Abstract
The recent advent of high-throughput sequencing and genotyping technologies makes it possible to produce, easily and cost effectively, large amounts of detailed data on the genotype composition of populations. Detecting locus-specific effects may help identify those genes that have been, or are currently, targeted by natural selection. How best to identify these selected regions, loci, or single nucleotides remains a challenging issue. Here, we introduce a new model-based method, called SelEstim, to distinguish putative selected polymorphisms from the background of neutral (or nearly neutral) ones and to estimate the intensity of selection at the former. The underlying population genetic model is a diffusion approximation for the distribution of allele frequency in a population subdivided into a number of demes that exchange migrants. We use a Markov chain Monte Carlo algorithm for sampling from the joint posterior distribution of the model parameters, in a hierarchical Bayesian framework. We present evidence from stochastic simulations, which demonstrates the good power of SelEstim to identify loci targeted by selection and to estimate the strength of selection acting on these loci, within each deme. We also reanalyze a subset of SNP data from the Stanford HGDP–CEPH Human Genome Diversity Cell Line Panel to illustrate the performance of SelEstim on real data. In agreement with previous studies, our analyses point to a very strong signal of positive selection upstream of the LCT gene, which encodes for the enzyme lactase–phlorizin hydrolase and is associated with adult-type hypolactasia. The geographical distribution of the strength of positive selection across the Old World matches the interpolated map of lactase persistence phenotype frequencies, with the strongest selection coefficients in Europe and in the Indus Valley.
Keywords: adaptation, genome scan, hierarchical Bayesian model, natural selection, population differentiation
IN the new era of population genomics, surveys of genetic polymorphism (“genome scans”) offer the opportunity to distinguish locus-specific from genome-wide effects at many loci (Black et al. 2001). Reliable inference of demography and phylogenetic history depends on being able to identify putative neutral regions of the genome, which are assumed to be influenced by genome-wide effects only (Ross et al. 1999). Conversely, detecting locus-specific effects may help identify those genes that have been, or still are, targeted by natural selection (Luikart et al. 2003). Such genes may be involved, for example, in the adaptation to new environments or in the arms race with pathogens (Nielsen 2005). The applications for population genomic analyses therefore cover a wide range of disciplines. Although some progress has been made (Nielsen 2001), the problem of how best to identify regions, loci, or single nucleotides that have been, or are currently, targets of selection remains challenging.
Tests of selective neutrality have been developed for samples drawn from single populations. Most of them are based on the comparison of some summary statistics of the site-frequency spectrum (i.e., the observed distribution of gene frequencies) to their expected distribution from diffusion theory under an infinitely many sites mutation model (Bustamante et al. 2001; Payseur et al. 2002; Nielsen et al. 2005b; Williamson et al. 2005). Accounting for different classes of markers (e.g., selected and neutral) is achieved by a Poisson random field (PRF) approximation (Sawyer and Hartl 1992), which assumes independent mutation and selection parameters across sites (see, e.g., Kim and Stephan 2002; Bustamante et al. 2003). In particular, Bustamante et al. (2003) developed a hierarchical PRF model that allows the estimation of selection coefficients at a set of DNA polymorphisms sampled in a single population (see also Nielsen et al. 2005a). Williamson et al. (2005) used a similar approach to infer selection in a nonequilibrium demographic model. Yet, they assume a priori which mutations are selectively neutral and which are not. The putatively neutral class of markers is then used to infer demographic parameters and, given these estimates, inferences regarding selection are performed on the other class of markers.
Other tests of selective neutrality are based on haplotype structure. Focusing on haplotypes at a locus of interest (referred to as “core haplotypes”), Sabeti et al. (2002) analyzed the decay of gene identity as a function of distance from the core, as measured by the extended haplotype homozygosity (EHH). Core haplotypes that have both a high population frequency and a high EHH are evidence of recent positive selection. Deriving the expected distribution of the EHH requires making strong assumptions about the underlying population history, however, which makes it difficult to evaluate the significance of observed values. Several extensions have therefore been proposed and adapted to genome-wide scans of single nucleotide polymorphism (SNP) data (reviewed in Gautier and Vitalis 2012), based on the empirical distribution of EHH-like statistics either for single populations (Voight et al. 2006) or for pairs of differentiated populations (Tang et al. 2007). Such approaches therefore rely on the assumption that most SNPs behave neutrally, so that the observed distribution of the statistics provides a proxy to the null distribution.
When markers are genotyped across multiple populations, it has been advocated that signatures of natural selection may simply be identified in the extreme tails of the empirical distribution of FST estimates (Goldstein and Chikhi 2002). The rationale is that loci that are involved in adaptation to local environmental conditions are expected to show unusually high levels of differentiation among populations. Conversely, loci that are under balancing selection are expected to show unusually low levels of differentiation. These model-free approaches are highly computationally efficient and, therefore, have early been applied to large data sets of tens to hundreds of thousands of SNPs, such as the Perlegen (Hinds et al. 2005) and the HapMap (International HapMap Consortium 2003, 2005) data in humans (see, e.g., Akey et al. 2002; Weir et al. 2005; Barreiro et al. 2008). Model-free approaches implicitly assume that most of the markers analyzed are selectively neutral, however, and the choice of a decision criterion to characterize “outlier loci” is fairly subjective. These methods are intended to be immune to arbitrary assumptions about the (unknown) demographic history of the sample. Dependence upon the unknown demography (including the geographic and historical relationship among populations) was indeed a severe criticism of Lewontin and Krakauer’s model-based test of selective neutrality (Lewontin and Krakauer 1973), which relies on the sampling distribution of the parameter FST (Robertson 1975; Nei and Maryuyama 1975).
Refinements of this controversial test (see, e.g., Beaumont and Nichols 1996) were based on the properties of gene genealogies in structured populations (Beaumont 2005), which, in many cases, tend toward a simple structure (Nordborg 1997; Wakeley 1999). In this simple structure, the recent genealogy of genes in each local population can be separated from the ancestral genealogy of the whole metapopulation (Wakeley 2004). If this separation-of-timescale approximation holds, the gene frequency distribution in each local population may be approximated as a Dirichlet-multinomial distribution (Balding and Nichols 1995; Balding 2003). Otherwise, it may be necessary to model demography explicitly (Nielsen et al. 2009), to restrict the analysis to pairwise comparisons (Vitalis et al. 2001), or to account for the covariance matrix of the population allele frequencies (Bonhomme et al. 2010; Coop et al. 2010; Frichot et al. 2013; Günther and Coop 2013; Guillot et al. 2014). Several likelihood-based approaches that take advantage of the Dirichlet-multinomial distribution of gene frequencies have been proposed, generally within a Bayesian framework. In particular, Beaumont and Balding (2004) proposed decomposing FST into locus-, population-, and locus-by-population components in a hierarchical model. Their model offered a new way to formulate the problem of inferring which loci are targets of selection. This framework was further extended by Riebler et al. (2008), Foll and Gaggiotti (2008), Guo et al. (2009), and Gompert and Buerkle (2011). Yet, these approaches (as most FST-based methods aimed at looking for locus-specific effects on FST estimates; see Beaumont and Nichols 1996) are typically not designed to identify population-specific selection. Furthermore, many species have a hierarchical structure, where a subset of populations share a recent ancestry, or exchange more migrants, relative to the full species range. For such complex structures, extensions of the Beaumont and Nichols (1996) test or the classical Lewontin and Krakauer (1973) test have been proposed (see Excoffier et al. 2009; Bonhomme et al. 2010, respectively). As an alternative to models based on the Dirichlet-multinomial distribution of gene frequencies, Coop et al. (2010) developed an extension to the truncated Gaussian model proposed by Nicholson et al. (2002), which accounts for the pattern of covariance in allele frequencies between populations (see also Frichot et al. 2013; Günther and Coop 2013; Guillot et al. 2014). The resulting multivariate normal distribution of gene frequencies allows testing a linear effect of some environmental variable on the allele frequency at some loci (Hancock et al. 2010, 2011; Frichot et al. 2013).
A major limitation of the methods based on comparisons among populations, however, is that they do not quantify selection. Rather, they are constructed as tests of departure from selective neutrality (Gautier et al. 2010). One exception, though, lies in Bazin et al. (2010), where the effective migration rate at a marker locus is expressed as a function of the selection coefficient at a positively selected locus and the recombination rate between the two (see Petry 1983). While neutrality is a convenient null hypothesis, a proper interpretation of the observed patterns of variability, in particular the extent to which the neutral model is applicable, requires methods that rely on nonneutral models (see, e.g., Donnelly et al. 2001). Furthermore, proper tests of selection should provide estimates of the parameters of interest, i.e., the strength and the type of selection acting on segregating polymorphisms.
Here, we provide a new method, called SelEstim, to distinguish neutral from selected polymorphisms and estimate the intensity of selection at the latter. Our model accounts explicitly for positive selection, and we suppose that all marker loci in the data set are responding to selection, to some extent. The method is based on a diffusion approximation for the distribution of allele frequency in a population subdivided in a number of demes that exchange migrants (i.e., an island model; see Wright 1931). The framework for statistical inference from this model consists in a hierarchical Bayesian model (see Gelman et al. 2004). We use a componentwise Markov chain Monte Carlo (MCMC) algorithm to sample from the joint posterior distribution of the model parameters. We then evaluate the performance of SelEstim, by means of stochastic simulations. Last, we reanalyze a subset of SNP data from the Centre d'Etude du Polymorphisme Humain (CEPH) Human Genome Diversity Panel (HGDP) (Cann et al. 2003) to illustrate the applicability of SelEstim on real data.
Model
Assumptions
We consider an infinite island model where the ith deme consists of Ni diploid individuals and receives immigrants from the whole population at rate mi. We define the scaled migration parameter in the ith deme as Mi ≡ 4Nimi. We consider biallelic markers, i.e., that only two alleles (denoted by A and a) may occur at a given locus. We denote by pij the frequency of allele A in deme i at locus j and by πj the frequency of allele A at the jth locus in the whole population. Since we consider that the population as a whole is made of an infinite number of islands, πj gives the frequency of allele A in the pool of migrant individuals. The following notations are used hereafter: the vector of allele frequencies in deme i at locus j is pij ≡ (pij, 1 − pij), and the vector of allele frequencies at locus j among migrants is πj ≡ (πj, 1 − πj). We consider a simple genic model of selection where, at each locus, the allele A provides a selective advantage. The homozygote individuals AA and the heterozygotes Aa have a relative increase of fitness of 1 + sij and 1 + sij/2, respectively, as compared to the aa homozygotes. We define the scaled coefficient of selection in deme i at locus j as σij ≡ 2Nisij. We define the indicator variable κij, which takes the value κij = 0 if allele A is selected for, and κij = 1 if allele a is selected for. Therefore, the frequency of the selected allele in deme i at locus j reads
The data consist of individuals collected in a set of nd demes and genotyped at L loci. We denote by nij the total number of genes sampled in the ith deme at the jth locus, out of which xij have allelic state A. The vector of allele counts in deme i at locus j therefore reads nij ≡ (xij, nij − xij).
Given the frequencies pij of allele A, the conditional distribution of allele counts nij in population i at locus j is binomial:
| (1) |
In the limit of large deme size, as Ni → ∞, and assuming that selection and random genetic drift are of comparable strength (i.e., that Mi and σij have a finite limit as Ni → ∞), the distribution of the pij may be approximated by the stationary density of a diffusion process, which has the form
| (2) |
(Wright 1949; Barton and Turelli 1987; Ethier and Nagylaki 1988; Bürger 2000). In Equation 2, C is the constant that ensures that the distribution integrates to 1. This constant can be evaluated as
| (3) |
where 1F1(a; b; z) is the confluent hypergeometric, or Kummer’s, function (see, e.g., Abramowitz and Stegun 1965, p. 504), and .
