Abstract
Background
Atopic dermatitis (AD) is a common disease, with an increasing prevalence. The primary pathogenesis of the disease is still elusive, resulting in the lack of specific treatments. AD is currently considered a biphasic disease, with Th2 predominating acute disease, and a switch to Th1 characterizing chronic disease. Elucidation of the molecular factors that participate in the onset of new lesions and maintenance of chronic disease is critical for the development of targeted therapeutics.
Objectives
We sought to characterize the mechanisms underlying onset and maintenance of AD.
Methods
We investigated intrapersonal sets of transcriptomes from non-lesional, acute and chronic lesions of ten AD patients through genomic, molecular and cellular profiling.
Results
Our study associated the onset of acute lesions with a striking increase in a subset of terminal differentiation proteins, specifically the cytokine-modulated S100A7, S100A8, and S100A9. Acute disease was also associated with significant increases in gene expression levels of major Th22- and Th2- cytokines, and smaller increases in IL-17. A lesser induction of Th1-associated genes was detected in acute disease, although some were significantly up-regulated in chronic disease. Further significant intensification of major Th22 and Th2 cytokines was observed between acute and chronic lesions.
Conclusions
Our data identified increased S100A7, S100A8 and S100A9 gene expression with AD initiation, and concomitant activation of Th2 and Th22 cytokines. Our findings support a model of progressive activation of Th2 and Th22 immune axes from acute to chronic phases, expanding the prevailing view of pathogenesis, with important therapeutic implications.
Keywords: atopic dermatitis, acute, chronic, Th2, Th22, Th17, IL-22, S100A7, S100A8, S100A9, terminal differentiation
INTRODUCTION
Atopic dermatitis (AD) is the most common inflammatory skin disease, with an increased prevalence in the past several decades.1,2 Together with asthma and allergic rhinitis, it constitutes the “atopic triad,” broadly encountered by the medical community.3 It is characterized by distinct acute and chronic stages; acute lesions are typically bright red, “wet” and flat, becoming dull red, dry and thick with chronicity.2,4
The pathogenesis of AD has been linked to both immune and barrier abnormalities.1,2,4 While an association between epidermal defects and genetic mutations in filaggrin was recently identified,5 the Th2 (IL-4, IL-13, IL-31) and Th22 (IL-22) cytokines implicated in AD6,7 were shown to suppress major terminal differentiation proteins (i.e. filaggrin and loricrin).2,8–11 Unlike psoriasis, another common inflammatory skin disease that is increasingly considered a Th17 disease, and for which an increased understanding of pathogenic mechanisms led to development of revolutionary targeted therapeutics, the pathogenesis of AD is still enigmatic, resulting in the lack of specific treatments.6,12–15 This is further complicated by the lack of data to address whether acute and chronic AD represent progressive stages across a continuum of inflammatory responses or if each has distinct immunologic mechanisms.1,2,4,15
Specifically, little is known about the factors that initiate AD lesions or sustain chronic inflammation.1,12,15 This stems from difficulties in obtaining uninvolved (non-lesional), acute and chronic skin lesions from single patients, ultimately resulting in very few reports, which offer comparisons on limited numbers of patients.16–20 Furthermore, a broad genomic analysis comparing acute and chronic disease is lacking.
The current model for acute AD largely originates from an experimental model that uses the atopy patch test (APT) with environmental allergens to simulate acute disease.21–24 Based on these studies, AD pathogenesis is characterized as a biphasic T-cell mediated disease; while a Th2 signal predominates the acute phase, a Th2 to Th1 switch promotes disease chronicity.2,21–23 As this is based on an experimental model, a characterization of spontaneous acute AD lesions and comparison to chronic lesions from the same patients is warranted.
We sought to characterize the mechanisms underlying the onset and maintenance of AD, based on paired comparisons between non-lesional, acute and chronic skin lesions from individual patients (Figure 1A).
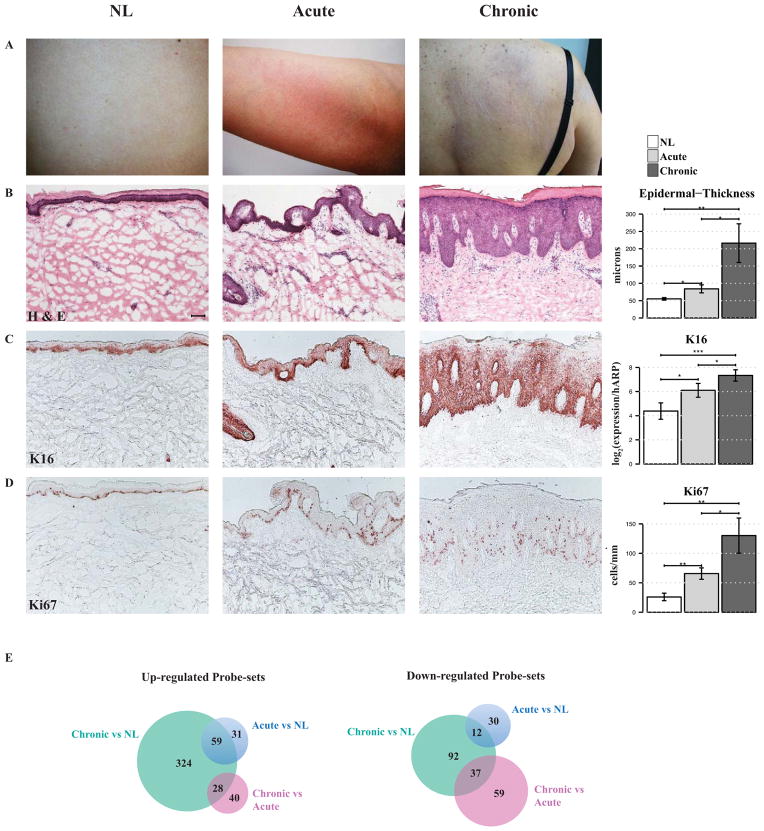
Figure 1. Clinical, histologic and genomic differences between non-lesional (NL), acute and chronic atopic dermatitis (AD).
A, Clinical images of NL, acute and chronic AD demonstrate bright erythema of acute and dullness and lichenification of chronic lesions. B–D, Representative immunohistochemistry stainings of B, H&E and the proliferation markers C, Keratin 16 (K16) and D, Ki67 in tissue sections of NL, acute and chronic AD skin, as well as B, quantification of epidermal thickness, C, K16 by real-time PCR (RT-PCR), represented in log2 [log2(expression/hARP)] and D, Ki67+ cell counts, showing significantly increased epidermal hyperplasia and abnormal proliferation in acute lesions, with further increases in chronic lesions. *P < .05, **P < .01, and ***P < .001, (n=10). Bar plots represent mean ± SEM. Scale bar = 100 μm. E, Venn diagrams of up- and down-regulated probe-sets in acute and chronic skin lesions, in comparison with NL skin, using criteria of fold change (FCH)>1.5 and p-value<0.1, (n=8).
