Abstract
When interacting with the CD4 receptor, the HIV gp120 envelope glycoprotein undergoes conformational changes that allow binding to the chemokine receptor. Receptor binding is proposed to lead to conformational changes in the gp41 transmembrane envelope glycoprotein involving the creation and/or exposure of a coiled coil consisting of three heptad repeat (HR) sequences. The subsequent interaction of the HR2 region of gp41 with this coiled coil results in the assembly of a six-helix bundle that promotes the fusion of the viral and target cell membranes. Here we show that CD4 binding to gp120 induces the formation and/or exposure of the gp41 HR1 coiled coil in a process that does not involve gp120 shedding and that depends on the proteolytic maturation of the gp160 envelope glycoprotein precursor. Importantly, BMS-806 and related HIV-1 entry inhibitors bind gp120 and block the CD4 induction of HR1 exposure without significantly affecting CD4 binding. Moreover, these compounds do not disrupt gp120-chemokine receptor binding or the HR1-HR2 interaction within gp41. These studies thus define a receptor-induced conformational rearrangement of gp120-gp41 that is important for both CD4-dependent and CD4-independent HIV-1 entry and is susceptible to inhibition by low-molecular-weight compounds.
The global epidemic of infection by HIV, the cause of AIDS (1, 2), has created an urgent need for new classes of antiretroviral agents. The entry of HIV-1 into target cells involves several potential targets for intervention. The HIV-1 envelope glycoprotein complex is a trimer consisting of three gp120 exterior envelope glycoproteins and three gp41 transmembrane glycoproteins (3-6). These are derived by cleavage of a gp160 precursor glycoprotein by host cell proteases. The mature envelope glycoprotein complex is expressed on the surface of infected cells and is incorporated into virion membranes. HIV-1 infection is initiated by gp120 binding to CD4 on the target cell surface (7, 8). CD4 binding induces conformational changes in the gp120 glycoprotein (9, 10), some of which allow gp120 to interact efficiently with one of the chemokine receptors, CCR5 and CXCR4, that serve as obligate coreceptors for HIV-1 (11-15). CD4 binding can also trigger the shedding of gp120, leading to functional inactivation of the viral envelope glycoprotein complex (16). Therefore, not every CD4-induced conformational change in the HIV-1 envelope glycoproteins is relevant or conducive to virus entry. The interaction of gp120 with its receptors is thought to promote conformational rearrangements in gp41 that drive the fusion of the viral and target cell membranes. The gp41 ectodomain can form a very stable six-helix bundle consisting in part of a trimeric coiled coil of heptad repeat (HR) sequences (4-6). An HR2 sequence in gp41 near the viral membrane can form an α helix that packs in the hydrophobic grooves of the HR1 coiled coil. This stable six-helix bundle is thought to represent the final, fusogenic conformation of gp41, and the favorable energy change associated with its formation may facilitate the apposition of the viral and cell membranes (4-6). That peptides mimicking the HR2 region inhibit HIV-1 infection in a dominant-negative fashion (17) suggests that, in some relevant conformation of gp41, the HR1 and HR2 regions are not associated. The HR2 region peptides and some gp41-directed monoclonal antibodies bind the HIV-1 envelope glycoproteins better after treatment with soluble CD4 (18-21), but it is uncertain whether this increase in binding results from an entry-related conformational change, gp120 shedding, or another process.
BMS-378806 (herein called BMS-806) and related compounds are low-molecular-weight inhibitors of HIV-1 entry that were recently identified by using a viral infection-based screen (22, 23). BMS-806 was shown to be specific for HIV-1, with no activity against HIV-2 or simian immunodeficiency virus. BMS-806 is active against HIV-1 isolates irrespective of chemokine receptor preference (23, 24). BMS-806 binds gp120, and changes in particular gp120 amino acid residues can alter the sensitivity of the virus to BMS-806 (23, 24). Here we study the mechanism of action of BMS-806 and related antiviral compounds.
Materials and Methods
Reagents. Compounds were synthesized as described (22), and their identity was confirmed (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Envelope glycoprotein-expressing plasmids (25, 26), soluble CD4, and anti-gp120 antibodies (27) are described in Supporting Materials and Methods.
C34-Ig consists of the HR2 region [amino acid residues 628-661, numbered according to current convention (28)] from the HXBc2 envelope glycoproteins, a GSGSG linker, and the Ig-constant region. By virtue of the Ig-constant region of the molecule, C34-Ig is dimeric. The production of C34-Ig is detailed in Supporting Materials and Methods.
Binding Assays. Assays to measure ligand-envelope glycoprotein binding are described in Supporting Materials and Methods.
HIV-1 Infectivity Assay. Recombinant HIV-1 expressing firefly luciferase was produced in 293T cells, incubated with varying concentrations of compound, and then used to infect either Cf2Th-CD4/CCR5 or Cf2Th-CCR5 target cells, as described (25). Details are provided in Supporting Materials and Methods.
