Abstract
F-box and WD repeat domain-containing 7 (FBW7), the substrate-binding subunit of E3 ubiquitin ligase SCFFBW7 (a complex of SKP1, cullin-1 and FBW7), plays important roles in various physiological and pathological processes. Although FBW7 is required for vascular development, its function in the endothelium remains to be investigated. In this study, we show that FBW7 is an important regulator of endothelial functions, including angiogenesis, leukocyte adhesion and the endothelial barrier integrity. Using RNA interference, we found that the depletion of FBW7 markedly impairs angiogenesis in vitro and in vivo. We identified the zinc finger transcription factor Krüppel-like factor 2 (KLF2) as a physiological target of FBW7 in endothelial cells. Knockdown of FBW7 expression resulted in the accumulation of endogenous KLF2 protein in endothelial cells. FBW7-mediated KLF2 destruction was shown to depend on the phosphorylation of KLF2 via glycogen synthase kinase-3 (GSK3) at two conserved phosphodegrons. Mutating these phosphodegron motifs abolished the FBW7-mediated degradation and ubiquitination of KLF2. The siRNA-mediated knockdown of FBW7 showed that KLF2 is an essential target of FBW7 in the regulation of endothelial functions. Moreover, FBW7-mediated KLF2 degradation was shown to be critical for angiogenesis in teratomas and in zebrafish development. Taken together, our study suggests a role for FBW7 in the processes of endothelial cell migration, angiogenesis, inflammation and barrier integrity, and provides novel insights into the regulation of KLF2 stability in vivo.
Keywords: angiogenesis, FBW7, KLF2, ubiquitin, degradation, cell migration
Introduction
The endothelium is the thin layer of cells that lines in the interior surface of all blood vessels and plays crucial roles in many aspects of the vascular system1. The loss of proper endothelial function is a hallmark of vascular diseases such as atherosclerosis, hypertension, blood clotting and inflammation2,3,4,5. The Krüppel-like factors (KLFs) are a family of zinc finger transcription factors with essential roles in cell growth, differentiation and tissue development6. Several KLFs, including KLF2, 4, 6, 10, and 15, have been reported to be involved in vascular functions7,8,9,10,11. Among them, KLF2 (also known as LKLF) is a central regulator of endothelial functions, including angiogenesis, inflammation, thrombosis and vascular tone7,12. A KLF2 deficiency impairs endothelial barrier integrity and increases endothelial leakage by regulating the expression of the key junction protein occludin7,13. The ectopic expression of KLF2 inhibits endothelial cell migration and angiogenesis by downregulating VEGFR214. KLF2 also regulates endothelial proinflammatory activation by upregulating eNOS expression through the NF-κB pathway15.
The regulation of KLF2 mRNA expression has been well studied in the endothelium16. Accumulating evidence shows that MEF2 transcription factors are the central regulators of KLF2 mRNA expression in endothelial cells. Laminar flow induces MEF2 phosphorylation through ERK5 activation and thus stimulates KLF2 mRNA expression17. Similarly, the anti-inflammation drugs statins also induce KLF2 expression via MEF2 activation18. Recent evidence shows that miRNA-92a regulates KLF2 mRNA in endothelial cells19. Moreover, proteasome inhibitors can also induce the expression of KLF2 mRNA in the endothelium and thus increase the expression of Thrombomodulin20. Although a number of studies have shown that KLF2 is tightly controlled at the transcriptional level, it is still unclear whether KLF2 protein is also regulated at the posttranslational level in endothelial cells.
Ubiquitin-dependent protein degradation is involved in various biological processes by regulation of protein abundance. The F-box and WD repeat domain-containing 7 (FBW7, also known as FBXW7, SEl-10, hCDC4, and hAgo) is the substrate recognition component of the SKP1-cullin-1-FBW7 (SCFFBW7) E3 ubiquitin ligase complex, which mediates the degradation of a variety of proteins, including c-Myc, Cyclin E, c-Jun, C/EBPα, MCL1, Notch and KLF521,22,23. FBW7 has been demonstrated to play important roles in various physiological and pathological processes, such as tumorigenesis, lipid metabolism, cell proliferation, stemness and differentiation21,24. Evidences from tissue-specific knockout mice indicate that FBW7 fine-tunes the substrate function in a tissue-specific manner25,26. For example, FBW7 regulates the proliferation and differentiation of hematopoietic stem cells through the modification of c-Myc abundance26, and orchestrates the differentiation of neural stem cells through regulation of Notch/c-Jun signaling pathways25,27. It has been reported that FBW7 is required for the formation of functional vasculature28. Fbw7- deficient mice show defects in vascular development and embryonic lethality at day E10.5 (E10.5)28. Recent studies indicate that FBW7 may regulate angiogenesis by targeting Notch, NF-1 or HIF129,30,31. However, the role of FBW7 in vascular biology remains to be investigated.
Results
Depletion of FBW7 impairs angiogenesis in vitro and in vivo
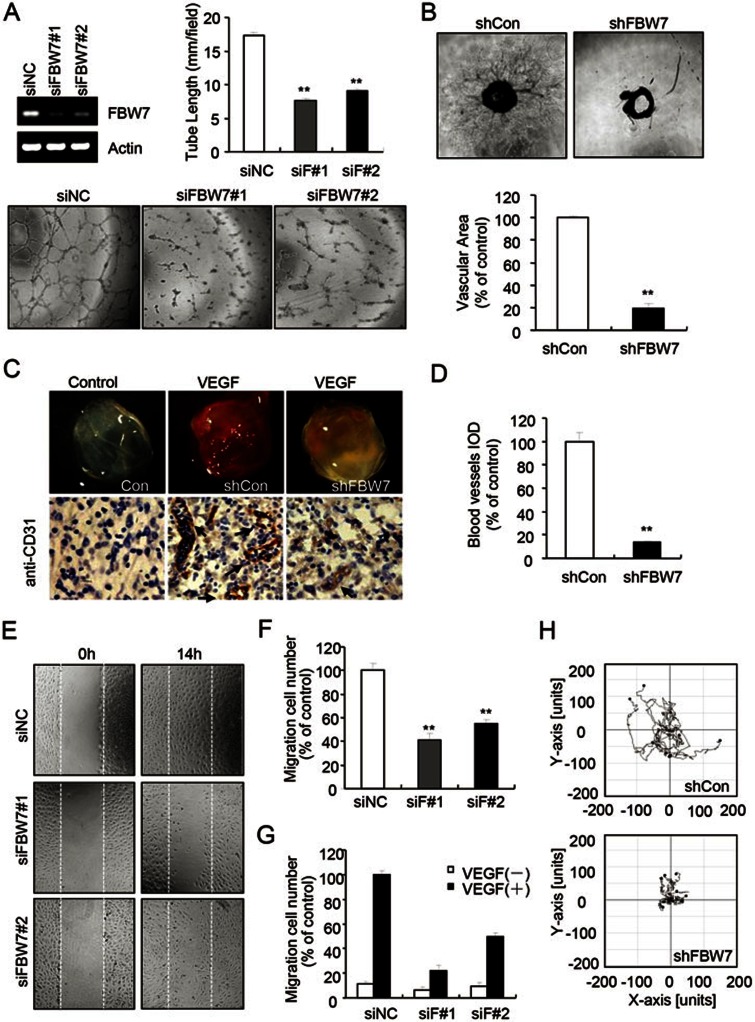
A previous study indicated that Fbw7 is notably expressed in the endothelial cell lineage of embryos28. Our data also indicated that FBW7 is highly expressed in HUVECs and aortic endothelial cells (Supplementary information, Figure S1A). Angiogenesis, which is the growth of new blood vessels from pre-existing vessels, is a vital process in development, wound healing, inflammation, ischemia and tumorigenesis3. To examine the potential role of FBW7 in angiogenesis, we investigated the effect of FBW7 siRNA on vascular endothelial tube formation in vitro. Three isoforms with different subcellular localization of FBW7 have been identified: FBW7α, β and γ32. HUVECs transfected with control or FBW7 siRNA targeting all three isoforms of FBW7 were plated on Matrigel in the presence of VEGF and capillary/tube-like-structure formation was examined. Transfection with FBW7 siRNA efficiently reduced the expression of FBW7 in HUVECs (Figure 1A and Supplementary information, Figure S1B) and significantly impaired the tube formation compared with the control cells (Figure 1A). Cells transfected with FBW7 siRNA formed partially connected networks. We measured the cumulative tube length and found that FBW7 siRNA reduced the tube length by ∼58% (Figure 1A). Similar results were obtained using FBW7 shRNA lentivirus, which significantly inhibited FBW7 expression in HUVECs (Supplementary information, Figure S2A-S2C). The effect of the FBW7 knockdown could be rescued using a shRNA-resistant mutant of FBW7 (Supplementary information, Figure S2D and S2E), indicating that the effect of FBW7 knockdown was specifically caused by the reduction of FBW7 expression. The effects of FBW7 knockdown on angiogenesis were also confirmed using a mouse aortic ring model. The infection of the mouse aortic ring with FBW7 shRNA lentivirus dramatically reduced the sprouting of endothelial tubules (Figure 1B). These results demonstrate that FBW7 plays a pivotal role in endothelial tube formation and sprouting.
Figure 1.
