Abstract
Chaperone-mediated autophagy (CMA) selectively degrades a subset of cytosolic proteins in lysosomes. A potent physiological activator of CMA is nutrient deprivation, a condition in which intracellular triglyceride stores or lipid droplets (LD) also undergo hydrolysis (lipolysis) to generate free fatty acids for energetic purposes. Here we report that LD-associated proteins perilipin 2 (PLIN2) and perilipin 3 (PLIN3) are CMA substrates and their degradation via CMA precedes lipolysis. In vivo studies revealed that CMA degradation of PLIN2 and PLIN3 was enhanced during starvation, concurrent with elevated levels of cytosolic adipose triglyceride lipase (ATGL) and macroautophagy proteins on LD. CMA blockage both in cultured cells and mouse liver or expression of CMA-resistant PLINs lead to reduced association of ATGL and macrolipophagy-related proteins with LD and the subsequent decrease in lipid oxidation and accumulation of LD. We propose a role of CMA in LD biology and in the maintenance of lipid homeostasis.
Keywords: Chaperones, Lipid droplets, Lysosomes, Lysosome-associated membrane protein 2A, Perilipins, Protein degradation
Introduction
Autophagy maintains cellular homeostasis by degrading proteins, lipids and organelles in lysosomes1, 2. Turnover of nonfunctional cellular components by autophagy ensures quality control and recycling of degraded products provides energy3. In chaperone-mediated autophagy (CMA) heat shock cognate protein of 70kDa (hsc70) recognizes proteins with a pentapeptide motif4 and delivers them to the lysosome surface5 for binding to lysosome-associated membrane protein 2A (LAMP-2A; L2A), the rate-limiting component of CMA6. L2A organizes into a multimeric complex7 and in cooperation with lysosomal hsc708 mediates lysosomal translocation of the unfolded substrates9 for degradation. CMA occurs in most mammalian cell-types10, 11 and is maximally activated following prolonged starvation12, mild oxidative stress13, hypoxia14 and lipogenic stressors15. Decrease in lysosomal stability of LAMP-2A results in reduced CMA activity in old organisms16, 17.
Added to the well-characterized turnover of proteins by autophagy, lipids can also undergo degradation by macroautophagy, by sequestration into double membrane vesicles (autophagosomes) that then fuse with lysosomes18, 19. Autophagy-mediated lipolysis (macrolipophagy) selectively targets lipid droplets (LD), intracellular lipid stores that serve as energy source through hydrolysis of triglycerides (TG) into free fatty acids (FFA). LD are surrounded by structural proteins of the perilipin (PLIN) family, with PLIN1 being primarily an adipocyte protein and PLIN2 and PLIN3 expressed ubiquitously20. Lipolysis can occur via cytosolic lipases, such as adipose triglyceride lipase (ATGL)21, or by lysosome luminal lipases when autophagosomes, formed through assembly of autophagy-related proteins (ATGs) on the LD surface, engulf portions of LD and target them to lysosomes18.
Although CMA can degrade only proteins and no lipids, mice with constitutive blockage of CMA in liver exhibit pronounced steatosis, even though non-selective macroautophagy was intact22. The discrete increase in lipogenesis, due to reduced degradation of lipogenic enzymes through CMA22, could not account alone for the massive hepatic lipid accumulation in these animals. This finding combined with the facts that, (i) in tissues, such as liver, lipid influx, lipolysis and CMA activation occur at maximal rates during starvation12, 23; (ii) lipogenic challenges initially upregulate CMA activity15; and (iii) conditions when CMA activity decreases such as aging16 often associate with intracellular lipid accumulation24, made us to further investigate the role of CMA in regulating lipid homeostasis.
Here, we demonstrate that blocking CMA both in cultured cells and in mouse liver in vivo decreases LD breakdown. Under conditions that promote lipolysis, CMA degrades LD proteins PLIN2 and PLIN3, and this facilitates the LD association of cytosolic lipase ATGL and of macroautophagy ATGs. Reduced CMA precludes recruitment of the lipolytic machinery to the LD, thereby positioning CMA as a critical upstream regulator of both macrolipophagy and cytosolic lipolysis.
Results
LAMP-2A-deficient cells accumulate LD
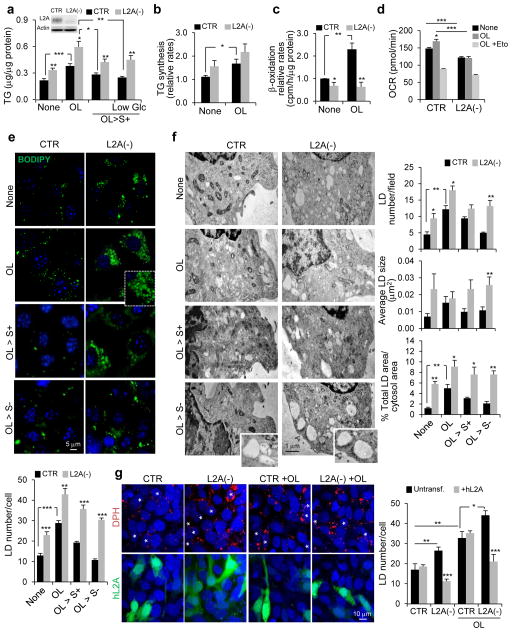
Using both, livers from mice conditionally knock-out for LAMP-2A (L2A) in hepatocytes22 (L2AKO) and mouse fibroblasts (NIH3T3 cells) knocked down for L2A (L2A(−)) to block CMA25 we confirmed that, despite lower dependence of fibroblasts on lipid metabolism when compared to hepatocytes, L2A-deficient fibroblasts accumulated significantly more triglycerides (TG) than control fibroblasts (Fig. 1a). These differences in TG content were even higher when intracellular lipid usage was forced by reducing glucose in the media or after a lipogenic stimulus (oleate; OL) (Fig. 1a).
Figure 1.
LAMP-2A-deficient cells accumulate LD. (a) Total triglycerides (TG) in control mouse fibroblasts (CTR) and in cells stably knocked down for LAMP-2A (L2A(−)) (inset) untreated or treated with OL, incubated with serum-supplemented regular media (OL>S+) or low glucose media (OL>S+ Low Glc) after OL treatment. n=4 (LowGlc), 6 (OL>S+) and 7 (all other conditions) independent experiments. (b) TG synthesis in cells as in (a). n=4 independent experiments. (c) Beta-oxidation rates in cells as in (a). n=6 independent experiments. (d) Oxygen consumption rates (OCR) in CTR and L2A(−) cells with the indicated treatments. Eto: etomoxir. n=5 time points from 8 independent experiments. (e) BODIPY493/503 staining in CTR and L2A(−) cells untreated or treated with OL, or incubated with serum-supplemented medium (OL>S+) or serum-deprived medium (OL>S−) after OL treatment. Graph: average LD number/cell (LD size shown in Supplementary Figure 1d). n=6 independent experiments with 40 cells per condition in each experiment. (f) Electron microscopy of cells treated as in (e). Graphs: area occupied by LD or average LD number/cell and LD size. n=3 independent experiments with 5 micrographs per condition. (g) DPH staining in CTR and L2A(−) cells transfected with hL2A, untreated or treated with OL. Asterisks: transfected cells. Graph: average LD number/cell calculated from orthoviews. n=5 independent experiments with 40 cells per condition in each experiment. Values are mean ± SEM. Differences are significant for *P<0.05, **P<0.01, ***P<0.001 using Student’s t-test. Source data is available in Supplementary Table 1.
As in the L2AKO mice22, we only found a slight trend towards higher TG synthesis in L2A(−) cells compared to control cells (Fig. 1b). In contrast, L2A(−) cells showed significantly reduced β-oxidation rates, an event downstream of TG hydrolysis, both under basal and lipogenic conditions (Fig. 1c), and failed to increase oxygen consumption rates (OCR) upon OL exposure (Fig. 1d; CTR cells: Δ20.9+1.7pmol/min; L2A(−) cells: Δ−1.3+0.2pmol/min OCR change). Decreased β-oxidation rates in L2A(−) cells was not due to defective mitochondria per se, since the fraction of mitochondria with intact membrane potential (Supplementary Figure 1a) and mitochondria turnover (detected with pH-sensitive MitoKeima reporter26; Supplementary Figure 1b) were comparable to CTR cells.. Differences in β-oxidation rates between control and L2A(−) cells were minimized in presence of etomoxir (inhibitor of mitochondrial FFA uptake) (Fig. 1d; upon OL addition, OCR in CTR cells was 48.4+2.1pmol/min higher than in L2A(−) cells, but after adding etomoxir the difference between both cell-types was reduced to 17.2+1.6pmol/min) suggesting a problem in lipid mobilization and not in its oxidation per se.
