Localized F-actin disassembly by ADF/cofilin drives invadopodial membrane recycling through endolysosomes, which promotes efficient cell transmigration through the basement membrane.
Abstract
Invadopodia are protrusive, F-actin–driven membrane structures that are thought to mediate basement membrane transmigration during development and tumor dissemination. An understanding of the mechanisms regulating invadopodia has been hindered by the difficulty of examining these dynamic structures in native environments. Using an RNAi screen and live-cell imaging of anchor cell (AC) invasion in Caenorhabditis elegans, we have identified UNC-60A (ADF/cofilin) as an essential regulator of invadopodia. UNC-60A localizes to AC invadopodia, and its loss resulted in a dramatic slowing of F-actin dynamics and an inability to breach basement membrane. Optical highlighting indicated that UNC-60A disassembles actin filaments at invadopodia. Surprisingly, loss of unc-60a led to the accumulation of invadopodial membrane and associated components within the endolysosomal compartment. Photobleaching experiments revealed that during normal invasion the invadopodial membrane undergoes rapid recycling through the endolysosome. Together, these results identify the invadopodial membrane as a specialized compartment whose recycling to form dynamic, functional invadopodia is dependent on localized F-actin disassembly by ADF/cofilin.
Introduction
Basement membrane is a thin, dense, sheet-like matrix that underlies or surrounds most tissues (Hohenester and Yurchenco, 2013). Cell invasion through basement membrane occurs in many normal and pathological processes, including embryo implantation, gastrulation, leukocyte trafficking, and cancer metastasis (Rowe and Weiss, 2008; Hagedorn and Sherwood, 2011). Because of its small pore size and cross-linked organization, cells are thought to require specialized mechanisms to transmigrate basement membrane (Madsen and Sahai, 2010). In particular, protrusive F-actin–based membrane structures, termed invadopodia, have been proposed to be critical for breaching basement membrane (Condeelis and Segall, 2003). Although extensively studied in transformed cells and cancer lines in vitro, the regulation of invadopodia in vivo has been difficult to establish due to the challenge of examining invasion in an intact organism (Beerling et al., 2011; Saltel et al., 2011).
Anchor cell (AC) invasion in Caenorhabditis elegans is a simple model of basement membrane transmigration that combines genetic analysis and imaging of the invasive cell–basement membrane interface (Hagedorn and Sherwood, 2011). The AC is a specialized uterine cell that invades through basement membrane to connect the developing uterine and vulval tissues during development (Sherwood and Sternberg, 2003). We have recently shown that F-actin–based AC invadopodia presage and then occupy the initial breach in the basement membrane (Hagedorn et al., 2013). Several actin regulators, including the C. elegans orthologue of Ena/VASP, UNC-34, and the membrane-associated Rac GTPases, MIG-2 and CED-10, localize to AC invadopodia. Animals harboring mutations in these genes have mild defects in invasion, suggesting these proteins are subtle modulators of invadopodia (Ziel et al., 2009).
Proteins of the actin-depolymerizing factor (ADF)/cofilin family regulate actin filament dynamics through their F-actin–severing activity, promotion of filament depolymerization, and ability to regulate nucleation (Carlier et al., 1999; Ichetovkin et al., 2002; Andrianantoandro and Pollard, 2006; Bernstein and Bamburg, 2010). In vitro studies have suggested that cofilin can promote either F-actin assembly or disassembly depending on its concentration relative to actin and other actin-binding proteins (Carlier et al., 1999; Van Troys et al., 2008). Alterations in ADF/cofilin activity affect formation of cellular protrusions, including neurite and lamellipodia extensions, and influence cell migration (Chen et al., 2001; Gungabissoon and Bamburg, 2003; Ghosh et al., 2004; Hotulainen et al., 2005; Kiuchi et al., 2007). Consistent with a possible role in cell invasion, cofilin is highly expressed in numerous metastatic cancers and tumor cell lines and promotes invadopodia stability in rat MTLn3 mammary tumor cells (Yamaguchi et al., 2005; Wang et al., 2007). Despite many studies, it remains unclear whether ADF/cofilin proteins primarily regulate actin assembly or disassembly during protrusive cellular activity (Van Troys et al., 2008). Moreover, it is unknown if ADF/cofilin proteins regulate invadopodia in vivo.
We find here that the C. elegans ADF/cofilin orthologue UNC-60A is expressed within the AC, localizes to AC invadopodia, and is required for AC invasion. RNAi-mediated depletion of the unc-60a transcript caused a reduction in F-actin disassembly, resulting in a pronounced expansion in F-actin at AC invadopodia. Strikingly, loss of unc-60a caused a specific disruption in the trafficking of the invadopodial membrane from the endolysosome to the invasive cell membrane, inhibiting invadopodia formation. Together our findings indicate that ADF/cofilin is a critical regulator of invadopodia in vivo, and support a model where ADF/cofilin directs localized F-actin filament turnover that promotes the trafficking of invadopodial membrane to the plasma membrane.
Results
UNC-60A (ADF/cofilin) is a critical cell-autonomous regulator of AC invasion
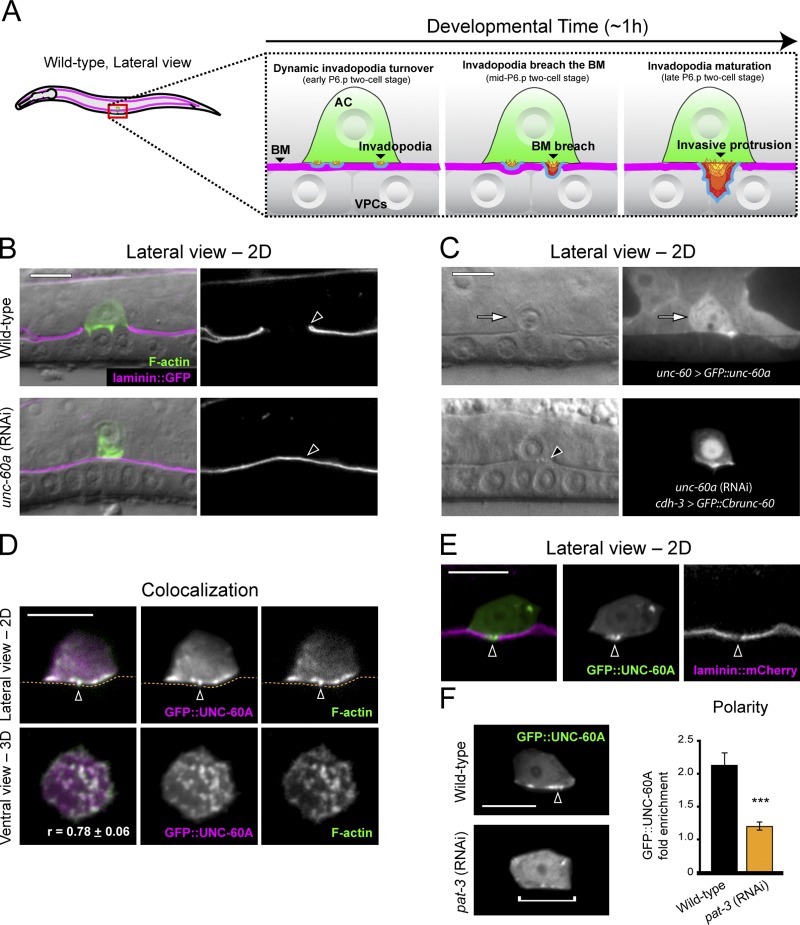
Recent work has shown that invadopodia form within the C. elegans uterine AC and mediate basement membrane penetration as the AC invades the vulval tissue (Fig. 1 A; Hagedorn et al., 2013). To identify direct regulators of AC invadopodia, we examined genes identified in a whole-genome RNAi screen for AC invasion (Matus et al., 2010). Of these, we noted that loss of unc-60a, one of two isoforms of the C. elegans orthologue of the ADF/cofilin gene (McKim et al., 1994; Anyanful et al., 2004), resulted in one of the most penetrant AC invasion defects. An expanded analysis of RNAi-mediated knockdown of unc-60a confirmed this finding, revealing a 90% AC invasion defect (Fig. 1 B; n = 14/140 [10%] normal invasion, 54/140 [39%] partial invasion, 72/140 [51%] no invasion; see Materials and methods for scoring details). unc-60 (su158) mutants, which are null for the muscle-specific isoform unc-60b, have normal AC invasion (Ono et al., 1999; Ono et al., 2003; Ziel et al., 2009), indicating that the unc-60a isoform regulates AC invasion.
