Abstract
In genome-based prediction there is considerable uncertainty about the statistical model and method required to maximize prediction accuracy. For traits influenced by a small number of quantitative trait loci (QTL), predictions are expected to benefit from methods performing variable selection [e.g., BayesB or the least absolute shrinkage and selection operator (LASSO)] compared to methods distributing effects across the genome [ridge regression best linear unbiased prediction (RR-BLUP)]. We investigate the assumptions underlying successful variable selection by combining computer simulations with large-scale experimental data sets from rice (Oryza sativa L.), wheat (Triticum aestivum L.), and Arabidopsis thaliana (L.). We demonstrate that variable selection can be successful when the number of phenotyped individuals is much larger than the number of causal mutations contributing to the trait. We show that the sample size required for efficient variable selection increases dramatically with decreasing trait heritabilities and increasing extent of linkage disequilibrium (LD). We contrast and discuss contradictory results from simulation and experimental studies with respect to superiority of variable selection methods over RR-BLUP. Our results demonstrate that due to long-range LD, medium heritabilities, and small sample sizes, superiority of variable selection methods cannot be expected in plant breeding populations even for traits like FRIGIDA gene expression in Arabidopsis and flowering time in rice, assumed to be influenced by a few major QTL. We extend our conclusions to the analysis of whole-genome sequence data and infer upper bounds for the number of causal mutations which can be identified by LASSO. Our results have major impact on the choice of statistical method needed to make credible inferences about genetic architecture and prediction accuracy of complex traits.
Keywords: genome-based prediction, variable selection, complex traits, genetic architecture, plant breeding populations, GenPred, shared data resources
GENOME-BASED prediction of genotypic values from marker or sequence information has been shown to be a valuable tool in plant and animal breeding (Meuwissen et al. 2001; Meuwissen and Goddard 2010; Albrecht et al. 2011). A series of statistical methods mainly differing in the extent of regularization and variable selection has been proposed in the literature (a review is in de los Campos et al. 2013). Simulation studies revealed clear differences between methods with respect to their predictive ability. Several factors affecting the prediction performance of these methods such as genetic trait architecture, span of linkage disequilibrium (LD), sample size, trait heritability, and marker density have been identified (Zhong et al. 2009; Daetwyler et al. 2010; Habier et al. 2010). However, how these methods account for the respective factors is still not fully understood, causing uncertainty about the best choice of method for a given population and trait.
High-throughput genotyping platforms deliver data sets where the number of available observations n is typically smaller than the number of markers p. In these high-dimensional data sets, strategies beyond the classical fixed linear regression model are required because the problem is underdetermined; i.e., there are more unknown parameters than observations (Hastie et al. 2009). Penalized regression techniques constrain the size of the regression coefficients by a penalty function to ensure stable estimates even when n < p. The form of the penalty function crucially affects properties of the respective methods. A frequently used method is ridge regression best linear unbiased prediction (RR-BLUP) (Meuwissen et al. 2001), where the penalty function is defined by the sum of the squared regression coefficients. Here, estimates of marker effects are strongly affected by collinearity between predictors through the so-called grouping effect (Ishwaran and Rao 2011). In RR-BLUP an upper bound exists for pairwise differences between estimated SNP effects, which is a function of their correlation coefficient and of the extent of regularization. All predictors are retained in the model, and marker effects within a block of correlated SNPs tend toward the same value, leading to estimates of low precision.
On the other hand, variable selection methods such as the least absolute shrinkage and selection operator (LASSO) (Tibshirani 1996) or BayesB (Meuwissen et al. 2001) attempt to bridge the gap between a small n and large p through variable selection in addition to regularization; i.e., some variables are effectively removed from the model. In theory, removing markers not in LD with a QTL through variable selection can help to improve prediction performance by reducing the prediction variance but at the expense of an increased estimation bias (Hastie et al. 2009). LASSO uses the sum of absolute values of the regression coefficients as a penalty function, which leads to a sparse solution with less than min(n, p) nonzero elements retained in the model (Hastie et al. 2009). BayesB uses as prior for the marker effects a mixture of a t-distribution and a point mass at zero to induce variable selection (de los Campos et al. 2013). Methods LASSO and BayesB do not exhibit the grouping effect and, therefore, should be able to tag QTL by individual SNPs. Zou and Hastie (2005) introduced the elastic net as a method that combines the properties of both LASSO and RR-BLUP. The elastic net performs variable selection but a potential advantage over LASSO is that for n < p it can retain more than n markers in the model. Moreover, the elastic net can select groups of correlated predictors while the LASSO is expected to select randomly one representative out of a group of correlated variables (Zou and Hastie 2005).
It has been hypothesized that methods employing variable selection are superior to RR-BLUP for traits that are influenced by a small number of QTL or when QTL effects are distributed nonuniformly across the genome (Daetwyler et al. 2010; Hayes et al. 2010). According to Meuwissen and Goddard (2010), the same should hold when predictions are based on whole-genome sequence data, where functional mutations affecting a trait of interest are included in the data with high probability. Furthermore, predictions across several selection cycles, as well as across breeds in animal genetics or across heterotic pools in hybrid plant breeding, may benefit from methods that return the genomic position of functional polymorphisms with higher accuracy rather than distributing effects across the genome. Results from simulation studies support this hypothesis and suggest a superiority of methods employing variable selection, such as BayesB or LASSO (Daetwyler et al. 2010; Meuwissen and Goddard 2010; Clark et al. 2011), over RR-BLUP for specific traits and populations.
Recently, method comparisons have been published for a number of plant populations (Heslot et al. 2012; Pérez-Rodríguez et al. 2012; Riedelsheimer et al. 2012). Using experimental data, only minor differences in prediction performance have been reported between methods with and without variable selection. Most authors concluded that this might reflect an infinitesimal genetic model underlying the traits under study. However, the ability of variable selection methods to identify a true model has not been well studied for plant breeding populations with a large extent of LD, potential substructure, and fairly small sample sizes relative to those in human genetics or animal breeding. It is known from statistical theory that a breakdown phenomenon determined by the number of true nonzero regression coefficients (p0), the number of predictors (p), and the number of observations (n) exists when a given variable selection method’s ability to recover the set of true nonzero coefficients breaks down (Donoho and Stodden 2006; Ishwaran and Rao 2011). In genome-based prediction true nonzero coefficients in the model can arise due to causal mutations, markers in LD with an unobserved QTL, or markers involved in epistatic interactions. To successfully perform variable selection with high-dimensional data (n < p) it is required that the model complexity level defined as ρ = p0/n is (much) smaller than 1.
The critical assumptions needed for successful variable selection have not been addressed in the context of genome-based prediction. However, these assumptions are of high relevance when determining the potential of a given method to remove predictors from the model without loss of information, as well as when making inferences about the genetic architecture underlying the trait of interest. We chose four frequently used methods in genome-based prediction, LASSO, the elastic net, BayesB, and RR-BLUP and investigated their performance in a computer simulation study. These methods can be ranked according to their variable selection intensity. While LASSO produces the sparsest solution, the elastic net and BayesB perform less stringent variable selection and RR-BLUP retains all markers in the model. Predictive performance of the four methods was also compared for a series of traits with presumably different genetic architecture as inferred from genome-wide association (GWA) studies in three experimental data sets of rice (Oryza sativa L.), wheat (Triticum aestivum L.), and the model plant Arabidopsis thaliana (L.) showing very distinct patterns of LD, population size, and stratification (Table 1).
Table 1. Description of experimental data sets.
| Species |
|||
|---|---|---|---|
| Ricea | Wheatb | Arabidopsisc | |
| No. observations n | 413 | 254 | 199 |
| No. SNPs p | 36,901 | 2,056 | 215,908 |
| No. chromosomes | 12 | 21 | 5 |
| Average distance of neighboring SNPs | 10.1 kb | NAd | 0.55 kb |
| Average minor allele frequency | 0.26 | 0.20 | 0.24 |
| Average r2 of neighboring SNPs | 0.39 | NAd | 0.26 |
| Traits analyzed (acronym) | Days to heading in Aberdeen (flowering time) | Yield | Flowering time in the field (flowering time) |
| Plant height | Thousand-kernel weight | Plant diameter at flowering (plant diameter) | |
| Panicle length | Days to heading | FRIGIDA (FRI) gene expression | |
| Length of seed with hull (seed length) | Plant diameter grown at 10° (plant width) | ||
Data sets previously described in Zhao et al. (2011).
Data sets previously described in Poland et al. (2012).
Data sets previously described in Atwell et al. (2010).
Marker positions not available.