Depletion of FBW7 impaired angiogenesis and endothelial cell migration. (A) Depletion of FBW7 inhibited the tube formation of HUVECs. HUVECs transfected with FBW7 or control siRNAs were placed in plates coated with Matrigel and tubular structures were photographed. Column, mean from three independent experiments with duplicates; bar, standard deviation (SD); *P < 0.05; **P < 0.01 vs control siRNA. (B) Knockdown of FBW7 reduced the sprouting of vessels. Aortic segments isolated from mice were infected with lentiviral vectors. The aortic segments were placed in Matrigel-covered wells for 6 days. Sprouts were recorded using Image-Pro Plus 6.0 software and were calculated as a percentage of the control groups. Column, mean; bar, SD. **P < 0.01 vs control. (C) The depletion of FBW7 inhibited VEGF-induced angiogenesis in vivo. Mice were injected with 0.5 ml of Matrigel containing the indicated lentivirus. The angiogenesis in the intact Matrigel plugs was examined after 7 days (n = 5). Plug sections of 5 μM were immunostained with a specific anti-CD31 antibody for blood vessels. The images were captured using a Leica DM 4000B photo microscope (magnification, 400×). Arrows, blood vessels. (D) Quantification of blood vessels from C. IOD, Integrated Optical Density; Column, mean; bar, SD. **P < 0.01 vs control. (E) The depletion of FBW7 inhibited HUVEC migration as measured by wound healing assay. HUVECs transfected with FBW7 or control siRNAs were treated with mitomycin C. Cells were scratched using a pipette tip and the wound margins were imaged. The migrated cells were quantified manually. (F) Quantification data of E. (G) Depletion of FBW7 inhibited HUVEC migration in a transwell assay. HUVECs were seeded in the upper chamber of a transwell. The bottom chamber was filled with ECGM supplemented with VEGF. (H) Depletion of FBW7 inhibited HUVEC motility. HUVECs were transfected with control or FBW7 siRNA and cell migration was recorded using time-lapse microscopy in the presence of VEGF. The motility was calculated using Image J software.
To determine whether FBW7 is important to angiogenesis in vivo, we studied the effect of FBW7 knockdown on the angiogenic response using an in vivo Matrigel plug assay in mice. Matrigel containing angiogenic factors (VEGF) mixed with control or Fbw7 shRNA virus was implanted subcutaneously into the abdomen of male mice. Without angiogenic factors, little to no red blood cells (RBCs) were observed in the Matrigel (Figure 1C). Matrigel plugs containing angiogenic factors mixed with control virus appeared red and were filled with RBCs 6 days after implantation, indicating that functional vascular structures were induced within the Matrigel (Figure 1C). In contrast, the addition of Fbw7 shRNA virus notably inhibited angiogenic factors-induced vascular growth and the Matrigel plugs were pale due to the lack of RBCs (Figure 1C). The angiogenesis was confirmed by immunohistochemistry using anti-CD31 antibody, which is a specific marker for endothelial cells. A ≥ 80% reduction of angiogenesis was observed in the presence of Fbw7 shRNA virus (Figure 1D). Collectively, these data suggest that FBW7 is a key regulator of angiogenesis in vivo.
FBW7 is involved in endothelial cell migration
Endothelial cell migration in response to angiogenic stimuli is a key step in angiogenesis33. Thus, we determined whether FBW7 affects endothelial cell migration. Our data showed that the transfection of FBW7 siRNA significantly reduced HUVECs migration in a wound-healing assay (Figure 1E and 1F). Similar results were obtained from a transwell assay (Figure 1G and Supplementary information, Figure S2F). The effect of FBW7 knockdown on HUVEC motility was also examined using time-lapse microscopy in the presence of VEGF. We observed that the FBW7 knockdown significantly reduced HUVEC cell motility (Figure 1H). Taken together, these data indicate that FBW7 is required for angiogenic factor-induced endothelial cell migration.
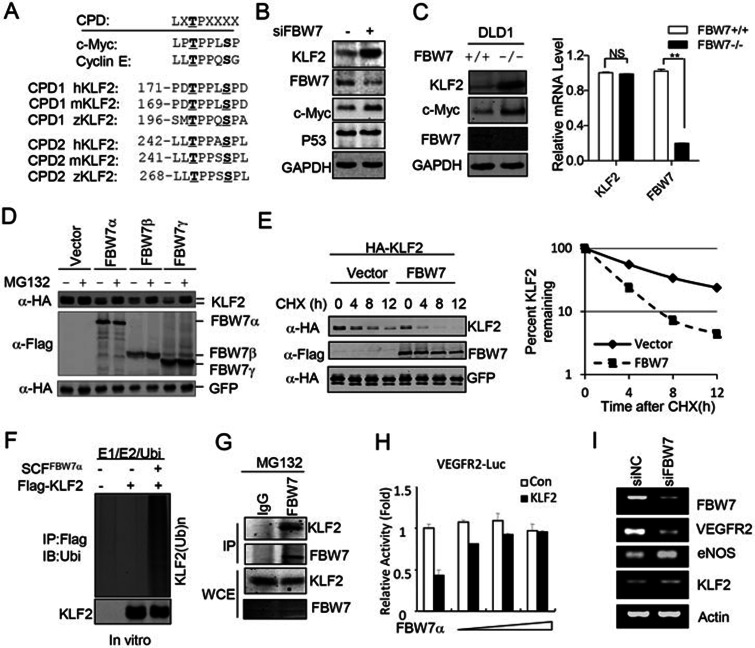
FBW7 targets KLF2 for degradation
As a key component of SCF E3 ubiquitin ligase, FBW7 primarily induces the ubiquitin-dependent degradation of proteins21. However, most of the FBW7 substrates contain a conserved phosphorylation site called the CDC4 phosphodegron (CPD)21. A global protein database search revealed that the transcription factor KLF2 contains two potential CPDs in its transcription inhibitory domain (Figure 2A). KLF2 has been well established as a central regulator of endothelial cell functions, including angiogenesis and cell migration. Therefore, we examined whether FBW7 regulates endothelial cell function through the destruction of KLF2. As shown in Figure 2B, KLF2 protein was significantly increased in the FBW7 siRNA-transfected HUVECs compared with the control cells. In contrast, the FBW7 knockdown showed no effect on p53 levels (Figure 2B). A cycloheximide (CHX) chase assay revealed that the knockdown of FBW7 reduced KLF2 turnover in HUVECs (Supplementary information, Figure S3A). Moreover, both the abundance and half-life of KLF2 protein were significantly increased in the FBW7-deficient DLD1 cell line compared with the parental cell line (Figure 2C and Supplementary information, Figure S3B). However, deficiency of FBW7 had no effect on the KLF2 mRNA level (Figure 2C). The depletion of FBW7 in HUVECs also markedly increased the expression of shear stress-induced KLF2 protein (Supplementary information, Figure S3C).These data suggest that FBW7 regulates endogenous KLF2 protein in vivo.
Figure 2.
FBW7 targeted KLF2 for degradation. (A) KLF2 contains putative CPD sequences. 'X' indicates any residue other than 'K' or 'R'. (B) Knockdown of FBW7 increased the protein level of endogenous KLF2 in HUVECs. GAPDH was used as loading control. (C) The KLF2 protein level, but not mRNA level, was increased in FBW7-null DLD1 cells. The protein levels were detected by western blotting. GAPDH was used as loading control. The mRNA levels were detected using Q-PCR. NS, not significant; **P < 0.01. (D) FBW7 mediated degradation of KLF2 in an ubiquitin-proteasome-dependent manner. HA-KLF2 was coexpressed with empty vector or different FBW7 isoforms. Transfected cells were treated with or without 10 μM MG132 for 6 h before harvesting. The expression of KLF2, FBW7, and GFP were analyzed using western blotting. (E) FBW7 promoted KLF2 turnover as measured using a CHX chase assay as indicated. (F) FBW7 promoted the ubiquitination of KLF2 in vitro. 293T cells were transfected with HA-KLF2 or Flag-FBW7 as indicated, followed by immunoprecipitated using an HA or Flag antibody and then subjected to in vitro ubiquitination. Ubi, Ubiquitin. (G) Interaction between endogenous KLF2 and FBW7 in DLD1 cells. DLD1 cells were treated with 10 μM MG132 for 6 h and lysed. FBW7 was immunoprecipitated using an anti-FBW7 antibody. The immunoprecipitates (IP) and the original whole-cell extracts (WCE) were analyzed by western blotting using an anti-KLF2 or anti-FBW7 antibody. (H) FBW7 reversed the inhibitory effect of KLF2 on VEGFR2 promoter. The VEGFR2 promoter reporter gene was transfected with KLF2 and FBW7 into 293Tcells as indicated. Promoter activity was analyzed using a Dual-Luciferase assay. (I) Depletion of FBW7 inhibited VEGFR2 expression. The mRNA expression was detected using RT-PCR.