Figure 1.
UNC-60A (ADF/cofilin) localizes to invadopodia and regulates invasion. (A) Diagram depicts normal anchor cell (AC) invasion (BM, basement membrane; VPCs, vulval precursor cells). AC invasion is precisely timed with underlying VPC development, and initiates breach of the basement membrane during the P6.p two-cell stage and completes basement membrane clearance by the P6.p four-cell stage (Hagedorn et al., 2013). AC invasion is initiated by F-actin and membrane-based invadopodia that breach the basement membrane and transform into a protrusion. (B) Fluorescence overlaid on DIC (left); grayscale of basement membrane fluorescence (right, visualized with laminin::GFP). Lateral-view images show a wild-type anchor cell (top, visualized with mCherry::moeABD) that has breached the basement membrane (arrowhead). Bottom panels show blocked AC invasion (arrowhead, intact basement membrane) unc-60a RNAi treatment. (C) unc-60 > GFP::UNC-60A (top) is up-regulated in the AC (arrow). AC-specific expression of the C. briggsae orthologue of ADF/cofilin (bottom) rescued invasion in animals depleted of the C. elegans unc-60a transcript (break in the phase-dense basement membrane line under the AC, arrowhead). (D) GFP::UNC-60A (overlay, left) colocalized with F-actin (mCherry::moeABD) at AC invadopodia (arrowheads, basement membrane position, dotted orange line). r-value reports mean Pearson’s correlation coefficient for colocalization of UNC-60A and F-actin within the AC ± SEM. (E) UNC-60A localized to the initial basement membrane breach (arrowhead). (F) RNAi-mediated knockdown of the β-integrin subunit pat-3 resulted in a loss of UNC-60A localization (bracket), which in wild-type animals is concentrated at the invasive cell membrane (arrowhead). The graph shows fold enrichment of GFP::UNC-60A at the invasive membrane. (P < 0.001, Student’s t test; n = 10 animals for each treatment, error bars report ±SEM). Bars, 5 µm.
To examine where unc-60a (ADF/cofilin) is expressed, we generated a translational GFP::UNC-60A fusion protein under the control of its endogenous promoter (unc-60 > GFP::unc-60a). At the time of invasion GFP::UNC-60A was present throughout the uterine tissue and at elevated levels in the AC (Fig. 1 C; 1.35-fold greater levels versus neighboring uterine cells; n = 11 animals examined). GFP::UNC-60A was also more enriched at the invasive cell membrane (1.29-fold enrichment vs. 0.75-fold for neighboring uterine cells; n = 11 animals examined). To determine if ADF/cofilin functions within the AC, we generated a construct in which the unc-60 (ADF/cofilin) transcript from the related nematode Caenorhabditis briggsae was expressed specifically in the AC (cdh-3 > GFP::Cbrunc-60). The C. briggsae unc-60 and C. elegans unc-60a genes share 83% nucleotide identity over the conserved first four exons (nucleotides 1–427 of C. elegans unc-60a), which is below the 95% threshold required for RNAi targeting (Rual et al., 2007). cdh-3 > GFP::Cbrunc-60 restored invasion in animals treated with C. elegans unc-60a RNAi (Fig. 1 C; n = 72/86 [84%] normal invasion, 12/86 [14%] partial invasion, 2/86 [2%] no invasion). Hence, UNC-60A promotes AC invasion cell autonomously.
UNC-60A (ADF/cofilin) localizes to AC invadopodia
To visualize the subcellular localization of UNC-60A, we generated animals with AC-specific GFP::UNC-60A expression (cdh-3 > GFP::unc-60a). GFP::UNC-60A localized in punctate structures at the AC–basement membrane interface before invasion (Fig. 1 D). To determine whether these structures were invadopodia, we examined their localization relative to F-actin and basement membrane breaching. We observed a tight correlation between F-actin localization and UNC-60A enrichment at the AC–basement membrane interface (Fig. 1 D), as well as localization to the invadopodium that breached the basement membrane (5/5 animals; Fig. 1 E). Further, UNC-60A localization to the AC–basement membrane interface was dependent on the integrin INA-1/PAT-3, whose activity is required for invasion and promotes the localization of other invadopodia constituents (Fig. 1 F; Hagedorn et al., 2009, 2013). Thus, UNC-60A is a component of AC invadopodia.
UNC-60A promotes F-actin turnover at AC invadopodia
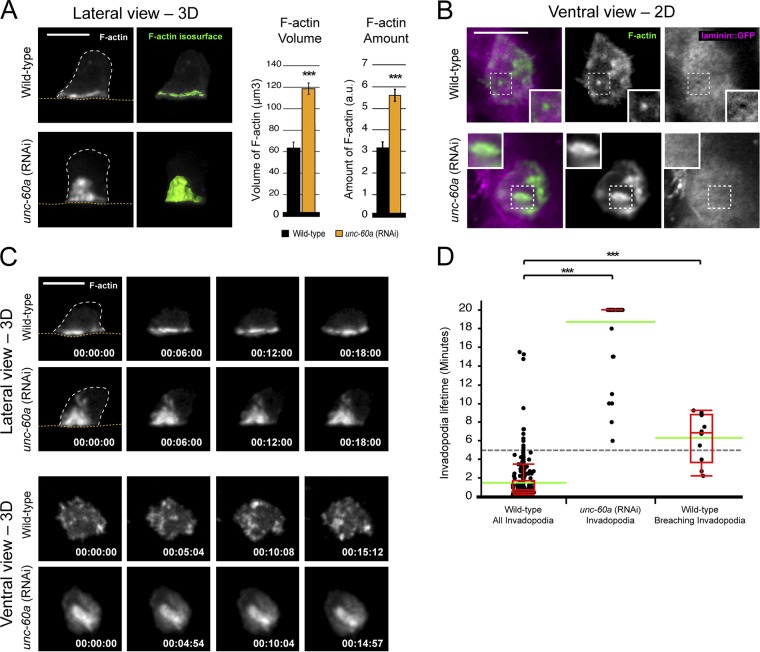
To determine whether UNC-60A regulates AC invadopodia, we examined F-actin within the AC after loss of unc-60a. In unc-60a (RNAi) animals we observed a twofold expansion in the total volume and amount of the F-actin network at the AC–basement membrane interface, suggesting UNC-60A promotes actin filament turnover (Fig. 2 A). Although F-actin remained polarized, it was no longer organized into discrete foci and instead formed larger structures that failed to penetrate basement membrane (Fig. 2 B). Compared with F-actin structures in wild-type ACs, which had a median diameter of 0.96 µm, in the absence of ADF/cofilin, F-actin structures had a median size of 1.90 µm (n = 30 structures from 10 animals for each; P < 0.0001). These larger F-actin structures were also more stable. In 20-min time-lapse movies of wild-type animals (n = 4 animals), the average lifetime of invadopodia was only 88 s (Fig. 2 D; n = 307 structures examined). In contrast, in unc-60a (RNAi) ACs (n = 10 animals), most F-actin structures (n = 43/51) persisted for the entire time-lapse (20 min; Fig. 2 C and Videos 1–3). This duration far exceeded that of invadopodia in wild-type animals that are associated with basement membrane breaching events (Fig. 2 D; average lifetime of 6.3 min; n = 10 invadopodia from 8 animals).
