Abstract
Photomorphogenesis is controlled by multiple signaling pathways, including the light and brassinosteroid (BR) pathways. BR signaling activates the BZR1 transcription factor, which is required for suppressing photomorphogenesis in the dark. We identified a suppressor of the BR hypersensitive mutant bzr1–1D and named it bzr1–1D suppressor1-Dominant (bzs1–D). The bzs1–D mutation was caused by overexpression of a B-box zinc finger protein BZS1, which is transcriptionally repressed by BZR1. Overexpression of BZS1 causes de-etiolation in the dark, short hypocotyls in the light, reduced sensitivity to BR treatment, and repression of many BR-activated genes. Knockdown of BZS1 by co-suppression partly suppressed the short hypocotyl phenotypes of BR-deficient or insensitive mutants. These results support that BZS1 is a negative regulator of BR response. BZS1 overexpressors are hypersensitive to different wavelengths of light and loss of function of BZS1 reduces plant sensitivity to light and partly suppresses the constitutive photomorphogenesis 1 (cop1) mutant in the dark, suggesting a positive role in light response. BZS1 protein accumulates at an increased level after light treatment of dark-grown BZS1–OX plants and in the cop1 mutants, and BZS1 interacts with COP1 in vitro, suggesting that light regulates BZS1 through COP1-mediated ubiquitination and proteasomal degradation. These results demonstrate that BZS1 mediates the crosstalk between BR and light pathways.
Keywords: photomorphogenesis, light signaling, Brassinosteroid, BZS1, Arabidopsis
INTRODUCTION
Wild-type Arabidopsis seedlings grown in the dark have long hypocotyls and closed cotyledons with undifferentiated chloroplast, a phenomenon termed etiolation or skotomorphogenesis. When exposed to light, seedlings undergo de-etiolation (also called photomorphogenesis) and thus display short hypocotyls, open cotyledons, and chloroplast differentiation (Wei and Deng, 1996). This light-induced developmental switch is controlled by the photoreceptor-mediated signaling transduction pathways, which act in part by inactivating the COP1 E3 ubiquitin ligase and stabilizing positive transcription factors. In addition to light signaling, brassinosteroid (BR) is another key signal that controls the switch between skotomorphogenesis and photomorphogenesis, as BR-deficient and insensitive mutants show de-etiolation phenotypes in the dark (Chory et al., 1991; Li et al., 1996; Szekeres et al., 1996; Song et al., 2009). Light and BR act antagonistically in photomorphogenesis. However, the molecular mechanisms of this antagonism are not fully understood.
BR signals are perceived by BRI1 receptor kinase and transduced through a well-defined signal transduction pathway to activate members of the BZR family transcription factors (Kim et al., 2009; Kim and Wang, 2010; Clouse, 2011). BZR1 and BZR2/BES1 transcription factors play central roles in BR regulation of plant growth and gene expression (Wang et al., 2002; Yin et al., 2002; He et al., 2005; Yin et al., 2005). The dominant bzr1–1D mutation causes constitutive BZR1 activation due to enhanced dephosphorylation by PP2A (Tang et al., 2011), and bzr1–1D fully suppresses the photomorphogenesis phenotype of bri1-116 and reverses the expression changes of about 80% of the genes affected in bri1-116 (Sun et al., 2010). A large number of light-induced genes are activated in the BR-deficient or insensitive mutants in the dark (Chory et al., 1991; Li et al., 1996; Szekeres et al., 1996; Song et al., 2009), suggesting that light and BR may regulate common transcription factors. Indeed, a recent study showed that a member of the GATA family transcription factors (GATA2) is transcriptionally repressed by BZR1 but posttranslationally activated by light signaling (Luo et al., 2010). GATA2 controls a subset of genes co-regulated by light and BR to promote photomorphogenesis. Many other transcription factors mediate light-regulated gene expression (Lau and Deng, 2010), but whether any of them are also regulated by BR remains unknown.
B-box proteins are a group of transcription factors containing one or more B-box domains that are stabilized by binding to zinc ions (Klug and Schwabe, 1995). There are 32 B-box containing proteins with diverse functions in Arabidopsis (Khanna et al., 2009). For example, CONSTANS (CO) is an important regulator of photoperiodic flowering (Putterill et al., 1995; Onouchi et al., 2000). The CO-like genes COL1 and COL2 are involved in regulating the circadian clock (Ledger et al., 2001). STH2, STH3, and COL3 are positive regulators of light responses (Datta et al., 2006, 2007, 2008), whereas STO and STH1 are negative regulators of photomorphogenesis (Khanna et al., 2006; Indorf et al., 2007).
In this study, we identified an activation-tagged bzr1–1D suppressor (bzs1–D), which showed dwarfism and suppressed bzr1–1D’s phenotypes of stem kink and insensitivity to BR biosynthesis inhibitor BRZ. The phenotypes of bzs1–D are caused by overexpression of a B-box protein we named BZS1 (also named BBX20), which is repressed at the transcription level by BR. Overexpression and loss-of-function experiments demonstrate that BZS1 plays a negative role in BR responses but positively regulates light responses. BZS1 protein level is increased upon light treatment of dark-grown seedlings and in the cop1 mutant. BZS1 interacts with COP1 in vitro, suggesting that light may regulate BZS1 accumulation through COP1. These data demonstrate that BZS1 acts downstream of both BR and light pathways. This study supports a mechanism of BR-light crosstalk through transcriptional repression and posttranslational activation of photomorphogenesis-promoting transcription factors.
