Abstract
BACKGROUND
Patients with ulcerative colitis (UC) are at risk of developing colorectal cancer. We have previously reported that cancer progression is associated with the presence of clonal expansions and shorter telomeres in non-dysplastic mucosa. We sought to validate these findings in an independent case-control study.
METHODS
This study included 33 patients with UC: 14 Progressors (patients with high-grade dysplasia or cancer) and 19 Non-Progressors. For each patient, a mean of 5 non-dysplastic biopsies from proximal, mid, and distal colon were assessed for clonal expansions, as determined by clonal length altering mutations in polyguanine tracts, and telomere length, as measured by Quantitative-PCR. Both parameters were compared with individual clinico-pathological characteristics.
RESULTS
Clonal expansions and shorter telomeres were more frequent in non-dysplastic biopsies from UC Progressors than Non-Progressors, but only for patients with early-onset of UC (diagnosis at less than 50 years of age). Late-onset Progressor patients had very few or no clonal expansions and longer telomeres. A few Non-Progressors exhibited clonal expansions, which were associated with older age and shorter telomeres. In Progressors, clonal expansions were associated with proximity to dysplasia. The mean percentage of clonally expanded mutations distinguished early-onset Progressors from Non-Progressors with 100% sensitivity and 80% specificity.
CONCLUSIONS
Early-onset Progressors develop cancer in a field of clonally expanded epithelium with shorter telomeres. The detection of these clones in a few random non-dysplastic colon biopsies is a promising cancer biomarker in early-onset UC. Curiously, late-onset UC patients appear to develop cancer without the involvement of such fields.
Keywords: inflammatory bowel disease, preneoplasia, field effect, cancer biomarker, age of onset
INTRODUCTION
Ulcerative colitis (UC) is a chronic inflammatory disease that affects approximately 593,000 individuals in the United States and its prevalence is steadily increasing (1, 2). It has two peaks of incidence: one between 25 and 35 years of age (early-onset) and another one between 55 and 65 years of age (late-onset) (2). It is widely accepted that patients with UC have an increased risk of colorectal cancer, although recent epidemiological studies suggest that the increased risk is limited to patients with early age of onset, extensive disease and long disease duration (3, 4) A variety of other important clinical differences between UC patients with early-onset and late-onset of disease have been reported. Late-onset patients appear to have a less aggressive clinical course (5) and better response to therapy (6). However, if neoplasia develops, it tends to arise in fewer years than in early-onset patients (7) and it tends to be localized instead of widespread (8). Given the aging of the population and the increasing prevalence of UC, it is crucial to determine the molecular pathways that underlie these differences in order to develop cancer biomarkers that can inform the clinical management of these patients. Tumorigenesis in UC is a multistep process that involves progression through low-grade dysplasia (LGD), high-grade dysplasia (HGD), and cancer (9). Current guidelines recommend that UC patients with more than 8 years of disease duration undergo colonoscopy every 1–2 years for early detection of dysplasia (10). Because UC-associated dysplasia is often flat and difficult to visually identify, 4-quadrant biopsies from every 10cm of colon are typically collected at colonoscopy for histological examination (10). This intensive surveillance protocol is time-consuming, expensive and uncomfortable for the patient. Moreover, it is biased towards identifying larger, more advanced neoplasia, given the probabilistic nature of arbitrary sampling. UC patients would greatly benefit from more sensitive biomarkers of cancer risk that could detect preneoplastic alterations that precede the appearance of histological changes.
We recently developed a technique that enables the detection of preneoplastic clonal expansions in histologically normal colon biopsies from UC patients with cancer elsewhere in the colon (11). The technique is based on the identification of clonal mutations in polyguanine tracts, which are short series of guanine repeats found mostly in intergenic and intronic areas of the genome. These repetitive tracts are highly prone to insertions and deletions, with a mutation rate on the order of ~10−4 per cell generation (12). Because they are mutated so readily, individual patterns of mutation can serve as a convenient marker of cell lineage. When a polyguanine mutation is detected in a colon biopsy, it indicates the presence of a clonal expansion, that is, an abnormal proliferation of cells. Most of these mutations do not confer any specific proliferative advantage, but rather, are simply inherited by daughter cells as ‘passengers’ on sweeps of clonal evolution driven by mutations and epimutations elsewhere in the genome. In our initial study, we found genetically related clonal expansions in non-dysplastic biopsies both immediately surrounding, and distant from cancer, suggesting that such clonal fields precede dysplasia in UC (11). Moreover, these preneoplastic clones were rarely found in patients without cancer, making them an attractive biomarker of concurrent, and potentially future, cancer in UC.
The presence of molecular alterations in histologically normal epithelium that surrounds cancer is known as ‘field cancerization’ or ‘field effect’ (13). We previously demonstrated that in UC, such fields comprise cells harboring aneuploidy, p53 mutations, and chromosomal instability (14–16), and can extend many centimeters along the colon (16). These fields are also characterized by shorter telomeres, increased DNA damage and increased senescence, which occurs in association with chronic inflammation (17). Our working hypothesis is that inflammation leads to telomere shortening through reactive oxygen species, which damage and shorten telomeres (18). Shorter telomeres lead to senescence in the presence of proficient checkpoints (p53, p16) (19) but, in the absence of checkpoints, lead to fusion-bridge-breakage cycles and self-perpetuating chromosomal instability (20). In this model, telomere shortening is an early event that might precede clonal outgrowth, but it is also likely that clonal expansion accelerates telomere shortening due to the end-replication problem. Regardless of the order of events, clonal expansions (11) and shorter telomeres (17, 20) are more frequent in non-dysplastic biopsies of UC patients with high grade dysplasia or cancer (UC Progressors) than UC patients without (UC Non-Progressors). The aim of this study is to validate those results in an independent case-control study and to explore the associations between clonal expansion, telomere shortening, and the clinico-pathological characteristics of UC patients to further characterize their potential as clinical biomarkers.
MATERIAL AND METHODS
Patients and biopsies
Thirty-three patients with UC who underwent colectomy at the Cleveland Clinic between 2005 and 2009 were included in this study. The clinico-pathological characteristics of the patients are presented in Table 1. Nineteen patients without HGD or cancer were classified as “Non-Progressors”. Fourteen patients with at least one biopsy with HGD or cancer were classified as “Progressors”. The indication for colectomy in UC Progressors was a diagnosis of HGD or cancer at colonoscopy, whereas for UC Non-Progressors was intractability of symptoms. All patients had documented pancolitis except for seven where information on extent of disease was unavailable. Only one patient presented with primary sclerosing colangitis. Biopsies obtained at colectomy were divided, with one half frozen at −70°C in Minimal Essential Medium with 10% DMSO and the other fixed and sectioned for histology. Diagnosis of dysplasia was performed by an expert GI pathologist (MPB). For each patient, 3–8 non-dysplastic biopsies (mean 5) obtained from distal, mid and proximal colon were analyzed, totaling 188 biopsies. For all biopsies, the distance to rectum and site of nearest HGD or cancer was recorded. For Progressors, an additional 9 dysplastic biopsies were collected for separate analysis. For all colons in the study, a muscularis sample was collected for defining normal germline genotype. All protocols were approved by Human Subjects Review Boards at the Cleveland Clinic and University of Washington.