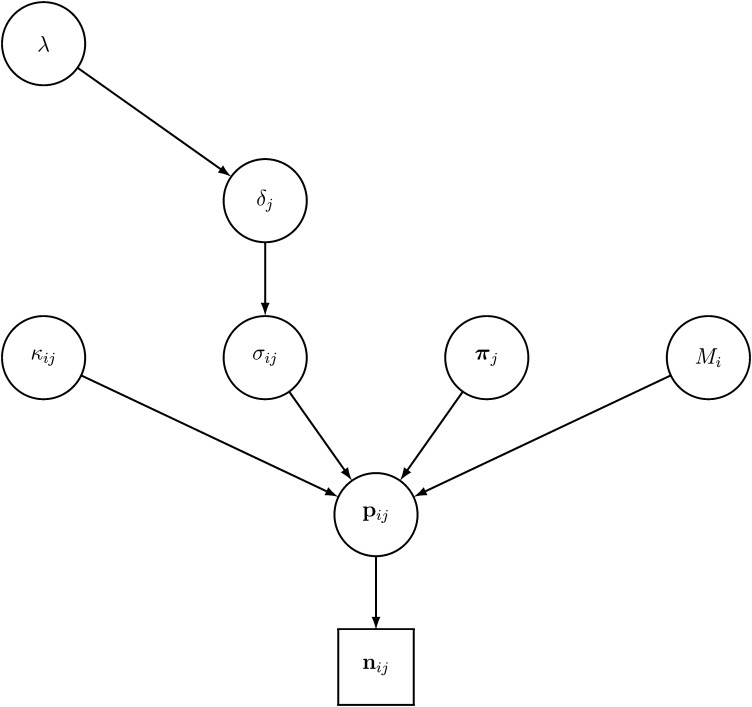
Given the model specified in Equations 1 and 2, we are interested in evaluating the parameters of interest , π ≡ (π1, …, πj, …, πL), , and , from the observed allele counts n over all sampled demes and loci. The directed acyclic graph (DAG) for this hierarchical-Bayesian model is shown in Figure 1.
Figure 1.
Directed acyclic graph (DAG) of the hierarchical Bayesian model.
We assume a Bernoulli prior distribution for the parameters κij, i.e., κij ∼ Ber(0.5), and a uniform prior for the πj’s, that is πj ∼ Beta(1, 1). We further assume a log-uniform prior for the Mi’s with support from 0.001 to 10,000; i.e., the priors of the Mi’s are uniform in log scale: log(Mi) ∼ u(log(10−3), log(104)). The prior distributions for the selection coefficients σij (at each locus, in each deme) are modeled hierarchically (see, e.g., Gelman et al. 2004, pp. 124–125). In particular, we assume that σij has an exponential prior distribution that depends upon the locus-specific hyperparameter δj, which represents the average effect of selection at locus j (over all demes). We further assume that this hyperparameter δj has an exponential prior distribution f(δj|λ) ∼ exp(λ−1) that depends, in turn, upon the hyperparameter λ, which represents the genome-wide effect of selection over all demes and loci. Last, we assume that the prior distribution of λ is f(λ) ∼ exp(Λ−1), with Λ = 1.0, in what follows. Assuming independence of allele frequencies among loci and populations, the posterior distribution of the parameters f(M, π, σ, κ, δ, λ| n), i.e., the conditional distribution of the parameters M, π, κ, σ, δ, and λ given the data n, depends upon the prior distributions of the parameters and the data as
| (4) |
Although in Equation 4 the likelihood from Equation 1 can be integrated analytically over the distribution of unknown population frequencies given by Equations 2 and 3, we found that it increases the computational burden significantly. This is so because additional gamma and confluent hypergeometric functions are then introduced in the likelihood function ℒ′(Mi, πj, κij, σij, nij).
We implemented a single-component Metropolis–Hastings (or Metropolis within Gibbs) algorithm (see, e.g., Ntzoufras 2009) to sample from the joint posterior distribution of f(M, π, κ, σ, δ, λ| n), which is specified by Equation 4. In practice, we therefore update one parameter at each time, iteratively, as detailed in Supporting Information, File S1. The proposal distributions for each of the M, π, κ, σ, δ, and λ parameters are adjusted by means of 25 short pilot runs of 1000 iterations to obtain acceptance rates between 0.25 and 0.40 (see, e.g., Gilks et al. 1996).
The software package implementing the model described here, called SelEstim, is available at http://www1.montpellier.inra.fr/CBGP/software/selestim. All the postprocessing statistical analyses were performed using the R software environment for statistical computing, v. 3.0.1 (R Core Team 2013).
Identifying loci under selection from the MCMC outputs
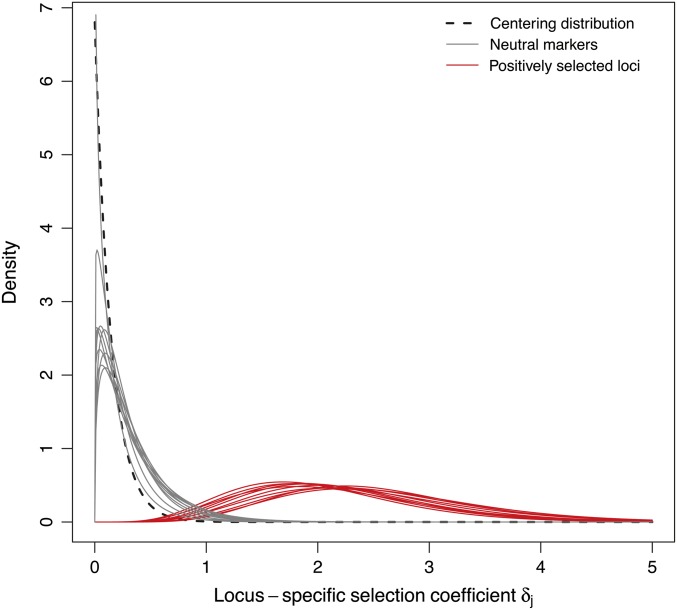
Because the model assumes that each and every locus in a data set is selected to a certain extent, we are particularly interested in the posterior densities of the locus-specific hyperparameters δj: we expect the density to be shifted toward zero for neutral markers and to positive values for (presumably) selected loci (see Figure 2). Yet, given the hierarchical structure of our model, it would not be sufficient to simply test whether, at a particular locus, the posterior distribution of δj departs from zero. This approach would neglect the genome-wide effects of selection. Since we assume in our model that the δj’s are drawn independently from a common hyperdistribution with parameter λ (which represents the genome-wide effect of selection), it is indeed more appropriate to compare the posterior distributions of the locus-specific coefficients of selection with the “centering” distribution derived from the hyperdistribution of the genome-wide effect of selection. This centering distribution is a good choice, rather than, for example, a point mass at zero, because it leads to consistent estimates (the hyperprior will shrink toward zero with more data). Furthermore, as becomes apparent, it provides some, albeit very weak, power to identify loci under balancing selection.
Figure 2.
Examples of posterior densities of the locus-specific hyperparameter δj for neutral markers (in gray) and positively selected loci (in red). The “centering” distribution (dashed line) is derived from the hyperdistribution of the genome-wide effect of selection, λ, and is defined as an exponential distribution , where is the posterior mean of λ, as estimated from the MCMC.
Following Guo et al. (2009), we consider the following steps to detect outlier loci in a data set: (i) approximate the posterior distributions of the locus-specific selection hyperparameters (δj); (ii) compare each of these distributions to a “centering” distribution derived from the hyperdistribution with parameter λ that describes the among-locus variation in the locus-specific effect of selection; and last (iii) measure the mean of the posterior distributions of σij for outlier loci over sampled populations to summarize the distribution of selection effects across sampling locations.
Our preliminary analyses suggested that the posterior distribution of the parameters δj is unimodal. Furthermore, its support is on [0, ∞), by definition of δj. For subsequent analyses, we approximate the posterior distributions of the δj’s by a gamma distribution with the same mean and variance as estimated from the MCMC sample. In doing so, we assume that slight discrepancies in higher-order moments will not affect the comparison of these distributions with their centering distribution. In the following, we therefore consider that the posterior distribution of δj is Γ(k0, θ0) with and , where and are the mean and the variance, respectively, of the posterior distribution of δj, as estimated from the MCMC outputs. Since we assumed an exponential prior distribution for the hyperparameters δj, i.e., f(δj) ∼ exp(λ−1), or equivalently f(δj) ∼ Γ(1, λ), we defined the centering distribution of δj as Γ(1, θ1), where is the posterior mean of λ, as estimated from the MCMC outputs. We expect that the stronger the intensity of selection at locus j, the larger the departure of the posterior distribution of δj from the centering distribution. We use the Kullback–Leibler divergence (KLD) to measure the distance of the posterior distribution of δj from the centering distribution. The KLD of a distribution with density f(x) from a distribution with density g(x) is defined as
| (5) |
With a little algebra, one can show that the KLD of a gamma distribution with shape and scale parameters (k0, θ0) from a gamma distribution with shape and scale parameters (1, θ1) is given by
| (6) |
where is the digamma function. Note that as an alternative to these computations, one might use other KLD estimators, such as those based on nearest neighbors (see, e.g., Pérez-Cruz 2008).
To provide a decision criterion for discriminating between neutral and selected markers, we calibrate the KLD using simulations from a predictive distribution based on the observed data set. The motivation here is to generate a set of loci equivalent to those that we observe in their levels of diversity and genetic variation. Note that generating a full posterior predictive distribution for KLD is unhelpful here because extreme P-values would indicate poor model fit rather than give evidence of selection per se. A key requirement in our calibration is that we generate a distribution of KLD under a null model in which the δj for each locus are drawn from their centering distribution. For this reason we cannot use a strictly neutral model (i.e., the beta-binomial parameterized by Mi). A further assumption that we make, for otherwise the approach would be computationally intractable, is that the loci are exchangeable. Thus the KLD computed for each locus in our data set is compared to the simulated distribution of KLD among all the loci generated from the predictive distribution. As described in File S2, we parameterize our predictive distribution using the estimated posterior means for Mi, πj, κij, and λ. We show below that, although the method is somewhat conservative, the calibration based on these posterior means is generally good. Thus, in practice, for each data set and each analysis, we use the algorithm detailed in File S2 to generate pseudo-observed data (POD). We then analyze that POD, using the same MCMC parameters (number and length of pilot runs, burn-in, chain length, etc.) as for the analysis of the original data set. The KLD values computed for each simulated locus are then combined to obtain an empirical distribution. The quantiles of this empirical distribution are computed and are used to calibrate the KLD observed for each locus in the original data: e.g., the 99% quantile of the KLD distribution from the POD analysis provides a 1% threshold KLD value, which is then used as a decision criterion to discriminate between selection and neutrality.
Materials and Methods
Simulation study
We evaluated the performance of the method by simulating artificial data sets for fixed parameter values. The simulations were performed according to an island model with 50 demes, each consisting of N = 250 diploid individuals. Following Beaumont and Balding (2004), we simulated allele counts data from a Wright–Fisher model with migration and selection.
