Abstract
The opportunistic human pathogen Acinetobacter baumannii is a concern to health care systems worldwide because of its persistence in clinical settings and the growing frequency of multiple drug resistant infections. To combat this threat, it is necessary to understand factors associated with disease and environmental persistence of A. baumannii. Recently, it was shown that a single biosynthetic pathway was responsible for the generation of capsule polysaccharide and O-linked protein glycosylation. Because of the requirement of these carbohydrates for virulence and the non-template driven nature of glycan biogenesis we investigated the composition, diversity, and properties of the Acinetobacter glycoproteome. Utilizing global and targeted mass spectrometry methods, we examined 15 strains and found extensive glycan diversity in the O-linked glycoproteome of Acinetobacter. Comparison of the 26 glycoproteins identified revealed that different A. baumannii strains target similar protein substrates, both in characteristics of the sites of O-glycosylation and protein identity. Surprisingly, glycan micro-heterogeneity was also observed within nearly all isolates examined demonstrating glycan heterogeneity is a widespread phenomena in Acinetobacter O-linked glycosylation. By comparing the 11 main glycoforms and over 20 alternative glycoforms characterized within the 15 strains, trends within the glycan utilized for O-linked glycosylation could be observed. These trends reveal Acinetobacter O-linked glycosylation favors short (three to five residue) glycans with limited branching containing negatively charged sugars such as GlcNAc3NAcA4OAc or legionaminic/pseudaminic acid derivatives. These observations suggest that although highly diverse, the capsule/O-linked glycan biosynthetic pathways generate glycans with similar characteristics across all A. baumannii.
Acinetobacter baumannii is an emerging opportunistic pathogen of increasing significance to health care institutions worldwide (1–3). The growing number of identified multiple drug resistant (MDR)1 strains (2–4), the ability of isolates to rapidly acquire resistance (3, 4), and the propensity of this agent to survive harsh environmental conditions (5) account for the increasing number of outbreaks in intensive care, burn, or high dependence health care units since the 1970s (2–5). The burden on the global health care system of MDR A. baumannii is further exacerbated by standard infection control measures often being insufficient to quell the spread of A. baumannii to high risk individuals and generally failing to remove A. baumannii from health care institutions (5). Because of these concerns, there is an urgent need to identify strategies to control A. baumannii as well as understand the mechanisms that enable its persistence in health care environments.
Surface glycans have been identified as key virulence factors related to persistence and virulence within the clinical setting (6–8). Acinetobacter surface carbohydrates were first identified and studied in A. venetianus strain RAG-1, leading to the identification of a gene locus required for synthesis and export of the surface carbohydrates (9, 10). These carbohydrate synthesis loci are variable yet ubiquitous in A. baumannii (11, 12). Comparison of 12 known capsule structures from A. baumannii with the sequences of their carbohydrate synthesis loci has provided strong evidence that these loci are responsible for capsule synthesis with as many as 77 distinct serotypes identified by molecular serotyping (11). Because of the non-template driven nature of glycan synthesis, the identification and characterization of the glycans themselves are required to confirm the true diversity. This diversity has widespread implications for Acinetobacter biology as the resulting carbohydrate structures are not solely used for capsule biosynthesis but can be incorporated and utilized by other ubiquitous systems, such as O-linked protein glycosylation (13, 14).
Although originally thought to be restricted to species such as Campylobacter jejuni (15, 16) and Neisseria meningitidis (17), bacterial protein glycosylation is now recognized as a common phenomenon within numerous pathogens and commensal bacteria (18, 19). Unlike eukaryotic glycosylation where robust and high-throughput technologies now exist to enrich (20–22) and characterize both the glycan and peptide component of glycopeptides (23–25), the diversity (glycan composition and linkage) within bacterial glycosylation systems makes few technologies broadly applicable to all bacterial glycoproteins. Because of this challenge a deeper understanding of the glycan diversity and substrates of glycosylation has been largely unachievable for the majority of known bacterial glycosylation systems. The recent implementation of selective glycopeptide enrichment methods (26, 27) and the use of multiple fragmentation approaches (28, 29) has facilitated identification of an increasing number of glycosylation substrates independent of prior knowledge of the glycan structure (30–33). These developments have facilitated the undertaking of comparative glycosylation studies, revealing glycosylation is widespread in diverse genera and far more diverse then initially thought. For example, Nothaft et al. were able to show N-linked glycosylation was widespread in the Campylobacter genus and that two broad groupings of the N-glycans existed (34).
During the initial characterization of A. baumannii O-linked glycosylation the use of selective enrichment of glycopeptides followed by mass spectrometry analysis with multiple fragmentation technologies was found to be an effective means to identify multiple glycosylated substrates in the strain ATCC 17978 (14). Interestingly in this strain, the glycan utilized for protein modification was identical to a single subunit of the capsule (13) and the loss of either protein glycosylation or glycan synthesis lead to decreases in biofilm formation and virulence (13, 14). Because of the diversity in the capsule carbohydrate synthesis loci and the ubiquitous distribution of the PglL O-oligosaccharyltransferase required for protein glycosylation, we hypothesized that the glycan variability might be also extended to O-linked glycosylation. This diversity, although common in surface carbohydrates such as the lipopolysaccharide of numerous Gram-negative pathogens (35), has only recently been observed within bacterial proteins glycosylation system that are typically conserved within species (36) and loosely across genus (34, 37).
In this study, we explored the diversity within the O-linked protein glycosylation systems of Acinetobacter species. Our analysis complements the recent in silico studies of A. baumannii showing extensive glycan diversity exists in the carbohydrate synthesis loci (11, 12). Employing global strategies for the analysis of glycosylation, we experimentally demonstrate that the variation in O-glycan structure extends beyond the genetic diversity predicted by the carbohydrate loci alone and targets proteins of similar properties and identity. Using this knowledge, we developed a targeted approach for the detection of protein glycosylation, enabling streamlined analysis of glycosylation within a range of genetic backgrounds. We determined that; O-linked glycosylation is widespread in clinically relevant Acinetobacter species; inter- and intra-strain heterogeneity exist within glycan structures; glycan diversity, although extensive results in the generation of glycans with similar properties and that the utilization of a single glycan for capsule and O-linked glycosylation is a general feature of A. baumannii but may not be a general characteristic of all Acinetobacter species such as A. baylyi.
MATERIALS AND METHODS
Bacterial Strains
Acinetobacter strains are provided in Table I. All Acinetobacter strains were grown in Luria Bertani (LB) (10g Tryptone, 5g yeast extract, and 10g of NaCl per 1L of dH2O supplemented with 15g of agar per liter of broth when needed) broth/agar at 37 °C with shaking at 200rpm. For protein purification (supplementary Methods) an additional 50 μg/ml of Kanamycin was added with 0.2% (w/v) l-arabinose also added for induction when required. For the generation of material for protein purification studies and ZIC-HILIC analysis 1L cultures were grown overnight as described above. Cells were harvested, washed twice in phosphate buffered saline and either used instantly for protein purification or freeze dried for ZIC-HILIC analysis preparation.
Table I. Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| A. baumannii | ||
| ATCC 17978 | Reference strain | [74] |
| ATCC 17978 ΔpglL | O-linked glycosylation negative strain | [14] |
| SDF | Body lice isolate | [75] |
| AYE | Human isolate | [75] |
| ATCC 19606 | Reference strain | [76] |
| 1224 | Clinical isolate, thigh | [77] |
| 1225 | Clinical isolate, coccyx | [77] |
| 1441 C1 | MDR Clinical isolate | This study |
| 1441 C3 | MDR Clinical isolate | This study |
| Arg1 | MDR Clinical isolate | This study |
| Arg2 | MDR Clinical isolate | This study |
| A. calcoaceticus 1217 | Clinical isolate, urine | [77] |
| A. calcoaceticus 1218 | Clinical isolate, leg | [77] |
| A. pittii 1219 | Clinical isolate, urine | [77] |
| A. baylyi ADP1 | Soil isolate | [78] |
| A. nosocomialis 1222 | Clinical isolate, iscial ulcer | [77] |
| E. coli | ||
| DH5α | General cloning and plasmid propagation | Invitrogen |
| Plasmids | ||
| pEXT20–A1S_1193His10X | Cloning and expression vector, IPTG inducible, Ampr, with A1S_1193 inserted at BamH1 and XbaI | [28] |
| pEC-A1S_1193His10X | Arabinose inducible, Kmr, plasmid used to express A1S_1193 (Inserted at HincII and SalI) in Acinetobacter for glycan fishing | [28] |
| pBAVMCS | Kmr pBAV1K-T5-gfp derivative with gfp ORF removed, Constitutive E. coli/Acinetobacter shuttle vector | [10] |
| pBAVMCS-A1S_1193His10X | Constitutive plasmid, Kmr, used to express A1S_1193 (inserted at BamHI and SalI) in Acinetobacter | This study |
Glycopeptide Identification from Purified A1S_1193
Purified proteins (Supplementary Methods) were resolved using 10% SDS-PAGE and stained with Coomassie blue to assess purity. In-gel trypsin digestion of the protein band corresponding to glycosylated A1S_1193 was performed according to Shevchenko et al. (2006) (38). Briefly, bands were washed with water and dehydrated with 100% acetonitrile (ACN), followed by repeated rehydration with 1:1 (v/v) water and ACN and dehydration with 100% ACN. Next disulfide bonds were reduced through treatment with 10 mm DTT in 50 mm NH4HCO3 for 60 min at 37 °C. Cysteine thiol groups were then alkylated with 50 mm iodoacetamide in 50 mm NH4HCO3 in the dark for 60 min at room temperature. Gel pieces were then washed with 50 mm NH4HCO3, dehydrated with 100% ACN and dried. A1S_1193 was then digested with 0.02 mg/ml trypsin in 50 mm NH4HCO3 (Promega, Madison, WI) at 37 °C for 16 h. Peptides were eluted from the gel through addition of 100% ACN and water, and lyophilized for mass spectrometry analysis. Peptides were resuspended in 0.1% trifluoroacetic acid and loaded onto a ZipTipC18 (Millipore, USA) column for desalting. Peptides were eluted with 60% ACN, dried down in a Speedvac and resuspended in 0.1% Formic acid (FA). The peptides were analyzed using a Q-TOF Premier (Waters, Manchester, UK) coupled to a nanoACQUITY (Waters) ultra-performance liquid chromatography system as briefly described (39) with MassLynx, v. 4.1 (Waters) employed for data analysis.