METHODS
Patient Population
Intrapersonal acute, chronic (> 72 hrs duration) and non-lesional (≥ 10 cm from active lesions) skin biopsies (Figure 1A) and blood samples were collected from each of 17 patients with moderate-to-severe AD under an Institutional Review Board-approved protocol. Biopsies were not taken from skin that was clinically judged as infected. No systemic or topical treatments were allowed for ≥4 weeks prior to biopsies. Ten patients (4 males and 6 females, age: 20–67 years; mean age: 44 years) (see Table E1 in the Online Repository) met the following criteria for distinguishing true acute from “acute on chronic” skin lesions: a) new lesions of <72 hours duration, as previously defined16; b) lack of skin lichenification; c) lack of regenerative hyperplasia, as defined by epidermal thickness ≤150μm (H&E) (Figure 1B) and basal or confluent supra-basal Keratin 16 (K16) positivity (Figure 1C). Serum IgE levels were increased in 7 of 10 patients (range: 27.9–3652 kU/L; mean: 1032 kU/L; reference range: 0–160 kU/L) and eosinophil counts in 1 patient (reference range: 0% to 7%). The Scoring of Atopic Dermatitis (SCORAD) index was used to evaluate disease severity (range: 40–63; mean: 50). No filaggrin gene mutations were found.
Skin biopsies from normal volunteers25 and psoriasis patients26 (n=15 in both) were used for comparisons (see Methods in the Online Repository).
Biopsies were frozen in OCT medium for immunohistochemistry (IHC) and liquid nitrogen for RNA extraction. Histologic and real-time PCR (RT-PCR) analyses were performed on non-lesional, acute, and chronic skin samples from all 10 patients. However, because of loss of tissue with laboratory processing, gene array analysis was performed on 8 patients.
Immunohistochemistry
IHC was performed on cryostat tissue sections using purified mouse anti-human monoclonal antibodies (see Table E2 and Methods in the Online Repository). Epidermal thickness was quantified and positive cells per millimeter were counted manually using computer-assisted image-analysis software (ImageJ 1.42; NIH, Bethesda, MD).
Sample preparation for RT-PCR and gene chip analysis
RNA was extracted for RT-PCR, which was performed with EZ-PCR Core Reagents (Life Technologies, Grand Island, NY), and custom primers were generated as previously described.27 We used human HGU133Plus2.0 GeneChip probe arrays (Affymetrix Inc, Santa Clara, Calif). Total RNA was extracted by Qiagen RNeasy Mini Kit (Valencia, Calif), and DNA was removed by Qiagen RNAse-free DNAse Set. Total RNA (50 ng) was reverse transcribed and amplified with Ovation Whole Blood Solution from NuGen (San Carlos, Calif). The labeled target was fragmented and hybridized to probe arrays, using Encore Biotin Module from NuGen (see Methods in the Online Repository).
Statistical analyses
Analysis of gene-array and RT-PCR data (log2 scale of normalized values) was through a linear mixed-effect model and of IHC cell counts was via the paired Wilcoxon Rank Sum Tests.
Affymetrix CEL files were scrutinized for spatial artifacts by Harshlight package.28 Expression values were obtained with the frozen robust multi-array analysis (fRMA) algorithm.29 Since samples were hybridized using different kits, the batch effect was removed using R’s package COMBAT.30 Raw data was deposited in NCBI’s Gene Expression Omnibus repository (accession no. GSE36842).
Expression values (in log2) were modeled by a linear mixed-effects model, with Group as the fixed effect and a random intercept for each patient, in the framework of Bioconductors’ limma package. The comparisons of interest were tested using moderated t-test,31 and resultant p-values were adjusted for multiples hypotheses by the Benjamini-Hochberg procedure. Since typical FDR cutoffs proved as very stringent for the small sample size, genes with fold change (FCH)>1.5 and p<0.01 were considered differentially expressed genes (DEGs) (see Methods and Table E3 in the Online Repository).
RESULTS
To identify the key pathways associated with acute skin lesions, we characterized the global cellular, molecular and gene-expression alterations that appear with the onset of AD lesions.
Acute disease triggers increased hyperplasia
We compared epidermal thickness and protein expression of the proliferation markers K16 and Ki67, among non-lesional, acute and chronic AD skin lesions (Figure 1B–D). A significant increase in epidermal thickness (52%, P=0.027) and proliferation (153% increase in Ki67+ cells, P=0.004) was observed from non-lesional to acute lesions, with additional respective increases of 141% (P=0.02) and 110% (P=0.02) from acute to chronic lesions (Figure 1B, D). K16 mRNA expression was also significantly up-regulated between non-lesional and acute (P=0.02, Figure 1C), with a further significant increase in chronic lesions (P=0.016, Figure 1C). While chronic lesions demonstrated continuous suprabasal K16 staining, acute lesions showed patchy K16 positivity (Figure 1C).
Genomic expression differences between acute, non-lesional and chronic AD
Gene-arrays were conducted to explore the characteristics of acute compared to chronic and non-lesional AD skin lesions, using the above criteria to define DEGs in acute compared to chronic and non-lesional AD (see Table E3 in the Online Repository).
Our study defined a set of intrapersonal transcriptomes from non-lesional, acute and chronic skin lesions of individual patients. 90 probe-sets (76 genes) were up-regulated and 42 (26 genes) were down-regulated in acute compared to non-lesional AD (Figure 1E). A higher number of DEGs were identified in chronic in comparison to non-lesional AD [411 probe-sets (310 genes) up- and 141 (110 genes) down-regulated]. A relatively small set of DEGs distinguished chronic and acute AD [68 probe-sets (47 genes) up-and 96 (75 genes) down-regulated] (Figure 1E and see Table E3 in the Online Repository).
Acute AD is associated with an abrupt increased expression of epidermal S100 proteins
As the top 50 DEGs between acute and non-lesional AD skin include many terminal differentiation genes (see Figure E1 in the Online Repository), we further analyzed this gene-group (Figure 2A).
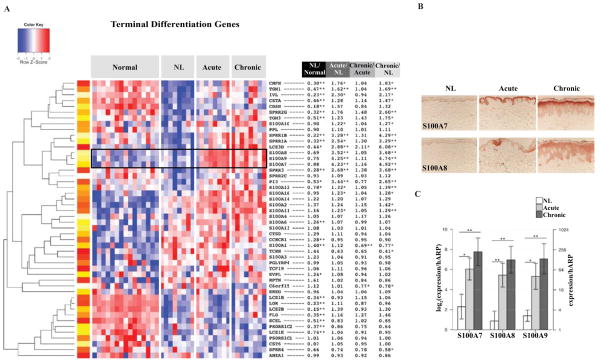
Figure 2. Onset of acute lesions is associated with coordinated increases in S100A7, S100A8 and S100A9.