Results
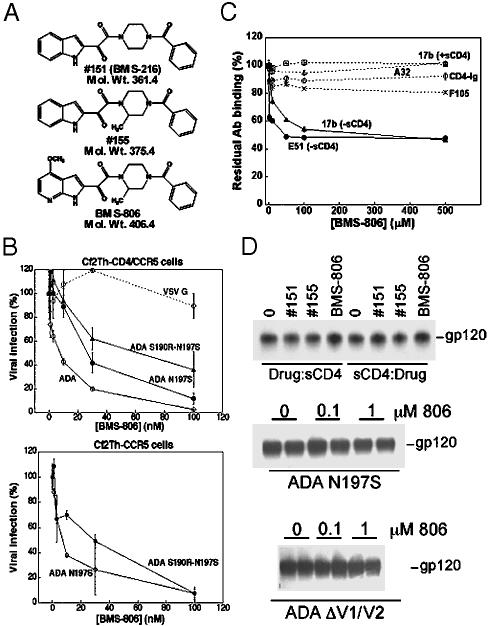
The Antiviral Activity of BMS-806 Is Not Due to Inhibition of Receptor Binding. BMS-806 and related compounds have been reported to block HIV-1 entry (23, 24). We synthesized BMS-806 and two analogues herein called #151 (previously reported as BMS-216) and #155 (Fig. 1A). All three compounds specifically inhibited a single-round infection by recombinant viruses with envelope glycoproteins derived from the X4 HXBc2, R5 YU2, or R5 ADA HIV-1 isolates (Fig. 1B Upper and data not shown). The IC50 values of BMS-806 for all three HIV-1 strains were between 1 and 10 nM. Thus, BMS-806 potently inhibited HIV-1 infection regardless of the particular chemokine receptor used. No inhibition of infection by viruses with the vesicular stomatitis virus glycoproteins was observed (Fig. 1B). We also examined the effect of BMS-806 on the infection of two CD4-independent viruses, one with the ADA N197S envelope glycoproteins and the other with the ADA S190R-N197S envelope glycoproteins. These viruses have previously been shown to infect CD4-negative target cells expressing CCR5 (29). CD4-independent infection of such cells by viruses with the ADA N197S and S190R-N197S envelope glycoproteins was inhibited by BMS-806 as efficiently as the infection of CD4-expressing cells by viruses with these envelope glycoproteins (Fig. 1B, compare Upper with Lower). Because BMS-806 potently inhibits HIV-1 infection of cells lacking CD4, we conclude that BMS-806 exerts its major antiviral effect on a step in the HIV-1 entry process other than CD4 binding.
Fig. 1.
BMS-806 does not inhibit HIV-1 receptor binding. (A) The structures of BMS-806 and related compounds #151 and #155 are illustrated. (B) Recombinant HIV-1 viruses expressing firefly luciferase and containing the indicated envelope glycoproteins were used to infect Cf2Th-CD4/CCR5 or Cf2Th-CCR5 cells in the presence of different concentrations of BMS-806. The percentage of luciferase measured in the target cells relative to that observed in the absence of drug is shown. The values shown represent the means and SDs of triplicate points in the assay. The results are typical of those obtained in three independent experiments. In separate experiments, BMS-806 did not inhibit the infection of Cf2Th-CCR5 cells by HIV-1 viruses pseudotyped by vesicular stomatitis virus glycoproteins (data not shown). (C) The YU2 gp120 glycoprotein was captured on ELISA plates coated with the D7324 antibody. Antibodies or CD4-Ig were incubated with the captured gp120 in the presence of the indicated concentrations of BMS-806, and, after washing, bound ligand was detected by peroxidase-conjugated goat anti-human IgG (Ig-constant region-specific) antibody. The values shown represent the optical densities obtained, relative to those observed for each ligand in the absence of BMS-806. (D)(Top) Radiolabeled ADA gp120 was incubated either first with BMS-806 followed by sCD4 or first with sCD4 followed by BMS-806. The mixtures were then incubated with Cf2Th-CCR5 cells; the bound gp120 glycoproteins are shown. (Middle and Bottom). Radiolabeled ADA N197S gp120 or ΔV1/V2 gp120 glycoproteins were incubated with Cf2Th-CCR5 cells without the addition of sCD4 and in the presence of the indicated concentrations of BMS-806. The bound gp120 variants from duplicate experiments are shown.
BMS-806 has been reported to bind the HIV-1 gp120 glycoprotein (23, 24). We confirmed the ability of BMS-806 to bind monomeric gp120 derived from the YU2 primary HIV-1 isolate by isothermal titration microcalorimetry, and determined a dissociation constant of ≈43 ± 15 nM for this gp120 glycoprotein (data not shown). Notably, the interaction of BMS-806 with gp120 was characterized by a binding enthalpy of -3.4 ± 0.5 kcal/mol (1 cal = 4.18 J) and a binding entropy of 22 ± 2 cal/(K × mol). The thermodynamics of BMS-806 binding to gp120 contrasts with the extremely large enthalpy and entropy changes associated with the binding of sCD4 or even CD4 miniproteins (30). The large negative entropy (approximately -167 cal/(K × mol) associated with the binding of soluble CD4 (sCD4) has been related to the significant structuring of gp120 residues that is necessary for the formation of the chemokine receptor-binding site (30). Apparently, BMS-806 does not induce such a large structuring of gp120; the measured entropy change resembles that of small hydrophobic molecules.