RESULTS
Isolation and Characterization of the bzr1–1D Suppressor Mutant
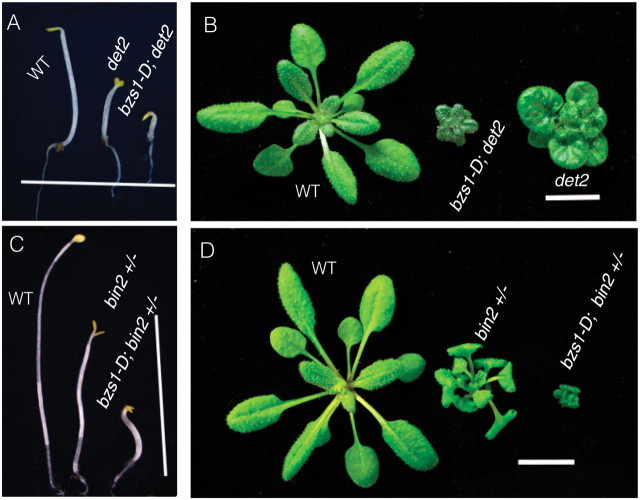
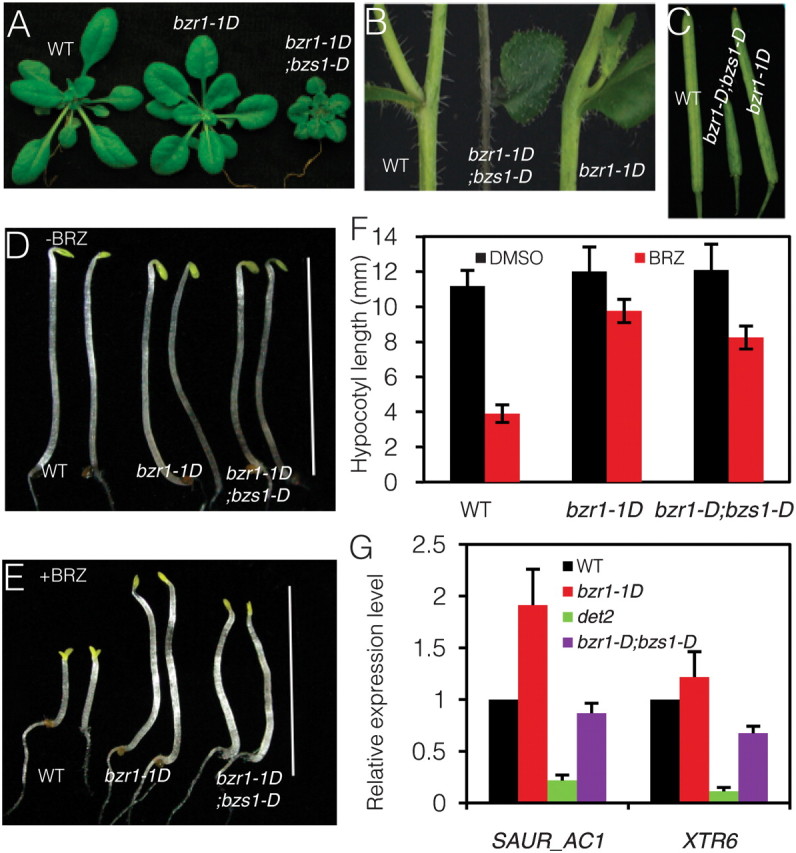
The activation tagging strategy has been successfully used in identification of novel BR signaling components, including BRS1, BAK1, BRL1, BSU1, BEN1, ATBS1, and TCP1 (Li et al., 2001, 2002; Mora-Garcia et al., 2004; Zhou et al., 2004; Yuan et al., 2007; Wang et al., 2009; Guo et al., 2010; Kang et al., 2010). To better understand the molecular mechanism of BZR1 function and identify additional components of the BR pathway, we previously carried out a large-scale activation tagging screen in bzr1–1D background (Cao et al., 2008). One of the mutant lines partially suppressed phenotypes of bzr1–1D; therefore, we named it bzr1–1D suppressor1-Dominant (bzs1–D). The bzr1–1D;bzs1–D mutant showed stronger BR-deficient phenotypes, such as dwarfism and extended lifecycle, compared to bzr1–1D itself (Figure 1A). The bzr1–1D;bzs1–D plants showed no stem kink, which is one of the most obvious phenotypes of light-grown bzr1–1D (Figure 1B), and a reduced degree of silique kink compared to bzr1–1D (Figure 1C).
Figure 1.
The bzs1–D Mutation Partly Suppresses bzr1–1D’s Phenotypes.
(A) Phenotypes of soil-grown 3-week-old plants.
(B) The stem kink phenotypes.
(C) The silique kink phenotype.
(D, E) Selective dark-grown seedlings on regular half-strength MS medium (D) or half-strength MS medium containing 2 μM BRZ (E).
(F) The average hypocotyl length of seedlings grown on medium with or without 2 μM BRZ. Error bars indicate SD (n = 40).
(G) Quantitative RT–PCR analysis of the expression levels of SAUR_AC1 and XTR6. UBC30 was used as internal control.
The bzr1–1D mutant is insensitive to brassinazole (BRZ), a BR biosynthesis inhibitor (Wang et al., 2002). BRZ inhibits hypocotyl elongation of wild-type in the dark but not of bzr1–1D (Figure 1D and 1E). While bzr1–1D;bzs1–D plants had similar hypocotyl length compared to wild-type and bzr1–1D when grown on medium without BRZ, their hypocotyls were shorter than bzr1–1D on the medium containing 2 μM BRZ, suggesting bzs1–D partially suppressed bzr1–1D’s BRZ-insensitive phenotype (Figure 1E and 1F).
To test whether bzs1–D affected the expression of BR-regulated genes, the expression levels of two BR-responsive genes, SAUR-AC1 and XTR6, were analyzed by quantitative reverse transcription (RT)–PCR. The expression levels of these two genes increased in bzr1–1D but decreased in the BR-deficient mutant det2. In bzr1–1D;bzs1–D, the expression levels of these two genes were reduced dramatically compared to bzr1–1D (Figure 1G). The results indicate that bzs1–D partially suppresses multiple developmental, physiological, and molecular phenotypes of bzr1–1D.
The bzr1–1D mutation suppresses the phenotypes of BR-deficient mutant det2 and BR-insensitive mutant bin2-1 (Wang et al., 2002). When bzs1–D was crossed into det2 and bin2-1, and the double mutants bzs1–D;det2 and bzs1–D;bin2-1(+/–) had shorter hypocotyls in the dark and more dramatic dwarf phenotypes in the light compared with det2 single mutant and bin2-1 heterozygote, respectively, while the segregated bzs1–D itself was phenotypically indistinguishable from wild-type (Figure 2A–2D). These genetic results suggest that bzs1–D has a negative effect on the BR pathway.
Figure 2.
The bzs1–D Mutation Aggravates Phenotypes of BR-Deficient Mutant det2 and Insensitive Mutant bin2-1.
(A, C) Five-day-old etiolated seedlings.