The objectives of this study were (1) to explore in silico the efficiency of variable selection methods under different levels of model complexity (ρ) and determinedness (n/p) in the context of genome-based prediction; (2) to investigate the influence of the LD structure, of the number of QTL, and of the trait heritability on the predictive ability of the different methods; (3) to elucidate contradictory results from computer simulations and experimental data with respect to the performance of variable selection methods compared to RR-BLUP; and (4) to assess prediction performance of LASSO, the elastic net, BayesB, and RR-BLUP in experimental data sets using traits of different genetic architecture. By combining computer simulations and experimental data we investigated the joint influence of factors such as genetic trait architecture and sample size and ensured that we explored scenarios relevant for real life experimental data. Finally, we discuss whether the assumptions required to benefit from variable selection are met in plant breeding populations employed in genome-based prediction.
Materials and Methods
Plant material
The rice (O. sativa L.) data set was recently analyzed by Zhao et al. (2011) and is publicly available. Data were downloaded from http://www.ricediversity.org/data/. The germplasm consists of a global diversity panel with 413 rice varieties from six subpopulations (Supporting Information, Figure S1). Individuals in the rice data were highly homozygous with a small fraction of residual heterozygosity (0.48%). Phenotypic data on 34 traits and genotypic data from an Affymetrix 44K SNP array were available. A final set of 36,901 high-quality SNPs was used for this study after quality control conducted by Zhao et al. (2011). Missing values in the marker matrices (4.3%) were reconstructed based on flanking markers, using Beagle (Browning and Browning 2009). Results from the GWA study using the mixed-model approach in Zhao et al. (2011) were used to identify four quantitative traits (flowering time, plant height, panicle length, and seed length) with contrasting genetic architectures.
The wheat (T. aestivum L.) data set was provided by Poland et al. (2012) and was downloaded from the corresponding supplemental material. It comprises 254 advanced breeding lines from the Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT) wheat breeding program. The F6 lines were derived from 122 crosses contributing from 1 to 12 lines, leading to familial substructure in the data (Figure S1). All lines were genotyped using a genotyping-by-sequencing approach, and 41,371 polymorphic SNPs were discovered (Poland et al. 2012). To minimize the number of missing genotypic scores, we selected the 2056 SNPs with the lowest number of missing values. In the absence of a reference genome, no physical positions can be assigned to the SNPs. Hence, remaining missing values (14%) were imputed based on the marginal allele frequencies. All lines were phenotyped in the year 2010 in Mexico in seven trials with three replications under irrigated and drought conditions. Here, we analyzed the adjusted means for the traits yield, thousand-kernel weight, and days to heading under irrigated conditions as provided by Poland et al. (2012).
The A. thaliana (L.) data set was previously described and analyzed by Atwell et al. (2010). Data were downloaded from https://cynin.gmi.oeaw.ac.at/home/resources/atpolydb and consisted of 199 accessions genotyped with a custom Affymetrix 250K SNP chip and phenotyped for a total of 107 traits. All individuals were fully homozygous inbred lines. SNPs were preselected for quality control according to the protocol in Atwell et al. (2010), resulting in 216,130 SNPs for the analysis. We updated the SNP positions to the current Arabidopsis Information Resource (TAIR) 10 assembly (http://www.arabidopsis.org) and removed 222 SNPs with mismatches to the reference genome, resulting in 215,908 high-quality SNPs used in this study. Atwell et al. (2010) conducted a GWA study, using the efficient mixed-model association (EMMA) program to test for associations of single SNPs after correcting for population stratification. From their results, we identified four quantitative traits [flowering time, plant diameter, FRIGIDA (FRI) gene expression, and plant width] with a contrasting genetic architecture that were analyzed in this study (Table 1). For the trait FRI gene expression, functional deletions at the FRI gene locus were included into the marker matrix by coding them as a SNP.
Linkage disequilibrium
LD between marker pairs was estimated with the software PLINK (Purcell et al. 2007, version 1.07) as the squared correlation (r2) between alleles at two loci (Hill and Robertson 1968). Short-range LD decay was measured by examining the average r2 of neighboring SNPs and values for the rice and Arabidopsis data sets are given in Table 1. Long-range LD decay was visualized with the network R package (Butts 2008). An LD network was constructed by connecting all SNP pairs with an r2 value above a certain threshold with an edge, while all other pairs were omitted from the network. To compare the LD in the experimental data sets based on a similar number of SNPs, we evaluated the 2056 SNPs of the wheat data set and 100 randomly selected subsets of 2000 SNPs for rice and Arabidopsis, respectively. The average density of a network was computed by the ratio of observed to potential edges (Butts 2008), using thresholds for r2 of 0.75, 0.50, and 0.25. For p SNPs, the number of potential edges in a network is given by p ⋅ (p − 1)/2. Extensive long-range LD will cause many edges between SNP pairs because their pairwise r2 value is above the threshold and the density will be large. The extent of long-range LD is visualized in a single network for each data set in Figure S2. The average densities for the wheat and 100 samples of the rice and Arabidopsis data set are presented in Table S1. The largest density was observed in the rice data, partly due to the admixture of subpopulations, with several SNPs being monomorphic within subpopulations but polymorphic across subpopulations. For the wheat data set, we recognize that the LD pattern will be influenced by both the imputing algorithm and the selected subset of markers. The observed density must be interpreted as a lower bound estimate because the extent of LD can be deflated by an imputing scheme that does not take into account flanking marker information.
Genome-based prediction methods
We used methods LASSO, the elastic net, BayesB, and RR-BLUP for genome-based prediction. All methods employ the same linear model, using training data with n individuals and p SNP markers,
| (1) |
with y denoting the n-dimensional vector of phenotypic values, 1 an n-dimensional vector of ones, μ the overall mean, W the n × p matrix of genotype scores coded as the number of copies of the minor allele using the synbreed R package (Wimmer et al. 2012), β the p-dimensional vector of marker effects, e the n-dimensional vector of residuals, I an n × n identity matrix, and σ2 the residual variance.
Estimates for LASSO, the elastic net, and RR-BLUP can be obtained from penalized regression by solving
| (2) |
where Pen(β) denotes the penalty function, which is defined by the squared L2 norm for RR-BLUP with , by the L1 norm with for LASSO, and by a mixture of both in the elastic net with (Hastie et al. 2009). Both RR-BLUP and LASSO can be seen as special cases of the elastic net with λ1 = 0 and λ2 = 0, respectively. An estimate for the regularization parameter λ in LASSO was obtained by scanning a grid of 100 values to select the that gives the minimum mean squared error within cross-validation (CV), using the glmnet R package (Friedman et al. 2010). For the elastic net, we used CV on a two-dimensional grid to optimize λ1 and λ2 simultaneously. For RR-BLUP, we calculated the noise-to-signal ratio with and being the residual and marker variance component estimates obtained by residual maximum likelihood (REML), using the ASReml software (Gilmour et al. 2009) according to Riedelsheimer et al. (2012).
For the BayesB method we followed Meuwissen et al. (2001). The prior for the marker effect βj for j = 1, …, p is given by the hierarchical prior
where δ0(⋅) denotes a point mass at zero that assigns zero variance to the effects of a fraction π of markers. A priori, only a fraction 1 − π of markers was selected to be in the model and a scaled inverted chi-square distribution χ−2(ν, S) was used as prior distribution for the variance of the marker effects with hyperparameters degrees of freedom ν and scale S. With π > 0 a variable selection feature is introduced in BayesB, and we used π = 0.7 for the rice and wheat data sets and 0.975 for the Arabidopsis data set. These prior values for π were chosen so that (1 − π) ⋅ p was in the same order of magnitude as the number of SNPs required to reach a plateau for the predictive ability evaluated with RR-BLUP and random marker subsets (Figure S3, Figure S4, and Figure S5). In the experimental data sets and computer simulations we chose ν = 5 for all traits and S according to Ober et al. (2012).
The BayesB method was fitted using the Metropolis–Hastings algorithm implemented in the GenSel software (Fernando and Garrick 2009, versions 4.0.1 and 4.36R, http://bigs.ansci.iastate.edu) for all experimental data sets. In all computer simulations, we employed an implementation of the algorithm in R (R Development Core Team 2012). For the analysis of the experimental data sets in GenSel, a chain of length 50,000 was generated with the first 10,000 samples declared as burn-in and the last 40,000 samples were used for posterior inference. For the computer simulations, we used 5000 iterations including a burn-in of 1000. The reduced chain length was found to be sufficient for the lower number of SNP markers that was used in the computer simulations. Differences compared to analyses with 13,000 iterations including a burn-in of 3000 were found to be small in selected scenarios representing the extremes of the simulation scheme. In addition, we observed only minor differences in the posterior distributions between two replicated chains and interpreted this as evidence that the algorithm converged.