Table 1.
Patient and biopsy information
| Patient number |
Patient ID |
NP/P | Higuest grade |
Gender | Age (years) |
Age onset disease |
Disease duration (years) |
Total biopsies non- dysplastic |
Biopsy location | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Distal colon |
Mid colon |
Proximal colon |
|||||||||
| 1 | PG10 | NP | NEG | F | 23 | 19 | 3.7 | 5 | 2 | 1 | 2 |
| 2 | PG19 | NP | NEG | M | 24 | 21 | 2.75 | 5 | 2 | 1 | 2 |
| 3 | PG1 | NP | NEG | M | 27 | 20 | 7 | 5 | 2 | 1 | 2 |
| 4 | PG14 | NP | NEG | M | 36 | 35 | 1.5 | 5 | 2 | 1 | 2 |
| 5 | PG7 | NP | NEG | F | 38 | 36 | 2 | 5 | 2 | 1 | 2 |
| 6 | PG16 | NP | NEG | F | 45 | 45 | 0.4 | 5 | 2 | 1 | 2 |
| 7 | PG18 | NP | NEG | M | 29 | 21 | 8 | 5 | 2 | 1 | 2 |
| 8 | PG41 | NP | NEG | M | 31 | 23 | 8 | 7 | 3 | 2 | 2 |
| 9 | PG36 | NP | NEG | F | 41 | 21 | 20 | 7 | 2 | 2 | 3 |
| 10 | PG13 | NP | NEG | M | 43 | 26 | 17 | 5 | 2 | 1 | 2 |
| 11 | PG42 | NP | NEG | M | 45 | 25 | 20 | 7 | 3 | 2 | 2 |
| 12 | PG30 | NP | NEG | M | 48 | 39 | 9 | 7 | 3 | 2 | 2 |
| 13 | PG48 | NP | NEG | F | 49 | 29 | 20 | 7 | 3 | 2 | 2 |
| 14 | PG26 | NP | NEG | F | 56 | 46 | 10 | 7 | 3 | 2 | 2 |
| 15 | PG37 | NP | NEG | M | 63 | 24 | 38.7 | 7 | 3 | 2 | 2 |
| 16 | PG22 | NP | NEG | M | 67 | 37 | 30 | 7 | 3 | 2 | 2 |
| 17 | PG4 | NP | NEG | F | 60 | 58 | 2 | 5 | 2 | 1 | 2 |
| 18 | PG2 | NP | NEG | M | 65 | 64 | 1.25 | 5 | 2 | 1 | 2 |
| 19 | PG24 | NP | NEG | F | 70 | 54 | 16 | 6 | 2 | 2 | 2 |
| 20 | PG28 | P | CA | M | 45 | 39 | 6 | 8 | 4 | 2 | 2 |
| 21 | PG29 | P | CA | M | 31 | 21 | 10 | 6 | 4 | 1 | 1 |
| 22 | PG9 | P | CA | F | 39 | 19 | 20 | 5 | 3 | 1 | 1 |
| 23 | PG12 | P | CA | M | 47 | 27 | 20 | 4 | 2 | 1 | 1 |
| 24 | PG25 | P | CA | F | 55 | 35 | 20 | 8 | 4 | 2 | 2 |
| 25 | PG8 | P | HGD | F | 57 | 12 | 45 | 5 | 3 | 1 | 1 |
| 26 | PG5 | P | HGD | F | 63 | 40 | 23 | 3 | 2 | 1 | NA |
| 27 | PG15 | P | CA | M | 66 | 19 | 47 | 5 | 3 | 1 | 1 |
| 28 | PG31 | P | CA | M | 67 | 37 | 30 | 8 | 4 | 2 | 2 |
| 29 | PG20 | P | CA | M | 66 | 53 | 13 | 5 | 2 | 1 | 2 |
| 30 | PG3 | P | HGD | M | 69 | 58 | 11 | 4 | 2 | 1 | 1 |
| 31 | PG17 | P | CA | F | 71 | 64 | 7.5 | 5 | 2 | 2 | 1 |
| 32 | PG6 | P | CA | F | 78 | 73 | 5 | 5 | 2 | 2 | 1 |
| 33 | PG11 | P | CA | M | 86 | 81 | 5 | 5 | 2 | 2 | 1 |
Abbreviations: NP: Non Progressor, P: Progressor, NEG: negative for dysplasia, CA: cancer, HGD: high-grade dysplasia, F: female, M: male, NA: non-available.
Epithelial cell isolation and DNA extraction
Epithelial cells from frozen biopsies were isolated by EDTA shake off as previously described (16). This method yields 90% enrichment for epithelial cells. DNA was extracted by silica filtration columns (Qiagen) and quantified by Nanodrop UV spectroscopy (Thermo Scientific).
Polyguanine Tract-Length Genotyping
Genotyping of the 17 polyguanine loci listed in Figure, Supplemental Digital Content 1, http://links.lww.com/IBD/A290 was carried out for each sample in a blinded fashion using the primers and protocol previously described (11). Briefly, 2 ng of DNA was used to seed PCR reactions with each fluorescently-labeled primer set on robotically loaded 384 well plates. PCR products of compatible amplicon lengths and labels were appropriately diluted and combined for multiplex capillary fragment length analysis on an ABI 3730xl instrument. Resulting electropherograms were analyzed through a combination of automated software and caller-blinded visual assessment. Mutations were defined as genotypes differing from a patient’s corresponding germline genotype obtained from a muscularis biopsy. Each genotyping reaction was carried out in triplicate and the small number of ungenotypable amplification or sequencing failures (1.7%) were marked as such and not included in subsequent mutation frequency calculations.
Telomere length quantification
Telomere length was measured by Quantitative PCR (Q-PCR) as previously described (21), with minor modifications. The analysis was performed on the same DNA previously used for polyguanine genotyping. For each sample, two PCRs were performed: the first to amplify the telomeric DNA and the second to amplify a single-copy control gene (36B4, acidic ribosomal phosphoprotein PO). A four-point standard curve (2–fold serial dilutions from 5 to 0.625ng of DNA) was included in all PCRs to allow the transformation of Ct (cycle threshold) into nanograms of DNA. All PCR reactions were performed in a Rotor-Gene Q (Qiagen) using 100-well discs and a reaction volume of 10ul. Each reaction included 1X Rotor Gene Sybrgreen PCR master mix (Qiagen), 2.5ng of DNA, and either 500nM of each telomere primer or 1uM of each control gene primer. All samples were run in triplicate and the median was used for subsequent calculations. The amount of telomeric DNA was divided by the amount of control-gene DNA, producing a relative measurement of the telomere length of the sample. Two control samples were run in each experiment to allow for normalization between experiments and periodic reproducibility experiments were performed to guarantee correct measurements. The inter-assay variability (coefficient of variation) for this assay was 7%.