A, Unsupervised hierarchical clustering of terminal differentiation genes across normal, non-lesional (NL), acute and chronic atopic dermatitis (AD) skin samples (red, up-regulated; blue, down-regulated). In contrast to the uniform down-regulation of well-characterized epidermal differentiation complex (EDC) genes such as filaggrin and loricrin throughout NL, acute and chronic skin lesions, there is a steep increase in expression of S100A7, S100A8 and S100A9 with the onset of acute disease, with further induction in chronic lesions, as indicated by the black highlighting. Probes with the largest fold change (FCH) were chosen when several probes represented single genes. *P < .05, **P < .01, and ***P < .001, (n=8 for AD; n=15 for normals). The significant induction of these S100 proteins has been validated by increases in both B, protein expression of S100A7 and S100A8 in representative immunohistochemistry stainings of acute and chronic lesions, compared to NL skin, and C, mRNA gene expression levels by real-time PCR (RT-PCR) represented in log2 [log2(expression/hARP)] and natural scale (expression/hARP) (~8 FCH, P<0.02). hARP, human acidic ribosomal protein. (n=10).
The transition from non-lesional to acute lesional skin was associated with a striking increase in expression of a subset of epidermal differentiation complex (EDC) genes, including S100A7, S100A8 and S100A9 (~4 FCH, Figure 2A, see Figure E1 and Table E3 in the Online Repository). These steep increases, detected by gene-arrays (~4 FCH), were confirmed by both strong protein expression of S100A7 and S100A8 in tissue sections of acute lesions, which localized to the upper spinous and granular layers of the epidermal keratinocytes (Figure 2B), and by comparatively large increases in gene expression levels by RT-PCR (~8-FCH, P=0.019, Figure 2C).
Compared to normal controls, the expression of other well-characterized EDC genes [i.e. filaggrin, loricrin and late cornified envelope 2B (LCE2B)] showed large reductions in non-lesional AD skin, and were unaltered with either acute or chronic disease (Figure 2A). This was further confirmed by RT-PCR, showing grossly similar mRNA expression of filaggrin and corneodesmosin across non-lesional and active disease (Figure 3).
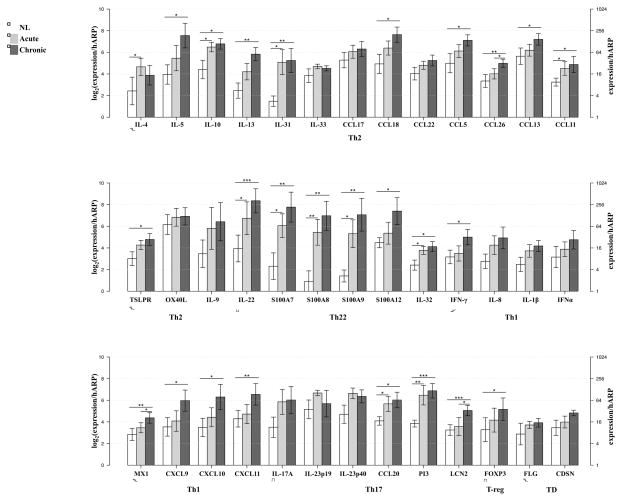
Figure 3. Marked activation of Th2 and Th22 immune pathways in acute disease with progressive activation in chronic skin lesions, by real-time PCR (RT-PCR).
Mean expression estimates normalized to human acidic ribosomal protein (hARP) are represented in log2 [log2(expression/hARP)] and natural scale (expression/hARP). The largest gene-expression increases are associated with Th2- and Th22-associated products, including IL-31, IL-22 and S100A7, S100A8, S100A9. Small increases in Th1-related products are evident in both acute and chronic lesions. In contrast with the immune mediators, the mRNA expression levels of filaggrin (FLG) and corneodesmosin (CDSN) showed similarity across the disease. Bar plots represent mean ± SEM. *P < .05, **P < .01, and ***P < .001, (n=10). FOXP3, forkhead box protein 3; LCN2, lipocalin 2; MX1, myxovirus resistance 1; OX40L, OX40 ligand; PI3, peptidase inhibitor 3; TD, terminal differentiation; TSLPR, thymic stromal lymphopoietin receptor.
Acute atopic dermatitis is associated with Th2 and Th22 cytokine activation
One hypothesis for the abrupt activation of the S100A7, S100A8 and S100A9 genes is that they are induced by T-cell derived cytokines, since these products were shown to be up-regulated in vitro by IL-22 and/or IL-17 cytokines.8,10,32,33 Thus, we broadly measured changes in activation of major cytokines and chemokines that define Th1, Th2, Th22 and Th17 T-cell subsets, as well as representative genes shown to be up-regulated in cytokine-treated keratinocytes6,34–41 using both gene-arrays and quantitative RT-PCR measures, since transcripts of primary cytokines are not always measured on gene-arrays (Figure 3 and see Figure E2 in the Online Repository).
The onset of acute lesions is associated with significant increases in gene expression levels of Th2- (i.e. IL-4, IL-13, IL-31) and Th22- (i.e. IL-22) defining cytokines (Figure 3 and see Figure E2A–B in the Online Repository). The largest quantitative gene-expression increases are associated with IL-31, IL-22, S100A7, S100A8 and S100A9, as demonstrated by both RT-PCR and gene-arrays (Figure 3 and see Figure E2B in the Online Repository). Acute disease showed a positive correlation between the SCORAD index and IL-22 mRNA expression (r=0.55, P=0.132). A lesser induction of Th1/interferon- [i.e. IFN-γ, myxovirus resistance 1 (MX-1), IL-1β, CXCL9-11] induced products was also evident in acute disease (Figure 3 and see Figure E2C in the Online Repository). Small increases in Th17 axis cytokines (IL-17, IL-23p19 and IL23p40) were detected in acute disease, accompanied by significant increases in IL-17-regulated products [CCL20, peptidase inhibitor 3 (PI3)/Elafin, and lipocalin 2 (LCN2)] (Figure 3 and see Figure E2D in the Online Repository).
Intensification of immune activation rather than an immune switch characterizes chronic disease
Progressive increases in IL-22 mRNA and its associated products (S100A7, S100A8, S100A9 and IL-32)42 were detected between acute and chronic lesions (Figure 3 and see Figure E2B in the Online Repository). Chronic lesions are also characterized by intensified Th2-related inflammation, with significant increases in most associated products (i.e. IL-5, IL-13, IL-10, IL-31, CCL5, CCL13, CCL18), with the exception of IL-4 and its receptor that decreased from acute-to-chronic lesions (Figure 3 and see Figure E2A in the Online Repository). Nevertheless, in chronic disease, changes in IL-4 mRNA expression correlate with the SCORAD index (r=0.833, P=0.008). Significant increases in Th1-related products (i.e. IFN-γ, MX1, CXCL9-11) were detected only in chronic skin lesions although small increases were evident in acute lesions. No significant differences were found in Th17-mediated products between acute and chronic disease (except for LCN2). Significant increases in several IL-17-regulated genes (CCL20, PI3, LCN2) were detected in chronic lesions, as compared to non-lesional skin (Figure 3 and see Figure E2D in the Online Repository). A significant increase in the regulatory T-cell marker, forkhead box protein 3 (FOXP3) was found in chronic disease alone (Figure 3).