Membrane Preparation for Glycopeptide Enrichment
Lipid free membranes were prepared according to Pessione et al. (40). Briefly, cells suspended in ice-cold 40 mm Tris (pH7.4) and lysed using three rounds of disruption at 30 kpsi using a cell disruptor (Constant System ltd, Kennesaw, GA). Lysates were then centrifuged at 100,000 × g for 70 min and the resulting pellet resuspended in 1 ml of 50 mm ammonium bicarbonate. Membrane pellets were delipidated using 4 ml of 2:1 v/v trifluoroethanol/chloroform that was allowed to incubate at 4 °C for 1 h and were mixed every 10 min. The delipidated samples were spun down at 10,000 × g at 4 °C for 10 min and the upper phase collected and dried before use.
Protease Digestion for Glycopeptide Enrichment
Dried membrane proteins were resuspended in 6 m urea, 2 m thiourea, and 40 mm NH4HCO3 and reduced/alkylated prior to digestion with Lys-C (1/200 w/w) and then trypsin (1/50 w/w) as previously described (30). All peptide digests were dialyzed against ultra-pure water overnight using a Mini Dialysis Kit with a molecular mass cut off of 1000 Da (Amersham Biosciences, Buckinghamshire, UK) and on completion were collected and lyophilized.
Identification of Glycopeptides using ZIC-HILIC Enrichment and Reversed Phase LC-MS/MS
ZIC-HILIC enrichment was performed according to (30) with minor modifications. Micro-columns composed of 10 μm ZIC-HILIC resin (Sequant, Umeå, Sweden) packed into p10 tips containing a 1 mm2 excised C8 Empore™ disc (Sigma) were packed to a bed length of 0.5 cm. Prior to use, the columns were washed with ultra-pure water, followed by 95% ACN and then equilibrated with 80% ACN and 5% FA. Samples were resuspended in 80% ACN, 5% FA and insoluble material removed by centrifugation at 20,000 × g for 5 min at 4 °C. Samples were adjusted to a concentration of 2 μg/μl and 100 μg of peptide material loaded onto a column and washed with 10 load volumes of 80% ACN, 5% FA. Peptides were eluted with three load volumes of ultra-pure water into low-bind tubes and concentrated using vacuum centrifugation. ZIC-HILIC fractions were resuspended in 0.5% formic acid and separated using EASY-nLC system (Thermo Scientific, San Jose, CA, USA) coupled to either an LTQ-Orbitrap XL with ETD, an LTQ-Orbitrap velos or an Orbitrap Elite (Thermo Scientific). Samples were eluted using a gradient from 100% buffer A (0.5% acetic acid) to 40% buffer B (0.5% acetic acid, 80% ACN) over 148 mins at a constant flow of 300 nL/min. The instrument was operated using Xcalibur v2.2 (Thermo Scientific) in a data-dependent mode automatically switching between MS and HCD/CID on the Orbitrap Elite and Velos whereas CID/ETD was used on the Orbitrap XL. ETD and CID scan events were analyzed with ITMS whereas HCD scans were analyzed using FTMS. On all instruments the 5 most abundant precursor ions were selected and dynamic exclusion of 30 s enabled. MS resolution was set to 60,000 with an ACG target of 1 × 106, maximum fill time of 500 ms and a mass window of 600 to 2000 m/z. HCD fragmentation (normalized collision energy 40) was carried out with an ACG of 2 × 105, maximum fill time of 250 ms, resolution set to 7500 and mass window 200 to 2000 m/z. CID (normalized collision energy 35), while CID fragmentation was carried out with an ACG target of 2 × 104 and maximum fill time of 100 ms. ETD fragmentation was carried out with an ACG target of 2 × 105, ETD reaction time of 100ms. Duplicate enrichments were generated for each glycopeptide analysis.
Glycopeptide Data Processing
The raw files were then processed within Proteome Discover version 1.3 (Thermo Scientific) to generate mgf files and searched using Sequest against a composite FASTA database of strain SDF, ATCC17978, and AYE (NCBI accession: NC_010400.1, NC_009085.1 and NC_010410.1 respectively, obtained from NCBI on 10/07/2012). Scan events that did not result in peptide identification from Sequest searches were exported to Excel (Microsoft, Redmond, WA, USA). To identify possible glycopeptides within this list, the “mgf graph” MS-MS module of GPMAW 8.2 was utilized to identified all scan events within the generated mgfs containing the diagnostic oxonium 301.104 or 204.086 m/z ion. Using Excel, all scan events that were not matched by Sequest and contained a predicted marker of glycosylation were identified. These events were manually inspected and identified as possible glycopeptides based on the presence of the glycan fragment within the CID scan. To facilitate glycopeptide assignments from HCD scans, the ions below the mass of the predicted deglycosylated peptides were extracted with Xcalibur v2.2 using the Spectrum list function. Ions with a deconvoluted mass above that of the deglycosylated peptide and ions corresponding to known carbohydrate oxoniums were removed in a similar approach to post-spectral processing of ETD data (41, 42). MASCOT v2.2 searches were using the Walter and Eliza Hall Institute Mascot server (https://sysbio-mascot.wehi.edu.au/mascot/home.html) of the proteobacteria taxonomy of the LugwigNR database. Searches were carried out with a parent ion mass accuracy of 20 ppm and a product ion accuracy of 0.02 Da with no protease specificity as well as the fixed modification carbamidomethyl (C) and variable modifications, oxidation (M), deamidation (N), and N-terminal formylation. The instrument setting of MALDI-QIT-TOF was chosen because of previous studies showing quadrupole-like fragmentation within HCD spectra (43) (generating a, b, and y ions) and our observation of internal cleavage products that are all included in this setting. All spectra were searched with the decoy option enabled and no matches to this database were detected (FDR 0%). To further validate glycopeptide matches, all spectra HCD spectra were annotated using the Expert Annotation tool (44) (http://www.biochem.mpg.de/mann/tools/) with a mass accuracy of 10ppm, whereas ETD data was annotated manually with a mass accuracy of 0.6 Da to ensure all major peaks were match providing further confidence of identity and localization. All annotated spectra are providing within supplemental Tables S1, S3, S4, S6A, and S7–S10. Isotopic distribution analysis was accomplished with the aid of the MS-Isotope module of Protein Prospector (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msisotope).
RESULTS
A. baumannii Strains Produced Diverse O-glycan Structures
Protein glycosylation and the carbohydrate synthesis loci thought to be responsible for the production of the O-linked glycan are ubiquitous in Acinetobacter (13, 14). As glycans are produced in a non-template driven manner we began exploring the potential O-glycan diversity by undertaking glycopeptide enrichment of commonly used sequenced A. baumannii strains as well as clinical isolates. These strains included; ATCC 19606, predicted to produce the same O-glycan as the previously characterized ATCC 17978 glycan (supplemental Fig. S1) (14), two strains thought to produce divergent glycans; AYE and SDF (supplemental Fig. S1); as well as clinical isolates of unknown glycan composition; Arg1, Arg2, 1441 C1, and 1441 C3. ZIC-HILIC enriched preparations of ATCC 19606 resulted in the identification of 50 unique glycopeptides (supplemental Table S1) corresponding to 13 unique glycoproteins. Of these glycoproteins, eight were previously unknown (supplemental Table S2). In agreement with their matching carbohydrate synthesis loci, the glycan moieties in ATCC 19606 and ATCC 17978 were identical, consisting of pentasaccharide β-GlcNAc3NAcA4OAc-4-(β-GlcNAc-6-)-α-Gal-6-β-Glc-3-β-GalNAc- (Fig. 1A).
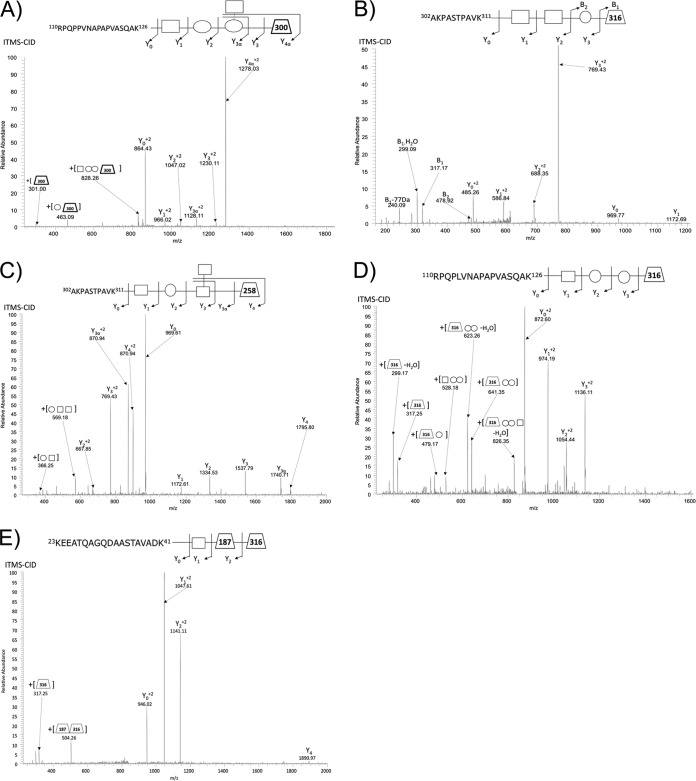
Fig. 1.
Major O-glycan structure identified using ZIC-HILIC enrichment. ITMS-CID fragmentation results in near exclusive glycan fragmentation of A. baumannii glycopeptides leading to the identification of A, the previously pentamer glycan (14) within strain ATCC 19606 (110RPQPPVNAPAPVASQAK126 of D0CDA9_ACIBA), B, a 4-mer glycan containing a 316 Da residue in strain SDF (302AKPASTPAVK311 of B0VKN6_ACIBS), C, a novel pentamer glycan within strain Arg1 containing a 258 residue (302AKPASTPAVK311 of A3M265_ACIBT), D, a novel 4-mer glycan within strain Arg2 (110RPQPLVNAPAPVASQAK126 of J5IPS4_ACIBA) containing a similar 316 Da residue to SDF, and E, the novel trisaccaride identified in strains 1441 C1 and C3 (23KEEATQAGQDAASTAVADK41 of A7FB63_ACIBT) containing the 316 Da residue of SDF and Arg 1.