Initialization was achieved by means of a Pólya urn scheme simulation of the coalescent (Donnelly and Tavaré 1995) with scaled mutation parameter θ ≡ 4Nμ = 1. This amounts to considering selection acting on standing variation and makes this simulation model similar in spirit to the models considered by Innan and Kim (2004) and Przeworsky et al. (2005). At each generation (generations were discrete and nonoverlapping), each individual produced a random number of offspring drawn from a Poisson distribution with mean 100. Mutations then occurred at rate 2 × 10−5. Dispersal of the (diploid) offspring then occurred at rate m, with dispersing individuals reaching necessarily a distinct deme. Selection of the offspring surviving to adulthood was then achieved, according to the scheme detailed below. A number N of adults was drawn from among the offspring, except if the number of offspring in a deme was <N, in which case all offspring survived. This life cycle was repeated for 25,000 generations. Samples were then taken, but only if the minimum allele frequency (the frequency of the least frequent allele) was >0.01. All loci were considered as independent, so that each multilocus data set was made of independent realizations of that process.
To account for the possibility of positive selection to local environmental conditions, the demes were arbitrarily provided with attributes (blue, red, or uncolored), which were assigned at random, independently for each selected locus. For positively selected loci, one allele B was considered as advantageous in a blue deme (and neutral in a red deme), while the other allele R was considered as advantageous in a red deme (and neutral in a blue deme). Both alleles were considered as neutral in uncolored demes. Therefore, BB homozygotes had fitness (1 + s) in blue demes and 1 in red and uncolored demes; RR homozygotes had fitness (1 + s) in red demes and 1 in blue and uncolored demes; BR heterozygotes had fitness 1 + s/2 in red and blue demes and 1 in uncolored demes. For loci under balancing selection, only the heterozygote genotypes were selected for in the blue and red demes, with relative fitness (1 + s). Homozygote genotypes were neutral (relative fitness 1) in all demes, as were the heterozygote genotypes in uncolored demes.
A total of 18 data sets were generated using the Wright–Fisher model described above (see Table 1). For each locus, 50 diploid individuals (100 genes) per deme were sampled. The details that distinguish the different data sets are given in Table 1. For example, for data sets 1–9, the samples were taken in 6 demes: 2 blue demes, 2 red demes, and 2 uncolored demes, and each simulated data set consisted in 10,000 SNPs, with 8000 neutral markers, 1000 positively selected loci, and 1000 loci under balancing selection. For data sets with selected loci (data sets 1–11), we assumed that 30% of all demes were blue demes, 30% were red demes, and 40% were uncolored demes. For data sets with neutral markers only (data sets 12–18), 50,000 SNPs were simulated and all demes were uncolored. Table 1 further gives the combinations of FST and σ values used for the simulations.
Table 1. Parameters of simulated data sets.
| Markers | Sampled demes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Data set | N | M ≡ 4Nm | FST | s | σ ≡ 2Ns | Pos. | Bal. | Neut. | No. | Categories |
| 1 | 250 | 18.067 | 0.05 | 0.02 | 10 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 2 | 250 | 18.067 | 0.05 | 0.05 | 25 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 3 | 250 | 18.067 | 0.05 | 0.10 | 50 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 4 | 250 | 8.664 | 0.10 | 0.02 | 10 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 5 | 250 | 8.664 | 0.10 | 0.05 | 25 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 6 | 250 | 8.664 | 0.10 | 0.10 | 50 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 7 | 250 | 3.858 | 0.20 | 0.02 | 10 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 8 | 250 | 3.858 | 0.20 | 0.05 | 25 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 9 | 250 | 3.858 | 0.20 | 0.10 | 50 | 1,000 | 1,000 | 8,000 | 6 | (2,2,2) |
| 10 | 250 | 8.664 | 0.10 | 0.05 | 25 | 1,000 | 1,000 | 8,000 | 3 | (1,1,1) |
| 11 | 250 | 8.664 | 0.10 | 0.05 | 25 | 1,000 | 1,000 | 8,000 | 12 | (4,4,4) |
| 12 | 250 | 84.604 | 0.01 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
| 13 | 250 | 44.619 | 0.02 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
| 14 | 250 | 18.067 | 0.05 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
| 15 | 250 | 8.664 | 0.10 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
| 16 | 250 | 5.468 | 0.15 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
| 17 | 250 | 3.858 | 0.20 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
| 18 | 250 | 2.888 | 0.25 | – | – | 0 | 0 | 50,000 | 6 | (0,0,6) |
All the simulations were performed according to an island model with nd = 50 demes, each made of N = 250 diploid individuals (500 genes). Fifty diploid individuals (100 genes) were sampled per deme. The migration rate m was fixed to achieve the desired value of FST, using Equation 6 in Rousset (1996). The number of loci simulated under positive selection (Pos.), balancing selection (Bal.) and neutrality (Neut.) is indicated. The total number of sampled demes is also given, together with the composition of the sample: (2,2,2) indicates that the sample of six demes consisted of two blue demes, two red demes, and two uncolored demes.
An additional set of four simulations was performed to test for the robustness to departure from the island model assumptions. To that end, a classical island model, a hierarchical island model (see, e.g., Excoffier et al. 2009), a stepping-stone model in one dimension, and a pure drift model (whereby nine populations diverge sequentially) were simulated (see Figure S1). The sample characteristics (number of individuals, number of sampled demes) were the same for all simulations, and the model parameters were tuned to achieve an overall realized FST of ≈ 0.24.
For each of these 22 data sets the MCMC algorithm was run to sample from the joint posterior distribution of the model parameters. For each Markov chain, 100,000 updating steps were completed after 25 short pilot runs of 1000 iteration and a burn-in of 25,000 steps. Samples were collected for all the model parameters every 25 steps (thinning) to reduce autocorrelations, yielding 4000 observations.
The data sets 1–11 (see Table 1) were analyzed using BayeScan v. 2.1 (Foll and Gaggiotti 2008) with default option values (except for the MCMC parameters, see below). BayeScan is based on the Dirichlet-multinomial model for allele frequencies in an island model of population structure. At each locus, the distribution of allele frequency in each subpopulation depends on the allele frequency in the common pool of migrants and a subpopulation-specific parameter. In BayeScan, as in Beaumont and Balding’s model (Beaumont and Balding 2004), the logit of is decomposed additively into a locus-specific component (αi) shared by all populations, and a population-specific component (βj) shared by all loci. Significantly positive (resp., negative) values of αi are taken as evidence of positive (resp., balancing) selection. BayeScan is based on a reversible-jump MCMC algorithm, which estimates the posterior probabilities of two alternative models, a purely neutral one (αi = 0), and one including selection (αi ≠ 0). Each Markov chain was run for 100,000 updating steps, after 25 short pilot runs of 1000 iteration each and a burn-in of 25,000 steps. Samples were collected every 25 steps (thinning), yielding 4000 observations. For each output, we computed the Bayes factor (BF) for the model including selection (αi ≠ 0), assuming prior odds of 10 for the neutral model (αi = 0).
The comparison of the relative efficiency of the two methods was achieved by means of receiver operating characteristic (ROC) analysis (see, e.g., Fawcett 2006, for further information), using the R software environment for statistical computing, v. 3.0.1 (R Core Team 2013).
Application to human data
We applied SelEstim on the Stanford HGDP–CEPH Human Genome Diversity Cell Line Panel (Cann et al. 2003) SNP genotyping data, which consist of genotypes at more than 650,000 SNPs (ftp://ftp.cephb.fr/hgdp_supp1). Because we were interested, for illustrative purpose, in measuring the genetic signature of selection in the lactase gene, we considered only the 53,765 SNPs mapping to chromosome 2 (HSA2) and incorporated the genotyping data of the two SNPs reported to be tightly associated with lactase persistence (−13910C → T and −22018G → A) as published by Bersaglieri et al. (2004). All the populations with <15 genotyped individuals were discarded from the data set. Furthermore, we removed seven populations from Oceania and Southern America, as well as three populations from Sub-Saharan Africa (the Biaka Pygmies, Mbuti Pygmies, and the Mandenka) that were absent from Bersaglieri et al.’s data set (Bersaglieri et al. 2004). The two Bantu populations (from Kenya and South Africa) were merged, as in Bersaglieri et al. (2004). Finally, we discarded all the SNPs with a minimum allele frequency (MAF) below 0.01 across the populations retained. The final data set consisted in 52,633 SNPs characterized in 23 populations from Africa and Eurasia.
Results
Evaluating the performance of SelEstim:
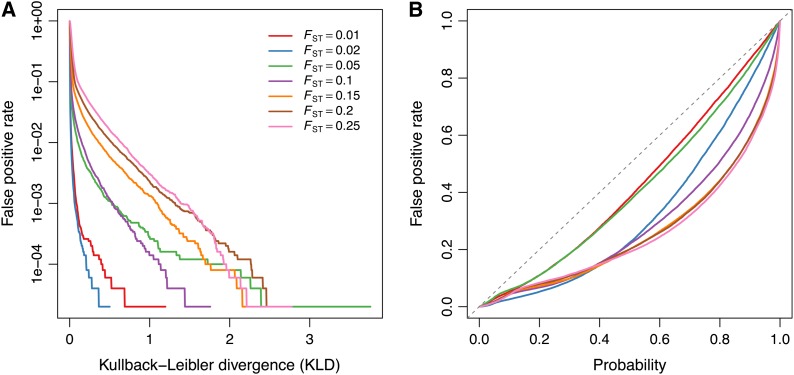
To asses the KLD calibration procedure, we first evaluate the method using simulated data sets made of neutral markers only (data sets 12–18; see Table 1). Figure 3A shows the false-positive rate, i.e., the proportion of markers that are incorrectly classified as under selection, as a function of various KLD thresholds. For each data set, KLD thresholds based on the quantiles of the KLD distributions from the POD analyses were computed and used as a decision criterion for discriminating between neutral and selected markers. Figure 3B represents the false-positive rate as a function of the quantile probability (comprised between 0 and 1). Figure 3B shows that, for the data sets considered here, the false-positive rate at any KLD threshold is always less than the corresponding quantile probability. This suggests that our calibration procedure is “conservative,” at least for the range of FST values considered here (ranging from 0.01 to 0.25).
Figure 3.
(A) False-positive rate (neutral loci detected as outliers) as a function of the Kullback–Leibler divergence (KLD) threshold, for data sets 12–18. (B) False-positive rate, as a function of the quantile probability. For each data set analysis, pseudo-observed data (POD) are generated from the inference model with hyperparameters λ, πj, and Mi set to their respective posterior means, using a rejection-sampling algorithm (see File S2). The POD is then analyzed, using the same MCMC parameters (number and length of pilot runs, burn-in, chain length, etc.) as for the analysis of the original data. Each quantile probability defines a KLD threshold, which is used as a decision criterion for discriminating between neutral and selected markers.