(B, D) Three-week-old soil-grown plants.
bzs1–D Overexpresses a B-box Protein
The population of segregated heterozygous bzr1–1D;bzs1–D lines showed a 3:1 (Basta+:Basta–) ratio, and the basta resistance co-segregated with the mutant phenotypes, indicating a single T-DNA insertion is responsible for the phenotypes of bzr1–1D;bzs1–D. Using thermal asymmetric interlaced PCR (TAIL–PCR), a T-DNA flanking sequence was identified at 1084 bp upstream of the translational start codon of gene At4g39070 (Figure 3A). As activation tagging usually activates the genes in the vicinity of the insert (Weigel et al., 2000), we investigated the expression levels of the genes near the T-DNA insertion site. Quantitative RT–PCR result showed that At4g39070 was overexpressed in mutant bzr1–1D;bzs1–D, but the other two flanking genes At4g39080 and At4g39090 were not significantly affected (Figure 3B). To confirm that the mutant phenotype is caused by overexpression of At4g39070, we overexpressed At4g39070 driven by the CaMV35S promoter in the bzr1–1D background. From 16 independent transgenic lines, 10 lines recapitulated bzr1–1D;bzs1–D mutant phenotype and the level of recapitulation was correlated to the transcript level (Figure 3C and 3D). Overexpression of At4g39070 in bzr1–1D also partly restored the sensitivity of bzr1–1D to BRZ (Supplemental Figure 1). RNA interference (RNAi) was also used to knock down the expression of At4g39070 in bzr1–1D;bzs1–D mutant. From 11 transgenic plants, 10 plants restored bzr1–1D phenotype, and the phenotypes correlated with reduced expression levels of At4g39070 (Figure 3E and 3F). Both overexpression and knockdown experiments confirmed that the phenotype of bzr1–1D;bzs1–D was due to overexpression of At4g39070, so we defined this gene as BZS1.
Figure 3.
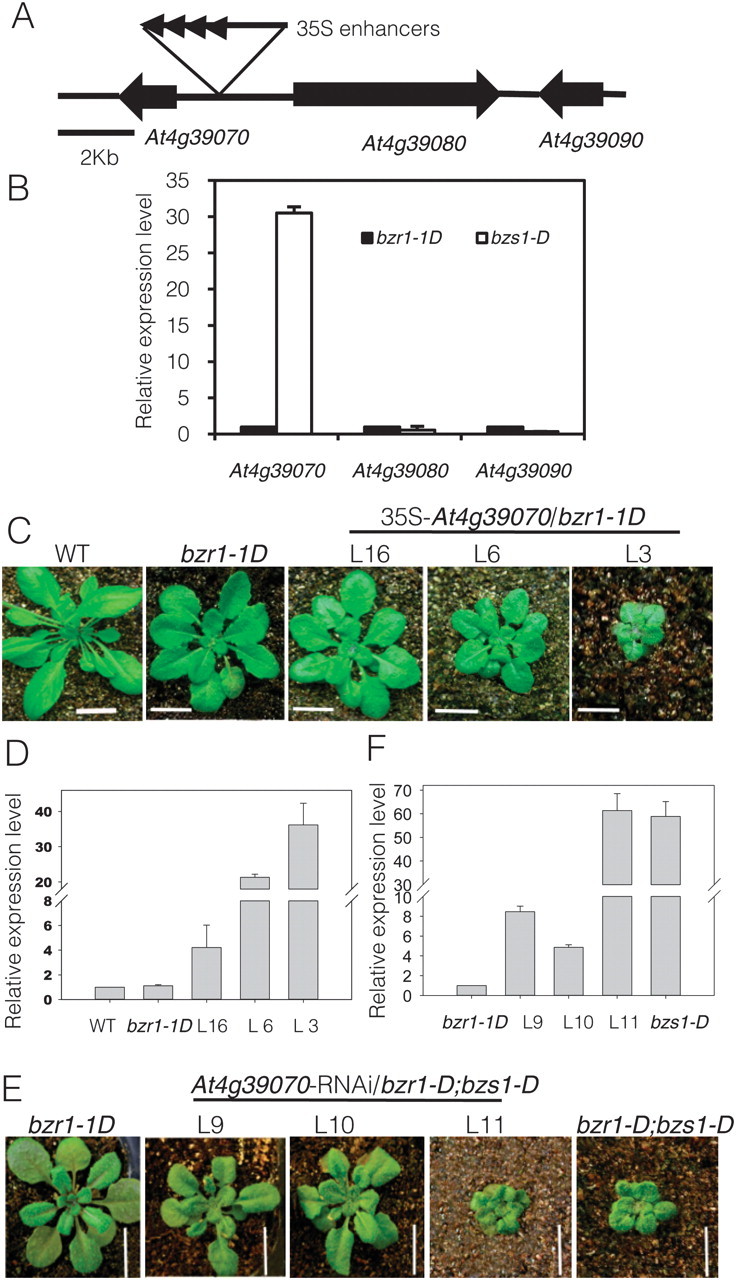
BZS1/BBX20 Is Responsible for the bzs1–D Phenotypes.
(A) A diagram of the genomic region flanking the T-DNA insertion site in bzr1–1D;bzs1–1D.
(B) Quantitative RT–PCR analysis of the genes flanking the T-DNA insertion.
(C) Overexpression of BZS1 in bzr1–1D (two independent lines: L6 and L3) recapitulates phenotype of bzr1–1D;bzs1–D.
(D) Quantitative RT–PCR analysis of the expression level of BZS1 in three independent overexpression lines.
(E) Suppressing the expression of BZS1 using RNAi in bzr1–1D;bzs1–D (two independent lines: L9 and L10) restores bzr1–1D phenotype.
(F) Quantitative RT–PCR analysis of the expression level of BZS1 in BZS1–RNAi plants. For all the qRT–PCR analysis, UBC30 was used as internal control.