Consistent with the pronounced hepatosteatosis of L2AKO mice (Supplementary Figure 1c), L2A(−) fibroblasts accumulate more and bigger lipid droplets (LD) visible with either BODIPY493/503 (Fig. 1e, Supplementary Figure 1d) or DPH (1,6-diphenylhexatriene27) (Supplementary Figure 1e,f), both under basal and OL-treated conditions. Unlike control cells, etomoxir-treated L2A(−) cells failed to increase LD supporting defective lipid mobilization (Supplementary Figure 1g). To further test this hypothesis, we used different experimental paradigms to force lipolysis in fibroblasts. OL-challenged control cells, upon removal of OL (OL>S+) showed a decrease in LD number and size, even more evident if maintained in serum-free media (OL>S−) (Fig. 1e, Supplementary Figure 1d–f). In contrast, OL-challenged L2A(−) cells maintained elevated LD number and size throughout. Similarly, culturing cells in low-glucose or in serum-free media, to facilitate cellular usage of lipid stores, revealed significantly reduced ability of L2A(−) cells to decrease their TG levels and LD content compared to control cells (Fig. 1a, Supplementary Figure 1h). Electron microscopy confirmed that LD number, average size, and occupied cellular area were significantly higher in CMA-deficient cells than in control cells in all conditions tested (Fig. 1f). LD accumulation in L2A(−) cells was a direct consequence of L2A loss since expressing L2A in these cells (Supplementary Figure 1i,j) was sufficient to reduce LD to control cell levels (Fig. 1g). Together, these results indicate that CMA blockage results in LD accumulation and this is primarily a consequence of decreased LD breakdown.
LD proteins PLIN2 and PLIN3 are CMA substrates
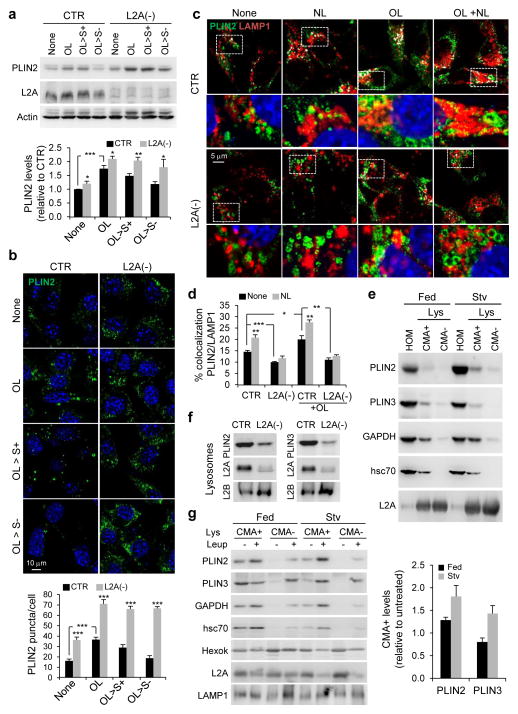
Since only proteins and not lipids can be CMA substrates, we next investigated a role of CMA in degradation of LD proteins, which could explain defective LD utilization in CMA-deficient cells. Immunoblotting (Fig. 2a, Supplementary Figure 2a) and immunofluorescence (Fig. 2b) for the ubiquitously expressed PLIN2 and PLIN3 showed significantly higher levels under basal, OL treatment and post-OL treatment in L2A(−) cells. Consistent with the described PLIN2 turnover by the ubiquitin-proteasome pathway28,29, proteasomal inhibitors increased LD number (Supplementary Figure 2b) and PLIN2 levels (Supplementary Figure 2c) in control cells but not in CMA-defective cells, although proteasomal degradation of ubiquitinated proteins and a degron-GFP reporter30 were preserved in these cells (Supplementary Figure 2c–e). Lipid accumulation upon proteasomal or lysosomal inhibition were comparable (Supplementary Figure 2b), leaving open the possibility that PLINs could also be amenable for lysosomal degradation.
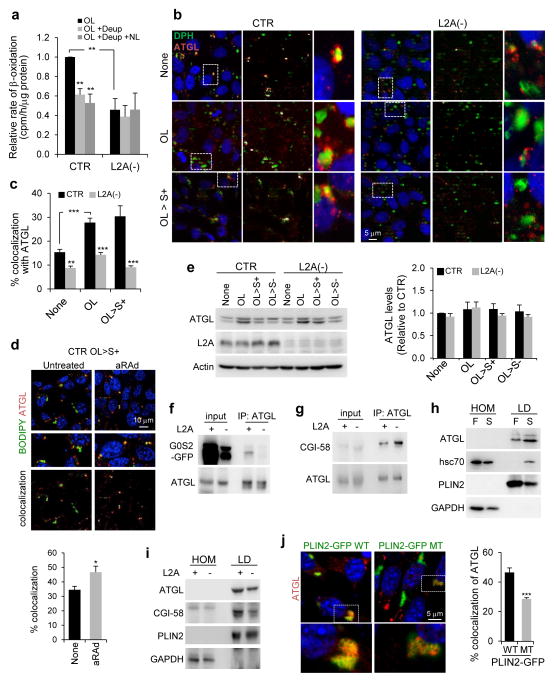
Figure 2.
PLIN2 and PLIN3 are CMA substrates. (a) Immunoblot for indicated proteins of total cell lysates from CTR and L2A(−) cells untreated or treated with OL or incubated with serum-supplemented medium (OL>S+) or serum-deprived medium (OL>S−) after OL treatment. Graph: PLIN2 levels relative to untreated CTR cells. n=7 independent experiments. (b) Immunostaining for PLIN2 in CTR and L2A(−) cells treated as in (a). Graph: average puncta/cell. n=5 independent experiments with 40 cells per condition in each experiment. (c) Coimmunostaining for PLIN2 and LAMP1 in CTR and L2A(−) cells treated or not with OL followed by treatment with lysosomal inhibitors, ammonium chloride and leupeptin (NL). Top: Colocalized pixels in white. Bottom: Merged image of the boxed area at higher magnification. (d) Graph: percentage colocalization of PLIN2 with LAMP1 from (c). n=5 independent experiments with 40 cells per condition in each experiment. (e) Immunoblot for indicated proteins of homogenates (HOM), lysosomes with high (CMA+) and low (CMA−) activity isolated from fed or starved (Stv) rat livers. Representative blots from 5 independent experiments. (f) Immunoblot for indicated proteins of isolated lysosome-enriched fractions from OL-treated control and L2A(−) cells. (g) Immunoblot for indicated proteins of HOM, CMA+ and CMA− lysosomes isolated from fed or starved (Stv) livers of rat untreated or treated with leupeptin (Leup). Graph: PLIN2 and PLIN3 levels in leupeptin-treated CMA+ lysosomes relative to untreated. Representative blots from 3 independent experiments (other two shown in Supplementary Figure 2h). Values are mean ± SEM. Differences are significant for *P<0.05, **P<0.01, ***P<0.001 using Student’s t-test. Uncropped images of blots are shown in Supplementary Figure 8. Source data is available in Supplementary Table 1.
To examine the possible contribution of CMA to PLIN2 and PLIN3 degradation, we first investigated their association with lysosomes. In support of PLINs lysosomal turnover, inhibiting lysosomal degradation with ammonium chloride and leupeptin in control cells significantly increased the colocalization of PLIN2 or PLIN3 with the lysosomal marker (LAMP1), and more so upon OL addition (Fig. 2c,d, Supplementary Figure 2f,g). In contrast, L2A(−) cells showed decreased association of PLIN2 and PLIN3 with lysosomes and impaired flux with lysosomal inhibitors (Fig. 2c,d, Supplementary Figure 2f,g). PLIN2 and PLIN3 were enriched in lysosomes active for CMA (CMA+, containing hsc70 and LAMP-2A31) as compared to lysosomes with lower CMA activity (CMA−, containing LAMP-2A but deficient in hsc7031) (Fig. 2e, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a CMA substrate32, is shown as a positive control). Lysosomal levels of both PLINs were reduced in L2A(−) cells (Fig. 2f). We confirmed that lysosomal association of PLIN2 and PLIN3 led to their degradation in vivo, since blocking lysosomal proteolysis in starved rats with leupeptin for 2h was sufficient to increase levels of these LD proteins in CMA-active lysosomes (Fig. 2g, Supplementary Figure 2h). CMA degradation of PLIN2 was also detected in fed mice, albeit at lower levels than in starved animals (Fig. 2g), whereas lysosomal degradation of PLIN3 in fed conditions was negligible (Fig. 2g). Some degradation of PLINs was observed in CMA- lysosomes, likely due to LD degradation by macrolipophagy, since these lysosomes preferentially fuse with autophagosomes33. Collectively, results from our in vitro and in vivo studies indicate that CMA degrades PLIN2 and PLIN3.