We examined the ability of BMS-806 to interfere with the binding of gp120 ligands to the YU2 gp120 glycoprotein captured on an ELISA plate. The antibodies tested (b12, F105, 15e, 17b, E51, 39F, 2G12, A32, 2/11c and C11) recognize a variety of gp120 epitopes (ref. 27 and Supporting Materials and Methods). We also included CD4-Ig in the assay. CD4-Ig consists of the amino-terminal two domains of CD4, which are sufficient for high-affinity gp120 binding, fused to dimeric Ig heavy-chain constant regions. BMS-806 had no effect on the binding of the divalent CD4-Ig molecule to the captured gp120 glycoprotein (Fig. 1C). Even at 500 μM concentrations, BMS-806 exerted little or no effect on the binding of monomeric sCD4 to the gp120 glycoproteins of the YU2 and JR-FL HIV-1 strains (Fig. 5, which is published as supporting information on the PNAS web site). At concentrations of up to 500 μM, BMS-806 partially inhibited the binding of only two antibodies, 17b and E51, to the YU2 gp120 glycoprotein (Figs. 1C and 5). The 17b and E51 antibodies recognize CD4-induced gp120 epitopes that overlap the binding site for the chemokine receptors (31, 32). When BMS-806, 17b antibody and sCD4 were simultaneously incubated with the captured gp120, no inhibition of 17b binding was observed, even at 500 μM concentrations of BMS-806 (Fig. 1C). A positive control, the 12p1 peptide, specifically inhibited CD4-Ig and 17b binding in this assay (data not shown), as expected from the previously reported activity of this 12-residue peptide (33). In parallel studies, high concentrations of BMS-806 decreased the binding of free gp120, but not gp120-sCD4 complexes, to the 17b antibody immobilized on a biosensor (data not shown). Thus, at high concentrations, BMS-806 can partially inhibit the binding of some CD4-induced antibodies to monomeric gp120, an effect that is not seen in the presence of sCD4.
For most HIV-1 isolates, CD4 binding creates and/or exposes conserved gp120 elements involved in both CXCR4 and CCR5 interaction (9, 10). The gp120 glycoproteins of CD4-independent HIV-1 isolates can bind CCR5 or CXCR4 directly, without the assistance of CD4 (29, 34). We investigated the ability of BMS-806 to inhibit CCR5 binding by gp120 glycoproteins from both CD4-dependent and CD4-independent viruses. Fig. 1D Top demonstrates that, at concentrations of up to 50 μM, BMS-806 did not inhibit the binding of the wild-type ADA gp120 glycoprotein to CCR5-expressing cells, regardless of whether the compound was incubated with the gp120 glycoprotein before or after the addition of sCD4. We conclude that BMS-806 does not block CD4-dependent binding of gp120 to CCR5.
When the binding of the CD4-independent ADA N197S gp120 to CCR5 was studied, ≈20% inhibition at the 1 μM concentration of BMS-806 was observed (Fig. 1D Middle). Further studies demonstrated that BMS-806, #151, and #155 inhibited CCR5 binding of the ADA N197S gp120 with IC50 values of ≈60 μM (data not shown). This inhibition was specific, given that it was not seen for CCR5 binding of another CD4-independent gp120, ADA ΔV1/V2 gp120 (Fig. 1D Bottom). The ADA ΔV1/V2 gp120 glycoprotein lacks the V1 and V2 variable loops and does not bind BMS-806 efficiently (unpublished observations). The observed inhibitory effects of BMS-806 on the binding of the CD4-independent N197S gp120 glycoprotein to CCR5 occur at drug concentrations ≈10,000-fold higher than those required to inhibit virus infection. Thus, even for this unusual HIV-1 variant, the major antiviral action of BMS-806 appears to involve inhibition of virus entry steps other than chemokine receptor binding.
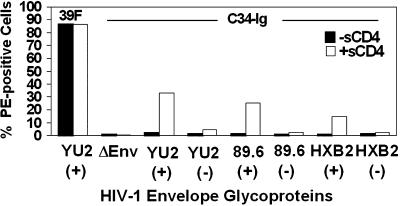
CD4 Induces HR2 Binding to the HIV-1 Envelope Glycoproteins. The data described above imply that BMS-806 might interfere with a process involved in HIV-1 entry other than receptor binding. It has been hypothesized that, for enveloped viruses such as HIV-1 that fuse viral and target cell membranes at neutral pH, receptor binding by the exterior envelope glycoprotein serves as the trigger that activates the fusogenic functions of the transmembrane envelope glycoprotein (3-6). Indeed, sCD4 binding to the HIV-1 envelope glycoproteins has been reported to result in the exposure of gp41 elements (18-20). However, as noted above, sCD4 binding can also induce gp120 shedding (16), allowing gp41 exposure by a process not obviously related to virus entry. Therefore, we investigated the ability of CD4 binding to induce HIV-1 envelope glycoprotein conformational changes that could be distinguished from gp120 shedding. To monitor the exposure of the hydrophobic groove in the HIV-1 gp41 HR1 trimeric coiled coil, we used a C34 peptide corresponding to the gp41 HR2 region linked to the Ig-constant region of an IgG molecule (C34-Ig). The C34-Ig fusion protein exhibited much less nonspecific association with cellular membranes than a construct, DP178-Ig, that contained a peptide corresponding to a more C-terminal segment of the gp41 HR2 region (data not shown). C34-Ig specifically inhibited HIV-1 infection (data not shown), indicating that it can access functionally relevant structures on the HIV-1 envelope glycoproteins. C34-Ig was incubated at 20°C with 293T cells expressing cytoplasmic tail-deleted YU2 envelope glycoproteins, in the absence or presence of sCD4. The envelope glycoproteins were derived from HIV-1 strains exhibiting different coreceptor preference: YU2 (R5), 89.6 (R5X4) and HXBc2 (X4). The addition of sCD4 significantly increased C34-Ig binding to the envelope glycoprotein-expressing cells (Fig. 2). By contrast, the binding of the 39F antibody, which recognizes the third variable (V3) loop of gp120 (27), and that of 2G12, which recognizes the gp120 outer domain (27), were minimally affected by the presence of sCD4 (Fig. 2 and Table 1, which is published as supporting information on the PNAS web site). We conclude that CD4 can induce the creation and/or exposure of a binding site for C34-Ig on the HIV-1 envelope glycoproteins, even in the apparent absence of gp120 shedding.
