Abstract
Combinatorial targeted therapies are more effective in treating cancer by blocking by-pass mechanisms or inducing synthetic lethality. However, their clinical application is hampered by resistance and toxicity. To meet this important challenge, we developed and tested a novel concept of biomarker-guided sequential applications of various targeted therapies using ErbB2-overexpressing/PTEN-low, highly aggressive breast cancer as our model. Strikingly, sustained activation of ErbB2 and downstream pathways drives trastuzumab resistance in both PTEN-low/trastuzumab-resistant breast cancers from patients and mammary tumors with intratumoral heterogeneity from genetically-engineered mice. Although lapatinib initially inhibited trastuzumab-resistant mouse tumors, tumors by-passed the inhibition by activating the PI3K/mTOR signaling network as shown by the quantitative protein arrays. Interestingly, activation of the mTOR pathway was also observed in neoadjuvant lapatinib-treated patients manifesting lapatinib resistance. Trastuzumab + lapatinib resistance was effectively overcome by sequential application of a PI3K/mTOR dual kinase inhibitor (BEZ235) with no significant toxicity. However, our p-RTK array analysis demonstrated that BEZ235 treatment led to increased ErbB2 expression and phosphorylation in genetically-engineered mouse tumors and in 3-D, but not 2-D, culture, leading to BEZ235 resistance. Mechanistically, we identified ErbB2 protein stabilization and activation as a novel mechanism of BEZ235 resistance, which was reversed by subsequent treatment with lapatinib + BEZ235 combination. Remarkably, this sequential application of targeted therapies guided by biomarker changes in the tumors rapidly evolving resistance doubled the life-span of mice bearing exceedingly aggressive tumors. This fundamentally novel approach of using targeted therapies in a sequential order can effectively target and reprogram the signaling networks in cancers evolving resistance during treatment.
Keywords: sequential therapy, tumor evolution, targeted therapy, trastuzumab resistance, BEZ235, ErbB2 stabilization
Introduction
In the era of personalized medicine, targeted cancer therapies have improved the clinical outcome in many cancer types including breast cancer. Almost 20% of breast cancers overexpress the HER2/ErbB2 receptor tyrosine kinase (RTK)1. Trastuzumab (also named Herceptin), a humanized monoclonal antibody against the extracellular domain of HER2/ErbB22, has been revolutionary in treating patients with ErbB2-overexpressing tumors in neoadjuvant3, adjuvant4 and metastatic settings5. After the success of trastuzumab, other drugs have been developed to target ErbB2 (e.g., pertuzumab, T-DM1) or to other members of the ErbB family of receptors (e.g., EGFR). Similarly, lapatinib, a dual tyrosine kinase inhibitor which targets the ATP-binding pockets of both ErbB2 and EGFR, has been approved by the FDA for treating ErbB2-overexpressing, trastuzumab-refractory breast cancers6. However, a major obstacle in the clinic for the success of trastuzumab and these other targeted agents is primary (or de novo) and secondary (or acquired) resistance. Therefore, it is essential to identify biomarkers and mechanisms of resistance to select subpopulations of patients who are likely responders and to develop new regimens or agents that target resistance pathways to reverse resistance.
Various mechanisms of resistance have been proposed for both de novo and acquired trastuzumab resistance, e.g., activation of other RTKs (e.g., ErbB3 or IGF-1R), steric hindrance of ErbB2-trastuzumab interaction by glycoproteins (e.g., MUC4), inhibition of trastuzumab binding to ErbB2 by cleavage of ErbB2 extracellular domain (p95ErbB2) and activation of PI3K pathway or SRC7,8. We previously demonstrated that loss of the phosphatase and tensin homolog (PTEN), a negative regulator of the PI3K/AKT pathway, confers de novo trastuzumab resistance in ErbB2-overexpressing breast cancer9. Later, PTEN was identified to be the only modulator of trastuzumab response in an RNA interference-based functional screen for genes involved in trastuzumab resistance in breast cancer10. Importantly, the tumor suppressor PTEN could be lost in up to 50% of breast cancers due to mutations, loss of heterozygosity or epigenetic modifications11. PTEN deficiency accelerates the development of multifocal and highly metastatic mammary tumors with ErbB2/neu overexpression in mice12 and the resulting tumors are highly aggressive and refractory to treatment with trastuzumab13. Therefore, novel therapies are urgently needed to overcome trastuzumab resistance in this patient subpopulation having highly aggressive disease.
To this end, combinatorial targeted therapies hold great promise through blocking the potential by-pass mechanisms or inducing synthetic lethality. We have recently shown that targeting SRC, a key node in the trastuzumab resistance network, by a SRC inhibitor (saracatinib) in combination with trastuzumab overcomes trastuzumab resistance from multiple resistance mechanisms, including PTEN loss14. Additionally, we demonstrated that in two different genetically-engineered mouse models of trastuzumab resistance (mammary gland-specific ErbB2/neu transgenic and PTEN knockout), an Akt inhibitor (triciribine) synergizes with trastuzumab in tumor inhibition13. Targeting the PI3K pathway in combination with trastuzumab has also been reported to overcome trastuzumab resistance in other models15. Although initially successful, these combinatorial regimens rapidly develop resistance and are often too toxic for clinical usage16. Therefore, innovative strategies of applying targeted agents with high efficacy but low toxicity are crucial in tackling the challenge.
Here, we developed and experimentally tested a novel strategy of employing targeted agents for potentially highly effective clinical cancer therapy: using arising biomarkers in an ErbB2-overexpressing cancer evolving therapy resistance to guide subsequent applications of various combinatorial targeted therapies in a sequential manner to inhibit the constantly changing resistant signals in the cancer during targeted therapies. We report that this strategy of sequential application of various combinatorial targeted therapies reprogrammed the signaling networks in cancer evolving therapy resistance and yielded cancer vulnerabilities compared to using all targeted agents at once. Remarkably, this fundamentally novel approach doubled the survival time of mice bearing highly aggressive, therapy-refractory ErbB2-overexpressing tumors. It is anticipated that this strategy can be rapidly translated into the clinic to benefit cancer patients who have highly aggressive and therapy-resistant tumors.
Results
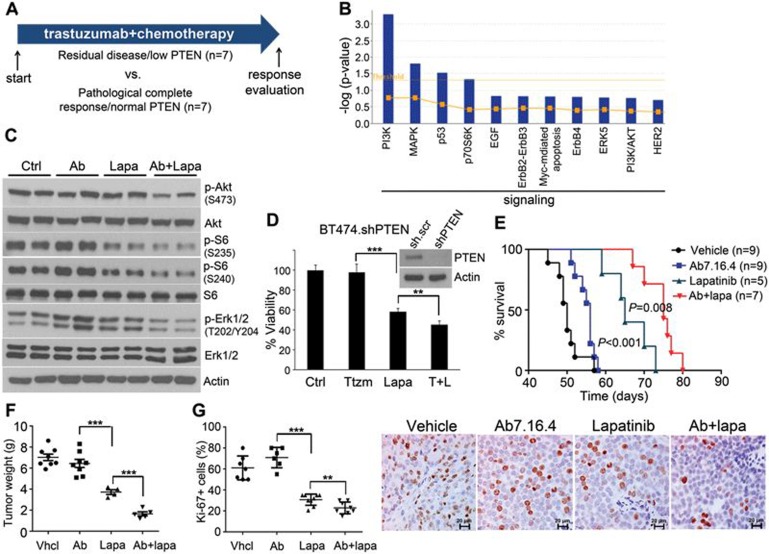
Biomarker analyses denote activation of multiple ErbB2 downstream pathways in PTEN-low/trastuzumab-resistant tumors that are inhibited by lapatinib
To explore novel strategies of employing targeted agents for highly effective cancer therapy that reverse resistance and reduce toxicity, we focused on HER2/ErbB2-overexpressing breast cancers. We previously found that PTEN loss confers resistance to trastuzumab in patients with ErbB2-overexpressing breast cancers9. Therefore, we initially compared the gene expression profiles and analyzed signaling events in breast cancers from PTEN-low/trastuzumab-resistant versus PTEN-normal/trastuzumab-sensitive patients who had neoadjuvant trastuzumab plus chemotherapy treatment (Figure 1A)17. First, we identified the differentially expressed genes from cDNA microarray of the fine-needle aspiration specimens between these two groups (Supplementary information, Table S1)17, and then determined the pathways in which these genes are enriched using Ingenuity Pathway Analysis (IPA) (Figure 1B and Supplementary information, Table S2). As expected, the PI3K pathway signature was significantly increased in PTEN-low/trastuzumab-resistant tumors. Additionally, we identified multiple pathways (e.g., MAPK, and p70S6K) significantly activated in PTEN-low/trastuzumab-resistant tumors (P < 0.05). Interestingly, we also observed a clear trend for increased expressions of EGF and ErbB2-ErbB3 signaling in this small cohort of PTEN-low/trastuzumab-resistant patients (Figure 1B).
Figure 1.