Statistical Analysis
Because multiple biopsies were analyzed for each patient, patients were first compared based on their mean values of percentage polyguanine mutations and telomere length. These associations were assessed using the t-test and linear regression. Second, patients were compared based on the individual values of all the biopsies analyzed. For this analysis, the generalized estimating equations (GEE) method was used for regression as it accounts for correlation among multiple biopsies from a given patient. The discriminatory ability of the percentage of mutations and telomere length to classify patients as Progressors or Non-Progressors was evaluated using receiver operating characteristic (ROC) curves. Classification accuracy was quantified using the area under the ROC curve (AUC) and percentile-based 95% confidence intervals were constructed by bootstrapping the data 1000 times. Age was controlled as a confounder by estimating covariate-adjusted ROC curves (22). Optimal thresholds were obtained as the values that maximized sensitivity for a specificity of 80% or greater. Statistical analyses were performed using Stata (version 10) and R statistical software (version 2.14.1). All reported p-values were two-sided at an alpha level of 0.05.
RESULTS
Clonal expansions and telomere length characterize different subgroups of UC Progressors and Non-Progressors
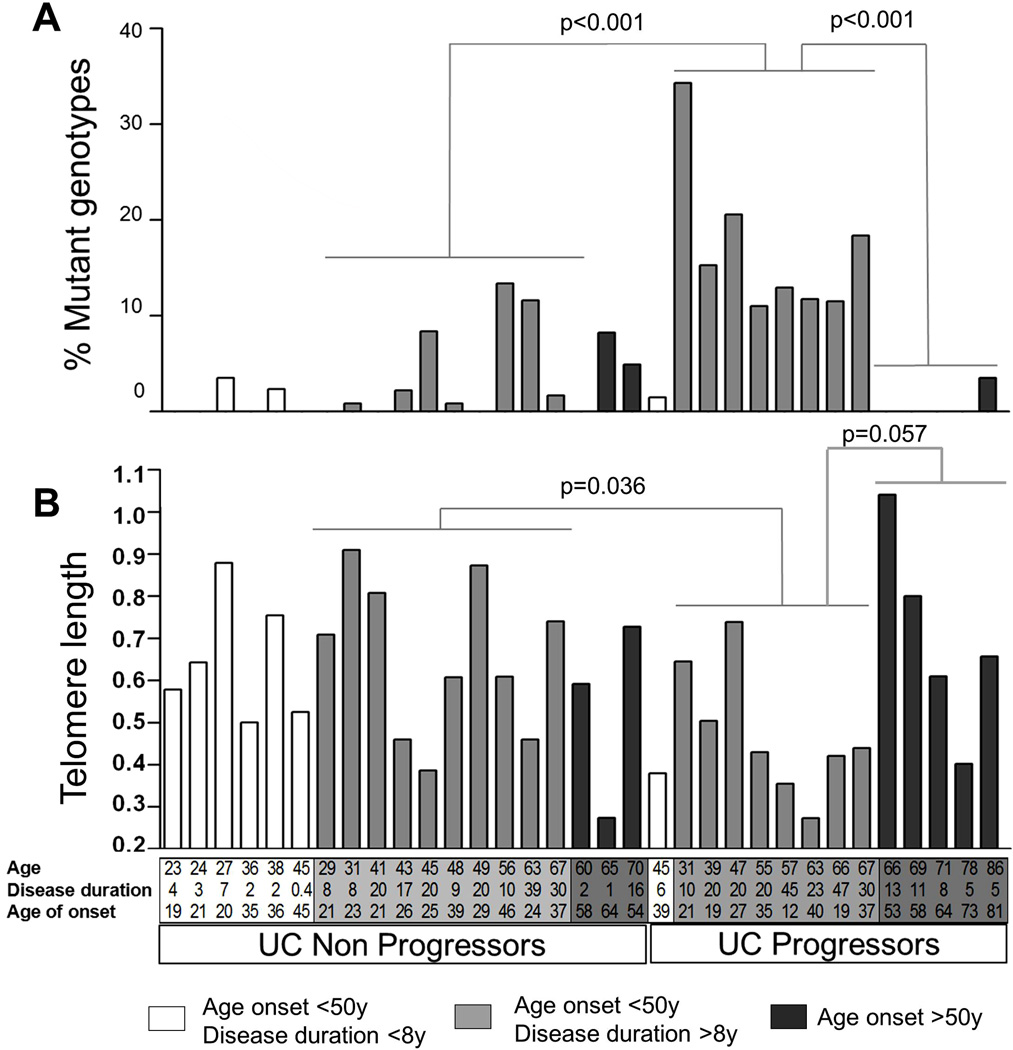
We previously observed many polyguanine mutation-defined clones in the non-dysplastic mucosa of UC Progressors, but not in UC Non-Progressors (11), and here sought to validate these results in an independent cohort. A complete listing of clonal mutations found at each of the 17 polyguanine loci analyzed in DNA from non-dysplastic colonic epithelium of 33 independent UC patients is tabulated in Figure, Supplemental Digital Content 1, http://links.lww.com/IBD/A290. The percentage of mutant genotypes for each biopsy was calculated as the number of loci mutated divided by the number of loci genotyped. Figure 1 summarizes the mean percentage of mutations (Fig 1A) and mean telomere length (Fig 1B), both calculated as the average of all non-dysplastic biopsies analyzed for from each patient. UC Progressors and Non-Progressors are shading-coded according to their age of onset of disease and years of disease duration. As is readily observed, mutations were most frequent in UC Progressor colons, but only in those with early-onset of disease and long disease duration. The cut off point of late-onset of disease at 50 years of age is based on recent data regarding UC incidence in a US cohort (2) and the study of Ha et al. (6), which reported important clinical differences between late-onset (>50 years at diagnosis) and early-onset UC patients. The cut off point for long disease duration is 8 years, which is the clinically established starting time for colonoscopic surveillance based on previous epidemiological studies (23). On average, early-onset UC Progressors with long disease duration displayed four times more mutations than early-onset UC Non-Progressors with long disease duration (17% vs 3.9%, t-test p<0.001, Fig 1A). Moreover, UC Progressors with late-onset disease displayed an average of only 0.71% mutations per biopsy, 24 times less than the Progressors with early-onset disease (t-test p<0.001, Fig 1A). These groups also differed in terms of their telomere length. Telomeres tended to be shorter in early-onset, long-disease duration UC Progressors than in both early-onset, long-disease duration Non-Progressors (0.476 vs. 0.656, t-test p=0.036), and late-onset UC Progressors (0.476 vs. 0.702, t-test p=0.057).
Figure 1. Clonal expansions and telomere length in UC patients.