The FBW7-induced degradation of KLF2 was confirmed in 293T cells. Our data showed that FBW7 specifically downregulated the level of KLF2 protein, which was not affected by other F-box proteins that we tested, including β-Trcp1, FBW2, FBW5 and FBW8 (Supplementary information, Figure S3D). Similar results were obtained in 293T cells from the coexpression of FBW7β and γ isoforms (Figure 2D). Interestingly, we found that expression of FBW7α and γ, but not β, reduced the protein levels of endogenous KLF2 in HUVECs (Supplementary information, Figure S3E). Consistently, using isoform-specific siRNA, we showed that knockdown of FBW7α and γ, but not β, resulted in the increased levels of endogenous KLF2 protein in HUVECs (Supplementary information, Figure S3F). These data indicate that endogenous KLF2 is mainly regulated by FBW7α and γ, but not β in HUVECs. The protease inhibitor MG132 blocked FBW7-mediated KLF2 destruction (Figure 2D), indicating that FBW7 degrades KLF2 via the proteasome-dependent pathway. Deletion of the F-box domain completely abolished FBW7-mediated KLF2 degradation (Supplementary information, Figure S3G). The FBW7-mediated KLF2 degradation was further confirmed using a CHX chase assay (Figure 2E). The coexpression of a dominant negative form of Cullin (dnCullin1) significantly blocked FBW7-mediated KLF2 destruction (Supplementary information, Figure S3H), indicating that the SCF complex is required for FBW7 to target KLF2 for degradation. Taken together, our data indicate that the SCFFBW7 complex targets KLF2 for degradation in a proteasome-dependent manner.
As a key component of the SCFFBW7 E3 ubiquitin ligase, FBW7 targets its substrates for degradation in a ubiquitination-dependent manner. The FBW7-mediated degradation of KLF2 prompted us to investigate whether FBW7 could target KLF2 for ubiquitination. Our data showed that the coexpression of FBW7 significantly increased the level of ubiquitinated KLF2 in vivo (Supplementary information, Figure S3I). The FBW7-mediated ubiquitination of KLF2 was also confirmed using an in vitro ubiquitination assay (Figure 2F). Taken together, these results demonstrate that KLF2 is a downstream target of the SCFFBW7 E3 ubiquitin ligase.
FBW7 interacts with KLF2
Binding to FBW7 is required for the destruction of substrates21. Thus, we investigated whether KLF2 interacts with FBW7 using a co-immunoprecipitation assay. Our data showed that KLF2 was only detected in the immunoprecipitated samples containing FBW7 but not in the samples containing other F-box proteins, including β-Trcp1, FBW2, FBW5 and FBW8 (Supplementary information, Figure S4A), suggesting that KLF2 specifically interacts with FBW7. Our data further showed that the FBW7 WD40 repeat was sufficient for its interaction with KLF2 (Supplementary information, Figure S4B and S4C). The results of an immunoprecipitation assay showed that endogenous KLF2 also interacted with endogenous FBW7 (Figure 2G). Immunofluorescence showed that KLF2 colocalized with FBW7α in the nucleus (Supplementary information, Figure S4D). Collectively, our data indicate that KLF2 binds to the substrate-binding domain of FBW7.
FBW7 regulates KLF2 transcriptional activity
We measured the effect of FBW7 on KLF2 transcriptional activity using a reporter gene assay. Consistent with the previous report34, the expression of KLF2 significantly increased S1P1 promoter activity. As expected, the coexpression of FBW7 eliminated the KLF2 effect (Supplementary information, Figure S4E). It was reported that KLF2 inhibited VEGFR2 expression by reducing the activity of the VEGFR2 promoter14. Therefore, we tested whether FBW7 affects the inhibitory effect of KLF2 on VEGFR2 promoter. As shown in Figure 2H, the coexpression of FBW7 completely abolished the KLF2-mediated inhibition of the VEGFR2 promoter.
Next, we tested whether the knockdown of FBW7 affected the expression of VEGFR2 in endothelial cells. We found that the knockdown of FBW7 significantly reduced VEGFR2 expression but increased eNOS expression (Figure 2I). This result is consistent with a previous report that both VEGR2 and eNOS are downstream targets of KLF27,33. Moreover, FBW7 has also been demonstrated as an efficient E3 ubiquitin ligase for Notch, which may regulate the expression of VEGFR2 in endothelial cells31,35. To exclude the effect of Notch, HUVECs transfected with control or FBW7 siRNA were treated with γ-secretase inhibitor DAPT for 24 h to block the Notch signaling. Then, the mRNA levels of VEGFR2 were analyzed using qRT-PCR. As shown in Supplementary information, Figure S4F, knockdown of FBW7 can still reduce the VEGFR2 expression in the presence of DAPT, suggesting that a Notch-independent pathway exists to inhibit the VEGFR2 expression upon FBW7 knockdown. Thus, our data suggest that FBW7 might regulate endothelial cell function by affecting KLF2 transcriptional activity in endothelial cells.
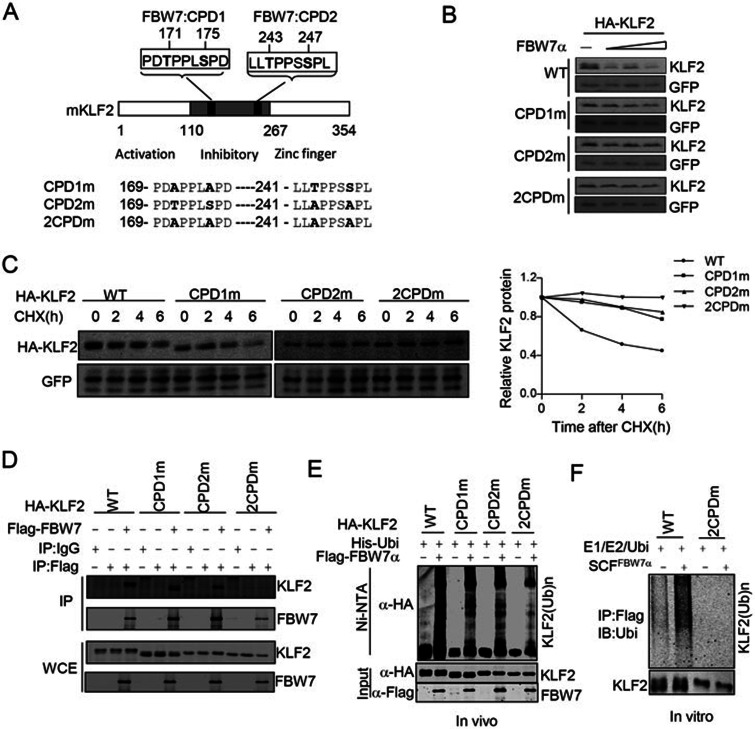
Identification of two conserved CPDs in KLF2
As described earlier, KLF2 contains two conserved putative CPDs (Figure 2A). To examine whether these CPDs are required for FBW7-mediated degradation, we made single or double CPD mutants of KLF2 (Figure 3A). Our data showed that individual mutations of either CPD or the simultaneous mutation of both CPDs dramatically reduced FBW7-mediated KLF2 degradation (Figure 3B and Supplementary information, Figure S5A). This result was confirmed using a CHX chase assay. The half-life of the CPD mutants was significantly extended compared with wild-type KLF2 (Figure 3C), indicating that both of the CPDs contributed to FBW7-mediated degradation. Moreover, the mutation of either CPD markedly reduced KLF2 binding to FBW7, and mutation of both CPDs completely abolished its interaction with FBW7 (Figure 3D). Consistent with these results, the CPD-null mutant of KLF2 was also resistant to FBW7-mediated ubiquitination both in vivo (Figure 3E) and in vitro (Figure 3F). These results strongly suggest that both CPD motifs are required for the SCFFBW7 E3 ubiquitin ligase-mediated ubiquitination and degradation of KLF2.
Figure 3.
Identification of CPDs in KLF2. (A) Schematic illustration of the various KLF2 mutants used in this study. CPD1m, T171A/S175A mutant; CPD2m, T243A/S247A mutant; 2CPDm, T171A/S175A/T243A/S247A mutant. (B) Mutation of CPDs impaired FBW7-mediated degradation. Wild-type or CPD mutated KLF2 was coexpressed with FBW7α and the degradation of KLF2 was monitored using western blotting analysis. (C) Mutation of CPDs impaired FBW7-mediated degradation. Wild-type or CPD mutant KLF2 was expressed in HEK293T cells and the half-life of KLF2 was examined by a CHX chase assay as indicated. (D) Mutation of CPDs impaired binding of KLF2 to FBW7. 293T cells were transfected with the indicated plasmids. 24 h post transfection, cells were subjected to co-immunoprecipition assay. *, IgG band; WCE, whole cell extract. (E) Mutation of CPDs impaired the FBW7-mediated ubiquitination of KLF2. The FBW7-mediated ubiquitination of KLF2 was examined using an in vivo ubiquitination assay. Ubi, Ubiquitin. (F) Ubiquitination of wild-type and the CPD mutant KLF2 in vitro. Ubi, Ubiquitin.
Previous data showed that FBW7 dimerization enhances the degradation of some FBW7 substrates36. The identification of the two CPDs in KLF2 prompted us to examine whether the dimerization of FBW7 is involved in KLF2 degradation. Consistent with a previous report36, the mutation of key residues required for FBW7 dimerization dramatically reduced the FBW7-mediated degradation of cyclin E, but had little effect on the degradation of c-Myc. Interestingly, the FBW7 L256E/I257E dimerization mutant36 was unable to degrade KLF2 (Supplementary information, Figure S5B), suggesting that dimerization of FBW7 is important for the efficient degradation of KLF2.