A multiple regression analysis identified the best set of SCORAD predictors in chronic AD as Th2-, Th1- and Th17-associated products, and Th2-derived CCL18 and CCL13 in acute AD (see Table E4 in the Online Repository).
Increased immune-cell infiltrates with disease onset by immunohistochemistry
Since these data associate the onset and chronicity of AD with progressive increases in cytokines and chemokines, we measured the infiltration of non-lesional, acute and chronic lesions by various inflammatory cells (Figure 4).
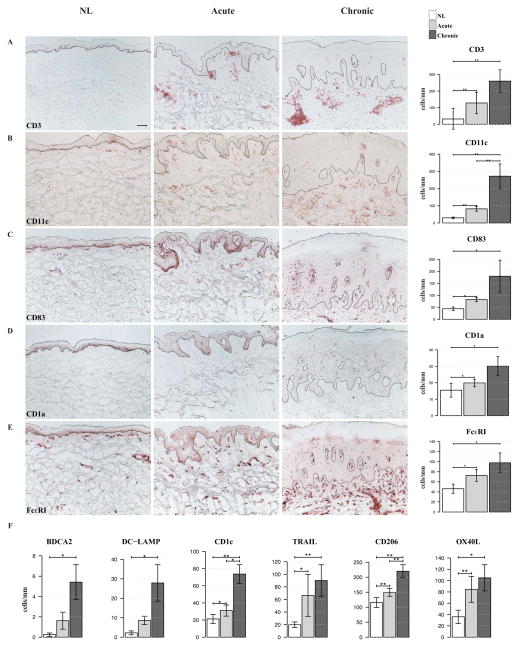
Figure 4. Significant increases in immune-cell infiltrates characterize acute disease onset and progression to chronic disease, as quantified by means of immunohistochemistry and cell counts.
A–E, Significant increases in (CD3+) T-cells, myeloid (CD11c+) dendritic cells (DCs), mature (CD83+) DCs, Langerhans (CD1a+) cells and inflammatory dendritic epidermal cells (IDECs) [Fc receptor for IgE (Fc ε RI+)] characterize acute disease. F, Cell counts of plasmacytoid [blood dendritic cell antigen 2 (BDCA2+)] DCs, mature [dendritic cell lysosome-associated membrane glycoprotein (DC-LAMP+)] DCs, resident (CD1c+) DCs, inflammatory [TNF-related-apoptosis-inducing ligand (TRAIL+)] DCs, IDECs (CD206+) and atopic [OX40 ligand (OX40L+)] DCs, increase from non-lesional (NL) through chronic atopic dermatitis (AD) skin. *P < .05, **P < .01, and ***P < .001, (n=10). Bar plots represent mean ± SEM. Scale bar = 100 μm.
Significant increases in infiltrating T (CD3+) cells (303%, P=0.004), myeloid (CD11c+) dendritic-cells (DCs) (178%, P=0.004), mature DCs (CD83+) (P=0.034) and inflammatory dendritic epidermal cells (IDECs) [as quantified by Fc receptor for IgE high affinity I (Fcε RI+) (P=0.038) and CD206+ (P=0.009) cells] were observed between non-lesional and acute AD skin lesions (Figure 4). Increases in cell counts were also noted in Langerhans cells (CD1a+) and other DC subsets, including: “inflammatory” [TNF-related-apoptosis-inducing ligand (TRAIL+)] DCs and “resident” (CD1c+) DCs.43 T-cell and DC infiltrates were further increased with disease chronicity (P<0.05 for most subsets, Figure 4).
Although scant numbers of eosinophils, neutrophils and mast cells were detected in skin lesions (as quantified by major basic protein+, neutrophil elastase+, and tryptase+ cell counts, respectively), we observed an increase in these infiltrates from non-lesional through chronic disease (P=0.036 and P=0.022, for eosinophils and neutrophils, respectively, see Figure E3 in the Online Repository).
Compared to psoriasis, IL-17-induced genes are not highly increased in AD
Th17/IL-17 up-regulates a large set of genes in keratinocytes that are coordinately up-regulated in psoriasis.10,44–46 Therefore, we examined disease profiles of acute AD, chronic AD and psoriasis (each compared to non-lesional skin) for genes induced by IL-17.10
In contrast to a strong up-regulation of Th17/IL-17-induced genes in psoriasis, this association is attenuated in both acute and chronic AD (Figure 5A–B, D). As visualized in Figure 5A–B and D, almost all of the IL-17-related genes (61/74, 82% in acute AD and 63/74, 85% in chronic AD) are within the psoriasis field, with few exceptions, including S100A7 and S100A8, which are synergistically induced by IL-17 and IL-22.8,10,33,43
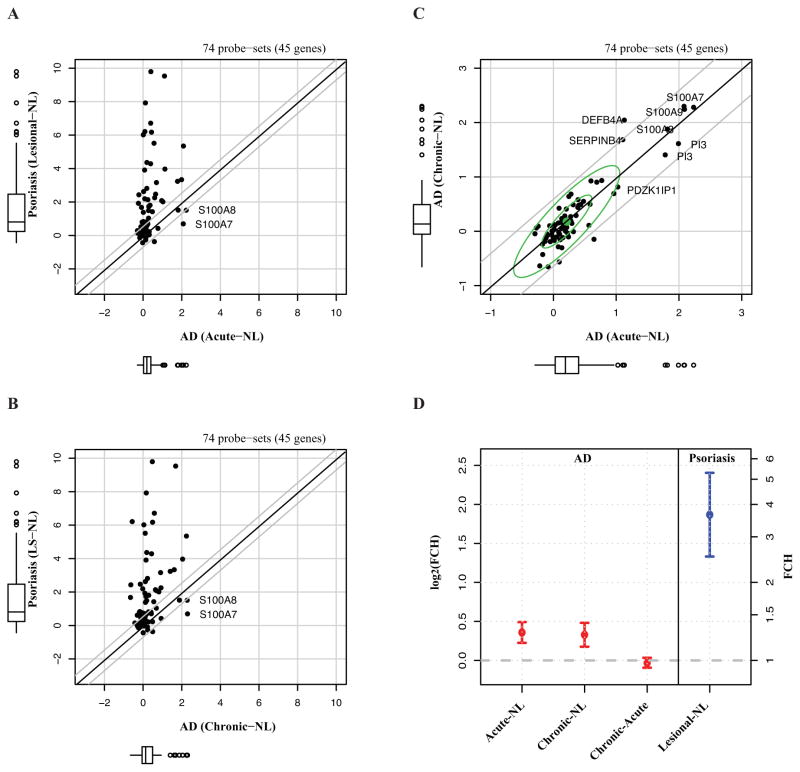
Figure 5. Disease profiles of IL-17-induced genes in acute and chronic atopic dermatitis (AD), as compared to psoriasis.
