Abstract
The cluster of human neuronal nicotinic receptor genes (CHRNA5/A3/B4) (15q25.1) has been associated with a variety of smoking and drug-related behaviors, as well as risk for lung cancer. CHRNA3/B4 intergenic single nucleotide polymorphisms (SNPs) rs1948 and rs8023462 have been associated with early initiation of alcohol and tobacco use, and rs6495309 has been associated with nicotine dependence and risk for lung cancer. An in vitro luciferase expression assay was used to determine whether these SNPs and surrounding sequences contribute to differences in gene expression using cell lines either expressing proteins characteristic of neuronal tissue or derived from lung cancers. Electrophoretic mobility shift assays (EMSAs) were performed to investigate whether nuclear proteins from these cell lines bind SNP alleles differentially. Results from expression assays were dependent on cell culture type and haplotype. EMSAs indicated that rs8023462 and rs6495309 bind nuclear proteins in an allele-specific way. Additionally, GATA transcription factors appeared to bind rs8023462 only when the minor/risk allele was present. Much work has been done to describe the rat Chrnb4/a3 intergenic region, but few studies have examined the human intergenic region effects on expression; therefore, these studies greatly aid human genetic research as it relates to observed nicotine phenotypes, lung cancer risk and potential underlying genetic mechanisms. Data from these experiments support the hypothesis that SNPs associated with human addiction-related phenotypes and lung cancer risk can affect gene expression, and are potential therapeutic targets. Additionally, this is the first evidence that rs8023462 interacts with GATA transcription factors to influence gene expression.
Keywords: luciferase, nicotinic receptor, EMSA, rs1948, rs6495309, rs8023462
1 Introduction
Nicotine dependence (ND) is the leading cause of preventable death in the world (Albuquerque et al., 2009; CDC, 2010) and is the major risk factor for chronic obstructive pulmonary disease (COPD) and lung cancer (Young et al., 2008). Genome Wide Association Studies (GWAS) have implicated the nicotinic acetylcholine receptor (nAChR) cluster of genes (CHRNA5/CHRNA3/CHRNB4) in risk for ND (Berrettini et al., 2008; Bierut et al., 2008; Saccone et al., 2009; Saccone et al., 2007; Thorgeirsson et al., 2008; Weiss et al., 2008), COPD and lung cancer (Amos et al., 2008; Hung et al., 2008; Liu et al., 2008; Thorgeirsson et al., 2008; Wang et al., 2009c). Recent meta-analyses sustain these associations (Bierut et al., 2010; Consortium, 2010; Thorgeirsson et al., 2010; Timofeeva et al., 2012). The most well studied single nucleotide polymorphism (SNP) within the cluster has been rs16969968, a missense variant that results in a change from aspartic acid (D) to asparagine (N) in the nAChR α5 subunit and decreased response to agonist when the risk (398N) allele is present (Berrettini and Doyle, 2012; Bierut et al., 2007; Bierut et al., 2008; Saccone et al., 2007). Yet, there are other known genetic influences on ND and lung cancer risk within this region, and variants in the cluster have a complex linkage disequilibrium (LD) structure, complicating identification of other functional polymorphism(s) (Pergadia et al., 2009).
Additional molecular experiments are necessary to understand other functional SNPs within the cluster (Berrettini and Doyle, 2012). At least one locus associated with increased risk for ND and lung cancer (locus 3 tagged by rs588765), is also associated with significantly increased α5 mRNA levels in frontal cortex of individuals harboring the risk allele (Wang et al., 2009c). However, many CHRNA3 promoter or intergenic variants investigated thus far (See Figure 1 for extent of investigation based on three previous studies) have not been shown to directly impact transcription (Doyle et al., 2011; Fornasari et al., 1997). A notable exception is the intergenic SNP rs6495309, which has been shown to influence luciferase expression in lung cancer cell lines (Wu et al., 2009). The Oct-1 transcription factor (TF) has been shown to bind the rs6495309 allele A (minor/protective) in the H1299 lung cancer cell line better than the G allele and decrease expression of a reporter gene in this and other cell lines (Wu et al., 2009). Thus, a biological mechanism for increased risk of early initiation of nicotine use and/or ND as well as lung cancer may involve changes in transcription and expression of the cluster genes influenced by additional genetic variants that have yet to be studied.
Figure 1. Constructs (A) and previous human expression studies (B).
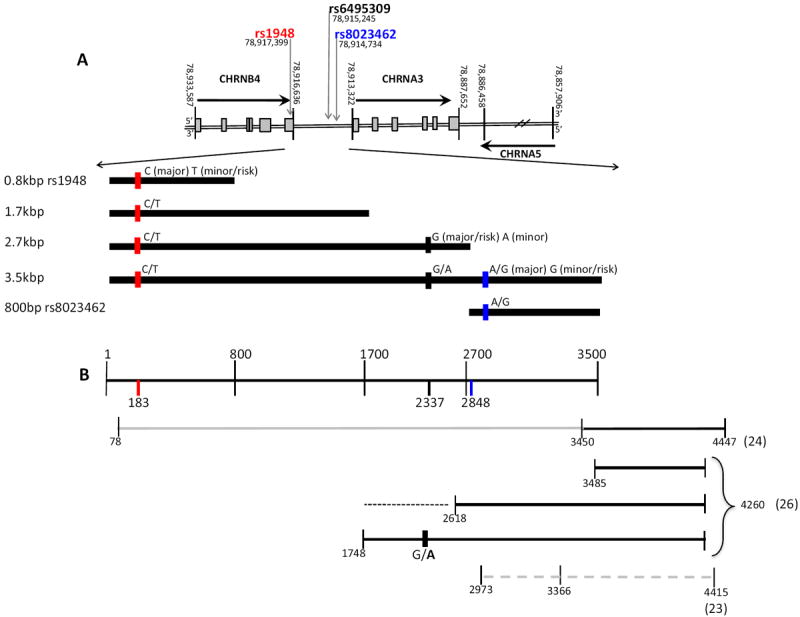
A. The CHRNA5/A3/B4 cluster with the reverse strand (coding strand for CHRNA3 and CHRNB4) of human chromosome 15 is shown on top, 5’ to 3’. Positions indicated from the Feb 2009 GRCh37/hg19 NCBI assembly database delineate borders of each gene and identify each SNP position. Grey boxes indicate exons. CHRNA5 is not to scale (indicated by hash marks) and exons are not identified. Bold arrows indicate direction of transcription. The five different length constructs, inserted upstream of the SV40 promoter in the promoter-reporter pGL3 luciferase expression vector (not shown), as well as each possible allele for each SNP, is shown below the cluster diagram.
B. Top line, indicating the 3.5kbp construct used here, describes nucleotide base pairs from relative position 1 to 3500. Numbers across the top denote nucleotide bps in the 3.5kbp construct, bottom numbers indicate relative SNP locations. Constructs shown below this metric are from 3 independent studies using luciferase reporter assays and human sequences upstream of CHRNA3 and inclusive of the promoter. Solid, dark lines indicate a significant effect of the sequence when placed upstream of the luciferase reporter (Fornasari et al., 1997; Wu et al., 2009). Grey solid line indicates no effect of fragment (no promoter used in construct) (Fornasari et al., 1997). Dotted black line indicates possible repressive region, and bold A at rs6495309 indicates repressed expression compared to G (Wu et al., 2009). Dashed grey line indicates no effect (inclusive of CHRNA3 promoter, numerous variants investigated) on expression (Doyle et al., 2011).
Our human genetic studies identified significant associations between age of initiation for alcohol and tobacco use and SNPs rs1948 and rs8023462 (minor/risk alleles, T and G respectively) in the intergenic region between CHRNA3 and CHRNB4 (Schlaepfer et al., 2008). These SNPs are in high LD with one another (r2>0.74 in European American samples; (Stephens et al., 2012)), but not with any of the other well-studied loci for ND. A recent meta-analysis also found that the SNP rs6495309 (in LD with locus 2 tagged by rs578776) is associated with early age of onset of regular smoking (Stephens et al, in preparation). Early age of initiation for both alcohol (Guerri and Pascual, 2010; Zucker et al., 2008) and tobacco use (Hu et al., 2006) is a strong predictor of future abuse and dependence, so understanding genetic risk factors for early drinking and smoking is important. Additionally, SNPs involved in early initiation may play a predisposing role by resulting in differentially susceptible brain neurochemistry that varies by allele. Thus, these initial studies are focused on possible allele-specific baseline differences in expression.
This paper investigates whether three SNPs, rs1948, rs6495309 and rs8023462, can influence gene expression in cell lines expressing neuronal proteins, or in small cell lung cancer (SCLC) cell lines, and whether nuclear proteins interact differentially with alleles of each SNP. Because these SNPs reside in a noncoding region upstream of CHRNA3 promoter and inclusive of the 3’UTR of CHRNB4, we sought to determine whether the minor and major alleles differentially affect gene expression in vitro when placed upstream of an SV40 promoter and luciferase gene. Use of a heterologous expression system was preferable in order to assess any potential repressive or enhancing effects of the region under investigation. Previous work by Fornasari et al. indicated that better expression can be achieved using the SV40 promoter than the CHRNA3 promoter (Doyle et al., 2011; Fornasari et al., 1997), and studies using the rat intergenic region have successfully employed similar heterologous constructs (McDonough and Deneris, 1997; McDonough et al., 2000). Luciferase promoter/reporter constructs were transfected into two cell lines expressing neuronal proteins (neuronal-like cell lines), human neuroblastoma SH-SY5Y and rat pheochromocytoma PC12 cells, under two different treatment conditions, untreated and differentiated, and two SCLC cells lines, H446 and H69. Gene expression from transfected cells was assessed at 96 hours (neuronal-like cells) or 48 hours (lung cancer cells) post-transfection. Only haplotypes known to exist in the current HapMap dataset were reported. Additionally, electrophoretic mobility shift assays (EMSAs) were performed for each SNP allele to determine if nuclear proteins interact, and whether any noted interactions between nuclear proteins and SNP allele might correlate with expression data.