Lapatinib alone or combined with anti-ErbB2 antibody Ab7.16.4 increases survival of PTEN−/−/NIC trastuzumab-resistant mice. (A) Treatment scheme of 14 ErbB2-positive breast cancer patients. Patients were classified into two groups: trastuzumab sensitive with high PTEN level (PCR/high PTEN) (n = 7) and trastuzumab resistant with normal PTEN level (RD/low PTEN) (n = 7). (B) Identification of significantly different canonical pathways between trastuzumab-resistant and -sensitive patients using IPA. Threshold is at P < 0.05 (dotted yellow line); y axis is −log of the P-value. (C) Western blot of PTEN−/−/NIC tumor lysates from mice treated with different drugs for 3 days. (D) WST-1 cell proliferation assay comparing proliferation of BT474.shPTEN cells after drug treatment for 72 h (Ttzm: trastuzumab and T + L: trastuzumab plus lapatinib). Inset shows western blot validation of PTEN knockdown. **P < 0.01, ***P < 0.001 by two-tailed t-test. (E) Survival analysis of the PTEN−/−/NIC mice treated with different drugs or combinations. P < 0.001 (by log-rank test) is between Ab7.16.4 and lapatinib; P = 0.008 is between lapatinib and Ab7.16.4 + lapatinib. (F) Left, tumor weight (mean ± SEM) of mice treated with vehicle, 7.16.4 mAb (Ab), lapatinib, or combination for 3 weeks. ***P < 0.001 by two-tailed t-test. (G) Ki-67 staining of tumor tissue obtained from mice treated with different drugs or their combination. Scale bar indicates 20 μm. **P < 0.01, ***P < 0.001 by two-tailed t-test.
We next examined whether these signatures detected in patients can be recapitulated in PTEN loss-mediated trastuzumab-resistant mouse models to be used for preclinical testing of new strategies of targeted therapies. We crossed HER2/neu-overexpressing (MMTV-neu-IRES-cre, or NIC) mice with PTENfl/fl mice (PTEN−/−/NIC), which developed highly aggressive neu-overexpressing/PTEN-deficient and trastuzumab-resistant mammary tumors with median survival of 45 days13. We applied a quantitative proteomics approach of reverse phase protein array (RPPA, containing > 150 antibodies)18 on lysates of mammary tumors from the PTEN−/−/NIC mice and compared it to the PTEN+/+/NIC (PTEN wild type) tumors. As with patient tumors, tumors of the PTEN−/−/NIC mice had activation of not only the PI3K pathway, but also ERK1/2, p70S6K, S6K, 4EBP1, Stat3 and SRC, as shown by RPPA and validated by western blot (Supplementary information, Figure S1A and S1B). Importantly, PTEN loss in mammary tumors of PTEN−/−/NIC mice also resulted in hyper-phosphorylation of the ErbB2/neu RTK (Supplementary information, Figure S1C), which resembles the trend in patients (Figure 1B). Based on (a) this enhanced ErbB2 and multiple ErbB2 downstream signaling pathways in tumors of PTEN−/−/NIC mice and PTEN-low/trastuzumab-resistant patients, and (b) the clinical use of lapatinib to treat trastuzumab-refractory breast cancers, we tested whether lapatinib can effectively inhibit activation of multiple ErbB2 downstream pathways and impede tumorigenesis of PTEN−/−/NIC mice. Indeed, lapatinib inhibited the phosphorylation of ErbB2 targets Akt, S6 and Erk1/2 compared to Ab7.16.4, the rodent equivalent of trastuzumab (Figure 1C)19. Significantly, combination of lapatinib with Ab7.16.4 inhibited the Akt, S6 and Erk1/2 phosphorylation more effectively than lapatinib alone. We also tested whether lapatinib inhibits the proliferation of an ErbB2-overexpressing BT474 human breast cancer cell line with stable PTEN knockdown (BT474.shPTEN), which is trastuzumab resistant14. As in the mouse model, lapatinib inhibited the BT474.shPTEN cell growth, and the combination of trastuzumab with lapatinib is more effective than lapatinib alone (Figure 1D). Importantly, lapatinib significantly increased survival of the Ab7.16.4-resistant PTEN−/−/NIC mice (P < 0.001), and lapatinib plus Ab7.16.4 further increased the survival of these mice than lapatinib alone (P = 0.008) (Figure 1E). Lapatinib alone or combination treatment reduced tumor weight after three weeks of treatment compared to vehicle or Ab7.16.4 (Figure 1F). Furthermore, lapatinib alone or combination treatment dramatically inhibited cell proliferation (Figure 1G), without affecting apoptosis of tumors in PTEN−/−/NIC mice (Supplementary information, Figure S2). Taken together, our biomarker analyses indicated that 1) PTEN loss leads to activation of multiple ErbB2 downstream pathways in PTEN-low/trastuzumab-resistant patient tumors and in PTEN−/−/NIC mouse mammary tumors with conferring trastuzumab resistance; 2) the use of lapatinib, alone or in combination with trastuzumab, targets these ErbB signaling events and overcomes PTEN loss-mediated trastuzumab resistance.
Activation of PI3K/mTOR pathways in tumors resistant to Ab7.16.4 + lapatinib combination
Although lapatinib was a rational choice with efficacy in treating PTEN-low/trastuzumab-resistant tumors based on the biomarker analysis (Figure 1), lapatinib in combination with trastuzumab provides only 4.5 months of survival advantage20, which is also reflected by the tumor relapse in PTEN−/−/NIC mice (Figure 1E). Therefore, we next aimed to identify biomarkers of resistance to lapatinib + trastuzumab treatment and test therapies to effectively overcome the resistance. To this end, we employed a tumor transplantation model by transplanting mammary tumors from the PTEN−/−/NIC donor mice to mammary fat pads (MFPs) of recipient syngeneic mice (in FVB background) to facilitate timely testing of different therapies (Supplementary information, Figure S3A). The tumor transplants faithfully recapitulated both molecular alterations (activation of signaling proteins, Supplementary information, Figure S3B) and biological properties (aggressive growth rate and trastuzumab resistance, Supplementary information, Figure S3C) of the donor PTEN−/−/NIC mice. Similar to the tumors of the donor mice, most transplants initially responded to Ab7.16.4 + lapatinib therapy with reduced cell proliferation (Supplementary information, Figure S3D and S3E) but no significant difference in apoptosis (Supplementary information, Figure S3F), whereas some were non-responding (Supplementary information, Figure S3D-S3F). Thus, this transplantation model is appropriate for testing biomarkers and targeted therapies for overcoming Ab7.16.4 + lapatinib resistance.
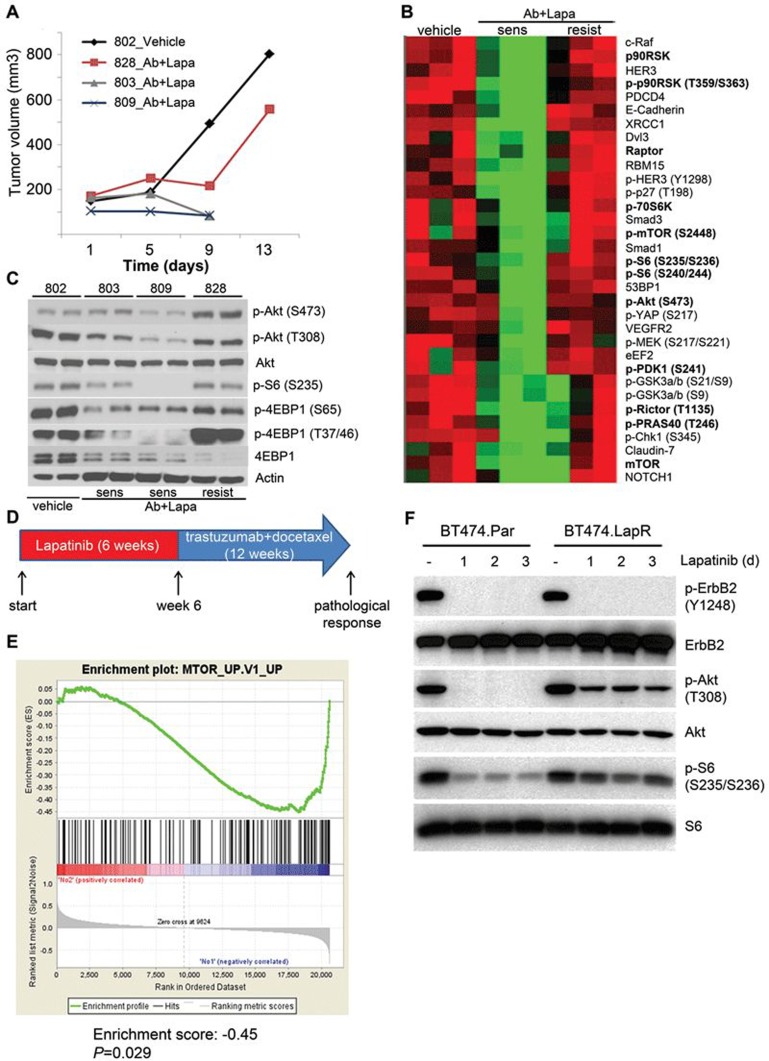
We treated mice bearing tumor transplants with Ab7.16.4 + lapatinib combinatorial therapy and followed their tumor growth. All transplants initially responded to therapy with reduced cell proliferation, but some acquired resistance later and tumors resumed growth (Figure 2A and Supplementary information, Figure S3D). Accordingly, we classified transplants into sensitive and resistant groups based on tumor volume change. We performed RPPA to compare tumor biomarkers in these two groups and the control-treated group. Clearly, Ab7.16.4 + lapatinib-sensitive tumors had reduced levels of p-AKT, p-PRAS40, p-mTOR, p-S6 and p-4EBP compared to the control group (Figure 2B and Supplementary information, Table S3). However, in Ab7.16.4 + lapatinib-resistant tumors, phosphorylation levels of all these proteins were similar to, or higher than, those in the control group, indicating that PI3K and downstream mTOR pathways remained active in resistant tumors. The RPPA data were further validated by western blots in tumor lysates from additional mice (Figure 2A-2C). These analyses indicated that the activation of the PI3K/mTOR pathway is potentially a critical molecular event (a biomarker) of Ab7.16.4 + lapatinib resistance in PTEN−/−/NIC tumors.