Figure 2.
UNC-60A promotes F-actin dynamics at AC invadopodia. (A) 3D projections (left) and isosurface renderings (green, right) of the normal F-actin network in wild-type (top; visualized with the F-actin binding probe mCherry::moeABD) and expanded F-actin in a unc-60a (RNAi) animal (bottom; dotted orange line is basement membrane; dotted white line is the AC’s apicolateral membrane). Graph reports the F-actin volume and amount in wild-type (black) and unc-60a (RNAi) animals (gray, n ≥ 10 animals each; P < 0.001, Student’s t test; error bars indicate ±SEM. (B) Ventral-view images (top) show an F-actin–rich AC invadopodium (green, mCherry::moeABD) at the initial basement membrane breach (inset). Bottom panels show a unc-60a (RNAi) animal that has enlarged F-actin foci that fail to invade. (C) Time-series show dynamic F-actin–rich AC invadopodia in a wild-type animal (top) and the enlarged, static F-actin structures in a unc-60a (RNAi) animal (bottom). Bars, 5 µm. (D) Scatterplots report invadopodia lifetime measurements for wild-type (left; invadopodia that were actively breaching the basement membrane are shown on the right) and unc-60a RNAi-treated animals (center). The interquartile range is shown in red; green bars denote the population mean (***, P < 0.0001; Wilcoxon rank-sum test).
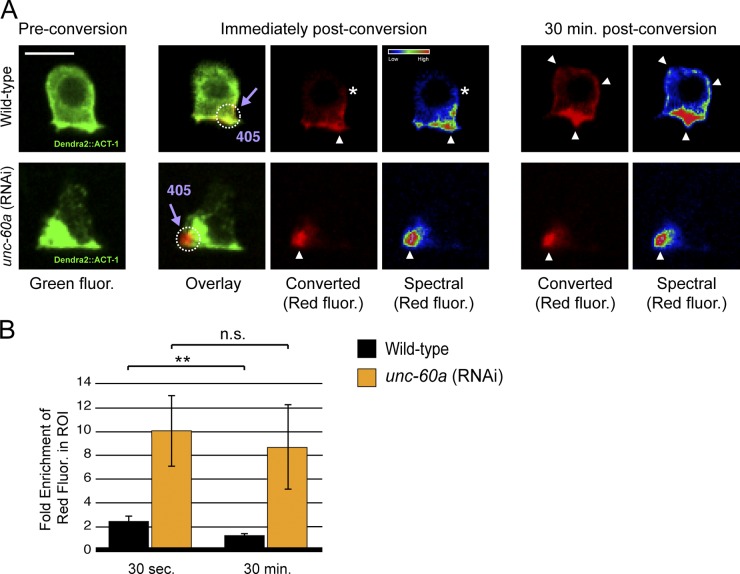
To test if the accumulation of F-actin resulted from reduced F-actin disassembly, we created animals that expressed an actin isoform tagged with Dendra2 in the AC. Dendra2 is a photoconvertible fluorophore that switches from green to red fluorescence after brief exposure to 405-nm light (Gurskaya et al., 2006). We treated the Dendra2::ACT-1 animals with either unc-60a RNAi or an empty vector control and then photoconverted a small region of actin (F-actin and likely a small portion of G-actin) at the invasive cell membrane where invadopodia reside (Fig. 3 A). In control animals, optically highlighted actin began redistributing immediately and spread throughout the AC by 30 min, consistent with rapid F-actin turnover (Fig. 3, A and B; n = 5/5 ACs). In contrast, after loss of unc-60a, optically highlighted actin did not disperse significantly, even after 30 min (Fig. 3, A and B; n = 4/4 ACs). Taken together, these results offer compelling evidence that UNC-60A (ADF/cofilin) regulates disassembly of F-actin within AC invadopodia.
Figure 3.
UNC-60A promotes F-actin disassembly at AC invadopodia. (A) Pre-conversion ACs expressing Dendra2::ACT-1 (a tagged form of the actin monomer found throughout the cytoplasm in monomeric form and incorporated within F-actin at invadopodia; left), immediately post-conversion (converted actin is shown in red and spectrally; middle), and 30 min after conversion (right). Optically highlighted regions of Dendra2::ACT-1 (dotted white circle, arrowhead) began to disperse immediately (asterisks) in wild-type animals (top) and were found throughout the AC by 30 min (arrowheads). In unc-60a (RNAi) animals (bottom), optically highlighted regions of actin failed to disperse, consistent with a lack of F-actin disassembly. (B) The fold enrichment of red signal (converted actin) in the optically highlighted region (dotted white circle) compared with the region outside the highlighted region in the cell is displayed for wild-type and unc-60a (RNAi) animals 30 s after conversion and 30 min after conversion (n = 5 ACs for wild-type animals, n = 4 for unc-60a RNAi animals; **, P = 0.005; Student’s t test; error bars indicate ±SEM. Bar, 5 µm.
UNC-60A regulates invadopodial membrane recycling from the endolysosome
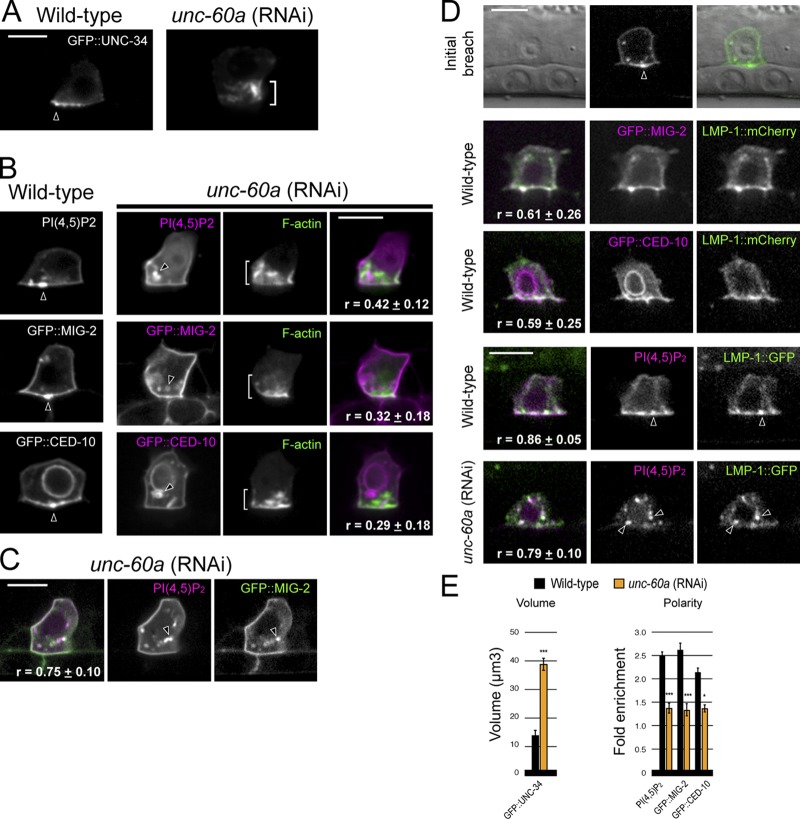
To further examine UNC-60A’s role in AC invadopodia, we analyzed the localization of other invadopodia components after loss of unc-60a. Consistent with localization to the actin cytoskeleton (Trichet et al., 2008), the actin regulator UNC-34 (Ena/VASP) was distributed throughout the large F-actin aggregates that formed after loss of unc-60a (Fig. 4, A and E). In contrast, the invadopodial membrane component PI(4,5)P2 and the membrane-associated Rac proteins MIG-2 and CED-10, which colocalize with F-actin at invadopodia in wild-type animals (Fig. S1), were not enriched at the plasma membrane or the expanded F-actin network. Instead, these molecules were localized within intracellular vesicles (Fig. 4, B and E). Colocalization analysis of PI(4,5)P2 and MIG-2 indicated that these molecules were within overlapping vesicles (Fig. 4 C), suggesting there is coordinated trafficking of invadopodial membrane to the plasma membrane regulated by UNC-60A.