A–B, A scatterplot of fold changes (FCHs) between lesional and non-lesional (NL) skin for each disease (in log2-scale) demonstrates that whereas in psoriasis, these genes are well associated with the genomic differences between NL and lesional skin, no such association was found for either acute or chronic AD. C, There is a similar distribution of the Th17-regulated genes between acute and chronic AD, both compared to NL. D, Means and 95% confidence intervals (CIs) of the log2(FCH) among IL-17 genes in psoriasis and AD transcriptomes (n=8 for AD; n=15 for psoriasis).
A similar magnitude of representation of IL-17-induced genes was found between acute and chronic AD lesions, demonstrating lack of major differences in this immune-axis between acute and chronic disease (Figure 5C–D).
Thus, while psoriasis has a strong Th17/IL-17 molecular fingerprint, this signature is less evident in both acute and chronic AD.
DISCUSSION
The pathogenesis of AD has been attributed to alternate mechanisms, including a) a defective formation of the epidermal barrier, with mutations in filaggrin that may contribute to this defect,5 or b) effects of immune cytokines such as IL-4, IL-13 and IL-31 that can suppress epidermal differentiation.2,8–11,47 In the first model, immune activation might arise as a consequence of abnormal growth/differentiation of the epidermis, whereas in the second model, disease-related barrier changes might be driven by underlying T-cell activation and cytokine production. While early models of immune activation in AD focused on the differential roles for Th2 and Th1 in disease initiation and maintenance, recently, roles for Th17 and Th22 T-cells have also been proposed.7,12,44,48
Within the context of alternate models of pathogenesis, it is extremely important to measure alterations in terminal epidermal differentiation and immune mediators that occur with the onset of acute skin lesions. However, it has been extremely difficult to perform studies on true acute AD lesions, since most AD patients present with only chronic lesions (defined as persisting for >72 hours).16 In turn, most studies of “acute” disease changes have examined skin inflammation induced by topical application of environmental allergens that is assumed to parallel immune changes in acute skin lesions.21–24 Measures made in these patch reactions have suggested the biphasic model, in which initiation of inflammation is associated with Th2 activation, and chronic disease is associated with a Th1 switch.21–23 The biphasic model is in marked contrast to the progressive cytokine activation that occurs in an IL-4 transgenic mouse model resembling AD inflammation.35
Our study represents the first comprehensive genomic and molecular comparison of intrapersonal non-lesional, acute and chronic skin lesions. With analysis of well-defined acute AD lesions, in comparison to chronic and clinically uninvolved AD skin, we have established detailed changes in epidermal growth/differentiation and immune activation that occur with both the onset of new lesions and persistence of disease.
While the expression of some epidermal differentiation gene products such as filaggrin and loricrin is largely reduced in non-lesional AD skin, further down-regulation does not occur with acute or chronic lesions. Thus, onset of clinically significant disease is not accompanied by decreases in epidermal differentiation genes, but is instead associated with marked activation of an EDC gene cluster on chromosome 1q, with the largest increases in expression of S100A7, S100A8, and S100A9. The increase in these gene products corresponds with the abrupt increased synthesis of S100 proteins in epidermal keratinocytes in acute and chronic AD skin lesions. Increased S100A7 in acute AD has been detected in only one past study, in which both a specific definition of acute AD, and a comparison to chronic lesions from the same patients were lacking.49 The S100A7, S100A8, and S100A9 proteins were shown to be highly expressed in chronic hyperproliferative epithelium, and have been associated with an alternative differentiation pathway.50–52 They have important functions in inflammation, such as chemotaxis of T-cells, monocytes and neutrophils as well as pro-inflammatory functions in many inflammatory diseases.8,53–55 The particular relevance of these proteins for inflammation was underlined with the identification of a new inflammatory disorder, whose hallmark is an abundance of S100A8 and S100A9.56 This disorder is characterized by recurrent skin infections and systemic inflammation, bearing similarities to AD.56 Yet, these proteins were also suggested to have antimicrobial activities, particularly against gram-negative bacteria such as E. coli. Indeed, AD skin is rarely infected with E. coli.51,57 However, since AD skin is characterized by an increased rate of S. aureus skin infections, even in the presence of up-regulated S100A7, S100A8 and S100A9 expression, these molecules may be more important as pro-inflammatory mediators in the pathogenesis of the disease. In acute AD, these proteins might contribute to chemotaxis of immune cells, and particularly T-cells, which are highly increased in acute disease.
The onset of acute AD skin lesions is also linked to increased skin infiltration by T-cells and DCs, along with significantly increased synthesis of cytokines that define activation of Th2 and Th22 T-cell subsets, as well as increased IL-17-related products (i.e. CCL20, PI3/Elafin and LCN2). S100A7, S100A8 and S100A9 were demonstrated to be regulated by IL-22 and IL-17 cytokines alone and in synergy.8,10,33,43 Thus, the marked induction of S100 genes in the epidermis could be explained by inductive effects of IL-22 and/or IL-17 cytokines on these gene products in epidermal keratinocytes, while the induction of epidermal hyperplasia at the onset of acute AD might result from a direct response of keratinocytes to IL-22.7 Although increased expression of IL-4, IL-13, and IL-31 has been hypothesized to decrease keratinocyte differentiation,9,47 we did not identify corresponding decreases in the expression of filaggrin, loricrin or other terminal differentiation genes from non-lesional through chronic skin lesions. A particularly dramatic induction of the itch-associated Th2 cytokine, IL-31, was measured in acute AD, corresponding to the marked pruritus experienced during development of new lesions.58,59
The sum of these observations suggests a new hypothesis, namely that onset of clinically evident acute AD lesions is linked to activation of a subset of epidermal differentiation genes, instead of a further suppression of terminal differentiation (Figure 6). This could be driven by a set of established activators of the S100 epidermal differentiation proteins,8,10,33,43 specifically IL-22 and IL-17 that are increased in acute lesions. While these cytokines are typically derived from Th17 and Th22 T-cell subsets, and corresponding increases in T-cell infiltrates are evident in acute lesions, further studies should still establish T-cells as the source of these cytokines. Furthermore, the S100 proteins could be modulated by additional factors, such as IL-1α, IL-6, oncostatin M, and TNF-α.60–65 We did not detect altered expression of these markers in our study, but cannot exclude the possibility that factors beyond increased IL-22 and/or IL-17 contribute to modulation of S100 proteins in acute disease.