Figure 2.
Resistance to Ab7.16.4 + lapatinib combination develops through activation of PI3K/mTOR pathways in PTEN−/−/NIC mice and mTOR gene signature is enriched in lapatinib-resistant patients. (A) Tumor volume measurements at different time points for transplant mice treated with Vehicle or Ab + lapatinib. Mice were sacrificed on day 9 (803 and 809) or day 13 (802 and 828). 803 and 809 were sensitive whereas 828 was resistant to Ab + lapatinib. (B) RPPA analysis showing expression or phosphorylation of proteins downregulated in Ab + lapatinib-sensitive tumors and upregulated or not affected in Ab + lapatinib-resistant tumors compared to control. PI3K/mTOR pathway components are bolded. (C) Western blot analysis of Ab + lapatinib-sensitive and -resistant tumors shown in A. (D) Treatment scheme of ErbB2-positive breast cancer patients treated with neoadjuvant lapatinib followed by trastuzumab + docetaxel. Gene expression profiling was done with RNA isolated from tumors before and after lapatinib treatment (“start” and “week 6”). (E) GSEA of tumors from lapatinib-resistant patients. Genes upregulated upon mTOR inhibition by everolimus were significantly downregulated. (F) Western blot analysis of parental (BT474.Par) and lapatinib-resistant (BT474.LapR) cells treated with DMSO or lapatinib (1 μM) over 3 days.
The mTOR pathway is upregulated in lapatinib-resistant patient tumors and activated in lapatinib-resistant human breast cancer cells
To test the clinical relevance of our findings from transplant models, we obtained patient tumor mRNAs isolated before and after 6 weeks of neoadjuvant single agent lapatinib treatment and examined their gene expression profiles21 (Figure 2D). The pathological response to treatment was evaluated after subsequent treatment with trastuzumab and docetaxel for 12 weeks and grouped into pathological complete response or near-complete response (pCR/pNCR) versus non-responders (NRs). After identifying differentially expressed genes between these two groups, we examined whether PI3K or mTOR pathway gene signatures were increased in the NR group. Gene set enrichment analysis (GSEA)22 revealed that tumors of NR patients had a significant downregulation of genes which were upregulated upon mTOR inhibition by everolimus (RAD001)23 (Figure 2E). This suggests that the mTOR gene signature is upregulated in resistant tumors of patients and may play an important role in lapatinib + trastuzumab resistance, similar to that observed in the transplant model (Figure 2B and 2C). To test whether the mTOR pathway is activated in lapatinib + trastuzumab-resistant human breast cancer cells, we selected lapatinib-resistant BT474 human breast cancer cell line (BT474.LapR) by continuous exposure of BT474 cells to lapatinib over 9 months. These BT474.LapR cells are also highly resistant to trastuzumab (Supplementary information, Figure S3G). We treated the cells with lapatinib for 1, 2, and 3 days and analyzed PI3K/mTOR pathway activation by western blot. Indeed, the lapatinib + trastuzumab-resistant BT474.LapR cell line showed a higher and sustained phosphorylation of Akt and S6 compared to the parental line (BT474.Par) (Figure 2F). Taken together, these data indicate that PI3K and/or its downstream mTOR pathway is activated in lapatinib + trastuzumab-resistant human breast cancers and cell lines, further supporting our findings from Ab7.16.4 + lapatinib-resistant transplant models and demonstrating their human pathological relevance.
Sequential application of Ab7.16.4 + BEZ235 following Ab7.16.4 + lapatinib is effective in reversing Ab7.16.4 + lapatinib resistance
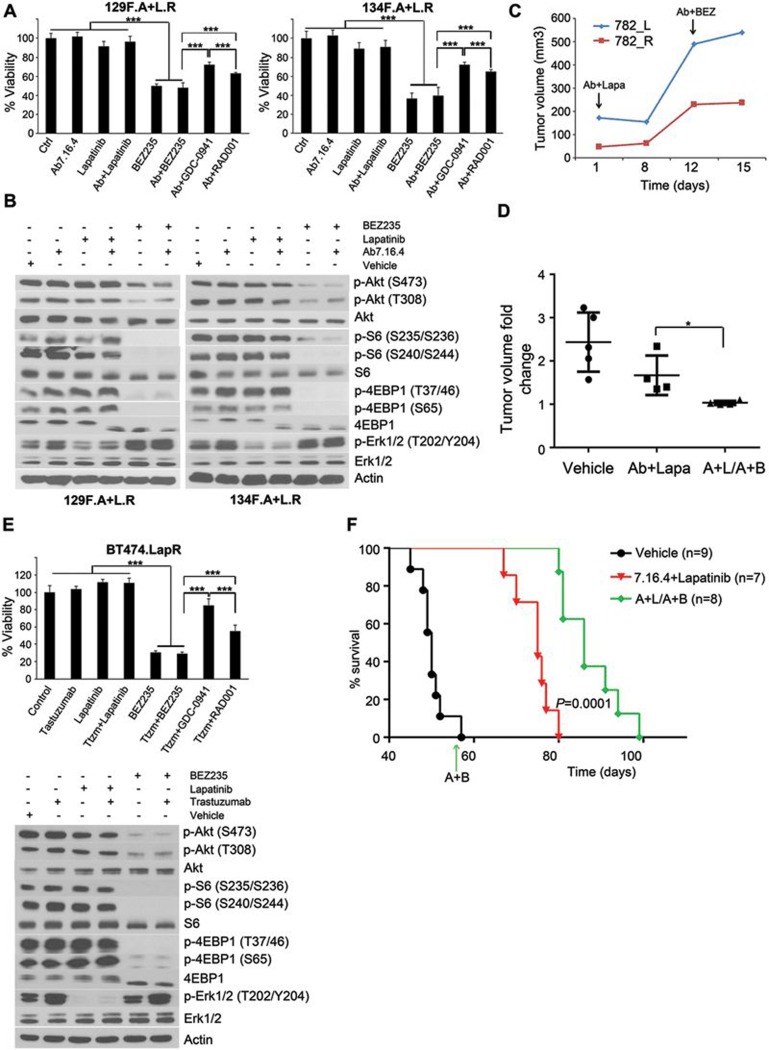
Guided by the above finding that PI3K/mTOR pathway is activated in lapatinib + trastuzumab-resistant breast cancers and Ab7.16.4 + lapatinib-resistant transplant tumors, we tested whether PI3K/mTOR inhibitors are effective in overcoming resistance. We first isolated mammary tumor cells from two PTEN−/−/NIC mice (129F and 134F) who acquired resistance after 6 weeks of Ab7.16.4 + lapatinib combination treatment (129F.A + L.R and 134F.A + L.R). These primary cultures were resistant to Ab7.16.4 and/or lapatinib similar to the original 129F.A + L.R and 134F.A + L.R tumors (Figure 3A). These primary culture cells were then treated with either GDC-0941 (PI3K P110α/β-inhibitor) or RAD001 (mTORC1 inhibitor) or BEZ235 (PI3K and mTOR dual inhibitor) in combination with Ab7.16.4. Clearly, BEZ235, alone or in combination with Ab7.16.4, was more effective than GDC-0941 + Ab7.16.4 or RAD001 + Ab7.16.4 in reducing cell viability, likely through simultaneous inhibition of p-AKT, p-S6 and p-4EBP (Figure 3A and 3B). Then, we tested whether BEZ235 + Ab7.16.4 may effectively inhibit Ab7.16.4 + lapatinib-resistant transplants in mice. Indeed, sequential application of Ab7.16.4 + BEZ235 (A + B) after acquired resistance to Ab7.16.4 + lapatinib (A + L) led to tumor stasis (Figure 3C and 3D) by reducing tumor cell proliferation (Figure 4C) but without significant changes in apoptosis (Supplementary information, Figure S4).
Figure 3.
Sequential application of Ab7.16.4 + BEZ235 combination overcomes Ab7.16.4 + lapatinib resistance. (A) WST-1 proliferation assay after 72 h of treatment with different drugs targeting PI3K and/or mTOR. Primary cells (129F.A + L.R and 134F.A + L.R) from PTEN−/−/NIC mice treated with Ab + lapatinib over 4 weeks were used. ***P < 0.001, by one-way ANOVA and ***P < 0.001, by two-tailed t-test. (B) Western blot analysis of primary cells treated with the indicated drugs for 3 h. (C) Tumor volume measurements at different time points for transplant mice (782_L and 782_R) treated with Ab + lapatinib first and then with Ab + BEZ235 on day 12 upon resistance development. (D) Tumor volume fold change of transplant mice treated with vehicle (ctrl) or Ab7.16.4 + lapatinib or A + L/A + B. *P < 0.05, by two-tailed t-test. (E) Upper panel, WST-1 proliferation assay after 72 h treatment with the indicated drugs targeting PI3K and/or mTOR pathways in BT474.LapR cells. ***P < 0.001, by two-tailed t-test. Lower panel, western blot analysis of signaling pathways in BT474.LapR cells upon treatment with different drugs or their combination for 3 h. (F) Survival analysis of the PTEN−/−/NIC mice treated with vehicle (ctrl) or Ab + lapatinib or A + L/A + B. The switch time for treatment was shown with a green arrow on the x-axis for the sequential therapy-treated mice. P-value was determined by log-rank test. P = 0.0001 is between Ab7.16.4 + lapatinib and A + L/A + B sequential therapy.