In contrast, A. baumannii strains SDF and AYE lack the genes required for the generation of GlcNAc3NAcA4OAc (supplemental S. S1), and consequently, no potential glycopeptides could be identified using the GlcNAc3NAcA4OAc oxonium ion. To enable the detection of divergent glycan attachments, we assessed the presence of the oxonium ion 204.086 m/z, generated by the presence of HexNAc moieties within ZIC-HILIC enriched samples of A. baumannii. Despite multiple attempts, glycosylation could not be identified within strain AYE on either the peptide or protein level, using Periodic Acid Schiff's (PAS) staining. Interestingly, PAS staining also failed to identify the present of capsular polysaccharide (supplemental Fig. S2) suggesting the absence of products from the AYE carbohydrate locus. As the capsular polysaccharide is required for complement resistant (13) we assess the resistance of AYE to complement-mediated killing. Consistent with the lack of the capsular polysaccharide AYE was highly sensitive to complement-mediated killing, compared with ATCC 17978 and all recent clinical isolates examined (supplemental Fig. S3). The levels of sensitivity were also consistent with that of the known capsule mutant ATCC 17978 ΔpglC (supplemental Fig. S3) (13), supporting the absence of carbohydrate locus products within AYE.
Within SDF the use of ZIC_HILIC enrichment lead to the identification of 13 unique glycopeptides containing a tetrasaccharide glycan composed of HexNAc2-Hex-NulO (mass 884.34 Da) (Fig. 1B), where NulO corresponds to a 316.13 Da nonulosonic acid sugar. The identification of this unique glycan attached to five unique proteins (Table II and supplemental Table S3) both confirmed SDF has an active general O-linked glycosylation system and produces a glycan different to the pentasaccharide characterized in ATCC 17978 and 19606. Analysis of glycan related fragments within identified glycopeptides of SDF revealed the mass and isotopic distribution of the 317.13 m/z oxonium ion (the MH+ of the 316.13 residue, supplemental Fig. S4A), was consistent with legionaminic acid (elemental composition C13H21O7N2) (45). To further assess the identity of the 316 Da moiety, analysis of the low mass region of the HCD spectra was undertaken (supplemental Fig. S4B) confirming fragmentation consistent with that of the legionaminic or pseudaminic acid oxonium ion (45). These observations are in line with the genetic analysis of the glycan locus of SDF, which contains homologs for legionaminic acid biosynthesis as noted previously (12) (supplemental Fig. S1 and S4C).
Table II. Glycans Identified within Acinetobacter species. The 15 strains investigated within this study are denoted with the total number of glycans identified, number of shared glycans and the number of unique glycan also indicated for each strain. Annotated examples of each unique glycoform are provided within Fig. 2 to 5, 7 to 9 and supplementary Figs. Major glycans (define by the most observed glycan species) are denoted with*.
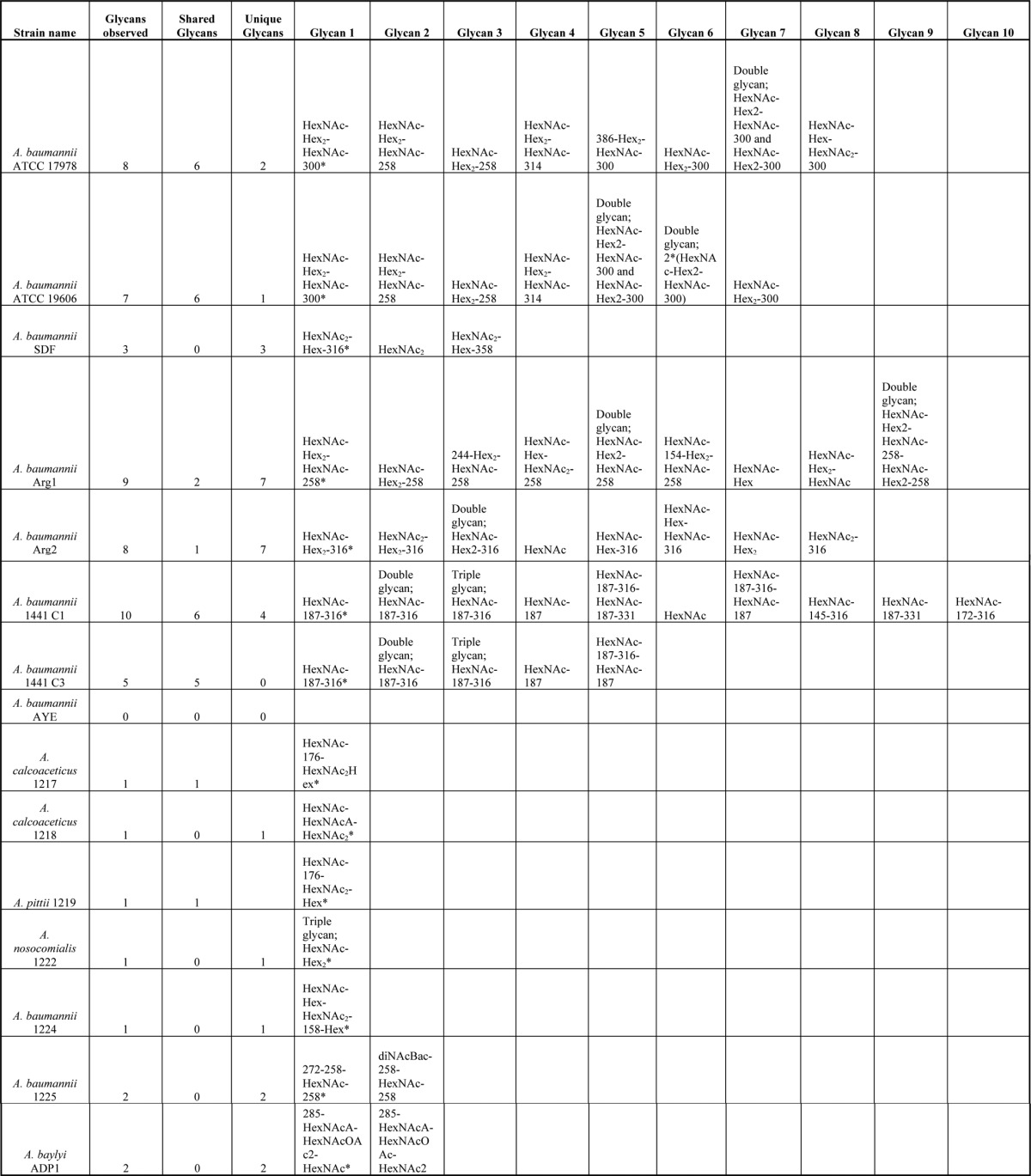
To further understand the O-linked glycosylation diversity and glycan characteristics within A. baumanii we analyzed four recent MDR clinical isolates (Arg1, Arg2, 1441 C1, and 1441 C3). As with SDF, the analysis of these strains revealed a diverse array of glycans with the major glycopeptides identified containing the glycans HexNAc-Hex2-HexNAc-258 (mass 988.34 Da), HexNAc-Hex2-NulO (mass 843.31 Da), and HexNAc-dHexNAc-NulO (mass 706.30 Da) within Arg1, Arg2, and 1441 C1 and 1441 C3 respectively (Fig. 1C–1E supplemental Tables S4–S7, and Table II). These glycopeptides corresponded to both previously identified O-glycosylation substrates and a multitude of novel protein acceptors (supplemental Tables S2 and S4–S7). Inspection of the mass and isotopic distribution of oxonium ions generated by these moieties (corresponding to the 258 and 316 Da residues) supported the assignments of these residues as a deaceylated form of GlcNAc3NAcA4OAc, a carbohydrate residue previously identified within the glycan of Acinetobacter lwoffii F78 (46), and legionaminic/pseudaminic acid (supplemental Fig. S5A, S5B). From these results, we confirm that diverse glycans are utilized in a range of strains, and that similar substrates are targeted for glycosylation across A. baumannii strains.
A. baumannii Strains Display Glycan Micro-heterogeneity Producing Multiple Related Glycan Structures
In addition to the major glycoform described above, at least three alternative glycoforms were unexpectedly identified within each A. baumannii strain, including SDF, Arg1, Arg2, 1441 C1, and 1441 C3 (Fig. 2A-J, Table II, supplemental Tables S3–S7). Glycan diversity appeared to be largely the result of chemical exchange and/or addition of functional groups such as acetyl and methyl groups to the bacterial specific residue observed within each strain, although examples of truncated glycan were also observed (Fig. 2A–2C). Inspection of the mass and isotopic distribution of oxonium ions generated by the modified bacterial specific residues (corresponding to the 331 and 358 Da residues) supported the assignments of these residues as methylated or acetylated forms of the nonulosonic sugar found in the major glycoform (supplemental Fig. S6). These modified forms although common and found on multiple peptide substrates appear less abundant, based on frequency of identification and ion intensity relative to the major glycoform within each strain (data not shown).
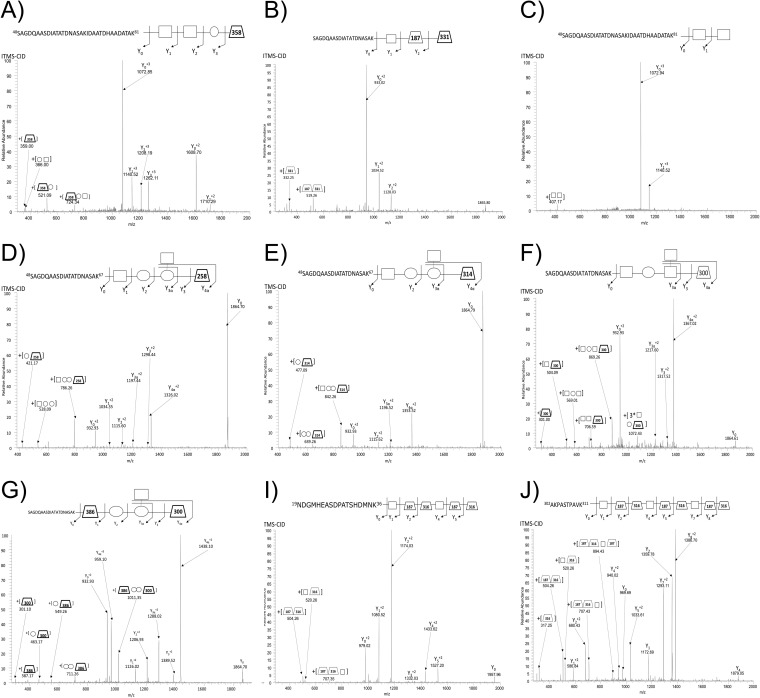
Fig. 2.