BZS1 encodes a 242-amino-acid B-box transcription factor that is a member of the BBX family and has been named BBX20 (Khanna et al., 2009). The expression pattern of BZS1 was examined using BZS1 promoter fused with GUS reporter gene. GUS stain results showed that BZS1 is expressed through the entire life circle, from etiolated seedlings, young seedlings, margins of adult leaves, flower buds, flowers, and siliques (Supplemental Figure 2A–2H). Consistently with BZS1 being repressed by BZR1 (Sun et al., 2010), the BZS1::GUS expression was reduced by BR treatment (Supplemental Figure 2B and 2D). In addition, the subcellular localization of BZS1 was examined using 35S::BZS1–YFP transgenic plants. Confocal microscope imaging results showed that BZS1 localized to both nucleolus and cytoplasm (Supplemental Figure 2I–2K).
BZS1 Is a Negative Regulator in BR Response
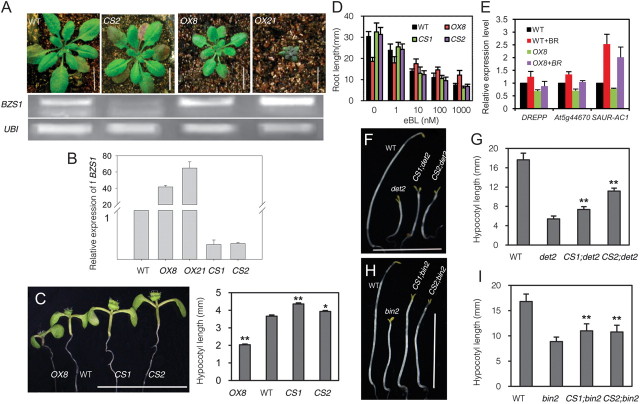
To further analyze the functions of BZS1, a BZS1 overexpression construct (35S::BZS1) was transformed into wild-type plants. Among 23 transgenic plants, 19 plants were smaller than wild-type control. RT–PCR analysis showed that the dwarf phenotype correlated with overexpression of BZS1, and two transgenic lines (BZS1–OX8 and BZS1–OX21) showing medium and strong phenotypes were selected for further analysis (Figure 4A and 4B). Two of the four transgenic lines showing no obvious phenotype when grown in soil (BZS1–CS1 and BZS1–CS2) showed lower expression levels of BZS1 than wild-type control, apparently due to co-suppression (Figure 4A and 4B).
Figure 4.
Overexpression of BZS1 Partly Inhibits BR Signaling.
(A) Soil-grown 3-week-old plants of wild-type and transgenic plants (upper panel); RT–PCR analysis of BZS1 expression (lower panel).
(B) Quantitative RT–PCR analysis of BZS1expression level in WT and BZS1–OX transgenic lines (OX8 and OX21) and co-suppressed lines (CS1 and CS2). UBC30 was used as internal control.
(C) Phenotypes of 7-day-old light-grown seedlings of wild-type, OX8, CS1, and CS2 (left panel) and the measured hypocotyl lengths (right panel). Representatives of the tallest seedlings of each population were chosen for photographs. Error bars indicate SD (n = 30), and significant differences from wild-type are marked (** P < 0.01, * P < 0.05).
(D) Quantification of the root length of seedlings grown on half-strength MS containing various amounts of BR.
(E) qRT–PCR analysis of downstream genes in wild-type and BZS1–OX8 plants with or without 100 nM eBL treatment (3 h). UBC30 was used as internal control.
(F, H) Dark-grown phenotypes of det2 (F) or bin2-1 (H) crossed with BZS1–CS lines (CS1 and CS2).
(G, I) Hypocotyl lengths of dark-grown seedlings in (F) and (H), respectively. Error bars indicate SD (n = 20), and significant differences from wild-type are marked (** P < 0.01, * P < 0.05).
When grown on half-strength Murashige-Skoog (MS) medium in the light, the BZS1–OX lines showed shorter hypocotyls, and the BZS1–CS lines showed significantly longer hypocotyls than wild-type (Figure 4C). While average hypocotyl lengths of the BZS1–CS lines were only slightly longer than wild-type, the tallest BZS1–CS individuals were obviously taller than the tallest wild-type. It seems that not all BZS1–CS seedlings maintain co-suppression. The BZS1–OX plants had shorter roots than wild-type when grown on medium without BR. Increasing concentration of BR inhibits root growth of wild-type plants but the BZS1–OX plants showed reduced response (Figure 4D), suggesting that accumulation of BZS1 reduces BR sensitivity. By contrast, two BZS1–CS lines had slightly longer roots than wild-type on medium without BR and had slightly shorter roots on medium with high concentration of BR, although the differences are not statistically significant. These results suggest that BZS1 reduces BR sensitivity (Figure 4D).
Quantitative RT–PCR results revealed that the expression levels of three BR-induced genes DREPP, At5g44670, and SAUR-AC1 were decreased in the BZS1–OX plants compared to wild-type (Figure 4E). To further analyze the possible role of BZS1 in BR-regulated responses, the BZS1–CS lines (CS1 and CS2) were crossed into BR mutants det2 and bin2-1. The double mutants BZS1–CS1;det2 and BZS1–CS2;det2 had longer hypocotyls than det2 in the dark (Figure 4F and 4G). Similarly, BZS1–CS1 and CS2 also increased the hypocotyl elongation of bin2+/– in the dark (Figure 4H and 4I). These results together demonstrate that BZS1 is a negative regulator of BR response.
BZS1 Promotes Photomorphogenesis
When grown in the dark, BZS1–OX lines showed de-etiolation phenotypes with short hypocotyls and open cotyledons (Figure 5A). The phenotypes of de-etiolation in the dark and hypersensitive to light of BZS1–OX plants indicated that BZS1 promotes photomorphogenesis and may be involved in light signaling. To investigate whether BZS1 is downstream of a specific photoreceptor, BZS1–OX plants and the two BZS1–CS lines were grown in different light conditions. BZS1–OX had shorter hypocotyls, while CS1 and CS2 showed slightly but significantly longer hypocotyls compared to wild-type in blue and red light (Figure 5B–5E and Supplemental Figure 3). Similarly, bzr1–1D;bzs1–D was hypersensitive to both red and blue light (Supplemental Figure 4). These results suggest that BZS1 is a common factor downstream of multiple photoreceptors and promotes photomorphogenesis.
Figure 5.