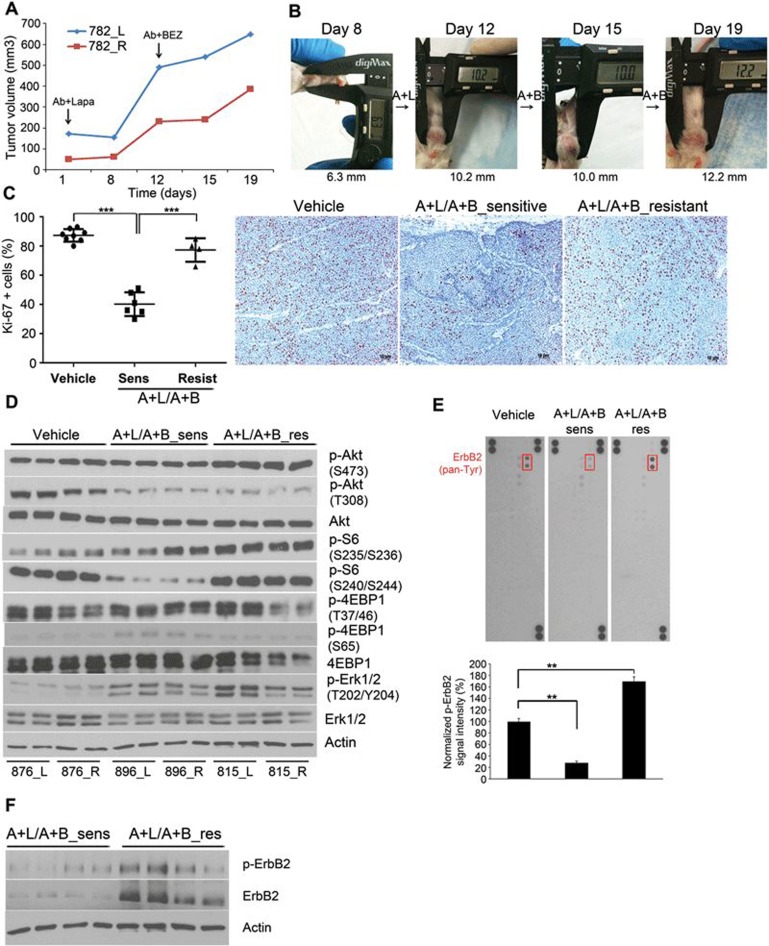
Figure 4.
Resistance to Ab7.16.4 + BEZ235 develops through activation of p-ErbB2 and downstream signaling. (A) Tumor volume measurements at different time points for transplant mice (782_L and 782_R) treated with Ab + lapatinib first and then with Ab + BEZ235 on day 12. Resistance to Ab + BEZ developed on day 19. (B) Representative pictures of tumors from mouse 782_L taken at shown time points after treatments indicated in A. (C) Ki-67 staining of the sensitive and resistant tumors treated with A + L/A + B sequential therapy. ***P < 0.001, by two-tailed t-test. (D) Western blot analysis of A + L/A + B-sensitive and -resistant tumors shown in C. (E) Top, phospho-RTK array comparing the phosphorylation of 39 RTKs in tumors from D. Bottom, quantification of p-ErbB2 signal intensity using Image J software. **P < 0.01, by two-tailed t-test. (F) Western blot analysis of A + L/A + B-sensitive and -resistant tumors shown in C. Actin was used as a loading control.
To test whether trastuzumab + BEZ235 also inhibits proliferation of the lapatinib + trastuzumab-resistant BT474.LapR human breast cancer cells, in which Akt and p-S6 are activated (Figure 2F), we treated the cells with lapatinib, BEZ235, GDC-0941, or RAD001 alone or in combination with trastuzumab. Although trastuzumab, lapatinib or their combination had no significant effect, BEZ235 or trastuzumab + BEZ235 inhibited BT474.LapR cell growth significantly and more effectively than GDC-0941 + BEZ235 or RAD001 + BEZ235 (Figure 3E, top). BEZ235 or trastuzumab + BEZ235 led to a complete blockage of Akt, S6 and 4EBP1 phosphorylation in BT474.LapR cells (Figure 3E, bottom) similar to its effect on Ab7.16.4 + lapatinib-resistant mouse mammary tumor cells (129F.A + L.R and 134F.A + L.R). This promoted us to further test the sequential treatment strategy in PTEN−/−/NIC mice. We first treated mice with Ab7.16.4 + lapatinib, which initially inhibited tumor growth. After 4 weeks of treatment, tumor started to regrow, and we switched to Ab7.16.4 + BEZ235 combination treatment. This sequential regimen further increased survival (median survival of 86 days) significantly (P = 0.0001) compared to mice treated continuously with Ab7.16.4 + lapatinib (median survival of 75 days) or vehicle control (median survival of 50 days) (Figure 3F). Altogether, these data indicated that trastuzumab + BEZ235 (or Ab7.16.4 + BEZ235 for mice) combination is effective in blocking proliferation of trastuzumab + lapatinib-resistant cells and Ab7.16.4 + lapatinib-resistant PTEN−/−/NIC tumors, and sequential application of Ab + lapatinib followed by Ab + BEZ235 (A + L/A + B) increases the survival of PTEN−/−/NIC mice with highly aggressive tumors.
BEZ235 enhances ErbB2 protein stability and activation
Although mice who had sequential (A + L/A + B) therapy had a significantly prolonged life-span, their tumors later developed resistance. To conquer resistance to the sequential (A + L/A + B) therapy, we sought to dissect the underlying resistance mechanism. To this end, we treated transplant PTEN−/−/NIC tumors with Ab7.16.4 + lapatinib until tumor growth increased (resistant) by day 12, then switched to Ab7.16.4 + BEZ235 (Ab + BEZ) treatment until resistance developed seven days later on day 19 (Figure 4A and 4B). Notably, the resistant tumors had increased Ki-67+ cells compared to sensitive ones (Figure 4C). Then, we examined the PI3K/mTOR and ERK1/2 pathway activities in resistant versus sensitive tumors. Phosphorylation of Akt (S473), S6 (S235/236 and S240/244) and ERK1/2 (T202/Y204) was higher in resistant tumors compared to that in the sensitive ones (Figure 4D). Interestingly, p-Akt T308 and p-4EBP1 were not significantly different between resistant and sensitive tumors (Figure 4D). The activation of multiple key signaling pathways led us to hypothesize that an upstream RTK could be involved in their activation and contribute to the resistance. Thus, we performed a phospho-RTK array to simultaneously detect the phosphorylation of 39 different mouse RTKs in tumor protein lysates. Remarkably, the only RTK with increased phosphorylation in resistant versus sensitive tumors was ErbB2/neu (Figure 4E). To verify the upregulation of ErbB2 phosphorylation and to test ErbB2 expression, we expanded our analyses of the ErbB2/p-ErbB2 levels in multiple A + L/A + B sequential treatment-resistant versus -sensitive mammary tumors using western blot. Indeed, both the expression and phosphorylation of ErbB2/neu were increased in dual sequential treatment-resistant tumors compared to that in sensitive tumors (Figure 4F).