Alterative O-glycan structure identified using ZIC-HILIC enrichment with A. baumannii strains. ITMS-CID fragmentation enabled the identification of A. baumannii glycopeptides containing alterative glycoforms within all strains examined including: A, A tetramer glycan composed of HexNAc2-Hex-NulOAc within A. baumannii SDF (48SAGDQAASDIATATDNASAKIDAATDHAADATAK81 of B0VLI0_ACIBS), B, A trisaccharide composed of HexNAc-dHexNAc-NulOCH3 within A. baumannii 1441 C1 (48SAGDQAASDIATATDNASAK67 of A7FB63_ACIBT), C, A disaccharide composed of HexNAc2 within A. baumannii SDF (48SAGDQAASDIATATDNASAK67 of B0VLI0_ACIBS), D, A pentamer glycan within A. baumannii 19606 composed of HexNAc-Hex2-HexNAc-GlcNAc3NAcA (48SAGDQAASDIATATDNASAK67 of D0CEI7_ACIBA), E, A pentamer glycan within A. baumannii 19606 and 17978 composed of HexNAc-Hex2-HexNAc-GlcNAc3NAcA4OAcCH3 (48SAGDQAASDIATATDNASAK67 of A7FB63_ACIBT), F, The known pentamer glycan of A. baumannii ATCC 17978 where a Hex has been exchanged for a HexNAc residue compared with the previously reported glycan (14) (48SAGDQAASDIATATDNASAK67 of A7FB63_ACIBT), G, The known pentamer glycan of A. baumannii ATCC 17978 where the linking HexNAc residue has been exchanged for the unique 386.11 Da residues compared with the previously reported glycan (14) (48SAGDQAASDIATATDNASAK67 of A7FB63_ACIBT), I, A dimer of the trisaccharide O-glycan of A. baumannii 1441 C1 (19NDGMHEASDPATSHDMNK36 of A7FB95_ACIBT), and J, A trimer of the trisaccharide O-glycan of A. baumannii 1441 C1 (302AKPASTPAVK311 of A3M265_ACIBT).
As glycan heterogeneity was not considered during the initial analysis A. baumannii ATCC 17978 and ATCC 19606, re-inspection of ZIC-HILIC enrichment datasets were carried out leading to the confirmation of glycan heterogeneity within both strains (Fig. 2D, 2E; supplemental Tables S1, S2, S8, and Table II). The dominant glycan structure produced by ATCC 17978 and ATCC 19606 is HexNAc-Hex2-HexNAc-GlcNAc3NAcA4OAc, however, alternative glycans composed of HexNAc-Hex2-HexNAc-258 and HexNAc-Hex2-HexNAc-314 were also identified (Fig. 2D, 2E) in both strains (supplemental Table S1 and S8). Examination of the low mass region of the HCD spectrum within these alterative glycans revealed the presence of two novel ions corresponding to 259 and 315 m/z (consistent with MH+ of the 258.09 and 314.12 Da residues respectively, supplemental Fig. S7). The observed masses and isotopic distributions of these oxonium ions were consistent with a methylated form and a deacetylated form of GlcNAc3NAcA4OAc, of 315.11733 Da and 259.09112 Da, respectively (supplemental Fig. S7). Within ATCC 17978 we also noted the existence of two unique glycoforms that differed in the internal carbohydrate of the glycan, one corresponding to the exchange of a HexNAc for a Hex residue (Fig. 2F) and the other to the exchange of the linking carbohydrate for a moiety of 386 Da (Fig. 2G). The analysis of the low m/z region of the HCD spectra confirmed the presence of a 387.11937 Da MH+ ion and provided insight into the identity of this moiety. By comparing spectra generated from modified and non-modified glycans (supplemental Fig. S8A, S8B) a putative characterization was possible suggesting this moiety corresponds to a composition of C13H24N1O10S1 that is consistent with the mass and isotopic pattern of this residue. (supplemental Fig. S8C). A molecule with this formula has not been previously reported within a bacterial glycopeptide. This predicted composition does not match any known carbohydrate and appears to represent a minor O-linked glycoform of ATCC 17978.
It should be noted that in addition to heterogeneity because of the exchange and/or addition of functional groups within the glycan, polymerized forms of the O-linked glycan, glycan oligomers, were also observed within all strains (Table II). These glycan oligomers were present on numerous peptides (supplemental Tables S1, S3–S7, Table II, and supplemental Figs. S9–S23) with dimeric glycans (Fig. 2I) identified in all strains. In addition to glycan dimers, trimers (Fig. 2J) were also readily detectible within strains 1441 C1 and 1441 C3, which produces the smallest A. baumannii O-glycan characterized to date, a linear trisaccharide of 706.39 Da.
Acinetobacter baumannii O-glycosylates Multiple Conserved Protein Substrates via Serine Residues in Low-complexity Regions
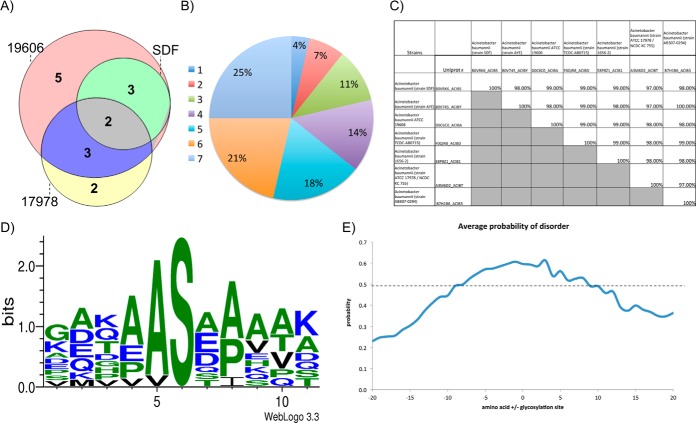
Examination of multiple A. baumannii strains expanded the repertoire of known glycosylation substrates adding 19 novel glycoproteins to the seven previously identified substrates (supplemental Table S2). Interestingly few glycoproteins identified were unique to a single strain; for example within the laboratory strains examined glycopeptides corresponding to the same eight glycoproteins were observed in SDF, ATCC 17978, and ATCC 19606 (Fig. 3A), whereas seven were unique to a single strain. This trend was seen across all A. baumannii strains examined where ∼70% of all glycoproteins were identified within at least two strains (Fig. 3B). These observations in combination with genetic analyses of multiple genome-sequenced strains of A. baumannii confirm that the identified glycoproteins are both conserved and targeted for glycosylation across divergent strains (supplemental Table S9). An example of this is the conserved, putative uncharacterized protein D0C6C0_ACIBA (Fig. 3C), which was identified in five out of the seven strains examined (supplemental Table S2). Interestingly, it was also noted that the observed glycoproteins are unique to Acinetobacter, with no homologs outside of this genus, and are highly conserved with >97% identity between strains (supplemental Table S9). No homologs were found in any other bacterium and therefore their function cannot be assigned based on homology.
Fig. 3.
A. baumannii prominently glycosylates Serine residues with Alanine residues in the −1 position. Analysis of glycoproteins and glycosylation sites identified of A. baumannii strains. A, Comparison of glycoproteins identified within each strain. Showing the high level of overlap between strains. B, Analysis of the overlap between strains glycoproteins identified in all seven A. baumannii strains. C, Comparison of the sequence identity of proteins between strains of A. baumannii demonstrating the high level of sequence identity between strains of the identified glycoproteins. D, Motif analysis of identified glycosylation sites showing a strong preference for the sequence AS. E, Comparison of the region of disorder around the identified sites of glycosylation, a disordered prediction >0.5 is consisted to be disordered according to PreDisorder, http://casp.rnet.missouri.edu/predisorder.html.
To accurately assess the local environment of glycosylation, based on the site of attachment, the sites of modification were characterized using ETD fragmentation (supplemental Table S10). Using this approach a total of seven sites of glycosylation could be localized, which in conjunction with four additional sites localized by the presence of only one hydroxyl containing amino acid in the sequence, lead to the localization of eleven glycosylation sites across the examined A. baumannii strains (supplemental Table 10). Within this dataset it was noted that only Serine appeared to be glycosylated within the identified glycopeptides, and that the sites of glycosylation seem to have a strong preference for alanine in the −1 position (Fig. 3D, supplemental Table S10). Because previous reports have suggested that glycosylation occurs at disordered regions in other bacterial O-linked glycosylation systems rather than at a specific sequon (47–49), we also examined the region surrounding the identified glycosylation sites. Similar to other bacterial O-linked systems, sites of A. baumannii O-linked glycosylation occur in low complexity regions, rich in proline, alanine, and serine (Fig. 3E), suggesting that the recognition of substrates by O-oligosaccharyltransferases is conserved in different bacteria.
Targeted Analysis of the Glycan Diversity in Acinetobacter Clinical Isolates
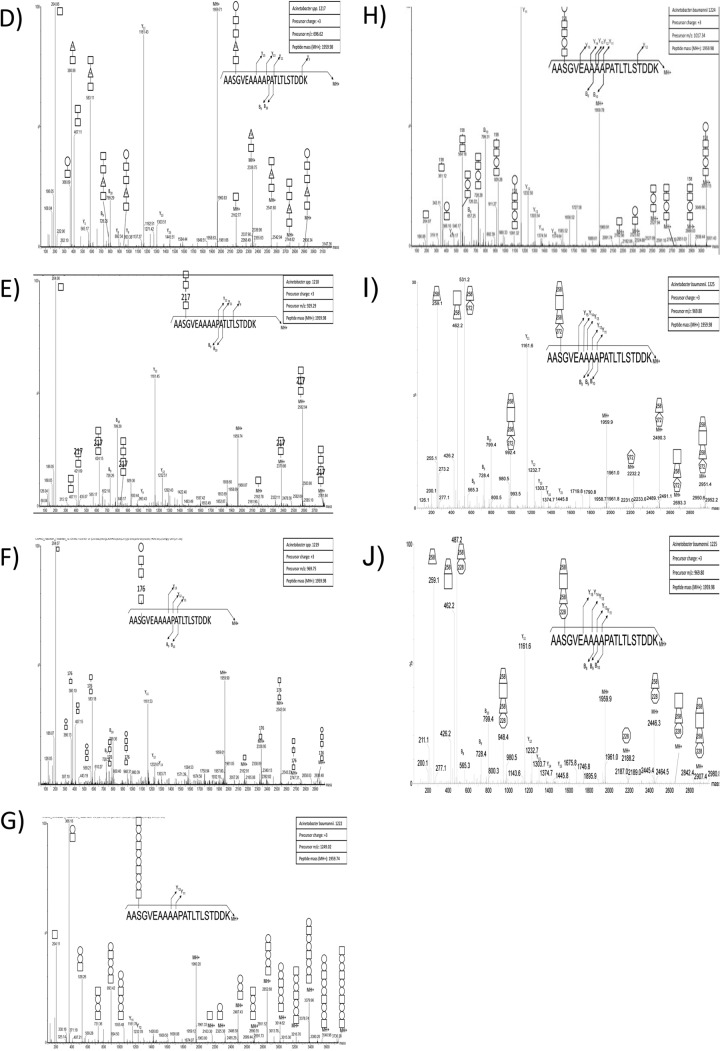
Although the results of the ZIC-HILIC enrichment confirmed our hypothesis of O-linked diversity within multiple strains, it also highlighted potential variability in the performance of ZIC-HILIC enrichment. As these methodologies rely on the ability to detect glycosylation based on the presence of diagnostic carbohydrate reporter ions, we reasoned that if strains possessed a glycan lacking known carbohydrate moieties or were ineffectively enriched with ZIC-HILIC, detection and determination of glycosylation would be compromised. In order to refine the analysis of glycan diversity and expand the number of strains analyzed, we developed a targeted approach to assess glycosylation. Based on the observation that the same proteins are glycosylated at sites with similar structural characteristics in multiple strains (Fig. 3A, 3D, and 3E), we reasoned that a His-tagged version of one of these conserved proteins could be expressed in other Acinetobacter strains and used as bait in order to isolate attached glycans. To achieve this goal we expressed the A. baumannii glycoprotein A1S_1193 in clinically relevant strains. Previous work from our lab showing the site of glycosylation within A1S_1193 is Serine-205 (28), contained within the tryptic peptide 203AASGVEAAAAPATLTLSTDDK223 facilitated characterization of the corresponding glycans (14). Expression of A1S_1193 leads to the decoration of the protein substrate with native glycosylation, enabling the isolation of the protein independent of the chemical properties of the sugars. Importantly, this approach does not require prior knowledge of the genomic sequence or glycan structure, enabling the assessment of glycosylation in unsequenced clinical strains.