PLINs interact with CMA chaperone hsc70
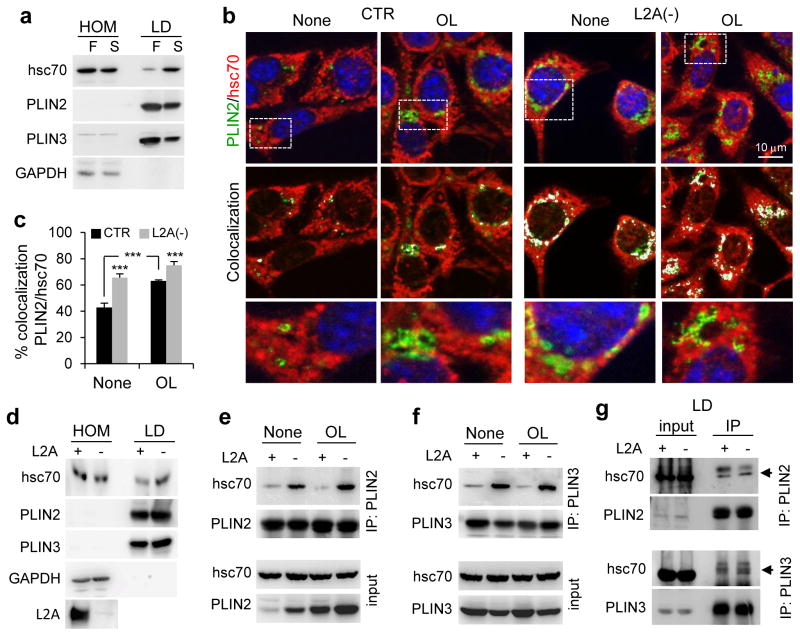
The first step in CMA is substrate interaction with hsc70 for subsequent lysosomal targeting. We found hsc70 in isolated rat liver LD and its levels increased during starvation, when hepatic lipolysis is highly active, coinciding with a decrease in LD levels of PLIN2 and PLIN3 (Fig. 3a). Immunofluorescence confirmed hsc70 colocalization with each PLIN on LD, which increased upon OL-challenge that induces lipolysis (Fig. 3b,c, Supplementary Figure 3a). Forcing lipid mobilization by placing cells in serum-free media post-OL challenge reduced association of hsc70 with LD (Supplementary Figure 3b). Remarkably, L2A(−) cells exhibited higher hsc70 colocalization with PLIN2 or PLIN3 in LD under all conditions (Fig. 3b,c, Supplementary Figure 3a,b). Similar higher abundance of hsc70 was also observed in LD isolated from livers of L2AKO mice compared to control littermates (Fig. 3d).
Figure 3.
PLIN2 and PLIN3 interact with CMA chaperone hsc70. (a) Immunoblot for indicated proteins of homogenates (HOM) and lipid droplets (LD) isolated from fed (F) or starved (S) rat livers. GAPDH is shown as a negative control for lack of cytosolic contamination in the LD fractions. Representative blots from 5 independent experiments. (b, c) Coimmunostaining for hsc70 and PLIN2 in CTR and L2A(−) cells treated or not with OL. Colocalized pixels are in white. Boxed areas are shown at higher magnification. Graph: percentage colocalization of PLIN2 with hsc70. n=5 independent experiments with 40 cells per condition in each experiment. (d) Immunoblot for indicated proteins of HOM and LD isolated from starved wild-type (+) or L2A knockout (−) mice livers. Representative blots from 3 independent experiments. (e, f) Coimmunoprecipitation (IP) of PLIN2 (e) and PLIN3 (f) in CTR (+) and L2A(−) (−) cells treated or not with OL. Extended blots are shown in Supplementary Figure 3f. Representative blots of from 5 (e) and 3 (f) independent experiments. (g) IP of PLIN2 (top) and PLIN3 (bottom) in LD-enriched fractions from mice as in (d). The experiment was repeated twice. Values are mean ± SEM. Differences are significant for ***P<0.001 using Student’s t-test. Uncropped images of blots are shown in Supplementary Figure 8. Source data is available in Supplementary Table 1.
We found direct interaction of hsc70 with PLIN2 and with PLIN3 in cultured cells. Hsc70 was recovered in PLIN2 and 3 pull-downs (Fig. 3e,f) and both PLINs were also detected in hsc70 pull-downs (Supplementary Figure 3c,d). For the same amount of PLINs pulled-down, we consistently observed higher levels of bound hsc70 in L2A(−) cells (Fig. 3e,f). Increased binding to hsc70 is characteristic of CMA substrates in these cells where disruption of CMA occurs at the level of the lysosomal receptor (L2A) while substrate recognition by hsc70 is intact (Supplementary Figure 3e shows the same effect in two other CMA substrates). This enhanced binding of hsc70 for multiple substrates explains why higher levels of PLINs (a single substrate) are not observed in hsc70 pull-downs (Supplementary Figure 3c,d). Upon treatment of L2A(−) cells with increasing OL concentrations previously shown to activate CMA15, hsc70 bound to PLIN2 or PLIN3 further increased whereas in control cells they remained constant, supporting continuous turnover of LD proteins by CMA (Fig. 3e,f, Supplementary Figure 3f). Since contrary to the almost exclusive LD location of PLIN2, PLIN3 can be present in other cellular compartments, we isolated mouse liver LD and confirmed that interaction of hsc70 with PLINs occurs in LD (Fig. 3g). These results are compatible with PLIN2 and PLIN3 being recognized by the CMA chaperone hsc70 at the LD surface before undergoing CMA-mediated degradation.
Modifications of CMA-targeted PLINs
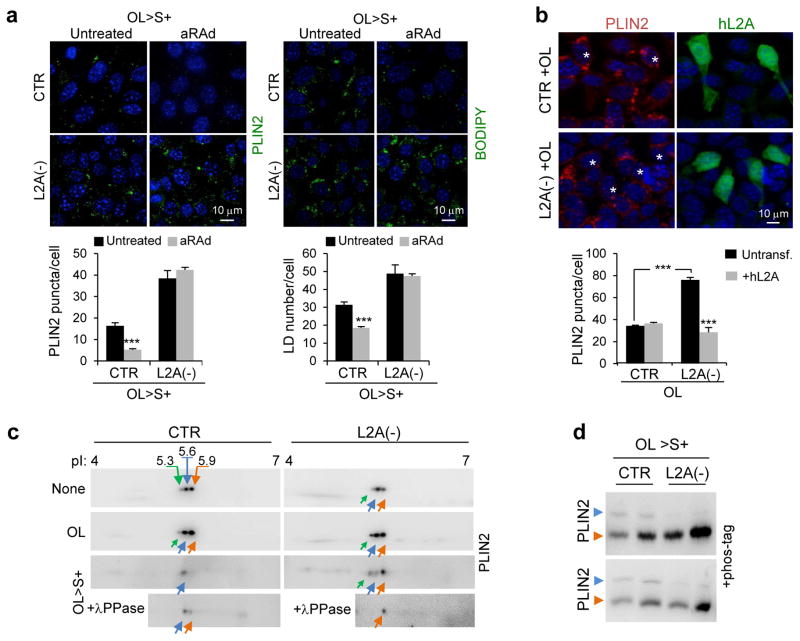
To further confirm the involvement of CMA in PLINs turnover, we activated CMA in OL-challenged cells using an atypical retinoic acid derivative (aRAd)34. Chemical activation of CMA significantly reduced PLIN2 levels (Fig. 4a) and LD content (Fig. 4b) in control cells but not in L2A(−) cells. In fact, restoration of L2A levels in L2A(−) cells through transient transfection was sufficient to bring PLIN2 levels close to control cell values (Fig. 4b).
Figure 4.
Posttranslational modifications of PLINs and their degradation by CMA. (a) Immunostaining for PLIN2 (left) or BODIPY493/503 staining (right) in CTR and L2A(−) cells incubated in serum-supplemented media after OL challenge (OL>S+) treated or not with atypical retinoic acid derivative (aRAd) shown to activate CMA34. Graphs: PLIN2 puncta or LD/cell. n=5 (PLIN2) and 6 (LD/cell) independent experiments with 40 cells per condition in each experiment. (b) Immunostaining for PLIN2 in CTR and L2A(−) cells transfected with hL2A and treated with OL. Asterisks: transfected cells. Graph: average PLIN2 puncta/cell calculated from orthoviews. n=5 independent experiments with 40 cells per condition in each experiment. (c) Bidimensional electrophoresis and immunoblot for PLIN2 in total cell lysates from CTR and L2A(−) cells untreated, treated with OL, or maintained in OL>S+ media. Bottom inset shows samples treated with lambda phosphatase (λPPase). Arrows: estimated isoelectric points. Representative blots of n=3 (OL>S+) and 6 (None and OL) independent experiments. (d) PLIN2 immunoblots of total cell lysates from CTR and L2A(−) cells subjected to indicated treatments run with Phos-tag to resolve phosphorylated proteins. Immunoblots from two independent experiments are shown. Arrows: orange (unphosphorylated PLIN2), blue (phosphorylated PLIN2). Values are mean ± SEM. Differences are significant for ***P<0.001 using Student’s t-test. Uncropped images of blots are shown in Supplementary Figure 8. Source data is available in Supplementary Table 1.