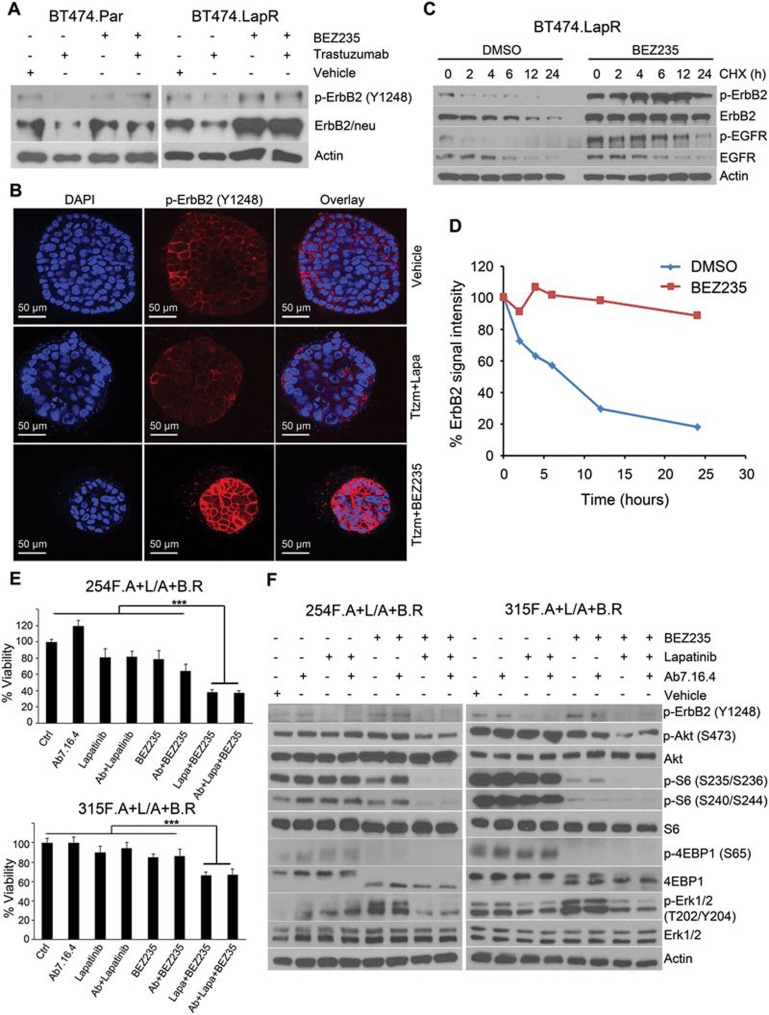
To elucidate how ErbB2 phosphorylation is increased upon Ab7.16.4 + BEZ235 treatment in tumor cells resistant to the A + L/A + B sequential targeted therapy, we treated BT474 parental (BT474.Par) and lapatinib + trastuzumab-resistant (BT474.LapR) cells with BEZ235 alone or in combination with trastuzumab over a week. Treatment had no significant effect on ErbB2 expression or phosphorylation levels in either parental or resistant lines under regular 2-D cell culture (Supplementary information, Figure S5). However, when the cells were cultured in 3-D with matrigel, ErbB2 expression and phosphorylation were dramatically increased upon BEZ235 or BEZ235 + trastuzumab treatment in BT474.LapR cells, but not in BT474.Par cells shown by western blot (Figure 5A). Immunofluorescence staining also revealed that lapatinib + trastuzumab reduced ErbB2 phosphorylation, but BEZ235 + trastuzumab led to increased ErbB2 phosphorylation despite effective inhibition of acinar proliferation (Figure 5B and Supplementary information, Figure S5B). Notably, ERBB2 mRNA level was not significantly increased in response to BEZ235 or BEZ235 + trastuzumab treatment in 3-D cultured BT474.LapR cells (Supplementary information, Figure S5C). To investigate how BEZ235 or BEZ235 + trastuzumab treatment increases ErbB2 protein level, we initially treated the BT474.LapR cells with DMSO or BEZ235 for a week in 3-D culture, then treated cells with cycloheximide (CHX) to block protein synthesis for different times up to 24 h and detected ErbB2 protein levels by western blot. ErbB2 protein decreased after 6 h of CHX treatment in DMSO-treated BT474.LapR cells, while it did not decrease after almost 24 h under CHX treatment in BEZ235-treated BT474.LapR cells (Figure 5C and 5D). ErbB2 phosphorylation had a similar pattern. Consistent with the transplant tumor RTK array data, the EGFR protein was similarly degraded in both DMSO- and BEZ235-treated BT474.LapR cells although EGFR phosphorylation was stable with BEZ235 treatment due to increased ErbB2 (Figure 5C). These data suggest that BEZ235 treatment specifically stabilizes ErbB2, leading to its increased phosphorylation and activation. Altogether, these data suggest that BEZ235 treatment-mediated ErbB2 protein stability and activation leads to the activation of multiple ErbB2 downstream pathways, thus conferring resistance to Ab + lapatinib/Ab + BEZ235 sequential treatment.
Figure 5.
BEZ235 stabilizes ErbB2 protein and lapatinib + BEZ235 combination overcomes resistance to Ab + lapatinib/Ab + BEZ235 in mouse primary cells. (A) Western blot analysis of cell lysates from BT474.Par and BT474.LapR cells grown in 3-D culture under different treatments for a week. (B) Immunofluorescence staining of BT474.LapR cells grown in 3-D culture under different treatment conditions for a week. Scale bar indicates 50 μm. (C) Protein stability assay in BT474.LapR cells treated with DMSO or BEZ235 (for a week) in combination with CHX over 24 h. (D) Quantification of ErbB2 signal intensity from C. (E) WST-1 proliferation assay after 72 h treatment with the indicated drugs or combinations in primary cells (254F.A + L/A + B.R and 315F.A + L/A + B.R) derived from PTEN−/−/NIC mice treated with Ab + lapatinib for 4 weeks and then with Ab + BEZ235 for 4 weeks, by which time resistance had developed. ***P < 0.001, by one-way ANOVA. (F) Western blot analysis of 254F.A + L/A + B.R and 315F.A + L/A + B.R primary cells after the indicated treatment for 3 h.
Marker-guided sequential treatment with A + L/A + B/A + B + L doubles the life-span of PTEN−/−/NIC mice bearing rapidly evolving aggressive mammary tumors
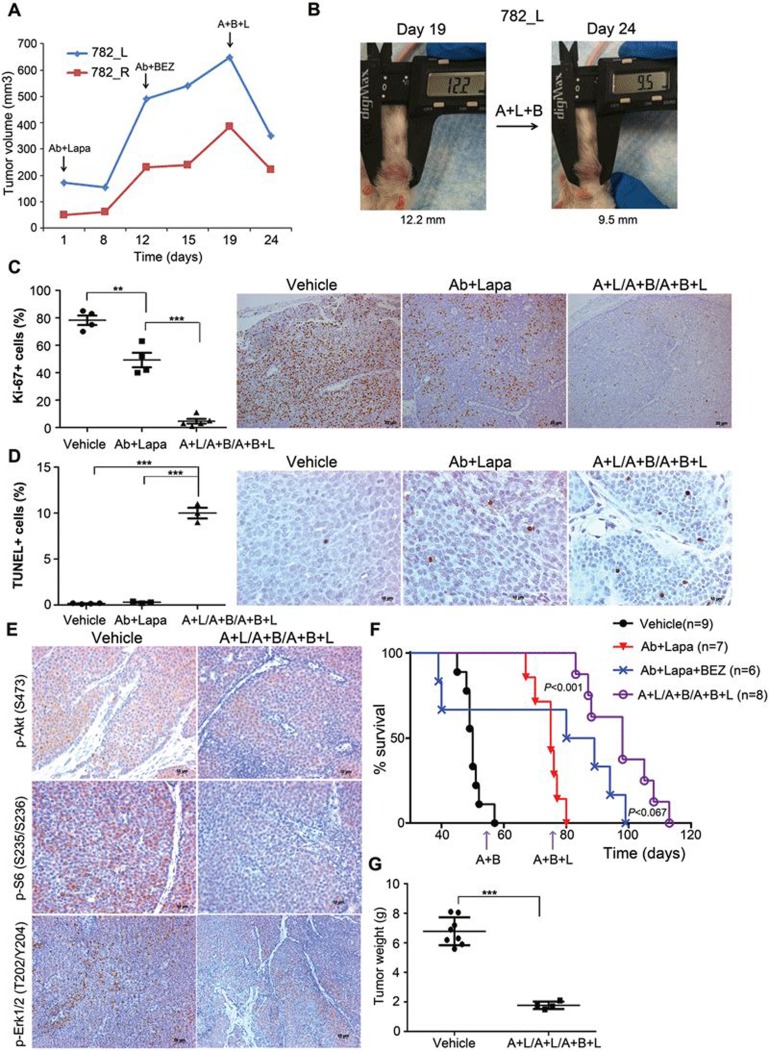
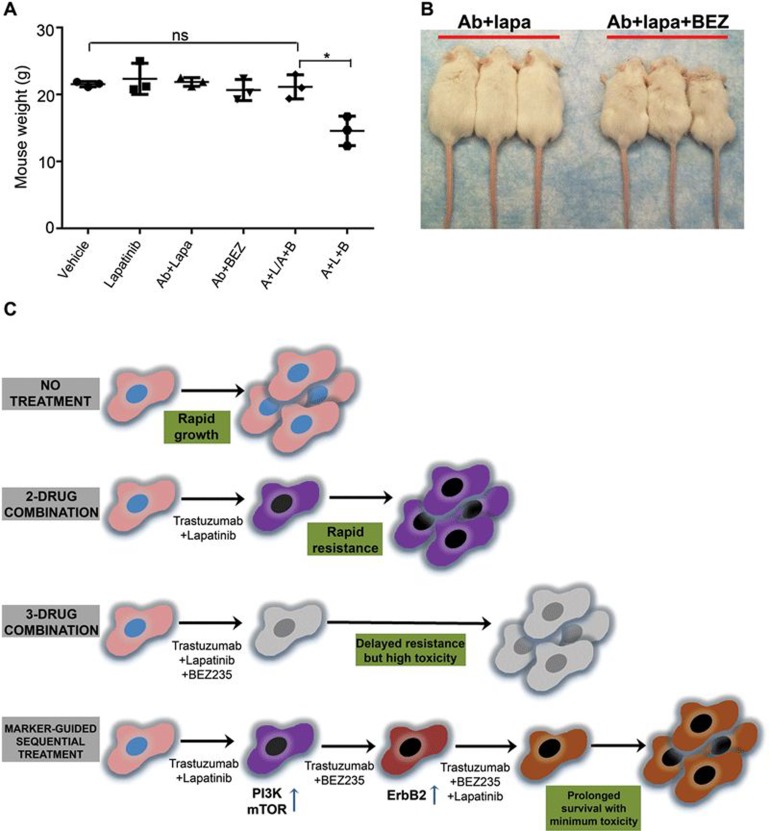
As ErbB2 expression and phosphorylation were increased in tumors resistant to Ab + lapatinib/Ab + BEZ235 dual sequential treatment, we next decided to add lapatinib, which inhibits ErbB2 kinase activity, back into the Ab + BEZ235 regimen to test whether the triple combination may overcome the resistance to sequential Ab + lapatinib/Ab + BEZ235 treatment. To this end, two donor PTEN−/−/NIC mice (254F and 315F) were first treated with Ab7.16.4 + lapatinib (A + L) for 4 weeks, followed by Ab7.16.4 + BEZ235 (A + B) treatment for another 4 weeks, and mammary tumor cells resistant to the sequential therapy (A + L/A + B.R) were isolated for primary culture (254F.A + L/A + B.R and 315F.A + L/A + B.R). Both primary cells were resistant to treatment with either lapatinib or BEZ235 alone or in combination with Ab7.16.4. However, combinatorial application of lapatinib and BEZ235 with or without Ab7.16.4 significantly inhibited proliferation of the A + L/A + B-resistant primary cells (Figure 5E). In both primary cells, although BEZ235 or Ab + BEZ treatment inhibited phosphorylation of S6 and 4EBP1 to a certain extent, Akt phosphorylation was not inhibited (Figure 5F). Furthermore, Erk1/2 phosphorylation, downstream of ErbB2, was increased upon BEZ235 treatment. Remarkably, combinatorial application of BEZ235 + lapatinib blocked Akt, S6, 4EBP1 and ERK1/2 signaling downstream of ErbB2 (Figure 5F). These results prompted us to apply the combination of Ab7.16.4, BEZ235, and lapatinib (A + B + L) in transplants resistant to Ab + lapatinib and Ab + BEZ235 sequential treatment. The A + B + L triple combination treatment effectively reduced tumor volume (Figure 6A and 6B) not only by inhibiting cell proliferation (Figure 6C), but also by increasing apoptosis (Figure 6D). We then tested the PI3K/mTOR and ERK1/2 pathway status in differently treated transplants and observed that sequential application of A + B + L after A + L/A + B treatment led to strong downregulation of Akt, S6 and ERK1/2 phosphorylation (Figure 6E). Finally, we applied the A + B + L after development of resistance to A + L/A + B sequential treatment in the original donor PTEN−/−/NIC mice. Triple sequential treatment (A + L followed by A + B, then followed by A + B + L, denoted as A + L/A + B/A + B + L) was highly effective in increasing the survival of the PTEN−/−/NIC mice (median survival: 98 days) than both A + L (median survival: 75 days, P < 0.0001 by log-rank test) or A + L/A + B (median survival: 86 days, P = 0.03 by log-rank test) treatments. The addition of the A + B + L triple combination after A + L/A + B sequential therapy doubled the life-span of mice bearing highly aggressive mammary tumors by increasing the median survival time from 45 days to 98 days (Figure 6F) and substantially reduced tumor size and multiplicity (Figure 6G). We also tested whether sequential treatment was superior to treatment with Ab7.16.4 + lapatinib + BEZ235 from the beginning (see Figure 6F, “A + L + B”). Sequential treatment showed a trend towards superior survival compared to A + L + B (P = 0.067; median survival 84.5 versus 98 days). Notably, two mice in the A + L + B triple combination had to be sacrificed around day 40 (Figure 6F) due to severe toxic responses including significant weight loss (Figure 7A and 7B). Thus sequential treatment maximized survival while minimizing toxicity compared to all other treatments tested (Figure 7C). Taken together, these data clearly demonstrate that biomarker-guided, sequential application of targeted therapies can effectively target and reprogram the signaling networks in cancer evolving resistance, create cancer vulnerabilities, and substantially increase the survival of mice bearing highly aggressive therapy-refractory tumors.