For each patient, mean clonal mutations (A) and mean telomere length (B) were calculated as the mean of all non-dysplastic biopsies analyzed per patient (mean 5, range 3–8). The same biopsies were analyzed in both assays. Age, disease duration (years), and age of onset are indicated for each patient on the X-axes. Patients are grouped by Non Progressor and Progressor status and shading-coded to facilitate group comparisons. White: patients with age of onset <50 years and disease duration <8 years; light gray: patients with age of onset <50 years and disease duration >8 years; dark grey: patients with age of onset >50 years. Patients without bars on Figure 1A do not have any mutant genotypes. p-values correspond to t- tests.
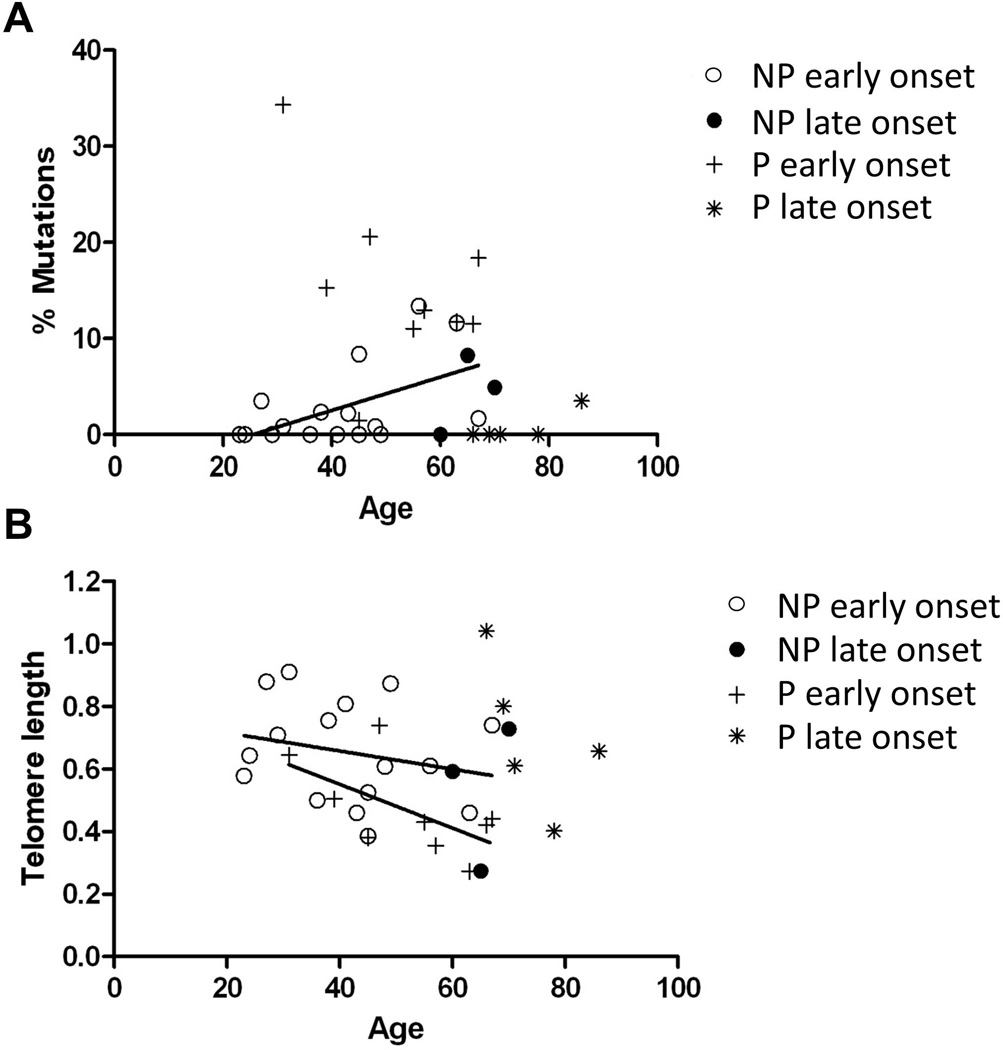
The presence of mutations in early-onset UC Non-Progressors was associated with older age (Fig 2A, Pearson r=0.520, p=0.039). This is in agreement with a variety of evidence indicating that, with age, mutations accumulate in various human organs and tissues, including colon (24). While aging is also expected to shorten telomeres, the observed trend was not statistically significant for either early-onset Non-Progressors or Progressors (Fig 2B). Interestingly, late-onset UC Progressors showed remarkably low levels of mutations and long telomeres in spite of being the eldest patients in the study.
Figure 2. Association between clonal expansions (A) and telomere length (B) with age.
The regression between percentage of mutations and age for Non-Progressors with early age of onset is significant (p=0.037 by Pearson test). The regressions between telomere length and age for Non-Progressors and Progressors with early-onset of disease had the expected trend (shorter telomeres with older age), but were not significant.
In order to eliminate any potential confounding by age, we used age-adjusted ROC curves to analyze the ability of the percentage of mutations and telomere length to distinguish UC Progressors and Non-Progressors (Table 2). Late-onset patients were excluded from this analysis because progression in these patients is not associated with clonal expansions or shorter telomeres, as noted above. We also excluded patients with less than 8 years disease duration because these patients have no epidemiologically increased risk of colorectal cancer (23) (accordingly, they showed very few clonal expansions) and were unfairly overrepresented in the Non Progressor group. Thus, the biomarker analysis was restricted to early-onset UC patients with more than 8 years of disease duration (10 Non-Progressors and 8 Progressors). These two groups of patients could be distinguished with 100% sensitivity (8/8) and 80% specificity (8/10) using a threshold of 10% average frequency of mutations. Telomere length was a weaker classifier, as there was no optimal threshold to maximize sensitivity without compromising specificity.
Table 2.