2 Results
2.1 Expression results from luciferase assays
Supplementary Table 1 shows Ns for each of the following transfections.
2.1.1 Neuronal-like cell lines
In the neuronal-like cell General Linear Model (GLM), all factors assessed, cell line, treatment condition and length had main effects (p≤0.001). Given these findings and biological justification, separate analyses were conducted within each cell line, treatment condition and construct length group. Follow-up analyses were conducted as described in section 4.6, statistical analysis.
2.1.1.1 SH-SY5Y untreated cells
Results for the untreated SH-SY5Y cells are shown in Figure 2. For 0.8kbp rs1948 constructs, the minor T allele showed decreased (nearing significance) gene expression in these cells compared to C (t(1,62)=1.9, p=0.06). In the 3.5kbp construct-transfected group a main effect of plasmid haplotype was detected (F(4,77)=11.48, p<0.001), and follow-up Tukey post-hoc analysis within the 3.5kbp group comparing all haplotypes indicated that 3.5kbp_CGG reduced expression compared to all other constructs (CAA, CGA, TGG each p<0.001, and CAG p=0.004). There were no differences in the 1.7kbp rs1948, 2.7kb and 800bp rs8023462 constructs.
Figure 2. Luciferase expression results from transfected SH-SY5Y cells.
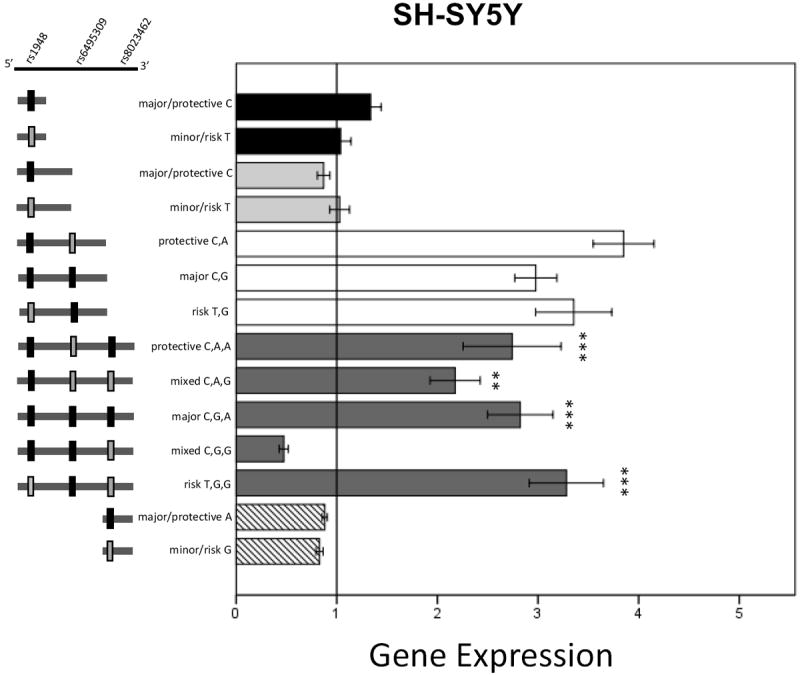
Data are mean +/- SEM. ** p<0.01,*** p<0.001. X-axis indicates luciferase gene expression normalized to the pGL3 empty vector (1.0). All comparisons are within length groups. Significant differences indicated are from post hoc Tukey analysis of the 3.5kbp constructs. Asterisks indicate significantly different from 3.5kbp_CGG.
2.1.1.2 SH-SY5Y differentiated cells
Results for differentiated SH-SY5Y cells are shown in Figure 3. Expression was increased for T compared to C for the 0.8kbp rs1948 length constructs (t(1,37)=-3.7, p=0.001). The short 800 bp rs8023462 constructs were also significantly different (F(1,66)=2.1, p=0.043), with the G allele decreasing expression. Analysis of expression results from 3.5kbp construct transfected cells indicated a main effect of plasmid haplotype, (F(4,72)=2.68, p=0.039). Follow-up Tukey post-hoc analysis within the 3.5kbp group comparing all haplotypes indicated that 3.5kbp_CGA decreased expression compared to TGG (nearing significance, p=0.055). There were no differences in the 1.7kbp rs1948 or 2.7kb constructs.
Figure 3. Luciferase expression results from transfected SH-SY5Y Differentiated cells.
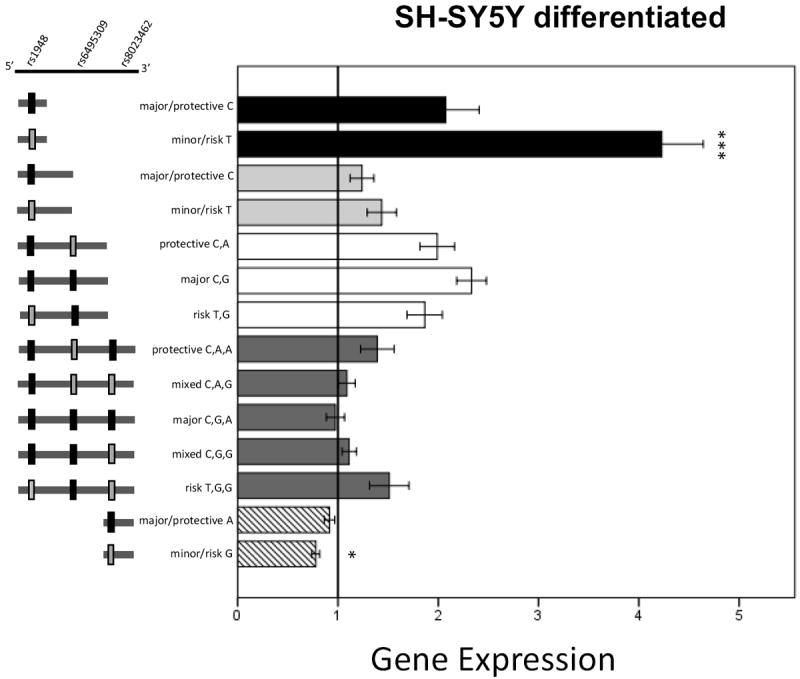
Data are mean +/- SEM. * p<0.05, *** p<0.001. 0.8kbp_T expression is significantly increased compared to C. 800bp rs8023462_G expression is significantly reduced compared to A.
2.1.1.3 PC12 untreated cells
Luciferase expression results for untreated PC12 cells are shown in Figure 4. Expression was significantly increased for 1.7kbp rs1948 T versus C (t(1,25)=-2.5, p=0.022). For the 3.5kbp length construct group a significant main effect of plasmid haplotype was detected (F(4,75)=5.85, p<0.001). Tukey post-hoc analysis within the 3.5kbp group comparing all 5 different construct haplotypes indicated that 3.5kbp_CGG reduced expression compared to both CAG (p=0.013) and TGG (p<0.001) and 3.5kbp_CAA reduced expression compared to TGG (p=0.048). There were no differences in the 0.8kbp rs1948, 2.7kb and 800bp rs8023462 constructs.
Figure 4. Luciferase expression results from transfected PC12 cells.
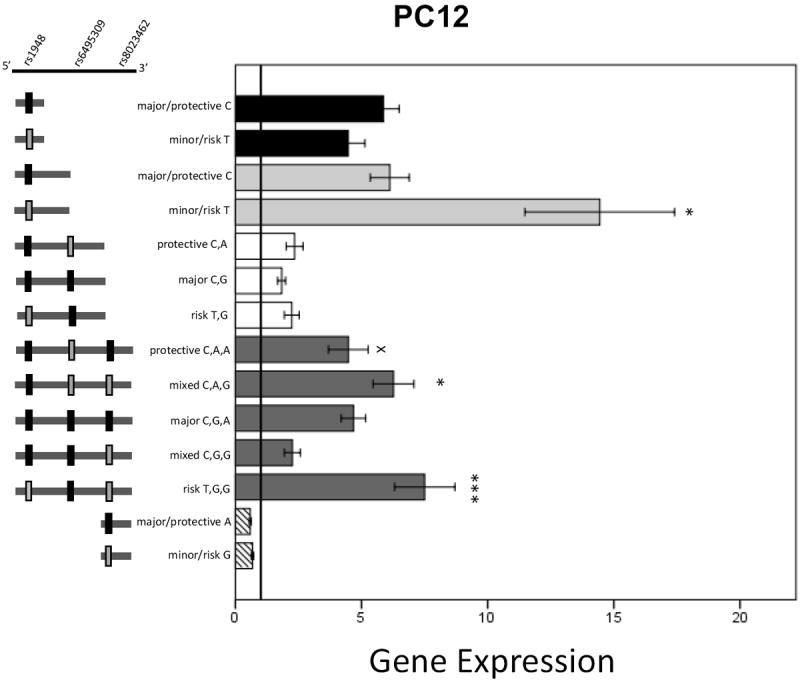
Data are mean +/- SEM. x or * p<0.05, *** p<0.001. 1.7kbp_T expression is significantly increased compared to 1.7kbp_C. In the 3.5kbp group, asterisks indicate significantly different from 3.5kbp_CGG, and X symbol indicates significantly different from 3.5kbp_TGG in the post hoc Tukey analysis for 3.5kbp length constructs.