Cross-validation and predictive ability
Fivefold CV was used to assess the prediction performance of the different statistical methods in the experimental data sets and computer simulations. Following Albrecht et al. (2011), the data set was divided into five mutually exclusive subsets; four of them formed the estimation set (ES) for fitting marker effects and the fifth subset was used as a test set (TS). We predicted the genotypic values in the TS according to
where the matrix WTS encodes the marker genotypes for the individuals in the TS, using the same reference alleles as in the ES, and are the estimates of the marker effects derived from the ES. Regularization parameters in LASSO, the elastic net, and RR-BLUP were derived from the ES.
Pearson’s correlation coefficient between predicted genotypic values and observed phenotypic values (yTS) in the TS describes the predictive ability of a method in experimental and simulated data. The accuracy of a method describes the correlation between predicted and true genotypic values and was available only for simulated data sets. In all scenarios, accuracies can be approximated from the predictive ability as , where h is the square root of the trait heritability (Legarra et al. 2008). For the experimental data sets, we report the average predictive ability from 10 replications of the CV scheme. Standard errors of the predictive abilities were calculated from the means of the replications.
Computer simulations
Computer simulations were employed to assess the performance of the statistical methods LASSO, the elastic net, BayesB, and RR-BLUP under scenarios differing in sample size, LD structure, genetic trait architecture, and trait heritability. First, we investigated the ability of the statistical methods to recover true models of varying complexity in an underdetermined system with more markers than observations. No correlation structure between predictor variables was simulated. Next, we used the marker information of the experimental data to simulate data sets that allowed us to investigate the influence of LD, trait architecture, and heritability on model performance. The following simulation procedures were conducted (see also the overview in Figure S6):
- Following Donoho and Stodden (2006), we generated 400 different scenarios varying for model complexity ρ = p0/n and determinedness level n/p. Values for ρ and n/p ranged from 0.05 to 1.00 with increments of 0.05. Although ρ can take any positive value, we focused on ρ ∈ [0, 1] because it was expected that the ability of the variable selection methods to recover the true model breaks down outside this interval (Donoho and Stodden 2006). In each scenario, we simulated p = 2000 independent biallelic SNP marker genotypes for n individuals according to the determinedness level. For each individual and SNP, the marker genotype was sampled from a Bernoulli distribution, taking the value of 2 with probability 0.3 and the value of 0 with probability 0.7, respectively. The marker genotypes were combined in the n × p marker matrix W. Next, a subset of p0 SNPs was declared to be true nonzero coefficients with effect sizes randomly sampled from a U(0, 100/p0) distribution. All p − p0 remaining markers were declared as true zero coefficients and hence the vector of the true marker effects is given as . True genotypic values were calculated as g = Wβ0 and phenotypic records were simulated as y = g + e with e ∼ N(0, Iσ2) and σ2 = Var(g) ⋅ (1 − h2)/h2 to obtain a trait heritability of h2. For each scenario, we estimated the marker effects in the whole data set, using LASSO, the elastic net, BayesB (π = 0.80), and RR-BLUP. These estimates were used to calculate the normalized L2 error according to Donoho and Stodden (2006) as a measure for the accuracy of estimated marker effects for each scenario as
between true (β0) and estimated marker effects, where ‖⋅‖2 denotes the L2 norm of a vector. The normalized L2 error loss function is ∈ [0, ∞) and evaluates the performance over true zero and nonzero coefficients. A small normalized L2 error indicates good agreement between estimated and simulated marker effects. A normalized L2 error of 1 implies that the sum of squared differences between estimated marker effects and simulated marker effects was equal to the sum of squared simulated marker effects. Normalized L2 errors of all 400 scenarios were visualized with heat maps. All scenarios were replicated four times for each method and results were presented as averages over replications. We also generated scenarios resembling the data structure of whole-genome sequence data, using p = 250,000 and n = 200. Here, we performed scenarios differing in level of model complexity (p0/n = 0.02, … , 0.20 with increments of 0.02) and trait heritability (h2 = 0.25, 0.50, 0.75, and 1.00). For each scenario, we performed 10 replications. For LASSO, we investigated the ability to detect the true nonzero coefficients by calculating the sensitivity according to Pepe (2004) as the empirical conditional probability that a true nonzero coefficient received a nonzero estimate; i.e., for all j = 1, … , p0. We evaluated scenarios in which the LD structure of the experimental data sets was reflected. These scenarios were generated to evaluate the predictive ability for different genetic trait architectures and heritabilities defined in silico but with data structures similar to those in the experimental data sets. Retaining original sample sizes from experimental data [rice (n = 413), wheat (n = 254), and Arabidopsis (n = 199)], we randomly selected 2000 SNPs from each data set to obtain LD structures in three simulation scenarios with large (rice), medium (wheat), and small (Arabidopsis) extents of LD, respectively (Table S1 and Figure S2). From these 2000 SNPs we randomly declared p0 SNPs (p0 = 1, 10, and 100) to be causal mutations under the restriction that the minor allele frequency (MAF) of the SNP was >0.05. All causal mutations were assigned additive genetic effects of equal magnitude and genotypic values were obtained by the sum of all mutation effects. Analogously to procedure 1, random Gaussian errors were simulated to obtain phenotypic records with h2 = 0.1, 0.5, and 0.9. For each combination of data set, p0, and h2, we performed 10 replications by sampling new sets of 2000 SNP markers from the experimental data sets and assigning new causal mutations. The predictive ability of both BayesB (π = 0.95) and RR-BLUP was assessed with fivefold CV within each replication.
- We assessed the joint influence of the model complexity and determinedness level on the normalized L2 error in scenarios with correlated markers. Here, it was not sufficient to sample SNP genotypes from the experimental data as in procedure 2 because these were fixed with respect to sample size. To obtain scenarios with varying sample sizes, we first simulated independent marker data W according to procedure 1. Next, we conveyed the LD structure of the rice, wheat, and Arabidopsis data sets to the simulated data. The LD structure from the experimental data was assessed by randomly selecting 2000 SNPs and computing their 2000 × 2000 empirical correlation matrix Σ. Because empirical correlation matrices are not necessarily positive definite, we used the nearPD function of the Matrix R package (Bates and Maechler 2012) to construct a positive definite matrix from Σ, denoted by Σ*. A Cholesky decomposition was used to construct a 2000 × 2000 upper-diagonal matrix U such that U′U = Σ*. By multiplying WU = W* we conveyed the correlation structure of each experimental data set to the simulated data. However, values in W* were not binary and hence we used component-wise thresholding for all i = 1, … , n and j = 1, … , 2000 to obtain a new matrix W** of the same dimension but entries were transformed using
with being the ijth element in matrix W* and the (1 − pj)th quantile of column . The value for pj equaled the MAF of marker j as observed in the experimental data set. Finally, the simulated data set W** consisted of n individuals and 2000 markers, where the MAF and correlation structure were similar to those of a specific experimental data set. To account for the variability in the sampling of the SNP markers, we evaluated each of the 400 scenarios with 10 different marker subsets and reported the average values across replications.
