Abstract
The relevance of changes to the coding sequence of the c-MYC oncogene to malignancy is controversial. Overexpression of a pristine form of MYC is observed in many cancers and is sufficient to drive tumorigenesis in most contexts. Yet missense changes to MYC are found in ~50% of Burkitt’s lymphomas, aggregate within an amino-terminal degron important for proteasomal destruction of MYC, and where examined profoundly enhance the tumorigenic properties of MYC in vitro and in vivo. Much of the controversy surrounding these mutants stems from the limited number of mutations that have been evaluated and their clustering within a single region of the MYC protein; the highly-conserved Myc box I (MbI) element. Here, by analysis of extant genomic data sets, we identify a previously-unrecognized hotspot for tumor-associated MYC mutations, located in a conserved central portion of the protein. We show that, despite their distal location in MYC, mutations in this region precisely phenocopy those in MbI in terms of stability, in vitro transformation, growth-promoting properties, in vivo tumorigenesis, and ability to escape p53-dependent tumor surveillance mechanisms. The striking parallels between the behavior of tumor-derived mutations in disparate regions of the MYC protein reveals that a common molecular process is disrupted by these mutations, implying an active role for these mutations in tumorigenesis and suggesting that different therapeutic strategies may be needed for treatment of lymphomas expressing wild-type versus mutant forms of MYC protein.
Keywords: MYC, lymphomagenesis, mutations
INTRODUCTION
c-MYC encodes an oncogene transcription factor that features prominently in cancer. Across the spectrum of malignancies, activation of MYC is typically driven by overexpression of the wild-type protein, but blood-borne tumors often possess changes to the MYC coding sequence. 1-12 Indeed, ~50% of Burkitt’s lymphomas (BL) harbor MYC mutations, the majority of which cluster at sites within the amino-terminus of the protein. 5 This region carries an expansive degron that signals MYC proteolysis, and we have reported that tumor mutations within this segment stabilize MYC, the most-pronounced effects being observed with mutations in a conserved element called Myc box I (MbI). 13 Subsequent studies showed that the core of MbI (residues 58–62) is a phosphorylation-dependent degron for the SCFFbw7 ubiquitin-ligase, 14 and that tumor mutations subvert proteolysis by disabling phosphorylation events within this region. Importantly, functional analyses of tumor-associated MbI mutations reveal that they render MYC profoundly oncogenic, and capable of driving lymphomagenesis without triggering Bim-dependent apoptosis and without selecting for loss of p53. 9 Although it is unclear how such mutations impact MYC’s transcriptional activity, it is clear that mutations in MbI induce both quantitative and qualitative changes in MYC that favor tumorigenesis.
Despite their widespread prevalence in BL, the relevance of tumor-associated MYC mutations to the etiology of the disease remains controversial. On one hand, the canonical t(8:14) translocation in BL is sufficient to drive high levels of MYC expression and places MYC in a hypermutable region of the genome, where random mutations could occur. On the other hand, these mutations do accumulate in specific regions of MYC and clearly enhance its tumorigenic functions, 9 implying that they confer a selective advantage to malignant cells. Much of the difficulty in understanding the significance of these mutations stems from the relatively small number of mutant alleles that have been sequenced, the often complex multi-residue nature of these mutations, and the fact that the best-characterized tumor-associated mutations localize to just a single region of MYC (MbI), leaving open the question of whether the handful of MbI mutations that have been studied to date reflect what occurs in BL patients. Clearly, resolution of this controversy requires analysis of additional tumor MYC alleles, with the most informative being those that lie outside of MbI.
Recent BL resequencing efforts 10-12 expanded the number of tumor-associated MYC alleles that have been characterized. Prompted by this work, we collated published reports of missense c-MYC mutations in BL and other lymphomas. We hypothesized that, as with MbI, functionally important mutations may cluster in critical regions of the MYC protein, and that identification of clustered mutations in novel regions of MYC would help address the relevance of these mutations to lymphomas. Here, we report the results of this analysis, and identify a novel hotspot for mutations in MYC, spanning residues 243–249 within the central portion of the protein. We show that mutations in this region disrupt a second phosphodegron within MYC, and that they precisely phenocopy effects of mutations within MbI in terms of stability, enhanced tumorigenesis, and immunity to p53-mediated tumor surveillance mechanisms. The remarkable similarity between the effects of tumor-associated mutations in disparate regions of MYC reveals a common molecular theme in MYC deregulation in lymphoma, and strongly implies that MYC mutations play an active role in the pathophysiology of the disease.