Fig. 2.
CD4 induces binding of C34-Ig to the HIV-1 envelope glycoproteins. The cleavage-competent [YU2(+), 89.6(+), and HXBc2(+)] and cleavage-defective [YU2(-), 89.6(-), and HXBc2(-)] envelope glycoproteins, all of which contain deletions of the cytoplasmic tail, were expressed on the surface of 293T cells and incubated with either the 39F or 2G12 anti-gp120 antibody or C34-Ig in the absence or presence of 20 μg/ml sCD4. The bound ligand was detected by a PE-conjugated goat anti-human IgG antibody, and the percentage of PE-positive cells was determined by fluorescence-activated cell sorter analysis. The results shown are typical of those obtained in three experiments. The effects of sCD4 on 2G12 binding are reported in Table 1.
To investigate the dependence of this phenomenon on proteolytic maturation of the HIV-1 envelope glycoproteins, we created cytoplasmic tail-deleted HXBc2, 89.6, and YU2 envelope glycoproteins that had altered cleavage sites so that processing of the gp160 precursor was abolished (35). All of the envelope glycoprotein variants were expressed at equivalent levels on the surface of transfected 293T cells and were able to bind soluble CD4 efficiently (35). Although the addition of sCD4 induced C34-Ig binding to the envelope glycoproteins that were competent for proteolytic processing, minimal induction of C34-Ig binding by sCD4 was observed for the cleavage-defective counterparts (Fig. 2). Thus, the sCD4 induction of C34-Ig binding to the HIV-1 envelope glycoproteins appears to require proteolytic processing of the gp160 envelope glycoprotein precursor.
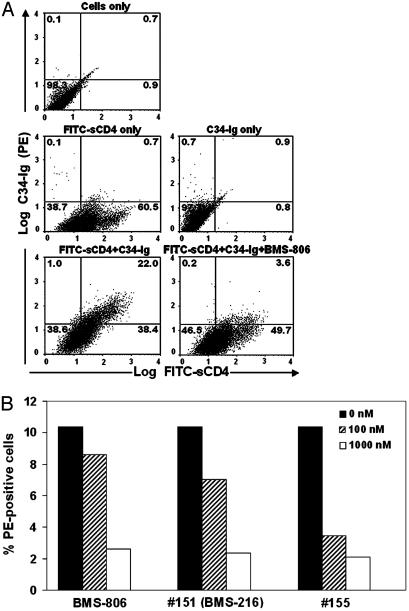
BMS-806 and Related Compounds Inhibit CD4-Induced C34-Ig Binding to the HIV-1 Envelope Glycoproteins. To understand better the relationship between the binding of CD4 and C34-Ig to the HIV-1 envelope glycoproteins, we labeled sCD4 with FITC so that the binding of both ligands could be simultaneously monitored by two-color flow cytometry. The envelope glycoprotein-expressing cells were readily stained with FITC-sCD4 (Fig. 3A). Incubation with C34-Ig alone resulted in little staining of the cells, consistent with the results obtained by single-color flow cytometry. Incubation with both FITC-sCD4 and C34-Ig resulted in a dramatic increase in double-positive cells (Fig. 3A). Essentially all of the cells positive for C34-Ig binding were also stained with FITC-sCD4, consistent with a cause-effect relationship between CD4 binding and C34-Ig binding.
Fig. 3.
BMS-806 and related compounds inhibit CD4-induced C34-Ig binding. (A) 293T cells expressing the cytoplasmic tail-deleted YU2 HIV-1 envelope glycoproteins were incubated with the indicated molecules and then analyzed for sCD4 (FITC) binding and C34-Ig (PE) binding by two-color flow cytometry. The concentration of BMS-806 was 1 μM. The percentage of cells in each quadrant is indicated. The results shown are typical of those obtained in four independent experiments. (B) 293T cells expressing the cytoplasmic tail-deleted YU2 HIV-1 envelope glycoproteins were incubated with FITC-sCD4 and C34-Ig in the presence of the indicated concentrations of BMS-806, #151, or #155. The percentage of FITC/PE-positive cells, which have bound both sCD4 and C34-Ig, was determined by two-color flow cytometry. The results shown are typical of those obtained in four independent experiments.