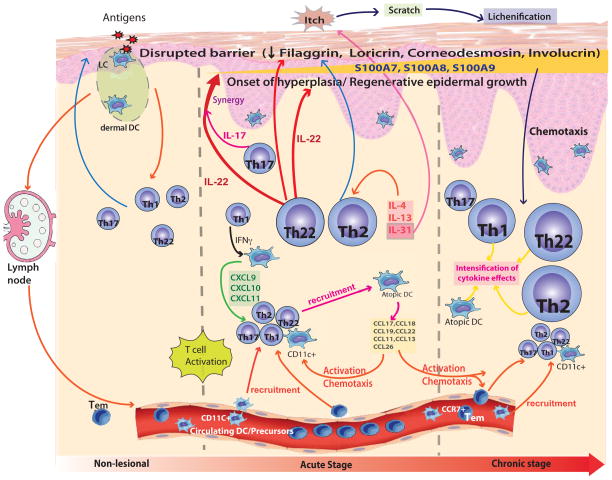
Figure 6. A schematic illustration of initiation of acute atopic dermatitis (AD) and progression to chronic skin lesions.
Non-lesional AD skin lesions show some immune infiltrates that produce inflammatory mediators, which might contribute to a defective epidermal barrier. Barrier defects lead to penetration by epicutaneous antigens that encounter Langerhans cells in the epidermis and dermal dendritic cells in the dermis, inducing marked immune activation and recruitment of inflammatory cells in acute AD lesions. Marked activation of Th2 and Th22 axes occurs in acute disease onset. Smaller increases in Th1 and Th17 immune axes were found in acute lesions. A progressive activation of Th2 and Th22, as well as Th1 pathways is characteristic of the chronic stage of AD. The relative induction of each T-cell subset, according to disease stage, is represented pictorially by their size, relative to the other T-cell subsets. Cytokines (i.e. IL-4, IL-13) and chemokines [i.e. CCL17, CCL18, CCL19, CXCL9, CXCL10, CXCL11] produced by various T-cells and DCs induce further activation and recruitment of additional immune cells. With the onset of acute disease, Th22 cells release IL-22, which induces epidermal hyperplasia, and synergistically with the Th17 cytokine, IL-17, drives an abrupt increase in a subset of terminal differentiation genes, specifically S100A7, S100A8 and S100A9 proteins. The increases in these barrier proteins contrast with the uniformly disrupted epidermal differentiation gene products (filaggrin, loricrin, corneodesmosin, etc.) throughout non-lesional, acute and chronic AD skin. The Th2 and Th22 cytokines contribute to inhibition of the terminal differentiation proteins. IL-31 is abruptly up-regulated in acute disease, potentially reflecting its role as an itch mediator in AD.
Chronic AD lesions are characterized by further increases in epidermal proliferation, skin infiltration by T-cells and DCs, and mRNA levels of IL-22 and S100 proteins, further supporting a probable link between expression of these transcriptional axes. The increase in Th1/IFNγ-related products in chronic lesions could either represent a pro-inflammatory stimulus for activation for Th17/Th22 T-cells,66 or a counter-regulatory signal to Th267 or Th1768 activation. Our findings indicate that progression to chronic disease is associated with intensification of the immune axes that are up-regulated in acute lesions, particularly Th22 and Th2, as well as significant increases in Th1-related products. Although less significant increases were detected in IL-17 cytokine expression (P=0.13), several IL-17 regulated products were also significantly increased in acute or chronic AD lesions (Figure 3 and see Figure E2D in the Online Repository). Furthermore, IL-17-related genes were among the set of best predictors of the SCORAD in chronic disease. Thus, IL-17 might still be biologically important in initiation and progression of AD lesions, and could activate or synergize with IL-22 for induction of S100A7, S100A8 and S100A9. However, the increases in IL-17-associated products in both acute and chronic versus non-lesional AD are much lower than in psoriasis (Figure 5A–B, D).46 The more modest contribution of the Th17 axis in AD might result from inhibition of this pathway by activated Th2 cytokines in both acute and chronic AD.42
Our data expands the prevailing view of pathogenic pathways16 and has important implications for the treatment of acute and chronic AD. One implication of these data is that potential antagonism of the Th2 and Th22 inflammatory pathways might not only reverse chronic AD lesions, but could also prevent the onset of new skin lesions.
Supplementary Material
Acknowledgments
JGK and MSF were supported by grant number 5UL1RR024143-02 from the National Center for Research Resources (NCRR), a component of the NIH, and NIH Roadmap for Medical Research. C.D.S. was supported by the NIH (R00AR055948). E.G.Y. was supported by the Dermatology Foundation Physician Scientist Career Development Award.
ABBREVIATIONS
- AD
Atopic dermatitis
- APT
Atopy patch test
- BDCA2
Blood dendritic cell antigen 2
- CDSN
Corneodesmosin
- CI
Confidence interval
- DC
Dendritic cell
- DC-LAMP
Dendritic cell lysosome-associated membrane glycoprotein
- DEG
Differentially expressed gene
- EDC
Epidermal differentiation complex
- Fcε RI
Fc receptor for IgE
- FCH
Fold change
- FLG
Filaggrin
- FOXP3
Forkhead box protein 3
- fRMA
Frozen robust multi-array analysis
- hARP
Human acidic ribosomal protein
- IDEC
Inflammatory dendritic epidermal cell
- IHC
Immunohistochemistry
- K16
Keratin 16
- LCE
Late cornified envelope
- LCN2
Lipocalin-2
- MX1
Myxovirus resistance 1
- NL
Non-lesional
- OX40L
OX40 ligand
- PI3
Peptidase inhibitor 3
- RT-PCR
Real-time PCR
- SCORAD
Scoring of Atopic Dermatitis
- TRAIL
TNF-related-apoptosis-inducing ligand
- TSLPR
Thymic stromal lymphopoietin receptor
Footnotes
Disclosures: The authors have declared that they have no conflict of interest.