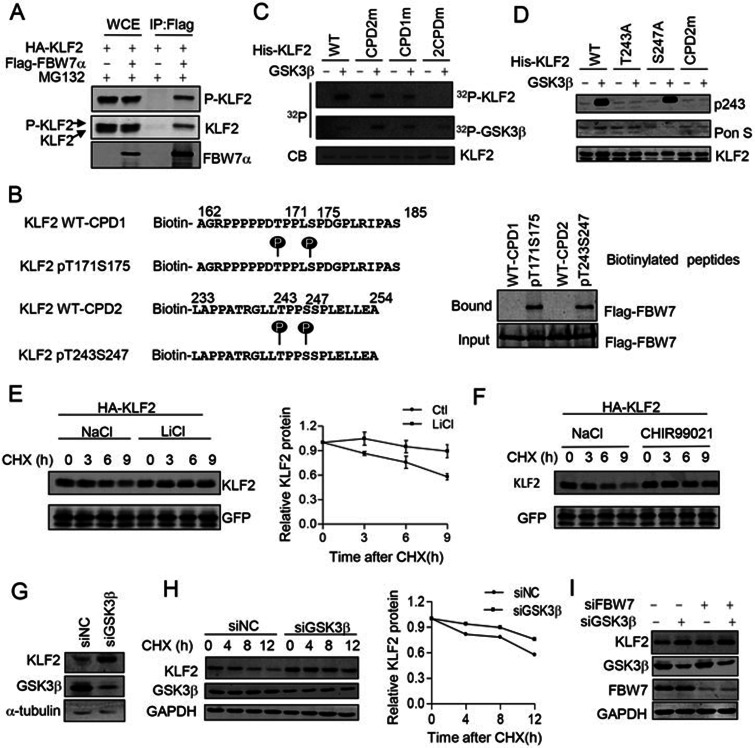
The phosphorylation of the Thr/Ser residue in the CPD motif is required for substrate recognition by FBW7. To test whether the CPDs of KLF2 are also phosphorylated, we generated an anti-KLF2 antibody that specifically recognized the phosphorylated T243, by using a synthesized phosphorylated peptide. The specificity of this antibody was characterized using T243A and S247A mutants. Our data showed that this antibody specifically recognized T243-phosphorylated KLF2, but not nonphosphorylated KLF2 (Supplementary information, Figure S6A). Treatment with protein phosphatase completely abolished the binding of the phospho-antibody to KLF2 immunoprecipitated from transfected 293T cells (Supplementary information, Figure S6B). To test whether FBW7 binds to phosphorylated or nonphosphorylated KLF2, HA-KLF2 was expressed in HEK293T cells in the presence or absence of Flag-FBW7. FBW7 was immunoprecipitated with an anti-Flag antibody, and the bound KLF2 was detected using both anti-HA antibody and the anti-pT243 antibody. As shown in Figure 4A, FBW7 specifically bound to the pT243-phosphorylated KLF2. Moreover, data from the biotin-labeled peptide pull-down assay showed that FBW7 bound to the phosphorylated CPD1 and CPD2, but not nonphosphorylated CPDs (Figure 4B). Thus, our data indicate that FBW7 binds to the KLF2 via the phosphodegron motifs.
Figure 4.
GSK3 mediated phosphorylation of KLF2. (A) FBW7 binds to the phosphorylated KLF2. (B) FBW7 bound to the phosphorylated but not nonphosphorylated KLF2 CPDs. Binding was examined using biotinylated peptide pull-down assay and analyzed using western blotting. (C) GSK3 mediated phosphorylation of KLF2. His-KLF2 fragments were purified from E.coli. The purified His-KLF2 fragment was subjected to in vitro kinase assay. CB, Coomassie blue. (D) The phosphorylation of WT or CPD mutated KLF2 was examined using a phospho-T243 antibody. CPD2m, KLF2 T243A/S247A mutant. (E) The inhibition of GSK3 stabilized KLF2. Cells expressing KLF2 were treated with 40 mM NaCl or LiCl. The half-life of KLF2 was examined using a CHX assay. The data are presented as the mean and standard error (n = 3). (F) GSK3 inhibitor CHIR99021 stabilized KLF2. Transfected cells were treated with 10 μM CHIR99021 and the half-life of KLF2 was examined using a CHX assay. (G) The GSK3β knockdown increased the protein levels of endogenous KLF2 in HUVECs. Tubulin was used as loading control. (H) The GSK3β knockdown in HUVECs prolonged the half-life of endogenous KLF2. HUVECs were transfected with the indicated siRNA and the half-life of KLF2 was examined using a CHX assay. Quantification was performed using GAPDH as a loading control. (I) The effect of GSK3 on KLF2 protein levels is FBW7 dependent. HUVECs were transfected with the indicated siRNA. 72 h after transfection, cells were lysed and analyzed by western blotting. GAPDH was used as loading control.
Phosphorylation of KLF2 by GSK3β
Next, we investigated the potential kinases required for KLF2 phosphorylation using various kinase inhibitors that might affect the KLF2 phosphorylation. Interestingly, we found that treatment with the glycogen synthase kinase-3 (GSK3) inhibitor LiCl reduced the level of phosphorylated T243; none of the other inhibitors tested, including SP600125, SB203580, U0126 and PD98059, reduced the phosphorylation of KLF2 at T243 (Supplementary information, Figure S6C). Thus, we further investigated whether GSK3 is the potential kinase involved in KLF2 phosphorylation using an in vitro kinase assay. As shown in Figure 4C, purified GSK3 robustly phosphorylated wild-type KLF2. The mutation of either CPD1 or CPD2 significantly reduced the GSK3-mediated phosphorylation. The double CPD mutation (2CPDm) completely abolished GSK3-mediated phosphorylation. The GSK3-mediated phosphorylation of KLF2 at T243 was confirmed using the phospho-KLF2 antibody (Figure 4D). Our data are consistent with a recent study showing that KLF2 is phosphorylated at T171 and T243 in mouse tissues using whole genome phosphorylation analysis (https://gygi.med.harvard.edu/phosphomouse) (Supplementary information, Figure S6D). We also investigated whether GSK3 activity is required for KLF2 degradation. Cells expressing HA-KLF2 were treated with or without GSK3 inhibitor LiCl or CHIR99021. KLF2 protein turnover was measured using a CHX chase assay. We found that both LiCl and CHIR99021 treatment significantly extended the half-life of KLF2 (Figure 4E and 4F). Moreover, knockdown of GSK3 significantly increased the protein level of KLF2 (Figure 4G) and extended the half-life of endogenous KLF2 (Figure 4H). The FBW7 knockdown abolished the increase in KLF2 protein induced upon GSK3 knockdown, and no significant differences were found between individual and double knockdown of FBW7 and GSK3β (Figure 4I); therefore, these results indicate that the effect of GSK3 on KLF2 protein levels is FBW7 dependent. Taken together, these data indicate that GSK3 phosphorylates KLF2 and consequently promotes the KLF2 degradation.
FBW7 inhibits KLF2 functions in endothelial cells
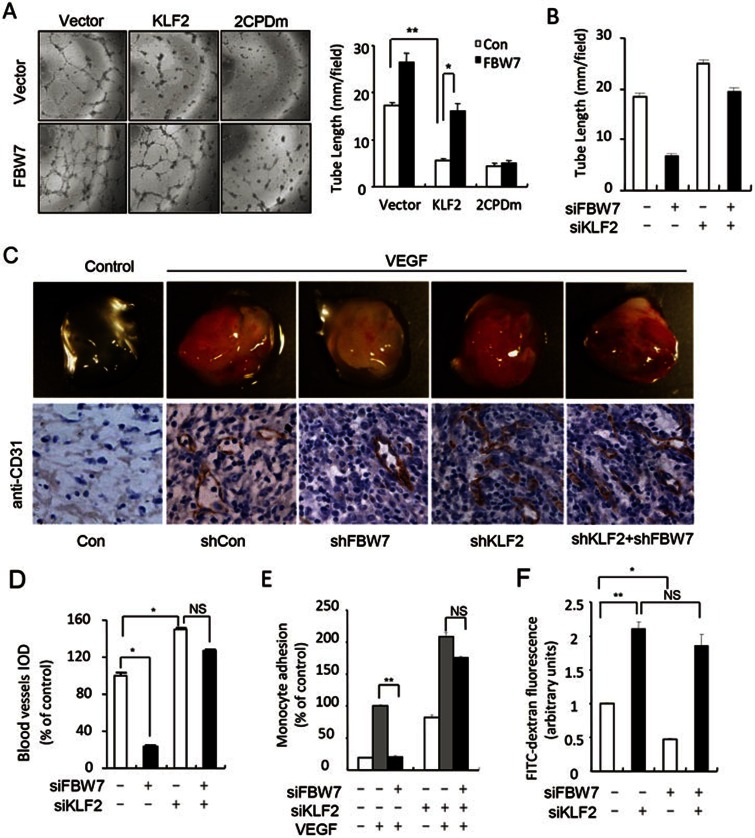
Next we investigated whether FBW7 regulates KLF2 function in endothelial cells. Consistent with a previous report14, the ectopic expression of KLF2 in HUVECs dramatically inhibited the tube formation and angiogenesis (Figure 5A and Supplementary information, Figure S7A). As expected, the co-expression of FBW7 significantly reduced the inhibitory effect of wild-type KLF2. In contrast, FBW7 had little effect on the CPD-null KLF2 mutant (Figure 5A). Moreover, the overexpression of KLF2 had similar effects on angiogenesis to the FBW7 knockdown (Supplementary information, Figure S7B-S7D), suggesting an important role for KLF2 in the effect of FBW7 on angiogenesis. Knockdown of KLF2 rescued the effect of FBW7 siRNA on the tube formation (Figure 5B and Supplementary information, Figure S8). The critical role of KLF2 in the FBW7-mediated regulation of angiogenesis was also confirmed using the Matrigel plug assay in vivo (Figure 5C and 5D). Together, these data indicate that KLF2 is involved in the negative effect of FBW7 knockdown on angiogenesis.