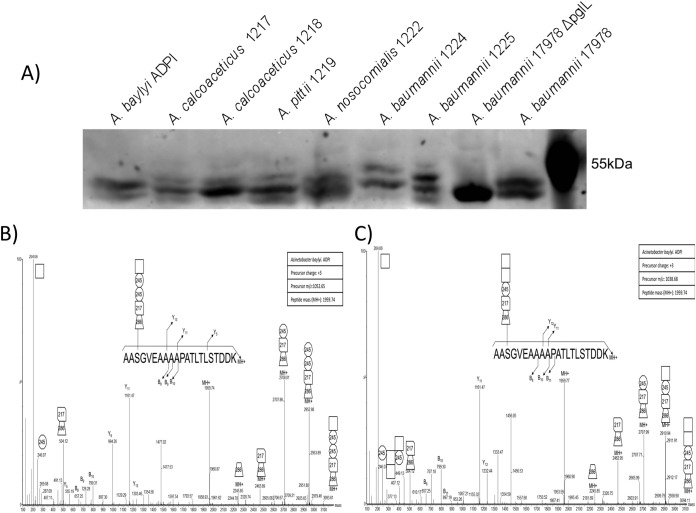
To investigate glycan diversity in the clinical setting, six clinical Acinetobacter isolates were selected for analysis using this targeted approach. The expression of A1S_1193 within these Acinetobacter clinical isolates lead to a detectible mass shift compared with the non-glycosylated control (Fig. 4A) suggesting glycosylation of the protein substrates. MS analysis confirmed the addition of glycan structures to A1S_1193 with a total of six novel glycan structures observed across the six strains (Fig. 4D–4J, Table II). From this analysis it was noted that all glycans were composed of four to six carbohydrates and were largely linear in nature with few branched sugars observed. As with SDF, ATCC 19606, and ATCC 17978, glycan heterogeneity was also observed within clinical isolates; for example within A. baumannii 1225 two glycans composed of 272–258-HexNAc-258 and 228–258-HexNAc-258 were identified (Fig. 5F, 5G). As with previously identified heterogeneity the alteration in A. baumannii 1225 occurred on the bacterial specific residues corresponding to the mass of diNAcBac (228 Da; bacillosamine). Interestingly the glycans of A. baumannii 1225 appear similar to the previously characterized glycans of A. lwoffii F78 and A. baumannii AB307–0294 where NMR was utilized to confirm the modification of the capsule with the deacetylated form of GlcNAc3NAcA4OAc (258 Da) or diNAcBac (228 Da) and BacNAc modified with 3-OH-butyrate (272 Da) respectively (46, 50). Furthermore sequencing of A. baumannii 1225 isolate has confirmed the presence of the genes necessary for GlcNAc3NAcA4OAc and diNAcBac biosynthesis supporting the assignment of these bacterial specific carbohydrates (supplemental Fig. S1C).
Fig. 4.
Western blot analysis used to resolve the mass difference between glycosylated and unglycosylated A1S_1193. A, Anti-Histidine Western blot analysis of Acinetobacter strains recombinantly expressing Acinetobacter glycoprotein A1S_1193 with a C-terminal Histidine tag. The slight increase in molecular weight indicates the protein has been post-translationally modified. B–J, ESI-QTOF MS/MS Analysis of the fished A. baumannii glycoprotein A1S_1193 to elucidate the glycan structure. ESI-QTOF-MS and MS/MS was carried out on purified A1S_1193, expressed in various Acinetobacter strains, to characterize the posttranslational modification. B–C, ESI-QTOF-MS/MS analysis of tryptic peptide 203AASGVEAAAAPATLTLSTDDK223 expressed in A. baylyi ADP1 revealed either the pentasaccharide 285-217–2452-HexNAc or 285-217–245-HexNAc2 attached to the glycopeptide. D, MS/MS fragmentation of 203AASGVEAAAAPATLTLSTDDK223 expressed in A. calcoaceticus 1217 displays modification with the pentasaccharide HexNAc-176-HexNAc2-Hex. E, A. calcoaceticus 1218 glycosylates the tryptic peptide 203AASGVEAAAAPATLTLSTDDK223 with the tetrasaccharide HexNAc-217-HexNAc2. F, Fragmentation of tryptic peptide 203AASGVEAAAAPATLTLSTDDK223 from A1S_1193 expressed in A. pittii 1219 reveals glycosylation with a pentasaccharide identical to 1217, HexNAc-176-HexNAc2-Hex. G, A. nosocomialis 1222 modifies glycopeptide 203AASGVEAAAAPATLTLSTDDK223 with the trisaccharide repeat unit HexNAc-Hex2. H, A. baumannii 1224 uses the hexasaccharide HexNAc-Hex-HexNAc2-158-Hex to glycosylate glycopeptide 203AASGVEAAAAPATLTLSTDDK223. I–J, A. baumannii 1225 modifies glycopeptide 203AASGVEAAAAPATLTLSTDDK223 with one of two tetrasaccharides, 272–258-HexNAc-258 or 228–258-HexNAc-258.
Not all O-glycan Structures within Acinetobacter Match the Predicted Carbohydrate Synthesis Locus: The Investigation of A. baylyi ADP1 Glycosylation
With the development of a targeted approach to investigate Acinetobacter O-linked glycosylation we also assessed its potential to characterize the O-linked glycan of the non-pathogenic model strain A. baylyi ADP1. This bacterium is frequently employed as a model Acinetobacter strain because of its amenability to genetic manipulation (51). Interestingly A. baylyi ADP1 was recently demonstrated to contain a functional glycosylation system (26), although the exact structure has not been elucidated. Using our targeted approach A. baylyi ADP1 was found to glycosylate A1S_1193 with a pentasaccharide composed of 285–217-2452-HexNAc or 285–217-245-HexNAc2, which is distinct from the capsule subunit of its parent strain A. baylyi BD4 (Fig. 4B, 4C, Table II). The residues of this glycan included two atypical sugars of mass 217 and 245 Da matching 2-acetamido-2-deoxy-d-hexuronic acid and O-acetyl-N-acetylhexosamine moieties respectively, which have also been recently identified within multiple Campylobacter species (31). Furthermore within A. baumannii AB307–0294 a form of hexuronic acid, galactosaminuronic acid, has been previously noted (50). This observation demonstrates the biosynthetic pathways required to generate the underivatized precursor of the 217 Da do exist in Acinetobacter. The identification of a shortened glycan composed of chemically unusual moieties demonstrated the importance of experimental analysis of glycan structure, rather than bioinformatic prediction and the convenience of our targeted based approach to rapidly identify glycosylation in the Acinetobacter genus.
DISCUSSION
Protein O-glycosylation is a common process in bacterial species, and is required for virulence and biofilm formation in A. baumannii ATCC 17978. However, the prevalence, diversity, and specific role of the O-glycan modification of A. baumannii has yet to be determined (14). Lees-Miller et al. showed that in the strain ATCC 17978 the building blocks employed for capsular polysaccharide and the O-linked glycan are identical and employ the same enzymatic machinery for their synthesis, which is encoded in a single glycan locus (13). Genetic comparison of the capsular loci revealed extensive variability in the predicted capsule structure (11). In this work, we analyzed the glycan structures attached to proteins in the strains most commonly used for molecular studies as well as ten clinical isolates and confirmed the presence of extensive O-linked glycan diversity. From this analysis a diverse array of glycans were observed across all 15 strains examined with a total of 11 unique main glycoforms identified (Tables II). These observations support the predictions of Hu et al. and the presence of extensive glycan diversity in A. baumannii. In addition to the prominent structures, alternative O-glycans were also identified demonstrating A. baumannii is capable of producing multiple glycoforms within a given strain. We observed that the majority of strains produced an array of unique glycans only found within that strain; exceptions to this are the reference strain ATCC 19606 that produced the identical O-linked pentasaccharide containing GlcNAc3NAcA4OAc of ATCC 17978 (Fig. 1A, Table II) and the closely related isolates 1441 C1 and 1441 C3 (Fig. 1E, Table II). Interestingly, the pentasacccharide of ATCC 17978 and ATCC 19606 has also been identified as the capsule-repeat unit of A. baumannii strain SMAL (52). Although the frequency of the observed GlcNAc3NAcA4OAc containing structure would suggest the commonality of this sugar in A. baumannii, both the recent bioinformatics analyses (11, 12) and this work suggest that the pentasacccharide of ATCC 17978 (13, 14), ATCC 19606, and SMAL (52) is just one of the multitude of glycans utilized by A. baumannii.