Figure 4.
UNC-60A (ADF/cofilin) regulates the invadopodial membrane. (A) In contrast to its tight localization at the invasive cell membrane in wild-type animals (arrowhead), GFP::UNC-34 (Ena/VASP) localizes to the expanded F-actin network (bracket) after unc-60a RNAi treatment. (B) The invadopodial membrane components PI(4,5)P2 (mCherry::PLCδPH) and the Racs GFP::MIG-2 and GFP::CED-10 were no longer localized to the invasive cell membrane after loss of unc-60a and were found within intracellular vesicles (arrowheads) independent of the expanded F-actin (brackets; r-value reports average Pearson’s correlation coefficient for colocalization within the AC ± SEM). (C) The invadopodial membrane components PI(4,5)P2 and GFP::MIG-2 colocalized within the same vesicles (arrowheads) after loss of unc-60a (r-value reports average Pearson’s correlation coefficient for colocalization within the AC ± SEM). (D) The endolysosome marker LMP-1::GFP localized to invadopodial structures and at the initial breach site (top, arrowhead). LMP-1::GFP colocalized with invadopodial membrane markers (GFP::MIG-2, GFP::CED-10, and PI(4,5)P2) in wild-type ACs (middle panels, arrowheads) and after loss of unc-60a within internalized vesicles (bottom, arrowheads). r-value reports average Pearson’s correlation coefficient for colocalization within the AC ± SEM. (E) Graphs show the volume of GFP::UNC-34 and the polarized localization of the invadopodial membrane components in wild-type (black) and unc-60a (RNAi) animals (orange; n ≥ 6 animals examined for each; *, P < 0.01; ***, P < 0.0001; Student’s t test; error bars indicate ±SEM). Bars, 5 µm.
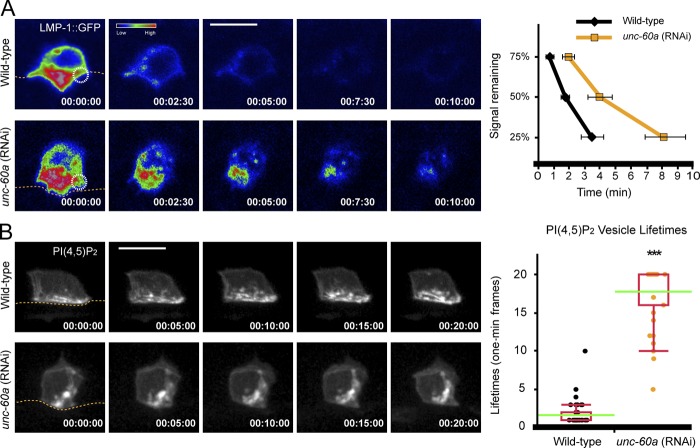
To better understand invadopodial membrane trafficking, we examined markers for the early (Rab5), late (Rab7), and recycling (Rab11) endosomes, and the endolysosome (LMP-1 and CUP-5; Balklava et al., 2007; Campbell and Fares, 2010; Humphries et al., 2011). This analysis revealed that the endolysosomal markers LMP-1 and CUP-5 were polarized at the invasive membrane (Fig. S2). Consistent with a normal role in trafficking through this compartment, the invadopodial membrane components PI(4,5)P2, CED-10, and MIG-2 colocalized strongly with LMP-1 at invadopodia and LMP-1 localized to the invadopodium at the site of breach (Fig. 4 D). Further, using fluorescence loss in photobleaching (FLIP), we found that repeated photobleaching of a small 1.0-µm region of the invasive cell membrane resulted in loss of LMP-1::GFP throughout the endolysosome within 5 min, indicating dynamic recycling of the invadopodial membrane through this compartment (Fig. 5 A, Fig. S3, and Video 4). Time-lapse analysis of PI(4,5)P2 confirmed active invadopodial membrane dynamics at the invasive cell membrane (Fig. 5 B and Video 5). Together, these results indicate that the invadopodial membrane recycles through the endolysosome during invadopodia formation.
Figure 5.
UNC-60A promotes invadopodial membrane recycling through the endolysosome. (A) Focused laser photobleaching of an ∼1.0-µm region at the invasive membrane (circle) in a wild-type AC (top row) resulted in loss of the endolysosome LMP-1::GFP signal throughout the AC shortly after two and a half minutes (time points, minutes). Note, these images were captured on a laser-scanning microscope optimized for FLIP analysis, which has reduced sensitivity for LMP-1::GFP signal compared with the spinning-disk confocal image captured in Fig. 4 D (see Materials and methods). In unc-60a (RNAi) animals the LMP-1::GFP signal persisted ∼10 min after initiation of photobleaching. Graphs report LMP-1::GFP signal remaining in neighboring nonbleached invasive membrane over time (n ≥ 5 animals each genotype; P < 0.01 for 25%, and P < 0.05 for 50 and 75% signaling remaining; Student’s t test). (B) Time-lapse analysis of PI(4,5)P2 (GFP::PLCδPH) at the invasive cell membrane in a wild-type AC (top row) and unc-60a (RNAi) animals (bottom row) revealed reduced membrane dynamics after loss of unc-60a. Graphs report lifetimes of internal vesicles (n ≥ 35 vesicles each genotype; ***, P < 0.0001; Wilcoxon rank-sum test). Bars, 5 µm.
The accumulation of invadopodial membrane in internal vesicles after loss of unc-60a and recycling through the endolysosome suggested that UNC-60A might regulate the trafficking of invadopodial membrane through this compartment. Consistent with this notion, after loss of unc-60a, the invadopodial membrane marker PI(4,5)P2 colocalized with the endolysosome marker LMP-1 within internalized vesicles (Fig. 4 D). FLIP analysis revealed over a twofold decrease in LMP-1::GFP recycling, indicating a reduction in trafficking (Fig. 5 A, Fig. S3, and Video 4). Time-lapse analysis of PI(4,5)P2 also confirmed these vesicles were more static (Fig. 5 B and Video 5). We conclude that UNC-60A promotes dynamic invadopodial membrane recycling from the endolysosome to the plasma membrane.