Figure 5.
FBW7 negatively regulated endothelial KLF2 function. (A) FBW7 reduced the inhibitory effect of KLF2 on angiogenesis. 2CPDm, KLF2 T171A/S175A/T243A/S247A mutant; Column, the means from three independent experiments with duplicates; bar, SD; *P < 0.05; **P < 0.01 vs control. (B) Knockdown of KLF2 reduced the inhibitory effect of FBW7 on the tube formation. (C) Knockdown of KLF2 reduced the inhibitory effect of FBW7 on angiogenesis in vitro. Mice were injected with 0.5 ml of Matrigel containing the indicated lentivirus. Angiogenesis in the intact Matrigel plugs was examined after 7 days (n = 5). Plug sections of 5 μM were immunostained with a specific anti-CD31 antibody for blood vessels. The images were captured using a Leica DM 4000B photo microscope (magnification, 400×). Arrows, blood vessels. (D) Quantification of blood vessels from C, and the images were captured. The data are presented as the mean and standard error (n = 3). IOD, Integrated Optical Density. *P < 0.05; **P < 0.01; NS, not significant. (E) FBW7 affected KLF2-mediated adhesion of U937 cells to HUVECs. U937 cells were labeled with CFSE and the adhesion of U937 cells to the transfected HUVECs was recorded using a microscope. **P < 0.01; NS, not significant. (F) The depletion of FBW7 decreased thrombin-induced endothelial leakage. HUVECs were transfected as indicated, treated with thrombin (1 U/mL), and the leakage assay was performed to measure the FITC-dextran passage. *P < 0.05; **P < 0.01; NS, not significant.
KLF2 has various functions in endothelial cells, such as anti-inflammatory activities and maintenance of the endothelial barrier7. Given that FBW7 is a negative regulator of KLF2, we investigated whether FBW7 regulates other KLF2-relevant endothelial functions. It has been reported that the upregulation of KLF2 in endothelial cells reduces the adhesion of leukocytes to the endothelium, which is critical for inflammation37. Consistent with a previous report, stimulation with VEGF significantly increased the adhesion of monocytes to HUVECs (Figure 5E and Supplementary information, Figure S9A). The knockdown of FBW7 markedly reduced the monocyte adhesion, while the depletion of KLF2 reduced the effect of FBW7 siRNA (Figure 5E). These data suggested that FBW7 affects leukocyte adhesion through KLF2. KLF2 is also a key regulator of endothelial barrier integrity13. The results of our dextran leakage assay showed that the knockdown of FBW7 reduced thrombin-induced endothelial leakage, which could be rescued by KLF2 knockdown (Figure 5F and Supplementary information, Figure S9B). We also examined the effect of FBW7 knockdown on the proliferation of HUVECs. Our data indicated that knockdown of FBW7 inhibited the proliferation of HUVECs, which could be rescued upon KLF2 knockdown (Supplementary information, Figure S10), suggesting that FBW7 is important for HUVEC proliferation via KLF2. Together, our data suggest that FBW7 is an important regulator of KLF2-relevant functions, including anti-inflammation, endothelial barrier integrity, and cell proliferation in endothelial cells.
The effect of FBW7 on the development of blood vessels in a teratoma assay
A previous study from knockout mice indicated that FBW7 is essential for vascular development28. Thus, we tested whether the FBW7-mediated degradation of KLF2 is also involved in the development of blood vessels during embryogenesis using a teratoma model, which has been demonstrated as a powerful gene-targeting method to study angiogenesis30,38. We first examined the expression of Fbw7 during mouse embryonic stem (ES) cell differentiation using an embryonic body (EB) differentiation model. Interestingly, our data showed that both Fbw7 and Vegfr2 mRNA levels were markedly increased during EB differentiation (Supplementary information, Figure S11A). To further test whether Fbw7 is expressed in endothelial lineage cells, the Vegfr2+ cells were sorted from the EB and the expression of Fbw7 in Vegfr2+ and Vegfr2– cells was examined using RT-PCR. Our data showed that the expression of Fbw7 in Vegfr2+ cells was higher than in Vegfr2– cells (Supplementary information, Figure S11B), suggesting that FBW7 might play roles in endothelial cell functions.
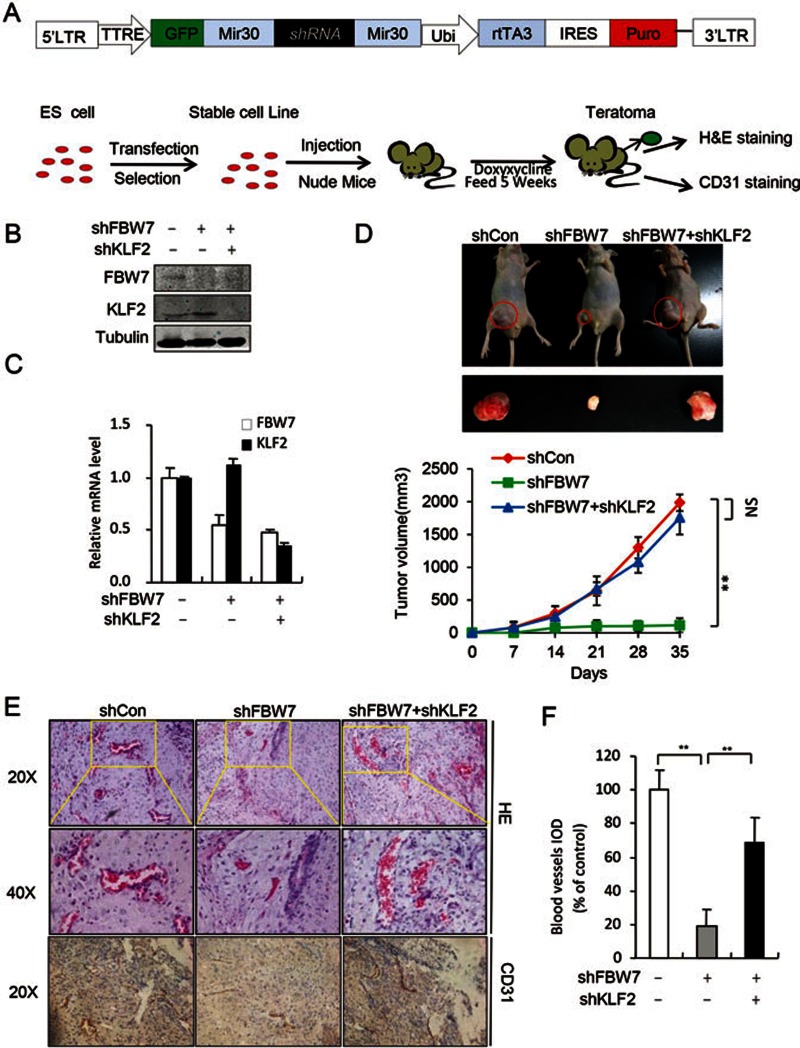
Subsequently, we generated a doxycycline-dependent knockdown stable ES cell line and the ES cells were injected into the nude mice as described39 (Figure 6A). Teratoma development was monitored by determination of endoderm, mesoderm and ectoderm (Supplementary information, Figure S11C). As shown in Figure 6B and 6C, both Fbw7 and Klf2 were efficiently depleted in mice fed with doxycycline for 5 weeks. Interestingly, knockdown of FBW7 dramatically inhibited teratoma growth, whereas knockdown of Klf2 rescued the effect of the FBW7 knockdown (Figure 6D). These data indicate that the FBW7-mediated degradation of KLF2 is involved in teratoma growth. We examined the development of blood vessels using anti-CD31 antibody staining. Our data showed that knockdown of FBW7 markedly reduced the number of blood vessels, which was rescued upon KLF2 knockdown (Figure 6E and 6F). Together, our data suggested that the FBW7-mediated KLF2 degradation is required for the angiogenesis in the development of teratoma from mouse ES cells.
Figure 6.
FBW7-mediated KLF2 degradation was required for teratoma growth and vascular development. (A) Procedure for the teratoma analysis. (B) FBW7-mediated KLF2 degradation is essential for the teratoma growth in nude mice. As indicated, the ES cells as indicated were injected subcutaneously into nude mice. Mice were fed with doxycycline for 5 weeks. The teratomas were harvested. (B and C) Teratomas were collected and the expression of FBW7 and KLF2 was analyzed using western blotting (B) and qRT-PCR (C). Tubulin was used as loading control. (D) The size of the tumors was measured every 7 days. Five mice were inoculated in each group, and data in the graph represent the mean ± SEM (n = 5, *P < 0.05). (E) Teratoma sections were stained with HE or CD31 for blood vessel. (F) Quantification of blood vessels in teratoma. IOD, Integrated Optical Density. *P < 0.05; **P < 0.01; NS, not significant.