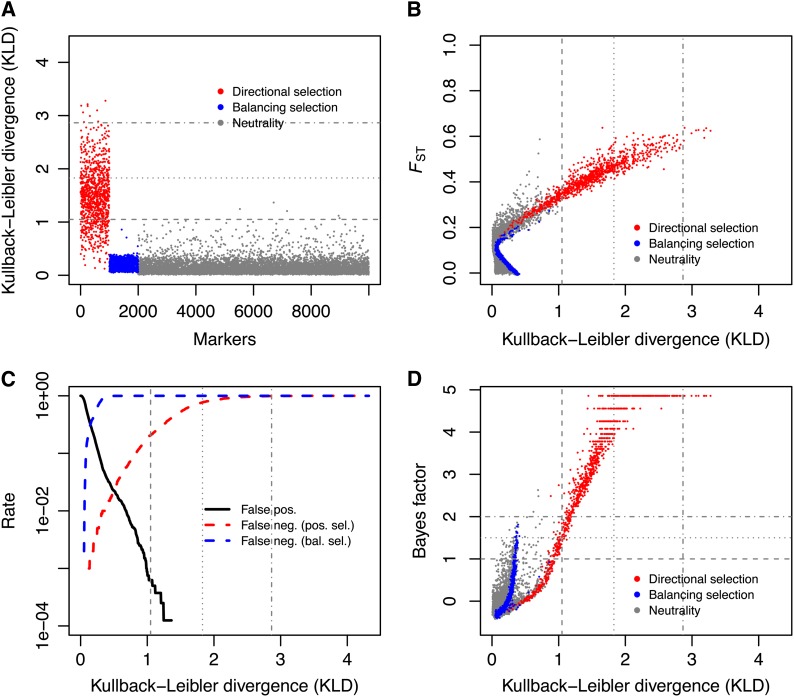
Figure 4 shows the performance of the method on data set 5 (see Table 1), which corresponds to FST = 0.10 and σ ≡ 2Ns = 25. Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, Figure S8, Figure S9, Figure S10, and Figure S11 provide the same outputs for simulated data sets 1–4 and 6–11. Figure 4A shows that the distribution of KLD measures for positively selected loci departs from that of the neutral markers and the loci under balancing selection. This is essentially true for the data sets for which FST ≥ 0.05 and σ ≥ 25, as can be seen from Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, Figure S8, and Figure S9. Not surprisingly, large KLD values correspond to large FST estimates (Figure 4B). This is so because for positively selected loci, one allele is selected for in blue populations and the other in red populations, which tends to exacerbate differentiation. A close examination of Figure 4C further shows that the false-positive rate (the proportion of neutral markers that exhibit a signature of selection) at any KLD threshold is always less than the corresponding KLD quantile probability (as in Figure 3B). This strengthens the notion that the KLD thresholds should not be interpreted as frequentist P-values. All else being equal, increasing the number of sampled demes improves the discrimination of neutral and positively selected markers (compare Figure 4, Figure S10, and Figure S11). Last, Figure 4 shows that SelEstim has some weak statistical power to identify loci under balancing selection. This arises because markers with very low divergence will tend to have posteriors for δ that are pushed further toward zero in comparison with the centering distribution and, hence, will also have a higher KLD. Yet, although the KLD for loci under balancing selection is indeed slightly higher, on average, than for neutral markers (Figure 4, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, Figure S8, Figure S9, Figure S10 and Figure S11), it remains very low. This result is not surprising, though, since the selection scheme considered in our model of inference accounts only for positive genic selection. Furthermore, previous simulation studies have also shown that, in the absence of an explicit model of selection, similar methods generally lack power to detect balancing selection (Beaumont and Balding 2004; Foll and Gaggiotti 2008; Riebler et al. 2008).
Figure 4.
Analysis of the allele count data from data set 5. (A) Kullback–Leibler divergence (KLD) measure between the posterior of δj and its centering distribution for all simulated loci. Loci under positive selection are depicted in red, loci under balancing selection in blue, and neutral markers are in gray. (B) FST as a function of the KLD measure for all loci. (C) False-positive (neutral loci detected as outliers) and false-negative (selected loci not detected as outliers) rates as a function of the KLD threshold. (D) Relationship between BayeScan Bayes factor log10(BF) and the KLD for all markers in data set 5. Positively selected markers are in red, loci under balancing selection are in blue, and neutral markers are in gray. The horizontal lines in A and the vertical lines in B–D indicate the KLD thresholds corresponding to the 95%, 99%, and 99.9% quantile of the KLD distribution from the POD analysis of data set 5. In D, the horizontal dotted lines indicate the log10(BF) = 1, log_10(BF) = 1.5 and log10(BF) = 2 thresholds, which correspond to strong, very strong and decisive support following Jeffreys’s (1961) scale of evidence for selection, respectively.
Comparison with BayeScan:
Figure 4D shows the relationship between BayeScan BF and the KLD for each and every locus from data set 5. According to Jeffreys’ rule (Jeffreys 1961; Kass and Raftery 1995), a BF >10 (log10(BF) ≥ 1) provides “strong” evidence for selection, and for a BF >100 (log10(BF) ≥ 2) the evidence is “decisive.” However, it should be noted that, by definition, the BF depends on the prior odds for the neutral model. For this set of simulated data, there is a good agreement between BayeScan BF criterion and the KLD, since markers with high KLD show decisive evidence of selection from BayeScan BF criterion. For BayeScan analyses, whenever the posterior probability of the model including selection (αi ≠ 0) was equal to 1, we arbitrarily defined the log10(posterior odd) as log10(3999.5/4000) − log10(0.5/4000) to account for the chain length (4000 iterations). By construction, the maximum value that the BF can take (log10(BF) = 4.857; see Figure 4D) therefore depends on the MCMC length and the prior odds for the neutral model (here equal to 10).
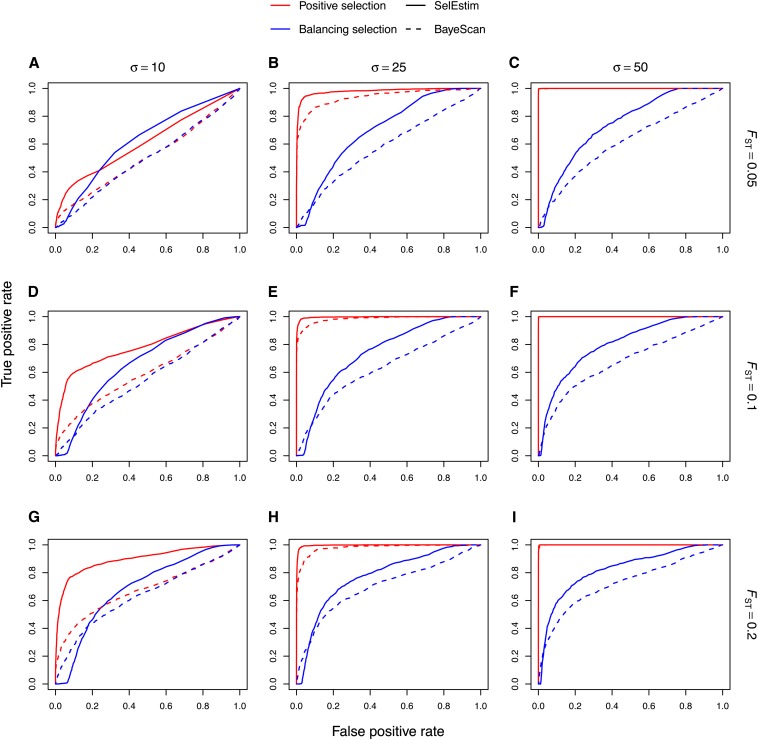
The ROC analysis (Figure 5) further shows a slight power gain of SelEstim over BayeScan (Foll and Gaggiotti 2008). In the ROC analysis, the proportion of false positives and true positives is computed for each possible value of the threshold that is used to classify a locus under selection (see, e.g., Fawcett 2006, for further information). For SelEstim, the classifying variable was the KLD of the posterior distribution of the locus-specific coefficient of selection δj from its centering distribution, while in the case of BayeScan it was the Bayes factor. The ROC analysis yields a monotonic curve with no positives (true or false) at one end and all positives at the other. If a method has no classification power, the curve should be linear with slope 1, and the area under the ROC curve (AUC) should be 0.5. If a method has perfect classification power, the curve should perfectly superimpose to the left-hand and top sides of the unit square, and the AUC should be 1. Considering positively selected loci first, the area under the ROC curve for SelEstim is slightly larger, and closer to 1, than that obtained for BayeScan (see Figure 5). In particular SelEstim appears to have progressively improved relative performance with decreasing values of σ. As for loci under balancing selection, SelEstim seems slightly better than BayeScan based on the ROC analysis, although both methods lack statistical power in these sets of simulated data.
Figure 5.
Receiver operating characteristic (ROC) analysis for the data sets 1 (A), 2 (B), 3 (C), 4 (D), 5 (E), 6 (F), 7 (G), 8 (H) and 9 (I). In the ROC analysis, the proportion of false positives and true positives is computed for each possible value of the threshold that is used to classify a locus under selection. For SelEstim, the classifying variable was the KLD between the posterior distribution of the locus-specific coefficient of selection δj and its centering distribution, while in the case of BayeScan it was the Bayes factor.
The analysis of data sets 1–11 (see Table 1) took 12,175 sec on average per data set (SD = 6921) with BayeScan and 12,633 sec (SD = 6149) with SelEstim. SelEstim is therefore 3.85% slower than BayeScan, based on the same MCMC parameters (number and length of pilot runs, burn-in, chain length, etc.) and using the same number of processor cores. Note, however, that the KLD calibration procedure of SelEstim comes at the cost of up to a doubled computing time, due to the additional analysis of the POD.
Robustness to model misspecification
We analyzed simulated data departing from the island model assumptions (see Figure S1). The rate of false positives detected by SelEstim at a given threshold was higher for data simulated from a hierarchical island model or a stepping-stone model, as compared to data simulated from a nonhierarchical model with the same overall FST (Figure S12). However, for all scenarios considered, the false-positive rate at any KLD threshold was less than or nearly equal to the corresponding quantile probability (Figure S12F), which suggests that our calibration procedure is “conservative,” even for strong departures from the island model assumptions. Furthermore, the rates of false positives for these scenarios were much lower as compared to BayeScan analyses of the same data (Figure S13).
Inference of selection coefficients
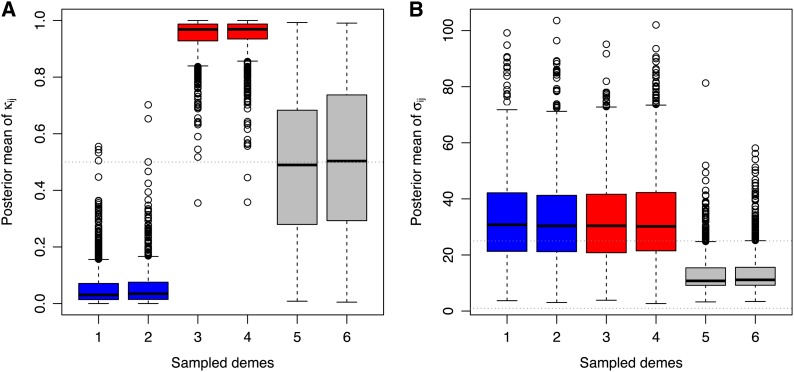
For data set 5 (see Table 1), we examined the distributions of the posterior means of the parameters κij (which indicate which allele is selected for), for the 1000 positively selected loci. Here, from the hypotheses of our simulation model, κij = 0 indicates that the blue allele is selected for, and κij = 1 indicates that the red allele is selected for. Figure 6A shows the distributions of the posterior means of κij in each sampled deme. Consistent with our expectation, it is apparent from Figure 6A that the posterior means of κij in demes 1 and 2 (blue demes) are shifted toward zero and that the posterior means of κij in demes 3 and 4 (red demes) are shifted toward one. Alleles of the right color are therefore selected for in the right deme. It is also reassuring to see that in demes 5 and 6 (uncolored demes), the posterior means of κij are centered around 0.5, which is consistent with the fact that neither allele should be selected for in these demes.
Figure 6.