2.1.1.4 PC12 differentiated cells
Results for the NGF-treated, differentiated PC12 cells are presented in Figure 5. A significant effect of the rs1948 allelic variants on luciferase expression was noted for both 0.8kbp and 1.7kbp constructs (t(1,27)=-4.1, p<0.001 and t(1,39)=4.4, p<0.001, respectively); the minor allele (T) showed increased gene expression in the 0.8kbp constructs, but decreased expression in the 1.7kbp constructs when compared to major (C) allele constructs. In the 3.5kbp construct-transfected group, a main effect of plasmid haplotype was detected (F(4,74)=7.65, p<0.001). Follow-up Tukey post-hoc analysis comparing all 5 haplotypes indicated that 3.5kbp_CGG reduced expression compared to all other constructs (CAA (p<0.001), CAG (p=0.003), CGA (p=0.008), and TGG (p=0.002)). There were no differences in 2.7kb or 800bp rs8023462 constructs.
Figure 5. Luciferase expression results from transfected PC12 Differentiated cells.
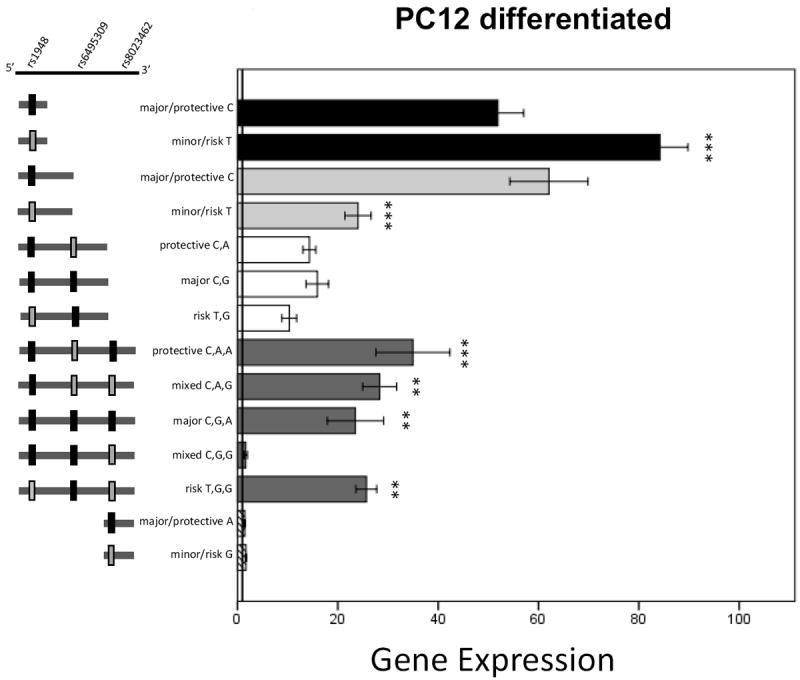
Data are mean +/- SEM. * p<0.05, xx or ** p<0.01, *** p<0.001. Note that the X-axis shows increments of 20 and ends at 100. 0.8kbp_T expression is significantly increased compared to 0.8kbp_C. 1.7kbp_T expression is significantly decreased compared to 1.7kbp_C. In the 3.5kbp group, asterisks indicate significantly increased from 3.5kbp_CGG in the post hoc Tukey analysis of the 3.5kbp constructs.
2.1.2 Lung cancer cell lines
In the lung cancer cell GLM, main effects of cell line and length (p<0.001) were detected. Given these findings and biological justification, separate analyses were conducted within each cell line, treatment condition and construct length group. Follow-up analyses were conducted as described in section 4.6, statistical analysis.
2.1.2.1 H446 cells
Both the 0.8kbp and the 1.7kbp length rs1948 constructs were significantly different (t(1,14)=-4.05, p=0.001 and t(1,11)=25.29, p<0.001, respectively), with the T allele (minor/risk) increasing expression relative to the C (major/protective) allele in the 0.8kbp experiments, but reducing expression in the 1.7kbp constructs. In the 3.5kbp length group a main effect of plasmid haplotype (F(4,39)=15.81, p<0.001) was detected and follow-up Tukey post-hoc analysis indicated that 3.5kbp_CGG had decreased expression compared to all other haplotypes (all p<0.001 except CAG p=0.020). Additionally, 3.5kbp_GAG had decreased expression compared to 3.5kbp_CGA (p=0.003). No other significant differences were noted in the 2.7kbp or 800bp rs8023462 groups, either between alleles or among haplotypes.
2.1.2.2 H69 cells
Luciferase gene expression results for the H69 cells are shown in Figure 7. In the 3.5kbp length group a main effect of plasmid haplotype (F(4,59)=3.95, p=0.007) was noted. Tukey post-hoc analysis within the 3.5kbp group comparing all 5 different haplotypes indicated that 3.5kbp_CGG had significantly reduced expression compared to TGG (p=0.004) and CGA (p=0.035). No other significant differences were noted in other length groups between alleles or among haplotypes.
Figure 7. Luciferase expression results from transfected H69 cells.
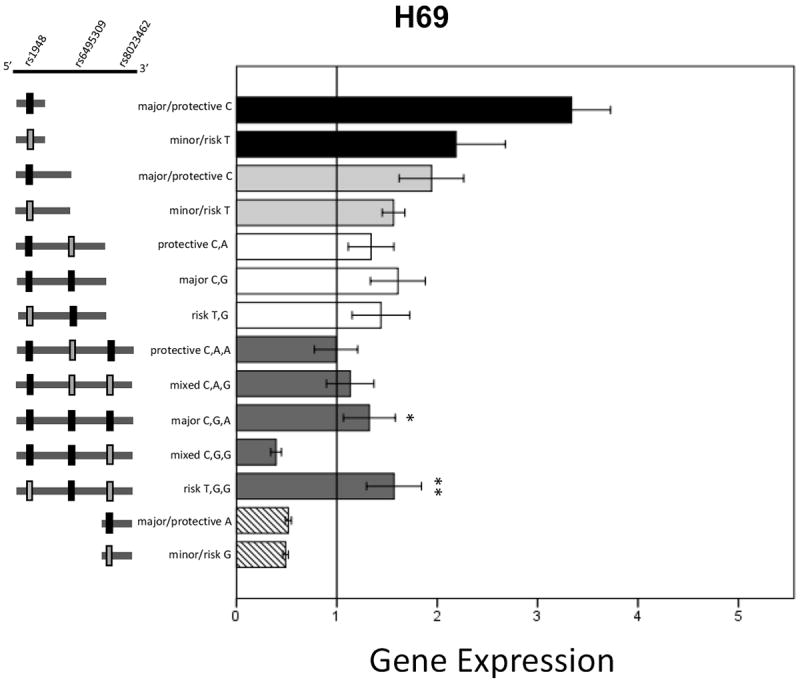
Data are mean +/- SEM. * p<0.05, ** p<0.01. Significant differences indicated are from post hoc Tukey analysis of the 3.5kbp constructs. Asterisks indicate significantly different from 3.5kbp_CGG.
2.2 EMSA Results
Both EMSAs for rs6495309 (Figure 8) and rs8023462 (Figure 9) had specific, shifted bands, as well as allele-dependent band differences using both neuronal-like and lung cancer cell nuclear extracts. EMSAs for rs1948 revealed no specific shifts.
Figure 8. rs6495309 EMSA: Biotin-labeled rs6495309_A shifts more nuclear extract than G.