Comparison of age-adjusted ROC models based on the analysis of polyguanine mutations and telomere length
| A. All Biopsies | ||||
|---|---|---|---|---|
| Biomarker | Comparison of Non-Progressors vs. Progressors (age of onset < 50 years, disease duration > 8 years) |
|||
| AUC* (95% CI) | Optimal threshold |
Sensitivity | Specificity | |
| % Polyguanine Mutations | ||||
| - Mean % mutation | 0.91 (0.60, 1.00) | 10% | 100.0% (8/8) | 80.0% (8/10) |
| - Maximum % mutation | 0.94 (0.66, 1.00) | 27.5% | 87.5% (7/8)** | 100.0% (10/10) |
| Telomere Length | ||||
| - Mean Telomere Length | 0.78 (0.45, 0.96) | 0.455† | 62.5% (5/8) | 90% (9/10) |
| - Minimum Telomere Length | 0.72 (0.40, 0.94) | 0.600† | 50.0% (4/8) | 80% (8/10) |
| B. 3 Biopsies (1 Proximal, 1 Mid, 1 Distal). If >1, selected one randomly | ||||
|---|---|---|---|---|
| Biomarker | Comparison of Non-Progressors vs. Progressors (age of onset < 50 years, disease duration > 8 years) |
|||
| AUC* (95% CI) | Optimal threshold |
Sensitivity | Specificity | |
| % Polyguanine Mutations | ||||
| - Mean % mutation | 0.86 (0.60, 1.00) | 9% | 87.5% (7/8) | 90.0% (9/10) |
| - Maximum % mutation | 0.86 (0.52, 1.00) | 15% | 75.0% (6/8) | 80.0% (8/10) |
| Telomere Length | ||||
| - Mean Telomere Length | 0.76 (0.48, 0.96) | 0.485† | 75.0% (6/8) | 80.0% (8/10) |
| - Minimum Telomere Length | 0.68 (0.36, 0.95) | 0.565† | 50.0% (4/8) | 80.0% (8/10) |
Adjusted for age as a covariate
Progressor undetected was only patient in the study with incomplete colon coverage
Sensitivity is estimated as the proportion of progressors with telomere length below optimal threshold and specificity as the proportion of non-progressors with telomere length above optimal threshold
Adjusted for age as a covariate
Sensitivity is estimated as the proportion of progressors with telomere length below optimal threshold and specificity as the proportion of non-progressors with telomere length above optimal threshold
Clonal expansions and short telomeres are present in most non-dysplastic biopsies from early-onset UC Progressors, but in few biopsies of Non-Progressors
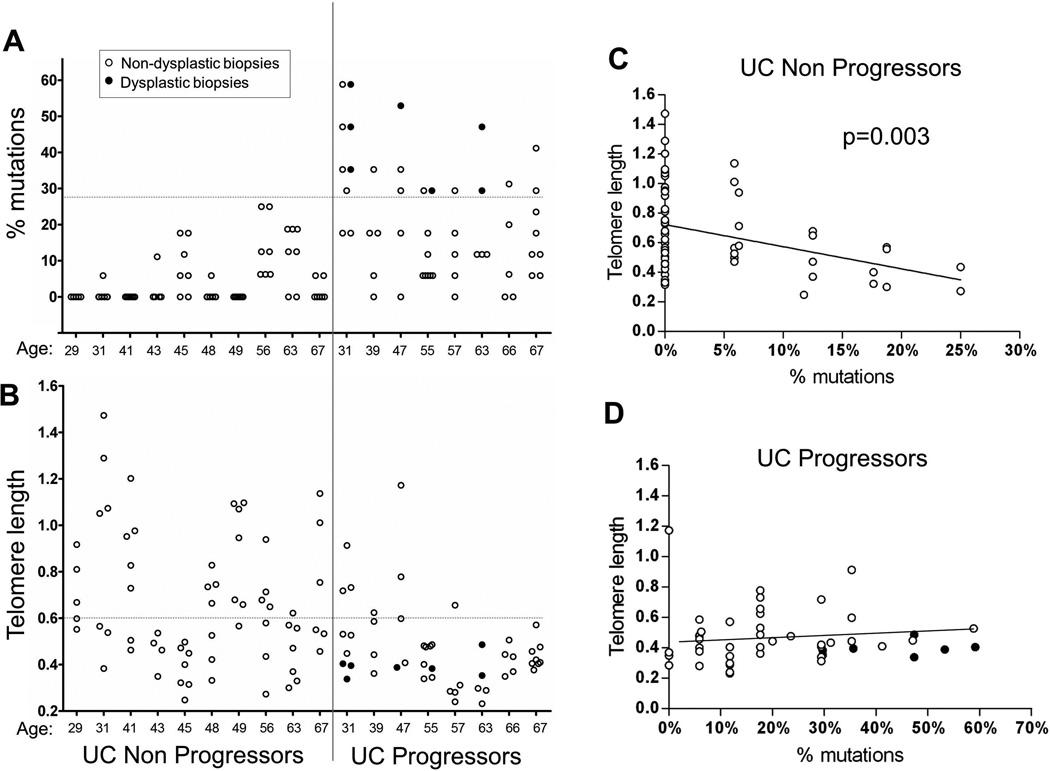
Within a given biopsy the presence of a single mutant locus is sufficient to define a clone and indicate abnormal proliferation of the cell population. The co-occurrence of multiple mutations, however, suggests that the clone has either accumulated more passengers through sequential rounds of clonal outgrowth, has been exposed to more mutagenic damage prior to expansion, or, perhaps, is derived from a genetically unstable population (25). To explore the clinical relevance of different degrees of mutation, we analyzed the percentage of mutant polyguanine genotypes for each biopsy of UC Non-Progressors and UC Progressors (only early-onset and >8 years disease duration, as discussed above, Fig 3A). Seven additional dysplastic biopsies from UC Progressors were also assessed.
Figure 3. Clonal expansions and telomeres in all biopsies analyzed for UC Non-Progressors and Progressors with early-onset of disease and more than 8 years disease duration.
Open circles: non-dysplastic biopsies; black circles: dysplastic biopsies. A–B: Age of patients is indicated in the X-axis. Vertical line separates Non-Progressors from Progressors. Horizontal dotted-lines indicate the thresholds that separate Non-Progressors from Progressors with optimal sensitivity and specificity. C–D: Association between telomere length and percentage of mutations for colon biopsies from UC Non-Progressors (C) and Progressors (D). Regression lines (excluding dysplastic biopsies) are shown and the statistically significant p-value for the GEE regression test in C is indicated.
As expected, dysplastic biopsies yielded the highest percentage of mutations (mean 40.4%, min 23.5%, max 58.8%). The average percentage of mutations in non-dysplastic biopsies was statistically significantly higher by 14.1% among Progressors compared to Non-Progressors [95% CI (6.5%, 22.1%), p-value <0.001]. The estimated odds of having at least one mutation was 14.7 times higher in Progressors compared to Non-Progressors [95% CI (3.8, 56.4), p-value <0.001]. In spite of the heterogeneity in the frequency of mutations, all but one Progressor had at least one non-dysplastic biopsy with more than 27.5% mutant genotypes whereas all Non-Progressor had all biopsies below that level (Fig 3A). This represents 100% specificity and 87.5% sensitivity (Table 2). The single Progressor without non-dysplastic biopsies above the 27.5% threshold was the only case in the study which lacked complete colon coverage (case PG5, Table 1). This patient had a proximal cancer and, unfortunately, no proximal non-dysplastic biopsies were available for analysis.
Telomere length could not distinguish UC Progressors from Non-Progressors as well as the percentage of polyguanine mutations (Fig 3B). Using a GEE model that adjusted for age, the mean telomere length was significantly shorter among Progressors compared to Non-Progressors [0.13 units, 95% CI (−0.25, −0.01), p-value = 0.036]. However, some Non Progressor biopsies showed relatively short telomeres, which compromised the discriminatory value of telomere length (Table 2A). Interestingly, a subset of these biopsies were the ones harboring polyguanine mutations, as shown by the significant negative association between telomere length and percentage of mutations [Fig 3C, slope=-0.010, 95% CI (−0.016, −0.003), p-value=0.003 from GEE model]. This indicates that clonal expansions are often accompanied by shorter telomeres, which might reflect telomere attrition due to multiple rounds of cell proliferation. A subset of biopsies, however, showed short telomeres without clonal expansions, which might be the consequence of telomere attrition secondary to oxidative damage (18). In contrast to UC Non-Progressors, among UC Progressors practically all biopsies had relatively short telomeres and frequent clonal expansions, hence the lack of association between the two (Fig 3D, slope=−0.002, 95% CI (−0.006, 0.002), p-value=0.361). Dysplastic biopsies showed very short telomeres and, as noted previously, the highest degree of mutations.