Analysis of the allele count data from data set 5. (A) Boxplot representation of the posterior means of the parameters κij (which indicate which allele is selected for) for the 1000 positively selected loci in blue demes (1–2), red demes (3–4), and uncolored demes (5–6). (B) Boxplot representation of the posterior means of the selection coefficients σij for positively selected loci in data set 5. For blue demes, the posterior means of the selection coefficients σij are conditional upon the blue allele being selected for (κij = 0). For red demes, the posterior means of the selection coefficients σij are conditional upon the red allele being selected for (κij = 1). For uncolored demes, the posterior means of the selection coefficients σij are unconditional. The horizontal dashed line gives the true (simulated) value of σij = 25.
For the same 1000 positively selected loci, we further examined the posterior means of the scaled coefficients of selection σij ≡ 2Nisij, conditionally on κij. By doing so, we estimate the coefficient of selection associated with the allele being effectively targeted by selection. Figure 6B shows that the posterior means of f(σij|κij = 0) in blue demes and the posterior means of f(σij|κij = 1) in red demes are very close to the simulated values (σ ≡ 2Ns =25 in data set 5; see Table 1). By contrast, in uncolored demes, the posterior means of σij that were not conditioned upon κij, are much lower and closer to the prior distribution of the hyperparameter λ (which represents the genome-wide effect of selection over all demes and loci). Figure S14, Figure S15, Figure S16, Figure S17, and Figure S18 reproduce the same outputs as in Figure 6 for data sets 1–4 and 6–11. The posterior means of the scaled coefficients of selection σij conditionally on κij are very close to the simulated values, for FST ≥ 0.05 and σ ≥ 25 (data sets 2–3, 5–6, and 8–9) and FST = 0.2 and σ = 10 (data set 7). All else being equal, increasing the number of sampled demes improves the estimation of the scaled coefficients of selection σij conditionally on κij (compare Figures 6 and Figure S18).
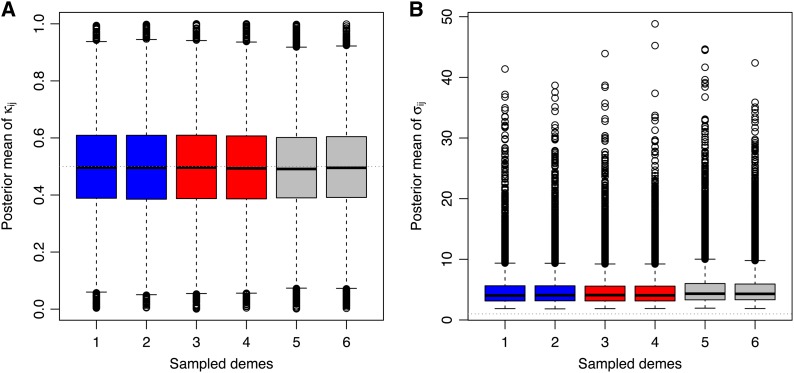
Last, we examined the distributions of the posterior means of κij for the 8000 neutral markers in data set 5. Figure 7A shows that the posterior means of κij, which do not depend on the color of the sampled demes, are all centered around 0.5. This result is consistent with the fact that neither allele should be selected for in these demes. Furthermore, the distributions of the posterior means of κij for neutral markers are narrower, as compared to the posterior means of κij for selected loci in uncolored demes (see Figure 6A). The distributions of the posterior means of κij for neutral markers are therefore closer to their prior distribution. Yet, the distributions are still wider than expected from the prior distribution, since the mean over 4000 independent samples (which corresponds to the MCMC length) from a Ber(0.5) distribution should be approximately normally distributed with mean 0.5 and standard deviation 0.008. This extra variance may be due to the hierarchical structure of the model, which produces a correlation between the parameters. In addition, Figure S19A shows that, in the absence of selection (data set 16; see Table 1), the distribution of the posterior means of κij is centered around 0.5 and narrower as compared to data sets that include positively selected loci (compare with Figure 7A). Therefore, the higher variance of the distributions of κij for selected loci in uncolored demes (as compared to neutral markers) certainly stems from the influence of selection occurring for the same loci in blue and red demes, through the prior on the locus-specific hyperparameter δj. The posterior means of σij for neutral markers (unconditionally upon κij) are very low and close to the posterior mean of the hyperparameter λ. In the absence of selection (data set 16), the posterior means of σij for neutral markers (unconditionally upon κij) are also very low and not different from the prior mean of the hyperparameter λ (Figure S19B).
Figure 7.
Analysis of the allele count data from data set 5. (A) Boxplot representation of the posterior means of the parameters κij (which indicate which allele is selected for) for the 8000 neutral markers in blue demes (1–2), red demes (3–4), and uncolored demes (5–6). (B) Boxplot representation of the posterior means of the selection coefficients σij for neutral markers in data set 5. The posterior means of the selection coefficients σij are unconditional.
Implicitly, Figure 6 and Figure 7 demonstrate that SelEstim is able to give accurate measures of the scaled coefficient of selection at one locus in different demes and therefore to provide evidence of local adaptation. This paves the way for the inference of the distribution of selection strength across populations in a landscape as is illustrated in the next section.
Last, we analyzed a set of 11 simulations using the same parameters as for data set 5 (see Table 1), but varying the proportion of selected loci from 10 to 5000 of 10,000 markers (hence, from 0.1 to 50%). Interestingly, we found a strong correlation between the posterior mean of the genome-wide coefficient of selection λ and the number of positively selected loci (see Figure S20). However, as the number of positively selected loci increases, the performance of SelEstim weakens (>20% of selected markers), although less markedly than BayeScan (see Figure S21).
Application to human data
We ran three independent MCMC analyses on a subset of the Stanford HGDP–CEPH Human Genome Diversity Cell Line Panel (Cann et al. 2003) SNP Genotyping Data. The data consisted in 52,631 SNPs from the HGDP–CEPH data, and two SNPs (−13910C → T and −22018G → A) known to be tightly associated with lactase persistence (Bersaglieri et al. 2004), genotyped in 23 populations from Africa and Eurasia. After 25 pilot runs of 1000 iterations, each MCMC was run for 100,000 updating steps, after a burn-in period of 25,000 steps. Samples were collected from the Markov chains for all the model parameters every 25 steps (thinning) to reduce autocorrelations, yielding 4000 samples for each parameter.
Convergence was assessed by computing the multivariate extension of Gelman–Rubin’s diagnostic (Brooks and Gelman 1998) on the three independent Markov chains. Gelman–Rubin’s diagnostic is based on the computation of the ratio of the pooled-chains variance over the within-chain variance and was calculated using the coda package, v. 0.16-1, (Plummer et al. 2006) as implemented for R (R Core Team 2013). Gelman–Rubin’s diagnostic was equal to 1.09 for the hyperparameter λ and to 1.07 for the parameters Mi, which indicates that the chains converge satisfactorily to the target distribution. The following analyses are based on the outputs from one of the three Markov chains.
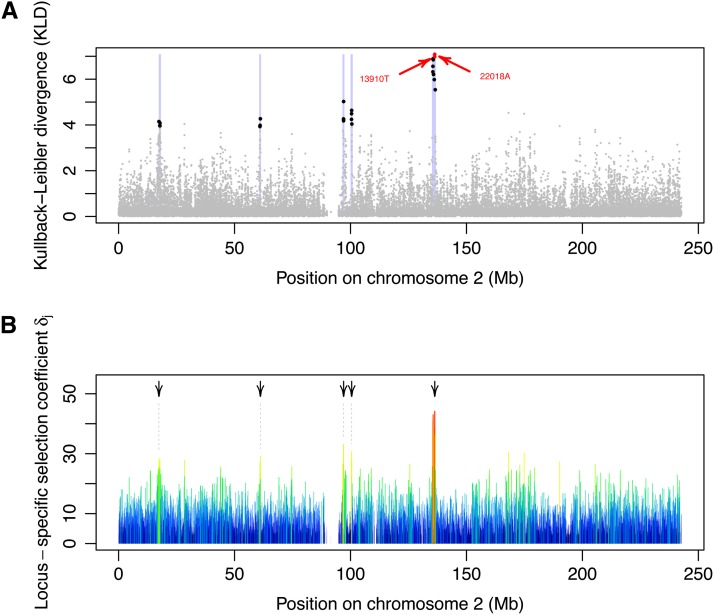
To identify the genomic regions showing the strongest signatures of selection in the HGDP–CEPH data, 1-Mb windows were constructed for each marker by including all markers that were ≤500 kb from that marker. The average number of markers per window was ∼248. Outstanding regions were then defined as the windows containing at least three SNPs above the critical KLD value of 3.924 (corresponding to the 99.9% quantile of the KLD distribution from the POD analysis). Figure 8A shows the distribution of the KLD for each SNP along HSA2. The two SNPs that are tightly associated with lactase persistence (−13910C → T and −22018G → A) are highlighted. These two SNPs have the two largest KLD values. Furthermore, the nine SNPs with the largest KLD values were located 3.7 kb and 1.0 Mb upstream of the LCT gene, at <805.2 kb from −13910C → T and <813.4 kb from −22018G → A. Figure 8B represents the distribution of the posterior means of the locus-specific selection parameter δj, along HSA2. This figure therefore represents the variation of the strength of selection along the chromosome and depicts a very strong signal of positive selection in the vicinity of the LCT gene (located from base pair 136,545,414 to 136,594,749), which encodes for the enzyme lactase–phlorizin hydrolase and is associated with adult-type hypolactasia. In addition to the LCT region, we found four other outstanding genomic regions, which are indicated by arrows in Figure 8B. The closest gene from each region was determined from the UCSC Genome Browser (http://genome.ucsc.edu/), using the Genome Reference Consortium GRCh37 assembly (hg19). Table 2 provides the list of these genes, along with their functions.
Figure 8.
Analysis of the HGDP–CEPH data. (A) KLD between the posterior of δj and its centering distribution. The genomic regions highlighted in light purple show the strongest signatures of selection in the HGDP–CEPH data. They correspond to sets of 1-Mb windows (centered around each marker) containing at least three SNPs above the critical KLD value of 3.924 (corresponding to the 99.9% quantile of the KLD distribution from the POD analysis). For each region, the SNPs with KLD ≥ 3.924 are shown in black. The alleles −13910T and −22018A associated with lactase persistence are shown in red. (B) Locus-specific selection coefficient δj along chromosome 2. The color (from blue to red) and the width of each segment is proportional to the strength of selection. The arrows highlight the five outstanding genomic regions highlighted in A. The closest gene from each of these regions is provided in Table 2.
Table 2. List of the four genomic regions from chromosome 2 (besides the LCT region) showing the strongest signatures of selection.
| Region | SNP ID | Position | KLD | Closest gene (position) | Function |
|---|---|---|---|---|---|
| 1 | rs7355461 | 17,538,316 | 4.148 | RAD51AP2 (chr2:17,691,986–17,699,706) | Unknown functiona |
| 2 | rs1177279 | 61,295,122 | 4.269 | KIAA1841 (chr2:61,293,006–61,316,639) | Unknown function |
| 3 | rs1256991 | 97,669,386 | 5.023 | FAM178B (chr2:97,541,619–97,652,301) | Unknown function |
| 4 | rs1519662 | 101,143,618 | 4.636 | NMS (chr2:101,086,944–101,099,742) | Neuropeptide signaling pathway |
To identify these regions (indicated by arrows in Figure 8B), 1-Mb windows were constructed for each marker by including all markers that were ≤500 kb from that marker. Outstanding regions were then defined as the windows containing at least three SNPs above the critical KLD value of 3.924 (corresponding to the 99.9% quantile of the KLD distribution from the POD analysis). For each region, the SNP with the highest KLD value is indicated, along with its position. The closest gene from each SNP, determined from the University of California—Santa Cruz Genome Browser (http://genome.ucsc.edu/) using the Genome Reference Consortium GRCh37 assembly (hg19), is provided along with its function.