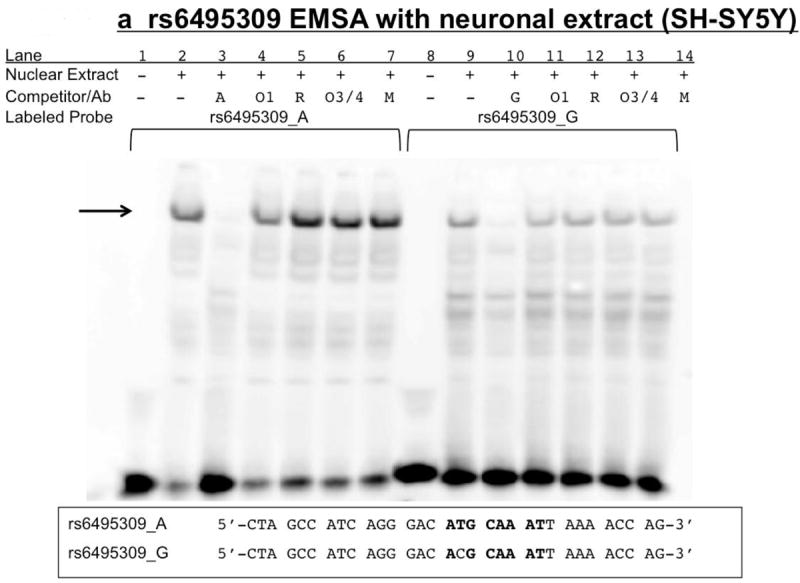
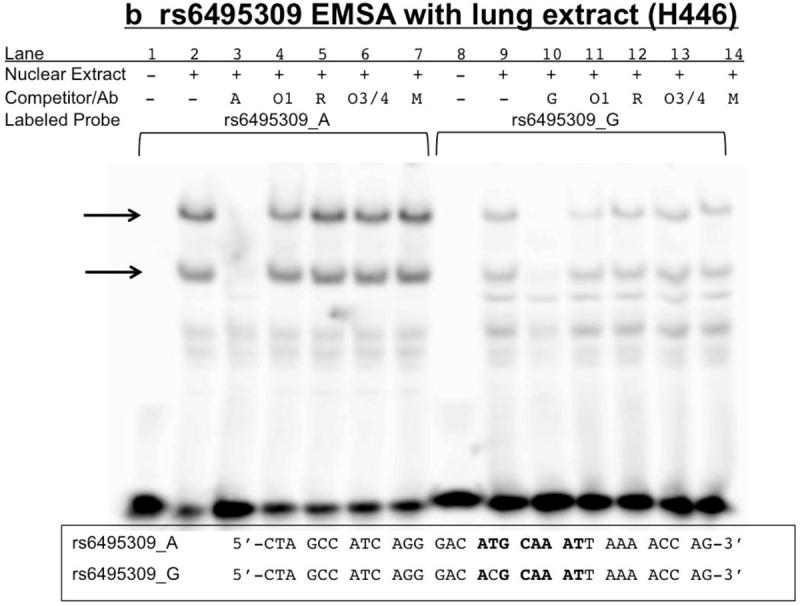
Nuclear extract (+) is either 15μg SH-SY5Y (8A) or H446 (8B) or no extract (-). Competitor/Ab indicates either none (-) or 100x molar excess, double-stranded, unlabeled rs6495309_A (A) or rs6495309_G (G), or antibodies specific to oct-1 (O1), oct-3/4 (O3/4) and their respective control antibodies (R=rabbit, M=mouse). Labeled probe is either 3’ biotin-labeled, double-stranded rs6495309_A or G. Arrows indicate specific bands of unknown protein composition (1 band for SH-SY5Y (8A) and 2 bands for H446 (8b). Sequences provided are for both the biotin labeled and unlabeled probes. Bold letters indicate putative Oct transcription factor binding site.
Figure 9. rs8023462 EMSAs: Biotin-labeled rs8023462_G yields a competitive supershift with antibodies to both GATA-4 and GATA-2/3 using SH-SY5Y nuclear extract.
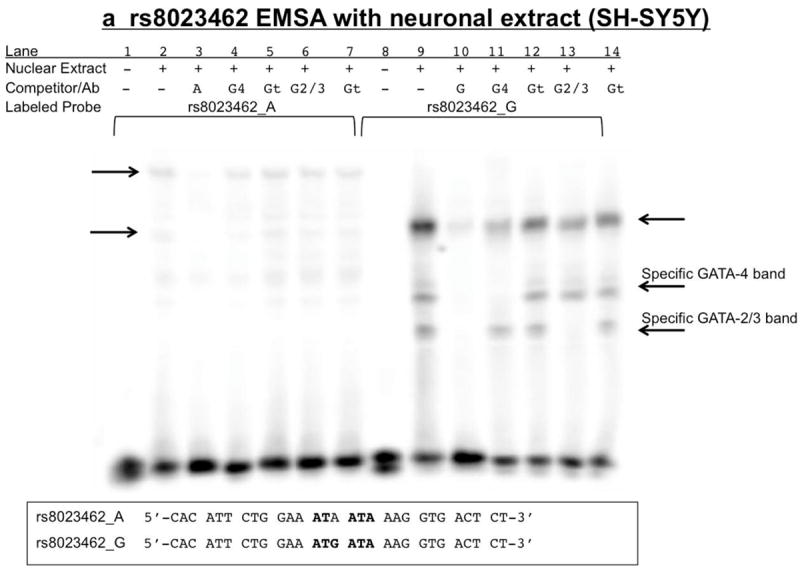
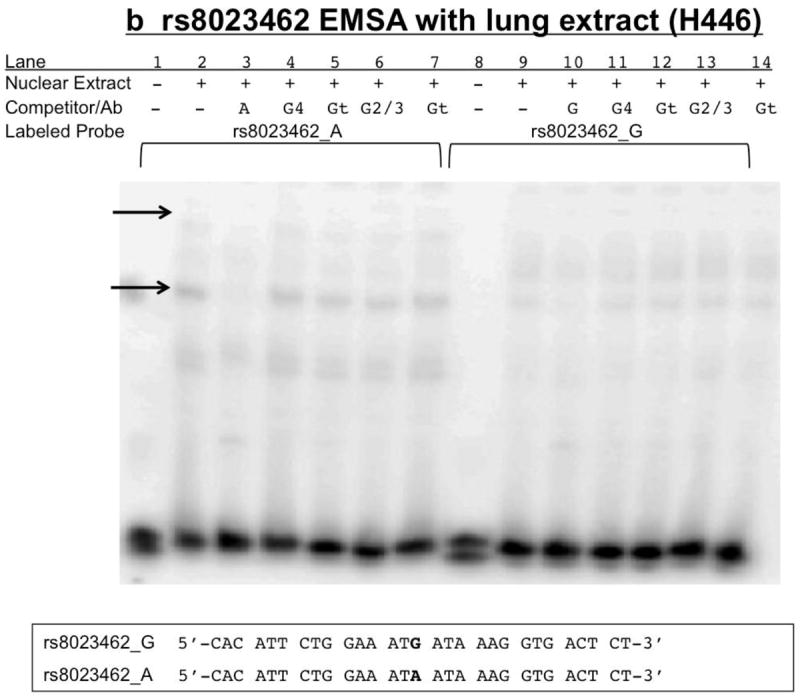
Nuclear extract (+) is either 15μg SH-SY5Y (8A) or H446 (8B) or no extract (-). Competitor/Ab indicates either none (-) or 100x molar excess, double-stranded, unlabeled rs8023462_G (G) or rs8023462_A (A), or antibodies specific to GATA-4 (G4), GATA 2/3 (G2/3) and the non-specific control antibody (Gt=goat). Labeled probe is either 3’ biotin-labeled, double-stranded rs8023462_G or A. Arrows indicate specific bands. Sequences provided are for both the biotin labeled and unlabeled probes. Bold letters indicate putative GATA transcription factor binding site. 9a) The upper right arrow as well as the upper left and lower left arrows indicate specific bands of unknown protein(s) binding specifically to the oligonucleotides. The rs8023462_A allele may bind a different protein (upper left arrow) than the rs8023462_G allele. 9b) GATA factors 2-4 do not appear to be present in the H446 extract.
2.2.1 rs6495309
2.2.1.1 Neuronal-like cell nuclear extracts and rs6495309 probes
For EMSAs using nuclear extract from SH-SY5Y untreated (Figure 8a), or differentiated cells, and EMSAs using untreated or differentiated PC12 cells (not shown), the rs6495309 A allele caused a more intense, shifted, specific band than the G allele, indicating a stronger/better association with nuclear protein when the A (minor/protective) allele is present than when the G (major/risk) allele is present (Figure 8a). This is supported by the lack of full competition when the unlabeled G oligonucleotide was used as a competitor, compared to the unlabeled A (100x molar excess) (Supplemental Figure 1a). The SH-SY5Y undifferentiated cell line was chosen for supershift experiments, as nuclear extracts from all neuronal-like cell lines tested yielded the same specific band, but EMSAs using the SH-SY5Y nuclear extract had the most intense band. Addition of antibodies specific for oct-1 or oct-3/4 did not result in a supershift (Figure 8a), whether added in 1 or 2 μL amounts, before or after probe and extract incubation. Addition of an unlabeled consensus oct-1 competitor at 450x molar excess also could not out-compete the shifted bands (Supplementary Figure 1a).
2.2.1.2 Lung cancer cell line nuclear extracts and rs6495309 probes
When combined with nuclear extract from either of the lung cancer cell lines tested, H446 (Figure 8b) or H69 (Supplemental Figure 1b), the biotin-labeled probes for rs6495309 bound specifically with nuclear protein(s), yielding two major bands. The rs6495309_A allele caused more intense, shifted, specific bands than the G allele, indicating a stronger/better association with nuclear protein(s) when the A allele was present than when the G allele was present. This was further demonstrated by the lack of full competition when the unlabeled G oligonucleotide was used as a competitor, compared to the unlabeled A (100x molar excess) (Supplementary Figure 1b). H446 nuclear extract was used for supershift experiments, since this cell line was investigated in work by Wu et al., which indicated Oct-1 binds rs6495309 in lung cancer cell lines (Wu et al., 2009), Addition of an antibody specific for Oct-1 did not result in a supershift (Figure 8b), whether added in 1 or 2 μL amounts, before or after probe and extract incubation.