Results and Discussion
Breakdown behavior of variable selection methods
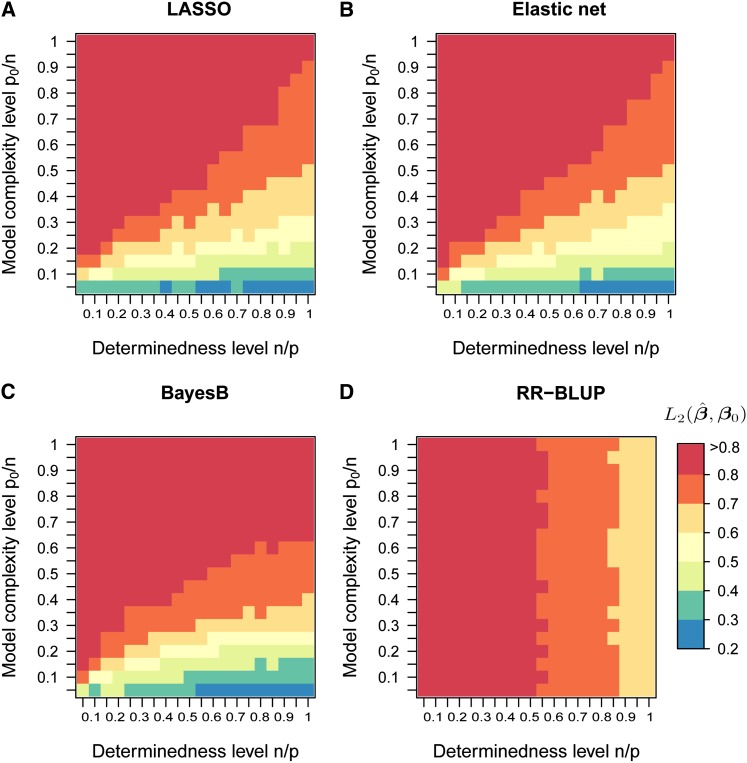
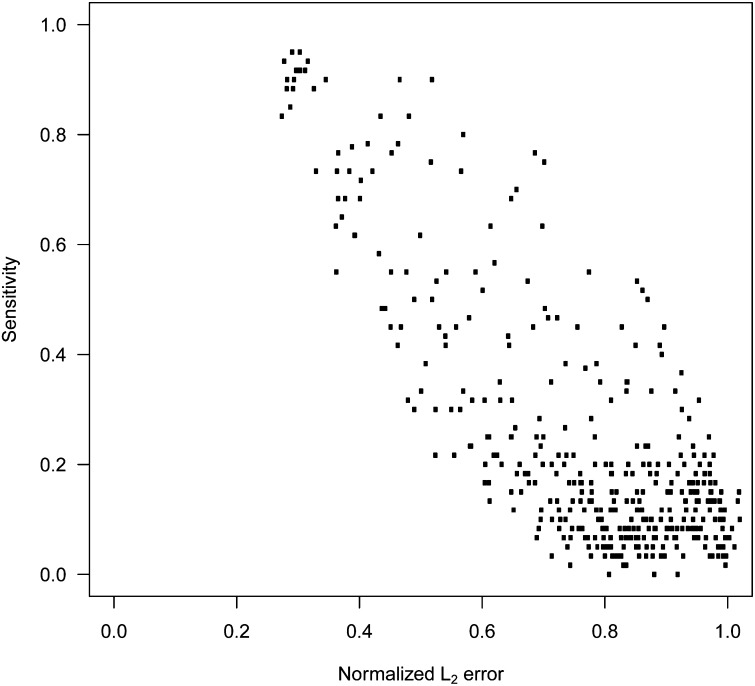
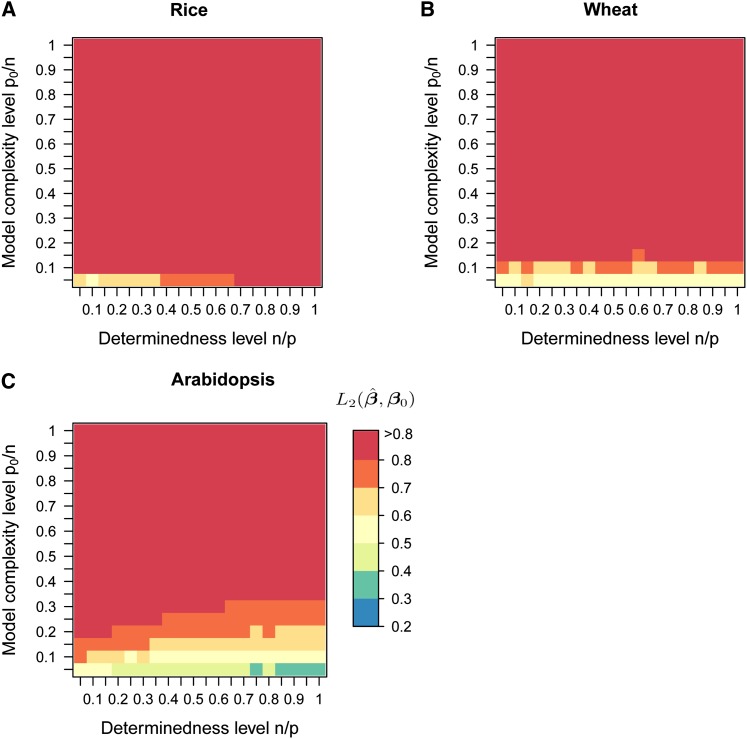
We evaluated the ability of the four methods to cope with high-dimensional data sets and models of varying complexity (simulation procedure 1, Figure S6). In Figure 1, method performance is given for 400 simulated scenarios differing in the number of observations n and the number of true nonzero coefficients p0 for LASSO, the elastic net, BayesB, and RR-BLUP (p = 2000 and h2 = 0.75). We observed a large influence of the model complexity level on the average normalized L2 error in LASSO, the elastic net, and BayesB, but not in RR-BLUP. The average normalized L2 error of all methods increased with decreasing determinedness level n/p, i.e., more markers per observation. To infer upper bounds for the number of true nonzero coefficients that can be accurately determined by the variable selection methods, we calculated the probability that the min(p0, 20) largest true nonzero effects were included in the model, for all 400 scenarios. For LASSO, a probability >0.8 was generally achieved in scenarios with a normalized L2 error <0.5 (Figure 2).
Figure 1.
(A–D) Accuracy of estimated marker effects in computer simulations using independent predictor variables. Simulations were conducted according to procedure 1 (Figure S6). In each scenario p = 2000 independent markers were simulated and h2 = 0.75 was used to simulate n phenotypic records. The normalized L2 errors of LASSO, the elastic net, BayesB, and RR-BLUP are displayed as heat maps for a grid of 20 values between 0.05 and 1.00 for the determinedness level n/p and model complexity level p0/n, respectively. The color key presents the normalized L2 error averaged over four replications for each scenario.
Figure 2.
Averaged normalized L2 error vs. the averaged sensitivity of LASSO across all 400 scenarios with four replications as in Figure 1. The sensitivity was evaluated as the empirical conditional probability that one of the min(p0, 20) largest true nonzero coefficients was identified.
We assume that a normalized L2 error of 0.5 is an upper bound for accurately estimated marker effects. Now, we can infer from Figure 1 the maximum level of model complexity where the normalized L2 error is <0.5 for each level of determinedness. For BayesB, a complexity level ρ = p0/n ≤ 0.2 is required to obtain an average normalized L2 error <0.5 for n/p = 1. Thus, n ≥ 5p0 are required, i.e., more than five phenotypic records per true nonzero coefficient, to accurately estimate marker effects. When the level of determinedness is decreased to n/p = 0.05, more than 20 phenotypic records per true nonzero coefficient are required for a normalized L2 error <0.5. These numbers are in good agreement with theoretical and empirical thresholds given by Donoho and Stodden (2006) for LASSO to perform comparably to an all-subset search. Over the 400 scenarios, the performance of the elastic net was more similar to that of LASSO than to that of RR-BLUP, which was expected for scenarios without pronounced correlation structure.
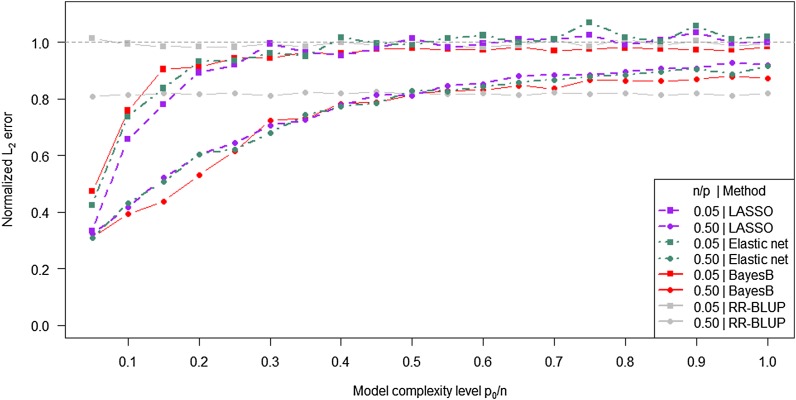
To investigate the influence of different model complexity levels on the accuracy of estimated marker effects more closely, we fixed the determinedness level n/p at 0.5 and 0.05, corresponding to scenarios of sample sizes n = 1000 and n = 100, respectively. The four methods revealed different curve characteristics for the normalized L2 error as a function of model complexity level (Figure 3). Even though there was no strong collinearity among markers, individual marker effects were estimated with low precision in RR-BLUP and the level of the normalized L2 error was >0.5 irrespective of the model complexity level. In contrast, LASSO, the elastic net, and BayesB benefited from a low model complexity level. As expected, the performance of all methods was higher for n = 1000 compared to n = 100. LASSO was found to perform better than the other variable selection methods for low levels of model complexity and n/p = 0.05. However, in general no method consistently dominated the other methods with respect to the normalized L2 error.
Figure 3.
Normalized L2 error for LASSO, the elastic net, BayesB, and RR-BLUP as a function of the model complexity level. Curves were extracted from the surfaces in Figure 1 by fixing the determinedness level n/p at 0.05 and 0.50, respectively.