The effect of BMS-806 on the assay was examined. The addition of 1 μM BMS-806 resulted in a dramatic decrease in the cells staining positive for C34-Ig, with minimal effects on the number of cells staining with FITC-sCD4 (Fig. 3A). In the absence of C34-Ig, BMS-806 exerted little effect on the binding of FITC-sCD4 to the envelope glycoprotein-expressing cells (Fig. 6, which is published as supporting information on the PNAS web site). The lack of an effect of BMS-806 on CD4 binding to the envelope glycoproteins was confirmed in a different assay in which a CD4-Ig fusion protein was incubated with envelope glycoprotein-expressing cells, and bound CD4-Ig was detected with a phycoerythrin (PE)-conjugated goat anti-human IgG antibody (data not shown). BMS-806, #151 and, #155 all decreased the sCD4-induced binding of C34-Ig to the HIV-1 envelope glycoproteins, with IC50 values in the 50-400 nM range (Fig. 3B). These values are consistent with the concentrations of BMS-806 and #155 required to inhibit the formation of syncytia between 293T cells expressing either full-length or cytoplasmic tail-deleted YU2 HIV-1 envelope glycoproteins and Cf2Th-CD4/CCR5 cells (Fig. 7, which is published as supporting information on the PNAS web site). The presence of the cytoplasmic tail of gp41 did not influence the efficacy of these drugs in inhibiting syncytia. We conclude that BMS-806 and related compounds inhibit the CD4-induced creation and/or exposure of the hydrophobic groove on the HIV-1 gp41 glycoprotein at concentrations comparable to those exerting suppressive effects on HIV-1 envelope glycoprotein function.
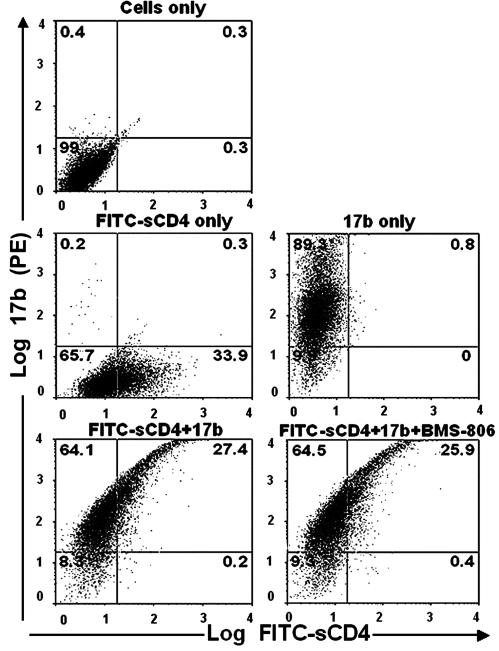
BMS-806 Does Not Inhibit 17b Binding to the Native YU2 HIV-1 Envelope Glycoproteins. The binding of CD4 to the HIV-1 envelope glycoproteins results in the exposure and/or the formation of gp120 epitopes recognized by CD4-induced antibodies (27, 31, 32). These epitopes overlap the binding sites for the chemokine receptors. Because this induction is also downstream of CD4 binding and has been shown to contribute to virus entry (9, 10, 36), we tested whether BMS-806 and related compounds affect the binding of a CD4-induced antibody, 17b, to the native HIV-1 envelope glycoprotein complex. Because 17b is a human monoclonal antibody, it can be conveniently substituted for C34-Ig in the two-color flow cytometry assay. As can be seen in Fig. 4, FITC-sCD4 and 17b individually bound envelope-expressing cells; ≈34% of the cells detectably bound CD4 under the conditions used. Simultaneous addition of FITC-sCD4 and 17b resulted in two major populations, one CD4- 17b+ and the other CD4+ 17b+. Staining with 17b was ≈10-fold brighter in the CD4+ subset of cells than in the CD4- subset, consistent with the ability of CD4 to induce the binding of 17b to gp120. In contrast to the effect seen above on C34-Ig binding, addition of BMS-806 had no significant effect on the pattern of fluorescence of the cells. Experiments with #151 and #155 yielded similar results (data not shown). We conclude that, at biologically relevant concentrations, BMS-806 and related compounds do not interfere with either CD4 or 17b binding to the native, trimeric HIV-1 envelope glycoproteins.
Fig. 4.
BMS-806 does not inhibit sCD4 or 17b antibody binding to the HIV-1 envelope glycoproteins. Cells (293T) expressing the cytoplasmic tail-deleted YU2 HIV-1 envelope glycoproteins were incubated with the indicated molecules and then analyzed for sCD4 binding (FITC) and 17b antibody binding (PE) by two-color flow cytometry. The concentration of BMS-806 was 1 μM. The percentage of cells in each quadrant is indicated. The experiment was repeated four times with similar results. The mean PE fluorescence in the absence of FITC-sCD4 was ≈250; in the presence of FITC-sCD4, the mean PE fluorescence was ≈150 in the FITC-negative population and ≈1,900 in the FITC-positive population.
BMS-806 Does Not Directly Interfere with HR2-HR1 Interactions. It is formally possible that BMS-806 could directly interfere with the interaction between the HR2 portion of C34-Ig and the exposed HR1 grooves in gp41. To investigate this possibility, sCD4 was absorbed on an ELISA plate and used to capture soluble YU2 gp130 envelope glycoprotein trimers. The soluble gp130 trimer lacks the HR2 region of gp41 and readily interacts with peptides corresponding to the HR2 region (26). BMS-806, #151, and #155 did not significantly inhibit the binding of C34-Ig to the soluble gp130 trimers at up to 10 μM concentrations (Fig. 8, which is published as supporting information on the PNAS web site). We conclude that these compounds do not directly interfere with HR2-HR1 interactions.