Although the majority of glycans were unique to specific strains, multiple glycans contained common bacterial specific sugars such as in the case of the reference strain SDF and clinical isolates Arg2, 1441 C1, and 1441 C3. Within these strains, multiple glycopeptides were identified decorated with residues matching the mass and fragmentation pattern of the negatively charged sugar, legionaminic or its stereoisomer pseudaminic acid (Fig. 1B, 1D, and 1E) (45). Bioinformatic analysis of the carbohydrate clusters of SDF as well as the unpublished clinical isolates A. baumannii 1441 C1 and C3 (Weber et al. unpublished data) supports the presence of the legionaminic acid biosynthesis pathway within these strains (supplemental Fig. S1B and S4C). This finding demonstrates A. baumannii is one of the many bacterial species now recognized to utilize legionaminic/pseudaminic acid or their derivatives within protein attached glycans (45, 53, 54). Within the best characterized of these systems, the O-linked glycosylation system of C. jejuni, these sialic acid analogs are required for ideal protein function where they are essential for autoagglutination, modulation of the hydrophobicity of the flagellin and dampening of the inflammation response by SigLec-10 binding (55–57). Interestingly, the presence of negatively charged sugars appears to be a common feature for most if not all the A. baumannii structures identified (Table II). From this trend it is tempting to speculate that these negative sugars may be advantages to A. baumannii biology and themselves may be important for virulence as seen for other bacterial species where negative surface carbohydrates can provide resistance to complement killing (58).
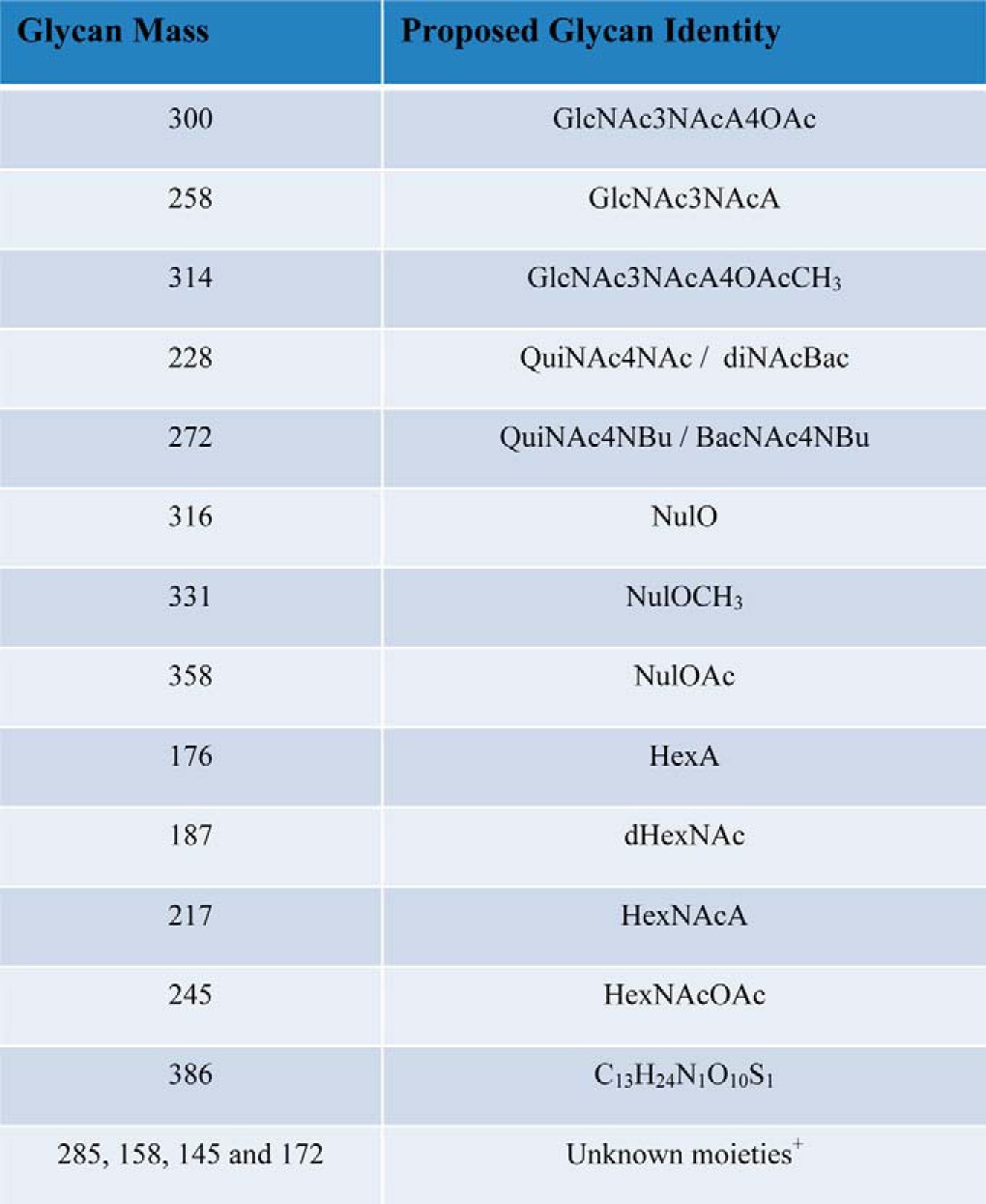
In addition to legionaminic acid, other carbohydrates such as the HexNAcA, HexNAcOAc, and diNAcBac residues, whose identity is assigned based on mass comparison to characterized glycans of bacterial glycosylation systems (31), were observed across multiple strains within this study (Table III). Residues with identical masses have been identified as components of glycans utilized within other bacterial protein glycosylation systems (47, 59–61) suggesting A. baumannii utilizes a similar carbohydrate repertoire as other Gram-negative protein glycosylation systems. Although the identities of these residues cannot be confirmed from the mass alone the convergence of similar residues, irrespective of their stereochemistry, is an observation of significant note. A similar concept of the utilization of a conserved carbohydrate repertoire has been noted within Neisseria and Campylobacter species where both systems utilize diBacNAc yet the enzymes responsible for the generation of diBacNAc represent two phylogenetically distinct clades (62–64). Widespread use of these unique carbohydrates by multiple bacterial glycosylation systems suggests preference for these sugars in protein modification, although the exact advantages of these residues are unknown. If specific carbohydrates are advantageous for virulence or glycosylation functionality this preference may be exploited by potential anti-microbial therapies and the generation of serological reagents, which may aid in the diagnosis and treatment of A. baumannii infections and is currently under investigation within our laboratory (65, 66).
Table III. Carbohydrates identified in the O-linked glycan of Acinetobacter species. Nominal mass of unique sugars identified within Acinetobacter O-linked glycans are provided as well as proposed glycan identities. +Unknown moieties correspond to residues where high mass accuracy measurements (<2 ppm) could not be obtained due to the 200 m/z HCD MWCO or data being acquired on a lower mass accuracy instrument.
With the exception of AYE, at least one glycan structure could be identified within each of the examined strains, with the presence of multiple alternative glycan forms being a common feature of A. baumannii strains. Although bacterial glycan heterogeneity is poorly understood it has been suggested that it could contribute to immune evasion (37) Similar to previously observed heterogeneity in the capsule structure of Acinetobacter strains (46, 52) glycan diversity appeared to be largely the result of chemical exchange and/or addition of functional groups such as acetyl and methyl groups to the bacterial specific residue observed within each strain, although examples of truncated glycan were also observed (Fig. 2A–2C). These alterations were not uniformly distributed on all carbohydrate residues but appeared to favor the alteration of bacterial specific carbohydrates related to diNAcBac, GlcNAc3NAcA4OAc, and legionaminic/pseudaminic acid. Within these sugars multiple alterations were associated with augmentation of the levels of acetylation (Table II). This observation is of key interest because of the association of acetylation levels in other capsule systems and resistance to complement mediated killing (58, 67, 68). As the capsule is essential in ATCC 17978 for complement resistance (13) variability in capsule structure may alter the levels of resistance, which may influence virulence. In addition to the exchange and addition of chemical functional groups, variations such as the addition or lack of sugars as well as changes in the order of the sugars (Fig. 2, supplemental Figs. S9–S23) were also observed across strains. These variations suggest that unlike the archetypical glycosylation system of C. jejuni, which only utilizes complete correctly formed glycans, A. baumannii, is more promiscuous with glycans utilized for protein modification. This promiscuous nature would be in line with other O-linked glycosylation systems such as that of Neisseria, which change the O-linked glycans because of phase variation of glycosyltransferases involved in the assembly of the glycan to aid in immune evasion (69).
In addition to the diversity resulting from the exchange/alteration of carbohydrates within glycans we also observed heterogeneity in the form of glycan oligomerization (Fig. 2I and 2J, Table II). This finding is in agreement with the previous work of Lees-Miller et al. (13) showing that polymerized capsule subunits can be attached to protein substrates in ATCC 17978, and support the notion that the shared glycan biosynthesis pathway for capsule and protein glycosylation is a general feature of A. baumannii. Interestingly the observed diversity within Acinetobacter glycans is consistent with the micro-heterogeneity recently noted in the glycosylation systems of other bacteria genera, such as Campylobacter, Burkholderia, and Francisella (32–39). These observations suggest that the majority of glycosylation systems utilize a range of related glycans with proteins substrates rarely decoration with a homogenous glycan. Although the physiological significance of the glycan micro-heterogeneity is still unknown, the extent of glycan microdiversity seen within Acinetobacter is among the highest reported to date (32–39).
Within this study glycosylation was detected in all strains examined except the multiple drug resistant strain A. baumannii AYE. As previously noted (11, 12), this strain carries a glycan cluster containing three glycosyltransferases and the genes required for the synthesis of diBacNAc, which has been demonstrated to lead to the generation of UDP-diBacNAc in heterogeneous expression systems (50). Because of the multiple drug resistant nature of AYE targeted glycosylation could not be undertaken leaving only enrichment of glycopeptides by ZIC-HILIC chromatography, which is a variable means to assess glycosylation. We initially reasoned that our inability to identify glycosylation within AYE using ZIC-HILIC enrichment may be the result of technical caveats including; the oligosaccharides may have failed to alter glycopeptides hydrophilicity sufficiently to enable partitioning to the pseudo-water of the ZIC-HILIC stationary phase (70); or the resulting glycan may have lacked diagnostics ions used to identify glycosylation within other strains. Conversely it is also possible that the glycosylation machinery in this strain is regulated, and only present under certain growth conditions or functionally inactive. However, the absence of PAS reactive capsular polysaccharide and glycoproteins in this strain, as well as its sensitivity to serum killing provides additional evidence that this strain does not produce surface carbohydrates under the conditions tested. The sensitivity of AYE to serum killing is in contrast to the resistance exhibited by recent clinical isolates (supplemental Fig. S3), suggesting without selective pressure A. baumannii may regulate or lose surface carbohydrate expression. The loss of surface carbohydrates in the laboratory has been noted in numerous bacterial species. For example, common E. coli K12 strains do not produce O antigen because of the mutation in a rhamnosyltransferase (71) Another example is the human pathogen Burkholderia cenocepacia strain J2315 that is extremely sensitive to human serum because of an insertion of IS402 within the glycosyl transferase wbxE (72). The requirement of capsular polysaccharide for serum resistance demonstrates the significant biological role for surface carbohydrates in the virulence and infective lifecycle of A. baumannii (13) and the importance for experimental elucidation of both glycans and there phenotypic roles.