Close to VSNL1 (chr2:17,720,393–17,838,285) in Pickrell et al. (2009) (top 10 XP–EHH signal).
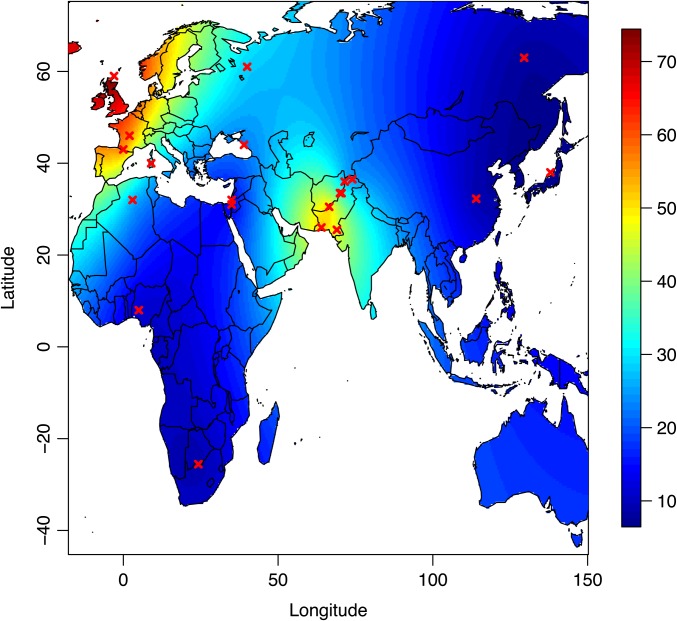
Figure 9 shows the distribution of the scaled coefficients of selection σij (conditionally on κij indicating allele −13910C → T to be targeted by selection) across African and Eurasian populations. The map from Figure 9 was extrapolated by kriging using the R package fields (Fields Development Core Team 2006), v. 6.8. It is obvious from Figure 9 that the intensity of selection is very strong in Europe and around the Indus valley and attains similar levels in both geographic regions.
Figure 9.
Extrapolated spatial distribution of the selection coefficient σij at locus −13910C → T, conditionally on allele −13910T being selected for, across African and Eurasian populations.
Discussion
We developed a hierarchical Bayesian model that considers explicitly the effect of genic selection on the distribution of allele frequencies at SNP loci. SelEstim extends previous methods based on the Dirichlet-multinomial distribution of allele frequencies (which reduces to the beta-binomial distribution for SNP data) that arises as the diffusion approximation of genetic drift in the migration-drift equilibrium island model (see, e.g., Beaumont and Balding 2004; Riebler et al. 2008; Foll and Gaggiotti 2008; Guo et al. 2009; Gautier et al. 2010). The beta-binomial model has been argued to be robust to the vagaries of demographic history (Beaumont and Nichols 1996; Beaumont 2005) because in many situations, the genealogy of genes in a metapopulation divides into a scattering phase, which represents the recent genealogy of each deme, and a collecting phase, which represents the ancestral genealogy of the whole metapopulation (Nordborg 1997; Wakeley 2004). With this separation of timescales, there are no mutations in the scattering phase and the distribution of gene frequencies in each deme depends upon the frequencies in the pool of migrants (i.e., the collecting phase) and the deme-specific FST. However, the assumption that each deme receives migrants from a unique migrant pool may not hold if populations share a history of successive divergences (Gaggiotti and Foll 2010). In that case indeed, gene frequencies may be correlated among closely related populations, which violates the assumption that populations are independent (Robertson 1975; Excoffier et al. 2009; Bonhomme et al. 2010; Gompert and Buerkle 2011). SelEstim should therefore be used with caution on populations that are known to be hierarchically structured (but see Figure S12). To conclude on a more positive note, we would argue that although violations from the island model assumptions certainly inflate the overall variance of the Mi parameters, it should not generate artificially correlated signals across closely linked SNPs as observed, e.g., in Figure 8. From a practical point of view, using sliding windows to identify genomic regions of interest may therefore constitute a valuable approach (see, e.g., Gautier et al. 2009). Accounting explicitly for the correlation of gene frequencies across populations due to shared history and gene flow might be achieved by considering the multivariate generalization of the Gaussian approximation of the gene frequency distribution (Coop et al. 2010; Günther and Coop 2013), which was recently extended to infer population splits and mixtures (Pickrell and Pritchard 2012). A Gaussian approximation for the distribution of allele frequencies (as suggested by Nicholson et al. 2002) can be justified whenever the deterministic equilibrium is located away from the boundaries (fixation of any one of the alleles). However, it is a poor approximation when the deterministic equilibrium is close to one of the boundaries (Barton and Rouhani 1987; Gautier and Vitalis 2013).
Our model introduces two major improvements over the methods based on the Dirichlet-multinomial or the beta-binomial distribution of gene frequencies. First, instead of being conceived as tests of departure from a neutral model (see, e.g., Gautier et al. 2010), SelEstim incorporates an explicit selection model, which allows selection strength to be inferred among a set of markers. Second, SelEstim provides the distribution of selection strength across populations, which allows identifying the local population(s) where selection is acting. This is so because the hierarchical structure of our model improves the estimation of locus- and population-specific coefficients of selection (σij) by borrowing strength across multiple populations.
Detecting selection among a set of markers
At a given SNP, j, the locus- and population-specific parameters of selection σij depend upon a locus-specific hyperparameter δj that gives the population-wide effect of selection at a particular locus. It is therefore natural to use the posterior distribution of the hyperparameters δj as a means to discriminate between neutral and selected markers. We indeed expect the posterior density of δj to be shifted toward zero if the jth marker is neutral and toward positive values if the jth marker is targeted by selection. To operate this classification, it would have been possible to follow Beaumont and Balding (2004) and adopt a simple informal criterion assuming that δj is significantly different from zero at some critical level P whenever its equal-tailed 100(1 − P)% credible interval excludes zero. Yet, this approach would neglect the genome-wide effect of selection, which in our model is driven by the hyperparameter λ. We therefore proposed comparing the posterior distributions of the locus-specific coefficients of selection δj with the centering distribution derived from the hyperdistribution with parameter λ. To that end, we used the KLD to measure the divergence between these two distributions. We calibrated this measure using simulations from a predictive distribution based on the observed data set. We found that the false-positive rate at any KLD threshold is always less than the corresponding quantile probability (Figure 3, Figure 4, Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, Figure S8, Figure S9, and Figure S10), which suggests that such calibration is conservative, even for strong departures from the island model assumptions (Figure S12F).
McCulloch (1989), Peng and Dey (1995), and Guo et al. (2009) proposed an alternative calibration, based on the following argument: consider flipping a “fair” coin with equal probability 0.5 for heads and tails vs. flipping a biased coin with probability ν for heads. Then the KLD between these two Bernoulli distributions denoted by Ber(0.5) and Ber(ν), respectively, can be computed and used as a threshold for any value of ν. In our case, however, we must divide ν in half, since we look for only unusually large KLD values. Therefore, the KLD threshold can be defined as KLD[Ber(0.5), Ber(ν)] = −log[ν(2 − ν)]/2. For example, the KLD between two Bernoulli distributions corresponding to flipping a fair coin and a biased coin that gives heads or tails with probability 0.05 (resp. 0.01) is equal to 1.164 (resp. 1.959). However, we found that this calibration procedure was overly conservative (see Table S1 and Table S2).
Last, we used ROC analyses, as in Riebler et al. (2008), to compare our model with BayeScan (Foll and Gaggiotti 2008). We found that SelEstim performed slightly better than BayeScan (Figure 5). Since BayeScan was shown to outperform Beaumont and Balding’s approach (Beaumont and Balding 2004), as well as some other popular moment-based methods using dominant markers (Pérez-Figueroa et al. 2010), we may therefore conclude that SelEstim represents an appreciable improvement to the population genomicist’s toolbox.
As an alternative to the KLD, it could have been possible to implement Bayesian model selection using, e.g., Bayes factors to discriminate between neutral and selected loci. Foll and Gaggiotti (2008) proposed using reversible-jump MCMC to estimate, for each marker, the posterior probabilities of two alternative models: a purely neutral one and one including selection. Riebler et al. (2008) also proposed an elegant reparameterization of Beaumont and Balding’s model (Beaumont and Balding 2004), by introducing a Bernoulli-distributed auxiliary variable to indicate whether a locus is targeted by selection. This parameterization was later shown to facilitate the computation of Bayes factors (Gautier et al. 2009). Both approaches (reversible-jump MCMC and auxiliary variable) are actually straightforward to implement in our hierarchical-Bayesian model (not shown). Nevertheless, the KLD has the practical advantage that, provided the MCMC sampler converges and a large enough sample is drawn from the δj and the λ posterior distributions, its computation does not depend upon the length of the MCMC. As an illustration example, we ran a BayeScan analysis of the 52,633 SNPs from the HGDP–CEPH data, using the same MCMC parameters (number and length of pilot runs, burn-in, chain length, etc.) as with SelEstim, assuming prior odds of 1000 for the neutral model (Figure S22A). It is clear from this figure that a substantial number of markers for which the Kullback–Leibler divergence provides no evidence of selection have log10(BF) ≥ 2 (Figure S22, B and C). This number increases with decreasing prior odds (not shown). Furthermore, since the maximum value that the BF can take is bounded by the MCMC length, we may observe a “saturation” effect with many of the outliers sharing the same evidence of selection (see Figure S22A). From a practical point of view, this may prevent the visual identification of genomic regions potentially targeted by selection (as, e.g., in Figure 8), unless very long MCMC are performed (to achieve an effective sample size of the order of the number of markers). In addition, we found that the outputs of BayeScan vary with the prior odds, which depend on the user’s prior belief for the proportion of presumably neutral SNPs. Our results therefore argue in favor of using KLD in empirical studies since it allows ranking the SNPs in order of the divergence between locus-specific and genome-wide selection strength, which indicates the degree of evidence that a locus is under selection. However, we concur with Coop et al. (2010) that making statements about the statistical significance of outlier loci might be hazardous. In particular, we refrained from defining P-values from KLD measures.
Inferring selection strength across populations
In an early analysis of population differentiation using the HapMap data set, Weir et al. (2005) already showed the utility of estimating population-specific FST values (Weir and Hill 2002). In particular, concentrating their analyses on chromosome 2, they did not find any outstanding peak of population average FST around the LCT gene, although there was a clear elevation of the population-specific FST values for Caucasians of European descent and European Americans. Yet, in Weir et al.’s study (Weir et al. 2005), the characterization of “exceptional regions” was based on the greatest difference between population-specific FST values (averaged over 5-Mb windows) being larger than 3 SD, which does not provide a definitive statistical criterion to decide which loci are outliers of the empirical, genome-wide distribution of FST.
Like Beaumont and Balding (2004), Riebler et al. (2008), Foll and Gaggiotti (2008), and Guo et al. (2009), who considered population-specific effects on FST, we considered in our model that the distribution of allele frequency depends upon population-specific parameters (Mi). Since we defined a parameter that indicates which allele is selected for, the selected allele need not to be the same in all the sampled demes. Furthermore, the strength of selection need not to be the same in all demes. SelEstim therefore accounts for situations where selection is acting in some populations, but not all, possibly in opposite direction (with alternative alleles being selected for in different environments). It is therefore particularly relevant to detect the signatures of local adaptation in subdivided populations.