Discussion
BMS-806 and related compounds bind the HIV-1 gp120 exterior envelope glycoprotein and specifically and potently inhibit HIV-1 entry. Our data agree with those of Lin et al. (23) and Guo et al. (24) on these important points. Several other gp120-directed inhibitors have been reported, but are either significantly larger than BMS-806 (e.g., CD4 miniproteins and 12p1 peptide) (33, 37) or bind multiple sites on gp120 because of electrostatic or lectin-carbohydrate interactions (e.g., sulfated polymers and cyanovirin) (38, 39). These considerations make BMS-806 much more attractive as a potential antiviral drug and as a probe to study HIV-1 entry.
Our results do not support the mechanistic model proposed by Lin et al. (23) and Guo et al. (24), suggesting that BMS-806 inhibits virus-CD4 interaction. In our hands, BMS-806 did not significantly block either CD4 or chemokine receptor binding, even at concentrations 100-fold higher than concentrations at which HIV-1 infection or syncytium formation were inhibited. In a key experiment addressing the mechanism of action, Lin et al. reported that BMS-806 inhibited infection by a CD4-independent HIV-1 only when CD4 was expressed on the target cells. By contrast, we observed potent BMS-806 inhibition of infection of CD4-expressing and CD4-negative target cells by viruses with an envelope glycoprotein variant identical to that used by Lin et al. (23). Importantly, we obtained similar results with another CD4-independent virus. The observed inhibition was specific, based on the lack of inhibitory effect on viruses with the vesicular stomatitis virus glycoprotein. Moreover, the IC50 values observed for these CD4-independent viruses were in the same range as those seen for other HIV-1 variants. Unlike the single-round viruses used in our study, the replication-competent and extensively passaged viruses used by Guo et al. (24) may have acquired env sequence changes that reduced sensitivity to BMS-806. Indeed, even in the CD4-expressing target cells, they achieved only 80% inhibition of their virus at very high (20 μM) concentrations of BMS-806. These considerations, in addition to the minimal effect of even high concentrations of BMS-806 on routine assays for CD4-gp120 interaction, make blockade of CD4 binding an extremely unlikely mechanism of drug activity.
CD4 binding is thought to lock the conformationally flexible free gp120 glycoprotein into a more rigid state that is competent for chemokine receptor binding (30). BMS-806 binding does not introduce a high degree of order into free gp120, nor does it preclude CD4 from doing so. Thus, the CD4-induced gp120 conformational adjustments required for coreceptor binding are not blocked by BMS-806 and related molecules.
The realization that BMS-806 blocks HIV-1 entry yet allows receptor binding implies that events critical to virus entry that follow the interaction of gp120 with CD4 and/or the chemokine receptor are disrupted by this compound. We established an assay that monitors CD4-induced conformational changes in the gp120-gp41 relationship, resulting in the creation and/or exposure of the gp41 HR1 region coiled coil. The exposure of this element after CD4 binding could allow the subsequent interaction with the gp41 HR2 region to form the six-helix bundle, a process that is thought to promote viral and target cell membrane fusion (4-6). Importantly, in contrast to previous studies (19, 20), we have designed our assay to rule out gp120 shedding as an explanation for CD4-induced exposure of gp41 moieties. The relevance of CD4-induced gp120 shedding to HIV-1 entry is doubtful, given the documented necessity of chemokine receptor binding to infection (11-15); gp120 shedding may represent an exaggerated or aberrant response to CD4 binding that ultimately inactivates the envelope glycoprotein spike (16). The use of subsaturating concentrations of sCD4 and room temperature incubation probably minimize the amount of gp120 shedding in our assay system.
The ability of small, potent inhibitors of HIV-1 entry to block the CD4-induced creation and/or exposure of the gp41 HR1 coiled coil supports the importance of this event for envelope glycoprotein function. We propose that CD4 binding results in two parallel pathways of conformational change in the HIV-1 envelope glycoproteins. One set of changes, involving the V2 variable loop and the core of gp120 (3, 36), results in competence for chemokine receptor binding. These changes do not depend on proteolytic cleavage of the gp160 envelope glycoprotein precursor and are not inhibited by BMS-806 and related compounds. A second set of changes involves the alteration of the gp120-gp41 relationship, with the end result being the exposure of the gp41 HR1 coiled coil. These changes depend on cleavage at the gp120-gp41 junction and are inhibited by BMS-806 and related molecules. The exposure of the hydrophobic groove of the gp41 HR1 coiled coil after CD4 binding is consistent with previous studies that have examined the ability of HR2-like peptides to inhibit HIV-1 entry and/or syncytium formation (17, 40). Additional studies on the binding site of BMS-806 should provide insights into the structural basis for the relevant conformational changes induced by CD4.
BMS-806 potently inhibited the infection of CCR5-expressing cells lacking CD4 by a CD4-independent virus. How might this observation be reconciled with a model in which BMS-806 inhibits a CD4-induced alteration in gp120-gp41 conformation? We suggest that negotiating this conformational transition is critical for successful HIV-1 entry, regardless of the involvement of CD4 in the process. Whereas CD4-dependent viruses rely on CD4 binding to promote this conformational change, CD4-independent viruses may employ chemokine receptor binding to trigger the change or may spontaneously undergo the change. The latter possibility would require that CD4-independent envelope glycoproteins have structural modifications that lower the activation barrier to the requisite conformational transitions.