Although all strains examined supported our hypothesis of diversity in the O-linked glycan, the agreement between the carbohydrate locus and the glycan structure was not always consistent. The large 34 ORF carbohydrate locus of A. baylyi ADPI encodes the TDP-rhamnose biosynthetic pathway, which is not necessary for production of the pentasaccharides we detected, that did not contain rhamnose. The locus encodes 10 glycosyltransferases, when only five would be needed for the pentasaccharide (supplemental Fig. S1 and Figs. 4B–4C). The O-linked glycan has an unusual sugar composition consisting of monosaccharides that match residues observed in other recently identified Campylobacter glycosylation systems as well as the A. baumannii capsule glycan of AB307–0294 (31, 50). As the sugars corresponding to the unusual masses are unknown, one possibility is that this strain generates one or multiple residues from a rhamnose based precursor. However, the capsule of the closely related strain A. baylyi BD4, which ADP1 is derived from (73), has been shown to contain l-rhamnose, d-glucose, d-glucuronic acid, and d-mannose (43). Therefore, it is tempting to speculate that in contrast to A. baumannii, ADP1 produces a capsule that is unrelated to the O-glycan. Consistent with this, ADP1 was the only strain that did not show evidence of higher oligomer glycan by Western blotting of the A1S_1193 bait (Fig. 4A). Additionally, the ADPI glycan cluster contains two initiating glycosyltransferase homologs suggesting the potential to produce two unique lipid-linked glycans, one for protein modification and one for capsule (supplemental Fig. S1). Additional work is required to further explore these observations to confirm the segregation of the capsule and O-glycan, as it will be interesting to know if and how the two pathways are compartmentalized to avoid possible crosstalk between them.
The ability to assess heterogeneity within this work was because of the use of both targeted and non-targeted technologies for bacterial glycosylation analysis. The ZIC-HILIC approach provides a non-targeted means to assess glycan diversity but requires significant experience in glycopeptide analysis to elucidate sugar structures as well as instrumentation capable of performing multiple fragmentation approaches. To overcome these shortcomings we developed a method to simplify glycosylation analysis that could be achieved on routine MS instrumentation. This method was developed based on the observation of conservation within proteins subjected to glycosylation and the structural properties of these proteins within Acinetobacter glycosylation. In this approach, a common tagged-glycosylation acceptor protein is introduced as “bait” into the strain of interest. The glycosylated protein is then purified via affinity chromatography, digested and subjected to MS analysis. This approach provides a scalable and optimizable means to produce glycosylated proteins without prior knowledge of the glycan structure, allowing analysis of un-sequence clinical strains. As the fragmentation pattern of the “bait” is known, the determination of the glycans attached to it is straightforward. This targeted approach was employed here to characterize the sugar composition of seven different Acinetobacter strains. We believe that in the future this strategy could be employed to simplify the characterization of other bacterial glycosylation systems. By using both targeted and non-targeted MS approaches we show extensive diversity exists in the Acinetobacter glycoproteome; that at least 26 proteins, most of which are unique to Acinetobacter, are subjected to O-linked glycosylation; and, further demonstrate that the use of a single glycan for both O-linked glycosylation and capsule production appears to be a general feature of A. baumannii.
Supplementary Material
Footnotes
Author contributions: N.E.S., R.L.K., and M.F.F. designed research; N.E.S., R.L.K., A.V.E., and S.M.D. performed research; M.R.L., S.M.D., J.S., and L.J.F. contributed new reagents or analytic tools; N.E.S., R.L.K., and M.F.F. analyzed data; N.E.S., R.L.K., and M.F.F. wrote the paper; N.E.S. and R.L.K. contributed equally to this work.
* This work was supported by funds provided by the Natural Sciences and Engineering Research Council of Canada (NSERC) to MFF. MFF is a CIHR Investigator. N.E.S. was supported by a National Health and Medical Research Council of Australia (NHMRC) Overseas (Biomedical) Fellow (APP1037373) and is currently a Michael Smith Foundation for Health Research Trainee Post-Doctoral Fellow (award # 5363). Operational funding support for N.E.S. and L.J.F. was provided by CIHR Operating grant # MOP-77688. RLK holds an NSERC fellowship.
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S10.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S10.
1 The abbreviations used are:
- MDR
- multiple drug resistant
- CID
- collision induced dissociation
- HCD
- higher energy collisional dissociation
- ETD
- electron transfer dissociation
- LC
- liquid chromatography
- MS
- mass spectrometry
- ZIC-HILIC
- zwitterionic hydrophilic interaction liquid chromatography.
REFERENCES
- 1. Gaynes R., Edwards J. R. (2005) Overview of nosocomial infections caused by gram-negative bacilli. Clin. Infect. Dis. 41, 848–854 [DOI] [PubMed] [Google Scholar]
- 2. Cisneros J. M., Rodriguez-Bano J. (2002) Nosocomial bacteremia due to Acinetobacter baumannii: epidemiology, clinical features, and treatment. Clin. Microbiol. Infect. 8, 687–693 [DOI] [PubMed] [Google Scholar]
- 3. Peleg A. Y., Seifert H., Paterson D. L. (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maragakis L. L., Perl T. M. (2008) Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 46, 1254–1263 [DOI] [PubMed] [Google Scholar]
- 5. Towner K. J. (2009) Acinetobacter: an old friend, but a new enemy. J. Hosp. Infect. 73, 355–363 [DOI] [PubMed] [Google Scholar]
- 6. Russo T. A., Luke N. R., Beanan J. M., Olson R., Sauberan S. L., MacDonald U., Schultz L. W., Umland T. C., Campagnari A. A. (2010) The K1 capsular polysaccharide of Acinetobacter baumannii strain 307–0294 is a major virulence factor. Infect. Immun. 78, 3993–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moffatt J. H., Harper M., Mansell A., Crane B., Fitzsimons T. C., Nation R. L., Li J., Adler B., Boyce J. D. (2013) Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect. Immun. 81, 684–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henry R., Vithanage N., Harrison P., Seemann T., Coutts S., Moffatt J. H., Nation R. L., Li J., Harper M., Adler B., Boyce J. D. (2012) Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 56, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reisfeld A., Rosenberg E., Gutnick D. (1972) Microbial degradation of crude oil: factors affecting the dispersion in sea water by mixed and pure cultures. Appl. Microbiol. 24, 363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakar D., Gutnick D. L. (2001) Analysis of the wee gene cluster responsible for the biosynthesis of the polymeric bioemulsifier from the oil-degrading strain Acinetobacter lwoffii RAG-1. Microbiology 147, 1937–1946 [DOI] [PubMed] [Google Scholar]
- 11. Hu D., Liu B., Dijkshoorn L., Wang L., Reeves P. R. (2013) Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PloS One 8, e70329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kenyon J. J., Hall R. M. (2013) Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PloS One 8, e62160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lees-Miller R. G., Iwashkiw J. A., Scott N. E., Seper A., Vinogradov E., Schild S., Feldman M. F. (2013) A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol. Microbiol. 89, 816–830 [DOI] [PubMed] [Google Scholar]
- 14. Iwashkiw J. A., Seper A., Weber B. S., Scott N. E., Vinogradov E., Stratilo C., Reiz B., Cordwell S. J., Whittal R., Schild S., Feldman M. F. (2012) Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 8, e1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 16. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St Michael F., Aberg E., Szymanski C. M. (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]
- 17. Stimson E., Virji M., Makepeace K., Dell A., Morris H. R., Payne G., Saunders J. R., Jennings M. P., Barker S., Panico M., Blench I., Moxon R.E. (1995) Meningococcal pilin: a glycoprotein substituted with digalactosyl 2,4-diacetamido-2,4,6-trideoxyhexose. Mol. Microbiol. 17, 1201–1214 [DOI] [PubMed] [Google Scholar]
- 18. Iwashkiw J. A., Vozza N. F., Kinsella R. L., Feldman M. F. (2013) Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol. Microbiol. 89, 14–28 [DOI] [PubMed] [Google Scholar]
- 19. Nothaft H., Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 20. Larsen M. R., Jensen S. S., Jakobsen L. A., Heegaard N. H. (2007) Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Mol. Cell. Proteomics 6, 1778–1787 [DOI] [PubMed] [Google Scholar]
- 21. Hagglund P., Bunkenborg J., Elortza F., Jensen O. N., Roepstorff P. (2004) A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3, 556–566 [DOI] [PubMed] [Google Scholar]
- 22. Zielinska D. F., Gnad F., Wisniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 23. Parker B. L., Thaysen-Andersen M., Solis N., Scott N. E., Larsen M. R., Graham M. E., Packer N. H., Cordwell S. J. (2013) Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J. Proteome Res. 2, 5791–5800 [DOI] [PubMed] [Google Scholar]
- 24. Kolarich D., Jensen P. H., Altmann F., Packer N. H. (2012) Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 7, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 25. Jensen P. H., Karlsson N. G., Kolarich D., Packer N. H. (2012) Structural analysis of N- and O-glycans released from glycoproteins. Nat. Protoc. 7, 1299–1310 [DOI] [PubMed] [Google Scholar]
- 26. Schulz B. L., Jen F. E., Power P. M., Jones C. E., Fox K. L., Ku S. C., Blanchfield J. T., Jennings M. P. (2013) Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PloS One 8, e62768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ding W., Nothaft H., Szymanski C. M., Kelly J. (2009) Identification and quantification of glycoproteins using ion-pairing normal-phase LC and MS. Mol. Cell. Proteomics 8, 2170–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Madsen J. A., Ko B. J., Xu H., Iwashkiw J. A., Robotham S. A., Shaw J. B., Feldman M. F., Brodbelt J. S. (2013) Concurrent automated sequencing of the glycan and peptide portions of O-linked glycopeptide anions by ultraviolet photodissociation mass spectrometry. Anal. Chem. 85, 9253–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Hojrup P., Packer N. H., Larsen M. R., Cordwell S. J. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, HCD and ETD-MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10.1074/mcp.M000031-MCP201-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Hojrup P., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by cid, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031MCP000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nothaft H., Scott N. E., Vinogradov E., Liu X., Hu R., Beadle B., Fodor C., Miller W. G., Li J., Cordwell S. J., Szymanski C. M. (2012) Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol. Cell. Proteomics 11, 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scott N. E., Nothaft H., Edwards A. V., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., Cordwell S. J. (2012) Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287, 29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lithgow K. V., Scott N. E., Iwashkiw J. A., Thomson E. L. S., Foster L. J., Feldman M. F., Dennis J. J. (2014) A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol. Microbiol. 92, 116–137 [DOI] [PubMed] [Google Scholar]