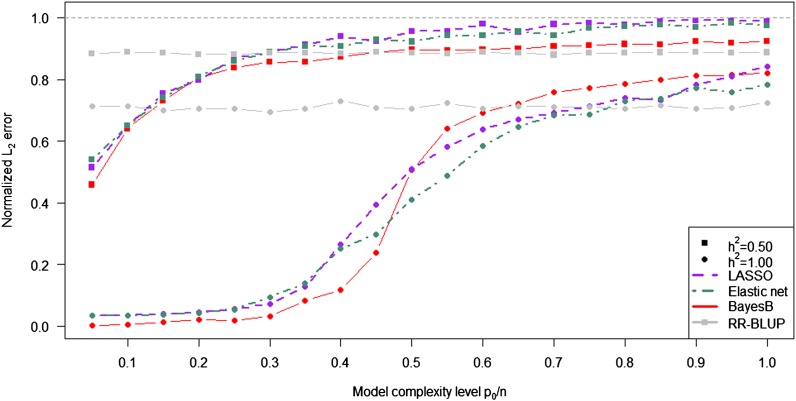
The effect of different levels of noise on the performance of the statistical methods was investigated in simulated scenarios with different trait heritabilities and fixed determinedness level n/p = 0.5 (Figure 4). As expected, differences in the performance of the variable selection methods and RR-BLUP disappeared earlier with h2 = 0.5 than with h2 = 1.0. The ability of LASSO, the elastic net, and BayesB to accurately recover and estimate the effects of true nonzero coefficients with h2 = 1.0 held until a complexity level of ρ = 0.3 was reached, a value similar to that observed for LASSO by Stodden (2006). The elastic net outperformed the other methods for medium levels of model complexity and h2 = 1.0 while BayesB and RR-BLUP gave the lowest normalized L2 error for h2 = 0.5 and high levels of model complexity.
Figure 4.
Normalized L2 error for LASSO, the elastic net, BayesB, and RR-BLUP as a function of the model complexity level for different trait heritabilities (h2 = 0.50 and 1.00) and n/p = 0.5. The simulations were conducted according to procedure 1 (Figure S6).
We conclude that the breakdown behavior of the variable selection methods with respect to recovering the true model was mainly dominated by dimensionality. Thus, we can use marker effects to learn about genetic trait architecture only if the trait has a sparse representation. In this case, methods such as LASSO, BayesB, and fixed-effect regression methods commonly used in GWA studies will lead to marker effects of higher precision than RR-BLUP because they do not exhibit the grouping effect. However, as soon as the trait architecture becomes more complex, the methods will not be successful in identifying SNP markers tagging a QTL with high probability. Recall that model complexity was presented as a function of n. With sample sizes commonly employed in plant breeding for GWA studies and genome-based prediction we tend to explore scenarios in the top left corner of the heat maps in Figure 1. Here, all methods lead to estimated marker effects of low precision and differences between methods melt. Only increasing the sample size n can alleviate this curse of dimensionality.
Because variable selection methods were favored over RR-BLUP with respect to the normalized L2 error of marker effect estimates given a low complexity level, we expected them to outperform RR-BLUP in genome-based prediction for traits of low complexity. To verify this hypothesis, we evaluated the predictive ability of LASSO, the elastic net, and RR-BLUP under different levels of model complexity and determinedness, using CV (simulation procedure 1, Figure S6). Results given in Table 2 show that differences between methods with respect to accuracy of marker effect estimates measured by the normalized L2 error translated into differences in predictive abilities. LASSO performed better than RR-BLUP when the level of model complexity was ρ = 0.1, but RR-BLUP outperformed LASSO in scenarios with ρ ≥ 0.5. The elastic net did improve prediction performance compared to RR-BLUP and LASSO in scenarios with intermediate determinedness and model complexity levels. However, the performance of the elastic net was dominated by one of its special cases in most scenarios. It was sufficient to use the L1 norm in LASSO for regularization in scenarios with low model complexity while in scenarios with high model complexity it was sufficient to use only the L2 norm for regularization within RR-BLUP. As expected, predictive ability of all methods increased with increasing determinedness level, i.e., more observations per marker. As was already evident from the heat map in Figure 1, prediction accuracies of the variable selection methods close to 1 could be achieved only when using >10 observations per true nonzero coefficient, i.e., for ρ = 0.1. Under this scenario, RR-BLUP was not optimal for prediction and differences between the predictive abilities of the methods were consistent with those in Meuwissen and Goddard (2010).
Table 2. Predictive abilities in computer simulations using LASSO, the elastic net, and RR-BLUP.
| Model complexity level ρ = p0/n |
|||||
|---|---|---|---|---|---|
| n/p | Method | 0.10 | 0.25 | 0.50 | 1.00 |
| 0.10 | LASSO | 0.69 ± 0.02 | 0.41 ± 0.02 | 0.20 ± 0.05 | 0.09 ± 0.04 |
| Elastic net | 0.62 ± 0.02 | 0.35 ± 0.03 | 0.19 ± 0.03 | 0.09 ± 0.03 | |
| RR-BLUP | 0.20 ± 0.03 | 0.21 ± 0.03 | 0.22 ± 0.03 | 0.22 ± 0.03 | |
| 0.25 | LASSO | 0.73 ± 0.01 | 0.46 ± 0.02 | 0.29 ± 0.03 | 0.13 ± 0.05 |
| Elastic net | 0.73 ± 0.01 | 0.48 ± 0.03 | 0.33 ± 0.02 | 0.19 ± 0.02 | |
| RR-BLUP | 0.38 ± 0.01 | 0.35 ± 0.01 | 0.34 ± 0.02 | 0.34 ± 0.01 | |
| 0.50 | LASSO | 0.77 ± 0.01 | 0.57 ± 0.02 | 0.39 ± 0.02 | 0.26 ± 0.02 |
| Elastic net | 0.76 ± 0.01 | 0.60 ± 0.01 | 0.43 ± 0.01 | 0.31 ± 0.01 | |
| RR-BLUP | 0.47 ± 0.01 | 0.48 ± 0.01 | 0.49 ± 0.01 | 0.48 ± 0.01 | |
| 1.00 | LASSO | 0.79 ± 0.003 | 0.68 ± 0.01 | 0.52 ± 0.01 | 0.42 ± 0.02 |
| Elastic net | 0.80 ± 0.004 | 0.67 ± 0.004 | 0.54 ± 0.01 | 0.45 ± 0.01 | |
| RR-BLUP | 0.61 ± 0.01 | 0.62 ± 0.01 | 0.61 ± 0.01 | 0.60 ± 0.01 | |
Average predictive abilities ± SEs were estimated using fivefold cross-validation with 10 replications for each scenario. All scenarios were simulated according to procedure 1 (Figure S6), using p = 2000 independent markers and h2 = 0.75.
Discrepancies in method comparisons between simulated and experimental data
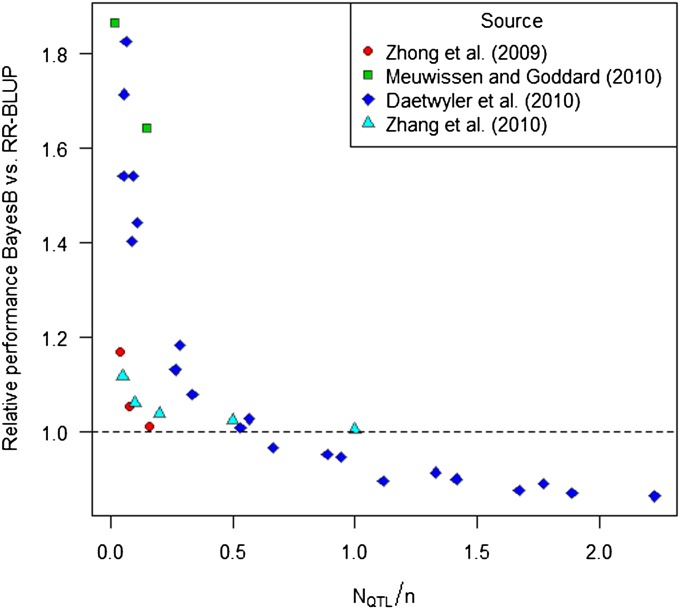
The hypothesis that genetic trait architecture has a strong influence on the relative performance of prediction methods is mainly supported by simulation studies comparing BayesB with RR-BLUP under scenarios differing in the number of simulated QTL (NQTL) and sample size n. Treating NQTL as a proxy for p0, we compared the relative performance of methods BayesB and RR-BLUP as a function of NQTL/n in four recently published simulation studies (Zhong et al. 2009; Daetwyler et al. 2010; Meuwissen and Goddard 2010; Zhang et al. 2010). BayesB outperformed RR-BLUP by up to 80% if the complexity level NQTL/n was smaller than 0.5 (Figure 5). However, with increasing complexity level, the relative superiority of BayesB over RR-BLUP sharply decreased in all studies and vanished for NQTL/n > 0.5. This decrease in relative performance of BayesB with increasing number of simulated QTL has been discussed earlier (e.g., Daetwyler et al. 2010 or de los Campos et al. 2013). However, it has not yet been related to the sample size and to the breakdown behavior of the respective methods.