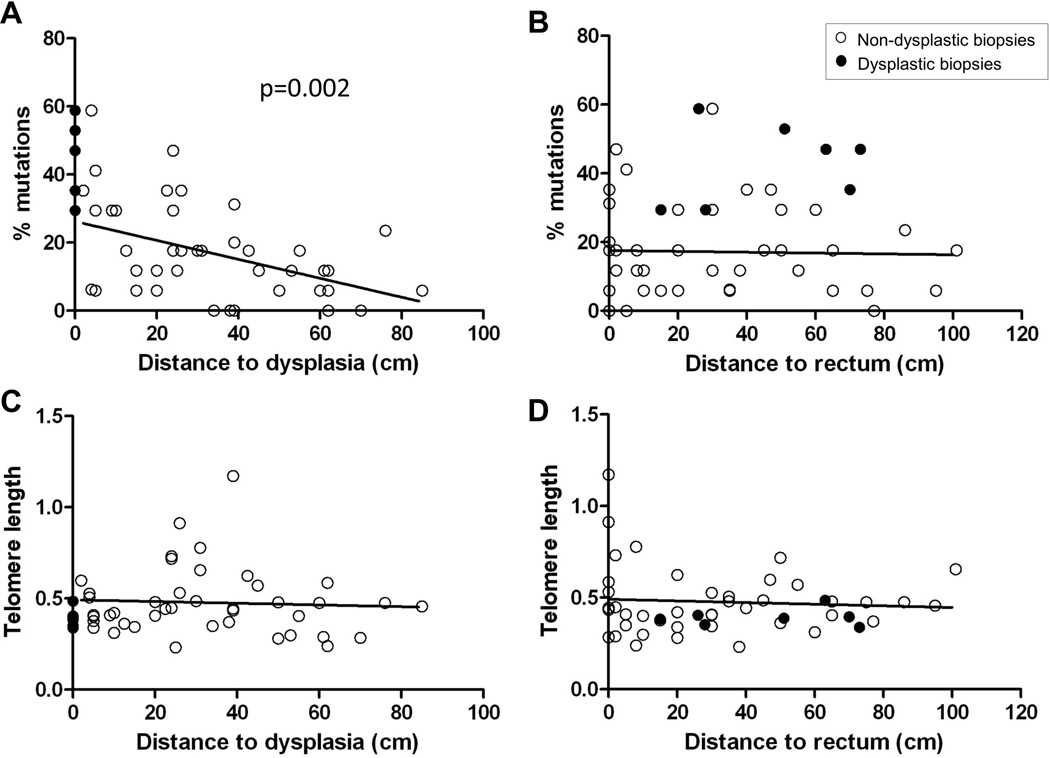
Biopsies closer to dysplasia show higher percentage of mutations
We next investigated if the variation in the percentage of mutant polyguanine genotypes of different biopsies was associated with position in the colon. Among non-dysplastic biopsies, a very strong association was found between the percentage of mutations in non-dysplastic biopsies and proximity to dysplasia (Fig 4A), but not between the percentage of mutations and the distance to rectum (Fig 4B). The average percentage of mutations was estimated to decrease by 2.7% for a 10 cm increase in distance from dysplasia [95% CI (−4.4, −1.0), p = 0.002 from GEE model). Dysplastic biopsies (located at distance=0 in Fig 4A) showed the highest percentage of mutations. We additionally plotted the association between percentage of mutations and distance to dysplasia for each patient (Fig., Supplemental Digital Content 2, http://links.lww.com/IBD/A291) and found that, while the percentage of mutations varied between patients, all but two patients showed a greater mutation frequency closer to dysplasia. Telomere length was not significantly associated with distance to dysplasia (Fig 4C) or distance to rectum (Fig 4D).
Figure 4. Associations of clonal expansions and telomere length with distance to dysplasia and rectum.
Regression plots of percentage of mutations and distance to dysplasia (A), percentage of mutations and distance to rectum (B), telomere length and distance to dysplasia (C), and telomere length and distance to rectum (D). Open circles: non-dysplastic biopsies; black circles: dysplastic biopsies. Dysplastic biopsies are not included in the regression tests. Statistically significant p-value for the GEE regression test in A is indicated.
Classification of UC Progressors and Non-Progressors based on the analysis of clonal expansions and telomere length using only 3 biopsies
With the goal of translating our clonal analysis into a clinically practical test to identify UC patients at risk of cancer, assay simplification and standardization are required. Thus, we compared the ability of clonal mutations to distinguish UC Progressors from Non-Progressors using all the biopsies in the study (Table 2A) versus only 3 randomly selected biopsies collected from distal, mid, and proximal colon (Table 2B). We reasoned that 3 biopsies along the colon might suffice because in spite of higher level of mutations closer to dysplasia, most biopsies from UC Progressors had at least one mutation, which was not the case for UC Non-Progressors (Fig 3A). The discriminatory performance of percentage of mutations and telomere length was found to be only slightly decreased when using 3 biopsies compared to all biopsies. The mean percentage of mutations showed the best discriminatory value with 87.5% sensitivity (as above, the patient missed was the one with incomplete colon coverage) and 90% specificity.
Clonal expansion analysis could be simplified with a reduced number of polyguanine markers
From the summary of mutations by marker (Fig. A, Supplemental Digital Content 3, http://links.lww.com/IBD/A292), it is readily apparent that certain polyguanine loci are more frequently informative than others. This suggests the possibility of simplifying the analysis by using only the most informative markers. In order to investigate the discriminatory power of each marker, we calculated the AUC for all possible combinations of all 17 markers. For combinations that fell into varying levels of performance based on AUC intervals, we calculated the proportion of combinations that contained each marker. Markers that were often present in combinations that had high AUC values and rarely present in combinations that had low AUC values would be potential candidates for simpler marker combinations (Fig. B, Supplemental Digital Content 3, http://links.lww.com/IBD/A292). We identified 24,419 different combinations of markers that produced an AUC higher than 0.95 (the best discriminatory value between Progressors and Non-Progressors).The marker with the best discriminatory power appeared in 90% of these combinations, with the next most frequent markers present in in 70% and 64% of the combinations, followed by several markers present in about half of the combinations. When calculating the percentage of mutations using only the 3 best discriminatory markers, we could, as expected, maximize discrimination between Progressors and Non-Progressors (100% sensitivity, 90% specificity, Fig. C, Supplemental Digital Content 3, http://links.lww.com/IBD/A292). While this limited marker panel remains to be validated in a larger study, it offers the possibility of a simpler assay by using only the most informative polyguanine loci.