Figure 6.
Marker-guided sequential treatment doubles the life-span of PTEN−/−/NIC mice bearing highly aggressive mammary tumors. (A) Tumor volume measurements at different time points for transplant mice treated with Ab + lapatinib (A + L) first, then with Ab + BEZ235 (A + B) on day 12, and Ab + lapatinib + BEZ235 (A + L + B) on day 19 upon resistance development. (B) Representative images of a tumor from transplant mouse 782_L taken before and after A + L + B treatment. (C) Ki-67 staining of transplant tumors treated with A + L/A + B/A + L + B sequential therapy compared to Ab + lapa treatment or vehicle. **P < 0.01, ***P < 0.001, by two-tailed t-test. (D) TUNEL staining of tumors from C. ***P < 0.001, by two-tailed t-test. (E) Immunohistochemical (IHC) staining of p-Akt, p-S6 and p-ERK1/2 of transplants treated with A + L/A + B/A + L + B sequential therapy compared to vehicle (Ctrl)-treated mice. (F) Survival analysis of the PTEN−/−/NIC mice treated with vehicle, Ab + lapatinib (A + L), Ab + lapatinib + BEZ235 (A + L + B) or A + L/A + B/A + B + L sequential therapy. P-value was determined by log-rank test. P < 0.001 is between Ab7.16.4 + lapatinib and A + L/A + B/A + B + L sequential therapy. (G) Tumor weight (mean ± SEM) of mice treated with vehicle (Ctrl) or A + L/A + B/A + L + B therapy. ***P < 0.001 by two-tailed t-test.
Figure 7.
The effects of treatments on mouse weight and appearance, and the model explaining the superiority of the biomarker-guided, sequential combinatorial targeted therapies. (A) Mouse weight measurements after treatment of the PTEN−/−/NIC mice with the indicated agents for 10 days (20 days in the case of sequential A + L/A + B). Vehicle, lapatinib, Ab7.16.4 + lapatinib (A + L), Ab7.16.4 + BEZ235 (A + B), Ab7.16.4 + lapatinib followed by Ab7.16.4 + BEZ235 (A + L/A + B), and Ab7.16.4 + lapatinib + BEZ235 (A + L + B). ns, not significant (P > 0.05) and *P < 0.05, by two-tailed t-test. A + L + B treatment led to weight loss. (B) Pictures of the mice treated with Ab7.16.4 + lapatinib (Ab + Lapa) or Ab7.16.4 + lapatinib + BEZ235 (Ab + Lapa + BEZ) from (A). (C) Model showing how biomarker-guided, sequential combinatorial targeted therapies maximize mouse survival in rapidly evolving tumors while reducing toxicity compared to current clinical practices.
Discussion
In the current metastatic or late-stage therapy settings even the most successful targeted agents can only prolong the survival of patients for a limited time. De novo or acquired resistance to therapies as well as over-treatment due to lack of robust biomarkers are the main problems associated with treatment failure. Trastuzumab, the mainstay therapy for ErbB2-positive patients, has an initial clinical response in only 12%-34% of patients, and the majority of these patients acquire resistance to the drug within one year24. Therefore, there is an urgent need for identification of biomarkers to choose patients with higher potential of response and novel targets to reverse resistance. Here, we designed a sequential targeted therapy strategy guided by identification of resistance mechanisms and resulting biomarkers. We tested the strategy in a highly aggressive trastuzumab-resistant transgenic animal model, ErbB2-overexpressing human breast cancer and primary culture mouse tumor cells. Here, we demonstrated that the biomarker-guided, sequential application of different targeted therapy combinations effectively targets the signaling networks in cancer evolving therapy resistance to better inhibit tumor progression than using a single targeted therapy combination throughout the course of the treatment.
The role of PTEN loss and the resulting activation of PI3K/Akt pathway has been well characterized in de novo resistance of breast cancer cells to trastuzumab9,10. However, PTEN's role in lapatinib resistance has been controversial. Loss of PTEN in BT474 cells by shRNAs was shown to lead to lapatinib resistance both in vitro and in vivo25. Similarly, activation of PI3K signaling resulting from PTEN loss or PI3K mutations has been reported for both trastuzumab and lapatinib resistance in patients26. On the other hand, the response to lapatinib of ErbB2-overexpressing PTEN-knockdown BT474 and AU565 cell lines, and of patients with inflammatory breast cancers is independent of PTEN status27. Interestingly, low PTEN level was associated with a higher pathological response to lapatinib in breast cancer patients21. Our study clearly demonstrates that PTEN loss does not confer lapatinib resistance in the PTEN−/−/NIC immune-competent mice with mammary gland-targeted ErbB2 overexpression and PTEN knockout (Figure 1E). Rather, when combined with trastuzumab, lapatinib increased the survival of mice by reducing the proliferation of cancer cells (Figure 1E-1G). Our data also showed that although PTEN loss confers de novo trastuzumab resistance, trastuzumab + lapatinib are more effective in tumor inhibition than lapatinib alone, indicating that trastuzumab should still be used with lapatinib to treat patients with PTEN-low tumors. Recently, a clinical study similarly found that lapatinib combined with trastuzumab was more effective (4.5 months overall survival advantage) than lapatinib alone for patients who had progressive disease on trastuzumab treatment20.
Our study shows that development of resistance to lapatinib involved the activation of the PI3K and mTOR pathways in the PTEN−/−/NIC mouse model, the human BT474.LapR cell lines with acquired resistance, and importantly, also in breast cancer patients treated with lapatinib. Several studies have indicated the role of the PI3K and/or mTOR pathways in acquired lapatinib resistance and have suggested using PI3K or mTOR inhibitors in combination with lapatinib25,28. However, these studies tested the combination either in cell culture or in xenograft models without considering toxicity, which is often a major obstacle in successful translation to the clinic. For example, combinatorial inhibition of PI3K and MAPK pathways in various advanced tumors led to better efficacy but higher toxicity compared to individual treatments16. Similarly, combinatorial treatment of mice bearing ovarian tumor xenografts with GNE493 (mTOR inhibitor) and ABT-737 (Bcl-2 inhibitor) led to 5% to 20% loss of body weight after 7 days of treatment29. We observed that triple combination treatment of Ab7.16.4 + BEZ235 + lapatinib for 10 days induced up to 20% loss of body weight and rough coat in some of the PTEN−/−/NIC mice (Figure 7A and 7B). However, sequential treatment of Ab7.16.4 + lapatinib for 10 days followed by Ab7.16.4 + BEZ235 for another 10 days did not induce significant weight loss. Clearly, sequential application of various targeted therapies may be a better strategy to manage both tumor growth and side-effects than simultaneous application of all drugs in the course of treatment. Accordingly, our strategy was to first apply Ab7.16.4 + lapatinib, then Ab7.16.4 + BEZ235, and finally Ab7.16.4 + BEZ235 + lapatinib only after PTEN−/−/NIC tumors developed resistance to Ab7.16.4 + BEZ235 for maximum tumor inhibition and minimum toxicity. Additionally, this sequential therapy can also generate cancer vulnerabilities to be effectively targeted with the subsequent agent. Supporting this idea, it was recently shown that sequential, but not simultaneous, application of an EGFR inhibitor and chemotherapy led to apoptosis of triple negative breast cancer cell lines by rewiring the DNA damage network resulting in activation of Caspase-8 in vitro30. Similarly, in our study, we showed for the first time that Ab7.16.4 + lapatinib combination reprogrammed the cancer cell signaling network and led to activation of the PI3K/mTOR pathway, which was successfully inhibited by subsequent application of Ab7.16.4 + BEZ235 in vivo.