Not surprisingly, we found that SelEstim has weak statistical power to identify loci under balancing selection (see Figure 5). Since our genic selection model allows for only positive selection, this was somewhat expected, but by using a centering distribution we are able, in principle, to identify loci with support for unusually low values of δ. Beaumont and Balding (2004) concluded from simulations that their method could not easily identify loci under balancing selection, even for very strong selection. Although Foll and Gaggiotti (2008) showed that microsatellites could be used to detect balancing selection, especially with data sets containing a large number of sampled populations, they needed 10 populations with SNPs to achieve the same rate of detection (Foll and Gaggiotti 2008). In principle, however, it should be possible to scan for SNPs targeted by balancing selection using a modified version of our model, in particular Equation 2, that would account for overdominance with population-specific selection pressures (see, e.g., Equation 13.60 in Wright 1969, p. 371). This strategy could be valuable for improving statistical power to identify loci under balancing selection.
Because our model accounts explicitly for positive selection, it cannot only be used to detect the genomic signatures of selection, but also to measure the strength of selection along the genome. As mentioned above, contrary to previous approaches that approximated selection as a locus-specific effect in a logistic regression model (Beaumont and Balding 2004) or a reduction in effective migration rate (see, e.g., Bazin et al. 2010), we introduced explicitly a scaled coefficient of selection σij ≡ 2Nisij for locus j in deme i, where sij represents the relative gain in fitness brought by a positively selected allele. We found that the posterior means of the scaled coefficients of selection σij (conditionally on κij) were close to the simulated value for positively selected loci, although slightly overestimated (Figure 6, Figure S14, Figure S15, Figure S16, Figure S17, and Figure S18). We also found that the variation of σij across populations with different selection regimes was remarkably well inferred, with selected loci exhibiting large coefficients of selection in the colored demes and small coefficients of selection in uncolored demes (Figure 6, Figure S14, Figure S15, Figure S16, Figure S17, and Figure S18).
The strong correlation between the posterior mean of the genome-wide coefficient of selection λ and the number of positively selected loci (Figure S20) would tend to suggest that the parameter λ provides some information about the extent of selection acting on the genome. This must be nuanced, however, at least for two reasons. First, as the number of positively selected loci increases, the performance of SelEstim weakens (see Figure S21). Second, we have observed that the parameter λ also depends on demography, and particularly on departures from the island model assumptions (not shown). This identifiability problem therefore prevents the comparison of λ estimates (to infer the overall effect of selection) across species with different population structures. We note that this identifiability problem is somewhat avoided in BayeScan with the Gaussian prior (zero mean and standard deviation of 1) put on the locus-specific component αi (which, therefore, provides no information whatsoever on the extent of selection acting on the genome).
Application example at the LCT gene
To illustrate how the inference of selection strength may provide new insights into the characterization of local adaptation, we investigated the well-studied and clear-cut example of the evolution of lactase persistence in humans (see Gerbault et al. 2011, for a review). The region around the LCT gene that allows lactose tolerance to persist into adulthood is indeed a very-well-known example of selection in humans (Sabeti et al. 2006). The first causative polymorphism described was the −13910C → T mutation (Enattah et al. 2002), which lies in the cis-acting regulatory element located in the 13th intron of a neighboring gene, MCM6. Although this single mutation of purported western Eurasian origin accounts for much of observed lactase persistence outside Africa, multiple independent mutations in the same region upstream of the LCT gene have been associated with this trait in pastoralists from Saudi Arabia (Enattah et al. 2008) and Africa (Tishkoff et al. 2007). The lactase persistence allele at the LCT locus lies on a haplotype that is common in Europeans but that extends largely undisrupted for >1 Mb, much farther than is typical for an allele of that frequency (Bersaglieri et al. 2004). More recently, Romero et al. (2012) found that the −13910C → T mutation also explains a substantial proportion of lactase persistence in the Indian subcontinent. Most interestingly, they showed that the −13910C → T mutation in India is identical by descent to the European allele and is associated with the same extended haplotype in both populations, which strongly suggests that the origin of the −13910C → T mutation is shared in Europe and India. These results are consistent with the high levels of present-day milk consumption in India and with archeological and genetic evidence for the independent domestication of cattle in the Indus valley ca. 7000 years ago (Romero et al. 2012).
In agreement with these studies, our analyses pointed to a very strong signal of positive selection between 3.7 kb and 1.0 Mb upstream of the LCT gene (Figure 8). The strongest evidence of selection (in terms of KLD) was found for the two SNPs that are tightly associated with lactase persistence (−13910C → T and −22018G → A). Building on the fact that our model is able to give accurate measures of the scaled coefficient of selection at each locus in different demes, we further examined the distribution of the strength of positive selection at the −13910C → T SNP across the 23 populations analyzed. We found the strongest selection coefficients in Europe and in the Indus Valley (Figure 9), which matches the interpolated map of lactase persistence phenotype frequencies in the Old World (Itan et al. 2010). More precisely, we found that the coefficients of selection (σ ≡ 2Ns) at the −13910T allele ranged from 8.73 (Sardinians) to 101.66 (Orcadians) in Europe and from 4.94 (Kalash) to 77.50 (Balochi) in Central/South Asia. There have been previous attempts to measure the strength of selection acting at the LCT gene, although most of them relied on strong assumptions on the demographic and adaptive history of the studied populations. For example, Aoki (1986) predicted that a selection coefficient s > 5% would be necessary to explain the observed allele frequency of the −13910T allele, assuming that this mutation appeared 6000 years ago in a population of effective size 500, which would give σ ≡ 2Ns = 50. Bersaglieri et al. (2004) estimated the coefficient of selection s to be 1–15% for a new mutation arising in a population of effective size of 500–5000. More recently, Tishkoff et al. (2007) estimated selection intensity by matching simulated data under a coalescent framework to the observed centimorgan span and the observed frequency of the allele targeted by selection. They found extremely recent and strong positive selection in many African populations (σ ≡ 2Ns ranging from 800 to 1940 assuming an effective population size N of 10,000). Modeling a geographical structuring of selection pressure by latitude, Gerbault et al. (2009) found selection coefficients in the range between 0.8 and 1.8% (Gerbault et al. 2011), also assuming a carrying capacity of 10,000. However, assuming an effective population size N of 10,000 may largely overestimate σ ≡ 2Ns (see Tenesa et al. 2007, for more accurate estimates of effective size based on measures of linkage disequilibrium). Last, using a spatially explicit model and approximate Bayesian computation (Beaumont et al. 2002), Itan et al. (2009) estimated coefficients of selection to lie in the range of 5.2–15.9%. The difficulty in comparing these values is that strong hypotheses about the effective population size need to be made. It is clear from the stationary density of the diffusion process in Equation 2 that the two parameters s and N are not identifiable. Estimating s therefore requires informative priors on N. Furthermore, the population size considered in our model is the local effective size of a deme, not the effective size of the total population. Therefore, considering the scaled coefficient of selection (σ ≡ 2Ns) might be more appropriate for interpreting the variation of the strength of selection exerted at different loci or at one locus in different populations.
Perspectives
SelEstim provides a new tool with which to detect signatures of selection from genome-wide scan studies and, perhaps most importantly, to infer the intensity of selection across loci and populations. However, as most FST-based methods aimed at looking for locus-specific effects on FST estimates, SelEstim assumes that molecular markers are independent from each other. There are few exceptions, though. For example Guo et al. (2009) introduced a conditional autoregressive model to incorporate the local correlation among SNPs. Gompert and Buerkle (2011) proposed an extension of the models developed by Beaumont and Balding (2004), Riebler et al. (2008), and Foll and Gaggiotti (2008), which incorporates genetic distances among haplotypes (φ-statistics; see Excoffier et al. 1992) in measures of genetic differentiation. More recently, Fariello et al. (2013) developed a haplotype-based method, which uses a multipoint linkage disequilibrium model (Scheet and Stephens 2006) that regroups individual chromosomes into local haplotype clusters. The reconstructed haplotypes are then used to measure differentiation between populations (see also Browning and Weir 2010). Handling conditional dependencies of markers along the genome would therefore be an essential step forward in future developments of SelEstim. In the meantime, we recommend potential users to view this method as a first step toward identify genomic regions of interest, which should then be characterized more specifically in further studies.
Supplementary Material
Acknowledgments
We acknowledge two anonymous reviewers for helpful comments, which led to a significantly improved version of this manuscript. We also warmly thank Pierre Pudlo and Christian P. Robert for insightful discussions on KLD calibration. This work was supported by a Biotechnology and Biological Sciences Research Council grant (BBS/B/12776) awarded to M.A.B. and K.J.D. M.A.B. was supported by a Natural Research Council Advanced Fellowship (NER/J/S/2001/00792). R.V. was supported by the French National Research Agency (ANR) programme NUTGENEVOL 07-BLAN-0064. Both R.V. and M.G. were supported by the ANR programme EMILE 09-BLAN-0145-01 and by the Institut National de la Recherche Agronomique, through the support of the starting group “IG-GiPop” (2010–2012).