RESULTS AND DISCUSSION
Identification of a novel hotspot for tumor-derived mutations in MYC
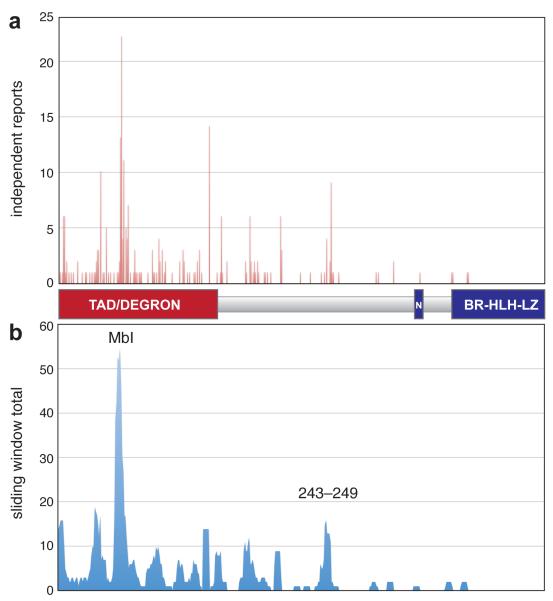
To generate a comprehensive view of c-MYC mutations in lymphoma, we compiled published reports 1-12 of mutations described in patient samples and cultured cell lines (Supplemental Table S1). Because MYC is subject to multiple mutations in ~50% of cases, we deconvoluted complex mutations and expressed the results as the frequency of independent reports of mutation at each residue in MYC (Figure 1a). To identify clusters of mutations that may impact a specific region of MYC, we also probed for mutations within a sliding six amino acid window along the protein (Figure 1b). These analyses showed that tumor-associated mutations in MYC are located predominantly in the amino-terminal degron, with the highest frequency found in MbI, confirming this site as the major mutational hotspot for MYC. Interestingly, however, we also identified a unique mutational cluster, spanning residues 243–249, that lies distal to the MYC degron. We selected the 243–249 region for analysis because this region is highly-conserved in vertebrate c-MYC proteins (Supplementary Figure S1), because the effects of tumor-derived mutations in this regions have not been reported, and because comparison with mutations in MbI would reveal whether common molecular processes are targeted by tumor-associated mutations in MYC.
Figure 1.
Distribution of tumor-derived mutations in c-MYC. (a) The graph illustrates the number of independent times mutations at each position in MYC have been reported, either in cell lines (n=17) or lymphoma patient samples (n=106). Raw data are presented in Supplemental Table S1. Beneath the graph is a scaled cartoon of MYC, showing key elements of the protein (‘TAD/DEGRON’; transcriptional activation domain and destruction element, ‘N’; nuclear localization sequence, ‘BR-HLH-LZ’; basic region helix-loop-helix leucine zipper DNA binding domain). (b) Sliding window analysis, showing the running totals for a sliding six amino acid residue along the MYC protein. The peaks corresponding to MbI and the 243–249 cluster (which summits in this analysis at residue 245) are indicated.
Identification of a central phosphodegron targeted by tumor-derived MYC mutations
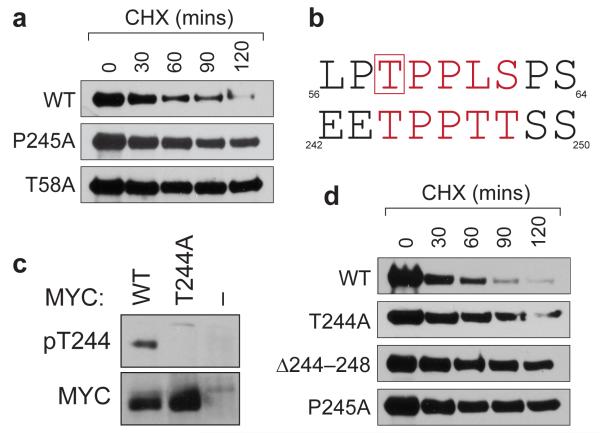
First, we asked whether mutations in this region impact MYC stability, as reported for those in MbI, such as T58A.13 We selected the P245A tumor mutant for this analysis, because it has been isolated as a single MYC mutation in BL patients, as well as in conjunction with other mutations (Supplemental Figure S1). For comparison, we stably expressed the wild-type (WT), T58A, and P245A forms of MYC in NIH3T3 cells, and used cycloheximide chase to infer their stability. This analysis (Figure 2a) revealed that the P245A mutation increases the half-life of MYC from ~40 to ~120 minutes, comparable to effects observed with the T58A mutation. Thus, like mutations in MbI, a tumor-derived mutation in the 243–249 segment of MYC subvert its rapid ubiquitin-mediated proteolysis.