Figure 5.
Meta-analysis of relative performance of BayesB compared to RR-BLUP with results from the literature. Results were extracted from the studies in Zhong et al. (2009), Daetwyler et al. (2010), Meuwissen and Goddard (2010), and Zhang et al. (2010), differing with respect to the number of QTL (NQTL) and sample size (n) of the training data set. Relative performance is defined as the ratio of accuracy or predictive ability of BayesB over RR-BLUP.
To investigate prediction performance of LASSO, the elastic net, BayesB, and RR-BLUP for experimental data with different levels of model complexity and determinedness we chose three experimental data sets differing with respect to sample size n, marker coverage, extent of LD, and substructure (Table 1). We selected traits for which prior knowledge about the genetic architecture based on GWA studies was available. The traits flowering time in the rice data set and FRI gene expression in the Arabidopsis data set were used as candidates for traits exhibiting a sparse genetic architecture (Atwell et al. 2010; Zhao et al. 2011) while the remaining traits were known to exhibit a more complex genetic architecture. Because heritability estimates are not comparable across data sets and traits, estimated predictive abilities and not accuracies are reported for the three populations and 11 traits. Thus, prediction performance can be compared across methods but not across traits. As in previous studies with experimental data (Heslot et al. 2012), we observed only minor interactions between genetic trait architecture and method with respect to prediction performance (Table 3). For traits assumed to have a low complexity level based on the GWA studies, variable selection did not consistently increase prediction performance, probably due to a combination of small sample sizes and medium trait heritabilities.
Table 3. Predictive abilities obtained with LASSO, the elastic net, BayesB, and RR-BLUP for three experimental data sets.
| Predictive ability |
||||||
|---|---|---|---|---|---|---|
| Data | Trait | n | LASSO | Elastic net | BayesB | RR-BLUP |
| Rice | Flowering time | 359 | 0.46 ± 0.013 | 0.52 ± 0.011 | 0.59 ± 0.006 | 0.59 ± 0.007 |
| Plant height | 383 | 0.71 ± 0.005 | 0.73 ± 0.003 | 0.76 ± 0.002 | 0.76 ± 0.002 | |
| Panicle length | 375 | 0.60 ± 0.004 | 0.61 ± 0.007 | 0.66 ± 0.006 | 0.66 ± 0.006 | |
| Seed length | 377 | 0.75 ± 0.003 | 0.78 ± 0.003 | 0.75 ± 0.003 | 0.75 ± 0.003 | |
| Wheat | Yield | 254 | 0.44 ± 0.010 | 0.43 ± 0.006 | 0.51 ± 0.006 | 0.51 ± 0.005 |
| TKWa | 254 | 0.48 ± 0.010 | 0.48 ± 0.011 | 0.55 ± 0.004 | 0.55 ± 0.004 | |
| Days to heading | 254 | 0.60 ± 0.005 | 0.61 ± 0.008 | 0.66 ± 0.005 | 0.66 ± 0.004 | |
| Arabidopsis | Flowering time | 180 | 0.71 ± 0.008 | 0.65 ± 0.004 | 0.75 ± 0.004 | 0.75 ± 0.004 |
| Plant diameter | 180 | 0.47 ± 0.012 | 0.49 ± 0.016 | 0.51 ± 0.006 | 0.51 ± 0.007 | |
| FRIb | 164 | 0.48 ± 0.020 | 0.36 ± 0.026 | 0.41 ± 0.014 | 0.41 ± 0.013 | |
| Plant width | 176 | 0.46 ± 0.024 | 0.49 ± 0.010 | 0.44 ± 0.009 | 0.43 ± 0.009 | |
Predictive abilities for all 11 traits were estimated using fivefold cross-validation and results were averaged over 10 replications ± SE.
TKW, thousand-kernel weight.
FRI, FRI gene expression.
For the rice data set, it should be noted that the estimated predictive abilities were inflated by the admixture of genetically and phenotypically diverse subpopulations. However, no method was consistently superior when CV was performed only within subpopulations (results not shown). We also investigated the influence of the marker density on prediction performance in the rice data set. Differences in predictive ability between the methods were not significant irrespective of the marker density (Figure S3). The predictive abilities obtained for the wheat data set were higher in this study compared to Poland et al. (2012) because we did not account for the family structure in our CV scheme.
BayesB did not enhance prediction performance compared to RR-BLUP but computational load was much higher. Predictive abilities obtained with LASSO were significantly reduced for 8 of the 11 traits compared to those with RR-BLUP. Recall that LASSO selects less than min(n, p) coefficients (the average numbers of selected markers for all traits are given in Table S2). Thus, for complex traits with a large number of QTL, we might have missed several nonzero coefficients in LASSO, leading to a loss of predictive ability. The inferiority of LASSO compared to RR-BLUP was most pronounced in the rice data that exhibit large long-range LD (Table S1). The elastic net improved prediction accuracies compared to LASSO for all four traits in the rice data set. However, in most cases the elastic net did not outperform RR-BLUP. For FRI gene expression in the Arabidopsis data set, a trait that was assumed to be influenced by a small number of loci, LASSO outperformed BayesB, the elastic net, and RR-BLUP. While LASSO was optimized for prediction using internal CV, in BayesB the fraction of selected markers was a priori the same for all traits within one data set. Hence, variable selection in BayesB was not as stringent as in LASSO and too many variables might have been selected for this trait. There are extensions of BayesB available such as BayesCπ, which addresses this issue by treating the fraction of selected markers as random (Habier et al. 2011) and might lead to a small improvement in predictive ability for traits such as FRI gene expression.
The question arose why predictive abilities observed in experimental data (Table 3) did not differ as much between methods as inferred from computer simulations (Table 2). In the literature, two sources of prediction accuracy have been discussed (Habier et al. 2007, 2010): accuracy due to marker–QTL LD and accuracy due to genetic relationships among individuals. In all experimental data sets, genetic relationships were observed (Figure S1). We hypothesize that in experimental data, a certain level of predictive ability was due to the presence of genetic relationships but this was not the case in computer simulations. With experimental data, all four methods seemed to exploit genetic relationships for prediction by assigning equal genotypic values to relatives sharing a large fraction of the genome (Hofheinz et al. 2012). Thus, for prediction of genomic breeding values, RR-BLUP is a good choice because this method is robust and computationally efficient. If there is prior knowledge that the trait has a sparse genetic architecture, as expected for metabolic data (Riedelsheimer et al. 2012), LASSO may be a good alternative. Moreover, variable selection might enhance the accuracy of predictions for genetic predisposition in humans with extremely large populations of nominally unrelated individuals (de Los Campos et al. 2010).
Influence of LD and trait heritability
To assess the influence of correlations among predictor variables on the normalized L2 error of estimated marker effects, we simulated three additional scenarios, imposing the correlation structure of each of the three experimental data sets on the 2000 SNP markers (simulation procedure 3, Figure S6). Performance of LASSO at different levels of model complexity, determinedness, and extent of LD is visualized in Figure 6. The normalized L2 error was larger throughout all three scenarios exhibiting LD compared to the scenarios with independent markers as depicted in Figure 1. The loss in efficiency was less pronounced in the scenarios where the correlations among markers were conveyed from the Arabidopsis data set, followed by the wheat and the rice data sets. Based on the extent of LD (Table S1), this order was expected. For a given combination of model complexity and determinedness level, the number of observations that were required to estimate one true nonzero coefficient with the same average normalized L2 error compared to the simulations with independent markers was at least doubled in the simulations where the LD structure was adopted from the Arabidopsis data set. LASSO tended to randomly select one variable from a group of correlated variables and hence a loss in efficiency can be expected when the variables exhibit strong correlations. This is especially true for highly selected populations where a large extent of LD is expected due to small effective population sizes. The elastic net was expected to overcome these problems of LASSO by allowing selection of groups of variables. However, based on the normalized L2 error, a consistent advantage of the elastic net could not be observed (Figure S7).
Figure 6.
(A–C) Heat maps for the normalized L2 error of LASSO with correlated predictor variables. The correlation structure of the simulated marker data was superimposed from the three experimental data sets (rice, wheat, and Arabidopsis). The simulations were conducted according to procedure 3 (Figure S6). Each of the 400 scenarios (p = 2000 and h2 = 0.75) was repeated 10 times and results were averaged over replications.