DISCUSSION
The goal of this study was to validate clonal expansions and telomere shortening as cancer biomarkers in UC. We previously reported that clonal expansions–defined by length altering mutations in polyguanine tracts–occur at much higher frequency in non-dysplastic biopsies of UC Progressors than Non-Progressors (11). We also demonstrated that non-dysplastic biopsies from UC Progressors show shorter telomeres than those from UC Non-Progressors (17, 20). In this report we independently confirmed these original findings but, intriguingly, discovered that they apply only to early-onset (disease arising prior to 50 years of age) UC patients. Late-onset UC Progressors lacked clonal expansions and displayed longer telomeres, suggesting that they might follow a different mechanism of tumor progression. For early-onset Progressors, clonal expansions were a significantly better marker of progression than short telomeres. Mean frequency of clonal expansions could discriminate early-onset Progressors from Non-Progressors with 100% sensitivity and 80% specificity, which is in close agreement with our original study (11). This confirms its value as a biomarker for the detection, and potentially prediction, of cancer risk in UC.
The age-of-onset dependent effect on clonal expansions could not have been observed in our first study because all Progressors in the initial cohort had early-onset of disease. Its discovery in this validation set was unanticipated, but lends biological substantiation to recent studies that have reported important clinical and epidemiological differences between early-onset and late-onset colitis. Late-onset UC patients do not appear to have an increased risk of colorectal cancer compared to the rest of the population (3) and, when they develop cancer, it frequently takes less than 8 years (7). This is reflected in the late-onset Progressors in our study, two of which had UC for less than 8 years. While the number of patients is small, our results provide the first molecular evidence that the mechanism of tumorigenesis in late-onset Progressors might be different from early-onset Progressors. Further investigations are needed to confirm these results, as they have significant clinical implications. If late-onset Progressors develop cancer in less than 8 years, then these patients should be enrolled earlier for colonoscopic surveillance. Moreover, if colorectal cancer does not arise as a consequence of underlying colitis and widespread preneoplastic clones, these patients might safely benefit from partial colon resection, such as is done with sporadic colorectal cancer patients, to spare them the morbidity of full colectomy.
A simple mechanistic explanation of why tumors in late-onset disease uniquely arise in the absence of preneoplastic fields and shortened telomeres is not obvious. It is possible that some of these cancers might simply be sporadic colorectal carcinomas (inherently more prevalent in the elderly (26)) that coincidentally arose in the background of UC. Sporadic colorectal cancers in non-IBD patients do not show a field effect of telomere shortening (27) like the colorectal cancers observed in the late-onset UC patients. Alternatively, UC might accelerate transformation of pre-existing age-associated adenomas into colorectal cancers, which could explain the clinical observation that progression to cancer appears to be more rapid in Iate-onset than in early-onset UC (7). Of greatest interest is the possibility that the nature of colitis and its progression to cancer is fundamentally different in late-onset patients compared to early-onset patients. In agreement with this notion, two different types of UC-associated colorectal cancer have been described: 1) widespread neoplasia, which exhibits co-existing diffusely scattered dysplasia and is associated with early-onset of disease, and 2) localized neoplasia, in which cancers tend to be unifocal and without accompanying dysplastic changes and is associated with late-onset of disease (8). Our results suggest that widespread neoplasia is likely to be the consequence of diffuse telomere shortening and clonal expansions, as observed in the colon of early-onset UC Progressors. In these patients, critically short telomeres may lead to chromosomal instability and tumor progression through cycles of chromosome bridge-breakage-fusion (20). In contrast, late-onset Progressors develop localized neoplasia in the absence of widespread genetic alterations, consistent with alternative mechanisms driving tumor development in this elderly population.
Consistent with our prior data, non-dysplastic clones remained a strong biomarker of concurrent cancer or HGD elsewhere in colon. In this validation study, we observed a higher percentage of clone-defining mutations than seen previously in both Progressors and Non-Progressors, which likely reflects the use of a smaller number of more informative markers and more uniform biopsy coverage of colons. Nevertheless, at an optimal ROC-defined mutation threshold, the sensitivity and specificity for distinguishing the two groups was almost identical to that previously reported (100% sensitivity and 80% vs. 88% specificity). That some Non-Progressors exhibit clones is to be expected when considered in the context that malignant transformation is a temporally graded process and carcinoma appears to arise from within pre-existing clones (11). A subset of Non-Progressors are destined to become future Progressors, and while the ability to predict concurrent malignancy will be clinically useful, a more significant possibility is that the abundance of mutant clones will predict risk of future progression. The ability to more precisely risk-stratify UC patients would allow redistribution of surveillance efforts to focus on the small subset at greatest risk, while potentially scaling back expensive and uncomfortable monitoring in the majority of patients who will never develop cancer. The expansion of preneoplastic clones has been documented by other techniques in different premalignant conditions of the gut (28). The analysis of polyguanine mutations provides a highly sensitive method to detect these clones and, as such, it has the potential for early cancer detection, not only in UC, but in other gastrointestinal neoplastic process. Future longitudinal studies will be necessary to address these questions.
With the goal of clinical translation, we explored several assay refinements to minimize laboratory processing time and sampling requirements. In this study, we halved the number of polyguanine loci interrogated in the last study to eliminate those that performed poorly. We also computationally simulated the most efficient subpanels of markers that could be used to further reduce the number of PCR reactions needed without compromising sensitivity or specificity. In addition, we examined the number of biopsies tested per person and found that three widely spaced samples performed nearly as well as a larger set for distinguishing UC Progressors from Non-Progressors.
While very effective, our current capillary-based genotyping approach remains considerably lower-throughput and more labor-intensive than contemporary massively-parallel sequencing platforms. In this study we examined 17 polyguanine tracts, yet there are more than 3000 comparable sequences throughout the genome that could be simultaneously interrogated on massively-parallel sequencing systems to obtain greater sensitivity. The digital nature of next generation sequencing additionally makes it possible to resolve mutant clones that have been substantially intermixed with unrelated cells. This feature might conceivably offer the possibility of collecting samples through less invasive means that ordinarily obscure clone homogeneity, such as colonic brushings or perhaps even DNA from stool. One concern is that the biochemical property which makes polyguanine homopolymers so mutable in vivo – and therefore such informative markers of cell lineage – also occurs during enzymatic manipulation in vitro and has traditionally confounded most next generation sequencers. We and our colleagues have recently developed biochemical error correction techniques that we anticipate will overcome this limitation (29, 30) and we are working to transition our current assay to this sequencing modality.