Figure 2.
The 244–248 segment of MYC is a phosphodegron that is inactivated by the P245A tumor mutation. (a) Site-directed mutagenesis was used to introduce the indicated mutations into the MSCV–MYC–IRES–GFP vector, 9 which expresses HA-epitope tagged MYC along with an IRES-driven green fluorescent protein (GFP). These constructs were retrovirally transduced into NIH3T3 cells. To infer MYC stability, stable transductants were treated with cycloheximide (CHX) for the indicated times to inhibit protein synthesis, 22 protein extracts prepared, and exogenous MYC levels determined by Western blotting with an anti-HA antibody. (b) Alignment of MYC residues 56–64 and 242–250. Core residues of the SCFFbw7 phosphodegron are in red. Residue T58, which must be phosphorylated to bind SCFFbw7, is boxed. (c) NIH3T3 cells expressing the indicated HA-tagged MYC proteins (or vector control ‘–’) were lysed, HA-tagged proteins recovered by immunoprecipitation, and either T244-phosphorylated, or total, MYC proteins detected by Western blotting. Characterization of the specificity of purified phospho-T244 antibodies is presented in Supplementary Figure S2. (d) As in (a) except that NIH3T3 cells expressed WT, T244A, Δ244–248, or P245A forms of MYC.
Inspection of the sequence surrounding P245 revealed that residues 244–248 of MYC bear significant similarity to the SCFFbw7 phosphodegron within MbI (Figure 2b), including a threonine corresponding to T58 (T244) that must be phosphorylated to promote MYC turnover. 14 The similarity between these sequences prompted us to ask whether residues 244–248 of MYC also constitute a phosphodegron. Three lines of evidence support this notion. First, residue T244 of MYC is phosphorylated in vivo (Figure 2c), as revealed by a phospho-specific antibody that we developed against this modification (Supplementary Figure S2). Second, mutation of T244 to alanine stabilizes MYC (Figure 2d), consistent with the idea that phosphorylation of this residue promotes MYC turnover. Finally, deletion of the entire 244–248 segment (Δ244–248) also retards the rate of MYC proteolysis (Figure 2d), arguing that point mutations in this region do not dominantly stabilize MYC, but rather inactivate an element that functions to drive MYC turnover. Based on these data, we conclude that the 244–248 region of MYC constitutes a tumor-disrupted phosphodegron that is required for rapid MYC proteolysis.
The P245A MYC mutant phenocopies T58A
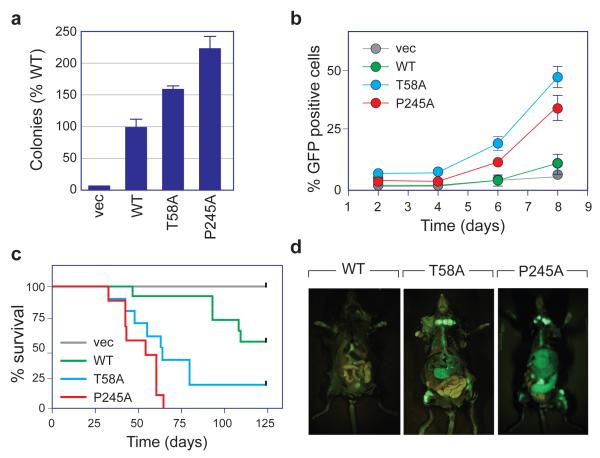
Previous analyses of the T58A mutation in MYC have revealed that it has an enhanced and very specific pattern of influence on MYC function. Not only is the T58A mutant increased in transformation potential in vitro 15 and in vivo, 9, 16 but it specifically fails to induce p53-dependent tumor surveillance mechanisms via the proapoptotic protein Bim 17 and does not require collaborating loss of p53 status to drive lymphomagenesis. 9 To determine whether a tumor-associated mutation in the 244–248 region of MYC leads to the same characteristic changes in MYC function, we compared the T58A and P245A mutants in a battery of functional assays. These analyses (Figure 3) revealed remarkable similarity between the two mutants. Both the T58A and P245A proteins showed increased activity in promoting anchorage-independent growth of NIH3T3 cells, compared to WT MYC (Figure 3a and Supplementary Figure S3a–b). Both proteins were considerably more potent than WT MYC at driving expansion of fetal liver cells grown in vitro under conditions that promote B-cell expansion 18 (Figure 3b and Supplementary Figure S3c). And both mutants behaved almost identically in an in vivo adoptive transfer assay of lymphomagenesis, 9 driving