Other polarized trafficking and secretion events are normal after loss of unc-60a
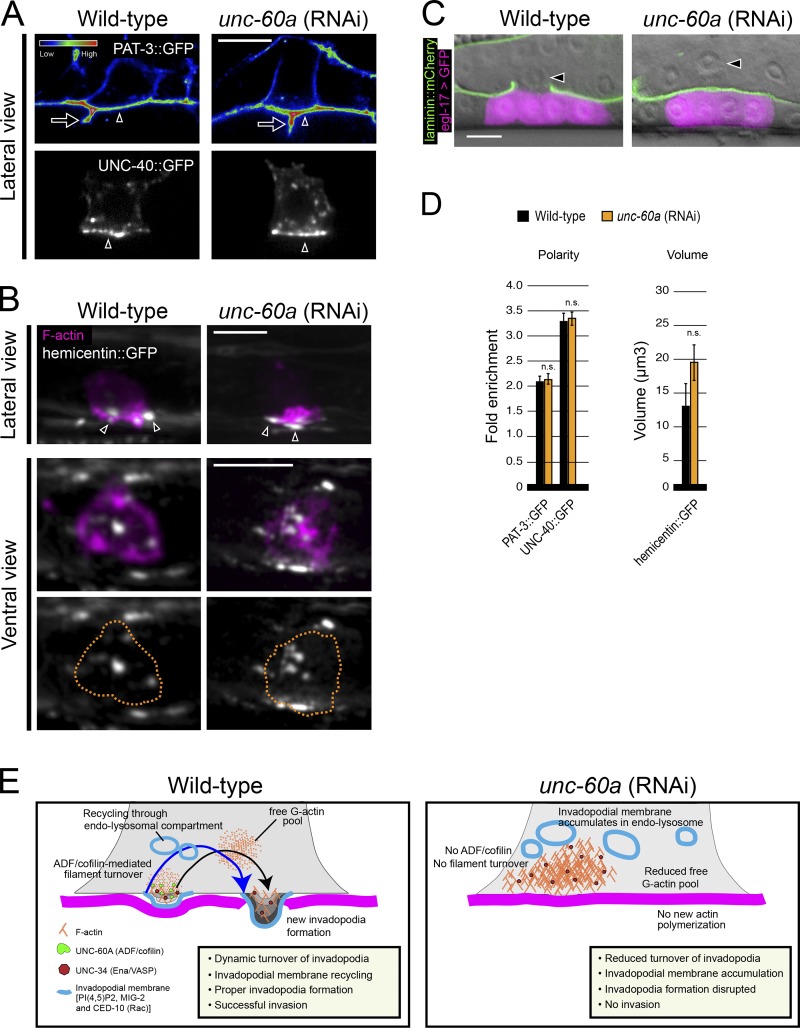
To determine whether the disruption in invadopodial membrane trafficking was an indirect effect from a general perturbation in vesicle trafficking or polarity, we examined markers of polarity and secretion. Notably, after depletion of unc-60a the integrin receptor INA-1/PAT-3 and the netrin receptor UNC-40 (DCC) were polarized normally to the AC’s invasive cell membrane (Fig. 6, A and D; Hagedorn et al., 2009; Ziel et al., 2009). Further, AC deposition of the matrix component hemicentin into the basement membrane (Fig. 6, B and D; Sherwood et al., 2005) and secretion of the EGF-like ligand LIN-3, which induces vulval development, were normal (Fig. 6 C). These results offer strong evidence that UNC-60A does not regulate general polarity or secretion in the AC.
Figure 6.
UNC-60A (ADF/cofilin) does not regulate other AC polarity and secretion functions. (A) The β-integrin subunit PAT-3::GFP (top, spectral representation of fluorescence intensity) and the netrin receptor UNC-40::GFP (bottom) localized to the invasive cell membrane in wild-type (left, arrowhead) and after loss of unc-60a (right, arrowhead). Note, the integrin indicated by the arrow is PAT-3::GFP that localizes between the central vulval precursor cells (it is not a protrusion from the AC). (B) Lateral- and ventral-view images show the normal deposition of the matrix component hemicentin (shown in grayscale, arrowheads) into the basement membrane below the AC (expressing the F-actin probe mCherry::moeABD, magenta) in wild-type (left) and after loss of unc-60a (right). (C) Vulval induction (indicated by egl-17::GFP, green) was normal after loss of unc-60a, indicating AC (arrowhead) secretion of the ligand LIN-3. (D) Graphs report the polarity of PAT-3::GFP and UNC-40::GFP, and the volume of hemicentin::GFP for wild-type (black) and unc-60a–depleted animals (orange; n ≥ 6 animals; n.s., not significant; Student’s t test; error bars indicate mean ± SEM). (E) UNC-60A drives F-actin disassembly at AC invadopodia, which is required for normal invadopodia dynamics, and the recycling of invadopodial membrane through endolysosome to the plasma membrane.
Discussion
Invadopodia, protrusive F-actin–based membrane structures capable of ECM remodeling, were identified in transformed cells and cancer lines in vitro over 20 years ago (Chen, 1989). Despite intense interest, the regulation and function of invadopodia in vivo has been elusive (Beerling et al., 2011; Murphy and Courtneidge, 2011). Our data indicate that the C. elegans ADF/cofilin orthologue UNC-60A is a critical and specific regulator of invadopodia during AC invasion. We find that UNC-60A mediates F-actin disassembly at AC invadopodia, which is critical for normal invadopodia dynamics and the trafficking of invadopodial membrane and associated membrane components to the plasma membrane (Fig. 6 E).
Our live-cell imaging and site-of-action studies indicate that UNC-60A specifically localizes to AC invadopodia and functions within the AC to promote breaching the basement membrane. The increased accumulation of F-actin and dramatically decreased actin turnover after loss of unc-60a indicate that UNC-60A promotes F-actin disassembly at invadopodia, rather than nucleating new actin filaments. Our data add weight to the notion that in most cell types ADF/cofilin contributes to actin dynamics primarily by mediating actin filament disassembly (Hotulainen et al., 2005; Kiuchi et al., 2007; Okreglak and Drubin, 2007; Lai et al., 2008). In the C. elegans AC, F-actin disassembly might be necessary for breaking down existing AC invadopodia and forming new invadopodia through the recycling of actin monomers. Dynamic F-actin turnover at AC invadopodia is also likely critical to generate the protrusive force required to breach basement membrane. Consistent with this idea, ADF/cofilin proteins are thought to generate actin monomers to promote lamellipodial extensions in Cos and MCF7 cell lines and migration in NIH 3T3 and B16F1 cells (Hotulainen et al., 2005; Kiuchi et al., 2007).
Ultrastructural analysis and TIRF microscopy have indicated that invadopodia in tumor cells have a dynamic, morphologically unique membrane (Baldassarre et al., 2003; Artym et al., 2011). Studies on the trafficking of invadopodial membrane components in cell culture have focused on the membrane-anchored matrix metalloproteinase MT1-MMP (Poincloux et al., 2009). Intriguingly, recent work in tumor cells has indicated that MT1-MMP is recycled from the endolysosomal compartment to the plasma membrane (Hoshino et al., 2012; Yu et al., 2012). Our studies indicate that the invadopodial membrane is a unique compartment, which is actively recycled through the endolysosome in an UNC-60A (ADF/cofilin)–dependent manner (Fig. 5 E). Trafficking through the endolysosome might facilitate dynamic membrane addition to invadopodia required for active protrusions, similar to roles of endolysosome membrane addition in wound healing and neurite outgrowth (Reddy et al., 2001; Arantes and Andrews, 2006). Active membrane movement through the endolysosome may also be required for delivery of proteases to AC invadopodia (Yu et al., 2012).