To identify possible posttranslational modifications that trigger PLIN2 for CMA degradation we performed bidimensional electrophoresis and immunoblot and found two very prominent forms of PLIN2 of 5.6 and 5.9 isoelectric point (pI) and a less abundant isoform of pI 5.3. When lipolysis and subsequent PLIN2 degradation was induced with OL, the less positively charged isoform was no longer observed in control cells but it persisted in L2A(−) cells (Fig. 4c). Phosphatase treatment confirmed that PLIN2 was phosphorylated during lipolysis in CTR cells but not in L2A(−) cells that accumulated a form of PLIN2 unchanged by the phosphatase (Fig. 4c). Analysis of LD by electrophoresis using Phos-tag (for resolution of phosphorylated variants) confirmed the presence of a phosphorylated form of PLIN2 in control cells almost completely absent in L2A(−) cells (Fig. 4d). Although further studies are needed to elucidate the reasons for reduced levels of phospho-PLIN2 in absence of functional CMA, our findings suggest that phosphorylation of PLIN2 may be required to trigger its CMA degradation.
CMA-resistant PLIN2 causes steatosis
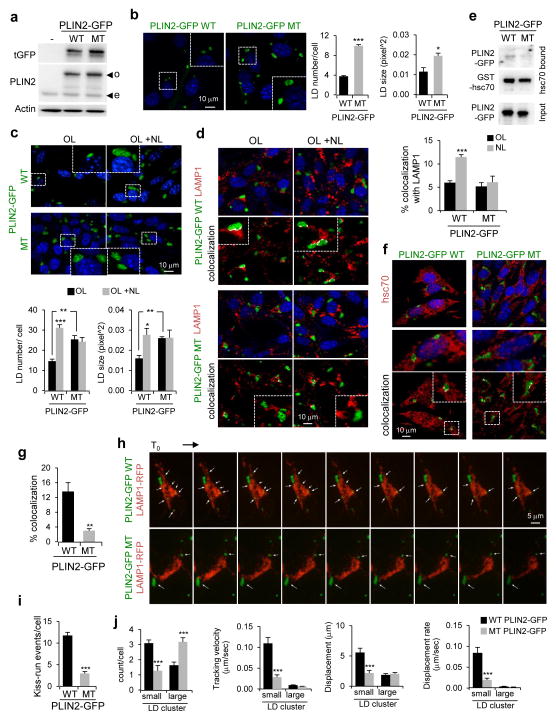
Hsc70 binds CMA substrates through a pentapeptide motif4. PLIN3 bears a canonical CMA-motif (100LDRLQ) and PLIN2 a putative motif (414SLKVQ). To analyze the consequences of preventing CMA degradation of PLIN2, independently from other changes that may occur during CMA blockage, we mutated the PLIN2 CMA-targeting motif to 414SLKAA. Expression of wild-type (WT) and CMA-mutant (MT) PLIN2-GFP in control cells led to higher levels of MT PLIN2-GFP (Fig. 5a) and a ~2.7-fold increase in LD content and size as compared to WT PLIN2-expressing cells under basal conditions (Fig. 5b) and upon OL-challenge (Fig. 5c). MT PLIN2-GFP accumulation was mainly due to its reduced lysosomal degradation as, treatment with lysosomal inhibitors (NL) increased WT but not MT PLIN2-GFP levels (Fig. 5c) and, unlike WT PLIN2, failed to increase colocalization of MT PLIN2 with the lysosomal marker LAMP1 (Fig. 5d).
Figure 5.
Cells expressing CMA-mutant PLIN2 accumulate LD. (a) Immunoblot for indicated proteins of total cell lysates from untransfected cells (−) or cells transfected with wild-type (WT) or CMA-mutant (MT) PLIN2-GFP. o: overexpressed protein; e: endogenous protein. Representative blots of n=3 independent experiments. (b) Cells expressing WT or MT PLIN2-GFP. Inset: Higher magnification of the boxed area. Graphs: average LD number or LD size. n=5 (LD number) and 3 (LD size) independent experiments with 40 cells per condition in each experiment. (c) Cells expressing WT or MT PLIN2-GFP treated with OL with or without lysosomal inhibitors ammonium chloride and leupeptin (NL). Inset: Higher magnification of the boxed area. Graphs: average LD number or LD size. n=5 (LD number) and 3 (LD size) independent experiments with 40 cells per condition in each experiment. (d) Immunostaining for LAMP1 in cells treated as in (c). Top: Merged images. Bottom: Colocalized pixels in white. Insets: higher magnification. Graph: percentage colocalization of PLIN2-GFP with LAMP1. n=5 independent experiments with 40 cells per condition in each experiment. (e) Binding of WT and MT PLIN2-GFP from cellular extracts to GST-hsc70 immobilized on agarose beads. Representative blots of n=3 independent experiments. (f, g) Immunostaining for hsc70 in OL-treated cells expressing WT or MT PLIN2-GFP. Colocalized pixels are in white. Insets: higher magnification. 3D-reconstruction shown in Supplementary Figure 4a. Graph: percentage colocalization of PLIN2-GFP with hsc70. n=5 independent experiments with 40 cells per condition in each experiment. (h–j) Live-cell imaging of cells cotransfected with WT or MT PLIN2-GFP and LAMP1-RFP and treated with OL. Sequential frames at 60s intervals are shown. Arrows: LD. Graphs: LD (i) kiss-run events/cell and (j) count/cell, tracking velocity, displacement and displacement rate. Small and large LD clusters defined as <1μm3 and >1μm3, respectively. 5i: n=24 cells from 3 independent experiments. 5j: n=103 (WT) and 85 (MT) LD clusters from 6 (WT) and 8 (MT) movies with >8 cells/movie from 2 independent experiments. Values are mean ± SEM. Differences are significant for *P<0.05, **P<0.01, ***P<0.001 using Student’s t-test. Uncropped images of blots are shown in Supplementary Figure 8. Source data is available in Supplementary Table 1.
We confirmed that disruption of the CMA-targeting motif in PLIN2 almost completely abrogates its binding to hsc70 (Fig. 5e), explaining the reduced colocalization of hsc70 with LD in cells expressing MT PLIN2 (Fig. 5f,g; 3D reconstruction of Z-sections shown in Supplementary Figure 4a and Supplementary Video 1). Persistence of MT PLIN2 on LD altered their cellular dynamics and their interplay with lysosomes. Thus, live-cell imaging of cells co-expressing WT PLIN2-GFP and LAMP1-RFP revealed that WT PLIN2-GFP LD were predominantly small and very mobile as reported earlier35, 36 (Fig. 5h–j, Supplementary Video 2 (green channel only)) and showed dynamic inter-LD associations and kiss-and-run events between LD and lysosomes (Fig. 5h–j, Supplementary Video 3 (merged channels)). In striking contrast, majority of LD in MT PLIN2-expressing cells formed larger, less dynamic clusters and the very few smaller LD rarely associated with lysosomes (Fig. 5h–j; Z-sections stack shown in Supplementary Figure 4b, Supplementary Video 2,3). In summary, preventing degradation of PLIN2 by CMA is sufficient to induce the intracellular lipid accumulation observed in CMA-deficient cells (Fig. 1 and Fig. 2) and to reduce LD/lysosome interactions.
Defective CMA of PLINs blocks lipolysis
TG in LD are broken down into FFA either by cytosolic lipases21, 23 or by lysosomal lipases that gain access to these cytosolic stores via macrolipophagy18. To assess which lipolytic pathway failed to mobilize LD in L2A(−) cells (Fig. 1), we treated cells with a lipase inhibitor, diethylumbelliferyl phosphate (Deup), and with lysosomal inhibitors (NL) and found that βoxidation decreased in control cells but L2A(−) cells remained unresponsive (Fig. 6a). Overall these findings suggested that blocking CMA results in impaired lipolysis by cytosolic lipases and macrolipophagy.
Figure 6.
Failure to remove PLINs by CMA blocks lipolysis. (a) Beta-oxidation rates in CTR and L2A(−) cells treated with OL followed by lipase inhibitor diethylumbelliferyl phosphate (Deup) and/or lysosomal inhibitors ammonium chloride and leupeptin (NL). n=3 independent experiments. (b, c) Costaining for DPH and ATGL in CTR and L2A(−) cells untreated or treated with OL or incubated with serum-supplemented medium (OL>S+) after OL treatment. Colocalized pixels are in white. Right: Higher magnification of boxed area. Graph: percentage colocalization of DPH with ATGL. n=6 independent experiments with 40 cells per condition in each experiment. (d) Costaining for BODIPY493/503 and ATGL in CTR cells incubated in OL>S+ treated or not with atypical retinoic acid derivative (aRAd). Middle: Higher magnification area. Bottom: Colocalized pixels. Graph: percentage colocalization of BODIPY with ATGL. n=5 independent experiments with 40 cells per condition in each experiment. (e) Immunoblot for indicated proteins in total cell lysates from CTR and L2A(−) cells untreated or treated with OL or incubated with serum-supplemented medium (OL>S+) or serum-deprived medium (OL>S−) after OL treatment. Graph: ATGL levels relative to untreated CTR cells. n=3 independent experiments. (f, g) Coimmunoprecipitation (IP) of ATGL in OL>S+ (f) or OL-treated (g) CTR (+) and L2A(−) cells. Cells in (f) are expressing G0S2-GFP. 1/10 input shown. Representative blots from n=3 (f) and 5 (g) independent experiments. (h, i) Immunoblot for indicated proteins of homogenates (HOM) and lipid droplets (LD) isolated from fed (F) or starved (S) rat livers (h), or starved wild-type (+) or L2A knockout (−) mice livers (i). Representative blots of n=4 (i; 3 additional blots shown in Supplementary Figure 5c) and 7(h) independent experiments. (j) Immunostaining for ATGL in OL-treated cells expressing WT or MT PLIN2-GFP. Inset: Higher magnification of the boxed area. Graph: percentage colocalization of ATGL with PLIN2-GFP. n=5 independent experiments with 40 cells per condition in each experiment. Values are mean ± SEM. Differences are significant for *P<0.05, **P<0.01, ***P<0.001 using Student’s t-test. Uncropped images of blots are shown in Supplementary Figure 8. Source data is available in Supplementary Table 1.