aggressive and rapidly fatal disease after reconstituting the bone marrow of sub-lethally-irradiated mice with either wild-type p53 or p53+/− hematopoietic stem cells (HSCs) expressing each protein (Figure 3c, d). For both mutants, recipient mice succumbed to pre-B cell lymphoma as judged by B220, Gr1, and Mac1 status (Supplementary Figures S4 and S5). Importantly, the P245A mutant mirrored T58A not only in disease phenotype, but in critical molecular characteristics, including the failure to induce Bim (Supplementary Figure S6a) and, in p53+/− cells, failure to select for loss of the wild-type copy of the p53 gene (Supplementary Figure S6b). 9 Thus the P245A mutant is able to drive lymphomagenesis in vivo without inducing Bim and without driving loss of p53 status, establishing that this mutant precisely phenocopies MbI MYC mutants such as T58A.
Figure 3.
The P245A tumor-mutation activates the oncogenic potential of MYC. (a) NIH3T3 cells, engineered to express the indicated MYC proteins, or vector control (vec), were grown in soft-agar. 23 After 21 days, colonies were counted, and colony numbers expressed relative to WT MYC. Data are presented as mean ± SEM (n=3). (b) Primary mouse fetal liver cells were transduced with MSCV–MYC–IRES–GFP vectors to express the indicated MYC proteins (or vector control), together with an IRES-driven green fluorescent protein (GFP). Transduced cells were then cultured under conditions that promote pre-B cell expansion, 18 harvested at the indicated times, and GFP-positive cells scored by FACS. Data are presented as mean ± SEM (n=6). Significance for (a) and (b) was established by comparing the effects of WT and mutant MYC proteins using Student’s t-test. (c) Kaplan–Meier survival curves of irradiated mice reconstituted with wild-type hematopoietic stem cells expressing the indicated MYC proteins (or vector control). 9 Kaplan–Meier curves were analyzed using the Log-rank test and statistical significance established via Chi-square analysis. The T58A (n=10) and P245A (n=9) MYC proteins drive tumorigenesis with a significantly (p<0.01) reduced latency (~50 days) compared to WT MYC (n=10, >100 days). The difference between T58A and P245A MYC is not significant. (d) Photos of representative mice from (c), imaging GFP-expressing cells. All mouse experiments were performed in compliance with US Federal laws and with approval of the Cold Spring Harbor Laboratory Animal Care and Use Committee.
Our characterization of the P245A MYC mutant establishes that naturally-occurring mutations outside of MbI enhance the oncogenicity of MYC. These findings demonstrate that activating mutations are not confined to a single region of MYC, and are therefore a larger part of the landscape of lymphomas than previously thought. Moreover, the near-identical behavior of the T58A and P245A mutants in our assays reveals that a common molecular process is affected by both sets of changes, arguing strongly that these changes are relevant to lymphomagenesis. Although mutations in both regions confer increased MYC stability, it is difficult to reconcile their behavior with the notion that they function solely by promoting MYC accumulation. Increased levels of MYC are associated with enhancement of its pro-apoptotic activities, 19 and although the T58A and P245A mutants are expressed at twice the levels of WT MYC (Supplementary Figure S3), they are defective at inducing Bim and do not apply selective pressure for loss of p53, demonstrating that these mutations selectively disable the ability of MYC to promote apoptosis. Thus these mutations confer distinct quantitative and qualitative changes in MYC that appear to provide a selective advantage to lymphoma cells.