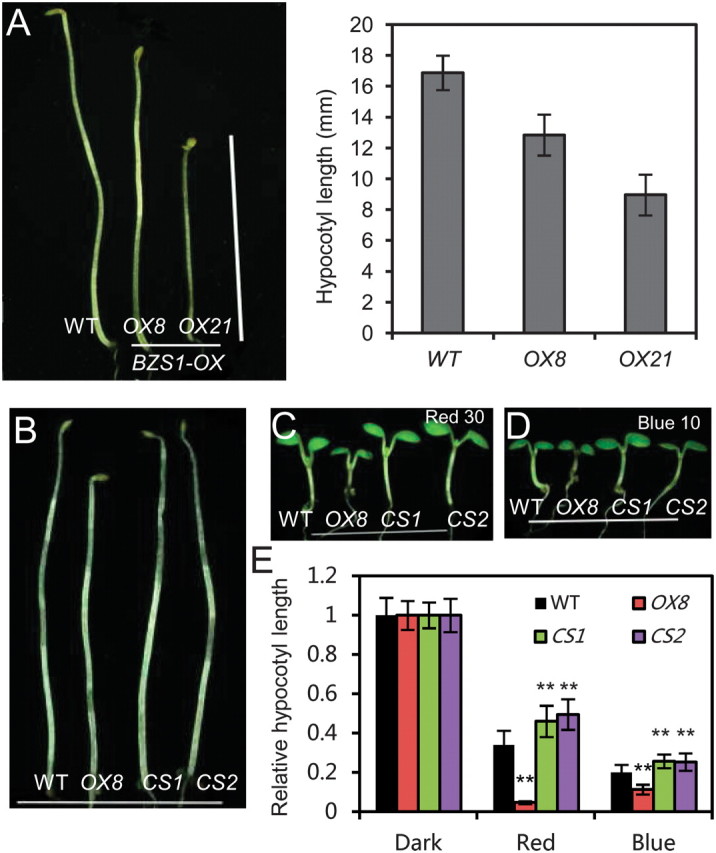
BZS1 Promotes Photomorphogenesis.
(A) Phenotypes of wild-type (WT) and two BZS1–OX transgenic lines (OX8 and OX21) grown in the dark. The hypocotyl length measurement of seedlings shown in (A) is in the right panel. Error bars indicate SD (n = 30).
(B–D) Phenotypes of BZS1–OX and BZS1–CS lines grown in the dark (B), red light (30 μmol m−2 s−1) (C), and blue light (10 μmol m−2 s−1) (D), for 7 d. Representatives of the tallest seedlings of each population were chosen for photographs.
(E) Relative hypocotyl length from (B), (C), and (D). Error bars indicate SD (n = 20), and significant differences from wild-type are marked (** P < 0.01, * P < 0.05).
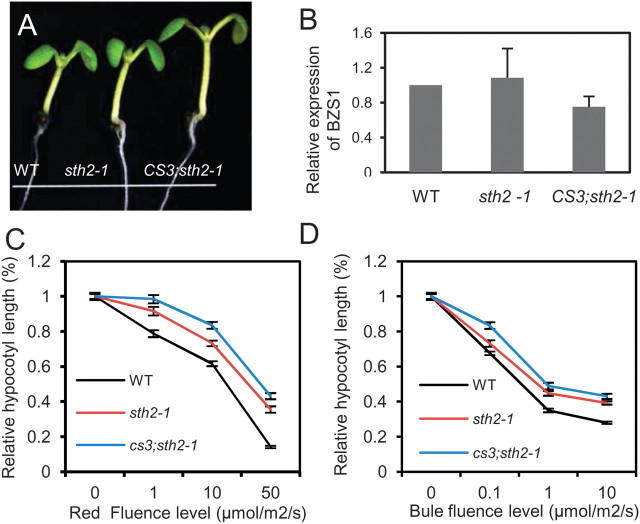
STH2, the closest homolog of BZS1, has been shown to play a positive role in light signaling. The loss-of-function mutant sth2-1 is hyposensitive to blue, red, and far-red light (Datta et al., 2007). We generated a BZS1 co-suppression line in sth2-1 background. Quantitative RT–PCR analysis showed the expression of BZS1 was reduced by about 30% in BZS1–CS3;sth2-1 plants (Figure 6B). Fluence response test showed that BZS1–CS3;sth2-1 plants had longer hypocotyls under various fluence rates of different lights than sth2-1 single mutant (Figure 6A, 6C, and 6D). These data suggest that BZS1 and STH2 play overlapping or redundant roles as positive regulators of light responses.
Figure 6.
BZS1 Functions Redundantly with STH2.
(A) Phenotypes of 7-day-old seedlings of wild-type, sth2-1, and BZS1–CS3;sth2-1 grown in blue light (10 μmol m−2 s−1).
(B) Expression level of BZS1 analyzed by quantitative RT–PCR.
(C, D) Fluence response of wild-type, sth2-1, and BZS1–CS3;sth2-1 measured as relative hypocotyl lengths under red light (C) or blue light (D). Error bars present standard error (n = 20).
BZS1 Affects Genes Responsive to Both Light and BR
To further understand the function of BZS1 in BR and light signaling pathways, we compared the transcriptomic changes caused by BZS1 overexpression, light treatment, and bri1 (Luo et al., 2010; Sun et al., 2010). Seven-day-old light-grown seedlings of BZS1–OX and wild-type (WT) were analyzed by microarray using the Arabidopsis ATH1 array (Affymetrix). The results showed that expression levels of 285 genes were altered in BZS1–OX plants, with 145 genes up-regulated and 140 down-regulated (fold ≥ 1.5 and q ≤ 0.05; Supplemental Table 1). About 66% (189 of the 285) of these genes were affected by light in at least one of the microarray analyses of light-responsive genes (Luo et al., 2010). Among these, about 89% (168) were affected in the same way by BZS1–OX and light treatment (Supplemental Figure 5 and Supplemental Table 2). confirming BZS1 as a positive regulator in the light signaling pathway. About 65 genes (23% of the 285) differentially expressed in BZS1–OX were also affected by bri1-116 mutation, and 83% of these 65 genes were also regulated by light (Supplemental Figure 5 and Supplemental Table 3). Among the genes co-regulated by BZS1 and light or bri1-116 mutation, about 89% or 77% were affected in the same way by BZS1–OX and light treatment or by bri1-116 mutation, respectively. The gene expression results indicate a role of BZS1 in mediating the antagonistic effects of BR and light on gene expression and photomorphogenesis.