Next, we investigated the joint influence of LD, trait heritability, and genetic trait architecture on the relative prediction performance of BayesB and RR-BLUP with simulation scenarios generated according to simulation procedure 2 (Figure S6). The performance of BayesB and RR-BLUP strongly depended on the values of h2, NQTL, and on the experimental and on the experimental data set used for the simulations (Table 4). For each data set, BayesB tended to perform better for medium to high heritabilities and sparse trait architectures. No superiority of BayesB over RR-BLUP was observed for h2 = 0.1 or NQTL = 100. Remarkable differences in the relative performance of BayesB and RR-BLUP were observed between the different data sets. The advantage of BayesB was most pronounced in the simulations based on the Arabidopsis genotypes, which was expected due to the low extent of LD. For the scenarios where the LD structure was adopted from rice, the superiority of BayesB over RR-BLUP had already disappeared with 10 QTL and h2 = 0.5. Presumably, the large extent of LD did not permit efficient variable selection in the scenario based on the rice data set.
Table 4. Predictive abilities from BayesB and RR-BLUP for simulations with different numbers of QTL and heritabilities.
| Rice (n = 413) |
Wheat (n = 254) |
Arabidopsis (n = 199) |
|||||
|---|---|---|---|---|---|---|---|
| h2 | No. QTL | BayesB | RR-BLUP | BayesB | RR-BLUP | BayesB | RR-BLUP |
| 0.1 | 1 | 0.16 ± 0.035 | 0.17 ± 0.033 | 0.17 ± 0.021 | 0.18 ± 0.020 | 0.08 ± 0.038 | 0.06 ± 0.030 |
| 10 | 0.26 ± 0.025 | 0.26 ± 0.021 | 0.14 ± 0.023 | 0.13 ± 0.018 | 0.07 ± 0.025 | 0.08 ± 0.032 | |
| 100 | 0.30 ± 0.012 | 0.30 ± 0.014 | 0.13 ± 0.034 | 0.22 ± 0.022 | 0.09 ± 0.031 | 0.09 ± 0.038 | |
| 0.5 | 1 | 0.68 ± 0.012 | 0.56 ± 0.032 | 0.68 ± 0.008 | 0.50 ± 0.021 | 0.64 ± 0.027 | 0.27 ± 0.035 |
| 10 | 0.65 ± 0.006 | 0.65 ± 0.008 | 0.50 ± 0.012 | 0.47 ± 0.014 | 0.33 ± 0.024 | 0.27 ± 0.015 | |
| 100 | 0.71 ± 0.004 | 0.71 ± 0.004 | 0.48 ± 0.027 | 0.55 ± 0.023 | 0.29 ± 0.030 | 0.27 ± 0.034 | |
| 0.9 | 1 | 0.94 ± 0.003 | 0.81 ± 0.018 | 0.94 ± 0.002 | 0.75 ± 0.020 | 0.94 ± 0.003 | 0.43 ± 0.039 |
| 10 | 0.94 ± 0.001 | 0.88 ± 0.014 | 0.93 ± 0.003 | 0.74 ± 0.008 | 0.92 ± 0.009 | 0.51 ± 0.029 | |
| 100 | 0.94 ± 0.002 | 0.94 ± 0.002 | 0.78 ± 0.019 | 0.83 ± 0.015 | 0.47 ± 0.030 | 0.50 ± 0.028 | |
The simulations were conducted according to procedure 2 (Figure S6). Predictive abilities were estimated with fivefold cross-validation for BayesB and RR-BLUP based on marker genotypes for 2000 randomly selected markers and 1, 10, and 100 simulated causal mutations. We report the average predictive ability ± SE of 10 replications for each scenario.
When considering the absolute values of the predictive abilities, we found a large influence of the data set on the performance of RR-BLUP. Across almost all scenarios, the largest predictive ability was observed in the scenarios where the LD structure and sample size were conveyed from the rice data set. Within each data set, the predictive ability of RR-BLUP increased with increasing number of QTL and increasing heritability. In contrast, the predictive ability of BayesB was similar across data sets in scenarios with medium to high heritabilities and true models of low complexity. With NQTL = 1 and h2 = 0.5 or 0.9, accuracies close to one were observed for BayesB. Interestingly, the predictive ability of BayesB increased with NQTL for h2 = 0.1 and h2 = 0.5 in the simulations based on the rice data set, while it decreased with NQTL in most of the other scenarios. Presumably, the large extent of LD in the rice data led to a situation where BayesB failed to identify the causal mutation with NQTL = 1. This confirms that both the sparsity of the true model and the absence of strong correlations in the marker matrix are crucial assumptions for successful variable selection.
We also compared the results from computer simulations in Table 4 with experimental results from Table 3 to interpret the absolute values found for predictive abilities in the experimental data sets. For FRI gene expression, a predictive ability of 0.41 was observed for BayesB. Based on results from the GWA study in Atwell et al. (2010), we assumed a sparse architecture for this trait. After considering the predictive abilities in Table 4 for NQTL = 1 and the Arabidopsis genotypes, we would expect a fairly low heritability between 0.1 and 0.5 for this trait. As can be seen from Table 4, BayesB was only better than RR-BLUP for prediction in the scenarios with h2 = 0.5 and h2 = 0.9. These findings explain why BayesB failed to outperform RR-BLUP for FRI gene expression. Although this trait was supposed to have a sparse architecture, no advantage of BayesB was observed with respect to prediction performance, because the trait has presumably a low heritability as expected for gene expression data.
Prediction with whole-genome sequence data
The promise of whole-genome sequence data is that causal variants will be included in the data with high probability (Meuwissen and Goddard 2010). Compared to SNP marker data we expect that p changes dramatically but not p0 and, hence, with whole-genome sequence data we explore experimental designs with a very low determinedness level. The limitations of the different methods to handle high levels of underdeterminedness are demonstrated in Figure 1. Thus, for predictions using whole-genome sequence data, we expect an advantage of variable selection only for traits with a sparse representation relative to the sample size. This hypothesis was recently supported by a first experimental study in Drosophila melanogaster predicting two quantitative traits with whole-genome sequence data (Ober et al. 2012). No differences in the predictive ability of BayesB and RR-BLUP were reported for the two traits.
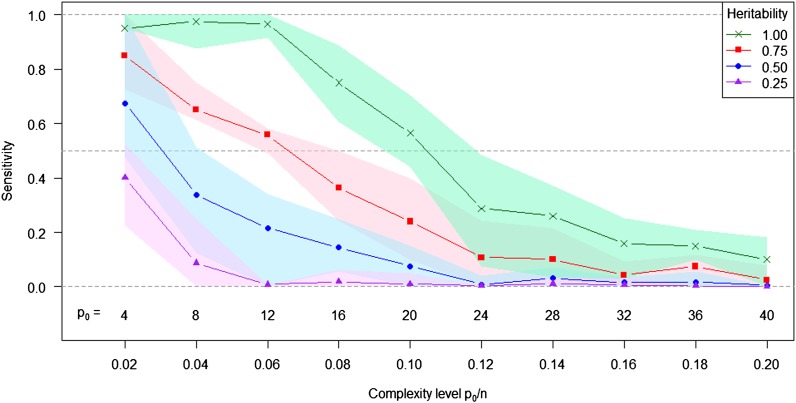
The ability of LASSO to identify the causal mutations in whole-genome sequence data was investigated in a scenario with p = 250,000 independent SNP markers and a small sample size of n = 200 (such as in Atwell et al. 2010; Ober et al. 2012). LASSO was selected because it produces a sparse solution with less than n nonzero elements. This is desirable if we want to pinpoint causal mutations. For different model complexity levels and heritabilities we evaluated the sensitivity of the method, i.e., the empirical conditional probability that a causal mutation was selected. As expected, the sensitivity of detecting the true nonzero coefficients decreased with increasing complexity level and decreasing heritability (Figure 7). With h2 = 0.5 and n = 200, the average sensitivity was >0.5 only for p0 ≤ 4. If more mutations underlay phenotypic variation, LASSO failed to pinpoint these and sensitivity was significantly reduced. However, even with h2 = 1.0, no more than 20 nonzero coefficients could be identified with an average sensitivity >0.5. It should also be noted that a considerable number of false positive nonzero coefficients were reported by LASSO (on average 39.7 and 19.4 for h2 = 1.0 and 0.5, respectively). The excess of false positives partly occurred because we tuned the regularization parameter in LASSO for prediction using CV. Avoiding false positives will require more severe regularization to obtain sparser solutions.
Figure 7.