Clinical Implications
Targeting Th2 and Th22 pathways in atopic dermatitis can be an effective therapeutic strategy regardless of disease stage.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–94. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 2.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–7. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol. 2003;112:S118–27. doi: 10.1016/j.jaci.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 4.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part I: clinical and pathologic concepts. J Allergy Clin Immunol. 2011;127:1110–8. doi: 10.1016/j.jaci.2011.01.053. [DOI] [PubMed] [Google Scholar]
- 5.Palmer CN, Irvine AD, Terron-Kwiatkowski A, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 6.Brandt EB, Sivaprasad U. Th2 Cytokines and Atopic Dermatitis. J Clin Cell Immunol. 2011:2. doi: 10.4172/2155-9899.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nograles KE, Zaba LC, Shemer A, et al. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J Allergy Clin Immunol. 2009;123:1244–52.e2. doi: 10.1016/j.jaci.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boniface K, Bernard FX, Garcia M, Gurney AL, Lecron JC, Morel F. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J Immunol. 2005;174:3695–702. doi: 10.4049/jimmunol.174.6.3695. [DOI] [PubMed] [Google Scholar]
- 9.Howell MD, Kim BE, Gao P, et al. Cytokine modulation of atopic dermatitis filaggrin skin expression. J Allergy Clin Immunol. 2009;124:R7–R12. doi: 10.1016/j.jaci.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Nograles KE, Zaba LC, Guttman-Yassky E, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159:1092–102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sehra S, Yao Y, Howell MD, et al. IL-4 regulates skin homeostasis and the predisposition toward allergic skin inflammation. J Immunol. 2010;184:3186–90. doi: 10.4049/jimmunol.0901860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman-Yassky E, Nograles KE, Krueger JG. Contrasting pathogenesis of atopic dermatitis and psoriasis--part II: immune cell subsets and therapeutic concepts. J Allergy Clin Immunol. 2011;127:1420–32. doi: 10.1016/j.jaci.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 13.Homey B, Steinhoff M, Ruzicka T, Leung DY. Cytokines and chemokines orchestrate atopic skin inflammation. J Allergy Clin Immunol. 2006;118:178–89. doi: 10.1016/j.jaci.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 14.Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med. 2009;361:496–509. doi: 10.1056/NEJMra0804595. [DOI] [PubMed] [Google Scholar]
- 15.Wilsmann-Theis D, Hagemann T, Jordan J, Bieber T, Novak N. Facing psoriasis and atopic dermatitis: are there more similarities or more differences? Eur J Dermatol. 2008;18:172–80. doi: 10.1684/ejd.2008.0357. [DOI] [PubMed] [Google Scholar]
- 16.Hamid Q, Boguniewicz M, Leung DY. Differential in situ cytokine gene expression in acute versus chronic atopic dermatitis. J Clin Invest. 1994;94:870–6. doi: 10.1172/JCI117408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamid Q, Naseer T, Minshall EM, Song YL, Boguniewicz M, Leung DY. In vivo expression of IL-12 and IL-13 in atopic dermatitis. J Allergy Clin Immunol. 1996;98:225–31. doi: 10.1016/s0091-6749(96)70246-4. [DOI] [PubMed] [Google Scholar]
- 18.Laberge S, Ghaffar O, Boguniewicz M, Center DM, Leung DY, Hamid Q. Association of increased CD4+ T-cell infiltration with increased IL-16 gene expression in atopic dermatitis. J Allergy Clin Immunol. 1998;102:645–50. doi: 10.1016/s0091-6749(98)70282-9. [DOI] [PubMed] [Google Scholar]
- 19.Taha RA, Leung DY, Ghaffar O, Boguniewicz M, Hamid Q. In vivo expression of cytokine receptor mRNA in atopic dermatitis. J Allergy Clin Immunol. 1998;102:245–50. doi: 10.1016/s0091-6749(98)70093-4. [DOI] [PubMed] [Google Scholar]
- 20.Taha RA, Minshall EM, Leung DY, et al. Evidence for increased expression of eotaxin and monocyte chemotactic protein-4 in atopic dermatitis. J Allergy Clin Immunol. 2000;105:1002–7. doi: 10.1067/mai.2000.106483. [DOI] [PubMed] [Google Scholar]
- 21.Grewe M, Walther S, Gyufko K, Czech W, Schopf E, Krutmann J. Analysis of the cytokine pattern expressed in situ in inhalant allergen patch test reactions of atopic dermatitis patients. J Invest Dermatol. 1995;105:407–10. doi: 10.1111/1523-1747.ep12321078. [DOI] [PubMed] [Google Scholar]
- 22.Thepen T, Langeveld-Wildschut EG, Bihari IC, et al. Biphasic response against aeroallergen in atopic dermatitis showing a switch from an initial TH2 response to a TH1 response in situ: an immunocytochemical study. J Allergy Clin Immunol. 1996;97:828–37. doi: 10.1016/s0091-6749(96)80161-8. [DOI] [PubMed] [Google Scholar]
- 23.Grewe M, Gyufko K, Schopf E, Krutmann J. Lesional expression of interferon-gamma in atopic eczema. Lancet. 1994;343:25–6. doi: 10.1016/s0140-6736(94)90879-6. [DOI] [PubMed] [Google Scholar]
- 24.Ring J, Darsow U, Behrendt H. Role of aeroallergens in atopic eczema: proof of concept with the atopy patch test. J Am Acad Dermatol. 2001;45:S49–52. doi: 10.1067/mjd.2001.117015.24. [DOI] [PubMed] [Google Scholar]
- 25.Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. J Allergy Clin Immunol. 2009;124:1235–44. e58. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 26.Suarez-Farinas M, Li K, Fuentes-Duculan J, et al. Expanding the psoriasis disease profile: Interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012 doi: 10.1038/jid.2012.184. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chamian F, Lowes MA, Lin SL, et al. Alefacept reduces infiltrating T cells, activated dendritic cells, and inflammatory genes in psoriasis vulgaris. Proc Natl Acad Sci U S A. 2005;102:2075–80. doi: 10.1073/pnas.0409569102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suarez-Farinas M, Pellegrino M, Wittkowski KM, Magnasco MO. Harshlight: a “corrective make-up” program for microarray chips. BMC Bioinformatics. 2005;6:294. doi: 10.1186/1471-2105-6-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–53. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Gutowska-Owsiak D, Schaupp AL, Salimi M, Taylor S, Ogg GS. Interleukin-22 downregulates filaggrin expression and affects expression of profilaggrin processing enzymes. Br J Dermatol. 2011;165:492–8. doi: 10.1111/j.1365-2133.2011.10400.x. [DOI] [PubMed] [Google Scholar]
- 33.Liang SC, Tan XY, Luxenberg DP, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–9. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cevikbas F, Steinhoff M. IL-33: A Novel Danger Signal System in Atopic Dermatitis. J Invest Dermatol. 2012;132:1326–9. doi: 10.1038/jid.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Martinez O, Overbergh L, Mathieu C, Prabhakar BS, Chan LS. Early up-regulation of Th2 cytokines and late surge of Th1 cytokines in an atopic dermatitis model. Clin Exp Immunol. 2004;138:375–87. doi: 10.1111/j.1365-2249.2004.02649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haider AS, Lowes MA, Suarez-Farinas M, et al. Identification of cellular pathways of “type 1,” Th17 T cells, and TNF- and inducible nitric oxide synthase-producing dendritic cells in autoimmune inflammation through pharmacogenomic study of cyclosporine A in psoriasis. J Immunol. 2008;180:1913–20. doi: 10.4049/jimmunol.180.3.1913. [DOI] [PubMed] [Google Scholar]