The role of the FBW7-KLF2 axis in in vivo angiogenesis in a zebrafish model
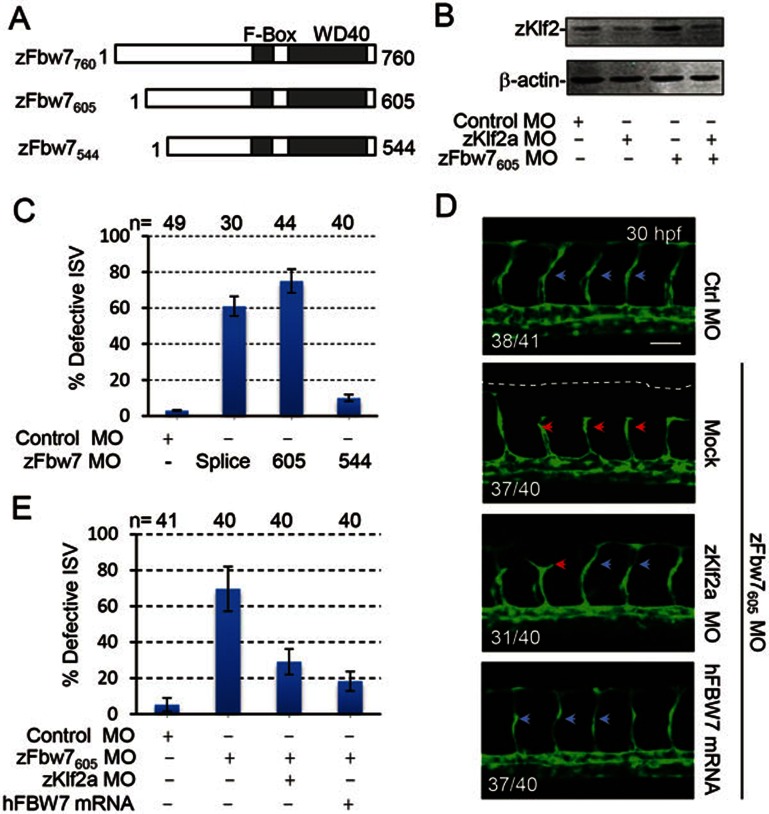
Furthermore, we employed a zebrafish model to study the role of FBW7 in vasculogenesis and angiogenesis during vertebrate embryogenesis. Three isoforms of zebrafish Fbw7 were identified through searching NCBI and Ensemble Genome Browser database (Figure 7A), and confirmed by RT-PCR and DNA sequencing (data not shown). We designed morpholino (MO) reagents targeting individual (labeled as zfbw7544 MO, or zfbw7605 MO) or all isoforms (labeled as zfbw7splice MO, blocking the splicing of exon4/intron5, shared by all zfbw7 isoforms) of Fbw7 (described in Materials and Methods), and further confirmed their efficacy through either coinjection with mRNA (Supplementary information, Figure S12A-S12D) or RT-PCR detection of morpholino-caused immature Fbw7 splicing products (Supplementary information, Figure S12E). Injection of zfbw7605 MO (Figure 7B) or zfbw7splice MO, but not zfbw7544 MO (data not shown), caused a remarkable increase in endogenous zKLF2 protein level in wild-type zebrafish embryos and kdrl:GFP-positive endothelial cells (Figure 7B and Supplementary information, Figure S12G), and a significant vascular morphogenesis defects in Tg(kdrl:EGFP)S843 transgenic zebrafish (Figure 7C and Supplementary information, Figure S12F). No gross morphological defects were evident in morphants and vasculogenic assembly of the trunk axial vessels also occurred normally (data not shown), thus, Fbw7 knockdown-induced angiogenic defects are specific to angiogenesis. At 30 hpf, these deficits on the growth of intersegmental vessels were grossly over 70% (Figure 7C), and could be significantly rescued by either expression of human FBW7 or co-injection of zklf2 morpholino (Figure 7D and 7E). These data indicate that zFbw7 indeed targets zKlf2, which in turn, controls normal angiogenesis in vivo.
Figure 7.
Fbw7-mediated Klf2 degradation was required for angiogenesis in zebrafish. (A) Three alternative splicing isoforms of Fbw7 have been identified in zebrafish. (B) The increase of endogenous zKlf2 protein level in zFbw7 morphants. β-actin:internal control. (C) Knockdown of Fbw7 caused angiogenic defects. Quantitation of intersegmental vessel (ISV) phenotype at 30 hpf in Tg(kdrl:EGFP)S843 transgenic zebrafish injected with zFbw7 morpholinos at one-cell stage. (D, E) Downregulating the expression of zKlf2 rescued the angiogenic defect in zFbw7 morphants. (D) Representative confocal fluorescence images for (E). Normal ISV growth (blue arrow); Defective ISV growth (red arrow). Scale Bar, 50 μm. (E) Quantitation of intersegmental vessel (ISV) phenotype at 30 hpf in Tg(kdrl: EGFP)S843 transgenic zebrafish injected with morpholino and mRNA, as indicated in C, at one-cell stage. Numbers of embryos (n =) counted for ISV phenotype were labeled on the top of panel C and E.
Discussion
In the present study, we provided the evidence showing that FBW7 plays an important role in endothelial functions, including cell migration, sprouting, tube formation, leukocyte adhesion and angiogenesis. Importantly, we identified that KLF2, a central endothelial transcription factor, is a functional target of FBW7.
We provided evidence showing that FBW7 is a physiological E3 ubiquitin ligase for KLF2. Although many studies have shown that KLF2 mRNA expression is regulated by various signals, including stress, growth factors and cytokines in endothelial cells, the mechanism that regulates the KLF2 abundance at the posttranslational level has remained largely unknown7. A previous study showed that the E3 ubiquitin ligase WWP1 inhibits KLF2 transcriptional activity and targets KLF2 for ubiquitination and degradation in a ubiquitin ligase activity-independent manner40,41. A recent study showed E3 ubiquitin ligase Smurf1 targets KLF2 for degradation in H1299 cells42. However, how KLF2 protein stability is regulated in endothelial cells has remained unknown. In the current study, we offer the first evidence to show that KLF2 is regulated through the ubiquitin-proteasome pathway at the posttranslational level in endothelial cells. Our results clearly demonstrate that SCFFBW7 E3 ubiquitin ligase negatively regulates KLF2 protein levels through ubiquitination and degradation. We also demonstrate that FBW7 is a critical negative regulator of KLF2-mediated endothelial functions, including angiogenesis, leukocyte adhesion and thrombin-induced leakage. Thus, our findings provide a novel mechanism for the regulation of KLF2 abundance and function in endothelial cells.
As an essential E3 ubiquitin ligase involved in cell proliferation, differentiation, tumorigenesis and development, FBW7 is regulated at both transcriptional and posttranslational level22. In this study, we identified FBW7 as an important regulator of endothelial functions. However, the mechanisms by which the FBW7 activity is regulated in endothelial cells are still unclear. Recent study indicates that PI3K-dependent phosphorylation stimulates the FBW7 activity to degrade its substrates43. Moreover, phosphorylation of FBW7 at T205 results in its binding to Pin1, which promotes the self-ubiquitination and degradation of FBW744. Thus, it will be interesting to examine whether angiogenic signals may regulate FBW7 activity through phosphorylation. Moreover, FBW7 has been demonstrated to be downstream target of p5345,46. Hypoxia or vascular injury may induce the expression of p53 in endothelial cells47. Thus, it is also possible that these signals may regulate FBW7 expression through the induction of p53.
Three splice-variant isoforms (α, β and γ) with different subcellular localization of FBW7 have been identified32,48. FBW7α is localized in the nucleoplasm, FBW7β is cytoplasmic and FBW7γ is nucleolar32. Despite their different subcellular localization, all of them have been shown to be able to degrade nuclear proteins49. Moreover, a recent study provides an interesting model that FBW7α and γ collaborate to target cyclin E for ubiquitination in the nucleolus in a Pin1-mediated isomerization-dependent manner50. KLF2 is a transcriptional factor that is mainly localized in the nucleoplasm. Although our data showed that overexpression of FBW7α, β and γ can promote the degradation of KLF2 in HEK293T cells, we found that FBW7α and γ, but not β, are involved in the degradation of endogenous KLF2 in HUVECs. Interestingly, we also noticed that KLF2 contains CPDs with double proline bond as cyclin E, thus it will be interesting to test whether KLF2 is ubiquitinated and degraded via a similar mechanism as cyclin E.
Although it is well established that FBW7 is an important tumor suppressor, which is mutated in various cancers, the pro-survival function has also been uncovered in multiple myeloid cells51. FBW7 is required for cell survival via targeting the p100 in the nucleus. Depletion of FBW7 significantly inhibits the NF-κB activation and cell survival. Moreover, FBW7 has also been demonstrated as an efficient E3 ubiquitin ligase for tumor suppressor NF1 and Nrf130,52. In our study, we reported that FBW7 is required for the growth of teratoma via targeting the KLF2 for degradation and inhibiting the angiogenesis. Thus, we conclude that FBW7 has a cell context-dependent E3 ubiquitin ligase activity by targeting different substrates.