Spatially restricted cofilin activity is necessary for vesicular trafficking of acetylcholine receptors to nascent postsynaptic sites as well as planar cell polarity proteins to apical membranes in the mouse embryo (Lee et al., 2009; Mahaffey et al., 2013). It is thought that localized F-actin turnover by ADF/cofilin facilitates vesicle targeting or fusion to the plasma membrane, perhaps by breaking the cortical actin barrier (Eitzen, 2003; Lee et al., 2009; Mahaffey et al., 2013). Thus, targeted F-actin turnover by UNC-60A at AC invadopodia might direct trafficking of invadopodial membrane from the endolysosome to discrete sites to form invadopodia. Cofilin has also been implicated in other trafficking events within cells, including sorting at the trans-Golgi network and endocytic trafficking to the yeast vacuole (Okreglak and Drubin, 2007; von Blume et al., 2009). In the trans-Golgi, cofilin is proposed to mediate trimming of F-actin, which allows the transmembrane Ca2+ ATPase SPCA1 to import Ca2+ into the lumen to direct proper sorting (von Blume et al., 2011). During endocytic delivery of vesicles to the yeast vacuole, cofilin is required to disassemble F-actin from the endocytic vesicles and regenerate the actin monomer pool, both of which might be required for late endocytic targeting or fusion with the vacuole (Okreglak and Drubin, 2007). It is thus possible that cofilin might regulate aspects of invadopodial membrane targeting, fusion, or sorting at the endolysosome itself that are required for trafficking back to the invasive membrane to form invadopodia. Given the tight localization of cofilin to invadopodia at the invasive cell membrane, however, we favor the notion that cofilin has a direct role in targeting invadopodial membrane to nascent invadopodia. Taken together, our results establish a unique organizing function for UNC-60A (cofilin) at invadopodia, coordinating both protrusive F-actin formation and invadopodial membrane addition.
Vertebrate ADF/cofilin proteins are overexpressed in many cancers and mediate invadopodia maturation in an adenocarcinoma cell line (Wang et al., 2007). The results of our functional in vivo studies here strongly support the notion that ADF/cofilin proteins are conserved regulators of invadopodia in both normal and pathological processes. Further, the specific defect in AC invadopodia formation and strong perturbation in ability to invade suggests that targeting ADF/cofilin activity might be an effective strategy to block invasion in diseases such as cancer.
Materials and methods
Strains and culture conditions
C. elegans were cultured as described previously (Brenner, 1974) and wild-type were strain N2. In the text and figures, linkage to a promoter is designated with “>” and linkages that fuse open reading frames with “::”. The following alleles and transgenes were used: qyEx411 [zmp-1 > lmp-1::mCherry], qyEx403 [cdh-3 > GFP::cup-5], qyIs61 [cdh-3 > GFP::unc-34], qyEx237 [unc-60 > GFP::unc-60a], qyIs222 [cdh-3 > GFP::unc-60a], qyIs223 [cdh-3 > GFP::unc-60a], qyIs221 [cdh-3 > GFP::ced-10], qyIs224 [cdh-3 > GFP::Cbrunc-60], qyIs219 [cdh-3 > GFP::PLCδPH], qyIs220 [cdh-3 > GFP::mig-2], qyIs67 [cdh-3 > unc-40::GFP], qyIs127 [laminin::mCherry], muIs27 [mig-2::GFP], qyIs211 [cdh-3 > lmp-1::GFP], qyIs205 [cdh-3 > mCherry::rab-11], qyIs252 [cdh-3 > mCherry::rab-7], qyIs256 [cdh-3 > mCherry::rab-5]; LGI: ayIs4 [egl-17 > GFP]; LGII: qyIs23 [cdh-3 > mCherry::PLCδPH]; LGIII: unc-119(ed4), rhIs23 [hemicentin::GFP]; LGIV: qyIs10 [laminin::GFP], qyIs42 [pat-3::GFP;genomic ina-1]; LGV: qyIs50 [cdh-3 > mCherry::moeABD]; LGX: qyIs7 [laminin::GFP], qyIs24 [cdh-3 > mCherry::PLCδPH], qyEx282 [cdh-3 > Dendra2::act-1].
Microscopy, image acquisition, processing, and analysis
Images were acquired using a spinning-disk confocal microscope (CSU-10; Yokogawa Corporation of America) mounted on a microscope (AxioImager; Carl Zeiss) with a 100× Plan Apochromat objective (1.4 NA) and controlled by iVision software (Biovision Technologies). The point spread function (PSF) reported as full width at half-maximum (FWHM) was measured by imaging fluorescent beads (210 nm; channel 488) on both microscopy systems used in this study: (1) spinning-disk confocal; XY (lateral) 287 nm, Z (axial) 571 nm; and (2) laser-scanning confocal; XY (lateral) 333 nm, Z (axial) 772.
Acquired images were processed using ImageJ 1.40g (National Institutes of Health) and Photoshop (CS6 Extended; Adobe Systems, Inc.). 3D reconstructions were built from confocal Z-stacks, analyzed, and exported as .MOV files using IMARIS 7.4 (Bitplane, Inc.). Figures and graphs were constructed using Illustrator (CS6 Extended; Adobe Systems, Inc.). Videos were annotated using Photoshop.
Scoring of AC invasion, fluorescence intensity, colocalization, and polarity
AC invasion was scored as previously described using DIC microscopy (Sherwood et al., 2005). In brief, animals were scored for invasion at the P6.p four-cell stage when basement membrane clearance is completed in wild-type animals. ACs were scored as “normal invasion” if the breach in the basement membrane was at least the width of the AC by the P6.p four-cell stage, “partial invasion” if there was a visible breach smaller than the width of the AC, and “no invasion” if there was no detectible breach in the basement membrane. For analysis of UNC-60A up-regulation in the AC, regions of interest were drawn in ImageJ around the AC and then a neighboring uterine cell in the same animal; mean fluorescence intensity of each cell was then determined. Up-regulation was calculated as the following ratio: [AC mean intensity – background]/[neighboring uterine cell mean intensity – background]. Colocalization analysis was performed on confocal z-sections using the “Coloc” module in Imaris. In brief, a mask was first used to exclude all signal outside of the AC. Pearson’s correlation coefficient was then calculated from the GFP and mCherry fluorescence inside the AC (n ≥ 5 animals for each comparison). All images were acquired using identical settings. Polarity in wild-type animals and animals treated with unc-60a RNAi was determined using the ratio of the mean fluorescence intensity from a 5-pixel-wide line scan drawn along the invasive and apicolateral membranes of ACs and in some cases neighboring uterine cells expressing unc-60 > GFP::unc-60a, GFP::MIG-2, GFP::CED-10, LMP-1::GFP, mCherry::PLCδPH, mCherry::RAB-5, mCherry::RAB-7, mCherry::RAB-11, GFP::CUP-5, PAT-3::GFP, and UNC-40::GFP. Numbers examined for each are indicated in the text or figure legends. Polarity was calculated as the following ratio: [invasive membrane mean intensity − background]/[apicolateral membrane mean intensity − background].
Scoring of F-actin volume and dynamics
Quantitative F-actin measurements were performed as described previously (Hagedorn et al., 2009). In brief, confocal Z-stacks were used to make 3D reconstructions of F-actin networks in ACs expressing the F-actin probe mCherry::moeABD using Imaris 6.0 (Bitplane, Inc.). Isosurface renderings of mCherry::moeABD were created setting a threshold that outlined the dense F-actin network at the invasive membrane in wild-type ACs. This same thresholding was used in cdh-3 > mCherry::moeABD-untreated and unc-60a RNAi-treated animals. Quantitative measurements were then made for the volume and amount of fluorescence intensity with these isosurface renderings (n ≥ 10 animals for each). Similar methods were used to quantify GFP::UNC-34 and hemicentin::GFP volumes. Quantitative measurements of the total (sum) fluorescence of mCherry::moeABD present in the AC showed no change after treatment with unc-60a RNAi (n = 10 animals each; P = 0.91; Student’s t test). AC invadopodia dynamics were quantified using the “Spots” module within Imaris, which facilitated the tracking of F-actin patches (cdh-3 > mCherry::moeABD) at the invasive membrane of the AC over time in wild-type and unc-60a RNAi-treated animals.