The rate-limiting cytosolic lipase in most cell-types, adipose triglyceride lipase (ATGL)21, colocalizes with LD (lipid- or protein-labeled) during lipogenic stimulus (OL) and in OL>S- or OL>low-glucose S+ conditions to induce lipolysis (Fig. 6b,c, Supplementary Figure 5a,b). Similarly, chemical activation of CMA increased colocalization of ATGL with LD (Fig. 6d). In contrast, CMA-defective cells displayed significantly reduced association of ATGL with LD in all conditions (Fig. 6b,c, Supplementary Figure 5a,b) despite total ATGL levels remaining unchanged (Fig. 6e), pointing towards defective localization of this lipase on LD. Reduced lipolytic activity of ATGL in CMA-deficient cells could not be attributed to changes in its association with endogenous modulators because, on the contrary, pull-down for ATGL in L2A(−) cells revealed reduced binding to its inhibitor G0/G1 switch gene 2 (G0S2) and enhanced association with its co-activator comparative gene identification 58 (CGI-58) (Fig. 6f,g), likely reflecting cellular compensatory efforts to sustain lipolysis. Similar changes occur in vivo as association of ATGL with rat liver LD was enhanced upon starvation, which activates both lipolysis and CMA (Fig. 6h), but LD isolated from livers of starved L2AKO mice22 displayed a discrete but consistent reduction in ATGL levels and a more pronounced decrease in levels of its co-activator CGI-58 when compared to LD isolated from control mice (Fig. 6i, Supplementary Figure 5c). Furthermore, expression of the CMA-resistant PLIN2, but not the WT variant, was sufficient to significantly reduce the association of ATGL with LD (Fig. 6j). Thus, the inability to remove PLIN2 from LD in CMA-deficient cells leads to reduced lipolysis, in part because it impairs the association of the cytosolic lipase ATGL with LD.
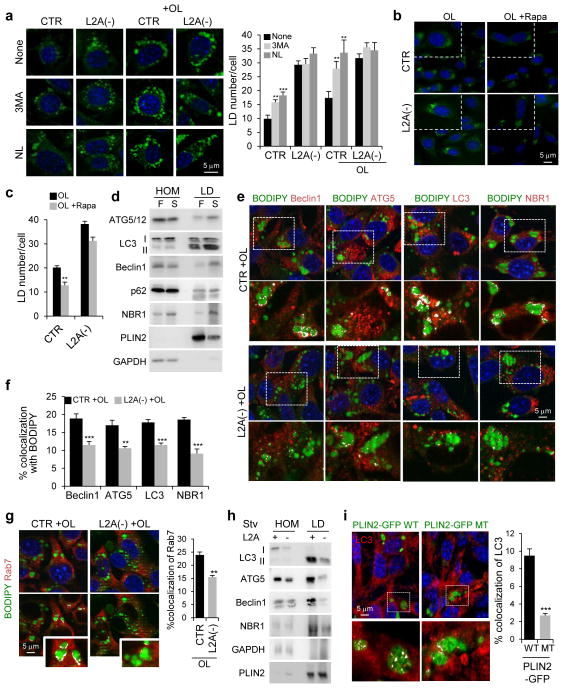
To investigate the reasons behind reduced lysosome-dependent lipophagy in CMA-defective cells (Fig. 6a), we first analyzed changes in LD abundance upon macroautophagy blockage (Fig. 7a, Supplementary Figure 5d). Addition of 3-methyladenine was efficient in reducing macroautophagy in both control and L2A(−) cells (Supplementary Figure 5e) but resulted in increased LD number only in control cells (Fig. 7a, Supplementary Figure 5d). Similarly, upregulation of macroautophagy with rapamycin resulted in a more pronounced decrease in LD content in control than in L2A(−) cells (Fig. 7b,c). This reduction in macrolipophagy in cells with defective CMA contrasts with the previously reported increase in non-selective macroautophagy in these cells25, that we also confirmed by LC3 staining (Supplementary Figure 5f). To investigate the reasons behind the macrolipophagy failure, we analyzed the recruitment of macroautophagy proteins (ATGs) to LD that occurs in liver when lipolysis is activated by prolonged starvation (Fig. 7d) and also in cultured fibroblasts upon OL challenge (Supplementary Figure 6a,b). Co-immunostaining in L2A(−) cells revealed a significant decrease in colocalization with LD of components of macroautophagy initiation complex (Beclin1, ATG14, vps15 and vps34), proteins involved in autophagosome elongation (ATG5) and the structural autophagosome component (LC3) (Fig. 7e,f, Supplementary Figure 6c,d). We also analyzed two cargo-recognition proteins and found that LD in L2A(−) cells show lower colocalization with NBR1 (Fig. 7e,f), the one that normally increases on LD during lipolysis (Fig. 7d, Supplementary Figure 6a,b), whereas colocalization with p62, that does not change during lipolysis, remained also unchanged in L2A(−) cells (Supplementary Figure 6e). Colocalization between LD and ATG7, the ligase that mediates elongation, remained unchanged whereas other ATG proteins such as ULK1, ATG9 and UVRAG did not colocalize with LD either in control or L2A(−) cells (Supplementary Figure 6e,f). Colocalization of Rab7 with LD, required for lysosome-mediated lipolysis37, was also significantly reduced in L2A(−) cells (Fig. 7g). In vivo analysis using isolated liver LD confirmed that levels of LC3, ATG5, Beclin1 and NBR1, were reduced in L2A-defective mice as compared to control mice (Fig. 7h). Furthermore, even in the context of functional CMA, expression of the CMA-resistant PLIN2 mutant was sufficient to significantly reduce the amount of LC3, ATG5 and Beclin1 associated to LD (Fig. 7i, Supplementary Figure 7a,b and Supplementary Video 4 and 3D reconstructions of Z-sections shown in Supplementary Figure 7c and Supplementary Video 5), confirming that inability to degrade LD-associated PLINs via CMA inhibited the association of the macroautophagy machinery to LD. We propose that failure to anchor ATGs at the LD surface prevents initiation of macrolipophagy. Ultrastructural analysis confirmed that the membranous structures observed in large LD in control cells upon induction of lipolysis (by OL challenge or starvation) and previously proposed as signature of macrolipophagy of portions of LD18, were almost completely absent in LD of L2A(−) cells (Supplementary Figure 7d). Collectively, these results indicate that removal of PLINs via CMA precedes macrolipophagy.
Figure 7.
Failure to remove PLINs by CMA blocks macrolipophagy. (a) BODIPY493/503 staining in CTR and L2A(−) cells untreated or treated with OL, or treated with 3-methyladenine (3MA) or lysosomal inhibitors ammonium chloride and leupeptin (NL). Full fields shown in Supplementary Figure 5c. Graph: average LD number/cell. n=9 fields with 1350 cells per condition from 3 independent experiments. (b, c) Cells stained as in (a) treated or not with rapamycin (Rapa). Insets: higher magnification. Graph: Average LD number/cell. n=9 fields with 1350 cells per condition from 3 independent experiments. (d) Immunoblot for indicated proteins in homogenates (HOM) and lipid droplets (LD) isolated from livers for fed (F) and starved (S) rats. Representative blots of n=5 independent experiments. (e, f) Costaining for indicated ATG proteins and BODIPY493/503 in CTR and L2A(−) cells treated with OL. Insets: Higher magnification of the boxed areas with colocalized pixels in white. Graph: percentage colocalization with BODIPY. n=6 independent experiments with 40 cells per condition in each experiment. (g) Costaining for Rab7 in the same cells as in (e). Graph: percentage colocalization with BODIPY. n=4 independent experiments with 40 cells per condition in each experiment. (h) Immunoblot for indicated proteins of HOM and LD isolated from starved wild-type (+) or L2A knockout (−) mice livers. (i) Immunostaining for LC3 in cells expressing WT or MT PLIN2-GFP treated with OL. Inset: Higher magnification of the boxed area with colocalized pixels in white. 3D-reconstruction shown in Supplementary Figure 7c. Graph: percentage colocalization of LC3 with PLIN2-GFP. n=4 independent experiments with 40 cells per condition in each experiment. Values are mean ± SEM. Differences are significant for **P<0.01, ***P<0.001 using Student’s t-test. Uncropped images of blots are shown in Supplementary Figure 8. Source data is available in Supplementary Table 1.