Light Regulates BZS1 Protein Accumulation Through COP1
Quantitative RT–PCR result showed that the transcript level of BZS1 was decreased in the light, but induced in the dark (Figure 7A and 7B). Similarly, BZS1 expression was inhibited in the cop1 mutants, suggesting light inhibits the transcription of BZS1 (Figure 7C). Such RNA expression patterns are opposite to the positive function of BZS1 in promoting photomorphogenesis. We thus examined the BZS1 protein level. Interestingly, the BZS1 protein level was increased in BZS1–OX plants after light treatment and in the cop1 mutant compared to WT (Figure 7D and 7E). The opposite changes of BZS1 RNA and protein by both light and cop1 mutation is likely due to negative feedback inhibition of BZS1 transcription by BZS1 protein or independent regulation at transcriptional and posttranslational levels. Suppression of BZS1 in cop1-4 and cop1-6 partly suppressed the photomorphogenic phenotypes of cop1 in the dark, confirming that accumulation of BZS1 contributes to the de-etiolation phenotype of cop1 mutant (Figure 7F and 7G).
Figure 7.
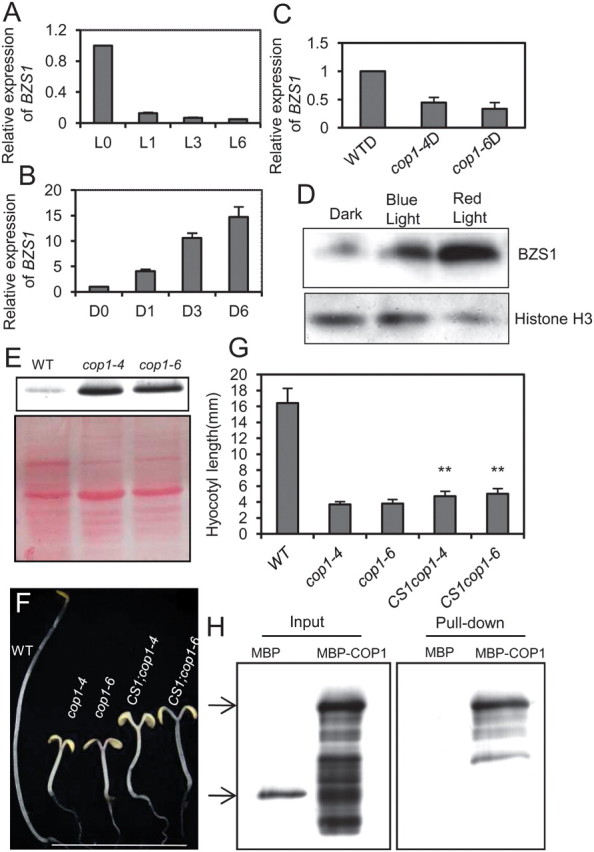
Light Regulates BZS1 Accumulation through COP1.
(A) Light represses BZS1 transcription levels. Etiolated wild-type seedlings were treated with white light for the indicated time and mRNA levels of BZS1 were analyzed by qRT–PCR.
(B) Dark induces BZS1 transcription levels. Light-grown wild-type seedlings were moved into dark for the indicated time and mRNA levels of BZS1 were analyzed by qRT–PCR.
(C) Quantitative RT–PCR analysis of BZS1 expression in the dark-grown wild-type and cop1 mutants.
(D) Immunoblot analysis of BZS1 protein in 3-day-old dark-grown OX8 line seedlings treated with blue and red light for 1 h.
(E) Immunoblot analysis of BZS1 protein levels in dark-grown wild-type and cop1 mutants.
(F) Dark-grown phenotypes of cop1 mutants crossed with BZS1–CS1.
(G) Hypocotyl lengths of wild-type, cop1 mutants, CS1;cop1-4, and CS1;cop1-6. Representatives of the tallest seedlings of each population were chosen for photographs. Error bars represent SD (n = 30) and significant differences from cop1 mutants are marked (** P < 0.01, * P < 0.05).
(H) In vitro pull-down assay showing the interaction between BZS1 and COP1.
COP1 is an E3 ubiquitin ligase that inhibits photomorphogenesis by promoting ubiquitination and degradation of positive regulators of light signal transduction pathways, such as HY5 and STH3 (Osterlund et al., 2000; Datta et al., 2008). The accumulation of BZS1 protein in cop1 mutants suggests that BZS1 might be a substrate of COP1. In vitro pull-down assay showed that BZS1 interacted physically with COP1 (Figure 7H). These results suggest that BZS1 is another positive transcription factor of photomorphogenesis that is directly regulated by COP1. Light-induced inactivation of COP1 leads to accumulation of BZS1 protein, which promotes photomorphogenesis, whereas BR inhibition of BZS1 transcription contributes to BR’s repression effects on photomorphogenesis.
DISCUSSION
Interactions between light and brassinosteroid signals are important for photomorphogenic development in higher plants. The underlying molecular network is not fully understood. In this study, we show that the B-box protein BZS1/BBX20 is another component of the network integrating BR and light signaling on gene expression and photomorphogenic development. BR represses the transcriptional level of BZS1/BBX20 through BZR1, whereas light promotes BZS1/BBX20 protein accumulation through inhibiting COP1’s activity. Our results provide further evidence for a general mode of BR-light crosstalk whereby photomorphogenesis-promoting transcription factors are activated by light-induced stabilization, through inhibition of the COP1-mediated ubiquitination/degradation, but are inhibited by BR signaling through BZR1-mediated transcriptional repression (Luo et al., 2010).
Genetic interactions with BR mutants support a negative role of BZS1/BBX20 in BR response. Overexpression of BZS1 suppressed the BR-activation phenotypes of bzr1–1D, but enhanced the dwarf phenotypes of BR-deficient det2-1 and BR-insensitive bin2-1. In contrast, knockdown of BZS1 expression partly suppressed the phenotypes of det2-1 and bin2-1. These results strongly support that BZS1/BBX20 plays a role in repressing BR-regulated response. BZS1/BBX20 has been shown in Chromatin-immunoprecipitation experiments to be a BZR1 target gene, and its expression is repressed by BR, suggesting that BR represses BZS1 expression through BZR1 (Sun et al., 2010). Consistently with BZS1/BBX20 function downstream of BZR1, bzs1–D had no effect on BZR1 accumulation and phosphorylation (Supplemental Figure 6). BZS1–OX suppressed multiple phenotypes of bzr1–1D, including BRZ-insensitivity and stem kink, suggesting that BZS1/BBX20 is a major component that mediates multiple BZR1-regulated developmental processes.