Footnotes
Communicating editor: W. Stephan
Literature Cited
- Abramowitz M., Stegun I. A., 1965. Handbook of Mathematical Functions. Dover, New York. [Google Scholar]
- Akey J. M., Zhang G., Jin L., Shriver M. D., 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K. A., 1986. Stochastic model of gene-culture coevolution suggested by the ‘culture historical hypothesis’ for the evolution of adult lactose absorption in humans. Proc. Natl. Acad. Sci. USA 83: 2929–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding D. J., 2003. Likelihood-based inference for genetic correlation coefficients. Theor. Popul. Biol. 63: 221–230. [DOI] [PubMed] [Google Scholar]
- Balding D. J., Nichols R. A., 1995. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica 96: 3–12. [DOI] [PubMed] [Google Scholar]
- Barreiro L. B., Laval G., Quach H., Patin E., Quintana-Murci L., 2008. Natural selection has driven population differentiation in modern humans. Nat. Genet. 40: 340–345. [DOI] [PubMed] [Google Scholar]
- Barton N., Rouhani S., 1987. The frequency of shifts between alternative equilibria. J. Theor. Biol. 125: 397–418. [DOI] [PubMed] [Google Scholar]
- Barton N., Turelli M., 1987. Adaptive landscapes, genetic distance and the evolution of quantitative characters. Genet. Res. 49: 157–173. [DOI] [PubMed] [Google Scholar]
- Bazin E., Beaumont M. A., Dawson K. J., 2010. Likelihood-free inference of population structure and local adaptation in a bayesian hierarchical model. Genetics 185: 587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont M. A., 2005. Adaptation and speciation: What can FST tell us? Trends Ecol. Evol. 20: 435–440. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Balding D. J., 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13: 969–980. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Nichols R. A., 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 263: 1619–1626. [Google Scholar]
- Beaumont M. A., Zhang W., Balding D. J., 2002. Approximate bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri T., Sabeti P., Patterson N., Vanderploeg T., Schaffner S., et al. , 2004. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74: 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black W. C., Baer C. F., Antolin M. F., DuTeau N. M., 2001. Population genomics: genome-wide sampling of insect populations. Annu. Rev. Entomol. 46: 441–469. [DOI] [PubMed] [Google Scholar]
- Bonhomme, M., C. Chevalet, B. Servin, S. Boitard, J. Abdallah et al., 2010 Detecting selection in population trees: the Lewontin and Krakauer test extended. Genetics 186: 241–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks S., Gelman A., 1998. General methods for monitoring convergence of iterative simulations. J. Comput. Graph. Statist. 7: 434–455. [Google Scholar]
- Browning S., Weir B., 2010. Population structure with localized haplotype clusters. Genetics 185: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger R., 2000. The Mathematical Theory of Selection, Recombination Mutation.Wiley, Chichester, England. [Google Scholar]
- Bustamante C. D., Wakeley J., Sawyer S. A., Hartl D. L., 2001. Directional selection and the site-frequency spectrum. Genetics 159: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C. D., Nielsen R., Hartl D. L., 2003. Maximum likelihood and bayesian methods for estimating the distribution of selective effects among classes of mutations using DNA polymorphism data. Theor. Popul. Biol. 63: 91–103. [DOI] [PubMed] [Google Scholar]
- Cann H. M., de Toma C., Cazes L., Legrand M.-F., Morel V., et al. , 2003. A human genome diversity cell line panel. Science 296: 261–262. [DOI] [PubMed] [Google Scholar]
- Coop G., Witonsky D., Di Rienzo A., Pritchard J. K., 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185: 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P., Tavaré S., 1995. Coalescents and genealogical structure under neutrality. Annu. Rev. Genet. 29: 401–421. [DOI] [PubMed] [Google Scholar]
- Donnelly P., Nordborg M., Joyce P., 2001. Likelihoods and simulation methods for a class of nonneutral population genetics models. Genetics 159: 853–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enattah N. S., Sahi T., Savilahti E., Terwilliger J. D., Peltonen L., et al. , 2002. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 30: 233–237. [DOI] [PubMed] [Google Scholar]
- Enattah N. S., Jensen T. G., Nielsen M., Lewinski R., Kuokkanen M., et al. , 2008. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 82: 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier S. N., Nagylaki T., 1988. Diffusion approximations of markov chains with two time scales and application to population genetics, II. Adv. Appl. Probab. 20: 525–545. [Google Scholar]
- Excoffier L., Smouse P. E., Quattro J. M., 1992. Analysis of molecular variance inferred from metric distances among dna haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Hofer T., Foll M., 2009. Detecting loci under selection in a hierarchically structured population. Heredity 103: 285–298. [DOI] [PubMed] [Google Scholar]
- Fariello, M. I., S. Boitard, H. Naya, M. SanCristobal, and B. Servin, 2013 Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 193: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T., 2006. An introduction to ROC analysis. Pattern Recognit. Lett. 27: 882–891. [Google Scholar]
- Fields Development Core Team, 2006 fields: Tools for Spatial Data. National Center for Atmospheric Research, Boulder, Colorado. [Google Scholar]
- Foll M., Gaggiotti O., 2008. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frichot E., Schoville S. D., Bouchard G., François O., 2013. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 30: 1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggiotti O., Foll M., 2010. Quantifying population structure using the F-model. Mol. Ecol. Res. 10: 821–830. [DOI] [PubMed] [Google Scholar]
- Gautier M., Vitalis R., 2012. rehh: An r package to detect footprints of selection in genome-wide snp data from haplotype structure. Bioinformatics 28: 1176–1177. [DOI] [PubMed] [Google Scholar]
- Gautier M., Vitalis R., 2013. Inferring population histories using genome-wide allele frequency data. Mol. Biol. Evol. 39: 654–668. [DOI] [PubMed] [Google Scholar]
- Gautier M., Flori L., Riebler A., Jaffrézic F., Laloé D., et al. , 2009. A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genomics 10: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M., Hocking T. D., Foulley J.-L., 2010. A Bayesian outlier criterion to detect SNPs under selection in large data sets. PLoS ONE 5: e11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Carlin J. B., Stern H. S., Rubin D. B., 2004. Bayesian Data Analysis, Ed. 2 Chapman & Hall, New York. [Google Scholar]
- Gerbault P., Moret C., Currat M., Sanchez-Mazas A., 2009. Impact of selection and demography on the diffusion of lactase persistence. PLoS ONE 4: e6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbault P., Liebert A., Itan Y., Powell A., Currat M., et al. , 2011. Evolution of lactase persistence: an example of human niche construction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366: 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks W. R., Richardson S., Spiegelhalter D. J., 1996. Markov Chain Monte Carlo in Practice, Ed. 2 Chapman & Hall, New York. [Google Scholar]
- Goldstein D. B., Chikhi L., 2002. Human migrations and population structure: what we know and why it matters. Annu. Rev. Genomics Hum. Genet. 3: 129–152. [DOI] [PubMed] [Google Scholar]
- Gompert Z., Buerkle C. A., 2011. A hierarchical bayesian model for next-generation population genomics. Genetics 187: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot, G., R. Vitalis, A. le Rouzic, and M. Gautier, 2014 Detecting correlation between allele frequencies and environmental variables as a signature of selection: a fast computational approach for genome-wide studies. Spatial. Stat. (in press).
- Günther T., Coop G., 2013. Robust identification of local adaptation from allele frequencies. Genetics 195: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Dey D. K., Holsinger K. E., 2009. A bayesian hierarchical model for analysis of single-nucleotide polymorphisms diversity in multilocus, multipopulation samples. J. Am. Stat. Assoc. 104: 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A. M., Witonsky D. B., Ehler E., Alkorta-Aranburu G., Beall C., et al. , 2010. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc. Natl. Acad. Sci. USA 107: 8924–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A. M., Witonsky D. B., Alkorta-Aranburu G., Beall C. M., Gebremedhin A., et al. , 2011. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 7: e1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds D. A., Stuve L. L., Nilsen G. B., Halperin E., Eskin E., et al. , 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307: 1072–1079. [DOI] [PubMed] [Google Scholar]
- Innan H., Kim Y., 2004. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl. Acad. Sci. USA 101: 10667–10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium , 2003. The international HapMap project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Powell A., Beaumont M. A., Burger J., Thomas M. G., 2009. The origins of lactase persistence in europe. PLOS Comput. Biol. 5: e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan Y., Jonesand B. L., Ingram C. J. E., Swallow D. M., Thomas M. G., 2010. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol. Biol. 10: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys H., 1961. Theory of Probability, Ed. 3 Oxford University Press, Oxford. [Google Scholar]
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795. [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C., Krakauer J., 1973. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphism. Genetics 74: 175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G., England P. R., Tallmon D., Jordan S., Taberlet P., 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4: 981–994. [DOI] [PubMed] [Google Scholar]
- McCulloch R., 1989. Local model influence. J. Am. Stat. Assoc. 84: 473–478. [Google Scholar]
- Nei M., Maryuyama T., 1975. Lewontin–Krakauer test for neutral genes. Genetics 80: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson G., Smith A. V., Jónsson F., Gústafsson Ó., Stefánsson K., et al. , 2002. Assessing population differentiation and isolation from single-nucleotide polymorphism data. J. R. Stat. Soc. Series B Stat. Methodol. 64: 695–715. [Google Scholar]
- Nielsen R., 2001. Statistical tests of selective neutrality in the age of genomics. Heredity 86: 641–647. [DOI] [PubMed] [Google Scholar]
- Nielsen R., 2005. Disclosure of variation. Nature 434: 288–289. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A. G., Glanowski S., Sackton T. B., et al. , 2005a A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., et al. , 2005b Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Hubisz M. J., Hellmann I., Torgerson D., Andrés A. M., et al. , 2009. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 19: 838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., 1997. Structured coalescent processes on different time scales. Genetics 146: 1501–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntzoufras I., 2009. Bayesian Modeling Using WinBugs. Wiley, Hoboken, NJ. [Google Scholar]
- Payseur B. A., Cutter A. D., Nachman M. W., 2002. Searching for evidence of positive selection in the human genome using patterns of microsatellite variability. Mol. Biol. Evol. 19: 1143–1153. [DOI] [PubMed] [Google Scholar]
- Peng F., Dey D. K., 1995. Bayesian analysis of outlier problems using divergence measures. Can. J. Stat. 23: 199–213. [Google Scholar]
- Pérez-Cruz, F., 2008 Kullback–Leibler divergence estimation of continuous distributions, pp. 1666–1670 in IEEE International Symposium on Information Theory (ISIT), Toronto, Ontario, Canada. [Google Scholar]
- Pérez-Figueroa A., García-Pereira M. J., Saura M., Rolán-Alvarez E., Caballero A., 2010. Comparing three different methods to detect selective loci using dominant markers. J. Evol. Biol. 23: 2267–2276. [DOI] [PubMed] [Google Scholar]
- Petry D., 1983. The effect on neutral gene flow of selection at a linked locus. Theor. Popul. Biol. 23: 300–313. [DOI] [PubMed] [Google Scholar]
- Pickrell J. K., Pritchard J. K., 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8: e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Coop G., Novembre J., Kudaravalli S., Li J. Z., et al. , 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19: 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M., Best N., Cowles K., Vines K., 2006. Coda: output analysis and diagnostics for MCMC. R News 6: 7–11. [Google Scholar]
- Przeworsky M., Coop G., Wall J., 2005. The signature of positive selection on standing variation. Evolution 59: 2312–2323. [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Riebler A., Held L., Stephan W., 2008. Bayesian variable selection for detecting adaptive genomic differences among populations. Genetics 178: 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A., 1975. Remarks on the Lewontin–Krakauer test. Genetics 80: 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero I. G., Mallick C. B., Liebert A., Crivellaro F., Chaubey G., et al. , 2012. Herders of indian and european cattle share their predominant allele for lactase persistence. Mol. Biol. Evol. 29: 249–260. [DOI] [PubMed] [Google Scholar]
- Ross K. G., Shoemaker D. D., Krieger M. J. B., DeHeer J., Keller L., 1999. Assessing genetic structure with multiple classes of molecular markers: a case study involving the introduced fire ant Solenopsis invicta. Mol. Biol. Evol. 16: 525–543. [DOI] [PubMed] [Google Scholar]
- Rousset F., 1996. Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics 142: 1357–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z. P., Richter D. J., et al. , 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., Schaffner S. F., Fry B., Lohmueller J., Varilly P., et al. , 2006. Positive natural selection in the human lineage. Science 312: 1614–1620. [DOI] [PubMed] [Google Scholar]
- Sawyer S. A., Hartl D. L., 1992. Population genetics of polymorphism and divergence. Genetics 132: 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M., 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K., Thornton K. R., Stoneking M., 2007. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5: e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenesa A., Navarro P., Hayes B. J., Duffy D. L., Clarke G. M., et al. , 2007. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 17: 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S. A., Reed F. A., Ranciaro A., Voight B. F., Babbitt C. C., et al. , 2007. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis R., Boursot P., Dawson K., 2001. Interpretation of variation across marker loci as evidence of selection. Genetics 158: 1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 1999. Nonequilibrium migration in human history. Genetics 153: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 2004. Metapopulation models for historical inference. Mol. Ecol. 13: 865–875. [DOI] [PubMed] [Google Scholar]
- Weir B. S., Hill W. G., 2002. Estimating F-statistics. Annu. Rev. Genet. 36: 721–750. [DOI] [PubMed] [Google Scholar]
- Weir B. S., Cardon L. R., Anderson A. D., Nielsen D. M., Hill W. G., 2005. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 15: 1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. H., Hernandez R. A. Fledel Alon, L. Zhu, R. Nielsen et al, 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA 102: 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1949. Adaptation and selection, pp. 365–389 in Genetics, Paleontology, and Evolution, edited byJepson G. L., Simpson G. G., Mayr E. University Press, Princeton, NJ. [Google Scholar]
- Wright, S., 1969 Evolution and the Genetics of Populations: Vol. II. The Theory of Gene Frequencies. University of Chicago Press, Chicago. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.