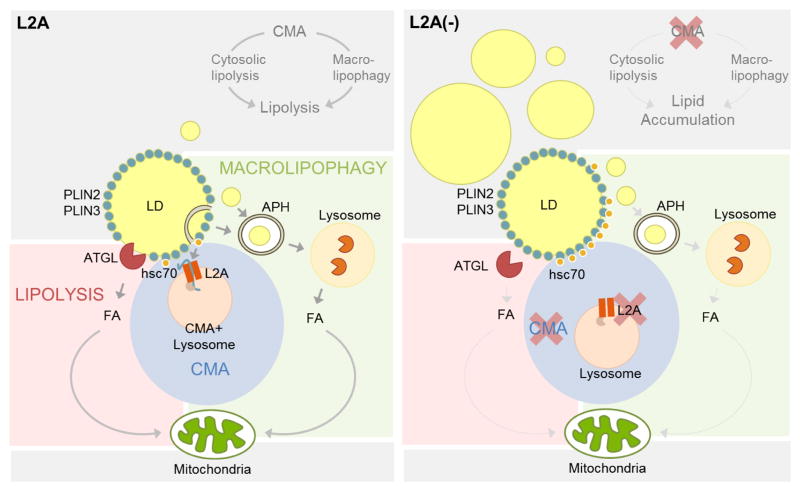
Altogether, we propose that degradation of LD proteins PLIN2 and PLIN3 by CMA is required for the LD association of both the cytosolic lipase and the macroautophagy effector proteins, and for subsequent lipolysis, explaining the intracellular lipid accumulation observed upon CMA compromise (Fig. 8).
Figure 8.
Scheme of the interplay of CMA with cytosolic and lysosomal lipolysis through PLIN2 and PLIN3 degradation. Left: Removal and degradation by CMA of lipid droplets (LD) associated proteins PLIN2 and PLIN3 facilitates access of the lipid core to the cytosolic lipase ATGL. Similarly, elimination of PLINs also allows for proteins involved in autophagosome formation (ATG) to anchor on the LD and initiate the sequestration of regions of LD in autophagosomes (APH) for delivery to lysosomes for degradation (macrolipophagy). Right: When CMA activity is compromised, the inability to remove PLINs from the surface of the LD prevents access of ATGL and ATGs to the lipid core and inhibits lipolysis by both pathways (cytosolic lipases and macrolipophagy).
Discussion
In this study, we demonstrate the role of CMA in regulating mobilization of intracellular lipid stores by degrading LD proteins PLIN2 and PLIN3. Removal of these LD proteins is a prerequisite for subsequent lipolysis to occur, since failure to remove these proteins via CMA results in decreased LD association of components of both arms of the lipolytic machinery – cytosolic lipases and macrolipophagy (Fig. 8).
TG in LD are broken down by cytosolic lipases to generate FFA that undergo β-oxidation in mitochondria21, 23 and by lysosomal lipases upon sequestration of complete or fractions of LD by macrolipophagy18, 19. ATGs and ATG-positive membranes are detected in LD during lipolysis, suggesting that the autophagosome limiting membrane forms in situ on LD surface. In fact, recent studies have shown that LD are a site for autophagosome biogenesis38. We have uncovered that CMA is required prior to the selective sequestration of LD by the autophagosome, suggesting that for ATGs to associate and initiate autophagosome biogenesis, LD peripheral proteins would have to be removed from the LD surface. Consequently, the only residual lipolysis observed in CMA-deficient cells could be attributable to in-bulk macroautophagy, shown upregulated in these cells25, and where cargo recognition is not required.
Cytosolic lipolysis is regulated by complex interactions between PLINs and lipases39. ATGL associates with LD under maximal lipolytic conditions39 and PLINs at the LD surface exert a ‘shielding effect’ that modulates the lipase’s accessibility to TG. In this regard, overexpression of PLIN2 reduces LD association of ATGL and hence, lipolysis39, 40. We propose that CMA facilitates lipolysis by removing PLINs from LD since we found decreased association of ATGL on LD in cells with elevated PLIN2 levels, either due to impaired CMA or due to expression of the CMA mutant. In addition, PLIN2 removal could also facilitate binding of the coactivator to ATGL already on LD, since CMA blockage reduced levels of CGI-58 in LD.
Although PLIN2 has been reported to be a proteasome substrate28, 29, both lysosomes and proteasomes can degrade the same protein depending on the cellular requirement or the cell-type, as reported, for example, for PLIN141–43. In our study, we observed both lysosomal and proteasomal degradation of PLIN2, but interestingly, CMA blockage also prevented PLIN2 proteasomal degradation. Although it is plausible that CMA might indirectly facilitate proteasomal degradation of PLIN2, our data disfavors this option since (i) PLIN2 directly interacts with hsc70; (ii) we detect PLIN2 in CMA-active lysosomes and its degradation in this compartment; and (iii) eliminating the CMA-targeting motif retains PLIN2 in LD. We propose thus that PLIN2 is directly identified by hsc70 in LD and targeted for CMA degradation. Proteasome may only be able to access LD PLIN2 once its removal has been initiated by CMA or alternatively, the PLIN2 amenable to proteasomal degradation could be located in a different compartment, as described before29.
We describe PLIN2 and PLIN3 as the first lipid-anchored proteins to be CMA substrates. Presence of hsc70 on LD, also described before44–47 and its persistence at the LD surface when CMA is blocked, together with the altered LD and lysosomal dynamics in these conditions, suggests the presence of a microenvironment at the LD surface, wherein lysosomes and LD are in close proximity. Dynamic and transient LD interactions, similar to the ones we propose here with lysosomes, have been described between LD and other organelles, such as mitochondria, and peroxisomes48–50, to channel FFA from lipolysis to sites of oxidation20. Our studies with live-cell microscopy support marked reduction in kiss-and-run events between LD and lysosomes in conditions that compromise PLIN degradation by CMA such as mutations in the CMA-targeting motif of PLIN2 that prevent hsc70 interaction with this protein.
We thus, propose that degradation of PLINs via CMA occurs in discrete areas on LD to facilitate punctual removal of PLINs from the LD surface for ATGL and ATGs to access the stored TG. ATGL rapidly cycles on and off of LD to catalyze lipolysis39 and similarly, association of LC3 with LD is polarized and occurs in discrete areas on the LD surface from where the limiting membrane starts forming18. Although the mechanisms that determine removal of only a fraction of PLINs in a localized region of LD require future investigation, we speculate that changes in PLINs phosphorylation may facilitate the release of a specific fraction of PLINs from LD. In fact, PLIN2 has been already identified as part of the phosphoproteome of LD51.
We propose that the pronounced hepatic lipid accumulation observed in L2AKO mice results from a combination of: increased levels of lipogenic enzymes degraded through CMA22, reduced VLDL secretion22, and reduced lipolysis due to the inability of cytosolic lipases and ATGs to bind to LD when PLINs are not degraded by CMA. We have previously shown that chronic lipid loading inhibits CMA activity15. In light of our current findings of regulation of lipid stores by CMA, it is conceivable that a compromise in CMA would set up a viscous cycle of lipid overloading in cells. CMA activity decreases with age16, and it is plausible that changes in CMA would impact cellular lipohomeostasis and the steatosis observed during aging, which in turn would further suppress CMA activity. Manipulating CMA could thus be a viable therapeutic option against excessive lipid accumulation.
Online Methods
Cell Culture and Treatments
NIH3T3 mouse fibroblasts (American Type Culture Collection) were cultured in complete Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich), 10% heat-inactivated newborn calf serum (HyClone), 1% penicillin/streptomycin/fungizone (Invitrogen) and maintained at 37°C/5% CO2. NIH3T3 clones stably RNA-interfered for LAMP-2A (L2A(−)) were generated as described previously25. All the cell lines were tested for mycoplasma contamination using DNA staining protocol with Hoechst 33258 dye. Oleic acid (Sigma-Aldrich) was conjugated to albumin as described52, and cells were treated with 0.06 mM oleate (OL) for 24 h unless otherwise stated. Where indicated, OL was washed off with Hanks’ Balanced Salt Solution (Invitrogen) and replaced by DMEM or low-glucose DMEM with or without serum (OL>S+, OL>S+ Low Glc, OL>S-, respectively) for 16 h. Cells were treated with lysosomal inhibitors ammonium chloride (20 mM; American Bioanalyticals) and leupeptin (100 μM; Fisher Scientific), macroautophagy activator rapamycin (100 nM; Sigma-Aldrich), proteasomal inhibitors lactacystin (5 μM; Enzo Life Sciences) and MG132 (5 μM; Sigma-Aldrich), mitochondrial depolarizer FCCP (2 μM; Sigma-Aldrich) for 16 h unless indicated otherwise. For imaging experiments, cells were treated with etomoxir (10 μM; Sigma-Aldrich) for 6 h.