Previous studies showed that the Fbw7 knockout in mice results in the embryonic lethality at E10.5 due to the impairment of vasculogenesis, suggesting that Fbw7 is required for proper endothelial functions28,53. In our current study, we provided several lines of evidence showing that the FBW7-mediated degradation of KLF2 positively regulates the endothelial functions both in vitro and in vivo: (1) knockdown of FBW7 in HUVECs impairs angiogenesis that can be rescued upon knockdown of KLF2; (2) inhibition of FBW7 expression impairs the blood vessel development in teratomas and knockdown of Klf2 rescued the phenotype; and (3) the depletion of Fbw7 in zebrafish impairs angiogenesis in vivo, which can be rescued upon knockdown of Klf2. However, our data cannot exclude the possibility that other FBW7-target proteins are also involved in the functions of FBW7 in endothelial cells. Previous studies using global knockout mice suggested that degradation of Notch by FBW7 is responsible for the development of normal blood vessels28,53. Results from a recent study using an endothelial cell-specific Fbw7 knockout mouse model support the notion that FBW7 is a potent positive regulator of angiogenesis by targeting Notch for degradation in endothelial cells31. Consistent with our study, they also showed that knockdown of FBW7 results in the inhibition of HUVEC proliferation31. Another study from Dr Sun's group identified neurofibromatosis type 1(NF1) as a novel target protein of SAG-CUL1-FBW7 E3 ubiquitin ligase. Disruption of SAG, a key component of SAG-CUL1-FBW7 E3 ubiquitin ligase, causes the accumulation of NF1, which abolishes the endothelial differentiation of mouse ES cells and blocks the angiogenesis and growth of teratomas30. Except the positive role of FBW7 in endothelial functions, FBW7 has also been shown to negatively regulate the cell migration and angiogenesis in HMEC-1 cells under both normoxic and hypoxic conditions through targeting HIF-1α for degradation29. The reason for this discrepancy is still unclear, but it is probably caused by the difference between HUVEC and HMEC-1. Unlike HUVECs, which are primary human umbilical vein endothelial cells, HMEC-1 is an immortal human EC line. Significant differences have been found between HUVEC and HMEC-1 cell lines54. Thus, together with our current study, we conclude that FBW7 is an indispensable regulator of endothelial functions by targeting multiple key factors, including KLF2, Notch, NF1 and HIF1-α, for degradation in a context-dependent manner.
KLF transcriptional factors, including KLF2, 4, 6, and 10, have been shown to regulate endothelial functions including angiogenesis and inflammation9,53,55. Among these factors, KLF2 has been identified as all essential regulator of endothelial function in mammals7. Overexpression of KLF2 in HUVECs significantly inhibits the angiogenesis14. Our study shows that degradation of KLF2 by FBW7 is indispensable for angiogenesis in mice. Moreover, Klf2 is essential for normal valve development56 and flow-induced angiogenesis during aortic arch development by regulating the expression of miR126 in zebrafish12. Our data showed that zebrafish Klf2 is also a target of zebrafish Fbw7. Depletion of Fbw7 results in the increase in KLF2 protein levels and inhibits the angiogenesis in zebrafish. Importantly, inhibition of Klf2a expression rescued the defective angiogenesis caused by Fbw7 depletion. Therefore, these data indicate that Fbw7-mediated Klf2 degradation is an evolutionally conserved pathway, which is important for angiogenesis in both zebrafish and mammal.
In addition to its essential roles in the endothelium, KLF2 is a critical regulator of lymphocyte trafficking, adipogenesis, erythropoiesis, development and ES cell self-renewal7. Thus, our study raises an interesting question of whether FBW7 regulates KLF2 function in other cell types. It has been established that FBW7 targets different substrates for degradation in a tissue-specific manner25,26. For example, FBW7 can target mTOR for degradation in a cancer cell line, but not in the liver55,57. In addition, FBW7 regulates the function of hematopoietic stem cells but not neural stem cells through c-Myc degradation25,26. As both KLF2 and FBW7 are widely expressed in many tissues other than the endothelium, their interaction in various physiological and pathological processes is worthy of further investigation using the recently developed tissue-specific FBW7 knockout mouse as a tool. Our findings are likely to have wider implications for the regulatory role of FBW7 in KLF2-dependent pathways beyond endothelial functions.
Materials and Methods
Animal studies
Mice were purchased from the National Rodent Laboratory Animal Resources, Shanghai Branch (Shanghai, China) and maintained in a laminar airflow cabinet under specific pathogen-free conditions according to the NIH standards established in the Guidelines for the Care. All the protocols were approved by East China Normal University.
In vivo Matrigel plug assay
The formation of new vessels in vivo was evaluated using a Matrigel plug assay. Briefly, lentiviruses expressing 1 × 109 pfu of lentiviral empty vector or shFbw7 were mixed in the Matrigel solution at 4 °C with 100 ng/ml VEGF165 (Peprotech), and 20 units of heparin. 500 μl of Matrigel containing lentiviruses was injected subcutaneously into the abdomen of 6-week-old C57BL/6 male mice. After 6 days, the skin of each mouse was pulled off to expose an intact Matrigel plug. Thereafter, hematoxylin and eosin (HE) staining was performed to identify the formation and infiltration of new functional microvessels. Functional microvessels with intact RBCs were quantified manually using a microscope (high-power field [HPF] 200×).
Fish care, morpholinos, microinjection and whole-mount mRNA in situ hybridization
Zebrafish maintenance, breeding and staging were performed as described previously58. The zebrafish facility and zebrafish study were approved by the Institutional Review Board of the Institute of Health Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. MO phosphorodiamidate oligonucleotides were designed by and purchased from Gene Tools (Carvalis, OR, USA). The sequences of Fbw7 MOs: Fbw7605: 5′-GGGAATAGGGCAATGCCGCAGCATT-3′ Fbw7544: 5′-TTAAAGTGCCGTAGAAACCCATGGT-3′ Fbw7-Splicing: 5′-AGCCAACTACAACAAGACAGAGACA-3′. The sequences of Klf2a MO: 5′-ATCGTTCCACTCAAAGCCATTTCCA-3′. The morpholinos were diluted in Danieu buffer. Capped mRNAs were synthesized using a Massage machine kit according to the manufacturer's instructions (Ambion). The zebrafish embryo microinjections were performed with a Harvard Apparatus microinjector at the one-cell stage. The dose of injected morpholinos/mRNA: 16 ng for Fbw7 splicing, Fbw7605 and Fbw7544 MO; 8 ng for KLF2a MO; and 0.1 ng for hFBW7. Confocal images were obtained using an Olympus Fluoview 1000 confocal microscope and data were analyzed as described59. All the experiments were repeated at least three times, and only representative results were shown.
Detailed methods are described in the Supplementary information, Data S1.
Acknowledgments
We thank Drs Mukesh K Jain (Case Western Reserve University), Michele Pagano (New York University School of Medicine), Steven I Reed (The Scripps Research Institute), Takashi Minami (The University of Tokyo), Wenyi Wei (Harvard Medical School), Feng Liu (Institute of Zoology, Chinese Academy of Sciences), Qunying Lei (Fudan University) and Jonathan Cherry (The Johns Hopkins University) for kindly providing reagents. We thank Min Deng , Bin Wei, Gang Pei from (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences), and Xiaoli Zhang, Jingjing Pan, Lufan Wang and Li Lai for technical help. We also thank Drs Martin A Schwartz (Yale School of Medicine) and Hardean E Achneck (Duke University) for help with the flow chamber. We thank other members of the Wang lab for their assistance. This work was supported by the National Basic Research Program of China (973 program; 2010CB529704, 2012CB910404, and 2013CB910900), the National Natural Science Foundation of China (30800587, 30971521, 31171338 and 31222037), and the Science and Technology Commission of Shanghai Municipality (11DZ2260300). P W is a scholar of the Program for New Century Excellent Talents in University (NCET-10-0387), the Shanghai Rising-Star Program from Science and Technology Commission of Shanghai Municipality (09QA1401900), the Dawn Program of Shanghai Education Commission (11SG27), the Thousand Talents Program for Distinguished Young Scholars, and the Hundred Talents Program of the Chinese Academy of Sciences.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
FBW7 expression in the vascular endothelium.
FBW7 shRNA inhibited the endothelial tube formation.
FBW7-mediated degradation of KLF2.
Interaction of KLF2 with FBW7.
KLF2 contains two CPDs.
Phosphorylation of KLF2 at T171 and T243.
Overexpression of KLF2 had a similar effect on the angiogenesis to the FBW7 knockdown.
KLF2 knockdown reduced the inhibitory effect of FBW7 on the tube formation.
FBW7 knockdown in HUVECs reduced the leukocyte adhesion.
FBW7 knockdown affected the cell proliferation and survival.
The expression of Fbw7 mRNA in Vegfr2-positive cells at day 6 of EB formation.
Knockdown of zFbw7 caused angiogenic defects in zebrafish.