Sensitivity of LASSO to identify true nonzero coefficients in simulated whole-genome sequence data. The lines indicate the average sensitivity over 10 replications, and the shaded area indicates the range between the 10% and 90% quantiles as a function of the model complexity level and trait heritability. The simulations were conducted according to procedure 1 (Figure S6) with the exception that n = 200 individuals and p = 250,000 markers were used. Heritabilities varied with h2 = 0.25, 0.50, 0.75, and 1.00.
We conclude that prediction based on whole-genome sequence data suffers dramatically from the curse of dimensionality. The question arises of whether we can cope better with the dimensionality of the data by a preselection of markers to increase the determinedness level n/p. We could preselect markers according to biological prior information; e.g., SNPs can be categorized according to plausible candidates based on gene ontology categories (Yu et al. 2012). Ideally, these approaches should remove only true zero coefficients to increase the efficiency of variable selection within the remaining subset of markers. However, currently no empirical results on preselecting markers are available and further research on strategies to preselect SNPs and their effect on prediction is urgently required.
Concluding remarks
In this study we investigated the performance of the statistical methods LASSO, the elastic net, and BayesB with respect to successful variable selection and compared them to RR-BLUP. Our most important finding is that variable selection methods can outperform RR-BLUP with respect to accuracy of marker effects and prediction in high-dimensional data sets, but only if crucial requirements are met. In scenarios where the number of causal mutations is small relative to the sample size, where markers do not exhibit strong LD, and where the trait heritability is high, LASSO, the elastic net, and BayesB can be advantageous over RR-BLUP with respect to prediction performance. However, if these requirements were not met, all three variable selection methods did not enhance prediction performance and marker effects were estimated with low precision. In the case of LASSO, model performance was even considerably derogated when model complexity and the level of underdeterminedness were high. As most traits of agronomic performance can be assumed to be controlled by a large number of segregating QTL with small effects (Schön et al. 2004) and because experimental settings in plant breeding generally suffer from a large extent of LD, medium trait heritabilities, and relatively small sample sizes, we recommend using RR-BLUP, which showed good performance in all experimental settings. The use of a variable selection method such as LASSO can be recommended for experimental settings with large effective population and sample sizes and prior knowledge that the trait is controlled by few genes of large effect (e.g., resistance traits).
In this study, we compared only additive models, recognizing that epistatic networks among loci are prevalent in nature. In first studies, nonlinear methods such as reproducing kernel Hilbert spaces regression or neural networks have been shown to perform better than linear methods in genome-based prediction in wheat (Pérez-Rodríguez et al. 2012). To the best of our knowledge, the breakdown behavior as a function of model complexity and determinedness level has been shown only for linear variable selection methods. Further research on how nonlinear models are influenced by data dimensionality would be highly desirable.
Supplementary Material
Acknowledgments
We thank Dorian Garrick for his help on running GenSel; Nicole Krämer, Ulrike Ober, and Daniel Gianola for discussions and comments; and Michael Lechermann for his help on running the cross-validation with GenSel. We thank the authors in Atwell et al. (2010), Zhao et al. (2011), and Poland et al. (2012) for making their data sets publicly available. The authors thank two anonymous reviewers for their constructive comments. This research was funded by the German Federal Ministry of Education and Research (BMBF) within the AgroClustEr Synbreed—Synergistic plant and animal breeding (FKZ 0315528A).
Footnotes
Communicating editor: F. Zou
Literature Cited
- Albrecht T., Wimmer V., Auinger H.-J., Erbe M., Knaak C., et al. , 2011. Genome-based prediction of testcross values in maize. Theor. Appl. Genet. 123: 339–350. [DOI] [PubMed] [Google Scholar]
- Atwell S., Huang Y., Vilhjálmsson B., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D., and M. Maechler, 2012 Matrix: sparse and dense matrix classes and methods. R Package Version 1.0–6.
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 846: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts C. T., 2008. Network: a package for managing relational data in R. J. Stat. Softw. 24: 1–36.18612375 [Google Scholar]
- Clark S., Hickey J., van der Werf J., 2011. Different models of genetic variation and their effect on genomic evaluation. Genet. Sel. Evol. 43: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Pong-Wong R., Villanueva B., Woolliams J. A., 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Allison D. B., 2010. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat. Rev. Genet. 11: 880–886. [DOI] [PubMed] [Google Scholar]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. L., 2013. Whole genome regression and prediction methods applied to plant and animal breeding. Genetics 193: 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho, D. L., and V. Stodden, 2006 Breakdown point of model selection when the number of variables exceeds the number of observations, pp. 1916–1921 in Proceedings of the International Joint Conference on Neural Networks. Vancouver, British Columbia. [Google Scholar]
- Fernando R., Garrick D., 2009. GenSel—User Manual of Genomic Selection Related Analyses, Iowa State University, Ames, Iowa. [Google Scholar]
- Friedman J., Hastie T., Tibshirani R., 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 30: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Gilmour A., Gogel B., Cullis B., Thompson R., 2009. ASReml User Guide Release 3.0. VSN International, Hemel Hempstead, UK. [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Tetens J., Seefried F., Lichtner P., Thaller G., 2010. The impact of genetic relationship information on genomic breeding values in German Holstein cattle. Genet. Sel. Evol. 42: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, T., R. Tibshirani, and J. Friedman, 2009 The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer Series in Statistics, Ed. 2). Springer-Verlag, Stanford, CA. [Google Scholar]
- Hayes B. J., Pryce J., Chamberlain A. J., Bowman P. J., Goddard M. E., 2010. Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk-fat percentage, and type in Holstein cattle as contrasting model traits. PLoS Genet. 6: e1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslot N., Yang H.-P., Sorrells M. E., Jannink J.-L., 2012. Genomic selection in plant breeding: a comparison of models. Crop Sci. 52: 146–160. [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 6: 226–231. [DOI] [PubMed] [Google Scholar]
- Hofheinz N., Borchardt D., Weissleder K., Frisch M., 2012. Genome-based prediction of test cross performance in two subsequent breeding cycles. Theor. Appl. Genet. 125: 1639–1645. [DOI] [PubMed] [Google Scholar]
- Ishwaran, H., and J. S. Rao, 2011 Generalized ridge regression: geometry and computational solutions when p is larger than n. Technical report. Cleveland Clinic and University of Miami, Miami. [Google Scholar]
- Legarra A., Robert-Granie C., Manfredi E., Elsen J., 2008. Performance of genomic selection in mice. Genetics 180: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Goddard M. E., 2010. Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U., Ayroles J. F., Stone E. A., Richards S., Zhu D., et al. , 2012. Using whole genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 8: e1002685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe M. S., 2004. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford University Press, New York. [Google Scholar]
- Pérez-Rodríguez, P., D. Gianola, J. M. González-Camacho, J. Crossa, Y. Manès et al., 2012 Comparison between linear and non-parametric regression models for genome-enabled prediction in wheat. G3 2: 1595–1605. [DOI] [PMC free article] [PubMed]
- Poland J., Endelman J., Dawson J., Rutkoski J., Wu S., et al. , 2012. Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genet. 5: 103–113. [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A. R., et al. , 2007. Plink: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team , 2012. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Riedelsheimer C., Technow F., Melchinger A., 2012. Comparison of whole-genome prediction models for traits with contrasting genetic architecture in a diversity panel of maize inbred lines. BMC Genomics 13: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön C.-C., Utz H. F., Groh S., Truberg B., Openshaw S., et al. , 2004. Quantitative trait locus mapping based on resampling in a vast maize testcross experiment and its relevance to quantitative genetics for complex traits. Genetics 167: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodden, V., 2006 Model selection when the number of variables exceeds the number of observations. Ph.D. Thesis, Department of Statistics, Stanford University, Stanford, CA. [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Wimmer V., Albrecht T., Auinger H.-J., Schön C.-C., 2012. synbreed: a framework for the analysis of genomic prediction data using R. Bioinformatics 28: 2086–2087. [DOI] [PubMed] [Google Scholar]
- Yu J., Li X., Zhu C., Yeh C. T., Wu W., et al. , 2012. Genic and non-genic contributions to natural variation of quantitative traits in maize. Genome Res. 12: 2436–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liu J., Ding X., Bijma P., De Koning D.-J., et al. , 2010. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS ONE 5: 0012648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., C.-W. Tung, G. C. Eizenga, M. H. Wright, M. L. Ali et al., 2011 Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2: 467. [DOI] [PMC free article] [PubMed]
- Zhong S., Dekkers J. C. M., Fernando R. L., Jannink J.-L., 2009. Factors affecting accuracy from genomic selection in populations derived from multiple inbred lines: a barley case study. Genetics 182: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67: 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.