At high concentrations, BMS-806 and related compounds partially inhibited the binding of CCR5 and CD4-induced antibodies to monomeric gp120. These effects occurred at concentrations significantly higher than those required for inhibition of envelope glycoprotein function, did not occur in the presence of CD4, and were not evident when native trimeric envelope glycoproteins were studied. Therefore, it is unlikely that modulation of chemokine receptor binding contributes to the anti-viral activity of BMS-806. However, BMS-806 binding in the absence of CD4 may exert subtle effects on the bridging sheet or adjacent β19 strand of gp120, which have been implicated in the interaction with chemokine receptors and CD4-induced antibodies (31, 32).
The search for virus-directed, low-molecular-weight inhibitors of HIV-1 receptor binding has been frustrating, despite extensive searches. Our results underscore the vulnerability of postreceptor binding conformational changes in the HIV-1 envelope glycoproteins to inhibition by small molecules. The development of chemical library screens that incorporate these conformational changes may allow the identification of novel inhibitors of HIV-1 entry.
Supplementary Material
Acknowledgments
We thank Ms. Yvette McLaughlin and Ms. Sheri Farnum for manuscript preparation. This work was supported by National Institutes of Health Grants AI24755, AI39420, and PO1-GM56550; the Bristol-Myers Squibb Foundation; the International AIDS Vaccine Initiative; and the late William F. McCarty-Cooper. Z.S. was supported in part by pilot project AI28691 from the Dana-Farber Cancer Institute/Children's Hospital Boston/Beth Israel Deaconess Medical Center Center for AIDS Research. N.M. was supported by National Research Service Award Postdoctoral Fellowship F32 NS43260M from the National Institutes of Health. J.M.C. was supported by American Cancer Society Postdoctoral Fellowship PF-03-005-01-CDD and the National Fisheries Institute.
Abbreviations: HR, heptad repeat; sCD4, soluble CD4; PE, phycoerythrin.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Barre-Sinoussi, F., Chermann, J. C., Rey, F., Nugeyre, M. T., Chamaret, S., Gruest, J., Dauguet, C., Axler-Blin, C., Vezinet-Brun, F., Rouzioux, C., et al. (1983) Science 220, 868-871. [DOI] [PubMed] [Google Scholar]
- 2.Gallo, R. C., Salahuddin, S. Z., Popovic, M., Shearer, G. M., Kaplan, M., Haynes, B. F., Palker, T. J., Redfield, R., Oleske, J., Safai, B., et al. (1984) Science 224, 500-503. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt, R. & Sodroski, J. (1998) Science 280, 1884-1888. [DOI] [PubMed] [Google Scholar]
- 4.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263-273. [DOI] [PubMed] [Google Scholar]
- 5.Tan, K., Liu, J., Wang, J., Shen, S. & Lu, M. (1997) Proc. Natl. Acad. Sci. USA 94, 12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. (1997) Nature 387, 426-430. [DOI] [PubMed] [Google Scholar]
- 7.Dalgleish, A. G., Beverley, P. C., Clapham, P. R., Crawford, D. H., Greaves, M. F. & Weiss, R. A. (1984) Nature 312, 763-767. [DOI] [PubMed] [Google Scholar]
- 8.Klatzmann, D., Champagne, E., Chamaret, S., Gruest, J., Guetard, D., Hercend, T., Gluckman, J. C. & Montagnier, L. (1984) Nature 312, 767-768. [DOI] [PubMed] [Google Scholar]
- 9.Wu, L., Gerard, N. P., Wyatt, R., Choe, H., Parolin, C., Ruffing, N., Borsetti, A., Cardoso, A. A., Desjardin, E., Newman, W., et al. (1996) Nature 384, 179-183. [DOI] [PubMed] [Google Scholar]
- 10.Trkola, A., Dragic, T., Arthos, J., Binley, J. M., Olson, W. C., Allaway, G. P., Cheng-Mayer, C., Robinson, J., Maddon, P. J. & Moore, J. P. (1996) Nature 384, 184-187. [DOI] [PubMed] [Google Scholar]
- 11.Feng, Y., Broder, C. C., Kennedy, P. E. & Berger, E. A. (1996) Science 272, 872-877. [DOI] [PubMed] [Google Scholar]
- 12.Choe, H., Farzan, M., Sun, Y., Sullivan, N., Rollins, B., Ponath, P. D., Wu, L., Mackay, C. R., LaRosa, G., Newman, W., et al. (1996) Cell 85, 1135-1148. [DOI] [PubMed] [Google Scholar]
- 13.Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., Di Marzio, P., Marmon, S., Sutton, R. E., Hill, C. M., et al. (1996) Nature 381, 661-666. [DOI] [PubMed] [Google Scholar]
- 14.Doranz, B. J., Rucker, J., Yi, Y., Smyth, R. J., Samson, M., Peiper, S. C., Parmentier, M., Collman, R. G. & Doms, R. W. (1996) Cell 85, 1149-1158. [DOI] [PubMed] [Google Scholar]