Optical highlighting and fluorescence loss in photobleaching (FLIP)
Imaging was conducted with a laser-scanning confocal microscope (LSM 510; Carl Zeiss) equipped with a 100× objective (1.4 NA). The PMT light detector used on the laser-scanning microscope is less sensitive in detecting fluorescence than the CCD camera on the spinning-disk confocal microscope (Murray et al., 2007). Furthermore, optimization to achieve numerical data for FLIP analysis on the laser-scanning confocal microscope resulted in a reduced signal-to-noise ratio. These features underlie the differences in the images shown in Fig. 3 A and Fig. 5 A and Fig. S3 captured using the laser-scanning microscope versus all other fluorescence images that were captured on a spinning-disk confocal microscope. Transgenic cdh-3 > Dendra2::act-1 worms were photoconverted by scanning regions of interest (ROIs; ∼1 µm) with a 405-nm laser at 2% power for 100 iterations. Images were captured before, immediately after, and 30 min after conversion. To quantify redistribution of optically highlighted actin (an estimate of F-actin turnover), ImageJ was used to measure the mean fluorescence intensity of the converted green-to-red ROI and the remaining area of the AC outside the ROI. The fold enrichment of red signal (561-nm excitation wavelength) was measured as follows: [ROI background]/[outside ROI background]. For FILP, selected ROIs (∼1 µm) of the AC in worms expressing cdh-3 > LMP-1::GFP were bleached every 30 s for 10 min at 100% power for 20 iterations, and images were taken before and after each beaching interval. Quantitative data were obtained with ImageJ by normalizing background-subtracted mean intensities to t = 0 of the LMP-1::GFP signal at the invasive membrane adjacent to the bleached ROI. For each technique, identical settings were used to acquire images at all time-points and ≥5 animals were analyzed in each group.
RNAi
RNAi targeting pat-3 and unc-60a was delivered by RNAi feeding (Sherwood et al., 2005). To bypass embryonic lethality, synchronized L1-stage larvae were plated on unc-60a RNAi, and synchronized L2-stage larvae were plated on pat-3 RNAi as described previously (Hagedorn et al., 2009). The empty RNAi vector L4440 was used as the wild-type control.
Construction of GFP protein fusions
The GFP::unc-60a fusions were constructed by placing GFP immediately after the unc-60a start codon and 1.9 kb downstream of the stop codon was included as the 3′ UTR. The GFP::UNC-60 amplicon was linked by PCR fusion to promoters, either endogenous (fragment encompassing 3.6 kb upstream of the unc-60 start codon, unc-60 > GFP::unc-60a) or AC specific (cdh-3 > promoter, cdh-3 > GFP::unc-60a). The Dendra2::act-1 fusion was constructed by cloning Dendra2 at the N terminus of a 1.7-kb genomic fragment of the C. elegans act-1 gene (fragment encompassing act-1 start codon and downstream 3′ UTR). The AC-specific cdh-3 > promoter was then cloned immediately upstream of the Dendra2 start codon. The lmp-1::GFP, GFP::cup-5, mCherry::rab-5, mCherry::rab-7, and mCherry::rab-11 reporters were amplified from plasmid DNA (see Table S2) and linked to the cdh-3 > promoter by PCR fusion. Constructs were co-injected with 50 ng/µl unc-119 rescue DNA, 50 ng/µl pBsSK, and 50 ng/µl EcoR1 cut salmon sperm DNA into unc-119(ed4) hermaphrodites. Extrachromosomal lines were established and integrated by gamma irradiation as described previously (Sherwood et al., 2005). See supplemental material for transgenic strains generated (Table S1) and primer sequences used (Table S2).
Site-of-action studies with C. briggsae unc-60a rescue
The GFP::Cbrunc-60a rescue construct was made using primers to amplify a 5.3-kb fragment of C. briggsae unc-60 gene from AF16 genomic DNA (fragment encompassing start codon and 2 kb downstream 3′ UTR). A three-step PCR fusion was used to link the C. briggsae amplicon to the C terminus of GFP and then the AC-specific cdh-3 > promoter. The presence of the GFP::CbrUNC-60 protein in the AC confirmed the C. briggsae unc-60 RNA was not depleted by C. elegans unc-60a RNAi.
Statistical analysis
All statistical analysis was performed in JMP version 9.0 (SAS Institute). Figure legends specify which test was used.
Online supplemental material
Online supplemental material includes three figures, five videos corresponding to the time-lapse data presented in the main text figures, and two tables: Table S1 for extrachromosomal arrays and integrated strains and Table S2 for primer sequences. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312098/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201312098.dv.
Supplementary Material
Acknowledgments
We thank G. Seydoux and H. Fares for reagents; S. Johnson of the Duke LMCF for imaging advice; the CGC for strains; and M. Morrissey, S. Ono, and L. Lohmer for helpful comments.
This work was supported by The Pew Scholars Program in the Biomedical Sciences, and National Institutes of Health grant GM100083 to D.R. Sherwood.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AC
- anchor cell
- ADF
- actin-depolymerizing factor
- FLIP
- fluorescence loss in photobleaching
References
- Andrianantoandro E., Pollard T.D. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Anyanful A., Ono K., Johnsen R.C., Ly H., Jensen V., Baillie D.L., Ono S. 2004. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J. Cell Biol. 167:639–647 10.1083/jcb.200407085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes R.M., Andrews N.W. 2006. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J. Neurosci. 26:4630–4637 10.1523/JNEUROSCI.0009-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym V.V., Matsumoto K., Mueller S.C., Yamada K.M. 2011. Dynamic membrane remodeling at invadopodia differentiates invadopodia from podosomes. Eur. J. Cell Biol. 90:172–180 10.1016/j.ejcb.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre M., Pompeo A., Beznoussenko G., Castaldi C., Cortellino S., McNiven M.A., Luini A., Buccione R. 2003. Dynamin participates in focal extracellular matrix degradation by invasive cells. Mol. Biol. Cell. 14:1074–1084 10.1091/mbc.E02-05-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Pant S., Fares H., Grant B.D. 2007. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 9:1066–1073 10.1038/ncb1627 [DOI] [PubMed] [Google Scholar]
- Beerling E., Ritsma L., Vrisekoop N., Derksen P.W., van Rheenen J. 2011. Intravital microscopy: new insights into metastasis of tumors. J. Cell Sci. 124:299–310 10.1242/jcs.072728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.W., Bamburg J.R. 2010. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20:187–195 10.1016/j.tcb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell E.M., Fares H. 2010. Roles of CUP-5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis. BMC Cell Biol. 11:40 10.1186/1471-2121-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., Ressad F., Pantaloni D. 1999. Control of actin dynamics in cell motility. Role of ADF/cofilin. J. Biol. Chem. 274:33827–33830 10.1074/jbc.274.48.33827 [DOI] [PubMed] [Google Scholar]
- Chen J., Godt D., Gunsalus K., Kiss I., Goldberg M., Laski F.A. 2001. Cofilin/ADF is required for cell motility during Drosophila ovary development and oogenesis. Nat. Cell Biol. 3:204–209 10.1038/35055120 [DOI] [PubMed] [Google Scholar]
- Chen W.T. 1989. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J. Exp. Zool. 251:167–185 10.1002/jez.1402510206 [DOI] [PubMed] [Google Scholar]
- Condeelis J., Segall J.E. 2003. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer. 3:921–930 10.1038/nrc1231 [DOI] [PubMed] [Google Scholar]
- Eitzen G. 2003. Actin remodeling to facilitate membrane fusion. Biochim. Biophys. Acta. 1641:175–181 10.1016/S0167-4889(03)00087-9 [DOI] [PubMed] [Google Scholar]
- Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D.S., Condeelis J.S. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746 10.1126/science.1094561 [DOI] [PubMed] [Google Scholar]
- Gungabissoon R.A., Bamburg J.R. 2003. Regulation of growth cone actin dynamics by ADF/cofilin. J. Histochem. Cytochem. 51:411–420 10.1177/002215540305100402 [DOI] [PubMed] [Google Scholar]
- Gurskaya N.G., Verkhusha V.V., Shcheglov A.S., Staroverov D.B., Chepurnykh T.V., Fradkov A.F., Lukyanov S., Lukyanov K.A. 2006. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 24:461–465 10.1038/nbt1191 [DOI] [PubMed] [Google Scholar]
- Hagedorn E.J., Sherwood D.R. 2011. Cell invasion through basement membrane: the anchor cell breaches the barrier. Curr. Opin. Cell Biol. 23:589–596 10.1016/j.ceb.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E.J., Yashiro H., Ziel J.W., Ihara S., Wang Z., Sherwood D.R. 2009. Integrin acts upstream of netrin signaling to regulate formation of the anchor cell’s invasive membrane in C. elegans. Dev. Cell. 17:187–198 10.1016/j.devcel.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn E.J., Ziel J.W., Morrissey M.A., Linden L.M., Wang Z., Chi Q., Johnson S.A., Sherwood D.R. 2013. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J. Cell Biol. 201:903–913 10.1083/jcb.201301091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester E., Yurchenco P.D. 2013. Laminins in basement membrane assembly. Cell Adhes. Migr. 7:56–63 10.4161/cam.21831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino D., Koshikawa N., Suzuki T., Quaranta V., Weaver A.M., Seiki M., Ichikawa K. 2012. Establishment and validation of computational model for MT1-MMP dependent ECM degradation and intervention strategies. PLOS Comput. Biol. 8:e1002479 10.1371/journal.pcbi.1002479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M.K., Lappalainen P. 2005. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 16:649–664 10.1091/mbc.E04-07-0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries W.H., IV, Szymanski C.J., Payne C.K. 2011. Endo-lysosomal vesicles positive for Rab7 and LAMP1 are terminal vesicles for the transport of dextran. PLoS ONE. 6:e26626 10.1371/journal.pone.0026626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichetovkin I., Grant W., Condeelis J. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84 10.1016/S0960-9822(01)00629-7 [DOI] [PubMed] [Google Scholar]
- Kiuchi T., Ohashi K., Kurita S., Mizuno K. 2007. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177:465–476 10.1083/jcb.200610005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F.P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H.G., Stradal T.E., Dunn G.A., Small J.V., Rottner K. 2008. Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27:982–992 10.1038/emboj.2008.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Han J., Bamburg J.R., Han L., Lynn R., Zheng J.Q. 2009. Regulation of acetylcholine receptor clustering by ADF/cofilin-directed vesicular trafficking. Nat. Neurosci. 12:848–856 10.1038/nn.2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen C.D., Sahai E. 2010. Cancer dissemination—lessons from leukocytes. Dev. Cell. 19:13–26 10.1016/j.devcel.2010.06.013 [DOI] [PubMed] [Google Scholar]
- Mahaffey J.P., Grego-Bessa J., Liem K.F., Jr, Anderson K.V. 2013. Cofilin and Vangl2 cooperate in the initiation of planar cell polarity in the mouse embryo. Development. 140:1262–1271 10.1242/dev.085316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus D.Q., Li X.Y., Durbin S., Agarwal D., Chi Q., Weiss S.J., Sherwood D.R. 2010. In vivo identification of regulators of cell invasion across basement membranes. Sci. Signal. 3:ra35 10.1126/scisignal.2000654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Matheson C., Marra M.A., Wakarchuk M.F., Baillie D.L. 1994. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242:346–357 10.1007/BF00280425 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., Courtneidge S.A. 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.M., Appleton P.L., Swedlow J.R., Waters J.C. 2007. Evaluating performance in three-dimensional fluorescence microscopy. J. Microsc. 228:390–405 10.1111/j.1365-2818.2007.01861.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178:1251–1264 10.1083/jcb.200703092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Parast M., Alberico C., Benian G.M., Ono S. 2003. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116:2073–2085 10.1242/jcs.00421 [DOI] [PubMed] [Google Scholar]
- Ono S., Baillie D.L., Benian G.M. 1999. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145:491–502 10.1083/jcb.145.3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux R., Lizárraga F., Chavrier P. 2009. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. J. Cell Sci. 122:3015–3024 10.1242/jcs.034561 [DOI] [PubMed] [Google Scholar]
- Reddy A., Caler E.V., Andrews N.W. 2001. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 106:157–169 10.1016/S0092-8674(01)00421-4 [DOI] [PubMed] [Google Scholar]
- Rowe R.G., Weiss S.J. 2008. Breaching the basement membrane: who, when and how? Trends Cell Biol. 18:560–574 10.1016/j.tcb.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Rual J.F., Klitgord N., Achaz G. 2007. Novel insights into RNAi off-target effects using C. elegans paralogs. BMC Genomics. 8:106 10.1186/1471-2164-8-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltel F., Daubon T., Juin A., Ganuza I.E., Veillat V., Génot E. 2011. Invadosomes: intriguing structures with promise. Eur. J. Cell Biol. 90:100–107 10.1016/j.ejcb.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Sherwood D.R., Sternberg P.W. 2003. Anchor cell invasion into the vulval epithelium in C. elegans. Dev. Cell. 5:21–31 10.1016/S1534-5807(03)00168-0 [DOI] [PubMed] [Google Scholar]
- Sherwood D.R., Butler J.A., Kramer J.M., Sternberg P.W. 2005. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 121:951–962 10.1016/j.cell.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Trichet L., Sykes C., Plastino J. 2008. Relaxing the actin cytoskeleton for adhesion and movement with Ena/VASP. J. Cell Biol. 181:19–25 10.1083/jcb.200710168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. 2008. Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87:649–667 10.1016/j.ejcb.2008.04.001 [DOI] [PubMed] [Google Scholar]
- von Blume J., Duran J.M., Forlanelli E., Alleaume A.M., Egorov M., Polishchuk R., Molina H., Malhotra V. 2009. Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J. Cell Biol. 187:1055–1069 10.1083/jcb.200908040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume J., Alleaume A.M., Cantero-Recasens G., Curwin A., Carreras-Sureda A., Zimmermann T., van Galen J., Wakana Y., Valverde M.A., Malhotra V. 2011. ADF/cofilin regulates secretory cargo sorting at the TGN via the Ca2+ ATPase SPCA1. Dev. Cell. 20:652–662 10.1016/j.devcel.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Wang W., Eddy R., Condeelis J. 2007. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer. 7:429–440 10.1038/nrc2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Lorenz M., Kempiak S., Sarmiento C., Coniglio S., Symons M., Segall J., Eddy R., Miki H., Takenawa T., Condeelis J. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol. 168:441–452 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zech T., McDonald L., Gonzalez E.G., Li A., Macpherson I., Schwarz J.P., Spence H., Futó K., Timpson P., et al. 2012. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J. Cell Biol. 199:527–544 10.1083/jcb.201203025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel J.W., Hagedorn E.J., Audhya A., Sherwood D.R. 2009. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat. Cell Biol. 11:183–189 10.1038/ncb1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.