The phenotypes of BZS1–OX and BZS1–CS plants grown under different light conditions indicate a role of BZS1 in photomorphogenesis. The BZS1–OX plants showed constitutive photomorphogenesis phenotypes in the dark, and hypersensitivity to blue, red, and white light. In contrast, BZS1–CS plants showed increased hypocotyl elongation under all light conditions. Furthermore, BZS1–CS partly suppresses the photomorphogenic cop1 mutant. These results support that BZS1 is a positive regulator of light response. The weak phenotype of the BZS1–CS plants is likely due to partial suppression of BZS1 and redundant function with homologous genes. The close homologs of BZS1/BBX20, namely STH2/BBX21, STH3/BBX22, and STO/BBX24, have been shown to positively regulate photomorphogenesis (Datta et al., 2007; Indorf et al., 2007). BZS1/BBX20 shares 71.82%, 54.13%, and 56.48% of amino-acid identity in the B-box domain to STH2/BBX21, STH3/BBX22, and STO/BBX24, respectively (Supplemental Figure 7). While sth2-1 and BZS1–CS plants each show weak long-hypocotyl phenotypes, the BZS1–CS/sth2-1 plants showed stronger phenotypes than each parental line, consistently with their redundant functions in promoting photomorphogenesis.
To mediate light-regulated development, BZS1/BBX20 protein level is increased by light signaling. Light regulation of BZS1/BBX20 protein is likely mediated by the COP1 ubiquitin ligase, as BZS1/BBX20 accumulates in the cop1 mutants and interacts with COP1 in vitro. Similarly, other members of the BBX family, including CO, COL3, STO, STH1, STH2, and STH3, have been shown to interact with COP1 (Holm et al., 2001; Datta et al., 2006, 2007, 2008; Liu et al., 2008). The triple mutant sth2-1 sth3 cop1 showed a stronger cop1-suppression phenotype compared to the sth3cop1 double mutant, suggesting that STH2 might also be the target of COP1 degradation (Datta et al., 2008). As such, interaction with COP1 seems a conserved feature of the B-box proteins consistently with their roles in light-regulated development.
Our microarray data of BZS1–OX plants provide further evidence for a positive role in regulating light-responsive gene expression. Although only a small set of genes are affected by BZS1 overexpression, the majority of these genes are light-responsive and the expression changes caused by BZS1–OX are similar to the effects of light. How BZS1 regulates gene expression remains unknown, but its homolog STH2/BBX21 has been shown to interact with HY5 (Datta et al., 2007; Holtan et al., 2011), a COP1-targeted b-ZIP transcription factor that positively regulates photomorphogenesis. It is likely that BZS1/BBX20 also interacts with HY5 or other light signaling transcription factors to control a subset of light-responsive genes.
Together, our results demonstrate that BZS1/BBX20 mediates the antagonistic interactions between light and BR signaling pathways. BZS1/BBX20 is oppositely regulated by BR and light through transcriptional and posttranscriptional mechanisms, respectively (Supplemental Figure 8). This is analogous to the GATA2 factor, which is also transcriptionally repressed by BZR1 and posttranslationally stabilized by light through a COP1-dependent mechanism (Luo et al., 2010). Therefore, it seems a general strategy of crosstalk that BR represses the expression and light increases accumulation of positive transcription factors that promote photomorphogenic gene expression and development. In addition, light inhibits BR response by increasing the expression of the Membrane Steroid Binding Protein 1 (MSBP1) (Shi et al., 2011). Furthermore, the BR signaling transcription factor BZR1 and the phytochrome-interacting factor (PIL5) co-regulate many common target genes (Sun et al., 2010). A recent study also showed that the HLH factor PAR1, which was previously identified as a negative regulator of shade-avoidance response (Bou-Torrent et al., 2008), binds to PIF4 to inhibit PIF4 DNA binding (Hao et al., 2012). The BR-activated HLH factor PRE1 interacts with and inhibits PAR1, freeing PIF4 to promote the response to shade and darkness (Hao et al., 2012). Therefore, the interactions between BR and light pathways appear to be mediated by a complex network of molecular interactions.
METHODS
Plant Material and Growth
Arabidopsis thaliana ecotype Columbia-0, various mutants, and transgenic plants obtained in this study were grown at 22°C under white light (16-h light/8-h dark cycles unless stated otherwise) either on half-strength MS medium or in the soil. Arabidopsis seeds were sterilized with 75% ethanol plus 0.01% Triton X-100 for 5 min, followed by three rinses with 95% ethanol, and dried in the hood. The surface-sterilized seeds were sowed on 0.7% phytoagar plates containing half-strength MS medium and 1% sucrose. The plates were kept at 4°C for 3 d and exposed to white light for 6 h before being transferred into the dark. Growth under red and blue light was carried out in an LED light chamber (E-30LEDL3, Percival) at 22°C. Seedlings were photographed and their hypocotyl lengths measured using ImageJ software (http://rsb.info.nih.gov/ij/).
Vector Construction and Transformation
A 1520-bp genomic fragment containing full-length BZS1 open reading frame was amplified by PCR and then cloned into the BamHI and KpnI sites of the pSN1301 binary vector to place BZS1 under the control of the CaMV 35S promoter. The primer sequence used was 5'-GGGGTACCCCAAGTTGGTTTGGATTCATG-3' and 5'-CGGGATCCCGAGAGTGAGAAGAAGCAAT-3'.
To knock down BZS1 in bzr1–1D;bzs1–D, a specific sequence was amplified from bzr1–1D;bzs1–D with primers 5'-CGAGCTCGGATCCTGGTCGGGTTTTGGCTCG-3' and 5'-GGACTAGTGGTACCTTGCCAAGAAGACAGAGC-3'. This fragment was first digested by BamHI and KpnI for the reverse insert to vector pTCK309. The forward insert was generated by SpeI and SacI digestion.