Experimental procedures
References
- Lee S, Chen TT, Barber CL, et al. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadini GP, Agostini C, Sartore S, Avogaro A. Endothelial progenitor cells in the natural history of atherosclerosis. Atherosclerosis. 2007;194:46–54. doi: 10.1016/j.atherosclerosis.2007.03.046. [DOI] [PubMed] [Google Scholar]
- Folkman J. Angiogenesis: an organizing principle for drug discovery. Nat Rev Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- Rao RM, Yang L, Garcia-Cardena G, Luscinskas FW. Endothelial-dependent mechanisms of leukocyte recruitment to the vascular wall. Cir Res. 2007;101:234–247. doi: 10.1161/CIRCRESAHA.107.151860b. [DOI] [PubMed] [Google Scholar]
- Sakao S, Tatsumi K, Voelkel NF. Endothelial cells and pulmonary arterial hypertension: apoptosis, proliferation, interaction and transdifferentiation. Respir Res. 2009;10:95. doi: 10.1186/1465-9921-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak L, Lin Z, Jain MK. "Go with the flow": how Kruppel-like factor 2 regulates the vasoprotective effects of shear stress. Antioxid Redox Signal. 2011;15:1449–1461. doi: 10.1089/ars.2010.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Martin EM, Blanco FJ, Roque M, et al. Vascular injury triggers Krüppel-like factor 6 mobilization and cooperation with Sp1 to promote endothelial activation through upregulation of the activin receptor-like kinase 1 gene. Cir Res. 2013;112:113–127. doi: 10.1161/CIRCRESAHA.112.275586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DH, Hsu CF, Lin CY, Guo JY, Yu WC, Chang VH. Krüppel-like factor 10 upregulates the expression of cyclooxygenase 1 and further modulates angiogenesis in endothelial cell and platelet aggregation in gene-deficient mice. Int J Biochem Cell Biol. 2012;45:419–428. doi: 10.1016/j.biocel.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Helbing T, Volkmar F, Goebel U, et al. Kruppel-like factor 15 regulates BMPER in endothelial cells. Cardiovasc Res. 2010;85:551–559. doi: 10.1093/cvr/cvp314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Hamik A, Nayak L, et al. Endothelial Krüppel-like factor 4 protects against atherothrombosis in mice. J Clin Invest. 2012;122:4727–4731. doi: 10.1172/JCI66056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Natesan V, Shi H, et al. Krüppel-like factor 2 regulates endothelial barrier function. Arterioscler Thromb Vasc Biol. 2010;30:1952–1959. doi: 10.1161/ATVBAHA.110.211474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, Senbanerjee S, Lin Z, et al. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the Kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- SenBanerjee S, Lin Z, Atkins GB, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–4873. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ha CH, Jhun BS, Wong C, Jain MK, Jin ZG. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood. 2010;115:2971–2979. doi: 10.1182/blood-2009-05-224824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen-Banerjee S, Mir S, Lin Z, et al. Krüppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112:720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- Wu W, Xiao H, Laguna-Fernandez A, et al. Flow-dependent regulation of Krüppel-like factor 2 is mediated by microRNA-92a. Circulation. 2011;124:633–641. doi: 10.1161/CIRCULATIONAHA.110.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi T, Deming CB, Zhao H, et al. Proteasome inhibitors enhance endothelial thrombomodulin expression via induction of Krüppel-like transcription factors. Arterioscler Thromb Vasc Biol. 2009;29:1587–1593. doi: 10.1161/ATVBAHA.109.191957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Wang Z, Inuzuka H, Zhong J, et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586:1409–1418. doi: 10.1016/j.febslet.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Li H, Li S, et al. The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J Biol Chem. 2010;285:18858–18867. doi: 10.1074/jbc.M109.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Inuzuka H, Fukushima H, et al. Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep. 2012;13:36–43. doi: 10.1038/embor.2011.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeck JD, Jandke A, Blake SM, et al. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci. 2010;13:1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- Reavie L, Della Gatta G, Crusio K, et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat Immunol. 2010;11:207–215. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Onoyama I, Sunabori T, Kageyama R, Okano H, Nakayama KI. Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J Biol Chem. 2011;286:13754–13764. doi: 10.1074/jbc.M110.194936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Yu W, Li M, et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc Natl Acad Sci USA. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flugel D, Gorlach A, Kietzmann T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood. 2012;119:1292–1301. doi: 10.1182/blood-2011-08-375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Zhao Y, Kim SJ, et al. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell. 2011;21:1062–1076. doi: 10.1016/j.devcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N, Helker C, Ehling M, Behrens A, Herzog W, Adams RH. Fbxw7 controls angiogenesis by regulating endothelial notch activity. PloS One. 2012;7:e41116. doi: 10.1371/journal.pone.0041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck CH, Strohmaier H, Sangfelt O, et al. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002;62:4535–4539. [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- Carlson CM, Endrizzi BT, Wu J, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- Williams CK, Li JL, Murga M, Harris AL, Tosato G. Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function. Blood. 2006;107:931–939. doi: 10.1182/blood-2005-03-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako K, Fukuhara S, Minami T, et al. Angiopoietin-1 induces Krüppel-like factor 2 expression through a phosphoinositide 3-kinase/AKT-dependent activation of myocyte enhancer factor 2. J Biol Chem. 2009;284:5592–5601. doi: 10.1074/jbc.M806928200. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang H, Boland P, et al. Embryonic stem cell tumor model reveals role of vascular endothelial receptor tyrosine phosphatase in regulating Tie2 pathway in tumor angiogenesis. Proc Natl Acad Sci USA. 2009;106:22399–22404. doi: 10.1073/pnas.0911189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CW, Yabuuchi A, Chen L, Viswanathan S, Kim K, Daley GQ. Ras-MAPK signaling promotes trophectoderm formation from embryonic stem cells and mouse embryos. Nat Genet. 2008;40:921–926. doi: 10.1038/ng.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Krüppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316:139–148. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Wani MA, Lingrel JB. Lung Kruppel-like factor contains an autoinhibitory domain that regulates its transcriptional activation by binding WWP1, an E3 ubiquitin ligase. J Biol Chem. 2001;276:29299–29306. doi: 10.1074/jbc.M103670200. [DOI] [PubMed] [Google Scholar]
- Xie P, Tang Y, Shen S, et al. Smurf1 ubiquitin ligase targets Kruppel-like factor KLF2 for ubiquitination and degradation in human lung cancer H1299 cells. Biochem Biophys Res Commun. 2011;407:254–259. doi: 10.1016/j.bbrc.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Schulein C, Eilers M, Popov N. PI3K-dependent phosphorylation of Fbw7 modulates substrate degradation and activity. FEBS Lett. 2011;585:2151–2157. doi: 10.1016/j.febslet.2011.05.036. [DOI] [PubMed] [Google Scholar]
- Min SH, Lau AW, Lee TH, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell. 2012;46:771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori T, Mimori K, Iwatsuki M, et al. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009;69:3788–3794. doi: 10.1158/0008-5472.CAN-08-2846. [DOI] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Stempien-Otero A, Karsan A, Cornejo CJ, et al. Mechanisms of hypoxia-induced endothelial cell death. Role of p53 in apoptosis. J Biol Chem. 1999;274:8039–8045. doi: 10.1074/jbc.274.12.8039. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- Zhao D, Zheng HQ, Zhou Z, Chen C. The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 2010;70:4728–4738. doi: 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- Bhaskaran N, van Drogen F, Ng HF, et al. Fbw7alpha and fbw7gamma collaborate to shuttle cyclin e1 into the nucleolus for multiubiquitylation. Mol Cell Biol. 2013;33:85–97. doi: 10.1128/MCB.00288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Millman SE, Scotto L, et al. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas M, Phan D, Watanabe M, Chan JY. The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J Biol Chem. 2011;286:39282–39289. doi: 10.1074/jbc.M111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, et al. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- Lidington EA, Moyes DL, McCormack AM, Rose ML. A comparison of primary endothelial cells and endothelial cell lines for studies of immune interactions. Transpl Immunol. 1999;7:239–246. doi: 10.1016/s0966-3274(99)80008-2. [DOI] [PubMed] [Google Scholar]
- Mao JH, Kim IJ, Wu D, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J, Forouhar AS, Liebling M, et al. Reversing blood flows act through klf2a to ensure normal valvulogenesis in the developing heart. PLoS Biol. 2009;7:e1000246. doi: 10.1371/journal.pbio.1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori T, Mimori K, Iwatsuki M, et al. Copy number loss of FBXW7 is related to gene expression and poor prognosis in esophageal squamous cell carcinoma. Int J Oncol. 2012;41:253–259. doi: 10.3892/ijo.2012.1436. [DOI] [PubMed] [Google Scholar]
- Fu YF, Du TT, Dong M, et al. Mir-144 selectively regulates embryonic alpha-hemoglobin synthesis during primitive erythropoiesis. Blood. 2009;113:1340–1349. doi: 10.1182/blood-2008-08-174854. [DOI] [PubMed] [Google Scholar]
- Pan W, Pham VN, Stratman AN, et al. CDP-diacylglycerol synthetase-controlled phosphoinositide availability limits VEGFA signaling and vascular morphogenesis. Blood. 2012;120:489–498. doi: 10.1182/blood-2012-02-408328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FBW7 expression in the vascular endothelium.
FBW7 shRNA inhibited the endothelial tube formation.
FBW7-mediated degradation of KLF2.
Interaction of KLF2 with FBW7.
KLF2 contains two CPDs.
Phosphorylation of KLF2 at T171 and T243.
Overexpression of KLF2 had a similar effect on the angiogenesis to the FBW7 knockdown.
KLF2 knockdown reduced the inhibitory effect of FBW7 on the tube formation.
FBW7 knockdown in HUVECs reduced the leukocyte adhesion.
FBW7 knockdown affected the cell proliferation and survival.
The expression of Fbw7 mRNA in Vegfr2-positive cells at day 6 of EB formation.
Knockdown of zFbw7 caused angiogenic defects in zebrafish.
Experimental procedures