2.2.2 rs8023462
2.2.2.1 Neuronal-like cell nuclear extracts and rs8023462 probes
For SH-SY5Y untreated (Figure 9A and Supplementary Figure 2A) and differentiated (Supplementary Figure 2B) cell nuclear extracts, rs8023462 probes yielded unique, allele-specific shifted bands, the biotin-labeled G allele causing a specific band overlaying a non-specific band with three secondary specific bands, and the A allele causing a more retarded, specific band in addition to the band overlaying a non-specific band. When the unlabeled A oligonucleotide was used as a 100x competitor for G, or when unlabeled G (100x) was used to compete with A, specific bands were slightly reduced (supplementary Figure 2A and 2B), but by no means eliminated. GATA antibodies caused competitive supershifts for only rs8023462_G (minor/risk), illustrated by disappearance of specific bands upon addition of the antibody (Lanes 11 and 13 Figure 9A, SH-SY5Y nuclear extract used). Biotin-labeled probes for rs8023462 did not cause a significant, specific shift in EMSAs using nuclear extracts from either untreated or differentiated PC12 cells.
2.2.2.2 Lung cancer cell nuclear extracts and rs8023462 probes
Nuclear extract from H69 (not shown) and H446 (Figure 9B and Supplementary Figure 2C) bound probes for rs8023462, and yielded one specific, shifted band. Band intensity was allele-dependent. The biotin-labeled A allele caused a more intense shifted band than the G allele, and competition with the alternate allele at 100x did not eliminate the specific band. (Supplementary Figure 2C). When H446 was tested for a supershift with GATA antibodies, no supershift was detected (Figure 9B).
2.2.3 rs1948
Biotin-labeled probes for rs1948, whether of the major C or minor/risk T variant, did not cause a significant specific shift in EMSAs using nuclear extracts from any of the neuronal-like or lung cancer cell lines tested, even after multiple attempts with various combinations of kit reagents and probe/extract concentrations to optimize the reaction.
3 Discussion
Data from the luciferase expression assays have shown that human CHRNB4/A3 SNPs associated with drug phenotypes contribute to differences in gene expression in vitro using both cells that express neuron-specific proteins and those from lung cancer specimens. The main effect of cell line noted in the overall GLM for neuronal-like cells may be due to a number of differences between PC12 and SH-SY5Y cells including species, tumor type and tissue. PC12 cells had enhanced expression when differentiated. Maturation-dependent expression changes may be involved in gene expression differences between untreated PC12 and differentiated cells. Results are consistent with the knowledge that expression of nicotinic receptor subunits is controlled within particular cell types during different stages of development or differentiation (Gotti et al., 2006), and provides some evidence that the human CHRNB4/A3 intergenic region may be regulated by developmental changes.
The effects of length of construct on expression are expected, given previous experiments indicating that multiple enhancing/repressive regions exist in the both the human CHRNA3 promoter (Battaglioli et al., 1998; Benfante et al., 2007; Doyle et al., 2011; Fornasari et al., 1997; Wu et al., 2009) and rat Chrnb4/a3 intergenic regions (Boyd, 1996; Improgo et al., 2010). A putative repressive region in the 2.7kbp construct but not in the 1.7kbp construct, previously identified by Wu et al. using lung cancer cell lines (Wu et al., 2009), does not appear to be globally repressive (Figures 2-7). The lack of effect on expression of rs8023462 in the smaller construct, compared to the 3.5kbp construct, could be due to required additional flanking sequences that may influence DNA structure or key regulatory sequences (Nakshatri et al., 1995). Alternately, the distance of SNP from the promoter may be a factor modifying transcription.
The constructs most routinely impacted by cell line and treatment condition appear to be the rs1948 0.8kbp and 1.7kbp constructs. Expression of the longer constructs in PC12 cells appears sensitive to effects of differentiation (Figures 4 and 5). The shorter rs1948 constructs had increased expression for the T allele versus the C allele in both differentiated neuronal-like cell lines (Figures 3 and 5). In the context of interpreting results from luciferase assays, it is important to recognize this approach does not establish the mechanism responsible for differences in gene expression. Given the negative EMSA results for rs1948, it appears that either a nuclear protein is not involved, or the assay is simply unable to detect nuclear protein binding which may require interactions with other DNA binding proteins not present in the short sequence of the oligos used for EMSAs. Though methylation may be an interesting epigenetic mechanism to investigate in regards to rs1948, to the best of our knowledge, there is not any evidence that epigenetic mechanisms can be effectively studied using transiently transfected constructs. Results reported here provide a definite platform from which to pursue additional questions, and support a functional contribution of rs1948.
EMSAs reveal, for the first time to our knowledge, that the minor./risk rs8023462_G allele (but not A) binds GATA transcription factors in SH-SY5Y nuclear extracts (Figure 9a). GATA TFs were chosen for investigation using the Transcription Element Search System (TESS) (URL: http://www.cbil.upenn.edu/tess). This family of TFs coordinates complex developmental processes (Bresnick et al., 2010), such as those involved in regulating receptor expression during critical windows in development, and GATA-4 has been identified through GWAS as a TF associated with alcohol dependence (Treutlein et al., 2009). Importantly, the supershift EMSA shown here in Figure 9a indicates that rs8023462_G binds both GATA 4 and GATA 2 or 3, while the A allele does not. It is interesting to note that 3.5kbp_CGG routinely has significantly lower expression in all cell lines tested in the luciferase assays, indicating that perhaps the context of the surrounding sequence can influence the presumably repressive effect of GATA binding at rs8023462_G.
As previously reported by Wu et al., the A allele of rs6495309 binds nuclear protein and causes a more intense band on EMSAs. However, we did not find support for Oct-1 binding rs6495309, particularly when using H446 (figure 8b), a cell line investigated by Wu et al. in their luciferase expression studies, though not in their EMSA studies, which instead used nuclear extract from H1299 cells. Perhaps H1299 but not H446 nuclear extract contains Oct-1. Indeed, the binding of another protein or proteins is indicated in Figure 8b using H446 extracts. Using TESS, nearly all POU TFs are suggested to bind this allele-containing region. Neuronal and lung cancer cell lines have high levels of N-Oct-3 (brain-2) (Schreiber et al., 1992), and so we investigated this TF as well and determined Oct-3 and Oct-4 do not appear to bind rs6495309 either. In the rat intergenic region, the POU family TF SCIP/Tst-1/Oct-6 has been identified as a cell-type specific regulator of transcription (Fyodorov and Deneris, 1996; Improgo et al., 2010; Yang et al., 1994). POU family TFs Brn-3a, b and c have also been shown to interact with the rat Chrna3 and Chrna2 promoters and to either activate or repress transcription, depending on surrounding sequence and subtype (Milton et al., 1996). It remains to be determined if these other POU TFs bind rs6495309.
While the clustering of CHRNA5/A3/B4 genes is well conserved across species (Battaglioli et al., 1998; Boulter et al., 1990; Couturier et al., 1990; Fornasari et al., 1997), only 7% appears to be conserved between rodent and human (Evolutionary Conserved Regions (ECR) browser) (Ovcharenko et al., 2004). Transfections using the rat PC12 cell line (Figures 4 and 5) were employed to begin to assess cross-species regulatory conservation in the context of previous findings for the rat. To begin to develop a model for the human sequence, Figure 10 provides a very preliminary model for the EMSA and gene expression effects noted here.
Figure 10. Model for SNP effects on gene expression based on EMSA results.
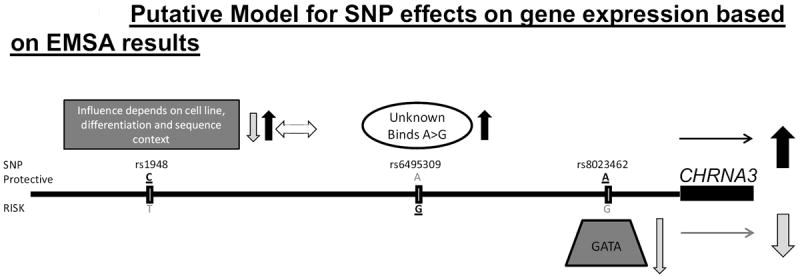
All 3 SNPs are indicated, along with very preliminary putative mechanisms for regulation of gene expression. Mechanisms that increase expression indicated on top, those that decrease expression on bottom. Bold/underline indicates the major allele for each SNP.
Though potential confounds exist related to co-morbidity (Young et al., 2008) as well as ethnicity (Shiraishi et al., 2009; Wu et al., 2009), it is apparent that variants within the cluster may be directly involved in carcinogenic events within the lung parenchyma where they are expressed. The risk of lung cancer that can be attributed to CHRNA5/A3/B4 variants is higher than what one could explain by simple effects of smoking quantity, supporting a gene-environment interaction (Thorgeirsson et al., 2008; Volkow et al., 2008). Some insist that receptors in the lung are not driving this interaction, rather that variants are primarily associated with increased intake of carcinogenic compounds like nitrosamine, even with cigarette numbers being equivalent. Individuals with the risk variants rs1051730 and rs16969968 have been shown to extract more carcinogens per cigarette (Le Marchand et al., 2008), as supported by increased tobacco metabolite (cotinine) levels in individuals with risk genotypes (Munafò et al., 2012).