A total 2042-bp genomic fragment containing the promoter region of BZS1 was amplified from WT genomic DNA and cloned into the vector pBI101 by enzymes BamHI and SalI. The primer sequence used was 5'-CGGGATCCACACCAAATCTTCATCTTTC-3' and 5'-ACGCGTCGACGTCCAGTAGTACATCCATG-3'.
The 35S::BZS1–YFP fusion construct was generated by inserting a full-length BZS1 cDNA without stop codon fused to the N-terminus of the pEZR–LNY vector. Primer sequence 5'-ACGCGTCGACAGAGAAGGGTTTGTGATCC-3' and 5'-CGGAATTCAGAGTGAGAAGAAGCAAT-3' was used.
The BZS1–OX, BZS1–RNAi, pBZS1::GUS, and 35S::BZS1–YFP binary constructs were transformed into the Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis thaliana plants via floral dip method.
Total RNA Extraction and Quantitative RT–PCR Analysis
Total RNA was extracted by Trizol (Invitrogen, USA). About 500 ng RNA was reverse-transcribed by AMV reverse transcriptase (Takara Biotechnology, Ltd) following the manufacture’s instruction. Quantitative RT–PCR analyses were carried out on ABI7500 (Applied Biosystems, USA) by using SYBR Green reagent (Toyobo, Japan). Two or three biological repeats and three technical repeats were performed in each treatment. The UBC30 gene was used as internal reference for all the qRT–PCR analysis.
Protein Expression and Antibody Preparation
The full-length BZS1 cDNA was cloned into the pGEX–6p-1 vector to express GST–BZS1 protein in E. coli BL21 cells (Novagen). The recombinant fusion protein was purified using glutathione-agarose beads (GE Healthcare) and used to immunize rabbit. The anti-BZS1 antibody was purified from the immune serum using immobilize un-conserved fragment (amino acids 101–242) of BZS1 tagged with maltose binding protein (MBP) (Amino link Immobilization Kit, Pierce Biotechnology). The anti-histone H3 antibody for loading control was from Sigma (catalog number H9289).
Protein Purification and Pull-Down Assay
The GST–BZS1 protein was expressed using the pGEX–6P-1 vector in E. coli BL21 cells. The recombinant fusion protein was purified using glutathione-agarose beads, and the protein was mostly full-length based on it size in SDS–PAGE gel. COP1 fused to MBP was purified using amylose resin (New England Biolabs). For pull-down assay, glutathione beads containing 1 μg GST–BZS1 were incubated with MBP and MBP–COP1. The mixture was rotated in a cold room for 1 h and the beads were washed at least five times with wash buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl). The proteins were eluted from the beads by boiling in equal volume of 2 SDS buffer and loaded onto a SDS–PAGE gel. Gel blots were analyzed using anti-MBP antibody (NEB).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This study was supported by grants from NSFC (30470169 and 30970253) and NIH (R01GM066258).
Supplementary Material
Acknowledgments
We thank Jia-Ying Zhu for help with editing the manuscript. No conflict of interest declared.
References
- Bou-Torrent J, Roig-Villanova I, Galstyan A, Martinez-Garcia JF. PAR1 and PAR2 integrate shade and hormone transcriptional networks. Plant Signal Behav. 2008;3:453–454. doi: 10.4161/psb.3.7.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao DM, Fan XY, Wang YS, Kang LF. Construction of an activation tagging library of Arabidopsis and mutants phenotype analysis. J. Agricult. Biotechnol. 2008;16:292–298. [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007;19:3242–3255. doi: 10.1105/tpc.107.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18:70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008;20:2324–2338. doi: 10.1105/tpc.108.061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell. 2010;22:1161–1173. doi: 10.1105/tpc.109.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Oh E, Choi G, Liang Z, Wang ZY. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant. 2012;5:688–697. doi: 10.1093/mp/sss011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001;20:118–127. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan HE, et al. BBX32, an Arabidopsis B-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 2011;156:2109–2123. doi: 10.1104/pp.111.177139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodriguez-Franco M. Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 2007;51:563–574. doi: 10.1111/j.1365-313X.2007.03162.x. [DOI] [PubMed] [Google Scholar]
- Kang B, Wang H, Nam KH, Li J. Activation-tagged suppressors of a weak brassinosteroid receptor mutant. Mol. Plant. 2010;3:260–268. doi: 10.1093/mp/ssp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang YS, Quail PH. Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell. 2006;18:2157–2171. doi: 10.1105/tpc.106.042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH. The Arabidopsis B-Box Zinc Finger Family. The Plant Cell. 2009;21:3416–3420. doi: 10.1105/tpc.109.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan SH, Sun Y, Deng ZP, Tang WQ, Shang JX, Sun Y, Burlingame AL, Wang ZY. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nature Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- Lau OS, Deng XW. Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 2010;13:571–577. doi: 10.1016/j.pbi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 2001;26:15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- Li J, Lease KA, Tax FE, Walker JC. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 2001;98:5916–5921. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role forbrassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo XM, et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Garcia S, Vert G, Yin Y, Cano-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol. 2000;124:1520–1524. doi: 10.1104/pp.124.4.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Shi QM, Yang X, Song L, Xue HW. Arabidopsis MSBP1 is activated by HY5 and HYH and is involved in photomorphogenesis and brassinosteroid sensitivity regulation. Mol. Plant. 2011;4:1092–1104. doi: 10.1093/mp/ssr049. [DOI] [PubMed] [Google Scholar]
- Song L, Zhou XY, Li L, Xue LJ, Yang X, Xue HW. Genome-wide analysis revealed the complex regulatory network of brassinosteroid effects in photomorphogenesis. Mol. Plant. 2009;2:755–772. doi: 10.1093/mp/ssp039. [DOI] [PubMed] [Google Scholar]
- Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim T-W, Zhou H-W, Deng Z, Gampala SS, Gendron JM, Jonassen EM, Lillo C, DeLong A, Burlingame AL, Sun Y, Wang Z-Y. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nature Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell. 2009;21:3781–3791. doi: 10.1105/tpc.109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW. The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Gou X, He K, Russell SD, Li J. BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J. 2007;51:220–233. doi: 10.1111/j.1365-313X.2007.03129.x. [DOI] [PubMed] [Google Scholar]
- Zhou A, Wang H, Walker JC, Li J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004;40:399–409. doi: 10.1111/j.1365-313X.2004.02214.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.