- 37.Hirahara K, Ghoreschi K, Laurence A, Yang XP, Kanno Y, O’Shea JJ. Signal transduction pathways and transcriptional regulation in Th17 cell differentiation. Cytokine Growth Factor Rev. 2010;21:425–34. doi: 10.1016/j.cytogfr.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirota T, Takahashi A, Kubo M, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–6. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma B, Liu W, Homer RJ, et al. Role of CCR5 in the pathogenesis of IL-13- induced inflammation and remodeling. J Immunol. 2006;176:4968–78. doi: 10.4049/jimmunol.176.8.4968. [DOI] [PubMed] [Google Scholar]
- 40.Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology. 2010;129:311–21. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang N, Pan HF, Ye DQ. Th22 in inflammatory and autoimmune disease: prospects for therapeutic intervention. Mol Cell Biochem. 2011;353:41–6. doi: 10.1007/s11010-011-0772-y. [DOI] [PubMed] [Google Scholar]
- 42.Eyerich S, Eyerich K, Pennino D, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–85. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, et al. Identification of TNF-related apoptosis-inducing ligand and other molecules that distinguish inflammatory from resident dendritic cells in patients with psoriasis. J Allergy Clin Immunol. 2010;125:1261–8. e9. doi: 10.1016/j.jaci.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol. 2008;181:7420–7. doi: 10.4049/jimmunol.181.10.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, et al. Integrative responses to IL-17 and TNF-alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131:677–87. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 46.Suarez-Farinas M, Lowes MA, Zaba LC, Krueger JG. Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA) PLoS One. 2010;5:e10247. doi: 10.1371/journal.pone.0010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cornelissen C, Marquardt Y, Czaja K, et al. IL-31 regulates differentiation and filaggrin expression in human organotypic skin models. J Allergy Clin Immunol. 2012;129:426–33. 33e1–8. doi: 10.1016/j.jaci.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 48.Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invst Dermatol. 2008;128:2625–30. doi: 10.1038/jid.2008.111. [DOI] [PubMed] [Google Scholar]
- 49.Harder J, Dressel S, Wittersheim M, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Invest Dermatol. 2010;130:1355–64. doi: 10.1038/jid.2009.432. [DOI] [PubMed] [Google Scholar]
- 50.Benoit S, Toksoy A, Ahlmann M, et al. Elevated serum levels of calcium-binding S100 proteins A8 and A9 reflect disease activity and abnormal differentiation of keratinocytes in psoriasis. Br J Dermatol. 2006;155:62–6. doi: 10.1111/j.1365-2133.2006.07198.x. [DOI] [PubMed] [Google Scholar]
- 51.Glaser R, Meyer-Hoffert U, Harder J, et al. The antimicrobial protein psoriasin (S100A7) is upregulated in atopic dermatitis and after experimental skin barrier disruption. J Invest Dermatol. 2009;129:641–9. doi: 10.1038/jid.2008.268. [DOI] [PubMed] [Google Scholar]
- 52.Martinsson H, Yhr M, Enerback C. Expression patterns of S100A7 (psoriasin) and S100A9 (calgranulin-B) in keratinocyte differentiation. Exp Dermatol. 2005;14:161–8. doi: 10.1111/j.0906-6705.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 53.Roth J, Vogl T, Sorg C, Sunderkotter C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 2003;24:155–8. doi: 10.1016/s1471-4906(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 54.Goyette J, Geczy CL. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids. 2011;41:821–42. doi: 10.1007/s00726-010-0528-0. [DOI] [PubMed] [Google Scholar]
- 55.Eckert RL, Broome AM, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. J Invest Dermatol. 2004;123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x. [DOI] [PubMed] [Google Scholar]
- 56.Sampson B, Fagerhol MK, Sunderkotter C, et al. Hyperzincaemia and hypercalprotectinaemia: a new disorder of zinc metabolism. Lancet. 2002;360:1742–5. doi: 10.1016/S0140-6736(02)11683-7. [DOI] [PubMed] [Google Scholar]
- 57.Reginald K, Westritschnig K, Werfel T, et al. Immunoglobulin E antibody reactivity to bacterial antigens in atopic dermatitis patients. Clin Exp Allergy. 2011;41:357–69. doi: 10.1111/j.1365-2222.2010.03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takaoka A, Arai I, Sugimoto M, Yamaguchi A, Tanaka M, Nakaike S. Expression of IL-31 gene transcripts in NC/Nga mice with atopic dermatitis. Eur J Pharmacol. 2005;516:180–1. doi: 10.1016/j.ejphar.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 59.Kim S, Kim HJ, Yang HS, Kim E, Huh IS, Yang JM. IL-31 Serum Protein and Tissue mRNA Levels in Patients with Atopic Dermatitis. Ann Dermatol. 2011;23:468–73. doi: 10.5021/ad.2011.23.4.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abtin A, Eckhart L, Glaser R, Gmeiner R, Mildner M, Tschachler E. The antimicrobial heterodimer S100A8/S100A9 (calprotectin) is upregulated by bacterial flagellin in human epidermal keratinocytes. J Invest Dermatol. 2010;130:2423–30. doi: 10.1038/jid.2010.158. [DOI] [PubMed] [Google Scholar]
- 61.Bai B, Yamamoto K, Sato H, Sugiura H, Tanaka T. Complex regulation of S100A8 by IL-17, dexamethasone, IL-4 and IL-13 in HaCat cells (human keratinocyte cell line) J Dermatol Sci. 2007;47:259–62. doi: 10.1016/j.jdermsci.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Guilloteau K, Paris I, Pedretti N, et al. Skin Inflammation Induced by the Synergistic Action of IL-17A, IL-22, Oncostatin M, IL-1{alpha}, and TNF-{alpha} Recapitulates Some Features of Psoriasis. J Immunol. 2010 doi: 10.4049/jimmunol.0902464. [DOI] [PubMed] [Google Scholar]
- 63.Hegyi Z, Zwicker S, Bureik D, et al. Vitamin D analog calcipotriol suppresses the Th17 cytokine-induced proinflammatory S100 “alarmins” psoriasin (S100A7) and koebnerisin (S100A15) in psoriasis. J Invest Dermatol. 2012;132:1416–24. doi: 10.1038/jid.2011.486. [DOI] [PubMed] [Google Scholar]
- 64.Johnston A, Gudjonsson JE, Aphale A, Guzman AM, Stoll SW, Elder JT. EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent manner. J Invest Dermatol. 2011;131:329–37. doi: 10.1038/jid.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nukui T, Ehama R, Sakaguchi M, et al. S100A8/A9, a key mediator for positive feedback growth stimulation of normal human keratinocytes. J Cell Biochem. 2008;104:453–64. doi: 10.1002/jcb.21639. [DOI] [PubMed] [Google Scholar]
- 66.Kryczek I, Bruce AT, Gudjonsson JE, et al. Induction of IL-17+ T cell trafficking and development by IFN-gamma: mechanism and pathological relevance in psoriasis. J Immunol. 2008;181:4733–41. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonder CS, Davies KV, Hosszu EK, Finlay-Jones JJ, Hart PH. IFN-gamma downregulates interleukin-4 functional activity on monocytes by multiple mechanisms. J Interferon Cytokine Res. 2002;22:287–93. doi: 10.1089/107999002753675703. [DOI] [PubMed] [Google Scholar]
- 68.Kalinke U, Prinz M. Endogenous, or therapeutically induced, type I interferon responses differentially modulate Th1/Th17-mediated autoimmunity in the CNS. doi: 10.1038/icb.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.