However, there is also evidence that nicotinic receptor activation within the lung parenchyma (Paliwal et al., 2010) (Dasgupta et al., 2006) (Xin and Deng, 2005) Catassi, Servent et al. 2008) Schuller 2009) can contribute to increased risk for lung cancer. Variants associated with increased risk for lung cancer may be influencing brain processes to sustain the activity of smoking AND also increasing pro-survival signals within the lung as a consequence of nicotinic receptor activation (West et al., 2003), providing a “double hit” in terms of conferring risk for lung cancer. Possible mechanisms include disregulation of mRNA (Falvella et al., 2009; Wang et al., 2009a; Wang et al., 2009b) and altered levels of gene expression given different haplotypes (Doyle et al., 2011; Falvella et al., 2010; Wu et al., 2009).
In conclusion, the work presented here supports the hypothesis that SNPs associated with human alcohol and tobacco behaviors may lead to allele-specific gene expression differences that are dependent on cell type and haplotype. These findings are consistent with previous work showing transcriptional regulation in this region is tightly controlled during development. Additionally, this is the first evidence that rs8023462 interacts with GATA transcription factors and influences gene expression.
4 Experimental Procedure
4.1 Plasmid constructs
A “construct” refers to the pGL3 promoter vector (Promega Corporation, Madison, WI, USA) with a sequence inserted between the 5’-MluI–NheI-3’ restriction sites (or 5’-Acc651-XhoI-3’ restriction sites for the 800 bp rs8023462 construct only), upstream of the SV40 promoter and firefly luciferase gene. Relative sizes of each construct with their respective SNPs are shown in Figure 1A.
Constructs using the positive/forward chromosomal strand of our region of interest (consensus sequence from NCBI assembly database GRCh37/hg19) were synthesized and placed upstream of the SV40 promoter in the pGL3 promoter vector at GenScript, Inc. (Piscataway, NJ, USA), and manipulated in our laboratory to excise and place the sequence in the correct, coding, orientation (the coding strand for CHRNA3/B4 is the negative/reverse chromosomal strand). Details of cut sites, sequences, and ligations are available upon request. The use of a viral promoter is important for determining the impact of these regions outside of the influence of the CHRNA3 promoter, and viral promoters have been used to study regulation of the rat cluster in a similar manner (McDonough et al., 2000). All constructs and control plasmids were transformed into Top10 cells (Invitrogen Corporation, Carlsbad, CA, USA), and prepared from bacterial cultures using FastPlasmid Mini Kit or the PerfectPrep Endofree Maxi Kit (5 Prime Inc., Gaithersburg, MD, USA). All constructs and control plasmids were sent for sequencing initially and re-sequenced halfway through the project at SeqWright Inc. (Houston, TX, USA) or GenScript (Piscataway, NJ, USA), for verification.
4.2 Cell culture and reagents
Cell culture conditions (growth, differentiation and maintenance) have been carried out as described previously by our group (Ehringer et al., 2010; Mexal et al., 2007). The PC12 and SH-SY5Y cell lines were chosen based on similarities such as the ability to transcribe nAChR subunit genes including those in the cluster (Dajas-Bailador et al., 2002; Lukas et al., 1993; Rogers et al., 1992; Wang et al., 1996), expression of genes characteristic of neurons (Biedler et al., 1978; Greene and Tischler, 1976), and the capacity to differentiate (Greene and Tischler, 1976; Påhlman et al., 1990; Rogers et al., 1992). H69 and H446 were chosen as SCLC is largely attributed to smoking. All cell lines were obtained from ATCC (Manassas, VA, USA), then passaged and seeded in the exponential growth phase. Cells were maintained in a humidified incubator with 5% CO2 at 37°C. Media was purchased from ATCC, unless otherwise indicated. Fetal bovine serum (FBS), horse serum (HS) and penicillin/streptomycin/amphotericin B (PSA) were purchased from Invitrogen Corporation (Carlsbad, CA, USA).
PC12 cells were grown in RPMI 1640 with 10% HS, 5% FBS and 1% PSA. Differentiated PC12 cells were cultured RPMI 1640 with nerve growth factor added and serum reduced (NGF 50 ng/mL, 1% HS, 0.5% FBS, 1% PSA) and seeded on plates pretreated with poly-L-lysine (PLL, 100 μg/mL). SH-SY5Y (human neuroblastoma) cells were grown in a 1:1 mixture of Ham’s F12 (Invitrogen) and Eagle’s Minimal Essential Medium (EMEM) with 10% FBS and 1% PSA. Differentiated SH-SY5Y cells were cultured in serum-reduced media with retinoic acid (RA 10μM, 1% FBS, 1% PSA) and seeded on plates pre-treated with PLL. H69 and H446 cells were grown in RPMI 1640 + 10% FBS and 1% PSA.
4.3 Cell transfection
100,000 cells/1mL (neuronal-like cells) or 150,000 cells/mL (lung cancer cells) were seeded in 12 or 24-well culture plates. Twenty-four hours after seeding (differentiation initiated on day of seeding), test plasmids (constructs or empty vector) (20 ng/μL final concentration) were transfected into cells using FuGene HD or X-tremeGene transfection reagent (H446 only) (Roche USA, Indianapolis, IN, USA) in GIBCO Opti-MEM ® 1 media (Invitrogen). A renilla luciferase control plasmid, (pRL-CMV, Promega Corporation) was co-transfected (0.002 ng/μL final concentration) with each construct or empty vector. Cells were maintained for another 96 hrs (neuronal-like cells) or 48 hrs (lung cancer cells) before harvesting and assaying for luciferase activity.
4.4 Promoter/reporter luciferase assay
The Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA) was used to assess gene expression following manufacturer’s instructions. Briefly, aliquots of 20 μL of lysate from each cell culture plate well were run in duplicate using 96-well plates. A PerkinElmer Victor 3V plate reader (Perkin Elmer, Wellesley, MA, USA) dispensed assay substrates and detected light generated by test (firefly luciferase; constructs and empty vector) and control (renilla luciferase) plasmids. A minimum of 2 separate maxi preps were tested with multiple replicates of each performed on at least 2 separate days.
4.5 Nuclear extracts and Electrophoretic Mobility Shift Assays (EMSAs)
Proteins were extracted from cell nuclei using the NE-PER nuclear and cytoplasmic extraction kit as directed, and measured using the BCA protein assay kit (both Thermo Fisher Scientific, Rockford, IL, USA) following the micro-assay protocol for the NanoDrop (Thermofisher Scientific, Welmington, DE, USA). Synthetic double-stranded and 3′ biotin–labeled oligonucleotides were prepared at IDT (Integrated DNA Technologies, Inc., San Diego, CA USA) and reconstituted in water. For competition experiments with the Oct-1 consensus probe, the Oct-1 EMSA Probe Set from Panomics was used (Panomics. Inc., Fremont. CA, USA). Biotin labeled probes (0.025 picoMoles) and cell nuclear extracts (10-15μg) were incubated together at room temperature for 30 min using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific, IL, USA) with 0.5 mM DTT and 5 mM MgCl2 in 1x binding buffer with 1 μg poly dIdC (for rs6495309) or with 0.5 mM DTT in 1x binding buffer with 1 μg poly dIdC (rs8023462). Reactions were electrophoresed using 5% polyacrylamide gels and transferred to Biodyne B Nylon membranes (Thermo Fisher Scientific, IL, USA). Bands were detected (Chemiluminescent Detection Kit, Thermo Fisher Scientific, IL, USA) and images were acquired using the GelLogic 2200Pro CCD-camera and accompanying software (Carestream Molecular Imaging, Woodbridge, CT, USA). Unlabeled oligonucleotides at 100-fold molar excess were added to the reaction for competition (15 minute pre-incubation with unlabeled oligos and nuclear extract, 15 minute further incubation following addition of biotin labeled oligos). To determine whether the DNA-binding protein in assays with rs6495309 was oct-1 or oct-3/4, we used antibodies specific to Oct-1 (sc-232x), Oct-3/4 (sc-5279x) or nonspecific rabbit IgG or mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA), as described by Wu et al. 2009 (Wu et al., 2009). For identification of protein in assays with rs8023462, the Transcription Element Search System (TESS) (URL: http://www.cbil.upenn.edu/tess) was used to determine possible TFs for investigation. GATA 4 and GATA 2/3 antibodies were used (sc-1237X and sc-1235X, respectively) with nonspecific goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
4.6 Statistical Analysis
Firefly luciferase activity of the test plasmids was divided by renilla luciferase activity of the control plasmid for each well, yielding a gene expression ratio. Duplicate readings were averaged to arrive at one ratio value per transfected cell-culture well. Values were then normalized to the average value of the empty pGL3 test plasmid (no insert) within the same cell line and experiment to yield the vector-normalized ratio, or relative luciferase activity, herein referred to as “gene expression”. Data were analyzed using IBM® SPSS® Statistics (Somers, NY, USA) version 19.0, after first discarding results with Z score values greater than 2.25. An initial analysis was performed separately for the neuronal-like cell data and the lung cancer cell data using a General Linear Model (GLM) Univariate Analysis, with gene expression level as the dependent variable and cell line (PC12/SH-SY5Y or H446/H69), treatment condition (differentiated or un-treated), and length as independent variables. In both the neuronal-like and lung cancer cell line GLMs, main effects were evident for cell line, treatment condition (neuronal-like) and length (p<0.001). Given these findings and biological justification, separate analyses were conducted within each cell line, treatment condition and construct length group. Follow-up analyses were conducted for each PC12, PC12-D, SH-SY5Y, SH-SY5Y-D, H446 and H69 groups using student’s t-tests (0.8kbp rs1948, 1.7kbp and 800bp rs8023462 length groups), or analysis of variance (ANOVA) with Tukey post-hoc analysis (2.7kbp and 3.5kbp length groups) to compare the variants or haplotypes, respectively.