What are the implications of our findings? The prevalence of these mutations in BL, and our demonstration of the profoundly altered activity of MYC tumor mutants, indicates that—even amidst a backdrop of MYC overexpression—MYC can act in fundamentally different ways in patients expressing WT versus mutant MYC. As Bim levels can predict the response of tumors to certain targeted therapies 20, the inability of MYC mutants described here to induce Bim may impact how BL patients respond to particular therapies. Burkitt’s lymphomas bearing mutant MYC proteins, for example, might be expected to be more sensitive to BH3 mimetics 21 than those expressing WT MYC, because the mutant MYC-expressing tumors, by virtue of attenuated Bim induction, have evolved without the need to lose pro-apoptotic cellular machinery by other means. We suggest that careful analysis of the form of MYC that is overexpressed in BL could inform not only basic mechanisms of disease progression and outlook, but also optimal therapeutic strategies.
Supplementary Material
ACKNOWLEDGEMENTS
For reagents we thank the CSHL Antibody Shared Resource and the Vanderbilt Antibody and Protein Shared Resource. We thank Simone Salghetti for technical assistance. This work was supported by the CSHL Cancer Center Support Grant CA45508, the Vanderbilt Ingram Cancer Center Support grant CA68485, and by US Public Health Service grant CA-13106 from the NCI. S.W.L is an investigator in the Howard Hughes Medical Institute.
Funding Sources: CSHL Cancer Center Support Grant CA45508; Vanderbilt Ingram Cancer Center Support grant CA68485; US Public Health Service grant CA-13106 from the NCI (WPT/SWL). Howard Hughes Medical Institute (SWL).
REFERENCES
- 1.Rabbitts TH, Hamlyn PH, Baer R. Altered nucleotide sequences of a translocated c-myc gene in Burkitt lymphoma. Nature. 1983;306:760–765. doi: 10.1038/306760a0. [DOI] [PubMed] [Google Scholar]
- 2.Westaway D, Payne G, Varmus HE. Proviral deletions and oncogene base-substitutions in insertionally mutagenized c-myc alleles may contribute to the progression of avian bursal tumors. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:843–847. doi: 10.1073/pnas.81.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Showe LC, Ballantine M, Nishikura K, Erikson J, Kaji H, Croce CM. Cloning and sequencing of a c-myc oncogene in a Burkitt’s lymphoma cell line that is translocated to a germ line alpha switch region. Mol Cell Biol. 1985;5:501–509. doi: 10.1128/mcb.5.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy W, Sarid J, Taub R, Vasicek T, Battey J, Lenoir G, et al. A translocated human c-myc oncogene is altered in a conserved coding sequence. Proc Natl Acad Sci U S A. 1986;83:2939–2943. doi: 10.1073/pnas.83.9.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 6.Bhatia K, Spangler G, Gaidano G, Hamdy N, Dalla-Favera R, Magrath I. Mutations in the coding region of c-myc occur frequently in acquired immunodeficiency syndrome-associated lymphomas. Blood. 1994;84:883–888. [PubMed] [Google Scholar]
- 7.Clark HM, Yano T, Otsuki T, Jaffe ES, Shibata D, Raffeld M. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- 8.Axelson H, Henriksson M, Wang Y, Magnusson KP, Klein G. The amino-terminal phosphorylation sites of C-MYC are frequently mutated in Burkitt’s lymphoma lines but not in mouse plasmacytomas and rat immunocytomas. Eur J Cancer. 1995;31A:2099–2104. doi: 10.1016/0959-8049(95)00449-1. [DOI] [PubMed] [Google Scholar]
- 9.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–5184. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. Embo J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welcker M, Orian A, Jin J, Grim JA, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conzen SD, Gottlob K, Kandel ES, Khanduri P, Wagner AJ, O’Leary M, et al. Induction of cell cycle progression and acceleration of apoptosis are two separable functions of c-Myc: transrepression correlates with acceleration of apoptosis. Mol Cell Biol. 2000;20:6008–6018. doi: 10.1128/mcb.20.16.6008-6018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer research. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaughlin J, Chianese E, Witte ON. In vitro transformation of immature hematopoietic cells by the P210 BCR/ABL oncogene product of the Philadelphia chromosome. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:6558–6562. doi: 10.1073/pnas.84.18.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer discovery. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 22.Tworkowski KA, Chakraborty AA, Samuelson AV, Seger YR, Narita M, Hannon GJ, et al. Adenovirus E1A targets p400 to induce the cellular oncoprotein Myc. Proc Natl Acad Sci U S A. 2008;105:6103–6108. doi: 10.1073/pnas.0802095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst A, Hemann MT, Tworkowski KA, Salghetti SE, Lowe SW, Tansey WP. A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO Rep. 2005;6:177–183. doi: 10.1038/sj.embor.7400333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.