Little is known about the mechanisms of resistance to BEZ235 treatment. Recently, two studies suggested the involvement of the MYC oncogene, which is downstream of the PI3K/mTOR pathway31. First, an unbiased screen showed that MYC conferred resistance to BEZ235 in the human mammary epithelial cell (HMEC) line MCF-10A, and that BEZ235-resistant human cell lines had higher c-MYC gene copy number compared to their sensitive counterparts32. Second, MYC and eIF4E amplification was identified in BEZ235-resistant HMECs in addition to genome-wide copy number alteration33. Here, we did not observe any change in MYC mRNA level in our BT474.Par or lapatinib-resistant BT474.Lap.R cells upon long-term BEZ235 treatment (Supplementary information, Figure S6A). c-Myc protein was also not increased after long-term treatment with BEZ235 in BT474 LapR cells (Supplementary information, Figure S6B). Rather, we observed a consistent increase in ErbB2 expression and phosphorylation in both mice and human breast cancer cells grown in 3-D culture with sequential lapatinib and BEZ235 treatment. The increase in ErbB2 phosphorylation led to an increase in downstream activation of Erk1/2, Akt and S6 which provided signals necessary for survival and proliferation of the cancer cells. As BEZ235 stabilizes ErbB2 and leads to its activation, the combination of ErbB2-targeting agents and BEZ235 reduced the phosphorylation of ErbB2 and downstream kinases and led to an increase in apoptosis and a decrease in cell proliferation at the expense of higher toxicity. Our findings bring novel insights into the mechanisms of BEZ235 resistance that may guide future development of strategies to reverse the resistance without increasing toxicity.
In conclusion, we propose a novel strategy of using biomarkers in cancer evolving therapy resistance to guide the application of various targeted therapies in a sequential manner, instead of simultaneously, to induce cancer vulnerability and effectively block tumor progression. We have validated this rational approach in multiple animal models and by using patient data. Our data demonstrate that sequential application of lapatinib and BEZ235 in combination with trastuzumab led to a doubling of the life-span of PTEN−/−/NIC mice bearing highly aggressive, treatment-refractory mammary tumors. Importantly, the mouse model that we used (PTEN−/−/NIC) reflects the intratumoral heterogeneity of breast cancer as PTEN loss is highly heterogeneous (determined by IHC), indicating that some parts of the tumor may be primarily driven by ErbB2/neu while others are driven by PTEN loss as well (Supplementary information, Figure S7). Similar heterogeneity has also been reported in other GEM models34,35. Additionally, our experimental approach of testing drugs in a time-efficient and robust manner using transplantation in immune-competent mice could be utilized for setting up synchronous mouse co-clinical trials to study the resistance mechanisms of patients in real time, similar to the elegant work done in lung cancer in which the mouse trial was used to predict the outcome of an on-going human clinical trial36. Notably, our findings may potentially change the current clinical management of patients having highly aggressive ErbB2-overexpressing trastuzumab-resistant breast cancers as the targeted therapies used in our study are either FDA approved or in clinical trials. More importantly, this strategy of using biomarker-guided, sequential, combinatorial targeted therapies in rapidly evolving tumors can also be rapidly translated into the clinic for patients with other cancer types receiving different targeted therapies to significantly reduce cancer deaths.
Materials and Methods
Transgenic animal model and syngeneic tumor transplantation
MMTV-NIC (Neu-IRES-Cre) mice37 were interbred with Flox-PTEN mice to generate HER2/neu-overexpressing PTEN homozygous loss (PTEN−/−/NIC) mice12. For tumor transplantation, the donor PTEN−/−/NIC tumors were cut into quasi-equal pieces of 3-5 mm in diameter after sacrificing donor mice. Recipient FVB mice were anesthetized, the skin was incised and donor tumors were transplanted into both left and right MFP of the recipient mice. Incisions were closed using wound clips which were removed after 7 days. Tumor size was measured 3-5 days later and treatment was started when tumors reached 4-5 mm. Tumors were measured using calipers and volume was calculated as length × width2/2. Tumor volume fold changes were calculated by taking the ratio of most recent measurements to previous measurements for the same tumor. All tumor measurements were included in the analysis. All animal studies were approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee.
Animal treatment
mAb7.16.4 hybridoma was a kind gift from Dr Mark Greene. PTEN−/−/NIC mice were randomized to treatment groups when the first palpable tumor was 3-5 mm in diameter. 7.16.4 mAb was administered intraperitoneally at 2 mg/kg body weight every 3 days in PBS as described previously13. We treated the original donor mice with lapatinib (LC Laboratories, Woburn, MA, USA) or BEZ235 (Novartis, Basel, Switzerland) daily at 100 mg/kg or 50 mg/kg, respectively, in vehicle (0.5% wt/vol hydroxypropyl-methylcellulose with 0.1% vol/vol Tween 20) via daily oral gavage. Mice were sacrificed either after treatment for 3 weeks or when the tumor size reached the institutional euthanasia criteria, and tumors were harvested and weighed.
Cell culture and primary cell preparation
BT474 m1 cells were a gift from Dr Dajun Yang. Lapatinib-resistant cells were established by exposure of the cells to 2.1 μM lapatinib for > 9 months. BT474.shPTEN cells were described previously14. Primary cells from mice were prepared by incubating minced tissue in growth medium containing collagenase A (Roche, Mannheim, Germany) and hyaluronidase (Sigma, St Louis, MO, USA) for 6 h at 37 °C with continuous mixing. After centrifugation and removal of debris and blood cells, cells were plated and grown for 48 h. These cells were then used for viability and western blot assays without further passaging.
In vitro drug treatment and cell viability assay
Primary mouse cells or human BT474.LapR or BT474.shPTEN cells were seeded in 96-well plates 24 h before the assay. After treatment of the cells with trastuzumab (2 μg/ml), Ab7.16.4 (5 μg/ml for mouse primary cells), lapatinib (1 μM), BEZ235 (500 nM), GDC-0941 (1 μM) or RAD001 (50 nM) for 72 h, cells were incubated with 20 μl of WST-1 (Roche, Penzberg, Germany) in 200 μl of final volume for 4 h. Absorbance at 450 nm was measured and percentage of viability was calculated.
3-D morphogenesis assay
BT474.Par and BT474.LapR cells were seeded into 8-chamber slides (BD Biosciences, San Jose, CA, USA) coated with 40 μl of growth factor-reduced matrigel (BD Biosciences). 3-D structures were fixed and stained as previously described38. For mRNA or protein analysis, cells were seeded in 2% Matrigel into 60-mm dishes coated with 4 ml of polyHEMA (Sigma, St Louis, MO, USA).
Protein stability sssay
BT474.LapR cells were grown in 3-D culture under DMSO or BEZ235 treatment over a week, and CHX (5 μg/ml; Calbiochem) was given for 0, 2, 4, 6, 12 or 24 h followed by western blot analysis.
Western blot and immunoprecipitation
Tumor protein extracts were prepared by homogenizing samples in tissue lysis buffer (10 mmol/l sodium phosphate (pH 7.3), 154 mmol/l NaCl, 5% sodium deoxycholate, 1% SDS) using a tissue grinder, followed by centrifugation. Western blot was done as previously described14,39. List of antibodies used in this study is given in Supplementary information, Table S4.
RPPA and p-RTK array
Protein lysate for RPPA was prepared according to MD Anderson RPPA Core instructions (available online) and run by the Core. Quantification of mouse phospho RTK arrays (ARY014, R&D, Minneapolis, MN, USA) was done with ImageJ (National Institutes of Health).
RNA extraction and quantitative real-time RT-PCR
RNA was isolated using Trizol (Invitrogen) and reverse transcribed with iScript™ cDNA synthesis kit (Biorad, Hercules, CA, USA). qRT-PCR was performed with iQ SYBR Green Supermix (Biorad, Hercules, CA) and StepOnePlus (Applied Biosystems, Grand Island, NY, USA) instrument. The genes analyzed were ERBB2, MYC and 18S (used as housekeeping control). Primer sequences were: ERBB2_forw_5′-GGGAAACCTGGAACTCACCT-3′ and ERBB2_rev_5′-CCCTGCACCTCCTGGATA-3′ MYC_forw_5′-CCTGGTCAAGAAGCATTTCAA-3′ and MYC_rev_5′-GCCCCAAAGATGAGGAGTATC-3′ and 18S_forw_5′-AACCCGTTGAACCCCATT-3′ and 18S_rev_5′-CCATCCAATCGGTAGTAGCG-3′. Data were analyzed with the delta-delta CT method.