Plasmids and Reagents
Mouse PLIN2-tGFP and G0S2-tGFP plasmids were from Origene, rat LAMP1-RFP plasmid from Addgene plasmid repository, human dsRed-LC3 plasmid53, mtKeima plasmid26, GFP-dgn/GFP-degFS30 plasmids as described before and human L2A (PGK-hL2A, plasmid includes the GFP transgene, to facilitate identification of transfected cells) was generated in our laboratory54. Mutagenesis was performed via the QuikChange site-directed mutagenesis kit (Agilent Biotechnologies). Cells were transfected with 1 μg DNA plasmid (or 0.5 μg of each plasmid in case of cotransfection) using Lipofectamine 2000 transfection reagent (Invitrogen) according to manufacturer's instructions. Sources of chemicals were as previously described18, 25, 55.
Antibodies
The dilutions and sources of antibodies used for immunoblot (IB) and immunofluorescence (IF) in this study are: PLIN2 (1:2000 (IB); 1:100 (IF); Progen Biotechnik; GP40), PLIN3 (1:2000 (IB); 1:100 (IF); ProSci Inc; 3881), L2A (1:5000 (1B); Life Technologies; 512200), mouse LAMP1 (1:3000 (IB); 1:100 (IF); Developmental Studies Hybridoma Bank; 1D4B), rat LAMP1 (1:3000 (IB); Enzo Life Sciences; Ly1C6), human LAMP2 (1:3000 (IB); 1:100 (IF); Developmental Studies Hybridoma Bank; H4B4), LAMP-2B (1:5000 (IB); generated in our laboratory), Hsc70 (1:5000 (IB); 1:100 (IF); Novus Biologicals; 13D3), GAPDH (1:3000 (IB); Abcam; 6C5), hexokinase (1:1000 (IB); Cell Signaling; C35C4), tGFP (1:2000 (IB); Origene; 2H8), ATGL (1:1000 (IB); 1:100 (IF); Cell Signaling; 30A4), CGI58 (1:3000 (IB); ProSci Inc; 45186), ATG5 (1:2000 (IB); 1:100 (IF); Novus Biologicals; 53818), Beclin1 (1:2000 (IB); 1:100 (IF); Novus Biologicals; 500249), LC3 (1:1000 (IB); 1:100 (IF); Cell Signaling; 2775), Atg14 (1:100 (IF); MBL; PD026), Atg7 (1:100 (IF); Cell Signaling; 2631), vps15 (1:100 (IF); Novus Biologicals; NBP130463), vps34 (1:2000 (IB); 1:100 (IF); Life Technologies; 382100), p62 (1:2000 (IB); Enzo Life Sciences; 9860), NBR1 (1:3000 (IB); 1:100 (IF); Abnova; A01), Rab7 (1:100 (IF); Abcam; EPR7589), ULK1-P (1:100 (IF); Cell Signaling; 5869), Atg9 (1:100 (IF); Novus Biologicals; 56893), UVRAG (1:100 (IF); Abcam; 174550), K48-specific ubiquitin (1:2000 (IB); Millipore; Apu2), IkB (1:5000 (IB); Santa Cruz Biotechnology; C21), and Actin (1:10,000 (IB); Abcam; AC15).
Animals and Treatments
Adult Wistar male rats (25) and C57BL/6 male mice (12) (Charles River Laboratories) were used. L2A knockout mice were generated as described22. Where indicated, rats or mice were starved for 48 h or 24 h before organelle isolation by removing food but maintaining water ad libitum. Animals were injected intraperitoneally with leupeptin (2 mg/100 g body weight) 2 h prior to organelle isolation. All animal studies were approved by the Institutional Animal Care and Use Committee. Randomization between control and L2AKO mice was not possible as animal group is based on genotype. In the case of starvation and treatment with vehicle or leupeptin, animals were randomly attributed to each group. The investigators were not blinded to allocation during experiments and outcome assessment. No statistical method was used to predetermine sample size.
TG Measurement
Triglycerides (TG) were measured by a commercial kit (Sigma-Aldrich) according to the manufacturer’s instructions.
TG Synthesis
TG synthesis was measured as described before18. Briefly, synthesis of triglycerides derived from [14C]-oleate (PerkinElmer) complexed with BSA was measured by incubation of untreated or OL-treated cells with [14C]-oleate-BSA (0.2 μCi, 4 h), followed by extraction of cellular lipids and analysis by thin-layer chromatography. Incorporation of radioactivity into TG was determined using a Storm Gel and Blot Imaging System (GE Healthcare) followed by densitometry using ImageJ. The rate of TG synthesis was calculated as the amount of incorporated [14C]-oleate in relative units per mg protein.
Beta-oxidation
Rates of fatty acid beta-oxidation were determined by metabolic radioactive labeling as described earlier18 and by respiratory measurements. Briefly, the rate of carbon dioxide production resulting from the oxidation of [14C]-oleate was measured. Cells pretreated with or without OL (0.06 mM) were treated with [14C]-oleate-BSA (0.8 μCi, 4 h). Where indicated inhibitors, ammonium chloride (20 mM) and leupeptin (100 μM), and diethylumbelliferyl phosphate (Deup) (100 μM) were added with the radiolabeled oleate for 4 h. The released [14C]-carbon dioxide was trapped at 37°C for 1 h onto filter paper pre-soaked in 100 mM sodium hydroxide. The rate of beta-oxidation was calculated as the amount of trapped [14C]-carbon dioxide in relative units produced per mg protein per hour. Equal numbers of cells were plated in XF96 plates (Seahorse Bioscience) in low-glucose DMEM (GIBCO) supplemented with serum. Cells were treated with OL (0.12 mM) for 16 h following which cell respiration was assayed as time-resolved measurements of oxygen consumption rate (OCR) in a respirometer (Seahorse Bioscience). Fatty acid beta-oxidation was calculated as specified by the manufacturer by injecting etomoxir (50 μM) in the buffered assay medium.
Cell Imaging
Immunofluorescence microscopy
Cells grown on coverslips were fixed with 4% paraformaldehyde (or pre-chilled methanol in case of staining for Hsc70, in order to highlight the membrane-associated protein) for 30 min, permeabilized and blocked with 1% BSA, 0.01% Triton X-100 in phosphate buffer saline (PBS). Incubation with primary and secondary antibody conjugated to Alexa Fluor 488 or Cy5 (Invitrogen) in 0.1% BSA in PBS was performed at room temperature for 1 h each. For LD staining, cells were incubated with BODIPY 493/503 (Invitrogen, 20 μg/ml) or 1,6-diphenyl-1,3,5-hexatriene (DPH, Sigma-Aldrich, 5 μM; pseudo-colored to red or green) for 20 min at 37°C prior to fixation. For mitochondria staining, cells were incubated with MitoTracker (50 mM) and MitoTracker CMXRos (50 nM) (Invitrogen) for 20 min 37°C prior to fixation. Coverslips were mounted in DAPI-Fluoromount-G (Southern Biotechnology) or Draq5 (Invitrogen; pseudo-colored to blue) to stain the nucleus. All images were acquired with Axiovert 200 fluorescence microscope (Carl Zeiss Microscopy) with ×63 objective and 1.4 numerical aperture, mounted with an ApoTome.2 slider, and prepared using Adobe Photoshop CS3 (Adobe Systems) and ImageJ (NIH). Where indicated, cells plated in glass-bottom 96-well plates were treated, stained with BODIPY493/503, fixed and images were acquired using a high-content microscope (Operetta, Perkin Elmer). Images of 9 different fields per well were captured, resulting in an average of 1500–2000 cells. Nuclei and puncta were identified using the manufacturer’s software. The number of particles/puncta per cell was quantified using ‘analyze particles’ function of ImageJ after thresholding in non-saturated images. The percentage of colocalization was determined by ‘JACoP’ plugin in ImageJ after thresholding of individual frames56. Where indicated, the colocalized pixels were highlighted using ‘colocalization’ plugin of ImageJ. 3D reconstruction images were modeled from Z-sections as mixed renderings using the Inside4D module for AxioVision Rel. 4.8. after applying the Nyquist-Shannon sampling theorem. Immunofluorescence and direct fluorescence quantifications of number of lipid droplets, number of puncta and percentage colocalization, were performed blinded.
Live-cell imaging
Live-cell imaging was performed in cells grown in glass-bottom dishes (MatTek) 24 h post-transfection (with 8 h OL treatment) at 37°C in a CO2-enriched atmosphere in recording medium (phenol red-free DMEM (GIBCO), 3.7 mg/ml NaHCO3, and 25 mM HEPES, pH 7.4) using a 4D spinning-disk confocal microscope (PerkinElmer, TE2000S) with 60x NA 1.4 oil objective and ORCA-ERA camera (Hamamatsu). Images were acquired as a stack of 50–50 Z optical planes (0.4 μm) encompassing the entire cell width at a fixed rate of maximum speed for ~20 min. Image sequences were processed with Volocity and ImageJ. Using Volocity, small and large LD clusters were identified as <1 μm3 and >1 μm3, respectively and were tracked using the software to calculate the displacement (straight line distance between the start and the end point of the track), displacement rate (displacement/(time of last time point in the track – time of the first time point of the track)) and tracking velocity (average speed over the whole track).