Supplementary Material
Figure 6. Luciferase expression results from transfected H446 cells.
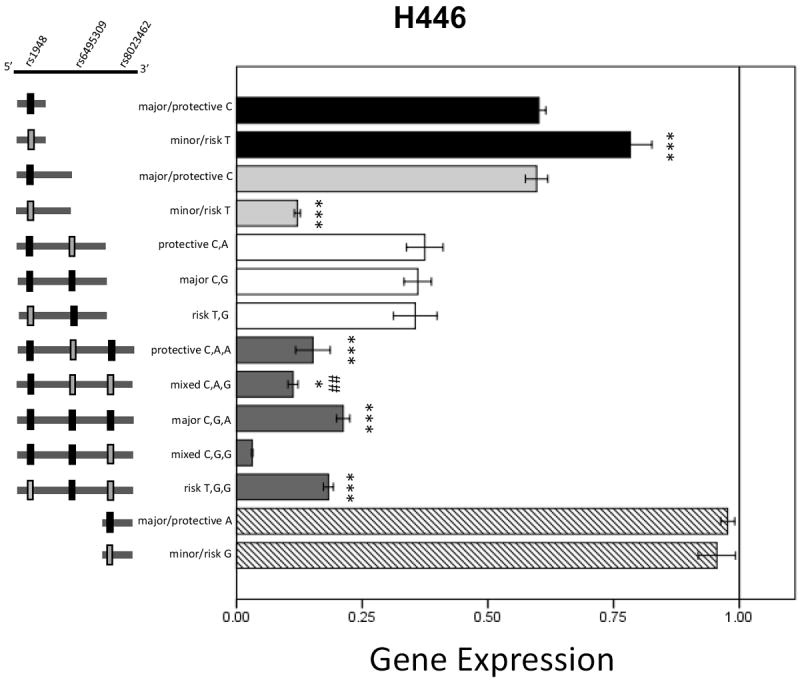
Data are mean +/- SEM. * p<0.05, ## p<0.01, *** p ≤ 0.001. Note that the X-axis shows increments of 0.25 and ends at 1.00. Asterisks in the 3.5kbp group indicate significantly different from 3.5kbp_CGG, pound symbol indicates significantly different from 3.5kbp_CGA in the post hoc Tukey analysis.
Acknowledgments
Support - This works was supported by Grants from National Institutes of Health (NIH R01 017889, R21 DA026901 and T32 AA007464) and the University of Colorado (Cancer Center IRG (Institutional Research Grant) 57-001-47) and P30 CA046934.M
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amber V Flora, Email: amber.flora@colorado.edu.
Cristian A Zambrano, Email: Cristian.Zambrano@colorado.edu.
Xavier Gallego, Email: francesc.gallegomoreno@colorado.edu.
Krista A Johnson, Email: Krista.A.Johnson@colorado.edu.
Katelyn A Cowan, Email: cowan.katelyn@gmail.com.
Jerry A Stitzel, Email: stitzel@colorado.edu.
References
- Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008 doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglioli E, Gotti C, Terzano S, Flora A, Clementi F, Fornasari D. Expression and transcriptional regulation of the human alpha3 neuronal nicotinic receptor subunit in T lymphocyte cell lines. J Neurochem. 1998;71:1261–70. doi: 10.1046/j.1471-4159.1998.71031261.x. [DOI] [PubMed] [Google Scholar]
- Benfante R, Flora A, Di Lascio S, Cargnin F, Longhi R, Colombo S, Clementi F, Fornasari D. Transcription Factor PHOX2A Regulates the Human 3 Nicotinic Receptor Subunit Gene Promoter. Journal of Biological Chemistry. 2007;282:13290–13302. doi: 10.1074/jbc.M608616200. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini WH, Doyle GA. The CHRNA5–A3–B4 gene cluster in nicotine addiction. Mol Psychiatry. 2012;17:856–866. doi: 10.1038/mp.2011.122. [DOI] [PubMed] [Google Scholar]
- Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38:3751–7. [PubMed] [Google Scholar]
- Bierut LJ, Madden PAF, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, Horton WJ, Breslau N, Budde J, Cloninger CR, Dick DM, Foroud T, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Kuperman S, Madden PAF, Mayo K, Nurnberger J, Pomerleau O, Porjesz B, Reyes O, Schuckit M, Swan G, Tischfield JA, Edenberg HJ, Rice JP, Goate AM. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–7. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter J, O’Shea-Greenfield A, Duvoisin RM, Connolly JG, Wada E, Jensen A, Gardner PD, Ballivet M, Deneris ES, McKinnon D. Alpha 3, alpha 5, and beta 4: three members of the rat neuronal nicotinic acetylcholine receptor-related gene family form a gene cluster. J Biol Chem. 1990;265:4472–82. [PubMed] [Google Scholar]
- Boyd RT. Transcriptional regulation and cell specificity determinants of the rat nicotinic acetylcholine receptor alpha 3 gene. Neurosci Lett. 1996;208:73–6. doi: 10.1016/0304-3940(96)12561-1. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Lee H-Y, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–93. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2010. pp. 1–727. C.f.D. Control. [PubMed] [Google Scholar]
- Consortium, T.a.G. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier S, Erkman L, Valera S, Rungger D, Bertrand S, Boulter J, Ballivet M, Bertrand D. Alpha 5, alpha 3, and non-alpha 3. Three clustered avian genes encoding neuronal nicotinic acetylcholine receptor-related subunits. J Biol Chem. 1990;265:17560–7. [PubMed] [Google Scholar]
- Dajas-Bailador FA, Mogg AJ, Wonnacott S. Intracellular Ca2+ signals evoked by stimulation of nicotinic acetylcholine receptors in SH-SY5Y cells: contribution of voltage-operated Ca2+ channels and Ca2+ stores. J Neurochem. 2002;81:606–14. doi: 10.1046/j.1471-4159.2002.00846.x. [DOI] [PubMed] [Google Scholar]
- Dasgupta P, Kinkade R, Joshi B, Decook C, Haura E, Chellappan S. Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc Natl Acad Sci U S A. 2006;103:6332–7. doi: 10.1073/pnas.0509313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle GA, Wang M-J, Chou AD, Oleynick JU, Arnold SE, Buono RJ, Ferraro TN, Berrettini WH. In vitro and ex vivo analysis of CHRNA3 and CHRNA5 haplotype expression. PLoS ONE. 2011;6:e23373. doi: 10.1371/journal.pone.0023373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, McQueen MB, Hoft NR, Saccone NL, Stitzel JA, Wang JC, Bierut LJ. Association of CHRN genes with “dizziness” to tobacco. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:600–9. doi: 10.1002/ajmg.b.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falvella FS, Galvan A, Frullanti E, Spinola M, Calabrò E, Carbone A, Incarbone M, Santambrogio L, Pastorino U, Dragani TA. Transcription deregulation at the 15q25 locus in association with lung adenocarcinoma risk. Clin Cancer Res. 2009;15:1837–42. doi: 10.1158/1078-0432.CCR-08-2107. [DOI] [PubMed] [Google Scholar]
- Falvella FS, Galvan A, Colombo F, Frullanti E, Pastorino U, Dragani TA. Promoter polymorphisms and transcript levels of nicotinic receptor CHRNA5. J Natl Cancer Inst. 2010;102:1366–70. doi: 10.1093/jnci/djq264. [DOI] [PubMed] [Google Scholar]
- Fornasari D, Battaglioli E, Flora A, Terzano S, Clementi F. Structural and functional characterization of the human alpha3 nicotinic subunit gene promoter. Mol Pharmacol. 1997;51:250–61. doi: 10.1124/mol.51.2.250. [DOI] [PubMed] [Google Scholar]
- Fyodorov D, Deneris E. The POU domain of SCIP/Tst-1/Oct-6 is sufficient for activation of an acetylcholine receptor promoter. Mol Cell Biol. 1996;16:5004–14. doi: 10.1128/mcb.16.9.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27:482–91. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–8. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerri C, Pascual M. Mechanisms involved in the neurotoxic, cognitive, and neurobehavioral effects of alcohol consumption during adolescence. Alcohol. 2010;44:15–26. doi: 10.1016/j.alcohol.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Hu MC, Davies M, Kandel DB. Epidemiology and correlates of daily smoking and nicotine dependence among young adults in the United States. Am J Public Health. 2006;96:299–308. doi: 10.2105/AJPH.2004.057232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcatova I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Improgo MRD, Scofield MD, Tapper AR, Gardner PD. The nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster: dual role in nicotine addiction and lung cancer. Prog Neurobiol. 2010;92:212–26. doi: 10.1016/j.pneurobio.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, Tiirikainen M, Wang H. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68:9137–40. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, Pinney SM, Yang P, de Andrade M, Petersen GM, Wiest JS, Fain PR, Gazdar A, Gaba C, Rothschild H, Mandal D, Coons T, Lee J, Kupert E, Seminara D, Minna J, Bailey-Wilson JE, Wu X, Spitz MR, Eisen T, Houlston RS, Amos CI, Anderson MW, You M. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326–30. doi: 10.1093/jnci/djn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas RJ, Norman SA, Lucero L. Characterization of Nicotinic Acetylcholine Receptors Expressed by Cells of the SH-SY5Y Human Neuroblastoma Clonal Line. Mol Cell Neurosci. 1993;4:1–12. doi: 10.1006/mcne.1993.1001. [DOI] [PubMed] [Google Scholar]
- McDonough J, Deneris E. beta43’: An enhancer displaying neural-restricted activity is located in the 3’-untranslated exon of the rat nicotinic acetylcholine receptor beta4 gene. J Neurosci. 1997;17:2273–83. doi: 10.1523/JNEUROSCI.17-07-02273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough J, Francis N, Miller T, Deneris ES. Regulation of transcription in the neuronal nicotinic receptor subunit gene cluster by a neuron-selective enhancer and ETS domain factors. J Biol Chem. 2000;275:28962–70. doi: 10.1074/jbc.M004181200. [DOI] [PubMed] [Google Scholar]