- 34. Nothaft H., Scott N. E., Vinogradov E., Liu X., Hu R., Beadle B., Fodor C., Miller W. G., Li J., Cordwell S. J., Szymanski C. M. (2012) Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol. Cell. Proteomics 11, 1203–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Whitfield C., Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem 83, 99–128 [DOI] [PubMed] [Google Scholar]
- 36. Szymanski C. M., Michael F. S., Jarrell H. C., Li J., Gilbert M., Larocque S., Vinogradov E., Brisson J. R. (2003) Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278, 24509–24520 [DOI] [PubMed] [Google Scholar]
- 37. Borud B., Viburiene R., Hartley M. D., Paulsen B. S., Egge-Jacobsen W., Imperiali B., Koomey M. (2011) Genetic and molecular analyses reveal an evolutionary trajectory for glycan synthesis in a bacterial protein glycosylation system. Proc. Natl. Acad. Sci. U. S. A. 108, 9643–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 39. Wang N., Mackenzie L., De Souza A. G., Zhong H., Goss G., Li L. (2007) Proteome profile of cytosolic component of zebrafish liver generated by LC-ESI MS/MS combined with trypsin digestion and microwave-assisted acid hydrolysis. J. Proteome Res. 6, 263–272 [DOI] [PubMed] [Google Scholar]
- 40. Pessione E., Pessione A., Lamberti C., Coisson D. J., Riedel K., Mazzoli R., Bonetta S., Eberl L., Giunta C. (2009) First evidence of a membrane-bound, tyramine, and beta-phenylethylamine producing, tyrosine decarboxylase in Enterococcus faecalis: a two-dimensional electrophoresis proteomic study. Proteomics 9, 2695–2710 [DOI] [PubMed] [Google Scholar]
- 41. Good D. M., Wenger C. D., Coon J. J. (2009) The effect of interfering ions on search algorithm performance for electron-transfer dissociation data. Proteomics 10, 164–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Good D. M., Wenger C. D., McAlister G. C., Bai D. L., Hunt D. F., Coon J. J. (2009) Post-acquisition ETD spectral processing for increased peptide identifications. J. Am. Soc. Mass Spectrom. 20, 1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 44. Neuhauser N., Michalski A., Cox J., Mann M. (2012) Expert system for computer assisted annotation of MS/MS spectra. Mol. Cell. Proteomics 11, 1500–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., Guerry P. (2001) Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870 [DOI] [PubMed] [Google Scholar]
- 46. Hanuszkiewicz A., Kaczyński Z., Lindner B., Goldmann T., Vollmer E., Debarry J., Heine H., Holst O. (2008) Structural analysis of the capsular polysaccharide from Acinetobacter lwoffii F78. Eur. J. Org. Chem. 2008, 6183–6188 [Google Scholar]
- 47. Vik A., Aas F. E., Anonsen J. H., Bilsborough S., Schneider A., Egge-Jacobsen W., Koomey M. (2009) Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 106, 4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Slynko V., Schubert M., Numao S., Kowarik M., Aebi M., Allain F. H. (2009) NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation. J. Am. Chem. Soc. 131, 1274–1281 [DOI] [PubMed] [Google Scholar]
- 49. Rangarajan E. S., Bhatia S., Watson D. C., Munger C., Cygler M., Matte A., Young N. M. (2007) Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni. Protein Sci. 16, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Russo T. A., Beanan J. M., Olson R., MacDonald U., Cox A. D., St Michael F., Vinogradov E. V., Spellberg B., Luke-Marshall N. R., Campagnari A. A. (2013) The K1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect. Immun. 81, 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Metzgar D., Bacher J. M., Pezo V., Reader J., Doring V., Schimmel P., Marliere P., de Crecy-Lagard V. (2004) Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 32, 5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fregolino E., Gargiulo V., Lanzetta R., Parrilli M., Holst O., Castro C. D. (2011) Identification and structural determination of the capsular polysaccharides from two Acinetobacter baumannii clinical isolates, MG1, and SMAL. Carbohydr. Res. 346, 973–977 [DOI] [PubMed] [Google Scholar]
- 53. McNally D. J., Aubrey A. J., Hui J. P., Khieu N. H., Whitfield D., Ewing C. P., Guerry P., Brisson J. R., Logan S. M., Soo E. C. (2007) Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem. 282, 14463–14475 [DOI] [PubMed] [Google Scholar]
- 54. Twine S. M., Paul C. J., Vinogradov E., McNally D. J., Brisson J. R., Mullen J. A., McMullin D. R., Jarrell H. C., Austin J. W., Kelly J. F., Logan S. M. (2008) Flagellar glycosylation in Clostridium botulinum. FEBS J. 275, 4428–4444 [DOI] [PubMed] [Google Scholar]
- 55. Ewing C. P., Andreishcheva E., Guerry P. (2009) Functional characterization of flagellin glycosylation in Campylobacter jejuni 81–176. J. Bacteriol. 191, 7086–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Howard S. L., Jagannathan A., Soo E. C., Hui J. P., Aubry A. J., Ahmed I., Karlyshev A., Kelly J. F., Jones M. A., Stevens M. P., Logan S. M., Wren B. W. (2009) Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect. Immun. 77, 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stephenson H. N., Mills D. C., Jones H., Milioris E., Copland A., Dorrell N., Wren B. W., Crocker P. R., Escors D., Bajaj-Elliott M. (2014) Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: a novel flagellin-host interaction. J. Infect. Dis. jiu 287 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lewis A. L., Nizet V., Varki A. (2004) Discovery and characterization of sialic acid O-acetylation in group B Streptococcus. Proc. Natl. Acad. Sci. U. S. A. 101, 11123–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morrison M. J., Imperiali B. (2013) Biosynthesis of UDP-N,N′-diacetylbacillosamine in Acinetobacter baumannii: Biochemical characterization and correlation to existing pathways. Arch. Biochem. Biophys. 536, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Anonsen J. H., Vik A., Egge-Jacobsen W., Koomey M. (2012) An extended spectrum of target proteins and modification sites in the general O-linked protein glycosylation system in Neisseria gonorrhoeae. J. Proteome Res. 11, 5781–5793 [DOI] [PubMed] [Google Scholar]
- 61. Jervis A. J., Butler J. A., Lawson A. J., Langdon R., Wren B. W., Linton D. (2012) Characterization of the structurally diverse N-linked glycans of Campylobacter species. J. Bacteriol. 194, 2355–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hartley M. D., Morrison M. J., Aas F. E., Borud B., Koomey M., Imperiali B. (2011) Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N′-diacetylbacillosamine. Biochemistry 50, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morrison M. J., Imperiali B. (2014) The renaissance of bacillosamine and its derivatives: pathway characterization and implications in pathogenicity. Biochemistry 53, 624–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nothaft H., Szymanski C. M. (2013) Bacterial protein N-glycosylation: new perspectives and applications. J. Biol. Chem. 288, 6912–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ciocchini A. E., Rey Serantes D. A., Melli L. J., Iwashkiw J. A., Deodato B., Wallach J., Feldman M. F., Ugalde J. E., Comerci D. J. (2013) Development and validation of a novel diagnostic test for human brucellosis using a glyco-engineered antigen coupled to magnetic beads. PLoS Negl. Trop. Dis. 7, e2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Iwashkiw J. A., Fentabil M. A., Faridmoayer A., Mills D. C., Peppler M., Czibener C., Ciocchini A. E., Comerci D. J., Ugalde J. E., Feldman M. F. (2012) Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb. Cell Fact. 11, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Berry D. S., Lynn F., Lee C. H., Frasch C. E., Bash M. C. (2002) Effect of O acetylation of Neisseria meningitidis serogroup A capsular polysaccharide on development of functional immune responses. Infect. Immun. 70, 3707–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kahler C. M., Lyons-Schindler S., Choudhury B., Glushka J., Carlson R. W., Stephens D. S. (2006) O-Acetylation of the terminal N-acetylglucosamine of the lipooligosaccharide inner core in Neisseria meningitidis. Influence on inner core structure and assembly. J. Biol. Chem. 281, 19939–19948 [DOI] [PubMed] [Google Scholar]
- 69. Borud B., Aas F. E., Vik A., Winther-Larsen H. C., Egge-Jacobsen W., Koomey M. (2010) Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192, 2816–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Buszewski B., Noga S. (2012) Hydrophilic interaction liquid chromatography (HILIC)–a powerful separation technique. Anal. Bioanal. Chem. 402, 231–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu D., Reeves P. R. (1994) Escherichia coli K12 regains its O antigen. Microbiology 140, 49–57 [DOI] [PubMed] [Google Scholar]
- 72. Ortega X., Hunt T. A., Loutet S., Vinion-Dubiel A. D., Datta A., Choudhury B., Goldberg J. B., Carlson R., Valvano M. A. (2005) Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187, 1324–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vaneechoutte M., Young D. M., Ornston L. N., De Baere T., Nemec A., Van Der Reijden T., Carr E., Tjernberg I., Dijkshoorn L. (2006) Naturally transformable Acinetobacter sp. strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72, 932–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Piechaud M., Second L. (1951) [Studies of 26 strains of Moraxella Iwoffi]. Ann. Inst. Pasteur 80, 97–99 [PubMed] [Google Scholar]
- 75. Vallenet D., Nordmann P., Barbe V., Poirel L., Mangenot S., Bataille E., Dossat C., Gas S., Kreimeyer A., Lenoble P., Oztas S., Poulain J., Segurens B., Robert C., Abergel C., Claverie J. M., Raoult D., Medigue C., Weissenbach J., Cruveiller S. (2008) Comparative analysis of Acinetobacters: three genomes for three lifestyles. PloS One 3, e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bouvet P. J. M. G., P. A. D. (1986) Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus, and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36, 228–240 [Google Scholar]
- 77. Weber B. S., Miyata S. T., Iwashkiw J. A., Mortensen B. L., Skaar E. P., Pukatzki S., Feldman M. F. (2013) Genomic and functional analysis of the type VI secretion system in Acinetobacter. PloS One 8, e55142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Juni E. (1972) Interspecies transformation of Acinetobacter: genetic evidence for a ubiquitous genus. J. Bacteriol. 112, 917–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.