Electron microscopy
Cells were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer pH 7.4 at 4°C for 3 h. Post-fixation with 1% osmium tetroxide was followed by dehydration with graded series of ethanol, infiltration and embedding in EMbed 812 (Electron Microscopy Sciences). 80 nm sections were cut with ultramicrotome (Leica), stained with uranyl acetate followed by lead citrate and viewed in JEM-1200EX transmission electron microscope (JEOL) at 80 kV. Morphometric analysis was done using ImageJ with LD identified as electron translucent structures lacking surrounding membrane. Micrographs were independently annotated by two people and the average of their scoring was used for each micrograph.
Cellular subfractionation
Lysosome isolation
Rat liver lysosomes were isolated from a light mitochondrial-lysosomal fraction by centrifugation in a discontinuous metrizamide density gradient57.
LD isolation
Rat and mouse liver LD were isolated as described previously18. Livers homogenized in 0.25 M sucrose were centrifuged at 6,800 g for 5 min at 4°C. The supernatant and the fatty layer were centrifuged at 17,000 g for 10 min at 4°C to pellet lysosomes. The supernatant was adjusted to 20% sucrose (LD-enriched fraction) and centrifuged in a discontinuous sucrose density gradient at 28,000 g for 60 min at 4°C. The LD fraction collected was delipidated, solubilized in SDS, and equal protein amount was analyzed by immunoblot.
Coimmunoprecipitation
Coimmunoprecipitation was performed in total cell lysates or LD-enriched fractions prepared in 25 mM Tris (pH 7.2), 150 mM NaCl, 5 mM MgCl2, 0.5% NP-40, 1 mM DTT, 5% glycerol and protease inhibitors using standard procedures. Equal protein amount of each fraction was incubated with primary antibody (1 μg) overnight followed by incubation with protein A-conjugated sepharose for 1 h, centrifugation and washes.
General Methods
Total cell lysates were prepared in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% NaDoc, 0.1% SDS, 50 mM Tris pH 8) containing protease and phosphatase inhibitors. Protein concentration was measured using bovine serum albumin as a standard58. Isoelectric focusing (IEF) was done using the Protean IEF Cell (Bio-Rad) at 20°C with rapid ramping to voltage 10,000 V at a current limit of 50 μA using IPG Strips with 4–7pH range (Bio-Rad). Where indicated total cellular fractions were treated with λ-phosphatase according to manufacturer’s instructions (New England BioLabs) before isoelectric focusing. Phos-tag PAGE was performed according to manufacturer’s instructions (Wako) using 100 μM Phos-tag. Samples were subjected to SDS-PAGE, transferred to nitrocellulose membrane, blocked with low-fat milk and incubated with primary antibody overnight. The proteins were visualized by using peroxidase-conjugated secondary antibodies and chemiluminescent reagent (PerkinElmer) in LAS-3000 Imaging System (Fujifilm). Densitometric quantification was performed on unsaturated images using ImageJ.
Sample size, Randomization and Statistical Analyses
In the studies performed in cell lines in culture, all experiments were repeated at least 3 times and often with duplicate blots in the same experiment. All numerical results are reported as mean ± standard error of the mean (SEM). Statistical significance of the difference between experimental groups was analyzed by two-tailed unpaired Student’s t-test using MS Excel. Differences were considered statistically significant for P<0.05. For those experiments in which an estimate of variation was possible based on previous studies or published work by others, we have confirmed that the variance is similar between the control and experimental groups compared. For those instances in which previous information was not available, we have compared with other conditions in which changes in cellular components resulted in changes in CMA.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health AG021904, AG031782, AG038072, DK098408 and the generous support of Robert and Renée Belfer. We thank Bindi Patel for performing the electron microscopy, Dr. Rajat Singh for assistance in biochemical lipid assays and the Analytical Imaging Facility for support with live-cell imaging. The authors declare no conflict of interest.
Footnotes
Author Contributions
SK designed and performed the experiments, analyzed and interpreted the data, and contributed to writing the manuscript; AMC coordinated the study, contributed to designing and interpretation of the experiments and to writing the manuscript.
Competing Financial Interest
The authors declare that they have no competing interests.
Supplementary Information is linked to the online version of the paper.
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 5.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 6.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 7.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275:27447–27456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 10.Arias E, Cuervo AM. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol. 2011;23:184–189. doi: 10.1016/j.ceb.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuervo AM, Knecht E, Terlecky SR, Dice JF. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 13.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohi E, et al. Hypoxic stress activates chaperone-mediated autophagy and modulates neuronal cell survival. Neurochem Int. 2012;60:431–442. doi: 10.1016/j.neuint.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Navarro JA, et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705–714. doi: 10.1073/pnas.1113036109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 17.Kiffin R, et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782–791. doi: 10.1242/jcs.001073. [DOI] [PubMed] [Google Scholar]
- 18.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quiroga AD, Lehner R. Liver triacylglycerol lipases. Biochim Biophys Acta. 2012;1821:762–769. doi: 10.1016/j.bbalip.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg AS, et al. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Ranall MV, Gabrielli BG, Gonda TJ. High-content imaging of neutral lipid droplets with 1,6-diphenylhexatriene. BioTechniques. 2011;51:35–36. 38–42. doi: 10.2144/000113702. [DOI] [PubMed] [Google Scholar]
- 28.Masuda Y, et al. ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J Lipid Res. 2006;47:87–98. doi: 10.1194/jlr.M500170-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Xu G, et al. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem. 2005;280:42841–42847. doi: 10.1074/jbc.M506569200. [DOI] [PubMed] [Google Scholar]
- 30.Greussing R, Unterluggauer H, Koziel R, Maier AB, Jansen-Durr P. Monitoring of ubiquitin-proteasome activity in living cells using a Degron (dgn)-destabilized green fluorescent protein (GFP)-based reporter protein. J Vis Exp. 2012;69 doi: 10.3791/3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 32.Aniento F, Roche E, Cuervo AM, Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem. 1993;268:10463–10470. [PubMed] [Google Scholar]
- 33.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anguiano J, et al. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9:374–382. doi: 10.1038/nchembio.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bostrom P, et al. Cytosolic lipid droplets increase in size by microtubule-dependent complex formation. Arterioscler Thromb Vasc Biol. 2005;25:1945–1951. doi: 10.1161/01.ATV.0000179676.41064.d4. [DOI] [PubMed] [Google Scholar]
- 36.Targett-Adams P, et al. Live cell analysis and targeting of the lipid droplet-binding adipocyte differentiation-related protein. J Biol Chem. 2003;278:15998–16007. doi: 10.1074/jbc.M211289200. [DOI] [PubMed] [Google Scholar]
- 37.Lizaso A, Tan KT, Lee YH. beta-adrenergic receptor-stimulated lipolysis requires the RAB7-mediated autolysosomal lipid degradation. Autophagy. 2013;9:1228–1243. doi: 10.4161/auto.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupont N, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24:609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011;286:15707–15715. doi: 10.1074/jbc.M110.207779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res. 2007;48:2751–2761. doi: 10.1194/jlr.M700359-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Kovsan J, Ben-Romano R, Souza SC, Greenberg AS, Rudich A. Regulation of adipocyte lipolysis by degradation of the perilipin protein: nelfinavir enhances lysosome-mediated perilipin proteolysis. J Biol Chem. 2007;282:21704–21711. doi: 10.1074/jbc.M702223200. [DOI] [PubMed] [Google Scholar]
- 42.Ogasawara J, et al. Oligonol-induced degradation of perilipin 1 is regulated through lysosomal degradation machinery. Nat Prod Commun. 2012;7:1193–1196. [PubMed] [Google Scholar]
- 43.Xu G, Sztalryd C, Londos C. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta. 2006;1761:83–90. doi: 10.1016/j.bbalip.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 45.Fujimoto T, Ohsaki Y. Cytoplasmic lipid droplets: rediscovery of an old structure as a unique platform. Ann N Y Acad Sci. 2006;1086:104–115. doi: 10.1196/annals.1377.010. [DOI] [PubMed] [Google Scholar]
- 46.Parent R, Qu X, Petit MA, Beretta L. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology. 2009;49:1798–1809. doi: 10.1002/hep.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turro S, et al. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- 48.Binns D, et al. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blanchette-Mackie EJ, Scow RO. Movement of lipolytic products to mitochondria in brown adipose tissue of young rats: an electron microscope study. J Lipid Res. 1983;24:229–244. [PubMed] [Google Scholar]
- 50.Wang H, et al. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res. 2011;52:2159–2168. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartz R, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 52.Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 53.Boland B, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- 55.Kaushik S, Massey AC, Cuervo AM. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006;25:3921–3933. doi: 10.1038/sj.emboj.7601283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 57.Kaushik S, Cuervo AM. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.