- Mexal S, Jenkins PM, Lautner MA, Iacob E, Crouch E, Stitzel JA. alpha 7 nicotinic receptor gene promoter polymorphisms in inbred mice affect expression in a cell type-specific fashion. J Biol Chem. 2007 doi: 10.1074/jbc.M610694200. [DOI] [PubMed] [Google Scholar]
- Milton NG, Bessis A, Changeux JP, Latchman DS. Differential regulation of neuronal nicotinic acetylcholine receptor subunit gene promoters by Brn-3 POU family transcription factors. Biochem J. 1996;317(Pt 2):419–23. doi: 10.1042/bj3170419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò MR, Timofeeva MN, Morris RW, Prieto-Merino D, Sattar N, Brennan P, Johnstone EC, Relton C, Johnson PCD, Walther D, Whincup PH, Casas JP, Uhl GR, Vineis P, Padmanabhan S, Jefferis BJ, Amuzu A, Riboli E, Upton MN, Aveyard P, Ebrahim S, Hingorani AD, Watt G, Palmer TM, Timpson NJ, Group ES, Davey Smith G. Association Between Genetic Variants on Chromosome 15q25 Locus and Objective Measures of Tobacco Exposure. J Natl Cancer Inst. 2012 doi: 10.1093/jnci/djs191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H, Nakshatri P, Currie RA. Interaction of Oct-1 with TFIIB. Implications for a novel response elicited through the proximal octamer site of the lipoprotein lipase promoter. J Biol Chem. 1995;270:19613–23. doi: 10.1074/jbc.270.33.19613. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Research. 2004;32:W280–6. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Påhlman S, Mamaeva S, Meyerson G, Mattsson ME, Bjelfman C, Ortoft E, Hammerling U. Human neuroblastoma cells in culture: a model for neuronal cell differentiation and function. Acta Physiol Scand Suppl. 1990;592:25–37. [PubMed] [Google Scholar]
- Paliwal A, Vaissière T, Krais A, Cuenin C, Cros M-P, Zaridze D, Moukeria A, Boffetta P, Hainaut P, Brennan P, Herceg Z. Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res. 2010;70:2779–88. doi: 10.1158/0008-5472.CAN-09-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergadia ML, Agrawal A, Loukola A, Montgomery GW, Broms U, Saccone SF, Wang JC, Todorov AA, Heikkila K, Statham DJ, Henders AK, Campbell MJ, Rice JP, Todd RD, Heath AC, Goate AM, Peltonen L, Kaprio J, Martin NG, Madden PA. Genetic linkage findings for DSM-IV nicotine withdrawal in two populations. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:950–9. doi: 10.1002/ajmg.b.30924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SW, Mandelzys A, Deneris ES, Cooper E, Heinemann S. The expression of nicotinic acetylcholine receptors by PC12 cells treated with NGF. J Neurosci. 1992;12:4611–23. doi: 10.1523/JNEUROSCI.12-12-04611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D, Johnson EO, Madden PAF, Swan GE, Wang JC, Goate AM, Rice JP, Bierut LJ. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–66. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, Swan GE, Goate AM, Rutter J, Bertelsen S, Fox L, Fugman D, Martin NG, Montgomery GW, Wang JC, Ballinger DG, Rice JP, Bierut LJ. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–46. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Himmelmann A, Malipiero U, Tobler A, Stahel R, Fontana A. Human small cell lung cancer expresses the octamer DNA-binding and nervous system-specific transcription factor N-Oct 3 (brain-2) Cancer Res. 1992;52:6121–4. [PubMed] [Google Scholar]
- Shiraishi K, Kohno T, Kunitoh H, Watanabe S-i, Goto K, Nishiwaki Y, Shimada Y, Hirose H, Saito I, Kuchiba A, Yamamoto S, Yokota J. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- Stephens SH, Hoft NR, Schlaepfer IR, Young SE, Corley RC, McQueen MB, Hopfer C, Crowley T, Stallings M, Hewitt J, Ehringer MA. Externalizing behaviors are associated with SNPs in the CHRNA5/CHRNA3/CHRNB4 gene cluster. Behavior genetics. 2012;42:402–14. doi: 10.1007/s10519-011-9514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsäter A, Flex A, Aben KKH, de Vegt F, Mulders PFA, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Mägi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tönjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Döring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen A-L, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PAF, Giegling I, Rujescu D, Järvelin M-R, Salomaa V, Stumvoll M, Spector TD, Wichmann H-E, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, Consortium E, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, Risch A, McKay JD, Wang Y, Dai J, Gaborieau V, McLaughlin J, Brenner D, Narod S, Caporaso NE, Albanes D, Thun M, Eisen T, Wichmann H-E, Rosenberger A, Han Y, Chen W, Zhu D, Spitz M, Wu X, Pande M, Zhao Y, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Krokan HE, Gabrielsen ME, Skorpen F, Vatten L, Njølstad I, Chen C, Goodman G, Lathrop M, Benhamou S, Vooder T, Välk K, Nelis M, Metspalu A, Raji O, Chen Y, Gosney J, Liloglou T, Muley T, Dienemann H, Thorleifsson G, Shen H, Stefansson K, Brennan P, Amos CI, Houlston R, Landi MT, Team fTR. Influence of Common Genetic Variation on Lung Cancer Risk: Meta-Analysis of 14,900 Cases and 29,485 Controls. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nöthen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–84. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N, Rutter J, Pollock JD, Shurtleff D, Baler R. One SNP linked to two diseases-addiction and cancer: a double whammy? Nicotine addiction and lung cancer susceptibility. Mol Psychiatry. 2008;13:990–2. doi: 10.1038/mp.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gerzanich V, Wells GB, Anand R, Peng X, Keyser K, Lindstrom J. Assembly of human neuronal nicotinic receptor alpha5 subunits with alpha3, beta2, and beta4 subunits. J Biol Chem. 1996;271:17656–65. doi: 10.1074/jbc.271.30.17656. [DOI] [PubMed] [Google Scholar]
- Wang JC, Bierut LJ, Goate AM. Variants weakly correlated with CHRNA5 D398N polymorphism should be considered in transcriptional deregulation at the 15q25 locus associated with lung cancer risk. Clin Cancer Res. 2009a;15:5599. doi: 10.1158/1078-0432.CCR-09-1108. [DOI] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, Duan W, Fox L, Grucza RA, Kern J, Mayo K, Reyes O, Rice J, Saccone SF, Spiegel N, Steinbach JH, Stitzel JA, Anderson MW, You M, Stevens VL, Bierut LJ, Goate AM. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009b;18:3125–35. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP, Duan W, Fox L, Grucza RA, Kern J, Mayo K, Reyes O, Rice J, Saccone SF, Spiegel N, Steinbach JH, Stitzel JA, Anderson MW, You M, Stevens VL, Bierut LJ, Goate AM collaborators, C.c.a.G. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009c;18:3125–35. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, Piper ME, Fiore MC, Scholand MB, Connett JE, Kanner RE, Gahring LC, Rogers SW, Hoidal JR, Leppert MF. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West KA, Brognard J, Clark AS, Linnoila IR, Yang X, Swain SM, Harris C, Belinsky S, Dennis PA. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111:81–90. doi: 10.1172/JCI16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Hu Z, Yu D, Huang L, Jin G, Liang J, Guo H, Tan W, Zhang M, Qian J, Lu D, Wu T, Lin D, Shen H. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–72. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- Xin M, Deng X. Nicotine inactivation of the proapoptotic function of Bax through phosphorylation. J Biol Chem. 2005;280:10781–9. doi: 10.1074/jbc.M500084200. [DOI] [PubMed] [Google Scholar]
- Yang X, McDonough J, Fyodorov D, Morris M, Wang F, Deneris ES. Characterization of an acetylcholine receptor alpha 3 gene promoter and its activation by the POU domain factor SCIP/Tst-1. J Biol Chem. 1994;269:10252–64. [PubMed] [Google Scholar]
- Young RP, Hopkins RJ, Hay BA, Epton MJ, Black PN, Gamble GD. Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur Respir J. 2008;32:1158–64. doi: 10.1183/09031936.00093908. [DOI] [PubMed] [Google Scholar]
- Zucker RA, Donovan JE, Masten AS, Mattson ME, Moss HB. Early developmental processes and the continuity of risk for underage drinking and problem drinking. Pediatrics. 2008;121(Suppl 4):S252–72. doi: 10.1542/peds.2007-2243B. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