- 15.Dragic, T., Litwin, V., Allaway, G. P., Martin, S. R., Huang, Y., Nagashima, K. A., Cayanan, C., Maddon, P. J., Koup, R. A., Moore, J. P. & Paxton, W. A. (1996) Nature 381, 667-673. [DOI] [PubMed] [Google Scholar]
- 16.Moore, J. P., McKeating, J. A., Weiss, R. A. & Sattentau, Q. J. (1990) Science 250, 1139-1142. [DOI] [PubMed] [Google Scholar]
- 17.Wild, C. T., Shugars, D. C., Greenwell, T. K., McDanal, C. B. & Matthews, T. J. (1994) Proc. Natl. Acad. Sci. USA 91, 9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuta, R. A., Wild, C. T., Weng, Y. & Weiss, C. D. (1998) Nat. Struct. Biol. 5, 276-279. [DOI] [PubMed] [Google Scholar]
- 19.Koshiba, T. & Chan, D. C. (2003) J. Biol. Chem. 278, 7573-7579. [DOI] [PubMed] [Google Scholar]
- 20.He, Y., Vassell, R., Zaitseva, M., Nguyen, N., Yang, Z., Weng, Y. & Weiss, C. D. (2003) J. Virol. 77, 1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattentau, Q. J., Moore, J. P., Vignaux, F., Traincard, F. & Poignard, P. (1993) J. Virol. 67, 7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, T., Zhang, Z., Wallace, O. B., Deshpande, M., Fang, H., Yang, Z., Zadjura, L. M., Tweedie, D. L., Huang, S., Zhao, F., et al. (2003) J. Med. Chem. 46, 4236-4239. [DOI] [PubMed] [Google Scholar]
- 23.Lin, P.-F., Blair, W., Wang, T., Spicer, T., Guo, Q., Zhou, N., Gong, Y.-F., Wang, H.-G. H., Rose, R., Yamanaka, G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 11013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, Q., Ho, H.-T., Dicker, I., Fan, L., Zhou, N., Friborg, J., Wang, T., McAuliffe, B. V., Wang, H.-G. H., Rose, R. E., et al. (2003) J. Virol. 77, 10528-10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si, Z., Cayabyab, M. & Sodroski, J. (2001) J. Virol. 75, 4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, X., Florin, L., Farzan, M., Kolchinsky, P., Kwong, P. D., Sodroski, J. & Wyatt, R. (2000) J. Virol. 74, 4746-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore, J. P. & Sodroski, J. (1996) J. Virol. 70, 1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korber, B., Foley, B., Kuiken, C., Pillai, S. & Sodroski, J. (1998) in Human Retroviruses and AIDS 1998 (Los Alamos Natl. Lab., Los Alamos, NM).
- 29.Kolchinsky, P., Mirzabekov, T., Farzan, M., Kiprilov, E., Cayabyab, M., Mooney, L. J., Choe, H. & Sodroski, J. (1999) J. Virol. 73, 8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myszka, D. G., Sweet, R. W., Hensley, P., Brigham-Burke, M., Kwong, P. D., Hendrickson, W. A., Wyatt, R., Sodroski, J. & Doyle, M. L. (2000) Proc. Natl. Acad. Sci. USA 97, 9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rizzuto, C. D., Wyatt, R., Hernandez-Ramos, N., Sun, Y., Kwong, P. D., Hendrickson, W. A. & Sodroski, J. (1998) Science 280, 1949-1953. [DOI] [PubMed] [Google Scholar]
- 32.Xiang, S.-H., Wang, L., Abreu, M., Huang, C.-C., Kwong, P. D., Rosenberg, E., Robinson, J. E. & Sodroski, J. (2003) Virology 315, 124-134. [DOI] [PubMed] [Google Scholar]
- 33.Ferrer, M. & Harrison, S. C. (1999) J. Virol. 73, 5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaBranche, C. C., Hoffman, T. L., Romano, J., Haggarty, B. S., Edwards, T. G., Matthews, T. J., Doms, R. W. & Hoxie, J. A. (1999) J. Virol. 73, 10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si, Z., Phan, N., Kiprilov, E. & Sodroski, J. (2003) AIDS Res. Hum. Retroviruses 19, 217-226. [DOI] [PubMed] [Google Scholar]
- 36.Sullivan, N., Sun, Y., Sattentau, Q., Thali, M., Wu, D., Denisova, G., Gershoni, J., Robinson, J., Moore, J. & Sodroski, J. (1998) J. Virol. 72, 4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vita, C., Drakopoulou, E., Vizzavona, J., Rochette, S., Martin, L., Menez, A., Roumestand, C., Yang, Y. S., Ylisastigui, L., Benjouad, A. & Gluckman, J. C. (1999) Proc. Natl. Acad. Sci. USA 96, 13091-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moulard, M., Lortat-Jacob, H., Mondor, I., Roca, G., Wyatt, R., Sodroski, J., Zhao, L., Olson, W., Kwong, P. D. & Sattentau, Q. J. (2000) J. Virol. 74, 1948-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esser, M. T., Mori, T., Mondor, I., Sattentau, Q. J., Dey, B., Berger, E. A., Boyd, M. R. & Lifson, J. D. (1999) J. Virol. 73, 4360-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wild, C., Greenwell, T. & Matthews, T. (1993) AIDS Res. Hum. Retroviruses 9, 1051-1053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.