Microarray data analysis of patient tumors
Neoadjuvant trastuzumab + chemotherapy (n = 45) patient gene expression data17 were classified into two groups: trastuzumab-sensitive/high PTEN (n = 7) and trastuzumab-resistant/low PTEN (n = 7) according to their pathological response and PTEN mRNA expression. Differentially expressed genes with fold change > 2 were analyzed by IPA of canonical pathways. Pre- and post-lapatinib treatment gene expression data from 13 patients (nine responders)21 were analyzed by GSEA22.
Immunohistochemistry and TUNEL staining
IHC and TUNEL staining were performed as described previously13 using antibodies for p-Akt (1:400), Ki-67 (DAKO, 1:200), p-S6 (1:400), and p-Erk (1:200).
Statistical analyses
Statistical differences were determined by 2-tailed t-test, one-way ANOVA or log-rank test. The GraphPad Prism 6 Program (GraphPad Software, La Jolla, CA, USA) was used to perform all statistical analyses. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Yu laboratory members for helpful discussions on the manuscript. We thank Novartis for providing BEZ235. We also thank Dr Mark Greene (University of Pennsylvania) for generously providing the hybridoma for 7.16.4 mAb. This study is supported by NIH grants P30-CA 16672 (MDACC), PO1-CA099031 project 4 (DY), RO1-CA112567-07 (DY), Susan G Komen Breast Cancer Foundation Promise Grant KG091020 (DY), Cancer Prevention Research Institute of Texas Grant RP100726 (DY), Career Development Award of MD Anderson Cancer Center Breast SPORE (OS), NIH/NCI pre-doctoral fellowship F31CA165819 (SB) and NIH grant U54CA149169 (STW). D Yu is the Hubert L & Olive Stringer Distinguished Chair in Basic Science at MD Anderson Cancer Center.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
List of differentially expressed probes between PTEN low/trastuzumab resistant (red) versus PTEN normal/trastuzumab sensitive (blue) breast cancer patients.
List of genes used for IPA to determine canonical pathways differentially enriched between PTEN normal/trastuzumab resistant vs. PTEN high/trastuzumab sensitive breast cancer patients.
List of proteins with downregulated expression/phosphorylation in Ab7.16.4+lapatinib sensitive PTEN−/−/NIC transplants compared to control and resistant tumors.
List of antibodies used in western blotting
PTEN loss leads to activation of ErbB2 and its downstream signaling in PTEN−/−/NIC mice.
TUNEL staining of the tumor tissue obtained from PTEN−/−/NIC mice treated with different drugs or their combination for determination of apoptosis.
Syngeneic tumor transplantation model recapitulates the both molecular and biological features of the original PTEN−/−/NIC mice.
TUNEL staining of the tumor tissue obtained from PTEN−/−/NIC mice treated with vehicle, Ab+Lapa or sequential A+L/A+B for determination of apoptosis.
BEZ235 does not stabilize ErbB2 in 2-D cell culture in BT474.Par or BT474.LapR cells and does not affect ErbB2 mRNA level in 3-D culture.
BEZ235 treatment does not significantly alter c-MYC level in 3-D culture.
PTEN loss is heterogeneous in Neu-driven, PTEN-loss mouse mammary tumors.
References
- Slamon D, Clark G, Wong S, Levin W, Ullrich A, McGuire W. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Carter P, Presta L, Gorman CM, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pelt AE, Mohsin S, Elledge RM, et al. Neoadjuvant trastuzumab and docetaxel in patients with breast cancer: preliminary results. Clin Breast Cancer. 2003;4:348–353. doi: 10.3816/cbc.2003.n.040. [DOI] [PubMed] [Google Scholar]
- Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable her2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- Geyer CE, Forster J, Lindquist D, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006;355:2733–2743. doi: 10.1056/NEJMoa064320. [DOI] [PubMed] [Google Scholar]
- Esteva FJ, Yu D, Hung MC, Hortobagyi GN. Molecular predictors of response to trastuzumab and lapatinib in breast cancer. Nat Rev Clin Oncol. 2010;7:98–107. doi: 10.1038/nrclinonc.2009.216. [DOI] [PubMed] [Google Scholar]
- Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Schade B, Rao T, Dourdin N, et al. PTEN deficiency in a luminal ErbB-2 mouse model results in dramatic acceleration of mammary tumorigenesis and metastasis. J Biol Chem. 2009;284:19018–19026. doi: 10.1074/jbc.M109.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li S-H, Wang H, et al. Concomitant targeting of tumor cells and induction of T-cell response synergizes to effectively inhibit trastuzumab-resistant breast cancer. Cancer Res. 2012;72:4417–4428. doi: 10.1158/0008-5472.CAN-12-1339-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Huang WC, Li P, et al. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A, Bhola NE, Sutton C, et al. Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors. Cancer Res. 2013;73:1190–1200. doi: 10.1158/0008-5472.CAN-12-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Tolcher AW, Papadopoulos KP, et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin Cancer Res. 2012;18:2316–2325. doi: 10.1158/1078-0432.CCR-11-2381. [DOI] [PubMed] [Google Scholar]
- Esteva FJ, Wang J, Lin F, et al. CD40 signaling predicts response to preoperative trastuzumab and concomitant paclitaxel followed by 5-fluorouracil, epirubicin, and cyclophosphamide in HER-2-overexpressing breast cancer. Breast Cancer Res. 2007;9:R87. doi: 10.1186/bcr1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel D, Brady B, Tabchy A, B. Mills G, Hennessy B. Proteomic classification of breast cancer. Curr Drug Targets. 2012;13:1495–1509. doi: 10.2174/138945012803530080. [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Q, Montone KT, et al. Shared antigenic epitopes and pathobiological functions of anti-p185her2/neu monoclonal antibodies. Exp Mol Pathol. 1999;67:15–25. doi: 10.1006/exmp.1999.2266. [DOI] [PubMed] [Google Scholar]
- Blackwell KL, Burstein HJ, Storniolo AM, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 Study. J Clin Oncol. 2012;30:2585–2592. doi: 10.1200/JCO.2011.35.6725. [DOI] [PubMed] [Google Scholar]
- Dave B, Migliaccio I, Gutierrez MC, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29:166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PK, Febbo PG, Bikoff R, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJA, Gili M, Scaltriti M, et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008;68:9221–9230. doi: 10.1158/0008-5472.CAN-08-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang Q, Zhang J, et al. PI3K pathway activation results in low efficacy of both trastuzumab and lapatinib. BMC Cancer. 2011;11:248. doi: 10.1186/1471-2407-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Husain I, Liu L, et al. Lapatinib antitumor activity is not dependent upon phosphatase and tensin homologue deleted on chromosome 10 in ErbB2-overexpressing breast cancers. Cancer Res. 2007;67:1170–1175. doi: 10.1158/0008-5472.CAN-06-2101. [DOI] [PubMed] [Google Scholar]
- Jegg AM, Ward TM, Iorns E, et al. PI3K independent activation of mTORC1 as a target in lapatinib-resistant ERBB2+ breast cancer cells. Breast Cancer Res Treat. 2012;136:683–692. doi: 10.1007/s10549-012-2252-9. [DOI] [PubMed] [Google Scholar]
- Muranen T, Selfors LM, Worster DT, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Blenis J, Yuan J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc Natl Acad Sci USA. 2008;105:6584–6589. doi: 10.1073/pnas.0802785105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muellner MK, Uras IZ, Gapp B V, et al. A chemical-genetic screen reveals a mechanism of resistance to PI3K inhibitors in cancer. Nat Chem Biol. 2011;7:787–793. doi: 10.1038/nchembio.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci USA. 2011;108:E699–E708. doi: 10.1073/pnas.1108237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng H, Santiago S, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17:1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milliken EL, Lozada KL, Johnson E, et al. Ovarian hyperstimulation induces centrosome amplification and aneuploid mammary tumors independently of alterations in p53 in a transgenic mouse model of breast cancer. Oncogene. 2008;27:1759–1766. doi: 10.1038/sj.onc.1210815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Cheng K, Walton Z, et al. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–617. doi: 10.1038/nature10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini-Siegel J, Hardy WR, Zuo D, et al. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. EMBO J. 2008;27:910–920. doi: 10.1038/emboj.2008.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Uhlmann S, Mannsperger H, Zhang JD, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of differentially expressed probes between PTEN low/trastuzumab resistant (red) versus PTEN normal/trastuzumab sensitive (blue) breast cancer patients.
List of genes used for IPA to determine canonical pathways differentially enriched between PTEN normal/trastuzumab resistant vs. PTEN high/trastuzumab sensitive breast cancer patients.
List of proteins with downregulated expression/phosphorylation in Ab7.16.4+lapatinib sensitive PTEN−/−/NIC transplants compared to control and resistant tumors.
List of antibodies used in western blotting
PTEN loss leads to activation of ErbB2 and its downstream signaling in PTEN−/−/NIC mice.
TUNEL staining of the tumor tissue obtained from PTEN−/−/NIC mice treated with different drugs or their combination for determination of apoptosis.
Syngeneic tumor transplantation model recapitulates the both molecular and biological features of the original PTEN−/−/NIC mice.
TUNEL staining of the tumor tissue obtained from PTEN−/−/NIC mice treated with vehicle, Ab+Lapa or sequential A+L/A+B for determination of apoptosis.
BEZ235 does not stabilize ErbB2 in 2-D cell culture in BT474.Par or BT474.LapR cells and does not affect ErbB2 mRNA level in 3-D culture.
BEZ235 treatment does not significantly alter c-MYC level in 3-D culture.
PTEN loss is heterogeneous in Neu-driven, PTEN-loss mouse mammary tumors.