon, J. I. (1998) J. Biol. Chem. 273, 25864-25874. [DOI] [PubMed] [Google Scholar]
- 28.Fisher, F., Jayaraman, P. S. & Goding, C. R. (1991) Oncogene 6, 1099-1104. [PubMed] [Google Scholar]
- 29.Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M., Boucherie, H., Toledano, M. B. & Labarre, J. (1998) J. Biol. Chem. 273, 22480-22489. [DOI] [PubMed] [Google Scholar]
- 30.Boy-Marcotte, E., Perrot, M., Bussereau, F., Boucherie, H. & Jacquet, M. (1998) J. Bacteriol. 180, 1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurvitz, A., Hiltunen, J. K., Erdmann, R., Hamilton, B., Hartig, A., Ruis, H. & Rottensteiner, H. (2001) J. Biol. Chem. 276, 31825-31830. [DOI] [PubMed] [Google Scholar]
- 32.Hedges, D., Proft, M. & Entian, K. D. (1995) Mol. Cell. Biol. 15, 1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozcan, S. & Johnston, M. (1995) Mol. Cell. Biol. 15, 1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, H. & Eide, D. J. (1997) Mol. Cell. Biol. 17, 5044-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyons, T. J., Gasch, A. P., Gaither, L. A., Botstein, D., Brown, P. O. & Eide, D. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa, N., DeRisi, J. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck, T. & Hall, M. N. (1999) Nature 402, 689-692. [DOI] [PubMed] [Google Scholar]
- 38.Kuruvilla, F. G., Shamji, A. F., Sternson, S. M., Hergenrother, P. J. & Schreiber, S. L. (2002) Nature 416, 653-657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.