In summary, we have validated that clonal expansions in non-dysplastic mucosa are a strong biomarker of concurrent cancer in early-onset UC and distinguish UC Progressors from Non-Progressors with high sensitivity and specificity. Cancer in early-onset UC arises in fields of expanded clones with short telomeres, which are absent in late-onset UC for unclear reasons that merit further elucidation. A lesser degree of clonal expansion also occurs in some Non-Progressors and the extent of this expansion may predict the subset of those patients at greatest risk for future progression. Future longitudinal studies using an assay adapted to higher throughput, massively parallel sequencing platforms will be helpful to confirm this compelling possibility.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Binkley and Calvin Ngo for technical assistance and Yasuko Tamura and Bonnie Shadrach for assistance with tissue de-archiving and preparation.
Grant support: This work was supported by the National Institutes of Health under award numbers K07 CA137136 to RAR, R01 CA160674 to LaAL, R01 CA068124 to TAB, P30 AG13280 to PSR, UL1 TR000423 to AB, and the Crohn’s and Colitis Foundation of America.
ABBREVIATIONS
- AUC
Area Under the Curve
- Ct
Cycle threshold
- HGD
High-Grade Dysplasia
- LGD
Low-Grade Dysplasia
- Q-PCR
Quantitative PCR
- ROC
Receiver Operating Characteristic
- UC
Ulcerative Colitis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors disclose no conflicts of interest.
Author contributions: JJS, TAB, LaAL, PSR and RAR designed the study. JJS, LiAL, and DAC conducted the experiments. AB, CHU, and RAR analyzed the data. MSH and MPB provided technical and material support. JJS and RAR interpreted the data and wrote the manuscript. All authors provided helpful discussion and critical reading of the manuscript.
REFERENCES
- 1.Kappelman MD, Moore KR, Allen JK, et al. Recent trends in the prevalence of Crohn's disease and ulcerative colitis in a commercially insured US population. Dig Dis Sci. 2013;58:519–525. doi: 10.1007/s10620-012-2371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou JK, Kramer JR, Richardson P, et al. The incidence and prevalence of inflammatory bowel disease among U.S. veterans: a national cohort study. Inflamm Bowel Dis. 2013;19:1059–1064. doi: 10.1097/MIB.0b013e31828028ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jess T, Simonsen J, Jorgensen KT, et al. Decreasing risk of colorectal cancer in patients with inflammatory bowel disease over 30 years. Gastroenterology. 2012;143:375–381. e371. doi: 10.1053/j.gastro.2012.04.016. quiz e313–374. [DOI] [PubMed] [Google Scholar]
- 4.Lutgens MW, van Oijen MG, van der Heijden GJ, et al. Declining risk of colorectal cancer in inflammatory bowel disease: an updated meta-analysis of population-based cohort studies. Inflamm Bowel Dis. 2013;19:789–799. doi: 10.1097/MIB.0b013e31828029c0. [DOI] [PubMed] [Google Scholar]
- 5.Charpentier C, Salleron J, Savoye G, et al. Natural history of elderly-onset inflammatory bowel disease: a population-based cohort study. Gut. 2013 doi: 10.1136/gutjnl-2012-303864. [DOI] [PubMed] [Google Scholar]
- 6.Ha CY, Newberry RD, Stone CD, et al. Patients with late-adult-onset ulcerative colitis have better outcomes than those with early onset disease. Clin Gastroenterol Hepatol. 2010;8:682–687. e681. doi: 10.1016/j.cgh.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baars JE, Kuipers EJ, van Haastert M, et al. Age at diagnosis of inflammatory bowel disease influences early development of colorectal cancer in inflammatory bowel disease patients: a nationwide, long-term survey. J Gastroenterol. 2012 doi: 10.1007/s00535-012-0603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brackmann S, Andersen SN, Aamodt G, et al. Two distinct groups of colorectal cancer in inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:9–16. doi: 10.1002/ibd.20542. [DOI] [PubMed] [Google Scholar]
- 9.Brentnall TA. Molecular underpinnings of cancer in ulcerative colitis. Curr Opin Gastroenterol. 2003;19:64–68. doi: 10.1097/00001574-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Ullman T, Odze R, Farraye FA. Diagnosis and management of dysplasia in patients with ulcerative colitis and Crohn's disease of the colon. Inflamm Bowel Dis. 2009;15:630–638. doi: 10.1002/ibd.20766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salk JJ, Salipante SJ, Risques RA, et al. Clonal expansions in ulcerative colitis identify patients with neoplasia. Proc Natl Acad Sci U S A. 2009;106:20871–20876. doi: 10.1073/pnas.0909428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer JC, Yamada NA, Roques CN, et al. Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Hum Mol Genet. 2002;11:707–713. doi: 10.1093/hmg/11.6.707. [DOI] [PubMed] [Google Scholar]
- 13.Braakhuis BJ, Tabor MP, Kummer JA, et al. A genetic explanation of Slaughter's concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 14.Brentnall TA, Crispin DA, Rabinovitch PS, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 15.Burmer GC, Rabinovitch PS, Haggitt RC, et al. Neoplastic progression in ulcerative colitis: histology, DNA content, and loss of a p53 allele. Gastroenterology. 1992;103:1602–1610. doi: 10.1016/0016-5085(92)91184-6. [DOI] [PubMed] [Google Scholar]
- 16.Rabinovitch PS, Dziadon S, Brentnall TA, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59:5148–5153. [PubMed] [Google Scholar]
- 17.Risques RA, Lai LA, Himmetoglu C, et al. Ulcerative colitis-associated colorectal cancer arises in a field of short telomeres, senescence, and inflammation. Cancer Res. 2011;71:1669–1679. doi: 10.1158/0008-5472.CAN-10-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 19.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 20.O'Sullivan JN, Bronner MP, Brentnall TA, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32:280–284. doi: 10.1038/ng989. [DOI] [PubMed] [Google Scholar]
- 21.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janes H, Pepe MS. Adjusting for covariates in studies of diagnostic, screening, or prognostic markers: an old concept in a new setting. Am J Epidemiol. 2008;168:89–97. doi: 10.1093/aje/kwn099. [DOI] [PubMed] [Google Scholar]
- 23.Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526–535. doi: 10.1136/gut.48.4.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 25.Salk JJ, Horwitz MS. Passenger mutations as a marker of clonal cell lineages in emerging neoplasia. Semin Cancer Biol. 20:294–303. doi: 10.1016/j.semcancer.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy G, Devesa SS, Cross AJ, et al. Sex disparities in colorectal cancer incidence by anatomic subsite, race and age. Int J Cancer. 2011;128:1668–1675. doi: 10.1002/ijc.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Sullivan J, Risques RA, Mandelson MT, et al. Telomere length in the colon declines with age: a relation to colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2006;15:573–577. doi: 10.1158/1055-9965.EPI-05-0542. [DOI] [PubMed] [Google Scholar]
- 28.Baker AM, Graham TA, Wright NA. Pre-tumour clones, periodic selection and clonal interference in the origin and progression of gastrointestinal cancer: potential for biomarker development. J Pathol. 2013 doi: 10.1002/path.4157. [DOI] [PubMed] [Google Scholar]
- 29.Hiatt JB, Pritchard CC, Salipante SJ, et al. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013 doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt MW, Kennedy SR, Salk JJ, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.